Abstract
Coronavirus disease 2019 (COVID-19) has become a global pandemic with a high rate of transmission. Currently, there is a lack of vaccines and specific drugs for this newly-emerged virus. Timely diagnosis and treatment, as well as isolation of patients and virus carriers, contribute to the effective prevention and control of this epidemic. This review focuses on early stage COVID-19 diagnosis methods and strategies, highlighting the guiding role of laboratory indicators on treatment strategy formulation, and prognosis assessments.
Keywords: COVID-19, laboratory examination, SARS-CoV-2, etiological examination, nucleic acid test
INTRODUCTION
Coronavirus disease 2019 (COVID-19) is creating dramatic and daily evolving changes with profound impacts on people’s lives and has become a global pandemic with a high rate of transmission. By April 13, 2020, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) had infected over 1.7 million people worldwide, resulting in 111 652 deaths[1]. The fact that coronavirus disease 2019 (COVID-19) cases has been reported in 202 countries, areas or territories in both hemispheres has increased the risk of a long-enduring pandemic, as multiple new centers of outbreaks have been reported in middle Asia, Europe and North America.
SARS-CoV-2 is high contagious via human-to-human transmission, although the exact route of transmission is unclear. The burden laid by SARS-CoV-2 on health and medical resources significantly outstrips that made by the other two newly emerged coronavirus diseases, severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS), particularly in the centers of outbreaks. While most infected patients don’t require respiratory support, the capacity to quickly control transmission and properly treat patients with severe symptoms, directly impact the mortality rate of COVID-19. Hubei Province, the first reported center of the COVID-19 outbreak, had a higher mortality rate than any other region in China (4.6% vs. 0.9%, average 4.01%)[2], due to a sudden shortage of medical resources in late January. Similar situations are ongoing at the current outbreak centers in Europe, where the infected population is over a hundred of thousand, and mortality has exceeded 10%[1].
While our understanding towards SARS-CoV-2 and COVID-19 increases every day, the development of SARS-CoV-2 specific antiviral drugs and vaccines has taken time. Currently, the most effective way of controlling this pandemic without overwhelming the medical system is to cut the chain of transmission in time, through quick diagnosis and effective separation of infected patients from the uninfected population. This review will provide an overview of currently available methods and techniques for COVID-19 diagnosis and their application under varied situations.
LABORATORY EXAMINATIONS
The standard COVID-19 diagnostic protocol assesses clinical symptoms (fever and respiratory symptoms), SARS-CoV-2 nucleic acid detection, serum-specific antibodies, lung imaging, comprehensive epidemiological history (including clustered onset) and other factors (Fig. 1). Dynamic observations of lung imaging and laboratory examinations (such as blood routine examination, biochemical parameters, cytokine levels), help to identify critically ill patients early in the disease.
Fig. 1.
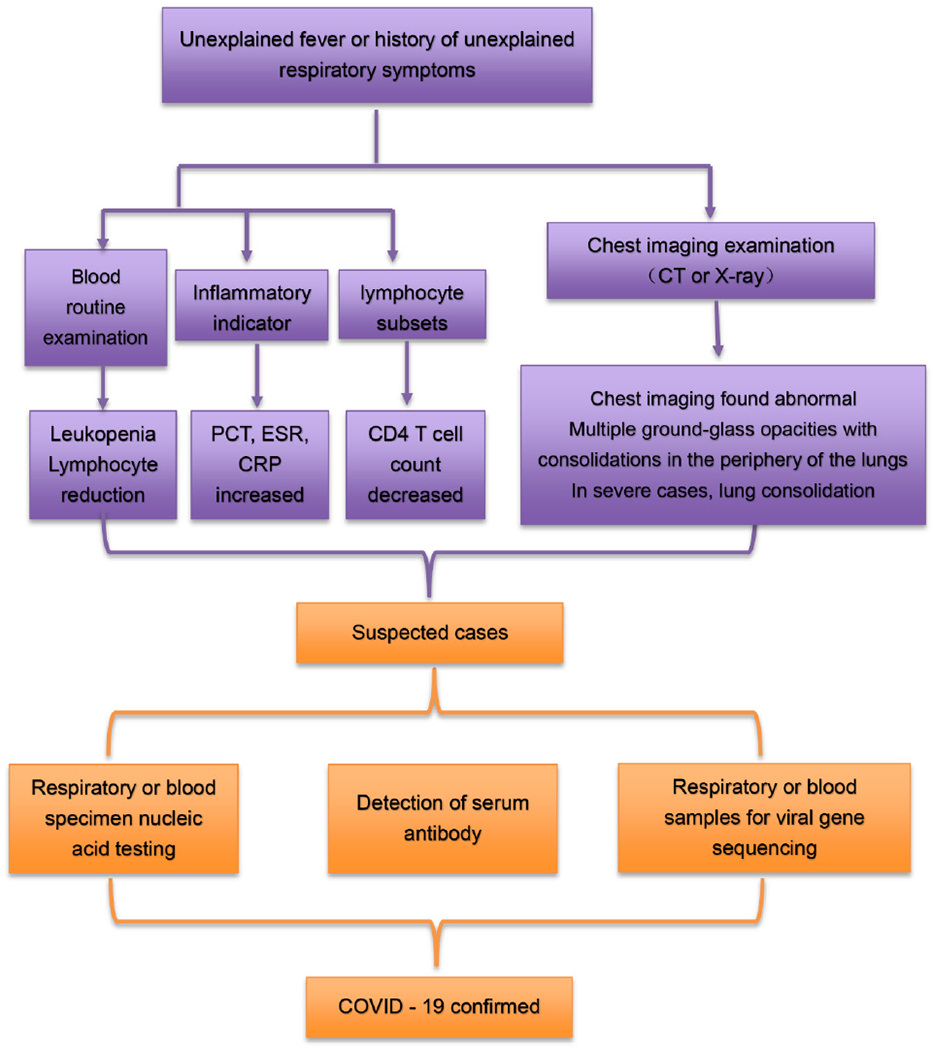
Diagnostic flow chart.
Blood routine examination has guiding significance for clinical treatment and early differential diagnosis
Although routine blood examination results are usually nonspecific, they can be used to distinguish COVID-19 from other types of pneumonia in the early stages. Laboratory results were combined with other studies which showed the characteristics of viral infections in the early stage of COVID-19: The blood cell examinations showed a normal leukocyte count in 246 of 338 patients (72.7%); the lymphocyte count (67.4%) was normal or low (67.4%, 228/338); forty-four of 51 (86.2%) patients’ neutrophil count was decreased (leukocyte count reference value: 4×109/L~10×109/L; lymphocyte count reference value: 0.8×109/L~4.0×109/L; neutrophil count reference value: 2×109/L~7×109/L) 3–6. Most patients had eosinopenia (52.9%, 73/138)7,8 (eosinophil count reference value: 0.02×109/L~0.52×109/L). As the disease progresses, it is more common to see the leukocyte and lymphocyte counts significantly decrease, accompanied by an increase in neutrophils[9]. In severe cases, the absolute value of lymphocytes decreased, showing a progressive reduction, while the number of neutrophils continued to increase in some infected patients. SARS-CoV-2 can directly invade the patient’s immunity system, destroy the lymphocytes and inhibit lymphocyte regeneration. In addition, COVID-19 patients showed normal or elevated neutrophil counts, but lymphocyte counts decreased, leading to the increase of NLR. NLR is further elevated as the illness progresses and remains decreased during convalescence. Therefore, NLR can serve as a visual clinical indicator[8].
In addition, patients’ blood routine examinations showed platelet counts had decreased (platelet counts reference value: 100×109/L ~300×109/L). This may be related to immune-related factors and other factors that caused secondary thrombocytopenia. Prof. Zhong conducted a retrospective study on the clinical characteristics of 1 099 COVID-19 patients in China, finding that about 36.2% of COVID-19 patients had thrombocytopenia, among which the higher incidence of severe cases was 57.7%, and that of non-severe cases was 31.6%[10–12]. However, one paper showed first an increased platelet peak, which then decreased in severe patients during treatment. This may be related to cytokine storms caused by SARS-CoV-2, which continued to stimulate platelet release. The cut-off value of platelet lymphocyte ratio difference (Δ PLR) as the patient’s condition progresses: If Δ PLR>126.7, medical staff must actively intervene to prevent further deterioration caused by the disease. This means the absolute value of platelet count and PLR dynamic changes are an important prognostic indicator for disease severity and prognosis[10,12].
Low CD4 T cell count is significantly associated with severe diseases. Some T-lymphocyte subset analysis research data showed CD4 and CD8 T-lymphocytes began to decrease at an early stage of COVID-19, but that the CD4/CD8 ratio was normal in 92.8% of the patients[13]. (CD4 reference value: 550/μL~1 600/μL; CD8 reference value: 320/μL~ 1 250/μL; CD4/CD8: 1.2~2.0). As the disease progresses, CD4 T cells have greater decrease in severe cases[14], meaning that the immunological function of COVID-19 patients is significantly suppressed. Thus, T-lymphocyte subset analysis would help improve the early diagnosis of COVID-19[8,15–16].
Abnormal changes in biochemical parameters suggest possible complications
In the laboratory’s biochemical indexes of COVID-19 patients, it was found that the indexes of some patients also changed with the development of the disease (Table 1). Liver function: alanine aminotransferase (ALT) and aspartate aminotransferase (AST) increased, albumin (ALB) was reduced, and the rate and extent of ALT and AST elevation in severe patients were higher than those in non-serious patients, suggesting that the patients had different degrees of liver damage[17]. However, from existing reports, patients’ liver enzymes were mostly mildly elevated [<3-4 times the upper limit of normal value (ULN)], and only very few patients had significant liver enzyme elevation (>10 ULN)[18]. Renal function: urea was generally reduced, serum iron was often reduced, and creatinine could be increased along with renal impairment. These suggest that novel coronavirus pneumonia combined with liver damage is more likely, owing to adverse drug reactions and systemic inflammatory responses in severely affected patients who are receiving medical treatment[19]. The abnormal biochemical indicators suggested that in addition to lung lesions, SARS-CoV-2 could also cause other complications such as liver or kidney damage[20].
Table 1.
Biomarkers and function of biochemical parameters
| Biomarkers | Function |
|---|---|
| Liver function: ALT, AST, TB, DB, LDH, ALB, SAA | When the liver cells of the human liver undergo inflammation, poisoning, and necrosis, the liver cells will be damaged, and the transaminase will be released into the blood, causing the transaminase in the serum to rise and turn abnormal. |
| Kidney function: CRE, UA | The body excretes metabolic waste from the kidneys to maintain the body’s electrolyte stability and acid-base balance. Kidney damage can cause a series of changes in serum indicators. |
| Myocardial enzymes: CK, CK-MB | Cardiac cell damage results in increased cell membrane permeability and release of cardiac enzymes into the blood. By detecting changes in myocardial enzymes, the degree of myocardial damage can be determined. |
Dynamic monitoring of the myocardial enzyme spectrum is also an important basis for the clinical assessment of COVID-19 patients with heart damage. Some COVID-19 patients had elevated creatine kinase (CK), lactate dehydrogenase (LDH), hydroxybutyrate dehydrogenase (HBDH) or aspartate aminotransferase (AST)[21]. It is speculated that as COVID-19 progressed, heart hypoxia will increase, myocardial cells will be damaged at varying degrees and cell membrane permeability will increase, leading to increased myocardial enzyme activity. The lactate dehydrogenase (LDH) and brain natriuretic peptide (BNP) in severe patients are both significantly higher than in ordinary patients[22]. This may be due to increased hypoxia leading to heart damage following disease progression. It is suggested that special care should be taken to protect heart function in COVID-19 patients[23]. The higher the ALT, AST, TB, DB, SAA, LDH, and CK-MB, the greater the risk of SARS-CoV-2 infection. It is suggested that these items can be used as early auxiliary diagnoses indicators of biochemical changes indicative of SARS-CoV-2[24].
Cytokine storm syndrome (CSS) is a very significant cause of COVID-19 patient death
Generally speaking, COVID-19 patients go through three stages of development. At the beginning of the infection, the virus begins to multiply in the body. In the second stage, the body desperately tries to remove the virus by releasing a large number of pro-inflammatory cytokines, while the excessive immune response causes damage to the body (Fig. 2). CSS causes rapid progression to acute respiratory distress syndrome, coagulopathy, and even multiple organ failure (Table 2). The present research found the following levels of pro-inflammatory cytokines: IL-2, IL-6, IL-7, IL-8, IL-9, IL-10, GM-CSF, MCP-1, TNF-α and INF-γ in the plasma of COVID-19 patients were increased[8,25]. Notably, expression levels of IL-6, IL-2R and IL-10 were greatly increased in severe patients. Monitoring the levels of IL-6, IL-2R and IL-10 in the disease’s early stages is helpful in assessing the risk of progression to a severe condition, and for the prognosis of patients. Therefore, this report recommends pro-inflammatory cytokines should be given more attention in the treatment of critically ill patients with COVID-19.
Fig. 2. Stage of COVID-19 development.
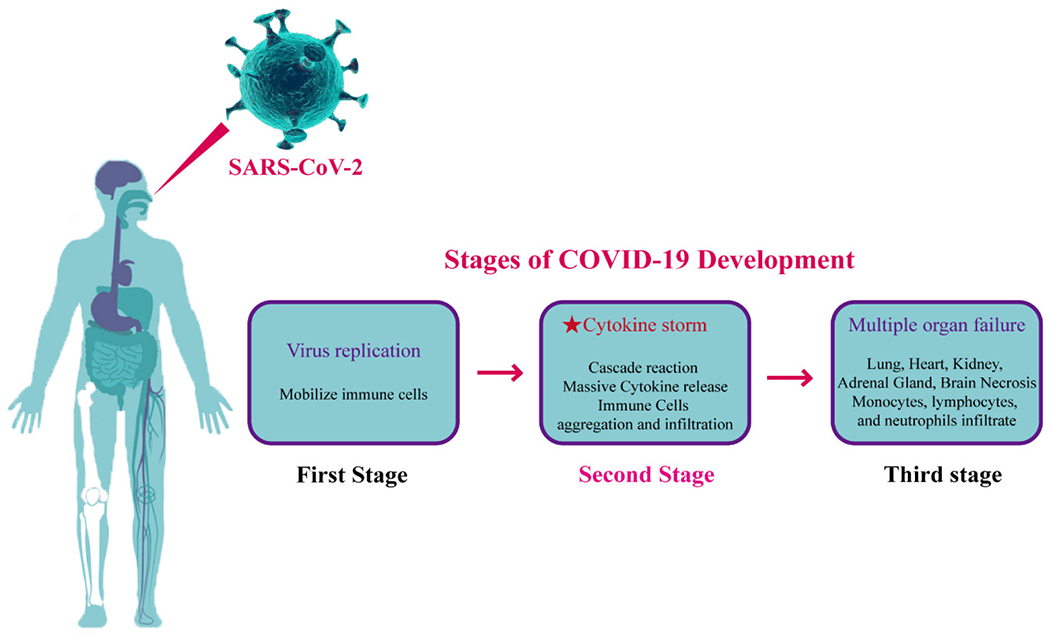
The most important factor is that a large number of cytokines are released in the process of clearing the viruses, causing cytokine storm.
Table 2.
Cytokine storm related cytokines and function
| Cytokins | Function |
|---|---|
| Interleukins (IL): IL-1β, IL-2 R, IL-6, IL-7, IL-8, IL-9, IL-10, IL-17 | Growth and differentiation of leukocytes |
| Interferons (IFN): IFN-γ | Activate antiviral properties and regulate innate immunity to play an antiproliferative role |
| Chemokines: MCP-1, IP10 | leukocyte recruitment |
| Colony-stimulating factors (CSF): G-CSF, GM-CSF | Stimulates hematopoietic progenitor cell proliferation and differentiation |
| Tumor necrosis factor (TNF) TNFα | Proinflammatory, activates cytotoxic T lymphocytes |
PCT, ESR and CRP are commonly used in the diagnosis of infectious diseases. Combined detection is a valuable indicator for the early diagnosis of COVID-19. While some evidence suggests that most patients with early stage COVID-19 have a normal level of procalcitonin (PCT), PCT was found significantly higher in severe patients than in non-severe patients. PCT is a specific inflammatory indicator of bacterial infection, and its significant increase indicates that patients are more susceptible to secondary bacterial infection and a poor prognosis[5,26]. In addition, erythrocyte-sedimentation rates (ESR) and levels of C-reactive protein (CRP) and hypersensitive C-reactive protein (HS-CRP) significantly increase in most COVID-19 patients[25–27] (ESR reference value: adult male 0-15mm/h, adult female 0-20mm/h; CRP reference value: <8mg/L; HS-CRP reference value: 1-10mg/L). Combined detection of CRP, hs-CRP, ESR and PCT levels can determine prognosis, combined with other indicators to analyze the severity of COVID-19[26].
The change of coagulation index is closely related to the development and prognosis of severe patients
Clotting function was normal in most patients, but coagulation dysfunction and the PT time was found longer in severe patients, even dead patients[28,29]. Which may be related to the large release of proinflammatory cytokines caused by severe neo-coronary pneumonia. These cytokines are mediators of atherosclerosis which directly lead to plaque rupture through local inflammation, induction of procoagulant factors, and changes in hemodynamics[30]. Current research indicates that disseminated intravascular coagulation (DIC) and pulmonary embolism may be one of the leading causes of death in patients with new crowns, however more research data is needed for inflammation (Fig. 3). This means that coagulation parameters are closely related to poor prognosis[31].
Fig. 3.
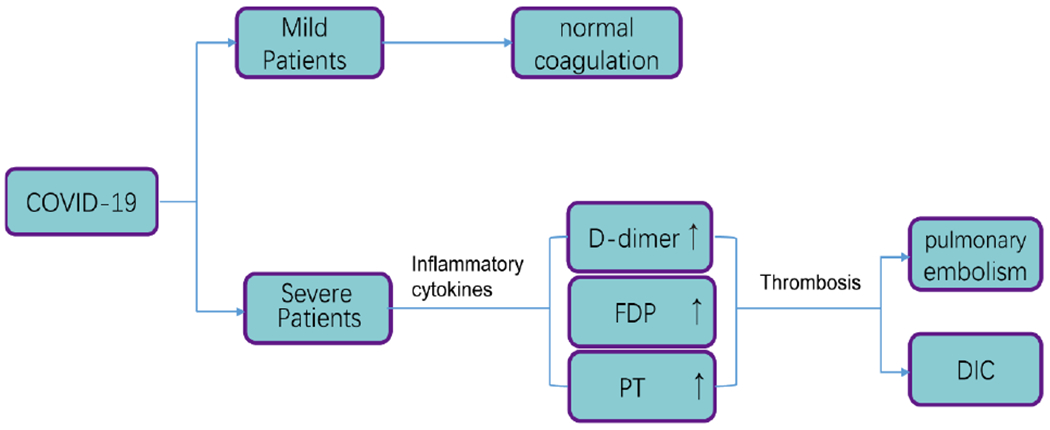
Coagulation index changes with the develop ment of the disease.
ETIOLOGICAL EXAMINATIONS
Molecular and phylogenetic characterizations of SARS-CoV-2
SARS-CoV-2 is a linear positive-sense single stranded RNA (ssRNA) virus, labeled under the subgenus Sarbecovirus of the genus Betacoronavirus[32]. Phylogenetic analysis has shown that SARS-CoV-2 has a closer relationship with two bat-derived coronaviruses (88% identity), bat-SL-CoVZC45 and bat-SL-CoVZXC21, than with SARS-CoV (~79% identity) and MERS-CoV (~50% identity) (Fig. 4)[33]. Over 2 000 SARS-CoV-2 genomes have been uploaded to public databases, providing a temporal resolution of nucleotide substitution rate (subs per site per year) of 8×10−4 without significant mutation[34–35], which is lower than that of Influenza A virus (1.43×10−3 to 11.62×10−3) [36] and SARS-CoV (0.80×10−3 to 2.38×10−3) [37], while being a little higher than MERS-CoV (4.81×10−4)[38]. According to recently published research, sequenced SARS-CoV-2 strains could be distinguished into 3 subtypes according to the amino acid changes. While A type was considered the ancestral type, B type was the most prevalent type in China and other areas in East Asia (Fig. 5)[39].
Fig. 4. Phylogenetic analysis of SARS-CoV-2 and representative viruses of the genus Betacoronavirus.
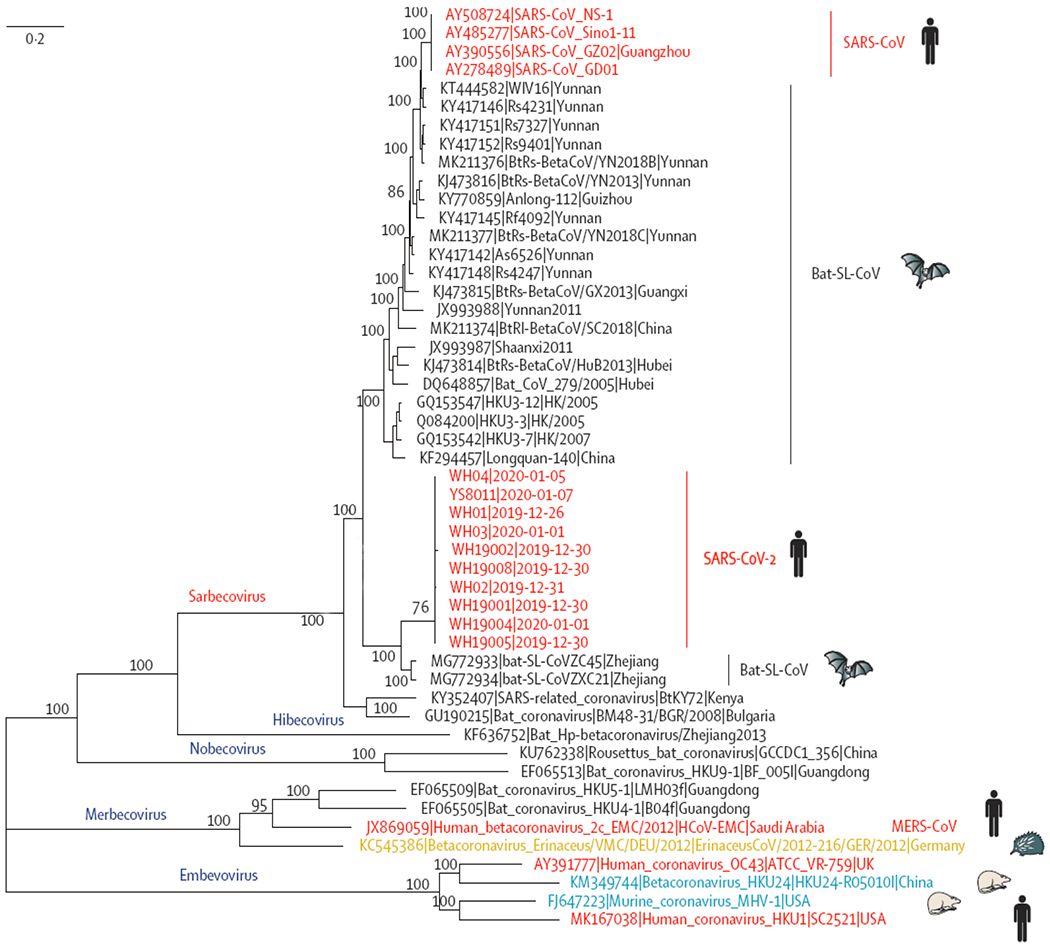
Adopted from Lu R. et al[33]. Phylogenetic analysis of SARS-CoV-2 and its closely related reference genomes revealed that SARS-CoV-2 belonged to the subgenus Sarbecovirus. The ten SARS-CoV-2 strains from Wuhan and the two bat-derived SARS-like strains from eastern China (bat-SL-CoVZC45 and bat-SL-CoVZXC21) formed a clade, while SARS-CoV-2 strains from humans and genetically similar SARS-like coronaviruses from bats of southwestern China formed another clade.
Fig. 5. Phylogenetic network of SARS-CoV-2 subtypes.
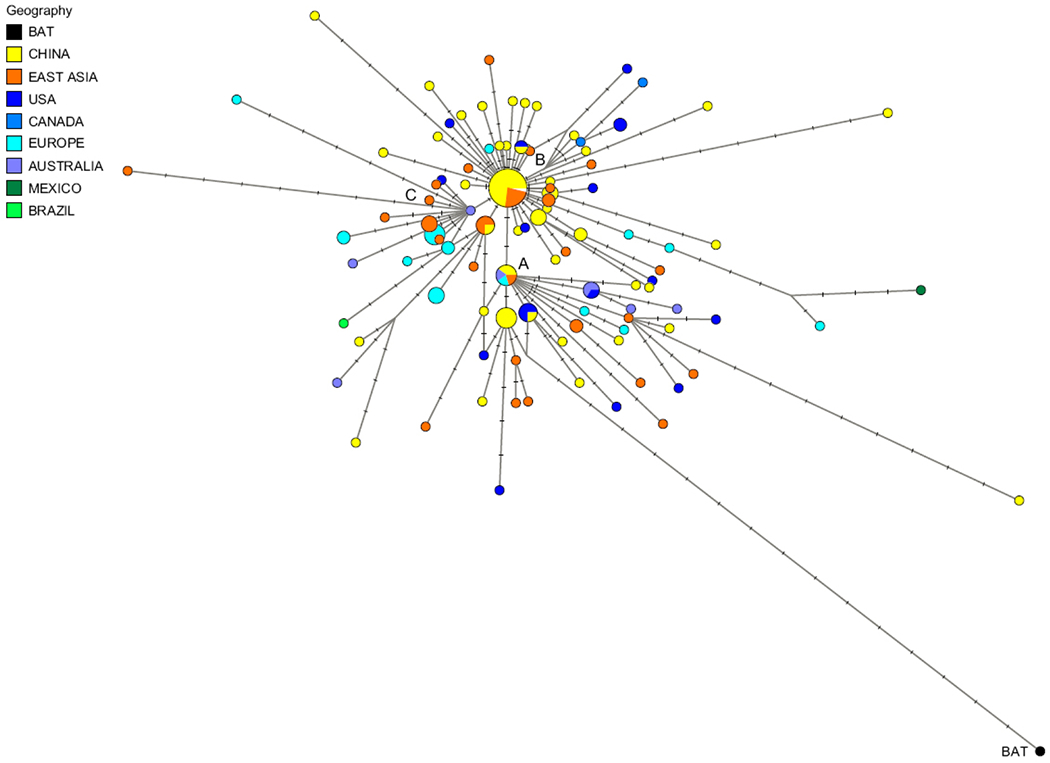
Adopted from Forster P. et al[39], using 160 SARS-CoV-2 genomes. The notch on the links represents a mutated nucleotide position, and the circle areas are proportional to the number of taxa. Using the bat virus as an outgroup resulted in the root of the network being placed in the “A” cluster of lineages. The network showed that ancestral viral genomes exist alongside their newly mutated daughter genomes. Cluster B is derived from A and most genomes in B cluster were sampled from Wuhan, from other parts of China, and from adjacent Asian countries. Cluster C derived from its parent Cluster B, but it contained no genomes from mainland Chinese samples.
The full-length genome of SARS-CoV-2 is about 29 903 nt, mainly encoding the non-structural protein replicase ORF1ab and four structural proteins, spike (S), envelope (E), membrane (M) and nucleocapsid (N) (Fig. 6) [32,40]. Seven conserved replication enzymes on ORF1ab are coronavirus-specific domains[41]. The S protein is responsible for receptor-binding and membrane fusion[42–43]; the M protein adapts a membrane region for virion assembly and structural proteins capture at the budding site[44–45]; the E protein is involved in the virus assembly and release[46–47]; and the N protein facilitate the binding of the viral RNA genome to the replication - transcription complex and virions packages[48–49]. Sequence identities between SARS-CoV-2 and other members of subgenus sarbecovirus vary from over 98% in the E gene to lower than 73% in the S gene[33]. So far, ORF1ab, E, N, RdRP and S genes have been used as molecular targets for SARS-CoV-2 nucleic acid tests[50].
Fig. 6. Schematic presentation of the SARS-CoV-2 genome organization.
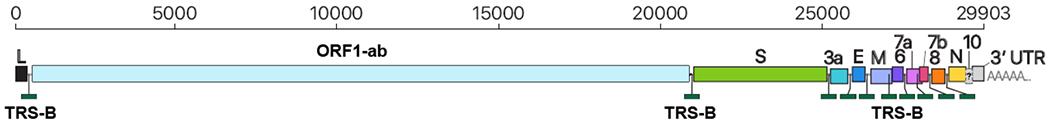
Adopted from Kim D. et al[40]. The representative SARS-CoV-2 full-length genomic RNA (29 903 nt) also serves as an mRNA. ORF1a and ORF1b are translated. Nine major subgenomic RNAs are produced. In order to provide better visualization, the sizes of the boxes representing small accessory proteins are bigger than the actual size of the ORF. The black box indicates the leader sequence.
Application of metagenomic high-throughput sequencing in SARS-CoV-2 detection
Metagenomic high-throughput sequencing is the core technology for obtaining the whole genome of SARS-CoV-2, which has laid the foundation for the development of diagnosis kits and epidemic analysis. Current major manufacturers of high-throughput sequencing platforms include Illumina, ThermoFisher, MGI, Oxford Nanopore and PacBio. Sequencing platforms produced by Illumina, MGI and ThermoFisher use next generation sequencing (NGS) technology, while single-molecule real-time sequencing (SMRT) techniques are applied by Oxford Nanopore and PacBio. NGS and SMRT have been reported in clini-cal use for pathogen detections, and both technologies were used in the de novo assembly of the first SARS-CoV-2 genomes[51].
The first complete genome of SARS-CoV-2 was published on the GISAID database on Jan 10th, 2020[52–53], 11 days after China reported clusters of pneumonia with unknown causes in association with the Huanan Seafood Market on Dec 31st, 2019[54]. The phylogenetic relation between SARS-CoV-2 and bat-derived coronaviruses was further confirmed by genetic analysis of genomes sequenced by DNBSEQ-T7, which is manufactured by MGI Shenzhen[33]. In China, high-throughput sequencing has been officially approved for COVID-19 diagnosis[55]. The DNBSEQ-T7 sequencing and analysis system has gone through emergency certification by the Chinese National Medical Products Administration (NMPA2020340061, NMPA2020340062), being the first certificated high-throughput sequencing system for SARS-CoV-2 detection and infection confirmation.
Despite the fact that high throughput sequencing has significantly contributed to our understanding of SARS-CoV-2, it is not as widely used as real-time reverse transcriptase polymerase chain reaction (rRT-PCR) and serology detection in clinical practice due to its higher cost and longer turn-around time. However, this technology is crucial not only in COVID-19 diagnosis, but also in origin tracing, infection mechanism exploration, mutations monitoring, and vaccine development.
Nucleic acid test for COVID-19 diagnosis
Nucleic acid detection is currently regarded as the golden standard for SARS-CoV-2/COVID-19 diagnosis. Reported nucleic acid detection methods for SARS-CoV-2 include rRT-PCR, metagenomic sequencing, isothermal amplification, digital PCR and CRISPR. As of 27 March 2020, there are at least 126 enterprises in China developing COVID-19 nucleic acid detection kits and 15 kits have been approved in China by the National Medical Products Administration (NMPA), including 10 rRT-PCR kits, 1 sequencing kit, 3 isothermal amplification kits and 1 Hybrid capture immunofluorescence kit[56]; the US FDA have given emergency use authorizations to 19 nucleic acid detection kits for diagnostic tests[57]. The WHO published a list of recommended target regions and primers from multiple national or regional health institutes (Table 3).
Table 3.
Recommended target region and primers for RT-PCR detection of SARS-CoV-2
| Institute | Gene targets | Primers and probe(s)(5′→3′) | PCR strategy | Sample/specimen type* |
|---|---|---|---|---|
| China CDC, China | ORF1ab and N |
Target 1: ORF1ab gene F: CCCTGTGGGTTTTACACTTAA R: ACGATTGTGCATCAGCTGA P: FAM–CCGTCTGCGGTATGTGGAAAGGTTATGG–BHQ1 Target 2: N gene F: GGGGAACTTCTCCTGCTAGAAT R: CAGACATTTTGCTCTCAAGCTG P: FAM–TTGCTGCTGCTTGACAGATT–TAMRA |
Real-time RT-PCR |
N/A |
| Institut Pasteur, Paris, France | Two targets in RdRP |
Target 1: RdRp gene / nCoV_IP2 nCoV_IP2–12669FW: ATGAGCTTAGTCCTGTTG nCoV_IP2–12759Rv: CTCCCTTTGTTGTGTTGT nCoV_IP2–12696bProbe(+): AGATGTCTTGTGCTGCCGGTA [5′]Hex [3′]BHQ–1 Target 2: RdRp gene / nCoV_IP4 nCoV_IP4–14059Fw GGTAACTGGTATGATTTCG nCoV 1 _IP4–14146Rv CTGGTCAAGGTTAATATAGG nCoV_IP4–14084Probe(+) TCATACAAACCACGCCAGG [5′]Fam [3′]BHQ–1 Target 3: E gene / E_Sarbeco E_Sarbeco_F1 ACAGGTACGTTAATAGTTAATAGCGT E_Sar 2 beco_R2 ATATTGCAGCAGTACGCACACA E_Sarbeco_P1 ACACTAGCCATCCTTACTGCGCTTCG [5′]Fam [3′]BHQ–1 |
RT-PCR | N/A |
| US CDC, USA | Three targets in N gene |
Target 1:2019–nCoV_N1-F/R/P F: GACCCCAAAATCAGCGAAAT R: TCTGGTTACTGCCAGTTGAATCTG Probe: FAM–ACCCCGCATTACGTTTGGTGGACC–BHQ1 Target 2:2019–nCoV_N2–F/R/P Fr: TTACAAACATTGGCCGCAAA R: GCGCGACATTCCGAAGAA Probe: FAM–ACAATTTGCCCCCAGCGCTTCAG–BHQ1 Target 3:2019–nCoV_N3–F/R/P 2019–nCoV_N3 Forward Primer: GGGAGCCTTGAATACACCAAA A 2019–nCoV_N3 Reverse Primer: TGTAGCACGATTGCAGCATTG 2019–nCoV_N3 Probe: FAM–AYCACATTGGCACCCGCAATCCTG–BHQ1 Target 4:RP–F/R/P RNAse P Forward Primer: AGATTTGGACCTGCGAGCG RNAse P Reverse Primer: GAGCGGCTGTCTCCACAAGT RNAse P Probe: FAM – TTCTGACCTGAAGGCTCTGCGCG–BHQ–1 |
RT-PCR | Upper respiratory specimen (Nasopharyngeal specimen, oropharyngeal (OP) specimen, nasal mid-turbinate (NMT) swab, anterior nares specimen) |
| National Institute of Infectious Diseases, Japan | Pancorona and multiple targets, Spike protein |
Target 1: ORF1a set 1 1st NIID_WH–1_F501 Sense TTCGGATGCTCGAACTGCACC 2 1st NIID_WH–1_R913 Antisense CTTTACCAGCACGTGCTAGAAGG 3 2nd NIID_WH–1_F509 Sense CTCGAACTGCACCTCATGG 4 2nd NIID_WH–1_R854 Antisense CAGAAGTTGTTATCGACATAGC Target 2: S set 7 1st WuhanCoV–spk1–f Sense TTGGCAAAATTCAAGACTCACTTT 8 1st WuhanCoV–spk2–r Antisense TGTGGTTCATAAAAATTCCTTTGTG 9 2nd NIID_WH–1_F24381 Sense TCAAGACTCACTTTCTTCCAC 10 2nd NIID_WH–1_R24873 Antisense ATTTGAAACAAAGACACCTTCAC |
Nested RT-PCR |
Pharyngeal swab |
| National Institute of Infectious Diseases, Japan | Pancorona and multiple targets, Spike protein |
Target 1: N protein gene region NIID_2019–nCOV_N_F2 AAATTTTGGGGACCAGGAAC NIID_2019–nCOV_N_R2 TGGCAGCTGTGTAGGTCAAC NIID_2019–nCOV_N_P2 FAM–ATGTCGCGCATTGGCATGGA–BHQ |
Real-time RT-PCR |
Pharyngeal swab |
| Charité, Germany | RdRP, E. N |
Target 1: RdRP gene RdRP_SARSr–F2 GTGARATGGTCATGTGTGGCGG RdRP_SARSr–R1 CARATGTTAAASACACTATTAGCATA RdRP_SARSr–P2 FAM–CAGGTGGAACCTCATCAGGAGATGC BBQ RdRP_SARSr–P1 FAMCCAGGTGGWACRTCATCMGGTGATGC BBQ Target 2: E gene E gene E_Sarbeco_F1 ACAGGTACGTTAATAGTTAATAGCGT E_Sarbeco_R2 ATATTGCAGCAGTACGCACACA E_Sarbeco_P1 FAM–ACACTAGCCATCCTTACTGCGCTTCGBBQ W is A/T; R is G/A; M is A/C ; FAM, 6-carboxyfluorescein; BBQ. blackberry quencher |
RT-PCR | Not mentioned |
| HKU, Hong Kong SAR | ORF1b-nsp14, N |
Assay 1 (Target: ORF1b–nsp14) Forward primer (HKU–ORF1b–nsp14F): TGGGGYTTTACRGGTAACCT Reverse primer (HKU– ORF1b–nsp14R): AACRCGCTTAACAAAGCACTC Probe (HKU–ORF1b-nsp141P): FAM-TAGTTGTGATGCWATCATGACTAG–TAMRA Assay 2 (Target: N) Forward primer (HKU–NF): TAATCAGACAAGGAACTGATTA Reverse primer (HKU–NR): CGAAGGTGTGACTTCCATG Probe (HKU–NP): FAM–GCAAATTGTGCAATTTGCGG–TAMRA |
RT-PCR | Upper respiratory and sputum samples. |
| National Institute of Health, Thailand | N | WH–NIC N–F CGTTTGGTGGACCCTCAGAT WH–NIC N–R CCCCACTGCGTTCTCCATT WH–NIC N–P FAM–CAACTGGCAGTAACCA– BQH1 |
RT-PCR | Not mentioned |
SARS-CoV-2 nucleic acid detection is not only critical for clinical diagnosis but is also important for precautionary screening inside the community and at borders. The experience in Hubei Province demon strated that the improvement of testing capacity is important for early diagnosis and outbreak control. At least 18 CDC centers, 66 hospitals, and 13 third-party independent institutions have been authorized for SARS-CoV-2 nucleic acid testing in Hubei Province[58]. In other areas outside Hubei Province, general screening projects have been launched for people with exposure histories or at high risk, in order to prevent potential community epidemics. For example, the COVID-19 Prevention Headquarter of Guangdong Province has ordered mandatory SARS-CoV-2 nucleic acid testing for all patients in the fever clinics[59]; and the Shanghai Municipal Health Commission requires all international travelers receive SARS-CoV-2 nucleic acid tests at the ports of entry[60].
rRT-PCR testing was the earliest and most widely applied nucleic acid detection method developed for SARS-CoV-2. The WHO guidelines recommend the collection of upper respiratory tract specimens (URT: nasopharyngeal and oropharyngeal) for SARS-CoV-2 rRT-PCR testing. If the URT detection results are negative in a suspected case, along with other clinical indications of SARS-CoV-2 (such as pneumonia), lower respiratory tract specimens (LRT: sputum, tracheal aspirate, or bronchoalveolar lavage fluid from mechanically ventilated patients) should be collected for viral nucleic acid examination[61]. Appropriate personal protective equipment (PPE) should be used during sample collection (droplets and contact precautions for URT samples and airborne precautions for LRT samples). Other WHO recommendations for URT sample collection include: ① use virus swabs (sterile polyester or rayon, not cotton wool swabs); ② promptly submit the test; ③ do not smear the nostrils or tonsils for sampling. Clinicians should avoid inducing sputum specimens as they may increase the risk of airborne transmission[61].
Besides specimens recommended by the WHO, the SARS-CoV-2 genome has been detected in other types of samples, such as fibrobronchoscopies, brush biopsies, feces and blood by rRT-PCR[62–63]. In a study conducted in Guangzhou, China, 55% (41/47) of COVID-19 patients had positive detection results in both their respiratory and fecal samples. In most patients, SARS-CoV-2 RNA was first detected in respiratory samples, but it remained longer in the fecal samples for up to a month after the respiratory samples became negative[63]. There are recommendations to recruit rRT-PCR test for fecal samples as an implementation to current discharge standards[64].
The detection efficiency of a nucleic acid test is impacted by the sample types, COVID-19 clinical stage and technological limitations of PCR. A few cases have been reported that, without disease relapse, recovered patients with two negative results tested positive again after days or with another type of specimens. Due to sensitivity limitations in nucleic acid detection technology, there may be false negative results in samples with low viral concentrations, including samples from patients during the incubation period and rehabilitation period. The reasons for false negatives include a failure in the collection of nucleic acid at sampling, improper storage, transportation or processing of specimens, or limitations in the technology itself, such as unpredicted impacts of virus mutations and PCR inhibition factors[55,65]. The fact that SARS-CoV-2 clearance requires such a long period requires comprehensive interpretation of the PCR results with precautions[63].
ANTIBODY TEST FOR COVID-19 DIAGNOSIS
Serology tests are widely applied in the diagnosis of respiratory infections and especially in rapid pathogen detection, due to shorter turn-around time and sampling convenience. After the development and certification of several nucleic acid test kits/systems, 8 antibody test kits have been certified for COVID-19 diagnosis in China until March 27, 2020[56], including ELISA, peptide-based magnetic chemiluminescence enzyme immunoassay (CLEIA), fluorescence immunochromatographic assay (FICA) and colloidal gold-immunochromatographic assay (GICA).
Real world clinical data on preprint papers indicated that antibody tests had a high specificity for detecting SARS-CoV-2, but that sensitivity for all test kits varied from 57.24% to 93.3%[66–71]. Test sensitivity was also impacted by the choice of antibody targets (Spike or nucleocapsid) and the type of immune globulin 5. It is generally considered that antibodies targeting the nucleocapsid provide higher sensitivity than those targeting the spike part 5. IgM and IgG are both considered sensitive in detecting SARS-CoV-2, however, sensitivity varies among kits from different manufacturers.
Unlike nucleic acid tests, which directly detect viral RNA, the target of antibody tests is lymphocytes following disease progression. For these cases, the sensitivity of antibody tests at the initial stage of infection may be quite low. According to the reported data, although antibodies could be detected early after fever onset, the positivity of IgG and IgM detection was higher at more advanced stage of infection[66,70]. In a study that involved 238 COVID-19 patients, although antibody tests had better sensitivity over rRT-PCR analysis (81.5% and 64.3%) in general, the unsteady detection results before the 11th day of disease onset rendered them unsuitable for initial screening[66]. A similar discovery was made in another study, where a high positivity of IgG and IgM detection was observed after 10 days of illness onset[44]. In conclusion, antibody tests have high specificity and sensitivity at later stages of COVID-19, making them reliable methods in infection confirmation especially in suspect patients with negative rRT-PCR results. However, due to their insufficient sensitivity at the latent period or initial stage of infection, they should not be the initial choice for precautionary screening.
CONCLUSIONS
Taken together, SARS-CoV-2 brought a serious risk to worldwide human health. Combined laboratory examinations and etiological examinations were important elements against the progress of COVID-19, including early diagnosis, therapeutic strategy formulation and prognosis assessment. Some reports suggested that there were significant changes: leukocyte count and lymphocyte count, CD4 T cells, have become the key to early diagnosis. Platelet count, NLR, PLR and some biochemical parameters such as ALT, AST, LDH, BNP and certain inflammatory factors such as IL-6, IL-2R and IL-10 appeared to be related to serious patients. CCS and abnormal blood coagulation caused rapid progression, which suggests a bad prognosis once happens.
Acknowledgments
This work was supported by the grant of National Science and Technology Major Project of China (No. 2018ZX10305409-001-001).
Footnotes
Conflict of interest: The authors have no conflict of interest to report.
References
- [1].World Health Organization. Coronavirus disease 2019 (COVID-19)-Situation Report-84[R/OL]. (2020–04–13) [2020–04–13]. https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200413-sitrep-84-covid-19.pdf?sfvrsn=44f511ab_2.
- [2].National Health Commission of the People’s Republic of China. National-wide Updates of COVID-19 Outbreaks[EB/OL]. (2020–04–13) [2020–04–13], http://www.nhc.gov.cn/2020.
- [3].Han R, Huang L, Jiang H, et al. Early clinical and CT manifestations of coronavirus disease 2019(COVID-19) pneumonia[J]. AJR Am J Roentgenol, 2020, 215(2): 338–343. [DOI] [PubMed] [Google Scholar]
- [4].Chen L, Liu HG, Liu W, et al. Analysis of clinical features of 29 patients with 2019 novel coronavi-rus pneumonia[J/OL]. Chinese Journal of Tuberculosis and Respiratory Diseases (in Chinese), 2020, 43:E005 10.3760/cma.j.issn.1001-0939.2020.0005. [DOI] [PubMed] [Google Scholar]
- [5].Zhang MQ, Wang XH, Chen YL, et al. Clinical features of 2019 novel coronavirus pneumonia in the early stage from a fever clinic in Beijing[J]. Chinese Journal of Tuberculosis and Respiratory Diseases (in Chinese), 2020, 43(3): 215–218. [DOI] [PubMed] [Google Scholar]
- [6].Song F, Shi N, Shan F, et al. Emerging 2019 novel coronavirus (2019-nCoV) pneumonia[J]. Radiology, 2020, 295(1): 210–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhang JJ, Dong X, Cao YY, et al. Clinical characteristics of 140 patients infected with SARS-CoV-2 in Wuhan, China[J]. Allergy, 2020, 75(7): 1730–1741. [DOI] [PubMed] [Google Scholar]
- [8].Qin C, Zhou L, Hu Z, et al. Dysregulation of immune response in patients with coronavirus 2019 (COVID-19) in wuhan, China[J]. Clin Infect Dis, 2020, 71(15): 762–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Li YX, Wu W, Yang T, et al. [Characteristics of peripheral blood leukocyte differential counts in patients with COVID-19][J]. Chinese Journal of Internal Medicine (in Chinese), 2020, 59(5): 372–374. [DOI] [PubMed] [Google Scholar]
- [10].Qu R, Ling Y, Zhang YH, et al. Platelet-to-lymphocyte ratio is associated with prognosis in patients with coronavirus disease-19[J/OL]. J Med Virol, 2020. 10.1002/jmv.25767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Liu Y, Gayle AA, Wilder-Smith A, et al. The reproductive number of COVID-19 is higher compared to SARS coronavirus[J/OL]. J Travel Med, 2020, 27(2): taaa021 10.1093/jtm/taaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Dhama K, Khan S, Tiwari R, et al. Coronavirus disease 2019–COVID–19[J]. Clin Microbiol Rev, 2020, 33(4): 00020–00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen J, Qi T, Liu L, et al. Clinical progression of patients with COVID-19 in Shanghai, China[J]. J Infect, 2020, 80(5): e1–e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chen N, Zhou M, Dong X, et al. Epidemiological and clinical characteristics of 99 cases of 2019 novel coronavirus pneumonia in Wuhan, China: a descriptive study[J]. Lancet, 2020, 395(20): 30211–30217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Yao XH, Li TY, He ZC, et al. [A pathological report of three COVID-19 cases by minimal invasive autopsies][J]. Chinese Journal of Pathology (in Chinese), 2020, 49(5): 411–417. [DOI] [PubMed] [Google Scholar]
- [16].Xu Z, Shi L, Wang Y, et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome[J]. Lancet Respir Med, 2020, 8(4): 420–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Holshue ML, Debolt C, Lindquist S, et al. First case of 2019 novel coronavirus in the United States[J]. N Engl J Med, 2020, 382(10): 929–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Chan KW, Wong VT, Tang S.COVID-19: an update on the epidemiological, clinical, preventive and therapeutic evidence and guidelines of integrative Chinese-Western medicine for the management of 2019 novel coronavirus disease[J]. Am J Chin Med, 2020, 48(3): 737–762. [DOI] [PubMed] [Google Scholar]
- [19].Liu C, Jiang ZC, Shao CX, et al. Preliminary study of the relationship between novel coronavirus pneumonia and liver function damage: a multicenter study[J]. Chinese Journal of Hepatology (in Chinese), 2020, 28(2): 107–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Yao N, Wang SN, Lian JQ, et al. Clinical characteristics and influencing factors of patients with novel coronavirus pneumonia combined with liver injury in Shaanxi region[J]. Chinese Journal of Hepatology (in Chinese), 2020, 28(3): 234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Liu M, He P, Liu HG, et al. Clinical characteristics of 30 medical workers infected with new coronavirus pneumonia[J]. Chinese Journal of Tuberculosis and Respiratory Diseases (in Chinese), 2020, 43(3): 209–214. [DOI] [PubMed] [Google Scholar]
- [22].Wu C, Hu X, Song J, et al. Heart injury signs are associated with higher and earlier mortality in coronavirus disease 2019 (COVID-19)[J/OL]. medRxiv, 2020. 10.1101/2020.02.26.20028589. [DOI] [Google Scholar]
- [23].Ma LK.What we know so far:COVID-19 current clinical knowledge and research[J]. Clin Med (Lond), 2020, 20(2): 124–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhang LP, Wang M, Wang Y, et al. Focus on the 2019 novel coronavirus (SARS-CoV-2)[J]. Future Microbiol, 2020, 15: 905–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Li G, Fan Y, Lai Y, et al. Coronavirus infections and immune responses[J]. J Med Virol, 2020, 92(4): 424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Lippi G, Plebani M. Procalcitonin in patients with severe coronavirus disease 2019 (COVID-19): A meta-analysis[J]. Clin Chim Acta, 2020, 505: 190–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Singhal T A review of coronavirus disease-2019(COVID-19)[J]. Indian J Pediatr, 2020, 87(4): 281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nishiga M, Wang DW, Han Y, et al. COVID-19 and cardiovascular disease: from basic mechanisms to clinical perspectives[J]. Nat Rev Cardiol, 2020, 17(9): 543–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Xiong Y, Sun D, Liu Y, et al. Clinical and High-Resolution CT features of the COVID-19 infection: comparison of the initial and follow-up changes[J]. Invest Radiol, 2020, 55(6): 332–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Tang N, Li D, Wang X, et al. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia[J]. J Thromb Haemost, 2020, 18(4): 844–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Zhou F, Yu T, Du R, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: a retrospective cohort study[J]. Lancet, 2020, 395(20): 30563–30566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wu F, Zhao S, Yu B, et al. A new coronavirus associated with human respiratory disease in China[J]. Nature, 2020, 579(7798): 265–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lu R, Zhao X, Li J, et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding[J]. Lancet, 2020, 395(20): 30251–30258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hadfield J, Megill C, Bell SM, et al. Nextstrain:real-time tracking of pathogen evolution[J]. Bioinformatics, 2018, 34(23): 4121–4123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].GISAID Initiative. Genomic epidemiology of hCoV-19 2020. [EB/OL], (2020–03–18)[2020–04–13], https://www.gisaid.org/.
- [36].Rejmanek D, Hosseini PR, Mazet JA, et al. Evolutionary dynamics and global diversity of influenza a virus[J]. J Virol, 2015, 89(21): 01515–01573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Zhao Z, Li H, Wu X, et al. Moderate mutation rate in the SARS coronavirus genome and its implications[J]. BMC Evol Biol, 2004, 4:21 10.1186/1471-2148-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Zhang Z, Shen L, Gu X. Evolutionary dynamics of MERS-CoV: potential recombination, positive selection and transmission[J/OL]. Sci Rep, 2016, 6: 25049 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Forster P, Forster L, Renfrew C, et al. Phylogenetic network analysis of SARS-CoV-2 genomes[J]. Proc Natl Acad Sci U S A, 2020, 117(17): 9241–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kim D, Lee JY, Yang JS, et al. The Architecture of SARS-CoV-2 Transcriptome[J]. Cell, 2020, 181(4): 914–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Chen Y, Liu Q, Guo D. Emerging coronaviruses: Genome structure, replication, and pathogenesis[J]. J Med Virol, 2020, 92(4): 418–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Aubriotdelmas B, Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression[J]. J Virol, 1990, 64(11): 5367–5375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Beniac DR, Andonov A, Grudeski E, et al. Architecture of the SARS coronavirus prefusion spike[J]. Nat Struct Mol Biol, 2006, 13(8): 751–752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Nal B, Chan C, Kien F, et al. Differential maturation and subcellular localization of severe acute respiratory syndrome coronavirus surface proteins S,M and E[J]. J Gen Virol, 2005, 86(5): 80670–80671. [DOI] [PubMed] [Google Scholar]
- [45].Neuman BW, Kiss G, Kunding AH, et al. A structural analysis of M protein in coronavirus assembly and morphology[J]. J Struct Biol, 2011, 174(1): 11–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis[J]. Methods Mol Biol, 2015, 1282: 1–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Chang CK, Sue SC, Yu TH, et al.Modular organization of SARS coronavirus nucleocapsid protein[J]. J Biomed Sci, 2006, 13(1): 59–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hurst KR, Koetzner CA, Masters PS. Identification of in vivo-interacting domains of the murine coronavirus nucleocapsid protein[J]. J Virol, 2009, 83(14): 7221–7234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cui L, Wang H, Ji Y, et al. The nucleocapsid protein of coronaviruses Acts as a viral suppressor of RNA silencing in mammalian cells[J]. J Virol, 2015, 89(17): 1315–1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].World Health Organization. Coronavirus disease (COVID-19) technical guidance: Laboratory testing for 2019-nCoV in humans[EB/OL], (2020–03–18)[2020–04–13], https://www.who.int/, 2020.
- [51].Zhu N, Zhang D, Wang W, et al. A novel coronavirus from patients with pneumonia in China, 2019[J]. N Engl J Med, 2020, 382(8): 727–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tan W, Zhao X, Wang W, et al. hCoV-19/Wuhan/IVDC-HB-01/2019. 01-10-2020 ed[EB/OL], (2020–03–18) [2020–04–13]. https://platform.gisaid.org/: Global Initiative on Sharing All Influenza Data, 2020. [Google Scholar]
- [53].Tan W, Ma X, Zhao X, et al. hCoV-19/Wuhan/IVDC-HB-05/2019. 01-10-2020 ed[EB/OL]. (2020–03–18) [2020–04–13]. https://platform.gisaid.org/: Global Initiative on Sharing All Influenza Data, 2020. [Google Scholar]
- [54].World Health Organization. Pneumonia of unknown cause-China: World Health Organization, 2020[R]. [Google Scholar]
- [55].National Health Commission of the People’s Republic of China. Protocol for prevention and control of COVID-19(6th ed)[R]. China CDC Weekly: Chinese Center for Disease Control and Prevention, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].National Medical Products Administration. Emergency Use Authorizations for COVID-19 Dignosis Kits[EB/OL]. (2020–03–18)[2020–04–13]. http://www.nmpa.gov.cn/WS04/CL2056/376095.htm, 2020.
- [57].US Food & Drug Administration. Coronavirus Disease 2019 (COVID-19) Emergency Use Authorizations for Medical Devices[EB/OL]. (2020–03–18)[2020–04–13], https://www.fda.gov/medical-devices/emergency-situations-medical-devices/emergency-use-authorizations-covid19ventilators, 2020. [PubMed]
- [58].Authorized Institution for Nucleic Acid Tests in Hubei Province [EB/OL]. (2020–3–10) [2020–04–13], http://wjw.hubei.gov.cn/bmdt/ztzl/fkxxgzbdgrfyyq/fkdt/202002/t20200210_2022611.shtml. [Google Scholar]
- [59].COVID-19 Prevention Headquarter of Guangdong Province. Next steps for COVID-19 control: mandatory nucleic acid tests for all patients in the fever clinics[EB/OL]. (2020–03–19) [2020–04–13]. http://wsjkw.gd.gov.cn/zwyw_gzdt/content/post_2887910.html: Health Commission of Guangdong province. [Google Scholar]
- [60].COVID-19 Prevention Headquarter of Shanghai. COVID-19 prevention at international ports of entry in Shanghai municipal health commission[R]. 2020. [Google Scholar]
- [61].World Health Organization. Clinical management of severe acute respiratory infection when novel coronavirus (nCoV) infection is suspected: Interim Guidance: Institutional Repository for Information Sharing [R]. 2020. [Google Scholar]
- [62].Wang W, Xu Y, Gao R, et al. Detection of SARS-CoV-2 in different types of clinical specimens[J/OL]. JAMA, 2020. 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wu Y, Guo C, Tang L, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples[J].Lancet Gastroenterol Hepatol,2020,20:30082–30083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Handbook of COVID-19 Prevention and Treatment[R]. Hangzhou, China: Jack Ma Foundation, Zhejiang University, The First Affiliated Hospital, Zhejiang University School of Medicine (FAHZU), 2020. [Google Scholar]
- [65].National Health Commission of the People’s Republic of China. Technical guide for laboratory testing of SARS-CoV-2(2nd ed)[R]. China CDC Weekly. Chinese Center for Disease Control and Prevention, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Liu L, Liu W, Zheng Y, et al. A preliminary study on serological assay for severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 238 admitted hospital patients[J]. Microbes Infect, 2020, 22(4–5): 206–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Cai XF, Chen J, Li Hu J, et al. A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of coronavirus disease 2019[J]. J Infect Dis, 2020, 222(2): 189–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Jia X, Zhang P, Tian Y, et al. Clinical significance of IgM and IgG test for diagnosis of highly suspected COVID-19 infection[J/OL]. medRxiv, 2020. 10.1101/2020.02.28.20029025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Xiang J, Yan M, Li H, et al. Evaluation of Enzyme-Linked Immunoassay and Colloidal Gold-Immunochromatographic Assay Kit for Detection of Novel Coronavirus (SARS-Cov-2) Causing an Outbreak of Pneumonia (COVID-19)[ J/OL]. medRxiv, 2020. 10.1101/2020.02.27.20028787. [DOI] [Google Scholar]
- [70].Li B, Feng F, Yang G, et al. Immunoglobulin G/M and Cytokines Detections in Continuous Sera from Patients with Novel Coronaviruses (2019-nCoV) Infection[J/OL]. Available at SSRN 3543609, 2020. 10.2139/ssrn.3543609. [DOI] [Google Scholar]
- [71].Zhang W, Du RH, Li B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes[J]. Emerg Microbes Infect, 2020, 9(1): 3. [DOI] [PMC free article] [PubMed] [Google Scholar]


