Abstract
Despite enormous progress in understanding the fundamentals of bacterial gene regulation, our knowledge remains limited when compared with the number of bacterial genomes and regulatory systems to be discovered. Derived from a small number of initial studies, classic definitions for concepts of gene regulation have evolved as the number of characterized promoters has increased. Together with discoveries made by new technologies, this knowledge has led to revised generalizations and principles. In this Expert Recommendation, we suggest precise, updated definitions that support a logical, consistent, conceptual framework of bacterial gene regulation, focusing on transcription initiation. The resulting concepts can be formalized by ontologies for computational modelling, laying the foundation for improved bioinformatics tools, knowledge-based resources and scientific communication. Thus, this work will help to construct better predictive models, with different formalisms, that will be useful in engineering, synthetic biology, microbiology and genetics.
Introduction
Gene expression, the transcription of DNA into RNA and the translation of RNA into a polypeptide chain, and its regulation encompass a collection of genetic and molecular programmes that underlie the major biological capabilities of eukaryotic and prokaryotic cellular differentiation and development. Gene expression is regulated in response to environmental conditions, which is critical for bacterial fitness and survival. Any step in the gene expression pathway can be regulated, from transcription initiation to mRNA stability and translation. The foundations of our understanding of gene expression regulation rely on terminology and models derived from research in Escherichia coli and bacteriophage λ1,2. These fundamental studies have been followed by decades of research in a number of regulated bacterial systems and more recently by studies using high-throughput genomic methodologies and advanced biophysical single-molecule approaches3–5, which have led to discoveries that could not have been imagined when the original concepts were proposed. Thus, some terms have acquired meanings that differ from their original ones. As experimental biology becomes a data science, mainly due to the advent of genomics, computational models that rigorously organize our knowledge become essential. Databases and ontologies are the two major tools that underpin the computational representation of knowledge. Because these tools are specified in formal language, they require definitions at a level of detail that is beyond what is common in communications among experts.
Here, we focus on transcription initiation, the most studied step in the regulation of gene expression. To discuss the limitations of existing definitions that constitute our core understanding of the regulation of transcription initiation in bacteria, the literature was searched for original and more recent definitions (Supplementary tables 1–5), as well as for examples of regulatory systems that do not conform to these definitions. A group of experts on the regulation of transcription initiation in bacteria organized a collective process of evaluating the necessity and sufficiency of the different features used to characterize the elements involved and their relation to other elements. More precisely, a feature X is not sufficient to define a class A, if there is an object that does not belong to A and has feature X. And, conversely, if a member of class A does not have feature X, then X is not necessary in the definition of A. Based on an initial draft, the authors engaged in systematic discussions, one concept at a time, on how to better expand each concept until final agreements were reached. Most of the reviewed concepts fall under the scope of the Sequence Ontology, which aims to define sequence features used in biological sequence annotation, to which final definitions were added or updated6. These updated definitions are also being incorporated into RegulonDB, a database that has curated knowledge on transcriptional regulation in E. coli7 for the past three decades and populates gene regulation data in EcoCyc8, a scientific database for E. coli K-12 MG1655.
Below, we begin with a brief overview of bacterial transcription initiation. Next, we contrast classic concepts and terms relating to bacterial transcription initiation and its regulation with their current use, in light of the current body of knowledge on transcriptional regulation in E. coli7 and other bacterial organisms. We aim to construct up-to-date and precise definitions that support a logically consistent conceptualization of gene regulation. We believe this will provide a reference for knowledge representation, for modelling and for future thinking about bacterial gene regulation. Indirectly, these concepts may also influence gene regulation frameworks beyond bacteria. Throughout the manuscript, terms followed by [G] can be found in the glossary at the end of the document.
Overview of transcription initiation
The first step in transcription is the formation of the RNA polymerase (RNAP) holoenzyme (Eσ)9, a molecular complex composed of the core RNAP plus a σ factor, which is capable of initiating gene transcription at specific DNA positions. Bacterial RNA polymerase (RNAP) is a multi-subunit enzyme. The core RNAP is comprised of five subunits: α2, β and β’, and ω. Core RNAP contains the active site that catalyses the formation of the phosphodiester bond of nascent RNA. RNAP α subunits interact with the upstream promoter (UP) element, which consists of two distinct subsites located upstream of the −35 element10. The RNAP core enzyme interacts in a sequence-specific manner with the template-strand positions –4 to +2, which constitute the core recognition element (CRE)11. To form the holoenzyme Eσ, RNAP is associated with a σ factor. There are two structurally and evolutionary distinct families of σ factors: σ54 and σ70. σ70-related factors contain up to four functional domains (σ1–4). The σ2 domain recognizes and interacts with the −10 element, and the σ4 domain interacts with the −35 element. The extended −10 element interacts with σ3, and this interaction is crucial in promoters whose −35 and −10 elements show a poor match to consensus sequences12. Some promoters, particularly the ones that respond to amino acid starvation, have an element called discriminator, which is recognized by and interacts with the σ2 domain (conserved region σ1.2)13,14. σ70 bound to the nontemplate strand captures the −10 region in an open complex and allows the single-strand template DNA to enter the active site. For this, σ70 does not need an energy source such as ATP or GTP15. σ interacts with different promoter elements to position the Eσ to unwind the double-stranded DNA in the region of the transcription start site (TSS), which corresponds to the first base of the transcript6. Most bacteria rely on different σ factors to take Eσ to different sets of promoters in response to changes in growth conditions16. Alternative σ factors are classified into two evolutionary distinct families: σ54 and σ70. σ54 is a single member family, whereas σ70 normally has several members, whose number varies among bacterial species. For example, σ24, σ28, σ32, σ38 and σ70 are members of the σ70 family in E. coli17.
Eσ initially binds promoters in a closed complex (RPc), whereby it covers DNA from approximately –55 bp to approximately +15 bp relative to the TSS (positive numbers represent bases downstream of the TSS, whereas negative ones represent upstream positions), according to DNA footprints18. This binding triggers a series of conformational changes both in the DNA and in Eσ that create an open complex (RPo), culminating in the separation of DNA strands from approximately positions –11 to +3 bp19. The region where the DNA strands are separated is often referred to as the ‘transcription bubble’, and it includes the base on the template strand, designated +1, that will act as the template for the first nucleotide of the transcript19,20. Transcription initiates with a short unstable region potentially subject to abortive transcription, in which Eσ synthesizes short products before transitioning to a stable elongation complex21. The transcription cycle then proceeds with elongation and termination steps, which have been reviewed in detail elsewhere22.
The role of promoter elements is primarily to interact with Eσ to dock the DNA–Eσ complex that is competent for the subsequent steps of transcription; hence, promoter elements determine the autonomous activity of the promoter23. The activity of different promoters varies by many orders of magnitude, ranging from promoters that produce less than one RNA copy per cell generation to promoters which generate tens of thousands of RNA copies24,25.
Essentially all promoters are subject to regulation, either indirectly, by changes in Eσ concentration26 or substrate concentrations27,28, or directly, by specific regulators. A subclass of these regulators includes activators [G] and repressors [G], collectively known as transcription factors (TFs), that act by binding to specific DNA targets. Promoters prone to activation are often intrinsically weak owing to their low affinity for Εσ, with activators compensating for this weakness by recruiting Εσ29. By contrast, repressors prevent Eσ from transcribing, often by directly occluding the promoter, or preventing some isomerization step30.
The activity of most DNA-binding transcription regulators is coupled to outside signals by a variety of mechanisms that facilitate quick responses and make gene expression sensitive to environmental changes31. Parallel mechanisms sensitive to internal and external changes support our molecular understanding of genetic developmental programmes in eukaryotes32,33.
Promoter
The original definition of promoter as defined by Jacob and Monod in 1964 is a sequence located between the operator [G] and the beginning of the operon [G] that is indispensable for operon initiation of gene expression34. This definition deserves to be revised; the following discussion shows that there is no well-defined sequence that can define any promoter.
Core RNAP transcription
Although core RNAP can transcribe single-stranded DNA in nicked regions or from DNA ends, and it has been shown that RNAP can initiate transcription from double-stranded, circular DNA35,36, sites of core RNAP transcription initiation are not promoters, because they are not specific. Thus, a promoter is not necessary for random transcription, although it is essential for transcript initiation at specific TSSs36.
Promoter sequence motifs
One of the most prominent characteristics of promoter sequences is the presence of Eσ recognition elements. There is a long history of promoter sequence comparisons and mutations, from which the base sequence consensus motifs [G] for −35 (Ref. 37), −10 (Ref.37–39), extended −10 (Refs. 12,40), −12 and −24 (Ref. 41), discriminator42, the Core Recognition Element (CRE)11, and the Upstream Promoter (UP)10,43 elements were identified. Instances of these motifs interact with the σ and α subunits of RNAP.
A variety of old and recent evidence points to the existence of sequences that conform to the motifs but does not support transcriptional activity, such as the presence of promoter-like sequences involved in transcriptional pausing [G]44, the presence of a high density of σ70 promoter-like sequences in intragenic regions45–47 and, in some bacteria, the overrepresentation of the –10 element in coding sequences48. Thus, the presence of motifs is not a sufficient condition for promoter activity.
Furthermore, the presence of a sequence in a DNA segment matching a motif is not necessary to regard a segment as a promoter. It is known that weak promoters may lack one or several functional elements, which can be compensated for by activators29. Furthermore, there may be sites that have poor matches to motifs but are recognized efficiently by Eσ; it has been shown that a single mutation over random 100 bp DNA stretches can generate a promoter49,50.
Eσ binding does not define a promoter
Eσ binding sites that drive transcription are defined as promoters. For both families of σ factors, there is evidence that Eσ binding is not sufficient to define a promoter. For example, there are sequences that conform to the motifs but do not support transcriptional activity, such as sequences leading to unproductive binding of Eσ70 (Ref. 51). Similarly, it has been suggested that Eσ70 binds in an inactive conformation under salicylic acid stress, because it was observed bound adjacent to strongly downregulated genes52.
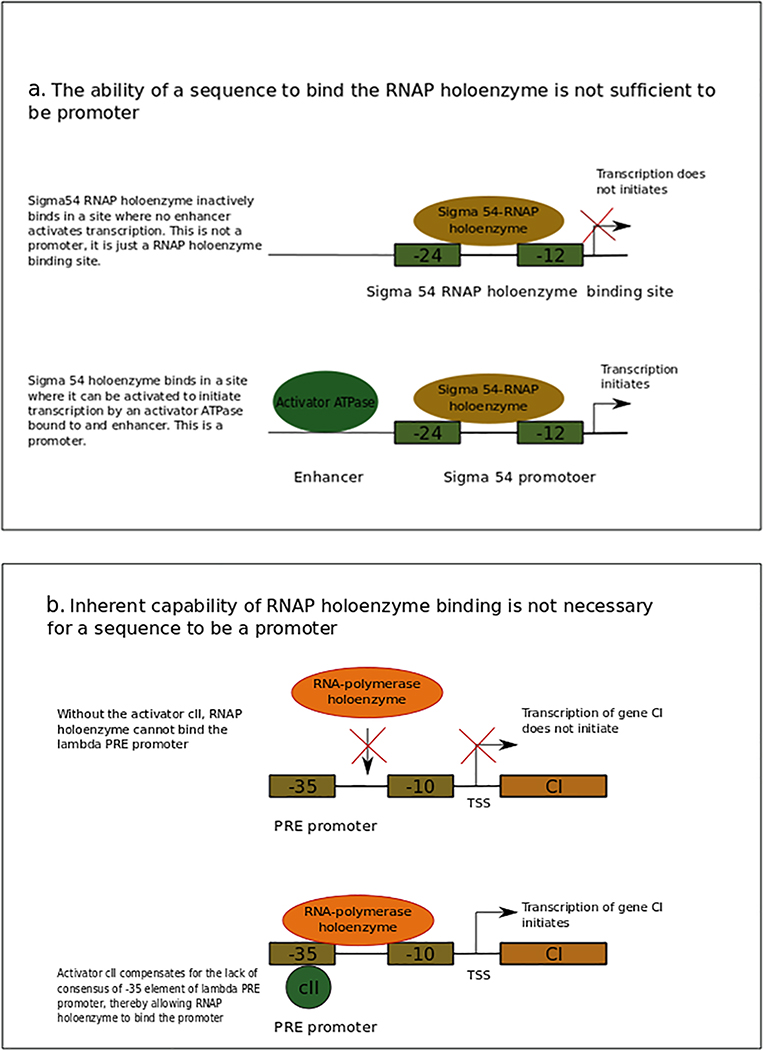
Eσ54 can bind promoters in a transcriptionally inactive state, and its activation requires an enhancer-binding protein (EBP)53. Genome-wide studies have greatly increased the number of known Eσ54 binding sites in E. coli54,55 and other bacteria56,57. Many of the newly found sites are intragenic, and most of them are not conserved in other species; thus, not all of them are likely to be functional. Hence, the binding of Eσ to a site is not a sufficient criterion to regard those sites as promoters (Fig. 1a).
Figure 1 |. RNAP holoenzyme (Eσ) intrinsic recognition by a sequence is neither necessary nor sufficient for that sequence to be a promoter.
a | σ54 is divided into three conserved regions. Region I (not shown) comprises a domain that inhibits polymerase isomerization and initiation in the absence of activation and stimulates initiation in response to activation. Due to this inhibiting element, σ54 requires an ATP-dependent activator (green oval)211. Region II (not shown) is variable and is implicated in DNA melting. Region III (brown oval) is the primary DNA-binding region and recognizes the −12 element and −24 element, to which an inactive Eσ54 (not shown) binds. Transcription initiates upon binding of an ATP-dependent activator to a sequence called enhancer. Binding sites of Eσ54 from which no transcription occurs, possibly because there is not an enhancer nearby, are not promoters. Thus, binding of the Eσ is not sufficient to define a promoter. To be a promoter, Eσ must bind and initiate transcription. b | A schematic representing the PRE promoter that does not have autonomous capability of binding Eσ. This promoter shows little to no interaction with the Eσ in the absence of activator cII (green bubble). When cII protein is bound to its site, which overlaps the −35 element, it compensates the lack of consensus of this element, thereby allowing the Eσ interaction at PRE and allowing transcription of the CI gene (brown box)210. Because there are promoters that are not bound autonomously by the Eσ, this is not a necessary feature in the definition of promoter.
Some promoters, comprising elements that are very different from the consensus sequence or that lack some recognition element, cannot bind Eσ alone; they require additional factors for their function. For example, the λ phage PRE promoter has −10 and −35 elements that differ from the consensus sequence, and it requires cII protein for Eσ binding12. Therefore, autonomous binding of Eσ to a sequence, that is, without the need for other molecules, is not a prerequisite for a sequence to be a promoter (Fig. 1b).
Although transcription initiation by Eσ may not lead to a functional RNA, it nevertheless requires a promoter. Some promoters generate short non-functional transcripts 2–15 bp long as a result of abortive transcription; this phenomenon plays a role in the regulation of transcription58. Non-functional RNAs result also from so-called TSS-associated RNAs of around 35–50 bp59 and other pervasively transcribed spurious RNAs60,61.
Regulatory binding sites are not part of the promoter
Some Eσ binding sites require the binding of activators to initiate transcription12,62, which raises the question of whether to annotate both the Eσ binding site and the required activator sites as part of the promoter. We propose not to do so, because not all promoters are activator-dependent, and it is thus not a necessary feature. In cases where the activator site overlaps the promoter, the annotated promoter will include the activator site; currently, RegulonDB contains at least 40 activator sites of 22 different TFs that overlap their corresponding promoter. However, the overlapped sequence should be annotated again independently as a regulatory site (discussed below). In cases where the activator site does not overlap the promoter, the region annotated as promoter should not include the activator site. Although this promoter sequence is not competent for transcription on its own, being annotated as a promoter indicates that it is the sequence recognized by RNAP, with the help of the activator to initiate transcription. Similarly, repressor sites and overlapping promoters are separate entities, since they are not necessary for promoter activity, although they are necessary for regulation. The existence of overlapping elements in DNA is a recurring theme in the modelling of transcriptional regulation.
Different σ factors initiating at the same TSS define different promoters
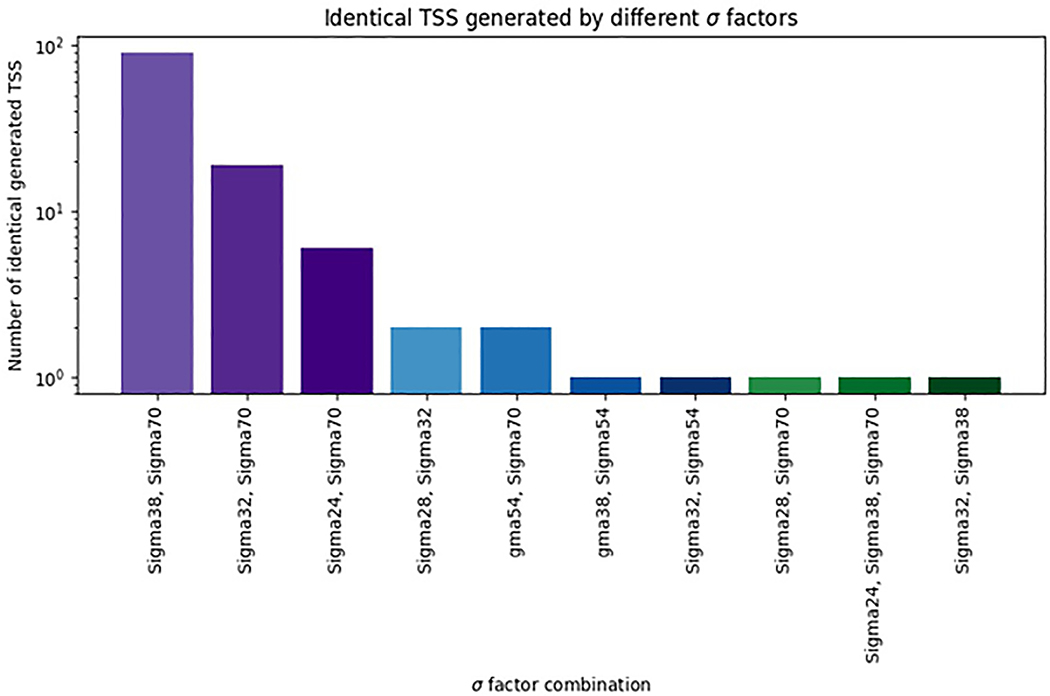
Overlapping promoters can initiate transcripts at the same TSS63,64. For example, glmY expression in E. coli is controlled by two overlapping promoters, one recognized by σ54 and the other by σ70 (Ref. 64). 5′-Rapid Amplification of cDNA Ends [G] (RACE) analyses showed that these promoters initiate transcription of the glmY gene at the same position. Mutations in the –10 element, recognized by σ70, and in the –24 element, recognized by σ54, abolished the corresponding Eσ activity while leaving the activity of the second Eσ unaffected64. This finding demonstrates that RNAP holoenzyme is using different recognition elements even when selecting the same base as TSS. Similarly, Eσ70 can initiate transcription from the majority of σ32 promoters at identical TSSs, and Eσ70 transcribes 40% of σ24 promoters65.
Fig. 2 shows the numbers of TSSs targeted by different σ factors, according to the information in RegulonDB7. We propose that overlapping binding sites targeted by different σ factors be considered separate promoters, even if the TSSs are the same. Certainly, such cases may be subject to different regulatory inputs, and different sequence elements will be used, according to the nature of the σ factor.
Figure 2 |.
Relative numbers of TSSs generated by two or more σ factors according to the information in RegulonDB.
Proposed definition for promoters
In summary, promoters are essential for transcription that is specific; an instance of a sequence motif is not mandatory; autonomous binding of Eσ is neither sufficient nor necessary; and promoters are σ factor-specific. Based on these considerations we define a promoter as a DNA segment essential for the specific initiation of transcription at a defined location in a DNA molecule, although this location might not be one single base. It is recognized by a specific Eσ, and this recognition is not necessarily autonomous.
Transcription factors
In the operon model, the product of a regulator gene, the cytoplasmic repressor, acts on the operator to affect the synthesis of a set of genes66,67. The chemical identity and mechanism of repressors were unknown until the lac and the λ phage repressors, proteins with high specificity to a site in the DNA, were isolated68,69. Later, an Eσ binding site was found to overlap λ repressor and lac repressor operator sites, which confirmed the mechanism for a repressor, which prevents RNAP from binding to the promoter70. It was assumed that gene regulation was mediated solely by repressors, until genes in the maltose and arabinose operons and in λ phage were found to be positively regulated71–73. Now, activators and repressors are collectively called TFs, and their sites of action are called transcription factor binding sites (TFBSs).
Many factors affect transcription
The term TF should not be confused with the more general class of factors that regulate transcription. Regulators that act on RNA, DNA or RNAP throughout the whole transcription cycle, including proteins, small peptides, noncoding RNAs and a variety of small ligands, have also been referred to as TFs20,74.
Originally, regulators of transcription initiation were thought to be proteins exclusively; however, there are other kinds of molecules that regulate transcription initiation, such as regulatory RNAs75,76 and small ligands, for example, ppGpp77. Here, we deal only with regulatory gene products as originally conceived by Jacob and Monod. For instance, E. coli 6S RNA regulates transcription initiation by directly binding to Eσ70 and preventing its binding to the promoter, leading to an increase in Eσ38 transcription78,79. However, the term TF has a traditionally well-established meaning in all domains of life: a protein that binds DNA to regulate transcription initiation80–85.
A large number of protein factors that bind directly to Eσ to regulate transcription initiation have been identified over the past 20 years86–89. Moreover, some proteins that regulate transcription initiation bind to both DNA and Eσ. Thus, the criterion of mere binding to a molecule, be it DNA or the holoenzyme, makes the definition of transcription factor imprecise.
By contrast, specificity — the ability to promote or repress the expression of a subset of genes — must be a feature of any regulator of gene expression, as it is the means to differentially respond to different conditions. This criterion can be used to decide whether a regulator that binds both the DNA and any component of the Εσ is a TF. If the specificity of the regulator is determined directly by the sequence of the DNA to which it binds, then it is a TF. If it is determined by its interaction with Eσ, then we propose to use the term Εσ-centered regulatory protein90.
To refine the TF definition, we focus on proteins that specifically bind to DNA and regulate transcription and asked if they should be covered by this term.
σ factors
Both TFs and σ factors bind DNA and lead Eσ to different sets of promoters in response to different environmental conditions. For example, the synthesis of σ29 is induced during sporulation in Bacillus subtilis, thereby enabling the transcription of the subset of genes required during sporulation91. Another example is E. coli σ32, whose expression is induced under heat-shock conditions; in turn, Eσ32 induces the expression of the heat-shock response genes92,93. σ factors could be regarded as activators, as they were initially discovered as factors that increase transcription activity in vitro94. However, σ factors can be defined as the proteins that regulate and are essential for specific transcription initiation while being part of the RNAP holoenzyme95. The features that make σ factors different from TFs are the ability to confer core RNAP promoter specificity, open duplex DNA and facilitate template strand entry into the RNAP active site96.
Some TFs have activities that are very similar to those of σ factors, such as forming a complex with RNAP in solution or stabilizing the open complex (RPo). For example, in E. coli, SoxS forms a complex with Eσ, which then scans DNA using SoxS to search for their cognate sites, called Sox boxes97,98. Although sequence specificity of SoxS has been demonstrated99, it does not aid in DNA opening and template strand entry to the active site; instead, this TF acts by pre-recruitment of Eσ. CarD and RbpA proteins bind both Eσ and the promoter just upstream of the –10 element to stabilize the unstable open complex (RPo) in Mycobacterium sp.100–102. Since specificity of CarD and RbpA have not yet been shown to be determined by the DNA sequence, these are RNAP-centred regulatory proteins90 that help σ to stabilize the open complex.
Nucleoid-associated proteins
The distinction between TFs and nucleoid-associated proteins (NAPs) is a perfect example of how a preconceived idea of two different types of molecules, based on genetics and function, led to the realization that TF and NAP functions are frequently performed by the same molecules103. NAPs are a group of DNA-binding proteins that are believed to play important roles in chromosome structure and compaction104,105. Some NAPs have been shown to function as site-specific regulators of transcription initiation104,106–114, similarly to other TFs, as well as acting at a distance from the target promoter by bending the intermediate DNA; integration host factor (IHF) is a well-known example at σ54 promoters115,116. Some NAPs tend to bind to many sites with fairly low sequence specificity117, and most global regulators [G] in E.coli are NAPs103,118. As NAPs have functions that overlap with those of TFs, for the purposes of the definitions we propose we do not consider them as a separate class.
Proposed definitions for TFs
A comprehensive terminology to describe all types of regulatory gene products that act on the different levels of transcription and/or other mechanisms of gene regulation is beyond the scope of this article; however, we outline terminology to designate different kinds of regulators. In the words of Jacob and Monod, we can begin by defining the general term ‘regulatory gene product’ as any gene product that increases or decreases the expression of a specific set of genes (note that gene product complexes such as heteromultimeric TFs are included, for example, IHF in E.coli).
We can distinguish two general kinds of regulatory gene products: regulatory RNA and regulatory proteins. These we can further subclassify according to the level at which they act to regulate gene expression or the specific mechanism of regulation. In particular, we have to define the class ‘DNA-binding regulatory protein of transcription initiation’ as the subclass of regulatory proteins that bind to specific DNA sequences to regulate transcription initiation. This class includes TFs, that is, DNA-binding regulatory proteins that bind near a promoter and affect transcript initiation at that promoter, and sigma factors, that is, DNA-binding regulatory proteins of transcription initiation that are part of the RNAP holoenzyme and are essential for specific initiation of transcription. Another subclass of regulatory proteins would be ‘RNAP-centred regulatory protein of transcription initiation’, defined as the proteins that regulate transcription initiation by interacting with Eσ, and whose specificity is not determined directly by recognition of specific DNA sequences.
Transcription factor binding sites
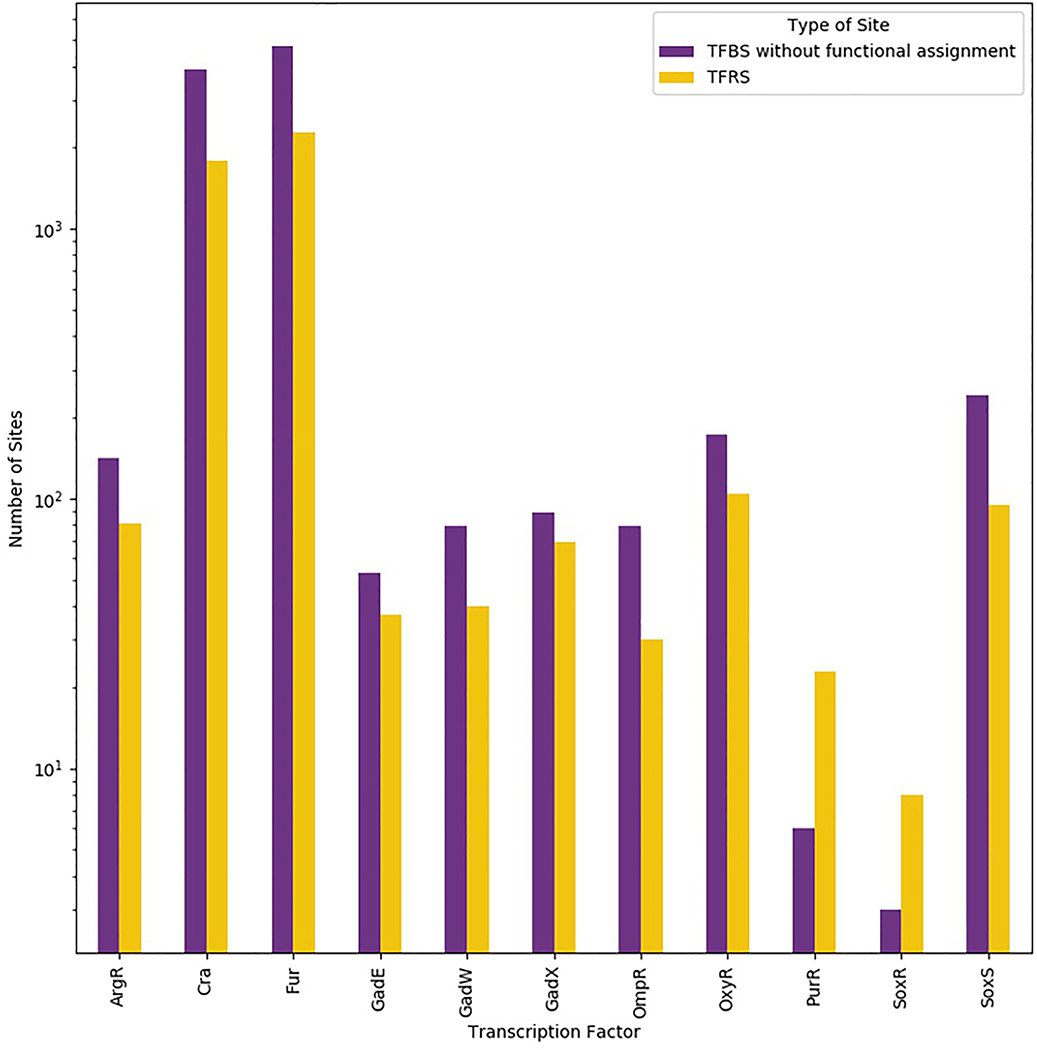
TFs bind specifically to their binding sites to activate or repress adjacent promoters. Recent genome-scale technologies capable of identifying TFBSs anywhere in the genome, such as chromatin immunoprecipitation followed by sequencing (ChIP–seq) or by identification using a DNA array, or chip (ChIP-chip) have shown that sites where TF binding has no direct effect on transcriptional regulation are common (Fig. 3). For instance, when analysing 12 studies of 9 different TFs in E. coli, only ~25% of 3,973 TF–gene interactions showed evidence of regulating local gene expression119. Similar observations have been made in other bacterial genomes, such as Mycobacterium tuberculosis4,120, Pseudomonas aeruginosa121,122, Salmonella enterica123, Listeria monocytogenes124, Helicobacter pylori125 and Shigella flexneri126. The diverse behaviour of TFs at the genomic level raises the question: which fraction of TFs has binding sites that are exclusively involved in transcriptional regulation and thus behaves like MelR127, OmpR128 and LexA129? Indeed, most TFs have sites that support other functions, such as contributing to the nucleoid structure of the genome, akin to NAPs130.
Figure 3 |. Number of transcription factor binding sites without functional assignment versus number of transcription factor regulatory sites in E. coli.
The sum of transcription factor binding sites (TFBSs) without functional assignment (purple) and transcription factor regulatory sites (TFRSs; yellow) represent the complete set of TFBSs. The figure was drawn using data from published chromatin immunoprecipitation (ChIP) experiments and data from classical experiments128,212–220. Only TFs with non-functional sites found by highthroughput methodologies were included. A logarithmic scale is used to help visualize the disparate numbers of sites known for Fur and Cra and other TFs, such as DpiA and TrpR.
It will be interesting to understand the different distribution of TFBSs in noncoding versus coding regions. For instance, two-thirds of 96 binding sites of the regulator of iron homeostasis Fur lie in intergenic regions131. Moreover, a genome-wide study of uncharacterized TFs in E. coli identified binding sites for 10 candidate TFs using ChIP-seq combined with exonuclease treatment (ChIP-exo) and showed that only 41% of 241 sites were in regulatory regions132. By contrast, as an extreme case, 70% of binding sites for RutR were found within coding regions in E.coli, a tendency conserved in other bacteria133.
Additionally, a more clear distinction of subclasses of binding sites is emerging, either owing to the contribution of additional TFs129,131,134 or the same TF working differently in varying conditions. For example, Fur was shown by ChIP–seq to bind to lower and higher numbers of TFBS under aerobic or anaerobic conditions, respectively131. In another study, ChIP-seq combined with RNA sequencing (RNA-seq) under nine physiological conditions showed that nutrient levels or growth phase affect the mechanism of action of the TF Lrp135.
Taken together, these studies distinguish between sites that affect transcription from those that do not. We thus propose the term TFBS to be defined as a DNA site where a TF binds specifically and that the term transcription factor regulatory sites (TFRS) be defined as the subset of TFBSs that are involved in transcription regulation (Fig. 3).
Architecture of regulatory regions
TFRSs, initially termed operator sites, were conceived as a single entity located near a cognate regulated promoter34,136. However, only 37% of all current promoters in RegulonDB are regulated by a single TFRS. Certainly, the steady increase of well characterized regulated promoters has led to what we now see as the rich architecture of regulatory regions, with promoters subject to the effect of one or several TFs binding to one or several TFRSs.
The diversity of TFRSs close to promoters became clear as a result of studies in the early 1990s in an initial review of 107 σ70 and 12 σ54-dependent regulated promoters3. A general principle emerging from this cohort of bacterial regulatory regions was the requirement for a proximal site, defined as an activator or repressor site located in positions that enable direct interaction of the TF in Eσ70 promoters. Only 4 out of 107 σ70 promoters lacked a proximal site3,137, a finding that has been sustained as more promoters have been characterized138. In one study, only 6.9% of σ70 promoters (48 out of 692) lacked a proximal site located between –95 to +20 relative to the TSS7. Note that promoters were counted individually even if multiple promoters coexisted in the same upstream region.
This principle does not apply to σ54-dependent promoters, the activator sites of which are more distal, with the activator often brought close to the promoter by DNA looping induced by IHF binding between the enhancer-like TF sites and the promoter139,140. This different organization is associated with the capability of Eσ54 to form stable but inactive closed complexes and the absolute requirement of an activator for transcript initiation, as opposed to the Eσ70 holoenzyme, which is competent to initiate transcription without activators and forms a transient closed complex3,141.
Regulatory modules or phrases
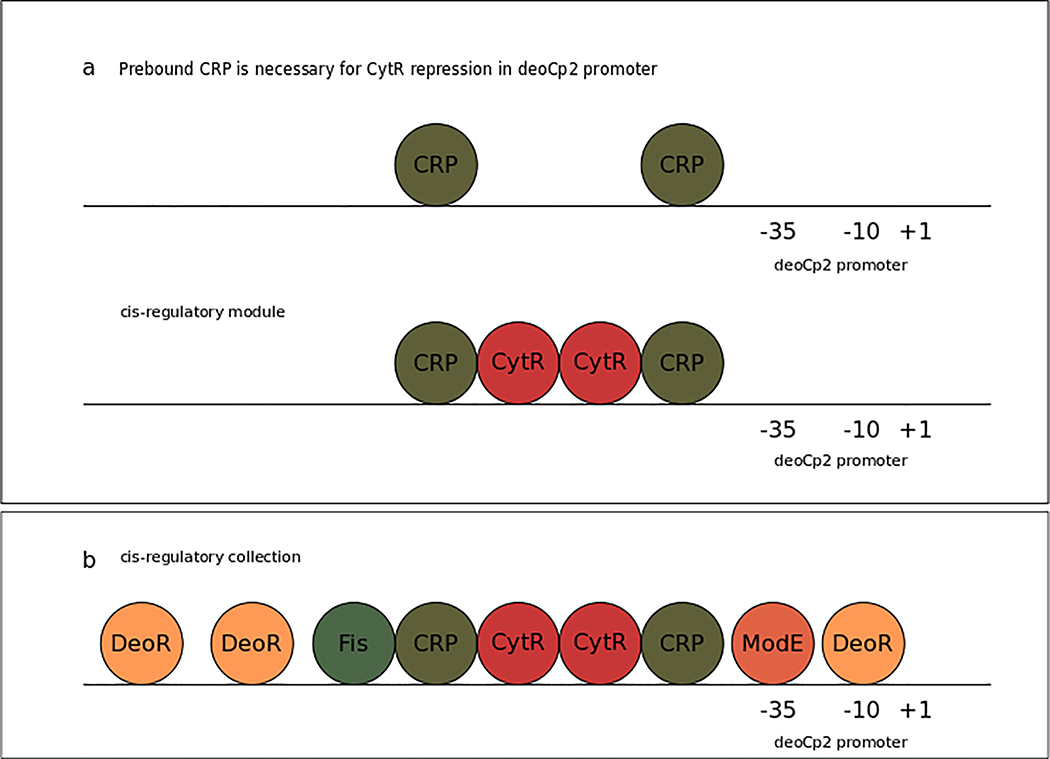
The collection of sites affecting a promoter can be partitioned into groups of sites that work together, similar to words in a sentence that form syntactic categories. These ‘regulatory phrases’ or ‘cis-modules’142 can be homotypic modules, grouping sites for the same TF, or heterotypic modules, grouping sites for different TFs that work jointly in the regulation of a promoter (Fig. 4). These modules constitute the building blocks of grammatical143, combinatorial logic143,144 and of quantitative thermodynamic models of regulated promoter transcriptional activity145–147. They are inferred in approaches searching to understand large amounts of gene expression data148 and aim at a combinatorial construction of all possible regulatory arrangements in bacterial genomics. For instance, a grammatical model was implemented with a reduced number of rules to generate the whole collection of regulatory architectures of σ70-regulated promoters143.
Figure 4 |. cis-regulatory architecture of the promoter deoCp2.
The line represents the deoCp2 promoter along with its transcription factor regulatory sites (TFRS). –35 and –10 elements and transcription start site (TSS; +1) are labelled, and the cis-regulatory architecture is represented by transcription factors (TFs; bubbles) bound to their corresponding TFRS. a | Cooperative regulatory interaction between CRP and CytR in the deoCp2 promoter. CytR is recruited as a corepressor by pre-bound CRP221. Since CytR necessarily requires pre-bound CRP to repress expression of the transcription unit downstream, these four sites form a TFRS module222. b | The TFRS collection of promoter deoCp2 is the complete set of TFRS known to regulate the deoCp2 promoter. Although there might be indirect regulatory interactions among DeoR, ModE, Fis, CRP and CytR, the only direct and necessary interaction is the one between CytR and CRP223–225. The other TFs act independently under different conditions on deoCp2, each with their own proximal sites.
We propose to define a bacterial TFRS module, or TFRS phrase, as a combination of one or several TFRSs whose bound TFs work jointly in the regulation of a promoter. A bacterial TFRS collection is defined as all the TFRSs that regulate a promoter.
Operon
The classic definition of an operon is the units of coordinated expression constituted by an operator and the group of structural genes coordinated by it136, thus requiring an operon to have one operator. However, multiple TFRSs organized in a module can regulate a single promoter. Furthermore, transcriptional regulation of some co-transcribed genes may be independent of TFs. For example, the genome of Mycoplasma genitalium encodes a limited number of TFs, and it has been suggested that this bacterium depends mostly on DNA supercoiling [G] for gene expression regulation149,150. For example, the –10 element along with DNA supercoiling induced by hyperosmolar conditions have been shown to be sufficient for the induction of expression of the MG_149 gene151. Because some cotranscribed genes may not be regulated by TFs, we propose TFRSs to be considered independent of these ‘units of coordinated expression’ and their connections captured by defining their regulatory relationships.
Cotranscribed genes are not limited to one pathway
The first operons studied coordinate the expression of genes whose products are involved in a common pathway (that is, lactose, maltose or arabinose). Hence, it was reasonable to expect that co-transcribed genes of an operon participate in the same biological pathway. However, not long after proposal of the operon model, an early example of co-transcribed genes involved in different pathways was found; in Bacillus subtilis, the tryptophan gene cluster is expressed coordinately with genes involved in histidine and tyrosine production under tryptophan-limiting conditions152. Comprehensive studies of functional classes of large numbers of transcripts in E. coli now provide us with numbers that underscore the diversity of functions of many co-transcribed genes153.
Although the operon concept was initially proposed to account for the discovery that units of transcriptional regulation in bacteria were not single genes, it was later extended to include monocistronic operons [G] by Jacob and Monod themselves154,155. Thus, we consider it to be unnecessary for an operon to have more than one gene.
Transcription units versus operons
The most general definition for both operon and transcription unit is a set of adjacent co-transcribed genes (Supplementary table 1), although there are ambiguities between the two concepts. We think that it is better to define transcription units as physical entities, whereas operons are more complex conceptual entities. At least in theory, every transcription unit in a cell can be determined by measurement (for example, cDNA sequencing). In the literature, there are two ways to define transcription units: they are segments of DNA that extend from the promoter to the terminator156,157, which are included, or they are DNA segments that begin at a TSS and end at a transcription termination site (TTS)158,159 (Supplementary table 1). The latter, and most common usage, is equivalent to defining transcription units as the DNA that corresponds to a primary transcript.
A promoter can have more than one TSS, and a terminator can have more than one TTS. Promoters were found to have on average 1.6 TSSs in an integrated genome-wide analysis of E. coli160. If we opt to define transcription units as the DNA sequences that begin at a TSS and end at a TTS, we would be representing multiple units that differ by a few nucleotides, but most of this microvariation can be considered functionally spurious. Studies using single-molecule fluorescence resonance energy transfer (FRET) supported a model in which this microvariation arises from the thermodynamics of transcription5. Thus, we prefer to think of these units of transcription as having variable ends and only consider the predominant ones for each promoter or terminator (Fig. 5a).
Figure 5 |. Transcription unit and operon schematic.
a | When different transcription start sites (TSSs) from the same promoter are not differentially regulated, they form a single transcription unit that is limited by, but not including, a single promoter and a single terminator. b | Schematic of the gadAXW complex operon. Several internal promoters and terminators enable different sets of genes to be co-transcribed in different combinations: gadAX by the gadAp promoter; gadX and gadXW by the gadXp promoter, and gadW by the gadWp1 and gadWp2 (not shown) promoters. Red wavy lines represent mRNAs. These promoters are subject to regulation by different sets of TFs (not shown), so they are not all subject to the same signals. There are at least four operons in RegulonDB with no evidence of a single polycistronic transcript including all the genes of the operon.
Nonetheless, some regulatory mechanisms alter TSS selection from a single promoter. For example, the TSSs of the pyrC and pyrD promoters of E. coli and Salmonella enterica are shifted by nucleotide concentration27,161. pyrC encodes a pyrimidine biosynthetic enzyme in E. coli and S. enterica serovar Typhimurium. Transcription of pyrC initiates at four adjacent sites named T6, C7, C8 and G9, each of which produces a different transcript. Under conditions of pyrimidine excess, the intracellular level of CTP is high, and position C7 is the dominant start site28,161. C7 transcripts are not translated, because they form a stable hairpin at their 5’ ends that blocks ribosome binding to the pyrC Shine-Dalgarno (SD) sequence. Under conditions of pyrimidine limitation, the GTP level is high, which makes the G9 start site dominant. G9 transcripts do not form the inhibitory hairpin and are readily translated28. Thus, C7 and G9 are different non-spurious TSSs from the same promoter (pyrCp), because they originate two transcripts that are differentially translated. Thus, these are two transcription units originating from the same promoter28.
In addition, some promoters can be primed by nanoRNAs in a growth phase-dependent manner, thereby altering TSS selection162. Regarding TTSs, it has long been known that Rho-dependent termination is diffuse, and elongation factors can conditionally allow bypass of terminators, which results in multiple termination points74,163–165 and complex patterns of expression166–168. To include microvariation subject to differential regulation, we propose to define transcription units as DNA regions delimited by different nonspurious TSS–TTS pairs.
A widely held distinction between transcription units and operons is that one gene can belong to one or more transcription unit but only to one operon. This distinction comes from the existence of promoters and terminators internal to operons. In 1967, dis-coordinated expression of the cluster of five tryptophan synthesis genes of Salmonella typhimurium was reported169. Deletions ranging from the operator to the second gene of the cluster suppressed expression of the first two genes only. The last three were silenced after the deletion reached the region between the second and third genes. Deletion of the operator upstream of the five genes deregulated all of them. These five genes were considered an operon regulated by a single operator but subdivided into two parts determined by promoter-like elements. Similarly, the glnALG operon is differentially transcribed in different transcription units depending on the nitrogen source by means of an internal terminator and alternative promoters170. Now, many systems in which subsets of genes of an operon are differentially expressed under different conditions due to alternative combinations of promoters and terminators are known158,160,168,171–175. This has led to the notion of operons with internal promoters and terminators as containing several transcription units.
However, internal promoters are often differentially regulated. Currently, 862 operons with known regulation are listed in RegulonDB. Of these, 143 operons consist of more than one transcription unit. Of the 143 multi-transcription unit operons, 92 are differentially regulated, whereby at least one pair of their constituent transcription units is regulated by different TFRS collections. Moreover, there are cases in which the genes of an operon are not all co-transcribed. This phenomenon is generally described as an operon containing genes ABC with two transcription units, AB and BC (Fig. 5b).
To preserve both the notion of operons being the set of genes in the same transcription unit and the notion of the set of genes coordinated in the maximal set of overlapping transcription units, we propose to use the term simple operon for the former, and complex operon for the latter. Although some complex operons may have genes that are not co-transcribed, there is ‘co-operation’ of their function (they use the same infrastructure); despite being differentially regulated, there is likely a complex functional interplay among the individual transcription units that makes it hard to consider them individually.
Whereas operons are defined in terms of genes, we think that transcription units do not necessarily bear genes or have a function. Transcriptome analysis has revealed the widespread production of non-canonical transcripts60,61, that is, transcripts that are noncoding and are often antisense; such ‘pervasive transcripts’ rarely have an assigned function176,177. Evolutionarily, we should not assume that transcriptional regulation has been selected as ‘optimal’, that is, to express exactly the right genes at the right moment in the right cells178–180. Non-functional transcripts may constitute raw material for evolution given novel mutations. Moreover, pervasive transcription may have a function in itself, that is, a basal level of pervasive transcription means that core RNAP or σ levels have to be higher, effectively buffering their levels in the cell181.
Co-expression may extend beyond operon limits. Significant coexpression has been observed in regions of 10 kb and even larger regions, which has been associated with transcriptional read-through [G], supercoiling and nearby regulons166,167,182. Read-through of terminators has been documented183,184, and a recent transcriptome obtained using single-molecule long-read sequencing has extended 34% of RegulonDB operons by at least one additional gene168.
Proposed definitions for operons
An operon is a set of adjacent genes whose transcription is coordinated by one or several mutually overlapping transcription units that are transcribed in the same direction and share at least one gene. A simple operon is an operon whose transcription is coordinated from a single transcription unit. A complex operon is an operon whose transcription is coordinated through several mutually overlapping transcription units that are transcribed in the same direction and share at least one gene.
Regulon
A regulon is a system in which the production of all enzymes (of a metabolic pathway) can be controlled by a single repressor substance; this substance may consist of several entities, but whatever its nature, it acts in a unitary fashion185.
Although regulons were originally proposed by studying the pathway of arginine biosynthesis, they must now be defined exclusively by regulation. From the 149 described E. coli regulons that include enzymes catalysing metabolic reactions, only 21% included enzymes for a single metabolic pathway whose inputs and outputs form a connected network186.
What kind of regulatory entity defines a regulon?
We suggest that the regulator that defines a regulon must be a regulatory gene product. Before this discussion, Sequence Ontology used to refer to the regulator as “regulatory signal”6. However, the term “signal” is used to refer to cues that are transformed into an effector that interacts with regulators, thereby providing information about environmental and physiological states so that the cell can adjust gene expression levels (see below). Although most TFs bind to one effector, there are documented cases where TFs allosterically bind several metabolites, for example, tryptophan, tyrosine and phenylalanine in the case of TyrR187. Conversely, small molecules may act as effectors for more than one TF, such as Zn+2 binding to ZntR and Zur, and tryptophan to TyrR and TrpR. Thus, regulation by an effector does not imply regulation by a specific TF. The initial Sequence Ontology definition of regulons corresponded to that of stimulons [G] 188.
Based on the original definition, a regulator entity should be either one regulator that can work independently (with one or multiple TFRSs) or any collection of regulators working in unity, such as complex heteromultimeric proteins. Currently, RegulonDB has documented 598 transcription units regulated by more than one TFRS; of these, 477 are regulated by different TFs. These data motivate the expansion of a regulon to include groups of genes subject to multiple regulators. They will not be acting in unity but will support a complex multiple input–output regulation. Such sets of multiple regulators may be, for instance, TFs comprising TFRS-modules anchored by a proximal site, or alternatively, the set of TFs binding the TFRS collection (Fig. 4). How these groupings will help to map mechanisms to physiology is an open question.
Regulated entity and regulated stage of gene expression
Regulator, regulated entity and the level at which gene expression is regulated are interdependent features. The concentration of products of genes transcriptionally coordinated may be uneven189, implying that the mapping of transcription units to translation units is complex. Most regulatory RNAs regulate gene expression post-transcriptionally by base pairing with mRNA. Thus, if the regulon definition is such that it includes RNAs as regulator entities, then regulated entities should include transcription units and transcribed coding sequences.
Proposed definition for regulons
In general, units of gene expression are defined as transcription units or transcribed coding sequences. A regulon is a set of units of gene expression directly regulated by a common set of one or more common regulatory gene products. A simple regulon is a regulon defined by considering one regulatory gene product, and a complex regulon is a regulon defined by considering the units of expression regulated by a specified set of regulatory gene products.
Signal and effector
Effectors were originally defined as compounds that bind specifically and reversibly to an allosteric site on a protein and bring about a discrete reversible alteration of its molecular structure that modifies its properties, changing one or several of its kinetic parameters154,190.
It is more difficult to trace the classic definition of a signal. As we want to define the term signal in the context of gene regulation, one appropriate definition to consider is a molecule originated from the environment or produced by metabolism to which a cellular response must be mounted191.
Difference between signal and effector
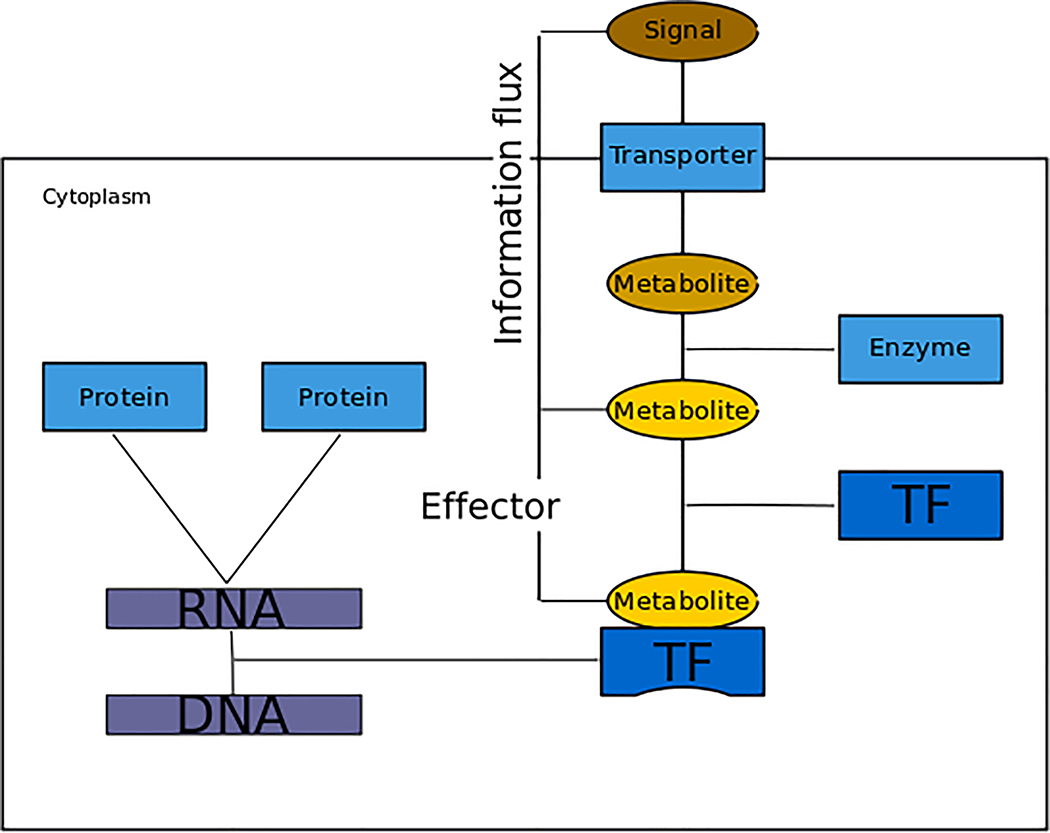
The concept of signal operates at the physiological level, whereas the effector is critical at the mechanistic level of gene expression. The signal is the starting point of an information flux that will use a variety of reactions and mechanisms ultimately reaching the regulatory machinery of cells. Effectors provide the necessary continuity to information flux by binding to the regulator that modifies gene expression (Fig. 6). Originally, effectors modulated protein activity. We propose to generalize the definition of targets of effectors to include all kinds of regulatory gene products. Some mRNAs have motifs to which small molecules bind to modify secondary structure, thereby regulating gene expression192.
Figure 6 |. Schematic of signal and effector.
A cell reacting to the environment is represented. The rectangle represents the cell membrane, separating cytoplasm and the environment. A signal is represented as an environmental molecule (brown external bubble) that elicits a cellular response. The environmental molecule is introduced to the cell (brown internal oval) through a membrane transporter (blue square) and transformed by an enzyme into another molecule (yellow oval) that plays the role of effector by binding a transcription factor (TF) and modifying its ability to recognize its DNA binding sites. The concept of genetic sensory response units (GENSOR units) captures all the elements from the signal via the effector and regulation to the final regulated gene products as the response186.
In this flux, an effector action has been considered a reversible allosteric transition that changes some chemical (kinetic or affinity) parameter of a regulatory molecule154. The effector concept must now be extended. Reversibility and allosteric features are no longer necessary, some effector-induced changes result in proteolysis, such as for E. coli LexA193,194, and covalently attached groups that irreversibly change TFs are also considered a chemical change produced by effectors. Phosphotriester and 6-O-Methylguanine DNA lesions irreversibly methylate Cys residues of Ada protein, a TF that induces its own expression195. Furthermore, the activation of this TF is not due to a conformational change but to changes in the electrostatic repulsion between the protein and the DNA196.
Signal and effector are not disjointed concepts. An example that fits perfectly with the 1963 definition of effector is that of an extracellular molecule that binds to a transmembrane sensor that reversibly modifies its conformation. This effector also acts as a signal, because the conformational change of the transmembrane sensor triggers an intracellular cascade of protein–protein interactions, that is, the signal transduction pathway, that ends in the activation or inactivation of a TF, which will in turn repress or activate the transcription of its target genes (Fig. 6).
What kinds of entities are signals and which are effectors?
Some signals are not material in nature. Environmental changes are mostly complex and elicit a plethora of changes in metabolic fluxes that generate internal signals in the form of changes in concentration and ratios of metabolites197.
We propose to extend Ptashne’s view, that any kind of molecule can be an effector33, to include other kinds of entities such as temperature and light, as they can induce conformational changes in DNA, in proteins and in RNA, thereby regulating gene expression. It has been proposed that changing from a cooler environment to a warm host triggers virulence factors in bacteria198–200. For example, in Salmonella typhimurium, high temperature unfolds the autorepressor TlpA, preventing dimerization201,202. TlpA monomers are unable to bind DNA. High temperature can directly melt mRNA-inhibitory structures that prevent binding of the ribosome, thereby inducing translation initiation201,202. Since RNA melting is a conformational change, temperature can play the role of effector (for example, in the induction of the heat-shock σ factors rpoH of E. coli and prfA of Listeria monocytogenes192). An example of a transcriptional regulator that is activated by light is the antirepressor AppA of Rhodobacter sphaeroides. Light sensed through the BLUF domain of AppA causes the reduction of affinity for DNA of the complex it forms with the repressor PpsR, thereby regulating expression of photosynthetic genes203,204.
To take into account non-material signals and effectors, we make use of the conceptualization of the Basic Formal Ontology (BFO), an upper-level domain-independent ontology that describes the most general classes under which domain-specific classes can be located. The BFO partitions reality into two general classes: continuants and occurrents. A continuant is defined as an entity that persists, endures or continues through time while maintaining its identity. It was defined in opposition to processes, or occurrents, which are entities that unfold in time205.
Proposed definitions for signals and effectors
Signals and effectors are any kind of continuant, where a signal is defined as a continuant that is the first step in a flow of information that causes a change in gene expression. An effector is defined as a continuant that produces a chemical change in a molecule and modifies its activity and/or specificity. More precisely, we propose the definition of an effector of gene expression regulator as an effector that acts on a regulatory component of a genetic switch.
Conclusions
Terms that historically made sense face limitations with new discoveries. As a consequence, ambiguities in their use emerge, requiring concepts to be revised and refined as natural science progresses. Problems arise when attempting to define classes of objects that include all and only the intended objects, given the abundance at all levels of unusual cases. Here, we analysed the adequacy of properties used to define elements involved in bacterial transcription initiation, considering possible extreme cases and generalizing definitions accordingly. We envision that an additional strategy to resolve ambiguities is to define terms that capture the object’s most general behaviour and to allow any member of a class to have a different role in specific circumstances. For example, RNAP can have a repressor role at a convergent adjacent promoter206.
Original concepts regarding sequence features were defined in genetic or physiological terms. The discovery of a functional sequence normally was followed by its characterization. However, sequence motifs cannot ensure the identity of a function, and not all functional sequences are motif compliant. Motifs are not obligatory elements in the new definitions.
Deriving universally robust functional definitions of sequences is complicated by the fact that evolution in bacteria happens rapidly, and many of the revealed features might serve no biological purpose. It is likely that some features are just pawns in the evolution game, as evidenced by the fact that nearly all DNA segments of a bacterial genome can be transcribed61,176,177,207. In fact, we suggest that the arrangement of DNA elements and regulatory architectures in a bacterial genome derive from the functional and anatomical properties of RNAPs, TFs and NAPs which are more conserved in evolution than DNA regulatory sequences. After analysing the different types of concepts, we can grasp some common themes. The distinction of functional versus spurious sequences is suggested as a guiding principle to define the level of signal versus noise at which sequences are to be classed as a functional element and which are not. Thus, promoters can be defined if their activity can be measured even if they transcribe a non-functional transcript. Similarly, when defining elements, a guiding principle to consider two entities as separate is whenever they are subject to different regulation. We have followed this guidance to describe variable TSSs or TTSs as one element, and to prefer modelling the same TSS transcribed by different sigma factors as different promoters. The same is true for the same binding site sequence being recognized by two different TFs. Certainly, differential regulation has to be captured in any model of gene regulation.
A recurrent theme is that overlapping of different DNA elements does not imply that these elements are a single entity. For example, different promoters can overlap and use the same TSS but use different sigma factors; similarly, activator and repressor sites can overlap a promoter region. In this sense, the correspondence between DNA and concepts is not a one-to-one relationship.
Overall, we have expanded definitions while trying to retain their essence. But, we are aware that different generalizations are feasible; for instance, we here focused on transcriptional regulators that are gene products, but they could be expanded to include small ligands such as ppGpp and consider the ppGpp regulon208.
The challenge in biology is that, experimentally, we may understand a few cases. However, evolution enables a much larger combination of possibilities. Ideally, a known corpus of well-known cases will generate principles that predict the universe of all possible combinations. For example, we can think of TFRS modules anchored at a proximal site in σ70-dependent promoters3,137 as an initial working hypothesis that restricts the range of possible promoter architectures that can be validated, or corrected, by bioengineering and synthetic approaches209. The challenge is to implement methods and strategies that test the validity of our current definitions on the one hand, and advance our quantitative and qualitative integrated understanding of microbial gene regulation on the other.
Supplementary Material
Acknowledgements
C.M.A is a doctoral student from the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México (UNAM) and has received CONACyT fellowship 576333. J.C.V. acknowledges funding by Universidad Nacional Autónoma de México (UNAM) and National Institutes of Health (NIH) [5R01GM110597-04, 1RO1GM131643-01A1 and R01GM077678]. J.C.V. acknowledges being on sabbatical leave at the Center for Genomic Regulation, Barcelona, Spain. B.O.P. acknowledges the support of the Galletti Endowment at UC San Diego. The authors acknowledge J. Soffer, S. Gama-Castro and H. Salgado for useful discussions, and David W. Sant for updating the definitions in Sequence Ontology. The authors also acknowledge the very valuable suggestions from the referees.
Glossary
- Operon
A set of adjacent cotranscribed genes
- Activators
Gene products that increase transcription, indicating their function is to enhance promoter activity
- Repressors
Gene products that decrease transcription, indicating their function is to hamper promoter activity
- Operator
A genetic entity adjacent to a group of genes that regulate their expression and that is sensitive to a repressor
- Motifs
Representations of a collection of binding sites that summarize their characteristics
- 5’-Rapid Amplification of cDNA Ends
(5’-RACE) is a method that amplifies mRNA between a defined internal site and its initiation site
- DNA supercoiling
The writhe of DNA over the double-stranded axis
- Monocistronic operons
Operons that encode a single gene product
- Transcriptional read-through
Transcription that allows RNA polymerase to continue transcription beyond termination sites
- Stimulons
Sets of genes (or sets of regulons) whose products are increased in response to a common environmental stimulus
- Global regulators
TFs that affect a large number of genes involved in many different functions
- Transcriptional pausing
A process in which the RNAP slows down transcription during elongation
Footnotes
Competing interests
The authors declare no competing interests.
Peer review information
Nature Reviews Genetics thanks A. S. Ribeiro, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
RegulonDB database: http://regulondb.ccg.unam.mx/
Ecocyc database: https://ecocyc.org
Sequence Ontology: http://www.sequenceontology.org
Basic Formal Ontology: https://basic-formal-ontology.org/
Supplementary information
Supplementary information is available for this paper at https://doi.org/10.1038/s41576-020-0254-8.
References
- 1.Miller JH The operon. 7, (Cold Spring Harbor Laboratory Pr, 1980). [Google Scholar]
- 2.Beckwith J The Operon: an Historical Account. in Escherichia coli and Salmonella: cellular and molecular biology (eds. Neidhardt F et al.) 1227–1231 (ASM Press, 1996). [Google Scholar]
- 3.Collado-Vides J, Magasanik B & Gralla JD Control site location and transcriptional regulation in Escherichia coli. Microbiol. Rev. 55, 371–394 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galagan JE et al. The Mycobacterium tuberculosis regulatory network and hypoxia. Nature (2013). doi: 10.1038/nature12337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robb NC et al. The transcription bubble of the RNA polymerase-promoter open complex exhibits conformational heterogeneity and millisecond-scale dynamics: Implications for transcription start-site selection. J. Mol. Biol. 425, 875–885 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eilbeck K et al. The Sequence Ontology: a tool for the unification of genome annotations. Genome Biol. 6, R44.1–R44.12 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Santos-Zavaleta A et al. RegulonDB v 10.5: Tackling challenges to unify classic and high throughput knowledge of gene regulation in E. coli K-12. Nucleic Acids Res. 47, D212–D220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karp PD et al. The ecocyc database. EcoSal Plus 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruff EF, Thomas Record M & Artsimovitch I Initial events in bacterial transcription initiation. Biomolecules 5, 1035–1062 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Estrem ST et al. Bacterial promoter architecture : subsite structure of UP elements and interactions with the carboxy-terminal domain of the RNA polymerase a subunit. Genes Dev. 13, 2134–2147 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y et al. Structural Basis of Transcription Initiation. Science (80-.). 338, 1076–1080 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keiltys S & Rosenberg M Constitutive Function of a Positively Regulated Promoter Reveals New Sequences Essential for Activity *. J. Biol. Chem. 262, 6389–6395 (1987). [PubMed] [Google Scholar]
- 13.Haugen SP et al. rRNA Promoter Regulation by Nonoptimal Binding of σ Region 1.2: An Additional Recognition Element for RNA Polymerase. Cell 125, 1069–1082 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Josaitis CA, Gaal T & Gourse RL Stringent control and growth-rate-dependent control have nonidentical promoter sequence requirements. Proc. Nati. Acad. Sci. USA 92, 1117–1121 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davis MC, Kesthely CA, Franklin EA & MacLellan SR The essential activities of the bacterial sigma factor. Can. J. Microbiol. 63, 89–99 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Losick R & Pero J Cascades of sigma factors. Cell 25, 582–584 (1981). [DOI] [PubMed] [Google Scholar]
- 17.Paget M & Helmann J Protein family review - The sigma(70) family of sigma factors. Genome Biol. 4, 1–6 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li X-Y & McClure WR Characterization of the closed complex intermediate formed during transcription initiation by Escherichia coli RNA polymerase. J. Biol. Chem. 273, 23549–23557 (1998). [DOI] [PubMed] [Google Scholar]
- 19.Saecker RM, Record MT & Dehaseth PL Mechanism of bacterial transcription initiation: RNA polymerase - Promoter binding, isomerization to initiation-competent open complexes, and initiation of RNA synthesis. J. Mol. Biol. 412, 754–771 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haugen SP, Ross W & Gourse RL Advances in bacterial promoter recognition and its control by factors that do not bind DNA. Nat. Rev. Microbiol. 6, 507–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Revyakin A, Liu C, Ebright RH & Strick TR Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science (80-.). 314, 1139–1143 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greive SJ & Von Hippel PH Thinking quantitatively about transcriptional regulation. Nat. Rev. Mol. Cell Biol. 6, 221–232 (2005). [DOI] [PubMed] [Google Scholar]
- 23.Helmann JD & Pieter L Protein - Nucleic Acid Interactions during Open Complex Formation Investigated by Systematic Alteration of the Protein and DNA Binding Partners †. 38, (1999). [DOI] [PubMed] [Google Scholar]
- 24.Schneider DA, Ross W & Gourse RL Control of rRNA expression in Escherichia coli. Curr. Opin. Microbiol. 6, 151–156 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Gourse RL et al. Strength and regulation without transcription factors: lessons from bacterial rRNA promoters. in Cold Spring Harbor symposia on quantitative biology 63, 131–140 (1998). [DOI] [PubMed] [Google Scholar]
- 26.Grigorova IL, Phleger NJ, Mutalik VK & Gross CA Insights into transcriptional regulation and σ competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl. Acad. Sci. U. S. A 103, 5332–5337 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sorensen KI, Baker KE, Kelln RA & Neuhard J Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J. Bacteriol. 175, 4137–4144 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Turnbough CL, Switzer RL & Sites TS Regulation of Pyrimidine Biosynthetic Gene Expression in Bacteria: Repression without Repressors. Microbiol. Mol. Biol. Rev. 72, 266–300 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adhya S, Gottesman M, Garges S & Oppenheim A Promoter resurrection by activators - a minireview. Gene 132, 1–6 (1993). [DOI] [PubMed] [Google Scholar]
- 30.Browning DF & Busby SJW The regulation of bacterial transcription initiation. Nat. Rev. Microbiol. 2, 1–9 (2004). [DOI] [PubMed] [Google Scholar]
- 31.Libis V, Delépine B & Faulon JL Sensing new chemicals with bacterial transcription factors. Curr. Opin. Microbiol. 33, 105–112 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Davidson EH The regulatory genome: gene regulatory networks in development and evolution. (Elsevier, 2010). [Google Scholar]
- 33.Ptashne M & Gann A Genes & Signals. (Cold Spring Laboratory Press, 2002). [Google Scholar]
- 34.Jacob F, Ullman A & Monod J Le promoteur, élément génétique nécesaire à l’expression d’un opéron. 258, 3125–3128 (1964). [PubMed] [Google Scholar]
- 35.Vogt V Breaks in DNA stimulate Transcription by Core RNA Polymerase. Nature 223, 854–855 (1969). [DOI] [PubMed] [Google Scholar]
- 36.Dausse J-P, Sentenac A & Fromageot P Interaction of RNA Polymerase from Escherichia coli with DNA: Influence of DNA Scissions on RNA-Polymerase Binding and Chain Initiation. Eur. J. Biochem. 31, 394–404 (1972). [DOI] [PubMed] [Google Scholar]
- 37.Takanami M, Sugimoto K, Sugisaki H & Okamoto T Sequence of promoter for coat protein gene of bacteriophage fd. Nature 260, 297–302 (1976). [DOI] [PubMed] [Google Scholar]
- 38.Schaller H, Gray C & Herrmann K Nucleotide Sequence of an RNA Polymerase Binding Site from the DNA of Bacteriophage fd. Proc. Natl. Acad. Sci. 72, 737–741 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pribnow D Nucleotide sequence of an RNA polymerase binding site at an early T7 promoter. Proc. Natl. Acad. Sci. U. S. A. 72, 784–788 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ponnambalam S, Webster C, Bingham A & Busby S Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal-35 region sequences. J. Biol. Chem. 261, 16043–16048 (1986). [PubMed] [Google Scholar]
- 41.Morett E & Buck M In vivo studies on the interaction of RNA polymerase-σ54 with the Klebsiella pneumoniae and Rhizobium meliloti nifH promoters. J. Mol. Biol. 210, 65–77 (1989). [DOI] [PubMed] [Google Scholar]
- 42.Travers AA Promoter Sequence for Stringent Control of Bacterial Ribonucleic Acid Synthesis. J. Bacteriol. 141, 973–976 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ross W et al. A Third Recognition Element in Bacterial Promoters: DNA Binding by the a Subunit of RNA Polymerase. Science (80-.). 262, 1407–1413 (1993). [DOI] [PubMed] [Google Scholar]
- 44.Harden TT et al. Bacterial RNA polymerase can retain σ70 throughout transcription. Proc. Natl. Acad. Sci. U. S. A. 113, 602–607 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun Z et al. Density of σ70 promoter-like sites in the intergenic regions dictates the redistribution of RNA polymerase during osmotic stress in Escherichia coli. Nucleic Acids Res. 47, 3970–3985 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huerta AM & Collado-Vides J Sigma70 promoters in Escherichia coli: specific transcription in dense regions of overlapping promoter-like signals. J. Mol. Biol. 333, 261–278 (2003). [DOI] [PubMed] [Google Scholar]
- 47.Huerta AM, Francino MP, Morett E & Collado-Vides J Selection for unequal densities of $σ$70 promoter-like signals in different regions of large bacterial genomes. PLoS Genet. 2, 1740–1750 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Froula JL & Francino MP Selection against Spurious Promoter Motifs Correlates with Translational Efficiency across Bacteria. PLoS One 2, 1–11 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yona AH, Alm EJ & Gore J Random sequences rapidly evolve into de novo promoters. Nat. Commun. 9, 1–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urtecho G et al. Genome-wide Functional Characterization of Escherichia coli Promoters and Regulatory Elements Responsible for their Function. bioRxiv (2020). doi: 10.1101/2020.01.04.894907 [DOI] [Google Scholar]
- 51.Jones BB, Chan H, Rothstein S, Wells RD & Reznikoff WS RNA polymerase binding sites in λplac5 DNA. Proc. Natl. Acad. Sci. U. S. A. 74, 4914–4918 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Grainger DC, Hurd D, Harrison M, Holdstock J & Busby SJW Studies of the distribution of Escherichia coli cAMP-receptor protein and RNA polymerase along the E. coli chromosome. Proc. Natl. Acad. Sci. 102, 17693–17698 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wigneshweraraj S et al. Modus operandi of the bacterial RNA polymerase containing the σ54 promoter-specificity factor. Mol. Microbiol. 68, 538–546 (2008). [DOI] [PubMed] [Google Scholar]
- 54.Bonocora RP, Smith C, Lapierre P & Wade JT Genome-Scale Mapping of Escherichia coli σ54 Reveals Widespread, Conserved Intragenic Binding. PLoS Genet. 11, 1–30 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schaefer J, Engl C, Zhang N, Lawton E & Buck M Genome wide interactions of wild-type and activator bypass forms of σ54. Nucleic Acids Res. 43, 7280–7291 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bono AC et al. Novel DNA Binding and Regulatory Activities for σ54 (RpoN) in Salmonella enterica Serovar Thyphimurium 14028s. J. Bacteriol. 199, 1–24 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shao X et al. RpoN-Dependent Direct Regulation of Quorum Sensing and the Type VI Secretion System in Pseudomonas aeruginosa PAO1. J. Bacteriol. 200, 1–17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Goldman SR, Ebright RH & Nickels BE Direct detection of abortive RNA transcripts in vivo. Science (80-.). 324, 927–928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yus E et al. Transcription start site associated RNAs in bacteria. Mol. Syst. Biol. 8, 1–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dornenburg JE, DeVita AM, Palumbo MJ & Wade JT Widespread antisense transcription in Escherichia coli. MBio 1, 1–4 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raghavan R, Sloan DB & Ochman H Pervasive transcription is widespread but rarely conserved in Enteric bacteria. MBio 3, 1–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sasse-dwight S & Gralla JAYD Probing the Escherichia coli glnALG upstream activation mechanism in vivo. Proc. Nati. Acad. Sci. USA 85, 8934–8938 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Domínguez-Cuevas P, Marín P, Ramos JL & Marqués S RNA polymerase holoenzymes can share a single transcription start site for the Pm promoter: Critical nucleotides in the −7 to −18 region are needed to select between RNA polymerase with σ38 or σ32. J. Biol. Chem. 280, 41315–41323 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Reichenbach B, Göpel Y & Görke B Dual control by perfectly overlapping σ54- and σ70-promoters adjusts small RNA GlmY expression to different environmental signals. Mol. Microbiol. 74, 1054–1070 (2009). [DOI] [PubMed] [Google Scholar]
- 65.Wade JT et al. Extensive functional overlap between σ factors in Escherichia coli. Nat. Struct. Mol. Biol. 13, 806–814 (2006). [DOI] [PubMed] [Google Scholar]
- 66.Jacob F & Monod J Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356 (1961). [DOI] [PubMed] [Google Scholar]
- 67.Jacob F, Monod J & Pasteur I The genetic control and cytoplasmic expression of “inducibility” in the synthesis of β-galactosidase by E. coli. J. Mol. Biol. 1, 165–178 (1959). [Google Scholar]
- 68.Gilbert W, Muller-Hill B & Müller-Hill B Isolation of the Lac Repressor. Proc. Natl. Acad. Sci. 56, 1891–1898 (1966). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ptashne M Specific Binding of the λ Phage Repressor to λ DNA. Nature 214, 232–234 (1967). [DOI] [PubMed] [Google Scholar]
- 70.Maniatis T et al. Recognition sequences of repressor and polymerase in the operators of bacteriophage lambda. Cell (1975). doi: 10.1016/0092-8674(75)90018-5 [DOI] [PubMed] [Google Scholar]
- 71.Englesberg E, Irr Joseph Power,’ And J & Lee’ N Positive Control of Enzyme Synthesis by Gene C in the L-Arabinose System. J. Bacteriol. 90, 946–957 (1965). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schwartz M Aspects biochimiques et génétiques du metabolisme du maltose chez Escherichia coli K12. COMPTES RENDUS Hebd. DES SEANCES L Acad. DES Sci. 260, 2613 (1965). [PubMed] [Google Scholar]
- 73.Thomas R Control of development in temperate bacteriophages. I. Induction of prophage genes following hetero-immune super-infection. J. Mol. Biol. 22, 79–95 (1966). [Google Scholar]
- 74.Borukhov S, Lee J & Laptenko O Bacterial transcription elongation factors: new insights into molecular mechanism of action. Mol. Microbiol. 55, 1315–1324 (2005). [DOI] [PubMed] [Google Scholar]
- 75.Storz G, Opdyke JA & Wassarman KM Regulating bacterial transcription with small RNAs. Cold Spring Harb. Symp. Quant. Biol. 71, 269–273 (2006). [DOI] [PubMed] [Google Scholar]
- 76.Schwenk S & Arnvig KB Regulatory RNA in Mycobacterium tuberculosis, back to basics. Pathog. Dis. 76, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Gourse RL et al. Transcriptional Responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wassarman KM 6S RNA: a small RNA regulator of transcription. Curr. Opin. Microbiol. 10, 164–168 (2007). [DOI] [PubMed] [Google Scholar]
- 79.Wassarman KM, Repoila F, Rosenow C, Storz G & Gottesman S Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15, 1637–1651 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ptashne M Specific Binding of the λ Phage Repressor to λ DNA. Nature 214, 232–234 (1967). [DOI] [PubMed] [Google Scholar]
- 81.Eckweiler D, Dudek C-A, Hartlich J, Brötje D & Jahn D PRODORIC2: the bacterial gene regulation database in 2018. Nucleic Acids Res. 46, D320–D326 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fornes O et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 48, D87–D92 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu H et al. AnimalTFDB 3.0: a comprehensive resource for annotation and prediction of animal transcription factors. Nucleic Acids Res. 47, D33–D38 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Jin J et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45, D1040–D1045 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teixeira MC et al. YEASTRACT: an upgraded database for the analysis of transcription regulatory networks in Saccharomyces cerevisiae. Nucleic Acids Res. 46, D348–D353 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paul BJ et al. DksA. Cell 118, 311–322 (2004). [DOI] [PubMed] [Google Scholar]
- 87.Gregory BD et al. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc. Natl. Acad. Sci. U. S. A. 101, 4554–4559 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pratt LA & Silhavy TJ Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29, 1225–1236 (1998). [DOI] [PubMed] [Google Scholar]
- 89.Srivastava DB et al. Structure and function of CarD, an essential mycobacterial transcription factor. Proc. Natl. Acad. Sci. U. S. A 110, 12619–12624 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Browning DF & Busby SJW Local and global regulation of transcription initiation in bacteria. Nat. Rev. Microbiol. (2016). doi: 10.1038/nrmicro.2016.103 [DOI] [PubMed] [Google Scholar]
- 91.Haldenwang WG, Lang N & Losick R A sporulation-induced sigma-like regulatory protein from b. subtilis. Cell 23, 615–624 (1981). [DOI] [PubMed] [Google Scholar]
- 92.Grossman AD, Erickson JW & Gross CA The htpR gene product of E. coli is a sigma factor for heat-shock promoters. Cell 38, 383–390 (1984). [DOI] [PubMed] [Google Scholar]
- 93.Taylor WE et al. Transcription from a heat-inducible promoter causes heat shock regulation of the sigma subunit of E. coli RNA polymerase. Cell 38, 371–381 (1984). [DOI] [PubMed] [Google Scholar]
- 94.Burgess RR & Travers AA Factor Stimulating Transcription by RNA Polymerase. Nature 221, 43–46 (1969). [DOI] [PubMed] [Google Scholar]
- 95.Feklístov A, Sharon BD, Darst SA & Gross CA Bacterial Sigma Factors: A Historical, Structural, and Genomic Perspective. Annu. Rev. Microbiol. 68, 357–376 (2014). [DOI] [PubMed] [Google Scholar]
- 96.Campagne S, Marsh ME, Capitani G, Vorholt JA & Allain FHT Structural basis for −10 promoter element melting by environmentally induced sigma factors. Nat. Struct. Mol. Biol. 21, 269–276 (2014). [DOI] [PubMed] [Google Scholar]
- 97.Griffith KL, Shah IM, Myers TE, O’Neill MC & Wolf RE Evidence for ‘pre-recruitment’ as a new mechanism of transcription activation in Escherichia coli: The large excess of SoxS binding sites per cell relative to the number of SoxS molecules per cell. Biochem. Biophys. Res. Commun. 291, 979–986 (2002). [DOI] [PubMed] [Google Scholar]
- 98.Shah IM & Wolf RE Novel protein-protein interaction between Escherichia coli SoxS and the DNA binding determinant of the RNA polymerase α subunit: SoxS functions as a co-sigma factor and redeploys RNA polymerase from UP-element-containing promoters to SoxS-dependent promot. J. Mol. Biol. 343, 513–532 (2004). [DOI] [PubMed] [Google Scholar]
- 99.Li Z & Demple B Sequence specificity for DNA binding by Escherichia coli SoxS and Rob proteins. Mol. Microbiol. 20, 937–945 (1996). [DOI] [PubMed] [Google Scholar]
- 100.Kaur G et al. Mycobacterium tuberculosis CarD, an essential global transcriptional regulator forms amyloid-like fibrils. Sci. Rep. 8, 1–13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Hubin EA et al. Structure and function of the mycobacterial transcription initiation complex with the essential regulator RbpA. Elife 6, 1–40 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Rammohan J, Manzano AR, Garner AL, Stallings CL & Galburt EA CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res. 43, 3272–3285 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dorman CJ, Schumacher MA, Bush MJ, Brennan RG & Buttner MJ When is a transcription factor a NAP? Curr. Opin. Microbiol. 55, 26–33 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Schneider R et al. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 29, 5107–5114 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Dillon SC & Dorman CJ Bacterial nucleoid-associated proteins, nucleoid structure and gene expression. Nat. Rev. Microbiol. 8, 185–195 (2010). [DOI] [PubMed] [Google Scholar]
- 106.Dame RT The role of nucleoid-associated proteins in the organization and compaction of bacterial chromatin. Mol. Microbiol. 56, 858–870 (2005). [DOI] [PubMed] [Google Scholar]
- 107.Blot N, Mavathur R, Geertz M, Travers A & Muskhelishvili G Homeostatic regulation of supercoiling sensitivity coordinates transcription of the bacterial genome. EMBO Rep. 7, 710–715 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Rimsky S, Zuber F, Buckle M & Buc H A molecular mechanism for the repression of transcription by the H-NS protein. Mol. Microbiol. 42, 1311–1323 (2001). [DOI] [PubMed] [Google Scholar]
- 109.Opel ML et al. Activation of transcription initiation from a stable RNA promoter by a Fis protein-mediated DNA structural transmission mechanism. Mol. Microbiol. 53, 665–674 (2004). [DOI] [PubMed] [Google Scholar]
- 110.Ihara K et al. Expression of the alaE gene is positively regulated by the global regulator Lrp in response to intracellular accumulation of L-alanine in Escherichia coli. J. Biosci. Bioeng. 123, 444–450 (2017). [DOI] [PubMed] [Google Scholar]
- 111.Finkel SE & Johnson RC The Fis protein: it’s not just for DNA inversion anymore. Mol. Microbiol. 7, 1023–1023 (1993). [DOI] [PubMed] [Google Scholar]
- 112.Brandi A, Giangrossi M, Giuliodori AM & Falconi M An interplay among FIS, H-NS, and guanosine tetraphosphate modulates transcription of the Escherichia coli cspA gene under physiological growth conditions. Front. Mol. Biosci. 3, 1–12 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Govantes F, Orjalo AV & Gunsalus RP Interplay between three global regulatory proteins mediates oxygen regulation of the Escherichia coli cytochrome d oxidase (cydAB) operon. Mol. Microbiol. 38, 1061–1073 (2002). [DOI] [PubMed] [Google Scholar]
- 114.Meenakshi S, Karthik M & Munavar MH A putative curved DNA region upstream of rcsA in Escherichia coli plays a key role in transcriptional regulation by H-NS. FEBS Open Bio 8, 1209–1218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Carmona M, Claverie-Martin F & Magasanik B DNA bending and the initiation of transcription at σ54-dependent bacterial promoters. Proc. Natl. Acad. Sci. 94, 9568–9572 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Ninfa AJ, Reitzer LJ & Magasanik B Initiation of transcription at the bacterial glnAp2 promoter by purified E. coli components is facilitated by enhancers. Cell 50, 1039–1046 (1987). [DOI] [PubMed] [Google Scholar]
- 117.Azam TA & Ishihama A Twelve species of the nucleoid-associated protein from Escherichia coli. J. Biol. Chem. 274, 33105–33113 (1999). [DOI] [PubMed] [Google Scholar]
- 118.Martinez-Antonio A & Collado-Vides J Identifying global regulators in transcriptional regulatory networks in bacteria. Curr. Opin. Microbiol. 6, 482–489 (2003). [DOI] [PubMed] [Google Scholar]
- 119.Santos-Zavaleta A et al. A unified resource for transcriptional regulation in Escherichia coli K-12 incorporating high-throughput-generated binding data into RegulonDB version 10.0. BMC Biol. 16, 1–12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Galagan J, Lyubetskaya A & Gomes A ChIP-Seq and the Complexity of Bacterial Transcriptional Regulation. 43–68 (2012). doi: 10.1007/82 [DOI] [PubMed] [Google Scholar]
- 121.Babin BM et al. SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 113, E597–E605 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jones CJ et al. ChIP-Seq and RNA-Seq Reveal an AmrZ-Mediated Mechanism for Cyclic di-GMP Synthesis and Biofilm Development by Pseudomonas aeruginosa. PLoS Pathog. 10, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Perkins TT et al. ChIP-seq and transcriptome analysis of the OmpR regulon of Salmonella enterica serovars Typhi and Typhimurium reveals accessory genes implicated in host colonization. Mol. Microbiol. 87, 526–538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Lobel L & Herskovits AA Systems Level Analyses Reveal Multiple Regulatory Activities of CodY Controlling Metabolism, Motility and Virulence in Listeria monocytogenes. PLoS Genet. 12, 1–27 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vannini A et al. Comprehensive mapping of the Helicobacter pylori NikR regulon provides new insights in bacterial nickel responses. Sci. Rep. 7, 1–14 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Vergara-Irigaray M, Fookes MC, Thomson NR & Tang CM RNA-seq analysis of the influence of anaerobiosis and FNR on Shigella flexneri. BMC Genomics 15, 1–22 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Grainger DC et al. Genomic studies with Escherichia coli MelR protein: applications of chromatin immunoprecipitation and microarrays. J. Bacteriol. 186, 6938–6943 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Seo SW et al. Revealing genome-scale transcriptional regulatory landscape of OmpR highlights its expanded regulatory roles under osmotic stress in Escherichia coli K-12 MG1655. Sci. Rep. 7, 1–10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wade JT, Reppas NB, Church GM & Struhl K Genomic analysis of LexA binding reveals the permissive nature of the Escherichia coli genome and identifies unconventional target sites. Genes Dev. 19, 2619–2630 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Visweswariah SS & Busby SJW Evolution of bacterial transcription factors : how proteins take on new tasks, but do not always stop doing the old ones. Trends Microbiol. 23, 463–467 (2015). [DOI] [PubMed] [Google Scholar]
- 131.Beauchene NA et al. Impact of anaerobiosis on expression of the iron-responsive Fur and RyhB regulons. MBio 6, e01947–15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Gao Y et al. Systematic discovery of uncharacterized transcription factors in Escherichia coli K-12 MG1655. Nucleic Acids Res. 46, 10682–10696 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Shimada T, Ishihama A, Busby SJW & Grainger DC The Escherichia coli RutR transcription factor binds at targets within genes as well as intergenic regions. Nucleic Acids Res. 36, 3950–3955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Myers KS et al. Genome-scale analysis of Escherichia coli FNR reveals complex features of transcription factor binding. PLoS Genet. 9, e1003565–e1003565 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Kroner GM, Wolfe MB & Freddolino PL Escherichia coli Lrp regulates one-third of the genome via direct, cooperative, and indirect routes. J. Bacteriol. 201, e00411–18 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Jacob F, Perrin D, Sanchez C & Monod J L’opéron: groupe de gènes à expression coordonnée par un opérateur. Comptes Rendus l’Academie des Sci. 250, 1727–1729 (1960). [PubMed] [Google Scholar]
- 137.Gralla JD & Collado-Vides J Organization and function of transcription regulatory elements. in Escherichia coli and Salmonella: Cellular and Molecular Biology (eds. Neidhardt F & Curtiss R) 1232–1245 (ASM Press, 1996). [Google Scholar]
- 138.Collado-Vides J et al. Bioinformatics resources for the study of gene regulation in bacteria. J. Bacteriol. 91, 23–31 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Reitzer LJ & Magasanik B Transcription of glnA in E. coli is stimulated by activator bound to sites far from the promoter. Cell 45, 785–792 (1986). [DOI] [PubMed] [Google Scholar]
- 140.Claverie-Martin F & Magasanik B Role of integration host factor in the regulation of the glnHp2 promoter of Escherichia coli. Proc. Natl. Acad. Sci. 88, 1631–1635 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gralla JD Promoter Recognition and mRNA Initiation by Escherichia coli Eσ70. Methods Enzymol. 185, 37–54 (1990). [DOI] [PubMed] [Google Scholar]
- 142.Hancock JM & Zvelebil MJ Concise Encyclopaedia of Bioinformatics and Computational Biology. (John Wiley & Sons, 2014). [Google Scholar]
- 143.Collado-Vides J The search for a grammatical theory of regulation is formally justified by showing the inadequacy of context-free grammars. Bioinformatics 7, 321–326 (1991). [DOI] [PubMed] [Google Scholar]
- 144.Buchler NE, Gerland U & Hwa T On schemes of combinatorial transcription logic. Proc. Natl. Acad. Sci. U. S. A. 100, 5136–5141 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Bintu L et al. Transcriptional regulation by the numbers: Applications. Curr. Opin. Genet. Dev. 15, 125–135 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Phillips R et al. Figure 1 Theory Meets Figure 2 Experiments in the Study of Gene Expression. Annu. Rev. Biophys. 48, 121–163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Bintu L et al. Transcriptional regulation by the numbers: models. Curr. Opin. Genet. Dev. 15, 116–124 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Segal E et al. Module networks: identifying regulatory modules and their condition-specific regulators from gene expression data. Nat. Genet. 34, 166–176 (2003). [DOI] [PubMed] [Google Scholar]
- 149.Fraser CM et al. The Minimal Gene Complement of Mycoplasma genitalium. Science (80-.). 270, 397–404 (1995). [DOI] [PubMed] [Google Scholar]
- 150.Dorman CJ Regulation of transcription by DNA supercoiling in mycoplasma genitalium: Global control in the smallest known self-replicating genome. Mol. Microbiol. 81, 302–304 (2011). [DOI] [PubMed] [Google Scholar]
- 151.Zhang W & Baseman JB Transcriptional regulation of MG_149, an osmoinducible lipoprotein gene from Mycoplasma genitalium. Mol. Microbiol. 81, 327–339 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Roth CW & Nester EW Co-ordinate control of tryptophan, histidine and tyrosine enzyme synthesis in Bacillus subtilis. J. Mol. Biol. 62, 577–589 (1971). [DOI] [PubMed] [Google Scholar]
- 153.Salgado H, Moreno-Hagelsieb G, Smith TF & Collado-Vides J Operons in Escherichia coli: Genomic analyses and predictions. Proc. Natl. Acad. Sci. 97, 6652–6657 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Monod J et al. Allosteric proteins and cellular control systems. J. Mol. Biol. 6, 306–329 (1963). [DOI] [PubMed] [Google Scholar]
- 155.Jacob F Genetics of the bacterial cell. Science (80-.). 152, 1470–1478 (1966). [DOI] [PubMed] [Google Scholar]
- 156.Bockhorst J et al. Predicting bacterial transcription units using sequence and expression data. 19, (2003). [DOI] [PubMed] [Google Scholar]
- 157.Mao X et al. DOOR 2.0: presenting operons and their functions through dynamic and integrated views. 42, 654–659 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Koide T et al. Prevalence of transcription promoters within archaeal operons and coding sequences. Mol. Syst. Biol. 5, 1–16 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Pray LA What is a gene? Colinearity and transcription units. Nat. Educ. 1, (2008). [Google Scholar]
- 160.Cho B et al. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 27, 1043–1049 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Liu J & Turnbough CL Effects of transcriptional start site sequence and position on nucleotide- sensitive selection of alternative start sites at the pyrC promoter in Escherichia coli. J. Bacteriol. 176, 2938–2945 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Goldman SR et al. NanoRNAs Prime Transcription Initiation In Vivo. Mol. Cell 42, 817–825 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Ciampi MS Rho-dependent terminators and transcription termination. Microbiology 152, 2515–2528 (2006). [DOI] [PubMed] [Google Scholar]
- 164.Lau LF & Roberts JW Rho-dependent transcription termination at lambda R1 requires upstream sequences. J. Biol. Chem. 260, 574–584 (1985). [PubMed] [Google Scholar]
- 165.Richardson LV & Richardson JP Rho-dependent Termination of Transcription Is Governed Primarily by the Upstream Rho Utilization (rut) Sequences of a Terminator. J. Biol. Chem. 271, 21597–21603 (1996). [DOI] [PubMed] [Google Scholar]
- 166.Jeong KS, Ahn J & Khodursky AB Spatial patterns of transcriptional activity in the chromosome of Escherichia coli. Genome Biol. 5, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Junier I, Unal EB, Yus E, Lloréns-Rico V & Serrano L Insights into the Mechanisms of Basal Coordination of Transcription Using a Genome-Reduced Bacterium. Cell Syst. 2, 391–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Yan B, Boitano M, Clark TA & Ettwiller L SMRT-Cappable-seq reveals complex operon variants in bacteria. Nat. Commun. 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Bauerle RH & Margolin P Evidence for two sites for initiation of gene expression in the tryptophan operon of Salmonella typhimurium. J. Mol. Biol. 26, 423–436 (1967). [DOI] [PubMed] [Google Scholar]
- 170.Ueno-Nishio S, Backman KC & Magasanik B Regulation at the glnL-operator-promoter of the complex glnALG operon of Escherichia coli. J. Bacteriol. 153, 1247–1251 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Conway T et al. Unprecedented High-Resolution View of Bacterial Operon Architecture Revealed by RNA Sequencing. MBio 5, e01442–14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Li S, Dong X & Su Z Directional RNA-seq reveals highly complex condition-dependent transcriptomes in E. coli K12 through accurate full-length transcripts assembling. BMC Genomics 14, 1–24 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mao X et al. Revisiting operons: An analysis of the landscape of transcriptional units in E. coli. BMC Bioinformatics 16, 1–9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Ju X, Li D & Liu S Full-length RNA profiling reveals pervasive bidirectional transcription terminators in bacteria. Nat. Microbiol. 4, 1907–1918 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Sharma CM et al. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464, 250–255 (2010). [DOI] [PubMed] [Google Scholar]
- 176.Lybecker M, Bilusic I & Raghavan R Pervasive transcription: Detecting functional RNAs in bacteria. Transcription 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Wade JT & Grainger DC Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat. Rev. Microbiol. 12, 647–653 (2014). [DOI] [PubMed] [Google Scholar]
- 178.Price MN et al. Indirect and suboptimal control of gene expression is widespread in bacteria. Mol. Syst. Biol. 9, 1–18 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Price MN, Wetmore KM, Deutschbauer AM & Arkin AP A comparison of the costs and benefits of bacterial gene expression. PLoS One 11, 1–22 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Shao W, Price MN, Deutschbauer AM, Romine MF & Arkin AP Conservation of transcription start sites within genes across a bacterial genus. MBio 5, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Wade JT & Grainger DC Spurious transcription and its impact on cell function. Transcription 9, 182–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Pannier L, Merino E, Marchal K & Collado-Vides J Effect of genomic distance on coexpression of coregulated genes in E. coli. PLoS One 12, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Stringer AM et al. Genome-scale analyses of Escherichia coli and Salmonella enterica AraC reveal noncanonical targets and an expanded core regulon. J. Bacteriol. 196, 660–671 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Chen Y-J et al. Characterization of 582 natural and synthetic terminators and quantification of their design constraints. Nat. Methods 10, 659 (2013). [DOI] [PubMed] [Google Scholar]
- 185.Maas WK & Clark AJ Studies on the mechanism of repression of arginine biosynthesis in Escherichia coli: II. Dominance of repressibility in diploids. J. Mol. Biol. 8, 365–370 (1964). [DOI] [PubMed] [Google Scholar]
- 186.Ledezma-Tejeida D, Altamirano-Pacheco L, Fajardo V & Collado-Vides J Limits to a classic paradigm: most transcription factors in E. coli regulate genes involved in multiple biological processes. Nucleic Acids Res. 47, 6656–6667 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Pittard J & Yang J Biosynthesis of the aromatic amino acids. EcoSal Plus 3, 1–39 (2008). [DOI] [PubMed] [Google Scholar]
- 188.Smith MW & Neidhardt FC Proteins induced by aerobiosis in Escherichia coli. J. Bacteriol. 154, 336–343 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Schaefer EM, Hartz D, Gold L & Simoni RD Ribosome-binding sites and RNA-processing sites in the transcript of the Escherichia coli unc operon. J. Bacteriol. 171, 3901–3908 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Monod J, Wyman J & Changeux J-P On the nature of allosteric transitions: a plausible model. J Mol Biol 12, 88–118 (1965). [DOI] [PubMed] [Google Scholar]
- 191.Hoch JA Two-component and phosphorelay systems Signal transduction systems. Curr. Opin. Microbiol. 3, 165–170 (2000). [DOI] [PubMed] [Google Scholar]
- 192.Grundy FJ & Henkin TM Regulation of gene expression by effectors that bind to RNA. Curr. Opin. Microbiol. 7, 126–131 (2004). [DOI] [PubMed] [Google Scholar]
- 193.Horii T et al. Regulation of SOS functions: purification of E. coli LexA protein and determination of its specific site cleaved by the RecA protein. Cell 27, 515–522 (1981). [DOI] [PubMed] [Google Scholar]
- 194.Jenal U & Hengge-Aronis R Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 6, 163–172 (2003). [DOI] [PubMed] [Google Scholar]
- 195.Uphoff S et al. Stochastic activation of a DNA damage response causes cell-to-cell mutation rate variation. Science (80-.). 351, 1094–1097 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Takinowaki H, Matsuda Y, Yoshida T, Kobayashi Y & Ohkubo T The solution structure of the methylated form of the N-terminal 16-kDa domain of Escherichia coli Ada protein. Protein Sci. 15, 487–497 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Kotte O, Zaugg JB & Heinemann M Bacterial adaptation through distributed sensing of metabolic fluxes. Mol. Syst. Biol. 6, 1–9 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Mekalanos JJ Environmental signals controlling expression of virulence determinants in bacteria. J. Bacteriol. 174, 1–7 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Maurelli AT Temperature regulation of virulence genes in pathogenic bacteria: a general strategy for human pathogens? Microb. Pathog. 7, 1–10 (1989). [DOI] [PubMed] [Google Scholar]
- 200.Miller JF, Mekalanos JJ & Falkow S Coordinate regulation and sensory transduction in the control of bacterial virulence. Science (80-.). 243, 1355–1362 (1989). [DOI] [PubMed] [Google Scholar]
- 201.Hurme R, Berndt KD, Normark SJ & Rhen M A proteinaceous gene regulatory thermometer in Salmonella. Cell 90, 55–64 (1997). [DOI] [PubMed] [Google Scholar]
- 202.Piraner DI, Abedi MH, Moser BA, Lee-Gosselin A & Shapiro MG Tunable thermal bioswitches for in vivo control of microbial therapeutics. Food, Pharm. Bioeng. Div. 2017 - Core Program. Area 2017 AIChE Annu. Meet. 2, 695–702 (2017). [DOI] [PubMed] [Google Scholar]
- 203.Lindner R et al. Photoactivation Mechanism of a Bacterial Light-Regulated Adenylyl Cyclase. J. Mol. Biol. 429, 1336–1351 (2017). [DOI] [PubMed] [Google Scholar]
- 204.Winkler A et al. A ternary AppA-PpsR-DNA complex mediates light regulation of photosynthesis-related gene expression. Nat. Struct. Mol. Biol. 20, 859–867 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Smith B, Kumar A & Bittner T Basic Formal Ontology for Bioinformatics. IFOMIS Reports (2005). [Google Scholar]
- 206.Strainic MG, Sullivan JJ, Collado-Vides J & DeHaseth PL Promoter interference in a bacteriophage lambda control region: Effects of a range of interpromoter distances. J. Bacteriol. 182, 216–220 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Scherrer K Primary transcripts: From the discovery of RNA processing to current concepts of gene expression - Review. Exp. Cell Res. 373, 1–33 (2018). [DOI] [PubMed] [Google Scholar]
- 208.Sanchez-Vazquez P, Dewey CN, Kitten N, Ross W & Gourse RL Genome-wide effects on Escherichia coli transcription from ppGpp binding to its two sites on RNA polymerase. Proc. Natl. Acad. Sci. 116, 8310–8319 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Browning DF, Butala M & Busby SJW Bacterial Transcription Factors: Regulation by Pick ‘N’Mix. J. Mol. Biol. 431, 4067–4077 (2019). [DOI] [PubMed] [Google Scholar]
- 210.Ho Y, Wulff DL & Rosenberg M Bacteriophage λ protein cII binds promoters on the opposite face of the DNA helix from RNA polymerase. Nature 304, 703–708 (1983). [DOI] [PubMed] [Google Scholar]
- 211.Buck M & Cannon W Specific binding of the transcription factor sigma-54 to promoter DNA. 358, 19–21 (1992). [DOI] [PubMed] [Google Scholar]
- 212.Ogasawara H, Shinohara S, Yamamoto K & Ishihama A Novel regulation targets of the metal-response BasS-BasR two-component system of Escherichia coli. Microbiology 158, 1482–1492 (2012). [DOI] [PubMed] [Google Scholar]
- 213.Yamamoto K et al. Anaerobic regulation of citrate fermentation by CitAB in Escherichia coli. Biosci. Biotechnol. Biochem. 72, 3011–3014 (2008). [DOI] [PubMed] [Google Scholar]
- 214.Cho BK, Federowicz S, Park YS, Zengler K & Palsson B Deciphering the transcriptional regulatory logic of amino acid metabolism. Nat. Chem. Biol. 8, 65–71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Cho BK et al. The PurR regulon in Escherichia coli K-12 MG1655. Nucleic Acids Res. 39, 6456–6464 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Kim D et al. Systems assessment of transcriptional regulation on central carbon metabolism by Cra and CRP. Nucleic Acids Res. 46, 2901–2917 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Shimada T, Yamamoto K & Ishihama A Involvement of the leucine response transcription factor LeuO in regulation of the genes for sulfa drug efflux. J. Bacteriol. 191, 4562–4571 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Seo SW et al. Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 5, 1–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Seo SW, Kim D, O’Brien EJ, Szubin R & Palsson BO Decoding genome-wide GadEWX-transcriptional regulatory networks reveals multifaceted cellular responses to acid stress in Escherichia coli. Nat. Commun. 6, 1–8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Seo SW, Kim D, Szubin R & Palsson BO Genome-wide Reconstruction of OxyR and SoxRS Transcriptional Regulatory Networks under Oxidative Stress in Escherichia coli K-12 MG1655. Cell Rep. 12, 1289–1299 (2015). [DOI] [PubMed] [Google Scholar]
- 221.Sogaard-Andersen L, Mellegaard NE, Douthwaite SR & Valentin-Hansen P Tandem DNA-bound cAMP-CRP complexes are required for transcriptional repression of the deoP2 promoter by the CytR repressor in Escherichia coli. Mol. Microbiol. 4, 1595–1601 (1990). [DOI] [PubMed] [Google Scholar]
- 222.Barnard A, Wolfe A & Busby S Regulation at complex bacterial promoters: How bacteria use different promoter organizations to produce different regulatory outcomes. Current Opinion in Microbiology 102–108 (2004). doi: 10.1016/j.mib.2004.02.011 [DOI] [PubMed] [Google Scholar]
- 223.Tao H, Hasona A, Do PM, Ingram LO & Shanmugam KT Global gene expression analysis revealed an unsuspected deo operon under the control of molybdate sensor, ModE protein, in Escherichia coli. Arch. Microbiol. 184, 225–233 (2005). [DOI] [PubMed] [Google Scholar]
- 224.González-Gil G, Bringmann P & Kahmann R FIS is a regulator of metabolism in Escherichia coli. Mol. Microbiol. 22, 21–29 (1996). [DOI] [PubMed] [Google Scholar]
- 225.Valentin-Hansen P, Albrechtsen B & Løve Larsen JE DNA-protein recognition: demonstration of three genetically separated operator elements that are required for repression of the Escherichia coli deoCABD promoters by the DeoR repressor. EMBO J. 5, 2015–2021 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.