Abstract
Efficient genome editing methods are essential for biotechnology and fundamental research. Homologous recombination (HR) is the most versatile method of genome editing, but techniques that rely on host RecA-mediated pathways are inefficient and laborious. Phage-encoded ssDNA annealing proteins (SSAPs) improve HR 1000-fold above endogenous levels; however, they are not broadly functional. Using Escherichia coli, Lactococcus lactis, Mycobacterium smegmatis, Lactobacillus rhamnosus, and Caulobacter crescentus we investigated the limited portability of SSAPs. We find that these proteins specifically recognize the C-terminal tail of the host’s single-stranded DNA-binding protein (SSB), and are portable between species if compatibility with this host domain is maintained. Furthermore, we find that co-expressing SSAPs with a paired SSB can significantly improve activity, in some species enabling SSAP functionality even without host-compatibility. Finally, we find that high-efficiency HR far surpasses the mutational capacity of commonly used random mutagenesis methods, generating exceptional phenotypes inaccessible through sequential nucleotide conversions.
Keywords: Genome engineering, Synthetic biology, Molecular evolution, Single-stranded DNA binding protein, Single-stranded DNA annealing protein
Genome modification is crucial to the study of microbes, and is used to investigate the effects of genetic variation in genes and regulatory sequences1,2, to explore the properties of human commensals and pathogens3,4,5,6,7,8, and to engineer strains for biotechnology9,10,11,12. Homologous recombination (HR) is the most versatile method of genome editing. These methods rely on flanking homology arms to incorporate DNA into a targeted locus, and in principle can introduce any genetic modification at any site on the genome. However, the endogenous pathway of RecA-mediated HR is inefficient and requires long homology arms, which makes the introduction of genetic changes unreliable and which limits the generation of genomic libraries13. Cas9, in contrast to its effects in mammalian cells, does not greatly improve the efficiency of HR in most prokaryotes14. Instead, since dsDNA breaks are often lethal, Cas9 is used as a tool to cut and select against non-edited cells14,15,16. Methods for high-throughput genome engineering in prokaryotes currently rely on overexpression of phage encoded single-stranded DNA annealing proteins (SSAPs), which integrate single-stranded oligonucleotide donors without nicking or breaking the genome (a method referred to as recombineering)17,13,18,19,20,21. Unfortunately, these proteins are not broadly portable, and have only been established in a small number of species.
The Red operon from Enterobacteria phage λ is the source of one of the most widely used SSAPs, λ-Red β. λ-Red β, along with the rest of the λ-Red operon, enables a suite of technologies that rely on efficient HR including gene knockouts using PCR products13, multiplex genome editing9, and state-of-the-art methods for synthetic genome assembly22,12. It also provides one of the most comprehensive methods for diversifying genetic loci1,23. Rather than expressing mutant genes from plasmids24, which introduce copy number effects, or performing random mutagenesis25, recombineering can be used to generate libraries of precisely defined genetic variants in their endogenous context. λ-Red β, however, only operates in a small set of phylogenetically related species (E. coli, Salmonella enterica and Citrobacter freundii, among others)26. In distantly related species such as Lactococcus lactis, homologs of RecT from the E. coli Rac prophage27, have been used for recombineering28, but these proteins also only function across a small host-range. The key features influencing the host-tropism exhibited by phage SSAPs is not well understood. In this work, we aimed to characterize limits to the portability of λ-Red β and homologs within its superfamily (RecT/Redβ proteins), as well as broaden the host-range of recombineering methods.
The most reliable strategy for establishing recombineering in species distantly related to E. coli has been to screen SSAPs from phages or prophages that infect the host of interest28,29,30. We therefore hypothesized that SSAPs may require a specific interaction with one or more bacterial proteins, limiting their functionality to species where this host-protein interaction is maintained. A key mechanistic step during recombineering is annealing of ssDNA to the genome at the lagging strand of the replication fork21,31,32. We thus focused our efforts on host proteins which reside or consistently interact with DNA on the lagging strand. Recently, λ-Red β was found to interact with a C-terminal peptide of E. coli SSB (single-stranded DNA-binding protein)33, which binds to lagging strand ssDNA. SSBs play important roles in phage recombination pathways34, and have been shown to influence recombineering efficiency35.
Here, we provide evidence that a specific interaction between SSAPs and the host bacterial SSB limits their portability. We find the majority of this interaction relies on recognition of SSB’s 7 C-terminal amino acids. We also find that, in some species, supplying an exogenous bacterial SSB significantly enhances SSAP activity and can make up for a lack of host-compatibility, allowing SSAPs to function in previously recalcitrant species. We then demonstrate the potential for high-efficiency HR in new species by optimizing recombineering in Lactococcus lactis. Specifically, we find that oligonucleotide recombination can be used to navigate genotypic landscapes that contain extensive epistatic effects and are inaccessible through error-prone diversification methods which primarily generate single-nucleotide conversions.
Results:
SSBs are key mediators of SSAP functionality
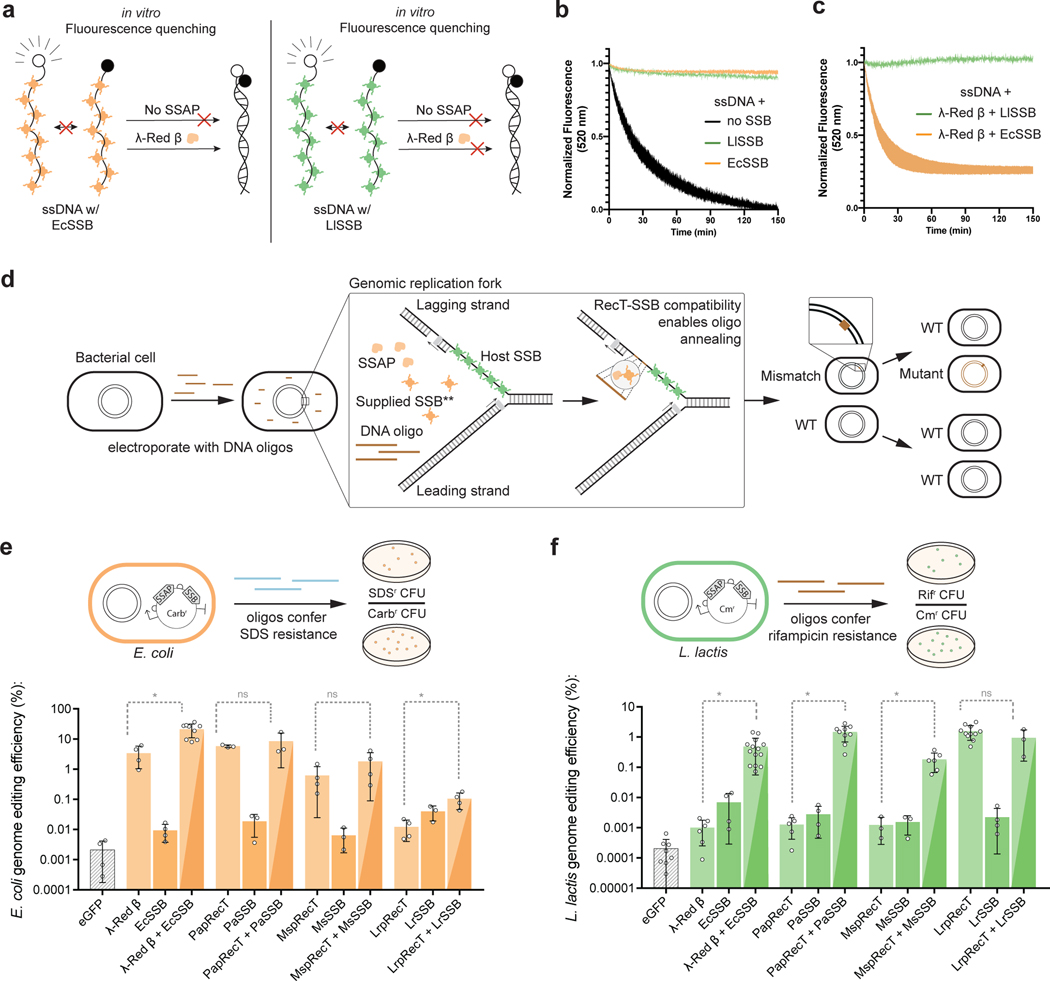
To understand the host-tropism displayed by SSAPs, we started by developing a simplified in-vitro model of oligonucleotide annealing that includes bacterial SSBs, the key host protein that coats ssDNA at the replication fork36. We first tested whether two 90bp oligos could anneal if they were pre-coated with SSB. We purified SSBs from E. coli (a gram-negative species), where most recombineering work has been performed, and L. lactis (a gram-positive species), a lactic acid bacterium distantly related to E. coli. Using fluorescence quenching to measure annealing, we found that while the free oligos annealed together slowly (Fig 1a,b), both E. coli SSB (EcSSB) and L. lactis SSB (LlSSB) completely inhibited oligonucleotide annealing. We then tested capacity of a SSAP to overcome this SSB-mediated inhibition of annealing. We thus purified λ-Red β, which is not broadly portable, but mediates efficient oligonucleotide annealing in E. coli. We found that adding λ-Red β overcame the inhibitory effect of EcSSB but not LlSSB, rapidly annealing the EcSSB-coated two oligos together (Fig 1a,c). These preliminary results indicate that while bacterial SSBs inhibit oligonucleotide annealing in vitro, SSAPs overcome the inhibitory effect in an SSB-specific manner.
Fig. 1: SSBs are key mediators of SSAP functionality.
(a) Model of ssDNA annealing inhibition by EcSSB or LlSSB, and ability of λ-Red β to overcome annealing inhibition by EcSSB. (b) In-vitro ssDNA annealing without SSB, pre-coated with EcSSB, or pre-coated with LlSSB. Shaded area represents the SEM of at least 2 replicates (c) In-vitro ssDNA annealing in the presence of λ-Red β when pre-coated with EcSSB or LlSSB. Shaded area represents the SEM of at least 2 replicates (d) Model for SSAP-mediated editing at the replication fork. An interaction between SSAP and the host SSB enables oligo annealing to the lagging strand. **Co-expressing an exogenous SSB that is compatible with the SSAP can in some species enable functionality even without host compatibility. (e),(f), The efficiency of editing in E. coli (e) and L. lactis (f) is compared using either SSAPs, SSBs, or “cognate pairs” (as described in the text). Error bars indicate SD from the mean of at least 3 biologically independent experiments. *: P value < .05; Welch’s two tailed t-test of Log-transformed data.
To validate this result in vivo, we developed an assay to measure the portability of SSAPs. We selected four variants known to enable high efficiency genome editing (λ-Red β and PapRecT in E. coli, LrpRecT in L. lactis, and MspRecT in M. smegmatis), and tested codon optimized versions of all four in E. coli and L. lactis. We use this RecT nomenclature as a way to specify the phylogenetic origin of each protein (PapRecT is a Pseudomonas aeruginosa prophage RecT, LrpRecT is a Lactobacillus reuteri prophage RecT, and MspRecT is a Mycobacterium smegmatis prophage RecT). We measured genome editing efficiency in E. coli and L. lactis by introducing oligos encoding known antibiotic resistance mutations (at TolC to generate SDS resistance in E. coli, and at RpoB to generate Rifampacin resistance in L. lactis), comparing the antibiotic resistant cell counts to the total number of viable cells in the population (Fig 1e, f). In E. coli, λ-Red β and PapRecT functioned well, and improved oligo incorporation 1600-fold and 2700-fold respectively, while MspRecT (290-fold improvement) and LrpRecT (5.6-fold improvement) were less effective (Fig 1e). In L. lactis, LrpRecT was the only functional homolog, and improved oligo incorporation 7,700-fold, while the three other SSAPs were nearly non-functional, improving oligo incorporation less than 7-fold (Fig 1f). No SSAP functioned well both in E. coli and L. lactis. This result matches those of previous studies, which have found that SSAPs are often not portable between distantly related bacterial species37.
If interaction with the bacterial SSB is required for SSAP functionality, one solution to establishing recombineering in a new species would be to replace the host SSB with one compatible with the chosen RecT. However, SSB proteins are essential, and mutations to SSB can result in severe growth defects36. We therefore evaluated if temporary overexpression of an exogenous SSB could supply the necessary requirements for recombineering and improve the activity of non-host compatible SSAPs. We synthesized bacterial SSB genes corresponding with each SSAP (See Methods) and tested the activity of all four cognate SSAP-SSB pairs in L. lactis and E. coli. Co-expression of a cognate bacterial SSB improved the genome editing efficiencies of all SSAPs with low host-compatibility (Fig 1e, f, Extended Data Fig. 1). The best performing pairs, λ-Red β + EcSSB and PapRecT + PaSSB demonstrated 483-fold and 1,168-fold improved editing efficiencies over the SSAPs alone in L. lactis, and still maintained high activity in E. coli (Fig 1e, f). In E. coli, λ-Red β + EcSSB also demonstrated 6.25-fold higher genome editing efficiency than λ-Red β alone (21.2% vs. 3.4%), and reduced the construct’s toxicity (Extended Data Fig. 2). These results, especially in L. lactis, demonstrate that expression of an exogenous cognate bacterial SSB can dramatically improve recombineering efficiency and overcome the host incompatibility of SSAPs.
The C-terminal tail of SSB affects SSAP compatibility
We next investigated domains on SSB that might mediate the SSAP interaction. An SSB domain-specific model for understanding SSAP portability would be more informative than previous models for portability, which relied on phylogenetic relationships between the host species37. SSAP have been shown to function in species with SSBs with relatively divergent sequences. For example, λ-Red β works well in E. coli, Salmonella enterica, and Citrobacter freundii which have SSBs with 88% identity, and PapRecT works in E. coli and Pseudomonas aeruginosa, which have SSBs of only 59% identity. To investigate the specific residues involved, we used our genome editing assay in L. lactis and evaluated the effect of co-expressing SSAPs with non-cognate or mutated SSBs.
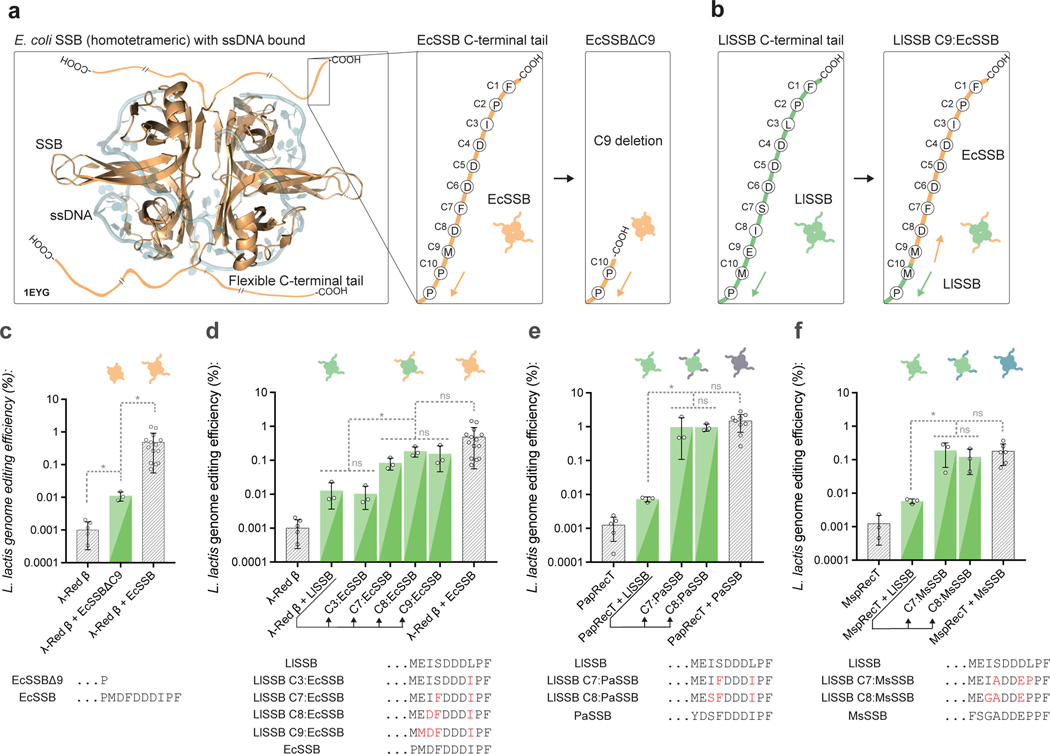
The C-terminal tail of E. coli SSB is known to be the binding domain for host proteins involved in DNA replication and repair36, and a 9-amino-acid EcSSB C-terminal tail peptide was shown to bind to λ-Red β in vitro33. We therefore evaluated the importance of the SSB C-terminal tail by co-expressing λ-Red β in L. lactis, along with a version of EcSSB that had a 9-amino-acid C-terminal deletion (EcSSBΔ9) (Fig 2a). In L. lactis, the genome editing efficiency of λ-Red β with EcSSBΔ9 was 44-fold lower than λ-Red β with EcSSB, indicating a key role for the C-terminal tail domain in the SSB-mediated efficiency improvement (Fig 2c). Next, we co-expressed λ-Red β with the L. lactis SSB (LlSSB), which we expect to have little-to-no compatibility. Co-expression of λ-Red β with LlSSB performed similarly to λ-Red β with EcSSBΔ9, and improved genome editing efficiency 38.5-fold less than λ-Red β with EcSSB (Fig 2d). We then co-expressed λ-Red β with chimeric versions of the LlSSB, where up to 9 amino acids of the LlSSB C-terminal tail were replaced with their corresponding residues from EcSSB (Fig 2b). Swapping the last 7 C-terminal residues (LlSSB C7:EcSSB) improved editing rates to within 5.9-fold of λ-Red β with EcSSB (Fig 2d), and swapping the last 8 C-terminal residues (LlSSB C8:EcSSB) improved editing rates within 2.6-fold of λ-Red β with EcSSB (Fig 2d). These results support a model where λ-Red β specifically recognizes at minimum the 7 C-terminal acids of E. coli SSB, but not that of L. lactis SSB.
Fig. 2: The C-terminal tail of SSB affects SSAP compatibility.
(a), A crystal structure of homotetrameric E. coli SSB bound to ssDNA (PDB-ID 1EYG)46. The amino acid sequence of the flexible C-terminal tail is diagramed in the right panel, along with the design of a 9AA C-terminal truncation to SSB. (b), The L. lactis SSB C-terminal tail is diagramed, along with an example of an SSB C-terminal tail replacement. In this case, the 9 C-terminal amino acids of the L. lactis SSB are replaced with the corresponding residues from E. coli SSB. The notation “LlSSB C9:EcSSB” is used as shorthand. (c), Editing efficiency in L. lactis of λ-Red β co-expressed with a 9AA C-terminally truncated EcSSB mutant. (d), Editing efficiency in L. lactis of λ-Red β co-expressed with LlSSB, or mutants of LlSSB with C3, C7, C8, or C9 terminal residues replaced with the corresponding residues from EcSSB. (e, f) Editing efficiency in L. lactis of PapRecT (e) or MspRecT (f) co-expressed with LlSSB, or mutants of LlSSB with the C7 or C8 terminal residues replaced with the corresponding residues from the cognate SSB. All experiments have at least 3 biologically independent replicates, error bars indicate SD from the mean. (c, d, e, f) *: P value < .05; ordinary one-way ANOVA of Log-transformed data, Holm-Sidak multiple comparisons test.
To evaluate if the SSB C-terminal 7 amino acids also affected the compatibility of the other two non-host compatible SSAPs, we performed similar SSB-chimera experiments with PapRecT and MspRecT. In L. lactis, the genome editing efficiency of PapRecT co-expressed with the L. lactis SSB was 135-fold less than when using the cognate pair (Fig 2e). However, this defect was completely recovered when PapRecT was co-expressed with L. lactis SSB chimeras where either the last 7 or 8 C-terminal residues were replaced (LlSSB C7:PaSSB, LlSSB C8:PaSSB) (Fig 2e). For MspRecT, the genome editing efficiency with LlSSB was 33-fold lower than when using the cognate pair (Fig 2f). Again, the defect was completely recovered when MspRecT was co-expressed with L. lactis SSB chimeras where either the last 7 or 8 C-terminal residues were replaced (LlSSB C7:MsSSB, LlSSB C8:MsSSB) (Fig 2f). Since the chimeric LlSSBs greatly improved the functionality of non-host compatible SSAPs, while the wild-type LlSSB did not, the SSAP-SSB interaction seems to be both specific and relatively modular, with the 7 C-terminal amino acids acting as the critical interaction domain.
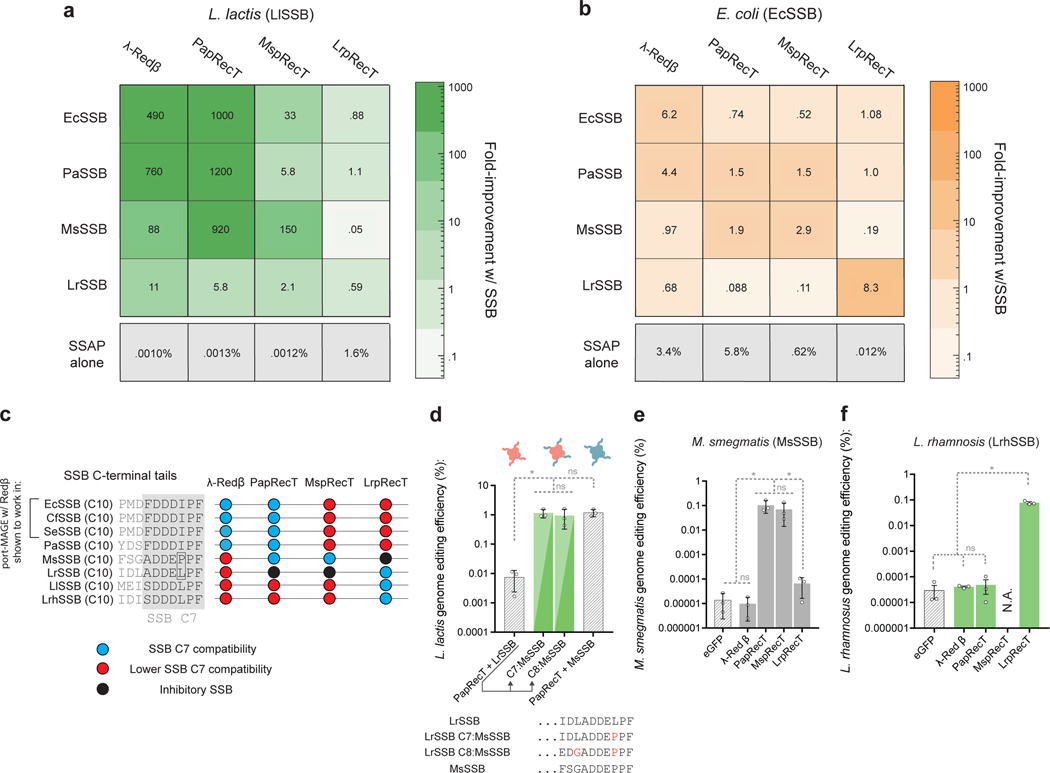
These results provide a molecular basis for the portability of SSAPs between species which have host SSBs with a conserved C-terminal tail. Specifically, although the SSBs have only 59% identity, the P. aeruginosa and E. Coli SSBs have a perfectly conserved 7 amino acid C-terminal tail domain (Fig 3c), supporting the functionality of PapRecT in E. coli. Additionally, E. coli, Salmonella enterica and Citrobacter freundii26 SSBs all have a perfectly conserved 7 amino acid C-terminal tails, supporting the portability of λ-Red β between these species (Fig 3c).
Fig. 3. SSAP-SSB interactions match SSAP portability across bacterial species.
(a, b) Heat map showing the fold improvement in editing efficiency due to SSB co-expression in (a) L. lactis or (b) E. coli of SSAP-SSB pairs as compared to the SSAP alone. (c), C-terminal sequences of SSBs as well as SSAP compatibility given by (a, b). (d), Editing efficiency in L. lactis of PapRecT co-expressed with LrSSB, MsSSB, or mutants of LrSSB which had the C7 or C8 terminal residues replaced with the corresponding residues from the MsSSB. (e), Editing efficiency in M. smegmatis of λ-Red β, PapRecT, MspRecT, and LrpRecT. (f), Editing efficiency in L. rhamnosus of λ-Red β, PapRecT, MspRecT, and LrpRecT. All experiments have at least 3 biologically independent replicates, error bars indicate SD from the mean. (d, e, f) *: P value < .05; ordinary one-way ANOVA of Log-transformed data, Holm-Sidak multiple comparisons test.
SSAP-SSB interactions match SSAP portability across bacterial species
Some SSAPs are known to be portable between species that have distinct SSB C-terminal tails. To better characterize the network of SSAP-SSB compatibility among the proteins analyzed here, we tested all possible pairs using the four SSAPs and SSBs in both E. coli and L. lactis (Fig 3a, b, S3, Extended Data Fig. 2). We found that EcSSB and PaSSB were relatively interchangeable, as might be expected since they share the same 7 amino acid C-terminal tail (Fig 3c). Interestingly, PapRecT displayed the characteristics of a more portable SSAP protein, and showed compatibility with EcSSB/PaSSB and MsSSB, even though their 7AA C-terminal tail sequences are distinct (Fig 3a, c). Importantly, co-expressing PapRecT with LrSSB did not provide a substantial improvement in genome editing efficiency in L. lactis, even though the 7 C-terminal tail amino acids of LrSSB differ only by a single residue from MsSSB (Fig 3a, c).
To test if PapRecT specifically interacts with the C-terminal tail of MsSSB, we co-expressed PapRecT with a chimeric version of LrSSB, with either the C7 or C8 amino acids matching that of MsSSB (Fig 3d). These constructs demonstrated the same editing efficiency as PapRecT + MsSSB, showing that a single amino acid change was sufficient to enable compatibility between the proteins (Fig 3d). The compatibility of PapRecT with the distinct EcSSB/PaSSB and MsSSB tails but not the LrSSB tail affirms that while the SSB C-terminal tail has a critical role in the SSAP-SSB interaction, there can be flexibility in the specific motif recognized.
We next evaluated if the interaction between PapRecT and the co-expressed MsSSB in L. lactis indicated that PapRecT would function in Mycobacterium smegmatis, where MsSSB is natively expressed. We tested all four SSAPs in this species, and used oligos targeting the M. smegmatis RpsL to produce streptomycin resistance. Indeed, we found that PapRecT enabled high efficiency editing in this host, where it incorporated oligos at the same rate as the host-associated protein MspRecT38, while the other two SSAPs had much lower efficiency (Fig 3e, S5, protein expression may play an additional effect).
Last, we used our model for SSAP portability to establish recombineering in Lactobacillus rhamnosus, a well-studied probiotic used to treat a variety of illnesses including diarrhea and bacterial vaginosis. Although the L. rhamnosus SSB and L. lactis SSB only have 47% identity, they share identical SSB C-terminal tail amino acids. Based on this we expected that LrpRecT (which functions in L. lactis) should be portable to L. rhamnosus, while the other SSAPs proteins would not be functional. We tested the 4 SSAPs in this species, and used oligos targeting L. rhamnosus RpoB to produce rifampicin resistance. This was indeed the case, and in L. rhamnosus, LrpRecT incorporated oligonucleotides over 2,500-fold above the background level, while λ-Red β and PapRecT had negligible activity and MspRecT was toxic.
SSAP-SSB pairs function across a broader host range than SSAPs alone
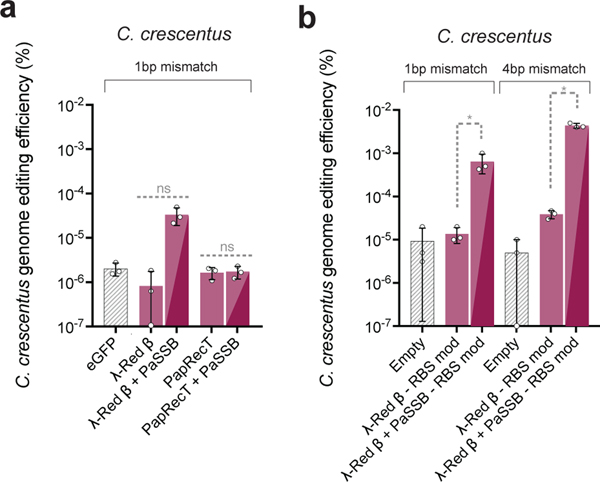
In L. lactis, the co-expression of PapRecT and λ-Red β with compatible SSB’s improved genome editing efficiency to a level comparable with the host-compatible LrpRecT. We hypothesized that for some species, SSAP-SSB pairs could provide functional recombineering capacity even if no functional SSAP had previously been identified. We therefore tested the two best-performing SSAP-SSB pairs (λ-Red β + PaSSB, and PapRecT + PaSSB) for activity in Caulobacter crescentus, a model organism for studying cell cycle regulation, replication, and differentiation.
In C. crescentus, we used oligos targeting RpoB to produce Rifampicin resistance, and did not detect any significant editing enhancement over the background with the SSAPs alone, or (surprisingly) with PapRecT + PaSSB. Since PapRecT and PaSSB are compatible, and PapRecT is well expressed in C. crescentus (Supplementary Fig. 4), we believe additional factors must contribute to the incompatibility of this pair in this species. However, using λ-Red β + PaSSB, we observed a 15-fold improvement over λ-Red β alone (Fig 4a). After expression optimization (Supplementary Fig. 4) and evasion of mismatch repair, λ-Red β + PaSSB demonstrated 873-fold improved editing efficiency over the background level, which was 112-fold higher than λ-Red β alone (Fig 4b). These results indicate that while SSAP-SSB pairs are not universally portable (Figure S7), the co-expression of a SSAP with a compatible bacterial SSB improves upon the editing efficiencies of SSAPs alone, and can be used to significantly enhance recombineering efficiency in species where no host-specific SSAP has been identified.
Fig. 4. SSAP-SSB pairs function across a broader host range than SSAPs alone.
(a), Editing efficiency in C. crescentus of two SSAP-SSB protein pairs, λ-Red β + PaSSB and PapRecT + PaSSB which had high genome editing efficiency in both E. coli and L. lactis. (b), Editing efficiency in C. crescentus of λ-Red β + PaSSB with ribosomal binding sites optimized for translation rate and using an oligo designed to evade mismatch repair. All experiments have at least 3 biologically independent replicates, error bars indicate SD from the men. (a, b) *: P value < .05; welch’s two tailed t-test of Log-transformed data.
Using SSAP-SSB pairs for genomic diversification
In E. coli, one of the unique capabilities of recombineering is the ability to generate rationally designed or high-coverage genomic libraries. Although this technique (termed MAGE) has been used for a variety of applications including optimizing metabolic pathways9, protein evolution39, and saturation mutagenesis1, it has only been used in a limited capacity in other species. We used L. lactis, a microbe distantly related to E. coli, to demonstrate how to optimize recombineering efficiency and perform high-coverage genomic mutagenesis after a functional SSAP has been identified.
To begin, we modified our construct in L. lactis to enable efficient incorporation of single, double, or triple nucleotide mutations, which are normally recognized and corrected by mismatch repair (MMR)31,40. We used the cognate pair PapRecT and PaSSB, and co-expressed either the dominant negative mismatch repair protein MutL.E32K from E. coli26, or took the host L. lactis MutL from the genome and made the equivalent mutation (LlMutL.E33K, Supplementary Fig. 6). While MutL.E32K from E. coli was nonfunctional, co-expression of LlMutL.E33K with PapRecT and PaSSB enabled the efficient introduction of 1bp pair changes (Extended Data Fig. 3). This indicates that while the E. coli dominant negative MutL is not broadly portable, a similarly mutated host MutL gene may be added to the recombineering construct to generate an inducible dominant negative MMR phenotype. Optimization of inducer and oligonucleotide concentrations further improved editing efficiency in L. lactis 26-fold to reach 23.9% for 1bp mutations at RpoB (Extended Data Fig. 3). Although we primarily focused on optimizing oligonucleotide recombination, we also found that PapRecT + PaSSB enabled the incorporation of long 1kb dsDNA constructs without the usual requirement for a cognate phage exonuclease (Extended Data Fig. 4).
Next, we used our optimized protocol to characterize the landscape of spectinomycin resistant variants at the ribosomal gene RpsE. Antibiotic resistant mutants at this locus were previously identified in E. coli using an efficient error-prone mutagenesis method, and the genotypes sampled reflect the capabilities of other error-prone mutagenesis techniques including hyper-mutator strains, PACE, and the amplification and cloning of genes using error-prone PCR. This previous approach found one antibiotic resistant amino acid mutation at RpsE (G30D in L. lactis), as well as a number of resistant mutants formed through amino-acid deletions25. However, error-prone mutagenesis imposes restrictions on the amino acids sampled due to primarily making single nucleotide conversions. The influence of amino acid mutations that require two or more nucleotide conversions, and the effects of combination amino-acid mutants are still not well understood.
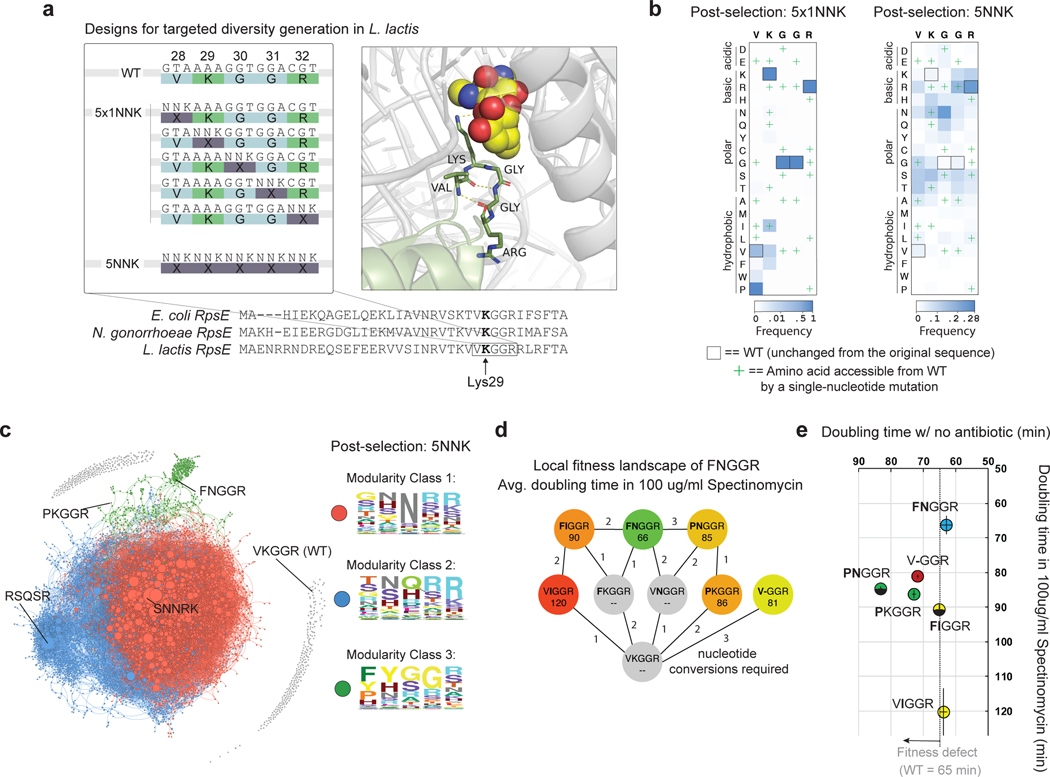
We began by targeting a 5-residue region near the spectinomycin binding pocket (L. lactis Lys 29), which is perfectly conserved between L. lactis, E. coli and N. gonorrhea, the species for which spectinomycin has been used clinically41 (Fig 5a, Supplementary Fig. 12). We first pooled 5 oligos, each of which mutates single codons, and sampled all possible single amino-acid conversions in the region (5×1NNK library, 100 variants) (Fig 5a). We also synthesized an oligo pool which mutates all 5 codons simultaneously, and generated an estimated 96.7% of the possible combination mutants between the 5 positions (5NNK library, 3.2 million variants, see Methods) (Fig 5a).
Fig. 5: Using SSAP-SSB pairs to interrogate complex phenotypic landscapes.
(a), Oligonucleotide design strategy. A sequence alignment of RpsE between E. coli, N. gonorrhoeae (a pathogen targeted by spectinomycin), and L. lactis shows a conserved 5AA region around the spectinomycin binding pocket (E. coli Lys26). 5 oligos can be pooled to introduce single degenerate codons at each amino acid position (5×1NNK), and a single oligo containing a fully degenerate sequence (5NNK) can be used to diversify the entire region. A crystal structure of E. coli RpsE bound to spectinomycin47 shows the approximate location of the antibiotic relative to the 5 targeted residues (Supplementary Fig. 12). (b), Normalized heat maps after selection and enrichment for the 5×1NNK single-amino acid saturation mutagenesis experiment vs. 5NNK combination mutagenesis experiment. (c), Force directed graph of all spectinomycin resistant combination mutants with at least 10 reads, lines connect variants that could be accessed through a single-nucleotide mutation, and the size of dots reflects the relative enrichment (d), Shortest paths to highly enriched double mutant FNGGR, as well as the nucleotide conversions required (e), Fitness of selected mutants in the presence and absence of antibiotic. Error bars represent the standard deviation from the mean of four biologically independent replicates.
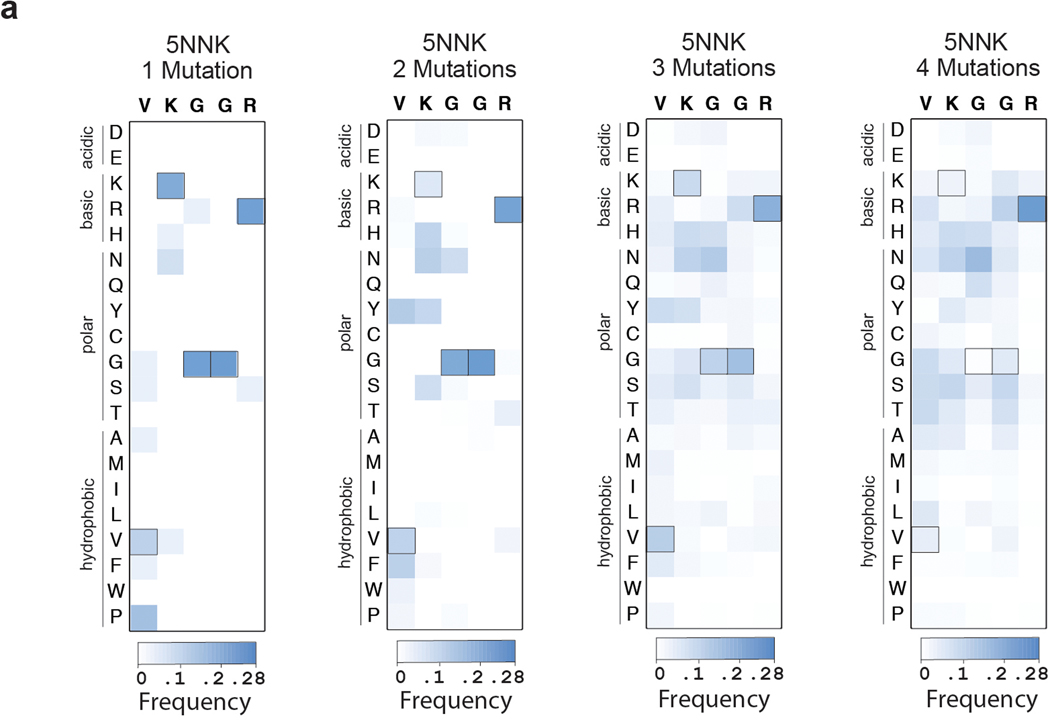
After enrichment of resistant cells by passaging, we used next-generation sequencing to identify the frequency of individual variants. Our saturation mutagenesis library (5×1NNK) revealed 15 new single amino acid variants, and 6 highly enriched mutants (Fig 5b). One mutant, PKGGR, was greatly over-represented, and required two simultaneous nucleotide conversions (Fig 5b). In the combination mutant library (5NNK), we found over 49,000 more candidate resistant mutants (Fig 5b). We generated heat maps of resistant variants containing 1, 2, 3 or 4 amino acid mutations relative to WT, and found that the enriched amino acids change at increasing mutational depth (Extended Data Fig 5), indicating substantial epistatic effects.
To analyze the combination mutant library holistically we generated a force-directed graph containing all mutants with at least 10 unique reads (8078 mutants, Fig 5c). We connected each mutant to every other mutant that was accessible through a single-nucleotide mutation, and found that none of the enriched combination mutants were accessible from WT (Fig 5c). Most of the resistant variants clustered into two closely related groups, in which all 5 amino acids are enriched with polar or charged residues. For a third cluster, WT residues were enriched at 3 of the 5 residues (“XX”GGR) (Fig 5c). We analyzed the most enriched mutant in this third cluster, FNGGR, and found that it is far more resistant than both the most resistant single mutant and a prototypical previously identified deletion mutant (Fig 5d). Furthermore, while the most resistant single mutants and deletion mutants exhibited fitness defects, FNGGR grew with no appreciable fitness defect (Fig 5e). Additionally, we found that FNGGR is formed by the epistatic combination of two mutations with no individual effect (Fig 5d). Here, efficient HR allowed the identification of new single mutants (PKGGR) and new clusters of antibiotic resistant genotypes (the high-fitness combination mutants) inaccessible through alternative approaches42. In this experiment, the high-editing efficiencies achievable via recombineering in L. lactis allowed us to simultaneously diversify 5 amino acid positions. In species with 100-fold (20^2) lower oligonucleotide incorporation efficiencies, similar experiments could be performed to diversify three neighboring amino-acid positions simultaneously.
Discussion
In this work we studied the host-tropism of phage SSAPs, which are used by synthetic biologists for genome engineering, and which form core components of phage recombination pathways34. Here, we discovered that an interaction between SSAPs and the host bacterial SSB largely determines their portability. Using chimeric SSBs, we found that this SSAP-SSB interaction relies on recognition of SSB C-terminal 7 amino acids. Although the SSAP-SSB interaction is specific, we found some SSAPs have a naturally broad host range (PapRecT) and interact with multiple distinct SSB tails. We also found that co-expression of an exogenous bacterial SSB broadens the host-range of SSAPs, and in certain species enables SSAP functionality even if there is no basal host compatibility. In E. coli, we found that co-expression of EcSSB significantly improved the recombineering efficiency of λ-Red β while reducing the construct’s toxicity. In other species, we used insights on the SSB interaction to develop new recombineering methods. In L. rhamnosus, we used the host-compatible LrpRecT to improve oligonucleotide incorporation frequency over 2,500-fold, while in C. crescentus used the co-expressed pair λ-Red β + PaSSB to improve oligonucleotide incorporation 870-fold.
We then demonstrated how SSAP-enabled recombineering surpasses error-prone methods of genome diversification by characterizing mutants of L. lactis RpsE that conferred spectinomycin resistance. The enriched variants would have been difficult to identify with even the most efficient nucleotide diversification methods due to extensive epistatic effects within the mutational landscape. We believe these methods will be useful for interrogating other protein-small molecule, or protein-protein interaction domains in gene’s native context. A follow-up study could use similar methods to identify mutations on the host-SSB’s C-terminal tail that are tolerated without a significant fitness penalty, since most of the currently identified SSB mutants are deleterious36. Similar methods might also be used to optimize the properties of enzymes within a host of interest39, or determine protein structures in-vivo by generating pairwise mutants43,44. While our work focused on a selectable phenotype, non-selectable phenotypes such as fitness can be examined as long as the sequencing depth is scaled by the efficiency of editing. In L. lactis with an efficiency of ~2% at the RpsE locus, 50x additional sequencing coverage would be needed to examine the same space of non-selected mutants.
There are many questions which remain about the mechanism of recombineering. First, there is no crystal structure of any full length SSAP, and a structural understanding of the steps involved in oligonucleotide binding and annealing in the presence of SSB should be characterized to better understand this mechanism. Second, while co-expression of an exogenous SSB can expand the host range of SSAPs, this is not a fully generalizable method, which raises questions about what other variables are relevant. Possible causes of SSAP-SSB portability failure may include: (1) an inhibitory interaction between SSAPs and the host SSB (there are a number of examples in Fig 3a, b where an SSB reduced the efficiency of an otherwise functional SSAP), (2) SSB-induced toxicity (neither PapRecT + PaSSB, or λ-Red β + PaSSB could be transformed and expressed in L. rhamnosus), and (3) limited protein expression in new hosts (λ-Red β is not expressed well in M. smegmatis). Finally, we are interested in examining the portability requirements of linear-linear homologous recombination (LLHR), since it is a distinct mechanism of recombineering that does not require replication45. With the long-term goal of reliably establishing high efficiency homologous recombination methods in all species, continuing research into the mechanisms of SSAPs will further help to expand and generalize these methods.
Methods:
Bacterial Strains and Culturing Conditions
The E. coli strain used was derived from EcNR2 with some modifications (EcNR2.dnaG_Q576A.tolC_mut.mutS::cat_mut.dlambda::zeoR)9. L. lactis strain NZ9000 (L. lactis subsp. cremoris MG1363 – pepN::nisRK) was provided as a kind gift from Jan Peter Van Pijkeren. M. smegmatis strain mc(2)155 was purchased from ATCC. The C. crescentus strain NA1000 and provided by Michael T Laub. L. rhamnosus GG was provided by Anik Debnath.
All chemicals were purchased from Sigma Aldrich, unless stated otherwise. E. coli and its derivatives were cultured in Lysogeny broth - Low sodium (Lb-L) (10 g/L tryptone, 5 g/L yeast extract (Difco), PH 7.5 with NaOH), in a roller drum at 34 °C. L. lactis was cultured in M17 broth (Difco, BD BioSciences) supplemented with 0.5% (w/v) D-glucose, static at 30 °C. M. smegmatis was cultured in Middlebrook 7H9 Broth (Difco, BD BioSciences) with AD Enrichment (10x stock: 50g/L BSA, 20g/L D-glucose, 8.5 g/L NaCl), supplemented with glycerol and Tween 80 to a final concentration of 0.2% (v/v) and 0.05% (v/v), respectively, in a roller drum at 37 °C. L. rhamnosus was cultured in MRS media (Difco, BD BioSciences) at 37 °C. C. crescentus was cultured in peptone-yeast extract (PYE) broth (2 g/L peptone, 1 g/L yeast extract (Difco), .3 g/L MgSO4, 1ml/L of 0.5M CaCl2), shaking at 30 °C.
Plating was done on petri dishes of LB agar for E. coli, M17 Agar (Difco, BD BioSciences) supplemented with 0.5% (w/v) D-glucose for L. lactis, 7H10 (Difco, BD BioSciences) supplemented with AD Enrichment and 0.2% (v/v) glycerol for M. smegmatis, MRS Agar (Difco, BD BioSciences) for L. rhamnosus, and PYE agar for C. crescentus. Antibiotics were added to the media for plasmid retention, at the following concentrations: 50 μg/mL carbenicillin for E. coli, 10 μg/mL chloramphenicol for L. lactis, and 20 μg/mL kanamycin for M. smegmatis, 10 μg/mL erythromycin for L. rhamnosus, 5 μg/ml kanamycin for C. crescentus. For the selective plates used to determine allelic recombination frequency, antibiotics were added as follows: 0.005% SDS for E. coli, 50 μg/mL rifampicin, and 100 μg/ml spectinomycin for L. lactis, 20 μg/mL streptomycin for M. smegmatis, 50 μg/mL rifampicin for L. rhamnosus, and 5 μg/ml rifampicin for C. crescentus.
Construction and Transformation of Plasmids
Plasmids were constructed using PCR fragments and Gibson Assembly. All primers and genes were obtained from Integrated DNA Technologies (IDT). Plasmids were derived from pARC8 for use in E. coli 48, pjp005 for use in L. lactis - a gift from Jan Peter Van Pijkeren28, pKM444 for use in M. smegmatis - a gift from Kenan Murphy (Addgene plasmid # 108319)8, and pBXMCS-2 for use in C. crescentus49. Genes were codon optimized for each of the host organisms using IDT’s online Codon Optimization Tool. E. coli and L. lactis plasmid constructs were Gibson assembled, then directly transformed into electrocompetent E. coli and L. lactis strains. M. smegmatis plasmids were first cloned in NEB 5-alpha Competent E. coli (New England Biolabs) for plasmid verification before transformation into electrocompetent M. smegmatis. All cloning was verified by Sanger sequencing (Genewiz).
Protein purification
To prepare λ-Red β for in vitro analysis, it was first cloned by Gibson cloning into pET-53-DEST, with a 6x poly-histidine tag followed by a glycine-serine linker and a TEV protease site (MHHHHHHGSGENLYFQG) appended to its N-terminus. After purification and treatment with TEV protease, this leaves only an N-terminal glycine before the start codon. Overnight cultures of E. coli BL21 (DE3) (NEB) with the expression construct were diluted 1:100 into Fernbach flasks, grown to an OD of ∼0.5, and induced with 1 mM IPTG at 37 °C for 4 h. Cultures were pelleted at 10,000 × g in a fixed angle rotor for 10 min and the supernatant decanted. Bacterial pellets were resuspended in 30 mL of lysis buffer (150 mM NaCl, 0.1% v/v Triton-X, 50 mM TRIS-HCl pH 8.0) and sonicated at 80% power, 50% duty cycle for 5 minutes on ice. The lysed cultures were again centrifuged for 10 min at 15,000 × g in a fixed angle rotor. The supernatant was then incubated for 30 minutes at room temperature with HisPur cobalt resin (Thermo) and column purified on disposable 25 ml polypropylene columns (Thermo). The protein-bound resin was washed with four column volumes of wash buffer (150 mM NaCl, 10 mM imidazole, 50 mM TRIS-HCl pH 8.0) and bound protein was eluted with two column volumes of elution buffer (150 mM NaCl, 250 mM imidazole, 50 mM TRIS-HCl pH 8.0). Protein eluates were dialyzed overnight against 25 mM TRIS-HCl pH 7.4 with 10,000 MWCO dialysis cassettes (Thermo), concentration was measured by Qubit (Thermo) and 1.5 mg of protein was cleaved in a 2 ml reaction with 240 Units of TEV protease (NEB) for two hours at 30 °C. The TEV cleavage reaction was re-purified with cobalt resin, except that in this case the flow-through was collected, as the His tag and the TEV protease were bound to the resin. Expression and successful TEV cleavage were confirmed by SDS-PAGE. Protein was concentrated in 10,000 MWCO Amicon protein concentrators (Sigma), protein concentration was assayed by Qubit, and an equal volume of glycerol was added to allow storage at −20 °C. E. coli and L. lactis SSBs were prepared according to previously published protocol (Lohman, Green, and Beyer, 1986) without the use of an affinity tag50.
Oligonucleotide annealing and quenching experiments
Fluorescent (tolC-r.null.mut-3’FAM) and quenching (tolC-f.null.mut-5’IBFQ) oligos were ordered from Integrated DNA Technologies. Unless otherwise indicated, 50 nM of each oligo was incubated in 25 mM TRIS-HCl pH 7.4 with 1.0 μM Ec_SSB or Ll_SSB at 30 °C for 30 minutes. 100 μl of each oligo mixture were then combined into a 96-well clear-bottom black assay plate (Costar), incubated a further 60 minutes at 30 °C, and annealing was tracked on a Synergy H4 microplate reader (BioTek) with fluorescence excitation set to 495 nm and emission set to 520 nm. After 60 minutes, 20 μl of a solution with or without 25μM λ-Red β and containing 100 mM MgCl2 was added to achieve a final reaction concentration of 2.5 μM λ-Red β and 10 mM MgCl2. The annealing was then tracked over 10 hours in a the Synergy H4 microplate reader with the setting indicated above.
Preparation of Electrocompetent E. coli
A single colony of E. coli was grown overnight to saturation. In the morning 30 μL of dense culture was inoculated into 3 mL of fresh media and grown for 1 hour. To induce gene expression of the pARC8 vector for recombineering experiments, L-arabinose was added to a final concentration of 0.2% (w/v) and the cells were grown an additional hour. 1 mL of cells were pelleted at 4 °C by centrifugation at 12,000 x g for 2.5 minutes and washed twice with 1 mL of ice-cold dH2O. Cells were resuspended in 50 μL ice-cold dH2O mixed with DNA and transferred to a pre-chilled 0.1 cm electroporation cuvette.
Preparation of Electrocompetent L. lactis
A single colony of L. lactis was grown overnight to saturation. 500 μL of dense culture was inoculated into 5 mL of fresh media, supplemented with 500mM sucrose and 2.5% (w/v) glycine, and grown for 3 hours. To induce gene expression of the pJP005 vector for recombineering experiments, the cells were grown for an additional 30 min after adding 1 ng/mL freshly diluted nisin, unless stated otherwise. For the optimized condition (Fig 4b), 10 ng/mL nisin was used. Cells were pelleted at 4 °C by centrifugation at 5,000 x g for 5 minutes and washed twice with 2 mL of ice-cold electroporation buffer (500mM sucrose containing 10% (w/v) glycerol) by centrifugation at 13,200 x g for 2.5 minutes. Cells were resuspended in 80 μL ice-cold electroporation buffer mixed with DNA and transferred to a pre-chilled 0.1 cm electroporation cuvette.
Preparation of Electrocompetent M. smegmatis
This method was adapted from: “Murphy, Kenan C., et al. “ORBIT: a new paradigm for genetic engineering of mycobacterial chromosomes.” MBio 9.6 (2018).”
A single colony of M. smegmatis was grown overnight to saturation. The next day 25 μL of dense culture was inoculated into 5 mL of fresh media in the evening and grown overnight. The next day, 500ng/ml ATC was added at an OD of .5 and grown for 3 hours. The cultures were placed on ice (swirling for 10 min), pelleted at 4 °C by centrifugation at 3,500 x g for 10 minutes and washed twice with 10 mL ice-cold 10% glycerol. Cells were resuspended in 350 μL ice-cold 10% glycerol mixed with DNA to a pre-chilled 0.2 cm electroporation cuvette.
Preparation of Electrocompetent L. rhamnosus
A single colony of L. rhamnosus was grown overnight at 37C in 5ml of media (stationary, screw-capped tube). The next day, cells were diluted back to an OD = 0.1, and supplemented with both 1ml 20% glycine (2% final concentration), and 5μg ATc (500 ng/ml final concentration) in 10ml total volume. Cells were incubated at 37C until an OD = 0.25. The culture was now supplemented with 10 μg/ml ampicillin, mixed using a 1ml pipette, and incubated at 37C until an OD = .5. The cells were pelleted by centrifugation at 5000 G for 15 min at ambient temperature. Cells were resuspended in 5ml “electroporation conditioner 1” (filter sterilized: .5M sucrose, 7mM potassium phosphate and 1mM magnesium chloride) and pelleted. This step was repeated one time. Cells were then resuspended in 100 μL “electroporation conditioner 1” mixed with DNA, and added to a pre-chilled .2cm cuvette.
Preparation of Electrocompetent C. crescentus
A single colony of C. crescentus was grown overnight. The next day cells were diluted back to OD ~0.001 in 25 mL PYE, and grown overnight. The next day, 250 μL of 30% xylose was added to cells at OD ~0.1. Cells were harvested at between OD = 0.5 and OD = 0.7, spun at 10,000 rpm for 10 min, and then washed twice in 12.5ml of ice-cold dH2O, washed once in 12.5ml of ice-cold 10% glycerol, then washed and resuspended in 2.5ml of ice-cold 10% glycerol mixed with DNA. 90 μL of cells and DNA were added to 0.1cm cuvettes and incubated on ice for 10 min.
Recombineering Experiments
90-mer oligos were added to electrocompetent cells at stock solution concentrations of: 1 μM (1.46μg in 50μL) for E. coli, 21.4 μM (50μg in 80μL) for L. lactis, 100μM (292μg in 100μL) for L. rhamnosus, and 10μM (26.3μg in 90μL) for C. crescentus. 70-mer oligos were used at 95.1 nM (1μg in 360μL) for M. smegmatis. All oligos were obtained from IDT and can be found under “Oligonucleotides for genome editing” in materials and methods. For dsDNA experiments L. lactis was electroporated with 1.5 μg purified linear dsDNA. Cells were electroporated using a Bio-Rad gene pulser set to 25 μF, 200 Ω, and 1.8 kV for E. coli, 2.0 kV for L. lactis, 1.7 kV for L. rhamnosus, and 1.5kV for C. crescentus and to 25 μF, 1000 Ω and 2.5 kV for M. smegmatis. Immediately after electroporation, cells were recovered in fresh media for 3 hours for E. coli, 1 hour for L. lactis, overnight for M. smegmatis, L. rhamnosus, and C. crescentus. L. lactis and L. rhamnosus recovery media was supplemented with 20mM MgCl2 and 2 mM CaCl2. For plasmid retention, E. coli recovery media was supplemented with carbenicillin, M. smegmatis recovery media was supplemented with kanamycin, L. rhamnosus recovery media was supplemented with erythromycin and C. crescentus recovery media was supplemented with 0.3% xylose and kanamycin. After recovery, the cells were serial diluted and plated on non-selective vs. selective agar plates to obtain approximately 50–500 CFU/plate. Colonies were counted using Fiji, and allelic recombination frequency was calculated by dividing the number of colonies on selective plates, with the number of colonies on non-selective plates.
Next-generation sequencing and analysis for L. lactis Spectinomycin resistant mutants
For the site-saturation experiment (1×5NNK) half of the cells recovered from a single transformation were used. For the combinatorial experiment (5NNK) cells recovered from 30 parallel transformations were used. After transformation, cells were recovered for 1hr, then plated on 100 μg/mL Spectinomycin GM17 plates. After 2 days of growth, cells were resuspended in 1 mL of culture per plate, and recovered overnight. The cultures were then passaged to enrich for resistant mutants. During passaging for 1×5NNK: 100 μL confluent cells were passaged into 10 mL culture and grown overnight for the single-amino acid library, and for 5NNK: 500 μL confluent cells were passaged into 50 mL culture and grown overnight for the combinatorial mutant library. Cultures were used as templates for initial amplification using the L. lactis RpsE locus primers given under “Next-generation sequencing of spectinomycin resistance mutants (L. lactis)”. Cultures were collected at various time points: 1 hr after electroporation and after overnight recovery in Spectinomycin for the 1NNK library, and 1 hr after electroporation as well as after 1, 2, and 3 overnight passages in Spectinomycin for the 5NNK library. The data from 3 overnight passages was used for subsequent analysis. 50 μL of cells at each timepoint were spun down to make a pellet and used in a 50 μL PCR reaction. Initial amplification was monitored by qPCR for ~20 cycles until late exponential phase. A second round of qPCR was used to add indexing primers for Illumina sequencing, and run for ~15 cycles until late exponential phase. Samples were quantified by Qubit, combined and sequenced using Illumina MiSeq with reagent kit V3. Sequencing was done using a single 150bp read using RpsE locus primer R. For each timepoint, sequencing reads were processed using a python script, filtered such that reads matched the genomic locus 21 bp upstream and 21 bp downstream of the target site.
E. coli SSAP-SSB Toxicity Assay
A single colony of each construct for E. coli was grown overnight in a 96-well plate containing 150 μL of media per well with carbenicillin. 1.5 μL overnight culture was inoculated into 150 μL fresh media and grown for 2 hours to reach exponential phage. 1.5 μL of exponential culture was re-inoculated into 150 μL fresh media containing .2% (v/v) L-arabinose to induce expression of the genes. OD600 was then measured every three minutes, for a time course of 7 hours in a BioTek Eon plate reader, set to 34 °C with double orbital shaking. Growth curves were analyzed in Matlab using a custom script to determine the doubling time.
L. lactis Spectinomycin and wildtype fitness assay
To assay spectinomycin resistance in L. lactis, single colonies were grown in 150 μL overnight with no antibiotic in 96 well plates. In the morning cells 1.5 μL confluent culture was then inoculated into 300 μL media containing no antibiotic or 100 μg/mL Spectinomycin in 96 well black assay plates with clear bottoms. These plates were covered with a “Breathe-Easy” sealing film, and cells were grown at 30C for 7 hours in a BioTek Eon plate reader, with OD600 measured every 3 minutes.
Generating the force-directed graph of spectinomycin resistance mutants
Using python, an edges matrix was generated between SpecR combination mutants that had more than 10 reads and were accessible through single nucleotide mutations. This matrix was uploaded to the network visualization software Gephi, and a separate node matrix of the SpecR mutants was uploaded to add the genotype labels and enrichment values. Using Gephi, a force directed graph was produced using Force Atlas 2 (scaling = 25, gravity = 250). Once the nodes reached steady-state locations, the modularity statistic was performed using a resolution of 3.0 to identify the major clusters.
Finding cognate SSBs for each SSAP:
Cognate SSBs corresponding to each of the phage SSAPs were selected as follows:
λ-Red β is found in an Escherichia coli phage. The selected strain E. coli K12 only has a single SSB protein annotated. (SSB tail motif: FDDDIPF)
PapRecT is found in a Pseudomonas aeruginosa phage. The selected strain P. aeruginosa PAO1 only has a single SSB protein annotated. (SSB tail motif: FDDDIPF)
MspRecT is found in a Mycobacterium smegmatis phage. The selected strain M. smegmatis mc(2)-155 has 3 annotated SSB proteins, however only one has a tail motif (DDEPPF) similar to the SSBs selected for PaSSB and EcSSB so we selected that variant.
LrpRecT is found in a Lactococcus reuteri prophage. The selected strain L. reuteri MM4–1A has two annotated SSB proteins, however only one has a tail motif (DDELPF) similar to the SSBs selected for PaSSB and EcSSB so we selected that variant.
Cutoffs for “SSB C7 compatibility”, Lower SSB C7 compatibility”, and “Inhibitory SSB” in Figure 3c:
Metrics for calculating “SSB C7 compatibility”, “An SSB C7 sequence was marked as “compatible” if the SSB caused at least a 5x increase in recombination efficiency above the SSAP alone, and was within 0.5x of the best performing SSB.
An SSB C7 sequence was marked as having “lower SSB compatibility” if the SSB caused between a 5x increase or 5x decrease in recombination efficiency, or contributed to an efficiency less then 0.5-fold of the best performing SSB.
An SSB C7 sequence was marked as “inhibitory” if the SSB caused a more than a 5x decrease in recombination efficiency.
Calculation of estimated number of edited cells and library coverage for 1NNK and 5NNK libraries:
5×1NNK:
We performed a single transformation and plated half of the recovered cells (235 million) on 100μg/mL spectinomycin plates (117.5 million). We estimate that a single doubling occurred during the hour recovery (~58.75 million unique cells electroporated). The average efficiency of known single-amino acid changes at that position is 3.2% (V28P, K29I), giving an approximate number of 1.88 million edited cells. We assume complete coverage of the 105 amino acid variants (including stop codons).
5NNK:
We performed 30 parallel transformations and plated all of the recovered cells (6.75 billion) on 100ug/ml spectinomycin plates. We estimate that a single doubling occurred during the 60min recovery (3.375 billion unique cells electroporated). The average efficiency of 5AA mutations is 1.54% (RTNAR, NGTRF, Supplementary Fig. 8), giving an approximate number of 52.1 million edited cells.
The likelihood of sampling of a particular variant is given by: 1 where represents the probability of each variant (i) and, L is the total number of edited cells. We calculate for each possible amino acid combination by assuming independence at each position, use the frequency of sampling each amino acid in the initial library (Supplementary Fig. 9), and take the product of the probabilities at each position.
For example:
To estimate total library coverage we then sum over all possible variants
Given L=52.1 million edited cells, our expected coverage is 3.95 million amino acid variants (including stop codons) or 96.7% of the total 4.08 million.
Protein structures
Protein structure images (Fig 2a) were downloaded from PyMOL: Schrodinger LLC, The PyMOL Molecular Graphics System, Version 1.8 (2015).
Data availability statement (DAS):
NGS data for the L. lactis spectinomycin experiment is deposited as a NCBI BioProject under accession number PRJNA648295. Source data is provided for all main text figures. All data and constructs are available from the authors upon reasonable request. For L. rhamnosus plasmids, contact the corresponding authors or anik@tenza.bio.
Code Availability
The scripts used to analyze doubling times in MATLAB, analyze Illumina sequencing data, and generate the adjacency matrix for the force-directed graph are available upon request.
Extended Data
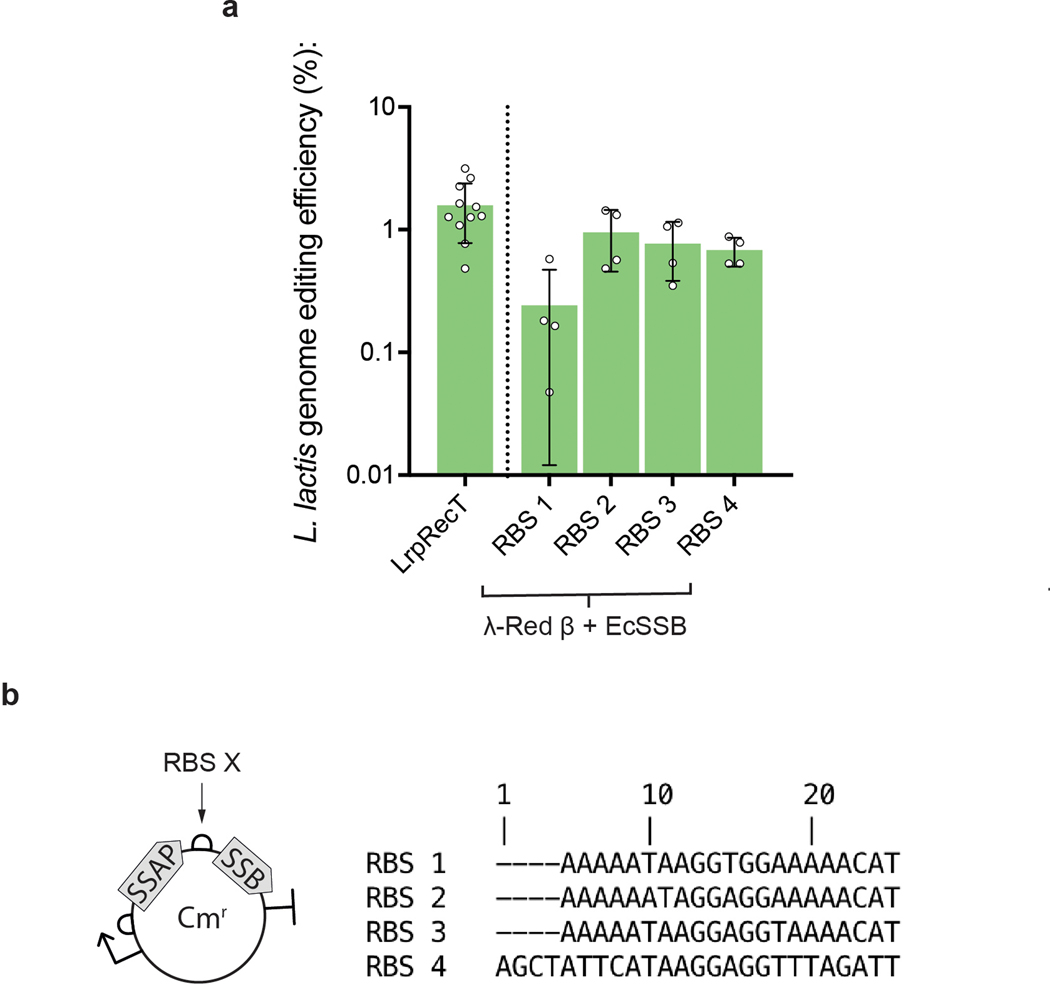
Extended Data Fig. 1. Bicistronic RBS optimization.
In L. lactis, the internal RBS sequence affected recombination efficiency using the bicistronic λ-Red β and EcSSB construct. (a, b) RBS 2, which enabled the highest efficiency genome editing in this experiment was selected used in all other constructs unless otherwise indicated. Error bars indicate SD from the mean of at least four biologically independent replicates.
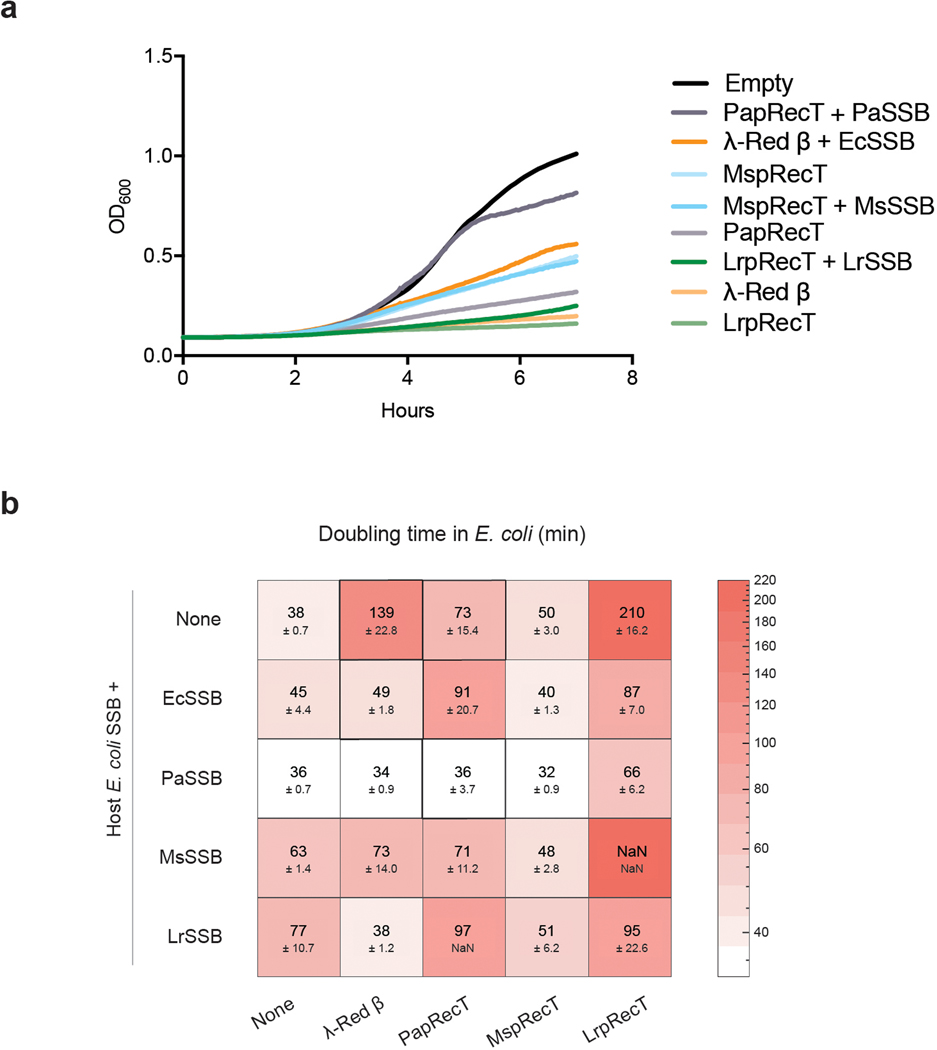
Extended Data Fig. 2. Doubling times in E. coli of constructs expressing SSAPs and SSBs reveal that co-expression of SSB can dramatically influence toxicity.
(a) Growth curve of cognate SSAP-SSB pairs and SSAPs alone in E. coli under constant induction (7hrs). (b) Doubling time measurements for all combinations of the 4 SSAPs and SSBs in E. coli under constant induction (7hrs) with mean and standard deviation presented for at least 3 biologically independent replicates. The SSAPs vary in toxicity, with λ-Red β showing considerable toxicity. The co-expression of SSBs reduces SSAP toxicity in a number of cases, especially for PaSSB. There are a number of constructs with low toxicity and high genome editing efficiency (λ-Red β + EcSSB, λ-Red β + PaSSB, PapRecT + PaSSB) showing that there is no direct correlation between toxicity and genome editing efficiency.
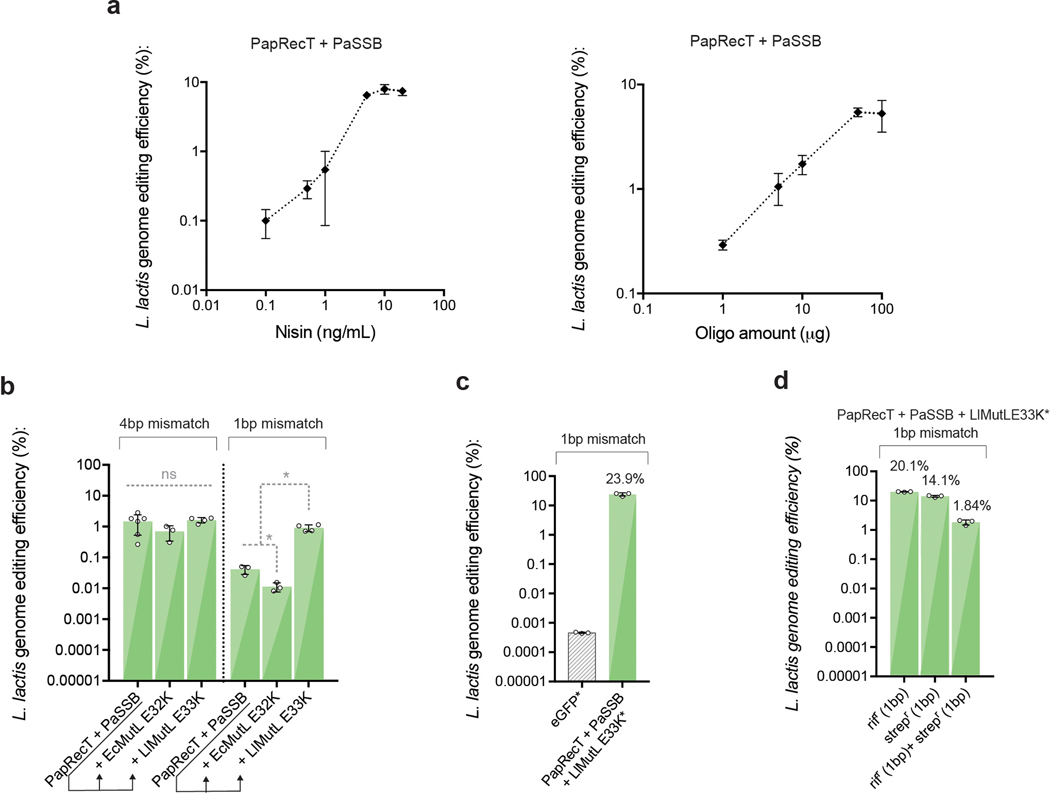
Extended Data Fig. 3. Optimization of recombineering efficiency in L. lactis.
(a) Optimization of nisin concentration to 10ng/ml contributed to a significant improvement in genome editing efficiency for PapRecT + PaSSB. (a) The optimal oligo amount plateaued at 50 μg of DNA, which corresponds 21.4 μM in 80 μL. (b) Expression of the L. lactis MutL variant E33K allowed the efficient introduction of 1bp mismatches at similar efficiency to 4bp mismatches which evade MMR. (c, d) After optimization from (a, b), PapRecT + PaSSB + LlMutLE33K enabled >20% editing efficiency at the Rif locus (c), and efficient multiplexed editing (d). Error bars indicate SD from the mean of at least three biologically independent replicates. (b) *: P value < .05; ordinary one-way ANOVA of Log-transformed data, Holm-Sidak multiple comparisons test.
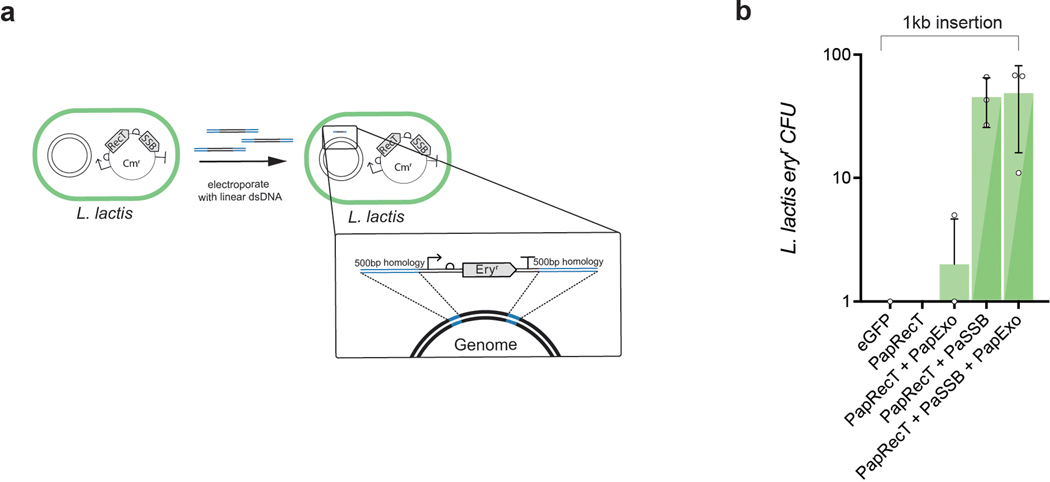
Extended Data Fig. 4. DsDNA recombineering with PapRecT and PaSSB in L. lactis.
Although this work mostly focused on ssDNA recombineering, dsDNA recombineering can be used to integrate larger constructs including genes and resistance markers, and usually requires the presence of a cognate phage exonuclease. These proteins are almost always found within the phage operon containing the SSAP, and can be readily co-expressed to enable dsDNA recombineering. Surprisingly, we find that PapRecT + PaSSB enabled dsDNA recombineering in L. lactis even without including a cognate phage exonuclease suggesting that the co-expressed SSB recruits an endogenous exonuclease, or the SSAP+SSB pair provides the sufficient requirements for dsDNA recombineering. (a) Gene knockins were performed in L. lactis using linear DNA with 500bp homology arms carrying an Erythromycin resistance cassette. (b) Co-expression of PapRecT + PaSSB enabled the efficient introduction of a 1kb selectable marker as dsDNA even without the addition of the cognate phage exonuclease. Error bars indicate SD from the mean of at least three biologically independent replicates.
Extended Data Fig. 5. Heat maps of RpsE mutagenesis at different mutational depths.
The 5NNK library diversification experiment (Fig 5) allow us to identify antibiotic resistant single, double, triple, or quintuple mutants when the other codons have WT amino acids. (a) heat maps showing the enrichment of amino acids in the 5NNK library, filtered to separately present those with 1, 2, 3, or 4 mutations vs. WT. The enriched amino acids change at increasing mutational depth. The “5NNK: 1 mutation” library has mutations enriched in the first 2 positions (28V, and 29K) similar to the 5×1NNK single-amino acid mutagenesis heat map, while the “5NNK: 4 mutation” library looks similar to the 5NNK library heat map (Fig 5b), with an enrichment to polar and charged residues at all 5 positions.
Supplementary Material
Acknowledgments:
We thank Gleb Kuznetsov and Georgia Squyres for reviewing the manuscript and providing helpful feedback. We thank Charles Bell for discussions about possible mechanisms of recombineering, and Jan Peter Van Pijkeren along with Jee-Hwan Oh for helping us setup initial protocols in L. lactis.
Funding: This work was supported through National Institute of General Medical Sciences under Grant No. 1U01GM110714-01 and the Department of Energy DE-FG02-02ER63445 to G.M.C.. This work was also supported by NIH grant R01GM082899 to M.T.L. who is an Investigator of the Howard Hughes Medical Institute. G.T.F. was supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE1745303. D.A.S was supported by a Landry Cancer Biology Research Fellowship. K.G. was supported by the National Science Foundation Graduate Research Fellowship under Grant No. DGE1745302.
Footnotes
Competing interests: G.M.C. is a founder of 64-x, Enevolv, and GRO Biosciences. G.T.F, T.M.W., and G.M.C. are named inventors on a patent application related to the technologies described in this article. Other potentially relevant financial interests are listed at http://arep.med.harvard.edu/gmc/tech.html.
References:
- 1.Kelsic ED et al. RNA Structural Determinants of Optimal Codons Revealed by MAGE-Seq. Cell Systems 3, 563–571.e6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sharon E et al. Functional Genetic Variants Revealed by Massively Parallel Precise Genome Editing. Cell 175, 544–557.e16 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Warrier T et al. N -methylation of a bactericidal compound as a resistance mechanism in Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences 113, E4523–E4530 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Riglar DT et al. Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nature Biotechnology 35, 653–658 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steidler L et al. Biological containment of genetically modified Lactococcus lactis for intestinal delivery of human interleukin 10. Nature Biotechnology 21, 785–789 (2003). [DOI] [PubMed] [Google Scholar]
- 6.Guo C-J et al. Depletion of microbiome-derived molecules in the host using Clostridium genetics. Science 366, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen PE & Shapiro BJ The advent of genome-wide association studies for bacteria. Current Opinion in Microbiology 25, 17–24 (2015). [DOI] [PubMed] [Google Scholar]
- 8.Murphy KC et al. ORBIT: a New Paradigm for Genetic Engineering of Mycobacterial Chromosomes. mBio 9, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang HH et al. Programming cells by multiplex genome engineering and accelerated evolution. Nature 460, 894–898 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garst AD et al. Genome-wide mapping of mutations at single-nucleotide resolution for protein, metabolic and genome engineering. Nat Biotechnol 35, 48–55 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Lajoie MJ et al. Genomically Recoded Organisms Expand Biological Functions. Science 342, 357–360 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fredens J et al. Total synthesis of Escherichia coli with a recoded genome. Nature 569, 514–518 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yu D et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proceedings of the National Academy of Sciences of the United States of America 97, 5978–83 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W, Bikard D, Cox D, Zhang F & Marraffini LA RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nature biotechnology 31, 233–9 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oh J-H & van Pijkeren J-P CRISPR–Cas9-assisted recombineering in Lactobacillus reuteri. Nucleic Acids Research 42, e131–e131 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corts AD, Thomason LC, Gill RT & Gralnick JA Efficient and Precise Genome Editing in Shewanella with Recombineering and CRISPR/Cas9-Mediated Counter-Selection. ACS Synth. Biol. 8, 1877–1889 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Murphy KC Use of Bacteriophage λ Recombination Functions To Promote Gene Replacement in Escherichia coli. Journal of Bacteriology 180, 2063–2071 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iyer LM, Koonin EV & Aravind L Classification and evolutionary history of the single-strand annealing proteins, RecT, Redbeta, ERF and RAD52. BMC genomics 3, 8 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Datsenko KA & Wanner BL One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. PNAS 97, 6640–6645 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y, Buchholz F, Muyrers JPP & Stewart AF A new logic for DNA engineering using recombination in Escherichia coli. Nature Genetics 20, 123–128 (1998). [DOI] [PubMed] [Google Scholar]
- 21.Ellis HM, Yu D, DiTizio T & Court DL High efficiency mutagenesis, repair, and engineering of chromosomal DNA using single-stranded oligonucleotides. Proc. Natl. Acad. Sci. U.S.A. 98, 6742–6746 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang K et al. Defining synonymous codon compression schemes by genome recoding. Nature 539, 59–64 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nyerges Á et al. Directed evolution of multiple genomic loci allows the prediction of antibiotic resistance. PNAS 115, E5726–E5735 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Podgornaia AI & Laub MT Protein evolution. Pervasive degeneracy and epistasis in a protein-protein interface. Science 347, 673–677 (2015). [DOI] [PubMed] [Google Scholar]
- 25.Halperin SO et al. CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window. Nature 560, 248–252 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Nyerges Á et al. A highly precise and portable genome engineering method allows comparison of mutational effects across bacterial species. Proceedings of the National Academy of Sciences 113, 2502–2507 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall SD, Kane MF & Kolodner RD Identification and characterization of the Escherichia coli RecT protein, a protein encoded by the recE region that promotes renaturation of homologous single-stranded DNA. Journal of Bacteriology 175, 277–287 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Pijkeren J-P & Britton RA High efficiency recombineering in lactic acid bacteria. Nucleic Acids Research 40, e76–e76 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Datta S, Costantino N, Zhou X & Court DL Identification and analysis of recombineering functions from Gram-negative and Gram-positive bacteria and their phages. Proc Natl Acad Sci U S A 105, 1626–1631 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Kessel JC & Hatfull GF Recombineering in Mycobacterium tuberculosis. Nat. Methods 4, 147–152 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Costantino N & Court DL Enhanced levels of λ Red-mediated recombinants in mismatch repair mutants. PNAS 100, 15748–15753 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomason LC, Costantino N & Court DL Examining a DNA Replication Requirement for Bacteriophage λ Red- and Rac Prophage RecET-Promoted Recombination in Escherichia coli. mBio 7, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Caldwell BJ et al. Crystal structure of the Redβ C-terminal domain in complex with λ Exonuclease reveals an unexpected homology with λ Orf and an interaction with Escherichia coli single stranded DNA binding protein. Nucleic Acids Research (2019) doi: 10.1093/nar/gky1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lo Piano A, Martínez-Jiménez MI, Zecchi L & Ayora S Recombination-dependent concatemeric viral DNA replication. Virus Research 160, 1–14 (2011). [DOI] [PubMed] [Google Scholar]
- 35.Yin J et al. Single-Stranded DNA-Binding Protein and Exogenous RecBCD Inhibitors Enhance Phage-Derived Homologous Recombination in Pseudomonas. iScience 14, 1–14 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shereda RD, Kozlov AG, Lohman TM, Cox MM & Keck JL SSB as an organizer/mobilizer of genome maintenance complexes. Critical reviews in biochemistry and molecular biology 43, 289–318 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sun Z et al. A high-efficiency recombineering system with PCR-based ssDNA in Bacillus subtilis mediated by the native phage recombinase GP35. Appl. Microbiol. Biotechnol. 99, 5151–5162 (2015). [DOI] [PubMed] [Google Scholar]
- 38.Murphy KC, Papavinasasundaram K & Sassetti CM Mycobacterial Recombineering. in Methods in molecular biology (Clifton, N.J.) vol. 1285 177–199 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Amiram M et al. Evolution of translation machinery in recoded bacteria enables multi-site incorporation of nonstandard amino acids. Nat Biotechnol 33, 1272–1279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawitzke JA et al. Probing cellular processes with oligo-mediated recombination; using knowledge gained to optimize recombineering. J Mol Biol 407, 45–59 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Unemo M et al. Neisseria gonorrhoeae Strain with High-Level Resistance to Spectinomycin Due to a Novel Resistance Mechanism (Mutated Ribosomal Protein S5) Verified in Norway. Antimicrobial Agents and Chemotherapy 57, 1057–1061 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miton CM & Tokuriki N How mutational epistasis impairs predictability in protein evolution and design. Protein Sci 25, 1260–1272 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmiedel JM & Lehner B Determining protein structures using deep mutagenesis. Nature Genetics 51, 1177–1186 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rollins NJ et al. Inferring protein 3D structure from deep mutation scans. Nature Genetics 51, 1170–1176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fu J et al. Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nature Biotechnology 30, 440–446 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Raghunathan S, Kozlov AG, Lohman TM & Waksman G Structure of the DNA binding domain of E. coli SSB bound to ssDNA. Nature Structural Biology 7, 648–652 (2000). [DOI] [PubMed] [Google Scholar]
- 47.Borovinskaya MA, Shoji S, Holton JM, Fredrick K & Cate JHD A Steric Block in Translation Caused by the Antibiotic Spectinomycin. ACS Chem. Biol. 2, 545–552 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only References:
- 48.Choe W, Chandrasegaran S & Ostermeier M Protein fragment complementation in M.HhaI DNA methyltransferase. Biochemical and Biophysical Research Communications 334, 1233–1240 (2005). [DOI] [PubMed] [Google Scholar]
- 49.Thanbichler M, Iniesta AA & Shapiro L A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res 35, e137–e137 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lohman TM, Green JM & Beyer RS Large-scale overproduction and rapid purification of the Escherichia coli ssb gene product. Expression of the ssb gene under .lambda. PL control. Biochemistry 25, 21–25 (1986). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
NGS data for the L. lactis spectinomycin experiment is deposited as a NCBI BioProject under accession number PRJNA648295. Source data is provided for all main text figures. All data and constructs are available from the authors upon reasonable request. For L. rhamnosus plasmids, contact the corresponding authors or anik@tenza.bio.