Abstract
TUG proteins bind and sequester GLUT4 glucose transporters intracellularly, and insulin stimulates TUG cleavage to translocate this GLUT4 to the cell surface and increase glucose uptake. This effect of insulin is independent of phosphatidylinositol-3-kinase, and its physiological relevance remains uncertain. Here, we show that this TUG cleavage pathway regulates both insulin-stimulated glucose uptake in muscle and organism-level energy expenditure. Using mice with muscle-specific TUG knockout and muscle-specific constitutive TUG cleavage, we show that, after GLUT4 release, the TUG C-terminal cleavage product enters the nucleus, binds PPARγ and PGC-1α, and regulates gene expression to promote lipid oxidation and thermogenesis. This pathway acts in muscle and adipose cells to upregulate sarcolipin and Ucp1, respectively. The PPARγ2 Pro12Ala polymorphism, which reduces diabetes risk, enhances TUG binding. The Ate1 arginyltransferase, which mediates a specific protein degradation pathway and controls thermogenesis, regulates the stability of the TUG product. We conclude that insulin-stimulated TUG cleavage coordinates whole-body energy expenditure with glucose uptake, that this mechanism might contribute to the thermic effect of food, and that its attenuation could promote obesity.
To stimulate glucose uptake into muscle and adipose cells, insulin causes the exocytic translocation of GLUT4 glucose transporters. This action requires the intersection of insulin signaling and membrane trafficking pathways1,2. Most work in this area has focused on signaling through Akt to specific Rab GTPases, but this pathway does not fully explain GLUT4 translocation or insulin resistance3–5. From a trafficking standpoint, GLUT4 is unusual not because of its targeting in the presence of insulin, but because of its targeting in unstimulated cells6–8. During fasting, intracellular sequestration of GLUT4 restricts glucose uptake; release of this trapped GLUT4 is the major effect of insulin. In insulin-resistant individuals, GLUT4 is mistargeted during fasting9,10. The GLUT4 sequestration compartment comprises preformed vesicles, termed “insulin-responsive vesicles” or “GLUT4 storage vesicles” (GSVs)11. Insulin mobilizes GSVs from the sequestered pool in a graded, dose-dependent manner5,6. How this occurs is not well understood.
First identified in a genetic screen, the TUG protein is a “Tether, containing a UBX domain, for GLUT4” that sequesters GSVs in unstimulated cells5,12,13. The TUG N-terminal region binds GLUT4 and IRAP, a transmembrane aminopeptidase present in GSVs8,14,15. The TUG C-terminal domain binds Golgi matrix proteins, including Golgin-160 and ACBD316,17. TUG may tether vesicles stably, or constrain a cycle of budding and fusion, to trap GSVs at the Golgi matrix. To mobilize these vesicles, insulin triggers site-specific endoproteolytic cleavage of TUG5,16–19. The N-terminal cleavage product is a ubiquitin-like modifier, called TUGUL (“TUG Ubiquitin-Like”). In adipocytes, TUGUL modifies kinesin motors to carry GLUT4 to the cell periphery5,20. Thus, TUG cleavage both liberates the trapped GSVs and also activates their microtubule-based movement to the plasma membrane.
Insulin stimulates TUG cleavage and GLUT4 movement by a phophatidylinositol-3-kinase (PI3K)-independent mechanism5,20–22. This requires the TC10α GTPase and its effector, PIST, which binds directly to TUG16. In muscle, targeted degradation of PIST causes constitutive TUG cleavage, GLUT4 translocation, and increased glucose uptake during fasting18. The data imply that PIST is a negative regulator of TUG cleavage, which is disinhibited by GTP-bound TC10α to promote TUG cleavage in fat and muscle8,16,18. The muscle isoform of the Usp25 protease, Usp25m, mediates TUG cleavage in adipocytes5. Insulin activates TC10α very transiently, likely because of an upstream feedforward circuit11,13,23, which may help explain equivocal data in muscle24–28. Such feedforward circuits enable cells to respond to the rate-of-change of an external signal29. Physiologically, TC10α activation may respond primarily to the rate-of-change of insulin concentration, so that TUG cleavage reflects glycemic load.
In addition to GLUT4, GSVs contain IRAP, which inactivates circulating vasopressin, and LRP1 and sortilin, which bind lipoproteins5,30. Transgenic mice with constitutive TUG cleavage in muscle exhibit not only increased fasting muscle glucose uptake, due to increased GLUT4 at the plasma membrane, but also increased water intake, due to increased IRAP at the plasma membrane and accelerated vasopressin degradation14. These mTUGUBX-Cter mice (here called “UBX mice”) also display increased energy expenditure, which remains unexplained18. Here, we show that TUG deletion in muscle mimics the effect of insulin to stimulate GLUT4 translocation, with consequent effects on glucose homeostasis. We further define an unanticipated action of the TUG C-terminal cleavage product, which enters the nucleus and regulates gene expression to control fatty acid oxidation and thermogenesis. Our results link insulin-stimulated glucose uptake with energy expenditure, and may have therapeutic implications for metabolic disease.
Results
TUG deletion in muscle increases glucose uptake but not energy expenditure.
In UBX mice, muscle-specific transgenic expression of an unstable TUG fragment recruits PIST for proteosomal degradation, and endogenous intact TUG proteins are cleaved constitutively in the absence of an insulin signal18. In fasting UBX mice, GSVs are translocated and TUG cleavage products are generated in muscle. Therefore, the 12–13% increase in energy expenditure observed in these mice18 could result from translocation of a GSV cargo protein or from effects of a cleavage product. To distinguish these possibilities, as well as to test effects on glucose homeostasis, we created muscle-specific TUG knockout (MTKO) mice. We predicted that in the absence of intact TUG, GSV cargoes would be translocated to the cell surface in muscle cells, as in adipocytes8, but no TUG cleavage products will be generated. With respect to production of the TUG C-terminal cleavage product, UBX and MTKO mice are gain- and loss-of-function models, respectively.
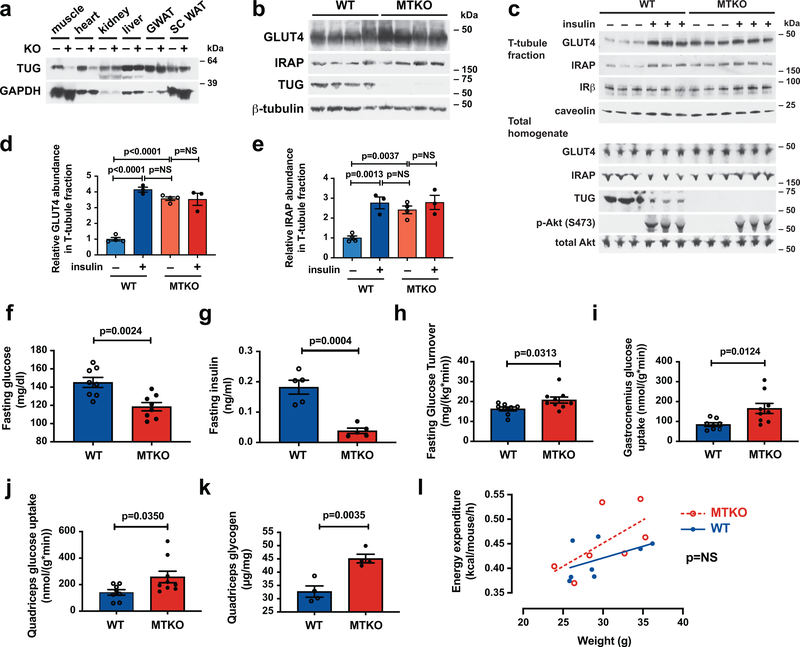
TUG was effectively deleted in skeletal muscle, and this did not affect GLUT4 or IRAP abundances (Fig. 1a,b; Extended Data Fig. 1a,b). TUG deletion caused the dramatic translocation of GLUT4 and IRAP to T-tubule membranes in quadriceps muscle (Fig. 1c, Extended Data Fig. 1c,d). The magnitude of this effect was indistinguishable from that of insulin stimulation in wildtype (WT) muscle, and insulin had no further effect in MTKO muscle (Fig. 1d,e). These results are similar to those previously observed after TUG knockdown in 3T3-L1 adipocytes and in quadriceps muscle of UBX mice8,16,18. In WT mice, insulin stimulation caused an ~80% decrease in the abundance of intact TUG (Fig. 1c, Extended Data Fig. 1e), consistent with TUG cleavage and similar to previous data in muscle and adipose tissues5,18. TUG destruction can account for the insulin-stimulated dissociation of GLUT4 from intact TUG observed in skeletal muscle31 and 3T3-L1 adipocytes5,12. TUG deletion had no effect on insulin-stimulated Akt phosphorylation (Fig. 1c), similar to results in 3T3-L1 adipocytes7. The data support the idea that intact TUG is required for intracellular sequestration of GSVs in muscle, as it is in 3T3-L1 adipocytes8,16.
Fig. 1. TUG deletion in muscle causes GLUT4 translocation and increased glucose uptake during fasting, without affecting energy expenditure, in mice fed regular chow.
a,b, Immunoblots were done as indicated on Muscle TUG Knockout (MTKO) and wildtype (WT) control mice. GWAT and SC WAT indicate gonadal and subcutaneous white adipose tissue, respectively. c, 12-week-old MTKO and WT mice were fasted 4–6 h, treated with intraperitoneal (IP) injection of insulin and glucose, or saline control, then euthanized 30 min. after IP injection. Quadriceps muscles were homogenized and T-tubule -enriched membrane fractions were purified and immunoblotted, as indicated. d,e, Abundances of GLUT4 (d) and IRAP (e) in T-tubule -enriched membranes were quantified using densitometry. N=4 in each unstimulated group, and N=3 in each stimulated group. f, Fasting glucose concentrations were measured in 10-week old mice. N=8 in each group. g, Fasting insulin concentrations were measured in 16-week old mice. N=5 in each group. h–j, Tracer infusions were used to measure whole-body glucose turnover (h), gastrocnemius muscle glucose uptake (i), and quadriceps muscle glucose uptake (j) in fasting 19-week-old mice. N=8 WT and 9 MKTO mice. k, Glycogen content was measured in quadriceps muscles of mice fasted for 2 h. N=4 in each group. l, Energy expenditure was measured using indirect calorimetry in 17-week old mice and linear regressions are plotted. N=8 WT and 7 MTKO mice. All data are presented as mean ± SEM of biologically independent samples, analyzed using ANOVA (d,e), two-tailed t-tests (f–k), and ANCOVA (l).
Like UBX mice18, MTKO mice had reduced fasting plasma glucose and insulin concentrations, compared to WT controls (Fig. 1f,g). Body weight and composition and heart weight was unchanged (Extended Data Fig. 1f–i). HOMA-IR, calculated from the product of fasting glucose and insulin concentrations, was markedly reduced in MTKO mice (Extended Data Fig. 1j,k). Dynamic measurements of glucose flux showed that in fasting MTKO mice, compared to controls, whole-body glucose turnover was increased by 27% (Fig. 1h, Extended Data Fig. 1l). Muscle-specific glucose uptake was increased 2.0-fold in gastrocnemius and 1.8-fold in quadriceps of MTKO mice, compared to controls (Fig. 1i,j). Heart-specific glucose uptake was unchanged (Extended Data Fig. 1m). In MTKO mice fasted for 2 h, quadriceps glycogen content was increased (Fig. 1k). We conclude that TUG deletion in muscle causes a marked increase in cell surface-targeting of GLUT4, and enhances muscle-specific glucose uptake and whole-body glucose turnover during fasting. These phenotypes are broadly similar to those observed in UBX mice18, and support the concept that TUG is required for intracellular sequestration of GLUT4 in muscle.
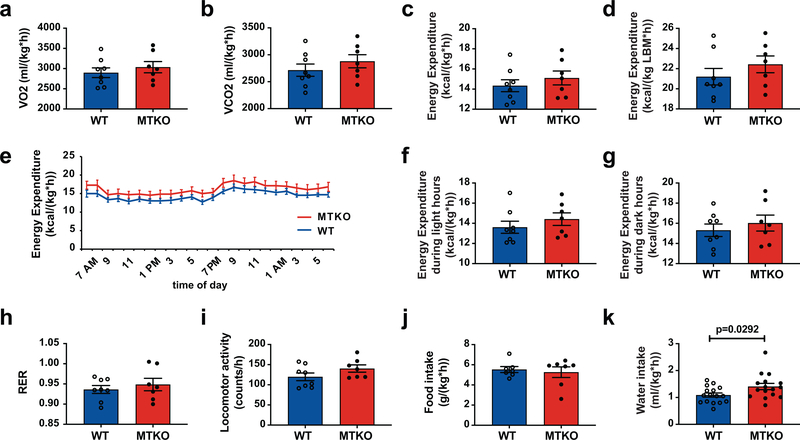
If the increased energy expenditure observed in UBX mice results from cell surface targeting of a GSV protein, then energy expenditure should be increased in the MTKO mice. This was not observed (Fig. 1l and Extended Data Fig. 2a–d). When light and dark hours were analyzed separately, energy expenditure remained similar in MTKO and WT mice (Extended Data Fig. 2e–g). There was no change in respiratory exchange ratio (RER), locomotor activity, or food intake (Extended Data Fig. 2h–j). In UBX mice, constitutive TUG cleavage results in increased targeting of IRAP to T-tubules, with consequently accelerated vasopressin degradation and increased water intake14. Water intake was increased by 29% in MTKO mice, compared to WT controls (Extended Data Fig. 2k), consistent with the observation that IRAP is translocated to T-tubule membranes (Fig. 1c,e). We conclude that in MTKO muscles lacking TUG, similar to UBX muscles with constitutive TUG cleavage, GSV cargoes are targeted to the cell surface and affect glucose metabolism and physiology. Yet, increased energy expenditure was observed only in UBX mice, and not in MTKO mice. The data suggest that the energy phenotype does not result from a GSV cargo protein, but that it may result from a TUG cleavage product.
On a high-fat diet, muscle TUG knockout mice have reduced energy expenditure.
When fed a high-fat diet (HFD), MTKO mice gained weight more rapidly than WT controls, and developed increased fasting plasma glucose and insulin concentrations (Fig. 2a–e and Extended Data Fig. 3a–c). This phenotype was opposite to that observed in regular chow (RC) -fed animals (Fig. 1f,g and Extended Data Fig. 1i,j). After 3 weeks on a HFD, MTKO mice had increased body weight, fat mass, and lean mass, with increased percent fat mass and decreased percent lean mass (Fig. 2f–j). By contrast, HFD-fed UBX mice do not gain excess weight compared to WT controls, despite a 14% increase in food intake, and they continue to have reduced fasting plasma glucose concentrations18. To study energy expenditure in MTKO mice, we used younger animals in order to make measurements prior to the development of significant differences in body weight and composition. In metabolic cages, HFD-fed MTKO mice had an overall 9% reduction in energy expenditure, which was more marked during the light (resting) hours than during the dark hours (Fig. 2k–n, Extended Data Fig. 3d–f). Similar changes were observed in VO2 and VCO2, and there was no change in RER (Extended Data Fig. 3g–k). No difference in locomotor activity or food intake was observed (Extended Data Fig. 3l–n). Intriguingly, on a per mouse basis, HFD-fed MTKO animals had a 22% increase in food intake (Extended Data Fig. 3o). When normalized to lean mass, energy expenditure was decreased by 6% in MTKO mice (Fig. 2o, Extended Data Fig. 3p,q). By contrast, UBX mice have a 13% increase in energy expenditure, based on previous data18 which are here normalized to lean mass (Fig. 2p). In linear regression analyses, we observed these opposite differences in MTKO and UBX per mouse energy expenditure across a range of body weights (Fig. 2q). We also observed increased weight gain and gonadal white adipose tissue (GWAT) mass in RC-fed male and female MTKO mice housed under thermoneutral conditions (Extended Data Fig. 4a–d). Thus, muscle-specific TUG deletion causes decreased energy expenditure, and muscle-specific constitutive TUG cleavage causes increased energy expenditure, despite both models having similar effects due to cell surface targeting of GSV cargoes. The concordant and divergent phenotypes in the two models are summarized in Table 1, which incorporates previous results from UBX mice14,18. The data support the idea that effects on energy expenditure do not result from translocation of a GSV cargo protein, but are due to action of a TUG cleavage product.
Fig. 2. On a high-fat diet, muscle TUG knockout mice have increased weight gain and reduced energy expenditure, which is opposite to effects in mice with constitutive TUG cleavage.
a, Mice were fed a high-fat diet (HFD) beginning at 15-weeks of age and weight gain was measured over time. N=10 WT and 12 MTKO mice. b,c, Weights of mice were measured before and after HFD feeding for 17 days. N=11 WT and 14 MTKO mice. d,e, Fasting plasma glucose and insulin concentrations were measured after 17 days on a HFD, in 17.5-week old mice. N=7 WT and 10 MTKO mice. f–j, Body weights and composition were measured in 18-week old mice that had been fed a HFD for 3 weeks. N=7 WT and 6 MTKO mice. k–n, Energy expenditure was measured in 14-week-old HFD-fed mice and is shown over time (k), during light hours (l), during dark hours (m), and averaged over 24 h (n). N=7 in each group. o, Energy expenditure in 14-week-old HFD-fed MTKO and WT mice was normalized to lean mass. N=7 in each group. p, Energy expenditure in 14-week-old HFD-fed UBX and WT mice was normalized to lean mass. N=6 in each group. q, Energy expenditure in UBX, WT, and MTKO mice are plotted as linear regressions vs. body weight. N=6 UBX, 13 WT, and 7 MTKO mice. Data are presented as mean ± SEM of biologically independent samples, analyzed using two-tailed t-tests (a–p), and ANCOVA (q). *p<0.05, **p<0.01, ***p<0.001.
Table 1. Summary of concordant and divergent phenotypes in mice with muscle-specific, constitutive TUG cleavage or muscle-specific TUG deletion.
Phenotypes for UBX mice are from Löffler, M.G., et al., J Biol Chem 2013 (ref. 18), and Habtemichael E.N., et al., J Biol Chem 2015 (ref. 14). Data for MTKO mice are presented here in Figs. 1 and 2 and in Extended Data Figs. 1–3.
| Constitutive TUG cleavage (UBX mice) | TUG deletion (MTKO mice) | |
|---|---|---|
| Concordant phenotypes attributable to translocation of vesicles containing GLUT4 and IRAP | ||
| Fasting plasma glucose | Reduced | Reduced |
| Fasting plasma insulin | Reduced | Reduced |
| Whole body glucose turnover (fasting) | 17% increase | 27% increase |
| Muscle glucose uptake | 2.7-fold increase in fasting glucose uptake in quadriceps; similar to insulin effect in WT; no further insulin effect in UBX | 2.0-fold increase fasting glucose uptake in gastroc.; 1.8-fold increase in fasting glucose uptake in quadriceps |
| Muscle glycogen | 1.7–2.1 -fold increase in quadriceps, gastroc., TA after 4–6 h fast | 38% increase in quadriceps after 2 h fast |
| T-tubule GLUT4 abundance | 3.6-fold increase during fasting | 3.6-fold increase during fasting, similar to insulin effect in WT; no further insulin effect in MTKO |
| T-tubule IRAP abundance | 3.2-fold increase during fasting | 2.4-fold increase during fasting; similar to insulin effect in WT; no further insulin effect in MTKO |
| Water intake | 55% increase | 29% increase |
| Divergent phenotypes attributable to action of the TUG C-terminal cleavage product | ||
| Energy expenditure | 12% increase | No change |
| Food intake | 14% increase (p=0.06) | No change |
| Energy expenditure per lean body mass | 14% increase | No change |
| Body weight after HFD | No change | Increased |
| Fat mass after HFD | No change | Increased |
| Fasting glucose after HFD | Reduced | Increased |
| Energy expenditure on HFD |
9% increase | 9% decrease |
| Food intake on HFD | 14% increase | No change |
| Energy expenditure per lean body mass on HFD | 13% increase | 6% decrease |
The TUG cleavage product acts with PPARγ–PGC-1α to control oxidative metabolism.
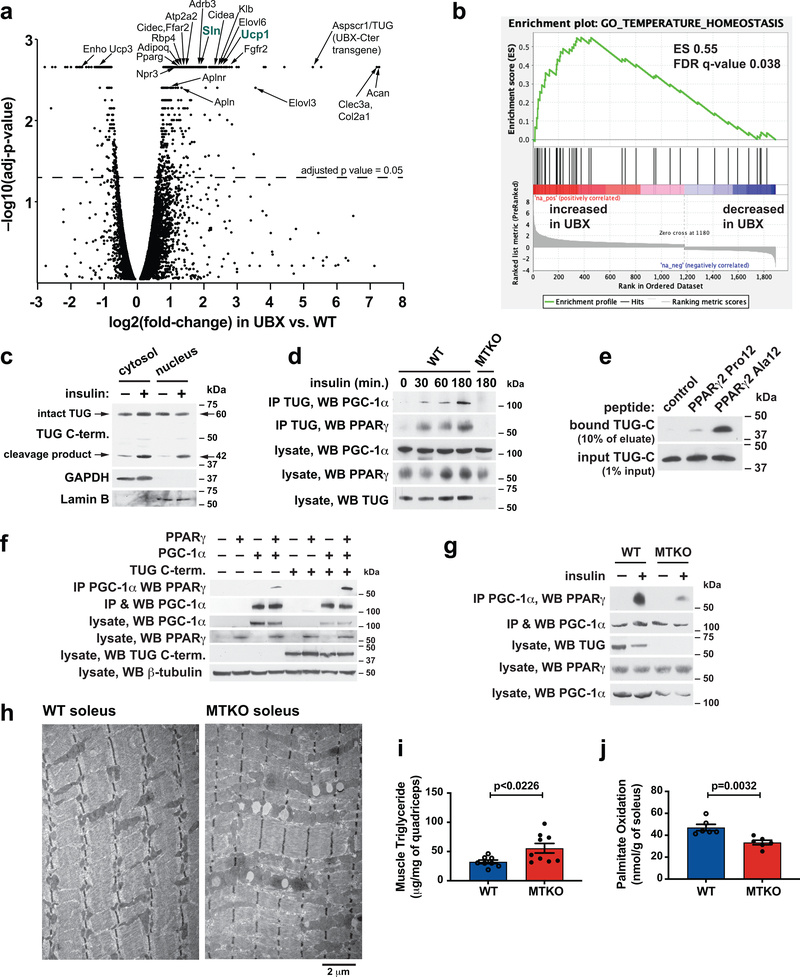
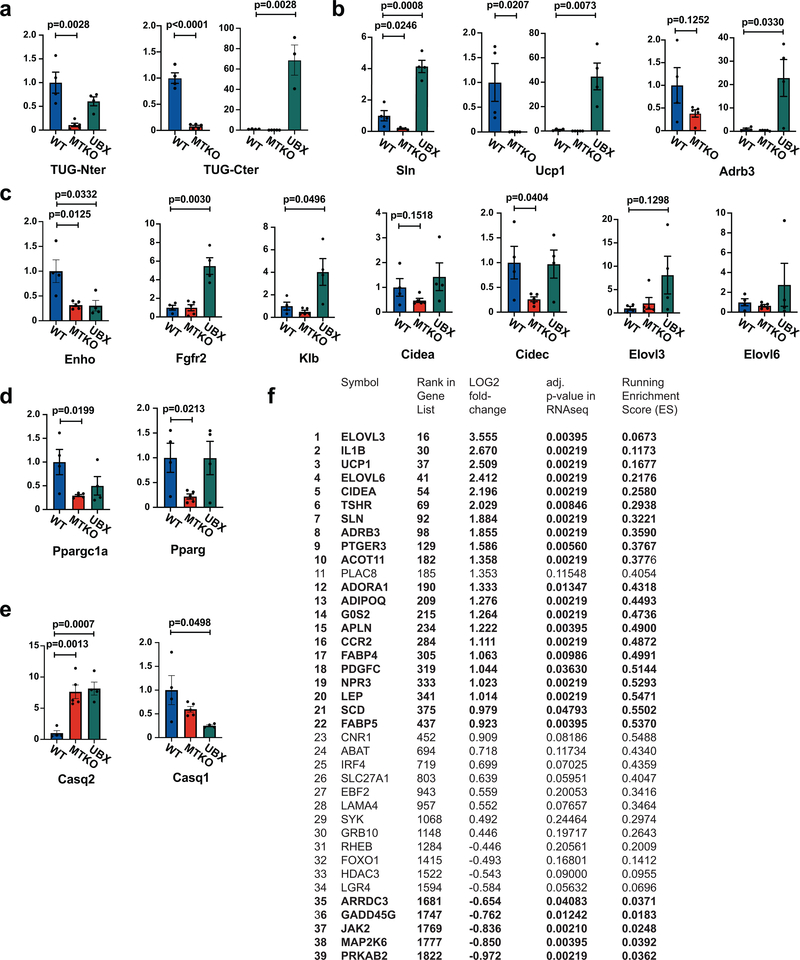
To learn how gene expression is altered in UBX vs. WT muscles, we analyzed transcriptomes by deep sequencing of RNA (RNA-seq). In UBX muscles, RNAs encoding several proteins for oxidative metabolism and thermogenesis were upregulated (Fig. 3a, Supplementary Table 1), including sarcolipin (Sln, 3.7-fold increase), Ucp1 (5.7-fold), β3-adrenergic receptor (Adrb3, 3.6-fold), and Atp2a2 (encoding SERCA2b, 2.6-fold)32–35. Quantitative PCR (qPCR) showed that there were decreased abundances of Sln and Ucp1 transcripts, and a trend toward decreased Adrb3 transcript, in MTKO muscles, and also confirmed the increased abundances of these transcripts in UBX muscles (Extended Data Fig. 5a,b). Effects on the abundances of several other differentially-regulated transcripts relevant to energy expenditure, lipid metabolism, and calcium handling were confirmed by qPCR (Extended Data Fig. 5c–e). Some of these (Fgfr2, Klb, Cidec, Elovl3) had changes consistent with possible regulation by the TUG C-terminal product, whereas others (Enho, Casq2) were altered similarly in MTKO and UBX muscle and may be regulated by translocation of a GSV cargo protein. Pathway analyses identified PPAR signaling (Supplementary Table 2) and temperature homeostasis (Fig. 3b and Extended Data Fig. 5f) as enriched programs of gene expression in UBX muscles, compared to WT controls.
Fig. 3. The TUG C-terminal cleavage product acts with PPARγ and PGC-1α to control oxidative metabolism.
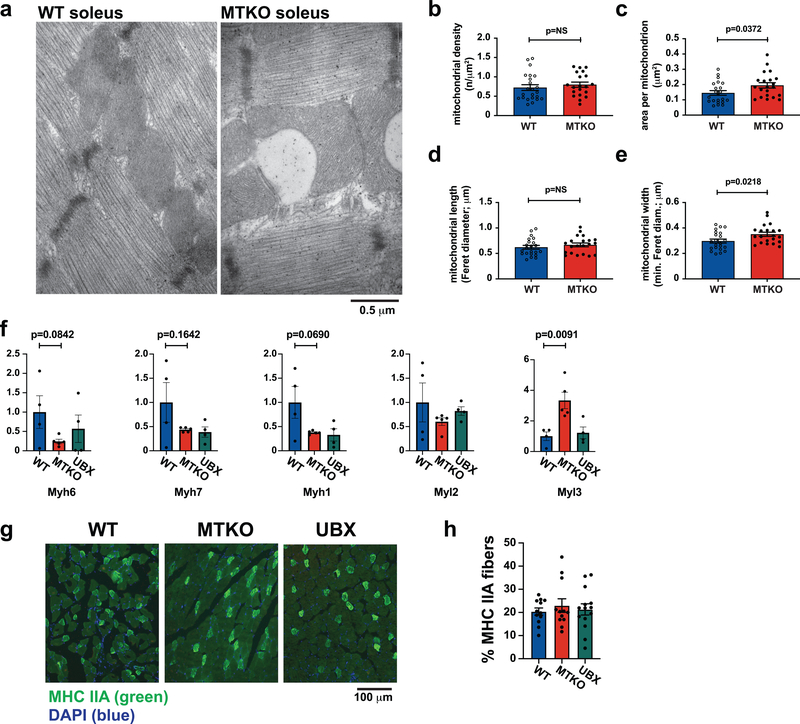
a, RNA-seq was used to analyze transcriptomes in quadriceps muscles of fasting UBX and WT mice (N=3 each), and changes in transcript abundance are presented using a volcano plot (see Methods). Identities of selected transcripts are indicated. b, Differentially expressed genes identified by RNA-seq were analyzed using Gene Set Enrichment Analysis, which revealed a significant enrichment for genes regulating temperature homeostasis. c, Wildtype mice were treated by intraperitoneal (IP) injection of insulin-glucose solution, or saline control, then cytosol and nuclear fractions were isolated from quadriceps muscle and immunoblotted as indicated. d, Mice were treated by IP injection of insulin-glucose solution, then lysates were prepared from hindlimb muscles at the indicated times. Immunoprecipitations were done using an antibody to the TUG C-terminus, and eluted proteins were immunoblotted as indicated. e, Peptides corresponding to the N-terminus of PPARγ2, containing Pro12 or Ala12, were immobilized on beads, then incubated with recombinant TUG C-terminal product. Immunoblots were done as indicated. f, Proteins were expressed by transfection, PGC-1α was immunoprecipitated, and immunoblots were done as indicated. g, The indicated mice were treated by IP injection of insulin-glucose, or saline control, then euthanized after 3 h. Lysates were prepared from quadriceps, PGC-1α was immunoprecipitated, and immunoblots were done as indicated. h, The indicated mice were fed a HFD for 2.5 weeks, then soleus muscles were imaged using electron microscopy. i, Intramyocellular triglyceride was measured in quadriceps from HFD-fed mice. N=8 WT and 9 MTKO mice. j, Palmitate oxidation was measured ex vivo in soleus muscles from mice raised at thermoneutrality. N=6 in each group. All data are biologically independent samples, analyzed using two-tailed t-tests and presented as mean ± SEM (i,j).
Previous data show that when overexpressed in transfected cells, TUG accumulates in the nucleus36. We observed the endogenous TUG C-terminal cleavage product in nuclear fractions of insulin-stimulated muscles (Fig. 3c and Extended Data Fig. 6a). The TUG product bound PPARγ and its cofactor, PGC-1α, in transfected cells, and we used recombinant proteins to show that these interactions are direct (Extended Data Fig. 6b–f). Immunoprecipitation of endogenous TUG copurified both PGC-1α and PPARγ from insulin-treated muscles, and not from unstimulated muscles, demonstrating that these interactions occur physiologically after insulin stimulation (Fig. 3d). The TUG product bound to peptides corresponding to the N-termini of PPARγ1 and PPARγ2 (Extended Data Fig. 6g). PPARγ2 contains a polymorphism, Pro12Ala, that predicts diabetes risk37. We observed increased binding of the TUG product to PPARγ2 peptides containing the protective Ala12 residue, compared to the Pro12 residue encoded by the risk allele (Fig. 3e and Extended Data Fig. 6h,i). The data suggest that increased binding of the TUG C-terminal product to PPARγ2 Ala12 may help to recruit PGC-1α and possibly other factors, resulting in enhanced expression of genes for oxidative metabolism and thermogenesis38.
To test whether the TUG product stabilizes a PPARγ–PGC-1α complex, we immunoprecipitated PGC-1α and immunoblotted PPARγ using transfected cells. Fig. 3f shows that a greater number of PPARγ–PGC-1α complexes were present when the TUG product was coexpressed. Conversely, fewer PPARγ–PGC-1α complexes were present after insulin stimulation in muscle from MTKO mice, compared to WT controls (Fig. 3g and Extended Data Fig. 6j). PGC-1α proteins have limited stability39–41, and insulin stimulation increased total PGC-1α protein abundance in muscle in WT, but not MTKO, mice (Extended Data Fig. 7a–f). Consistent with reduced PGC-1α action in MTKO mice, electron microscopy revealed enlarged, swollen mitochondria with disorganized cristae in muscles of HFD-fed MTKO animals, compared to WT controls; lipid droplets were also observed and triglyceride content was increased in muscle lacking TUG (Fig. 3h,i, and Extended Data Fig. 8a–e). Ex vivo palmitate oxidation was reduced in soleus muscle of MTKO mice, compared to controls (Fig. 3j). No large effects on muscle fiber type were detected by qPCR of myosin isoforms or by immunohistochemistry (Extended Data Fig. 8f–h). The data show that the TUG product binds and stabilizes a PGC-1α–PPARγ complex, increases overall PGC-1α abundance, and promotes fatty acid oxidation.
Insulin acts through TUG to stimulate the production of thermogenic proteins.
We next examined whether the TUG C-terminal product regulates the abundance of sarcolipin, which mediates thermogenesis in muscle by uncoupling ATP hydrolysis from Ca2+ transport into the sarcoplasmic reticulum33. Insulin stimulated an increase in sarcolipin abundance in WT muscle, which was abrogated by TUG deletion (Fig. 4a,b, and Extended Data Fig. 9a). Conversely, in UBX muscle, sarcolipin was increased during fasting (Fig. 4c and Extended Data Fig. 9b), consistent with Sln transcript abundance (Fig. 3a and Extended Data Fig. 5b) and calorimetry18. After insulin stimulation, the TUG product associated with the sarcolipin promoter (Fig. 4d). Insulin also recruited PGC-1α and, to a lesser extent, PPARγ to this promoter, and this effect was largely independent of TUG (Extended Data Fig. 9c,d). The TUG C-terminal product enhanced the action of PPARγ and PGC-1α to activate transcription from the sarcolipin promoter (Fig. 4e). In muscle of mice fed a HFD, TUG processing was attenuated and the abundance of the TUG protease, Usp25m, was reduced (Fig. 4f,g and Extended Data Fig. 9e,f). As well, TUG binding at the sarcolipin promoter was reduced (Fig. 4h) and the insulin-stimulated increase in sarcolipin protein abundance was greatly attenuated (Fig. 4i,j). We conclude that the effects on energy expenditure we observe are mediated, in part, by action of the TUG product to increase sarcolipin abundance, and that this effect is inhibited when TUG cleavage is impaired in HFD-induced insulin resistance.
Fig. 4. Insulin acts through TUG to enhance production of the thermogenic protein, sarcolipin.
a, Mice were treated with IP insulin-glucose, or saline control, and hindlimb muscles were immunoblotted at the indicated times. b, Studies similar to (a) were repeated in mice raised at 30° C, and sarcolipin abundance was quantified and plotted. N=7 WT unstimulated, 8 WT insulin-stimulated, 3 MTKO unstimulated, and 5 MTKO insulin-stimulated mice. c, The indicated mice were fasted for 4–6 h, then hindlimb muscles were immunoblotted to quantify sarcolipin abundance. N=4 WT, 5 UBX, and 5 MTKO mice. d, The indicated mice were treated with insulin, and hindlimb muscles were used for chromatin immunoprecipitation with a TUG C-terminus antibody. PCR detected sequences at the indicated sites upstream of the sarcolipin (Sln) transcription start site. e, A reporter construct containing the sarcolipin promoter was used to measure transcription activity in transfected in 293 cells. N=3 in each group. f, WT mice were fed regular chow (RC) or a high-fat diet (HFD) for 3 weeks, then treated for 30 min. with IP insulin-glucose, or saline control. Quadriceps muscles were immunoblotted to detect intact TUG and the 42 kDa C-terminal product. The ratio of cleavage product to intact TUG is plotted, normalized to basal control samples. N=3 in each group. g, Usp25m abundance was measured by densitometry of immunoblots from replicate samples, as in (f). Data are plotted relative to RC-fed controls. N=5 in each group. h, RC- or HFD-fed mice were fasted, treated with IP insulin-glucose for 30 min. as indicated, then sacrificed. Chromatin immunoprecipitation was done in hindlimb muscles using a TUG C-terminus antibody and PCR to detect sites upstream of the sarcolipin transcription start site. i, RC- or HFD- fed mice were treated with IP insulin-glucose or saline control, then quadriceps muscles were immunoblotted as indicated. j, Replicates of data in (i) were quantified. N=3 RC unstimulated, 4 RC insulin-stimulated, 4 HFD-fed unstimulated, and 5 HFD-fed insulin-stimulated mice. All data are presented as mean ± SEM of biologically independent samples, analyzed using a two-tailed t-tests (g) or ANOVA with adjustment for multiple comparisons (b,c,e,f,j).
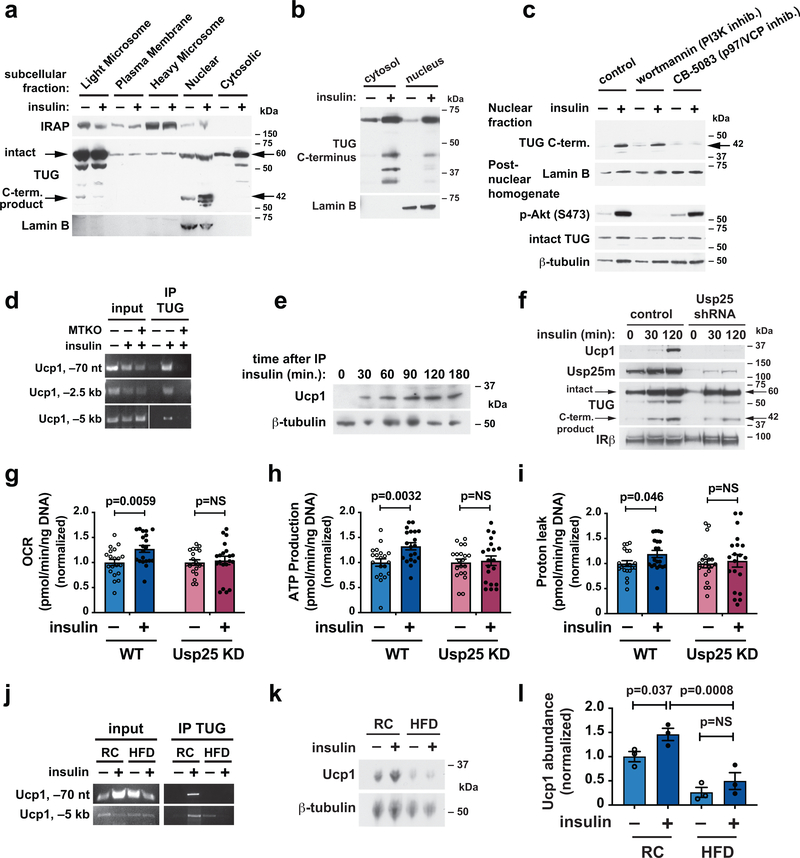
To learn whether the TUG C-terminal product acts by a similar mechanism in adipocytes, we used both primary adipocytes and roscovitine-treated 3T3-L1 cells, which adopt a brown adipocyte -like phenotype42. In adipocytes, as in muscle, insulin caused entry of the TUG product into the nucleus (Fig. 5a,b). We hypothesized that the TUG cleavage product may be extracted from a protein complex by the action of p97/VCP ATPases, which bind TUG36,43,44. This process can expose a nuclear localization signal36, so that no additional insulin signal may be required for nuclear translocation. Supporting this idea, the insulin-stimulated entry of the TUG C-terminal product into the nucleus was independent of PI3K, but was blocked by the p97 inhibitor CB-5083 (Fig. 5c). Chromatin immunoprecipitation showed that after insulin stimulation, the TUG C-terminus associates with the Ucp1 promoter in muscle (Fig. 5d). In adipose tissue, insulin stimulated an increase in Ucp1 protein abundance (Fig. 5e); this effect was also observed in roscovitine-treated 3T3-L1 adipocytes, and was abrogated by shRNA-mediated depletion of the TUG protease, Usp25m (Fig. 5f). To corroborate these data, we studied mitochondrial function by measuring cellular oxygen consumption rates. Insulin caused an increase in cellular O2 consumption, ATP production, and uncoupled respiration in control cells, and these effects were abrogated in cells in which TUG cleavage was inhibited by depletion of Usp25m (Fig. 5g–i). Previous data show that in mice fed a HFD, TUG proteolytic processing is reduced in adipose5. Accordingly, in HFD-fed mice, binding of TUG at the Ucp1 promoter was decreased and the insulin-stimulated increase in Ucp1 abundance was abolished (Fig. 5j–l). The data support the idea that the TUG C-terminal cleavage product acts in adipose, as it does in muscle, to enhance the expression of thermogenic proteins, and that this effect is reduced because of attenuated TUG cleavage in HFD-induced insulin resistance.
Fig. 5. Insulin acts through TUG to enhance production of Ucp1 in adipocytes.
a, Subcellular fractions from basal and insulin-treated 3T3-L1 adipocytes were immunoblotted as indicated. b, Mice were fasted, treated by IP injection of insulin-glucose solution or saline control for 30 min., then gonadal white adipose tissue (GWAT) was isolated. Cytosolic and nuclear fractions were immunoblotted as indicated. c, 3T3-L1 adipocytes were serum starved, treated with wortmannin or CB-5083, then stimulated with insulin as indicated. Homogenates and nuclear fractions were immunoblotted as indicated. d, WT and MTKO mice were fasted, treated by IP insulin-glucose or saline control for 30 min., then hindlimb muscles were isolated. Chromatin immunoprecipitations were done using an antibody to the TUG C-terminus. PCR detected sequences at the indicated locations upstream of the Ucp1 transcription start site. e, Mice were treated with IP insulin-glucose as indicated and GWAT was immunoblotted as shown. f, Control 3T3-L1 adipocytes and cells containing a shRNA to deplete Usp25 protease were differentiated using roscovitine to induce a brown-like phenotype, then stimulated with insulin and immunoblotted as indicated. g–i, 3T3-L1 adipocytes as in (f) were treated with or without insulin for 3 h, then used for analyses of mitochondrial function. Oxygen consumption rate (OCR) (g), and rates of ATP production (h) and proton leak (i) were normalized to measurements in unstimulated cells. N=20 independent measurements in each group. Data are presented as mean ±SEM and analyzed using a two-tailed t-test. j, Chromatin immunoprecipitation was done as in (d) using GWAT from mice maintained on regular chow (RC) or fed a high-fat diet (HFD) for 3 weeks. Mice were treated with insulin-glucose or saline for 30 min. as indicated prior to euthanasia. k, Immunoblots were done on GWAT from RC or HFD-fed mice, treated prior to euthanasia with IP saline or glucose-insulin solution, as indicated. l, Quantification of relative Ucp1 protein abundances in replicates of the experiment shown in (k). Data are presented as mean ± SEM of biologically independent samples (N=3 in each group), analyzed using ANOVA with adjustment for multiple comparisons.
The TUG C-terminal cleavage product is degraded by an Ate1-dependent mechanism.
The stability of C-terminal cleavage products is controlled by N-degron (formerly, “N-end rule”) pathways, which commonly target proteins for degradation by recognizing their N-terminal residues45. We considered that a particular N-degron pathway, requiring the Ate1 arginyltransferase, may regulate the TUG product. Acute, whole-body deletion of Ate1 in mice results in increased energy expenditure, reduced fat mass, and upregulation of Ucp1 in adipose tissue46. Ate1 attaches an arginine to specific N-terminal residues to create a degradation signal, but proteins that are stabilized upon Ate1 deletion to enhance energy expenditure remain unknown. The TUG product contains an N-terminal serine residue (S165 of intact TUG). Serine is not known to be a physiologic substrate of Ate1, and we used ubiquitin fusion proteins to test whether the TUG product is subject to this pathway (Fig. 6a). Deubiquitylase-mediated cleavage of such fusions generates the C-terminal product in an insulin- and cell type- independent manner. As controls, we used fusions that contained mutations predicted to stabilize, destabilize, or confer Ate1-dependent stability upon the TUG product. We expressed these fusions in WT and Ate1-deficient (KO) murine embryonic fibroblasts (MEFs) and studied effects on TUG product abundance.
Fig. 6. The TUG C-terminal cleavage product stabilizes PGC-1α and is degraded by an Ate1-dependent mechanism.
a, A diagram is shown to indicate how the TUG C-terminal product is produced with different residues at its N-terminus. b, The indicated constructs were stably expressed in wildtype (WT) and Ate1 knockout (KO) MEFs using retroviruses. Lysates were immunoblotted as indicated. c, WT and Ate1 KO cells expressing the wildtype TUG C-terminal product were treated with cycloheximide for the times indicated and samples were immunoblotted. d, HeLa cells were transfected to express PGC-1α and PPARγ, with or without the TUG C-terminal product (beginning with Met), as indicated. Cycloheximide was added for the times indicated and samples were immunoblotted. e, PGC-1α was expressed, with or without coexpressed HA-Ub-TUG C-term. (WT) fusion protein, in WT and Ate1 KO cells, as indicated. Cycloheximide was added for the times indicated and samples were immunoblotted. The approximate PGC-1α half-life for each condition is indicated at bottom.
In Ate1 KO MEFs, compared to WT control cells, there was marked accumulation of the wildtype TUG product, as shown in Fig. 6b (compare lane 4 to lane 3; numbering at bottom). Immunoblots of HA-tagged ubiquitin showed that the constructs were expressed at similar levels, implying that the product accumulates because of slowed degradation, not increased production. The Ser165 residue was not fully destabilizing in WT control MEFs, because TUG products containing an N-terminal Asp residue, a known Ate1 substrate, did not accumulate; this is shown most clearly on a darker exposure of the TUG immunoblot (second panel, compare lanes 3 and 9). As well, TUG products containing a destabilizing N-terminal Leu residue did not accumulate, regardless of whether these fusions were expressed in WT or Ate1 KO MEFs (lanes 7, 8). The accumulation of the wildtype TUG product in Ate1 KO MEFs was similar to that of TUG products containing a stabilizing N-terminal Met residue, produced by the S165M mutant (lanes 4, 5, 6). The data support the idea that Ate1 controls the main pathway for degradation of the wildtype TUG C-terminal product. Accumulation of the TUG C-terminal product consistently caused an increase in the abundance of endogenous, intact TUG, which may reflect the formation of oligomeric complexes36. To further test the effect of Ate1 deletion on TUG C-terminal product stability, we treated cells acutely with cycloheximide to block protein synthesis, and followed the disappearance of the TUG product over time. In these “cycloheximide chase” experiments, the half-life of the TUG product was greatly prolonged in Ate1 KO MEFs, compared to control cells (Fig. 6c). The data show that Ate1 deletion increased the half-life of the wildtype TUG product from ~2 h to ≥16 h. We conclude that the stability of the TUG C-terminal product is regulated by an Ate1-dependent degradation pathway.
To test whether the half-life of PGC-1α is prolonged by coexpression of the TUG product, we used transfected cells. Cycloheximide chase experiments showed that the half-life of PGC-1α in cells not expressing the TUG product was ~12 min., similar to previous results39–41, and this half-life was substantially prolonged (to ~60 min.) by coexpression of the TUG product (Fig. 6d). These data are consistent with results, above, showing that the TUG product stabilizes PGC-1α-PPARγ complexes and controls overall PGC-1α abundance in muscle (Fig. 3f,g, and Extended Data Fig. 7a–d). To test whether Ate1 may regulate PGC-1α stability, we performed cycloheximide chase experiments using WT or Ate1-deficient MEFs. Ate1 deletion had a small effect to stabilize PGC-1α in cells not containing the TUG product, and coexpression of the TUG product both stabilized PGC-1α in WT cells and conferred a marked increase in PGC-1α stability in Ate1 KO cells (Fig. 6e). Thus, not only does the TUG C-terminal product bind and stabilize PGC-1α, but it also confers Ate1-dependent stability upon PGC-1α. Together with previous results46 and data above, we conclude that an Ate1-regulated N-degron pathway acts through the TUG C-terminal cleavage product and bound PGC-1α to control overall energy expenditure.
Discussion
Here we describe a novel action of insulin to stimulate energy expenditure. This action is mechanistically coupled to the stimulation of glucose uptake, insofar as insulin-stimulated TUG endoproteolytic cleavage mediates both effects (Fig. 7). Previous data support a model in which insulin triggers TUG cleavage to release GSVs that are trapped at the Golgi matrix5,11,18. The N-terminal cleavage product, TUGUL, links these vesicles to kinesin motors to promote microtubule-based GLUT4 movement to the plasma membrane in adipocytes5,20,47. Data here show that the C-terminal TUG cleavage product enters the nucleus, binds and stabilizes a PGC-1α–PPARγ complex, and promotes the expression of genes to enhance oxidative metabolism. Specific genes that are regulated include sarcolipin and Ucp1, in muscle and adipose, respectively. These thermogenic proteins can account for the divergent energy expenditure phenotypes observed in UBX and MTKO mice. The TUG cleavage product has a limited half-life and controls PGC-1α stability, so that both proteins are subject to an Ate1-regulated protein degradation pathway. Thus, the thermogenic effect resulting from the mechanism we describe normally occurs after nutrient intake and has limited duration. This duration is likely hours-long, and repeated insulin stimulation of this pathway can account for ongoing effects to increase total energy expenditure.
Fig. 7. A model for the coordinated regulation of glucose uptake and energy expenditure.
Insulin acts through a phosphaditylinositol-3-kinase -independent pathway to stimulate site-specific TUG cleavage, which is mediated by the Usp25m protease. Cleavage releases GLUT4 from the Golgi matrix. The N-terminal cleavage product, TUGUL, facilitates GLUT4 translocation to the plasma membrane by linking it to kinesin motors. The TUG C-terminal cleavage product is extracted from the Golgi matrix and enters the nucleus, where it binds PPARγ and PGC-1α. The TUG product stabilizes a complex containing these proteins, which activates the transcription of genes, including sarcolipin and Ucp1, that promote oxidative metabolism and thermogenesis. This action of the TUG C-terminal product is terminated by an Ate1-dependent degradation pathway, which limits the duration of the thermogenic effect.
The thermic effect of food is a transient increase in energy expenditure that occurs after meals, which in humans lasts ~6 h and can account for ~10% of total energy expenditure48,49. The mechanism we describe can account, at least in part, for this phenomenon. Signaling upstream of TUG cleavage is independent of PI3K-Akt, so that this mechanism for insulin-regulated gene expression is complementary to those involving FOXO or SREBP proteins50. As well, this signaling pathway is proposed to contain a feed-forward circuit, so that TUG cleavage and the increased thermogenesis that results may be proportional to glycemic load5,13. Such a feed-forward circuit may also release a proportional amount of sequestered GLUT4 into a cell surface recycling pathway, as is physiologically appropriate to dispose of the glycemic load. Attenuated TUG processing may contribute to the reduced thermic effect of food observed in insulin-resistant individuals51, and to reduced expression of PGC-1α-responsive genes in type 2 diabetes38,52.
The differential binding of the TUG product to PPARγ2 Pro12 and Ala12 peptides may elucidate how these PPARγ2 variants can modulate diabetes risk37. We propose that increased binding of the TUG product to the protective Ala12 variant enhances the stability of protein complexes containing TUG, PPARγ2 and PGC-1α. Individuals having PPARγ2 Ala12 proteins may then have an increased extent or duration of food-induced thermogenesis. Such an effect may amount to only a small fraction of total energy expenditure, yet the cumulative effect over time may cause altered fat mass and diabetes risk37. Estimates of the population attributable risk of the Pro12 variant imply that it accounts for as much as 25% of type 2 diabetes in the population. Thus, understanding the mechanistic basis of this effect is important for public health. Our data imply that the TUG cleavage pathway is a major biochemical mechanism that can influence type 2 diabetes pathogenesis.
Although the TUG product caused increased the abundance of PGC-1α, its metabolic effects cannot be attributed solely to this effect. PGC-1α is increased after exercise and acts together with several proteins to increase mitochondrial biogenesis and uncoupling53–56. Transgenic mice with increased PGC-1α in muscle have increased mitochondrial density and ATP synthesis, but not increased energy expenditure57. Paradoxically, these mice are prone to fat-induced insulin resistance, reflecting increased de novo lipogenesis without increased oxidation; this is improved by exercise-induced lipid oxidation58,59. By contrast, unexercised UBX mice have increased energy expenditure and are resistant to diet-induced weight gain, and HFD-fed MTKO mice had reduced energy expenditure and increased weight gain. Possibly, the TUG C-terminal product recruits PGC-1α to specific transcription factors, such as PPARγ, and not to others that are involved in lipogenesis. We do not know whether TUG is cleaved in response to exercise. Data show that TUG is cleaved in cardiac muscle after ischemia-reperfusion60 and cleavage may be stimulated by an obscurin-TC10α pathway in skeletal muscle25. If so, this may contribute to increased PGC-1α abundance after exercise, together with enhanced fatty acid oxidation and energy expenditure, and to exercise-induced glucose uptake.
Our data support the idea that TUG regulates the insulin-responsive vesicles containing GLUT4, GSVs, and that this is a major site of insulin action in muscle. TUG deletion caused a dramatic translocation of GLUT4 and IRAP to T-tubule membranes in unstimulated muscle. This mimicked the effect of insulin stimulation, and insulin had no further effect on GLUT4 targeting in muscles lacking TUG. These results are consistent with live cell imaging of adipocytes, which showed that TUG depletion causes an increased rate of GSV exocytosis that is indistinguishable from that observed after insulin stimulation7. Live cell imaging data further show that during ongoing insulin exposure, GSV cargoes bypass a TUG-regulated compartment and return to the cell surface directly from endosomes. This effect is due to insulin signaling through Akt and Rab proteins, which redirects the targeting of endocytosed GLUT41,7,61. Thus, the data support a role for TUG cleavage at the transition from fasting to fed states. In muscles lacking TUG, muscle-specific glucose uptake was increased ~2-fold during fasting, whereas T-tubule GLUT4 abundance was increased ~3.6-fold. This difference may reflect effects of other transporters (e.g. GLUT1), limitations in the assays that were used, or potential effects of insulin on other regulatory mechanisms62. Of note, in UBX mice, quadriceps-specific glucose uptake was increased 2.7-fold during fasting; the magnitude of this effect was similar to that of insulin, and no further effect of insulin to increase uptake was observed in hyperinsulinemic clamps18. Thus, the data support the concept that insulin action through TUG accounts for a large fraction of its overall effect to stimulate glucose uptake in muscle.
Our data show that insulin-stimulated TUG cleavage is attenuated in muscle of HFD-fed mice. We previously observed a similar effect of HFD feeding in adipose5. In both cases, reduced TUG proteolytic processing was accompanied by reduced Usp25m protein abundance5. The data are consistent with the idea that in the setting of insulin resistance, the targeting of GLUT4 and other GSV cargoes is altered during fasting9,10. The result is that a complex containing intact TUG, Usp25m, and other proteins cannot trap GSVs at the Golgi matrix5. It seems likely that TUG cleavage and GSV mobilization are inter-dependent, and that Usp25m is destabilized when it is not incorporated into such a complex to retain the GSVs in an insulin-responsive configuration. Altered targeting of GSV proteins may occur in parallel to attenuated Akt signaling, and may account for insulin resistance not attributable to attenuated Akt signaling4. Of note, altered targeting of IRAP proteins during fasting may alter vasopressin dynamics to contribute to hypertension in the setting of insulin resistance14,30. The data presented here suggest that these alterations may also reduce energy expenditure, since the generation and subsequent action of the TUG C-terminal product is decreased. Thus, in addition to the effect of obesity to promote insulin resistance, insulin resistance may, potentially, promote obesity.
Our data imply that the Ate1-mediated degradation pathway for the TUG C-terminal cleavage product may be targeted therapeutically. Stabilization of the TUG product, and of protein complexes in which it participates, is predicted to enhance energy expenditure. This effect may be functionally similar to that of the PPARγ Ala12 variant. We do not know whether Ate1 acts directly to arginylate the TUG Ser165 residue, which is newly exposed at the N-terminus of the TUG cleavage product. Ate1 acts on acidic residues (Asp, Glu) and on oxidized Cys residues, but has not been described to act physiologically on Ser63. One possibility is that the Ser may need to be oxidized or otherwise modified to make it susceptible to Ate1-mediated arginylation. If so, this may serve as part of a regulatory mechanism to control oxidative metabolism.
Many questions remain. We do not know whether the TUG product modifies the transcriptome that is regulated by PGC-1α–PPARγ complexes. As well, the TUG product may act with other transcriptional regulators, which have not yet been defined. We also do not know whether this pathway contributes to cross-tissue effects, or if stimuli other than insulin may affect TUG cleavage to control glucose uptake and fatty acid oxidation. Nonetheless, our data implicate the TUG C-terminal product as an important regulator of energy expenditure, and suggest that further elucidation of this pathway will be significant for understanding metabolic disease.
Materials and methods
Animals.
Muscle TUG Knockout (MTKO) mice were produced using a targeting construct obtained from the NIH Knock-out Mouse Program (KOMP; CSD30881). Homologous recombination inserts loxP sites flanking exon 5 of the Aspscr1 gene, which encodes TUG protein. The construct was electroporated into 129Sv ES cells, recombination was obtained using positive-negative selection, and cells were injected into C57BL/6 blastocysts and implanted into pseudopregnant females. After germline transmission, mice were crossed with Frt transgenic deleter strain to remove the selection cassette. Mice were backcrossed to C57BL/6J (Jax stock #000664) for at least 10 generations. To delete TUG in muscle, mice homozygous for floxed TUG allele (TUGfl/fl) were bred with TUGfl/fl mice containing a MCK-Cre transgene (Tg(Ckmm-cre)5Khn/J; Jax stock #006475). Most experiments compared TUGfl/fl (WT) and TUGfl/fl + MCK-Cre (MTKO) mice, and littermates were used as controls. Of note, this strategy facilitated comparison of MTKO and UBX mice, since a MCK promoter was used on a C57BL/6J background in UBX mice18. Mice were maintained on at 12 h light/dark cycle (7 AM – 7 PM) and had ad libitum access to food and water. Mice were housed at 22° C and at 30 – 70% relative humidity, except for the described experiments done using mice house under thermoneutral conditions. For these, mice were housed at 30° C, at ~50% relative humidity, from the time of weaning using Memmert climate chambers. Male mice were used except where indicated. The standard regular chow (RC) diet was Harlan-Teklad 2018S and the high-fat diet (HFD) was Research Diets D12492 (60% kcal from fat). The Yale Institutional Animal Care and Use Committee approved all procedures.
For genotyping, genomic DNA was retrieved by overnight proteinase K digestion of tail biopsies, and was used in PCR to assess the presence of both the floxed TUG allele and the MCK-Cre transgene. For the floxed TUG allele, PCR used 39 cycles with 95° C for 30 seconds, 67° C for 30 seconds, and 72° C for 2.5 minutes, together with the following primer pair: 5’-AGGGCACTGCTCTCATTCTTTG-3’ and 5’-GCCCGCCCAGCTCAGGACAC-3’. For the MCK-Cre transgene, PCR used 38 cycles with 95° C for 30 seconds, 55.5° C for 30 seconds, and 72° C for 2 minutes with the following primer pair: 5’-GCCTTCTCTACACCTGCGG-3’ and 5’-GGTTCGCAAGAACCTGATGG-3’. Alternatively, a touchdown protocol was used (JAX Protocol 23304) together with the following primer pair: 5’-GTGAAACAGCATTGCTGTCACTT-3’ and 5’-TAAGTCTGAACCCGGTCTGC-3’. Genotyping of UBX mice (previously called mTUGUBX-Cter mice) was described previously18.
Cell Culture.
3T3-L1, MEF, HEK293, and HeLa cells were cultured in high glucose DMEM GlutaMAX medium (Invitrogen) containing 10% EquaFETAL bioequivalent serum (Atlas Biologicals), antibiotic antimycotic solution (Sigma), and plasmocin (Invivogen). The 3T3-L1, HEK293, and HeLa cells were obtained from ATCC or Zen-Bio, Inc. or from the Lodish, Toomre, or Kandror laboratories. MEFs were derived from C57BL/6J mice by serial passaging. Ate1 knockout (KO) and wildtype (WT) control murine embryonic fibroblasts (MEFs) were a gift of Dr. Anna Kashina and were described previously64. 3T3-L1 adipocytes were differentiated in 10% fetal bovine serum (FBS) or EquaFETAL serum with supplements, essentially as described previously17. Briefly, cells were allowed to become confluent for at least 2 days prior to induction of differentiation, and media was changed the day before inducing differentiation. Differentiation was induced (on day 0) with media as above supplemented with 0.25 mM dexamethasone, 160 nM insulin, and 500 mM methylisobutylxanthine, and 2 mM rosiglitazone (Cayman chemical). After 3 days, media was changed to DMEM containing FBS, as above, without supplements. Cells were typically used between days 8 and 14 after induction of adipocyte differentiation. Where indicated, 3T3-L1 adipocytes were differentiated in the presence of 5 μM roscovitine to induce brown adipocyte -like phenotype, as described42. For stable expression of exogenous proteins or Usp25 shRNA, MEFs and 3T3-L1 cells were infected with retroviruses and selected using puromycin or FACS5,65. Control cells containing empty vector were also subjected to puromycin selection, which together with plasmocin helped to maintain cells free of mycoplasma.
Metabolic and tissue analyses.
Fasting blood glucose and insulin concentrations were measured using a handheld glucometer (Onetouch UltraMini, Lifescan) and by an ultrasensitive ELISA (ALPCO, 80-INSMSU-E01). For tail vein glucose measurements, mice were fasted for 4 h in separate cages with unrestricted access to water. Mice were restrained by hand, and an 18 g needle was used to create a single venipuncture site on the tail vein. Blood was expressed through manual milking of the tail and one large drop (~10 μl) was applied to a glucometer. For measurement of simultaneous insulin and glucose concentrations, mice were fasted 4 – 6 h, then cardiac puncture was performed using isoflurane anesthesia. Mice were euthanized and used for post-mortem analysis of tissues. HOMA-IR was calculated as [glucose]*[insulin] / 22.5, where glucose is given in mmol/l and insulin is in mU/l.
Fat and lean mass were measured using 1H NMR (Minispec, Bruker Biospin)18. Rates of oxygen consumption (VO2) and carbon dioxide production (VCO2), energy expenditure, respiratory exchange ratio, locomotor activity, food consumption, and water intake were measured using CLAMS metabolic cages (Columbus Instruments), as previously18, and were analyzed as described66. Control groups for UBX and MTKO mice had similar weight and body composition and were studied at the same facility and ambient temperature; therefore these groups were pooled in Fig. 2q to increase statistical power, based on recent data67. Dynamic measurements of glucose flux were done as previously, using a Beckman Glucose Analyzer II18,68,69. Briefly, a catheter was placed in the right jugular vein 6–7 days before turnover studies were done. After a 6 h fast, 3-[3H]glucose (Perkin-Elmer Life Sciences) was infused at a rate of 0.05 μCi/min for 120 min for basal glucose turnover measurement. Next, to measure muscle-specific fasting glucose uptake, 10 μCi of 2-deoxy-D-[1-14C] glucose was infused over 20 min., without insulin and with monitoring of plasma glucose and care to minimize any increases.
Blood samples were drawn from the tail vein at 5, 15, 25, 35, 45, and 55 min after initiation of 2-deoxyglucose infusion. At the end of the study, mice were treated with intravenous pentobarbital sodium injection (150 mg/kg), tissues were quickly excised, snap frozen in liquid nitrogen, and stored at −80° C for subsequent analysis. Intracellular (6-phosphorylated) 2-deoxyglucose was measured and used to calculate tissue-specific glucose transport as described previously18,70.
Muscle glycogen and triglyceride measurements were done on quadriceps from 4–5 h fasted WT and MTKO mice. For glycogen measurements, chow-fed mice were analyzed using a glycogen assay kit (Biovision, Cat. No. K648–100). For intramyocellular triglyceride, determinations were performed on HFD-fed mice essentially as described 71. Triglycerides were extracted from 80–130 mg quadriceps tissue from each mouse. Tissues were homogenized in ice cold 2:1 chloroform:methanol, and lipids were extracted with shaking at room temperature for 3–4 hours. H2SO4 was added to ~100 mM, samples were vortexed, then centrifuged to achieve phase separation, and the organic phase was collected. Aliquots were dried and resuspended in Sekisui Triglyceride-SL reagent (Sekisui) for spectrophotometric determination of triglyceride content. The standard curve was generated using the DC-Cal multi analyte calibrator (Sekisui).
Palmitate oxidation was measured ex vivo in soleus muscles from regular chow -fed 20-week old MTKO and WT mice, which had been housed under thermoneutral conditions (30° C) from weaning. Oxidation was measured by collecting released CO2 72,73. Briefly, soleus muscles were quickly removed and attached to stainless steel clips to maintain resting tension. Muscles were preincubated in 1.5 ml Krebs-Ringer bicarbonate buffer (KRBB) containing 10 mM glucose and 0.5% BSA, pH 7.4, pregassed for 30 min with O2, at 35° C. Muscles were then transferred to new vials containing 1.5 ml of the same buffer, but with 0.1 mM palmitic acid and 0.2 μCi/mL [1-14C]palmitic acid added. NaOH (0.3 ml at 2 N) was added to an open microtube inside these vials for 14CO2 adsorption. Incubation was performed for 1 h under the same conditions. At the end of the incubation, muscles were removed, 0.5 ml of 2 N HCl was added to the KRBB, and the incubation was continued for 2 h longer at 35° C. Finally, the NaOH solution (0.3 ml) containing the adsorbed CO2 was added to scintillation vials containing scintillation cocktail for radioactivity determination.
Electron microscopy and immunohistochemistry.
For electron microscopy, soleus muscles from HFD-fed WT and MTKO mice, fasted 4–5 h prior to sacrifice, were fixed in 2.5% glutaraldehyde, 2% paraformaldehyde, 0.1 M sodium cacodylate buffer (pH 7.4) at room temperature for 1 h. After rinsing in the same buffer twice, tissue was post-fixed in 1% OsO4 at room temperature for 1 h. Specimens were stained en bloc using 2% aqueous uranyl acetate for 30 min, dehydrated in a graded series of ethanol to 100%, substituted with propylene oxide, and embedded in EMbed 812 resin. Sample blocks were polymerized in an oven at 60° C overnight. Thin sections (60 nm) were cut using a Leica ultramicrotome (UC7) and post-stained with 2% uranyl acetate and lead citrate. Sections were examined with a FEI Tecnai transmission electron microscope at 80 kV accelerating voltage, and digital images were recorded with an Olympus Morada CCD camera and iTEM imaging software. Image analysis was done blindly using ImageJ/FIJI, and mitochondria were traced manually for measurement of mitochondrial density (number of mitochondria per cross-sectional area, in μm2), area per mitochondrion, mitochondrial length (Feret diameter), and mitochondrial width (minimum Feret diameter). Quantification was done for three mice of each genotype, and mitochondria were quantified on 5–9 images (similar to Fig. 3h) from each mouse, so that data from 20 WT and 23 MTKO images were quantified. Averages of the above parameters from each cross-sectional image are plotted in Extended Data Fig. 8b–e.
Immunohistochemistry was done essentially as described74,75. Briefly, quadriceps muscles were removed, embedded in OCT compound, and rapidly frozen in liquid nitrogen -cooled isopentane, then sectioned and stored at −80°C. For staining, the slides were air dried for 10 min. and all procedures were performed at room temperature. Two of three sections were blocked with 10% normal goat serum, then primary antibodies (BA-F8, 1:180 and SC-71, 1:3000) were applied for a two-hour incubation period. The third section was incubated with isotype control antibodies. After a brief wash using PBS, sections were stained with secondary antibodies (AlexaFluor555-conjugated anti-mouse IgG2b, 1:500 and AlexaFluor488-conjugated anti-mouse IgG1, 1:500). After washing, coverslips were applied using Prolong Gold antifade reagent. Images were acquired using a Zeiss Axiovert 200M wide field microscope equipped with a Colibri LED light source and driven by AxioVision 4.7 imaging software. Image analysis was done using ImageJ/FIJI and Photoshop. In quadriceps, few fibers stained for myosin heavy chain type I (BA-F8). The percentage of fibers staining for myosin heavy chain type IIA (SC-71) was quantified by two independent blinded observers.
Reagents and transfection.
Antibodies directed to TUG, GLUT4, and IRAP were described previously 5,8,12,17. Other antibodies were purchased, including those directed to GAPDH (Millipore MAB374), β-tubulin (Developmental Studies Hybridoma Bank (DSHB) at Univ. of Iowa, clone E7-b), LaminB1 (Cell Signaling Technology (CST) clone D4Q4Z, 12586S), PGC-1α (Thermo Fisher Invitrogen PA5–72948), PPARγ (Santa Cruz Biotechnology clone E-8, sc-7273), Sarcolipin (Millipore ABT13), Usp25 (Novus NBP180631 and Abcam ab187156), HA epitope tag (Biolegend clone HA.11, 901502; CST clone C29F4, 3724S), Ate1 (Millipore clone 6F11, MABS436), β-actin (Thermo Scientific PIMA515739), flag epitope tag (CST clone D6W5B, 14793S and Sigma clone M2, F3165 and A2220), GST (CST clone 91G1, 2625S), insulin receptor β-subunit (Millipore 07–724 and CST clone 4B8, 3025S), caveolin (CST clone D46G3, 3267S), Akt (CST clone C67E7, 4691S), phospho-Akt (Ser473) (CST clone D9E, 4060S), Ucp1 (Abcam ab10983 and ab209483), DHPR α-subunit (DSHB, clone IIID5E1), myosin heavy chain type 1 (DSHB, BA-F8-c), and myosin heavy chain type IIA (DSHB, SC-71-c). For immunoblots, antibodies were used at 1:1000 dilution or at ~2 μg/ml unless otherwise noted.
Plasmids to express flag-tagged PPARγ, PGC-1α, and PGC-1β were gifts of Dr. Bruce Spiegelman and were obtained from Addgene (plasmids #8895, 1026, 1031, respectively)76–78, as were plasmids for recombinant production of PGC-1α (Addgene #1028 and 1029)54. pGEX TUG plasmids were previously described36. The pGEX 4T-1-PPARγ2 plasmid was a gift of Dr. J. Song (Addgene #78773)79. A plasmid encoding HA-tagged ubiquitin was a gift of Dr. Edward Yeh (Addgene #18712)80. A plasmid containing the sarcolipin promotor driving luciferase (pTR.SLN.Luc) was a gift of Dr. Dongtak Jeong81.
Lipofectamine 2000 (Invitrogen) was used for transient transfection of HEK293 and HeLa cells. For electroporation of MEFs, cells were grown to 90% confluence and resuspended in 1 ml of 0.25% trypsin. Cells were pelleted by spinning at 5000 x g and pellet was resuspended in 1 ml of OptiMEM (Gibco). 100 μl of resuspended cells and 2–4 μg of desired plasmid DNA in 5 μL of volume were pipetted into electroporation cuvettes (2 mm gap, Bulldog Bio 12358–346) and electroporated using a NEPA21 electroporation system. Settings used were Poring pulse: V=150, Length=5 ms, Interval=50 ms, Number of pulses=2, Decay rate=10%, Polarity= +. Transfer pulse: V=20, Length=50 ms, Interval=50 ms, Number of pulses=5, Decay rate=40%, Polarity= +/−. After electroporation, cells were diluted with 20 mL of DMEM and plated in 6 well dishes.
Immunoblots and Immunoprecipitations.
For immunoblots of basal and insulin-stimulated tissues, lysates were prepared from mice that had been fasted for 4–6 hours, then treated with intraperitoneal (IP) injection of insulin (8 U/kg) and glucose (1 g/kg) in phosphate buffered saline (PBS), or with an equivalent volume (0.3 ml) of PBS alone, as previously18. After 30 min (or other indicated durations), mice were anesthetized and sacrificed by cervical dislocation.
Glucometer measurements confirmed that no hypoglycemia occurred during the 3 h after IP injections using this protocol. Gonadal white adipose tissue (GWAT), quadriceps, gastrocnemius, and soleus muscles, and other tissues were collected and flash frozen in liquid nitrogen and stored at −80° C. For experiments using basal and insulin stimulated 3T3-L1 adipocytes, cells were typically serum starved for 3 h prior to insulin stimulation. Insulin was used at 80–160 nM for 15–30 min unless otherwise specified.
For immunoblots, tissues were quickly thawed and 200 mg of each tissue were weighed and mixed with lysis buffer (1% IGEPAL CA-630 (Sigma), 20 mM Tris, pH 7.4, 150 mM NaCl, 2 mM EDTA with Complete (Roche) protease inhibitors). A Qiagen TissueLyser II was used to grind the tissue for 3 min. at 30 cycles/sec. To remove insoluble debris, lysates were centrifuged 10 min at 13,000 rpm in a tabletop centrifuge (Eppendorf 5424R) at 4° C. Supernatants were analyzed by SDS-PAGE using Invitrogen NuPAGE gels, transferred to nitrocellulose membranes using a semidry apparatus, and imaged using peroxidase conjugated secondary antibodies and detection on film, as previously5,16,17. Quantification was done on exposures within the linear range of the film and used transillumination (Epson Perfection V700 flatbed scanner) together with Silverfast 8 (Lasersoft Imaging, version 8.8.0) and ImageJ (FIJI, version 2.1.0/1.53c) software. Figures were prepared using Adobe Photoshop (version 20.0.6) and Illustrator (version 23.1.1; Creative Cloud 2019), and Graphpad Prism (version 9.0 for MacOS) software.
Immunoprecipitations both tissue and cell lysates were done using the above buffer, and were allowed to proceed overnight at 4° C after addition of the immunoprecipitating antibody. Protein A sepharose (CL-4B, GE LifeSciences) was added and incubations were continued an additional 4 h at 4° C. For immunoprecipitations using epitope tags, affinity matrices were incubated overnight with cell lysates. After pelleting in a benchtop microfuge, beads were washed 6 times with 1% or 0.5% IGEPAL CA-630 buffer and transferred to new tubes. Samples were eluted by heating (5 min., 95° C) in SDS-PAGE sample buffer with 15% 2-mercaptoethanol or without heat using glycine buffer (pH 2.5) with neutralization by Tris base (pH 9). Samples were separated on 4–12% NuPAGE bis-tris gels and immunoblotted as above.
Fractionations and cycloheximide chase experiments.
To prepare nuclear and cytosolic fractions of muscle and adipose tissues, 100 mg of tissue was homogenized using 10 strokes in a 2 ml ground glass dounce-type tissue grinder in the buffers provided in the NE-PER nuclear and cytoplasmic extraction kit from ThermoFisher Scientific. Nuclear and cytoplasmic fractions were then prepared according to kit instructions and used for SDS-PAGE and immunoblotting as above.
Subcellular fractionation of 3T3-L1 adipocytes was performed as described previously8,17,82. Briefly, for each sample, five 10 cm plates of 3T3-L1 adipocytes were homogenized in 5 ml of an ice-cold TES buffer (250 mM sucrose, 10 mM Tris pH 7.4, 0.5 mM EDTA, protease inhibitor cocktail, and 20 mM iodoacetamide) using a glass dounce-type homogenizer. Plasma membrane, light microsome, heavy microsome, nuclear, and cytosolic fractions were isolated by differential centrifugation8,17,82. Pellets were resuspended in SDS-PAGE sample buffer with 10% 2-mercaptoethanol. Samples were heated for 5 minutes at 95 °C, separated on 4–12% NuPAGE bis-tris gels, transferred to nitrocellulose membranes, and immunoblotted as above. For experiments in which cells were pre-treated with pharmacologic inhibitors, wortmannin (Sigma) was used at 100 nM and was added 30 min. prior to insulin treatment, and CB-5083 (Cayman Chemical) was used at 5 μM and was added 1.5 h prior to insulin treatment. Cells were treated with 80 nM insulin for 60 min. prior to homogenization, fractionation, and immunoblotting as above.
T-tubule-enriched membrane fractions were isolated from quadriceps muscles using a protocol similar to that described previously14,18. Briefly, 12-week old mice were fasted 4–6 h, then treated by IP injection of insulin and glucose solution, or saline control, as above. After 30 min., mice were anesthetized and euthanized by cervical dislocation. One entire quadriceps muscle (~100 mg) was minced in 2 ml of ice-cold Buffer A (20 mM Na4P2O7, 20 mM NaH2PO4, 1 mM MgCl2, 0.3 M sucrose, 0.5 mM EDTA, 20 mM iodoacetamide, and two protease inhibitor tablets (Roche Applied Science) per 50 ml). Samples were homogenized using a Polytron tissue grinder for 10–15 sec. at 8000 rpm and then centrifuged at 13,000 rpm for 20 min. at 4 °C using a SS-34 rotor (Sorvall). Pellets were resuspended in 1.7 ml of Buffer A, homogenized again using a Polytron (13,500 rpm for 45 sec.), and then centrifuged at 11,000 rpm for 20 min. at 4 °C using an SS-34 rotor. The supernatant was placed in a new tube and centrifuged in a TLA-120.2 rotor (Beckman) at 18,000 rpm for 10 min. at 4 °C. Supernatant was removed carefully and pellets were resuspended in 200 μL of Buffer A. To strip myofibrillar proteins, 500 μL of Buffer B (0.3 M sucrose, 20 mM Tris, pH 7.0) and 300 μL of 4M KCl were added to make a final KCl concentration of 1.2 M. Samples were incubated at 4 °C for 1 h with gentle agitation. Samples were then centrifuged at 57,000 rpm in a TLA-120.2 rotor for 10 min at 4 °C. The pellets representing the T-tubule-enriched membrane fraction were resuspended in 150 μL of Buffer B, mixed with 4x LDS loading buffer (Invitrogen) and analyzed by SDS-PAGE and immunoblotting.
For initial tests of protein stability, retroviruses were used to express HA-tagged ubiquitin fused to the TUG C-terminal product (residues 165–550), containing various residues at position 165 as indicated. These fusion constructs were expressed in Ate1 KO MEFs and WT control cells using the pBICD2 vector, and cells with similar levels of expression were selected using FACS of cell surface CD2 8,12,65. The relative abundances of HA-tagged ubiquitin were further tested using immunoblots. To assess the rate of TUG C-terminal product degradation, 500 μM cycloheximide was added acutely, cells were lysed at various times after cycloheximide addition, and immunoblots were performed.
To assess effects of TUG and of Ate1 knockout on PGC-1α stability, cycloheximide chase experiments were using HeLa cells or MEFs. Cells cultured in 10 cm dishes transfected with indicated plasmids using Lipofectamine 2000 or by electroporation, as above. Two days later, cycloheximide was added in 1 ml of prewarmed media to a final concentration of 500 μM. Cells were maintained in cycloheximide for time periods indicated and washed with cold PBS twice prior to lysis, SDS-PAGE, and immunoblotting. Replicate experiments were quantified using densitometry, and protein half-lives were estimated based on a least-squares fit to a first-order exponential decay.
Transcriptome analysis and chromatin immunoprecipitation.
Total RNA was prepared from quadriceps muscles of 11-week old, 4–6 h fasted UBX and WT mice (N=3 each). A NucleoSpin RNA preparation kit (Macherey-Nagel) was used with tissues that had been flash frozen and stored at −80° C. 100 mg of each sample was defrosted on ice and lysed using 10 strokes in a 2 ml glass dounce-type tissue grinder in the buffers provided. For deep sequencing, rRNA were removed using Ribo-Zero (Illumina). Six strand-specific sequencing libraries, 3 replicates per condition, were produced from purified total RNA samples by the Illumina TruSeq stranded protocol. The libraries underwent 76bp single-end sequencing using Illumina HiSeq 2500 according to Illumina protocols, generating between 40–55 million reads per sample. For each read, we trimmed the first 6 nucleotides and the last nucleotides at the point where the Phred score of an examined base fell below 20 using in-house scripts. If, after trimming, the read was shorter than 45-bp, the whole read was discarded. Trimmed reads were mapped to the mouse reference genome (mm10) with a known transcriptome index (UCSC Known Gene annotation) with Tophat v2.1.183 using the very-sensitive preset, first strand library type, and providing the corresponding gene model annotation. Only the reads that mapped to a single unique location within the genome, with a maximum of two mismatches in the anchor region of the spliced alignment, were reported in these results. We used the default settings for all other Tophat options. Tophat alignments were then processed by Cuffdiff Cufflinks v2.2.1, 84 to obtain differential gene expression using first strand library type, providing gene model annotation and the genome sequence file for detection and correction of sequence-specific bias that random hexamer can cause during library preparation. Expression between UBX and WT quadriceps was significantly different for 674 transcripts after genome-wide adjustment using Benjamini (to adjusted p<0.05). Of these, 467 had increased and 207 had decreased expression. Pathway analyses were done using DAVID85 (Supplementary Table 2), GOrilla86, Ingenuity Pathway Analysis (Qiagen), MetaCore (Clarivate), and GSEA38,87. For GSEA, transcripts that were altered between UBX and WT mice were ranked in order of significance, and preranked data were analyzed. Fig. 3b and Extended Data Fig. 5f present an analysis of the top 2000 preranked transcripts using the GO Biological Process ontology gene set from MSigDB v7.2.
For quantitative PCR (qPCR), RNA was extracted using the Qiagen RNA-easy preparation kit as directed. qPCR was done as previously5. Briefly, 670 nmol of RNA was used with a high capacity cDNA reverse transcriptase kit (Applied Biosystems). Real time PCR was done using a StepOnePlus system and Power Sybr Green Master Mix (Applied Biosystems). mRNA abundances were normalized to that for 18S RNA or β-actin (Actb). Primers are listed in Supplementary Table 3. Two or three technical replicates were done for each biological replicate and were averaged. Biological replicates (from separate mice) are plotted and were used to compare mice.
Chromatin immunoprecipitation was carried out using a ChIP-IT Express Enzymatic Kit (Active Motif). Tissues were quickly thawed and 200 mg of each was weighed, minced, and placed in an Eppendorf tube. Samples were fixed in PBS containing 1.5% formaldehyde for 15 minutes with rotation, then washed three times using PBS. Cleaned fixed tissues were then homogenized using 10–20 strokes in a 2 ml ground glass dounce-type tissue grinder in buffers provided by the ChIP-IT Kit. Precipitations were done using antibodies to the TUG C-terminus, PGC-1α, or PPARγ antibody, together with magnetic beads. Chromatin was washed, eluted, and cross links were reversed per protocol instructions. Eluted DNA as well as pre-IP controls were amplified using GoTaq Green Master Mix (Promega) with primers designed for the promoter regions of Ucp1 and Sarcolipin. Primers used are listed in Supplementary Table 3.
Recombinant protein expression and pulldowns.
GST-tagged constructs were expressed in BL21(DE3)pLys GOLD E. coli (Agilent Technologies). 30 ml bacterial starter cultures were grown at 37° C overnight and then added to 1.5 liters of LB media at 37° C. Protein expression was induced with isopropyl-β-D-thiogalactopyranoside at a final concentration of 1 mM once an OD of 0.8 was reached. After 3–4 h, bacteria were lysed in 50 mM Tris, pH 8.0, 300 mM NaCl, 1% Triton X-100 and 1mM PMSF in a French Press. Insoluble debris was pelleted at 16,000 x g for 20 min. GST-tagged protein from supernatant were purified using glutathione-Sepharose 4B (GE Healthcare). Sepharose bound proteins were used as the protein columns. GST-free eluates of protein were prepared by using 1 unit of biotinylated thrombin (Millipore 69672–3) per 100 μl of beads and letting samples incubate on a rotator at 4° C overnight. Thrombin was removed by treating the supernatant with 36 μl of 50% neutravidin bead slurry per 1 unit of thrombin used. Thrombin-cleaved eluted proteins were incubated with protein-bound columns overnight at 4° C, then samples were washed 4–6 times using 0.25% IGEPAL CA-630 buffer. Eluates were analyzed by SDS-PAGE and immunoblotting as above.
For peptide pulldown experiments, biotin-containing synthetic peptides were dissolved to a concentration of 2 mg/mL in Dulbecco’s phosphate buffered saline and 500 μl of peptide solution was incubated with 200 μl of Pierce NeutrAvidin Agarose at 4°C overnight on a rotator. After overnight incubation, beads were washed with 1% IGEPAL CA-630 buffer as above three times, and beads were collected by centrifugation for 1 minute at maximum speed in a benchtop centrifuge. Biotin alone at a concentration of 1 mg/mL was used to generate negative control beads. These pre-incubated beads were then stored at 4°C prior to use. The synthetic peptides used were obtained from LifeTein and were: mmPPARG2, NH2-GETLGDSPVDPEHGAFADALPMSTSQEITMVDTEMPF-Ahx-Ahx-Lys(biotin)-COOH; mmPPARG1, NH2-VDTEMPFWPTNFGISSVDLSVMEDHSHSFDIKPFTTV-Ahx-Ahx-Lys(biotin)-COOH; hsPPARG2P, NH2-GETLGDSPIDPESDSFTDTLSANISQEMT-Ahx-Ahx-Lys(biotin)-COOH; hsPPARG2A, NH2-GETLGDSPIDAESDSFTDTLSANISQEMT-Ahx-Ahx-Lys(biotin)-COOH. Ahx denotes aminohexanoic acid, which was used as a spacer to extend the peptide away from the biotin binding site on the neutravidin beads.
Beads with bound peptides (or biotin only, as a control) were incubated with cell lysates or with recombinant proteins, in different experiments. For cell lysates, 3T3-L1 adipocytes, MEFs, or HEK293 cells were used. Lysates were typically prepared using 0.5% IGEPAL CA-630, although in some experiments this detergent was used at 0.25% or at 1%. For recombinant proteins, 1% IGEPAL CA-630 was used. Whole cell lysates or recombinant proteins were added to 100 μl of beads, prepared as above, and incubated at 4°C overnight on a rotator. Beads were then washed with the same buffer four times, collected by centrifugation in a benchtop centrifuge. Bound proteins were eluted using 100 microliters of sample buffer and heating at 95°C for 5 min. Samples were analyzed by SDS-PAGE and immunoblotting as above.
Transcription activation assays.
A pTR-SLN-Luc plasmid, containing 1029 nt from the human SLN promoter upstream of firefly luciferase, was a gift of Dr. Dongtak Jeong81. The pRL-SV40P plasmid, containing the SV40 promoter driving Renilla luciferase, was a control and was a gift of Ron Prywes (Addgene plasmid #27163)88. HEK293 cells were plated on 10 cm dishes and were transfected with 1 μg of each of the indicated plasmids. Controls were transfected with pTR-SLN-Luc or pRL-SV40P alone or with both of these plasmids. An empty vector pBICD4 plasmid was used to equalize the amount of DNA transfected in each sample16. In total, 5 μg of plasmid was suspended in 250 μl of DMEM and allowed to equilibrate for 5 min. Lipofectamine (25 μl) was combined with 225 μl of DMEM in the different tube for 5 min. Lipofectamine mixture was then added to the plasmid mixture and allowed to mix for 20 minutes before being added to the 10 cm dishes. 72 h after plasmid addition, cells were scraped in 1 ml of 1x lysis buffer from the Promega dual luciferase kit. Cells were then placed at −20° C overnight for a freeze/thaw cycle. 20 μl of lysate was then added to each well of a 96 well plate and combined with 100 μl of firefly Luciferase substrate. Measurements were made in triplicate. Plates were read using the luminescence protocol on a Victor3 plate reader with a 3 second read time per well. 100 μl of Stop and Glo buffer was then added to each well and Renilla activity was measured using the same Victor protocol. SLN-luc light units were normalized to Renilla light units, and data were plotted and analyzed using ANOVA with Tukey post-hoc correction for comparison of each sample with every other sample.
Cell Oximetry.
Cell metabolic analyses were done using a Seahorse Analyzer. Mitochondrial respiration and proton leak-linked oxygen consumption rates (OCR) were measured in 3T3-L1 adipocytes, differentiated as above using roscovitine to induce a brown like phenotype42. OCR was measured using the Seahorse XF Cell Mito Stress Test Kit (Agilent) according to the manufacturer’s instructions. Briefly, Control 3T3-L1 adipocytes and cells containing a shRNA to knockdown Usp25 (shRNA #1) were used5. Cells were differentiated on Seahorse XF24 V7 Culture Microplates that had been pre-coated with gelatin (Geltrex, Gibco). Following overnight serum starvation, cells were treated in the presence or absence of 160 nM insulin for 3 h. Cells were then washed twice with 1 ml XF24 Assay Media (DMEM Base containing 1.0 mM pyruvate, 2 mM glutamine and 5.5 mM glucose, pH 7.4). 500 μl of XF24 Assay Media was added to each well and plates were equilibrated at 37° C for 1 h in a non-CO2 incubator. Cells were then assayed on Seahorse XFe24 Analyzer (Agilent) following a 12-min equilibration period. Respiration rates were measured using an instrument protocol of 3-minute mix, 2-minute wait, and 3-minute measure. Following three baseline measurements, oligomycin (1.5 μM final concentration), FCCP (0.75 μM final concentration) and Rote/AA (1 μM final concentration) were sequentially injected and oxygen consumption rates were recorded using the same instrument protocol. Flux rates were normalized to DNA content following cell lysis using the Quant-iT™ PicoGreen® dsDNA Reagent (Invitrogen) according to the manufacturer’s instructions. Five separate experiments were performed, and four sets of cells under each condition were analyzed in each experiment.
Statistics and Reproducibility.
All data were replicated in at least two independent experiments, and usually three or more replicates were performed using biologically independent samples. On bar graphs showing individual replicates, data are presented as mean ±S.E.M. Biological replicates indicate that data were obtained using different mice, different plates of cultured cells, or cells that were induced separately to undergo adipocyte differentiation. Significance was assessed using unpaired, two-tailed t-tests or using one-way ANOVA with Bonferroni adjustment for multiple comparisons of preselected pairs. Exact p values are indicated in the figures, except for p<0.0001. Differences were considered significant at p<0.05. No statistical tool was used to pre-determine sample sizes; rather, the availability of materials and estimates of variances based on previous experience determined the number of biological replicates that were used.
Reporting Summary.
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Characterization of glucose homeostasis in MTKO mice.
a,b, Relative TUG abundance was quantified in quadriceps (a) and heart (b) using densitometry of immunoblots of tissues from 12-week old MTKO and WT control mice. N=5 in each group. c, Quadriceps muscles from fasted MTKO and WT mice were homogenized and T-tubule -enriched membrane fractions were purified and immunoblotted, as indicated. The first three samples in each group are the same as the unstimulated samples shown in Fig. 1c. All four unstimulated samples shown here were included in the quantification shown in Figs. 1d, e. d, Control immunoblots of T-tubule fractions and total homogenates from quadriceps of fasted mice were done as indicated to demonstrate the purity of the fractions. e, The relative abundance of intact TUG in quadriceps of WT mice was quantified by densitometry of the immunoblot in Fig. 1c. N=3 in each group. f–h, Body weights and composition were measured in 17-week old WT and MTKO mice. N=8 in each group. i, Heart weights were measured in 12-week old WT and MTKO mice. N=9 WT, 8 MTKO mice. j, Fasting glucose concentrations were measured in blood obtained by cardiac puncture of 16-week old mice. N=9 WT and 11 MTKO mice. k, HOMA-IR was calculated from paired insulin and glucose measurements plotted individually in Fig. 1g and in (j), in 4–6 h fasted 16-week old mice. N=5 in each group. l, Basal plasma glucose was measured prior to turnover studies in fasting 19-week old mice. N=8 WT and 9 MKTO mice. m, Heart-specific glucose uptake was measured in fasting 19-week old mice. N=8 WT and 9 MKTO mice. All data are presented as mean ± SEM of biologically independent samples, analyzed using two-tailed t-tests.
Extended Data Fig. 2. Characterization of energy expenditure in MTKO mice.
a–d, The indicated parameters were measured in 17-week old WT and MTKO mice in metabolic cages. Energy expenditure was normalized to total weight (c) and lean body mass (LBM; d). N=8 WT and 7 MTKO mice. e–g, Energy expenditure measurements are plotted vs. time of day (e) and during light (f) and dark (g) hours. N=8 WT and 7 MTKO mice. h–j, Respiratory exchange ratio (RER; h), locomotor activity (i), and food intake (j) are plotted. N=8 WT and 7 MTKO mice. k, Water intake was measured in 22-week old WT and MTKO mice in metabolic cages. N=16 in each group. All data are presented as mean ± SEM of biologically independent samples, analyzed using two-tailed t-tests.
Extended Data Fig. 3. MTKO mice are susceptible to obesity and have reduced energy expenditure on a high-fat diet.
a, Mice were fed a high-fat diet (HFD) beginning at 15 weeks of age, and the percentage weight gain from baseline is plotted. N=10 WT and 12 MTKO mice. b, After the HFD, HOMA-IR was calculated from paired measurements of plasma glucose and insulin. N=7 WT and 10 MTKO mice. c, Immunoblots were done as indicated on hindlimb muscles from mice fed a HFD for 3 weeks. d–f, Body weights and composition of 14-week old MTKO and WT mice fed a HFD for 3 weeks prior to measurements done in metabolic cages. N=7 in each group. g–q, HFD-fed 14-week old MTKO and WT mice were housed in metabolic cages and the indicated parameters were measured. N=7 in each group (g–l, p, q); N=7 WT and 6 MTKO mice (m–o). RER, respiratory exchange ratio; LBM, lean body mass. All data are presented as mean ± SEM of biologically independent samples, analyzed using two-tailed t-tests. *p<0.05, **p<0.01, ***p<0.001.
Extended Data Fig. 4.
a, Body weights were measured in male MTKO and WT mice housed at 30° C from weaning. All mice were maintained on regular chow. N = 11 WT and 9 MTKO mice. b, Body weights were measured in female MTKO and WT mice housed at 30° C from weaning. All mice were maintained on regular chow. N = 12 WT and 8 MTKO mice. c,d, Gonadal white adipose tissues from 20-week old mice used for data in (a) and (b) were weighed. N=5 WT and 3 MTKO males, and 10 WT and 5 MTKO females. All data are presented as mean ± SEM of biologically independent samples, analyzed using two-tailed t-tests. *p<0.05, **p<0.01, ***p<0.001.
Extended Data Fig. 5. Expression of specific genes involved in energy expenditure is reduced in MTKO mice and increased in UBX mice.
a–e, RNA was prepared from quadriceps muscles of ad lib fed 10-week old WT, MTKO, and UBX mice, and qPCR was used to measure the relative abundances of selected transcripts. N=4 WT, 5 MTKO, and 4 UBX mice except for N=3 for UBX TUG-Cter data (a). a, Control reactions were used to verify knockout of TUG in MTKO mice and expression of the UBX-Cter transgene in UBX mice. b, Relative abundances of transcripts for Sarcolipin (Sln), Ucp1, and β3-adrenergic receptor (Adrb3) are shown. c, Relative abundances of the indicated transcripts involved in energy expenditure and lipid metabolism are shown. d, Relative abundances of transcripts encoding PGC-1α (Ppargc1a) and PPARγ (Pparg) are shown. e, Relative abundances of transcripts encoding the indicated calsequestrin proteins are shown. Data are presented as mean ± SEM of biologically independent samples, analyzed using two-tailed t-tests (a–e). f, Gene set enrichment analysis was done on transcripts that were differentially expressed in quadriceps of 11-week old, 4–6 h fasted UBX mice, compared to WT controls (N=3 mice in each group; transcripts were analyzed using CuffDiff as described in the Methods section). Transcripts were ranked in order of significance, and the top 2000 transcripts were analyzed using the GO Biological Process ontology gene set. The gene set corresponding to “Temperature Homeostasis” was significantly enriched (Fig. 3b; False Discovery Rate q value = 0.038)). Here, specific genes in this set that were differentially expressed in UBX vs. WT muscles are listed, together with their rank in the gene list, the fold-change in expression in UBX muscles compared to controls, the adjusted p-value in the RNAseq data set (text is bold if p<0.05 after adjustment for multiple comparisons using the Benjamini method), and the running enrichment score calculated by GSEA software.
Extended Data Fig. 6. The TUG C-terminal product enters the nucleus and binds PPARγ and PGC-1α.
a, Nuclear fractions were prepared from quadriceps of WT, UBX, and MTKO mice that had been fasted, treated with IP injection of insulin-glucose solution or saline control, and euthanized 30 min. after injection. Immunoblots were done as indicated. b,c, Proteins were expressed by transient transfection of HEK293 cells, and immunoprecipitations (IP) and western blots (WB) were performed, as indicated. d, Recombinant proteins were produced as GST fusions, immobilized on glutathione beads, and incubated with recombinant TUG C-terminal cleavage product (residues 165–550). Bound TUG protein was eluted and western blots were performed as indicated. e, Recombinant proteins were immobilized and incubated with soluble recombinant PPARγ2 protein. Bound PPARγ2 was eluted and western blots were performed as indicated. f, Truncated forms of TUG were produced as GST fusions and the GST was cleaved off to yield soluble TUG fragments. These were incubated with immobilized GST, PGC-1α, and PPARγ2 as indicated. Bound proteins were eluted and immunoblotted as indicated. g, Peptides corresponding to the 37 residues at the N-termini of PPARγ1 or PPARγ2 were immobilized on streptavidin beads. The TUG C-terminal product (beginning with a Met residue) was expressed stably in MEFs using a retrovirus, and lysates from these cells were incubated with the beads. Bound proteins were eluted and immunoblotted. h, Peptides corresponding to the 29 N-terminal residues of PPARγ2, containing Pro12 or Ala12 residues, were immobilized on beads and incubated with lysates of MEFs expressing the TUG C-terminal product. Bound proteins were eluted and immunoblotted as indicated. i, Peptides used in (h) were incubated with HEK293 lysates, and bound endogenous (human) intact TUG was eluted and immunoblotted. j, WT and MTKO mice were treated by IP injection of insulin-glucose solution, then euthanized after 3 h. PGC-1α was immunoprecipitated from lysates of quadriceps, and immunoblots were done to detect bound PPARγ, as in Fig. 3g. The relative abundances of PPARγ in replicate experiments were quantified. N=3 biologically independent samples, presented as mean ±SEM, analyzed using a two-tailed t-test.
Extended Data Fig. 7. TUG controls PGC-1α protein abundance.
a, WT and MTKO mice were treated by IP injection of insulin-glucose, or saline control, then sacrificed at the indicated times after injection. Quadriceps muscles were immunoblotted as indicated. b,c, WT and MTKO mice were treated by IP injection of insulin-glucose, or saline control, then sacrificed after 3 h. Lysates were prepared from quadriceps muscles, PGC-1α was immunoblotted, and the relative abundances in each sample were quantified using densitometry. Data in (c) are presented as mean ±SEM of biologically independent samples (N=3 in each group), analyzed using ANOVA with adjustment for multiple comparisons. d, WT and MTKO mice were treated with IP insulin-glucose, or saline control, then sacrificed 3 h later. RNA was prepared from quadriceps muscles, and Q-PCR was used to measure PGC-1α (Ppargc1a) mRNA abundance. Data are presented as mean ±SEM of biologically independent samples (N=3 in each group), analyzed using ANOVA with adjustment for multiple comparisons.
Extended Data Fig. 8. TUG regulates mitochondrial function and morphology and has no large effect on muscle fiber type.
a, WT and MTKO soleus muscles from mice that had been fed a HFD for 2.5 weeks were imaged using electron microscopy. Lipid droplets were noted in MTKO muscles, but not WT muscles, and were adjacent to mitochondria, as shown. b–e, Images of soleus muscles from HFD-fed WT and MTKO mice (N=3 each) were obtained by electron microscopy and were analyzed to quantify mitochondrial density (b), area (c), length (d), and width (e). Mitochondria were traced manually on 5–9 images from each mouse. Each data point represents the average of the measurements from a single image. N=22 WT and 21 MTKO images were quantified. Data are presented as mean ±SEM and analyzed using two-tailed t-tests. f, Relative abundances of the indicated transcripts in quadriceps muscles were measured using qPCR. N=4 WT, 5 MTKO, and 4 UBX mice. Data are plotted as mean ±SEM and pairwise analyses were done using two-tailed t-tests. g, Cross-sections of quadriceps muscles were stained to detect myosin heavy chain type IIA using immunohistochemistry. h, The percentage of muscle fibers that stained for MHC type IIA was quantified from 2–3 images from each of 5 separate mice of each genotype. N=12 WT, 12 MTKO, and 14 UBX images were quantified, and data are plotted as mean ±SEM.
Extended Data Fig. 9. Sarcolipin abundance is regulated by TUG and is decreased in diet-induced insulin resistance.
a, WT and MTKO mice were housed at 30° C from the time of weaning. At age 20 weeks, mice were fasted, treated with IP insulin-glucose or saline control, and sacrificed 3 h later. Hindlimb muscle lysates were immunoblotted as indicated. Data from replicates of this experiment were quantified and are plotted in Fig. 4b. b, WT and UBX mice were treated with IP insulin-glucose, or saline control, sacrificed after 30 min., and quadriceps muscles were immunoblotted as indicated. c,d, WT and MTKO mice were treated with IP insulin-glucose, or saline control, and sacrificed after 30 min. Hindlimb muscles were used for chromatin immunoprecipitation using PGC-1α (c) and PPARγ (d) antibodies, as indicated. PCR was used to detect an amplicon at −200 nucleotides relative to the sarcolipin transcription start site. e, WT mice were fed regular chow (RC) or a high-fat diet (HFD) for 3 weeks, then treated for 30 min. with IP insulin-glucose, or saline control. Quadriceps lysates were immunoblotted to detect intact TUG and the C-terminal cleavage product. Replicates were quantified using densitometry and are plotted in Fig. 4f. f, WT mice were fed RC or a HFD for 3 weeks, fasted, and sacrificed. Hindlimb muscles were isolated and immunoblotted as indicated. Replicates were quantified using densitometry and are plotted in Fig. 4g.
Supplementary Material
Acknowledgments
We thank Michael G. Löffler, Abel Alcázar-Román, Omar Julca-Zevallos, Jacob Culver, Eleanor Zagoren, Shrikant Mane, Whitney Harris Brown, Eric Li, Roland Calia-Bogan, Anna Kashina, Dongtak Jeong, and Derek Toomre for advice, reagents, and assistance. This work used Core Facilities of the Yale Diabetes Research Center (DRC; NIH P30 DK045735) and services of the CCMI EM facility and Keck Biotechnology Resource at Yale University. This work was supported by NIH R01 DK092661 and R56 DK092661 (to J.S.B.), by American Diabetes Association 1-17-IBS-40 (to J.S.B.), and by R01 DK116774 and R01 DK114793 (to G.I.S.). D.T.L. was supported by T32 GM136651 and F30 DK115037, E.N.H. was supported by a Yale DRC Pilot, under P30 DK45735, and D.F.V. was supported by R01 DK124272. L.G. was supported by K99 HL150234. J.-P.C. was supported by São Paulo Research Foundation (FAPESP) grant 2018/04956-5. H.L. was supported by the Natural Science Foundation of Zhejiang Province, China, No. LY18H070004. S.H. was supported by the Brazilian Coordination for the Improvement of Higher Education Personnel (CAPES/PVEX-88881.170862/2018-01) and Postgraduate and Research Dean/Cruzeiro do Sul (PRPGP/UNICSUL-0708/2019).
Footnotes
Competing interests
The authors declare no competing interests.
Data Availability
All data are available in the manuscript or extended data, or in the associated source data files. Additional source data are available from the corresponding author upon reasonable request. RNA-seq data have been deposited in Gene Expression Omnibus (GEO) under the accession number GSE134846.
References
- 1.Bogan JS Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem 81, 507–532, doi: 10.1146/annurev-biochem-060109-094246 (2012). [DOI] [PubMed] [Google Scholar]
- 2.Leto D & Saltiel AR Regulation of glucose transport by insulin: traffic control of GLUT4. Nat Rev Mol Cell Biol 13, 383–396, doi: 10.1038/nrm3351 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Xu Y, Nan D, Fan J, Bogan JS & Toomre D Optogenetic activation reveals distinct roles of PIP3 and Akt in adipocyte insulin action. J Cell Sci 129, 2085–2095, doi: 10.1242/jcs.174805 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Czech MP Insulin action and resistance in obesity and type 2 diabetes. Nat Med 23, 804–814, doi: 10.1038/nm.4350 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Habtemichael EN et al. Usp25m protease regulates ubiquitin-like processing of TUG proteins to control GLUT4 glucose transporter translocation in adipocytes. J Biol Chem 293, 10466–10486, doi: 10.1074/jbc.RA118.003021 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Govers R, Coster AC & James DE Insulin increases cell surface GLUT4 levels by dose dependently discharging GLUT4 into a cell surface recycling pathway. Molecular and cellular biology 24, 6456–6466, doi: 10.1128/MCB.24.14.6456-6466.2004 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu Y et al. Dual-mode of insulin action controls GLUT4 vesicle exocytosis. J Cell Biol 193, 643–653, doi: 10.1083/jcb.201008135 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu C, Cresswell J, Loffler MG & Bogan JS The glucose transporter 4-regulating protein TUG is essential for highly insulin-responsive glucose uptake in 3T3-L1 adipocytes. J Biol Chem 282, 7710–7722, doi: 10.1074/jbc.M610824200 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Garvey WT et al. Evidence for defects in the trafficking and translocation of GLUT4 glucose transporters in skeletal muscle as a cause of human insulin resistance. J Clin Invest 101, 2377–2386 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maianu L, Keller SR & Garvey WT Adipocytes exhibit abnormal subcellular distribution and translocation of vesicles containing glucose transporter 4 and insulin-regulated aminopeptidase in type 2 diabetes mellitus: implications regarding defects in vesicle trafficking. J Clin Endocrinol Metab 86, 5450–5456 (2001). [DOI] [PubMed] [Google Scholar]
- 11.Li DT et al. GLUT4 Storage Vesicles: Specialized Organelles for Regulated Trafficking. Yale J Biol Med 92, 453–470 (2019). [PMC free article] [PubMed] [Google Scholar]
- 12.Bogan JS, Hendon N, McKee AE, Tsao TS & Lodish HF Functional cloning of TUG as a regulator of GLUT4 glucose transporter trafficking. Nature 425, 727–733, doi: 10.1038/nature01989 (2003). [DOI] [PubMed] [Google Scholar]
- 13.Belman JP, Habtemichael EN & Bogan JS A proteolytic pathway that controls glucose uptake in fat and muscle. Reviews in endocrine & metabolic disorders 15, 55–66, doi: 10.1007/s11154-013-9276-2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habtemichael EN et al. Coordinated Regulation of Vasopressin Inactivation and Glucose Uptake by Action of TUG Protein in Muscle. J Biol Chem 290, 14454–14461, doi: 10.1074/jbc.C115.639203 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kandror KV & Pilch PF The sugar is sIRVed: sorting Glut4 and its fellow travelers. Traffic 12, 665–671, doi: 10.1111/j.1600-0854.2011.01175.x (2011). [DOI] [PubMed] [Google Scholar]
- 16.Bogan JS et al. Endoproteolytic cleavage of TUG protein regulates GLUT4 glucose transporter translocation. J Biol Chem 287, 23932–23947, doi: 10.1074/jbc.M112.339457 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belman JP et al. Acetylation of TUG protein promotes the accumulation of GLUT4 glucose transporters in an insulin-responsive intracellular compartment. J Biol Chem 290, 4447–4463, doi: 10.1074/jbc.M114.603977 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Löffler MG et al. Enhanced fasting glucose turnover in mice with disrupted action of TUG protein in skeletal muscle. J Biol Chem 288, 20135–20150, doi: 10.1074/jbc.M113.458075 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantley JL et al. Targeting sterol receptor coactivator-1 with antisense oligonucleotides increases insulin stimulated skeletal muscle glucose uptake in male rats. Am J Physiol Endocrinol Metab, ajpendo 00148 02014, doi: 10.1152/ajpendo.00148.2014 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Semiz S et al. Conventional kinesin KIF5B mediates insulin-stimulated GLUT4 movements on microtubules. Embo J 22, 2387–2399 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiang SH et al. Insulin-stimulated GLUT4 translocation requires the CAP-dependent activation of TC10. Nature 410, 944–948 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Baumann CA et al. CAP defines a second signalling pathway required for insulin-stimulated glucose transport. Nature 407, 202–207, doi: 10.1038/35025089 (2000). [DOI] [PubMed] [Google Scholar]
- 23.Chang L, Chiang SH & Saltiel AR TC10alpha is required for insulin-stimulated glucose uptake in adipocytes. Endocrinology 148, 27–33 (2007). [DOI] [PubMed] [Google Scholar]
- 24.JeBailey L et al. Skeletal muscle cells and adipocytes differ in their reliance on TC10 and Rac for insulin-induced actin remodeling. Mol Endocrinol 18, 359–372 (2004). [DOI] [PubMed] [Google Scholar]
- 25.Coisy-Quivy M et al. TC10 controls human myofibril organization and is activated by the sarcomeric RhoGEF obscurin. J Cell Sci 122, 947–956, doi: 10.1242/jcs.040121 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupte A & Mora S Activation of the Cbl insulin signaling pathway in cardiac muscle; dysregulation in obesity and diabetes. Biochem Biophys Res Commun 342, 751–757, doi: 10.1016/j.bbrc.2006.02.023 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Bernard JR, Reeder DW, Herr HJ, Rivas DA & Yaspelkis BB 3rd. High-fat feeding effects on components of the CAP/Cbl signaling cascade in Sprague-Dawley rat skeletal muscle. Metabolism 55, 203–212, doi: 10.1016/j.metabol.2005.08.013 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Bernard JR, Saito M, Liao YH, Yaspelkis BB 3rd & Ivy JL Exercise training increases components of the c-Cbl-associated protein/c-Cbl signaling cascade in muscle of obese Zucker rats. Metabolism 57, 858–866, doi: 10.1016/j.metabol.2008.01.035 (2008). [DOI] [PubMed] [Google Scholar]
- 29.Hart Y & Alon U The Utility of Paradoxical Components in Biological Circuits. Molecular Cell 49, 213–221, doi: 10.1016/j.molcel.2013.01.004 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Li DT, Habtemichael EN & Bogan JS Vasopressin inactivation: Role of insulin-regulated aminopeptidase. Vitam Horm 113, 101–128, doi: 10.1016/bs.vh.2019.08.017 (2020). [DOI] [PubMed] [Google Scholar]
- 31.Schertzer JD et al. A transgenic mouse model to study glucose transporter 4myc regulation in skeletal muscle. Endocrinology 150, 1935–1940, doi: 10.1210/en.2008-1372 (2009). [DOI] [PubMed] [Google Scholar]
- 32.Nedergaard J & Cannon B The browning of white adipose tissue: some burning issues. Cell Metab 20, 396–407, doi: 10.1016/j.cmet.2014.07.005 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Bal NC, Sahoo SK, Maurya SK & Periasamy M The Role of Sarcolipin in Muscle Non-shivering Thermogenesis. Front Physiol 9, 1217, doi: 10.3389/fphys.2018.01217 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ikeda K et al. UCP1-independent signaling involving SERCA2b-mediated calcium cycling regulates beige fat thermogenesis and systemic glucose homeostasis. Nat Med 23, 1454–1465, doi: 10.1038/nm.4429 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reinisch I, Schreiber R & Prokesch A Regulation of thermogenic adipocytes during fasting and cold. Mol Cell Endocrinol 512, 110869, doi: 10.1016/j.mce.2020.110869 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Orme CM & Bogan JS The ubiquitin regulatory X (UBX) domain-containing protein TUG regulates the p97 ATPase and resides at the endoplasmic reticulum-golgi intermediate compartment. J Biol Chem 287, 6679–6692, doi: 10.1074/jbc.M111.284232 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altshuler D et al. The common PPARgamma Pro12Ala polymorphism is associated with decreased risk of type 2 diabetes. Nat Genet 26, 76–80, doi: 10.1038/79216 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Mootha VK et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34, 267–273, doi: 10.1038/ng1180 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Trausch-Azar J, Leone TC, Kelly DP & Schwartz AL Ubiquitin proteasome-dependent degradation of the transcriptional coactivator PGC-1{alpha} via the N-terminal pathway. J Biol Chem 285, 40192–40200, doi: 10.1074/jbc.M110.131615 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adamovich Y et al. The protein level of PGC-1alpha, a key metabolic regulator, is controlled by NADH-NQO1. Mol Cell Biol 33, 2603–2613, doi: 10.1128/MCB.01672-12 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sano M et al. Intramolecular control of protein stability, subnuclear compartmentalization, and coactivator function of peroxisome proliferator-activated receptor gamma coactivator 1alpha. J Biol Chem 282, 25970–25980, doi: 10.1074/jbc.M703634200 (2007). [DOI] [PubMed] [Google Scholar]
- 42.Wang H, Liu L, Lin JZ, Aprahamian TR & Farmer SR Browning of White Adipose Tissue with Roscovitine Induces a Distinct Population of UCP1(+) Adipocytes. Cell Metab 24, 835–847, doi: 10.1016/j.cmet.2016.10.005 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Banchenko S et al. Common Mode of Remodeling AAA ATPases p97/CDC48 by Their Disassembling Cofactors ASPL/PUX1. Structure 27, 1830–1841 e1833, doi: 10.1016/j.str.2019.10.001 (2019). [DOI] [PubMed] [Google Scholar]
- 44.van den Boom J & Meyer H VCP/p97-Mediated Unfolding as a Principle in Protein Homeostasis and Signaling. Mol Cell 69, 182–194, doi: 10.1016/j.molcel.2017.10.028 (2018). [DOI] [PubMed] [Google Scholar]
- 45.Varshavsky A N-degron and C-degron pathways of protein degradation. Proc Natl Acad Sci U S A 116, 358–366, doi: 10.1073/pnas.1816596116 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brower CS & Varshavsky A Ablation of arginylation in the mouse N-end rule pathway: loss of fat, higher metabolic rate, damaged spermatogenesis, and neurological perturbations. PLoS One 4, e7757, doi: 10.1371/journal.pone.0007757 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cui J et al. Adipose-specific deletion of Kif5b exacerbates obesity and insulin resistance in a mouse model of diet-induced obesity. FASEB J 31, 2533–2547, doi: 10.1096/fj.201601103R (2017). [DOI] [PubMed] [Google Scholar]
- 48.Lowell BB & Spiegelman BM Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660, doi: 10.1038/35007527 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Reed GW & Hill JO Measuring the thermic effect of food. Am J Clin Nutr 63, 164–169 (1996). [DOI] [PubMed] [Google Scholar]
- 50.Haeusler RA, McGraw TE & Accili D Biochemical and cellular properties of insulin receptor signalling. Nat Rev Mol Cell Biol 19, 31–44, doi: 10.1038/nrm.2017.89 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.de Jonge L & Bray GA The thermic effect of food is reduced in obesity. Nutr Rev 60, 295–297; author reply 299–300 (2002). [DOI] [PubMed] [Google Scholar]
- 52.Patti ME et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A 100, 8466–8471, doi: 10.1073/pnas.1032913100 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Finck BN & Kelly DP PGC-1 coactivators: inducible regulators of energy metabolism in health and disease. J Clin Invest 116, 615–622, doi: 10.1172/JCI27794 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Puigserver P et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science 286, 1368–1371, doi: 10.1126/science.286.5443.1368 (1999). [DOI] [PubMed] [Google Scholar]
- 55.Wu Z et al. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98, 115–124, doi: 10.1016/S0092-8674(00)80611-X (1999). [DOI] [PubMed] [Google Scholar]
- 56.St-Pierre J et al. Bioenergetic analysis of peroxisome proliferator-activated receptor gamma coactivators 1alpha and 1beta (PGC-1alpha and PGC-1beta) in muscle cells. J Biol Chem 278, 26597–26603, doi: 10.1074/jbc.M301850200 (2003). [DOI] [PubMed] [Google Scholar]
- 57.Choi CS et al. Paradoxical effects of increased expression of PGC-1alpha on muscle mitochondrial function and insulin-stimulated muscle glucose metabolism. Proceedings of the National Academy of Sciences of the United States of America 105, 19926–19931, doi: 10.1073/pnas.0810339105 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Summermatter S, Baum O, Santos G, Hoppeler H & Handschin C Peroxisome proliferator-activated receptor {gamma} coactivator 1{alpha} (PGC-1{alpha}) promotes skeletal muscle lipid refueling in vivo by activating de novo lipogenesis and the pentose phosphate pathway. J Biol Chem 285, 32793–32800, doi: 10.1074/jbc.M110.145995 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Summermatter S et al. PGC-1alpha improves glucose homeostasis in skeletal muscle in an activity-dependent manner. Diabetes 62, 85–95, doi: 10.2337/db12-0291 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quan N et al. Sestrin2 prevents age-related intolerance to ischemia and reperfusion injury by modulating substrate metabolism. FASEB J 31, 4153–4167, doi: 10.1096/fj.201700063R (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Y et al. Rab10 and myosin-Va mediate insulin-stimulated GLUT4 storage vesicle translocation in adipocytes. Journal of Cell Biology 198, 545–560, doi: 10.1083/jcb.201111091 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wasserman DH, Kang L, Ayala JE, Fueger PT & Lee-Young RS The physiological regulation of glucose flux into muscle in vivo. Journal of Experimental Biology 214, 254–262, doi: 10.1242/jeb.048041 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hu RG et al. The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986, doi: 10.1038/nature04027 (2005). [DOI] [PubMed] [Google Scholar]
- 64.Karakozova M et al. Arginylation of beta-actin regulates actin cytoskeleton and cell motility. Science 313, 192–196, doi: 10.1126/science.1129344 (2006). [DOI] [PubMed] [Google Scholar]
- 65.Liu X et al. Generation of mammalian cells stably expressing multiple genes at predetermined levels. Anal Biochem 280, 20–28, doi: 10.1006/abio.2000.4478 (2000). [DOI] [PubMed] [Google Scholar]
- 66.Tschop MH et al. A guide to analysis of mouse energy metabolism. Nat Methods 9, 57–63, doi: 10.1038/nmeth.1806 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Corrigan JK et al. A big-data approach to understanding metabolic rate and response to obesity in laboratory mice. Elife 9, doi: 10.7554/eLife.53560 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Camporez JP et al. Mechanism by which arylamine N-acetyltransferase 1 ablation causes insulin resistance in mice. Proc Natl Acad Sci U S A 114, E11285–E11292, doi: 10.1073/pnas.1716990115 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Camporez JP et al. Cellular mechanism by which estradiol protects female ovariectomized mice from high-fat diet-induced hepatic and muscle insulin resistance. Endocrinology 154, 1021–1028, doi: 10.1210/en.2012-1989 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Choi CS et al. Continuous fat oxidation in acetyl-CoA carboxylase 2 knockout mice increases total energy expenditure, reduces fat mass, and improves insulin sensitivity. Proceedings of the National Academy of Sciences of the United States of America 104, 16480–16485, doi: 10.1073/pnas.0706794104 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vatner DF et al. Thyroid hormone receptor-beta agonists prevent hepatic steatosis in fat-fed rats but impair insulin sensitivity via discrete pathways. Am J Physiol Endocrinol Metab 305, E89–100, doi: 10.1152/ajpendo.00573.2012 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chaudry IH & Gould MK Kinetics of glucose uptake in isolated soleus muscle. Biochim Biophys Acta 177, 527–536, doi: 10.1016/0304-4165(69)90315-8 (1969). [DOI] [PubMed] [Google Scholar]
- 73.Cuendet GS, Loten EG, Jeanrenaud B & Renold AE Decreased basal, noninsulin-stimulated glucose uptake and metabolism by skeletal soleus muscle isolated from obese-hyperglycemic (ob/ob) mice. J Clin Invest 58, 1078–1088, doi: 10.1172/JCI108559 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wang C, Yue F & Kuang S Muscle Histology Characterization Using H&E Staining and Muscle Fiber Type Classification Using Immunofluorescence Staining. Bio Protoc 7, doi: 10.21769/BioProtoc.2279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Bloemberg D & Quadrilatero J Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One 7, e35273, doi: 10.1371/journal.pone.0035273 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hauser S et al. Degradation of the peroxisome proliferator-activated receptor gamma is linked to ligand-dependent activation. J Biol Chem 275, 18527–18533, doi: 10.1074/jbc.M001297200 (2000). [DOI] [PubMed] [Google Scholar]
- 77.Lin J, Puigserver P, Donovan J, Tarr P & Spiegelman BM Peroxisome proliferator-activated receptor gamma coactivator 1beta (PGC-1beta ), a novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem 277, 1645–1648, doi: 10.1074/jbc.C100631200 (2002). [DOI] [PubMed] [Google Scholar]
- 78.Monsalve M et al. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol Cell 6, 307–316 (2000). [DOI] [PubMed] [Google Scholar]
- 79.Kim JH et al. Suppression of PPARgamma through MKRN1-mediated ubiquitination and degradation prevents adipocyte differentiation. Cell Death Differ 21, 594–603, doi: 10.1038/cdd.2013.181 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kamitani T, Kito K, Nguyen HP & Yeh ET Characterization of NEDD8, a developmentally down-regulated ubiquitin-like protein. J Biol Chem 272, 28557–28562, doi: 10.1074/jbc.272.45.28557 (1997). [DOI] [PubMed] [Google Scholar]
- 81.Yoo J, Kohlbrenner E, Kim O, Hajjar RJ & Jeong D Enhancing atrial-specific gene expression using a calsequestrin cis-regulatory module 4 with a sarcolipin promoter. J Gene Med 20, e3060, doi: 10.1002/jgm.3060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bogan JS, McKee AE & Lodish HF Insulin-responsive compartments containing GLUT4 in 3T3-L1 and CHO cells: regulation by amino acid concentrations. Mol Cell Biol 21, 4785–4806, doi: 10.1128/MCB.21.14.4785-4806.2001 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Trapnell C, Pachter L & Salzberg SL TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111, doi: 10.1093/bioinformatics/btp120 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Trapnell C et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28, 511–515, doi: 10.1038/nbt.1621 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Huang da W, Sherman BT & Lempicki RA Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols 4, 44–57, doi: 10.1038/nprot.2008.211 (2009). [DOI] [PubMed] [Google Scholar]
- 86.Eden E, Navon R, Steinfeld I, Lipson D & Yakhini Z GOrilla: a tool for discovery and visualization of enriched GO terms in ranked gene lists. BMC Bioinformatics 10, 48, doi: 10.1186/1471-2105-10-48 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Subramanian A et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102, 15545–15550, doi: 10.1073/pnas.0506580102 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chen X & Prywes R Serum-induced expression of the cdc25A gene by relief of E2F-mediated repression. Mol Cell Biol 19, 4695–4702, doi: 10.1128/mcb.19.7.4695 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the manuscript or extended data, or in the associated source data files. Additional source data are available from the corresponding author upon reasonable request. RNA-seq data have been deposited in Gene Expression Omnibus (GEO) under the accession number GSE134846.