Abstract
Epithelia are structurally integral elements in the fabric of oral mucosa with significant functional roles. Similarly, the gingival epithelium performs uniquely critical tasks in responding to a variety of external stimuli and dangers through the regulation of specific built-in molecular mechanisms in a context-dependent fashion at cellular levels. Gingival epithelial cells form an anatomic architecture that confers defense, robustness, and adaptation toward external aggressions, most critically to colonizing microorganisms, among other functions. Accordingly, recent studies unraveled previously uncharacterized response mechanisms in gingival epithelial cells that are constructed to rapidly exert biocidal effects against invader pathobiotic bacteria, such as Porphyromonas gingivalis, through small danger molecule signaling. The host-adapted bacteria, however, have developed adroit strategies to 1) exploit the epithelia as privileged growth niches and 2) chronically target cellular bactericidal and homeostatic metabolic pathways for successful bacterial persistence. As the overgrowth of colonizing microorganisms in the gingival mucosa can shift from homeostasis to dysbiosis or a diseased state, it is crucial to understand how the innate modulatory molecules are intricately involved in antibacterial pathways and how they shape susceptibility versus resistance in the epithelium toward pathogens. Thus, in this review, we highlight recent discoveries in gingival epithelial cell research in the context of bacterial colonizers. The current knowledge outlined here demonstrates the ability of epithelial cells to possess highly organized defense machineries, which can jointly regulate host-derived danger molecule signaling and integrate specific global responses against opportunistic bacteria to combat microbial incursion and maintain host homeostatic balance. These novel examples collectively suggest that the oral epithelia are equipped with a dynamically robust and interconnected defense system encompassing sensors and various effector molecules that arrange and achieve a fine-tuned and advanced response to diverse bacteria.
Keywords: gingival epithelia, opportunistic bacteria, epithelial defense mechanisms, danger signaling, chronic infection, epithelial homeostasis
Introduction
The epithelial tissues, apart from the well-known protective barrier functions, are a dynamic ecosystem where host cells are constantly exposed to environmental cues and stressors while managing to sustain cellular and microbial homeostasis (Larsen et al. 2020). To take on this unique challenge, the epithelial cells are excellent sentinels capable of recognizing perturbations in the environment, initiating cascades of intracellular events, and secreting immune effectors such as cytokines/chemokines and defensins (Ross and Herzberg 2016). Thus, the underlying tissues below the epithelial layer are provided with resistance to any potential mechanical and external insults. Fundamentally different from professional immune cells, the epithelial cells are somatic cells that not only provide structural integrity of the organs but also are critical couriers for efficient signaling between the internal and external environment. As the first line of defense in the oral cavity and other mucosal areas of the body, epithelial cells are an important part of the host immune system, undertaking proper molecular and tactile recognition and responses against invading bacteria (Gunther and Seyfert 2018).
While epithelial cells often maintain a well-concerted relationship with resident microflora composed of indigenous bacteria, some microorganisms have developed adaptive strategies to colonize inside the epithelial cells and permeate to deeper tissues (Ribet and Cossart 2015; Lee, Spooner, et al. 2020). Oral opportunistic bacteria, especially the fastidious anaerobes Porphyromonas gingivalis and Filifactor alocis, can invade the nutrient-rich gingival epithelium to forge ahead a persistence reservoir in the oral mucosa and later systemic spread (Lee, Spooner, et al. 2020). Once inside the epithelial cells, P. gingivalis can produce a series of well-orchestrated adaptive responses for survival (Lee et al. 2017; Lee, Roberts, Atanasova, et al. 2018; Lee, Roberts, Choi, et al. 2018). In recent years, pathogenic intrusions to the epithelial cells have been shown to elicit release of small danger molecules from the host cells, including extracellular ATP (eATP) and adenosine (Alam et al. 2015; Okumura and Takeda 2017). These “danger signal” molecules can prime the innate and adaptive immune arms to respond to tissue threat via purinergic signaling. Similarly, when the danger signaling becomes excessive or prolonged, chronic inflammatory conditions such as periodontitis might ensue (Burnstock 2014; Ramos-Junior et al. 2020). Thus, accumulation of eATP and adenosine beyond physiologic levels can shift the ecophysiologic homeostasis to ecologic upheaval, eventually leading to cellular pathophysiology and microbial dysbiosis (Spooner et al. 2014; Ramos-Junior et al. 2020). In face of the host defenses designed to reign microbial colonization, the ability of some bacterial organisms to modulate the intensity of danger signals can become a key feature to securing prolonged infection within the epithelia and can lead to the formation of chronic inflammation (Spooner et al. 2014; Binderman et al. 2017; Bhalla et al. 2020; Lee, Chowdhury, et al. 2020).
To illustrate recent discoveries of gingival epithelial cell (GEC) and bacteria interaction that can shape host resistance to opportunistic colonization, this review 1) presents newly unraveled dynamic antibacterial and homeostatic machineries of the gingival epithelial system associated with endogenous danger molecule signaling, 2) highlights the novel molecular aspects of cellular autophagy and apoptosis as specific host defenses against oral bacteria for maintaining epithelial barrier function, and 3) underlines molecular underpinnings of the hidden cellular plasticity of GECs toward pathogenic versus commensal bacteria in a species-specific manner. Thus, decoding the molecular handles of eclectically complex epithelial cell and bacteria interplay can define gradual switching from health to disease states in the oral tissues.
A New Line of Progress in Sensing and Responding Mechanisms of the Gingival Epithelia
Introduction to Extracellular ATP, Adenosine, and HMGB1
Classical studies of the epithelial mucosa have identified antimicrobial molecules such as calprotectin and β-defensins as key built-in defenses majorly contributing to epithelial barrier function (Hans and Madaan Hans 2014; Ross and Herzberg 2016). Developing research in human primary GECs has lately provided increasingly new mechanistic insights on sensing and responding mechanisms to colonizing bacteria via release of small molecules such as eATP. Growing evidence supports that the select danger signals outlined here, released in a controlled manner in the gingival epithelial mucosa, can contextually affect modulation of bacterial establishment and cellular homeostasis (Roberts et al. 2017; Lee, Chowdhury, et al. 2020).
Increased levels of eATP are found in various pathologic conditions (Burnstock 2014). The release of ATP to the extracellular milieu occurs in a regulated manner via mechanisms primarily associated with pannexin hemichannels (Dahl 2015). After release, eATP can activate cell surface purinergic receptors, most notably P2X7, in specialized immune cells (Di Virgilio et al. 2017). Interestingly, human primary GECs have emerged as one of the key cell types to have active purinergic signaling as a way to respond to danger, including bacteria (Yilmaz et al. 2010; Hung et al. 2013). In infected GECs, eATP release occurs through pannexin 1, which functions as an extracellular translocation pathway for a virulence factor for P. gingivalis: nucleoside-diphosphate kinase (NDK; Atanasova et al. 2016). ATP acting via P2X7 is the second signal to the assembly of NLRP3 inflammasome, which induces maturation and secretion of proinflammatory cytokines IL-1β and IL-18, as well as reactive oxygen species (ROS) production. Furthermore, eATP/P2X7 coupling is involved in caspase 1 activation and induction of cellular apoptosis (Choi et al. 2013; Di Virgilio et al. 2017).
Although the predominantly proinflammatory nature of eATP in epithelial cells is a critical action of the cellular innate defense machinery upon sensing microbial danger, sustained abundance of eATP may lead to undesirable tissue damage due to an overly exuberant immune response (Antonioli et al. 2013; Burnstock 2014). Therefore, eATP becomes rapidly hydrolyzed by cell surface–bound ectonucleotidases such as CD73 to anti-inflammatory adenosine for maintenance of purine balance (Antonioli et al. 2013). The extracellular adenosine can activate G protein–coupled adenosine receptors in the GECs, including A2a that can change adenylate cyclase activity, followed by intracellular cyclic AMP (cAMP) levels as a second messenger (Spooner et al. 2014). The A2a receptor is sensitive to extracellular adenosine and suppresses inflammation through cAMP-dependent activation of Akt, CREB, and NF-kB (Jacobson and Gao 2006). The functional presence of the A2a receptor has been confirmed in various cells in the gingiva, including GECs (Murakami et al. 2002; Spooner et al. 2014).
High mobility group box 1 protein (HMGB1) is an endogenous homeostatic molecule traditionally associated with chromatin, which can mediate specific responses to stress or infection when secreted into the cytosol (Lu et al. 2012). The release of HMGB1 to the extracellular milieu can act as a ligand for toll-like receptors 2 and 4 (TLR2 and TLR4) and the receptor for advanced glycation end products (RAGE; Das et al. 2016). HMGB1 has been shown to synergize with eATP, stimulating the P2X7 receptors to potent immunomodulatory functions, including inflammasome activation that leads to secretion of antibacterial, proinflammatory cytokines (e.g., IL-1β; Kang et al. 2014). Interestingly, HMGB1 was recently studied in the GECs in the context of oral bacterial infection (Johnson et al. 2015; Bui et al. 2016). Both opportunistic periodontopathic bacteria P. gingivalis and Fusobacterium nucleatum can cause release of HMGB1 from the nucleus to the cytosol in the GECs. However, P. gingivalis solely induces HMGB1 release to the extracellular space in the presence of eATP as a secondary stimulus (Johnson et al. 2015), whereas F. nucleatum does not require any secondary stimulus to translocate HMGB1 out of the cell (Bui et al. 2016). A homolog of NDK in P. gingivalis was shown to attenuate eATP-induced caspase 1 activation, IL-1β secretion, and, importantly, release of HMGB1 in GECs (Johnson et al. 2015). F. nucleatum lacks the NDK gene homolog with an essential metabolic activity (Wolf et al. 2004).
Collectively, these findings suggest that differential regulation of the endogenous danger molecules by various bacteria can shift the homeostatic equilibrium of pro- versus anti-inflammatory mediators and lead to tissue damage.
Recently Characterized Antibacterial Mechanisms of GECs
eATP-Induced ROS Generation through a Distinct NADPH Oxidase Is a Novel Gingival Epithelial Antibacterial Mechanism against Opportunistic Bacteria
With the rising importance of danger signaling pathways in the oral cavity (Kay et al. 2019), recent studies show that epithelial cells execute a variety of immunologic functions against potential pathogens as effectively as traditional immune cells (Roberts et al. 2017; Groeger and Meyle 2019). Stimulation by eATP can induce multiple immune responses, such as powerful antibacterial ROS production in the GECs (Choi et al. 2013; Hung et al. 2013). Increased cellular ROS generation and the resulting oxidative stress over the physiologic levels are typically regarded as a potent host defense response that can lead to elimination of intracellular pathogens (Boyum et al. 2010). The generation of ROS in response to eATP signaling can originate by 2 main sources, mitochondrion and membrane-associated NADPH oxidase (NOX), which have synergistic crosstalk to augment ROS generation upon increasing danger molecule sensing (Fang 2011).
A recent study showed that human primary GECs have robust NOX expression and that, while most of the isoforms are present, not all are functional upon eATP treatment (Roberts et al. 2017). Among the 7 members of the NOX family of proteins, NOX2 is known to have antibacterial functions in specialized in immune cells, and it appears to play a dynamically specific role in GECs following increased levels of eATP. However, during P. gingivalis infection in GECs, NOX2 complex formation was disrupted in multiple levels, resulting in declined eATP-induced ROS and enhanced bacterial survival in GECs. Moreover, the epithelial cells in the presence of danger signal eATP revealed substantially increased activity of myeloperoxidase (MPO), an enzyme known to generate biocidal hypochlorous acid in neutrophils (Roberts et al. 2017). Strong expression of MPO was also implicated in the gingival epithelia of patients with periodontal inflammation (Kuzenko et al. 2015). Interestingly, while increased MPO activity was fully present during infection in GECs, the cellular hypochlorous acid generation was rapidly neutralized through the induction of a major host antioxidant glutathione, which has been shown to increase with P. gingivalis infection in GECs (Choi et al. 2013).
Treponema denticola, one of the key bacteria in chronic periodontitis, has been shown in GECs to suppress human β-defensin expression by inhibiting TLR2 signaling (Shin et al. 2010). TLR2 is a critical mediator for innate immunity and can significantly contribute to shaping the composition of microbiota and modulating the homeostasis of neutrophils in gingival epithelium (Chang et al. 2019). Interestingly, another study showed that T. denticola can attenuate cellular ROS production in immortalized GECs (Shin and Choi 2012). Similarly in neutrophils, which are often in close interaction with epithelial cells in the oral mucosa, the antimicrobial ROS formation was specifically attenuated by F. alocis (Edmisson et al. 2018), a newly appreciated pathogen in chronic periodontitis. These studies demonstrate a steady role of cellular ROS as an antimicrobial response being targeted by various oral bacteria. Currently, no published study has yet confirmed whether the putative ROS modulation by F. alocis or T. denticola in the epithelial cells is mediated via the eATP-induced NOX pathways. New clinical evidence pointing the in situ dual-species colonization of F. alocis and P. gingivalis in the human gingival epithelium prompts speculation that the NOX signaling might be a functional antibacterial mechanism also targeted by F. alocis in the epithelial mucosa (Lee, Spooner, et al. 2020).
Homeostatic Adenosine Signaling Can Modulate Gingival Epithelia and Oral Bacteria Interaction
Extracellular purine homeostasis is a critical regulatory pathway that is controlled by a series of surface-bound enzymes maintaining the balance in extracellular concentration of danger molecules ATP and adenosine (Bours et al. 2006). Ectonucleotidase-CD73, an integral cell membrane enzyme catalyzing the irreversible conversion of adenosine monophosphate (AMP) into adenosine, has been identified as an anti-inflammatory modulator in immune cells (Alam et al. 2015). However, there remains a knowledge gap on CD73 and its potential role in epithelial cells. A latest study suggested a novel host-pathogen adaptation mechanism where active CD73 functions as a switch for oral bacterial survival and growth in epithelial cells through the regulation of local purine levels (Lee, Chowdhury, et al. 2020). The activity and surface expression of CD73 were increased when the AMP-pretreated GECs were infected with P. gingivalis and the enhanced CD73 resulted in abrogating eATP-induced ROS production in GECs and aiding P. gingivalis to grow to higher numbers intracellularly. The inhibition of CD73 via siRNA showed that CD73 is a negative regulator of IL-6 secretion and ROS production from GECs during infection by P. gingivalis. In addition, this study delineated that exogenous treatment of antibacterial IL-6 negatively affects intracellular bacterial replication, which can be significantly reversed by CD73 overexpression and stimulation by exogenous AMP. These novel findings underline the key role of local extracellular purine metabolism and CD73 in modulation of bacterial colonization in epithelial cells.
The concentration of CD73-generated adenosine is an important factor for maintaining the purine balance in the extracellular space. High availability of adenosine over physiologic levels is required to activate various adenosine receptors (i.e., A1, A2a, A2b, and A3; Hasko and Cronstein 2013). An earlier study showed that GECs express all forms of adenosine receptors and that A2a receptor activation results in enhanced intracellular growth of P. gingivalis partially via elevating levels of an anti-inflammatory mediator, cAMP in GECs (Spooner et al. 2014). The A2a receptor was shown to inhibit inflammation caused by the T-cell lymphocytes while aiding persistence of Helicobacter pylori in the gastric epithelial mucosa (Alam et al. 2009), suggesting a shared mechanism of A2a in gingival and gastric mucosa with opportunistic bacteria. Interestingly, a recent study demonstrated that adenosine signaling via the A1 receptor can significantly reduce the ability of Streptococcus pneumoniae to bind human pulmonary epithelial cells by decreasing the expression of the platelet-activating factor receptor, a host protein used by S. pneumoniae to adhere to host cells (Bhalla et al. 2020). These examples underscore the potential conserved specificity in the repertoire of adenosine receptors and their nonredundant functions in epithelial cells for controlling opportunistic bacterial infection and inflammation.
Active Innate Molecular Aspects of Autophagy and Apoptosis in GECs
Autophagy, a homeostatic intrinsic process that recycles dysfunctional cytoplasmic material mainly through lysosomal degradation pathways, can serve in detecting, sequestering, and removing intracellular pathogens (Jiao and Sun 2019). Xenophagy, which is a variant of select autophagy induced upon microbial cellular invasion, forms specific cargos containing the pathogens that are marked by ubiquitin markers (e.g., NDP52 and p62) and targeted to autophagosomes for selective degradation (Sharma et al. 2018). While NDP52 and p62 can function as autophagy adapter proteins, they also perform as cytosolic defense sensors of the innate system in professional phagocytic cells.
A recent study performed in GECs revealed that P. gingivalis, which is a persistent colonizer of the epithelium, can evade the antibacterial ubiquitin marking designed to deliver pathogens to autophagolysosomes for intracellular killing and can induce a select autophagy that is probacterial (Lee, Roberts, Choi, et al. 2018). By using high-resolution 3-dimensional tomographic transmission electron microscopy accompanied with advanced approaches, the study characterized that after invasion of GECs, P. gingivalis becomes highly associated with endoplasmic reticulum–rich autophagic structures that display intact replicating bacteria. Only a small subset of P. gingivalis was detected in the cytosol and captured by the ubiquitin adapters NDP62 and p62. These findings depicted the operational existence of multiple innate autophagy pathways in the GECs that can be selectively modulated by host-acclimatized pathogenic bacteria (Lee, Roberts, Choi, et al. 2018). Interestingly, the gingival expression profile of autophagy-related genes examined in a recent large-animal study suggested that bacteria-orchestrated impairment of autophagy could increase the risk for persistent infection and periodontal inflammation (Ebersole et al. 2021).
Newer studies on Aggregatibacter actinomycetemcomitans, an oral pathogen associated with rapid progression of periodontal destruction, showed that the microorganism employs selective autophagy for internalization to macrophages resulting in attenuation of antimicrobial IL-1β and ROS production (Lee, Park, et al. 2020) and sidesteps the autophagic ubiquitin marking via a select pathway to survive in transformed GECs (Appendix, reference 12, Vicencio et al. 2020). Interestingly,T. denticola can invade transformed GECs and remain within the epithelial cells by resisting endolysosomal degradation (Shin and Choi 2012). However, it remains to be determined whether T. denticola employs an autophagic machinery to extend its colonization in the GECs. Furthermore, autophagy can suppress the danger signals–induced activation (eATP and HMGB1) of the NLRP3 inflammasome by eliminating damaged cellular debris, which prevents excess production of biocidal ROS (Saitoh and Akira 2016). This observation implies that multiple host-protective machineries likely crosstalk and counterbalance each other to maintain the immune equilibrium under cellular stress of microbial aggression.
Similarly, apoptosis—programmed cell death that is tightly controlled for the proper maintenance of host cells—can participate in the host cellular defense systems to control intracellular pathogenic bacteria (Weinrauch and Zychlinsky 1999). Major functions of the apoptosis for epithelial cells can be exploited by opportunistic pathogens. For example, oral bacterium P. gingivalis can modulate a variety of host apoptotic pathways in a context-dependent fashion for successful survival in human primary GECs (Yilmaz 2008; Lee, Roberts, Atanasova, et al. 2018). Heat shock protein 27 (HSP27) is a small antistress phosphoprotein known to activate antiapoptotic events when phosphorylated (Garrido et al. 2006). HSP27 protects the cell from programmed cell death via the intrinsic apoptosis pathway, acting as a negative regulator of cytochrome c release, a key participant of ATP synthesis in the mitochondria (Garrido et al. 2006). A recent study on the putative interaction between HSP27 and P. gingivalis revealed that the effector of P. gingivalis, NDK, can directly phosphorylate HSP27 on selected serine residues and activate an antistress pathway that specifically participates in blocking the intrinsic apoptosis in GECs (Lee, Roberts, Atanasova, et al. 2018). Furthermore, sequestration of cytochrome c in the mitochondria of GECs was shown to depend on the presence of P. gingivalis NDK. Since P. gingivalis NDK can disrupt NOX-mediated ROS production via ATP scavenging (Choi et al. 2013), these findings suggest that antistress molecules such as HSP27 in the antiapoptotic pathway can be targeted for intracellular survival of opportunistic bacteria in GECs.
Unlike P. gingivalis, which is capable of bringing a prosurvival phenotype in GECs for prolonged intracellular life, other oral pathogens (F. nucleatum, A. actinomycetemcomitans, and Streptococcus gordonii) are proposed to associate with a sustained increase in apoptosis of the epithelial cells (Dickinson et al. 2011). Given the chronically prominent presence of P. gingivalis in the gingival epithelia, cellular autophagy and apoptosis might be integral homeostatic processes of epithelial cells that are orchestrated by the specific inside-out signals to deter immediate development of pathogenic states in the mucosa, while the variants of autophagy and apoptosis can be usurped by the host-adapted pathogens (Mostowy and Cossart 2012).
Epithelial Cell Barrier Behavior Is Switched by Distinct Oral Bacteria
Commensal Bacteria–Induced Re-epithelization in the Gingival Epithelia
As opposed to opportunistic bacteria with high potential for inducing pathogenesis, some bacteria (also known as commensals) that surround epithelial mucosa have evolved to benefit the host at immunophysiologic levels (Engevik and Versalovic 2017). Currently, there is a general lack of understanding of which cellular changes occur and/or which host molecules are targeted in the oral mucosa by the beneficial bacteria. However, a recent study described advantageous cellular events in the oral epithelia induced by Streptococcus salivarius, traditionally considered a commensal bacterium (Fernandez-Gutierrez et al. 2017). S. salivarius is one of the earliest colonizers in the oral cavity, remaining present as a major inhabitant of the normal microflora (Kaci et al. 2014). Re-epithelialization, the resurfacing of a wound with new epithelium, is one of the key phases in acute wound repair regulated by complex signaling events. A failed healing process for acute wound may result in chronic wounds highly predisposed to colonization by opportunistic bacteria. By using an improved scratch assay through live-automated fluorescence microscopy and image segmentation to model the kinetics of re-epithelialization in the GEC line Ca9-22, various lactic acid bacteria were screened for their ability to stimulate re-epithelialization, and S. salivarius strains showed significantly improved re-epithelization performance of the cells as compared with other lactic acid bacteria (Fernandez-Gutierrez et al. 2017). It remains to be determined by what epithelial molecular mechanisms the wound closure was achieved in the presence of commensal bacteria. Furthermore, detailed mechanistic studies to identify potential effector proteins produced by S. salivarius could aid in the development of novel strategies to support epithelial integrity in the oral mucosa.
Transdifferentiation of Epithelial Cells into Motile Mesenchymal-like Cells by Oral Bacteria
One of the most studied oral bacteria, P. gingivalis, is equipped with host-adaptive strategies by invading epithelial cells and facilitating its long-term persistence in the gingival mucosa (Roberts et al. 2017; Lee, Roberts, Atanasova, et al. 2018; Lee, Roberts, Choi, et al. 2018). Several studies recently pointed to the ability of P. gingivalis to induce an epithelial-mesenchymal transition (EMT)–like phenotype, which is a central event in epithelial wound repair/regeneration and can associate with neoplastic development (Sztukowska et al. 2016; Lee et al. 2017). Specifically, differential expression and activation of principal EMT markers were observed during >5 d of infection with P. gingivalis in primary GECs (Lee et al. 2017). Phosphorylation of GSK3β, an important EMT regulator, in GECs was significantly increased, and the EMT-associated transcription factors, such as Zeb1, Slug, and Snail, were significantly regulated. There were further phenotypic changes in the host cells, as evidenced by decreased E-cadherin, an integral membrane protein for epithelial architecture and attachment by bacteria (Lee et al. 2017). Interestingly, the study elucidated higher EMT of GECs in the presence of P. gingivalis and F. nucleatum. Since the migration of the cells induced by P. gingivalis was significantly more than that of F. nucleatum alone, this suggests that chronic infection of P. gingivalis may play a predominant role in the initial transdifferentiation of the epithelial cells with early migration. Another study demonstrated that F. nucleatum and S. gordonii were not able to reprogram Zeb1 expression in the immortalized GECs, which suggests that the specific EMT induction shown with P. gingivalis is not merely due to the tissue destruction by bacterial challenge but likely associated with slow-rate neoplastic transformation (Sztukowska et al. 2016). A recent study utilizing oral primary rat epithelial cells similarly illustrated that prolonged exposure of the cells to P. gingivalis and F. nucleatum generated EMT-like features (Abdulkareem et al. 2018). Overall, these findings could aid in unveiling specific synergistic participation of opportunistic pathogens to the development of pathologies associated with the oral epithelia, such as periodontal disease, chronic mucositis, and cancer.
Concluding Remarks
Epithelial tissues are fundamentally core structural components that provide physical integrity and protection to environmental aggressions. More important, the novel knowledge described here illustrates that epithelial cells carry specifically robust systems to interact with surrounding or invading bacteria via a variety of context-dependent mechanisms, which can have an impact on overall host tissue homeostasis as well as bacterial host colonization (see Table for a summary of presented gingival epithelial mechanisms). This embodies the intrinsically complex nature of host-bacteria interactions in the oral epithelium. From the standpoint of the epithelial cells, the present host-protective machineries appear indispensable for these cells to coexist and/or combat against the bacteria, resulting in preservation of the tissue homeostasis intricately regulated by various external cues. To summarize, the discussed antibacterial host defense machineries include inducible biocidal pathways, specific cytokine production, selective autophagy, targeting of select antiapoptotic pathways, and tight regulation of redox systems. It is tempting to speculate that these systems are crucial epithelial cell homeostatic processes that can shift not only healthy physiology of epithelia to pathogenic states with excessive levels of activation by danger signals but also the mucosal microbial ecology to the pathogenic state (see Figure for a summary of current knowledge and developing concepts in the GEC system).
Table.
Newly Recognized Actions of Host-Derived Danger Signal Molecules and Putative Cellular Innate Responses with Important Pathophysiologic Roles During Epithelial Cell and Bacteria Interaction.
| Endogenous Danger Molecule | Newly Characterized Innate Mechanisms for Epithelial Cell and Bacteria Interaction | Oro- and Aerodigestive Bacteria | Cell Type |
|---|---|---|---|
| Extracellular Adenosine | A) Disrupted purine homeostasis [1, 2] B) CD73 expression and activity [1, 2] C) Regulation of cellular ROS [1] D) Modulation of NOX activity [1] E) Regulation of proinflammatory IL-6 via activation of CD73 [1] |
Porphyromonas gingivalis(A to E)Salmonella species(A, B) | Primary gingival epithelial cellsIntestinal epithelial cells |
| A) Adenosine receptor activation modulating host resistance to infection [3 to 5] B) cAMP dependent CREB activation [3] C) Regulation of proinflammatory IL-6 via adenosine receptor [4] |
Streptococcus pneumoniae (A)Clostridium difficile (A, C)Porphyromonas gingivalis (A, B) | Pulmonary epithelial cellsCecum epithelial cellsPrimary gingival epithelial cells | |
| A) Pannexin-1 mediated release of bacterial effector [6] B) eATP-evoked ROS modulation by CD73 [1] C) NLRP3 inflammasome activation [7, 8] D) Modulation of proinflammatory cytokine secretion (IL-1β and/or IL-6) [1, 7] |
Porphyromonas gingivalis (A to D) | Primary gingival epithelial cells | |
| Fusobacterium nucleatuma (C, D) | Primary gingival epithelial cells | ||
| ExtracellularATP | A) Mitochondria and NOX2-generated ROS [9] B) Induction/activation of NOX2 [9] C) Activation of bactericidal NOX2/MPO pathway [9] D) Induction/production of glutathione [9] |
Porphyromonas gingivalis (A to D) | Primary gingival epithelial cells |
| A) Cas-1 and/or Cas-4 activation [8, 10] B) Induction of eATP release [6, 11] C) Infection leading to induction of host danger signals (e.g., eATP) and pyroptosis [10] D) Induction of selective autophagy [12, 13] |
Porphyromonas gingivalis (A, B, D)Aggregatibacter actinomycetemcomitans (B, D)Tannerella forsythia (A, C)Treponema denticola (A, C) | Primary gingival epithelial cellsPeriodontal fibroblasts gingival epithelial cell linebMacrophagesbMacrophagesb | |
| HMGB1 | A) Infection leading to initiation of multiple host danger signals [7, 14] B) Cas-1 activation [14] C) IL-1β, HMGB1 interplay [14] |
Fusobacterium nucleatum (A, B)Porphyromonas gingivalis (A to C) | Primary gingival epithelial cellsPrimary gingival epithelial cells |
| Calprotectin | A) p38, JNK MAPK, and NF-κB signaling induces Calprotectin via RAGE, TLR2 [15] B) Modulation of ROS [16] C) IL-1α regulation [17] |
Porphyromonas gingivalis (A)Salmonella typhimurium (B)Listeria monocytogenes (C) | Gingival epithelial cell lineIntestinal epithelial cellsBuccal epithelial cell line |
Table references are cited by numbers in brackets; see Appendix.
cAMP, cyclic AMP; Cas, caspase; CREB, cAMP response element-binding protein; eATP, extracellular ATP; HMGB1, high mobility group box 1 protein; JNK, c-Jun NH2-terminal kinase; MAPK, mitogen-activated protein kinase; MPO, myeloperoxidase; NF-κB, nuclear factor kappa B; NOX, NADPH oxidase; NOX2, NADPH oxidase 2; RAGE, receptor for advanced glycation end products; ROS, reactive oxygen species; TLR2, toll-like receptor 2.
F. nucleatum does not require eATP signal to activate NLRP3 inflammasome and subsequent IL-1β secretion, which is contrary to P. gingivalis, which needs ATP as a secondary stimulus.
Macrophages and fibroblasts were included in the table since their related mechanisms were specifically highlighted in this review as potentially shared but differentially modulated innate machineries reserved in epithelial cells.
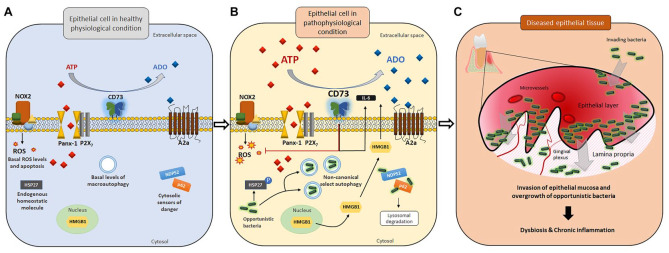
Figure.
Disrupted homeostatic environment in gingival epithelial cells upon invasion by opportunistic bacteria may lead to bacterial dysbiosis in tissues. In gingival epithelial cells, various host defense machineries are in place to combat potential bacterial offenses. (A) At the physiologic level, a minimal amount of ATP is released to the extracellular space via the pannexin 1 (Panx-1) hemichannel without activating P2X7-mediated purinergic signaling and ROS-generating NADPH oxidase 2 (NOX2), resulting in a basal level of apoptosis. The equilibrium in extracellular purine concentration between ATP and adenosine is well balanced. Other danger signal molecules (e.g., HMGB1) and ubiquitination markers/cytosolic sensors of danger (including NDP52 and p62) remain mostly inactive. Antiapoptotic protein HSP27 is largely unphosphorylated. (B) Upon pathogenic bacterial insults, the extracellular ATP (eATP) release drastically increases, which then activates the P2X7 receptor and NOX2 as antibacterial responses. HMGB1, which requires high eATP levels as a second stimulus, becomes translocated from the nucleus to the cytosol and extracellular space. However, opportunistic bacteria such as Porphyromonas gingivalis can modulate multiple antibacterial host machineries (NOX2, P2X7, HMGB1, and ubiquitination marking) and stimulate probacterial host machineries (probacterial autophagy, CD73 activation, HSP27 phosphorylation, and A2a receptor signaling) for successful long-term colonization. (C) Opportunistic pathogens can invade host epithelial tissue and establish chronic colonization while penetrating deeper into the lamina propria. Uncontrolled growth of such pathogenic bacteria and further host tissue invasion lead to bacterial dysbiosis in the epithelia, ultimately initiating the formation of chronic inflammation and diseased epithelial tissues with increased formation of microvasculature.
Various bacterial species show distinct outcomes while interacting with the epithelia: some commensal bacteria have a beneficial effect, whereas some bacteria disturb the typical epithelial physiologic state, promoting undesirable plasticity. As many more host mechanisms remain to be unveiled, future research endeavors are warranted to appreciate the complexity of synchronously ongoing epithelial defenses that are crucial for the health of the epithelial mucosa.
Author Contributions
J.S. Lee, contributed to design, drafted and critically revised the manuscript; Ö. Yilmaz, contributed to conception and design, drafted and critically revised the manuscript. Both authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520973012 for Key Elements of Gingival Epithelial Homeostasis upon Bacterial Interaction by J.S. Lee and Ö. Yilmaz in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by funding from the National Institutes of Health (grants R01DE016593, R56DE016593, R01DE030313, F30DE029103, and T32DE017551).
ORCID iDs: J.S. Lee  https://orcid.org/0000-0001-6937-9416
https://orcid.org/0000-0001-6937-9416
Ö. Yilmaz  https://orcid.org/0000-0003-3487-7217
https://orcid.org/0000-0003-3487-7217
References
- Abdulkareem AA, Shelton RM, Landini G, Cooper PR, Milward MR. 2018. Potential role of periodontal pathogens in compromising epithelial barrier function by inducing epithelial-mesenchymal transition. J Periodontal Res. 53(4):565–574. [DOI] [PubMed] [Google Scholar]
- Alam MS, Costales MG, Cavanaugh C, Williams K. 2015. Extracellular adenosine generation in the regulation of pro-inflammatory responses and pathogen colonization. Biomolecules. 5(2):775–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alam MS, Kurtz CC, Wilson JM, Burnette BR, Wiznerowicz EB, Ross WG, Rieger JM, Figler RA, Linden J, Crowe SE, et al. 2009. A2A adenosine receptor (AR) activation inhibits pro-inflammatory cytokine production by human CD4+ helper T cells and regulates Helicobacter-induced gastritis and bacterial persistence. Mucosal Immunol. 2(3):232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonioli L, Pacher P, Vizi ES, Hasko G. 2013. CD39 and CD73 in immunity and inflammation. Trends Mol Med. 19(6):355–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanasova K, Lee J, Roberts J, Lee K, Ojcius DM, Yilmaz Ö. 2016. Nucleoside-diphosphate-kinase of P. gingivalis is secreted from epithelial cells in the absence of a leader sequence through a pannexin-1 interactome. Sci Rep. 6:37643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhalla M, Hui Yeoh J, Lamneck C, Herring SE, Tchalla EYI, Heinzinger LR, Leong JM, Bou Ghanem EN. 2020. A1 adenosine receptor signaling reduces Streptococcus pneumoniae adherence to pulmonary epithelial cells by targeting expression of platelet-activating factor receptor. Cell Microbiol. 22(2):e13141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binderman I, Gadban N, Yaffe A. 2017. Extracellular ATP is a key modulator of alveolar bone loss in periodontitis. Arch Oral Biol. 81:131–135. [DOI] [PubMed] [Google Scholar]
- Bours MJ, Swennen EL, Di Virgilio F, Cronstein BN, Dagnelie PC. 2006. Adenosine 5′-triphosphate and adenosine as endogenous signaling molecules in immunity and inflammation. Pharmacol Ther. 112(2):358–404. [DOI] [PubMed] [Google Scholar]
- Boyum A, Skrede KK, Myhre O, Tennfjord VA, Neurauter CG, Tolleshaug H, Knudsen E, Opstad PK, Bjoras M, Benestad HB. 2010. Calprotectin (S100A8/S100A9) and myeloperoxidase: co-regulators of formation of reactive oxygen species. Toxins (Basel). 2(1):95–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui FQ, Johnson L, Roberts J, Hung SC, Lee J, Atanasova KR, Huang PR, Yilmaz O, Ojcius DM. 2016. Fusobacterium nucleatum infection of gingival epithelial cells leads to NLRP3 inflammasome-dependent secretion of IL-1β and the danger signals ASC and HMGB1. Cell Microbiol. 18(7):970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnstock G. 2014. Purinergic signalling in the gastrointestinal tract and related organs in health and disease. Purinergic Signal. 10(1):3–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AM, Liu Q, Hajjar AM, Greer A, McLean JS, Darveau RP. 2019. Toll-like receptor-2 and -4 responses regulate neutrophil infiltration into the junctional epithelium and significantly contribute to the composition of the oral microbiota. J Periodontol. 90(10):1202–1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi CH, Spooner R, DeGuzman J, Koutouzis T, Ojcius DM, Yilmaz Ö. 2013. Porphyromonas gingivalis-nucleoside-diphosphate-kinase inhibits ATP-induced reactive-oxygen-species via P2X7 receptor/NADPH-oxidase signalling and contributes to persistence. Cell Microbiol. 15(6):961–976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl G. 2015. ATP release through pannexon channels. Philos Trans R Soc Lond B Biol Sci. 370(1672):20140191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das N, Dewan V, Grace PM, Gunn RJ, Tamura R, Tzarum N, Watkins LR, Wilson IA, Yin H. 2016. HMGB1 activates proinflammatory signaling via TLR5 leading to allodynia. Cell Rep. 17(4):1128–1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson BC, Moffatt CE, Hagerty D, Whitmore SE, Brown TA, Graves DT, Lamont RJ. 2011. Interaction of oral bacteria with gingival epithelial cell multilayers. Mol Oral Microbiol. 26(3):210–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Virgilio F, Dal Ben D, Sarti AC, Giuliani AL, Falzoni S. 2017. The P2X7 receptor in infection and inflammation. Immunity. 47(1):15–31. [DOI] [PubMed] [Google Scholar]
- Ebersole JL, Kirakodu S, Novak MJ, Dawson D, Stromberg AJ, Orraca L, Gonzalez-Martinez J, Burgos A, Gonzalez OA. 2021. Gingival tissue autophagy pathway gene expression profiles in periodontitis and aging. J Periodontal Res. 56(1):34–45. [DOI] [PubMed] [Google Scholar]
- Edmisson JS, Tian S, Armstrong CL, Vashishta A, Klaes CK, Miralda I, Jimenez-Flores E, Le J, Wang Q, Lamont RJ, et al. 2018. Filifactor alocis modulates human neutrophil antimicrobial functional responses. Cell Microbiol. 20(6):e12829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engevik MA, Versalovic J. 2017. Biochemical features of beneficial microbes: foundations for therapeutic microbiology. Microbiol Spectr. 5(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang FC. 2011. Antimicrobial actions of reactive oxygen species. MBio. 2(5):e00141-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Gutierrez MM, Roosjen PPJ, Ultee E, Agelink M, Vervoort JJM, Keijser B, Wells JM, Kleerebezem M. 2017. Streptococcus salivarius MS-oral-D6 promotes gingival re-epithelialization in vitro through a secreted serine protease. Sci Rep. 7(1):11100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrido C, Brunet M, Didelot C, Zermati Y, Schmitt E, Kroemer G. 2006. Heat shock proteins 27 and 70: anti-apoptotic proteins with tumorigenic properties. Cell Cycle. 5(22):2592–2601. [DOI] [PubMed] [Google Scholar]
- Groeger S, Meyle J. 2019. Oral mucosal epithelial cells. Front Immunol. 10:208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunther J, Seyfert HM. 2018. The first line of defence: insights into mechanisms and relevance of phagocytosis in epithelial cells. Semin Immunopathol. 40(6):555–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hans M, Madaan Hans V. 2014. Epithelial antimicrobial peptides: guardian of the oral cavity. Int J Pept. 2014:370297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasko G, Cronstein B. 2013. Regulation of inflammation by adenosine. Front Immunol. 4:85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung SC, Choi CH, Said-Sadier N, Johnson L, Atanasova KR, Sellami H, Yilmaz O, Ojcius DM. 2013. P2X4 assembles with P2X7 and pannexin-1 in gingival epithelial cells and modulates ATP-induced reactive oxygen species production and inflammasome activation. PLoS One. 8(7):e70210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobson KA, Gao ZG. 2006. Adenosine receptors as therapeutic targets. Nat Rev Drug Discov. 5(3):247–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, Sun J. 2019. Bacterial manipulation of autophagic responses in infection and inflammation. Front Immunol. 10:2821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson L, Atanasova KR, Bui PQ, Lee J, Hung SC, Yilmaz O, Ojcius DM. 2015. Porphyromonas gingivalis attenuates ATP-mediated inflammasome activation and HMGB1 release through expression of a nucleoside-diphosphate kinase. Microbes Infect. 17(5):369–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaci G, Goudercourt D, Dennin V, Pot B, Dore J, Ehrlich SD, Renault P, Blottiere HM, Daniel C, Delorme C. 2014. Anti-inflammatory properties of Streptococcus salivarius, a commensal bacterium of the oral cavity and digestive tract. Appl Environ Microbiol. 80(3):928–934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R, Chen R, Zhang Q, Hou W, Wu S, Cao L, Huang J, Yu Y, Fan XG, Yan Z, et al. 2014. HMGB1 in health and disease. Mol Aspects Med. 40:1–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay JG, Kramer JM, Visser MB. 2019. Danger signals in oral cavity–related diseases. J Leukoc Biol. 106(1):193–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzenko Y, Romanyuk A, Politun A, Karpenko L. 2015. S100, bcl2 and myeloperoxid protein expirations during periodontal inflammation. BMC Oral Health. 15:93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen SB, Cowley CJ, Fuchs E. 2020. Epithelial cells: liaisons of immunity. Curr Opin Immunol. 62:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HA, Park MH, Song Y, Na HS, Chung J. 2020. Role of Aggregatibacter actinomycetemcomitans–induced autophagy in inflammatory response. J Periodontol. 91(12):1682–1693. [DOI] [PubMed] [Google Scholar]
- Lee J, Roberts JS, Atanasova KR, Chowdhury N, Han K, Yilmaz O. 2017. Human primary epithelial cells acquire an epithelial-mesenchymal-transition phenotype during long-term infection by the oral opportunistic pathogen, Porphyromonas gingivalis. Front Cell Infect Microbiol. 7:493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Roberts JS, Atanasova KR, Chowdhury N, Yilmaz O. 2018. A novel kinase function of a nucleoside-diphosphate-kinase homologue in Porphyromonas gingivalis is critical in subversion of host cell apoptosis by targeting heat-shock protein 27. Cell Microbiol. 20(5):e12825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Chowdhury N, Roberts JS, Yilmaz O. 2020. Host surface ectonucleotidase-CD73 and the opportunistic pathogen, Porphyromonas gingivalis, cross-modulation underlies a new homeostatic mechanism for chronic bacterial survival in human epithelial cells. Virulence. 11(1):414–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JS, Spooner R, Chowdhury N, Pandey V, Wellslager B, Atanasova K, Evans Z, Yilmaz O. 2020. In situ intraepithelial localizations of opportunistic pathogens, Porphyromonas gingivalis and Filifactor alocis, in human gingiva. Curr Res Microb Sci. 1(1):7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Roberts JS, Choi CH, Atanasova KR, Yilmaz O. 2018. Porphyromonas gingivalis traffics into endoplasmic reticulum-rich-autophagosomes for successful survival in human gingival epithelial cells. Virulence. 9(1):845–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Nakamura T, Inouye K, Li J, Tang Y, Lundback P, Valdes-Ferrer SI, Olofsson PS, Kalb T, Roth J, et al. 2012. Novel role of PKR in inflammasome activation and HMGB1 release. Nature. 488(7413):670–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostowy S, Cossart P. 2012. Bacterial autophagy: restriction or promotion of bacterial replication? Trends Cell Biol. 22(6):283–291. [DOI] [PubMed] [Google Scholar]
- Murakami S, Yoshimura N, Koide H, Watanabe J, Takedachi M, Terakura M, Yanagita M, Hashikawa T, Saho T, Shimabukuro Y, et al. 2002. Activation of adenosine-receptor-enhanced iNOS mRNA expression by gingival epithelial cells. J Dent Res. 81(4):236–240. [DOI] [PubMed] [Google Scholar]
- Okumura R, Takeda K. 2017. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp Mol Med. 49(5):e338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos-Junior ES, Pedram M, Lee RE, Exstrom D, Yilmaz O, Coutinho-Silva R, Ojcius DM, Morandini AC. 2020. CD73-dependent adenosine dampens interleukin-1β-induced CXCL8 production in gingival fibroblasts: association with heme oxygenase-1 and adenosine monophosphate-activated protein kinase. J Periodontol. 91(2):253–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribet D, Cossart P. 2015. How bacterial pathogens colonize their hosts and invade deeper tissues. Microbes Infect. 17(3):173–183. [DOI] [PubMed] [Google Scholar]
- Roberts JS, Atanasova KR, Lee J, Diamond G, Deguzman J, Hee Choi C, Yilmaz Ö. 2017. Opportunistic pathogen Porphyromonas gingivalis modulates danger signal ATP-mediated antibacterial NOX2 pathways in primary epithelial cells. Front Cell Infect Microbiol. 7:291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross KF, Herzberg MC. 2016. Autonomous immunity in mucosal epithelial cells: fortifying the barrier against infection. Microbes Infect. 18(6):387–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Akira S. 2016. Regulation of inflammasomes by autophagy. J Allergy Clin Immunol. 138(1):28–36. [DOI] [PubMed] [Google Scholar]
- Sharma V, Verma S, Seranova E, Sarkar S, Kumar D. 2018. Selective autophagy and xenophagy in infection and disease. Front Cell Dev Biol. 6:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Choi Y. 2012. The fate of Treponema denticola within human gingival epithelial cells. Mol Oral Microbiol. 27(6):471–482. [DOI] [PubMed] [Google Scholar]
- Shin JE, Kim YS, Oh JE, Min BM, Choi Y. 2010. Treponema denticola suppresses expression of human {beta}-defensin-3 in gingival epithelial cells through inhibition of the toll-like receptor 2 axis. Infect Immun. 78(2):672–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spooner R, DeGuzman J, Lee KL, Yilmaz O. 2014. Danger signal adenosine via adenosine 2a receptor stimulates growth of Porphyromonas gingivalis in primary gingival epithelial cells. Mol Oral Microbiol. 29(2):67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sztukowska MN, Ojo A, Ahmed S, Carenbauer AL, Wang Q, Shumway B, Jenkinson HF, Wang H, Darling DS, Lamont RJ. 2016. Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cell Microbiol. 18(6):844–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinrauch Y, Zychlinsky A. 1999. The induction of apoptosis by bacterial pathogens. Annu Rev Microbiol. 53(1):155–187. [DOI] [PubMed] [Google Scholar]
- Wolf M, Muller T, Dandekar T, Pollack JD. 2004. Phylogeny of Firmicutes with special reference to Mycoplasma (Mollicutes) as inferred from phosphoglycerate kinase amino acid sequence data. Int J Syst Evol Microbiol. 54(Pt 3):871–875. [DOI] [PubMed] [Google Scholar]
- Yilmaz O. 2008. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology (Reading). 154(Pt 10):2897–2903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yilmaz O, Sater AA, Yao L, Koutouzis T, Pettengill M, Ojcius DM. 2010. ATP-dependent activation of an inflammasome in primary gingival epithelial cells infected by Porphyromonas gingivalis. Cell Microbiol. 12(2):188–198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520973012 for Key Elements of Gingival Epithelial Homeostasis upon Bacterial Interaction by J.S. Lee and Ö. Yilmaz in Journal of Dental Research