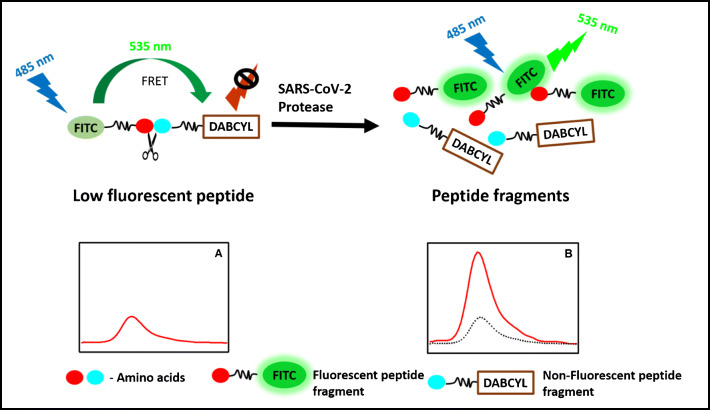
Abstract
The novel corona (SARS-CoV-2) virus causes a global pandemic, which motivates researchers to develop reliable and effective methods for screening and detection of SARS-CoV-2. Though there are several methods available for the diagnosis of SARS-CoV-2 such as RT-PCR and ELSIA, nevertheless, these methods are time-consuming and may not apply at the point of care. In this study, we have developed a specific, sensitive, quantitative and fast detection method for SARS-CoV-2 by fluorescence resonance energy transfer (FRET) assay. The total extracellular protease proteolytic activity from the virus has been used as the biomarker. The specific peptide sequences from the library of 115 dipeptides were identified via changes in the fluorescence signal. The fluorogenic dipeptide substrates have the fluorophore and a quencher at the N- and the C- terminals, respectively. When the protease hydrolyzes the peptide bond between the two specific amino acids, it leads to a significant increase in the fluorescence signals. The specific fluorogenic peptide (H-d) produces a high fluorescence signal. A calibration plot was obtained from the changes in the fluorescence intensity against the different concentrations of the viral protease. The lowest limit of detection of this method was 9.7 ± 3 pfu/mL. The cross-reactivity of the SARS-CoV-2-specific peptide was tested against the MERS-CoV which does not affect the fluorescence signal. A significant change in the fluorescence signal with patient samples indicates that this FRET-based assay might be applied for the diagnosis of SARS-CoV-2 patients.
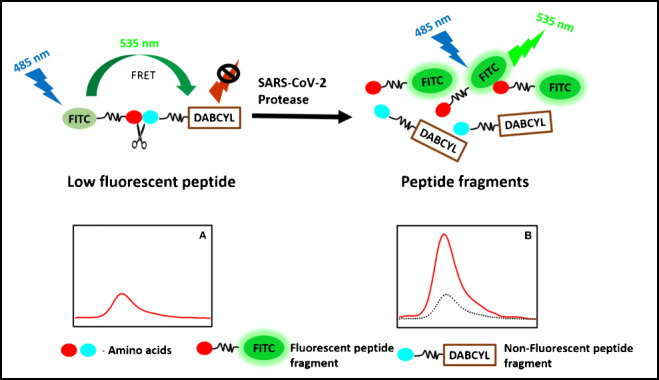
Graphical abstract
Keywords: SARS-CoV-2 detection, COVID-19 FRET substrate, Detection of Covid-19, Fluorogenic peptide sequences
Introduction
The outbreak of coronavirus disease 2019 (COVID-19) caused by the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2/2019-nCoV) poses a serious threat to global public health and local economies [1]. It causes hundreds of thousands of serious illnesses and human life losses all over the world. Human respiratory tract infections caused by coronaviruses such as HCoV-229E and HCoVOC43 are known since the 1960s [2], and later SARS-CoV-1, HCoV-NL63, HCoV-HKU1, and MERS-CoV have been discovered [3]. Severe respiratory illness in humans due to COVID-19 was first reported in December 2019 in China. Since then, 81,187,266 confirmed cases of infection and 1,772,853 deaths of 28 December 2020 [1, 4, 5]. People with chronic diseases such as diabetes, renal failure, chronic lung disease, and immunocompromised patients are at high risk to die due to COVID-19 [4]. The incubation period of COVID-19 varies between 2 and 14 days and is susceptible to all age groups [5]. The virus can be transmitted from human to human and might cause mild illnesses, severe pneumonia, severe respiratory syndrome, and multi-organs failure [6]. Other reported symptoms are fever, anorexia, nausea, vomiting, abdominal pain, diarrhea, and disseminated intercellular coagulation. The WHO and Center for Disease Control (CDC) recommended that COVID-19 cases must be confirmed by RT-PCR via targeting three genes: RdRP gene, E gene, and N gene [7]. Different methods have been developed for the diagnosis and detection of COVID-19 such as molecular-based assays, serological based tests, point of care devices, radiology-based tests, and viral cell culture-based methods [8]. The molecular diagnostic assays rely on the identification and amplification of the viral RNA by real-time reverse transcriptase-polymerase chain reaction (rRT-PCR) [9] obtained from the suspected individuals. The WHO and CDC have established a standard protocol with a set of primers that can bind to the specific region of RNA and facilitate the amplification process [10]. When SARS-CoV-2 infects human host cell, it produces a specific antibody against SARS-CoV-2. Enzyme-linked Immunosorbent Assay (ELISA) is used for the qualitative and quantitative detection of SARS-CoV-2 antibody. Chemiluminescent immunoassay (CLIA) is used for the quantitative measurement of SARS-CoV-2 antibodies such as IgG, IgM, and IgA [11]. Western blots have been employed to detect specific SARS-CoV-2 proteins, and Northern blot hybridization has been applied for targeting specific SARS-CoV-2 genes [12]. Nucleic acid amplification techniques such as PCR [7, 13], nucleic acid sequence-based amplification (NASBA) [14], next-generation sequencing (NGS) [15], and Lawrence Livermore microbial detection array (LMDA) [16] were developed and optimized for the identification of SARS-CoV-2 genome from clinical samples. Although these methods are sensitive, the mutation rate in SARS-CoV-2 genome is high, which may lead to false results. Alternatively, detection of SARS-CoV-2 in short time with low cost has been the focus of this study by the screening of protease using the cleavage of a specific peptide bond from a substrate library. Recently, protease-based nanobiotechnology plays a potential role in the clinical application in the detection and identification of emerging viruses [17–19]. FRET peptides are the potential candidate to follow the enzymatic activity in crude biological fluids and culture media. FRET-based assay is rapid, extremely sensitive, and straightforward. The increased fluorescence signal is directly proportional to the quantity of the protease from SARS-CoV-2.
In this study, we used the proteases as diagnostic markers for the detection of SARS-CoV-2. Several proteases are acting as host receptors for the coronavirus entry including aminopeptidase N and angiotensin-converting enzyme 2 (hACE2). We developed a simple, low cost, and reliable assay for the detection of the SARS-CoV-2 using the activity of the SARS-CoV-2 secreted proteases. We used short peptide containing the cleavage site of the SARS-CoV-2 protease and it was labeled with a fluorophore and a quencher at both ends. The peptide is cleaved in the presence of protease; therefore, the fluorophore and the quencher are separated from each other which leads to high fluorescence. The specific peptide sequence cleaves by the SARS-CoV-2 protease indicates the presence of SARS-CoV-2.
Materials and methods
Materials
Vero E6 cells (ATCC® number 1568) were purchased from the American Type Culture Collection (ATCC®). Dulbecco’s Eagle medium (DMEM) and fetal bovine serum (FBS), streptomycin and penicillin, HEPES, agarose, paraformaldehyde, crystal violet, PBS, and DMSO were purchased from Sigma-Aldrich (Gillingham, UK). Dipeptide library was purchased from Pepmic Co Ltd. (Suzhou, China).
Experimental
Cell line and SARS-CoV-2 propagation
Vero E6 cells (ATCC® number 1568) were maintained and grown in Dulbecco’s Eagle medium (DMEM) contained 10% fetal bovine serum (FBS) as described before [20]. SARS-CoV-2/human/SAU/85791C/2020 (Gene accession number MT630432.1) was isolated from a human nasopharyngeal swab confirmed positive by RT-PCR. IRP number H-02-K-076-00520-298 was obtained from the Saudi Ministry of Health to use patient samples. All experiments involved live SARS-CoV-2 were performed following the international recommended safety measures and precautions in Biosafety Level 3 Facility at the Special Infectious Agent Unit, King Fahd Medical Research Center, King Abdulaziz University, Saudi Arabia.
SARS-CoV-2 was propagated and titrated using Median Tissue Culture Infectious Dose (TCID50). In brief, SARS-CoV-2 was inoculated on 90–95% confluent Vero E6 cells in a T175 tissue culture flask and incubated at 37 °C for 1 h in a humidified 5% CO2 incubator with shaking every 15 min. Then, 25 mL of viral inoculation medium (DMEM supplemented with 10 mmol/L HEPES, 1% streptomycin and penicillin, and 2% FBS) was used to replace the inoculum. The cells were then incubated at 37 °C in a humidified 5% CO2 incubator for 72 h or until 90% of the cells illustrated CPE (cytopathic effect). The supernatant was then harvested and centrifuged at 500×g for 5 min at room temperature. Ultimately, SARS-CoV-2 was aliquoted and stored at − 80 °C, and the plaque assay was used to determine the virus titer and TCID50. MERS-CoV was isolated from human nasopharyngeal swab confirmed positive by RT-PCR. The MERS-CoV-positive sample was inoculated on the 95% confluent Vero E6 cells, and finally, the virus was harvested as described above in the propagation of SARS-CoV-2.
Plaque assay
Plaque assay was conducted as described previously [21]. In brief, DMEM medium was used to grow 1 × 105/mL Vero E6 cells. Every well of six-well tissue culture plates were seeded with 2 mL Vero E6 cells and incubated at 37 °C overnight. Serial dilution for each sample (starting from 10−1) was performed in the inoculated DMEM, and 200 μL of each dilution were transferred to the Vero E6 cell monolayers and incubated at 37 °C for 1 h with shaking every 15 min. Overlay DMEM with 0.8% agarose was then added to replace the inoculum and incubated at 37 °C for 3–4 days. Subsequently, the overlay was removed, and 4% paraformaldehyde was used for 15 min to fix the cells. Crystal violet was used to stain the cells, and plaques were counted to determine SARS-CoV-2 titer as plaque-forming units (pfu/mL). TCID50 for SARS-CoV-2 was calculated as 3.16 × 105.
Peptide library design
The peptide library consisted of 115 substrates with two amino acids of the same type or different types including D-amino acids. The library substrates were FRET-based fluorogenic peptides, each peptide had two amino acids of two L-amino acids, or C-terminal l-amino acid and N-terminal d-amino acid, or two d-amino acids. The upper-case letters represent L-amino acids, and the lower-case letter represents D-amino acids. The FITC (fluorescein isothiocyanate) and the dabcyl, [4-((4-(dimethylamino)phenyl)azo)benzoic acid] are the most efficient FRET pairs; therefore, they were introduced at both ends of all the substrates. 6-aminohexanoic acid, Ahx linker was introduced between the FITC fluorophore and the N- terminal amino acids. Lysine was introduced between the C-terminal amino acids and the dabcyl quencher molecule. The combination of different dipeptide substrates has been validated [22, 23].
Identification of SARS-CoV-2-specific substrate by FRET assay
SARS-CoV-2-specific peptide was identified from the library of 115 FRET substrates by high-throughput screening. All the fluorogenic peptides with FITC and the dabcyl were added to the wells of 96 well fluorescence microtiter plate. The proteolytic activity of the COVID-19 protease on the individual substrates was monitored from the changes in the fluorescence of the FITC at its emission maxima. 0.5 μL of each peptide from 800 μM solution, and 50 μl PBS were mixed with 50 μl of SARS-CoV-2 total protease (the supernatant of the overnight culture media). Culture media was used as a negative control. The changes in the fluorescence intensity of FITC was monitored every 2 min for 2.5 h at 37 °C. The samples were excited at 485 nm and the emission was recorded at 535 nm. The relative fluorescence unit (RFU) change in each sample with time was calculated by subtracting the sample fluorescence values from the negative control values. The protease activity on the substrate reflects the changes in the RFU per minute (RFU/min). The specific peptide with the highest RFU/m was the most specific to SARS-CoV-2. L-Histidine-D-aspartic acid FITC-Ahx-H-d-K(Dabcyl) was found to be the most active peptide in the presence of SARS-CoV-2 protease. FITC-Ahx-H-d-K(Dabcyl) dipeptide was used for further studies.
Real-time fluorescence measurement from the SARS-CoV-2-specific FRET substrate
The proteolytic activity of SARS-CoV-2 protease on the fluorescence of the FITC-Ahx-H-d-K(Dabcyl) fluorogenic peptides has been studied. A constant amount (4 μM) of the H-d peptide in PBS buffer was added to nine different wells of the microtiter plate. Live SARS-CoV-2 viral solution containing 108 pfu/mL to 101 pfu/mL was added to each well containing the specific peptide. The changes in the fluorescence were monitored at 535 nm with the excitation at 495 nm. The fluorescence of FITC in each well was recorded every 2 min for 3 h. The well with the only dipeptide without the live virus was considered as a negative control. The fluorescence values were obtained by subtracting negative control values from the sample values. The raw plot of fluorescence signal change against time for individual samples is presented in Fig. 1a. The percentage change was calculated by (F-F0/F0)×100. Figure 1b represents the exponential fluorescence signal growth of FITC-Ahx-H-d-K(Dabcyl) peptide in the presence of 108 pfu/mL at different time intervals.
Fig. 1.
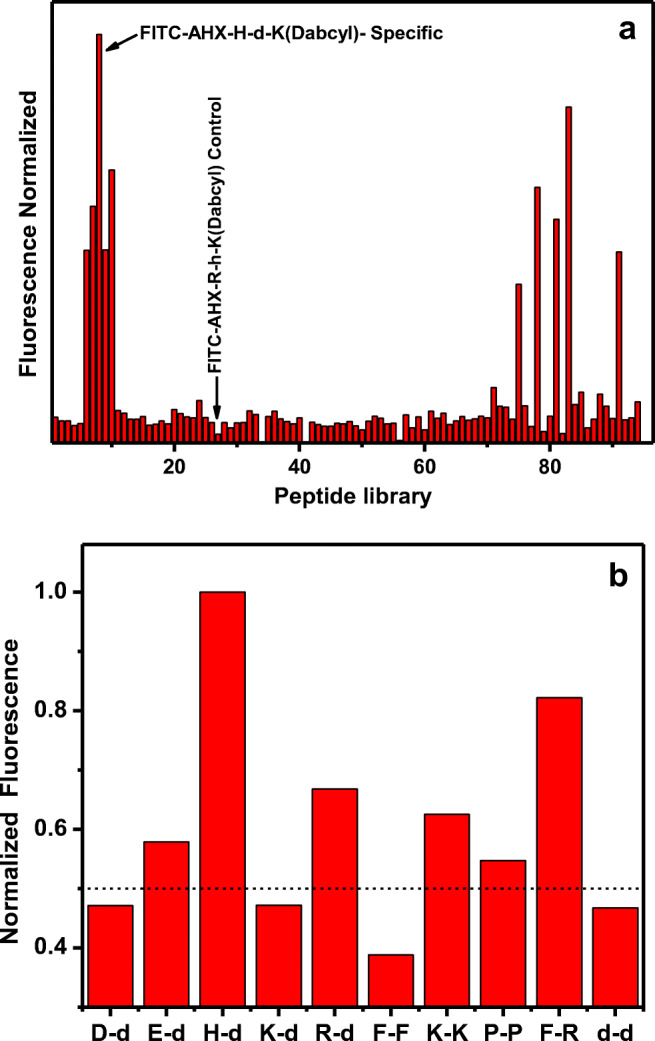
a The changes in the fluorescence signal of 115 different fluorogenic substrates presence of 108 pfu/mL at 37 °C. b The FRET substrates with significant increase in the fluorescence signal upon incubation with 108 pfu/mL of Covid-19 viral particles at 37 °C. The dotted line represents the threshold fluorescence. The samples were excited at 485 nm and the fluorescence was observed at 535 nm
Detection limit and specificity
The limit of detection (LOD) of a developed assay determines the successful development of a sensor platform. The sensitivity of the SARS-COV-2 protease-specific substrate: FITC-Ahx-H-d-K(Dabcyl) was determined from the calibration plot, obtained from the fluorescence of the substrate with different concentrations of the SARS-CoV-2 live virus in the dynamic range of 101–108 pfu/mL. Each sample with variable concentrations of the protease and their respective fluorescence intensities are represented as the linear plot in Fig. 4. The specificity of the peptide substrate against the other closely associated virus such as MERS-CoV has been tested under the same conditions. No significant change in the fluorescence intensity is observed with MER-CoV protease (Fig. 5). Changes in the fluorescence signal have been observed in the presence of SARS-CoV-2-positive patient sample, and the corresponding pfu/mL was calculated from the standard calibration plot.
Fig. 4.
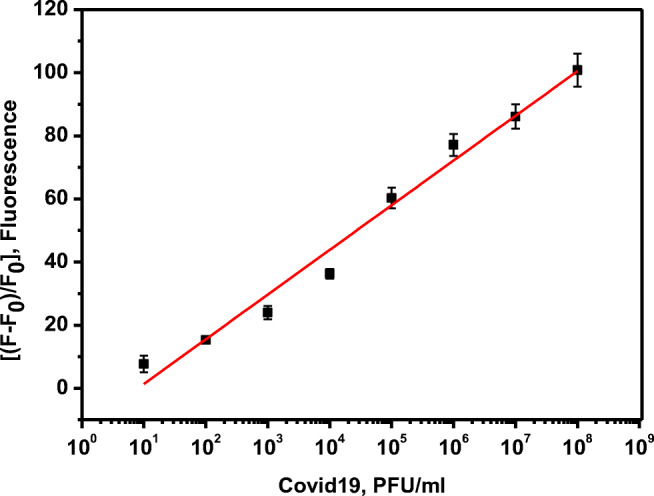
The plot of change in the fluorescence of FITC-Ahx-H-d-(Dabcyl)peptide substrate against the various concentrations of Covid-19 live virus ( 108–10 pfu/mL) after 3 hours of incubation at 37°C. The samples were excited at 485 nm and the fluorescence was observed at 535 nm. The standard errors were calculated from the three different measurements
Fig. 5.
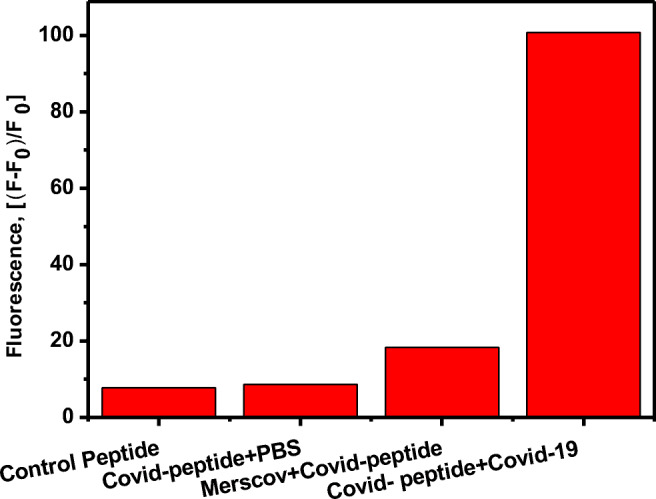
Changes in the fluorescence of FITC-Ahx-H-d-(Dabcyl) peptide substrate in the presence Covid-19 live virus, Mers-CoV (108 pfu/mL) and PBS buffer after 3 hours of incubation at 37°C. The samples were excited at 485 nm and the fluorescence was observed at 535 nm. The standard errors were calculated from the three different measurements
Real-time RT-PCR for SARS-CoV-2 patient samples
PowerChek™ 2019-nCoV Real-time PCR Kit (Cat No. R6900TD–100 Samples) was used to perform the RT-PCR test for nasopharyngeal swabs collected from SARS-CoV-2-infected patients. The kit was purchased from Kogenebiotech Co., Ltd. Republic of Korea, and it was approved by the FDA. The kit was utilized according to the manufacturer’s instructions, and it targets the RdRp gene for 2019-nCoV in nasopharyngeal swab and sputum. In brief, ExiPrep™ 96 Lite was used with ExiPrep™ 96 Viral DNA/RNA Kit (BioNEER Corp) to extract SARS-CoV-2 RNA. PowerChek™ provides a one-step real-time RT-PCR premix with specific primers and probes. PCR mixture was prepared by adding 11 μL of RT-PCR Premix, 4 μL of each primer/probe mix, and 5 μL of RNA sample to reach a total reaction volume of 20 μL. The real-time RT-PCR reaction was conducted by programing the LightCycler® 480 Instrument II using the following temperature profile: 50 °C for 30 min (1 cycle), 95 °C for 10 min (1 cycle), 95 °C for 15 s (40 cycles), and 60 °C for 1 min (40 cycles). The fluorescence curves were analyzed on FAM fluorescence detection channel for the RdRp gene-2019-nCoV and JOE (VIC or HEX) for the internal control. The result was considered positive if the corresponding fluorescence accumulation curve crosses the threshold line. Negative, positive, and internal controls were used, and their results must be passed for the run to be accepted.
Results and discussions
Enzymes can hydrolyze the peptide bond between the naturally occurring amino acids. The composition of the amino acid pair determines the specificity of the substrate for the enzyme of interest. The favorable unique conformation of amino acids and their chemical structure with their side chains between the peptide links are the most important parameters for the hydrolysis of the peptide bond by the enzymes. Under pathological conditions, the proteolytic activity of the enzymes plays a major role in many pathological processes. Therefore, the identification of these enzymes is important for understanding the mechanism and retarding the pathological enzymatic process. Determination of the hydrolysis of the specific peptide bond would lead to the development of therapeutic inhibition of the process by throughput screening. The proteolytic activity of the proteases in pathogens draws attention to develop sensitive and specific methods to detect pathogens. The fluorogenic (FRET) peptides have a fluorescent donor group attached to one of the amino acids that transfer the excited energy to the quencher acceptor group attached to another amino acid in the substrate by FRET processes [24, 25] (Scheme 1). In general, the donor and the acceptor are in very close proximity in the fluorogenic peptides and show low fluorescence due to the internal quenching (Scheme 1, A). However, the hydrolysis of the peptide bond between the donor and the acceptor leads to the physical separation of the FRET pairs; subsequently, the fluorescence increases (Scheme 1, B). An increase in the fluorescence intensity can be monitored at the emission peak of the fluorophore, which is proportional to the enzyme activity. The fluorescence intensity implies the quantity of the enzyme activity and hydrolysis rate of the specific peptide bond by the protease.
Scheme 1.
Schemtaic for the FRET assay used for screening the peptides library and the assay development. (A) Fluorescent molecule and quencher attached to the peptide seqeunce (low fluorescence signal); (B) protease cleaves the peptide seqeunce and separate the fluorecent molecules from the quencher, which results in increase in the fluorescence signal
Screening for SARS-CoV-2-specific peptide
SARS-CoV-2 protease proteolytic activity was tested against 115 different fluorogenic peptides to identify the specific substrate from the fluorescence signal. We used dipeptide of L-amino acids, D-amino acids, and the combination of both. L-amino acids present in the major natural proteins. Some bacterial species produce D-isomers in milli-molar range in the cell wall [26]. Therefore, it is assumed that the protease produced from bacteria may have the combination of both L and D-amino acids [27], and it might digest D-amino acid substrates [22]. Listeria monocytogenes protease specifically cleaves the D-amino acid peptides faster than the L-amino acids of the same peptide [28]. SARS-CoV-2 main protease (Mpro) specific substrate and 3L protease ( 3Lpro) substrate have been identified using FRET assay by high-throughput screening assay [18, 19]. More number of amino acids in the peptide substrate would lead to the complication in the identification of the specific peptide bond in the substrate. Hence, we used dipeptide with a single peptide bond between the amino acids which is the only peptide bond that undergoes hydrolysis and induces the fluorescence signal [23]. SARS-CoV and SARS-CoV-2 main protease-specific substrates have been screened by a high-throughput screening method using both natural and unnatural amino acids [19, 29, 30]. In this screening process, the SARS-CoV-2 total protease was incubated with the individual FRET substrate for 2 h at 37 °C. Change in the fluorescence intensity of each peptide was compared to find the efficient hydrolysis of the peptide bond by SARS-CoV-2 protease on the specific peptide substrate. The fluorescence signal response generated by substrate hydrolysis is illustrated in Fig. 1a. We selected ten substrates that showed considerable fluorescence signal change in the presence of SARS-CoV-2 protease (D-d, E-d, H-d, K-d, R-d, F-F, P-P, F-R, and d-d). The relative fluorescence intensity of the substrates is summarized in Table 1. The highest fluorescence change was observed from FITC-Ahx-H-d-K(Dabcyl dipeptide), and thus, we considered this the relative scale of 100%. FITC-Ahx-F-R-K(Dabcyl) confirmed 82% increase in the fluorescence intensity compared to H-d peptide (Fig. 1b). Interestingly, most of the active peptide has a combination of L- and D-amino acids. More specifically, the L-amino acids at the N-terminal and D-amino acids in the C-terminals with the exception of F-R and d-d. The two highly sensitive peptides, FITC-Ahx-H-d-K(Dabcyl) and FITC-Ahx-F-R-K(Dabcyl), have the L-amino acids with conjugated cyclic side chains, and both peptides comprise the positive charge in one of the amino acids. The results indicated that SARS-CoV-2-specific dipeptide substrate has either the combination of both L- and D-amino acids or only L-amino acids with cyclic side chain and/or positive charge in one of the amino acids. More than 50% of the active peptides have at least one dicarboxylic acid amino acids (aspartic acid or glutamic acid) implies that the presence of dicarboxylic acid amino acids might be one of the favorable factors for SARS-CoV-2 protease proteolytic activity. SARS-CoV-2 protease digests the FITC-Ahx-H-d-K(Dabcyl) efficiently and showed maximum fluorescence intensity after 3 h of incubation at 37 °C. Interestingly, the fluorogenic dipeptide FITC-Ahx-D-d-K(Dabcyl) and FITC-Ahx-d-d-K(Dabcyl) had the same effect on the cleavage of peptide bond independently of L- or D-amino acids (Fig. 1b), which indicates the presence of specific amino acid in the protease binding site and their orientations were important for the peptide bond cleavage in the substrate. The FITC-Ahx-R-h-K(Dabcyl) was very stable in the presence of SARS-CoV-2 protease incubated at 37 °C and confirmed insignificant increase in the fluorescence intensity after 3 h. Based upon the fluorescence results, we selected FITC-Ahx-H-d-K(Dabcyl) for further studies, and we used FITC-Ahx-R-h-K(Dabcyl) as a control.
Table 1.
Changes in the fluorescence signal of different substrates in the presence of SARS-CoV-2 protease
| FRET substrate | Relative fluorescence |
|---|---|
| FITC-Ahx-H-d-K(Dabcyl) | 100 |
| FITC-Ahx-F-R-K(Dabcyl) | 82 |
| FITC-Ahx-R-d-K(Dabcyl) | 67 |
| FITC-Ahx-K-K-K(Dabcyl) | 62 |
| FITC-Ahx-E-d-K(Dabcyl) | 58 |
| FITC-Ahx-P-P-K(Dabcyl) | 55 |
| FITC-Ahx-K-d-K(Dabcyl) | 47 |
| FITC-Ahx-D-d-K(Dabcyl) | 47 |
| FITC-Ahx-d-d-K(Dabcyl) | 47 |
Proteolytic activity of SRAS-CoV-2 protease on FITC-Ahx-H-d-K(Dabcyl) substrate
The rate of cleavage for the H-D substrate in the presence of variable concentrations of SARS-CoV-2 protease was monitored when incubated with 101–108 pfu/mL of SARS-CoV-2 supernatant. The fluorescence intensity of the FRET substrate increases rapidly in the presence of 108 pfu/mL of protease (Fig. 2, black curve). The rate of hydrolysis of the FITC-Ahx-H-d-K(Dabcyl) peptide reduces with a reduction in the protease concentrations as illustrated in Fig. 2 curves from top to bottom. There was no significant change in the fluorescence signal of the substrate with 101 pfu/mL of the protease. The fluorescence value is close to the control sample after 3 h of incubation at 37 °C (Fig. 2 dotted line). The fluorescence signal of FITC-Ahx-H-d-K(Dabcyl) peptide substrate was increasing linearly with time in the presence of 108 pfu/mL of the protease, indicating that the rate of hydrolysis was very fast in the initial stage of the reaction. The rate decreased over the period of time, and the curve reached a plateau after 1.5 h. When the protease and the FRET substrates mixed in the wells at the early stage, more number of substrates were available for the protease to digest the H-d peptide link in the FITC-Ahx-H-d-K(Dabcyl) substrate. The protease to substrate ratio decreased with time, and fewer substrate numbers were available for the hydrolysis, and thus, the increase in the fluorescence intensity was not linear anymore. The kinetics curves of the FITC-Ahx-H-d-K(Dabcyl) with different concentrations are illustrated in Fig. 2a. At the initial stage of the reaction, a greater number of FITC-Ahx-H-d-K(Dabcyl) substrates were available for the protease to cleave and the reaction was extremely fast. However, there was less relative fluorescence units (rfu/min) after 30 min due to the limited number of substrates available for the protease to interact with. The changes in the protease amount after the reaction was insignificant due to the high protease concentration in this reaction. Therefore, the rate of the reaction was only dependent on the concentration of the substrate.
Fig. 2.
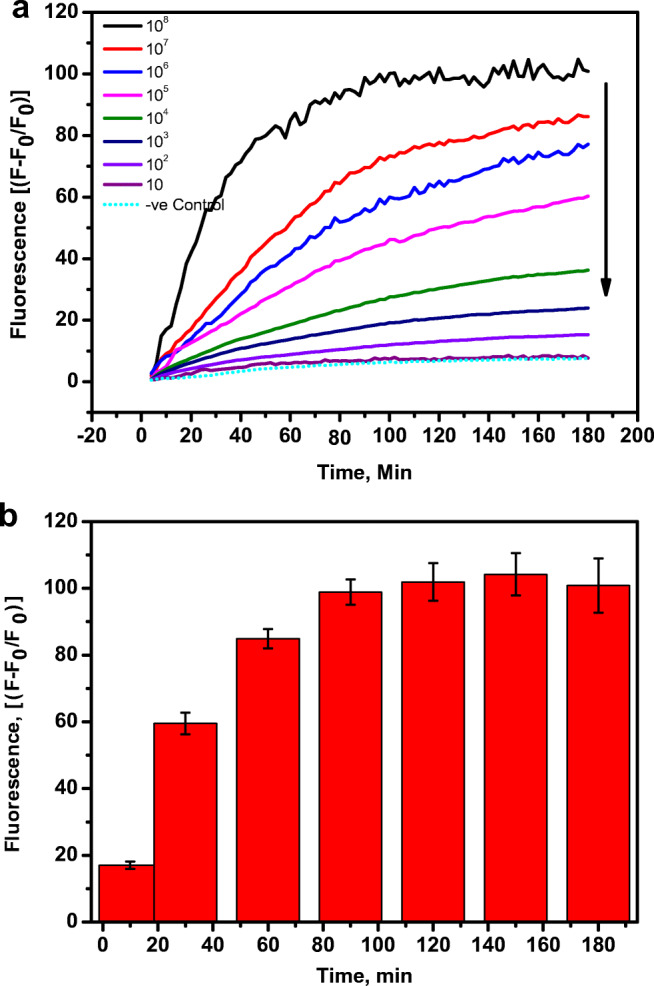
(a) The real time fluorescence change in FITC-Ahx-H-d-(Dabcyl)peptide in the presence of various concentrations of Covid-19 live virus in the dynamic range of 108–10 pfu/mL at 37 °C. (b) The increase in the fluorescence signal of FITC-Ahx-H-d-(Dabcyl) incubation with 108 pfu/mL of Covid-19 viral particles at 37 °C. The samples were excited at 485 nm and the fluorescence was observed at 535 nm. The standard errors were calculated from the three different measurements
SARS-CoV-2 sensing using FRET substrate
The sensitive detection of SARS-CoV-2 was performed by the hydrolysis of the specific peptide bond between H- and d-amino acids in FITC-Ahx-H-d-K(Dabcyl) substrate by the total protease. When the substrate was incubated with live SARS-CoV-2 viral particles, the protease produced as a metabolic by-product hence, specifically digest the H-d bond rapidly and the FITC donor and the dabcyl acceptor were separated from each other. Therefore, when FITC was excited at 485 nm, the emitted photons were not absorbed by the dabcyl acceptor, because the donor and the acceptor were not in close vicinity and there was no fluorescence quenching. All the emitted photons were monitored as fluorescence emission. The quantitative detection of SARS-CoV-2 was achieved by incubating a constant amount of FRET fluorogenic peptide with different concentrations of SARS-CoV-2 viral protease ranging from 10 to 108 pfu/mL at 37 °C for 3 h. Then, the fluorescence of FITC in all the samples is monitored, and the relative change in the fluorescence is represented in Fig. 3. The black and red bars represent the change in the fluorescence intensity after 180 min and 30 min, respectively. A significant amount of the reaction was completed within 30 min. For example, in the presence of 108 pfu/mL of SARS-CoV-2, more than 50% of the reaction was completed within 30 min. Figure 4 illustrates the plot of the endpoint of fluorescence values after 3 h against the respective concentrations of the viral particle. Further, it shows the calibration curve which was obtained by plotting the fluorescence signal change against the logarithm of SARS-CoV-2 particle (protease) concentration (pfu/mL). The minimum SARS-CoV-2 sensing ability of FITC-Ahx-H-d-K(Dabcyl), (limit of detection or LOD), was calculated according to the following formula: 3SD/m, where (SD) is the standard deviation of the fluorescence signal in the absence of SARS-CoV-2 particles and (m) is the slope of the straight line obtained from the linear fitting. The calculated LOD of FITC-Ahx-H-d-K(Dabcyl) substrate was found to be 9 ± 3 pfu/mL. Our FRET-based results were compared with the RT-PCR method, which is the gold standard for SARS-CoV-2 diagnosis. Table 2 represents the comparison of eight different patient samples (high CT, medium CT, and near the cutoff CT). The samples with rfu of 77 to 100 showed high CT values, rfu of 25 to 60 had medium CT values, and rfu of 25 and less had a weak CT values (within the cutoff values). The results confirmed that our FRET-based assay is comparable with the standard methods such as RT-PCR. Corman et al. developed RT-PCR method for the detection of SARS-CoV-2 and quantified the limit of detection in the range of 2 to 12 copies per reaction [7]. Van Kasteren et al. compared the commercially available RT-PCR kits with their in-house developed method for the detection of SARS-CoV-2. The detection limit of the evaluated kits was in the range of 3 to 10 copies/mL; however, their in-house PCR method has the LOD of 0.91 copies/mL [30]. A field-effect transistor (FET)-based biosensor has been developed for the sensitive detection of SARS-CoV-2 in clinical sample. The sensing platform was fabricated by coating graphene sheets of the FET with anti-SARS-CoV-2 spike protein antibody. The sensor was tested for antigen protein, cultured virus, and specimen from the SARS-CoV-2-positive patients. The sensor was able to detect as low as 16 pfu/mL [31]. Two high-affinity aptamers against the receptor-binding domain (RBD) of SARS-CoV-2 spike glycoprotein have been selected with the affinity of 5.8 nM and 19.9 nM, respectively. The aptamer-based recognized receptor is an ideal tool for the development of sensitive and selective aptasensor for the detection of SARS-CoV-2 [32]. Various methods used for the detection of SARS-CoV-2 are compared with our developed assay and summarized in Table 3. The previously reported RT-PCR method is more sensitive than other methods due to amplification processes. However, the other methods are comparable or less sensitive than the reported values by our FRET assay. Although our developed assay is highly sensitive, it has some limitations. The turbid sample leads to fluorescence scattering; therefore, samples should be clear. The presence of any molecule in the sample absorbs in the region of the FITC emission which reduces the fluorescence intensity due to the fluorescence quenching. The fluorogenic substrates have to be protected from the light as it may undergo photobleaching when exposing to light.
Fig. 3.
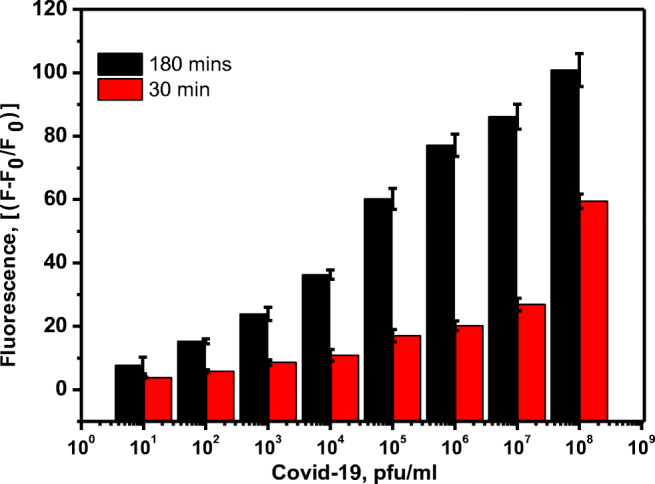
Changes in the fluorescence of FITC-Ahx-H-d-(Dabcyl) peptide substrate in the presence of various concentrations of Covid-19 live virus (108–10 pfu/mL) after 30 minutes and 3 hours of incubation at 37 °C. The samples were excited at 485 nm and the fluorescence was observed at 535 nm. The standard errors were calculated from the three different measurements
Table 2.
Comparison of FRET assay with RT-PCR
| Patient samples | Fluorescence (relative percentage) | RT-PCR (CT value) |
|---|---|---|
| Patient 1 | 71–100 | High |
| Patient 2 | High | |
| Patient 3 | High | |
| Patient 4 | 31–70 | Medium |
| Patient 5 | Medium | |
| Patient 6 | Medium | |
| Patient 7 | <30 | Weak(~Cut off) |
| Patient 8 | Weak (~Cut off) |
Table 3.
Comparison of different methods used for the detection of SARS-CoV-2
| Method | Sample type | LOD | Reference | |
|---|---|---|---|---|
| 1 | RT-PCR | Viral -RNA | 0.91 copies/mL | [30] |
| 2 | RT-PCR | RdRp gene | 3.6 copies/reaction | [7] |
| 3 | Field effect transistor | S-protein antibody | 16 copies/mL | [31] |
| 4 | RT-LAMP | mRNA | 50 copies/μL | [33] |
| 5 | RT-q(PCR-LAMP) | mRNA | 5 copies/reaction | [34] |
| 6 | ITP-Crisper | cDNA of mRNA | 10 copies/μL | [35] |
| 7 | Molecular POC | mRNA | 10 copies/μL | [36] |
| 8 | Electrochemical | Viral antigen | 0.8 pg/mL | [37] |
| 9 | Lateral flow immunoassay | IgM/IgG | 1 pg/mL | [38] |
| 10 | Fluorescence | Protease | 0.9 copies/mL | [This study] |
Cross-reactivity of FRET substrate
The selective hydrolysis of the H-D peptide bond by SARS-CoV-2 was validated using the protease obtained from closely associated viruses, such as MERS-CoV. The same concentration of MERS-CoV viral particles were incubated with FITC-Ahx-H-d-K(Dabcyl), under the similar condition as SARS-CoV-2 as mentioned in Section 2.5, and presented in Fig. 5. There was no significant increase in the fluorescence intensity (less than 20%) of FITC-Ahx-H-d-K(Dabcyl) in the presence of MERS-CoV virus. The MERS-CoV protease did not cleave the H-d peptide link in the FITC-Ahx-H-d-K(Dabcyl) substrate. The change in the fluorescence intensity was close to the FITC-Ahx-H-d-K(Dabcyl) peptide in the absence of SARS-CoV-2 virus and control peptide, FITC-Ahx-R-h-K(Dabcyl) which confirmed very minimum fluorescence change in the presence of SARS-CoV-2. The results indicated that the fluorogenic substrate, FITC-Ahx-H-d-K(Dabcyl), is very specific to SARS-CoV-2. Further, it could specifically digest H-D peptide bond in the mixture of other closely associated viral particles. The specific substrate was validated by incubating SARS-CoV-2-positive patient sample with FITC-Ahx-H-d-K(Dabcyl), and it showed the value of 34,304 pfu/mL from the standard calibration plot. The results suggest that FITC-Ahx-H-d-K(Dabcyl) could be used for the diagnosis of SARS-CoV-2-positive samples in bulk in a short time using simple spectrofluorometric measurements.
Conclusions
We successfully developed a high-throughput peptide screening for the sensitive detection of SARS-CoV-2 as a diagnostic marker. FRET assay has been carried out using fluorogenic dipeptides (L, D, or both amino acids in the dipeptide) with a fluorophore and a quencher at both ends. SARS-CoV-2 protease-specific dipeptide substrates were identified from the quantity of fluorescence signal change. FITC-Ahx-H-d-(Dabcyl) fluorogenic dipeptide is considered as the most specific peptide substrate as it showed the highest fluorescence signal in the presence of SARS-CoV-2 protease. The cross-reactivity of the substrate with other closely associated viral proteases such as MERS-CoV has been confirmed from insignificant fluorescence signal change. Clinical patient samples have been used for the validation of the FRET assay, and the values were highly comparable to the standard RT-PCR methods. Therefore, our FRET assay is straightforward, low cost, and highly sensitive for the analysis of clinical patient samples.
Funding
The author acknowledges the financial support provided by King Abdulaziz City for Science and Technology (General Directorate for Research & Innovation Support) (GDRIS) (King Abdulaziz University) to implement. This work is through fast track program for COVID-19 Research Project No. 5-20-01-009-0002.
Declarations
Conflict of interest
This work is protected under USPTO filing No. 17108263.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hani A. Alhadrami and Raja Chinnappan contributed equally to this work.
Contributor Information
Hani A. Alhadrami, Email: hanialhadrami@kau.edu.sa
Esam I. Azhar, Email: eazhar@kau.edu.sa
Mohammed Zourob, Email: mzourob@alfaisal.edu.
References
- 1.Rothan HA, Byrareddy SN. The epidemiology and pathogenesis of coronavirus disease (COVID-19) outbreak. J Autoimmun. 2020;109:102433. doi: 10.1016/j.jaut.2020.102433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hamre D, Procknow JJ. A new virus isolated from the human respiratory tract. Proc Soc Exp Biol Med. 1966;121:190–193. doi: 10.3181/00379727-121-30734. [DOI] [PubMed] [Google Scholar]
- 3.Chen Y, Chan KH, Kang Y, Chen H, Luk HKH, Poon RWS, Chan JFW, Yuen KY, Xia N, Lau SKP, Woo PCY. A sensitive and specific antigen detection assay for Middle East respiratory syndrome coronavirus. Emerg Microbes Infect. 2015;4:e26. doi: 10.1038/emi.2015.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Williamson EJ, Walker AJ, Bhaskaran K, BaconS BC, Morton CE, Curtis HJ, Mehrkar A, Evans D, Inglesby P, Cockburn J, McDonald HI, MacKenna B, Tomlinson L, Douglas IJ, Rentsch CT, Mathur R, Wong AYS, Grieve R, Harrison D, Forbes H, Schultze A, Croker R, Parry J, Hester F, Harper S, Perera R, Evans SJW, Smeeth L, Goldacre B. Factors associated with COVID-19-related death using OpenSAFELY. Nature. 2020;584:430–436. doi: 10.1038/s41586-020-2521-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tan WYT, Wong LY, Leo YS, Toh MPHS. Does incubation period of COVID-19 vary with age? A study of epidemiologically linked cases in Singapore. Epidemiol Infect. 2020;148:e197. doi: 10.1017/S0950268820001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaim S, Chong JH, Sankaranarayanan V, Harky A. COVID-19 and multiorgan response. Curr Probl Cardiol. 2020;45:100618. doi: 10.1016/j.cpcardiol.2020.100618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Corman VM, Landt O, Kaiser M, Molenkamp R, Meijer A, Chu DKW, Bleicker T, Brünink S, Schneider J, Schmidt ML. Detection of 2019 novel coronavirus (2019-nCoV) by real-time RT-PCR. Eurosurveillance. 2020;25:2000045. doi: 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adhikari SP, Meng S, Wu YJ, Mao YP, Ye RX, Wang QZ, Sun C, Sylvia S, Rozelle S, Raat H. Epidemiology, causes, clinical manifestation and diagnosis, prevention and control of coronavirus disease (COVID-19) during the early outbreak period: a scoping review. Infect Dis Poverty. 2020;9(1):1–12. doi: 10.1186/s40249-020-00646-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan WJF, Yip CCY, To KKW, Tang THC, Wong SCY, Leung KH, Fung AYF, Ng ACK, Zou Z, Tsoi HW (2020) Improved molecular diagnosis of COVID-19 by the novel, highly sensitive and specific COVID-19-RdRp/Hel real-time reverse transcription-PCR assay validated in vitro and with clinical specimens. J Clin Microbiol 58(5) [DOI] [PMC free article] [PubMed]
- 10.World Health Organization . Coronavirus disease (COVID-19) technical guidance: laboratory testing for 2019-nCoV in humans. Geneva: Switzerland; 2020. [Google Scholar]
- 11.Cai X, Chen J, Hu J, Long Q, Deng H, Fan K, Liao P, Liu B, Wu G, Chen Y (2020)A peptide-based magnetic chemiluminescence enzyme immunoassay for serological diagnosis of corona virus disease 2019 (COVID-19), medRxiv [DOI] [PMC free article] [PubMed]
- 12.Guo L, Ren L, Yang S, Xiao M, Chang D, Yang F, Dela Cruz CS, Wang Y, Wu C, Xiao Y, Zhang L, Han L, Dang S, Xu Y, Yang QW, Xu SY, Zhu HD, Xu YC, Jin Q, Sharma L, Wang L, Wang J. Profiling early humoral response to diagnose novel coronavirus disease (COVID-19) Clin Infect Dis. 2020;71:778–785. doi: 10.1093/cid/ciaa310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Corman VM, Muller MA, Costabel U, Timm J, Binger T, Meyer B, Kreher P, Lattwein E, Eschbach-Bludau M, Nitsche A, Bleicker T, Landt O, Schweiger B, Drexler JF, Osterhaus AD, Haagmans BL, Dittmer U, Bonin F, Wolff T, Drosten C (2012) Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill 17(49) http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20334 [DOI] [PubMed]
- 14.Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, Chen H, Mubareka S, Gubbay JB, Chan WCW. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14:3822–3835. doi: 10.1021/acsnano.0c02624. [DOI] [PubMed] [Google Scholar]
- 15.Kustin T, Ling G, Sharabi S, Ram D, Friedman N, Zuckerman N, Bucris ED, Glatman-Freedman A, Stern A, Mandelboim M. A method to identify respiratory virus infections in clinical samples using next-generation sequencing. Sci. Rep. 2019;9:2606–2606. doi: 10.1038/s41598-018-37483-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keightley MC, Sillekens P, Schippers W, Rinaldo C, George KS. Real-time NASBA detection of SARS-associated coronavirus and comparison with real-time reverse transcription-PCR. J Med Virol. 2005;77:602–608. doi: 10.1002/jmv.20498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campos EVR, Pereira AES, de Oliveira JL, Carvalho LB, Guilger-Casagrande M, de Lima R, Fraceto LF. How can nanotechnology help to combat COVID-19? Opportunities and urgent need. J Nanobiotechnol. 2020;18:125. doi: 10.1186/s12951-020-00685-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu W, Xu M, Chen CZ, Guo H, Shen M, Hu X, Shinn P, Klumpp-Thomas CMichael SG, Zheng W. Identification of SARS-CoV-2 3CL protease inhibitors by a quantitative high-throughput screening. ACS Pharmacol Transl Sci. 2020;3:1008–1016. doi: 10.1021/acsptsci.0c00108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rut W, Groborz K, Zhang L, Sun X, Zmudzinski M, Hilgenfeld R, Drag M (2020) Substrate specificity profiling of SARS-CoV-2 Mpro protease provides basis for anti-COVID-19 drug design. Biorxiv. 10.1101/2020.03.07.981928
- 20.Al-Amri SS, Abbas AT, Siddiq LA, Alghamdi A, Sanki MA, Al-Muhanna MK, Alhabbab RY, Azhar EI, Li X, Hashem AM. Immunogenicity of candidate MERS-CoV DNA vaccines based on the spike protein. Sci Rep. 2017;7:44875. doi: 10.1038/srep44875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coleman CM, Frieman MB. Growth and quantification of MERS-CoV infection. Curr Protoc Microbiol. 2015;37:15E-2. doi: 10.1002/9780471729259.mc15e02s37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kaman WE, Voskamp-Visser I, de Jongh DMC, Endtz HP, van Belkum A, Hays JP, Bikker FJ. Evaluation of a D-amino-acid-containing fluorescence resonance energy transfer peptide library for profiling prokaryotic proteases. Anal Biochem. 2013;441:38–43. doi: 10.1016/j.ab.2013.06.015. [DOI] [PubMed] [Google Scholar]
- 23.Kaman WE, Hulst AG, van Alphen PTW, Roffel S, van der Schans MJ, Merkel T, van Belkum A, Bikker FJ. Peptide-based fluorescence resonance energy transfer protease substrates for the detection and diagnosis of Bacillus species. Anal Chem. 2011;83:2511–2517. doi: 10.1021/ac102764v. [DOI] [PubMed] [Google Scholar]
- 24.Chinnappan R, Dubé A, Lemay J-F, Lafontaine DA. Fluorescence monitoring of riboswitch transcription regulation using a dual molecular beacon assay. Nucleic Acids Res. 2013;41(10):e106–e106. doi: 10.1093/nar/gkt190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Raja C, Ferner J, Dietrich U, Avilov S, Ficheux D, Darlix J-L, de Rocquigny H, Schwalbe H, Mély Y. A tryptophan-rich hexapeptide inhibits nucleic acid destabilization chaperoned by the HIV-1 nucleocapsid protein. Biochemistry. 2006;45(30):9254–9265. doi: 10.1021/bi052560m. [DOI] [PubMed] [Google Scholar]
- 26.Lam H, Oh DC, Cava F, Takacs CN, Clardy J, de Pedro MA, Waldor MK. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science. 2009;325:1552–1555. doi: 10.1126/science.1178123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Aliashkevich A, Alvarez L, Cava F. New insights into the mechanisms and biological roles of D-amino acids in complex eco-systems. Front Microbiol. 2018;9:683. doi: 10.3389/fmicb.2018.00683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alhogail S, Suaifan GARY, Zourob M. Rapid colorimetric sensing platform for the detection of Listeria monocytogenes foodborne pathogen. Biosens Bioelectron. 2016;86:1061–1066. doi: 10.1016/j.bios.2016.07.043. [DOI] [PubMed] [Google Scholar]
- 29.Van de Plassche MAT, Barniol-Xicota M, Verhelst S. Peptidyl acyloxymethyl ketones as activity-based probes for the main protease of SARS-CoV-2. Chembiochem. 2020;21:3383–3388. doi: 10.1002/cbic.202000371. [DOI] [PubMed] [Google Scholar]
- 30.van Kasteren PB, van der Veer B, van den Brink S, Wijsman L, de Jonge J, van den Brandt A, Molenkamp R, Reusken CBEM, Meijer A. Comparison of commercial RT-PCR diagnostic kits for COVID-19. J Clin Virol. 2020;128:104412. doi: 10.1016/j.jcv.2020.104412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seo G, Lee G, Kim MJ, Baek SH, Choi M, Ku KB, Lee CS, Jun S, Park D, Kim HG, Kim SJ, Lee JO, Kim BT, Park EC, Kim SI. Rapid detection of COVID-19 causative virus (SARS-CoV-2) in human nasopharyngeal swab specimens using field-effect transistor-based biosensor. ACS Nano. 2020;14:5135–5142. doi: 10.1021/acsnano.0c02823. [DOI] [PubMed] [Google Scholar]
- 32.Song Y, Song J, Wei X, Huang M, Sun M, Zhu L, Lin B, Shen H, Zhu Z, Yang C. Discovery of aptamers targeting the receptor-binding domain of the SARS-CoV-2 spike glycoprotein. Anal Chem. 2020;92:9895–9900. doi: 10.1021/acs.analchem.0c01394. [DOI] [PubMed] [Google Scholar]
- 33.Ganguli A, Mostafa A, Berger J, Aydin MY, Sun F, de Ramirez SA, Valera E, Cunningham BT, King WP, Bashir R. Rapid isothermal amplification and portable detection system for SARS-CoV-2. Proc Natl Acad Sci. 2020;117:22727–22735. doi: 10.1073/pnas.2014739117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Varlamov DA, Blagodatskikh KA, Smirnova EV, Kramarov VM, Ignatov KB. Combinations of PCR and isothermal amplification techniques are suitable for fast and sensitive detection of SARS-CoV-2 viral RNA. Front Bioeng Biotechnol. 2020;8:1294. doi: 10.3389/fbioe.2020.604793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ramachandran A, Huyke DA, Sharma E, K. Sahoo M K, Huang C, Banaei N, Pinsky BA, Santiago J G. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. PNAS. 2020;117:29518–29525. doi: 10.1073/pnas.2010254117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, Sasso JM, Gregg AC, Soares DJ, Beskid TR. JerveySR, Liu C. ACS Cent Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eissa S, Zourob M. Development of a low-cost cotton-tipped electrochemical immunosensor for the detection of SARS-CoV-2. Anal Chem. 2021;93(3):1826–1833. doi: 10.1021/acs.analchem.0c04719. [DOI] [PubMed] [Google Scholar]
- 38.Liu H, Dai E, Xiao R, Zhou Z, Zhang M, Bai Z, Shao Y, Qi K, Tu J, Wang C, Wang S. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens Actuators B Chem. 2021;329:129196. doi: 10.1016/j.snb.2020.129196. [DOI] [PMC free article] [PubMed] [Google Scholar]