Abstract
Background
Laboratory-based methods for SARS-CoV-2 antibody detection vary widely in performance. However, there are limited prospectively-collected data on assay performance, and minimal clinical information to guide interpretation of discrepant results.
Methods
Over a 2-week period, 1080 consecutive plasma samples submitted for clinical SARS-CoV-2 IgG testing were tested in parallel for anti-nucleocapsid IgG (anti-N, Abbott) and anti-spike IgG (anti-S1, EUROIMMUN). Chart review was conducted for samples testing positive or borderline on either assay, and for an age/sex-matched cohort of samples negative by both assays. CDC surveillance case definitions were used to determine clinical sensitivity/specificity and conduct receiver operating characteristics curve analysis.
Results
There were 52 samples positive by both methods, 2 positive for anti-N only, 34 positive for anti-S1 only, and 27 borderline for anti-S1. Of the 34 individuals positive for anti-S1 alone, 8 (24%) had confirmed COVID-19. No anti-S1 borderline cases were positive for anti-N or had confirmed/probable COVID-19. The anti-N assay was less sensitive (84.2% [95% CI 72.1-92.5%] vs 94.7% [95% CI 85.4-98.9%]) but more specific (99.2% [95% CI 95.5-100%] vs 86.9% [95% CI 79.6-92.3%]) than anti-S1. Abbott anti-N sensitivity could be improved to 96.5% with minimal effect on specificity if the index threshold was lowered from 1.4 to 0.6.
Conclusion
Real-world concordance between different serologic assays may be lower than previously described in retrospective studies. These findings have implications for the interpretation of SARS-CoV-2 IgG results, especially with the advent of spike antigen-targeted vaccination, as a subset of patients with true infection are anti-N negative and anti-S1 positive.
Keywords: SARS-CoV-2, COVID-19, serology
Introduction
At the conclusion of 2019, the emergence of a novel coronavirus, severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), brought about the coronavirus disease 2019 (COVID-19) pandemic, resulting in devastating loss of life and disruption in the fabric of global societies. Over the course of the pandemic, substantial progress has been made in understanding the humoral response to infection with SARS-CoV-2, with IgM, IgG, and IgA antibodies specific for various SARS-CoV-2 antigens. These include the S1 domain and receptor binding domain (RBD) of the spike protein, as well as nucleocapsid protein (N), which become detectable at a median of approximately 2 weeks after onset of symptoms (1, 2). Antibody titers peak at 1 month post symptom onset, with levels directly correlating with severity of illness. Titers then begin to decrease, relatively rapidly for IgM and IgA, and more gradually for IgG, although the ultimate duration of SARS-CoV-2 antibody responses remains an area of active investigation (3). SARS-CoV-2 vaccines for which data are available elicit robust antibody responses, and licensing studies so far have demonstrated short-term protection from natural infection (4–6).
SARS-CoV-2 antibody testing plays an important complementary role in COVID-19 diagnosis. Specifically, antibody testing is used to evaluate patients with a high clinical suspicion of infection and repeatedly negative nucleic acid amplification tests (NAATs), as well as in the assessment of suspected multisystem inflammatory syndrome in children (7, 8). SARS-CoV-2 antibody testing is also a critical public health tool, enabling surveillance efforts to characterize seroprevalence and inform policy decisions. Finally, SARS-CoV-2 antibody testing may be used to monitor the humoral response to vaccines, and potentially to differentiate humoral responses due to natural infection (i.e., anti-N and anti-S) from those due to vaccination with spike-only vaccines (anti-S only), though the United States Centers for Disease Control and Prevention (CDC) does not currently recommend use of serologic assays for this purpose.
Laboratory-based methods for SARS-CoV-2 antibody detection include enzyme-linked immunosorbent assays (ELISA) and chemiluminescent immunoassays (CLIA). Among the most common of these assays are the Abbott anti-nucleocapsid antigen IgG CLIA (anti-N) and EUROIMMUN anti-S1 domain spike protein IgG ELISA (anti-S1) (9, 10). While numerous diagnostic accuracy studies have evaluated the comparative performance of the EUROIMMUN and Abbott SARS-CoV-2 IgG immunoassays (11–20), these studies rely on retrospective, nonconsecutive samples, collected prior to the pandemic or selected from individuals with known SARS-CoV-2 NAAT results. Furthermore, they offer minimal clinical information to guide laboratorians and clinicians in the interpretation of discordant results.
To address the limitations of these retrospective studies, this work prospectively assessed 1080 consecutive clinical samples tested in parallel with the Abbott anti-N and EUROIMMUN anti-S1 assays. A case-control study design was used to clinically characterize individuals with discrepant results. Diagnostic accuracy was then determined based on the CDC surveillance case definitions (21).
Materials and Methods
Clinical Specimens
The Stanford Clinical Virology and Special Chemistry Laboratories receive specimens from tertiary-care academic hospitals and affiliated outpatient facilities in the San Francisco Bay Area, California. From August 3 to August 15, 2020, all plasma samples submitted for clinical SARS-CoV-2 IgG testing were prospectively tested in parallel on 2 different SARS-CoV-2 IgG testing platforms. The purpose of this parallel testing was to assess concordance in anticipation of switching assays used in our clinical practice. Venipuncture blood samples submitted for testing were collected in lithium heparin-coated vacutainers from asymptomatic, symptomatic, and convalescent inpatients and outpatients, either for clinical care, or in the context of COVID-19 related epidemiologic surveillance studies and drug trials at our institution. Generally, asymptomatic individuals with no history of exposure were offered serologic testing in addition to NAAT prior to oncologic care or planned procedures, or as part of employer/school-mandated screening protocols. Symptomatic individuals and those with history of exposure may have been initially offered serologic testing in addition to NAAT as an adjunctive diagnostic tool, and in the weeks to months following diagnosis for the purpose of immunologic surveillance. No individuals were known to be enrolled in vaccine trials, and only 1 individual received convalescent plasma prior to serologic testing.
Commercially Available Serologic Assays
After centrifugation, plasma samples were tested for both anti-N IgG and anti-S1 IgG antibodies on commercially available Architect (Abbott) and EUROLabWorkstation (EUROIMMUN) platforms, respectively. The Abbott anti-N IgG assay is an automated 2-step CLIA conducted and interpreted according to manufacturer guidelines (22). A sample-to-calibrator relative light unit index of ≥1.4 is considered positive, while an index value of <1.4 is considered negative. The EUROIMMUN anti-S1 IgG assay is an ELISA conducted and interpreted according to manufacturer guidelines (23). A sample-to-calibrator optical density (OD) ratio of ≥1.1 is considered positive, while a ratio of ≥0.8 to <1.1 is considered borderline, and a ratio of <0.8 is considered negative. Samples were tested on both platforms independently and in parallel within 24 h of each other, without prior knowledge of results on the other platform.
Laboratory-Developed Serologic Assays
Specimens with sufficient volume that were positive or borderline for either anti-N or anti-S1 IgG antibodies were further evaluated using a laboratory-developed ELISA designed to detect human IgG antibodies to the SARS-CoV-2 spike protein RBD. The ELISA was performed on the ESP 600 ELISA instrument (Inova Diagnostics) using 96-well Corning Costar high binding plates (Thermo Fisher) coated with recombinant SARS-CoV-2 RBD proteins produced and purified as previously described (1). In brief, plates are coated with RBD protein at a concentration of 0.1 µg per well overnight at 4 °C and incubated with plasma at a 1:100 dilution, with secondary detection by horseradish peroxidase conjugated goat anti-human IgG at a 1:6,000 dilution (Thermo Fisher). An OD at 450 nm of ≥0.3 is interpreted as positive.
These specimens were additionally evaluated for the ability of anti-RBD IgG present in the sample to block binding between purified RBD protein and recombinant human angiotensin-converting enzyme 2 (ACE2) receptor using a laboratory-developed competition ELISA, performed on the ESP 600 instrument as previously described (1). In brief, RBD-coated plates are incubated with plasma at a 1:10 dilution, recombinant ACE2 joined to a mouse IgG2a Fc (ACE2-mFc) is added at 0.5 µg/mL, and secondary detection is performed using horseradish peroxidase conjugated goat anti-mouse IgG at a 1:10,000 dilution. Samples are run in duplicate, and average OD at 450 nm is used to calculate the sample-to-negative calibrator ratio. Blocking activity is reported as a percentage as follows: (1-ratio) x 100, with higher percentages corresponding to lower levels of RBD-ACE2 binding.
SARS-CoV-2 anti-RBD IgM testing was only performed when ordered for clinical purposes. Testing was conducted on the ESP 600 instrument in an identical manner to anti-RBD IgG testing as described above, but with secondary detection by horseradish peroxidase conjugated goat anti-human IgM at a 1:6,000 dilution (Sigma) (1). Due to the observation of nonspecific plastic-binding antibodies in some individuals, a PBS-coated control plate was run in tandem with the RBD-coated plate. Samples were only reported as positive for IgM if the OD at 450 nm was ≥0.4, and the OD was higher than that of the uncoated control plate.
Respiratory Sample Nucleic Acid Amplification Tests
Respiratory sample NAAT results reported in this study were performed as part of routine clinical care. These tests were conducted using a variety of methods including a lab-developed reverse transcription quantitative polymerase chain reaction (RT-qPCR) targeting the envelope gene on the Rotor-Gene Q (Qiagen), as well as commercially available RT-qPCR or transcription mediated amplification methods on the Panther Fusion or Aptima platforms, respectively, (Hologic) targeting open reading frame 1ab (24–28). Cycle threshold (Ct) values are reported only for RT-qPCR testing.
Clinical Data and Statistical Analysis
Five physicians conducted retrospective electronic medical record review, each for a different subset of cases (individuals with samples testing positive or borderline on either IgG assay), as well as for a selected cohort of matched controls (individuals testing negative on both assays). Each positive or borderline case was matched to a single negative control on the basis of age, sex, and availability of demographic data using an optimal matching algorithm through R package “MatchIt” (29). Standardized differences were used to assess balance between matched cohorts.
The Research Electronic Data Capture (REDCap) platform was used to collect and manage demographic data, history of present illness, unplanned hospital or intensive care unit (ICU) admission, and relevant laboratory information for each case that were entered into the electronic medical record between March 1, 2020 and the date of sample collection. CDC COVID-19 case surveillance definitions were used to classify cases as confirmed, probable, or suspect (21). Per these definitions, symptomatic individuals showed either 1) at least 2 of the following: fever, chills, rigors, myalgia, headache, sore throat, nausea or vomiting, diarrhea, fatigue, congestion or runny nose, 2) any one of the following: cough, shortness of breath, difficulty breathing, anosmia, ageusia, or 3) clinical or radiographic evidence of pneumonia or acute respiratory distress. Exposures were defined as close contact with an individual with confirmed or probable COVID-19 documented in the medical record. Confirmed cases were defined as having had a positive NAAT. Probable cases were defined as those without positive NAATs who were both symptomatic without a more likely alternate diagnosis and had evidence of exposure. Suspect cases were defined as those positive or borderline by either antibody assay without documented positive NAAT, symptoms, or evidence of exposure as outlined above. This study was conducted with Stanford institutional review board approval (protocol 48973) and individual consent was waived.
Agreement, sensitivity, specificity, and accuracy statistics were reported with exact (Clopper-Pearson) 95% confidence intervals (CI) (30). Positive and negative likelihood ratio 95% CIs were calculated using the log method (30). Receiver operating characteristic (ROC) curve analysis was conducted using R package “pROC” (31). Clinical performance characteristics and ROC curve analysis use the CDC surveillance case definitions of confirmed or probable cases as “disease positive,” with assumption of 100% seroconversion.
Results
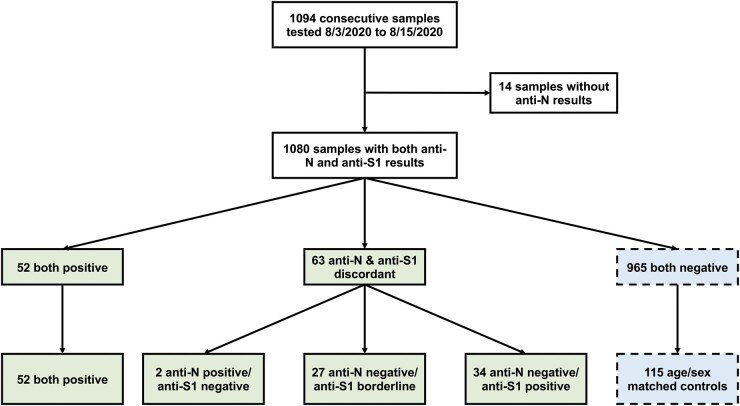
From August 3 to August 15, 2020, 1094 plasma samples were tested for presence of anti-SARS-CoV-2 IgG. Fourteen samples with insufficient volume to perform testing for both anti-N (Abbott Architect) and anti-S1 (EUROIMMUN) were excluded from subsequent analysis. There were 52 samples positive by both methods, 63 samples with discordant anti-N and anti-S1 results, and 965 samples negative by both methods (Fig. 1). Anti-N was negative in 40% (34/86) of cases considered positive for anti-S1 (index ≥1.1), and in 100% (27/27) of the cases considered borderline by anti-S1 (index between 0.8 and 1.1) (Supplemental Table 1). In contrast, anti-S1 was negative in only 4% (2/54) of cases considered positive by anti-N, as reflected by the change in positive percent agreement depending on which assay was considered the reference method (Table 1).
Fig. 1.
Flow chart of consecutive plasma samples tested for SARS-CoV-2 IgG. The 115 samples positive/borderline for either or both Abbott anti-nucleocapsid IgG (anti-N) and EUROIMMUN anti-spike protein IgG (anti-S1) were all included as cases. Out of the 965 samples negative by both assays, 115 controls were selected for chart review based on one-to-one matching for availability of demographic data, age, and sex (see Supplemental Table 2).
Table 1.
Agreement between Abbott’s anti-N and EUROIMMUN anti-S1 IgG assays from August 3 to August 15, 2020 (n = 1080).
| Statistic | Treating anti-S1 borderline as negative | Treating anti-S1 borderline as positive |
|---|---|---|
| PPA (anti-S1 as reference) | 60.5% (49.3%–70.9%) | 46.0% (36.6%–55.7%) |
| PPA (anti-N as reference) | 96.3% (87.3%–99.6%) | 96.3% (87.3%–99.6%) |
| NPA (anti-S1 as reference) | 99.8% (99.3%–100.0%) | 99.8% (99.3%–100.0%) |
| NPA (anti-N as reference) | 96.7% (95.4%–97.7%) | 94.1% (92.4%–95.4%) |
| Overall % Agreement | 96.7% (95.4%–97.7%) | 94.2% (92.6%–95.5%) |
| Cohen’s Kappa | 0.73 (0.64–0.81) | 0.60 (0.51–0.68) |
95% confidence intervals are provided in parentheses.
PPA, positive percent agreement; NPA, negative percent agreement; S1, spike protein; N, nucleocapsid protein.
Individuals dual positive by anti-N and anti-S1 were more likely to have history of exposure, symptoms, hospital admission, positive respiratory NAAT, and positive IgM, when compared to matched dual negative controls (Table 2, Supplemental Table 2). Among samples with sufficient volume, our laboratory-developed anti-spike RBD IgG assay was positive in 100% (31/31) of tested samples dual positive for anti-N and anti-S1, and in 4% (1/25) of samples positive only for anti-S1, but negative in both samples positive only for anti-N (Supplemental Fig. 1). The single patient receiving convalescent plasma was dual positive. Of the 52 total dual anti-N/anti-S1 positive specimens, 14 had ≥50% blocking activity of ACE2 receptor binding; these individuals had a median anti-N IgG index of 6.9 [interquartile range (IQR) 5.8–7.2] and median anti-S1 IgG index of 12.4 (IQR 10.7–15.2). In contrast, for the 38 dual anti-N/anti-S1 positive individuals with <50% blocking activity, the median anti-N index was 3.6 (1.9–5.6), and median anti-S1 index was 5.3 (2.8-7.8).
Table 2.
Epidemiologic, clinical, and laboratory data for cases vs controls (n = 230).
| Both positive | Anti-N only | Anti-S1 only |
Both negative | ||
|---|---|---|---|---|---|
| Positive | Positive | Borderline | |||
| Clinical data available | 73.1% (38/52) | 100% (2/2) | 67.6% (23/34) | 66.7% (18/27) | 77.4% (89/115) |
| Any exposure | 42.1% (16/38) | 100% (2/2) | 13.0% (3/23) | 22.2% (4/18) | 7.9% (7/89) |
| Household exposure | 21.1% (8/38) | 0.0% (0/2) | 8.7% (2/23) | 16.7% (3/18) | 1.1% (1/89) |
| Symptomatica | 76.3% (29/38) | 50.0% (1/2) | 56.5% (13/23) | 27.8% (5/18) | 28.1% (25/89) |
| Symptomatic no alt dxb | 63.2% (24/38) | 50.0% (1/2) | 34.8% (8/23) | 11.1% (2/18) | 13.5% (12/89) |
| Days from symptom onsetc | 34 (25-48) | 12 | 82.5 (32-149) | 121 | 144 (60-147) |
| Hospital admission | 23.7% (9/38) | 0.0% (0/2) | 13.0% (3/23) | 0.0% (0/18) | 0.0% (89/89) |
| ICU admission | 10.5% (4/38) | 0.0% (0/2) | 4.4% (1/23) | 0.0% (0/18) | 0.0% (89/89) |
| NAAT performed | 100.0% (52/52) | 100.0% (2/2) | 100.0% (34/34) | 81.5% (22/27) | 85.2% (98/115) |
| NAAT positive | 86.5% (45/52) | 100.0% (2/2) | 23.5% (8/34) | 0.0% (0/22) | 1.0% (1/98) |
| Days from NAATd | 31 (25-45) | 21 (11-31) | 30 (14-118) | … | 22 |
| RT-qPCR Ct valuee | 22.4 (16.9-32.6) | 24.1 | 19.9 (15.7-28.4) | … | 38.2 |
| RBD performed | 59.6% (31/52) | 100.0% (2/2) | 73.5% (25/34) | 92.6% (25/27) | 0.0% (0/115) |
| RBD positive | 100.0% (31/31) | 0% (0/2) | 4.0% (1/25) | 4.0% (1/25) | … |
| RBD IgG indexf | 1.2 (0.7-1.7) | … | 0.4 | 0.5 | … |
| Blocking performed | 100.0% (52/52) | 100.0% (2/2) | 29.4% (10/34) | 3.7% (1/27) | 0.0% (0/115) |
| Percent blockingg | 9% (1%-51%) | 3% (1.0-4.0%) | 4% (0-11%) | 0% | … |
| IgM performed | 32.7% (17/52) | 0.0% (0/2) | 50.0% (17/34) | 40.7% (11/27) | 53.0% (61/115) |
| IgM positive | 52.9% (9/17) | … | 0.0% (0/17) | 0.0% (0/11) | 0.0% (0/61) |
| IgM indexf | 1.1 (0.6-1.7) | … | … | … | … |
| Confirmed or probable case | 97.9% (46/47) | 100.0% (2/2) | 34.8% (8/23) | 0.0% (0/18) | 1.1% (1/89) |
Categorical data presented as % (numerator/denominator); continuous data presented as median (interquartile range).
S1, spike protein; N, nucleocapsid protein; ICU, intensive care unit; NAAT, nucleic acid amplification test; RT-qPCR, reverse-transcription quantitative polymerase chain reaction; Ct, cycle threshold; RBD, spike protein receptor-binding domain; ACE2, angiotensin converting enzyme 2.
Individuals with symptoms satisfying CDC clinical criteria, without consideration of whether symptoms were most likely due to COVID-19 vs other causes (21).
Individuals with symptoms satisfying CDC clinical criteria for COVID-19 in the absence of a more likely diagnosis (21).
Symptom onset date available for n = 23, 1, 8, 1, and 8 individual(s) from left to right columns.
Days from first positive respiratory NAAT.
Ct value data available for n = 32, 1, 5, 0, and 1 individual(s) from left to right columns.
IgG and IgM index value summary statistics are calculated from positive results only.
Indicates the percentage of blocking activity the RBD IgG antibody has against the ACE2 receptor. Higher values correspond to a stronger blocking response.
The 2 individuals positive only for anti-N both had positive NAATs. One was symptomatic with serology sample drawn 12 days after symptom onset. The other was asymptomatic, with serology sample drawn 31 days after positive NAAT. In contrast, among 34 individuals positive for anti-S1 alone, only 8 (24%) had confirmed COVID-19. Specimen collection for these 8 individuals was more likely to have occurred further from symptom onset than those dual-positive, with median of 83 days (IQR 32–149) vs 34 days (IQR 25–48). Most of these individuals had anti-N IgG index values approaching the threshold value of 1.4 (Fig. 2). None of the individuals with borderline anti-S1 index values were NAAT-positive or met probable case criteria.
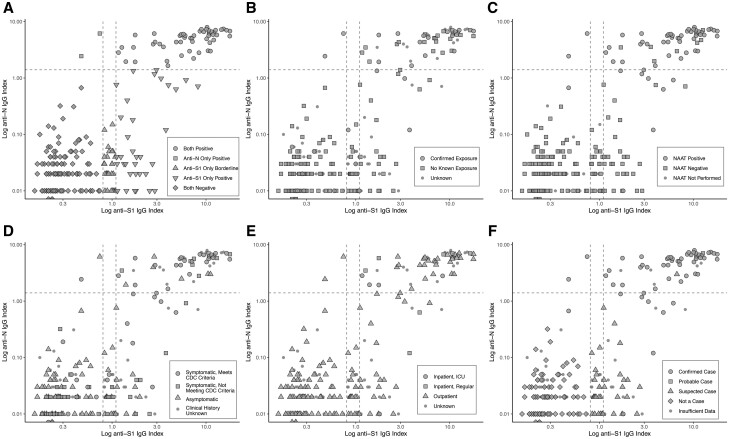
Fig. 2.
Log-transformed Abbott anti-nucleocapsid IgG (anti-N) index value vs EUROIMMUN anti-spike protein IgG (anti-S1) optical density ratio for the age and sex-matched cases/controls (n = 230). The horizontal dashed line represents the anti-N threshold of positivity (1.4); the 2 vertical dashed lines represent the anti-S1 threshold for borderline (0.8) and positive (1.1), respectively. The top right corner contains dual-positive samples, the bottom left corner contains dual-negative samples, and the remaining points represent samples with discordant results. The different colors/shapes represent A) IgG results, B) exposure history, C) respiratory nucleic acid amplification test (NAAT) results, D) symptom history according to Centers for Disease Control (CDC) clinical case criteria, E) inpatient and/or intensive care unit (ICU) admission history, F) case classification based on CDC surveillance case definitions.
There were 179 individuals with adequate available clinical information to classify as confirmed (n = 56), probable (n = 1), suspect (n = 34), or noncases (n = 88) using CDC criteria. The single probable case was a NAAT-negative individual who was symptomatic, had exposure, and was later found to be IgM-positive. Among the confirmed and probable COVID-19 cases, the 2 patients positive only for anti-N were observed within the first 31 days (Supplemental Fig. 2A–C), while the 8 patients positive only for anti-S1 were observed 0–145 days after diagnosis. Among symptomatic patients not meeting confirmed or probable CDC criteria, none were positive for anti-N only, whereas 7 and 5 were positive and borderline for anti-S1 only, respectively (Supplemental Fig. 2D–F).
When considering only confirmed and probable cases as “disease positive,” the Abbott anti-N assay was less sensitive (84.2% vs 94.7%), but more specific (99.2% vs 86.9%) than the EUROIMMUN anti-S1 assay if borderline cases were interpreted as negative (Table 3). Considering borderline anti-S1 results as positive decreased the test specificity (from 86.9% to 72.1%) without any gain in sensitivity. ROC curve analysis demonstrated that decreasing the Abbott anti-N assay index value threshold from 1.4 to 0.6 would increase the sensitivity (from 84.2% to 96.5%) without appreciably decreasing specificity (from 99.2% to 98.4%) (Fig. 3). If this decreased threshold of 0.6 were applied to the 1080 individuals in this study, 14 (1.3%) would be reclassified from anti-N negative to positive, with 8 (0.7%) additional true positives and 3 (0.3%) false positives, and 3 (0.3%) with insufficient clinical data to assess CDC criteria (2 of 3 had positive EUROIMMUN IgG indices ≥4).
Table 3.
Clinical sensitivity and specificity of Abbott’s anti-N and EUROIMMUN’s anti-S1 IgG assays for confirmed or probable COVID-19 case definitions by CDC criteria (n = 179).
| Statistic | Anti-N | Anti-S1, borderline as negative | Anti-S1, borderline as positive |
|---|---|---|---|
| Sensitivity | 84.2% (72.1%–92.5%) | 94.7% (85.4%–98.9%) | 94.7% (85.4%–98.9%) |
| Specificity | 99.2% (95.5%–100%) | 86.9% (79.6%–92.3%) | 72.1% (63.3%–79.9%) |
| Positive likelihood ratio | 102.7 (14.5–725.9) | 7.2 (4.6–11.5) | 3.4 (2.5–4.6) |
| Negative likelihood ratio | 0.16 (0.09–0.29) | 0.06 (0.02–0.18) | 0.07 (0.02–0.22) |
95% confidence intervals are provided in parentheses.
N, nucleocapsid protein; S1, spike protein.
Fig. 3.
Receiver operating curve characteristics for A-B) Abbott anti-nucleocapsid IgG (anti-N) and C-D) EUROIMMUN anti-spike protein IgG (anti-S1). While the 2 assays have similar areas under the curve (AUCs) with overlapping 95% confidence intervals (95% CI), the anti-N assay has a more favorable sensitivity (solid red line) vs specificity (dashed blue line) profile as compared to the anti-S1 assay. The Abbott anti-N index threshold for positivity could be lowered from 1.4 to 0.6 to improve sensitivity without a substantial decrement to specificity.
Discussion
There are currently over 60 available FDA emergency use authorized antibody assays for the diagnosis of prior COVID-19 infection. These assays use a variety of methodologies, differing antigen targets, and have widely variable performance characteristics (18). However, little evidence exists to guide laboratorians and clinicians in reporting and interpreting discordant results.
In this prospective study of 1080 consecutive plasma samples concurrently tested by the Abbott anti-N and EUROIMMUN anti-S1 assays, almost 6% of samples had discordant results. These 2 platforms were selected for clinical testing and comparison in our laboratory based upon their high-throughput capacity, widespread availability in the United States, and differing methodology (ELISA and CLIA). Notably, it was more common for a sample to be positive/borderline on only 1 assay than to be positive by both commercial assays. This finding contrasts with a prior study which reported higher agreement (Cohen’s Kappa of 0.83) between these 2 specific platforms (17). Similarly, many studies have reported relatively high concordance between anti-N and anti-S1 assays (3, 12, 16, 32).
These differences in reported interassay concordance may be due to our prospective study design, which allowed for unbiased evaluation of consecutive samples from patients whose physicians had initiated serologic testing. In contrast, almost all prior validation, seroprevalence, and method comparison studies on antibody assays have used nonconsecutive, selected known NAAT-positive or pre-pandemic samples (3, 12, 13, 16, 19, 33–35). Such samples are more likely to be drawn from patients with well-characterized states of health or infection, and are less likely to include patients with ambiguous presentations or nonspecific symptoms that are included in routine clinical testing. Accordingly, previously-reported interassay concordance from selected nonconsecutive specimens may be falsely high and less generalizable to the clinically-tested population.
We also considered the possibility that our inclusion of samples drawn more than 1 month after diagnosis might have impacted concordance; however, our data showed no clear time-dependent relationship between concordance and days from diagnosis. Prior studies have also reported mixed results regarding anti-S1 vs anti-N persistence (17, 36–38). Further studies will be required to determine why some individuals mount only an anti-S1 or anti-N response, or whether the ratio of such a response is clinically important.
Although several method comparison studies have reported lower agreement between anti-N and anti-S1 serologic assays, no clinical data for discrepant samples were provided (13, 19). In this study, only 2 individuals had positive anti-N with negative anti-S1, and both had prior positive NAATs. This is consistent with previous reports of high specificity (>99%) for the Abbott anti-N assay (33, 34). Additionally, we observed that decreasing the Abbott anti-N index value threshold from 1.4 to 0.6 offered increased sensitivity (from 84.2% to 96.5%) with minimal decrement in specificity (from 99.2% to 98.4%). This finding corroborates prior studies reporting an optimal cutoff between 0.55–0.8 (15, 33, 39). Increased serologic sensitivity may be justified in the diagnosis of patients with late symptomatic presentation and negative NAAT during times of high prevalence. However, even small decrements in specificity have the potential to misclassify patients, and current Infectious Diseases Society of America and CDC guidelines for serologic testing recommend assays with ≥99.5% specificity in low-prevalence settings.
In contrast, only one-quarter of the individuals positive for anti-S1 IgG and negative for anti-N IgG represented confirmed or probable cases of COVID-19 per CDC criteria. Accordingly, the EUROIMMUN anti-S1 assay specificity observed was lower than in prior retrospective studies, even when interpreting borderline specimens as negative (17, 35). However, a non-negligible proportion of individuals positive for only anti-S1 were confirmed or probable COVID-19 cases. As such, qualitative detection of anti-S1 without anti-N should not be considered a specific marker for immunization. The CDC does not currently recommend serologic testing for assessment or confirmation of vaccination status, and our findings demonstrate that such testing with these 2 methods would have low specificity. This is particularly relevant as vaccines become more widely available (4, 5).
An area of uncertainty is the degree to which anti-S1 and anti-N antibodies confer immunity, as median percent blocking was low even in concordant specimens. Previous data suggest that anti-RBD antibody titers are correlated with neutralization and ACE2 blocking activity (1); in the present study, 100% of concordant specimens were positive for anti-RBD, whereas 0% and 4% of anti-N and anti-S1 only specimens were positive for anti-RBD, respectively. However, it is unclear how our laboratory-developed anti-RBD assay might perform against other commercially available anti-N or anti-S assays.
One major strength of this study is the mitigation of selection biases associated with comparing specimens from well-characterized disease states through concurrent anti-N and anti-S1 clinical serologic testing on a large consecutive cohort of patients. Additional strengths include the incorporation of descriptive clinical/exposure data and use of combined clinical, epidemiologic linkage, and laboratory evidence to define cases. These findings may be of practical interest to laboratories switching serologic assays or offering both anti-N and anti-S1 assays as vaccines become more widely available.
Limitations of this study include the comparison of only 2 assays over a relatively short duration, where antibodies to 2 different antigens (N and S1) were tested with different assay methods (CLIA vs ELISA), limiting our ability to determine whether differences in assay performance are related to the antigen or to the assay method. Indeed, these findings may not be generalizable to other assays, including OrthoClinical or DiaSorin, which were used by approximately twice as many labs as EUROIMMUN according to a 2020 College of American Pathologists survey (9). For example, heterogeneity in performance has been reported among anti-S assays, with EUROIMMUN generally demonstrating lower sensitivity and/or specificity than its counterparts in prior retrospective studies of well-characterized specimens (16, 20, 40).
An additional limitation of our approach is that it does not fully address the challenge posed by potential asymptomatic infection cases, which would have led to an underestimation of anti-S1 assay sensitivity. The individuals who were positive for anti-S1 and negative for anti-N but did not meet CDC case definitions could have, in fact, truly been infected with SARS-CoV-2, and either have been asymptomatic, or had poor documentation in the electronic medical record. Lastly, this consecutive cohort of tested patients received care at a single high-resource tertiary-care institution with a relatively high prevalence of complex medical comorbidities, which may impact clinician ordering practices and serologic status. These findings may therefore not be generalizable to health systems with differing serologic testing policies or patient populations. We expect that the advent of vaccination will also markedly impact assay concordance.
In this large single-institutional case-control study of consecutive specimens tested concurrently with the EUROIMMUN anti-S1 ELISA and the Abbott Architect anti-N CLIA, we observed that discordant results were more common than concordant positive results, in contrast to many methodological comparison studies with selected well-characterized specimens. Based on our findings, low-positive EUROIMMUN anti-S1 results should be interpreted with caution by laboratorians, whereas borderline EUROIMMUN anti-S1 results should be considered negative. The Abbott anti-N assay threshold could be lowered to 0.6 if maximization of sensitivity is desired. Review of clinical data in conjunction with serologic testing is advisable for adjudication, and laboratorians should be prepared to interpret discrepant results to minimize patient and clinician uncertainty. These findings may be particularly relevant with the availability of spike-targeted vaccination, as laboratories may be asked to distinguish between past infection and vaccine-derived immunity.
Supplemental Material
Supplemental material is available at Clinical Chemistry online.
Supplementary Material
Nonstandard Abbreviations:
- SARS-CoV-2
severe acute respiratory syndrome coronavirus-2
- COVID-19
coronavirus disease 2019
- RBD
receptor binding domain
- N
nucleocapsid protein
- S
spike protein
- NAAT
nucleic acid amplification test
- ELISA
enzyme-linked immunosorbent assay
- CLIA
chemiluminescent immunoassay
- anti-N
anti-nucleocapsid antigen IgG
- anti-S1
anti-S1 domain spike protein IgG
- CDC
United State Centers for Disease Control and Prevention
- ACE2
human angiotensin-converting enzyme 2
- RT-qPCR
reverse transcription quantitative polymerase chain reaction
- Ct
cycle threshold
- REDCap
Research Electronic Data Capture platform
- ICU
intensive care unit
- PPA
positive percent agreement
- NPA
negative percent agreement
- CI
confidence interval
- ROC
receiver operating characteristic
- IQR
interquartile range
Author Contributions
All authors confirmed they have contributed to the intellectual content of this paper and have met the following 4 requirements: (a) significant contributions to the conception and design, acquisition of data, or analysis and interpretation of data; (b) drafting or revising the article for intellectual content; (c) final approval of the published article; and (d) agreement to be accountable for all aspects of the article thus ensuring that questions related to the accuracy or integrity of any part of the article are appropriately investigated and resolved.
H. Wang, statistical analysis; R.Z. Shi, administrative support, provision of study material or patients; S.D. Boyd, financial support.
Authors’ Disclosures or Potential Conflicts of Interest
Upon manuscript submission, all authors completed the author disclosure form. Disclosures and/or potential conflicts of interest:
Employment or Leadership
None declared.
Consultant or Advisory Role
S.D. Boyd, Regeneron, Sanofi, Novartis.
Stock Ownership
S.D. Boyd, AbCellera.
Honoraria
None declared.
Research Funding
The Stanford REDCap platform (http://redcap.stanford.edu) is developed and operated by Stanford Medicine Research IT team. The REDCap platform services at Stanford are subsidized by a) Stanford School of Medicine Research Office, and b) the National Center for Research Resources and the National Center for Advancing Translational Sciences, National Institutes of Health, through grant UL1 TR001085. Portions of this work were supported by NIH/NIAID R01AI127877 (S.D. Boyd), NIH/NIAID R01AI130398 (S.D. Boyd), NIH 1U54CA260517 (S.D. Boyd and B.A. Pinsky), an endowment from the Crown Family Foundation (S.D. Boyd), and a Coulter COVID-19 Rapid Response Award (S.D. Boyd). B.A. Pinsky, Abbott Diagnostics.
Expert Testimony
None declared.
Patents
S.D. Boyd, provisional patent applications for COVID-19 antibody tests, no patents awarded.
Role of Sponsor
The funding organizations played no role in the design of study, choice of enrolled patients, review and interpretation of data, preparation of manuscript, or final approval of manuscript.
Acknowledgments:
We are grateful to the Stanford Clinical Virology and Special Chemistry Laboratory staff for their hard work on the front lines and resilience in the face of the unprecedented challenges presented by the COVID-19 pandemic.
References
- 1. Röltgen K, Powell AE, Wirz OF, Stevens BA, Hogan CA, Najeeb J, et al. Defining the features and duration of antibody responses to SARS-CoV-2 infection associated with disease severity and outcome. Sci Immunol 2020;5:eabe0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Long Q-X, Liu B-Z, Deng H-J, Wu G-C, Deng K, Chen Y-K, et al. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat Med 2020;26:845–8. [DOI] [PubMed] [Google Scholar]
- 3. Gudbjartsson DF, Norddahl GL, Melsted P, Gunnarsdottir K, Holm H, Eythorsson E, et al. Humoral immune response to SARS-CoV-2 in Iceland. N Engl J Med 2020;383:1724–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. ; C4591001 Clinical Trial Group. Safety and efficacy of the BNT162b2 mRNA Covid-19 vaccine. N Engl J Med 2020;383:2603–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, et al. ; Oxford COVID Vaccine Trial Group. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet 2021;397:99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Widge AT, Rouphael NG, Jackson LA, Anderson EJ, Roberts PC, Makhene M, et al. ; mRNA-1273 Study Group. Durability of responses after SARS-CoV-2 mRNA-1273 vaccination [Letter]. N Engl J Med 2021;384:80–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hanson KE, Caliendo AM, Arias CA, Englund JA, Lee MJ, Loeb M, et al. Infectious Diseases Society of America Guidelines on the Diagnosis of COVID-19: serologic testing. Clin Infect Dis 2020;doi:10.1093/cid/ciaa1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Theel ES, Slev P, Wheeler S, Couturier MR, Wong SJ, Kadkhoda K.. The role of antibody testing for SARS-CoV-2: is there one? [Commentary]. J Clin Microbiol 2020;58:e00797–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.College of American Pathologists. SARS-CoV-2 Serology Survey Results. CAP Proficiency Testing Program. 2020.
- 10. Haselmann V, Özçürümez MK, Klawonn F, Ast V, Gerhards C, Eichner R, et al. Results of the first pilot external quality assessment (EQA) scheme for anti-SARS-CoV2-antibody testing. Clin Chem Lab Med 2020;58:2121–30. [DOI] [PubMed] [Google Scholar]
- 11. Jääskeläinen A, Kuivanen S, Kekäläinen E, Ahava M, Loginov R, Kallio-Kokko H, et al. Performance of six SARS-CoV-2 immunoassays in comparison with microneutralisation. J Clin Virol 2020;129:104512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Prince HE, Givens TS, Lapé-Nixon M, Clarke NJ, Schwab DA, Batterman HJ, et al. Detection of SARS-CoV-2 IgG targeting nucleocapsid or spike protein by four high-throughput immunoassays authorized for emergency use. J Clin Microbiol 2020;58:e01742–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Perkmann T, Perkmann-Nagele N, Breyer MK, Breyer-Kohansal R, Burghuber OC, Hartl S, et al. Side-by-side comparison of three fully automated SARS-CoV-2 antibody assays with a focus on specificity. Clin Chem 2020;66:1405–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nicol T, Lefeuvre C, Serri O, Pivert A, Joubaud F, Dubée V, et al. Assessment of SARS-CoV-2 serological tests for the diagnosis of COVID-19 through the evaluation of three immunoassays: Two automated immunoassays (Euroimmun and Abbott) and one rapid lateral flow immunoassay (NG Biotech). J Clin Virol 2020;129:104511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lau CS, Oh HML, Hoo SP, Liang YL, Phua SK, Aw TC.. Performance of an automated chemiluminescence SARS-CoV-2 IG-G assay. Clin Chim Acta 2020;510:760–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Theel ES, Harring J, Hilgart H, Granger D.. Performance characteristics of four high-throughput immunoassays for detection of IgG antibodies against SARS-CoV-2. J Clin Microbiol 2020;58:e01243–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tang MS, Hock KG, Logsdon NM, Hayes JE, Gronowski AM, Anderson NW, et al. Clinical performance of two SARS-CoV-2 serologic assays. Clin Chem 2020;66:1055–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lisboa Bastos M, Tavaziva G, Abidi SK, Campbell JR, Haraoui L-P, Johnston JC, et al. Diagnostic accuracy of serological tests for COVID-19: systematic review and meta-analysis. BMJ 2020;370:m2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grzelak L, Temmam S, Planchais C, Demeret C, Tondeur L, Huon C, et al. A comparison of four serological assays for detecting anti–SARS-CoV-2 antibodies in human serum samples from different populations. Sci Transl Med 2020;12:eabc3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Charlton CL, Kanji JN, Johal K, Bailey A, Plitt SS, MacDonald C, et al. Evaluation of six commercial mid- to high-volume antibody and six point-of-care lateral flow assays for detection of SARS-CoV-2 antibodies. J Clin Microbiol 2020;58:e01361–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.U.S. Centers for Disease Control and Prevention. Coronavirus Disease 2019 (COVID-19) 2020 Interim Case Definition, Approved August 5, 2020. https://wwwn.cdc.gov/nndss/conditions/coronavirus-disease-2019-covid-19/case-definition/2020/08/05/ (Accessed November 2020).
- 22.Abbott. Food and Drug Administration Emergency Use Authorization: SARS-CoV-2 IgG Architect. https://www.fda.gov/media/137383/download (Accessed November 2020).
- 23.EUROIMMUN. Food and Drug Administration Emergency Use Authorization: Anti-SARS-CoV-2 ELISA (IgG). https://www.fda.gov/media/137609/download (Accessed November 2020).
- 24. Bulterys PL, Garamani N, Stevens B, Sahoo MK, Huang CH, Hogan CA, et al. Comparison of a laboratory-developed test targeting the envelope gene with three nucleic acid amplification tests for detection of SARS-CoV-2. J Clin Virol 2020;129:104427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, et al. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance 2020;25:2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stanford Health Care Clinical Virology Laboratory. Food and Drug Administration Emergency Use Authorization: SARS-CoV-2 RT-PCR Assay. https://www.fda.gov/media/136818/download (Accessed November 2020).
- 27.Hologic. Food and Drug Administration Emergency Use Authorization: SARS-CoV-2 Assay (Panther Fusion System). https://www.fda.gov/media/136156/download (Accessed November 2020).
- 28.Hologic. Food and Drug Administration Emergency Use Authorization: Aptima SARS-CoV-2. https://www.fda.gov/media/138096/download (Accessed November 2020).
- 29. Ho DE, Imai K, King G, Stuart EA.. MatchIt: nonparametric preprocessing for parametric causal inference. J Stat Softw 2011;42:1–28. [Google Scholar]
- 30. Altman D, Machin D, Bryant T, Gardner M, editors. Statistics with Confidence. 2nd Ed. London: British Medical Journal Books; 2000. [Google Scholar]
- 31. Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ainsworth M, Andersson M, Auckland K, Baillie JK, Barnes E, Beer S, et al. Performance characteristics of five immunoassays for SARS-CoV-2: a head-to-head benchmark comparison. Lancet Infect Dis 2020;20:1390–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bryan A, Pepper G, Wener MH, Fink SL, Morishima C, Chaudhary A, et al. Performance characteristics of the Abbott Architect SARS-CoV-2 IgG assay and seroprevalence in Boise, Idaho. McAdam AJ, editor. J Clin Microbiol 2020;58:e00941–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chew KL, Tan SS, Saw S, Pajarillaga A, Zaine S, Khoo C, et al. Clinical evaluation of serological IgG antibody response on the Abbott Architect for established SARS-CoV-2 infection. Clin Microbiol Infect 2020;26:1256.e9–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meyer B, Torriani G, Yerly S, Mazza L, Calame A, Arm-Vernez I, et al. ; Geneva Center for Emerging Viral Diseases. Validation of a commercially available SARS-CoV-2 serological immunoassay. Clin Microbiol Infect 2020;26:1386–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fenwick C, Croxatto A, Coste AT, Pojer F, André C, Pellaton C, et al. Changes in SARS-CoV-2 spike versus nucleoprotein antibody responses impact the estimates of infections in population-based seroprevalence studies. J Virol 2020;95:e01828–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grandjean L, Saso A, Ortiz AT, Lam T, Hatcher J, Thistlethwayte R, et al. Long-term persistence of spike antibody and predictive modeling of antibody dynamics following infection with SARS-CoV-2. Preprint at https://www.medrxiv.org/content/10.1101/2020.11.20.20235697v1 (2020).
- 38. Bruni M, Cecatiello V, Diaz-Basabe A, Lattanzi G, Mileti E, Monzani S, et al. Persistence of anti-SARS-CoV-2 antibodies in non-hospitalized COVID-19 convalescent health care workers. JCM 2020;9:3188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Perkmann T, Perkmann-Nagele N, Ozsvar-Kozma M, Koller T, Breyer M-K, Breyer-Kohansal R, et al. Increasing both specificity and sensitivity of SARS-CoV-2 antibody tests by using an adaptive orthogonal testing approach. Preprint at https://www.medrxiv.org/content/10.1101/2020.11.05.20226449v2 (2020).
- 40.United States Food and Drug Administration. Independent Evaluations of COVID-19 Serological Tests. https://open.fda.gov/apis/device/covid19serology/ (Accessed February 2021).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.