Abstract
Objective
To evaluate the performance of a DNA methylation-based digital droplet polymerase chain reaction (ddPCR) assay to detect aberrant DNA methylation in cell-free DNA (cfDNA) and to determine its application in the detection of hepatocellular carcinoma (HCC).
Methods
The present study recruited patients with liver-related diseases and healthy control subjects. Blood samples were used for the extraction of cfDNA, which was then bisulfite converted and the extent of DNA methylation quantified using a ddPCR platform.
Results
A total of 97 patients with HCC, 80 healthy control subjects and 46 patients with chronic hepatitis B/C virus infection were enrolled in the study. The level of cfDNA in the HCC group was significantly higher than that in the healthy control group. For the detection of HCC, based on a cut-off value of 15.7% for the cfDNA methylation ratio, the sensitivity and specificity were 78.57% and 89.38%, respectively. The diagnostic accuracy was 85.27%, the positive predictive value was 81.91% and the negative predictive value was 87.20%. The positive likelihood ratio of 15.7% in HCC diagnosis was 7.40, while the negative likelihood ratio was 0.24.
Conclusions
A sensitive methylation-based assay might serve as a liquid biopsy test for diagnosing HCC.
Keywords: Circulating tumour DNA methylation, hepatocellular carcinoma diagnosis, digital droplet PCR
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common tumour type and the third most common cause of cancer-related deaths worldwide according to the World Health Organization report in 2020.1 In China, HCC is the third most common cause of cancer-related deaths in male and female patients.2 The majority of patients are diagnosed and treated at the late stages of HCC, which leads to poor prognoses. Although abdominal ultrasonography and serum alpha-fetoprotein (AFP) measurement is widely accepted, the sensitivity and specificity of existing tumour biomarker tests are relatively low when screening for HCC.3,4 In recent years, several protein biomarkers, gene mutations and epigenetic modifications have been analysed for the molecular diagnosis of HCC.5,6 As a non-invasive liquid biopsy analyte, circulating cell-free DNA (cfDNA) has gained considerable attention for the diagnosis of various cancer diseases.7,8 Circulating nucleic acid is derived from apoptotic cells, and typically, cfDNA is lower in healthy individuals.9 cfDNA, released from tumour cells, is termed circulating tumour DNA (ctDNA).10 It exhibits primary tumoral heterogeneity, which is a potential biomarker for cancer diagnosis, mostly detectable methylated ctDNA at the beginning of tumorigenesis.10,11 Specifically, the detection of ctDNA, genetic material from cancer cells naturally found in the bloodstream, has also been utilized for cancer detection and disease monitoring.12 As a new biomarker, obtained by analysing cancer-associated gene mutations,13 aberrant DNA methylation,14 copy number variation,15 microsatellite instability,16 and other epigenetic alternations in ctDNA, bioinformative ctDNA in peripheral circulation implies dynamic changes in the tumour in cancer patients. These alterations not only indicate potential responses to treatment but also predict the prognosis of patients in many cancers.17–19
DNA methylation is a crucial regulator of epigenetic modification in cell physiology.20 It has been found to be associated with the occurrence and development of tumours by silencing tumour suppressor gene promoter regions and activating oncogene expression.21 Owing to the tissue specificity, the DNA methylation signature is considered as the fingerprint of the tumour, with characteristics of primary tumorigenesis.22 These modifications in a single cell precede tumour formation, which could help identify tumorigenesis and manage cancer patients.23 Recent studies showed that the ctDNA methylation pattern occurring early in carcinogenesis is a novel robust biomarker that improves the clinical utilization of liquid biopsy in cancer diagnosis.24,25 It has been demonstrated that specific DNA methylation patterns are reliable for early diagnosis, surveillance and prognosis of HCC.26 Currently, digital droplet polymerase chain reaction (ddPCR) is utilized for the quantitation of DNA mutations. However, the DNA methylation pattern in the circulating DNA of HCC remains unclear and a limited number of methylations are arranged on the ddPCR platform.27–30 In contrast, although many methylated genes have been shown to play a major role in HCC,6 a specific indicator has not been confirmed in HCC diagnosis.
The epigenetic alternation plays a significant role in a variety of disease processes, including cancer and other common diseases.14 DNA methylation regulates the expression of tumour suppressor genes.31 An increased number of methylated tumour suppressor genes are detected at the beginning of tumorigenesis events, which makes monitoring cancer based on DNA methylation patterns possible.32 Currently, there are several strategies for the testing and validation of DNA methylation biomarkers. The majority of these approaches are based on quantitative real-time polymerase chain reaction (qPCR) and bisulfite genomic sequencing (BSP). The qPCR-based technique for methylation analysis identifies the methylation status by relative calibration curve analysis based on that generated by a reference for the standard.33–35 BSP is used to read uracil from cytosine directly after bisulfite conversion, rendering it as an important method in cfDNA analysis.36 It provides robust and comprehensive genetic information on cancer monitoring.37 Sensitive technologies, such as digital PCR,38 droplet digital PCR (ddPCR),39 an BEAMing,40 are required for ctDNA analysis. These PCR-based and NGS analytical methodologies are complicated, whereas ddPCR can be used to evaluate the methylation status by directly counting the number of methylated and unmethylated copies.28
This current study validated the potential of a non-invasive DNA methylation test to detect the presence of cancer and provide quantitative data using plasma samples via the ddPCR platform.
Patients and methods
Patient samples
The present study recruited consecutive patients with liver cancer and liver-related chronic diseases and healthy control subjects in the Department of Clinical Laboratory Medicine, Xijing Hospital, Air Force Medical University, Xi'an, Shaanxi Province, China, between March 2018 and May 2019. The eligibility criteria for HCC patients were as follows: (i) age >18 years; (ii) without other malignant tumours; (iii) with liver function test results; (iv) with pathological or imaging evidence of HCC or non-HCC or diagnosis of HCC based on the diagnostic criteria from the ‘Evidence-based practice guidelines for the standardized pathological diagnosis guidelines of primary liver cancer in 2015’.41 Patients that presented with HCC of Barcelona-Clinic Liver Cancer (BCLC) stage A–D were defined in this study. The HCC stage was categorized by the criteria of the BCLC staging system.42 Patients with chronic hepatitis were confirmed by clinicians. A group of healthy volunteers was enrolled as negative controls in addition to those without liver diseases and their serum AFP level was <7 ng/ml. Healthy individuals were local residents that came to Xijing Hospital for routine physical screening. All participants were selected for routine physical body examination, liver-related function testing and imagining screening. Patients with chronic hepatitis with an abnormal AFP level and long-term hepatitis virus infection, as well as HCC patients with BCLC A and B, without clinical liver-related symptoms, were enrolled for the evaluation of diagnostic efficiency.
The study was approved by the Ethics Committee, the First Affiliated Hospital (Xijing Hospital) of the Air Force Military Medical University (no. KY20182078-C-1). All study participants provided written informed consent.
Sample preparation
A 5-ml sample of peripheral whole blood was collected from each participant by venipuncture. The blood samples were stored in K2-ethylenediaminetetra-acetic acid tubes, followed by centrifugation at 2000 g for 10 min at room temperature in a low-speed centrifuge (TDL-80-2C; Shanghai Anting Scientific Factory, Shanghai, China). Isolated plasma (2 ml) was transferred into a new sterile tube and preserved at 4°C for approximately 4 h, followed by centrifugation at 16 000 g at 4°C for 10 min in a Z 32 HK centrifuge (HERMLE Labortechnik GmbH, Wehingen, Germany) before storage at –80°C prior to subsequent use.
Quantification of cfDNA by qPCR
The amount of cfDNA in the plasma samples was quantified using qPCR. The initial cfDNA sample template was used at a dilution of 1:100. A volume of 7.5 μl reaction buffer was mixed with 2.5 μl diluted plasma sample and amplified in a 96-well optical reaction plate on the CFX96 Real-Time PCR System (Bio-Rad, Hercules, CA, USA). The reaction buffer contained: 2.8 µl 5× HemoKlenTaq Reaction Buffer (New England Biolabs, Ipswich, MA, USA); 0.2 µl Deoxynucleotide (dNTP) Solution Mix (New England Biolabs); 0.1 µl SYBR™Green I Nucleic Acid Gel Stain (Thermo Fisher Scientific, Waltham, MA, USA); 0.4 µl primers (Guangzhou Youze Biological Pharmaceutical Technology Company Ltd., Guangzhou, China) and 4.0 µl DNase-free H2O. The PCR protocol was as follows: preliminary denaturation at 95°C for 3 min, followed by 35 cycles of denaturation at 95°C for 10 s, annealing at 60°C for 30 s, and elongation at 65°C for 5 s, followed by a final elongation step at 95°C for 10 min. Standard samples were prepared using a Qubit™ dsDNA HS Assay Kit (Thermo Fisher Scientific), using 1 ng/ml cfDNA as the initial concentration and five serial dilutions for the standard curve. The relative cfDNA concentration of the sample was further calculated by Cq value, slopes and y-intercepts derived from the calibration curves of qPCR. The primers for the qPCR reaction were selected and designed as described previously.43
cfDNA extraction and measurement
The cfDNA was extracted from the plasma using the EliteHealth cfDNA extraction kit (EliteHealth, Guangzhou Youze, China) following the manufacturer’s instructions. Briefly, 1.2–1.5 ml plasma was mixed with 0.2 ml proteinase K. Then, 1.6 ml of buffer ACL was added and the reaction incubated at 60°C for 30 min, followed by the addition of 3.2 ml of buffer ACB and incubation on ice for 5 min. Subsequently, the mixture was filtered through the column. The bound cfDNA was consecutively washed with 600 µl of buffer DCW1, 600 µl of buffer DCW2 and 600 µl of ethanol, with centrifugation at 13000 g for 60 s in a Z 32 HK centrifuge (HERMLE Labortechnik GmbH) at room temperature after every washing step. To elute the cfDNA, 20 µl of nuclease-free water was added to the tube and incubated for 3 min at room temperature before centrifugation at 13000 g for 1 min in a Z 32 HK centrifuge (HERMLE Labortechnik GmbH) at room temperature. The concentration of the extracted cfDNA in the plasma was estimated by DNA fluorometric quantitation (Qubit™ dsDNA HS Assay Kit; Thermo Fisher Scientific).
Bisulfite conversion
The extracted cfDNA was treated with bisulfite using a cfDNA methylation kit (EliteHealth, Guangzhou Youze, China) according to the manufacturer’s instructions. Briefly, 130 μl of bisulfite reagent was mixed with 19 μl of purified cfDNA and incubated at 98°C for 8 min, 54°C for 60 min and then at 4°C for 20 h. Then, 600 μl M-binding buffer was mixed with a hybridized solution, followed by centrifugation at 12000 g for 30 s in a Z 32 HK centrifuge (HERMLE Labortechnik GmbH) at room temperature, washed with 100 μl M-wash buffer and centrifuged at 3000 g for 5 min in a Z 32 HK centrifuge (HERMLE Labortechnik GmbH) at room temperature. Subsequently, 200 μl of L-desulfonation buffer was added and the mixture incubated for 20 min at room temperature until centrifugation at 12000 g for 3 min in a Z 32 HK centrifuge (HERMLE Labortechnik GmbH) at room temperature. After washing, the extracted bisulfite DNA was eluted in 20 μl elution buffer for 5 min. The converted DNA was quantified by absolute DNA fluorometric quantitation (Qubit™ ssDNA HS Assay Kit; Thermo Fisher Scientific).
ddPCR analysis
All ddPCR analyses were performed using the QX100 Droplet Digital PCR System according to the manufacturer’s instructions (Bio-Rad). Bisulfide cfDNA subsequently determined the methylation ratio using the markers provided by the Guangzhou Youze Biological Pharmaceutical Technology Company Ltd. (Guangzhou, China). The assay was designed to measure the methylation status at target sites within specific genes of cfDNA extracted from blood samples by using a ddPCR platform. These target sites are hypermethylated in specific cancers.19 The target region was amplified by the primer pair, cg23612220-Forward: 5′-GTAATGGTGGTAGAGGAAT, cg23612220-Reverse: 5′-AAAACTAAACTAAACTCTACAAAAA; fluorescent probe for methylated allele detection, cg23612220-M 5'/6-FAM/TGTGAAATTTTCGTTTGTATAATTTTTGG/BHQ1/-3'; probe for unmethylated allele detection, cg23612220-NM5'/HEX/TGTGAAATTTTTGTTTGTATAATTTTTGGG/BHQ1/-3'. An equivalent of 10 ng bisulfite cfDNA was utilized for ddPCR, which amplified the target region under the following conditions: 95°C for 10 min, followed by 45 cycles at 94°C for 30 s and 54°C for 60 s, 98°C for 10 min, and maintained at 12°C. Subsequently, the droplet plate was read using the QuantaSoft™ Analysis Pro 1.0.596 software (Bio-Rad) and the data analysed. The test results were reported as a quantitative methylation percentage, which indicated the methylation status of the target sites. For each sample, ddPCR analytic data were presented as copies in a 20 μl reaction system. The total copies were calculated as the methylation copies plus non-methylation copies. The cfDNA methylation ratio was calculated as the methylation copies/(methylation copies +unmethylation copies).
Statistical analyses
All statistical analyses were performed using IBM SPSS Statistics for Windows, Version 22.0 (IBM Corp., Armonk, NY, USA). The correlation analysis of relative and absolute cfDNA quantitation methods between qPCR and DNA fluorometric quantitation was undertaken using Spearman’s correlation coefficient. Mann–Whitney U-test was used to compare the HCC patients with or without liver-related clinical symptoms. Student’s t-test was used to compare the mean value differences of cfDNA amount between the tumour, chronic hepatitis and healthy control groups. The methylation ratio of each sample was calculated. Kruskal–Wallis test and post-hoc Dunn test were performed to assess the variations in cfDNA amount, methylation copies and methylation rates in the BCLC subgroups. Youden index was generated from the area under the receiver operating characteristic (ROC) curve (AUC) in order to determine the optimal cut-off value of cfDNA methylation patterns. The figures were generated using GraphPad Prism for Windows 8.0.1 (GraphPad Software Inc., San Diego, CA, USA). A P-value < 0.05 was considered statistically significant. Venn diagrams were generated using Venny 2.1 (Oliveros, J.C. (2007-2015) Venny) An interactive tool for comparing lists with Venn's diagrams.44
Results
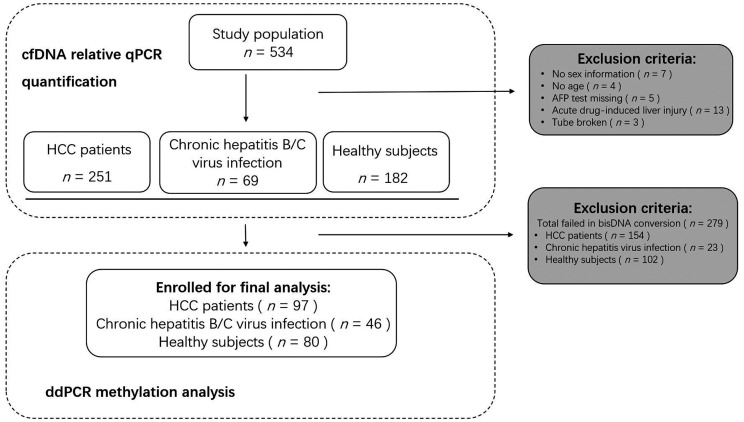
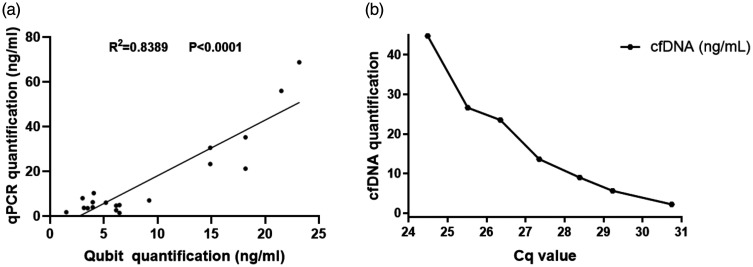
Optimized qPCR was performed to quantify the cfDNA concentration at the beginning of the study. Of the 534 participants enrolled for the relative cfDNA quantitation, 97 were HCC patients, 46 were patients with chronic hepatitis B/C virus infection and 80 were healthy control subjects, with well-integrated clinical information and reliable statistics for further methylation analysis (Figure 1). The remaining 311 participants were excluded from the study as they met the exclusion criteria. The cfDNA quantification results showed optimized qPCR as assessed by Qubit™ (Figure 2a; r2 = 0.8389, P < 0.0001). The Cq values generated from the relative qPCR test represented the cfDNA amount in the study population (Figure 2b).
Figure 1.
Workflow showing sample selection and inclusion in a study designed to determine the potential of a non-invasive DNA methylation test to detect the presence of cancer and provide quantitative data using plasma samples via the ddPCR platform. HCC, hepatocellular carcinoma; AFP, alpha-fetoprotein; cfDNA, cell-free DNA; qPCR, quantitative real-time polymerase chain reaction; ddPCR, digital droplet polymerase chain reaction.
Figure 2.
Correlation analysis of cell-free DNA (cfDNA) quantification. (a) Relative (optimized quantitative real-time polymerase chain reaction [qPCR]) and absolute (Qubit™) calculation; (b) Relative cfDNA quantification and its Cq value in healthy individualized by optimized qPCR.
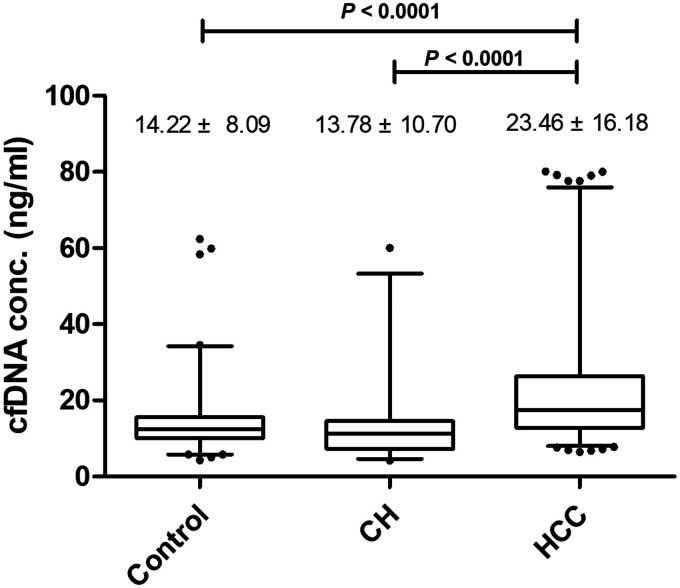
The mean ± SD plasma cfDNA level in the healthy control group was 14.22 ± 8.09 ng/ml compared with 23.46 ± 16.18 ng/ml in the HCC group and 13.78 ± 10.70 ng/ml in the group with chronic hepatitis B/C virus infection (Figure 3). The plasma cfDNA level in the HCC group was significantly higher than that in the group with chronic hepatitis B/C virus infection and the healthy control subjects (P < 0.0001 for both comparisons). There was no significant difference in the cfDNA level between the healthy control subjects and the group with chronic hepatitis B/C virus infection.
Figure 3.
Cell-free DNA (cfDNA) analysis in all study participants (n = 502). Data presented as mean ± SD. Between-group comparisons undertaken using Student’s t-test. The central black horizontal line in each group is the median; the upper extremity of the box is the 25th percentile and the lower extremity of the box is the 75th percentile; the error bars represent minimum and maximum outliers; and the circles above and below the error bars represent extreme outliers. Control, healthy subjects; CH, patients with chronic hepatitis B/C virus infection; HCC, patients with hepatocellular carcinoma.
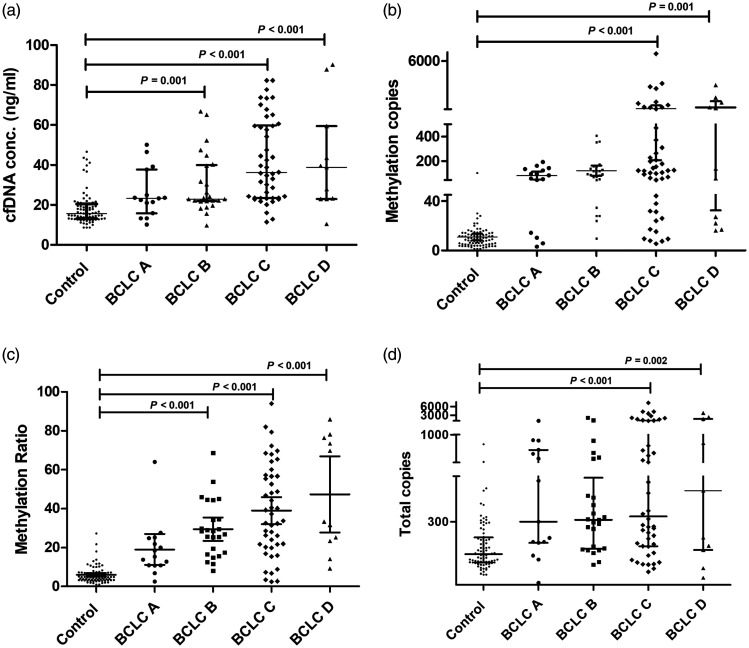
Demographic, clinical and cfDNA data of the 223 study participants are presented in Table 1. The cfDNA methylation ratio reflected the percentage of methylated alleles of CpG. The HCC subgroup analysis in which patients were stratified according to the BCLC staging system classification demonstrated significantly higher cfDNA levels and methylation ratio in patients with clinical symptoms and a heavy tumour burden (the symptomatic groups BCLC C and BCLC D) compared with the asymptomatic groups (groups BCLC A and BCLC B) (P < 0.05 for both comparisons) (Table 2). When considering the methylation patterns, the asymptomatic patients (groups BCLC A and BCLC B) carried a low number of cfDNA fragments and presented with a low methylation status (Figure 4a). Both the methylated copies (Figure 4b) and methylation ratio (Figure 4c) in HCC patients with BCLC C and BCLC D were significantly higher than in those patients with BCLC A and BCLC B (P < 0.001 for all comparisons). The methylation ratio was significantly lower in the healthy control subjects compared with each of the four subgroups of HCC patients (P < 0.05 for all comparisons) (Figure 4c). Methylated copies (Figure 4b) and total methylated copies (Figure 4d) were significantly higher in patients with late-stage BCLC (C and D) compared with the healthy control subjects (P < 0.05 for all comparisons).
Table 1.
Demographic and clinical characteristics of all study participants (n = 223).
| Characteristic | Healthy control groupn = 80 | Patients with chronic hepatitis B/C virus infectionn = 46 | Patients with hepatocarcinoman = 97 |
|---|---|---|---|
| Age, years | 42 (32–58) | 49 (43–55) | 54 (43–64) |
| cfDNA, ng/ml | 15.61 (13.10–20.43) | 9.97 (6.96–12.60) | 28.62 (22.33–47.51) |
| AFP, ng/ml | 3.20 (2.20–4.89) | 3.43 (2.72–5.01) | 40.00 (5.71–547.00) |
| Methylation copies, copies/20 μl | 9 (5–14) | 29 (15–44) | 104 (36–218) |
| Total copies, copies/20 μl | 156 (119–230) | 247 (162–373) | 311 (190–825) |
| Methylation ratio, % | 4.8 (3.31–7.22) | 10.78 (8.44–14.31) | 28.35 (16.68–48.44) |
Data represented as median (interquartile range, IQR).
cfDNA, cell-free DNA; AFP, alpha-fetoprotein.
Table 2.
Comparison of demographic and clinical characteristics in patients with hepatocellular carcinoma (HCC) stratified according to the presence or absence of clinical symptoms.
| HCC patients n = 97 |
Statistical significancea | ||||
|---|---|---|---|---|---|
| Asymptomatic |
Symptomatic |
||||
| BCLC A | BCLC B | BCLC C | BCLC D | ||
| Patients, n | 15 | 25 | 46 | 11 | |
| Age, years | NS | ||||
| Median | 49 | 58 | 53 | 54 | |
| Interquartile range | 42–61 | 46–65 | 39–64 | 43–64 | |
| cfDNA, ng/ml | P = 0.001 | ||||
| Median | 23.28 | 22.91 | 36.11 | 38.78 | |
| Interquartile range | 15.90–37.71 | 21.87–39.98 | 23.42–59.79 | 22.92–59.43 | |
| AFP, ng/ml | NS | ||||
| Median | 39.96 | 8.15 | 53.11 | 39.14 | |
| Interquartile range | 8.59–338.20 | 2.83–1239.50 | 13.02–694.88 | 2.95–81.31 | |
| Methylation copies, copies/20 μl | NS | ||||
| Median | 84 | 92 | 122 | 138 | |
| Interquartile range | 14–134 | 49–146 | 37–490 | 22–1260 | |
| Total copies, copies/20 μl | NS | ||||
| Median | 300 | 308 | 323 | 436 | |
| Interquartile range | 206–726 | 180–493 | 191–1087 | 175–1714 | |
| Methylation ratio, % | P = 0.002 | ||||
| Median | 15.25 | 28.35 | 35.56 | 33.33 | |
| Interquartile range | 10.88–24.91 | 16.99–39.50 | 21.27–56.83 | 23.40–76.47 | |
aBetween-group comparisons undertaken using Kruskal–Wallis test and post-hoc Dunn test; NS, no significant between-group difference (P ≥ 0.05).
BCLC, Barcelona-Clinic Liver Cancer; cfDNA, cell-free DNA; AFP, alpha-fetoprotein.
Figure 4.
DNA methylation parameters in healthy control subjects (n = 80) and hepatocellular carcinoma (HCC) groups (n = 97; subgroups: BCLC A, BCLC B, BCLC C, BCLC D). (a) Comparison of cell-free DNA (cfDNA) concentration; (b) comparison of methylated copies; (c) comparison of methylation ratio; (d) comparison of total copies. Data presented as median ± interquartile range. Between-group comparisons undertaken using Kruskal–Wallis test and post-hoc Dunn test. BCLC, Barcelona-Clinic Liver Cancer.
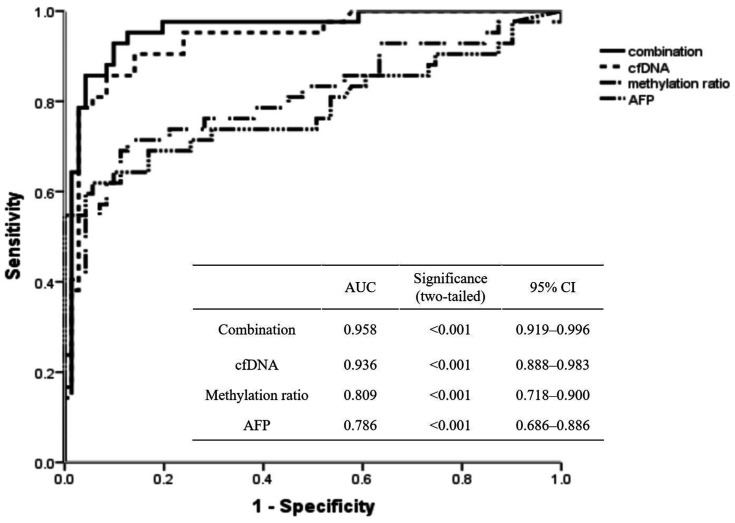
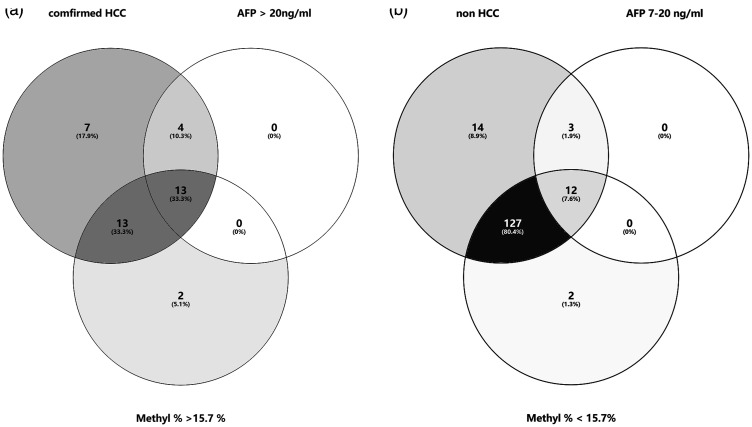
Patients with (i) chronic hepatitis B/C virus infection as well as an abnormal AFP level and (ii) asymptotic HCC patients were enrolled in an AUC analysis. The efficiency of relevant tumour markers for HCC diagnosis was assessed (Figure 5). The ROC curve analysis determined the AUCs as follows: 0.786 (95% confidence interval [CI] 0.686, 0.886) for the AFP level and 0.809 (95% CI 0.718, 0.900) for the methylation ratio to differentiate HCC from non-cancer participants. The optimal cut-off value of the methylation ratio was 15.7%, which was obtained using the ROC curve analysis (AUC of 0.809 (95% CI 0.718, 0.900) for HCC diagnosis. A series of laboratory diagnostic indexes were then analysed using the cut-off value of 15.7% for the methylation ratio (Table 3). Based on the cut-off value of 15.7% in HCC screening, the sensitivity and specificity of HCC detection were 78.57% and 89.38%, respectively. The diagnostic accuracy was 85.27%, the positive predictive value was 81.91% and the negative predictive value was 87.20%. The positive likelihood ratio of 15.7% in HCC diagnosis was 7.40, while the negative likelihood ratio was 0.24. The combination of AFP, cfDNA and methylation ratio can distinguish asymptomatic HCC in the population with an AUC of 0.958 (95% CI 0.919, 0.996) (Figure 5). By combining AFP and methylation ratio, 13 new cases were confirmed to have HCC (Figure 6a). When screening the population, 12 individuals with a positive AFP level (range, 7–20 ng/ml; reference range, 0–7 ng/ml), but with a methylation ratio < 15.7%, were later confirmed by ultrasound examination without HCC tumours (Figure 6b).
Figure 5.
Receiver operating characteristic curve analysis of the relative markers in a diagnosis of hepatocellular carcinoma. cfDNA, cell-free DNA; AFP, alpha-fetoprotein.
Table 3.
Use of cell-free DNA methylation ratio for predicting hepatocellular carcinoma.
| Pathological evidence |
|||||||
|---|---|---|---|---|---|---|---|
| Positive | Negative | Total | PPV | NPV | +LR | –LR | |
| Cut-off value 15.7% | |||||||
| Positive | 77 | 17 | 94 | 0.8191 | |||
| Negative | 21 | 143 | 164 | 0.8720 | |||
| Total | 98 | 160 | 258* | ||||
| Sensitivity | 0.7857 | 7.3983 | 0.2398 | ||||
| Specificity | 0.8938 | 0.8527 | |||||
*Study individuals selected from: (i) healthy control group (n = 80); (ii) patients with chronic hepatitis B/C virus infection (n = 46); (iii) patients with hepatocarcinoma (n = 97); (iv) new chronic hepatitis out-patient cases (n = 35).
PPV, positive predictive value; NPV, negative predictive value; +LR, positive likelihood ratio; –LR, negative likelihood ratio.
Figure 6.
Venn diagrams representing the diagnostic ability of various clinical markers in hepatocellular carcinoma (HCC) diagnosis. (a) Combined with a methylation ratio ≥ 15.7% in screening asymptomatic individuals. (b) Combined with a methylation ratio < 15.7% in screening individuals suspected of having HCC. AFP, alpha-fetoprotein; M%, methylation ratio.
Discussion
This current study quantified cfDNA variations under different clinical conditions. Subsequently, the study validated the dynamic changes in the peripheral DNA methylation ratio that corresponded with HCC progression. Next, the diagnostic value of the methylation ratio for HCC diagnosis was investigated. This current study confirmed the potential application of tissue-specific methylation markers, combined with AFP and cfDNA analysis, in the investigation of susceptible HCC patients differentiated from the asymptomatic population. The differential methylation status between HCC patients, patients with chronic hepatitis B/C virus infection and healthy control subjects was investigated using a cut-off value of 15.7% for the methylation ratio combined with the AFP level and cfDNA level. This yielded a sensitivity of 78.57%, a specificity of 89.38% and a diagnostic accuracy of 85.27%. It was possible to distinguish symptomatic individuals from the population. Moreover, this current study demonstrated that cfDNA methylation analysis was a reliable and robust method for the early diagnosis of HCC. The DNA methylation patterns were highly correlated with the pathological subgroups based on the BCLC classification system. This current study also demonstrated that the ddPCR method efficiently detects trace amounts of DNA methylation. This relatively inexpensive approach is generalizable and could be easily used for personalized medicine, which would potentially improve health management.
The current study used the BCLC classification system, instead of the AJCC/TNM stage, to define the status of the HCC patients. The reason is because BCLC shows a strong ability to classify and predict prognosis, especially in high-risk populations in which it can identify early liver cancer patients for diagnosis and treatment.45 It was interesting to explore if the methylation ratio had the same power as the BCLC system to predict prognosis. However, the results of this current study were not as expected due to lost follow-up data. It has been reported that using a combination of several methylation patterns would increase prognostic ability.26 This study focused on the diagnostic performance of the explored DNA methylation markers. In cancer patients, increased cfDNA levels indicate pathological progression.46,47 According to the BCLC classification system, patients with stages C and D are defined as having tumours that have started to spread via the blood vessels or there has been extrahepatic spread.48,49 The current study demonstrated higher DNA methylation ratios and cfDNA levels in patients with advanced (BCLC C) and late (BCLC D) stages of HCC.
Alpha-fetoprotein is a conventional biomarker that presents as an increased level in pregnant women and in patients with acute liver inflammatory diseases.50 The American College of Radiology Appropriateness Criteria® Chronic Liver Disease guidelines do not recommend screening populations for AFP levels because it is not associated with a statistically significant improvement in HCC detection in the USA.3,4,51 An AFP level > 20 ng/ml provides a sensitivity of 65% and a specificity of 94% for HCC screening.4 A previous study analysed a HCC index, which included age, cfDNA and AFP level; and it demonstrated a sensitivity of 87% and a specificity of 100% for the diagnosis of HCC.49 The current study demonstrated an AUC for AFP in asymptomatic population screening of 0.786; and this was improved to 0.958 by combining AFP with the DNA methylation ratio and cfDNA level.
In conclusion, these current findings emphasize the potential utility of a DNA methylation-based ddPCR platform for the minimally invasive, blood-based early detection of cancer, including HCC. The combined detection of the methylation of multiple genes might improve the diagnostic efficiency.26 This current study also confirmed that ddPCR technology is a robust methodology that can be used in a clinical laboratory and that the analysis of peripheral DNA methylation using the ddPCR platform can be easily implemented in a clinical setting. In addition, it showed great potential for the diagnostic, prognostic and evaluation of therapeutic efficacy during individualized cancer management.
Acknowledgements
We gratefully acknowledge all of the participants in Youze Biological Pharmaceutical Technology Company Ltd. for their technical assistance. We thank Dr Dhruvajyoti Roy from the Laboratory for Advanced Medicine (West Lafayette, IN, USA) for his lively, helpful discussions.
Authors' contributions: Juan Wang contributed to the data analysis and drafted the manuscript. Liu Yang, Jiayun Liu and Yueyun Ma helped to critically revise the manuscript. Yanjun Diao contributed to acquisition and analysis of data. Jinjie Li and Rui Li participated in the interpretation and analysis of data. Lianghong Zheng contributed to the design and critically revised the manuscript. Kang Zhang contributed to the conception and critically revised the manuscript. Xiaoke Hao contributed to the conception, design and critically revised the manuscript. All authors read and approved the final manuscript.
Declaration of conflicting interest: The authors declare that there are no conflicts of interest.
Funding: This research was funded by a grant from the Key Research and Development Plan of Shaanxi Province, China (no. 2019SF-092).
ORCID iD: Xiaoke Hao https://orcid.org/0000-0002-3680-4882
References
- 1.International Agency for Research on Cancer, World Health Organization. Cancer Factor Sheet, https://gco.iarc.fr/today/fact-sheets-cancers (2020, accessed 21 February 2021).
- 2.Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin 2016; 66: 115–132. DOI: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Forner A, Reig M, Bruix J. Alpha-fetoprotein for hepatocellular carcinoma diagnosis: the demise of a brilliant star. Gastroenterology 2009; 137: 26–29. DOI: 10.1053/j.gastro.2009.05.014. [DOI] [PubMed] [Google Scholar]
- 4.Gonzalez SA, Keeffe EB. Diagnosis of hepatocellular carcinoma: role of tumor markers and liver biopsy. Clin Liver Dis 2011; 15: 297–306, vii-x. DOI: 10.1016/j.cld.2011.03.012. [DOI] [PubMed] [Google Scholar]
- 5.Ye Q, Ling S, Zheng S, et al. Liquid biopsy in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. Mol Cancer 2019; 18: 114. DOI: 10.1186/s12943-019-1043-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li J, Han X, Yu X, et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J Exp Clin Cancer Res 2018; 37: 213. DOI: 10.1186/s13046-018-0893-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Oxnard GR, Klein EA, Seiden MV, et al. Simultaneous multi-cancer detection and tissue of origin (TOO) localization using targeted bisulfite sequencing of plasma cell-free DNA (cfDNA). Ann Oncol 2019; 30(suppl 5): v912. DOI: 10.1200/JGO.2019.5.suppl.44. [Google Scholar]
- 8.Liu MC, Jamshidi A, Venn O, et al. Genome-wide cell-free DNA (cfDNA) methylation signatures and effect on tissue of origin (TOO) performance. J Clin Oncol 2019; 37(suppl 15): 3049. DOI: 10.1200/JCO.2019.37.15_suppl.3049. [Google Scholar]
- 9.Jahr S, Hentze H, Englisch S, et al. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res 2001; 61: 1659–1665. [PubMed] [Google Scholar]
- 10.Corcoran RB and, Chabner BA. Application of cell-free DNA analysis to cancer treatment. N Engl J Med 2018; 379: 1754–1765. DOI: 10.1056/NEJMra1706174. [DOI] [PubMed] [Google Scholar]
- 11.Chan KC, Jiang P, Chan CW, et al. Noninvasive detection of cancer-associated genome-wide hypomethylation and copy number aberrations by plasma DNA bisulfite sequencing. Proc Natl Acad Sci U S A 2013; 110: 18761–18768. DOI: 10.1073/pnas.1313995110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis G, Stein S. Circulating Cell-Free Tumour DNA in the Management of Cancer. Int J Mol Sci 2015; 16: 14122–14142. DOI: 10.3390/ijms160614122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veldore VH, Choughule A, Routhu T, et al. Validation of liquid biopsy: plasma cell-free DNA testing in clinical management of advanced non-small cell lung cancer. Lung Cancer (Auckl) 2018; 9: 1–11. DOI: 10.2147/lctt.s147841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egger G, Liang G, Aparicio A, et al. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004; 429: 457–463. DOI: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 15.Chan KC, Jiang P, Zheng YW, et al. Cancer genome scanning in plasma: detection of tumor-associated copy number aberrations, single-nucleotide variants, and tumoral heterogeneity by massively parallel sequencing. Clin Chem 2013; 59: 211–224. DOI: 10.1373/clinchem.2012.196014. [DOI] [PubMed] [Google Scholar]
- 16.Niu B, Ye K, Zhang Q, et al. MSIsensor: microsatellite instability detection using paired tumor-normal sequence data. Bioinformatics 2014; 30: 1015–1016. DOI: 10.1093/bioinformatics/btt755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kamat AA, Bischoff FZ, Dang D, et al. Circulating cell-free DNA: a novel biomarker for response to therapy in ovarian carcinoma. Cancer Biol Ther 2006; 5: 1369–1374. DOI: 10.4161/cbt.5.10.3240. [DOI] [PubMed] [Google Scholar]
- 18.Schwarzenbach H, Stoehlmacher J, Pantel K, et al. Detection and monitoring of cell-free DNA in blood of patients with colorectal cancer. Ann N Y Acad Sci 2008; 1137: 190–196. DOI: 10.1196/annals.1448.025. [DOI] [PubMed] [Google Scholar]
- 19.Ren N, Ye QH, Qin LX, et al. Circulating DNA level is negatively associated with the long-term survival of hepatocellular carcinoma patients. World J Gastroenterol 2006; 12: 3911–3914. DOI: 10.3748/wjg.v12.i24.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reik W, Surani MA. Germline and Pluripotent Stem Cells. Cold Spring Harb Perspect Biol 2015; 7: a019422. DOI: 10.1101/cshperspect.a019422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toh TB, Lim JJ, Chow EK. Epigenetics in cancer stem cells. Mol Cancer 2017; 16: 29. DOI: 10.1186/s12943-017-0596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hao X, Luo H, Krawczyk M, et al. DNA methylation markers for diagnosis and prognosis of common cancers. Proc Natl Acad Sci U S A 2017; 114: 7414–7419. DOI: 10.1073/pnas.1703577114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes . Science 2013; 339: 1546–1558. DOI: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Widschwendter M, Evans I, Jones A, et al. Methylation patterns in serum DNA for early identification of disseminated breast cancer. Genome Med 2017; 9: 115. DOI: 10.1186/s13073-017-0499-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Widschwendter M, Zikan M, Wahl B, et al. The potential of circulating tumor DNA methylation analysis for the early detection and management of ovarian cancer. Genome Med 2017; 9: 116. DOI: 10.1186/s13073-017-0500-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu RH, Wei W, Krawczyk M, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater 2017; 16: 1155–1161. DOI: 10.1038/nmat4997. [DOI] [PubMed] [Google Scholar]
- 27.Postel M, Roosen A, Laurent-Puig P, et al. Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev Mol Diagn 2018; 18: 7–17. DOI: 10.1080/14737159.2018.1400384. [DOI] [PubMed] [Google Scholar]
- 28.Yu M, Carter KT, Makar KW, et al. MethyLight droplet digital PCR for detection and absolute quantification of infrequently methylated alleles. Epigenetics 2015; 10: 803–809. DOI: 10.1080/15592294.2015.1068490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Wesenbeeck L, Janssens L, Meeuws H, et al. Droplet digital PCR is an accurate method to assess methylation status on FFPE samples. Epigenetics 2018; 13: 207–213. DOI: 10.1080/15592294.2018.1448679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Barault L, Amatu A, Bleeker FE, et al. Digital PCR quantification of MGMT methylation refines prediction of clinical benefit from alkylating agents in glioblastoma and metastatic colorectal cancer. Ann Oncol 2015; 26: 1994–1999. DOI: 10.1093/annonc/mdv272. [DOI] [PubMed] [Google Scholar]
- 31.Feinberg AP, Koldobskiy MA and, Göndör A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression. Nat Rev Genet 2016; 17: 284–299. DOI: 10.1038/nrg.2016.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Baylin SB, Jones PA. Epigenetic Determinants of Cancer. Cold Spring Harb Perspect Biol 2016; 8: a019505. DOI: 10.1101/cshperspect.a019505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee HS, Hwang SM, Kim TS, et al. Circulating methylated septin 9 nucleic Acid in the plasma of patients with gastrointestinal cancer in the stomach and colon. Transl Oncol 2013; 6: 290–296. DOI: 10.1593/tlo.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fujii M, Fujimoto N, Hiraki A, et al. Aberrant DNA methylation profile in pleural fluid for differential diagnosis of malignant pleural mesothelioma. Cancer Sci 2012; 103: 510–514. DOI: 10.1111/j.1349-7006.2011.02180.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kisiel JB, Klepp P, Allawi HT, et al. Analysis of DNA Methylation at Specific Loci in Stool Samples Detects Colorectal Cancer and High-Grade Dysplasia in Patients With Inflammatory Bowel Disease. Clin Gastroenterol Hepatol 2019; 17: 914–921.e5. DOI: 10.1016/j.cgh.2018.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y and, Tollefsbol TO. DNA methylation detection: bisulfite genomic sequencing analysis . Methods Mol Biol 2011; 791: 11.–. DOI: 10.1007/978-1-61779-316-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu MC, Oxnard GR, Klein EA, et al. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann Oncol 2020; 31: 745.–. DOI: 10.1016/j.annonc.2020.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbreteau G, Vallée A, Knol AC, et al. Quantitative monitoring of circulating tumor DNA predicts response of cutaneous metastatic melanoma to anti-PD1 immunotherapy. Oncotarget 2018; 9: 25265–25276. DOI: 10.18632/oncotarget.25404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takayama Y, Suzuki K, Muto Y, et al. Monitoring circulating tumor DNA revealed dynamic changes in KRAS status in patients with metastatic colorectal cancer. Oncotarget 2018; 9: 24398–24413. DOI: 10.18632/oncotarget.25309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.García-Foncillas J, Alba E, Aranda E, et al. Incorporating BEAMing technology as a liquid biopsy into clinical practice for the management of colorectal cancer patients: an expert taskforce review. Ann Oncol 2017; 28: 2943–2949. DOI: 10.1093/annonc/mdx501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Evidence-based practice guidelines for standardized pathological diagnosis of primary liver cancer in China: 2015. Zhonghua Gan Zang Bing Za Zhi 2015; 23: 321–327 [Article in Chinese]. [PubMed] [Google Scholar]
- 42.Llovet JM, Brú C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis 1999; 19: 329–338. DOI: 10.1055/s-2007-1007122. [DOI] [PubMed] [Google Scholar]
- 43.Breitbach S, Tug S, Helmig S, et al. Direct quantification of cell-free, circulating DNA from unpurified plasma. PLoS One 2014; 9: e87838. DOI: 10.1371/journal.pone.0087838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Oliveros JC. Venny. An interactive tool for comparing lists with Venn's diagrams, https://bioinfogp.cnb.csic.es/tools/venny/index.html (2007–2015, accessed 1 March 2021).
- 45.Zhang JF, Shu ZJ, Xie CY, et al. Prognosis of unresectable hepatocellular carcinoma: comparison of seven staging systems (TNM, Okuda, BCLC, CLIP, CUPI, JIS, CIS) in a Chinese cohort. PLoS One 2014; 9: e88182. DOI: 10.1371/journal.pone.0088182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chang Y, Tolani B, Nie X, et al. Review of the clinical applications and technological advances of circulating tumor DNA in cancer monitoring. Ther Clin Risk Manag 2017; 13: 1363–1374. DOI: 10.2147/tcrm.s141991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piciocchi M, Cardin R, Vitale A, et al. Circulating free DNA in the progression of liver damage to hepatocellular carcinoma. Hepatol Int 2013; 7: 1050–1057. DOI: 10.1007/s12072-013-9481-9. [DOI] [PubMed] [Google Scholar]
- 48.Huang A, Zhang X, Zhou SL, et al. Plasma Circulating Cell-free DNA Integrity as a Promising Biomarker for Diagnosis and Surveillance in Patients with Hepatocellular Carcinoma. J Cancer 2016; 7: 1798–1803. DOI: 10.7150/jca.15618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yan L, Chen Y, Zhou J, et al. Diagnostic value of circulating cell-free DNA levels for hepatocellular carcinoma. Int J Infect Dis 2018; 67: 92–97. DOI: 10.1016/j.ijid.2017.12.002. [DOI] [PubMed] [Google Scholar]
- 50.Mizejewski GJ. Alpha-fetoprotein structure and function: relevance to isoforms, epitopes, and conformational variants. Exp Biol Med (Maywood) 2001; 226: 377–408. DOI: 10.1177/153537020122600503. [DOI] [PubMed] [Google Scholar]
- 51.Horowitz JM, Kamel IR, Arif-Tiwari H, et al. ACR Appropriateness Criteria® Chronic Liver Disease. J Am Coll Radiol 2017; 14: S391–S405. DOI: 10.1016/j.jacr.2017.08.045. [DOI] [PubMed] [Google Scholar]