Abstract
Background
There is an urgent need for accurate, rapid, inexpensive biomarkers that can differentiate coronavirus disease 2019 (COVID-19) from bacterial pneumonia. We assess the role of the ferritin-to-procalcitonin (F/P) ratio to classify pneumonia cases into those due to COVID-19 vs those due to bacterial pathogens.
Methods
This multicenter case–control study compared patients with COVID-19 with those with bacterial pneumonia, admitted between March 1 and May 31, 2020. Patients with COVID-19 and bacterial pneumonia co-infection were excluded. The F/P in patients with COVID-19 vs with bacterial pneumonia were compared. Receiver operating characteristic curve analysis determined the sensitivity and specificity of various cutoff F/P values for COVID-19 vs bacterial pneumonia.
Results
A total of 242 COVID-19 pneumonia cases and 34 bacterial pneumonia controls were included. Patients with COVID-19 pneumonia had a lower mean age (57.1 vs 64.4 years; P = .02) and a higher body mass index (30.74 vs 27.15 kg/m2; P = .02) compared with patients with bacterial pneumonia. Cases and controls had a similar proportion of women (47% vs 53%; P = .5), and COVID-19 patients had a higher prevalence of diabetes mellitus (32.6% vs 12%; P = .01). The median F/P was significantly higher in patients with COVID-19 (4037.5) compared with the F/P in bacterial pneumonia (802; P < .001). An F/P ≥877, used to diagnose COVID-19, resulted in a sensitivity of 85% and a specificity of 56%, with a positive predictive value of 93.2% and a likelihood ratio of 1.92. In multivariable analyses, an F/P ≥877 was associated with greater odds of identifying a COVID-19 case (odds ratio, 11.27; 95% CI, 4–31.2; P < .001).
Conclusions
An F/P ≥877 increases the likelihood of COVID-19 pneumonia compared with bacterial pneumonia.
Keywords: COVID-19, ferritin, pneumonia, procalcitonin, SARS-CoV-2
Coronavirus disease 2019 (COVID-19) was declared a global pandemic by the World Health Organization on March 11, 2020 [1]. It is challenging to differentiate COVID-19 pneumonia from bacterial pneumonia at the time of clinical presentation. The clinical similarity between COVID-19 and bacterial pneumonia has resulted in concomitant use of antibiotics in up to 100% of COVID-19 patients [2–4]. In 4267 COVID-19 patients in New York City, co-infections were diagnosed in 3.6% of patients (mostly bacterial), yet antibiotics were given in 71% of these patients [5]. This mismatch between true co-infection and clinically presumed co-infection at the time of presentation emphasizes the difficulty in separating COVID-19 from bacterial infection. Early diagnosis enables expeditious use of appropriate antiviral or antibacterial treatments, minimizes adverse effects and costs associated with empirical therapies targeting both viral and bacterial infections, and facilitates infection control measures designed to prevent COVID-19 transmission. Real-time reverse transcriptase polymerase chain reaction (RT-PCR), used to diagnose COVID-19, typically requires at least 24 hours to return and can take more than 72 hours in low-resource settings. The clinical utility of other rapid antigen or molecular-based tests has not been established [6]. Thus, investigation of biomarkers has emerged as an important area of study in efforts to discriminate COVID-19 pneumonia from bacterial pneumonia.
Several laboratory markers have been studied for diagnosing COVID-19 and for assessing the severity of COVID-19 at the time of clinical presentation. We created the ferritin/procalcitonin (F/P) index using a model-based approach to biomarker development. A scientific model is a simplified description of a phenomenon (disease) that represents the disease to learn about the mechanism of disease production (how it arises) and suggests treatments [7, 8]. A common example of this activity is the use of animal models to explore pathogenesis and therapy. In our model, we use simple components that include respiratory cells and the pathogen severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). The model specifies interactions between pathogen and respiratory cells derived from the literature. In this model, SARS-CoV-2 produces disease by virion replication and obligate cytolysis in the respiratory epithelium, which results in respiratory dysfunction. The magnitude of virus replication and cytolysis reflects disease severity. There is also an associated inhibition of systemic inflammation caused by products of viral replication [9, 10]. In contrast, in bacterial pneumonia, most organisms do not require intracellular replication and cytolysis and thus produce less respiratory cell destruction compared with SARS-CoV-2. Procalcitonin (PCT) is a marker of systemic inflammation that is consistently higher in bacterial infections compared with viral infections [11]. This model focuses on significant differences in the mechanism of pathogen replication and host inflammation during infection, and we sought biomarkers that best represented these 2 model components. We restricted candidate biomarkers to those widely available, rapidly obtainable, and inexpensive. This model-based approach led us to focus on PCT and the iron-storage protein ferritin.
Our model predicts the ratio of circulating ferritin level (ng/mL) and PCT (ng/mL), called the F/P ratio, would be higher in COVID-19 pneumonia than in bacterial pneumonia.
METHODS
Ethics Statement
This investigation was approved by the Colorado Multiple Institutional Review Board (COMIRB) at the University of Colorado Anschutz Medical Campus (UCHealth), the Institutional Review Board (IRB) at University of Georgia in Athens, Georgia (USA), and the IRB at Yichang Central People’s Hospital of China Three Gorges University. Clinical data were obtained under a protocol reviewed and approved by IRBs and ethics committees at UCHealth (COMIRB Protocol 20–0690), University of Georgia (PROJECT00002679), and Yichang Central People’s Hospital (HEC-KYJJ-2020-026-01).
Patients and Data Collection
COVID-19 patients admitted to UCHealth, Yichang Central People’s Hospital, or Phoebe Putney Memorial Hospital from March 1 to May 31, 2020, were considered for inclusion. These sites were included based on collaborative efforts and the adoption of similar diagnostic criteria. The diagnosis of COVID-19 pneumonia was established by positive RT-PCR, clinical signs and symptoms, and imaging findings suggesting pneumonia. Exclusion criteria included patients younger than age 18 years, pregnancy, death on admission, missing baseline data, or transfer to another hospital. We excluded patients with concurrent COVID-19 and bacterial pneumonia at the time of hospital presentation. Based on our hypothesis of cytolysis-mediated lung injury, we anticipated that the co-infected patients with COVID-19 and bacterial pneumonia would also have an elevated F/P ratio, making differentiation more difficult. Patients diagnosed with bacterial pneumonia in the absence of COVID-19 served as controls. Bacterial pneumonia was diagnosed by clinical signs and symptoms of pneumonia, radiologic evidence of pneumonia on chest imaging, microbiologic documentation of a bacterial pathogen, and a negative COVID-19 PCR test.
Electronic medical records were manually abstracted for clinical and laboratory variables. At UCHealth and Phoebe Putney Memorial Hospital, data were retrospectively collected and stored in REDCap (electronic capture tools). At Yichang Central People’s Hospital, data were transferred manually into an Excel file. The following data were collected: gender, race/ethnicity, age, weight, body mass index (BMI), oxygen saturation in the emergency department, recorded preexisting medical conditions including diabetes mellitus (DM) and hypertension (HTN), and laboratory results including lactate dehydrogenase (LDH), ferritin, and PCT. LDH level was extracted as it is an intracellular enzyme that is elevated in the setting of organ damage. Based on our hypothesis that SARS-CoV-2 causes damage by cytolytic lysis, we predict LDH to be more elevated in COVID-19 patients compared with bacterial pneumonia patients.
Real-time Reverse Transcriptase Polymerase Chain Reaction
At UCHealth, upper respiratory tract samples were subjected to reverse transcription PCR amplification for SARS-CoV-2 (Roche Cobas 6800 EUA, Roche Diagnostics Corporation, Indianapolis, IN, USA).
At Phoebe Putney Memorial Hospital, several PCR platforms were used, including tests employed at Labcorp, Quest, and the Cepheid Xpert Xpress SARS-CoV-2 (Cepheid, Sunnyvale, CA, USA).
At Yichang Central People’s Hospital, RT-PCR was performed using the Detection Kit for SARS-CoV-2 RNA (PCR-Fluorescence Probing, Da An Gene Co., Ltd., of Sun Yat-sen University, Guandong, China) to detect SARS-CoV-2 virus nucleic acid.
Statistical Analysis
Statistical analyses were performed using STATA software, version 12.1 (StataCorp, College Station, TX, USA). Continuous variables were summarized using means (± SDs) or medians (± interquartile ranges). Frequencies (percentages) were used to describe categorical variables. Dichotomous outcomes were compared using chi-square analysis. The Fisher exact test was used to compare nominal independent variables, and a t test or Mann-Whitney U test was employed for interval-independent variables. Co-linear variables or variables missing significant data were excluded. We excluded from analysis 102 cases with COVID-19 and bacterial pneumonia co-infections. We compared F/P in patients diagnosed with COVID-19 with F/P in patients with bacterial pneumonia using a logistic regression controlling for age, sex, BMI, DM, HTN, and cohort site. A receiver operating characteristic (ROC) curve was used to compare true-positive rates (sensitivity) with false-positive rates (1- specificity) for various F/P values. The optimal F/P value that separated patients with COVID-19 pneumonia from patients with bacterial pneumonia was determined. Statistical significance was set at the .05 alpha level.
We analyzed F/P in the UCHealth cohort and separately analyzed F/P in the other 2 hospitals combined. An ROC curve was constructed for F/P in UCHealth patients and separately in the 2 hospital cohorts combined, and areas under the ROC curves were calculated.
Data Access
The corresponding author had full access to data in the study and had final responsibility for the decision to submit the manuscript for publication. The data sets used in the current study are available from the corresponding author upon reasonable request.
RESULTS
Clinical Characteristics and Laboratory Data in the 3 Participating Sites
Table 1 shows the clinical characteristics and laboratory data obtained in study subjects in each participating hospital. COVID-19 patients in the UCHealth cohort were Hispanic (43.2%), followed by African American (21.4%), White (18.2%), Asian (10%), and other (7.2%). In the Phoebe Putney Memorial Hospital cohort, most patients were African American (77.4%), followed by White (19.4%) and Hispanic (3.2%). In Yichang Central People’s Hospital, all patients were Asian (100%). DM and HTN were most prevalent in the Phoebe Putney Memorial Hospital group. A significantly larger percentage of patients from UCHealth and Phoebe Putney Memorial Hospitals were treated with intubation and mechanical ventilation compared with patients from the Yichang Central People’s Hospital. The median ferritin and procalcitonin levels were higher in Phoebe Putney Memorial Hospital patients compared with patients at UCHealth and Yichang Central People’s Hospital. The median F/P was lower in the UCHealth cohort compared with the cohorts from Phoebe Putney Memorial Hospital and Yichang Central People’s Hospital.
Table 1.
Characteristics of Patients Included at the 3 Participating Sites
| UCHealth (n = 209) | Phoebe Putney Memorial Hospital (n = 31) | Yichang Central People’s Hospital (n = 36) | |
|---|---|---|---|
| Cohort, No. (%) | |||
| Cases | 192 (92) | 27 (87) | 23 (64) |
| Controls | 17 (8) | 4 (13) | 13 (36) |
| Race, % | |||
| African American | 21.4 | 77.4 | 0 |
| Hispanic | 43.2 | 3.2 | 0 |
| White | 18.2 | 19.4 | 0 |
| Asian | 10 | 0 | 100 |
| Others | 7.3 | 0 | 0 |
| Age, mean (SD), y | 56.77 (18.3) | 62 (13.4) | 62.08 (15.3) |
| BMI, mean (SD), kg/m2 | 30.57 (7.1) | 36 (12) | 23.5 (2.8) |
| Female gender, No. (%) | 101 (48) | 19 (61) | 11 (31) |
| Comorbidities, No. (%) | |||
| Hypertension | 95 (46) | 21 (68) | 18 (50) |
| Diabetes mellitus | 64 (31) | 11 (36) | 8 (22) |
| Intubated, No. (%) | 32 (15.3) | 9 (29) | 4 (11) |
| LDH, median [IQR], IU/L | 324.5 [261–429.5] | 379 [345–562] | 248 [191–327] |
| Procalcitonin, median [IQR], ng/mL | 0.12 [0.05–0.24] | 0.18 [0.08–0.72] | 0.1 [0.05–0.24] |
| Ferritin, median [IQR], ng/mL | 353 [168–608] | 786.4 [385.10–1059] | 543.8 [285.15–1306.18] |
| F/P, median [IQR] | 3088.46 [1231.82–6600] | 6205.56 [877.75–12 961.11] | 6829.08 [2491.63–9658.67] |
| F/P ≥877, No. (%) | 166 (79) | 24 (77) | 30 (83) |
Abbreviations: BMI, body mass index; F/P, ferritin/procalcitonin ratio; IQR, interquartile range; LDH, lactate dehydrogenase.
Ferritin, Procalcitonin, and F/P in the Derivation Cohort (UCHealth)
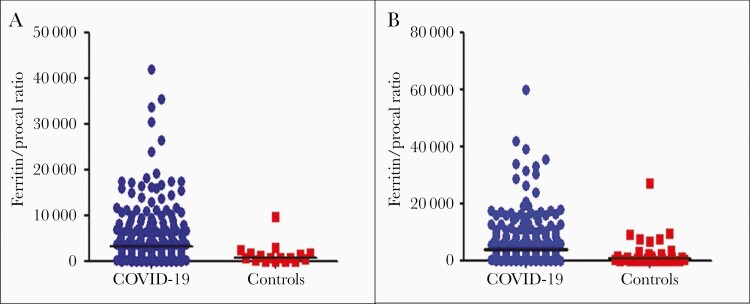
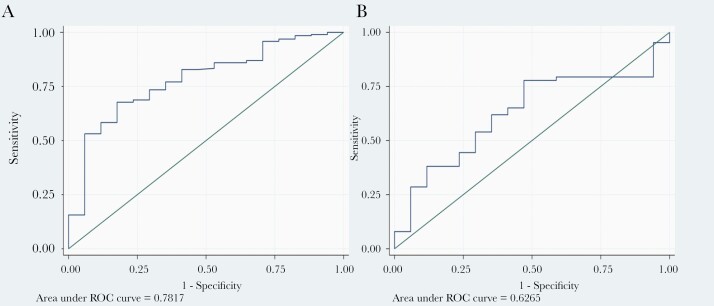
A total of 192 patients with COVID-19 and 17 patients with bacterial pneumonia were diagnosed at UCHealth. The median F/P in the UCHealth group was significantly higher in patients with COVID-19 pneumonia than in the bacterial pneumonia group (3195 vs 860, respectively; P < .001) (Figure 1A). An F/P cutoff point of ≥1250 generated a sensitivity of 78% and a specificity of 59% to separate COVID-19 cases from bacterial pneumonia cases, with an area under the curve (AUC) of 0.78 (Figure 2A). Adjusted for age, gender, BMI, DM, and HTN, an F/P ≥1250 associates with COVID-19 (odds ratio [OR], 4.9; 95% CI, 1.5–16.1; P = .009).
Figure 1.
A, Ferritin to procalcitonin ratios of patients with COVID-19 pneumonia and patients with bacterial pneumonia (controls) at UC Health. B, Ferritin to procalcitonin ratios in patients with COVID-19 pneumonia and patients with bacterial pneumonia (controls) in the 3 cohorts combined. Abbreviation: COVID-19, coronavirus disease 2019.
Figure 2.
A, Receiver operating characteristic analysis of ferritin-to-procalcitonin ratio cutoff values predicting COVID-19 diagnosis for UCHealth. B, Receiver operating characteristic analysis of ferritin-to-procalcitonin ratio cutoff values predicting COVID-19 diagnosis in the Putney Memorial and Yichang Central People’s Hospital Cohorts. Abbreviations: COVID-19, coronavirus disease 2019; ROC, receiver operating characteristics.
Ferritin, Procalcitonin, and F/P in the Validation Cohort
The F/P was then assessed for consistency using data from the Phoebe Putney Memorial Hospital group and Yichang Central People’s Hospital group as validation cohorts. A total of 50 patients with COVID-19 and 17 patients with bacterial pneumonia were diagnosed at Phoebe Putney Memorial Hospital and Yichang Central People’s Hospital. The median F/P in these cohorts was significantly higher in patients with COVID-19 pneumonia than in the bacterial pneumonia group (7461.1 vs 414, respectively; P < .001). A cutoff F/P of 1250 would correctly classify 68% of the COVID-19 cases for patients in the Phoebe Putney Memorial Hospital and Yichang Central People’s Hospital cohorts combined. With a cutoff of 877, this percentage increases to 73%. The ROC curve is shown in Figure 2B, with an AUC of 0.63.
Characteristics of Patients With COVID-19 and Patients With Bacterial Pneumonia
Our combined cohort comprised 242 hospitalized patients diagnosed with COVID-19 and 34 hospitalized patients diagnosed with bacterial pneumonia in the 3 participating hospitals combined, and patient characteristics are shown in Table 2 (none of these patients were co-infected). Patients with COVID-19 pneumonia were significantly younger and had a significantly higher mean BMI than patients with bacterial pneumonia. The groups had similar proportions of women. DM was significantly more common in the COVID-19 group compared with the group with bacterial pneumonia. Mean oxygen saturation in COVID-19 patients upon presentation to the emergency department was lower than the mean in patients with bacterial pneumonia, although the difference was not statistically significant. Patients with COVID-19 pneumonia were more likely to have a course complicated by intubation compared with patients with bacterial pneumonia, without reaching statistical significance.
Table 2.
Characteristics of Patients With COVID-19 Pneumonia and Bacterial Pneumonia
| COVID-19 Pneumonia (n = 242) | Bacterial Pneumonia (n = 34) | P Value | |
|---|---|---|---|
| Age, mean (SD), y | 57.11 (17.36) | 64.4 (17.72) | .02 |
| BMI, mean (SD), kg/m2 | 30.74 (8.10) | 27.15 (7.98) | .02 |
| Female gender, No. (%) | 113 (47) | 18 (53) | .50 |
| Comorbidities, No. (%) | |||
| Hypertension | 117 (48.3) | 17 (50) | .86 |
| Diabetes mellitus | 79 (32.6) | 4 (12) | .01 |
| Intubated, No. (%) | 42 (17.36) | 3 (9) | .98 |
| ED O2 saturation, mean (SD) | 89.6 (7.87) | 95.8 (4.21) | .08 |
| LDH, median [IQR], IU/L | 324.5 [258.5–423] | 262 [197–466.5] | .15 |
| Procalcitonin, median [IQR], ng/mL | 0.11 [0.05–0.22] | 0.33 [0.11–1.26] | <.001 |
| Ferritin, median [IQR], ng/mL | 427 [220.5–873] | 287.5 [124–468] | .01 |
| F/P | 4037.5 [1508.86–8118.89] | 802.09 [128.57–2597.81] | <.001 |
| F/P ≥877, No. (%) | 205 (84.7) | 15 (44.1) | <.001 |
| F/P ≥1250, No. (%) | 192 (79.34) | 15 (44.1) | <.001 |
Abbreviations: BMI, body mass index; COVID-19, coronavirus disease 2019; F/P, ferritin/procalcitonin ratio; IQR, interquartile range; LDH, lactate dehydrogenase.
Streptococcus pneumoniae, Escherichia coli, Serratia marcescens, and Pseudomonas aeruginosa were among the bacteria isolated in some of the controls included in the study. There was no predominant type of bacteria.
The median ferritin level was significantly higher in the COVID-19 pneumonia group compared with the bacterial pneumonia group. Patients with bacterial pneumonia had a significantly higher median procalcitonin level compared with patients with COVID-19 pneumonia. The median F/P was significantly higher in patients with COVID-19 (Table 2, Figure 1B).
Patients with COVID-19/bacterial pneumonia co-infection had a median F/P ratio of 5018, compared with 4037.5 (P = .58) among COVID-19 patients without coexisting bacterial pneumonia.
Predictors of COVID-19 Pneumonia
Adjusting for age, gender, BMI, DM, HTN, and admission to the hospital, an F/P ≥877 was associated with an 11-fold increased odds of COVID-19 compared with bacterial pneumonia (OR, 11.27; 95% CI, 4.07–31.20; P < .001) (Table 3).
Table 3.
Predictors of COVID-19 Pneumonia in Multivariate Logistic Regression
| Parameter | OR (95% CI) | P Value |
|---|---|---|
| F/P ≥877 | 11.27 (4.07–31.20) | <.001 |
| Age | 0.98 (0.95–1.01) | .16 |
| Gender | 0.36 (0.14–0.98) | .05 |
| BMI | 1.06 (0.99–1.13) | .07 |
| DM type 2 | 2.66 (0.78–9.04) | .12 |
| Hypertension | 0.47 (0.16–1.38) | .17 |
| Hospital | 0.45 (0.25–0.80) | .006 |
Abbreviations: BMI, body mass index; COVID-19, coronavirus disease 2019; F/P, ferritin/procalcitonin ratio; OR, odds ratio.
DISCUSSION
In this retrospective multicenter study, an F/P cutoff point of ≥877 generated a sensitivity of 85% and a specificity of 56% to differentiate COVID-19 pneumonia from bacterial pneumonia. We found similar F/P performance in patients from UCHealth and the validation cohort, suggesting that the diagnostic value of the F/P generalizes to other patient populations.
The F/P may have substantial clinical applicability. Circulating ferritin and procalcitonin levels can be easily obtained on admission blood testing, with results available within minutes to a few hours and providing real-time information on whether patients are more likely to have COVID-19 compared with bacterial pneumonia. This information may guide antimicrobial administration, patient isolation until the diagnosis of COVID-19 is confirmed, and specific treatment considerations. Unnecessary antimicrobial therapy and associated harms may be reduced. The prevalence of bacterial co-infections among patients with COVID-19 in our study was 15.4%, and we excluded these patients from this report. In a large cohort of 1705 COVID-19 patients, the prevalence of community-onset bacterial co-infections was only 3.5%. Despite the low rate of bacterial co-infections, the rate of empiric antimicrobial use was 56.6% [12]. In another cohort of 4267 COVID-19 patients in New York City, co-infections were diagnosed in 3.6% of cases [5]. The larger percentage of co-infected patients in our study in the total patient group reflects patient population variability. The disproportionately high rate of antimicrobial use in COVID-19 patients despite the low prevalence of co-infections is uniformly reported across studies [13]. We realize that the sensitivity of F/P in our cohort is too low to accurately rule out SARS-CoV-2 infection. We believe this tool is complementary to clinical judgment and other laboratory tests that help in the distinction between COVID-19 and bacterial infection. Its utility might be best in low-resource settings or in situations where the results of the PCR are delayed. It is also best used in settings where the PCR is falsely negative and the clinical suspicion for COVID-19 is high. Although not addressed in this report, we are evaluating F/P for use in discerning co-infected patients from patients infected with COVID-19 alone.
There are limited data available on serum ferritin levels in swine H1N1– and avian H5N1–infected patients to tell whether this ratio can be applied to COVID-19 [14]. A recent study [15] showed that hyperferritinemia is associated with poor outcomes in influenza A infection. As lung injury in influenza is mediated by cytolysis—in a similar manner as SARS-CoV-2—we hypothesize that the ratio may have some utility in influenza pneumonia as well.
Significant cytolysis caused by SARS-CoV-2 replication is suggested by in vitro observations in the closely related virus SARS-CoV, where prodigious viral replication within human primary ciliated respiratory epithelial cells caused cytopathy and cell shedding from tissues [16]. Ferritin was chosen as the marker for cytolysis during viral infection. Ferritin stores iron within cells and protects cell components from iron-induced free radical generation [17]. Circulating ferritin is often mischaracterized as an acute-phase reactant and likely originates from cell injury or cytolysis. Observations supporting this concept include the absence of a mechanism for cell secretion of ferritin [18]. Serum ferritin levels are unaffected by elevated plasma levels of the acute phase–inducing cytokines tumor necrosis factor–alpha (TNF) and interleukin-6 (IL-6) in healthy individuals. In contrast, there is a correlation between serum ferritin levels and circulating cell-free DNA (cfDNA) in hemodialysis patients [19]. As cfDNA is a cytolysis footprint, this report indicates that cytolysis is mirrored by elevated ferritin [20]. A significant role for cytolysis during COVID-19 is indicated by the presence of elevated cfDNA in COVID-19, and increased COVID-19 cfDNA associates with increased disease severity [21]. The association of viral infection of cells with cytolysis and liberation of intracellular ferritin into the blood is consistent with the substantial elevation of blood ferritin reported in COVID-19 patients [3, 22, 23]. As the exact source of circulating cfDNA and ferritin during COVID-19 is unknown, lysis of nonrespiratory epithelial cells may be an additional source for these cytolysis products. Regardless of the source, the magnitude of total cytolysis in COVID-19 patients likely reflects total viral replication activity and indicates disease severity. In contrast to COVID-19, bacterial pathogenesis does not necessitate host cell lysis, as most typical bacterial pneumonia pathogens do not require intracellular invasion to replicate. We believe it is useful to think of circulating ferritin as a biomarker indicating cytolysis and suggest an analogy with circulating levels of cardiac isoenzymes (often troponin proteins) as indicators of cardiomyocyte lysis or damage.
TNF and IL-1β (IL-1) are the prototype pro-inflammatory mediators involved in the host immune response. TNF levels are significantly lower in patients with COVID-19 and ARDS compared with patients with bacterial septic shock with ARDS [24], suggesting a less robust inflammatory response in COVID-19 compared with bacterial infections. Procalcitonin (PCT) is an established marker of inflammation and has been shown to increase in response to TNF and IL-1. Moreover, PCT synthesis is suppressed by interferon-gamma (IFN), which is produced in response to viral infections and may participate in lowering PCT levels during viral infections [25]. A review comparing PCT levels in viral and bacterial pneumonia in pediatric patients showed lower PCT concentrations in viral pneumonia [26]. For adult pneumonia, a systematic review and meta-analysis revealed that circulating PCT >0.05 µg/mL used to diagnose bacterial infection is associated with sensitivity and specificity of 0.55 and 0.76, respectively (considered a moderate degree of discrimination) [27]. Considered together, it appears that viral pneumonia—like COVID-19—produces lower systemic inflammation compared with bacterial pneumonia, and this is reflected in lower PCT concentrations in viral pneumonia. Given its widespread availability, lower cost, and fast turnaround time compared with levels of TNF or IL-1, we chose PCT as a surrogate marker of inflammation.
This study has several limitations. Proper use of F/P requires obtaining ferritin and PCT simultaneously as soon as possible after clinical presentation. However, our data included the first available levels of ferritin and PCT, and these levels were often not obtained simultaneously or immediately after initial presentation. The diagnostic characteristics of F/P may improve with the use of early and simultaneous acquisition of ferritin and PCT. Another limitation is the low specificity of this metric in either including or excluding bacterial pneumonia. Therefore, it might not be helpful in the decision-making of initiating or withholding antibiotics. Our study is likely underpowered to detect differences between COVID-19 pneumonia cases and bacterial pneumonia controls as we included only controls with microbiologic documentation of a bacterial infection, whereas most cases of community-acquired pneumonia do not have a positive bacterial culture. Additionally, without a matching strategy due to the relatively small number of patients, the sample size might be underpowered, limiting the reliability of the results presented. Our total patient cohort from all 3 hospitals included only 34 patients with bacterial pneumonia, and more bacterial pneumonia cases may provide a more accurate estimate of the utility of F/P. The F/P ratio is also dichotomized using ROC inflection points, and this dichotomization likely led to decreased statistical power. The retrospective study design makes it susceptible to several biases, and the applicability of F/P to other patient populations is uncertain. Due to limitations of available data, we were unable to assess the F/P association with important outcomes like mortality, intensive care unit stay, and mechanical ventilation. Although we adjusted for several confounders, there may be confounders unaccounted for in our analysis. Furthermore, F/P overlap occurred at low values for F/P (Figure 1), and our model predicts that this is likely due to COVID-19 patients with mild disease. Low F/P in COVID-19 likely associates with smaller amounts of cytolysis and increased inflammation, a combination associated with bacterial infection. It may therefore be difficult to separate mild COVID-19 cases from cases of bacterial pneumonia. If our model accurately reflects disease mechanism, serial calculation of F/P could be used as a tool to quantify the severity of infection over time and may predict clinical outcomes like mortality. This report introduces this novel index, and it is our belief F/P will perform better when ferritin and PCT are obtained as soon as possible after hospital presentation.
In summary, the novel F/P may assist with the early identification of COVID-19 patients. It is best used in combination with clinical judgment and other laboratory tools, where it could assist in the decision-making to withhold antibiotics in noncritical cases until the results of PCR and other laboratory markers and cultures are out. Further studies should evaluate its potential to quantify COVID-19 severity and prognosis.
Acknowledgments
We acknowledge the support of the University of Colorado medical students who contributed significantly to the chart abstraction.
Financial support. Dr. Leland Shapiro is supported by The Emily Foundation. Dr. Erlandson is funded by the National Institute on Aging (AG054366-05S1) and has received research and consultative funds from Gilead Sciences (paid to the University of Colorado).
Potential conflicts of interest. None declared. All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
Author contributions. The manuscript was read and approved by all authors. All authors contributed to the work of this report. K.J., J.H., P.H., J.F., F.M., and S.O. collected the clinical data. A.G. wrote the initial draft of the manuscript. A.H.M. and L.S. assisted in the study conception and design. A.H.M. analyzed the data. D.C., W.Z., J.M., K.E., S.S., C.F.P., A.H.M., and L.S. edited and reviewed the final manuscript.
Patient consent. The design of the study was approved by the Institutional Review Board at the University of Colorado Anschutz Medical Campus, University of Georgia, and Yichang Central People’s Hospital. Due to the retrospective nature of this study, the need for patient consent was waived.
References
- 1.World Health Organization. Timeline: WHO’s COVID-19 response. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/interactive-timeline/. Accessed 26 August 2020. 2020.
- 2. Huang C, Wang Y, Li X, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020; 395:497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study. Lancet 2020; 395:507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Li L, Zhang W, Hu Y, et al. Effect of convalescent plasma therapy on time to clinical improvement in patients with severe and life-threatening COVID-19: a randomized clinical trial. JAMA 2020; 324:460–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nori P, Cowman K, Chen V, et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect Control Hosp Epidemiol. 2021; 42:84–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dinnes J, Deeks JJ, Adriano A, et al. ; Cochrane COVID-19 Diagnostic Test Accuracy Group . Rapid, point-of-care antigen and molecular-based tests for diagnosis of SARS-CoV-2 infection. Cochrane Database Syst Rev 2020; 8:CD013705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bailer-Jones DM. Scientific Models in Philosophy of Science. Pittsburgh: University of Pittsburgh Press;2009. [Google Scholar]
- 8. Gerlee P, Lundh T.. Scientific Models. Red Atoms, White Lies and Black Boxes in a Yellow Book. Switzerland: Springer; 2016. [Google Scholar]
- 9. Schubert K, Karousis ED, Jomaa A, et al. SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 2020; 27:959–66. [DOI] [PubMed] [Google Scholar]
- 10. Schubert K, Karousis ED, Jomaa A, et al. Author correction: SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat Struct Mol Biol 2020; 27:1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lubell Y, Blacksell SD, Dunachie S, et al. Performance of C-reactive protein and procalcitonin to distinguish viral from bacterial and malarial causes of fever in Southeast Asia. BMC Infect Dis 2015; 15:511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vaughn VM, Gandhi T, Petty LA, et al. Empiric antibacterial therapy and community-onset bacterial co-infection in patients hospitalized with COVID-19: a multi-hospital cohort study. Clin Infect Dis. 2021; 72:e533–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rawson TM, Moore LS, Zhu N, et al. Bacterial and fungal co-infection in individuals with coronavirus: a rapid review to support COVID-19 antimicrobial prescribing. Clin Infect Dis. 2021; 71:2459–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kappert K, Jahić A, Tauber R. Assessment of serum ferritin as a biomarker in COVID-19: bystander or participant? Insights by comparison with other infectious and non-infectious diseases. Biomarkers. 2021; 25:616–25. [DOI] [PubMed] [Google Scholar]
- 15. Lalueza A, Ayuso B, Arrieta E, et al. ; INFLUDOC group . Elevation of serum ferritin levels for predicting a poor outcome in hospitalized patients with influenza infection. Clin Microbiol Infect 2020; 26:1557.e9–15. [DOI] [PubMed] [Google Scholar]
- 16. Sims AC, Baric RS, Yount B, et al. Severe acute respiratory syndrome coronavirus infection of human ciliated airway epithelia: role of ciliated cells in viral spread in the conducting airways of the lungs. J Virol 2005; 79:15511–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knovich MA, Storey JA, Coffman LG, et al. Ferritin for the clinician. Blood Rev 2009; 23:95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kell DB, Pretorius E. Serum ferritin is an important inflammatory disease marker, as it is mainly a leakage product from damaged cells. Metallomics 2014; 6:748–73. [DOI] [PubMed] [Google Scholar]
- 19. Cichota LC, Bochi GV, Tatsch E, et al. Circulating double-stranded DNA in plasma of hemodialysis patients and its association with iron stores. Clin Lab 2015; 61:985–90. [DOI] [PubMed] [Google Scholar]
- 20. Butt AN, Swaminathan R. Overview of circulating nucleic acids in plasma/serum. Ann N Y Acad Sci 2008; 1137:236–42. [DOI] [PubMed] [Google Scholar]
- 21. Cheng AP, Cheng MP, Gu W, et al. Cell-free DNA in blood reveals significant cell, tissue and organ specific injury and predicts COVID-19 severity. medRxiv 2020.07.27.20163188 [Preprint]. 29 July 2020. Available at: https://doi.org/10.1101/2020.07.27.20163188. Accessed 1 October 2020. [Google Scholar]
- 22. Wu C, Chen X, Cai Y, et al. Risk factors associated with acute respiratory distress syndrome and death in patients with coronavirus disease 2019 pneumonia in Wuhan, China. JAMA Intern Med 2020; 180:934–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Richardson S, Hirsch JS, Narasimhan M, et al. ; the Northwell COVID-19 Research Consortium . Presenting characteristics, comorbidities, and outcomes among 5700 patients hospitalized with COVID-19 in the New York City area. JAMA 2020; 323:2052–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kox M, Waalders NJB, Kooistra EJ, et al. Cytokine levels in critically ill patients with COVID-19 and other conditions. JAMA. 2021; 324:1565–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gilbert DN. Neglected variables in the interpretation of serum procalcitonin levels in patients with septic shock. J Infect Dis 2020; 222:S96–102. [DOI] [PubMed] [Google Scholar]
- 26. Thomas J, Pociute A, Kevalas R, et al. Blood biomarkers differentiating viral versus bacterial pneumonia aetiology: a literature review. Ital J Pediatr 2020; 46:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kamat IS, Ramachandran V, Eswaran H, et al. Procalcitonin to distinguish viral from bacterial pneumonia: a systematic review and meta-analysis. Clin Infect Dis 2020; 70:538–42. [DOI] [PubMed] [Google Scholar]