Abstract
Nonalcoholic steatohepatitis (NASH) is an aggressive liver disease threatening human health, yet no medicine is developed to treat this disease. In this study, we first discovered that Leptin mutant rats (LepΔI14/ΔI14) exhibit characteristic NASH phenotypes including steatosis, lymphocyte infiltration, and ballooning after postnatal week 16. We then examined NASH progression by performing an integrated analysis of hepatic transcriptome in Leptin-deficient rats from postnatal 4 to 48 weeks. Initially, simple steatosis in LepΔI14/ΔI14 rats were observed with increased expression of the genes encoding for rate-limiting enzymes in lipid metabolism such as acetyl-CoA carboxylase and fatty acid synthase. When NASH phenotypes became well developed at postnatal week 16, we found gene expression changes in insulin resistance, inflammation, reactive oxygen species and endoplasmic reticulum stress. As NASH phenotypes further progressed with age, we observed elevated expression of cytokines and chemokines including C-C motif chemokine ligand 2, tumor necrosis factor ɑ, interleukin-6, and interleukin-1β together with activation of the c-Jun N-terminal kinase and nuclear factor-κB pathways. Histologically, livers in LepΔI14/ΔI14 rats exhibited increased cell infiltration of MPO+ neutrophils, CD8+ T cells, CD68+ hepatic macrophages, and CCR2+ inflammatory monocyte-derived macrophages associated with macrophage polarization from M2 to M1. Subsequent cross-species comparison of transcriptomes in human, rat, and mouse NASH models indicated that Leptin-deficient rats bear more similarities to human NASH patients than previously established mouse NASH models. Taken together, our study suggests that LepΔI14/ΔI14 rats are a valuable pre-clinical rodent model to evaluate NASH drug safety and efficacy.
Keywords: Nonalcoholic steatohepatitis, transcriptome, hepatic inflammation, animal model
Impact statement
Nonalcoholic steatohepatitis (NASH) is an aggressive liver disease affecting human health. In order to develop effective drugs for NASH, we need to develop useful animal NASH models. Previously, Leptin-deficient ob/ob mice were widely used as nonalcoholic fatty liver disease models. However, they do not develop spontaneous NASH. Here we found that Leptin-deficient rats (LepΔI14/ΔI14) exhibit unique NASH phenotypes at postnatal week 16. We therefore analyzed disease progression in LepΔI14/ΔI14 rats via transcriptomic analysis of hepatic gene expression. Our work reveals that NASH progression is coupled with sequential changes in gene expression that are linked to inflammatory signaling pathways and immune cell infiltration. We conclude that LepΔI14/ΔI14 rats will serve as a valuable pre-clinical rodent model to screen potential drug targets and biomarkers for NASH drug development.
Introduction
Nonalcoholic fatty liver disease (NAFLD) is the most common liver disease worldwide, affecting approximately 24% of the population.1 This disease can be categorized into nonalcoholic fatty liver (NAFL) and the more advanced stage of nonalcoholic steatohepatitis (NASH) with inflammation and hepatocyte damage.2 NAFLD, especially NASH, will eventually develop into liver fibrosis, cirrhosis, and hepatocellular carcinoma.3 Historically, the severity of NAFLD has been assessed through many diagnostic modalities, including examination of demographic record, serum parameters, and histopathological characteristics of liver biopsies. Recently, non-invasive imaging techniques such as magnetic resonance imaging-proton density fat fraction and magnetic resonance elastography have been used.4 However, the severity level only provides a snapshot of the temporal progression of the disease. The pathogenesis of NAFLD/NASH in human beings is still poorly understood due to its very slow progression rate.5
To study the pathogenesis and progression of NAFLD/NASH, many animal models were generated by way of gene mutations and/or dietary induction.6–9 Among them, Leptin-deficient ob/ob mice are widely used as a NAFLD model to mirror the human disease with obesity, hyperglycemia, and hyperinsulinemia.9 Leptin is an adipokine secreted by white adipocyte tissues that regulates food intake, glucose and lipid metabolism, energy homeostasis, the immune system, reproduction, and more.10,11 However, it should be noted that ob/ob mice do not develop spontaneous NASH.6,7 Moreover, it is still unclear how leptin functions during NASH progression in humans.12 13 We previously generated Leptin-deficient (LepΔI14/ΔI14) rats to study diabetes by CRISPR/Cas9 technology.14 In the current study, we found that LepΔI14/ΔI14 rats also display distinctive NASH features in adulthood, providing us with an ideal rodent model to study NASH. We therefore performed integrated transcriptomic analysis, identifying temporal changes of hepatic gene expression in association with NASH progression in LepΔI14/ΔI14 rats.
Materials and methods
Animals
Rats and mice were kept in a 12-h light, 12-h dark cycle with ad libitum access to regular chow food and water. The littermates were used as the controls. B6/JNju-Lepem1Cd25/Nju mice were purchased from Nanjing Biomedical Research Institute of Nanjing University (Cat#T001461). All the protocols were in accordance with the guidelines of Tongji University’s Committee on Animal Care and Use. All the experimental procedures as described below were approved by the animal experiment administration committee of Tongji University (# TJLAC-016-021).
Serum biochemical analyses
The animals were fasted for 16 h and serum was obtained after centrifugation. Serum alanine aminotransferase (ALT) and aspartate aminotransferase (AST) were determined by using the spectrometric kits (Sigma Diagnostics, St. Louis, MO, USA). Triglyceride, high-density lipoprotein cholesterol (HDL-C), and low-density lipoprotein cholesterol (LDL-C) were examined by Adicon Central Lab. Serum IL1β was detected by ELISA kit (Boster Biological Technology, Wuhan, China).
Histological analyses
Liver samples of the animals were collected and fixed according to the routine methods for hematoxylin and eosin (HE) staining, Oil red O staining, Sirius red staining, and immunohistochemistry (IHC), respectively. Detailed information of the antibodies for IHC are summarized in Table S1. To quantify the IHC signals, we stained the liver sections with antibodies against immune-cell markers for three LepΔI14/ΔI14 rats and three WT controls at different timepoints. The positively stained cells were counted in three random pictures of each animal. Then we determined the immune cell infiltration by statistics described in “Statistical analyses” section.
RT-PCR
Total RNA was extracted from snap-frozen liver samples using TRIzol (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instructions. The complementary DNA (cDNA) was generated by RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, USA). Quantitative polymerase chain reaction (qPCR) was performed using TB GreenTM Premix Ex TaqTM II (TaKaRa, RR820A, Kusatsu, Shiga, Japan) on QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, Carlsbad, CA, USA). PCR primers were designed to specifically amplify genes of interest (Table S2). qPCR was performed with β-tubulin as internal control.
Western blot
Total protein was extracted from snap-frozen liver samples using radioimmunoprecipitation lysis buffer (EpiZyme, PC104, Shanghai, China) with protease inhibitors (EpiZyme, GRF101, Shanghai, China) and phosphatase inhibitors (EpiZyme, GRF102, Shanghai, China). Western blot was performed following the routine method. The antibodies are listed in Table S1. The expression level of the genes was normalized to that of β-tubulin for each sample in all Western blots.
RNA-seq
Total RNA was extracted from snap-frozen liver samples using TRIzol (Life Technologies, Carlsbad, CA, USA) according to the manufacturer’s instruction. RNA degradation and contamination were monitored on 1% agarose gels. RNA purity was checked using the Nano Photometer spectrophotometer (IMPLEN, München, Germany). RNA concentration was measured by Qubit 2.0 Fluorometer (Life Technologies, Carlsbad, CA, USA). RNA integrity was assessed using RNA Nano 6000 Assay Kit of the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). A total amount of 3 μg RNA per sample was used as input material for the RNA sample preparations. Ribosomal RNA was removed by Ribo-zero rRNA Removal Kit (Epicentre Biotechnologies, Madison, WI, USA). Library construction was done using NEBNext Ultra Directional RNA Library Prep Kit for Illumina (NEB, Ipswich, MA, USA) following manufacturer’s protocols. After cluster generation (TruSeq PE Cluster Kit v3-cBot-HS [Illumina, San Diego, CA, USA]), the libraries were sequenced on an Illumina Hiseq X platform and 150 bp paired-end reads were generated in the facilities of Novogene (Beijing, China).
RNA-seq data analysis. Clean data were obtained by removing reads containing adapters, reads containing ploy-N, and low-quality reads from raw reads. Meanwhile, Q20, Q30, and GC content of the clean data were calculated. All the analyses were based on the clean data with high quality. Using Hisat2 (v2.1.0), the rat RNA-seq reads were mapped to the rn6 reference genome (RefSeq). For transcriptome quantification, we used featureCounts (v1.6.1) to generate read counts for all genes. Raw counts were normalized by DESeq2 (v1.20.0) using rlog method and normalized counts of replicates were averaged. The differentially expressed genes (DEGs) between two samples were identified by DESeq2 (v1.20.0) with a Benjamini-Hochberg-adjusted P value <0.05 and a fold change >1.5. Principal component analysis (PCA) on samples were plotted with R package ggplot2 on read counts normalized with R package DESeq2.
Gene ontology (GO) analysis. We performed functional annotation of gene sets using R package clusterProfiler. P values for clusters were plotted to show the significance of functional clusters. Heatmaps were generated with R package pheatmap (https://CRAN.R-project.org/package=pheatmap) and RColorBrewer (https://CRAN.R-project.org/package=RColorBrewer).
Time-series analysis of fat accumulation and inflammation geneset. Genes representing each feature were collected as described in the main text. Their gene expression levels were normalized by Z-score and presented as scatter plots.
Species comparison analysis. Homologous genes of three species (human, rat, mouse) were downloaded from the Ensembl genome browser Release 91 (http://dec2017.archive.ensembl.org/index.html). The significantly DEGs and pathways in human beings with healthy obesity, NAFLD, and NASH were retained from the previous study.15 Specifically, we screened the mouse and rat DEGs and re-annotated to match the human genes. Then we used gene set enrichment analysis to pool the enriched KEGG pathways (P < 0.05). Pre-defined set of genes were collected from the official Molecular Signatures Database-C2-KEGG section. The statistical test was done by a phenotypic-based permutation test to produce a null distribution for the enrichment score. The P value was determined by comparison to the null distribution. Similarly, we obtained the enriched KEGG pathways in human beings with healthy obesity, NAFLD, and NASH. The common pathway alteration was determined by the overlap between different species.
Accession numbers. The accession number for the rat RNA-seq data generated in this study is GSE124002.
Statistical analyses
At each timepoint, the data from LepΔI14/ΔI14 rats were compared to the WT controls by t test. The P value of <0.05 was defined as the level of significance, and all graphs and analyses were performed using GraphPad Prism (version 6; GraphPad Software Inc., La Jolla, CA, USA).
Results
Leptin deficiency leads to NASH phenotype in rats but not in mice
LepΔI14/ΔI14 rats share many similar phenotypes with ob/ob mice, such as obesity, hyperglycemia, hyperinsulinemia, NAFLD, and infertility.14 Our previous histopathological examination showed that the NAFLD feature in LepΔI14/ΔI14 rats arises at postnatal week 8.14 In the present work, we did hematoxylin-eosin (HE) and Oil Red O staining of liver sections prepared at different timepoints during disease progression (Figure 1(a)). As expected, the WT controls developed simple steatosis during aging (Figure 1(a)). Compared to the controls, LepΔI14/ΔI14 rats showed earlier and more severe NAFLD phenotypes (Figure 1(a)). In line with HE and Oil Red O staining, body weight and relative liver weight of LepΔI14/ΔI14 rats were also significantly higher than those of their wild-type (WT) littermates after eight postnatal weeks (Figure S1(a)). We also observed an increase in serum triglyceride, HDL-C, and LDL-C (Figure S1(a)). Interestingly, ALT and AST levels dramatically increased and reached a plateau at postnatal week 16 (Figure S1(a)). The plateau of these NAFLD markers made us wonder how NASH phenotypes progress in LepΔI14/ΔI14 rats after week 16. We re-examined the liver sections at different time points, looking particularly for NASH features such as steatosis, lymphocyte infiltration and ballooning. Indeed, NASH phenotypes such as steatosis, lymphocyte infiltration, and ballooning were only detected in LepΔI14/ΔI14 rats after week 16 (Figure 1(b), (c), and (d)). Compared to the controls, myeloperoxidase positive (MPO+) neutrophil infiltration was also significantly increased after week 16 (Figure 1(e)). Using the NASH scoring system (Table S3),16 we found that LepΔI14/ΔI14 rats reached the significance of NASH score (≥5) from week 16 (Figure 1(f)). We did not see fibrosis in LepΔI14/ΔI14 rats at any timepoints through 48 weeks postnatally, which was the latest time point we could carry out experiments with LepΔI14/ΔI14 rats (Figure S1(b)). In short, our results showed that LepΔI14/ΔI14 rats develop the NASH phenotype of hepatic inflammation from postnatal week 16 on.
Figure 1.
Unique NASH phenotypes of LepΔI14/ΔI14 rats. (a) HE and Oil red O staining of liver sections showing that NASH phenotypes can be observed at postnatal week 16. (b to d) Magnified views of HE staining in (a) showing NASH phenotype at postnatal week 16 (b), 32 (c), and 48 (d). Arrow heads indicate hepatocyte ballooning; dashed rectangles indicate lymphocyte infiltration. (e) IHC showing MPO positive neutrophil infiltration. Left panel shows a representative image at postnatal week 16, and right panel is the statistical analysis at each time point (n = 6). (f) The NASH scores at each time point. ***P < 0.001, ****P < 0.0001. (A color version of this figure is available in the online journal.)
It has been reported that Leptin-deficient ob/ob mice do not develop spontaneous NASH.6,7 To determine whether species differences between mice and rats are real, we also examined Leptin-deficient mice in our own breeding colony with similar breeding conditions to LepΔI14/ΔI14 rats. Leptin-deficient ob/ob mice in our breeding conditions also showed significantly higher body weight, elevated liver/body weight ratio, and higher AST and ALT levels (Figure S2(a), (b), (c), and (d)) at postnatal week 16, confirming the presence of NAFLD. However, HE and Oil Red O staining showed that Leptin-deficient mice had simple steatosis without NASH-like histological phenotypes at week 16 and 32 (Figure S2(e) and (g)). Nor did they show any difference in immune cell infiltration by LY6G+ neutrophils, F4/80+ macrophages, CD11C+ M1 macrophages, CD163+ M2 macrophages or CCR2+ inflammatory monocyte-derived macrophages (Figure S2(f) and (h)). Taken together, Leptin deficiency leads to NASH phenotypes uniquely in rats but not in mice.
The temporal progression of NASH is accompanied with distinct hepatic gene expression patterns
To understand the mechanism of disease progression, previous studies have investigated the transcriptome of liver biopsies from NAFLD patients at mild, medium, and severe stages.17,18 However, the heterogeneities in different patients cannot reflect the temporal progression of the disease in a particular person. Therefore, we decided to examine transcriptomic changes during NASH progression in this unique LepΔI14/ΔI14 rat model. We performed RNA-sequencing (RNA-seq) of liver samples from littermate WT and LepΔI14/ΔI14 rats at postnatal 4-week, 8-week, 16-week, 32-week, and 48-week timepoints. Overall, the hepatic transcriptome showed dynamic changes at different timepoints (Figure 2(a)). At week 4, the expression profiles of WT controls and LepΔI14/ΔI14 rats were very similar and clustered together. From week 8 to week 32, the expression profiles of WT controls and LepΔI14/ΔI14 rats clustered separately, implying a temporally regulated different expression pattern during NAFLD progression. The expression profiles of LepΔI14/ΔI14 rats at week 48 were still clearly separated from that of WT controls (Figure 2(a)), although differences in gene expression patterns may have been reduced during the aging process. We then identified the DEGs between LepΔI14/ΔI14 and WT rats for each timepoint (Table S4) and annotated their functions with enriched GO terms. We found that two metabolic pathways (fatty acid metabolic process [GO:0006631] and regulation of small molecule metabolic processes [GO:0062012]) were constantly de-regulated in all timepoints analyzed (Figure 2(b), Table S5). Further analysis of both upregulated DEGs and downregulated DEGs indicated that gradual changes of gene expression occurred from postnatal week 4 to 48 (Figure S3, Tables S4 and S6). This suggested that NAFLD progresses stepwisely with activation/inactivation of different sets of genes. Indeed, when we examined the enriched GO terms (LepΔI14/ΔI14 vs. WT) at different timepoints, we found that GO terms related to lipid synthesis initially appeared at week 4, which was followed in turn by those related to insulin resistance, inflammation, reactive oxygen species (ROS), endoplasmic reticulum (ER) stress, and mitochondrial function (Figure 2(c), Table S5). We did not observe more enriched GO terms at week 48 than week 16 and 32 (Figure 2(c)). While looking into DEGs within the pathways described above (Figure S4), we found that many genes in fatty acid metabolic processes such as Gk, Fabp4, Fasn, and Acaca were upregulated (Figure S4). Interestingly, some insulin-resistance related genes such as Ppara, Irs1, and Irs2 were downregulated, while others such as Prkcd and Pkm were upregulated (Figure S4). Many inflammation related genes were upregulated at week 32, including Mapk9, Fasn, Ikbkb, Nfkb1, Nos2, Ccl2, and Mmp9 (Figure S4). Nevertheless, many genes related to cellular respiration were downregulated, especially at week 32 (Figure S4). The gene expression patterns at week 4 show very minor differences between LepΔI14/ΔI14 rats and WT controls. The difference in gene expression patterns in WT and LepΔI14/ΔI14 rats became obvious at week 8 (Figure S4). Overall, our results show that NASH progresses in LepΔI14/ΔI14 rats stepwisely with sequential activation/inactivation of genes in lipid metabolism, insulin resistance, inflammation, ROS, ER stress, and mitochondrial function.
Figure 2.
Hepatic transcriptome alteration during NASH progression in LepΔI14/ΔI14 rats. (a) PCA of the RNA-seq data in LepΔI14/ΔI14 rats and the WT controls from postnatal week 4 to 48 (n = 3 for samples at each time point except n = 2 for LepΔI14/ΔI14 rats at week 32). (b) Venn plot showing the overlap of enriched GO terms among different time points. (c) Heatmap showing a stepwise enrichment of different pathways during NASH progression. Followed by lipid synthesis at week 4, the pathways related to insulin resistance, inflammation, reactive oxygen species (ROS), endoplasmic reticulum (ER) stress, and mitochondrial function are gradually enriched at different time points. (A color version of this figure is available in the online journal.)
The expression of rate-limiting enzymes is upregulated in LepΔI14/ΔI14 rats during NASH progression
As shown in Figure 2, progression of NASH in LepΔI14/ΔI14 rats was accompanied with temporal gene expression related to lipid metabolism, inflammation, and mitochondrial function. We then asked if the trend of gene expression related to lipid accumulation and inflammation can explain the plateau of inflammation at week 16 during NASH progression. We first profiled the dynamic expression of genes with functions related to lipid metabolism and inflammation, respectively (fatty acid biosynthetic process [GO:0006633] and positive regulation of inflammatory response [GO:0050729]). As expected, the inflammation-related genes started to be expressed significantly higher in LepΔI14/ΔI14 rats than WT at week 16 (P < 0.05) and maintained stably at the elevated expression (P < 0.05) all the way to week 48 (Figure 3(a)). In contrast, many lipid-metabolism related genes showed no significant difference between LepΔI14/ΔI14 and the WT controls across all timepoints (Figure 3(a)). As liver steatosis is a morbid accumulation of triglyceride (TAG) in hepatocytes, we selected nine genes which encode well-known essential enzymes including acetyl-CoA carboxylase (ACC) and fatty acid synthase (FASN) for TAG synthesis. As steatosis developed during aging (Figure 1), the expression of these nine genes displayed an elevating trend in both WT controls and LepΔI14/ΔI14 rats. Again, LepΔI14/ΔI14 rats showed much higher expression than in WT controls (Figure 3(b) and (c)). The increase in Cd36, Scd1, and Fasn in LepΔI14/ΔI14 rat liver was confirmed by qRT-PCR (Figure 3(d)). Our results indicate that liver steatosis is correlated with the elevated expression of a small number of rate-limiting enzymes, which can serve as therapeutic targets of NASH by reducing fat accumulation in the liver.
Figure 3.
The inflammation related genes and pathways during NASH progression in LepΔI14/ΔI14 rats. (a) The expression trend of DEGs in GO terms “fatty acid biosynthetic process” (GO:0006633) and “positive regulation of inflammatory response” (GO:0050729). (b) The expression trend of nine genes encoding rate-limiting enzymes for TAG synthesis. (c) Heatmap showing expression levels of the genes in (b). log2 (foldchange) is shown as normalized gene expression. (d) qRT-PCR showing the relative expression (LepΔI14/ΔI14 rats vs. WT controls) of Cd36, Scd1 and Fasn using Actb as internal control. Data are presented as mean ± SD. (A color version of this figure is available in the online journal.)
Restricted hepatic inflammation in LepΔI14/ΔI14 rats is associated with the inflammation related genes, pathways, and immune cell infiltration
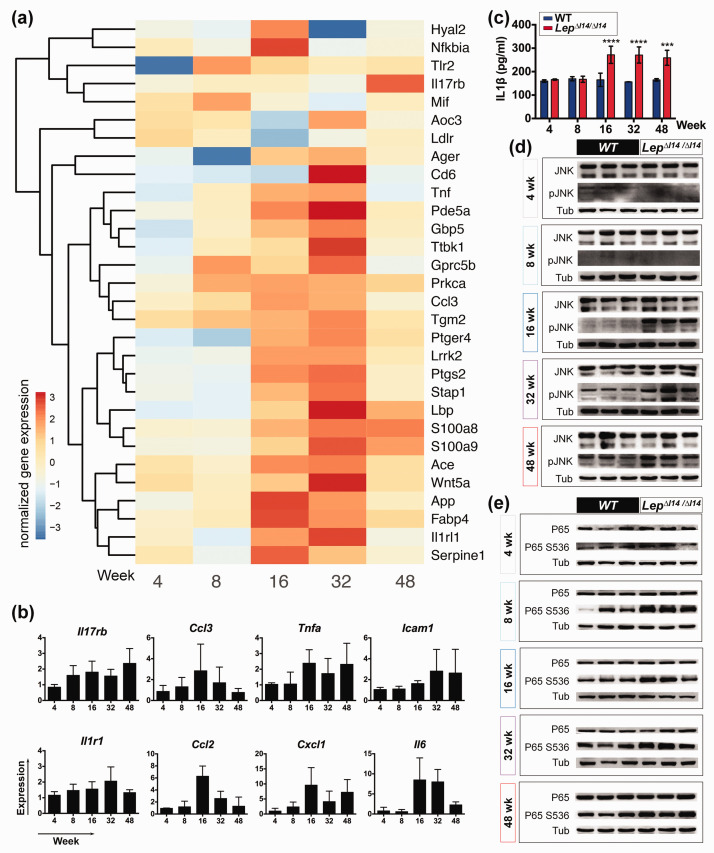
As demonstrated above (Figure 3(a)), we saw a unique expression trend of genes with function in positive regulation of inflammatory response (GO:0050729) in LepΔI14/ΔI14 rats. Indeed, these pro-inflammation genes started to be expressed with a high level at week 16 in LepΔI14/ΔI14 rats (Figure 4(a)). We also performed qRT-PCR to confirm the upregulation of these genes including Ccl2, Tnfa, and Il6 at week 16 (Figure 4(b)). Serum IL1β level was elevated at week 16 and remained at a steady level afterwards (Figure 4(c)). It is known that inflammatory processes are largely regulated by conserved signaling pathways through the phosphorylation of several key molecules such as c-Jun N-terminal kinase (JNK) and nuclear factor-κB (NF-κB).19,20 So, we examined the expression and post-translational modification of these molecules at different time points by Western blot (Figure 4(d) and (e)). We found that the activated (phosphorylated) form of JNK (Figure 4(d)) was markedly higher after week 16 in LepΔI14/ΔI14 rats compared to the WT controls. The elevation of phosphorylated P65 was observed as early as week 8 (Figure 4(e)), which is in line with the fact that NF-κB mediated signaling is required for the activation of pro-inflammation gene expression.
Figure 4.
The inflammation related genes and pathways during NASH progression in LepΔI14/ΔI14 rats. (a) Heatmap of the relative gene expression related to inflammation in LepΔI14/ΔI14 rats. log2 (foldchange) is shown as normalized gene expression. (b) qRT-PCR verification of the expression of inflammation related genes (n = 3) such as Il17rb, Tnfa, Ccl2, Icam1, Ccl3, Il1r1, Cxcl1, and Il6. Shown is the relative expression (LepΔI14/ΔI14 rats vs. WT controls) using Actb as internal control. Data are presented as mean ± SD. (c) Serum IL1β levels at different time points by ELISA (n = 3). Data are presented as mean ± SD. ***P < 0.001, ****P < 0.0001. (d) and (e) Western blot showing the levels of phosphorylated JNK (d) and p65 (e) at different time points (n = 3). (A color version of this figure is available in the online journal.)
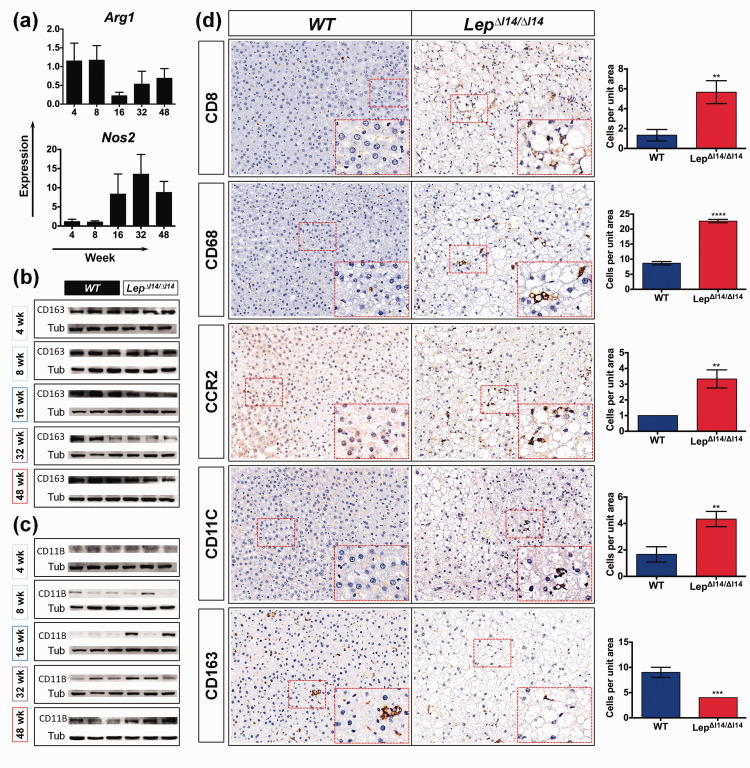
Macrophage polarization of pro-inflammatory M1 (classically activated macrophages) and anti-inflammatory M2 (alternatively activated macrophages) contributes to liver inflammation and injury.21 Interestingly, we found that the M2 related gene Arg1 was downregulated in the livers of LepΔI14/ΔI14 rats at week 16, while M1 related gene Inos (Nos2) was upregulated, which was validated by qRT-PCR (Figure 5(a)). At the protein level, M2 marker CD163 was reduced at week 16 (Figure 5(b)), while M1 marker CD11B22,23 was increased (Figure 5(c)). Since inflammation in the liver involves infiltration of distinct types of immune cells,24,25 this suggests that NASH progression in LepΔI14/ΔI14 rats also involves immune cell infiltration. Indeed, we observed an increase of CD8+ T cells and CD68+ hepatic macrophages (Figure 5(d)) in addition to MPO+ neutrophil infiltration (Figure 1(e)). M2 to M1 polarization was evidenced by the results that the number of inflammatory monocyte-derived macrophages (CCR2+) and M1 macrophages/dendritic cells (CD11C+) were significantly higher in the livers of LepΔI14/ΔI14 rats after week 16, while that of M2 macrophages (CD163+) was less than the controls (Figure 5(d), Figure S5). We also found immune cell infiltration from week 4 to week 48 (Figure S5). As stated above, such phenotypes were not observed in Leptin-deficient mice raised with the same conditions (Figure S2). Taken together, hepatic inflammation in LepΔI14/ΔI14 rats is evoked but restricted at a certain level after postnatal week 16, which is associated with altered gene expression, elevated inflammatory signaling pathways, and immune cell infiltration.
Figure 5.
Immune cell infiltration during NASH progression in LepΔI14/ΔI14 rats. (a) Dynamic changes (relative expression (LepΔI14/ΔI14 rats vs. WT controls)) of Arg1 and Nos2 expression verified by qRT-PCR (n = 3). (b) and (c) Western blot showing the expression of CD163 (b) and CD11B (c) (n = 3). (d) Representative images of IHC using antibodies against CD8, CD68, CCR2, CD11C, and CD163 in liver sections of week-16 LepΔI14/ΔI14 rats compared to WT controls. Right panel shows the statistics of immune cell infiltration (n = 3). Data are presented as mean ± SD. **P < 0.01, ***P < 0.001, ****P < 0.0001. (A color version of this figure is available in the online journal.)
LepΔI14/ΔI14 rats share many evolutionarily conserved DEGs and pathways with human patients and may serve as a useful model for drug evaluation
As described above, LepΔI14/ΔI14 rats develop NASH with a well-defined progression pattern. However, is the molecular etiology of NASH in LepΔI14/ΔI14 rats evolutionarily conserved with human beings? To answer this question, we compared the DEGs in LepΔI14/ΔI14 rats across stages to those in human beings during disease progression with healthy obese, NAFLD, and NASH from the previous study.15 Forty-two DEGs were significantly and concordantly regulated in both LepΔI14/ΔI14 rats at week 16 and human NASH (Figure 6(a)). In contrast, there were at most 18 DEGs shared between the mouse model (MCD8 mice26) and human NASH.15 Comparative analysis found that 20 pathways (Table S7) were significantly and concordantly regulated in both LepΔI14/ΔI14 rats at week 16 and human NASH (Figure 6(b)).
Figure 6.
NASH progression in LepΔI14/ΔI14 rats is evolutionarily conserved to human beings. (a) and (b) Counts of the significantly differentially regulated genes (a) and pathways (b) in LepΔI14/ΔI14 rats that are also significantly and concordantly regulated in healthy obese human, NAFLD, and NASH, respectively. (c) Heatmap showing expression levels of the drug target genes in clinical trials. log2 (foldchange) is shown as normalized gene expression. Blue squares indicate antagonist targets; red circles indicate agonist targets; check marks indicate that gene expression is correlated with the targeted drug effect. (A color version of this figure is available in the online journal.)
Many drug candidates for treatment of NASH are still in clinical trials.27,28 So, we asked if LepΔI14/ΔI14 rats could be useful to evaluate the efficacy of drug candidates. We generated a list of hepatic genes27,28 that are known to be targeted by either agonist or antagonist drugs. Indeed, we found that Fxr (Nr1h4) and Pparα/δ/γ were downregulated at many timepoints in Lep ΔI14/ΔI14 rats. Similarly, Scd1, Acc (Acaca), Aoc3, Casp3, Fgf21,and Galectin3 (Lgals3) were upregulated in mutant rats that would be useful to evaluate antagonist drugs (Figure 6(c)).27–29 Thus, pharmacologic agents targeting the list of dys-regulated genes in Figure 6(c) can be tested for drug efficacy in LepΔI14/ΔI14 rats.
Discussion
For the past several decades, many animal models have been created to study NASH.6–9 Many NASH animal models are based on the ‘‘two hit’’ model30 or “the multiple parallel hits hypothesis”.31 Dietary induction animal models are widely used; however, human beings rarely consume such experimental diets, suggesting a limitation in the use of dietary induction models for NASH.26 In mice, chronic liver injury in ob/ob mice can lead to development of the NASH phenotypes such as liver fibrosis,13 even though ob/ob mice themselves cannot spontaneously exhibit NASH phenotypes with regular diet. This supports the “two hit” hypothesis.
In the present study, we discovered LepΔI14/ΔI14 rats as a novel animal model to study human NASH. We found NASH features in LepΔI14/ΔI14 rats including inflammation and immune cell infiltration when LepΔI14/ΔI14 rats age over 16 weeks postnatally (Figure S6). Analysis of hepatic transcriptome profiles indicate that NASH in LepΔI14/ΔI14 rats progresses by multiple hits in a stepwise manner. As known, Leptin deficiency causes excess food intake. At postnatal week 8, TAG accumulation in the liver triggers simple steatosis (NAFL), which is evidenced by elevated expression of the genes encoding rate-limiting enzymes in lipid metabolism. Then, the pressure of excess nutrition alters the gene expression related to pathways such as insulin resistance, inflammation, ROS and ER stress, which induces NASH phenotypes at postnatal week 16. Accompanied with NASH progression, JNK and NF-κB pathways are activated by protein phosphorylation. Meanwhile, distinct immune cell infiltration appears to promote inflammation in the liver. For example, CD8+ T cells, DC, and neutrophils are increased. Macrophages in the liver undergo polarization from M2 to M1 as well. It should be pointed out that our work focused on hepatic gene expression. We perceive that many other tissues and organs must be involved in hepatic phenotypes of NASH,32,33 and this should be further addressed in the future.
Nowadays, drug development usually targets specific molecules. Although there are still no approved therapies for NASH, many drug candidates are in clinical trials and target important pathways related to metabolism, inflammation, fibrosis, etc. Recently, obeticholic acid, a farnesoid X receptor agonist, achieved promising results in a phase 3 trial.34 Other phase 3 trial drugs include Elafibranor (a peroxisome proliferator activated receptor-α/δ dual agonist) and Selonsertib (an ASK-1 inhibitor). Interestingly, Pparα/δ was downregulated in LepΔI14/ΔI14 rats (Figure 6(c)). Of note, Scd1, Acc (Acaca), Casp3, and Galectin3 (Lgals3) were upregulated in LepΔI14/ΔI14 rats, and their antagonists are in phase 2 trials (Figure 6(c)). We suggest that LepΔI14/ΔI14 rats will be useful for screening potential biomarkers and target genes of NASH. Given that rats are widely used in pharmacology and toxicology,35 our characterization of NASH in LepΔI14/ΔI14 rats will greatly facilitate their usage in evaluating drug efficacy during NASH drug development.
Supplemental Material
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220976530 for Characterizing disease progression of nonalcoholic steatohepatitis in Leptin-deficient rats by integrated transcriptome analysis by Ping Lu, Guang Yang, Lichun Jiang, Wen He, Wanwan Wu, Lingbin Qi, Shijun Shen, Junhua Rao, Peng Zhang, Zhigang Xue, Cizhong Jiang, Guoping Fan and Xianmin Zhu in Experimental Biology and Medicine
Supplemental material, sj-xlsx-2-ebm-10.1177_1535370220976530 for Characterizing disease progression of nonalcoholic steatohepatitis in Leptin-deficient rats by integrated transcriptome analysis by Ping Lu, Guang Yang, Lichun Jiang, Wen He, Wanwan Wu, Lingbin Qi, Shijun Shen, Junhua Rao, Peng Zhang, Zhigang Xue, Cizhong Jiang, Guoping Fan and Xianmin Zhu in Experimental Biology and Medicine
Supplemental material, sj-xlsx-3-ebm-10.1177_1535370220976530 for Characterizing disease progression of nonalcoholic steatohepatitis in Leptin-deficient rats by integrated transcriptome analysis by Ping Lu, Guang Yang, Lichun Jiang, Wen He, Wanwan Wu, Lingbin Qi, Shijun Shen, Junhua Rao, Peng Zhang, Zhigang Xue, Cizhong Jiang, Guoping Fan and Xianmin Zhu in Experimental Biology and Medicine
Supplemental material, sj-xlsx-4-ebm-10.1177_1535370220976530 for Characterizing disease progression of nonalcoholic steatohepatitis in Leptin-deficient rats by integrated transcriptome analysis by Ping Lu, Guang Yang, Lichun Jiang, Wen He, Wanwan Wu, Lingbin Qi, Shijun Shen, Junhua Rao, Peng Zhang, Zhigang Xue, Cizhong Jiang, Guoping Fan and Xianmin Zhu in Experimental Biology and Medicine
ACKNOWLEDGMENTS
The authors thank Dr Wei Wang from Biomedical Big Data Platform of SIAIS, ShanghaiTech University for assisting bioinformatics analysis and helpful discussions. The authors thank Dr Felice Elefant at Drexel University for revising the text.
AUTHORS’ CONTRIBUTIONS: XZ, CJ, and GF designed research; PL, HW, WW, LQ, and JR performed experiments; GY, LJ, and SS did bioinformatic analysis; ZX and PZ coordinated project; PL, XZ, and GF wrote the article. All authors approved the final version of the manuscript for its submission.
DECLARATION OF CONFLICTING INTERESTS: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
FUNDING: The work was supported by National Program on Key Basic Research Project (973 Program 2015CB964702, 2015CB964601); National Natural Science Foundation of China (81570521, Key Program 81430026); Experimental Animal Research Fund, and Science and Technology Commission of Shanghai Municipality (15140903900).
ORCID iD: Guoping Fan https://orcid.org/0000-0001-5235-6410
SupplementAL MATERIAL: Supplemental material for this article is available online.
References
- 1.Younossi ZM, Koenig AB, Abdelatif D, Fazel Y, Henry L, Wymer M. Global epidemiology of nonalcoholic fatty liver disease-meta-analytic assessment of prevalence, incidence, and outcomes. Hepatology 2016; 64:73–84 [DOI] [PubMed] [Google Scholar]
- 2.Chalasani N, Younossi Z, Lavine JE, Diehl AM, Brunt EM, Cusi K, Charlton M, Sanyal AJ. The diagnosis and management of non-alcoholic fatty liver disease: practice guideline by the American Association for the Study of Liver Diseases, American College of Gastroenterology, and the American Gastroenterological Association. Hepatology 2012; 55:2005–23 [DOI] [PubMed] [Google Scholar]
- 3.Anstee QM, Targher G, Day CP. Progression of NAFLD to diabetes mellitus, cardiovascular disease or cirrhosis. Nat Rev Gastroenterol Hepatol 2013; 10:330–44 [DOI] [PubMed] [Google Scholar]
- 4.Younossi ZM, Loomba R, Anstee QM, Rinella ME, Bugianesi E, Marchesini G, Neuschwander-Tetri BA, Serfaty L, Negro F, Caldwell SH, Ratziu V, Corey KE, Friedman SL, Abdelmalek MF, Harrison SA, Sanyal AJ, Lavine JE, Mathurin P, Charlton MR, Goodman ZD, Chalasani NP, Kowdley KV, George J, Lindor K. Diagnostic modalities for non-alcoholic fatty liver disease (NAFLD), non-alcoholic steatohepatitis (NASH) and associated fibrosis. Hepatology 2018; 68:349–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rowe IA. Too much medicine: overdiagnosis and overtreatment of non-alcoholic fatty liver disease. Lancet Gastroenterol Hepatol 2018; 3:66–72 [DOI] [PubMed] [Google Scholar]
- 6.Lau JK, Zhang X, Yu J. Animal models of non-alcoholic fatty liver disease: current perspectives and recent advances. J Pathol 2017; 241:36–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kohli R, Feldstein AE. NASH animal models: are we there yet? J Hepatol 2011; 55:941–3 [DOI] [PubMed] [Google Scholar]
- 8.Varela-Rey M, Embade N, Ariz U, Lu SC, Mato JM, Martinez-Chantar ML. Non-alcoholic steatohepatitis and animal models: understanding the human disease. Int J Biochem Cell Biol 2009; 41:969–76 [DOI] [PubMed] [Google Scholar]
- 9.Santhekadur PK, Kumar DP, Sanyal AJ. Preclinical models of non-alcoholic fatty liver disease. J Hepatol 2018; 68:230–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Y, Proenca R, Maffei M, Barone M, Leopold L, Friedman JM. Positional cloning of the mouse obese gene and its human homologue. Nature 1994; 372:425–32 [DOI] [PubMed] [Google Scholar]
- 11.Allison MB, Myers MG., Jr. 20 years of leptin: connecting leptin signaling to biological function. J Endocrinol 2014; 223:T25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naylor C, Petri WA., Jr. Leptin regulation of immune responses. Trends Mol Med 2016; 22:88–98 [DOI] [PubMed] [Google Scholar]
- 13.Leclercq IA, Farrell GC, Schriemer R, Robertson GR. Leptin is essential for the hepatic fibrogenic response to chronic liver injury. J Hepatol 2002; 37:206–13 [DOI] [PubMed] [Google Scholar]
- 14.Xu S, Zhu X, Li H, Hu Y, Zhou J, He D, Feng Y, Lu L, Du G, Hu Y, Liu T, Wang Z, Ding G, Chen J, Gao S, Wu F, Xue Z, Li Y, Fan G. The 14th ile residue is essential for leptin function in regulating energy homeostasis in rat. Sci Rep 2016; 6:28508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teufel A, Itzel T, Erhart W, Brosch M, Wang XY, Kim YO, von Schonfels W, Herrmann A, Bruckner S, Stickel F, Dufour JF, Chavakis T, Hellerbrand C, Spang R, Maass T, Becker T, Schreiber S, Schafmayer C, Schuppan D, Hampe J. Comparison of gene expression patterns between mouse models of nonalcoholic fatty liver disease and liver tissues from patients. Gastroenterology 2016; 151:513–25 e0 [DOI] [PubMed] [Google Scholar]
- 16.Kleiner DE, Brunt EM, Van Natta M, Behling C, Contos MJ, Cummings OW, Ferrell LD, Liu YC, Torbenson MS, Unalp-Arida A, Yeh M, McCullough AJ, Sanyal AJ. Nonalcoholic steatohepatitis clinical research N. design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology 2005; 41:1313–21 [DOI] [PubMed] [Google Scholar]
- 17.Murphy SK, Yang H, Moylan CA, Pang H, Dellinger A, Abdelmalek MF, Garrett ME, Ashley-Koch A, Suzuki A, Tillmann HL, Hauser MA, Diehl AM. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology 2013; 145:1076–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moylan CA, Pang H, Dellinger A, Suzuki A, Garrett ME, Guy CD, Murphy SK, Ashley-Koch AE, Choi SS, Michelotti GA, Hampton DD, Chen Y, Tillmann HL, Hauser MA, Abdelmalek MF, Diehl AM. Hepatic gene expression profiles differentiate presymptomatic patients with mild versus severe nonalcoholic fatty liver disease. Hepatology 2014; 59:471–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hotamisligil GS. Inflammation, metaflammation and immunometabolic disorders. Nature 2017; 542:177–85 [DOI] [PubMed] [Google Scholar]
- 20.Cai D, Yuan M, Frantz DF, Melendez PA, Hansen L, Lee J, Shoelson SE. Local and systemic insulin resistance resulting from hepatic activation of IKK-beta and NF-kappaB. Nat Med 2005; 11:183–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tacke F, Zimmermann HW. Macrophage heterogeneity in liver injury and fibrosis. J Hepatol 2014; 60:1090–6 [DOI] [PubMed] [Google Scholar]
- 22.Kinoshita M, Uchida T, Sato A, Nakashima M, Nakashima H, Shono S, Habu Y, Miyazaki H, Hiroi S, Seki S. Characterization of two F4/80-positive Kupffer cell subsets by their function and phenotype in mice. J Hepatol 2010; 53:903–10 [DOI] [PubMed] [Google Scholar]
- 23.McGettigan B, McMahan R, Orlicky D, Burchill M, Danhorn T, Francis P, Cheng LL, Golden-Mason L, Jakubzick C, Rosen H. Dietary lipids differentially shape NASH progression and the transcriptome of Kupffer cells and infiltrating macrophages. Hepatology 2019; 70:67–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heymann F, Tacke F. Immunology in the liver–from homeostasis to disease. Nat Rev Gastroenterol Hepatol 2016; 13:88–110 [DOI] [PubMed] [Google Scholar]
- 25.Gadd VL, Skoien R, Powell EE, Fagan KJ, Winterford C, Horsfall L, Irvine K, Clouston AD. The portal inflammatory infiltrate and ductular reaction in human nonalcoholic fatty liver disease. Hepatology 2014; 59:1393–405 [DOI] [PubMed] [Google Scholar]
- 26.Rinella ME, Green RM. The methionine-choline deficient dietary model of steatohepatitis does not exhibit insulin resistance. J Hepatol 2004; 40:47–51 [DOI] [PubMed] [Google Scholar]
- 27.Friedman SL, Neuschwander-Tetri BA, Rinella M, Sanyal AJ. Mechanisms of NAFLD development and therapeutic strategies. Nat Med 2018; 24:908–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Konerman MA, Jones JC, Harrison SA. Pharmacotherapy for NASH: current and emerging. J Hepatol 2018; 68:362–75 [DOI] [PubMed] [Google Scholar]
- 29.Solon-Biet SM, Cogger VC, Pulpitel T, Heblinski M, Wahl D, McMahon AC, Warren A, Durrant-Whyte J, Walters KA, Krycer JR, Ponton F, Gokarn R, Wali JA, Ruohonen K, Conigrave AD, James DE, Raubenheimer D, Morrison CD, Le Couteur DG, Simpson SJ. Defining the nutritional and metabolic context of FGF21 using the geometric framework. Cell Metab 2016; 24:555–65 [DOI] [PubMed] [Google Scholar]
- 30.Day CP, James OF. Steatohepatitis: a tale of two "hits"? Gastroenterology 1998; 114:842–5 [DOI] [PubMed] [Google Scholar]
- 31.Tilg H, Moschen AR. Evolution of inflammation in nonalcoholic fatty liver disease: the multiple parallel hits hypothesis. Hepatology 2010; 52:1836–46 [DOI] [PubMed] [Google Scholar]
- 32.Byrne CD, Targher G. NAFLD: a multisystem disease. J Hepatol 2015; 62:S47–64 [DOI] [PubMed] [Google Scholar]
- 33.Gehrke N, Schattenberg JM. Metabolic inflammation – a role for hepatic inflammatory pathways as drivers of comorbidities in nonalcoholic fatty liver disease? Gastroenterology 2020; 158:1929–47 e6 [DOI] [PubMed] [Google Scholar]
- 34.Younossi ZM, Ratziu V, Loomba R, Rinella M, Anstee QM, Goodman Z, Bedossa P, Geier A, Beckebaum S, Newsome PN, Sheridan D, Sheikh MY, Trotter J, Knapple W, Lawitz E, Abdelmalek MF, Kowdley KV, Montano-Loza AJ, Boursier J, Mathurin P, Bugianesi E, Mazzella G, Olveira A, Cortez-Pinto H, Graupera I, Orr D, Gluud LL, Dufour JF, Shapiro D, Campagna J, Zaru L, MacConell L, Shringarpure R, Harrison S, Sanyal AJ, Investigators RS. Obeticholic acid for the treatment of non-alcoholic steatohepatitis: interim analysis from a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 2019; 394:2184–96 [DOI] [PubMed] [Google Scholar]
- 35.Jacob HJ. Functional genomics and rat models. Genome Res 1999; 9:1013–6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-pdf-1-ebm-10.1177_1535370220976530 for Characterizing disease progression of nonalcoholic steatohepatitis in Leptin-deficient rats by integrated transcriptome analysis by Ping Lu, Guang Yang, Lichun Jiang, Wen He, Wanwan Wu, Lingbin Qi, Shijun Shen, Junhua Rao, Peng Zhang, Zhigang Xue, Cizhong Jiang, Guoping Fan and Xianmin Zhu in Experimental Biology and Medicine
Supplemental material, sj-xlsx-2-ebm-10.1177_1535370220976530 for Characterizing disease progression of nonalcoholic steatohepatitis in Leptin-deficient rats by integrated transcriptome analysis by Ping Lu, Guang Yang, Lichun Jiang, Wen He, Wanwan Wu, Lingbin Qi, Shijun Shen, Junhua Rao, Peng Zhang, Zhigang Xue, Cizhong Jiang, Guoping Fan and Xianmin Zhu in Experimental Biology and Medicine
Supplemental material, sj-xlsx-3-ebm-10.1177_1535370220976530 for Characterizing disease progression of nonalcoholic steatohepatitis in Leptin-deficient rats by integrated transcriptome analysis by Ping Lu, Guang Yang, Lichun Jiang, Wen He, Wanwan Wu, Lingbin Qi, Shijun Shen, Junhua Rao, Peng Zhang, Zhigang Xue, Cizhong Jiang, Guoping Fan and Xianmin Zhu in Experimental Biology and Medicine
Supplemental material, sj-xlsx-4-ebm-10.1177_1535370220976530 for Characterizing disease progression of nonalcoholic steatohepatitis in Leptin-deficient rats by integrated transcriptome analysis by Ping Lu, Guang Yang, Lichun Jiang, Wen He, Wanwan Wu, Lingbin Qi, Shijun Shen, Junhua Rao, Peng Zhang, Zhigang Xue, Cizhong Jiang, Guoping Fan and Xianmin Zhu in Experimental Biology and Medicine