Abstract
The N501Y mutation in SARS-CoV-2 variants found in several strains from the UK, South Africa and Brazil has been linked to increased transmission.
In order to discriminate N501Y variants quickly, a single nucleotide polymorphism (SNP) discrimination assay was designed and validated. It was then deployed prospectively in 757 nasopharyngeal swabs.
Validation of the novel variant discrimination assay corroborated the results in all validation panel samples (n = 63) through sequencing.
This novel variant discrimination assay was then deployed prospectively in 757 clinical nasopharyngeal swabs during the last week of January 2021. N501Y was found in 206 (27.4 %) of the samples: 94 (28.2 %) men and 112 (26.85 %) women (p = 0.73). The patients in whom it was identified had a mean age of 47.8 ± 25.8 (0–96) years, similar to that of patients without this variant: 51.7 ± 25.9 (0–104) years (p = 0.06).
501Y variant was confirmed in 34 samples by sequence method and 501 N wild type was confirmed in 67.
This method is sensitive, specific, and simple to apply in any microbiology lab.
Keywords: SARS-CoV-2, Mutation, Variant discrimination assay, N501Y, Sequencing, New generation sequencing
1. Introduction
During the evolution and adaptation of a virus it is very common for variants to be seen, particularly in RNA viruses like SARS-CoV-2 (van Dorp et al., 2020). The worrying thing about these variants is their potential capacity to elude the defense response (natural defense mechanisms as well as a vaccine).
In the case of SARS-CoV-2, the scientific community must be extremely alert to the development of variants of the spike protein, which can alter the virus binding mechanism and result in the virus escaping the action of natural/vaccine-induced antibodies. The spike D614 G mutation of SARS-CoV-2 emerged in Europe in February 2020 and was linked to more serious infection and increased transmissibility. However, vaccinated subjects produce antibodies capable of neutralizing this variant (Korber et al., 2020; Zhang et al., 2020; Volz et al., 2021).
At the end of 2020, three new variants with increased transmission and potential clinical importance appeared. The first was the variant B.1.1.7 which initially appeared in the United Kingdom in September (ECDC, 2020; Leung et al., 2021). Later, the B.1.351 variant was identified in South Africa and B.1.128.1 in Brazil (WHO, 2020; Weisblum et al., 2020; Faria et al., 2021). All three have the N501Y mutation, which is involved in the receptor-binding mechanism and may have clinical impacts (Leung et al., 2021; Makowski et al., 2021).
The detection of SARS-CoV-2 variants has principally been through the use of the traditional Sanger sequencing. In order to achieve a faster and cheaper method a new one-step variant discrimination real time PCR (VD RT-PCR) assay to detect the N501Y SARS-CoV-2 mutation was designed and developed, and then assayed in clinical samples.
2. Materials and methods
2.1. Nasopharyngeal samples
A total of 64 SARS-CoV-2 positive nasopharyngeal swabs collected in UTM media (Copan, Italy) between December 2020-January 2021 and previously characterized by sequencing (35 of which had the N501Y mutation) were used to validate the method. Ct for amplification of these samples was below 30.
Next, the novel N501Y VD RT-PCR method was used on 757 nasopharyngeal swabs collected prospectively between January 20 and 27 2021, under the same conditions as previous samples. Patients were 335 males (mean age 48.2 ± 24.2, range 0–99, years) and 422 females (mean age 52.5 ± 27.1 range 1–104 years).
Amplification of these samples gave Ct values of between 15 and 35.
Following extraction, all samples were stored at 4 °C until further use.
2.2. Processing of nasopharyngeal samples and SARS-CoV-2 detection
In the first step of the initial SARS-CoV-2 detection from the nasopharyngeal swabs, RNA was isolated using a MagNA Pure 96 System (Roche Diagnostics, Switzerland) following the manufacturer’s protocol. Amplification and detection were then carried out using in-house real time (RT)-PCR developed to detect the Orf1ab and nucleoprotein genes of SARS-CoV-2.
Viral genomes were amplified using TaqMan® Fast Virus 1-Step Master Mix (Life Technologies, CA) and the primers and FAM/VIC-labelled MGB (minor groove binding) probes shown in Table 1 . Amplifications and data analysis were performed using either a 7500 or a QS5 Real Time PCR System (Applied Biosystems, CA) under the following conditions: retrotranscription at 50 °C for 15 min; denaturation at 95 °C for 5 min; 40 cycles at 95 °C for 10 s followed by 60 °C for 20 s.
Table 1.
Primers and probes used: for RT-PCR, for N501Y detection and for sequencing.
| Use | Position | Name | Sequence (5’-3’) | Gen |
|---|---|---|---|---|
| Sense primer | CoV-2-OVI-S | ATCAAGTTAATGGTTACCCTAACATGT | SARS-CoV-2 ORF1ab |
|
| Antisense primer | CoV-2-OVI-A | AACCTAGCTGTAAAGGTAAATTGGTACC | ||
| Probe MGB FAM | CoV-2-OVI-FAM | CCGCGAAGAAGCTA | ||
| SARS-CoV-2 | Sense primer1 | 2019-nCoV_N1-F | GACCCCAAAATCAGCGAAAT | SARS-CoV-2 Gen N |
| detection | Antisense primer1 | 2019-nCoV_N1-R | TCTGGTTACTGCCAGTTGAATCTG | |
| Probe MGB VIC1 | 2019-nCoV_N1-P-VIC | CCGCATTACGTTTGGT2 | ||
| Sense primer | Beta-TR-S | ACACAACTGTGTTCACTAGC | Human β-globin gene |
|
| Antisense primer | Beta-TR-A | CCAACTTCATCCACGTTCACC | ||
| Probe Cy5.0 | Beta-Cy5.0 | TGCATCTGACTCCTGAGGA | ||
| Sense primer | CoV-N501Y-Fwd2 | AATCTATCAGGCCGGTAGCACA | SARS-CoV-2 Gen S |
|
| N501Y | Antisense primer | CoV-N501Y-Rev | ACAGTTGCTGGTGCATGTAGA | |
| detection | Probe MGB FAM | CoV-WT-N501-FAM | CCACTAATGGTGTTGGT3 | |
| Probe MGB VIC | CoV-Mut-Y501-VIC | CCACTTATGGTGTTGGT3 | ||
| N501Y | Sense primer | CoV-N501Y-Fwd2 | AATCTATCAGGCCGGTAGCACA | SARS-CoV-2 Gen S |
| Sequencing | Antisense primer | N501-Seq-R2 | CAGGGACTTCTGTGCAGTTAACA |
US Centers for Disease Control and Prevention (2020) 2019-Novel coronavirus (2019-nCoV) real-time rRT-PCR panel primers and probes. Available in: https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html.
Probe sequence has been shortened as it is a MGB probe.
The AAT (Asn) and TAT (Tyr) codons are underlined.
In addition, the human ß-globin gene was quantified in each sample in order to evaluate sample quality and to calculate normalized viral load (Alvarez-Argüelles et al, 2015; Yahav et al., 2021).
2.3. Variant N501 discrimination assay
Sequence-specific forward and reverse oligonucleotide primers were used, along with two TaqMan MGB variant specific probes, each with a different reporter dye at the 5’ end and a non-fluorescent quencher (NFQ) at the 3’ end (Table 1): one labelled with VIC dye to detect the Allele 1 (501Y) sequence and one with FAM to detect the Allele 2 (501 N) sequence.
To design the primers and probes for the one step VD RT-PCR assay, sequences of the variants of the SARS-CoV-2 spike gene deposited in the NCBI (National Center of Biotechnology Information) were aligned using the ClustalW program. The oligonucleotide primers and probes used in the study were provided by Applied Biosystems, Life Technologies Corporation (Warrington, UK). The final concentrations of primers and probes used were 400 nM and 100 nM, respectively.
The reaction was carried out using 2.5 μL of extracted sample in a 2.5 μL reaction volume of TaqMan® Fast Virus 1-Step Master Mix (Life Technologies, CA) in either a StepOnePlus or a QS5 Real-Time PCR System (Applied Biosystems, Life Technologies Corporation, Foster City, CA, USA) under the following conditions: 50 °C for 15 min; 95 °C for 2 min; 40 cycles at 95 °C for 5 s and 60 °C for 30 s. Pre- and post-readings were carried out at 50 °C and acquisition of other data was obtained during the annealing/extension stage of each cycle in both FAM and VIC filters. The total duration of the process was approximately 46 min.
As negative controls, four non-template controls (water) were included in each VD RT-PCR assay run. Three positive controls were also tested in each run: a confirmed N501 N variant sequence, a confirmed N501Y variant sequence and a mixture of the two.
2.4. Sequencing
For the identification of the SARS-CoV-2 N501 variants, a fragment of 450 bp from the spike gene (Nt 1412-1862; aa 472-620) was amplified with primers designed in our laboratory (Table 1). The PCR mix contained 25 pmol of each primer, 0.2 mM of each deoxynucleotide triphosphate, 1 × PCR buffer, 2 mM MgCl2, Titan One tube RT-PCR System (Roche Diagnostics, Mannheim, Germany), and 5 μL of extracted sample in a 25 μL reaction volume. The thermal profile was: 50° for 15 min; denaturation at 94 °C for 5 min followed by 45 cycles of 94 °C for 30 s; 55 °C for 45 s; 72 °C for 3 min; a final extension of 10 min at 72 °C.
The PCR products were purified using the Montage PCR purification kit (DNA Gel Extraction Kit, Bedford, MA). Sequencing was performed in a 5 μL reaction volume with the same primers using a BigDye Terminator v1.1 Cycle Sequencing Ready Reaction kit (Applied Biosystems, Life Technologies Corporation, Austin, TX, USA) and then removes unincorporated reaction components with the BigDye Xterminator purification kit (Life Technologies Corporation, Bedford, USA). Genome sequences were obtained with an ABI PRISM 3130X Genetic Analyzer Avant Sequencer (Applied Biosystems, Life Technologies Corporation, Foster City, CA, USA). The chromatogram files were edited and assembled using Chromas Pro 2.1.6). The nucleotide sequence data were examined using MEGA (version 7.0).
NGS (new generation sequencing) was performed on 11 of 63 strains from the validation panel with the 501Y mutation using the Ion GeneStudio™ S5 System (Thermo Fisher Scientific) following the manufacturer’s protocol. Libraries were prepared using the Ion AmpliSeq SARS-CoV-2 Research Panel designed by Thermo Fisher Scientific for complete viral genome sequencing.
From the 750 specimens collected, with conclusive results sequencing method was performed in 101 cases, 67 using Sanger sequencing and 34 using NGS.
2.5. Data and statistical analysis and ethical approval
All statistical tests were performed using GraphPad InStat version 3.00 for Windows 95 (GraphPad Software, San Diego, CA) following the manufacturer’s instructions.
This study was approved by the Local Ethical Committee.
3. Results
3.1. Validation panel
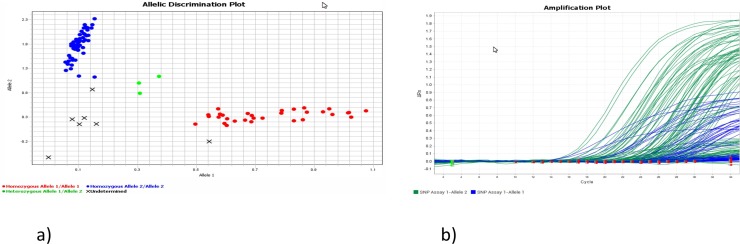
The validation panel was assembled using 29 SARS-CoV-2 501 N (wild) strains and 35 501Y variant strains. One wild variant did not amplify, but in the other 63, complete concordance was observed between the results of the RNA sequencing and the novel VD RT-PCR assay (Fig. 1).
Fig. 1.
Variant discrimination assay. a) Variant discrimination assay plot: samples with sequence confirmed SARS-CoV-2 wild strain (N501) in blue, N501Y variants in either red, and mixed variants in green. b) Graph of variant amplification assay: amplification of wild type variant in green and N501Y variants in blue (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
NGS was performed on 11 of these 63 strain N501Y variants to ascertain the specific genotypes and linages. All of them where characterized like SARS-CoV-2 B.1.1.7 strain by Pangolin program (https://pangolin.cog-uk.io/).
3.2. Results for prospective determination of variant
Between January 20 and 27 2021, variant discrimination assays were performed on 757 nasopharyngeal swab samples. Conclusive results were achieved in 750 cases (99.07 %).
The N501Y mutation was present in 206 samples (27.4 %). These variants were found in 94 (28.2 %) men and 112 (26.85 %) women (p = 0.73).
Mean age of patients with N501Y was 47.8 ± 25.8 years (range 0–96) compared to 51.7 ± 25.9 years (range 0–104) for patients without this mutation (p = 0.06).
With the method described above, amplification of samples resulted in Cts of between 15 and 35. The N501Y mutation was present in 12 samples (5.8 %) with Ct of over 30 and wild type in 34 (6.25 %) such samples (p = 0.9).
In 101 sequenced, variants was present in 67 (36 by Sanger and 31 by NGS) and no present in 34 (31 by Sanger and 3 by NGS).
4. Discussion
Mutations in the SARS-CoV-2 spike protein could be linked to loss of natural/vaccine-induced immune response and increased virulence, which could have significant impacts on public health provisions. To this end, it is important internationally for health organizations to detect these variants quickly in order to take actions and adopt specific measures to minimize its community transmission (ECDC, 2021; WHO, 2021).
In 2020, the SARS-CoV-2 spike protein variant D614 G which replaced the original strains identified, was found to be associated with increased transmissibility and more serious pathology (Korber et al., 2020; Zhang et al., 2020; Volz et al., 2021). Variants with this mutation are now globally dominant (Conti et al., 2020).
Recently, the newly identified SARS-CoV-2 variants B.1.1.7, B1.351, and B.1.1.28.1 have come under scrutiny because they can increase transmissibility and reduce neutralization (Leung et al., 2021; WHO, 2021; Weisblum et al., 2020; Conti et al., 2020).All of these variants share the spike protein mutation N501Y, which is involved in virus binding, and is principally found in mutations associated with virus transmission (ECDC, 2020; Leung et al., 2021; WHO, 2021; Weisblum et al., 2020; Virological, 2021; Makowski et al., 2021).
The method designed was able to discriminate the variant in all but one instance: a wild type SARS-CoV-2 which was collected 20 days before the assay and had a Ct of 32. These characteristics could have influenced in the result.
In the other 63 samples (35 of them the N510Y variant), the correlation between the novel VD RT-PCR method and the sequencing method was complete.
Importantly, this VD RT-PCR method can be performed and provide results in less than one hour. What is more, this work indicates that it would be possible to design and develop a real time RT-PCR system that includes both diagnosis and variant detection. This would be a fast and simple system, the drawback being that more reagents would be needed.
The eleven samples analyzed by NGS confirmed that the N501Y variant found was most similar to strain B.1.1.7.
This method was implemented in clinical samples collected prospectively over 7 days at the end of January 2021. Results were possible in close to 100 % of cases. Only in 7 (0.9 %) samples with Ct over 30 was amplification not possible. N501Y variant was present in almost 30 % of these samples. Since the presence of these variants in our locality only began in early January, these results suggest that the N501Y variant can now replace circulating wild-type strains very early. No differences in the presence of the N501Y variant were found in terms gender or mean age.
As before, in all samples sequenced, the correlation with this method was complete.
The method developed in this work to identify N501Y SARS-CoV-2 variants is fast and simple, and can be performed in any basic laboratory in less than one hour.
Author statement
Conceptualization; Santiago Melón García, Jose Antonio Boga, Marta Elena Alvarez Argüelles, Susana Rojo Alba, Santiago Melón García.
Data curation; Marta Sandoval Torrientes, Jose Antonio Boga, Marta Elena Alvarez Argüelles, Susana Rojo Alba, Santiago Melón García.
Formal analysis; Marta Sandoval Torrientes, Marta Elena Alvarez Argüelles, Susana Rojo Alba, Santiago Melón García.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
We thank Ronnie Lendrum for her help with correcting the English, and ASCOL for his technical support.
References
- Álvarez-Argüelles M.E., de Oña-Navarro M., Rojo-Alba S., Torrens-Muns M., Junquera-Llaneza M.L., Antonio-Boga J., Pérez-Castro S., Melón-García S. Quantification of human papilloma virus (HPV) DNA using the Cobas 4800 system in women with and without pathological alterations attributable to the virus. J. Virol. Methods. 2015;222(September 15):95–102. doi: 10.1016/j.jviromet.2015.05.016. doi: 0.1016/j.jviromet.2015.05.016. Epub 2015 Jun 6.PMID: 26057221. [DOI] [PubMed] [Google Scholar]
- Conti P., Caraffa A., Gallenga C.E., Kritas S.K., Frydas S.K., Younes A., Emidio P.D., Tetè G., Pregiasco F., Ronconi G. The British variant of the new coronavirus-19 (Sars-Cov-2) should not create a vaccine problem. J. Biol. Regul. Homeost. Agents. 2020;35(December 30 (1)) doi: 10.23812/21-3-E. [DOI] [PubMed] [Google Scholar]
- European Centre for Disease Prevention and Control . 2020. Threat Assessment Brief: Rapid Increase of a SARS-CoV-2 Variant With Multiple Spike Protein Mutations Observed in the United Kingdom. [Google Scholar]
- ECDC . 2021. Sequencing of SARS-Cov-2 [internet]: European Centre for Disease Prevention and Control. [January 13th, 2021]. https//www.ecdc.europa.eu/en/publications-data/sequencing-sars-cov-2. [Google Scholar]
- Faria N.R., Morales I., Candido D., Moyses L.A., Andrade P.S., Coleti T.M., Silva C.A., Sales F.C., Manuli E.R., Aguiar R.S., Gaburo N., Camilo C., Fraiji N.A., Esashika M.A., Carvalho M.P., Rambaut A., Loman N., Pybus O.G., Sabino E.C., Virological . 2021. Genomic Characterisation of an Emergent SARS-CoV-2 Lineage in Manaus: Preliminary Findings.https://virological.org/t/genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-manaus-preliminary-findings [Google Scholar]
- Korber B., Fischer W.M., Gnanakaran S., Yoon H., Theiler J., Abfalterer W., Hengartner N., Giorgi E.E., Bhattacharya T., Foley B., Hastie K.M., Parker M.D., Partridge D.G., Evans C.M., Freeman T.M., de Silva T.I., Sheffield COVID-19 Genomics Group, McDanal C., Perez L.G., Tang H., Moon-Walker A., Whelan S.P., LaBranche C.C., Saphire E.O., Montefiori D.C. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. 2020;182(August 20 (4)):812–827.e19. doi: 10.1016/j.cell.2020.06.043. Epub 2020 Jul 3. PMID: 32697968; PMCID: PMC7332439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung K., Shum M., Leung G., Lam T., Wu J. Early transmissibility assessment of the N501Y mutant strains of SARS-Cov2 in the United Kingdom, October to November 2020. Euro Surveill. 2021;26(Jan. (1)) doi: 10.2807/1560-7917.ES2020.26.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makowski L., Olson-Sidford W., W-Weisel J. Biological and clinical consequences of integrin binding via a rogue RGD motif in the SARS CoV-2 spike protein. Viruses. 2021;13(January 20 (2)):E146. doi: 10.3390/v13020146. PMID: 33498225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dorp L., Richard D., Tan C.C.S., Shaw L.P., Acman M., Balloux F. No evidence for increased transmissibility from recurrent mutations in SARS-CoV-2. Nat. Commun. 2020;11(November 25 (1)):5986. doi: 10.1038/s41467-020-19818-2. PMID: 33239633; PMCID: PMC7688939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E., Hill V., McCrone J.T., Price A., Jorgensen D., O’Toole Á, Southgate J., Johnson R., Jackson B., Nascimento F.F., Rey S.M., Nicholls S.M., Colquhoun R.M., da Silva Filipe A., Shepherd J., Pascall D.J., Shah R., Jesudason N., Li K., Jarrett R., Pacchiarini N., Bull M., Geidelberg L., Siveroni I., COG-UK Consortium, Goodfellow I., Loman N.J., Pybus O.G., Robertson D.L., Thomson E.C., Rambaut A., Connor T.R. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell. 2021;184(January 7 (1)):64–75.e11. doi: 10.1016/j.cell.2020.11.020. Epub 2020 Nov 19.PMID: 33275900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisblum Y., Schmidt F., Zhang F., DaSilva J., Poston D., Lorenzi J.C., Muecksch F., Rutkowska M., Hoffmann H.H., Michailidis E., Gaebler C., Agudelo M., Cho A., Wang Z., Gazumyan A., Cipolla M., Luchsinger L., Hillyer C.D., Caskey M., Robbiani D.F., Rice C.M., Nussenzweig M.C., Hatziioannou T., Bieniasz P.D. Escape from neutralizing antibodies by SARS-CoV-2 spike protein variants. Elife. 2020;9(October 28):e61312. doi: 10.7554/eLife.61312. PMID: 33112236; PMCID: PMC7723407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SARS-CoV-2 Variants. WHO. https://www.who.int/csr/don/31-december-2020-sars-cov2-variants/en/.
- WHO . 2021. Genomic Sequencing of SARS-Cov-2: A Guide to Implementation for Maximum Impact on Public Healt. [January 13th, 2021]. https//www.who.int/publications-detail-redirect/9789240018440. [Google Scholar]
- Yahav D., Yelin D., Eckerle I., Eberhardt C.S., Wang J., Cao B., Kaiser L. Definitions for coronavirus disease 2019 reinfection, relapse and PCR re-positivity. Clin. Microbiol. Infect. 2021;27(March (3)):315–318. doi: 10.1016/j.cmi.2020.11.028. Epub 2020 Dec 5. PMID: 33285276; PMCID: PMC7718119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Jackson C.B., Mou H., Ojha A., Rangarajan E.S., Izard T., Farzan M., Choe H. The D614G mutation in the SARS-CoV-2 spike protein reduces S1 shedding and increases infectivity. bioRxiv [Preprint]. 2020;(Jun 12) doi: 10.1101/2020.06.12.148726. 2020.06.12.148726, Update in: Nat Commun. 2020 Nov 26;11(1):6013. PMID: 32587973; PMCID: PMC7310631. [DOI] [Google Scholar]