Abstract
Studies of large proteins, protein complexes, and membrane protein complexes pose new challenges, most notably the need for increased ion mobility (IM) and mass spectrometry (MS) resolution. This review covers evolutionary developments in IM-MS in the authors’ and key collaborators’ laboratories with specific focus on developments that enhance the utility of IM-MS for structural analysis. IM-MS measurements are performed on gas phase ions, thus “structural IM-MS” appears paradoxical—do gas phase ions retain their solution phase structure? There is growing evidence to support the notion that solution phase structure(s) can be retained by the gas phase ions. It should not go unnoticed that we use “structures” in this statement because an important feature of IM-MS is the ability to deal with conformationally heterogeneous systems, thus providing a direct measure of conformational entropy. The extension of this work to large proteins and protein complexes has motivated our development of Fourier-transform IM-MS instruments, a strategy first described by Hill and coworkers in 1985 (Anal Chem, 1985, 57, pp. 402–406) that has proved to be a game-changer in our quest to merge drift tube (DT) and ion mobility and the high mass resolution orbitrap MS instruments. DT-IMS is the only method that allows first-principles determinations of rotationally averaged collision cross sections (CSS), which is essential for studies of biomolecules where the conformational diversities of the molecule precludes the use of CCS calibration approaches. The Fourier transform-IM-orbitrap instrument described here also incorporates the full suite of native MS/IM-MS capabilities that are currently employed in the most advanced native MS/IM-MS instruments.
Keywords: drift tube ion mobility, orbitrap MS, 1st principles CCS, native MS/IM-MS, conformational heterogeneity, cryogenic ion-mobility MS
I. INTRODUCTION
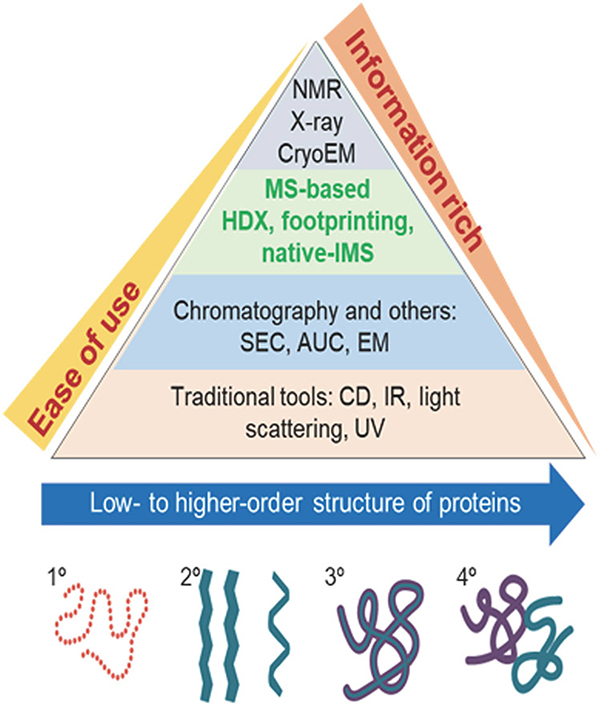
Structural mass spectrometry (MS) has evolved from the early art form of interpreting bonding configurations of atoms/functional groups of small molecules ionized by electron impact ionization (Turecek & Mclafferty, 1993) to the method of choice for determining the amino acid sequences of proteins, to studies of higher-order protein structure (2°, 3°, and 4°). These evolutionary developments have expanded the experimental versatility, mass range, and resolution of MS instruments, thereby opening new vistas for studies of large biomolecules (Dyachenko et al., 2013), membrane protein complexes (Laganowsky et al., 2014), and even viruses (Lutomski et al., 2018). These more advanced instruments incorporate multiple MS-based techniques, for example, MS/MS, ion mobility mass spectrometry (IM-MS), and MS-IM-MS, that facilitate more integrative approaches as well as substantial gains in terms of sensitivity and resolution, for both IM and MS (Zinnel, Pai, & Russell, 2012). More importantly, the versatility of these new technologies allows users to design and implement novel experimental strategies that can then be employed to carry out new untested measurement strategies which oftentimes provide solutions to previously intractable problems (Chen, Chen, & Russell, 2014). These modern instruments are increasingly used for characterizing the structure, stabilities, and dynamics of proteins (Dixit, Polasky, & Ruotolo, 2018) as well as the stoichiometry and topology (4°) of biomolecular complexes (Zhou & Wysocki, 2014), including membrane protein complexes (Vimer et al., 2020) as illustrated by the following reports. Tian and Ruotolo (2018) have described comprehensive integrated MS strategies, viz. proteomics, chemical labeling MS, native MS and IM-MS, and their use in the discovery and development of therapeutic antibodies. Similar strategies are now providing new insights related to protein structure and function, including allostery associated with lipid binding to membrane proteins (Patrick et al., 2018). Variable-temperature high-resolution native MS has been used to determine the thermodynamics of individual lipid binding events to the ammonia channel membrane (AmtB), which revealed characteristic thermodynamic signatures for the binding of different lipids as well as distinct changes in these signatures for AmtB mutants (Cong et al., 2016). In a similar study, the kinetics and thermodynamics of the intrinsic hydrolysis of K-RAS and its oncogenic mutants were investigated by high-resolution native MS, and native IM-MS measurements of K-RAS and its oncogenic mutants indicated that a native-like conformation is preserved in the IM-MS instrument (Moghadamchargari et al., 2019). Native IM-MS has also been used to directly monitor small molecule (toxin tertiapin Q) binding to the mammalian potassium channel GIRK2. This study revealed that phosphatidylinositides enhance the binding selectivity relative to other phospholipids, and CIU IM-MS was used to show a similar increase in binding affinities (Liu et al., 2019). It is interesting to consider areas of structural biology on which native MS and native IM-MS will be the most impactful. The traditional structural analysis techniques, that is, nuclear magnetic resonance (NMR), X-ray diffraction (XRD) crystallography, cryogenic electron microscopy (cryo-EM), and optical spectroscopy, are well-established and broadly used by the structural biology community. However, MS-based approaches are only slowly being accepted by this community owing to questions related to the “IMS Paradox” (vide infra)…do gas-phase ions retain their solution-phase structure? It is important to realize, however, that not all structure-based MS methods are based solely on the native MS data. The structural information obtained by hydrogen/deuterium exchange (Masson et al., 2019), hydroxyl radical footprinting (Kiselar & Chance, 2018; Niu & Gross, 2019), and covalent labeling (Rajabi, Ashcroft, & Radford, 2015) samples the solution-phase ion structure. Gross (Mass spectrometry-based structural proteomics solves problems in biochemistry and bio-therapeutics. Award Address (ACS Award in Analytical Chemistry sponsored by the Battelle Memorial Institute). Abstracts of Papers, 255th ACS National Meeting and Exposition, New Orleans, LA, United States, March 18–22, 2018, ANYL-368) addressed this question using the diagram shown in Figure 1, where the major structure analysis techniques are compared in terms of “ease of use” and “information-richness.” NMR, XRD, and cryo-EM are capable of providing the highest resolution structural information; but when the complete suite of native MS/IM-MS structural tools, for example, collision-induced unfolding (CIU; Polasky et al., 2019), collisional-induced dissociation (CID; Donor, Shepherd, & Prell, 2020), surface-induced dissociation (SID; Zhou & Wysocki, 2014), ultraviolet photodissociation (UVPD; Sipe & Brodbelt, 2019), and infrared multiphoton dissociation (Pagel et al., 2009; Seo et al., 2017), are factored into the equation, native-MS offers an unprecedented advantage in the field of structural biology.
FIGURE 1.
Comparison of native IM-MS to other the more conventional structural biology techniques used for structural studies. First presented by M. L. Gross as part of 2018 Symposium to honor his ACS Award in Analytical Chemistry. IM, ion mobility; MS, mass spectrometry.
This review focuses on the development of native MS with conditions allowing for retention of solution phase structure, which when combined with MS-MS and IMS, open new paradigms for studies of chemical processes that occur in solution. Here, we focus on protein folding-unfolding, changes in structure, stability and dynamics, and aggregation as the solution conditions (i.e., pH, temperature, mutations and/or post-translational modification, presence of ligands, metal ions (Dong, Wagner, & Russell, 2018), small molecules (Gault et al., 2018), and even other peptide/proteins (Yewdall et al., 2018)), change. There are, however, two salient issues that remain questionable: (i) Buffers and sample preparation used for native MS are designed to produce low charge states, but whether low charge states are better correlated to solution-phase structure(s)/conformations is still debated. (ii) There also exists an “IMS Paradox,” “…for how long, under what conditions, and to what extent is (solution) structure retained in the absence of solvent” (Breuker & McLafferty, 2008; Hewitt et al., 2014; Rolland & Prell, 2019). This question refers to issues related to how both hydrophobic and hydrophilic solute/solvent interactions influence the conformational preferences of the ions as they transition from bulk solution, to nanodroplets, and then to solvent-free (SF) gas phase ions.
It is important to note, however, that a similar paradox exists for all structural characterization techniques. Crystallography requires high quality crystals, NMR requires relatively high concentrations of high purity samples, and both report ensemble-averaged responses, which limit their utility for studies of conformationally heterogeneous samples. Pochapsky and Pochapsky (2019) specifically noted that “…crystallization only captures conformers that fit into the growing (crystal) lattice…,” thus crystallization is a purification step. Meisburger and Ando (2017) noted in their review that “the quality of diffraction cannot correlate with the visual appearance of crystals,” whereas small-angle X-ray scattering better reveals “…what is hidden…,” noting the “next holy grail of crystallography is to embrace imperfection toward a dynamic picture of enzymes.” Pricer et al. note that conformational heterogeneity enables the “context-specific function to emerge in response to changing cellular conditions…” that ultimately gives rise to a greater diversity of function for single structural motifs (Pricer, Gestwicki, & Mapp, 2017). There is a greater realization that dynamics or “conformational entropy” are fundamental to biological processes including cellular signaling, molecular recognition, and protein-ligand interactions (Frederick et al., 2007; Wand & Sharp, 2018).
In the following sections, studies are described that illustrate the unique capabilities of IM-MS for investigations of conformationally heterogeneous systems, whether the heterogeneity is inherent to chemical species or due to changes in the solution environment. While it is challenging to design experiments to unequivocally address the “IMS Paradox,” the studies that are discussed herein all produce ions under conditions that involve rapid evaporative cooling of charged nanodroplets. Beauchamp and coworkers described this evaporative drying process as “freeze-drying” (Lee et al., 1998) owing to the effects of evaporative cooling where temperatures of the dehydrated ions are estimated at 130–150 K. Cryogenic ion mobility-mass spectrometry (cryo-IM-MS) (vida infra), takes advantage of “freeze-drying” to generate cold-hydrated ions that are then cooled further (80 K), thereby preserving a relatively high level of hydration. It is interesting to compare the freeze-drying and cryo-IM-MS experiments with cryo-EM. Cryo-EM structures of large molecules/complexes are obtained from water droplets that are rapidly cooled to cryogenic temperatures (186 K in liquid ethane or 230 K in propane), thereby trapping the sample molecules in vitreous ice. There exist important parallels between cryo-EM and freeze-drying electrospray ionization (ESI)-MS/cryo-IM-MS (vide infra) in that both techniques take advantage of kinetic-trapping of molecules as they exist in solution.
A holy grail of structural biology lies in structural characterization of the peptides/proteins that comprise the proteome, including identification of structural changes introduced by modifications, changes in concentration, or presence of osmolytes (Clemmer, Russell, & Williams, 2017). While such objectives may seem intractable, recent advances in technologies for native MS/IM-MS are now poised to establish feasible strategies. The following sections of this review describe selected IM-MS studies aimed at determining conformational preferences of relatively simple peptides to more complex soluble proteins, and finally to protein complexes. Emphasis is placed on results obtained using IM-MS, but in nearly all cases these studies integrate molecular dynamics (MD) simulations (Jarrold, 2000), bottom-up and/or top-down proteomics strategies, and techniques such as CIU.
Moreover, despite several studies attempting to correlate the ion mobility measurements to exact shape of ions (Kaldmaee et al., 2019; Landreh et al., 2020), a priori assignment of detailed structure on the basis of rotationally averaged collision cross section (CCS) is not possible; therefore, it is common practice to compare the measured CCS to calculated CCS for structures obtained from NMR, circular dichroism (CD), crystallography, and/or MD simulations.
II. CONFORMATIONAL HETEROGENEITY OF MODEL PEPTIDES
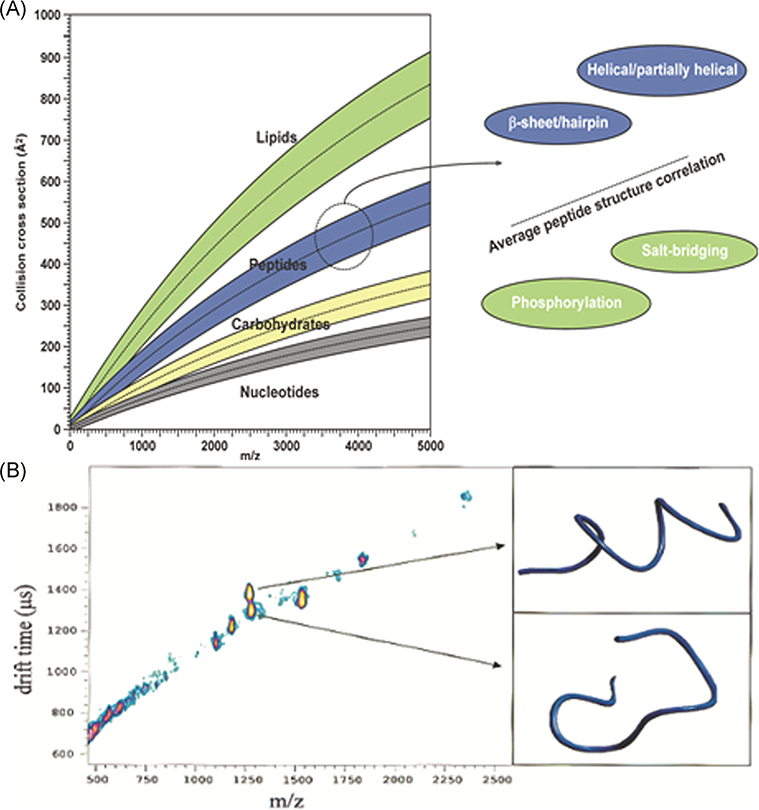
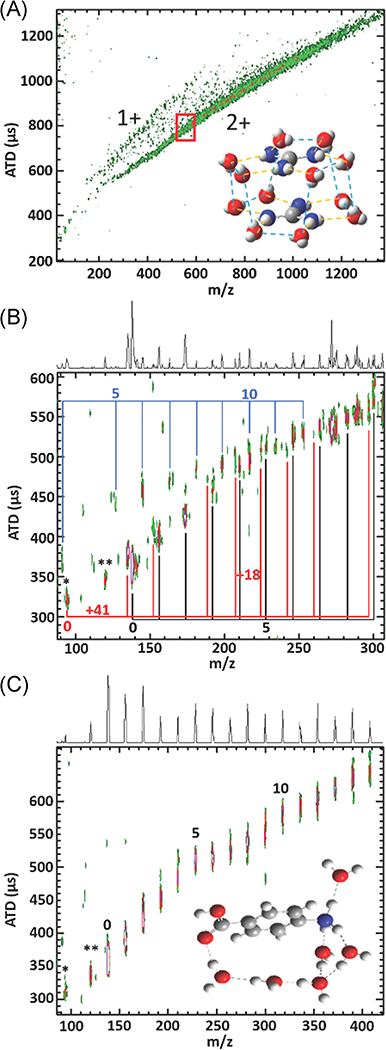
Initial developments in IM-MS occurred in parallel with proteomic MS and tandem MS; consequently, much of our early IM-MS work focused on peptides and small protein ions generated by matrix-assisted laser desorption ionization (MALDI; Ruotolo et al., 2002a). MALDI is readily compatible with IM-TOF-MS, and cocrystallization of the peptides with MALDI matrix formed inclusion complexes (Lehmann et al., 2000) that might yield structures that are similar to those obtained by crystallography. While size/shape-to-charge and mass-to-charge (m/z) ratios are not highly orthogonal, separations based on these metrics do provide information on ion structure/conformation as well as compound class (Fig. 2A; McLean et al., 2005). Note that mobility (plotted as CCS or drift time) vs m/z plots can be used to distinguish different classes of compounds as well as conformers that have different shapes and/or sizes. As an example, Figure 2B contains well-resolved signals for two peptides, one a random coil and the other a helix, that differ by only ~9 Da, but these ions separate on the basis of their different CCS (>10%) (Ruotolo et al., 2002a). Later studies showed that posttranslational modifications (PTMs) that alter the conformations of the ion can also be separated from the native peptides (Ruotolo et al., 2002b, 2004), and cyclic conformers can be separated from their linear analogs when complexed with alkali metal ions (Ruotolo, Tate, & Russell, 2004).
FIGURE 2.
(A) 2-D plots of mobility (arrival-time distributions or CCS) vs m/z illustrating of separation on the basis of compound class and “conformation space.” (B) Mobility (ATD) vs m/z plots for the hemoglobin tryptic peptide fragment ions (residues 104–115, LLGNVLVVVLAR (m/z 1275.5) and residues 30–39, LLVVYPWTQR m/z 1284.5). Reproduced from McLean et al. (2005) and Ruotolo et al. (2002a). ATD, arrival time distributions; CCS, collision cross section.
III. SOLVENT-DEPENDENT CONFORMATIONAL PREFERENCES OF MODEL PEPTIDES
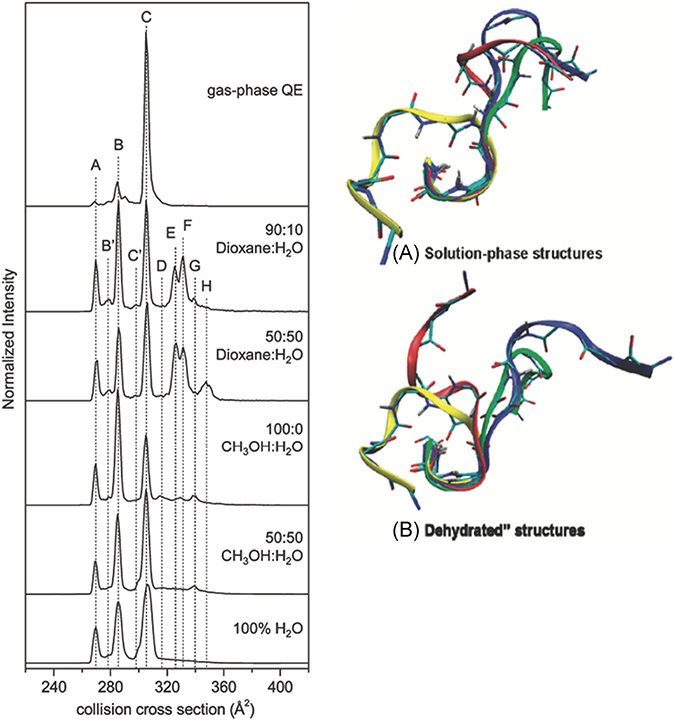
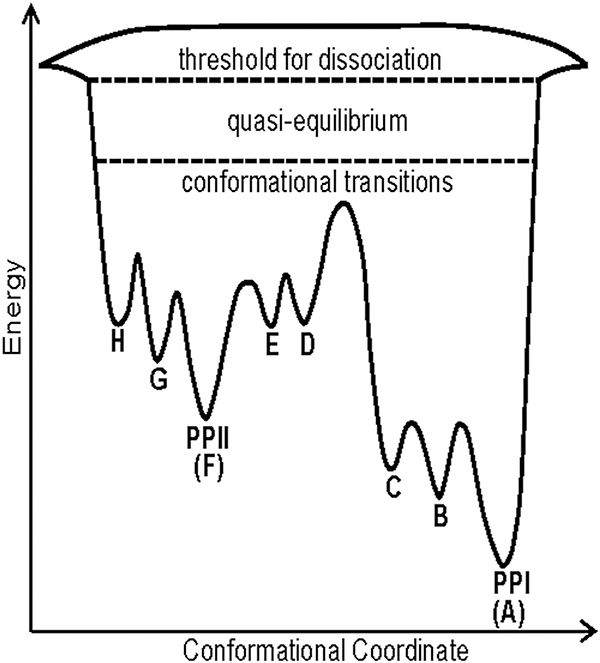
Ion mobility complements other structural MS approaches, but quite possibly the most important figures-of-merit are sensitivity, large dynamic range for measurements of ion abundance, and the ability to measure the conformational heterogeneity of states at equilibrium under different solution environment conditions. Structure and dynamics regulate functions of peptides and proteins, and interaction with the solvent is intrinsic to stabilization of protein structure. In addition to solvent, there are many parameters associated with IM-MS that may also affect the conformational preferences of the ions under investigation. McLean et al. (2010) showed that multiple potential charge sites have significant effects on the helical preferences of singly charged peptide ions and a study by Xiao et al. (2015) observed that charge state, charge site, and side-chain interactions also affect conformational preference of peptide ions. More recently, the effects of solvent composition (mixtures of methanol/water and dioxane/water) on the conformational preferences of the nonapeptide bradykinin (BK) provided evidence for 10 independent populations of conformers (Pierson et al., 2011). In some solutions (90:10 dioxane/water) as many as eight different conformers were found to coexist (Fig. 3). Later studies showed these conformational preferences are strongly linked to cis/trans configurations of the proline residues at positions 2, 3, and 7 (RPPGFSPFR) (Fuller et al., 2018); similar behavior had been observed for BK fragments 1–5 (RPPGF), 1–7 (RPPGFSP), and 1–8 (RPPGFSPF) (Sawyer et al., 2005; Fernandez-Lima et al., 2009). The Clemmer and Russell groups have extended these solution manipulation methods to monitor conformational entropy using kinetics and equilibrium measurements as a function of both solvent and solution temperature, respectively. A series of papers describe in great detail the effects of solvent (propanol/water) on cis-trans configuration changes for a series of polyprolines (Shi et al., 2014, 2016; El-Baba et al., 2016). A particularly interesting configurationally coupled proton transfer reaction was observed for polyproline-7 (PPPPPPP, denoted Pro7) (Shi et al., 2015). It is now well established from this study as well as prior work that cis configurations of proline are preferred in less polar solvents, whereas trans configurations are preferred in polar solvents. For example, Pro7 is initially observed as the all-cis [PPI + H]+ ion in propanol, and when transferred to 40:60 propanol/water under acidic conditions, a very slow protonation reaction coupled with configuration conversion of the PPI helix to the PPII helix is observed to produce all-trans [PPII + 2H]2+ ions. Kinetic and equilibrium measurements as a function of temperature were used to determine the thermochemistry of the proton transfer reaction, which provided evidence that the configurationally coupled reaction is subject to large entropic effects, ~190 J·mol−1·K−1. Two potential mechanisms were considered to rationalize the observed reaction: (i) a mechanism wherein the second proton adds to the C-terminus of the PPI chain to produce [PPI + 2H]2+ ions, or (ii) a mechanism involving migration of the proton initially located on the N-terminus down the peptide chain, possibly as H3O+. Presumably this reaction would occur in concert with the cis-to-trans configurational change of each proline. As this migration of the proton proceeds, the N-terminus can accept a second proton to form the [PPII + 2H]2+ ions. It was also noted that the pore size (~3–4 Å) of the PPI helix is sufficiently large and hydrophilic to allow for H3O+ ions to migrate through the pore (Fig. 4).
FIGURE 3.
Solvent-dependent CCS profiles and examples of four low-energy structures for bradykinin 3+ ions in solution phase and “dehydrated” states. The solvent composition used for each solvent system is shown to the right of each plot. Reproduced from Pierson et al., 2011. CCS, collision cross section.
FIGURE 4.
A qualitative folding landscape for PPI/PPII transitions for Pro13 in vacuo. system. The barrier height is estimated from collision voltages rather than more accurate solution phase measurements or well-defined single collision energies. Note that the reaction D→C varies with H2O and is highest for a “dry” environment. Adapted from Shi et al. (2016b).
Pro13 was also found to fold through a number (est. ~6) of sequential long-lived intermediate states as it converts from an all-cis configuration to an all-trans configuration upon transfer from propanol to water. It was also found that the induction period for this reaction is shifted to longer times at lower temperatures (Shi et al., 2014). HisPro13, however, behaves quite differently from Pro13 in that the intermediate conformers are no longer observed and folding occurs by a cooperative two-state transition having a long temperature-dependent induction period (El-Baba et al., 2016). It appears that the overall reaction occurs by an entropically favored endothermic mechanism that involves 13–17 long-lived intermediate states. It was proposed that the reaction is initiated by hydration of the protonated N-terminus followed by hydration along the peptide backbone. The fact that intermediates are not observed for the reaction PPIPrOH→PPIIaq is attributed to the weakening of the helix dipole, thereby promoting the intermediate structure to revert to the dipole stabilized PPI conformation. An alternative explanation that may be linked to the kinetic trapping effects of “freeze-drying” has not yet been fully explored. Both explanations point to the potential importance of developing better understanding of the role of the confined environments of cold nanodroplets.
A similar conformationally coupled reaction was observed for bradykinin (BK; RPPGFSPFR) and the neuropeptide substance P (SP; RPKPQQFFGLM). At elevated temperatures (65°C) BK undergoes a proton transfer reaction [BK + 2H]2++ H+→[BK + 3H]3+ that is coupled with trans→cis configuration change that then leads to cleavage of the Pro2-Pro3 bond (Fuller et al., 2018). Interestingly, Pro2-Pro3 bond cleavage does not occur by any human enzyme. Similar reactions were also observed for the neuropeptide SP (RPKPQQFFGLM) (Conant et al., 2019). In this case however, the product of the penultimate proline reaction is observed for both the intact SP precursor as well as the formed product ions, viz. [KPQQFFGLM + 2H]2+→[QQFFGLM + 2H]2+.
All of the above reactions occur in solution, but it is interesting to consider a specific example where solution- and gas-phase chemistries are compared. The cis/trans configuration preferences for “wet” and “dry” PPI/PPII were examined for Pro13 using tandem IMS-IMS-MS (Shi et al., 2014). In these experiments, all-trans PPII or the all-cis PPI conformer were formed from water or propanol:water solutions, and specific conformers were selected by IMS-1. The ions were then subjected to mild collisional activation (CA), and the products of CA were then analyzed by IMS-2. For example, SF PPII intermediates efficiently (>90%) refold to form the all cis PPI upon CA, and these refolding reactions follow a similar pathway, that is, the CCS of the intermediates in the reactions PPII→PPI transitions are similar to those observed using the mixed solvent (propanol;water) experiment. A later study showed that SF environments strongly favor the cis (PPI) conformer (Shi et al., 2016b). In addition, the intermediates involved in the PPIISF→PPISF transitions are similar, in terms of CCS, to those observed in solution, thus some transitions observed in water, that is, wet folding conditions, are accessible in vacuo. Conversely, the SF PPI→PPII transition does not occur, which underscores the role of water as the major factor promoting trans proline.
IV. EFFECTS OF HYDRATION ON CONFORMER PREFERENCES: TRACKING HYDRATION DURING THE TRANSITION FROM SOLUTION TO THE GAS PHASE
Experimental and theoretical studies underscore the importance of both long- and short-range interactions between ligands/solutes and biological molecules (Ladbury, 1996; Bogan & Thorn, 1998; Levy & Onuchic, 2004; Papoian et al., 2004; Levy & Onuchic, 2006). However, a detailed understanding of the effects of water on protein structures, stabilities, dynamics, and functions is largely confined to the effects of bulk solvent and specific cases where a limited number of localized water molecules, referred to as “biological water” (Nandi & Bagchi, 1997; Pal, Peon, & Zewail, 2002), interact with the protein backbone and/or specific amino acid side chains (Halle, 2004; Rodier et al., 2005; Nucci, Pometun, & Wand, 2011; Duboue-Dijon & Laage, 2014). The ability to experimentally probe the effects of solvation on peptide and protein structure across the range of “bulk-like” water (where species are hydrated beyond the first solvent shell) to a small number of water molecules has the potential to provide new insights as to the role water plays in biological structure and functionality. Cryo-IM-MS is uniquely suited to explore these considerations as the “kinetically-trapped” structures which have been evaporatively “freeze-dried” in various hydration states by ESI are preserved by its 80 K drift tube (DT; Fig. 5), allowing the analysis of hydrated ions (May & Russell, 2011; Silveira et al., 2013a, 2013b; Servage et al., 2015c), including the determination of their CCS from their arrival time distributions (ATD) after traveling through the DT.
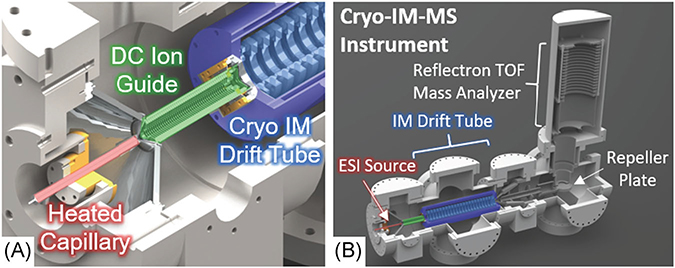
FIGURE 5.
Solidworks rendering of (A) the cryo-IM-MS source and (B) instrument with major components labeled. Ions generated by electrospray ionization are transferred to the heated capillary (red), which is heated to control the degree of hydration. Ions are then passed through the DC ion guide (green) and into the cryogenic IM drift tube (light blue). The kinetically trapped, hydrated ions are preserved as they pass through the drift tube by using drift gas precooled by liquid nitrogen circulating through the Dewar jacket (dark blue). After leaving the drift tube, the ions are detected by TOF mass analyzer. Instrument details are described in further detail elsewhere (Silveira et al., 2013b). TOF, time-of-flight.
Recent studies bring to the forefront two key roles water plays in affecting ions in confined nanodroplets. First, water has been shown to play a stabilizing role in forming like-charged ion complexes. Scheraga et al. (Magalhaes et al., 1994; No, Nam, & Scheraga, 1997) showed that protonated guanidine (GdmH+) side-chains of surface arginine residues were interacting, implying water plays a role in stabilizing like-charge ion pairing; Vazdar et al. (2011) showed with MD that as few as 12 water molecules were required to stabilize the ion pair. Cryo-IM-MS shows a distinct fall-off in relative abundance of the like-charged GdmH+-GdmH+(H2O)n ion pairs with decreasing numbers (below ~n = 55) of water molecules adducted, highlighted by the red box in Figure 6A, suggesting that water plays a stabilizing role (Hebert & Russell, 2019) likely by screening the excess charge and supplying vital H-bonding to stabilize the clusters, as suggested by Vazdar et al. (2011). These results imply that water can mitigate coulombic repulsions, suggesting a mechanism for the stabilization of highly charged, albeit covalently bound peptides and proteins seen in previous cryo-IM-MS experiments, which would support the long-held belief that coulombic repulsion causes biomolecules to unfold (Servage et al., 2016). For instance, stepwise dehydration of hydrated SP (RPKPQQGLM) [SP + 3H]3+ ions produces a compact conformer, denoted A, that has been shown to unfold to a more extended conformer only after complete dehydration of its highly charged (NH3-RPK)3+ region to form the extended B conformer (Silveira et al., 2013a). Likewise, noncovalent ubiquitin (Ubq) dimers dissociate only after near-active complete dehydration (vide infra) (Servage et al., 2015b). The second role water plays, which is related to the first, is the facilitation of charge delocalization. A recent investigation of hydrated 4-aminobenzoic acid (4-ABAH+) hydrated ions revealed that water plays a key role in transferring the proton originally located on the –NH3+ to the –COHOH+ group in sparsely hydrated systems, as shown in Figure 6B and C (Hebert & Russell, 2020). The proton transfer through the water bridge can be inhibited by the presence of polar molecules (such as acetonitrile) that directly interact with the charged region to block the water bridge. In the absence of water, biomolecules must rely only on rearrangement of charge carriers, which can introduce very high charge transfer barriers for sterically hindered regions. Intramolecular proton transfer reactions negatively impact the formation of native-like structures (via different salt bridges, hydration structures, and intramolecular charge solvation) formed by ESI, altering observed gas-phase structures (Wyttenbach, Liu, & Bowers, 2005; Wyttenbach & Bowers, 2009; Xiao, Perez, & Russell, 2015). The following section displays results from cryo-IM-MS experiments that further illustrate these points; these examples demonstrate the utility of kinetic trapping using freeze-drying ESI and cryo-IM-MS to study peptide and protein ions.
FIGURE 6.
(A) ATD is plotted as a function of m/z for a mixture of GdmH+(H2O)n (denoted 1+) with GdmH+(H2O)n and GdmH+-GdmH+(H2O)n (denoted 2+). The red box highlights where the ion abundance begins to decrease with decreasing hydration numbers. The inset shows a proposed structure of GdmH+-GdmH+(H2O)12. (B) ATD vs m/z plot of 4-ABAH+(H2O)n sprayed from 0.1% formic acid in H2O showing the change in the trendline at the n = 6 proton transfer from –NH3+ to –COHOH+. (C) ATD vs m/z plot of a mixture of H+(H2O)n, 4-ABAH+(H2O)n, and NH3 +C6H5(ACN)1(H2O)n clusters labelled in blue, black, and red, respectively, sprayed from 0.1% formic acid in 1:1 ACN/H2O. This plot shows no clear change in the trendline suggesting the –COHOH+ conformer persists. *denotes 94 m/z (CO2 loss) and **denotes 120 m/z (H2O loss). Adapted from Hebert and Russell (2019) and Hebert and Russell (2020). ATD, arrival time distribution.
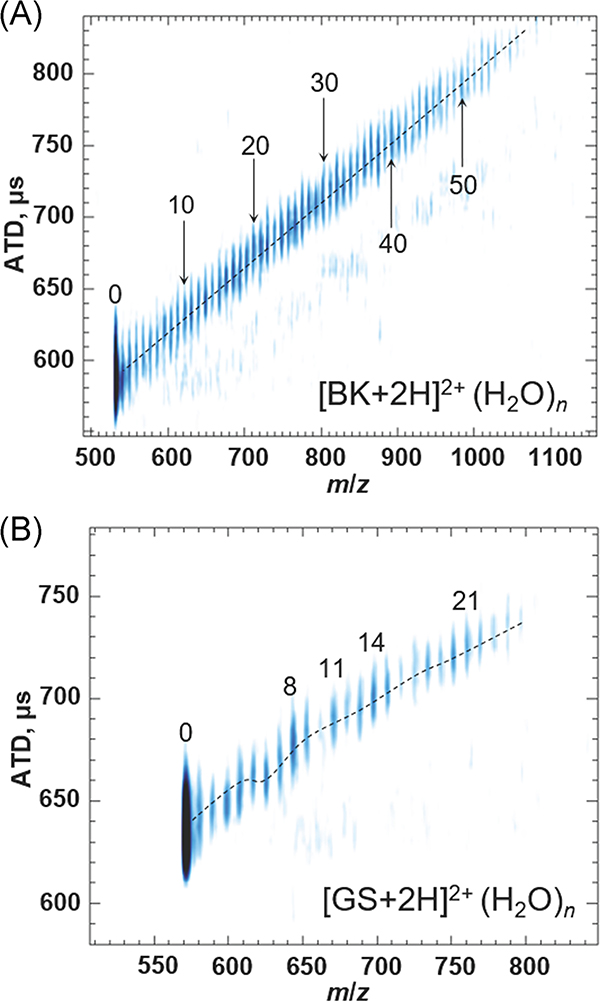
Hydration-dependent conformer preferences of model peptides illustrate the unique capabilities of ESI-cryo-IM-MS for studies of water and its effects on conformational preferences of biomolecules. Such studies also complement results obtained by cold-ion spectroscopy (Papadopoulos et al., 2012; Nagornova, Rizzo, & Boyarkin, 2012). Cold-ion spectroscopy measures spectroscopic signatures of the solute, whereas cryo-IM-MS provides direct measurements of the 3-D shapes as well as conformational heterogeneity of the ion population. The various effects of hydration on such conformational heterogeneity are illustrated in Figure 7 which contains hydration data for BK (RPPGFSPFR) and gramicidin S (GS; cyclic-(VOLFP)2) (Silveira et al., 2013b; Servage et al., 2016). Although both peptides are detected as [M + 2H]2+ ions, BK2+ hydrates are observed for n > ~60 whereas the GS2+ hydrate ions drop off sharply at n = ~26 (Rodriguez-Cruz, Klassen, & Williams, 1997). ATDs increase monotonically for BK2+ ions, suggesting the structure of BK2+ does not change as a function of n. However, the ATD for GS2+ ions shows significant variations that are correlated with the previously reported “magic numbers” (n = 8, 11, 14) ions, which are likely affected by the structures formed with the charged ammonium groups (Rodriguez-Cruz, Klassen, & Williams, 1997; Nagornova, Rizzo, & Boyarkin, 2012). These magic number clusters for GS2+ have also been reported by Beauchamp and coworkers (Lee et al., 1998). The n = 14 may represent the maximum water:charge ratio of 7, as indicated by gas-phase peptide hydrations reported by Liu et al. (2003). Several decreases in ATD suggest reoccurring linkages of the cyclic peptide, probably through water bridging of the ornithine groups first (n = ~4–6) followed by other functional groups (n = ~10–11). CCS characterization of this region will enable a better understanding of the currently circumstantial evidence of magic number cluster formation (Silveira et al., 2013b; Hebert & Russell, 2020). BK2+ ions can have a more complex charge distribution, viz. a protonated N-terminus, two arginine ions (RH+), and a deprotonated C-terminus, or two charges may reside on the two RH+ groups owing to high proton affinities of the guanidine groups (Strittmatter & Williams, 2000). The extra charge sites may lead to formation of a stable structure, although the charge distribution might shift as a result of hydration. In contrast, the charges on GS2+ ion reside exclusively on ornithine (O) side chains. Since the –NH3+ ions of GS2+ are small and the solvent shell is expected to be well ordered, variations in ATD vs m/z plots are attributed to different structures for the water molecules solvating the –NH3+ ions (vide infra).
FIGURE 7.
Cryo-IM-MS plots of (A) bradykinin peptide and (B) gramicidin S. BK displays a typical example of the linear desolvation patterns of larger molecules, whereas GS shows distinct shifts in the mobility and magic number clusters. Adapted from Servage et al. (2016) and Silveira et al. (2013b). BK, bradykinin; Cryo-IM-MS, cryogenic ion mobility-mass spectrometry; GS, gramicidin S.
V. Water-mediated conformer preferences of hydrated SP3+ and SP3+ mutant ions
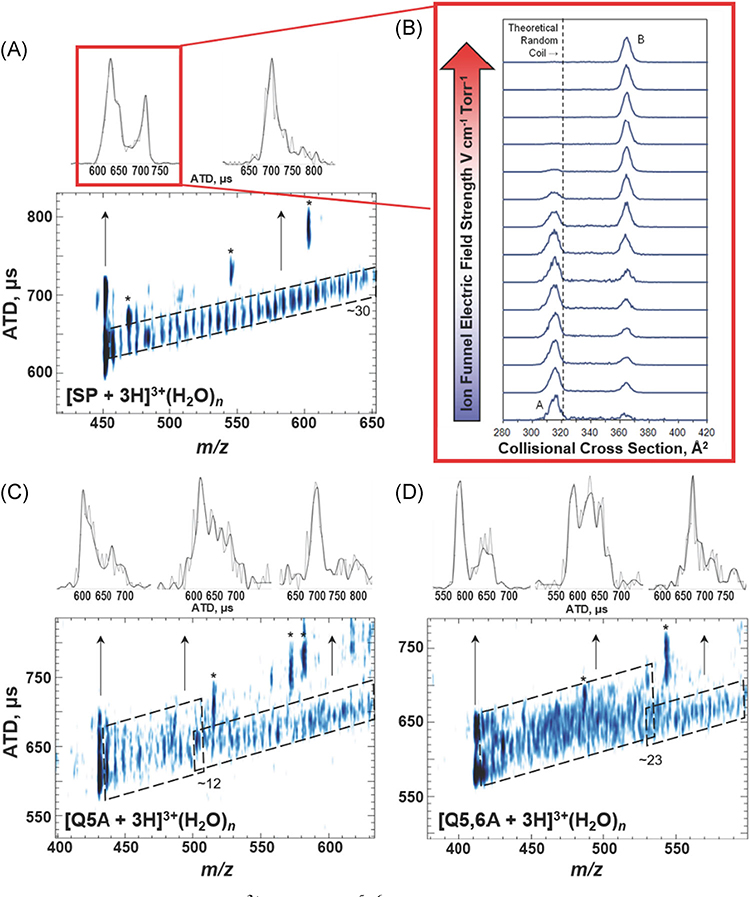
It is interesting to compare hydration of BK2+ and GS2+ with that observed for SP3+ ions (amino acid sequence RPKPQQFFGLM) owing to differences in the numbers of hydrophilic side chains. The N-terminus of SP3+ is quite hydrophilic (RPKPQQ) whereas the C-terminus (FFGLM) is hydrophobic. Figure 8A contains the first experimental data that trace ion formation from a bulk-like solvent environment to a “solvent-free” gas-phase ion (Silveira et al., 2013a). SP3+ ions are detected as [M + 3H]3+(H2O)n, where n > ~30, and the hydrated ions undergo stepwise loss of H2O monomer to yield a compact [M + 3H]3+ ion, denoted conformer A, which upon collisional heating rearranges to conformer B (see Fig. 8B). Thus, conformer A is a “kinetically-trapped” solution-like state of SP, and conformer B is the thermodynamically favored gas-phase conformer (Silveira et al., 2013a; Fort et al., 2014) MD simulations and IM-MS ATDs of SP mutant ions (SPM) show that intramolecular interactions involving the N-terminal charge sites (N-terminus, R and K side chains) and side chains of Q at position 5 and 6, and to a lesser degree F at position 7 and 8 (pi-cation interactions), facilitate “kinetic trapping” of conformer A (Fort et al., 2014).
FIGURE 8.
(A) ATD vs m/z for SP3+ ions (RPKPQ5Q6FFGLM). (B) Plot of CCS as a function of the electric field strength of two [SP + 3H]3+ conformers CCS, illustrating the conversion of species A to B under collision induced unfolding conditions. The theoretical random coil trendline (321 Å2) is shown with a dashed line. (C) and ATD vs m/z for SP mutant Q5A and (D) Q5,6A. All peaks labeled with an asterisk correspond to fragment ions observed at higher capillary temperatures. The upper panels contain extracted ATD for specific m/z ranges as indicated by the arrows. The black ATD lines are the result of plotting every other data point while the full data set is plotted in gray. Note that the differences in the peak widths of the extracted ATDs reveal conformational heterogeneity for the ions at each m/z ratio. While the plots of ATD vs m/z were collected using an 80 K drift tube, the plot of ion funnel electric field strength versus CCS was collected under ambient conditions. Adapted from Servage et al. (2015a) and Silveira et al. (2013b). ATD, arrival time distributions; CCS, collision cross section.
The hydration data for SP Q5A and Q5,6A mutants are quite different from that for SP3+ ions seen in Figure 8A which closely resemble the data from BK2+. In Figure 7C, while the ATD for hydrated SPM3+ Q5A ions is relatively narrow for n > ~12, the ATD for SP Q5A with n < ~12 are scattered, indicating a more heterogeneous conformer population, comparable to what is observed for the GS2+ hydration trendline. A similar change in the ATDs is observed for the Q5,6A mutant for n < ~23 in Figure 7A (Servage et al., 2015a). These results are interpreted as an “order” to “disorder” transition that occurs at a specific number of water molecules, which is taken as evidence that the conformation of the hydrated peptide is dictated by solute/solvent interactions. The dependence on the numbers of H2O molecules is attributed to restrictions on the motion of the peptide backbone imposed by hydration of the hydrophilic peptide side chains, viz. the N-terminus, side chains of R and K as well as stabilizing interactions of Q at position 5 and Q at position 6 (Halle, 2004; Mattea, Qvist, & Halle, 2008; Halle & Nilsson, 2009). In addition, the narrowness of the ATD for the hydrated ions in the range n < 12 of Q5A and n < 23 of Q5,6A suggest that these ions may also have ordered conformers.
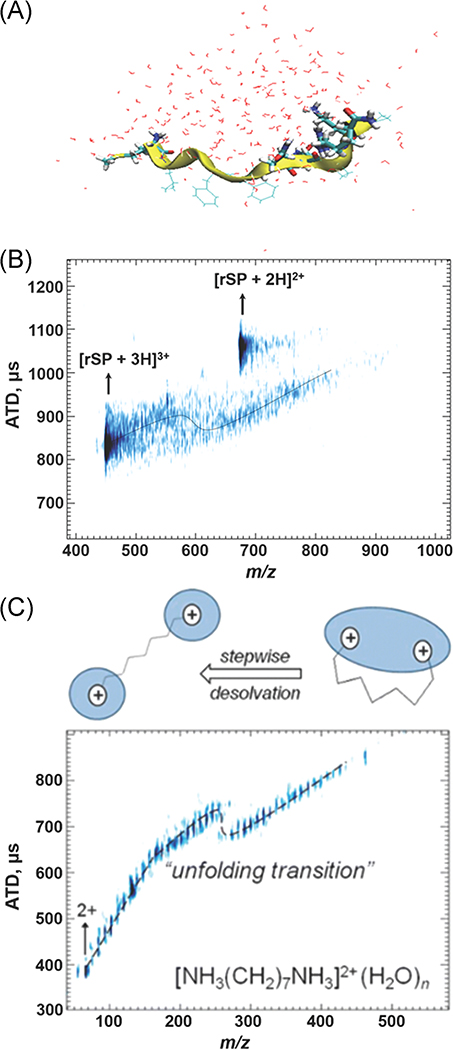
Unpublished data from the Russell group for retro-sequence SP (rSP; MLGFFQQPKPR-NH2) shown in Figure 9A is compared to SP, where influences of coulombic repulsion are quite different; the protonated N-terminus is well separated from the protonated side chains of R and K. The shift in the ATD of the hydrated rSP3+ ion that is observed at n ~ 30 is indicative of a significant change in the conformation of this system (Fig. 9B).
FIGURE 9.
(A) MDS of hydrated rSP. (B) m/z vs ATD plot of rSP showing the conformational change undergone by increased desolvation. MDS of solvated rSP ion (C) m/z vs ATD plot of 1,7-diammoniumheptane demonstrates the impact water has on structure on solvated ions, with inset schematic of changes in solvation motif with dehydration. Adapted from Kim et al. (2017); Servage et al. (2015a). ATD, arrival time distributions; rSP, retro-sequence substance P.
Results from MD simulations are consistent with the solvation and water bridging of the charge centers rather than the backbone (Fig. 9A). In the case of the diammonium alkyl cation, a similar phenomenon is seen with both ammonium ions contained in a single water droplet for n > 18, but at n < 18 the two –NH3+ groups are contained in individual droplets (Servage et al., 2015a). This transition, seen in Figure 9C, is driven by coulombic repulsion, analogous to the case of GdnH+-GdnH+(H2O)n. Here, however, since the charge centers are connected, a resultant shift in the ATD vs m/z for [rSP + 3H+]3+ hydrated ions is observed. This is indicative of a transition from a more compact state to a more extended conformer, that is, from a single water network hydrating the charged N-terminus and the charged K and R residues to separate water networks hydrating each charged region.
Kim et al. (2017) used MD simulations to model changes in the conformational preferences of SP3+ ions during ESI, specifically conformational changes that occur during the transition from a charged nanodroplet to the SF gas phase ion as shown in Figure 10. The initial charged droplet contained ~2,400 water molecules, 22 hydronium ion, 10 chloride ions, and a single SP peptide. Droplet shrinkage involved water evaporation and loss of excess charge by the ejection of hydronium ions. Further droplet shrinkage occurred by evaporation followed by fission events as well as the loss of Cl− ions. The conformation of the SP3+ ions responded to changes in droplet size by small changes in conformation and migration of the hydrophobic C-terminus to regions of the droplet near the surface. The hydrophilic N-terminus remained buried in the droplet interior by water-mediated interactions with the H3O+ and Cl− ions. The final conformations obtained from the simulations are shown in Figure 10A-iii and B-iii. The simulations suggest that interactions between SP3+ and Cl− are retained until the final stages of ESI droplet evaporation; however, evidences of these adduct ions are not observed in the mass spectra. Presumably, Cl− is lost owing to mild collisional heating or by elimination of neutral HCl, that is, charge-reduction reactions (Mirza & Chait, 1994). Subsequent studies present additional evidence that Cl− is involved in preserving compact conformers. Although CCS profiles for both protonated SP and melittin electrosprayed from water and water/0.1% HCl solution are indistinguishable, the abundances of more compact conformers are higher for melittin-Cl adduct ions (Servage et al., 2015a; Kim et al., 2017).
FIGURE 10.
(A, B) Plots of the evolution of two fully elongated structures showing the (raw data, black line; smoothed, yellow line) CCS of SP3+ ions and (blue points) numbers of water molecules vs time extracted from selected simulations. The experimentally determined CCS of SP3+ (316 Å2 orange dash and 368 Å2, green dash) is also shown for reference. Structures labeled as (i−iii) depict representative snapshots of the simulations (A, B): (i) the post-fission compact structures observed at 2000 ps, (ii) the elongated structures observed later in the simulation, and (iii) the final frame of the desolvation simulation. Blue dots represent water molecules, purple spheres represent Cl−, and H3O+ are shown in green. Figure is from Kim et al. (2017). CCS, collision cross section.
Insight into the interactions of hydrated proteins with Cl− may be gleaned from a follow-up study by Wagner et al. which showed that ESI of Ubq from acidified (HCl) solutions produced abundant ions having the general formula, [M + nH + xCl](n − x)+, and these ions eliminate HCl when subjected to mild CA, Figure 11A–C (Wagner, Kim, & Russell, 2016; Kim et al., 2017; Oh & Consta, 2017). Moreover, increased activation energy was required to unfold anion-protein complexes to their more stable gas-phase structures. This suggested that introducing anion interactions may cause solution-phase structures to be more easily trapped in the gas phase due to reduced coulombic repulsion. More importantly, the CCS of the product ions of chloride adduction are similar to those of the “native state” of the protein. Another prominent series of adduct ions, viz. [M + nH + xCl + yH2O](n − x)+, was also observed. While the relative abundances of these ions increased with increasing numbers of Cl− anions, the water adducts had no measurable effect on the CCS profiles. Thus, it was proposed that the water molecules interact with the anions rather than the protein.
FIGURE 11.
(A–C) Heat maps showing the effect of collisional activation on the CCS profiles of [M + nH + xCl](n − x)+ ubiquitin ions, with a total charge of 5+, 6+, and 7+ (A–C, respectively). The CCS profiles observed using a collision voltage of 5 V is shown to the left of each map. Adapted from Wagner et al. (2016). CCS, collision cross section.
Ignoring the electrostatic role of anions has vastly simplified positive-ESI-IM-MS spectra, but anions interact directly with proton sites, playing a key role in reducing the activation of proteins/protein complexes. These studies are most relevant to contemporary issues frequently encountered in native MS, most notably peak broadening associated with large proteins and protein complexes where the high m/z tails of the peaks can be trimmed by CA (Benesch, 2009). The results noted above potentially explain why the CCS for these signals are not sensitive to the presence of adducted species, that is, salts and water. Additional studies addressing these issues are needed, especially studies utilizing high-resolution IM and MS measurements (vide infra).
VI. WATER-MEDIATED DIMERIZATION OF Ubq
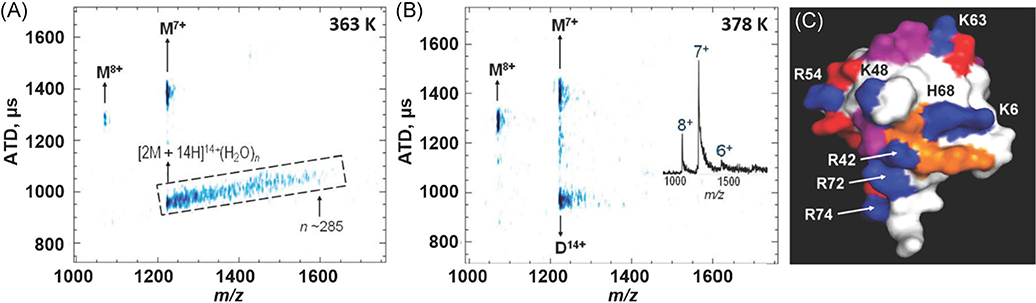
The extent of hydration of model peptides described above shows a dependence on the number of charged functional groups, and it is expected that larger peptides and proteins would have a greater capacity for hydration. The question is raised whether ESI-cryo-IM-MS can be used to directly measure, or at a minimum provide a more accurate estimate, of the solvent-accessible surface area (SASA; Servage et al., 2015b). Results from MD simulations on Ubq (~8 kDa) estimated that the first solvation shell is composed of ~187 water molecules (Tompa et al., 2009). Surprisingly, cryo-IM-MS detected relatively few water molecules (~10) associated with Ubq [M + 7H]7+ monomer ions, whereas hydrated Ubq dimer ions ([2 M + 14H]14+(H2O)n) with n ~ 285 were detected, which is considerably less than 374 assuming the SASA of two monomers. The decreased numbers of water molecules observed for the dimer versus the number of water molecules based on the SASA of two monomers is consistent with a dimer formed by face-to-face contacts involving the hydrophobic patch (Nucci, Pometun, & Wand, 2011).
The formation of the Ubq dimer has been described as water-mediated, meaning that water plays a key role in this process. The hydrophobic patch of Ubq formed by the side chains of Leu-8, Ile-44, and Val-70 (shown in orange in Fig. 12) meet the necessary criteria for a protein-protein binding “hot spot,” including the required occlusion of water to nearby hydrophilic sites (R42, K48, H68, R72, and R74; see Fig. 12). A similar rationale was described by Liu et al. (2012) and Wand and coworkers have shown that the water molecules solvating these hydrophobic sites exhibit restricted hydration dynamics (Nucci, Pometun, & Wand, 2011). Further evidence supporting the proposed mechanism for water-mediated formation of the Ubq dimer comes from studies on the CIU of the noncovalent dimer and the K-6, K-11, K-48, and K-63 covalent dimers. The K-48 dimer is known to contain strong interactions involving the I-44 hydrophobic patch, and the CIU heat maps for the K-48 and noncovalent dimer are almost identical (Wagner & Russell, 2016).
FIGURE 12.
ATD vs m/z plot for ubiquitin at heated capillary temperatures of (A) 363 K and (B) 378 K, and (C) an annotated structure of the native state of ubiquitin. The hydrophobic patch (orange) and surrounding basic (blue), acidic (red), and glutamine (purple) residues are shown. The hydrophobic patch (L8, I44, V70) is surrounded by K6, K11 (not visible), R42, K48, H68, R72, and R74 residues, which are more solvent accessible and may serve as initiators of dimer formation. Adapted from Servage et al. (2015b).
Invoking formation of an occlusion complex to explain water-mediated dimers of Ubq raises questions about possible coulombic repulsion within confined regions where a substantial fraction of the hydrophilic side chains (R42, K48, H68, R72, and R74) are charged. As described, this is an archetypal model for a protein “hot spot” in which a hydrophobic core is surrounded by a hydrophilic, charged peripheral region. A similar “hot spot” also appears in the GdmH+-GdmH+(H2O)n complex described earlier. Loss of hydration is what causes dissociation in both cases; the occlusion of water forms the thermodynamic impetus for dimerization (hydrophobic effect), but the hydrophilic peripheral regions appear necessary to maintain these dimers.
Cryo-IM-MS excels at probing the effects of solvation on the structure of ions. This unique approach reveals the crucial role of water in the regulation of structure, and thus function. However, many other physiological factors, for example, crowding, mutation, temperature, protein interaction, and ligand can influence the structure which is the main focus of the following section. Thermal changes, as well as interactions with and between these solutes, can not only define adduction but also affect whether the bound protein undergoes structural and thus functional change. The CIU technique is uniquely suited to investigations of these phenomena. In the following section, this technique is used to investigate structural changes based on metal binding interactions and the effects of tagging and dimerization linkages.
VII. STUDIES OF NATIVE AND NON-NATIVE PROTEIN FOLDING: TRACKING INTERMEDIATES DURING THE UNFOLDING PROCESS
In recent years, there has been growing interest in the development of MS-based methods for studies on the folding/unfolding of biomolecules and biomolecular ions. In the following subsections, two MS approaches for probing protein folding are discussed: (i) CIU (Ruotolo et al., 2007; Dixit, Polasky, & Ruotolo, 2018) and (ii) thermal melting of proteins in solution through variable-temperature electrospray ionization (VT-ESI; Benesch, Sobott, & Robinson, 2003). CIU reports on the energies required to unfold gas-phase ions as reported in a recent paper that found quantitative barriers for CIU can be obtained using approaches previously developed for blackbody infrared radiative dissociation (Donor, Shepherd, & Prell, 2020). On the other hand, thermal melting of a protein in solution is better described as a “folding/refolding” equilibrium reaction that occurs upon changes in the chemical potential that serve as driving forces for thermal unfolding/refolding reactions. For solution-phase melting the driving forces for refolding are the changes in the dielectric constant, ranging from ~78 at 25°C to 56 at 100°C for water (Malmberg & Maryott, 1956) as compared to ~33 at 25°C for methanol (Cunningham, Vidulich, & Kay, 1967; Shirke et al., 2001). As the protein responds to these changing forces, the new state formed under these conditions is the native state. For CIU studies, the folding/unfolding terminology is correct as the products sampled in this reaction are under kinetic control; thus they are not in equilibrium with the local environment and cannot refold. The CIU results provide a means for determining the energetics of the unfolding reactions limited by the time frame on which the measurements are sampled. Combining results of VT-ESI and CIU studies provides information regarding the behavior of protein structure in non-native states which are thermodynamically unfolded/refolded into equilibrium states and/or collisionally excited in kinetically trapped states.
VIII. STABILITIES OF GAS-PHASE PROTEIN IONS: CIU
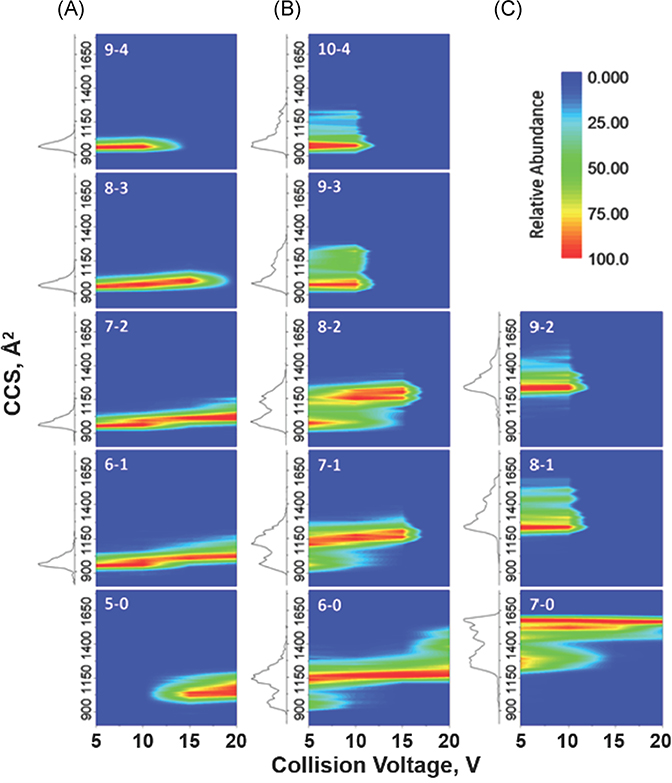
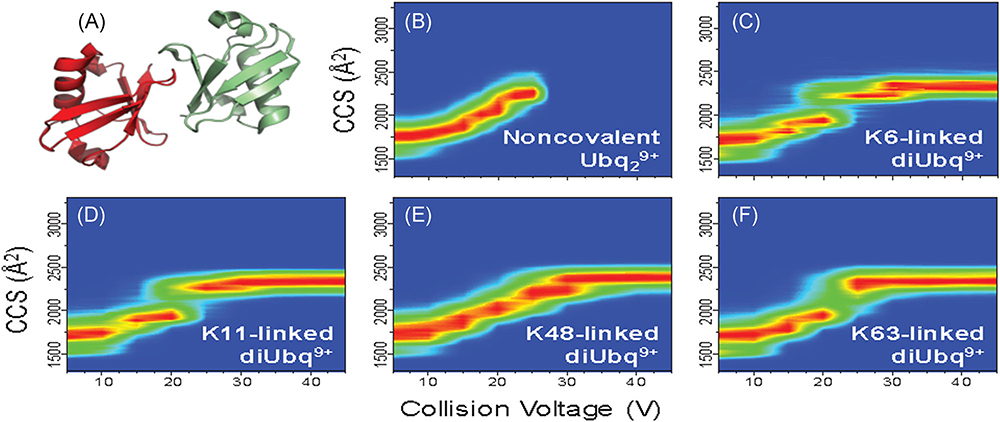
A major aim for native IM-MS is to characterize biomolecules and biomolecular ions based on conformational preferences, that is, secondary and tertiary structure. CIU, which measures changes in CCS of the ions as a function of collisional heating, can be used to compare the stabilities of the gas-phase ions (Dong et al., 2020). Ubiquitylation, a posttranslational modification, is an important cellular process that encodes an array of cellular functions (Komander & Rape, 2012). Native IM-MS and CIU were employed to better understand differences in subunit interfacial interactions for noncovalent Ubq dimers by studying conformational preferences for K6-, K11-, K48-, and K63-linked covalent ubiquitin dimers (diUbq; Wagner, Clemmer, & Russell, 2017). The four diUbq are not easily differentiated based on IM CCS as they all have strong IM features with CCS of ~1,700 Å2 when sampled under minimally activating instrument conditions (Chen & Russell, 2015a). However, the CIU heat maps for the diUbq ions show distinct differences (Fig. 13B–F) and unfolding pathways with unique linkage-dependent features. Comparison of the CIU heat map of noncovalent ubiquitin dimers (Ubq29+; Fig. 13B) to those of the lysine-linked covalent ubiquitin dimers (diUbq9+; Fig. 13C–F) revealed similarities between unfolding of noncovalent Ubq dimer and K48-linked diUbq (Fig. 13A), suggesting similar gas-phase structures and dimer interfacial interactions.
FIGURE 13.
CIU heatmaps of ubiquitin dimers [2M + 9H]9+ from acidified solutions (0.1% formic acid). (A) NMR structure of K48-linked ubiquitin dimer (PDB 2PEA). (B) CIU of the noncovalent ubiquitin dimer shows unfolding from a from ~1,750 A2 to ~2,300 A2 before dissociation at a collision voltage of 25 V. CIU heatmaps of (C) K6-, (D) K11-, (E) K48-, and (F) K63-linked covalent ubiquitin dimers show similar conformer distributions at low collision voltages yet distinct linkage-dependent unfolding. Similarities between the unfolding pathways of noncovalent and K48-linked ubiquitin dimers (B and E, respectively) suggest the noncovalent dimer adopts similar subunit interfacial interactions to the K48-linked covalent ubiquitin dimer. CIU heatmaps are reproduced from references Wagner and Russell (2016); Wagner et al. (2017). CIU, collision-induced unfolding; NMR, nuclear magnetic resonance.
IX. FOLDING/REFOLDING PROTEINS IN SOLUTION USING VT-ESI
Protein melting (El-Baba et al., 2018) or thermal heating of the solution (Wang, Bondarenko, & Kaltashov, 2018; Kohler et al., 2019) within the ESI emitter provide new approaches for determining the thermal stabilities of peptides, proteins, and even protein complexes. The combination of VT-ESI with IM-MS affords the ability to investigate structural transitions (refolded states) of biomolecules by kinetically trapping folding intermediates in the gas phase. VT-ESI-IM-MS has been used primarily to study soluble proteins and protein complexes but has also been used for thermodynamic studies of lipid binding to membrane protein complexes (Cong et al., 2016).
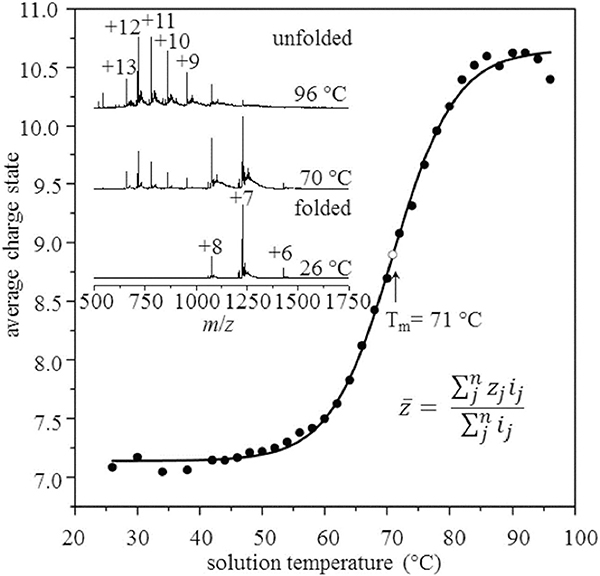
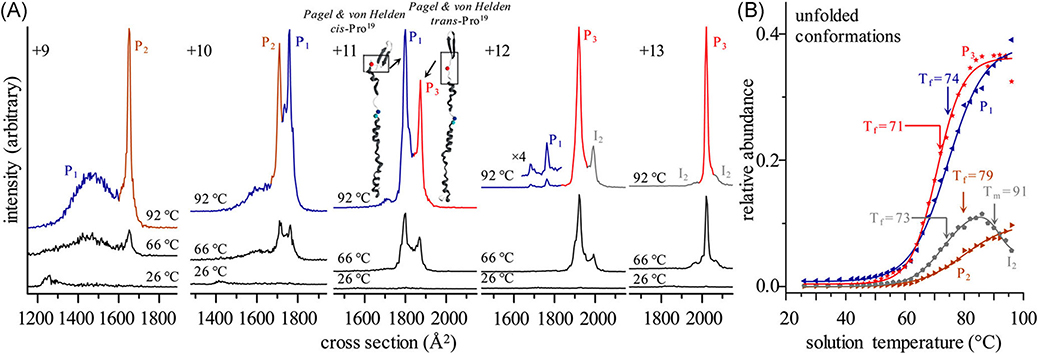
The utility of VT-ESI for studies of protein folding/refolding was initially illustrated using monomeric ubiquitin (Ubq; El-Baba et al., 2017). Melting curves shown in Figure 14 are consistent with a two-state, cooperative unfolding transition having a melting temperature of ~71°C (Wintrode, Makhatadze, & Privalov, 1994) however, the IM-MS data shown in Figure 15 reveal the presence of several intermediate states (i.e., conformers) involved in the thermal melting reaction. As examples, [Ubq + 9H]9+ ions form two conformers, labeled as P1 (blue trace) and P2 (brown trace), with P2 increasing in abundance as the temperature is increased from 26°C to 96°C. For the [Ubq + 11H]11+ state, the broad signal for the P1 (blue trace) is still observed, but more importantly, a new conformer is observed that is assigned to a different configuration of the proline at position 19 (P3, red trace); consistent with cis/trans Pro19 previously assigned by Pagel and von Helden (Warnke et al., 2014). Careful analysis of all the IM-MS data, showed that thermal unfolding of the various charge states of Ubq occurs by “refolding” reactions involving at least nine unique conformer states: three native or native-like states, four states at higher temperatures, and two states that differ based on cis/trans configuration about the Glu18-Pro19 peptide bond.
FIGURE 14.
The average charge state for ubiquitin (10 μM in acetic acid, pH 3.0) was determined from the spectra shown in the upper left by taking the weighted average of charge state as function of solution temperature. The midpoint at Tm = 71°C is in excellent agreement with 71 ± 2°C reported by Wintrode et al. (1994). Reproduced from El-Baba et al. (2017).
FIGURE 15.
(A) CCS distributions for [M + 9H]9+-[M + 13H]13+ions of ubiquitin at various temperatures. Traces are shown in different colors when IMS peaks for different charge states show indistinguishable temperature profiles. (B) Relative abundance profiles as a function of temperature for each configuration reveal three distinct solution products (P1, P2, P3) and one high-temperature equilibrium intermediate (I2). Relative abundances of these identified conformers show distinct freezing curves as these conformers form from the unfolding/refolding of the compact, native ubiquitin ions at elevated temperatures. Reproduced from El-Baba et al. (2017). CCS, collision-induced unfolding.
Melting curves for myohemerythrin obtained with VT-ESI (shown in Fig. 16) are very different from any other system. The native state favors a four-helix bundle motif and a bridging diiron oxo cofactor that binds oxygen, and at ~35°C some bound oxygen dissociates. A melting transition (loss of cofactor) is observed at ~66°C giving rise to both folded and unfolded apoprotein. At ~85°C the folded apoprotein dominates and the IM-MS data reveal evidence (shift in the CCS profile and a mass loss of 2 Da) of the formation of a non-native disulfide bond at high temperatures. The Tm values obtained by VT-ESI are in excellent agreement with those obtained by CD spectroscopy (64.5 and 67.0°C at λ222nm and λ209nm, respectively) (Woodall et al., 2019). This example illustrates the increased chemical information obtained from VT-ESI coupled with IM-MS: (i) the thermally induced loss of oxygen and the cofactor were not observed using CD spectroscopy, and (ii) the stabilization of the final product results from the formation of a non-native disulfide bond.
FIGURE 16.
(A) Melting curves for myohemerythrin (20 μM in 30 mM ammonium acetate, pH 6.8) show a unique unfolding and refolding pathway dictated by a structural rearrangement and formation of a non-native disulfide bond. Inset mass spectra show shifts towards higher charge and transition from holoprotein (filled circles) to apoprotein (open circles) with increasing temperature, followed by a shift towards lower charge state following the formation of the nonnative disulfide bond at high temperature. (B) Structures of the products formed by melting are shown along with respective CCS profiles and MS spectra. Reproduced from Woodall et al. (2019).
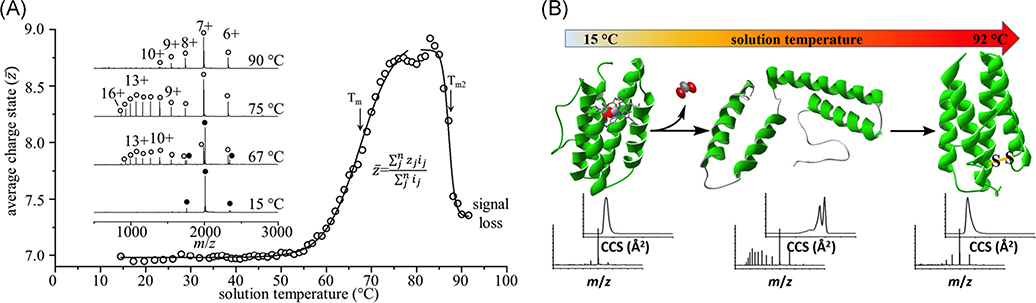
While VT-ESI methods have been used to explore monomeric protein folding/unfolding and infer details regarding 2° and 3° structure dynamics, recent reports have shifted focus towards VT-ESI of protein complexes to study the effects of temperature on the quaternary structure using hemoglobin and concanavalin A as examples. In the case of hemoglobin, VT-ESI-IM-MS was used to track the melting transition of the heterotetramer (α2β2) to the heterodimer (αβ), which ultimately forms aggregates at temperatures above 60°C. New insights for the melting were observed by VT-ESI-IM-MS, viz. both the tetramers and dimers form less compact conformers at elevated temperatures before temperatures where dissociation dominates (Woodall et al., 2020). VT-ESI-IM-MS was also used to study the folding/unfolding of concanavalin A, a homotetrameric protein complex. The use of VT-ESI-IM-MS to study the effects of thermal unfolding of concanavalin A revealed four conformers of the tetrameric complex with distinct melting temperatures, a detail that could not be observed by conventional, solution-averaged melting measurements (El-Baba & Clemmer, 2019). In addition to the expected shifts in charge states and tetramer dissociation at elevated temperature, Gibbs-Helmholtz analyses of VT-ESI-IM-MS results suggest that changes in specific heat (ΔCp) of identified concanavalin A tetrameric conformers result from solvation of nonpolar amino acids (positive ΔCp) found in conformers stable at low temperature or solvation of hydrophilic residues (negative ΔCp) found in conformers stable at high temperature (El-Baba & Clemmer, 2019). These results suggest that amino acid hydration and changes in SASA are driving forces for protein folding even for protein complexes and provide a means for deeper fundamental thermodynamic analyses of structural dynamics and conformer preference of proteins and protein complexes.
X. COMBINING CIU AND VT-ESI-IM-MS TO PROBE STABILITIES OF PROTEIN COMPLEX: THE EFFECTS OF SEQUENCE TAGS
The breadth of the studies described above clearly illustrate the unique potential for CIU, VT-ESI-IM-MS, and MS/MS for studies of complex protein systems; however, incorporating additional MS-based structural approaches adds to the biophysical characterization of proteins and protein complexes and their labeling with covalent and/or noncovalent modifiers. As examples, our current level of understanding of the structure(s) of partially metalated metallothionein comes from combining covalent labeling of the cysteine side chains with N-ethylmaleimide (Kohler et al., 2019) with top-down and bottom-up proteomics approaches (Chen, Russell, & Russell, 2013; Chen & Russell, 2015b).
A similar case exists for our studies on transthyretin (TTR), a homotetrameric protein complex whose subunits (when partially unfolded) are implicated in fibril formation. TTR aggregation has been shown to be involved in several diseases (Gertz et al., 2019) but the detailed mechanism(s) for aggregation have been elusive (Dunn, 2005), owing to the instrumental inabilities to identify and structurally characterize the transient intermediates (kinetic and/or thermodynamic) that are involved. While X-ray crystallography and NMR have provided an atomistic-level structure for TTR and even the TTR complexes with bound thyroxine (T4), these structural techniques are not well-suited for studies of structurally heterogeneous samples, especially those where low abundance transient intermediates are responsible for misfunction (Woods, Radford, & Ashcroft, 2013).
Another factor that complicates studies of TTR is related to the presence of endogenous metals. For example, TTR is complexed by Zn(II) and functions as a metallopeptidase where three distinct sites have been determined (de C. Palmieri et al., 2010) and prior studies have shown that excess Zn(II) binding decreases retinol transport function and increased rates of fibril formation (Castro-Rodrigues et al., 2011). Moreover, changes in structure/stability occur upon Zn(II) binding which might ultimately influence T4 and/or retinol-binding. New evidence for metal-induced oxidation observed using native IM-MS underscores the importance of rigorous analytical measurements to understand protein behavior (vide infra) (Poltash et al., 2019).
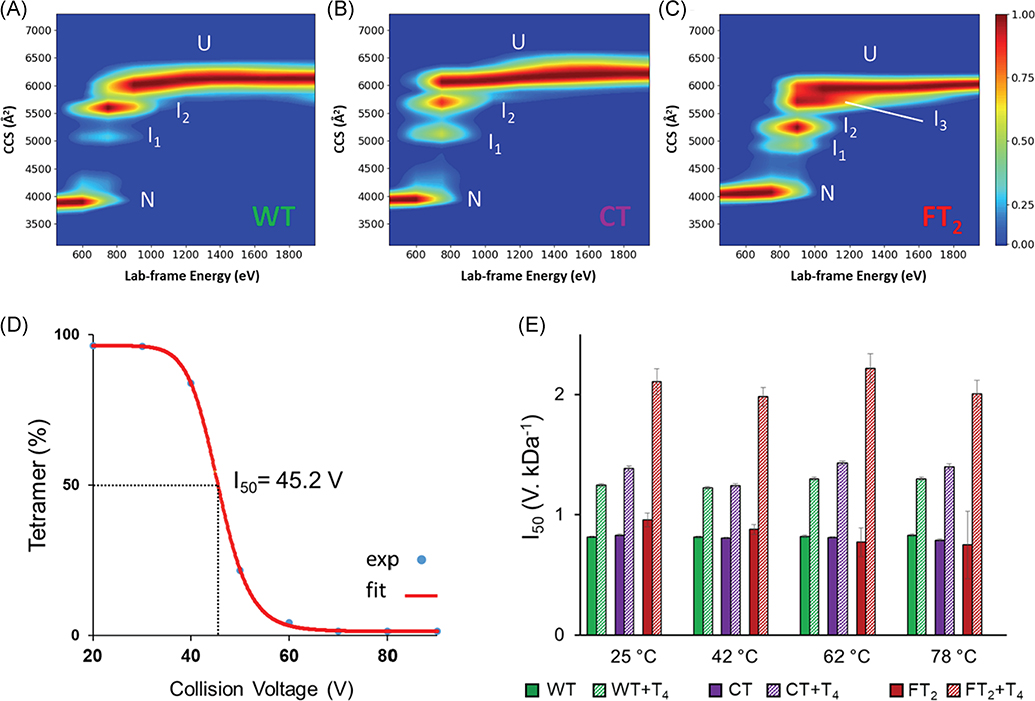
Recombinant TTR is routinely used to study physiological function and structure, a common practice for many other proteins and antibodies. This practice has revolutionized the field of biochemistry, and massive production of recombinant proteins paved the way for the development and commercialization of many protein-based therapeutics. However, despite their invaluable contribution to science and biology to achieve the desired sample quality, the purification requires specific tags that can impact the structure and function of the expressed protein. Cleavage of covalent tags is often time consuming thus the necessity of tag removal remains a challenge for many laboratories. Recently, CIU experiments were utilized to analyze the conformation space the gas-phase stability of two sequence-based tags, C-terminal tag (CT) and dual FLAG-tag (FT2) (Shirzadeh et al., 2020) used in previous TTR subunit exchange studies (Shirzadeh et al., 2019). CT-TTR exhibited similar unfolding energetics to wild type (WT) as well as gas-phase unfolding pathway (two intermediates), whereas FT2-TTR showed a third intermediate and higher gas-phase stability (Fig. 17A–C) (Shirzadeh et al., 2020).
FIGURE 17.
CIU and melting data for WT-, CT-, and FT2-TTR. CIU heatmaps of (A) WT, (B) C-terminal tagged (CT), and (C) dual flag tagged (FT2) show that FT2 tag increases gas- and solution-phase stability of TTR and alters its unfolding pathway in the gas phase as a third intermediate was observed in CIU plot. (D) Solution stability was measured using I50 values corresponding to the energy required to dissociate 50 percent of the tetramer. T4 was used as a control as it is known that it enhances the solution stability of TTR. (E) I50 values for WT-, CT-, and FT2-TTR were plotted as a function of temperature, demonstrating slightly higher stability of FT2-TTR at room temperature which diminishes at higher temperatures. Reproduced from Shirzadeh et al. (2020). CIU, collision-induced unfolding; TTR, transthyretin; WT, wild-type.
Top-down experiments also confirmed a backbone cleavage (Lys9-Cys10) in FT2-TTR at ambient temperature which increased in abundance with temperature (Shirzadeh et al., 2020). Previous studies have shown high thermostability of TTR (Tm > 98°C) (Shnyrov et al., 2000); thus, the secondary and tertiary structure of TTR cannot be responsible for observed cleavage. To dissect the effect of temperature on the quaternary structure of TTR, CID of the tetrameric complex was performed at various solution temperatures using VT-ESI (Fig. 17D). These results revealed a decrease in the stability of FT2 tag with increasing temperature (Figure 17E) but not significant enough to justify the observed cleavage. Detailed analysis of these variants linked the origin of backbone degradation to the metalloprotease activity of TTR which can be inhibited by removing metals. Both CIU and VT-ESI-CID experiments point to structural impact of FT2 tag on TTR which was hidden in previous studies (Rappley et al., 2014; Robinson & Reixach, 2014).
All together, these unfolding studies point to potential applications of native IM-MS and tandem MS to interrogate conformation space and stability of biomolecules. The example provided for backbone cleavage of FT2-TTR further emphasizes that this detailed information cannot be achieved with other traditional techniques such as differential scanning calorimetry, which measures stability analysis indirectly by the heat change in response to protein unfolding.
XI. NEXT-GENERATION IM-MS TECHNOLOGIES FOR STUDIES OF LARGE PROTEINS AND PROTEIN COMPLEXES
Instrument development for native-MS is largely driven by needs to understand the structural details of large protein complexes, viz. solvent manipulation (Shi et al., 2014), effects of protein-protein interactions (Cong et al., 2016), PTMs (Fornelli et al., 2020), stoichiometry (Kaltashov & Mohimen, 2005; Zhou & Wysocki, 2014; Shirzadeh et al., 2019; Stiving et al., 2019; VanAernum et al., 2019), and ligand binding events (Hyung, Robinson, & Ruotolo, 2009; Allison et al., 2015; Cong et al., 2016; Marchand et al., 2018), investigations of which are made possible by recent advances in MS instrumentation (Giles et al., 2019 Poltash et al., 2020). Advances in IM-MS technology and experiments were accelerated by the readily adopted Waters SYNAPT platform. The versatile SYNAPT platform allows for pre-IM ion mass selection allowing for tandem MS experiments that can be further interrogated by the novel traveling-wave IM with additional fragmentation, that is, CID (Mitchell Wells & McLuckey, 2005), SID (Zhou & Wysocki, 2014; Stiving et al., 2019; VanAernum et al., 2019), and electron transfer/capture dissociation (Zhurov et al., 2013). However, perturbations in the structure of native proteins are difficult to study, because the changes in structure may lead to small changes in CCS that are unresolvable on commercial instrumentation (Poltash et al., 2020). To understand the relatively minute structural perturbations for large proteins and protein complexes, new instrumentation needs to be developed to overcome the limitations of commercial instrumentation in terms of insufficient resolving power (RP), that is, peak centroid/separation of two closely spaced peaks) in the IM and m/z domains (RPIM and RPm/z, respectively; Poltash et al., 2020). For example, as shown for TTR, a RPm/z of 840 is needed to resolve Zn binding to the intact tetrameric protein that was previously hidden. Traditional uniform-field drift-tube (UF-DT) IMS systems lack the necessary radial confinement of ions as they traverse the DT (Silveira et al., 2010). This poor confinement results in the diffusional broadening of the ion beam during IM analysis resulting in decreased sensitivity and signal loss. Increased ion transmission has been achieved namely by radially focusing the ions, viz. post-IMS focusing funnels (Bush et al., 2010; Allen & Bush, 2016; Garimella et al., 2017; Ibrahim et al., 2017; Stiving et al., 2020), magnetic fields (Bluhm, Gillig, & Russell, 2000; Bluhm, Gillig, & Russell, 2000; Bluhm, North, & Russell, 2001), segmented quadrupoles (Javahery & Thomson, 1997), and/or periodic-focusing (PF)-DT (Gillig et al., 2004; Verbeck et al., 2004; Silveira et al., 2010; Blase et al., 2011; Gamage et al., 2011; Silveira et al., 2012). Periodic focusing drift tube (PF-DT) radially focuses ions using a distance-dependent waveform (~pseudo kHz) as ions traverse axially through the DT (Gillig et al., 2004; Verbeck et al., 2004; Silveira et al., 2010; Blase et al., 2011; Gamage et al., 2011; Silveira et al., 2012). The distance-dependent electric field leads to an oscillatory ion trajectory increasing the drift times relative to UF-DT for ions of the same size and shape (Verbeck et al., 2004; Gillig et al., 2004; Silveira et al., 2010; Blase et al., 2011; Gamage et al., 2011; Silveira et al., 2012). PF-DT drift times deviate from typical UF-DT, but the mobility term, K, can be accurately accounted for by applying a calculated mobility dampening term, α, from first-principles measurements, as shown by Silveira et al. (2012).
Typically, IMS is coupled to time-of-flight (TOF) MS because the duty cycle of the TOF operates on the microsecond time scale, and the typical mobility separation occurs on the millisecond time scale, allowing for nested MS analysis of ions as they exit the DT. Current commercial IM-MS instruments suffer from low MS resolution (Rm/z) in the TOF with only recent implementations of high-resolution TOFs (e.g., Waters SELECT Series Cyclic IMS or Agilent 6545XT Q-TOF MS, but the latter lacks IM capabilities) (Giles et al., 2019). Alternatively, IM can be coupled to a high-resolution orbitrap platform, but this introduces a duty-cycle mismatch since the orbitrap operates on a timescale of hundreds of milliseconds (Clowers & Hill, 2005; Szumlas, Ray, & Hieftje, 2006; Morrison, Siems, & Clowers, 2016; Poltash et al., 2018). This problem may be mitigated by supplementing the gate at the entrance of the DT with a second gate implemented post-IM to isolate a single drift time of interest to be transferred for mass analysis; this process is then iterated over the entire drift time window. Alternatively, Fourier transform IM (FT-IM) uses a linear chirp waveform to modulate both gates by the same pulse which sweeps a user-defined frequency range over a designated period (Knorr et al., 1985; Morrison, Siems, & Clowers, 2016; Poltash et al., 2018; Poltash et al., 2019). Applying the same pulse pattern to both gates converts the DT into a frequency filter outputting to the orbitrap mass analyzer. FT-IM thus overcomes the duty cycle mismatch by synchronizing the period of the IM frequency sweep with a multiple of the orbitrap duty cycle. The frequency of modulation the ions experience via the FT-IM mode is encoded into their respective time-dependent m/z signals which can be deconvoluted into an ATD.
The combination of the aforementioned PF-DT coupled to the high-resolution capabilities of an orbitrap MS allows for the interrogation of protein structure to a level that has not been previously achieved. The first-generation native FT-IM-PF-DT orbitrap EMR (Fig. 18A), has sufficient RPm/z to resolve apo-TTR ions from the Zn- and thyroxine-containing ions, m/z differences of 4.5 and 55.4, respectively, for TTR (~56 kDa tetrameric protein complex (Fig. 18B-a). Studies performed using low mass resolution instruments report composite IM profiles for all three species (apo, Zn-bound, and T4-bound), whereas IM data obtained using high mass RP yields IM profiles for each species (Fig. 18B-b and B-c). In addition, the high-resolution ion mobility capability of the FT-IM-PF-DT orbitrap EMR provides evidence for structural perturbation of TTR upon metal-induced oxidation (Poltash et al., 2019). As shown in Figure 19A, time-dependent oxidation of TTR can be resolved on EMR indicated by several stepwise mass shifts of 64 Da. The utility of ion mobility coupled to high-resolution mass analyzer allows the detection of two extended conformers of TTR upon oxidation (Fig. 19B). While the mass resolution offered by this platform is superior compared to that of a commercial IM-TOF instrument (Fig. 20C), the resolution, in both mass and mobility domains, is not sufficient to resolve the sequential oxidation of subunits for the intact tetramer, as they are evident on ejected monomers produced from SID of TTR (Fig. 19D and E).
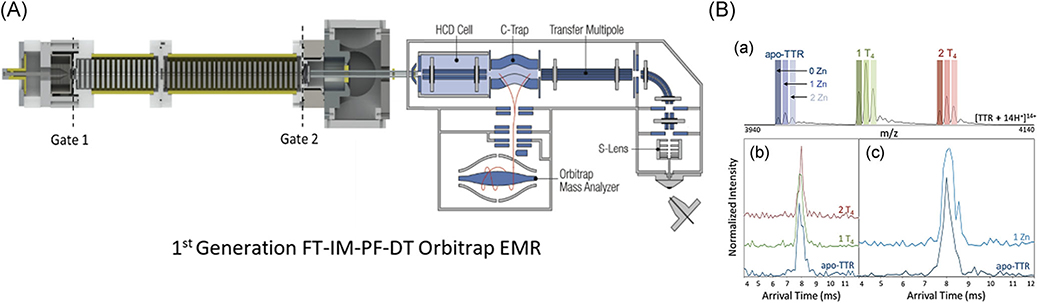
FIGURE 18.
(A) Solidworks schematic of first generation FT-IM-PF-DT coupled to a Thermo Scientific Exactive Plus orbitrap MS with EMR that was used for the following section of experimentation Briefly, ion is generated via static-spray nano-ESI into a heated capillary at ~100°C. Ions are then transmitted into an RF ion funnel at (250 Vpp). The ion beam is modulated at both the Gate 1 and Gate 2 by a linear frequency chirp of 5–5,005 Hz over 8 min to overcome the duty-cycle mismatch of IM separation and MS analysis. (B) Resolved peaks for T4 and endogenous zinc binding to TTR and corresponding ATDs obtained from instrument shown in panel A. Figure is adapted from Poltash et al. (2018). ATD, arrival time distributions; DT, drift-tube; EMR, extended mass range; ESI, electrospray ionization; FT, Fourier-transform; IM, ion-mobility; PF, periodic-focusing; TTR, transthyretin.
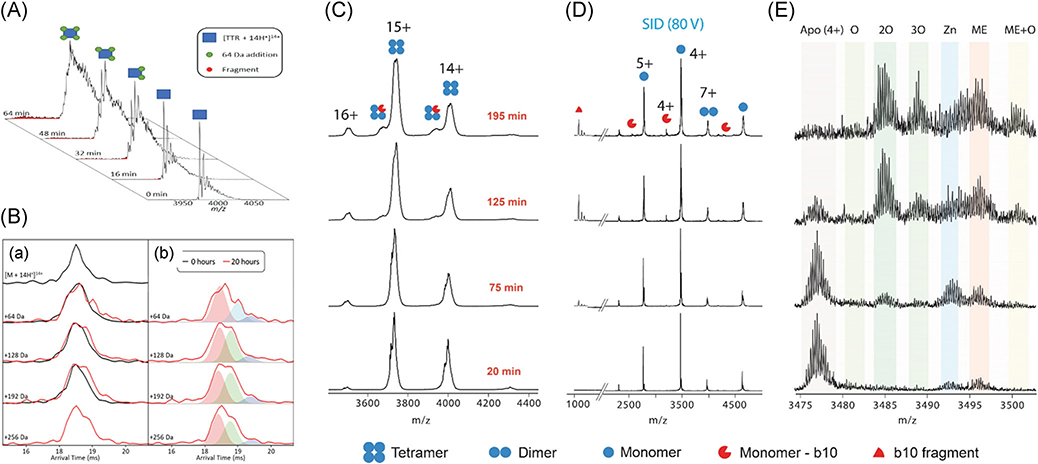
FIGURE 19.
(A) Stepwise increase in the mass of TTR upon electrospray ionization. (B) ATDs of TTR14+ (a) just loaded and (b) after 20 hr continuous ESI showing the unfolding of TTR due to oxidation (multiple extended conformers shown with green and purple peaks). (C) Repeated experiment as panel (A) but using Synapt G2 (Waters) without sufficient resolving power to detect stepwise 64 Da mass shift on tetramer, and (D) corresponding SID spectra showing ejected monomers. (E) SID spectra for M4+ showing several oxidations on monomeric TTR as well as zinc binding. Reproduced from Poltash et al. (2019). ESI, electrospray ionization; SID, surface-induced dissociation; TTR, transthyretin.
FIGURE 20.
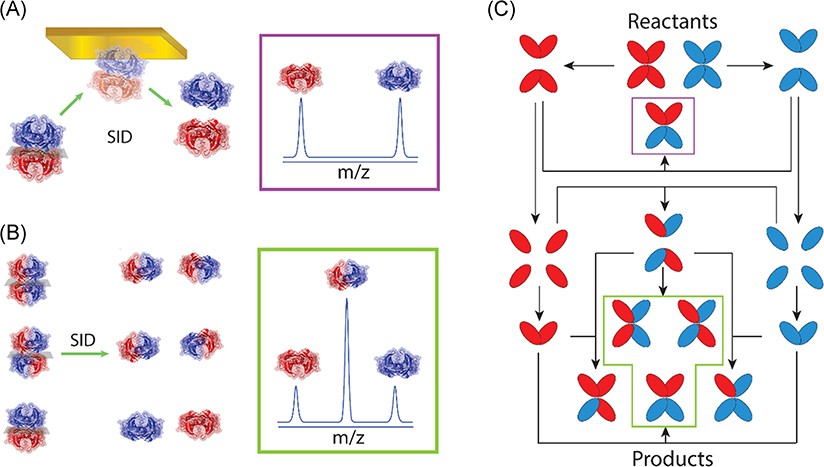
SID dissects topology of TTR products of SUE and provides a detailed mechanism of TTR disassembly in solution. (A) SID of 2:2 heterotetramer consisting of light/light and heavy/heavy dimers yields a mass spectrum for two homodimers with equal ion abundance. (B) For SID of an equimolar mixture of 2:2 heterotetramers, a ratio of 1:4:1 (LL/LH/HH) is obtained for dimers. (C) TTR disassembly mechanism supported by SID of SUE exchange products. After mixing the reactants, dissociation to dimers results in the production of the first product, shown in purple box and panel (A). Following dissociation of dimers to monomers yields all three topologies of 2:2 heterotetramer at equilibrium, shown in green box and panel (B). Reproduced from Shirzadeh et al. (2019). HH, light/light; LL, heavy/heavy; SUE, subunit exchange; TTR, transthyretin.
IM-MS results are augmented by techniques such as SID (Ma, Loo, & Wysocki, 2015; Quintyn, Harvey, & Wysocki, 2015; Quintyn et al., 2015; Song et al., 2015) which yields folded subunits which preserve ligand binding and reports on the topology of the complex (Fig. 20A, B) (Shirzadeh et al., 2019). In the case of ligand binding, SID of TTR reveals zinc-binding (Fig. 19E) while it is obscured in the CID experiment (Poltash et al., 2019). Recent native MS-SID-IM-MS studies on TTR using commercial instruments illustrate the diversity and wealth of information that can be gleaned using SID for topology analysis (Shirzadeh et al., 2019). The SID results show definitive data that TTR disassembly in solution occurs by a two-step reaction, tetramer→dimer→monomer (Foss, Wiseman, & Kelly, 2005), followed by self-assembly of the monomers (Fig. 20C); however, there remain open questions as to the conditions in which the monomers are properly folded, partially folded, or unfolded (Palhano et al., 2009).
One challenging aspect of any gas-phase analysis is conformational rearrangement upon transition from solution to the gas phase as discussed earlier. Thus, CCS of gas-phase ions is routinely compared with solution-phase structures to ensure the validity of data obtained by native IM-MS. Recently, first-principles CCS values of proteins and protein complexes, ranging from 8 to 810 kDa, were calculated and benchmarked against previously reported literature values using the FT-IM-PF-DT orbitrap EMR. The CCS was calculated via a Mason-Schamp equation modified by incorporating the mobility dampening term, α, determined for native complexes at varying reduce electric field strengths. The resulting CCS values fall within 5.5% difference of previous literature values with increasing deviation as the molecular weight (MW) of the proteins increase (McCabe et al., 2020, Anal Chem, submitted).
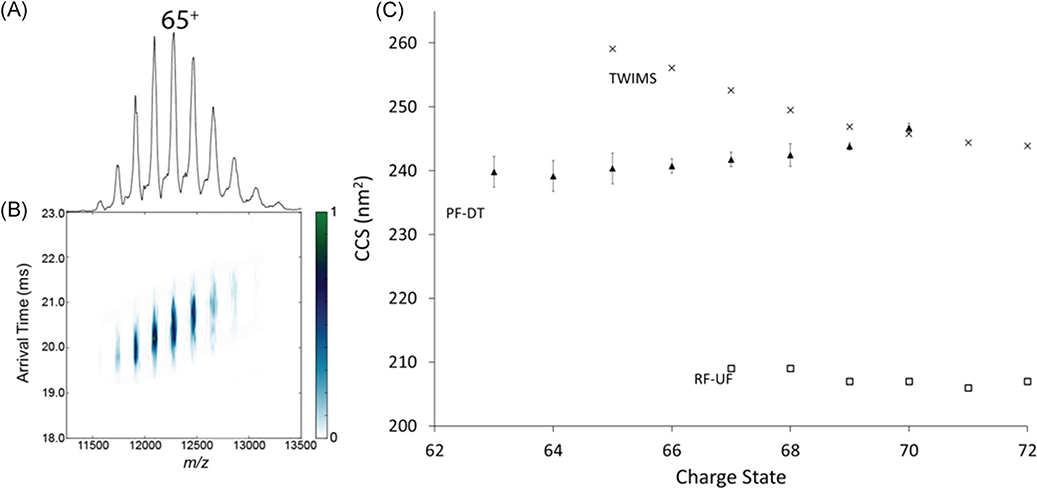
GroEL, a homotetradecameric chaperonin protein complex, was analyzed under native conditions to evaluate the performance of the FT-IM-PF-DT orbitrap platform for large protein complexes (Weaver et al., 2017). Figure 21 contains the IM-MS data for GroEL obtained via the FT-IM-PF-DT orbitrap EMR together with literature values of traveling wave ion mobility spectrometry (TWIMS; van Duijn et al., 2009) and of radiofrequency confining uniform field (Bush et al., 2010) for comparison. CCS obtained via the FT-IM-PF-DT orbitrap EMR, is well bracketed by both TWIMS, and RF-confining UF shows that the FT-IM-PF-DT orbitrap platform is capable of determining first-principles CCS measurements of proteins that approach 1 MDa. The utility of the FT-IM-PF-DT orbitrap is the ability to determine CCS values for protein and protein complexes over large MW range while exploiting the high-resolution capabilities of the IM and orbitrap mass analyzer.
FIGURE 21.
(A) Mass spectra, (B) ATD vs m/z plots, and (C) CCS vs CSD of native GroEL (810 kDa) with comparisons to PF-DT, TWIMS, RF-UF in 200 mM ammonium acetate. ATD, arrival time distributions; CCS, collision cross section; RF-UF, radio-frequency confining uniform field; PF-DT, periodic focusing drift tube; TWIMS, traveling-wave ion mobility.
While the current instrumentation shown in Figure 18A allows for high-resolution measurements of ions in the gas phase, additional instrument development is needed to gain sufficient RP to distinguish the signals contributing to peak heterogeneity. The current iteration of the FT-IM-PF-DT orbitrap EMR lacks the capabilities to do various pre-IM experiments, such as previously mentioned CIU, CID, VT-ESI, and SID. To increase the utility of the current instrument, CA is needed to clean up ions, for example, adduct removal from membrane proteins or inherently heterogeneous analytes. Soluble proteins typically do not require activation to remove the nonspecific adducts to the protein, whereas membrane proteins are solubilized in detergents to prevent precipitation in the solution (Laganowsky et al., 2014). The current FT-IM-PF-DT technology is adaptable to the Thermo Fisher Scientific Q Exactive Ultra-High Mass Resolution (UHMR) orbitrap MS, allowing for faster MS scan speeds and a m/z range of up to 80,000 Thomson (Th), where the previous upper m/z limit on the EMR platform was 20,000 Th (Fort et al., 2018), by increasing the scan frequency to 12 Hz (at a resolution of 12.5 k). With the commercial UHMR ion optics and software, the user has more control of various potentials and timing events, allowing for further optimization of the sensitivity of the instrument at a level was previously unavailable to users.
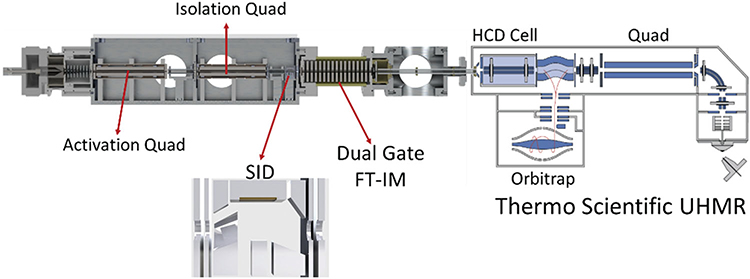
A modular vacuum chamber can be implemented pre-IM to exploit the abilities of IM as a size to charge separations technique, as shown in Figure 22. This platform allows for the implementation of two quadrupoles and SID to trap, isolate, fragment, dissociate, unfold, and/or manipulate ions before entering the PF-DT for IM separation. In the proposed design, the first quadrupole will be used as an activation quadrupole (denoted q), while the second quadrupole will be used as an isolation quadrupole (denoted Q) before SID. Development of SID modules in TOFs, orbitraps, and ion cyclotron resonance MS has been mainly driven by the Wysocki Group. While IMS is standard on commercial TOFs, commercially, this is not the case for oribtraps and to confirm that native structures are obtained opens an avenue for IM development on these platforms.
FIGURE 22.
Solidworks schematic of the implementation of a modular qQ-SID platform with FT-IM-PF-DT on the UHMR platform. The design of the SID cell is similar to that described by Zhou and Wysocki (2014). SID, surface-induced dissociation; UHRM, ultra-high mass resolution.
XII. CONCLUSION
Structural IM-MS has evolved from a technique developed by chemical physicists and physical chemists through various stages with uncertain potential for analytical applications, but now it is poised to take a lead role in areas of protein biophysical chemistry and structural biology. A major step in the evolutionary development of structural IM-MS was the introduction of the traveling-wave IM-MS SYNAPT series, an instrument that was geared for applications and fundamental studies. More important, the TWIMS SYNAPT was user friendly and adaptable to a broad range of studies. The major limitation of this technology is the limited IMS resolution and the limitations of CCS to the use of calibration standards. The limitations of using calibration standards is clearly illustrated in Figure 2A; the range of “conformation space” for large biomolecules which varies for different structural motifs, that is, disordered regions, random coils, sheets and helices, necessitates CCS determinations based on first-principles determinations.
The increased resolution and ion transmission for large, multiply charged ions afforded by the PF-DT combined with increased sample throughput of FT-IM-EMR orbitrap instrument (Fig. 18A) is already advancing our structural biology and native MS studies. The ability to incorporate ancillary MS techniques, specifically VT-ESI, CIU, SID, and native IM-MS on a single instrument is a game changer for MS-based structural studies. The qQ-SID-FT-IM-PF-DT UHMR orbitrap instrument (Fig. 22) creates an unrivaled advantage for studies of conformational diversity of proteins, protein complexes, and membrane protein complexes with the ancillary structural tools in the biophysical toolbox.
ACKNOWLEDGMENTS
Current funding for research in the Russell Lab is provided by the National Institutes of Health (P41GM128577 and R01GM121751) and the National Science Foundation (CHE-1707675). This review/perspective is written in honor of Professor Michael L. Gross for his many contributions to fundamental and applied mass spectrometry research, his 25 years as Editor-in-Chief of the Journal of the American Society for Mass Spectrometry, and his many contributions to the education of the next generation of MS scientists.
ABBREVIATIONS
- ATD
arrival time distributions
- CA
collisional activation
- CCS
collision cross section
- CD
circular dichroism
- CID/CIU
collision-induced dissociation/unfolding
- Cryo-EM
cryogenic electron microscopy
- Cryo-IM-MS
cryogenic ion mobility-mass spectrometry
- DT
drift tube
- ESI
electrospray ionization
- ETD/ECD
electron transfer/capture dissociation
- FT
Fourier transform
- ICR
ion cyclotron resonance
- IM-MS
ion mobility mass spectrometry
- m/z
mass-to-charge ratio
- MALDI
matrix-assisted laser desorption ionization
- MD
molecular dynamic simulations
- NMR
nuclear magnetic resonance
- PF-DT
periodic focusing drift tube
- PTM
posttranslational modification
- R
resolution
- RP
resolving power
- SASA
solvent-accessible surface area
- SAXS
small angle X-ray scattering
- SID
surface induced dissociation
- TOF
time-of-flight
- TTR
transthyretin
- TWIMS
traveling-wave ion mobility
- UF-DT
uniform-field drift-tube
- UVPD
ultraviolet photodissociation
- VT-ESI
variable-temperature electrospray ionization
- XRD
X-ray diffraction crystallography
Biography
Jacob W. McCabe received his BS in Chemistry from Baylor University in 2015 and MS in Chemistry from Texas A&M University—Commerce in 2017. He is a PhD degree candidate in Professor David Russell’s research group at Texas A&M University, developing high-resolution ion-mobility instrumentation coupled to the orbitrap platform.
Michael J. Hebert received his BS in Chemistry from University of North Carolina in 2015 and is currently a PhD degree candidate in Professor Russell’s group at Texas A&M University. His research focuses on hydrated like-charged ion pair interactions and late-stage ESI structural changes via cryo-IM-MS.
Mehdi Shirzadeh received his master’s degree from the University of Tehran and his PhD from Texas A&M University under Professor David Russell. His doctoral research focused on studies on the mechanism of transthyretin aggregation using IM-MS and surface-induced dissociation. He will continue working on applications of surface-induced dissociation for structural biology as a postdoctoral research assistant in the Russell group.
Christopher S. Mallis is a PhD degree candidate in the David H. Russell research group at Texas A&M University. His research interests include applications of mass spectrometry and ion mobility-mass spectrometry towards the characterization of protein complexes and intrinsically disordered proteins.
Joanna K. Denton is a postdoctoral researcher with Professor David H. Russell. She received her PhD from Yale University examining small atmospheric ions’ metal binding and microhydration behavior with gas-phase cold IR spectroscopy. Her research interests include application and development of IMS-MS to study the effects of solvent conditions on protein structure.
Thomas E. Walker is a graduate student in the David Russell Lab at Texas A&M University. His research interests include development of mass spectrometry instrumentation as well as study of water structure and its impact on biomolecule conformations using cryogenic ion mobility-mass spectrometry.
David H. Russell is a Professor of Chemistry and MDS-Sciex Professor of Mass Spectrometry at Texas A&M University. His research interests focus on fundamentals of mass spectrometry and ion mobility-mass spectrometry, bioanalytical chemistry, and biophysical chemistry. The major focus of his most recent research is on proteins and protein complexes and the development of novel, integrated instrumentation for biomolecule structural analysis.
REFERENCES
- Allen SJ, Bush MF. 2016. Radio-frequency (Rf) confinement in ion mobility spectrometry: Apparent mobilities and effective temperatures. J Am Soc Mass Spectrom 27(12):2054–2063. [DOI] [PubMed] [Google Scholar]
- Allison TM, Reading E, Liko I, Baldwin AJ, Laganowsky A, Robinson CV. 2015. Quantifying the stabilizing effects of protein-ligand interactions in the gas phase. Nat Commun 6:8551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benesch JL, Sobott F, Robinson CV. 2003. Thermal dissociation of multimeric protein complexes by using nanoelectrospray mass spectrometry. Anal Chem 75(10):2208–2214. [DOI] [PubMed] [Google Scholar]
- Benesch JLP. 2009. Collisional activation of protein complexes: Picking up the pieces. J Am Soc Mass Spectrom 20(3):341–348. [DOI] [PubMed] [Google Scholar]
- Blase RC, Silveira JA, Gillig KJ, Gamage CM, Russell DH. 2011. Increased ion transmission in IMS: A high resolution, periodic-focusing DC ion guide ion mobility spectrometer. Int J Mass Spectrom 301(1-3):166–173. [Google Scholar]
- Bluhm BK, Gillig KJ, Russell DH. 2000. Development of a Fourier-transform ion cyclotron resonance mass spectrometer-ion mobility spectrometer. Rev Sci Instrum 71(11):4078–4086. [Google Scholar]
- Bluhm BK, North SW, Russell DH. 2001. Separation of spin-orbit coupled metastable states of Kr+ and Xe+ by ion mobility. J Chem Phys 114(4):1709–1715. [Google Scholar]
- Bogan AA, Thorn KS. 1998. Anatomy of hot spots in protein interfaces. J Mol Biol 280(1):1–9. [DOI] [PubMed] [Google Scholar]
- Breuker K, McLafferty FW. 2008. Stepwise evolution of protein native structure with electrospray into the gas phase, 10(−12) to 10(2) S. Proc Natl Acad Sci USA 105(47):18145–18152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bush MF, Hall Z, Giles K, Hoyes J, Robinson CV, Ruotolo BT. 2010. Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal Chem 82(22):9557–9565. [DOI] [PubMed] [Google Scholar]
- Castro-Rodrigues AF, Gales L, Saraiva MJ, Damas AM. 2011. Structural insights into a zinc-dependent pathway leading to Leu55pro transthyretin amyloid fibrils. Acta Crystallogr D Biol Crystallogr 67(Pt 12):1035–1044. [DOI] [PubMed] [Google Scholar]
- Chen S-H, Russell WK, Russell DH. 2013. Combining chemical labeling, bottom-up and top-down ion-mobility mass spectrometry to identify metal-binding sites of partially metalated metallothionein. Anal Chem 85(6):3229–3237. [DOI] [PubMed] [Google Scholar]
- Chen SH, Chen L, Russell DH. 2014. Metal-induced conformational changes of human metallothionein-2a: A combined theoretical and experimental study of metal-free and partially metalated intermediates. J Am Chem Soc 136(26):9499–9508. [DOI] [PubMed] [Google Scholar]
- Chen SH, Russell DH. 2015a. How closely related are conformations of protein ions sampled by IM-MS to native solution structures? J Am Soc Mass Spectrom 26(9):1433–1443. [DOI] [PubMed] [Google Scholar]
- Chen SH, Russell DH. 2015b. Reaction of human Cd7 metallothionein and N-ethylmaleimide: Kinetic and structural insights from electrospray ionization mass spectrometry. Biochemistry 54(39):6021–6028. [DOI] [PubMed] [Google Scholar]
- Clemmer DE, Russell DH, Williams ER. 2017. Characterizing the conformationome: Toward a structural understanding of the proteome. Acc Chem Res 50(3):556–560. [DOI] [PubMed] [Google Scholar]
- Clowers BH, Hill HH Jr. 2005. Mass analysis of mobility-selected ion populations using dual gate, ion mobility, quadrupole ion trap mass spectrometry. Anal Chem 77(18):5877–5885. [DOI] [PubMed] [Google Scholar]
- Conant CR, Fuller DR, Zhang Z, Woodall DW, Russell DH, Clemmer DE. 2019. Substance P in the gas phase: Conformational changes and dissociations induced by collisional activation in a drift tube. J Am Soc Mass Spectrom 30(6):932–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong X, Liu Y, Liu W, Liang X, Russell DH, Laganowsky A. 2016. Determining membrane protein-lipid binding thermodynamics using native mass spectrometry. J Am Chem Soc 138(13):4346–4349. [DOI] [PubMed] [Google Scholar]
- Cunningham GP, Vidulich GA, Kay RL. 1967. Several properties of acetonitrile-water acetonitrile-methanol and ethylene carbonate-water systems. J Chem Eng Data 12(3):336–337. [Google Scholar]
- de C. Palmieri L, Lima LMTR, Freire JBB, Bleicher L, Polikarpov I, Almeida FCL, Foguel D. 2010. Novel Zn2+-binding sites in human transthyretin: Implications for amyloidogenesis and retinol-binding protein recognition. J Biol Chem 285(41):31731–31741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixit SM, Polasky DA, Ruotolo BT. 2018. Collision induced unfolding of isolated proteins in the gas phase: Past, present, and future. Curr Opin Chem Biol 42:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Shirzadeh M, Fan L, Laganowsky A, Russell DH. 2020. Ag+ ion binding to human metallothionein-2A is cooperative and domain specific [published online ahead of print June 9, 2020]. Anal Chem 10.1021/acs.analchem.0c00829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong S, Wagner ND, Russell DH. 2018. Collision-induced unfolding of partially metalated metallothionein-2a: Tracking unfolding reactions of gas-phase ions. Anal Chem 90(20):11856–11862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donor MT, Shepherd SO, Prell JS. 2020. Rapid determination of activation energies for gas-phase protein unfolding and dissociation in a Q-IM-TOF mass spectrometer. J Am Soc Mass Spectrom 31(3):602–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duboue-Dijon E, Laage D. 2014. Comparative study of hydration shell dynamics around a hyperactive antifreeze protein and around ubiquitin. J Chem Phys 141(22):22D529. [DOI] [PubMed] [Google Scholar]
- Dunn MF. 2005. Zinc–ligand interactions modulate assembly and stability of the insulin hexamer—A review. Biometals 18(4):295–303. [DOI] [PubMed] [Google Scholar]
- Dyachenko A, Gruber R, Shimon L, Horovitz A, Sharon M. 2013. Allosteric mechanisms can be distinguished using structural mass spectrometry. Proc Natl Acad Sci USA 110(18):7235–7239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Baba TJ, Kim D, Rogers DB, Khan FA, Hales DA, Russell DH, Clemmer DE. 2016. Long-lived intermediates in a cooperative two-state folding transition. J Phys Chem B 120(47):12040–12046. [DOI] [PubMed] [Google Scholar]
- El-Baba TJ, Woodall DW, Raab SA, Fuller DR, Laganowsky A, Russell DH, Clemmer DE. 2017. Melting proteins: Evidence for multiple stable structures upon thermal denaturation of native ubiquitin from ion mobility spectrometry-mass spectrometry measurements. J Am Chem Soc 139(18):6306–6309. [DOI] [PubMed] [Google Scholar]
- El-Baba TJ, Fuller DR, Woodall DW, Raab SA, Conant CR, Dilger JM, Toker Y, Williams ER, Russell DH, ClemmerE D. 2018. Melting proteins confined in nanodroplets with 10.6 μm light provides clues about early steps of denaturation. Chem Commun 54(26):3270–3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Baba TJ, Clemmer DE. 2019. Solution thermochemistry of concanavalin a tetramer conformers measured by variable-temperature ESI-IMS-MS. Int J Mass Spectrom 443:93–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Lima FA, Wei H, Gao YQ, Russell DH. 2009. On the structure elucidation using ion mobility spectrometry and molecular dynamics. J Phys Chem A 113(29):8221–8234. [DOI] [PubMed] [Google Scholar]
- Fornelli L, Srzentic K, Toby TK, Doubleday PF, Huguet R, Mullen C, Melani RD, Seckler HDS, DeHart CJ, Weisbrod CR, Durbin KR, Greer JB, Early BP, Fellers RT, Zabrouskov V, Thomas PM, Compton PD, Kelleher NL. 2020. Thorough performance evaluation of 213 nm ultraviolet photodissociation for top-down proteomics. Mol Cell Proteomics 19(2):405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort KL, Silveira JA, Pierson NA, Servage KA, Clemmer DE, Russell DH. 2014. From solution to the gas phase: Factors that influence kinetic trapping of substance P in the gas phase. J Phys Chem B 118(49): 14336–14344. [DOI] [PubMed] [Google Scholar]
- Fort KL, van de Waterbeemd M, Boll D, Reinhardt-Szyba M, Belov ME, Sasaki E, Zschoche R, Hilvert D, Makarov AA, Heck AJR. 2018. Expanding the structural analysis capabilities on an orbitrap-based mass spectrometer for large macromolecular complexes. Analyst 143(1):100–105. [DOI] [PubMed] [Google Scholar]
- Foss TR, Wiseman RL, Kelly JW. 2005. The pathway by which the tetrameric protein transthyretin dissociates. Biochemistry 44(47):15525–15533. [DOI] [PubMed] [Google Scholar]
- Frederick KK, Marlow MS, Valentine KG, Wand AJ. 2007. Conformational entropy in molecular recognition by proteins. Nature 448(7151):325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller DR, Conant CR, El-Baba TJ, Brown CJ, Woodall DW, Russell DH, Clemmer DE. 2018. Conformationally regulated peptide bond cleavage in bradykinin. J Am Chem Soc 140(30):9357–9360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamage CM, Silveira JA, Blase RC, Russell DH. 2011. Gas-phase ion dynamics in a periodic-focusing DC ion guide (part II): Discrete transport modes. Int J Mass Spectrom 303(2-3):154–163. [Google Scholar]
- Garimella SVB, Webb IK, Prabhakaran A, Attah IK, Ibrahim YM, Smith RD. 2017. Design of a TW-SLIM module for dual polarity confinement, transport, and reactions. J Am Soc Mass Spectrom 28(7):1442–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gault J, Lianoudaki D, Kaldmae M, Kronqvist N, Rising A, Johansson J, Lohkamp B, Laín S, Allison TM, Lane DP, Marklund EG, Landreh M. 2018. Mass spectrometry reveals the direct action of a chemical chaperone. J Phys Chem Lett 9(14):4082–4086. [DOI] [PubMed] [Google Scholar]
- Gertz MA, Mauermann ML, Grogan M, Coelho T. 2019. Advances in the treatment of hereditary transthyretin amyloidosis: A review. Brain and Behavior 9(9):e01371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giles K, Ujma J, Wildgoose J, Pringle S, Richardson K, Langridge D, Green M. 2019. A cyclic ion mobility-mass spectrometry system. Anal Chem 91(13):8564–8573. [DOI] [PubMed] [Google Scholar]
- Gillig KJ, Ruotolo BT, Stone EG, Russell DH. 2004. An electrostatic focusing ion guide for ion mobility-mass spectrometry. Int J Mass Spectrom 239(1):43–49. [Google Scholar]
- Halle B 2004. Protein hydration dynamics in solution: A critical survey. Philos Trans R Soc London, Ser B 359(1448):1207–1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halle B, Nilsson L. 2009. Does the dynamic stokes shift report on slow protein hydration dynamics? J Phys Chem B 113(24):8210–8213. [DOI] [PubMed] [Google Scholar]
- Hebert MJ, Russell DH. 2019. Hydration of guanidinium ions: An experimental search for like-charged ion pairs. J Phys Chem Lett 10(6):1349–1354. [DOI] [PubMed] [Google Scholar]
- Hebert MJ, Russell DH. 2020. Tracking the structural evolution of 4-aminobenzoic acid in the transition from solution to the gas phase. J Phys Chem B 124(11):2081–2087. [DOI] [PubMed] [Google Scholar]
- Hewitt D, Marklund E, Scott DJ, Robinson CV, Borysik AJ. 2014. A hydrodynamic comparison of solution and gas phase proteins and their complexes. J Phys Chem B 118(29):8489–8495. [DOI] [PubMed] [Google Scholar]
- Hyung S-J, Robinson CV, Ruotolo BT. 2009. Gas-phase unfolding and disassembly reveals stability differences in ligand-bound multiprotein complexes. Chem Biol 16(4):382–390. [DOI] [PubMed] [Google Scholar]
- Ibrahim YM, Hamid AM, Deng L, Garimella SV, Webb IK, Baker ES, Smith RD. 2017. New frontiers for mass spectrometry based upon structures for lossless ion manipulations. Analyst 142(7):1010–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrold MF. 2000. Peptides and proteins in the vapor phase. Annu Rev Phys Chem 51(1):179–207. [DOI] [PubMed] [Google Scholar]
- Javahery G, Thomson B. 1997. A segmented radiofrequency-only quadrupole collision cell for measurements of ion collision cross section on a triple quadrupole mass spectrometer. J Am Soc Mass Spectrom 8(7):697–702. [Google Scholar]
- Kaldmaee M, Sahin C, Saluri M, Marklund EG, Landreh M. 2019. A strategy for the identification of protein architectures directly from ion mobility mass spectrometry data reveals stabilizing subunit interactions in light harvesting complexes. Protein Sci. 28(6):1024–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltashov IA, Mohimen A. 2005. Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal Chem 77(16):5370–5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Wagner N, Wooding K, Clemmer DE, Russell DH. 2017. Ions from solution to the gas phase: A molecular dynamics simulation of the structural evolution of substance P during desolvation of charged nanodroplets generated by electrospray ionization. J Am Chem Soc 139(8):2981–2988. [DOI] [PubMed] [Google Scholar]
- Kiselar J, Chance MR. 2018. High-resolution hydroxyl radical protein footprinting: Biophysics tool for drug discovery. Annu Rev Biophys 47:315–333. [DOI] [PubMed] [Google Scholar]
- Knorr FJ, Eatherton RL, Siems WF, Hill HH Jr. 1985. Fourier transform ion mobility spectrometry. Anal Chem 57(2):402–406. [DOI] [PubMed] [Google Scholar]
- Kohler M, Marchand A, Hentzen NB, Egli J, Begley AI, Wennemers H, Zenobi R. 2019. Temperature-controlled electrospray ionization mass spectrometry as a tool to study collagen homo- and heterotrimers. Chem Sci 10(42):9829–9835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komander D, Rape M. 2012. The ubiquitin code. Annu Rev Biochem 81(1):203–229. [DOI] [PubMed] [Google Scholar]
- Ladbury JE. 1996. Just add water! The effect of water on the specificity of protein-ligand binding sites and its potential application to drug design. Chem Biol 3(12):973–980. [DOI] [PubMed] [Google Scholar]
- Laganowsky A, Reading E, Allison TM, Ulmschneider MB, Degiacomi MT, Baldwin AJ, Robinson CV. 2014. Membrane proteins bind lipids selectively to modulate their structure and function. Nature 510(7503):172–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landreh M, Sahin C, Gault J, Sadeghi S, Drum CL, Uzdavinys P, Drew D, Allison T, Degiacomi M, Marklund EG. 2020. Predicting the shapes of protein complexes through collision cross section measurements and database searches. ChemRxiv. Preprint. 10.26434/chemrxiv.12275057.v1 [DOI] [PubMed] [Google Scholar]
- Lee S-W, Freivogel P, Schindler T, Beauchamp JL. 1998. Freeze-dried biomolecules: FT-ICR studies of the specific solvation of functional groups and clathrate formation observed by the slow evaporation of water from hydrated peptides and model compounds in the gas phase. J Am Chem Soc 120(45):11758–11765. [Google Scholar]
- Lehmann E, Diederich F, Zenobi R, Salih B, Gómez-López M. 2000. Do matrix-assisted laser desorption/ionization mass spectra reflect solution-phase formation of cyclodextrin inclusion complexes? Analyst 125(5):849–854. [Google Scholar]
- Levy Y, Onuchic JN. 2004. Water and proteins: A love- hate relationship. Proc Natl Acad Sci USA 101(10):3325–3326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy Y, Onuchic JN. 2006. Water mediation in protein folding and molecular recognition. Annu Rev Biophys Biomol Struct 35:389–415. [DOI] [PubMed] [Google Scholar]
- Liu D, Wyttenbach T, Barran PE, Bowers MT. 2003. Sequential hydration of small protonated peptides. J Am Chem Soc 125(28):8458–8464. [DOI] [PubMed] [Google Scholar]
- Liu Y, LoCaste CE, Liu W, Poltash ML, Russell DH, Laganowsky A. 2019. Selective binding of a toxin and phosphatidylinositides to a mammalian potassium channel. Nat Commun 10(1):1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Zhang W-P, Xing Q, Ren X, Liu M, Tang C. 2012. Noncovalent dimerization of ubiquitin. Angew Chem Int Ed 51(2):469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutomski CA, Lyktey NA, Pierson EE, Zhao Z, Zlotnick A, Jarrold MF. 2018. Multiple pathways in capsid assembly. J Am Chem Soc 140(17):5784–5790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Loo JA, Wysocki VH. 2015. Surface induced dissociation yields substructure of methanosarcina thermophila 20S proteasome complexes. Int J Mass Spectrom 377:201–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes A, Maigret B, Hoflack J, Gomes JNF, Scheraga HA. 1994. Contribution of unusual arginine-arginine short-range interactions to stabilization and recognition in proteins. J Protein Chem 13(2):195–215. [DOI] [PubMed] [Google Scholar]
- Malmberg CG, Maryott AA. 1956. Dielectric constant of water from 0°C to 100°C. J Res Nat Bur Stand 56(1):1–8. [Google Scholar]
- Marchand A, Rosu F, Zenobi R, Gabelica V. 2018. Thermal denaturation of DNA G-quadruplexes and their complexes with ligands: Thermodynamic analysis of the multiple states revealed by mass spectrometry. J Am Chem Soc 140(39):12553–12565. [DOI] [PubMed] [Google Scholar]
- Masson GR, Burke JE, Ahn NG, Anand GS, Borchers C, Brier S, Bou-Assaf GM, Engen JR, Walter Englander S, Faber J, Garlish R, Griffin PR, Gross ML, Guttman M, Hamuro Y, Heck AJR, Houde D, Iacob RE, Jørgensen TJD, Kaltashov IA, Klinman JP, Konermann L, Man P, Mayne L, Pascal BD, Reichmann D, Skehel M, Snijder J, Strutzenberg TS, Underbakke ES, Wagner C, Wales TE, Walters BT, Weis DD, Wilson DJ, Wintrode PL, Zhang Z, Zheng J, Schriemer DC, Rand KD. 2019. Recommendations for performing, interpreting and reporting hydrogen deuterium exchange mass spectrometry (HDX-MS) experiments. Nat Methods 16(7):595–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattea C, Qvist J, Halle B. 2008. Dynamics at the protein-water interface from 17O spin relaxation in deeply supercooled solutions. Biophys J 95(6):2951–2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JC, Russell DH. 2011. A mass-selective variable-temperature drift tube ion mobility-mass spectrometer for temperature dependent ion mobility studies. J Am Soc Mass Spectrom 22(7):1134–1145. [DOI] [PubMed] [Google Scholar]
- McLean JA, Ruotolo BT, Gillig KJ, Russell DH. 2005. Ion mobility-mass spectrometry: A new paradigm for proteomics. Int J Mass Spectrom 240(3):301–315. [Google Scholar]
- McLean JR, McLean JA, Wu Z, Becker C, Perez LM, Pace CN, Scholtz JM, Russell DH. 2010. Factors that influence helical preferences for singly charged gas-phase peptide ions: The effects of multiple potential charge-carrying sites. J Phys Chem B 114(2):809–816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisburger SP, Ando N. 2017. Correlated motions from crystallography beyond diffraction. Acc Chem Res 50(3):580–583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirza UA, Chait BT. 1994. Effects of anions on the positive ion electrospray ionization mass spectra of peptides and proteins. Anal Chem 66(18):2898–2904. [DOI] [PubMed] [Google Scholar]
- Mitchell Wells J, McLuckey SA. 2005. Collision-induced dissociation (CID) of peptides and proteins. Methods Enzymol 402(148):18. [DOI] [PubMed] [Google Scholar]
- Moghadamchargari Z, Huddleston J, Shirzadeh M, Zheng X, Clemmer DE, Raushel FM, Russell DH, Laganowsky A. 2019. Intrinsic gtpase activity of K-RAS monitored by native mass spectrometry. Biochemistry 58(31):3396–3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison KA, Siems WF, Clowers BH. 2016. Augmenting ion trap mass spectrometers using a frequency modulated drift tube ion mobility spectrometer. Anal Chem 88(6):3121–3129. [DOI] [PubMed] [Google Scholar]
- Nagornova NS, Rizzo TR, Boyarkin OV. 2012. Interplay of intra- and intermolecular H-bonding in a progressively solvated macrocyclic peptide. Science 336(6079):320–323. [DOI] [PubMed] [Google Scholar]
- Nandi N, Bagchi B. 1997. Dielectric relaxation of biological water. J Phys Chem B 101(50):10954–10961. [Google Scholar]
- Niu B, Gross ML. MS-based hydroxyl radical footprinting: Methodology and application of fast photochemical oxidation of proteins (FPOP). Paper presented at: Mass Spectrometry-Based Chemical Proteomics Conference; Online Computer File 2019. [Google Scholar]
- No KT, Nam K-Y, Scheraga HA. 1997. Stability of like and oppositely charged organic ion pairs in aqueous solution. J Am Chem Soc 119(52):12917–12922. [Google Scholar]
- Nucci NV, Pometun MS, Wand AJ. 2011. Mapping the hydration dynamics of ubiquitin. J Am Chem Soc 133(32):12326–12329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh MI, Consta S. 2017. What factors determine the stability of a weak protein-protein interaction in a charged aqueous droplet? Phys Chem Chem Phys 19(47):31965–31981. [DOI] [PubMed] [Google Scholar]
- Pagel K, Kupser P, Bierau F, Polfer NC, Steill JD, Oomens J, Meijer G, Koksch B, von Helden G. 2009. Gas-phase IR spectra of intact alpha-helical coiled coil protein complexes. Int J Mass Spectrom 283(1-3):161–168. [Google Scholar]
- Pal SK, Peon J, Zewail AH. 2002. Biological water at the protein surface: Dynamical solvation probed directly with femtosecond resolution. Proc Natl Acad Sci USA 99(4):1763–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palhano FL, Leme LP, Busnardo RG, Foguel D. 2009. Trapping the monomer of a non-amyloidogenic variant of transthyretin: Exploring its possible use as a therapeutic strategy against transthyretin amyloidogenic diseases. J Biol Chem 284(3):1443–1453. [DOI] [PubMed] [Google Scholar]
- Papadopoulos G, Svendsen A, Boyarkin OV, Rizzo TR. 2012. Conformational distribution of bradykinin [Bk + 2H]2+ revealed by cold ion spectroscopy coupled with FAIMS. J Am Soc Mass Spectrom 23(7):1173–1181. [DOI] [PubMed] [Google Scholar]
- Papoian GA, Ulander J, Eastwood MP, Luthey-Schulten Z, Wolynes PG. 2004. Water in protein structure prediction. Proc Natl Acad Sci USA 101(10):3352–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick JW, Boone CD, Liu W, Conover GM, Liu Y, Cong X, Laganowsky A. 2018. Allostery revealed within lipid binding events to membrane proteins. Proc Natl Acad Sci USA 115(12):2976–2981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierson NA, Chen L, Valentine SJ, Russell DH, Clemmer DE. 2011. Number of solution states of bradykinin from ion mobility and mass spectrometry measurements. J Am Chem Soc 133(35):13810–13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pochapsky TC, Pochapsky SS. 2019. What your crystal structure will not tell you about enzyme function. Acc Chem Res 52(5):1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polasky DA, Dixit SM, Fantin SM, Ruotolo BT. 2019. Ciusuite 2: Next-generation software for the analysis of gas-phase protein unfolding data. Anal Chem 91(4):3147–3155. [DOI] [PubMed] [Google Scholar]
- Poltash ML, McCabe JW, Shirzadeh M, Laganowsky A, Clowers BH, Russell DH. 2018. Fourier transform-ion mobility-orbitrap mass spectrometer: A next-generation instrument for native mass spectrometry. Anal Chem 90(17):10472–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltash ML, Shirzadeh M, McCabe JW, Moghadamchargari Z, Laganowsky A, Russell DH. 2019. New insights into the metal-induced oxidative degradation pathways of transthyretin. Chem Commun (Camb) 55(28):4091–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltash ML, McCabe JW, Shirzadeh M, Laganowsky A, Russell DH. 2020. Native IM-orbitrap MS: Resolving what was hidden. Trends Anal Chem 124:115533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pricer R, Gestwicki JE, Mapp AK. 2017. From fuzzy to function: The new frontier of protein-protein interactions. Acc Chem Res 50(3):584–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintyn RS, Harvey SR, Wysocki VH. 2015. Illustration of SID-IM-SID (surface-induced dissociation-ion mobility-SID) mass spectrometry: Homo and hetero model protein complexes. Analyst 140(20):7012–7019. [DOI] [PubMed] [Google Scholar]
- Quintyn RS, Zhou M, Yan J, Wysocki VH. 2015. Surface-induced dissociation mass spectra as a tool for distinguishing different structural forms of gas-phase multimeric protein complexes. Anal Chem 87(23):11879–11886. [DOI] [PubMed] [Google Scholar]
- Rajabi K, Ashcroft AE, Radford SE. 2015. Mass spectrometric methods to analyze the structural organization of macromolecular complexes. Methods 89:13–21. [DOI] [PubMed] [Google Scholar]
- Rappley I, Monteiro C, Novais M, Baranczak A, Solis G, Wiseman RL, Helmke S, Maurer MS, Coelho T, Powers ET, Kelly JW. 2014. Quantification of transthyretin kinetic stability in human plasma using subunit exchange. Biochemistry 53(12):1993–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson LZ, Reixach N. 2014. Quantification of quaternary structure stability in aggregation-prone proteins under physiological conditions: The transthyretin case. Biochemistry 53(41):6496–6510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodier F, Bahadur RP, Chakrabarti P, Janin J. 2005. Hydration of protein-protein interfaces. Proteins: Struct, Funct, Bioinf 60(1):36–45. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Cruz S, Klassen J, Williams E. 1997. Hydration of gas-phase gramicidin S (M + 2H) ions formed by electrospray: The transition from solution to gas-phase structure. J Am Soc Mass Spectrom 8(5):565–568. [Google Scholar]
- Rolland AD, Prell JS. 2019. Computational insights into compaction of gas-phase protein and protein complex ions in native ion mobility-mass spectrometry. Trac-Trend Anal Chem 116:282–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruotolo BT, Verbeck GF, Thomson LM, Gillig KJ, Russell DH. 2002a. Observation of conserved solution-phase secondary structure in gas-phase tryptic peptides. J Am Chem Soc 124(16):4214–4215. [DOI] [PubMed] [Google Scholar]
- Ruotolo BT, Verbeck GFIV, Thomson LM, Woods AS, Gillig KJ, Russell DH. 2002b. Distinguishing between phosphorylated and nonphosphorylated peptides with ion mobility-mass spectrometry. J Proteome Res 1(4):303–306. [DOI] [PubMed] [Google Scholar]
- Ruotolo BT, Gillig KJ, Woods AS, Egan TF, Ugarov MV, Schultz JA, Russell DH. 2004. Analysis of phosphorylated peptides by ion mobility-mass spectrometry. Anal Chem 76(22):6727–6733. [DOI] [PubMed] [Google Scholar]
- Ruotolo BT, Tate CC, Russell DH. 2004. Ion mobility-mass spectrometry applied to cyclic peptide analysis: Conformational preferences of gramicidin S and linear analogs in the gas phase. J Am Soc Mass Spectrom 15(6):870–878. [DOI] [PubMed] [Google Scholar]
- Ruotolo BT, Hyung SJ, Robinson PM, Giles K, Bateman RH, Robinson CV. 2007. Ion mobility-mass spectrometry reveals long-lived, unfolded intermediates in the dissociation of protein complexes. Angew Chem Int Ed 46(42):8001–8004. [DOI] [PubMed] [Google Scholar]
- Sawyer HA, Marini JT, Stone EG, Ruotolo BT, Gillig KJ, Russell DH. 2005. The structure of gas-phase bradykinin fragment 1-5 (RPPGF) Ions: An ion mobility spectrometry and H/D exchange ion-molecule reaction chemistry study. J Am Soc Mass Spectrom 16(6):893–905. [DOI] [PubMed] [Google Scholar]
- Seo J, Hoffmann W, Warnke S, Huang X, Gewinner S, Schoellkopf W, Pagel K, Bowers MT, von Helden G, Page K. 2017. An infrared spectroscopy approach to follow β-sheet formation in peptide amyloid assemblies. Nat Chem 9(1):39–44. [DOI] [PubMed] [Google Scholar]
- Servage KA, Fort KL, Silveira JA, Shi L, Clemmer DE, Russell DH. 2015a. Unfolding of hydrated alkyl diammonium cations revealed by cryogenic ion mobility-mass spectrometry. J Am Chem Soc 137(28):8916–8919. [DOI] [PubMed] [Google Scholar]
- Servage KA, Silveira JA, Fort KL, Clemmer DE, Russell DH. 2015b. Water-mediated dimerization of ubiquitin ions captured by cryogenic ion mobility-mass spectrometry. J Phys Chem Lett 6(24):4947–4951. [DOI] [PubMed] [Google Scholar]
- Servage KA, Silveira JA, Fort KL, Russell DH. 2015c. From solution to gas phase: The implications of intramolecular interactions on the evaporative dynamics of substance P during electrospray ionization. J Phys Chem B 119(13):4693–4698. [DOI] [PubMed] [Google Scholar]
- Servage KA, Silveira JA, Fort KL, Russell DH. 2016. Cryogenic ion mobility-mass spectrometry: Tracking ion structure from solution to the gas phase. Acc Chem Res 49(7):1421–1428. [DOI] [PubMed] [Google Scholar]
- Shi L, Holliday AE, Shi H, Zhu F, Ewing MA, Russell DH, Clemmer DE. 2014. Characterizing intermediates along the transition from polypro-line I to polyproline II using ion mobility spectrometry-mass spectrometry. J Am Chem Soc 136(36):12702–12711. [DOI] [PubMed] [Google Scholar]
- Shi L, Holliday AE, Khanal N, Russell DH, Clemmer DE. 2015. Configurationally-coupled protonation of polyproline-7. J Am Chem Soc 137(27):8680–8683. [DOI] [PubMed] [Google Scholar]
- Shi L, Holliday AE, Glover MS, Ewing MA, Russell DH, Clemmer DE. 2016. Ion mobility-mass spectrometry reveals the energetics of intermediates that guide polyproline folding. J Am Soc Mass Spectrom 27(1):22–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi L, Holliday AE, Bohrer BC, Kim D, Servage KA, Russell DH, Clemmer DE. 2016b. “Wet” versus “Dry” folding of polyproline. J Am Soc Mass Spectrom 27(6):1037–1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirke RM, Chaudhari A, More NM, Patil PB. 2001. Temperature dependent dielectric relaxation study of ethyl acetate-alcohol mixtures using time domain technique. J Mol Liquids 94(1):27–36. [Google Scholar]
- Shirzadeh M, Boone CD, Laganowsky A, Russell DH. 2019. Topological analysis of transthyretin disassembly mechanism: Surface-Induced dissociation reveals hidden reaction pathways. Anal Chem 91(3):2345–2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirzadeh M, Poltash ML, Laganowsky A, Russell DH. 2020. Structural analysis of the effect of a dual-flag tag on transthyretin. Biochemistry 59(9):1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shnyrov VL, Villar E, Zhadan GG, Sanchez-Ruiz JM, Quintas A, Saraiva MJ, Brito RM. 2000. Comparative calorimetric study of non-amyloidogenic and amyloidogenic variants of the homotetrameric protein transthyretin. Biophys Chem 88(1-3):61–67. [DOI] [PubMed] [Google Scholar]
- Silveira JA, Gamage CM, Blase RC, Russell DH. 2010. Gas-phase ion dynamics in a periodic-focusing DC ion guide. Int J Mass Spectrom 296(1-3):36–42. [Google Scholar]
- Silveira JA, Jeon J, Gamage CM, Pai PJ, Fort KL, Russell DH. 2012. Damping factor links periodic focusing and uniform field ion mobility for accurate determination of collision cross sections. Anal Chem 84(6):2818–2824. [DOI] [PubMed] [Google Scholar]
- Silveira JA, Fort KL, Kim D, Servage KA, Pierson NA, Clemmer DE, Russell DH. 2013a. From solution to the gas phase: Stepwise dehydration and kinetic trapping of substance P reveals the origin of peptide conformations. J Am Chem Soc 135(51):19147–19153. [DOI] [PubMed] [Google Scholar]
- Silveira JA, Servage KA, Gamage CM, Russell DH. 2013b. Cryogenic ion mobility-mass spectrometry captures hydrated ions produced during electrospray ionization. J Phys Chem A 117(5):953–961. [DOI] [PubMed] [Google Scholar]
- Sipe SN, Brodbelt JS. 2019. Impact of charge state on 193 nm ultraviolet photodissociation of protein complexes. Phys Chem Chem Phys 21(18):9265–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Nelp MT, Bandarian V, Wysocki VH. 2015. Refining the structural model of a heterohexameric protein complex: Surface induced dissociation and ion mobility provide key connectivity and topology information. ACS Cent Sci 1(9):477–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiving AQ, VanAernum ZL, Busch F, Harvey SR, Sarni SH, Wysocki VH. 2019. Surface-induced dissociation: An effective method for characterization of protein quaternary structure. Anal Chem 91(1):190–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiving AQ, Jones BJ, Ujma J, Giles K, Wysocki VH. 2020. Collision cross sections of charge-reduced proteins and protein complexes: A database for collision cross section calibration. Anal Chem 92(6):4475–4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strittmatter EF, Williams ER. 2000. Structures of protonated arginine dimer and bradykinin investigated by density functional theory: Further support for stable gas-phase salt bridges. J Phys Chem A 104(25):6069–6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szumlas AW, Ray SJ, Hieftje GM. 2006. Hadamard transform ion mobility spectrometry. Anal Chem 78(13):4474–4481. [DOI] [PubMed] [Google Scholar]
- Tian Y, Ruotolo BT. 2018. The growing role of structural mass spectrometry in the discovery and development of therapeutic antibodies. Analyst 143(11):2459–2468. [DOI] [PubMed] [Google Scholar]
- Tompa K, Bánki P, Bokor M, Kamasa P, Lasanda G, Tompa P. 2009. Interfacial water at protein surfaces: Wide-line NMR and DSC characterization of hydration in ubiquitin solutions. Biophys J 96(7):2789–2798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turecek F, Mclafferty FW. 1993. Interpretation of mass spectra. 4th ed. Mill Valley, CA: University Science Books. [Google Scholar]
- van Duijn E, Barendregt A, Synowsky S, Versluis C, Heck AJR. 2009. Chaperonin complexes monitored by ion mobility mass spectrometry. J Am Chem Soc 131(4):1452–1459. [DOI] [PubMed] [Google Scholar]
- VanAernum ZL, Gilbert JD, Belov ME, Makarov AA, Horning SR, Wysocki VH. 2019. Surface-induced dissociation of noncovalent protein complexes in an extended mass range orbitrap mass spectrometer. Anal Chem 91(5):3611–3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazdar M, Vymětal J, Heyda J, Vondrášek J, Jungwirth P. 2011. Like-charge guanidinium pairing from molecular dynamics and ab initio calculations. J Phys Chem A 115(41):11193–11201. [DOI] [PubMed] [Google Scholar]
- Verbeck GF, Ruotolo BT, Gillig KJ, Russell DH. 2004. Resolution equations for high-field ion mobility. J Am Soc Mass Spectrom 15(9):1320–1324. [DOI] [PubMed] [Google Scholar]
- Vimer S, Ben-Nissan G, Morgenstern D, Kumar-Deshmukh F, Polkinghorn C, Quintyn RS, Vasil’ev YV, Beckman JC, Elad N, Wysocki VH, Sharon M. 2020. Comparative structural analysis of 20S proteasome ortholog protein complexes by native mass spectrometry. ACS Cent Sci 6(4):573–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner ND, Kim D, Russell DH. 2016. Increasing ubiquitin ion resistance to unfolding in the gas phase using chloride adduction: Preserving more “native-like” conformations despite collisional activation. Anal Chem 88(11):5934–5940. [DOI] [PubMed] [Google Scholar]
- Wagner ND, Russell DH. 2016. Defining noncovalent ubiquitin homodimer interfacial interactions through comparisons with covalently linked diubiquitin. J Am Chem Soc 138(51):16588–16591. [DOI] [PubMed] [Google Scholar]
- Wagner ND, Clemmer DE, Russell DH. 2017. ESI-IM-MS and collision-induced unfolding that provide insight into the linkage-dependent interfacial interactions of covalently linked diubiquitin. Anal Chem 89(18):10094–10103. [DOI] [PubMed] [Google Scholar]
- Wand AJ, Sharp KA. 2018. Measuring entropy in molecular recognition by proteins. Annu Rev Biophys 47(1):41–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Bondarenko PV, Kaltashov IA. 2018. Multi-step conformational transitions in heat-treated protein therapeutics can be monitored in real time with temperature-controlled electrospray ionization mass spectrometry. Analyst 143(3):670–677. [DOI] [PubMed] [Google Scholar]
- Warnke S, Baldauf C, Bowers MT, Pagel K, von Helden G. 2014. Photodissociation of conformer-selected ubiquitin ions reveals site-specific cis/trans isomerization of proline peptide bonds. J Am Chem Soc 136(29):10308–10314. [DOI] [PubMed] [Google Scholar]
- Weaver J, Jiang M, Roth A, Puchalla J, Zhang J, Rye HS. 2017. Groel actively stimulates folding of the endogenous substrate protein pepq. Nat Commun 8(1):15934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wintrode PL, Makhatadze GI, Privalov PL. 1994. Thermodynamics of ubiquitin unfolding. Proteins 18(3):246–253. [DOI] [PubMed] [Google Scholar]
- Woodall DW, El-Baba TJ, Fuller DR, Liu W, Brown CJ, Laganowsky A, Russell DH, Clemmer DE. 2019. Variable-temperature ESI-IMS-MS analysis of myohemerythrin reveals ligand losses, unfolding, and a non-native disulfide bond. Anal Chem 91(10):6808–6814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodall DW, Brown CJ, Raab SA, El-Baba TJ, Laganowsky A, Russell DH, Clemmer DE. 2020. Melting of hemoglobin in native solutions as measured by IMS-MS. Anal Chem 92(4):3440–3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods LA, Radford SE, Ashcroft AE. 2013. Advances in ion mobility spectrometry-mass spectrometry reveal key insights into amyloid assembly. Biochim Biophys Acta, Proteins Proteomics 1834(6): 1257–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyttenbach T, Liu D, Bowers MT. 2005. Hydration of small peptides. Int J Mass Spectrom 240(3):221–232. [Google Scholar]
- Wyttenbach T, Bowers MT. 2009. Hydration of biomolecules. Chem Phys Lett 480(1–3):1–16. [Google Scholar]
- Xiao C, Perez LM, Russell DH. 2015. Effects of charge states, charge sites and side chain interactions on conformational preferences of a series of model peptide ions. Analyst 140(20):6933–6944. [DOI] [PubMed] [Google Scholar]
- Yewdall NA, Pearce FG, Yewdall NA, Gerrard JA, Allison TM, Robinson CV, Gerrard JA. 2018. Self-assembly of toroidal proteins explored using native mass spectrometry. Chem Sci 9(28):6099–6106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou M, Wysocki VH. 2014. Surface induced dissociation: Dissecting noncovalent protein complexes in the gas phase. Acc Chem Res 47(4):1010–1018. [DOI] [PubMed] [Google Scholar]
- Zhurov KO, Fornelli L, Wodrich MD, Laskay ÜA, Tsybin YO. 2013. Principles of electron capture and transfer dissociation mass spectrometry applied to peptide and protein structure analysis. Chem Soc Rev 42(12):5014–5030. [DOI] [PubMed] [Google Scholar]
- Zinnel NF, Pai PJ, Russell DH. 2012. Ion mobility-mass spectrometry (IM-MS) for top-down proteomics: Increased dynamic range affords increased sequence coverage. Anal Chem 84(7):3390–3397. [DOI] [PubMed] [Google Scholar]