Abstract
Osteosarcoma is the most common malignant bone tumor in adolescents and young adults, and identifying biomarkers for prognosis and therapy is necessary. Bone morphogenetic protein receptor 2 (BMPR2) is involved in various cellular functions, including cell adhesion, proliferation and invasion, inflammation, apoptosis and metastatic spread. However, the correlation between BMPR2 expression levels and prognosis and tumor-infiltrating immune cells in osteosarcoma is not well understood. In the present study, the expression level of BMPR2 was investigated using the Oncomine and R2 databases. The association between the expression level of BMPR2 and the clinical prognosis of patients with cancer was analyzed using the R2 database. The relationship between the expression level of BMPR2 and immune cell infiltration in the stroma of osteosarcoma was assessed using the Tumor Immune Estimation Resource (TIMER) and CIBERSORT. The correlations between BMPR2 expression level and infiltrated immune cell gene marker sets in osteosarcoma were validated in the TIMER and R2 databases. Analysis of a cohort of patients with osteosarcoma revealed that BMPR2 expression was significantly higher in osteosarcoma compared with in normal tissue and was correlated with poor prognosis. M0 macrophages, M2 macrophages, resting mast, γ δ T and CD8+ T cells were the top five immune cells with the highest degrees of infiltration in osteosarcoma. In addition, BMPR2 expression level showed a significant negative correlation with the gene markers of CD8+ T cells, monocytes and M2 macrophages. Low levels of infiltrating CD8+ T cells, monocytes or M2 macrophages in osteosarcoma was significantly associated with poor survival. These data suggested that CD8+ T cells, monocytes and M2 macrophages play significant roles in the establishment of the immune microenvironment of osteosarcoma. High BMPR2 expression was associated with poor prognosis and low infiltration of CD8+ T cells, monocytes and M2 macrophages in osteosarcoma. Hence, BMPR2 can be considered a biomarker of the immune infiltration, metastasis and prognosis of osteosarcoma.
Keywords: BMPR2, immune infiltration, metastasis, prognosis, osteosarcoma
Introduction
Osteosarcoma is the most common malignant bone tumor that occurs predominantly in adolescents and young adults (1,2). Despite significant progress in the management of osteosarcoma, no significant improvement in the survival rate has been achieved over the past three decades (3). Despite the continuous improvement of surgical techniques and the application of neoadjuvant chemotherapy, the 5-year non-invasive survival rate has remained <70% for decades (4,5). Patients with metastatic or recurrent osteosarcoma have a poor prognosis, with a 5-year event-free survival of <30% (6–8). Hence, more effective treatment strategies are needed to improve the prognosis of these patients.
Recently, a number of studies have aimed to elucidate the interaction between the immune system and tumors, and to identify potential targets within this interaction as therapeutic interventions (9–11). In numerous malignant tumors, the composition of the tumor microenvironment (TME) requires the participation of innate and adaptive immune cells, which interact with lymphocytes, natural killer cells (NK) and antigen-presenting cells, such as dendritic cells (DC) and macrophages, to effectively control the tumor (12). Abnormal immune cell function can result in poor tumor monitoring and clearance, can exacerbate local inflammation, and can establish a conducive environment for tumor growth, thus facilitating tumor progression (13,14). Although previous studies have indicated that immune cells may act on osteosarcoma (15,16), the precise mechanisms underlying the function of the immune microenvironment in osteosarcoma are not well known. Therefore, it is important to elucidate the immunophenotype of the interaction between tumors and immune cells, and to identify novel immune-related therapeutic targets for osteosarcoma.
Bone morphogenetic protein receptor 2 (BMPR2) is a member of the TGF-β superfamily, which is reported to play a dual role in regulating tumor growth. In some tumors, BMPR2 acts as a tumor suppressor gene. For example, a lack of BMPR2 expression in the colonic matrix has been found to lead to colonic epithelial hyperplasia and polyps (17). In addition, deficient BMPR2 expression is more likely in bladder transitional cell carcinoma tissues compared with in normal tissues (18). Meanwhile, in some other tumors, BMPR2 functions as an oncogene. For example, in colorectal cancer where Smad4 is absent, BMPR2 can bind to LIM domain kinase 1 to activate the Rho/Rho-associated protein kinase (ROCK) pathway, thereby promoting tumor invasion and metastasis (19). In addition, BMPR2-overexpression is associated with poor prognosis in patients with resected osteosarcoma (20). Notably, a recent study revealed that BMP4 secreted by bladder cancer cells induced M2 macrophage polarization in a tumor-promoting immune environment, favoring tumor progression (21). However, the potential role of BMPR2 in tumor progression and its exact mechanism in osteosarcoma is not clear.
The present study comprehensively analyzed BMPR2 expression levels and their relationship with the prognosis of patients with osteosarcoma in databases such as Gene Expression Omnibus (GEO), Oncomine and R2. Furthermore, the relationship between BMPR2 and immune tumor-infiltrating cells in the osteosarcoma microenvironment was investigated using the Tumor Immune Estimation Resource (TIMER) and CIBERSORT. The study demonstrated the important role of BMPR2 in osteosarcoma and showed latent mechanisms in the BMPR2 and tumor-immune interactions. These findings will improve our knowledge about the of tumor immuno-microenvironment in osteosarcoma.
Materials and methods
Oncomine database analysis
Data for the expression level of the BMPR2 gene in different types of tumors was gathered from the Oncomine cancer database (https://www.oncomine.org/resource/login.html) (22). The thresholds for significance were P<0.05, fold-change of 2.0.
R2 database analysis
The difference in the BMPR2 expression level between sarcoma and normal tissue, as well as the relationship between BMPR2 expression and survival in osteosarcoma, was calculated using the R2 database (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Since the sample size of the sarcoma dataset in the Oncomine database was small, the expression level of BMPR2 was further analyzed in osteosarcoma and rhabdomyosarcoma using the R2 platform. R2 searches a large number of publicly available cancer microarray datasets (23–26) (for relationships between gene expression and patient prognosis, such as the time period for overall survival (OS) and metastasis-free survival. The threshold was set to a Cox P<0.05. The correlation coefficient was calculated by Spearman's method. Unpaired t-tests were applied to identify the differences of two groups, and P<0.05 was considered to indicate a statistically significant difference.
Sample information
The normalized expression data for patients with osteosarcoma were obtained from the GEO database (GSE33382). GSE33382 is one of the osteosarcoma datasets of the R2 platform (24). The clinical information including age, sex, tumor site, metastasis within 5 years, histological subtype and Huvos classification (27) was also extracted. Information from a total of 83 cases of osteosarcoma was obtained, including expression data and clinical data.
The Cancer Genome Atlas (TCGA) database analysis
The gene expression quantification data of 88 osteosarcoma samples were downloaded from TCGA (cancer.gov/tcga) on May 22, 2020. Updated clinical data about these osteosarcoma samples, such as age, sex, overall survival time and vital status, were also downloaded from the TARGET database (https://ocg.cancer.gov/programs/target) on May 22, 2020. Finally, information on a total of 85 cases of osteosarcoma was obtained, including expression data and clinical data.
TIMER database analysis
The TIMER Database (https://cistrome.shinyapps.io/timer/), a comprehensive database for the systematic analysis of immune invasion, was used to investigate the infiltrating pattern of immune cells in osteosarcoma (28). The links between BMPR2 expression and the proportions of infiltrated immune cells, including CD4+ T, CD8+ T and B cells, macrophages, neutrophils and dendritic cells, were investigated using gene modules. Furthermore, the relationship between the expression level of BMPR2 and tumor infiltrating immune cell gene markers was investigated through correlation modules. Genetic markers for tumor infiltrating lymphocytes included markers of B cells, CD8+ T cells, T cells (general), monocytes, tumor-associated macrophages (TAMs), M1 macrophages, M2 macrophages, dendritic cells (DCs), neutrophils, natural killer (NK), follicular helper T (Tfh), regulatory T (Tregs), T helper 1 (Th1), Th2, Th17 and exhausted T cells. These genetic markers were taken from previous studies (29–31). The correlation module analyzed and generated a scatter plot of expression between a pair of user-defined genes in a certain cancer type, as well as Spearman's correlation and estimated statistical significance. BMPR2 was used for the y-axis with gene symbols, and related marker genes are represented on the x-axis as gene symbols. The gene expression level was displayed with log2 RNA-Seq by Expectation-Maximization.
CIBERSORT analysis
CIBERSORT (https://cibersort.stanford.edu/index.php) uses a deconvolution algorithm to evaluate the pattern of tumor infiltrating lymphocytes in the TME. According to the standardized expression data in the tumor, the abundance of 22 types of infiltrating immune cells was analyzed by the CIBERSORT method (32). The immune cell profiles of all osteosarcoma samples were analyzed using CIBERSORT, and the number of permutations was set to 100. Samples of osteosarcoma with a P<0.05 were included.
Gene set enrichment analysis (GSEA)
To identify the potential biological mechanisms between two biological states, GSEA was performed (33). The gene sets were collected from the Molecular Signatures Database (33) (‘c5.bp.v7.0.symbols.gmtb (Gene Ontology)’). GSEA (3.0) was used to analyze the potential biological processes and signaling pathways associated with BMPR2 that affect the prognosis of osteosarcoma. The normalized enrichment score was obtained by performing gene set permutations with 1,000 times. The nominal (NOM) P<0.05 was used to quantify statistically significant enrichment. GSEA was performed (false discovery rate <0.25, nominal P<0.05) in the enrichment of ‘c5.bp.v7.0.symbols.gmtb (Gene Ontology)’.
Statistical analysis
Kaplan-Meier analysis with log-rank was used. The results from the Oncomine database analysis are displayed as P-values, fold-changes and ranks. Survival curves were generated by the R2 Platform. Spearman's correlation was used to assess the correlation of gene expression, P<0.05 was considered to indicate a statistically significant difference. The strength of the correlation was also determined by rs-value correlation with rs-value >0.3 or <-0.3 was considered as significant and between −0.3 to 0.3 was considered as weak or negligible correlation. The relationships between BMPR2 expression and tumor-infiltrating immune cells were analyzed using SPSS software (version 20). The correlations between BMPR2 expression and the clinicopathological features of osteosarcoma were evaluated by logistic regression analysis. The matrix data of gene expression levels were normalized with the limma package of R software (version 3.5.2) (34) and R studio (version 1.2.5001) (35). The specificity and sensitivity of survival prediction according to BMPR2 expression levels were analyzed by time-dependent receiver operating characteristic (ROC) curves, whose area under the curve (AUC) values quantified with the pROC package (https://cran.rstudio.com/bin/macosx/contrib/4.0/pROC_1.16.2.tgz).
Results
BMPR2 mRNA expression levels in different types of human cancer
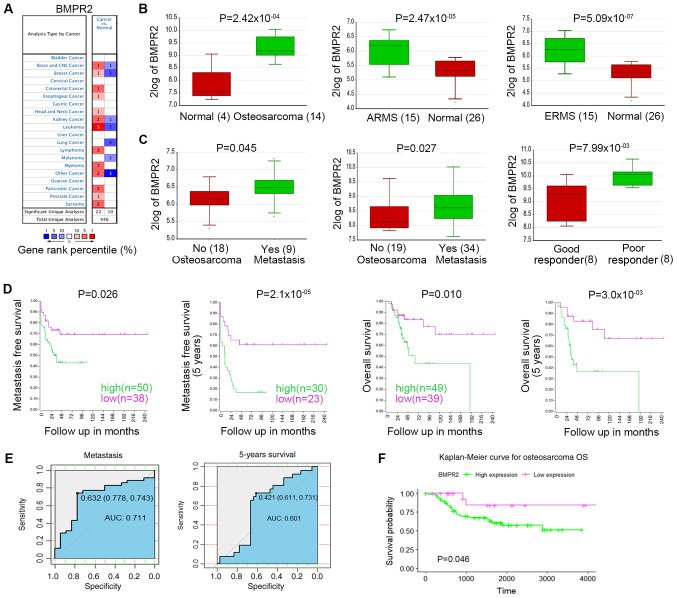
To verify the difference in the BMPR2 expression level between tumors and normal tissues, the Oncomine database was used to analyze the levels of BMPR2 mRNA in different types of tumors and the corresponding normal tissues. The results showed that BMPR2 expression was higher in brain and CNS (oligodendroglioma), breast (invasive ductal breast carcinoma), colorectal (rectal mucinous adenocarcinoma), esophageal (esophageal adenocarcinoma) and kidney cancer (clear cell sarcoma of the kidney and renal oncocytoma), leukemia (chronic lymphocytic leukemia), head and neck cancer (floor of the mouth carcinoma), lymphoma tumors (follicular lymphoma and germinal center B-cell-like diffuse large B-cell lymphoma), pancreatic (pancreatic ductal adenocarcinoma) and prostate cancer (prostate carcinoma), sarcomas (clear cell sarcoma of the kidney) and others, including skin basal cell carcinoma and teratoma, compared with in normal tissues (Fig. 1A; Table I). Meanwhile, BMPR2 expression was lower in glioblastoma, lung cancer (large cell lung carcinoma, lung adenocarcinoma and squamous cell lung carcinoma), invasive lobular breast carcinoma, chromophobe renal cell carcinoma, T cell prolymphocytic leukemia, skin basal cell carcinoma and myeloma compared with in normal tissues. Detailed results of BMPR2 expression in various types of cancer are listed in Table I.
Figure 1.
BMPR2 expression levels in different types of human cancer. (A) Abnormal expression of BMPR2 in data sets of different cancer types compared with normal tissues in the Oncomine database. (B) BMPR2 expression was higher in osteosarcoma (14 samples) compared with in normal tissues (four samples). BMPR2 expression level was higher in AMRS (15 samples) and ERMS (15 samples) compared with in the normal tissues (26 samples). (C) BMPR2 expression was significantly higher in metastatic osteosarcomas than in non-metastatic osteosarcomas. BMPR2 expression was significantly higher in sarcomas that did not respond well to chemotherapy compared with in those that did. (D) High expression level of BMPR2 was associated with worse OS and metastasis-free OS. (E) AUC values for the expression level of BMPR2 in predicting metastatic and 5-year survival. (F) High expression level of BMPR2 was associated with worse OS. ARMS, acinar rhabdomyosarcoma; ERMS, embryonal rhabdomyosarcoma; BMPR2, bone morphogenetic protein receptor 2; OS, overall survival; AUC, area under the curve.
Table I.
BMPR2 expression in cancer vs. normal tissue in the Oncomine database.
| Cancer | P-value | Fold-change | Rank, % | Sample size | (Refs.) |
|---|---|---|---|---|---|
| Brain and CNS cancer | |||||
| Oligodendroglioma | 5.19×10−8 | 2.132 | 5 | 73 | Sun et al (70) |
| Glioblastoma | 6.73×10−4 | −3.216 | 7 | 15 | TCGAa |
| Breast cancer | |||||
| Invasive ductal breast carcinoma | 1.60×10−2 | 2.896 | 6 | 25 | Turashvili et al (71) |
| Invasive lobular breast carcinoma | 1.40×10−2 | −2.075 | 3 | 25 | Turashvili et al (71) |
| Colorectal cancer | |||||
| Rectal mucinous adenocarcinoma | 2.10×10−4 | 2.171 | 5 | 9 | Kaiser et al (72) |
| Esophagus cancer | |||||
| Esophageal adenocarcinoma | 6.11×10−4 | 2.529 | 9 | 5 | Hao et al (73) |
| Head and neck cancer | |||||
| Floor of the mouth carcinoma | 1.61×10−4 | 3.473 | 7 | 5 | Pyeon et al (74) |
| Kidney cancer | |||||
| Clear cell sarcoma | 4.44×10−4 | 2.479 | 4 | 17 | Cutcliffe et al (75) |
| Renal oncocytoma | 1.00×10−3 | 2.37 | 6 | 9 | Yusenko et al (76) |
| Chromophobe renal cell carcinoma | 7.00×10−3 | −2.961 | 6 | 9 | Yusenko et al (76) |
| Leukemia | |||||
| Chronic lymphocytic leukemia | 2.96×10−13 | 3.465 | 1 | 12 | Alizadeh et al (77) |
| Chronic lymphocytic leukemia | 1.69×10−10 | 2.589 | 1 | 39 | Rosenwald et al (78) |
| Chronic lymphocytic leukemia | 1.23×10−92 | 3.446 | 1 | 448 | Haferlach et al (79) |
| Chronic lymphocytic leukemia | 1.00×10−3 | 2.188 | 7 | 3 | Rosenwald et al (80) |
| Chronic lymphocytic leukemia | 1.50×10−6 | 3.205 | 5 | 34 | Basso et al (81) |
| T cell prolymphocytic leukemia | 6.04×10−4 | −2.972 | 3 | 6 | Dürig et al (82) |
| Lung cancer | |||||
| Lung adenocarcinoma | 5.66×10−17 | −2.281 | 3 | 43 | Hou et al (83) |
| Large cell lung carcinoma | 1.08×10−12 | −2.775 | 3 | 19 | Hou et al (83) |
| Squamous cell lung carcinoma | 6.86×10−17 | −3.47 | 3 | 27 | Hou et al (83) |
| Lymphoma | |||||
| Follicular lymphoma | 7.00×10−3 | 2.425 | 5 | 6 | Basso et al (81) |
| Germinal center B cell-like diffuse large | 3.31×10−6 | 2.934 | 3 | 9 | Compagno et al (84) |
| B cell lymphoma | |||||
| Follicular lymphoma | 3.99×10−9 | 3.239 | 9 | 38 | Compagno et al (84) |
| Myeloma | |||||
| Non-neoplastic nevus | 4.00×10−3 | −2.388 | 7 | 9 | Haqq et al (85) |
| Pancreatic cancer | |||||
| Pancreatic ductal adenocarcinoma | 1.82×10−12 | 2.055 | 2 | 39 | Badea et al (86) |
| Pancreatic adenocarcinoma | 1.82×10−12 | 3.078 | 6 | 12 | Iacobuzio-Donahue et al (87) |
| Prostate cancer | |||||
| Prostate carcinoma | 2.70×10−2 | 4.768 | 8 | 15 | Luo et al (88) |
| Sarcoma | |||||
| Clear cell sarcoma of the kidney | 4.44×10−4 | 2.479 | 4 | 14 | Cutcliffe et al (75) |
| Gastrointestinal stromal tumor | 3.69×10−4 | 2.62 | 7 | 6 | Cho et al (89) |
| Other | |||||
| Skin basal cell carcinoma | 2.51×10−5 | 2.144 | 3 | 15 | Riker et al (90) |
| Teratoma, NOS | 2.48×10−08 | 2.606 | 4 | 14 | Korkola et al (91) |
| Skin basal cell carcinoma | 5.08×10−06 | −2.996 | 1 | 15 | Riker et al (90) |
TCGA, The Cancer Genome Atlas, No Associated Paper, http://tcga-data.nci.nih.gov/tcga/; CNS, central nervous system; NOS, not otherwise specified.
The results revealed that BMPR2 expression was higher in osteosarcoma, acinar rhabdomyosarcoma (ARMS) and embryonal rhabdomyosarcoma (ERMS) compared with in normal tissues (Fig. 1B). BMPR2 expression was significantly higher in metastatic osteosarcomas compared with in non-metastatic osteosarcomas. Its expression was also significantly higher in sarcomas that did not respond well to chemotherapy compared with in those that did (Fig. 1C). These data suggested that BMPR2 is a protumor gene in osteosarcoma and rhabdomyosarcoma.
High BMPR2 expression impacts the prognosis of patients with osteosarcoma
To improve our understanding of the role and underlying mechanism of BMPR2 expression in osteosarcoma, the relationship between BMPR2 expression and the clinicopathological features of patients with osteosarcoma was examined in the R2 database. High expression levels of BMPR2 indicated worse OS and metastasis-free survival (Fig. 1D). In addition, patients with osteosarcoma with high BMPR2 expression had a 2.794 times greater risk of metastasis within 5 years compared with those without metastasis (Table II). The specificity and sensitivity of metastatic and survival prediction according to the expression level of BMPR2, as shown by time-dependent ROC curves, were analyzed to evaluate its ability to predict metastasis and survival in patients. The results suggested that the AUC for the expression level of BMPR2 in predicting metastatic and 5-year survival were 0.711 and 0.601, respectively (Fig. 1E). The relationship between BMPR2 expression level and the prognosis of osteosarcoma was also analyzed using TCGA database. The results showed that patients with high expression levels of BMPR2 had worse OS (Fig. 1F). These data indicated that a high BMPR2 expression level serves as an adverse factor in the prognosis of osteosarcoma.
Table II.
Association between high bone morphogenetic protein receptor 2 expression with clinicopathological characteristics of osteosarcoma.
| Factors | OR | CI | P-value | Value, n |
|---|---|---|---|---|
| Sex, female vs. male | 0.908 | 0.638–1.291 | 0.629 | 71 |
| Age, <16 vs. ≥16 years | 1.047 | 0.743–1.473 | 0.825 | 83 |
| Tumor site, femur vs. tibia/fibula | 0.856 | 0.577–1.268 | 0.466 | 68 |
| Metastasis within 5 years, yes vs. no | 2.794 | 1.282–6.089 | 0.001 | 53 |
| Histological subtype | ||||
| Osteolastic vs. chondroblastic | 0.985 | 0.637–1.523 | 1.000 | 68 |
| Osteolastic vs. fibroblastic | 1.436 | 0.594–3.469 | 0.427 | 59 |
| Huvos grade, 1 and 2 vs. 3 and 4 | 0.908 | 0.638–1.291 | 0.629 | 71 |
OR, odds ratio; CI, confidence interval.
Infiltrating immune cells in osteosarcoma
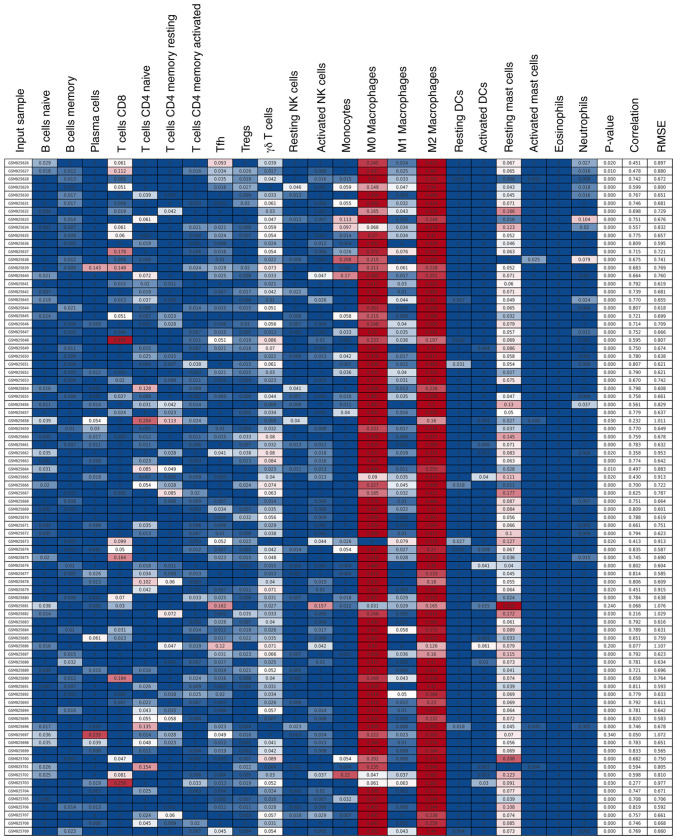
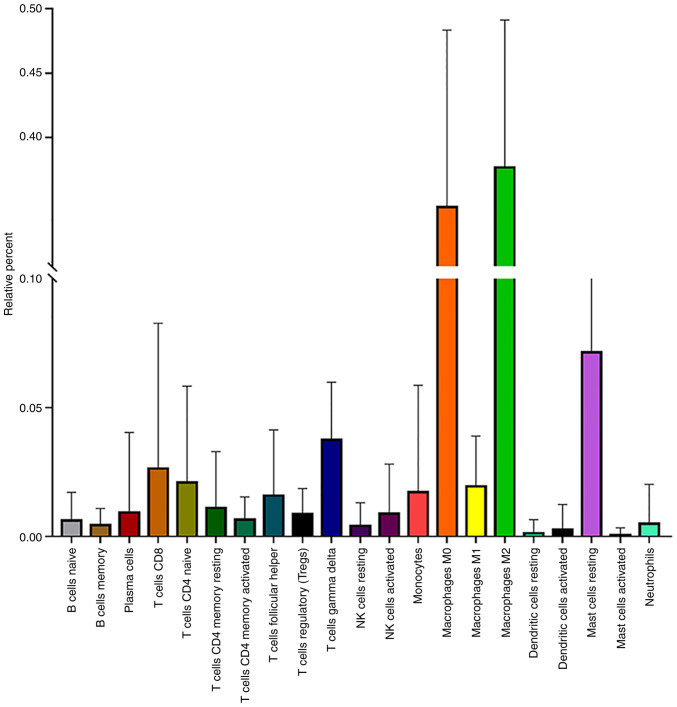
It has been reported that malignant tumors tend to construct an immunosuppressive microenvironment by interacting with their stromal contents, including infiltrating immune cells (35). However, the function and mechanism of infiltrating immune cells in regulating the microenvironment of osteosarcoma remain unclear. Therefore, 81 osteosarcoma samples with a P<0.05 were obtained by the CIBERSORT method (Fig. 2). Among 22 immune cell types, M2 macrophages, M0 macrophages, resting mast, γ δ T and CD8+ T cells were the top five immune cells with the highest degree of infiltration in osteosarcoma (Fig. 3). These results indicated that these five immune cells play important roles in maintaining the immune microenvironment of osteosarcoma.
Figure 2.
Infiltrating immune cells in osteosarcoma analyzed using the CIBERSORT method. Color from blue to red represents the infiltration abundance of the immune cells from low to high RNA-Seq by Expectation-Maximization.
Figure 3.
Infiltration of 22 types of immune cells in osteosarcoma. NK, natural killer.
Correlation analysis between BMPR2 expression and immune marker sets
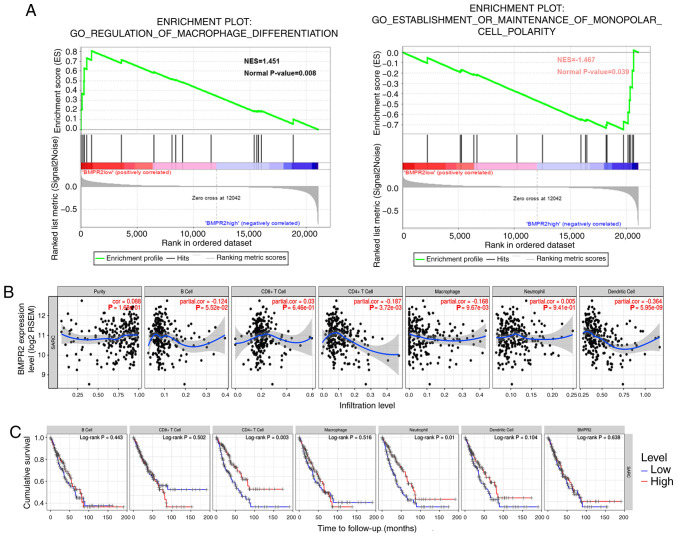
To investigate the signaling pathways activated by BMPR2 in osteosarcoma cells, GSEA was performed. The significant results are displayed in Figs. S1, S2A and B (NOM P<0.05) in the enrichment of ‘c5.bp.v7.0.symbols.gmtb (Gene Ontology)’. Notably, the data showed that the ‘regulation of the macrophage differentiation pathway’ and ‘establishment or maintenance of the monopolar cell polarity pathway’ were modulated by BMPR2 (Fig. 4A).
Figure 4.
Correlation analysis between BMPR2 expression and the infiltrating immune cells. (A) GSEA was performed (false discovery rate <0.25, nominal P<0.05) in the enrichment of ‘c5.bp.v7.0.symbols.gmtb (Gene Ontology)’. The regulation of the macrophage differentiation pathway and establishment or maintenance of the monopolar cell polarity pathway were mediated by BMPR2 in osteosarcoma. (B) BMPR2 expression level in sarcoma was statistically significant with CD4+ T cell, and negligibly correlated with macrophage and dendritic cell infiltration. (C) Low infiltration of CD4+ T cells and neutrophil cells was correlated with worse survival in sarcoma. BMPR2, bone morphogenetic protein receptor 2; NES, normalized enrichment score.
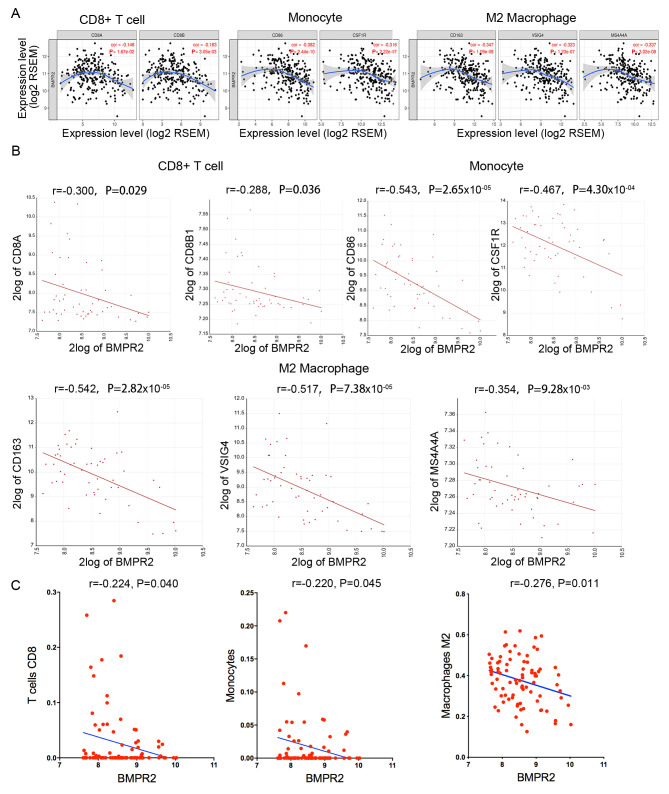
Additionally, the correlation between BMPR2 expression and immune cell infiltration in sarcoma was studied in the TIMER database. The results suggested that the BMPR2 expression level was inversely correlated with the concentrations of DCs. CD4+ T cells (P=3.72×10−3) and macrophages (P=9.67×10−3) was statistically significant with P-values, while the strength of the correlation was weak or negligibly according to the aforementioned cut-offs (Fig. 4B). In addition, low infiltration of CD4+ T cells and neutrophils were associated with worse survival (Fig. 4C).
To further explore the correlations between the expression level of BMPR2 and the degrees of infiltration of different types of immune cells, the relationship between the expression of BMPR2 and the marker genes of various lymphocytes in sarcomas was investigated using the TIMER database. The detailed results about the relationships between BMPR2 expression and immune marker sets of various types of immune cells, including T (general), B and CD8+ T cells, monocytes, TAMs, M1 and M2 macrophages, NKs, DCs, neutrophils, Th1, Th2, Th17 and Tfh cells, Tregs and exhausted T cells in sarcoma, are shown (Table III, Figs. 4, 5A and S3). Notably, the data reported that the majority of marker sets of the immune cells, including gene markers of T (general), B and CD8+ T cells, monocytes, TAMs, M1 and M2 macrophages, DCs, neutrophils, Th1, Th2 and Th17 cells, Tregs and exhausted T cells, had significant negative correlations with BMPR2 expression in sarcoma (Table III).
Table III.
Correlation analysis between bone morphogenetic protein receptor 2 and related genes and markers of immune cells in R2 and Tumor Immune Estimation Resource databases.
| OS | SARC | |||
|---|---|---|---|---|
| Gene markers | Cor | P-value | Cor | P-value |
| CD8+ T cell | ||||
| CD8A | −0.300 | 0.029 | −0.148 | 0.017 |
| CD8B | −0.288 | 0.036 | −0.183 | 0.003 |
| T cell (general) | ||||
| CD3D | −0.109 | 0.437 | −0.267 | 0.001a |
| CD3E | −0.355 | 0.009 | −0.196 | 0.001 |
| CD2 | −0.273 | 0.048 | −0.200 | 0.001 |
| B cell | ||||
| CD19 | −0.133 | 0.342 | −0.288 | 0.001a |
| CD79A | −0.099 | 0.479 | −0.162 | 0.009 |
| Monocyte | ||||
| CD86 | −0.543 | 0.001a | −0.382 | 0.001a |
| CD115 (CSF1R) | −0.467 | 0.001a | −0.316 | 0.001 |
| TAM | ||||
| CCL2 | −0.099 | 0.48 | −0.197 | 0.001 |
| CD68 | 0.312 | 0.023 | −0.453 | 0.001a |
| IL10 | 0.012 | 0.933 | −0.345 | 0.001a |
| M1 Macrophage | ||||
| INOS (NOS2) | −0.043 | 0.761 | 0.129 | 0.037 |
| IRF5 | −0.385 | 0.004 | −0.325 | 0.001a |
| COX2 (PTGS2) | 0.248 | 0.073 | 0.149 | 0.016 |
| M2 Macrophage | ||||
| CD163 | −0.542 | 0.001a | −0.347 | 0.001a |
| VSIG4 | −0.517 | 0.001a | −0.323 | 0.001a |
| MS4A4A | −0.354 | 0.009 | −0.337 | 0.001a |
| Neutrophils | ||||
| CD66b (CEACAM8) | −0.183 | 0.190 | −0.126 | 0.043 |
| CD11b (ITGAM) | −0.224 | 0.106 | −0.327 | 0.001a |
| CCR7 | −0.228 | 0.101 | −0.082 | 0.185 |
| Natural killer cell | ||||
| KIR2DL1 | −0.131 | 0.349 | −0.155 | 0.012 |
| KIR2DL3 | −0.169 | 0.228 | −0.142 | 0.022 |
| KIR2DL4 | 0.014 | 0.921 | −0.312 | 0.001a |
| KIR3DL1 | −0.365 | 0.007 | −0.066 | 0.289 |
| KIR3DL2 | −0.167 | 0.233 | −0.041 | 0.507 |
| KIR3DL3 | −0.258 | 0.062 | −0.119 | 0.055 |
| KIR2DS4 | 0.219 | 0.115 | −0.140 | 0.024 |
| Dendritic cell | ||||
| HLA-DPB1 | −0.201 | 0.150 | −0.294 | 0.001a |
| HLA-DQB1 | −0.259 | 0.061 | −0.274 | 0.001a |
| HLA-DRA | −0.484 | 0.001a | −0.265 | 0.001a |
| HLA-DPA1 | −0.451 | 0.001a | −0.238 | 0.001a |
| BDCA-1 (CD1C) | 0.235 | 0.090 | 0.020 | 0.752 |
| BDCA-4 (NRP1) | 0.183 | 0.189 | 0.167 | 0.007 |
| CD11c (ITGAX) | −0.331 | 0.015 | −0.325 | 0.001a |
| Th1 | ||||
| T-bet (TBX21) | 0.197 | 0.157 | −0.204 | 0.001a |
| STAT4 | −0.031 | 0.827 | −0.181 | 0.003 |
| STAT1 | −0.166 | 0.234 | 0.064 | 0.303 |
| IFN-γ (IFNG) | −0.216 | 0.120 | −0.216 | 0.001a |
| TNF-α (TNF) | −0.314 | 0.022 | −0.203 | 0.001 |
| Th2 | ||||
| GATA3 | −0.181 | 0.195 | −0.162 | 0.009 |
| STAT6 | 0.092 | 0.514 | 0.253 | 0.001a |
| STAT5A | −0.483 | 0.001a | −0.183 | 0.003 |
| IL13 | −0.320 | 0.019 | −0.030 | 0.632 |
| Tfh | ||||
| BCL6 | 0.099 | 0.479 | 0.018 | 0.771 |
| IL21 | 0.031 | 0.825 | −0.113 | 0.070 |
| Th17 | ||||
| STAT3 | −0.545 | 0.001a | 0.448 | 0.001a |
| IL17A | −0.529 | 0.001a | 0.003 | 0.961 |
| Treg | ||||
| FOXP3 | −0.171 | 0.221 | −0.126 | 0.042 |
| CCR8 | −0.072 | 0.608 | −0.025 | 0.689 |
| STAT5B | 0.158 | 0.257 | 0.365 | 0.001a |
| TGFβ (TGFB1) | −0.045 | 0.751 | −0.4 | 0.001a |
| T cell exhaustion | ||||
| PD-1 (PDCD1) | −0.133 | 0.341 | −0.237 | 0.001a |
| CTLA4 | −0.112 | 0.424 | −0.2 | 0.001 |
| LAG3 | −0.209 | 0.133 | −0.357 | 0.001a |
| TIM-3 (HAVCR2) | −0.599 | 0.001a | −0.407 | 0.001a |
| GZMB | −0.324 | 0.018 | −0.409 | 0.001a |
P<0.001. OS, osteosarcoma; SARC, sarcoma; TAM, tumor-associated macrophage; Th, T helper cell; Tfh, follicular helper T cell; Treg, regulatory T cell; Cor, coefficient value of Spearman's correlation.
Figure 5.
Association of BMPR2 expression and immune marker sets in sarcoma and osteosarcoma. (A) BMPR2 expression in sarcoma was correlated with CD8+ T cell, monocyte and M2 macrophage markers, such as CD8A, CD8B, CD86, CD115, CD163, VSIG4 and MS4A4A. (B) BMPR2 expression in osteosarcoma was correlated with CD8+ T cell, monocyte and M2 macrophage markers, such as CD8A, CD8B, CD86, CD115, CD163, VSIG4 and MS4A4A. (C) High expression of BMPR2 in osteosarcoma was correlated with low infiltration of CD8+ T cells, monocytes and M2 macrophages. BMPR2, bone morphogenetic protein receptor 2; RSEM, RNA-Seq by Expectation-Maximization.
In addition, the relationship between BMPR2 expression and marker genes of various immune cells in osteosarcoma was examined using the R2 database (Table III and Figs. S5 and 6). The results revealed that CD8+ T cell, monocyte and M2 macrophage markers, such as CD8A, CD8B, CD86, CD115 (CSF1R), CD163, VSIG4 and MS4A4A, showed statistical significance with BMPR2 expression (Table III and Fig. 5B).
Next, whether BMPR2 expression is associated with the immune infiltration pattern was investigated by assessing the abundance of 22 types of immune cells in 81 osteosarcoma samples. As displayed in Table IV and Fig. 5C, BMPR2 expression was statistical negatively correlated with CD8+ T, γ δ T cells, monocytes, M2 macrophages and neutrophil cells. Meanwhile, BMPR2 was statistically positively correlated with the concentrations of naïve B cells, naïve CD4+T, memory resting CD4+T and M0 macrophages. Taken considering of the rs-values, the correlation was defined as weak or negligible correlation according to the aforementioned cut-offs.
Table IV.
Relationship between bone morphogenetic protein receptor 2 and infiltration of immune cells according to CIBERSORT analysis.
| Cell type | Cor | P-value |
|---|---|---|
| B cells naive | 0.237 | 0.030 |
| B cells memory | −0.196 | 0.074 |
| Plasma cells | 0.169 | 0.124 |
| T cells CD8 | −0.224 | 0.040 |
| T cells CD4 naive | 0.260 | 0.017 |
| T cells CD4 memory resting | 0.487 | 0.001 |
| T cells CD4 memory activated | −0.059 | 0.593 |
| T cells follicular helper | 0.023 | 0.836 |
| T cells regulatory | −0.165 | 0.133 |
| T cells γ δ | −0.254 | 0.020 |
| NK cells resting | 0.212 | 0.053 |
| NK cells activated | 0.150 | 0.174 |
| Monocytes | −0.220 | 0.045 |
| Macrophages M0 | 0.233 | 0.033 |
| Macrophages M1 | −0.138 | 0.212 |
| Macrophages M2 | −0.276 | 0.011 |
| Dendritic cells resting | −0.156 | 0.157 |
| Dendritic cells activated | 0.010 | 0.930 |
| Mast cells resting | 0.091 | 0.411 |
| Mast cells activated | −0.103 | 0.352 |
| Neutrophils | −0.251 | 0.021 |
NK, natural killer.
Taken together, the aforementioned results indicated that high expression of BMPR2 was correlated with low infiltration of CD8+ T cells, monocytes and M2 macrophages in osteosarcoma.
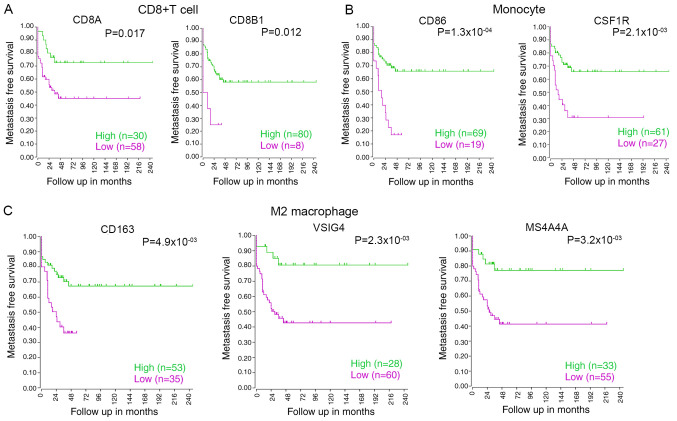
The relationship between the survival of patients with osteosarcoma and the markers of CD8+ T cells, monocytes and M2 macrophages, such as CD8A, CD8B, CD86, CD115, CD163, VSIG4 and MS4A4A, were analyzed using the R2 database. The results showed that low expression levels of CD8A, CD8B1, CD86, CSF1R, CD163, VSIG4 or MS4A4A were correlated with poor survival in osteosarcoma (Fig. 6A-C). Hence, these data indicated that low infiltration of CD8+ T cells, monocytes or M2 macrophages in osteosarcoma is significantly associated with poor prognosis.
Figure 6.
Relationship between the infiltration of immune cells and osteosarcoma survival. (A) Low expression of the CD8+ T cell markers CD8A and CD8B was associated with low metastasis-free survival in osteosarcoma. (B) Low expression of the monocyte cell markers CD86 and CSF1R was correlated with low metastasis-free survival in osteosarcoma. (C) Low expression of M2 macrophage cell markers CD163, VSIG4 and MS4A4A was related to low metastasis-free survival in osteosarcoma.
Discussion
BMPs and receptors (BMPR1 and BMPR2) are members of the TGF-β superfamily. BMPR2 patriciates in numerous cellular functions, including cell adhesion, proliferation and invasion, inflammation, apoptosis and metastatic spread (36,37). In recent years, studies have reported the expression and roles of BMPR2 in various types of cancer malignant tumors, such as colorectal cancer, prostate cancer and osteosarcoma (17,19,20,38). However, the function of BMPR2 in cancer immunology is unclear. The present study reported the differential expression of BMPR2 in different types of cancer. A high level of BMPR2 expression was associated with metastasis and poor prognosis in osteosarcoma, indicating the predictive value of BMPR2 for risk of metastasis in osteosarcoma. In addition, the results showed that the level of BMPR2 expression in sarcomas and osteosarcomas was negligibly correlated with different sets of gene markers of immune cells. Low infiltration of CD8+ T cells, monocytes and M2 macrophages was associated with poor survival in osteosarcoma. Consequently, the results indicated the influential role of BMPR2 in tumor immunology and its potential application as a tumor biomarker. As aforementioned, correlation with a rs -value between −0.3 to 0.3 was considered as weak or negligible correlation. Therefore, the current results do not present powerful statistical evidence to demonstrate the correlation between the gene markers of T (general) and B cells, TAMs, M1 macrophages, DCs, neutrophils, Th1, Th2 and Th17 cells, Tregs and exhausted T cells (Table III) and CD8+ T, γ δ T cells, monocytes, M2 macrophages, neutrophils, naïve B, naïve CD4+T, and memory resting CD4+T cells and M0 macrophages (Table IV) with BMPR2 expression. Further basic experiments and clinical studies focused on tumor infiltration immune cells and tumor microenvironment are needed to identify whether this statistical difference is true or negligible correlation.
Over the past few decades, immunosuppression in TME has received increasing attention in cancer research. Myeloid cells (macrophages, monocytes and DCs) and T lymphocytes act as key members of the immune microenvironment in osteosarcoma (39). Osteosarcoma cells can regulate the recruitment and induce the differentiation of infiltrating immune cells by secreting cytokines, such as CCL2, CCL4 or IFN-γ, thus establishing an immune-tolerant or immune-resistant local microenvironment (39). The TME causes CD8+ T cell dysfunction or exhaustion, removing the cytotoxic effect of CD8+ T cells towards tumor cells (40,41). Previous studies have shown that functional damage to CD8+ T cells is present in a variety of malignancies, including lung cancer (42), melanoma (43), prostate cancer (44) and osteosarcoma (45). The CD8+/forkhead box protein P3+/- ratio has been reported to be a powerful prognostic indicator for patients with osteosarcoma (46). It was also found that TAMs are correlated with a positive prognosis (47,48). The current study demonstrated that M2 and M0 macrophages, resting mast, γ δ T and CD8+ T cells were the top five immune cells with the highest degrees of infiltration in osteosarcoma. Zhang et al (49) analyzed the immune infiltration of osteosarcoma samples in TCGA database. Their results showed that the two most common types of immune cells in osteosarcoma tissue are macrophages and T lymphocytes, in line with the present findings. According to the current understanding of the TME, low infiltration levels of CD8+ T cells, monocytes and M2 macrophages in osteosarcoma are associated with poor prognosis. It can be concluded that CD8+ T cells, monocytes and M2 macrophages play critical roles in regulating the immune microenvironment of osteosarcoma.
In the adult immune system, the BMP signaling pathway actively participates in dominating the fate of the immune lineage and the development of immune cells (50). BMP ligands secreted by tumor cells into the extracellular matrix act on immune cells, thereby regulating the functions of immune cells (21,51,52). Studies have suggested that activated Bone Morphogenic Protein Receptor 1α (BMPR1α)-deficient CD4+ T cells produce a large amount of IFN-γ and enhance tumor proliferation, indicating the adverse effect of BMP signaling on the adaptive immune response (53). Abrogation of BMPR1α signaling during CD4+ T cell activation induced inflammatory effector cells to express various of cytokines such as IL-17, IFN-γ, and TNF family and transcription factors defining the Th17 cell lineage (54). BMPs and BMPRs also serve an important role during the process of B cell activation (55). The current data revealed that the expression levels of BMPR2 in chronic lymphocytic leukemia, follicular lymphoma and germinal center B cell-like diffuse large B cell lymphoma were significantly higher compared with those in normal tissues. These results suggested that BMPR2 plays a pivotal role in the progression of some cancer types. Moreover, CD8+ T cell, monocyte and M2 macrophage markers, such as CD8A, CD8B, CD86, CSF1R, CD163, VSIG4 or MS4A4A, were negatively correlated with BMPR2 expression. These results indicated the importance of BMPR2 in the suppression and regulation of infiltrating immune cells in the osteosarcoma microenvironment. However, the detailed mechanism by which BMPR2 inhibits the infiltration of CD8+T cells, monocytes and M2 macrophages remains unclear. Activated BMPR2 can sequentially activate the downstream p38 MAPK/ERK, RHO/ROCK/LIMK and JNK signaling pathways through a non-classical pathway (56). The p38 MAPKs proteins are key therapeutic targets in Rheumatoid arthritis, Crohn's disease and other inflammatory diseases (57). It has been revealed that p38 MAPKs can inhibit expression of pro-inflammatory cytokines, such as IL-6, TNF-α, MCP-1 and IL-1β, and promote expression of anti-inflammatory cytokines, such as IL-10 and TGF-β (58). This signaling pathway has been shown to be important to the immune microenvironment of numerous tumor types (59,60). Zhang et al demonstrated the mechanisms of crosstalk between myeloid cells and tumor cells through EGFR/MAPK signaling pathway to restore CD8+ mediated anti-tumor immunity in pancreatic cancer (59). MAPK-mutant tumors are the only CD8+ T cell-inflamed tumors with high immunoreactivity and a constitutively cytolytic tumor microenvironment. The MAPK-mutant head and neck squamous-cell carcinoma model with immune function shows active cell death and extensive CD8+ T cell recruitment (61,62). We infer that BMPR2 regulates the immune microenvironment of osteosarcoma tissue through the MAPK/ERK pathway. In the future, more clinical and experimental studies are expected to future clarify the role of BMPR2 in regulating the infiltration of immune cells in osteosarcoma.
Several studies have shown that BMPR2 mutation and methylation affect BMPR2 expression levels in disease such as pulmonary hypertension disease (63–66) and tumor (67). Kodach et al (67) demonstrated that BMPR2 was mutated in the microsatellite-unstable of colorectal cancer cells, which decreased the BMPR2 expression level. In addition, the mutation of the BMPR2 gene has been positively correlated with the abundance of CD8+ T cells and neutrophils, suggesting that the mutation of the BMPR2 gene can promote the infiltration of lymphocytes in colon adenocarcinoma (68). Previous studies have shown that patients with osteosarcoma with high expression levels of BMPR2 have a worse prognosis compared with patients expressing low levels of BMPR2 (20,69). This is in line with the current results. However, the present results revealed that AUC for the expression level of BMPR2 in predicting 5-year survival was 0.601, suggesting that BMPR2 was not a good indicator of 5-year survival. This may be because the GEO dataset that was analyzed only collected gene expression data from high-grade osteosarcoma diagnostic biopsies. In addition, our previous study demonstrated that the depletion of BMPR2 in osteosarcoma cells markedly reduced the invasive capacity in vitro and metastatic potential in vivo. Mechanistically, it was revealed that BMPR2 could active LIMK2 through the RhoA/ROCK pathway and could also interact with LIMK2 directly in osteosarcoma cells (20,69). Similarly, the present study reported that the AUC for the expression level of BMPR2 in predicting metastatic was 0.711, indicating that BMPR2 was a good indicator to predict metastasis. However, whether BMPR2 is mutated before the development of osteosarcoma and its role in lymphocyte infiltration is unknown.
Above all, the present study demonstrated that CD8+ T cells, monocytes and M2 macrophages play critical roles in constructing the immune microenvironment of osteosarcoma. Increased BMPR2 expression was associated with decreased infiltration of osteosarcoma tissues by CD8+ T cells, monocytes and M2 macrophages, and was also correlated with poor prognosis in patients with osteosarcoma. Therefore, BMPR2 has potential as a prognostic biomarker for immune infiltration, metastasis and prognosis in osteosarcoma.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- BMPR2
bone morphogenetic protein receptor 2
- TIMER
Tumor Immune Estimation Resource
- OS
overall survival
- NK
natural killer cells
- DC
dendritic cells
- ARMS
acinar rhabdomyosarcoma
- ERMS
embryonal rhabdomyosarcoma
- TAMs
tumor-associated macrophages
Funding Statement
This study was supported by The National Natural Science Foundation of China (grant nos. 81602361 and 81702261), and The China Postdoctoral Science Foundation (grant no. 205425).
Funding
This study was supported by The National Natural Science Foundation of China (grant nos. 81602361 and 81702261), and The China Postdoctoral Science Foundation (grant no. 205425).
Availability of data and materials
The datasets generated and/or analyzed during the current study are available in the Oncomine repository (https://www.oncomine.org/resource/main.html), R2 repository, (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi), TARGET repository, (https://ocg.cancer.gov/programs/target); The Cancer Genome Atlas repository (http://tcga-data.nci.nih.gov/tcga/) and TIMER repository, (https://cistrome.shinyapps.io/timer/).
Authors' contributions
HC participated in the analysis of the data and was a major contributor in writing the manuscript. SQ and LZ helped collect and interpretate the data. YC participated in designing the study. GJ analyzed the data and revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hameed M, Mandelker D. Tumor syndromes predisposing to osteosarcoma. Adv Anat Pathol. 2018;25:217–222. doi: 10.1097/PAP.0000000000000190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Otoukesh B, Boddouhi B, Moghtadaei M, Kaghazian P, Kaghazian M. Novel molecular insights and new therapeutic strategies in osteosarcoma. Cancer Cell Int. 2018;18:158. doi: 10.1186/s12935-018-0654-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison DJ, Geller DS, Gill JD, Lewis VO, Gorlick R. Current and future therapeutic approaches for osteosarcoma. Expert Rev Anticancer Ther. 2018;18:39–50. doi: 10.1080/14737140.2018.1413939. [DOI] [PubMed] [Google Scholar]
- 4.Harting MT, Blakely ML. Management of osteosarcoma pulmonary metastases. Semin Pediatr Surg. 2006;15:25–29. doi: 10.1053/j.sempedsurg.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 5.Carrle D, Bielack S. Osteosarcoma lung metastases detection and principles of multimodal therapy. Cancer Treat Res. 2009;152:165–184. doi: 10.1007/978-1-4419-0284-9_8. [DOI] [PubMed] [Google Scholar]
- 6.Link MP, Goorin AM, Miser AW, Green AA, Pratt CB, Belasco JB, Pritchard J, Malpas JS, Baker AR, Kirkpatrick JA, et al. The effect of adjuvant chemotherapy on relapse-free survival in patients with osteosarcoma of the extremity. N Engl J Med. 1986;314:1600–1606. doi: 10.1056/NEJM198606193142502. [DOI] [PubMed] [Google Scholar]
- 7.Meyers PA, Healey JH, Chou AJ, Wexler LH, Merola PR, Morris CD, Laquaglia MP, Kellick MG, Abramson SJ, Gorlick R. Addition of pamidronate to chemotherapy for the treatment of osteosarcoma. Cancer. 2011;117 doi: 10.1002/cncr.25744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirabello L, Troisi RJ, Savage SA. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, Epidemiology, and end results program. Cancer. 2009;115:1531–1543. doi: 10.1002/cncr.24121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Locy H, de Mey S, de Mey W, De Ridder M, Thielemans K, Maenhout SK. Maenhout, Immunomodulation of the tumor microenvironment: Turn Foe into friend. Front Immunol. 2018;9:2909. doi: 10.3389/fimmu.2018.02909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumauthioz N, Labiano S, Romero P. Tumor resident memory T cells: New players in immune surveillance and therapy. Front Immunol. 2018;9:2076. doi: 10.3389/fimmu.2018.02076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Blanc C, Hans S, Tran T, Granier C, Saldman A, Anson M, Oudard S, Tartour E. Targeting resident memory T cells for cancer immunotherapy. Front Immunol. 2018;9:1722. doi: 10.3389/fimmu.2018.01722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gajewski TF, Schreiber H, Fu YX. Innate and adaptive immune cells in the tumor microenvironment. Nat Immunol. 2013;14:1014–1022. doi: 10.1038/ni.2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schreiber RD, Old LJ, Smyth MJ. Cancer immunoediting: Integrating immunity's roles in cancer suppression and promotion. Science. 2011;331:1565–1570. doi: 10.1126/science.1203486. [DOI] [PubMed] [Google Scholar]
- 15.Lussier DM, O'Neill L, Nieves LM, McAfee MS, Holechek SA, Collins AW, Dickman P, Jacobsen J, Hingorani P, Blattman JN. Enhanced T-cell immunity to osteosarcoma through antibody blockade of PD-1/PD-L1 interactions. J Immunother. 2015;38:96–106. doi: 10.1097/CJI.0000000000000065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen JK, Cote GM, Choy E, Yang P, Harmon D, Schwab J, Nielsen GP, Chebib I, Ferrone S, Wang X, et al. Programmed cell death ligand 1 expression in osteosarcoma. Cancer Immunol Res. 2014;2:690–698. doi: 10.1158/2326-6066.CIR-13-0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beppu H, Mwizerwa ON, Beppu Y, Dattwyler MP, Lauwers GY, Bloch KD, Goldstein AM. Stromal inactivation of BMPRII leads to colorectal epithelial overgrowth and polyp formation. Oncogene. 2008;27:1063–1070. doi: 10.1038/sj.onc.1210720. [DOI] [PubMed] [Google Scholar]
- 18.Kim IY, Lee DH, Lee DK, Kim WJ, Kim MM, Morton RA, Lerner SP, Kim SJ. Restoration of bone morphogenetic protein receptor type II expression leads to a decreased rate of tumor growth in bladder transitional cell carcinoma cell line TSU-Pr1. Cancer Res. 2004;64:7355–7360. doi: 10.1158/0008-5472.CAN-04-0154. [DOI] [PubMed] [Google Scholar]
- 19.Voorneveld PW, Kodach LL, Jacobs RJ, Liv N, Zonnevylle AC, Hoogenboom JP, Biemond I, Verspaget HW, Hommes DW, de Rooij K, et al. Loss of SMAD4 alters BMP signaling to promote colorectal cancer cell metastasis via activation of Rho and ROCK. Gastroenterology. 2014;147:196–208 e113. doi: 10.1053/j.gastro.2014.03.052. [DOI] [PubMed] [Google Scholar]
- 20.Wang S, Ren T, Huang Y, Bao X, Sun K, Shen D, Guo W. BMPR2 and HIF1-α overexpression in resected osteosarcoma correlates with distant metastasis and patient survival. Chin J Cancer Res. 2017;29:447–454. doi: 10.21147/j.issn.1000-9604.2017.05.09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martínez VG, Rubio C, Martínez-Fernández M, Segovia C, López-Calderón F, Garín MI, Teijeira A, Munera-Maravilla E, Varas A, Sacedón R, et al. BMP4 induces M2 macrophage polarization and favors tumor progression in bladder cancer. Clin Cancer Res. 2017;23:7388–7399. doi: 10.1158/1078-0432.CCR-17-1004. [DOI] [PubMed] [Google Scholar]
- 22.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: Genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones KB, Salah Z, Del Mare S, Galasso M, Gaudio E, Nuovo GJ, Lovat F, LeBlanc K, Palatini J, Randall RL, et al. miRNA signatures associate with pathogenesis and progression of osteosarcoma. Cancer Res. 2012;72:1865–1877. doi: 10.1158/0008-5472.CAN-11-2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuijjer ML, Rydbeck H, Kresse SH, Buddingh EP, Lid AB, Roelofs H, Bürger H, Myklebost O, Hogendoorn PC, Meza-Zepeda LA, Cleton-Jansen AM. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer. 2012;51:696–706. doi: 10.1002/gcc.21956. [DOI] [PubMed] [Google Scholar]
- 25.Kobayashi E, Masuda M, Nakayama R, Ichikawa H, Satow R, Shitashige M, Honda K, Yamaguchi U, Shoji A, Tochigi N, et al. Reduced argininosuccinate synthetase is a predictive biomarker for the development of pulmonary metastasis in patients with osteosarcoma. Mol Cancer Ther. 2010;9:535–544. doi: 10.1158/1535-7163.MCT-09-0774. [DOI] [PubMed] [Google Scholar]
- 26.Schafer W. Rhabdomyosarcoma vs normal muscle. R2 database. 2000-01-01 ed., 2000. p. Available on R2 since: 2000-01-01. [Google Scholar]
- 27.Huvos AG, Rosen G, Marcove RC. Primary osteogenic sarcoma: Pathologic aspects in 20 patients after treatment with chemotherapy en bloc resection, and prosthetic bone replacement. Arch Pathol Lab Med. 1977;101:14–18. [PubMed] [Google Scholar]
- 28.Li T, Fan J, Wang B, Traugh N, Chen Q, Liu JS, Li B, Liu XS. TIMER: A web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77:e108–e110. doi: 10.1158/0008-5472.CAN-17-0307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siemers NO, Holloway JL, Chang H, Chasalow SD, Ross-MacDonald PB, Voliva CF, Szustakowski JD. Genome-wide association analysis identifies genetic correlates of immune infiltrates in solid tumors. PLoS One. 2017;12:e0179726. doi: 10.1371/journal.pone.0179726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Danaher P, Warren S, Dennis L, D'Amico L, White A, Disis ML, Geller MA, Odunsi K, Beechem J, Fling SP. Gene expression markers of tumor Infiltrating leukocytes. J Immunother Cancer. 2017;5:18. doi: 10.1186/s40425-017-0215-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sousa S, Maatta J. The role of tumour-associated macrophages in bone metastasis. J Bone Oncol. 2016;5:135–138. doi: 10.1016/j.jbo.2016.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gentles AJ, Newman AM, Liu CL, Bratman SV, Feng W, Kim D, Nair VS, Xu Y, Khuong A, Hoang CD, et al. The prognostic landscape of genes and infiltrating immune cells across human cancers. Nat Med. 2015;21:938–945. doi: 10.1038/nm.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.R Core Team, corp-author. R Foundation for Statistical Computing; Vienna, Austria: 2012. R: A language and environment for statistical computing. ISBN 3-900051-07-0, URL. [Google Scholar]
- 35.Binnewies M, Roberts EW, Kersten K, Chan V, Fearon DF, Merad M, Coussens LM, Gabrilovich DI, Ostrand-Rosenberg S, Hedrick CC, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24:541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bami M, Mavrogenis AF, Angelini A, Milonaki M, Mitsiokapa E, Stamoulis D, Soucacos PN. Bone morphogenetic protein signaling in musculoskeletal cancer. J Cancer Res Clin Oncol. 2016;142:2061–2072. doi: 10.1007/s00432-016-2149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Siegel PM, Massague J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat Rev Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 38.Kim IY, Lee DH, Lee DK, Ahn HJ, Kim MM, Kim S, Morton RA. Morton: Loss of expression of bone morphogenetic protein receptor type II in human prostate cancer cells. Oncogene. 2004;23:7651–7659. doi: 10.1038/sj.onc.1207924. [DOI] [PubMed] [Google Scholar]
- 39.Heymann MF, Lézot F, Heymann D. The contribution of immune infiltrates and the local microenvironment in the pathogenesis of osteosarcoma. Cell Immunol. 2019;343:103711. doi: 10.1016/j.cellimm.2017.10.011. [DOI] [PubMed] [Google Scholar]
- 40.Apetoh L, Smyth MJ, Drake CG, Abastado JP, Apte RN, Ayyoub M, Blay JY, Bonneville M, Butterfield LH, Caignard A, et al. Consensus nomenclature for CD8(+) T cell phenotypes in cancer. Oncoimmunology. 2015;4:e998538. doi: 10.1080/2162402X.2014.998538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Speiser DE, Utzschneider DT, Oberle SG, Münz C, Romero P, Zehn D. T cell differentiation in chronic infection and cancer: Functional adaptation or exhaustion? Nat Rev Immunol. 2014;14:768–774. doi: 10.1038/nri3740. [DOI] [PubMed] [Google Scholar]
- 42.Mittal R, Chen CW, Lyons JD, Margoles LM, Liang Z, Coopersmith CM, Ford ML. Murine lung cancer induces generalized T-cell exhaustion. J Surg Res. 2015;195:541–549. doi: 10.1016/j.jss.2015.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baitsch L, Baumgaertner P, Devêvre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P, et al. Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest. 2011;121:2350–2360. doi: 10.1172/JCI46102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Japp AS, Kursunel MA, Meier S, Mälzer JN, Li X, Rahman NA, Jekabsons W, Krause H, Magheli A, Klopf C, et al. Dysfunction of PSA-specific CD8+ T cells in prostate cancer patients correlates with CD38 and Tim-3 expression. Cancer Immunol Immunother. 2015;64:1487–1494. doi: 10.1007/s00262-015-1752-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng W, Xiao H, Liu H, Zhou Y. Expression of programmed death 1 is correlated with progression of osteosarcoma. APMIS. 2015;123:102–107. doi: 10.1111/apm.12311. [DOI] [PubMed] [Google Scholar]
- 46.Fritzsching B, Fellenberg J, Moskovszky L, Sápi Z, Krenacs T, Machado I, Poeschl J, Lehner B, Szendrõi M, Bosch AL, et al. CD8+/FOXP3+-ratio in osteosarcoma microenvironment separates survivors from non-survivors: A multicenter validated retrospective study. Oncoimmunology. 2015;4:e990800. doi: 10.4161/2162402X.2014.990800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Qian BZ, Pollard JW. Macrophage diversity enhances tumor progression and metastasis. Cell. 2010;141:39–51. doi: 10.1016/j.cell.2010.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dumars C, Ngyuen JM, Gaultier A, Lanel R, Corradini N, Gouin F, Heymann D, Heymann MF. Dysregulation of macrophage polarization is associated with the metastatic process in osteosarcoma. Oncotarget. 2016;7:78343–78354. doi: 10.18632/oncotarget.13055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang C, Zheng JH, Lin ZH, Lv HY, Ye ZM, Chen YP, Zhang XY. Profiles of immune cell infiltration and immune-related genes in the tumor microenvironment of osteosarcoma. Aging (Albany NY) 2020;12:3486–3501. doi: 10.18632/aging.102824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eixarch H, Calvo-Barreiro L, Montalban X, Espejo C. Bone morphogenetic proteins in multiple sclerosis: Role in neuroinflammation. Brain Behav Immun. 2018;68:1–10. doi: 10.1016/j.bbi.2017.02.019. [DOI] [PubMed] [Google Scholar]
- 51.Singla DK, Singla R, Wang J. BMP-7 treatment increases M2 macrophage differentiation and reduces inflammation and plaque formation in Apo E-/- mice. PLoS One. 2016;11:e0147897. doi: 10.1371/journal.pone.0147897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Limmer A, Wirtz DC. Osteoimmunology: Influence of the immune system on bone regeneration and consumption. Z Orthop Unfall. 2017;155:273–280. doi: 10.1055/s-0043-100100. [DOI] [PubMed] [Google Scholar]
- 53.Kuczma M, Kurczewska A, Kraj P. Modulation of bone morphogenic protein signaling in T-cells for cancer immunotherapy. J Immunotoxicol. 2014;11:319–327. doi: 10.3109/1547691X.2013.864736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Browning LM, Pietrzak M, Kuczma M, Simms CP, Kurczewska A, Refugia JM, Lowery DJ, Rempala G, Gutkin D, Ignatowicz L, et al. TGF-β-mediated enhancement of TH17 cell generation is inhibited by bone morphogenetic protein receptor 1α signaling. Sci Signal. 2018;11:eaar2125. doi: 10.1126/scisignal.aar2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huse K, Bakkebø M, Oksvold MP, Forfang L, Hilden VI, Stokke T, Smeland EB, Myklebust JH. Bone morphogenetic proteins inhibit CD40L/IL-21-induced Ig production in human B cells: Differential effects of BMP-6 and BMP-7. Eur J Immunol. 2011;41:3135–3145. doi: 10.1002/eji.201141558. [DOI] [PubMed] [Google Scholar]
- 56.Gámez B, Rodriguez-Carballo E, Ventura F. BMP signaling in telencephalic neural cell specification and maturation. Front Cell Neurosci. 2013;7:87. doi: 10.3389/fncel.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kumar S, Boehm J, Lee JC. p38 MAP kinases: Key signalling molecules as therapeutic targets for inflammatory diseases. Nat Rev Drug Discov. 2003;2:717–726. doi: 10.1038/nrd1177. [DOI] [PubMed] [Google Scholar]
- 58.Liang X, Wang L, Wang M, Liu Z, Liu X, Zhang B, Liu E, Li G. MicroRNA-124 inhibits macrophage cell apoptosis via targeting p38/MAPK signaling pathway in atherosclerosis development. Aging (Albany NY) 2020;12:13005–13022. doi: 10.18632/aging.103387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang Y, Velez-Delgado A, Mathew E, Li D, Mendez FM, Flannagan K, Rhim AD, Simeone DM, Beatty GL, Pasca di Magliano M. Myeloid cells are required for PD-1/PD-L1 checkpoint activation and the establishment of an immunosuppressive environment in pancreatic cancer. Gut. 2017;66:124–136. doi: 10.1136/gutjnl-2016-312078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wu M, Chen X, Lou J, Zhang S, Zhang X, Huang L, Sun R, Huang P, Wang F, Pan S. TGF-β1 contributes to CD8+ Treg induction through p38 MAPK signaling in ovarian cancer microenvironment. Oncotarget. 2016;7:44534–44544. doi: 10.18632/oncotarget.10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liao P, Song K, Zhu Z, Liu Z, Zhang W, Li W, Hu J, Hu Q, Chen C, Chen B, et al. Propranolol suppresses the growth of colorectal cancer through simultaneously activating autologous CD8(+) T cells and inhibiting tumor AKT/MAPK pathway. Clin Pharmacol Ther. 2020;108:606–615. doi: 10.1002/cpt.1894. [DOI] [PubMed] [Google Scholar]
- 62.Ngan HL, Liu Y, Fong AY, Poon PHY, Yeung CK, Chan SSM, Lau A, Piao W, Li H, Tse JSW, et al. MAPK pathway mutations in head and neck cancer affect immune microenvironments and ErbB3 signaling. Life Sci Alliance. 2020;3:e201900545. doi: 10.26508/lsa.201900545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pousada G, Baloira A, Valverde D. Methylation analysis of the BMPR2 gene promoter region in patients with pulmonary arterial hypertension. Arch Bronconeumol. 2016;52:293–298. doi: 10.1016/j.arbr.2016.03.020. (In English, Spanish) [DOI] [PubMed] [Google Scholar]
- 64.Liu D, Yan Y, Chen JW, Yuan P, Wang XJ, Jiang R, Wang L, Zhao QH, Wu WH, Simonneau G, et al. Hypermethylation of BMPR2 promoter occurs in patients with heritable pulmonary arterial hypertension and inhibits BMPR2 expression. Am J Respir Crit Care Med. 2017;196:925–928. doi: 10.1164/rccm.201611-2273LE. [DOI] [PubMed] [Google Scholar]
- 65.Gräf S, Haimel M, Bleda M, Hadinnapola C, Southgate L, Li W, Hodgson J, Liu B, Salmon RM, Southwood M, et al. Identification of rare sequence variation underlying heritable pulmonary arterial hypertension. Nat Commun. 2018;9:1416. doi: 10.1038/s41467-018-03672-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhu N, Pauciulo MW, Welch CL, Lutz KA, Coleman AW, Gonzaga-Jauregui C, Wang J, Grimes JM, Martin LJ, He H, et al. Novel risk genes and mechanisms implicated by exome sequencing of 2572 individuals with pulmonary arterial hypertension. Genome Med. 2019;11:69. doi: 10.1186/s13073-019-0685-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kodach LL, Wiercinska E, de Miranda NF, Bleuming SA, Musler AR, Peppelenbosch MP, Dekker E, van den Brink GR, van Noesel CJ, Morreau H, et al. The bone morphogenetic protein pathway is inactivated in the majority of sporadic colorectal cancers. Gastroenterology. 2008;134:1332–1341. doi: 10.1053/j.gastro.2008.02.059. [DOI] [PubMed] [Google Scholar]
- 68.Shi J, Jiang D, Yang S, Sun Y, Wang J, Zhang X, Liu Y, Lu Y, Yang K. Molecular profile reveals immune-associated markers of lymphatic invasion in human colon adenocarcinoma. Int Immunopharmacol. 2020;83:106402. doi: 10.1016/j.intimp.2020.106402. [DOI] [PubMed] [Google Scholar]
- 69.Wang S, Ren T, Jiao G, Huang Y, Bao X, Zhang F, Liu K, Zheng B, Sun K, Guo W. BMPR2 promotes invasion and metastasis via the RhoA-ROCK-LIMK2 pathway in human osteosarcoma cells. Oncotarget. 2017;8:58625–58641. doi: 10.18632/oncotarget.17382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun L, Hui AM, Su Q, Vortmeyer A, Kotliarov Y, Pastorino S, Passaniti A, Menon J, Walling J, Bailey R, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9:287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 71.Turashvili G, Bouchal J, Baumforth K, Wei W, Dziechciarkova M, Ehrmann J, Klein J, Fridman E, Skarda J, Srovnal J, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kaiser S, Park YK, Franklin JL, Halberg RB, Yu M, Jessen WJ, Freudenberg J, Chen X, Haigis K, Jegga AG, et al. Transcriptional recapitulation and subversion of embryonic colon development by mouse colon tumor models and human colon cancer. Genome Biol. 2007;8:R131. doi: 10.1186/gb-2007-8-7-r131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hao Y, Triadafilopoulos G, Sahbaie P, Young HS, Omary MB, Lowe AW. Gene expression profiling reveals stromal genes expressed in common between Barrett's esophagus and adenocarcinoma. Gastroenterology. 2006;131:925–933. doi: 10.1053/j.gastro.2006.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Pyeon D, Newton MA, Lambert PF, den Boon JA, Sengupta S, Marsit CJ, Woodworth CD, Connor JP, Haugen TH, Smith EM, et al. Fundamental differences in cell cycle deregulation in human papillomavirus-positive and human papillomavirus-negative head/neck and cervical cancers. Cancer Res. 2007;67:4605–4619. doi: 10.1158/0008-5472.CAN-06-3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cutcliffe C, Kersey D, Huang CC, Zeng Y, Walterhouse D, Perlman EJ, Renal Tumor Committee of the Children's Oncology Group Clear cell sarcoma of the kidney: Up-regulation of neural markers with activation of the sonic hedgehog and Akt pathways. Clin Cancer Res. 2005;11:7986–7994. doi: 10.1158/1078-0432.CCR-05-1354. [DOI] [PubMed] [Google Scholar]
- 76.Yusenko MV, Kuiper RP, Boethe T, Ljungberg B, van Kessel AG, Kovacs G. High-resolution DNA copy number and gene expression analyses distinguish chromophobe renal cell carcinomas and renal oncocytomas. BMC Cancer. 2009;9:152. doi: 10.1186/1471-2407-9-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 78.Rosenwald A, Alizadeh AA, Widhopf G, Simon R, Davis RE, Yu X, Yang L, Pickeral OK, Rassenti LZ, Powell J, et al. Relation of gene expression phenotype to immunoglobulin mutation genotype in B cell chronic lymphocytic leukemia. J Exp Med. 2001;194:1639–1647. doi: 10.1084/jem.194.11.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Haferlach T, Kohlmann A, Wieczorek L, Basso G, Kronnie GT, Béné MC, De Vos J, Hernández JM, Hofmann WK, Mills KI, et al. Clinical utility of microarray-based gene expression profiling in the diagnosis and subclassification of leukemia: Report from the International Microarray Innovations in Leukemia Study Group. J Clin Oncol. 2010;28:2529–2537. doi: 10.1200/JCO.2009.23.4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, et al. The use of molecular profiling to predict survival after chemotherapy for diffuse large-B-cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 81.Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, Califano A. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. doi: 10.1038/ng1532. [DOI] [PubMed] [Google Scholar]
- 82.Dürig J, Bug S, Klein-Hitpass L, Boes T, Jöns T, Martin-Subero JI, Harder L, Baudis M, Dührsen U, Siebert R. Combined single nucleotide polymorphism-based genomic mapping and global gene expression profiling identifies novel chromosomal imbalances, mechanisms and candidate genes important in the pathogenesis of T-cell prolymphocytic leukemia with inv(14)(q11q32) Leukemia. 2007;21:2153–2163. doi: 10.1038/sj.leu.2404877. [DOI] [PubMed] [Google Scholar]
- 83.Hou J, Aerts J, den Hamer B, van Ijcken W, den Bakker M, Riegman P, van der Leest C, van der Spek P, Foekens JA, Hoogsteden HC, et al. Gene expression-based classification of non-small cell lung carcinomas and survival prediction. PLoS One. 2010;5:e10312. doi: 10.1371/journal.pone.0010312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Compagno M, Lim WK, Grunn A, Nandula SV, Brahmachary M, Shen Q, Bertoni F, Ponzoni M, Scandurra M, Califano A, et al. Mutations of multiple genes cause deregulation of NF-kappaB in diffuse large B-cell lymphoma. Nature. 2009;459:717–721. doi: 10.1038/nature07968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Haqq C, Nosrati M, Sudilovsky D, Crothers J, Khodabakhsh D, Pulliam BL, Federman S, Miller JR, III, Allen RE, Singer MI, et al. The gene expression signatures of melanoma progression. Proc Natl Acad Sci USA. 2005;102:6092–6097. doi: 10.1073/pnas.0501564102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Badea L, Herlea V, Dima SO, Dumitrascu T, Popescu I. Combined gene expression analysis of whole-tissue and microdissected pancreatic ductal adenocarcinoma identifies genes specifically overexpressed in tumor epithelia. Hepatogastroenterology. 2008;55:2016–2027. [PubMed] [Google Scholar]
- 87.Iacobuzio-Donahue CA, Maitra A, Olsen M, Lowe AW, van Heek NT, Rosty C, Walter K, Sato N, Parker A, Ashfaq R, et al. Exploration of global gene expression patterns in pancreatic adenocarcinoma using cDNA microarrays. Am J Pathol. 2003;162:1151–1162. doi: 10.1016/S0002-9440(10)63911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Luo JH, Yu YP, Cieply K, Lin F, Deflavia P, Dhir R, Finkelstein S, Michalopoulos G, Becich M. Gene expression analysis of prostate cancers. Mol Carcinog. 2002;33:25–35. doi: 10.1002/mc.10018. [DOI] [PubMed] [Google Scholar]
- 89.Cho JY, Lim JY, Cheong JH, Park YY, Yoon SL, Kim SM, Kim SB, Kim H, Hong SW, Park YN, et al. Gene expression signature-based prognostic risk score in gastric cancer. Clin Cancer Res. 2011;17:1850–1857. doi: 10.1158/1078-0432.CCR-10-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Riker AI, Enkemann SA, Fodstad O, Liu S, Ren S, Morris C, Xi Y, Howell P, Metge B, Samant RS, et al. The gene expression profiles of primary and metastatic melanoma yields a transition point of tumor progression and metastasis. BMC Med Genomics. 2008;1:13. doi: 10.1186/1755-8794-1-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Korkola JE, Houldsworth J, Chadalavada RS, Olshen AB, Dobrzynski D, Reuter VE, Bosl GJ, Chaganti RS. Down-regulation of stem cell genes, including those in a 200-kb gene cluster at 12p13.31, is associated with in vivo differentiation of human male germ cell tumors. Cancer Res. 2006;66:820–827. doi: 10.1158/0008-5472.CAN-05-2445. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Oncomine repository (https://www.oncomine.org/resource/main.html), R2 repository, (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi), TARGET repository, (https://ocg.cancer.gov/programs/target); The Cancer Genome Atlas repository (http://tcga-data.nci.nih.gov/tcga/) and TIMER repository, (https://cistrome.shinyapps.io/timer/).