Abstract
TAR DNA-binding protein 43 (TDP-43) inclusions are a pathological hallmark of frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS), including cases caused by G4C2 repeat expansions in the C9orf72 gene (c9FTD/ALS). Providing mechanistic insight into the link between C9orf72 mutations and TDP-43 pathology, we demonstrated that a glycine-arginine repeat protein [poly(GR)] translated from expanded G4C2 repeats was sufficient to promote aggregation of endogenous TDP-43. In particular, toxic poly(GR) proteins mediated sequestration of full-length TDP-43 in an RNA-independent manner to induce cytoplasmic TDP-43 inclusion formation. Moreover, in GFP-(GR)200 mice, poly(GR) caused the mislocalization of nucleocytoplasmic transport factors and nuclear pore complex proteins. These mislocalization events resulted in the aberrant accumulation of endogenous TDP-43 in the cytoplasm where it co-aggregated with poly(GR). Last, we demonstrated that treating G4C2 repeat–expressing mice with repeat-targeting antisense oligonucleotides lowered poly(GR) burden, which was accompanied by reduced TDP-43 pathology and neurodegeneration, including lowering of plasma neurofilament light (NFL) concentration. These results contribute to clarification of the mechanism by which poly(GR) drives TDP-43 proteinopathy, confirm that G4C2-targeted therapeutics reduce TDP-43 pathology in vivo, and demonstrate that alterations in plasma NFL provide insight into the therapeutic efficacy of disease-modifying treatments.
INTRODUCTION
Frontotemporal dementia (FTD) is characterized clinically by changes in personality, behavior, and/or language, whereas amyotrophic lateral sclerosis (ALS) is characterized by motor neuron signs. Nevertheless, these two fatal neurodegenerative diseases share pathologic features and genetic causes, and frequently co-occur within the same individuals. FTD and ALS exist on the same disease spectrum and are caused by common pathogenic mechanism(s). A hallmark feature of the vast majority of ALS cases and about half of FTD cases is TAR DNA-binding protein 43 (TDP-43) pathology. TDP-43, a predominantly nuclear DNA/RNA-binding protein with a prion-like domain, plays many roles in RNA metabolism. In patients with FTD/ALS-TDP-43, however, TDP-43 forms cytoplasmic aggregates, which result in its nuclear depletion (1, 2). The underlying factors that initiate these events are unknown, but the discovery of a G4C2 repeat expansion in chromosome 9 open reading frame 72 (C9orf72) as the most common genetic cause of FTD and ALS (c9FTD/ALS) (3, 4) has provided important insight into potential mechanisms that drive TDP-43 pathology.
In addition to the aggregation of TDP-43, pathological features uniquely derived from the C9orf72 repeat expansion are observed in c9FTD/ALS. These include C9orf72 haploinsufficiency, as well as the accumulation of nuclear RNA foci composed of sense G4C2 and antisense G2C4 repeat transcripts (3, 5, 6) and dipeptide repeat (DPR) proteins [poly(GA), poly(GP), poly(GR), poly(PR), or poly(PA)] translated from these transcripts through unconventional means (7–11). Several lines of evidence suggest that TDP-43 pathology in c9FTD/ALS results from repeat expansion products rather than loss of C9orf72. For instance, knockout of C9orf72 in mice does not lead to TDP-43 pathology, neurodegeneration, or motor deficits (12). In contrast, mice that express G4C2 repeat expansions not only develop RNA foci and express DPR proteins but also exhibit phosphorylated and aggregated endogenous TDP-43 (13–16), indicating that the TDP-43 pathology is likely triggered by repeat RNA and/or DPR proteins produced from the expanded G4C2 repeats. Repeat expansion products, especially arginine-rich poly(GR) and poly(PR) proteins, have been implicated in causing nucleocytoplasmic transport defects (17–22) and impairing stress granule (SG) dynamics (13, 23–26), two pathomechanisms linked to TDP-43 mislocalization and aggregation, in c9FTD/ALS. For instance, poly(GR) immunoprecipitates with TDP-43, nucleocytoplasmic transport factors, and SG-associated proteins (26, 27). These findings, along with evidence that essential protein components and key regulators of SG assembly, such as ataxin-2 and T cell intracellular antigen-1 (TIA-1), colocalize with poly(GR) and phosphorylated TDP-43 (pTDP-43) in (G4C2)149 mice (13), suggest that poly(GR) plays a key role in inducing TDP-43 pathology through the disruption of nucleocytoplasmic transport and SG biology. That poly(GR) colocalizes with TDP-43 inclusions in the motor cortex of patients with c9ALS supports this notion (28). Nonetheless, the mechanistic links between poly(GR) and TDP-43 remain unclear. Here, we used pure protein biochemistry, cell and animal models, as well as human postmortem brain tissue to investigate the relationship between poly(GR) and TDP-43 proteinopathy.
RESULTS
Poly(GR) accelerates and enhances TDP-43 aggregation
To investigate the relationship between poly(GR) and TDP-43, we first assessed their behavior at the pure protein level. Thus, we incubated purified recombinant maltose-binding protein (MBP)–tagged TDP-43 with or without (GR)20 (a protein with 20 GR repeats) or (GA)20 (a protein with 20 GA repeats). When tagged with MBP, wild-type TDP-43 (TDP-43WT) remained soluble in the absence or presence of (GR)20 or (GA)20 (Fig. 1A). However, after selective removal of the MBP tag by tobacco etch virus (TEV) protease, TDP-43WT formed aggregates in a time-dependent manner (Fig. 1A). Sedimentation analysis at the end point of the turbidity measurements confirmed that the increased absorbance in our aggregation assay is due to the formation of TDP-43WT aggregates (fig. S1A). Upon near completion of MBP cleavage, TDP-43WT in the absence or presence of (GR)20 or (GA)20 fully sedimented into the pellet fraction, whereas free MBP remained soluble (fig. S1A). Of note, co-incubation with (GR)20, but not (GA)20, significantly accelerated and enhanced TDP-43WT aggregation (P < 0.0001), although (GR)20 and (GA)20 alone did not aggregate (Fig. 1, A and B). More specifically, (GR)20 elicited the rapid formation of visible, solid-like TDP-43 aggregates at times where no TDP-43 aggregates formed in the presence of buffer or (GA)20 (Fig. 1, A and C). Transmission electron microscopy (TEM) revealed that whereas uncleaved TDP-43WT showed no aggregate formation in the absence or presence of (GR)20 or (GA)20 (fig. S1B), (GR)20 induced the formation of very dense TDP-43WT aggregates upon MBP cleavage (Fig. 1D). In contrast, (GA)20 had no obvious effect on TDP-43, which displayed a typical granulo-filamentous morphology (Fig. 1D). Quantitative analysis revealed that the total area and intensity of TDP-43 aggregates induced by (GR)20 are significantly higher compared to TDP-43 alone (P < 0.05) or with (GA)20 (P = 0.0027) (Fig. 1E). Collectively, our pure protein studies established that (GR)20 enhances and accelerates the aggregation of TDP-43 into dense condensates, whereas (GA)20 has no effect.
Fig. 1. Poly(GR) directly accelerates and enhances TDP-43 aggregation.
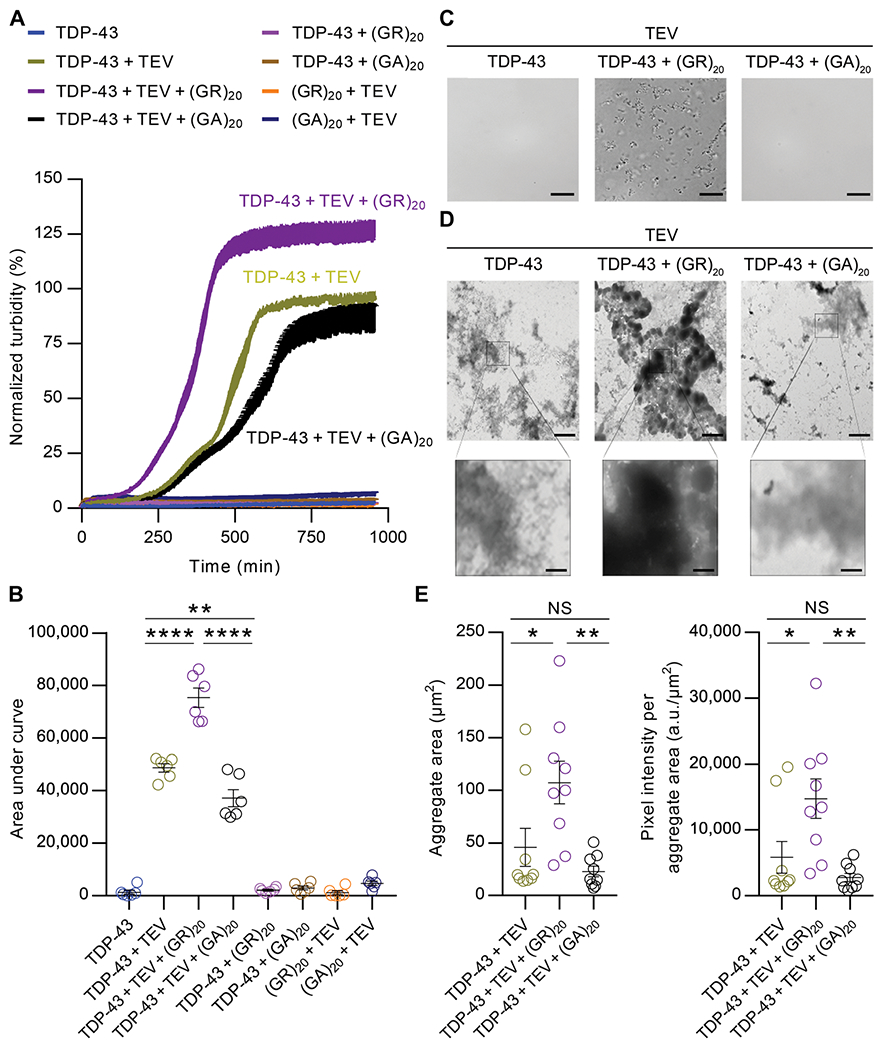
(A) TDP-43–MBP (5 μM) was incubated with buffer, 2 μM poly(GR), or 2 μM poly(GA) in the presence or absence of TEV protease (1 μg/ml). Aggregation was assessed by turbidity measured at an absorbance of 395 nm. Values are normalized mean ±SEM (n = 6). (B) Quantification of the area under the curve in turbidity for each condition in the in vitro aggregation assay (n = 6). (C) TDP-43 aggregation was initiated as described above, with the addition of TEV protease (10 μg/ml) with or without co-incubation with poly(GR) or poly(GA) for 30 min. Aggregation was visualized by differential interference contrast microscopy. Scale bars, 10 μm. (D) Representative electron micrographs of TDP-43 with or without poly(GR) or poly(GA). Samples were processed for EM at the end point of the turbidity assay. Scale bars, 2 μm (top) and 0.4 μm (bottom). (E) EM quantification of TDP-43 aggregate area and intensity per aggregate area with or without poly(GR) or poly(GA) (n = 9). Data shown are the mean ± SEM, one-way ANOVA, Tukey’s post hoc analysis. In (B), **P = 0.0027 and ****P < 0.0001. In (E), * (left to right) P = 0.0300 and P = 0.0263, ** (left to right) P = 0.0027 and P = 0.0027, NS (not significant; left to right) P = 0.5707 and P = 0.6080. a.u., arbitrary units.
Poly(GR) recruits cytoplasmic TDP-43 and SG-associated proteins to inclusions
To investigate the effects of poly(GR) aggregates on TDP-43, we first examined the morphology of poly(GR) aggregates using immuno-electron microscopy (IEM). We observed gold labeling of poly(GR) antibody on both granular and filamentous materials within cytoplasmic aggregates (Fig. 2A). Following confirmation that poly(GR) forms filaments and granular deposits in transfected cells, human embryonic kidney (HEK) 293T cells expressing green fluorescent protein (GFP) or GFP-tagged (GR)100 were cotransfected with various Myc-tagged TDP-43 constructs. Consistent with our previous findings (29), we observed that cytoplasmic GFP-(GR)100 inclusions were immunopositive for the SG markers TIA-1, eukaryotic translation initiation factor 3η (eIF3η), and ataxin-2 (Fig. 2B and fig. S2A). TDP-43WT, however, was absent from poly(GR)/TIA-1-positive inclusions, instead remaining in the nucleus (Fig. 2, B and C). In contrast, TDP-43 with a mutated nuclear localization signal (TDP-43NLSm) was confined to the cytosol where it co-aggregated with poly(GR) and TIA-1 (Fig. 2, B and C). To confirm that co-aggregation was not merely caused by TDP-43NLSm overexpression, we examined the distribution and expression of TDP-43NLSm in the absence of poly(GR) (fig. S2BD). TDP-43NLSm remained diffusely distributed throughout the cytosol in GFP-transfected cells (Fig. 2C and fig. S2B), despite the fact that TDP-43NLSm expression was higher in GFP–expressing cells than in GFP-(GR)100–expressing cells (fig. S2, C and D) [likely because poly(GR) inhibits translation (26, 29)]. Unlike poly(GR), poly(GA) inclusions in GFP-(GA)100–expressing cells failed to sequester cytoplasmic TDP-43NLSm (fig. S2E). Together, these data indicate that both the cytoplasmic localization of TDP-43 and its specific recruitment to poly(GR) inclusions promote TDP-43 aggregation.
Fig. 2. Poly(GR) mediates sequestration of cytosolic full-length TDP-43 into the inclusions.
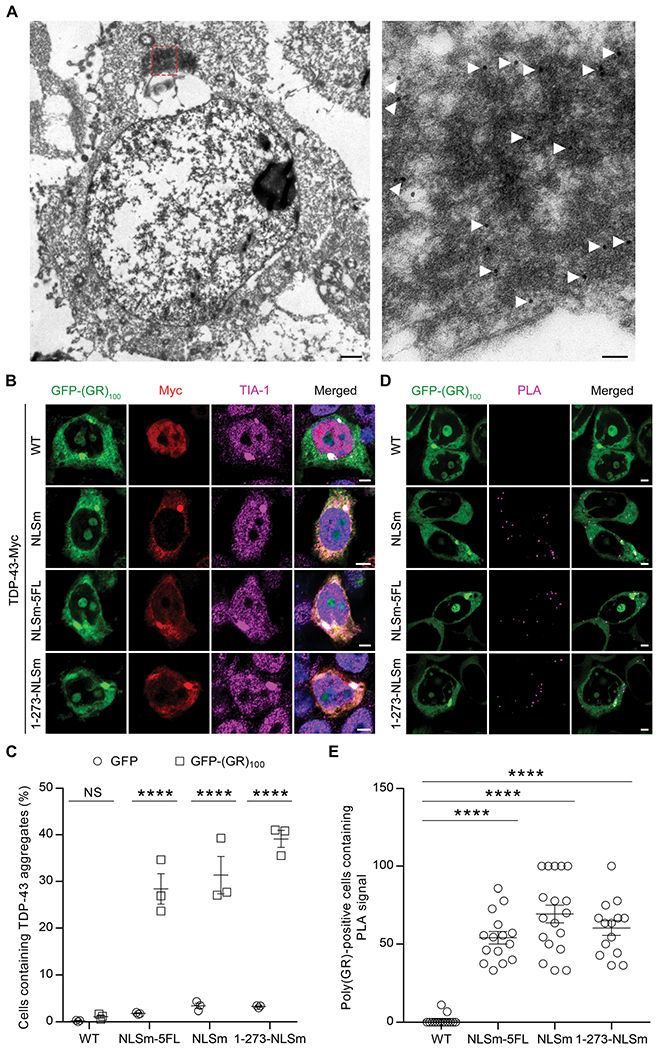
(A) IEM using an anti-poly(GR) antibody labeled with gold particles (18 nm) in HEK293T cells expressing GFP(GR)100. The selected region in the low-magnification image (left) is shown at high magnification (right). Scale bars, 1 μm (left) and 0.1 μm (right). (B) Triple-immunofluorescence staining for poly(GR), TDP-43, and TIA-1 in HEK293T cells expressing GFP-(GR)100 and various Myc-tagged TDP-43 species. Scale bars, 5 μm. (C) Quantification of the number of cytoplasmic TDP-43 aggregates in HEK293T cells co-expressing various Myc-tagged TDP-43 constructs with either GFP or GFP-(GR)100 (n = 3 independent experiments). (D) Proximity ligation assay (PLA) for GFP-(GR)100 and Myc-tagged TDP-43 species in HEK293T cells, with the PLA signal being indicative of their interaction. Scale bars, 5 μm. (E) Quantification of the PLA signal in HEK293T cells expressing GFP-(GR)100 and Myc-tagged TDP-43 species (n = 14 to 18 images). Data shown are the mean ± SEM. In (C), ****P < 0.0001 and NS P = 0.9959, two-way ANOVA, Tukey’s post hoc analysis. In (E), ****P < 0.0001, one-way ANOVA, Tukey’s post hoc analysis. WT, wild type.
Given that TDP-43 is an RNA-binding protein, we examined whether cytoplasmic TDP-43 recruitment to GFP-(GR)100 inclusions was RNA dependent by mutating five phenylalanine residues within the RNA-binding motifs of TDP-43NLSm to leucine residues (TDP-43NLSm-5FL). Although these mutations disrupt the RNA-binding ability of TDP-43 (30), TDP-43NLSm-5FL nonetheless colocalized to poly(GR) aggregates (Fig. 2, B and C). Next, because the C-terminal prion-like domain of TDP-43 is prone to aggregate (31, 32), we co-expressed TDP-43 C-terminal fragments (CTFs) or TDP-43NLSm lacking the C-terminal domain (TDP-431-273-NLSm) with GFP-(GR)100. Whereas TDP-431-273-NLSm maintained the ability to be recruited to poly(GR) inclusions (Fig. 2, B and C), TDP-43 CTF formed cytoplasmic aggregates devoid of poly(GR) (fig. S2F). To study the poly(GR)–TDP-43 interaction in more detail, we used a proximity ligation assay (PLA) in which a fluorescent PLA signal is indicative of GFP-(GR)100 binding to a given Myc-tagged TDP-43 species. Consistent with a lack of colocalization, no PLA signal was detected in cells expressing GFP-(GR)100 and TDP-43WT. In contrast, poly(GR) did interact with cytoplasmic TDP-43NLSm, TDP-43NLSm-5FL, and TDP-431-273-NLSm as revealed by the positive PLA signal within cytoplasmic poly(GR) inclusions (Fig. 2, D and E). Collectively, our results indicate that poly(GR) interacts with and recruits TDP-43 into inclusions, independent of RNA-binding ability or the aggregation-prone C-terminal prion-like domain of TDP-43.
Poly(GR) aggregates sequester endogenous TDP-43 and SG-resident proteins in vivo
Our cell culture studies indicate that poly(GR) inclusions sequester TDP-43NLSm but not TDP-43WT. This difference raises the possibility that the short experimental period of this cell culture system fails to meet the temporal requirement for poly(GR)-mediated recruitment of TDP-43WT. We therefore examined whether the extended time frame that in vivo studies permit would allow for TDP-43WT to be recruited to poly(GR) aggregates. We recently demonstrated that GFP-(GR)100 expression in the brains of mice causes neurodegeneration and translational repression through the binding of poly(GR) to ribosomal subunits and translation initiation factors (29). However, no inclusions of endogenous mouse TDP-43 were observed, which we speculate was due to the lack of GFP-(GR)100 aggregation in this model. To overcome this issue, we generated an adeno-associated viral (AAV) construct encoding 200 GR DPRs, which was injected into the lateral ventricles of postnatal day 0 mice. Although poly(GR) predominantly showed a diffuse intracellular distribution in the brains of these mice, poly(GR) aggregates were nonetheless observed in about 7.9 and 3.5% of poly(GR)-positive cells in 2-week-old and 3-month-old mice, respectively (Fig. 3, A and B, and fig. S3A). We detected a significant reduction in the number of diffuse (63.1% decrease, P < 0.0001) and aggregated (90.4% decrease, P = 0.0001) poly(GR)-positive cells in the cortex from 2 weeks to 3 months of age (Fig. 3B), likely attributed to toxicity of poly(GR) expression and subsequent neuronal loss. Ataxin-2 and TDP-43 aggregates were also observed in brains of GFP-(GR)200 mice, but not in GFP mice (Fig. 3, C and D, and fig. S3B). Furthermore, we found that poly(GR) aggregates, but not diffuse poly(GR), colocalized with ataxin-2, eIF3η, TDP-43, and pTDP-43 (Fig. 3, E to G). Of note, about 95.2 and 78.1% of poly(GR) inclusions in GFP-(GR)200 mice were positive for TDP-43 and pTDP-43, respectively (Fig. 3H and fig. S3C). Moreover, consistent with the nuclear depletion of TDP-43 in cells bearing cytoplasmic TDP-43 inclusions in patients with FTD and ALS, significant (P < 0.0001) nuclear depletion of TDP-43 was observed in cells containing poly(GR) aggregates compared to nontransduced and diffuse poly(GR)-positive cells (Fig. 3, B, F, and H). Quantitative analysis revealed that about 30.9% of cells containing TDP-43–positive inclusions display TDP-43 nuclear reduction (Fig. 3H), likely reflecting different stages of inclusion formation. We observed poly(GR) inclusions immunopositive for TDP-43 and eIF3η in c9FTD/ALS patient tissues (Fig. 3I and fig. S3D), and these were accompanied by varying degrees of nuclear TDP-43 depletion. Unlike poly(GR) inclusions in GFP-(GR)200 mice, poly(GA) inclusions formed in GFP-(GA)50 mice were negative for TDP-43 and eIF3η (Fig. 3J), which is consistent with the inability of poly(GA) alone to stimulate TDP-43 aggregation in vitro (Fig. 1) or in cultured cells (fig. S2C). Although poly(GA) is detected within TDP-43 inclusions in c9FTD/ALS postmortem brain tissue (fig. S3E), there is greater colocalization between TDP-43 and poly(GR) (fig. S3F). These results might suggest that poly(GA) colocalizes with TDP-43 in the presence of other G4C2-associated pathologies, whereas poly(GR) alone exhibits a unique ability to recruit TDP-43 to inclusions. Together, our data demonstrate that poly(GR) aggregation, in the absence of other G4C2-associated pathologies, is sufficient to promote the aggregation of endogenous, wild-type TDP-43 in vivo.
Fig. 3. Poly(GR) aggregation is sufficient to induce mislocalization and aggregation of endogenous TDP-43 in vivo.
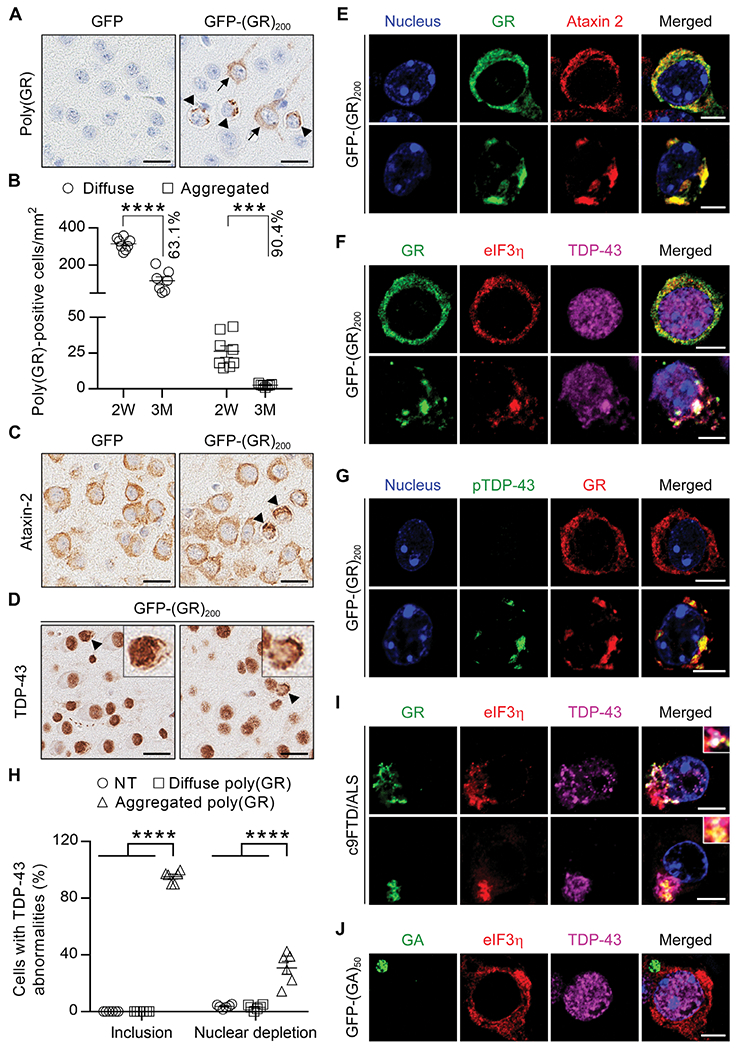
(A) Representative images of immunohistochemical analysis of poly(GR) in the cortex of 2-week-old GFP mice or GFP-(GR)200 mice (diffuse labeling noted by black arrows, aggregates indicated by black arrowheads). Scale bars, 20 μm. (B) Quantification of the number of poly(GR)-positive cells with either diffuse or aggregated poly(GR) in 2-week-old (n = 8) or 3-month-old (n = 7) GFP-(GR)200 mice. (C) Representative images of immunohistochemical analysis of ataxin-2 (inclusions indicated by black arrowheads) in the cortex of 2-week-old GFP or GFP-(GR)200 mice. (D) Representative images of immunohistochemical analysis of TDP-43 in the cortex of 2-week-old GFP-(GR)200 mice. Arrowheads in GFP-(GR)200 mice indicate cells with TDP-43–positive inclusions and either normal (left panel) or reduced (right panel) nuclear TDP-43. Scale bars, 20 μm. (E) Double-immunofluorescence staining for poly(GR) and ataxin-2 in the cortex of 2-week-old GFP-(GR)200 mice (n = 6). Scale bars, 5 μm. (F) Triple-immunofluorescence staining for eIF3η, TDP-43, and poly(GR) in the cortex of 2-week-old GFP-(GR)200 mice (n = 6). Scale bars, 5 μm. (G) Double-immunofluorescence staining for poly(GR) and pTDP-43 in the cortex of 2-week-old GFP-(GR)200 mice (n = 6). Scale bars, 5 μm. (H) Quantification of the percentage of cells that are either nontransduced (NT) cells or transduced cells with either diffuse or aggregated poly(GR) that contain TDP-43 inclusions and exhibit depleted nuclear TDP-43 in 2-week-old GFP-(GR)200 mice (n = 6). (I) Triple-immunofluorescence staining for poly(GR), eIF3η, and TDP-43 in the hippocampus of patients with c9FTD/ALS [case #1 (top), case #2 (bottom); see table S3 for patient information]. (J) Triple-immunofluorescence staining for poly(GA), eIF3η, and TDP-43 in the cortex of 3-month-old GFP-(GA)50 mice (n = 3). Scale bars, 5 μm. Data shown are the mean ± SEM. In (B), ***P = 0.0001 and ****P < 0.0001, two-tailed unpaired t test. In (H), **** P < 0.0001, one-way ANOVA, Tukey’s post hoc analysis.
Poly(GR) aggregates sequester nucleocytoplasmic transport factors and nuclear pore complex proteins in vivo
Because the cytoplasmic localization of TDP-43 is required for its sequestration to poly(GR) inclusions, we next determined how endogenous TDP-43 becomes mislocalized in GFP-(GR)200 mice. Given that poly(GR) disrupts nucleocytoplasmic transport in yeast, flies, and cultured cells (17, 18, 20), which could disrupt TDP-43 transport to and from the nucleus, we evaluated whether poly(GR) influences nucleocytoplasmic transport factors. We first examined importin α5 and karyopherin α2 (KPNA2), two nuclear import factors that mediate TDP-43 nuclear import (33). Co-immunofluorescence staining demonstrated that the colocalization of poly(GR) and importin α5 or KPNA2 was observed in both diffuse and aggregated poly(GR)-positive cells (Fig. 4A). However, compared to nontransduced cells, aggregated but not diffuse poly(GR) affected the distribution pattern of importin α5 and KPNA2 by recruiting these proteins in poly(GR) inclusions (Fig. 4, A and B). We next assessed various components of the nuclear pore complex (NPC), which consists of about 30 nucleoporins. Using an NPC antibody that detects four nucleoporins (NUP62, NUP153, NUP214, and NUP358), we observed that NPCs were homogeneously distributed throughout the nuclear envelope in nontransduced and diffuse poly(GR)-positive cells (Fig. 4C). In contrast, in poly(GR) inclusion–bearing cells, NPCs were irregularly distributed around the nuclear envelope and partially colocalized with poly(GR) aggregates, and this was similarly observed for the NPC components NUP98 and pore membrane protein of 121 kDa (POM121) (Fig. 4C and fig. S4). Quantitative analysis revealed that more than 90% of poly(GR) inclusion–bearing cells contained abnormal KPNA2 and NUP98 staining (Fig. 4, B and D). Last, we evaluated whether TDP-43 or eIF3η distribution was altered in poly(GR)-positive cells with NPC abnormalities. As anticipated, aggregated poly(GR) was associated with aberrant NPC distribution, loss of nuclear TDP-43, and recruitment of cytoplasmic TDP-43 and eIF3η to poly(GR) inclusions (Fig. 4, E to H). These results provide in vivo evidence that poly(GR) expression alone is sufficient to disturb nucleocytoplasmic transport factors and NPC proteins, contributing to the mislocalization of endogenous, wild-type TDP-43 and ultimately driving TDP-43 proteinopathy.
Fig. 4. Poly(GR) aggregates sequester nucleocytoplasmic transport factors and NPC proteins in vivo.
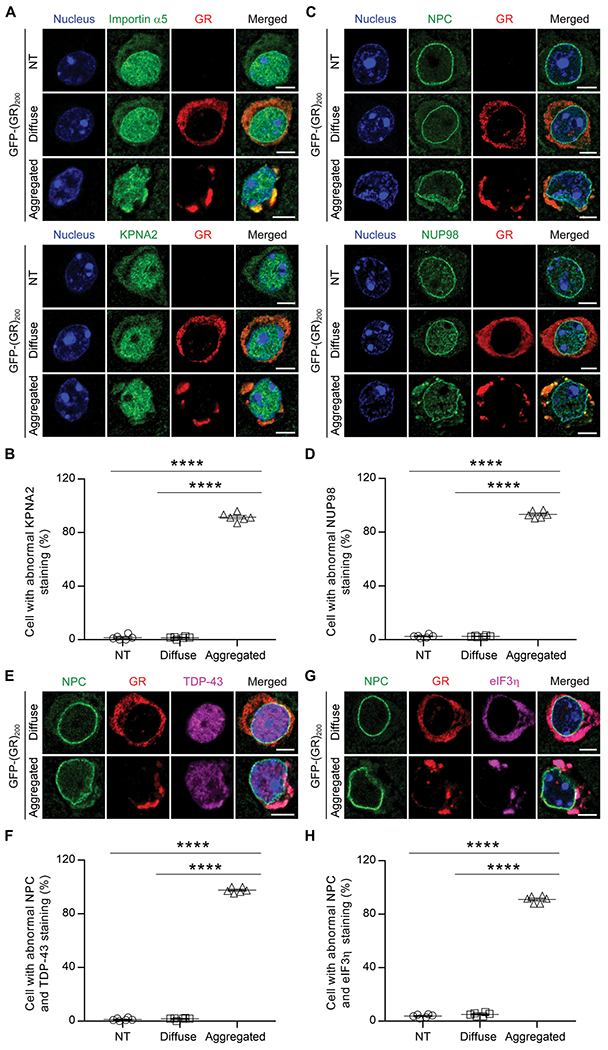
(A) Double-immunofluorescence staining for poly(GR) and importins in nontransduced (NT), diffuse, or aggregated poly(GR) cells, including importin α5 (top three panels) and karyopherin α2 (KNPA2; bottom three panels) in the cortex of 2-week-old GFP-(GR)200 mice (n = 6). Scale bars, 5 μm. (B) Quantification of the percentage of NT cells or of transduced cells with either diffuse or aggregated poly(GR) that contain abnormal KPNA2 in 2-week-old GFP-(GR)200 mice (n = 6). (C) Double-immunofluorescence staining for poly(GR) and several NPC proteins, including NPC (top three panels) and NUP98 (bottom three panels) in the cortex of 2-week-old GFP-(GR)200 mice (n = 6). Scale bars, 5 μm. (D) Quantification of the percentage of NT cells or of transduced cells with either diffuse or aggregated poly(GR) that contain abnormal NUP98 in 2-week-old GFP-(GR)200 mice (n = 6). (E) Triple-immunofluorescence staining for NPC, poly(GR), and TDP-43 in the cortex of GFP-(GR)200 mice (n = 6). Scale bars, 5 μm. (F) Quantification of the percentage of NT cells or of transduced cells with either diffuse or aggregated poly(GR) that contain abnormal NPC and TDP-43 in 2-week-old GFP-(GR)200 mice (n = 6). (G) Triple-immunofluorescence staining for NPC, poly(GR), and eIF3η in the cortex of 2-week-old GFP-(GR)200 mice (n = 6). Scale bars, 5 μm. (H) Quantification of the percentage of NT cells or of transduced cells with either diffuse or aggregated poly(GR) that contain abnormal NPC and eIF3η in 2-week-old GFP-(GR)200 mice (n = 6). Data shown are the mean ± SEM; ****P < 0.0001, one-way ANOVA, Tukey’s post hoc analysis.
A G4C2-targeted therapeutic mitigates poly(GR) and TDP-43 aggregation and rescues neurodegeneration in vivo
We previously reported that expressing expanded G4C2 repeats in the brain of mice leads to the accumulation of repeat-containing transcripts, DPR proteins, and pTDP-43 pathology (13, 14). Data from the present study strongly suggest that poly(GR) is directly responsible for inducing the TDP-43 pathology detected in (G4C2)66 and (G4C2)149 mice, and this role of poly(GR) is further supported by our previous observation that poly(GR) aggregates in (G4C2)149 mice are immunopositive for pTDP-43 and TIA-1 (13). Although we have shown that treating (G4C2)66 mice with antisense oligonucleotides targeting G4C2 repeats (c9ASO) decreases G4C2 repeat RNA and DPR protein expression (34), it remains unclear whether c9ASOs, which are currently being tested in clinical trials, will also alleviate pTDP-43 pathology. This issue is critical as it is likely imperative to combat TDP-43 proteinopathy and restore functional TDP-43 to the nucleus to rescue degenerating neurons. To investigate this issue, c9ASOs were injected into the brain of 3-month-old (G4C2)149 mice, which develop robust RNA foci and DPR protein pathology by this age (13). As anticipated, G4C2 repeat–containing RNA expression, assessed by both fluorescence in situ hybridization (FISH) and quantitative reverse transcription polymerase chain reaction (qRT-PCR), was reduced 3 months after treatment (Fig. 5A and fig. S5, A and B). To assess DPR burden, we used both immunohistochemistry of fixed brain tissue and immunoassays of cortical brain lysates, which confirmed that poly(GR) was reduced by c9ASO treatment, along with the other sense DPR proteins poly(GA) and poly(GP) (Fig. 5B and fig. S5C). Moreover, the number of inclusions immunopositive for pTDP-43 or ataxin-2 was also significantly reduced (Fig. 5, C to E; P < 0.0001 and P = 0.0407 for pTDP-43 and ataxin-2, respectively), validating that reduced poly(GR) and other G4C2-associated pathologies are associated with diminished TDP-43 pathology.
Fig. 5. A G4C2-targeting therapeutic reduces poly(GR) and alleviates TDP-43 aggregation and neurodegeneration in vivo.
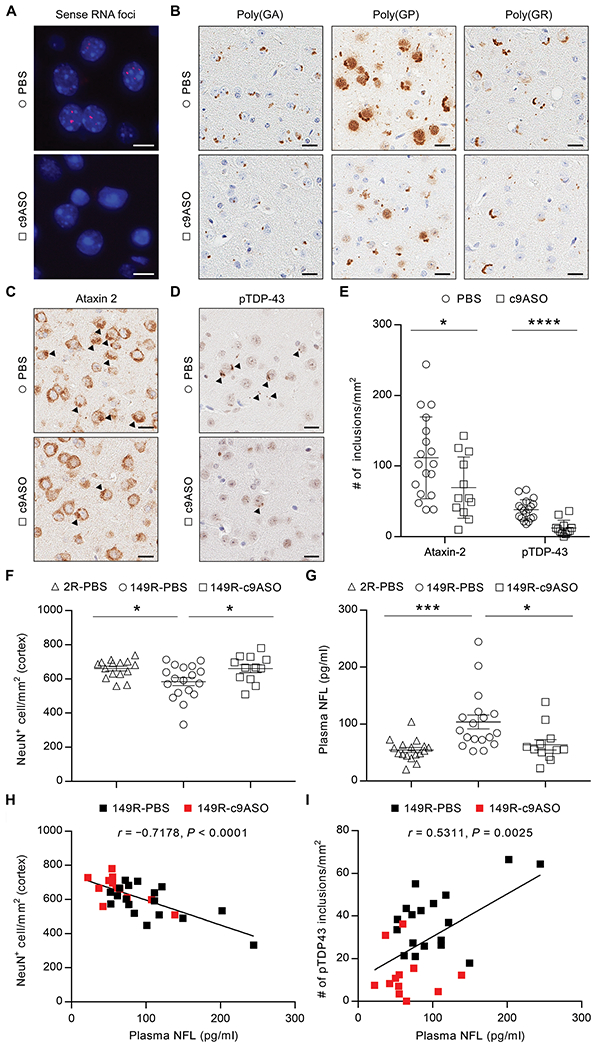
(A) Representative images of RNA FISH for the detection of sense RNA foci in (G4C2)149 mice treated with PBS or c9ASO. Scale bars, 10 μm. (B) Immunohistochemical labeling of poly(GA), poly(GP), and poly(GR) in PBS- or c9ASO-treated (G4C2)149 mice. Scale bars, 20 μm. (C and D) Representative images of immunohistochemical analysis of ataxin-2 (C) and pTDP-43 (D) in PBS- or c9ASO-treated (G4C2)149 mice, with inclusions indicated by black arrowheads. Scale bars, 20 μm. (E) Quantitative analysis of the number of ataxin-2 or pTDP-43 inclusions per square millimeter of cortex in (G4C2)149 mice treated with PBS (n = 18) or c9ASO (n = 12). (F) Quantitative analysis of the number of NeuN-positive neurons per square millimeter of cortex in (G4C2)149 mice treated with PBS (149R-PBS, n = 18) or c9ASO (149R-c9ASO, n = 12) relative to (G4C2)2 control mice treated with PBS (2R-PBS, n = 15). (G) Plasma neurofilament light (NFL) concentrations in mice measured using the Simoa HD-1 Analyzer (2R-PBS, n = 17; 149R-PBS, n = 18; 149R-c9ASO, n = 12). (H) The number of NeuN-positive neurons per square millimeter of cortex negatively correlated with plasma NFL concentrations in (G4C2)149 mice (PBS- and c9ASO-treated mice indicated by black and red squares, respectively). (I) The number of pTDP-43 inclusions per square millimeter of cortex positively correlated with plasma NFL concentrations in (G4C2)149 mice. Data represent the mean ± SEM. In (E), *P=0.0407 and ****P < 0.0001, unpaired two-tailed t test. In (F), * (left to right) P=0.0293 and P=0.0426, one-way ANOVA, Tukey’s post hoc analysis. In (G), *P=0.0166 and ***P=0.0009, one-way ANOVA, Tukey’s post hoc analysis.
To assess the neuroprotective effect of c9ASO-mediated alleviation of the abovementioned pathologies, we examined both neuronal number and amount of plasma neurofilament light (NFL), a biomarker of neuronal injury (35–37). c9ASO treatment prevented the reduction in NeuN-positive cortical neurons otherwise observed in (G4C2)149 mice (Fig. 5F). In addition, we observed increased plasma NFL concentrations in (G4C2)149 mice compared to control (G4C2)2 mice, which were attenuated in c9ASO-treated (G4C2)149 mice (Fig. 5G). To examine the pathological meaning of alterations in plasma NFL in the (G4C2)149 model, we evaluated the relationship between plasma NFL and neuronal counts, as well as TDP-43 pathology. Of note, plasma NFL negatively correlated with the number of NeuN-positive neurons (Fig. 5H) and positively correlated with pTDP-43 pathology (Fig. 5I) in (G4C2)149 mice, suggesting that plasma NFL concentrations are reflective of neuronal viability and TDP-43 burden in the brain. Together, these findings demonstrate that c9ASO treatment mitigates TDP-43 proteinopathy, which is associated with a reduction in neuronal loss and stabilization of plasma NFL concentrations in vivo.
DISCUSSION
In this study, we uncovered a key role for G4C2 repeat–derived poly(GR) in promoting TDP-43 proteinopathy in vitro and in vivo. Specifically, we established that poly(GR) directly accelerates and enhances TDP-43 aggregation. Moreover, poly(GR) sequesters cytoplasmic full-length TDP-43 through an RNA-independent mechanism. In doing so, poly(GR) induces the formation of dense TDP-43 aggregates in vitro. The formation of poly(GR) inclusions immunopositive for TDP-43 and SG-resident proteins in GFP-(GR)200 mice demonstrates that poly(GR) aggregation, in the absence of other G4C2-associated pathologies, is sufficient to induce endogenous TDP-43 aggregation. Combined with corroborating findings of inclusions containing both poly(GR) and TDP-43 in patients with c9FTD/ALS, these results provide in vivo validation in both mouse and human postmortem brain tissues that poly(GR) accumulation directly induces TDP-43 proteinopathy.
A key feature of TDP-43 pathology in humans is that TDP-43 inclusions are accompanied by the loss of nuclear TDP-43. Thus, our observation that a subset of neurons with cytoplasmic poly(GR)/TDP-43–positive inclusions exhibited reduced nuclear TDP-43 in GFP-(GR)200 mice increases pathological meaning. Given that importins and NPC proteins interact with poly(GR) (20, 26, 27), we asked whether the aberrant cytoplasmic distribution of endogenous TDP-43 in poly(GR)-positive cells might be caused by poly(GR)-induced nucleocytoplasmic transport defects. In support of this idea, we present evidence that several importins and nucleoporins are abnormally distributed in poly(GR)-positive cells and partially colocalize with poly(GR) inclusions in GFP-(GR)200 mice. These features were accompanied by both the loss of nuclear TDP-43 and its recruitment to cytoplasmic poly(GR) inclusions. These results support a temporal mechanism in which poly(GR) impairs nucleocytoplasmic transport by sequestering key nucleocytoplasmic transport factors and nucleoporins, which subsequently drives cytoplasmic accumulation and ultimately co-aggregation of TDP-43 with poly(GR) through protein-protein interactions.
Given our observations that poly(GR) alone is sufficient to induce endogenous TDP-43 mislocalization and aggregation, it is noteworthy that both TDP-43 and poly(GR) burden correlate with neurodegeneration in patients (28, 38, 39). Consistent with this notion, we observed an age-dependent loss of poly(GR)-positive cells in GFP-(GR)200 mice, suggesting that expression of poly(GR) is toxic to neurons. Moreover, the more marked reduction in aggregated poly(GR) compared to diffuse poly(GR) is likely due to the combination of poly(GR) and TDP-43 abnormalities in cells with poly(GR) inclusions. Therefore, therapeutics targeting poly(GR), TDP-43, or both may be needed to modulate neurotoxicity in c9FTD/ALS. Although the current study demonstrates that c9ASOs reduce TDP-43 pathology in an AAV model of c9FTD/ALS, future studies will be needed to explore the benefit of therapeutic strategies specifically targeting poly(GR) rather than all G4C2 repeat–associated pathologies. Although our results indicate that poly(GA) alone does not cause TDP-43 aggregation at either the pure protein level, in cultured cells, or in the mouse brain, it remains possible that additional products generated from the C9orf72 repeat expansion (RNA foci and other DPR proteins), either on their own or in combination, contribute to disease pathogenesis. TDP-43 has been shown to colocalize with poly(GA) and poly(GP) inclusions in patients with c9ALS, albeit less frequently than with poly(GR) (28). These findings are consistent with our observations in patients with c9FTD/ALS, suggesting that poly(GR) plays a more direct role in driving TDP-43 inclusion formation. In contrast to poly(GR), poly(GA)-induced TDP-43 pathology may require the presence of other C9orf72-associated pathology. It also bears mentioning that because poly(PR) shares similar biophysical properties and functions as poly(GR) (26), it too may also cause TDP-43 aggregation. However, poly(PR) inclusions are rare in patients with c9FTD/ALS (10, 40) and thus unlikely to be a major driver of TDP-43 pathology.
Although our findings reveal a new mechanism to explain TDP-43 pathology in poly(GR)-positive cells, it is important to note that there are other mechanisms underpinning TDP-43 proteinopathy. For example, our results indicate that the aggregation of pathological TDP-43 CTFs is independent of poly(GR) in cultured cells. Moreover, despite the abundance of TDP-43 pathology in the spinal cord of patients with c9ALS, poly(GR) aggregates in this region are rare (41). In addition, two recent studies provide insight into the underlying causes of TDP-43 pathology in sporadic ALS and FTD and/or poly(GR)-negative cells by demonstrating that cytoplasmic TDP-43 aggregation is mediated by aberrant phase transitions and occurs independently of SGs (42, 43). These data suggest that the observed recruitment of SG proteins to poly(GR) aggregates in GFP-(GR)200 mice occurs through their direct interaction with poly(GR) rather than by binding TDP-43. This mechanism is also consistent with the identification of the SG proteins RasGAP SH3 domain binding protein 1 (G3BP1) and ataxin-2 as poly(GR)-interacting proteins (26, 27), and with our finding that endogenous TIA-1 is recruited to poly(GR) inclusions in cultured cells expressing nuclear TDP-43WT. Confined to the nucleus, TDP-43WT remains absent from these poly(GR) aggregates. Last, the fact that blocking the RNA-binding ability of TDP-43 disrupts its interaction with ataxin-2 (44) and its localization to SGs (43), but does not prevent its sequestration to poly(GR)-induced aggregates immunopositive for SG proteins, provides compelling evidence that the recruitment of TDP-43 to poly(GR) inclusions is mediated by a direct interaction, rather than its co-aggregation, with cytoplasmic SGs.
Considering that c9ASOs targeting G4C2 repeat transcripts are currently being tested in clinical trials, our data showing that c9ASO treatment in (G4C2)149 mice mitigates poly(GR) burden, TDP-43 pathology, the aberrant SG response, and neurodegeneration are especially relevant. Moreover, we demonstrate that plasma NFL, which is abnormally elevated in (G4C2)149 mice and reduced with c9ASO treatment, also correlates with neuronal loss and TDP-43 pathology in (G4C2)149 mice. These results indicate that alterations in plasma NFL concentrations are both reflective of the severity of endogenous TDP-43 pathology in the (G4C2)149 model and responsive to treatment.
Along with the insights provided by our findings, it is important to acknowledge the limitations of the current study. Although we and others have observed substantial colocalization between TDP-43 and poly(GR) in patients with c9FTD/ALS using TDP-43 antibodies targeting the N-terminal region and phosphorylated serine-409/410 sites (28), additional studies using an array of antibodies targeting different epitopes of the TDP-43 protein in multiple brain regions from a large cohort of c9FTD/ALS patients are warranted. Next, we demonstrated the promising effects of c9ASO treatment in (G4C2)149 mice. However, as all G4C2 repeat–associated pathologies are decreased by c9ASO treatment, additional studies are needed to determine whether the protective effect is mediated by reductions in poly(GR), poly(GA), or G4C2 repeat–containing RNA transcripts. In addition, considering that c9ASOs were administered relatively early in the disease course, before substantial pTDP-43 deposition in the (G4C2)149 mouse model (13), it is unclear whether similar treatment efficacy would be observed if c9ASOs were delivered at a later time point.
In conclusion, we established that poly(GR) potently promotes TDP-43 aggregation through protein-protein interactions (independent of RNA binding and its C-terminal region), sequesters SG-resident proteins and nucleocytoplasmic transport factors, and thus drives TDP-43 proteinopathy in vivo. These pathological features, as well as neuronal loss, can be alleviated by c9ASO treatment. Together, the results of this study establish poly(GR) as a mechanistic link between the C9orf72 repeat expansion and TDP-43 proteinopathy, and show that decreasing poly(GR) with G4C2-targeted therapeutics is associated with reduced TDP-43 pathology and neurodegeneration in vivo. Moreover, our discovery that plasma NFL in (G4C2)149 mice correlates with neuronal loss, combined with our previous findings that NFL concentrations in patients with c9FTD correlate with disease severity and brain atrophy (35), indicates that plasma NFL might be used to predict the degree of neurodegeneration. It may also be used to assess efficacy of disease-modifying therapeutics in preclinical models to ultimately guide and facilitate the successful translation of future therapies to humans.
MATERIALS AND METHODS
Study design
The goals of this study were to (i) investigate the role of poly(GR) in driving the cytosolic mislocalization and aggregation of TDP-43 and (ii) determine whether antisense oligonucleotides targeting the G4C2 repeat would affect TDP-43 pathology and neurodegeneration in a c9FTD/ALS mouse model. To thoroughly evaluate the mechanistic relationship between poly(GR) and TDP-43, we included in vitro experiments to demonstrate a direct interaction at the pure protein level, cell culture experiments to interrogate cytosolic co-aggregation and the recruitment of SG-associated proteins, and in vivo experiments to assess the ability of poly(GR) to drive accumulation of endogenous mouse TDP-43. To determine whether a therapeutic strategy targeting the G4C2 repeat would alleviate TDP-43 proteinopathy and neurodegeneration, we used an AAV-based model that expresses either 2 or 149 G4C2 repeats, which we previously demonstrated recapitulates key pathologic features of c9FTD/ALS. At 3 months of age, (G4C2)149 mice were randomly assigned to either the phosphate-buffered saline (PBS) or c9ASO group, with littermates being assigned to separate groups, and also controlling for gender between groups. Poly(GR) aggregates, TDP-43 inclusions, and C9orf72-associated pathologies, as well as nucleocytoplasmic transport factors and NPC protein abnormalities were examined by biochemistry, electron microscopy, immunochemistry, immunofluorescence, FISH, PLA, and immunoassays. Mouse and human samples were randomly selected for analyses. Image analysis was performed in a blinded or unblinded fashion, as detailed in individual experiments. Quantification was performed in a blinded manner. Data from cell culture studies are based on three independent experiments. Group sizes for in vivo studies vary, but are noted in the figure legend. Sample sizes were adequately powered to observe the effects based on previous reports (13, 14, 45–47).
Statistics
All statistical analyses were performed in GraphPad Prism. Data are presented as mean ± SEM. Data were analyzed by one-way or two-way analysis of variance (ANOVA) followed by Tukey’s post hoc analysis, except analyses with two groups were analyzed with two-tailed unpaired t test (Figs. 3B and 5E and fig. S3F), and association analyses were performed by determining the Pearson’s correlation coefficient (Fig. 5, H and I). Specific tests are also noted in each figure legend. P < 0.05 is considered statistically significant.
Supplementary Material
Data file S1. Raw data for all the quantitative figures where n < 20.
Materials and Methods
Fig. S1. Poly(GR) accelerates and enhances TDP-43 aggregation.
Fig. S2. Poly(GR) mediates sequestration of cytosolic full-length TDP-43 into the inclusions.
Fig. S3. TDP-43 and DPR burden in mouse and human.
Fig. S4. Poly(GR) aggregates sequester nuclear pore POM121 protein in vivo.
Fig. S5. c9ASO reduces G4C2 repeat–containing RNA and sense DPR protein burden.
Table S1. cDNA sequence of the GFP-(GR)200 and mCherry-(GR)100 plasmids.
Table S2. Primary antibodies for Western blot, immunohistochemistry, and immunofluorescence staining.
Table S3. Characteristics of patients with c9FTD/ALS.
Table S4. Primers for qPCR.
Acknowledgments:
We are grateful to all patients who agreed to donate postmortem tissue. We thank W.-L. Lin for IEM study. We thank S. Boeynaems for providing the (GR)20 and (GA)20 peptides.
Funding: This work was supported by the NIH (R35NS097273 to L.P.; P01NS084974 to D.W.D., T.F.G., Y.-J.Z., and L.P.; P01NS099114 to T.F.G. and L.P.; R01NS088689 to L.P.; and R21AG065854 and R01GM099836 to J.S.), Mayo Clinic Foundation (to L.P.), Amyotrophic Lateral Sclerosis Association (to T.F.G., L.P., J.S., and Y.-.J.Z.), Robert Packard Center for ALS Research at Johns Hopkins (to L.P. and J.S.), Target ALS Foundation (to T.F.G., L.P., J.S., and Y.-J.Z.), Biogen Idec (to L.P.), and AstraZeneca postdoctoral fellowship (to H.M.O.).
Footnotes
SUPPLEMENTARY MATERIALS
View/request a protocol for this paper from Bio-protocol.
Competing interests: B.O. has consulted for Biogen, Medicinova, Mitsubishi, Amylyx, and Tsumura. F.R. is a paid employee of Ionis Pharmaceuticals. G.S.T. and A.M. are paid employees of Biogen Idec. L.P. is a consultant for Expansion Therapeutics. Other authors declare no competing interests.
Data and materials availability: c9ASOs were provided by Ionis Pharmaceuticals (F.R.) and are covered under a material transfer agreement (MTA). All data associated with this study are available in the main text or the Supplementary Materials. All requests for reagents should be made to the corresponding authors upon reasonable request. They will be made available through appropriate administrative channels (MTA).
REFERENCES AND NOTES
- 1.Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, Bruce J, Schuck T, Grossman M, Clark CM, McCluskey LF, Miller BL, Masliah E, Mackenzie IR, Feldman H, Feiden W, Kretzschmar HA, Trojanowski JQ, Lee VM-Y, Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science 314, 130–133 (2006). [DOI] [PubMed] [Google Scholar]
- 2.Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, Mann D, Tsuchiya K, Yoshida M, Hashizume Y, Oda T, TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun 351, 602–611 (2006). [DOI] [PubMed] [Google Scholar]
- 3.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, Kouri N, Wojtas A, Sengdy P, Hsiung G-YR, Karydas A, Seeley WW, Josephs KA, Coppola G, Geschwind DH, Wszolek ZK, Feldman H, Knopman DS, Petersen RC, Miller BL, Dickson DW, Boylan KB, Graff-Radford NR, Rademakers R, Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron 72, 245–256 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Renton AE, Majounie E, Waite A, Simón-Sánchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, Kalimo H, Paetau A, Abramzon Y, Remes AM, Kaganovich A, Scholz SW, Duckworth J, Ding J, Harmer DW, Hernandez DG, Johnson JO, Mok K, Ryten M, Trabzuni D, Guerreiro RJ, Orrell RW, Neal J, Murray A, Pearson J, Jansen IE, Sondervan D, Seelaar H, Blake D, Young K, Halliwell N, Callister JB, Toulson G, Richardson A, Gerhard A, Snowden J, Mann D, Neary D, Nalls MA, Peuralinna T, Jansson L, Isoviita V-M, Kaivorinne A-L, Hölttä-Vuori M, Ikonen E, Sulkava R, Benatar M, Wuu J, Chiò A, Restagno G, Borghero G, Sabatelli M; The ITALSGEN Consortium, Heckerman D, Rogaeva E, Zinman L, Rothstein JD, Sendtner M, Drepper C, Eichler EE, Alkan C, Abdullaev Z, Pack SD, Dutra A, Pak E, Hardy J, Singleton A, Williams NM, Heutink P, Pickering-Brown S, Morris HR, Tienari PJ, Traynor BJ, A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron 72, 257–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lagier-Tourenne C, Baughn M, Rigo F, Sun S, Liu P, Li H-R, Jiang J, Watt AT, Chun S, Katz M, Qiu J, Sun Y, Ling S-C, Zhu Q, Polymenidou M, Drenner K, Artates JW, McAlonis-Downes M, Markmiller S, Hutt KR, Pizzo DP, Cady J, Harms MB, Baloh RH, Vandenberg SR, Yeo GW, Fu X-D, Bennett CF, Cleveland DW, Ravits J, Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. U.S.A 110, E4530–E4539 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mizielinska S, Lashley T, Norona FE, Clayton EL, Ridler CE, Fratta P, Isaacs AM, C9orf72 frontotemporal lobar degeneration is characterised by frequent neuronal sense and antisense RNA foci. Acta Neuropathol. 126, 845–857 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ash PEA, Bieniek KF, Gendron TF, Caulfield T, Lin W-L, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW III, Rademakers R, Boylan KB, Dickson DW, Petrucelli L, Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron 77, 639–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mori K, Arzberger T, Grässer FA, Gijselinck I, May S, Rentzsch K, Weng S-M, Schludi MH, van der Zee J, Cruts M, Van Broeckhoven C, Kremmer E, Kretzschmar HA, Haass C, Edbauer D, Bidirectional transcripts of the expanded C9orf72 hexanucleotide repeat are translated into aggregating dipeptide repeat proteins. Acta Neuropathol. 126, 881–893 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Mori K, Weng S-M, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, Haass C, Edbauer D, The C9orf72 GGGGCC repeat is translated into aggregating dipeptide-repeat proteins in FTLD/ALS. Science 339, 1335–1338 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Gendron TF, Bieniek KF, Zhang YJ, Jansen-West K, Ash PE, Caulfield T, Daughrity L, Dunmore JH, Castanedes-Casey M, Chew J, Cosio DM, van Blitterswijk M, Lee WC, Rademakers R, Boylan KB, Dickson DW, Petrucelli L, Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 126, 829–844 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zu T, Liu Y, Bañez-Coronel M, Reid T, Pletnikova O, Lewis J, Miller TM, Harms MB, Falchook AE, Subramony SH, Ostrow LW, Rothstein JD, Troncoso JC, Ranum LPW, RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. U.S.A 110, E4968–E4977 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koppers M, Blokhuis AM, Westeneng H-J, Terpstra ML, Zundel CAC, Vieira de Sá R, Schellevis RD, Waite AJ, Blake DJ, Veldink JH, van den Berg LH, Pasterkamp RJ, C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann. Neurol 78, 426–438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chew J, Cook C, Gendron TF, Jansen-West K, del Rosso G, Daughrity LM, Castanedes-Casey M, Kurti A, Stankowski JN, Disney MD, Rothstein JD, Dickson DW, Fryer JD, Zhang Y-J, Petrucelli L, Aberrant deposition of stress granule-resident proteins linked to C9orf72-associated TDP-43 proteinopathy. Mol. Neurodegener 14, 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chew J, Gendron TF, Prudencio M, Sasaguri H, Zhang Y-J, Castanedes-Casey M, Lee CW, Jansen-West K, Kurti A, Murray ME, Bieniek KF, Bauer PO, Whitelaw EC, Rousseau L, Stankowski JN, Stetler C, Daughrity LM, Perkerson EA, Desaro P, Johnston A, Overstreet K, Edbauer D, Rademakers R, Boylan KB, Dickson DW, Fryer JD, Petrucelli L, C9ORF72 repeat expansions in mice cause TDP-43 pathology, neuronal loss, and behavioral deficits. Science 348, 1151–1154 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J, Zhu Q, Gendron TF, Saberi S, McAlonis-Downes M, Seelman A, Stauffer JE, Jafar-Nejad P, Drenner K, Schulte D, Chun S, Sun S, Ling S-C, Myers B, Engelhardt J, Katz M, Baughn M, Platoshyn O, Marsala M, Watt A, Heyser CJ, Ard MC, De Muynck L, Daughrity LM, Swing DA, Tessarollo L, Jung CJ, Delpoux A, Utzschneider DT, Hedrick SM, de Jong PJ, Edbauer D, Van Damme P, Petrucelli L, Shaw CE, Bennett CF, Da Cruz S, Ravits J, Rigo F, Cleveland DW, Lagier-Tourenne C, Gain of toxicity from ALS/FTD-linked repeat expansions in C9ORF72 is alleviated by antisense oligonucleotides targeting GGGGCC-containing RNAs. Neuron 90, 535–550 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Pattamatta A, Zu T, Reid T, Bardhi O, Borchelt DR, Yachnis AT, Ranum LPW, C9orf72 BAC mouse model with motor deficits and neurodegenerative features of ALS/FTD. Neuron 90, 521–534 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee K-H, Badders N, Valentine M, Miller BL, Wong PC, Petrucelli L, Kim HJ, Gao F-B, Taylor JP, GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature 525, 129–133 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jovičić A, Mertens J, Boeynaems S, Bogaert E, Chai N, Yamada SB, Paul III JW, Sun S, Herdy JR, Bieri G, Kramer NJ, Gage FH, Van Den Bosch L, Robberecht W, Gitler AD, Modifiers of C9orf72 dipeptide repeat toxicity connect nucleocytoplasmic transport defects to FTD/ALS. Nat. Neurosci 18, 1226–1229 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, Gupta S, Thomas MA, Hong I, Chiu S-L, Huganir RL, Ostrow LW, Matunis MJ, Wang J, Sattler R, Lloyd TE, Rothstein JD, The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature 525, 56–61 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang K, Daigle JG, Cunningham KM, Coyne AN, Ruan K, Grima JC, Bowen KE, Wadhwa H, Yang P, Rigo F, Taylor JP, Gitler AD, Rothstein JD, Lloyd TE, Stress granule assembly disrupts nucleocytoplasmic transport. Cell 173, 958–971.e17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang YJ, Gendron TF, Grima JC, Sasaguri H, Jansen-West K, Xu Y-F, Katzman RB, Gass J, Murray ME, Shinohara M, Lin W-L, Garrett A, Stankowski JN, Daughrity L, Tong J, Perkerson EA, Yue M, Chew J, Castanedes-Casey M, Kurti A, Wang ZS, Liesinger AM, Baker JD, Jiang J, Lagier-Tourenne C, Edbauer D, Cleveland DW, Rademakers R, Boylan KB, Bu G, Link CD, Dickey CA, Rothstein JD, Dickson DW, Fryer JD, Petrucelli L, C9ORF72 poly(GA) aggregates sequester and impair HR23 and nucleocytoplasmic transport proteins. Nat. Neurosci 19, 668–677 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y-J, Guo L, Gonzales PK, Gendron TF, Wu Y, Jansen-West K, O’Raw AD, Pickles SR, Prudencio M, Carlomagno Y, Gachechiladze MA, Ludwig C, Tian R, Chew J, DeTure M, Lin W-L, Tong J, Daughrity LM, Yue M, Song Y, Andersen JW, Castanedes-Casey M, Kurti A, Datta A, Antognetti G, McCampbell A, Rademakers R, Oskarsson B, Dickson DW, Kampmann M, Ward ME, Fryer JD, Link CD, Shorter J, Petrucelli L, Heterochromatin anomalies and double-stranded RNA accumulation underlie C9orf72 poly(PR) toxicity. Science 363, eaav2606 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Boeynaems S, Bogaert E, Kovacs D, Konijnenberg A, Timmerman E, Volkov A, Guharoy M, De Decker M, Jaspers T, Ryan VH, Janke AM, Baatsen P, Vercruysse T, Kolaitis RM, Daelemans D, Taylor JP, Kedersha N, Anderson P, Impens F, Sobott F, Schymkowitz J, Rousseau F, Fawzi NL, Robberecht W, Van Damme P, Tompa P, Van Den Bosch L, Phase separation of C9orf72 dipeptide repeats perturbs stress granule dynamics. Mol. Cell 65, 1044–1055.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cheng W, Wang S, Mestre AA, Fu C, Makarem A, Xian F, Hayes LR, Lopez-Gonzalez R, Drenner K, Jiang J, Cleveland DW, Sun S, C9ORF72 GGGGCC repeat-associated non-AUG translation is upregulated by stress through eIF2α phosphorylation. Nat. Commun 9, 51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green KM, Glineburg MR, Kearse MG, Flores BN, Linsalata AE, Fedak SJ, Goldstrohm AC, Barmada SJ, Todd PK, RAN translation at C9orf72-associated repeat expansions is selectively enhanced by the integrated stress response. Nat. Commun 8, 2005 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee K-H, Zhang P, Kim HJ, Mitrea DM, Sarkar M, Freibaum BD, Cika J, Coughlin M, Messing J, Molliex A, Maxwell BA, Kim NC, Temirov J, Moore J, Kolaitis R-M, Shaw TI, Bai B, Peng J, Kriwacki RW, Taylor JP, C9orf72 dipeptide repeats impair the assembly, dynamics, and function of membrane-less organelles. Cell 167, 774–788.e17 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayes LR, Duan L, Bowen K, Kalab P, Rothstein JD, C9orf72 arginine-rich dipeptide repeat proteins disrupt karyopherin-mediated nuclear import. eLife 9, e51685 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saberi S, Stauffer JE, Jiang J, Garcia SD, Taylor AE, Schulte D, Ohkubo T, Schloffman CL, Maldonado M, Baughn M, Rodriguez MJ, Pizzo D, Cleveland D, Ravits J, Sense-encoded poly-GR dipeptide repeat proteins correlate to neurodegeneration and uniquely co-localize with TDP-43 in dendrites of repeat-expanded C9orf72 amyotrophic lateral sclerosis. Acta Neuropathol. 135, 459–474 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y-J, Gendron TF, Ebbert MTW, O’Raw AD, Yue M, Jansen-West K, Zhang X, Prudencio M, Chew J, Cook CN, Daughrity LM, Tong J, Song Y, Pickles SR, Castanedes-Casey M, Kurti A, Rademakers R, Oskarsson B, Dickson DW, Hu W, Gitler AD, Fryer JD, Petrucelli L, Poly(GR) impairs protein translation and stress granule dynamics in C9orf72-associated frontotemporal dementia and amyotrophic lateral sclerosis. Nat. Med 24, 1136–1142 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buratti E, Baralle FE, Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem 276, 36337–36343 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Zhang Y-J, Xu Y-F, Cook C, Gendron TF, Roettges P, Link CD, Lin W-L, Tong J, Castanedes-Casey M, Ash P, Gass J, Rangachari V, Buratti E, Baralle F, Golde TE, Dickson DW, Petrucelli L, Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl. Acad. Sci. U.S.A 106, 7607–7612 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson BS, Snead D, Lee JJ, McCaffery JM, Shorter J, Gitler AD, TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem 284, 20329–20339 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishimura AL, Župunski V, Troakes C, Kathe C, Fratta P, Howell M, Gallo J-M, Hortobágyi T, Shaw CE, Rogelj B, Nuclear import impairment causes cytoplasmic trans-activation response DNA-binding protein accumulation and is associated with frontotemporal lobar degeneration. Brain 133, 1763–1771 (2010). [DOI] [PubMed] [Google Scholar]
- 34.Gendron TF, Chew J, Stankowski JN, Hayes LR, Zhang Y-J, Prudencio M, Carlomagno Y, Daughrity LM, Jansen-West K, Perkerson EA, O’Raw A, Cook C, Pregent L, Belzil V, van Blitterswijk M, Tabassian LJ, Lee CW, Yue M, Tong J, Song Y, Castanedes-Casey M, Rousseau L, Phillips V, Dickson DW, Rademakers R, Fryer JD, Rush BK, Pedraza O, Caputo AM, Desaro P, Palmucci C, Robertson A, Heckman MG, Diehl NN, Wiggs E, Tierney M, Braun L, Farren J, Lacomis D, Ladha S, Fournier CN, McCluskey LF, Elman LB, Toledo JB, McBride JD, Tiloca C, Morelli C, Poletti B, Solca F, Prelle A, Wuu J, Jockel-Balsarotti J, Rigo F, Ambrose C, Datta A, Yang W, Raitcheva D, Antognetti G, McCampbell A, Van Swieten JC, Miller BL, Boxer AL, Brown RH, Bowser R, Miller TM, Trojanowski JQ, Grossman M, Berry JD, Hu WT, Ratti A, Traynor BJ, Disney MD, Benatar M, Silani V, Glass JD, Floeter MK, Rothstein JD, Boylan KB, Petrucelli L, Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci. Transl. Med 9, eaai7866 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meeter LHH, Gendron TF, Sias AC, Jiskoot LC, Russo SP, Donker Kaat L, Papma JM, Panman JL, van der Ende EL, Dopper EG, Franzen S, Graff C, Boxer AL, Rosen HJ, Sanchez-Valle R, Galimberti D, Pijnenburg YAL, Benussi L, Ghidoni R, Borroni B, Laforce R Jr., del Campo M, Teunissen CE, van Minkelen R, Rojas JC, Coppola G, Geschwind DH, Rademakers R, Karydas AM, Oijerstedt L, Scarpini E, Binetti G, Padovani A, Cash DM, Dick KM, Bocchetta M, Miller BL, Rohrer JD, Petrucelli L, van Swieten JC, Lee SE, Poly(GP), neurofilament and grey matter deficits in C9orf72 expansion carriers. Ann. Clin. Transl. Neurol 5, 583–597 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gaetani L, Blennow K, Calabresi P, Di Filippo M, Parnetti L, Zetterberg H, Neurofilament light chain as a biomarker in neurological disorders. J. Neurol. Neurosurg. Psychiatry 90, 870–881 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Shahim P, Zetterberg H, Tegner Y, Blennow K, Serum neurofilament light as a biomarker for mild traumatic brain injury in contact sports. Neurology 88, 1788–1794 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sakae N, Bieniek KF, Zhang Y-J, Ross K, Gendron TF, Murray ME, Rademakers R, Petrucelli L, Dickson DW, Poly-GR dipeptide repeat polymers correlate with neurodegeneration and clinicopathological subtypes in C9ORF72-related brain disease. Acta Neuropathol. Commun 6, 63 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mackenzie IR, Arzberger T, Kremmer E, Troost D, Lorenzl S, Mori K, Weng S-M, Haass C, Kretzschmar HA, Edbauer D, Neumann M, Dipeptide repeat protein pathology in C9ORF72 mutation cases: Clinico-pathological correlations. Acta Neuropathol. 126, 859–879 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Mackenzie IRA, Frick P, Grässer FA, Gendron TF, Petrucelli L, Cashman NR, Edbauer D, Kremmer E, Prudlo J, Troost D, Neumann M, Quantitative analysis and clinico-pathological correlations of different dipeptide repeat protein pathologies in C9ORF72 mutation carriers. Acta Neuropathol. 130, 845–861 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Gomez-Deza J, Lee Y.-b., Troakes C, Nolan M, Al-Sarraj S, Gallo J-M, Shaw CE, Dipeptide repeat protein inclusions are rare in the spinal cord and almost absent from motor neurons in C9ORF72 mutant amyotrophic lateral sclerosis and are unlikely to cause their degeneration. Acta Neuropathol. Commun 3, 38 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gasset-Rosa F, Lu S, Yu H, Chen C, Melamed Z, Guo L, Shorter J, Da Cruz S, Cleveland DW, Cytoplasmic TDP-43 de-mixing independent of stress granules drives inhibition of nuclear import, loss of nuclear TDP-43, and cell death. Neuron 102, 339–357.e7 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mann JR, Gleixner AM, Mauna JC, Gomes E, DeChellis-Marks MR, Needham PG, Copley KE, Hurtle B, Portz B, Pyles NJ, Guo L, Calder CB, Wills ZP, Pandey UB, Kofler JK, Brodsky JL, Thathiah A, Shorter J, Donnelly CJ, RNA binding antagonizes neurotoxic phase transitions of TDP-43. Neuron 102, 321–338.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Elden AC, Kim H-J, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, Padmanabhan A, Clay-Falcone D, McCluskey L, Elman L, Juhr D, Gruber PJ, Rüb U, Auburger G, Trojanowski JQ, Lee VM-Y, Van Deerlin VM, Bonini NM, Gitler AD, Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–1075 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guo L, Kim HJ, Wang H, Monaghan J, Freyermuth F, Sung JC, O–Donovan K, Fare CM, Diaz Z, Singh N, Zhang ZC, Coughlin M, Sweeny EA, DeSantis ME, Jackrel ME, Rodell CB, Burdick JA, King OD, Gitler AD, Lagier-Tourenne C, Pandey UB, Chook YM, Taylor JP, Shorter J, Nuclear-import receptors reverse aberrant phase transitions of RNA-binding proteins with prion-like domains. Cell 173, 677–692.e20 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang A, Conicella AE, Schmidt HB, Martin EW, Rhoads SN, Reeb AN, Nourse A, Ramirez Montero D, Ryan VH, Rohatgi R, Shewmaker F, Naik MT, Mittag T, Ayala YM, Fawzi NL, A single N-terminal phosphomimic disrupts TDP-43 polymerization, phase separation, and RNA splicing. EMBOJ. 37, e97452 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang Y-J, Caulfield T, Xu Y-F, Gendron TF, Hubbard J, Stetler C, Sasaguri H, Whitelaw EC, Cai S, Lee WC, Petrucelli L, The dual functions of the extreme N-terminus of TDP-43 in regulating its biological activity and inclusion formation. Hum. Mol. Genet 22, 3112–3122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Prudencio M, Jansen-West KR, Lee WC, Gendron TF, Zhang Y-J, Xu Y-F, Gass J, Stuani C, Stetler C, Rademakers R, Dickson DW, Buratti E, Petrucelli L, Misregulation of human sortilin splicing leads to the generation of a nonfunctional progranulin receptor. Proc. Natl. Acad. Sci. U.S.A 109, 21510–21515 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gendron TF, van Blitterswijk M, Bieniek KF, Daughrity LM, Jiang J, Rush BK, Pedraza O, Lucas JA, Murray ME, Desaro P, Robertson A, Overstreet K, Thomas CS, Crook JE, Castanedes-Casey M, Rousseau L, Josephs KA, Parisi JE, Knopman DS, Petersen RC, Boeve BF, Graff-Radford NR, Rademakers R, Lagier-Tourenne C, Edbauer D, Cleveland DW, Dickson DW, Petrucelli L, Boylan KB, Cerebellar c9RAN proteins associate with clinical and neuropathological characteristics of C9ORF72 repeat expansion carriers. Acta Neuropathol. 130, 559–573 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1. Raw data for all the quantitative figures where n < 20.
Materials and Methods
Fig. S1. Poly(GR) accelerates and enhances TDP-43 aggregation.
Fig. S2. Poly(GR) mediates sequestration of cytosolic full-length TDP-43 into the inclusions.
Fig. S3. TDP-43 and DPR burden in mouse and human.
Fig. S4. Poly(GR) aggregates sequester nuclear pore POM121 protein in vivo.
Fig. S5. c9ASO reduces G4C2 repeat–containing RNA and sense DPR protein burden.
Table S1. cDNA sequence of the GFP-(GR)200 and mCherry-(GR)100 plasmids.
Table S2. Primary antibodies for Western blot, immunohistochemistry, and immunofluorescence staining.
Table S3. Characteristics of patients with c9FTD/ALS.
Table S4. Primers for qPCR.


