A tumor-suppressive circular RNA brings in the proteasome to destroy the foothold of liver cancer cells.
Abstract
Circular RNAs (circRNAs) have emerged as important regulators of various cellular processes and have been implicated in cancer. Previously, we reported the discovery of several dysregulated circRNAs including circPABPC1 (polyadenylate-binding protein 1) in human hepatocellular carcinoma (HCC), although their roles in HCC development remained unclear. Here, we show that circPABPC1 is preferentially lost in tumor cells from clinical samples and inhibits both intrahepatic and distant metastases in a mouse xenograft model. This tumor-suppressive function of circPABPC1 can be attributed to its inhibition of cell adhesion and migration through down-regulating a key member of the integrin family, ITGB1 (β1 integrin). Mass spectrometry and biochemical evidence demonstrate that circPABPC1 directly links ITGB1 to the 26S proteasome for degradation in a ubiquitination-independent manner. Our data have revealed an uncanonical route for integrin turnover and a previously unidentified mode of action for circRNAs in HCC that can be harnessed for anticancer treatment.
INTRODUCTION
Hepatocellular carcinoma (HCC) is the sixth most common malignancy and the fourth leading cause of cancer-related death globally (1). Despite a comprehensive regimen available including surgery, chemotherapy, intervention, targeted therapy, and immune therapy, the overall survival of patients with advanced HCC remains poor (1). Genetic and phenotypic heterogeneity is a hallmark of HCC and also poses a daunting challenge for diagnosis, treatment, and patient care (2). A deeper understanding of molecular mechanisms underlying HCC progression is urgently needed.
Most cancer lethality results from metastasis, for which integrin signaling is indispensable. Integrins are heterodimeric cell surface molecules mediating the physical interaction between cells and the extracellular matrix (ECM). An integrin molecule consists of an α subunit and a β subunit, each encoded by multiple genes in human cells. As single-pass transmembrane proteins, integrins bind to ECM components such as collagen, fibronectin, and laminin as well as other molecules in the extracellular microenvironment and transduce chemical or mechanical signals to the interior of cells through their short cytoplasmic tails. Integrins are not only essential for cell-ECM adhesion but also important regulators of cell survival, differentiation, migration, invasion, and drug resistance in cancer cells via intricate cross-talk with other signaling pathways (3–5). As a common subunit, β1 integrin (ITGB1) can pair with multiple α integrins and plays a central role in intrahepatic metastasis, a serious problem for HCC treatment and prognosis in the clinic (6).
Several strategies have been devised to target integrins for disease treatment. Monoclonal antibodies and small-molecule binding blockers are commonly used in research but have not been successful in the cancer clinic due to complicated reasons (3, 5). Recent years have witnessed an important paradigm shift in protein targeting approaches represented by various degrader technologies (7). The best studied and most promising example is proteolysis targeting chimera (PROTAC), which recruits the protein of interest to an E3 ubiquitin ligase, leading to its ubiquitination and proteasomal degradation (8). The 26S proteasome is a multisubunit complex consisting of one or two 19S regulatory particles (RPs) and a 20S core particle (CP). Proteasomal degradation of ubiquitinated and nonubiquitinated substrates accounts for the majority of selective proteolysis in eukaryotic cells (9). ITGB1, a classical type I transmembrane protein, is generally thought to be degraded via the lysosomal pathway as the other integrins (10, 11). However, proteasome inhibition has been shown to stabilize ITGB1, while the underlying mechanisms were not investigated (12, 13). Another approach to targeting ITGB1 is using small interfering RNAs (siRNAs) delivered in nanoparticles, which has been effective in suppressing HCC in mice (14).
Circular RNAs (circRNAs) are noncoding RNAs usually produced from back-splicing of exons of pre-mRNAs. They are usually present in the cytoplasm and relatively stable, functioning as microRNA (miRNA) sponges or protein scaffolds to facilitate protein-protein interactions (15). Increasing evidence supports a considerable role of circRNAs during cancer initiation and progression, including in HCC (16–26). Our previous circRNA microarray analysis revealed that multiple circRNAs hosted by the PABPC1 (polyadenylate-binding protein 1) gene were markedly suppressed by activated androgen receptor (AR) in HCC, a unique feature compared to circRNAs from other host genes (27). Among them, circPABPC1 (hsa_circ_0085154) was the only one that appeared to have antitumor activities.
Here, we performed in-depth analyses of circPABPC1 from human patient samples and in a mouse xenograft model and confirmed its role as a tumor suppressor in HCC. circPABPC1 negatively regulates HCC cell adhesion and migration by directly binding and down-regulating ITGB1. Unexpectedly, we found that ITGB1, of which degradation is normally handled by the lysosome, could be degraded by the 26S proteasome in HCC cells in a ubiquitination-independent manner, and circPABPC1 could serve as a bridge between the proteasome and ITGB1 to promote the degradation of the latter both in cells and in vitro. Our work has defined an important tumor-suppressive circRNA in HCC, revealed a previously unrecognized mechanism for substrate recognition by the proteasome, and suggested a potential means to induce ITGB1 degradation for anticancer treatment.
RESULTS
Decreased circPABPC1 expression in HCC correlates with poor prognosis
Our previous study identified circPABPC1 (hsa_circ_0085154) as a 91-base circRNA suppressed by AR, hence its former name circARSP91 (Fig. 1A) (27). To explore the function of circPABPC1 in HCC, we began by examining its expression in clinical samples from a small group of 22 patients (cohort 1) by quantitative real-time quantitative polymerase chain reaction (RT-qPCR). Confirming our previous microarray results, circPABPC1 level was significantly lower in the tumor samples than in matched peritumor or nontumor tissues (fig. S1A). We then expanded our survey to another 92 pairs of tumor/nontumor samples (cohort 2), and 75% of them (69 of 92) exhibited circPABPC1 down-regulation in HCC tumor tissues as compared to the surrounding tissues (Fig. 1B and fig. S1, B and C). In addition, three independent HCC datasets from the Gene Expression Omnibus (GEO) database were analyzed (GSE97332, GSE94508, and GSE78520). Although the differences between tumor and nontumor tissues did not reach statistical significance due to small sample sizes (n = 3, 5, and 7, respectively), an overall decrease in circPABPC1 in tumor tissues was discernable (table S1). In a third cohort of 91 patients with HCC (cohort 3) with follow-up records (table S2), low circPABPC1 expression in tumors strongly correlated with shortened overall survival and disease-free survival (DFS) (Fig. 1, C and D). These findings point to a tumor-suppressive function of circPABPC1 in HCC.
Fig. 1. circPABPC1 is a tumor suppressor in HCC.

(A) Schematic of circPABPC1 formation. (B) The expression of circPABPC1 was measured by RT-qPCR in matched tumor and nontumor tissues from 92 patients with HCC (cohort 2, means ± SD, ***P < 0.001, paired Student’s t test). (C and D) Kaplan-Meier curves showing overall survival (C) and DFS (D) of 91 patients with HCC (cohort 3) followed up to 60 months. Patients were separated by the median expression level of circPABPC1. (E) Top: Cartoon depicting the timeline of xenograft tumor growth in this study. Bottom: In vivo imaging system (IVIS) images of LM3 tumors at 6 weeks after transplantation, when both local growth and intrahepatic metastasis became discernable (blue arrowheads). (F) Intrahepatic tumor foci in (E) were quantified. *P < 0.05, unpaired Student’s t test. (G) IVIS images of gastrointestinal (GI) tract metastases at 12 weeks after transplantation (orange arrowheads). (H) Metastatic foci in the GI tract (G) were quantified. *P < 0.05, unpaired Student’s t test.
circPABPC1 suppresses HCC migration and metastasis in vivo
We next investigated the impact of circPABPC1 on HCC progression in vivo using an orthotopic xenograft model. A highly aggressive human HCC cell line LM3 derived from lung metastasis (28) was chosen because of its ability to form both intrahepatic and distant metastases in our pilot experiments (Fig. 1E), recapitulating the stepwise spreading of HCC that we observe in the clinic. LM3 cells were stably transduced with an empty vector or circPABPC1 (27) and labeled with luciferase to allow for tracking by the in vivo imaging system (IVIS). Cells were first injected subcutaneously into immunocompromised mice to form tumors, which were then surgically transplanted into the liver of recipient mice to avoid leakage of free cancer cells into the abdominal cavity. At the sixth week after transplantation, orthotopic tumor growth became readily detectable by IVIS. Almost all mice (six of seven) receiving the control tumors developed multifocal lesions, a typical sign of intrahepatic metastasis of HCC. However, this was rarely seen with the circPABPC1-overexpressing tumors (one of seven), suggesting that circPABPC1 effectively suppressed this local spreading at the early stage of HCC development (Fig. 1, E and F). By the 12th week after transplantation, control tumors had widely spread to the gastrointestinal (GI) tract and the lung of most mice (six of seven), representing metastasis via implantation and blood circulation, respectively. In contrast, GI and pulmonary metastases were largely inhibited by circPABPC1 (Fig. 1, G and H, and fig. S1, D to H). At this point, the primary tumors with circPABPC1 overexpression were also significantly smaller than the control tumors, although both groups of tumors had grown quite large, requiring the mice to be euthanized (fig. S1, I and J). These data demonstrate that circPABPC1 could function as a suppressor of HCC cell growth and metastasis in vivo.
circPABPC1 destabilizes cancer cell adhesion to ECM
To investigate the cellular function of circPABPC1, we manipulated circPABPC1 levels in three HCC cell lines by stable overexpression or siRNA-mediated knockdown (fig. S2, A to C). We noticed that, during routine cell culture, high-level circPABPC1 markedly accelerated cell detachment after trypsinization, while circPABPC1 depletion rendered cells more resistant to trypsin digestion (Fig. 2A and fig. S2, D and E). Similar results were observed in all three HCC cell lines, suggesting an important role of circPABPC1 in regulating cell adhesion. To test this idea, we performed cell adhesion assays by seeding the above HCC cells in Matrigel or on plates coated with defined components of ECM, namely, fibronectin, laminin, collagen VI, and vitronectin, all of which are known ligands of ITGB1-containing integrins (5). The results clearly showed that circPABPC1 inhibited cell adhesion under all tested conditions (Fig. 2, B to D, and fig. S2, F to H). Consistently, stress fiber formation was suppressed by circPABPC1 (Fig. 2E and fig. S2, I and J). Furthermore, circPABPC1 also retarded cell mobility in transwell migration assays (Fig. 2F). Because cell adhesion is crucial for cancer cell survival, proliferation, migration, and metastasis, these in vitro results provided a reasonable explanation for the tumor-suppressive function of circPABPC1 in vivo.
Fig. 2. circPABPC1 inhibits cell adhesion.

(A) Trypsin digestion assay on HA22T cells with circPABPC1 overexpression or knockdown. Cells without (−) or with (+) trypsinization were fix-stained with crystal violet and photographed. (B) HA22T cells as in (A) were seeded in Matrigel-coated plates and allowed to adhere for 60 to 120 min. Attached cells were stained and imaged. (C and D) HA22T cells with circPABPC1 overexpression (C) or knockdown (D) were seeded in plates coated with the indicated ECM components. Adhered cells were stained with crystal violet, followed by measurement of OD540nm (optical density at 540 nm) (means ± SD, *P < 0.05, **P < 0.01, and ***P < 0.001, unpaired Student’s t test). (E) Stress fiber formation in HA22T cells expressing vector control or circPABPC1 was shown by phalloidin staining of F-actin. Scale bar, 10 μm. (F) Transwell migration assays with HA22T and LM3 cells. Representative fields of the porous membranes are shown (left), and cell numbers per field are quantified (right) (means ± SD, *P < 0.05, **P < 0.01, and ***P < 0.001, n = 3 biological replicates, unpaired Student’s t test).
ITGB1 is a functionally relevant binding protein and target of circPABPC1
To dissect how circPABPC1 regulates cell adhesion and migration, we first determined the subcellular localization of circPABPC1 by RNA fluorescence in situ hybridization (FISH) and cell fractionation, followed by RT-qPCR. In all three HCC cell lines, circPABPC1 predominantly localized in the cytoplasm (Fig. 3, A and B, and fig. S3, A to D) as has been reported for most circRNAs. However, the small size of circPABPC1 (91 bases) makes it unlikely to be a miRNA sponge (15). No polypeptide-coding sequence can be found in circPABPC1, and it had no influence on the expression of its host gene, PABPC1 (fig. S3, E and F). Therefore, we reasoned that circPABPC1 might function as a protein scaffold in the cytoplasm.
Fig. 3. Identification of ITGB1 as a circPABPC1-interacting protein.

(A) Cytoplasmic localization of circPABPC1 in HA22T cells revealed by FISH. Cell nuclei were labeled with 4′,6-diamidino-2-phenylindole (DAPI). Scale bars, 10 μm. (B) RT-qPCR detection of circPABPC1 from the indicated compartments of HA22T cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA and U6 RNA were used as reference RNAs from the cytoplasm and nucleus, respectively. (C) Flowchart of identifying circPABPC1-binding proteins by RNA pull-down and mass spectrometry (MS). (D) circPABPC1-binding proteins identified by MS. ITGB1, ITGA3, and PSMA4 are highlighted. (E) Gene ontology (GO) analysis of circPABPC1-interacting proteins. Cell adhesion–related terms are marked by arrowheads. (F) HA22T and Huh7 cell extracts were incubated with a control oligo or circPABPC1-specific probe. circPABPC1-ITGB1 binding was confirmation by RNA pull-down and immunoblotting (IB), and a representative result is shown (top). Successful pull-down of circPABPC1 was confirmed by RT-qPCR (bottom) (***P < 0.001 from three independent repeats, unpaired Student’s t test). (G) circPABPC1 FISH and anti-ITGB1 immunofluorescence staining showing colocalization of these two molecules (arrowheads) in HA22T cells. Note that circPABPC1 overexpression reduced the intensity of endogenous ITGB1. Hoechst, nuclear marker. Scale bars, 4 μm.
We took a circRNA pull-down approach to examining whether circPABPC1 could bind proteins (Fig. 3C). By incubating HA22T cell extracts with a biotinylated probe complementary to circPABPC1, we successfully enriched circPABPC1 and coisolated a number of its potential binding proteins, 37 of which were identified by mass spectrometry (MS) (Fig. 3D). Gene ontology (GO) analysis showed that these proteins strongly correlate with cell adhesion (Fig. 3E), consistent with the role of circPABPC1 observed in the in vitro adhesion assays. In particular, α3 integrin (ITGA3) and ITGB1, two integrin family members crucial for cell-ECM interaction, were found in circPABPC1 pull-down (Fig. 3D). Western blot results confirmed the binding between circPABPC1 and endogenous ITGB1 (Fig. 3F). No ITGB1 was pulled down by the same probe in circPABPC1 knockdown cells or by probes toward two unrelated circRNAs (29, 30), demonstrating the specificity of interaction between circPABPC1 and ITGB1 (fig. S3, G and H). circPABPC1 and ITGB1 colocalized intracellularly as shown by fluorescence microscopy (Fig. 3G and fig. S3I). Moreover, overexpression of circPABPC1 led to a decrease in ITGB1 protein in three HCC cell lines, while ITGA3 expression was not affected (fig. S3, E, F, J, and K). These findings prompted us to further investigate the involvement of ITGB1 in circPABPC1-mediated regulation of cell adhesion.
Not only circPABPC1 down-regulated ITGB1 in HCC cell lines (Fig. 4, A and B) but its expression also negatively correlated with ITGB1 protein level in HCC patient samples (cohort 1, Pearson r = −0.5754, P < 0.01; fig. S4A). Integrin engagement with ECM is well known to activate focal adhesion kinase (FAK), transducing signals to further downstream prosurvival factors such as phosphatidylinositol 3-kinase (PI3K) and Akt (31). As expected, phosphorylation/activity of FAK, PI3K, and Akt was all negatively controlled by circPABPC1 in the same way as ITGB1 (Fig. 4, A and B). In trypsin digestion assays, circPABPC1 overexpression facilitated cell detachment as seen earlier, while ectopic expression of ITGB1 restored FAK activation and strengthened cell adhesion, completely blocking the effect of circPABPC1 (Fig. 4, C and D). Then, we gauged cell adhesion to the different ECM components. circPABPC1 overexpression again weakened cell attachment, which was fully rescued by ITGB1 (Fig. 4E and fig. S4, B and C). On the other hand, knockdown of circPABPC1 promoted cell adhesion, and this effect was fully reversed by partial deletion of ITGB1 using CRISPR-Cas9–mediated gene targeting (Fig. 4F and fig. S4, B to D). Furthermore, circPABPC1 impeded cell migration in transwell assays as seen before, which was dependent on and could be reversed by ITGB1 (Fig. 4G and fig. S4, D and E). Together, these epistasis analyses strongly suggest that circPABPC1 inhibits HCC cell adhesion and mobility through repressing its downstream effector ITGB1.
Fig. 4. circPABPC1 regulates cell adhesion through down-regulating ITGB1.
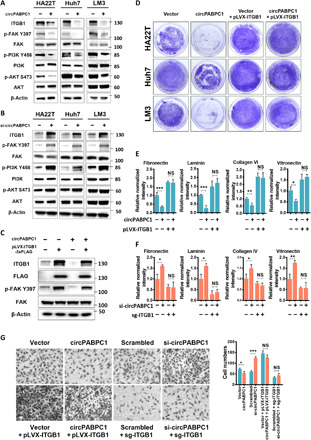
(A and B) HCC cells with circPABPC1 overexpression (A) or knockdown (B) were analyzed for ITGB1 expression and its downstream signaling by immunoblotting with the indicated antibodies. (C) HCC cells with or without circPABPC1 overexpression were transduced with ITGB1-3xFLAG. ITGB1 expression and FAK activation were determined by Western blot. (D and E) Ectopic expression of ITGB1 rescued circPABPC1-induced phenotypes in trypsin digestion assays (D) and cell adhesion assays (E). OD540nm values were normalized against the control sample (first bar, set to 1.0) in each experiment. *P < 0.05, **P < 0.01, and ***P < 0.001, one-way analysis of variance (ANOVA). n.s., not significant. (F) Depletion of endogenous ITGB1 in HA22T cells by CRISPR-Cas9 eliminated the effect of circPABPC1 knockdown on cell adhesion. Data were analyzed and presented as in (E). (G) Transwell migration assay with the indicated HA22T cells. Representative images of migrated cells (left) and the quantified results (right) are shown (means ± SD, *P < 0.05 and ***P < 0.001, n = 3 biological replicates, one-way ANOVA).
circPABPC1 promotes proteasomal degradation of ITGB1
Next, we examined the mechanism by which circPABPC1 regulates ITGB1 level. Down-regulation of ITGB1 did not seem to result from changes in its mRNA expression in circPABPC1-overexpressing cells (fig. S3K). Rather, circPABPC1 evidently promoted ITGB1 protein degradation when the HCC cell lines were treated with cycloheximide (CHX) to block protein synthesis (Fig. 5, A and B, and fig. S5, A and B). ITGB1 is generally thought to be degraded through the canonical lysosomal pathway (10, 11). Unexpectedly, we found that circPABPC1-induced ITGB1 decline was not much affected by the lysosome inhibitor chloroquine (CQ) but could be fully prevented by proteasome inhibitors MG-132 and bortezomib (Fig. 5C and fig. S5C). The same effect was seen with partial depletion of a 20S proteasome subunit PSMA4 (also known as α3) (fig. S5D). Although ubiquitination has been implicated in the sorting and degradation of integrins (10, 11), ITGB1 ubiquitination was barely detectable in our HCC cells even in the presence of MG-132, and it was not affected by circPABPC1 (fig. S5E). Moreover, treating HCC cells with the ubiquitin-activating enzyme (UBA1/E1) inhibitor PYR-41 substantially reduced total ubiquitination level but did not prevent circPABPC1-induced ITGB1 degradation (Fig. 5C). These data indicate that, in HCC cells, circPABPC1 promotes ubiquitin-independent proteasomal degradation of ITGB1.
Fig. 5. circPABPC1 promotes ITGB1 degradation by the proteasome.

(A and B) HA22T cells ± circPABPC1 overexpression were treated with CHX (5.0 μg/ml) as indicated. Endogenous ITGB1 was determined by Western blot (A) and quantified (B). (C) HA22T cells were treated with dimethyl sulfoxide, MG-132 (10 μM, 4 hours), bortezomib (BTZ) (10 μM, 4 hours), CQ (10 μM, 20 hours), or PYR-41 (20 μM, 20 hours). Endogenous ITGB1, ubiquitin (Ub), and LC3 levels were probed. (D and E) Simultaneous detection of endogenous ITGB1 and proteasome subunits/chaperone following circPABPC1 pull-down from HA22T cells. (F) RNA-FISH and immunostaining showing colocalization of circPABPC1 and PSMA4 in HA22T cells. Scale bars, 4 μm. (G and H) ITGB1-3xFlag coimmunoprecipitated with endogenous PSMA4 and circPABPC1 as determined by Western blot (G) and RT-qPCR (H), respectively. (I and J) Endogenous PSMA4 coimmunoprecipitated with endogenous ITGB1 and circPABPC1 as determined by Western blot (I) and RT-qPCR (J), respectively. (K) A mammalian two-hybrid assay with 293T cells expressing the indicated proteins with or without circPABPC1. Normalized luciferase reporter activity is shown (top), and protein expression was confirmed by Western blot (bottom). Data in (H), (J), and (K) are means ± SD. ***P < 0.001, unpaired Student’s t test.
circPABPC1 physically connects ITGB1 with the proteasome
In the aforementioned MS results, the proteasome subunit PSMA4 was found to be a circPABPC1-interacting protein (Fig. 3D). Therefore, we wondered whether circPABPC1 could directly recruit ITGB1 to the proteasome. We first confirmed by Western blot that circPABPC1 could simultaneously pull down ITGB1, PSMA4, as well as other 20S and 19S subunits from cells (Fig. 5, D and E, and fig. S5F). This suggests that circPABPC1 can associate with the entire 26S proteasome, which is probably mediated by its direct interaction with PSMA4 as shown in the in vitro binding assays (fig. S5H), consistent with the MS finding (Fig. 3D). Similar to ITGB1, PSMA4 colocalized with circPABPC1 in the cytoplasm (Fig. 5F and fig. S5G). Moreover, reciprocal coimmunoprecipitation assays showed that ITGB1 physically interacted with PSMA4, circPABPC1, and other proteasome subunits (Fig. 5, G to J, and fig. S5, I and J). Last, we adopted a mammalian two-hybrid assay using pBIND-PSMA4 and pACT-ITGB1 fusion proteins as bait and prey, respectively. Neither fusion protein alone nor circPABPC1 itself was able to promote reporter expression. However, coexpression of both proteins induced a fivefold increase in the luciferase reporter activity over control, and introduction of circPABPC1 further potentiated the interaction between PSMA4 and ITGB1 (Fig. 5K). On the contrary, knockdown of circPABPC1 disrupted the synergy between PSMA4 and ITGB1 (fig. S5K). Together, these results strongly suggest that circPABPC1, ITGB1, and PSMA4/proteasome can form a macromolecular complex.
Bivalent binding of circPABPC1 to ITGB1 and PSMA4 is required for its function
To further verify the role of circPABPC1 in mediating ITGB1 proteasomal degradation, we predicted the binding sites of ITGB1 and PSMA4 on circPABPC1 using structural modeling. On the basis of the secondary structure of circPABPC1 with the lowest free energy (fig. S6, A and B), we generated three-dimensional (3D) models of circPABPC1 in complex with ITGB1 and PSMA4, respectively (see Materials and Methods for details and fig. S6, C and D). According to the simulation, ITGB1 and PSMA4 bind different regions of circPABPC1, compatible with a quaternary structure formed by the three molecules. The structural simulation also returned bases of circPABPC1 potentially important for ITGB1 and PSMA4 binding. We mutated these sites separately and confirmed circularization of each mutant (Fig. 6A and fig. S6, E to G). In line with our models, mutation of key bases for ITGB1 binding (circ-mu-ITGB1) completely abrogated circPABPC1-ITGB1 interaction, while the association between circPABPC1 and PSMA4 was spared (Fig. 6B). Conversely, the circ-mu-PSMA4 mutant showed much reduced binding with PSMA4 but maintained the interaction with ITGB1 (Fig. 6B). We then compared wild-type and mutant circPABPC1 for their function in HCC cells. In all three cell lines, either of the circPABPC1 mutations abolished its effect on ITGB1 degradation and FAK activation (Fig. 6, C and D). Consistently, both circPABPC1 mutants failed to down-regulate cell-ECM adhesion (Fig. 6E and fig. S6, H and I) and had no impact on cell detachment in the trypsin digestion assay (Fig. 6F). These loss-of-function phenotypes further demonstrate the importance of circPABPC1 as a bridge between ITGB1 and the proteasome.
Fig. 6. Simultaneous interaction with ITGB1 and PSMA4 is necessary for circPABPC1 function.
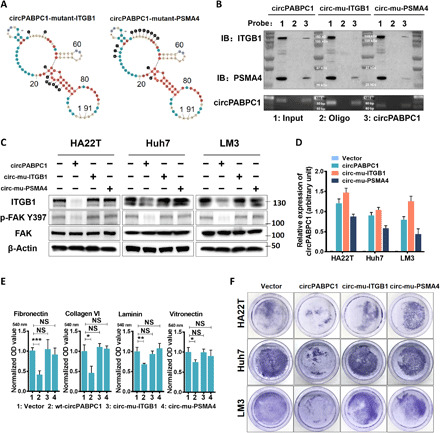
(A) circPABPC1 mutations that selectively block its binding with ITGB1 (left) or PSMA4 (right). (B) Wild-type circPABPC1 and mutants expressed in HA22T cells were isolated by RNA pull-down, and copurified ITGB1 and PSMA4 were probed. (C) Endogenous ITGB1 level and FAK activation in HCC cells expressing wild-type or mutant circPABPC1 were determined by Western blot. (D) circPABPC1 expression in HCC cells (C) was measured by RT-qPCR. (E) HA22T cells expressing the indicated circPABPC1 variants were examined for adhesion to the ECM substrates. OD540nm of crystal violet-stained cells was measured (means ± SD, *P < 0.05, **P < 0.01, and ***P < 0.001, one-way ANOVA). (F) Trypsin digestion assay of HCC cells expressing the indicated circPABPC1 variants.
circPABPC1 promotes ITGB1 degradation by the proteasome in vitro
We then tested whether purified proteasomes could directly interact with and degrade recombinant ITGB1 in vitro. The ubiquitin-like domain (UBL) of Rad23 was fused to glutathione S-transferase (GST) and immobilized on the glutathione resin, which captured intact endogenous proteasomes from HCC cells (fig. S7, A and B) (32). Recombinant ITGB1 protein was then added to the proteasome, and GST pull-down was performed to determine ITGB1-proteasome interaction. ITGB1 alone showed little binding with the proteasome, while the addition of circPABPC1 greatly enhanced their association (Fig. 7A). The same system was then supplemented with adenosine 5′-triphosphate (ATP) to determine proteasome-mediated ITGB1 degradation. Consistent with the above results, ITGB1 alone was essentially impervious to proteasomal digestion even after prolonged incubation. In contrast, the proteasome effectively degraded ITGB1 in the presence of circPABPC1, which could be blocked by MG-132 (Fig. 7, B and C). Furthermore, ATP hydrolysis was required in this process, as the nonhydrolyzable ATP analog, ATPγS, could not support in vitro degradation of ITGB1 by the proteasome (Fig. 7D). Given the lack of ubiquitination of recombinant ITGB1 (generated from wheat germ cell-free expression system), these results substantiate our hypothesis that circPABPC1 can directly promote ITGB1 degradation by the 26S proteasome in a ubiquitin-independent manner and that the proteasome ATPase activity probably plays a necessary role in the engaging, unfolding, and translocation of ITGB1.
Fig. 7. In vitro proteasome binding and degradation of ITGB1 are facilitated by circPABPC1.
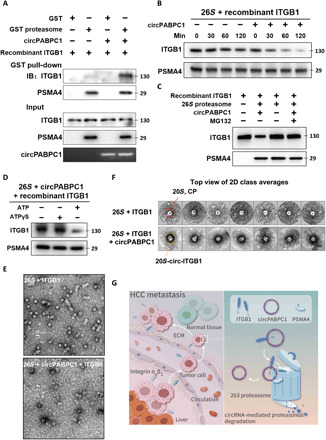
(A) For in vitro binding assay, 26S proteasomes immobilized on GST-UBL resin were incubated with recombinant ITGB1 in the presence or absence of circPABPC1. MG-132 (10 μM) was added to prevent ITGB1 degradation. After GST pull-down, proteasome-bound ITGB1 was detected by Western blot. (B) Active 26S proteasomes were purified as in (A), eluted, and incubated with recombinant ITGB1 in the presence or absence of circPABPC1. ATP was supplemented. Samples were taken at the indicated time points for Western blot analysis. (C) In vitro degradation of ITGB1 was measured as in (B) with or without MG-132 (10 μM). (D) In vitro degradation of ITGB1 was measured as in the presence or absence of ATP or ATPγS. (E) Representative negative-stained micrographs of purified 26S proteasomes after incubation with ITGB1 in the absence (top) or presence (bottom) of circPABPC1. Scale bar, 20 nm. (F) 2D class averages of samples in (E). The typical ring-shaped appearance of 20S CP (top) was altered in the presence of circPABPC1 and ITGB1 (bottom). (G) ITGB1 plays a key role in HCC metastasis. Frequent loss of circPABPC1 correlates with increased ITGB1 level in patients with advanced HCC. As a tumor suppressor, circPABPC1 physically links ITGB1 to the proteasome and promotes ubiquitin-independent ITGB1 degradation, thereby reducing tumor cell adhesion, migration, and metastatic spreading.
Last, we wondered whether the circPABPC1-mediated ITGB1-proteasome interaction could be visualized by transmission electron microscopy (EM). The proteasome was purified using the Rpn11-TBHA system (33) and incubated with recombinant ITGB1 in the presence of MG-132, with/without circPABPC1. The entire complexes were collected for negative-staining EM analysis (Fig. 7E). 2D class averages of negative stain images clearly showed the overall architectures of different proteasome assemblies (RP2CP, RP1CP, and CP; fig. S7C). Only when circPABPC1 was provided could extra densities consistent with the size of ITGB1 be observed that attached to the periphery of 20S (Fig. 7F). This finding further supports our model that circPABPC1 assists in the delivery of ITGB1 to the proteasome for ubiquitin-independent degradation, thereby down-regulating tumor cell adhesion to ECM and inhibiting HCC metastasis (Fig. 7G).
DISCUSSION
The production, function, and regulation of circRNAs have become a topic of intensive research in recent years, and numerous studies have reported their link with cancer (15–17). Both oncogenic and tumor-suppressive functions of circRNAs have been proposed, although cancer cells often exhibit a reduction in global circRNA abundance, which may result from dilution by rapid cell proliferation (34). Our previous research on gender disparity in HCC uncovered a mechanism for down-regulation of circRNAs by the AR, which also led to the discovery of circPABPC1 as a clinically relevant molecule in HCC (27). We found that circPABPC1 attenuated cancer cell proliferation, survival, and in vivo growth as xenograft tumors (27), consistent with it being a tumor suppressor. However, the role of circPABPC1 in HCC metastasis was not addressed in that study, and the molecular function of this circRNA has remained unknown.
Several other circRNAs have been implicated in HCC including circSMARCA5 (SWI/SNF Related, matrix associated, actin dependent regulator of chromatin, subfamily A, member 5), circMAT2B (methionine adenosyltransferase 2B), and circASAP1 (adenosine diphosphate ribosylation factor guanylate kinase 1), which have distinct roles in HCC progression but were all suggested to function as competing endogenous RNAs (18) that antagonize their cognate miRNAs (35–37). However, this sponging function of circRNAs has been controversial, given the endogenous miRNA:target ratios and the high numbers of additional targets required to achieve measurable consequences (38). The small size of circPABPC1 makes it even less likely to act as a miRNA sponge, and we found no change in the expression level of its host gene, PABPC1. On the other hand, mounting evidence indicates that circRNAs, especially those relatively small in size, can serve as scaffolds to facilitate interactions between proteins that may or may not be recognized as regular RNA binding proteins (19–25, 39). These circRNAs can be viewed as a molecular glue that enables binding and biochemical reactions between proteins that are otherwise physically separated, as we have demonstrated for circPABPC1 in regulating ITGB1 degradation. In addition to ITGB1 and proteasome subunits, our MS results identified dozens of other proteins that can interact with circPABPC1. Some of them may also be involved in circPABPC1-mediated tumor suppression in ways yet to be uncovered.
Integrins are essential for the survival of normal cells and participate in every step of cancer dissemination and metastasis (3, 40, 41). ITGB1 is particularly relevant in HCC as it partners with different α integrins to form receptors of most ECM components in the liver, including integrin α1β1 (receptor of laminin and collagen), integrin α5β1 (receptor of fibronectin), and integrin α9β1 (receptor of tenascin) (4). Our data showed that down-regulation of ITGB1 is a major mechanism by which circPABPC1 inhibits HCC cell adhesion and metastasis. AR was also reported to influence HCC metastasis by increasing ITGB1 expression (42). Our previous (27) and current studies on circPABPC1 have thus provided a mechanistic explanation for that finding. Very recently, another circRNA, circSKA3 (spindle and kinetochore associated complex subunit 3), was shown to bind ITGB1 and promote ITGB1-dependent invadopodia formation in breast cancer (26). Although circPABPC1 and circSKA3 operate in completely different ways, circRNA-mediated regulation of integrin signaling may be a common phenomenon in cancer.
ITGB1, as well as its partners (such as ITGA5), has been shown to be ubiquitinated, internalized, and degraded by the lysosome (10, 11), a typical degradation pathway for many proteins in the plasma membrane. Unexpectedly, however, circPABPC1-induced ITGB1 was selectively blocked by proteasome inhibitors but not CQ. Moreover, circPABPC1 had no obvious effects on the ubiquitination status of ITGB1 and could still promote ITGB1 degradation in the presence of ubiquitin E1 inhibitor, PYR-41. These data strongly argue that circPABPC1 shunts ITGB1 to a distinct pathway, namely, ubiquitin-independent proteasomal degradation. The molecular basis for this degradation is the proteasome-ITGB1 complex assembled by circPABPC1. To the best of our knowledge, this has been the first example of circRNA directly shuttling protein substrate to the proteasome. In addition to PSMA4 identified in the current study, several other proteasome subunits have been shown to be potential RNA binding proteins, although the biological meanings thereof remain uninvestigated (43). It is tempting to speculate that the associated RNAs may represent an unusual mechanism for substrate recognition by the proteasome.
Supporting the above model, purified 26S proteasome could directly degrade ITGB1 in vitro in the presence of circPABPC1. The recombinant full-length ITGB1 protein used as substrate was produced from a wheat germ expression system and free of ubiquitination. Therefore, the cytoplasmic tail of ITGB1 is likely to serve as a degron and/or initiation region (44) that engages with the proteasome. An intriguing question is how ITGB1 is extracted from the membrane for proteasomal degradation. Despite the potential role of p97/VCP (valosin-containing protein) complex in endoplasmic reticulum–associated degradation pathway (45, 46), ITGB1 might be extracted by the membrane-anchored proteasome itself (47, 48), with the assistance of circPABPC1. This is also consistent with the requirement for ATP hydrolysis (i.e., participation of the 19S RP) in ITGB1 degradation assays.
The tumor-suppressive function of circPABPC1 suggests potential therapeutic value of this circRNA for HCC treatment, which may be technically possible with recent advances in RNA-delivering methods (49, 50). The “matchmaker” property of circPABPC1 in ITGB1 regulation bears considerable similarity with that of PROTACs, except that circPABPC1 can directly target ITGB1 to the proteasome, bypassing the E3 ubiquitin ligase.
Recent single-cell sequencing studies have provided critical information about intrinsic alterations of tumor cells and their dynamic interactions with the tumor microenvironment (TME) in HCC (51, 52). A single clone of cancer cells with sufficient growth advantage can form multinodular HCC through intrahepatic spreading (52), which was recapitulated in our orthotopic xenograft model. A better understanding of the interactions between HCC cells and the TME is obviously required from both biological and clinical viewpoints. By binding with integrins and other proteins (including those yet to be identified), circPABPC1 may influence numerous components within the TME. The expression profiles and functional roles of circPABPC1 warrant further investigation in other cell types. Findings from these studies will provide important and necessary guiding principles for translational research on circPABPC1 and other circRNAs.
MATERIALS AND METHODS
Reagents
Detailed information about antibodies, chemicals, purified proteins, primer sequences, and plasmids can be found in the Supplementary Materials.
Patient samples
Three independent cohorts including a total of 205 patients with HCC were used in this study. All clinical samples were collected from Sir Run Run Shaw Hospital, Hangzhou, China. Cohort 1 contained a small group of 22 randomly selected HCC samples collected since August 2018, which were used for determining the expression levels of circPABPC1 and ITGB1. Cohort 2 consisted of 92 randomly selected samples collected since October 2017, which were used for circPABPC1 detection. Cohort 3 included 91 randomly selected samples collected since February 2006 with 5-year follow-up information. The follow-up period was defined as the interval from the date of surgery to the date of recurrence or death. Patients alive at the end of follow-up were censused. DFS was defined as the interval from the date of surgery until the detection of tumor recurrence. Patients without signs of recurrence at the end of follow-up were censused. Patients who died from diseases other than HCC or unexpected events were excluded from the study cohort. The low and high circPABPC1 expressions were cut off by median expression.
This study conformed to the principles of the Declaration of Helsinki and was approved by the Institutional Review Board of the Sir Run Run Shaw Hospital. All patients signed informed consent before surgery for the use of their tissues for scientific research.
Mouse xenograft studies
Male 6- to 8-week-old BALB/c nude mice were used for experiments. LM3 cells were engineered to stably express the luciferase reporter by transfection of pcDNA3.0 luciferase, followed by selection with G418 (27). circPABPC1-overexpressing or control LM3 cells were first injected into nude mice to form subcutaneous tumors. For orthotopic xenograft, 1 mg of tumor tissue (LM3-circPABPC1 or LM3-vector) was surgically transplanted into the left lobe of mouse liver, seven mice per group. Two weeks later, tumor formation and metastasis were routinely monitored with a fluorescent imager (IVIS Spectrum, Caliper Life Sciences, Hopkinton, MA) after intraperitoneal injection of d-luciferin (150 mg/kg). Mice were euthanized at 12 weeks after xenograft, and tumor/organ samples were isolated for further testing.
Cell culture
HA22T, LM3, 293T Rpn11-TBHA cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen, Grand Island, NY) with 10% fetal bovine serum (FBS) and 1% glutamine. Huh7 cell was maintained in minimum essential medium (Invitrogen, Grand Island, NY) with 10% FBS and 1% glutamine. Cell line sources are described in the Supplementary Materials.
Lentiviral infection and plasmid transfection
Cells with stable expression were established on the basis of a previous report (27). Briefly, human embryonic kidney 293 cells were transfected with the core plasmid (LentiV2, pLVX, or pCDH), with the psPAX2 packaging plasmid, and pMD2.G envelope plasmid, for 48 hours to obtain the lentivirus supernatant, which was frozen at −80°C for later infection to produce stable polyclonal populations. For the plasmid and siRNA transfection, we used Lipofectamine 3000 (Invitrogen), according to the manufacturer’s instructions. In addition, single guide RNAs for CRISPR were generated by following the LentiV2 manufacturer’s instructions (53).
Quantitative real-time PCR
Total RNAs were isolated using TRIzol reagent (Invitrogen), either from cultured cells or frozen tissue of primary tumors and adjacent nontumor liver tissues. One microgram of total RNA was subjected to reverse transcription using SuperScript III transcriptase (Invitrogen). RT-qPCR was conducted using a Bio-Rad CFX96 system (Bio-Rad, Hercules, CA) with SYBR green to determine the expression level of the RNA of interest, which was compared to that of GAPDH as an internal reference. For detection and quantification of circRNAs, qPCR primers were all designed toward the unique head-to-tail junction of circRNAs (27). Circularity of RNA was confirmed by ribonuclease R treatment that digests linear RNAs, before any further analysis.
Cell adhesion and migration assays
Cell adhesion assay was performed by seeding HCC cells in Matrigel-, fibronectin-, laminin-, collagen IV– or vitronectin-coated plates. Optimal conditions for attachment were determined for each cell line and application. Briefly, pure Matrigel was diluted with phosphate-buffered saline (PBS) at 1:1 before coating. ECM components were prepared according to the manufacturer’s instructions and diluted into PBS solution. The coating concentration was listed as fibronectin (1 to 2 μg/cm2), collagen type IV (0.1 to 0.2 mg/cm2), laminin (1 to 2 μg/cm2), and vitronectin (0.1 μg/cm2). Tissue culture plates were coated with the above protein solutions and air-dried overnight. HCC cells were seeded on coated plates and washed with PBS after partially attached. The remained cells were stained by crystal violet after 4% paraformaldehyde fixing. Images were captured, and absorbance at 540 nm was measured after dissolving crystal violet in 100% ethanol.
For transwell migration assay, HCC cells were seeded with serum-free medium into the upper chamber at 1 × 105 cells per well, and the bottom chamber of the apparatus contained culture medium with 10% FBS and then incubated for 24 hours at 37°C. Following incubation, the migrated cells attached to the lower surface of the membrane were fixed by 4% paraformaldehyde and stained with 0.1% crystal violet for observation.
For trypsin digestion assay, the same amounts of the indicated HCC cells were seeded in each well and allowed for attachment overnight. The next day, cells were treated with or without 0.05% trypsin/EDTA (Thermo Fisher Scientific) for 1 to 2 min at 37°C. After removal of detached cells, the remaining cells were fix-stained as above and imaged.
Cell lysis, immunoprecipitation, and immunoblotting
Cells were homogenized in radioimmunoprecipitation assay or immunoprecipitation (IP) lysis buffer supplemented with protease inhibitor cocktail, phosphatase inhibitor cocktail, panobinostat, and methylstat. Lysates were cleared by centrifugation at 13,000 rpm for 15 min at 4°C. Supernatants were used for immunoblotting or IP with the indicated antibodies. For IP, the prepared cell lysates were precleared using 10 μl of Protein A + G agarose beads by rotating at 4°C for 3 hours. The indicated antibodies or control immunoglobulin G (IgG) were added to precleared lysates, to the precleared lysates, followed by adding 20 μl of Protein A + G agarose beads to the mixture with rotating the tubes at 4°C for 3 to 5 hours for each process. The beads were washed with lysis buffer three times and eluted with 50 μl of SDS loading buffer, and the eluted protein or protein complexes were detected by immunoblot. Proteins were separated on 10 to 15% SDS–polyacrylamide gel electrophoresis gel and then transferred onto polyvinylidene difluoride membranes (Millipore, Billerica, MA). After blocking, membranes were incubated with appropriate dilutions of specific primary antibodies, followed by incubation with horseradish peroxidase–conjugated secondary antibodies and visualization using ECL system (Bio-Rad) (54).
RNA FISH assay and immunofluorescence staining
RNA localization was determined using a FISH kit from RiboBio according to the manufacturer’s protocol. Briefly, cells in chamber slides were fixed in 4% formaldehyde for 15 min. The fixed cells were further treated with pepsin (1% in 10 mM HCl) and subsequently dehydrated with 70, 90, and 100% ethanol. The air-dried cells were to incubated with 40 nM Cy3-conjugated FISH probe (RiboBio) in the hybridization buffer [dextran sulfate (100 mg/ml) and 10% formamide in 2× SSC] at 80°C for 2 min. Hybridization was performed at 55°C for 2 hours, and the slide was washed with 0.1× SSC at 65°C, followed by dehydration with 70, 90, and 100% ethanol. The slides were mounted using the ProLong Gold Antifade Reagent with 4′,6-diamidino-2-phenylindole (DAPI) for confocal fluorescence microscopy (54).
For immunofluorescence, cells were cultured in chamber slides overnight and fixed with 3.7% formaldehyde in PBS for 10 min at 4°C, followed by permeabilization with 0.5% Triton X-100 in PBS for 10 min. Cells were then blocked for nonspecific binding with 10% goat serum in PBS and 0.1% Tween 20 overnight and incubated with the indicated antibodies for 1 hour at room temperature, followed by incubation with a goat anti-mouse or anti-rabbit IgG (H + L) antibody (Alexa Fluor 488, Thermo Fisher Scientific) for 30 min at room temperature (54). For stress fiber staining, the Rhodamine Phalloidin F-actin Stain Kit from Thermo Fisher Scientific was used. Coverslips were mounted on slides using antifade mounting medium with DAPI or Hoechst. Immunofluorescence images were acquired on a Zeiss Axio Observer Z1 fluorescence microscope. For each channel, all images were acquired with the same settings.
circRNA pull-down and purification
The Magnetic RNA-Protein Pull-Down Kit (Pierce) was used for circRNA pull-down assays. Cells lysates prepared in the IP lysis buffer (Beyotime) were precleared by incubation with washed streptavidin magnetic beads at 4°C for 1 hour. Biotin-labeled circRNA probes immobilized on the streptavidin magnetic beads were then added to the cell lysates. After overnight incubation at 4°C, beads were magnetically separated and washed five times. For Western blot detection, the beads were boiled in SDS buffer for protein elution. For MS assays, the beads were incubated with nonionic water at 70°C for 5 min. For RT-qPCR assays and circRNA purification, TRIzol reagent was used to extract bound RNA from the beads.
MS analysis
The peptide samples collected from HA22T cells were analyzed on a Q Exactive mass spectrometer (Thermo Fisher Scientific, Bremen, Germany) by KangChen Bio-tech (Shanghai, China). The full mass and subsequent MS/MS analyses were performed in an Orbitrap analyzer with a resolution of 70,000 for MS1 [at 200 mass/charge ratio (m/z)] and 17,500 for MS2, respectively. The automatic gain control target for MS1 was set to 3.0 ×10+6 with max injection time (IT) of 50 ms and 5.0 × 10+4 for MS2 with max IT of 100 ms. The top 20 most intense ions were fragmented by higher-energy collisional dissociation (HCD) with normalized collision energy of 27% and isolation window of 2 m/z. The dynamic exclusion was set at 30 s. Raw data were processed with MaxQuant software (version 1.5.6.0). The intracellular pathway analysis was performed using clusterProfiler in R package to search GO and Kyoto Encyclopedia of Genes and Genomes database.
RNA immunoprecipitation and RNA extraction
To prepare antibody-coated beads, 20 μl of Protein A + G agarose beads were incubated with 1 to 5 μg of antibody or control IgG in 500 μl of lysis buffer supplemented with protease inhibitor cocktail, phosphatase inhibitor cocktail, panobinostat, and methylstat at 4°C overnight. Then, the beads were washed twice with lysis buffer and kept on ice. Cell lysates were precleared using 10 μl of Protein A + G agarose beads by rotating at 4°C for 3 hours. The precleared lysates were transferred to tubes with antibody-coated beads and then incubated at 4°C for 3 to 5 hours. Beads were washed with lysis buffer three times. RNA was collected from the beads using TRIzol. Nuclear and cytoplasmic fractionation of cells was performed using the PARIS Kit (Thermo Fisher Scientific) (54).
Mammalian two-hybrid assays
Assays were performed using the CheckMate Mammalian Two-Hybrid System (Promega) after transfection of the indicated plasmids. PSMA4 and ITGB1 complementary DNAs (cDNAs) were cloned into the pBIND and pACT vectors, respectively, and transfected along with a firefly luciferase vector (pG5luc, Promega). Reporter activities were measured 48 hours later using the Dual-Luciferase Reporter Assay Kit (Promega) and normalized to the Renilla luciferase activity.
Proteasome purification, in vitro degradation, and GST pull-down assay
Endogenous 26S proteasomes were isolated using the Human Proteasome Isolation Kit (Merck). HA22T cells were lysed in 50 mM tris-HCl (pH 7.5), 0.5% NP-40, 125 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, and 5 mM ATP, and lysates were incubated with UBL-GST-agarose beads at 4°C for 4 hours. The beads were washed at least three times before the immobilized 26S proteasomes were used for further assays.
For in vitro degradation assays, we followed the steps of a previous report (55). Briefly, 26S proteasomes purified from 1 × 107 HA22T cells as above were incubated with recombinant ITGB1 protein (Abcam) in 20 μl of buffer A [50 mM tris-HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, and 10% glycerol] with 10 mM dithiothreitol, 10× ATP regenerating system, and 5 mM ATP (or 5 mM ATPγS) added. The degradation reaction was allowed to proceed at 37°C for 1 hour in the presence or absence of circPABPC1 (purified from 1 × 107 HA22T-circPABPC1 cells). The reaction was terminated by boiling in SDS sample buffer and used for immunoblotting directly.
In vitro GST pull-down assays were set up with the same ingredients as above in the presence of 10 μM MG-132. The mixture was centrifuged, and the beads were washed at least three times. Bound proteins were eluted by boiling in SDS sample buffer and used for immunoblotting.
For large-scale proteasome purification, 293T Rpn11-TBHA cells (5 × 106) were used as described previously (33). Cell lysates were incubated with High Capacity Streptavidin Agarose (Thermo Fisher Scientific, USA) at 4°C for 2 hours. Beads were washed three times, and 26S proteasomes were cleaved off the beads by tobacco etch virus (TEV) protease (Thermo Fisher Scientific, USA). This yields high-quality proteasomes that will be used for negative stain EM.
PSMA4, PSMA5, PSMB5, and PSMD9 proteins were produced from pcDNA3-based plasmids using the TNT T7 Coupled Wheat Germ Extract System (Promega). GST-PSMD2 and GST-PSMD4 were expressed and purified from Escherichia coli as previously described (56).
RNA-protein interaction simulation and analysis
We used Mfold (version2.3, http://unafold.rna.albany.edu/?q=mfold) to calculate the lowest theoretical value of free energy for circPABPC1 (57). Dot-bracket notation was the filed used for RNAComposer (http://rnacomposer.cs.put.poznan.pl/) to generate the 3D model (58). Next, the 3D model of circPABPC1 was put in HDOCK software (http://hdock.phys.hust.edu.cn/) together with PSMA4 [Protein Data Bank (PDB) no. 5VFO, entity ID: 3] and ITGB1 (PDB no. 4DX9, entity ID: 2) for RNA/protein interaction simulation (59). Distance-based approach was used to identify the binding site residues/nucleotides for the protein-RNA complexes using a specific cutoff value. Two atoms (one in RNA and another in protein) were considered to be interacting with each other if the distance between them was <4.5 Å.
In the secondary structures of circRNAs, the first and last bases form the junction site, and they keeps unpaired with each other (60). To determine the secondary structures with lowest free energy, we used Mfold (version2.3) to calculate minimum free energy (57). Mfold uses energy minimization to identify optimal folding of a nucleic acid sequence within a specified energy increment to simulate the ensemble of possible structures. In addition, the secondary structure with lowest theoretical value of free energy would be chosen for 3D modeling. Dot-bracket notation was the filed used for RNAComposer to generate the 3D model (58). The latter is a web-based software for RNA folding and 3D structure prediction.
According to Mfold’s calculation, the best secondary structure for circPABPC1 would have a free energy of −15 kcal/mol, with −10.93, −10.20, −9.90, and −9.84 (kcal/mol) for the rest of top five models. Then, we used RNAComposer to analyze the tertiary structure and generated the 3D model of circPABPC1 in HDOCK software. HDOCK is a web server for in silico protein-protein and protein-DNA/RNA docking based on a hybrid strategy (59). Combined with protein structure data from PDB, we simulated the molecular interaction of circPABPC1, PSMA4 (PDB no. 5VFO, entity ID: 3) and ITGB1 (PDB no. 4DX9, entity ID: 2), respectively. The molecular simulation results confirmed that these three molecules could form a complex without a spatial conformation conflict.
Bases that might be responsible for interaction were also acquired from HDOCK, according to the distance to target protein. Next, we generated mutations in circPABPC1 for binding to ITGB1 and PSMA4, respectively, namely, circ-mu-ITGB1/circ-mu-PSMA4. The principle of mutation we adopted is avoiding changing the bases near conjunction site to ensure the ring formation. Since the mutation sites were not completely continuous, we recomposed the whole 91 bases of circPABPC1 according to mutation blueprint. After reconstructing the plasmid, we verified the ring formation and mutation by specifically designed primers and sequencing.
Negative stain EM grid preparation
Uranyl formate solution (0.75%) for negative stain EM was prepared as reported (61). Three hundred–mesh copper grids with carbon film (Electron Microscopy Sciences) were glow-discharged (PELCO easiGlow Glow Discharge Cleaning System) for 1 min at 25 mA before 3.5 μl of purified complex was applied to the grids and incubated for 30 s. After blotting the sample using filter paper, the grid surface was touched on two drops of 30 μl of 0.75% uranyl formate, and then the grids were stained on the third drop of uranyl formate with gentle stirring for 40 s. Stained grids were blotted to remove excess stain.
Image acquisition and data processing
Negative stain data collection was carried out on a Tecnai G2 Spirit transmission EM (Thermo Fisher Scientific, FEI) operating at 120 kV. Images were collected at a nominal magnification of 105,000 (3.1-Å pixel size) within a −0.5- to −3.0-μm defocus range. For 26S proteasome alone, 100 micrographs were collected, and particle selection was performed using autopicking command in RELiON-3.0-beta2 (62), yielding 29,585 particle projections. For 26S proteasome complexed with circPABPC1 and ITGB1, 100 micrographs were collected, and 33,465 particles were autopicked in RELiON. Each dataset was subjected to reference-free 2D classification.
Quantification and statistical analysis
The experiment was set up to use three samples/repeats per experiment/group/condition to detect a twofold difference with power of 80% and at the significance level of 0.05 by a two-sided test for significant studies. Results are reported as means ± SD. For protein half-life simulation, nonlinear regression with model of one-phase decay in GraphPad Prism v6.01 was performed. Comparisons were performed using two tailed paired Student’s t test or one-way analysis of variance (ANOVA), as indicated in individual figures. For survival analysis, the expression of indicated targets was treated as a binary variant and divided into “high” and “low” level. Kaplan-Meier survival curves were compared using the Gehan-Breslow test in GraphPad Prism v6.01.
Acknowledgments
We thank Y. Sun (University of Rochester, USA) for suggestions. We thank the Center of Cryo-Electron Microscopy (CCEM), Zhejiang University for technical assistance on transmission microscopy. All cDNAs of human proteasome subunits with N- or C-terminal Flag tag were provided by S. Murata from University of Tokyo. Funding: L.S. was supported by Postdoctoral Science Fund of China (2019tq0280 and 2019 M662079) and Zhejiang Natural Science Foundation (LQ20H160024). L.H. was supported by Zhejiang Natural Science Foundation (LQ20C010005). Yan Zhang was supported by the National Natural Science Foundation of China (81922071), the National Key Basic Research Program of China (2019YFA0508800), Zhejiang Province Natural Science Fund for Excellent Young Scholars (LR19H310001), and the Fundamental Research Funds for the Central Universities (2019XZZX001-01-06). X.G. was funded by Zhejiang Natural Science Foundation (LR18C050001) and Natural Science Foundation of China (31671391 and 31870762). X.C. was supported by the Natural Science Foundation of China (81772546). Author contributions: X.C. and X.G. conceived and designed the research. X.G. and Y.W. provided administrative support and revision suggestion. L.S. and B.L. performed most of the biochemical and molecular experiments, with the assistance from P.Y., Yanan Zhang, Y.T., L.H., G.J., Y. Zhu., Y.L., and B.S. D.-d.S. designed, performed, and analyzed the negative-staining EM experiments and participated in figure preparation under the supervision of Yan Zhang X.L. and H.Y. collected and provided clinical samples and data. L.S. and X.G. wrote the manuscript with input from B.L. All authors read and approved this version of manuscript. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Profiling by circRNA Array in our previous work has been deposited in the GEO database as GSE94520. RNA pull-down–MS data in this paper have been deposited on Mendeley Data (https://data.mendeley.com/datasets/bg2fcctfms/1). Additional data related to this paper may be requested from the authors.
SUPPLEMENTARY MATERIALS
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/13/eabe5043/DC1
REFERENCES AND NOTES
- 1.Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Llovet J. M., Zucman-Rossi J., Pikarsky E., Sangro B., Schwartz M., Sherman M., Gores G., Hepatocellular carcinoma. Nat. Rev. Dis. Primers. 2, 16018 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Desgrosellier J. S., Cheresh D. A., Integrins in cancer: Biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hamidi H., Ivaska J., Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper J., Giancotti F. G., Integrin signaling in cancer: Mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 35, 347–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang C., Zeisberg M., Lively J. C., Nyberg P., Afdhal N., Kalluri R., Integrin α1β1 and α2β1 are the key regulators of hepatocarcinoma cell invasion across the fibrotic matrix microenvironment. Cancer Res. 63, 8312–8317 (2003). [PubMed] [Google Scholar]
- 7.Ding Y., Fei Y., Lu B., Emerging new concepts of degrader technologies. Trends Pharmacol. Sci. 41, 464–474 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schapira M., Calabrese M. F., Bullock A. N., Crews C. M., Targeted protein degradation: Expanding the toolbox. Nat. Rev. Drug Discov. 18, 949–963 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Finley D., Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78, 477–513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lobert V. H., Brech A., Pedersen N. M., Wesche J., Oppelt A., Malerød L., Stenmark H., Ubiquitination of α5β1 integrin controls fibroblast migration through lysosomal degradation of fibronectin-integrin complexes. Dev. Cell 19, 148–159 (2010). [DOI] [PubMed] [Google Scholar]
- 11.Böttcher R. T., Stremmel C., Meves A., Meyer H., Widmaier M., Tseng H. Y., Fässler R., Sorting nexin 17 prevents lysosomal degradation of β1 integrins by binding to the β1-integrin tail. Nat. Cell Biol. 14, 584–592 (2012). [DOI] [PubMed] [Google Scholar]
- 12.Ueki Y., Saito K., Iioka H., Sakamoto I., Kanda Y., Sakaguchi M., Horii A., Kondo E., PLOD2 is essential to functional activation of integrin β1 for invasion/metastasis in head and neck squamous cell carcinomas. iScience 23, 100850 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wujak L., Böttcher R. T., Pak O., Frey H., el Agha E., Chen Y., Schmitt S., Bellusci S., Schaefer L., Weissmann N., Fässler R., Wygrecka M., Low density lipoprotein receptor-related protein 1 couples β1 integrin activation to degradation. Cell. Mol. Life Sci. 75, 1671–1685 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogorad R. L., Yin H., Zeigerer A., Nonaka H., Ruda V. M., Zerial M., Anderson D. G., Koteliansky V., Nanoparticle-formulated siRNA targeting integrins inhibits hepatocellular carcinoma progression in mice. Nat. Commun. 5, 3869 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L. L., The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21, 475–490 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Dong Y., He D., Peng Z., Peng W., Shi W., Wang J., Li B., Zhang C., Duan C., Circular RNAs in cancer: An emerging key player. J. Hematol. Oncol. 10, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fu L., Jiang Z., Li T., Hu Y., Guo J., Circular RNAs in hepatocellular carcinoma: Functions and implications. Cancer Med. 7, 3101–3109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hansen T. B., Jensen T. I., Clausen B. H., Bramsen J. B., Finsen B., Damgaard C. K., Kjems J., Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Li H., Yang F., Hu A., Wang X., Fang E., Chen Y., Li D., Song H., Wang J., Guo Y., Liu Y., Li H., Huang K., Zheng L., Tong Q., Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol. Med. 11, e10835 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yang F., Hu A., Li D., Wang J., Guo Y., Liu Y., Li H., Chen Y., Wang X., Huang K., Zheng L., Tong Q., Circ-HuR suppresses HuR expression and gastric cancer progression by inhibiting CNBP transactivation. Mol. Cancer 18, 158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang H., Li X., Meng Q., Sun H., Wu S., Hu W., Liu G., Li X., Yang Y., Chen R., CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol. Cancer 19, 13 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fang L., du W. W., Lyu J., Dong J., Zhang C., Yang W., He A., Kwok Y. S. S., Ma J., Wu N., Li F., Awan F. M., He C., Yang B. L., Peng C., MacKay H. J., Yee A. J., Yang B. B., Enhanced breast cancer progression by mutant p53 is inhibited by the circular RNA circ-Ccnb1. Cell Death Differ. 25, 2195–2208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang S., Li X., Zheng H., Si X., Li B., Wei G., Li C., Chen Y., Chen Y., Liao W., Liao Y., Bin J., Loss of super-enhancer-regulated circRNA Nfix induces cardiac regeneration after myocardial infarction in adult mice. Circulation 139, 2857–2876 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li Q., Wang Y., Wu S., Zhou Z., Ding X., Shi R., Thorne R. F., Zhang X. D., Hu W., Wu M., CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 30, 157–173.e7 (2019). [DOI] [PubMed] [Google Scholar]
- 25.Chen R. X., Chen X., Xia L. P., Zhang J. X., Pan Z. Z., Ma X. D., Han K., Chen J. W., Judde J. G., Deas O., Wang F., Ma N. F., Guan X., Yun J. P., Wang F. W., Xu R. H., Dan Xie, N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 10, 4695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du W. W., Yang W., Li X., Fang L., Wu N., Li F., Chen Y., He Q., Liu E., Yang Z., Awan F. M., Liu M., Yang B. B., The circular RNA circSKA3 binds integrin β1 to induce invadopodium formation enhancing breast cancer invasion. Mol. Ther. 28, 1287–1298 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shi L., Yan P., Liang Y., Sun Y., Shen J., Zhou S., Lin H., Liang X., Cai X., Circular RNA expression is suppressed by androgen receptor (AR)-regulated adenosine deaminase that acts on RNA (ADAR1) in human hepatocellular carcinoma. Cell Death Dis. 8, e3171 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang R. Y., Chen L., Chen H.-Y., Hu L., Li L., Sun H.-Y., Jiang F., Zhao J., Liu G.-M.-Y., Tang J., Chen C.-Y., Yang Y.-C., Chang Y.-X., Liu H., Zhang J., Yang Y., Huang G., Shen F., Wu M.-C., Zhou W.-P., Wang H.-Y., MUC15 inhibits dimerization of EGFR and PI3K-AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology 145, 1436–1448.e12 (2013). [DOI] [PubMed] [Google Scholar]
- 29.Xu J., Wan Z., Tang M., Lin Z., Jiang S., Ji L., Gorshkov K., Mao Q., Xia S., Cen D., Zheng J., Liang X., Cai X., N6-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol. Cancer 19, 163 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shen P., Yang Y., Liu G., Chen W., Chen J., Wang Q., Gao H., Fan S., Shen S., Zhao X., CircCDK14 protects against Osteoarthritis by sponging miR-125a-5p and promoting the expression of Smad2. Theranostics 10, 9113–9131 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parsons J. T., Focal adhesion kinase: The first ten years. J. Cell Sci. 116, 1409–1416 (2003). [DOI] [PubMed] [Google Scholar]
- 32.Besche H. C., Goldberg A. L., Affinity purification of mammalian 26S proteasomes using an ubiquitin-like domain. Meth. Mol. Biol. 832, 423–432 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Guo X., Wang X., Wang Z., Banerjee S., Yang J., Huang L., Dixon J. E., Site-specific proteasome phosphorylation controls cell proliferation and tumorigenesis. Nat. Cell Biol. 18, 202–212 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bachmayr-Heyda A., Reiner A. T., Auer K., Sukhbaatar N., Aust S., Bachleitner-Hofmann T., Mesteri I., Grunt T. W., Zeillinger R., Pils D., Correlation of circular RNA abundance with proliferation—Exemplified with colorectal and ovarian cancer, idiopathic lung fibrosis, and normal human tissues. Sci. Rep. 5, 8057 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yu J., Xu Q.-G., Wang Z.-G., Yang Y., Zhang L., Ma J.-Z., Sun S.-H., Yang F., Zhou W.-P., Circular RNA cSMARCA5 inhibits growth and metastasis in hepatocellular carcinoma. J. Hepatol. 68, 1214–1227 (2018). [DOI] [PubMed] [Google Scholar]
- 36.Li Q., Pan X., Zhu D., Deng Z., Jiang R., Wang X., Circular RNA MAT2B promotes glycolysis and malignancy of hepatocellular carcinoma through the miR-338-3p/PKM2 axis under hypoxic stress. Hepatology. 70, 1298–1316 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Hu Z. Q., Zhou S.-L., Li J., Zhou Z.-J., Wang P.-C., Xin H.-Y., Mao L., Luo C.-B., Yu S.-Y., Huang X.-W., Cao Y., Fan J., Zhou J., Circular RNA sequencing identifies CircASAP1 as a key regulator in hepatocellular carcinoma metastasis. Hepatology 72, 906–922 (2019). [DOI] [PubMed] [Google Scholar]
- 38.Thomson D. W., Dinger M. E., Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 17, 272–283 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Du W. W., Yang W., Liu E., Yang Z., Dhaliwal P., Yang B. B., Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dongre A., Weinberg R. A., New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 41.Friedl P., Gilmour D., Collective cell migration in morphogenesis, regeneration and cancer. Nat. Rev. Mol. Cell Biol. 10, 445–457 (2009). [DOI] [PubMed] [Google Scholar]
- 42.Ma W. L., Jeng L. B., Lai H. C., Liao P. Y., Chang C., Androgen receptor enhances cell adhesion and decreases cell migration via modulating β1-integrin-AKT signaling in hepatocellular carcinoma cells. Cancer Lett. 351, 64–71 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Castello A., Fischer B., Eichelbaum K., Horos R., Beckmann B. M., Strein C., Davey N. E., Humphreys D. T., Preiss T., Steinmetz L. M., Krijgsveld J., Hentze M. W., Insights into RNA biology from an atlas of mammalian mRNA-binding proteins. Cell 149, 1393–1406 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Yu H., Matouschek A., Recognition of client proteins by the proteasome. Annu. Rev. Biophys. 46, 149–173 (2017). [DOI] [PubMed] [Google Scholar]
- 45.van den Boom J., Meyer H., VCP/p97-mediated unfolding as a principle in protein homeostasis and signaling. Mol. Cell 69, 182–194 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Liu Y., Ye Y., Proteostasis regulation at the endoplasmic reticulum: A new perturbation site for targeted cancer therapy. Cell Res. 21, 867–883 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ramachandran K. V., Fu J. M., Schaffer T. B., Na C. H., Delannoy M., Margolis S. S., Activity-dependent degradation of the nascentome by the neuronal membrane proteasome. Mol. Cell 71, 169–177.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramachandran K. V., Margolis S. S., A mammalian nervous-system-specific plasma membrane proteasome complex that modulates neuronal function. Nat. Struct. Mol. Biol. 24, 419–430 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhaskaran V., Yao Y., Bei F., Peruzzi P., Engineering, delivery, and biological validation of artificial microRNA clusters for gene therapy applications. Nat. Protoc. 14, 3538–3553 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang L., Han B., Zhang Z., Wang S., Bai Y., Zhang Y., Tang Y., du L., Xu L., Wu F., Zuo L., Chen X., Lin Y., Liu K., Ye Q., Chen B., Li B., Tang T., Wang Y., Shen L., Wang G., Ju M., Yuan M., Jiang W., Zhang J. H., Hu G., Wang J., Yao H., Extracellular vesicle–mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation 142, 556–574 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Hohmann T., Dehghani F., The cytoskeleton-A complex interacting meshwork. Cells 8, 362 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duan M., Hao J., Cui S., Worthley D. L., Zhang S., Wang Z., Shi J., Liu L., Wang X., Ke A., Cao Y., Xi R., Zhang X., Zhou J., Fan J., Li C., Gao Q., Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 28, 359–373 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sanjana N. E., Shalem O., Zhang F., Improved vectors and genome-wide libraries for CRISPR screening. Nat. Methods 11, 783–784 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sang L.-J., Ju H.-Q., Liu G.-P., Tian T., Ma G.-L., Lu Y.-X., Liu Z.-X., Pan R.-L., Li R.-H., Piao H.-L., Marks J. R., Yang L.-J., Yan Q., Wang W., Shao J., Zhou Y., Zhou T., Lin A., LncRNA CamK-A regulates Ca2+-signaling-mediated tumor microenvironment remodeling. Mol. Cell 72, 71–83.e7 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Li Y., Tomko R. J. Jr., Hochstrasser M., Proteasomes: Isolation and activity assays. Curr. Protoc. Cell Biol. 67, 3.43.1–3.43.20 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu X., Xiao W., Zhang Y., Wiley S. E., Zuo T., Zheng Y., Chen N., Chen L., Wang X., Zheng Y., Huang L., Lin S., Murphy A. N., Dixon J. E., Xu P., Guo X., Reversible phosphorylation of Rpn1 regulates 26S proteasome assembly and function. Proc. Natl. Acad. Sci. U.S.A. 117, 328–336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zuker M., Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Popenda M., Szachniuk M., Antczak M., Purzycka K. J., Lukasiak P., Bartol N., Blazewicz J., Adamiak R. W., Automated 3D structure composition for large RNAs. Nucleic Acids Res. 40, e112 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan Y., Zhang D., Zhou P., Li B., Huang S. Y., HDOCK: A web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 45, W365–W373 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hofacker I. L., Stadler P. F., Memory efficient folding algorithms for circular RNA secondary structures. Bioinformatics 22, 1172–1176 (2006). [DOI] [PubMed] [Google Scholar]
- 61.Booth D. S., Avila-Sakar A., Cheng Y., Visualizing proteins and macromolecular complexes by negative stain EM: From grid preparation to image acquisition. J. Vis. Exp., (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Scheres S. H., RELION: Implementation of a Bayesian approach to cryo-EM structure determination. J. Struct. Biol. 180, 519–530 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hou L., Zhao J., Gao S., Ji T., Song T., Li Y., Wang J., Geng C., Long M., Chen J., Lin H., Cai X., Cang Y., Restriction of hepatitis B virus replication by c-Abl–induced proteasomal degradation of the viral polymerase. Sci. Adv. 5, eaau7130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material for this article is available at http://advances.sciencemag.org/cgi/content/full/7/13/eabe5043/DC1


