Abstract
The utility of reporter genes has gained significant momentum over the last three decades. Reporter genes are used to understand the transcriptional activity of a gene both in vitro and in vivo, and in pathway analysis and drug screening for diseases involving protozoan parasites, and in anti-cancer drug developments. Here, using a human prostate cancer xenograft model (PC3), we describe a method to construct and validate hypoxia reporter genes with different half-lives. Using molecular biology and optical imaging techniques, we have validated the expression of long half-life enhanced green fluorescence protein (EGFP) expression and short half-life luciferase gene expression to report on the spatial and temporal evolution of hypoxia in vivo.
Keywords: Bioluminescence, Lentivirus, Luciferase assay, Hypoxia, Hypoxia response elements (HRE), Reporter gene
1. Introduction
Our understanding of gene function has primarily come from the successful introduction and transfer of genes into cells. Detecting and screening the transformants for phenotypic changes in the transformed cells has advanced from the first use of chloramphenicol acetyltransferase (CAT) as a reporter of enzyme activity [1]. Reporter genes reliably measure gene expression and help in understanding the transcriptional activity of genes in response to a specific signaling event. Subsequently, reporter genes such as acid phosphatase and alkaline phosphatase detected by colorimetry [2, 3] and firefly and renilla luciferase detected by bioluminescence imaging (BLI) have been successfully used as reporters [4-7]. Visualization and localization of successful gene transfer, and corresponding reporter activity has become less challenging since the purification and cloning of green fluorescent protein (GFP) [8, 9]. Colorimetric, BLI, and fluorescence-based reporter genes have both advantages and disadvantages. The GFP protein is a single chain 238 amino acid polypeptide with a compact structure that is stable under various conditions. GFP is minimally toxic and can be noninvasively detected in live cells without cell disruption or addition of an external co-factor, such as luciferin required for BLI. Modifications in the GFP protein to encode a destabilized red-shift variant (d2EGFP) led to brighter fluorescence signals in mammalian cells (excitation max = 488 nm, emission max = 507 nm) and fusion of a sequence rich in proline (P), glutamic acid (E), serine (S), threonine (T) (PEST sequence) on the C-terminus region enabled the degradation of the protein decreasing its half-life in mammalian cells to 9.8 h [10].
Luciferase has been used as a reporter since it was cloned in 1985 [7]. This 550 amino acid, 62 kDa single polypeptide chain enzyme catalyzes the substrate d-luciferin, to emit bioluminescence that is visualized at 562 nm. The disadvantage of luciferase is the need for an additional substrate/co-factor, as well as oxygen and ATP, to detect the reporter. The need to destroy the cell/tissue to determine luciferase reporter activity was considered a disadvantage. However, with the advent of sensitive imaging instruments, bioluminescence can be detected noninvasively. Although the half-life of luciferase is only 2 h, the quantum yield is very high compared to fluorescent reporters, making it a useful reporter gene.
The transcription factor hypoxia inducible factor (HIF) is a heterodimer formed by a constitutively expressed protein subunit HIF-1β and an oxygen regulated protein subunit HIF-1α. In response to hypoxia, HIF binds to response element on the promoter regions of many genes to activate transcription [11]. In a proof-of-principle study, we have cloned five tandem repeats of hypoxia response elements (HREs) to create an artificial promoter and drive the transcription of EGFP and luciferase [12]. This chapter provides in-depth details on the construction, detection, and validation of short and long half-life reporters that are activated in response to hypoxia both in vitro and in vivo.
2. Materials
2.1. Antibodies
EGFP, GAPDH.
2.2. Antibiotics
Neomycin (G418-400 μg/mL from a 50 mg/mL stock). Puromycin (1 μg/mL from a stock of 10 mg/mL).
2.3. Cell Culture Related Materials
Fetal bovine serum (FBS).
Hanks balanced salt solution (HBSS).
OPTI-MEM medium.
RPMI medium.
PBS/DPBS- Dulbecco’s phosphate-buffered saline (Calcium and Magnesium free).
Petri dishes (60 mm and 100 mm).
Tissue culture Flasks (175 mm).
2.4. Cells
293 T cells for virus generation.
PC3 cells.
2.5. Chemicals
d-Luciferin (150 μg/mL for in vitro, 3 mg/mouse).
Dithiothreitol (DTT)—1 μL of DTT (1 M) per 1 mL of RIPA buffer.
Luciferase assay reagent (Promega) diluted according to the manufacturer’s instructions.
Orthovanadate (0.2 M stock).
Passive lysis buffer (1×)—Dilute 5× stock to 1× working stock with water.
Poly-d-lysine.
Phenylmethylsulfonyl fluoride (PMSF, 0.2 M).
Polybrene—8 μg/mL (Stock-8 mg/mL).
Protease Inhibitor mixture.
Radioimmunoprecipitation assay buffer (RIPA).
Tween 20–0.05%.
2.6. Instruments
PCR—polymerase chain reaction machine.
Plate reader.
2.7. Plasmids
EGFP plasmid with minimal Cytomegalovirus (CMV) promoter.
Lentiviral packaging plasmids (deltaR8.2 for accessory proteins, VSV-G for envelope protein).
2.8. Transfection Reagent
Lipofectamine 2000 or similar.
3. Construction of Hypoxia Regulated Long-Life EGFP Reporter Expression Plasmid Vector
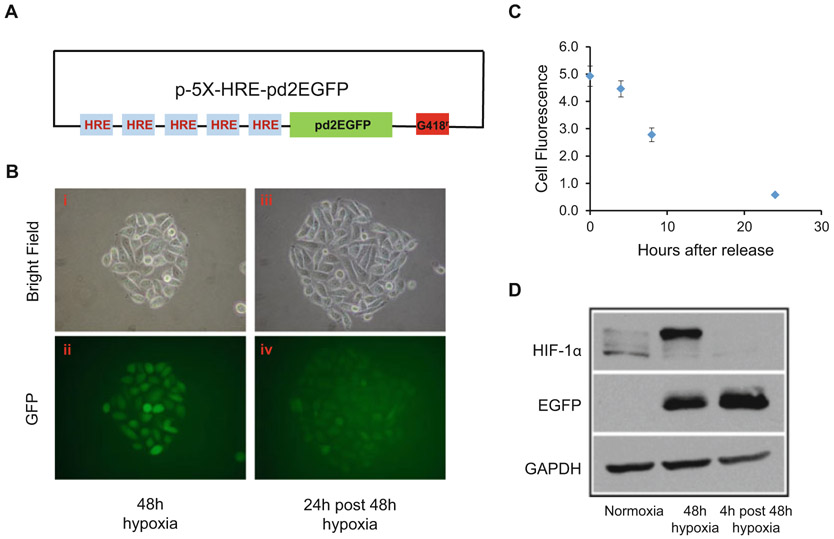
Five tandem copies of hypoxia response element (5XHRE) generation obtained by polymerase chain reaction (PCR) amplification from vascular endothelial growth factor (VEGF) promoter and their cloning into an expression vector are described by Shibata et al. [13]. PCR amplified 5XHRE promoter was cloned in frame on the 5′-end of the CMV-EGFP sequence to obtain the plasmid pd2-5X-HRE-CMV min-EGFP (HRE-EGFP) as shown in Fig. 1a. This mammalian expression vector also contains a geneticin resistant gene as a selection marker. By adding antibiotic G418, which is titrated depending upon the cell line used, cells expressing HRE-EGFP are selected as detailed below.
Fig. 1.
Long half-life reporter gene activity. (a) Box and line schematic showing the long half-life GFP reporter construct under the control of five tandem repeats of HRE. (b) Representative photomicrograph of PC3 cells stably expressing the plasmid containing the long half-life GFP reporter construct. Bright field and fluorescent images of a colony of PC3 cells stably expressing the HRE-GFP construct exposed to hypoxia for 48 h (i and ii). The same colony was photographed 24 h post hypoxia release (iii and iv). (c) Graph showing decrease in fluorescence intensity following incubation at various times after hypoxia release. Using ImageJ software, fluorescence intensity was calculated from photographs taken on a microscope equipped with a camera, from at least 3 to 5 colonies for each time point. Values represent mean ± S.E from three biological replicates. (d) Representative immunoblots showing expression of HIF-1α and EGFP in PC3 cells expressing HRE-GFP culture under normoxia, exposed to hypoxia for 48 h or maintained under normoxia for 4 h after 48 h hypoxia treatment
3.1. Establishing Stable Cell Line-Expressing HRE-EGFP
Prior to transduction, perform a “kill curve” to determine sensitivity of the cell line to G418. The example below uses PC-3 cells, a human prostate cancer cell line [14].
Plate 5 × 105 PC3 prostate cancer cells in a 60 mm dish 24 h before transfection.
In an eppendorf tube, dilute 1 μg of 5 × HRE-EGFP in 250 μL of OPTI-MEM medium In a second eppendorf tube with 250 μL of optimum medium, add 2 μL lipofectamine 2000 (Invitrogen, Carlsbad, CA) and mix well.
Add diluted plasmid DNA to diluted lipofectamine dropwise and incubate the complex for 20 min. Meanwhile, replace the media on the plate containing PC3 cells with low serum (1% FBS) containing RPM1640. At the end of incubation time, add the plasmid-lipofectamine complex dropwise on the cells and gently swirl the plate to evenly mix with the medium on the plate and distribute the complex evenly.
Twenty-four hours post transfection, change the media on the plate and replace with RPMI-1640 containing 10% FBS. For selecting PC-3 cells expressing the expression plasmid, add G418 (500 μg/mL).
Change the media on the plate every 4 days with fresh G418-containing medium till most of the non-transfected cells die. Once cells form colonies on the plate, pick healthy colonies with a sterile pipet tip, and transfer them to a 6-well culture dish (Note 1). Expand each colony separately and test the cells for GFP expression.
3.1.1. Detection and Validation of Hypoxia Regulated GFP Reporter Gene Expression
Reporter gene expression can be detected optically and validated for protein expression by western blot analysis of various PC3-HRE-GFP clones as follows.
Plate 300,000 PC3-HRE-GFP cells in three 60 mm petri dish.
Twenty-four hours later, change the media in both the plates. Place one plate in a 37 °C CO2 incubator. Place the other two plate in a modular incubator chamber (hypoxia chamber) and flush at 2 psi for 3 min with a gas mixture composition of 1% O2/5% CO2 and 94%N2, close the chamber and place it in the 37 °C CO2 incubator for 48 h (Note 2).
At the end of the 48-h period, using a 20× objective, EGFP expression can be detected using a fluorescence microscope equipped with a digital camera and a broadband halogen source (Fig. 1b, image ii). The excitation filter to detect EGFP is set to 500 nm and the emission filter for the detection at 500–540 nm.
To determine the half-life of EGFP expression, incubate the EGFP-expressing PC3 cells to normoxia and detect green fluorescence at various time points. As shown in Fig. 1b (image iv), GFP fluorescence is detected at low levels even after 24 h of reoxygenation and diminishes after 30 h (Fig. 1c).
To further validate reporter gene expression, perform immunoblotting for GFP and HIF-1α protein using antibodies specific for each component (Fig. 1d).
Briefly, seed 1 × 106 PC3-HRE-GFP cells in three 100 mm petri dish. On day 2, change media on the plates and place two plates (Plate #2, #3) in the hypoxia chamber and flush with a gas mixture as mentioned above to create hypoxia. Seal the incubator and place it in a 37 °C CO2 incubator for 48 h. Place the 100 mm petri dish (plate #1) under normoxic condition in a 37 °C CO2 incubator. At the end of incubation time, open the hypoxia chamber and place one dish (Plate #3) in the CO2 incubator to reoxygenate the cells for an additional 4 h. Place plate #1 and #2 on ice and aspirate the media. Wash the cells once with ice-cold DPBS and add 1 mL of ice-cold PBS and scrape the cells from the plate using a cell scraper. Collect the cells in an eppendorf tube and spin the tube in a centrifuge at 0.2 × g for 5 min at 4 °C to pellet. Aspirate the PBS and resuspend the pellet in radio immuno precipitation assay buffer (RIPA) fortified with protease inhibitor mixture, serine protease inhibitor-phenylmethylsulfonyl fluoride (PMSF, 0.2 M), 1 μL/mL of 1 M DTT, 0.2 M of sodium orthovanadate and 0.5 M sodium fluoride and incubate at 4 °C on ice for 30 min. Spin the contents at 16.1 × g for 30 min in a centrifuge set at 4 °C. Collect the supernatant and measure the concentration of the protein.
To detect the reporter activity, resolve the 100 μg of protein by polyacrylamide gel electrophoresis (PAGE) and transfer the separated protein to a nitrocellulose membrane by applying 40 mA of current overnight at 4 °C (Note 3).
Once the transfer of protein is complete, cut the membrane around 80–130 kDa and 32–50 kDa. Block the membrane with 5% nonfat dry milk in tris-buffered saline with Tween-20 (TBST) for 2 h. Probe for HIF-1α and EGFP expression using monoclonal antibodies against HIF-1α (1:1000 dilution, HIF-1α-67, Novus Biological, Littleton, Co) and anti-EGFP (1:2000 dilution, BD Biosciences, San Jose, CA). After the incubation with the primary antibody, wash the membranes three times with TBST and add horseradish peroxidase (HRP) conjugated goat anti-mouse secondary (1:2000). Once EGFP is detected, strip the EGFP bound antibody signal by soaking the membrane in stripping buffer at room temperature for 5 min and wash with phosphate-buffered saline (PBS), and probe for a loading control protein such as GAPDH (Fig. 1d).
3.2. Construction of Hypoxia Regulated Short Half-Life Luciferase Reporter Expression Plasmid Vector:
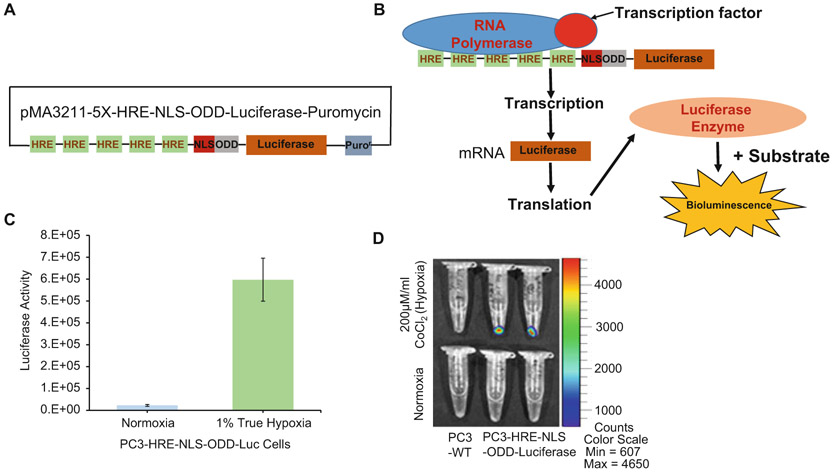
A plasmid containing five tandem copies of hypoxia response element (5×HRE) with oxygen-dependent degradation domain (ODD), nuclear localization signal (NLS), and firefly luciferase gene was kindly provided by Dr. Harada [15]. 5×HRE-NLS-ODD-Luc insert was later sub-cloned into a lentiviral vector as shown in Fig. 2a. This lentiviral vector also has the puromycin resistant gene as a selection marker.
Fig. 2.
Short half-life reporter gene activity. (a) Box and line schematic showing the short half-life luciferase reporter construct under the control of five tandem repeats of HRE. (b) Schematic showing binding of the transcription factor to the HRE and activating luciferase gene transcription that can be visualized as bioluminescence when the supplied substrate is converted by luciferase. (c) Luciferase assay quantifying the activity of the enzyme in PC3-HRE-NLC-ODD-Luc cells in response to 48 h of true hypoxia (1% oxygen). Values represent mean ± SEM from three independent experiments. *** p < 0.0005. (d) Representative image showing bioluminescence in PC3-HRE-NLS-ODD-Luc cells in response to 48 h treatment with hypoxia mimetic CoCl2. Images were captured using IVIS Spectrum Xenogen system
3.3. Generation of Virions-Expressing 5XHRE-NLS-ODD-Luciferase
To generate lentiviral particles plate 5 × 106 HEK 293-T cells on a 100 mm petri dish that has previously been coated with polylysine-D (Sigma, St. Louis, MO).
Twenty-four hours later, (day 2) change the medium to DMEM with 1% fetal bovine serum and transfect 293-T cells with a combination of lentiviral vector-expressing gene of interest, a packaging construct (ΔR8.2) and plasmid construct to pseudo type the envelop express vesicular stomatitis virus-G protein (VSVG) in the ratio of 12 μg:6 μg:1.5 μg using lipofectamine 2000, or similar reagent.
Forty-eight hours post transfection, collect the supernatant, and spin at 0.8 × g for 30 min. Transfer the viral supernatant carefully to a new tube (Note 4).
Seed PC3-HRE-GFP cells at 0.3 × 106 in a 60 mm dish. Twenty-four hours later, add 2 μL of polybrene to 2 mL of viral supernatant-expressing HRE-NLS-ODD-Luc virions, gently mix, and add it on to PC3-HRE-GFP cells. Allow the transduction to occur for 24 h and repeat this process again.
After a two-time infection, start selecting for clones that express both HRE-GFP and HRE-NLS-ODD-Luciferase by adding G418 and puromycin as described in the earlier section.
3.4. Detection and Validation of Hypoxia Regulated Luciferase Reporter Activity
Quantification of reporter activity in vitro can be performed by luciferase assay of various clones expressing HRE-NLS-ODD-Luciferase gene (Fig. 2). In the luciferase assay, firefly luciferase, the monomeric 61 kDa enzyme, catalyzes luciferin (substrate) oxidation by using ATP-Mg2+ as a cosubstrate. During this process a flash of light is generated that decays rapidly after the enzyme and substrate is combined and the bioluminescence is read at peak emission of 560 nm (yellow-green light) (Fig. 2b).
Briefly, plate PC3-HRE-NLS-ODD-Luc-expressing cells in two 100 mm petri dishes.
Twenty-four hours later, change the media in both the plates. Place one plate in a 37 °C CO2 incubator. Place the other plate in a modular incubator chamber and flush at 2 psi for 3 min with a gas mixture composition of 1% O2/5% CO2 and 94% N2. Close the chamber and place in the 37 °C CO2 incubator for 48 h.
At the end of the 48-h period, wash the cells with ice-cold PBS. Add 100 μL of 1× lysis buffer to lyse the cells. For the assay, add 10 μL of lysate in a 96-well plate with a solid bottom. Place the plate on a plate reader and pump 100 μL of luciferase assay reagent. The amount of light emitted reflects luciferase activity (Fig. 2c).
Short half-life luciferase reporter activity can also be detected optically and validated for protein expression in vitro conditions qualitatively by measuring BLI using optical instruments such as the Xenogen IVIS® series (Perkin Elmer).
To detect reporter activity in vitro, plate 300,000 HRE-NLS-ODD-Luciferase-expressing cells in two 60 mm petri dishes.
Twenty-four hours later, change the media in both the plates. In one set, add 200 μM cobalt chloride which is a hypoxia mimetic. Place both the plates in the 37 °C CO2 incubator for 48 h.
At the end of incubation, wash the cells with ice-cold phosphate-buffered saline (PBS), and scrape the cells in a small volume of buffer. Spin the content to pellet the cells and place them on ice. Switch on the optical imaging equipment (IVIS spectrum) and adjust the setting as follows: exposure is set to 30 s, field of view (FOV) to position (C): 12.8 × 12.8 cm, F at 1 and binning to 8, add 50 μL of the substrate d-luciferin to the pellet and mix and place the tubes on the stage inside the detection unit and acquire the image. Figure 2d clearly shows increase in bioluminescence in response to hypoxia in PC3 cells expressing HRE-NLS-ODD-Luciferase reporter cells and not in the control cells.
For the in vivo detection of reporter activity, appropriate mouse models should be used depending upon the cell line you choose to study. In the present example, for the in vivo detection of reporter gene activity, PC3 clones expressing both HRE-GFP and HRE-NLS-ODD-Luc construct were validated for EGFP expression and luciferase activity as mentioned above. We used 4–6 weeks old severely combined immunodeficient (SCID) male mice for this study (Note 5).
3.5. Tumor Implantation, Monitoring, and Imaging of Reporter Gene Activity
Grow PC3-HRE-GFP + HRE-NLS-ODD-Luciferase cells in T175 flasks till they reach 80–90% confluence. Trypsinize the cells and resuspend in Hanks balanced salt solution (HBSS) such that the concentration is 2 × 106 cells per 50 μL. Prepare enough for at least twice the number of animals that one is planning to inject. Monitor the tumor growth by measuring the tumor size with a caliper.
Prepare anesthetic according to the guidelines formulated by respective institutional Animal Care and Use Committee to temporarily immobilize the animal and minimize any pain.
Once the animal is anesthetized, mix the cell suspension and inject 50 μL of cell suspension subcutaneously in the flank.
Monitor tumor growth regularly. Once the tumor reaches 200 mm3, prepare for in vivo optical imaging using In Vivo Imaging System (IVIS)-Xenogen (Perkin Elmer) or a similar imaging instrument.
Place the mice into a clear plexiglass anesthesia box and pass gas-based anesthetic (2.5–3.5% isofluorane) through the tubing until anesthetized (Note 6).
Once anesthetized, transfer the animals from anesthesia box to the imaging chamber, and place with the nose inserted into the nose cones attached to the manifold in the imaging chamber. Continue to pass anesthetic at 2.5% isoflurane/O2.
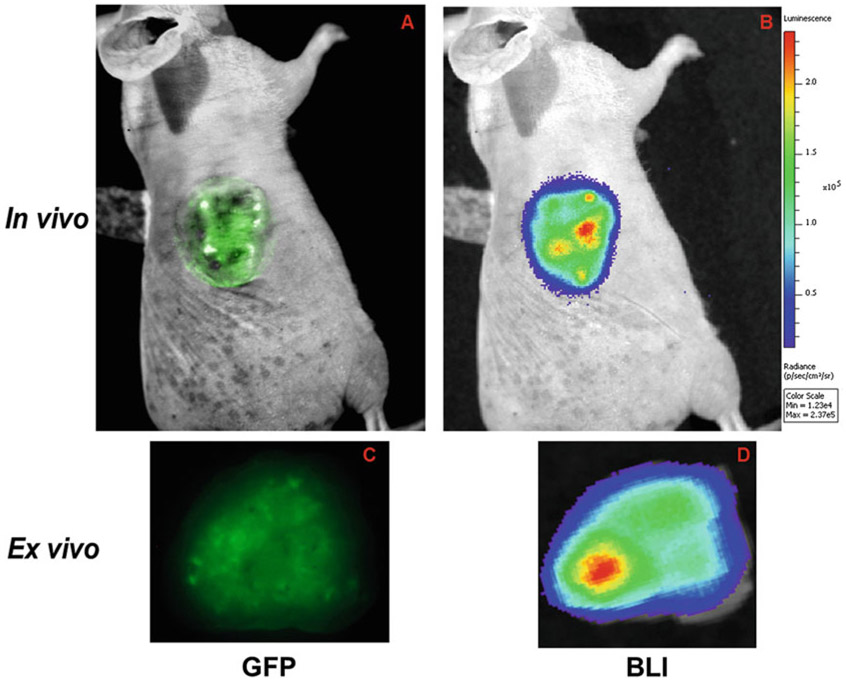
For reporter activity of long half-life protein, i.e., HRE-GFP, the following parameters are set for the GFP detection. Set the Channel to: 465/520, 465/540, 500/540 (excitation/emission) (Note 7). Set the exposure time to 1 s and FOV to (B): 12.8 × 12.8 cm. Set the f-stop to position 2 and binning to 8. Acquire the GFP image and save it (Fig. 3a).
Without moving the animal, inject d-luciferin and wait for 15 min before imaging the short half-life reporter using the following parameter: Exposure 30 seconds, FOV set to position (C): 12.8 × 12.8 cm, F = 1 and binning at 8 (Fig. 3b). (Note 8)
To minimize autofluorescence in the GFP channel and accurately detect both long half-life GFP and short half-life luciferase reporter activity, ex vivo imaging of the tumor slice can be performed.
Remove the tumor from the euthanized mouse. Make sure to remove the skin from the tumor.
Place the tumor on a tissue slicer to make 1 mm thick slices and place on a glass slide.
Place the glass slide with tissue slices on the imaging stage of the Xenogen scanner and capture GFP images using the parameters mentioned before.
Without changing the orientation, perform bioluminescence (BLI) imaging using the previously mentioned parameters.
Fig. 3.
Hypoxia reporter gene activity in tumors. Representative fluorescence image of a tumor derived from PC3 cells stably expressing both long half-life GFP and short half-life luciferase reporter under HRE control in vivo (a, b) and ex vivo (c, d)
Acknowledgments
This work was supported by National Institutes of Health R01 CA73850, R01 CA82337, and P50 CA103175.
Footnotes
Alternatively, cells can be plated in a 100 mm petri dish and subjected to selection pressure as described previously. As a precaution, dislodge single colonies one at a time and do not mix them if you want to select a clone with highest GFP expression. Before you start the antibiotic selection, it is advisable to perform a survival curve with different concentrations of antibiotics using wild-type cells.
The maximum psi (pounds per square inch) the modular incubator can hold/withstand is 2 psi. Therefore, do not increase the force gas mixture above 2 psi. This can be monitored and regulated by adjusting the knob on the regulator attached to the gas tank. When the knob is released at the end of the hypoxia treatment, the sound of gas escaping from the chamber will be heard.
Acrylamide is a hazardous chemical. Avoid contact with skin. Precast gels may also be used. However, make sure a large volume can be loaded into the wells, especially if the protein concentration is very low. When performing wet transfer of proteins on the gel to nitrocellulose membrane, make sure that there are no air bubbles.
Production and handling of lentiviral vectors and viral particles need prior approval from the institutional biosafety committee and environmental health and safety officials. All tissue culture laminar cabinets should comply with BSL 2 certification. The laboratory should be equipped for handling and storing viral particles. Always wear double gloves with sleeves covering the forearm. Have 10% bleach ready in the event of any spill. Clean the bench and all equipment used for viral production with 10% bleach. Dispose used tips and pipettes after soaking them in a bottle containing 10% bleach. 293 T cells are weakly adherent to tissue culture plates. Make sure that the plates are well coated with poly-d-lysine before plating 293 T cells.
All surgical procedures and animal handling should be carried out in accordance with protocols approved by the Institutional Animal Care and Use Committee.
Ensure the vaporizer is filled with the specific anesthetic agent for which it is designed and certified for use by the manufacturer. Fill the vaporizer using an anti-spill bottle adaptor. Check for leaks, defects, and damage in anesthesia equipment (including hoses and valves) and scavenging system. Always use a sealed chamber. Any isoflurane waste should be disposed of as hazardous waste.
Choose the best excitation/emission combination for your study depending upon the expression of the fluorescent protein.
Incubation with d-Luciferin and BLI must be standardized for each separate experiment. Conditions may vary depending upon the tumor type and tumor model.
References
- 1.Gorman CM, Moffat LF, Howard BH (1982) Recombinant genomes which express chloramphenicol acetyltransferase in mammalian cells. Mol Cell Biol 2:1044–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddy SV, Takahashi S, Haipek C, Chirgwin JM, Roodman GD (1993) Tartrate-resistant acid phosphatase gene expression as a facile reporter gene for screening transfection efficiency in mammalian cell cultures. BioTechniques 15:444–447 [PubMed] [Google Scholar]
- 3.Henthorn P, Zervos P, Raducha M, Harris H, Kadesch T (1988) Expression of a human placental alkaline phosphatase gene in transfected cells: use as a reporter for studies of gene expression. Proc Natl Acad Sci U S A 85:6342–6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Wet JR, Wood KV, DeLuca M et al. (1987) Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol 7:725–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gould SJ, Subramani S (1988) Firefly luciferase as a tool in molecular and cell biology. Anal Biochem 175:5–13 [DOI] [PubMed] [Google Scholar]
- 6.de Wet JR, Wood KV, Helinski DR et al. (1986) Cloning firefly luciferase. Methods Enzymol 133:3–14 [DOI] [PubMed] [Google Scholar]
- 7.de Wet JR, Wood KV, Helinski DR et al. (1985) Cloning of firefly luciferase cDNA and the expression of active luciferase in Escherichia coli. Proc Natl Acad Sci U S A 82:7870–7873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prasher D, McCann RO, Cormier MJ (1985) Cloning and expression of the cDNA coding for aequorin, a bioluminescent calciumbinding protein. Biochem Biophys Res Commun 126:1259–1268 [DOI] [PubMed] [Google Scholar]
- 9.Shimomura O, Johnson FH, Saiga Y (1962) Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan, Aequorea. J Cell Comp Physiol 59:223–239 [DOI] [PubMed] [Google Scholar]
- 10.Li X, Zhao X, Fang Y et al. (1998) Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 273:34970–34975 [DOI] [PubMed] [Google Scholar]
- 11.Wang GL, Jiang BH, Rue EA et al. (1995) Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci 92:5510–5514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danhier P, Krishnamachary B, Bharti S et al. (2015) Combining optical reporter proteins with different half-lives to detect temporal evolution of hypoxia and reoxygenation in tumors. Neoplasia 17:871–881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shibata T, Giaccia AJ, Brown JM (2000) Development of a hypoxia-responsive vector for tumor-specific gene therapy. Gene Ther 7:493–498 [DOI] [PubMed] [Google Scholar]
- 14.Kaighn ME, Narayan KS, Ohnuki Y et al. (1979) Establishment and characterization of a human prostatic carcinoma cell line (PC-3). Investig Urol 17:16–23 [PubMed] [Google Scholar]
- 15.Harada H, Kizaka-Kondoh S, Itasaka S et al. (2007) The combination of hypoxia-response enhancers and an oxygen-dependent proteolytic motif enables real-time imaging of absolute HIF-1 activity in tumor xenografts. Biochem Biophys Res Commun 360:791–796 [DOI] [PubMed] [Google Scholar]