Abstract
Delivery systems for controlled release of RNA interference (RNAi) molecules, including small interfering (siRNA) and microRNA (miRNA), have the potential to direct stem cell differentiation for regenerative musculoskeletal applications. To date, localized RNA delivery platforms in this area have focused predominantly on bulk scaffold-based approaches, which can interfere with cell-cell interactions important for recapitulating some native musculoskeletal developmental and healing processes in tissue regeneration strategies. In contrast, scaffold-free, high density human mesenchymal stem cell (hMSC) aggregates may provide an avenue for creating a more biomimetic microenvironment. Here, photocrosslinkable dextran microspheres (MS) encapsulating siRNA-micelles were prepared via an aqueous emulsion method and incorporated within hMSC aggregates for localized and sustained delivery of bioactive siRNA. siRNA-micelles released from MS in a sustained fashion over the course of 28 days, and the released siRNA retained its ability to transfect cells for gene silencing. Incorporation of fluorescently labeled siRNA (siGLO)-laden MS within hMSC aggregates exhibited tunable siGLO delivery and uptake by stem cells. Incorporation of MS loaded with siRNA targeting green fluorescent protein (siGFP) within GFP-hMSC aggregates provided sustained presentation of siGFP within the constructs and prolonged GFP silencing for up to 15 days. This platform system enables sustained gene silencing within stem cell aggregates and thus shows great potential in tissue regeneration applications.
Keywords: Mesenchymal stem cells, aggregates, sustained delivery, gene therapy, RNA interference
Introduction
RNA interference (RNAi) has come to the forefront of tissue regeneration as a strategy to control cell fate at the messenger RNA (mRNA) level [1]. RNAi molecules, which include both microRNAs (miRNAs) and small interfering RNAs (siRNAs), are short double-stranded RNA (dsRNA) molecules that act to post-transcriptionally silence gene expression by translational repression or cleavage of homologous mRNA [2]. miRNAs are transcribed endogenously, and typically target multiple genes by binding with partial complementarity to several mRNA targets [3]. In contrast, siRNA can be engineered to target a single, specific gene for silencing. Reports have highlighted the capacity for RNAi to act as both a negative and positive regulator of stem cell chondrogenic [4–6] and osteogenic [7–10] differentiation with applications in musculoskeletal tissue engineering. However, delivery of naked RNAi molecules that are not complexed with a transfection agent is limited by their susceptibility to RNAses in the microenvironment and their hydrophilic, anionic nature, which prevents cellular uptake [11, 12]. To overcome these limitations, numerous transfection agents have been developed, including micelles, liposomes, and cationic polymers, to enable formation of nanoscale particles for RNA cellular uptake and protection [13, 14]. Despite improved transfection efficiency and protection from RNAses, these nanocomplexes are readily dispersed upon systemic injection, often lack cellular targeting, and typically result in transient knockdown lasting less than 1 week [15–17], making sustained and localized release of RNAi molecules an important therapeutic strategy for further development [18–20].
Incorporation of naked RNA or RNA nanoparticles within hydrogels, 3D water-swollen, crosslinked polymer networks, is often employed in tissue engineering strategies as it allows for long-term, controlled presentation of RNA to co-encapsulated cells or those in the surrounding microenvironment [21–32] and can replicate important properties of native extracellular matrix (ECM) [33]. For example, sustained release of bioactive siRNA has been achieved from collagen [19, 34], alginate [19], poly(amidoamine)-dextran aldehyde [30], and poly(organophosphazene) [31] hydrogels. In addition, our group reported on the fine-tuned control of RNA release from hydrolysable methacrylated dextran (DEX) hydrogels via affinity interaction [35] or chemical tethering to the hydrogel network via hydrolysable linkages [36], polyethylene glycol (PEG) hydrogels via controlled hydrolytic degradation [26], and photodegradable PEG-based hydrogels upon UV application [22–24].
Despite the promising results achieved with bulk hydrogels, scaffold-based tissue engineering must meet the challenge of matching scaffold degradation rate to that of new tissue formation [37] to avoid compromising tissue function. While hydrogel scaffold-based approaches haven been successfully used for sustained siRNA delivery in in vivo applications, such as for regenerating critical-sized bone defects [38, 39], a scaffold can interfere with critical cell-cell interactions necessary for differentiation [40]. Scaffold-free tissue engineering approaches, including self-assembled human mesenchymal stem cell (hMSC) aggregates, which are formed in vitro typically via spontaneous cell aggregation following gravity-driven sedimentation or centrifugation, bypass the need for a scaffold and provide a more biomimetic microenvironment with abundant cell-cell and cell-ECM interactions. These cell aggregates may more closely recapitulate native developmental and healing processes involving high-density cell aggregates [40–42], and have application in the engineering of many tissue types including cartilage [43, 44] and bone [45–47]. Conventional culture of these aggregates requires repeated supplementation with exogenous growth factors in the medium to drive differentiation, but diffusion limitations and uptake by cells at the periphery may deplete supply for cells within the interior. Alternatively, incorporation of growth factor-loaded polymeric microparticles or microspheres (MS) within hMSC aggregates can deliver these bioactive factors locally to circumvent diffusion limitations and provide a sustained supply of inductive cues to drive both cartilage and bone formation without lengthy in vitro culture time prior to implantation in a defect [42, 48–59]. Similarly, and plasmid DNA encoding for specific growth factors can be loaded into MSs for delivery to cells within porcine MSC aggregates to drive, for example, bone formation [60]. Depending on the biomaterial, unloaded microparticles or MS can also serve as microenvironmental regulators of differentiation or act as ECM spacers within cells aggregates to improve oxygen and nutrient permeability, potentially allowing for engineering of large constructs [49, 61].
Given the emergence of RNAi as a powerful tool for programing regenerative cells, delivery of RNAi molecules has the potential to direct differentiation, alone or in combination with growth factors or pDNA, within cell aggregates. However, current high cell density technologies utilizing RNAi gene therapy typically transfect monolayer or suspended cells prior to aggregate formation [5, 62], which offers little control over the duration of gene silencing due to the transient nature of one-time miRNA or siRNA dosing. Encapsulation of RNAi mediators within MS has the capacity to incorporate these therapeutic molecules within stem cell aggregates and also package, protect, and deliver them for localized, controlled, and sustained gene knockdown. While delivery of bioactive RNA from MS incorporated within cell aggregates would be beneficial in tissue engineering strategies, a system with this capability has yet to be demonstrated.
In this work, an aqueous-based and phase-separated emulsion method was employed to engineer DEX-based hydrogel MS encapsulating siRNA complexed with endosmolytic micellar nanoparticles (siRNA-micelles), which were then incorporated within hMSC aggregates to permit localized delivery of siRNA-micelles for sustained gene silencing. The size of MS, siRNA encapsulation efficiency (EE), siRNA release behavior from MS, and bioactivity of released siRNA were examined. In addition, the ability of the siRNA-laden MS incorporated within hMSC aggregates to control the presentation of bioactive siRNA for sustained cellular gene silencing was examined. This approach has potential applications in the engineering of replacement tissues using high cell density aggregates to partially recreate native development and healing processes.
Materials and Methods
siRNA-micelle synthesis and characterization
The siRNA delivery vehicles used in this work are endosmolytic siRNA-micelles, previously shown to allow for effective cellular uptake and delivery of siRNA to the cytosol [27, 28, 63]. The siRNA-micelles are assembled from a reversible addition-fragmentation chain transfer (RAFT)-polymerized diblock copolymer comprising 2-(dimethylamino)ethyl methacrylate (DMAEMA), 2-propylacrylic acid (PAA), and butyl methacrylate (BMA). The diblock copolymer structure is poly{DMAEMA66-b-(DMAEMA86-co-BMA140-co-PAA56)} (Mn = 50.5 kDA, PDI = 1.32). The detailed chemical structure of the diblock copolymer is presented in Fig. S1 in Supporting Information (SI). Micelles (1 mg/mL) were assembled by dissolving the diblock copolymer in 100% ethanol (Sigma-Aldrich; 10 μL per 1 mg polymer) overnight followed by addition of RNAse-free diH2O at a rate of 8 mL/h, which triggers formation of micelles with cationic poly(DMAEMA) on the surface, amenable to RNA loading, and an ampholytic, pH-responsive poly(DMAEMA-co-BMA-co-PAA) core. siRNA in RNAse-free diH2O (50 μM) was electrostatically loaded on the micelle surface for 45 min at an N/P ratio of 6 in RNAse-free microcentrifuge tubes, where N and P refer to the number of amine and phosphate groups in the copolymer and siRNA, respectively [64]. To preserve bioactivity of siRNA during the lyophilization process, a solution of trehalose (Sigma-Aldrich) in RNAse-free diH2O (50 mg/mL) was then added to the siRNA-micelle nanoparticle solution (10% of the volume of the nanoparticle solution), and the mixture was allowed to stabilize at RT for 30 min, aliquoted, frozen, and lyophilized for at least 12 h before storing at −20 °C for future experiments. The sizes of siRNA-micelles pre- and post-lyophilization were measured using number-weighted dynamic light scattering (DLS, 90 Plus Particle Size Analyzer, Brookhaven Instrument Corporation, Holtsville, NY) at a concentration of 0.1 mg/mL micelle in diH2O (N=3 separate preparations per condition). The following sequences of siRNAs were used in this work: siGLO (fluorescently tagged siRNA; proprietary sequence by Dharmacon, Lafayette, CO), siRNA against green fluorescent protein (siGFP, 5’- GCA AGC UGA CCC UGA AGU UC-3’, Dharmacon), and negative control siRNA (siNC; 5’-UUC UCC GAA CGU GUC ACG UTT-3’; Shanghai GenePharma; Shanghai, China).
Synthesis of DEX-HEMA Microspheres
The MS polymer backbone chosen was DEX modified with the photocrosslinkable moiety hydroxyethyl methacrylate (DEX-HEMA; chemical structure in Fig. 1A), which was synthesized as previously reported [25, 35] and is described in more detail in SI (Fig. S2). MS with 6 or 10% (w/w) DEX-HEMA (MS6 and MS10, respectively) were synthesized using a modification of a previously described aqueous emulsion method [65, 66], which is summarized in Fig. 1B. DEX-HEMA solution (6 or 10%, w/w) in sterile RNAse-free diH2O containing 0.05% (w/v) Irgacure D-2959 photoinitiator (PI) (0.3 mL) was rapidly added to 1.5 mL solution of 20% (w/w) PEG (MW 10,000; Sigma-Aldrich) in RNAse-free diH2O containing 0.05% (w/v) PI in a 15 mL RNAse-free conical tube (Fisher Scientific). The mixture was vortexed for 10 s to achieve emulsification followed by immediate UV crosslinking in a sterile 50 mL beaker using Omnicure S100 UV Spot Cure System (Lumen Dynamics Group, Mississauga, Ontario, Canada) at 16–20 mW/cm2 for 30 s. The MS were then washed twice with diH2O (5 mL total) and collected by centrifugation at 1000 g (Survall Legend RT+ Centrifuge, Fisher Scientific) for 5 min followed by freezing at −80°C and lyophilization for at least 12 h before storing in lyophilized form at −20°C for use in future experiments. To prepare siRNA-loaded MS, lyophilized siRNA-micelles (40.00 μg siRNA) were suspended in 0.3 mL DEX-HEMA solutions before preparing the MS. Forty μg siRNA were loaded per batch of MS unless otherwise noted. Unloaded MS are termed empty MS, and MS encapsulating siRNA are termed siRNA-MS.
Figure 1.
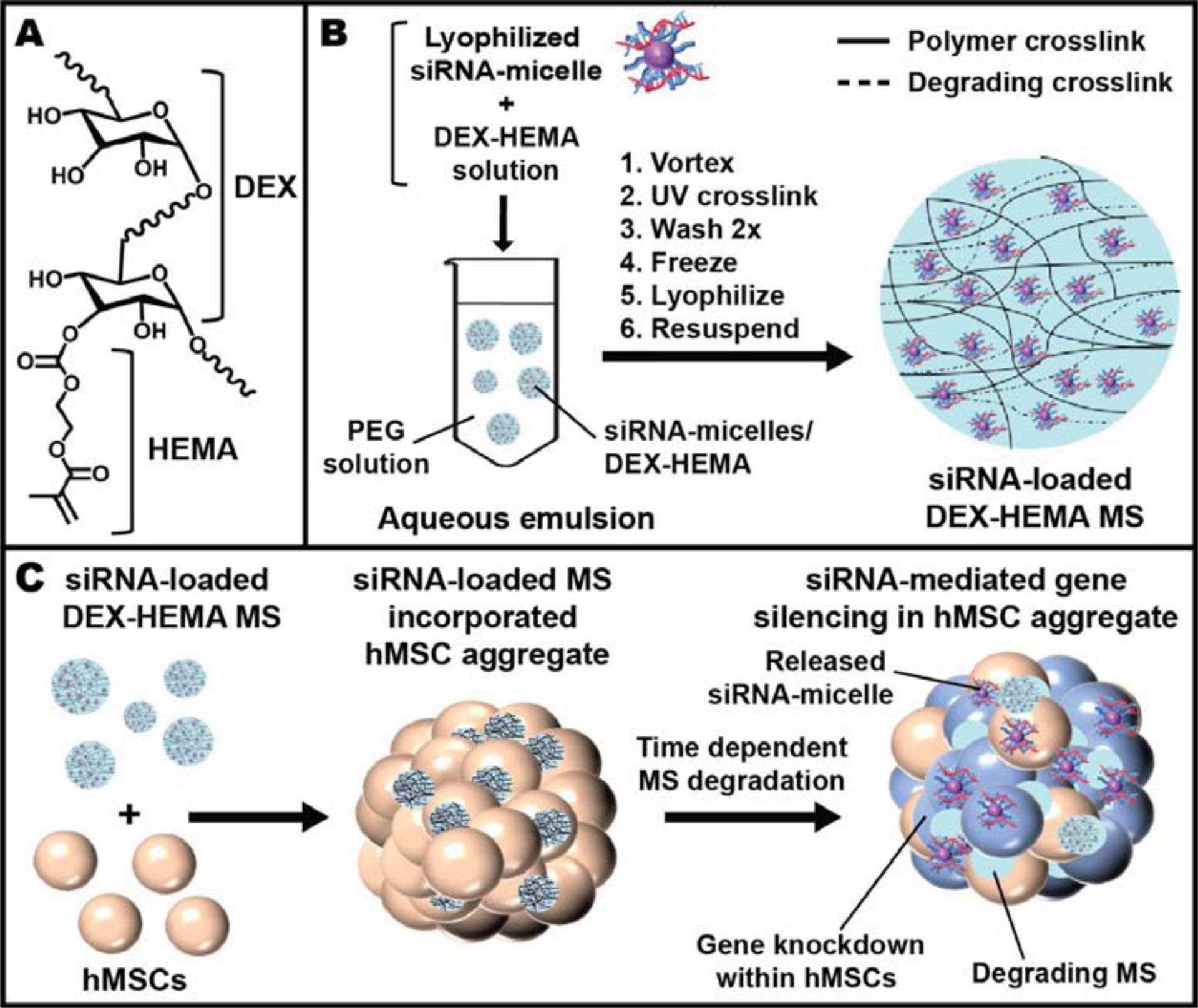
A) Chemical structure of DEX-HEMA. B) Schematic showing the fabrication of DEX-based MS encapsulating siRNA-micelles. C) Schematic depicting the incorporation of siRNA-MS into an hMSC aggregate for localized and sustained siRNA presentation and subsequent sustained gene silencing within a stem cell aggregate. Graphics are not to scale.
Light microscopy images of synthesized MS suspended in diH2O prior to lyophilization were acquired on an Olympus BX61VS microscope (Olympus, Center Valley, PA, USA) with a Pike F-505 camera (Allied Vision Technologies, Stadtroda, Germany), and MS diameters were measured using Image J analysis software (NIH, Washington, DC; N = 300 MS per batch). The reported values are the average values from three different MS fabrication batches per condition.
siRNA encapsulation efficiency (EE)
To determine the EE of siRNA in MS, lyophilized MS encapsulating siGLO-micelles (siGLO-MS) were prepared as previously described (40.00 μg siRNA per MS batch). Lyophilized siGLO-MS (2.0 mg) were dissolved in 0.5 mL 10 N NaOH (Fisher Scientific) overnight at 37°C with rotation to allow for complete degradation of crosslinked DEX-HEMA and dissociation of siGLO from micelles. The obtained solutions were then diluted 3-fold with a mixture of diH2O and Dulbecco’s Modified Eagles Medium (DMEM, Sigma-Aldrich) phenol red-free media (1/1, v/v) followed by siGLO quantification. A series of standard samples with known siGLO concentrations were used to establish a standard curve to calculate the concentration of siGLO in the samples. To prepare the standard samples, 8.00 μg siGLO in the form of lyophilized siGLO-micelles were suspended in 1 mL RNAse-free diH2O containing 0.05% (w/v) PI, vortexed, and UV applied for 30 s to mimic UV applied during the formation of MS. The solution was then frozen, lyophilized, and resuspended with a mixture of 10N-NaOH/diH2O/DMEM-phenol red-free medium (1/1/1, v/v/v). The amount of siGLO in the standards and samples was then quantified via recording of fluorescence intensity of 90 μL of samples or standards in duplicate wells of black 96-well microplates on a microplate reader (Synergy H1 hybrid reader, Biotek, Winooski, VT) set for excitation at 480 nm and emission at 520 nm. The samples from empty MS were measured in parallel and the average fluorescence signal from empty MS was subtracted from corresponding values of siGLO-MS to account for potential effects of degraded polymers in the samples. EE was calculated by EE (%) = WsiGLO-A/WsiGLO-T*100, where WsiGLO-A and WsiGLO-T refer to the actual measured amount and the theoretical amount of siGLO in 2.0 mg siGLO-MS, respectively. The WsiGLO-T values for 2.0 mg of MS6 and MS10 are 4.44 and 2.66 μg, respectively (i.e. WsiGLO-T 6 w/w MS = 2.0 mg MS *(40.00 μg siRNA)/(18.0 mg DEX-HEMA per batch MS) = 4.44 μg siRNA; WsiGLO-T 10 w/w MS = 2.0 mg MS *(40.00 μg siRNA)/(30.0 mg DEX-HEMA per batch MS) = 2.66 μg siRNA; 0.3 mL of 6 and 10% (w/w) DEX-HEMA solutions contain 18.0 and 30.0 mg of DEX-HEMA, respectively). We assumed that RNA encapsulation efficiency for all subsequent experiments in this work was similar to that for siGLO in the MS.
siRNA release kinetics from MS
To examine the release kinetics of siRNA from MS, release of siGLO from 3 batches of siGLO-MS was performed. Lyophilized siGLO-MS (2.0 mg) were prepared as described previously and suspended in release medium consisting of 0.5 mL DMEM-phenol red-free medium in 1.7 mL RNAse-free microcentrifuge tubes (N=3), which were then incubated at 37°C with rotation. At predetermined time points, the samples were centrifuged at 1000 g for 5 min (Survall Legend RT+ Centrifuge) and releasates were collected and stored at −20°C prior to assaying samples. Fresh release medium (0.5 mL) was added to the sample tubes to continue the release. To quantify the siGLO concentration in the releasates, a series of standard samples with known siGLO concentrations was used to establish a standard curve. To prepare the standard samples, 8.00 μg siGLO in the form of lyophilized siGLO-micelles was suspended in 1.5 mL RNAse-free diH20 containing 0.05% (w/v) PI, vortexed, and UV was applied for 30 s prior to freezing, lyophilizing, and resuspending in DMEM-phenol red-free media. Mixtures of samples or standards were mixed with diH2O and a solution of heparin (10 mg/mL; Sigma-Aldrich) in diH2O (1/1/2, v/v/v) and added in duplicate to wells of black 96-well microplates. The dissociation of siRNA from nanoparticles in the presence of heparin in solution was confirmed (Fig. S3 in SI). The microplates were then placed on a shaker at RT for 1 h before recording fluorescence intensity on a microplate reader set for excitation at 480 nm and emission at 520 nm. The releasates from empty MS were measured in parallel and the average fluorescence signal of empty MS was subtracted from corresponding values of siGLO-MS to account for potential effects of degraded polymers in the results. Percentage cumulative siRNA release was calculated based on WsiGLO-A from 2.0 mg MS.
Cell culture
hMSCs were isolated from bone marrow aspirates from the posterior iliac crest of healthy human donors as previously described [26], and details are presented in SI. Cells were expanded in expansion medium consisting of DMEM-Low Glucose (DMEM-LG, Sigma-Aldrich) supplemented with pre-screened 10% (v/v) FBS (Sigma-Aldrich), 1% (v/v) penicillin/streptomycin (P/S, Corning Inc, Corning, NY), and 10 ng/mL fibroblast growth factor-2 (FGF-2, R&D Systems, Minneapolis, MN). Passage 3 or 4 hMSCs were used for monolayer and aggregate experiments.
To prepare GFP-hMSCs, vesicular stomatitis virus vector particles (VSV-G) expressing GFP pseudotyped lentiviruses were fabricated and transduced into hMCSs. To fabricate VSV-G expressing GFP pseudotyped lentiviruses, 5 μg pLenti-CMV-MCS-GFP-SV-Puro-Addgene plasmid #73582 (a generous gift from Dr. Paul Odgren, Department of Cell and Developmental Biology, University of Massachusetts Medical School, Worcester, MA) [67], 5 μg pCMV delta R8.2-Addgene plasmid #12263, and 2 μg pMD2.G-Addgene plasmid #12259 (generous gifts from Dr. Didier Trono, School of Life Sciences, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland) vectors were transiently transfected with PEI (Sigma-Aldrich) into HEK 293-T cells (ATCC, Manassas, VA) cultured in a T150 flask. Viruses were collected at 36 and 48 h after transfection and filtered through a 0.22 μm-filter. VSV-G expressing GFP pseudotyped lentiviruses were then transduced into hMSCs (passage 3) by spinoculating at 3480 rpm (centrifuge 5810R, Eppendorf) for 1.5 h at room temperature followed by culture in T25 flasks (2.5 × 104 cells/flask) for 2 days [68, 69]. The transduced hMSCs were isolated by culture in medium composed of DMEM-LG supplemented with 10% (v/v) characterized FBS, 1% (v/v) P/S and puromycin (2 μg/mL; Invitrogen, Carlsbad, CA) for 3 days before harvesting to confirm GFP expression via flow cytometry and expanding to increase cell number. The GFP-hMSCs were cultured in expansion medium, as defined above, and passage 5 and 6 GFP-hMSCs were used for monolayer experiments and aggregate formation.
Destabilized GFP (deGFP)-expressing HeLa cells (generously gifted by Dr. Matthew Levy, Albert Einstein College of Medicine, Bronx, NY) were cultured in growth medium of DMEM-High Glucose (DMEM-HG, Sigma-Aldrich) supplemented with 5% (v/v) characterized FBS (Corning), 1% (v/v) P/S, and 0.5% G-418 (500 μg/mL; Fisher Scientific). G-418 was used in expansion culture to select for cells expressing GFP to ensure that cells expressed the target gene prior to use in the experimental studies. All cells in this study were cultured in a humidified incubator at 37°C and 5% CO2.
Bioactivity of released siRNA-micelles from MS
To examine the bioactivity of encapsulated and released siRNA, empty MS, siNC-MS, or siGFP-MS were UV-sterilized in the cell culture hood for 20 min before performing the release experiment, which was performed in a sterile manner, and collected releasates were applied to deGFP-expressing HeLa cells in monolayer culture. deGFP-expressing HeLa cells were plated at a density of 5 × 104 cells/well in 24-well plates (Fisher Scientific) containing 0.5 mL growth medium for 24 h prior to treating with 0.5 mL releasates (N=3 wells per condition per time point). Untreated cells were cultured with DMEM-phenol red-free media containing no RNA (N=3 wells) during the transfection period. Positive and negative controls were treated with 0.33 μg siNC in the form of lyophilized siNC-micelles ((−) ctrl, N=3 wells) and 0.33 μg siGFP in the form of lyophilized siGFP-micelles ((+) ctrl, N=3 wells), respectively, suspended in 0.5 mL DMEM-phenol red-free media. After 6 h of transfection, the medium was replaced with growth medium without G-418. Cells were then cultured for 2 days followed by harvesting for flow cytometry analysis (BD Leica LSR II, Becton Dickinson and Company, Franklin Lakes, NJ; 10,000 cells per sample). Untreated cells were assigned 100% GFP expression, and GFP expression in all groups was normalized to the untreated group.
Distribution of MS within hMSC aggregates and siRNA delivery to hMSCs
To examine the incorporation and distribution of MS within cell aggregates, rhodamine-labeled MS were prepared and incorporated into hMSC aggregates, followed by imaging of the rhodamine fluorescence signal. To prepare rhodamine-labeled MS, DEX-HEMA was dissolved in a solution of methacrylated-rhodamine (0.2 mg/mL; Polysciences, Warrington, PA) in RNAse-free diH2O containing 0.05% (w/v) PI, and the MS were fabricated as previously described. Empty rhodamine-labeled MS were UV-sterilized in the cell culture hood for at least 20 min before suspending in chondrogenic pellet medium (CPM), consisting of DMEM-HG supplemented with 1% ITS+ Premix (Corning Inc), 37.5 μg/mL ascorbate-2-phosphate (Wako USA, Richmond, VA), 10−7 M dexamethasone (MP Biomedicals, Solon OH), 1% nonessential amino acids (NEAA; Lonza Group, Basel, Switzerland), 1% sodium pyruvate (GE Healthcare Life Sciences, Logan, UT), 1% P/S, and 10 ng/mL TGF-β1 (PeproTech, Rocky Hill, NJ), at a concentration of 0.75 mg/mL (0.15 mg/aggregate) or 1.5 mg/mL (0.30 mg/aggregate). Harvested hMSCs were then suspended in CPM containing MS (1.25 × 106 cells/mL), and 200 μL cell suspension solution was dispensed per well into sterile 96-well V-bottom polypropylene microplates (Fisher Scientific), which were then centrifuged at 500 g for 5 min to form aggregates. The aggregates were cultured for 3 days, and then harvested, embedded in optimal cutting temperature (OCT; Fisher Scientific), flash-frozen in liquid nitrogen, and stored at −20°C prior to cryosectioning (10 μm thick sections). Slides with mounted tissues were fixed in acetone for 20 min prior to application of DAPI solution (1 μg/mL; Thermo Scientific) for 20 min, and then they were mounted with cover slips (Fluoromount; Sigma-Aldrich). Images of DAPI and rhodamine-labeled MS distribution in aggregates were acquired on a fluorescence microscope (ECLIPSE TE 300; Nikon, Tokyo, Japan) equipped with a digital camera (Retiga-SRV; Qimaging, Burnaby, BC, Canada) (N=3 aggregates).
To determine the ability of MS to deliver siRNA to cells in aggregates, UV-sterilized rhodamine-labeled MS encapsulating no RNA or lyophilized siGLO-micelles (16.00 μg siRNA in 0.3 mL DEX-HEMA solution) were used. The DAPI, rhodamine, and GLO signals of the sectioned aggregates were acquired on a Leica TCS SP8 confocal microscope (Buffalo Grove, IL) (N=2 aggregates per time point).
Encapsulation of siGFP-MS within GFP-hMSC aggregates for GFP silencing
Prior to encapsulating siGFP-MS within GFP-hMSC aggregates, the ability of lyophilized, siGFP-micelles to silence GFP expression in monolayer cultured GFP-hMSCs was confirmed (Fig. S7 in SI). GFP-hMSC aggregates (passage 6) incorporating empty, siNC-, or siGFP-MS (0.30 mg MS/aggregate) were fabricated as previously described, cultured, and harvested at predetermined time points for quantification of GFP mRNA expression, visualization of GFP expression, and analysis of DNA content, the latter of which is presented in SI.
To quantify the expression of GFP reporter gene in aggregates, the harvested GFP-hMSC aggregates (N=3 aggregates per condition per time point) were suspended in 1 mL of TriReagent (Sigma-Aldrich) followed by homogenization (35,000 rpm, TH Homogenizer, Omni International, Marietta, GA) for 30 s on ice. RNA was then isolated according to the manufacturer’s instructions and quantified spectrophotometrically based on A260 measurement using a NanoDrop spectrophotometer (Labtech International, Uckfield, UK). Following first strand cDNA synthesis (Applied Biosystems High Capacity Reverse Transcription Kit, Foster City, CA), fold changes in mRNA transcripts were determined by quantitative real-time polymerase chain reaction (qRT-PCR) using SYBR® Premix Ex Taq™ kit and RT-PCR detection system (Takara, St. Louis, MO). Relative levels of GFP mRNA were calculated using the ΔΔCt method and normalized to relative mRNA levels of the constitutively expressed 18S gene. Fold changes were calculated relative to cells only aggregates at each time point. The following primer sets (Invitrogen) were used in this work: 18S Forward: GTA ACC CGT TGA ACC CCA TT; 18S Reverse: CCA TCC AAT CGG TAG TAG CG; GFP Forward: CTG GTC GAG CTG GAC GGC G; and GFP Reverse: CTC GTC CAT GCC GAG AGT G.
To visualize GFP expression, the aggregates were embedded in OCT (N=3 aggregates per condition per time point) for subsequent sectioning, DAPI staining (Fig. S8 in SI), and confocal imaging, as described previously.
Statistical analysis
Results are presented as means ± standard deviation. Statistical analyses of siRNA release curves were performed using two-way ANOVA with post-hoc Tukey analysis using GraphPad Prism software (La Jolla, CA). All other statistical analyses were performed using one-way ANOVA with post-hoc Tukey analysis. p<0.05 were considered statistically significant.
Results
Characterization of siRNA-micelles
Delivery carriers are typically required to facilitate cellular uptake and release of unmodified siRNA into the cytoplasm. The endosmolytic micelle nanoparticles used in this work (chemical structure shown in Fig. S1 in SI) were previously shown to destabilize at pH 5 to allow for pH-sensitive micelle destabilization and endosomal rupture for effective delivery of siRNA to the cell cytosol [28]. DLS analysis revealed that both micelles alone and siRNA-micelles increased in size post-lyophilization (Fig. 2). The micelle particle average diameter pre-lyophilization was 53 ± 14 nm and post-lyophilization was 179 ± 151 nm, an approximate 3.4-fold increase in size. Similarly, the average diameter of siRNA-micelles pre-lyophilization was 60 ± 20 and post-lyophilization was 140 ± 56 nm, an approximate 2.4-fold increase in size.
Figure 2.
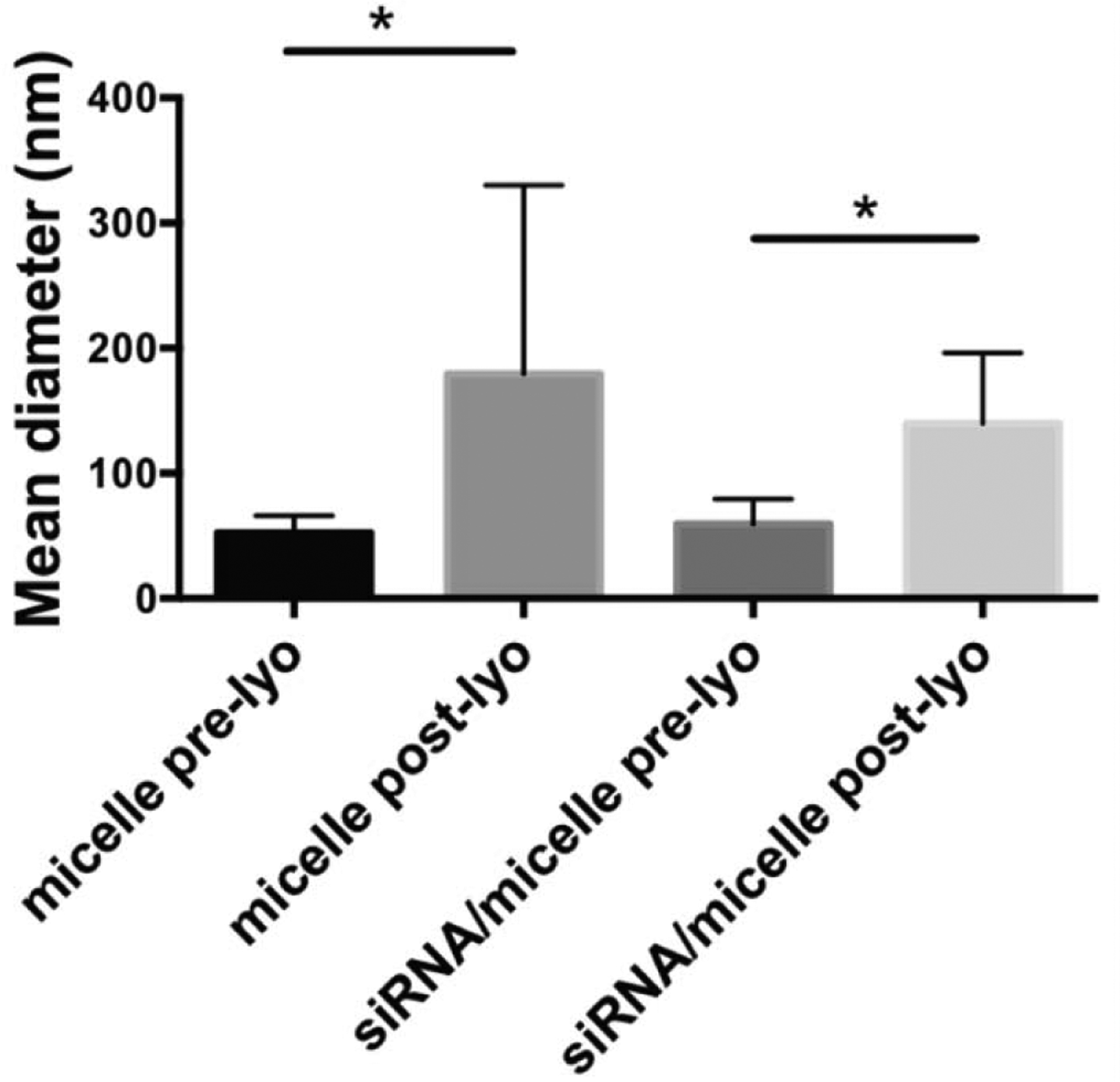
DLS characterization of micelle size shows the influence of siRNA loading lyophilization (lyo), pre- and post-lyo, on particle size in the presence of trehalose. * p<0.05.
MS fabrication, siRNA encapsulation, and MS characterization
DEX-HEMA was fabricated by conjugating HEMA to the backbone of DEX, a bacterial polysaccharide [70] with a history of use in FDA-approved products [71, 72], to enable creation of a biodegradable, cytocompatible hydrogel which has been used for sustained siRNA delivery to encapsulated and surrounding cells [25, 35]. DEX-HEMA with 6.7% actual degree of HEMA modification, determined by 1H-NMR (Fig. S2 in SI), was used to prepare MS with and without loaded siRNA-micelles by an aqueous emulsion technique based on immiscibility of DEX in PEG, as depicted in Fig. 1B. Two formulations of MS, MS6 and MS10, were fabricated to examine the role of polymer concentration on degradation rate and resultant siRNA release kinetics. The characteristics of fabricated MS are summarized in Table 1. Light microscopy images of MS (Fig. 3) reveal a spherical shape with mean diameter ranging from 3.8 to 6.5 μm depending on polymer concentration and the presence or absence of encapsulated RNA. Changing hydrogel polymer concentration and/or encapsulating siRNA-micelle nanoparticles within MS did not significantly affect the mean diameter of the fabricated MS. The EEs were calculated from 3 different batches of siGLO into MS6 and MS10 (84 ± 7 % and 85 ± 9 %, respectively) and were not significantly different.
Table 1:
Characteristics of fabricated microspheres
| Formulation | Mean diameter (μm) | Theoretical siRNA (2.0 mg MS) | Avg. actual siRNA (2.0 mg MS) | Avg. EE (%) |
|---|---|---|---|---|
| Empty-MS6 | 5.4 ± 2.0 | -- | -- | -- |
| siRNA-MS6 | 5.0 ± 1.7 | 4.44 μg | 3.75 μg | 84 ± 7 |
| Empty-MS10 | 6.5 ± 2.9 | -- | -- | -- |
| siRNA-MS10 | 3.8 ± 1.8 | 2.67 μg | 2.13 μg | 85 ± 9 |
Figure 3.
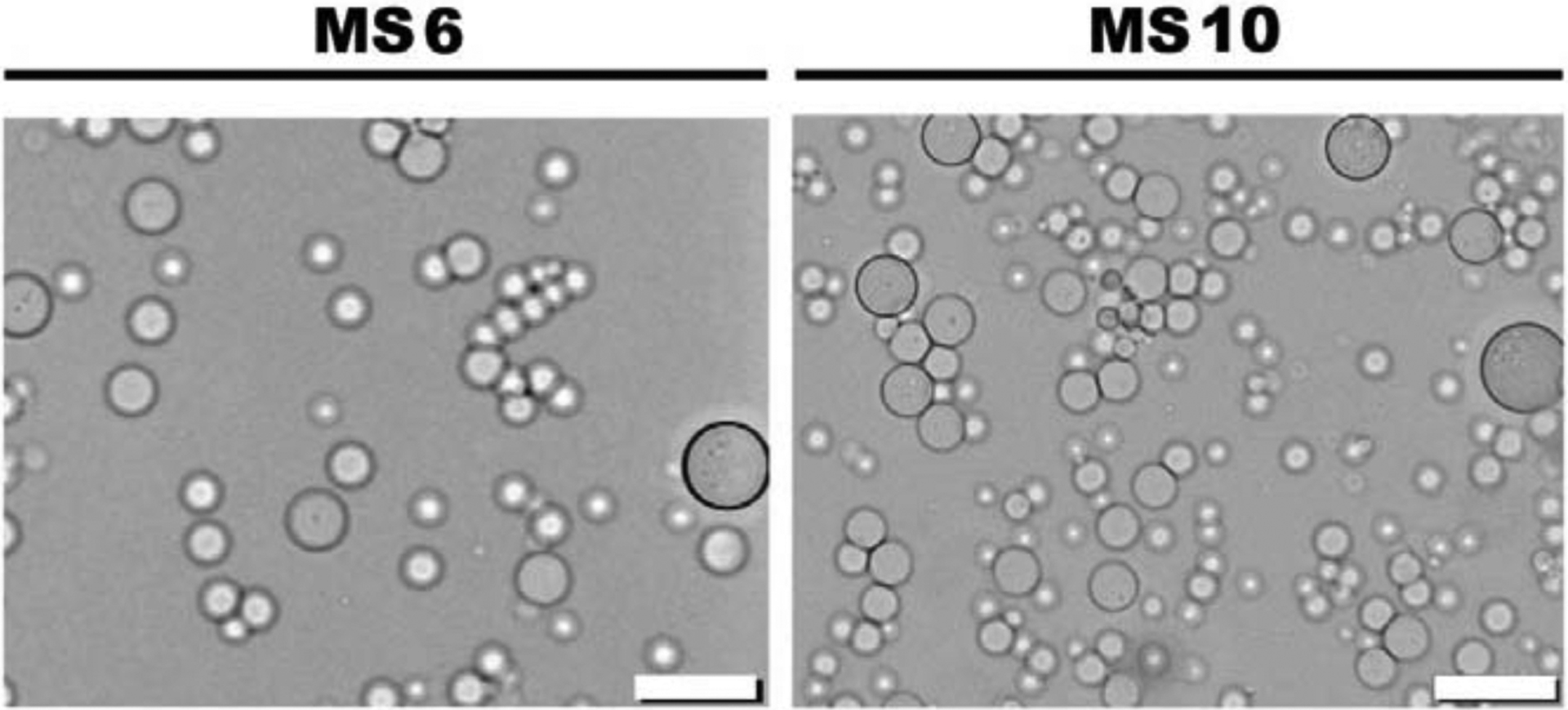
Light microscopy images of siGLO-MS6 and siGLO-MS10. Scale bars indicate 20 μm.
siRNA release kinetics from MS
To determine the effect of MS crosslinking density on the release kinetics of siRNA-micelles, the release of siGLO from MS6 and MS10 was performed using 3 different MS batches per formulation, and the data was plotted separately (Fig. 4). The cumulative siRNA release profiles demonstrate similar initial release rates from both MS6 (Fig. 4A) and MS10 (Fig. 4B) formulations, with approximately 30% payload released by day 8, and sustained siRNA release up to 28 days from both formulations. MS6 release rate then increased relative to MS10 with linear release observed in MS6 releases 2 and 3, which released approximately 80% siRNA by day 28. MS6 release 1 showed significantly higher release rate with most siRNA released by day 18 (Fig. 4A). Release from MS10 demonstrated similar release profiles between the three MS batches, resulting in sustained release profiles with approximately 58% siRNA release by 28 days (Fig. 4B).
Figure 4.
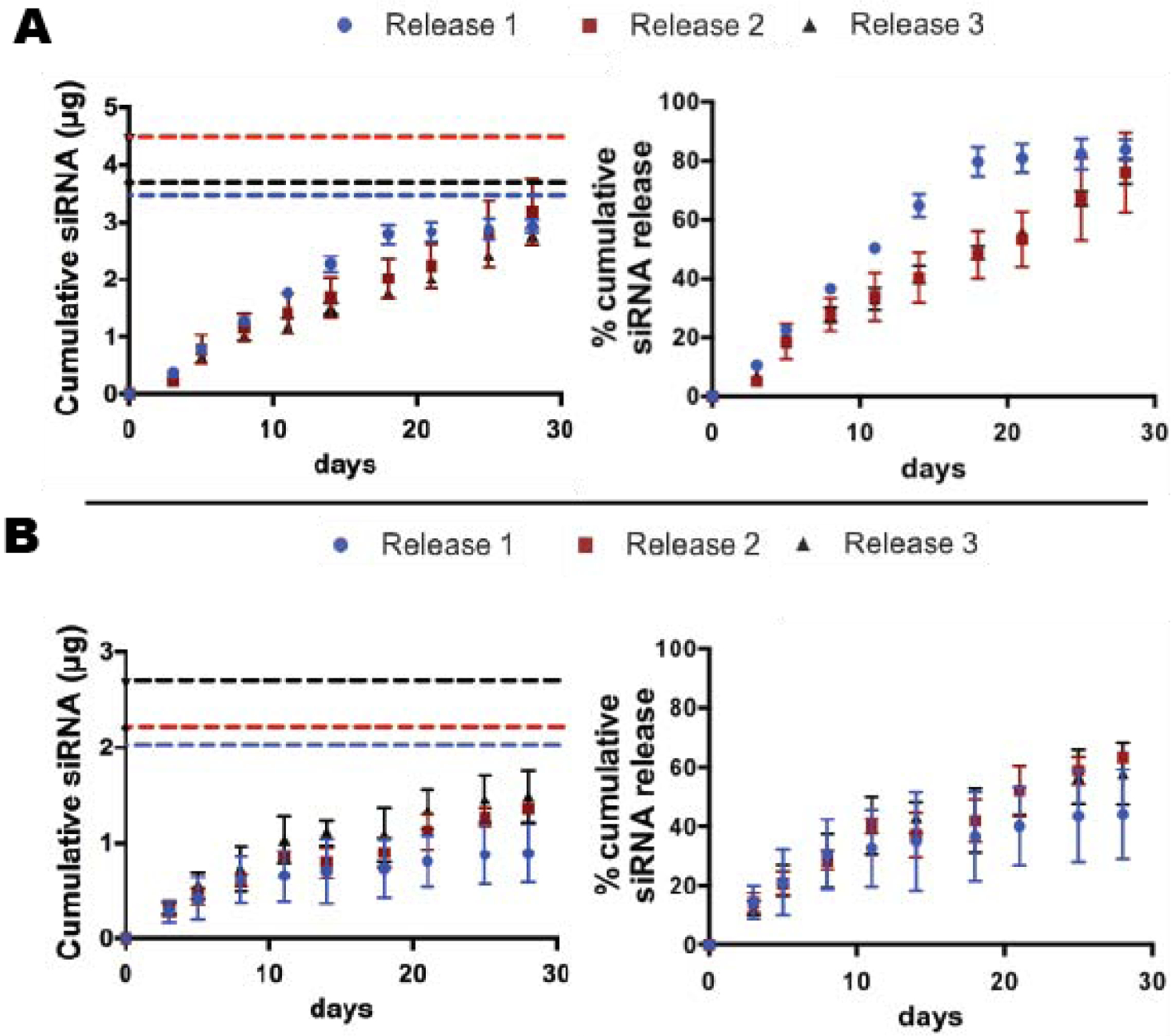
siGLO release profiles from A) MS6 and B) MS10 over the course of 28 days. Cumulative siRNA (left) and % cumulative siRNA (right) release data from 3 different MS batches are plotted separately. The dashed lines (left) represent the actual incorporated siRNA content in 2.0 mg MS for each batch. MS6 release 1 was significantly different from the other two MS6 release profiles, p<0.05.
Bioactivity of released siRNA-micelles from MS
It was then important to determine the capacity of the siRNA-micelles to maintain target gene knockdown following lyophilization, MS fabrication, and the release process. Released siGLO-micelles from MS retained their capacity for internalization by monolayer hMSCs (Fig. S4 in SI). To validate the bioactivity of released siRNA, the capacity of siGFP-micelles released from MS6 and MS10 to mediate gene silencing of the reporter gene, deGFP, was assessed. Monolayer deGFP-HeLa cells were used to take advantage of the destabilized nature of the GFP expressed by this cell population, which is an efficient RNAi reporter target due to the shorter half-life of deGFP relative to wild-type GFP protein [19]. Cells treated with siNC-micelles ((−) ctrl) or releasates from empty MS showed similar GFP expression compared to untreated cells (100% GFP expression), while GFP expression in cells treated with siGFP-micelles ((+) ctrl) was completely knocked-down (Fig. 5, Fig. S5A in SI). Cellular GFP expression levels were significantly decreased with treatment of releasates collected between days 0–5 from MS10 and releasates between days 6–10 from both siGFP-loaded MS formulations compared to no treatment or treatment with releasates from empty or siNC-MS (Fig. 5). No GFP knockdown was observed when cells were treated with releasates collected between days 11–15 from either siGFP-MS formulations. The bioactivity of siGFP released from MS6 and MS10 was also assessed using additional MS batches for each formulation, and the data is plotted separately and presented in Fig. S5 in SI. Results from experiments using different MS batches demonstrated a similar trend in which GFP knockdown was achieved with treatment of releasates collected between days 6–10 from both siGFP-MS formulations. In addition, both UV application in the presence of PI and DEX degradation byproducts reduced the capacity of siGFP to silence GFP expression in monolayer deGFP-HeLa cells, whereas incubation of siRNA-micelles in media, which could potentially result in nanoparticle aggregation, had no effect on siRNA bioactivity (Fig. S6 in SI).
Figure 5.
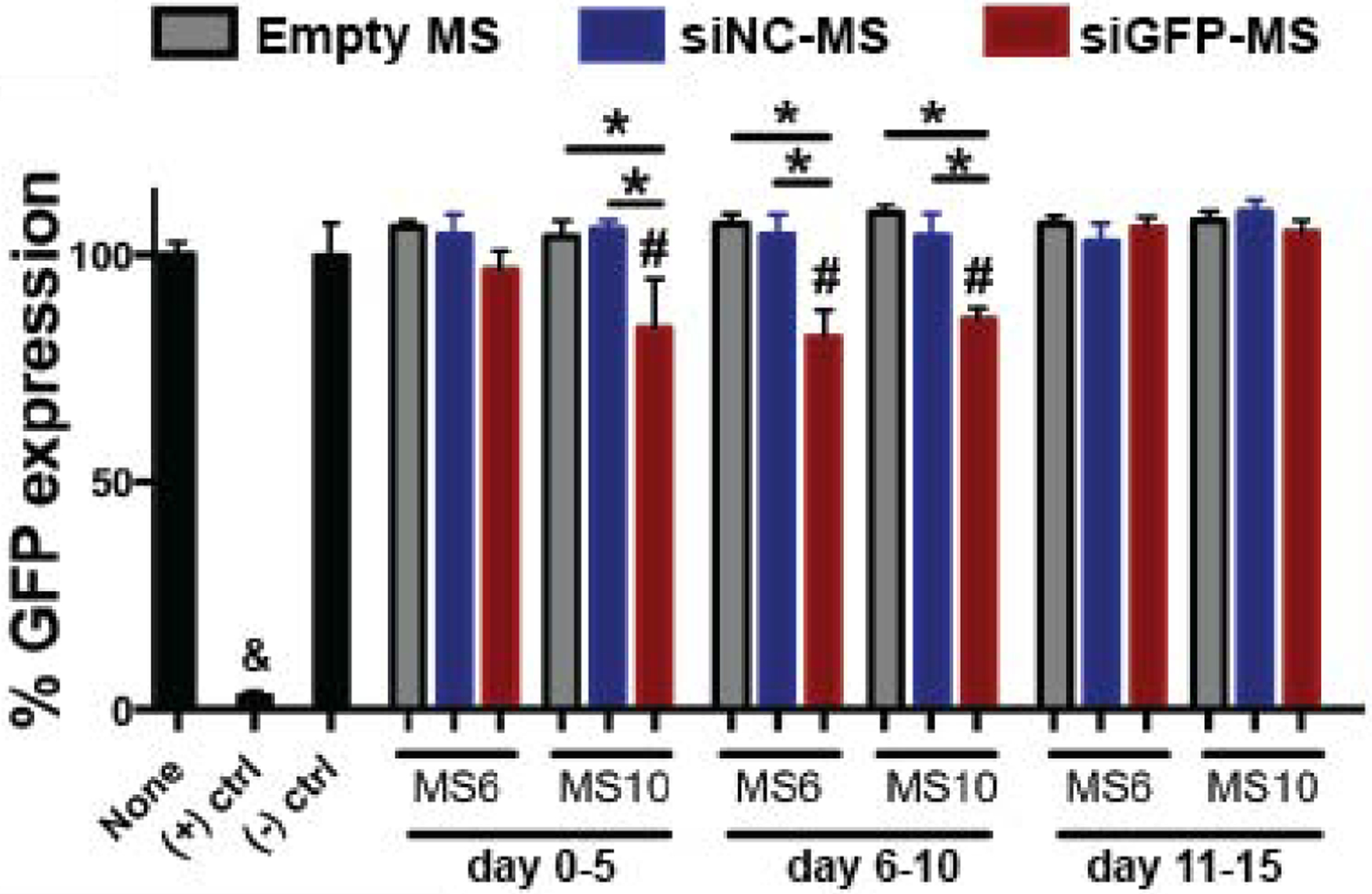
GFP expression of monolayer deGFP-Hela cells treated with MS releasates. & p<0.05 compared to all other groups, # p<0.05 compared to None and (−) ctrl, * p<0.05. None = untreated cells.
MS distribution and siRNA delivery to cells in aggregates
To investigate spatial distribution of MS within hMSC aggregates, MS labeled with rhodamine, a fluorescent molecule, were incorporated within hMSC aggregates and the distribution of rhodamine was examined. Incorporation of both MS formulations at 2 concentrations (i.e., 0.15 and 0.30 mg MS/aggregate) within aggregates did not disrupt aggregate formation, and the MS were distributed throughout the 3D cell constructs (Fig. 6A). In addition, siGLO-micelles were also loaded into rhodamine-labelled MS before incorporation into aggregates for visualization of siGLO cellular uptake over time. The localized and sustained delivery of siGLO to stem cells in 3D aggregates was achieved in both MS formulations, with decreasing siGLO signal over time potentially due to diffusion of siGLO into the surrounding environment, siRNA dilution through cell division, siRNA degradation, or increased matrix production leading to a higher distribution of microspheres and resultant decrease in local siRNA concentration (Fig. 6B). Increased intensity of siGLO fluorescence was visualized in hMSC aggregates embedded with MS6 compared to MS10 at day 14, potentially due to the higher amount of siGLO in MS6 loaded aggregates (WsiGLO-A = 0.11 and 0.07 μg siGLO in MS6 and MS10 incorporated aggregates, respectively). MS10 showed sustained presentation of siGLO within the aggregates up to day 20 (Fig. 6B).
Figure 6.
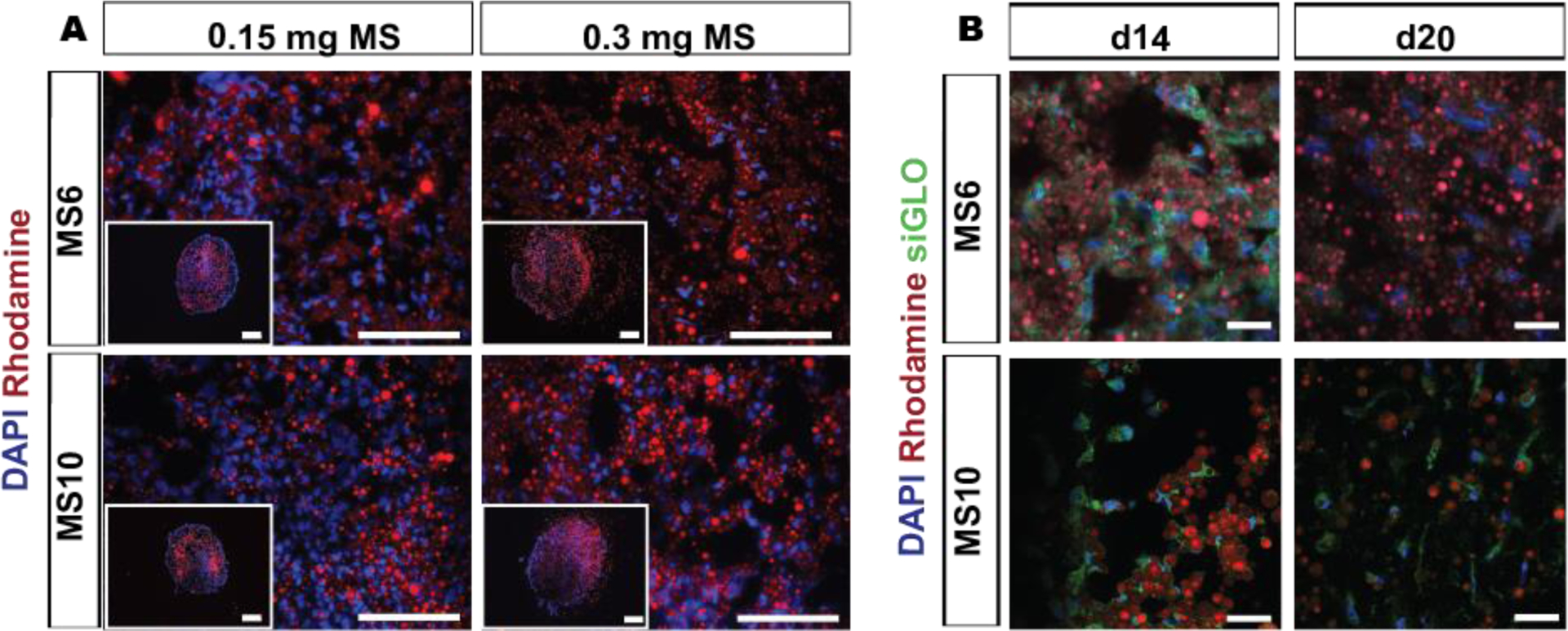
Distribution of incorporated MS in hMSC aggregates for sustained siRNA presentation. A) Fluorescence photomicrographs of rhodamine-labeled MS (red) incorporated into hMSC aggregates (0.15 and 0.30 mg MS per aggregate) and DAPI-stained cell nuclei (blue) after 3 days in vitro culture. Scale bars indicate 200 μm. B) Fluorescence confocal photomicrographs of rhodamine-labeled (red) siGLO-MS incorporated into hMSC aggregates to visualize siGLO uptake (green) and DAPI-stained hMSC nuclei (blue) in 3D aggregates after different culture periods. Scale bars indicate 30 μm.
GFP silencing within GFP-hMSC aggregates
Having demonstrated that the MS homogenously incorporated within hMSC aggregates (Fig. 6A) and lyophilized siGFP-micelles or MS-released siGFP-micelles suppressed GFP expression in monolayer GFP-hMSCs (Fig. S7 in SI) or deGFP-HeLa cells (Fig. 5, Fig. S5 in SI), respectively, the capacity of siRNA-MS incorporated into hMSC aggregates to regulate gene expression was investigated. GFP mRNA expression of GFP-hMSC aggregates embedded with siGFP-MS was quantified by qRT-PCR analysis after 5, 10, 15 and 20 days of culture. When siGFP-micelle-loaded MS6 were incorporated into the GFP-hMSC aggregates (0.56 μg siGFP in 0.30 mg siRNA-MS6, based on EE of siGLO, incorporated per aggregate), significant reduction in GFP expression was achieved in a sustained fashion for up to 15 days. Specifically, 77% and 95% knockdown of GFP mRNA occurred at days 5 and 10, respectively, which was significantly greater than cells only, empty MS6, and siNC-MS6 conditions, and 42% knockdown was attained at day 15, which was significantly greater than the siNC-MS6 condition (Fig. 7A–C). No significant reduction in GFP expression was observed at day 20 (Fig. 7D). The incorporation of siGFP-MS10 into the GFP-hMSC aggregates (0.32 μg siGFP in 0.30 mg siRNA-MS10, based on EE of siGLO, incorporated per aggregate) resulted in 38% and 35% reduction in GFP expression after 5 and 10 days, respectively, with significant GFP knockdown achieved at day 5 relative to siNC-MS10 (Fig. 7). Knockdown was not statistically significant at day 10 or later time points. Interestingly, incorporation of empty MS10 or siRNA-MS10 into the GFP-hMSC aggregates resulted in increased GFP expression compared to cells only aggregates at day 20.
Figure 7.
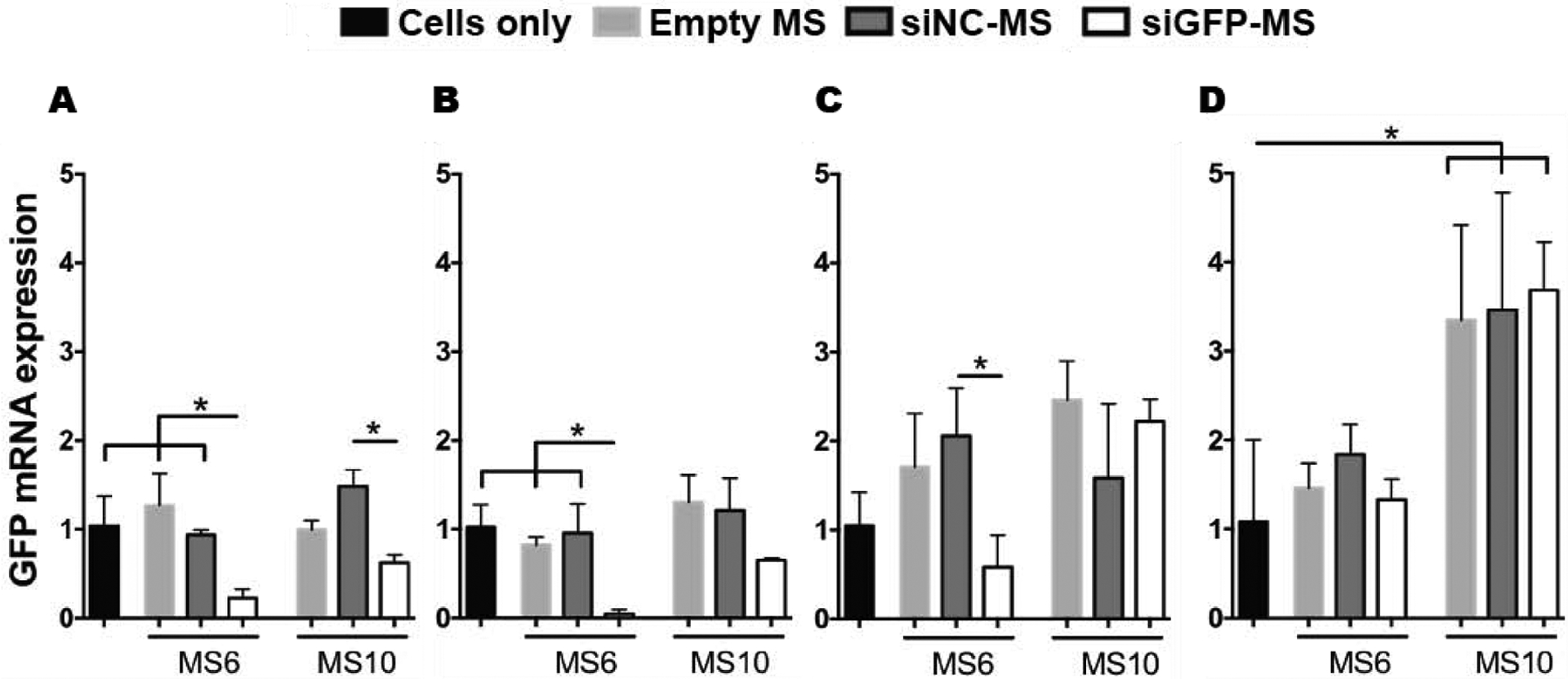
siGFP-mediated knockdown of GFP mRNA expression within GFP-hMSC aggregates. Analysis of fold changes in GFP mRNA expression of GFP-hMSC aggregates embedded with different MS formulations compared to cells only aggregates at A) 5, B) 10, C) 15 and D) 20 days in vitro culture. * p<0.05.
The capacity of encapsulated siRNA-MS to regulate gene expression within the hMSC aggregates was investigated via visualization of cellular GFP expression in sectioned aggregates with confocal microscopy. Fluorescence images of GFP-hMSCs aggregates corroborated the qRT-PCR analysis (Fig. 8). A similar or slightly increased degree of GFP signal was observed in aggregates incorporating empty and siNC-MS6 compared to cells only aggregates at all experimental time points. In contrast, when siGFP-MS6 were incorporated into aggregates, a substantial decrease in GFP signal was observed at all experimental time points (Fig. 8) while the DAPI signal remained similar to siNC-MS6 group (Fig. S8 in SI), indicating siGFP-mediated GFP silencing. In addition, incorporation of MS6s into the aggregates did not adversely affect cell number by day 20 (Fig. S9 in SI).
Figure 8.
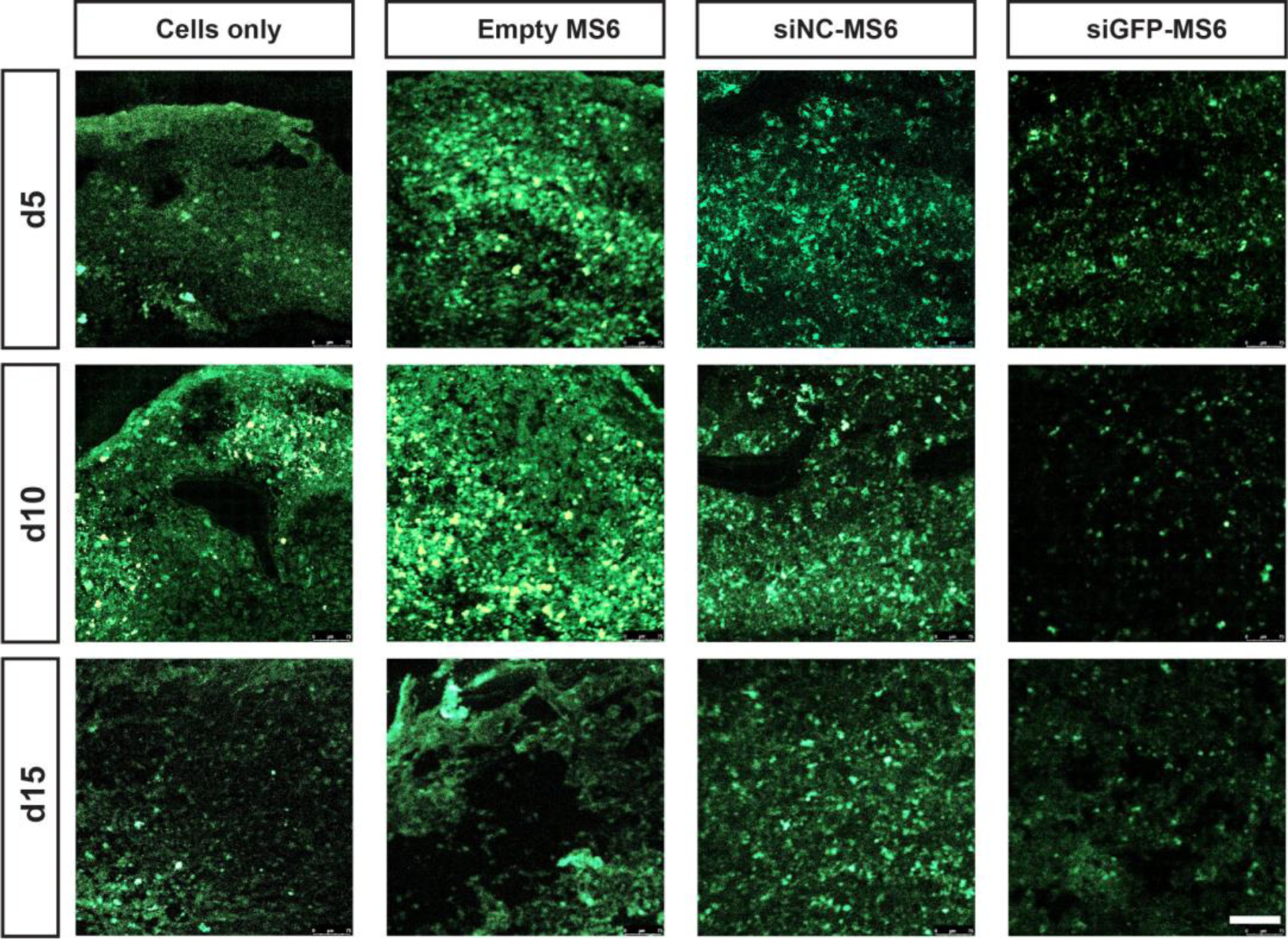
siGFP-mediated silencing of GFP expression within GFP-hMSC aggregates. Tissue sections of aggregates show GFP signal (green) of GFP-hMSC aggregates incorporated with cells only, and empty-, siNC- and siGFP-MS6 at different culture times. Scale bar indicates 100 μm.
Discussion
Here, we pursued the concept that partially recapitulating aspects of native musculoskeletal development and/or healing processes with high density cell aggregates and controlling the presentation of RNAi molecules within these aggregates is a promising strategy for tissue engineering. Since RNAi has been demonstrated to have a widespread role in the formation and/or regeneration of tissue such as cartilage [4–6] and bone [8–10], bulk hydrogel-mediated delivery systems have been employed to achieve RNAi-mediated enhanced tissue formation [23, 26] and to address issues associated with RNA instability and short half-life [15, 17]. This work differs from current scaffold-based approaches by mediating gene transfer from siRNA-loaded hydrogel MS within cellular aggregates. This confers the advantage of scaffold-based delivery systems, namely, localized and controlled presentation of siRNA, while also allowing for fabrication of a more biomimetic microenvironment that permits ample cell-cell interactions. The overall purpose of this present study was to provide a new strategy for temporally controlled gene silencing within stem cell aggregates for potential application in the engineering of orthopedic tissues.
Despite the great potential of siRNA in tissue regeneration strategies, delivery remains the major hurdle to more widespread use in the clinic [73]. Numerous transfection reagents have been developed to shield siRNA from RNAses in the microenvironment and allow for cellular uptake [11, 12]. Here, highly effective endosomolytic micellar nanoparticles, whose therapeutic potential has been previously demonstrated when delivered from PEG-based hydrogels to heal bone fractures [29] and from porous scaffolds to promote angiogenesis or improve wound healing [27, 28, 64, 74], were used as carriers to protect siRNA from RNAse degradation, enhance cellular uptake, and allow for endo-lysosomal escape [63, 75]. The electrostatic interactions between lyophilized anionic siRNA and cationic polymer micelles is strong enough to prevent separation under strong electric field, as demonstrated by the gel retardation assay (Fig. S3 in SI). The siRNA-micelles were fabricated in lyophilized form to allow for long-term storage and to concentrate the therapeutic payload. Trehalose was added to minimize loss of bioactivity during the lyophilization process [27, 76, 77]. Despite an increase in size after lyophilization, the siRNA-micelles maintained an average diameter below 200 nm (Fig. 2), allowing for efficient cellular uptake [78] (Fig. S4 in SI) and targeted gene silencing in monolayer (Fig. 5, Fig. S5, S7 in SI). The increase in size of the post-lyo siRNA-micelles is likely due to disruption of siRNA and micelle interactions during the lyophilization process. siRNA loaded micelles post-lyo may be smaller in size than those without siRNA due to the compacted arrangement of particles caused by interaction between negatively charged RNA and positively charged polymer.
siRNA-laden poly-lactic-glycolic acid (PLGA) MS have been used as injectable platforms for treatment of rheumatoid arthritis [79, 80] or incorporated within scaffolds to control siRNA release in bone regeneration strategies [39]. However, to the best of our knowledge, no reports have demonstrated encapsulation of siRNA within hydrogel microparticles for tunable delivery, which may be beneficial for tissue engineering applications. Photocrosslinked DEX-HEMA MS were synthesized in this study for encapsulation and subsequent prolonged delivery of siRNA-micelles within cell aggregates using a modification of a previously described aqueous-based, single emulsion procedure [65, 66], which has been reported to fabricate DEX-based MS without the use of harsh organic solvents typically employed in phase-separation MS synthesis approaches to minimize potential damage to encapsulated therapeutic molecules. UV-crosslinking aimed to circumvent the use of cytototoxic crosslinking agents (tetramethyl ethylenediamine and ammonium persulfate) used in these previous reports [81], which, unlike with UV-mediated radical formation, are not consumed during the reaction process. The sizes of fabricated DEX-HEMA MS were not significantly different between conditions and average diameter ranged from 3.8 to 6.5 μm, depending on the hydrogel polymer concentration and encapsulation of siRNA-micelles (Table 1). At this size, immediate cellular uptake may be avoided [82, 83] and MS can incorporate within cell aggregates without interfering with cell-cell interactions, as demonstrated previously with incorporation of microparticles between 5–8 μm within hMSC aggregates [48]. siRNA-micelles were loaded into the developed MS with relatively high EEs of 84% and 85% for MS6 and MS10, respectively (Table 1), avoiding extensive loss of unencapsulated siRNA. Variability in initial loading between batches may have resulted from small batch-to-batch differences that can occur during steps involved in MS fabrication, including vortexing, UV application, and washing. siRNA release kinetics from both MS formulations demonstrated sustained release over 28 days with no burst release associated with rapid diffusion-based release profiles, indicating that release kinetics are likely predominantly dictated by the degradation rate of the MS (Fig. 4). The faster release rates observed in siRNA-MS6 compared to MS10 (Fig. 4) were likely a result of lower crosslinking density in the MS6, which would increase hydrogel pore size and degradation rate for accelerated diffusional release. Decreasing hydrogel MS crosslinking density by lowering the hydrogel concentration in MS6 vs MS10 also increased total siRNA incorporated per mass of MS, providing additional ability to control siRNA dosage given a constant amount of MS incorporated per aggregate (3.75 and 2.13 μg siRNA in 2.0 mg of MS6 and MS10, respectively). Of note, the higher siRNA loading in the same mass of MS6 vs MS10 may have increased the diffusion gradient of siRNA from the MS6, and this may have also potentially contributed to the faster observed release kinetics. In addition, although not explored here, release in this system might be altered by varying the degree of modification of the photocrosslinkable linkage in the DEX backbone [32].
The goal of this delivery system was not only to provide local, sustained siRNA presentation, but also to protect the siRNA from degradation and maintain the capacity for nanocomplexes to regulate gene expression. siRNA-micelles released from the fabricated hydrogel MS was internalized by cells (Fig. S4 in SI) and subsequently silenced gene expression (Fig. 5, Fig. S5 in SI). Monolayer hMSCs treated with releasates collected between days 6–10 from two siGLO MS formulations exhibited enhanced siGLO uptake compared to untreated cells (Fig. S4 in SI), which indicates higher amounts of bioactive siRNA-micelles during this time frame and is consistent with bioactivity results from monolayer deGFP-HeLa cells which showed significant GFP silencing with treatment of siGFP-releasates collected between days 6–10 from both MS formulations (Fig. 5, Fig. S5 in SI). Although, released siGFP from MS significantly silenced GFP expression in monolayer cultured deGFP-HeLa cells at certain time points (Fig. 5, Fig. S5 in SI), the gene silencing ability of released siRNA was decreased compared to (+) ctrl of siGFP-micelles alone. For example, although average day 5 releasates from both MS formulations contained equal or higher amounts of siRNA (based on siGLO release, 0.75 and 0.56 μg siRNA in releasates from MS6 and MS10, respectively; Fig. 4) compared to (+) ctrl (0.50 μg siRNA), cells treated with siGFP-containing releasates showed significantly lower degree of GFP silencing (<21%) compared to (+) ctrl (>95%) (Fig. 5), a trend that was observed with different MS batches (Fig. S5). The decrease in gene knockdown capacity of released siRNA-micelles may be a due to a combination of both UV radiation in the presence of PI and interactions of negatively charged carboxylic acids at the ends of the degraded polymer backbone with cationic siRNA-micelles, resulting in decreased transfection efficiency (Fig. S6A and C in SI), the latter of which has been previously reported [23]. However, incorporation of siRNA-MS within cell aggregates may benefit from substrate-mediated transfection, in which cells can uptake payload complexes directly from a biomaterial substrate. This substrate-mediated transfection has been reported to improve siRNA transfection efficiency compared to conventional 2D transfection, potentially by concentrating the payload [84, 85]. The juxtaposition of cells and siRNA-MS within 3D stem cell aggregates may also prevent or decrease interactions between hydrogel degradation byproducts and cationic siRNA-micelles due to the competition of surface charge between cell membranes and the degraded polymer byproducts. Therefore, MS may serve to package, protect, and sustain delivery of RNA as well as enhance efficacy of transfection in 3D cell culture.
Upon incorporation within hMSC aggregates, MS distributed throughout the cell constructs (Fig. 6) without disrupting aggregate formation, and the MS did not negatively impact cell number after 20 days of culture (Fig. S9 in SI), indicating biocompatibility of the developed delivery system. In addition, the sustained presentation of siRNA-micelles in aggregates was visualized for up to 14 and 20 days (Fig. 6B), resulting in extended gene silencing in aggregates (Fig. 7 and 8) which is supported by siRNA release kinetics (Fig. 4) and quantification of gene knockdown in monolayer cells treated with released siRNA (Fig. 5, Fig. S5 in SI). Analysis of GFP mRNA expression confirmed that sustained delivery of siGFP from MS within GFP-hMSC aggregates significantly reduced GFP expression (Fig. 7). A higher dose of siRNA, faster delivery, or combination of the two in GFP-hMSC aggregates incorporating siGFP-MS6 (0.56 μg siGFP per aggregate) led to a time course of GFP silencing compared to limited silencing upon delivery of a lower siRNA dose and/or more delayed release with incorporation of siGFP-MS10 (0.32 μg siGFP per aggregate) (Fig. 7). Specifically, loading siGFP-MS6 into GFP-hMSC aggregates resulted in a sustained reduction of GFP mRNA for at least 10 days compared to all other groups with 77% and 95% GFP silencing at days 5 and 10, respectively (Fig. 7A–C). The sustained knockdown of GFP mRNA expression within GFP-hMSC aggregates incorporating siGFP-MS6 was further confirmed by GFP fluorescent images, which showed substantially decreased GFP fluorescence signal for up to 15 days compared to controls (Fig. 8) with similar DAPI signal (Fig. S8). In contrast, when siGFP-MS10 were incorporated into the GFP-hMSC aggregates, significant silencing of GFP mRNA expression was only achieved at day 5 with respect to siNC-MS aggregates (Fig. 7A), demonstrating differential degree of gene knockdown in aggregate with incorporation of different MS formulations. It is not clear why there was an increase in GFP expression in empty MS10 or siRNA-MS10 incorporated aggregates compared to cells only aggregates at day 20. From the overall findings of this work, embedding of siRNA-MS within stem cell aggregates may offer many advantages for tissue regeneration, including preservation of critical cell-cell interactions, prolonged presentation of RNAi molecules for sustained gene silencing, substrate-mediated enhancement in transfection efficacy, and tunable siRNA release profiles to control the time course of gene expression.
Conclusion
This work presents the first description of controlled release of bioactive RNA from hydrogel microparticles capable of incorporation within 3D stem cell aggregates for sustained RNAi-mediated gene silencing. As siRNA is transient in nature, its sustained delivery from microparticles allows for long-term gene silencing, circumventing the need for repeat treatment in vitro to create readily implantable constructs due to the self-contained nature of the system. Importantly, the developed MS are versatile as different formulations were shown to regulate siRNA release kinetics and resultant gene knockdown in aggregate, supporting the potential for tailoring siRNA release for specific applications. The system can also accommodate the delivery of a multiple different siRNAs, potentially with different release profiles and/or doses. In addition, this delivery approach can be used with other cell types (e.g., mature differentiated cells, embryonic stem cells, induced pluripotent stem cells) and cellular aggregates (e.g., embryoid bodies, organoid cultures). This spatiotemporally controlled RNA delivery strategy may have great utility in enhancing cell aggregate-based formation, development and function of tissues and organs, addressing basic biological questions, and disease therapeutics.
Supplementary Material
significance statement.
This work presents a new strategy to deliver RNA-nanocomplexes from photocrosslinked dextran microspheres for tunable presentation of bioactive RNA. These microspheres were embedded within scaffold-free, human mesenchymal stem cell (hMSC) aggregates for sustained gene silencing within three-dimensional cell constructs while maintaining cell viability. Unlike exogenous delivery of RNA within culture medium that suffers from diffusion limitations and potential need for repeated transfections, this strategy provides local and sustained RNA presentation from the microspheres to cells in the constructs. This system has the potential to inhibit translation of hMSC differentiation antagonists and drive hMSC differentiation toward desired specific lineages, and is an important step in the engineering of high-density stem cell systems with incorporated instructive genetic cues for application in tissue regeneration.
Acknowledgements
The authors gratefully acknowledge funding from the National Institutes of Health (R01AR063194, R01AR069564, R01EB023907, T32GM007250, R01 EB019409), National Science Foundation (CAREER BMAT 1349604), and the Department of Defense Congressionally Directed Medical Research Programs (OR110196). We also thank Rui Tang for help with NMR analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of interests
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- [1].Raisin S, Belamie E, Morille M Non-viral gene activated matrices for mesenchymal stem cells based tissue engineering of bone and cartilage, Biomaterials 104 (2016) 223–37. [DOI] [PubMed] [Google Scholar]
- [2].Whitehead KA, Langer R, Anderson DG Knocking down barriers: advances in siRNA delivery, Nat Rev Drug Discov 8 (2) (2009) 129–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Beavers KR, Nelson CE, Duvall CL MiRNA inhibition in tissue engineering and regenerative medicine, Adv Drug Deliv Rev 88 (2015) 123–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hong E, Reddi AH MicroRNAs in chondrogenesis, articular cartilage, and osteoarthritis: implications for tissue engineering, Tissue Eng Part B Rev 18 (6) (2012) 445–53. [DOI] [PubMed] [Google Scholar]
- [5].Bobick BE, Matsche AI, Chen FH, Tuan RS The ERK5 and ERK1/2 signaling pathways play opposing regulatory roles during chondrogenesis of adult human bone marrow-derived multipotent progenitor cells, J Cell Physiol 224 (1) (2010) 178–86. [DOI] [PubMed] [Google Scholar]
- [6].Gibson G, Asahara H microRNAs and cartilage, J Orthop Res 31 (9) (2013) 1333–44. [DOI] [PubMed] [Google Scholar]
- [7].Aleckovic M, Kang Y Bone marrow stroma-derived miRNAs as regulators, biomarkers and therapeutic targets of bone metastasis, Bonekey Rep 4 (2015) 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Elangovan S, Khorsand B, Do AV, Hong L, Dewerth A, Kormann M, Ross RD, Sumner DR, Allamargot C, Salem AK Chemically modified RNA activated matrices enhance bone regeneration, J Control Release 218 (2015) 22–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Rose L, Uludag H Realizing the potential of gene-based molecular therapies in bone repair, J Bone Miner Res 28 (11) (2013) 2245–62. [DOI] [PubMed] [Google Scholar]
- [10].Ghadakzadeh S, Mekhail M, Aoude A, Tabrizian M, Hamdy RC Small Players Ruling the Hard Game: siRNA in Bone Regeneration, J Bone Miner Res 31 (7) (2016) 1481. [DOI] [PubMed] [Google Scholar]
- [11].Turner JJ, Jones SW, Moschos SA, Lindsay MA, Gait MJ MALDI-TOF mass spectral analysis of siRNA degradation in serum confirms an RNAse A-like activity, Mol Biosyst 3 (1) (2007) 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wang J, Lu Z, Wientjes MG, Au JL Delivery of siRNA therapeutics: barriers and carriers, AAPS J 12 (4) (2010) 492–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kanasty R, Dorkin JR, Vegas A, Anderson D Delivery materials for siRNA therapeutics, Nat Mater 12 (11) (2013) 967–77. [DOI] [PubMed] [Google Scholar]
- [14].Oh YK, Park TG siRNA delivery systems for cancer treatment, Adv Drug Deliv Rev 61 (10) (2009) 850–62. [DOI] [PubMed] [Google Scholar]
- [15].Bartlett DW, Davis ME Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging, Nucleic Acids Res 34 (1) (2006) 322–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Lee JY, Kim KS, Kang YM, Kim ES, Hwang SJ, Lee HB, Min BH, Kim JH, Kim MS In vivo efficacy of paclitaxel-loaded injectable in situ-forming gel against subcutaneous tumor growth, Int J Pharm 392 (1–2) (2010) 51–6. [DOI] [PubMed] [Google Scholar]
- [17].Chiu YL, Rana TM RNAi in human cells: basic structural and functional features of small interfering RNA, Mol Cell 10 (3) (2002) 549–61. [DOI] [PubMed] [Google Scholar]
- [18].Sarett SM, Nelson CE, Duvall CL Technologies for controlled, local delivery of siRNA, J Control Release 218 (2015) 94–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Krebs MD, Jeon O, Alsberg E Localized and sustained delivery of silencing RNA from macroscopic biopolymer hydrogels, J Am Chem Soc 131 (26) (2009) 9204–6. [DOI] [PubMed] [Google Scholar]
- [20].Krebs MD, Alsberg E Localized, targeted, and sustained siRNA delivery, Chemistry 17 (11) (2011) 3054–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Huynh CT, Liu F, Cheng Y, Coughlin KA, Alsberg E Thiol-Epoxy “Click” Chemistry to Engineer Cytocompatible PEG-Based Hydrogel for siRNA-Mediated Osteogenesis of hMSCs, ACS Appl Mater Interfaces 10 (31) (2018) 25936–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Huynh CT, Nguyen MK, Tonga GY, Longe L, Rotello VM, Alsberg E Photocleavable Hydrogels for Light-Triggered siRNA Release, Adv Healthc Mater 5 (3) (2016) 305–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Huynh CT, Nguyen MK, Naris M, Tonga GY, Rotello VM, Alsberg E Light-triggered RNA release and induction of hMSC osteogenesis via photodegradable, dual-crosslinked hydrogels, Nanomedicine (Lond) 11 (12) (2016) 1535–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Huynh CT, Zheng Z, Nguyen MK, McMillan A, Yesilbag Tonga G, Rotello VM, Alsberg E Cytocompatible Catalyst-Free Photodegradable Hydrogels for Light-Mediated RNA Release To Induce hMSC Osteogenesis, ACS Biomater Sci Eng 3 (9) (2017) 2011–23. [DOI] [PubMed] [Google Scholar]
- [25].Hill MC, Nguyen MK, Jeon O, Alsberg E Spatial control of cell gene expression by siRNA gradients in biodegradable hydrogels, Adv Healthc Mater 4 (5) (2015) 714–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Nguyen MK, Jeon O, Krebs MD, Schapira D, Alsberg E Sustained localized presentation of RNA interfering molecules from in situ forming hydrogels to guide stem cell osteogenic differentiation, Biomaterials 35 (24) (2014) 6278–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Nelson CE, Gupta MK, Adolph EJ, Guelcher SA, Duvall CL siRNA Delivery from an Injectable Scaffold for Wound Therapy, Adv Wound Care (New Rochelle) 2 (3) (2013) 93–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nelson CE, Kim AJ, Adolph EJ, Gupta MK, Yu F, Hocking KM, Davidson JM, Guelcher SA, Duvall CL Tunable delivery of siRNA from a biodegradable scaffold to promote angiogenesis in vivo, Adv Mater 26 (4) (2014) 607–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Wang Y, Malcolm DW, Benoit DSW Controlled and sustained delivery of siRNA/NPs from hydrogels expedites bone fracture healing, Biomaterials 139 (2017) 127–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Segovia N, Pont M, Oliva N, Ramos V, Borros S, Artzi N Hydrogel doped with nanoparticles for local sustained release of siRNA in breast cancer, Adv Healthc Mater 4 (2) (2015) 271–80. [DOI] [PubMed] [Google Scholar]
- [31].Kim YM, Park MR, Song SC Injectable polyplex hydrogel for localized and long-term delivery of siRNA, ACS Nano 6 (7) (2012) 5757–66. [DOI] [PubMed] [Google Scholar]
- [32].Nguyen MK, McMillan A, Huynh CT, Schapira DS, Alsberg E Photocrosslinkable, biodegradable hydrogels with controlled cell adhesivity for prolonged siRNA delivery to hMSCs to enhance their osteogenic differentiation, J Mater Chem B 5 (3) (2017) 485–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Geckil H, Xu F, Zhang X, Moon S, Demirci U Engineering hydrogels as extracellular matrix mimics, Nanomedicine (Lond) 5 (3) (2010) 469–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Vinas-Castells R, Holladay C, di Luca A, Diaz VM, Pandit A Snail1 down-regulation using small interfering RNA complexes delivered through collagen scaffolds, Bioconjug Chem 20 (12) (2009) 2262–9. [DOI] [PubMed] [Google Scholar]
- [35].Nguyen K, Dang PN, Alsberg E Functionalized, biodegradable hydrogels for control over sustained and localized siRNA delivery to incorporated and surrounding cells, Acta Biomater 9 (1) (2013) 4487–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Nguyen MK, Huynh CT, Gilewski A, Wilner SE, Maier KE, Kwon N, Levy M, Alsberg E Covalently tethering siRNA to hydrogels for localized, controlled release and gene silencing, Sci Adv 5 (8) (2019) eaax0801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Alsberg E, Kong HJ, Hirano Y, Smith MK, Albeiruti A, Mooney DJ Regulating bone formation via controlled scaffold degradation, J Dent Res 82 (11) (2003) 903–8. [DOI] [PubMed] [Google Scholar]
- [38].Nguyen MK, Jeon O, Dang PN, Huynh CT, Varghai D, Riazi H, McMillan A, Herberg S, Alsberg E RNA interfering molecule delivery from in situ forming biodegradable hydrogels for enhancement of bone formation in rat calvarial bone defects, Acta Biomater 75 (2018) 105–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Zhang X, Li Y, Chen YE, Chen J, Ma PX Cell-free 3D scaffold with two-stage delivery of miRNA-26a to regenerate critical-sized bone defects, Nat Commun 7 (2016) 10376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Athanasiou KA, Eswaramoorthy R, Hadidi P, Hu JC Self-organization and the self-assembling process in tissue engineering, Annu Rev Biomed Eng 15 (2013) 115–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].DuRaine GD, Brown WE, Hu JC, Athanasiou KA Emergence of scaffold-free approaches for tissue engineering musculoskeletal cartilages, Ann Biomed Eng 43 (3) (2015) 543–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Solorio LD, Vieregge EL, Dhami CD, Alsberg E High-density cell systems incorporating polymer microspheres as microenvironmental regulators in engineered cartilage tissues, Tissue Eng Part B Rev 19 (3) (2013) 209–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells, Exp Cell Res 238 (1) (1998) 265–72. [DOI] [PubMed] [Google Scholar]
- [44].Mackay AM, Beck SC, Murphy JM, Barry FP, Chichester CO, Pittenger MF Chondrogenic differentiation of cultured human mesenchymal stem cells from marrow, Tissue Eng 4 (4) (1998) 415–28. [DOI] [PubMed] [Google Scholar]
- [45].Muraglia A, Corsi A, Riminucci M, Mastrogiacomo M, Cancedda R, Bianco P, Quarto R Formation of a chondro-osseous rudiment in micromass cultures of human bone-marrow stromal cells, J Cell Sci 116 (Pt 14) (2003) 2949–55. [DOI] [PubMed] [Google Scholar]
- [46].Farrell E, van der Jagt OP, Koevoet W, Kops N, van Manen CJ, Hellingman CA, Jahr H, O’Brien FJ, Verhaar JA, Weinans H, van Osch GJ Chondrogenic priming of human bone marrow stromal cells: a better route to bone repair?, Tissue Eng Part C Methods 15 (2) (2009) 285–95. [DOI] [PubMed] [Google Scholar]
- [47].Freeman FE, Haugh MG, McNamara LM Investigation of the optimal timing for chondrogenic priming of MSCs to enhance osteogenic differentiation in vitro as a bone tissue engineering strategy, J Tissue Eng Regen Med 10 (4) (2016) E250–62. [DOI] [PubMed] [Google Scholar]
- [48].Dang PN, Dwivedi N, Phillips LM, Yu X, Herberg S, Bowerman C, Solorio LD, Murphy WL, Alsberg E Controlled Dual Growth Factor Delivery From Microparticles Incorporated Within Human Bone Marrow-Derived Mesenchymal Stem Cell Aggregates for Enhanced Bone Tissue Engineering via Endochondral Ossification, Stem Cells Transl Med 5 (2) (2016) 206–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Solorio LD, Dhami CD, Dang PN, Vieregge EL, Alsberg E Spatiotemporal regulation of chondrogenic differentiation with controlled delivery of transforming growth factor-beta1 from gelatin microspheres in mesenchymal stem cell aggregates, Stem Cells Transl Med 1 (8) (2012) 632–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Solorio LD, Phillips LM, McMillan A, Cheng CW, Dang PN, Samorezov JE, Yu X, Murphy WL, Alsberg E Spatially organized differentiation of mesenchymal stem cells within biphasic microparticle-incorporated high cell density osteochondral tissues, Adv Healthc Mater 4 (15) (2015) 2306–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Solorio LD, Vieregge EL, Dhami CD, Dang PN, Alsberg E Engineered cartilage via self-assembled hMSC sheets with incorporated biodegradable gelatin microspheres releasing transforming growth factor-beta1, J Control Release 158 (2) (2012) 224–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Dikina AD, Strobel HA, Lai BP, Rolle MW, Alsberg E Engineered cartilaginous tubes for tracheal tissue replacement via self-assembly and fusion of human mesenchymal stem cell constructs, Biomaterials 52 (2015) 452–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Solorio LD, Fu AS, Hernandez-Irizarry R, Alsberg E Chondrogenic differentiation of human mesenchymal stem cell aggregates via controlled release of TGF-beta1 from incorporated polymer microspheres, J Biomed Mater Res A 92 (3) (2010) 1139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Dang PN, Dwivedi N, Phillips LM, Bowerman C, Murphy WL, Alsberg E Guiding chondrogenesis and osteogenesis with mineral-coated hydroxyapatite and BMP-2 incorporated within high-density hMSC aggregates for bone regeneration ACS Biomater Sci Eng 2 (1) (2016) 30–42. [DOI] [PubMed] [Google Scholar]
- [55].Dikina AD, Alt DS, Herberg S, McMillan A, Strobel HA, Zheng Z, Cao M, Lai BP, Jeon O, Petsinger VI, Cotton CU, Rolle MW, Alsberg E A Modular Strategy to Engineer Complex Tissues and Organs, Adv Sci (Weinh) 5 (5) (2018) 1700402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Dang PN, Herberg S, Varghai D, Riazi H, McMillan A, Awadallah A, Phillips LM, Jeon O, Nguyen MK, Dwivedi N, Yu X, Murphy WL, Alsberg E Endochondral Ossification in Critical-Sized Bone Defects via Readily Implantable Scaffold-Free Stem Cell Constructs, Stem Cells Transl Med 6 (7) (2017) 1644–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].McDermott AM, Herberg S, Mason DE, Collins JM, Pearson HB, Dawahare JH, Tang R, Patwa AN, Grinstaff MW, Kelly DJ, Alsberg E, Boerckel JD Recapitulating bone development through engineered mesenchymal condensations and mechanical cues for tissue regeneration, Sci Transl Med 11 (495) (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Herberg S, McDermott AM, Dang PN, Alt DS, Tang R, Dawahare JH, Varghai D, Shin JY, McMillan A, Dikina AD, He F, Lee YB, Cheng Y, Umemori K, Wong PC, Park H, Boerckel JD, Alsberg E Combinatorial morphogenetic and mechanical cues to mimic bone development for defect repair, Sci Adv 5 (8) (2019) eaax2476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Herberg S, Varghai D, Cheng Y, Dikina AD, Dang PN, Rolle MW, Alsberg E High-density human mesenchymal stem cell rings with spatiotemporally-controlled morphogen presentation as building blocks for engineering bone diaphyseal tissue, Nanotheranostics 2 (2) (2018) 128–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].McMillan A, Nguyen MK, Gonzalez-Fernandez T, Ge P, Yu X, Murphy WL, Kelly DJ, Alsberg E Dual non-viral gene delivery from microparticles within 3D high-density stem cell constructs for enhanced bone tissue engineering, Biomaterials 161 (2018) 240–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wang Y, Yu X, Baker C, Murphy WL, McDevitt TC Mineral particles modulate osteochondrogenic differentiation of embryonic stem cell aggregates, Acta Biomater 29 (2016) 42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Shim MS, Bhang SH, Yoon K, Choi K, Xia Y A bioreducible polymer for efficient delivery of Fas-silencing siRNA into stem cell spheroids and enhanced therapeutic angiogenesis, Angew Chem Int Ed Engl 51 (47) (2012) 11899–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Convertine AJ, Benoit DS, Duvall CL, Hoffman AS, Stayton PS Development of a novel endosomolytic diblock copolymer for siRNA delivery, J Control Release 133 (3) (2009) 221–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Nelson CE, Gupta MK, Adolph EJ, Shannon JM, Guelcher SA, Duvall CL Sustained local delivery of siRNA from an injectable scaffold, Biomaterials 33 (4) (2012) 1154–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Stenekes RJ, Franssen O, van Bommel EM, Crommelin DJ, Hennink WE The use of aqueous PEG/dextran phase separation for the preparation of dextran microspheres, Int J Pharm 183 (1) (1999) 29–32. [DOI] [PubMed] [Google Scholar]
- [66].Franssen O, Hennink WE A novel preparation method for polymeric microparticles without the use of organic solvents, International Journal of Pharmaceutics 168 (1) (1998) 1–7. [Google Scholar]
- [67].Witwicka H, Hwang SY, Reyes-Gutierrez P, Jia H, Odgren PE, Donahue LR, Birnbaum MJ, Odgren PR Studies of OC-STAMP in Osteoclast Fusion: A New Knockout Mouse Model, Rescue of Cell Fusion, and Transmembrane Topology, PLoS One 10 (6) (2015) e0128275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Nguyen K, Das B, Dobrowolski C, Karn J Multiple Histone Lysine Methyltransferases Are Required for the Establishment and Maintenance of HIV-1 Latency, MBio 8 (1) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wang GG, Calvo KR, Pasillas MP, Sykes DB, Häcker H, Kamps MP Quantitative production of macrophages or neutrophils ex vivo using conditional Hoxb8, Nat Methods 3 (4) (2006) 287–93. [DOI] [PubMed] [Google Scholar]
- [70].Van Tomme SR, Hennink WE Biodegradable dextran hydrogels for protein delivery applications, Expert Rev Med Devices 4 (2) (2007) 147–64. [DOI] [PubMed] [Google Scholar]
- [71].Castaneda RT, Khurana A, Khan R, Daldrup-Link HE Labeling stem cells with ferumoxytol, an FDA-approved iron oxide nanoparticle, J Vis Exp (57) (2011) e3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Wu M, Sun D, Tyner K, Jiang W, Rouse R Comparative Evaluation of U.S. Brand and Generic Intravenous Sodium Ferric Gluconate Complex in Sucrose Injection: In Vitro Cellular Uptake, Nanomaterials (Basel) 7 (12) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Tatiparti K, Sau S, Kashaw SK, Iyer AK siRNA Delivery Strategies: A Comprehensive Review of Recent Developments, Nanomaterials (Basel) 7 (4) (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Martin JR, Nelson CE, Gupta MK, Yu F, Sarett SM, Hocking KM, Pollins AC, Nanney LB, Davidson JM, Guelcher SA, Duvall CL Local Delivery of PHD2 siRNA from ROS-Degradable Scaffolds to Promote Diabetic Wound Healing, Adv Healthc Mater 5 (21) (2016) 2751–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Sarett SM, Kilchrist KV, Miteva M, Duvall CL Conjugation of palmitic acid improves potency and longevity of siRNA delivered via endosomolytic polymer nanoparticles, J Biomed Mater Res A 103 (9) (2015) 3107–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Kaushik JK, Bhat R Why is trehalose an exceptional protein stabilizer? An analysis of the thermal stability of proteins in the presence of the compatible osmolyte trehalose, J Biol Chem 278 (29) (2003) 26458–65. [DOI] [PubMed] [Google Scholar]
- [77].Yadava P, Gibbs M, Castro C, Hughes JA Effect of lyophilization and freeze-thawing on the stability of siRNA-liposome complexes, AAPS PharmSciTech 9 (2) (2008) 335–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Rejman J, Oberle V, Zuhorn IS, Hoekstra D Size-dependent internalization of particles via the pathways of clathrin- and caveolae-mediated endocytosis, Biochem J 377 (Pt 1) (2004) 159–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].De Rosa G, Salzano G PLGA microspheres encapsulating siRNA, Methods Mol Biol 1218 (2015) 43–51. [DOI] [PubMed] [Google Scholar]
- [80].Presumey J, Salzano G, Courties G, Shires M, Ponchel F, Jorgensen C, Apparailly F, De Rosa G PLGA microspheres encapsulating siRNA anti-TNFalpha: efficient RNAi-mediated treatment of arthritic joints, Eur J Pharm Biopharm 82 (3) (2012) 457–64. [DOI] [PubMed] [Google Scholar]
- [81].Desai ES, Tang MY, Ross AE, Gemeinhart RA Critical factors affecting cell encapsulation in superporous hydrogels, Biomed Mater 7 (2) (2012) 024108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chithrani BD, Ghazani AA, Chan WC Determining the size and shape dependence of gold nanoparticle uptake into mammalian cells, Nano Lett 6 (4) (2006) 662–8. [DOI] [PubMed] [Google Scholar]
- [83].Zheng M, Yu J The effect of particle shape and size on cellular uptake, Drug Deliv Transl Res 6 (1) (2016) 67–72. [DOI] [PubMed] [Google Scholar]
- [84].Adler AF, Leong KW Emerging links between surface nanotechnology and endocytosis: impact on nonviral gene delivery, Nano Today 5 (6) (2010) 553–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Fujita S, Ota E, Sasaki C, Takano K, Miyake M, Miyake J Highly efficient reverse transfection with siRNA in multiple wells of microtiter plates, J Biosci Bioeng 104 (4) (2007) 329–33. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


