Abstract
One of the central goals in molecular evolutionary biology is to determine the sources of variation in the rate of sequence evolution among proteins. Gene expression level is widely accepted as the primary determinant of protein evolutionary rate, because it scales with the extent of selective constraints imposed on a protein, leading to the well-known negative correlation between expression level and protein evolutionary rate (the E–R anticorrelation). Selective constraints have been hypothesized to entail the maintenance of protein function, the avoidance of cytotoxicity caused by protein misfolding or nonspecific protein–protein interactions, or both. However, empirical tests evaluating the relative importance of these hypotheses remain scarce, likely due to the nontrivial difficulties in distinguishing the effect of a deleterious mutation on a protein’s function versus its cytotoxicity. We realized that examining the sequence evolution of viral proteins could overcome this hurdle. It is because purifying selection against mutations in a viral protein that result in cytotoxicity per se is likely relaxed, whereas purifying selection against mutations that impair viral protein function persists. Multiple analyses of SARS-CoV-2 and nine other virus species revealed a complete absence of any E–R anticorrelation. As a control, the E–R anticorrelation does exist in human endogenous retroviruses where purifying selection against cytotoxicity is present. Taken together, these observations do not support the maintenance of protein function as the main constraint on protein sequence evolution in cellular organisms.
Keywords: protein evolutionary rate, avoidance of cytotoxicity, maintenance of protein function, the E–R anticorrelation, gene expression level, protein homeostasis
Significance
Understanding variation in the rate of sequence evolution among proteins encoded by the same genome has always been one of the central goals in molecular evolutionary biology. Two evolutionary mechanisms have been proposed to explain the widely observed negative correlation between the expression level and the protein evolutionary rate (the E–R anticorrelation) in cellular organisms: 1) the maintenance of protein function and/or 2) the avoidance of cytotoxicity. However, empirical tests evaluating the relative importance of these mechanisms on the rate of protein sequence evolution remain scarce. We estimated the E–R correlation in ten virus species and observed a complete absence of any E–R anticorrelation. The observation does not support function maintenance as the primary selective constraint on protein sequence evolution.
Introduction
The rate of protein sequence evolution varies by orders of magnitude among proteins encoded by a single genome (Zuckerkandl and Pauling 1965; Wilson et al. 1977; Li et al. 1985; Koonin and Wolf 2010), and understanding sources of such variation is a central goal in the molecular evolution research field (Kimura 1968; Wilson et al. 1977; Pal et al. 2006; Rocha 2006; Zhang and Yang 2015; Echave et al. 2016). The protein evolutionary rate is determined primarily by the level of the selective constraint—the factor(s) reducing the extent of divergence for a protein sequence relative to a neutral evolutionary process, owing to the operation of purifying selection. The selective constraint has been found to scale with a given protein’s gene expression level: The occurrence of a deleterious mutation in a more highly expressed gene will result in a larger number of improperly formed proteins. As a consequence, the expression level (E) is negatively correlated with protein evolutionary rate (R) in cellular organisms of all three domains of life (Pal et al. 2001; Zhang and Yang 2015). This phenomenon is known as the E–R anticorrelation.
Two mutually nonexclusive explanations have been proposed for the ubiquitous E–R anticorrelation reported in cellular organisms (Pal et al. 2001; Zhang and Yang 2015). First, the function maintenance hypothesis explained the E–R anticorrelation based on the idea that the observed optimal expression level of a protein represents a stable equilibrium between the benefit of protein function and the biochemical cost of protein expression (Cherry 2010; Gout et al. 2010). Therefore, around the optimal expression level, the functional benefit of having one more protein molecule can be approximated by the biochemical cost of expressing this molecule. Similarly, the functional decrease induced by a slightly deleterious mutation can be approximated by the loss of the cellular resources synthesizing nonfunctional protein molecules (or more accurately, the functionally impaired protein components), which is proportional to both the effect size of the deleterious mutation and the expression level of the gene (Cherry 2010; Gout et al. 2010). Although the function maintenance hypothesis in theory can explain the E–R anticorrelation, it has not been extensively tested using empirical data (Zhang and Yang 2015).
A second explanation for the E–R anticorrelation is the cytotoxicity avoidance hypothesis. Here, the term “cytotoxicity” is used in its broad sense: the negative consequence caused by the misfolding of a protein (Drummond et al. 2005; Drummond and Wilke 2008; Vavouri et al. 2009; Yang et al. 2010) or by its nonspecific interaction (i.e., misinteraction) with other proteins in the cell (Zhang et al. 2008; Vavouri et al. 2009; Levy et al. 2012; Yang et al. 2012). Such cytotoxicity also scales with the gene expression level as a deleterious mutation will render more misfolded or misinteracted proteins if it occurs in a more highly expressed gene. Thus, the term selective constraint—in addition to the intuitive meaning of “maintaining the function of a protein”—has been hypothesized to entail the selective pressure for avoiding cytotoxicity (Drummond et al. 2005). Although the role of cytotoxicity avoidance in protein sequence evolution has been supported by results from many studies (reviewed in Zhang and Yang 2015), there has been considerable debate over its validity in recent years (Plata and Vitkup 2018; Razban 2019; Biesiadecka et al. 2020; Usmanova et al. 2021).
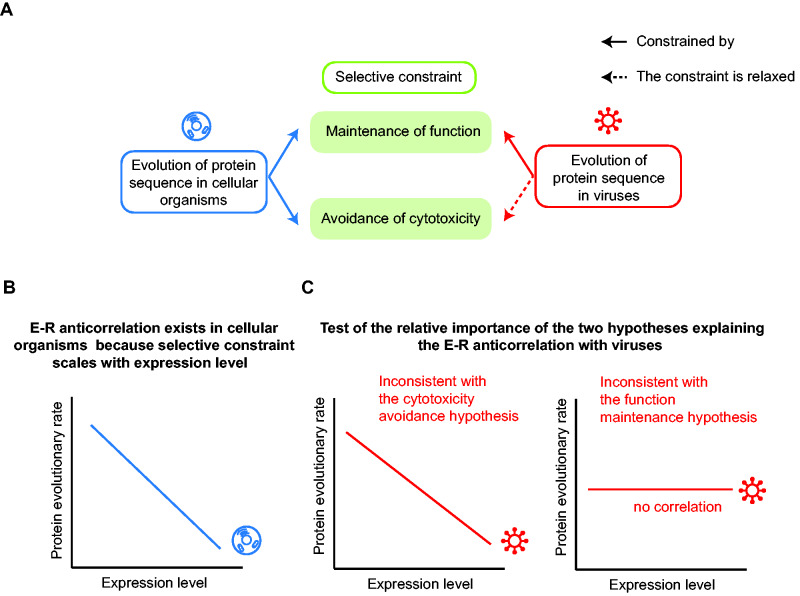
In principle, both the function maintenance hypothesis and the cytotoxicity avoidance hypothesis can be used to explain the E–R anticorrelation observed in cellular organisms (fig. 1A, left). However, a rigorous evaluation of the relative importance of these hypotheses using empirical data remains absent, likely because it is highly complicated and challenging to experimentally decipher, precisely, whether a deleterious mutation disrupts the function of a protein, induces cytotoxicity, or both.
Fig. 1.
The virus is an excellent system to distinguish two hypotheses explaining the E–R anticorrelation in cellular organisms. (A) Selective constraints imposed on the evolution of a protein sequence are hypothesized to entail the maintenance of protein function, the avoidance of cytotoxicity caused by protein misfolding and misinteraction, or both. Protein sequence evolution of cellular organisms may be constrained by both mechanisms, but the selection against cytotoxicity is relaxed for viral proteins (red dashed arrow). (B) The E–R anticorrelation has been observed in all three domains of cellular organisms. (C) Viruses can be used to test the relative importance of the two hypotheses explaining the E–R anticorrelation. If the E–R anticorrelation remains strong among viral ORFs, the cytotoxicity avoidance hypothesis is not supported (left), because the avoidance of cytotoxicity appears to be unnecessary for creating an E–R anticorrelation. In contrast, if the E–R anticorrelation is missing in viruses, the function maintenance hypothesis is not supported (right), because the maintenance of protein function per se is insufficient to create an E–R anticorrelation.
We speculated that viruses might provide an excellent opportunity for evaluating the relative importance of these two hypotheses (fig. 1A, right). On the one hand, viruses do not have cell structures where cytotoxicity can play a direct role. Even if viral proteins cause some host cell cytotoxicity, such cytotoxicity may not significantly reduce the proliferation rate of the virus, because it is likely masked (at least to some extent) by the rapid cytopathogenic damage that the regular viral infect-and-replicate life cycle causes to the host cells. Consider that viruses hijack the machinery of host cells, assemble new viruses, then burst out from and kill host cells, doing so within a rapid time frame (El-Sayed et al. 2016; Bojkova et al. 2020). In sharp contrast, consider that cytotoxicity plays a role mainly in a chronic manner because it may take up to years for misfolded proteins to accumulate and aggregate to an effective level (Gáspári and Perczel 2010; Bergh et al. 2015). Consistently, protein aggregation-associated diseases often occur in the long-lived cells, such as neurons (Chiti and Dobson 2017), in elder animals.
On the other hand, it is conceivable that slightly deleterious mutations that impair viral protein functions should remain being selected against, because the impaired infect-and-replicate functions should reduce the proliferation efficiency of viruses. Furthermore, the extent of such selective constraint is still proportional to the viral gene expression level: The equilibrium between the functional benefit and the biochemical cost of protein expression should remain effective in determining the optimal expression level of viral proteins. The reason is explained as follows. Although viruses do not have their own protein synthesis machinery, they often shut off transcription and translation of the host cell genes (Walsh and Mohr 2011) and regard the current protein synthesis resources of the infected host cells as their own to express viral proteins. Consequently, viruses incur a fitness cost for the production of viral proteins of impaired functions, as such resources could have been used to synthesize fully functional viral proteins.
Taken together, we reasoned that a mutation in the viral genome would likely evolve under relaxed purifying selection or even neutrally if it merely causes cytotoxicity to the host cell; in contrast, the evolution of viral genes remains constrained by the maintenance of protein function. Therefore, it should be informative to determine whether the E–R anticorrelation exists among viral proteins. The cytotoxicity avoidance hypothesis is not supported if the E–R anticorrelation persists in viruses, and the function maintenance hypothesis is not supported if the E–R anticorrelation is missing in viruses (fig. 1B and C).
Intriguingly, a previous study reported that the E–R anticorrelation exists among ten Mononegavirales virus species (Pagán et al. 2012) although the aim of the study was not to use viruses as a system to determine the molecular mechanism(s) of selective constraints. The authors ultimately reported the apparent existence of the E–R anticorrelation in these viruses only after they excluded ten “outlier” open reading frames (ORF) from a total of 82 ORFs in their analysis. We were deeply interested in these findings, and we suspect that excluding these “outlier” ORFs which were not specified a priori may have masked the real situation. We also suspect that another limitation of their study was the relatively low number of sequenced variants for the virus species they examined (the median number was 18 for a virus species), which would have limited the accuracy in the estimation of the protein evolutionary rate.
Given that the current SARS-CoV-2 pandemic is occurring in the world nearly ubiquitously equipped with the capacity for high-throughput sequencing, the thousands of confirmed variants for this virus provide us with the opportunity to rigorously evaluate the relative explanatory power of the function maintenance hypothesis and the cytotoxicity avoidance hypothesis. SARS-CoV-2 is a positive-sense single-stranded RNA virus, which replicates using an RNA-dependent RNA polymerase and does not integrate into the genome of its host’s cells (Wang et al. 2020; Wu et al. 2020; Zhou et al. 2020). Note that SARS-CoV-2 often lyses host cells within only dozens of hours after infecting them (Bojkova et al. 2020). We evaluated the presence of E–R anticorrelation in SARS-CoV-2 and in nine other virus species. Fundamentally, our finding that these viral proteins do not exhibit any E–R anticorrelation does not support the role of function maintenance as the main determining factor in protein sequence evolution.
Results
No E–R Anticorrelation Observed in SARS-CoV-2
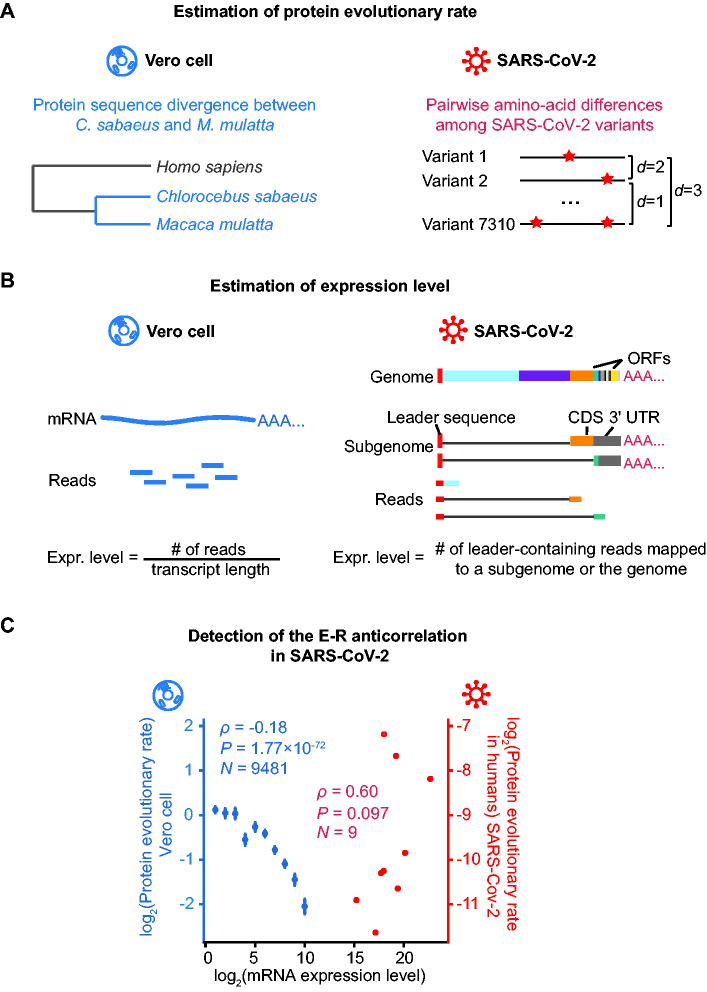
To determine the E–R correlation coefficient in the SARS-CoV-2 genome, we estimated the protein evolutionary rate for each of the nine ORFs from the reported sequences of 7,310 SARS-CoV-2 variants isolated from patients. Specifically, the protein evolutionary rate for an ORF was defined as the average pairwise amino acid differences among variants, normalized by the ORF length (fig. 2A and table 1). To determine whether these rates are correlated with mRNA expression levels, we further estimated the respective expression levels of these ORFs from high-throughput RNA sequencing data (Kim et al. 2020) for Vero cells infected by SARS-CoV-2 (fig. 2B and table 1).
Fig. 2.
Detection of the E–R anticorrelation in SARS-CoV-2. (A) A schematic shows how the protein evolutionary rate is estimated for Vero cell and SARS-CoV-2. The red stars on the right panel denote amino acid alternations. (B) A schematic shows how expression level is estimated for Vero cell and SARS-CoV-2. On the left penal, the expression level for each Vero cell gene is estimated as the normalized reads abundance aligned to it. On the right penal, the genome and two out of the eight subgenomes of SARS-CoV-2 are shown as examples. The gray lines denote the regions skipped by the leader-to-body fusion during the discontinuous transcription in coronaviruses. Note that the coding sequence (CDS) of a subgenome could be the 3′-UTR of another subgenome. Therefore, the expression level for each ORF in SARS-CoV-2 was estimated as the number of lead-containing reads of the corresponding subgenome. (C) The E–R correlation in the Vero cell (in blue) and in SARS-CoV-2 (in red). For Vero cells, expressed protein-coding genes are split into ten bins according to their expression levels: (−∞, 0.5), [0.5, 1.5), [1.5, 2.5), [2.5, 3.5), [3.5, 4.5), [4.5, 5.5), [5.5, 6.5), [6.5, 7.5), [7.5, 8.5), [8.5, 9.5), [9.5, +∞). The mean and standard errors of evolutionary rates are shown for each bin. Spearman’s correlation coefficients were calculated from the unbinned data.
Table 1.
The Protein Evolutionary Rates and the Expression Levels of SARS-CoV-2 ORFs.
| ORF | Protein Evolutionary Rate (R) |
Expression Level (E) |
|||
|---|---|---|---|---|---|
| In Humansa | In Animal Hostsb | Vs. SARS-CoVc | mRNA Level (×104)d | Protein Levele | |
| ORF1ab | 0.43 | 0.031 | 0.049 | 25.38 | 1.09 |
| S | 0.54 | 0.043 | 0.023 | 114.42 | 7.54 |
| ORF3a | 2.45 | 0.081 | 0.172 | 60.38 | 3.13 |
| E | 0.16 | 1.0 × 10−4 | 0.179 | 14.44 | 0.04f |
| M | 0.40 | 0.013 | 0.081 | 21.25 | 5.00 |
| ORF6 | 0.26 | 0.057 | 0.097 | 3.80 | 0.25 |
| ORF7a | 0.31 | 0.091 | 0.109 | 68.35 | 0.92 |
| ORF8 | 3.44 | 0.052 | 0.004 | 26.22 | 1.57 |
| N | 1.72 | 0.097 | 0.142 | 662.10 | 35.49 |
The average pairwise amino acid differences per 1,000 amino acids among 7,310 SARS-CoV-2 variants.
The average dN/dS ratio in the phylogenetic tree of SARS-CoV-2 and three related viruses, RaTG13 isolated from bats in Yunnan, GX-P5E from pangolins in Guangxi, and GD-1 from pangolins in Guangdong.
The dN/dS ratio estimated between SARS-CoV-2 and SARS-CoV.
The number of leader-containing reads mapped to the SARS-CoV-2 genomic RNA (ORF1ab) or a subgenomic RNA (ORFs other than ORF1ab).
Protein levels were calculated as the number of PSMs (retrieved from Davidson et al. [2020]) of an ORF, normalized by the number of its theoretical peptides.
PSM was not detected for E protein and the number was arbitrarily assigned to 0.5, which is half of the minimum nonzero value.
We detected no E–R anticorrelation in SARS-CoV-2 (ρ = 0.60, P = 0.097, N = 9, Spearman’s correlation; fig. 2C). As a control, we estimated whether E–R anticorrelation exists in Vero cells; these cells were originally isolated from the kidneys of green monkey Chlorocebus sabaeus and are widely used to culture viruses (Rhim et al. 1969). Specifically, we estimated the protein sequence divergence using a comparison between green monkey (C. sabaeus) and its close relative, macaque (Macaca mulatta, fig. 2A), and correlated the protein sequence divergence values with the mRNA levels of nuclear genes for Vero cells from the same RNA sequencing data set (fig. 2B). We found that expression level and evolutionary rate were negatively correlated in Vero cells (ρ = −0.18, P = 1.77 × 10−72, N = 9,481, Spearman’s correlation; fig. 2C), confirming as expected that these host cells do exhibit the E–R anticorrelation.
Presumably, the absence of the E–R anticorrelation in SARS-CoV-2 could be caused by the narrower range in which the ORFs of SARS-CoV-2 are expressed (174 times between the most highly and the most lowly expressed ORFs, N and ORF6, respectively, table 1). To test whether this relatively narrow range of gene expression has disabled the detection of the E–R anticorrelation in SARS-CoV-2, we restricted the detection of the E–R correlation coefficient in the Vero cell to a subset of endogenous genes. The expression levels of the top 20% endogenous genes in the Vero Cell are within a range of 150 times, which is smaller than 174 times as in SARS-CoV-2. Among these genes, expression level remained negatively correlated with evolutionary rate (ρ = −0.08, P = 4.4 × 10−4, N = 1,915, Spearman’s correlation; supplementary fig. S1, Supplementary Material online), suggesting that the expression range of 174 times in SARS-CoV-2 does provide sufficient statistical power for the detection of the E–R anticorrelation.
Protein Synthesis Dynamics Do Not account for the Absence of the E–R Anticorrelation in SARS-CoV-2
The lack of E–R anticorrelation in SARS-CoV-2 suggests that function maintenance unlikely plays the primary role in determining the protein evolutionary rate for cellular genes. However, there are some other possible explanations for our finding that SARS-CoV-2 does not exhibit the E–R anticorrelation. One possible explanation is the potential impact of specific SARS-CoV-2 protein synthesis dynamics (Brierley et al. 1989; Wu et al. 2020). That is, the translation of ORF1ab is known to be subject to a programmed ribosomal frameshift between ORF1a and ORF1b, and the encoded polypeptides are known to be subject to further proteolysis in host cells; this ORF thus encodes for a total of 11 or 15 nonstructural proteins (nsps, fig. 3A). As a consequence, although these nsps have the same mRNA abundance, they can vary in protein abundance, owing for example to the programmed ribosomal frameshift, as well as potential variation in translation elongation rate (resulted from codon usage bias or other cis-regulatory elements) (Chen et al. 2020; Zhao et al. 2020, 2021) and protein stability (Chen et al. 2019).
Fig. 3.
The E–R anticorrelation remains absent in SARS-CoV-2 when nsps are considered individually. (A) A schematic shows nsps derived from the proteolysis of pp1ab, which is encoded by ORF1ab. The black arrows denote the cleavage sites. (B) The absence of the E–R anticorrelation among 23 proteins encoded by the SARS-CoV-2 genome. (C) Similar to (B), but three proteins (S, E, and M) known to be packaged for secretion at the endoplasmic reticulum are excluded.
To evaluate this possibility, we tested whether the E–R anticorrelation exists for the 23 proteins encoded by the SARS-CoV-2 genome (15 nsps by ORF1ab plus eight proteins by the other eight ORFs). Specifically, we evaluated the expression level for each protein based on mass spectrometry data for SARS-CoV-2-infected Vero cells (Davidson et al. 2020). As our analysis based on RNA expression data, this protein data-based analysis found no evidence for the E–R anticorrelation (ρ = 0.36, P = 0.09, N = 23, Spearman’s correlation; fig. 3B and supplementary table S1, Supplementary Material online), suggesting that the use of RNA expression data in our analysis does not account for the undetected E–R anticorrelation in SARS-CoV-2.
Three SARS-CoV-2 proteins (S, E, and M) are known to be packaged for secretion at the endoplasmic reticulum. Secreted proteins—especially those that undergo N-linked glycosylation—and membrane proteins have been postulated to be under relatively weak selective constraints for avoiding cytotoxicity; this thinking is based on the stringent quality control mechanisms of the endoplasmic reticulum or on the impacts of their subcellular locations. Indeed, there have been reports indicating that some of these proteins exhibit a weaker E–R anticorrelation (Feyertag et al. 2017) or even a positive E–R correlation (Feyertag et al. 2019). To control for impacts specific to confounding factors from protein secretion, we performed an additional analysis that excluded proteins S, E, and M from the 23 proteins encoded by the SARS-CoV-2 genome. The E–R anticorrelation remained absent (ρ = 0.28, P = 0.24, N = 20, Spearman’s correlation; fig. 3C), suggesting that these three proteins do not account for the undetected E–R anticorrelation in SARS-CoV-2.
Host-Jumping Does Not Account for the Absence of the E–R Anticorrelation in SARS-CoV-2
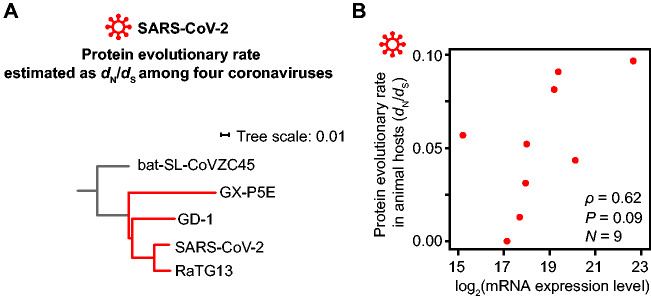
Another possible explanation for our finding that SARS-CoV-2 does not exhibit the E–R anticorrelation is the potential impact from its host-jumping into humans (Zhou et al. 2020). That is, the evolutionary rate estimated from the sequencing data for SARS-CoV-2 variants in human hosts may not genuinely reflect its long-term evolutionary rate (Longdon et al. 2014), due to for example relaxed purifying selection in a subset of viral ORFs in human hosts. Seeking to exclude this possibility, we estimated the protein evolutionary rates in a phylogenetic tree of SARS-CoV-2 and three related coronaviruses that have not achieved zoonotic transfer into humans (RaTG13 isolated from bats and GX-P5E and GD-1 from pangolin samples; fig. 4A).
Fig. 4.
The E–R anticorrelation remains absent in SARS-CoV-2 when the protein evolutionary rates in animal hosts are used. (A) The phylogenetic tree (red branches) from which the protein evolutionary rate (dN/dS) of individual SARS-CoV-2 ORFs was estimated. The scale bar represents 0.01 substitutions per nucleotide. (B) The absence of an anticorrelation between expression level, estimated by the number of leader-containing reads, and the protein evolutionary rate, estimated as the dN/dS ratio among four coronaviruses shown in (A).
For each ORF, we calculated the number of nonsynonymous substitutions per nonsynonymous site (dN) and the number of synonymous substitutions per synonymous site (dS). We used their ratio (dN/dS) to infer the protein evolutionary rate in an effort to control for the genomic variation on mutation rates (table 1). As in our analysis based on virus sequence data for human hosts, in this animal host-based analysis we found no evidence for the E–R anticorrelation among the nine SARS-CoV-2 ORFs (ρ = 0.62, P = 0.09, N = 9, Spearman’s correlation; fig. 4B). It is worth noting that the saturation of synonymous changes could have led to an inaccurate estimation of dS, and consequently, the dN/dS. Nevertheless, we also detected no E–R anticorrelation in SARS-CoV-2 when dN was used as the protein evolutionary rate (supplementary fig. S2A, Supplementary Material online). All these observations suggest that the estimation of protein evolutionary rate using variants collected in human hosts does not account for the undetected E–R anticorrelation in SARS-CoV-2.
No E–R Anticorrelation Observed in Nine Other Virus Species
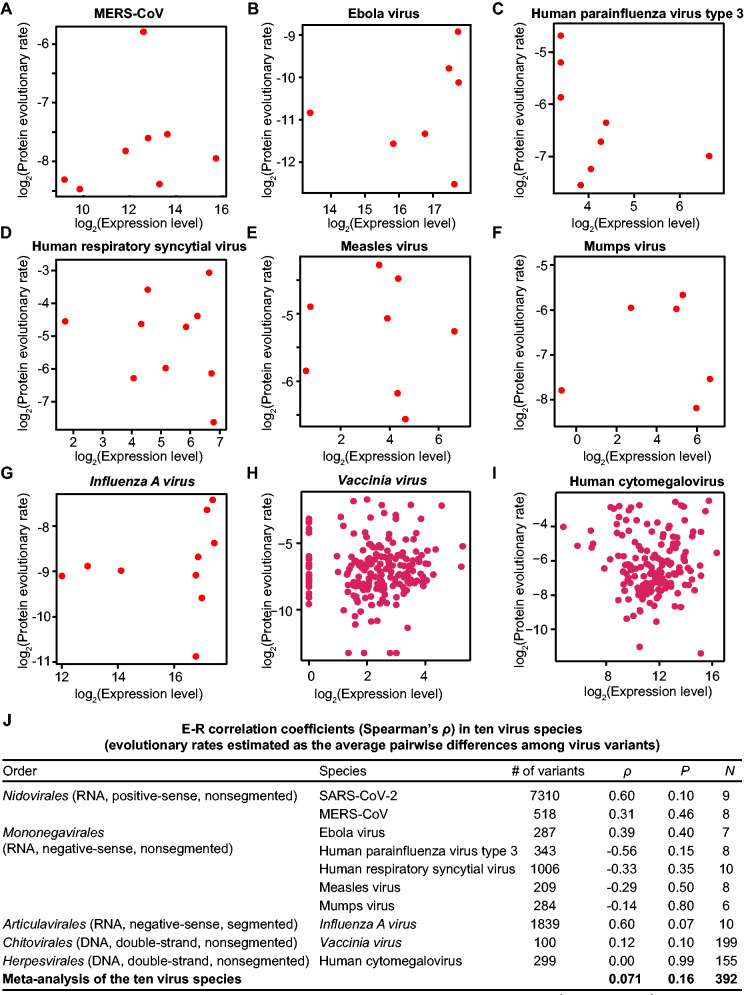
So far we have shown that in SARS-CoV-2, a positive-sense nonsegmented virus, no E–R anticorrelation was detected, contradicting the prediction of the function maintenance hypothesis. To test whether the E–R anticorrelation exists in virus species of other types, we assessed E–R anticorrelation for nine other virus species across five virus orders, including MERS-CoV (Middle East respiratory syndrome related coronavirus), Ebola virus (Zaire ebolavirus), human parainfluenza virus type 3 (Human respirovirus 3), human respiratory syncytial virus (Human orthopneumovirus), Influenza A virus subtype H1N1, measles virus (Measles morbillivirus), mumps virus (Mumps orthorubulavirus), Vaccinia virus, and human cytomegalovirus (Human betaherpesvirus 5). Vaccinia virus and human cytomegalovirus are double-stranded DNA viruses, MERS-CoV is a positive-sense single-stranded RNA virus, and the other six are negative-sense single-stranded RNA viruses. Also note that Influenza A virus is segmented whereas the others (including SARS-CoV-2) are nonsegmented (https://viralzone.expasy.org/, last accessed March 6, 2021). There have been hundreds of variants isolated for each of these virus species (Hatcher et al. 2017; Shu and McCauley 2017).
For each virus species, we estimated the evolutionary rate as the average pairwise sequence difference among isolated variants for each ORF (normalized by the ORF length). Gene expression levels were estimated from the RNA sequencing or genome tiling array data of cell cultures infected by the corresponding viruses (Assarsson et al. 2008; Cheng et al. 2017; Albarino et al. 2018; Blanco-Melo et al. 2020; Zhang et al. 2020) or were retrieved from studies that estimated mRNA levels based on the band densities from electrophoresis gels (Cattaneo et al. 1987; De et al. 1990; Barik 1992; Takeuchi et al. 1993). We did not detect the E–R anticorrelation for any of these nine virus species (Spearman’s correlation; fig. 5A–I).
Fig. 5.
The absence of E–R anticorrelation in nine other virus species. (A–I) The E–R correlation in individual virus species. The number of variants used for the estimation of protein evolutionary rate for each virus species is shown in (J). For Vaccinia virus (H), a constant number (one) has been added to all expression data prior to applying the logarithm transform, to avoid the logarithm of zero. (J) A meta-analysis of E–R correlation for ten virus species. Spearman’s correlation coefficient for individual viruses is shown. The combined correlation coefficient is given by meta-analysis of correlation coefficients (metacor function in the “meta” package of R, which combines correlation coefficients from ten individual viruses into one pooled correlation estimate). Here, the random-effects model with Sidik–Jonkman estimator is used for the estimation of between-study heterogeneity τ2, and z-transformed correlations are used for the meta-analysis. The results of the fixed-effect model is presented because the test of heterogeneity gave a P value = 0.16 (I2 = 21.5%). The effective sample size N is also shown for the meta-analysis.
Seeking to identify whether a common trend exists between the expression level and evolutionary rate of these viruses, we tested whether the E–R correlation coefficients of the ten viruses (including SARS-CoV-2) were with the same sign. Six positive and four negative correlation coefficients (regardless of the statistical significance) were observed (fig. 5J), which does not support a common trend of the E–R correlation among these viruses (P = 0.75, the binomial test with the hypothesized probability equal to 50% for both positive and negative correlation coefficients). But recalling that the relatively small number of ORFs (and therefore, limited statistical power) for each virus species could prevent robust trend detection, we attempted to synthesize the data across all ten virus species examined in our study. We understand that the evolutionary rates and expression levels among these viruses are not directly comparable; therefore, we performed a meta-analysis to combine the correlation coefficients calculated for the individual virus species. Briefly, Fisher’s z-transformation was performed to obtain the weight for each virus species, and the generic inverse-variance pooling method was applied to estimate a pooled correlation coefficient. The pooled E–R correlation coefficient ρ was equal to 0.08 (P = 0.16, N = 392; fig. 5J), with 95% confidence intervals [−0.03, 0.17], a finding supporting the lack of the E–R anticorrelation for these viruses.
No E–R Anticorrelation Observed in the Long-Term Virus Evolution
It is worth noting that the evolutionary rate estimated from the genomic sequences of virus variants is polymorphic and therefore, may not genuinely reflect the substitution rate in the long-term evolution. Seeking to exclude this possibility, we further estimated the evolutionary rate from the orthologous genes of sister virus species or subspecies. Specifically, we estimated the evolutionary rates as the dN/dS ratio for each orthologous ORF pair between SARS-CoV-2 and SARS-CoV; the latter caused the severe acute respiratory syndrome in 2003. Again, we detected no E–R anticorrelation (ρ = −0.07, P = 0.88, N = 9, Spearman’s correlation; fig. 6A).
Fig. 6.
The absence of E–R anticorrelation when the protein evolutionary rates are estimated as the dN/dS ratio of orthologous genes. (A–C) The E–R correlation coefficients in individual virus groups. The expression level ORFs of SARS-CoV-2 (A), human cytomegalovirus (B), or Vaccinia virus (C) were used; the protein evolutionary rates were estimated as the dN/dS ratio between SARS-CoV-2 and SARS-CoV (A), between human and chimpanzee cytomegalovirus (B), or among five Orthopoxvirus species (C). The scale bar of the phylogenetic tree in (C) represents 0.01 substitutions per nucleotide. For Vaccinia virus, a constant number (one) has been added to all expression data prior to applying the logarithm transform, to avoid the logarithm of zero. (D) Similar to figure 5J, showing a meta-analysis of E–R correlation for three groups of viruses in figure 6A–C. The results of the fixed-effect model is presented because the test of heterogeneity gave a P value = 0.88 (I2 = 0.0%). The effective sample size N is also shown for the meta-analysis.
Note that SARS-CoV-2 and SARS-CoV are often not considered to be two virus species. The evolutionary rates between SARS-CoV-2 and its sister species (MERS-CoV) were not presented because their sequence divergence was too high to warrant precise sequence alignment. For example, the protein similarity of the ORF N was only 43% between SARS-CoV-2 and MERS-CoV in the alignable region, and that of the ORF S was only 35%. Such extensive sequence divergence, even compared with the sister species, was also common for some other virus species used in figure 5. Nevertheless, we successfully estimated the dN/dS ratios between human cytomegalovirus (Human betaherpesvirus 5) and Chimpanzee cytomegalovirus (Panine betaherpesvirus 2), and among five Orthopoxvirus species (Vaccinia virus, Variola virus, Cowpox virus, Monkeypox virus, and Skunkpox virus).
We detected no E–R anticorrelation for cytomegalovirus or Orthopoxvirus (fig. 6B and C) or in a meta-analysis that estimated the pooled Spearman’s correlation coefficient from three groups of viruses (ρ = 0.13, P = 0.11, N = 158; fig. 6D). Keeping in mind the possibility of saturated synonymous changes, we also estimated the E–R correlation with dN instead of dN/dS being used to estimate the protein evolutionary rate. We again found no evidence for the E–R anticorrelation (supplementary fig. S2B–D, Supplementary Material online). Taken together, the E–R anticorrelation was not evident in virus evolution in the short term (evolutionary rates estimated from pairwise sequence difference among variants) or in the long term (evolutionary rates estimated as dN or dN/dS of orthologous genes of virus species).
The E–R Anticorrelation Exists among ORFs of Endogenous Retroviruses
No E–R anticorrelation was detected for any of the ten virus species we examined in this study, indicating that an E–R anticorrelation will not be evident when protein sequence evolution is constrained only by the maintenance of protein function. Therefore, this observation contradicts the function maintenance hypothesis but is consistent with the cytotoxicity avoidance hypothesis.
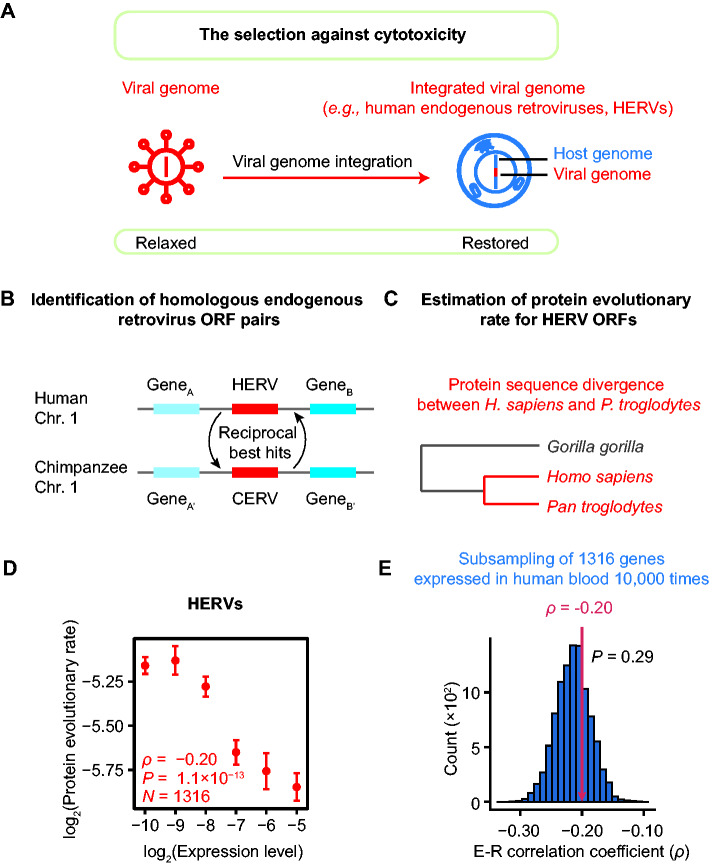
The cytotoxicity avoidance hypothesis further predicts that the E–R anticorrelation should be present among viral ORFs if the selection against cytotoxicity exists (fig. 7A). We realized that this prediction could be tested with human endogenous retroviruses (HERVs), which are the remnants of ancient integration of exogenous retroviruses that occurred in the germline (Griffiths 2001). There are tens of thousands of HERVs in the human genome (Belshaw et al. 2004), each including some or all of the four ORFs: gag (encoding core proteins), pro (protease), pol (reverse transcriptase), and env (envelope proteins). The vast majority of HERVs are presumably neither infectious nor functional, but numerous HERVs retained their protein expression activity in human cells (Andersson et al. 2002; Seifarth et al. 2005). The cytotoxicity induced by HERVs can reduce the fitness of the host organisms, which will, in turn, compromise the propagation of these viral elements. Consequently, any purifying selection against cytotoxicity could reasonably be expected to play a role in the sequence evolution of HERVs (fig. 7A). Therefore, the cytotoxicity avoidance hypothesis can be tested with HERVs; the hypothesis is not supported if the E–R anticorrelation among ORFs of HERV is not detected.
Fig. 7.
The presence of E–R anticorrelation among HERV ORFs. (A) The selection against cytotoxicity is restored when a viral genome is integrated into a host genome. (B) The syntenic reciprocal best BLAST hits endogenous retrovirus pairs were identified as human–chimpanzee orthologous pairs. (C) A schematic shows the estimation of the protein evolutionary rate for each HERV ORF. (D) The E–R correlation in HERVs (ρ = −0.20, P = 1.1 × 10−13, N = 1,316, Spearman’s correlation). The expression levels of HERV ORFs are estimated from RNA-seq data for human peripheral blood mononuclear cells from six healthy individuals. The plot shows the expression level in healthy #1 (replicate #1). Replicates #2–#6 are shown in supplementary figure S3, Supplementary Material online. The HERV ORFs are split into six bins according to their expression levels: (−∞, −9.5), [−9.5, −8.5), [−8.5, −7.5), [−7.5, −6.5), [−6.5, −5.5), and [−5.5, +∞). The mean (±standard errors) of the evolutionary rate is shown for each bin. Spearman’s correlation coefficient is calculated from the unbinned data. (E) The E–R anticorrelation among HERVs (indicated by the red arrow) was not significantly different from that among human nonvirus protein-coding genes (ρ = −0.21, P = 6.2 × 10−182, N = 17,410). The one-tailed P value (0.29) was calculated from a subsampling test. The histogram shows the distributions of 10,000 E–R correlation coefficients estimated from individual subset of 1,316 genes.
To test whether the E–R anticorrelation exists among HERVs, we identified orthologous endogenous retrovirus pairs between humans (Homo sapiens) and chimpanzees (Pan troglodytes, fig. 7B) and estimated their protein evolutionary rate from their sequence divergence (fig. 7C). We further estimated the expression level for each HERV ORF from RNA sequencing data (Tokuyama et al. 2018). We found that expression level was negatively correlated with protein evolutionary rate among HERV ORFs (ρ = −0.20, P = 1.1 × 10−13, N = 1,316, Spearman’s correlation; fig. 7D and supplementary fig. S3, Supplementary Material online), indicating the co-occurrence between the E–R anticorrelation and purifying selection against cytotoxicity. Furthermore, the correlation was not significantly different from the E–R anticorrelation trend among human endogenous genes (P = 0.29, subsampling test; fig. 7E). Taken together, these observations confirm that the E–R anticorrelation can be evident in viruses when purifying selection against cytotoxicity is present.
Discussion
In this study, we show that the widely reported E–R anticorrelation in cellular organisms is missing in viruses, where selection against cytotoxicity is relaxed but selection against impaired function remained (figs. 2–6). Furthermore, this correlation exists when selection against cytotoxicity is present as viral sequences are integrated into the host genome (fig. 7). These observations suggest that, whereas the maintenance of protein function definitely plays some roles in constraining protein sequence evolution, it is not likely the main component of the selective constraint that generates the E–R anticorrelation. Instead, the selective constraint may be better interpreted as the consequence of the avoidance of cytotoxicity or other mechanisms not yet known. Nevertheless, we realize that the lack of E–R anticorrelation observed for the viruses in our study is subject to at least three caveats, which we discuss below.
First, the E–R anticorrelation is expected under the assumption that positive selection does not play a role, because beneficial mutations are considered too rare to affect the protein evolutionary rate (Zhang and Yang 2015). In principle, the absence of the E–R anticorrelation in SARS-CoV-2 could result from the violation of this assumption; that is, it is conceivable that positive selection could have extensively driven the sequence evolution of viral proteins, due to for example the escape from the host immune systems. However, this is unlikely for several reasons. First, there are multiple reports of strong signals for purifying selection among the SARS-CoV-2 variants isolated from patients (Shen et al. 2020; Tang et al. 2020). Moreover, to the best of our knowledge there are only a few examples of signals suggesting positive selection of SARS-CoV-2 in human hosts.
Although not highlighted in our results, note that we did conduct additional analyses to examine this caveat possibility that positive selection could have driven the evolution of SARS-CoV-2. We estimated the dN/dS ratio on the phylogenetic tree of SARS-CoV-2 and three related coronaviruses (shown in fig. 4A) using Phylogenetic Analysis by Maximum Likelihood (PAML) (Yang 2007). The dN/dS ratio was smaller than 0.1 and significantly smaller than 1 (P < 0.001, likelihood ratio tests) for each of the nine SARS-CoV-2 ORFs (supplementary table S2, Supplementary Material online). Further, the proportion of sites showing a signal for positive selection (dN/dS > 1) was equal to zero for each ORF (supplementary table S2, Supplementary Material online). Similar results have also been reported in previous studies of Influenza A virus and other RNA viruses (Suzuki 2006; Pybus et al. 2007). All of these observations suggest that positive selection is rare and is unlikely to account for the observed absence of the E–R anticorrelation in viruses.
A second caveat is that the avoidance of cytotoxicity could still exert some role in the evolution of viral genomes. For example, if cytotoxicity from a viral protein reduces the health of a host cell, then this reduced health could in turn reduce the proliferation rates for the infecting viruses; this scenario would cause selection-based avoidance of this type of cytotoxicity on viral genome evolution. However, we suspect that such an effect is unlikely to be strong, as SARS-CoV-2 kills the host cell within a time window of only dozens of hours (Bojkova et al. 2020), whereas cytotoxicity is known to act mainly in long-lived cells, such as in neurons (David et al. 2010; Chiti and Dobson 2017). More obviously, if such avoidance of cytotoxicity in viral genome evolution is still effective to some extent, our data-driven challenge of the function maintenance hypothesis remains: The selective constraint from function maintenance would not be sufficient to create any trend suggesting the E–R anticorrelation.
Third, our study so far tested the function maintenance and cytotoxicity avoidance as two competing hypotheses because they are the two most popular theories used to explain the E–R anticorrelation. We do realize the possibility that the E–R anticorrelation observed in cellular organisms is explained by the additional mechanisms not yet known. Nevertheless, the identification of such novel mechanisms will not affect our conclusion that the function maintenance hypothesis is not well supported by the empirical data because we have reported that the maintenance of function (alone, or together with additional unknown mechanisms that operate in viruses) is not sufficient to generate a significant E–R anticorrelation in protein sequence evolution.
We noted in the introduction that Pagán et al. (2012) claimed the detection of the E–R anticorrelation in ten negative-stranded Mononegavirales virus species. However, there are at least two major limitations of their study. First, the detection of the E–R anticorrelation in Pagán et al. (2012) was dependent on excluding ten “outlier” ORFs from a total of 82 ORFs. We believe that this procedure was inappropriate because these “outliers” were not specified a priori. As a matter of fact, no E–R anticorrelation (P = 0.17) was detected by Pagán et al. before these “outliers” were removed, which is actually consistent with our results. Furthermore, it is notable that the number of sequenced variants for each virus species in Pagán et al. was much smaller than that of our data sets. Specifically, in Pagán et al., the number of isolated and sequenced variants for each virus species used to estimate the evolutionary rate varied between 6 (Sendai virus) and 110 (Newcastle disease virus), with the median equal to 18. In fact, five out of their ten virus species have also been included in our analysis (Ebola virus, human parainfluenza virus type 3, human respiratory syncytial virus, measles virus, and mumps virus; fig. 5). In sharp contrast to their work, the variant numbers we assessed for these five virus species were between 209 (measles virus) and 1,006 (human respiratory syncytial virus). This much-enlarged number of sequenced variants per virus species in our study enabled a significant improvement in accuracy for estimation of protein evolutionary rate, which bolstered our confidence for a complete absence of any E–R anticorrelation in viruses.
Some viruses can infect multiple cellular species, leading to symptoms in some hosts (symptomatic hosts) but not in others (asymptomatic hosts). It is conceivable that reducing virus-associated cytotoxicity in the asymptomatic host can help propagate and spread the virus (Chen et al. 2020). By this logic, it should be informative to test whether the E–R anticorrelation is present in asymptomatic hosts but is absent in symptomatic hosts for the same virus. However, we realized that technical hurdles in partitioning the evolutionary histories between the symptomatic and asymptomatic hosts for the same virus could hinder the investigation in this vein. Taking the Zika virus for example, the partition between the viral genome evolution in its symptomatic host (humans) versus in its asymptomatic host (mosquitos) is not trivial. It is because the Zika virus has been reported transmitted from mosquitos to humans time and again (Gutierrez-Bugallo et al. 2019). The sequence difference between the virus variants isolated from two humans can arise from the evolutionary history either in humans or in mosquitos. Nevertheless, comparing the E–R correlation coefficients for the same virus between its asymptomatic and symptomatic hosts deserves further investigation once the technical hurdle is overcome in the future.
In sum, our results provide an empirical test for evaluating the relative importance between the function maintenance hypothesis and the cytotoxicity avoidance hypothesis. Our results indicate that a given mutation may not confer its impacts through a specific loss of function, instead, through other mechanisms such as cytotoxicity. In addition to this theoretical purport, our findings also have medical implications. That is, the currently widespread default thinking from experimental biologists and medical scientists that disease-associated mutations are likely to impact some specific molecular function should be expanded: The potentially high likelihood that a given mutation confers its effects via cytotoxicity per se should be taken into consideration. Consistent with this idea, 83% of disease-causing missense polymorphisms observed in human genomes have been estimated to affect protein stability, potentially influencing the misfolding propensity of proteins (Wang and Moult 2001). This new insight will help to computationally predict disease-causing mutations from genome-wide sequencing data, an ongoing research direction that has been widely valued in recent years (Cooper and Shendure 2011; Wu et al. 2014), by pointing to the direction to identify nonsynonymous mutations that can reduce the structural stability of proteins.
Materials and Methods
Estimation of Gene Expression Levels
RNA Sequencing Data Sets Used in This Study
The expression levels of Vero cell genes and SARS-CoV-2 ORFs were estimated from the nanoball-based RNA sequencing data of Vero cells infected by SARS-CoV-2 BetaCoV/Korea/KCDC03/2020 (Kim et al. 2020). The expression levels of MERS-CoV ORFs were estimated from RNA sequencing data of human lung adenocarcinoma epithelial (Calu-3) cells harvested at 6 h after infected by MERS-CoV HCoV-EMC/2012 (Zhang et al. 2020). The expression levels of Influenza A virus ORFs were estimated from RNA sequencing data of primary human lung epithelium (NHBE) infected with the subtype H1N1 Influenza A virus A/Puerto Rico/8/1934 (Blanco-Melo et al. 2020). The gene expression levels of Ebola virus ORFs were estimated from RNA sequencing data of human liver cells (Huh7) infected by the Ebola virus Makona isolate (Albarino et al. 2018). The expression levels of HERV ORFs were estimated from the RNA sequencing data for human peripheral blood mononuclear cells from six healthy individuals (Tokuyama et al. 2018).
Reference Genomes
The genomes of C. sabaeus (Ensembl, release 99), H. sapiens (Ensembl, release 100), SARS-CoV-2 (GenBank: MN908947.3), MERS-CoV (GenBank: NC_019843.3), Influenza A virus (GenBank: NC_002023.1, NC_002021.1, NC_002022.1, NC_002017.1, NC_002019.1, NC_002018.1, NC_002016.1, and NC_002020.1), and Ebola virus (GenBank: KT589389) were used as references for mapping the sequencing reads, respectively.
Expression Levels in SARS-CoV-2 and MERS-CoV
The mRNA level of an ORF of coronaviruses cannot be estimated from the abundance of mapped reads in RNA sequencing data due to the nested nature of the genome and subgenomes (Kim et al. 2020). Nevertheless, such nested subgenomes were produced by the discontinuous negative-strand RNA synthesis (i.e., leader-to-body fusion); therefore, the abundance of a subgenome can be estimated from the number of the leader-to-body junction reads (fig. 2B). The reason is briefly described as follows. There are two types of transcription-regulating sequences (TRS): TRS-L is downstream to the leader sequence, and the body TRSs (TRS-B) are upstream to individual ORFs. During the synthesis of the negative-strand RNA, the negative-strand TRS-B may hybridize with the TRS-L in the positive-sense genomic RNA, and the synthesis continues using the leader sequence as the template, resulting in a leader-to-body junction. For each subgenome, only the most upstream (5′-) ORF is translated (when leaky scanning of start codons is not under consideration). Therefore, the mRNA level of an ORF in a subgenome can be inferred from the number of leader-containing reads. ORF1ab is translated from the genomic RNA. Therefore, although there is no leader-to-body junction, leader-containing reads can still be used to estimate its mRNA level.
The RNA sequencing reads of coronavirus-infected cell lines were aligned to the references with STAR 2.7.3a (Dobin et al. 2013) using the parameters as follows: --outFilterTypeBySJout --outFilterMultimapNmax 20 --align SJoverhangMin 8 --outSJfilterOverhangMin 12 12 12 12 --outSJfilterCountUniqueMin 1 1 1 1 --outSJfilterCount TotalMin 1 1 1 1 --outSJfilterDistToOtherSJmin 0 0 0 0 --outFilterMismatchNmax 999 --outFilterMismatchNover ReadLmax 0.04 --scoreGapNoncan -4 --scoreGapATAC -4 --chimOutType WithinBAM HardClip --chimScoreJunction NonGTAG 0 --alignSJstitchMismatchNmax -1 -1 -1 -1 --alignIntronMin 20 --alignIntronMax 1000000 --alignMates GapMax 1000000.
The TRS-L of SARS-CoV-2 is the 70th–75th nucleotides in its genome, and that of MERS-CoV is the 62th–67th nucleotides. Therefore, only the reads with the 5′-junction site located between the 55th and 85th nucleotides of the SARS-Cov-2 genome (or between 45th and 80th nucleotides of the MERS-CoV genome) were counted as they represented subgenomes generated by canonical leader-to-body fusions. A leader-to-body junction read was assigned to the subgenome according to the ORF sequence immediately downstream of its 3′-junction site; a leader-to-body junction read will be discarded if its 3′-junction site is beyond 50 nucleotides upstream of the start codon of any ORFs. The mRNA level of ORF1ab was calculated from the number of the leader-containing reads aligned to the genome RNA without gaps. ORF10 in SARS-CoV-2 was excluded in this study because it does not have a TRS-B, nor was its subgenome detected (Kim et al. 2020). Zhang et al. (2020) provided three replicate RNA-seq data sets for MERS-CoV-infected Calu-3 cells. Considering that the ranks of mRNA level among ORFs were consistent in the three replicates, replicate #3 was used in this study.
Expression Levels in Influenza A virus and Ebola Virus
The expression level of an ORF was estimated from the abundance of sequencing reads mapped to it. The RNA sequencing data were aligned to the corresponding references with STAR using default parameters. The expression level of an ORF was given by RSEM (Li and Dewey 2011). It is worth noting that the Influenza A virus subtype H1N1 strain used for estimating expression level (A/Puerto Rico/8/1934) is not directly related to the virus variants used for estimating evolutionary rate, the viruses from the 2009 pandemic. Nevertheless, we surmised that the relative expression levels of ORFs unlikely changed dramatically among Influenza A virus variants as a stoichiometric balance among viral proteins has to be largely maintained to ensure the correct packaging of the viral particle. Albarino et al. (2018) provided three RNA-seq data sets at different time points after Huh7 cells were infected by Ebola viruses; the average expression level at three time points was used for each ORF.
Expression Levels in Vaccinia virus and Human Cytomegalovirus
The expression levels of Vaccinia virus ORFs were retrieved from the genome tiling array data of Vaccinia virus-infected HeLa cells (Assarsson et al. 2008); most of the viral ORFs had initiated transcription at 4 h after infection, and therefore, the expression levels at this time point were used for further analyses. The expression levels of human cytomegalovirus ORFs were retrieved from a previous study that performed RNA sequencing experiments on human cytomegalovirus-infected CD34+ hematopoietic stem cells (Cheng et al. 2017). The RNA sequencing data of cells harvested at 2 days after infection, when the viral ORFs were most actively transcripted, were used in this study.
Expression Levels in Human Parainfluenza Virus Type 3, Human Respiratory Syncytial Virus, Measles Virus, and Mumps Virus
The expression level of an ORF was retrieved from the band densities from electrophoresis gels estimated in previous studies (Cattaneo et al. 1987; De et al. 1990; Barik 1992; Takeuchi et al. 1993; Pagán et al. 2012).
The Estimation of Expression Levels for Overlapping ORFs
Some viral ORFs overlap with each other, and their encoded proteins are translated with various mechanisms such as leaky scanning, ribosomal frameshifting, and RNA editing. Three scenarios were considered when we estimate the expression level of overlapping ORFs. First, overlapping ORFs share the same start codon (e.g., ORF1a/ORF1ab in SARS-CoV-2). Second, overlapping ORFs are translated in different frames due to leaky scanning (e.g., ORF3a/ORF3b in SARS-CoV-2). In both scenarios, we assigned the estimated mRNA level to the longest ORF (e.g., ORF1ab or ORF3a) for simplicity; the evolutionary rate was also estimated from this ORF. In the third scenario, for instance P/V/C in measles virus and in human parainfluenza virus type 3, there are overlapping ORFs sharing the same start codon (P and V) as well as overlapping ORFs in different translation frames (C and P) in the same genomic region; we split the total estimated mRNA level into individual ORFs according to the reported relative protein abundance (ViralZone, https://viralzone.expasy.org/857, retrieved on June 18, 2020).
Expression Levels of Monkey and Human Genes
The RNA sequencing data of SARS-CoV-2-infected Vero cells were aligned to the genome of C. sabaeus with STAR using default parameters. Similarly, the RNA sequencing data for human peripheral blood mononuclear cells were aligned to the human genome with STAR using default parameters. Each read was assigned to its aligned gene under the parameter --quantMode TranscriptomeSAM. Considering that multiple splicing isoforms may exist for a gene, the abundance of each isoform was estimated, and the total abundance of all isoforms of a gene was used as its expression level.
Expression Levels of HERV ORFs
The RNA sequencing data for human peripheral blood mononuclear cells were aligned to the human genome with STAR using the parameters as follows: --outSAMtype BAM SortedByCoordinate --outFilterMultimapNmax 30. The annotation file of HERV ORFs was downloaded from gEVE on August 5, 2020 (genome-based endogenous viral element database, http://geve.med.u-tokai.ac.jp) (Nakagawa and Takahashi 2016). Noted that some HERV ORFs were annotated as multiple truncated regions. For each annotated ORF or ORF region, the expression level was defined as the ratio between the aligned reads number, which was estimated with Telescope 1.0.3 (Bendall et al. 2019) with the default parameters, and the length of ORF or ORF region. As annotated regions of the same HERV ORF can have different estimated expression levels, we used the average expression level of all expressed annotated ORF regions of the same ORF to represent the expression level of this ORF.
Protein Levels in SARS-CoV-2
The abundance of proteins encoded by the SARS-CoV-2 genome was estimated from the number of peptide-spectrum matches (PSMs) of a protein, normalized by the number of its theoretical peptides. The number of PSMs was retrieved from a previous proteomic analysis on SARS-CoV-2-infected Vero cells using tandem mass spectrometry (Davidson et al. 2020). The theoretical peptides of proteins digested by trypsin were in silico generated using in-house python script, in which the maximum number of missed arginine or lysine during digestion was set to 2. Only the peptide with a length between 7 and 140 amino acids was counted. The estimated protein abundances of the nine ORFs of SARS-CoV-2 were highly correlated with the corresponding mRNA abundances (r = 0.99, P = 6 × 10−7, Pearson’s correlation; table 1).
Estimation of Protein Evolutionary Rates
Retrieval of the Genomic Sequence of Virus Variants
Complete genome sequences of variants of SARS-CoV-2 and Influenza A virus were downloaded from GISAID (Global Initiative on Sharing All Influenza Data, https://www.gisaid.org/) (Shu and McCauley 2017); those of MERS-CoV, Ebola virus, measles virus, mumps virus, human parainfluenza virus type 3, human respiratory syncytial virus, Vaccinia virus, and human cytomegalovirus were downloaded from NCBI Virus (https://www.ncbi.nlm.nih.gov/labs/virus/vssi/) (Hatcher et al. 2017). Specifically, for SARS-CoV-2, 13,556 high-coverage genome sequences were downloaded on May 12, 2020. Genome sequences isolated from bat, pangolin, tiger, or mink, and those with over 50 bases not determined during sequencing (labeled as Ns in the genome sequence) were filtered, and the 7,310 remaining sequences were used for the downstream analysis. For MERS-CoV, genome sequences with over 200 nucleotides mismatches against the reference genome were filtered, and the 518 remaining genome sequences (from both human and animal hosts) were used for the downstream analysis. For Ebola virus, 287 genome sequences collected in Sierra Leone and Guinea during the 2013–2016 Western African outbreak were downloaded. For Influenza A virus subtype H1N1, 1,839 genome sequences collected during the 2009 pandemic were downloaded. For measles virus and mumps virus, 209 and 284 genome sequences were downloaded, respectively. For human parainfluenza virus type 3, a total of 353 complete genome sequences were downloaded. For human respiratory syncytial virus, 1,006 complete genome sequences of subgroup A were downloaded. For Vaccinia virus, a total of 103 complete genome sequences were downloaded. Two of the sequences (NC_006998 and AY243312) were identical to each other, and we kept only the reference genome NC_006998 for further analyses. In addition, two of the virus variants (MG012795 and MG012796) that exhibited exceptionally long external branches in the phylogenetic tree were excluded for further analyses. As a consequence, a total of 100 remaining sequences were used for the estimation of protein evolutionary rate in Vaccinia virus. For human cytomegalovirus, a total of 299 complete genome sequences were downloaded.
Estimation of Protein Evolutionary Rate in Viruses
The protein evolutionary rate of ten virus species examined in this study was inferred from the average pairwise difference among variants. Specifically, for each virus species, the reference sequence of each ORF was aligned against the genomes of individual virus variants by EMBOSS needle (Rice et al. 2000). The aligned sequences (without gaps) were in silico translated to protein sequences. The number of different amino acids was counted for each variant pair, and the average was estimated from all possible variant pairs, which was further normalized by the length of the peptide sequence.
Estimation of the Protein Evolutionary Rate for SARS-CoV-2 Before Zoonotic Transfer
The rate of SARS-CoV-2 protein sequence during the evolution in animal hosts was estimated from the dN/dS ratio among SARS-CoV-2 and three related coronaviruses isolated from bats or pangolins. The sequences of SARS-CoV-2 (Wu et al. 2020), RaTG13 (GenBank: MN996532.1) isolated from bats in Yunnan (Zhou et al. 2020), GD-1 (GISAID: EPI_ISL_410721) isolated from pangolins in Guangdong (Xiao et al. 2020), and GX-P5E (GISAID: EPI_ISL_410541) isolated from pangolins in Guangxi (Lam et al. 2020) were aligned by MUSCLE (Edgar 2004); the phylogenetic tree was constructed using the neighbor-joining method in MEGA X (Kumar et al. 2018) with maximum composite likelihood as the substitution model. A coronavirus (bat-SL-CoVZC45, GenBank: MG772933.1) isolated from Zhejiang (Hu et al. 2018) was used to root the phylogenetic tree. The homologous ORFs among SARS-CoV-2, RaTG13, GD-1, and GX-P5E were aligned by MUSCLE. The average dN, dS, and dN/dS ratio on the phylogenetic tree were estimated for each ORF by codeml in PAML (Yang 2007) under the parameters model = 0, NSsites = 0, fix_omega = 0, and clock = 0.
Estimation of the Cross-species dN/dS Ratio in Three Groups of Virus Species/Subspecies
The dN/dS ratios were also estimated across virus species, for example, among five Orthopoxvirus species, including Vaccinia virus (GenBank: NC_006998), Variola virus (GenBank: NC_001611), Cowpox virus (GenBank: NC_003663), Monkeypox virus (GenBank: NC_003310), and Skunkpox virus (GenBank: NC_031038). Orthologous ORFs of Vaccinia virus in a second Orthopoxvirus species were identified by the reciprocal best BLAST hits and were aligned by MUSCLE. The phylogenetic tree was constructed from the concatenated protein coding sequences of four ORFs (A5R, A10L, A24R, and E6R) using the neighbor-joining method in MEGA X with maximum composite likelihood as the substitution model. The average dN, dS, and dN/dS on this phylogenetic tree were estimated for each ORF by codeml in PAML under the parameters model = 0, NSsites = 0, fix_omega = 0, and clock = 0. The dN, dS, and dN/dS of ORFs of SARS-CoV-2 and human cytomegalovirus (GenBank: NC_006273) were similarly estimated using their respective sister species, SARS-CoV (GenBank: NC_004718.3) and chimpanzee cytomegalovirus (GenBank: NC_003521).
Estimation of the Evolutionary Rate of Proteins Encoded by the Nuclear Genome of Primates
We estimated the evolutionary rate of proteins encoded in the genome of Vero cells from the protein divergence between C. sabaeus and M. mulatta. For each gene, the fractions of identical amino acids in the sequence alignment between the two species, “C. sabaeus % ID” and “M. mulatta % ID,” were retrieved from Ensembl (release 99). The homologous gene pair was filtered if the two fractions differ by greater than 5%. The mean of the two factions was estimated and its difference from 100% was used to infer the protein evolutionary rate. The evolutionary rate of human proteins was similarly estimated as the average fraction of identical amino acids in the sequence alignment between the human and the chimpanzee, “H. sapiens % ID” and “P. troglodytes % ID.”
Estimation of the Evolutionary Rate for HERV ORFs
The endogenous retrovirus protein sequences in humans and chimpanzees were downloaded from gEVE. A human–chimpanzee orthologous endogenous retrovirus pair for an annotated ORF region was identified following three criteria: 1) The orthologous pair presents the syntenic reciprocal best BLAST hits; 2) the identity of the aligned sequence was greater than 85%; and 3) the number of aligned amino acids was no less than the 85% of full protein length in either annotated region. The evolutionary rate of each annotated ORF region was estimated from the protein divergence between human and chimpanzee, defined as the number of varied amino acids divided by the average protein length of the orthologous pair. Similar to the estimation of expression levels, we used the average evolutionary rate of all annotated ORF regions to represent the evolutionary rate of the ORF when multiple regions are annotated for the same ORF.
Code Availability
All scripts used to analyze the data and to generate the figures are available at https://github.com/ChangshuoWei/E-R_anticorrelation-associated-hypotheses.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
We acknowledge the authors and laboratories for generating and submitting the sequences to GISAID Database on which this research is based. The list is detailed in supplementary tables S3 and S4, Supplementary Material online. We thank Dr Jian-Rong Yang and Dr Xionglei He at Sun Yat-sen University, Dr Lucas Carey at Peking University, Dr Jianzhi George Zhang at University of Michigan, Dr Katarzyna Tomala at Jagiellonian University for comments and suggestions, and Dr Zhang Zhang at Beijing Institute of Genomics CAS for technical supports. This work was supported by grants from the National Key R&D Program of China (2019YFA0508700 to W.Q.) and the National Natural Science Foundation of China (31922014 to W.Q.).
Author Contributions
W.Q. designed the study; C.W. and Y.-M.C. performed data analyses; Y.C. and W.Q. wrote the manuscript.
Data Availability
All data that were used to support the findings of this study are available in the public databases (NCBI, https://www.ncbi.nlm.nih.gov/; GISAID, https://www.gisaid.org/; Ensembl, https://www.ensembl.org/; gEVE, http://geve.med.u-tokai.ac.jp).
Literature Cited
- Albarino CG, Wiggleton Guerrero L, Chakrabarti AK, Nichol ST.. 2018. Transcriptional analysis of viral mRNAs reveals common transcription patterns in cells infected by five different filoviruses. PLoS One 13(8):e0201827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson AC, et al. 2002. Developmental expression of HERV-R (ERV3) and HERV-K in human tissue. Virology 297(2):220–225. [DOI] [PubMed] [Google Scholar]
- Assarsson E, et al. 2008. Kinetic analysis of a complete poxvirus transcriptome reveals an immediate-early class of genes. Proc Natl Acad Sci U S A. 105(6):2140–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barik S. 1992. Transcription of human respiratory syncytial virus genome RNA in vitro: requirement of cellular factor(s). J Virol. 66(11):6813–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, et al. 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci U S A. 101(14):4894–4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall ML, et al. 2019. Telescope: characterization of the retrotranscriptome by accurate estimation of transposable element expression. PLoS Comput Biol. 15(9):e1006453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergh J, et al. 2015. Structural and kinetic analysis of protein-aggregate strains in vivo using binary epitope mapping. Proc Natl Acad Sci U S A. 112(14):4489–4494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biesiadecka MK, Sliwa P, Tomala K, Korona R.. 2020. An overexpression experiment does not support the hypothesis that avoidance of toxicity determines the rate of protein evolution. Genome Biol Evol. 12(5):589–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Melo D, et al. 2020. Imbalanced host response to SARS-CoV-2 drives development of COVID-19. Cell 181(5):1036–1045.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bojkova D, et al. 2020. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature 583(7816):469–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brierley I, Digard P, Inglis SC.. 1989. Characterization of an efficient coronavirus ribosomal frameshifting signal: requirement for an RNA pseudoknot. Cell 57(4):537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo R, et al. 1987. Altered transcription of a defective measles virus genome derived from a diseased human brain. EMBO J. 6(3):681–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, et al. 2020. Dissimilation of synonymous codon usage bias in virus-host coevolution due to translational selection. Nat Ecol Evol. 4(4):589–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, et al. 2019. Overdosage of balanced protein complexes reduces proliferation rate in aneuploid cells. Cell Syst. 9(2):129–142.e5. [DOI] [PubMed] [Google Scholar]
- Cheng S, et al. 2017. Transcriptome-wide characterization of human cytomegalovirus in natural infection and experimental latency. Proc Natl Acad Sci U S A. 114(49):E10586–E10595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JL. 2010. Expression level, evolutionary rate, and the cost of expression. Genome Biol Evol. 2:757–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiti F, Dobson CM.. 2017. Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu Rev Biochem. 86:27–68. [DOI] [PubMed] [Google Scholar]
- Cooper GM, Shendure J.. 2011. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 12(9):628–640. [DOI] [PubMed] [Google Scholar]
- David DC, et al. 2010. Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8(8):e1000450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson AD, et al. 2020. Characterisation of the transcriptome and proteome of SARS-CoV-2 reveals a cell passage induced in-frame deletion of the furin-like cleavage site from the spike glycoprotein. Genome Med. 12(1):68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De BP, Galinski MS, Banerjee AK.. 1990. Characterization of an in vitro system for the synthesis of mRNA from human parainfluenza virus type 3. J Virol. 64(3):1135–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, et al. 2013. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29(1):15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Bloom JD, Adami C, Wilke CO, Arnold FH.. 2005. Why highly expressed proteins evolve slowly. Proc Natl Acad Sci U S A. 102(40):14338–14343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond DA, Wilke CO.. 2008. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell 134(2):341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echave J, Spielman SJ, Wilke CO.. 2016. Causes of evolutionary rate variation among protein sites. Nat Rev Genet. 17(2):109–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sayed I, Bassiouny K, Nokaly A, Abdelghani AS, Roshdy W.. 2016. Influenza A virus and influenza B virus can induce apoptosis via intrinsic or extrinsic pathways and also via NF-kappaB in a time and dose dependent manner. Biochem Res Int. 2016:1738237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyertag F, Berninsone PM, Alvarez-Ponce D.. 2017. Secreted proteins defy the expression level-evolutionary rate anticorrelation. Mol Biol Evol. 34(3):692–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feyertag F, Berninsone PM, Alvarez-Ponce D.. 2019. N-glycoproteins exhibit a positive expression level-evolutionary rate correlation. J Evol Biol. 32(4):390–394. [DOI] [PubMed] [Google Scholar]
- Gáspári Z, Perczel A.. 2010. Chapter 2 - Protein dynamics as reported by NMR. In: Webb GA, editor. Annual reports on NMR spectroscopy. Cambridge (MA): Academic Press. p. 35–75. [Google Scholar]
- Gout JF, Kahn D, Duret L, Paramecium Post-Genomics C; Paramecium Post-Genomics Consortium. 2010. The relationship among gene expression, the evolution of gene dosage, and the rate of protein evolution. PLoS Genet. 6(5):e1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths DJ. 2001. Endogenous retroviruses in the human genome sequence. Genome Biol. 2(6):REVIEWS1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Bugallo G, et al. 2019. Vector-borne transmission and evolution of Zika virus. Nat Ecol Evol. 3(4):561–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatcher EL, et al. 2017. Virus Variation Resource - improved response to emergent viral outbreaks. Nucleic Acids Res. 45(D1):D482–D490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu D, et al. 2018. Genomic characterization and infectivity of a novel SARS-like coronavirus in Chinese bats. Emerg Microbes Infect. 7(1):154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, et al. 2020. The architecture of SARS-CoV-2 transcriptome. Cell 181(4):914–921.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. 1968. Evolutionary rate at the molecular level. Nature 217(5129):624–626. [DOI] [PubMed] [Google Scholar]
- Koonin EV, Wolf YI.. 2010. Constraints and plasticity in genome and molecular-phenome evolution. Nat Rev Genet. 11(7):487–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K.. 2018. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 35(6):1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam TT, et al. 2020. Identifying SARS-CoV-2-related coronaviruses in Malayan pangolins. Nature 583(7815):282–285. [DOI] [PubMed] [Google Scholar]
- Levy ED, De S, Teichmann SA.. 2012. Cellular crowding imposes global constraints on the chemistry and evolution of proteomes. Proc Natl Acad Sci U S A. 109(50):20461–20466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN.. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Wu CI, Luo CC.. 1985. A new method for estimating synonymous and nonsynonymous rates of nucleotide substitution considering the relative likelihood of nucleotide and codon changes. Mol Biol Evol. 2(2):150–174. [DOI] [PubMed] [Google Scholar]
- Longdon B, Brockhurst MA, Russell CA, Welch JJ, Jiggins FM.. 2014. The evolution and genetics of virus host shifts. PLoS Pathog. 10(11):e1004395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa S, Takahashi MU.. 2016. gEVE: a genome-based endogenous viral element database provides comprehensive viral protein-coding sequences in mammalian genomes. Database (Oxford) 2016:baw087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán I, Holmes EC, Simon-Loriere E.. 2012. Level of gene expression is a major determinant of protein evolution in the viral order Mononegavirales. J Virol. 86(9):5253–5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Hurst LD.. 2001. Highly expressed genes in yeast evolve slowly. Genetics 158(2):927–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal C, Papp B, Lercher MJ.. 2006. An integrated view of protein evolution. Nat Rev Genet. 7(5):337–348. [DOI] [PubMed] [Google Scholar]
- Plata G, Vitkup D.. 2018. Protein stability and avoidance of toxic misfolding do not explain the sequence constraints of highly expressed proteins. Mol Biol Evol. 35(3):700–703. [DOI] [PubMed] [Google Scholar]
- Pybus OG, et al. 2007. Phylogenetic evidence for deleterious mutation load in RNA viruses and its contribution to viral evolution. Mol Biol Evol. 24(3):845–852. [DOI] [PubMed] [Google Scholar]
- Razban RM. 2019. Protein melting temperature cannot fully assess whether protein folding free energy underlies the universal abundance-evolutionary rate correlation seen in proteins. Mol Biol Evol. 36(9):1955–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhim JS, Schell K, Creasy B, Case W.. 1969. Biological characteristics and viral susceptibility of an African green monkey kidney cell line (Vero). Proc Soc Exp Biol Med. 132(2):670–678. [DOI] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A.. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16(6):276–277. [DOI] [PubMed] [Google Scholar]
- Rocha EP. 2006. The quest for the universals of protein evolution. Trends Genet. 22(8):412–416. [DOI] [PubMed] [Google Scholar]
- Seifarth W, et al. 2005. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J Virol. 79(1):341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Z, et al. 2020. Genomic diversity of severe acute respiratory syndrome-coronavirus 2 in patients with coronavirus disease 2019. Clin Infect Dis. 71(15):713–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y, McCauley J.. 2017. GISAID: global initiative on sharing all influenza data – from vision to reality. Euro Surveill. 22(13):30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki Y. 2006. Natural selection on the influenza virus genome. Mol Biol Evol. 23(10):1902–1911. [DOI] [PubMed] [Google Scholar]
- Takeuchi K, Tanabayashi K, Okazaki K, Hiahiyama M, Yamada A.. 1993. In vitro transcription and replication of the mumps virus genome. Arch Virol. 128(1–2):177–183. [DOI] [PubMed] [Google Scholar]
- Tang XL, et al. 2020. On the origin and continuing evolution of SARS-CoV-2. Natl Sci Rev. 7(6):1012–1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuyama M, et al. 2018. ERVmap analysis reveals genome-wide transcription of human endogenous retroviruses. Proc Natl Acad Sci U S A. 115(50):12565–12572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usmanova DR, Plata G, Vitkup D.. 2021. The relationship between the misfolding avoidance hypothesis and protein evolutionary rates in the light of empirical evidence. Genome Biol Evol. doi:10.1093/gbe/evab006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B.. 2009. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell 138(1):198–208. [DOI] [PubMed] [Google Scholar]
- Walsh D, Mohr I.. 2011. Viral subversion of the host protein synthesis machinery. Nat Rev Microbiol. 9(12):860–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Horby PW, Hayden FG, Gao GF.. 2020. A novel coronavirus outbreak of global health concern. Lancet 395(10223):470–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Moult J.. 2001. SNPs, protein structure, and disease. Hum Mutat. 17(4):263–270. [DOI] [PubMed] [Google Scholar]
- Wilson AC, Carlson SS, White TJ.. 1977. Biochemical evolution. Annu Rev Biochem. 46:573–639. [DOI] [PubMed] [Google Scholar]
- Wu F, et al. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579(7798):265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Li Y, Jiang R.. 2014. Integrating multiple genomic data to predict disease-causing nonsynonymous single nucleotide variants in exome sequencing studies. PLoS Genet. 10(3):e1004237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao K, et al. 2020. Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins. Nature 583(7815):286–289. [DOI] [PubMed] [Google Scholar]
- Yang JR, Liao BY, Zhuang SM, Zhang J.. 2012. Protein misinteraction avoidance causes highly expressed proteins to evolve slowly. Proc Natl Acad Sci U S A. 109(14):E831–E840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JR, Zhuang SM, Zhang J.. 2010. Impact of translational error-induced and error-free misfolding on the rate of protein evolution. Mol Syst Biol. 6:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24(8):1586–1591. [DOI] [PubMed] [Google Scholar]
- Zhang J, Maslov S, Shakhnovich EI.. 2008. Constraints imposed by non-functional protein-protein interactions on gene expression and proteome size. Mol Syst Biol. 4:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Yang JR.. 2015. Determinants of the rate of protein sequence evolution. Nat Rev Genet. 16(7):409–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, et al. 2020. Competing endogenous RNA network profiling reveals novel host dependency factors required for MERS-CoV propagation. Emerg Microbes Infect. 9(1):733–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, et al. 2021. Disome-seq reveals widespread ribosome collisions that promote cotranslational protein folding. Genome Biol. 22(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao TL, Zhang S, Qian WF.. 2020. [Cis-regulatory mechanisms and biological effects of translation elongation]. Yi Chuan 42(7):613–631. [DOI] [PubMed] [Google Scholar]
- Zhou P, et al. 2020. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuckerkandl E, Pauling L.. 1965. Evolutionary divergence and convergence in proteins. In: Bryson V, Vogel HJ, editors. Evolving genes and proteins. New York: Academic Press. p. 97–166. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data that were used to support the findings of this study are available in the public databases (NCBI, https://www.ncbi.nlm.nih.gov/; GISAID, https://www.gisaid.org/; Ensembl, https://www.ensembl.org/; gEVE, http://geve.med.u-tokai.ac.jp).