Abstract
Background
Chest x-ray is a relatively accessible, inexpensive, fast imaging modality that might be valuable in the prognostication of patients with COVID-19. We aimed to develop and evaluate an artificial intelligence system using chest x-rays and clinical data to predict disease severity and progression in patients with COVID-19.
Methods
We did a retrospective study in multiple hospitals in the University of Pennsylvania Health System in Philadelphia, PA, USA, and Brown University affiliated hospitals in Providence, RI, USA. Patients who presented to a hospital in the University of Pennsylvania Health System via the emergency department, with a diagnosis of COVID-19 confirmed by RT-PCR and with an available chest x-ray from their initial presentation or admission, were retrospectively identified and randomly divided into training, validation, and test sets (7:1:2). Using the chest x-rays as input to an EfficientNet deep neural network and clinical data, models were trained to predict the binary outcome of disease severity (ie, critical or non-critical). The deep-learning features extracted from the model and clinical data were used to build time-to-event models to predict the risk of disease progression. The models were externally tested on patients who presented to an independent multicentre institution, Brown University affiliated hospitals, and compared with severity scores provided by radiologists.
Findings
1834 patients who presented via the University of Pennsylvania Health System between March 9 and July 20, 2020, were identified and assigned to the model training (n=1285), validation (n=183), or testing (n=366) sets. 475 patients who presented via the Brown University affiliated hospitals between March 1 and July 18, 2020, were identified for external testing of the models. When chest x-rays were added to clinical data for severity prediction, area under the receiver operating characteristic curve (ROC-AUC) increased from 0·821 (95% CI 0·796–0·828) to 0·846 (0·815–0·852; p<0·0001) on internal testing and 0·731 (0·712–0·738) to 0·792 (0·780–0 ·803; p<0·0001) on external testing. When deep-learning features were added to clinical data for progression prediction, the concordance index (C-index) increased from 0·769 (0·755–0·786) to 0·805 (0·800–0·820; p<0·0001) on internal testing and 0·707 (0·695–0·729) to 0·752 (0·739–0·764; p<0·0001) on external testing. The image and clinical data combined model had significantly better prognostic performance than combined severity scores and clinical data on internal testing (C-index 0·805 vs 0·781; p=0·0002) and external testing (C-index 0·752 vs 0·715; p<0·0001).
Interpretation
In patients with COVID-19, artificial intelligence based on chest x-rays had better prognostic performance than clinical data or radiologist-derived severity scores. Using artificial intelligence, chest x-rays can augment clinical data in predicting the risk of progression to critical illness in patients with COVID-19.
Funding
Brown University, Amazon Web Services Diagnostic Development Initiative, Radiological Society of North America, National Cancer Institute and National Institute of Biomedical Imaging and Bioengineering of the National Institutes of Health.
Introduction
As of Feb 28, 2021, there were more than 113·4 million confirmed cases of COVID-19 worldwide, with daily increases in the number of new cases per day in the USA,1 so it is imperative that health-care providers efficiently triage patients with COVID-19. An early prediction of disease severity could be helpful in allocating resources in a timely manner to patients who are severely ill or who will progress to require critical care.
This prognostication can be made possible with medical imaging. For example, chest CT could be used for early diagnosis and determination of prognosis in patients with COVID-19.2, 3 Despite the high sensitivity and three-dimensional nature of CT, chest x-rays might be more useful in the COVID-19 pandemic due to their relative speed, low cost, portability, and accessibility, especially in low-resource settings, and with high patient volumes and critically ill patients whose transport for CT might be physically challenging.4 Chest x-rays have been shown to be efficacious in predicting the deterioration of patients with severe acute respiratory syndrome to critical status.5 In the context of COVID-19, chest x-rays have been analysed to predict the risk for hospital admission, length of hospitalisation, and risk of critical outcomes.6, 7, 8 However, there is a scarcity of studies that integrate artificial intelligence into chest x-ray analysis for time-to-event progression risk, demonstrate the incremental value of chest x-rays on prediction model based on clinical data alone, and compare the efficacy of these models with radiologist-derived severity scores.6, 7, 8, 9
Research in context.
Evidence before this study
We searched PubMed for articles published from database inception to Sept 1, 2020, with the search terms (“COVID-19” OR “coronavirus disease 2019”) AND (“artificial intelligence” OR “machine learning” OR “deep learning”) AND (“chest x-ray” OR “chest radiograph”), with no language restrictions, and found ten publications. Eight of ten studies used chest x-rays for COVID-19 diagnosis. Two of ten studies used chest x-rays to predict the severity of COVID-19 lung infection. This search indicated that there is a scarcity of studies related to artificial intelligence based on chest x-rays, especially for the prognostication of patients with COVID-19. To our knowledge, there are no published studies that predict the progression of patients with COVID-19 or the time until their deterioration using artificial intelligence based on chest x-rays.
Added value of this study
We used artificial intelligence based on chest x-rays to predict the severity of disease in patients with COVID-19 and their risk of disease progression during hospitalisation. The proposed model was trained and internally tested on a multicentre cohort of 1834 patients with COVID-19, who presented to hospital through the emergency department. The model was then externally tested on an independent cohort of 475 patients with COVID-19 who presented through a separate institution. The disease severity prediction has a binary outcome of critical or non-critical. A critical severity was defined as requiring ventilation or admission to the intensive care unit, or leading to death, during hospitalisation. The progression prediction has a time-to-event outcome, allowing clinicians to predict when the patient will deteriorate to a critical event. The model performance was evaluated and compared with manually derived severity scores by the radiologists.
Implications of all the available evidence
Efficient and effective prognostication of patients with COVID-19 is necessary for improved triaging of care and resources. Artificial intelligence has an auxiliary role in medicine to further improve the clinical workflow. Using the proposed artificial intelligence models, it might be possible to take advantage of readily available imaging data like chest x-rays to identify high-risk patients early and improve outcomes.
Artificial intelligence using chest x-rays has been used to assist in diagnosis and prognosis for patients with COVID-19.9, 10 By contrast, we aimed to implement a deep-learning artificial intelligence model using initial chest x-rays and clinical data to predict disease severity and risk of progression to critical illness in patients with COVID-19. We also aimed to compare the performance of the artificial intelligence model to that of radiologist-derived severity scores and clinical data.
Methods
Study design and participants
We did a retrospective study and identified patients with COVID-19 who presented to the emergency department of hospitals within the University of Pennsylvania Health System in Philadelphia, PA, USA, who we randomly assigned (7:1:2) to model training, validation, and internal testing sets, and patients who presented to Brown University affiliated hospitals in Providence, RI, USA, who were assigned to external testing (appendix p 2).
All patients included in the study presented to the hospital through the emergency department; from there they were either discharged or admitted to inpatient services for further monitoring and investigations, or died. Another criterion for inclusion in the study was a confirmatory RT-PCR for COVID-19 in the emergency department. The RT-PCR (COVID-19 RT-PCR test; Laboratory Corporation of America, Burlington, NC, USA) results were extracted from their electronic medical records. For patients with more than one RT-PCR assay, the earliest positive result from the emergency department was used. In addition, during the same hospitalisation, patients were required to have had at least one chest x-ray in anteroposterior view done in the emergency department or inpatient services (if admitted). If they had multiple chest x-rays from the admission, then the x-ray from closest to the time of initial presentation to the emergency department was downloaded from the hospital picture archiving and communications systems and included in the study.
The institutional review boards of all hospital institutions included in this study provided ethical approval. The requirement for written informed consent was waived. To avoid any potential breach of patient confidentiality, the data were deidentified and had no linkage to the researchers.
Procedures
For each patient, clinical data including age, sex, temperature, oxygen saturation on room air, white blood cell count, lymphocyte count, creatinine, C-reactive protein, and comorbidities such as cardiovascular disease, hypertension, chronic obstructive pulmonary disease, diabetes, chronic liver disease, chronic kidney disease, cancer, and HIV status were collected.11, 12 The vital signs and laboratory values were taken at the initial time of presentation. Additional information such as date and time of utilisation of mechanical ventilation, admission to the intensive care unit (ICU), and progression to death or discharge from the hospital were recorded. Disease outcome severity was defined as critical if the patient had any of the following outcomes: utilisation of mechanical ventilation, admission to the ICU, or death. If the patient did not have any of these critical outcomes, the disease severity was defined as non-critical. For patients with critical disease, the time to progression to a critical event was found by measuring the timeframe between their chest x-ray and their first critical outcome. For example, if the patient was on mechanical ventilation and later died, then their time to progression was defined as the time between their chest x-ray and mechanical ventilation.
The chest x-rays included in the study were independently scored for disease severity by two radiologists with 3 years of experience, with the supervision of a senior radiologist (W-HL). Additional information on chest x-ray severity scoring is provided in the appendix (pp 4–5).
For the severity prediction model, the chest x-rays were first segmented by a deep-learning model using a trained U-Net architecture.13 A pretrained visual geometry group architecture (VGG-11) was used as a feature extractor. This was followed by five encoder blocks and five decoder blocks to learn the transformation from input images and the corresponding binary masks. All images and masks were resized to 512 × 512 pixels size and normalised to the range 0–1 before being input to the segmentation U-Net. Negative log-likelihood loss was used to train network. Softmax operation was applied to model output. An Adam optimiser was used with a learning rate of 0·0005. Additional information on chest x-ray segmentation is provided in the appendix (pp 6–7).
An artificial intelligence model was built to predict the binary outcome of disease severity (critical or non-critical). For the image-based prediction, chest x-rays were preprocessed by normalising them to the range 0–1. The images were rescaled to 512 × 512 pixels. The processed images and their lung segmentations were used to generate masked images. The masked images were then combined with the feature-representation layers of the EfficientNet-B0 architecture pretrained on ImageNet14 and four prediction layers.
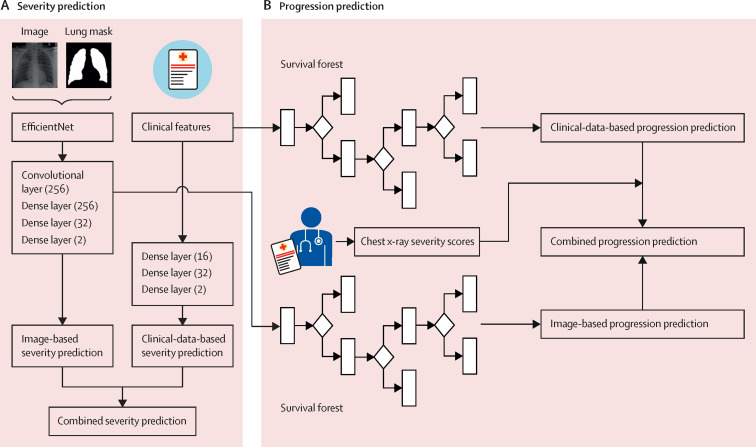
For the clinical-data-based prediction, a model with dense layers (16, 32, and 2) was trained to distinguish critical disease from non-critical disease on the basis of 16 collected clinical variables. Lastly, the combined severity prediction model was derived from the weighted sum of the image-based and clinical-data-based prediction models (figure 1A ). Additional information on the severity prediction model is provided in the appendix (pp 8–9).
Figure 1.
Illustration of our analysis pipeline
Severity prediction (A) and progression prediction (B).
For the progression prediction model, time-to-event models were built to predict the risk of progression to first critical outcome in patients with COVID-19 (figure 1B). For the image-based prediction, a series of 256-dimensional deep-learning image features were extracted from dense layer (256) of the aforementioned severity prediction model, and for the clinical-data-based model the input features were the same 16 clinical variables. The features were input to a survival forest model to derive image-based risk scores. The weighted sum of the image-based and clinical-data-based risk scores acts as the combined progression risk for each patient. For comparison, the chest x-ray severity scores were regarded as the progression risk measure to calculate the corresponding time-to-event evaluation and used to predict progression risks of patients in combination with clinical data. Additional information on the progression prediction model is provided in the appendix (p 10).
Statistical analysis
Area under the receiver operating characteristic curve (ROC-AUC) was calculated for the binary disease classification of critical or non-critical. 95% CI were determined using the adjusted Wald method.15 The concordance index (C-index) for right-censored data was applied to evaluate the performance of progression prediction models16 by comparing the progression information (positive labels and progression days) with the ranks of predicted risk scores. The Kaplan-Meier method was used to further stratify patients into high-risk and low-risk subgroups according to the median of progression risk scores.17 The stratification performance was evaluated using a log-rank test based on the predicted risk scores and critical progression information of the stratified subgroups. The time-dependent ROC-AUC was calculated for the progression prediction model.18 The precision-recall curves and F-scores were used to evaluate the severity and progression prediction models.19 Different prediction models were compared using a binomial test to show differences in performance. A p value of less than 0·05 was considered significant.
The lung segmentation model and severity prediction model were implemented with PyTorch (version 1.3.0) and trained with NVIDIA Titan V graphics processing units (NVIDIA, Santa Clara, CA, USA). The image-based and clinical-data-based progression prediction models were implemented with scikit-learn (version 0.21.3). The C-index and ROC-AUC were calculated using the Python package of scikit-learn (version 0.21.3). The binomial test was calculated using the Python package of scipy (version 1.5.0). The Kaplan-Meier curve was calculated using the R package of survminer (version 0.4.8). The model files, model parameters, and codes of our segmentation, severity, and progression prediction models are publicly available on GitHub. It is recognised that the development of a highly effective artificial intelligence model requires a multidisciplinary approach, so a web-based application of our severity prediction model is publicly available for use by other researchers and clinicians.
Role of the funding source
The funders of the study had no role in data collection, data analysis, data interpretation, or writing of the report.
Results
1834 patients who presented via the University of Pennsylvania Health System between March 9 and July 20, 2020, were identified and randomly divided into model training (n=1285), validation (n=183), and testing (n=366) sets. The internal test set was strictly separated from the training and validation sets at the patient level to avoid data leakage. 475 patients who presented via the Brown University affiliated hospitals between March 1 and July 18, 2020, were identified for external testing of the models (table 1 , appendix p 3).
Table 1.
Patient characteristics
| Patients from University of Pennsylvania Health System (n=1834) | Patients from Brown University affiliated hospitals (n=475) | p value | ||
|---|---|---|---|---|
| Age, years | .. | .. | <0·001 | |
| Median (IQR) | 55 (31·0) | 60 (26·5) | .. | |
| <20 | 28 (2%) | 6 (1%) | .. | |
| 20–39 | 493 (27%) | 66 (14%) | .. | |
| 40–59 | 539 (29%) | 160 (34%) | .. | |
| 60–79 | 577 (31%) | 168 (35%) | .. | |
| ≥80 | 197 (11%) | 75 (16%) | .. | |
| Sex | .. | .. | <0·001 | |
| Male | 854 (47%) | 278 (59%) | .. | |
| Female | 980 (53%) | 197 (41%) | .. | |
| Body temperature | .. | .. | 0·92 | |
| Elevated (>37°C) | 1186 (65%) | 311 (65%) | .. | |
| Not elevated (≤37°C) | 632 (34%) | 164 (35%) | .. | |
| Oxygen saturation on room air | .. | .. | <0·001 | |
| Not decreased (≥94%) | 1505 (82%) | 345 (73%) | .. | |
| Decreased (<94%) | 283 (15%) | 118 (25%) | .. | |
| White blood cell count | .. | .. | 0·0010 | |
| Elevated (>11 × 109/L) | 240 (13%) | 100 (21%) | .. | |
| Not elevated (≤11 × 109/L) | 1339 (73%) | 363 (76%) | .. | |
| Lymphocyte count | .. | .. | <0·001 | |
| Not decreased (≥1·0 × 109/L) | 914 (50%) | 195 (41%) | .. | |
| Decreased (<1·0 × 09/L) | 650 (35%) | 268 (56%) | .. | |
| Creatinine | .. | .. | 0·0040 | |
| Elevated (≥1·27 mg/dL) | 481 (26%) | 113 (24%) | .. | |
| Not elevated (<1·27 mg/dL) | 1062 (58%) | 353 (74%) | .. | |
| C-reactive protein | .. | .. | 0·16 | |
| Elevated (≥1·0 mg/dL) | 425 (23%) | 299 (63%) | .. | |
| Not elevated (<1·0 mg/dL) | 41 (2%) | 40 (8%) | .. | |
| Comorbidities | .. | .. | .. | |
| Cardiovascular disease | 390 (21%) | 124 (26%) | 0·021 | |
| Hypertension | 682 (37%) | 201 (42%) | <0·001 | |
| COPD | 90 (5%) | 32 (7%) | 0·11 | |
| Diabetes | 395 (22%) | 114 (24%) | 0·23 | |
| Chronic liver disease | 50 (3%) | 12 (3%) | 0·82 | |
| Chronic kidney disease | 215 (12%) | 40 (8%) | 0·043 | |
| Malignant tumour | 92 (5%) | 24 (5%) | 0·96 | |
| HIV | 27 (1%) | 9 (2%) | 0·50 | |
| COVID-19 disease severity | .. | .. | 0·15 | |
| Critical | 425 (23%) | 125 (26%) | .. | |
| Non-critical | 1409 (77%) | 350 (74%) | .. | |
| Outcomes | .. | .. | .. | |
| Inpatient admission* | 1082 (59%) | 412 (87%) | <0·0001 | |
| ICU admission | 360 (20%) | 92 (19%) | 0·91 | |
| Mechanical ventilation | 243 (13%) | 70 (15%) | 0·40 | |
| Death | 138 (8%) | 51 (11%) | 0·022 | |
| Discharge | 1690 (92%) | 412 (87%) | <0·0001 | |
| Progression from chest x-ray to critical event | .. | .. | 0·11 | |
| Median time to progression, days (IQR) | 0·61 (2·44) | 0·76 (2·91) | .. | |
| Day 1 | 221 (12%) | 62 (13%) | .. | |
| Day 2 | 51 (3%) | 13 (3%) | .. | |
| Day 3 | 28 (2%) | 11 (2%) | .. | |
| After day 3 | 87 (5%) | 33 (7%) | .. | |
| Censored† | 52 (3%) | 6 (1%) | .. | |
Data are n (%) unless otherwise stated. COPD=chronic obstructive pulmonary disease. ICU=intensive care unit.
Includes ICU admission.
Includes patients whose chest x-ray and clinical data were taken during or after a critical event.
Of the combined total of 2309 patients with a chest x-ray in anteroposterior view, 550 patients (24%) had a critical outcome. The median age of patients with critical outcomes was higher than that of patients with non-critical outcomes (67 years vs 51 years; p<0·0001). The median time from chest x-ray to critical event was 0·63 days (IQR 2·61). Additional information on the patient cohort is provided in the appendix (pp 11–16).
The image-based severity prediction model had an ROC-AUC of 0·803 (95% CI 0·773–0·817) and an F score of 0·792 (0·776–0·807) on the internal test set and an ROC-AUC of 0·753 (0·746–0·772) and an F score of 0·688 (0·676–707) on the external test set. The clinical-data-based severity prediction had an ROC-AUC of 0·821 (0·796–0·828) and an F score of 0·799 (0·785–0·815) on the internal test set and an ROC-AUC of 0·731 (0·712–0·738) and an F score of 0·721 (0·708–0·737) on the external test set. When the image-based severity prediction was combined with clinical data, the ROC-AUC improved to 0·846 (0·815–0·852) and the F score improved to 0·830 (0·813–0·847) on the internal test set, and the ROC-AUC improved to 0·792 (0·780–0·803) and the F score improved to 0·792 (0·775–0·802) on the external test set (table 2 ). The combined severity prediction model had a significant improvement (p<0·0001) compared with the image-based prediction and clinical-data-based prediction. Detailed evaluation of the severity prediction model performance is provided in the appendix (pp 17–20).
Table 2.
Performance of severity prediction models
| ROC-AUC (95% CI) | F score (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | p value | |
|---|---|---|---|---|---|
| Internal test set | |||||
| Image-based model | 0·803 (0·773–0·817) | 0·792 (0·776–0·807) | 0·671 (0·660–0·696) | 0·819 (0·815–0·824) | <0·0001 |
| Clinical-data-based model | 0·821 (0·796–0·828) | 0·799 (0·785–0·815) | 0·683 (0·669–0·709) | 0·827 (0·823–0·831) | <0·0001 |
| Image and clinical data combined model | 0·846 (0·815–0·852) | 0·830 (0·813–0·847) | 0·738 (0·727–0·761) | 0·853 (0·850–0·856) | ref |
| Severity-score-based model | 0·723 (0·710–0·762) | 0·752 (0·719–0·781) | 0·611 (0·601–0·633) | 0·777 (0·769–0·790) | <0·0001 |
| Severity score and clinical data combined model | 0·837 (0·820–0·849) | 0·806 (0·790–0·817) | 0·724 (0·712–0·739) | 0·820 (0·810–0·830) | 0·067 |
| External test set | |||||
| Image-based model | 0·753 (0·746–0·772) | 0·688 (0·676–0·707) | 0·662 (0·639–0·676) | 0·696 (0·691–0·706) | <0·0001 |
| Clinical-data-based model | 0·731 (0·712–0·738) | 0·721 (0·708–0·737) | 0·632 (0·609–0·641) | 0·688 (0·680–0·695) | <0·0001 |
| Image and clinical data combined model | 0·792 (0·780–0·803) | 0·792 (0·775–0·802) | 0·728 (0·711–0·739) | 0·701 (0·695–0·709) | ref |
| Severity-score-based model | 0·655 (0·617–0·685) | 0·658 (0·638–0·667) | 0·621 (0·609–0·632) | 0·643 (0·638–0·660) | <0·0001 |
| Severity score and clinical data combined model | 0·736 (0·717–0·754) | 0·690 (0·674–0·703) | 0·625 (0·615–0·639) | 0·687 (0·679–0·702) | <0·0001 |
A larger ROC-AUC represents better severity prediction performance. The p value from binomial test measures the difference in performance between the image and clinical data combined model and other prediction models; a smaller p value represents greater likelihood of a difference between the combined model and other models. ROC-AUC=area under the receiver operating characteristic curve.
The image-based progression prediction model had a C-index of 0·737 (95% CI 0·713–0·773) and an F score of 0·790 (0·776–0·808) on the internal test set and a C-index of 0·721 (0·700–0·727) and an F score of 0·795 (0·779–0·813) on the external test set. The clinical-data-based progression prediction model had a C-index of 0·769 (0·755–0·786) and an F score of 0·811 (0·803–0·836) on the internal test set and a C-index of 0·707 (0·695–0·729) and an F score of 0·769 (0·756–0·780) on the external test set. When the deep-learning features extracted from chest x-rays were combined with clinical data, the progression prediction model performance improved to a C-index of 0·805 (0·800–0·820) and an F score of 0·843 (0·836–0·863) on the internal test set and a C-index of 0·752 (0·739–0·764) and an F score of 0·805 (0·791–0·825) on the external test set. The combined progression prediction model had a significant improvement (p<0·0001) compared with the image-based prediction and clinical-data-based prediction (table 3 ).
Table 3.
Performance of progression prediction models
| C-index (95% CI) | F score (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | Binomial p value* | Log-rank p value† | χ2 (95% CI)† | |
|---|---|---|---|---|---|---|---|
| Internal test set | |||||||
| Image-based model | 0·737 (0·713–0·773) | 0·790 (0·776–0·808) | 0·696 (0·664–0·718) | 0·775 (0·769–0·782) | <0·0001 | <0·0001 | 17·33 (13·73–22·02) |
| Clinical-data-based model | 0·769 (0·755–0·786) | 0·811 (0·803–0·836) | 0·656 (0·631–0·674) | 0·811 (0·801–0·817) | <0·0001 | <0·0001 | 31·77 (24·58–36·56) |
| Image and clinical data combined model | 0·805 (0·800–0·820) | 0·843 (0·836–0·863) | 0·720 (0·700–0·749) | 0·845 (0·840–0·850) | ref | <0·0001 | 26·51 (21·65–33·56) |
| Severity-score–based model | 0·696 (0·676–0·711) | 0·761 (0·752–0·775) | 0·656 (0·635–0·669) | 0·743 (0·736–0·752) | <0·0001 | <0·0001 | 18·15 (9·45–23·70) |
| Severity score and clinical data combined model | 0·781 (0·755–0·787) | 0·805 (0·798–0·832) | 0·678 (0·666–0·700) | 0·798 (0·793–0·807) | 0·0002 | <0·0001 | 42·23 (33·63–49·59) |
| External test set | |||||||
| Image-based model | 0·721 (0·700–0·727) | 0·795 (0·779–0·813) | 0·633 (0·606–0·662) | 0·791 (0·788–0·796) | <0·0001 | <0·0001 | 39·17 (28·62–48·58) |
| Clinical-data-based model | 0·707 (0·695–0·729) | 0·769 (0·756–0·780) | 0·602 (0·583–0·621) | 0·753 (0·751–0·762) | <0·0001 | <0·0001 | 31·72 (26·41–42·94) |
| Image and clinical data combined model | 0·752 (0·739–0·764) | 0·805 (0·791–0·825) | 0·667 (0·643–0·698) | 0·798 (0·791–0·803) | ref | <0·0001 | 52·04 (46·50–66·14) |
| Severity-score–based model | 0·606 (0·584–0·627) | 0·720 (0·704–0·733) | 0·528 (0·512–0·541) | 0·695 (0·686–0·701) | <0·0001 | <0·0001 | 11·65 (6·84–15·43) |
| Severity score and clinical data combined model | 0·715 (0·704–0·721) | 0·778 (0·757–0·795 | 0·667 (0·649–0·677) | 0·759 (0·756–0·765) | <0·0001 | <0·0001 | 37·62 (26·68–46·95) |
C-index for right-censored data measures the model performance by comparing the progression information (critical labels and progression days) with predicted risk scores; a larger C-index correlates with better progression prediction performance. C-index=concordance index.
Measures the difference in performance between the image and clinical data combined model and other prediction models; a smaller p value represents greater likelihood of a difference between the combined model and other models.
Shows a comparison of stratification performance of different models; a smaller p value and larger χ2 correlate with better risk stratification performance.
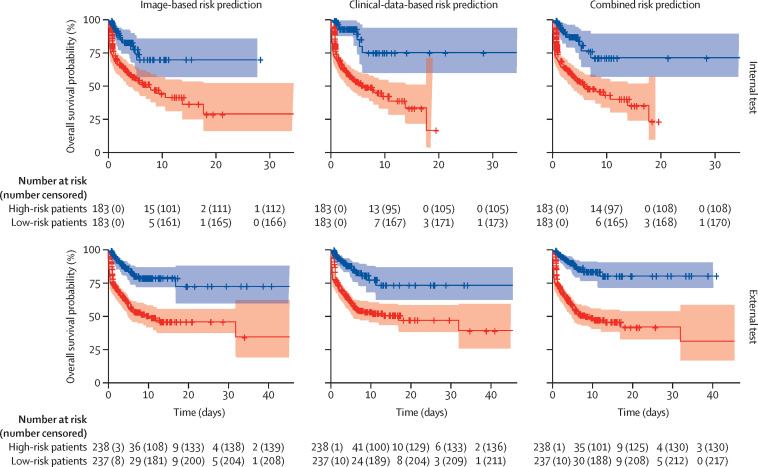
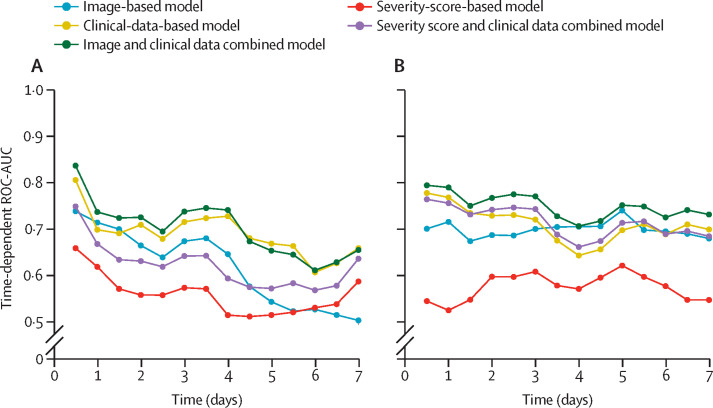
Kaplan-Meier curves for risk stratification are shown in figure 2 . As shown, the combined image-based and clinical-data-based model was discriminative in stratifying patients into high-risk and low-risk subgroups on both internal testing (p<0·0001) and external testing (p<0·0001). The time-dependent ROC-AUCs are shown in figure 3 . Detailed evaluation of the progression prediction model performance is provided in the appendix (pp 21–27).
Figure 2.
Kaplan-Meier curves for progression risk prediction
Figure 3.
Time-dependent ROC-AUCs of progression prediction
Time-dependent ROC-AUCs on internal testing (A) and external testing (B). ROC-AUC=area under the receiver operating characteristic curve.
The progression prediction model based on chest x-ray severity scores had a C-index of 0·696 (95% CI 0·676–0·711) on the internal test set and a C-index of 0·606 (0·584–0·627) on the external test set. On the subset of patients who had a severity score of 0, the model had a C-index of 0·821 (0·805–0·841) on internal testing and 0·824 (0·799–0·836) on external testing. Compared with combined chest x-ray severity score and clinical data, the combined model using deep-learning features extracted from chest x-rays and clinical data had significantly better performance on both internal testing (C-index 0·805 vs 0·781; p=0·0002) and external testing (C-index 0·752 vs 0·715; p<0·0001). The C-index values of the progression prediction models and their risk stratification performance are summarised in table 3.
Discussion
The prediction of disease severity and progression in patients with COVID-19 is important, as early intervention has been shown to reduce mortality.20, 21 Chest x-ray is a versatile imaging modality that has shown promise in aiding diagnosis and prognosis during the COVID-19 pandemic. However, the added value of chest x-rays to clinical data in disease prognostication needs further evaluation. In this study, an artificial intelligence model based on the initial chest x-ray and clinical data of patients with COVID-19 who presented to the emergency department was implemented and shown to incrementally improve the prognostic ability of clinical variables. Furthermore, the artificial intelligence model, using deep-learning features extracted from chest x-rays, performed significantly better in prognostication as compared with radiologist-derived severity scores.
Medical imaging and clinical data could have great utility in prognostication of patients with COVID-19. For example, a previous study developed an automated system to predict if patients with COVID-19 would die or require mechanical ventilation using radiomics features from CT scans, data on six clinical variables (age, sex, high blood pressure, diabetes, lymphocyte count, and C-reactive protein level), and two image indexes (disease extent and fat ratio).22 Similarly, in our study we explored the value of medical imaging in combination with clinical data, but chest x-rays and data on 16 clinical variables were utilised for a binary severity prediction (critical vs non-critical) and a time to critical event model. Knowing which patients will progress to a critical level and the timespan from a patient's first chest x-ray to a critical event will permit health-care providers to appropriately triage and manage these patients while optimising the use of resources.
This study has several differences compared with previous studies. Earlier literature in this area utilised a small cohort and focused on diagnostics or binary outcomes for severity prediction, which do not take into account the time of progression to a critical event.5, 6, 8, 10, 23, 24 By contrast, our study shows the incremental value of artificial intelligence based on chest x-rays in improving the utility of clinical variables for predicting progression to a critical event from the time the chest x-ray was done. One of the largest previous studies on the relation of clinical variables to COVID-19 severity found chest x-ray abnormality as an independent predictor of progression to severe disease in a multicentre cohort of 1590 patients, but did not discuss the added value of chest x-ray to clinical variables in making this prediction, nor did it discuss time to development of severe disease.11 By contrast with a previous artificial intelligence study, which utilised quantitative lung lesion features from chest CT scans of 442 patients with COVID-19 to determine prognosis from the time of admission,25 the model in our study extracted deep-learning features from chest x-rays and measured the time interval from chest x-ray to critical event in building the time to event model, rather than from time of admission. Another relevant chest x-ray study that integrated artificial intelligence into severity prediction only included 314 patients and found a correlation between high severity score and risk of critical outcome within 3 days, but did not take into account time to development of critical outcome or utilise clinical variables for its prediction model.9 Chest x-ray severity scores are powerful tools that have been shown to have value in predicting the risk of hospitalisation, ICU admission, or intubation in patients with COVID-19,6, 7, 8 but they have limitations such as intrarater and inter-rater variability, and inability to capture all the information contained within the image. Our study is unique and clinically relevant because it shows COVID-19 severity and specific time-to-critical-event windows can be predicted using clinical variables, and that using deep-learning features extracted from chest x-rays can incrementally increase the strength of those predictions and outperform the prediction by radiologist-derived severity scores.
This study has several limitations. First, the artificial intelligence model showed decreased performance on the external testing set relative to the internal testing set, indicating that generalisation might not be possible. This finding could have been due to several factors, including heterogeneous data and image acquisition between the different hospital systems. Although a lower performance on external testing is a common finding in deep-learning studies using multi-institutional data,26, 27 including our study, the addition of chest x-rays to clinical data improved the artificial performance on both internal and external testing. Second, several patients had a timeframe of less than 1 day for progression to a critical event from the time of their initial chest x-ray. The clinicians might have known or anticipated the clinical deterioration when they requested the chest x-ray. However, the distribution of time for these patients is widespread, as shown in the appendix (p 16), with the largest number of patients having a timeframe of 4 h, suggesting that the effect of this potential bias on the time-to-event analysis is likely to be minimal. Third, the disease severity and progression of patients were determined by critical events that occurred during the hospitalisation via the emergency department. This patient inclusion and exclusion criteria could have contributed to selection bias because it does not include any patients who presented to the outpatient services, or patients who had a critical event during another hospitalisation after their discharge from the emergency department. Fourth, although this study had a large number of patients from two separate hospital systems, the sample size is still smaller than for artificial intelligence models trained on ImageNet.28 Further improvements in model performance can be achieved with a larger cohort size, along with further validation and refinement by other researchers using our publicly available model files and codes. In addition to addressing these limitations, future studies could benefit from an end-to-end deep-learning model that simultaneously addresses severity and progression predictions, and from an incorporation of serial chest x-rays taken at different timepoints during a hospitalisation, to further improve the model performance.
In conclusion, artificial intelligence based on chest x-rays augmented the performance of clinical data in predicting risk of progression to critical illness in patients with COVID-19 who presented to hospital through the emergency department, and outperformed radiologist-derived severity scores. Using this approach, it might be possible to take advantage of this readily available imaging data to identify high-risk patients early and improve outcomes.
This online publication has been corrected. The corrected version first appeared at thelancet.com/digital-health on March 31, 2021
Data sharing
All data collected for the study are not publicly available for download in respect of patient confidentiality and consent. However, the corresponding author can be contacted for academic inquiries. As stated in the Methods, the artificial intelligence code and model are available for public use at https://github.com/ZhichengJiao/COVID-19-prognostic-model.
Declaration of interests
HXB reports grants from Brown University, Amazon Web Service, Radiological Society of North America, and National Cancer Institute of the National Institute of Health, during the conduct of the study. XF currently works in Carina Medical, a for-profit organisation that develops clinical products, outside of the submitted work. KC reports grants from National Institute of Biomedical Imaging and Bioengineering and National Cancer Institute of the National Institute of Health, during the conduct of the study. YF reports grants from National Institute of Health, during the conduct of the study. All other authors declare no competing interests.
Acknowledgments
Acknowledgments
This study was funded by the Brown University COVID-19 Research Seed Award, Amazon Web Services Diagnostic Development Initiative, Radiological Society of North America Research Scholar grant, and the National Cancer Institute of the National Institutes of Health R03 grant (R03CA249554 to HXB); National Institutes of Health grants (CA223358, DK117297, MH120811, and EB022573 to YF); and a training grant from the National Institute of Biomedical Imaging and Bioengineering (5T32EB1680) and the National Cancer Institute of the National Institutes of Health grant (F30CA239407 to KC). TL was partially supported by Institutional Development Award (U54GM115677) from the National Institute of General Medical Sciences of the National Institutes of Health.
Contributors
HXB, W-HL, and YF designed the study. JWC coordinated research efforts including data gathering and writing of the manuscript. ZJ led the training of artificial intelligence models and performing of the analyses. YF, TTH, SL, MKA, MF, and PJLZ provided the data. ZJ, JWC, and HXB accessed and verified the data. JWC, KH, TMLT, BH, FE, SAC, and RW performed data collection and organisation. ZJ and KC trained the algorithm. XF provided advice on lung segmentation. ZJ, JWC, and JWu performed the analyses. W-HL and DW organised the efforts for severity scoring. TTY and ATD implemented the algorithm. TL, JWa, and LY gave advice on algorithm development or data analyses. RW, TL, and BH generated the figures. All authors read and reviewed the final manuscript. All authors had full access to all the data in the study and had final responsibility for the decision to submit for publication.
Supplementary Material
References
- 1.WHO Weekly epidemiological update—2 March 2021. March 2, 2021. https://www.who.int/publications/m/item/weekly-epidemiological-update-2-march-2021
- 2.Bai HX, Hsieh B, Xiong Z. Performance of radiologists in differentiating COVID-19 from non-COVID-19 viral pneumonia at chest CT. Radiology. 2020;296:e46–e54. doi: 10.1148/radiol.2020200823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bai HX, Wang R, Xiong Z. Artificial intelligence augmentation of radiologist performance in distinguishing COVID-19 from pneumonia of other origin at chest CT. Radiology. 2020;296:e156–e165. doi: 10.1148/radiol.2020201491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang W, Sirajuddin A, Zhang X. The role of imaging in 2019 novel coronavirus pneumonia (COVID-19) Eur Radiol. 2020;30:4874–4882. doi: 10.1007/s00330-020-06827-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chau T-N, Lee P-O, Choi K-W. Value of initial chest radiographs for predicting clinical outcomes in patients with severe acute respiratory syndrome. Am J Med. 2004;117:249–254. doi: 10.1016/j.amjmed.2004.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toussie D, Voutsinas N, Finkelstein M. Clinical and chest radiography features determine patient outcomes in young and middle-aged adults with COVID-19. Radiology. 2020;297:e197–e206. doi: 10.1148/radiol.2020201754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cozzi D, Albanesi M, Cavigli E. Chest x-ray in new coronavirus disease 2019 (COVID-19) infection: findings and correlation with clinical outcome. Radiol Med (Torino) 2020;125:730–737. doi: 10.1007/s11547-020-01232-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim HW, Capaccione KM, Li G. The role of initial chest x-ray in triaging patients with suspected COVID-19 during the pandemic. Emerg Radiol. 2020;27:617–621. doi: 10.1007/s10140-020-01808-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li MD, Arun NT, Gidwani M. Automated assessment of COVID-19 pulmonary disease severity on chest radiographs using convolutional Siamese neural networks. medRxiv. 2020 doi: 10.1101/2020.05.20.20108159. published online May 26. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khuzani AZ, Heidari M, Shariati SA. COVID-classifier: an automated machine learning model to assist in the diagnosis of COVID-19 infection in chest x-ray images. medRxiv. 2020 doi: 10.1101/2020.05.09.20096560. published online May 18. (preprint). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang W, Liang H, Ou L. Development and validation of a clinical risk score to predict the occurrence of critical illness in hospitalized patients with COVID-19. JAMA Intern Med. 2020;180:1–9. doi: 10.1001/jamainternmed.2020.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kermali M, Khalsa RK, Pillai K, Ismail Z, Harky A. The role of biomarkers in diagnosis of COVID-19—a systematic review. Life Sci. 2020;254 doi: 10.1016/j.lfs.2020.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ronneberger O, Fischer P, Brox T. U-Net: convolutional networks for biomedical image segmentation. In: Navab N, Hornegger J, Wells W, Frangi A, editors. Medical image computing and computer-assisted intervention—MICCAI 2015. Springer; Cham: 2015. pp. 243–245. [Google Scholar]
- 14.Tan M, Le QV. EfficientNet: rethinking model scaling for convolutional neural networks. Sept 11, 2020. http://arxiv.org/abs/1905.11946
- 15.Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. Am Stat. 1998;52:119. [Google Scholar]
- 16.Harrell FE, Jr, Lee KL, Mark DB. Multivariable prognostic models: issues in developing models, evaluating assumptions and adequacy, and measuring and reducing errors. Stat Med. 1996;15:361–387. doi: 10.1002/(SICI)1097-0258(19960229)15:4<361::AID-SIM168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 17.Heagerty PJ, Zheng Y. Survival model predictive accuracy and ROC curves. Biometrics. 2005;61:92–105. doi: 10.1111/j.0006-341X.2005.030814.x. [DOI] [PubMed] [Google Scholar]
- 18.Cai T, Pepe MS, Zheng Y, Lumley T, Jenny NS. The sensitivity and specificity of markers for event times. Biostatistics. 2006;7:182–197. doi: 10.1093/biostatistics/kxi047. [DOI] [PubMed] [Google Scholar]
- 19.Saito T, Rehmsmeier M. The precision-recall plot is more informative than the ROC plot when evaluating binary classifiers on imbalanced datasets. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun Q, Qiu H, Huang M, Yang Y. Lower mortality of COVID-19 by early recognition and intervention: experience from Jiangsu Province. Ann Intensive Care. 2020;10:33. doi: 10.1186/s13613-020-00650-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyal DK, Mansab F, Iqbal A, Bhatti S. Early intervention likely improves mortality in COVID-19 infection. Clin Med (Lond) 2020;20:248–250. doi: 10.7861/clinmed.2020-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chassagnon G, Vakalopoulou M, Battistella E. AI-driven quantification, staging and outcome prediction of COVID-19 pneumonia. Med Image Anal. 2021;67 doi: 10.1016/j.media.2020.101860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vancheri SG, Savietto G, Ballati F. Radiographic findings in 240 patients with COVID-19 pneumonia: time-dependence after the onset of symptoms. Eur Radiol. 2020;30:6161–6169. doi: 10.1007/s00330-020-06967-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Holshue ML, DeBolt C, Lindquist S. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang K, Liu X, Shen J. Clinically applicable AI system for accurate diagnosis, quantitative measurements, and prognosis of COVID-19 pneumonia using computed tomography. Cell. 2020;181:1423. doi: 10.1016/j.cell.2020.04.045. 33.e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zech JR, Badgeley MA, Liu M, Costa AB, Titano JJ, Oermann EK. Variable generalization performance of a deep learning model to detect pneumonia in chest radiographs: a cross-sectional study. PLoS Med. 2018;15 doi: 10.1371/journal.pmed.1002683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.AlBadawy EA, Saha A, Mazurowski MA. Deep learning for segmentation of brain tumors: impact of cross-institutional training and testing. Med Phys. 2018;45:1150–1158. doi: 10.1002/mp.12752. [DOI] [PubMed] [Google Scholar]
- 28.Gozes O, Frid-Adar M, Greenspan H. Rapid AI development cycle for the coronavirus (COVID-19) pandemic: initial results for automated detection & patient monitoring using deep learning CT image analysis. March 24, 2020. http://arxiv.org/abs/2003.05037
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data collected for the study are not publicly available for download in respect of patient confidentiality and consent. However, the corresponding author can be contacted for academic inquiries. As stated in the Methods, the artificial intelligence code and model are available for public use at https://github.com/ZhichengJiao/COVID-19-prognostic-model.