Introduction
Endothelins are peptide ligands belonging to the family of 21-amino acid isopeptides. They get primarily secreted by the endothelium and are crucially involved in vasomodulation. There are three types of endothelins, i.e., EDN1 (Endothelin 1), EDN2 (Endothelin 2), and EDN3 (Endothelin 3) encoded by chromosomes 6, 1 and, 20, which are produced by sequential cleavage of the respective endothelin precursors. The endothelins are secreted as an inactive pre-pro-endothelins mainly in vascular endothelial cells, which then undergoes subsequent proteolytic cleavage to produce pro-endothelins. The proteolysis of pro-endothelins into big-endothelins, which in the case of big-EDN1, is carried out by furin-like proteases (Barton and Yanagisawa 2008). The conversion of the inactive precursor peptide, i.e., big-endothelin to the active endothelin, is carried out by endothelin converting enzymes (ECEs) or secreted soluble endopeptidases (Barton et al. 2003). Among the endothelins, EDN1 is the most abundant, primarily secreted by the endothelial cells and other cell types including, airway epithelial cells, macrophages, fibroblasts cardiomyocytes, neurons, and vascular smooth muscle cells (Hynynen and Khalil 2006, Sugo et al. 2001). EDN2 is secreted by ovary, intestinal epithelial cells, whereas EDN3 is expressed in endothelial cells, brain neurons, renal tubular epithelial cells and intestinal epithelial cells (Kedzierski and Yanagisawa 2001, Matsumoto et al. 1989). The endothelins are known to interact with endothelin receptors, i.e., endothelin receptor A (EDNRA) and endothelin receptor B (EDNRB), both of which belong to the family of G protein-coupled receptors (GPCRs). EDNRA shows the highest affinity to EDN1 and EDN2 more than that of EDN3, whereas EDNRB has equal affinities for all endothelin peptides (Barton and Yanagisawa 2008). The distribution pattern of endothelins varies extensively in different organs, which predict their role in both physiological and pathophysiological functions. The endothelin-mediated signaling is reported to differ mainly in normal and pathological conditions (Miyauchi and Goto 1999). The EDNRA receptor is known to mediate cellular proliferation, growth, vasoconstriction and inflammation, resulting in adverse vascular effects. In contrast, the EDNRB receptor is involved in vasodilation and inhibition of growth and inflammation (Luscher and Barton 2000). The opposing roles of ENDRA and EDNRB reiterate their importance in normal and pathophysiological conditions. Thereby it necessitates a better understanding of the cellular response to endothelin receptor signaling.
There is vast literature available on information related to the molecular cascades involved in endothelin-mediated signalling however, the information is scattered throughout the literature. In this regard, we have attempted to bring all the information related to the endothelin mediated signaling pathway together in a comprehensive manner. The information on molecular reactions was categorized into six different types based on NetPath criteria, based on which we have developed an extensive map of the endothelin signaling pathway. Previously, we published pathways including IL-18, serotonin, IL-33, corticotrophin, oncostatin M signaling (Rex et al. 2020, Pinto et al. 2018, Verma et al. 2016, Subbannayya et al. 2013, Dey et al. 2013). Endothelin mediated signaling pathway is made accessible through the WikiPathways Database (https://www.wikipathways.org/index.php/Pathway).
Methodology
An extensive literature search was carried out on PubMed using an appropriate keywords and analysis was performed over the reports hitherto published the downstream reactions of all endothelin ligands binding to the endothelin receptors. Articles were fetched using the following search terms “endothelin receptor type B “ OR “EDNRB” OR “ABCDS” OR “ET-B” OR “ET-BR” OR “ETB” OR “ETB1” OR “ETBR” OR “ETRB” OR “HSCR” OR “HSCR2” OR “WS4A” OR “endothelin receptor type A” OR “EDNRA” OR “ET-A” OR “ETA” OR “ETA-R” OR “ETAR” OR “ETRA” OR “MFDA” OR “hET-AR” OR “endothelin-1″ “ET-1” OR “endothelin-2” OR “ET-2” OR “ET-3” OR “endothelin-3″) AND (“pathway” OR “signaling” OR “signalling”. A systematic manual screening of research articles was done to organize the endothelin ligand-receptor downstream signaling reactions comprehensively. Only the original articles, except reviews and short communications, were considered for annotation according to the principles described in the previously published NetPath annotation criteria (Kandasamy et al. 2010). The articles were manually annotated for downstream signaling reactions of endothelin ligand-receptor binding into the following categories; 1) post-translational modification (PTM), 2) activation/inhibition, 3) gene regulation at the mRNA level (both up- and down-regulation), 4) induced protein expression, 5) molecular association (protein-protein interactions), 6) translocation/transport of proteins between subcellular compartments. The annotation principles were strictly followed as per the criteria described earlier in NetPath (Kandasamy et al. 2010). The information regarding the details of the type of cell lines/tissue/animals used, PTM residue and site, gene regulation type (upregulation/downregulation), along with the experiment type, were also curated during the annotation. The annotated reactions were utilized for mapping the endothelin pathway using the PathVisio tool (Version 3.2.2). The annotated reactions and pathway map were further extended for an external review by a pathway authority, who is working in the field of endothelin pathway. All the suggestions from the pathway authority were incorporated in the signaling pathway map. Individual reactions annotated in the endothelin signaling pathway were hyperlinked to the respective original PubMed article, from which the reactions have been acquired.
Results and discussion
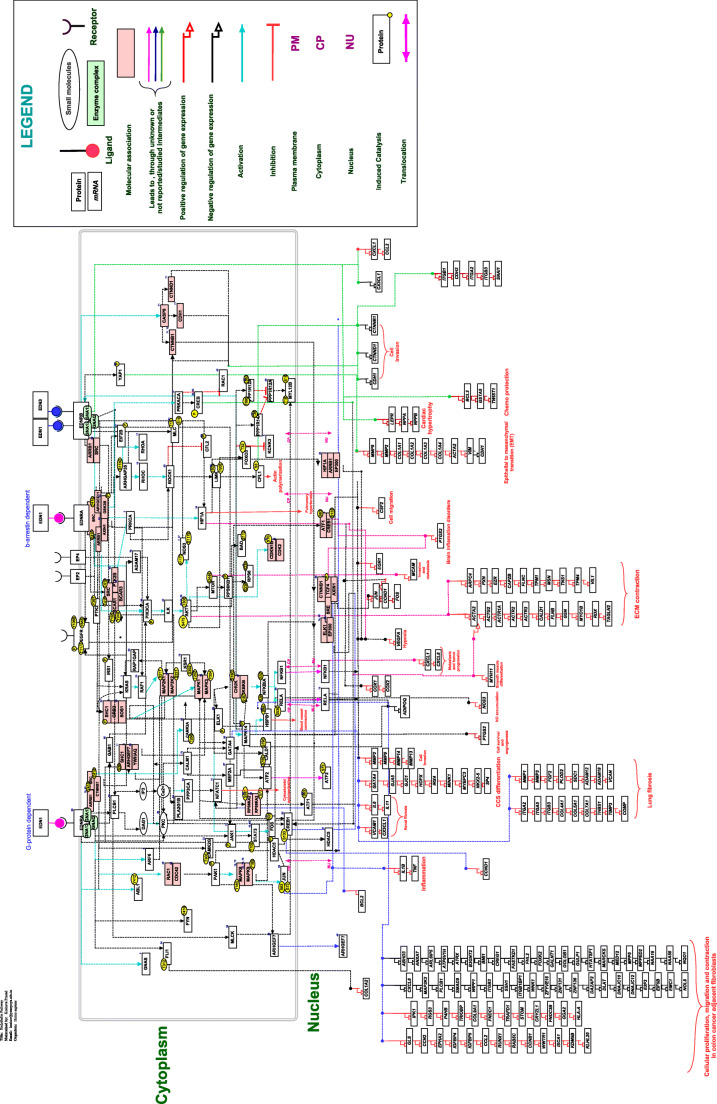
In the present study, the search term retrieved 2800 papers in Pubmed, which were used for initial screening. Of these, 640 research articles were reviewed for endothelin-mediated reactions, and out of these, 332 articles had information that was compatible with endothelin-mediated signaling and was selected for further curation. The manual curation of selected articles documented 87 activation/inhibition events,102 PTM’s or catalysis events, 54 molecular associations, 262 gene regulation events, 150 types of protein expression, and 29 protein translocation events. We reported PTMs, including phosphorylation and dephosphorylation. We could not annotate the site and residue of PTM for 26 and 16 molecules, respectively, due to the non-availability of such experimental data in the literature. A total of 262 differential gene regulation events were catalogued, in different mammalian systems, and of these, 198 and 64 genes were found to be upregulated and downregulated, respectively. These molecular reactions were represented in the signaling map of endothelin (Fig. 1). The pathway datasheet describes the molecular reactions involved in endothelin ligand and receptor-mediated signaling (Supplementary material). We submitted the complete pathway map of the endothelin-mediated signaling pathway to the WikiPathways database with the ID WP4857. It can be freely accessible to the scientific community by using following the URL:https://www.wikipathways.org/index.php/Pathway:WP4857.
Fig. 1.
Schematic representation of endothelin mediated signaling pathway. Schematic representation of endothelin mediated signaling reactions. The signaling pathway map represents molecules involved in ligand-receptor interactions of endothelin activated downstream molecular events including molecular association, enzyme catalysis, translocation, and gene regulation events. Information regarding the post-translational modification site and the residue is also shown in the pathway.
A summary of endothelin signaling pathway
The events mediated by endothelin ligand-receptor interaction are involved in many physiological as well as pathophysiological conditions such as vasoconstriction and dilation, cardiovascular remodelling, cell proliferation, cell differentiation, extracellular matrix production, and control of water and sodium secretion (Unic et al. 2011). The endothelin signaling pathway depicts the activation of EDNRA to be dependent on either G protein or β-arrestin. The binding of endothelin ligands to EDNRA and EDNRB receptor induces the activation of G-proteins- including G protein subunit alpha Q (GNAQ), G protein subunit alpha 12 and 13 (GNA12 and GNA13). Overexpression of EDN1 and subsequent stimulation of EDNRA induces the upregulation of genes involved in the repair of the extracellular matrix (ECM) of normal lung fibroblasts (Shi-Wen et al. 2004). In human airway smooth muscle cells, EDN1 induces hypertrophy and anti-apoptosis by stimulating both EDNRA and EDNRB through the activation of JAK1/STAT-3 and MAPK1/3 cell signaling pathways (McWhinnie et al. 2007). Its stimulation also induces the over expression of cytokines such as IL-6 and IL-11in an EDNRA-dependent manner, via the activation of map kinases, regulating proliferation and fibrosis in human lung fibroblasts (Gallelli et al. 2005). Endothelin also triggers cardiac hypertrophy through the activation of PLC, PKC, and MAP kinases via the activation of both EDNRA and EDNRB receptors (Li et al. 2014, Takahashi et al. 2005, Hautala et al. 2002). Similarly, the role of EDN1 is also implicated in renal interstitial fibrosis by induction of the inflammatory NF-kappaB signaling pathway (Gerstung et al. 2007). Activation of EDNRB by EDN1 in human monocytes triggers the cytokine storm which is critical for the development and maintenance of several inflammatory diseases (Juergens et al. 2008). Epithelial to mesenchymal transition is involved in tubulointerstitial fibrosis via Rho-kinase and YAP pathways (Seccia et al. 2016).
In most of the human cancers, endothelin activates MAPK, NF-kB, β-catenin, PI3K/AKT, and Rho GTPase pathways, regulating the expression of genesessential for cell survival, proliferation, drug resistance, angiogenesis, osteogenesis, immune modulation, invasion and metastasis (Johnson et al. 2017, Seccia et al. 2016, Tanja et al. 2015, Knowles et al. 2012, Nelson et al. 2005).Activation of EDNRA by EDN1 induces MAPK activation, PGE2 and VEGF secretion via Src- mediated EGFR transactivation which are responsible for ovarian cancer cell proliferation, survival and invasiveness (Rosano et al. 2007, Spinella et al. 2004). Our attempt to map the endothelin ligand-receptor pathway helps to consolidate the various signaling events that mediate cellular response. The pathway can be a resource for cellular network information to understand the activities involved in pathological responses of endothelin signaling.
Conclusions
The endothelin signaling map made available in the WikiPathway resource can be helpful for the scientific community to understand the association of various proteins in both physiological and pathophysiological conditions by overlaying the data on gene expression dynamics obtained from various experimental as well as clinical conditions. We believe that the updated endothelin pathway map will help the researchers to understand the role of endothelin signaling in disease progression that will assist them to design the better near future therapeutics.
Electronic supplementary material
(XLSX 383 kb)
Acknowledgments
We thank Karnataka Biotechnology and Information Technology Services (KBITS), Government of Karnataka, for the support to Center for Systems Biology and Molecular Medicine at Yenepoya (Deemed to be University) under the Biotechnology Skill Enhancement Programme in Multiomics Technology (BiSEP GO ITD 02 MDA 2017). RDAB is a recipient of the Senior Research Fellowship from the Indian Council of Medical Research (ICMR), Government of India. Varshasnata Mohanty is a recipient of the Women Scientist-A award from the Department of Science & Technology (DST), Government of India.
Abbreviations
- EDN1
Endothelin 1
- EDN2
Endothelin 2
- EDN3
Endothelin 3
- EDNRA
Endothelin Receptor A
- EDNRB
Endothelin Receptor B
- GPCRs
G protein-coupled receptors
- PPI
Protein-protein interaction
Compliance with ethical standards
Conflict of interest
The authors report no conflicts of interest.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Shobha Dagamajalu and D.A.B. Rex contributed equally to this work.
Contributor Information
Shobha Dagamajalu, Email: shobha_d@yenepoya.edu.in.
D.A.B. Rex, Email: rexprem@yenepoya.edu.in
Lathika Gopalakrishnan, Email: lathika@ibioinformatics.org.
Gayathree Karthikkeyan, Email: gayathreek@yenepoya.edu.in.
Sumrati Gurtoo, Email: sumrati@yenepoya.edu.in.
Prashant Kumar Modi, Email: prashantmodi@yenepoya.edu.in.
Varshasnata Mohanty, Email: varsham@yenepoya.edu.in.
M. Mujeeburahiman, Email: mujeeburahiman@gmail.com
Sowmya Soman, Email: sowmya.soman@gmail.com.
Rajesh Raju, Email: rajrrnbt@gmail.com.
Vinod Tiwari, Email: vtiwari.phe@itbhu.ac.in.
T.S. Keshava Prasad, Email: keshav@yenepoya.edu.in.
References
- Barton M, Traupe T, Haudenschild CC. Endothelin, hypercholesterolemia and atherosclerosis. Coron Artery Dis. 2003;14:477–490. doi: 10.1097/00019501-200311000-00002. [DOI] [PubMed] [Google Scholar]
- Barton M, Yanagisawa M. Endothelin: 20 years from discovery to therapy. Can J Physiol Pharmacol. 2008;86:485–498. doi: 10.1139/Y08-059. [DOI] [PubMed] [Google Scholar]
- Dey G, Radhakrishnan A, Syed N, Thomas JK, Nadig A, Srikumar K, Mathur PP, Pandey A, Lin SK, Raju R, Prasad TS. Signaling network of Oncostatin M pathway. J Cell Commun Signal. 2013;7:103–108. doi: 10.1007/s12079-012-0186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallelli L, Pelaia G, D’agostino B, Cuda G, Vatrella A, Fratto D, Gioffre V, Galderisi U, De Nardo M, Mastruzzo C, Salinaro ET, Maniscalco M, Sofia M, Crimi N, Rossi F, Caputi M, Costanzo FS, Maselli R, Marsico SA, Vancheri C. Endothelin-1 induces proliferation of human lung fibroblasts and IL-11 secretion through an ET(A) receptor-dependent activation of MAP kinases. J Cell Biochem. 2005;96:858–868. doi: 10.1002/jcb.20608. [DOI] [PubMed] [Google Scholar]
- Gerstung M, Roth T, Dienes HP, Licht C, Fries JW. Endothelin-1 induces NF-kappaB via two independent pathways in human renal tubular epithelial cells. Am J Nephrol. 2007;27:294–300. doi: 10.1159/000101999. [DOI] [PubMed] [Google Scholar]
- Hautala N, Tenhunen O, Szokodi I, Ruskoaho H. Direct left ventricular wall stretch activates GATA4 binding in perfused rat heart: involvement of autocrine/paracrine pathways. Pflugers Arch. 2002;443:362–369. doi: 10.1007/s004240100699. [DOI] [PubMed] [Google Scholar]
- Hynynen MM, Khalil RA. The vascular endothelin system in hypertension--recent patents and discoveries. Recent Pat Cardiovasc Drug Discov. 2006;1:95–108. doi: 10.2174/157489006775244263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson MG, Konicke K, Kristianto J, Gustavson A, Garbo R, Wang X, Yuan B, Blank RD. Endothelin signaling regulates mineralization and posttranscriptionally regulates SOST in TMOb cells via miR 126-3p. Physiol Rep. 2017;5:e13088. doi: 10.14814/phy2.13088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juergens UR, Racke K, Uen S, Haag S, Lamyel F, Stober M, Gillissen A, Novak N, Vetter H. Inflammatory responses after endothelin B (ETB) receptor activation in human monocytes: new evidence for beneficial anti-inflammatory potency of ETB-receptor antagonism. Pulm Pharmacol Ther. 2008;21:533–539. doi: 10.1016/j.pupt.2007.12.005. [DOI] [PubMed] [Google Scholar]
- Kandasamy K, Mohan SS, Raju R, Keerthikumar S, Kumar GS, Venugopal AK, Telikicherla D, Navarro JD, Mathivanan S, Pecquet C, Gollapudi SK, Tattikota SG, Mohan S, Padhukasahasram H, Subbannayya Y, Goel R, Jacob HK, Zhong J, Sekhar R, Nanjappa V, Balakrishnan L, Subbaiah R, Ramachandra YL, Rahiman BA, Prasad TS, Lin JX, Houtman JC, Desiderio S, Renauld JC, Constantinescu SN, Ohara O, Hirano T, Kubo M, Singh S, Khatri P, Draghici S, Bader GD, Sander C, Leonard WJ, Pandey A. NetPath: a public resource of curated signal transduction pathways. Genome Biol. 2010;11:R3. doi: 10.1186/gb-2010-11-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedzierski RM, Yanagisawa M. Endothelin system: the double-edged sword in health and disease. Annu Rev Pharmacol Toxicol. 2001;41:851–876. doi: 10.1146/annurev.pharmtox.41.1.851. [DOI] [PubMed] [Google Scholar]
- Knowles JP, Shi-Wen X, Haque SU, Bhalla A, Dashwood MR, Yang S, Taylor I, Winslet MC, Abraham DJ, Loizidou M. Endothelin-1 stimulates colon cancer adjacent fibroblasts. Int J Cancer. 2012;130:1264–1272. doi: 10.1002/ijc.26090. [DOI] [PubMed] [Google Scholar]
- Li H, Gao S, Ye J, Feng X, Cai Y, Liu Z, Lu J, Li Q, Huang X, Chen S, Liu P. COX-2 is involved in ET-1-induced hypertrophy of neonatal rat cardiomyocytes: role of NFATc3. Mol Cell Endocrinol. 2014;382:998–1006. doi: 10.1016/j.mce.2013.11.012. [DOI] [PubMed] [Google Scholar]
- Luscher TF, Barton M. Endothelins and endothelin receptor antagonists: therapeutic considerations for a novel class of cardiovascular drugs. Circulation. 2000;102:2434–2440. doi: 10.1161/01.CIR.102.19.2434. [DOI] [PubMed] [Google Scholar]
- Matsumoto H, Suzuki N, Onda H, Fujino M. Abundance of endothelin-3 in rat intestine, pituitary gland and brain. Biochem Biophys Res Commun. 1989;164:74–80. doi: 10.1016/0006-291X(89)91684-7. [DOI] [PubMed] [Google Scholar]
- Mcwhinnie R, Pechkovsky DV, Zhou D, Lane D, Halayko AJ, Knight DA, Bai TR. Endothelin-1 induces hypertrophy and inhibits apoptosis in human airway smooth muscle cells. Am J Physiol Lung Cell Mol Physiol. 2007;292:L278–L286. doi: 10.1152/ajplung.00111.2006. [DOI] [PubMed] [Google Scholar]
- Miyauchi T, Goto K. Heart failure and endothelin receptor antagonists. Trends Pharmacol Sci. 1999;20:210–217. doi: 10.1016/S0165-6147(99)01297-3. [DOI] [PubMed] [Google Scholar]
- Nelson JB, Udan MS, Guruli G, Pflug BR. Endothelin-1 inhibits apoptosis in prostate cancer. Neoplasia. 2005;7:631–637. doi: 10.1593/neo.04787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto SM, Subbannayya Y, Rex DAB, Raju R, Chatterjee O, Advani J, Radhakrishnan A, Keshava Prasad TS, Wani MR, Pandey A. A network map of IL-33 signaling pathway. J Cell Commun Signal. 2018;12:615–624. doi: 10.1007/s12079-018-0464-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rex DAB, Agarwal N, Prasad TSK, Kandasamy RK, Subbannayya Y, Pinto SM (2020) A comprehensive pathway map of IL-18-mediated signalling. J Cell Commun Signal 14:257–266 [DOI] [PMC free article] [PubMed]
- Rosano L, Di Castro V, Spinella F, Tortora G, Nicotra MR, Natali PG, Bagnato A. Combined targeting of endothelin A receptor and epidermal growth factor receptor in ovarian cancer shows enhanced antitumor activity. Cancer Res. 2007;67:6351–6359. doi: 10.1158/0008-5472.CAN-07-0883. [DOI] [PubMed] [Google Scholar]
- Seccia TM, Caroccia B, Gioco F, Piazza M, Buccella V, Guidolin D, Guerzoni E, Montini B, Petrelli L, Pagnin E, Ravarotto V, Belloni AS, Calo LA, Rossi GP (2016) Endothelin-1 drives epithelial-Mesenchymal transition in hypertensive Nephroangiosclerosis. J Am Heart Assoc 5 [DOI] [PMC free article] [PubMed]
- Shi-Wen X, Chen Y, Denton CP, Eastwood M, Renzoni EA, Bou-Gharios G, Pearson JD, Dashwood M, Du Bois RM, Black CM, Leask A, Abraham DJ. Endothelin-1 promotes myofibroblast induction through the ETA receptor via a rac/phosphoinositide 3-kinase/Akt-dependent pathway and is essential for the enhanced contractile phenotype of fibrotic fibroblasts. Mol Biol Cell. 2004;15:2707–2719. doi: 10.1091/mbc.e03-12-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinella F, Rosano L, Di Castro V, Natali PG, Bagnato A. Endothelin-1-induced prostaglandin E2-EP2, EP4 signaling regulates vascular endothelial growth factor production and ovarian carcinoma cell invasion. J Biol Chem. 2004;279:46700–46705. doi: 10.1074/jbc.M408584200. [DOI] [PubMed] [Google Scholar]
- Subbannayya T, Balakrishnan L, Sudarshan G, Advani J, Kumar S, Mahmood R, Nair B, Sirdeshmukh R, Mukherjee KK, Umathe SN, Raju R, Prasad TS. An integrated map of corticotropin-releasing hormone signaling pathway. J Cell Commun Signal. 2013;7:295–300. doi: 10.1007/s12079-013-0197-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugo S, Minamino N, Shoji H, Isumi Y, Nakao K, Kangawa K, Matsuo H. Regulation of endothelin-1 production in cultured rat vascular smooth muscle cells. J Cardiovasc Pharmacol. 2001;37:25–40. doi: 10.1097/00005344-200101000-00004. [DOI] [PubMed] [Google Scholar]
- Takahashi H, Takeishi Y, Seidler T, Arimoto T, Akiyama H, Hozumi Y, Koyama Y, Shishido T, Tsunoda Y, Niizeki T, Nozaki N, Abe J, Hasenfuss G, Goto K, Kubota I. Adenovirus-mediated overexpression of diacylglycerol kinase-zeta inhibits endothelin-1-induced cardiomyocyte hypertrophy. Circulation. 2005;111:1510–1516. doi: 10.1161/01.CIR.0000159339.00703.22. [DOI] [PubMed] [Google Scholar]
- Tanja TT, Outi N, Sakari S, Jarmo L, Kaisa P, Leila K. Preliminary Finnish measures of eating competence suggest association with health-promoting eating patterns and related psychobehavioral factors in 10–17 year old adolescents. Nutrients. 2015;7:3828–3846. doi: 10.3390/nu7053828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unic A, Derek L, Hodak N, Ceprnja M, Serdar T, Krhac M, Romic Z (2011) Endothelins -- clinical perspectives. Biochem Med (Zagreb) 21(3):231–242 [DOI] [PubMed]
- Verma R, Balakrishnan L, Sharma K, Khan AA, Advani J, Gowda H, Tripathy SP, Suar M, Pandey A, Gandotra S, Prasad TS, Shankar S. A network map of Interleukin-10 signaling pathway. J Cell Commun Signal. 2016;10:61–67. doi: 10.1007/s12079-015-0302-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX 383 kb)