Abstract
The adhesion GPCR latrophilin 3 (ADGRL3) has been associated with increased risk of attention-deficit/hyperactivity disorder (ADHD) and substance use in human genetic studies. Knockdown in multiple species leads to hyperlocomotion and altered dopamine signaling. Thus, ADGRL3 is a potential target for treatment of neuropsychiatric disorders that involve dopamine dysfunction, but its basic signaling properties are poorly understood. Identification of adhesion GPCR signaling partners has been limited by lack of tools to acutely activate these receptors in living cells. Here, we designed a novel acute activation strategy to characterize ADGRL3 signaling by engineering a receptor construct in which we could trigger acute activation enzymatically. Using this assay, we found that ADGRL3 signals through G12/G13 and Gq, with G12/13 the most robustly activated. Gα12/13 is a new player in ADGRL3 biology, opening up unexplored roles for ADGRL3 in the brain. Our methodological advancements should be broadly useful in adhesion GPCR research.
Introduction
Adhesion G protein-coupled receptors (aGPCRs) are crucial regulators of diverse functions in the nervous, immune, cardiac, and musculoskeletal systems, and their dysregulation has been linked to a variety of diseases and cancers1,2. The aGPCR latrophilin 3 (ADGRL3, human homolog) was recently associated with an increased risk of attention-deficit/hyperactivity disorder (ADHD) and substance use in human genetic studies3,4. Gene knockdown of ADGRL3 homologs in flies5, fish6,7, rat8 and mouse9 has been associated with a pan-species hyperlocomotor phenotype, as well as dysregulation of dopamine signaling. Thus, ADGRL3 may offer a novel target for modulating dopamine signaling, with important therapeutic implications for the treatment of ADHD and other neuropsychiatric disorders that involve dopamine dysfunction, such as schizophrenia. However, the identity of the G protein partners and downstream effector pathways controlled by ADGRL3 are poorly understood. Therefore, mapping these basic signaling properties remains a crucial first step in understanding how this receptor modulates dopaminergic neurotransmission.
Adhesion GPCRs form the second largest, yet most enigmatic class of the human GPCR superfamily. These atypical and complex GPCRs take part in cell-cell interactions through their enormous extracellular N-terminal domain and intracellular signaling via their canonical heptahelical transmembrane domain (Fig. 1a). During biosynthesis, the aGPCR is cleaved at the conserved GPCR autoproteolysis site (GPS), but the resulting N-terminal fragment (NTF) and C-terminal fragment (CTF) remain attached to each other and incorporate into the plasma membrane as a non-covalently bound unit10,11. Early studies in other aGPCRs showed that truncating the receptor by removal of the NTF up to the GPS cleavage site enhanced signaling12, suggesting that the NTF acts to suppress the intrinsic activity of the CTF13. The peptide segment immediately following the GPS is critical for enhanced signaling, suggesting that this peptide stretch (also known as the tethered agonist (TA), Stachel, or stalk peptide) acts as an internal agonist in a manner analogous to that of protease-activated GPCRs (PARs)14,15.
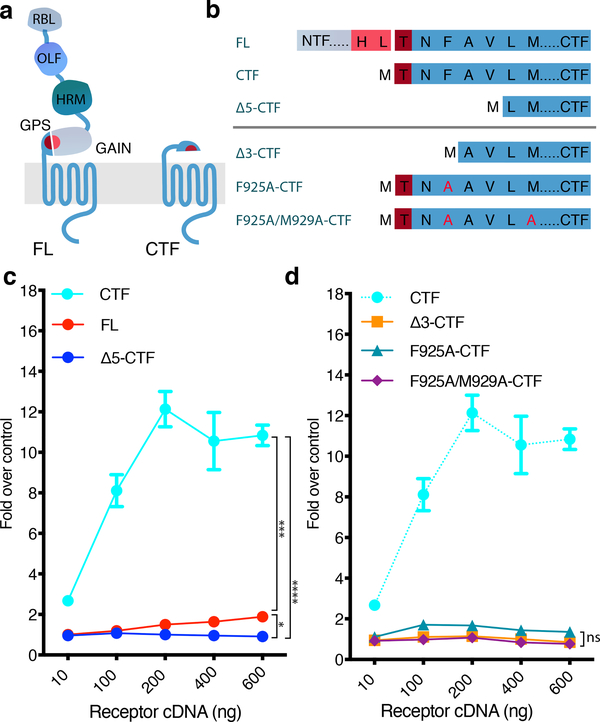
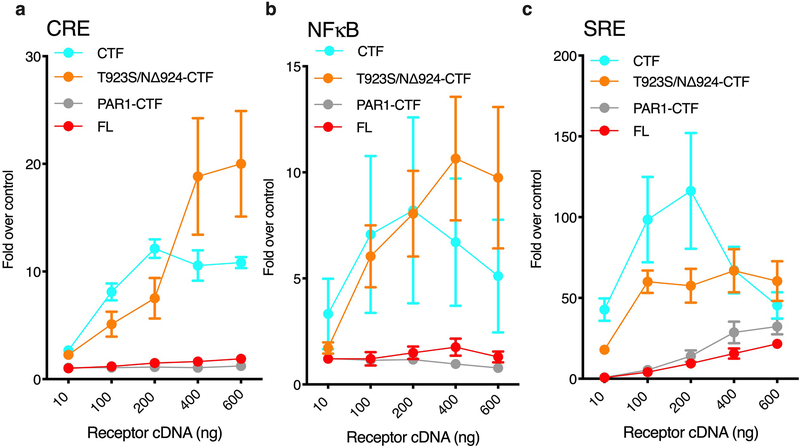
Figure 1. Exposure of the Adgrl3 TA promotes intracellular signaling.
(a) Schematic outlining the tertiary architecture of full-length (FL) and TA-exposed (CTF) Adgrl3 constructs. Adgrl3 FL is composed of a transmembrane GPCR fold (CTF) and a large N terminus (NTF) comprising 4 protein domains. Proteolysis occurs at the GPS cleavage site, which is buried in the GAIN domain. The peptide stretch (TA) immediately following the GPS is involved in regulating signaling. RBL: Rhamnose-binding lectin. OLF: Olfactomedin. HRM: Hormone receptor motif. GPS: GPCR proteolytic site. GAIN: GPCR autoproteolysis-inducing domain. GPCR: 7 transmembrane helix domain. (b) Schematic outlining the sequences of Adgrl3 constructs used in c and d. Proteolysis is marked in FL by a break in sequence between HL and T. (c) FL Adgrl3 constitutively enhances CRE and this signaling is increased when the entire N-terminal fragment (NTF) up to the GPS cleavage site (CTF) is removed. Further truncating 5 amino acids from the GPS site (Δ5-CTF) abolish es signaling (d) Truncating the first three amino acids following the GPS abolish CTF signaling (Δ3-CTF). Mutating the conserved phenylalanine in the TA (F943A-CTF) almost eliminates CTF activity, and two TA point mutations (F925A/M929A-CTF) abolish CTF signaling. The cyan dashed line is reprinted from c for direct comparison. All data points are normalized to empty vector control. In all panels data are presented as mean ±SEM from 3 independent experimental replicates. Unpaired two-tailed t-tests were performed to compare the conditions indicated by horizontal brackets using the 600 ng datapoints (a, *p=0.0325, ***p=0.0002, ****p<0.0001) (b, F925A-CTF vs. Δ3-CTF ns, p= 0.0660, F925A/M929A-CTF vs. Δ3-CTF ns, p=0.6227, F925A/M929A-CTF vs. F925A-CTF ns, p=0.0522).
Adhesion GPCR signaling is difficult to study because there have been no controlled means to acutely expose the TA in a live cell system. Signaling studies have been largely restricted to comparing the constitutive activity of full-length (FL) receptors and truncated CTF constructs at the second messenger level, and upon the addition of synthetic TA peptides14,15. Signaling reports for the ADGRL1–3 family have arrived at different conclusions regarding second messenger engagement16–22, and the full set of potential G protein-coupling pathways remain to be directly elucidated. To overcome these limitations, we designed a novel acute activation strategy to characterize ADGRL3 signaling in living cells by engineering a receptor construct where we replaced the GPS cleavage motif with a thrombin-recognition sequence that can be activated by treatment with thrombin to trigger acute TA exposure. We used this approach to study Adgrl3 (mouse homolog, ~90% identity with human, UniProt) signaling at the level of G protein activation in living cells.
We first systematically screened Adgrl3 FL and truncated CTF constructs for the full set of potential G protein interaction partners using gene reporter readouts in CRISPR knockout cell lines lacking the relevant Gα subunits. We optimized the assays to be dependent on the reintroduction of each Gα subunit individually so we could definitively identify the activated G proteins. Armed with the identified coupling partners we applied our new acute activation strategy in a panel of energy transfer assays to characterize acute Adgrl3 signaling at the plasma membrane. We found that TA-exposed Adgrl3 signals through G12/13 and Gq, with G12/13 being by far the most robustly activated. The Gα12/13 interaction partner is a new player in Adgrl3 biology, opening up a whole panel of unexplored roles for Adgrl3 in the nervous system. In addition, we anticipate that these methodological advancements will be broadly useful in aGPCR research as well as for orphan GPCRs in general.
Results
The Adgrl3 tethered agonist enhances signaling activity
To determine the signaling pathways engaged by Adgrl3 we first sought to establish whether the tethered agonist (TA) functions to activate downstream intracellular signaling, as has been reported for several other aGPCRs23. We truncated Adgrl3 full-length receptor (FL) at the GPS cleavage site (HL/T, where / indicates the point of autoproteolytic cleavage) to expose the TA (construct called CTF) and, in a parallel construct, also removed the first five amino acids subsequent to the GPS (construct called Δ5-CTF) ( Fig. 1a–b). We used a CRE luciferase reporter-gene assay that has been successfully used for orphan class A and other aGPCRs to characterize signaling activity24. In this assay, constitutive signaling can be measured as a change in luciferase expression under control of the CRE promoter, which is activated by CREB (cAMP response element-binding protein) downstream of protein kinase A. We verified that the assay worked as expected for constitutive activity of two well-characterized Gs and Gi/o coupled receptors (Supplementary Fig. 1). Notably, while this reporter provides a readout of canonical Gs and Gi/o signaling, CRE activity is distal to G protein activation and can also reflect more complex signaling crosstalk, as we discuss below.
We co-transfected HEK293T cells with a CRE-luciferase plasmid and increasing concentrations of the three Adgrl3 receptor constructs (FL, CTF, Δ5-CTF) and measured the level of expressed luciferase using luminescence as a readout (Fig. 1c and Supplementary Fig. 2). We observed elevated activity with increasing concentrations of the FL receptor, suggesting some level of constitutive activity. Signaling was greatly enhanced (~12 fold) by removal of the Adgrl3 NTF (CTF), but this signal was suppressed by removal of the first 5 residues of the TA (Δ5-CTF). The FL, CTF, and?5-CTF constructs are expressed at the cell surface at comparable levels (Supplementary Fig. 3). These results support a role for Adgrl3 in intracellular signaling, and establish that exposure of the TA greatly enhances signaling.
To explore the influence of the TA residues on the enhanced signaling activity, we performed the CRE gene expression assay for a construct in which the three amino acids subsequent to the GPS site were removed (Δ3-CTF) and for two constructs with alanine substitutions for the phenylalanine and methionine residues (at the P3’ and P7’ positions in the TA, where PN’ is used to denote the amino acid position downstream of the point of proteolysis25) that are highly conserved across the aGPCR family (constructs F925A-CTF and F925A/M929A-CTF) ( Fig. 1b). CTF CRE activity was abolished for the Δ3-CTF construct. Furthermore, mutating the highly conserved phenylalanine alone nearly eliminated CTF activity, and the combination of mutating the two conserved amino acids completely abolished CTF signaling, indicating that these key amino acids in the TA are crucial for enhancing signaling (Fig. 1d).
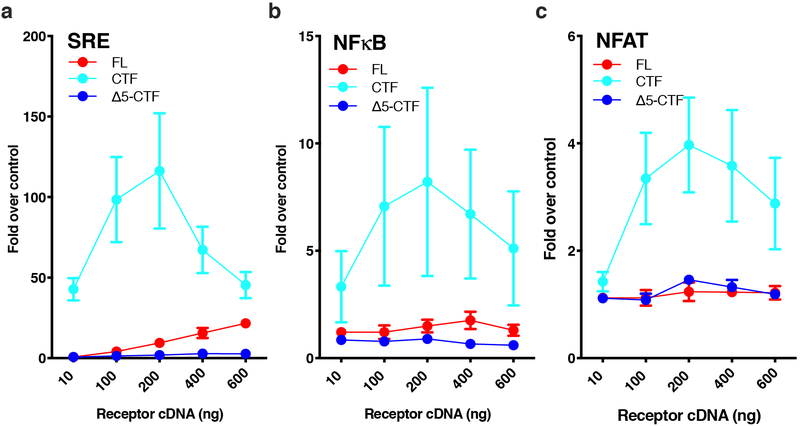
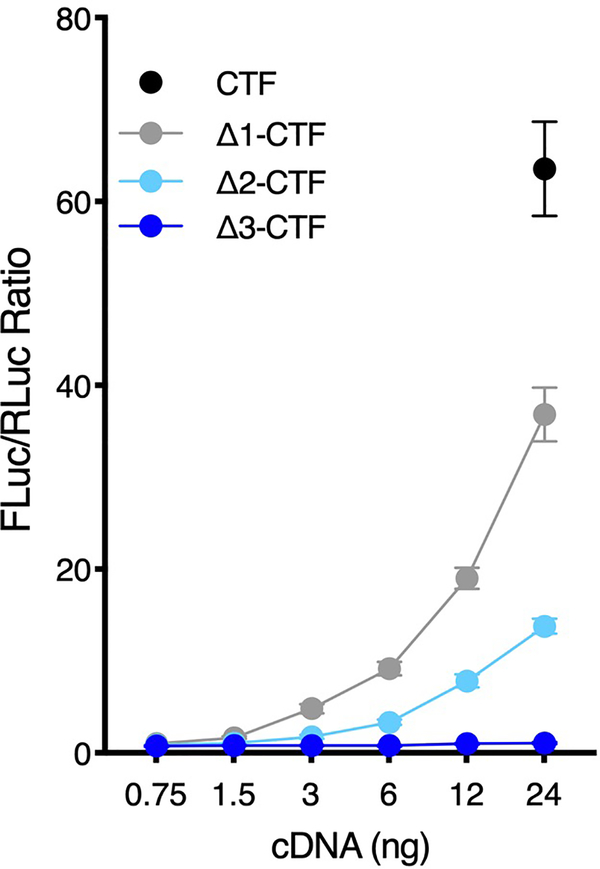
We next tested other luminescence-based gene expression assays (SRE, NFκB, and NFAT) that have previously been used for orphan GPCRs to infer about activity in other G protein signaling pathways (Gq and G12/G13). Like for CRE gene expression (Fig. 1), we found that the CTF greatly enhanced signaling over FL and Δ5-CTF in all these assays, indicating that activated Adgrl3 potentially couples to multiple G protein signaling pathways (Extended Data Fig. 1). Using the serum response element (SRE) luciferase reporter, we found that systematically truncating each TA residue abolished signaling after removing 3 amino acids subsequent to the GPS (Extended Data Fig. 2), consistent with our observations in the CRE assay (Fig. 1d).
Activated Adgrl3 couples to several Gα subtypes
To delineate which of the four main G protein signaling pathways is engaged by Adgrl3, we used a HEK293 knockout (KO) CRISPR cell line simultaneously lacking Gαs/Gαolf, Gαz, Gαq/Gα11, and Gα12/Gα13 (HEKΔ7)26. The only G proteins expressed by this cell line are those in the Gαi/o family. ADGRL3 has been observed to decrease cAMP, and a quantification of GTPγS exchange in detergent-treated membrane homogenates suggested that the truncated receptor can activate Gi21. While a recent study found that an ADGRL3 brain isoform did not modulate cAMP activity, pancreatic ADGRL3 isoforms decreased cAMP22. We failed to observe a decreases in CRE signals for FL, CTF, or Δ5-CTF in the HEKΔ7 cells which contain Gαi/o, even when we raised basal cAMP levels (Supplementary Fig. 4). In contrast, we observed a clear inhibition curve for the dopamine D2 receptor (D2R), which is known to signal through Gi/o.
We next developed a scheme for testing GPCR signaling by re-introducing one Gα protein isoform at a time in the CRE, SRE, and NFκB assays. For each Gα, we titrated the amount of cDNA to find an optimal level that restored signaling by defined receptors known to couple to the targeted isoform without substantially elevating the baseline signal (Supplementary Fig. 5). This insured that the expression level of each Gα subtype was set experimentally at a level that facilitates screening orphan receptors for Gα coupling. We first verified our approach for several well characterized class A GPCRs (Fig. 2a–c and Supplementary Fig. 5). For β2AR we observed a significant luminescence signal in CRE only when Gαs was expressed (Fig. 2a), and for the Gq/G12/13-coupled endothelin receptor type A (ETA) we observed an NFκB-dependent luminescence signal only in the presence of co-transfected Gαq or Gα13 (Fig. 2b–c, for Gα12 see Supplementary Fig. 5). Our approach thus allowed us to monitor Gα subtype-specific signals, thereby providing a platform to screen orphan GPCRs for G protein signaling partners while minimizing the confounding effect of signaling crosstalk.
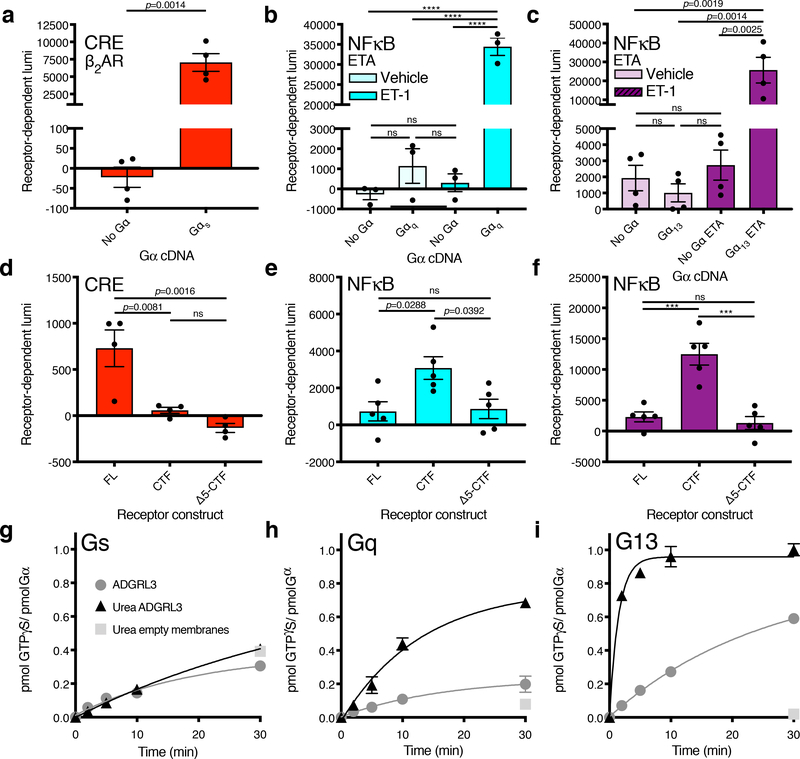
Figure 2. Adgrl3 CTF signals through Gq and G13.
Screen of Adgrl3 signaling in the major G protein signaling pathways utilizing a CRISPR knockout cell line (HEKΔ7) and a panel of gene expression assays. (a-c) Assay controls showing that the Gαs-coupled β2AR signals in CRE only when Gαs is reintroduced (a) and that ETA signals in NFκB only when Gαq (b) or Gα13 (c) is reintroduced. In (a) an unpaired two-tailed t-test was used to determine statistical significance between the No Gα and Gαs conditions; in (b,c) One-way ANOVA was employed with Tukey’s multiple-comparison post-hoc test (b, ****p<0.0001). (d-f) Gene expression signals for Adgrl3 constructs FL, CTF, and Δ5-CTF. (d) CRE with Gαs (e) NFκB with Gαq (f) NFκB with Gα13. Each Gα protein species was reintroduced at an optimized cDNA concentrations (Supplementary Fig. 5). In (d-f) One-way ANOVA with Tukey’s multiple-comparison post-hoc test was performed to determine statistical significance between the FL, CTF and Δ5-CTF conditions (ns p>0.05, ***p<0.001, ****p<0.0001). In panels (a-f) the baseline signal of empty vector was subtracted to show receptor-dependent luminescence (lumi). The full screen for Adgr3 in HEKΔ7 is shown in Extended Data Fig. 3. (g-i) ADGRL3 N-terminal dissociation induced by urea enhances G13 and Gq activation. Mock and urea-treated ADGRL3 membranes or empty High-Five membranes were reconstituted with purified Gαs (g) Gαq (h) Gα13 (i) and Gβ1Gγ2 heterodimer and receptor-stimulated [35S]-GTPγS binding kinetics were measured15,51,52. In panels (a-f) bars are presented as mean ±SEM from (a) n=4 (b-c) n=3 (d) n=4 (e-f) n=5 independent experimental replicates. In panels (g-i) data are from one representative experiment performed 3 times. Error bars mean ±SD from three technical replicates. See Supplementary Data for the full set of p-values.
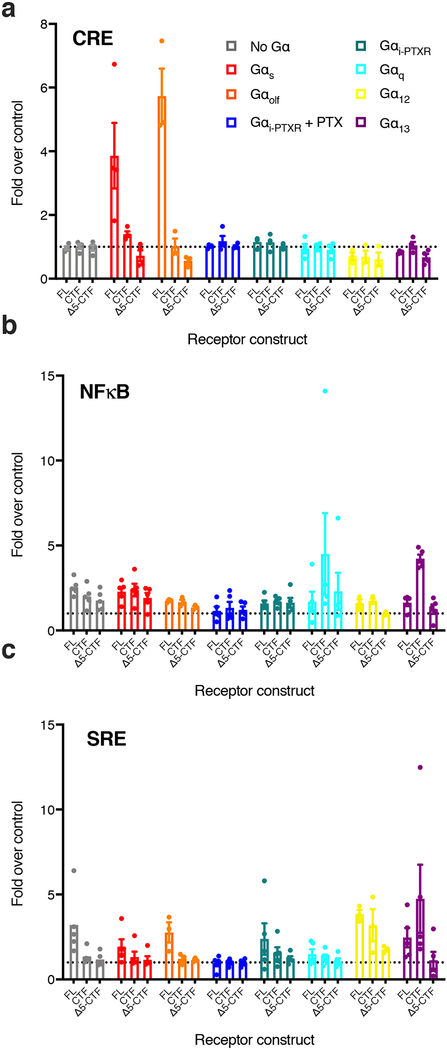
We screened Adgrl3 FL, CTF, and Δ5-CTF constructs for the three gene expression signals using an array of Gα subtypes (Extended Data Fig. 3). To our surprise, in contrast to our findings in Fig. 1 with wild type HEK cells, we found that compared to empty vector control, FL produced a small but significant Gαs-dependent CRE signal, whereas neither CTF nor Δ5-CTF impacted the CRE signal (Fig. 2d and Extended Data Fig. 3). In contrast, CTF greatly enhanced Gαq- and Gα13-dependent NFκB-dependent luciferase expression compared to FL and Δ5-CTF levels (~4 and ~6-fold respectively) (Fig. 2e–f). This result suggests that Adgrl3, upon activation and TA exposure, signals through Gq and G13.
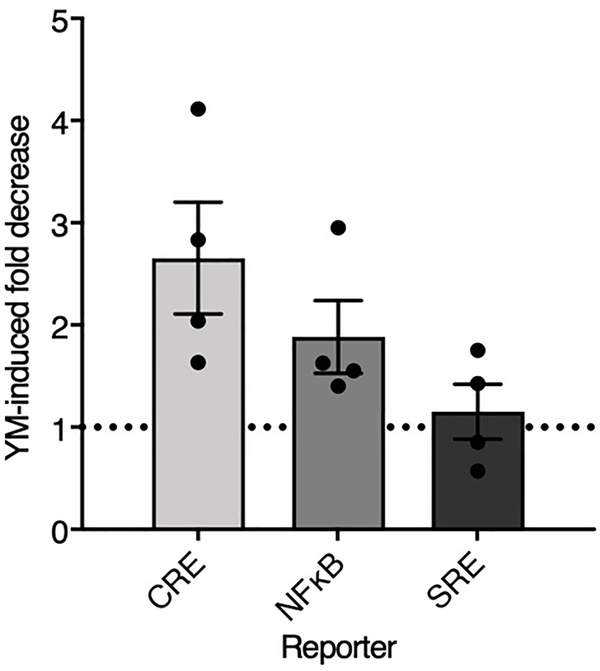
Given these findings and the hits from the Gα subunit screen, we revisited the enhanced CTF signaling in regular HEK293T cells (Fig. 1) using a selective Gαq inhibitor (YM-254890). We verified that the inhibitor suppresses Gq-mediated gene expression but does not affect Gs or G13 signals (Supplementary Fig. 6). The CTF-enhanced CRE and NFκB signals were both reduced by the Gαq inhibitor (Extended Data Fig. 4). These results suggest that Gq activation contributes to signals not only in the NFκB assay but also in the CRE assay, which is widely thought to be a readout of only Gs and Gi family activation, the isoforms that directly regulate adenylyl cyclase.
To validate our findings with an independent approach, we directly tested ADGRL3-mediated G protein activation by monitoring the kinetics of G protein [35S]-GTPγS binding in membranes reconstituted with purified Gβ1γ2 and either Gαs, Gαq or Gα13. In agreement with our Gα-subunit screen, ADGRL3 membranes treated with urea to remove the NTF and expose the TA (Extended Data Fig. 5) robustly activated both G13 and Gq, with the [35S]-GTPγS binding kinetics (0.67 ± 0.13 min−1 and 0.08 ± 0.02 min−1 respectively) being ~9-fold faster for G13 (Fig. 2g–i). For Gs we failed to observe any ADGRL3 activation above background with or without urea treatment, consistent with our finding that CTF did not produce any Gαs-dependent CRE signal in the KO HEKΔ7 cells, but seemingly inconsistent with our findings that the FL construct produced a small Gαs- and Gαolfdependent CRE signal (Fig. 2d and Extended Data Fig. 3). Notably, in the absence of Gαs and Gαolf, adenylyl cyclase cannot be activated by forskolin (Supplementary Fig. 7)27,28. Thus, co-expression of Gαs might “enable” another pathway to adenylyl cyclase activation, thereby leading to CRE signal without direct activation of Gs by FL. To explore further if FL Adgrl3 CRE signaling is dependent on direct Gs activation, we used a truncated Gαs (GαsΔ10), which has been shown to abolish β2AR coupling (Supplementary Fig. 7), but which still works to complement adenylyl cyclase activity. With co-expression of GαsΔ10 we still observed a FL concentration-dependent increase in CRE in the HEKΔ7 cells, and we therefore conclude that this baseline activity does not result from direct Gs activation by the receptor but rather through activation of another effector pathway that somehow indirectly activates adenylyl cyclase (Supplementary Fig. 7). The origin of the CRE signal generated by FL Adgrl3 thus remains an open question for future study.
Acute TA exposure leads to direct G protein activation
Several energy transfer approaches (BRET and FRET) would be useful for studying aGPCR signaling events at the plasma membrane of living cells, but these techniques are not well suited to constitutive activation and require acute receptor activation, typically with a ligand, to induce a change in protein-protein interactions that can be monitored within the dynamic range of the assays. To achieve this goal, w e modeled the TA-exposure mechanism found in protease-activated GPCRs (PAR1–4) and fused the PAR1 N terminus with its cleavage site LDPR/SF (/ marks the point of cleavage) to the CTF construct (PAR1-CTF). In the PAR1 cleavage site the P2’ sidechain phenylalanine was determined to have a major influence on cleavage rate whereas the P3’ sidechain was less important25. For aGPCRs in general, the GPS P1’ residue is either a threonine or a serine and the consensus TA sequence is TXFAVLMXX. Therefore, to maintain P2’ as phenylalanine we designed our activation construct so that thrombin cleaves to expose a TA with the sequence SFAVLM. Thus, in comparison to the endogenous Adgrl3 TA (TNFAVLM) the first amino acid, threonine, is replaced by a serine and the TA is shortened by deletion of the non-conserved asparagine normally at position P2’. Our finding that truncating the TA by two amino acids did not abolish the enhanced CTF signaling activity (Extended Data Fig. 2) supported this approach. To confirm that this engineered TA is able to signal similarly to the endogenous TA, we created a TA-exposed control construct, M-SFAVLM-CTF (construct called T923S/ΔN924-CTF), and verified that this construct signals at levels similar to that of the CTF construct in all the gene expression assays tested (Extended Data Fig. 6). Since expressing a FLAG epitope tag N terminal to the CTF construct abolished signaling to levels comparable to FL (Supplementary Fig. 8), presumably by masking the TA, we hypothesized that placing the N terminus of PAR1 N terminal to the CTF would also abolish signaling activity. Indeed, PAR1-CTF gene expression activity was not distinguishable from that of FL receptor (Extended Data Fig. 6). Both the PAR1-CTF and T923S/ΔN924-CTF constructs expressed at the cell surface at levels comparable to the FL and CTF constructs (Supplementary Fig. 3). Thus, when its N terminus is intact, the PAR1-CTF fusion construct recapitulates the basal levels of FL Adgrl3 signaling, but when we simulate thrombin cleavage, the newly exposed TA behaves like the endogenous Adgrl3 TA.
To determine if we could generate a G protein-activation signal by acute thrombin-induced cleavage, we first tested the PAR1-CTF construct in a Gβγ release BRET assay, monitoring energy transfer between a membrane-anchored luminescent donor and a fluorescent acceptor fused to the Gγ subunit of the G protein heterotrimer29 (Fig. 3a). We systematically tested this BRET scheme in the HEKΔ7 cell line with each of the key Gα subunits in question (Gαs/Gαq/Gα12/Gα13) (Fig. 3b). For both Gα12 and Gα13 we observed a robust response with thrombin activation of Adgrl3 PAR1-CTF and PAR1, the latter serving as a positive control. As expected, we did not observe any thrombin-induced effects for the T923S/ΔN924-CTF construct lacking the thrombin cleavage site. This result supports our finding that Adgrl3 TA exposure leads to G12/13 activation. As expected, we did not observe significant signals for conditions without co-transfection of Gα subunits. Neither did we observe any PAR1-CTF Gs activity, in agreement with our findings from the Gα-subunit screen and GTPγS binding assays shown in Fig. 2. Curiously, we also failed to observe significant Adgrl3 CTF Gq-dependent activation, which we attribute to the fact that Gq activation is less efficient30, consistent with the nearly 10-fold lower GTPγS binding rate compared to that for G13, measured using the G protein reconstituted membranes (Fig. 2h).
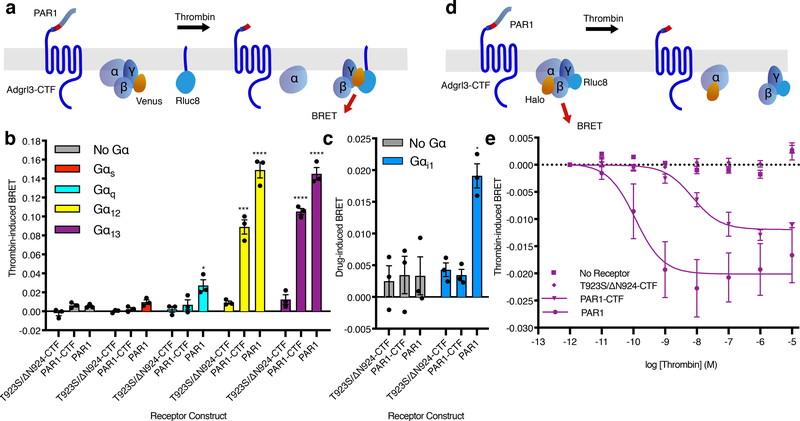
Figure 3. Adgrl3 couples to Gα12/13 upon acute exposure of the TA.
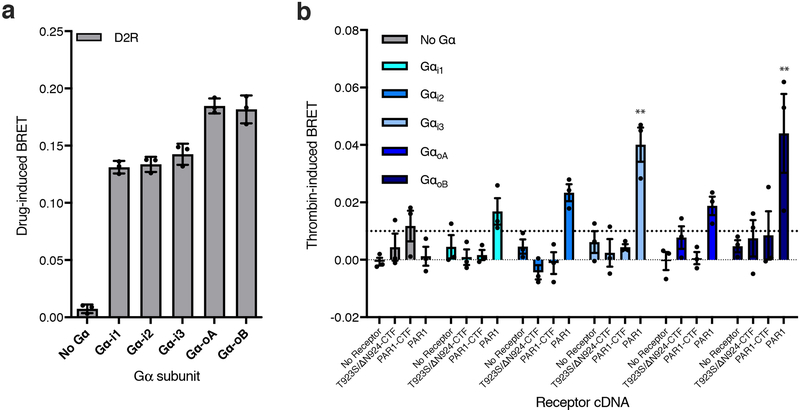
(a) Cartoon outlining the principle of the Gβγ release BRET assay. Drug-induced BRET occurs when Gβγ-Venus is released from the G protein to interact with the C-terminal fragment of the G protein receptor kinase 3 fused to Rluc8. (b) Gβγ release assay testing the T923S/ΔN924-CTF, PAR1-CTF, and PAR1 activation of Gs, Gq, G12, and G13 in HEKΔ7 cells. Unpaired two-tailed t-tests were performed for T923S/ΔN924-CTF, PAR1-CTF, and PAR1 individually, to determine statistical significance between the No Gα control and each Gα subtype (*p<0.05, ***p<0.001, ****p<0.0001). (c) Gβγ release assay testing the T923S/ΔN924-CTF, PAR1-CTF, and PAR1 activation of Gαi in HEK full G KO cells. Experimental conditions identical to b. An unpaired two-tailed t-test was performed for each cDNA construct to determine statistical significance between the No Gα control and Gαi conditions (*p<0.05). (d) Cartoon outlining the principle of the inter-molecular heterotrimer α-γ BRET assay. BRET occurs between the heterotrimer subunits Gα-Halo (labelled by JF-525) and Gγ-Rluc8. When the heterotrimer is activated the BRET signal is decreased. (e) Thrombin dose-response curves for empty vector control, T923S/ΔN924-CTF, PAR1-CTF, and PAR1 receptor constructs in the Gα-γ BRET assay. In b-e Luminescence was read after 10 min of stimulation with 1 μM thrombin. Data are normalized to buffer controls and show the BRET effect induced by thrombin. For panels (b-c) and (e) error bars represent ±SEM from 3 independent experimental replicates. See Supplementary Data for the full set of p-values in b and c.
As noted above, Adgr13 has previously been inferred to cpuple to Gαi21. While we failed to see inhibition of cAMP in the CRE gene expressoin assay, signal inhibition can be complicated to interpret at the level of gene expression. We therefore souhgt to investigate if we could detect Adgrl3 coupling to the Gαi/o family of G protein subtypes by acute TA exposure in the Gβγ release assay. For this purpose, we used a HEK293 CRISPR cell line lacking all Gαi/o subtypes in addition to the other main Gα families in HEKΔ7 (HEK full Gα KO). We verified that the assay worked for D2R when each of the five Gαi/o subtypes (αi1/αi2/αi3/αOA and αOB) were reinroduced independently (Extended Data Fig. 7a). However, consistent with the results of the CRE gene expressions assay, we failed to observe any Adgrl3-dependebt Gβγ release for the PAR1-CTF construct when any of the Gαi/o subtypes were co-expressed (Fig. 3c and Extended Data Fig. 7b).
To further establish that CTF indeed activates G13 in a dose-dependent manner, we also optimized an inter-molecular heterotrimer Gα-γ BRET assay (Fig. 3d) that reads out energy transfer within the heterotrimer itself. We introduced a Halo-tag in Gα13 and labelled it with the acceptor dye Janelia fluorophore 525 (JF-525) which is highly membrane permeant and can serve as a suitable acceptor for BRET. Therefore, upon TA exposure we expect to observe a decrease in drug-induced BRET as the receptor activates the heterotrimer. Indeed, both wild type PAR1 and PAR1-CTF led to a thrombin-concentration-dependent loss of BRET (Fig. 3e). The PAR1 response to thrombin is more potent and efficacious than PAR1-CTF, likely because PAR1 has, in addition to its cleavage site, a hirudin-like sequence that binds the thrombin anion exosite region and affects cleavage potency31. Importantly, without transfected receptor or with the T923S/ΔN924-CTF construct expressed, there was no response to thrombin. Our measured EC50 for PAR1 was consistent with literature values31 and the dose-dependent behavior, which is expected for a ligand-stimulated receptor response, is recapitulated for PAR1-CTF, highlighting that CTF activates Gα13 upon TA exposure.
TA-exposed Adgrl3 recruits Arrestin
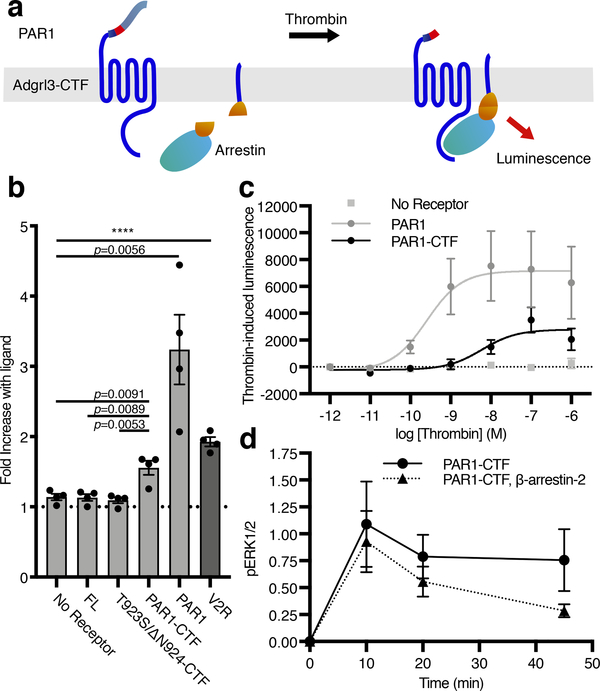
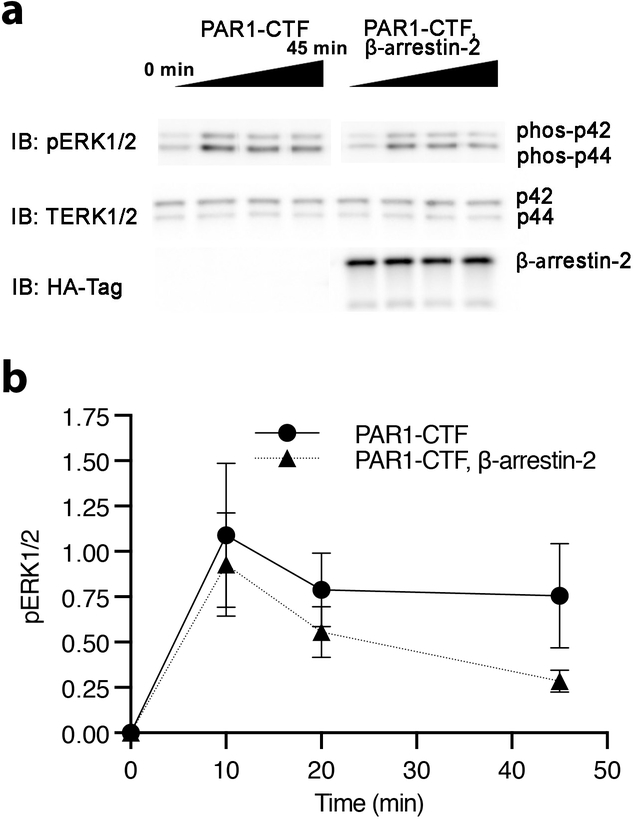
Having established that Adgrl3 engages in TA-dependent G protein signaling, we were interested in determining if TA exposure also leads to β-arrestin recruitment. We used a split complementation luminescence assay (Hauge, M., Pham, J., Mancebo, H., Inoue, A. & Javitch, J., manuscript in revision), in which β-arrestin fused to a C-terminal fragment of NanoLuc (nluc) is recruited to the membrane to complement an N-terminal fragment of nluc fused to a membrane anchor, in response to receptor activation. Thus, a luminescence signal is only produced by the reconstitution of a functional nluc when β-arrestin translocates to the plasma membrane (Fig. 4a). As a positive control we showed that vasopressin acted at the vasopressin receptor 2 (V2R) to recruit β-arrestin-2 (Fig. 4b, dark gray bar). Similarly, thrombin-mediated activation of both PAR1-CTF and PAR1 led to robust concentration-dependent recruitment of β-arrestin-232 (Fig. 4c). In contrast, no increase in luminescence was detected for the three negative controls that were not expected to respond to thrombin (empty vector, FL, and T923S/ΔN924-CTF) (Fig. 4c). Finally, we addressed the effect of arrestin on Adgrl3-mediated activation of extracellular signal-regulated kinase 1/2 (ERK1/2) in a HEK cell line devoid of β-arrestin-1/2 (HEK Δβarr1/2). We found that thrombin-induced TA exposure mediated ERK1/2 phosphorylation in a G protein-dependent manner and that β-arrestin-2 recruitment inhibited this response (Fig. 4d and Extended Data Fig. 8).
Figure 4. Adgrl3 recruits β-arrestin to the plasma membrane in living cells.
(a) Cartoon outlining the principle of the split complementation luminescence β-arrestin assay. After receptor activation (and potentially phosphorylation) β-arrestin-C-nluc is recruited to the membrane to complement a membrane anchored N-nluc. Upon β-arrestin-translocation and reconstitution of a functional nluc, a luminescence signal is produced. (b) β-Arrestin-2 membrane-recruitment complementation assay for negative controls of empty vector, FL, and T923S/ΔN924-CTF constructs as well as for PAR1-CTF, PAR1 in response to 1 μM thrombin, and the vasopressin receptor V2R in response to 1 uM Vasopressin (AVP), which is a high affinity β-arrestin binder. Data are shown as fold over control (buffer). Unpaired two-tailed t-tests were performed to determine statistical significance between the PAR1-CTF and ‘No receptor’, FL and T923S/ΔN924-CTF constructs, as well as ‘No receptor’ and controls PAR1 and the vasopressin receptor V2R. (c) Dose-response curves for a negative control (empty vector), PAR1-CTF, and PAR1. Bars in (b) and data points in (c) are presented as mean ±SEM from 4 independent experimental replicates. (d) β-arrestin-2 decreased PAR1-CTF ERK1/2 phosphorylation. HEK Δβarr1/2 cells were transfected with PAR1-CTF, or PAR1-CTF with β-arrestin-2. After 48 hr, the cells were acutely activated with 1 μM thrombin. The level of phospho-ERK1/2 was normalized to total ERK and the baseline at 0 min was subtracted to produce the time-dependent change in pERK1/2. Data in (d) represent mean ±SEM from 3 (PAR1-CTF) and 4 (PAR1-CTF, β-arrestin-2) independent experimental replicates.
Discussion
The remarkable convergence of human genetics findings and behavioral data from four different animal species implicating ADGRL3 in the pathophysiology of ADHD and substance use makes it a critical new target. To begin to understand Adgrl3 biology, we sought to identify the basic signaling properties of this aGPCR. Here we show that TA exposure in Adgrl3 leads to robust activation of G12/13, as well as a weaker activation of Gq.
In contrast to family A GPCRs where potent small molecule agonists are usually available, the tools to study aGPCRs have been limited, and there has been no method to acutely expose the endogenous TA from a FL aGPCR in a controlled manner in a live cell system. For a handful of receptors synthetic peptides mimicking the TA sequence have been used to activate signaling when applied to both FL and CTF constructs14,15,33. A hurdle in using TA peptides for acute activation is that for many aGPCRs (including Adgrl3) such peptides are highly hydrophobic and difficult to solubilize1. Because they are not covalently tethered to the CTF, very high concentrations are typically required to evoke a significant change in second messenger levels14,33. To overcome these limitations, we engineered a receptor construct that allows for controllable acute proteolytic TA exposure by thrombin and verified our findings from the Gα-subunit gene reporter assays at the level of acute activation of G proteins in the plasma membrane of living cells. We anticipate that this strategy will be broadly useful for the aGPCR field.
A typical feature for most class A GPCRs that signal to G12/13 is that they also activate Gq/11 pathways34. This fits with our observations of Adgrl3 as well as for aGPCRs in other families that have been implicated in the G12/13 pathway, for example, ADGRG1 (commonly known as GPR56)35,36, ADGRG237,38, and ADGRB1–2 (commonly known as BAI1–2)39,40. The most common action of G12/13 is the direct regulation of a group of Rho guanine nucleotide exchange factors (RhoGEFs) that activate the Ras-family small GTPase RhoA, which is involved in several cellular functions such as shape changes, migration, adhesion, and contraction34. Next steps will be to explore the downstream functions mediated by Adgrl3-dependent activation of G12/13, as well as to determine how, and in which subset of neurons in the brain, the receptor is activated. Adgrl3 has been shown to stabilize and shape synapse morphology and formation via its transsynaptic interactions with two single-pass transmembrane protein ligands, fibronectin leucine-rich transmembrane proteins (FLRTs) and teneurins18,41–43. Also a recent study implicates Adgrl3 in influencing actin cytoskeleton dynamics44. Intriguingly, these reports and our observation of G12/13 signaling raise the exciting possibility that ligand interaction with the FL receptor might also activate these pathways, although this has yet to be explored.
We also found that Adgrl3 recruits β-arrestin. Several other aGPCRs (ADGRG1–3 and ADGRB1/3) have been reported to bind β-arrestin using co-immunoprecipitation strategies37,39 and cell-based screening platforms with modified receptors45. Here, we show acute TA-dependent β-arrestin recruitment to Adgrl3 and establish that recruitment of β-arrestin inhibits Adgrl3-mediated G protein-dependent ERK phosphorylation, the first such example for an aGPCR. Whether β-arrestin recruitment to Adgrl3 and other aGPCRs also functions to internalize receptors and plays a role in signaling, as has been shown for multiple Family A GPCRs46, will also be an important area of future study47.
Adhesion GPCRs encompass a vital duality, combining cell-cell adhesion interactions and metabotropic signaling, and it remains an open question how this receptor class is activated in vivo. Recent advances in the field suggest that NTF interactions and G protein-coupling might be functionally integrated through mechanical stress20,48, which leads to the proposal of two potential signaling mechanisms that are not necessarily mutually exclusive. Mechanical force exerted on the extracellular region of aGPCRs could 1) remove the NTF altogether to expose the TA and activate G protein signaling or 2) alter the conformation of the extracellular protein domains in a tunable manner to either expose the TA and/or affect signaling in a TA-independent manner. Removal of the entire NTF would result in an ‘all or none’ activation mechanism; however, animal studies of ADGRL3 homologs (flies, larvae and mouse) have shown that GPS cleavage is not essential for at least some of its actions20,42,49,50. For Adgrl3 it remains an open question how the TA is exposed in vivo, but, a crucial first step to building a mechanistic understanding of its signal activation is to understand which intracellular G protein partners/signaling pathways to assay in response to physiological stimuli such as mechanical forces. Here we discovered a robust activation of G12/13, as well as a weaker activation of Gq, and therefore G12/13 should be the natural pathway to monitor in future studies dissecting potential mechanical activation of the receptor.
Online Methods
Materials
Dulbecco’s modified Eagle’s medium (DMEM), Hank’s Balanced Salt Solution, and Penicillin-Streptomycin were from Gibco. Fetal Bovine Serum (FBS), 0.05% Trypsin, and Dulbecco’s Phosphate Buffered Saline (DPBS) were from Corning. Lipofectamine 2000 was from Invitrogen. Firefly D-Luciferin was from NanoLight Technology. Endothelin 1 (ET-1) was from Tocris. YM-254890 was from AdipoGen Life Sciences. Enzyme free cell dissociation solution, Thrombin, and Bovine Serum Albumin were from Millipore Sigma. FluoroBrite DMEM was from Thermo Fischer Scientific.
Plasmid DNA Constructs
Adgrl3 (NM_198702, mouse homolog) cDNA was used as a PCR template to make the various Adgrl3 constructs used in this study followed by insertion into pCDNA3.1 (Supplementary Table 1). Plasmid construction was done either by restriction enzyme digestion followed by ligation or by Gibson Assembly using the NEBuilder HiFi DNA Assembly Master Mix (NEB). All sequences were confirmed with Macrogen’s DNA sequencing service.
Cell culture
HEK-293 cells with targeted deletion via CRISPR-Cas9 of GNAS, GNAL (HEKΔGs), HEK-293 cells with targeted deletion via CRISPR-Cas9 of GNAS, GNAL, GNAQ, GNA11, GNA12, GNA13, and GNAZ (HEKΔ7), as well as HEK-293 cells with targeted deletion via CRISPR-Cas9 of GNAS, GNAL, GNAQ, GNA11, GNA12, GNA13, GNAZ, GNAI1, GNAI2, GNAI3, GNAO1, GNAT1, and GNAT2 (HEK full G KO), and HEK-293 cells with targeted deletion via CRISPR-Cas9 of ARRB1 and ARRB2 (HEK Δγarr1/2) were used26,53,54. HEK293T, HEKΔGs, HEKΔ7, HEK full G KO, and HEK Δγarr1/2 cells were cultured in DMEM supplemented with 10% FBS and 1% Penicillin-Streptomycin at 37° in a 5% CO2 humidified incubator. For the HEKΔ7 and HEKΔ13 cells, FBS was heat-inactivated at 56° Celsius for 30 minutes.
Gene-expression assays
Cells were plated in 12-well culture plates at a density of 3–4 × 105 cells per well and incubated overnight. 24 hours after seeding, cells were transfected using Lipofectamine 2000 (2.3 μL/1 μg cDNA). For assays in HEK293T, 600 ng of reporter (CRE-luc/SRE-luc/NFκB-luc) was co-transfected with varied amounts of receptor (10–600 ng) and pcDNA5/FRT cDNA to balance the total amount of DNA up to 1200 ng. For assays in HEKΔ7, 300 ng of reporter (CRE-luc/SRE-luc/NFκB-luc) was co-transfected with 300 ng of receptor, a varying amount of Ga subunit, and pcDNA5/FRT cDNA to balance the total amount of DNA up to 800 ng. For the Gα subunit screens in HEKΔ7, the amounts of Gα plasmid used were as follows: Gαs 10 ng, Gαolf 100 ng, Gαi 100 ng, Gαq 200 ng, Gα12 1 ng, Gα13 10 ng, pertussis toxin 100 ng.
Twenty-four hours after transfection, cells were washed with DPBS and detached in enzyme free. Cells were centrifuged at 500 rcf for 3 minutes, the supernatant was aspirated, and the cells were resuspended in 200 μL assay buffer (1× HBSS, 20 mM HEPES, 0.1% w/v BSA, pH 7.5). Cells were distributed into a 96 -well Black/White Isoplate (Perkin Elmer Life Sciences) in triplicates at a volume of 60 μL per well. 30 μL of D-Luciferin dissolved in assay buffer was added to each well to a final concentration of 2 mM, and emission was read at 525 nm after 30 min incubation using a PHERAstar FS (BMG LABTECH). For assays using the SRE-Luc reporter, media was exchanged to serum-free DMEM 6 hours after transfection. For assays using the Gαq-inhibitor YM-254890, media was exchanged to DMEM containing 1 μM YM-254890 6 hours after transfection. For the assays using ET-1, media was exchanged to DMEM containing 100 nM ET-1 5 hours prior to lifting the cells for assay measurement.
For the SRE gene expression assay presented in Extended Data Fig. 2 a dual luciferase assay was used. HEK239T cells (105 cells per well) in 24 -well format were transfected using PEI reagent with varying amounts of ADGRL3 CTF pcDNA3.1 constructs (1–24 ng), 100 ng of SRE-Luciferase plasmid, 1 ng of Renilla luciferase plasmid, and balancer pcDNA3.1 to equal 350 ng of total DNA. At 24 hours post-transfection, cells were washed and incubated with serum-free DMEM for 8–10 hours. Cells were washed with Tyrode’s solution and incubated with 350 μl of the Dual Luciferase Firefly reagent (Promega). Cell lysate (100 μl) was pipetted in triplicate into 96 well gray optiplates (Perkin Elmer) and Firefly Luciferase signal was read using a Berthold TriStar2 plate reader. Dual Luciferase Renilla assay reagent (100 μl) was then added to each well, the plates were incubated for 5 min, shaken in the TriStar2 and Renilla Luciferase signal was measured.
BRET assays
For single point measurements cells were plated in 12-well culture plates at a density of 3–4 × 105 cells per well. For dose-response experiments, cells were plated in 6-well culture plates at a density of 9 × 105 cells per well. For the βγ-release BRET assay, cells were co-transfected 24 hours after seeding using Lipofectamine 2000 (2.3 μL/1 μg cDNA), receptor (200 ng), Gα (720 ng), Gβ1 (250 ng), Gγ2-Venus (250 ng), membrane-anchored GRK3ct-Rluc8 (50 ng), and pcDNA5/FRT to balance the total amount of DNA (1470 ng).
For the Gα-γ BRET assay, cells were co-transfected 24 hours after seeding using Lipofectamine 2000 (2.3 μL/1 μg cDNA) with receptor (1000 ng), Gα-Halo (1000 ng), Gγ2-Rluc8 (100 ng), and pcDNA5/FRT plasmid to balance the total amount of DNA (2100 ng). For dose-response experiments these values were scaled up by 2.7×, and two 6-well plates were combined for each curve. Gα-Halo was labelled before proceeding with the BRET assay by incubating cells with 250 nM JF-525 in DMEM for 30 min at 37° in a 5% CO2 and allowed to recover in fresh DMEM for 1 hour. Janelia Fluor 525 dye (JF-525) was generously provided by Luke Lavis and Jonathan B. Grimm (Janelia Research Campus).
The BRET assays were performed 24 hours after transfection. Cells were washed with DPBS and BRET buffer (DPBS containing 5 mM glucose). After washing, the cells were resuspended in either 400 μL or 1200 μL of BRET buffer (for 12-well and 6-well cell culture plates, respectively) and distributed into 96-well OptiPlates (Perkin Elmer Life Sciences) at 45 μl per well. Cells were incubated for 10 min with 5 μM coelenterazine H (NanoLight Technologies) before ligand addition to reach a final well volume of 100 μl. The donor (Rluc8) and acceptor (mVenus or JF-525) emission was collected at 485 nm and 525 nm, respectively. BRET intensities were measured on a Pherastar FS plate reader (BMG) 10 min after ligand addition. The BRET signal was calculated as the ratio of light emitted at 525 nM over that emitted at 485 nM. The drug-induced BRET ratio was obtained by subtracting baseline BRET (buffer) for each condition. Dose-response curves were fit by nonlinear regression to a sigmoidal dose-response relationship using GraphPad Prism.
For the BRET experiments presented in Supplementary Fig. 7 permeabilized cells were used to deplete GTP. HEK293 cells lacking Gαs-family subunits (HEKΔGs) were transiently transfected with β2AR-Rluc8, a Gα subunit (or vector control), Venus-1–155-Gγ2, and Venus-155–239-Gβ1 in a (1:3:1:1) ratio. After 24–48 hours cells were washed twice with permeabilization buffer (KPS) (140 mM KCl, 10 mM NaCl, 1 mM MgCl2, 0.1 mM KEGTA, 20 mM NaHEPES, pH 7.2), harvested by trituration, permeabilized in KPS buffer containing 10 μg ml−1 high purity digitonin, and transferred to opaque black 96-well plate (Perkin Elmer Life Sciences). BRET measurements were made from permeabilized cells supplemented with either 0.5 mM GDP or 2U ml−1 apyrase, with either agonist or inverse agonist.
Arrestin Recruitment Assay
Cells were plated in 12-well culture plates at a density of 3–4 × 105 cells per well and incubated overnight. Cells were co-transfected 24 hours after seeding using lipofectamine 2000 (2.3 μL/1 μg cDNA), receptor (300 ng), membrane-anchored N-terminal of Nluc (50 ng), β-Arrestin fused to the C-terminal of Nluc (50 ng), GRK2 (300 ng), and pcDNA5/FRT to balance the total amount of DNA (1200 ng). For dose-response experiments these values were scaled up by 2.7× and two 6-wells were combined for each curve. 24 hours after transfection cells were washed with DPBS and BRET buffer and resuspended in either 400 μL or 1200 μL BRET buffer (for single-point and dose-response curves respectively) before distributing 45 μL per well into a 96-well reading plate. Cells were incubated for 10 min with 5 μM coelenterazine H (NanoLight Technologies) before ligand addition to reach a final well volume of 100 μl. Nluc emission at 485 nM was measured on a Pherastar FS plate reader (BMG) 10 min after ligand addition. Fold increase with ligand was calculated by dividing the average luminescence signal by baseline (buffer). Thrombin-induced luminescence was obtained by subtracting baseline BRET (buffer) for each condition. Dose-response curves were fit by nonlinear regression to a sigmoidal dose-response relationship using GraphPad Prism.
UREA stripping and [35S]-GTPγS binding assay
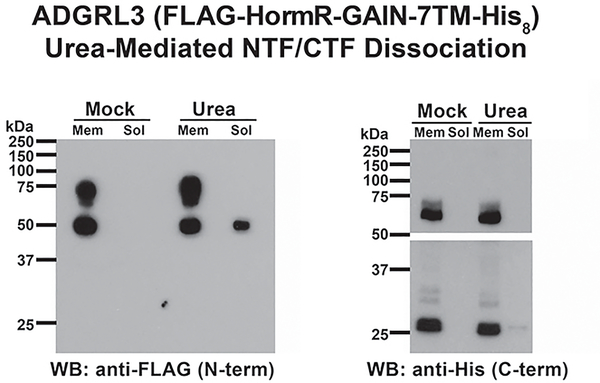
An ADGRL3 truncation that comprises the FLAG-tagged extracellular HormR and GAIN domains and C-terminal His8-tagged 7TM domain was expressed in High-Five insect cells. Prepared membranes from the cells were treated with buffer (20 mM Hepes pH 7.4, 1 mM EGTA) (Mock) or buffer containing 7M urea (Urea) at 4 ºC. The membranes were precipitated and washed and the solubilized material was collected. Both preparations were western blotted with mouse anti-FLAG (sigma, F1804, 1:5,000 in milk BLOTTO) and anti-mouse-HRP (GE Healthcare, NA931, 1:5,000 in milk BLOTTO) to detect the N-terminal and mouse anti-penta-His (Qiagen, 34660, 1:5,000 in 5% BSA in PBS due to its known reactivity with milk protein) and anti-mouse-HRP (GE Healthcare, NA931, 1:5,000 in milk BLOTTO) to detect the 7TM domain. Mock- and urea-treated ADGRL3 membranes or empty High-Five membranes were reconstituted with 100 nM purified Gα subunit (Gα13, Gαq, or Gαs) and 250 nM purified Gβ1Gγ2 heterodimer. 1 μM [35S]-GTPγS (~20,000 cpm/pmol) was added to initiate kinetic G protein GTPγS binding reactions at 25 ºC. At the indicated time points, Gα-[35S]-GTPγS was precipitated onto nitrocellulose filters, the filters were washed and subjected to scintillation counting to quantify the amount of GTPγS bound to G protein.
ERK1/2 Phophosphorylation Assay
HEK293 cells lacking β-arrestin-1/2 (HEK Δβarr1/2) were plated in 12-well culture plates at a density of 3.5 × 105 cells/well. Cells were transfected 24 hours after seeding using lipofectamine 2000 (2 μL/1 μg cDNA) with the corresponding receptor (200 ng), β-arrestin-2 (400 ng), and pcDNA5 to balance the total amount of DNA (600 ng). Forty-eight hours after transfection, the cells were incubated with 1 μM thrombin over a time course of 0, 10, 20, or 45 min. To stop the reaction, the media was immediately removed and replaced with 200 μL RIPA buffer (Sigma Aldrich) for 5 min. The cells were then centrifuged at 15,000xg for 10 min and the supernatant was placed into 200 μL Laemmli 2X Concentrate (Sigma Aldrich). For each condition, 3.5 μL of sample was loaded onto a 12% SDS-PAGE gel, transferred onto a PDVF membrane, and incubated with primary antibodies against phosphoERK1/2 (Cell Signaling Technology #9101S, 1:1,000 dilution), total ERK1/2 (CST #9102S, 1:1,000 dilution), or HA (CST #2367S, 1:1,000 dilution) at 4°C overnight. The membranes were then washed and incubated with the corresponding HRP-conjugated secondary antibodies (1:10,000 dilution) for 1 hr at room temperature. Bands were detected using the Azure 600 Imaging System with enhanced chemiluminescence (ECL, Pierce). PhosphoERK1/2 expression was normalized to total-ERK1/2 expression and the baseline at 0 min was subtracted to produce the time-dependent change in pERK1/2. The density of each time-point was quantified using ImageJ 1.5255.
Microscopy
Cells were plated in 6-well culture plates at a density of 9 × 105 cells per well. Cells were co-transfected 24 hours after seeding with 800 ng Halo-tagged receptor constructs using 6 μL of lipofectamine 2000. 24 hours after transfection cells were incubated with 250 nM JF-646, generously provided by Luke Lavis and Jonathan B. Grimm (Janelia Research Campus), in DMEM containing 10% fetal bovine serum for 30 min at 37° in 5% CO2 and subsequently in fresh DMEM for 1 hr, before being washed in DPBS and resuspended in enzyme free. After labelling, cells were washed 3X in PBA (PBS with 0.1% w/v BSA) and diluted in microscopy buffer (FluoroBrite DMEM, 20 mM HEPES, pH 7.5). Cells were seeded on a fibronectin-coated glass cover slip (dimensions 22 × 22 mm, thickness 0.17 nm, SCHOTT Nexterion) and incubated in FluoroBrite DMEM for 2–4 hours at 37 °C in 5% CO2. Finally, cells were fixed in a 5% paraformaldehyde solution overnight at 4 °C for next day imaging on a Leica SP8 using the 640 nm laser line.
Data availability
All cDNA constructs and data are available on request from the authors. Statistical source data are provided for all main figures, extended data figures and supplementary figures. Unprocessed full scans are provided for the immunoblots shown in Extended Data Fig. 5 and 8.
Extended Data
Extended Data Fig. 1. TA-enhanced signaling effect is also observed in SRE, NFAT, and NFκB gene expression assays.
(a) SRE (b) NFκB and (c) NFAT. For all gene response elements (SRE, NFAT, and NFκB) signaling was increased significantly when the entire N-terminal fragment up to the GPS cleavage site (CTF) was removed; FL receptor also showed some activity in SRE (comparable to CRE in Figure 1). Luminescence in (a-c) was measured for a range of increased receptor cDNA concentrations ~24 hours after transfection in HEK293T cells. All data points are normalized to an empty vector control. Data are presented as mean values ±SEM from 3 independent experimental replicates.
Extended Data Fig. 2. Successive truncation of the first three ADGRL3 tethered agonist residues dramatically blunts SRE-Luciferase gene reporter activation.
SRE gene expression assay for ADGRL3 (human homolog) CTF, Δ1-CTF, Δ2-CTF and Δ3-CTF. Luminescence was measured for a range of increased receptor cDNA amounts (ng) ~24 hours after transfection in HEK293T cells. For the dual luciferase assay, data are presented as Firefly/Renilla luciferase units, and all data points are normalized to the corresponding ratio for the empty vector control15,51–52. Data are from one representative experiment performed 3 times. Data are presented as mean ±SD from triplicate technical replicates.
Extended Data Fig. 3. Screen of Adgrl3 (FL, CTF, and Δ5-CTF constructs) signaling in the 4 major G protein signaling pathways utilizing a HEK-293 CRISPR knockout cell line (HEKΔ7) and a panel of gene expression assays.
(a) CRE (b) NFκB (c) SRE. Each Gα protein species was reintroduced one at a time (see color legend for specification) at optimized cDNA concentrations and luminescence signals were evaluated for empty vector control and receptor constructs ~24 hours after transfection. All data points are normalized to corresponding empty vector control. Bars indicate mean values ±SEM from 4 (a) and 5 (b-c) independent experimental replicates. Bars for Gαolf and Gα12 are presented as mean values ±SEM from 3 independent experimental replicates.
Extended Data Fig. 4. CTF Gαq signaling is detected both in CRE and NFκB.
CTF signaling in CRE, NFκB, and SRE was evaluated after 18 hours of treatment with either vehicle or a potent Gαq inhibitor (YM-254890, 1 μM). Data was collected in regular HEK293T cells. Data points are normalized to empty vector control and displayed as the fold decrease with YM-254890. Bars show mean ±SFM from 4 independent experimental replicates
Extended Data Fig. 5. Urea-mediated ADGRL3 N-terminal Fragment dissociation.
For the membrane urea treatment experiments presented in this figure, a FLAG- (N-terminal) and His8- (C-terminal) tagged ADGRL3 construct that was truncated N-terminally to the HormR domain was used21 (See Fig. 1 for Adgrl3 architecture). Insect cell membranes (High-Five) with expressed ADGRL3 were mock treated or extracted with urea. The presence of the ADGRL3 NTF and CTF in the membrane (Mem) and extract (Soluble, Sol) fractions was determined by immunoblotting with an anti-FLAG antibody to detect the NTF and an anti-penta-His antibody to detect the CTF. The NTF (apparent MW ~50 kDa) was partially solubilized with the urea, whereas the CTF (apparent MW ~27 kDa) was not. The penta-His blot panels are from one contiguous blot, but broken to avoid oversaturation of the ~70 kDa band (unprocessed receptor) and to show a higher exposure of low MW panel (~27 kDa CTF). Data from one representative experiment that was repeated three times.
Extended Data Fig. 6. CRE, NFκB, and SRE gene expression assays for PAR1-CTF and corresponding T923S/ΔN924-CTF control construct.
CRE (a), NFκB (b) and SRE (c) signaling was increased significantly for T923S/ΔN924-CTF to levels comparable with CTF, whereas PAR1-CTF signals were comparable to FL levels. CTF and FL are replotted from Figure 1c and Extended Data Fig. 1 for direct comparison. Luminescence was measured for a range of receptor cDNA concentrations ~24 hours after transfection in HEK293T cells. All data points are normalized to an empty vector control. Data are shown as mean ±SEM from 3 independent experimental replicates.
Extended Data Fig. 7. TA-exposed Adgrl3 does not activate the Gαi/o family.
(a) Gβγ release assay testing D2R activation of Gαi1, Gαi2, Gαi3, GαoA and GαoB in HEK full G protein KO cells. In comparison to the HEKΔ7 CRISPR knockout, this cell line also lacks the full Gαi/o family. Luminescence was read 10 min after stimulation with 10 jliM quinpirole. (b) Gβγ release assay testing the T923S/ΔN924-CTF, PAR1-CTF, and PAR1 activation of Gαi1, Gαi2, Gαi3, GαoA and GαoB in HEK full G protein KO cells. Luminescence was read 10 min after stimulation with 1 μM thrombin. All data are normalized to buffer controls and show the BRET effect induced by ligands. Bars show mean ±SEM from 3 independent experimental replicates. One-way ANOVA with Dunnett’s multiple-comparison post-hoc test was performed for each cDNA construct individually, (no receptor (empty vector), T923S/ΔN924-CTF, PAR1-CTF, and PAR1) to determine statistical significance between the No Gα control and each Gα subtype (For Gαi3**p=0.0064, for GαoB **p=0.0032). See Supplementary Data for the full set of p-values.
Extended Data Fig. 8. β-arrestin-2 decreases G protein-dependent ERK1/2 phosphorylation.
HEK Δarr1/2 cells were transfected with PAR1-CTF or PAR1-CTF with β-arrestin-2. After 48 hr, the cells were acutely activated with 1 μM thrombin over a time course of 45 min. (a) Representative immunoblotting analysis with antibodies against phosphoERK1/2 (#9101S), total ERK1/2 (#9102S), and HA (#2367S). Each sample was derived from the same experiment and the blots were processed in parallel. The HA blot was used as a sample processing control to ensure uniform β-arrestin-2 expression. (b) The level of phospho-ERK1/2 was normalized to total ERK and the baseline at 0 min was subtracted to produce the time-dependent change in pERK1/2. Data are presented as mean ±SEM from 3 (PAR1-CTF) 4 (PAR1-CTF, β-Arrestin-2) independent experimental replicates.
Supplementary Material
Acknowledgements
This work was supported by NIH grants MH112156 (J.A.J.), GM130142 (N.A.L.), GM131672 (N.O.), and T32-GM007315 (A.V.) and by the Hope for Depression Research Foundation (J.A.J.). A.I. was funded by the PRIME 18gm5910013 and the LEAP 18gm0010004 from the Japan Agency for Medical Research and Development (AMED) and KAKENHI 17K08264 from by the Japan Society for the Promotion of Science (JSPS). The authors thank L. Lavis and J. B. Grimm (Janelia Research Campus) for generously providing the JF-525 and JF-646 fluorophores.
Footnotes
Competing Interests statement
The authors declare no competing financial interests.
References main text
- 1.Purcell RH & Hall RA Adhesion G Protein-Coupled Receptors as Drug Targets. Annu Rev Pharmacol Toxicol 58, 429–449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamann J, Aust G, Arac D, Engel FB,Formstone C,Fredriksson R,Hall RA,Harty BL, Kirchhoff C, Knapp B, Krishnan A, Liebscher I, Lin HH, Martinelli DC, Monk KR, Peeters MC, Piao X, Promel S, Schoneberg T, Schwartz TW, Singer K, Stacey M, Ushkaryov YA, Vallon, Wolfrum U, Wright MW, Xu L, Langenhan T & Schioth HB. International Union of Basic and Clinical Pharmacology. XCIV. Adhesion G protein-coupled receptors. Pharmacol Rev 67, 338–367 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arcos-Burgos M, Castellanos F.x., Pineda D, Lopera F, Palacio JD, Palacio LG, Rapoport JL, Berg K, Bailey-Wilson JE & Muenke M Attention-deficit/hyperactivity disorder in a population isolate: Linkage to loci at 4q13.2, 5q33.3, 11q22, and 17p11. Am J Hum Genet 75, 998–1014 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arcos-Burgos M, Velez JI, Martinez AF, Ribases M, Ramos-Quiroga JA, Sanchez-Mora C, Richarte V, Roncero C, Cormand B, Fernandez-Castillo N, Casas M, Lopera F, Pineda DA, Palacio JD, Acosta-Lopez JE, Cervantes-Henriquez ML, Sanchez-Rojas MG, Puentes-Rozo PJ, Molina BSG, Group MTAC, Boden MT, Wallis D, Lidbury B, Newman S, Easteal S, Swanson J, Patel H, Volkow N, Acosta MT, Castellanos FX, de Leon J, Mastronardi CA & Muenke M ADGRL3 (LPHN3) variants predict substance use disorder. Transl Psychiatry 9, 42 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van de Voet M, Harich B, Franke B & Schenck A ADHA-asoociated dopamine transporter, latrophilin and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol Psychiatry (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange M, Norton W, Coolen M, Chaminade M, Merker S, Proft F, Schmitt A, Vernier P, Lesch KP & Bally-Cuif L The ADHD-susceptibility gene lphn3.1 modulates dopaminergic neuron formation and locomotor activity during zebrafish development. Mol Psychiatry 17, 946–954 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Lange M, Froc C, Grunwald H, Norton WHJ & Bally-Cuif L Pharmacological analysis of zebrafish lphn3.1 morphant larvae suggests that saturated dopaminergic signaling could underlie the ADHD-like locomotor hyperactivity. Prog Neuropsychopharmacol Biol Psychiatry 84, 181–189 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Regan SL, Hufgard JR, Pitzer EM, Sugimoto C, Hu YC, Williams MT & Vorhees CV Knockout of latrophilin-3 in Sprague-Dawley rats causes hyperactivity, hyper-reactivity, under-response to amphetamine, and disrupted dopamine markers. Neurobiol Dis 130, 104494 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wallis D, Hill DS, Mendez IA, Abbott LC, Finnell RH, Wellman PJ & Seltow B Initial characterization of mice null for Lphn3, a gene implicated in ADHD and addiction. Brain Res 1463, 85–92 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Arac D, Boucard AA, Bolliger MF, Nguyen J, Soltis SM, Sudhof TC & Brunger AT A novel evolutionarily conserved domain of cell-adhesion GPCRs mediates autoproteolysis. Embo Journal 31, 1364–1378 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin HH, Chang GW, davies JQ, Stacey M, Harris J & Gordon S Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. Journal of Biological Chemistry 279, 31823–31832 (2004). [DOI] [PubMed] [Google Scholar]
- 12.Paavola KJ, Stephenson JR, Ritter SL, Alter SP & Hall RA The N terminus of the adhesion G protein-coupled receptor GPR56 controls receptor signaling activity. J Biol Chem 286, 28914–28921 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Paavola KJ & Hall RA Adhesion G protein-coupled receptors: signaling, pharmacology, and mechanisms of activation. Mol Pharmacol 82, 777–783 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liebscher I, Schon J, Petersen SC, Fisher L, Auerbach N, Demberg LM, Mogha A, Coster M, Simon KU, Rothemund S, Monk KR & Schoneberg T A Tethered Agonist within the Ectodomain Activates the Adhesion G Protein-Coupled Receptors GPR126 and GPR133. Cell Rep 9, 2018–2026 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stoveken HM, Hajduczok AG, Xu L & Tall GG Adhesion G protein-coupled receptors are activated by exposure of a cryptic tethered agonist. Proc Natl Acad Sci U S A 112, 6194–6199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lelianova VG, Davletov BA, Sterling A, Rahman MA, Grishin EV, Totty NF & Ushkaryov YA Alpha-latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J Biol Chem 272, 21504–21508 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Rahman MA, Ashton AC, Meunier FA, Davletov BA, Dolly JO & Ushkaryov YA Norepinephrine exocytosis stimulated by alpha-latrotoxin requires both external and stored Ca2+ and is mediated by latrophilin, G proteins and phospholipase C. Philos Trans R Soc Lond B Biol Sci 354, 379–386 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Silva JP, Lelianova VG, Ermolyuk YS, Vysokov N, Hitchen PG, Berninghausen O, Rahman MA, Zangrandi A, Fidalgo S, Tonevitsky AG, Dell A, Volynski KE & Ushkaryov YA Latrophilin 1 and its endogenous ligand Lasso/teneurin-2 form a high-affinity transsynaptic receptor pair with signaling capabilities. Proc Natl Acad Sci U S A 108, 12113–12118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller A, Winkler J, Fiedler F, Sastradihardja T, Binder C, Schnabel R, Kungel J, Rothemund S, Hennig C, Schoneberg T & Promel S Oriented Cell Division in the C. elegans Embryo Is Coordinated by G-Protein Signaling Dependent on the Adhesion GPCR LAT-1. Plos Genet 11(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholz N, Guan C, Nieberler M, Grotemeyer A, Maiellaro I, Gao S, Beck S, Pawlak M, Sauer M, Asan E, Rothemund S, Winkler J, Promel S, Nagel G, Langenhan T & Kittel RJ Mechanodependent signaling by Latrophilin/CIRL quenches cAMP in proprioceptive neurons. eLife 6(2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nazarko O, Kibrom A, Winkler J, Leon K, Stoveken H, Salzman G, Merdas K, Lu Y, Narkhede P, Tall G, Promel S & Arac D A Comprehensive Mutagenesis Screen of the Adhesion GPCR Latrophilin-1/ADGRL1. iScience 3, 264–278 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rothe J, Thor D, Winkler J, Knierim AB, Binder C, Huth S, Kraft R, Rothemund S, Schoneberg T & Promel S Involvement of the Adhesion GPCRs Latrophilins in the Regulation of Insulin Release. Cell Rep 26, 1573–1584 e1575 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Liebscher I & Schoneberg T Tethered Agonism: A Common Activation Mechanism of Adhesion GPCRs. Handb Exp Pharmacol 234, 111–125 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Martin AL, Steurer MA & Aronstam RS Constitutive Activity among Orphan Class-AG Protein Coupled Receptors. Plos One 10(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LeBonniec BF, Myles T, Johnson T, Knight CG, Tapparelli C & Stone SR Characterization of the P2' and P3' specificities of throm bin using fluorescence-quenched substrates and mapping of the subsites by mutagenesis. Biochemistry 35, 7114–7122 (1996). [DOI] [PubMed] [Google Scholar]
- 26.Okashah N, Wan Q, Ghosh S, Sandhu M, Inoue A, Vaidehi N & Lambert NA Variable G protein determinants of GPCR coupling selectivity. Proc Natl Acad Sci U S A 116, 12054–12059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Green DA & Clark RB Direct evidence for the role of the coupling proteins in forskolin activation of adenylate cyclase. J Cyclic Nucleotide Res 8, 337–346 (1982). [PubMed] [Google Scholar]
- 28.Downs RW Jr. & Aurbach GD The effects of forskolin on adenylate cyclase in S49 wild type and cyccells. J Cyclic Nucleotide Res 8, 235–242 (1982). [PubMed] [Google Scholar]
- 29.Hollins B, Kuravi S, Digby GJ & Lambert NA The c-terminus of GRK3 indicates rapid dissociation of G protein heterotrimers. Cell Signal 21, 1015–1021 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuho I, Ostrovskaya O, Kramer GM, Jones CD, Xie KQ & Martemyanov KA Distinct profiles of functional discrimination among G proteins determine the actions of G protein-coupled receptors. Science Signaling 8(2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vu TK, Wheaton VI, Hung DT, Charo I & Coughlin SR Domains specifying thrombin-receptor interaction. Nature 353, 674–677 (1991). [DOI] [PubMed] [Google Scholar]
- 32.Soh UJ & Trejo J Activated protein C promotes protease-activated receptor-1 cytoprotective signaling through beta-arrestin and dishevelled-2 scaffolds. Proc Natl Acad Sci U S A 108, E1372–1380 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demberg LM, Rothemund S, Schoneberg T & Liebscher I Identification of the tethered peptide agonist of the adhesion G protein-coupled receptor GPR64/ADGRG2. Biochem Biophys Res Commun 464, 743–747 (2015). [DOI] [PubMed] [Google Scholar]
- 34.Worzfeld T, Wettschureck N & Offermanns S G(12)/G(13)-mediated signalling in mammalian physiology and disease. Trends in pharmacological sciences 29, 582–589 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Iguchi T, Sakata K, Yoshizaki K, Tago K, Mizuno N & Itoh H Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem 283, 14469–14478 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Ohta S, Sakaguchi S, Kobayashi Y, Mizuno N, Tago K & Itoh H Agonistic antibodies reveal the function of GPR56 in human glioma U87-MG cells. Biol Pharm Bull 38, 594–600 (2015). [DOI] [PubMed] [Google Scholar]
- 37.Zhang DL, Sun YJ, Ma ML, Wang YJ, Lin H, Li RR, Liang ZL, Gao Y, Yang Z, He DF, Lin A, Mo H, Lu YJ, Li MJ, Kong W, Chung KY, Yi F, Li JY, Qin YY, Li J, Thomsen ARB, Kahsai AW, Chen ZJ, Xu ZG, Liu M, Li D, Yu X & Sun JP Gq activity- and beta-arrestin-1 scaffolding-mediated ADGRG2/CFTR coupling are required for male fertility. eLife 7(2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peeters MC, Fokkelman M, Boogaard B, Egerod KL, van de Water B, AP IJ & Schwartz TW The adhesion G protein-coupled receptor G2 (ADGRG2/GPR64) constitutively activates SRE and NFkappaB and is involved in cell adhesion and migration. Cell Signal 27, 2579–2588 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Kishore A, Purcell RH, Nassiri-Toosi Z & Hall RA Stalk-dependent and Stalk-independent Signaling by the Adhesion G Protein-coupled Receptors GPR56 (ADGRG1) and BAI1 (ADGRB1). J BiolChem 291, 3385–3394 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Okajima D, Kudo G & Yokota H Brain-specific angiogenesis inhibitor 2(BAI2) may be activated by proteolytic processing. J Recept Sig Transd 30, 143–153 (2010). [DOI] [PubMed] [Google Scholar]
- 41.O'Sullivan ML, de Wit J, Savas JN, Comoletti D, Otto-Hitt S, Yates JR 3rd & Ghosh A FLRT proteins are endogenous latrophilin ligands and regulate excitatory synapse development. Neuron 73, 903–910 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sando R, Jiang X & Sudhof TC Latrophilin GPCRs direct synapse specificity by coincident binding of FLRTs and teneurins. Science 363(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li J, Shalev-Benami M, Sando R, Jiang X, Kibrom A,Wang J, Leon K, Katanski C, Nazarko O, Lu YC, Sudhof TC, Skiniotis G & Arac D Structural Basis for Teneurin Function in Circuit-Wiring: A Toxin Motif at the Synapse. Cell 173, 735–748 e715 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cruz-Ortega JS & Boucard AA Actin cytoskeleton remodeling defines adistinct cellular function for adhesion G protein-coupled receptors ADGRL/latrophilins 1, 2 and 3. Biol Open 8(2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Southern C, Cook JM, Neetoo-Isseljee Z, Taylor DL, Kettleborough CA, Merritt A, Bassoni DL, Raab WJ, Quinn E, Wehrman TS, Davenport AP, Brown AJ, Green A, Wigglesworth MJ & Rees S Screening beta-arrestin recruitment for the identification of natural ligands for orphan G-protein-coupled receptors. J Biomol Screen 18, 599–609 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Lefkowitz RJ Arrestins come of age: a personal historical perspective. Prog Mol Biol Transl Sci 118,3–18 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Spiess K, Bagger SO, Torz LJ, Jensen KHR, Walser AL, Kvam JM, Mogelmose AK, Daugvilaite V, Junnila RK, Hjorto GM & Rosenkilde MM Arrestin-independent constitutive endocytosis of GPR125/ADGRA3. Ann N Y Acad Sci 1456, 186–199 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petersen SC, Luo R, Liebscher I, Giera S, Jeong SJ, Mogha A, Ghidinelli M, Feltri ML, Schoneberg T, Piao X & Monk KR The adhesion GPCR GPR126 has distinct, domain-dependent functions in Schwann cell development mediated by interaction with laminin-211. Neuron 85, 755–769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Scholz N, Gehring J, Guan C, Ljaschenko D, Fischer R, Lakshmanan V, Kittel RJ & Langenhan T The adhesion GPCR latrophilin/CIRL shapes mechanosensation. Cell Rep 11, 866–874 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Promel S, Frickenhaus M, Hughes S, Mestek L, Staunton D, Woollard A, Vakonakis I, Schoneberg T, Schnabel R, Russ AP & Langenhan T The GPS motif is a molecular switch for bimodal activities of adhesion class G protein-coupled receptors. Cell Rep 2, 321–331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
References online methods
- 51.Stoveken HM, Bahr LL, Anders MW, Wojtovich AP, Smrcka AV & Tall GG Dihydromunduletone Is a Small-Molecule Selective Adhesion G Protein-Coupled Receptor Antagonist. Molecular Pharmacology 90, 214–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stoveken HM, Larsen SD, Smrcka AV & Tall GG Gedunin- and Khivorin-Derivatives Are Small-Molecule Partial Agonists for Adhesion G Protein-Coupled Receptors GPR56/ADGRG1 and GPR114/ADGRG5. Molecular Pharmacology 93, 477–488 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Grundmann M, Merten N, Malfacini D, Inoue A, Preis P, Simon K, Ruttiger N, Ziegler N, Benkel T, Schmitt NK, Ishida S, Muller I, Reher R, Kawakami K, Inoue A, Rick U, Kuhl T, Imhof D, Aoki J, Konig GM, Hoffmann C, Gomeza J, Wess J & Kostenis E Lack of beta-arrestin signaling in the absence of active G proteins. Nat Commun 9, 341 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stallaert W, van der Westhuizen ET, Schonegge AM, Plouffe B, Hogue M, Lukashova V,Inoue A, Ishida S, Aoki J, Le Gouill C & Bouvier M Purinergic Receptor Transactivation by the beta2-Adrenergic Receptor Increases Intracellular Ca(2+) in Nonexcitable Cells. Mol Pharmacol 91, 533–544 (2017). [DOI] [PubMed] [Google Scholar]
- 55.Schneider CA, Rasband WS & Eliceiri KW NIH Image to Image J: 25 yearsof image analysis. Nat Methods 9, 671–675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All cDNA constructs and data are available on request from the authors. Statistical source data are provided for all main figures, extended data figures and supplementary figures. Unprocessed full scans are provided for the immunoblots shown in Extended Data Fig. 5 and 8.