Abstract
In breast cancer, undetected lymph node metastases can spread to distal parts of the body for which the 5-year survival rate is only 27%, making accurate nodal metastases diagnosis fundamental to reducing the burden of breast cancer, when it is still early enough to intervene with surgery and adjuvant therapies. Currently, breast cancer management entails a time consuming and costly sequence of steps to clinically diagnose axillary nodal metastases status. The purpose of this study is to determine whether preoperative, clinical DCE MRI of the primary tumor alone may be used to predict clinical node status with a deep learning model. If possible then many costly steps could be eliminated or reserved for only those with uncertain or probable nodal metastases. This research develops a data-driven approach that predicts lymph node metastasis through the judicious integration of clinical and imaging features from preoperative 4D dynamic contrast enhanced (DCE) MRI of 357 patients from 2 hospitals. Innovative deep learning classifiers are trained from scratch, including 2D, 3D, 4D and 4D deep convolutional neural networks (CNNs) that integrate multiple data types and predict the nodal metastasis differentiating nodal stage N0 (non metastatic) against stages N1, N2 and N3. Appropriate methodologies for data preprocessing and network interpretation are presented, the later of which bolster radiologist confidence that the model has learned relevant features from the primary tumor. Rigorous nested 10-fold cross-validation provides an unbiased estimate of model performance. The best model achieves a high sensitivity of 72% and an AUROC of 71% on held out test data. Results are strongly supportive of the potential of the combination of DCE MRI and machine learning to inform diagnostics that could substantially reduce breast cancer burden.
Keywords: Breast cancer, Nodal metastases, DCE MRI, Deep learning
1. Introduction
Breast cancer is the most common cancer among women in many countries including the USA and causes more premature deaths than any cancer other than lung cancer. Among women with undetected breast lymph node metastasis, the 5 year survival rate is only 27% [3]. The presence of lymph node metastasis is the single most important prognostic factor in breast cancer [1]. Beyond prognosis, the detection of nodal metastases is used for cancer staging and to determine the course of surgical treatment, and is an important index for postoperative chemotherapy and radiotherapy.
The management of breast cancer entails a time consuming sequence of costly steps to diagnose whether the patient has axillary nodal metastases. In many hospitals this process entails: (1) ultrasound (US) imaging of the axilla along with tumor diagnosis costing $750, (2) breast DCE MRI (if US is negative MRI may still be positive) at $3,500, (3) axillary US again to reidentify the sentinel node $750, (4) US guided biopsy at $1,500, (5) pathology evaluation of the biopsy specimen costing $1,200. The earlier steps, typically (1) and (2), are used towards the clinical diagnosis of the lymph node status, cNode, which has 4 levels:N0 (no nodal metastasis), and increasing levels of nodal metastasis: N1, N2, and N3.
The purpose and clinical value of this study is to determine whether preoperative, clinical DCE MRI of the primary tumor may be used to predict node status with a deep learning model. If possible then steps 1,3 and 4 could be eliminated or reserved for only those patients with uncertain or probable metastases. Relying upon clinical MRI (1.5T) enables research results to have potential for direct clinical impact without costly upgrades to 3.0T. Furthermore, methods developed using the primary tumor will be applicable to most patients given the current standard protocol that images the tumor, while axillary nodes may not be in the field of view (FOV).
Dynamic contrast enhanced (DCE) MRI contains abundant information about the vascularity and structure of the tumor and is valuable to quantify cancer aggressiveness. Recently, preliminary results demonstrated that hand crafted tumoral features from 2D DCE MRI are can help predict nodal metastasis using classical machine learning SVM [6]. While promising these results relied upon research grade 3.0T MRI and were restricted to data from just 100 subjects acquired at one hospital. Furthermore validation performance is reported rather than more rigorous test set performance.
Deep learning has been shown to find useful patterns for image analysis [4]. It has been used to detect breast cancer with near expert radiologist accuracy [7]. However to the best of our knowledge, deep learning has not been used to predict axillary nodal metastasis from DCE MRI.
The strengths and contributions of this work are four-fold: (1) A 4D CNN model is proposed that automatically learns to fuse information from 4D DCE MRI (3D over time) and non-imaging clinical information. (2) The model relies exclusively on the primary tumor and does not require nodes to be within the FOV nor high field strength imaging which may not be available. (3) The model achieves a promising 72% accuracy while using rigorous nested cross-fold validation and while training and testing upon an extensive dataset of 357 subjects from two hospital sites using two distinct image acquisition protocols and hardware. (4) Saliency mapping demonstrates that the proposed model correctly learns to utilize primary tumor voxels when identifying both metastatic and non-metastatic subjects.
2. Materials and Methods
2.1. Materials
Clinical 1.5T DCE MRI was obtained from 357 breast cancer patients, whose characteristics are summarized in Table 1. Data for each subject includes a single precontrast and four serial dynamic image volumes acquired at a temporal resolution of 90s/phase obtained before and immediately after intravenous bolus infusion of a contrast agent. 221 subjects were obtained from Parkland Hospital, Dallas, Texas where dynamic VIBRANT sagittal images were acquired with a GE Optima MR450w 1.5T scanner using 0.1 mmol/kg gadopentetate dimeglumine contrast medium. The remaining 136 subjects came from UT Southwestern Medical Center, Dallas, Texas where dynamic FSPGR (THRIVE) axial images are acquired with a Philips Intera 1.5T scanner using 0.1 mmol/kg Gadavist contrast medium. Additionally, four clinical features were obtained including age (yrs), estrogen receptor status (ER), human epidermal growth factor receptor-2 (HER2), and a marker for proliferation (Ki-67). Clinical node status (cNode) ground truth was determined by one of 13 board-certified radiologists, fellowship-trained in breast imaging and breast MRI, who assess the 4D DCE MRI and ultrasound imaging information, clinical measures, clinical history available at the time of the image reading.
Table 1.
Demographics and disease characteristic of subjects included in this analysis.
| Variable | Age | cNode status | Tumor stage | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Category | 21–30 | 31–40 | 41–50 | 51–60 | 61–70 | 71–80 | 81–90 | N0 | N1 | N2 | N3 | T1 | T2 | T3 | T4 |
| Percentage | 2 | 16 | 34 | 23 | 19 | 5 | 1 | 62 | 28 | 4 | 6 | 30 | 44 | 19 | 7 |
| Number of patients | 7 | 57 | 121 | 82 | 68 | 18 | 4 | 221 | 101 | 13 | 22 | 107 | 157 | 68 | 25 |
2.2. Methods
Each subject has five 3D MRI volumes which are denoted as time1, time2, time3, time4, time5. Board certified radiologists traced the boundary of the primary tumor of each subject on the time3 volume. Then a 3D cuboidal bounding box encompassing the tumor region of interest (ROI) and peri-tumoral area was defined and used to crop each subject data to consistently-sized 3D volumes. Three difference images were then defined for each subject by subtracting voxelwise the cropped time3-time1, time4-time1 and time5-time1. These difference volumes are used to train the deep learning model and the processing steps are illustrated in Fig. 1. Next the intensities for the difference images from each hospital are harmonized by: (1) clipping the values of the lowest and highest 0.5% intensities, and (2) computing the mean and standard deviation of the intensities per hospital and transforming the intensities to have zero mean and unit variance.
Fig. 1.
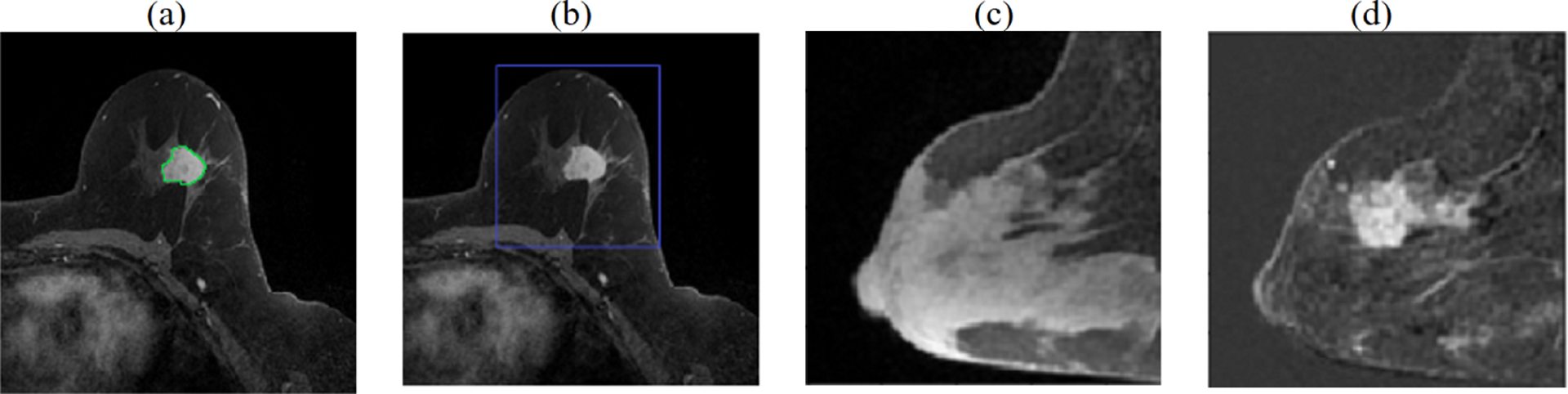
Preprocessing the volumetric DCE MRI. (a) primary tumor is radiologist delineated at time3 in each slice (green contour), (b) MRI is cropped to a cuboidal volume around tumor, (c) sagittal view showing breast at time1, (d) tumor is enhanced by computing difference images, shown here: time3-time1.
The data is partitioned using nested, stratified group 10-fold cross-validation with the test data held out and not used during training nor validation. This ensured a subject’s data appears in only 1 fold and each fold has the same ratio of patients from each hospital, and the same ratio of negative (cNode = N0) and positive (N1, N2, N3) labels. Data is augmented 27× by small random translation and rotation of the cropping volume.
In this work, we develop two-category classifiers to predict whether or not there is lymph node metastasis. Our classifiers are predominantly CNNs [2] because they automatically learn a hierarchy of intensity features from difference volumes. For brevity we explain the most complex 4D hybrid CNN model that we build which takes as input a set of difference volumes and the clinical data. The remaining models (e.g. 2D CNN, 3D CNN) are simpler and their architectural details can be found in the supplemental file. The 4D hybrid CNN model consist of four 4D convolutional layers and three fully connected layers. Each convolutional layer is followed by max-pooling. Then the output of the convolutional layers are concatenated with the clinical features to form the final classifier and all layers use batch normalization [5], except the last which outputs the metastasis diagnosis. Figure 2 visualizes the model architecture and how the data dimensions change in each layer.
Fig. 2.
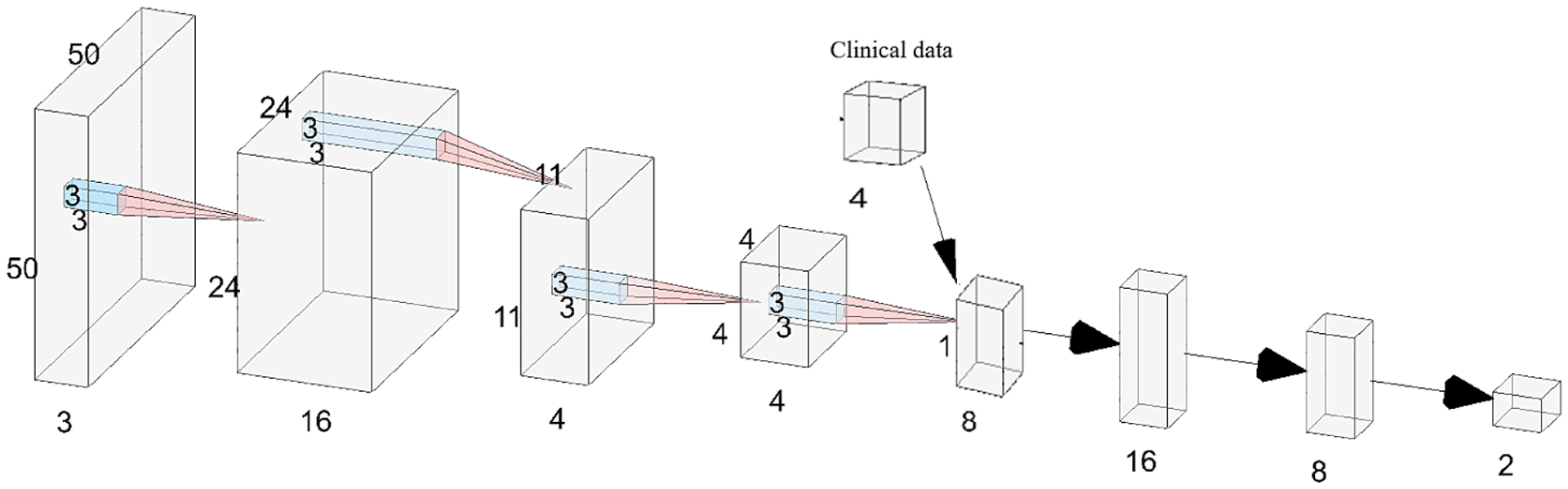
The 4D CNN model architecture. The model consists of 4 convolutional layers (red pyramids) followed by 3 fully connected layers (horz. arrows). Input and feature maps are 4D tensors; to visualize them they are rendered as 3D volumes by omitting one dimension, e.g. the input layer (left) is 50 × 50× 50 ×3 but rendered as 50 × 50 × 3. Four clinical features are concatenated with the 4 outputs of last conv. layer, creating an 8-vector as input to the dense layers. One-hot encoded output layer predicts probability of nodal metastasis and no metastasis. Illustration generated using [12].
The output of the model can be summarized in Eq. (1)
| (1) |
where xk is the set of 3D difference images for the kth patient, ck is their clinical data, and 2-dimensional p is the predicted probabilities that the model f() assigns to the two output categories. The threshold is >= 0.5 for positive prediction and < 0.5 for negative prediction.
Since the dataset has moderate imbalance (62% of subjects are non-metastatic) and we would like the model to focus more on positive than negative cases, we apply a weighted cost function where the cost for positive cases is twice that of the negative cases:
| (2) |
where E is the cost function, N is the number of subjects, lk and pk are the label and prediction of the kth patient respectively, and w is the weight assigned according to class label where: w1 = 1 and w2 = 2. This MSE cost function works well on classification tasks when using a softmax output [10]. Model fitting is trained from scratch and model weights are learned with the Adam [9] optimizer using: an adaptive learning rate initialized to 0.001, beta1 = 0.9, beta2 = 0.999, and a batch size of 36. Models are implemented in Tensorflow and trained on a Linux workstation with 2 NVIDIA V100s GPUs.
3. Results
Five types of input feature sets were tested: clinical features alone, 2D images alone, 3D images alone, 4D images (3D + time), and 4D with clinical features. Results are summarized in Table 2. With clinical data only, an XGBoost classifier [11] attained a performance just above chance accuracy with an AUC = 0.55. Dense feedforward neural networks did not perform better. The relative importance of the 4 clinical features computed as f scores, are shown in Fig. 3. Ki67 and HER2 are top ranked and have approximately equal importance while age and ER status are less important. Using 2D difference images and a deep 2D CNN, performance improved to an AUC = 0.61. Using the additional context of 3D difference images performance further improved to AUC = 0.66. Each 3D difference image is input independently, however group stratification is used so that a subject’s data appears in either train, validation or test. Subsequently, all three 3D difference images of a patient (time3-time1, 4–1, and 5–1) are concatenated forming an input 4D tensor to the 4D CNN. This produced the best results including an AUC = 0.67 and, when combined with clinical data an AUC of 0.71 and true positive rate of 0.72. This compares favorably with the results of a related study [13] in which texture features were manually specified.
Table 2.
Comparative performance across input feature sets on the held out test set. Increasing prediction performance was observed across the five sets of inputs evaluated including: only clinical features, 2D images, 3D images, 4D images (3D + time), and 4D with clinical features.
| Clinical only | 2D image | 3D image | 4D image | 4D img & clinical | |||||
|---|---|---|---|---|---|---|---|---|---|
| AUC | TPR | AUC | TPR | AUC | TPR | AUC | TPR | AUC | TPR |
| 0.55 | 0.24 | 0.61 | 0.35 | 0.66 | 0.84 | 0.67 | 0.72 | 0.71 | 0.72 |
Fig. 3.
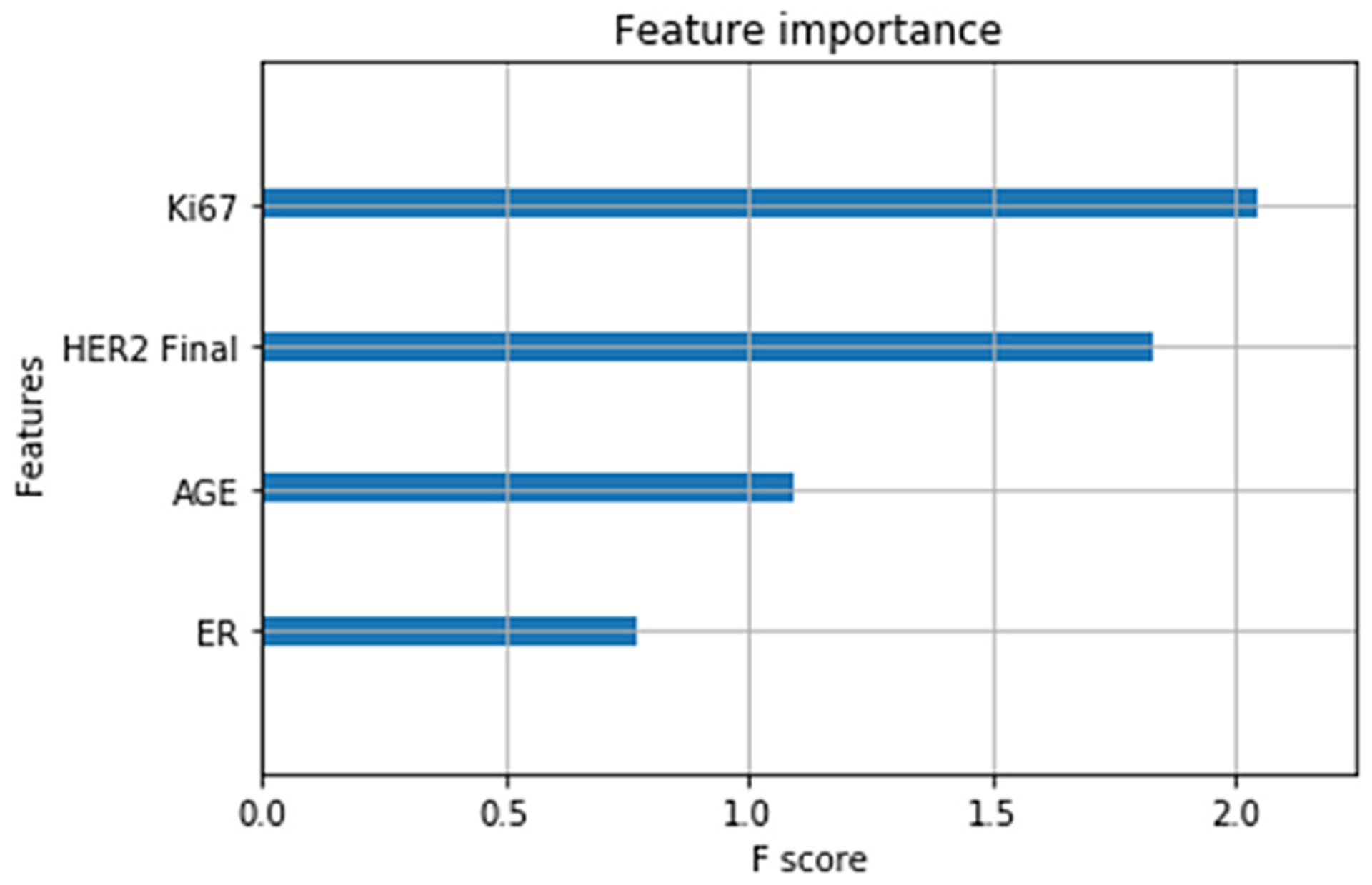
Relative importance of each clinical feature. Features with higher scores are more valuable for predicting metastasis.
4. Discussion
The results (Table 2) suggest the more data the model has the better its performance. This makes sense: 4D provides spatiotemporal context that 3D and 2D lack, while clinical features provide biomolecular information not accessible through MRI. To reveal which regions the top model learned as important Grad-CAM [8] was used to generate saliency maps. The most important voxels to predict metastasis free (Fig. 4a) or high probability of metastasis (Fig. 4b) are the primary tumor and its surround. More distal voxels are less important. That healthy breast tissue and non-breast tissue are of less value, makes sense biologically, and since the algorithm was not explicitly provided the non-linear tumor boundary this saliency map results suggests the model learned the segmentation on its own. Additional benefits are observed when adding clinical data to the 4D model including improving both the AUC and the true negative rate (TNR), while simultaneously reducing the variance across folds (Table 3), which further increases confidence in these performance estimates.
Fig. 4.
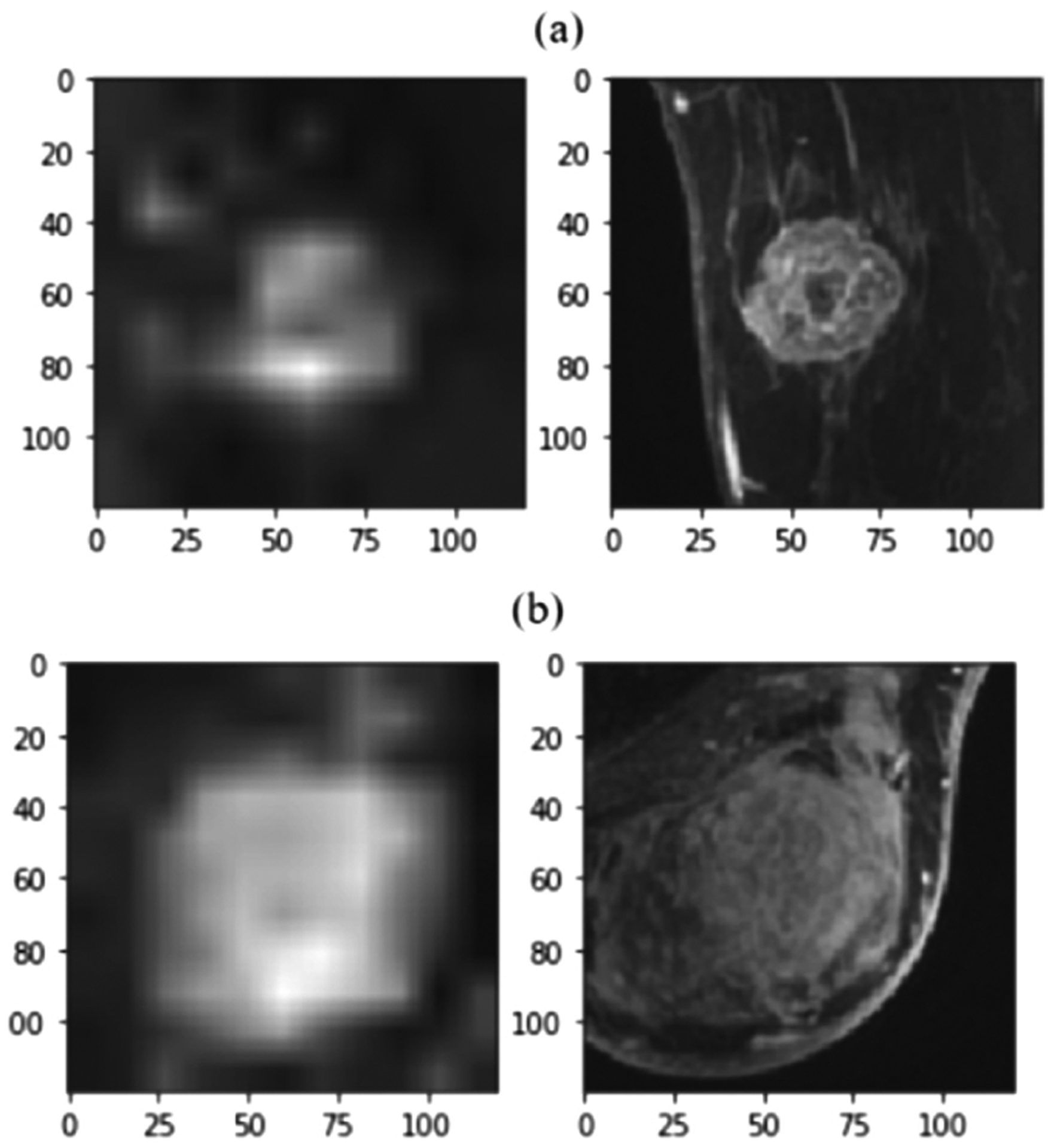
Important voxels revealed through saliency mapping. Saliency mapping with Grad-CAM (left column) demonstrates that the primary active tumor voxels in MRI (right column) are those most valuable for predicting boht (a) non-metastatic cancer, and (b) axillary metastatic cancer.
Table 3.
Comparison prediction accuracy and stability across feature sets. Both accuracy and stability increases when clinical features are added to the 4D CNN.
| Fold | 4D image | 4D & clinical | ||||
|---|---|---|---|---|---|---|
| AUC | TPR | TNR | AUC | TPR | TNR | |
| 0 | 0.61 | 0.71 | 0.45 | 0.77 | 0.71 | 0.73 |
| 1 | 0.78 | 0.79 | 0.68 | 0.85 | 0.71 | 0.76 |
| 2 | 0.71 | 0.93 | 0.23 | 0.83 | 0.86 | 0.64 |
| 3 | 0.82 | 0.71 | 0.77 | 0.67 | 1.00 | 0.00 |
| 4 | 0.37 | 1.00 | 0.00 | 0.58 | 0.29 | 0.87 |
| 5 | 0.65 | 0.64 | 0.59 | 0.62 | 0.71 | 0.55 |
| 6 | 0.82 | 1.00 | 0.00 | 0.8 | 0.83 | 0.61 |
| 7 | 0.62 | 0.38 | 0.64 | 0.54 | 0.62 | 0.41 |
| 8 | 0.75 | 0.62 | 0.82 | 0.8 | 0.92 | 0.48 |
| 9 | 0.55 | 0.46 | 0.55 | 0.56 | 0.54 | 0.52 |
| mean | 0.67 | 0.72 | 0.47 | 0.71 | 0.72 | 0.56 |
| std | 0.14 | 0.21 | 0.3 | 0.11 | 0.21 | 0.24 |
Our approach has some room for further improvement. Our reported AUC is 0.72 is very promising, but still needs improvement. We expect to improve the AUC to at least 0.8 to facilitate clinical adoption, while 0.9 is a long term target. There are several steps that can lead us there. Differences between hospital data is currently mitigated through preprocessing, however adversarial domain adaption could directly learn a model agnostic to these differences and may improve performance. Second, unsupervised pretraining on external data could improve performance. Third, other data combination methods beyond concatenation could be beneficial. Fourth, our preprocessing assumes no motion between MRI time frames, however patient movement can occur and could impede model performance. In the future we will apply motion correction to suppress any difference image artifact.
5. Conclusions
This work demonstrates that a deep 4D CNN has the potential to learn to predict axillary nodal metastases with high accuracy through the judicious fusion of spatiotemporal features of the primary tumor visible in DCE MRI, and that this further improves with the addition of clinical measures. Such high precision non-invasive methods that utilize standard clinical MRI (1.5T) and do not require complete imaging of the axillary nodes would fit well with clinical practices and could with further refinement, help patients avoid the costs associated with unnecessary lymph node surgery. The proposed method was tested on an extensive dataset of 357 subjects from 2 hospitals with distinct imaging protocols. Saliency mapping demonstrated that the model used tumor voxels to predict nodal metastasis which agrees with the expectation that aggressive tumors that spread to the nodes have appearance distinct from less invasive tumors. Such diagnostic methods hold the potential to streamline time consuming and costly steps currently used to clinically diagnose nodal metastases, improve doctor efficiency, and help select safe and effective treatments that reduce postoperative complications.
Supplementary Material
Footnotes
Electronic supplementary material The online version of this chapter (https://doi.org/10.1007/978-3-030-59713-9_32) contains supplementary material, which is available to authorized users.
References
- 1.Fisher B, et al. : Relation of number of positive axillary nodes to the prognosis of patients with primary breast cancer. An NSABP update. Cancer 52(9), 1551–1557 (1983) [DOI] [PubMed] [Google Scholar]
- 2.Soffer S, Ben-Cohen A, Shimon O, Amitai MM, Greenspan H, Klang E: Convolutional neural networks for radiologic images: a radiologist’s guide. Radiology 290(3), 590–606 (2019) [DOI] [PubMed] [Google Scholar]
- 3.American Cancer Society Homepage, March 2020. https://www.cancer.org/cancer/breast-cancer/understanding-a-breast-cancer-diagnosis/breast-cancer-survival-rates.html
- 4.LeCun Y, Bengio Y, Hinton G: Deep learning. Nature 521(7553), 436–444 (2015) [DOI] [PubMed] [Google Scholar]
- 5.Ioffe S, Szegedy C: Batch normalization: accelerating deep network training by reducing internal covariate shift (2015). arXiv preprint arXiv:1502.03167 [Google Scholar]
- 6.Cui X, et al. : Preoperative prediction of axillary lymph node metastasis in breast cancer using radiomics features of DCE-MRI. Sci. Rep 9(1), 1–8 (2019) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhou J, et al. : Weakly supervised 3D deep learning for breast cancer classification and localization of the lesions in MR images. J. Magn. Reson. Imaging 50(4), 1144–1151 (2019) [DOI] [PubMed] [Google Scholar]
- 8.Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D: Grad-CAM: visual explanations from deep networks via gradient-based localization. In: Proceedings of the IEEE International Conference on Computer Vision, pp. 618–626 (2017) [Google Scholar]
- 9.Kingma DP, Ba J: Adam: a method for stochastic optimization (2014). arXiv preprint arXiv:1412.6980 [Google Scholar]
- 10.Tyagi K, Nguyen S, Rawat R, Manry M: Second order training and sizing for the multilayer perceptron. Neural Process. Lett 51(1), 963–991 (2019). 10.1007/s11063-019-10116-7 [DOI] [Google Scholar]
- 11.Chen T, Guestrin C: XGBoost: a scalable tree boosting system. In: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, pp. 785–794, August 2016 [Google Scholar]
- 12.LeNail A: NN-SVG: publication-ready neural network architecture schematics. J. Open Source Softw 4(33), 747 (2019). 10.21105/joss.00747 [DOI] [Google Scholar]
- 13.Kulkarni S, Xi Y, Ganti R, Lewis M, Lenkinski R, Dogan B: Contrast texture-derived MRI radiomics correlate with breast cancer clinico-pathological prognostic factors. In: Radiological Society of North America 2017 Scientific Assembly and Annual Meeting, Chicago, IL, 26 November–1 December 2017. archive.rsna.org/2017/17002173.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


