Abstract
Background:
Tamoxifen (TAM) is the eminent first-line drug for endocrine therapy of hormone receptor positive premenopausal breast cancer and reduces the risk of recurrence by ∼50%. However, many patients developed TAM resistance and their diseases recurred. Our previous study on transcriptome profile of TAM resistant breast cancer cells revealed that the TMEM47 is one of the most significantly differentially expressed genes. The mechanism of how TMEM47 is involved in TAM resistance was not known.
Methods:
We constructed a mammal breast cancer cell line, in which TMEM47 was stably overexpressed (TMEM47-OE/MCF-7), to further verify the role of TMEM47 in TAM resistance. siRNA targeting TMEM47 was transfected into TAMR / MCF-7 cells by Liposome. TMEM47 expression was validated on mRNA and protein level by qRT-PCR and western blotting. We tested the cytotoxicity of TAM in the cells. Apoptosis was detected by flow cytometry.
Results:
Compared to the MCF7 cells, TMEM47 mRNA was significantly up regulated more than 6 folds in the TAMR/MCF7 cells and so its protein. TMEM47 expression level in TMEM47-OE/MCF-7 was similar as in the TAMR/MCF-7 cells. The 50% inhibitory concentration (IC50) value (mean ± SD) of TAM in MCF-7, TAMR/MCF-7 and TMEM47-OE/MCF-7 cells was 1.58 ± 0.19, 2.74 ± 0.24 and 3.12 ± 0.32 µγ/mL, respectively. The apoptosis rates of TAMR/MCF-7 and TMEM47-OE/MCF-7 cell lines were significantly lower than that of MCF-7 cells. After 24 and 48 hours TAM treatments, cell viability was significantly inhibitied in TMEM47 knockdown TAMR/MCF7 cells (P < 0.01). Consistant with the decreased cell viability, the apoptosis rate in TMEM47 knockdown TAMR/MCF-7 cells was significantly increased.
Conclusions:
Our results suggest that overexpression of TMEM47 in MCF-7 cells acquired TAM resistance to those cells, and knockdown of TMEM47 in TAMR/MCF-7 cells reversed their resistance to TAM. TMEM47 might confer TAM resistance on MCF-7 cells through the inhibition of apoptosis.
Keywords: tamoxifen (TAM), resistance, TMEM47, overexpression, breast cancer
Introduction
Breast cancer remains one of the most common cancers that affect women’s health globally. Breast cancers are classified into 3 subgroups based on the molecular biomarkers, the estrogen receptor-positive, HER2-positive and triple-negative, of which the estrogen receptor-positive patients account for up to 70% cases. Endocrine therapy which targets to the estrogen receptor-signaling pathway is the major pharmacotherapy for ER+ breast cancers. Clinically, Tamoxifen (TAM) is the first-line drug for endocrine therapy of hormone receptor positive premenopausal breast cancer patients. Unfortunately, drug resistance develops in approximately 40% patients after TAM treatment, which significantly affects the prognosis of patients. Emerging studies are performed to address endocrine-resistance, but the mechanism of TAM resistance remains unclear. Therefore, unraveling the molecular mechanism(s) that underlies TAM-resistance may extend the use of TAM and provide benefits for patients.
To understand the molecular mechanism that might contribute to TAM resistance, we previous generated a TAM-resistant MCF-7 cell line (TAMR/MCF-7) and preformed RNA sequencing (RNA-seq) to compare the transcriptome profiles between TAMR/ MCF-7 cells and its parent MCF-7 cells. In that study, we identified several differentially-expressed genes (DEGs). One of the “top” DEGs is transmembrane protein 47 (TMEM47), which is significantly up-regulated in TAMR/MCF-7 cells (FDR = 1.67E-15; log2 fold change = 6.86)1 TMEM47 encodes a tetraspan protein in the PMP22/EMP/Claudin family, which is involved in the maintenance of cell junction organization. Although the function of TMEM47 is unknown, other TMEM family proteins are known to be critical components on various biological membrane and play roles in many physiological and pathological process.2 It has been reported that TMEM family members, such as TMEM97 and TMEM48, may serve as biomarkers for cancer prognosis.3-8 Some of TMEM members, such as TMEM158, TMEM14A, TMEM97 and TMEM88, are known involved in carcinogenesis.9 Of note, several TMEM family members such as TEME45A, TMEM88 and TMEM205, have been demonstrated that they could contribute to chemotherapy resistance.10-12 Since we have observed that TMEM47 mRNA is significantly increased in TAM-resistance MCF-7 cells,1 we hypothesized that TMEM47 might contribute to TAM resistance in breast cancer cells.
In the present study, we tested our hypothesis by measuring TAM cytotoxicity in MCF-7 breast cancer cells with manipulation of TMEM47 expression. In addition to the TAMR/MCF-7 cell line, we generated a stable TMEM47-overexpression MCF-7 cell line (TMEM47-OE/MCF-7). TAM cytotoxicity in both cell lines were measured and compared with their parent MCF-7 cells. Cell viability and apoptosis in those cell lines after TAM exposure were assayed. Our study demonstrated that TMEM47 confers TAM resistance on MCF-7 breast cancer cells, which might provide a biomarker for TAM resistance in breast cancers.
Materials and Methods
Construction of Human TMEM47 Overexpression in MCF-7 Cell Lines (TMEM47-OE/MCF-7)
The primers that were used to amplify TMEM47 cDNA were designed based on the TMEM47 gene sequence in the NCBI database (https://www.ncbi.nlm.nih.gov/gene/83604). The primer sequences are following,
Forward primer: 5’-accgagctcggatccATGGCTTCGGCGGGCAGCG-3’, and reverse primer: 5’-cggttcattgctagcCTAGTAGTAGTCTTCATAGT-3’. PCR amplicon was inserted to the pLOC vector after NheI and BamHI digestion. The plasmid was transformed into DH5 α, and the positive clone was selected to extract the plasmid. The constructed plasmids with target fragments and lentivirus packaging plasmids were transfected into T293 cells for packaging of lentivirus, and the supernatant of cells was collected to obtain virus particles, which were then transfected into the target cell line MCF-7. The stable transfected cells expressing TMEM47 were obtained by drug screening.
TMEM47 Knockdown (KD) in TAMR/MCF-7 Cells
Small interfering RNAs (siRNAs) targeting TMEM47 mRNA were designed and synthesized. TMEM47 siRNAs and non-target control (NC) siRNA were transfected into TAMR/MCF-7 cells by liposome regent to knock down TMEM47 expression.
mRNA Quantification by qRT-PCR
TMEM47 expression was quantified on mRNA level by using qRT-PCR. TRIZOL Reagent was used to extract total RNA from the cells according to the standardized operation process as recommended. Nanodrop spectrophotometer was used to measure the concentration of total RNA. RNA Nano 6000 Assay Kit with the Agilent Bioanalyzer 2100 system was used to assess the integrity of total RNA extracted.
RNA extracted was used to synthesize cDNA subsequently. PrimeScript™ RT reagent Kit with the gDNA Eraser Kit were used to reverse-transcribe mRNA according to the standardized operation process as manufacturer recommended. The target gene was detected by qRT PCR based on SYBR green dye method. With GAPDH as endogenous control, all reactions were carried out in 3 times. The relative gene expression was calculated by comparing cycle threshold (2-ΔΔ CT).
Western Blot
Cultured cells were collected to extract the total protein of cells. SDS gel PAGE gel was prepared for electrophoresis to separate the target protein and then imprinted onto PVDF membrane. Western blot was performed with the appropriate primary antibody and the secondary antibody conjugated with horseradish peroxidase. The expression of TMEM47 was validated on protein level.
Drug Cytotoxicity
Cells were collected and plated in 96-well plates at the density of 1 × 104. 4-hydroxy-tamoxifen (4OH-TAM), an active TAM metabolite, was diluted into DMEM medium according to the designed concentration gradient (0-100 μM) and the drug-containing media were added into 96 well plates. The cells were cultured at 37°C for 48 h in a 5% CO2 humidified incubator. Cell counting kit-8 (CCK-8) was used to detect cell proliferation under the condition of drug toxicity.
The cytotoxicity of 4OH-TAM in MCF-7, Leti-control/MCF-7, TAMR/MCF-7 and TMEM47-OE/MCF-7 cells was tested using the method described above. The experiment was carried out at least 3 times. Graphpad Prism 8.0 was used to calculate the 50% inhibitory concentration (IC50) value (mean ± SD) of TAM and plot the dose-response growth inhibitory curve. Student’s t-test (two-tailed) was used to perform statistical evaluations. A p-value <0.01 was considered to denote a significant difference.
Cell Viability
In order to detect the drug sensitivity of TAMR/MCF-7 cell lines to TAM after TMEM47 knockdown, CCK-8 method was used to detect the cell viability. A total of 5000 cells per well were plated into 96-well plates, cultured at 37°C for 48 h in a 5% CO2 humidified incubator overnight; The cells were treated with 1 μM 4OH-TAM for 24 h. After 24 h for cell culture, Cell counting kit-8 (CCK-8) was used to detect cell proliferation under the condition of drug toxicity. Three duplication wells were set and each experiment was duplicated for at least 3 times.
The percentage of cell viability was measured as follows:
Viability = (A treatment − A blank)/(A control − A blank) × 100% (A = absorbance). The histogram was plotted according to the average value of each experimental well.
Detection of Apoptosis by Flow Cytometry
After digesting the cells in a 100 × 20 mm cell culture dish, 2 × 104 cells were evenly inoculated into a well of 6-well cell culture plate. After incubation overnight, the serum-free DMEM medium containing different concentrations of 4OH-TAM was replaced and incubated for 24 hours. After cells were detached with 200 μ? of trypsin without EDTA, the cells were collected and centrifuged at 1000 rpm for 10 min.Cells were then washed with DPBS buffer for 3 times. After absorbing and discarding the supernatant, the cells were fully suspended with 500μ? of binding buffer solution. 5μ? of annexin V-FITC and propidum lodide (PI) solutions were added to the cells for blowing and mixing. The cells were placed at room temperature for 5-20 min in dark. After filtering and dispersing into single cells, the cells were placed at 488 nm of flow cytometer to detect apoptosis.
Results
TMEM47 Was Up-Regulated in TAMR/MCF-7 Cells
In our previous study, we generated a TAM-resistant MCF-7 breast cancer cell line (TAMR/MCF-7) and compared the transcriptome profiles between the TAMR/MCF-7 cells and their parent MCF-7 cells by using RNA-Seq. TMEM47 was one of the “top” differentially-expressed genes. TMEM47 mRNA level was significantly up-regulated (6.8 times) in the TAMR/MCF7 cells.1 (Figure 1a) To further confirm this observation, we performed qRT-PCR and Western blot to validate the TMEM47 mRNA and protein levels in the TAMR/MCF-7 cells, respectively. The qRT-PCR results again showed that TMEM47 mRNA expression was dramatically up-regulated in TAMR/MCF-7 cells (Log2FC = 7.12) (Figure 1b). Consistent with the mRNA level, TMEM47 protein level was significantly increased in TAMR/MCF-7 cells as shown by Western blot assays (Figure 1c).
Figure 1.
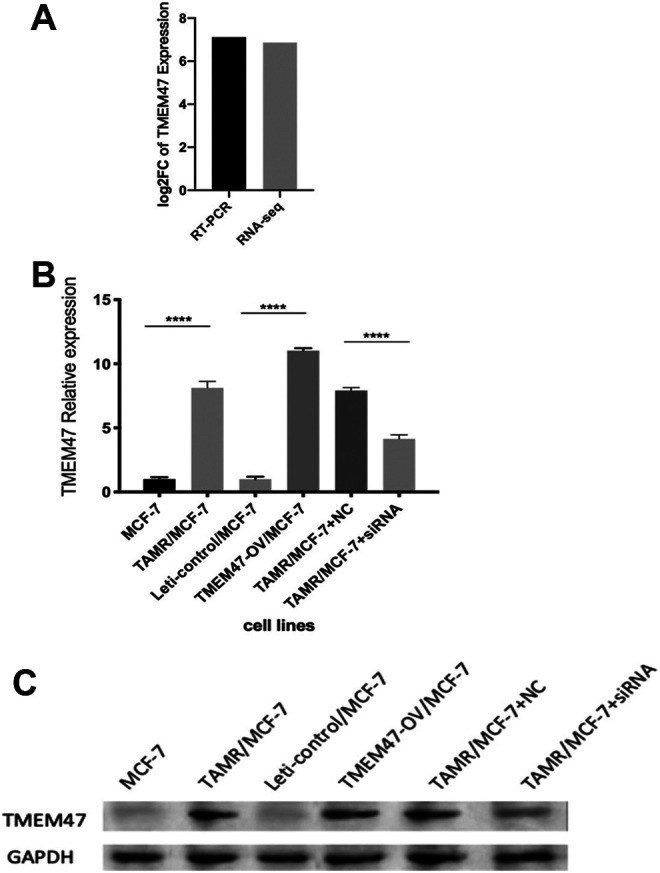
Validation of TMEM47 expression on mRNA and protein level. (a) The relative expression of TMEM47 in TAMR/MCF-7 was validated by qRT-PCR and RNA Seq. (b) qRT-PCR was performed to validate TMEM47 expression in the 6 cell lines. The expression level of each gene was normalized to the level in MCF-7 cells. (****P < 0.0001) Results are representative of 3 independent experiments. (c) From 1 to 6 was TMEM47 expression in MCF-7, TAMR/MCF-7, Leti-control/MCF-7, TMEM47-OE/MCF-7, TAMR/MCF-7 transfected with NC and TAMR/MCF-7 transfected with siRNA. TMEM47 could not be detected in MCF-7 and Leti-control/MCF-7.
Generation of TMEM47-Overexpression Cell Line (TMEM47-OE/MCF-7)
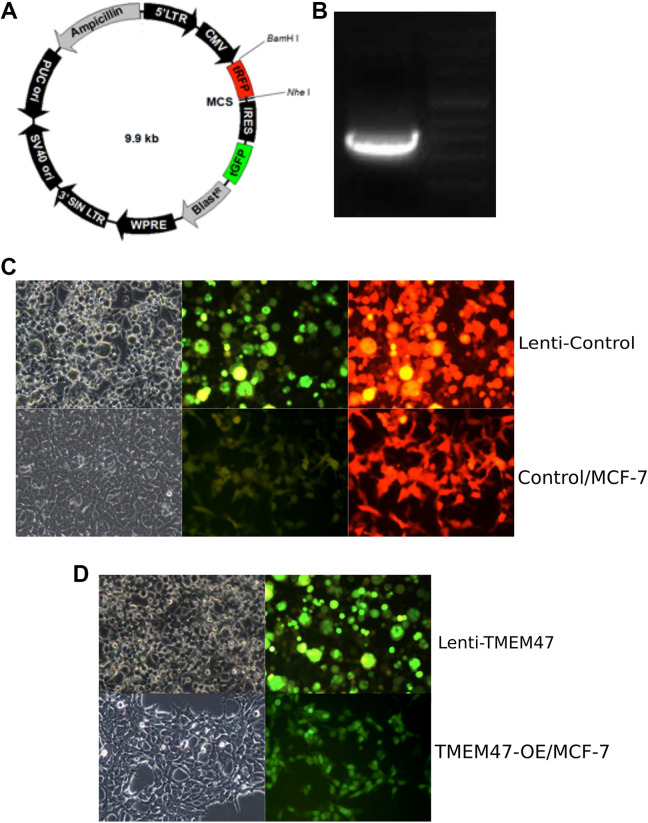
To further investigate whether TMEM47 alone could cause TAM resistance in TAMR/MCF-7 cells, we generated stable TMEM47 overexpression MCF-7 cell line (TMEM47-OE/MCF-7) to study TAM cytotoxicity. The map of pLOC vector was presented in Figure 2a. As schematically depicted in Figure 2b, the sequence of TMEM47 was cloned into pLOC vector. Then the plasmid was treated with restriction enzyme and the 550 bp target fragment was analyzed using agarose gel electrophoresis. The result showed that the target fragment is intact and could be detected. Next, TMEM47 plasmid and lentivirus packaging vector were co-transfected into 293 T cells. As shown in Figure 2c, the packaging lentivirus was efficiently expressed in 293 T cells. Next, the lentivirus was collected from the conditioned medium of 293 T and infected into MCF-7 cell line. TMEM47-overexpressing stable cell line (TMEM47-OE/MCF-7) was generated upon the selection by blasticidin. Compared to directly Lentivirus infected MCF-7, TMEM47-OE/MCF-7 exhibited the similar levels of GFP signals, indicating the lentivirus vector were successfully integrated into the genome (Figure 2d). Next, we investigate the expression of TMEM47 in TMEM47-OE/MCF-7 cells. The result of qRT-PCR (Figure 1b) showed that the mRNA level of TMEM47 was up regulated in TMEM47-OE/MCF-7 (Log2FC = 10.03) compared to that in vehicle infected MCF-7 (Lenti-control/MCF-7). The result of Western Blotting (Figure 1c) showed that the protein levels of TMEM47 were significantly increased in TMEM47-OE/MCF-7. Taken together, TMEM47-overexpressing stable cell line were generated successfully.
Figure 2.
Construction of human TMEM47 overexpression in MCF-7 cell line (a) the map of pLOC vector with the restriction site NheI and Bam HI (b) The insert fragment was 550 bp after plasmid digestion. (c) Packaging efficiency in 293 T cells of lentivirus in control group and leti-control/MCF-7 cells after Blasticidin screening. Leti-control group was the vector without any fragment insert packaged in 293 T cells. Green fluorescence and red fluorescence could be detected in the control group. Blasticidin was used to screen for stable expression MCF-7 cells. (d) Packaging efficiency in 293 T cells of lentivirus in TMEM47 group and TMEM47-OE/MCF-7 cells after Blasticidin screening. TMEM47 was inserted into the pLOC vector; TMEM47 lentivirus plasmid and packaging plasmid were co-transferred to 293 T cells; TMEM47-OE/MCF-7 cells were obtained after Blasticidin screening. Green fluorescence could be detected.
Knockdown of TMEM47 in TAMR/MCF-7 Cell Lines
To further verify the role of TMEM47 in TAM resistance, we knocked down the expression of TMEM47 by transfecting siRNA into TAMR/MCF-7 cells. The result of qRT-PCR showed that the relative expression of TMEM47 in TAMR/MCF-7 was Log2FC = 6.92 (transfected with NC) and Log2FC = 3.14 (transfected with siRNA) compared with MCF-7. (Figure 2b) TMEM47 expression was significantly down regulated in TAMR/MCF-7 cells transfected with siRNA. (P < 0.01) The result of Western Blot in Figure 2c showed that the protein levels of TMEM47 were significantly decreased in TAMR/MCF-7.
TMEM47 Dampens the Sensitivity of MCF-7 to TAM
In order to verify the role of TMEM47 on the sensitivity of cancer cells to TAM, we performed cell cytotoxicity analysis on MCF-7 treated with different concentrations of 4OH-TAM for 24 hours. Then the cell activity was analyzed using CCK-8 method and IC50 was calculated. The dose-response inhibition curves showed that, compared to TAM-sensitive cells (MCF-7), the sensitivity of TMEM47-OE/MCF-7 cells to TAM was attenuated, reaching the same extent to that of TAMR/MCF-7 cells (Figure 3a). Importantly, TMEM47-OE/MCF-7 cells exhibited a slightly higher IC50 value than that of TAMR/MCF-7 cells (Table 1), indicating that up-regulation of TMEM47 is a major contributor to TAM resistance.
Figure 3.
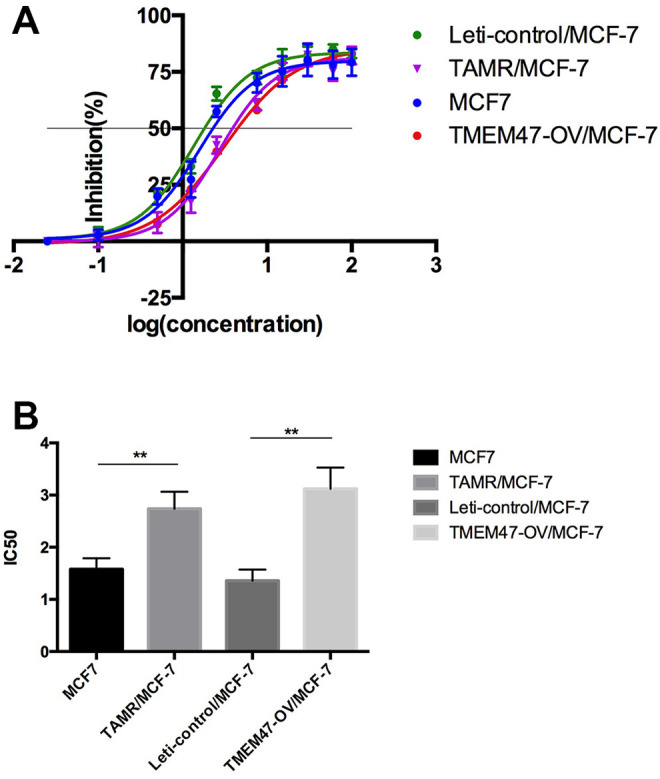
TMEM47 overexpression enhanced the resistance of MCF-7 to TAM (a). The inhibitory effects of different concentrations of TAM on the 4 cells; cell viability, as assessed by the CCK-8 assay, was determined after exposure amounts of TAM for 48 h. Results represent the average of triplicate wells and are representative of 3 independent experiments. (b) The IC50 value (mean ± SD) of TAM in the cells was calculated (**P < 0.01). Results are representative of 3 independent experiments.
Table 1.
The IC50 Value (mean ± SD) of TAM in the Cells.
| samples | treat with TAM | |
|---|---|---|
| IC50 | RI* | |
| MCF-7 | 1.58 ± 0.19 | / |
| TAMR/MCF-7 | 2.74 ± 0.24 | 1.74 |
| Leti-control/MCF-7 | 1.36 ± 0.27 | / |
| TMEM47-OV/MCF-7 | 3.12 ± 0.32 | 2.30 |
* resistant index.
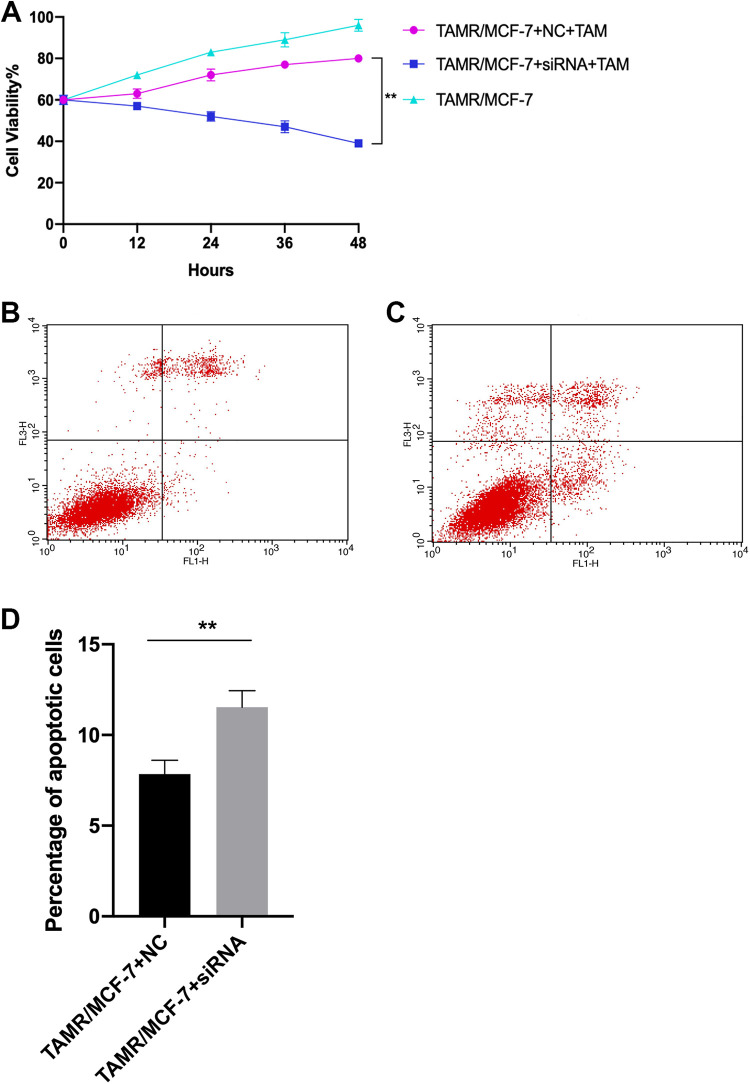
In order to further verify effect of TMEM47 knockdown, siRNA targeting TMEM47 was transfected into TAMR / MCF-7 cells by Liposome. Cell viability and apoptosis were detected by CCK-8 and flow cytometry. The cell viability TAMR/MCF-7 knocked down of TMEM47 was 52% and 39%(treated with TAM for 24 and 48 hours); while the cell viability of TAMR/MCF-7 cells transfected with NC was 72% and 80% (P < 0.01), (Figure 4a) indicating that knockdown of TMEM47 in TAMR / MCF-7 cells enhanced the sensitivity to TAM.
Figure 4.
Knockdown of TMEM47 in TAMR / MCF-7 cells enhanced the sensitivity to TAM(a). The cell viability TAMR/MCF-7 knocked down of TMEM47 was 52% and 39%(treated with TAM for 24 and 48 hours); while the cell viability of TAMR/MCF-7 cells transfected with NC was 72% and 80%. (**P < 0.01). (b-c) Fig4b and fig4c were presented to show apoptosis rate detection by Flow cytometry in TAMR/MCF-7 transfected with NC and siRNA targeting TMEM47 respectively.(d) The apoptosis rate of the 4 cell lines was presented. Each experiment was performed in triplicate. (**P < 0.01).
TMEM47 Suppresses Apoptosis of MCF-7 Cells
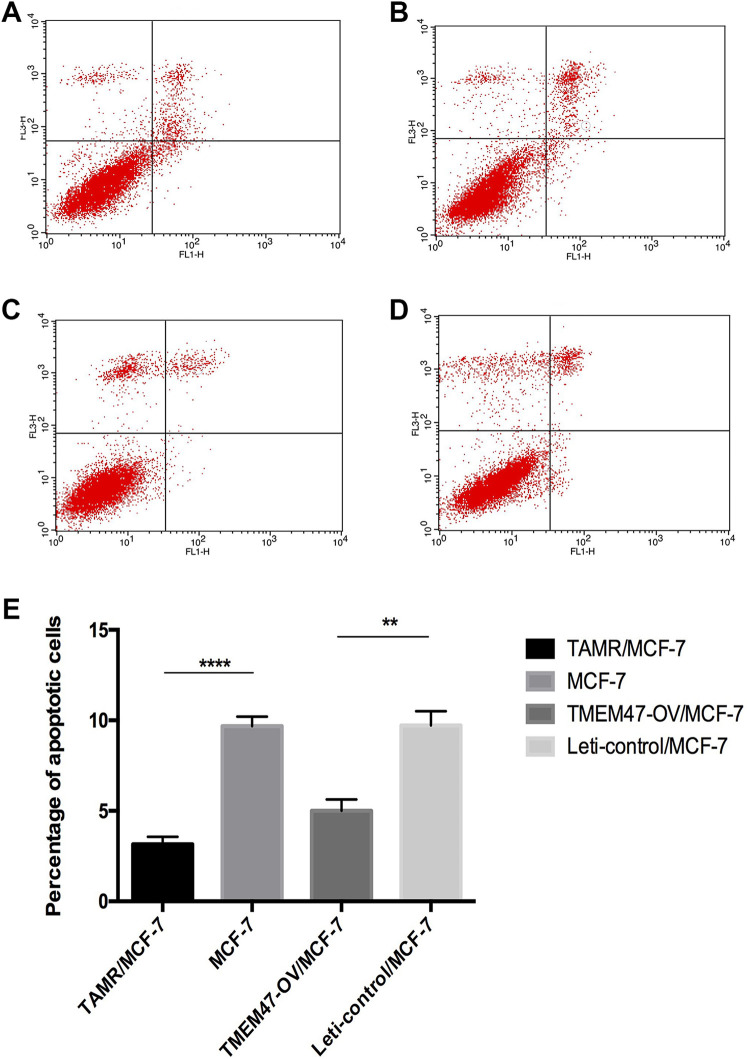
In order to further verify the effect of overexpression of TMEM47 on apoptosis, we performed flow cytometry using Annexin V-FITC/PI double staining As shown in Table 2 and Figure 5, compared to MCF-7, TMEM47-OE/MCF-7 and TAMR/MCF-7 cells showed significantly decreased apoptosis rate in both early phase and late phase upon treatment of 4OH-TAM. Of note, TMEM47-OE/MCF-7 and TAMR/MCF-7 cells showed the similar extend of apoptosis, indicating that TMEM47 is a main candidate gene for apoptosis.
Table 2.
The Apoptosis Rate of the Cells Treated With TAM.
| samples | apoptosis rate | ||
|---|---|---|---|
| early phase | late phase | total | |
| MCF-7 | 6.63 | 3.05 | 9.68 |
| TAMR/MCF-7 | 2.80 | 0.36 | 3.16 |
| Leti-control/MCF-7 | 7.92 | 1.80 | 9.72 |
| TMEM47-OE/MCF-7 | 3.99 | 1.01 | 5.00 |
| TAMR/MCF-7 (transfected with NC) | 6.65 | 1.20 | 7.85 |
| TAMR/MCF-7 (transfected with siRNA) |
5.45 | 6.08 | 11.53 |
Figure 5.
Overexpression of TMEM47 decreased the apoptosis rate(a-d) fig5a-fig5d were presented to show apoptosis rate detection by flow cytometry in MCF-7, Leti-control/MCF-7, TAMR/MCF-7 and TMEM47-OE/MCF-7 respectively.(e) The apoptosis rate of the 4 cell lines was presented. Each experiment was performed in triplicate. (**P < 0.01; **** P < 0.0001).
The apoptosis rate of TAMR/MCF-7 transfected with siRNA was significantly increased (Figure 4b-d), compared to TAMR/MCF-7 transfected with NC. It was indicating that knockdown of TMEM47 in TAMR / MCF-7 cells promoted cell apoptosis after 4OH-TAM treatment.
Discussion
TMEM family is closely related to the occurrence and development of cancer. TMEM family has many members, and these proteins are located on different biomembranes. With the further development of research in recent years, some of them are involved in the process of tumor development, invasion and metastasis, others are related to the poor prognosis of clinical patients. These results indicate that TMEMs can be used as a biomarker of prognosis.
Different regulation of TMEMs expression has been observed in many cancers. For example, TMEM176 has been found to be differentially expressed in the development of lymphoma,13-15 TMEM25 is also found to be closely related to colorectal cancer,16 the same as TMEM7 to hepatic cancer17 and TMEM48 to lung cancer.4 Many TMEMs such as TMEM45A, TMEM116, TMEM207, TMEM213 located in ER predictably have been implicated as potential prognostic biomarkers of cancer grade in renal cancers.18 Besides, many TMEMs have also been found to play an important role in tumorigenesis and drug resistance. These studies show that TMEM family is an important gene family that cannot be ignored in tumor research.
Qiao W. et al performed a research on 60 non-small cell lung carcinoma patients to explore the biological function of TMEM48. The result showed that TMEM48 was overexpression in cancer tissues compared to healthy tissues. It implicated that TMEM48 overexpression was closely related to poor prognosis, tumor metastasis and short survival time.4 Through the case study of breast cancer and cervical lesions, Flamant L. et al suggested that TMEM45A is essential for hypoxia-induced chemoresistance in breast cancer cells; Manawapat-Klopfer A. et al indicated that TMEM45A transcript levels are predictive for development of high-grade cervical lesions. These results showed that TMEM45A can be regarded as a potential biomarker of progressive breast cancer and cervical lesions, and the high expression level of tmem45a is related to the low level of overall survival rate.10,19 All these studies demonstrated that TMEM expression could act an significant role in the prognosis of many cancer patients including liver cancer, glioma, renal cancer, head and neck cancer, ovarian cancer and ductal breast cancer.10,18-23
Tamoxifen (TAM) is the first-line drug for endocrine therapy of hormone receptor positive premenopausal breast cancer patients. However, it is reported that patients will develop drug resistance after an average of 3 to 5 years. This will cause progressive disease in patients, which is not conducive to the prognosis of patients. It is of great significance to explore the mechanism of TAM resistance and find the target of drug resistance for the development of new drugs and the strategy of clinical treatment in the future.
In the process of TAM resistance, the expression of many genes will be up regulated or down regulated in varying degrees, participating in different metabolic pathways, affecting cell proliferation and apoptosis, migration and invasion. We found many key differentially expressed genes based on the analysis of expression profile sequencing in previous study. These DEGs involved in the pathways of Axonal Guidance Signaling, Noradrenaline and Adrenaline Degradation, Hepatic Fibrosis/Hepatic Stellate Cell Activation and ER related regulate many biological activities such as cell proliferation and survival, motility and migration, and tumor cell invasion. These genes affect the sensitivity of cells to TAM through a series of pathways, leading to the resistance of cells to TAM. TMEM47 is one of them. It was reported that the expression of TMEM47 in progressive disease patients was higher than that in complete response patients, a number of genes involved in TMEM family were reported as prognostic biomarkers for matastasis.24 But few evidence has been reported about the relationship between TMEM family and TAM resistance in breast cancer.
The sequencing analysis of our previous expression profile also showed that TMEM47 in TAMR / MCF-7 cell line was 6.86 times higher than that in MCF-7 cell line.1 All of these indicated that TMEM47 plays an important role in the development of TAM resistance.
In this study, we constructed a lentiviral plasmid of TMEM47 that was overexpressed in MCF-7, and screened a stable TMEM47 expression cell line TMEM-OE/MCF-7. RT-PCR and Western blot showed that TMEM47 was stably expressed in TMEM-OE/MCF-7 at mRNA and protein levels. Then we tested the sensitivity of 3 cell lines to TAM, and found that the IC50 of TMEM-OE/MCF-7 was significantly higher than that of MCF-7 (P < 0.01), Overexpression of TMEM47 decreased the sensitivity to TAM by 50% and increased the tolerance. (P < 0.01). This result suggests that the up regulation of TMEM47 may be a cause of drug resistance.
Detection of apoptosis rate by flow cytometry showed that after being treated with the same concentrate of 4OH-TAM, the apoptosis rate in TMEM-OE/MCF-7 and TAMR/MCF-7 was lower than MCF-7 and Leti-control/MCF-7. It revealed that overexpression of TMEM47 increased TAM tolerance through reduce apoptosis and promote cell proliferation. It is worth mentioning that there is no linear relationship between the degree of TAM resistance of cell lines and the overexpression of TMEM47, while there is a positive correlation between them. That is to say, overexpression of TMEM47 is an important cause of TAM resistance, but not the only one. As we found in previous studies, the occurrence of drug resistance is a regulatory process involving many key genes and pathways, many genes as well as mi RNA and lnc RNA is involved. We selected some genes from nearly 3000 DEGs for further verification according to log2FC and key pathways as well as some research reported in the reference literature. Many DEGs have been shown to be unrelated to TAM resistance after verification. TMEM47 is one of the most effective DEGs for TAM resistance.
These conclusions are based on experiment in cell lines. In the previous study, we used high-dose shock method to induce and construct MCF-7 resistant cell line TAMR/MCF-7. We tried to construct TAMR/T47D in the same way but failed. After several cell passages, the drug resistance of TAMR/T47D cells was lost and the sensitivity to tamoxifen was resumed. Therefore, we cannot carry out subsequent cell proliferation activity and apoptosis analysis. We believe that drug resistance is caused by mutation or differential expression of some genes under the pressure of drug selection. This is the stress response of cells to environmental pressure. The mutation or expression difference is random. Maybe the cells that can produce stable drug resistance were not been screened, or some other mutations occurred, which lead to the reversal of drug resistance. This might be a very complicated process. Although drug-resistant cell lines are useful models to understand drug resistance in vitro to search for prognostic or predictive biomarkers, the complexity of tumor microenvironment is much higher than that of cell line. So we need more animal experimental data and clinical data to further verify and support our conclusions. Some researchers applied Weighted gene co-expression network analysis (WGCNA) to explore the differences between aggressive and non-aggressive breast cancer cell lines to predict the clinical outcome of patients, they found that TMEM47 was involved in a co-expression network to be associated with an aggressive phenotype.25 In the CCLE database, According to the dataset of GSE66495(PRJNA277145;), SLITRK2, TMEM47, and LYPD1 were found to be specifically overexpressed by more than 5 fold in 231-BR cells compared with cells isolated from other sites of metastasis. Ingenuity pathway analysis of differentially expressed genes revealed activation of pathways that would enable cancer cells to adapt to organs of metastasis such as drug detoxification. Actually, in previous study, In order to find the DEGs related to TAM resistance in the tissues of breast cancer patients with acquired TAM resistance, to predict the DEGs related to TAM resistance in BRCA patients after treatment with TAM, TCGA RNA-seq dataset was used to screen gene expression data from 22 unique breast cancer samples. When we examined the association of the DEGs with complete response and progressive disease of BRCA, 1669 and 2404 DEGs were identified in PD compared with normal samples and CR compared with normal samples respectively. Considering the breast cancer (BRCA) samples data obtained from TCGA were limited (22 cases), even TMEM47 was found to be down regulated in Progressive Disease (PD) groups, we hope to collect more clinical samples with sufficient information to prove our results in the next study and we will design animal experiments to further verify the conclusion.
In conclusion, we explored the effort of TMEM47 on TAM resistance in MCF-7 cell line in this study. The results revealed that overexpression of TMEM47 could be the reason to TAM resistance through reducing the cell apoptosis rate. TMEM47 could be a prognostic biomarker to TAM resistance. In any case, the results in this study is a good basis for further exploring the mechanism of TAM resistance, finding prognostic biomarkers for clinical diagnosis and providing targets for new drug development.
Supplemental Material
Supplemental Material, sj-jpeg-1-tct-10.1177_15330338211004916 for Overexpression of TMEM47 Induces Tamoxifen Resistance in Human Breast Cancer Cells by Xin Men, Mengyang Su, Jun Ma, Yueyang Mou, Penggao Dai, Chao Chen and Xi An Cheng in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-1-tct-10.1177_15330338211004916 for Overexpression of TMEM47 Induces Tamoxifen Resistance in Human Breast Cancer Cells by Xin Men, Mengyang Su, Jun Ma, Yueyang Mou, Penggao Dai, Chao Chen and Xi An Cheng in Technology in Cancer Research & Treatment
Footnotes
Author Contribution: Men X., Chen C. and Dai P.G. contributed conception and design of the study; Men X. Su M.Y. and Mou Y.Y. performed the experiment, Men X. and Ma J. performed the statistical analysis; Men X. wrote the first draft of the manuscript; Men X., Chen C. and Cheng X.A. contributed to manuscript major revision, read and approved the submitted version.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Statement: Our study did not require an ethical board approval because it did not contain human or animal trials.
Funding: The author(s) received no financial support for the research, authorship, and/or publication of this article.
ORCID iD: Chao Chen  https://orcid.org/0000-0003-2197-1992
https://orcid.org/0000-0003-2197-1992
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Men X, Ma J, Wu T, et al. Transcriptome profiling identified differentially expressed genes and pathways associated with tamoxifen resistance in human breast cancer. Oncotarget. 2018;9(3):4074–4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Marx S, Dal Maso T, Chen JW, et al. Transmembrane (TMEM) protein family members: poorly characterized even if essential for the metastatic process. Semin Cancer Biol. 2020;60:96–106. [DOI] [PubMed] [Google Scholar]
- 3. Akkafa F, Koyuncu I, Temiz E, Dagli H, Dilmec F, Akbas H. miRNA-mediated apoptosis activation through TMEM 48 inhibition in A549 cell line. Biochem Biophys Res Commun. 2018;503(1):323–329. [DOI] [PubMed] [Google Scholar]
- 4. Qiao W, Han Y, Jin W, et al. Overexpression and biological function of TMEM48 in non-small cell lung carcinoma. Tumour Biol. 2016;37(2):2575–2586. [DOI] [PubMed] [Google Scholar]
- 5. Cantonero C, Camello PJ, Abate C, et al. NO1, a New sigma 2 receptor/TMEM97 fluorescent ligand, downregulates SOCE and promotes apoptosis in the triple negative breast cancer cell lines. Cancers. 2020;12(2):257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oyer HM, Sanders CM, Kim FJ. Small-molecule modulators of sigma1 and sigma2/TMEM97 in the context of cancer: foundational concepts and emerging themes. Front Pharmacol. 2019;10:1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Qu T, Zhao Y, Chen Y, et al. Down-regulated MAC30 expression inhibits breast cancer cell invasion and EMT by suppressing Wnt/beta-catenin and PI3K/Akt signaling pathways. Int J Clin Exp Pathol. 2019;12(5):1888–1896. [PMC free article] [PubMed] [Google Scholar]
- 8. Schmidt HR, Kruse AC. The molecular function of sigma receptors: past, present, and future. Trends Pharmacol Sci. 2019;40(9):636–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Karagiannis GS, Pastoriza JM, Wang Y, et al. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci Transl Med. 2017;9(397):eaan0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Flamant L, Roegiers E, Pierre M, et al. TMEM45A is essential for hypoxia-induced chemoresistance in breast and liver cancer cells. BMC Cancer. 2012;12(1):391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Shen DW, Gottesman MM. RAB8 enhances TMEM205-mediated cisplatin resistance. Pharm Res. 2012;29(3):643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen DW, Ma J, Okabe M, Zhang G, Xia D, Gottesman MM. Elevated expression of TMEM205, a hypothetical membrane protein, is associated with cisplatin resistance. J Cell Physiol. 2010;225(3):822–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zuccolo J, Bau J, Childs SJ, Goss GG, Sensen CW, Deans JP. Phylogenetic analysis of the MS4A and TMEM176 gene families. PLoS One. 2010;5(2):e9369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zuccolo J, Deng L, Unruh TL, et al. Expression of MS4A and TMEM176 genes in human B lymphocytes. Front Immunol. 2013;4:195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuajungco MP, Podevin W, Valluri VK, Bui Q, Nguyen VH, Taylor K. Abnormal accumulation of human transmembrane (TMEM)-176A and 176B proteins is associated with cancer pathology. Acta Histochem. 2012;114(7):705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hrasovec S, Hauptman N, Glavac D, Jelenc F, Ravnik-Glavac M. TMEM25 is a candidate biomarker methylated and down-regulated in colorectal cancer. Dis Markers. 2013;34(2):93–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhou X, Popescu NC, Klein G, Imreh S. The interferon-alpha responsive gene TMEM7 suppresses cell proliferation and is downregulated in human hepatocellular carcinoma. Cancer Genet Cytogenet. 2007;177(1):6–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wrzesinski T, Szelag M, Cieslikowski WA, et al. Expression of pre-selected TMEMs with predicted ER localization as potential classifiers of ccRCC tumors. BMC Cancer. 2015;15(1):518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Manawapat-Klopfer A, Thomsen LT, Martus P, et al. TMEM45A, SERPINB5 and p16INK4A transcript levels are predictive for development of high-grade cervical lesions. Am J Cancer Res. 2016;6(7):1524–1536. [PMC free article] [PubMed] [Google Scholar]
- 20. Lee S, Stewart S, Nagtegaal I, et al. Differentially expressed genes regulating the progression of ductal carcinoma in situ to invasive breast cancer. Cancer Res. 2012;72(17):4574–4586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guo J, Chen L, Luo N, Yang W, Qu X, Cheng Z. Inhibition of TMEM45A suppresses proliferation, induces cell cycle arrest and reduces cell invasion in human ovarian cancer cells. Oncol Rep. 2015;33(6):3124–3130. [DOI] [PubMed] [Google Scholar]
- 22. Sun W, Qiu G, Zou Y, et al. Knockdown of TMEM45A inhibits the proliferation, migration and invasion of glioma cells. Int J Clin Exp Pathol. 2015;8(10):12657–12667. [PMC free article] [PubMed] [Google Scholar]
- 23. Burnett RM, Craven KE, Krishnamurthy P, et al. Organ-specific adaptive signaling pathway activation in metastatic breast cancer cells. Oncotarget. 2015;6(14):12682–12696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Entenberg D, Oktay MH, D’Alfonso T, et al. Validation of an automated quantitative digital pathology approach for scoring TMEM: a prognostic biomarker for metastasis. Cancers. 2020;12(4):846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guo L, Zhang K, Bing Z. Application of a coexpression network for the analysis of aggressive and nonaggressive breast cancer cell lines to predict the clinical outcome of patients. Mol Med Rep. 2017;16(6):7967–7978. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Material, sj-jpeg-1-tct-10.1177_15330338211004916 for Overexpression of TMEM47 Induces Tamoxifen Resistance in Human Breast Cancer Cells by Xin Men, Mengyang Su, Jun Ma, Yueyang Mou, Penggao Dai, Chao Chen and Xi An Cheng in Technology in Cancer Research & Treatment
Supplemental Material, sj-pdf-1-tct-10.1177_15330338211004916 for Overexpression of TMEM47 Induces Tamoxifen Resistance in Human Breast Cancer Cells by Xin Men, Mengyang Su, Jun Ma, Yueyang Mou, Penggao Dai, Chao Chen and Xi An Cheng in Technology in Cancer Research & Treatment