Abstract
The etiology and pathogenesis of craniofacial birth defects are multifactorial and include both genetic and environmental factors. Despite the identification of numerous genes associated with congenital craniofacial anomalies, our understanding of their etiology remains incomplete, and many affected individuals have an unknown genetic diagnosis. Here, we show that conditional loss of a Mediator complex subunit protein, Med23 in mouse neural crest cells (Med23fx/fx;Wnt1-Cre), results in micrognathia, glossoptosis, and cleft palate, mimicking the phenotype of Pierre Robin sequence. Sox9 messenger RNA and protein levels are both upregulated in neural crest cell–derived mesenchyme surrounding Meckel’s cartilage and in the palatal shelves in Med23fx/fx;Wnt1-Cre mutant embryos compared to controls. Consistent with these observations, we demonstrate that Med23 binds to the promoter region of Sox9 and represses Sox9 expression in vitro. Interestingly, Sox9 binding to β-catenin is enhanced in Med23fx/fx;Wnt1-Cre mutant embryos, which, together with downregulation of Col2a1 and Wnt signaling target genes, results in decreased proliferation and altered jaw skeletal differentiation and cleft palate. Altogether, our data support a cell-autonomous requirement for Med23 in neural crest cells, potentially linking the global transcription machinery through Med23 to the etiology and pathogenesis of craniofacial anomalies such as micrognathia and cleft palate.
Keywords: neural crest cells, micrognathia, cleft palate, Mediator, Pierre Robin sequence, cleidocranial dysplasia
Introduction
The craniofacial complex comprises specialized tissues and organs, including the primary sense organs, central and peripheral nervous systems, and musculoskeletal components of the head and neck. Anatomically, it is the most complex region of the human body and is developmentally prone to genetic and environmental insult, resulting in craniofacial anomalies. Jaw, palate, and skull malformations are among the most common human birth defects and have considerable functional, aesthetic, and social consequences. Despite advances in phenotyping and genotyping, many affected individuals have an unknown genetic diagnosis. Therefore, it is imperative to identify novel genetic and environmental factors and the cellular mechanisms they regulate during normal craniofacial development and in disease.
Neural crest cells (NCCs) comprise a migratory progenitor cell population essential for vertebrate development. Born in the neuroepithelium during early embryogenesis, NCCs undergo an epithelial to mesenchymal transition, delaminate, migrate, and differentiate into most of the cartilage, bone, and connective tissues of the head and face. Most craniofacial disorders are associated with defects in NCC development, but depending on which phase of NCC formation, migration, or differentiation is disrupted, distinct malformations such as micrognathia, cleft palate, or cleidocranial dysplasia can occur.
Mediator is a multiprotein complex that plays an indispensable role in regulating gene transcription (Conaway and Conaway 2013). Recently, Mediator has emerged as an integral coordinator of development and cell lineage determination with evidence for roles in development, metabolism, and cancer (Yin and Wang 2014). Mediator may, therefore, function as a centralized hub that integrates multiple aspects of transcriptional regulation to ensure the proper pattern, timing, and intensity of global gene expression during development and in the etiology and pathogenesis of disease (Malik and Roeder 2000; Carlsten et al. 2013).
We previously described a novel allele in the Mediator complex subunit, Med23, termed snouty (sn) (Sandell et al. 2011; Dash et al. 2020). Med23sn/sn mutant mice exhibit craniofacial defects such as a shortened frontonasal prominence and hypoplastic pharyngeal arches, and they are embryonic lethal around E10.5. To bypass this lethality and demonstrate a cell-autonomous function for Med23 in NCC and craniofacial development, we generated a conditional knockout of Med23, which resulted in micrognathia, glossoptosis, cleft palate, and cleidocranial dysplasia. We subsequently determined Med23 to be a direct upstream repressor of Sox9 in mice. Loss of Med23 results in increased Sox9 expression and Sox9–β-catenin protein interactions, which leads to decreased Col2a1 and downstream components of Wnt signaling, as well as perturbation of NCC-derived palatal mesenchyme proliferation and osteochondroprogenitor differentiation. Thus, Med23 potentially links the global transcription machinery in NCC to cartilage and bone differentiation during craniofacial development.
Materials and Methods
Details of the methods are provided in the Appendix.
Results
NCC-Specific Deletion of Med23 Results in Defects in Meckel’s Cartilage Development and Cleft Palate
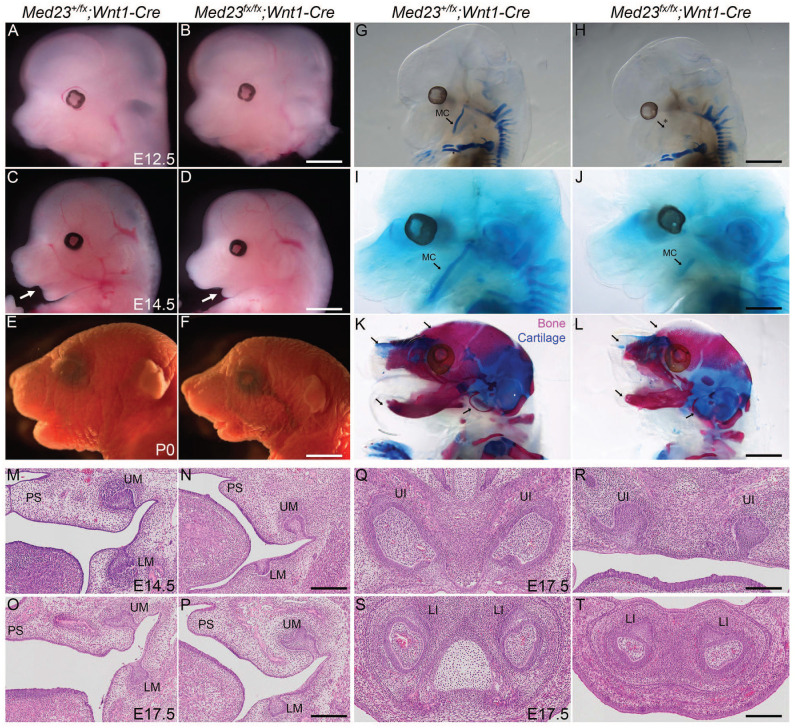
To investigate the cell-autonomous function of Med23, we generated NCC-specific mutants of Med23 by intercrossing Med23fx/fx mice with Wnt1-Cre mice. Med23fx/fx;Wnt1-Cre mutant embryos are morphologically indistinguishable from Med23+/fx; Wnt1-Cre control littermates until E12.5 (Fig. 1A, B). Furthermore, NCC migration and differentiation into neurons of the peripheral nervous system are comparable between mutant and control littermates at E10.5 as observed by Sox10 in situ hybridization (Appendix Fig. 1) and Tuj1 immunostaining (Dash et al. 2020). However, by E14.5, Med23fx/fx;Wnt1-Cre embryos exhibit severe craniofacial defects, including micrognathia (small jaw) and glossoptosis (abnormally positioned tongue) (Fig. 1C, D). By postnatal day (P)0, the severity of micrognathia and glossoptosis is further evident, and difficulty in breathing and feeding causes neonatal lethality (Fig. 1E, F). To better define the cranioskeletal defects, we performed bone and cartilage staining beginning at E12.5 and observed a lack of Meckel’s cartilage (MC) development in Med23fx/fx;Wnt1-Cre mutants compared to controls (Fig. 1G and H). By E14.5, Med23fx/fx;Wnt1-Cre mutants exhibit severely hypoplastic MCs (Fig. 1I, J), and because MC provides a scaffold for mandible development, its smaller size likely contributes to a smaller mandible at P0. In addition, Med23fx/fx;Wnt1-Cre mutants exhibit agenesis of the palatine bones and nasal cartilage and bones, together with abnormal development of the tympanic ring and skull bones, which result in cleidocranial dysplasia (Fig. 1K, L; Appendix Fig. 2).
Figure 1.
Neural crest cell–specific deletion of Med23 causes craniofacial defects. (A–F) Bright-field images of Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre embryos indicate that the mutant embryos have no overt phenotype at E12.5. By E14.5, the mutant embryos have a smaller lower jaw. At P0, the lower jaw is still hypoplastic and the tongue is abnormally positioned in the mutants compared to controls. (G–L) Skeletal staining of Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre embryos indicates that Meckel’s cartilage development is delayed at E12.5 and hypoplastic at E14.5 in Med23fx/fx;Wnt1-Cre embryos. At P0, the mandible is smaller in mutants compared to controls. Furthermore, the incisors are underdeveloped, the nasal cartilage is absent, and the otic bones are hypoplastic in Med23fx/fx;Wnt1-Cre embryos (indicated by arrows). (M–T) Histology of Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre embryos suggests that development of the upper and lower molars as well as upper and lower incisors is delayed in the Med23fx/fx;Wnt1-Cre embryos compared to Med23+/fx;Wnt1-Cre control embryos. LI, lower incisor; LM, lower molar; MC, Meckel’s cartilage; PS, palatal shelf; UI, upper incisor; UM, upper molar. Scale bar for A, B is 200 mm; C, D is 250 mm; E, F is 350 mm; G–J is 200 mm; K, L is 300 mm; M, N is 70 mm; and O–T is 45 mm.
Med23fx/fx;Wnt1-Cre embryos also present with defects in teeth development. While the molars in control embryos differentiate to the cap stage by E14.5 and late bell stage by E17.5, the molars in Med23fx/fx;Wnt1-Cre embryos are developmentally delayed at the bud stage at E14.5 and early bell stage at E17.5. Similarly, development of the upper incisors is also delayed in Med23fx/fx;Wnt1-Cre embryos having only reached the bell stage, compared to control embryos where the upper incisors are transitioning from the late bell to eruption stage. Furthermore, the lower incisors are also considerably smaller in Med23fx/fx;Wnt1-Cre mutants compared to controls (Fig. 1M–T).
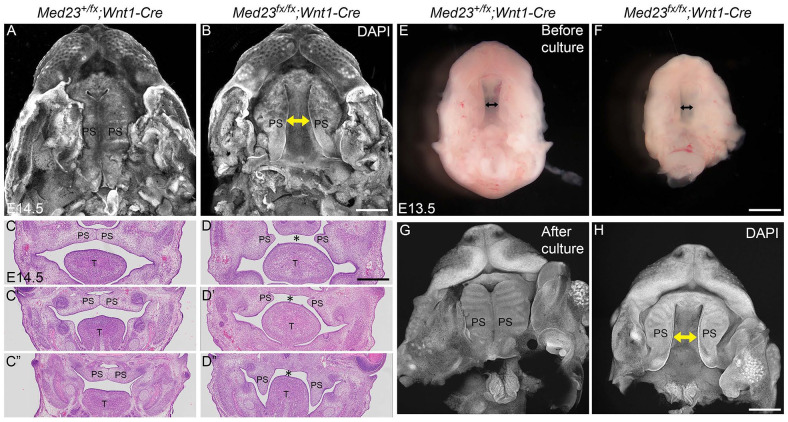
The absence of the palatine bones in Med23fx/fx;Wnt1-Cre mutant embryos is indicative of cleft palate and is clearly observed by pseudo–scanning electron microscopy (SEM)imaging (Sandell et al. 2012) in E14.5 embryos (Fig. 2A, B). The presence of cleft palate is also evident in histological sections wherein the palatal shelves remain separated by the tongue in association with apparent failure of palatal shelf elevation and fusion (Fig. 2C, D′′). By E17.5, the palatal shelves in the anterior of mutant embryos have elevated above the tongue but still remain far apart. The posterior palatal shelves fail to elevate and remain well separated by the tongue (Appendix Fig. 3). This indicates that palatal shelf elevation is incomplete, which contributes to the failure of palatal shelf fusion and thus cleft palate in Med23fx/fx;Wnt1-Cre embryos.
Figure 2.
Med23fx/fx;Wnt1-Cre embryos exhibit cleft palate. (A, B) Pseudo–scanning electron microscopy imaging of the palate of Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre embryos at E14.5 shows that the palatal shelves in Med23fx/fx;Wnt1-Cre embryos remain far apart, which is indicative of cleft palate. (C′, D′′) Histological analysis of Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre palatal shelves at E14.5 shown anterior (C, D) to posterior (C′′, D′′) revealed that the palatal shelves in Med23fx/fx;Wnt1-Cre embryos are hypoplastic in the anterior region, while in the posterior region, palatal shelf growth is vertical beside the tongue. Asterisk denotes cleft palate. (E, F) Bright-field images of the palatal shelves at E13.5 prior to ex vivo culture illustrate that the palatal shelves are not fused at this stage in Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre embryos. (G, H) Seventy-two hours after ex vivo roller culture, the palatal shelves of Med23+/fx;Wnt1-Cre have fused, while those of Med23fx/fx;Wnt1-Cre embryos remain unfused. Double-ended arrows indicate the unfused palatal shelves. PS, palatal shelf; T, tongue. Scale bar for A, B is 200 µm; C–D′′ is 180 µm; E–H is 200 µm.
Failure of Palate Fusion in Med23fx/fx;Wnt1-Cre Embryos Is Independent of Tongue Development
We hypothesized that the presence of the tongue between the palatal shelves in Med23fx/fx;Wnt1-Cre embryos may serve as a physical impediment to palatal shelf fusion. To test this hypothesis, we dissected the maxillary apparatus from control and mutant embryos at E13.5 and cultured them ex vivo in roller culture. Prior to culture, the palatal shelves were not fused in either Med23+/fx;Wnt1-Cre control or Med23fx/fx;Wnt1-Cre mutant embryos (Fig. 2E, F). However, after 72 h culture, the control palatal shelves developed rugae and fused. In contrast, although rugae formed in Med23fx/fx;Wnt1-Cre embryos, the palatal shelves remained separated and unfused (Fig. 2G, H). This indicates that cleft palate in Med23fx/fx;Wnt1-Cre mutant embryos is independent of tongue development or its position.
Sox9 Is Upregulated in Med23fx/fx;Wnt1-Cre Palatal Shelf Mesenchyme
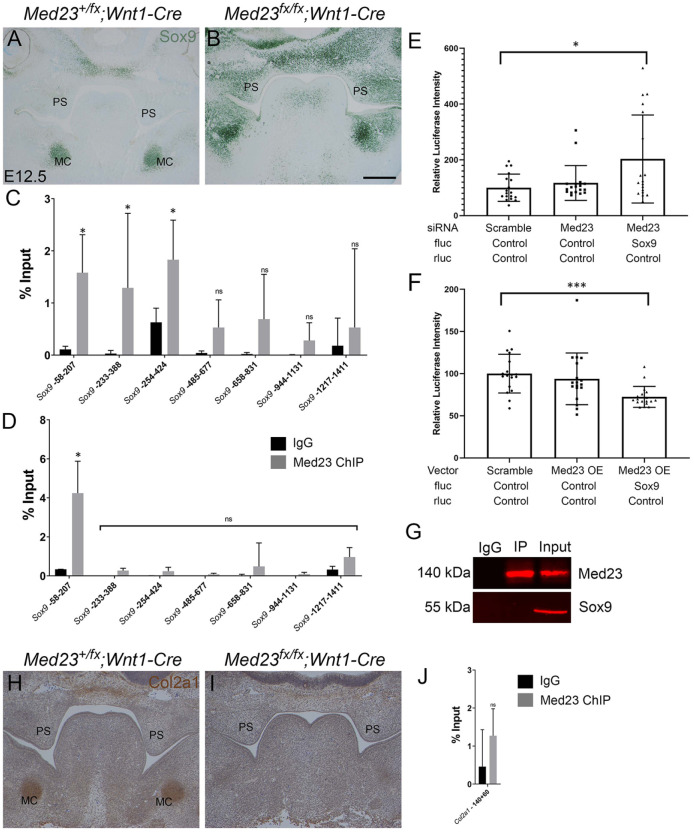
The micrognathia, cleft palate, and glossoptosis observed in Med23fx/fx;Wnt1-Cre mutants resemble the characteristics of Pierre Robin sequence (PRS) in humans, which is associated with mutations in an enhancer of SOX9 (Benko et al. 2009; Selvi and Mukunda Priyanka 2013). SOX9 is an early marker of migratory NCC and a master regulator of chondrogenesis (Lefebvre et al. 1997; Cheung and Briscoe 2003). SOX9 directly binds to and activates chondrocyte-specific genes, including COL2A1 (Bell et al. 1997; Lefebvre et al. 1997; Ng et al. 1997). Therefore, we examined the expression of Sox9 in Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre embryos by in situ hybridization and immunostaining, which revealed that both Sox9 RNA and protein are upregulated in the mutants, especially in the frontonasal prominence, and NCCs colonizing the first pharyngeal arch between E9.5 and E11.5 (Appendix Fig. 4A–H). The upregulation of Sox9 is also clearly evident at E12.5 and E14.5, when Sox9 protein is ectopically expressed in the palatal shelves and in the region that condenses to form MC in Med23fx/fx;Wnt1-Cre embryos (Fig. 3A, B; Appendix Fig. 4I–L, O, P). Alcian blue counterstaining revealed that MC does not properly condense and is loosely packed in mutant embryos compared to controls (Appendix Fig. 4K′, L′).
Figure 3.
Sox9 is misexpressed in Med23fx/fx;Wnt1-Cre embryos. (A, B) Sox9 is overexpressed in the palatal shelves and presumptive Meckel’s cartilages of Med23fx/fx;Wnt1-Cre embryos, as observed by immunostaining. (C) Chromatin immunoprecipitation performed on E13.5 wild-type palatal shelves using a Med23 antibody demonstrates that Med23 binds to the minimal promoter (−58 to −207) of Sox9 as well as to a region −233 to −424 bp upstream of the Sox9 transcription start site. (D) Chromatin immunoprecipitation performed on E13.5 wild-type limbs revealed Med23 binds to the minimal promoter of Sox9 but not to the −233-bp to −424-bp region upstream of the Sox9 transcription start site. (E) Luciferase activity is increased in 3T3-Swiss albino cells when the cells are cotransfected with small interfering RNA (siRNA) against Med23 and Sox9 enhancer-Luc vector compared to controls that are cotransfected with scramble siRNA and PGL3-enhancer vector and siRNA against Med23 and Sox9 enhancer-Luc vector. (F) On the other hand, luciferase activity is decreased when cells are cotransfected with Sox9 enhancer-Luc vector and Med23 overexpression vector compared to controls. (G) Coimmunoprecipitation of Med23 with Sox9 shows that Med23 and Sox9 do not form a complex. (H, I) Col2a1 immunostaining in the palatal shelves demonstrates that Col2a1 is significantly reduced in Meckel’s cartilages in Med23fx/fx;Wnt1-Cre embryos compared to Med23+/fx;Wnt1-Cre controls. (J) Chromatin immunoprecipitation performed on wild-type palatal shelves using a Med23 antibody, followed by quantitative polymerase chain reaction of the Sox9 binding region of the Col2a1 promoter, indicates that Med23 does not bind to Col2a1 promoter. MC, Meckel’s cartilage; PS, palatal shelf. Asterisk denotes statistical significance with P < 0.05. Scale bar for A–F is 120 mm.
The upregulation of both Sox9 transcript and protein in Med23fx/fx;Wnt1-Cre embryos suggests that Med23 either directly or indirectly regulates Sox9 transcription. To test this hypothesis, we performed chromatin immunoprecipitation (ChIP) on wild-type E13.5 palatal tissue using a Med23-specific antibody. We observed Med23 binding to 2 regions upstream of the Sox9 transcription start site (TSS) (Fig. 3C). The −58-bp to −207-bp region upstream of the Sox9 TSS has previously been identified to contain the minimal promoter of Sox9 (Kanai and Koopman 1999). However, Med23 also binds to −233 bp to −424 bp upstream of the Sox9 TSS, which is an unannotated region. To identify if Med23 binds to this region in a tissue that expresses Sox9, other than palatal mesenchyme, we performed ChIP on E13.5 limbs. We observed that Med23 binds only the minimal promoter of Sox9 and not the −233 to −424 region (Fig. 3D), suggesting that binding to this region might be required for tissue-specific regulation of Sox9 in palatal mesenchyme.
To determine if Med23 binding to the Sox9 promoter region is functional, we performed a luciferase assay in Swiss-3T3 cells. When Med23 is knocked down by 50% using small interfering RNA (siRNA), luciferase activity under control of the Sox9 promoter region increased (Fig. 3E). Conversely, a 50% increase in Med23 expression resulted in decreased luciferase activity (Fig. 3F). This suggests that Med23 binds to the promoter region of Sox9 and represses Sox9.
Sox9 positively regulates Col2a1 transcription (Bell et al. 1997; Lefebvre et al. 1997) and we, therefore, hypothesized that increased Sox9 expression should result in Col2a1 upregulation. Col2a1 is expressed in MC and nasal cartilage in E12.5 Med23+/fx;Wnt1-Cre embryos. However, Col2a1 was surprisingly downregulated in the presumptive MC of Med23fx/fx; Wnt1-Cre mutants (Fig. 3H, I). The downregulation of Col2a1 in the presence of elevated Sox9 is suggestive of an abrogation of Sox9 function and cranioskeletal maturation in Med23fx/fx; Wnt1-Cre embryos.
To determine if Med23 directly regulates Col2a1 expression, we performed ChIP–quantitative polymerase chain reaction (qPCR) on the Sox9 binding region of Col2a1 promoter but observed no binding between Med23 and the Col2a1 promoter (Fig. 3J). We also performed coimmunoprecipitation of Med23 and Sox9 in E12.5 wild-type palatal shelves to determine if Med23 facilitated Sox9 binding at the Col2a1 promoter but did not identify Sox9 to be a protein binding partner of Med23 (Fig. 3G). This suggests that Med23 is not directly required for Sox9-mediated transcriptional control of Col2a1 and that the decrease in Col2a1 occurs via another mechanism.
Med23 Binds to Runx2 Protein but Does Not Regulate Its Expression
Our data revealed that Med23 loss of function in NCC results in micrognathia, cleft palate, and cleidocranial dysplasia. Runx2 is a master regulator of osteoblast differentiation (Ducy et al. 1997; Komori et al. 1997), and mutations in RUNX2 are associated with cleidocranial dysplasia in humans (Mundlos et al. 1997). We therefore examined the differentiation of NCC-derived mesenchymal cells into primary osteoblasts at E12.5 via Runx2 immunostaining and observed the spatiotemporal patterns of Runx2 expression to be similar in the mesenchymal cells surrounding MC in Med23+/fx;Wnt1-Cre and Med23fx/fx; Wnt1-Cre embryos (Appendix Fig. 5A–C). In addition, ChIP-qPCR performed on the P1 promoter of Runx2 indicated that Med23 does not directly bind to the Runx2 promoter (Appendix Fig. 5D) and thus does not directly regulate Runx2 expression.
However, to further understand potential defects in osteoblast differentiation, we performed alkaline phosphatase staining and observed that while the mesenchyme had begun differentiating in E12.5 Med23+/fx;Wnt1-Cre controls, there were no differentiating cells observed in Med23fx/fx;Wnt1-Cre mutants (Appendix Fig. 5E, F). Reduced endogenous alkaline phosphatase staining was observed in the mesenchyme surrounding MC in E14.5 Med23fx/fx;Wnt1-Cre mutants compared to controls (Appendix Fig. 4K, L). This suggests that while Runx2 expression is not affected in Med23fx/fx;Wnt1-Cre mutants, early osteogenic differentiation is perturbed. Because Runx2 and Med23 proteins are known to interact in mouse preosteoblast cell lines (Liu et al. 2016), we performed Med23-Runx2 coimmunoprecipitation using E12.5 wild-type palatal shelves and observed that Med23 indeed binds to Runx2 (Appendix Fig. 5G). This suggests that Med23 may function as a cofactor for Runx2 in mediating transcriptional control of its target genes.
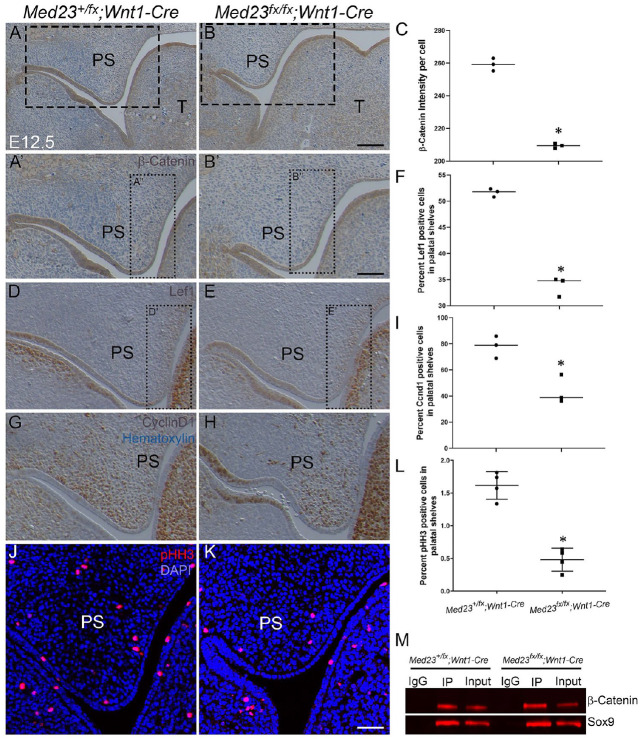
β-Catenin Is Downregulated in the Palatal Mesenchyme of Med23fx/fx;Wnt1-Cre Embryos
Sox9 upregulation has previously been shown to result in cleft palate in mice in association with downregulated Wnt signaling (Mori-Akiyama et al. 2003; Akiyama et al. 2004). Briefly, Sox9 overexpression leads to a dose-dependent increase in binding to β-catenin, which results in inactivation of both Sox9 and β-catenin, diminished Sox9 transcriptional activation of Col2a1, and downregulation of β-catenin target genes. We therefore immunostained sections of E12.5 Med23fx/fx; Wnt1-Cre and Med23+/fx; Wnt1-Cre embryos with β-catenin and observed that β-catenin is downregulated in the palatal mesenchyme in association with upregulated Sox9 expression (Fig. 4A–C, A′, B′; Appendix Fig. 6A, B). Furthermore, we determined that Lef1 and Ccnd1, which are downstream targets of β-catenin, were also downregulated in the palatal shelves of Med23fx/fx;Wnt1-Cre mutant embryos compared to controls (Fig. 4D–I; Appendix Fig. 6C, D). To test if Med23 directly regulates Lef1 and Ccnd1, we performed ChIP-qPCR for known transcription factor binding sites of Lef1 and Ccnd1. We observed no binding of Med23 to the promoters of Lef1 and Ccnd1 (Appendix Fig. 6E, F), suggesting that the downregulation of these genes is a consequence of diminished β-catenin activity independent of Med23.
Figure 4.
β-Catenin is downregulated in Med23fx/fx;Wnt1-Cre palatal shelves. (A–C) β-Catenin is downregulated specifically in the palatal shelves of Med23fx/fx;Wnt1-Cre embryos as observed by immunostaining with an antibody against β-catenin. The sections are counterstained with hematoxylin to visualize nuclei. Immunostaining with downstream targets of β-catenin, Lef1 (D–F), and Ccnd1 (G–I) demonstrates the number of palatal shelf mesenchymal cells expressing Lef1 and Ccnd1 is severely reduced in the Med23fx/fx;Wnt1-Cre embryos. (J–L) The number of proliferating cells is also significantly reduced in the Med23fx/fx;Wnt1-Cre palatal shelves compared to Med23+/fx;Wnt1-Cre. (M) Immunoprecipitation with Sox9 and immunoblotting with β-catenin using palatal shelves from E12.5 Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre embryos indicates that the binding of Sox9 and β-catenin is higher in Med23fx/fx;Wnt1-Cre palatal shelves, despite β-catenin itself being downregulated as observed by band size in input lanes. The concentration of total protein in the input lanes is 10% of the concentration used for the immunoprecipitation experiment. PS, palatal shelf. Asterisk denotes statistical significance with P < 0.05. Scale bar for A, B is 160 mm; A′, B′, D, E, G, H, J, K is 70 mm.
Jaw, orofacial clefts, and skull vault disorders have each been attributed to mutations in WNT signaling component genes (Manocha et al. 2019; Reynolds et al. 2019). Since Wnt signaling is a critical regulator of cell proliferation through Ccnd1, we hypothesized that decreased proliferation may underpin the pathogenesis of cleft palate in Med23fx/fx;Wnt1-Cre mutant embryos. Phospho-histone H3 staining revealed fewer proliferating cells in the palatal shelves of E12.5 Med23fx/fx; Wnt1-Cre mutants compared to controls, further corroborating our β-catenin and Ccnd1 immunostaining data (Fig. 4J–L). Although the number of proliferating cells was similar between controls and mutants at E14.5, by this stage, the palatal shelves of Med23fx/fx;Wnt1-Cre mutants were considerably smaller compared to controls, which affects their ability to meet in the midline (Appendix Fig. 7G–I). We also tested for apoptosis during palatogenesis via Cleaved-Caspase3 immunostaining and observed a similar number of apoptotic cells in controls and mutants (Appendix Fig. 7A–F). This suggests that reduced proliferation at E12.5 results in lower cell density by E14.5 and that fewer proliferating cells in the palatal mesenchyme limit the ability of the palatal shelves to grow and fuse to form a normal palate in Med23fx/fx;Wnt1-Cre embryos.
Sox9 and β-Catenin Have Increased Interaction in Med23fx/fx;Wnt1-Cre Palatal Shelves
To test if Sox9 upregulation results in increased Sox9 and β-catenin interaction, we performed immunoprecipitation with a Sox9-specific antibody on E12.5 Med23+/fx;Wnt1-Cre and Med23fx/fx;Wnt1-Cre palatal shelves. We found that β-catenin pulldown via Sox9 immunoprecipitation is higher in Med23fx/fx; Wnt1-Cre mutants compared to controls (Fig. 4M; Appendix Fig. 6G). Altogether, our data suggest that Med23 is required to repress Sox9 transcription, which helps to maintain optimal Sox9 expression in the palatal shelves. In the absence of Med23, Sox9 expression increases, resulting in elevated Sox9–β-catenin interaction and decreased Wnt signaling. Collectively, this leads to perturbation of palatal shelf mesenchyme proliferation and osteoprogenitor differentiation, which underpins the pathogenesis of micrognathia and cleft palate in Med23fx/fx; Wnt1-Cre embryos.
Discussion
Med23 is a tail subunit protein of the Mediator complex, which is required for enabling transcription by RNA polymerase II (Monté et al. 2018). Along with other tail complex proteins, Med23 connects the Mediator complex with sequence-specific transcription factors such as Elk1 (Stevens et al. 2002), Runx2 (Liu et al. 2016), and Irf7 (Griffiths et al. 2013). In addition, Med23 has been shown to function during transcript elongation (Wang et al. 2013), splicing (Huang et al. 2012), and chromatin modification (Yao et al. 2015). Here, we demonstrate that Med23 is required for craniofacial development, and its deletion in NCC results in micrognathia, cleft palate, glossoptosis, teeth defects, and cleidocranial dysplasia.
In humans, mutations in the enhancer and exons of SOX9 are associated with PRS (Jakobsen et al. 2007; Benko et al. 2009; Selvi and Mukunda Priyanka 2013). The SOX9 enhancer region, denoted as HCNE-F2, can drive the expression of LacZ in craniofacial tissues in E11.5 transgenic reporter mouse embryos and contains binding sites for transcription factors such as MSX1, EN1, and ZNF628 (Benko et al. 2009). Since SOX9 is required for chondrogenesis in the craniofacial and appendicular skeleton, mutations in this tissue-specific enhancer region could explain craniofacial bone and cartilage defects without limb defects in patients with PRS.
Both overexpression and downregulation of Sox9 result in cleft palate and severe chondrodysplasia in mice, indicating that Sox9 expression needs to be tightly regulated for proper chondrogenesis (Mori-Akiyama et al. 2003; Akiyama et al. 2004). We hypothesize a similar mechanism in Med23fx/fx; Wnt1-Cre embryos where absence of Med23 lifts transcriptional repression of Sox9, resulting in its overexpression. Sox9, when overexpressed, has previously been shown to increase its binding with β-catenin, thereby reducing the pool of free Sox9 and β-catenin, resulting in reduced expression of their respective downstream genes such as Col2a1 and Ccnd1. Furthermore, the physical interaction of Sox9 and β-catenin triggers the ubiquitination/proteasome pathway, resulting in inactivation and degradation of both proteins (Akiyama et al. 2004). Similar to Sox9, β-catenin is essential for palatal development and growth, and it functions by interacting with cadherins that are required during chondrogenic mesenchymal condensation (Delise and Tuan 2002). β-Catenin controls cell proliferation, migration, and differentiation during embryonic development, and its ablation results in orofacial clefts (He et al. 2011). Thus, in Med23fx/fx;Wnt1-Cre embryos, a diminished pool of freeβ-catenin reduces the expression of downstream targets ofβ-catenin that are required for proliferation of palatal mesenchymal cells. Decreased cell proliferation at this crucial developmental stage results in smaller palatal shelves, contributing to the pathogenesis of cleft palate (Fig. 5).
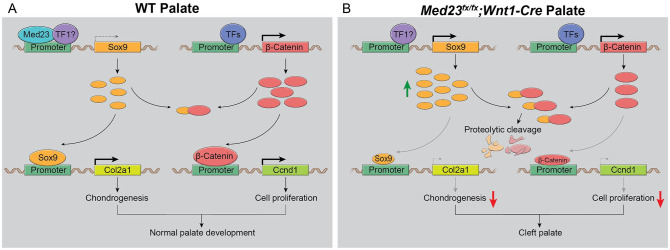
Figure 5.
Sox9 and β-catenin bind to each other in the palatal shelves. (A) In wild-type palatal shelves, Med23 represses the transcription of Sox9 (denoted by dashed arrow), probably in concert with other transcription factors. Sox9, when expressed in moderation, enables the transcription of its downstream targets such as Col2a1, thus regulating chondrogenesis. Meanwhile, β-catenin is also expressed in palatal shelf mesenchymal cells and enhances the expression of targets such as Ccnd1, thereby promoting cell proliferation. Together, Sox9 and β-catenin promote normal palate development. (B) In Med23fx/fx;Wnt1-Cre mutant palatal shelves, the absence of Med23 leads to upregulation of Sox9 (indicated by green arrow), which exhibits increased binding to β-catenin. Sox9 and β-catenin interactions can promote mutual proteolytic cleavage and inactivation of both proteins. However, Sox9 and β-catenin binding can also inhibit Sox9 from activating Col2a1 transcription. Collectively, this leads to downregulation of downstream targets of both Sox9 and β-catenin, such as Col2a1 and Ccnd1, respectively (indicated by dashed arrow), in the pathogenesis of cleft palate.
While we expected upregulation of the Sox9 downstream target Col2a1, and consequently increased chondrogenesis in Med23fx/fx;Wnt1-Cre embryos, instead we observed downregulation of Col2a1 and delayed chondrogenesis. Interestingly, Sox9–β-catenin binding has been shown to diminish the amount of Sox9 freely available in progenitor cells, as well as inhibit Sox9’s ability to transcriptionally activate Col2a1, which collectively can account for the reduction in Col2a1 activity in Med23fx/fx;Wnt1-Cre embryos. However, we cannot rule out that Med23 may regulate other factors critical for Col2a1 expression. Nonetheless, mutations in COL2A1 result in Stickler syndrome in humans (Richards et al. 2000; Liberfarb et al. 2003; Hoornaert et al. 2010; Higuchi et al. 2017), which is characterized by cleft palate together with ocular, auditory, and skeletal defects. Thus, downregulation of Col2a1 in Med23fx/fx;Wnt1-Cre mice, in addition to misregulation of Sox9 and β-catenin activity, is also consistent with the pathogenesis of cleft palate.
Med23fx/fx;Wnt1-Cre mutants exhibit cleidocranial dysplasia similar to the phenotypes observed following mesenchymal stem cell–specific knockout of Med23 (Med23fx/fx;Prx1-Cre) (Liu et al. 2016) and in Runx2+/− mutant mice (Mundlos et al. 1997). Although Runx2 expression was not reduced in Med23fx/fx; Wnt1-Cre embryos, Med23 is known to physically associate with Runx2 and influence the expression of transcriptional targets of Runx2. Thus, in the absence of Med23, Runx2 transcriptional activity is diminished, contributing to reduced bone growth in association with micrognathia and cleidocranial dysplasia. The Med23-Sox9 and Med23-Runx2 interactions, together with increased Sox9–β-catenin binding and decreased transcription of Sox9 and β-catenin targets, molecularly account for the phenotypes observed in Med23fx/fx;Wnt1-Cre embryos and suggest that Med23 functions in concert with multiple transcription factors to regulate gene expression during craniofacial development.
In support of our findings, 2 other Mediator complex proteins, Med12 and Med25, have also been shown to bind to Sox9 protein and regulate craniofacial and limb development, respectively (Rau et al. 2006; Nakamura et al. 2011). This provides a precedent for the Mediator complex in regulating chondrogenesis as well as craniofacial development through Sox9. It is important to note, however, that in contrast to Med12, Med25, and Med31, which activate Sox9 activity in mice (Rau et al. 2006; Risley et al. 2010; Nakamura et al. 2011), Med23 represses Sox9. Collectively, this implies that regulation by Mediator is context dependent and that certain subunits such as Med23 may only be present as part of the complex at specific times to repress or activate osteochondrogenesis during craniofacial development. Therefore, it will be imperative in the future to identify the full repertoire of Med23 binding partners that govern its transcriptional regulation output.
Author Contributions
S. Dash, P.A. Trainor, contributed to conception, design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; S. Bhatt, L.L. Sandell, contributed to conception, design, data acquisition, analysis, or interpretation, critically revised the manuscript; K.T. Falcon, contributed to data acquisition, analysis, and interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, DS_10.1177_0022034520969109 for Med23 Regulates Sox9 Expression during Craniofacial Development by S. Dash, S. Bhatt, K. T. Falcon, L.L. Sandell and P.A. Trainor in Journal of Dental Research
Acknowledgments
The authors thank members of the Trainor lab for their insights and discussions. We greatly appreciate Melissa Childers for her expertise in breeding, maintaining, and caring for the mice used in this project, and we acknowledge Rodney McCay, Lacey Ellington, and the Stowers Institute ES Cell and Transgenic Core for generating Med23bgeo chimeric mice. We also thank Mark Miller for illustrating Figure 5.
Footnotes
A supplemental appendix to this article is available online.
Declaration of Conflicting Interests: The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Stowers Institute for Medical Research (P.A. Trainor) and a Kirschstein-NRSA F31 predoctoral fellowship (DE027860) from the National Institute for Dental and Craniofacial Research (K.T. Falcon). Original data underlying this article can be accessed from the Stowers Original Data Repository at http://www.stowers.org/research/publications/LIBPB-1527.
ORCID iDs: S. Dash  https://orcid.org/0000-0002-9030-3341
https://orcid.org/0000-0002-9030-3341
P.A. Trainor  https://orcid.org/0000-0003-2774-3624
https://orcid.org/0000-0003-2774-3624
References
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, et al. 2004. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 18(9):1072–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell DM, Leung KK, Wheatley SC, Ng LJ, Zhou S, Ling KW, Sham MH, Koopman P, Tam PP, Cheah KS. 1997. SOX9 directly regulates the type-II collagen gene. Nat Genet. 16(2):174–178. [DOI] [PubMed] [Google Scholar]
- Benko S, Fantes JA, Amiel J, Kleinjan D-J, Thomas S, Ramsay J, Jamshidi N, Essafi A, Heaney S, Gordon CT, et al. 2009. Highly conserved non-coding elements on either side of SOX9 associated with Pierre Robin sequence. Nat Genet. 41(3):359–364. [DOI] [PubMed] [Google Scholar]
- Carlsten JOP, Zhu X, Gustafsson CM. 2013. The multitalented Mediator complex. Trends Biochem Sci. 38(11):531–537. [DOI] [PubMed] [Google Scholar]
- Cheung M, Briscoe J. 2003. Neural crest development is regulated by the transcription factor Sox9. Development. 130(23):5681–5693. [DOI] [PubMed] [Google Scholar]
- Conaway RC, Conaway JW. 2013. The Mediator complex and transcription elongation. Biochim Biophys Acta. 1829(1):69–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Bhatt S, Sandell LL, Siedel C, Ahn Y, Krumlauf R, Trainor PA. 2020. The Mediator subunit, Med23 is required for embryonic survival and regulation of canonical WNT signaling during cranial ganglia development. Front Physiol. 11:531933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delise AM, Tuan RS. 2002. Analysis of N-cadherin function in limb mesenchymal chondrogenesis in vitro. Dev Dyn. 225(2):195–204. [DOI] [PubMed] [Google Scholar]
- Ducy P, Zhang R, Geoffroy V, Ridall AL, Karsenty G. 1997. Osf2/Cbfa1: a transcriptional activator of osteoblast differentiation. Cell. 89(5):747–754. [DOI] [PubMed] [Google Scholar]
- Griffiths SJ, Koegl M, Boutell C, Zenner HL, Crump CM, Pica F, Gonzalez O, Friedel CC, Barry G, Martin K, et al. 2013. A systematic analysis of host factors reveals a Med23-interferon-λ regulatory axis against herpes simplex virus type 1 replication. PLoS Pathog. 9(8):e1003514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He F, Xiong W, Wang Y, Li L, Liu C, Yamagami T, Taketo MM, Zhou C, Chen Y. 2011. Epithelial Wnt/β-catenin signaling regulates palatal shelf fusion through regulation of Tgfβ3 expression. Dev Biol. 350(2):511–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y, Hasegawa K, Yamashita M, Tanaka H, Tsukahara H. 2017. A novel mutation in the COL2A1 gene in a patient with Stickler syndrome type 1: a case report and review of the literature. J Med Case Rep. 11(1):237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoornaert KP, Vereecke I, Dewinter C, Rosenberg T, Beemer FA, Leroy JG, Bendix L, Björck E, Bonduelle M, Boute O, et al. 2010. Stickler syndrome caused by COL2A1 mutations: genotype–phenotype correlation in a series of 100 patients. Eur J Hum Genet. 18(8):872–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Li W, Yao X, Lin Q-J, Yin J-W, Liang Y, Heiner M, Tian B, Hui J, Wang G. 2012. Mediator complex regulates alternative mRNA processing via the MED23 subunit. Mol Cell. 45(4):459–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakobsen LP, Ullmann R, Christensen SB, Jensen KE, Mølsted K, Henriksen KF, Hansen C, Knudsen MA, Larsen LA, Tommerup N, et al. 2007. Pierre Robin sequence may be caused by dysregulation of SOX9 and KCNJ2.J Med Genet. 44(6):381–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Koopman P. 1999. Structural and functional characterization of the mouse Sox9 promoter: implications for campomelic dysplasia. Hum Mol Genet. 8(4):691–696. [DOI] [PubMed] [Google Scholar]
- Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao Y-H, Inada M, et al. 1997. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 89(5):755–764. [DOI] [PubMed] [Google Scholar]
- Lefebvre V, Huang W, Harley VR, Goodfellow PN, de Crombrugghe B. 1997. SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene. Mol Cell Biol. 17(4):2336–2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberfarb RM, Levy HP, Rose PS, Wilkin DJ, Davis J, Balog JZ, Griffith AJ, Szymko-Bennett YM, Johnston JJ, Francomano CA. 2003. The Stickler syndrome: genotype/phenotype correlation in 10 families with Stickler syndrome resulting from seven mutations in the type II collagen gene locus COL2A1. Genet Med. 5(1):21–27. [DOI] [PubMed] [Google Scholar]
- Liu Z, Yao X, Yan G, Xu Y, Yan J, Zou W, Wang G. 2016. Mediator MED23 cooperates with RUNX2 to drive osteoblast differentiation and bone development. Nat Commun. 7:11149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik S, Roeder RG. 2000. Transcriptional regulation through Mediator-like coactivators in yeast and metazoan cells. Trends Biochem Sci. 25(6):277–283. [DOI] [PubMed] [Google Scholar]
- Manocha S, Farokhnia N, Khosropanah S, Bertol JW, Santiago J, Fakhouri WD. 2019. Systematic review of hormonal and genetic factors involved in the nonsyndromic disorders of the lower jaw. Dev Dyn. 248(2):162–172. [DOI] [PubMed] [Google Scholar]
- Monté D, Clantin B, Dewitte F, Lens Z, Rucktooa P, Pardon E, Steyaert J, Verger A, Villeret V. 2018. Crystal structure of human Mediator subunit MED23. Nat Commun. 9(1):3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori-Akiyama Y, Akiyama H, Rowitch DH, de Crombrugghe B. 2003. Sox9 is required for determination of the chondrogenic cell lineage in the cranial neural crest. Proc Natl Acad Sci USA. 100(16):9360–9365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, Cole WG, Henn W, Knoll JH, et al. 1997. Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell. 89(5):773–779. [DOI] [PubMed] [Google Scholar]
- Nakamura Y, Yamamoto K, He X, Otsuki B, Kim Y, Murao H, Soeda T, Tsumaki N, Deng JM, Zhang Z, et al. 2011. Wwp2 is essential for palatogenesis mediated by the interaction between Sox9 and mediator subunit 25. Nat Commun. 2:251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LJ, Wheatley S, Muscat GE, Conway-Campbell J, Bowles J, Wright E, Bell DM, Tam PP, Cheah KS, Koopman P. 1997. SOX9 binds DNA, activates transcription, and coexpresses with type II collagen during chondrogenesis in the mouse. Dev Biol. 183(1):108–121. [DOI] [PubMed] [Google Scholar]
- Rau MJ, Fischer S, Neumann CJ. 2006. Zebrafish Trap230/Med12 is required as a coactivator for Sox9-dependent neural crest, cartilage and ear development. Dev Biol. 296(1):83–93. [DOI] [PubMed] [Google Scholar]
- Reynolds K, Kumari P, Rincon LS, Gu R, Ji Y, Kumar S, Zhou CJ. 2019. Wnt signaling in orofacial clefts: crosstalk, pathogenesis and models. Dis Model Mech. 12(2):dmm037051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards AJ, Martin S, Yates JR, Scott JD, Baguley DM, Pope FM, Snead MP. 2000. COL2A1 exon 2 mutations: relevance to the Stickler and Wagner syndromes. Br J Ophthalmol. 84(4):364–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risley MD, Clowes C, Yu M, Mitchell K, Hentges KE. 2010. The Mediator complex protein Med31 is required for embryonic growth and cell proliferation during mammalian development. Dev Biol. 342(2):146–156. [DOI] [PubMed] [Google Scholar]
- Sandell LL, Iulianella A, Melton KR, Lynn M, Walker M, Inman KE, Bhatt S, Leroux-Berger M, Crawford M, Jones NC, et al. 2011. A phenotype-driven ENU mutagenesis screen identifies novel alleles with functional roles in early mouse craniofacial development. Genesis. 49(4):342–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Kurosaka H, Trainor PA. 2012. Whole mount nuclear fluorescent imaging: convenient documentation of embryo morphology. Genesis. 50(11):844–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvi R, Mukunda Priyanka A. 2013. Role of SOX9 in the etiology of Pierre-Robin syndrome. Iran J Basic Med Sci. 16(5):700–704. [PMC free article] [PubMed] [Google Scholar]
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ. 2002. Transcription control by E1A and MAP kinase pathway via Sur2 mediator subunit. Science. 296(5568):755–758. [DOI] [PubMed] [Google Scholar]
- Wang W, Yao X, Huang Y, Hu X, Liu R, Hou D, Chen R, Wang G. 2013. Mediator MED23 regulates basal transcription in vivo via an interaction with P-TEFb. Transcription. 4(1):39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X, Tang Z, Fu X, Yin J, Liang Y, Li C, Li H, Tian Q, Roeder RG, Wang G. 2015. The Mediator subunit MED23 couples H2B mono-ubiquitination to transcriptional control and cell fate determination. EMBO J. 34(23):2885–2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Wang G. 2014. The Mediator complex: a master coordinator of transcription and cell lineage development. Development. 141(5):977–987. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, DS_10.1177_0022034520969109 for Med23 Regulates Sox9 Expression during Craniofacial Development by S. Dash, S. Bhatt, K. T. Falcon, L.L. Sandell and P.A. Trainor in Journal of Dental Research