Abstract
Termini often determine the fate of RNA molecules. In recent years, 3′ ends of almost all classes of RNA species have been shown to acquire nontemplated nucleotides that are added by terminal nucleotidyltransferases (TENTs). The best‐described role of 3′ tailing is the bulk polyadenylation of messenger RNAs in the cell nucleus that is catalyzed by canonical poly(A) polymerases (PAPs). However, many other enzymes that add adenosines, uridines, or even more complex combinations of nucleotides have recently been described. This review focuses on metazoan TENTs, which are either noncanonical PAPs or terminal uridylyltransferases with varying processivity. These enzymes regulate RNA stability and RNA functions and are crucial in early development, gamete production, and somatic tissues. TENTs regulate gene expression at the posttranscriptional level, participate in the maturation of many transcripts, and protect cells against viral invasion and the transposition of repetitive sequences.
This article is categorized under:
RNA Interactions with Proteins and Other Molecules > Protein‐RNA Recognition
RNA Processing > 3′ End Processing
RNA Turnover and Surveillance > Regulation of RNA Stability
Keywords: polyadenylation, TENT5, terminal nucleotidyltransferase, TUT4/7, uridylation
Terminal nucleotidyltransferases are a diverse group of enzymes that regulate many processes in metazoan cells. Here, we comprehensively summarize recent work on the role of posttranscriptional 3′ end RNA extensions in metabolism, organism development, and disease progression.
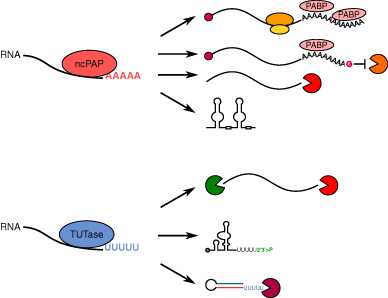
1. INTRODUCTION
Gene expression is precisely regulated at multiple levels to ensure the correct function of cells under normal conditions and rapidly and correctly respond to continually changing environmental conditions. A substantial part of such regulation is provided by tight coordination between transcriptional and posttranscriptional mechanisms. The posttranscriptional addition of nontemplated nucleotides to the 3′ end of various types of RNA molecules, also known as RNA tailing, is a highly conserved process that modulates their stability and activity in multiple organisms and plays a crucial role in regulating gene expression. The 3′ ends of RNA molecules, especially their tails, are a powerful regulatory location that can determine the fate of target RNAs. Almost all classes of RNA species can be subjected to some kind of 3′ end tailing at various steps of their existence. Thus, enzymes that are responsible for these extensions, namely terminal nucleotidyltransferases (TENTs), play an especially important role among RNA metabolism machinery.
The best‐described role of 3′ tailing is the bulk polyadenylation of messenger RNAs (mRNAs) in the cell nucleus that is catalyzed by canonical poly(A) polymerases (PAPs), which also belong to the TENT superfamily (Aravind & Koonin, 1999; Martin & Keller, 2007). Canonical PAPs are, however, outside the scope of the present review, which focuses on other TENTs that play more specific regulatory roles. Various genomes encode different numbers of TENTs per organism. In humans, 11 TENTs are grouped into 6 subfamilies (TENT1–TENT6), based on their phylogenetic analysis (Figure 1a). The nomenclature of TENTs can be somewhat confusing because the same enzymes have often been described under different names. The updated nomenclature that was approved by the HUGO Gene Nomenclature Consortium advises referring to all vertebrate noncanonical PAPs (ncPAPs) and terminal uridylyltransferases (TUTases) by their respective TENT subfamily name. However, for some enzymes (e.g., mitochondrial poly(A) RNA polymerase [MTPAP], TUT4, and TUT7), their well‐recognized functional names can still be used. The invertebrate orthologs are named according to nomenclature guidelines for the respective organism, and their names do not always correspond to the human orthologs. To aid readers of the present review, Table 1 summarizes information about the nomenclature and functional diversity of TENTs in vertebrates, Caenorhabditis elegans and Drosophila melanogaster.
FIGURE 1.
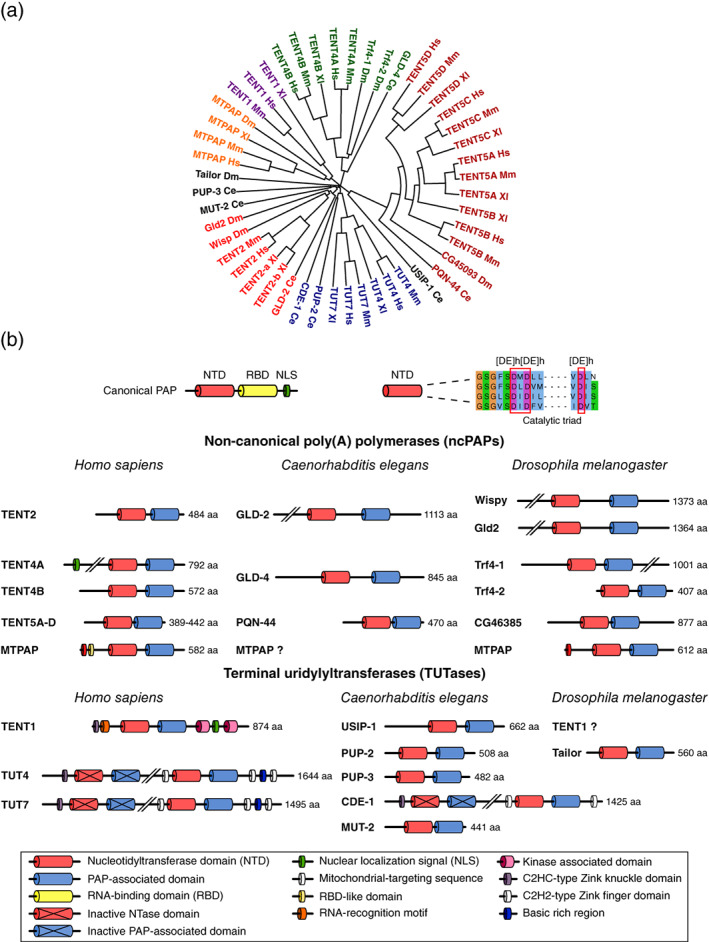
(a) A phylogenetic relationship of TENTs from vertebrates, worms, and fruit fly. (b) Schematic representation of terminal nucleotidyltransferases (TENTs) from the human, worm, and fruit fly genomes. Canonical nuclear poly(A) polymerase (PAP) is a highly evolutionarily conserved PAP. It comprises an N‐terminal nucleotidyltransferase (NTase) catalytic domain, a central RNA‐binding domain (RBD), and a C‐terminal domain with a nuclear localization signal (NLS). Catalytic activity relies on the highly conserved aspartate triad ([DE]h[DE]h [DE]h in the NTase domain. All TENTs share similar catalytic domain architecture but lack the RBD. The domain architecture of TENTs is diverse, and all additional domains and regions are indicated for each TENT. The length (aa) of each enzyme is indicated on the right
TABLE 1.
Summary of current knowledge about metazoan TENTs
| Subfamily | Enzyme (synonyms) | Activity | RNA substrate | Effect on target RNAs | Reference |
|---|---|---|---|---|---|
| Vertebrates | |||||
| TENT1 | TENT1 (TUT1, U6 TUTase, Star‐PAP, PAPD2) | Oligouridylation | U6 snRNA | Maturation | Trippe et al. (2006) |
| Polyadenylation (controversial) | Pre‐mRNA | Stabilization | Laishram and Anderson (2010), Mellman et al. (2008) | ||
| TENT2 | TENT2 (GLD‐2, PAPD4, TUT‐2) | Polyadenylation | mRNA | Activation of translationally dormant mRNA | Barnard et al. (2004), Rouhana et al. (2005), Swanger et al. (2013), Udagawa et al. (2012), Yamagishi et al. (2016) |
| Monoadenylation | miRNA | Stabilization | D'Ambrogio et al. (2012), Hojo et al. (2020), Katoh et al. (2009), Kim et al. (2016) | ||
| TENT3 |
TUT4 (TENT3A, PAPD3, ZCCHC11, KIAA0191) TUT7 (TENT3B, PAPD6, ZCCHC6, KIAA1711) |
Oligouridylation |
mRNA Histone mRNA LINE‐1 mRNA Pre‐miRNA miRNA Pre‐rRNA Pol III‐ncRNAs Viral RNA TSS RNA |
Degradation | Chang et al. (2014, 2018), Faehnle et al. (2014), Gutiérrez‐Vázquez et al. (2017), Heo et al. (2009), Jones et al. (2012), Łabno et al. (2016), Lackey et al. (2016), Le Pen et al. (2018), Lim et al. (2014), Morgan et al. (2019, 2017), Mullen and Marzluff (2008), Pirouz et al. (2016, 2019), Piskounova et al. (2011), Thornton et al. (2014), Ustianenko et al. (2018, 2013, 2016), Warkocki, Krawczyk, et al. (2018) |
| Monouridylation | Pre‐miRNA | Processing | Heo et al. (2012), Kim et al. (2015) | ||
| TENT4 | TENT4A (PAPD7, POLS, TRF4‐1, TUT5, POLK) | Polyadenylation (tethering assay) | mRNA | Not defined | Ogami et al. (2013) |
| Mixed A/G tailing (with TENT4B) |
mRNA Viral RNA |
Stabilization | Hyrina et al. (2019), Kim et al. (2020), Lim et al. (2018), Mueller et al. (2019) | ||
| TENT4B (PAPD5, TRF4‐2, GLD‐4, TUT3) | Polyadenylation |
Pre‐rRNA rRNA Y RNA hTR |
Degradation | Fasken et al. (2011), Lubas et al. (2011), Preker et al. (2011), Shcherbik et al. (2010), Sinturel et al. (2017), Sudo et al. (2016) | |
| mRNA | Stabilization | Burns et al. (2011), Shin et al. (2017) | |||
| Oligoadenylation |
snoRNA scaRNA miRNA |
Maturation Degradation |
Berndt et al. (2012), Son et al. (2018) Shukla et al. (2019) |
||
| Monoadenylation | miRNA | Degradation | (Boele et al. (2014) | ||
| Mixed A/G tailing (with TENT4A) |
mRNA Viral RNA |
Stabilization | Hyrina et al. (2019), Kim et al. (2020), Lim et al. (2018), Mueller et al. (2019) | ||
| TENT5 |
TENT5A (FAM46A) TENT5B (FAM46B) TENT5C (FAM46C) TENT5D (FAM46D) |
Polyadenylation | mRNA | Stabilization (shown for TENT5C and TENT5B) | Bilska et al. (2020), Herrero et al. (2020), Hu et al. (2020), Mroczek et al. (2012) |
| TENT6 | MTPAP (TENT6, PAPD1, SPAX4) | Polyadenylation | Mt‐mRNA | Stop codon generation, stabilization | Anderson et al. (1981), Nagaike et al. (2005), Tomecki et al. (2004), Wilson et al. (2014) |
| Oligoadenylation | Mt‐tRNA | Maturation | Fiedler et al. (2015), Pearce et al. (2017) | ||
| Caenorhabditis elegans | |||||
| TENT1 | USIP‐1 | Oligouridylation | U6 snRNA |
Stabilization Recycling |
Rüegger et al. (2015) |
| TENT2 | GLD‐2 | Polyadenylation | Oocyte mRNA |
Activation Stabilization |
Kim et al. (2010), Nousch et al. (2014), Schmid et al. (2009), Suh et al. (2006) |
| TENT3 | CDE‐1 (PUP‐1, CID‐1) | Uridylation |
siRNA Viral RNA |
Degradation | Le Pen et al. (2018), van Wolfswinkel et al. (2009) |
| PUP‐2 | Uridylation | Pre‐miRNA | Processing | Lehrbach et al. (2009) | |
| PUP‐3 | Uridylation | Preston et al. (2019) | |||
| MUT‐2 (RDE‐3) | Tandem UG repeats addition |
siRNA Transposons |
Trigger gene silencing | Preston et al. (2019), Shukla et al. (2020) | |
| TENT4 | GLD‐4 | Polyadenylation | Oocyte mRNA | Not defined | Nousch et al. (2014) |
| TENT5 | PQN‐44 (TENT5) | Putative ncPAP | |||
| Drosophila melanogaster | |||||
| TENT2 | Gld2 | Polyadenylation | mRNA | Activation | Jae et al. (2008), Sartain et al. (2011) |
| Wispy | Polyadenylation | mRNA |
Activation Stabilization |
Benoit et al. (2008), Coll et al. (2018), Cui et al. (2008, 2013), Dufourt et al. (2017), Eichhorn et al. (2016), Lim et al. (2016) | |
| Oligoadenylation |
miRNA sisRNAs |
Degradation Stabilization |
Lee et al. (2014), Jia Ng et al. (2018) | ||
| TENT3 | Tailor | Oligouridylation |
Mirtrons 5S rRNA tRNA snRNA snoRNA RNase MRP |
Degradation | Bortolamiol‐Becet et al. (2015), Reimão‐Pinto et al. (2015), Reimão‐Pinto et al. (2016) |
| TENT4 | Trf4‐1 | Polyadenylation |
snRNA mRNA |
Degradation | Harnisch et al. (2016), Nakamura et al. (2008) |
| Trf4‐2 | Inactive | Nakamura et al. (2008) | |||
| TENT5 | CG46385 | Putative ncPAP | |||
Both TENTs and canonical PAPs belong to the nucleotidyltransferase superfamily of DNA polymerase β, which is conserved in eukaryotes. Canonical PAP comprises three domains: an N‐terminal nucleotidyltransferase (NTase) catalytic domain with the signature helix‐turn motif, a central domain, and a C‐terminal RNA‐binding domain (RBD) with a nuclear localization signal and multiple regulatory sites (Martin, Keller, & Doublié, 2000; Yang, Nausch, Martin, Keller, & Doublié, 2014). Three conserved aspartates in the NTase domain are essential for catalytic activity. TENTs share a similar catalytic domain architecture and the mechanism of catalysis with canonical PAP, but they also have some notable differences (Figure 1b). TENTs carry the conserved NTase domain but lack a typical RBD (with the exception of TENT1). The NTase domain of TENTs is accompanied by a PAP‐associated domain that is absent in canonical PAP. TENTs could also carry some additional regulatory elements, such as sequences that are responsible for subcellular localization or accessory domains to perform their diverse functions (Martin & Keller, 2007). Based on their preference for adenosine triphosphate (ATP) or uridine triphosphate (UTP) incorporation, TENTs are divided into ncPAPs and TUTases. However, some TENTs may exhibit broader specificity for the incorporated ribonucleotide, depending on the cellular context, and can incorporate more than one type of ribonucleotide, thereby leading to so‐called mixed tailing.
Despite the fact that all TENTs belong to one superfamily and share a conserved catalytic domain, they have remarkable functional diversity that affects RNA maturation, stability, and decay and the activation of translationally dormant deadenylated mRNAs (Martin, Doublié, & Keller, 2008). Moreover, TENTs from different organisms have been shown to play specific regulatory roles in the nucleus, cytoplasm, and mitochondria. Finally, TENTs add mono‐, oligo‐, and polynucleotides that are homo‐ or heteroribonucleotide extensions to a variety of RNA substrates, including mRNA, small nuclear RNA (snRNA), micro RNA (miRNA), and aberrant rRNA. Such variables (i.e., substrates, localization, added extensions, and functional outcome) make RNA tailing a potent instrument of the posttranscriptional regulation of gene expression. In recent years, many groundbreaking discoveries of the role of TENTs in many important processes in different organisms have been made, and the development of new high‐throughput methods (Box 1) has allowed our understanding of RNA 3′ end modifications to substantially expand. Many excellent review articles have described the role of mammalian TENTs in the regulation of gene expression (De Almeida, Scheer, Zuber, & Gagliardi, 2018; Menezes, Balzeau, & Hagan, 2018; Warkocki, Liudkovska, Gewartowska, Mroczek, & Dziembowski, 2018; Yu & Kim, 2020). However, these enzymes also act in other organisms. Many crucial findings have been reported using invertebrate model organisms, the most studied of which are C. elegans and D. melanogaster.
BOX 1. HIGH‐THROUGHPUT METHODS TO STUDY RNA 3′ END EXTENSIONS.
TAIL‐seq allows researchers to determine the exact 3′ end of RNA molecules, measure the poly(A) tail length, and capture noncanonical tail modifications, such as uridylation and guanylation (Chang, Lim, Ha, & Kim, 2014). To prepare sequencing libraries, total RNA is subjected to the size‐fractionation (>200 nt) and depletion of ribosomal RNA (rRNA). Next, the biotinylated 3′ adaptor is ligated, and RNA is treated with a low concentration of RNase T1, which partially digests the RNA body but leaves poly(A) tails intact. The fragmented RNA is pulled down with streptavidin beads, phosphorylated at the 5′ end, and gel purified (500–1,000 nt). Following ligation of the 5′ adaptor, RNA is reverse transcribed, amplified by polymerase chain reaction (PCR), and sequenced in the pair‐end mode. A dedicated algorithm is used to detect signals from long thymine stretches (Chang et al., 2014). TAIL‐seq is a powerful method, but it is technically challenging and expensive. In a modified version of TAIL‐seq (mRNA TAIL‐seq [mTAIL‐seq]), splint ligation of the 3′ hairpin adaptor is used to significantly increase sensitivity of the method to mRNAs and decrease costs (Lim, Lee, Son, Chang, & Kim, 2016). The main drawback of mTAIL‐seq is its limited ability to capture very short tails (<8 nt) and tails with non‐adenosine terminal nucleotides. Although enrichment for specific types of the RNA terminus is possible by changing the design of splint ligation adaptors, mTAIL‐seq is still less effective for comprehensively studying all types of RNA termini than the original method and is mostly used to analyze the poly(A) tail length (Lim et al., 2016).
PAL‐seq (poly(A) tail length profiling by sequencing) accurately measures the length of poly(A) tails (Subtelny, Eichhorn, Chen, Sive, & Bartel, 2014). A splint 3′ adaptor is specifically ligated to 3′ ends of polyadenylated RNA after partial digestion with RNase T1. The gel‐purified mRNA fragments are bound to streptavidin beads before phosphorylating the 5′ ends for adaptor ligation, reverse transcribed, and released from the beads. The sequencing clusters are generated by annealing the primers to the 3′ end of the poly(A) sequence and extended using deoxythymidine triphosphate and biotinylated deoxyuridine triphosphate (dUTP). Next, fluorescent streptavidin tags are attached to biotin‐dUTP, and their signal intensity is used to quantify the length of adenosine homopolymers. One of the main limitations of PAL‐seq is that it offers information about RNA 3′ ends that consist only of adenosines and does not reveal non‐adenosine tailings. PAL‐seq also requires a highly elaborate and customized setup that can be executed only on the discontinued Illumina Genome Analyzer II sequencer. The newest version of the method, PAL‐seq v2, combines the advantages of TAIL‐seq and PAL‐seq and allows researchers to measure poly(A) tails with non‐adenosine termini using more readily accessible contemporary Illumina sequencing equipment. For further technical details, see Eisen et al. (2020).
FLAM‐seq (full‐length poly(A) and mRNA sequencing) and PAIso‐seq (poly(A) inclusive RNA isoform sequencing) enable the global and accurate determination of the full‐length sequence of endogenous mRNAs, including their poly(A) tails along with non‐adenosine residues at all positions within the tails (Legnini, Alles, Karaiskos, Ayoub, & Rajewsky, 2019; Liu, Nie, Liu, & Lu, 2019). The principle of both methods is very similar and includes an RNA 3′ end extension, template‐switching, cDNA amplification from full‐length mRNA, and PCR amplification following PacBio sequencing. The main difference between FLAM‐seq and PAIso‐seq lies in the approach to add a 3′ adaptor to the end of poly(A) tails. FLAM‐seq uses G/I tailing, whereas PAIso‐seq employs template extension. FLAM‐seq and PAIso‐seq have comparable accuracy, but PAIso‐seq is possibly more sensitive and suitable for single‐cell analysis.
Oxford nanopore direct RNA sequencing allows long‐read full RNA molecule sequencing and accurate quantification of the poly(A) tail length without a need for RNA fragmentation or amplification, thus significantly reducing bias that can be introduced by the PCR amplification of long adenosine tracts within tails (for review, see Feng, Zhang, Ying, Wang, & Du, 2015). The 3′ sequencing primer that carries the motor protein is attached to the RNA molecule or RNA–DNA hybrid after optional reverse transcription. The motor protein passes the RNA strand through the pore at a consistent rate and in an ATP‐dependent manner. Sequencing then proceeds in the 3′–5′ direction, starting from the adaptor primer. Although this technology is one of the best for analyzing the poly(A) tail length, it does not yet allow the identification of non‐adenosine tail insertions or tailing. Moreover, a large amount of purified mRNA is currently still required for this method.
TRAID‐seq (tethered rNTase activity identified by high‐throughput sequencing) is a screening approach to analyze nontemplated 3′ end extensions that are added to reporter RNA by a tethered known or putative TENT (Preston et al., 2019). TRAID‐seq allows both the assessment of candidate enzyme activity and single‐nucleotide resolution analysis of the identity of the added tailing. A specific TENT is fused to MS2 coat protein and expressed in yeast together with a reporter RNA that carries high‐affinity MS2‐binding sites. The interaction between MS2 and its binding site in the reporter brings the fused TENT to the reporter RNA, and its 3′ end modifications are then assessed by Illumina sequencing.
2. NONCANONICAL POLYADENYLATION
The role of poly(A) tails that are added by canonical nuclear PAP is well‐described. They are crucial for regulating mRNA stability, export to the cytoplasm, the regulation of mRNA translation, and decay. Noncanonical nuclear and cytoplasmic polyadenylation may have varying effects on target RNA molecules, depending on the particular enzyme or subcellular localization. This section summarizes our current knowledge about all metazoan families of ncPAPs.
2.1. TENT2, GLD‐2, Wispy, and dmGLD2
Germline development defective‐2 (GLD‐2) was first identified in C. elegans as a gene that controls the germline mitosis‐to‐meiosis decision, progression through meiosis, and gamete production (Kadyk & Kimble, 1998; Wang, Eckmann, Kadyk, Wickens, & Kimble, 2002). GLD‐2 is evolutionary conserved, and its homologs are also active PAPs in other species, including D. melanogaster, Xenopus laevis, and mammals (Barnard, Ryan, Manley, & Richter, 2004; Benoit, Papin, Kwak, Wickens, & Simonelig, 2008; Cui, Sackton, Horner, Kumar, & Wolfner, 2008; Kwak, Wang, Ballantyne, Kimble, & Wickens, 2004; Rouhana et al., 2005; Sartain, Cui, Meisel, & Wolfner, 2011). Similar to the majority of all TENTs, GLD‐2/TENT2 lacks the RNA‐recognition motif and is recruited to its various substrate RNAs by other RNA‐binding proteins. In vitro, GLD‐2 itself has low catalytic activity; in the presence of its co‐factors, however, it can effectively elongate mRNA tails (Wang et al., 2002). At least in worms, all known molecular and biological functions of GLD‐2 may depend on its enzymatic activity (Nousch, Minasaki, & Eckmann, 2017).
In C. elegans, GLD‐2 influences multiple aspects of germline development. High‐throughput analysis and case‐specific studies identified over 500 germline‐enriched mRNAs that are potential substrates for GLD‐2, the expression levels of which are significantly downregulated in gld‐2‐defective worms (Kim, Wilson, & Kimble, 2010; Nousch, Yeroslaviz, Habermann, & Eckmann, 2014; Schmid, Küchler, & Eckmann, 2009; Suh, Jedamzik, Eckmann, Wickens, & Kimble, 2006). Although some of these mRNAs may have been indirectly downregulated because of the developmental defect of gld‐2 mutants, the number of direct GLD‐2 substrates was still very high. The loss of gld‐2 led to the shortening of mRNA poly(A) tails and a decrease in mRNA abundance, suggesting that GLD‐2 promotes the stability and expression of translationally repressed germline mRNAs (Kim et al., 2010; Nousch et al., 2014). GLD‐2, together with its cofactor GLD‐3 (an RNA‐binding protein of the Bicaudal‐C family), plays an important role in entry and progression through the meiosis of germ cells (Figure 2a) (Eckmann, Crittenden, Suh, & Kimble, 2004). One of the critical targets of GLD‐2 is gld‐1 mRNA that encodes the repressor of mitosis‐promoting mRNAs (Suh et al., 2006). Translationally repressed gld‐1 mRNA ensures mitosis in the germline (for review, see Kimble & Crittenden, 2007; Wang & Voronina, 2020). However, upon the GLD‐2/GLD‐3‐mediated polyadenylation of gld‐1 mRNA, newly translated GLD‐1 protein promotes the germ cell mitosis‐to‐meiosis transition by repressing mitosis‐specific mRNAs. Moreover, GLD‐2 may also activate other mRNAs that are required for meiosis. Other processes that are regulated by GLD‐2 include gamete identity determination, progression through spermatogenesis and oogenesis, and oocyte development (Kim et al., 2009, 2010). These functions of GLD‐2 are facilitated by its selective interactions with two distinct RNA‐binding cofactors, RRM domain‐containing protein 8 (RNP‐8) and GLD‐3 (Figure 2b). During gamete production in hermaphrodites, GLD‐3 and RNP‐8 compete with each other and form separate complexes with GLD‐2 to promote its activity toward distinct pools of mRNA targets (Eckmann et al., 2004; Kim et al., 2009, 2010; Nakel et al., 2016; Nakel, Bonneau, Eckmann, & Conti, 2015). Gamete production in worms starts with spermatogenesis and later switches to oogenesis. Through differential interactions with GLD‐3 and RNP‐8, GLD‐2 controls gamete sex specification. GLD‐2/GLD‐3 promotes spermatogenesis, whereas GLD‐2/RNP‐8 specifies oogenesis (Kim et al., 2009; Nakel et al., 2016). Interestingly, instead of antagonizing each other, RNP‐8 and GLD‐3 have been shown to act synergistically during oocyte development (Kim et al., 2010).
FIGURE 2.
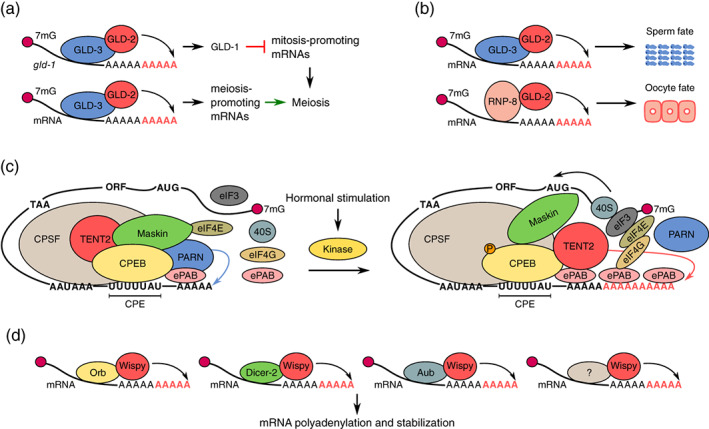
Selected aspects of polyadenylation by GLD‐2, TENT2, and Wispy in worms, frogs, and flies, respectively. (a) Polyadenylation by GLD‐2/GLD‐3 plays an important role in the mitosis‐to‐meiosis decision in the adult worm germline through the stabilization of gld‐1 mRNA, which encodes a repressor of mitosis‐promoting mRNAs. At the same time, GLD‐2/GLD‐3 activates repressed meiosis‐related mRNAs. Both events result in the switch from mitosis to meiosis. (b) In worms, GLD‐2 forms two separate complexes with RNA‐binding proteins. GLD‐2/GLD‐3 specifies spermatogenesis, and GLD‐2/RNP‐8 specifies oogenesis. (c) One of the proposed mechanisms of mRNA reactivation in Xenopus oocytes. In immature Xenopus oocytes, CPE‐containing mRNAs (UUUUUAU motif) are bound by CPEB and form a complex with CPSF, TENT2, PARN, ePABP, and Maskin that prevents assembly of the translation initiation complex and thus keeps mRNAs translationally repressed. Upon hormonal stimulation, CPEB is phosphorylated that leads to substantial rearrangements. PARN dissociates from the complex and allows TENT2 to extend poly(A) tails. As a result, ePABP binds to the newly elongated tails. eIF4G displaces Maskin from eIF4E and promotes formation of the translation initiation complex, initiating the translation of mRNAs. 40S, small subunit of the cytoplasmic ribosome; CPE, cytoplasmic polyadenylation element; CPEB, CPE‐binding protein; CPSF, cleavage and polyadenylation specificity factor; eIF3, eukaryotic initiation factor 3; eIF4, eukaryotic initiation factor 4; ePABP, embryonic poly(A) binding protein; PARN, poly(A)‐specific ribonuclease. (d) In the fruit fly, Wispy is recruited to its target mRNAs via interactions with various RNA‐binding proteins
The role of TENT2/GLD‐2 in gamete production is conserved among many species. Cytoplasmic polyadenylation has been studied in great detail throughout oocyte maturation and early embryo development in the frog X. laevis (for a comprehensive review, see Charlesworth, Meijer, & De Moor, 2013; Ivshina, Lasko, & Richter, 2014; Reyes & Ross, 2016). In frog oocytes that are arrested at the end of the first meiotic prophase, numerous maternal mRNAs are stored with short poly(A) tails to prevent their translation. These mRNAs acquire their dormant state through the association of their U‐rich cytoplasmic polyadenylation element (CPE) that is located in 3′ untranslated regions with CPE‐binding protein (CPEB). mRNA‐CPEB further interacts with cleavage and polyadenylation specificity factor, and the whole complex is exported to the cytoplasm. Two models have been proposed for the mechanism of the CPEB‐mediated inhibition of mRNA translation and its subsequent activation (for review, see Kang & Han, 2011; Radford, Meijer, & de Moor, 2008). However, both require the activity of TENT2 for mRNA readenylation after hormonal stimulation of the oocytes. In the first model, CPEB recruits the poly(A)‐specific ribonuclease PARN which competes with TENT2 activity to keep poly(A) tails short. Additionally, CPBE interacts with Maskin, which binds cap‐binding protein eukaryotic initiation factor 4E (eIF4E) and prevents translation. The phosphorylation of CPEB upon the hormonal stimulation of oocytes causes the dissociation of PARN, elongation of the poly(A) tail by TENT2, and consequently the release of Maskin from the masked mRNA (Figure 2c). Notably, however, the mammalian ortholog of Maskin, known as transforming acidic coiled‐coil‐containing protein 3 (TACC3), is a nonmotor microtubule‐associated protein that plays an important role in mitotic spindle stability and organization (O'Brien et al., 2005; Yao, Natsume, & Noda, 2007). The second model of CPEB action does not require Maskin, which interaction with CPEB was shown to be weak and probably not conserved (Minshall, Reiter, Weil, & Standart, 2007). Instead, CPEB is proposed to recruit the RNA helicase Xp54, RNA‐associated protein 55 (RAP55), and 4E‐T protein to interact with eIF4E1b. The presence of this complex is proposed to stop the translation of targeted mRNAs (Minshall et al., 2007).
The D. melanogaster genome encodes two homologs of TENT2/GLD‐2. The first homolog, Gld2 (CG5732), is highly enriched in the testes and acts during postmeiotic spermatogenesis (Sartain et al., 2011). Gld2‐deficient males do not produce mature sperm and are sterile (Sartain et al., 2011). Consistent with its role in fly spermatogenesis, Gld2 was found to interact with the GLD‐3 homolog BIC‐C in a yeast two‐hybrid assay (Sartain et al., 2011). Surprisingly, the enzymatic activity of Gld2 is also essential for long‐term memory formation in neurons (Jae et al., 2008). The second homolog, Wispy (CG15737), is expressed in the female germline and is required for fruit fly oogenesis (Benoit et al., 2008; Cui et al., 2008; Cui, Sartain, Pleiss, & Wolfner, 2013). Wispy polyadenylates the vast majority of maternal mRNAs, with the exception of ribosomal protein transcripts, during late oogenesis and upon egg activation (Benoit et al., 2008; Cui et al., 2008, 2013; Eichhorn et al., 2016; Lim et al., 2016). Interestingly, Wispy may not only activate these mRNAs but also protect their poly(A) tails from deadenylation. In wispy mutants, poly(A) tail lengths are shortened during late oogenesis (Lim et al., 2016). Although the homolog of CPEB, Orb, interacts with Wispy (Benoit et al., 2008), not all maternal mRNAs carry Orb‐binding sequences, suggesting that Orb is not the only regulator of cytoplasmic polyadenylation during oogenesis in flies (Figure 2d) (Cui et al., 2013; Ivshina et al., 2014). Indeed, Wispy polyadenylates the innate immunity mRNA Toll in a Dicer‐2‐dependent manner. Dicer‐2 is implicated in RNA interference (RNAi), but its role in cytoplasmic polyadenylation is independent of RNAi machinery (Coll et al., 2018). The PIWI protein Aubergine also acts as a Wispy‐recruiting factor to stabilize mRNAs in the Drosophila germ plasm (Dufourt et al., 2017). Wispy polyadenylates and stabilizes so‐called stable intronic sequence RNAs (sisRNAs) during fly oocyte development (Jia Ng, Zheng, Osman, & Pek, 2018). sisRNAs are long intronic sequences that are excised during splicing and present as extremely stable linear or lariat molecules with high regulatory potential (for review, see Chan & Pek, 2019). The exact function of sisRNAs in fruit flies is only beginning to be known, but some have been proposed to be required for proper fly larva development via the clearance of long noncoding RNAs (Jia Ng et al., 2018). The mechanism of Wispy recruitment to sisRNAs remains to be established.
In contrast to invertebrates and Xenopus, the involvement of mammalian TENT2 in gametogenesis and early embryo development is still not precisely known, since the disruption of the TENT2 gene has no effect on poly(A) tail elongation in mouse oocytes or on oocyte maturation (Nakanishi et al., 2007). Furthermore, TENT2‐deficient male and female mice are fertile and healthy (Nakanishi et al., 2007), raising the possibility that other TENTs might be involved in mRNA polyadenylation in the mammalian germline (Kashiwabara et al., 2002). TENT2 function has been better described in the mouse brain. TENT2 is ubiquitously expressed in the cerebellum and hippocampus and proposed to regulate synaptic plasticity (Rouhana et al., 2005). TENT2 activates over 100 mRNAs in mouse hippocampal dendrites in a mode that is similar to frog oocytes, in which synaptic stimulation induces the phosphorylation of CPEB, leading to protein rearrangement on masked mRNAs and the polyadenylation of these mRNAs by TENT2 (Udagawa et al., 2012). Among the most prominent targets of TENT2 in the hippocampus are mRNAs that encode the NR2A and GluN2A subunits of N‐methyl‐d‐aspartate receptors that are necessary for long‐term synaptic plasticity (Swanger, He, Richter, & Bassell, 2013; Udagawa et al., 2012). TENT2 knockout animals do not exhibit any behavioral abnormalities.
In somatic cells, other RNA‐binding proteins may recruit TENT2 to target mRNAs for polyadenylation and the activation of translation. For example, QKI‐7 specifically targets heterogeneous nuclear ribonucleoprotein A1 (hnRNPA1), p27, and β‐catenin mRNAs for TENT2 polyadenylation in human embryonic kidney 293 (HEK293) cells (Yamagishi, Tsusaka, Mitsunaga, Maehata, & Hoshino, 2016).
In addition to their role in mRNA expression, TENT2 and Wispy have been implicated in regulating the stability of certain mature miRNAs. TENT2 gene deletion in a human fibroblast cell line significantly reduced a fraction of monoadenylated miRNAs (D'Ambrogio, Gu, Udagawa, Mello, & Richter, 2012). The liver‐specific miRNA‐122 plays an essential role in liver development and metabolism and has been shown to be 3′ monoadenylated in both human hepatocytes and the mouse liver (Katoh et al., 2009). The level of miRNA‐122 is significantly lower in TENT2 knockout mice, suggesting that TENT2 monoadenylation enhances its stability (Katoh et al., 2009). Moreover, miRNA‐122 stabilization in hepatocytes is promoted by QKI‐7. Similar to HEK293 cells, QKI‐7 in hepatocytes recruits TENT2 for miRNA‐122 monoadenylation (Hojo et al., 2020). Another example of a TENT2‐stabilizing effect on miRNA‐122 was the observation that the hepatitis C virus core protein promotes miRNA‐122 decay by inhibiting TENT2 (Kim et al., 2016). Interestingly, the depletion of TENT2 adenylation has also been shown to did not affect miRNA stability (Burroughs et al., 2010; Mansur et al., 2016). Furthermore, an opposite effect of maternal miRNA adenylation was reported in fly early embryos (Lee et al., 2014). The deletion of wispy resulted in the accumulation of miRNAs, whereas its overexpression increased adenylation and reduced miRNA abundance, suggesting that Wispy‐mediated adenylation may promote miRNA decay and clearance during the maternal‐to‐zygotic transition. miRNA adenylation is proposed to be mediated by an interaction between Wispy and Argonaute 1, presenting yet another selective recruiting factor (Lee et al., 2014). Still unclear is what exactly specifies the adenylation effect on miRNA stability. One speculation is that the addition of one adenosine may stabilize miRNA, whereas a longer oligo(A) tail may recruit exonucleases and result in miRNA degradation. The processivity of TENT2 and adenylation effect on miRNA stability may also depend on the cell type, molecular context, and particular miRNA species.
2.2. TENT4A, TENT4B, GLD‐4, and Trf4‐1
Human TENT4A and TENT4B are orthologs of the yeast ncPAPs Trf4p and Trf5p (Houseley & Tollervey, 2006; San Paolo et al., 2009). In yeast, Trf4p, together with the zinc knuckle protein Air2p and RNA helicase Mtr4p, forms the Trf4/Air2/Mtr4p polyadenylation (TRAMP) complex, which recruits the nuclear exosome to degrade various RNA species (LaCava et al., 2005; Vaňáčová et al., 2005). TENT4A and TENT4B have a high level of similarity, but there is only partial overlap between their described functions (Burroughs et al., 2010; Lim et al., 2018; Lubas et al., 2011; Shcherbik, Wang, Lapik, Srivastava, & Pestov, 2010). TENT4A exists in two isoforms that differ in their N‐terminal region: TENT4A short (S) and the more prevalent TENT4A long (L) (Lim et al., 2018; Ogami, Cho, & Hoshino, 2013). Only TENT4A (L) can polyadenylate reporter mRNA in the tethering assay. TENT4A (L) localizes predominantly in the nucleus, and its unique N‐terminal region is crucial for the nuclear localization and activity. In contrast, inactive TENT4A (S) is distributed evenly in the cell (Ogami et al., 2013). The functions of TENT4A have not been extensively studied. TENT4A has not been shown to be a component of the human TRAMP complex, and unknown is whether it can form TRAMP‐like complexes (Fasken et al., 2011). The functional relevance of TENT4B is much closer to its yeast ortholog (Figure 3). TENT4B localizes to the nucleus, accumulates in the nucleolus, and forms a conserved TRAMP complex with the Air1/Air2p homolog ZCCHC7 and Mtr4p homolog SKIV2L2 (Fasken et al., 2011; LaCava et al., 2005; Lubas et al., 2011; Rammelt, Bilen, Zavolan, & Keller, 2011; Sudo, Nozaki, Uno, Ishida, & Nagahama, 2016; Weick et al., 2018). As a part of the TRAMP complex, TENT4B adds tails and promotes either the decay or maturation of various RNA species.
FIGURE 3.
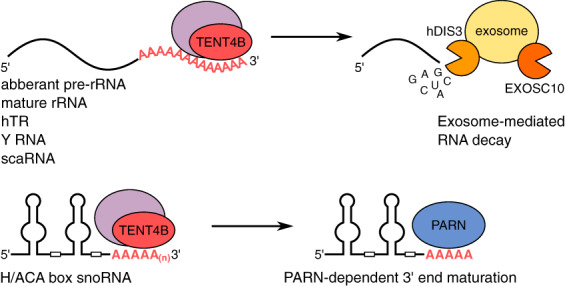
In the nucleus, polyadenylation by TENT4B/A, acting alone or in complex with other proteins, induces the exosome‐mediated decay of various RNA species (upper panel). TENT4B/A also cooperates with the poly(A)‐specific ribonuclease PARN to promote H/ACA box snoRNA maturation (bottom panel)
The role of TENT4B/Trf4p in rRNA surveillance is highly conserved between yeast and mammals (Dez, Houseley, & Tollervey, 2006; LaCava et al., 2005). TENT4B participates in the adenylation of aberrant and superfluous precursor rRNA (pre‐rRNA) fragments, leading to their degradation by the nucleolar exosome (Lubas et al., 2011; Shcherbik et al., 2010; Sudo et al., 2016). Additionally, TENT4B‐mediated polyadenylation contributes to the degradation of mature rRNAs in mouse hepatocytes. In mice, daily oscillations in liver mass and size result from regulated changes in the number and activity of ribosomes (Sinturel et al., 2017). During a diurnal cycle, pre‐rRNAs are continually synthesized, whereas ribosomal proteins are produced rhythmically, leading to a situation in which not all rRNAs are packed into mature ribosomes. Excess 18S and 28S rRNAs that are not incorporated into ribosomes are polyadenylated by TENT4B and degraded by the nuclear exosome (Sinturel et al., 2017). Another conserved function of TENT4B as part of the TRAMP complex involves formation of the mature 3′ end of small nucleolar RNAs (snoRNAs) (Grzechnik & Kufel, 2008; LaCava et al., 2005). In humans, TENT4B participates in the maturation of H/ACA box snoRNAs by adding oligo(A) tails to the processing intermediate after initial 3′–5′ exonucleolytic trimming of the excised intron that contains the snoRNA precursor (Figure 3). The oligo(A) tails, together with remaining intronic nucleotides of snoRNA, are then trimmed by PARN, which generates mature and stable snoRNAs (Berndt et al., 2012; Son, Park, & Kim, 2018).
The adenylation by TENT4B may influence the targeting of some miRNAs and, together with PARN and other 3′–5′ exoribonucleases, affect their stability (Burroughs et al., 2010; Rammelt et al., 2011; Shukla, Bjerke, Muhlrad, Yi, & Parker, 2019; Wyman et al., 2011). Similar to the TENT2‐mediated adenylation of miRNA, adenylation by TENT4 may have either a positive or negative impact on miRNA stability that likely depends on the cellular context and length of the added tail. Monoadenylation of the 3′ end of oncogenic miRNA‐21 marks it for direct degradation by PARN (Boele et al., 2014). Intriguingly, this observation is opposite to the monoadenylation effect of TENT2 that instead stabilizes miRNAs (D'Ambrogio et al., 2012; Hojo et al., 2020; Katoh et al., 2009). In a more complicated pathway, the oligoadenylation of miRNA‐25 and miRNA‐92 by TENT4B poises regulation of their stability to enable the tight and rapid modulation of their levels (Shukla et al., 2019). In such a scenario, PARN removes oligo(A) tails that protect these miRNAs from complete degradation by the 3′–5′ exonucleases DIS3‐like exonuclease 1 (DIS3L) and DIS3L2. As a result, these two ribonuclease activities of PARN versus DIS3L/DIS3L2 have an opposing outcome with regard to levels of miRNA that are sensitized by TENT4B (Shukla et al., 2019). Similar competition for the TENT4B‐oligoadenylated substrate between PARN and other exoribonucleases has been described for the human telomerase RNA (hTR) precursor (Roake et al., 2019; Shukla, Schmidt, Goldfarb, Cech, & Parker, 2016). The telomerase ribonucleoprotein component Dyskerin binds and stabilizes the hTR precursor, thereby promoting its maturation by the exosome. Dyskerin also controls mature hTR levels by recruiting TENT4B and PARN with the accumulation of oligoadenylated hTR that leads to its exosome‐mediated degradation (Roake et al., 2019; Shukla et al., 2016). TENT4B is also implicated in the removal of abundant RNA polymerase III (Pol III)‐transcribed Y RNAs (Shukla & Parker, 2017; Son et al., 2018). In humans, Y RNAs play a role in DNA replication, the DNA damage response, histone mRNA processing, and RNA quality control (Kowalski & Krude, 2015). Again, PARN can remove oligo(A) tails that are added by TENT4B, leading to the stabilization of these RNAs that are otherwise completely degraded by DIS3L (Shukla & Parker, 2017; Son et al., 2018). Finally, in a similar scenario, PARN and target of EGR1 protein 1 have also been shown to promote the maturation of small Cajal body‐specific RNAs by trimming their 3′ ends that contain oligo(A) tails that are added by TENT4B (Son et al., 2018). In summary, in all of the aforementioned cases, adenylation by TENT4B sensitizes the RNA molecule for both PARN and exosome activities but with opposing outcomes on its fate (i.e., stabilization and degradation, respectively). Such a mechanism probably allows better regulation of the levels of different RNA species.
Intriguingly, TENT4B may have a positive regulatory impact on mRNAs. Polyadenylation by TENT4B has been shown to stabilize TP53 mRNA (also known as p53) and modulate its translational competence. In the proposed model, TENT4B is recruited to its target mRNA through its interaction with CPEB (Burns, D'Ambrogio, Nottrott, & Richter, 2011). TENT4B may also mediate the CPEB‐dependent stabilization of several mRNAs that are involved in carbohydrate metabolism, including GLUT1 mRNA, which encodes the major glucose transporter (Shin, Paek, Ivshina, Stackpole, & Richter, 2017). However, other studies do not provide evidence of TENT4B‐CPEB interactions.
Another function that has been attributed to both TENT4B and TENT4A is the regulation of mRNA stability through mixed A/G tailing (Figure 4) (Lim et al., 2018). TENT4 ncPAPs have promiscuous nucleotide specificity. Although ATP is their predominant substrate, they could also incorporate non‐adenosine residues, with the strongest preference for guanosine triphosphate over UTP and cytidine triphosphate. TENT4 ncPAPs incorporate single guanosine residues along with adenosines while extending long poly(A) tails of mRNAs, protecting them from the deadenylating complex CCR4‐NOT, because this complex is unable to efficiently remove the guanidine residue (Lim et al., 2018). The depletion of either TENT4A or TENT4B does not affect guanylation, whereas the depletion of both led to its strong decrease, suggesting that TENT4A and TENT4B are redundantly responsible for these additions (Lim et al., 2018). However, the ways in which TENT4A and TENT4B select their mRNA targets, presumably in the nucleus, remain to be established. Consistent with the potential stabilizing role of TENT4A/B is the observation that TENT4A and TENT4B stabilize viral RNA that is transcribed from hepatitis B virus (HBV) and human cytomegalovirus (HCMV) through the mixed tailing of their poly(A) (Hyrina et al., 2019; Kim et al., 2020; Mueller et al., 2019). This process occurs independently of other components of the TRAMP complex (i.e., zinc finger CCHC domain‐containing protein 7 [ZCCHC7] and hMTR4). Instead, TENT4A/B forms a complex with ZCCHC14 (Hyrina et al., 2019; Kim et al., 2020). Transcripts of both HBV and HCMV carry the posttranscriptional regulatory element (PRE). ZCCHC14 recognizes and binds to the CNGGN pentaloop within the PRE of viral transcripts and recruits TENT4A/B (Kim et al., 2020). Mixed tailing prevents the rapid deadenylation of viral RNA and results in its stabilization (Kim et al., 2020).
FIGURE 4.
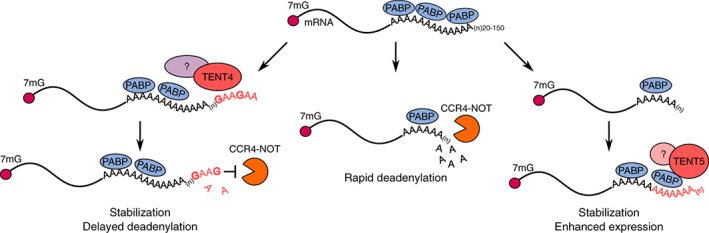
Stabilization of polyadenylated mRNAs. During their lifetime, the mRNA poly(A) tails become gradually shortened through CCR4‐NOT‐mediated deadenylation (center) that later leads to complete mRNA degradation from both ends (not shown). TENT4A/B enhances the stability of mRNAs by the mixed A/G tailing of their poly(A) tails (on the left). The guanosine residues in the terminal positions of poly(A) tails protect mRNA from deadenylation by CCR4‐NOT because CCR4‐NOT is unable to efficiently remove the guanidine residue. Proteins from the TENT5 family elongate mRNA poly(A) tails in the cytoplasm, ultimately extending the mRNA lifespan and enhancing its expression (on the right). Remaining unknown are the ways in which TENT4 and TENT5 ncPAPs recognize their target RNA and whether other protein cofactors facilitate their function
Drosophila has two homologs of yeast Trf4/5 ncPAPs that are expressed with the same pattern during development (Nakamura et al., 2008). Although both Trf4‐1 and Trf4‐2 bear the NTase domain, only Trf4‐1 has PAP activity in vitro. The knockdown or overexpression of trf4‐1 is lethal, whereas the disruption of trf4‐2 expression has no effect on the fly's viability. Trf4‐1, together with dmRrp6, participates in the polyadenylation‐mediated degradation of snRNAs in the nucleus (Nakamura et al., 2008). Additionally, Trf4‐1 has been proposed to facilitate exosomal 3′ mRNA decay in the cytoplasm (Harnisch et al., 2016).
In C. elegans, GLD‐4 is an ortholog of human TENT4B and TENT4A but appears to functionally diverge from these ncPAPs. GLD‐4 is expressed predominantly in the germline and localizes to the cytoplasm and P granules where its PAP activity relies on interactions with germline survival defective 1 (GLS‐1), an ortholog of human ZCCHC14 (Minasaki, Rudel, & Eckmann, 2014; Schmid et al., 2009). GLD‐4 is implicated in the regulation of some aspects of germline development through the cytoplasmic polyadenylation of mRNAs. However, in contrast to GLD‐2, the impact of GLD‐4 on bulk polyadenylation in the germline is mild, in which worms that lack gld‐4 exhibit only minor changes in mRNA poly(A) tail length (Nousch et al., 2014). The depletion of gld‐4 leads to no alterations of mRNA abundance, suggesting that GLD‐4 neither stabilizes nor promotes the degradation of mRNAs (Nousch et al., 2014). However, gld‐4 mutant worms exhibit fewer polysomes, and both GLD‐4 and GLS‐1 proteins co‐purify with them (Nousch et al., 2014). Based on these observations, the catalytic activity of GLD‐4 has been hypothesized to be required for efficient polysome formation. Alternatively, GLD‐4/GLS‐1 might assist with polysome formation independent of the enzymatic activity of GLD‐4 (Nousch et al., 2014). The role of GLD‐4 in nuclear RNA surveillance has not yet been assessed. However, the wide evolutionary aspects of this function and fewer polysomes in GLD‐4 mutants suggest that it may also be the case in worms.
In summary, multiple functions have been attributed to the TENT4 family of PAPs. Mechanistically, the best understood role is nuclear RNA surveillance with TENT4 as a part of the TRAMP complex that attracts the RNA‐degrading exosome complex. The stabilizing function of mixed tailing by TENT4A/B is a very intriguing possibility. However, more data are needed to understand this phenomenon and its role in mRNA metabolism.
2.3. TENT5 family
In mammals, TENT5, also known as family with sequence similarity 46 (FAM46), is a family of four highly similar (>40% overall sequence identity) proteins—TENT5A, TENT5B, TENT5C, and TENT5D—that are differentially expressed in tissues and organs (Kuchta et al., 2016). TENT5 proteins are conserved in metazoa, and variable numbers of TENT5 homologs are present in the genome of all animals (Kuchta et al., 2016). TENT5 proteins have been shown to act as cytoplasmic ncPAPs in functional in vitro and in vivo studies (Hu et al., 2020; Mroczek et al., 2017). The recently solved structure of TENT5B in Xenopus tropicalis revealed that TENT5 proteins share a two‐domain architecture that is comparable to other TENTs, although with some distinguishable properties (Hu et al., 2020). The N‐terminal NTase and C‐terminal helical domain of TENT5B form a large cleft where the highly conserved hG[GS] pattern and three catalytic residues [DE]h[DE]h and h[DE]h are located. The N‐terminal catalytic domain of xtTENT5B is prominently larger and contains more secondary elements compared with other PAPs. TENT5 proteins have been proposed to have the largest catalytic domain among all known PAPs. Another interesting characteristic of TENT5 proteins is that their overall fold resembles prokaryotic PAP/CCA adding enzymes. However, this similarity is only limited to the protein fold and not reflected by TENT5 activity, in which TENT5 proteins do not have CCA‐adding activity in vitro (Hu et al., 2020). Importantly, human TENT5B has almost exclusive preference for ATP incorporation, suggesting that TENT5 proteins are highly unlikely to perform mixed RNA tailing. Biochemical analysis revealed that human TENT5B prefers substrate RNAs that are adenosine‐rich close to the very 3′ end, suggesting that TENT5 proteins preferentially act on already polyadenylated mRNAs (Hu et al., 2020). The functional relevance of cytoplasmic polyadenylation by TENT5 ncPAP is only beginning to emerge because it is a relatively newly discovered family of TENTs (Figure 4) (Kuchta et al., 2016). Notably, despite a high level of similarity between TENT5 paralogs, they may have distinct functions.
2.3.1. TENT5A
In mice, TENT5A has strong expression in embryonic and adult tissues that undergo mineralization, suggesting its role in bone development and postnatal bone homeostasis (Diener et al., 2016). TENT5A‐deficient mice exhibit growth delay, a shorter body length, and severe skeletal abnormalities (Diener et al., 2016). Furthermore, variable number of tandem repeat polymorphisms in the second exon of the TENT5A gene are associated with a higher risk of large‐joint osteoarthritis in humans (Etokebe et al., 2015). Loss‐of‐function mutations of the human TENT5A gene have been reported in patients who suffer from an autosomal recessive form of osteogenesis imperfecta bone disease (Doyard et al., 2018). The mechanism of TENT5A function in bones is currently unknown. TENT5A has been reported to be a potential binding partner of SMAD proteins, which are important components of signaling pathways that are involved in bone development (Colland et al., 2004). However, the functional relevance of this potential interaction to bone development in mammals has not been assessed. Interestingly, TENT5A has been shown to interact with Smad1/Smad4 and regulate the bone morphogenetic protein signaling pathway during early differentiation in Xenopus embryos (Watanabe et al., 2018). TENT5A is expressed in the frog preplacodal ectoderm, a specialized region that differentiates into the anterior pituitary gland and sensory organs, and TENT5A knockdown causes abnormalities in eye formation and body pigmentation (Watanabe et al., 2018). TENT5A was previously shown to be expressed in ameloblast nuclei of tooth buds and potentially involved in the development of teeth and enamel production in mice (Etokebe et al., 2009). Moreover, TENT5A has been described as a negative regulator of leptin during a low‐calorie diet (Carayol et al., 2017). Leptin is a hormone that regulates energy balance by inhibiting hunger and diminishing fat storage in human adipocytes. TENT5A knockdown does not influence leptin gene expression but leads to the higher secretion of leptin (Carayol et al., 2017). Additionally, mutations of the TENT5A gene are associated with the human autosomal recessive eye disease retinis pigmentosa (Barragán et al., 2008; Lagali, Kakuk, Griesinger, Wong, & Ayyagari, 2002; Schulz, Goetz, Kaschkoetoe, & Weber, 2004). A variable number of tandem repeat polymorphism in TENT5A was connected with susceptibility to tuberculosis (Etokebe, Bulat‐Kardum, Munthe, Balen, & Dembic, 2014). The molecular mechanism of action of TENT5A in these processes and pathogenesis of these diseases is unknown.
2.3.2. TENT5B
As mentioned above, the structure of xtTENT5B protein has been solved, and the biochemical properties of TENT5B have recently been reported (Hu et al., 2020). However, few studies have described the functional relevance of this ncPAP. TENT5B has been proposed to serve as a potential molecular marker that may be useful for the diagnosis and prognosis of refractory lupus nephritis (Benjachat et al., 2015). The expression level of TENT5B is downregulated in patients with prostate cancer, and overexpression of the TENT5B gene inhibits prostate cancer cell proliferation in vitro and tumor growth in vivo (Liang et al., 2018).
Large‐scale transcriptomic analyses revealed that TENT5B is specifically expressed in human embryonic stem cells (hESCs) and human induced pluripotent stem cells, and its level is sharply downregulated during differentiation (Hu et al., 2020). In hESCs, TENT5B knockout is lethal, whereas knockdown leads to a lower abundance of newly synthesized proteins and the severely compromised viability of hESCs. A tempting speculation is that TENT5B is involved in the cytoplasmic polyadenylation of mRNA in the early stage of human embryogenesis (Hu et al., 2020).
2.3.3. TENT5C
TENT5C is one of the most frequently mutated genes in multiple myeloma (MM), which occurs in approximately 20% of all cases. MM is a malignant proliferation of bone marrow plasma cells, and the loss of TENT5C is associated with limited survival (Barbieri et al., 2016; Boyd et al., 2011; Chapman et al., 2011; Hu, Chen, & Wang, 2019; Lohr et al., 2014; Vikova et al., 2019; Walker et al., 2018). Further work revealed that TENT5C is a B‐cell lineage‐specific growth suppressor. Several MM cell lines that carried mutations of the TENT5C gene exhibited significantly lower growth and lower survival rates upon the expression of wild‐type TENT5C (Mroczek et al., 2017; Zhu et al., 2017). Primary B cells that were isolated from TENT5C knockout mice proliferated faster than B cells that were isolated from wild‐type animals (Mroczek et al., 2017). TENT5C overexpression decreased levels of the transcription factors interferon regulatory factor 4, CCAAT/enhancer‐binding protein, and Myc proto‐oncogene protein and upregulated the expression of immunoglobulin (Ig) light chain and genes that are involved in the unfolded protein response (Zhu et al., 2017). The most crucial observation is that the reintroduction of active TENT5C into MM cell lines leads to the broad polyadenylation and stabilization of mRNAs, with a strong preference for those that encode endoplasmic reticulum (ER)‐targeted secreted proteins (Mroczek et al., 2017). It appears that TENT5C plays a broad role in B cell biology. Cytoplasmic polyadenylation by TENT5C was recently shown to stimulate the humoral immune response (Bilska et al., 2020; Herrero et al., 2020). A global analysis of poly(A) tail distribution by nanopore direct RNA sequencing revealed that mRNAs that encode Igs carry shorter poly(A) tails and have lower steady‐state levels in B cells that are isolated from TENT5C knockout mice compared with wild‐type mice (Bilska et al., 2020). TENT5C‐deficient B cells exhibit a reduction of Ig mRNA and protein abundance and secrete fewer antibodies (Bilska et al., 2020; Herrero et al., 2020).
TENT5C is also associated with other tumors (Tanaka et al., 2017; Wan et al., 2017; Zhang, Yue, Jiang, Han, & Xin, 2017). TENT5C expression is significantly lower in hepatocellular carcinoma (HCC) than in normal liver tissue (Zhang et al., 2017). TENT5C has been reported to serve as a biomarker of HCC recurrence and mediate the antimetastatic effects of norcantharidin on HCC cells (Wan et al., 2017; Zhang et al., 2017). A significant reduction of TENT5C levels was observed in gastric cancer, in which low TENT5C levels were associated with a poor prognosis and a higher risk of metastasis or recurrence (Tanaka et al., 2017). TENT5C also acts as a tumor suppressor gene in squamous cell carcinoma of the lungs (Xia et al., 2018) and oral cells (Zhuang & Lu, 2018).
TENT5C has been shown to be strongly expressed in spermatids in the mouse testes and play an essential role in spermiogenesis (Zheng et al., 2019). TENT5C knockout in mice results in male sterility, characterized by the production of headless spermatozoa or spermatozoa with severely compromised fertilization ability (Zheng et al., 2019). The exact mechanism of TENT5C function during spermiogenesis is unknown.
2.3.4. TENT5D
TENT5D has strong polyadenylation activity in vitro, but only little data are available on the functional relevance of TENT5D. It has been proposed to be a testis cancer antigen, in which the TENT5D gene has a restricted expression pattern in the normal testis but is aberrantly expressed in testicular cancer (Bettoni et al., 2009). TENT5D is also overexpressed in the cerebral cortex in a mouse model of autism spectrum disorder (Hamilton et al., 2011).
2.4. MTPAP (TENT6) and DmMTPAP
MTPAP is the only known TENT that acts in mitochondria. It has an N‐terminal mitochondrial targeting sequence (residues 1–37) and localizes exclusively to this organelle where it contributes to the regulation of gene expression via the polyadenylation of mitochondrial RNAs (Nagaike, Suzuki, Katoh, & Ueda, 2005; Tomecki, Dmochowska, Gewartowski, Dziembowski, & Stepien, 2004). Structural and biochemical analyses indicate that MTPAP has strong substrate preference for ATP and acts as a dimer. Although it is rather self‐sufficient for substrate recognition, some proteins can stimulate its activity (Bai, Srivastava, Chang, Manley, & Tong, 2011; Chujo et al., 2012; Lapkouski & Hällberg, 2015; Wang et al., 2014). For example, leucine‐rich pentatricopeptide repeat‐containing protein (LRPPRC) stimulates the polyadenylation activity of MTPAP on sense mitochondrial mRNAs (mt‐mRNAs). Upon LRPPRC silencing, mt‐mRNAs have shorter poly(A) tails (Bratic et al., 2011; Chujo et al., 2012; Pajak et al., 2019; Wilson et al., 2014). Other mitochondrial enzymes, such as the RNA helicase SUV3 and RNA exoribonuclease PNPase, cooperate with MTPAP to regulate RNA metabolism in mitochondria (Wang et al., 2014). C. elegans appears to lack an ortholog of MTPAP. None of its closest homologs have been shown to reside in worm mitochondria, with no information about polyadenylation in C. elegans mitochondria. In contrast, the D. melanogaster genome encodes an ortholog of human MTPAP, DmMTPAP (CG11418). Despite only 30% homology between human and fly proteins, their functions and some properties are similar (Bratic et al., 2016). In the fruit fly, DmMTPAP is essential for mitochondrial function, fly development, and survival (Bratic et al., 2016). In humans, MTPAP is responsible for the polyadenylation of mitochondrial tRNAs and mRNAs. In D. melanogaster, mitochondrial rRNAs (mt‐rRNAs) have been shown to carry poly(A) tails. However, their functional relevance is not entirely clear (Bratic et al., 2016).
MTPAP is important for the processing and maturation of mt‐tRNAs in both humans and flies (Figure 5a). In human mitochondria, the genes that encode tRNATyr and tRNACys overlap by one nucleotide that results in a tRNATyr precursor that is devoid of 3′‐terminal adenosines. MTPAP is able to extend the tRNATyr transcript 3′ end with a few adenosines. In the case of monoadenylation, the tRNATyr precursor immediately becomes a substrate for the addition of CCA and further acetylation. If more adenosines are present, then either the deadenylase 2′,5′‐phosphodiesterase 12 (PDE12) or endonuclease RNase Z removes the excessive nucleotides, producing a substrate for the addition of CCA (Fiedler, Rossmanith, Wahle, & Rammelt, 2015; Pearce et al., 2017). Interestingly, in addition to the maturation of these two tRNAs, a spurious poly(A) tail could also be added by MTPAP to all other mt‐tRNAs, thus preventing their aminoacylation and leading to mitochondrial ribosome stalling and consequently a defect of the oxidative phosphorylation (OXPHOS) complex. PDE12 has been proposed to remove the spurious polyadenylation of mt‐tRNAs and restore the properly matured pool of mt‐tRNAs that are available for aminoacylation and correct translation (Pearce et al., 2017). In D. melanogaster, although genes that encode these tRNAs do not overlap, the loss of dmmtpap results in disturbances of maturation, a reduction of steady‐state levels of tRNACys, and incorrect CCA modification, indicating that polyadenylation by MTPAP is involved in the processing or stability of at least some mt‐tRNAs in flies (Bratic et al., 2016).
FIGURE 5.
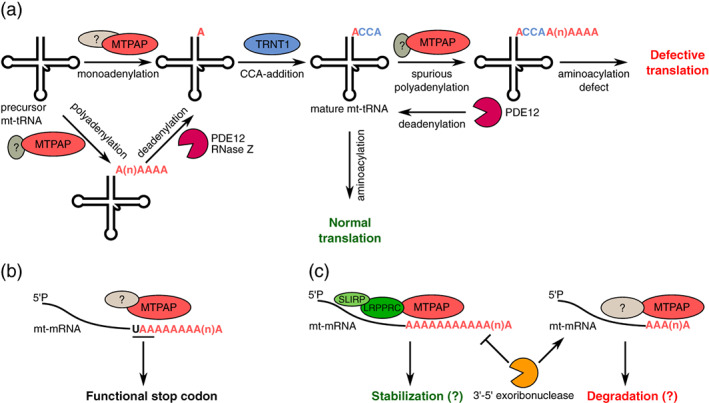
Mitochondrial poly(A) RNA polymerase (MTPAP) is the only TENT that acts in mitochondria. (a) MTPAP plays a role in the maturation of mt‐tRNAs through adenylation of the tRNA precursor, which generates a substrate for the addition of CCA and further acetylation. MTPAP also polyadenylates mature mt‐tRNAs, leading to defective translation. In both cases, the deadenylase PDE12 removes excessive adenosines. (b) MTPAP generates a complete UAA stop codon and functional ORFs for mt‐mRNA, for which full termination codons are not encoded in the mitochondrial genome. (c) The effect of polyadenylation on mt‐mRNA stability and turnover may be cofactor‐ and/or transcript‐dependent and is not completely understood. In humans and flies, the leucine‐rich pentatricopeptide‐repeat containing protein (LRPPRC)/stem‐loop‐interacting RNA‐binding protein (SLIRP) complex stimulates the polyadenylation activity of MTPAP and protects mRNA from degradation
In humans, 7 of 13 mitochondrial genome‐encoded mRNAs (ND1, ND2, ND3, ND4, CytB, COIII, and ATP6) have incomplete stop codons with either a terminal U or UA because full termination codons are not encoded in the mitochondrial genome. For these mt‐mRNAs, adenylation by MTPAP is essential to generate a complete UAA stop codon and thus functional ORFs (Figure 5b) (Anderson et al., 1981; for review, see Borowski, Szczesny, Brzezniak, & Stepien, 2010; Gagliardi, Stepien, Temperley, Lightowlers, & Chrzanowska‐Lightowlers, 2004; Nagaike, Suzuki, & Ueda, 2008). Importantly, these mt‐mRNAs are essential, and in the case of MTPAP deficiency, these mt‐mRNAs would encode defective proteins that are unable to integrate into the OXPHOS complex. Apart from the generation of complete stop codons of mt‐mRNAs, MTPAP also polyadenylates mature mRNAs in mitochondria (Tomecki et al., 2004). The effect of poly(A) tails on mt‐mRNA stability, turnover, and translation has been a topic of discussion (for review, see Borowski et al., 2010; Gagliardi et al., 2004; Nagaike et al., 2008; Wilson et al., 2014). The downregulation of MTPAP leads to the shortening of mt‐mRNA poly(A) tails but also results in diverse effects on mt‐mRNAs, with some transcripts that are upregulated, some that are unaffected, and some that are downregulated (Nagaike et al., 2005; Rorbach, Nicholls, & Minczuk, 2011; Tomecki et al., 2004). These results suggest that the effects of MTPAP are mt‐mRNA‐specific, and individual transcripts may be differentially regulated, likely through additional factors (Figure 5c).
In fruit fly larvae, deletion of the dmmtpap gene leads to a severe reduction of mitochondrial polyadenylation, with all mt‐mRNAs having trimmed 3′ ends (Bratic et al., 2016). Surprisingly, despite the lack of functioning 3′ ends of mt‐mRNAs, these mRNAs were translated but led to the production of incomplete and defective proteins that could not be integrated into the correct complexes (Bratic et al., 2016). Interestingly, the loss of SUV3, PNPase, or MTPAP leads to the accumulation of mitochondrion‐derived antisense double‐stranded RNA in the cytoplasm of cells, which is associated with alterations of the immune response in flies (Pajak et al., 2019). However, whether polyadenylation is required for the degradation of these antisense RNAs and other RNA species in D. melanogaster remains to be established.
MTPAP has also been implicated in mitophagy, a process that involves the selective autophagic degradation of defective or damaged mitochondria (Furuya et al., 2018). The autophagy receptor NDP52 enters depolarized mitochondria and, together with MTPAP, forms an oligomeric autophagy receptor complex that presumably detects damaged mitochondria and facilitates their elimination. The MTPAP‐NDP52 interaction itself and not the catalytic activity of MTPAP has been proposed to be important for mitophagy (Furuya et al., 2018).
The disruption of RNA polyadenylation in human mitochondria that is caused by mutations of MTPAP has been linked to some genetic disorders. Notably, all disease‐related mutations have been identified within the fingers domain, which is a highly conserved region of MTPAP. Importantly, none of the identified mutations appears to affect the expression, oligomeric state, or cellular localization of MTPAP. The N478D mutation of MTPAP is associated with an autosomal‐recessive disease (i.e., severe progressive spastic ataxia with optic atrophy (Crosby et al., 2010). In individuals who carry the homozygous N478D mutation, the poly(A) tails of mt‐mRNAs are shorter compared with healthy individuals. Although the effect of tail shortening on mt‐mRNA stability is transcript‐dependent, it leads to alterations of mitochondrial protein expression and an OXPHOS defect (Wilson et al., 2014). The same homozygous N478D mutation causes a distinct cellular phenotype that is characterized by DNA damage, impairments in DNA repair kinetics, and an increase in cell death following exposure to ionizing radiation (Martin et al., 2014). Other rare missense mutations of MTPAP (i.e., heterozygous I428T and R523W mutations and the homozygous I385F mutation) have been associated with autosomal recessive perinatal lethal encephalopathy (Van Eyck et al., 2019). For N478D, these mutations also lead to a decrease in the polyadenylation of mitochondrial transcripts and alterations of the expression of mitochondrially encoded proteins.
3. URIDYLATION
RNA uridylation, although discovered much later than noncanonical polyadenylation, is a widespread posttranscriptional modification that targets various coding and noncoding RNAs in mammals, plants, trypanosomes, fission yeast, worms, fruit flies, and other organisms. TENT1/TUT1, TUT4/TENT3A, and TUT7/TENT3B are mammalian TUTases that mediate template‐independent uridylation. The functional ortholog of TENT1 in worms is USIP‐1, but its counterpart in flies is unknown. Tailor is a TUTase in fruit flies, which has functions that are reminiscent of TUT4/7 (Bortolamiol‐Becet et al., 2015). Worms also have two orthologs of TUT4/7, also known as poly(U) polymerases (PUPs): CDE‐1 (also known as PUP‐1 and CID‐1) and its paralog PUP‐2. The third TUTase, PUP‐3, is only distantly related to TUT4/7 (Jae & Wickens, 2007). C. elegans also has MUT‐2, the first enzyme that was shown to have the ability to add UG tails. This section summarizes our current knowledge about all known metazoan enzymes that drive RNA uridylation.
3.1. TENT1 (TUT1) and USIP‐1
In mammals, TENT1 is widely expressed in all tissues and essential for cell proliferation and survival (Hart et al., 2015; Trippe et al., 2006). TENT1 has a complex structure that consists of different domains that are distinct from those that are present in other TENTs. It comprises an N‐terminal zinc finger (ZF), an RNA‐recognition motif (RRM) followed by a palm domain that contains a proline‐rich region, fingers, a kinase‐associated‐1 (KA‐1) domain, and a C‐terminal nuclear localization signal (Yamashita, Takagi, Nagaike, & Tomita, 2017). Notably, TENT1 is the only member of the TENT family that carries the RRM and possibly does not rely on other RNA‐binding proteins for substrate specificity. TENT1 is localized to the nucleolus, nucleoplasm, and nuclear speckles (Mellman et al., 2008; Trippe et al., 2006).
Uridylation by TENT1 plays an essential role in the maturation of spliceosomal U6 snRNA that is responsible for the catalysis of pre‐mRNA splicing (Figure 6) (Hirai, Lee, Natori, & Sekimizu, 1988; Trippe et al., 2006; Trippe, Richly, & Benecke, 2003). U6 snRNA is transcribed by Pol III, which uses a short stretch of thymine nucleotides in DNA as a termination signal that results in four encoded uridines at the 3′ end of the U6 transcript (Lund & Dahlberg, 1992). Terminal uridines attract the chaperone‐like La protein, which stabilizes the U6 precursor (Pannone, Xue, & Wolin, 1998). To become fully functional, U6 snRNA undergoes further 3′ end maturation that starts with the removal of La protein and oligouridylation by TENT1 (Hirai et al., 1988; Trippe et al., 2003, 2006). Oligo(U) tails are subsequently trimmed by the 3′–5′ exoribonuclease USB1 until the tail contains only five uridines (Didychuk et al., 2017; Mroczek et al., 2012; Shchepachev, Wischnewski, Missiaglia, Soneson, & Azzalin, 2012; Shchepachev, Wischnewski, Soneson, Arnold, & Azzalin, 2015). USB1 also has very specific exoribonuclease activity that generates a 2′,3′‐cyclic phosphate at the RNA 3′ end—a hallmark of U6 snRNA that has a stabilizing effect by protecting it from oligouridylation‐mediated decay (Didychuk et al., 2017; Lund & Dahlberg, 1992; Mroczek et al., 2012; Shchepachev et al., 2012, 2015). The uridylation of U6 snRNA also plays an indirect important role in pre‐mRNA splicing by recruiting the LSM2‐8 protein complex to the 3′ end of U6 snRNA that facilitates the annealing of U6 snRNA with U4 snRNA and formation of the snRNP complex that is crucial for the splicing reaction (for review, see Didychuk, Butcher, & Brow, 2018).
FIGURE 6.
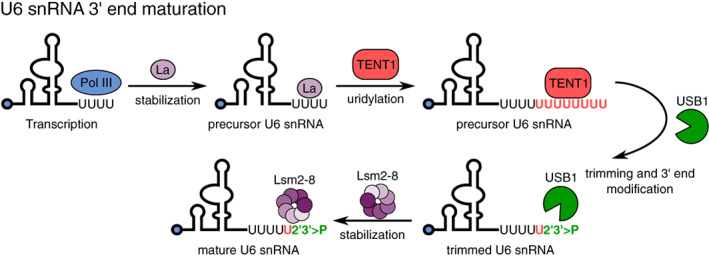
Maturation of U6 snRNA. Following transcription by Pol III, La protein protects the U6 snRNA precursor by binding the four uridines at the 3′ end. La protein is later replaced by TENT1, which uridylates the U6 snRNA 3′ end. The 3′‐5′ exoribonuclease and phosphodiesterase USB1 removes uridines, leaving only five of them, and a terminal 2′,3′ cyclic phosphate (2′3′ > P). The U6 snRNA is further protected by recruitment of the Lsm2‐8 protein complex. The blue dot at the 5′ end represents the γ‐monomethylguanosine triphosphate (meGTP) cap
The function of TENT1 is highly conserved among vertebrates, with some preservation of the degree of functional conservation among invertebrates. Similar to humans, U6 snRNAs in worms have four or more uridines at the 3′ end (depending on the U6 snRNA gene isoform, in which four to five uridines are encoded in the DNA), and these ends are also protected at their 3′ termini. Intriguingly, the identity of the protective moiety/modification remains unknown, in which U6 snRNAs in worms do not carry 2′,3′‐cyclic‐, 3′‐monophosphate, or 2′‐O‐methylation blocking groups (Lund & Dahlberg, 1992; Rüegger, Miki, Hess, & Großhans, 2015). In C. elegans, the closest ortholog of TENT1 is USIP‐1 (U6 snRNA‐interacting protein‐1, ZK863.4), which is involved in regulating U6 snRNA stability and function (Rüegger et al., 2015). In usip‐1‐deficient mutants, the level of U6 snRNA is significantly decreased, in which molecules that have 3′ ends that cannot be extended to their normal 4–5 uridines are rapidly degraded (Rüegger et al., 2015). However, unclear is whether USIP‐1 directly participates in U6 snRNA biogenesis and stabilization because USIP‐1 interacts both physically and genetically with the bona fide U6 snRNA recycling factor squamous cell carcinoma antigen recognized by T‐cells 3.
There is some controversy about the substrate preference and function of TENT1. It was reported to function as a highly processive nuclear ncPAP called speckle targeted phosphatidylinositol‐4,5‐bisphosphate regulated PAP (Star‐PAP) (Mellman et al., 2008). Star‐PAP has been proposed to affect the expression of oxidative stress and apoptotic response genes by regulating the 3′ end formation of their mRNAs (Laishram & Anderson, 2010; Li et al., 2012; Li, Laishram, & Anderson, 2013; Mellman et al., 2008). Such a duality of TENT1 action could be explained by TENT1 substrate specificity and activity that are tightly regulated by its subnuclear localization, the presence of specific signals in RNA substrates, or the regulation of TENT1 through posttranslational modifications. Importantly, however, recent structural and biochemical studies revealed higher preference of TENT1 for uridylation over adenylation (Yamashita et al., 2017).
3.2. TUT4/7, CDE‐1, PUP‐2, PUP‐3, and Tailor
Human TUT4 and TUT7 and their homologs are major mediators of uridylation that target a wide variety of coding and noncoding RNAs and RNA decay intermediates (De Almeida et al., 2018; Menezes et al., 2018). Uridylation is mostly linked to RNA surveillance and decay. However, depending on the substrate RNAs, cofactors, and the cellular context, uridylation may also be a crucial step of RNA biogenesis and maturation (Table 1 and Figure 7). TUT4 and TUT7 are proposed to act redundantly, and their functions are almost always described together, referring to them as TUT4/7. Human TUT4/7 and worm CDE‐1 are large multidomain enzymes, whereas other worm TUTases, such as PUP‐2 and PUP‐3, and the D. melanogaster TUTase Tailor are smaller and less complex (Figure 1).
FIGURE 7.
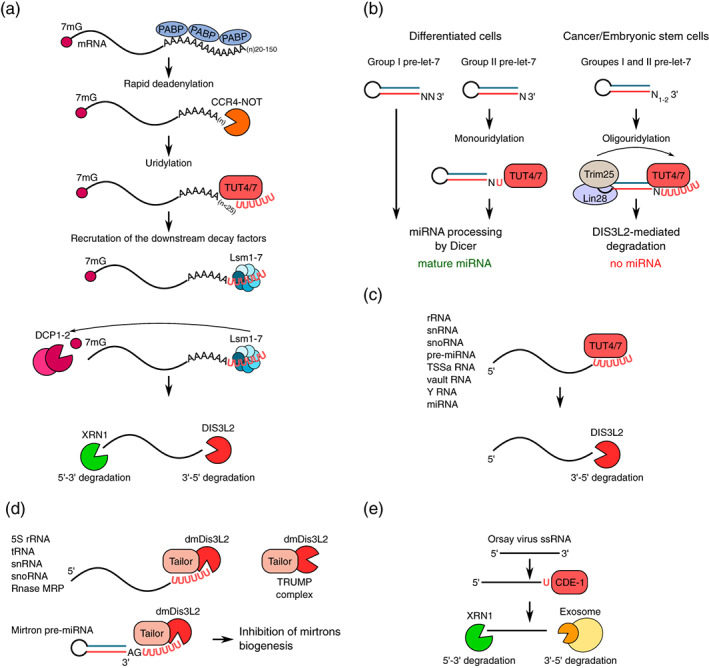
Selected uridylation‐dependent processes in humans, worms, and flies. (a) Uridylation‐mediated degradation of polyadenylated mRNAs in humans. After deadenylation, mRNAs with poly(A) tails that are shorter than 25 nt are uridylated by TUT4/7, leading to the recruitment of downstream RNA decay factors. The LSM1‐7 complex first interacts with the U‐tail and promotes mRNA decapping by the DCP1‐2 complex. mRNAs that are devoid of a cap are then degraded by the 5′–3′ exonuclease XRN1. Additionally, mRNA can be degraded by the exosome and/or DIS3L2 exonuclease in the 3′‐5′ direction. (b) Pre‐miRNA let‐7 uridylation by TUT4/7. Following their transcription and initial processing, the precursor miRNA (pre‐miRNA) are exported into the cytoplasm (not shown). In differentiated cells where Lin28 is absent, group II pre‐miRNAs that carry a one‐nucleotide 3′ overhang are monouridylated by TUT4/7, thus enabling further processing by Dicer. In cancer and embryonic stem cells, Lin28 binds both groups of let‐7 pre‐mRNAs and recruits TUT4/7. Dicer cannot process oligouridylated pre‐miRNAs, and they are degraded by the 3′‐5′ exonuclease DIS3L2. (c) Various RNA species that are transcribed by Pol I, Pol II, and Pol III can be targeted to the 3′–5′ exosome and/or DIS3L2 exonuclease degradation in a uridylation‐mediated manner. (d) In Drosophila, the decay of multiple RNAs relies on uridylation by Tailor, which forms a complex with the 3′–5′ exonuclease dmDis3L2 (a so‐called TRUMP complex). The stability of mirtrons is particularly controlled by uridylation. Mirtrons arise during splicing and lariat debranching of introns and carry AG at their 3′ ends. The 3′‐AG is uridylated by Tailor, which inhibits their biogenesis and leads to degradation by dmDis3L2. (e) Antiviral RNA uridylation in worms. Upon infection, Orsay virus RNAs are monouridylated by CDE‐1, which promotes their degradation by the 5′–3′ exonuclease XRN1 and 3′–5′ exonucleases of the exosome
Uridylation has been studied in great detail in the context of pre‐miRNA processing in both worms and humans (Figure 7b). In C. elegans, let‐7 miRNA regulates several transcription factors in the progression from larvae to adults; thus, its expression is tightly controlled to ensure a proper transition (Pasquinelli et al., 2000; Reinhart et al., 2000). The conserved RNA‐binding protein LIN‐28 binds pre‐let‐7, preventing its processing by Dicer. PUP‐2 uridylates pre‐let‐7 in a LIN‐28‐dependent manner, tagging it for degradation (Lehrbach et al., 2009). The mechanism is conserved in other species. In humans, the let‐7 miRNA family suppresses cell proliferation and promotes differentiation (Lee, Han, Kwon, & Lee, 2016). Furthermore, in differentiated cells, let‐7 suppresses several oncogenes and thus is repressed in some cancers (Balzeau, Menezes, Cao, & Hagan, 2017; Menezes et al., 2018). TUT4/7 plays a dual role in controlling let‐7 levels, either promoting the maturation of pre‐let‐7 or marking it for degradation. In humans, only 3 of 12 members of the let‐7 family carry the typical two‐nucleotide 3′ overhang in their precursors (group I pre‐miRNAs) and are directly processed by Dicer, whereas the rest have one nucleotide 3′ overhang (group II pre‐miRNAs) and require an additional step for their maturation (Bartel, 2018). In differentiated cells, monouridylation by TUT4/7 restores the two‐nucleotide 3′ overhang of group II pre‐let‐7, making them a substrate for Dicer and leading to effective let‐7 maturation (Heo et al., 2012; Kim et al., 2015). Importantly, this distributive activity of TUT4/7 at least partially arises from the absence of LIN28a in differentiated cells because LIN28a is a TUT4/7 positive processivity factor (Faehnle, Walleshauser, & Joshua‐Tor, 2017). In nondifferentiated cells and some cancers, LIN28a blocks let‐7 biogenesis to ensure pluripotency and self‐renewal (Hagan, Piskounova, & Gregory, 2009; Heo et al., 2009; Thornton, Chang, Piskounova, & Gregory, 2012). In this case, TUT4/7 oligouridylates pre‐let‐7, leading to its degradation by DIS3L2 (Faehnle, Walleshauser, & Joshua‐Tor, 2014; Hagan et al., 2009; Heo et al., 2009; Piskounova et al., 2011; Ustianenko et al., 2013, 2018). In addition to LIN28a, the E3 ligase tripartite motif‐containing protein 25 (Trim25) may stimulate the TUT4‐mediated oligouridylation of pre‐let‐7 (Choudhury et al., 2014).
In D. melanogaster, TUTase Tailor prevents the processing and promotes the degradation of mirtrons (Figure 7d) (Bortolamiol‐Becet et al., 2015; Reimão‐Pinto et al., 2015). Mirtrons are miRNA precursors that are located in introns of mRNA‐coding genes. Mirtrons rely on splicing and intron‐debranching machinery to generate short hairpins that end in AG, which are later processed by Dicer to produce mature miRNAs (Westholm & Lai, 2011). In fruit flies, mirtrons with AG‐ending hairpins are a preferred substrate for Tailor (Cheng et al., 2019; Kroupova, Ivascul, Reimao‐Pinto, Ameres, & Jinek, 2019). Uridylation promotes their degradation by dmDis3L2 and prevents the accumulation of mature miRNAs (Bortolamiol‐Becet et al., 2015; Reimão‐Pinto et al., 2015). Although mirtrons are also present in worms and humans, unknown is whether uridylation is involved in their decay.
Unclear are whether and how uridylation influences the activity and abundance of mature miRNAs. TUT4/7 has been reported to participate in the uridylation‐mediated degradation of mature miRNAs during differentiation and in response to environmental changes (Gutiérrez‐Vázquez et al., 2017; Jones et al., 2012; Thornton et al., 2014). Uridylation by TUT4/7 can also modulate target recognition, enhancing the repertoire of mRNAs that are regulated by a particular miRNA (Yang et al., 2019).
TUT4/7 also targets other RNA species for DIS3L2‐mediated decay, including aberrant snRNA, snoRNA, rRNA, 7SL, tRNA, Y and vault RNA, and Pol II transcription start site‐associated short RNA that are generated from bidirectional promotors (Figure 7c) (Łabno et al., 2016; Pirouz, Du, Munafò, & Gregory, 2016; Pirouz, Munafò, Ebrahimi, Choe, & Gregory, 2019; Ustianenko et al., 2016). This function is conserved in flies. Tailor, together with dmDis3L2, forms the cytoplasmic terminal RNA uridylation‐mediated processing complex that mediates cytoplasmic quality control and the degradation of various Pol III transcripts, such as 5S rRNA, tRNA, snRNA, snoRNA, and RNase MRP (Figure 7d) (Reimão‐Pinto et al., 2016).
In humans, TUT4/7 also regulates the lifespan of mRNAs. The TUT4/7‐mediated uridylation of replication‐dependent histone mRNAs recruits exosome to carry their degradation (for review, see Marzluff & Koreski, 2017). These mRNAs carry a stabilizing 3′ stem‐loop instead of a poly(A) tail. At the end of the S‐phase or during the inhibition of replication, histone mRNAs are no longer needed and are rapidly degraded in the TUT7‐mediated process. Interestingly, the activity of TUT4 in this process appears to play only a minor role, providing a rare example that TUT4 and TUT7 do not always act together (Lackey, Welch, & Marzluff, 2016). In addition to histone mRNAs, uridylation also initially targets polyadenylated mRNAs (Chang et al., 2014; Lim et al., 2014; Rissland & Norbury, 2008). In the cytoplasm, the poly(A) tails of mRNAs gradually become shortened by deadenylases until they can no longer accommodate PABPC (less than 25 nt). Such unprotected, short poly(A) tails are selectively uridylated by TUT4/7, triggering mRNA degradation from both ends (Figure 7a) (Chang et al., 2014; Lim et al., 2014; Malecki et al., 2013). The same mechanism occurs for mRNAs that are targeted for nonsense‐mediated decay in eukaryotes (Kurosaki, Miyoshi, Myers, & Maquat, 2018). Recent work with a conditional knockout mouse model strongly suggests that the TUT4/7‐mediated regulation of mRNA stability is particularly important during gametogenesis and early embryonic development (Chang et al., 2018; Morgan et al., 2019, 2017). The deletion of TUT4 and TUT7 leads to infertility and alterations of the transcriptome of germ cells, whereas the effect on somatic cells is minimal. Interestingly, in D. melanogaster, mRNAs and their degradation intermediates appear to not be frequently uridylated, suggesting that, in contrast to mammals, uridylation may not be the major trigger for their decay (Chang et al., 2014).
Finally, TUT4/7 also plays a role in the restriction of human LINE‐1 retrotransposition by the uridylation‐dependent degradation of LINE‐1 mRNA (Warkocki, Krawczyk, et al., 2018). LINE‐1 is a group of abundant vertebrate retrotransposons that proliferate through a “copy and paste” mechanism (Faulkner & Garcia‐Perez, 2017). LINE‐1 proliferation involves transcription and reintegration into a new site within genomic DNA by a so‐called target‐primed reverse transcription (TPRT) mechanism, potentially leading to de novo mutations of the germline, neurons, and cancers (Beck, Garcia‐Perez, Badge, & Moran, 2011; Scott et al., 2016). The uridylation of LINE‐1 mRNA enhances its degradation and blocks the initiation of reverse transcription during TPRT (Warkocki, Krawczyk, et al., 2018).
In C. elegans, uridylation by CDE‐1 is important in the regulation of various processes. In embryos, CDE‐1 localizes to mitotic chromosomes, where it interacts with the RNA‐dependent RNA polymerase EGO‐1 to produce endogenous small‐interfering RNAs (endo‐siRNAs) that are incorporated into the Argonaute (Ago) protein CSR‐1. CDE‐1 uridylates these siRNAs to limit their accumulation (van Wolfswinkel et al., 2009). Worms that lack cde‐1 exhibit an increase in endo‐siRNA levels and defects in both mitotic and meiotic chromosome segregation, likely because of the inappropriate loading of siRNAs into pathways that are mediated by other Ago proteins. Thus, CDE‐1 protects the transcriptome against excessive EGO‐1‐generated siRNAs and restricts them to the CSR‐1‐mediated RNAi pathway (van Wolfswinkel et al., 2009). A related process has been demonstrated in the fission yeast Schizosaccharomyces pombe, in which spurious small RNAs are eliminated to prevent the unintended silencing of euchromatin genes (Pisacane & Halic, 2017). CDE‐1 was also identified in the forward genetic screen as a gene that is required for RNAi inheritance, a phenomenon in which the progeny of animals that are exposed to double‐stranded RNA continue to silence genes that are targeted by these RNAs in previous generations (Spracklin et al., 2017). The role of CDE‐1 in long‐term gene silencing needs further investigation, but CDE‐1 likely uridylates siRNAs that are associated with the Ago protein WAGO‐4 that promotes the transmission of siRNAs for RNAi inheritance (Spracklin et al., 2017; Xu et al., 2018).
In C. elegans, the balance between CDE‐1, PUP‐2, and PUP‐3 activity is essential for germline development (Li & Maine, 2018). The lack of CDE‐1 and PUP‐2 allows the expression of soma‐specific genes in the germline, leading to severe defects in development and reproduction. Moreover, under temperature stress, cde‐1/pup‐2 double‐mutant worms do not maintain the identity of the germline and exhibit transgenerational germ cell loss. However, the identity of direct targets of CDE‐1 and PUP‐2 is unknown. Interestingly, PUP‐3 levels increased in the cde‐1/pup‐2 double‐mutant germline, indicating that CDE‐1 and PUP‐2 may regulate PUP‐3 levels. The loss of pup‐3 partially rescues the germline phenotypes in the cde‐1/pup‐2 mutant. PUP‐3 targets are most likely distinct from CDE‐1 and PUP‐2 substrates, and PUP‐3 likely exerts an opposite effect on germline development. The authors proposed that the coordinated activity of these three enzymes guarantees the proper abundance of their RNA targets, which impacts gene expression in the developing germline (Li & Maine, 2018).
CDE‐1 and human TUT4/7 play a critical role in antiviral immunity (Le Pen et al., 2018). In worms during infection with Orsay virus, CDE‐1 uridylates the 3′ end of the viral RNA genome and promotes its degradation (Figure 7e). The depletion of diverse RNA decay enzymes, such as those that are encoded by dis‐3, exos‐2, and xrn‐2, led to the accumulation of uridylated viral RNAs, indicating their contribution to viral RNA removal. Human TUT4/7 enzymes mark influenza A virus mRNAs for degradation, thus decreasing the rate of infection (Le Pen et al., 2018). In worms, the uridylation pathway acts in parallel with RNAi to fight RNA viruses. In mammals, the uridylation pathway appears to complement innate and adaptive immune responses. In other organisms, many exogenous RNAs of viral origin are oligouridylated, suggesting that uridylation is a conserved antiviral mechanism (Huo et al., 2016; Yeo & Kim, 2018).
3.3. MUT‐2
MUT‐2 (also known as RDE‐3) is a TENT that is homologous to GLD‐2, MTPAP, and CDE‐1. It was first identified in a screen of mutants with a higher frequency of transposon Tc1 excision in the C. elegans germline (Collins, Saari, & Anderson, 1988). Since then, it has been implicated in transposon silencing and the enhancement of RNAi (Chen et al., 2005; Gu et al., 2009; Tsai et al., 2015). The latest findings show that MUT‐2 can add long stretches that contain tandem UG repeats (pUG tails) both in vitro and when tethered to mRNA in heterologous expression systems (Preston et al., 2019). Furthermore, in vivo, MUT‐2 has been reported to add pUG tails to transposon RNAs and to other targets of RNAi (Shukla et al., 2020). Such extension leads to the recruitment of RNA‐dependent RNA polymerase that directly synthesizes siRNAs. In turn, these small RNAs help target MUT‐2 to its substrates, which creates a self‐reinforcing loop that enables RNA‐based transgenerational inheritance (Shukla et al., 2020).
4. OUTLOOK
As summarized in the present review, metazoan TENTs are crucial for homeostasis in animals through the regulation of gene expression at the posttranscriptional level. RNA tailing clearly participates in many biological processes, although we are only at the beginning of exploring its significance. Thus, much more needs to be discovered.
An important aspect of TENT function that is critical for further progress in the field is to understand the molecular mechanisms that underlie the specificity of TENT enzymes. TENTs often rely on interacting RNA‐binding proteins to identify their target mRNAs (Martin & Keller, 2007). In worms, GLD‐2 substrate selectivity is provided by interactions with the RNA‐binding proteins GLD‐3 and RNP‐8. Interestingly, the GLD‐2/GLD‐3 and GLD‐2/RNP‐8 complexes modify distinct pools of mRNAs and control gamete identity by promoting spermatogenesis or oogenesis, respectively (Kim et al., 2009, 2010; Nakel et al., 2016). In Drosophila, Wispy relies on the CPEB homolog Orb and other RNA‐binding proteins, such as Dicer‐2 and Aubergine, for target RNA recognition (Benoit et al., 2008; Coll et al., 2018; Cui et al., 2013; Dufourt et al., 2017). In Xenopus, cytoplasmic polyadenylation by TENT2 is CPEB‐dependent (for review, see Radford et al., 2008; Reyes & Ross, 2016). In mammals, the situation is more complicated. Although TENT2 knockout mice have no discernible phenotype, knockouts of CPEB family members result in a plethora of different phenotypes, including gametogenesis defects (Ivshina et al., 2014; Maillo et al., 2017). Thus, two alternative scenarios are possible. CPEBs may cooperate with other TENTs or CPEB performs biological functions that are not directly related to cytoplasmic polyadenylation. Further research is needed to resolve this issue. TENT4A/B acts in complex with other proteins (e.g., TRAMP) to target RNA for exosome‐mediated degradation (Fasken et al., 2011; LaCava et al., 2005) but can also be recruited to other substrates, such as viral RNAs, by ZCCHC14 (Hyrina et al., 2019; Kim et al., 2020). TUT4/7 substrate specificity is highly dependent on the molecular context. For the uridylation of mRNA, a reduction of poly(A) tail length to less than 20 nt appears to be a sufficient trigger (Lim et al., 2014). In the case of the LINE‐1 retrotransposon, this process requires cooperation with the helicase Moloney leukemia virus 10 (MOV10) (Warkocki, Krawczyk, et al., 2018). However, for other RNA species, TUT4/7 relies on many RNA binding cofactors and other target selectivity mechanisms (Choudhury et al., 2014; Faehnle et al., 2014; Hagan et al., 2009; Heo et al., 2009). Finally, MTPAP does not appear to require any interaction partner to polyadenylate mRNAs. The same is true for the uridylation of U6 snRNA by TENT1. For other TENT families, such as TENT5, the mechanism of substrate selection remains to be established.
Another issue is the nucleotide specificity and processivity of enzymes. Despite quite intensive efforts, the molecular mechanisms that underlie the selection of nucleotides by TENTs are not fully understood. Some explanations have been proposed for uridines over adenosines selection by TUTases, such as TUT7, Tailor, and lower eukaryotic counterparts (Kobylecki, Kuchta, Dziembowski, Ginalski, & Tomecki, 2017; Munoz‐Tello, Gabus, & Thore, 2012, 2014; Yates et al., 2012). Still unknown, however, is why TENT4 family members incorporate guanosines with relative frequency. The best way to predict specificity is currently based on overall sequence similarity with the best biochemically characterized members. However, there are peculiarities. For example, the worm NPOL‐1, paralogous to GLD‐2 and orthologous to human TENT2, has broad nucleotide specificity with a slight preference for uridines/adenosines (Preston et al., 2019) and does not promote mRNA translation once tethered to reporter mRNA, unlike its other family members (Jae & Wickens, 2007). The basis of TENT processivity and the switch between processive and distributive modes remain largely unknown. Some enzymes, such as TUT4/7, are processive in vitro for unstructured substrates and distributive for structured substrates (Faehnle et al., 2017; Warkocki, Krawczyk, et al., 2018). Based on biochemical experiments in TENT4 orthologs in yeast, the RNA helicase subunit of the TRAMP complex has been proposed to control the processivity of PAP (Jia et al., 2011). Although the exact mechanisms that modulate other TENT modes of action await further investigation, one hypothesis could be that structural properties of RNA substrates and TENT cofactors are important in this regard.
Finally, TENT activity is known to play an important role in some biological processes, such as the activation of dormant mRNAs during oogenesis, and in some neuronal processes. In mammals, however, the identity of the enzyme that is responsible for this process remains to be established.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest for this article.
AUTHOR CONTRIBUTIONS
Vladyslava Liudkovska: Visualization; writing‐original draft; writing‐review and editing. Andrzej Dziembowski: Conceptualization; funding acquisition; supervision; writing‐original draft; writing‐review and editing.
RELATED WIREs ARTICLES
Specificity factors in cytoplasmic polyadenylation
Deadenylation: Enzymes, regulation, and functional implications.
Poly(A) RNA‐binding proteins and polyadenosine RNA: New members and novel functions.
Exonucleases and endonucleases involved in polyadenylation‐assisted RNA decay
ACKNOWLEDGMENTS
The authors thank Michal Krzyszton for critically reading the manuscript and helpful comments and Michael Arends for proofreading the manuscript.
Liudkovska V, Dziembowski A. Functions and mechanisms of RNA tailing by metazoan terminal nucleotidyltransferases. WIREs RNA. 2021;12:e1622. 10.1002/wrna.1622
Funding information ERA Chair European Union's Horizon 2020 programme, Grant/Award Number: 810425; National Science Center Poland, Grant/Award Number: UMO‐2019/33/B/NZ2/01773
REFERENCES
- Anderson, S. , Bankier, A. T. , Barrell, B. G. , De Bruijn, M. H. L. , Coulson, A. R. , Drouin, J. , … Young, I. G. (1981). Sequence and organization of the human mitochondrial genome. Nature, 290(5806), 457–465. 10.1038/290457a0 [DOI] [PubMed] [Google Scholar]
- Aravind, L. , & Koonin, E. V. (1999). DNA polymerase β‐like nucleotidyltransferase superfamily: Identification of three new families, classification and evolutionary history. Nucleic Acids Research, 27(7), 1609–1618. 10.1093/nar/27.7.1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai, Y. , Srivastava, S. K. , Chang, J. H. , Manley, J. L. , & Tong, L. (2011). Structural basis for dimerization and activity of human PAPD1, a noncanonical poly(A) polymerase. Molecular Cell, 41(3), 311–320. 10.1016/j.molcel.2011.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balzeau, J. , Menezes, M. R. , Cao, S. , & Hagan, J. P. (2017). The LIN28/let‐7 pathway in cancer. Frontiers in Genetics, 8(March), 1–16. 10.3389/fgene.2017.00031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbieri, M. , Manzoni, M. , Fabris, S. , Ciceri, G. , Todoerti, K. , Simeon, V. , … Lionetti, M. (2016). Compendium of FAM46C gene mutations in plasma cell dyscrasias. British Journal of Haematology, 174(4), 642–645. 10.1111/bjh.13793 [DOI] [PubMed] [Google Scholar]
- Barnard, D. C. , Ryan, K. , Manley, J. L. , & Richter, J. D. (2004). Symplekin and xGLD‐2 are required for CPEB‐mediated cytoplasmic polyadenylation. Cell, 119(5), 641–651. 10.1016/j.cell.2004.10.029 [DOI] [PubMed] [Google Scholar]
- Barragán, I. , Borrego, S. , Abd El‐Aziz, M. M. , El‐Ashry, M. F. , Abu‐Safieh, L. , Bhattacharya, S. S. , & Antiñolo, G. (2008). Genetic analysis of FAM46A in Spanish families with autosomal recessive retinitis pigmentosa: Characterisation of novel VNTRs. Annals of Human Genetics, 72(1), 26–34. 10.1111/j.1469-1809.2007.00393.x [DOI] [PubMed] [Google Scholar]
- Bartel, D. P. (2018). Metazoan microRNAs. Cell, 173(1), 20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck, C. R. , Garcia‐Perez, J. L. , Badge, R. M. , & Moran, J. V. (2011). LINE‐1 elements in structural variation and disease. Annual Review of Genomics and Human Genetics, 12(1), 187–215. 10.1146/annurev-genom-082509-141802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjachat, T. , Tongyoo, P. , Tantivitayakul, P. , Somparn, P. , Hirankarn, N. , Prom‐On, S. , … Townamchai, N. (2015). Biomarkers for refractory lupus nephritis: A microarray study of kidney tissue. International Journal of Molecular Sciences, 16(6), 14276–14290. 10.3390/ijms160614276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benoit, P. , Papin, C. , Kwak, J. E. , Wickens, M. , & Simonelig, M. (2008). PAP‐ and GLD‐2‐type poly(A) polymerases are required sequentially in cytoplasmic polyadenylation and oogenesis in Drosophila. Development, 135(11), 1969–1979. 10.1242/dev.021444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berndt, H. , Harnisch, C. , Rammelt, C. , Stöhr, N. , Zirkel, A. , Dohm, J. C. , … Wahle, E. (2012). Maturation of mammalian H/ACA box snoRNAs: PAPD5‐dependent adenylation and PARN‐dependent trimming. RNA, 18(5), 958–972. 10.1261/rna.032292.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettoni, F. , Filho, F. C. , Grosso, D. M. , Galante, P. A. F. , Parmigiani, R. B. , Geraldo, M. V. , … Camargo, A. A. (2009). Identification of FAM46D as a novel cancer/testis antigen using EST data and serological analysis. Genomics, 94(3), 153–160. 10.1016/j.ygeno.2009.06.001 [DOI] [PubMed] [Google Scholar]
- Bilska, A. , Kusio‐kobia, M. , Krawczyk, P. S. , Gewartowska, O. , Tarkowski, B. , Koby, K. , … Mroczek, S. (2020). Immunoglobulin expression and the humoral immune response is regulated by the non‐canonical poly(A) polymerase TENT5C. Nature Communications, 11(2031), 1–17. 10.1038/s41467-020-15835-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boele, J. , Persson, H. , Shin, J. W. , Ishizu, Y. , Newie, I. S. , Søkilde, R. , … De Hoon, M. J. L. (2014). PAPD5‐mediated 3′ adenylation and subsequent degradation of miR‐21 is disrupted in proliferative disease. Proceedings of the National Academy of Sciences of the United States of America, 111(31), 11467–11472. 10.1073/pnas.1317751111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski, L. S. , Szczesny, R. J. , Brzezniak, L. K. , & Stepien, P. P. (2010). RNA turnover in human mitochondria: More questions than answers? Biochimica et Biophysica Acta – Bioenergetics, 1797(6–7), 1066–1070. 10.1016/j.bbabio.2010.01.028 [DOI] [PubMed] [Google Scholar]
- Bortolamiol‐Becet, D. , Hu, F. , Jee, D. , Wen, J. , Okamura, K. , Lin, C. J. , … Lai, E. C. (2015). Selective suppression of the splicing‐mediated microRNA pathway by the terminal uridyltransferase tailor. Molecular Cell, 59(2), 217–228. 10.1016/j.molcel.2015.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd, K. D. , Ross, F. M. , Walker, B. A. , Wardell, C. P. , Tapper, W. J. , Chiecchio, L. , … Morgan, G. J. (2011). Mapping of chromosome 1p deletions in myeloma identifies FAM46C at 1p12 and CDKN2C at 1p32.3 as being genes in regions associated with adverse survival. Clinical Cancer Research, 17(24), 7776–7784. 10.1158/1078-0432.CCR-11-1791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic, A. , Clemente, P. , Calvo‐Garrido, J. , Maffezzini, C. , Felser, A. , Wibom, R. , … Wredenberg, A. (2016). Mitochondrial polyadenylation is a one‐step process required for mRNA integrity and tRNA maturation. PLoS Genetics, 12(5), e1006028. 10.1371/journal.pgen.1006028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratic, A. , Wredenberg, A. , Grönke, S. , Stewart, J. B. , Mourier, A. , Ruzzenente, B. , … Larsson, N. G. (2011). The bicoid stability factor controls polyadenylation and expression of specific mitochondrial mRNAs in Drosophila melanogaster . PLoS Genetics, 7(10), 1–14. 10.1371/journal.pgen.1002324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns, D. M. , D'Ambrogio, A. , Nottrott, S. , & Richter, J. D. (2011). CPEB and two poly(A) polymerases control miR‐122 stability and p53 mRNA translation. Nature, 473(7345), 105–108. 10.1038/nature09908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs, A. M. , Ando, Y. , de Hoon, M. J. L. , Tomaru, Y. , Nishibu, T. , Ukekawa, R. , … Daub, C. O. (2010). A comprehensive survey of 3′ animal miRNA modification events and a possible role for 3′ adenylation in modulating miRNA targeting effectiveness. Genome Research, 20(10), 1398–1410. 10.1101/gr.106054.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carayol, J. , Chabert, C. , Di Cara, A. , Armenise, C. , Lefebvre, G. , Langin, D. , … Hager, J. (2017). Protein quantitative trait locus study in obesity during weight‐loss identifies a leptin regulator. Nature Communications, 8(1), 1–14. 10.1038/s41467-017-02182-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, S. N. , & Pek, J. W. (2019). Stable intronic sequence RNAs (sisRNAs): An expanding universe. Trends in Biochemical Sciences, 44(3), 258–272. 10.1016/j.tibs.2018.09.016 [DOI] [PubMed] [Google Scholar]
- Chang, H. , Lim, J. , Ha, M. , & Kim, V. N. (2014). TAIL‐seq: Genome‐wide determination of poly(A) tail length and 3′ end modifications. Molecular Cell, 53(6), 1044–1052. 10.1016/j.molcel.2014.02.007 [DOI] [PubMed] [Google Scholar]
- Chang, H. , Yeo, J. , Kim, J. g. , Kim, H. , Lim, J. , Lee, M. , … Kim, V. N. (2018). Terminal uridylyltransferases execute programmed clearance of maternal transcriptome in vertebrate embryos. Molecular Cell, 70(1), 72–82.e7. 10.1016/j.molcel.2018.03.004 [DOI] [PubMed] [Google Scholar]
- Chapman, M. A. , Lawrence, M. S. , Keats, J. J. , Cibulskis, K. , Sougnez, C. , Schinzel, A. C. , … Golub, T. R. (2011). Initial genome sequencing and analysis of multiple myeloma. Nature, 471(7339), 467–472. 10.1038/nature09837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth, A. , Meijer, H. A. , & De Moor, C. H. (2013). Specificity factors in cytoplasmic polyadenylation. WIREs: RNA, 4(4), 437–461. 10.1002/wrna.1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. C. G. , Simard, M. J. , Tabara, H. , Brownell, D. R. , McCollough, J. A. , & Mello, C. C. (2005). A member of the polymerase β nucleotidyltransferase superfamily is required for RNA interference in C. elegans . Current Biology, 15(4), 378–383. 10.1016/j.cub.2005.01.009 [DOI] [PubMed] [Google Scholar]
- Cheng, L. , Li, F. , Jiang, Y. , Yu, H. , Xie, C. , Shi, Y. , & Gong, Q. (2019). Structural insights into a unique preference for 3′ terminal guanine of mirtron in Drosophila TUTase tailor. Nucleic Acids Research, 47(1), 495–508. 10.1093/nar/gky1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury, N. R. , Nowak, J. S. , Zuo, J. , Rappsilber, J. , Spoel, S. H. , & Michlewski, G. (2014). Trim25 is an RNA‐specific activator of Lin28a/TuT4‐mediated uridylation. Cell Reports, 9(4), 1265–1272. 10.1016/j.celrep.2014.10.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo, T. , Ohira, T. , Sakaguchi, Y. , Goshima, N. , Nomura, N. , Nagao, A. , & Suzuki, T. (2012). LRPPRC/SLIRP suppresses PNPase‐mediated mRNA decay and promotes polyadenylation in human mitochondria. Nucleic Acids Research, 40(16), 8033–8047. 10.1093/nar/gks506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll, O. , Guitart, T. , Villalba, A. , Papin, C. , Simonelig, M. , & Gebauer, F. (2018). Dicer‐2 promotes mRNA activation through cytoplasmic polyadenylation. RNA, 24(4), 529–539. 10.1261/rna.065417.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colland, F. , Jacq, X. , Trouplin, V. , Mougin, C. , Groizeleau, C. , Hamburger, A. , … Gauthier, J. M. (2004). Functional proteomics mapping of a human signaling pathway. Genome Research, 14(7), 1324–1332. 10.1101/gr.2334104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, J. , Saari, B. , & Anderson, P. (1988). Activation of a transposable element in the germ line but not the soma of Caenorhabditis elegans . Nature, 328(6132), 726–728. 10.1038/328726a0 [DOI] [PubMed] [Google Scholar]
- Crosby, A. H. , Patel, H. , Chioza, B. A. , Proukakis, C. , Gurtz, K. , Patton, M. A. , … Lightowlers, R. N. (2010). Defective mitochondrial mRNA maturation is associated with spastic ataxia. American Journal of Human Genetics, 87(5), 655–660. 10.1016/j.ajhg.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J. , Sackton, K. L. , Horner, V. L. , Kumar, K. E. , & Wolfner, M. F. (2008). Wispy, the Drosophila homolog of GLD‐2, is required during oogenesis and egg activation. Genetics, 178(4), 2017–2029. 10.1534/genetics.107.084558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J. , Sartain, C. V. , Pleiss, J. A. , & Wolfner, M. F. (2013). Cytoplasmic polyadenylation is a major mRNA regulator during oogenesis and egg activation in Drosophila. Developmental Biology, 383(1), 121–131. 10.1016/j.ydbio.2013.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Ambrogio, A. , Gu, W. , Udagawa, T. , Mello, C. C. , & Richter, J. D. (2012). Specific miRNA stabilization by Gld2‐catalyzed monoadenylation. Cell Reports, 2(6), 1537–1545. 10.1016/j.celrep.2012.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Almeida, C. , Scheer, H. , Zuber, H. , & Gagliardi, D. (2018). RNA uridylation: A key posttranscriptional modification shaping the coding and noncoding transcriptome. WIREs: RNA, 9(1), e1440. 10.1002/wrna.1440 [DOI] [PubMed] [Google Scholar]
- Dez, C. , Houseley, J. , & Tollervey, D. (2006). Surveillance of nuclear‐restricted pre‐ribosomes within a subnucleolar region of Saccharomyces cerevisiae . EMBO Journal, 25(7), 1534–1546. 10.1038/sj.emboj.7601035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didychuk, A. L. , Butcher, S. E. , & Brow, D. A. (2018). The life of U6 small nuclear RNA, from cradle to grave. RNA, 24(4), 437–460. 10.1261/rna.065136.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didychuk, A. L. , Montemayor, E. J. , Carrocci, T. J. , Delaitsch, A. T. , Lucarelli, S. E. , Westler, W. M. , … Butcher, S. E. (2017). Usb1 controls U6 snRNP assembly through evolutionarily divergent cyclic phosphodiesterase activities. Nature Communications, 8(1), 497. 10.1038/s41467-017-00484-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diener, S. , Bayer, S. , Sabrautzki, S. , Wieland, T. , Mentrup, B. , Przemeck, G. K. H. , … Lorenz‐Depiereux, B. (2016). Exome sequencing identifies a nonsense mutation in Fam46a associated with bone abnormalities in a new mouse model for skeletal dysplasia. Mammalian Genome, 27(3–4), 111–121. 10.1007/s00335-016-9619-x [DOI] [PubMed] [Google Scholar]
- Doyard, M. , Bacrot, S. , Huber, C. , Di Rocco, M. , Goldenberg, A. , Aglan, M. S. , … Cormier‐Daire, V. (2018). FAM46A mutations are responsible for autosomal recessive osteogenesis imperfecta. Journal of Medical Genetics, 55(4), 278–284. 10.1136/jmedgenet-2017-104999 [DOI] [PubMed] [Google Scholar]
- Dufourt, J. , Bontonou, G. , Chartier, A. , Jahan, C. , Meunier, A. C. , Pierson, S. , … Simonelig, M. (2017). piRNAs and Aubergine cooperate with Wispy poly(A) polymerase to stabilize mRNAs in the germ plasm. Nature Communications, 8(1), 1–12. 10.1038/s41467-017-01431-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckmann, C. R. , Crittenden, S. L. , Suh, N. , & Kimble, J. (2004). GLD‐3 and control of the mitosis/meiosis decision in the germline of Caenorhabditis elegans . Genetics, 168(1), 147–160. 10.1534/genetics.104.029264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichhorn, S. W. , Subtelny, A. O. , Kronja, I. , Kwasnieski, J. C. , Orr‐Weaver, T. L. , & Bartel, D. P. (2016). mRNA poly(A)‐tail changes specified by deadenylation broadly reshape translation in Drosophila oocytes and early embryos. eLife, 5(July), 1–24. 10.7554/eLife.16955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, T. J. , Eichhorn, S. W. , Subtelny, A. O. , Lin, K. S. , McGeary, S. E. , Gupta, S. , & Bartel, D. P. (2020). The dynamics of cytoplasmic mRNA metabolism. Molecular Cell, 77, 1–14. 10.1016/j.molcel.2019.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etokebe, G. E. , Bulat‐Kardum, L. , Munthe, L. A. , Balen, S. , & Dembic, Z. (2014). Association of variable number of tandem repeats in the coding region of the FAM46A gene, FAM46A rs11040 SNP and BAG6 rs3117582 SNP with susceptibility to tuberculosis. PLoS One, 9(3), e91385. 10.1371/journal.pone.0091385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etokebe, G. E. , Jotanovic, Z. , Mihelic, R. , Jericevic, B. M. , Nikolic, T. , Balen, S. , … Dembic, Z. (2015). Susceptibility to large‐joint osteoarthritis (hip and knee) is associated with BAG6 rs3117582 SNP and the VNTR polymorphism in the second exon of the FAM46A gene on chromosome 6. Journal of Orthopaedic Research, 33(1), 56–62. 10.1002/jor.22738 [DOI] [PubMed] [Google Scholar]
- Etokebe, G. E. , Küchler, A. M. , Haraldsen, G. , Landin, M. , Osmundsen, H. , & Dembic, Z. (2009). Family‐with‐sequence‐similarity‐46, member A (Fam46a) gene is expressed in developing tooth buds. Archives of Oral Biology, 54(11), 1002–1007. 10.1016/j.archoralbio.2009.08.005 [DOI] [PubMed] [Google Scholar]
- Faehnle, C. R. , Walleshauser, J. , & Joshua‐Tor, L. (2014). Mechanism of Dis3l2 substrate recognition in the Lin28‐let‐7 pathway. Nature, 514(7521), 252–256. 10.1038/nature13553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faehnle, C. R. , Walleshauser, J. , & Joshua‐Tor, L. (2017). Multi‐domain utilization by TUT4 and TUT7 in control of let‐7 biogenesis. Nature Structural and Molecular Biology, 24(8), 658–665. 10.1038/nsmb.3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasken, M. B. , Leung, S. W. , Banerjee, A. , Kodani, M. O. , Chavez, R. , Bowman, E. A. , … Corbett, A. H. (2011). Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5‐Air1/2‐Mtr4 polyadenylation (TRAMP) RNA quality control complex. Journal of Biological Chemistry, 286(43), 37429–37445. 10.1074/jbc.M111.271494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faulkner, G. J. , & Garcia‐Perez, J. L. (2017, November). L1 Mosaicism in mammals: Extent, effects, and evolution. Trends in Genetics, 33, 802–816. 10.1016/j.tig.2017.07.004 [DOI] [PubMed] [Google Scholar]
- Feng, Y. , Zhang, Y. , Ying, C. , Wang, D. , & Du, C. (2015). Nanopore‐based fourth‐generation DNA sequencing technology. Genomics, Proteomics and Bioinformatics, 13, 4–16. 10.1016/j.gpb.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler, M. , Rossmanith, W. , Wahle, E. , & Rammelt, C. (2015). Mitochondrial poly(A) polymerase is involved in tRNA repair. Nucleic Acids Research, 43(20), 9937–9949. 10.1093/nar/gkv891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya, N. , Kakuta, S. , Sumiyoshi, K. , Ando, M. , Nonaka, R. , Suzuki, A. , … Hattori, N. (2018). NDP 52 interacts with mitochondrial RNA poly(A) polymerase to promote mitophagy. EMBO Reports, 19(12), e46363. 10.15252/embr.201846363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gagliardi, D. , Stepien, P. P. , Temperley, R. J. , Lightowlers, R. N. , & Chrzanowska‐Lightowlers, Z. M. A. (2004). Messenger RNA stability in mitochondria: Different means to an end. Trends in Genetics, 20, 260–267. 10.1016/j.tig.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Grzechnik, P. , & Kufel, J. (2008). Polyadenylation linked to transcription termination directs the processing of snoRNA precursors in yeast. Molecular Cell, 32(2), 247–258. 10.1016/j.molcel.2008.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, W. , Shirayama, M. , Conte, D. , Vasale, J. , Batista, P. J. , Claycomb, J. M. , … Mello, C. C. (2009). Distinct Argonaute‐mediated 22G‐RNA pathways direct genome surveillance in the C. elegans germline. Molecular Cell, 36(2), 231–244. 10.1016/j.molcel.2009.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutiérrez‐Vázquez, C. , Enright, A. J. , Rodríguez‐Galán, A. , Pérez‐García, A. , Collier, P. , Jones, M. R. , … Sánchez‐Madrid, F. (2017). 3′ Uridylation controls mature microRNA turnover during CD4 T‐cell activation. RNA, 23(6), 882–891. 10.1261/rna.060095.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagan, J. P. , Piskounova, E. , & Gregory, R. I. (2009). Lin28 recruits the TUTase Zcchc11 to inhibit let‐7 maturation in mouse embryonic stem cells. Nature Structural and Molecular Biology, 16(10), 1021–1025. 10.1038/nsmb.1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, S. M. , Spencer, C. M. , Harrison, W. R. , Yuva‐Paylor, L. A. , Graham, D. F. , Daza, R. A. M. , … Paylor, R. (2011). Multiple autism‐like behaviors in a novel transgenic mouse model. Behavioural Brain Research, 218(1), 29–41. 10.1016/j.bbr.2010.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harnisch, C. , Cuzic‐Feltens, S. , Dohm, J. C. , Götze, M. , Himmelbauer, H. , & Wahle, E. (2016). Oligoadenylation of 3′ decay intermediates promotes cytoplasmic mRNA degradation in Drosophila cells. RNA, 22(3), 428–442. 10.1261/rna.053942.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, T. , Chandrashekhar, M. , Aregger, M. , Steinhart, Z. , Brown, K. R. , MacLeod, G. , … Moffat, J. (2015). High‐resolution CRISPR screens reveal fitness genes and genotype‐specific cancer liabilities. Cell, 163(6), 1515–1526. 10.1016/j.cell.2015.11.015 [DOI] [PubMed] [Google Scholar]
- Heo, I. , Ha, M. , Lim, J. , Yoon, M. J. , Park, J. E. , Kwon, S. C. , … Kim, V. N. (2012). Mono‐uridylation of pre‐microRNA as a key step in the biogenesis of group II let‐7 microRNAs. Cell, 151(3), 521–532. 10.1016/j.cell.2012.09.022 [DOI] [PubMed] [Google Scholar]
- Heo, I. , Joo, C. , Kim, Y. K. , Ha, M. , Yoon, M. J. , Cho, J. , … Kim, V. N. (2009). TUT4 in concert with Lin28 suppresses microRNA biogenesis through pre‐MicroRNA uridylation. Cell, 138(4), 696–708. 10.1016/j.cell.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Herrero, A. B. , Quwaider, D. , Corchete, L. A. , Mateos, M. V. , García‐Sanz, R. , & Gutiérrez, N. C. (2020). FAM46C controls antibody production by the polyadenylation of immunoglobulin mRNAs and inhibits cell migration in multiple myeloma. Journal of Cellular and Molecular Medicine, 00, 1–12. 10.1111/jcmm.15078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai, H. , Lee, D. I. , Natori, S. , & Sekimizu, K. (1988). Uridylation of U6 RNA in a nuclear extract of Ehrlich ascites tumor cells. Journal of Biochemistry, 104(6), 991–994. 10.1093/oxfordjournals.jbchem.a122597 [DOI] [PubMed] [Google Scholar]
- Hojo, H. , Yashiro, Y. , Noda, Y. , Ogami, K. , Yamagishi, R. , Okada, S. , … Suzuki, T. (2020). The RNA‐binding protein QKI‐7 recruits the poly(A) polymerase GLD‐2 for 3′ adenylation and selective stabilization of microRNA‐122. Journal of Biological Chemistry, 295(2), 390–402. 10.1074/jbc.RA119.011617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley, J. , & Tollervey, D. (2006). Yeast Trf5p is a nuclear poly(A) polymerase. EMBO Reports, 7(2), 205–211. 10.1038/sj.embor.7400612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, J. L. , Liang, H. , Zhang, H. , Yang, M. Z. , Sun, W. , Zhang, P. , … Gao, S. (2020). FAM46B is a prokaryotic‐like cytoplasmic poly(A) polymerase essential in human embryonic stem cells. Nucleic Acids Research, 48(5), 2733–2748. 10.1093/nar/gkaa049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu, Y. , Chen, W. , & Wang, J. (2019). Progress in the identification of gene mutations involved in multiple myeloma. Oncotargets and Therapy, 12, 4075–4080. 10.2147/OTT.S205922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huo, Y. , Shen, J. , Wu, H. , Zhang, C. , Guo, L. , Yang, J. , & Li, W. (2016). Widespread 3′‐end uridylation in eukaryotic RNA viruses. Scientific Reports, 6(1), 25454. 10.1038/srep25454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrina, A. , Jones, C. , Chen, D. , Clarkson, S. , Cochran, N. , Feucht, P. , … Holdorf, M. (2019). A genome‐wide CRISPR screen identifies ZCCHC14 as a host factor required for hepatitis B surface antigen production. Cell Reports, 29(10), 2970–2978.e6. 10.1016/j.celrep.2019.10.113 [DOI] [PubMed] [Google Scholar]
- Ivshina, M. , Lasko, P. , & Richter, J. D. (2014). Cytoplasmic polyadenylation element binding proteins in development, health, and disease. Annual Review of Cell and Developmental Biology, 30(1), 393–415. 10.1146/annurev-cellbio-101011-155831 [DOI] [PubMed] [Google Scholar]
- Jae, E. K. , Drier, E. , Barbee, S. a. , Ramaswami, M. , Yin, J. C. P. , & Wickens, M. (2008). GLD2 poly(A) polymerase is required for long‐term memory. Proceedings of the National Academy of Sciences of the United States of America, 105(38), 14644–14649. 10.1073/pnas.0803185105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jae, E. K. , & Wickens, M. (2007). A family of poly(U) polymerases. RNA, 13(6), 860–867. 10.1261/rna.514007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, H. , Wang, X. , Liu, F. , Guenther, U. P. , Srinivasan, S. , Anderson, J. T. , & Jankowsky, E. (2011). The RNA helicase Mtr4p modulates polyadenylation in the TRAMP complex. Cell, 145(6), 890–901. 10.1016/j.cell.2011.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Ng, S. S. , Zheng, R. T. , Osman, I. , & Pek, J. W. (2018). Generation of Drosophila sisRNAs by independent transcription from cognate introns. IScience, 4, 68–75. 10.1016/j.isci.2018.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, M. R. , Blahna, M. T. , Kozlowski, E. , Matsuura, K. Y. , Ferrari, J. D. , Morris, S. A. , … Mizgerd, J. P. (2012). Zcchc11 uridylates mature miRNAs to enhance neonatal IGF‐1 expression, growth, and survival. PLoS Genetics, 8(11), e1003105. 10.1371/journal.pgen.1003105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadyk, L. C. , & Kimble, J. (1998). Genetic regulation of entry into meiosis in Caenorhabditis elegans . Development, 125(10), 1803–1813 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/9550713 [DOI] [PubMed] [Google Scholar]
- Kang, M. K. , & Han, S. J. (2011). Post‐transcriptional and post‐translational regulation during mouse oocyte maturation. BMB Reports, 44(3), 147–157. 10.5483/BMBRep.2011.44.3.147 [DOI] [PubMed] [Google Scholar]
- Kashiwabara, S. I. , Noguchi, J. , Zhuang, T. , Ohmura, K. , Honda, A. , Sugiura, S. , … Baba, T. (2002). Regulation of spermatogenesis by testis‐specific, cytoplasmic poly(A) polymerase TPAP. Science, 298(5600), 1999–2002. 10.1126/science.1074632 [DOI] [PubMed] [Google Scholar]
- Katoh, T. , Sakaguchi, Y. , Miyauchi, K. , Suzuki, T. , Suzuki, T. , Kashiwabara, S. I. , & Baba, T. (2009). Selective stabilization of mammalian microRNAs by 3′ adenylation mediated by the cytoplasmic poly(A) polymerase GLD‐2. Genes and Development, 23(4), 433–438. 10.1101/gad.1761509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, B. , Ha, M. , Loeff, L. , Chang, H. , Simanshu, D. K. , Li, S. , … Kim, V. N. (2015). TUT 7 controls the fate of precursor micro‐RNAs by using three different uridylation mechanisms. The EMBO Journal, 34(13), 1801–1815. 10.15252/embj.201590931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, D. , Lee, Y. s. , Jung, S. J. , Yeo, J. , Seo, J. J. , Lee, Y. Y. , … Kim, V. N. (2020). Viral hijacking of the TENT4–ZCCHC14 complex protects viral RNAs via mixed tailing. Nature Structural and Molecular Biology, 27, 581–588. 10.1038/s41594-020-0427-3 [DOI] [PubMed] [Google Scholar]
- Kim, G. W. , Lee, S. H. , Cho, H. , Kim, M. , Shin, E. C. , & Oh, J. W. (2016). Hepatitis C virus Core protein promotes miR‐122 destabilization by inhibiting GLD‐2. PLoS Pathogens, 12(7), e1005714. 10.1371/journal.ppat.1005714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K. W. , Nykamp, K. , Suh, N. , Bachorik, J. L. , Wang, L. , & Kimble, J. (2009). Antagonism between GLD‐2 binding partners controls gamete sex. Developmental Cell, 16(5), 723–733. 10.1016/j.devcel.2009.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, K. W. , Wilson, T. L. , & Kimble, J. (2010). GLD‐2/RNP‐8 cytoplasmic poly(A) polymerase is a broad‐spectrum regulator of the oogenesis program. Proceedings of the National Academy of Sciences of the United States of America, 107(40), 17445–17450. 10.1073/pnas.1012611107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimble, J. , & Crittenden, S. L. (2007). Controls of germline stem cells, entry into meiosis, and the sperm/oocyte decision in Caenorhabditis elegans . Annual Review of Cell and Developmental Biology, 23(1), 405–433. 10.1146/annurev.cellbio.23.090506.123326 [DOI] [PubMed] [Google Scholar]
- Kobylecki, K. , Kuchta, K. , Dziembowski, A. , Ginalski, K. , & Tomecki, R. (2017). Biochemical and structural bioinformatics studies of fungal CutA nucleotidyltransferases explain their unusual specificity toward CTP and increased tendency for cytidine incorporation at the 3′‐terminal positions of synthesized tails. RNA, 23(12), 1902–1926. 10.1261/rna.061010.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski, M. P. , & Krude, T. (2015). Functional roles of non‐coding Y RNAs. International Journal of Biochemistry and Cell Biology, 66, 20–29. 10.1016/j.biocel.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroupova, A. , Ivascul, A. , Reimao‐Pinto, M. M. , Ameres, S. L. , & Jinek, M. (2019). Structural basis for acceptor RNA substrate selectivity of the 3 terminal uridylyl transferase Tailor. Nucleic Acids Research, 47(2), 1030–1042. 10.1093/nar/gky1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuchta, K. , Muszewska, A. , Knizewski, L. , Steczkiewicz, K. , Wyrwicz, L. S. , Pawlowski, K. , … Ginalski, K. (2016). FAM46 proteins are novel eukaryotic non‐canonical poly(A) polymerases. Nucleic Acids Research, 44(8), 3534–3548. 10.1093/nar/gkw222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosaki, T. , Miyoshi, K. , Myers, J. R. , & Maquat, L. E. (2018). NMD‐degradome sequencing reveals ribosome‐bound intermediates with 3′‐end non‐templated nucleotides. Nature Structural and Molecular Biology, 25(10), 940–950. 10.1038/s41594-018-0132-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak, J. E. , Wang, L. , Ballantyne, S. , Kimble, J. , & Wickens, M. (2004). Mammalian GLD‐2 homologs are poly(A) polymerases. Proceedings of the National Academy of Sciences of the United States of America, 101(13), 4407–4412. 10.1073/pnas.0400779101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Łabno, A. , Warkocki, Z. , Kulínski, T. , Krawczyk, P. S. , Bijata, K. , Tomecki, R. , & Dziembowski, A. (2016). Perlman syndrome nuclease DIS3L2 controls cytoplasmic non‐coding RNAs and provides surveillance pathway for maturing snRNAs. Nucleic Acids Research, 44(21), 10437–10453. 10.1093/nar/gkw649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCava, J. , Houseley, J. , Saveanu, C. , Petfalski, E. , Thompson, E. , Jacquier, A. , & Tollervey, D. (2005). RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell, 121(5), 713–724. 10.1016/j.cell.2005.04.029 [DOI] [PubMed] [Google Scholar]
- Lackey, P. E. , Welch, J. D. , & Marzluff, W. F. (2016). TUT7 catalyzes the uridylation of the 3′ end for rapid degradation of histone mRNA. RNA, 22(11), 1673–1688. 10.1261/rna.058107.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagali, P. S. , Kakuk, L. E. , Griesinger, I. B. , Wong, P. W. , & Ayyagari, R. (2002). Identification and characterization of C6orf37 a novel candidate human retinal disease gene on chromosome 6q14. Biochemical and Biophysical Research Communications, 293(1), 356–365. 10.1016/S0006-291X(02)00228-0 [DOI] [PubMed] [Google Scholar]
- Laishram, R. S. , & Anderson, R. a. (2010). The poly A polymerase Star‐PAP controls 3‐2‐end cleavage by promoting CPSF interaction and specificity toward the pre‐mRNA. EMBO Journal, 29(24), 4132–4145. 10.1038/emboj.2010.287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapkouski, M. , & Hällberg, B. M. (2015). Structure of mitochondrial poly(A) RNA polymerase reveals the structural basis for dimerization, ATP selectivity and the SPAX4 disease phenotype. Nucleic Acids Research, 43(18), 9065–9075. 10.1093/nar/gkv861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Pen, J. , Jiang, H. , Di Domenico, T. , Kneuss, E. , Kosałka, J. , Leung, C. , … Miska, E. A. (2018). Terminal uridylyltransferases target RNA viruses as part of the innate immune system. Nature Structural and Molecular Biology, 25(9), 778–786. 10.1038/s41594-018-0106-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, H. , Han, S. , Kwon, C. S. , & Lee, D. (2016). Biogenesis and regulation of the let‐7 miRNAs and their functional implications. Protein and Cell, 7, 100–113. 10.1007/s13238-015-0212-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, M. , Choi, Y. , Kim, K. , Jin, H. , Lim, J. , Nguyen, T. A. , … Kim, V. N. (2014). Adenylation of maternally inherited microRNAs by Wispy. Molecular Cell, 56(5), 696–707. 10.1016/j.molcel.2014.10.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legnini, I. , Alles, J. , Karaiskos, N. , Ayoub, S. , & Rajewsky, N. (2019). FLAM‐seq: Full‐length mRNA sequencing reveals principles of poly(A) tail length control. Nature Methods, 16(9), 879–886. 10.1038/s41592-019-0503-y [DOI] [PubMed] [Google Scholar]
- Lehrbach, N. J. , Armisen, J. , Lightfoot, H. L. , Murfitt, K. J. , Bugaut, A. , Balasubramanian, S. , & Miska, E. A. (2009). LIN‐28 and the poly(U) polymerase PUP‐2 regulate let‐7 microRNA processing in Caenorhabditis elegans . Nature Structural and Molecular Biology, 16(10), 1016–1020. 10.1038/nsmb.1675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Laishram, R. S. , & Anderson, R. A. (2013). The novel poly(A) polymerase Star‐PAP is a signal‐regulated switch at the 3′‐end of mRNAs. Advances in Biological Regulation, 53(1), 64–76. 10.1016/j.jbior.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W. , Laishram, R. S. , Ji, Z. , Barlow, C. A. , Tian, B. , & Anderson, R. A. (2012). Star‐PAP control of BIK expression and apoptosis is regulated by nuclear PIPKIα and PKCδ signaling. Molecular Cell, 45(1), 25–37. 10.1016/j.molcel.2011.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. , & Maine, E. M. (2018). The balance of poly(U) polymerase activity ensures germline identity, survival and development in Caenorhabditis elegans . Development (Cambridge), 145(19), dev165944. 10.1242/dev.165944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, T. , Ye, X. , Liu, Y. , Qiu, X. , Li, Z. , Tian, B. , & Yan, D. (2018). FAM46B inhibits cell proliferation and cell cycle progression in prostate cancer through ubiquitination of β‐catenin. Experimental and Molecular Medicine, 50(12), 1–12. 10.1038/s12276-018-0184-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. , Ha, M. , Chang, H. , Kwon, S. C. , Simanshu, D. K. , Patel, D. J. , & Kim, V. N. (2014). Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell, 159(6), 1365–1376. 10.1016/j.cell.2014.10.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim, J. , Kim, D. , Lee, Y. S. , Ha, M. , Lee, M. , Yeo, J. , … Kim, V. N. (2018). Mixed tailing by TENT4A and TENT4B shields mRNA from rapid deadenylation. Science, 361(6403), 701–704. 10.1126/science.aam5794 [DOI] [PubMed] [Google Scholar]
- Lim, J. , Lee, M. , Son, A. , Chang, H. , & Kim, V. N. (2016). mTAIL‐seq reveals dynamic poly(A) tail regulation in oocyte‐to‐embryo development. Genes and Development, 30(14), 1671–1682. 10.1101/gad.284802.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y. , Nie, H. , Liu, H. , & Lu, F. (2019). Poly(A) inclusive RNA isoform sequencing (PAIso‐seq) reveals wide‐spread non‐adenosine residues within RNA poly(A) tails. Nature Communications, 10(1), 5292. 10.1038/s41467-019-13228-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr, J. G. , Stojanov, P. , Carter, S. L. , Cruz‐Gordillo, P. , Lawrence, M. S. , Auclair, D. , … Golub, T. R. (2014). Widespread genetic heterogeneity in multiple myeloma: Implications for targeted therapy. Cancer Cell, 25(1), 91–101. 10.1016/j.ccr.2013.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubas, M. , Christensen, M. S. , Kristiansen, M. S. , Domanski, M. , Falkenby, L. G. , Lykke‐Andersen, S. , … Jensen, T. H. (2011). Interaction profiling identifies the human nuclear exosome targeting complex. Molecular Cell, 43(4), 624–637. 10.1016/j.molcel.2011.06.028 [DOI] [PubMed] [Google Scholar]
- Lund, E. , & Dahlberg, J. E. (1992). Cyclic 2′,3′‐phosphates and nontemplated nucleotides at the 3′ end of spliceosomal U6 small nuclear RNA's. Science, 255(5042), 327–330. 10.1126/science.1549778 [DOI] [PubMed] [Google Scholar]
- Maillo, C. , Martín, J. , Sebastián, D. , Hernández‐Alvarez, M. , García‐Rocha, M. , Reina, O. , … Méndez, R. (2017). Circadian‐ and UPR‐dependent control of CPEB4 mediates a translational response to counteract hepatic steatosis under ER stress. Nature Cell Biology, 19(2), 94–105. 10.1038/ncb3461 [DOI] [PubMed] [Google Scholar]
- Malecki, M. , Viegas, S. C. , Carneiro, T. , Golik, P. , Dressaire, C. , Ferreira, M. G. , & Arraiano, C. M. (2013). The exoribonuclease Dis3L2 defines a novel eukaryotic RNA degradation pathway. EMBO Journal, 32(13), 1842–1854. 10.1038/emboj.2013.63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansur, F. , Ivshina, M. , Gu, W. , Schaevitz, L. , Stackpole, E. , Gujja, S. , … Richter, J. D. (2016). Gld2‐catalyzed 3′ monoadenylation of miRNAs in the hippocampus has no detectable effect on their stability or on animal behavior. RNA, 22(10), 1492–1499. 10.1261/rna.056937.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. , Doublié, S. , & Keller, W. (2008). Determinants of substrate specificity in RNA‐dependent nucleotidyl transferases. Biochimica et Biophysica Acta – Gene Regulatory Mechanisms, 1779(4), 206–216. 10.1016/j.bbagrm.2007.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. , & Keller, W. (2007). RNA‐specific ribonucleotidyl transferases. RNA, 13(11), 1834–1849. 10.1261/rna.652807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. , Keller, W. , & Doublié, S. (2000). Crystal structure of mammalian poly(A) polymerase in complex with an analog of ATP. The EMBO Journal, 19(16), 4193–4203. 10.1093/emboj/19.16.4193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, N. T. , Nakamura, K. , Paila, U. , Woo, J. , Brown, C. , Wright, J. A. , … Concannon, P. (2014). Homozygous mutation of MTPAP causes cellular radiosensitivity and persistent DNA double‐strand breaks. Cell Death and Disease, 5(3), e1130. 10.1038/cddis.2014.99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff, W. F. , & Koreski, K. P. (2017). Birth and death of histone mRNAs. Trends in Genetics, 33(10), 745–759. 10.1016/j.tig.2017.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellman, D. L. , Gonzales, M. L. , Song, C. , Barlow, C. A. , Wang, P. , Kendziorski, C. , & Anderson, R. A. (2008). A PtdIns4,5P2‐regulated nuclear poly(A) polymerase controls expression of select mRNAs. Nature, 451(7181), 1013–1017. 10.1038/nature06666 [DOI] [PubMed] [Google Scholar]
- Menezes, M. R. , Balzeau, J. , & Hagan, J. P. (2018). 3′ RNA uridylation in epitranscriptomics, gene regulation, and disease. Frontiers in Molecular Biosciences, 5, 1–20. 10.3389/fmolb.2018.00061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasaki, R. , Rudel, D. , & Eckmann, C. R. (2014). Increased sensitivity and accuracy of a single‐stranded DNA splint‐mediated ligation assay (sPAT) reveals poly(A) tail length dynamics of developmentally regulated mRNAs. RNA Biology, 11(2), 111–123. 10.4161/rna.27992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minshall, N. , Reiter, M. H. , Weil, D. , & Standart, N. (2007). CPEB interacts with an ovary‐specific eIF4E and 4E‐T in early Xenopus oocytes. Journal of Biological Chemistry, 282(52), 37389–37401. 10.1074/jbc.M704629200 [DOI] [PubMed] [Google Scholar]
- Morgan, M. , Kabayama, Y. , Much, C. , Ivanova, I. , Di Giacomo, M. , Auchynnikava, T. , … O'Carroll, D. (2019). A programmed wave of uridylation‐primed mRNA degradation is essential for meiotic progression and mammalian spermatogenesis. Cell Research, 29(3), 221–232. 10.1038/s41422-018-0128-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan, M. , Much, C. , DiGiacomo, M. , Azzi, C. , Ivanova, I. , Vitsios, D. M. , … O'Carroll, D. (2017). mRNA 3′ uridylation and poly(A) tail length sculpt the mammalian maternal transcriptome. Nature, 548(7667), 347–351. 10.1038/nature23318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek, S. , Chlebowska, J. , Kuliński, T. M. , Gewartowska, O. , Gruchota, J. , Cysewski, D. , … Dziembowski, A. (2017). The non‐canonical poly(A) polymerase FAM46C acts as an onco‐suppressor in multiple myeloma. Nature Communications, 8(1), 619. 10.1038/s41467-017-00578-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek, S. , Krwawicz, J. , Kutner, J. , Lazniewski, M. , Kuciński, I. , Ginalski, K. , & Dziembowski, A. (2012). C16orf57, a gene mutated in poikiloderma with neutropenia, encodes a putative phosphodiesterase responsible for the U6 snRNA 3′ end modification. Genes and Development, 26(17), 1911–1925. 10.1101/gad.193169.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, H. , Lopez, A. , Tropberger, P. , Wildum, S. , Schmaler, J. , Pedersen, L. , … Javanbakht, H. (2019). PAPD5/7 are host factors that are required for hepatitis B virus RNA stabilization. Hepatology, 69(4), 1398–1411. 10.1002/hep.30329 [DOI] [PubMed] [Google Scholar]
- Mullen, T. E. , & Marzluff, W. F. (2008). Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes and Development, 22(1), 50–65. 10.1101/gad.1622708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munoz‐Tello, P. , Gabus, C. , & Thore, S. (2012). Functional implications from the Cid1 poly(U) polymerase crystal structure. Structure, 20(6), 977–986. 10.1016/j.str.2012.04.006 [DOI] [PubMed] [Google Scholar]
- Munoz‐Tello, P. , Gabus, C. , & Thore, S. (2014). A critical switch in the enzymatic properties of the Cid1 protein deciphered from its product‐bound crystal structure. Nucleic Acids Research, 42(5), 3372–3380. 10.1093/nar/gkt1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaike, T. , Suzuki, T. , Katoh, T. , & Ueda, T. (2005). Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria‐specific poly(A) polymerase and polynucleotide phosphorylase. Journal of Biological Chemistry, 280(20), 19721–19727. 10.1074/jbc.M500804200 [DOI] [PubMed] [Google Scholar]
- Nagaike, T. , Suzuki, T. , & Ueda, T. (2008). Polyadenylation in mammalian mitochondria: Insights from recent studies. Biochimica et Biophysica Acta – Gene Regulatory Mechanisms, 1779(4), 266–269. 10.1016/j.bbagrm.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Nakamura, R. , Takeuchi, R. , Takata, K. ‐i. , Shimanouchi, K. , Abe, Y. , Kanai, Y. , … Sakaguchi, K. (2008). TRF4 is involved in polyadenylation of snRNAs in Drosophila melanogaster . Molecular and Cellular Biology, 28(21), 6620–6631. 10.1128/mcb.00448-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakanishi, T. , Kumagai, S. , Kimura, M. , Watanabe, H. , Sakurai, T. , Kimura, M. , … Baba, T. (2007). Disruption of mouse poly(A) polymerase mGLD‐2 does not alter polyadenylation status in oocytes and somatic cells. Biochemical and Biophysical Research Communications, 364(1), 14–19. 10.1016/j.bbrc.2007.09.096 [DOI] [PubMed] [Google Scholar]
- Nakel, K. , Bonneau, F. , Basquin, C. , Habermann, B. , Eckmann, C. R. , & Conti, E. (2016). Structural basis for the antagonistic roles of RNP‐8 and GLD‐3 in GLD‐2 poly(A)‐polymerase activity. RNA, 22(8), 1139–1145. 10.1261/rna.056598.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakel, K. , Bonneau, F. , Eckmann, C. R. , & Conti, E. (2015). Structural basis for the activation of the C. elegans noncanonical cytoplasmic poly(A)‐polymerase GLD‐2 by GLD‐3. Proceedings of the National Academy of Sciences of the United States of America, 112(28), 8614–8619. 10.1073/pnas.1504648112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch, M. , Minasaki, R. , & Eckmann, C. R. (2017). Polyadenylation is the key aspect of GLD‐2 function in C. elegans . RNA, 23(8), 1180–1187. 10.1261/rna.061473.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nousch, M. , Yeroslaviz, A. , Habermann, B. , & Eckmann, C. R. (2014). The cytoplasmic poly(A) polymerases GLD‐2 and GLD‐4 promote general gene expression via distinct mechanisms. Nucleic Acids Research, 42(18), 11622–11633. 10.1093/nar/gku838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Brien, L. L. , Albee, A. J. , Liu, L. , Tao, W. , Dobrzyn, P. , Lizarraga, S. B. , & Wiese, C. (2005). The Xenopus TACC homologue, maskin, functions in mitotic spindle assembly. Molecular Biology of the Cell, 16(6), 2836–2847. 10.1091/mbc.E04-10-0926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogami, K. , Cho, R. , & Hoshino, S. i. (2013). Molecular cloning and characterization of a novel isoform of the non‐canonical poly(A) polymerase PAPD7. Biochemical and Biophysical Research Communications, 432(1), 135–140. 10.1016/j.bbrc.2013.01.072 [DOI] [PubMed] [Google Scholar]
- Pajak, A. , Laine, I. , Clemente, P. , El‐Fissi, N. , Schober, F. A. , Maffezzini, C. , … Wredenberg, A. (2019). Defects of mitochondrial RNA turnover lead to the accumulation of double‐stranded RNA in vivo. PLoS Genetics, 15(7), e1008240. 10.1371/journal.pgen.1008240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannone, B. K. , Xue, D. , & Wolin, S. L. (1998). A role for the yeast La protein in U6 snRNP assembly: Evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO Journal, 17(24), 7442–7453. 10.1093/emboj/17.24.7442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquinelli, A. E. , Reinhart, B. J. , Slack, F. , Martindale, M. Q. , Kuroda, M. I. , Maller, B. , … Ruvkun, G. (2000). Conservation of the sequence and temporal expression of let‐7 heterochronic regulatory RNA. Nature, 408(6808), 86–89. 10.1038/35040556 [DOI] [PubMed] [Google Scholar]
- Pearce, S. F. , Rorbach, J. , Van Haute, L. , D'Souza, A. R. , Rebelo‐Guiomar, P. , Powell, C. A. , … Minczuk, M. (2017). Maturation of selected human mitochondrial tRNAs requires deadenylation. eLife, 6, e27596. 10.7554/eLife.27596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirouz, M. , Du, P. , Munafò, M. , & Gregory, R. I. (2016). Dis3L2‐mediated decay is a quality control pathway for noncoding RNAs. Cell Reports, 16(7), 1861–1873. 10.1016/j.celrep.2016.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirouz, M. , Munafò, M. , Ebrahimi, A. G. , Choe, J. , & Gregory, R. I. (2019). Exonuclease requirements for mammalian ribosomal RNA biogenesis and surveillance. Nature Structural and Molecular Biology, 26(6), 490–500. 10.1038/s41594-019-0234-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisacane, P. , & Halic, M. (2017). Tailing and degradation of Argonaute‐bound small RNAs protect the genome from uncontrolled RNAi. Nature Communications, 8, 15332. 10.1038/ncomms15332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskounova, E. , Polytarchou, C. , Thornton, J. E. , Lapierre, R. J. , Pothoulakis, C. , Hagan, J. P. , … Gregory, R. I. (2011). Lin28A and Lin28B inhibit let‐7 microRNA biogenesis by distinct mechanisms. Cell, 147(5), 1066–1079. 10.1016/j.cell.2011.10.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preker, P. , Almvig, K. , Christensen, M. S. , Valen, E. , Mapendano, C. K. , Sandelin, A. , & Jensen, T. H. (2011). PROMoter uPstream transcripts share characteristics with mRNAs and are produced upstream of all three major types of mammalian promoters. Nucleic Acids Research, 39(16), 7179–7193. 10.1093/nar/gkr370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston, M. A. , Porter, D. F. , Chen, F. , Buter, N. , Lapointe, C. P. , Keles, S. , … Wickens, M. (2019). Unbiased screen of RNA tailing activities reveals a poly(UG) polymerase. Nature Methods, 16(5), 437–445. 10.1038/s41592-019-0370-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford, H. E. , Meijer, H. A. , & de Moor, C. H. (2008). Translational control by cytoplasmic polyadenylation in Xenopus oocytes. Biochimica et Biophysica Acta – Gene Regulatory Mechanisms, 1779(4), 217–229. 10.1016/j.bbagrm.2008.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rammelt, C. , Bilen, B. , Zavolan, M. , & Keller, W. (2011). PAPD5, a noncanonical poly(A) polymerase with an unusual RNA‐binding motif. RNA, 17(9), 1737–1746. 10.1261/rna.2787011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimão‐Pinto, M. M. , Ignatova, V. , Burkard, T. R. , Hung, J. H. , Manzenreither, R. A. , Sowemimo, I. , … Ameres, S. L. (2015). Uridylation of RNA hairpins by Tailor confines the emergence of MicroRNAs in Drosophila . Molecular Cell, 59(2), 203–216. 10.1016/j.molcel.2015.05.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimão‐Pinto, M. M. , Manzenreither, R. A. , Burkard, T. R. , Sledz, P. , Jinek, M. , Mechtler, K. , & Ameres, S. L. (2016). Molecular basis for cytoplasmic RNA surveillance by uridylation‐triggered decay in Drosophila . The EMBO Journal, 35(22), 2417–2434. 10.15252/embj.201695164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhart, B. J. , Slack, F. J. , Basson, M. , Pasquienelll, A. E. , Bettlnger, J. C. , Rougvle, A. E. , … Ruvkun, G. (2000). The 21‐nucleotide let‐7 RNA regulates developmental timing in Caenorhabditis elegans . Nature, 403(6772), 901–906. 10.1038/35002607 [DOI] [PubMed] [Google Scholar]
- Reyes, J. M. , & Ross, P. J. (2016). Cytoplasmic polyadenylation in mammalian oocyte maturation. WIREs: RNA, 7(1), 71–89. 10.1002/wrna.1316 [DOI] [PubMed] [Google Scholar]
- Rissland, O. S. , & Norbury, C. J. (2008). The Cid1 poly(U) polymerase. Biochimica et Biophysica Acta – Gene Regulatory Mechanisms, 1779(4), 286–294. 10.1016/j.bbagrm.2008.03.003 [DOI] [PubMed] [Google Scholar]
- Roake, C. M. , Chen, L. , Chakravarthy, A. L. , Ferrell, J. E. , Raffa, G. D. , & Artandi, S. E. (2019). Disruption of telomerase RNA maturation kinetics precipitates disease. Molecular Cell, 74(4), 688–700.e3. 10.1016/j.molcel.2019.02.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach, J. , Nicholls, T. J. J. , & Minczuk, M. (2011). PDE12 removes mitochondrial RNA poly(A) tails and controls translation in human mitochondria. Nucleic Acids Research, 39(17), 7750–7763. 10.1093/nar/gkr470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouhana, L. , Wang, L. , Buter, N. , Jae, E. K. , Schiltz, C. A. , Gonzalez, T. , … Wickens, M. (2005). Vertebrate GLD2 poly(A) polymerases in the germline and the brain. RNA, 11(7), 1117–1130. 10.1261/rna.2630205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rüegger, S. , Miki, T. S. , Hess, D. , & Großhans, H. (2015). The ribonucleotidyl transferase USIP‐1 acts with SART3 to promote U6 snRNA recycling. Nucleic Acids Research, 43(6), 3344–3357. 10.1093/nar/gkv196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- San Paolo, S. , Vanacova, S. , Schenk, L. , Scherrer, T. , Blank, D. , Keller, W. , & Gerber, A. P. (2009). Distinct roles of non‐canonical poly(A) polymerases in RNA metabolism. PLoS Genetics, 5(7), e1000555. 10.1371/journal.pgen.1000555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartain, C. V. , Cui, J. , Meisel, R. P. , & Wolfner, M. F. (2011). The poly(A) polymerase GLD2 is required for spermatogenesis in Drosophila melanogaster . Development, 138(8), 1619–1629. 10.1242/dev.059618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid, M. , Küchler, B. , & Eckmann, C. R. (2009). Two conserved regulatory cytoplasmic poly(A) polymerases, GLD‐4 and GLD‐2, regulate meiotic progression in C. elegans . Genes and Development, 23(7), 824–836. 10.1101/gad.494009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz, H. L. , Goetz, T. , Kaschkoetoe, J. , & Weber, B. H. F. (2004). The retinome – Defining a reference transcriptome of the adult mammalian retina/retinal pigment epithelium. BMC Genomics, 5(1), 50. 10.1186/1471-2164-5-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, E. C. , Gardner, E. J. , Masood, A. , Chuang, N. T. , Vertino, P. M. , & Devine, S. E. (2016). A hot L1 retrotransposon evades somatic repression and initiates human colorectal cancer. Genome Research, 26(6), 745–755. 10.1101/gr.201814.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shchepachev, V. , Wischnewski, H. , Missiaglia, E. , Soneson, C. , & Azzalin, C. M. (2012). Mpn1, mutated in poikiloderma with neutropenia protein 1, is a conserved 3′‐to‐5′ RNA exonuclease processing U6 small nuclear RNA. Cell Reports, 2(4), 855–865. 10.1016/j.celrep.2012.08.031 [DOI] [PubMed] [Google Scholar]
- Shchepachev, V. , Wischnewski, H. , Soneson, C. , Arnold, A. W. , & Azzalin, C. M. (2015). Human Mpn1 promotes post‐transcriptional processing and stability of U6atac. FEBS Letters, 589(18), 2417–2423. 10.1016/j.febslet.2015.06.046 [DOI] [PubMed] [Google Scholar]
- Shcherbik, N. , Wang, M. , Lapik, Y. R. , Srivastava, L. , & Pestov, D. G. (2010). Polyadenylation and degradation of incomplete RNA polymerase I transcripts in mammalian cells. EMBO Reports, 11(2), 106–111. 10.1038/embor.2009.271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin, J. , Paek, K. Y. , Ivshina, M. , Stackpole, E. , & Richter, J. D. (2017). Essential role for non‐canonical poly(A) polymerase GLD4 in cytoplasmic polyadenylation and carbohydrate metabolism. Nucleic Acids Research, 45(11), 6793–6804. 10.1093/nar/gkx239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, A. , Yan, J. , Pagano, D. J. , Dodson, A. E. , Fei, Y. , Gorham, J. , … Kennedy, S. (2020). Poly(UG)‐tailed RNAs in genome protection and epigenetic inheritance. Nature, 582, 1–6. 10.1038/s41586-020-2323-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S. , Bjerke, G. A. , Muhlrad, D. , Yi, R. , & Parker, R. (2019). The RNase PARN controls the levels of specific miRNAs that contribute to p53 regulation. Molecular Cell, 73(6), 1204–1216.e4. 10.1016/j.molcel.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S. , & Parker, R. (2017). PARN modulates Y RNA stability and its 3′‐end formation. Molecular and Cellular Biology, 37(20), e00264–e00217. 10.1128/mcb.00264-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla, S. , Schmidt, J. C. , Goldfarb, K. C. , Cech, T. R. , & Parker, R. (2016). Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nature Structural and Molecular Biology, 23(4), 286–292. 10.1038/nsmb.3184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinturel, F. , Gerber, A. , Mauvoisin, D. , Wang, J. , Gatfield, D. , Stubblefield, J. J. , … Schibler, U. (2017). Diurnal oscillations in liver mass and cell size accompany ribosome assembly cycles. Cell, 169(4), 651–663.e14. 10.1016/j.cell.2017.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, A. , Park, J. E. , & Kim, V. N. (2018). PARN and TOE1 constitute a 3′ end maturation module for nuclear non‐coding RNAs. Cell Reports, 23(3), 888–898. 10.1016/j.celrep.2018.03.089 [DOI] [PubMed] [Google Scholar]
- Spracklin, G. , Fields, B. , Wan, G. , Becker, D. , Wallig, A. , Shukla, A. , & Kennedy, S. (2017). The RNAi inheritance machinery of Caenorhabditis elegans . Genetics, 206(3), 1403–1416. 10.1534/genetics.116.198812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtelny, A. O. , Eichhorn, S. W. , Chen, G. R. , Sive, H. , & Bartel, D. P. (2014). Poly(A)‐tail profiling reveals an embryonic switch in translational control. Nature, 508(7494), 66–71. 10.1038/nature13007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudo, H. , Nozaki, A. , Uno, H. , Ishida, Y. i. , & Nagahama, M. (2016). Interaction properties of human TRAMP‐like proteins and their role in pre‐rRNA 5′ETS turnover. FEBS Letters, 590(17), 2963–2972. 10.1002/1873-3468.12314 [DOI] [PubMed] [Google Scholar]
- Suh, N. , Jedamzik, B. , Eckmann, C. R. , Wickens, M. , & Kimble, J. (2006). The GLD‐2 poly(A) polymerase activates gld‐1 mRNA in the Caenorhabditis elegans germ line. Proceedings of the National Academy of Sciences of the United States of America, 103(41), 15108–15112. 10.1073/pnas.0607050103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanger, S. A. , He, Y. A. , Richter, J. D. , & Bassell, G. J. (2013). Dendritic GluN2A synthesis mediates activity‐induced NMDA receptor insertion. Journal of Neuroscience, 33(20), 8898–8908. 10.1523/JNEUROSCI.0289-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka, H. , Kanda, M. , Shimizu, D. , Tanaka, C. , Kobayashi, D. , Hayashi, M. , … Kodera, Y. (2017). FAM46C serves as a predictor of hepatic recurrence in patients with resectable gastric Cancer. Annals of Surgical Oncology, 24(11), 3438–3445. 10.1245/s10434-016-5636-y [DOI] [PubMed] [Google Scholar]
- Thornton, J. E. , Chang, H. M. , Piskounova, E. , & Gregory, R. I. (2012). Lin28‐mediated control of let‐7 microRNA expression by alternative TUTases Zcchc11 (TUT4) and Zcchc6 (TUT7). RNA, 18(10), 1875–1885. 10.1261/rna.034538.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton, J. E. , Du, P. , Jing, L. , Sjekloca, L. , Lin, S. , Grossi, E. , … Gregory, R. I. (2014). Selective microRNA uridylation by Zcchc6 (TUT7) and Zcchc11 (TUT4). Nucleic Acids Research, 42(18), 11777–11791. 10.1093/nar/gku805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomecki, R. , Dmochowska, A. , Gewartowski, K. , Dziembowski, A. , & Stepien, P. P. (2004). Identification of a novel human nuclear‐encoded mitochondrial poly(A) polymerase. Nucleic Acids Research, 32(20), 6001–6014. 10.1093/nar/gkh923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe, R. , Guschina, E. , Hossbach, M. , Urlaub, H. , Lührmann, R. , & Benecke, B. J. (2006). Identification, cloning, and functional analysis of the human U6 snRNA‐specific terminal uridylyl transferase. RNA, 12(8), 1494–1504. 10.1261/rna.87706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trippe, R. , Richly, H. , & Benecke, B. J. (2003). Biochemical characterization of a U6 small nuclear RNA‐specific terminal uridylyltransferase. European Journal of Biochemistry, 270(5), 971–980. 10.1046/j.1432-1033.2003.03466.x [DOI] [PubMed] [Google Scholar]
- Tsai, H. Y. , Chen, C. C. G. , Conte, D. , Moresco, J. J. , Chaves, D. A. , Mitani, S. , … Mello, C. C. (2015). A ribonuclease coordinates siRNA amplification and mRNA cleavage during RNAi. Cell, 160(3), 407–419. 10.1016/j.cell.2015.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udagawa, T. , Swanger, S. A. , Takeuchi, K. , Kim, J. H. , Nalavadi, V. , Shin, J. , … Richter, J. D. (2012). Bidirectional control of mRNA translation and synaptic plasticity by the cytoplasmic polyadenylation complex. Molecular Cell, 47(2), 253–266. 10.1016/j.molcel.2012.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko, D. , Chiu, H. S. , Treiber, T. , Weyn‐Vanhentenryck, S. M. , Treiber, N. , Meister, G. , … Zhang, C. (2018). LIN28 selectively modulates a subclass of Let‐7 microRNAs. Molecular Cell, 71(2), 271–283.e5. 10.1016/j.molcel.2018.06.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko, D. , Hrossova, D. , Potesil, D. , Chalupnikova, K. , Hrazdilova, K. , Pachernik, J. , … Vanacova, A. (2013). Mammalian DIS3L2 exoribonuclease targets the uridylated precursors of let‐7 miRNAs. RNA, 19(12), 1632–1638. 10.1261/rna.040055.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ustianenko, D. , Pasulka, J. , Feketova, Z. , Bednarik, L. , Zigackova, D. , Fortova, A. , … Vanacova, S. (2016). TUT‐DIS3L2 is a mammalian surveillance pathway for aberrant structured non‐coding RNAs. The EMBO Journal, 35(20), 2179–2191. 10.15252/embj.201694857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Eyck, L. , Bruni, F. , Ronan, A. , Briggs, T. A. , Roscioli, T. , Rice, G. I. , … Crow, Y. J. (2019). Biallelic mutations in MTPAP associated with a lethal encephalopathy. Neuropediatrics, 51, 178–184. 10.1055/s-0039-3400979 [DOI] [PubMed] [Google Scholar]
- van Wolfswinkel, J. C. , Claycomb, J. M. , Batista, P. J. , Mello, C. C. , Berezikov, E. , & Ketting, R. F. (2009). CDE‐1 affects chromosome segregation through uridylation of CSR‐1‐bound siRNAs. Cell, 139(1), 135–148. 10.1016/j.cell.2009.09.012 [DOI] [PubMed] [Google Scholar]
- Vaňáčová, Š. , Wolf, J. , Martin, G. , Blank, D. , Dettwiler, S. , Friedlein, A. , … Keller, W. (2005). A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biology, 3(6), 986–997. 10.1371/journal.pbio.0030189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vikova, V. , Jourdan, M. , Robert, N. , Requirand, G. , Boireau, S. , Bruyer, A. , … Moreaux, J. (2019). Comprehensive characterization of the mutational landscape in multiple myeloma cell lines reveals potential drivers and pathways associated with tumor progression and drug resistance. Theranostics, 9(2), 540–553. 10.7150/thno.28374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, B. A. , Mavrommatis, K. , Wardell, C. P. , Cody Ashby, T. , Bauer, M. , Davies, F. E. , … Morgan, G. J. (2018). Identification of novel mutational drivers reveals oncogene dependencies in multiple myeloma. Blood, 132(6), 587–597. 10.1182/blood-2018-03-840132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan, X. Y. , Zhai, X. F. , Jiang, Y. P. , Han, T. , Zhang, Q. Y. , & Xin, H. L. (2017). Antimetastatic effects of norcantharidin on hepatocellular carcinoma cells by up‐regulating FAM46C expression. American Journal of Translational Research, 9(1), 155–166 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/28123642 [PMC free article] [PubMed] [Google Scholar]
- Wang, D. D. H. , Guo, X. E. , Modrek, A. S. , Chen, C. F. , Chen, P. L. , & Lee, W. H. (2014). Helicase SUV3, polynucleotide phosphorylase, and mitochondrial polyadenylation polymerase form a transient complex to modulate mitochondrial mRNA polyadenylated tail lengths in response to energetic changes. Journal of Biological Chemistry, 289(24), 16727–16735. 10.1074/jbc.M113.536540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, L. , Eckmann, C. R. , Kadyk, L. C. , Wickens, M. , & Kimble, J. (2002). A regulatory cytoplasmic poly(A) polymerase in Caenorhabditis elegans . Nature, 419(6904), 312–316. 10.1038/nature01039 [DOI] [PubMed] [Google Scholar]
- Wang, X. , & Voronina, E. (2020). Diverse roles of PUF proteins in germline stem and progenitor cell development in C. elegans . Frontiers in Cell and Developmental Biology, 8(29), 1–12. 10.3389/fcell.2020.00029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki, Z. , Krawczyk, P. S. , Adamska, D. , Bijata, K. , Garcia‐Perez, J. L. , & Dziembowski, A. (2018). Uridylation by TUT4/7 restricts retrotransposition of human LINE‐1s. Cell, 175, 1537–1548. 10.1016/j.cell.2018.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warkocki, Z. , Liudkovska, V. , Gewartowska, O. , Mroczek, S. , & Dziembowski, A. (2018). Terminal nucleotidyl transferases ( TENTs ) in mammalian RNA metabolism. Philosophical Transactions of the Royal Society B: Biological Sciences, 373(1762), 20180162. 10.1098/rstb.2018.0162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe, T. , Yamamoto, T. , Tsukano, K. , Hirano, S. , Horikawa, A. , & Michiue, T. (2018). Fam46a regulates bmp‐dependent pre‐placodal ectoderm differentiation in xenopus. Development (Cambridge), 145(20), dev166710. 10.1242/dev.166710 [DOI] [PubMed] [Google Scholar]
- Weick, E.‐M. , Puno, M. R. , Januszyk, K. , Zinder, J. C. , Dimattia, M. A. , & Lima, C. D. (2018). Helicase‐dependent RNA decay illuminated by a cryo‐EM structure of a human nuclear RNA exosome‐MTR4 complex. Cell, 173(7), 1–15. 10.1016/j.cell.2018.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westholm, J. O. , & Lai, E. C. (2011). Mirtrons: MicroRNA biogenesis via splicing. Biochimie, 93(11), 1897–1904. 10.1016/j.biochi.2011.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, W. C. , Hornig‐Do, H. T. , Bruni, F. , Chang, J. H. o. , Jourdain, A. A. , Martinou, J. C. , … Lightowlers, R. N. (2014). A human mitochondrial poly(A) polymerase mutation reveals the complexities of post‐transcriptional mitochondrial gene expression. Human Molecular Genetics, 23(23), 6345–6355. 10.1093/hmg/ddu352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman, S. K. , Knouf, E. C. , Parkin, R. K. , Fritz, B. R. , Lin, D. W. , Dennis, L. M. , … Tewari, M. (2011). Post‐transcriptional generation of miRNA variants by multiple nucleotidyl transferases contributes to miRNA transcriptome complexity. Genome Research, 21(9), 1450–1461. 10.1101/gr.118059.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia, E. , Kanematsu, S. , Suenaga, Y. , Elzawahry, A. , Kondo, H. , Otsuka, N. , … Yokoi, S. (2018). MicroRNA induction by copy number gain is associated with poor outcome in squamous cell carcinoma of the lung. Scientific Reports, 8(1), 1–10. 10.1038/s41598-018-33696-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, F. , Feng, X. , Chen, X. , Weng, C. , Yan, Q. , Xu, T. , … Guang, S. (2018). A cytoplasmic Argonaute protein promotes the inheritance of RNAi. Cell Reports, 23(8), 2482–2494. 10.1016/j.celrep.2018.04.072 [DOI] [PubMed] [Google Scholar]
- Yamagishi, R. , Tsusaka, T. , Mitsunaga, H. , Maehata, T. , & Hoshino, S. I. (2016). The STAR protein QKI‐7 recruits PAPD4 to regulate post‐transcriptional polyadenylation of target mRNAs. Nucleic Acids Research, 44(6), 2475–2490. 10.1093/nar/gkw118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita, S. , Takagi, Y. , Nagaike, T. , & Tomita, K. (2017). Crystal structures of U6 snRNA‐specific terminal uridylyltransferase. Nature Communications, 8, 15788. 10.1038/ncomms15788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, A. , Bofill‐De Ros, X. , Shao, T. J. , Jiang, M. , Li, K. , Villanueva, P. , … Gu, S. (2019). 3′ Uridylation confers miRNAs with non‐canonical target repertoires. Molecular Cell, 75(3), 511–522.e4. 10.1016/j.molcel.2019.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Q. , Nausch, L. W. M. , Martin, G. , Keller, W. , & Doublié, S. (2014). Crystal structure of human poly(A) polymerase gamma reveals a conserved catalytic core for canonical poly(A) polymerases. Journal of Molecular Biology, 426(1), 43–50. 10.1016/j.jmb.2013.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, R. , Natsume, Y. , & Noda, T. (2007). TACC3 is required for the proper mitosis of sclerotome mesenchymal cells during formation of the axial skeleton. Cancer Science, 98(4), 555–562. 10.1111/j.1349-7006.2007.00433.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates, L. A. , Fleurdépine, S. , Rissland, O. S. , De Colibus, L. , Harlos, K. , Norbury, C. J. , & Gilbert, R. J. C. (2012). Structural basis for the activity of a cytoplasmic RNA terminal uridylyl transferase. Nature Structural and Molecular Biology, 19(8), 782–787. 10.1038/nsmb.2329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo, J. , & Kim, V. N. (2018). U‐tail as a guardian against invading RNAs. Nature Structural and Molecular Biology, 25(10), 903–905. 10.1038/s41594-018-0139-0 [DOI] [PubMed] [Google Scholar]
- Yu, S. , & Kim, V. N. (2020). A tale of non‐canonical tails: Gene regulation by post‐transcriptional RNA tailing. Nature Reviews Molecular Cell Biology. 10.1038/s41580-020-0246-8. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- Zhang, Q. Y. , Yue, X. Q. , Jiang, Y. P. , Han, T. , & Xin, H. L. (2017). FAM46C is critical for the anti‐proliferation and pro‐apoptotic effects of norcantharidin in hepatocellular carcinoma cells. Scientific Reports, 7(1), 396. 10.1038/s41598-017-00313-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, C. , Ouyang, Y. C. , Jiang, B. , Lin, X. , Chen, J. , Dong, M. Z. , … Han, C. (2019). Non‐canonical RNA polyadenylation polymerase FAM46C is essential for fastening sperm head and flagellum in mice†. Biology of Reproduction, 100(6), 1673–1685. 10.1093/biolre/ioz083 [DOI] [PubMed] [Google Scholar]
- Zhu, Y. X. , Shi, C. X. , Bruins, L. A. , Jedlowski, P. , Wang, X. , Kortüm, K. M. , … Stewart, A. K. (2017). Loss of FAM46C promotes cell survival in myeloma. Cancer Research, 77(16), 4317–4327. 10.1158/0008-5472.CAN-16-3011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang, X. , & Lu, M. (2018). The potential functions of FAM46C in oral squamous cell carcinoma. Oncotargets and Therapy, 11, 8915–8923. 10.2147/OTT.S185244 [DOI] [PMC free article] [PubMed] [Google Scholar]


