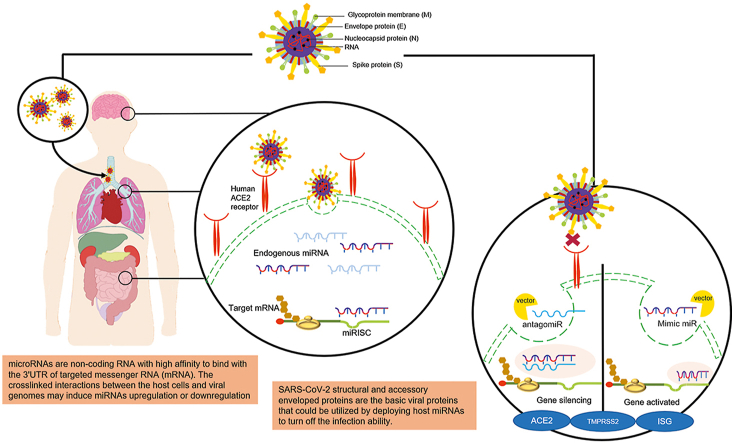
Abstract
Since the World Health Organization (WHO) declared COVID-19, the disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), as a pandemic in March 2020, and more than 117 million people worldwide have been confirmed to have been infected. Scientists, medical professionals, and other stakeholders are racing against time to find and develop effective medicines for COVID-19. However, no drug with high efficacy to treat SARS-CoV-2 infection has been approved. With the increasing popularity of gene therapy, scientists have explored the utilization of small RNAs such as microRNAs (miRNAs) as therapeutics. miRNAs are non-coding RNAs with high affinity for the 3′-UTRs of targeted messenger RNAs (mRNAs). Interactions between host cells and viral genomes may induce the upregulation or downregulation of various miRNAs. Therefore, understanding the expression patterns of these miRNAs and their functions will provide insights into potential miRNA-based therapies. This review systematically summarizes the potential targets of miRNA-based therapies for SARS-CoV-2 infection and examines the viability of possible transfection methods.
Keywords: microRNA, SARS-CoV-2, COVID-19, Targeted therapy
Graphical abstract
Highlights
-
•
Currently there is no single approved treatments for SARS-CoV-2.
-
•
ACE2 and structural proteins of SARS-CoV-2 are the main targets for recent drug development.
-
•
Gene therapy especially the advantageous of microRNA (miRNA/miR) has gained popularity even for infectious disease.
-
•
MiRNA based therapeutics in a form of miR antagonist and mimic approach has potential to answer the challenge.
1. Introduction
MicroRNAs (miRNAs) are short non-coding RNAs, 19–25 nucleotides in length. After their biogenesis, mature miRNAs interact with certain messenger RNAs (mRNAs) in the cytoplasm during post-transcriptional regulation in a multiprotein RNA-induced silencing complex (RISC) [1,2]. These miRNA-induced silencing complexes (miRISCs) interact with target mRNAs primarily at complementary sequences in the 3′-untranslated region (3′-UTR) and mainly decrease mRNA stability or promote mRNA degradation [3,4]. Perfect complementary binding results in mRNA cleavage; however, imperfect binding results in aborted translation [5]. miRNAs were first observed in phenotype variants of Caenorhabditis elegans; through several experimental approaches, a negative regulatory element (NRE) located at the 3′-UTR of the lin-14 gene was found to be responsible for the observed abnormal morphology, and it was shown that the element produced small RNA molecules. However, protein levels were not affected [3]. This surprising finding was later discussed, and interest in the phenomenon began to increase [6]. Although miRNAs are non-coding RNAs, more than 500 miRNAs have been identified in humans, and a single miRNA can modulate hundreds of different mRNAs [7]. Several studies have confirmed that 30%–60% of mammalian mRNAs can be regulated by a single miRNA [8,9]. Studies of miRNAs have become quite popular, and much of the focus has been on identifying and characterizing miRNAs for their potential as either biomarkers or in targeted therapies not only in animal cells but also in human cells [6,10].
Epigenetic factors and dysregulation during miRNA biogenesis could alter their expression and can lead to abnormalities in various cellular activities, such as proliferation, growth, differentiation, apoptosis, cell cycle, and tissue development [11,12]. Moreover, several miRNAs have been reported to mediate shifts in host defense mechanisms related to cellular immunity [13].
2. Roles of miRNAs in viral infections
Immunity is a complex process. During an immune reaction, several molecular pathways, which include miRNAs, have long been known to mediate the process of inflammation primarily via immune cell modulation. Like their hosts, most pathogens, such as viruses, also encode miRNAs to promote their survival [14]. Due to their small size, lack of immunogenicity, and excellent functional flexibility, miRNAs are susceptible to host miRNA-mRNA co-modulation expression during infection [9]. To ensure their survival, viruses must replicate their genetic material by using the host's cellular machinery. During this process, crosstalk can occur between host miRNAs and the viral genome [8,15,16]. These interactions can occur via three different pathways: (i) mRNA blocking, (ii) mRNA destabilization, and (iii) mRNA degradation via protein complexes [17]. However, viruses can also mediate changes in cellular miRNAs prior to infection, to create an advantageous environment for pathogenesis. These miRNAs mostly bind to the 5′ and 3′ non-translated regions (NTRs), although some bind in the coding regions of viral proteins [18,19].
Some studies have suggested that host miRNA-viral genome interactions are beneficial to virus sustainability [20]. Wang et al. [21] observed that oncogenic human papillomavirus virus 16 HPV16 and HPV18 infection could suppress the expression of the host miRNA miR-34 and destabilize the expression of the tumor suppressor p53 to induce tumor formation. Poxvirus, which causes smallpox in humans, has been reported to facilitate the degradation of host miRNAs. The further investigation showed that the ability of poxvirus to evade the host miRNA defense system and the binding of host miRNAs in the 3′-UTR of the viral genome massively downregulated host miRNAs to enhance the proliferation of infected cells [16]. Cell degradation after viral infection was also shown after an miRNA from the influenza PB1 gene coding region bound to the host miRNAs miR-323, miR-491, miR-485, miR-654, and miR-3145 [19]. Human epithelial type 2 (HEp-2) cells infected with respiratory syncytial virus (RSV)-green fluorescent protein (GFP) contained lower miRNA levels when compared to the levels in non-infected cells, and the NF-κB monomers p65, p50, and p52 were significantly altered. Although RSV does not encode any miRNA in its genome, its binding to the host miRNA miR-146a-5p reduced its expression and downregulated miR-345–5p, let-7c-5p, and miR-221. Furthermore, these cells did not appropriately respond to and kill the virus because the expression of p21 and the 70 kDa heat shock protein (HSP-70) was suppressed [9].
In contrast, some host miRNAs block virus accumulation. The presence of miRNA response elements (MREs) within the 5′- and 3′-UTRs of the viral genome has been suggested as the main way that miRNAs respond to pathogenic viral activities [22]. MiR-32 can interfere with further spread of the retrovirus primate foamy virus type 1 (PFV-1) in normal human cells by activating the RNA-mediated silencing machinery to act on the viral proteins Tas and Bet [23]. In a mouse model, miR-144 targeted the tumor necrosis factor (TNF) receptor-associated factor 6 (TRAF6)/IRF7 signaling axis and further decreased type-1 interferon (IFN–I) expression during H1N1 influenza virus infection [24]. Peng et al. [25] investigated the contribution of hsa-miR-1-3p to the inhibition of viral replication by targeting the supportive host factor ATP6VIA after infection with two main influenza A virus (IAV) subtypes H1N1 (PR8) and H3N2 in HEK293T cells.
The growing interest in miRNA-related studies is due to in part to its role in unraveling the mysteries of RNA and because they can also be used as a tool to discover targets for alternative therapies for various diseases, such as COVID-19, the disease caused by severe acute respiratory syndrome-corona virus-2 (SARS-CoV-2). The latest epidemiological data from the WHO reported [26] that SARS-CoV-2 has spread to more than 200 countries and has infected over 65.8 million people, resulting in nearly 1.5 million deaths globally. SARS-CoV-2, which like SARS-CoV and Middle East respiratory syndrome coronavirus (MERS-CoV), belongs to the family Coronaviridae (subfamily Coronavirinae) but has lower morbidity and mortality rates than the two previous viruses [27,28]. SARS-CoV-2 is an enveloped, non-segmented, positive-sense single-stranded RNA virus [27,29] and is of the same lineage as SARS-CoV, which caused the SARS epidemic in 2003. SARS-CoV-2 is one of the seven members of the CoV family that can infect humans and cause diseases with a wide spectrum of clinical manifestations, by attacking the respiratory epithelial barrier and causing serious acute lung injury (ALI), sometimes acute respiratory distress syndrome (ARDS), and even death from multi-organ failure (MOF) due to massive inflammatory cell infiltration and proinflammatory cytokine/chemokine release, which is sometime referred to as a “cytokine storm” [28,30,31].
3. Host-pathogen interactions in SARS-CoV-2 infection
SARS-CoV-2 shares many of the same characteristics as most coronavirus family viruses, including a pleomorphic or spherical shaped capsid envelope 150–160 nm in size, a polycistronic mRNA with a 5′ cap and 3′ poly-A-tail, and an unsegmented, positive, single-stranded RNA genome (30 kb) [[32], [33], [34]]. SARS-CoV-2 has two large polyproteins, ORF1a and ORF1b; four structural proteins: spike (S), envelope (E), membrane (M), and nucleocapsid (N); and eight accessory proteins: ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, and ORF9b. It has also been determined that SARS-CoV-2 lacks a hemagglutinin esterase gene, which is strongly related to the betacoronavirus human coronavirus (hCoV) HKU1 [32,34]. ORF1ab expresses a total of 16 non-structural proteins (NSP) named NSP1–16 with the help of papain-like protease (PLpro) and 3C-like protease (3CLpro), which cleave the replicase polyprotein. Each NSP has a particular role in disrupting the host cell environment while keeping the virus secure. Some of these NSPs also encode proteins with essential features, such as PLpro (nps3), 3CLpro (nsp5), and RNA-dependent RNA polymerase/RdRp (nsp12) [32,35]. RdRp plays a major role in the viral life cycle, and scientists believe that one of the active sites of RdRp could be used as a potential target for drug treatment [36]. The structural proteins S, E, M, and N are essential for virus assembly and have the primary function of inducing host cell infection [34].
Like other viruses, SARS-CoV-2 creates various problems in host cells, ranging from triggering abnormal gene expression to global cell destruction, which are related to the host immune response. The molecular mechanism is still observable because it involves complex mechanisms throughout the system [37]. Type II pneumocytes and mucosal ciliated cells in the lung are thought to be the major targets of SARS-CoV and IAV infection because these epithelia are relatively vulnerable to respiratory infection due to their high risk of pathogen exposure [38]. The main target cells of SARS-CoV-2 in the airway epithelial, alveolar epithelial, and vascular endothelial cells are angiotensin-converting enzyme 2 (ACE2) receptor and transmembrane protease serine 2 (TMPRSS2) [39]. The receptor binding domain (RBD) in the S1 subunit of the surface-expressed S protein of SARS-CoV-2 binds to ACE2 receptors on the target host cells, and this binding initiates the process of infection. The S1 subunit is cleaved, while the S2 subunit, which consists of a fusion peptide (FP) and two heptad repeat regions, HR1 and HR2, folds in on itself, bringing the two heptad repeat regions together. Then, the viral genome is unpackaged in the host cell cytoplasm [39].
Studies have revealed that during SARS-CoV-2 infection, several inflammatory pathways can be activated [40]. When the innate immune system is activated, pattern recognition receptors (PRRs), especially Toll-like receptor type 3 (TLR3), respond to pathogen-associated molecular patterns (PAMPS), resulting in robust intercellular signaling through the Toll/IL-1R domain-containing adaptor-inducing interferon-beta (IFN-β) (TRIF)-dependent pathways [14]. Through these pathways, the transcription factor nuclear factor kappa B (NF-κB) may be activated to stimulate further immune responses [14,29,41]. Activation of NF-κB can lead to the induction of downstream factors including proinflammatory cytokines, such as interleukin-6 (IL-6), interferon-alpha (IFN-α) and IFN-β, IL-1b, IL-6, IL-12, IL-18, IL-33, TNF-α, and tumor growth factor-beta (TGF-β); several chemokines, including C–C motif chemokine ligands CCL2, CCL3, and CCL5, and C-X-C motif chemokine ligands CXCL8, CXCL9, and CXCL10 [29,42]. Activation of these cytokines can lead to a “cytokine storm”. In patients with moderate to severe COVID-19, Yang et al. [43] found that elevated concentrations of three proteins, IL-1 receptor antagonist, CCL7, and CXCL10, were correlated with viral load and lung damage. In such cases, poor outcomes, including death, were often unavoidable.
The presence of immunoglobulin M (IgM) and immunoglobulin G (IgG) antibodies was detected at days 6–8 after exposure [44], indicating that adaptive immunity was also activated during the infection; IgM was reported to peak at 3 weeks and decline slowly, whereas IgG continued to increase and may persist beyond 7 weeks [45]. Infected cells are recognized by cytotoxic T lymphocytes (CTLs) or CD8+ T lymphocytes through major histocompatibility complex class 1 (MHC class 1) complexes on the cell surface. This recognition attracts immune molecules to diminish cells. As reported by Baruah and Bose [46], there are five different CTL epitopes, three sequential B cell epitopes, and five discontinuous B cell epitopes in the SARS-CoV-2 surface glycoprotein. However, samples collected from SARS-CoV-2-infected subjects suggested that CD3+, CD4+, and CD8+ levels were mostly below the normal range as both B cells and NK cells were significantly decreased. This finding paralleled those of studies showing direct viral killing of lymphocytes as a reason for the massive reductions in various immune cell populations, which caused lymphopenia in COVID-19 patients [39,47]. Xiong et al. [48] reported the presence of several altered genes in bronchoalveolar lavage fluid, with one surprising result, an increase of TP53I3, which is known to be vital during p53 signaling-induced programmed immune cell death.
Since a trial of recombinant IFN showed remarkable results as a SARS-CoV treatment, a similar trial is currently under way for SARS-CoV-2 [49]. Various pharmacological interventions that showed significant improvement in IAV, SARS, and MERS infection are now being applied to SARS-CoV-2 cases. However, specific, effective therapies for patients with COVID-19 have not yet received clinical approval [50].
4. miRNAs in SARS-CoV-2
The discovery of miRNAs as part of RNAi with antiviral ability was observed quite well but still remained unclear. Attachment of SARS-CoV-2 to host receptors enables the virus to release its genome into the cytoplasm, where it employs the host machinery for replication. Once in the cytoplasm, an antiviral defense, in the form of host endogenous miRNAs, is activated that it recognizes small viral RNA (svRNA) [51,52]. The RNA-ase III endonuclease Dicer, the enzyme that processes mature miRNAs, allows the host RNAi machinery to control viral RNA replication by specifically recognizing the viral double-stranded RNA (dsRNA) through complementary sequence recognition. This allows the virus to evade the host antigen-specific response to viral proteins [53].
An in silico study suggested that numerous cellular miRNAs could target the SARS-CoV-2 genome, with each miRNA having up to 10 target sites. Thus, these miRNAs have the potential as therapeutic agents. However, as shown in an miRNA analysis by Fulzele et al. [20], miRNA activity in patients infected with SARS-CoV-2 is age-dependent. Younger patients have more host miRNAs with viral complementary binding sites than aged individuals, and thus may be better able to prevent viral replication. This finding is also supported by a study by Huan et al. [54], which showed that miRNA levels in peripheral blood are related to age.
4.1. miRNA-based therapeutic approaches in SARS-CoV-2
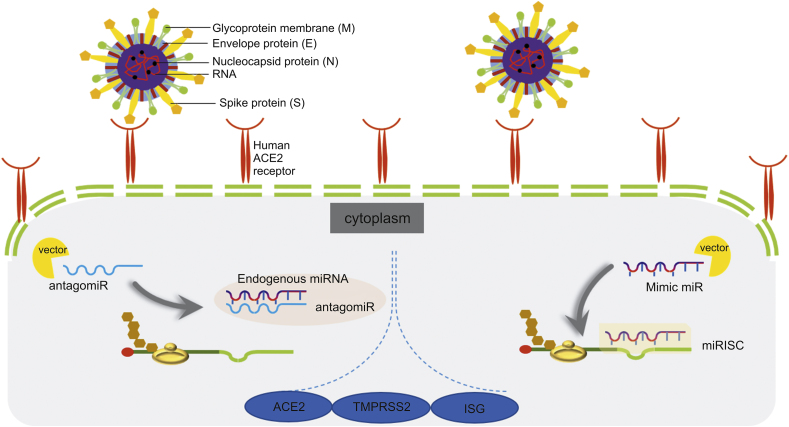
The advantage of identifying the hallmarks of any disease, such as those caused by viral infections, is that such information helps scientists develop therapies. Nucleosides, nucleotides, viral nucleic acids, and enzymes/proteins involved in the replication and transcription of SARS-CoV-2 are potential treatment targets [49]. As mentioned briefly above, miRNAs have a significant relationship with viral infection, as they can either enhance viral activity or stop replication through numerous pathways. Several studies of miRNA-based therapies have been conducted, and in cancer, the use of miRNAs has successfully increased the survival rate [12]. Current miRNA-based therapies focus on suppressing or enhancing miRNA activity. Strategies that inhibit miRNA expression utilize modified miRNA inhibitors or oligomers, such as miRNA antisense therapy, small molecule inhibitors, locked nucleic acids (LNAs), peptide nucleic acids (PNAs), morpholinos, miRNA sponges, and miRNA binding site silencing using gene-targeted single-stranded RNA (ssRNA) sequences. In contrast, miRNA replacement therapy usually refers to augmenting cellular miRNA activity by transfecting synthetic mimics or miRNA-loaded lentiviral vectors [55]. An antagomir is a small synthetic RNA with the ability to bind to certain genes through complementary sequences [56]. Antagomir miR-122 transfection was reported as the first synthetic oligonucleotide therapy, and it was shown to cure hepatitis C virus (HCV) infection. Thereafter, several studies have been conducted using the same strategy. In contrast to antagomirs, miRNA mimics interact with the target by assembling with the non-distinguishable sense of true endogenous miRNAs to induce changes in mRNA expression through miRISC (Fig. 1) [5,22]. The main purpose of administering miRNA mimics is to restore the functional level of an underexpressed intracellular miRNA that is related to disease. Several miRNA mimic therapies have shown potential, especially in cancer, and one clinical trial for hepatocellular carcinoma has been conducted. However, due to five cases of immune-related side effects, the study was halted in September 2016 [4,22].
Fig. 1.
The illustration shows how transfected antagomir and miR mimic to host cell being processed intracellular. MiRNA: microRNA; antagomir: antagonist microRNA; ACE2: angiotensin converting enzyme-2; TMPRSS2: transmembrane protease serine 2; ISG: interferon stimulated gene; miRISC: miRNA-induced silencing complex.
4.2. miRNAs that target viral proteins related to host binding
Among the structural proteins that envelop SARS-CoV-2, the spike (S) glycoprotein is considered to bind to host cells through the ACE2 receptor [29]. ACE2 receptors are abundantly expressed in the lungs and heart, and are also expressed in the digestive tract, testes, and kidneys. This distribution of ACE2 receptors explains the wide variation in the symptoms and clinical signs of SARS-CoV-2 infection [57]. In the circulation, ACE2, which is an ACE homolog, plays a role in regulating blood pressure and electrolyte levels [58,59]. Similar to SARS-CoV, after the S1 subunit binds to ACE2, the S2 subunit of the S protein is likely to undergo conformational changes, leading to viral-to-host attachment, which promotes viral entry. Internalization causes SARS-CoV-2 to release its viral RNA genome into the cytoplasm (Fig. 1). The attachment process is peptidase-activity independent, but several transmembrane proteinases may be involved in binding [59,60]. RNA interference (RNAi) has been widely utilized to silence certain mRNAs in SARS-CoV infection since the SARS/MERS pandemics more than 10 years ago [61]. For example, Qin et al. [60] transfected the sense and antisense templates of small interfering RNAs (siRNAs) into SARS CoV-infected HEK293T cells and tested the ability of the small non-coding RNAs to bind with the SARS-CoV S gene and modulate its degradation, resulting in the failure of host cell receptor binding.
Like siRNA, miRNA-mediated RNAi has attracted much interest. During a respiratory infection, ACE2 is thought to function to protect the lungs from excess inflammation that would trigger ARDS. As shown in Table 1 [[62], [63], [64], [65], [66]], the results of one study targeting the ACE2 receptors in the lungs during H5N1 influenza infection led to acute lung injury or ARDS. This study showed that the virus massively disrupted ACE2 expression upon binding. In a mouse model, plasma miR-200c-3p levels were significantly elevated and were correlated with ARDS symptoms. Administration of antagomir miR-200c-3p successfully ameliorated lung edema in the mouse model by upregulating ACE2 expression, which induced NF-κB activation [62]. This study was also supported by the findings of Nersisyan et al. [67], who identified several miRs, such as hsa-let-7e/hsa-miR-125a and hsa-miR-141/hsa-miR-200 that target ACE2 and TMPRSS2 genes and are modulated by lysine-specific demethylase 5 B (JARID18). Surprisingly, Sacconi et al. [68] demonstrated through bioinformatics and experimental analyses that patients with head and neck cancer might have higher resistance to SARS-CoV-2 infection due to downregulated expression of TMPRSS2. Further observation showed a strong correlation with the upregulation of miR-31–3p and miR-503–5p. However, this finding requires confirmation in studies targeting these miRNAs in SARS-CoV-2-infected cells. Another study investigated several host miRNA targets within SARS-CoV that were expressed in bronchoalveolar stem cells (BASCs) (Table 1 [63]). Downregulated expressions of miR-17, miR-574–5p, and miR-214 in BASCs were found to enhance the activities of not only the S glycoprotein but also the nucleotide (N) protein, matrix (M) protein, small envelope (E) protein, and ORF1a primarily by acting upon host receptor binding. Therefore, suppressing the expressed viral proteins by upregulating these miRs is likely to inhibit viral internalization. Similar results were observed for miR-223 and miR-98; downregulation of these miRs (by 8.7- and 5.6-fold, respectively) enabled the N protein and S glycoprotein to engage in viral-to-host transmission [63].
Table 1.
Examples of miRNA-based therapies for respiratory infectious diseases.
| miRNA therapy | Virus | Targets | Effects | Refs. |
|---|---|---|---|---|
| Antagomir-miR-200c-3p | H5N1 influenza | ACE2, NF-κB upregulation | Decrease lung edema in severe pneumonia in mice model | [62] |
| Antagomir-200c-5p | H5N1 influenza | DUSP1 | Attenuate pulmonary inflammatory responses and lung injury | [62] |
| miR-17 mimic | SARS-CoV | Viral S, N, E, M protein and ORF1a | Deactivate viral proteins based on microarray analysis (BASCGAP consortium) in bronchoalveolar stem cells (BASC) | [63] |
| miR-574–5p mimic | ||||
| miR-214 mimic | ||||
| miR-223 mimic | SARS-CoV | N and S protein | Viral entry based on microarray analysis (BASCGAP consortium) in BASC | [63] |
| miR-98 mimic | ||||
| miR-155 mimic | VSV | STAT1, STAT2 | Phosphorylate STAT1/2 to activate ISGs transcription in murine-VSV infected ex vivo model | [64] |
| Inhibitor hsa-let-7e-5p | Human-Adv | NF-κB, SOC, STAT, and IFN | Viral replication in human primary lung fibroblasts (IMR-90) cells infected with HAdV5 | [65,66] |
Abbreviation: Antagomir: antagonist miRNA; ACE-2: angiotensin converting enzyme-2; DUSP1: dual-specificity phosphatase 1; BASC: bronchoalveolar stem cells; BASCGAP: bronchoalveolar stem cells genome anatomy projects; STAT1/STAT2: signal transducers and activators of transcription 1 and 2; VSV: vesicular stomatitis virus; ISG: interferon stimulated gene; Human-Adv: human adenovirus; SOC: suppressor of cytokine; IFN: interferon; HAdV5: human adenovirus-5.
In silico studies have shown remarkable results in identifying 3CLpro and RdRp as potential target candidates. However, no miRNA-related studies have confirmed this association. In a study conducted by He et al. [69], it was shown that six designed 21-mer siRNAs were able to target various regions of the SARS-CoV replicase gene (ORF1a). Although this in vitro experiment showed promising results, its clinical usefulness remains to be determined [69,70].
4.3. Role of miRNAs in accelerating interferon-induced antiviral activity
Thus far, three studies have been published that were aimed to identify miRNAs significantly expressed in lung epithelial cells from SARS-CoV-2 patients. Chow and Salmena [71] found 11 miRNAs that were common in all the studies: hsa-miR-5047, hsa-miR-1301–3p, hsa-miR-125a-3p, hsa-miR-19b-2-5p, hsa-miR-141–3p, hsa-miR-19b-1-5p, hsa-miR-1301–3p, hsa-miR-196a-5p, hsa-miR-4758–5p, hsa-miR-15b-3p, and hsa-miR-153–5p [20,72]. Analyses such as these will enable scientists to develop miRNA-based therapies for patients with SARS-CoV-2 infection. Although no existing and clinically approved antagomirs or mimics have been used for SARS-CoV-2, some studies have shown positive results against other viral infectious diseases, such as influenza A [25,71].
Genome wide analyses have revealed host miRNAs that function as either viral enhancers or inhibitors. As antiviral agents, miRNAs mainly act by inhibiting viral RNA production, blocking viral replication, suppressing pro-viral proteins, or inducing the virus to enter a latent phase [73]. Khan et al. [74] found that dysregulation of host miRNAs caused by viral genome interaction in SARS-CoV-2 infection primarily disrupts TLR signaling, which is vital to the activation of antiviral activities, such as IFN regulation and cytokine production, and it also disrupts IL, TRAF6, S1P1, estrogen receptor, protease-activated receptor (PAR), and bone morphogenetic (BMP) signaling. When viral-host recognition occurs, the host innate immunity immediately responds by activating the NF-κB transcription factor to modulate the expression of interferon-stimulated genes (ISGs). ISGs exert antiviral effects by expressing the genes that encode type 1 IFNs. Several miRNAs have been identified that enhance modulation during this process [75]. A study by Wang et al. [64] showed that delivery of miR-155 mimics could alter the phosphorylation of signal transducer and activator of transcription 1 and 2 (STAT1 and STAT2) in murine macrophages after ex vivo vesicular stomatitis virus (VSV) infection. STAT1/2 is characterized by its ability to attach to DNA in the regions that encode ISG proteins. Thus, they act as promoters of transcription [73,76]. Using chromatin immunoprecipitation (ChiP) analysis of miR-21 in the DU145 cell line, Yang et al. [77] identified the binding site for IFN-induced NF-κB in the miR-21 promoter of a complex that showed to contain STAT3 and the p65 NF-κB subunit. Downregulation of miR-200a in the host increased ISG protein expression by targeting IFN-α receptors and STAT2/4, while upregulation of miR-223 induced apoptosis in influenza virus-infected patients by repressing CREB activity and modulating NF-κB activity, thereby supporting proinflammatory responses [78,79]. Lai et al. [80] investigated numerous miRNAs with elevated expression during human coronavirus OC43 infection that suppressed the virus by binding to its nucleocapsid, and miR-9 was one of the miRs that also upregulated NF-κB expression and synergized cell sensitivity to TNF-α.
Activation of T and B lymphocytes in SARS-CoV-2 infection could also be mediated by miRNA. There have been several studies on the protective function of various miRs in adaptive immune responses, such as miR-17-92, which is considered to be a key regulator of T cell development and supports T cell survival through BCL-2 and phosphatase and homolog-mediated tensin (PTEN) [81]. In addition, in vitro and in vivo studies of miR-155 have suggested that its upregulation is correlated with larger numbers of active human CD8+ T cells. Conversely, an miR-155 deficiency resulted in increased type I IFN signaling and reduced proliferation of CD8+ T cells [82].
Recent studies using computational approaches have suggested that SARS-CoV-2 encodes miRNAs that may play a role in evading host immune surveillance through several signaling pathways that may be targeted indirectly, such as the TLR, IL, TRAF6, autophagy, IFN-I, Wnt, and mammalian target of rapamycin (mTOR) signaling pathways [74]. This finding was also supported by a study conducted by Chow and Salmena [71], which showed that SARS-CoV-2 might have evolved to form a complex secondary structure involving the coding region, and as a result, could evade the effects of endogenous RNAi. However, this conclusion did not finalize the hypothesis, and more studies were needed to confirm these findings. The first study to observe the miRNAs in a virus was an investigation of bovine leukemia virus (BLV). In this virus, replication can occur only when miRNAs bind to the mRNA of transcription factors. Ebola virus (EBOV) has also been reported to encode several miRNAs based on observations obtained using HEK293T cells transfected with a plasmid harboring an EBOV pre-miRNA, and the host cell machinery induced viral miRNA production upon interaction [17].
5. MiRNA delivery systems
Systemic miRNA delivery still faces significant challenges since delivered miRNAs could access many cytosolic mRNAs. In infections, immunity acts as a double-edged sword, and it has been reported that miRNAs may activate the innate immune response or even be rejected by cellular immunity [12,22]. In addition, the physiological characteristics of miRNAs, such as their short half-life, make them difficult to handle. Before an miRNA reaches its target, it may be degraded [83], and delivery of naked miRNA is impossible. Therefore, each miRNA needs to be conjugated to a special vehicle. Several crucial points must be considered when constructing vectors for miRNA delivery, including possible toxicity and formulation of the effective dose to maximize bioavailability [12]. To minimize possible toxicity, nanocarriers may be the answer. Recently, well-developed nanocarriers based on organic, inorganic, lipid, protein, glycan, or even synthetic components have been developed [84,85]. Some carriers used in clinical settings are liposome-based, nanocrystal-based, emulsion-based, and iron-carbohydrate complex-based nanocarriers [86]. Nanocarriers are often chemically modified with peptide-like antibodies, proteins, or small molecules to ensure high transfection through cells [87]. To minimize possible toxicity and immunogenicity risks of viral vectors, nanocarriers may be the answer. Nucleic acid delivery systems usually require a nanocarrier or vector. The use of a viral-capsid vector is still a popular notion; however, they often trigger serious immune responses in the host [88,89].
6. Conclusions
Understanding the role of miRNAs in SARS-CoV-2 pathobiology has been gaining interest. Although the overall mechanism of viral infection for SARS-CoV-2 has been well clarified based on those of the related viruses, SARS-CoV and MERS-CoV, many questions remain regarding the exact molecular signaling pathways involved. Various miRNA-related studies have attempted to explore the association between small non-coding RNAs and the expression of numerous mRNAs. The structural and accessory envelope proteins of SARS-CoV-2 are the basic viral proteins that can be targeted by host miRNAs to block infection. For instance, miR-17, miR-574–5p, and miR-214 are shown to induce degradation of the S glycoprotein, N and M proteins, and ORF1a. Upon binding to ACE2 receptors, host immunity is activated in an attempt to kill the virus and stop further replication. Several miRNAs, such as miR-155, would enable us to target both the innate and adaptive immunity of the host. It has also been shown that miR-9 plays a role in upregulating NF-κB expression and boosting immunity. Some miRNAs could be used as diagnostic tools to screen patients or even as therapies. The results of the miRNAs imbalance through screenings can be developed into either miR-inhibitor or miR-mimics for therapeutic strategies. However, several challenges remain, especially in the safe delivery of miRNAs. Various solutions are being developed to manage this problem. We do know that some non-viral vectors can be used to transfect miRNAs with fewer immunogenic and toxic effects. However, further studies are required in the near future to develop suitable and precise delivery of miRNAs for the treatment of SARS-CoV-2 infection.
Declaration of competing interest
The authors declare that there are no conflicts of interest.
Footnotes
Peer review under responsibility of Xi'an Jiaotong University.
References
- 1.Mitash N., Donovan J.E., Swiatecka-Urban A. The role of microrna in the airway surface liquid homeostasis. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21113848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nishioka C., Ikezoe T., Pan B. MicroRNA-9 plays a role in interleukin-10-mediated expression of E-cadherin in acute myelogenous leukemia cells. Canc. Sci. 2017;108:685–695. doi: 10.1111/cas.13170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pager C.T., Wehner K.A., Fuchs G. Chapter 5 MicroRNA-mediated gene silencing. In: Hershey J.W.B., editor. first ed. Elsevier Inc.; 2009. pp. 187–210. (Progress in Molecular Biology and Translational Science). [DOI] [PubMed] [Google Scholar]
- 4.Mekuria A.N., Abdi A.D., Mishore K.M. MicroRNAs as a potential target for cancer therapy. J. Canc. Sci. Ther. 2018;10:152–161. [Google Scholar]
- 5.Hosseinahli N., Aghapour M., Duijf P.H.G. Treating cancer with microRNA replacement therapy: a literature review. J. Cell. Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 6.Bhaskaran M., Mohan M. MicroRNAs: history, biogenesis, and their evolving role in animal development and disease. Vet. Pathol. 2014;51:759–774. doi: 10.1177/0300985813502820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Umbach J.L., Cullen B.R. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23:1151–1164. doi: 10.1101/gad.1793309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eilam-Frenkel B., Naaman H., Berkic G. MicroRNA 146-5p, miR-let-7c-5p, miR-221 and miR-345-5p are differentially expressed in Respiratory Syncytial Virus (RSV) persistently infected HEp-2 cells. Virus Res. 2018;251:34–39. doi: 10.1016/j.virusres.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 10.O’Brien J., Hayder H., Zayed Y. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018;9 doi: 10.3389/fendo.2018.00402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan G., Tang X., Tang F. The role of microRNAs in nasopharyngeal carcinoma. Tumor Biol. 2014;36:69–79. doi: 10.1007/s13277-014-2847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mollaei H., Safaralizadeh R., Rostami Z. MicroRNA replacement therapy in cancer. J. Cell. Physiol. 2019;234:12369–12384. doi: 10.1002/jcp.28058. [DOI] [PubMed] [Google Scholar]
- 13.Saini S., Saini A., Thakur C.J. Genome-wide computational prediction of miRNAs in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) revealed target genes involved in pulmonary vasculature and antiviral innate immunity. Mol. Biol. Res. Commun. 2020;9:83–91. doi: 10.22099/mbrc.2020.36507.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandan K., Gupta M., Sarwat M. Role of host and pathogen-derived MicroRNAs in immune regulation during infectious and inflammatory diseases. Front. Immunol. 2020;10 doi: 10.3389/fimmu.2019.03081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scaria V., Hariharan M., Maiti S. Host-virus interaction: a new role for microRNAs. Retrovirology. 2006;3 doi: 10.1186/1742-4690-3-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guo Y.E., Steitz J.A. Virus meets host MicroRNA: the destroyer, the booster, the hijacker. Mol. Cell Biol. 2014;34:3780–3787. doi: 10.1128/MCB.00871-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.González Plaza J.J. Current roles of microRNAs in infectious diseases – advancing into healthcare. Croat. J. Infect. 2016;36:5–15. [Google Scholar]
- 18.Trobaugh D.W., Gardner C.L., Sun C. RNA viruses can hijack vertebrate microRNAs to suppress innate immunity. Nature. 2014;506:245–248. doi: 10.1038/nature12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Trobaugh D.W., Klimstra W.B. MicroRNA regulation of RNA virus replication and pathogenesis. Trends Mol. Med. 2017;23:80–93. doi: 10.1016/j.molmed.2016.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fulzele S., Sahay B., Yusufu I. COVID-19 virulence in aged patients might be impacted by the host cellular MicroRNAs abundance/profile. Aging Dis. 2020;11:509–522. doi: 10.14336/AD.2020.0428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang X., Wang H.-K., McCoy J.P. Oncogenic HPV infection interrupts the expression of tumor-suppressive miR-34a through viral oncoprotein E6. RNA. 2009;15:637–647. doi: 10.1261/rna.1442309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wong R.R., Abd-Aziz N., Affendi S. Role of microRNAs in antiviral responses to dengue infection. J. Biomed. Sci. 2020;27 doi: 10.1186/s12929-019-0614-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lecellier C.H., Dunoyer P., Arar K. A cellular microRNA mediates antiviral defense in human cells. Science. 2005;308:557–560. doi: 10.1126/science.1108784. [DOI] [PubMed] [Google Scholar]
- 24.Nguyen T.H., Liu X., Su Z.Z. Potential role of MicroRNAs in the regulation of antiviral responses to influenza infection. Front. Immunol. 2018;9 doi: 10.3389/fimmu.2018.01541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peng S., Wang J., Wei S. Endogenous cellular MicroRNAs mediate antiviral defense against influenza A virus. Mol. Ther. Nucleic Acids. 2018;10:361–375. doi: 10.1016/j.omtn.2017.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.World Health Organization Weekly epidemiological update-8 December 2020. 2020. https://www.who.int/publications/m/item/weekly-epidemiological-update-8-december-2020 (Accessed 13 December 2020)
- 27.Machhi J., Herskovitz J., Senan A.M. The natural history, pathobiology, and clinical manifestations of SARS-CoV-2 infections. J. Neuroimmune Pharmacol. 2020;15:359–386. doi: 10.1007/s11481-020-09944-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dhama K., Khan S., Tiwari R. Coronavirus disease 2019–COVID-19. Clin. Microbiol. Rev. 2020;33 doi: 10.1128/CMR.00028-20. e00028-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Astuti I., Ysrafil Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): an overview of viral structure and host response. Diabetes Metab. Syndr. Clin. Res. Rev. 2020;14:407–412. doi: 10.1016/j.dsx.2020.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bradley B.T., Bryan A. Emerging respiratory infections: the infectious disease pathology of SARS, MERS, pandemic influenza, and Legionella. Semin. Diagn. Pathol. 2019;36:152–159. doi: 10.1053/j.semdp.2019.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crimi E., Benincasa G., Figueroa-Marrero N. Epigenetic susceptibility to severe respiratory viral infections: pathogenic and therapeutic implications: a narrative review. Br. J. Anaesth. 2020:1002–1017. doi: 10.1016/j.bja.2020.06.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoshimoto F.K. The proteins of severe acute respiratory syndrome coronavirus-2 (SARS CoV-2 or n-COV19), the cause of COVID-19. Protein J. 2020;39:198–216. doi: 10.1007/s10930-020-09901-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kannan S., Ali P.S.S., Sheeza A. COVID-19 (Novel Coronavirus 2019) – recent trends. Eur. Rev. Med. Pharmacol. Sci. 2020;24:2006–2011. doi: 10.26355/eurrev_202002_20378. [DOI] [PubMed] [Google Scholar]
- 34.Wu J., Yuan X., Wang B. Severe acute respiratory syndrome coronavirus 2: from gene structure to pathogenic mechanisms and potential therapy. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aftab S.O., Ghouri M.Z., Masood M.U. Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J. Transl. Med. 2020;18 doi: 10.1186/s12967-020-02439-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia H., Gong P. A structure-function diversity survey of the rna-dependent rna polymerases from the positive-strand RNA viruses. Front. Microbiol. 2019;10 doi: 10.3389/fmicb.2019.01945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arisan E.D., Dart A., Grant G.H. The prediction of miRNAs in SARS-CoV-2 genomes: host responses and virus pathogenicity-related KEGG pathways significant for comorbidities. Viruses. 2020;12 doi: 10.3390/v12060614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Totura A.L., Whitmore A., Agnihothram S. Toll-like receptor 3 signaling via TRIF contributes to a protective innate immune response to severe acute respiratory syndrome coronavirus infection allison. mBio. 2015;6 doi: 10.1128/mBio.00638-15. e00638-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tay M.Z., Poh C.M., Rénia L. The trinity of COVID-19: immunity, inflammation and intervention. Nat. Rev. Immunol. 2020;20:363–374. doi: 10.1038/s41577-020-0311-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paces J., Strizova Z., Smrz D. COVID-19 and the immune system. Physiol. Res. 2020;69:379–388. doi: 10.33549/physiolres.934492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vidya M.K., Kumar V.G., Sejian V. Toll-like receptors: significance, ligands, signaling pathways, and functions in mammals. Int. Rev. Immunol. 2018;37:20–36. doi: 10.1080/08830185.2017.1380200. [DOI] [PubMed] [Google Scholar]
- 42.Neufeldt C.J., Cerikan B., Cortese M. BioRxiv; 2020. SARS-CoV-2 Infection Induces a Pro-inflammatory Cytokine Response through cGAS-STING and NF-kB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Y., Shen C., Li J. MedRxiv; 2020. Exuberant Elevation of IP-10, MCP-3 and IL-1ra during SARS-CoV-2 Infection Is Associated with Disease Severity and Fatal Outcome. [DOI] [Google Scholar]
- 44.World Health Organization, What we know about the COVID-19 immune response, 2020, https://www.who.int/docs/default-source/coronaviruse/risk-comms-updates/update-34-immunity-2nd.pdf?sfvrsn=8a488cb6_2. (Accessed 29 January 2021).
- 45.Liu C., Yu X., Gao C. Characterization of antibody responses to SARS-CoV-2 in convalescent COVID-19 patients. J. Med. Virol. 2021;93:2227–2233. doi: 10.1002/jmv.26646. [DOI] [PubMed] [Google Scholar]
- 46.Baruah V., Bose S. Immunoinformatics-aided identification of T cell and B cell epitopes in the surface glycoprotein of 2019-nCoV. J. Med. Virol. 2020;92:495–500. doi: 10.1002/jmv.25698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allegra A., Di Gioacchino M., Tonacci A. Immunopathology of SARS-CoV-2 infection: immune cells and mediators, prognostic factors, and immune-therapeutic implications. Int. J. Mol. Sci. 2020;21 doi: 10.3390/ijms21134782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xiong Y., Liu Y., Cao L. Transcriptomic characteristics of bronchoalveolar lavage fluid and peripheral blood mononuclear cells in COVID-19 patients, Emerg. Microb. Infect. 2020;9:761–770. doi: 10.1080/22221751.2020.1747363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khan S., Siddique R., Shereen M.A. Emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus 2: biology and therapeutic options. J. Clin. Microbiol. 2020;58:e00187–e00220. doi: 10.1128/JCM.00187-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bhatti J.S., Bhatti G.K., Khullar N. Therapeutic strategies in the development of anti-viral drugs and vaccines against SARS-CoV-2 infection. Mol. Neurobiol. 2020;57:4856–4877. doi: 10.1007/s12035-020-02074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bartoszewski R., Dabrowski M., Jakiela B. SARS-CoV-2 may regulate cellular responses through depletion of specific host miRNAs. Am. J. Physiol. Lung Cell Mol. Physiol. 2020;319:L444–L455. doi: 10.1152/ajplung.00252.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mishra P.K., Tandon R., Byrareddy S.N. Diabetes and COVID-19 risk: an miRNA perspective. Am. J. Physiol. Heart Circ. Physiol. 2020;319:H604–H609. doi: 10.1152/ajpheart.00489.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou R., Rana T. RNA-based mechanisms regulating host-virus interactions. Immunol. Rev. 2013;253:97–111. doi: 10.1111/imr.12053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huan T., Chen G., Liu C. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell. 2018;17 doi: 10.1111/acel.12687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gambari R., Brognara E., Spandidos D.A. Targeting oncomiRNAs and mimicking tumor suppressor miRNAs: Ew trends in the development of miRNA therapeutic strategies in oncology (Review) Int. J. Oncol. 2016;49:5–32. doi: 10.3892/ijo.2016.3503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ji W., Sun B., Su C. Targeting microRNAs in cancer gene therapy. Genes. 2017;8 doi: 10.3390/genes8010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wicik Z., Eyileten C., Jakubik D. ACE2 interaction networks in COVID-19: a physiological framework for prediction of outcome in patients with cardiovascular risk factors. J. Clin. Med. 2020;9:3743. doi: 10.3390/jcm9113743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lu D., Chatterjee S., Xiao K. MicroRNAs targeting the SARS-CoV-2 entry receptor ACE2 in cardiomyocytes. J. Mol. Cell. Cardiol. 2020;148:46–49. doi: 10.1016/j.yjmcc.2020.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ni W., Yang X., Yang D. Role of angiotensin-converting enzyme 2 (ACE2) in COVID-19. Crit. Care. 2020;24 doi: 10.1186/s13054-020-03120-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin Z.L., Zhao P., Zhang X.L. Silencing of SARS-CoV spike gene by small interfering RNA in HEK 293T cells. Biochem. Biophys. Res. Commun. 2004;324:1186–1193. doi: 10.1016/j.bbrc.2004.09.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y., Li T., Fu L. Silencing SARS-CoV Spike protein expression in cultured cells by RNA interference. FEBS Lett. 2004;560:141–146. doi: 10.1016/S0014-5793(04)00087-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu Q., Du J., Yu X. MiRNA-200c-3p is crucial in acute respiratory distress syndrome. Cell Discov. 2017;3 doi: 10.1038/celldisc.2017.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mallick B., Ghosh Z., Chakrabarti J. MicroRNome analysis unravels the molecular basis of SARS infection in bronchoalveolar stem cells. PloS One. 2009;4 doi: 10.1371/journal.pone.0007837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang P., Hou J., Lin L. Inducible microRNA-155 feedback promotes type I IFN signaling in antiviral innate immunity by targeting suppressor of cytokine signaling 1. J. Immunol. 2010;185:6226–6233. doi: 10.4049/jimmunol.1000491. [DOI] [PubMed] [Google Scholar]
- 65.Huang F., Zhang J., Yang D. MicroRNA expression profile of whole blood is altered in adenovirus-infected pneumonia children. Mediat. Inflamm. 2018 doi: 10.1155/2018/2320640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Makkoch J., Poomipak W., Saengchoowong S. Human microRNAs profiling in response to influenza A viruses (subtypes pH1N1, H3N2, and H5N1) Exp. Biol. Med. 2016;241:409–420. doi: 10.1177/1535370215611764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Nersisyan S., Shkurnikov M., Turchinovich A. Integrative analysis of miRNA and mRNA sequencing data reveals potential regulatory mechanisms of ACE2 and TMPRSS2. PloS One. 2020;15 doi: 10.1371/journal.pone.0235987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sacconi A., Donzelli S., Pulito C. TMPRSS2, a SARS-CoV-2 internalization protease is downregulated in head and neck cancer patients. J. Exp. Clin. Canc. Res. 2020;39 doi: 10.1186/s13046-020-01708-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.He M.L., Zheng B., Peng J. Inhibition of SARS-associated coronavirus infection and replication by RNA interference. J. Am. Med. Assoc. 2003;290:2665–2666. doi: 10.1001/jama.290.20.2665. [DOI] [PubMed] [Google Scholar]
- 70.Tong T.R. Therapies for coronaviruses. Part 2: inhibitors of intracellular life cycle. Expert Opin. Ther. Pat. 2009;19:415–431. doi: 10.1517/13543770802600698. [DOI] [PubMed] [Google Scholar]
- 71.Chow J.T.-S., Salmena L. Prediction and analysis of SARS-CoV-2-targeting microRNA in human lung epithelium. Genes. 2020;11 doi: 10.3390/genes11091002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Saçar Demirci M.D., Adan A. Computational analysis of microRNA-mediated interactions in SARS-CoV-2 infection. PeerJ. 2020;8 doi: 10.7717/peerj.9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Barbu M.G., Condrat C.E., Thompson D.C. MicroRNA involvement in signaling pathways during viral infection. Front. Cell Dev. Biol. 2020;8 doi: 10.3389/fcell.2020.00143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Khan M.A.A.K., Sany M.R.U., Islam M.S. Epigenetic regulator miRNA pattern differences among SARS-CoV, SARS-CoV-2, and SARS-CoV-2 world-wide isolates delineated the mystery behind the epic pathogenicity and distinct clinical characteristics of pandemic COVID-19. Front. Genet. 2020;11 doi: 10.3389/fgene.2020.00765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leon-Icaza S.A., Zeng M., Rosas-Taraco A.G. microRNAs in viral acute respiratory infections: immune regulation, biomarkers, therapy, and vaccines. ExRNA. 2019;1 doi: 10.1186/s41544-018-0004-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.de Veer M.J., Holko M., Frevel M. Functional classification of interferon-stimulated genes identified using microarrays. J. Leukoc. Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 77.Yang C.H., Li K., Pfeffer S.R. The type I IFN-Induced miRNA, miR-21. Pharmaceuticals. 2015;8:836–847. doi: 10.3390/ph8040836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y., Chan E.Y., Li J. MicroRNA expression and virulence in pandemic influenza virus-infected mice. J. Virol. 2010;84:3023–3032. doi: 10.1128/JVI.02203-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wen A.Y., Sakamoto K.M., Miller L.S. The role of the transcription factor CREB in immune function. J. Immunol. 2017;185:6413–6419. doi: 10.4049/jimmunol.1001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Lai F.W., Stephenson K.B., Mahony J. Human coronavirus OC43 nucleocapsid protein binds MicroRNA 9 and potentiates NF- B activation. J. Virol. 2014;88:54–65. doi: 10.1128/JVI.02678-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Głobińska A., Pawełczyk M., Kowalski M.L. MicroRNAs and the immune response to respiratory virus infections. Expet Rev. Clin. Immunol. 2014;10:963–971. doi: 10.1586/1744666X.2014.913482. [DOI] [PubMed] [Google Scholar]
- 82.Gracias D.T., Stelekati E., Hope J.L. MicroRNA-155 controls CD8+ T cell responses by regulating interferon signaling. Nat. Immunol. 2013;14:593–602. doi: 10.1038/ni.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chaudhary V., Jangra S., Yadav N.R. Nanotechnology based approaches for detection and delivery of microRNA in healthcare and crop protection. J. Nanobiotechnol. 2018;16 doi: 10.1186/s12951-018-0368-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wicki A., Witzigmann D., Balasubramanian V. Nanomedicine in cancer therapy: challenges, opportunities, and clinical applications. J. Contr. Release. 2015;200:138–157. doi: 10.1016/j.jconrel.2014.12.030. [DOI] [PubMed] [Google Scholar]
- 85.Bahrami B., Hojjat-Farsangi M., Mohammadi H. Nanoparticles and targeted drug delivery in cancer therapy. Immunol. Lett. 2017;190:64–83. doi: 10.1016/j.imlet.2017.07.015. [DOI] [PubMed] [Google Scholar]
- 86.Flühmann B., Ntai I., Borchard G. Nanomedicines: the magic bullets reaching their target? Eur. J. Pharmaceut. Sci. 2019;128:73–80. doi: 10.1016/j.ejps.2018.11.019. [DOI] [PubMed] [Google Scholar]
- 87.Xu X., Ho W., Zhang X. Cancer nanomedicine: from targeted delivery to combination therapy. Trends Mol. Med. 2015;21:223–232. doi: 10.1016/j.molmed.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Santos-Carballal B., Aaldering L.J., Ritzefeld M. Physicochemical and biological characterization of chitosan- microRNA nanocomplexes for gene delivery to MCF-7 breast cancer cells. Sci. Rep. 2015;5 doi: 10.1038/srep13567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bai Z., Wei J., Yu C. Non-viral nanocarriers for intracellular delivery of microRNA therapeutics. J. Mater. Chem. B. 2019;7:1209–1225. doi: 10.1039/c8tb02946f. [DOI] [PubMed] [Google Scholar]