To the Editor
Convolutional neural networks (CNNs) are a class of deep-learning systems that are highly effective for classifying and analyzing image data (Krizhevsky et al., 2012). For skin cancer diagnosis, it has been claimed that CNNs can perform at a level of accuracy approaching that of a dermatologist (Brinker et al., 2019; Esteva et al., 2017). As a consequence, there is an impetus to apply these approaches in a clinical setting. However, the misclassification of skin cancer as a benign skin lesion can have serious clinical consequences, and it is therefore imperative that we have a full understanding of potential failure modes for CNN classifiers.
There is an increasing realization that in comparison with the human visual system, there are limitations of the CNN architecture in the ability to generalize from training images to novel data (Sabour et al., 2017). In keeping with this observation, CNNs can be misled into incorrect classifications by artificially perturbing natural-world images. For example, a subtle perturbation to an input image of a panda that is imperceptible to the human visual system is sufficient to cause misclassification as a gibbon (Goodfellow et al., 2015). This type of manipulation of an input image with the goal of deceiving the network into an incorrect classification is called an adversarial attack. Adversarial attacks are of interest for understanding the functional basis of neural networks and highlighting the important differences to the human visual system. In this letter, we report two, to our knowledge, previously unreported classes of relatively simple adversarial attacks that could arise inadvertently in a clinical setting: (i) alterations in color balance and (ii) alterations in rotation and/or translation of the input image that lead to misclassification of melanoma as a benign nevus. We additionally show that systematic alterations in image color balance and rotation affect the overall accuracy of a CNN classifier.
We first implemented a CNN classifier for melanoma versus benign melanocytic nevi. In line with previous work (Esteva et al., 2017), we fine-tuned a pretrained CNN (Inception, version 3) on a dataset of skin lesion images compiled by the International Skin Imaging Collaboration Challenge dataset 2018 (NCF Codella unpublished data, 2018; M Combalia, unpublished data, 2019; Tschandl et al., 2018) (Supplementary Materials and Methods). Sensitivity, specificity, and area under the curve were comparable or superior to other published models (Supplementary Table S1).
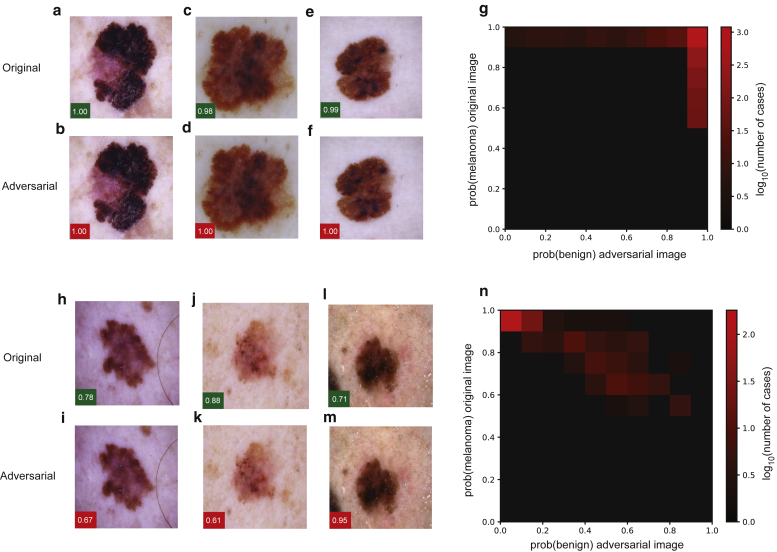
To facilitate a comparison with previous studies of adversarial attack on medical imaging systems (SG Finlayson, unpublished data, 2019; X Ma, unpublished data, 2020), we began by implementing the fast gradient sign method attack (Supplementary Materials and Methods), which makes subtle adjustments to the red, blue, and green values for each pixel in the input image according to the magnitude (gradient) of that pixel’s effect on the final classification with the goal of switching the output of the classifier to an incorrect classification (Goodfellow et al., 2015). As previously published, we found that adversarial attacks could be created with a high degree of success (Figure 1a–g). Next, we implemented a three-pixel attack (Figure 1h–m; Supplementary Materials and Methods), which modifies only three pixels within the input image leaving all others unchanged. We found that this method also led to successful adversarial attacks; however, in comparison with the fast gradient sign method, there was a lower success rate, and attacks were only successful when the initial confidence of the network in predicting melanoma was lower for a particular image (Figure 1n).
Figure 1.
FGSM and three-pixel attacks on deep-learning systems for skin cancer diagnosis. (a–g) Attacks were implemented against a pretrained Inception, version 3, network that was fine tuned for the differentiation of melanoma from benign melanocytic nevi. Adversarial attack with the FGSM. Examples of (a, c, e) original and (b, d, f) perturbed images are shown. Green boxes indicate the confidence (i.e., the output of the network in favor of this class after softmax transformation) of the network in predicting melanoma for the original images, and red boxes indicate the confidence in the prediction of a benign nevus for the adversarial images. (g) Image illustrates the dependency of successful adversarial attacks on initial classification by the network. For each image in the validation set, after the softmax transformation, the output of the final classification layer of the network is plotted for the original image (y-axis) versus the adversarial image (x-axis). (h–n) Adversarial attack through modification of three pixels within the input image. Examples of (h, j, l) original and (i, k, m) perturbed images are shown along with the dependency of the successful adversarial attacks on the original classification by the (n) network plotted as in g. FGSM, fast gradient sign method.
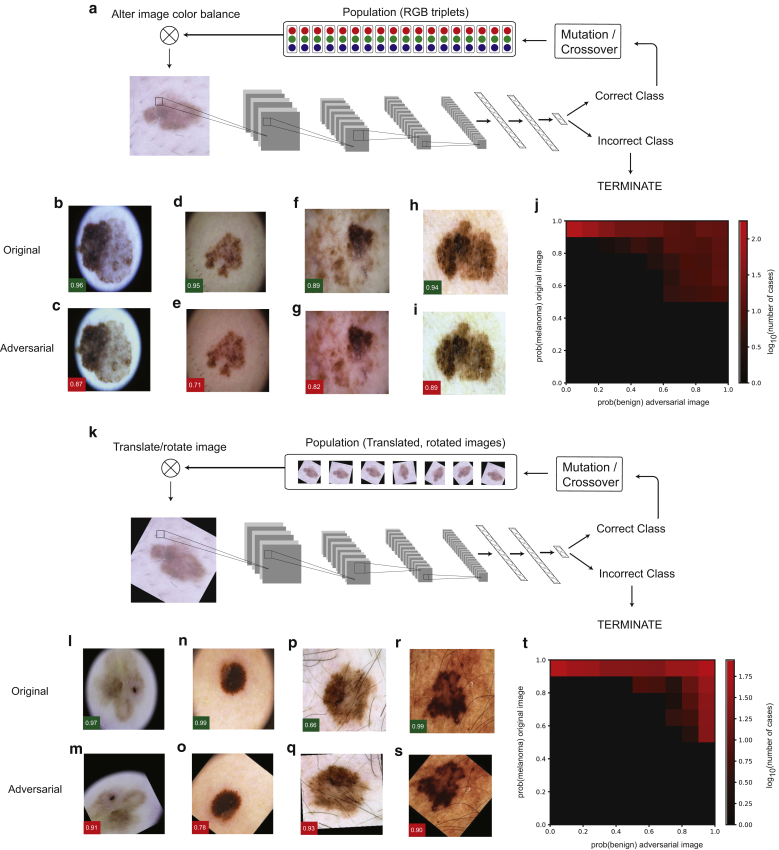
Although the methods described previously can deceive a CNN into misclassifying melanoma as a benign nevus, it is highly improbable that these complex image perturbations would arise by chance in a clinical context. We therefore wondered whether it would be possible to craft additional classes of adversarial attacks with greater clinical relevance. Deep-learning systems for skin cancer diagnosis are typically trained with images obtained using epiluminescence skin surface microscopy (dermoscopy). The color balance of these images is influenced by skin pigmentation, dermoscopic illumination, and image capture and processing. To explore whether alterations in the overall image color balance could influence the accuracy of skin cancer lesion classification by a CNN, we employed a differential evolution algorithm to search for subtle perturbations of global color balance that could lead to misclassification (Figure 2a). Differential evolution is a population-based optimization method that does not require information about the internal state of the neural network (Supplementary Materials and Methods). Remarkably, for numerous melanoma images, there existed subtle alterations in the red, blue, and green color balance that resulted in misclassification as a benign nevus (Figure 2b–i). These adversarial attacks were successful even when the initial confidence of the network in the diagnosis of melanoma was high (Figure 2j). To understand whether this could be mitigated by exposing the network to variation in color balance during training, we retrained the network with each training image subjected to random color variation in each training batch (Supplementary Materials and Methods). This resulted in a 33% decrease in the number of successful attacks from 15.35% (120 of 782 test images) to 10.23% (80 of 782 test images); however, it did not eliminate them. To assess further the effects of color-balance alterations, we systematically introduced minor changes to the color balance of all images in the test data set (Supplementary Figure S1). Even barely perceptible perturbations of image color balance had a large impact on the rate of missed melanoma diagnosis (Supplementary Figure S1); for example, subtracting 10 units from the green channel (maximum 255) led to a 235% increase in false negatives (from 68 to 160 cases).
Figure 2.
Clinically relevant adversarial attacks on deep-learning systems for skin cancer diagnosis. Attacks were implemented against a pretrained Inception, version 3, network that was fine tuned for the differentiation of melanoma from benign melanocytic nevi. (a–j) Differential evolution-based adversarial attack through the modification of global color balance. (a) A schematic illustration of the differential evolution algorithm in addition to examples of (b, d, f, h) original and (c, e, g, i) perturbed images are shown. Green boxes indicate the confidence (i.e., the output of the network in favor of this class after softmax transformation) of the network in predicting melanoma for the original images, and red boxes indicate the confidence in the prediction of a benign nevus for the adversarial images. (j) Image illustrates the dependency of the successful adversarial attacks on initial classification by the network. For each image in the validation set, after the softmax transformation, the output of the final classification layer of the network is plotted for the original image (y-axis) versus the adversarial image (x-axis). (k–t) Differential evolution-based adversarial attack through the modification of image translation and rotation. (k) A schematic illustration of the differential evolution algorithm in addition to examples of (l, n, p, r) original and (m, o, q, s) perturbed images are shown along with the dependency of the successful adversarial attacks on the original classification by the (t) network plotted as in j. RGB, red green blue.
Because there is no correct orientation for a dermoscopic image and there is some variability in lesion positioning within an image, we investigated whether perturbations involving rotation or translation would affect diagnostic accuracy. We employed a similar differential evolution-based optimization method, allowing arbitrary combinations of rotation (up to 360°) and translation (up to 50 pixels; input image size of 299 × 299 pixels) in both horizontal and vertical orientations (Figure 2k, Supplementary Materials and Methods). We found that for a large proportion (45.6%) of tested images, simple rotation and translation deceived the network into classifying melanoma as a benign nevus (Figure 2l–s). As for the color-balance attack, successful adversarial examples could be achieved even when the initial confidence of the network in favor of melanoma was high (Figure 2t). This reduction in accuracy occurred despite data augmentation with a random rotation of images during the training of the network. We also examined the consequences of systematic rotation of the test images by either 45° or 180°. We found that 45° rotation and 180° rotation both increased the incidence of missed melanoma diagnosis (false negative rate) by 11% (from 68 to 76; Supplementary Figure S1).
To ensure that the attacks described above represent a general vulnerability of CNNs rather than a specific weakness of the network that we tested, we retrained our model with a different split of train and/or test data and showed that this did not prevent adversarial attacks (Supplementary Table S2). Next, we obtained a pretrained model that has been extensively characterized in a previous publication (Han et al., 2018). Using our differential evolution-based attacks with test images of melanoma that were released along with this publication, we showed that this alternative CNN architecture was also vulnerable to the attacks that we have described (Supplementary Table S1; Supplementary Materials and Methods).
Finally, having established that CNN architectures are vulnerable to perturbations in image color balance and rotation, we wished to establish whether the human visual system was susceptible to similar perturbations. To test whether similar image perturbations affected the ability of dermatologists to correctly diagnose skin cancer, we tested four dermatologists (two consultants and two experienced residents) with a set of 204 images, half of which were unmodified and half were adversarial images, which had defeated the CNN. In contrast to the CNN, we observed no statistically significant differences in accuracy for human dermatologists (Supplementary Figure S2) when tested with adversarial versus unaltered images.
In summary, we have shown that CNNs for the diagnosis of melanoma are susceptible to two, to our knowledge, previously unreported forms of relatively simple and clinically relevant adversarial attack: (i) subtle alterations in color balance and (ii) rotation and/or translation of the input image. Alterations such as these mimic variations and inconsistencies in images and image capture methods in clinical settings, which are currently not standardized. It is possible that variance in these factors may explain degradation in classification accuracy when CNNs are tested on imaging data that were not represented in the training data (Narla et al., 2018). Interestingly, a recent prospective real-world study of CNN accuracy in skin cancer diagnosis found substantial differences in classification according to whether the image was taken with an iPhone, Samsung phone, or digital single-lens reflex, which may, at least in part, be explained by variations in color balance (Phillips et al., 2019). Another recent publication reported that the presence of blue marker ink in dermoscopic images also has a negative impact on CNN classification accuracy (Winkler et al., 2019). These studies demonstrate the importance of investigating failure modalities and testing CNNs rigorously to ensure that they are robust before unsupervised clinical deployment. Limitations of our study include the evaluation of a limited number of CNN network architectures and models, and it will be interesting to evaluate a wider range in the future.
Almost all recent successful applications of CNNs to medical imaging data have fine-tuned pretrained, off-the-shelf CNNs (Esteva et al., 2017; Gulshan et al., 2016). There is an increasing awareness that there are limitations of CNN architectures—which were originally developed for the classification of natural images—in the ability to generalize to novel data, and this has stimulated the development of novel network architectures (Sabour et al., 2017). It will be essential to ensure that CNN skin cancer classifiers are robust to alterations in image color balance and translation and/or rotation. In addition, it will be important to explore strategies (in addition to random variation in color balance jitter and rotation during the training process as described in this letter) to mitigate these classes of adversarial attacks; this could include the retraining of models with generated adversarial images. Finally, it is essential to verify that other applications of artificial intelligence in medical imaging are robust to similar perturbations both through adversarial challenges as described earlier and standardization of image acquisition in the clinical setting.
Data availability statement
Deep-learning models and adversarial attacks were implemented in PyTorch and were trained on an NVidia 2080ti GPU. Training images were obtained from publicly available International Skin Imaging Collaboration Challenge dataset. Code for the training of models, generation of adversarial challenges, and testing of dermatologists is available at https://github.com/thelynchlab/adversarial.
ORCIDs
Xinyi Du-Harpur: http://orcid.org/0000-0002-5738-8734
Callum Arthurs: http://orcid.org/0000-0001-8508-3754
Clarisse Ganier: http://orcid.org/000-0003-1143-7905
Rick Woolf: http://orcid.org/0000-0003-3200-0075
Zainab Laftah: http://orcid.org/0000-0001-6219-9041
Manpreet Lakhan: http://orcid.org/0000-0002-8121-489X
Amr Salam: http://orcid.org/0000-0002-0907-2372
Bo Wan: http://orcid.org/0000-0002-8109-1850
Fiona M. Watt: http://orcid.org/0000-0001-9151-5154
Nicholas M. Luscombe: http://orcid.org/0000-0001-5293-4778
Magnus D. Lynch: http://orcid.org/0000-0001-7586-4338
Conflict of Interest
XDH is a clinical advisor to Skin Analytics. FMW is currently on secondment as executive chair of the Medical Research Council. The remaining authors state no conflict of interest.
Acknowledgments
XDH is the recipient of an Accelerator Award from Cancer Research United Kingdom. FMW gratefully acknowledges financial support from the United Kingdom Medical Research Council (MR/PO18823/1), Biotechnology and Biological Sciences Research Council (BB/M007219/1), and the Wellcome Trust (206439/Z/17/Z). This work was supported by the Francis Crick Institute, which receives its core funding from Cancer Research United Kingdom (FC010110), the United Kingdom Medical Research Council Journal of Investigative Dermatology (FC010110), and the Wellcome Trust (FC010110). NML is a Winton Group leader in recognition of the Winton Charitable Foundation’s support toward the establishment of the Francis Crick Institute. NML is also funded by a Wellcome Trust Joint Investigator Award (103760/Z/14/Z), the Medical Research Council eMedLab Medical Bioinformatics Infrastructure Award (MR/L016311/1), and core funding from the Okinawa Institute of Science and Technology Graduate University. MDL gratefully acknowledges financial support from the Wellcome Trust (211276/E/18/Z).
Author Contributions
Conceptualization: XDH, CA, CG, FMW, NML, MDL; Formal Analysis: CG, RW, ZL, ML, AS, BW; Software: XDH, CA, MDL; Supervision: FMW, NML, MDL; Validation: XDH, CA, RW, ZL, ML, AS, BW, FMW, NML, MDL; Visualization: XDH, CA, FMW, NML, MDL; Writing - Original Draft Preparation: XDH, CA, CG, FMW, NML, MDL
Accepted manuscript published online 12 September 2020; corrected proof published online 26 November 2020
Footnotes
Supplementary material is linked to the online version of the paper at www.jidonline.org, and at https://doi.org/10.1016/j.jid.2020.07.034.
Supplementary Material
References
- Brinker T.J., Hekler A., Enk A.H., Klode J., Hauschild A., Berking C. A convolutional neural network trained with dermoscopic images performed on par with 145 dermatologists in a clinical melanoma image classification task. Eur J Cancer. 2019;111:148–154. doi: 10.1016/j.ejca.2019.02.005. [DOI] [PubMed] [Google Scholar]
- Esteva A., Kuprel B., Novoa R.A., Ko J., Swetter S.M., Blau H.M. Dermatologist-level classification of skin cancer with deep neural networks. Nature. 2017;542:115–118. doi: 10.1038/nature21056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodfellow IJ, Shlens J, Szegedy C. Explaining and harnessing adversarial examples. Paper presented at: ICLR 2015. 7–9 May 2015; San Diego, CA.
- Gulshan V., Peng L., Coram M., Stumpe M.C., Wu D., Narayanaswamy A. Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA. 2016;316:2402–2410. doi: 10.1001/jama.2016.17216. [DOI] [PubMed] [Google Scholar]
- Han S.S., Kim M.S., Lim W., Park G.H., Park I., Chang S.E. Classification of the clinical images for benign and malignant cutaneous tumors using a deep learning algorithm. J Invest Dermatol. 2018;138:1529–1538. doi: 10.1016/j.jid.2018.01.028. [DOI] [PubMed] [Google Scholar]
- Krizhevsky A., Sutskever I., Hinton G.E. ImageNet classification with deep convolutional neural networks. In: Pereira F., Burges C.J.C., Bottou L., Weinberger K.Q., editors. Advances in Neural Information Processing Systems 25. Curran Associates, Inc.; New York: 2012. pp. 1097–1105. [Google Scholar]
- Narla A., Kuprel B., Sarin K., Novoa R., Ko J. Automated classification of skin lesions: from pixels to practice. J Invest Dermatol. 2018;138:2108–2110. doi: 10.1016/j.jid.2018.06.175. [DOI] [PubMed] [Google Scholar]
- Phillips M., Marsden H., Jaffe W., Matin R.N., Wali G.N., Greenhalgh J. Assessment of accuracy of an artificial intelligence algorithm to detect melanoma in images of skin lesions. JAMA Netw Open. 2019;2 doi: 10.1001/jamanetworkopen.2019.13436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabour S., Frosst N., Hinton G. 31st Conference on Neural Information Processing Systems (NIPS 2017) CA, USA; Long Beach: 2017. Dynamic routing between capsules. [Google Scholar]
- Tschandl P., Rosendahl C., Kittler H. The HAM10000 dataset, a large collection of multi-source dermatoscopic images of common pigmented skin lesions. Sci Data. 2018;5:180161. doi: 10.1038/sdata.2018.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler J.K., Fink C., Toberer F., Enk A., Deinlein T., Hofmann-Wellenhof R. Association between surgical skin markings in dermoscopic images and diagnostic performance of a Deep Learning Convolutional Neural Network for Melanoma Recognition. JAMA Dermatol. 2019;155:1135–1141. doi: 10.1001/jamadermatol.2019.1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Deep-learning models and adversarial attacks were implemented in PyTorch and were trained on an NVidia 2080ti GPU. Training images were obtained from publicly available International Skin Imaging Collaboration Challenge dataset. Code for the training of models, generation of adversarial challenges, and testing of dermatologists is available at https://github.com/thelynchlab/adversarial.