Abstract
Immunoglobulin A (IgA) is the most abundant antibody at mucosal surfaces and has been the subject of many investigations involving microbiota research in the last decade. While the classic functions of IgA include neutralization of harmful toxins, more recent investigations have highlighted an important role for IgA in regulating the composition and function of the commensal microbiota. Multiple reviews have comprehensively covered the literature that describes recent, novel mechanisms of action of IgA and development of the IgA response within the intestine. Here we focus on how the interaction between IgA and the microbiota promotes homeostasis with the host to prevent disease.
Introduction
While Immunoglobulin M (IgM) and IgG are also found at mucosal sites, the human body produces several grams of IgA daily, making IgA the most abundant antibody in mucosal secretions (Pabst, 2012). IgA is predominantly produced in a dimeric form at mucosal sites, where the two monomers are held together by the joining protein (J chain). Upon binding to the polymeric Ig receptor (pIgR) in intestinal epithelia, IgA is transcytosed through the cell and secreted into the lumen of the gut where a piece of pIgR, called secretory component (SC) remains covalently linked to the dimer to produce secretory IgA (SIgA) (Pabst and Slack, 2020). While IgA has the typical Fab portion for antigen binding, the J chain and SC are highly glycosylated and also serve to bind microbes.
Mucosal sites can be divided into type 1 and type 2 mucosa, which differ significantly based on their cell types and immunoglobulin profile (Iwasaki, 2016). Type 1 mucosa, which includes gastrointestinal, respiratory, and the upper female reproductive tract, contains columnar cells that express pIgR and the IgG receptor, FcRn. Thus, type I mucosal sites possess the ability to secrete both IgA and IgG; however, in these tissues, IgA secretion dominates and high luminal levels of IgG are mainly observed during inflammation. Type 2 mucosal sites, such as oral, esophageal and the lower female reproductive tract, however, contain mostly stratified squamous epithelia which do not express pIgR, but do retain FcRn, and therefore lack the ability to transport IgA. This review will focus primarily on the antibody-microbiota interactions within the type I mucosal tissue of the intestine.
Characterizing IgA interactions with the microbiota
Using diverse scientific approaches, researchers have been able to identify what organisms and epitopes are bound by IgA and how IgA reactivity might influence interactions between the host and the microbiota. These techniques have been reviewed elsewhere (Round and Palm, 2018). Based on this literature, SIgA responses can be categorized by their reactivity or based on their functional impact on the microorganisms they physically bind to. These aspects of SIgA biology are important to consider when thinking about ways to harness IgA for therapeutic benefit. In the following section we review the literature on IgA reactivity and function, and begin to categorize IgA responses against the microbiota based on these properties.
Reactivity of antibody responses against the microbiota
From the most current literature, homeostatic IgA responses can be placed into three different categories based on their reactivity (Figure 1). The first is cross-species reactive, defined by Pabst and Slack as SIgA complexes that are able to bind to multiple distinct taxonomic groups of the microbiota (Pabst and Slack, 2020). The second can be considered species-specific, meaning SIgA molecules that bind only to members of a particular species of microbe. Here we propose the use of the term strain-specific IgA to describe the third category, which will refer to SIgA that discriminate between distinct strains of the same species. Based on the current available techniques, it difficult to quantify the proportion of these three different types of antibody categories. For example, strains of microbes will differ across individuals and the amount of antigen placed in any given assay can alter the experimental outcome, thus different categories of antibodies may be missed or mis-categorized. However, the literature clearly supports the presence of these three categories which are described in detail below.
Figure 1: Depiction of cross-species reactive IgA, species-specific IgA and strain-specific IgA responses against bacteria in the intestinal mucosa.
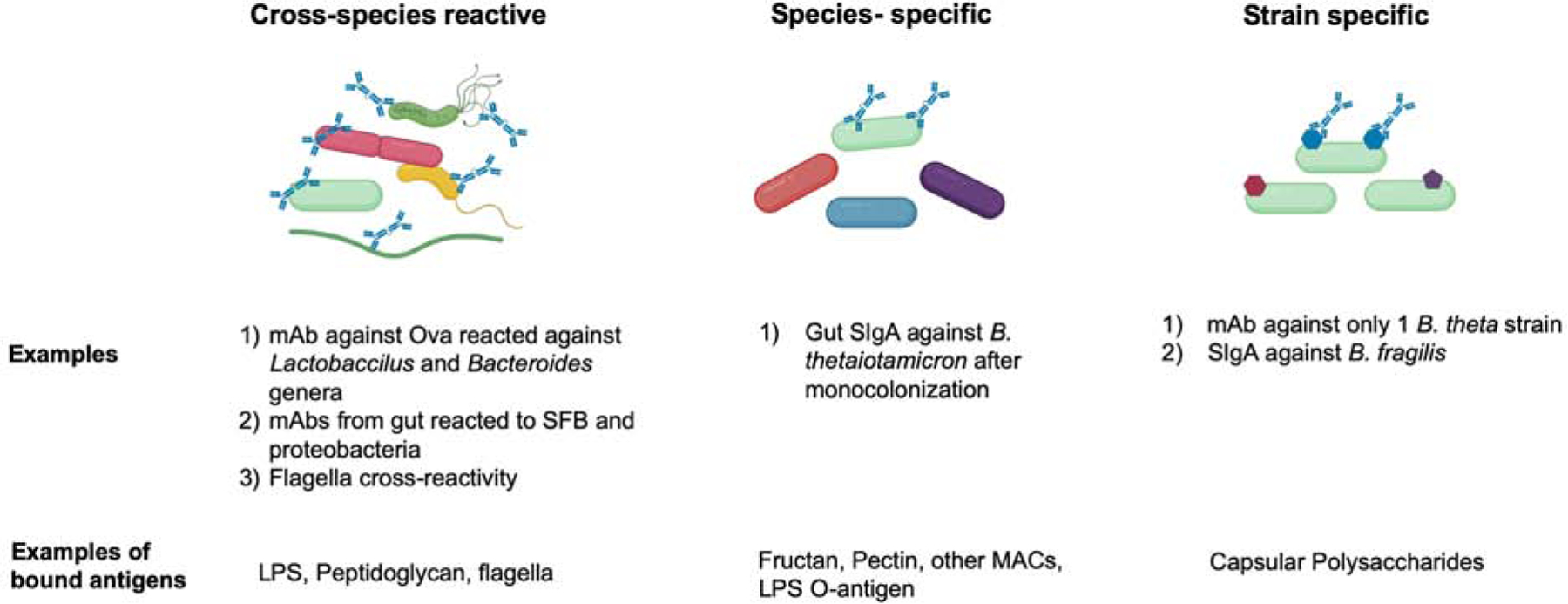
The left column shows known examples of cross-species reactive SIgA (in blue) binding many different species of commensal bacteria within the gut to various known and unknown antigens. This can be Fab -dependent or independent. Center column depicts species-specific SIgA binding to just one species within a genera, for example B. theta among other Bacteroides. Right column shown a strain specific SIgA binding to one strain of bacteria among other strains of the same species, for example a B. theta strain among other B. theta strains with specificity for one surface molecule such as a capsular polysaccharide.
Cross-species reactive IgA responses
Current data indicate that a proportion of the IgA responses against the microbiota can be cross-species reactive, a term proposed in a recent review whereby IgA can bind to members across a range of different taxa (Pabst and Slack, 2020). To clarify, this is distinct from the term poly-reactive, which defines individual SIgA molecules that can bind to a variety of distinct antigens such as LPS and DNA. In many cases, cross-species reactive antibodies are also polyreactive. Many of the targets of cross-species reactive IgA are highly glycosylated moieties on the cell surface that can be either Fab fragment dependent or independent. IgA itself is heavily glycosylated with N and O-glycans present on both the Fc and Fab fragment as well as on the secretory component (SC) and J chain. In a recent study, a heavily glycosylated monoclonal antibody against ovalbumin (OVA), which is not a bacterial antigen, called 7–6IgA, readily reacted with multiple members of the Bacteroides and Lactobacillus genera (Nakajima et al., 2018). The binding of 7–6IgA was Fab - independent as pre-absorption of OVA did not disrupt bacterial binding. Additionally, a different clone of antibody that maintained OVA specificity, but was less glycosylated, did not react with Bacteroides species. This study further demonstrated that 7–6IgA interacted with the LPS component of these gram-negative bacteria. Similar studies carried out on gram-positive bacteria revealed that glycosylation of the secretory component (SC) and polymeric IgA were responsible for binding to the peptidoglycan within gram positive bacteria such as Lactobacillus (Mathias and Corthesy, 2011). Thus, in these examples, cross-species reactive antibody interactions can occur independently of the Fab fragment and instead bind to multiple glycans present within the Fc portion of IgA or the highly glycosylated SC component. The functional consequences of cross-species reactive antibodies and their potential contribution to homeostasis will be discussed in later sections.
Cross-species reactive antibodies can also be Fab dependent. Bendelac and coworkers harvested and cloned monoclonal antibodies (mAbs) from single IgA+ plasma B cells or naïve B cells from the lamina propria of mice (Bunker et al., 2017). These mAbs were cloned and expressed as monomeric human IgG1-Igκ chimeras to allow for a focused analysis of Fab specificities. Interestingly, ~30% of the mAbs isolated from naïve B cells, splenic B cells, and human anti-influenza plasmablasts showed high reactivity against the microbiota. Screening 157 mAbs against a panel of seven structurally different antigens that included DNA, insulin, LPS, flagellin, albumin, cardiolipin, and keyhole limpet hemocynanin (KLH) revealed that while only 30% of the mAbs from naïve B cells were poly-reactive, 60% of the mAbs isolated from IgA+ gut plasmablasts were polyreactive. Many of those monomeric mAbs reacted with multiple members of the microbiota such as segmented filamentous bacteria (SFB) and Proteobacteria indicating that cross-species antibody production can be Fab dependent as well. Thus, to date, the epitopes that mediate cross-species IgA interactions are basic cell wall components, such as LPS and peptidoglycan (PGN), that are possessed by many different types of bacteria.
Cross-species antibodies have also been documented between viruses and bacteria. In a recent example, IgA was isolated from germfree mice mono-associated with norovirus and was used to probe different Lactobacillus isolates. Indeed, IgA from norovirus infected germfree animals reacted with several Lactobacillus isolates, albeit at a fairly low level (Huus et al., 2020). These data support that a high amount of cross-species reactive antibodies are present within the normal intestinal repertoire and the functional consequences of these antibodies are discussed in later sections.
Species-specific IgA responses
While the characterization of specific epitopes from the microbiota that are bound by IgA is still in its infancy, several studies have identified that gut IgA can be species-specific. For clarification, the term species has been variously defined, however we utilize the definition as proposed in Yan et. al 2020 to be where genotype is considered a trait (phenotype) by which different isolates can be classified into self-similar groups (Yan et al., 2020). Thus, this definition takes into account the genomic information that is now available using high-throughput sequencing techniques as well as more classic parameters such as various phenotypic traits. The term strain here will refer to a single isolate that has a specific microbial genome, however, the definition of strain level differences is an evolving field with important recent contributions.
Demonstrating that IgA can be species-specific, Peterson and colleagues showed that IgA antibodies can react with a number of strains from the same species but do not react with a variety of non-related species (Joglekar et al., 2019). This group first established an ex vivo system to harvest gut IgA. Following mono-association of germfree mice with Bacteroides thetaiotaomicron (B. theta) VPI-5482, the small intestinal (SI) lamina propria was harvested and cultured to isolate culture supernatants as a source of B. theta IgA. Initial characterization of isolated IgA against antigens from three distinct strains of B. theta, five other Bacteroides species including B. ovatus, B. finegoldii, B. fragilis, B. vulgatus and B. caccae and nine other un-related bacteria such as Lactobacillus, Bifidobacteria and Akkermansia species, revealed that while the IgA reacted strongly with all three strains of B. theta tested, it did not cross-react with other Bacteroides or other taxa. Much of the IgA response generated in the intestine targeted one or more of the eight high molecular weight B. theta capsular polysaccharides, as has been reported prior to this study (Peterson et al., 2007). However, in this study, protease digestion revealed that the relevant epitope for this species-specific reaction was a protein.
The same group used a different approach to study antibody specificity. B cells were isolated from the lamina propria of germfree mice mono-associated with B. theta and fused with a myeloma cell line to make hybridomas (Peterson et al., 2015). One of these hybridomas, termed 260.8, produced a mAb that was tested against 16 distinct strains of hospital and community acquired B. theta and 13 different Bacteroides species by ELISA. The results revealed that mAb 260.8 was highly specific for B. theta as it did not recognize other Bacteroides species (B. fragilis, B. ovatus, etc.), however it did react with multiple isolates and strains of B. theta. Whole genome mutagenesis of B. theta identified a 19 gene locus involved in LPS O-antigen polysaccharide synthesis that is required for the reactivity against mAb 260.8. Thus, proteins and carbohydrates that are necessary for colonization and likely conserved across various strains of the same species can be targets of host IgA.
Strain-specific IgA responses
Anyone who has worked on some of the more commonly studied microbes, such as E. coli, appreciates that there are many microbes that are taxonomically classified as the same species based on genomic data, but have completely different biological outcomes depending on what strain is under study. Indeed, several E. coli strains are clearly pathogenic and produce various toxins that are associated with colorectal cancer, urinary tract infections or sepsis. However, other E. coli strains (such as Nissle) can have beneficial effects on their hosts. Thus, it is very important in microbiology to distinguish what strain is being used in any particular investigation. The mammalian immune system also seems to have the capacity to distinguish between distinct strains of the same species. One of the first studies to characterize an IgA epitope from the commensal organism B. theta created hybridomas from B cells isolated from the gut of germfree mice mono-associated with B. theta strain VPI-5482 (Peterson et al., 2007). Further characterization of one of the mAbs, called 225.4, produced from one of these hybridomas, identified an antibody against B. theta that was highly specific for this strain. Indeed, probing 225.4 mAb against antigens isolated from 25 distinct strains of B. theta and four other Bacteriodes species demonstrated that not only was this antibody specific for B. theta, as it did not react with any other Bacteriodes tested, but this antibody also did not react with 25 other B. theta strains. To identify the epitope recognized by this mAb, a library of 4600 transposon mutants was screened for loss of 225.4 antibody reactivity. B. theta possesses eight capsular polysaccharides, and 225.4 reacted specifically against just one of these surface moieties.
Another study from Palm et. al characterized IgA coated microbes in individuals with inflammatory bowel disease (IBD) using IgA-Seq (cell sorting to isolate IgA-bound bacteria vs unbound bacteria followed by 16S rDNA sequencing) (Palm et al., 2014). This study identified members of the same species that were differentially coated in IgA. In particular, they found that some strains of B. fragilis were targeted by IgA while others were not. Carefully curated culture collections from various human samples isolated IgA bound versus unbound strains of B. fragilis and tested them in an animal model of IBD. IgA bound strains of B. fragilis induced worsened disease when compared to unbound strains of B. fragilis, demonstrating that IgA was targeting strains of B. fragilis that could be harmful. These studies demonstrate the remarkable specificity of the mammalian immune response that is capable of distinguishing between distinct strains of highly related species that likely possess similar surface architecture. Further, strain-specific IgA responses could be used to identify specific bacterial effectors, pathogenic or commensal, that drive functional differences between strains. Thus, immune-microbiota interactions might be useful diagnostic tools for a variety of diseases.
Functional effects of IgA on the microbiota
Current basic immunology textbooks highlight two major functions of IgA in the gut based on extensive studies performed on pathogenic species. Firstly, IgA can bind to microbes or their secreted toxins, either within the lumen, epithelial cells, or in the lamina propria. Binding of IgA can block the microbe from invading or prevent toxins from binding receptors on host cells, so that the complex can ultimately be “flushed” out of the body. This function will not be discussed here as it has been covered by many other reviews. The second function is uptake of IgA bound complexes by dendritic cells for presentation to T cells to elicit antigen specific responses, which also ultimately works to clear pathogens. When IgA targeted organisms are placed back into a host with disease, several studies have shown that IgA bound bacteria exacerbate disease (Kau et al., 2015; Palm et al., 2014; Viladomiu et al., 2017). This has led to the notion that IgA primarily targets organisms that warrant exclusion from the intestine. However, recent literature has highlighted other mechanisms by which IgA can exert effects on microbial organisms. This newer data reveals that the function of IgA is not solely eradication of microorganisms, but rather a communication between host and microbe to ensure homeostasis. Collectively, these data allow us to divide the functional effects of IgA into elimination, neutralization, colonization, and sculpting functions (Figure 2).
Figure 2: Mechanisms by which SIgA affects bacteria.

Mechanisms by which SIgA influence gut microbes. The current mechanisms by which SIgA can affect microbes can either be divided into ways in which SIgA causes Elimination or Neutralization of bacteria or ways in which SIgA can alter gene expression patterns or functions of microbes that might actually aid in their colonization. (A) SIgA eliminates toxins and/or neutralizes microbial molecules by direct binding. This binding prevents binding of the microbial molecule/toxin to the host target receptor so that it will eventually be cleared by peristalsis. (B) by directly binding to the pathogen and by limiting motility and likely invasion. For instance, SIgA can bind to flagella and prevent organisms from accessing the epithelia. These organisms are likely to be eliminated by peristalsis. (C) Aggregation of rapidly dividing bacteria by enchained growth prevents over-population by proliferation while also limiting access to the epithelia. These organisms are likely eliminated by peristalsis. (D) Biofilms are often mechanisms by which microbes can adhere to surfaces and allow colonization. SIgA can prevent biofilm formation which would be predicted to prevent some organisms from colonizing surfaces. It is also possible that that this mechanism could prevent the expression of genes required for biofilm formation. Therefore, this could be a mechanism for both Elimination and Sculpting. (E) SIgA can preferentially bind to surface microbial molecules that anchor the microbe to the epithelial surface. In this way, SIgA might positively select for organisms that express these molecules. (F) Microbes are known for the ability to sense environmental cues and respond by changing their gene expression patterns. SIgA binding of specific molecules on microbes might be sensed by the microbe such that binding results in down-regulation of that surface molecule. In this way, SIgA could fine-tune microbial gene expression to prevent the production of proteins that may be harmful to the host.
IgA can aggregate bacteria for removal
IgA can facilitate aggregation of bacteria by crosslinking and agglutinating bacteria together. This mechanism leads to aggregation of the bacteria into deactivated clumps in the mucosa, precluding them from antigen presentation and further immune recognition and priming, in what is termed immune exclusion (Mantis et al., 2011). Classically, aggregated bacteria within the mucus are thought to become entrapped as an agglutinated complex and these pathogen-IgA complexes are cleared from the intestinal mucosa via peristalsis. This has been considered a mechanism by which IgA helps to eliminate bacteria. However, bacterial aggregation has more recently been shown to enhance intestinal colonization of some commensals and therefore might be a mechanism for maintenance within the gut and is discussed later (Castagliuolo et al., 2005; Tareb et al., 2013; Voltan et al., 2007).
Aggregation by IgA followed by expulsion via peristalsis is a well-studied phenomenon. Dating back to 1972, Williams and Gibbons showed that SIgA could inhibit bacterial adherence by binding and agglutinating pathogenic bacteria, and that this interaction could be blocked in vitro with anti-IgA antisera (Williams and Gibbons, 1972). While that interpretation is limited due to its lack of in vivo data, a recent example of this demonstrated that IgA aggregates bacteria for removal by binding to bacteria as they are currently dividing, called enchained growth. In the mouse model of Salmonella enterica Typhimurium (STM) intestinal infection, Moor et al investigated the way in which IgA against STM LPS O-antigen interacts with STM at varying concentrations, with the logic that likely agglutination only occurs at high bacterial densities (Moor et al., 2017). STM vaccinated mice were able to block STM colonization and infection and the bacteria were found clumped together in the center of the lumen. The same result occurred using strains of STM without flagella or type III secretion systems, indicating that the clumping and clearance occurred independently of epithelial cell virulence factor recognition, indicative of aggregation or immune exclusion. However, using live microscopy and two different fluorescently labeled strains of STM, the authors were able to discern that clumping was typically confined to one color or another, indicating clonal populations suggestive of enchained growth. A follow up study demonstrated that bacteria with high replication rates could divide before the SIgA linkage is broken and face greater chances of entrapment, whereas bacteria that replicate at a slower rate have a greater chance for the IgA crosslinking to fail prior to formation of a new daughter cell. Thus, pathogens like STM in log phase would be the most susceptible to enchained growth (Bansept et al., 2019). This suggests that slower growing commensal bacteria may evade enchained growth based on these dynamics and that may play an important role in maintaining long lasting host colonization and is discussed later.
SIgA has been shown to bind to other surface proteins such as bacterial pili and fimbriae (Wold et al., 1990). Indeed, one study found that SIgA could bind specifically to the commensal E. coli type 1 pili and drive aggregation of the bacteria, independent of other important adhesion factors in what is likely species or strain-specific IgA binding (Orndorff et al., 2004). As pili are important for E. coli adherence and invasion of intestinal epithelial cells, this immunoglobulin mediated aggregation could serve both to prevent invasion and as a mode of immune exclusion. Other work points to SIgA being important for breaking up biofilms. Indeed, SIgA has been shown to diminish biofilm formation of the pathogen Vibrio cholera in WT vs IgA−/− mouse and human SIgA was shown to block V. cholera binding to cultured epithelial cells. Together these data suggest that IgA can mediate bacterial aggregation and removal and that it seems to depend largely upon specific recognition of bacterial molecules.
SIgA aid colonization of some commensals
In many cases, the induction of an IgA response does not lead to complete elimination of a microorganism. Indeed, in the homeostatic gut, IgA responses are detected against multiple commensal species that are still quite abundant. This suggests that IgA targeting can function beyond brute force elimination of offending organisms and instead might act through more elegant mechanisms that have been speculated to enhance colonization of some commensal species (Kubinak and Round, 2016). A few recent studies have now experimentally demonstrated this phenomenon.
Recent work from the Mazmanian lab showed that a human commensal, Bacteroides fragilis, colonized differentially based upon its capsular polysaccharides, regulated by the commensal colonization factor (CCF) system (Donaldson et al., 2018). When the authors analyzed how IgA interacted with WT B. fragilis vs capsule mutants, they found that IgA did not bind as well to the mutants without CCF driven capsule polysaccharides. Further, in an IgA−/− mouse, mucosal colonization of wild type B. fragilis was reduced, indicating that IgA may be used by B. fragilis to help anchor itself into the gut mucosa and promote colonization. In this work other commensal species such as Rikenellaceae, Blautia sp., and SFB were also highly IgA-coated. They further found that Blautia sp and SFB were increased in the mucosa, or even more closely associated with tissue in the absence of IgA, whereas Rikenellaceae was reduced in the absence of functional IgA much like B. fragilis, indicating a more widespread phenomena across multiple commensal organisms.
Supporting this, several studies have demonstrated that aggregation of specific commensals aids in host colonization. Studies using the commensal, Lactobacillus crispatus, demonstrated that the ability to aggregate enhanced colonization and tempered immune responses within the intestine. An isogenic mutant of L. crispatus that lacked the ability to aggregate, was completely out-competed by the strain that could auto-aggregate (Voltan et al., 2007). Additionally, while the aggregating L. crispatus strain was capable of protecting from an animal model of colitis, the non-aggregating L. crispatus strain failed to protect (Castagliuolo et al., 2005), suggesting that the ability to aggregate can not only aid in commensal colonization, but also confers benefits to the host. While the effect of IgA on aggregation of L. crispatus was not examined within these studies, a recent study from Brett Finlay’s group demonstrated that IgA interactions with commensal strains of Lactobacillus induces aggregation of these organisms (Huus et al., 2020), suggesting that SIgA mediated aggregation of specific microbes might allow them to outcompete non-aggregating strains. While the level of specificity of these antibodies was not tested in these studies, one might speculate that this mechanism could utilize strain- or species-specific antibody responses, as this would enhance competition between organisms that occupy similar niches.
While IgA is thought to disrupt biofilms, it has also been shown to enhance them. Indeed, in a series of in vitro experiments, work from Bollinger and colleagues first showed that the presence of SIgA facilitated commensal E. coli biofilm formation on cultured human epithelial cell surfaces (Bollinger et al., 2003). In follow up studies, the same team showed that, in a range of environmental E. coli isolates, expression of the type 1 pili was necessary for IgA-mediated biofilm formation on polystyrene and live epithelial cells (Bollinger et al., 2006). This work still needs to be validated in vivo, but suggests that classic functions of IgA that were originally thought to eliminate bacteria might also function to enhance them.
Retrotranscytosis of IgA within the Peyers Patch
Another well documented function of SIgA is retrotranscytosis through microfold (M) cells within Peyer’s patches (PPs) allowing interactions with underlying dendritic cells (DCs) in both free antibody form or in complexes with bound antigen. This mechanism is important for responses against enteric pathogens as well as commensals. Using the inflammatory human pathogen Shigella flexneri, it was identified that that a subset of subepithelial dome DCs can be differentially conditioned by sIgA. Cross-species antibodies were identified to be bound by Dectin-1 and SIGNR3 receptors on DCs, that are known to recognize carbohydrate motifs (Mikulic et al., 2017a). This recognition induced internalization of the sIgA-S. flexneri complex into DCs. While DCs exposed to S. flexneri alone, expressed high levels of the inflammatory cytokines IL-12, TNF-α and KC, DCs incubated with sIgA-S. flexneri complexes had reduced inflammatory cytokine production and instead produced high levels of the anti-inflammatory mediator, TGF-β. Thus, SIgA binding to pathogens can dampen harmful inflammatory processes while still neutralizing the offending microbe.
This process also functions against common commensal organisms. In a similar study, using Lactobacillus rhamnosus, PP or MLN derived DCs were incubated with either L. rhamnosus alone or in complex with SIgA (Mikulic et al., 2017b). Incubation with L. rhamnosus alone induced the expression of several negative regulators of TLR signaling including Tollip, Sigirr and Socs1, suggesting that this commensal can prevent inflammatory TLR signaling. Interestingly, incubation of DCs with sIgA-L. rhamnosus complexes significantly enhanced this effect, indicating that the presence of SIgA can further upregulate anti-inflammatory signaling. Incubation of SIgA-L. rhamnosus conditioned DCs with T cells also induced the conversion of naïve T cells to Foxp3+ T regulatory cells. Thus, uptake of SIgA complexes can enhance the anti-inflammatory effect of some commensal species. It is important to note that these studies were conducted with isolated intestinal cells in vitro and therefore the in vivo relevance of these findings is still unknown. Recent studies have uncovered the importance of these same DCs in directly interacting with B cells in the PP to promote IgA production against commensals (Reboldi et al., 2016) . As TGF-β production is imperative for IgA class switching, these studies also suggest that uptake of SIgA complexes might function to further induce IgA responses in the gut.
Regulation of bacterial expression
Much of this review has focused on SIgA binding to bacteria, those epitopes, and its immunological implications. An exciting outstanding question in the field centers around how SIgA binding actually affects the microbe that it’s bound to by altering gene expression. The examples we highlight here demonstrate that IgA can cause downregulation of the expression of surface epitopes or mobile elements (such as flagella) in microbial species that allow them to be retained within the gut. This phenomenon is often referred to as sculpting or pruning, as this process does not lead to eradication of the microbial population but rather a trimming of molecules that may present harm to the host. While this possibility requires further experimental investigation, there is enough evidence to highlight it here.
The seminal suggestion of this possibility was work from Petersen et al in 2007, mentioned previously (Peterson et al., 2007). The authors utilized GeneChip to profile B. theta that was colonizing Rag−/− mice that were injected with a hybridoma expressing the monoclonal IgA 225.4 against the capsular polysaccharide 4 (CPS4). IgA binding was associated with down-regulation of CPS4, which in turn induced a more robust inflammatory response from the host. Antibody binding does not always down-regulate a surface epitope. In similar studies, the same group characterized a species-specific antibody against B. theta, termed 260.8 (Peterson et al., 2015) . This particular antibody did not impact expression of the epitope nor did it change the population density of B. theta in the gut, however, it did impact B. theta gene expression that reduced its capacity to elicit inflammation within the gut.
Other work from this same group demonstrated that IgA can influence B. theta’s metabolic capacity. Bacterial antigens bound by SIgA were identified using an ex vivo small intestine culture assay from mice monocolonized with B. theta. The authors found that SIgA targeted B. theta capsular polysaccharides, lipopolysaccharides, and proteins, and the proteins most targeted were those from the polysaccharide utilization locus (PUL) that are necessary for utilization of fructan (Joglekar et al., 2019). Further, they found that in the presence of dietary polysaccharides (i.e. fructan), PUL specific-SIgA responses increased, recognizing the machinery responsible for utilization of the dietary sugars, which subsequently drove downregulation of B. theta expression of those same PUL proteins. This suggests that SIgA binding of the PUL proteins results in a change in B. theta gene expression, offering a mechanism by which B. theta adapts to immune pressures within the gut in a metabolically dependent fashion.
Work from the Martens lab has also provided evidence that gut bacteria, as modeled by B. theta, can change capsule polysaccharide (CPS) expression in response to immune pressures. The authors interrogated the CPS variations by competing single over-expressing CPS mutants in germ free animals. One CPS, CPS5, was particularly successful and its presence on the B. theta surface correlated with higher amounts of SIgA in the gut. The most successful B. theta strains were ones that had all CPS options available to them (WT), and after antibiotic perturbation, WT B. theta dominantly displayed CPS5 on its surface in a mouse with adaptive immunity intact (Porter et al., 2017). These data suggest a dynamic role in B. theta CPS expression leading to enhanced fitness that is guided by immune pressures of SIgA.
Supporting this, Nakajima et al. showed that a cross-species monoclonal antibody to OVA, 7–6IgA, enhanced the colonization fitness of B. theta by modulating gene expression (Nakajima et al., 2018). Gene expression patterns of B. theta were analyzed in animals using the backpack model described previously. Several PUL genes, found in multiple Bacteroides species, were upregulated in the presence of mAb 7–6IgA. Disruption of those PUL genes reduced the ability of B. theta to colonize, and increased colitis in a mouse. Interestingly, changes in B. theta gene expression also impacted other members of the microbiota. In a complex community, the expression of PUL genes in WT B. theta induced an expansion of Clostridiales, which was correlated with protection from colitis. Taken together this demonstrates that IgA targeting of one organism can influence the community composition and host health.
However, because all of these studies were conducted in vivo, it is difficult to distinguish between bacterial down-regulation of gene expression versus elimination of bacteria expressing the antibody binding epitope. A study from Ruth Ley’s group, supports that bacterial gene regulation can directly be influenced by the presence of SIgA. In mice, lacking TLR5, there is a reduction in cross-species anti-flagella immunoglobulins in the gut, resulting in an increase of microbial flagella gene expression from a wide diversity of gut bacteria including Roseburia, Eubacterium, and Clostridium. Further, in the TLR5 knockout mouse, gut commensals were found closer to the small intestine villi, and flagellated bacteria were able to breach the colonic barrier (Cullender et al., 2013). This was associated with worsened colitis and metabolic syndrome (Carvalho et al., 2012; Vijay-Kumar et al., 2010). To demonstrate that antibodies might directly influence bacterial gene expression, an E. coli strain was engineered to express GFP under the control of the flagella (FliC) promoter to allow real time monitoring of flagella gene expression. Addition of this anti-flagella antibodies to an in vitro culture of E. coli-FliC-GFP caused a rapid down regulation of flagella gene expression. As this experiment was conducted in vitro, elimination of flagella expressing bacteria by SIgA was ruled out, supporting that bacteria may respond to antibody binding directly by modulating gene expression Taken together, these studies demonstrate that host antibodies can function in a variety of ways to influence the commensal microbiota. However, the field has only begun to fully understand these interactions and novel mechanisms will likely be revealed as more investigation is conducted in other commensal organisms, including non-bacterial organisms such as fungi.
IgA associations with disease
IgA deficiency is the most common primary immunodeficiency disease in humans and often develops later in life. While many IgA-deficient individuals are asymptomatic, many with this immunodeficiency develop recurrent infections, have metabolic diseases, gastrointestinal diseases and are at greater risk for developing colorectal cancer (Ludvigsson et al., 2014). That IgA deficient individuals do not have more pronounced disease is likely due to compensatory expression of other Igs such as IgM and IgG, which can both be transported across the intestinal layer. Supporting this, a recent study analyzed fifteen human subjects with secretory IgA deficiencies and found that, as compared with healthy controls, they exhibited much higher levels of secretory IgM in their feces as well as expanded levels of IgM coated bacteria (Catanzaro et al., 2019). However, despite elevated levels of IgM, the authors note altered gut microbial communities between the two groups and differentially targeted bacteria between immunoglobulins further supporting an important role for antibody mediated sculpting of the microbiota. This effect was also observed by Fadlallah and colleagues, where they observed mild dysbiosis of gut microbial communities in patients with IgA deficiencies (Fadlallah et al., 2018). Since IgA has been shown to strongly influence the gene expression patterns of commensal microorganisms, meta-transcriptomic analysis on the microbiota of IgA deficient individuals might reveal even more dramatic effects. However, subtleties in the way IgA is induced or even slight changes in IgA selection or abundance may have important consequences on human health, that have yet to be tested. Here we review the literature that supports that defective IgA selection can lead to disease.
Inflammatory Bowel Disease
Inflammatory bowel disease (IBD), which includes Crohn’s Disease (CD) and ulcerative colitis (UC) are known to be influenced by the microbiota during inflammation and disease progression (Huttenhower et al., 2014; Maloy and Powrie, 2011). Many investigators have employed IgA-Seq to identify microorganisms of interest in these diseases. Commonalities in IgA targets have begun to emerge from these collective studies (Table 1). Work from Flavell and colleagues has shown that IgA preferentially targets pro-colitogenic bacteria. In this study, IgA-Seq within the homeostatic gut revealed that Lactobacillus, Bacteroidales (S24–7), SFB and a member of Erysipelotrichaceae were all differentially coated by IgA. However, when this technique was employed on animals that had inflammasome-mediated colitis, a member of the Prevotellaceae family and Helicobacter sp. flexispira became highly enriched in the IgA-bound population (Palm et al., 2014). To investigate the human relevance of these findings, IgA-Seq was performed on human fecal samples from patients with IBD and healthy controls. Patients with IBD had a greater total amount of IgA coating, and many bacterial species from patients with IBD were also more highly coated, including Streptococcus luteciae and Haemophilus parainfluenza. Organisms that were uniquely IgA coated in IBD patients included Lactobacillus mucosae, Allobaculum sp., Bulleidia, and Weissella sp. Transfer of IgA-bound bacteria to animals followed by colitis induction resulted in worsened disease when compared to transfer of unbound bacteria. In a similar study, IgA-bound bacteria were isolated from individuals with CD that had also developed spondyloarthritis (SpA), a common extra-intestinal manifestation of CD. In these individuals, IgA coated a large number of adherent invasive E. coli species. When these isolates were placed into animal models of colitis or arthritis, the severity of disease was significantly worsened (Viladomiu et al., 2017).
Table 1:
Examples of bacteria that are differentially bound by sIgA during disease
| Bacteria | Disease associated | Host | IgA binding change |
|---|---|---|---|
| Prevotellaceae | |||
| Helicobacter sp. flexispira | |||
| Segmented Filamentous | Mouse | Increased | |
| Bacteria | |||
| Lactobacillus | |||
| Streptococcus luteciae | IBD | ||
| Haemophilus parainfluenza | (UC and CD) | ||
| Lactobacillus mucosae | |||
| Allobaculum sp. | Human | ||
| Ruminococcaceae | Increased | ||
| Blautia | |||
| Bacteroides fragilis | |||
| Clostridium methylpentosum | |||
| Bifidobacterium animalis | |||
| Bilophila | |||
| Oscillospira | Decreased | ||
| Adherent invasive E. coli | Spondyloarthritis | Human | increased |
| Fusobacteria nucleatum | Colorectal Cancer | Human | increased |
| Enterobacteriaceae | Malnutrition | Human | increased |
|
Lactobaccillus sp. Bifidobacterium Eubacterium |
Undernutrition | Mouse | Decreased |
| Clostridia | Metabolic disease | Mouse | increased |
A new study by Shapiro et al. analyzed IgA-bound bacteria using IgA-Seq in 184 IBD patients and also found that many taxa that were highly coated in IgA were associated with IBD. Organisms that were highly coated in IgA and differentially abundant in IBD patients included B. fragilis, Haemophilus parainfluenzae, Clostridium methylpentosum, Bifidobacterium animalis, and a Bilophila sp. Further, they found that low IgA coating of one taxon of Oscillospira was predictive of disease progression to surgical intervention (Shapiro et al., 2021). Other studies have been conducted on human and mouse samples and will continue to be important in identifying disease relevant organisms and pathways that influence IgA targeting (Kubinak et al., 2015a; Rengarajan et al., 2020; Wang et al., 2015). These studies together have led to the notion that IgA targets organisms that are harmful to the host.
Colorectal Cancer
One co-morbidity that is correlated with IgA-deficient humans is an increase in colorectal cancer (CRC). Unlike with IBD, transfer of IgA coated microbes into disease models of CRC has yet to be done, making conclusions about IgA coating during CRC unclear. Since IBD is a major risk factor for CRC, it is reasonable to hypothesize that IgA-bound microbes might also increase CRC severity. However, as of yet few studies have found support for this hypothesis.
In a human study of 190 participants, 150 of which had CRC, total abundance of IgA was surveyed in fecal contents pre-and post-surgery. The presence of tumors positively correlated with an increase in IgA, among other markers, and postoperative IgA levels largely return to normal after tumor removal (Chalkias et al., 2011). This data could suggest that either the tumor itself or, bacteria that are commonly associated with the tumor, could induce IgA responses. Fusobacterium nucleatum, enterotoxigenic Bacteroides fragilis and genotoxin producing E. coli have recently been demonstrated to be found within CRC tumors (Garrett, 2019). In particular, F. nucleatum has been described by many as a biomarker for disease, and several studies have found elevated F. nucleatum specific IgA and IgG in the serum of CRC patients compared to healthy controls (Wang et al., 2016), suggesting that serum anti-F. nucleatum antibodies could be used diagnostically. Since many antibody reactions against gut bacteria are also found within the serum (Zeng et al., 2016), this also supports a role for gut antibody responses in CRC.
As the interleukin (IL) 33 signaling pathway has been associated with IBD, Malik et al. recently determined whether disruption of IL-33 could lead to colitis associated CRC (Malik et al., 2016). IL-33 deficient animals had increased colitis and CRC. Interestingly, mice lacking IL-33 had a decrease in intestinal IgA, which led to an altered microbiota. Cohousing IL-33−/− mice with their WT counterparts abrogated the IgA loss, microbial shifts, and decreased colitis and CRC severity. Whether altered IgA promotes tumor severity or was the result of microbial shifts was not fully addressed. However, this study provides another example that IgA changes might influence CRC. Future work centering on the interaction between IgA and the microbiota during CRC might reveal novel diagnostic tools and mechanisms that influence the development of this cancer.
Metabolic disease
A staggering 820 million individuals are malnourished worldwide and at increased risk of diarrheal diseases. A severe form of undernutrition is kwashiorkor, which is caused by sufficient caloric intake but a lack of protein that leads to edema and enlarged liver with fatty infiltrates. As the microbiota is an important part of promoting host immunity and digestion of food, studies to understand the microbiota’s role during malnutrition might lead to novel interventions. To address this, Kau et al analyzed IgA targeting in mice colonized with human microbiotas from Malawian twins that were discordant for kwashiorkor (Kau et al., 2015). Animals were either placed on a Malawian diet (macro- and micro-nutrient deficient) or a nutrient sufficient standard diet. Despite its presence within all the animal groups, only animals that were colonized with the kwashiorkor microbiota and receiving the Malawian diet developed an IgA response toward multiple members of Enterobacteriaceae. IgA-bound bacteria were sorted and transferred to germfree recipients receiving a Malawian diet. IgA-bound bacteria isolated from the kwashiorkor microbiota, but not from the healthy co-twin microbiota, resulted in 50% mortality after only 1 week and severe weight loss in animals that did survive. This result was dependent on diet as even animals with the IgA-bound bacteria from the individual with kwashikor, but maintained on a standard diet, thrived. Interestingly, inoculation of kwashikor animals with the bacteria, A. muciniphila and Clostridia scindens, present in the healthy twin, was sufficient to reduce mortality. IgA analysis in a larger cohort of individuals revealed that IgA targeting of Enterobacteriaceae was indicative of a pathogenic community, demonstrating that IgA targeting could potentially be used as a diagnostic to identify individuals that will succumb to disease.
Further supporting a role between diet and IgA-microbiota interactions, the Finlay groups demonstrated Lactobacillus johnsonii/gasseri, as well as the Bifidobacterium and Eubacterium generas, failed to become IgA targeted in a mouse model of undernutrition. Sequencing of Lactobacillus isolates that were either IgA bound or not revealed that the limitation of available nutrients in a mouse diet led to genomic mutations in carbohydrate and metabolism genes, that altered bacterial surface glycosylation patterns and IgA binding (Huus et al., 2020). Changes in Lactobacillus glycan expression were replicated in Rag−/− animals that lack B and T cells, indicating that SIgA itself was not the main selective pressure for cell surface glycosylation, but rather nutrient availability drove this phenotype. While the functional consequences of the SIgA targeting of Lactobacillus were not addressed in this study, others have demonstrated that L. johnsonii can enhance body weight in undernourished mice, suggesting that SIgA targeting of this organism might be a marker of nutritional health (Kaburagi et al., 2007).
Our lab has identified that animals lacking MyD88 within the T cell compartment develop an age dependent spontaneous obesity and metabolic syndrome on normal chow (Petersen et al., 2019). The most notable immunological phenotype in these animals was a reduction in TFH cells within the PP and reductions in IgA binding of the microbiota (Kubinak et al., 2015a). Sequencing of IgA coated microbes in obesogenic animals demonstrated an inappropriate targeting of members of the Clostridia family, accompanied by an overall expansion in the amount of Desulfovibrio. The development of obesity was associated with T cell dependent IgA responses as the transfer of WT TFH to obesity prone animals was sufficient to prevent obesity and restore appropriate IgA binding of the microbiota. Supporting a role for the relevance of IgA targeting of the microbiota in humans, individuals with obesity and/or metabolic syndrome have reduced mucosal IgA responses, are more prone to recurrent infections and have reduced responses to vaccination (Must et al., 1999; Pallaro et al., 2002). Together, these data suggest that immune system control of the microbiota is a critical component of metabolic health that requires further exploration.
Therapeutic manipulation of host-microbe interactions
While much basic work needs to be done, now that the microbiota has been implicated in a variety of disease settings, how might we utilize this data to improve human health? Prevention of intestinal infections, treatment of under or over nutrition, boosting immune responses, preventing autoimmunity, or even expediting the microbiota recovery after common procedures such as colonoscopy or antibiotic treatment, are potential places where microbiota-based therapies might be relevant. The following section will propose ways in which our knowledge of microbiota-IgA interactions might be employed therapeutically and is summarized in Figure 3.
Figure 3: Potential therapeutic strategies based on sIgA.

Therapeutic strategies include A) cross-species antibody responses against flagella, LPS, murine lipoprotein, and other outer membrane structures where a commensal can prime against a pathogen B) Blocking invasion and stimulating immune responses to assist in anti-bacterial clearance, for example, blocking SFB invasion while also increasing Th17 responses. Induction of an SIgA response blocking bacteriophages may weaken bacteria fitness and protect against IBD (C). Therapeutic strategies could include utilizing SIgA to facilitate seeding of the gut with beneficial microbes, such as B. fragilis (D). Vaccinations using lyophilized bacteria could be useful for inducing SIgA responses that can be passed through breastmilk to protect neonates (E).
Use of cross-species antibody responses
Control of gene expression
In several examples discussed above, both pathogenic and commensal flagellated bacteria can gain access to the host epithelia and cause damage and disease. Indeed, there are several species of flagellated commensal Enterobacteriaceae that have been associated with intestinal disease (Carvalho et al., 2012; Vijay-Kumar et al., 2010; Vijay-Kumar et al., 2007). IgA targeting of mobility elements such as flagella could be an effective means of preventing organisms from getting too physically close to host tissue to cause damage and inflammation. Supporting that, IgA responses against flagella are important during human disease, obese subjects have increased flagella expression in their microbiota and reduced fecal anti-flagellin IgA, while individuals with colitis have increased serum anti-flagellin IgA (Tran et al., 2019; Wallis et al., 2013). As some of these elements are conserved across multiple species, eliciting a cross-species antibody response might aid in the protection from disease globally. A recent example of this has been tested in animal models.
Tran et al. administered purified Salmonella derived flagellin via an intraperitoneal injection once a week for 10 weeks in animals (Tran et al., 2019). This regimen elicited a robust cross-species serum and fecal anti-flagellin specific IgA and IgG response that was associated with downregulation of flagellin within the microbiota and reduced adherence of microbes to the intestinal epithelia. Importantly, immunization against flagella protected animals from high fat diet induced obesity and a mouse model of colitis. These data provide the proof of principle that a cross-species antibody response against commensals expressing flagella can protect against disease and highlight the importance of an appropriate immune response against the microbiota for the prevention of a variety of diseases.
Prevention of systemic infection
Commensal driven cross-species antibody responses have been shown to react against pathogenic organisms, suggesting that having appropriate commensal antibodies can protect from pathogen invasion. Nunez and colleagues demonstrated a large proportion of the serum IgG and IgA is reactive to the gut microbiota. This study demonstrated that the pre-existing microbiota IgG responses were specific to murein-lipoprotein (MLP), found on many gram-negative bacteria (Zeng et al., 2016). Indeed, the commensal MLP specific antibodies protected animals from sepsis when challenged with two distinct pathogens, Salmonella and E. coli. Thus, this could be exploited in two ways to help bolster immunity against pathogens. The first is to identify commensal organisms that elicit protective cross-species antibody responses and subsequently colonize individuals with these organisms. A second possibility is to immunize individuals with the epitopes that elicit cross-species antibody responses. Continued identification of organisms that elicit the induction of cross-species antibody responses against common bacteria, viral, or fungal epitopes could be used to repeal the colonization of pathogenic species. These types of therapeutic or preventative measures might be particularly useful in individuals about to undergo chemotherapy or have taken antibiotics.
Protection of neonates
A long-appreciated function of IgA and IgG is that they are delivered through breastmilk and helps protect neonates that have yet to develop their own effective immunity from pathogenic insult. In a recent study, the Kasper and Mekalanos groups demonstrated that induction of IgG responses against common commensal organisms within the mouse gut was passed through the breast milk and protected the neonate from enteric infection (Zheng et al., 2020). Isolation of the organisms that were associated with protection uncovered a Pantoea-1 Proteobacteria. When the Pantoea-1 isolate was pre-incubated with the cross-reactive sera, the titer of cross-species reactive antibody was significantly diminished, indicating that the antibody recognized epitopes on Pantoea-1. When germfree mothers were immunized with formalin-killed Pantoea-1, their neonates were protected from infection with enterotoxigenic E. coli, confirming its protective role. Interestingly, the cross-species reactive antibodies were only specific for other members of the Enterobacteriacae and no cross-reactivity was observed with Bacteroides or Staphylococcus species, indicating some level of specificity. While humans and mice differ in their dominant immunoglobulins passed through breastmilk (with mice largely passing IgG and humans IgA), these results open up the exciting possibility that colonization with certain species or immunization approaches within mothers could protect newborns from a variety of childhood infections. Colonization of mothers with commensal organisms that can elicit protective cross-species reactive antibodies against organisms associated with diarrheal diseases might be particularly useful in countries that are under-developed. Indeed, while diarrheal diseases are preventable and treatable in industrialized countries, over 450,000 young children a year die across the globe from these diseases. Given that bacterial organisms can be lyophilized, packaged in easy to take pills, and are shelf stable, this strategy could be a viable option for countries that lack resources to vaccinate.
Use of species and strain-specific antibody responses
Control of harmful gene expression
There are many examples, some of which were highlighted above, where a normal resident of the commensal community causes damage and disease. In these cases, boosting IgA responses against particular species that can control potentially pathogenic interactions through mucosal immunization strategies might be an effective therapeutic intervention.
We don’t fully understand why some commensals can suddenly cause damage in a host, and since many of these organisms might also have beneficial effects, complete eradication of the offending microbe might not always be the best strategy. Since binding of IgA has been shown to cause downregulation of specific genes within the microbe, an immunization strategy against a specific epitope on a microbe might be an effective way to maintain colonization while preventing damage. Adhesion, mobility and invasion are all attributes of organisms that can elicit an IgA response. Adhesion of microbes to epithelia provides them access to invade host tissue and possibly disseminate to more sensitive sites. To date, there have only been a few organisms that reside within the commensal population that are capable of direct invasion, including Helicobacter species, SFB, and some Enterobacteraceae such as E. coli. SFB is known to initiate Th17 responses that aggravate chronic models of inflammation including colitis, multiple sclerosis and arthritis (Lee et al., 2011; Stepankova et al., 2007; Wu et al., 2010). Induction of this pro-inflammatory response requires invasion of epithelial cells by SFB (Atarashi et al., 2015). In diseases where chronic inflammation drives disease progression, it might be useful to initiate an IgA response to specific epitopes on SFB that allow for this invasion. A curated IgA response to those epitopes could cause downregulation of its invasion machinery and prevent the harmful inflammation that fuels colitis. SFB, however, has not just been associated with detrimental consequences to the host. Indeed, SFB can prevent and cure rotavirus infection and Citrobacter infection in animals (Ivanov et al., 2009; Shi et al., 2019). Protection from rotavirus infection was found to be independent of the Th17 inducing capacity of SFB, but instead was dependent on the ability of SFB to elicit epithelial turnover so that virus was excluded from the epithelia. While SFB has not been definitively identified as a human commensal, several human associated commensals are known to induce Th17 responses (Tan et al., 2016). Therefore, based on these studies, species-specific immunization strategies that target the invasive, Th17 driven responses of microbes like SFB, might help retain their protective effects while eradicating their chronic inflammatory consequences. As we begin to uncover the more mechanistic underpinnings of specific commensal effects on the host, this strategy could extend to many disease settings and allow us to sculpt personalized microbial communities.
Aid in beneficial commensal colonization
It has been well-documented that probiotic strains are usually not able to stably colonize in the gut (Sanders, 2011). This could be due to colonization resistance, that is, where niches within the gut are already filled by specific organisms such that new organisms cannot compete for nutrients or space. However, another possibility is that the host immune system must ‘select’ for certain organisms within the intestinal tract. We and others have reasoned that since commensal and pathogenic microbes share many of the same motifs recognized by TLRs, that this is not the primary means by which the immune system discriminates between benign and harmful organisms. Antigen specific responses, mediated by major histocompatibility complex (MHC), are capable of using microbe specific epitopes to mount fine-tuned immune responses and have been recently demonstrated to be an important component of commensal host-reactions. Using congenic mice that only differed at their MHC locus, we have shown that unique IgA signatures were associated with distinct microbial communities depending on the MHC locus (Kubinak et al., 2015b). Similarly, Silverman et. al showed that a protective MHC allele shapes the intestinal microbiota, and this protects from Type I diabetes (Silverman et al., 2017). Thus, MHC directing of IgA responses against the microbiota governs the microbial community that takes form within the gut. These data, coupled with the study demonstrating that IgA is necessary for B. fragilis colonization (Donaldson et al., 2018)(Donaldson et al., 2018)(Donaldson et al., 2018)(Donaldson et al., 2018)(Donaldson et al., 2018)(Donaldson et al., 2018), suggests that immune responses could be harnessed to enhance colonization of beneficial organisms. Based on this, a combination therapy that involves eliciting a strain-specific IgA response followed by seeding of the gut with the beneficial strain could help facilitate long-term colonization. As strains of B. fragilis that encode toxins are associated with colorectal cancer, it would be important that this type of approach be strain specific.
While there have been many studies that have uncovered important mechanisms that mediate IgA-microbiota interactions, much work still needs to be done. Understanding how species-specific versus cross-species specific antibody responses are elicited will be important as we begin to employ these mechanisms therapeutically. One important consideration in using cross-species and strain-specific antibody responses for protection comes from an evolutionary perspective. It may be important, in an effort to mitigate selecting for immune evasion by pathogens, to simultaneously apply selective pressure on both the species and a particular strain to aid in the specificity and strength of the protective effect. Therefore, the use of both cross-species and strain-specific antibodies in a combination therapy approach might be important therapeutically. Moreover, identifying specific epitopes, many of which will likely be carbohydrates or glycoproteins, will be another critical endeavor. To further complicate this, it is becoming clear that individuals might harbor distinct strains of bacteria with differing functions, therefore uncovering mechanisms that differ between important strains of commensal bacteria will be necessary when thinking about the use of strain-specific antibody responses. A greater understanding of the microbial metagenome will aid in this endeavor. As the medical field continues to employ more personalized medical approaches, there is no doubt that the interactions between IgA and the microbiota will need to be considered.
ACKNOWLEDGMENTS
We would like to thank Zac Stephens, Jennifer Hill, Kyla Ost, and Tyson Chiaro for critically reading the manuscript. A.M.W. is supported by the NIH NRSA Postdoctoral Fellowship through NCI F32CA243501. J.R. is supported by a Burrough’s Wellcome Investigator in Pathogenesis Grant, the Helmsley Foundation, The W.M. Keck Foundation, The MS Society, The Margolis Foundation and NIDDK R01DK124336 and R01DK124317.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Atarashi K, Tanoue T, Ando M, Kamada N, Nagano Y, Narushima S, Suda W, Imaoka A, Setoyama H, Nagamori T, et al. (2015). Th17 Cell Induction by Adhesion of Microbes to Intestinal Epithelial Cells. Cell 163, 367–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansept F, Schumann-Moor K, Diard M, Hardt WD, Slack E, and Loverdo C (2019). Enchained growth and cluster dislocation: A possible mechanism for microbiota homeostasis. PLoS Comput Biol 15, e1006986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger RR, Everett ML, Palestrant D, Love SD, Lin SS, and Parker W (2003). Human secretory immunoglobulin A may contribute to biofilm formation in the gut. Immunology 109, 580–587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollinger RR, Everett ML, Wahl SD, Lee YH, Orndorff PE, and Parker W (2006). Secretory IgA and mucin-mediated biofilm formation by environmental strains of Escherichia coli: role of type 1 pili. Mol Immunol 43, 378–387. [DOI] [PubMed] [Google Scholar]
- Bunker JJ, Erickson SA, Flynn TM, Henry C, Koval JC, Meisel M, Jabri B, Antonopoulos DA, Wilson PC, and Bendelac A (2017). Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho FA, Koren O, Goodrich JK, Johansson ME, Nalbantoglu I, Aitken JD, Su Y, Chassaing B, Walters WA, Gonzalez A, et al. (2012). Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell host & microbe 12, 139–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castagliuolo I, Galeazzi F, Ferrari S, Elli M, Brun P, Cavaggioni A, Tormen D, Sturniolo GC, Morelli L, and Palu G (2005). Beneficial effect of auto-aggregating Lactobacillus crispatus on experimentally induced colitis in mice. FEMS Immunol Med Microbiol 43, 197–204. [DOI] [PubMed] [Google Scholar]
- Catanzaro JR, Strauss JD, Bielecka A, Porto AF, Lobo FM, Urban A, Schofield WB, and Palm NW (2019). IgA-deficient humans exhibit gut microbiota dysbiosis despite secretion of compensatory IgM. Sci Rep 9, 13574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalkias A, Nikotian G, Koutsovasilis A, Bramis J, Manouras A, Mystrioti D, and Katergiannakis V (2011). Patients with colorectal cancer are characterized by increased concentration of fecal hb-hp complex, myeloperoxidase, and secretory IgA. Am J Clin Oncol 34, 561–566. [DOI] [PubMed] [Google Scholar]
- Cullender TC, Chassaing B, Janzon A, Kumar K, Muller CE, Werner JJ, Angenent LT, Bell ME, Hay AG, Peterson DA, et al. (2013). Innate and adaptive immunity interact to quench microbiome flagellar motility in the gut. Cell host & microbe 14, 571–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson GP, Ladinsky MS, Yu KB, Sanders JG, Yoo BB, Chou WC, Conner ME, Earl AM, Knight R, Bjorkman PJ, et al. (2018). Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fadlallah J, El Kafsi H, Sterlin D, Juste C, Parizot C, Dorgham K, Autaa G, Gouas D, Almeida M, Lepage P, et al. (2018). Microbial ecology perturbation in human IgA deficiency. Sci Transl Med 10. [DOI] [PubMed] [Google Scholar]
- Garrett WS (2019). The gut microbiota and colon cancer. Science 364, 1133–1135. [DOI] [PubMed] [Google Scholar]
- Huttenhower C, Kostic AD, and Xavier RJ (2014). Inflammatory bowel disease as a model for translating the microbiome. Immunity 40, 843–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huus KE, Bauer KC, Brown EM, Bozorgmehr T, Woodward SE, Serapio-Palacios A, Boutin RCT, Petersen C, and Finlay BB (2020). Commensal Bacteria Modulate Immunoglobulin A Binding in Response to Host Nutrition. Cell Host Microbe 27, 909–921 e905. [DOI] [PubMed] [Google Scholar]
- Ivanov II, Atarashi K, Manel N, Brodie EL, Shima T, Karaoz U, Wei D, Goldfarb KC, Santee CA, Lynch SV, et al. (2009). Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A (2016). Exploiting Mucosal Immunity for Antiviral Vaccines. Annu Rev Immunol 34, 575–608. [DOI] [PubMed] [Google Scholar]
- Joglekar P, Ding H, Canales-Herrerias P, Pasricha PJ, Sonnenburg JL, and Peterson DA (2019). ntestinal IgA Regulates Expression of a Fructan Polysaccharide Utilization Locus in Colonizing Gut Commensal Bacteroides thetaiotaomicron. mBio 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaburagi T, Yamano T, Fukushima Y, Yoshino H, Mito N, and Sato K (2007). Effect of Lactobacillus johnsonii La1 on immune function and serum albumin in aged and malnourished aged mice. Nutrition 23, 342–350. [DOI] [PubMed] [Google Scholar]
- Kau AL, Planer JD, Liu J, Rao S, Yatsunenko T, Trehan I, Manary MJ, Liu TC, Stappenbeck TS, Maleta KM, et al. (2015). Functional characterization of IgA-targeted bacterial taxa from undernourished Malawian children that produce diet-dependent enteropathy. Sci Transl Med 7, 276ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinak JL, Petersen C, Stephens WZ, Soto R, Bake E, O’Connell RM, and Round JL (2015a). MyD88 signaling in T cells directs IgA-mediated control of the microbiota to promote health. Cell Host Microbe 17, 153–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinak JL, and Round JL (2016). Do antibodies select a healthy microbiota? Nat Rev Immunol 16, 767–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubinak JL, Stephens WZ, Soto R, Petersen C, Chiaro T, Gogokhia L, Bell R, Ajami NJ, Petrosino JF, Morrison L, et al. (2015b). MHC variation sculpts individualized microbial communities that control susceptibility to enteric infection. Nat Commun 6, 8642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YK, Menezes JS, Umesaki Y, and Mazmanian SK (2011). Proinflammatory T-cell responses to gut microbiota promote experimental autoimmune encephalomyelitis. Proc Natl Acad Sci U S A 108 Suppl 1, 4615–4622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludvigsson JF, Neovius M, and Hammarstrom L (2014). Association between IgA deficiency & other autoimmune conditions: a population-based matched cohort study. J Clin Immunol 34, 444–451. [DOI] [PubMed] [Google Scholar]
- Malik A, Sharma D, Zhu Q, Karki R, Guy CS, Vogel P, and Kanneganti TD (2016). IL-33 regulates the IgA-microbiota axis to restrain IL-1alpha-dependent colitis and tumorigenesis. J Clin Invest 126, 4469–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy KJ, and Powrie F (2011). Intestinal homeostasis and its breakdown in inflammatory bowel disease. Nature 474, 298–306. [DOI] [PubMed] [Google Scholar]
- Mantis NJ, Rol N, and Corthesy B (2011). Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal immunology 4, 603–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathias A, and Corthesy B (2011). Recognition of gram-positive intestinal bacteria by hybridoma- and colostrum-derived secretory immunoglobulin A is mediated by carbohydrates. J Biol Chem 286, 17239–17247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikulic J, Bioley G, and Corthesy B (2017a). SIgA-Shigella Immune Complexes Interact with Dectin-1 and SIGNR3 to Differentially Regulate Mouse Peyer’s Patch and Mesenteric Lymph Node Dendritic Cell’s Responsiveness. J Mol Biol 429, 2387–2400. [DOI] [PubMed] [Google Scholar]
- Mikulic J, Longet S, Favre L, Benyacoub J, and Corthesy B (2017b). Secretory IgA in complex with Lactobacillus rhamnosus potentiates mucosal dendritic cell-mediated Treg cell differentiation via TLR regulatory proteins, RALDH2 and secretion of IL-10 and TGF-beta. Cell Mol Immunol 14, 546–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moor K, Diard M, Sellin ME, Felmy B, Wotzka SY, Toska A, Bakkeren E, Arnoldini M, Bansept F, Co AD, et al. (2017). High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544, 498–502. [DOI] [PubMed] [Google Scholar]
- Must A, Spadano J, Coakley EH, Field AE, Colditz G, and Dietz WH (1999). The disease burden associated with overweight and obesity. JAMA 282, 1523–1529. [DOI] [PubMed] [Google Scholar]
- Nakajima A, Vogelzang A, Maruya M, Miyajima M, Murata M, Son A, Kuwahara T, Tsuruyama T, Yamada S, Matsuura M, et al. (2018). IgA regulates the composition and metabolic function of gut microbiota by promoting symbiosis between bacteria. J Exp Med 215, 2019–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orndorff PE, Devapali A, Palestrant S, Wyse A, Everett ML, Bollinger RR, and Parker W (2004). Immunoglobulin-mediated agglutination of and biofilm formation by Escherichia coli K-12 require the type 1 pilus fiber. Infect Immun 72, 1929–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst O (2012). New concepts in the generation and functions of IgA. Nat Rev Immunol 12, 821–832. [DOI] [PubMed] [Google Scholar]
- Pabst O, and Slack E (2020). IgA and the intestinal microbiota: the importance of being specific. Mucosal Immunol 13, 12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pallaro A, Barbeito S, Taberner P, Marino P, Franchello A, Strasnoy I, Ramos O, and Slobodianik N (2002). Total salivary IgA, serum C3c and IgA in obese school children. J Nutr Biochem 13, 539. [DOI] [PubMed] [Google Scholar]
- Palm NW, de Zoete MR, Cullen TW, Barry NA, Stefanowski J, Hao L, Degnan PH, Hu J, Peter I, Zhang W, et al. (2014). Immunoglobulin a coating identifies colitogenic bacteria in inflammatory bowel disease. Cell 158, 1000–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen C, Bell R, Klag KA, Lee SH, Soto R, Ghazaryan A, Buhrke K, Ekiz HA, Ost KS, Boudina S, et al. (2019). T cell-mediated regulation of the microbiota protects against obesity. Science 365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson DA, McNulty NP, Guruge JL, and Gordon JI (2007). IgA response to symbiotic bacteria as a mediator of gut homeostasis. Cell host & microbe 2, 328–339. [DOI] [PubMed] [Google Scholar]
- Peterson DA, Planer JD, Guruge JL, Xue L, Downey-Virgin W, Goodman AL, Seedorf H, and Gordon JI (2015). Characterizing the interactions between a naturally primed immunoglobulin A and its conserved Bacteroides thetaiotaomicron species-specific epitope in gnotobiotic mice. J Biol Chem 290, 12630–12649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter NT, Canales P, Peterson DA, and Martens EC (2017). A Subset of Polysaccharide Capsules in the Human Symbiont Bacteroides thetaiotaomicron Promote Increased Competitive Fitness in the Mouse Gut. Cell Host Microbe 22, 494–506 e498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reboldi A, Arnon TI, Rodda LB, Atakilit A, Sheppard D, and Cyster JG (2016). IgA production requires B cell interaction with subepithelial dendritic cells in Peyer’s patches. Science 352, aaf4822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rengarajan S, Vivio EE, Parkes M, Peterson DA, Roberson EDO, Newberry RD, Ciorba MA, and Hsieh CS (2020). Dynamic immunoglobulin responses to gut bacteria during inflammatory bowel disease. Gut Microbes 11, 405–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Round JL, and Palm NW (2018). Causal effects of the microbiota on immune-mediated diseases. Sci Immunol 3. [DOI] [PubMed] [Google Scholar]
- Sanders ME (2011). Impact of probiotics on colonizing microbiota of the gut. J Clin Gastroenterol 45 Suppl, S115–119. [DOI] [PubMed] [Google Scholar]
- Shapiro JM, de Zoete MR, Palm NW, Laenen Y, Bright R, Mallette M, Bu K, Bielecka AA, Xu F, Hurtado-Lorenzo A, et al. (2021). Immunoglobulin A Targets a Unique Subset of the Microbiota in Inflammatory Bowel Disease. Cell Host Microbe 29, 83–93 e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Z, Zou J, Zhang Z, Zhao X, Noriega J, Zhang B, Zhao C, Ingle H, Bittinger K, Mattei LM, et al. (2019). Segmented Filamentous Bacteria Prevent and Cure Rotavirus Infection. Cell 179, 644–658 e613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman M, Kua L, Tanca A, Pala M, Palomba A, Tanes C, Bittinger K, Uzzau S, Benoist C, and Mathis D (2017). Protective major histocompatibility complex allele prevents type 1 diabetes by shaping the intestinal microbiota early in ontogeny. Proc Natl Acad Sci U S A 114, 9671–9676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepankova R, Powrie F, Kofronova O, Kozakova H, Hudcovic T, Hrncir T, Uhlig H, Read S, Rehakova Z, Benada O, et al. (2007). Segmented filamentous bacteria in a defined bacterial cocktail induce intestinal inflammation in SCID mice reconstituted with CD45RBhigh CD4+ T cells. Inflamm Bowel Dis 13, 1202–1211. [DOI] [PubMed] [Google Scholar]
- Tan TG, Sefik E, Geva-Zatorsky N, Kua L, Naskar D, Teng F, Pasman L, Ortiz-Lopez A, Jupp R, Wu HJ, et al. (2016). Identifying species of symbiont bacteria from the human gut that, alone, can induce intestinal Th17 cells in mice. Proc Natl Acad Sci U S A 113, E8141–E8150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tareb R, Bernardeau M, Gueguen M, and Vernoux JP (2013). In vitro characterization of aggregation and adhesion properties of viable and heat-killed forms of two probiotic Lactobacillus strains and interaction with foodborne zoonotic bacteria, especially Campylobacter jejuni. J Med Microbiol 62, 637–649. [DOI] [PubMed] [Google Scholar]
- Tran HQ, Ley RE, Gewirtz AT, and Chassaing B (2019). Flagellin-elicited adaptive immunity suppresses flagellated microbiota and vaccinates against chronic inflammatory diseases. Nat Commun 10, 5650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, and Gewirtz AT (2010). Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328, 228–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijay-Kumar M, Sanders CJ, Taylor RT, Kumar A, Aitken JD, Sitaraman SV, Neish AS, Uematsu S, Akira S, Williams IR, et al. (2007). Deletion of TLR5 results in spontaneous colitis in mice. The Journal of clinical investigation 117, 3909–3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viladomiu M, Kivolowitz C, Abdulhamid A, Dogan B, Victorio D, Castellanos JG, Woo V, Teng F, Tran NL, Sczesnak A, et al. (2017). IgA-coated E. coli enriched in Crohn’s disease spondyloarthritis promote TH17-dependent inflammation. Sci Transl Med 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voltan S, Castagliuolo I, Elli M, Longo S, Brun P, D’Inca R, Porzionato A, Macchi V, Palu G, Sturniolo GC, et al. (2007). Aggregating phenotype in Lactobacillus crispatus determines intestinal colonization and TLR2 and TLR4 modulation in murine colonic mucosa. Clin Vaccine Immunol 14, 1138–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis D, Asaduzzaman A, Weisman M, Haroon N, Anton A, McGovern D, Targan S, and Inman R (2013). Elevated serum anti-flagellin antibodies implicate subclinical bowel inflammation in ankylosing spondylitis: an observational study. Arthritis Res Ther 15, R166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HF, Li LF, Guo SH, Zeng QY, Ning F, Liu WL, and Zhang G (2016). Evaluation of antibody level against Fusobacterium nucleatum in the serological diagnosis of colorectal cancer. Sci Rep 6, 33440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Charbonnier LM, Noval Rivas M, Georgiev P, Li N, Gerber G, Bry L, and Chatila TA (2015). MyD88 Adaptor-Dependent Microbial Sensing by Regulatory T Cells Promotes Mucosal Tolerance and Enforces Commensalism. Immunity 43, 289–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RC, and Gibbons RJ (1972). Inhibition of bacterial adherence by secretory immunoglobulin A: a mechanism of antigen disposal. Science 177, 697–699. [DOI] [PubMed] [Google Scholar]
- Wold AE, Mestecky J, Tomana M, Kobata A, Ohbayashi H, Endo T, and Eden CS (1990). Secretory immunoglobulin A carries oligosaccharide receptors for Escherichia coli type 1 fimbrial lectin. Infect Immun 58, 3073–3077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HJ, Ivanov II, Darce J, Hattori K, Shima T, Umesaki Y, Littman DR, Benoist C, and Mathis D (2010). Gut-residing segmented filamentous bacteria drive autoimmune arthritis via T helper 17 cells. Immunity 32, 815–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Nguyen LH, Franzosa EA, and Huttenhower C (2020). Strain-level epidemiology of microbial communities and the human microbiome. Genome Med 12, 71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng MY, Cisalpino D, Varadarajan S, Hellman J, Warren HS, Cascalho M, Inohara N, and Nunez G (2016). Gut Microbiota-Induced Immunoglobulin G Controls Systemic Infection by Symbiotic Bacteria and Pathogens. Immunity 44, 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng W, Zhao W, Wu M, Song X, Caro F, Sun X, Gazzaniga F, Stefanetti G, Oh S, Mekalanos JJ, et al. (2020). Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 577, 543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]


