Abstract
In the era of precision oncology, major strides are being made to use individual tumor information for clinical decision-making. Differing from traditional biopsy methods, the emerging practice of liquid biopsy provides a minimally invasive way of obtaining tumor cells and derived molecules. Liquid biopsy provides a means to detect and monitor disease progression, recurrence, and treatment response in a noninvasive way, and to potentially complement classical biopsy. Uveal melanoma (UM) is a unique malignancy, with diagnosis heavily reliant on imaging, few repeat biopsies, and a high rate of metastasis, which occurs hematogenously and often many years after diagnosis. In this disease setting, a noninvasive biomarker to detect, monitor, and study the disease in real time could lead to better disease understanding and patient care. While advances have been made in the detection of tumor-disseminated components, sensitivity and specificity remain important challenges. Ambiguity remains in how to interpret current findings and in how liquid biopsy can have a place in clinical practice. Related publications in UM are few compared to other cancers, but with further studies we may be able to uncover more about the biology of disseminated molecules and the mechanisms involved in the progression to metastasis.
Keywords: Liquid biopsy, Uveal melanoma, Biomarkers
Introduction
Uveal melanoma (UM) is the most common intraocular malignancy in adults and the second most common form of melanoma. It has an incidence rate of 4.9 per million in the US [1] and 3.75 per million in Canada [2]. In Europe, the incidence rate varies according to latitude, with <2 per million in southern regions and >8 per million in northern countries like Norway and Denmark [3]. UM arises from melanocytes of the uveal tract in the eye, which is composed of the iris, ciliary body, and the choroid, where 90% of the tumors are found [1].
Our understanding of the biology of UM has increased tremendously with the discovery of initiating mutations in GNAQ/11, which are found in >80% of patients and code for the α-subunit of heterotrimeric G proteins [4, 5]. GNAQ/11 play a role in activating G protein signaling cascade via IP3, DAG, and cAMP, which activates MAP kinase/AKT signaling [5, 6]. Mutations in GNAQ or GNA11 are often utilized as a marker of melanocytic tumor origin. While 15–20% of tumors do not possess a mutation in GNAQ or GNA11, many have a mutation in cysteinyl leukotriene receptor 2 (CYSLTR2) or phospholipase C beta 4 (PLCB4), which both also lead to activation of GNAQ signaling [7, 8]. A mutually exclusive mutation in GNAQ, GNA11, CYSLTR2, and PLCB4 is found in almost all cases and thus thought to be an initiating event in UM [9].
Despite significant improvements in local treatments of primary tumors, the prognosis for UM patients remains unchanged, with up to 50% developing metastasis, which occurs primarily in the liver. With a lack of efficient treatment options, the average overall patient survival after diagnosis of metastatic UM is less than a year. The risk of developing metastasis in UM is traditionally estimated by clinical tumor features such as tumor thickness, diameter, location, and extraocular extension, as well as histopathology. Yet, genetic and molecular markers of the tumor that have been identified in recent years are more reliable at predicting the prognosis of UM patients.
Although biopsies are less needed for the purpose of confirming the diagnosis (which can now be done with assistance of various imaging techniques), obtaining a sample of primary tumor remains important for its prognostic value. In recent years, gene expression profiling of UM has been used for the prognostication of tumors through classification into class 1 or 2, which translate into low and high risk, respectively, for developing metastasis [10, 11]. Chromosome 3 monosomy remains the strongest single cytogenetic factor to predict UM metastasis [12, 13, 14].
Alongside chromosome 3 monosomy, mutations associated with risk of metastasis have emerged in UM. BAP1, which encodes a nuclear ubiquitin carboxy-terminal hydrolase with deubiquitinase activity, acts as a tumor suppressor gene in UM. Somatic mutation in BAP1, resulting in loss of protein expression, is associated with increased risk of metastasis, and approximately 84% of metastatic UM patients maintain only the mutated allele of BAP1 [15]. Additionally, mutations in SF3B1 and EIF1AX may confer differing risk of UM metastasis. BAP1, SF3B1, and EIF1AX mutations are mutually exclusive and are thought to be associated with early (BAP1), late (SF3B1), or no (EIF1AX) metastasis [16]. In tumors with wild-type SF3B1 but similar structural chromosomal aberrations, mutations in another gene of the spliceosome complex serine and arginine rich splicing factor 2 (SRSF2), have been found [17]. Similarly to SF3B1, SRSF2 has been associated with class 1B gene expression profiling, disomy 3, and intermediate risk of metastasis [18]. Many additional gene mutations have been discovered that could occur sequentially after the canonical mutations [16], although it has been hypothesized that the mutations essential for malignancy occur an early time point and additional mutations are neutral to tumor evolution [19]. Certain genetic aberrations and their correlations with prognosis have been summarized into a 15-gene assay developed by Harbour and Chen [10], which has been validated for its prognostic efficiency.
Although biopsy of the primary tumor can provide valuable prognostic information, biopsies of intraocular tumors are invasive procedures, with rare but severe complications, including hemorrhage, retinal detachment, and cataract. Additionally, although it has been disproved empirically, some are still concerned about the possibility of intraocular biopsies provoking unwanted systemic tumor cell dissemination. Furthermore, biopsies have been reported to show discordant profiles depending on where tissue is sampled [20]. As such, single biopsies may inadequately sample disease and provide only a snapshot of the tumor; repeated biopsy to monitor genomic changes and heterogeneity is simply not possible. Hence, an easier and less invasive biopsy method to sample the ocular tumor is warranted in UM. This ideal method should (1) reflect both the status of the primary tumor and the risk or presence of metastasis, (2) monitor the molecular changes that occur as part of the natural history of disease and in response to therapy, (3) be easy to repeat, and (4) be well tolerated and minimally invasive.
Liquid Biopsy
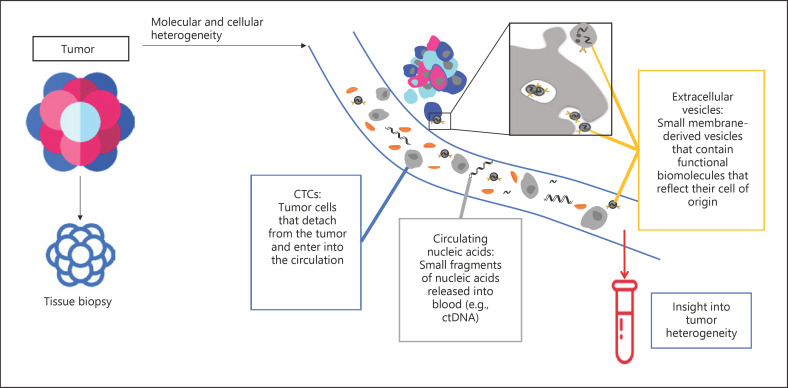
Meeting all these criteria, liquid biopsy is now emerging as an alternative approach to analyze the molecular features of tumors in individual patients. Liquid biopsy involves the sampling of tumor-derived molecules in body fluids such as blood. There are various components for analysis in liquid biopsy, including circulating tumor cells (CTCs), circulating tumor DNA (ctDNA), cell-free microRNAs (miRNAs), as well as tumor-derived extracellular vesicles (EVs), that can be informative of genetic and molecular characteristics of the tumor (Fig. 1). Liquid biopsy provides a means to detect and monitor disease progression, recurrence, and response to treatment in a noninvasive way and to potentially complement classical biopsy. Detection of CTCs, ctDNA, and EVs like exosomes may contribute to early diagnosis and management. It is also a valuable tool for longitudinal assessment of the genetic heterogeneity of cancer: recurrent or metastatic lesions may exhibit a different molecular profile than the primary lesion, and these changes may dictate the most effective treatment. While traditional biopsies are limited to obtaining only one portion of the primary tumor and do not reflect the status of any unrecognized secondary or metastatic tumors, another major advantage of liquid biopsy lies in its potential to capture the heterogeneous nature of cancer.
Fig. 1.
Main types of liquid biopsy analytes. Schematic representation of tumor-derived molecules that can be found in liquid biopsy (circulating tumor cells, circulating nucleic acids, and extracellular vesicles). CTCs, circulating tumor cells; ctDNA, circulating tumor DNA.
Although most studies in liquid biopsy focus on blood, other body fluids can also be utilized for analysis, including urine, cerebrospinal fluid, ascites, saliva, as well as aqueous and vitreous humor. The rationale of which body fluid to use can depend on the type and anatomical location of the cancer being studied. For instance, saliva may be used for head and neck cancers, urine may be used for bladder cancers, and cerebrospinal fluid may be better suited for brain tumors [21]. In ocular diseases, the vitreous and aqueous can also be important sources of material. In UM, where there is an exclusively hematogenous dissemination of tumor cells, blood makes an ideal candidate for analysis and, due to the ease of its sampling, has been the most studied analyte. Moreover, aqueous humor is also under investigation by our group and others as a source of liquid biopsy analyte. Aqueous humor should be investigated because it could provide a more concentrated source of tumor-derived molecules. The aqueous is also currently under investigation as an important liquid biopsy bioanalyte for retinoblastoma [22].
CTCs in Liquid Biopsy
Cells released from the primary tumor are believed to contribute to metastatic disease by acting as seeding cells that disseminate and eventually form colonies at metastatic sites [23]. Over the past decade, many studies have focused on CTC counts and prognosis in cancer, and CTCs have been demonstrated to have strong prognostic value in breast, colorectal, and prostate cancer [23]. Considering the high risk of metastasis through hematogenous dissemination, CTCs in the blood are of particular interest in UM.
CTC detection can be difficult given the rarity of CTCs in the circulation, with an estimated concentration in peripheral blood ranging from 1 to 10 CTCs per 10 mL of blood [24]. For this reason, enrichment steps to concentrate CTCs in the sample are necessary. Such methods can be biological, based on positive and/or negative selection for protein markers, or based on physical properties like size and electric charges. In UM, CTCs have been isolated using different technical approaches, based on either RT-PCR, size filtration, or immunodetection [25, 26, 27, 28]. However, the prognostic value of CTCs in UM remains unclear, and comparison between studies is difficult due to varying methodologies [29]. Moreover, the clinical use of CTCs is constrained by the difficulty in differentiating tumor-specific CTCs from other cell types in the circulation (Table 1).
Table 1.
Previous studies analyzing the value of CTCs in UM
| Title | Reference | Patient number | Technique used | Major findings |
|---|---|---|---|---|
| Detection of melanocytes from uveal melanoma in peripheral blood using the polymerase chain reaction | Tobal et al. [31], 1993 | UM patients (n = 6) | RT-PCR of tyrosinase mRNA | CTCs detected in 3 of 6 patients; CTC positive result preceded overt metastatic disease in 1 patient; limit of detection sensitivity of 10 cells/5 mL of blood |
| The detection of melanoma cells in peripheral blood by reverse transcription-polymerase chain reaction | Foss et al. [32], 1995 | UM patients (n = 36) | RT-PCR of tyrosinase mRNA | No patient sample was positive for CTCs; limit of detection sensitivity of 1 cell/mL of blood; CTCs detected in 2 of 31 control blood samples |
| Quantitative detection of circulating tumor cells in cutaneous and ocular melanoma and quality assessment by real-time reverse transcriptase-polymerase chain reaction | Keilholz et al. [37], 2004 | high-risk primary UM (n = 21); metastatic UM (n = 26) | real-time PCR of tyrosinase, MelanA/MART1, and gp 100 mRNA | mRNA of tyrosinase, MelanA/MART1, and gp 100 expressed in higher quantities and frequency in stage IV UM patients compared to earlier stages |
| Five-year results of prognostic value of tyrosinase in peripheral blood of uveal melanoma patients | Boldin et al. [34], 2005 | UM patients (n = 41); no metastasis at diagnosis | RT-PCR of tyrosinase mRNA | CTCs detected in 16 of 41 patients; 11 of the 16 CTC-positive patients became CTC-negative after therapy; CTC positivity correlated with 5-year survival; limit of detection sensitivity of 3 cells/5 mL of blood |
| Circulating tumor cells as prognostic factor for distant metastases and survival in patients with primary uveal melanoma | Schuster et al. [35], 2007 | high-risk primary UM (n = 110) | real-time PCR of tyrosinase and MelanA/MART1 mRNA | CTCs detected in 11 of 110 patients, with no associations with clinical features; presence of tyrosinase or MelanA/MART1 transcripts was prognostic for metastasis and survival |
| Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study | Callejo et al. [36], 2007 | primary UM (n = 30) | nested RT-PCR of tyrosinase and MelanA mRNA | CTCs detected in 29 of 30 patients during the follow-up period; CTC presence was detected regardless of tumor size or time after treatment; limit of detection sensitivity of 10 cells/2 mL of blood |
| Tyrosinase mRNA levels in the blood of uveal melanoma patients: correlation with the number of circulating tumor cells and tumor progression | Pinzani et al. [33], 2010 | UM patients (n = 41) | real-time PCR of tyrosinase mRNA | 20 of 41 patients were positive for tyrosinase mRNA; correlation found between tyrosinase mRNA levels and CTC counts suggest that tyrosinase could be used as an indirect measurement for CTC level |
| Visualization of circulating melanoma cells in peripheral blood of patients with primary uveal melanoma | Ulmer et al. [38], 2008 | primary UM (n = 52) | immunostaining based on MCSP expression | CTCs detected in 10 of 52 patients, and not in the controls; CTC presence associated with ciliary body invasion, advanced tumor stage, and anterior tumor location; limit of detection sensitivity of 1 cell/mL of blood |
| Circulating melanoma cells in peripheral blood of patients with uveal melanoma before and after different therapies and association with prognostic parameters: a pilot study | Suesskind et al. [39], 2011 | UM patients (n = 81) | immunostaining based on MCSP expression | various primary tumor therapies showed no significant impact on CTC number in circulation |
| Circulating tumor cells detection and counting in uveal melanomas by a filtration-based method | Mazzini et al. [27], 2014 | primary UM (n = 31); choroidal nevi (n = 10) | isolation by size of epithelial tumor cells | single CTCs or clusters of cells detected in 17 of 31 UM patients, while no CTCs detected in choroidal nevi patients; patients with<10 CTCs/10 mL blood had significantly worse DFS and OS; limit of detection sensitivity of 1 cell/mL of blood |
| Identification of circulating melanoma cells in uveal melanoma patients by dual-marker immunoenrichment | Tura et al. [26], 2014 | primary UM patients (n = 31) | immunomagnetic enrichment of NKI/C3- and NKI/beteb-positive cells; followed by immunostaining for NKI/C3 or MCSP | CTCs detected in 29 of 31 patients using dual-immunomagnetic enrichment assay for NKI/C3 and NKI/beteb; suggest that a combination of two antibodies instead of single antibody may be more efficient at detecting CTCs |
| Immunomagnetic detection of micro metastatic cells in bone marrow of uveal melanoma patients: a paradox | Eide et al. [40], 2015 | UM patients (n = 328) | immunodetection of MCSP | CTCs detected in 2% of the patient cohort |
| Analysis of monosomy-3 in immunomagnetically isolated circulating melanoma cells in uveal melanoma patients | Tura et al. [43], 2016 | primary UM patients (n = 44) | immuno-FISH assay to detect chromosome 3 | CTCs detected in 91% of the patients; 58% of detected CTCs were positive for monosomy 3, associated with advanced tumor stage |
| Detection of circulating melanoma cells in choroidal melanocytic lesions | Bande et al. [41], 2015 | choroidal nevus (n = 4); primary UM (n = 8) | CellSearch | 50% of UM patients had<1 CTC detected in their blood sample, while no CTCs were found in the nevi patients; no significant correlation was found between CTC detection and clinical parameters (largest basal diameter, tumor height) |
| Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma | Bidard et al. [23], 2014 | metastatic UM (n = 40) | CellSearch | CTCs detected in 12 of 40 patients; CTC levels associated with presence of miliary hepatic metastasis, metastasis volume, PFS, and OS |
| Arterial blood, rather than venous blood, is a better source for circulating melanoma cells | Terai et al. [44], 2015 | metastatic UM (n = 17) | CellSearch | CTCs detected with greater frequency (100%) and quantities in arterial compared to venous blood; no significant association was found between CTC count and tumor burden within the liver |
| Pilot study of circulating tumor cells in early-stage and metastatic uveal melanoma | Anand et al. [42], 2019 | primary UM (n = 20); metastatic UM (n = 19) | CellSearch | OS at 24 months significantly lower in CTC-positive vs.-negative early-stage UM; mean CTC count was higher in the metastatic group |
CTCs, circulating tumor cells; DFS, disease-free survival; OS, overall survival; PFS, progression-free survival; UM, uveal melanoma.
CTCs have a half-life in bloodstream of about 1–2.4 h [30]. They carry information from the tumor at the DNA, RNA, and protein levels. In fact, the first groups to research CTCs in UM took an indirect approach at detecting CTC presence by measuring RNA transcripts of melanoma-associated proteins with RT-PCR. The earliest reports of CTCs in UM were published in the 1990s, with UM cells detected in the circulation through PCR. In 1993, Tobal et al. [31] reported the feasibility of detecting as few as 10 CTCs in 5 mL of blood in 3 of 6 patients and indicated that a positive CTC result preceded overt metastatic disease in 1 patient. Following this, Foss et al. [32] attempted to detect CTCs in 36 patients, but found none to be positive. Several groups have investigated the presence of mRNA of tyrosinase [33, 34, 35, 36], an enzyme involved in the biosynthesis of melanin, which is not usually detected in the blood of healthy subjects [35]. By testing 64 UM samples, Keilholz et al. [37] simultaneously quantified RNA levels of tyrosinase, MelanA/MART1, and gp100 and found all three markers to be expressed in higher quantities and frequencies in stage IV UM patients compared to earlier-stage patients. However, the detection rates of CTCs based on this indirect method were highly variable, ranging from 0 to 97% [31, 32, 34, 35, 36].
Around the same time, other groups (Ulmer et al. [38], Suesskind et al. [39], and Eide et al. [40]) took a more direct way of detecting CTCs through an immunomagnetic enrichment technique based on expression of melanoma-associated chondroitin sulfate proteoglycan. Later groups have taken advantage of CellSearch, an FDA-approved CTC detection approach, to directly detect CTCs in UM [23]. Using the CellSearch system, cells are first enriched for CD146 using immunomagnetic isolation. Then UM CTCs are selected for expression of the high-molecular-weight melanoma-associated antigen and stained for CD45 and CD34 to distinguish from leukocytes and endothelial cells [23]. Using CellSearch, Bande et al. [41] detected CTCs in the blood of patients with primary nonmetastatic UM tumors and patients with nevus. In that study, 50% of the UM patients had more than 1 CTC detected in their blood sample, while no CTCs were found in the nevus patients. Also using CellSearch in a prospective study, the detection rate and prognostic value of both CTCs and ctDNA were evaluated in 40 patients with metastatic UM [23]. The results showed that 12 patients (30%) had at least one CTC detected in blood and negatively correlated CTCs with overall survival. However, CTC detection was not found to be an independent prognostic factor. A main limitation of CTCs is reflected in this study, for as many as 70% of enrolled patients, all of whom were metastatic, had no detectable CTCs, highlighting an insufficient sensitivity of detection [23].
The relationship between CTC positivity and disease prognosis has also been reported by Mazzini et al. [27], utilizing a different detection method of size filtration. Single CTCs or clusters of cells were found in 17 of 31 UM patients, while no CTCs were detected in choroidal nevus patients. Patients with more than 10 CTCs per 10 mL blood were found to have significantly worse disease-free and overall survival. Finally, a very recent pilot study conducted by Anand et al. [42] in 40 UM patients (including early-stage and metastatic patients) also reported a negative correlation between CTC positivity and patient outcome. The study suggests the potential of CTC presence in early-stage UM to predict metastatic risk and confirms its association with poor disease outcome.
Many of these studies also explored the relationship between CTC presence and various clinical parameters such as larger basal diameter and tumor height. Bande et al. [41] showed no correlation with larger basal diameter and tumor height, but suggested a correlation with extrascleral extension, although the latter observation was based on 1 patient only. Similarly, Mazzini et al. [27] also failed to correlate CTC presence to clinical parameters of the tumor, such as larger basal diameter and tumor height as well as TNM. However when the CTC-positive samples were stratified according to CTC counts, the number of CTCs significantly correlated with larger basal diameter and tumor height.
In contrast with other studies, Callejo et al. [36] reported detectable CTCs in almost all 30 UM patients in their study. In that study, CTCs were identified at diagnosis and even after treatment, including enucleation. To further explore CTCs as prognostic for metastatic risk, another group showed that instead of merely considering the presence of CTCs, it would be more valuable prognostically to analyze the detected CTCs for genomic aberrations such as monosomy 3 [43].
Conflicting data from previous reports may be improved with better detection efficiency for CTCs. For example, a combination of two antibodies instead of a single one may be more efficient at detecting CTCs [26]. Interestingly, with regards to the source of analysis sample, one study showed that arterial blood may be a better sample source than venous blood and may help achieve higher detection rates [44]. The authors infer that the difference could be attributed to increased degradation and fragmentation of CTCs following circulation in the peripheral venous system. As more studies are conducted that improve CTC detection, along with advances in techniques, the clinical value of CTCs will become apparent. These cells contain information from the tumor of origin and likely contribute actively to systemic disease. Detecting and characterizing CTCs will allow us to exploit them as both biomarkers and therapeutic targets.
Circulating Tumor DNA
ctDNA are small fragments of DNA that are released by tumor cells and enter the circulation. Since blood, irrespective of health state, contains circulating free DNA (cfDNA) derived from the physiologic apoptosis of hematopoietic and other normal cells, evaluation of ctDNA is often done via detection of a tumor-specific mutation that distinguishes cfDNA from DNA of tumor origin (ctDNA). As such, malignancies with defined mutations are good candidates for assessing the value of ctDNA to monitor disease. The large amount of cfDNA in patients compared to ctDNA makes detection of ctDNA a major challenge for liquid biopsy. The challenge is made greater when patients undergo cancer therapies such as radiotherapy or chemotherapy, which cause significant amounts of cfDNA to be released as a result of tissue damage [30].
Mutations found in cfDNA from patient plasma likely embody both primary tumor and metastatic sites, making it representative of systemic disease. Evidence suggests that the half-life of cfDNA in the circulation is between 16 min and several hours [45, 46], allowing it to provide information about tumor heterogeneity and subclonal changes in real time [47, 48]. Further work is needed to elucidate the exact mechanism of ctDNA release from cells, but it is currently believed that the majority of ctDNA in blood results from apoptosis [49], with necrosis and active release mechanisms also accounting for a proportion of ctDNA. It is still unclear whether ctDNA has biological roles on other cells, but studies have shown ctDNA uptake by host cells, and in vitro cells have been reported to be transformed by cfDNA from cancer patients [30]. The quantity of ctDNA found in the blood has been correlated to tumor burden and cell turnover [50]. Recent studies have shown a concordance rate of 80% (mutations) between tissue and ctDNA in liquid biopsies. Thus, liquid biopsy serves as a reliable proxy for conventional tissue biopsy. In 2016, the FDA approved the first ctDNA test for EGFR mutations in non-small-cell lung cancer patients (cobas® Mutation Test version 2), highlighting the clinical utility of ctDNA (Table 2).
Table 2.
Previous studies analyzing the value of ctDNA in UM
| Title | Reference | Patient number | Major findings |
|---|---|---|---|
| Pyrophosphorolysis-activated polymerization detects circulating tumor DNA in metastatic uveal melanoma | Madic et al. [51], 2012 | metastatic UM patients (n = 21) | ctDNA detected in 20 of 21 patients (range 1.3–2,125 copies/mL of plasma); ctDNA levels correlated with tumor burden; sensitivity: 1 ctDNA copy per reaction; specificity: 104 copies of normal DNA were not detected |
| Ultradeep sequencing detects GNAQ and GNA11 mutations in cell-free DNA from plasma of patients with uveal melanoma | Metz et al. [52], 2013 | metastatic UM patients (n = 28; amplicon sequencing performed on 22 patients) | ctDNA detected in 9 of 22 patients; no association between primary tumor characteristics (tumor height, largest basal diameter) and ctDNA detection; specificity: 0.1% false-positive results in healthy blood |
| Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma | Bidard et al. [23], 2014 | metastatic UM patients (n = 40; 26 with ctDNA measurements) | ctDNA detected in 22 of 26 patients (range 4–11,421 copies/mL); ctDNA levels associated with presence of miliary hepatic metastasis, metastasis volume, PFS, and OS; multivariate analyses showed ctDNA to be a better prognostic marker than CTCs; sensitivity: <1 copy/mL; no false-positives in healthy controls (n = 20) |
| Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study | Cabel et al. [53], 2017 | metastatic UM patients (n = 3; 15 patients in total) | patients with undetectable ctDNA by week 8 of treatment had a significantly better PFS and OS than patients with persistently detectable ctDNA; ctDNA levels could be valuable in assessing response to anti-PD1 therapy; sensitivity:<0.1% mutant copies in normal DNA |
| Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors | Rodrigues et al. [97], 2018 | metastatic UM patients (n = 42) | one patient responded to pembrolizumab treatment; therapeutic response was measured by ctDNA levels and imaging, which showed consistent results |
ctDNA, circulating tumor DNA; OS, overall survival; PFS, progression-free survival; UM, uveal melanoma.
GNAQ and GNA11 are two genes of particular interest in the detection of UM ctDNA, since mutations in these genes can be exploited to differentiate tumor-derived DNA from cfDNA. Bidard et al. [23] investigated the level of ctDNA in metastatic UM patients based on point mutations in GNAQ and GNA11 using the bidirectional pyrophosphorolysis-activated polymerization (Bi-PAP) technique. ctDNA was found in 22 of 26 patients, and a correlation was made between higher levels of ctDNA and hepatic miliary metastases, tumor volume, and CTC count. ctDNA was also shown to be an independent prognostic factor for progression-free and overall survival. Also using Bi-PAP and similar techniques, Madic et al. [51] showed the potential of using ctDNA to detect patients with minimal metastatic mass and enable early adjuvant therapy intervention.
Using a different technique, ultradeep amplicon sequencing, Metz et al. [52] similarly looked to detect GNAQ and GNA11 mutations in ctDNA and found positive results in 9 of 22 patients, with no correlations to clinical parameters. The advantage of this approach over Bi-PAP is not needing prior knowledge of the specific mutational information.
As mentioned before, one of the anticipated utilities of liquid biopsy is to monitor treatment response, which would allow clinicians to modify treatment according to this biomarker. This clinical utility was validated in a proof-of-principle study that monitored ctDNA levels during anti-PD1 immunotherapy in patients with non-small-cell lung cancer, microsatellite-unstable colorectal cancer, and UM [53]. ctDNA was detected in 10 of 15 patients at baseline measurement (of whom 2 were UM patients). After 8 weeks, a change in ctDNA level correlated significantly with change in tumor size, and ctDNA detection at week 8 was prognostic of progression-free survival. Such results need to be validated in a larger patient cohort, especially since only 3 UM patients were included. Of the 3 UM patients included, 1 had undetectable ctDNA levels throughout the study, while the other 2 patients showed mild changes in ctDNA levels at week 8.
Our group focused on using digital droplet PCR, a highly sensitive technique, to detect ctDNA for mutations in GNAQ, GNA11, PLCB4, and CYSLTR2. In our study of 40 patients with UM, patients with nevus, and healthy individuals, we saw a strong correlation between plasma-based ctDNA detection and disease, with highest levels seen in patients with UM. In our nevus cohort, only patients with risk factors had detectable levels of ctDNA (unpublished data). Interestingly, using an animal model, we also showed an enrichment for ctDNA in the blood as well as aqueous humor of animals, which supports the exploration of these analytes as potentially valuable liquid biopsy (unpublished data).
ctDNA is a powerful biomarker because of its specificity in identifying tumor-derived nucleic acids. With advances in the sensitivity of detection technologies such as sequencing and digital PCR, ctDNA has the potential to be used for monitoring treatment response and disease course.
Circulating miRNA
miRNAs are small noncoding RNAs of around 22 nucleotides [54]. They function as regulators of gene expression that impact many important biological processes, including cancer development. miRNAs found in the circulation are termed circulating cell-free miRNA, and have a relatively long half-life [55]. In liquid biopsy, there are currently three major techniques for their detection: quantitative RT-PCR, microarray analyses, and deep sequencing [54] (Table 3).
Table 3.
Previous studies analyzing the value of miRNAs in UM
| Title | Reference | Patient number | Major findings |
|---|---|---|---|
| Circulating immune cell and microRNA in patients with uveal melanoma developing metastatic disease | Achberger et al. [58], 2014 | UM patients (n = 6) | plasma levels of miR-20a, miR-125b, miR-146a, miR-155, miR-181a, and miR-233 higher in UM patients compared to healthy controls; upon development of metastasis, all miRs with the exception of miR-181a increased from the time of diagnosis |
| miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: pathological and diagnostic implications | Ragusa et al. [55], 2015 | UM patients (n = 6) | 32 miRNAs found to be differentially expressed in UM patients compared to healthy controls; vitreous humor circulating miRNA profile only partially overlaps that in serum; miR-146a was found to be upregulated in serum of UM patients, a potential circulating marker |
| Increased levels of miRNA-146a in serum and histologic samples of patients with uveal melanoma | Russo et al. [56], 2016 | UM patients (n = 14) | SAM analysis showed 8 serum miRNAs to be differentially expressed between patients and controls; when singularly validated with TaqMan assays, only significant overexpression of miRNA-146a was found |
| A panel of circulating microRNAs detects uveal melanoma with high precision | Stark et al. [57], 2019 | uveal nevus (n = 10); localized UM (n = 50); metastatic UM (n = 5) | 6 miRNAs (miR-16, miR-145, miR-146a, miR-204, miR-211, and miR-363-3p) were differentially expressed between uveal nevi compared to localized or metastatic UM; miR-211 had the ability to distinguish metastatic from localized UM; the 6 miRNAs together had 93% sensitivity and 100% specificity in identifying UM |
miRNA, microRNA; UM, uveal melanoma.
A previous study found that 32 miRNAs are differentially expressed in UM patients compared to healthy controls [55]. The study compared miRNA profiles from vitreous humor, vitreous humor-derived exosomes, as well as serum from 6 UM patients and 6 control participants. miR-146a, which is thought to play an important role in the survival of melanocytes in UM, was found to be upregulated in the serum of UM patients. The significance of miR-146a as a serum marker was subsequently confirmed by another group [56]. Recently, a panel of six miRNAs − miR-16, miR-145, miR-146a, miR-204, miR-211, and miR-363-3p − was reported to distinguish between patients with UM and uveal nevi [57]. Immune regulatory miRNAs were also studied, measured in 6 UM patients from the time of diagnosis up until development of metastasis. Plasma levels of miR-20a, miR-125b, miR-146a, miR-155, miR-181a, and miR-233 were all higher in UM patients compared to healthy controls, and upon development of metastasis, all miRNAs with the exception of miR-181a had increased compared to the time of diagnosis [58].
Extracellular Vesicles
Emitted by both normal and cancer cells, EVs are nanoparticles with a lipid bilayer membrane and range from 50 nm to several micrometers in size. EVs are highly heterogeneous and can be categorized mainly into microvesicles, apoptotic bodies, and exosomes based on size, shape, and origin [59, 60]. The process of EV emission from cells is conserved throughout evolution from prokaryotes to eukaryotes [61, 62]. They can transport cargo such as RNA, DNA, and proteins and act as a messenger between cells, playing a vital role in intercellular communication [63]. However, the underlying mechanism involved in EV formation, delivery of cargo inside EVs, and ultimately emission is still not clear.
Tumor-derived EVs have been implicated in promoting cancer cell proliferation, migration, invasion, and disease progression [64, 65, 66, 67]. EVs have been shown to play a major role in mediating metastasis, ranging from oncogenic reprogramming of recipient cells to formation of the premetastatic niche through uptake by normal cells and subsequent priming of the microenvironment for colonization by CTCs [68, 69, 70, 71]. Indeed, evidence has emerged suggesting that EVs released by cancer cells can transfer their oncogenic properties to recipient cells, which constitutes a novel mechanism of cancer dissemination [72, 73]. The contents of EVs are thus informative of their originating tumor, making them promising candidates for liquid biopsy analysis. Double-stranded DNA cargo could reflect specific mutations of the originating tumor. The lipid bilayer also conveniently protects the RNA and miRNA contents from degrading due to RNase catalytic actions [74]. There are some advantages of using EVs as a liquid biopsy target. Compared to using CTCs as targets, there are currently more isolation protocols already developed for harvesting EVs, including commercially available kits. Additionally, EVs express specific markers that allow for identification. Markers like HSP70 and Alix allow their separation from other subcellular vesicles, while they also express markers that allow identification of their originating cells as well as target cells [74]. An immunoaffinity-based isolation protocol has been developed to distinguish melanoma-derived exosomes in patient plasma, based on the chondroitin sulphate peptidoglycan 4 (CSPG4) tumor antigen [75]. Finally, EVs are relatively stable while in circulation [74] (Table 4).
Table 4.
Previous studies analyzing the value of extracellular vesicles in UM
| Title | Reference | Patient number | Major findings |
|---|---|---|---|
| MicroRNA in exosomes isolated directly from the liver circulation in patients with metastatic uveal melanoma | Eldh et al. [76], 2014 | metastatic UM patients (n = 12) | metastatic UM releases exosomes into the liver circulation; patients have more circulating exosomes in systemic circulation compared to healthy controls; miRNA profiles of exosomes are similar between patients, but different compared with the cell line controls |
| miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: pathological and diagnostic implications | Ragusa et al. [55], 2015 | UM patients (n = 6) | 90% of miRNAs were common between vitreous humor and vitreal exosomes; the major source of miRNA in vitreous humor could be from exosomes |
| Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma | Sharma et al. [75], 2018 | ocular melanoma (n = 2) | immunoaffinity-based isolation protocol based on antibody for CSPG4 could isolate melanoma-derived exosomes in patient plasma with good reproducibility |
| Oncosuppressor-mutated cells as a liquid biopsy test for cancer-screening | Abdouh et al. [78], 2019 | dysplastic melanocytic nevus (n = 1); choroidal melanoma (n = 1) | exosomes from cancer patient sera were able to transform OMCs; suggested that OMCs could be used as a biological liquid biopsy test for early cancer screening |
miRNA, microRNA; OMCs, oncosuppressor-mutated cells; UM, uveal melanoma.
Recent studies on EVs in UM have focused on blood exosomes, which are small EVs (30–100 nm) secreted by cells that contain functional biomolecules that reflect their cell of origin. Using plasma from liver perfusates from metastatic UM patients, Eldh et al. [76] reported tumor-originated exosomes in the hepatic circulation, and although comparison with healthy controls failed due to insufficient exosome level in healthy blood, it was shown that they contain a different miRNA profile compared to other tumor cell lines. In particular, a cluster of six miRNAs (miR-370, miR-210, miR-320a, miR-124, miR-107, and miR-486-5p) is shared between UM patients and likely indicates a melanoma origin. A higher overall concentration of exosomes in systemic blood, measured by total exosomal protein, was also found in metastatic UM patients (median 75.6 µg/mL plasma) compared to healthy controls (median 13.8 µg/mL plasma) [76]. Exosomes produced by metastatic melanoma cells have been reported to contribute to premetastatic niche formation, preparing the environment for colonization by CTCs [77]. In a study on the use of oncosuppressor-mutated cell-based liquid biopsy tests for cancer screening, EVs from cancer patients of many tumor types, including choroidal melanoma, were capable of transforming oncosuppressor-mutated cells to form tumors in vivo, suggesting the horizontal transfer of malignant traits [78]. In-depth studies on UM-derived exosomes are limited, and characterization of EVs in this disease setting is lacking. It would be relevant to further evaluate mechanisms to inhibit the synthesis and uptake of tumor exosomes as an additional alternative to prevent melanoma progression and develop new adjuvant therapies.
Other Biomarkers
Currently, liver function tests (LFTs) are conventionally used for detecting and monitoring UM metastasis to the liver. LFTs assess levels of various liver enzymes including alanine aminotransferase, aspartate aminotransferase, alkaline phosphatase, gamma-glutamyl transpeptidase, lactic dehydrogenase, and bilirubin. However, LFTs often have low sensitivity and only detect metastases at later stages of the disease. As a result, attention has been given to tumor serum markers, many of which were first utilized in the context of cutaneous melanoma, such as calcium-binding protein B (S100β) and melanoma inhibitory activity (MIA) protein. S100β, MIA, osteopontin, and tissue polypeptide-specific antigen have been shown to have prognostic potential in predicting liver metastasis [79, 80]. In one study, while up to 96% of studied patients showed elevated levels of serum tumor markers at the time of metastasis diagnosis, only 52% showed elevated levels of LFT enzymes [81], indicating that serum tumor markers may have higher prognostic potential compared to LFT. Another group recently developed a 7-plex immunoassay for serum UM markers that expanded to include CEACAM-1, MIC-1, SPON1, POSTN, and HSP27 in addition to osteopontin and MIA [82].
Other serum markers include DJ-1, a 189-amino-acid protein that has been identified as an oncogene and has been shown to be a promising biomarker for the detection of metastatic UM [83]. Another emerging target is tumor-educated platelets, which are platelets that have been influenced by tumor cells through the uptake of transferred tumor-originating biomolecules such as RNAs [54]. mRNA sequencing of tumor-educated platelets in other cancer types has been shown to identify malignancies with extremely high accuracy [84]. Further studies are needed to assess their significance in UM.
Aqueous and Vitreous Humor in Liquid Biopsy
Although not as minimally invasive as blood samples, ocular fluids like the aqueous and vitreous humor can also be sampled for intraocular diseases. In retinoblastoma, ctDNA in the aqueous humor has been explored as a potential biomarker [22]. Aqueous humor samples from 26 retinoblastoma patients were evaluated for somatic chromosomal copy number alterations, and 6p gain in aqueous humor ctDNA was found to correlate with poorer clinical outcome [85].
In UM, aqueous and vitreous have also been sampled from patients in various contexts, although far less so than blood samples. Sakuma et al. [86] described a case of choroidal melanoma where diagnosis made after a modified Shorr stain on a sample of vitreous humor revealed cancer cells associated with melanin granules. The aqueous and vitreous have also been investigated as a source of other biomarkers. High levels of S-100 protein were found in both vitreous and aqueous humors in patients with ocular melanoma [87]. Levels of vascular endothelial growth factor (VEGF) were also observed in the aqueous humor of UM patients, with aqueous VEGF-A concentration correlating with largest basal tumor diameter and tumor height [88, 89]. Additionally, the level of soluble HLA in aqueous humor was found to be an unfavorable prognostic factor that correlates with larger tumors and worse survival [90].
EVs have also been detected in ocular fluids. In a study by Ragusa et al. [55], 32 miRNAs were found to be differentially expressed in UM patients compared to healthy controls, and in particular, miR-146a, miR-21, and miR-34a were significantly upregulated in both the vitreous humor and vitreal exosomes, while miR-618 was downregulated in the vitreous humor and upregulated in vitreal exosomes. It was concluded that since 90% of miRNAs were common between the vitreous humor and vitreal exosomes, exosomes are likely a major source of miRNA in the vitreous humor.
Clinical Utility of Liquid Biopsy
Screening and Early Detection
Tumor-originating biomarkers such as CTCs and ctDNA have potential for early tumor detection. Evidence suggests that many patients have disseminated tumor cells at the time of diagnosis. Indeed, it has been estimated that at the point of primary UM tumor treatment, CTCs and micrometastases may have already been present for 2–3 years [91]. However, designing a clinically useful liquid biopsy-based screening test with high enough specificity and sensitivity remains an obstacle. Our group recently showed a correlation between positive plasma ctDNA and choroidal nevus with risk factors for growth and malignancy, suggesting that ctDNA may help diagnose small melanomas (unpublished data).
Predicting Prognosis and Metastatic Risk
Given the lack of effective systemic therapies available for metastatic disease, doubt exists as to the clinical utility of prognostic indicators for UM patients. Despite this, identifying patients who are at high risk of metastasis would allow closer monitoring of disease progression. Moreover, albeit limited, there are data suggesting that systemic therapies can prolong survival [92, 93], and it is probable that early initiation of treatment at the micrometastasis stage will translate to better response due to lower disease burden. Moreover, surgical intervention can, in certain instances, be an option when metastatic disease is limited to few foci. With many clinical trials underway and our growing understanding of UM biology, more effective systemic therapies are likely to emerge, which will make determining metastatic risk clinically relevant.
Interestingly, the 2010 TNM classification system has already included CTC count in its criteria, denoted by “i+” [30]. However, it has not been widely used due to very low CTC counts in early tumor stages, limiting its clinical value [30].
Monitoring Cancer Evolution
Tumors are in constant evolution throughout the disease course as both part of their natural history and in response to selective pressure from treatment. New mutations and mechanisms of resistance can emerge as a result of clonal selection. A safe and noninvasive method of monitoring cancer evolution that can be done repeatedly is needed, and liquid biopsy is an ideal way of tracking such changes. Through analysis of ctDNA, for example, mutations that arise in patients can be monitored and used to design more personalized treatment plans that take into account any emergent drug-resistant clones [30].
Monitoring Therapeutic Response and Minimal Residual Disease
Studies have shown the validity of using CTC quantification to assess therapeutic effects in some cancers, including metastatic breast, colorectal, and prostate cancer [94, 95, 96]. Following therapeutic intervention, patients can be analyzed for persisting high levels of CTCs or ctDNA as an indication of minimal residual disease and any need to adjust treatment. Such studies in the context of systemic UM therapies are lacking. Nevertheless, Suesskind et al. [39] did explore CTC levels in UM patients before and after various primary tumor therapies and found no significant difference in the number of blood CTCs. This provides evidence against the concern of primary therapy causing further tumor cell dissemination into the bloodstream. Although UM patients generally do not respond to immune checkpoint therapies such as anti-PD1, an outlier response was reported for a patient who responded unusually well, presumably due to a hypermutated tumor status. The study used ctDNA to monitor therapeutic response, which correlated well with imaging results [97].
Stratification for Therapeutic Intervention
In the era of targeted treatment, patient stratification is pivotal to determine the most effective therapy. New clinical trials are being conducted in UM that target specific molecular profiles of tumors. Liquid biopsy could potentially help stratify patients according to the genetic and molecular characteristics of their tumor. In fact, in 2016, the FDA approved the first ctDNA analysis test to detect EGFR mutations in lung cancer patients who could be eligible for EGFR-targeted therapy [98], indicating that such approaches are clinically useful through liquid biopsy analysis.
Discussion
Liquid biopsy has already been incorporated into clinical practice for certain cancers, yet insufficient detection sensitivity and consistency limit its current widespread utility. Liquid biopsy in UM remains a developing but exciting field, and more studies are warranted. Common challenges encountered by previous studies include limitation of detection sensitivity and small patient sample size. Recent advances in technology have already brought significant progress to the field. For example, ultrasensitive technologies such as digital PCR now allow for consistent detection of ctDNA in the circulation. New approaches, better markers of disseminated cells and molecules, and commercially available extraction kits are making liquid biopsy studies more consistent and valuable. Thus, it is reasonable to expect more accurate detection in the near future with further technological developments. Moreover, despite the rarity of UM, multicentric studies will help overcome limitations in sample size.
Studies on CTCs make up the majority of the current published literature on liquid biopsy in UM, but ctDNA is quickly attracting growing interest. While CTCs and ctDNA may appear as two competing targets for liquid biopsy, these two biomarkers provide different and complementary information regarding the tumor [30]. Bidard et al. [23] compared the prognostic value of CTC and ctDNA and concluded that only ctDNA was an independent prognostic factor for survival. However, the two markers were shown to correlate with each other, as reported in other cancers as well [23].
There is inconsistency in the detection rate of CTCs in published studies of UM. Particularly for RT-PCR-based studies, they showed insufficient accuracy and reproducibility, and thus proved not yet ready for clinical use in guiding treatment decisions [28]. While there is some controversy on whether the presence or absence of CTCs is informative regarding a patient's metastatic risk, a better correlation is likely to be found if we also consider the molecular traits of CTCs detected rather than only the total count. Studies to date on CTCs in UM generally showed that CTCs can be detected in UM patients from a very early time point. Knowing that not all UM patients will develop metastasis, it is proposed that the presence of CTCs may be needed but not sufficient for metastasis. Studying and characterizing CTCs could provide insight to the specific events that result in successful seeding of disseminated cells into clinically evident metastatic tumors. Thus, gene expression profiling of CTCs may be an important direction of research in the future, and liquid biopsy would be an attractive noninvasive way of collecting CTC samples even after primary tumor treatment. Moreover, understanding the role of tumor cell-derived EVs will likely shed considerable light on the communication that mediates metastasis. While few studies exist in EVs in UM, this field is growing rapidly, assisted by new EV isolation, detection, characterization, and visualization tools.
Importantly, liquid biopsy provides us not only with biomarkers of disease, but also with tumor-derived molecules that can be exploited noninvasively to better understand the disease. While CTCs, ctDNA, and EVs may be clinically valuable as biomarkers, these disseminated molecules are likely to play important active roles in driving systemic disease. CTCs may travel to and seed site specifically in distant organs, making understanding their molecular characteristics pivotal. Moreover, these cells release nucleic acids into the circulation, such as miRNAs and ctDNA, and these molecules can provide real-time insight into genomic alterations behind disseminated disease. Finally, EVs may be actively contributing to preparing organs such as the liver for colonization, and thus are an important new focus of therapeutics. While the biological mechanisms leading to metastasis in UM remain largely unknown, future efforts in clarifying this process may be key to the development of new effective adjuvant therapies. Such new insight may help explain why despite primary control, patients can develop metastasis, and further demonstrate that to improve patient survival, a systemic approach is needed to target tumor-derived molecules.
Conflict of Interest Statement
The authors have no conflict of interest to disclose.
Funding Sources
The authors have no funding to report.
Author Contributions
E. Jin and J.V. Burnier carried out the literature search and wrote and reviewed the manuscript.
References
- 1.McLaughlin CC, Wu XC, Jemal A, Martin HJ, Roche LM, Chen VW. Incidence of noncutaneous melanomas in the U.S. Cancer. 2005 Mar;103((5)):1000–7. doi: 10.1002/cncr.20866. [DOI] [PubMed] [Google Scholar]
- 2.Ghazawi FM, Darwich R, Le M, Rahme E, Zubarev A, Moreau L, et al. Uveal melanoma incidence trends in Canada: a national comprehensive population-based study. Br J Ophthalmol. 2019 Dec;103((12)):1872–6. doi: 10.1136/bjophthalmol-2018-312966. [DOI] [PubMed] [Google Scholar]
- 3.Virgili G, Gatta G, Ciccolallo L, Capocaccia R, Biggeri A, Crocetti E, et al. EUROCARE Working Group Incidence of uveal melanoma in Europe. Ophthalmology. 2007 Dec;114((12)):2309–15. doi: 10.1016/j.ophtha.2007.01.032. [DOI] [PubMed] [Google Scholar]
- 4.Van Raamsdonk CD, Bezrookove V, Green G, Bauer J, Gaugler L, O'Brien JM, et al. Frequent somatic mutations of GNAQ in uveal melanoma and blue naevi. Nature. 2009 Jan;457((7229)):599–602. doi: 10.1038/nature07586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Van Raamsdonk CD, Griewank KG, Crosby MB, Garrido MC, Vemula S, Wiesner T, et al. Mutations in GNA11 in uveal melanoma. N Engl J Med. 2010 Dec;363((23)):2191–9. doi: 10.1056/NEJMoa1000584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen X, Wu Q, Tan L, Porter D, Jager MJ, Emery C, et al. Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene. 2014 Sep;33((39)):4724–34. doi: 10.1038/onc.2013.418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moore AR, Ceraudo E, Sher JJ, Guan Y, Shoushtari AN, Chang MT, et al. Recurrent activating mutations of G-protein-coupled receptor CYSLTR2 in uveal melanoma. Nat Genet. 2016 Jun;48((6)):675–80. doi: 10.1038/ng.3549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Johansson P, Aoude LG, Wadt K, Glasson WJ, Warrier SK, Hewitt AW, et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget. 2016 Jan;7((4)):4624–31. doi: 10.18632/oncotarget.6614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amaro A, Gangemi R, Piaggio F, Angelini G, Barisione G, Ferrini S, et al. The biology of uveal melanoma. Cancer Metastasis Rev. 2017 Mar;36((1)):109–40. doi: 10.1007/s10555-017-9663-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harbour JW, Chen R. The DecisionDx-UM Gene Expression Profile Test Provides Risk Stratification and Individualized Patient Care in Uveal Melanoma. PLoS Curr. 2013 Apr;5:5. doi: 10.1371/currents.eogt.af8ba80fc776c8f1ce8f5dc485d4a618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Onken MD, Worley LA, Ehlers JP, Harbour JW. Gene expression profiling in uveal melanoma reveals two molecular classes and predicts metastatic death. Cancer Res. 2004 Oct;64((20)):7205–9. doi: 10.1158/0008-5472.CAN-04-1750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Horsman DE, Sroka H, Rootman J, White VA. Monosomy 3 and isochromosome 8q in a uveal melanoma. Cancer Genet Cytogenet. 1990 Apr;45((2)):249–53. doi: 10.1016/0165-4608(90)90090-w. [DOI] [PubMed] [Google Scholar]
- 13.Prescher G, Bornfeld N, Hirche H, Horsthemke B, Jöckel KH, Becher R. Prognostic implications of monosomy 3 in uveal melanoma. Lancet. 1996 May;347((9010)):1222–5. doi: 10.1016/s0140-6736(96)90736-9. [DOI] [PubMed] [Google Scholar]
- 14.Damato B, Duke C, Coupland SE, Hiscott P, Smith PA, Campbell I, et al. Cytogenetics of uveal melanoma: a 7-year clinical experience. Ophthalmology. 2007 Oct;114((10)):1925–31. doi: 10.1016/j.ophtha.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 15.Harbour JW, Onken MD, Roberson ED, Duan S, Cao L, Worley LA, et al. Frequent mutation of BAP1 in metastasizing uveal melanomas. Science. 2010 Dec;330((6009)):1410–3. doi: 10.1126/science.1194472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shain AH, Bagger MM, Yu R, Chang D, Liu S, Vemula S, et al. The genetic evolution of metastatic uveal melanoma. Nat Genet. 2019 Jul;51((7)):1123–30. doi: 10.1038/s41588-019-0440-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Poppelen NM, Drabarek W, Smit KN, Vaarwater J, Brands T, Paridaens D, et al. SRSF2 Mutations in Uveal Melanoma: A Preference for In-Frame Deletions? Cancers (Basel) 2019 Aug;11((8)):1200. doi: 10.3390/cancers11081200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Park JJ, Diefenbach RJ, Joshua AM, Kefford RF, Carlino MS, Rizos H. Oncogenic signaling in uveal melanoma. Pigment Cell Melanoma Res. 2018 Nov;31((6)):661–72. doi: 10.1111/pcmr.12708. [DOI] [PubMed] [Google Scholar]
- 19.Field MG, Durante MA, Anbunathan H, Cai LZ, Decatur CL, Bowcock AM, et al. Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat Commun. 2018 Jan;9((1)):116. doi: 10.1038/s41467-017-02428-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Augsburger JJ, Corrêa ZM, Augsburger BD. Frequency and implications of discordant gene expression profile class in posterior uveal melanomas sampled by fine needle aspiration biopsy. Am J Ophthalmol. 2015 Feb;159((2)):248–56. doi: 10.1016/j.ajo.2014.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.De Mattos-Arruda L, Mayor R, Ng CK, Weigelt B, Martínez-Ricarte F, Torrejon D, et al. Cerebrospinal fluid-derived circulating tumour DNA better represents the genomic alterations of brain tumours than plasma. Nat Commun. 2015 Nov;6((1)):8839. doi: 10.1038/ncomms9839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berry JL, Xu L, Murphree AL, Krishnan S, Stachelek K, Zolfaghari E, et al. Potential of Aqueous Humor as a Surrogate Tumor Biopsy for Retinoblastoma. JAMA Ophthalmol. 2017 Nov;135((11)):1221–30. doi: 10.1001/jamaophthalmol.2017.4097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bidard FC, Madic J, Mariani P, Piperno-Neumann S, Rampanou A, Servois V, et al. Detection rate and prognostic value of circulating tumor cells and circulating tumor DNA in metastatic uveal melanoma. Int J Cancer. 2014 Mar;134((5)):1207–13. doi: 10.1002/ijc.28436. [DOI] [PubMed] [Google Scholar]
- 24.Alix-Panabières C, Pantel K. Challenges in circulating tumour cell research. Nat Rev Cancer. 2014 Sep;14((9)):623–31. doi: 10.1038/nrc3820. [DOI] [PubMed] [Google Scholar]
- 25.Charitoudis G, Schuster R, Joussen AM, Keilholz U, Bechrakis NE. Detection of tumour cells in the bloodstream of patients with uveal melanoma: influence of surgical manipulation on the dissemination of tumour cells in the bloodstream. Br J Ophthalmol. 2016 Apr;100((4)):468–72. doi: 10.1136/bjophthalmol-2015-306955. [DOI] [PubMed] [Google Scholar]
- 26.Tura A, Lüke J, Merz H, Reinsberg M, Lüke M, Jager MJ, et al. Identification of circulating melanoma cells in uveal melanoma patients by dual-marker immunoenrichment. Invest Ophthalmol Vis Sci. 2014 Jun;55((7)):4395–404. doi: 10.1167/iovs.14-14512. [DOI] [PubMed] [Google Scholar]
- 27.Mazzini C, Pinzani P, Salvianti F, Scatena C, Paglierani M, Ucci F, et al. Circulating tumor cells detection and counting in uveal melanomas by a filtration-based method. Cancers (Basel) 2014 Feb;6((1)):323–32. doi: 10.3390/cancers6010323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Torres V, Triozzi P, Eng C, Tubbs R, Schoenfiled L, Crabb JW, et al. Circulating tumor cells in uveal melanoma. Future Oncol. 2011 Jan;7((1)):101–9. doi: 10.2217/fon.10.143. [DOI] [PubMed] [Google Scholar]
- 29.Fernandes BF, Belfort RN, Di Cesare S, Burnier MN., Jr Circulating uveal melanoma cells: should we test for them? Can J Ophthalmol. 2008 Apr;43((2)):155–8. doi: 10.3129/i08-011. [DOI] [PubMed] [Google Scholar]
- 30.Alix-Panabières C, Pantel K. Clinical Applications of Circulating Tumor Cells and Circulating Tumor DNA as Liquid Biopsy. Cancer Discov. 2016 May;6((5)):479–91. doi: 10.1158/2159-8290.CD-15-1483. [DOI] [PubMed] [Google Scholar]
- 31.Tobal K, Sherman LS, Foss AJ, Lightman SL. Detection of melanocytes from uveal melanoma in peripheral blood using the polymerase chain reaction. Invest Ophthalmol Vis Sci. 1993 Aug;34((9)):2622–5. [PubMed] [Google Scholar]
- 32.Foss AJ, Guille MJ, Occleston NL, Hykin PG, Hungerford JL, Lightman S. The detection of melanoma cells in peripheral blood by reverse transcription-polymerase chain reaction. Br J Cancer. 1995 Jul;72((1)):155–9. doi: 10.1038/bjc.1995.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pinzani P, Mazzini C, Salvianti F, Massi D, Grifoni R, Paoletti C, et al. Tyrosinase mRNA levels in the blood of uveal melanoma patients: correlation with the number of circulating tumor cells and tumor progression. Melanoma Res. 2010 Aug;20((4)):303–10. doi: 10.1097/CMR.0b013e32833906e3. [DOI] [PubMed] [Google Scholar]
- 34.Boldin I, Langmann G, Richtig E, Schwantzer G, Ardjomand N, Wegscheider B, et al. Five-year results of prognostic value of tyrosinase in peripheral blood of uveal melanoma patients. Melanoma Res. 2005 Dec;15((6)):503–7. doi: 10.1097/00008390-200512000-00004. [DOI] [PubMed] [Google Scholar]
- 35.Schuster R, Bechrakis NE, Stroux A, Busse A, Schmittel A, Scheibenbogen C, et al. Circulating tumor cells as prognostic factor for distant metastases and survival in patients with primary uveal melanoma. Clin Cancer Res. 2007 Feb;13((4)):1171–8. doi: 10.1158/1078-0432.CCR-06-2329. [DOI] [PubMed] [Google Scholar]
- 36.Callejo SA, Antecka E, Blanco PL, Edelstein C, Burnier MN., Jr Identification of circulating malignant cells and its correlation with prognostic factors and treatment in uveal melanoma. A prospective longitudinal study. Eye (Lond) 2007 Jun;21((6)):752–9. doi: 10.1038/sj.eye.6702322. [DOI] [PubMed] [Google Scholar]
- 37.Keilholz U, Goldin-Lang P, Bechrakis NE, Max N, Letsch A, Schmittel A, et al. Quantitative detection of circulating tumor cells in cutaneous and ocular melanoma and quality assessment by real-time reverse transcriptase-polymerase chain reaction. Clin Cancer Res. 2004 Mar;10((5)):1605–12. doi: 10.1158/1078-0432.ccr-0610-3. [DOI] [PubMed] [Google Scholar]
- 38.Ulmer A, Beutel J, Süsskind D, Hilgers RD, Ziemssen F, Lüke M, et al. Visualization of circulating melanoma cells in peripheral blood of patients with primary uveal melanoma. Clin Cancer Res. 2008 Jul;14((14)):4469–74. doi: 10.1158/1078-0432.CCR-08-0012. [DOI] [PubMed] [Google Scholar]
- 39.Suesskind D, Ulmer A, Schiebel U, Fierlbeck G, Spitzer B, Spitzer MS, et al. Circulating melanoma cells in peripheral blood of patients with uveal melanoma before and after different therapies and association with prognostic parameters: a pilot study. Acta Ophthalmol. 2011 Feb;89((1)):17–24. doi: 10.1111/j.1755-3768.2009.01617.x. [DOI] [PubMed] [Google Scholar]
- 40.Eide N, Faye RS, Høifødt HK, Sandstad B, Qvale G, Faber R, et al. Immunomagnetic detection of micrometastatic cells in bone marrow of uveal melanoma patients: a paradox. Acta Ophthalmol. 2015 Feb;93((1)):59–66. doi: 10.1111/aos.12462. [DOI] [PubMed] [Google Scholar]
- 41.Bande MF, Santiago M, Muinelo-Romay L, Blanco MJ, Mera P, Capeans C, et al. Detection of circulating melanoma cells in choroidal melanocytic lesions. BMC Res Notes. 2015 Sep;8((1)):452. doi: 10.1186/s13104-015-1420-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Anand K, Roszik J, Gombos D, Upshaw J, Sarli V, Meas S, et al. Pilot Study of Circulating Tumor Cells in Early-Stage and Metastatic Uveal Melanoma. Cancers (Basel) 2019 Jun;11((6)):856. doi: 10.3390/cancers11060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tura A, Merz H, Reinsberg M, Lüke M, Jager MJ, Grisanti S, et al. Analysis of monosomy-3 in immunomagnetically isolated circulating melanoma cells in uveal melanoma patients. Pigment Cell Melanoma Res. 2016 Sep;29((5)):583–9. doi: 10.1111/pcmr.12507. [DOI] [PubMed] [Google Scholar]
- 44.Terai M, Mu Z, Eschelman DJ, Gonsalves CF, Kageyama K, Chervoneva I, et al. Arterial Blood, Rather Than Venous Blood, is a Better Source for Circulating Melanoma Cells. EBioMedicine. 2015 Sep;2((11)):1821–6. doi: 10.1016/j.ebiom.2015.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo YM, Zhang J, Leung TN, Lau TK, Chang AM, Hjelm NM. Rapid clearance of fetal DNA from maternal plasma. Am J Hum Genet. 1999 Jan;64((1)):218–24. doi: 10.1086/302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008 Sep;14((9)):985–90. doi: 10.1038/nm.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murtaza M, Dawson SJ, Pogrebniak K, Rueda OM, Provenzano E, Grant J, et al. Multifocal clonal evolution characterized using circulating tumour DNA in a case of metastatic breast cancer. Nat Commun. 2015 Nov;6((1)):8760. doi: 10.1038/ncomms9760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.De Mattos-Arruda L, Weigelt B, Cortes J, Won HH, Ng CK, Nuciforo P, et al. Capturing intra-tumor genetic heterogeneity by de novo mutation profiling of circulating cell-free tumor DNA: a proof-of-principle. Ann Oncol. 2014 Sep;25((9)):1729–35. doi: 10.1093/annonc/mdu239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jiang P, Lo YM. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet. 2016 Jun;32((6)):360–71. doi: 10.1016/j.tig.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 50.Stroun M, Lyautey J, Lederrey C, Mulcahy HE, Anker P. Alu repeat sequences are present in increased proportions compared to a unique gene in plasma/serum DNA: evidence for a preferential release from viable cells? Ann N Y Acad Sci. 2001 Sep;945((1)):258–64. doi: 10.1111/j.1749-6632.2001.tb03894.x. [DOI] [PubMed] [Google Scholar]
- 51.Madic J, Piperno-Neumann S, Servois V, Rampanou A, Milder M, Trouiller B, et al. Pyrophosphorolysis-activated polymerization detects circulating tumor DNA in metastatic uveal melanoma. Clin Cancer Res. 2012 Jul;18((14)):3934–41. doi: 10.1158/1078-0432.CCR-12-0309. [DOI] [PubMed] [Google Scholar]
- 52.Metz CH, Scheulen M, Bornfeld N, Lohmann D, Zeschnigk M. Ultradeep sequencing detects GNAQ and GNA11 mutations in cell-free DNA from plasma of patients with uveal melanoma. Cancer Med. 2013 Apr;2((2)):208–15. doi: 10.1002/cam4.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabel L, Riva F, Servois V, Livartowski A, Daniel C, Rampanou A, et al. Circulating tumor DNA changes for early monitoring of anti-PD1 immunotherapy: a proof-of-concept study. Ann Oncol. 2017 Aug;28((8)):1996–2001. doi: 10.1093/annonc/mdx212. [DOI] [PubMed] [Google Scholar]
- 54.Cayrefourcq L, Alix-Panabières C. In: CTCs as liquid biopsy: where are we now? Molecular Medicine, editor. IntechOpen; 2019. p. p. 84366. Available from: https://www.intechopen.com/books/molecular-medicine/ctcs-as-liquid-biopsy-where-are-we-now- [Google Scholar]
- 55.Ragusa M, Barbagallo C, Statello L, Caltabiano R, Russo A, Puzzo L, et al. miRNA profiling in vitreous humor, vitreal exosomes and serum from uveal melanoma patients: pathological and diagnostic implications. Cancer Biol Ther. 2015;16((9)):1387–96. doi: 10.1080/15384047.2015.1046021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Russo A, Caltabiano R, Longo A, Avitabile T, Franco LM, Bonfiglio V, et al. Increased Levels of miRNA-146a in Serum and Histologic Samples of Patients with Uveal Melanoma. Front Pharmacol. 2016 Nov;7:424. doi: 10.3389/fphar.2016.00424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stark MS, Gray ES, Isaacs T, Chen FK, Millward M, McEvoy A, et al. A Panel of Circulating MicroRNAs Detects Uveal Melanoma With High Precision. Transl Vis Sci Technol. 2019 Nov;8((6)):12–12. doi: 10.1167/tvst.8.6.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Achberger S, Aldrich W, Tubbs R, Crabb JW, Singh AD, Triozzi PL. Circulating immune cell and microRNA in patients with uveal melanoma developing metastatic disease. Mol Immunol. 2014 Apr;58((2)):182–6. doi: 10.1016/j.molimm.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018 Apr;19((4)):213–28. doi: 10.1038/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- 60.Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015 Jun;25((6)):364–72. doi: 10.1016/j.tcb.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 61.Deatherage BL, Cookson BT. Membrane vesicle release in bacteria, eukaryotes, and archaea: a conserved yet underappreciated aspect of microbial life. Infect Immun. 2012 Jun;80((6)):1948–57. doi: 10.1128/IAI.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson DG, Ding Y, Jiang L. Unconventional protein secretion in plants: a critical assessment. Protoplasma. 2016 Jan;253((1)):31–43. doi: 10.1007/s00709-015-0887-1. [DOI] [PubMed] [Google Scholar]
- 63.Zaborowski MP, Balaj L, Breakefield XO, Lai CP. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience. 2015 Aug;65((8)):783–97. doi: 10.1093/biosci/biv084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Skog J, Würdinger T, van Rijn S, Meijer DH, Gainche L, Sena-Esteves M, et al. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat Cell Biol. 2008 Dec;10((12)):1470–6. doi: 10.1038/ncb1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Al-Nedawi K, Meehan B, Micallef J, Lhotak V, May L, Guha A, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008 May;10((5)):619–24. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 66.Keller S, König AK, Marmé F, Runz S, Wolterink S, Koensgen D, et al. Systemic presence and tumor-growth promoting effect of ovarian carcinoma released exosomes. Cancer Lett. 2009 Jun;278((1)):73–81. doi: 10.1016/j.canlet.2008.12.028. [DOI] [PubMed] [Google Scholar]
- 67.Aga M, Bentz GL, Raffa S, Torrisi MR, Kondo S, Wakisaka N, et al. Exosomal HIF1α supports invasive potential of nasopharyngeal carcinoma-associated LMP1-positive exosomes. Oncogene. 2014 Sep;33((37)):4613–22. doi: 10.1038/onc.2014.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, et al. Tumour exosome integrins determine organotropic metastasis. Nature. 2015 Nov;527((7578)):329–35. doi: 10.1038/nature15756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dos Anjos Pultz B, Andrés Cordero da Luz F, Socorro Faria S, Peixoto Ferreira de Souza L, Cristina Brígido Tavares P, Alonso Goulart V, et al. The multifaceted role of extracellular vesicles in metastasis: priming the soil for seeding. Int J Cancer. 2017 Jun;140((11)):2397–407. doi: 10.1002/ijc.30595. [DOI] [PubMed] [Google Scholar]
- 70.Chin AR, Wang SE. Cancer-derived extracellular vesicles: the ‘soil conditioner’ in breast cancer metastasis? Cancer Metastasis Rev. 2016 Dec;35((4)):669–76. doi: 10.1007/s10555-016-9639-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Becker A, Thakur BK, Weiss JM, Kim HS, Peinado H, Lyden D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell. 2016 Dec;30((6)):836–48. doi: 10.1016/j.ccell.2016.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee TH, Chennakrishnaiah S, Meehan B, Montermini L, Garnier D, D'Asti E, et al. Barriers to horizontal cell transformation by extracellular vesicles containing oncogenic H-ras. Oncotarget. 2016 Aug;7((32)):51991–2002. doi: 10.18632/oncotarget.10627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horibe S, Tanahashi T, Kawauchi S, Murakami Y, Rikitake Y. Mechanism of recipient cell-dependent differences in exosome uptake. BMC Cancer. 2018 Jan;18((1)):47. doi: 10.1186/s12885-017-3958-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Halvaei S, Daryani S, Eslami-S Z, Samadi T, Jafarbeik-Iravani N, Bakhshayesh TO, et al. Exosomes in Cancer Liquid Biopsy: A Focus on Breast Cancer. Mol Ther Nucleic Acids. 2018 Mar;10:131–41. doi: 10.1016/j.omtn.2017.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sharma P, Ludwig S, Muller L, Hong CS, Kirkwood JM, Ferrone S, et al. Immunoaffinity-based isolation of melanoma cell-derived exosomes from plasma of patients with melanoma. J Extracell Vesicles. 2018 Feb;7((1)):1435138. doi: 10.1080/20013078.2018.1435138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Eldh M, Olofsson Bagge R, Lässer C, Svanvik J, Sjöstrand M, Mattsson J, et al. MicroRNA in exosomes isolated directly from the liver circulation in patients with metastatic uveal melanoma. BMC Cancer. 2014 Dec;14((1)):962. doi: 10.1186/1471-2407-14-962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Peinado H, Alečković M, Lavotshkin S, Matei I, Costa-Silva B, Moreno-Bueno G, et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat Med. 2012 Jun;18((6)):883–91. doi: 10.1038/nm.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abdouh M, Gao ZH, Arena V, Arena M, Burnier MN, Arena GO. Oncosuppressor-Mutated Cells as a Liquid Biopsy Test for Cancer-Screening. Sci Rep. 2019 Feb;9((1)):2384. doi: 10.1038/s41598-019-38736-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barak V, Kaiserman I, Frenkel S, Hendler K, Kalickman I, Pe'er J. The dynamics of serum tumor markers in predicting metastatic uveal melanoma (part 1) Anticancer Res. 2011 Jan;31((1)):345–9. [PubMed] [Google Scholar]
- 80.Haritoglou I, Wolf A, Maier T, Haritoglou C, Hein R, Schaller UC. Osteopontin and ‘melanoma inhibitory activity’: comparison of two serological tumor markers in metastatic uveal melanoma patients. Ophthalmologica. 2009;223((4)):239–43. doi: 10.1159/000206139. [DOI] [PubMed] [Google Scholar]
- 81.Hendler K, Pe'er J, Kaiserman I, Baruch R, Kalickman I, Barak V, et al. Trends in liver function tests: a comparison with serum tumor markers in metastatic uveal melanoma (part 2) Anticancer Res. 2011 Jan;31((1)):351–7. [PubMed] [Google Scholar]
- 82.Song J, Merbs SL, Sokoll LJ, Chan DW, Zhang Z. A multiplex immunoassay of serum biomarkers for the detection of uveal melanoma. Clin Proteomics. 2019 Mar;16((1)):10. doi: 10.1186/s12014-019-9230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chen LL, Tian JJ, Su L, Jing Y, Zhang SC, Zhang HX, et al. DJ-1: a promising marker in metastatic uveal melanoma. J Cancer Res Clin Oncol. 2015 Feb;141((2)):315–21. doi: 10.1007/s00432-014-1804-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, et al. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell. 2015 Nov;28((5)):666–76. doi: 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Berry JL, Xu L, Kooi I, Murphree AL, Prabakar RK, Reid M, et al. Genomic cfDNA Analysis of Aqueous Humor in Retinoblastoma Predicts Eye Salvage: The Surrogate Tumor Biopsy for Retinoblastoma. Mol Cancer Res. 2018 Nov;16((11)):1701–12. doi: 10.1158/1541-7786.MCR-18-0369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sakuma T, Iseki R, Mimura A, Tanigawa N, Takamizu R. Rapid cytologic diagnosis of choroidal malignant melanoma by vitreous smear. J Fr Ophtalmol. 2012 Sep;35((7)):535.e1–4. doi: 10.1016/j.jfo.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 87.Cochran AJ, Holland GN, Saxton RE, Damato BE, Foulds WR, Herschman HR, et al. Detection and quantification of S-100 protein in ocular tissues and fluids from patients with intraocular melanoma. Br J Ophthalmol. 1988 Nov;72((11)):874–9. doi: 10.1136/bjo.72.11.874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Missotten GS, Notting IC, Schlingemann RO, Zijlmans HJ, Lau C, Eilers PH, et al. Vascular endothelial growth factor a in eyes with uveal melanoma. Arch Ophthalmol. 2006 Oct;124((10)):1428–34. doi: 10.1001/archopht.124.10.1428. [DOI] [PubMed] [Google Scholar]
- 89.Boyd SR, Tan D, Bunce C, Gittos A, Neale MH, Hungerford JL, et al. Vascular endothelial growth factor is elevated in ocular fluids of eyes harbouring uveal melanoma: identification of a potential therapeutic window. Br J Ophthalmol. 2002 Apr;86((4)):448–52. doi: 10.1136/bjo.86.4.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wierenga AP, Gezgin G, van Beelen E, Eikmans M, Spruyt-Gerritse M, Brouwer NJ, et al. Soluble HLA in the Aqueous Humour of Uveal Melanoma Is Associated with Unfavourable Tumour Characteristics. Cancers (Basel) 2019 Aug;11((8)):1202. doi: 10.3390/cancers11081202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Eskelin S, Pyrhönen S, Summanen P, Hahka-Kemppinen M, Kivelä T. Tumor doubling times in metastatic malignant melanoma of the uvea: tumor progression before and after treatment. Ophthalmology. 2000 Aug;107((8)):1443–9. doi: 10.1016/s0161-6420(00)00182-2. [DOI] [PubMed] [Google Scholar]
- 92.Carvajal RD, Sosman JA, Quevedo JF, Milhem MM, Joshua AM, Kudchadkar RR, et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: a randomized clinical trial. JAMA. 2014 Jun;311((23)):2397–405. doi: 10.1001/jama.2014.6096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Luke JJ, Callahan MK, Postow MA, Romano E, Ramaiya N, Bluth M, et al. Clinical activity of ipilimumab for metastatic uveal melanoma: a retrospective review of the Dana-Farber Cancer Institute, Massachusetts General Hospital, Memorial Sloan-Kettering Cancer Center, and University Hospital of Lausanne experience. Cancer. 2013 Oct;119((20)):3687–95. doi: 10.1002/cncr.28282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, et al. Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med. 2004 Aug;351((8)):781–91. doi: 10.1056/NEJMoa040766. [DOI] [PubMed] [Google Scholar]
- 95.Cohen SJ, Punt CJ, Iannotti N, Saidman BH, Sabbath KD, Gabrail NY, et al. Relationship of circulating tumor cells to tumor response, progression-free survival, and overall survival in patients with metastatic colorectal cancer. J Clin Oncol. 2008 Jul;26((19)):3213–21. doi: 10.1200/JCO.2007.15.8923. [DOI] [PubMed] [Google Scholar]
- 96.de Bono JS, Scher HI, Montgomery RB, Parker C, Miller MC, Tissing H, et al. Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin Cancer Res. 2008 Oct;14((19)):6302–9. doi: 10.1158/1078-0432.CCR-08-0872. [DOI] [PubMed] [Google Scholar]
- 97.Rodrigues M, Mobuchon L, Houy A, Fiévet A, Gardrat S, Barnhill RL, et al. Outlier response to anti-PD1 in uveal melanoma reveals germline MBD4 mutations in hypermutated tumors. Nat Commun. 2018 May;9((1)):1866. doi: 10.1038/s41467-018-04322-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Brown P. The Cobas® EGFR Mutation Test v2 assay. Future Oncol. 2016 Feb;12((4)):451–2. doi: 10.2217/fon.15.311. [DOI] [PubMed] [Google Scholar]