Abstract
Hepatocellular carcinoma (HCC) is the fourth leading cause of cancer-associated mortality worldwide. Thus, there is an urgent requirement to identify novel diagnostic and prognostic biomarkers for this disease. The present study aimed to identify the hub genes associated with the progression and prognosis of patients with HCC. A total of three expression profiles of HCC tissues were extracted from the Gene Expression Omnibus (GEO) database, followed by the identification of differentially expressed genes (DEGs) using the GEO2R method. The identified DEGs were assessed for survival significance using Kaplan-Meier analysis. Among the 15 identified DEGs in HCC tissues [cytochrome P450 family 39 subfamily A member 1, cysteine rich angiogenic inducer 61, Fos proto-oncogene, forkhead transcription factor 1 (FOXO1), growth arrest and DNA damage inducible β, Inhibitor of DNA binding 1, interleukin-1 receptor accessory protein, metallothionein-1M, pleckstrin homology-like domain family A member 1, Rho family GTPase 3, serine dehydratase, suppressor of cytokine signaling 2 (SOCS2), tyrosine aminotransferase (TAT), S100 calcium-binding protein P and serine protease inhibitor Kazal-type 1 (SPINK1)]. Low expression levels of FOXO1, SOCS2 and TAT and high SPINK1 expression indicated poor survival outcomes for patients with HCC. In addition, SOCS2 was associated with distinct stages of HCC progression in patients and presented optimal diagnostic value. In vitro functional experiments indicated that overexpression of SOCS2 inhibited HCC cell proliferation and migration. Taken together, the results of the present study suggest that SOCS2 may act as a valuable prognostic marker that is closely associated with HCC progression.
Keywords: hepatocellular carcinoma, bioinformatics analysis, differentially expressed genes, prognosis
Introduction
Hepatocellular carcinoma (HCC) is one of the most highly malignant and fatal cancers worldwide (1). It is the fourth leading cause of cancer-associated mortality worldwide, with ~841,000 new cases and 782,000 mortalities per year (1). The risk factors of HCC include hepatitis B virus or hepatitis C virus infection, consumption of aflatoxin contaminated food, alcohol abuse, obesity and smoking (2). Several strategies have been used for HCC treatment, such as surgical resection, chemotherapy, radiotherapy and targeted therapy (2). However, for patients with end-stage HCC, the 5-year survival rate remains <10% (3). HCC is a neoplastic disease with complex molecular mechanisms, which are affected by genetic or epigenetic mutations, genomic instability and environmental factors (3,4). Chronic inflammation, dysregulation of angiogenesis, changes in cellular metabolism and abnormal endocrine hormones may also be involved in the tumorigenesis of HCC (5). Recently, several studies have revealed key signaling pathways and genes that play critical roles in HCC (6,7). However, the underlying molecular mechanisms of HCC onset and progression remain unclear. Thus, it is important to investigate the molecular mechanisms of HCC pathogenesis to identify key molecular targets for the early diagnosis and treatment of patients with HCC.
Recently, the development of high-throughput sequencing and microarray technologies have provided a novel platform for studies of gene expression profiles and identification of key factors associated with tumor development. Microarray technique is a method used to analyze general genetic alterations, which has been extensively applied in the investigations of tumorigenicity to identify promising biomarkers for cancer diagnosis, treatment and prognosis (6,8). For example, analysis of gene expression profiles of 64 primary prostate tumors and 24 metastatic samples revealed that patients with metastasis had 415 upregulated and 364 downregulated genes, indicating high heterogeneity of the metastatic samples (9). Systematic analysis of publicly available sequencing data using integrated bioinformatics methods may be an efficient way to overcome limitations, such as the use of different sequencing platforms or small sample sizes, and can provide further insight for identifying novel diagnostic markers and therapeutic targets in different types of tumor tissues, such as endometrial cancer (10), osteosarcoma (11), non-small cell lung cancer (12) and gastric cancer (13).
The present study analyzed three independent sequencing datasets of HCC tissues and identified differentially expressed genes (DEGs) using a series of bioinformatics analysis methods. A protein-protein interaction (PPI) network was constructed, and function enrichment and survival analyses were performed to thoroughly investigate the molecular features of the DEGs. In addition, the key targets affecting the tumorigenesis of HCC cells were identified via biological function studies.
Materials and methods
Data source acquisition
The gene microarray expression datasets, GSE22058 (14,15), GSE57957 (16) and GSE14323 (17), including 186 HCC tissue samples and 150 adjacent tumor tissue samples, were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). The microarray datasets included in the present study satisfied the following selection criteria: i) They included human HCC tissues and adjacent tumor tissues; ii) the number of cases in the HCC and adjacent tumor groups was at least 10 and iii) they had intact RNA expression profiles for further analysis. The data acquisition and application methods in the present study complied with the guidelines and policies of the GEO database (18).
Identification of DEGs
The GSE22058, GSE57957 and GSE14323 expression profiles were normalized and analyzed using the GEO2R tool (https://www.ncbi.nlm.nih.gov/geo/geo2r). The criteria of P<0.05 and |logFC|>1 was applied to screen for the DEGs. The volcano plot of each dataset was constructed using the ‘volcano R’ package (version 3.2.0; R Foundation). The overlap of DEGs between the GSE22058, GSE57957 and GSE14323 datasets were categorized as common DEGs, which were retained for further studies.
PPI network and module analysis
The PPI network was constructed using Cytoscape software (version 3.4.0; National Resource for Network Biology). Associations between the DEG-encoded proteins were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/cgi/input.pl). PPIs with a confidence score ≥0.4 were reserved.
Functional enrichment analysis of the DEGs
Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses (19) were performed using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf.gov) to identify the biological processes, molecular functions and cellular components, and signaling pathways associated with the DEGs. P<0.05 was considered to indicate statistical significance.
Survival analysis
Overall survival (OS) and disease-free survival (DFS) analyses of the integrated DEGs were performed using the cBioPortal database (http://www.cbioportal.org). A total of five HCC studies, including MSK, Clin Cancer Res 2018 (20); INSERM, Nat Genet 2015 (21); AMC, Hepatology 2014 (22); RIKEN, Nat Genet 2012 (23) and The Cancer Genome Atlas (TCGA) (24), which included 1,000 patients with clinical information, were selected. OS analysis was performed using the Gene Expression Profiling Interactive Analysis (GEPIA) database (http://gepia.cancer-pku.cn). A total of 80 patients with HCC from the 920th Hospital were classified into the high expression group (n=40) or the low expression group (n=40), based on the median expression value (22.178) to determine the prognosis of suppressor of cytokine signaling 2 (SOCS2) using the ‘survival R’ package (version 3.2.0; R Foundation). P<0.05 was considered to indicate prognostic significance. Detailed information of the 80 patients is listed in Table SV.
Expression levels of SOCS2, TAT, FOXO1 and SPINK in TCGA dataset
The mRNA expression and clinical data were downloaded from TCGA-Liver Hepatocellular Carcinoma database (TCGA-LIHC, http://tcga-data.nci.nih.gov/tcga), including 50 healthy individuals and 371 patients with HCC. The expression levels of four prognosis-related genes (SOCS2, TAT, FOXO1 and SPINK) were detected in healthy individuals and different pathological stages of patients with HCC.
Receiver operating characteristic (ROC) curve
ROC curves of SOCS2, TAT, FOXO1 and SPINK were plotted based on their expression and sample feature (tumor vs. normal) in the GSE22058 dataset to determine their diagnostic values. The ROC curves were plotted using SPSS software (version 21; IBM Corp.).
Cell lines and clinical tissues
The HCC cell line, Huh7, was purchased from the Chinese Academy of Sciences Cell Bank and maintained in DMEM medium supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher Scientific, Inc.), at 37°C in 5% CO2.
A total of 12 pairs of HCC tissues and adjacent normal tissues (5 cm away from HCC tissues) were collected from patients with HCC who received surgical resection at the 920th Hospital between June 2018 and October 2019. The patients included 9 men and 3 women (age range, 53–65 years; mean age, 60.17 years). The histopathologic features of tumor tissues and adjacent normal tissues were confirmed by H&E staining. Fresh clinical samples were stored at −80°C until subsequent experimentation. The present study was approved by the Ethics Committee of the 920th Hospital (Kunming, China; approval no. 2018-020-01) and written informed consent was provided by all patients prior to the study start.
Reverse transcription-quantitative (RT-q)PCR
Total RNA was extracted from Huh7 cells using TRIzol® reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. A total of 1 µg RNA was reverse transcribed into cDNA using the Hifair® II 1st Strand cDNA Synthesis SuperMix kit (Yeasen Biotech Co.), under the following conditions: 25°C for 5 min, 55°C for 15 min and 85°C for 5 min. qPCR was subsequently performed using the Hieff® qPCR SYBR Green Master Mix kit (Yeasen Biotech Co.) in QuantStudio™ 5 System (Thermo Fisher Scientific, Inc.). The following thermocycling conditions were used for qPCR: 95°C for 5 min, followed by 40 cycles at 95°C for 10 sec, 60°C for 30 sec and elongation at 72°C for 2 min. The following primer sequences were used for qPCR: SOCS2 forward, 5′-GAGCCGGAGAGTCTGGTTTC-3′ and reverse, 5′-ATCCTGGAGGACGGATGACA-3′; and GAPDH forward, 5′-GGTCTCCTCTGACTTCAACA-3′ and reverse, 5′-GTGAGGGTCTCTCTCTTCCT-3′. Relative mRNA levels were calculated using the 2−ΔΔCq method (25) and normalized to the internal reference gene GAPDH.
Plasmid construction and cell transfection
To generate the SOCS2 overexpression construct, the SOCS2 ORF sequence was amplified via RT PCR and cloned into a pcDNA3.1 vector (Addgene) (OE). An empty pcDNA3.1 vector was used as the negative control (NC). SOCS2-specific small interfering (si)RNA and control siRNA were designed by Shanghai GenePharma Co., Ltd. The primer sequences used for vector construction and siRNA sequences were as follows: SOCS2 forward, 5′-GGATCCATGACCCTGCGGTGCCTTGAG-3′ and reverse, 5′-CTCGAGTTATACCTGGAATTTATATTCTTC-3′; control siRNA, 5′-GGATCAACTAACTTCCGAA-3′; and SOCS2 siRNA, 5′-GGACCAACTAATCTTCGAA-3′. Cells were transfected with the plasmids (2.5 µg) or siRNAs (50 nM) using Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific, Inc.) in 6-well plates, according to the manufacturer's protocol. Following incubation for 48 h at 37°C, transfected cells were harvested for subsequent experimentation.
Western blotting
Total protein was extracted from Huh7 cells using RIPA lysis buffer (Beyotime Institute of Biotechnology), containing ×100 protease inhibitor cocktail (Bio-Rad Laboratories, Inc.). Protein concentrations of lysates were detected using the bicinchoninic acid (BCA) assay (Beyotime Institute of Biotechnology). Equal amounts of protein lysates (20 µg/well) were separated by 10% SDS-PAGE, transferred onto polyvinylidene difluoride membranes (Bio-Rad Laboratories, Inc.) and blocked with 5% skim milk solution for 1 h at room temperature. The membranes were incubated with primary antibodies against SOCS2 (cat. no. A5703) and GAPDH (cat. no. AC001) (both 1:1,000 and purchased from ABclonal Biotech Co., Ltd.) overnight at 4°C. Following the primary incubation, membranes were incubated with horseradish peroxidase-conjugated Goat Anti-Rabbit IgG secondary antibody (1:2,000; cat. no. AS014; ABclonal Biotech Co., Ltd.) for 1 h at room temperature. Protein bands were visualized using BeyoECL Plus regent (Beyotime Institute of Biotechnology) in ImageQuant LAS4000 (GE Healthcare). GAPDH was used as the loading control.
Cell proliferation assay
HCC cells were transfected with the indicated siRNAs or plasmids for 24 h. Subsequently, cells were seeded into 96-well culture plates at a density of 4,000 cells/well. Cell proliferation was assessed via the Cell Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies, Inc.) at 0, 12, 24, 36 and 48 h following cell culture. The cell proliferation curve at each time point was plotted using the values of relative absorbance. EdU immunofluorescence staining was performed using the EdU kit (Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The results were quantified using ImageJ software (version 1.8.0; National Institutes of Health).
Wound healing assay
HCC cells were seeded into 6-well plates at a density of 4×105 cells/well and cultured until they reached 90% confluency. The cell monolayers were subsequently scratched using 200 µl pipette tips to create a gap. Cells were washed with phosphate buffered saline and cultured in fresh DMEM serum-free medium (Gibco; Thermo Fisher Scientific, Inc.). Images were acquired at 0 and 24 h using the Olympus IX73 light microscope (Olympus Corporation) to assess cell migration. Wounded areas between the cells were analyzed using ImageJ software (version 1.8.0; National Institutes of Health).
Statistical analysis
Statistical analysis was performed using GraphPad Prism 6 software (GraphPad Software, Inc.). All in vitro experiments were performed in triplicate and data are presented as the mean ± standard deviation. A two-tailed unpaired or paired Student's t-tests were used to compare differences between two groups. One-way ANOVA followed by Dunnett's test were used to compare differences between multiple groups. Pearson's correlation analysis was performed to determine the correlation between SOCS2 and FOXO1 expression. Survival curves were obtained via Kaplan-Meier analysis and the log-rank test between patients in the high and low expression groups, and Landmark analysis was performed when the survival curves cross. Age, gender, stage and SOCS2 expression level of 80 HCC patients were made for univariate and multivariate Cox analyses using SPSS software (version 21; IBM Corp.). P<0.05 was considered to indicate a statistically significant difference.
Results
Identification of DEGs
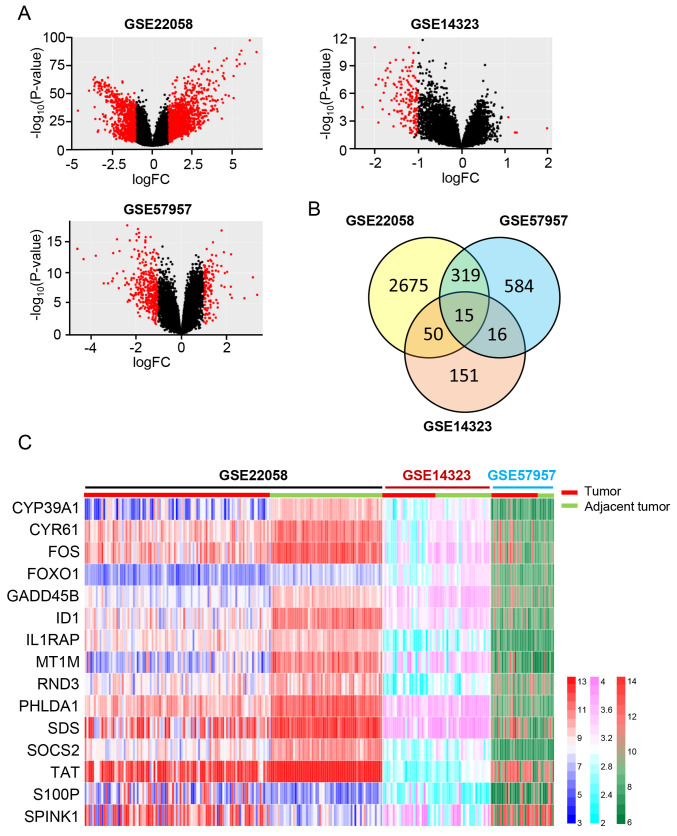
To identify the key DEGs in a large cohort of HCC samples, three sequencing datasets from the GEO database were selected, including HCC samples and adjacent normal tumor samples. Detailed information of the three datasets are presented in Table I. According to the screening criteria, a total of 2,657 DEGs were identified between the HCC tissues and adjacent normal tumor tissues in the GSE22058 dataset, which included 981 upregulated genes and 1,694 downregulated genes. A total of 584 DEGs were identified in the GSE57957 dataset, which consisted of 256 upregulated genes and 328 downregulated genes in HCC tissues compared with adjacent normal tumor tissues. The DEGs obtained from the GSE14323 dataset included five upregulated genes and 146 downregulated genes in HCC tissues. The volcano plots of the three datasets are presented in Fig. 1A. A total of 15 overlapping DEGs (13 downregulated genes and two upregulated genes) were identified by intersecting the three datasets (Fig. 1B and Table II). The heat map displays the detailed expression data of these 15 DEGs in each tissue sample of the three sequencing datasets (Fig. 1C).
Table I.
Gene Expression Omnibus datasets used in the present study.
Figure 1.
Identification of DEGs. (A) Volcano plots of the GSE22058, GSE14323 and GSE57957 datasets. Red dots represent DEGs with |logFC| >1 and P<0.05, black dots represent genes without significant changes. (B) Venn diagrams of overlapping DEGs between the GSE22058, GSE14323 and GSE57957 datasets. (C) A heat map of overlapping DEGs. Each column represents a sample, and each row represents one gene. The gradient color represents the gene expression value. DEGs, differentially expressed genes; FC, fold change.
Table II.
Detailed information of the 15 key differentially expressed genes.
| GSE22058 | GSE57957 | GSE14323 | |||||
|---|---|---|---|---|---|---|---|
| Gene symbol | Gene title | P-value | logFC | P-value | logFC | P-value | logFC |
| CYP39A1 | Cytochrome P450 family 39 subfamily A member 1 | 6.33×10−29 | −2.93 | 1.55×10−13 | −1.55 | 0.000307 | −1.11 |
| CYR61 | Cysteine rich angiogenic inducer 61 | 3.11×10−29 | −1.66 | 1.23×10−07 | −1.40 | 0.000624 | −1.12 |
| FOS | Fos proto-oncogene, AP-1 transcription factor subunit | 2.75×10−29 | −2.05 | 1.17×10−13 | −2.54 | 0.00240 | −1.21 |
| FOXO1 | Forkhead transcription factor 1 | 4.62×10−22 | −1.25 | 9.36×10−10 | −1.12 | 3.51×10−04 | −1.22 |
| GADD45B | Growth arrest and DNA damage inducible β | 3.86×10−35 | −1.79 | 6.63×10−12 | −1.58 | 0.000124 | −1.06 |
| ID1 | Inhibitor of DNA binding 1, HLH protein | 3.89×10−41 | −2.54 | 2.06×10−08 | −1.42 | 0.000219 | −1.03 |
| IL1RAP | Interleukin-1 receptor accessory protein | 2.24×10−14 | −1.02 | 4.64×10−11 | −1.34 | 0.000798 | −1.09 |
| MT1M | Metallothionein-1M | 2.23×10−53 | −3.97 | 4.54×10−10 | −2.75 | 0.00324 | −1.22 |
| PHLDA1 | Pleckstrin homology-like domain family A member 1 | 4.83×10−21 | −1.67 | 5.45×10−05 | −1.09 | 1.28×10−04 | −1.40 |
| RND3 | Rho family GTPase 3 | 2.45×10−42 | −1.94 | 1.94×10−13 | −1.64 | 1.32×10−04 | −1.03 |
| SDS | Serine dehydratase | 2.16×10−15 | −2.25 | 8.1×10−07 | −1.95 | 0.00144 | −1.04 |
| SOCS2 | Suppressor of cytokine signaling 2 | 2.95×10−34 | −2.13 | 3.66×10−10 | −1.24 | 0.000347 | −1.00 |
| TAT | Tyrosine aminotransferase | 1.64×10−12 | −1.38 | 6.28×10−08 | −2.10 | 1.40×10−09 | −1.40 |
| S100P | S100 calcium-binding protein P | 6.41×10−08 | 1.73 | 0.000373 | 1.25 | 0.00182 | 1.27 |
| SPINK1 | Serine protease inhibitor Kazal-type 1 | 6.06×10−13 | 2.67 | 1.1×10−08 | 3.38 | 0.001 | 1.93 |
Negative values represent downregulated genes, while positive values represent upregulated genes in hepatocellular carcinoma. FC, fold change.
PPI network and functional enrichment analysis of DEGs
To determine the associations between DEG-encoded proteins, a PPI network was constructed using the STRING database. In total, 34 proteins, 14 DEGs and 20 neighbor genes were obtained, and 251 edges were included in the PPI network (Fig. S1A). In the network, a large protein node indicated a strong ability to interact with other proteins. The top five highest value nodes were serine/threonine-protein kinase 1, V-Jun avian sarcoma virus 17 oncogene homolog, mitogen-activated protein kinase 8, mammalian target of rapamycin and early growth response 1, all of which have been reported to be involved in the development of multiple tumors (26–29). The top five highest value DEGs were Fos proto-oncogene (FOS), SOCS2, forkhead transcription factor 1 (FOXO1), growth arrest and DNA damage inducible β (GADD45B) and cysteine rich angiogenic inducer 61 (CYR61), which are also considered to play critical roles in tumor progression (30–34).
GO and KEGG enrichment analyses of the DEGs were performed using DAVID. The results demonstrated that the DEGs were predominantly enriched in ‘nucleoplasm’ and ‘transcription factor AP-1 complex’. Furthermore, they were significantly enriched in multiple biological processes and molecular functions associated with response to ‘abiotic stimuli’, ‘regulation of cell death’, ‘transcription factor binding’ and ‘activity of DNA-binding transcription activator’ (Fig. S1B-D and Tables SI–III). According to KEGG pathway enrichment analysis, the DEGs were significantly involved in ‘colorectal cancer’, ‘FOXO signaling pathway’, ‘osteoclast differentiation’, ‘insulin signaling pathway’ and ‘prolactin signaling pathway’ (Fig. S1E and Table SIV).
Survival analysis
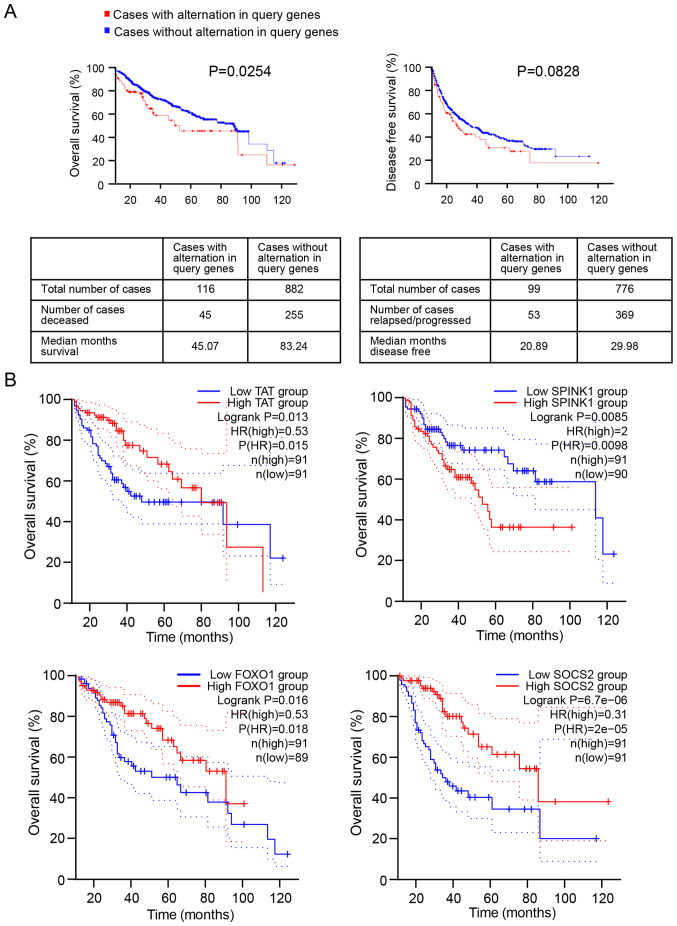
According to the clinical information of patients with HCC in the cBioPortal database, five HCC studies; MSK, Clin Cancer Res 2018 (20); INSERM, Nat Genet 2015 (21); AMC, Hepatology 2014 (22); RIKEN, Nat Genet 2012 (23) and TCGA (24), were selected to determine the prognostic values of the 15 DEGs. As presented in Fig. 2A, expression alteration of the 15 DEGs was significantly associated with a shorter OS time of patients with HCC (P=0.0254). The median survival time of 116 patients with DEG expression changes was 45.07 months, and the median survival time of 882 patients without alterations in DEG expression was 83.24 months. However, no significant difference was observed between the alteration of DEG levels and the DFS time of patients with HCC (P=0.0828).
Figure 2.
Prognostic values of the DEGs in patients with HCC. (A) Overall survival analysis (left) and disease-free survival analysis (right) of the 15 integrated DEGs in patients with HCC. (B) Overall survival analysis indicated that FOXO1, SPINK1, SOCS2 and TAT are prognostic markers in patients with HCC. DEGs, differentially expressed genes; HCC, hepatocellular carcinoma; FOXO1, forkhead transcription factor 1; SPINK1, serine protease inhibitor Kazal-type 1; SOCS2, suppressor of cytokine signaling 2; TAT, tyrosine aminotransferase.
To further investigate the association between the 15 DEGs and OS time of patients with HCC, Kaplan-Meier survival analysis of each DEG was performed using the GEPIA database. The results demonstrated that only four DEGs were significantly associated with the OS time of patients with HCC, including FOXO1 (P=0.016), serine protease inhibitor Kazal-type 1 (SPINK1, P=0.0085), SOCS2 (P=6.7×10−06) and tyrosine aminotransferase (TAT, P=0.013) (Figs. 2B and S2). Notably, low expression levels of FOXO1, SOCS2 and TAT, and high SPINK1 expression were associated with poor prognosis of patients with HCC.
SOCS2 is associated with HCC stages and demonstrates good diagnostic ability for HCC
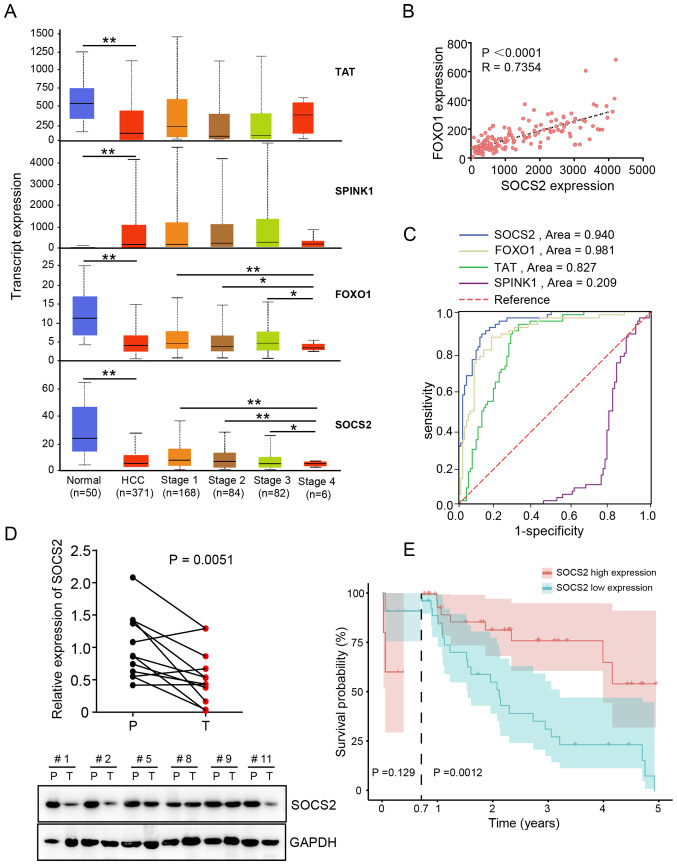
The expression levels of the four prognosis-related DEGs in TCGA dataset were assessed. The results demonstrated that the expression levels of FOXO1, SOCS2 and TAT were significantly downregulated in HCC tissues, whereas SPINK1 expression was significantly upregulated in HCC tissues compared with adjacent normal tissues. In addition, FOXO1 and SOCS2 expression were associated with HCC progression, whereby lower expression levels were observed in stage 4 patients with HCC (Fig. 3A). Furthermore, FOXO1 expression was positively correlated with SOCS2 in the GSE22058 dataset (Fig. 3B). ROC curve analysis demonstrated that SOCS2 presented a good diagnostic ability for HCC (Fig. 3C), and its area under the curve (AUC) value was 0.940 [95% confidence interval (CI), 0.906–0.975]. The AUC value of TAT, FOXO1 and SPINK1 were 0.827 (95% CI, 0.764–0.891), 0.891 (95% CI, 0.838–0.944) and 0.209 (95% CI, 0.138–0.280), respectively (Table SV).
Figure 3.
SOCS2 is a potential diagnostic marker for HCC. (A) The expression levels of the four key DEGs in The Cancer Genome Atlas HCC dataset. (B) FOXO1 expression was positively correlated with SOCS2 expression in the GSE22058 dataset. (C) Receiver operating characteristic curves of the four key DEGs were used to determine their diagnostic values. (D) SOCS2 mRNA (above, n=12 pairs) and protein (below, n=6 pairs) expression levels in clinical tissues. (E) Overall survival analysis of 80 patients with HCC from the 920th Hospital, based on SOCS2 expression. *P<0.05, **P<0.01 SOCS2, suppressor of cytokine signaling 2; HCC, hepatocellular carcinoma; DEGs, differentially expressed genes; FOXO1, forkhead transcription factor 1; SPINK1, serine protease inhibitor Kazal-type 1; TAT, tyrosine aminotransferase; P, normal tissue; T, HCC tissue.
SOCS2 expression was further validated in 12 pairs of clinical HCC tissues and adjacent normal tissues. The results demonstrated that SOCS2 mRNA and protein expression levels were downregulated in HCC tissues compared with adjacent normal tissues (Fig. 3D). Based on the median SOCS2 expression values, 80 patients with HCC from the 920th Hospital were classified into the high expression group (n=40) or the low expression group (n=40) (Table SVI). Long-term follow-up (0.7–5.0 years) showed that HCC cases in the low expression group were associated with a poor prognosis compared with the high expression group (P=0.0012), while there was no significant difference between the two groups in the short-term follow-up (0.0–0.7 years) (Fig. 3E). Furthermore, univariate and multivariate Cox analyses demonstrated that HCC clinical stage 1 and SOCS2 levels were independent prognostic factors for patients with HCC (Table III).
Table III.
Univariate and multivariate Cox analyses of hepatocellular carcinoma clinical characteristics and SOCS2 expression.
| Univariate analysis | Multivariate analysis | |||
|---|---|---|---|---|
| Variable | HR (95% CI) | P-value | HR (95% CI) | P-value |
| Age | 1.005 (0.986–1.024) | 0.593 | – | – |
| Gender | 1.033 (0.565–1.888) | 0.916 | – | – |
| Stage | ||||
| I | 2.338 (1.245–4.392) | 0.008 | 2.295 (1.093–4.821) | 0.028 |
| II | 0.657 (0.347–1.245) | 0.198 | – | – |
| III/IV | 0.517 (0.270–0.989) | 0.046 | 1.128 (0.512–2.485) | 0.765 |
| SOCS2 | 0.968 (0.942–0.994) | 0.018 | 0.968 (0.941–0.996) | 0.024 |
SOCS2, suppressor of cytokine signaling 2; HR, hazard ratio; CI, confidence interval; -, not applicable.
SOCS2 is a tumor suppressor for HCC progression
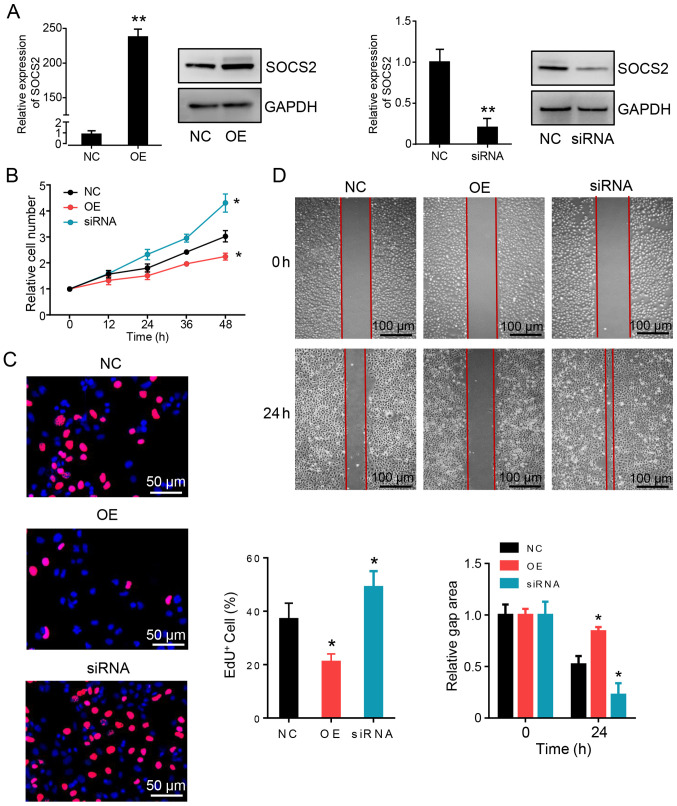
To determine the biological function of SOCS2 in HCC cells, SOCS2 expression in Huh7 cells was exogenously changed using recombinant expression plasmids and siRNAs. Both the overexpression and knockdown phenotypes of SOCS2 were confirmed via RT-qPCR and western blot analyses (Fig. 4A). Ectopic overexpression of SOCS2 inhibited HCC cell proliferation, as measured by the CCK-8 assay (Fig. 4B) and EdU staining (Fig. 4C). The wound healing assay demonstrated that overexpression of SOCS2 inhibited HCC cell metastasis compared with cells in the control group (Fig. 4D). Notably, SOCS2 knockdown promoted HCC cell proliferation and metastasis. Taken together, these results suggest that SOCS2 inhibits tumorigenesis in HCC cells.
Figure 4.
Overexpression of SOCS2 inhibits hepatocellular carcinoma tumorigenesis. (A) SOCS2 mRNA and protein expression levels in Huh7 cells. (B) The Cell Counting Kit-8 assay was performed to assess proliferation in SOCS2 overexpressed and suppressed Huh7 cells. (C) EdU staining was performed to assess proliferation in SOCS2 overexpressed and suppressed Huh7 cells. Scale bar, 50 µm. (D) The wound healing assay was performed to assess the migratory ability in SOCS2 overexpressed and suppressed Huh7 cells. Scale bar, 100 µm. *P<0.05, **P<0.01. SOCS2, suppressor of cytokine signaling 2; OE, SOCS2 overexpression; NC, negative control; si, small interfering.
Discussion
HCC carcinogenesis is a sophisticated and complex pathological process associated with specific tumor genes, multiple signaling cascades and epigenetic modifications (35,36). Recently, bioinformatics analyses have been extensively performed to identify novel diagnostic markers and therapeutic targets for different types of cancer (37,38), thus providing useful tools in tumor research.
The present study performed bioinformatics analyses to investigate the DEGs between HCC tissues and adjacent normal tumor tissues, based on three independent GEO expression datasets. A total of 15 overlapping DEGs were identified, including cytochrome P450 family 39 subfamily A member 1, CYR61, FOS, FOXO1, GADD45B, inhibitor of DNA binding 1, interleukin-1 receptor accessory protein, metallothionein-1M, pleckstrin homology-like domain family A member 1, Rho family GTPase 3, serine dehydratase, SOCS2, TAT, S100 calcium-binding protein P and SPINK1, all of which exhibited consistent expression patterns in the three sequencing datasets. A total of four potentially prognostic DEGs (FOXO1, SPINK1, SOCS2 and TAT) were further identified via Kaplan-Meier analysis.
FOXO1 is one of the forkhead family transcription factors, which participates in several processes of tumor development (30,39). It is well-known that FOXO1 expression is downregulated in the early stages of human pancreatic ductal adenocarcinoma, and may function as a valuable diagnostic marker (39). Sequencing analyses have demonstrated that FOXO1 and paired box 3 (PAX3) expression are upregulated in alveolar rhabdomyosarcoma; double knockdown of PAX3 and FOXO1 significantly inhibits tumor cell proliferation, survival and migration by targeting interleukin-24 (30). SOCS2 is involved in the inhibition of signal transduction (40). It has been reported that SOCS2 regulates the immune response during acute liver injury caused by acetaminophen (41). SOCS2 also plays a critical role in the development of prostate cancer (42). TAT is a tyrosine transaminase that is predominantly expressed in liver tissues, and its deficiency can lead to tyrosinemia (43). Low TAT expression has been observed in HCC and is associated with tumor progression (44). Previous studies have reported that SPINK1 is associated with the development of different types of cancer (8,45,46). Microarray analysis has demonstrated that SPINK1 expression is upregulated in HCC, which promotes the proliferation, migration and invasion of HCC cells (45). SOCS2 was selected as a hub gene in the present study due to its pathological stage association and good diagnostic ability in patients with HCC. Functional studies demonstrated that overexpression of SOCS2 inhibited HCC cell proliferation and migration, whereas SOCS2 knockdown promoted HCC tumorigenesis, suggesting that SOCS2 may function as a tumor suppressor in HCC development.
The present study was not without limitations. First, mechanistic experiments for the antitumor role of SOCS2 are required to determine the molecular mechanism of SOCS2 in HCC progression. Secondly, the present study only investigated the function of SOCS2 in HCC cells. Further biological experiments are required to validate the roles of other DEGs in the diagnosis and treatment of HCC. Thirdly, the quality and heterogeneity of the public data that were uploaded by other researchers and used in the present study cannot be accurately determined.
In conclusion, the present study identified four key DEGs from the HCC gene expression profile datasets using integrated bioinformatics analyses. The PPI network, and functional enrichment and prognostic analyses suggest that these DEGs may be involved in the pathogenesis and prognosis of HCC. In addition, the antitumor role of SOCS2 was investigated, and the results demonstrated that SOCS2 may serve as a potential diagnostic marker in patients with HCC. Collectively, these results provide further insight on the prognostic prediction and molecular targeting therapy for patients with HCC.
Supplementary Material
Acknowledgements
Not applicable.
Funding Statement
The present study was supported by the research projects fund of 920th Hospital (grant no. 2019YGB20).
Funding
The present study was supported by the research projects fund of 920th Hospital (grant no. 2019YGB20).
Availability of data and materials
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request. GSE22058, GSE57957, GSE14323 datasets are available in the GEO (https://www.ncbi.nlm.nih.gov/geo).
Authors' contributions
JL, ZL and SZ conceived the study. JL, ZL and WL collected data. ZL and WL performed the experiments. JL and ZL analyzed the data. JL, ZL, WL and SZ drafted the initial manuscript. SZ supervised the study. JL and SZ confirmed the authenticity of all raw data. JL and SZ revised the manuscript for important intellectual content. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of 920th Hospital (Kunming, China; grant no. 2018-020-01). Written informed consent was provided by all patients prior to the study start.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clini. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156:477–491.e1. doi: 10.1053/j.gastro.2018.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang H, Lu Z, Zhao X. Tumorigenesis, diagnosis, and therapeutic potential of exosomes in liver cancer. J Hematol Oncol. 2019;12:133. doi: 10.1186/s13045-019-0806-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dhanasekaran R, Felsher DW. A tale of two complications of obesity: NASH and hepatocellular carcinoma. Hepatology. 2019;70:1056–1058. doi: 10.1002/hep.30649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kudo M. Systemic therapy for hepatocellular carcinoma: Recent advances and future perspective. Nihon Shokakibyo Gakkai Zasshi. 2019;116:8–17. doi: 10.11405/nisshoshi.116.8. (In Japanese) [DOI] [PubMed] [Google Scholar]
- 6.Weng Q, Chen M, Li M, Zheng YF, Shao G, Fan W, Xu XM, Ji J. Global microarray profiling identified hsa_circ_ 0064428 as a potential immune-associated prognosis biomarker for hepatocellular carcinoma. J Med Genet. 2019;56:32–38. doi: 10.1136/jmedgenet-2018-105440. [DOI] [PubMed] [Google Scholar]
- 7.Lin J, Lin W, Ye Y, Wang L, Chen X, Zang S, Huang A. Kindlin-2 promotes hepatocellular carcinoma invasion and metastasis by increasing Wnt/β-catenin signaling. J Exp Clin Cancer Res. 2017;36:134. doi: 10.1186/s13046-017-0603-4. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Bhatia V, Yadav A, Tiwari R, Nigam S, Goel S, Carskadon S, Gupta N, Goel A, Palanisamy N, Ateeq B. Epigenetic silencing of miRNA-338-5p and miRNA-421 drives SPINK1-positive prostate cancer. Clin Cancer Res. 2019;25:2755–2768. doi: 10.1158/1078-0432.CCR-18-3230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chandran UR, Ma C, Dhir R, Bisceglia M, Lyons-Weiler M, Liang W, Michalopoulos G, Becich M, Monzon FA. Gene expression profiles of prostate cancer reveal involvement of multiple molecular pathways in the metastatic process. BMC Cancer. 2007;7:64. doi: 10.1186/1471-2407-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu L, Lin J, He H. Identification of potential crucial genes associated with the pathogenesis and prognosis of endometrial cancer. Front Genet. 2019;10:373. doi: 10.3389/fgene.2019.00373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hu G, Cheng Z, Wu Z, Wang H. Identification of potential key genes associated with osteosarcoma based on integrated bioinformatics analyses. J Cell Biochem. 2019;120:13554–13561. doi: 10.1002/jcb.28630. [DOI] [PubMed] [Google Scholar]
- 12.Ni M, Liu X, Wu J, Zhang D, Tian J, Wang T, Liu S, Meng Z, Wang K, Duan X, et al. Identification of candidate biomarkers correlated with the pathogenesis and prognosis of non-small cell lung cancer via integrated bioinformatics analysis. Front Genet. 2018;9:469. doi: 10.3389/fgene.2018.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X, Wu J, Zhang D, Bing Z, Tian J, Ni M, Zhang X, Meng Z, Liu S. Identification of potential key genes associated with the pathogenesis and prognosis of gastric cancer based on integrated bioinformatics analysis. Front Genet. 2018;9:265. doi: 10.3389/fgene.2018.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burchard J, Zhang C, Liu AM, Poon RT, Lee NP, Wong KF, Sham PC, Lam BY, Ferguson MD, Tokiwa G, et al. microRNA-122 as a regulator of mitochondrial metabolic gene network in hepatocellular carcinoma. Mol Syst Biol. 2010;6:402. doi: 10.1038/msb.2010.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu AM, Yao TJ, Wang W, Wong KF, Lee NP, Fan ST, Poon RT, Gao C, Luk JM. Circulating miR-15b and miR-130b in serum as potential markers for detecting hepatocellular carcinoma: A retrospective cohort study. BMJ Open. 2012;2:e000825. doi: 10.1136/bmjopen-2012-000825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mah WC, Thurnherr T, Chow PK, Chung AY, Ooi LL, Toh HC, Teh BT, Saunthararajah Y, Lee CG. Methylation profiles reveal distinct subgroup of hepatocellular carcinoma patients with poor prognosis. PLoS One. 2014;9:e104158. doi: 10.1371/journal.pone.0104158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mas VR, Maluf DG, Archer KJ, Yanek K, Kong X, Kulik L, Freise CE, Olthoff KM, Ghobrial RM, McIver P, Fisher R. Genes involved in viral carcinogenesis and tumor initiation in hepatitis C virus-induced hepatocellular carcinoma. Mol Med. 2009;15:85–94. doi: 10.2119/molmed.2008.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edgar R, Barrett T. NCBI GEO standards and services for microarray data. Nat Biotechnol. 2006;24:1471–1472. doi: 10.1038/nbt1206-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harding JJ, Nandakumar S, Armenia J, Khalil DN, Albano M, Ly M, Shia J, Hechtman JF, Kundra R, El Dika I, et al. Prospective genotyping of hepatocellular carcinoma: Clinical implications of next-generation sequencing for matching patients to targeted and immune therapies. Clin Cancer Res. 2019;25:2116–2126. doi: 10.1158/1078-0432.CCR-18-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schulze K, Imbeaud S, Letouze E, Alexandrov LB, Calderaro J, Rebouissou S, Couchy G, Meiller C, Shinde J, Soysouvanh F, et al. Exome sequencing of hepatocellular carcinomas identifies new mutational signatures and potential therapeutic targets. Nat Genet. 2015;47:505–511. doi: 10.1038/ng.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahn SM, Jang SJ, Shim JH, Kim D, Hong SM, Sung CO, Baek D, Haq F, Ansari AA, Lee SY, et al. Genomic portrait of resectable hepatocellular carcinomas: Implications of RB1 and FGF19 aberrations for patient stratification. Hepatology. 2014;60:1972–1982. doi: 10.1002/hep.27198. [DOI] [PubMed] [Google Scholar]
- 23.Fujimoto A, Totoki Y, Abe T, Boroevich KA, Hosoda F, Nguyen HH, Aoki M, Hosono N, Kubo M, Miya F, et al. Whole-genome sequencing of liver cancers identifies etiological influences on mutation patterns and recurrent mutations in chromatin regulators. Nat Genet. 2012;44:760–764. doi: 10.1038/ng.2291. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez-Vega F, Mina M, Armenia J, Chatila WK, Luna A, La KC, Dimitriadoy S, Liu DL, Kantheti HS, Saghafinia S, et al. Oncogenic signaling pathways in the cancer genome atlas. Cell. 2018;173:321–337.e10. doi: 10.1016/j.cell.2018.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Chen DG, Zhu B, Lv SQ, Zhu H, Tang J, Huang C, Li Q, Zhou P, Wang DL, Li GH. Inhibition of EGR1 inhibits glioma proliferation by targeting CCND1 promoter. J Exp Clin Cancer Res. 2017;36:186. doi: 10.1186/s13046-017-0656-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xu Z, Xu M, Liu P, Zhang S, Shang R, Qiao Y, Che L, Ribback S, Cigliano A, Evert K, et al. The mTORC2-Akt1 cascade is crucial for c-Myc to promote hepatocarcinogenesis in mice and humans. Hepatology. 2019;70:1600–1613. doi: 10.1002/hep.30697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yoon H, Kim M, Jang K, Shin M, Besser A, Xiao X, Zhao D, Wander SA, Briegel K, Morey L, et al. p27 transcriptionally coregulates cJun to drive programs of tumor progression. Proc Natl Acad Sci USA. 2019;116:7005–7014. doi: 10.1073/pnas.1817415116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xu P, Zhang G, Hou S, Sha LG. MAPK8 mediates resistance to temozolomide and apoptosis of glioblastoma cells through MAPK signaling pathway. Biomed Pharmacother. 2018;106:1419–1427. doi: 10.1016/j.biopha.2018.06.084. [DOI] [PubMed] [Google Scholar]
- 30.Lacey A, Hedrick E, Cheng Y, Mohankumar K, Warren M, Safe S. Interleukin-24 (IL24) is suppressed by PAX3-FOXO1 and is a novel therapy for rhabdomyosarcoma. Mol Cancer Ther. 2018;17:2756–2766. doi: 10.1158/1535-7163.MCT-18-0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim JH, Lee MJ, Yu GR, Kim SW, Jang KY, Yu HC, Cho BH, Kim DG. Alterations in the p53-SOCS2 axis contribute to tumor growth in colon cancer. Exp Mol Med. 2018;50:1–10. doi: 10.1038/s12276-018-0193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Senapati P, Dey S, Sudarshan D, Das S, Kumar M, Kaypee S, Mohiyuddin A, Kodaganur GS, Kundu TK. Oncogene c-fos and mutant R175H p53 regulate expression of Nucleophosmin implicating cancer manifestation. FEBS J. 2018;285:3503–3524. doi: 10.1111/febs.14625. [DOI] [PubMed] [Google Scholar]
- 33.Huang H, Wang Q, Du T, Lin C, Lai Y, Zhu D, Wu W, Ma X, Bai S, Li Z, et al. Matrine inhibits the progression of prostate cancer by promoting expression of GADD45B. Prostate. 2018;78:327–335. doi: 10.1002/pros.23469. [DOI] [PubMed] [Google Scholar]
- 34.Habel N, Stefanovska B, Carene D, Patino-Garcia A, Lecanda F, Fromigue O. CYR61 triggers osteosarcoma metastatic spreading via an IGF1Rβ-dependent EMT-like process. BMC Cancer. 2019;19:62. doi: 10.1186/s12885-019-5282-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Z, Li W, Pang Y, Zhou Z, Liu S, Cheng K, Qin Q, Jia Y, Liu S. SF3B4 is regulated by microRNA-133b and promotes cell proliferation and metastasis in hepatocellular carcinoma. EBioMedicine. 2018;38:57–68. doi: 10.1016/j.ebiom.2018.10.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toh TB, Lim JJ, Chow EK. Epigenetics in cancer stem cells. Mol Cancer. 2017;16:29. doi: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cantini L, Bertoli G, Cava C, Dubois T, Zinovyev A, Caselle M, Castiglioni I, Barillot E, Martignetti L. Identification of microRNA clusters cooperatively acting on epithelial to mesenchymal transition in triple negative breast cancer. Nucleic Acids Res. 2019;47:2205–2215. doi: 10.1093/nar/gkz016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liang Y, Zhang C, Dai DQ. Identification of differentially expressed genes regulated by methylation in colon cancer based on bioinformatics analysis. World J Gastroenterol. 2019;25:3392–3407. doi: 10.3748/wjg.v25.i26.3392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Al-Zoughbi W, Schauer S, Pichler M, Hoefler G. Early loss of forkhead transcription factor, O subgroup, member 1 protein in the development of pancreatic ductal adenocarcinoma. Pathobiology. 2018;85:342–347. doi: 10.1159/000492433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Quentmeier H, Geffers R, Jost E, Macleod RA, Nagel S, Röhrs S, Romani J, Scherr M, Zaborski M, Drexler HG. SOCS2: Inhibitor of JAK2V617F-mediated signal transduction. Leukemia. 2008;22:2169–2175. doi: 10.1038/leu.2008.226. [DOI] [PubMed] [Google Scholar]
- 41.Monti-Rocha R, Cramer A, Gaio Leite P, Antunes MM, Pereira RVS, Barroso A, Queiroz-Junior CM, David BA, Teixeira MM, Menezes GB, Machado FS. SOCS2 is critical for the balancing of immune response and oxidate stress protecting against acetaminophen-induced acute liver injury. Front Immunol. 2018;9:3134. doi: 10.3389/fimmu.2018.03134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Das R, Gregory PA, Fernandes RC, Denis I, Wang Q, Townley SL, Zhao SG, Hanson AR, Pickering MA, Armstrong HK, et al. MicroRNA-194 promotes prostate cancer metastasis by inhibiting SOCS2. Cancer Res. 2017;77:1021–1034. doi: 10.1158/0008-5472.CAN-16-2529. [DOI] [PubMed] [Google Scholar]
- 43.Gokay S, Kendirci M, Ustkoyuncu PS, Kardas F, Bayram AK, Per H, Poyrazoğlu HG. Tyrosinemia type II: Novel mutations in TAT in a boy with unusual presentation. Pediatr Int. 2016;58:1069–1072. doi: 10.1111/ped.13062. [DOI] [PubMed] [Google Scholar]
- 44.Fu L, Dong SS, Xie YW, Tai LS, Chen L, Kong KL, Man K, Xie D, Li Y, Cheng Y, et al. Down-regulation of tyrosine aminotransferase at a frequently deleted region 16q22 contributes to the pathogenesis of hepatocellular carcinoma. Hepatology. 2010;51:1624–1634. doi: 10.1002/hep.23540. [DOI] [PubMed] [Google Scholar]
- 45.Huang K, Xie W, Wang S, Li Q, Wei X, Chen B, Hua Y, Li S, Peng B, Shen S. High SPINK1 expression predicts poor prognosis and promotes cell proliferation and metastasis of hepatocellular carcinoma. J Invest Surg. 2020;33:1–10. doi: 10.1080/08941939.2020.1805654. [DOI] [PubMed] [Google Scholar]
- 46.Xu L, Lu C, Huang Y, Zhou J, Wang X, Liu C, Chen J, Le H. SPINK1 promotes cell growth and metastasis of lung adenocarcinoma and acts as a novel prognostic biomarker. BMB Rep. 2018;51:648–653. doi: 10.5483/BMBRep.2018.51.12.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the present study are available from the corresponding author upon reasonable request. GSE22058, GSE57957, GSE14323 datasets are available in the GEO (https://www.ncbi.nlm.nih.gov/geo).