Abstract
SARS-CoV-2 infects humans through the binding of viral S-protein (spike protein) to human angiotensin I converting enzyme 2 (ACE2). The structure of the ACE2-S-protein complex has been deciphered and we focused on the 27 ACE2 residues that bind to S-protein. From human sequence databases, we identified nine ACE2 variants at ACE2–S-protein binding sites. We used both experimental assays and protein structure analysis to evaluate the effect of each variant on the binding affinity of ACE2 to S-protein. We found one variant causing complete binding disruption, two and three variants, respectively, strongly and mildly reducing the binding affinity, and two variants strongly enhancing the binding affinity. We then collected the ACE2 gene sequences from 57 nonhuman primates. Among the 6 apes and 20 Old World monkeys (OWMs) studied, we found no new variants. In contrast, all 11 New World monkeys (NWMs) studied share four variants each causing a strong reduction in binding affinity, the Philippine tarsier also possesses three such variants, and 18 of the 19 prosimian species studied share one variant causing a strong reduction in binding affinity. Moreover, one OWM and three prosimian variants increased binding affinity by >50%. Based on these findings, we proposed that the common ancestor of primates was strongly resistant to and that of NWMs was completely resistant to SARS-CoV-2 and so is the Philippine tarsier, whereas apes and OWMs, like most humans, are susceptible. This study increases our understanding of the differences in susceptibility to SARS-CoV-2 infection among primates.
Keywords: : COVID-19, ACE2, S-protein, resistant to SARS-CoV-2
Introduction
SARS-CoV-2, the cause of COVID-19, was first found in Wuhan, China, in late 2019. It infects humans at a higher rate than the 2002–2003 SARS-CoV (Lee et al. 2003; Peiris et al. 2003; Chen et al. 2020; Fung et al. 2020; Singhal 2020; Zhou et al. 2020) and has caused the most widespread pandemic in written human history. SARS-CoV-2, like SARS-CoV, infects humans mainly through the binding of its S-protein to human angiotensin I converting enzyme 2 (ACE2) (Shang et al. 2020; Walls et al. 2020; Wang et al. 2020; Yan et al. 2020; Zhang et al. 2020; Zhou et al. 2020). Thus, it is interesting to know whether there exist ACE2 variants in humans that confer resistance to SARS-CoV-2 infection. This question has been investigated before (Cao, Li, Feng, et al. 2020; Damas et al. 2020), but as described later, there are far more human sequence data available for identifying human ACE2 variants. ACE2 variants have also been identified in nonhuman primates (Damas et al. 2020; Melin et al. 2020), but we have collected ACE2 sequence data from many more nonhuman primate species (57 vs. 38 in Damas et al. 2020 and 28 in Melin et al. 2020).
The structural basis for the binding between ACE2 and S-protein has been deciphered (Shang et al. 2020; Walls et al. 2020; Wang et al. 2020; Yan et al. 2020). As S-protein variants have been extensively studied, we focus on ACE2 variants at the binding interface between ACE2 and S-protein. For all ACE2 variants found at binding residue sites, we conduct experiments to evaluate their effects on the binding affinity of ACE2 to S-protein. Moreover, we also use computational structural biology tools to infer their mutational effects. This inference provides a structural view of how a mutation affects the binding affinity. This combination of extensive ACE2 sequence data analysis, structural biology inference, and experimental assessment should provide a good understanding of how the susceptibility of primates to SARS-CoV-2 has evolved from the common ancestor of primates to extant species. Many human ACE2 variants have been produced by deep mutagenesis and their mutational effects on ACE2’s binding to S-protein have been assayed (Chan et al. 2020). Those data are compared with our data.
Results
ACE2 Variants in Humans
As ACE2 is an angiotensin converting enzyme, which controls blood pressure, and also the receptor for both SARS-CoV and SARS-CoV-2, it is interesting to identify its variants in humans. From the human ACE2 DNA sequence data in gnomAD (Karczewski et al. 2020), dbSNP (Sherry et al. 2001), ChinaMap (Cao, Li, Xu, et al. 2020), UK10K (Consortium 2015 b), 3.5KJPNv2 (Tadaka et al. 2019), 1KGP (1000 Genomes Project), the Korean Genome Project (Jeon et al. 2020), the Human Genome Diversity Project (Bergström et al. 2020), DiscovEHR (Dewey et al. 2016), and the NHLBI Exome Sequencing Project (Fu et al. 2013), we infer the nonsynonymous variants (supplementary fig. S1 and data 1, Supplementary Material online). In total, we find 407 nonsynonymous single nucleotide polymorphisms (SNPs), 9 of which have a premature stop codon and will not be discussed further. The remaining 398 missense SNPs lead to 396 amino acid variants and their allele counts are given in supplementary data 1, Supplementary Material online. Thus, about half of the 805 residue sites of ACE2 are variable in humans. However, there is only one variant, N720D (AAC → GAC), with allele frequency >0.01 in gnomAD, UK10K, DiscovEHR, and the NHLBI Exome Sequencing Project (supplementary data 1, Supplementary Material online). All other variants have frequencies lower than 1%, suggesting that ACE2 is under purifying selection in humans.
Among the 398 missense ACE2 variants, 8 are in the region from residue 1–18 (8/18 = 0.44), which is prior to the start of the protease domain (PD), 284 variants (284/597 = 0.47) are in PD, 59 variants (59/111 = 0.53) are in the collectrin-like domain (CLD), 16 variants (16/30 = 0.53) are in the transmembrane region, 20 variants (20/37 = 0.54) are on the cytosolic side, and 11 variants (11/30 = 0.36) are in the region 727–738, which lies in between CLD and the transmembrane region (supplementary fig. S1, Supplementary Material online). Although the proportions of variant sites vary considerably among regions, they are not statistically different (P = 0.52, prop. test). Interestingly, the catalytic active sites, the zinc binding sites, and the substrate binding sites in the PD all have amino acid variants in humans. However, these sites are not more variable than the rest of the whole protein (P = 0.72, prop. test).
The 27 ACE2 residues on the interface between ACE2 and viral S-protein are the major focus of this study; they are called key residues in this study. Eight of these residues show variants in humans (table 1 and fig. 1). As two variants (E35K and E35D) are found at residue 35, and M82I actually represents two nonsynonymous mutations (ATG → ATT and ATG → ATA), there are in total ten nucleotide variants observed at these eight residue sites. The 27 binding residues, which show only 8 variable residues (8/27 = 0.30), are on average better conserved than the remaining 778 residues of ACE2, which show 388 variable residues (388/778 = 0.50) (P < 0.02, prop. test).
Table 1.
Primate ACE2 Variants at Key Residues with Respect to the Human Wild-Type Amino Acid Sequence.a
| AA Position | Human Variants | Apes(6 sp.) | Old World Monkeys(20 sp.) | New World Monkeys(11 sp.) | Philippine Tarsier(1 sp.) | Prosimians(19 sp.) | |
|---|---|---|---|---|---|---|---|
| Endregion B | S19 | P | Ab | Fb | |||
| Q24 | Eb | ||||||
| T27 | A | Ab | Ab | Ab, Ib | |||
| D30 | Eb | ||||||
| K31 | Eb | Nb | |||||
| L79 | Rb | I | Rb, Ib | ||||
| M82 | I | Tc | S | Nd, Td | |||
| P84 | T | ||||||
| Middle | H34 | Q | Qb, Nb, Rb | ||||
| E35 | K, D | ||||||
| E37 | K | ||||||
| D38 | E | E | |||||
| Y41 | Hc | H | Hb | ||||
| Q42 | Lb | Ec | |||||
| Endregion A | Q325 | Rb | |||||
| N330 | Kb | ||||||
| K353 | N | Qb | |||||
| G354 | Qc | S | Db | ||||
| D355 | N |
Binding residues F28, L45, Y83, N90, T324, F356, R357, and R393 show no variants among the primates studied and so are not listed in the table.
Found in “only one or a few species of the group.”
Found in all the New World monkey species studied.
T is found at residue position 82 in 18 of the 19 prosimian species studied, whereas N is found in only 1 species.
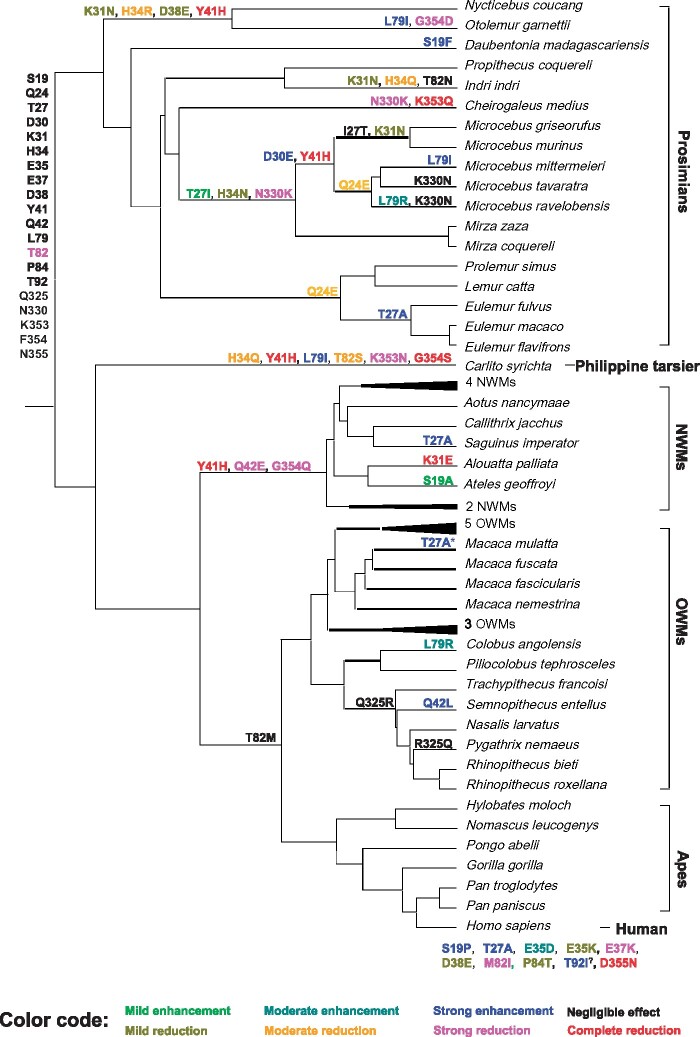
Fig. 1.
Cladogram of the primates studied and changes at key ACE2 residues during primate evolution. The inferred amino acids at the key ACE2 residues in the common ancestor of primates are shown at the root of the cladogram. The nine human ACE2 variants identified in this study are shown at the bottom of the figure, whereas the nonhuman primate variants are shown on the tree branches. The effect of each variant on binding affinity was evaluated by RBD attachment assay (see fig. 3) and is indicated by a color according to the color code at the bottom of this figure. Although site 92 is not a key binding residue, it is close to the N90 glycosylation site and the T92I mutation we identified in humans was found by Chan et al. (2020) to increase the binding affinity of ACE2 to S-protein; however, as its effect on binding affinity was not evaluated in this study, it is marked by “?” The T27A substitution in rhesus macaque (Macaca mulatta) is marked by an * because it only appeared in one individual we collected (supplementary data 4, Supplementary Material online).
ACE2 Variants in Nonhuman Primates
In the six ape species studied, no ACE2 variant at S-protein binding sites is found; that is, the amino acids at these residues are the same as those in human (table 1 and fig. 1) (the variants are annotated using the human ACE2 sequence, [table 1] or the primate ancestral ACE2 sequence [fig. 1] as the reference). In the 20 Old World monkey (OWM) species studied, 3 variants (T27A, Q42L, and L79R) each are found in only 1 species, whereas 1 variant (Q325R) is found in 5 species (fig. 1). In the 11 New World monkeys (NWMs) studied, the 4 variants Y41H, Q42E, M82T (i.e., T82), and G354Q are found in all 11 species, whereas the 3 variants S19A, T27A, and K31E each are found in only one species (fig. 1). The Philippine tarsier shows six variants (H34Q, Y41H, L79I, M82S, K353N, and G354S). In the 19 prosimians studied, 21 variants at binding residues are found; most of them are in only one or a few species (fig. 1). The variant M82T (i.e., T82) is found in 18 of the 19 prosimians studied and so T is likely the ancestral amino acid of prosimians at residue 82, whereas T82N is found in the remaining one species studied (Indri indri) (fig. 1). The E35, E37, P84, and D355 residue sites are found to have variants only in humans and the Q24 and N330 sites are variable only in prosimians, whereas the other sites in table 1 are found to have variants in multiple primate families (fig. 1), which apparently represent repeated mutations.
Functional Assays of RBD Attachment to ACE2 Variants
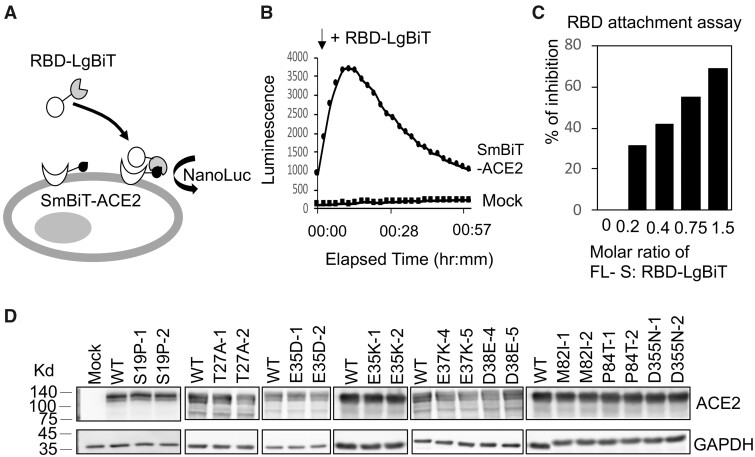
We established a cell-based S-protein attachment assay to evaluate the change in binding affinity due to a given ACE2 variant (fig. 2A). We used NanoLuc Binary Technology (NanoBiT), which splits NanoLuc luciferase into two parts, a large BiT (LgBiT) subunit and a small complimentary peptide with only 11 amino acids (SmBiT) (Dixon et al. 2016). Specifically, we first produced a recombinant LgBiT fusion protein with the receptor binding domain (RBD, amino acids 330–521) of S-protein (Wrapp et al. 2020) and generated expression constructs of human ACE2 with SmBiT tagged at the N-terminus. The attachment of RBD to the cell surface ACE2 receptor was measured by detecting luciferase activity when LgBiT and SmBiT were brought into close proximity upon RBD attachment (fig. 2B). RBD attachment was reduced when full-length S-protein (FLS) was included as a competitor (fig. 2C), implying that RBD-LgBiT and FLS competed for the same binding site on human ACE2. To investigate whether an ACE2 variant may affect host susceptibility to SARS-CoV-2, we tested RBD attachment on HeLa cells expressing the ACE2 variant. ACE2 variants were constructed by site-directed mutagenesis from the wild-type (WT) human SmBiT-ACE2. Expressions of these ACE2 variants were confirmed by western blotting following transient transfection into HeLa cells. ACE2 was detected with a molecular mass of ∼130 kDa for all human variants (fig. 2D).
Fig. 2.
The interaction between RBD and ACE2 variants measured by the RBD attachment assay. (A) Schematic illustration of the RBD attachment assay. Complementation of an active NanoLuc is achieved when the LgBiT and Sm-BiT are brought together upon interaction of RBD and ACE2. (B) HeLa cells transfected with mock construct or SmBiT-ACE2 were treated with RBD-LgBiT and subjected to bioluminescence detection for 1 h. Peak bioluminescence signal was detected at 10 min after the addition of RBD-LgBiT. (C) FLS was mixed with RBD-LgBiT at different molar ratios before incubation with HeLa cells transfected with SmBiT-ACE2. Bioluminescence signal was reduced by increasing the molar ratio of FLS. (D) Expression levels of the wild-type and human ACE2 variants were confirmed by western blotting. Two separate clones were tested for each variant transfection. The GAPDH expression served as an internal control for each experiment.
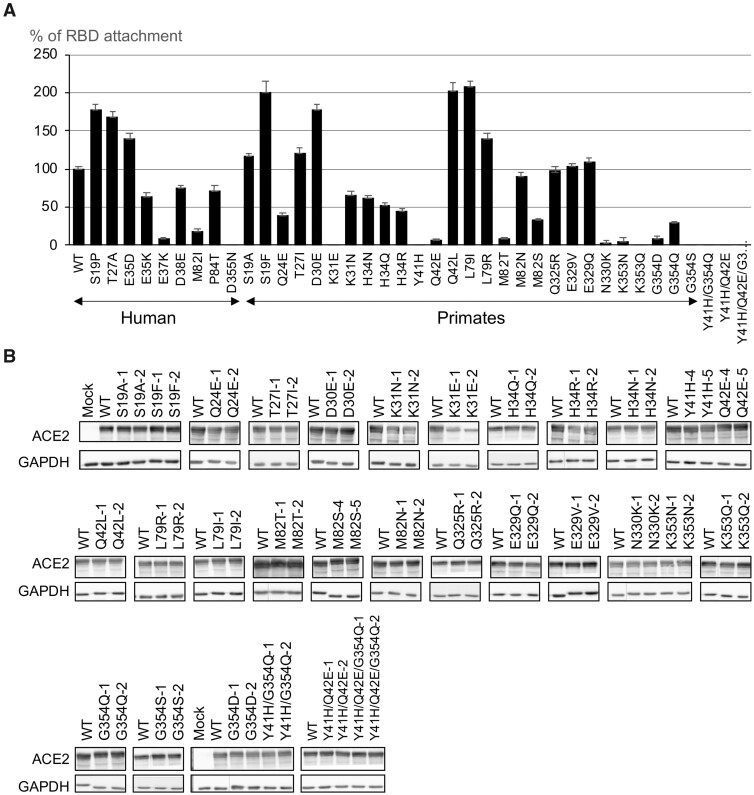
We next applied the RBD attachment assay to measure interactions between RBD and ACE2 variants. For each RBD attachment assay, 15,000 transfected cells were incubated with 250 ng RBD-LgBiT for 10 min before bioluminescence detection. We used the human WT ACE2 as the control and classified the effect of a variant as follows: 1) an increase in binding affinity is said to be strong if the observed binding affinity is >150% of that for WT, moderate if it is 125–150%, mild if it is 110–125%, and negligible if it is 100–110% and 2) a reduction is said to be negligible if the observed binding affinity is 90–100% of that for WT, mild if it is 60–90%, moderate if it is 30–60%, strong if it is <30%, and complete if it is 0%.
Among the nine human variants, S19P and T27A strongly enhanced (>150%) the RBD attachment and E35D moderately enhanced (>125%) the RBD attachment. On the other hand, RBD attachment was strongly reduced (<25%) in E37K and M82I and mildly reduced (<75%) in E35K, D38E, and P84T. Notably, RBD attachment was completely lost in the variant D355N (fig. 3A). For nonhuman primate variants, K31E, Y41H, K353Q, and G354S completely lost RBD attachment (fig. 3A). RBD attachment activity was moderately reduced in Q24E, H34R, M82S, and G354Q and strongly reduced in Q42E, M82T, N330K, K353N, and G354D. We detected a mild reduction of RBD attachment for variants K31N, H34Q, and H34N. Variants M82N and Q235R did not significantly reduce RBD attachment. On the other hand, S19A and T27I mildly and L79R moderately increased the binding affinity. Notably, variants S19F, D30E, Q42L, and L79I enhanced RBD attachment by approximately 1.5–2-fold. Except for K31E, all variants expressed equally in HeLa cells as shown by western blotting (fig. 3B).
Fig. 3.
Measuring RBD attachment on primate ACE2 variants. (A) RBD attachment activity measured on 9 human and 27 nonhuman primate ACE2 variants. The maximum bioluminescence signal measured in the RBD attachment assay for each variant was normalized by that of wild-type ACE2. Data are mean ± SEM of at least three replicates. (B) Intracellular protein expression levels of 27 single primate ACE2 variants, 2 double mutants, and 1 triple mutant were confirmed by western blotting. Expressions of two independent constructs for each variant were shown.
Interaction between S1 and ACE2 Variants
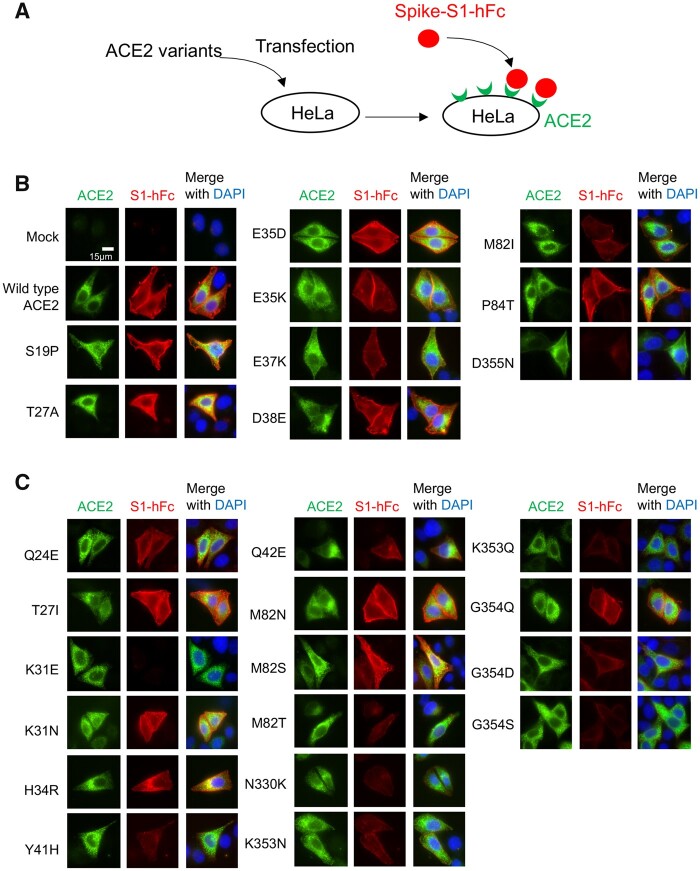
To confirm the interaction between S-protein and ACE2, we incubated recombinant spike-S1 protein containing a human Fc tag (S1-hFc) with HeLa cells expressing different ACE2 variants and detected cell surface-bound S1-hFc by immunofluorescence staining (fig. 4A). As expected, recombinant S1 protein was detected in HeLa cells expressing WT ACE2, but not in mock-transfected cells (fig. 4B). By comparing S1 protein interaction with nine different human ACE2 variants, we found that S1 binding was enhanced in cells expressing variant S19P, T27A, or E35D, mildly reduced in variant E35K, D38E, or P84T, and severely reduced in variant E37K or M82I. Notably, we detected no S1 signal in cells expressing variant D355N (fig. 4B). We then investigated the interaction between S1-hFc and 16 primate variants. As expected, variants K31E, Y41H, M82T, N330K, and G354S showed severely reduced S1 binding ability. A reduction in the S1 binding was also detected for variants Q42E, K353N, G354Q, and G354D. Besides, S1 binding ability was not affected for variants T27I, K31N, H34R, M82S, and M82N (fig. 4C).
Fig. 4.
Binding of S1-hFc to cell surface ACE2 variants. (A) Schematic illustration of the binding assay. HeLa cells transfected with ACE2 expression constructs were incubated 1 h with recombinant S1-hFc and cell surface bound S1-hFc was detected by immunofluorescence staining. (B) Representative images of S1-hFc (red) and HeLa cells expressing human ACE2 variants (green). (C) Representative images of S1-hFc (red) and HeLa cells expressing nonhuman primate ACE2 variants (green).
Structural Evaluation of ACE2 Variants
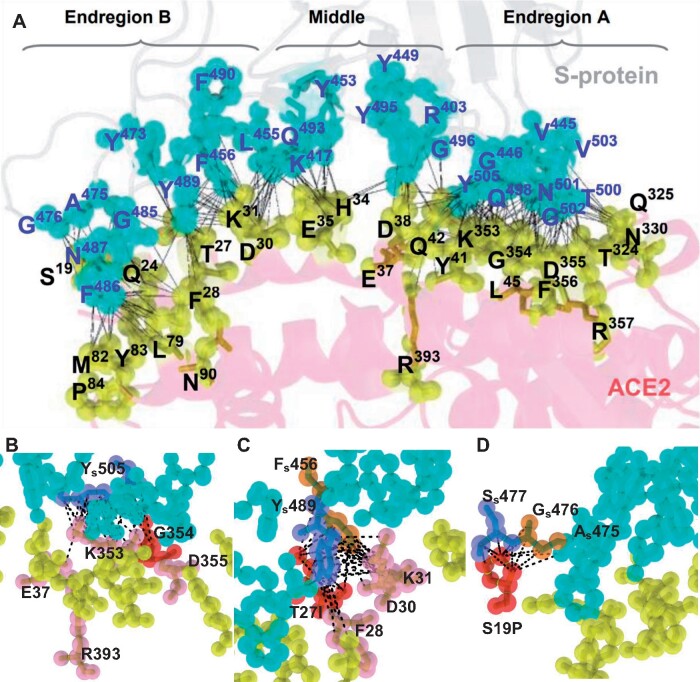
Using the X-ray crystal structure and cryo-EM structure of the ACE2–S-protein complex (Lan et al. 2020; Wrapp et al. 2020; Yan et al. 2020) and using topology theory (Edelsbrunner and Mucke 1994; Edelsbrunner et al. 1995, 1998) and geometric computations (Edelsbrunner et al. 1995, 1998; Liang, Edelsbrunner, Fu, et al. 1998; Tseng and Li 2009), we classify the 27 ACE2 residues on the binding interface into Endregion A (9 residues: L45, T324, Q325, N330, K353, G354, D355, F356, and R357), Middle (7 residues: H34, E35, E37, D38, Y41, Q42, and R393) and Endregion B (11 residues: S19, Q24, T27, F28, D30, K31, L79, M82, Y83, P84, and N90) (fig. 5A). We assess the mutational effect of a residue variant in terms of geometric measurements (Tseng, Dundas, et al. 2009; Tseng, Dupree, et al. 2009; Tseng and Li 2009, 2011), topographic properties of surfaces including solvent accessible area, number of atomic contacts (Liang, Edelsbrunner, Fu, et al. 1998; Liang, Edelsbrunner and Woodward 1998) and electrostatic potential (see Materials and Methods). It is clear from figure 5A that the density of atomic contacts is highest in Endregion A and lowest in Middle. We have evaluated all 27 variants but describe below only a number of variants with a strong effect on binding affinity.
Fig. 5.
The binding interface and atomic contacts between ACE2 and S-protein. (A) Binding between S-protein and ACE2 (in cyan and yellow spheres, respectively). The ACE2 binding residues (in yellow spheres) on the interface are classified into Endregion A, Middle, and Endregion B. Each contact between an ACE2 atom and an S-protein atom is analytically calculated and represented by a black dotted line. K353 of ACE2 has 42 atomic contacts, the strongest binding residue on the ACE2–S-protein interface. The highest density of atomic contacts between ACE2 and S-protein is detected at K353, Y41, and K31 of ACE2. (B–D) Close-up views of atomic contacts on the interface. (B) G354 in Endregion A is situated in the middle of the surface patch of K353, G354, and D355 (in red spheres), which strongly binds to the most crucial residue Ys505 (in slate) of S-protein. The amide backbone of G354 is replaced by a bulky polar side chain in G354Q (in red), leading to the removal of all five atomic contacts to Ys505 and reducing the binding affinity of K353, R393, and E37 (in pink) to Ys505. (C) The aromatic rings of Ys489 (in slate) and Fs456 (in orange) are docked into the groove of I27, F28, D30, and K31 (in pink). The polar side chain of T27 is replaced by a hydrophobic side-chain in the T27I mutation (in red), adding 3 and 5 atomic contacts to Ys489 and Fs456, respectively. (D) The S19P mutation on Endregion B gains 6 and 3 atomic contacts to Ss477 and Gs476, respectively, conferring a large increase in binding affinity to S-protein.
Variant D355N (Endregion A)
D355 has a total of 15 atomic contacts with S-protein, including 9 with T500, 4 with G502, and 2 with N501 of S-protein (fig. 5A); the pattern is represented by 9: T500, 4: G502, 2: N501. The D355N mutation removes the negative charge of D355 and changes the atomic contact pattern to 7: T500, 4: G502, 1: N501. D355N also affects the crucial residues T500, N501, and Y505 of S-protein on the binding interface. For instance, it removes 2 and 1 (−2 and −1) atomic contacts with T500 and N501 of S-protein. Moreover, it alters the atomic contacts of E37, K353, G354, A386, and R393 with Y505 of S-protein by −5, +4, −5, −1, and −2, respectively, causing a reduction of nine atomic contacts. The above analysis may explain why D355N abolishes the interaction between ACE2 and S-protein (figs. 3A and 4B).
Variants K353N/Q (Endregion A)
K353 is the strongest binding residue because it interacts with six S-protein residues (20: Y505, 13: N501, 3: G496, 2: Q498, 2: G502, 2: Y495) with a total of 42 atomic contacts. The removal of a positively charged side chain by K353N or K353Q influences many binding residues on the interface, including Y41, E37, D38, K353, G354, D355, A386, and R393 of ACE2. This structural analysis predicts that both K353N and K353Q abolish the binding between ACE2 and S-protein, in agreement with the RBD attachment assay (fig. 3A) and the S1 binding assay (fig. 4C).
Variants G354Q/S/D (Endregion A)
G354Q provides an example that the replacement of a small residue by a bulky polar side chain amino acid may greatly reduce the binding affinity to S-protein (fig. 5B). G354 lies in the middle of the surface patch of K353, G354, and D355 of ACE2 that strongly binds to the most crucial residue Y505 of S-protein. Its contact pattern includes 11 atomic contacts with S-protein (6: G502, 5: Y505). The replacement of the amide backbone of G354 by a large polar side chain in the G354Q mutation disrupts all five atomic contacts to Y505 of S-protein in the network of K353 and D355 and perturbs the binding of K353 and E37 to Y505 of S-protein on the interface. Thus, G354Q likely causes a strong reduction in binding affinity. G354S and G354D represent even stronger changes in physicochemical properties, so each of them likely causes an even stronger reduction on binding affinity than G354Q. These predictions are qualitatively in agreement with the RBD attachment assay data (fig. 3A).
Variant N330K (Endregion A)
N330 has seven atomic contacts with the crucial residue T500 of S-protein (fig. 5A). The N330K mutation adds a positive charge and alters the binding of ACE2 to T500 of S-protein, which also binds to the key residues D355 and R357 of ACE2. Therefore, N330K would cause a severe reduction in the binding affinity of ACE2 to S-protein, qualitatively in agreement with our experimental evaluation (figs. 3A and 4C)
Variant Y41H (Middle region)
K353, K31, and Y41 are the top three key residues on the ACE2-S-protein interface (supplementary table S1, Supplementary Material online). Y41 has the atomic contact pattern 11: Q498, 8: N501, 5: T500, so it has a total of 24 atomic contacts with S-protein. In Y41H, the phenol of site chain of Y41 is replaced by the imidazole side chain of H resulting in the pattern 9: Q498, 4: N501, 3: T500. As the atomic contact number is reduced from 24 to 16, Y41H would greatly reduce the binding affinity of ACE2 to S-protein, qualitatively in agreement with our experimental evaluation (figs. 3A and 4C).
Variants Q42E/L (Middle region)
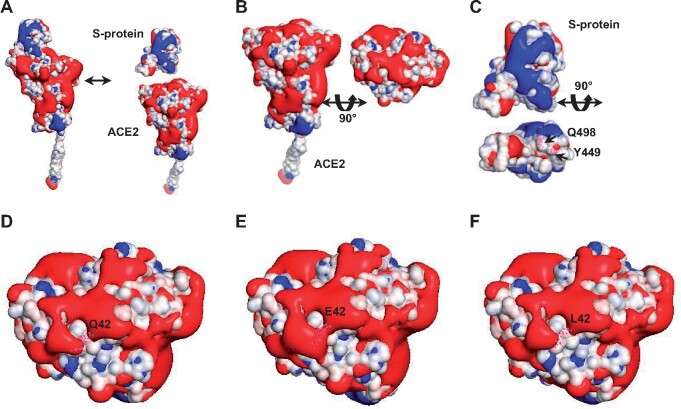
Q42 lies in the middle of the binding network of D38, Y41, Q42, and K353 of ACE2 that binds the crucial residues Q498, Y505, and Y449 of S-protein. It has the contact pattern: 2: Q498, 2: Y449, 1: G446. The Q42E and Q42L mutations alter the contact numbers in the bindings of D38, Y41, Q42, and K353 to Q498 of S-protein by −3, −2, +1, +1, and −2, −3, +1, +1, respectively. The Q42E mutation adds a negatively charged side change and thus perturbs the electrostatic surface. To gain a better understanding of the mutational effects of Q42E and Q42L on the binding affinity of ACE2 to S-protein, it requires an analysis of their effects on the electrostatic potential (Baker et al. 2001) of the neighboring residues in the binding network. As shown in figure 6A, the binding interface of ACE2 exhibits mostly negatively charged surface areas (red) (fig. 6B), whereas that of S-protein includes mostly hydrophobic areas (white) (fig. 6C). Specifically, the side chain of Q42 (fig. 6D) displays only a mild negative charge with a hydrophobic area of 26.04 Å2, whereas that of E42 significantly expands into a negatively charged surface with its neighboring residues (fig. 6E), effectively inhibiting the binding of S-protein to ACE2. In contrast, the Q42L mutation displays a 2.7-fold increase in hydrophobic area (i.e., 69.06 Å2 = 95.10–26.04 Å2, fig. 6F), providing a favorable condition for binding the counterpart surface at Q498 and Y449 of S-protein (fig. 6C). This analysis of perturbations in electrostatic potential due to residue changes predicts strikingly opposite effects of Q42E (a large reduction) and Q42L (a large increase) on the binding affinity of ACE2 to S-protein. These findings from electrostatic potential calculations qualitatively agree with the RBD attachment assay data (fig. 3A).
Fig. 6.
Electrostatic potential analysis of the binding interface of ACE2 for the wild-type Q42 and mutants E42 and L42. The atoms on the binding interface of ACE2 are colored according to their charges. The ACE binding interface displays charged surfaces with iso-surfaces drawn at 1.0 (blue) and −1.0 (red) kT/e, where k, T, and e denote the Boltzmann constant, the temperature, and the charge units, respectively. (A) ACE2 and S-protein display two distinctive patterns of electrostatic surfaces. (B) ACE2 is rotated to show that its binding interface includes mostly negatively charged areas (red). (C) After rotation, the S-protein interface shows mostly hydrophobic areas (white) with negatively charged patches (red). As indicated, Q498 and Y449 (in pink dots that represent atoms) are situated on the hydrophobic surface, interacting with Q42 of ACE2. (D) The wild-type Q42 (in pink dots) of ACE2 exhibits a mildly negatively charged surface. In comparison, the Q42E (in pink dots) mutation enhances the negatively charged surface with its neighboring residues (E), which may largely reject the interaction of ACE2 with S-protein, whereas the Q42L (in pink dots) mutation expands into a larger hydrophobic surface (F), strongly promoting the interaction of ACE2 with the hydrophobic surface of S-protein in (C).
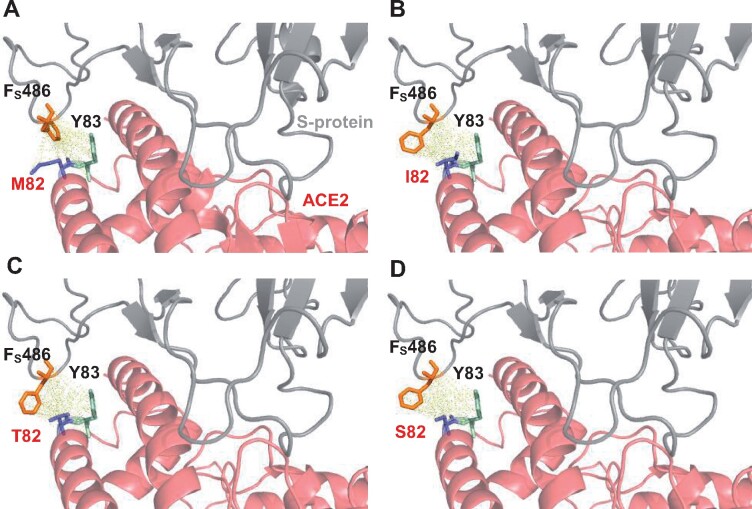
Variants M82I/S/T (Endregion B)
In the ACE2-S-protein complex, M82, Y83, and L79 of ACE2 form a subgroup (fig. 5A). Upon binding to S-protein, the cluster is oriented in the direction facing F486 on the flexible loop of S-protein, so that M82 might be a starting point of the binding between ACE2 and S-protein. M82 has 10 atomic contacts with F486 (10: F486) of S-protein (supplementary table S1, Supplementary Material online), and it teams up with Y83, which has 9 atomic contacts with F486 (9: F486) of S-protein (fig. 7A). We remodel the structure of M82I (Melo et al. 2002) and compute all atomic contacts and distances at position 82 (supplementary table S2, Supplementary Material online). The M82I mutation eliminates four contacts with F486 (6: F486) of S-protein. The accessible area and volume on the interface occupied by M82 and I82 are (30.35 Å2, 16.43 Å3) and (22.50 Å2, 1.53 Å3), respectively, so that the accessible area and volume on the interface are reduced by 7.85 (30.35–22.50) Å2 and 14.9 (16.43–1.53) Å3. The large alterations in accessible area (7.85/30.35 = 25.8%) and volume (14.9/16.43 = 90.7%) on the interface would largely block Y83’s binding to F486 of S-protein and eliminate four atomic contacts of L79 with F486 of S-protein. While the triad of L79, M82, and Y83 is oriented on the ACE2-S-protein interface, M82I modifies the structural conformation and stability of binding sites. Indeed, F486 of S-protein would not totally fit into the spatial position of I82 (fig. 7B). It further blocks the ring-stacking between F486 of S-protein and Y83 of ACE2 and the hydrophobic interaction between F486 of S-protein and L79 of ACE2. Taken together, our structural analysis indicates that M82I would strongly reduce the binding affinity between ACE2 and S-protein. M82S and M82T also reduce the number of atomic contacts with F486 of S-protein and prevent Y83 of ACE2 from binding to F486 of S-protein (fig. 7C and D). Specifically, the ten atomic contacts of M82 with F486 of S-protein are reduced to 3, 4, and 6 in M82S, M82T, and M82I, respectively. Furthermore, all three variants completely block Y83 of ACE2 from stacking with F486 of S-protein. This structural evaluation is supported by geometric analysis. Thus, the binding affinity to S-protein would be somewhat more strongly reduced in M82S and M82T than M82I. Our predictions for M82I and M82T are qualitatively in agreement with the RBD attachment assay data while give a larger reduction in binding affinity for M82S than the RBD attachment assay data (fig. 3A).
Fig. 7.
The effects of the M82I, M82T, and M82S mutations on the binding interface between ACE2 and S-protein. The shortest distance is 2.44 Å between M82 of ACE2 and Fs486 of S-protein and 5.85 Å between Y83 of ACE2 and Fs486 of S-protein. (A) The phenyl ring of Fs486 is oriented into the groove between M82 and Y83 of human wild-type ACE2 (PDB6m17). Pairwise atomic distances are displayed as dashed lines in yellow. (B) The residue packing at the local region of the M82I-mutant ACE2 is significantly altered. A prominent modification is that the phenyl ring of Fs486 is reoriented and situated away from the hydroxyphenyl ring of Y83 of the M82I mutant. (C–D) The replacement of the bulky hydrophobic methyl side chain in M82 by a polar side chain in T82 (C) or S82 (D) severely inhibits direct atomic contacts with the phenyl ring of Fs486, weakening the interaction between Fs486 and Y83 of ACE2 in M82T and M82S.
Variants T27A/I (Endregion B)
T27 contacts multiple residues of S-protein, including F456, Y473, A475, and Y489 and is situated in the compressed middle of the subgroup of T27, F28, D30, and K31 of ACE2 (fig. 5C). The T27A mutation causes a change from a polar to a short hydrophobic residue, leading to a smaller solvent accessible area. Upon binding, T27A induces a fit on the surface of F456 and Y489 of S-protein, strongly enhancing the binding affinity between ACE2 and S-protein. The T27I mutation replaces the polar side chain of T27 by a larger hydrophobic side chain and gains nine atomic contacts, eight of which are directly linked to the aromatic rings (5: F456, 3: Y489) of S-protein. Thus, T27I would enhance the binding affinity between ACE2 and S-protein. These two predictions are largely in agreement with the RBD attachment data (fig. 3A) and the S1 binding assay (fig. 4C).
Variants S19P/A/F (Endregion B)
S19 is on the border of the interface of the ACE2-S-protein complex (PDB: 6m0j) (fig. 5A). It has a simple atomic contact pattern (3: G476, 1: A475). The S19P mutation (fig. 5D) gains nine atomic contacts, six of which directly connect to S477 of S-protein (6: S477, 5: G476, 2: A475), implying a large increase in binding affinity. The S19A mutation only gains three additional atomic contacts with S477 of S-protein (3: G476, 3: S477, 1: A475), so it would confer only a mild increase in binding affinity. The S19F mutation replaces the polar side chain of S19 by a phenyl ring and changes the atomic contact pattern from 3: G476, 1: A475 to 10: A475, 8: G476, 5: S477, 3: Q474, 2: Y473, gaining 24 atomic contacts with S-protein. Moreover, it expands into a larger hydrophobic area and more effectively enhances affinity than the pyrrolidine side change of S19P. Thus, the S19F mutation would greatly increase the binding affinity of ACE2 to S-protein. These three predictions are qualitatively in agreement with the RBD attachment assay data (fig. 3A).
Discussion
This study used structural analysis and experimental assays to evaluate each observed ACE2 variant’s mutational effect on its binding to S-protein. We found that the two approaches usually gave similar results. For example, our structural analysis predicted that the D355N mutation would abolish the binding between ACE2 and S-protein, and our RBD attachment assay indeed supported this prediction. Moreover, our structural analysis predicted the importance of K353, K31, Y41, Q42, T27, and H34 in the binding to S-protein because each of these residues has >20 atomic contacts with the S-protein (supplementary table S1, Supplementary Material online). Our RBD attachment assays indeed showed that mutations at these residues strongly reduced or increased the binding affinity of ACE2 to S-protein. For instance, our RBD attachment assays showed a 100% reduction in binding affinity by K353Q and a > 90% reduction by K353N. As another example, our structural analysis predicted that S19F gains 24 atomic contacts with S-protein. It dramatically increases the binding affinity of ACE2 to S-protein, and our RBD attachment assay showed a 100% increase in binding affinity. However, as predicting interactions between two proteins is a complex problem, in some cases, including T27A, Q42L, and M82S, the predicted effect was different from the experimental evaluation. Our first structural analysis of Q42L was conducted solely in terms of the pattern of atomic contacts, and it predicted a mild reduction in binding affinity. However, taking its effect on electrostatic potential into account predicted a large increase in binding affinity, which was qualitatively consistent with the experimental assay.
Recently, Damas et al. (2020) proposed a set of rules for classifying the risks for SARS-CoV-2 infection in vertebrates. Their rules classify amino acid changes into conservative, semiconservative, and nonconservative and consider the number of identical key residues between a sequence and human ACE2 but with a particular emphasis on four key residues K31, E35, M82, and K353 and three glycosylation sites N53, N90, and N322. Our study confirmed that the four residues K31, E35, M82, and K353 indeed play important roles in the binding of ACE2 to SARS-CoV-2. In addition, we found other variants, including E37K, Y41H, Q42E, G354S, G354D, and D355N with a very strong effect on binding affinity (fig. 3). Moreover, we found that the effects of conservative residue substitutions (with similar physicochemical properties) can be strong. For example, Damas et al. (2020) classified the Q24E and Q42E mutations as “conservative,” implying no substantial effect on the binding affinity of ACE2 to S-protein. However, our structural analysis predicted that Q24E strongly hinders (data not shown), whereas Q42E largely disrupts the interactions between ACE2 and S-protein. The RBD attachment assay validated both predictions. Thus, our structural analysis can facilitate the understanding of why the same mutation at two different residue sites (e.g., Q24E vs. Q42E) and two different mutations at the same site (e.g., Q42E vs. Q42L) can have strikingly opposite effects on binding affinity (fig. 3A).
Melin et al. (2020) identified ACE2 variants at 12 amino acid residues critical for binding of ACE2 to S-protein. They studied the effect of amino acid change at ACE2 critical residues on the susceptibility of the host by estimating the binding free energy change (). Their study included 28 nonhuman primates among which apes and OWMs were inferred to have the same set of 12 amino acid residues as humans and so were equally susceptible to SARS-CoV-2 infection as humans. Their set of 12 critical amino acid residues is nested within our set of 27 residues and their inferences are largely consistent with our findings from 57 nonhuman primate species. Moreover, their inference of 400-fold reduction in SARS-CoV-2 susceptibility of NWMs compared with humans is also consistent with our evaluation by RBD attachment assays that “NWMs are completely resistant to SARS-CoV-2.” However, we found that the values for the five variants (Y41H, Q42E, M82T, D38E, and D30E) they studied and our RBD attachment assay data are only weakly correlated (r = 0.58) (supplementary fig. S2A, Supplementary Material online). The correlation coefficient became considerably lower (r = 0.20), when we compared our RBD attachment assay data to the values, we obtained using the SSIPe webserver (supplementary fig. S2B, Supplementary Material online). Thus, one should be cautious when using binding free energy change to infer the effect of mutation on binding affinity.
In an effort to identify ACE2 variants with a high binding affinity to S-protein, Chan et al. (2020) generated a large number of ACE2 variants by deep mutagenesis. Supplementary figure S3, Supplementary Material online, shows the comparison of their enrichment ratios and our RBD attachment assay data with a good correlation (r = 0.89), suggesting a qualitative agreement for the majority of mutations. For example, substantial reductions in the S-protein binding ability were reported for K31E, Y41H, N330K, K353Q, G354S, and D355N by Chan et al. The RBD attachment activities of these variants were mostly lost in our study (fig. 3A).
We collected extensive human sequence data to identify human ACE2 variants. Among the 398 missense variants, there were 9 variants located at ACE2–S-protein binding sites. These nine variants include the eight variants identified by Damas et al. (2020) and one novel variant (P84T). In addition, we identified T92I (supplementary fig. S1, Supplementary Material online), which is not on the ACE2–S-protein binding interface, but, according to Chan et al. (2020), it disrupts the glycosylation site N90 and strongly reduces the binding affinity of ACE2 to S-protein. From 57 nonhuman primate species, we identified all of the 26 nonhuman primate ACE2 variants identified by Damas et al. (2020), except S19Q, which was said to be found in Carlito syrichta, Microcebus murinus, and Otolemur garnettii, but we found S (i.e., no mutation) at residue 19 of these three species. Moreover, we identified a novel variant Q42L in OWM (fig. 1), which doubles the binding affinity of ACE2 to S-protein (fig. 3A).
This study provided the first evidence for changes in the host cell susceptibility by different human and primate ACE2 variants. We showed that attachment to SARS-CoV-2 S-protein was increased in cells expressing S19P/T27A/E35D, reduced in E37K and M82I, and drastically reduced in cells expressing variant D355N (figs. 3 and 4). Notably, the allele frequency of susceptible variant S19P is the highest among human variants studied in this study, with an allele frequency of 2,572 per million (m) in the African population and 3,911/m in African Americans (supplementary data 1, Supplementary Material online). As S19P, T27A, and E35D increase the binding affinity to S-protein (figs. 3A and 4B), they represent genetic risk factors for SARS-CoV-2. On the other hand, individuals carrying variants E37K, M82I, and D355N are likely moderately or strongly resistant to SARS-CoV-2. Variant E37K was found in three different data sets with an allele frequency of 112/m in Africans, 782/m in African Americans, and 319/m in Europeans. M82I was found in one data set, with an allele frequency of 202/m in Africans. Finally, the most resistant variant D355N was mainly found in Europeans, although the allele frequency was as low as 26/m. Despite the generally low allele frequencies of resistant ACE2 variants in the current human population, the status quo may change with the outbreak of COVID-19.
The identification of D355N as a strong resistant variant may provide an opportunity to study the evolution of a beneficial genetic variant following a pandemic outbreak over time. The human CC-type chemokine receptor 5 (CCR5) is a coreceptor of human immunodeficiency virus type-1 (HIV-1), and a 32-bp deletion in the coding region (CCR5-Δ32) confers resistance to HIV-1 (Samson et al. 1996). This finding has contributed to the clinical breakthrough for long-term control of HIV by stem-cell transplantation (Hutter et al. 2009). A recent study of the allele frequency of CCR5 in 1.3 million individuals in 87 countries found that CCR5-Δ32 allele frequencies ranged from 16.4% in the Norwegian sample to 0% in Ethiopia (Solloch et al. 2017). Similarly, it will be interesting to study the allele frequencies of ACE2-D355N, E37K, and M82I in human populations in the future. Presently, it is not clear if a chronic infection of SARS-CoV-2 may be established in patients. Evidence from the previous SARS coronavirus epidemic suggests that systemic and long-term tissue damage can last for years. Months after the COVID-19 outbreak, some patients are still battling crushing fatigue, lung damage and other ‘long COVID’ symptoms (Marshall 2020). Increasing cases of SARS-CoV-2 reinfection indicate that the protective immunity may be short-term (Tillett et al. 2020) and cannot eliminate the virus from the human body. In this regard, the identification of ACE2–D355N as a strongly resistant variant may confer a natural selective advantage against SARS-CoV-2.
Our data allow us to infer how the susceptibility to SARS-CoV-2 in primates has evolved. In essence, the evolution of primate susceptibility to SARS-CoV-2 can be captured by the evolution of four key residue sites: H41, Q42, M82, and G354. First, at the 27 ACE2 binding residues, the common ancestor of primates and human differed only at residue 82: T versus M (fig. 1). Compared with M82, T82 strongly reduces the binding affinity of ACE2 to S-protein. Therefore, while human is susceptible to SARS-CoV-2, the common ancestor of primates would be strongly resistant or only weakly susceptible. Second, like the common ancestor of primates, the common ancestor of prosimians and that of the tarsiers, NWMs, OWMs, apes, and human would be strongly resistant because their ACE2 sequences were identical to that of the common primate ancestor at the 27 binding residues. Third, the Philippine tarsier is likely completely resistant because its ACE2 includes the two mutations: Y41H and G354S, both of which strongly reduce the binding affinity of ACE2 to S-protein. Fourth, the common ancestor of NWMs should be completely resistant because it possessed H, E, T, and Q at residue sites 41, 42, 82, and 354. Indeed, our experimental assays showed that the combination of H41, Q42, and Q354 completely disrupts the binding (fig. 3). Moreover, it had T at residue 82. Fifth, the common ancestor of OWMs, apes, and human was susceptible because like humans, it had M at residue site 82. In summary, the common ancestor of primates was strongly resistant to and that of NWMs was completely resistant to SARS-CoV-2, whereas apes and OWMs, like most humans, are susceptible.
Besides the evolutionary changes at residue sites 41, 42, 82, and 354, some of the remaining episodic changes have changed the susceptibility to SARS-CoV-2 in some primate lineages. First, the three OWM species Macaca mulatta, Colobus angolensis, and Semnopithecus entellus have the substitutions T27A, L79R, and Q42L that should have greatly increased their susceptibility to SARS-CoV-2 (fig. 1). Second, among the NWMs, some individuals of Alouatta palliata should have stronger resistance compared with the other NWMs because of the K31E substitution, while some individuals of Saguinus imperator and Ateles geoffroyi might have relative weaker resistance compared with the other NWMs because they carry T27A and S19A, respectively (fig. 1). Third, although Philippine tarsier carries the L97I substitution, which might enhance the interaction between its ACE2 protein and S-protein, it has many other substitutions, such as H34Q, Y41H, T82S, K353N, G354S, which reduce the interaction (fig. 1). Therefore, it should be resistant to SARS-CoV-2. Last, some prosimian lineages have undergone evolutionary changes at ACE2 binding residues. There are several lineages that should be strongly resistant to SARS-CoV-2, such as I. indri that harbors N31, Q34, and T82, Cheirogaleus medius that harbors T82, K330, and Q353, and the common ancestor of Prolemur spp., Lemur spp., and Eulemur spp. that harbored E24 and T82 (fig. 1).
Recently, three new SARS-CoV-2 variants (table 2), namely B.1.1.7 (United Kingdom), B.1.351 (South Africa), and P.1 (Japan, a descendant of B.1.1.28), have received much attention because they appear to have a higher transmission rate and an increased viral burden due to mutations on the viral S-protein (Tegally et al. 2020; Sabino et al. 2021). According to the dynamic nomenclature classification of SARS-CoV-2 lineages (Rambaut et al. 2020), they all descended from B.1, which carries the D614G mutation outside the RBD of S-protein (Plante et al. 2020; Volz et al. 2021). This mutation has spread worldwide, so it likely has a selective advantage over D614 (Plante et al. 2020; Volz et al. 2021). In addition to this mutation, B.1.1.7, B.1.351, and P.1 carry 9, 11, and 10 other mutations in their S-protein, respectively (table 2). Here, we computationally assess the effect of each nonsynonymous mutation in the RBD of the S-protein on the binding affinity of S-protein variants to ACE2. Specifically, B.1.1.7 carries the N501Y mutation, whereas B.1.351 and P.1 carry N501Y, K417N, and E484K on the RBD of their S-protein. The mutations N501Y, K417N, and E484K are mapped, respectively, onto Endregion A, Middle, and the neighborhood of Endregion B of the binding interface between S-protein and ACE2 (see fig. 5). As we have already pointed above, N501 of S-protein is one of the top key binding residues of S-protein because it tightly interacts with K355, Y41, D355, and G326 of ACE2; our shape analysis reveals the atomic contact pattern 13: K353, 8: Y41, 2: D355, 1: G326. The N501Y mutation changes the atomic contact pattern to 18: K353, 12: Y41, 3: D355, 1: D38, increasing the number of atomic contacts from 24 to 34. As B.1.1.7, B.1.351, and P.1 all carry the N501Y mutation, they would all exhibit a higher binding affinity and thus probably also a higher transmission rate. For the B.1.351 and P.1 variants, the K417N mutation reduces three atomic contacts with D30 and H34 but enhances the binding affinity of the neighboring residue L455 by gaining six additional atomic contacts with D30 of ACE2. Thus, the K417N mutation would mildly increase the binding affinity of S-protein to ACE2. The E484K mutation perturbs the electrostatic potential on the surface of S-protein by the charge inversion from negatively charged E to positively charged K. The positively charged surface of E484K hinders the binding of S-protein to the positively charged K31 and K353 of ACE2, but overall mildly increases the binding affinity of S-protein to ACE2 because the binding interface of ACE2 mostly exhibits negatively charged surface areas (see fig. 6B). In summary, B.1.351 and P.1 likely mildly enhance the binding affinity of S-protein to ACE2, compared with B.1.1.7.
Table 2.
The Amino Acid Changes in the S-proteins of SARS-CoV-2 Variants (Tegally et al. 2020; Sabino et al. 2021).
| SARS-CoV-2 Variant Lineages | Mutations in S-Protein | Amino Acid Change on the RBD of S-Protein |
|---|---|---|
| B.1.1.7 (United Kingdom) | D614G, del H69, delV70, del Y144, N501Y, A570D, P681H, T716I, S982A, D1118H | N501Y |
| B.1.351 (South Africa) | D614G, L18F, D80A, D215G, R246I, K417N, E484K, N501Y, A701V, del L242, delA243, del L244 | N501Y, E484K, K417N |
| P.1 (Japan) | D614G, L18F, T20N, P26S, D138Y, R190S, K417T/N, E484K, N501Y, H655Y, T1027I | N501Y, E484K, K417T/N |
In conclusion, our combination of bioinformatics analysis of primate ACE2 sequences and RBD attachment assay has identified 15 ACE2 variants, each of which strongly reduces or completely disrupts the binding of ACE2 to S-protein, and 6 variants, each of which strongly enhances the binding affinity. Our computational protein structural analysis provided a basis for connecting structural changes in binding residues to changes in the binding affinity of ACE2 to S-protein. Complementing this, we established a novel, in vivo NanoLuc reporter assay to evaluate the effect on the binding of ACE2 variants to SARS-CoV-2 pseudovirus carrying the viral S-protein. From these findings, we propose a scenario for ACE2 sequence evolution in primates and how this affected the resistance or susceptibility of primates to SARS-CoV-2 infection.
Materials and Methods
Data Collection
To identify human ACE2 variants, we downloaded the human reference genome GRCh38 (hg38) and the following databases on July 12, 2020: the dbSNP (v154) (Sherry, et al. 2001), the NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium (132,345 individuals, obtained from dbSNP) (Kowalski et al. 2019), the Genome Aggregation Database (gnomAD) v3 (71,702 individuals) (Karczewski et al. 2020), UK10K (3,781 individuals) (The UK10K Consortium 2015), 3.5KJPNv2 (3,552 individuals) (Tadaka et al. 2019), 1KGP (The 1000 Genome Project Consortium 2015) (1000 Genomes Project phase 3, 2,504 individuals), Korean Genome Project (1,094 individuals) (Jeon et al. 2020), ChinaMap (10,588 individuals) (Cao, Li, Xu, et al. 2020) and Human Genome Diversity Project (929 individuals) (Bergström et al. 2020) and the exome sequencing data of gnomAD v.2.1.1 (125,748 individuals), DiscovEHR (50,726 individuals) (Dewey et al. 2016), and NHLBI Exome Sequencing Project (6,503 individuals) (Fu et al. 2013). The human variants we obtained are available as supplementary data 1, Supplementary Material online.
To have a comprehensive identification of nonhuman primate ACE2 variants, we downloaded all nonhuman primate genomes available on August 21, 2020, which include 13 ape genomes, 31 OWM genomes, 17 NWM genomes, 1 tarsier genome, and 21 prosimian genomes. In addition, we also downloaded all available ACE2 gene sequences of nonhuman primates on August 21, 2020, which include the ACE2 gene sequences of 26 rhesus macaques (M. mulatta) obtained by Chen et al. (2008) and 1 grivet (Chlorocebus aethiops) from GenBank. Moreover, we obtained the available ACE2 gene annotation of the downloaded genomes from GenBank (Sayers et al. 2020), RefSeq (Rajput et al. 2019), and Ensembl (Yates et al. 2020). The genomes and gene sequences we downloaded covered human, 6 apes, 20 OWMs, 11 NWMs, the Philippine tarsier, and 19 prosimians. We also downloaded the genome of Galeopterus variegatus (Sunda flying lemur, order Dermoptera) and its ACE2 coding sequences from GenBank (Sayers et al. 2020) to serve as an outgroup for inferring the ancestral ACE2 sequences of all extant primates.
Identification of Human ACE2 Variants
The ACE2 sequence of the human reference genome GRCh38 (hg38) was the same as the consensus sequence of all human ACE2 sequences we collected and was used as the reference for the identification of human ACE2 variants. The downloaded variants were validated using Ensembl variant effect predictor (McLaren et al. 2016). We only considered the nonsynonymous variants in subsequent analyses (supplementary data 1, Supplementary Material online). The variants along with their allele counts were plotted against their respective amino acid residues along the reference human ACE2 protein sequence (supplementary fig. S1, Supplementary Material online).
We used the R functionality prop. test (R Core Team 2020) for testing the null hypothesis that the proportions of ACE2 variants in different functional regions of the human ACE2 protein sequence were not different.
Search of Primate ACE2 Sequences
From the available ACE2 gene annotations of the downloaded genomes and gene sequences, we obtained a reference coding sequence set from 31 of the downloaded genomes (including one genome from each of Homo sapiens, Pan troglodytes, P. paniscus, Gorilla gorilla, Hylobates moloch, Mandrillus leucophaeus, Cercocebus atys, Papio anubis, Theropithecus gelada, Macaca nemestrina, Chlorocebus sabaeus, Piliocolobus tephrosceles, Callithrix jacchus, Aotus nancymaae, Saimiri boliviensis, Cebus capucinus, Sapajus apella, C. syrichta, Otolemur garnettii, Propithecus coquereli, Prolemur simus, and two genomes from each of Pongo abelii, Nomascus leucogenys, M. mulatta, Macaca fascicularis, and Rhinopithecus roxellana) and 27 of the nonhuman ACE2 gene sequences we collected. Each of the reference sequences includes the stop codon and has a length of 2,418 nucleotides. We then used the reference coding sequence set to search against all of the 56 nonhuman primate genomes without ACE2 gene annotation by BlastN of the Blast+ suite (version 2.9.0) (Camacho et al. 2009) to find the ACE2 coding sequences of all primate genomes. The ACE2 coding sequence of each primate genome was recovered based on the best BlastN hits. For the cases with incomplete coding regions, we inserted N’s in the missing regions; the number of N’s inserted was estimated from the best BlastN hits. After the search, we have 111 primate ACE2 coding sequences in total (supplementary data 2, Supplementary Material online).
Alignment of ACE2 Sequences and Inference of Ancestral Primate ACE2 Sequences
The 111 ACE2 coding sequences obtained above were aligned using MUSCLE (Edgar 2004) in MEGA X (Kumar et al. 2018). The alignment was 2,418 bps long and is available as supplementary data 3, Supplementary Material online. The amino acid alignment based on the nucleotide sequence alignment is presented in supplementary data 4, Supplementary Material online.
To determine the likely history of the amino acid substitutions at the key residues along different lineages during the course of primate evolution, we conducted ancestral sequence reconstruction as follows. We used the species name of the 58 primates we studied and the Sunda flying lemur (G. variegatus) to search the TimeTree database (Kumar et al. 2017) for their reference species tree and obtained a tree that covered 57 of the primates (except Microcebus sp. 3 GT-2019) studied and the Sunda flying lemur. We then selected one representative ACE2 nucleotide coding sequence for each of the 57 primates and the Sunda flying lemur. For human, the ACE2 coding sequence in GRCh38 was selected as the representative nucleotide coding sequence. For a nonhuman primate species with more than one ACE2 coding sequence, we first generated the consensus of all its ACE2 coding sequences; we then selected the coding sequence that is closest to the consensus as the representative sequence because we prefer not to include a product that is not supported by any of the genomes or gene sequences we obtained. The codon-based multiple sequence alignment of the representative sequences was obtained from the codon-based multiple sequence alignment of all ACE2 sequences we have. The best nucleotide substitution model was determined as the general time reversible model (Tavaré 1986) with five rate categories (GTR + G) considering both the codon-based multiple sequence alignment and the species tree using MEGA X (Kumar et al. 2018). Finally, the ancestral sequence reconstruction based on maximum-likelihood model was done using MEGA X (Kumar et al. 2018), considering the nucleotide sequence alignment, the nucleotide substitution model (GTR + G), and the reference tree.
Identification of ACE2 Variants of Nonhuman Primates
We used the human consensus ACE2 sequence as the reference sequence to identify the ACE2 variants in the alignment of primate ACE2 sequences obtained above. Thus, our primate ACE2 variants are variants with respect to the human ACE2 consensus sequence (supplementary data 4, Supplementary Material online).
Evaluating the Effect of Each ACE Variant on the Binding Affinity of ACE2 to S-Protein
The 3D complex structures (PDB ID: 6m0j, Lan et al. 2020 and 6m17, Yan et al. 2020) of human ACE2 and viral S-protein were used for structural analysis. We first computed the binding interface between ACE2 and S-protein using the 3D alpha-shape theory (Edelsbrunner and Mucke 1994; Edelsbrunner et al. 1995, 1998). We then used the Volbl package (Liang, Edelsbrunner, Fu, et al. 1998; Liang, Edelsbrunner, and Woodward 1998) to conduct structural analysis as described below.
To assess the effect of a residue change on the binding affinity between ACE2 and S-protein, we used the Modeller homology modeling tool (Melo et al. 2002) to construct a model of the complex of the ACE2 variant and S-protein for simulating mutational effects. We selected a target residue of ACE2 for mutagenesis and performed a molecular dynamics optimization at a fixed temperature (293.0 K) by Modeller with default parameters. We then conducted geometric calculations using Volbl (Liang, Edelsbrunner, Fu, et al. 1998; Liang, Edelsbrunner and Woodward 1998) to determine the binding interface between the WT ACE2 and S-protein and that between a mutant ACE2 and S-protein. In shape analysis (Tseng, Dundas, et al. 2009; Tseng, Dupree, et al. 2009; Tseng and Li 2009, 2011), we computed the atomic contacts between the WT residue of ACE2 and S-protein and those between a residue variant of ACE2 and S-protein, using the weighted Delaunay triangulation (Edelsbrunner and Mucke 1994; Edelsbrunner et al. 1995, 1998) (an atomic contact is defined as a link (edge) between one atom of ACE2 and one atom of S-protein in the weighted Delaunay triangulation of the ACE2–S-protein complex.). We then used the data to infer the atomic contact pattern for each selected residue of ACE2 (supplementary table S1, Supplementary Material online). Removing or adding atomic contacts to a pattern alters the interaction between the two proteins. A reduction in binding affinity is deemed severe if more than five atomic contacts are removed and is moderate or mild if fewer than five atomic contacts are removed. The smallest number of atomic contacts in this interface is 2. Thus, an increase in binding affinity is deemed strong if more than seven atomic contacts are added but moderate or mild if fewer than seven atomic contacts are added.
We also used Volbl to calculate solvent assessable area and volume of a site-specific residue. We further computed the polar and nonpolar solvent accessible area and volume of a residue. In analyzing electrostatic potential, we used the adaptive Poisson–Boltzmann solver (APBS) (Baker et al. 2001) with default parameters to assess the charge modification on protein surface. We then evaluated the mutational effect by comparing the atomic contact pattern, solvent accessible area, and electrostatic potential of each observed variant residue to those of the WT residue. The APBS and PDB2PQR (Dolinsky et al. 2007) software packages were used for electrostatics calculations. An input structure is reconstructed by adding hydrogens, assigning atomic charges, radii, and force field and repairing missing heavy atoms by PDB2PQR for electrostatic analysis in APBS. The resulting electrostatic potential map is displayed as isosurface using PyMOL apbsplugin (https://pymolwiki.org/index.php/Apbsplugin, last accessed January, 10, 2021). The isosurface is visualized as a color-coded electrostatic surface at 1.0 (blue) and −1.0 (red) kT/e, where k, T, and e represent the Boltzmann constant, the temperature, and the charge units, respectively. Structural representations in figures are prepared by PyMOL (https://github.com/schrodinger/pymol-open-source, last accessed January, 10, 2021).
To assess the effect of a residue mutation on the binding free energy change () between ACE2 and S-protein, we utilized the SSIPe webserver (https://zhanglab.ccmb.med.umich.edu/SSIPe/, last accessed January, 10, 2021) (Huang et al. 2020) with the default settings, using the 3D complex structure (PDB ID: 6m0j) (Lan et al. 2020) as the reference.
ACE2 Mutagenesis Experiment Design
The human ACE2 coding gene was obtained from Addgene (Plasmid #1786). To ectopically express SmBiT-hACE2 in mammalian cells, full-length hACE2 gene was subcloned into an EF-1α promoter-driven mammalian expression vector (which is flanked with PiggyBac transposon inverted repeat sequence), SmBiT (VTGYRLFEEIL from Promega)-Ala-Gly-Ala was used for site-directed insertion between hACE2 amino acid 17th and 18th residues. We used high-fidelity polymerase (CloneAmp HiFi polymerase; Takara Bio) and gene-specific primers (supplementary table S3, Supplementary Material online) to clone the WT human ACE2 gene into pJET1.2 vector (Thermo Scientific), which was then used as a template for mutagenesis. To construct each human ACE2 point mutation, we used high-fidelity polymerase and site-directed mutagenesis primers (supplementary table S3, Supplementary Material online) to amplify the entire plasmid in a polymerase chain reaction (Liu and Naismith 2008), generating a circular, mutant DNA product. The template DNA, carrying the WT allele, was digested with DpnI. All the DNA products were transformed into Escherichia coli DH5α competent cells and incubated overnight. Then, we picked a colony and sequenced it to confirm that it contained the desired mutation and no other mutation. All the mutated human ACE2 fragments were released by digestion with HpaI and KpnI and were cleaned using the Gel extraction kit (QIAGEN). T7 DNA ligase (New England Biolabs) was used to ligate the cleaned ACE2 fragments with the vector (containing SmBiT), which was treated with HpaI and KpnI. The QIAGEN plasmid midi kit was used to prepare plasmids for the attachment and binding affinity assays.
Cell-Based RBD Attachment Assay
RBD attachment assay was established to monitor the binding between recombinant RBD-LgBiT and ACE2 on a cell-based assay platform (manuscript in preparation). In short, 3 × 105 HeLa cells were plated overnight before transient transfection with 1 µg of the ACE2 expression construct. Transfection reagents were removed from culture at 24-h posttransfection and replaced with fresh culture medium. At 48-h posttransfection, transfected cells were removed from culture dish and seeded into a white 96-well plate at a density of 1.5 × 104 cells per well (in triplicate). The residual cells were collected for checking recombinant ACE2 expression by western blotting with rabbit anti-ACE2 (Novus biologicals, clone SN0754). For each attachment assay, cell culture medium was removed and rinsed once with phosphate-buffered saline (PBS). Following the removal of PBS, a 50 µl reaction mixture (containing 250 ng recombinant RBD-LgBiT, 0.5 µl of Nano-Glo luciferase assay substrate, and 9.5 µl of luciferase assay diluent) was added into each well, and luminescence was measured every 2 min and continuously for 1 h. For the competition assay, recombinant FLS (kindly provided by Danny Hsu, Academia Sinica, Taiwan) was included in the reaction mixture. The recombinant RBD-LgBiT protein was kindly provided by SMOBIO Inc., Taiwan.
S1 Binding Assay
To characterize the binding affinity between ACE2 and S-protein, 3 × 105 HeLa cells were preseeded on coverslips and transfected with plasmids containing the WT ACE2 or a variant. At 48-h posttransfection, transfected cells were washed three times with PBS, fixed with 4% paraformaldehyde for 10 min. The cells were then incubated in 100 µl of PBS containing 120 ng of Fc-tagged spike S1 recombinant protein (Cat: 40591-V02H, Sino Biological) for 1 h. The cells were washed three times with PBS and incubated with rabbit anti-ACE2 antibody, then incubated with goat anti-rabbit antibody conjugated with Alexa Fluor 488 (Molecular Probes) and goat anti-human antibody conjugated with Alexa Fluor 594, and counterstained with DAPI. The cells were then visualized on an epifluorescence microscope (Leica DMI2000).
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Supplementary Material
Acknowledgments
We thank John Wang and Soojin Yi for valuable suggestions. We appreciate the R&D team of SMOBIO Inc., Taiwan for supporting the mass production of recombinant protein RBD-LgBiT. This study was supported by Academia Sinica (AS-SUMMIT-109 and AS-KPQ-110-EIMD) and by Ministry of Science and Technology, Taiwan (MOST 107-2311-B-001-016-MY3, 107-2221-E-007-107-MY3, and 109-2327-B-007-002) and National Tsing Hua University (109Q2808E1). Y.Y.T. was supported by National Cancer Institute (RO1CA204962) and National Institute of Diabetes, Digestive and Kidney Diseases (RO1DK105963 and RO1DK76629), National Institutes of Health.
Data Availability
All data generated in this article are shown as tables and figures and as supplementary materials.
References
- The 1000 Genome Project Consortium. 2015. A global reference for human genetic variation. Nature 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA.. 2001. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 98(18):10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström A, McCarthy SA, Hui R, Almarri MA, Ayub Q, Danecek P, Chen Y, Felkel S, Hallast P, Kamm J, et al. 2020. Insights into human genetic variation and population history from 929 diverse genomes. Science 367(6484):eaay5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL.. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10(1):421–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li L, Feng Z, Wan S, Huang P, Sun X, Wen F, Huang X, Ning G, Wang W.. 2020. Comparative genetic analysis of the novel coronavirus (2019-nCoV/SARS-CoV-2) receptor ACE2 in different populations. Cell Discov. 6(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Li L, Xu M, Feng Z, Sun X, Lu J, Xu Y, Du P, Wang T, Hu R.. 2020. The ChinaMAP analytics of deep whole genome sequences in 10,588 individuals. Cell Res. 30:717–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KK, Dorosky D, Sharma P, Abbasi SA, Dye JM, Kranz DM, Herbert AS, Procko E.. 2020. Engineering human ACE2 to optimize binding to the spike protein of SARS coronavirus 2. Science 369(6508):1261–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu L, Wei Q, Zhu H, Jiang H, Tu X, Qin C, Chen Z.. 2008. Rhesus angiotensin converting enzyme 2 supports entry of severe acute respiratory syndrome coronavirus in Chinese macaques. Virology 381(1):89–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Liu Q, Guo D.. 2020. Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol. 92(4):418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damas J, Hughes GM, Keough KC, Painter CA, Persky NS, Corbo M, Hiller M, Koepfli K-P, Pfenning AR, Zhao H, et al. 2020. Broad host range of SARS-CoV-2 predicted by comparative and structural analysis of ACE2 in vertebrates. Proc Natl Acad Sci U S A. 117:22311–22322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey FE, Murray MF, Overton JD, Habegger L, Leader JB, Fetterolf SN, O’Dushlaine C, Van Hout CV, Staples J, Gonzaga-Jauregui C, et al. 2016. Distribution and clinical impact of functional variants in 50,726 whole-exome sequences from the DiscovEHR study. Science 354(6319):aaf6814. [DOI] [PubMed] [Google Scholar]
- Dixon AS, Schwinn MK, Hall MP, Zimmerman K, Otto P, Lubben TH, Butler BL, Binkowski BF, Machleidt T, Kirkland TA, et al. 2016. NanoLuc complementation reporter optimized for accurate measurement of protein interactions in cells. ACS Chem Biol. 11(2):400–408. [DOI] [PubMed] [Google Scholar]
- Dolinsky TJ, Czodrowski P, Li H, Nielsen JE, Jensen JH, Klebe G, Baker NA.. 2007. PDB2PQR: expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35(Web Server Issue):W522–W525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelsbrunner H, Facello M, Fu P, Liang J.. 1995. Measuring proteins and voids in proteins. Proceedings of the 28th Hawaii International Conference System Sciences. Vol. 5. Wailea (HI): IEEE. p. 256–264. [Google Scholar]
- Edelsbrunner H, Facello M, Liang J.. 1998. On the definition and the construction of pockets in macromolecules. Discrete Appl Math. 88(1–3):83–102. [PubMed] [Google Scholar]
- Edelsbrunner H, Mucke E.. 1994. Three-dimensional alpha shapes. ACM Trans Graph. 13(1):43–72. [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu W, O’Connor TD, Jun G, Kang HM, Abecasis G, Leal SM, Gabriel S, Rieder MJ, Altshuler D, Shendure J, et al. 2013. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 493(7431):216–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung SY, Yuen KS, Ye ZW, Chan CP, Jin DY.. 2020. A tug-of-war between severe acute respiratory syndrome coronavirus 2 and host antiviral defence: lessons from other pathogenic viruses. Emerg Microbes Infect. 9(1):558–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Zheng W, Pearce R, Zhang Y.. 2020. SSIPe: accurately estimating protein–protein binding affinity change upon mutations using evolutionary profiles in combination with an optimized physical energy function. Bioinformatics 36(8):2429–2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutter G, Nowak D, Mossner M, Ganepola S, Mussig A, Allers K, Schneider T, Hofmann J, Kucherer C, Blau O, et al. 2009. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 360(7):692–698. [DOI] [PubMed] [Google Scholar]
- Jeon S, Bhak Y, Choi Y, Jeon Y, Kim S, Jang J, Jang J, Blazyte A, Kim C, Kim Y, et al. 2020. Korean Genome Project: 1094 Korean personal genomes with clinical information. Sci Adv. 6(22):eaaz7835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karczewski KJ, Francioli LC, Tiao G, Cummings BB, Alföldi J, Wang Q, Collins RL, Laricchia KM, Ganna A, Birnbaum DP, et al. 2020. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 581(7809):434–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski MH, Qian H, Hou Z, Rosen JD, Tapia AL, Shan Y, Jain D, Argos M, Arnett DK, Avery C, et al. 2019. Use of >100,000 NHLBI Trans-Omics for Precision Medicine (TOPMed) Consortium whole genome sequences improves imputation quality and detection of rare variant associations in admixed African and Hispanic/Latino populations. PLoS Genet. 15(12):e1008500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Suleski M, Hedges SB.. 2017. TimeTree: a Resource for Timelines, Timetrees, and Divergence Times. Mol Biol Evol. 34(7):1812–1819. [DOI] [PubMed] [Google Scholar]
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, et al. 2020. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature 581(7807):215–220. [DOI] [PubMed] [Google Scholar]
- Lee N, Hui D, Wu A, Chan P, Cameron P, Joynt GM, Ahuja A, Yung MY, Leung CB, To KF, et al. 2003. A major outbreak of severe acute respiratory syndrome in Hong Kong. N Engl J Med. 348(20):1986–1994. [DOI] [PubMed] [Google Scholar]
- Liang J, Edelsbrunner H, Fu P, Sudhakar PV, Subramaniam S.. 1998. Analytical shape computation of macromolecules: I. Molecular area and volume through alpha shape. Proteins 33(1):1–17. [PubMed] [Google Scholar]
- Liang J, Edelsbrunner H, Woodward C.. 1998. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 7(9):1884–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Naismith JH.. 2008. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall M. 2020. The lasting misery of coronavirus long-haulers. Nature 585(7825):339–341. [DOI] [PubMed] [Google Scholar]
- McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, Flicek P, Cunningham F.. 2016. The Ensembl variant effect predictor. Genome Biol. 17(1):122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melin AD, Janiak MC, Marrone F, Arora PS, Higham JP.. 2020. Comparative ACE2 variation and primate COVID-19 risk. Commun Biol. 3(1):641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo F, Sanchez R, Sali A.. 2002. Statistical potentials for fold assessment. Protein Sci. 11(2):430–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris JSM, Lai ST, Poon LLM, Guan Y, Yam LYC, Lim W, Nicholls J, Yee WKS, Yan WW, Cheung MT, et al. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361(9366):1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plante JA, Liu Y, Liu J, Xia H, Johnson BA, Lokugamage KG, Zhang X, Muruato AE, Zou J, Fontes-Garfias CR, et al. 2020. Spike mutation D614G alters SARS-CoV-2 fitness and neutralization susceptibility. bioRxiv. doi:10.1101/2020.09.01.278689. [DOI] [PMC free article] [PubMed]
- R Core Team. 2020. R: a language and environment for statistical computing. Vienna (Austria: ): R Foundation for Statistical Computing. Available from: https://www.R-project.org/. Accessed January 10, 2021. [Google Scholar]
- Rajput B, Pruitt KD, Murphy TD.. 2019. RefSeq curation and annotation of stop codon recoding in vertebrates. Nucleic Acids Res. 47(2):594–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A, Holmes EC, O’Toole A, Hill V, McCrone JT, Ruis C, du Plessis L, Pybus OG.. 2020. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nat Microbiol. 5(11):1403–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabino EC, Buss LF, Carvalho MPS, Prete CA J, Crispim MAE, Fraiji NA, Pereira RHM, Parag KV, da Silva Peixoto P, Kraemer MUG, et al. 2021. Resurgence of COVID-19 in Manaus, Brazil, despite high seroprevalence. Lancet. 397(10273):452–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson M, Libert F, Doranz BJ, Rucker J, Liesnard C, Farber CM, Saragosti S, Lapoumeroulie C, Cognaux J, Forceille C, et al. 1996. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature 382(6593):722–725. [DOI] [PubMed] [Google Scholar]
- Sayers EW, Cavanaugh M, Clark K, Ostell J, Pruitt KD, Karsch-Mizrachi I.. 2020. GenBank. Nucleic Acids Res. 48(D1):D84–D86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F.. 2020. Structural basis of receptor recognition by SARS-CoV-2. Nature 581(7807):221–224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward M-H, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K.. 2001. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 29(1):308–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal T. 2020. A review of coronavirus disease-2019 (COVID-19). Indian J Pediatr. 87(4):281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solloch UV, Lang K, Lange V, Bohme I, Schmidt AH, Sauter J.. 2017. Frequencies of gene variant CCR5-Delta32 in 87 countries based on next-generation sequencing of 1.3 million individuals sampled from 3 national DKMS donor centers. Hum Immunol. 78(11–12):710–717. [DOI] [PubMed] [Google Scholar]
- Tadaka S, Katsuoka F, Ueki M, Kojima K, Makino S, Saito S, Otsuki A, Gocho C, Sakurai-Yageta M, Danjoh I, et al. 2019. 3.5 KJPNv2: an allele frequency panel of 3552 Japanese individuals including the X chromosome. Hum Genome Var. 6(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavaré S. 1986. Some probabilistic and statistical problems in the analysis of DNA sequences. Lect Math Life Sci. 17:57–86. [Google Scholar]
- Tegally H, Wilkinson E, Giovanetti M, Iranzadeh A, Fonseca V, Giandhari J, Doolabh D, Pillay S, San EJ, Msomi N.. 2020. Emergence and rapid spread of a new severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) lineage with multiple spike mutations in South Africa. medRxiv.
- Tillett RL, Sevinsky JR, Hartley PD, Kerwin H, Crawford N, Gorzalski A, Laverdure C, Verma SC, Rossetto CC, Jackson D, et al. 2020. Genomic evidence for reinfection with SARS-CoV-2: a case study. Lancet Infect Dis. 21:P52–P58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Dundas J, Liang J.. 2009. Predicting protein function and binding profile via matching of local evolutionary and geometric surface patterns. J Mol Biol. 387(2):451–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Dupree C, Chen ZJ, Li WH.. 2009. SplitPocket: identification of protein functional surfaces and characterization of their spatial patterns. Nucleic Acids Res. 37(Web Server Issue):W384–W389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng YY, Li WH.. 2009. Identification of protein functional surfaces by the concept of a split pocket. Proteins 76(4):959–976. [DOI] [PubMed] [Google Scholar]
- Tseng YY, Li WH.. 2011. Evolutionary approach to predicting the binding site residues of a protein from its primary sequence. Proc Natl Acad Sci U S A. 108(13):5313–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The UK10K Consortium. 2015. The UK10K project identifies rare variants in health and disease. Nature 526:82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volz E, Hill V, McCrone JT, Price A, Jorgensen D, O’Toole A, Southgate J, Johnson R, Jackson B, Nascimento FF, et al. 2021. Evaluating the effects of SARS-CoV-2 spike mutation D614G on transmissibility and pathogenicity. Cell 184(1):64–75.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walls AC, Park Y-J, Tortorici MA, Wall A, McGuire AT, Veesler D.. 2020. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 181(2):281–292.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Zhang Y, Wu L, Niu S, Song C, Zhang Z, Lu G, Qiao C, Hu Y, Yuen K-Y, et al. 2020. Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181(4):894–904.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS.. 2020. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science 367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q.. 2020. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 367(6485):1444–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates AD, Achuthan P, Akanni W, Allen J, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Azov AG, Bennett R, et al. 2020. Ensembl 2020. Nucleic Acids Res. 48(D1):D682–D688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Kang Z, Gong H, Xu D, Wang J, Li Z, Cui X, Xiao J, Meng T, Zhou W, et al. 2020. The digestive system is a potential route of 2019-nCov infection: a bioinformatics analysis based on single-cell transcriptomes. bioRxiv. doi:10.1101/2020.01.30.927806.
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, et al. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this article are shown as tables and figures and as supplementary materials.