Abstract
Exosomes are a class of cell-secreted, nano-sized extracellular vesicles with a bilayer membrane structure of 30–150 nm in diameter. Their discovery and application have brought breakthroughs in numerous areas, such as liquid biopsies, cancer biology, drug delivery, immunotherapy, tissue repair, and cardiovascular diseases. Isolation of exosomes is the first step in exosome-related research and its applications. Standard benchtop exosome separation and sensing techniques are tedious and challenging, as they require large sample volumes, multi-step operations that are complex and time-consuming, requiring cumbersome and expensive instruments. In contrast, microfluidic platforms have the potential to overcome some of these limitations, owing to their high-precision processing, ability to handle liquids at a microscale, and integrability with various functional units, such as mixers, actuators, reactors, separators, and sensors. These platforms can optimize the detection process on a single device, representing a robust and versatile technique for exosome separation and sensing to attain high purity and high recovery rates with a short processing time. Herein, we overview microfluidic strategies for exosome isolation based on their hydrodynamic properties, size filtration, acoustic fields, immunoaffinity, and dielectrophoretic properties. We focus especially on advances in label-free isolation of exosomes with active biological properties and intact morphological structures. Further, we introduce microfluidic techniques for the detection of exosomal proteins and RNAs with high sensitivity, high specificity, and low detection limits. We summarize the biomedical applications of exosome-mediated therapeutic delivery targeting cancer cells. To highlight the advantages of microfluidic platforms, conventional techniques are included for comparison. Future challenges and prospects of microfluidics towards exosome isolation applications are also discussed. Although the use of exosomes in clinical applications still faces biological, technical, regulatory, and market challenges, in the foreseeable future, recent developments in microfluidic technologies are expected to pave the way for tailoring exosome-related applications in precision medicine.
Keywords: exosomes, extracellular vesicles, microfluidics, isolation, detection, tumor-targeted drug delivery
Graphical abstract
1. Introduction
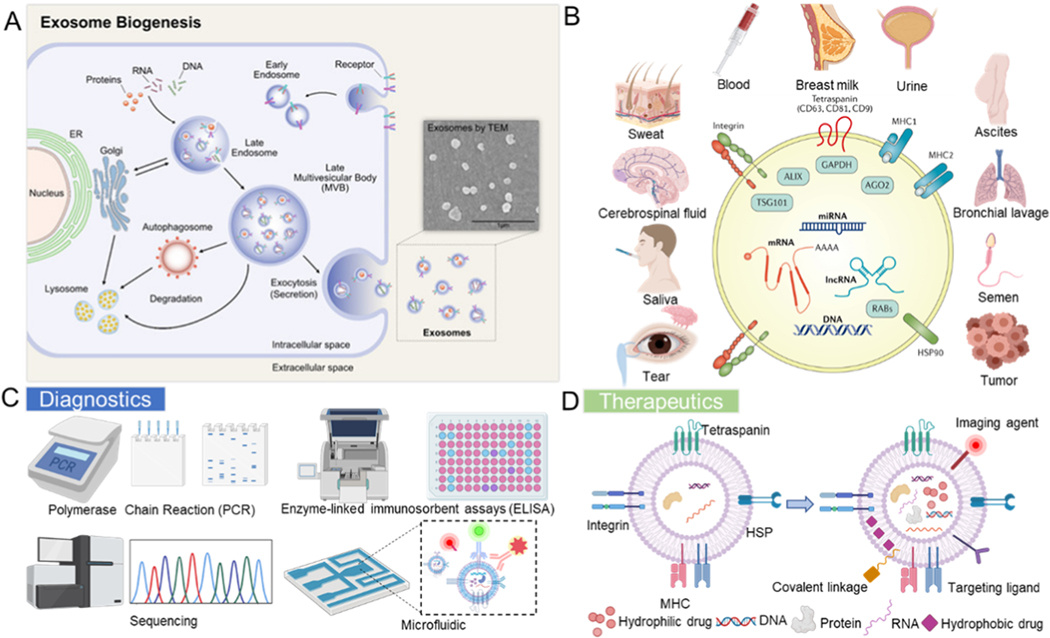
Exosomes are endogenous, nano-sized extracellular vesicles (EVs) composed of proteins, cholesterol, and phospholipids. Significant interest in exosomes stems from their unique biological functions as intercellular messengers and their clinical potential in disease diagnostics, while their potential as natural drug delivery carriers for cancer treatment is gaining increasing recognition [1–3]. Exosomes are the most investigated and characterized subtype of EVs, which have three main subpopulations based on differences in biogenesis mechanisms and size: exosomes, apoptotic bodies and microvesicles (MVs) [4]. Apoptotic bodies are vesicles with diameters ranging from 1 μm to 5 μm, formed by cells to expel toxic components from apoptotic cells during apoptosis or mechanical stress [5]. MVs are membranous vesicles (0.1–1 μm in diameter), which are released from the cytoplasmic membrane. Exosomes were reported in the early 1980s, and their diameters range from 30 nm to 150 nm [6]. Initially, they were only considered to be merely the waste disposed of by cells. Many studies have now shown that exosomes have essential biological functions and significance. They contain a variety of cargo, including nucleic acids [24, 63] and functional and structural proteins [7, 8], of which tetraspanins are currently the largest family of proteins, including CD9 and CD63 [9]. Exosomes are generated by the efflux of the multivesicular body (MVB) in the process of endocytosis (Figure 1A) [10, 11]. They exist widely in a variety of bodily fluids, including blood, saliva, cerebrospinal fluid, semen, breast milk, and urine (Figure 1B) [12, 13].
Figure 1.
(A) Illustration of the exosome biogenesis mechanism. Reprinted with permission from Ref. [11]. Copyright 2019, Multidisciplinary Digital Publishing Institute. (B) Schematic of a typical exosomal composition. Reprinted with permission from Ref. [13]. Copyright 2017, Nature Publishing Group. Exosomes exist in various biological fluids such as saliva and urine. (C) Diagnostic and (D) therapeutic applications of exosomes.
Exosomes are released by most types of living cells in both normal and pathological conditions and carry their secreted active biomolecules, including nucleic acids and proteins. They can reflect the physiological states of the parental cells and their local microenvironment and regulate intercellular communication in multicellular biological systems [14, 15]. Exosomes can also transfer their cargo, such as messenger RNAs, microRNAs, long noncoding RNAs (lncRNAs), and proteins, to recipient cells directly as functional biomolecules to alter the function and phenotype of the recipient cells [16–19]. Importantly, exosomes have been explored as novel biomarkers for early disease diagnosis (Figure 1C) [20, 21]. They also play functional roles in an extensive range of applications, including regulating disease processes, such as in cancer [22], liver diseases [21], neurodegeneration [23], and cardiopulmonary disorders [24, 25]. More recently, by virtue of their non-immunogenic characteristics due to their composition being similar to cells in the body and their stable lipid bilayer membrane structure, they can also be utilized as natural drug delivery carriers [20, 26–28]. Exosomes can stably maintain the encapsulated drug, especially nucleic acid or protein drugs, and improve the solubility of hydrophobic drugs (Figure 1D).
2. Challenges Associated with Exosomes
The structures and functions of exosomes are complex and variable, and the biogenesis and functional characteristics of exosomes are influenced by many factors, such as cell type, confluency or density of cells, cell culture conditions, and stimulation of cells with exogenous compounds [29]. Different cell types may give different exosome yields, and immature dendritic cells, for instance, can only secrete a limited number of exosomes, while mesenchymal stem cells may secrete much larger quantities of exosomes [151].
As exosomes are lipid vesicles released by various cell types, they vary in size and composition [30]. Different sizes and components enable exosomes to serve diverse biological functions. However, cell debris and the complex composition of biological fluids can affect the analysis of exosomal functions. The presence of impurities and other EV subtypes can also interfere with the molecular profiling of exosomes, including the quality of protein and nucleic acid yields, as well as downstream proteomic and transcriptomic analyses.
For EV diagnostic analysis and drug delivery applications, isolation and detection techniques should give a high yield of exosomes of high purity and specificity. These techniques should ensure that the composition, biological activity, and structure of exosomes are preserved intact, as their biological cargo can offer prognostic information for various diseases. Additionally, the integrity and stability of the exosomal membrane are of utmost importance for drug delivery. Intact exosomes can be loaded with drugs and can facilitate uptake by recipient cells. The integrity of the membrane structure can also help ensure exosome preservation from degradation or clearance by the immune system and avoid their uptake by the mononuclear phagocytic system.
Current conventional exosome isolation techniques, such as differential ultracentrifugation (the most commonly used method for the purification of exosomes), are time-consuming and require large volumes of sample, rigorous isolation steps, and expensive instruments, but they result in relatively low exosome recovery and purity. Techniques based on microfluidics have advantages of precise processing of fluids, tracing of valuable samples at the microscale level, and greater speed of detection of analytes in a sample by accelerated reactions via the large surface-to-volume ratio in microchannels. Optimized and integrated microfluidic platforms are expected to further improve exosome separation and detection performance. In the following section, we present an overview and comparison of existing and emerging microfluidic approaches for exosome detection and isolation.
3. Conventional Separation Techniques for Exosomes
There are several approaches for isolating exosomes from cell culture media and complex bodily fluids and mainly consist of differential centrifugation and bead-based, filtration-based, and precipitation-based isolation technologies. A comparison of these various conventional exosome separation methods is presented in Table 1.
Table 1.
Current conventional exosome isolation methods.
| Isolation technique | Principle | Time | Advantages | Disadvantages |
|---|---|---|---|---|
Differential ultracentrifugation
|
Particles with various size and density demonstrate various sediment speed under centrifugation | 5–18 hours | ✓ Suitable for large volume preparation | ➣Require expensive equipment ➣Potential damage because of the high-speed centrifugation ➣Time-consuming |
Size
|
Separate with specific molecular size exclusion | 2–4 hours | ✓ Isolate native exosomes | ➣ Exosomes and proteins clogging on nanopores ➣ Potential isolate other nanovesicles in similar size with exosomes |
Bead
|
On the basis of interaction between antibodies/ligan ds and exosome markers | 2–6 hours | ✓ High-purity ✓ Useful for isolating specific exosomes from target origin |
➣Expensive antibodies functionalization ➣Low yields and processing volume ➣Exosome markers must be optimized |
Polymer
|
High hydrophilic water-excluding polymers changes the solubility of exosomes | 0.5–12 hours | ✓ Available for all types of samples ✓ Easy to use |
➣Potential polymeric, protein aggregates, and other extracellular vesicles contaminants. |
3.1. Differential Ultracentrifugation-Based Exosome Isolation
Differential ultracentrifugation (UC) is commonly used to isolate exosomes [31, 32]. After removing cells and their debris by low-speed centrifugation (300×g, 2000×g, and 10,000×g), vesicles such as exosomes are precipitated and purified from the soluble molecules, such as free proteins and protein complexes, by another centrifugation (100,000×g) for at least 70 mins or longer [33]. Exosomes are subsequently washed at least once with PBS or serum-free growth medium to remove free residual proteins. Ultracentrifugation is also typically used in combination with a sucrose density gradient (a continuous distribution of density from low to high) or a sucrose cushion (typically 30% sucrose cushion) to purify exosomes in the sucrose density range of 1.13–19.19 g/mL [34]. Also, all centrifugation steps must be performed at 4 °C to keep proteases, DNases, and RNases in an inactive state during the long centrifugation. This method is widely used for isolating EVs from various biological samples. However, the process of ultracentrifugation is time-consuming, with multiple manual, labor-intensive steps and requires expensive equipment. The isolation efficiency varies from sample to sample and operator to operator and also depends on rotor type, acceleration, and sample viscosity. Moreover, the repeated centrifugation steps can damage exosome structure and reduce their quality. Further, soluble proteins in the sample can also aggregate and clump together with exosomes, causing contamination and adversely affecting purity. As an additional challenge that comes with the centrifugal forces during UC, this method cannot distinguish among exosome subpopulations or other particles with similar density and size, such as protein aggregates, lipids, and miscellaneous nucleic acid complexes, resulting in an impure product.
3.2. Size-Based Exosome Isolation
Exosomes can be separated by their size [35], and there are various size-based exosome isolation methods, including ultrafiltration [36], sequential filtration [37], size-exclusion chromatography [38], fractionation [39], hydrostatic filtration dialysis [40], and commercially available isolation products such as the ExoMir kit (Bioo Scientific; Austin, TX, USA). Ultrafiltration is the selective separation of samples through using ultrafiltration membranes, which have various molecular weight cut-offs (MWCO). The smaller insolvent components, based on molecular size, can transit through the membrane [41]. However, components of larger molecular size are retained because they are bigger than the size of nanopores in the membrane. Therefore, this is a simple and efficient method of separating exosomes without affecting their biological functions or cargo. This has also been reported as one of the best methods for studying exosomal RNA as it produces a higher RNA yield than other conventional methods. On the other hand, it can isolate other nanovesicles of a similar size to exosomes, such as viruses, and cause erroneous detection results. Size-exclusion chromatography is another robust approach that can perform exosome separation, which relies on the size of exclusion pores of a resin-packed column [42]. Molecules smaller than the pores diffuse into the column pores, while larger molecules are blocked out of the pores and then eluted from the column. While size-exclusion chromatography can achieve isolation of exosomes with high yield and purity, it is not suitable for processing large volumes of samples.
3.3. Bead-Based Exosome Isolation
Many receptors and proteins are found on the exosomal membrane. This characteristic is beneficial for the development of targeted techniques for exosome isolation by leveraging the immunoaffinity interactions between exosome surface proteins and their cognate antibodies. Among them, tetraspanin CD63 is a common exosome surface marker that is used to functionalize beads to specifically isolate exosomes [43]. The bead-based approach is easy to apply and compatible with a variety of downstream applications, including electron microscopy, flow cytometry, immunoblotting, and qRT-PCR. However, bead-based methods require multiple steps of centrifugation, which can be time-consuming. Moreover, the heterogeneity of expression of exosomal surface markers limits the reliability and purity of isolated exosomes [44]. Although the use of nano-sized magnetic beads [45] can help to isolate specific subpopulations of exosomes by targeting their surface markers, it is not optimal for separating exosomes from large volumes of samples.
3.4. Polymer-Based Exosome Isolation
Waterproof polymers such as PEG (polyethylene glycol) can facilitate hydrophobic protein and lipid molecule precipitation. They can reduce exosomal solubility to achieve exosome separation through combing their use with low-speed centrifugation or filtration. Precipitation using these polymers has multiple advantages, including ease of use and under neutral pH conditions without potentially affecting biological activity [35, 46].
ExoQuickTM (System Biosciences, Mountain View, CA, USA) is among the commercially available, polymer-based exosome isolation kits [47] and results in a high yield of exosomes when combined with ultracentrifugation. However, contamination by materials that are not exosomal (including polymeric materials, proteins, and protein aggregates) remains an issue in polymer-based exosome separation methods. Also, the presence of polymeric materials can interfere with downstream analysis. For example, several contaminants such as immunoglobulin, albumin, and residue polymer molecules have been detected after analyzing the polymer precipitation using mass spectrometry [48].
4. Advancement of Microfluidic-Based Exosome Isolation
Exosome isolation is a fundamental first step for their application in various biomedical disciplines. In microfluidics, micro- and nano-fabricated channels and structures are used for manipulating small volumes of fluids, and this method offers unique advantages of minimal consumption of reagents and biological samples and efficient isolation of high purity exosomes with accelerated separation and detection speed. Currently, microfluidic-based separation techniques can be placed in two categories: (i) immunoaffinity-based isolation utilizing specific biological markers such as antibodies, (ii) combining microfluidics with acoustic waves and dielectric electrophoresis, to achieve label-free separation of exosomes on the basis of their electrical and physical properties (Table 2).
Table 2.
Microfluidic platforms for label-free separation of exosomes.
| Exosome Isolation Approach | Sample | Isolated Size (nm) | Isolation Capacity (μL) | Isolation Throughput (μL/min) | Recovery Yield(%) | Reference |
|---|---|---|---|---|---|---|
| Filtration-based separation | ||||||
| Vesicle trapping on an array of ciliated nanowires | Liposome, beads | ~83–120 | 100 | 10 | ~10 | [63] |
| Nanowire-induced electrostatic collection | Urine | ~30–200 | 1000 | 50 | N/A | [64] |
| ZnO nanowires-coated 3D PDMS scaffold | Blood | ~40–200 | 100 | 5 | N/A | [61] |
| Double filtration | Urine | ~155 | 8000 | 33 | 74.2 | [62] |
| Exodisc: double filtration | Urine | ~20–600 | 1000 | 36 | >95 | [60] |
| ExoTIC:multi-membranes | Cell culture supernatant, Plasma, Urine | ~30–100 | 5000 | ~83 | >90 | [65] |
| Pressure-driven filtration | Mouse whole blood | ~150 | 3 | 0.075 | >1.5 | [58] |
| Hydrodynamic properties-based separation | ||||||
| Nano-DLD sorting using pillar array | Urine-derived | <100 | 0.72 | 0.0001–0.0002 | N/A | [70] |
| Nano-DLD sorting | Urine, Serum | ~30–200 | 900 | 15 | ~50 | [74] |
| Continuous viscoelasticity-based and field-free microfluidic sorting | Fetal bovine serum | <200 | 100 | 10 | 93.6 | [85] |
| Mild external force separation | Cell culture supernatant | ~30–100 | 5 | ~5 | ~80 | [66] |
| Wavy microchannel structures within viscoelastic fluids sorting | Cell culture supernatant | ~30–200 | N/A | 44.9 | >81 | [69] |
| Acoustic field-based separation | ||||||
| Acoustic purification | Cell culture supernatant, stored red blood cell products | <200 | 10 | 1.68 | >90 | [94] |
| Acousticfluidic collection | Human whole blood | ~100 | 500 | 10 | 99 | [92] |
| Acoustic trapping isolation | Cell culture supernatant, urine, human whole blood | 110–141 | 300 | 15 | 44.4 | [87] |
| Acoustofluidic separation | Human plasma | 20–600 | N/A | 0.5 | N/A | [88] |
| Acoustofluidic isolation | Saliva | ~22–200 | 100 | 11 | N/A | [93] |
| Electrical based separation | ||||||
| Electrophoresis-driven filtration | Mouse whole blood | ~150 | 240 | 2 | 1.5 | [58] |
| Electrophoretic isolation on nanoporous membrane | Mouse plasma |
~10–400 | 1000 | 20 | 65 | [59] |
| Alternating current electrokinetic (ACE) microarray chip | Undiluted human plasma samples | 50–150 | 30–50 | ~2–3.3 | N/A | [115] |
| Electric field driven filtration | Cell culture supernatant, serum | <150 | 50–66 | 2.5–3.5 | 60–80 | [95] |
| Electrokinetic concentration isolation | Cell culture supernatant | ~50–75 | 30 | 1 | N/A | [96] |
| Ion-depletion zone sorting | Cell culture supernatant | ~30–200 | N/A | 1 | 98 | [97] |
4.1. Microfluidic-Based Separation of Exosomes by Immunoaffinity
Exosome isolation using immobilized antibodies on microfluidic surfaces is a strategy to pre-enrich exosomes from cell cultures or blood samples [49–53]. Kanwar et al. designed a device called ExoChip to specifically isolate CD63-specific exosomes from serum in an hour [54]. This platform is composed of narrow channels interconnected with circular capture chambers to increase the retention time of exosomes in the channels to increase surface interaction time of exosomes. The surface of the chip is functionalized with antibodies against CD63, a representative exosomal marker protein as discussed above. This method facilitates simultaneous separation, quantification, and characterization of exosomes directly from blood serum. However, isolation throughput is only 4 μL/min. Further improvements in surface topography and chemistry of the microfluidic channels can enhance the separation sensitivity by increasing the surface area on which the interaction between microfluidic channels and exosomes occurs.
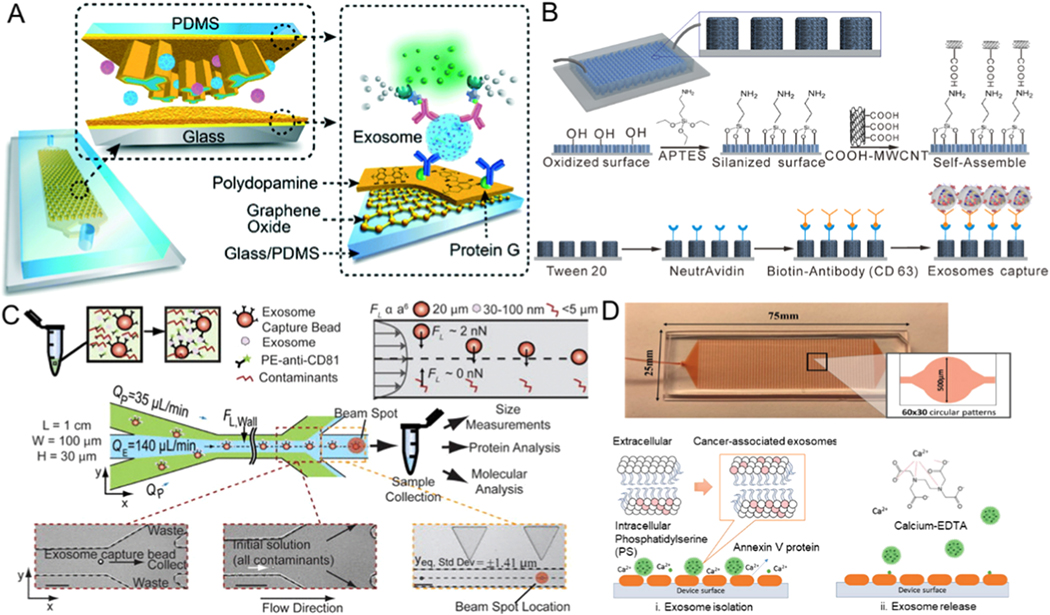
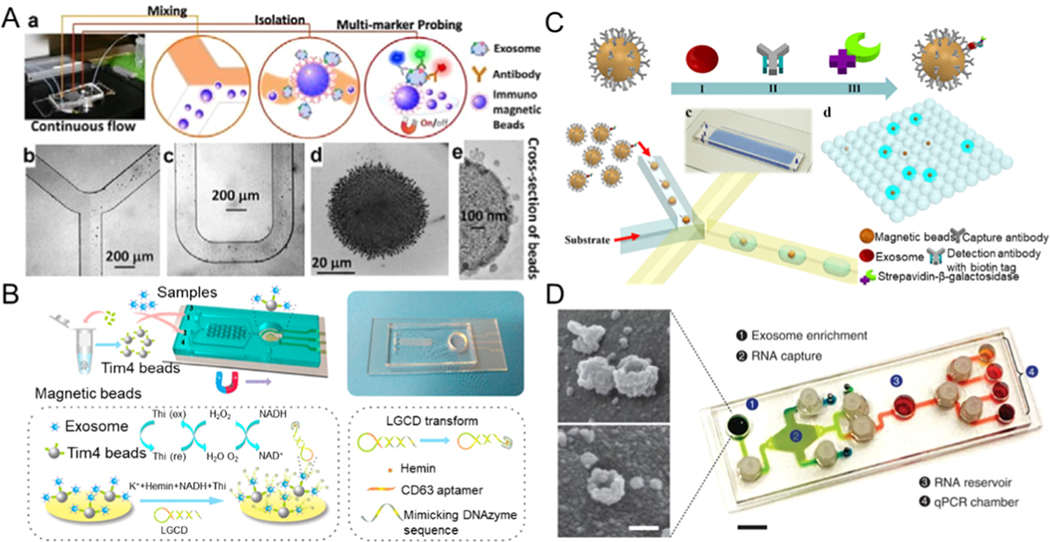
For example, a nano-IMEX device composed of Y-shaped micropillar arrays has been demonstrated to isolate exosomes. Micropillars were functionalized using graphene oxide and by further coating polydopamine on these micropillar surface interfaces to enlarge the capture surface and to improve mixing of the sample in the device (Figure 2A) [50]. This platform can separate exosomes from 2 μL plasma with a sensitivity of 50 μL−1 (80 aM). To further increase the separation efficiency and shorten the time of isolation, a three-dimensional microfluidic chip that is composed of multi-walled carbon nanotubes (MWCNTs) and functionalized arrays of polydimethylsiloxane (PDMS) pillars has been reported (Figure 2B) [52]. Through the use of anti-CD63 antibodies on MWCNTs surfaces to immobilize exosomes and optimizing the geometry of the chip for isolating exosomes, effective separation of exosomes can be achieved from a small quantity (400 μL) of cell culture medium at a flow rate of one milliliter per hour.
Figure 2.
Microfluidic exosome isolation based on immunoaffinity. (A) Capturing of exosomes on a GO/PDA nanoroughness interface in the nano-IMEX platform. Reprinted with permission from Ref. [50]. Copyright 2016, Royal Society of Chemistry. (B) Schematic illustration of the process of chemical modification with anti-CD63 antibody on 3D MWCNTs functionalized PDMS micropillars for immunocapturing of exosomes. Reprinted with permission from Ref. [52]. Copyright 2017, American Chemical Society. (C) Exosomes captured using magnetic beads and combining the complex with inertial lift forces in a microfluidic channel to further purify the isolated exosomes from other cellular contaminants. Reprinted with permission from Ref. [55]. Copyright 2015, American Institute of Physics. (D) 3D ripple-like structure chip for the immunocapture and release of exosomes based on the interaction between annexin V and phosphatidylserine. Reprinted with permission from Ref. [57]. Copyright 2019,Wiley.
Dudani et al. demonstrated a microfluidic chip for exosome analysis where the exosomes were immunocaptured by beads (Figure 2C) [55]. Polystyrene beads were functionalized with anti-CD63 antibodies to isolate exosomes, and the exosome-bead complex was injected into the optimized microfluidic device that can achieve rapid exosome purification through inertial force-induced solution exchange. Although the bead-based method does not have a limit on sample volume, achieving the efficient release of captured exosomes from the antibody-functionalized beads requires dissociation of the strong interaction between the antibody and the antigen. This is generally accomplished by trypsinization to break the cross-links between proteins, which compromises the structure of the exosome by damaging its surface membrane proteins. Cleavable link-based antibody immobilization approaches, such as chemical modification of dithiobis (sulfosuccinimidylpropionate) between the antibody and the capture area, allow exosome release for downstream studies and functional applications [56].
In addition to modifying the chip surfaces or beads with CD63 to capture specific exosomes, alternative methods to target specific exosomal lipids were also investigated recently. Kang et al. presented an annexin V immobilized microfluidic device integrated with alternating 3D ripple-like structures to enhance the contraction of the surface between exosomes and phosphatidylserine-targeting molecules (Figure 2D) [57]. The device achieved high (90%) separation efficiency for cancer cell-derived exosomes isolated in spiked experiments, compared to 38% separation efficiency for normal cell-derived exosomes. Further, the immune-captured exosomes were quickly released by chelation of Ca2+ at a flow rate of 1mL/h for downstream analysis. Although immunoaffinity-based microfluidic platforms allow specific isolation of exosomes at high purity, only those exosomes that contain specific protein markers or targeted phosphatidylserine expression on their surfaces can be separated by this method. The process to separate exosomes by flow from surfaces further dilutes the exosome concentration. Moreover, the hydrophobic surface of the PDMS surface can also cause the non-specific adsorption of proteins and exosomes, which might reduce isolation performance. The rapid depletion of reagents owing to the high surface-to-volume ratio of microfluidic platforms should also be taken into account for microchannel surface chemistry.
To overcome the limitations of affinity-based isolation methods, increasing research effort is now focused on label-free separation approaches on microfluidic devices without the use of an antibody or biological markers. These methods are based on physical and mechanical properties, size, electrical characteristics, and deformability of exosomes. Label-free microfluidic strategies for exosome separation show promise for isolating exosomes without compromising the morphological structure and biological composition or activity of the exosomes. In the following sections, we highlight several exciting and representative label-free microfluidic devices for exosome separation and their promising biomedical and clinical applications.
4.2. Microfluidic Devices for Label-free Separation of Exosomes
4.2.1. Microfluidic-Based Separation of Exosomes by Filtration
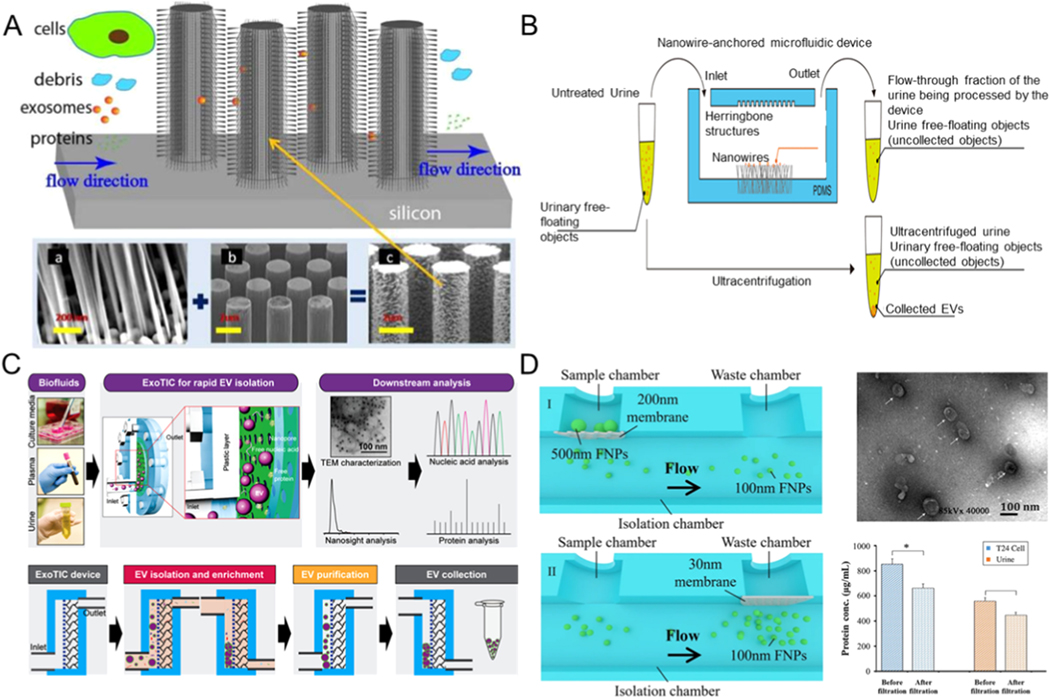
Similar to the traditional ultrafiltration approach, the filtration method of exosome separation based on microfluidics also involves the use of nanomembranes or nanowires [58–61]. The nanoporous membrane or nanowires permits particles that are smaller than the nanopores or the spacing in nanowires to be filtered while keeping other particles out [62]. Wang et al. applied a novel structure of porous silicon nanowires to isolate exosomes (Figure 3A) [63]. The fabricated microfluidic equipment captured exosomes between 40 and 100 nm in diameter while filtering out other extracellular vesicles, proteins, and cell debris. With a sample volume of 30 μL, the retention rate of 83 nm lipid vesicles was sixty percent, while that of larger vesicles of 500 nm diameter was only ten percent. The captured exosomes could be recovered after the porous silicon nanowires were dissolved in PBS buffer. However, when samples of larger volumes were used, the recovery rate declined due to the saturation effect on the surfaces. Using a similar strategy, a three-dimensional PDMS anchored with ZnO nanowires was designed to achieve exosome capture in the device (Figure 3B) [64]. It could collect exosomes with a diameter of ~ 30–200 nm from 1mL urine, providing a robust tool to improve the early diagnosis of urine-related diseases.
Figure 3.
Microfluidic exosome isolation based on filtering mechanisms. (A) Exosome separation by microfluidic device composed of ciliated micropillars. Reprinted with permission from Ref. [63]. Copyright 2013, Royal Society of Chemistry. (B) Schematic illustrations for the exosome isolation from urine using a microfluidic device anchored with ZnO/Al2O3 core-shell nanowire. Exosomes can be captured using these nanowires through electrostatic interactions. Reprinted with permission from Ref. [64]. Copyright 2017, American Association for the Advancement of Science. (C) Schematic description of the ExoTIC platform for exosomes isolation. Reprinted with permission from Ref. [65]. Copyright 2017, American Chemical Society. (D) Schematic of exosome isolation using microfluidic device combined with double-filtration. Reprinted with permission from Ref. [62]. Copyright 2017, Nature Publishing Group.
For label-free separation of exosomes, Liu et al. proposed an Exosome Total Isolation Chip (abbreviated as ExoTIC) to isolate exosomes on the basis of their size differences [60]. This chip is simple, modularized, and can easily handle various types of samples, including cell culture media, saliva, serum, urine, and plasma. The platform is macrofluidic in scale and sealed with plastic gaskets and metal screws to ensure leak-free connections and permit high throughput processing of the sample at 5 mL/h. This device can isolate exosomes from finger prick volumes of blood (10–100 μL) with yields that are around 4- to1000-fold higher than that of ultracentrifugation, making the ExoTIC ideal for point-of-care exosome-based clinical testing (Figure 3C) [65]. Another double-filtration microfluidic system for isolating exosomes of a certain size from urine samples was developed by Liang et al. [62]. They used a 200 nm nanopore membrane for preserving larger impurities and vesicles and removing soluble proteins utilizing another membrane with 30 nm pores (Figure 3D).
Physically capturing exosomes using nanowires and nanopores appears to be a prospective label-free method, especially owing to its relatively higher isolation efficiency and faster processing than ultracentrifugation and the ability to retain the native state of exosomes. However, further off-chip downstream analysis is necessary because exosomes can easily be separated together with microvesicles having similar membrane surface and size features. Moreover, exosome aggregates may block the nanopores of the filter, affecting membrane life and reducing the efficiency of exosome separation. Increasing the active filtering area will address some of these challenges to optimize the device operation with various samples and exosome concentrations.
4.2.2. Microfluidic-Based Separation of Exosomes by Hydrodynamic Properties
Continuous-flow sorting depends on the physical constraints of a microfluidic platform to enable exosomes to follow the streamline to flow continuously through the channels. The hydrodynamic constraints can be produced by arrays of nano-pillars or mechanical characteristics in the device [66]. Centrifugal effects result from dean flow in curved microchannels and will influence the position of suspended particles with surroundingforces [67, 68]. Currently, there are mainly two major microfluidic techniques that utilize hydrodynamic properties to separate exosomes: viscoelastic-flow sorting and deterministic lateral displacement (DLD) [69].
Deterministic lateral displacement is a technology that utilizes the specific arrays of nanopillars within a microfluidic channel to generate an asymmetric laminar fluid bifurcation. Particle flows through the DLD array are affected by both the laminar flow forces and the effect of nanopillar obstacles. When the particle is positioned at the gap of nanopillars, particles with a radius smaller than the first width of the streamline will follow a zigzag path with laminar flow, while particles that are larger than the first width of the streamline will be displaced laterally across the array of nanopillars. Therefore, the first streamline width is a critical diameter (Dc), and the Dc can be geometrically determined by placing an array of nanopillars in microchannels by photolithography. The DLD technique can precisely and continuously control trajectory and separate particles larger than the critical diameter (Dc) from particles smaller than Dc with high resolution [70].
Utilizing the DLD technique, Wunsch et al. designed an exosome separation method based on lateral displacement to isolate exosomes from urine, as shown in Figure 4A [70]. By optimizing the parameters of the array of nano-pillars, exosomes (diameter <100 nm) can be separated with a resolution as low as 10 nm. This label-free exosome separation approach is non-destructive and provides an ideal and novel way to separate and quantitatively analyze exosomes in trace samples (0.72 μL) [71, 72]. However, the flow rate of this device with a single array is very low (0.1–0.2 nL/min) owing to its high hydrodynamic resistance [73]. To increase the flowrate, Smith et al. reported an integrated platform with over 1000 nano-DLD parallel arrays and achieved higher throughputs (15 μL/min), applying technology more appropriate to the targeted exosome separation capacity (Figure 4B) [74]. It can isolate exosomes with a diameter of ~ 30–120 nm from urine and serum samples, and the recovery yield is ~ 50%. This strategy exploits the possibility for high-resolution analysis of exosomes within a selected size range. However, the versatility of the device in use with various bodily fluids needs further evaluation because its fabrication involves a complex photolithographic process.
Figure 4.
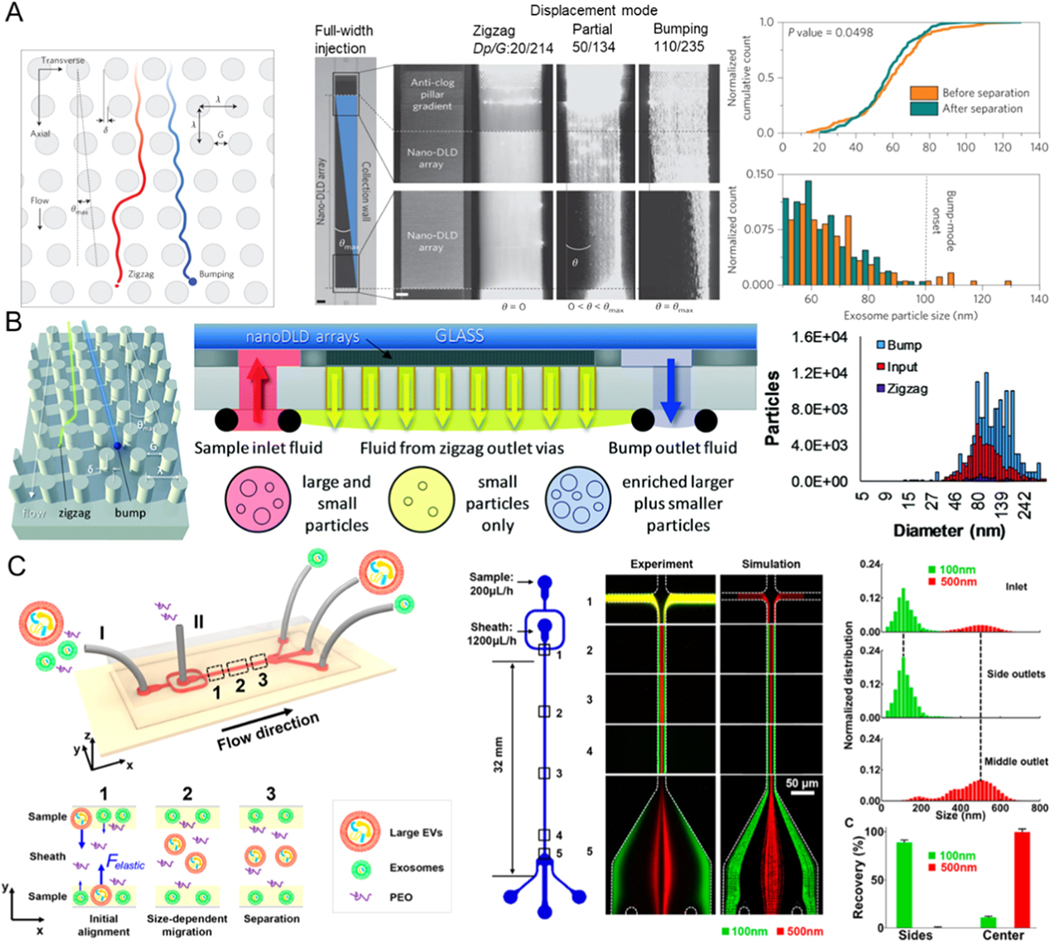
Microfluidic exosome isolation based on hydrodynamic properties. (A) Exosome isolation by microchip is composed of arrays of the DLD pillar. Reprinted with permission from Ref. [70]. Copyright 2016, Nature Publishing Group. (B) Separation of exosomes using microfluidic platforms combined with nanoscale DLD arrays. Reprinted with permission from Ref. [74]. Copyright 2018, Royal Society of Chemistry. (C) Exosome separation based on viscoelasticity on a chip. Poly(oxyethylene) as a sheath fluid is introduced from inlet II, while the sample is introduced from inlet I. Exosomes are collected at the side outlet, while larger vesicles are flowed to the middle outlet. Reprinted with permission from Ref. [85]. Copyright 2017, American Chemical Society.
For viscoelastic flow sorting, separation of different sizes of particles is achieved by utilizing different actions of elastic lift flow forces [75]. This technique has been used for label-free, non-sophisticated manipulation and separation of micro/nanoparticles such as microspheres [76–78], tumor cells [79–81], droplets [82], bacteria [83, 84], and blood cells [79]. Although the viscoelastic-based microfluidic separation method has many advantages, it cannot generate a sufficiently strong viscoelastic power on the nanoparticles.
To overcome this limitation, Liu et al. proposed to use the polymer polyoxyethylene (PEO) as an additive to the medium to change the viscoelasticity of the fluid (Figure 4C) [85]. PEO can increase the microfluidic viscoelasticity and thereby increase the viscoelastic force acting on exosomes. EVs of larger size are guided to the channel center by a more significant viscoelastic force. In contrast, exosomes are moved to the sides of the microchannel by controlling the sheath flow rate of the inlet during the separation process. The smaller exosomes are subject to limited viscoelastic effects, and their displacement is small so that exosomes and larger EVs can move to different outlets. This device attained high purity of exosomes up to 90% and high recovery efficiency up to 80% when used with fetal bovine serum or cell culture media. Sun et al. reported a microfluidic co-flow platform with Newtonian sheath fluid and viscoelastic sample fluid to isolate exosomes based on size within 30 min. They analyzed the surface protein profiles (HER2 and EpCAM) of their isolated exosome subtypes [86]. This system provides a robust tool to determine the heterogeneity of exosomes at the single exosome level. The exosome separation efficiency of this device can reach 96%, and the recovery efficiency can reach 91%. For their high-throughput and simplicity, viscoelastic microfluidics may offer a promising avenue for different types of exosome-related biomedical and clinical applications, where further verification with clinical samples are required. Moreover, the current viscoelastic microfluidic separation techniques require the use of a cleanroom and complicated fabrication processes. For example, high-resolution photolithography is needed to fabricate nanopillar arrays for DLD-based microfluidic platforms.
4.2.3. Microfluidic-Based Separation of Exosomes by Acoustic Fields
Acoustic waves possess a high level of biocompatibility and can be controlled with precision [87]. Therefore, acoustic waves are a well-recognized means for size-based separation of particles [88]. Standing acoustic waves are generated using an interdigitated microelectrode that converts electrical signals into propagating mechanical stress that travels along the surface of the piezoelectric substrate material. The standing acoustic waves inside the microfluidic channel can generate a series of pressure nodes. The principle for acoustic-based separation is that particles flowing through the channel will be encountering the pressure node. The force will guide the particles slightly off the center of the channel. The distance of the particle displacement depends on its properties, including compressibility differences and particle size. Therefore, particles of different sizes can be moved to different outlets to achieve particle separation [89–91]. Wu et al. presented a separation platform that integrated surface acoustic waves (SAW) with microfluidic elements to separate exosomes (~ 100 nm) from 500 μL whole blood samples without any contact and label (Figure 5A) [92]. It is composed of a module for cell-removal and a module for exosome-separation. In the first separation module, cell-sized particles are removed to enrich EVs. After this, EVs are flowed into the second acoustic isolation module and are further purified by removing the other nano-sized vesicles, including apoptotic bodies and microvesicles (MVs). This device that combines acoustics and microfluidics can, to a great extent, simplify the pre-processing of a complex blood sample and achieve high efficiency (99%) with an isolation throughput of 10 μL/min in exosome separation in a biocompatible manner, as the sound waves used in the process are mild. Exosomes only need to be subjected to the sound waves for separation for several seconds. Since this method uses size and the characteristics of the acoustic impedance of the objects for their separation, the other components in the plasma of size and acoustic impedance characteristics similar to those of the exosomes would inevitably interfere with the separation. Moreover, concentrating the exosomes in a small sample volume is difficult. The authors then used this platform to isolate exosomes (~ 22–200 nm) from 100 μL of saliva with 11 μL/min isolation throughput (Figure 5B) [93].
Figure 5.
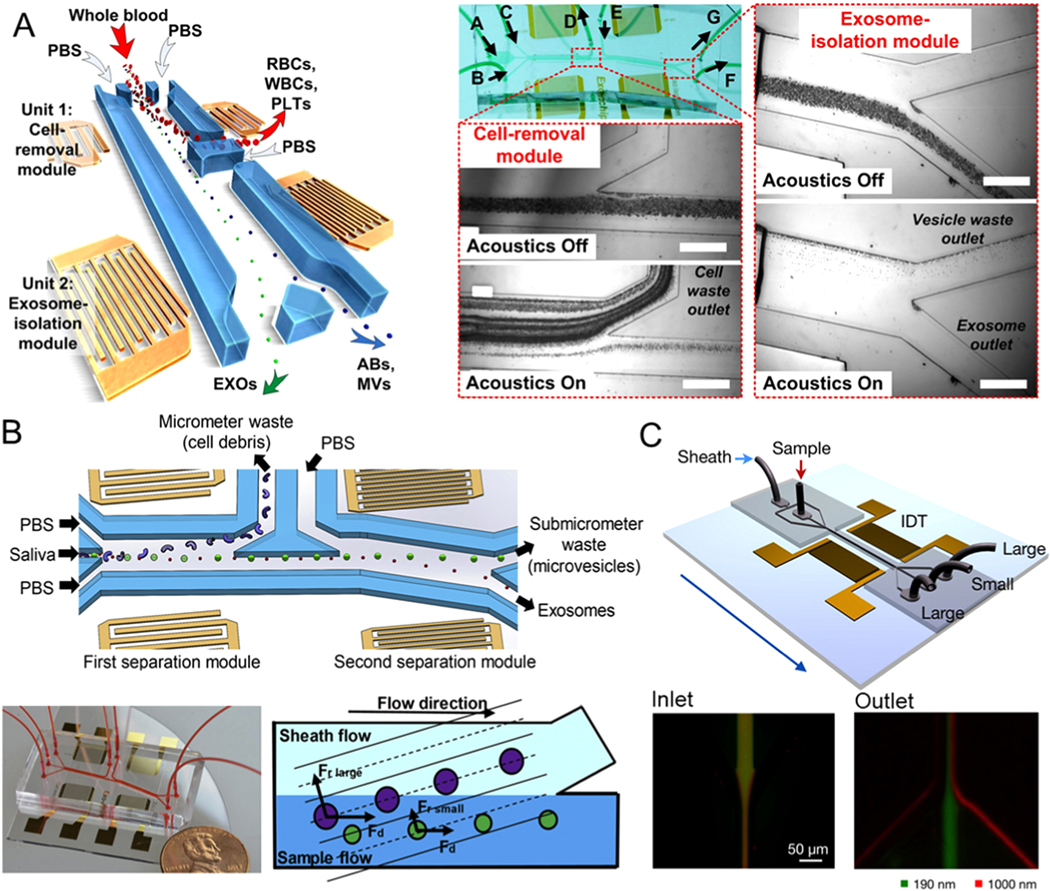
Microfluidic exosome isolation based on the acoustic field. (A) Schematic of the microfluidic device integrated with acoustic mode for label-free isolation of exosomes. Poly(oxyethylene) as a sheath fluid is introduced from inlet II, while the sample is introduced from inlet I. Exosomes are collected at the side outlet, while larger vesicles are flowing to the middle outlet. Reprinted with permission from Ref. [92]. Copyright 2017, National Academy of Sciences. (B) Schematic and optical image of the acoustic fluidic device for salivary exosome separation. Poly(oxyethylene) as a sheath fluid is introduced from inlet II, while the sample is introduced from inlet I. Exosomes are collected at the side outlet, while larger vesicles are flowing to the middle outlet. Reprinted with permission from Ref. [93]. Copyright 2020, Elsevier. (C) Illustration of the microfluidic device combined with acoustic nano-filter to label-free and size-specifically isolate exosomes. The standing acoustic waves are generated by the interdigitated transducer electrodes. Larger microvesicles are collected at the two side outlets, while smaller exosomes are collected at the middle outlets. Reprinted with permission from Ref. [94]. Copyright 2015, American Chemical Society.
Lee et al. presented a microfluidic platform using ultrasonic transducers as well as electronics to isolate and analyze exosome size, specifically in a contact-free and continuous manner (Figure 5C) [94]. Interdigital transducer (IDT) electrodes were uniformly patterned on the platform and used to produce surface acoustic waves across the direction of the flow. To acquire binary isolation for specific exosomes, a theoretical analysis model to adjust the cutoff size is further established. A higher force of radiation was exerted on particles larger than the size threshold, and these were redirected at the acoustic pressure modules, while the remaining smaller particles were enriched at the outlet. The exosome (diameter <200 nm) separation efficiency of this system is up to ∼90%, and the isolation throughput is around 1.68 μL/min. It is expected that the system would be integrated with other techniques for the separation of a particular subpopulation of exosomes, as well as the design of various SAW-fluid interaction geometries. However, complex fabrication processes are one of the limitations of this method, and the isolation throughput, especially for SAW-based microfluidic techniques, would need to be improved.
4.2.4. Microfluidic-Based Separation of Exosomes by Electrical Properties
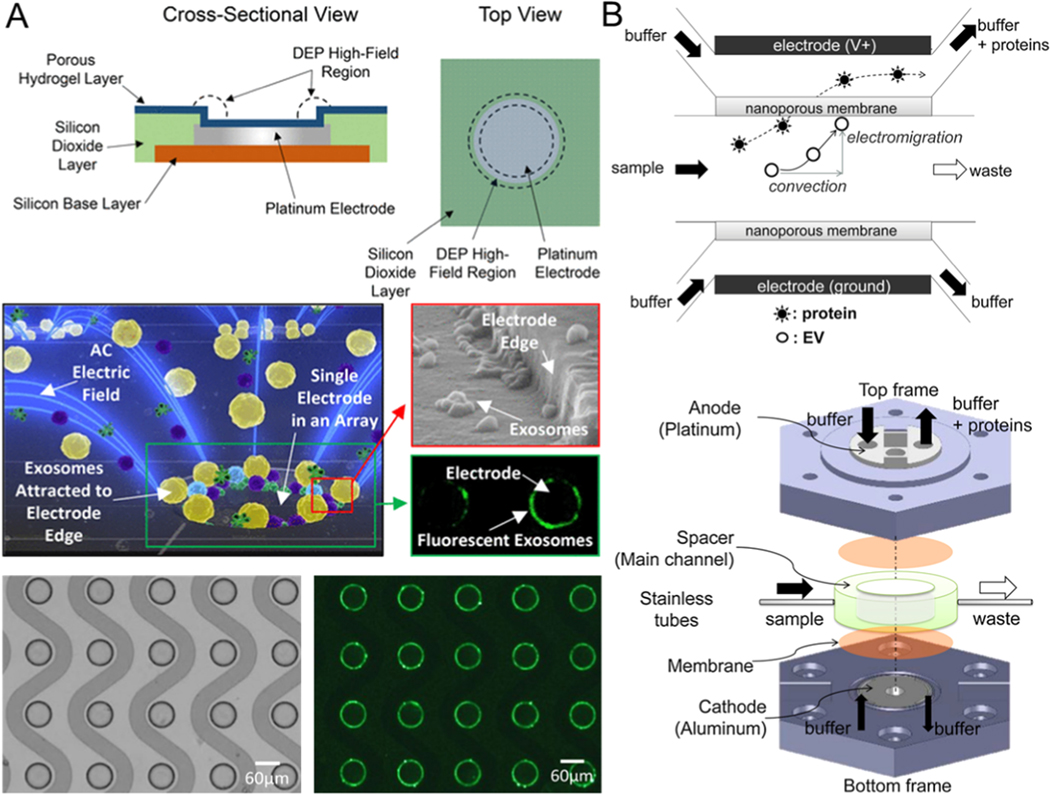
The application of AC voltage to the microelectrode placed in the solution can produce electro-hydraulic dynamic phenomena such as electroosmotic flow, dielectrophoresis, and electrothermal flow on the electrode’s surface [95–97]. This electro-hydraulic phenomenon on the electrode surface is widely used in microfluidic mixing [98–101], micro- and nano-particle manipulation [102–104], and other applications [105–109], because the phenomenon can be flexibly controlled by altering the amplitude and the frequency of the AC voltage. AC electroosmosis is suitable for working with low-conductivity solutions because the electric double layer formed on the surface of the electrode has a small density if the solution has a relatively high conductivity. This weakens the effect of AC electroosmosis, and exosome isolation from cell culture and serum becomes difficult [110–112]. Dielectric particles can be polarized in the non-uniform electric field and interact with this field to produce dielectrophoretic (DEP) force that causes the directional movement of polarized particles [113]. Since the DEP force is correlated with the dielectric properties and the size of the manipulated particles, a specific electrode structure needs to be designed to apply dielectrophoresis for the separation and purification of exosomes from blood. Heineck et al. observed that the planar electrode array model was not affected by electric heating flow under high conductance conditions. This suggested a direction for isolating exosomes from highly conductive biological fluids such as blood [114].
Ibsen et al. designed an AC motorized microarray chip (Figure 6A) [115] based on dielectrophoresis technology. It can separate and recover glioblastoma exosomes quickly from an undiluted blood sample. Exosome separation is achieved because the exosomes and other components in plasma have different dielectric properties so that they are subject to varying dielectrophoretic forces. Exosomes are attracted to the area around the edges of a microelectrode where the electrophoretic field is robust, whereas cells and macromolecular proteins are pushed into the areas of weak dielectrophoretic field between the electrodes. The dielectrophoretic force separation gives a high yield of exosomes (50–150 nm) from undiluted human plasma samples (30–50 μL) with ~ 2–3.3 μL/min isolation throughput in a short time (within 15 min). However, a potential disadvantage of this method is that the structure of exosomes and the exosomal content may be affected because exosomes come in direct contact with the electrodes. One possible solution to this problem is to build a porous hydrogel layer on the electrode array so that the exosomes do not come in direct contact with the electrode.
Figure 6.
Microfluidic exosome isolation based on dielectrophoretic. (A) Dielectrophoresis-driven exosomes isolation by the microfluidic device composed of the microarray of ACE. Reprinted with permission from Ref. [115]. Copyright 2017, American Chemical Society. (B) Schematic of microfluidics combined with a filtration system to isolate exosomes by electrophoretic force. Proteins smaller than the membrane pore size can be propelled through the membrane while EVs larger than the membrane pore size cannot diffuse into the membrane and are captured on the nanoporous membrane. Reprinted with permission from Ref. [59]. Copyright 2016, Elsevier.
Another example of an electrical property-based exosome isolation technique is by leveraging electro-kinetic filtration to allow electrophoretic sorting based on charge-mass ratios and different sizes of exosomes. This eliminates the need for any external pumps to move the fluid. Davies et al. reported a microfluidic nano-porous membrane filtration system driven by electrophoresis that can separate exosomes [58]. Negatively charged exosomes in this device can be driven through the membrane by electrophoretic force under the application of bias voltage across the membrane. It can prevent the clogging of the pores in the membrane because the continuous crossflow parallel to the filter membrane can wash away larger particles and improve the purity of the isolate. On the other hand, the electrophoretic force can push the positively charged proteins in the opposite direction. Unlike in conventional filtration methods, this device controls the size of nanopores in the membrane by altering the concentration of pore-forming solvent to prepolymer solution so that exosomes of the required size are separated and extracted. To further improve isolated exosome purity, DC electrophoresis was utilized to push particles through the membrane and achieve increasing separation efficiency of the vesicles from the free-floating proteins in solution. It can eliminate the interference of some soluble proteins and improve purity by using electrophoresis-driven filtration. Cho et al. integrated microfluidics with nanoporous membranes to isolate larger exosomes by electrophoretically migrating proteins of molecules sizes smaller than the exosomes through the membrane (Figure 6B) [59]. Compared to conventional ultracentrifugation, the recovery rate of this platform achieved 65% on the basis of the quantities of RNA obtained (around 8X better than that achieved with ultracentrifugation), and the separation efficiency was up to 83.6% based on the quantities of protein removal. However, the major limitation of this approach is the formation of gas bubbles on the electrodes when the voltage of the electrical field is greater than 7 V per centimeter (the layer of bubbles narrows the distance between the electrodes, hindering the flow of the solution through the channel in this platform). Moreover, heavy benchtop instrumentation is required for the generation of electric fields, and possible heating of the solution is also a limitation of this electrical property-based exosome separation technology.
5. Integrated Microfluidic Platforms for Exosome Detection and Analysis
Liquid biopsy analysis of tumors provides an ideal diagnostic and prognostic tool for molecular cancer diagnosis and for choosing treatment options. This promises a shift in medical paradigms towards personalized medicine. For use in liquid biopsies of tumors, exosome detection shows promise of revolutionizing prediction and treatment applications. It is a promising technique for diagnosing various other illnesses, including neurodegenerative diseases, cancer, infections, and autoimmune diseases [16–18]. Several recently reported technologies can be applied successfully to quantify the concentration as well as size distributions of exosomes, such as ELISA (Enzyme-linked immunosorbent assay), nanoparticle tracking analysis (NTA), and flow cytometry technologies [116–119]. However, when detecting highly heterogeneous exosomes in complex body fluids, it is hard to avoid “biological noise” (i.e., non-specific adsorption of biomolecules). Moreover, slow throughput, extensive and complex sample preparation, high cost of equipment, and operating cost are the disadvantages of these conventional approaches.
The growing field of microfluidics is transforming current conventional, quantitative methods of detecting exosomes and their molecular characterization into portable, integrated platforms because of their unique advantages of high throughput and sensitivity, use of small quantities of reagents and sample volumes, and low cost (Table 3) [35, 51, 120, 121].
Table 3.
Microfluidic devices for exosome detection and analysis.
| Microfluidic Detection | Sample | Throughput (μL/min) | Limit of Detection | Measurement | Disease | Ref |
|---|---|---|---|---|---|---|
| Colorimetric based exosome detection | ||||||
| Sequential Exodisc: -Double filtration isolation -Colorimetric ELISA |
Urine | 16.7 | N/A | Overall levels of CD81 and CD9 | Bladder cancer | [60] |
| Sequential stages: -Double filtration isolation -Colorimetric on-chip ELISA |
Urine | 17.2 | N/A | Overall levels of CD63 | Bladder cancer | [62] |
| Electrohydro-dynamic flow assisted immuno-capture stages: -Electric field driven analyte transport -Colorimetric ELISA |
Cell culture media | 7 | 2760 exosomes/μL | Overall levels of CD9 and HER2 | Breast cancer | [122 |
| Magnetic bead-based exosome detection | ||||||
| Sequential immunomagnetic stages: -immunomagnetic isolation -ELISA of intra-vesicular protein |
Plasma | 0.3 | 0.281 pg/mL | Phosphoryl ation levels of IGF-1R | Non-small-cell lung cancer | [51] |
| μNMR device -Immunomagnetic tagging -miniaturized micronuclear magnetic resonance system detection |
Blood | N/A | >104 exosomes/μL | Overall levels of CD63 | Glioblastoma tumor | [129] |
| ExoPCD-chip -Immunomagnetic enrichment - In situ electrochemical analysis |
Serum | ~0.14 | 4.39 × 106 exosomes/μL | Overall levels of CD63 | Liver cancer | [130] |
| Droplet digital ExoELISA -Immunomagnetic tagging -Fluorogenic ELISA |
Plasma | N/A | ~5 exosomes/μL | Protein levels of Glypican-1 | Breast cancer | [22] |
| iMER device - Immunomagnetic isolation - Exosome RNA analysis |
Blood | ~0.83 | ~1011 exosomes/μL | miRNA levels of EPHA2, EGFR, PDPN | Glioblastoma tumor | [131] |
| Surface plasmon resonance (SPR) based exosome detection | ||||||
| SPRi device - antibody microarray isolation - surface plasmon resonance imaging and detection |
Ascites | 300 | ∼4.87 × 107 exosomes/cm2 | Protein levels of CD9, CD81, CD82, and E-cadherin | Ovarian cancer | [144] |
| nPLEX biosensor - exosomes bind to antibody functionalized nanoholes - Detect the spectral shifts and intensity changes induced by exosomes binding |
Ascites | 10 | ~3,000 exosomes | Protein levels of CD 63, mRNA levels of GADPH | Ovarian cancer | [145] |
| Electrochemical property-based exosome detection | ||||||
| iMEX platform - Immunomagnetic enrichment - profile through the electrochemical reaction |
Plasma | ~0.16 | <105 exosomes | Protein levels of CD63, EpCAM, CD24, and CA125 | Ovarian cancer | [158] |
| Aptasensor for Electrochemical detection | Cell culture media | ~10–400 | ~1012 exosomes/μL | Protein levels of CD63 | Liver cancer | [159] |
| Signal amplified electrochemical aptasensor | Plasma | N/A | 9.54 × 105 exosomes/μL | Protein levels of MUC1 | Gastric cancer | [169] |
| Aptamer recognition-induced multi-DNA release and cyclic enzymatic amplification-based electrochemical detection | ultra centrifuged fetal bovine serum | N/A | 70 exosomes/μL | Amount of multimessenger DNAs (mDNAs) | Prostate cancer | [170] |
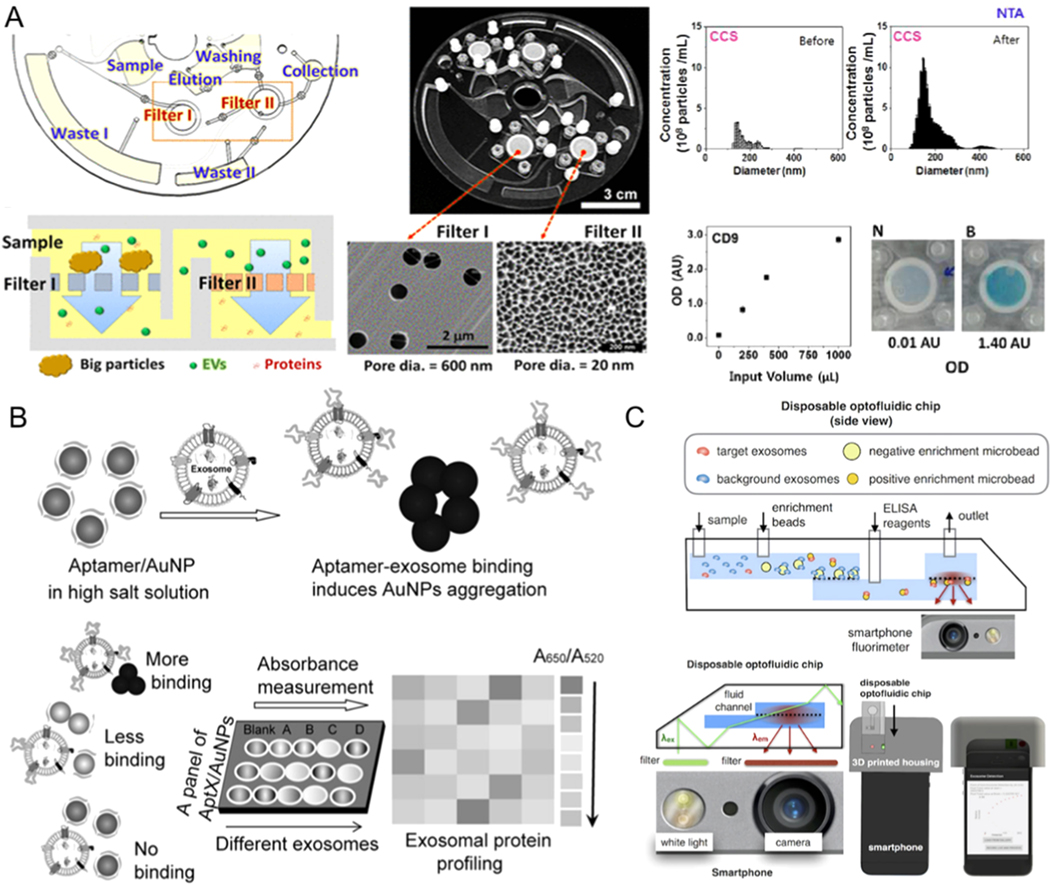
5.1. Colorimetric-Based Exosome Detection
Microfluidic methods that simplify the equipment with the use of direct colorimetric detection of exosomes have been developed in recent years [50, 61, 62, 122–124]. ELISA-on-chip can process immunological assays at high-throughput without the need for extensive training, electrical power, regular maintenance, and costly assays required for conventional ELISA diagnostic platforms, which makes them suitable for resource-limited or point-of-care settings. Integrated ELISA with microfluidics for exosome detection from a bladder cancer sample is shown in Figure 7A [60]. After the separation with filter, a lab on chip (LOC) ELISA was performed, and the OD values of samples at 450 nm were tested. The LOC offered an effective device to automate the modification, sequential capture, and detection operations. It can detect exosomes from urine samples with 16.7 μL/min throughput.
Figure 7.
Microfluidic exosome detection based on colorimetric mechanisms. (A) The microfluidic device combined with filter for exosome isolation and detection using ELISA. Reprinted with permission from Ref. [60]. Copyright 2017, American Chemical Society. (B)Biosensor for colorimetric profiling of exosomal proteins. The binding of exosomes with aptamer/gold nanoparticles complex in the high salt solution can induce the aggregation of gold nanoparticles and cause the absorbance displacement. Reprinted with permission from Ref. [125]. Copyright 2017, Wiley. (C) Schematic illustration of microfluidic-based mobile exosome detector (μMED). The quantitative data can be read with the camera of the smartphone. Reprinted with permission from Ref. [127]. Copyright 2016, Nature Publishing Group.
Jiang et al. demonstrated a multiplexed sensor platform, which was generated through assembling gold nanoparticles with a panel of aptamers to target exosome surface proteins that are either putative or ubiquitous (Figure 7B) [125]. The complexation of aptamers can protect AuNPs so that they do not aggregate in the high salt solution. However, the presense of exosomes can break the weaker, non-specific binding balance between AuNPs and the aptamers, resulting in AuNPs aggregation and changing the solution color from red to blue. This process can be monitored through absorption spectroscopy. By combining this process with a microfluidic chip composed of gold pattern arrays, different proteins on various kinds of cancer cell-derived exosomes (6.4 μg/mL) can be visually and quantitatively detected [121, 126].
Ko et al. fabricated a portable optofluidic platform and combined it with a smartphone to detect and profile exosomes secreted from the brain. The quantitative data can be read with the camera of a smartphone within an hour, which is nearly 10 times faster than conventional exosome detection methods (Figure 7C) [127]. The limit of detection of this device is about 107 exosomes/mL with a sensitivity of 73% and specificity of 71% for the detection of exosomes from mouse blood. Smartphone-based exosome biomarker readout optical platforms enable exosome detection from serum samples without using expensive equipment, which is suitable for remote settings.
5.2. Magnetic Bead-Based Exosome Detection
Exosomes can be trapped and labeled with beads and analyzed on a microfluidic sensor at high sensitivity [4, 51, 117, 128, 129]. Zhao et al. presented a microfluidic platform via continuous-flow, i.e., ExoSearch chip [53]. ExoSearch chip provides enrichment of plasma exosomes for an in situ analysis of exosomes through immune-magnetic beads (Figure 8A). They used three tumor markers (CD 24, CA-125, and EpCAM) combined with an ExoSearch chip to diagnose ovarian cancer in a non-invasive manner. Its accuracy and diagnostic ability tested with the standard Bradford assay proved to be very high (p = 0.001, a.u.c. = 1.0). However, to elute exosomes from bound antibodies in a mild elution condition is extremely difficult on account of strong antibody-antigen interactions. Phosphatidylserine (PS) on exosomal membranes can be identified easily through the PS-binding receptor Tim 4, which is seemingly necessary for exosomal germination from late endosomes. The capture of exosomes with Tim 4 immobilized magnetic beads is Ca2+-dependent, so exosomes can remain intact without their structure being easily destroyed by the use of a chelator for elution. Inspired by the newly found affinity method and considering the need for portability and sensitivity, Xu et al. proposed an ExoPCD chip. ExoPCD chip integrates the separation of exosomes in blood samples and their in situ electrochemical analysis [130]. The microchip consists of a series of Y-shaped micropillars and cascaded indium tin oxide (ITO) electrodes. PDMS micropillar arrays were designed in a repeating cross-mixed channel to achieve a better chance of entrapping the Tim 4 modified magnetic beads on the exosome membrane. It could detect exosomes from serum with a throughput of ~ 0.14 μL/min, and the limit of detection can reach up to 4.39 × 106 exosomes/μL. Exosomes can be detected and captured sensitively by the novel strategy of enriching magnetic particles on the top layer of the ITO electrode for the transduction of signals. Xu et al. demonstrated an electrochemical biosensor without the need for immobilization. The platform consists of mimicking the DNAzyme sequence with a hairpin structure and CD63 aptamer (named as LGCD transform). The hairpin of DNA opens and forms a G-quadruplex with hemin when the target CD63-positive exosomes are present. This G-quadruplex / hemin can be utilized with the HRP-mimicking DNAzyme and NADH oxidase at the same time. NADH oxidation generates H2O2, which can be continuously catalyzed to produce significant signal enhancement. (Figure 8B) [130]. This differs from the reported approaches of using protein-modified electrodes for detecting exosomes; these newly developed exosomal probes can be used in a microfluidic chip without the need for high-cost nucleic acid modifications, which also involve complex processes of immobilization and signal amplification. The current research shows that the ExoPCD chip effectively captures tumor-derived exosomes from a small 30 microliter sample. Liu et al. proposed an immunomagnetic droplet method to detect a single exosome to quantify the number of exosomes (Figure 8C) [22]. Exosomes were anchored to the magnetic micro-beads through a sandwich ELISA with a specific kind of enzymatic reporter with the capability of producing fluorescent signals. Next, the beads were separated and encapsulated with droplets. The volume of fluid formed into a droplet is just sufficient to encapsulate a single bead. Such a droplet-based digital ELISA method gives results of unprecedented accuracy in absolute measurement of cancer-specific exosomes. The apparatus can detect ten enzyme-tagged complexes of exosomes in every microliter (∼10–17 M).
Figure 8.
Microfluidic exosome detection based on magnetic beads. (A) A microfluidic platform for exosome detection combining exosome capture and fluorescent analysis. Reprinted with permission from Ref. [53]. Copyright 2016, Royal Society of Chemistry. (B) A microfluidic chip that features Y-shaped microcolumns for enhancing exosome labeling to detect exosomes. Reprinted with permission from Ref. [130]. Copyright 2018, American Chemical Society. (C) A droplet-based single-exosome-counting enzyme-linked immunoassay. Reprinted with permission from Ref. [22]. Copyright 2018, American Chemical Society. (D) A microfluidic platform that contains exosome RNA analysis and exosome enrichment units. Reprinted with permission from Ref. [131]. Copyright 2015, Nature Publishing Group.
Microfluidics also can be leveraged for non-invasive diagnostics. Recently, a microfluidic platform called iMER for immuno-magnetic in situ exosome RNA analysis has been reported. Following the separation of exosomes using magnetic beads, a lysis buffer was injected to release exosomal RNA. The magnetic beads transported the lysed RNA into another chamber for reverse transcription and finished the analysis of the target mRNAs by using RT-qPCR after adding the reverse transcribed DNA. This platform is not only comprehensive but also sensitive since it can detect mRNA amounts of glioblastoma multiforme markers in tumor exosomes isolated from blood using the magnetic beads. The limit of detection is reported as ~ 1011 exosomes/μL, and the throughput is ~ 0.83 μL/min. The analysis result of iMER is consistent with the conventional system test result (R2 = 0.986) (Figure 8D) [131]. This system combines into one microfluidic chip format the enrichment of exosomes by magnetic beads, lysis, the collection of RNA, and real-time PCR.
5.3. Surface Plasmon Resonance (SPR)-Based Exosome Detection
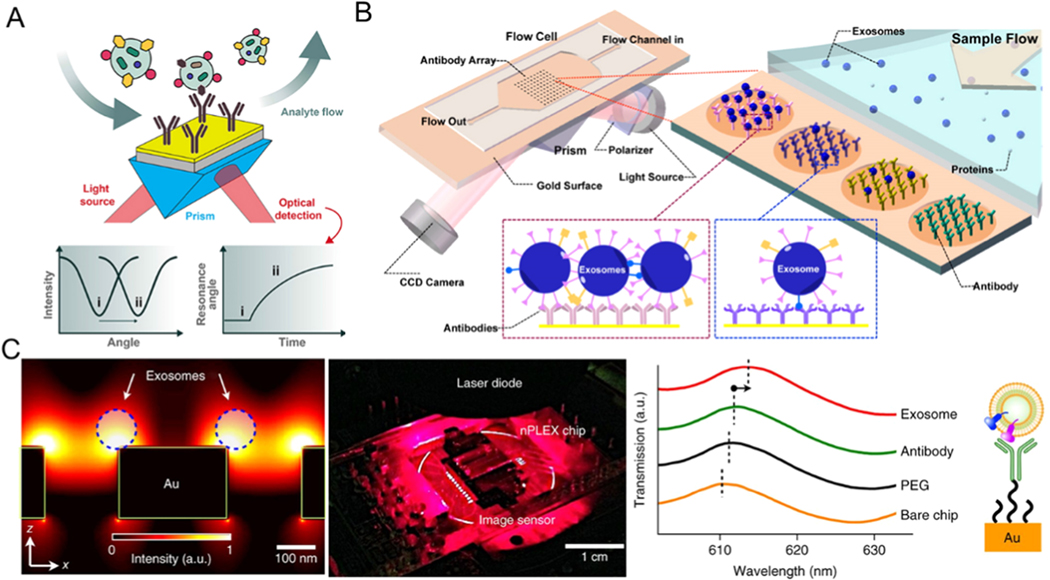
Surface Plasmon Resonance (SPR) is an optical-physical phenomenon. If polarized light enters the interface of two media with different refractive indices (such as silver and gold plating on a glass surface) at a critical angle, it can cause resonance of free electrons in the metal film, which dramatically reduces the reflected light at a specific angle [132–136]. The reflected light can completely disappear at a certain angle, and this angle is called the SPR angle. By monitoring the dynamic changes in the SPR angle during the biological response, specific signals for binding and interaction between biological molecules can be obtained (Figure 9A) [137].
Figure 9.
Surface plasmon resonance (SPR) based microfluidic exosome detection. (A) A typical SPR biosensor set-up. Reprinted with permission from Ref. [137]. Copyright 2019, Frontiers Media. (B) Schematic diagram of the detection of cancerous exosomes using SPR imaging with the combining use of antibody arrays. Reprinted with permission from Ref. [144]. Copyright 2014, American Chemical Society. (C) A nano-plasmonic sensor for detection and profiling of exosomes. Reprinted with permission from Ref. [145]. Copyright 2014, Nature Publishing Group.
In recent years, biomolecules (target molecules) have been coupled to a microfluidic surface, which is then injected with a solution containing another biomolecule (analyte) to flow over a functionalized surface [138–142]. By detecting the SPR angle change, the information about the analyte concentration, affinity of exosomes and functionalized surface, kinetic constant, and specificity can be obtained in real-time [143]. The combination of SPR and microfluidics has the advantages of high specificity, stability, reliability, and label-free detection.
Zhu et al. reported a microfluidic platform composed of exosome-specific antibody functionalized microarrays to capture exosomes from biological fluids. They utilized SPR imaging for label-free quantitative detection of exosomes and real-time monitoring of the exosome secretion (Figure 9B) [144]. The limit of detection can achieve up to ∼4.87 × 107 exosomes/cm2 with a throughput of 300 μL/min. Im et al. proposed a nanoplasmonic microfluidic platform, which contains nanopores functionalized with specific antibodies to capture exosomes (Figure 9C) [145] and can simultaneously profile various exosomal proteins based on the transmitted SPR changes on periodic nanohole arrays. This SPR-based microfluidic device provides a solution to the problem of identifying exosomes from different samples at the same time. The limit of detection can achieve ∼ 3000 exosomes with a throughput of 10 μL/min. SPR technology has laid the foundation for developing a simple, quick, highly sensitive, inexpensive, and highly specific tool to detect exosomes and profile them at a molecular level [146–151].
5.4. Electrochemical Property-Based Exosome Detection
Electrochemical assays have shown great promise for detecting trace amounts of biomolecules with high sensitivity and specificity in complex biological matrices by measuring changes in electrical signals of substances [152–157].
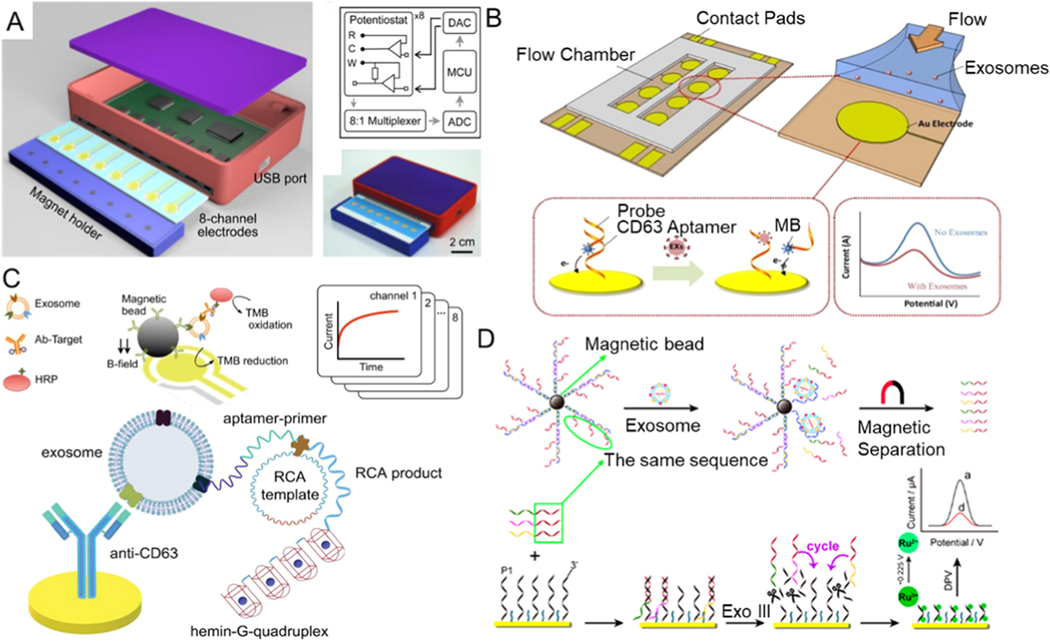
Recently, Jeong et al. designed a compact portable sensor that can identify exosomes in plasma within an hour and only needs a 10 μL sample (Figure 10A) [158]. It integrates a large magnet integrated with electrochemical analysis. A sensor captures exosomes from patient blood samples by an immunomagnetic bead-based sorting. Then, exosomes are detected through signal amplification with redox-active reporters and quantified by measuring the electrical currents at high-throughput. The limit of detection can achieve up to <105 exosomes with a throughput of ~ 0.16 μL/min.
Figure 10.
Microfluidic exosome detection based on electrochemical. (A) Schematic representation of the integrated iMEX platform. Reprinted with permission from Ref. [158]. Copyright 2016, American Chemical Society. (B) Schematic diagram of exosome detection by aptamer-based microfluidic. Reprinted with permission from Ref. [159]. Copyright 2016, Elsevier. (C) Schematic diagram of a sensitive aptasensor to detect cancerous exosomes. Reprinted with permission from Ref. [169]. Copyright 2019, Wiley. (D) Schematic illustration of an aptamer amplification strategy to detect exosomes. Reprinted with permission from Ref. [170]. Copyright 2018, American Chemical Society.
Zhou et al. designed an electrochemical biosensor that is aptamer-based to detect exosomes quantitatively (Figure 10B) [159]. Aptamers, as short ssDNA or RNA molecules, can bind to specific targets [160–163]. Exosomal transmembrane protein-specific nucleic acid aptamers and antibodies are often used as detection probes for exosomes, but for clinical practice, exosomes are still quite challenging to detect. The majority of detection probes that researchers use at present are CD63-specific aptamers or anti-CD63 antibodies [164–168]. Through immobilizing CD63-specific aptamers on gold electrode surfaces and integrating the functionalized electrodes with a microfluidic device, exosomes can be detected from cell culture media. The limit of detection can achieve ~ 1012 exosomes/μL with a throughput of ~ 10–400 μL/min.
Electrochemical assays have a wide range of advantages in the exosome detection field because of their small sample volume needed, low cost, and simplicity of these sensors. Electrochemical sensing can be combined with aptamers to detect exosomes using various signal generation and amplification strategies, such as DNA nano-tetrahedrons, metal nanoparticles, and nucleic acid-based amplification analysis. In some cases, the amplification analysis of nucleic acid needs higher temperatures and the heat can influence the activity of the exosomal content. Huang et al. reported a label-free electrochemical aptamer sensor that combines the hemin/G-quadruplex system with rolling circle amplification (RCA) to analyze exosomes of gastric cancer selectively and sensitively (Figure 10C) [169]. Researchers designed gastric cancer exosome-specific aptamers, and chemically functionalized them on the gold electrodes. Captured exosomes from gastric cancer samples produce a large number of G-quadruplexes through RCA. This product of the RCA reaction is then incubated with hemin to generate a hemin/G-quadruplex structure and eventually catalyzes H2O2 to create an electrochemical signal. Such an aptamer sensor is highly sensitive and selective for exosomes in gastric cancer. It can detect as few as 9.54 × 102 exosomes per milliliter, with a linear response range of 4.8 × 103 − 4.8 × 106 exosomes per milliliter, showing great potential to be leveraged as an effective device to diagnose gastric cancer at an early stage.
Dong et al. used a similar strategy to detect tumor-specific exosomes (Figure 10D) [170]. This strategy is on the basis of the release of multi-DNA and cyclic enzymatic amplification induced by aptamer recognition. First, nucleic acid aptamer magnetic bead biomolecules were utilized to capture exosomes, which resulted in messenger DNA release. The released messenger DNAs were hybridized with the probe DNAs after magnetic separation. The electroactive ruthenium(iii) hexamine (Ru(NH3)6 3+) was utilized as a signal reporter for its ability to attract DNA electrostatically. The concentration of Ru(NH3)63+ can modify the electrochemical signal relative to the level of messenger DNAs. The level of the released messenger DNAs is correlated with exosome concentration. Therefore, exosomes of the tumor are detected through the analysis of variations in the Ru(NH3)63+ peak current. The limit of detection can achieve down to 70 exosomes per milliliter.
5.5. Thermophoretic-based Exosome Detection
Thermophoresis is a phenomenon about the drift of suspended particles along an applied thermal gradient [171]. It occurs because the momentum transferred to the particles is different between gas molecules with high thermal velocity and gas molecules with low thermal velocity. The thermophoresis of particles is sensitive to the size of particles and the surface interactions with the surrounding medium [172]. Recent studies have demonstrated that thermophoretic effects can be utilized to manipulate and analyze particles, vesicles, and molecules in aqueous solutions [173]. Sun et al. presented a thermophoretic sensor combined with gold nanoflares to achieve exosome enrichment to amplify the fluorescence signal and realize in situ detection of exosomal miRNA [174]. It allows for direct quantification of exosomal miRNAs without additional RNA extraction, and the detection limit of exosomal miRNAs was 0.36 fM in 0.5 μL serum samples. Exosomal miRNA-375 was chosen as a well-recognized breast cancer biomarker, as their detection target. The result showed this approach could detect early stages of breast cancer (stage I and stage II) with 83% specificity and 88% sensitivity. The thermophoresis can also be leveraged to profile exosome surface proteins from 0.1 μL volume of clinical serum sample [175]. Aptamer-conjugated exosomes were rapidly accumulated (1400-fold) under thermophoretic force within 10 min. The enrichment of exosomes can also further lead to fluorescence signal amplification. The concentration of the target surface protein of exosomes can be detected via measuring the intensity of the fluorescent signal without any pre-isolation exosome steps. The overall detection can be finished within 3 hours, and the cost is reported as less than a dollar, offering promising potential for point of care testing.
6. Exosome-based Therapeutic Delivery for Cancer Therapy
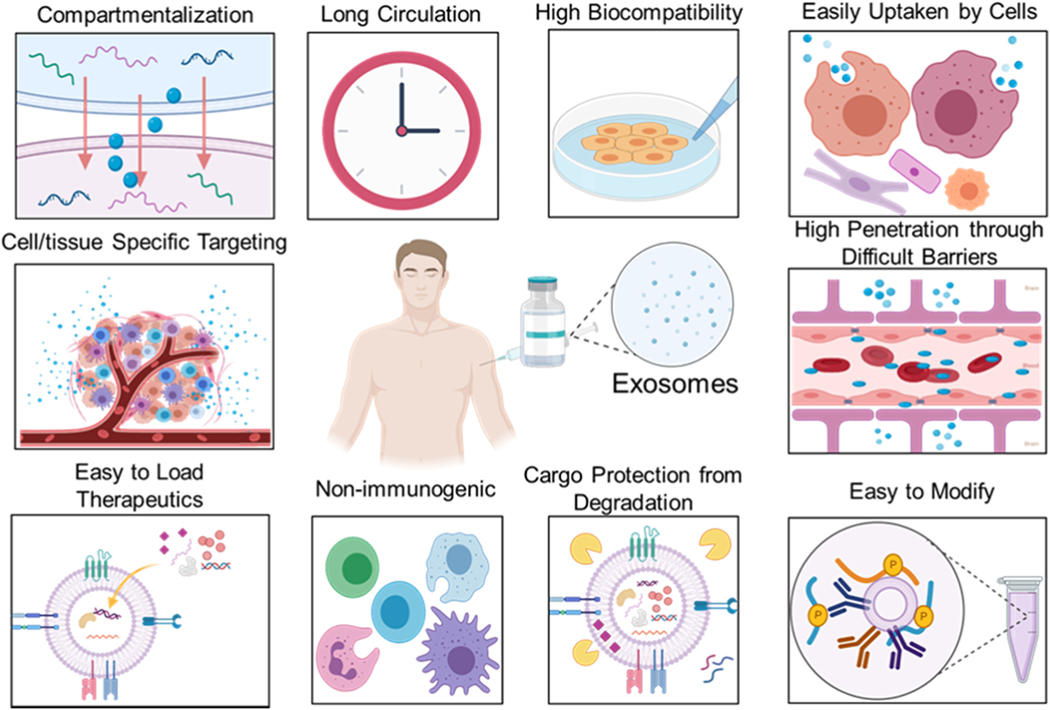
Exosome-based therapies include immunotherapy and tissue regeneration, but drug delivery is one of the most promising and investigated exosome-based applications [176]. With the development of modern medicine, more and more nanomaterials are being developed for use as drug delivery carriers to improve the stability of the encapsulated chemical or molecular drug, targeting ability, and therapeutic efficacy [177–179]. Exosomes, as natural nanocarriers released by cells, have favorable stability because of their phospholipid bilayer structure and satisfactory biocompatibility [180]. By their homing characteristics, exosomes can traverse long distance to deliver the encapsulated cargo to specific targets. Moreover, their enormous value derives from their other advantages that are difficult to create in synthetic nanomaterials, including their extensive distribution [181, 182], easy availability [183, 184], ability to participate in cellular communication [29, 185], cross-barrier transportability (plasma membrane and the blood/brain barrier) [186–189], and the inability of the reticuloendothelial system to clear them out (Figure 11) [190, 191].
Figure 11.
Advantages of exosome as a natural nano-carrier for therapeutic delivery.
Next, we will outline application of exosomes as drug delivery carriers for tumor treatment. So far, the drugs that are loaded into exosomes mainly include genes (mRNA, siRNA, miRNA) [189, 192–194], chemotherapy agents (e.g., doxorubicin, paclitaxel) [180, 184, 190, 195, 196], anti-inflammatory medications, antigens, proteins that can enhance immunity, and other types of drugs [197–200]. The current techniques for loading drugs into exosomes mainly include electroporation, sonication, chemical transfection, and cellular engineering (Table 4) [201].
Table 4.
Comparison of the strengths and limitations of therapeutic-loading nanotechnologies.
| Techniques | Advantages | Disadvantages |
|---|---|---|
| Electroporation | ✓Vector free ✓Effective for loading of hydrophilic cargos |
➣May affect surface zeta potential damage ➣Manufacturing challenges for scaling up |
| Sonication | ✓ small RNAs are easily encapsulation |
➣Drugs may simply adhere to the extraluminal surface ➣Risk of membrane deformation |
| Direct transfection | ✓ Effective for encapsulation of drugs with versatile potential |
➣ Transfection reagents may affect the membrane of exosome |
| Cellular engineering | ✓ Effective for gene modification and chemical editing in vitro | ➣ Risk of genotoxicity and cause adverse host immune response ➣ Time-consuming and hard to scalable production |
6.1. Techniques for Loading Drugs into Exosomes
6.1.1. Electroporation
Cells/exosomes can be processed with high-voltage and short pulses to produce temporary openings on the cell/exosome membranes to accelerate drug uptake efficiency. The electroporation method is easy to control because its parameters (electric voltage) are adjustable, and the challenges with the chemical transfection method that we describe below are avoided because this method doesn’t require any carriers [202–204]. The types of cells and exosomes and properties of other exogenous materials have a minimal effect on the drug loading efficiency. After setting the parameters for the equipment and electroporation conditions, a large number of cells/exosomes can be loaded with drugs quickly and steadily in a reproducible manner.
Wang et al. used electroporation to load siRNA/microRNA into the nucleic acid aptamer AS1411 (AS1411 can recognize the nucleolin that is highly expressed on breast cancer cells) modified vesicles, and, then carry out targeted delivery of siRNA/microRNA cargo to breast cancer tissue via exosomes [205]. They presented that this engineered exosome could be delivered precisely to human breast cancer cells as well as greatly inhibit tumor growth without any significant non-specific side effects or immune response.
Hadla et al. loaded doxorubicin into exosomes by electroporation and obtained better anti-tumor effects than free drug [206]. Although doxorubicin, as an established chemotherapeutic drug, is widely used for the treatment of tumors, it still has unavoidable toxic side effects (e.g., cardiotoxicity). Compared with pure exosomes and free doxorubicin, exosomes loaded with doxorubicin partially limit the passage of doxorubicin through cardiovascular endothelial cells and reduce the accumulation of doxorubicin in the heart. Thus, this technique reduces cardiac toxicity. However, some studies have shown that electroporation can cause the aggregation of exosomes [207]. Hod et al. found that using trehalose pulsed medium in the electroporation process can effectively address this aggregation problem [208].
6.1.2. Sonication
Sonication is a recently developed, effective method for loading drugs into exosomes [209]. Hydrophilic or lipophilic drugs can enter into the exosome phospholipid bilayer by this method [191, 210–212]. Lamichhane et al. demonstrated that small RNA could be packed into exosomes efficiently by sonication [213]. However, the drug may only simply be adsorbed on the exosomal extraluminal layer, which would hamper its subsequent release. Microfluidic sonication could also be used to assemble exosome membrane onto polymeric nanoparticles for homotypic targeting with better biocompatibility, prolonged blood circulation, and enhanced immune evasion capability [209]. The microfluidic sonication platform composed of an ultrasonic bath and a microfluidic chip enables producing strong micro-vortices. The high flow velocity within the microchannel can enhance the hydrodynamic mixing of exosomes and nanoparticles and facilitate uniform coating of the exosome membrane onto the nanoparticles. The vigorous pressure produced by the microfluidic sonication platform enabled simultaneous assembly and coating of different kinds of membranes onto the nanoparticles with high efficiency (up to 93%).
6.1.3. Chemical Transfection
This technique involves chemical transfection of donor cells or incubation of drugs with donor cells so that medications can be packed in exosomes that the donor cells secrete. The chemical transfection of the donor cells is usually done with a commercial transfection agent to modify the donor cells to facilitate the expression of a specific gene in the donor cells, and the secreted exosomes would also contain the particular gene [27, 193, 214, 215].
Morishita et al. proposed an active exosome-based system with the ability of co-delivering the tumor antigen-adjuvant [216]. By transfection of murine melanocytes with the plasmid vector that encodes the protein that fuses streptavidin, and with the exosome promoting protein, tumor donor cells can produce genetically engineered exosomes that contain these two molecules to ensure their effective delivery to murine dendritic cells to effectively activate the receptor cells to enhance their ability to present tumor antigen, and thus realize the immunotherapy of cancer. However, due to the limitation of safe and effective gene carriers, the transfection efficiency and gene expression effect of this method should be improved by developing efficient gene delivery vectors to overcome these systemic barriers [207]. Moreover, it is difficult to separate exosomes from transfection agentsin this method, so that it is hard to tell whether exosomes or transfection reagent is working as the treatment of the tumor. Also, this method cannot guarantee that all the gene drugs will enter the exosomes and not attach to their surface.
6.2. Exosome Functionalization Strategy for Tumor-Targeting Drug Delivery
6.2.1. Membrane Transfection
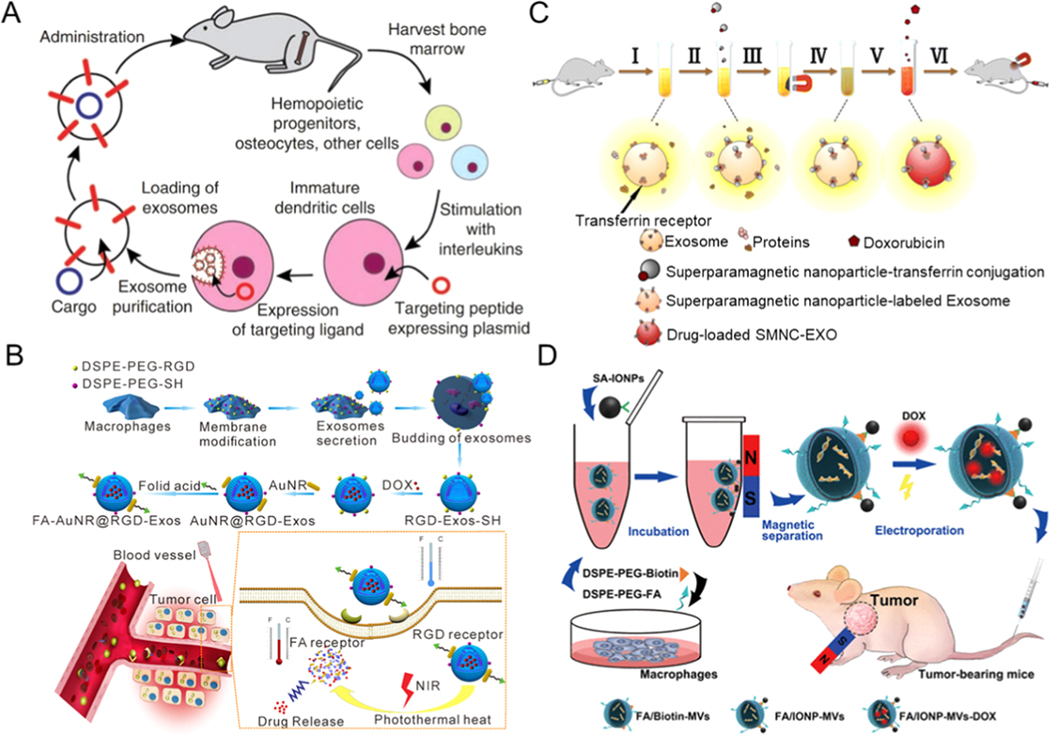
Through the transfection of donor cells, secreted exosomes can carry particular proteins, which endows the secreted exosomes with targeted therapeutic abilities. Alvarez-Erviti et al. presented the delivery of siRNA by using a vector that transfers RNA between human cells (Figure 12A) [189]. Individually, by expressing neuron-targeting proteins on the surface of exosomes and injecting these engineered exosomes into the bloodstream of mice after loading siRNA, they achieved specific gene knockdowns in the brain.
Figure 12.
Engineered exosomes for targeted drug delivery. (A) Schematic diagram of production, acquisition, and re-administration of targeted self-exosomes for the delivery of genes. Reprinted with permission from Ref. [189]. Copyright 2011, Nature Publishing Group. (B) Schematic diagram of how the engineered exosomes are delivered and the process of chemo-photothermal synergistic tumor therapy. Reprinted with permission from Ref. [218]. Copyright 2018, Wiley. (C) Schematic diagram of construction and delivery of drug-loaded superparamagnetic nanoparticles-labeled exosomes. Reprinted with permission from Ref. [224]. Copyright 2016, American Chemical Society. (D) Schematic illustration of the design of functionalized exosomes for drug targeted delivery. Reprinted with permission from Ref. [225]. Copyright 2017, American Chemical Society.
Based on this work, Tian et al. used mouse immature dendritic cells as exosomal donor cells and allowed the dendritic cells and their secreted exosomes to express Lamp 2b protein by transfection. Lamp 2b protein and RGD peptide can show outstanding tumor-targeting ability. They proved that the drug-loaded exosomes combined with RGD peptide could target the delivery of chemotherapy drugs to the site of the tumor, which demonstrated the great potential for the use of this technique in clinical applications [190]. However, the transfection procedure is demanding, requires skilled technicians to operate, and the efficiency of transfection is also easily affected by the environment.
6.2.2. Membrane Chemical Modification
Exosomal phospholipid bilayer structure and its fluid properties allow its phospholipids to easily self-assemble with phospholipid-polymer conjugates. Wang et al. self-assembled the functionalized DSPE-PEG (1,2-Distearoyl-sn-glycero-3-phospho-ethanol-amine-poly (ethylene glycol)) to endow exosomes with tumor-targeting ability [52]. They also applied this membrane chemical modification strategy to allow exosomes with blood vessel targeting capabilities to provide highly selective angiogenesis treatment [217]. Exosomal tumor release strategies were proposed by modifying donor cell phospholipids. They cultured donor cells in a medium containing DSPE-PEG-SH and DSPE-PEG-RGD to permit the modification of the membranes of the secreted exosomes with sulfhydryl groups and RGD (Figure 12B) [218]. Gold rods were stably adsorbed on the surface of the modified exosomes by the Au-S bonds. To further enable the exosomes to target tumors more efficiently, they functionalized another tumor-targeting ligand (folate) on the gold rod by forming a covalent bond with the gold rod. They proved that due to the synergistic effect of the dual ligands, exosomes could be concentrated at the tumor site. Since the gold rods, when exposed to near-infrared rays, could locally generate heat and destabilize the exosome membrane, the instability of the exosome membrane further resulted in the rapid release of the encapsulated drug. They realized tumor-targeted chemical-photothermal combination therapy with reduced toxic side effects.
6.2.3. Membrane Engineered with Magnetic Materials
Magnetic materials can combine with nanocarriers through chemical modification and help exosomes to be rapidly isolated from blood samples under a magnetic field, as well as re-dispersed well after the magnetic field is turned off in vitro. After being injected into an animal, drug delivery carriers functionalized with magnetic materialscan target the tumor site under the magnetic field and release the encapsulated drugs at the tumor site [219–223].
Qi et al. designed functional exosomes combined with magnetic nanoparticle clusters as drug delivery vehicles for targeted tumor treatment (Figure 12C) [224]. Exosomes were first isolated from the blood by using transferrin-modified magnetic beads, and then doxorubicin was loaded into the membrane of exosomes using electroporation. This cluster of exosomes loaded with superparamagnetic nanoparticle exhibits a more robust superparamagnetic behavior than a superparamagnetic nanoparticle alone. The researchers also presented that exosomes with magnetic beads attached to their membranes could effectively target tumor sites under the influence of external magnetic fields to deliver the drug to inhibit tumor growth. However, the magnetic targeting strategy is difficult to use in deep tissues accurately. The efficiency of magnetic bead-based targeting is affected not only by the magnetic bead itself but also by the blood flow velocity in the body.
To further improve the targeting efficiency of the exosomes, Zhang et al. not only functionalized exosomes with magnetic beads but also modified the surface of exosomes with chemical ligands (folate) (Figure 12D) [225]. This passive and active targeting combinatorial strategy dramatically improved the enrichment efficiency of exosomes at the tumor site.
7. Outlook
The versatility of microfluidics has demonstrated extraordinary capabilities in advanced exosome-related research for clinically precise applications in medicine. An optimized microfluidic platform can facilitate highly efficient exosome separation from complex and heterogeneous biological fluids and their analysis. It is of primary significance in various exosome-related biomedical applications, including diagnostics and treatment. Nevertheless, there are still many biological, technical, regulatory, financial, and market challenges to be addressed before exosomes can be widely used in clinical medicine.
7.1.1. Biological Challenges
Exosomes are comprised of a complex repertoire of biomolecular cargo that is still being elucidated. Cutting-edge genomic technologies are being deployed to comprehensively characterize the nucleic acid contents of exosomes, which will help realize their potential for cancer diagnostics. The Extracellular RNA Communication Consortium [226], funded by the National Institutes of Health, has extensively characterized small extracellular RNAs from human biofluids, including plasma, saliva, and urine. However, other classes of extracellular RNA are present in exosomes and will require additional in-depth genomic studies to fully profile these potentially important biomarker RNAs. Recent work by Reggiardo et al. using RNA sequencing demonstrated that exosomes are enriched with lncRNAs in human airway epithelial cells that are transformed by mutant KRAS [25]. Moreover, these exosomes also contain significant amounts of repetitive noncoding RNAs derived from transposable elements, which are specifically upregulated by oncogenic RAS signaling. Additional studies will be needed to provide biological insights into how specific mutations alter exosomal RNA profiles in the context of various cancer types, which would enable the development of more precise liquid biopsy approaches for cancer diagnostics, monitoring and early detection approaches.
7.1.2. Technical and Regulatory Challenges
To transform the use of exosomes from scientific research into clinical applications, the potential of microfluidics for exosome separation and analysis should be fully exploited. More bio-friendly chemical or gene functionalization strategies can potentially be developed to further improve the therapeutic performance of exosomes.
For exosome isolation and detection, future research will benefit from focusing on the following areas: (i) The current microfluidic platforms still produce a low number of exosomes than appropriate for clinical studies; thus, increasing separation yields, especially in drug delivery applications is important. Scaling up devices with a multi-channel format to process multiple samples simultaneously, so as to increase the throughput, is one of the potential means for surmounting this challenge. (ii) Numerous existing microfluidic-based exosome isolation techniques rely on immunoaffinity, which involves the binding and dissociation between antibody and antigen on the exosome surface using non-physiological elution and non-neutral pH buffers. These techniques may affect the biological functions of exosomes. Therefore, more label-free microfluidic technologies to separate exosomes without affecting their native structure and biological activity are urgently needed. (iii) Exosomes are fingerprints of their originating cells. One of the challenges for exosome detection is distinguishing exosome origin and accurately selecting exosome surface targets. The heterogeneity of antigen expression, especially for cancer cells, may produce false-negative results [42, 227, 228]. Moreover, other proteins or biomolecules are likely to non-specifically bind on the surface of exosomes and cause interference with the analysis of exosome composition. Therefore, more effort is needed for identifying reliable and specific exosomal markers for a given cell type and combining them with microfluidic technologies to provide more reliable and accurate information for non-invasive disease diagnosis. (iv) Different subtypes of exosomes possess various bio-functions. Considering the inherent heterogeneity of exosomes, it is an exceptionally difficult technical challenge to isolate the desired subtype, and then differentiate it from other subtypes in complex bodily fluids. (v) Also, in order to develop diagnostics that are minimally invasive, biofluids such as saliva and sweat should be taken into account, as they are easily accessible sources for isolation and analysis of exosomes. Sweat analysis, for example, can be performed non-invasively compared with blood, but the concentration of exosomes in sweat is lower than in blood. Additionally, sweat exosome proteomic profiling has physiological significance in immune homeostasis [229], but it is highly challenging to isolate/detect exosomes from sweat samples, and the use of microfluidics to isolate/detect exosomes from sweat samples needs further studies. The development of techniques for microfluidic-based exosome isolation/detection from other body fluids is thus an important direction worth pursuing. (vi) Another impediment for translating exosome research into clinical applications is the lack of rigorous, reproducible, and standardized approaches for the separation of a pure vesicular population. Microfluidic platforms for exosome isolation and detection developed possess different operating parameters and performance (e.g., purity, yield, efficiency, assay time, productivity, limit of detection). The amount and cargo of exosomes are affected by many factors, such as collection approaches, storage conditions and duration, and patient-related biological factors. Developing standard, reproducible operation protocols for separation and detection of exosomes can improve the reproducibility and optimize the technology through unbiased cross-platform comparison. It is also an important parameter to optimize the kits used downstream for exosome cargo extraction for the reproducibility and rigor of the results.
Although there has been significant progress in fundamental research of exosome-based drug delivery, there are still many challenges that need to be overcome to replicate these successes in the clinic: (i) Safety is the first and foremost consideration for translation of the scientific research into the clinic. Here the choice of optimal cell source is of utmost importance. For example, mesenchymal stem cell- and macrophage/dendritic cell-derived exosomes have proven to be safe for use in clinical settings because they have preferable immunomodulatory properties [230]. Other cell sources could be suitable for clinical applications, and they need to be thoroughly characterized and tested. Moreover, therapeutic exosomes should be further assessed for their long-term safety and therapeutic effect (pharmacokinetic and pharmacodynamic profiles) in relevant preclinical models to support predictions of clinical dose. (ii) Although the electroporation method is commonly used for loading therapeutics into exosomes, it may affect the integrity and stability of exosomes [231]. Microfluidic technology has potential to overcome this limitation owing to its unique characteristics of continuous flow mode, miniaturization, and precise controllability. The continuous flow working mode of microfluidics can be precisely controlled with electric pulses and electric fields, spatially and temporally, to permit electroporation with high-efficacy and minimal side effects at the microscale, such as pH variation, bubble formation, and Joule heating that are often observed in conventional static cuvettes [232]. Although microfluidic platforms have exhibited excellent performance over the past two decades using electroporation for loading exogenous molecules into cells [233], there is no report of the use of microfluidics for exosome electroporation, which is a critical area of exploration. (iv) Although exosomes combined with synthetic nanomaterials have shown promising advantages in cancer treatment (e.g., by providing exosomes with a more robust targeting efficiency) [234], the potential disadvantages, including unwanted immune responses, should be thoroughly investigated in future studies.
7.1.3. Market Challenges
Exosomes show great potential for clinical applications, including liquid biopsy, diagnostics and therapeutics [61, 235, 236]. Therefore, they represent tremendous commercial opportunities and have attracted considerable attention. During the past few years, numerous biotechnology companies are working on applications of exosomes in clinical trials. Some of these companies have collaborated with universities and research hospitals to conduct small-scale, first-in-class clinical trials on humans. For example, some of these companies have been committed to converting technology patents into new highly-sensitive diagnostic products in various pathologies, including prostate, bladder, kidney, breast cancers, and glioblastoma [237]. For instance, a urine-based product, ExoDx® Prostate (IntelliScore) (EPI), aims to track exosomes and genomic markers released from prostate cancer cells to help clinicians determine whether patients with unclear test results of PSA (prostate-specific antigen) need to undergo prostate biopsy [238].
Although microfluidic platforms, as promising laboratory tools, have the potential for effective isolation and detection of exosome samples at reduced cost and increased efficiency as compared to conventional benchtop equipment-based methods, their market penetration is so far limited. To improve the end-user experience and market acceptance, efforts are needed in the following areas: (i) Microfluidic devices with less complexity that can be mass-produced to isolate exosomes in batches need to be designed. This is a critical step in subsequent industry-scale development, especially for use in point-of-care settings. (ii) The commercial applications and acceptance in the market should be considered in the early design stages. For example, currently, most microfluidic devices for exosome isolation and detection require external pumps and experienced operators to manipulate these devices and analyze their results. Further, microfluidic platforms for exosome isolation and detection require off-chip sample pre-treatment, for example, centrifuging the sample at low speeds to remove the cells and staining the exosomes with a fluorescent dye and washing them repeatedly. Microfluidic devices with pretreatment of samples, purification of exosomes, and in-situ detection integrated into a single chip need to be developed. (iii) The display of results can be designed in a simple-readout manner to minimize the need for readout instruments and minimize subjective user interpretation. For example, distance-based visual readouts, quantitative detection methods, which are often used in microfluidics, could be integrated into the microfluidic platform for exosome detection [239]. End-users would only need to interact with a user-friendly system and could read visible signal bars to get the detection results directly for simplicity depending on the application. (iv) Microfluidic devices can be combined with robotics and deep learning to fully realize the automation of exosome detection capabilities. A robotic operation can eliminate “human errors” caused by the tediousness of manual procedures, and improve the experience of users who lack prior microfluidic expertise [240]. Deep learning models such as convolutional neural networks can minimize the reliance on highly trained clinicians to analyze results, which would be of great clinical market value, both with potential impact in developed countries as well as in resource-limited settings. (v) Academic efforts could synchronize and collaborate with the rapid developments in industry. Researchers from diverse disciplines (engineering, medicine, business) could work together to investigate and validate the need for exosomes to address urgent needs in the pre-clinical and clinical markets. For example, industrial partners with marketing, sales management capabilities, and established sales channels can participate in determining the product potential for success in the market and provide marketing strategies early on for successful translation of emerging technologies. Clinicians and healthcare experts can give valuable input on user needs and expectations in terms of integrating these new assays and tools into the clinical workflow and disease management strategies. Further, analysis on socioeconomic benefit will inform reimbursement strategies for emerging assays and treatments. Therefore, these multiple needs and strategies need to be integrated to design exclusiveand innovative products that are likely to attain success on the translational pipeline.
8. Conclusion
Microfluidic platforms have shown unique capabilites as versatile tools for exosome isolation and detection. Microfluidic technologies offer customizable opportunities for developing label-free separation techniques that can keep exosomal biological contents active and intact. These extraordinary features of microfluidics will continue to contribute to the advancement of exosome-based drug delivery applications. While the utilization of microfluidic technologies for exosome separation and analysis is still in its infancy, many advanced designs discussed above would potentially address existing challenges, and hence, bring breakthroughs in broader fields of clinical science, including liquid biopsies and therapeutic nanomedicine. Microfluidic technologies are poised to facilitiate the rapid development of exosome-based theranostic nanoplatforms for clinical use with broad applications that are likely to emerge with commercialization of these tools enabling new beneficial capabilities, high-impact applications and assays in the clinic and the marketplace.
Highlights.
Exosomes have been identified to hold exceptional value in clinical diagnostics and tumor therapy.
A short overview of conventional methods of exosome isolation is summarized.
The recent advancements of microfluidic strategy for exosomes isolation and detection are overviewed.
A brief overview of exosome-based drug delivery for tumor therapy is provided.
The current challenges and outlook of these fields are assessed.
Acknowledgments
This work was supported by the Canary Foundation seed grant, National Cancer Institute (NCI) Center for Cancer Nano-technology Excellence for Translational Diagnostics (NCI-U54CA199075), and the National Institutes of Health grant (R01 EB029805).
Biography

Dr. Jie Wang is a postdoctoral researcher at Stanford University School of Medicine, with a research focus in developing micro/nanomaterials technologies. She has expertise in microfabrication, chemistry, material science with a focus on biomedical engineering applications. She has been focused on developing microfluidic technology to isolate and detect rare cell populations and exosomes from whole blood, as well as engineered the isolated exosomes as a drug delivery carrier for tumor diagnostic and therapy.

Peng Ma is a Ph.D. candidate at Huazhong University of Science and Technology. He is a visiting student at Stanford University School of Medicine from 2019 to 2020. He is mainly engaged in research on microfluidic biochips and biomedical engineering to explore molecular biology, cell biology, biomedical engineering, and related frontier scientific issues, research, and development of application-oriented portable medical testing instruments.
Dr. Daniel H. Kim is currently an Assistant Professor of Biomolecular Engineering at the University of California Santa Cruz and an Associate Member at the Canary Center at Stanford for Cancer Early Detection. His research is focused on understanding the functions of noncoding RNAs in stem cells and cancer, and his lab is also developing RNA-based liquid biopsy approaches to enable highly sensitive and specific early detection of cancer and other diseases using genomic approaches. His research has been recognized by awards from the Damon Runyon Cancer Research Foundation and the National Academy of Sciences.

Dr. Bifeng Liu is currently a Professor at the College of Life Science and Technology, Huazhong University of Life Science and Technology. His research mainly focused on combining biomedical engineering, biochemistry, and molecular biology, analytical chemistry, etc., on developing new microfluidic technology for system biomedicine study, including (i) organ chip technology; (ii) single-cell omics technology; (iii) exosomal discovery and analysis technology; (iv) In vitro diagnostic technology. His recent research results have been published in advanced international academic journals such as Nature Communications, Advanced Materials, Advanced Functional Materials, ACS Nano, Angewandte Chemie International Edition and so on.

Dr. Demirci is currently a Professor at Stanford University School of Medicine with tenure at the Canary Center for Early Cancer Detection. Prior to his Stanford appointment, he was an Associate Professor of Medicine at Brigham and Women’s Hospital, Harvard Medical School and at Harvard-MIT Division of Health Sciences and Technology. He leads a group of 20+ researchers focusing on micro- and nano-scale technologies. He published over 150 peer-reviewed journal publications, 24 book chapters, 4 edited books, over 200 abstracts and proceedings, over 25 issued and pending patents and disclosures. (H-index, 73).
Footnotes
Declaration of Competing Interest
Prof. Utkan Demirci (UD) is a founder of and has an equity interest in: (i) DxNow Inc., a company that is developing microfluidic IVF tools and imaging technologies for point-of-care diagnostic solutions, (ii) Koek Biotech, a company that is developing microfluidic technologies for clinical solutions, (iii) Levitas Inc., a company focusing on developing microfluidic products for sorting rare cells from liquid biopsy in cancer and other diseases, and (iv) Hillel Inc., a company bringing microfluidic cell phone tools to home settings. UD’s interests were viewed and managed in accordance with the conflict of interest policies.
Author statement
Jie Wang, Bifeng Liu and Utkan Demirci: conception and design, manuscript writing, final approval of manuscript. Peng Ma: literature review and manuscript writing. Daniel Kim: manuscript writing.
Declaration of interests
☒ The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Thery C, Zitvogel L, Amigorena S, Nat. Rev. Immunol 2 (2002) 569–579. [DOI] [PubMed] [Google Scholar]
- [2].Yanez-Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro-da Silva A, Fais S, Falcon-Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj-Iglic V, Kramer-Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek-Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte-’t Hoen EN, Nyman TA, O’Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez-Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH, De Wever O, Extracell J. Vesicles 4 (2015) 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Kilchert C, Wittmann S, Vasiljeva L, Nat. Rev. Mol. Cell Biol 17 (2016) 227–239. [DOI] [PubMed] [Google Scholar]
- [4].Contreras-Naranjo JC, Wu HJ, Ugaz VM, Lab Chip 17 (2017) 3558–3577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Thery C, Ostrowski M, Segura E, Nat. Rev. Immunol 9 (2009) 581–593. [DOI] [PubMed] [Google Scholar]
- [6].Trams EG, Lauter CJ, Salem N Jr., Heine U, Biochim. Biophys. Acta 645 (1981) 63–70. [DOI] [PubMed] [Google Scholar]
- [7].Gross JC, Chaudhary V, Bartscherer K, Boutros M, Nat. Cell Biol 14 (2012) 1036–1045. [DOI] [PubMed] [Google Scholar]
- [8].Hoshino A, Kim HS, Bojmar L, Gyan KE, Cioffi M, Hernandez J, Zambirinis CP, Rodrigues G, Molina H, Heissel S, Mark MT, Steiner L, Benito-Martin A, Lucotti S, Di Giannatale A, Offer K, Nakajima M, Williams C, Nogues L, Pelissier Vatter FA, Hashimoto A, Davies AE, Freitas D, Kenific CM, Ararso Y, Buehring W, Lauritzen P, Ogitani Y, Sugiura K, Takahashi N, Aleckovic M, Bailey KA, Jolissant JS, Wang H, Harris A, Schaeffer LM, Garcia-Santos G, Posner Z, Balachandran VP, Khakoo Y, Raju GP, Scherz A, Sagi I, Scherz-Shouval R, Yarden Y, Oren M, Malladi M, Petriccione M, De Braganca KC, Donzelli M, Fischer C, Vitolano S, Wright GP, Ganshaw L, Marrano M, Ahmed A, DeStefano J, Danzer E, Roehrl MHA, Lacayo NJ, Vincent TC, Weiser MR, Brady MS, Meyers PA, Wexler LH, Ambati SR, Chou AJ, Slotkin EK, Modak S, Roberts SS, Basu EM, Diolaiti D, Krantz BA, Cardoso F, Simpson AL, Berger M, Rudin CM, Simeone DM, Jain M, Ghajar CM, Batra SK, Stanger BZ, Bui J, Brown KA, Rajasekhar VK, Healey JH, de Sousa M, Kramer K, Sheth S, Baisch J, Pascual V, Heaton TE, La Quaglia MP, Pisapia DJ, Schwartz R, Zhang H, Liu Y, Shukla A, Blavier L, DeClerck YA, LaBarge M, Bissell MJ, Caffrey TC, Grandgenett PM, Hollingsworth MA, Bromberg J, Costa-Silva B, Peinado H, Kang Y, Garcia BA, O’Reilly EM, Kelsen D, Trippett TM, Jones DR, Matei IR, Jarnagin WR, Lyden D, Cell 182 (2020) 1044–1061 e1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].I. Mager ELAS, Breakefield XO, Wood MJ, Nat. Rev. Drug Discov 12 (2013) 347–357. [DOI] [PubMed] [Google Scholar]
- [10].Tian T, Zhu YL, Zhou YY, Liang GF, Wang YY, Hu FH, Xiao ZD, J. Biol. Chem 289 (2014) 22258–22267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Gurunathan S, Kang MH, Jeyaraj M, Qasim M, Kim JH, Cells 8 (2019) 307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT, Nat. Cell Biol 13 (2011) 423–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Szabo G, Momen-Heravi F, Nat. Rev. Gastroenterol. Hepatol 14 (2017) 455–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Corrado C, Raimondo S, Chiesi A, Ciccia F, De Leo G, Alessandro R, Int. J. Mol. Sci 14 (2013) 5338–5366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chandy M, Rhee JW, Ozen MO, Williams DR, Pepic L, Liu C, Zhang H, Malisa J, Lau E, Demirci U, Wu JC, Circulation 142 (2020) 1794–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gibbings DJ, Ciaudo C, Erhardt M, Voinnet O, Nat. Cell Biol 11 (2009) 1143–1149. [DOI] [PubMed] [Google Scholar]
- [17].Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO, Nat. Cell Biol 9 (2007) 654–659. [DOI] [PubMed] [Google Scholar]
- [18].Mittelbrunn M, Gutierrez-Vazquez C, Villarroya-Beltri C, Gonzalez S, Sanchez-Cabo F, Gonzalez MA, Bernad A, Sanchez-Madrid F, Nat. Commun 2 (2011) 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yuan K, Shamskhou EA, Orcholski ME, Nathan A, Reddy S, Honda H, Mani V, Zeng Y, Ozen MO, Wang L, Demirci U, Tian W, Nicolls MR, de Jesus Perez VA, Circulation 139 (2019) 1710–1724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].He C, Zheng S, Luo Y, Wang B, Theranostics 8 (2018) 237–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Hoshino A, Costa-Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont-Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Labori KJ, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J, Lyden D, Nature 527 (2015) 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Liu C, Xu X, Li B, Situ B, Pan W, Hu Y, An T, Yao S, Zheng L, Nano Lett. 18 (2018) 4226–4232. [DOI] [PubMed] [Google Scholar]
- [23].ACS JH Appl Mater InterfacesLee, Choi JH, Chueng SD, Pongkulapa T, Yang L, Cho HY, Choi JW, Lee KB, ACS Nano 13 (2019) 8793–8803. [DOI] [PubMed] [Google Scholar]
- [24].Saha P, Sharma S, Korutla L, Datla SR, Shoja-Taheri F, Mishra R, Bigham GE, Sarkar M, Morales D, Bittle G, Gunasekaran M, Ambastha C, Arfat MY, Li D, Habertheuer A, Hu R, Platt MO, Yang P, Davis ME, Vallabhajosyula P, Kaushal S, Sci. Transl. Med 11 (2019) eaau1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Reggiardo RE, Velandi Maroli S, Halasz H, Ozen M, Carrillo D, LaMontagne E, Whitehead L, Kim E, Malik S, Fernandes J, Marinov G, Collisson E, Demirci U, Kim DH, BioRixv (2020) 10.1101/2020.11.04.367771 (accessed 4 November 2020). [DOI] [Google Scholar]
- [26].Vader P, Mol EA, Pasterkamp G, Schiffelers RM, Adv. Drug Deliv. Rev 106 (2016) 148–156. [DOI] [PubMed] [Google Scholar]
- [27].Yim N, Ryu SW, Choi K, Lee KR, Lee S, Choi H, Kim J, Shaker MR, Sun W, Park JH, Kim D, Heo WD, Choi C, Nat. Commun 7 (2016) 12277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Armstrong JPK, Stevens MM, Adv. Drug Deliv. Rev 130 (2018) 12–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mathieu M, Martin-Jaular L, Lavieu G, Thery C, Nat. Cell Biol 21 (2019) 9–17. [DOI] [PubMed] [Google Scholar]
- [30].Abels ER, Breakefield XO, Cell Mol. Neurobiol 36 (2016) 301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Jeppesen DK, Hvam ML, Primdahl-Bengtson B, Boysen AT, Whitehead B, Dyrskjot L, Orntoft TF, Howard KA, Ostenfeld MS, Extracell J. Vesicles 3 (2014) 25011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte-’t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH, Hochberg F, Extracell J. Vesicles 2 (2013) 20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Cvjetkovic A, Lotvall J, Lasser C, Extracell J. Vesicles 3 (2014) 23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Szatanek R, Baran J, Siedlar M, Baj-Krzyworzeka M, Int. J. Mol. Med 36 (2015) 11–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Lobb RJ, Becker M, Wen SW, Wong CS, Wiegmans AP, Leimgruber A, Moller A, Extracell J. Vesicles 4 (2015) 27031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS, Star RA, Am. J. Physiol. Renal Physiol 292 (2007) F1657–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Heinemann ML, Ilmer M, Silva LP, Hawke DH, Recio A, Vorontsova MA, Alt E, Vykoukal J, Chromatogr J. A 1371 (2014) 125–135. [DOI] [PubMed] [Google Scholar]
- [38].Nordin JZ, Lee Y, Vader P, Mager I, Johansson HJ, Heusermann W, Wiklander OP, Hallbrink M, Seow Y, Bultema JJ, Gilthorpe J, Davies T, Fairchild PJ, Gabrielsson S, Meisner-Kober NC, Lehtio J, Smith CI, Wood MJ, El Andaloussi S, Nanomedicine 11 (2015) 879–883. [DOI] [PubMed] [Google Scholar]
- [39].Kang D, Oh S, Ahn SM, Lee BH, Moon MH, J. Proteome Res 7 (2008) 3475–3480. [DOI] [PubMed] [Google Scholar]
- [40].Musante L, Tataruch D, Gu D, Benito-Martin A, Calzaferri G, Aherne S, Holthofer H, Sci. Rep 4 (2014) 7532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL, Immunol J. Methods 411 (2014) 55–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Yang D, Zhang W, Zhang H, Zhang F, Chen L, Ma L, Larcher LM, Chen S, Liu N, Zhao Q, Tran PHL, Chen C, Veedu RN, Wang T, Theranostics 10 (2020) 3684–3707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kim DK, Nishida H, An SY, Shetty AK, Bartosh TJ, Prockop DJ, Proc. Natl. Acad. Sci. U. S. A 113 (2016) 170–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Suarez H, Gamez-Valero A, Reyes R, Lopez-Martin S, Rodriguez MJ, Carrascosa JL, Cabanas C, Borras FE, Yanez-Mo M, Sci. Rep 7 (2017) 11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kabe Y, Sakamoto S, Hatakeyama M, Yamaguchi Y, Suematsu M, Itonaga M, Handa H, Anal. Bioanal. Chem 411 (2019) 1825–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Saenz-Cuesta M, Arbelaiz A, Oregi A, Irizar H, Osorio-Querejeta I, Munoz-Culla M, Banales JM, Falcon-Perez JM, Olascoaga J, Otaegui D, Front. Immunol 6 (2015) 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, Seremwe M, Dismuke WM, Bieberich E, Stamer WD, Hamrick MW, Liu Y, PLoS One 12 (2017) e0170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, Hulett MD, Mathivanan S, Proteomics 13 (2013) 3354–3364. [DOI] [PubMed] [Google Scholar]
- [49].Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M, Lab Chip 15 (2015) 2388–2394. [DOI] [PubMed] [Google Scholar]
- [50].Zhang P, He M, Zeng Y, Lab Chip 16 (2016) 3033–3042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].He M, Crow J, Roth M, Zeng Y, Godwin AK, Lab Chip 14 (2014) 3773–3780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Wang J, Li W, Zhang L, Ban L, Chen P, Du W, Feng X, Liu BF, ACS Appl. Mater. Interfaces 9 (2017) 27441–27452. [DOI] [PubMed] [Google Scholar]
- [53].Zhao Z, Yang Y, Zeng Y, He M, Lab Chip 16 (2016) 489–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kanwar SS, Dunlay CJ, Simeone DM, Nagrath S, Lab Chip 14 (2014) 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Dudani JS, Gossett DR, Tse HT, Lamm RJ, Kulkarni RP, Carlo DD, Biomicrofluidics 9 (2015) 014112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Kang YT, Kim YJ, Bu J, Cho YH, Han SW, Moon BI, Nanoscale 9 (2017) 13495–13505. [DOI] [PubMed] [Google Scholar]
- [57].Kang YT, Purcell E, Palacios-Rolston C, Lo TW, Ramnath N, Jolly S, Nagrath S, Small 15 (2019) e1903600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Davies RT, Kim J, Jang SC, Choi EJ, Gho YS, Park J, Lab Chip 12 (2012) 5202–5210. [DOI] [PubMed] [Google Scholar]
- [59].Cho S, Jo W, Heo Y, Kang JY, Kwak R, Park J, Sens. Actuators B Chem 233 (2016) 289–297. [Google Scholar]
- [60].Woo HK, Sunkara V, Park J, Kim TH, Han JR, Kim CJ, Choi HI, Kim YK, Cho YK, ACS Nano 11 (2017) 1360–1370. [DOI] [PubMed] [Google Scholar]
- [61].Chen Z, Cheng SB, Cao P, Qiu QF, Chen Y, Xie M, Xu Y, Huang WH, Biosens. Bioelectron 122 (2018) 211–216. [DOI] [PubMed] [Google Scholar]
- [62].Liang LG, Kong MQ, Zhou S, Sheng YF, Wang P, Yu T, Inci F, Kuo WP, Li LJ, Demirci U, Wang S, Sci. Rep 7 (2017) 46224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wang Z, Wu HJ, Fine D, Schmulen J, Hu Y, Godin B, Zhang JX, Liu X, Lab Chip 13 (2013) 2879–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Yasui T, Yanagida T, Ito S, Konakade Y, Takeshita D, Naganawa T, Nagashima K, Shimada T, Kaji N, Nakamura Y, Thiodorus IA, He Y, Rahong S, Kanai M, Yukawa H, Ochiya T, Kawai T, Baba Y, Sci. Adv 3 (2017) e1701133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Liu F, Vermesh O, Mani V, Ge TJ, Madsen SJ, Sabour A, Hsu EC, Gowrishankar G, Kanada M, Jokerst JV, Sierra RG, Chang E, Lau K, Sridhar K, Bermudez A, Pitteri SJ, Stoyanova T, Sinclair R, Nair VS, Gambhir SS, Demirci U, ACS Nano 11 (2017) 10712–10723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Shin S, Han D, Park MC, Mun JY, Choi J, Chun H, Kim S, Hong JW, Sci. Rep 7 (2017) 9907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ramachandraiah H, Ardabili S, Faridi AM, Gantelius J, Kowalewski JM, Martensson G, Russom A, Biomicrofluidics 8 (2014) 034117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yeo JC, Kenry Z Zhao P Zhang Z Wang CT Lim, Biomicrofluidics 12 (2018) 024103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Zhou Y, Ma Z, Tayebi M, Ai Y, Anal. Chem 91 (2019) 4577–4584. [DOI] [PubMed] [Google Scholar]
- [70].Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, Austin RH, Stolovitzky G, Astier Y, Nat. Nanotechnol 11 (2016) 936–940. [DOI] [PubMed] [Google Scholar]
- [71].Huang LR, Cox EC, Austin RH, Sturm JC, Science 304 (2004) 987–990. [DOI] [PubMed] [Google Scholar]
- [72].Zeming KK, Thakor NV, Zhang Y, Chen CH, Lab Chip 16 (2016) 75–85. [DOI] [PubMed] [Google Scholar]
- [73].Huang LR, Cox EC, Austin RH, Sturm JC, Science 304 (2004) 987–990. [DOI] [PubMed] [Google Scholar]
- [74].Smith JT, Wunsch BH, Dogra N, Ahsen ME, Lee K, Yadav KK, Weil R, Pereira MA, Patel JV, Duch EA, Papalia JM, Lofaro MF, Gupta M, Tewari AK, Cordon-Cardo C, Stolovitzky G, Gifford SM, Lab Chip 18 (2018) 3913–3925. [DOI] [PubMed] [Google Scholar]
- [75].Tian F, Feng Q, Chen Q, Liu C, Li T, Sun J, Microfluid. Nanofluid 23 (2019) 68. [Google Scholar]
- [76].D’Avino G, Greco F, Maffettone PL, Annu. Rev. of Fluid Mech 49 (2017) 341–360. [Google Scholar]
- [77].Nam J, Tan JK, Khoo BL, Namgung B, Leo HL, Lim CT, Kim S, Biomicrofluidics 9 (2015) 064117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Yuan D, Zhao Q, Yan S, Tang SY, Alici G, Zhang J, Li W, Lab Chip 18 (2018) 551–567. [DOI] [PubMed] [Google Scholar]
- [79].Tian F, Cai L, Chang J, Li S, Liu C, Li T, Sun J, Lab Chip 18 (2018) 3436–3445. [DOI] [PubMed] [Google Scholar]
- [80].Wang G, Crawford K, Turbyfield C, Lam W, Alexeev A, Sulchek T, Lab Chip 15 (2015) 532–540. [DOI] [PubMed] [Google Scholar]
- [81].Luo YN, Chen DY, Zhao Y, Wei C, Zhao XT, Yue WT, Long R, Wang JB, Chen J, Sens. Actuators B Chem 202 (2014) 1183–1189. [Google Scholar]
- [82].Hatch AC, Patel A, Beer NR, Lee AP, Lab Chip 13 (2013) 1308–1315. [DOI] [PubMed] [Google Scholar]
- [83].Hassanpourfard M, Ghosh R, Thundat T, Kumar A, Lab Chip 16 (2016) 4091–4096. [DOI] [PubMed] [Google Scholar]
- [84].Kim KP, Kim YG, Choi CH, Kim HE, Lee SH, Chang WS, Lee CS, Lab Chip 10 (2010) 3296–3299. [DOI] [PubMed] [Google Scholar]
- [85].Liu C, Guo J, Tian F, Yang N, Yan F, Ding Y, Wei J, Hu G, Nie G, Sun J, ACS Nano 11 (2017) 6968–6976. [DOI] [PubMed] [Google Scholar]
- [86].Liu C, Zhao J, Tian F, Chang J, Zhang W, Sun J, J. Am. Chem. Soc 141 (2019) 3817–3821. [DOI] [PubMed] [Google Scholar]
- [87].Ku A, Lim HC, Evander M, Lilja H, Laurell T, Scheding S, Ceder Y, Anal. Chem 90 (2018) 8011–8019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Wu M, Chen C, Wang Z, Bachman H, Ouyang Y, Huang PH, Sadovsky Y, Huang TJ, Lab Chip 19 (2019) 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Li P, Mao Z, Peng Z, Zhou L, Chen Y, Huang PH, Truica CI, Drabick JJ, El-Deiry WS, Dao M, Suresh S, Huang TJ, Proc. Natl. Acad. Sci. U. S. A 112 (2015) 4970–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Laurell T, Petersson F, Nilsson A, Chem. Soc. Rev 36 (2007) 492–506. [DOI] [PubMed] [Google Scholar]
- [91].Wu M, Mao Z, Chen K, Bachman H, Chen Y, Rufo J, Ren L, Li P, Wang L, Huang TJ, Adv. Funct. Mater 27 (2017) 1606039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH, Chen C, Li H, Li P, Quinn D, Dao M, Suresh S, Sadovsky Y, Huang TJ, Proc. Natl. Acad. Sci. U. S. A 114 (2017) 10584–10589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Wang Z, Li F, Rufo J, Chen C, Yang S, Li L, Zhang J, Cheng J, Kim Y, Wu M, Abemayor E, Tu M, Chia D, Spruce R, Batis N, Mehanna H, Wong DTW, Huang TJ, J. Mol. Diagn 22 (2020) 50–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Lee K, Shao H, Weissleder R, Lee H, ACS Nano 9 (2015) 2321–2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Marczak S, Richards K, Ramshani Z, Smith E, Senapati S, Hill R, Go DB, Chang HC, Electrophoresis (2018) 2029–2038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Cheung LS, Sahloul S, Orozaliev A, Song YA, Micromachines (Basel) 9 (2018) 306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Mogi K, Hayashida K, Yamamoto T, Anal. Sci 34 (2018) 875–880. [DOI] [PubMed] [Google Scholar]
- [98].Viefhues M, Eichhorn R, Fredrich E, Regtmeier J, Anselmetti D, Lab Chip 12 (2012) 485–494. [DOI] [PubMed] [Google Scholar]
- [99].Gadish N, Voldman J, Anal. Chem 78 (2006) 7870–7876. [DOI] [PubMed] [Google Scholar]
- [100].Li Y, Xu F, Liu C, Xu Y, Feng X, Liu BF, Analyst 138 (2013) 4475–4482. [DOI] [PubMed] [Google Scholar]
- [101].Li Y, Liu C, Feng X, Xu Y, Liu BF, Anal. Chem 86 (2014) 4333–4339. [DOI] [PubMed] [Google Scholar]
- [102].Wiklund M, Gunther C, Lemor R, Jager M, Fuhr G, Hertz HM, Lab Chip 6 (2006) 1537–1544. [DOI] [PubMed] [Google Scholar]
- [103].Yafouz B, Kadri NA, Ibrahim F, Sensors (Basel) 14 (2014) 6356–6369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Yang SM, Yu TM, Huang HP, Ku MY, Hsu L, Liu CH, Opt. Lett 35 (2010) 1959–1961. [DOI] [PubMed] [Google Scholar]
- [105].Ou X, Chen P, Huang X, Li S, Liu BF, J. Sep. Sci 43 (2020) 258–270. [DOI] [PubMed] [Google Scholar]
- [106].Kuczenski RS, Chang HC, Revzin A, Biomicrofluidics 5 (2011) 32005–3200515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Thomas RS, Mitchell PD, Oreffo RO, Morgan H, Biomicrofluidics 4 (2010) 022806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Chung C-C, Cheng IF, Lin C-C, Chang H-C, Microfluid. Nanofluid 10 (2010) 311–319. [Google Scholar]
- [109].Liu C, Li Y, Li Y, Chen P, Feng X, Du W, Liu BF, Talanta 149 (2016) 237–243. [DOI] [PubMed] [Google Scholar]
- [110].Chiriaco MS, Bianco M, Nigro A, Primiceri E, Ferrara F, Romano A, Quattrini A, Furlan R, Arima V, Maruccio G, Sensors (Basel) 18 (2018) 3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Ayala-Mar S, Perez-Gonzalez VH, Mata-Gomez MA, Gallo-Villanueva RC, Gonzalez-Valdez J, Anal. Chem 91 (2019) 14975–14982. [DOI] [PubMed] [Google Scholar]
- [112].Salafi T, Zeming KK, Zhang Y, Lab Chip 17 (2016) 11–33. [DOI] [PubMed] [Google Scholar]
- [113].Sin ML, Gao J, Liao JC, Wong PK, J. Biol. Eng 5 (2011) 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Heineck DP, Lewis JM, Heller MJ, Electrophoresis 38 (2017) 1475–1482. [DOI] [PubMed] [Google Scholar]
- [115].Ibsen SD, Wright J, Lewis JM, Kim S, Ko SY, Ong J, Manouchehri S, Vyas A, Akers J, Chen CC, Carter BS, Esener SC, Heller MJ, ACS Nano 11 (2017) 6641–6651. [DOI] [PubMed] [Google Scholar]
- [116].Boriachek K, Islam MN, Moller A, Salomon C, Nguyen NT, Hossain MSA, Yamauchi Y, Shiddiky MJA, Small 14 (2018) 1702153. [DOI] [PubMed] [Google Scholar]
- [117].Im H, Lee K, Weissleder R, Lee H, Castro CM, Lab Chip 17 (2017) 2892–2898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Lee JH, Kim JA, Kwon MH, Kang JY, Rhee WJ, Biomaterials 54 (2015) 116–125. [DOI] [PubMed] [Google Scholar]
- [119].Lee JH, Kim JA, Jeong S, Rhee WJ, Biosens. Bioelectron 86 (2016) 202–210. [DOI] [PubMed] [Google Scholar]
- [120].Zhang P, Zhou X, He M, Shang Y, Tetlow AL, Godwin AK, Zeng Y, Nat. Biomed. Eng 3 (2019) 438–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ko J, Carpenter E, Issadore D, Analyst 141 (2016) 450–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Vaidyanathan R, Naghibosadat M, Rauf S, Korbie D, Carrascosa LG, Shiddiky MJ, Trau M, Anal. Chem 86 (2014) 11125–11132. [DOI] [PubMed] [Google Scholar]
- [123].Gholizadeh S, Shehata Draz M, Zarghooni M, Sanati-Nezhad A, Ghavami S, Shafiee H, Akbari M, Biosens. Bioelectron 91 (2017) 588–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Boriachek K, Masud MK, Palma C, Phan HP, Yamauchi Y, Hossain MSA, Nguyen NT, Salomon C, Shiddiky MJA, Anal. Chem 91 (2019) 3827–3834. [DOI] [PubMed] [Google Scholar]
- [125].Jiang Y, Shi M, Liu Y, Wan S, Cui C, Zhang L, Tan W, Angew. Chem. Int. Ed 56 (2017) 11916–11920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Choi J, Seong TW, Jeun M, Lee KH, Adv. Healthc. Mater 6 (2017) 1700796. [DOI] [PubMed] [Google Scholar]
- [127].Ko J, Hemphill MA, Gabrieli D, Wu L, Yelleswarapu V, Lawrence G, Pennycooke W, Singh A, Meaney DF, Issadore D, Sci. Rep 6 (2016) 31215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Rho J, Chung J, Im H, Liong M, Shao H, Castro CM, Weissleder R, Lee H, ACS Nano 7 (2013) 11227–11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Shao H, Chung J, Balaj L, Charest A, Bigner DD, Carter BS, Hochberg FH, Breakefield XO, Weissleder R, Lee H, Nat. Med 18 (2012) 1835–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Xu H, Liao C, Zuo P, Liu Z, Ye BC, Anal. Chem 90 (2018) 13451–13458. [DOI] [PubMed] [Google Scholar]
- [131].Shao H, Chung J, Lee K, Balaj L, Min C, Carter BS, Hochberg FH, Breakefield XO, Lee H, Weissleder R, Nat. Commun 6 (2015) 6999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Hassani A, Skorobogatiy M, Opt. Express 14 (2006) 11616–11621. [DOI] [PubMed] [Google Scholar]
- [133].Ouellet E, Lausted C, Lin T, Yang CW, Hood L, Lagally ET, Lab Chip 10 (2010) 581–588. [DOI] [PubMed] [Google Scholar]
- [134].Wang DS, Fan SK, Sensors (Basel) 16 (2016) 1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Luo Y, Yu F, Zare RN, Lab Chip 8 (2008) 694–700. [DOI] [PubMed] [Google Scholar]
- [136].Malic L, Veres T, Tabrizian M, Biosens. Bioelectron 24 (2009) 2218–2224. [DOI] [PubMed] [Google Scholar]
- [137].Rojalin T, Phong B, Koster HJ, Carney RP, Front. Chem 7 (2019) 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Kurita R, Yokota Y, Sato Y, Mizutani F, Niwa O, Anal. Chem 78 (2006) 5525–5531. [DOI] [PubMed] [Google Scholar]
- [139].Lee KH, Su YD, Chen SJ, Tseng FG, Lee GB, Biosens. Bioelectron 23 (2007) 466–472. [DOI] [PubMed] [Google Scholar]
- [140].Hoa XD, Kirk AG, Tabrizian M, Biosens. Bioelectron 23 (2007) 151–160. [DOI] [PubMed] [Google Scholar]
- [141].Ansari MIH, Hassan S, Qurashi A, Khanday FA, Biosens. Bioelectron 85 (2016) 247–260. [DOI] [PubMed] [Google Scholar]
- [142].Malic L, Veres T, Tabrizian M, Biosens. Bioelectron 26 (2011) 2053–2059. [DOI] [PubMed] [Google Scholar]
- [143].Mataji-Kojouri A, Ozen MO, Shahabadi M, Inci F, Demirci U, ACS Nano 14 (2020) 8518–8527. [DOI] [PubMed] [Google Scholar]
- [144].Zhu L, Wang K, Cui J, Liu H, Bu X, Ma H, Wang W, Gong H, Lausted C, Hood L, Yang G, Hu Z, Anal. Chem 86 (2014) 8857–8864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Im H, Shao H, Park YI, Peterson VM, Castro CM, Weissleder R, Lee H, Nat. Biotechnol 32 (2014) 490–495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Psaltis D, Quake SR, Yang C, Nature 442 (2006) 381–386. [DOI] [PubMed] [Google Scholar]
- [147].Inci F, Tokel O, Wang S, Gurkan UA, Tasoglu S, Kuritzkes DR, Demirci U, ACS Nano 7 (2013) 4733–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Kim J, Lab Chip 12 (2012) 3611–3623. [DOI] [PubMed] [Google Scholar]
- [149].Shafiee H, Lidstone EA, Jahangir M, Inci F, Hanhauser E, Henrich TJ, Kuritzkes DR, Cunningham BT, Demirci U, Sci. Rep 4 (2014) 4116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Tokel O, Inci F, Demirci U, Chem. Rev 114 (2014) 5728–5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [151].Tokel O, Yildiz UH, Inci F, Durmus NG, Ekiz OO, Turker B, Cetin C, Rao S, Sridhar K, Natarajan N, Shafiee H, Dana A, Demirci U, Sci. Rep 5 (2015) 9152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [152].Hsieh K, Patterson AS, Ferguson BS, Plaxco KW, Soh HT, Angew. Chem. Int. Ed 51 (2012) 4896–4900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Fang TH, Ramalingam N, Xian-Dui D, Ngin TS, Xianting Z, Lai Kuan AT, Peng Huat EY, Hai-Qing G, Biosens. Bioelectron 24 (2009) 2131–2136. [DOI] [PubMed] [Google Scholar]
- [154].Kwakye S, Goral VN, Baeumner AJ, Biosens. Bioelectron 21 (2006) 2217–2223. [DOI] [PubMed] [Google Scholar]
- [155].Safavieh M, Ahmed MU, Tolba M, Zourob M, Biosens. Bioelectron 31 (2012) 523–528. [DOI] [PubMed] [Google Scholar]
- [156].Sanghavi BJ, Moore JA, Chavez JL, Hagen JA, Kelley-Loughnane N, Chou CF, Swami NS, Biosens. Bioelectron 78 (2016) 244–252. [DOI] [PubMed] [Google Scholar]
- [157].Wang Y, Xu H, Luo J, Liu J, Wang L, Fan Y, Yan S, Yang Y, Cai X, Biosens. Bioelectron 83 (2016) 319–326. [DOI] [PubMed] [Google Scholar]
- [158].Jeong S, Park J, Pathania D, Castro CM, Weissleder R, Lee H, ACS Nano 10 (2016) 1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Zhou Q, Rahimian A, Son K, Shin DS, Patel T, Revzin A, Methods 97 (2016) 88–93. [DOI] [PubMed] [Google Scholar]
- [160].Sefah K, Shangguan D, Xiong X, O’Donoghue MB, Tan W, Nat. Protoc 5 (2010) 1169–1185. [DOI] [PubMed] [Google Scholar]
- [161].Pusuluri A, Krishnan V, Lensch V, Sarode A, Bunyan E, Vogus DR, Menegatti S, Soh HT, Mitragotri S, Angew. Chem. Int. Ed 58 (2019) 1437–1441. [DOI] [PubMed] [Google Scholar]
- [162].Wang J, Yu J, Yang Q, McDermott J, Scott A, Vukovich M, Lagrois R, Gong Q, Greenleaf W, Eisenstein M, Ferguson BS, Soh HT, Angew. Chem. Int. Ed 56 (2017) 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [163].Wang S, Zhang L, Wan S, Cansiz S, Cui C, Liu Y, Cai R, Hong C, Teng IT, Shi M, Wu Y, Dong Y, Tan W, ACS Nano 11 (2017) 3943–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [164].Zhou YG, Mohamadi RM, Poudineh M, Kermanshah L, Ahmed S, Safaei TS, Stojcic J, Nam RK, Sargent EH, Kelley SO, Small 12 (2016) 727–732. [DOI] [PubMed] [Google Scholar]
- [165].Cheng N, Du D, Wang X, Liu D, Xu W, Luo Y, Lin Y, Trends. Biotechnol 37 (2019) 1236–1254. [DOI] [PubMed] [Google Scholar]
- [166].Chen J, Xu Y, Lu Y, Xing W, Anal. Chem 90 (2018) 14207–14215. [DOI] [PubMed] [Google Scholar]
- [167].Hong SL, Wan YT, Tang M, Pang DW, Zhang ZL, Anal. Chem 89 (2017) 6535–6542. [DOI] [PubMed] [Google Scholar]
- [168].Zhou J, Meng L, Ye W, Wang Q, Geng S, Sun C, Anal. Chim. Acta 1022 (2018) 124–130. [DOI] [PubMed] [Google Scholar]
- [169].Huang R, He L, Xia Y, Xu H, Liu C, Xie H, Wang S, Peng L, Liu Y, Liu Y, He N, Li Z, Small 15 (2019) e1900735. [DOI] [PubMed] [Google Scholar]
- [170].Dong H, Chen H, Jiang J, Zhang H, Cai C, Shen Q, Anal. Chem 90 (2018) 4507–4513. [DOI] [PubMed] [Google Scholar]
- [171].Yu LH, Chen YF, Anal. Chem 87 (2015) 2845–2851. [DOI] [PubMed] [Google Scholar]
- [172].Niether D, Wiegand S, J. Phys. Condens. Matter 31 (2019) 503003. [DOI] [PubMed] [Google Scholar]
- [173].Lee H, Castro CM, Nat. Biomed. Eng 3 (2019) 163–164. [DOI] [PubMed] [Google Scholar]
- [174].Zhao J, Liu C, Li Y, Ma Y, Deng J, Li L, Sun J, J. Am. Chem. Soc 142 (2020) 4996–5001. [DOI] [PubMed] [Google Scholar]
- [175].Liu C, Zhao J, Tian F, Cai L, Zhang W, Feng Q, Chang J, Wan F, Yang Y, Dai B, Cong Y, Ding B, Sun J, Tan W, Nat. Biomed. Eng 3 (2019) 183–193. [DOI] [PubMed] [Google Scholar]
- [176].Garcia-Manrique P, Matos M, Gutierrez G, Pazos C, Blanco-Lopez MC, Extracell J. Vesicles 7 (2018) 1422676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Barreto JA, O’Malley W, Kubeil M, Graham B, Stephan H, Spiccia L, Adv. Mater 23 (2011) H18–40. [DOI] [PubMed] [Google Scholar]
- [178].Park JH, von Maltzahn G, Xu MJ, Fogal V, Kotamraju VR, Ruoslahti E, Bhatia SN, Sailor MJ, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Abbas M, Zou Q, Li S, Yan X, Adv. Mater 29 (2017) 1605021. [DOI] [PubMed] [Google Scholar]
- [180].Batrakova EV, Kim MS, Control J. Release 219 (2015) 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [181].Pascucci L, Cocce V, Bonomi A, Ami D, Ceccarelli P, Ciusani E, Vigano L, Locatelli A, Sisto F, Doglia SM, Parati E, Bernardo ME, Muraca M, Alessandri G, Bondiolotti G, Pessina A, Control J. Release 192 (2014) 262–270. [DOI] [PubMed] [Google Scholar]
- [182].Yeo RW, Lai RC, Zhang B, Tan SS, Yin Y, Teh BJ, Lim SK, Adv. Drug Deliv. Rev 65 (2013) 336–341. [DOI] [PubMed] [Google Scholar]
- [183].Munagala R, Aqil F, Jeyabalan J, Gupta RC, Cancer Lett. 371 (2016) 48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [184].Saari H, Lazaro-Ibanez E, Viitala T, Vuorimaa-Laukkanen E, Siljander P, Yliperttula M, Control J. Release 220 (2015) 727–737. [DOI] [PubMed] [Google Scholar]
- [185].Regev-Rudzki N, Wilson DW, Carvalho TG, Sisquella X, Coleman BM, Rug M, Bursac D, Angrisano F, Gee M, Hill AF, Baum J, Cowman AF, Cell 153 (2013) 1120–1133. [DOI] [PubMed] [Google Scholar]
- [186].Wood MJ, O’Loughlin AJ, Samira L, Ther. Deliv 2 (2011) 1095–1099. [DOI] [PubMed] [Google Scholar]
- [187].El Andaloussi S, Lakhal S, Mager I, Wood MJ, Adv. Drug Deliv. Rev 65 (2013) 391–397. [DOI] [PubMed] [Google Scholar]
- [188].Zhao Z, Zlokovic BV, Cell Res. 27 (2017) 849–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ, Nat. Biotechnol 29 (2011) 341–345. [DOI] [PubMed] [Google Scholar]
- [190].Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J, Nie G, Biomaterials 35 (2014) 2383–2390. [DOI] [PubMed] [Google Scholar]
- [191].Haney MJ, Klyachko NL, Zhao Y, Gupta R, Plotnikova EG, He Z, Patel T, Piroyan A, Sokolsky M, Kabanov AV, Batrakova EV, Control J. Release 207 (2015) 18–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Tan A, Rajadas J, Seifalian AM, Adv. Drug Deliv. Rev 65 (2013) 357–367. [DOI] [PubMed] [Google Scholar]
- [193].Wahlgren J, De LKT, Brisslert M, Vaziri Sani F, Telemo E, Sunnerhagen P, Valadi H, Nucleic Acids Res. 40 (2012) e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [194].Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM, Proc. Natl. Acad. Sci. U. S. A 107 (2010) 6328–6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [195].Jang SC, Kim OY, Yoon CM, Choi DS, Roh TY, Park J, Nilsson J, Lotvall J, Kim YK, Gho YS, ACS Nano 7 (2013) 7698–7710. [DOI] [PubMed] [Google Scholar]
- [196].Au Yeung CL, Co NN, Tsuruga T, Yeung TL, Kwan SY, Leung CS, Li Y, Lu ES, Kwan K, Wong KK, Schmandt R, Lu KH, Mok SC, Nat. Commun 7 (2016) 11150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [197].Chen G, Huang AC, Zhang W, Zhang G, Wu M, Xu W, Yu Z, Yang J, Wang B, Sun H, Xia H, Man Q, Zhong W, Antelo LF, Wu B, Xiong X, Liu X, Guan L, Li T, Liu S, Yang R, Lu Y, Dong L, McGettigan S, Somasundaram R, Radhakrishnan R, Mills G, Lu Y, Kim J, Chen YH, Dong H, Zhao Y, Karakousis GC, Mitchell TC, Schuchter LM, Herlyn M, Wherry EJ, Xu X, Guo W, Nature 560 (2018) 382–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [198].Poggio M, Hu T, Pai CC, Chu B, Belair CD, Chang A, Montabana E, Lang UE, Fu Q, Fong L, Blelloch R, Cell 177 (2019) 414–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [199].Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, Barnes S, Grizzle W, Miller D, Zhang HG, Mol. Ther 18 (2010) 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [200].Alexander M, Hu R, Runtsch MC, Kagele DA, Mosbruger TL, Tolmachova T, Seabra MC, Round JL, Ward DM, O’Connell RM, Nat. Commun 6 (2015) 7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [201].Syn NL, Wang L, Chow EK, Lim CT, Goh BC, Trends. Biotechnol 35 (2017) 665–676. [DOI] [PubMed] [Google Scholar]
- [202].Melo SA, Sugimoto H, O’Connell JT, Kato N, Villanueva A, Vidal A, Qiu L, Vitkin E, Perelman LT, Melo CA, Lucci A, Ivan C, Calin GA, Kalluri R, Cancer Cell 26 (2014) 707–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [203].Kamerkar S, LeBleu VS, Sugimoto H, Yang S, Ruivo CF, Melo SA, Lee JJ, Kalluri R, Nature 546 (2017) 498–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [204].Kojima R, Bojar D, Rizzi G, Hamri GC, El-Baba MD, Saxena P, Auslander S, Tan KR, Fussenegger M, Nat. Commun 9 (2018) 1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [205].Wang Y, Chen X, Tian B, Liu J, Yang L, Zeng L, Chen T, Hong A, Wang X, Theranostics 7 (2017) 1360–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [206].Hadla M, Palazzolo S, Corona G, Caligiuri I, Canzonieri V, Toffoli G, Rizzolio F, Nanomedicine 11 (2016) 2431–2441. [DOI] [PubMed] [Google Scholar]
- [207].Johnsen KB, Gudbergsson JM, Skov MN, Pilgaard L, Moos T, Duroux M, Biochim. Biophys. Acta 1846 (2014) 75–87. [DOI] [PubMed] [Google Scholar]
- [208].Hood JL, Scott MJ, Wickline SA, Anal. Biochem 448 (2014) 41–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [209].Liu C, Zhang W, Li Y, Chang J, Tian F, Zhao F, Ma Y, Sun J, Nano Lett. 19 (2019) 7836–7844. [DOI] [PubMed] [Google Scholar]
- [210].Kim MS, Haney MJ, Zhao Y, Mahajan V, Deygen I, Klyachko NL, Inskoe E, Piroyan A, Sokolsky M, Okolie O, Hingtgen SD, Kabanov AV, Batrakova EV, Nanomedicine 12 (2016) 655–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [211].Arteaga CL, Sliwkowski MX, Osborne CK, Perez EA, Puglisi F, Gianni L, Nat. Rev. Clin. Oncol 9 (2011) 16–32. [DOI] [PubMed] [Google Scholar]
- [212].Baselga J, Swain SM, Nat. Rev. Cancer 9 (2009) 463–475. [DOI] [PubMed] [Google Scholar]
- [213].Lamichhane TN, Jeyaram A, Patel DB, Parajuli B, Livingston NK, Arumugasaamy N, Schardt JS, Jay SM, Cell Mol. Bioeng 9 (2016) 315–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [214].Lin Y, Wu J, Gu W, Huang Y, Tong Z, Huang L, Tan J, Adv. Sci 5 (2018) 1700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [215].El-Andaloussi S, Lee Y, Lakhal-Littleton S, Li J, Seow Y, Gardiner C, Alvarez-Erviti L, Sargent IL, Wood MJ, Nat. Protoc 7 (2012) 2112–2126. [DOI] [PubMed] [Google Scholar]
- [216].Morishita M, Takahashi Y, Matsumoto A, Nishikawa M, Takakura Y, Biomaterials 111 (2016) 55–65. [DOI] [PubMed] [Google Scholar]
- [217].Wang J, Li W, Lu Z, Zhang L, Hu Y, Li Q, Du W, Feng X, Jia H, Liu BF, Nanoscale 9 (2017) 15598–15605. [DOI] [PubMed] [Google Scholar]
- [218].Wang J, Dong Y, Li Y, Li W, Cheng K, Qian Y, Xu G, Zhang X, Hu L, Chen P, Du W, Feng X, Zhao Y-D, Zhang Z, Liu B-F, Adv. Funct. Mater 28 (2018) 1707360. [Google Scholar]
- [219].Kumar CS, Mohammad F, Adv. Drug Deliv. Rev 63 (2011) 789–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [220].Ji T, Zhao Y, Ding Y, Nie G, Adv. Mater 25 (2013) 3508–3525. [DOI] [PubMed] [Google Scholar]
- [221].Silva AK, Di Corato R, Pellegrino T, Chat S, Pugliese G, Luciani N, Gazeau F, Wilhelm C, Nanoscale 5 (2013) 11374–11384. [DOI] [PubMed] [Google Scholar]
- [222].Liang R, Wei M, Evans DG, Duan X, Chem. Commun 50 (2014) 14071–14081. [DOI] [PubMed] [Google Scholar]
- [223].Wang Y, Huang R, Liang G, Zhang Z, Zhang P, Yu S, Kong J, Small 10 (2014) 109–116. [DOI] [PubMed] [Google Scholar]
- [224].Qi H, Liu C, Long L, Ren Y, Zhang S, Chang X, Qian X, Jia H, Zhao J, Sun J, Hou X, Yuan X, Kang C, ACS Nano 10 (2016) 3323–3333. [DOI] [PubMed] [Google Scholar]
- [225].Zhang W, Yu ZL, Wu M, Ren JG, Xia HF, Sa GL, Zhu JY, Pang DW, Zhao YF, Chen G, ACS Nano 11 (2017) 277–290. [DOI] [PubMed] [Google Scholar]
- [226].Das S, Extracellular RNACC, Ansel KM, Bitzer M, Breakefield XO, Charest A, Galas DJ, Gerstein MB, Gupta M, Milosavljevic A, McManus MT, Patel T, Raffai RL, Rozowsky J, Roth ME, Saugstad JA, Van Keuren-Jensen K, Weaver AM, Laurent LC, Cell 177 (2019) 231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [227].Ludwig N, Whiteside TL, Reichert TE, Int. J. Mol. Sci 20 (2019) 4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [228].Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP, Biomed Res. Int 2018 (2018) 8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [229].Wu CX, Liu ZF, J. Invest. Dermatol 138 (2018) 89–97. [DOI] [PubMed] [Google Scholar]
- [230].Baek G, Choi H, Kim Y, Lee HC, Choi C, Stem Cells Transl. Med 8 (2019) 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [231].Johnsen KB, Gudbergsson JM, Skov MN, Christiansen G, Gurevich L, Moos T, Duroux M, Cytotechnology 68 (2016) 2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [232].Geng T, Lu C, Lab Chip 13 (2013) 3803–3821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [233].Wang S, Lee LJ, Biomicrofluidics 7 (2013) 11301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [234].Zhang Z, Dombroski JA, King MR, Cell Mol. Bioeng 13 (2020) 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [235].Su W, Li H, Chen W, Qin J, Trends Anal. Chem 118 (2019) 686–698. [Google Scholar]
- [236].Shao H, Im H, Castro CM, Breakefield X, Weissleder R, Lee H, Chem. Rev 118 (2018) 1917–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [237].Zipkin M, Nat. Biotechnol 37 (2019) 1395–1400. [DOI] [PubMed] [Google Scholar]
- [238].Tutrone R, Donovan MJ, Torkler P, Tadigotla V, McLain T, Noerholm M, Skog J, McKiernan J, Prostate Cancer Prostatic Dis. 23 (2020) 607–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [239].Tian T, Li J, Song Y, Zhou L, Zhu Z, Yang CJ, Lab Chip 16 (2016) 1139–1151. [DOI] [PubMed] [Google Scholar]
- [240].Vuong QH, Ho MT, Vuong TT, La VP, Ho MT, Nghiem KP, Tran BX, Giang HH, Giang TV, Latkin C, Nguyen HT, Ho CSH, Ho RCM, J. Clin. Med 8 (2019) 168. [DOI] [PMC free article] [PubMed] [Google Scholar]