Abstract
The clinical spectrum of hypertensive disorders of pregnancy (HDP) is determined by the interplay between environmental and genetic factors, most of which remains unknown. ERAP1, ERAP2 and LNPEP genes code for multifunctional aminopeptidases involved with antigen processing and degradation of small peptides such as angiotensin II (Ang II), vasopressin and oxytocin. We aimed to test for associations between genetic variants in aminopeptidases and HDP. A total of 1282 pregnant women (normotensive controls, n = 693; preeclampsia, n = 342; chronic hypertension with superimposed preeclampsia, n = 61; eclampsia, n = 74; and HELLP syndrome, n = 112) were genotyped for variants in LNPEP (rs27300, rs38034, rs2303138), ERAP1 (rs27044, rs30187) and ERAP2 (rs2549796 rs2927609 rs11135484). We also evaluated the effect of ERAP1 rs30187 on plasma Ang II levels in an additional cohort of 65 pregnant women. The genotype C/C, in ERAP1 rs30187 variant (c.1583 T > C, p.Lys528Arg), was associated with increased risk of eclampsia (OR = 1.85, p = 0.019) whereas ERAP2 haplotype rs2549796(C)–rs2927609(C)–rs11135484(G) was associated with preeclampsia (OR = 1.96, corrected p-value = 0.01). Ang II plasma levels did not differ across rs30187 genotypic groups (p = 0.895). In conclusion, ERAP1 gene is associated with eclampsia whereas ERAP2 is associated with preeclampsia, although the mechanism by which genetic variants in ERAPs influence the risk of preeclampsia and eclampsia remain to be elucidated.
Subject terms: Genetic association study, Genetics research, Hypertension
Introduction
Hypertensive disorders of pregnancy (HDP) account for 14% of all maternal deaths1 and contribute to increase the cardiovascular risk in both mothers2 and offspring3. As other complex diseases, HDP have a broad clinical spectrum ranging from mild hypertension without proteinuria to severe proteinuria, and eventual seizures (i.e. eclampsia), or with hemolysis elevated liver enzymes and low platelet liver disease and severe inflammation (i.e. HELLP syndrome). Risk factors for preeclampsia, such as pre-gestational body mass index, nulliparity, change in partners, and advanced maternal age have been reported for different populations4. Regarding the genetics of preeclampsia, genes INHBP5, FLT16 and PLEKHG17 were identified from genome wide association studies, however, the genetic architecture underlying the disease mechanism remains largely unknown8.
Endoplasmic reticulum aminopeptidases -1 (ERAP1), -2 (ERAP2) and leucyl/cystinyl aminopeptidase (LNPEP), also known as A-LAP, L-RAP and P-LAP, respectively, are multifunctional enzymes belonging to the M1 family of aminopeptidases9. These aminopeptidases act in concert to trim peptides to be presented by the major histocompatibility complex (MHC) class I molecules10 and, in addition, they cleave a variety of bioactive peptides, including angiotensins, bradykinin, kallidin and oxytocin11. Not surprisingly, these enzymes are involved in several biological processes such as immune and inflammatory responses, blood pressure regulation and pregnancy maintenance12,13. There is also increasing evidence that LNPEP is involved with preterm delivery due to its oxytocinase activity14.
Johnson and colleagues identified a quantitative trait locus (QTL) for preeclampsia on chromosome 5q, in a region harboring the aminopeptidases genes15 and, subsequently, confirmed the genetic association between ERAP2 and preeclampsia16. The missense genetic variants in ERAP1, rs27044 and rs30187, have consistently been reported as associated with ankylosing spondylitis, psoriasis, multiple sclerosis and Crohn’s disease17. Lastly, maternal LNPEP variants were reported as associated with increased risk of preterm birth18. Thus, the present study aimed to evaluate genetic variants in ERAP1, ERAP2 and LNPEP for association with the full clinical spectrum of HDP. For the first time, eclampsia and HELLP phenotypes, which are the most severe and rare phenotypes, were tested for these genes.
Methods
Population and study design
Our study population was recruited from Maternidade Escola Januário Cicco, a tertiary center for women’s health, located in Natal, Rio Grande do Norte state, Brazil. A total of 1693 women were recruited from 2002 to 2010, as part of a broader study aiming to investigate clinical, epidemiological and genetic aspects of hypertensive disorders of pregnancy. Clinical data as well as blood samples were collected at the time of enrollment. For the current study, we retrospectively selected 1282 women based on their pregnancy outcome: 693 normotensive women (control), 342 preeclampsia (PE), 61 superimposed preeclampsia (PEsuper), 74 eclampsia, and 112 HELLP syndrome cases. All Methods were performed in accordance with the Declaration of Helsink and followed the Brazilian ethical standards of scientific research. The research protocol was reviewed and approved by the Federal University of Rio Grande do Norte (CEP-UFRN 88) and Brazilian National Ethical Committee (CONEP 5059). All research participants or their legal guardian provided informed consent.
Phenotype definition
The diagnostic criteria followed the recommendations from the American College of Obstetrician and Gynecologists19. Preeclampsia was defined as the new onset hypertension (SBP ≥ 140 mmHg or DBP ≥ 90 mmHg) and proteinuria (≥ + 1 on dipstick) after 20 weeks of gestation. Superimposed preeclampsia occurred when the woman had a previous diagnosis of chronic hypertension and developed proteinuria after 20 weeks of gestation. Eclampsia was defined by the presence of seizure, while HELLP syndrome diagnosis was based on Mississippi Class III system (AST > 40 IU/L and LDH > 600 IU/L and platelets < 150,000/μL)20. Controls were healthy pregnant women with no history of hypertension. Women with multiple pregnancies, diabetes or other chronic diseases were excluded from study.
Genetic variants
The variants in ERAP2 (rs2549796, rs2927609, rs11135484) and LNPEP (rs27300, rs38034, rs2303138) were all tag-variants, identified through a pairwise selection strategy with an r2 threshold ≥ 0.8 in Haploview 4.221 using the HapMap CEU population genotype data (HapMap Rel 27 phase II + III). Variants rs30187 and rs27044, in ERAP1, were chosen based on their effect on protein function22,23 as well as their implication in other diseases17.
Genotyping
DNA extraction was carried out as previously described24. Samples were genotyped by SNaPshot technique and the capillary electrophoresis performed on ABI PRISM 3100 Avant Genetic Analyzer (Applied Biosystems). Technique standardization was carried out according to Lins and colleagues25. GeneMapper software (Applied Biosystems, CA, USA) was used for the genotype calling.
Population stratification assessment
To avoid confounding by ethnicity we used a panel with 27 ancestry informative markers (AIMs) particularly designed for the Brazilian population26. A sub-sample of 756 women randomly selected was used to assess the genetic ancestry of our study population (n = 1282) using principal component analysis in SNPRelate R package27. Samples from The 1000 Genomes Project28 of European (IBS), African (ASW, MSL, YRI) and American (CLM) origins were used as reference populations.
Functional validation
Variant effects on mRNA and protein levels were assessed from GTEx (dbGaP Accession phs000424.v8.p2)29 and single nucleotide polymorphisms annotator (SNiPA)30 databases. Aiming to functionally validate the ERAP1 genetic finding, we recruited an additional cohort of 65 pregnant women, including 29 normotensive controls and 36 severe preeclampsia cases, that had their Ang II plasma concentration measured by ELISA commercial kit (MyBioSource, San Diego, CA, USA, Cat.Num. MBS453098). Briefly, blood samples were systematically collected between 7 and 9 am in EDTA tubes and immediately centrifuged. The obtained plasma was stored at − 80 °C until assay.
Statistical analysis
Clinical and demographic data were analyzed through chi-squared and t-test for categorical and quantitative variables, respectively. With regard to the genetic data, allele frequencies were compared by Fisher exact test, whereas genotype and haplotype association tests were performed through logistic regression models including maternal age and parity (primigesta vs others) as covariates. Haplotype frequencies were estimated by Expectation–Maximization algorithm with a minor haplotype frequency threshold of 0.03. The p-values were corrected for family-wise error rate by permutation procedures (10,000 ×) implemented in PLINK31. All analyses were performed by comparing each case phenotype (i.e. PE, PEsuper, eclampsia and HELLP) against the normotensive control group.
Results
Demographics and clinical characteristics
Table 1 summarizes the main clinical characteristics and demographics for our study population. Women with eclampsia and HELLP were the youngest and oldest, respectively, when compared to the control group, whereas the proportion of primiparas was higher in the preeclampsia and eclampsia groups. Women with HELLP syndrome delivered their babies earlier in pregnancy (mean gestational age = 34.3 weeks), followed by eclampsia (mean gestational age = 36.2 weeks) and preeclampsia (37.2 weeks) groups. Overall, the frequency of family members affected by chronic hypertension was higher in the case groups when compared to control group, suggesting shared genetic components between essential hypertension and hypertensive disorders of pregnancy (Table 1). Of note, the prevalence of chronic hypertension and eclampsia in family members was much higher in superimposed preeclampsia (PEsuper) and eclampsia groups, respectively.
Table 1.
Demographics and clinical characteristics.
| Characteristics | Control | PE | PEsuper | Eclampsia | HELLP |
|---|---|---|---|---|---|
| Sample size, n | 693 | 342 | 61 | 74 | 112 |
| Maternal age, y, mean (± SD) | 24.4 (± 6.2) | 25.2 (± 6.8) | 31.1 (± 6.9)a | 20.5 (± 6.1)a | 27.0 (± 6.7)a |
| SBP, mmHg, mean (± SD) | 117 (± 12) | 156 (± 19)a | 166 (± 28)a | 159 (± 21)a | 154 (± 21)a |
| DBP, mmHg, mean (± SD) | 75 (± 9) | 105 (± 13)a | 108 (± 14)a | 108 (± 16)a | 101 (± 13)a |
| Proteinuria, n (%) | |||||
| Negative | 693 (100) | 0 (0.0) | 0 (0.0) | 2 (2.7) | 7 (6.3) |
| 1 + | – | 104 (30.4) | 28 (45.9) | 11 (14.9) | 13 (11.6) |
| ≥ 2 + | – | 238 (69.6) | 16 (26.2) | 43 (58.1) | 85 (75.8) |
| Missing | – | 0 (0.0) | 17 (27.9) | 18 (24.3) | 7 (6.3) |
| Primigestas, n (%) | 300 (43.3) | 180 (52.8)a | 14 (23.3) a | 54 (73.0)a | 46 (41.4) |
| Gestational age at delivery, w, mean (± SD) | 38.8 (± 3.1) | 37.2 (± 3.0)a | 35.8 (± 3.5)a | 36.2 (± 3.6)a | 34.3 (± 4.0)a |
| Number of antenatal care visits, mean (± SD) | 5.9 (± 2.8) | 6.2 (± 2.3) | 6.1 (± 2.9) | 4.5 (± 2.5)a | 5.3 (± 2.3)a |
| Family history of | |||||
| Chronic hypertensionb | 238 (35.5) | 178 (56.9)a | 46 (80.7)a | 32 (46.4) | 55 (49.6)a |
| Preeclampsiac | 22 (3.8) | 41 (15.9)a | 5 (10.9)a | 12 (21.0)a | 8 (8.3)a |
| Eclampsiac | 7 (1.2) | 11 (4.4)a | 3 (6.5)a | 5 (8.8)a | 2 (2.1) |
SBP/DBP systolic blood pressure/diastolic blood pressure.
PEsuper: chronic hypertension with superimposed preeclampsia.
ap < 0.05 for comparison with the control group.
bIf at least one first degree relative has the disease.
cIf the mother had had the disease.
Genetic analysis
All genetic variants achieved standard quality control thresholds (i.e. genotyping error rates < 0.05, minor allele frequency > 0.01, and p-value > 0.05 for Hardy–Weinberg equilibrium test). In addition, there was no evidence of population stratification, since cases and controls were equally distributed across the reference ethnical groups (Supplementary Fig. 1).
Figure 1 shows the linkage disequilibrium (LD) pattern across the genomic region encompassing the studied variants. ERAP1 variants were moderately correlated (r2 = 0.65) while ERAP2 and LNPEP variants seemed to belong to the same haploblock.
Figure 1.
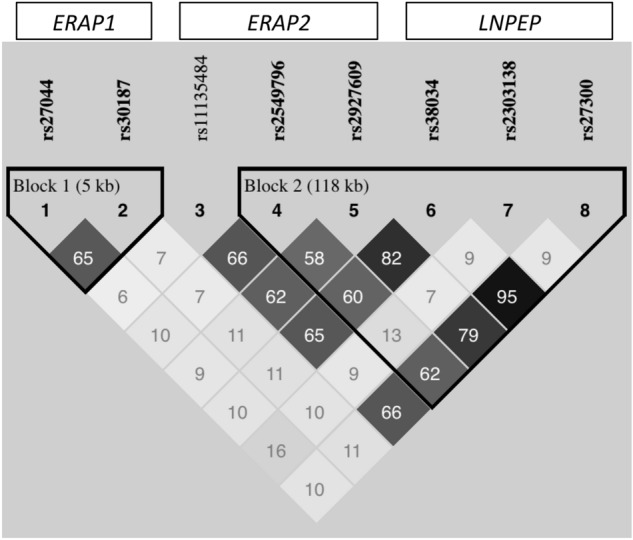
Linkage disequilibrium (LD) pattern among the studied markers. In the LD plot, the inside-square number represents the correlation coefficient value (r2). The genomic organization and LD pattern suggest ERAP2 and LNPEP markers as belonging to the same haploblock.
In order to analyze the combined effect of variants on disease risk, we performed haplotype-based tests (Table 2). As result, ERAP2 haplotype rs2549796–rs2927609–rs11135484 was associated with preeclampsia (corrected p = 0.0109). There was no haplotype associated with the remaining case groups (corrected p > 0.05).
Table 2.
Haplotype tests of association showing ERAP2 C–C–G haplotype associated with preeclampsia.
| Haplotypes | Haplotype frequency | ||||
|---|---|---|---|---|---|
| Control | PE | PEsuper | Eclampsia | HELLP | |
| ERAP1 | |||||
| T–G | 0.354 | 0.358 | 0.328 | 0.341 | 0.323 |
| T–C | 0.105 | 0.097 | 0.121 | 0.051 | 0.074 |
| C–C | 0.541 | 0.545 | 0.552 | 0.609 | 0.602 |
| ERAP2 | |||||
| C–T–A | 0.362 | 0.361 | 0.356 | 0.301 | 0.352 |
| C–C–A | 0.064 | 0.066 | 0.060 | 0.065 | 0.079 |
| C–C–G | 0.045 | 0.076a | 0.050 | 0.045 | 0.065 |
| T–C–G | 0.488 | 0.482 | 0.515 | 0.523 | 0.452 |
| LNPEP | |||||
| T–C–A | 0.123 | 0.130 | 0.105 | 0.132 | 0.135 |
| C–T–G | 0.398 | 0.376 | 0.375 | 0.345 | 0.397 |
| T–C–G | 0.479 | 0.494 | 0.520 | 0.521 | 0.468 |
ERAP1: rs30187–rs27044.
ERAP2: rs2549796–rs2927609–rs11135484.
LNPEP: rs27300–rs38034–rs2303138.
aPE versus Control (Uncorrected p = 0.0013; p-value corrected for family-wise error rates by running 10,000 permutations: p = 0.0109).
There was no difference regarding allele frequencies between control and case groups (Supplementary Table 1), although the genotype distribution for ERAP1 variants in eclampsia group seemed to differ, when compared to controls (Table 3). The frequency of genotype C/C (rs30187) was notably higher in eclampsia group (40.9%), what would be consistent with a recessive genetic model.
Table 3 .
Genotype distribution for ERAP1 variants across phenotypic groups.
| SNP | Genotypes | Genotype distribution, n (%) | ||||
|---|---|---|---|---|---|---|
| Control | PE | PEsuper | Eclampsiaa | HELLP | ||
| rs30187 | T/T | 139 (20.5) | 68 (20.5) | 12 (19.7) | 16 (22.5) | 16 (14.6) |
| C/T | 343 (50.7) | 167 (50.5) | 32 (52.5) | 26 (36.6) | 56 (50.9) | |
| C/C | 195 (28.8) | 96 (29.0) | 17 (27.8) | 29 (40.9) | 38 (34.6) | |
| rs27044 | G/G | 86 (12.7) | 48 (14.5) | 9 (15.5) | 14 (19.2) | 12 (10.9) |
| G/C | 307 (45.4) | 141 (42.6) | 20 (34.5) | 23 (31.5) | 48 (43.6) | |
| C/C | 283 (41.9) | 142 (42.9) | 29 (50.0) | 36 (49.3) | 50 (45.5) | |
aChi-squared test of genotypic association for Control vs Eclampsia: p = 0.055 for rs30187 and p = 0.079 for rs27044.
Of note, rs30187 C allele codes for Arg528 (instead of Lys528), resulting in an enzyme type characterized by lower peptidase activity against Ang II32. Therefore, we defined a recessive genetic model with C/C as the risk genotype for eclampsia. The model was implemented through logistic regression with genotype (T/T + T/C vs C/C) as the main explanatory variable and maternal age, and parity as covariates. As result, women homozygotes for rs30187 C/C risk genotype were more likely to develop eclampsia (OR = 1.85, p = 0.019) (Table 4).
Table 4.
Genetic effect of rs30187 on eclampsia risk under recessive genetic model.
| Genotypic group | Controla | Eclampsiaa | Eclampsia riskb | |
|---|---|---|---|---|
| n (%) | n (%) | OR (95% CI) | p-value | |
| T/T + C/T | 482 (71.2) | 40 (58.0) | 1.0 | 0.019 |
| C/C | 195 (28.8) | 29 (42.0) | 1.85 (1.11–3.11) | |
aChisquared test of association for genotype distribution between Control and Eclampsia (p = 0.022).
bOdds ratio (OR) and confidence interval (CI) estimated by logistic regression model adjusted for maternal age and parity.
Functional validation
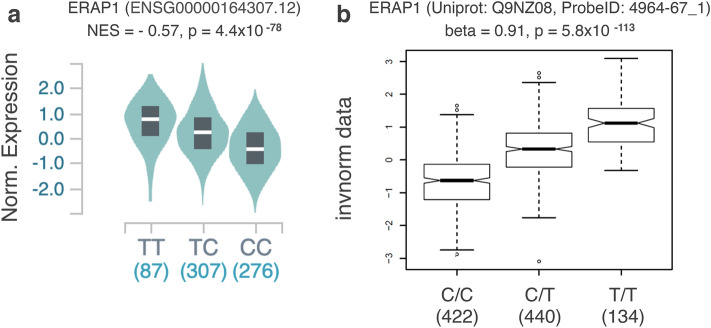
According to GTEx and SNiPa data, rs30187 has a significant effect on both ERAP1 mRNA and protein levels in blood with C/C genotype associated with the lowest expression levels (Fig. 2).
Figure 2.
Effect of ERAP1 rs30187 variant on mRNA (Data source: GTEx) and protein levels (Data source: SNiPA) in blood. ERAP1 rs30187 CC genotype is associated with the lowest expression level. Each T allele additivelly increases ERAP1 mRNA in whole blood cells (a) and Erap1 protein concentration in plasma (b). NES: Normalized effect size; Norm. Expression: normalized expression; invnorm data: inverse-normal scaled data, (b) was kindly provided by Karsten Suhre from SNiPA team.
Given the qualitative and quantitative effect of rs30187 on ERAP1 expression, we hypothesized that women homozygotes for C/C genotype have higher circulating levels of Ang II. In order to test that, plasma Ang II concentrations were determined in an additional cohort of women with severe preeclampsia (n = 36) and normotensive pregnant controls (n = 29). We rejected this hypothesis (p = 0.895) since no difference was detected between genotypic groups (Fig. 3). The intra-group analysis (i.e. cases-only and controls-only) did not detect any difference in Ang II levels across genotypic groups as well (data not shown).
Figure 3.
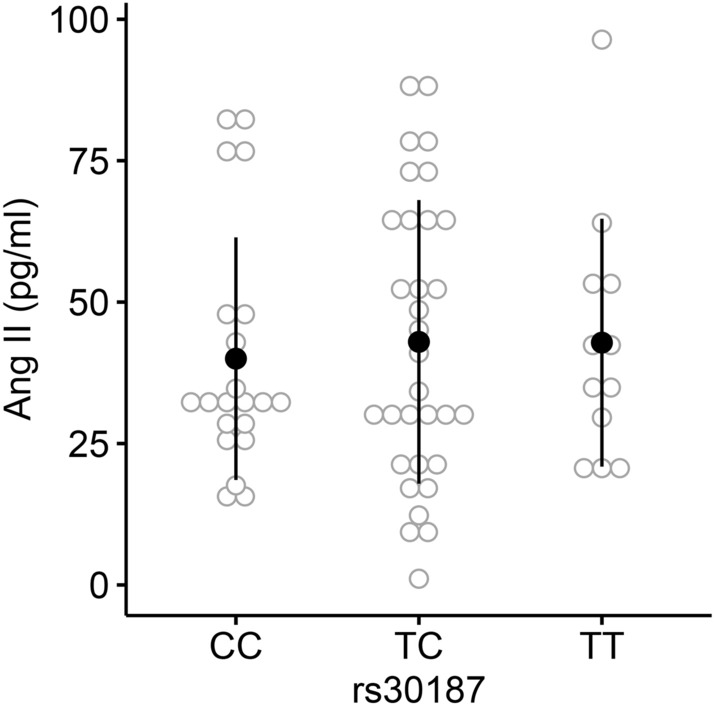
Plasma Ang II levels across rs30187 genotypic groups. Analysis of plasma Ang II concentration in pregnant women (n = 65). No difference was detected among genotypic groups (ANOVA, p = 0.895).
Discussion
Endoplasmic reticulum aminopeptidases (ERAPs), as well as leucyl/cystinyl aminopeptidase (LNPEP), play roles in antigen processing, inflammatory response, blood pressure regulation and angiogenesis, all processes potentially implicated in preeclampsia pathophysiology. The present study confirmed a genetic association between ERAP2 and preeclampsia, and, for the first time, reported an association between ERAP1 and eclampsia. These findings may help to disentangle the intricate association between the correlated phenotypes preeclampsia/eclampsia and the functionally and physically connected genes ERAP1/ERAP2. In addition, our results are consistent with the hypothesis of distinct genetic bases for preeclampsia and eclampsia, reinforcing the importance to separate the two phenotypes when designing genetic association studies.
Johnson et al., tested ERAP1, ERAP2 and LNPEP for associations with preeclampsia in Australian and Norwegian populations, and identified ERAP2 variants (rs2549782, rs2548538, rs2287988 and rs17408150) associated with preeclampsia16. In the same study, rs27044 and rs30187 (ERAP1) were not associated with disease although borderline association with preeclampsia was found for markers rs3734016 and rs34750, both within ERAP1 gene. It is important to highlight that the Australian cohort contained both preeclampsia and eclampsia cases, but they were analyzed as a unique group16, while our study treated the two phenotypes as different entities. A recent study with 148 preeclamptic women and 133 controls from Iran investigated four variants in ERAP1 (including rs30187) and three variants in ERAP2. None of the variants were associated with disease, but a haplotype encompassing the seven variants was associated with preeclampsia33. In another Iranian independent study, ERAP2 variants (rs2549782 and rs17408150) were also associated with preeclampsia34. In our study, the ERAP2 variants rs2549796, rs2927609 and rs11135484 were not associated with preeclampsia in a single marker analysis but the haplotype C–C–G was overrepresented in the preeclampsia group (Table 2). Interestingly, the fetal minor allele for variant rs2549782 (ERAP2) was associated with preeclampsia in African American population35. Besides the genetic association findings, Founds et al. showed ERAP2 was differentially expressed in the first trimester placentas of women who later developed preeclampsia36.
The ERAP1 rs30187 C allele codes an enzyme with Arg528 that causes a reduction on peptidase activity for angiotensin II degradation by approximately 60%, when compared to the enzyme with Lys528, coded by the T allele22,32. We failed to confirm the hypothesis that women carrying two copies of the C allele have increased levels of Ang II in their blood, which in turn could cause blood pressure elevation and seizure. However, we cannot rule out a potential effect of rs30187 on local RAS (e.g. brain and kidney). While Arg528 variant is associated with hypertensive disease37, the Lys528 variant is strongly associated with susceptibility to ankylosing spondylitis38 and other autoimmune diseases17. Since both Arg528 and Lys528 alleles are associated with bad outcomes, it is likely that ERAP genes would be subject to balancing selection, a process where heterozygous individuals are more adaptive than either of the two types of homozygous39. In addition, these genes play key role in the maintenance of immunotolerance to self-peptides as well as protecting against infectious agents, such as HIV40.
The small sample size for some of our case groups represents an important limitation for the present study, even though we should consider that eclampsia and HELLP are extremely rare phenotypes. On the other hand, the marker associated with eclampsia (rs30187) has been well characterized as affecting the protein function, what strengthens the biological plausibility for the genetic association reported here. Furthermore, we accounted for important confounders such as age, parity and ethnicity.
The mechanism by which endoplasmic reticulum aminopeptidases (ERAPs) influence the risk of preeclampsia/eclampsia remains to be elucidated. In addition to Ang II degradation and peptide trimming for antigen presentation via MHC-1, ERAPs also play role in inflammatory response by shedding cytokine receptors (e.g. IL-6R, IL-1R2 and TNFR)41–43. Lastly, ERAP1 plays crucial role in VEGF-stimulated proliferation and migration of endothelial cells, as well as angiogenesis, via the binding and modification of PDk144. All the above-mentioned mechanisms are potentially involved in the causal pathway of preeclampsia/eclampsia.
Conclusions
In conclusion, we identified genetic variants in ERAP1 and ERAP2 associated with eclampsia and preeclampsia, respectively. Sequencing and functional studies are needed in order to elucidate the mechanisms underlying these genetic associations.
Supplementary Information
Acknowledgements
We thank the staff of Maternidade Escola Januário Cicco for their help in ascertaining patients’ clinical classification and sample collection. We do appreciate the collaboration of SNiPA team (Karsten Suhre and Arnold Matthias) that kindly generated rs30187 pQTL plot (Figure 2b) and Victor Lima (on behalf of Laboratório de Análises Clínicas DNA Center) to perform capillary electrophoresis needed to genotype the additional cohort (n = 65) of pregnant women.
Author contributions
L.C.F. worked on the study design, carried out the genotyping and the genetic analysis, and the manuscript writing. C.E.M.G. contributed with the study design and genotyping. P.D. contributed with the genetic analysis and revising the manuscript. I.P.H. and P.R.P.N. carried out sample processing and Ang II measurement. A.S.L. contributed with the study participant recruitment and phenotype ascertainment. S.M.B.J. contributed with the study design, recruitment of subjects and manuscript revision.
Funding
Funding was provided by Conselho Nacional de Desenvolvimento Científico e Tecnológico, Coordenação de Aperfeiçoamento de Pessoal de Nível Superior and Federal University of Rio Grande do Norte (PIBIC-UFRN).
Data availability
The genetic data used in the present study is available from the corresponding author on reasonable request.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-86240-z.
References
- 1.Say L, et al. Global causes of maternal death: a WHO systematic analysis. Lancet Glob. Health. 2014;2:e323–e333. doi: 10.1016/S2214-109X(14)70227-X. [DOI] [PubMed] [Google Scholar]
- 2.Benschop L, Duvekot JJ, van Lennep JER. Future risk of cardiovascular disease risk factors and events in women after a hypertensive disorder of pregnancy. Heart. 2019;105:1273–1278. doi: 10.1136/heartjnl-2018-313453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andraweera PH, Lassi ZS. Cardiovascular risk factors in offspring of preeclamptic pregnancies—systematic review and meta-analysis. J. Pediatr. 2019 doi: 10.1016/j.jpeds.2018.12.008. [DOI] [PubMed] [Google Scholar]
- 4.Duckitt K, Harrington D. Risk factors for pre-eclampsia at antenatal booking: systematic review of controlled studies. BMJ. 2005;330:565. doi: 10.1136/bmj.38380.674340.E0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson MP, et al. Genome-wide association scan identifies a risk locus for preeclampsia on 2q14, near the inhibin Beta B gene. PLoS ONE. 2012;7:e33666. doi: 10.1371/journal.pone.0033666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGinnis R, et al. Variants in the fetal genome near FLT1 are associated with risk of preeclampsia. Nat. Genet. 2017;49:1255–1260. doi: 10.1038/ng.3895. [DOI] [PubMed] [Google Scholar]
- 7.Kathryn JG, et al. Gene-centric analysis of preeclampsia identifies maternal association at PLEKHG1. Hypertension. 2018;72:408–416. doi: 10.1161/HYPERTENSIONAHA.117.10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Staines-Urias E, et al. Genetic association studies in pre-eclampsia: systematic meta-analyses and field synopsis. Int. J. Epidemiol. 2012;41:1764–1775. doi: 10.1093/ije/dys162. [DOI] [PubMed] [Google Scholar]
- 9.Tsujimoto M, Hattori A. The oxytocinase subfamily of M1 aminopeptidases. Biochim. Biophys. Acta. 2005;1751:9–18. doi: 10.1016/j.bbapap.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 10.Saveanu L, et al. IRAP identifies an endosomal compartment required for MHC class I cross-presentation. Science. 2009;325:213–217. doi: 10.1126/science.1172845. [DOI] [PubMed] [Google Scholar]
- 11.Mitsui T, Nomura S, Itakura A, Mizutani S. Role of aminopeptidases in the blood pressure regulation. Biol. Pharm. Bull. 2004;27:768–771. doi: 10.1248/bpb.27.768. [DOI] [PubMed] [Google Scholar]
- 12.Cifaldi L, Romania P, Lorenzi S, Locatelli F, Fruci D. Role of endoplasmic reticulum aminopeptidases in health and disease: from infection to cancer. Int. J. Mol. Sci. 2012;13:8338–8352. doi: 10.3390/ijms13078338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haroon N, Inman RD. Endoplasmic reticulum aminopeptidases: biology and pathogenic potential. Nat. Rev. Rheumatol. 2010;6:461–467. doi: 10.1038/nrrheum.2010.85. [DOI] [PubMed] [Google Scholar]
- 14.Mizutani S, Wright JW, Kobayashi H. Placental leucine aminopeptidase- and aminopeptidase a-deficient mice offer insight concerning the mechanisms underlying preterm labor and preeclampsia. J. Biomed. Biotechnol. 2011;2011:286947. doi: 10.1155/2011/286947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson MP, et al. Identification of two novel quantitative trait loci for pre-eclampsia susceptibility on chromosomes 5q and 13q using a variance components-based linkage approach. Mol. Hum. Reprod. 2006;13:61–67. doi: 10.1093/molehr/gal095. [DOI] [PubMed] [Google Scholar]
- 16.Johnson MP, et al. The ERAP2 gene is associated with preeclampsia in Australian and Norwegian populations. Hum. Genet. 2009;126:655–666. doi: 10.1007/s00439-009-0714-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fierabracci A, Milillo A, Locatelli F, Fruci D. The putative role of endoplasmic reticulum aminopeptidases in autoimmunity: Insights from genomic-wide association studies. Autoimmun. Rev. 2012;12:281–288. doi: 10.1016/j.autrev.2012.04.007. [DOI] [PubMed] [Google Scholar]
- 18.Kim J, et al. Sequence variants in oxytocin pathway genes and preterm birth: a candidate gene association study. BMC Med. Genet. 2013;14:77. doi: 10.1186/1471-2350-14-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.American College of Obstetricians and Gynecologists Hypertension in pregnancy. Report of the American College of Obstetricians and Gynecologists’ task force on hypertension in pregnancy. Obstet. Gynecol. 2013;122:1122–1131. doi: 10.1097/01.AOG.0000437382.03963.88. [DOI] [PubMed] [Google Scholar]
- 20.Joshi D, James A, Quaglia A, Westbrook RH, Heneghan MA. Liver disease in pregnancy. Lancet. 2010;375:594–605. doi: 10.1016/S0140-6736(09)61495-1. [DOI] [PubMed] [Google Scholar]
- 21.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 22.Kochan G, et al. Crystal structures of the endoplasmic reticulum aminopeptidase-1 (ERAP1) reveal the molecular basis for N-terminal peptide trimming. Proc. Natl. Acad. Sci. 2011;108:7745–7750. doi: 10.1073/pnas.1101262108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nguyen TT, et al. Structural basis for antigenic peptide precursor processing by the endoplasmic reticulum aminopeptidase ERAP1. Nat. Struct. Mol. Biol. 2011;18:604–613. doi: 10.1038/nsmb.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grimberg J, et al. A simple and efficient non-organic procedure for the isolation of genomic DNA from blood. Nucl. Acids Res. 1989;17:8390. doi: 10.1093/nar/17.20.8390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lins TCL, et al. A multiplex single-base extension protocol for genotyping Cdx2, FokI, BsmI, ApaI, and TaqI polymorphisms of the vitamin D receptor gene. Genet. Mol. Res. GMR. 2007;6:316–324. [PubMed] [Google Scholar]
- 26.Lins TC, Vieira RG, Abreu BS, Grattapaglia D, Pereira RW. Genetic composition of Brazilian population samples based on a set of twenty-eight ancestry informative SNPs. Am. J. Hum. Biol. NA-NA. 2009 doi: 10.1002/ajhb.20976. [DOI] [PubMed] [Google Scholar]
- 27.Zheng X, et al. A high-performance computing toolset for relatedness and principal component analysis of SNP data. Bioinforma Oxf. Engl. 2012;28:3326–3328. doi: 10.1093/bioinformatics/bts606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.GTEx Consortium The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suhre K, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat. Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goto Y, Hattori A, Ishii Y, Tsujimoto M. Reduced activity of the hypertension-associated Lys528Arg mutant of human adipocyte-derived leucine aminopeptidase (A-LAP)/ER-aminopeptidase-1. FEBS Lett. 2006;580:1833–1838. doi: 10.1016/j.febslet.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 33.Dargahi H, et al. Association study of single nucleotide polymorphisms of endoplasmic reticulum aminopeptidase 1 and 2 genes in Iranian women with preeclampsia. Iran. J. Public Health. 2019;48:531–540. [PMC free article] [PubMed] [Google Scholar]
- 34.Soltani S, Nasiri M. Association of ERAP2 gene variants with risk of pre-eclampsia among Iranian women. Int. J. Gynecol. Obstet. 2019;145:337–342. doi: 10.1002/ijgo.12816. [DOI] [PubMed] [Google Scholar]
- 35.Hill LD, et al. Fetal ERAP2 variation is associated with preeclampsia in African Americans in a case-control study. BMC Med. Genet. 2011;12:64. doi: 10.1186/1471-2350-12-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Founds SA, et al. Altered global gene expression in first trimester placentas of women destined to develop preeclampsia. Placenta. 2009;30:15–24. doi: 10.1016/j.placenta.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamamoto N, et al. Identification of 33 polymorphisms in the adipocyte-derived leucine aminopeptidase (ALAP) gene and possible association with hypertension. Hum. Mutat. 2002;19:251–257. doi: 10.1002/humu.10047. [DOI] [PubMed] [Google Scholar]
- 38.Evans DM, et al. Interaction between ERAP1 and HLA-B27 in ankylosing spondylitis implicates peptide handling in the mechanism for HLA-B27 in disease susceptibility. Nat. Genet. 2011;43:761–767. doi: 10.1038/ng.873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andrés AM, et al. Balancing selection maintains a form of ERAP2 that undergoes nonsense-mediated decay and affects antigen presentation. PLoS Genet. 2010;6:e1001157. doi: 10.1371/journal.pgen.1001157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cagliani R, et al. Genetic diversity at endoplasmic reticulum aminopeptidases is maintained by balancing selection and is associated with natural resistance to HIV-1 infection. Hum. Mol. Genet. 2010;19:4705–4714. doi: 10.1093/hmg/ddq401. [DOI] [PubMed] [Google Scholar]
- 41.Cui X. An aminopeptidase, ARTS-1, is required for interleukin-6 receptor shedding. J. Biol. Chem. 2003;278:28677–28685. doi: 10.1074/jbc.M300456200. [DOI] [PubMed] [Google Scholar]
- 42.Cui X, Rouhani FN, Hawari F, Levine SJ. Shedding of the type II IL-1 decoy receptor requires a multifunctional aminopeptidase, aminopeptidase regulator of TNF receptor type 1 shedding. J. Immunol. 2003;171:6814–6819. doi: 10.4049/jimmunol.171.12.6814. [DOI] [PubMed] [Google Scholar]
- 43.Cui X, et al. Identification of ARTS-1 as a novel TNFR1-binding protein that promotes TNFR1 ectodomain shedding. J. Clin. Invest. 2002;110:515–526. doi: 10.1172/JCI0213847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yamazaki T. Puromycin-insensitive leucyl-specific aminopeptidase (PILSAP) binds and catalyzes PDK1, allowing VEGF-stimulated activation of S6K for endothelial cell proliferation and angiogenesis. Blood. 2004;104:2345–2352. doi: 10.1182/blood-2003-12-4260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genetic data used in the present study is available from the corresponding author on reasonable request.