Abstract
Glutamine (gln) metabolism has emerged as a cancer therapeutic target in past few years, however, the effect of gln-deprivation of bCSCs remains elusive in breast cancer. In this study, effect of glutamine on stemness and differentiation potential of bCSCs isolated from MCF-7 and MDAMB-231 were studied. We have shown that bCSCs differentiate into CD24+ epithelial population under gln-deprivation and demonstrated increased expression of epithelial markers such as e-cadherin, claudin-1 and decreased expression of mesenchymal protein n-cadherin. MCF-7-bCSCs showed a decrease in EpCAMhigh population whereas MDAMB-231-bCSCs increased CD44high population in response to gln-deprivation. The expression of intracellular stem cell markers such sox-2, oct-4 and nanog showed a drastic decrease in gene expression under gln-deprived MDAMB-231-bCSCs. Finally, localization of β-catenin in MCF-7 and MDAMB-231 cells showed its accumulation in cytosol or perinuclear space reducing its efficiency to transcribe downstream genes. Conclusively, our study demonstrated that gln-deprivation induces differentiation of bCSCs into epithelial subtypes and also reduces stemness of bCSCs mediated by reduced nuclear localization of β-catenin. It also suggests that basal and luminal bCSCs respond differentially towards changes in extracellular and intracellular gln. This study could significantly affect the gln targeting regimen of breast cancer therapeutics.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12079-020-00603-1.
Keywords: Breast cancer stem cells, Glutamine, 2‐Diazo‐norleucine (DON), Epithelial mesenchymal transition, β-Catenin
Introduction
Tumor microenvironment plays an important role in the growth and development of cancer. It involves various components such as cells present in the periphery of cancer cells, pH, extracellular matrix and nutrients. It is well established that cancer cells have a high uptake of various nutrients such as glucose, amino acid (essential/conditionally essential) to meet their biosynthetic needs and proliferation (Lyssiotis and Kimmelman 2017). Glutamine is a conditionally essential amino acid involved in various metabolic processes. It serves as an important source of nitrogen for anabolic reactions, acts as a carbon donor in TCA cycle and is an important precursor for nucleotide biosynthesis. Despite of being conditionally essential its uptake is increased in tumors of different tissue origins (Altman et al. 2016). Requirement for glutamine varies among the type of cancer and also the subtype, tissue of origin etc. Breast cancer is a heterogeneous disease with different subtypes which may vary in their metabolic requirements (Demas et al. 2019; El Ansari et al. 2018) e.g. luminal subtype is reported to grow independent of glutamine (Gln) while the basal subtype grow is dependent on gln due to absence of lineage specific expression of glutamine synthetase in later (Kung et al. 2011). Anti-metabolite drugs have served as useful anticancer tools in mitigation of the disease. Number of genomic mutation such as p53, kras, c-myc in various cancers have shown toxicity with either intracellular or extracellular gln-deprivation (Choi and Park 2018; Jariyal et al. 2019). Diazo-O-norleucine (DON) was the earliest glutaminase inhibitor due to its analogy with glutamine (Seltzer et al. 2010; Shapiro et al. 1991; Unger et al. 2005). Subsequently, number of glutamine metabolism inhibitors was designed in past year and few of them such as BPTES, CB-839are in clinical trials for their safety and efficacy evaluation (Song et al. 2018; Wu et al. 2018; Xu et al. 2018). CB-839 is a glutaminase inhibitor which is also reported to show anti-proliferative effect in triple-negative breast cancer cells. Earlier studies have also shown that gln is important for the survival and differentiation of hematopoietic cells to erythroid lineage (Oburoglu et al. 2014). Glutamine is reported to directly prevent the degradation of oct4 by protecting the oxidation of cysteine residue which is essential for its stability (Marsboom et al. 2016). However the role of gln in cancer stem cell studies is still controversial. Few reports suggesting that in the core of tumors (CSCs niche), the low concentrations of gln are responsible for maintaining stemness by inducing hypermethylation leading to dedifferentiation of tumor cells to CSCs (Pan et al. 2016) while other study reports that although gln is not essential for growth of murine embryonic stem cells but its uptake is required for maintaining high levels of α-KG needed for demethylation and maintaining pluripotency (Carey et al. 2015). A recent study with lung and pancreatic cell lines showed that gln is required for the maintenance of redox balance, the gln-deprivation leads to an imbalance in redox state of the cell which further downregulates the β-catenin pathway and therefore decrease the side population which were depicted CSCs-like population (Liao et al. 2017). Similarly in hepatocellular carcinoma the inhibition of main catabolic enzyme of glutamine i.e. glutaminase1 leads to decrease in stemness following increased ROS accumulation and inhibition of wnt/β-catenin pathway (Li et al. 2019).The increased ROS levels due to gln-deprivation are reported to sensitize pancreatic ductal carcinoma stem cells towards radiotherapy (Li et al. 2015). In colorectal cancer, the gln-deprivation is reported to sensitize the metformin resistant CSCs. The treatment with glutaminase C inhibitor (Compound 968) and metformin sensitization of the cells have demonstrated high expression of gln transporters with CSC properties (Kim et al. 2018). Gln is also required for the maintenance of redox balance in breast cancer when cells are grown in non-adherent conditions i.e. to avoid detachment induced cell death known as anoikis. Similarly, the CSCs are also found to be anoikis resistant as they are enriched in tumorsphere assay in the low/non-adherent conditions. Recently, Stanley et al. have shown that glutamine-deprivation leads to decrease in the ganglioside (GD2+) bCSCs population in MDAMB-231 and SUM159 cells (Ly et al. 2020). Further, the CSCs in breast cancer are identified with CD24−/low/CD44high/+/CD326high/+ expression on the surface. However, how glutamine-deprivation affects the proliferation and differentiation capacity of bCSCs population still needs to be evaluated. In this study, we have aimed to understand the effect of gln-deprivation in bCSCs (CD24−/low/CD44high/+/CD326high/+ population) isolated from MCF-7 and MDAMB-231 cells. We have also used DON as an inhibitor of glutaminase enzyme which inhibits the conversion of gln to glutamate therefore, depriving intracellular gln. We found that gln-deprivation differentially affects the bCSCs population isolated from MCF-7 and MDAMB-231 cells. We have also checked the changes in intracellular stem cell markers and demonstrated that the degradation of β-catenin may be involved in reducing the stem cell population.
Materials and methods
Cell culture
MDA-MB-231 and MCF-7 human breast cancer cell line were obtained from American Type Culture Collection (ATCC). MCF 7 were grown in Dulbecco’s Modified Eagle Medium (DMEM, Life Technology, USA) while MDA-MB-231 were cultured in Leibovitz’s L15 medium (Life Technology, USA) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Life technology, USA), 2 mM glutamine, 1% penicillin and streptomycin. Cells were cultured in tissue culture flasks (Falcon, USA) and were kept in CO2 incubator at 37 °C in a humidified atmosphere with 5% CO2. MDA-MB-231 cells were cultured at 37 °C under humid environment in a carbon dioxide free incubator. We have performed most of experiments for 72 h, however to check the long term effect of gln deprivation on breast cancer cell lines we have consider 8 days time point where we were able to observe morphological changes and surface expression changes prominently. The proliferation assay was also performed for 10 days and data of alternate days was represented.
Proliferation assay
Cells were harvested and seeded at a density of 5000 cells/well in a 24-well culture plate for 10 days. After 24 h, cells were treated with complete glutamine (2 mM) and glutamine-deprived media (0 mM). Cell proliferation was measured using Alamar blue (10 µg/ml) and cells were maintained at 37 °C with 5% CO2 for 4 h to detect its metabolic activity. Fluorescence intensity was measured using microplate reader with excitation/emission ant 560/590 nm at 2nd, 4th, 6th, 8th and 10th day.
Tumorspheres assay
Single cell suspensions were plated in non-adherent 6-well tissue culture plates coated with poly-2-hydroxyethylmethacrylate (Sigma Aldrich, USA) at a density of 0.1× 105 cells in serum free media with 1% penicillin/streptomycin, 1X B27 (Gibco, Life Technology, USA), 20 ng/ml EGF (Sigma Aldrich, USA) and 20 ng/ml bFGF (Sigma Aldrich, USA) to check the formation of tumorspheres. The number of spheres formed in each well was examined after 8 days. For secondary tumorsphere formation primary spheres were centrifuged at 300 g and washed with PBS. Tumorspheres were trypsinized and seeded again in fresh media and were allowed to for secondary spheres. The sphere formation was analysed by measuring the size of each sphere using ImageJ software.
Immunocytochemistry
MCF-7 and MDAB-231 cells were seeded in density of 2 × 104 cells/ well on Millicell EZ slide 8-well glass, sterile (Merck, Millipore) and grown to subconfluency. Cells were treated for 8 days with glutamine-deprived media and DON. After completion of treatment, medium was discarded and cells were washed with PBS followed by fixation with 4% paraformaldehyde for 10 min at room temperature. Permeabilization was done using 0.25% Triton X-100 for 2 min at room temperature. Permeabilized cells were then blocked with 1% BSA for 30 min, followed by incubation with primary antibody anti-beta catenin (ab16051-abcam), E-cadherin (ab15148-abcam), N-cadherin (sc-59,987- Santa Cruz-Biotechnology) and claudin-1 (71-7800-Invitrogen) for 1 h at room temperature (RT). After complete incubation, the cells were washed thrice with PBS and incubated with secondary antibody Goat anti-Rabbit IgG H&L alexa fluor 488 (ab150077) or Goat anti-mouse IgG H&L alexa fluor 647 (ab150115). Finally, the CD24-RPE (MCA1379PET—Bio-Rad) was added Cells were again washed thrice with PBS and nuclei were stained with DAPI. Finally, images were taken under confocal microscope (Leica SP8) and analyzed.
Flow cytometry analysis
The cells were harvested by trypsinization and washed with PBS. Cells were kept for blocking using 10% fetal bovine serum for 1 h in ice cold condition and then stained with anti-CD44-Alexaflour 647 (Bio-Rad), anti-CD24-FITC (Bio-Rad), anti-CD326-PeCy (Bio-Rad) in PBS for 1 h at 4 °C. The samples were washed with PBS and finally re-suspended in 300 µl PBS. Flow cytometry analysis was performed on a Prosort™ Flow Cytometer (Bio-Rad).
Isolation of breast cancer stem cells
MDA-MB 231 and MCF 7 cells were seeded in T-75cm2 flasks to reach confluency. Cells were harvested, trypsinized and resuspended in 1X PBS. BCSC’s were isolated based on expression of CD44+, CD24− and CD326+ (Supplementary Table 1). Briefly cells were stained with anti-CD44-Alexaflour 647, Anti-CD24-FITC and Anti-CD326-PeCy7 and BCSC’s was identified by the flow cytometry. Cells were sorted into 5 ml tubes containing 1 ml culture medium supplemented with 10% FBS and 1% penicillin and streptomycin. The sorted cells were seeded on a 24-well plate treated with Glutamine-deprived media and DON containing media. The untreated sorted cells were taken as control. The effect of glutamine on stem cells was analyzed after 72 h using Flow cytometry.
Apoptotic assay
The cells were seeded in 6 well plates and allowed to attach overnight. After 24 h, cells were treated with complete glutamine, glutamine-deprived media and 25 µM DON. Cells were harvested by trypsinization and incubated with Annexin V-Alexafluor 488 (Invitrogen, Life Technology, USA) and propidium iodide (Life technology, USA) in Annexin binding buffer for 15 min in dark. Stained cells were immediately subjected to flow cytometry and studied after 72 h and 8 days of treatment.
Quantitative real time‐polymerase chain reaction (qRT-PCR)
Total RNA was extracted using Trizol (Ambion, USA) according to the manufacturer’s protocol. Quantification of RNA concentration was done using the Nano Drop 2000 Spectrophotometer (Nano Drop Technologies, Thermo, USA). cDNA preparation was done using cDNA Synthesis Kit (Bio-Rad) according to the manufacturer’s instruction. ITaq™ RT-PCR SYBR Green kit (Bio-Rad, USA) was used to prepare the reaction mix according to instructions of the manufacturer. The primers for target genes such as Sox2, Nanog, Pou5F-1/oct-4 and housekeeping (18 s rRNA) genes were designed to amplify only the cDNAs compatible to mRNA sequence but not any genomic sequences. (Supplementary Table 2) shows oligonucleotide sequence used for this study. The analysis and quantification of gene expression were carried out using the Step one software, version 2.0 (Applied Bio systems, USA).
Western blot analysis
MDAMB-231 and MCF-7 cells were grown in gln-deprived condition and were treated with DON for 72 h. Cells were washed thrice with PBS and protein content was isolated using RIPA buffer. Protein was quantified using BCA reagent kit (Invitrogen). Nuclear and cytosolic extract of cells were extracted using nuclear extraction kit from abcam (ab113474). An equal amount of protein from each experimental condition was loaded to perform gel electrophoresis in 10–12% acrylamide. Proteins were transferred on PVDF membrane and were probed against β-catenin, sox-2 and oct-4. Data for total cell protein expression was normalized using total protein stained with commassie blue.
For nuclear and cytosolic extracts β-actin and PCNA were used as internal controls respectively.
Results
Intracellular and extracellular glutamine‐deprivation sensitizes breast cancer cells towards apoptosis
MDAMB-231 and MCF-7 cells were grown in gln-deprived media to analyse the effect of gln-deprivation on these cell types. Gln-deprivation in media significantly decreases the proliferation rate of MCF-7 and MDAMB-231 cells (supplementary Fig. 1a). The MDAMB-231 cells showed a more pronounced effect of gln-deprivation and did not reach confluency in gln-deprived conditions. Glutamine is a conditionally essential amino acid which is also required for nucleotide synthesis, therefore apoptotic assay was conducted to find out whether cells were dividing slowly or were undergoing apoptosis. Results of cell staining with annexin-V and PI showed that the percentage of apoptotic cells in MDAMB-231 cells significantly increased in gln-deprived conditions as compared to cells grown in gln containing complete media (Supplementary Fig. 1b). Furthermore, MDAMB-231 cells were more sensitive towards the withdrawn of extracellular gln. DON is an analogue of glutamine and act as an inhibitor of enzymes utilizing glutamine in biochemical reactions. Glutaminase (GLS) is an enzyme with two isoforms (GLS1 and GLS2) found in cytosol and mitochondria. GLS is involved in the conversion of glutamine to glutamate, consequently important for utilization of extracellular and intracellular gln by cell. Therefore, to check if hampering intracellular utilization of gln sensitizes MCF-7 cells which are utilizing intracellular gln (Kung et al. 2011). Treatment of 25 µM DON in complete media significantly increased the apoptotic population in MCF-7 cells as compare to control cells and cells grown in gln-deprived media (Supplementary Fig. 1b). However, in MDAMB-231 cells the intracellular and extracellular gln deficiency did not showed any significant difference in cell viability (Supplementary Fig. 1a). Conclusively, MCF-7 cells utilize intracellular gln whereas MDAMB-231 cells are more dependent on extracellular supply of gln and therefore are sensitive to intracellular and extracellular-deprivation of gln respectively.
Glutamine‐deprivation alters the morphology of MDAMB-231 cells
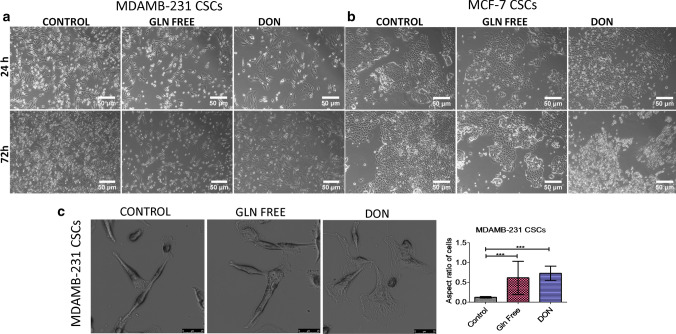
MDAMB-231 cells grown in gln-deprived media undergo cell death, however few cells were viable when grown in gln deficiency but undergone morphological changes. Also, the cells treated with DON were also undergone drastic change in morphology as compare to control cells. The cells become more flattened showing widened cytoplasm and oval nuclei in treated group while the untreated cells showed elongated cells with spindle shaped morphology (Supplementary Fig. 2a). The cells also showed a significant increase in their size in treated groups (Supplementary Fig. 3a). The morphological changes were more pronounced in cells treated with DON as compared to cells grown in gln-deprivation. However in MCF-7 cells no morphological changes were observed after depriving extracellular or intracellular gln (supplementary Fig. 2b). Further, freshly isolated breast cancer stem cells (bCSCs) population response was studied in gln-deprived conditions. We isolated CD24−/CD44+ and EpCAM+ cell population from MCF-7 and MDAMB-231 cells. CD24−/CD44+ and EpCAM+ cells are known to have bCSCs properties (Ghebeh et al. 2013; Xie et al. 2016). bCSCs isolated from MDAMB-231 cells showed similar changes in cell morphology and in increase in size as shown by MDAMB-231 cell line. bCSCs grown in gln-deprived media and DON changed their morphology towards epithelial like cells (Fig. 1a, c) (Supplementary Fig. 3b). Surprisingly, bCSCs isolated from MCF-7 cells were highly sensitive to gln-deprivation in contrast to the MCF-7 cells line. bCSCs treated with DON showed stress in cells within 24 h and cells undergo apoptosis within 72 h (Fig. 1b). Conclusively, it has been shown that bCSCs isolated from MCF-7 cells are very sensitive to gln-deprivation as compare to whole cell population of MCF-7 cells and bCSCs isolated from MDAMB-231 cells.
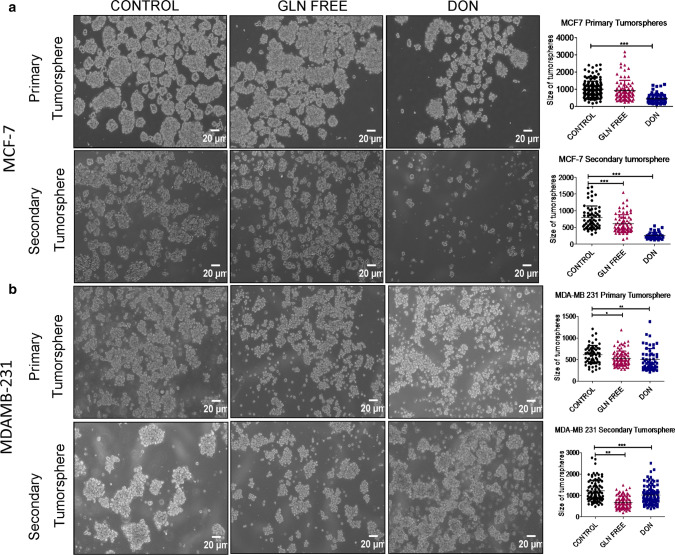
Fig. 1.
Morphological changes in breast cancer cells after glutamine depletion (a) and (b). Representative images of MDAMB231-bCSCs and MCF-7-bCSCs showing morphological changes in cells grown in gln-deprived media and DON containing complete media. (c). Higher magnification images showing morphological changes in MDAMB-231-CSCs
Glutamine‐deprivation changes the surface expression of stem cell markers in breast cancer cell population
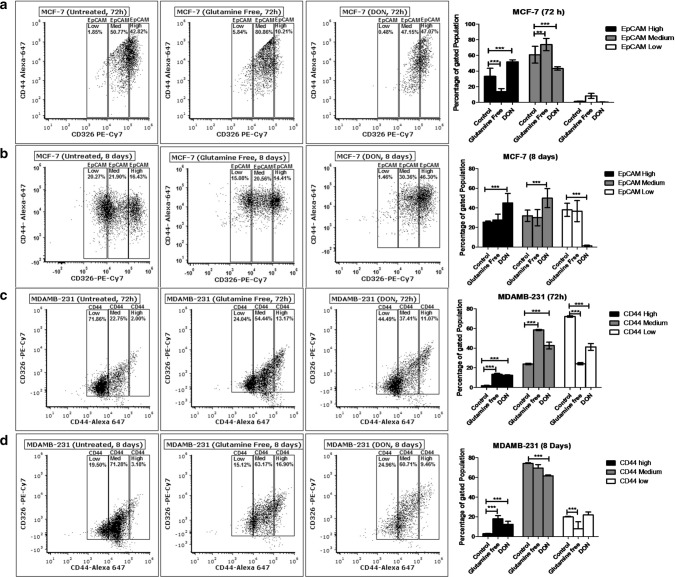
We analysed the surface expression of stem cell markers CD24, CD44 and EpCAM in MCF-7 and MDAMB-231 after gln-deprivation. The study was conducted for 72 h and 8 days to check the short term and long term effect of gln depletion on breast cancer cells. After 72 h of gln-deprivation and DON treatment, MCF-7 cells showed a significant decrease in population with high surface expression of EpCAM (Fig. 2a). However, no significant change in the EpCAM expressing population was observed after deprivation of gln for 8 days. Notably, 8 days treatment of DON showed a significant increase in EpCAMmedium and EpCAMhigh population in MCF-7 cells (Fig. 2b). There was no significant change in the expression of CD44 was observed after deprivation of gln in MCF-7 cells. Furthermore, MDAMB-231 cell showed a significant increase in the population with CD44medium and CD44high expression after 72 h and 8 days of gln-deprivation (Fig. 2c, d). Similarly, treatment of DON for 72 h and 8 days have shown to increase the population of CD44high cells in MDAMB-231 cells (Fig. 2c, d).
Fig. 2.
Stem cell marker modulations after glutamine-deprivation (a) and (b). Flow cytometry data shows the effect of gln-deprived media and DON containing complete media on EpCAM in MCF-7 after 72 and 8 days of treatment. (c) and (d) Flow cytometry data showing effect of gln-deprived media and DON containing complete media on CD44 in MDAMB-231 cells after 72 and 8 days of treatment. Data was analyzed using two-way ANOVA, each bar represents n = 3, ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
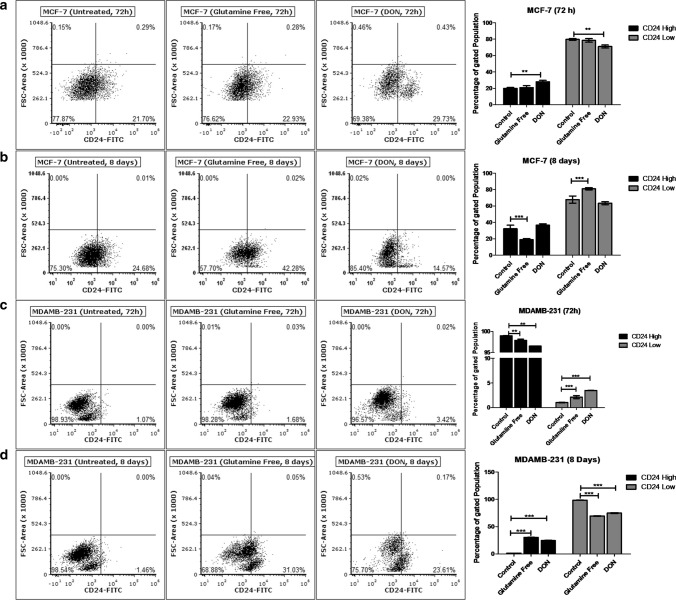
Effect of gln-deprivation on surface expression of CD24 was also observed in MCF-7 and MDAMB-231 cells. Gln-deprivation in media did not show any changes in the surface expression of CD24 for 72 h (Fig. 3a). However, after long term deprivation of gln for 8 days, MCF-7 cells with CD24+ve population doubled as compared to cells grown in complete media (Fig. 3b). In contrary, treatment of DON for 72 h increased the cells with CD24+ve population and after 8 days, decreased the CD24−ve population compared to control cells was observed. In MDAMB-231 cells, deprivation of gln did not changed CD24+ ve population (Fig. 3c), but the long-term deprivation of gln and treatment of DON significantly increased the CD24+ ve population compared to control cells (Fig. 3d). Conclusively, gln-deprivation changed the cells from CD24−ve to CD24+ve phenotype in MCF-7 and MDAMB-231 cells. EpCAM expression of MCF-7 cells was changed by gln-deprivation for short-term while the DON significantly increased the population of EpCAMhigh cells. Also, MDAMB-231 cells responded to gln-deprivation and DON treatment by increasing the CD44high population. Therefore, it has been shown that surface expression of stem cell markers is responsive to extracellular and intracellular deprivation of gln.
Fig. 3.
Glutamine depletion changes CD24 surface expression (a) and (b). Flow cytometry data showing effect of gln-deprived media and DON containing complete media on CD24 expressing population in MCF-7 after 72 and 8 days of treatment. (c) and (d) Flow cytometry data showing effect of gln-deprived media and DON containing complete media on CD24 expressing population in MDAMB-231 cells after 72 and 8 days of treatment. Data was analyzed using two-way ANOVA, each bar represents n = 3, ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
Glutamine‐deprivation changed the surface expression of stem cell markers on breast cancer stem cell population
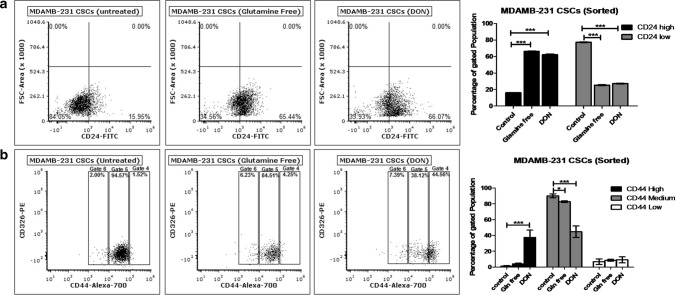
Freshly isolated bCSCs (CD24−/CD44+ and EpCAM+) population from MDAMB-231 cells were incubated in gln-deprived culture media and the surface expression of CD24, CD44 and EpCAM was further evaluated using flow cytometry. MDAMB-231-bCSCs grown in gln-deprived conditions and DON containing media have shown significant increase in the CD24+ population from 16 to 66% within 72 h (Fig. 4a). MDAMB-231-bCSCs population with CD44 expression showed a shift from CD44mediumto CD44high in gln-deprived conditions but a more prominent and significant population shift was observed in DON treated group (Fig. 4b). These results suggest that MDAMB-bCSCs may have differentiated into cancer cells in the absence of gln due to increase in the population of CD24+ cells.
Fig. 4.
Glutamine modulates stemness of bCSCs: (a) Flow cytometry data showing effect of gln-deprived media and DON containing complete media on CD24 expressing population in MDAMB-231-bCSCs after 72 of treatment. (b) Flow cytometry data showing effect of gln-deprived media and DON containing complete media on CD44 expressing population in MDAMB-231-bCSCs cells after 72 of treatment. Data was analyzed using two-way ANOVA, each bar represents n = 3, ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
Furthermore, we also checked the tumorsphere formation of MDAMB-231 and MCF-7 cells either in the absence of extracellular gln or presence of DON. To analyse the self -renewal properties of cells in tumorspheres we have also performed secondary tumorsphere formation in gln-deprived and DON treated conditions. Gln-deprivation and DON treatment in MCF-7 significantly decreased the size of primary tumorspheres. However the sphere forming capability was drastically reduced during secondary tumorsphere formation when cells were deprived of gln and were treated with DON (Fig. 5a). MDAMB-231 cells also showed a decrease in the size of tumorsphere in gln-deprived conditions. However, the DON treatment surprisingly has shown to increase the size of tumorspheres. Secondary tumorspheres of MDAMB-231 cells also showed a decrease in size of spheres when gln was deprived in media. Similar to primary spheres DON also formed larger secondary spheres. Furthermore, the spheres formed in gln-deprived or DON treated group were less compact as compared to untreated spheres (Fig. 5b).
Fig. 5.
Effect of gln deprivation on tumorsphere formation: (a) Representative images of primary and secondary tumorsphere formation in MCF-7 grown in gln-deprived media and DON containing complete media. (b) Representative images of primary and secondary tumorsphere formation in MCF-7 grown in gln-deprived media and DON containing complete media. Statistical analysis was done using one-way ANOVA, each bar represents n = 3, ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
Intracellular stem cell markers in breast cancer cells and breast cancer stem cells
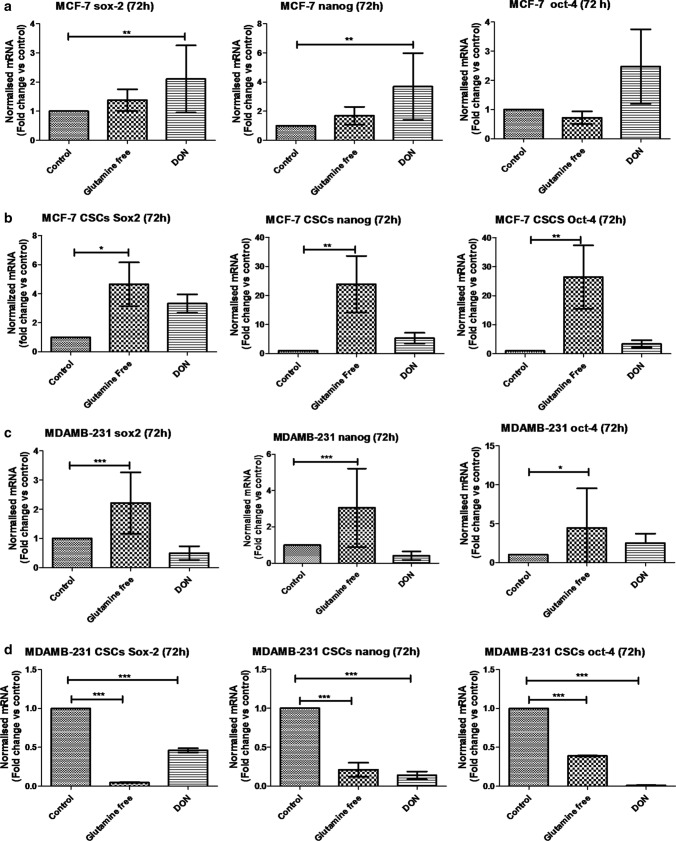
To analyse the effect of gln-deprivation on breast cancer cell lines, MCF-7 and MDAMB-231 cells were either grown in gln-deprived media or in DON containing media for 72 h. The gene expression analysis of MCF-7 cells revealed a significant increase in expression of sox-2 and nanog in DON containing media (Fig. 6a). However, there was no significant difference found in stem cell markers expression in gln-deprived conditions as compare to control. We have also checked the protein levels of sox-2 and oct-4 in MCF-7 and MDAMB-231 cells. The protein expression of sox-2 was found to be increased after gln-deprivation and DON treatment. The oct-4 protein levels remained unchanged after gln-deprivation in media while it was significantly reduced after DON treatment (Supplementary Fig. 4). Furthermore, similar experiment was conducted on the isolated bCSCs from MCF-7 cells. The mRNA expression showed that MCF-7-bCSCs grown in gln-deprived conditions showed a significant increase in the expression of sox2, nanog and oct-4, suggesting that these cells may not require extracellular gln supply and enhance stem cell gene expression in gln-deprived conditions (Fig. 6b). Nevertheless, as shown earlier the MCF-7 bCSCs were highly sensitive to the DON treatment and were not able to increase their stem cell gene expression (Fig. 6b).
Fig. 6.
qPCR analysis of stem cell markers: (a) and (b) mRNA expression of stem cell markers Sox-2, oct-4 and Nanog in MCF-7 and MCF-7-bCSCs after growing cells in gln-deprived media and DON-containing complete media for 72 h. (c) and (d) mRNA expression of stem cell markers Sox-2, oct-4 and Nanog in MDAMB-231 and MDAMB-231-bCSCs after growing cells in gln-deprived media and DON containing complete media for 72 h. Statistical analysis: one-way-ANOVA, each bar represents n = 3, ± SD, *p < 0.05, **p < 0.01, ***p < 0.001
Glutamine-deprivation in MDAMB-231 cells for 72 h have demonstrated significant increase in the mRNA expression of sox-2, oct-4 and nanog while the treatment with DON did not show any significant change in the expression of stem cell markers (Fig. 6c). Breast CSCs isolated from MDAMB-231 were also grown in gln-deprived media and DON containing media. mRNA expression of MDAMB-231-bCSCs showed a significant decrease in stem cell markers such as sox2, oct-4 and nanog (Fig. 6d) after availability of gln was hindered. In summary, these results demonstrated that the extracellular gln-deprivation could lead to an increase in stem cell markers in whole MDAMB-231 cell population while in whole MCF-7 cell population restraining intracellular gln supply by DON showed the similar effect. In contrary to normal cells, MCF-7-bCSCs demonstrated increase in stem cell gene expression in the absence of gln in growth media; however, MDAMB-231 bCSCs have shown decrease in the stem cell gene expression in absence of intracellular as well as extracellular gln.
Gln‐deprivation induces changes in wnt/β-catenin signalling
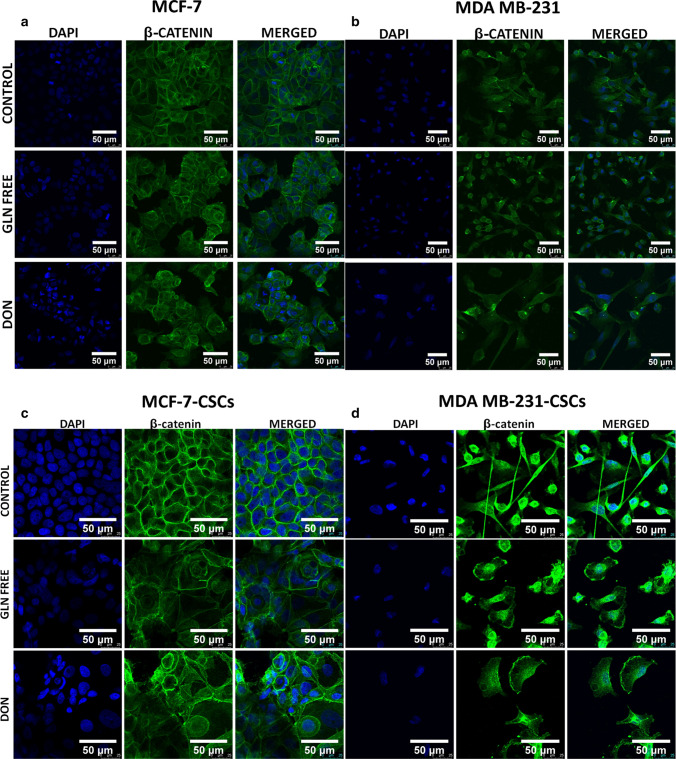
We have demonstrated the localization of catenin in MCF-7 and MDAMB-231 cells upon gln-deprivation. MCF-7 cells grown under gln deficiency have shown the expression of β-catenin in cytosol and on nuclear boundaries (perinuclear space) (Fig. 7a). The cells grown in DON showed cytosolic catenin also, deformities in nuclei of these cells were observed (Fig. 7a). However, MCF-7 cells grown in complete media with glutamine clearly showed β-catenin on the cell membrane only (Fig. 7a). In MDAMB-231 cells β-catenin was located in the cytosol of the normal cells while cells grown in gln-deprived media showed catenin localization near the nuclei of cells (Fig. 7b). The cells treated with DON showing morphological changes as discussed earlier have demonstrated aggregation of β-catenin in cytosol (Fig. 7b). For further confirmation of localization of β-catenin, nuclear and cytosolic fractions from MCF-7 and MDAMB-231 cells were extracted and its expression was analysed. MDAMB-231 cells showed very low expression of β-catenin as compared to MCF-7 cells. Although the cytosolic expression of β-catenin remain unaffected by gln-deprivation of DON treatment there was a significant decrease in its nuclear localization was observed (Supplementary Fig. 5).
Fig. 7.
β-catenin localization in breast cancer cells and breast cancer stem cells. (a) MCF-7 cells showing localization of β-catenin on cell membrane in control cells. β-catenin is localized in cytosol and perinuclear space after MCF-7 cells were grown in gln derived media DON containing complete media for 8 days. (b) MDAMB-231 cells showing localization of β-catenin in cytosol in control cells. β-catenin is localization increases in perinuclear space after MDAMB-231 cells were grown in gln derived media. Accumulation/aggregation of β-catenin in cytosol of MDAMB-231 cells after treatment with DON containing complete media for 8 days. (c) MCF-7-CSCs showing localization of β-catenin on cell membrane. β-catenin is localized in cytosol and perinuclear space after MCF-7-CSCs were grown in gln derived media DON containing complete media for 72 h. (d) MDAMB-231-CSCs cells showing localization of β-catenin in cytosol in control cells. β-catenin is localization is observed in nucleus and plasma membrane after MDAMB-231-CSCS were grown in gln derived media. Cells were stained with primary antibody against β-catenin, secondary antibody Alexaflour-488 tagged and counter stained with DAPI
Furthermore, the β-catenin expression was also analysed in MCF-7-CSCs and MDAMB-231 CSCs. Similar to MCF-7 cells MCF-7-CSCs clearly showed the pericellular accumulation of β-catenin in gln-deprived and DON treated conditions (Fig. 7c). MCF-7-CSCs grown in complete media showed β-catenin localization on the cell membrane. However in MDAMB-231-CSCs a clear difference from MDAMB-231 cells was observed in β-catenin localization. MDAMB-231-CSCs grown in Gln-deprived and DON conditions showed β-catenin distribution on cell membrane and nucleus (Fig. 7d).
Gln‐deprivation induces epithelial differentiation of breast cancer stem cells
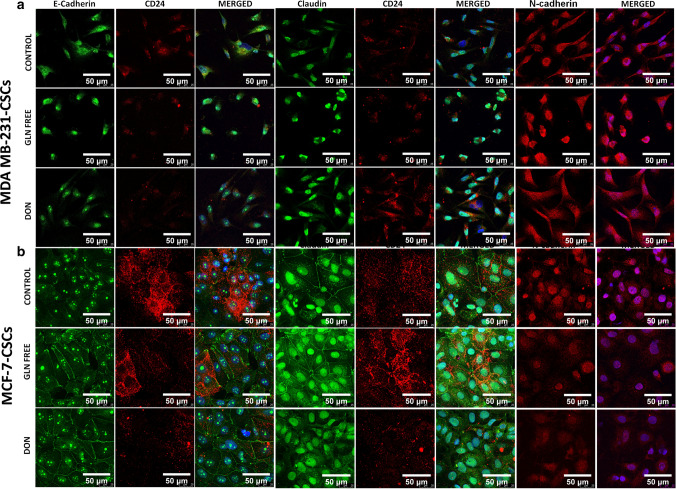
To identify if gln-deprivation could affect the differentiation of CSCs we have checked the expression of epithelial and mesenchymal protein after growing cells in gln-deprived conditions. Interestingly, gln-deprivation increased the expression of epithelial protein such as e-cadherin and claudin-1 in MDAMB-231-CSCs and MCF-7-CSCs (Fig. 8). Gln-deprivation in the media leads to intense nuclear localization of e-cadherin in MDAMB-231-CSCs while in cells grown in complete media showed cytosolic expression of the protein (Fig. 8a). Claudin-1 was found to be located in nucleus of MDAMB-231-CSCs however gln-deprivation significantly increased its expression (Fig. 8a and Supplementary Fig. 6b). Similarly, n-cadherin was also translocated to nucleus after gln-deprivation while in DON treated cells it was localized in cytosol. However n-cadherin expression was found to be increased significantly in cells deprived of gln (Fig. 8a and supplementary Fig. 6b). Notably, the expression of CD24 was also found to be increased after CSCs were grown in gln-deprived conditions (Supplementary Fig. 3b) as shown earlier by flow cytometery (Fig. 4b).
Fig. 8.
Expression of epithelial and mesenchymal markers in MDAMB-231-CSCs and MCF-7-CSCs: (a) MDAMB-231-CSCs stained with e–cadherin showed cytosolic expression in control cells and intense nuclear localization in gln-deprived and DON treated conditions. Claudin-1 showed nuclear localization of protein in control, gln-deprived and DON treated conditions and n–cadherin showed cytosolic expression in control and DON treated cells while the protein was nuclear localized in gln-deprived conditions. (b) MCF-7-CSCs stained with e-cadherin showed nuclear expression in control cells and nuclear and plasma membrane localization in gln-deprived and DON treated conditions. Claudin-1 showed nuclear and plasma membrane localization of protein in control, gln-deprived and DON treated conditions. N-cadherin showed nuclear expression in control, gln-deprived and DON treated cells
MCF-7-CSCs also showed a clear distribution of e-cadherin on cell membrane after gln-deprivation and DON treatment however the cells grown in complete media the protein was more intensely located in the nucleus (Fig. 8b). Similarly, the claudin-1 was also localized to plasma membrane after gln-deprivation notably, the nuclear expression of protein was also visible in control and gln-deprived cells (Fig. 8b). Expression of e- cadherin and claudin-1 was significantly increased after gln-deprivation and in DON treated cells (Supplementary Fig. 6c). In contrary, the expression of mesenchymal marker n-cadherin was significantly decreased after growing cells in gln-deprived and DON containing conditions as compared to control cells (Fig. 8b and Supplementary Fig. 6c). Also, the CD24 expression was found to be decreased in MCF-7 CSCs after gln-deprivation and DON treatment (Fig. 8b).
Discussion
In last decade glutamine metabolism has emerged as a potential anticancer target due to dependency of various tumors on gln (Bott et al. 2019). After glucose, gln is used as secondary source of carbon by cells and it is the only and major source of nitrogen for the cells. Gln is involved in TCA cycle through anaplerosis, proliferation of cells by de novo synthesis of nucleotides and is also required for synthesis of glutathione to maintain redox balance in the cells (Bott et al. 2019). Although, role of gln in various cancers has been explored widely but very few studies have investigated its contribution in cancer stem cell maintenance (Chae and Kim 2018; Peiris-Pagès et al. 2016). In breast cancer, only a recent study has shown that bCSCs identified by GD2+ cells were sensitive towards gln-deprivation and to glutaminase inhibitor CB839(Ly et al. 2020). Here, we have determined the effect of gln on bCSCs (CD24−/low/CD44high/+/CD326high/+ population) isolated form MCF-7 and MDAMB-231 cell lines as in vitro model. Firstly, we have shown that gln-deprivation decelerate proliferation of MCF-7 and MDAMB-231 cells. MDAMB-231 cells were also found to be more sensitive to the gln deficiency than MCF-7 cells. We have also observed drastic changes in the morphology of MDAMB-231 cells after gln-deprivation. As shown earlier that basal cells are more dependent on extracellular supply of gln (Kung et al. 2011), we have used DON, a glutaminase inhibitor to inhibit the intracellular conversion of gln to glu. Treatment with DON has significantly decreased the viability of MCF-7 cells (Supplementary Fig. 1). These results suggest that intracellular deprivation of gln sensitize both basal and luminal cells. Thereafter, similar experiment was conducted with MCF-7-bCSCs and MDAMB-231-bCSCs. Surprisingly, MCF-7-bCSCs were highly sensitive to intracellular deprivation of gln whereas MDAMB-231-bCSCs showed similar morphological changes as observed in MDAMB-231 cells (Fig. 1). A recent study suggested that gln-deprivation is detrimental for the survival of soft tissue sarcomas particularly due to their mesenchymal origin (Lee et al. 2020). Earlier studies have shown that gln metabolism is necessary for the differentiation of haematopoietic stem cells into erythroid lineage (Oburoglu et al. 2014). Also, it has been shown that human induced pluripotent stem cells (iPSCs) require gln oxidation for maintaining their naive undifferentiated state (Tohtama et al. 2016). Ramirez-Peña Esmeralda et al. has shown that epithelial mesenchymal transitions (EMT) cause cells to become independent of gln by suppressing the expression of GLS2 (Ramirez-Peña et al. 2019). Therefore, if the gln is unavailable to MDAMB-231-bCSCs they are only able to differentiate into epithelial phenotype and in case of MDAMB-231 cells alter their mesenchymal phenotype to epithelial in absence of gln.
Furthermore, the analysis of bCSCs surface stem cell marker analysis revealed an increase in CD24+ population in MCF-7 and MDAMB-231 cells after gln-deprivation, it is worth mentioning here that the conversion of CD24− population into CD24+ population was only observed after deprivation of the glutamine for longer period of time (8 days) but not in 72 h (Fig. 3). CD24 is small mucin-like GPI anchor found on the surface of various cells and is associated with distant metastasis in breast cancer (Abraham et al. 2005). In breast cancer, CD24−/low/CD44high/+/CD326high/+ population have been linked with cancer stem cell properties (Al-Hajj et al. 2003; Ghebeh et al. 2013; Xie et al. 2016). Staining of mouse mammary gland cells has revealed that CD24−, CD24low and CD24high population was representing non-epithelial, myoepithelial/basal and luminal epithelial cells respectively (Sleeman et al. 2006). Similarly, the number of EpCAMhigh population decreased in exogenous gln-deprivation, however long term deprivation merely showed any changes in EpCAM expression in MCF-7 cells (Fig. 2a, b). Previously, it has been shown that knockdown of EpCAM in MCF-7 cells inhibit their invasion properties (Martowicz et al. 2012). Also, the increase in the EpCAM expression has been correlated to poor survival rate in breast cancer (Agboola et al. 2012; Gao et al. 2017; Martowicz et al. 2012; Ohashi et al. 2016; Schnell et al. 2013; Ye et al. 2015). These results suggest that gln-deprivation was leading breast cancer cells towards epithelial phenotype by changing CD24 and EpCAM expression in MCF-7 cells. Contrarily, the number of CD44high population was increased after gln-deprivation in MDAMB-231 cells (Fig. 2) but has also shown morphological changes of epithelial phenotype. Further, isolated MDAMB-231-bCSCs grown in gln-deprived medium showed similar increase in CD24+ population but did not show any changes in the CD44high population after gln-deprivation (Fig. 4). However, DON treatment significantly increased the CD44high population in MDAMB-231 and MDAMB-231-bCSCs.We studied tumorsphere formation in gln-deprived condition and in the presence of DON to understand whether the increase in CD44high population is related to increase in sphere forming capability of cells. Although MCF-7 cells grow well in adherent condition in gln-deprived condition but they were unable to form spheres in non-adherent conditions. Notably, MDAMB-231 formed larger spheres in DON treated group but did not show any significant changes in gln-deprived conditions (Fig. 5). Earlier studies have explored the role of CD44 in increasing the aggregation of cells in the presence of intracellular hyaluronic acid (Cooper and Dougherty 1995; Goodison et al. 1999; Green et al. 1988; Underhill and Dorfman 1978). Therefore, the large spheres observed in DON treated group were perhaps the cell aggregates formed due to increased CD44 expression and do not depict actual cancer stem cell population. We have further checked the intracellular stem cell markers such as sox-2, oct-4 and nanog in gln-deprived conditions to demonstrate the stemness. It was observed that MCF-7 cells grown in gln-deprived media does not showed any changes in the expression of sox2 and nanog but DON treatment have significantly increased the expression of sox-2 and nanog in MCF-7 cells. However, the MCF-7-bCSCs have shown increased expression of stem cell markers in gln-deprived conditions and were very sensitive to DON treatment. MDAMB-231 cells grown in gln-deprived medium demonstrated increase in the expression of stem cell markers (Fig. 6). Earlier, such observations have been reported in melanoma and MDAMB-231 derived xenograft mice models, subsequent to hypermethylation of DNA due to decreased activity of Jumonji-domain-containing histone demethylases owing to depletion in α-KGA derived from gln (Pan et al. 2016).These results suggest that intracellular stem cells markers in MCF-7-bCSCs and MDAMB-231 cells may respond to gln-deprivation in a similar way. Furthermore, MDAMB-231-bCSCs decreased the expression of stem cell markers in response to gln-deprivation supporting the earlier observation of morphological changes and differentiation towards epithelial lineage. Our study has suggested that luminal and basal bCSCs differ in their response towards gln-deprivation in their microenvironment. Studies in human breast cancer and murine models has shown important role of Wnt/β-catenin signalling in development, morphogenesis and tumorogenesis of mammary glands. Wnt signalling is also involved in self renewal of normal and cancer stem cells (Valkenburg et al. 2011). Up regulation of β-catenin expression in breast cancer is found to mediate cancer stem cell like phenotype (Chen et al. 2007; Valkenburg et al. 2011; Woodward et al. 2007). Therefore, we have checked the β-catenin expression in MCF-7 and MDAMB-231 cells after deprivation of gln and treatment with DON. Immunocytochemistry staining have demonstrated cytosolic, perinuclear and membrane localization of β-catenin in MCF-7 cells grown in gln-deprived conditions while in DON treated cells showed only cytosolic localization of β-catenin. DON treated MCF-7 cells also showed nuclear deformities as shown by DAPI (Fig. 7). MDAMB-231 staining with β-catenin showed accumulation of protein in perinuclear space whereas in DON treated group the protein aggregation was observed in cytosol. Earlier the studies have shown that overexpression of β-catenin leads to its aggregation in nucleus or cytosol making it unavailable for transcriptional activation of genes (Giannini et al. 2000; Jazi and Najafi 2017).Furthermore, the differentiation of MCF-7-CSCs and MDAMB-231-CSCs into epithelial phenotype was shown by increased expression of epithelial markers such as e-cadherin and claudin-1 and decreased expression of n-cadherin (Fig. 8 and Supplementary Fig. 3). A recent study suggests that e-cadherin localization into nucleus inhibits β-catenin induced signalling by competing with its binding to transcription factors and is inversely correlated to metastasis and cancer stem cell phenotype (Su et al. 2015). In MDAMB-231-CSCs gln-deprivation leads to nuclear localization of e-cadherin (Fig. 8) and a corresponding change in morphology and membrane expression of β-catenin (Fig. 7) was observed. Similarly, e-cadherin was also located in the nucleus and membrane of MCF-7-CSCs in gln-deprived conditions (Fig. 8) and absence of β-catenin in nucleus was observed (Fig. 7). Therefore, it is postulated that e-cadherin could be inhibiting the β-catenin induced gene modulation in these cells.
Conclusion
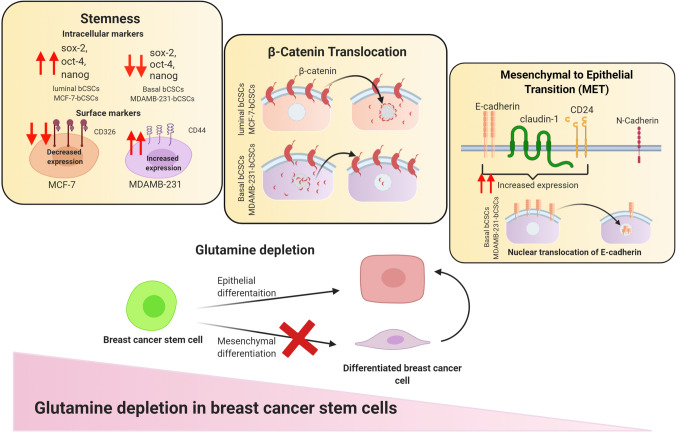
The present study has explored the role of gln metabolism in maintenance of bCSCs isolated from luminal and basal breast cancer cell lines (Fig. 9). We have demonstrated that deprivation of gln could reduce the stemness of bCSCs and drive them towards epithelial differentiation. It was found that that luminal-bCSCs were very sensitive to intracellular gln-deprivation and differentiated into non-CSCs upon gln-deprivation. While basal breast cancer stem cells differentiated to show an epithelial phenotype in gln-deprived conditions. Furthermore, the intracellular inhibition of gln by glutaminase inhibitor has profoundly changed the phenotype of basal bCSCs. More, elaborative studies with other gln inhibitors and in vivo evaluation of this study will elucidate the therapeutic potential gln metabolism in breast cancer stem cells.
Fig. 9.
Schematic representing the effect of glutamine-deprivation of luminal and basal breast cancer and breast cancer stem cells
Supplementary Information
Below is the link to the Supplementary Information.
Supplementary Information 1 (DOCX 2162 kb)
Acknowledgements
Mrs. Monika Seervi for their technical support for flow cytometry experiments.
Author contribution
H.J. and A.S. designed the manuscript. H.J., C.G., S.A. S.G. performed the experiments. H.J. and A.S analysed the data. H.J, drafted the manuscript and A.S. edited the manuscript. All authors have read and approved the manuscript.
Funding
National Institute of Pharmaceuticals Education and Research-Ahmedabad, Department of Pharmaceutics, Ministry of Chemicals and Fertilizers, Government of India. DST-SERB Grant (ECR/2016/002038), DBT supported Ramalingaswami Grant (BT/HRD/35/02/2006).
Compliance with ethical standards
Conflict of interest
The authors declare they have no such interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Abraham BK, Fritz P, McClellan M, Hauptvogel P, Athelogou M, Brauch H. Prevalence of CD44+/CD24–/low cells in breast cancer may not be associated with clinical outcome but may favor distant metastasis. Clin Cancer Res. 2005;11:1154–1159. [PubMed] [Google Scholar]
- Agboola AJ, Paish EC, Rakha EA, Powe DG, Macmillan RD, Ellis IO, Green AR. EpCAM expression is an indicator of recurrence in basal-like breast cancer. Breast Cancer Res Treat. 2012;133:575–582. doi: 10.1007/s10549-011-1813-7. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman BJ, Stine ZE, Dang CV. From Krebs to clinic: glutamine metabolism to cancer therapy. Nat Rev Cancer. 2016;16:619–634. doi: 10.1038/nrc.2016.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bott AJ, Maimouni S, Zong W-X. The pleiotropic effects of glutamine metabolism in cancer. Cancers. 2019;11:770. doi: 10.3390/cancers11060770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature. 2015;518:413. doi: 10.1038/nature13981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae YC, Kim JH. Cancer stem cell metabolism: target for cancer therapy. BMB Rep. 2018;51:319. doi: 10.5483/BMBRep.2018.51.7.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MS, Woodward WA, Behbod F, Peddibhotla S, Alfaro MP, Buchholz TA, Rosen JM. Wnt/β-catenin mediates radiation resistance of Sca1 + progenitors in an immortalized mammary gland cell line. J Cell Sci. 2007;120:468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- Choi Y-K, Park K-G. Targeting glutamine metabolism for cancer treatment. Biomol Therap. 2018;26:19. doi: 10.4062/biomolther.2017.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DL, Dougherty GJ. To metastasize or not? Selection of CD44 splice sites. Nat Med. 1995;1:635–637. doi: 10.1038/nm0795-635. [DOI] [PubMed] [Google Scholar]
- Demas DM, Demo S, Fallah Y, Clarke R, Nephew KP, Althaouse S, Sandusky G, He W, Shajahan-Haq AN. Glutamine metabolism drives growth in advanced hormone receptor positive breast cancer. Front Oncol. 2019;9:686. doi: 10.3389/fonc.2019.00686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Ansari R, McIntyre A, Craze ML, Ellis IO, Rakha EA, Green AR. Altered glutamine metabolism in breast cancer; subtype dependencies and alternative adaptations. Histopathology. 2018;72:183–190. doi: 10.1111/his.13334. [DOI] [PubMed] [Google Scholar]
- Gao S, Sun Y, Liu X, Zhang D, Yang X. EpCAM and COX-2 expression are positively correlated in human breast cancer. Mol Med Rep. 2017;15:3755–3760. doi: 10.3892/mmr.2017.6447. [DOI] [PubMed] [Google Scholar]
- Ghebeh H, Sleiman GM, Manogaran PS, Al-Mazrou A, Barhoush E, Al-Mohanna FH, Tulbah A, Al-Faqeeh K, Adra CN. Profiling of normal and malignant breast tissue show CD44 high/CD24 low phenotype as a predominant stem/progenitor marker when used in combination with Ep-CAM/CD49f markers. BMC Cancer. 2013;13:289. doi: 10.1186/1471-2407-13-289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannini A, Vivanco M, Kypta R. Analysis of β-catenin aggregation and localization using GFP fusion proteins: nuclear import of α-catenin by the β-catenin/Tcf complex. Exp Cell Res. 2000;255:207–220. doi: 10.1006/excr.1999.4785. [DOI] [PubMed] [Google Scholar]
- Goodison S, Urquidi V, Tarin D. CD44 cell adhesion molecules. Mol Pathol. 1999;52:189. doi: 10.1136/mp.52.4.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SJ, Tarone G, Underhill CB. Aggregation of macrophages and fibroblasts is inhibited by a monoclonal antibody to the hyaluronate receptor. Exp Cell Res. 1988;178:224–232. doi: 10.1016/0014-4827(88)90393-x. [DOI] [PubMed] [Google Scholar]
- Jariyal H, Weinberg F, Achreja A, Nagarath D, Srivastava A. Synthetic lethality: a step forward for personalized medicine in cancer. Drug Discov Today. 2019;25:305–320. doi: 10.1016/j.drudis.2019.11.014. [DOI] [PubMed] [Google Scholar]
- Jazi MS, Najafi SMA. Beta-catenin forms protein aggregation at high concentrations in HEK293TCells. Iran J Med Sci. 2017;42:66. [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Lee KJ, Seo Y, Kwon JH, Yoon JP, Kang JY, Lee HJ, Park SJ, Hong SP, Cheon JH. Effects of metformin on colorectal cancer stem cells depend on alterations in glutamine metabolism. Sci Rep. 2018;8:409. doi: 10.1038/s41598-017-18762-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kung H-N, Marks JR, Chi J-T. Glutamine synthetase is a genetic determinant of cell type-specific glutamine independence in breast epithelia. PLoS Genet. 2011;7:e1002229. doi: 10.1371/journal.pgen.1002229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee P, Malik D, Perkons N, Huangyang P, Khare S, Rhoades S, Gong YY, Burrows M, Finan JM, Nissim I. Targeting glutamine metabolism slows soft tissue sarcoma growth. Nat Commun. 2020;11:1–15. doi: 10.1038/s41467-020-14374-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Fu Z, Chen R, Zhao X, Zhou Y, Zeng B, Yu M, Zhou Q, Lin Q, Gao W. Inhibition of glutamine metabolism counteracts pancreatic cancer stem cell features and sensitizes cells to radiotherapy. Oncotarget. 2015;6:31151. doi: 10.18632/oncotarget.5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Cao Y, Meng G, Qian L, Xu T, Yan C, Luo O, Wang S, Wei J, Ding Y. Targeting glutaminase 1 attenuates stemness properties in hepatocellular carcinoma by increasing reactive oxygen species and suppressing Wnt/beta-catenin pathway. EBioMedicine. 2019;39:239–254. doi: 10.1016/j.ebiom.2018.11.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao J, Liu PP, Hou G, Shao J, Yang J, Liu K, Lu W, Wen S, Hu Y, Huang P. Regulation of stem-like cancer cells by glutamine through β-catenin pathway mediated by redox signaling. Mol Cancer. 2017;16:51. doi: 10.1186/s12943-017-0623-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ly S, Nguyen K, Andreeff M, Battula VL. Abstract P3-02-05: targeting glutamine metabolism inhibits GD2 + breast cancer stem cell function in triple negative breast cancer. Philadelphia: AACR; 2020. [Google Scholar]
- Lyssiotis CA, Kimmelman AC. Metabolic interactions in the tumor microenvironment. Trends Cell Biol. 2017;27:863–875. doi: 10.1016/j.tcb.2017.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsboom G, Zhang G-F, Pohl-avila N, Zhang Y, Yuan Y, Kang H, Hao B, Brunengraber H, Malik AB, Rehman J. Glutamine metabolism regulates the pluripotency transcription factor OCT4. Cell Rep. 2016;16:323–332. doi: 10.1016/j.celrep.2016.05.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martowicz A, Spizzo G, Gastl G, Untergasser G. Phenotype-dependent effects of EpCAM expression on growth and invasion of human breast cancer cell lines. BMC Cancer. 2012;12:501. doi: 10.1186/1471-2407-12-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oburoglu L, Tardito S, Fritz V, De Barros SC, Merida P, Craveiro M, Mamede J, Cretenet G, Mongellaz C, An X. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell. 2014;15:169–184. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Ohashi R, Kawahara K, Fujii T, Takei H, Naito Z. Higher expression of EpCAM is associated with poor clinical and pathological responses in breast cancer patients undergoing neoadjuvant chemotherapy. Pathol Int. 2016;66:210–217. doi: 10.1111/pin.12404. [DOI] [PubMed] [Google Scholar]
- Pan M, Reid MA, Lowman XH, Kulkarni RP, Tran TQ, Liu X, Yang Y, Hernandez-davies JE, Rosales KK, Li H. Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat Cell Biol. 2016;18:1090. doi: 10.1038/ncb3410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris-Pagès M, Martinez-Outschoorn UE, Pestell RG, Sotgia F, Lisanti MP. Cancer stem cell metabolism. Nat Cell Biol. 2016;18:55. doi: 10.1186/s13058-016-0712-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Peña E, Arnold J, Shivkumar V, Joseph R, Vidya Vijay G, Den Hollander P, Bhangre N, Allegakoen P, Prasad R, Conley Z. The epithelial to mesenchymal transition promotes glutamine independence by suppressing GLS2 expression. Cancers. 2019;11:1610. doi: 10.3390/cancers11101610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell U, Cirulli V, Giepmans BN. EpCAM: structure and function in health and disease. Biochim Biophys Acta Biomembr. 2013;1828:1989–2001. doi: 10.1016/j.bbamem.2013.04.018. [DOI] [PubMed] [Google Scholar]
- Seltzer MJ, Bennett BD, Joshi AD, Gao P, Thomas AG, Ferraris DV, Tsukamoto T, Rojas CJ, Slusher BS, Rabinowitz J. Inhibition of glutaminase preferentially slows growth of glioma cells with mutant IDH1. Cancer Res. 2010;70:8981–8987. doi: 10.1158/0008-5472.CAN-10-1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro RA, Farrell L, Srinivasan M, Curthoys N. Isolation, characterization, and in vitro expression of a cDNA that encodes the kidney isoenzyme of the mitochondrial glutaminase. J Biol Chem. 1991;266:18792–18796. [PubMed] [Google Scholar]
- Sleeman KE, Kendrick H, Ashworth A, Isacke CM, Smalley MJ. CD24 staining of mouse mammary gland cells defines luminal epithelial, myoepithelial/basal and non-epithelial cells. Breast Cancer Res. 2006;8:R7. doi: 10.1186/bcr1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Kim S-H, Im CY, Hwang H-J. Recent development of small molecule glutaminase inhibitors. Curr Top Med Chem. 2018;18:432–443. doi: 10.2174/1568026618666180525100830. [DOI] [PubMed] [Google Scholar]
- Su Y, Chang Y, Lin W, Liang C, Lee J. An aberrant nuclear localization of E-cadherin is a potent inhibitor of Wnt/β-catenin-elicited promotion of the cancer stem cell phenotype. Oncogenesis. 2015;4:e157–e157. doi: 10.1038/oncsis.2015.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohtama S, Fujita J, Hishiki T, Matsuura T, Hattori F, Ohno R, Kanazawa H, Seki T, Nakajima K, Kishino Y. Glutamine oxidation is indispensable for survival of human pluripotent stem cells. Cell Metab. 2016;23:663–674. doi: 10.1016/j.cmet.2016.03.001. [DOI] [PubMed] [Google Scholar]
- Underhill C, Dorfman A. The role of hyaluronic acid in intercellular adhesion of cultured mouse cells. Exp Cell Res. 1978;117:155–164. doi: 10.1016/0014-4827(78)90438-x. [DOI] [PubMed] [Google Scholar]
- Unger C, Harzmann R, Muller C, Witt C, Roberts J, Sethuraman N. Phase I dose escalating study of PEG-PGA and DON: a new amino acid depleting anti cancer drug approach. J Clin Oncol. 2005;23:3130–3130. [Google Scholar]
- Valkenburg KC, Graveel CR, Zylstra-Diegel CR, Zhong Z, Williams BO. Wnt/β-catenin signaling in normal and cancer stem cells. Cancers. 2011;3:2050–2079. doi: 10.3390/cancers3022050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward WA, Chen MS, Behbod F, Alfaro MP, Buchholz TA, Rosen JM. WNT/β-catenin mediates radiation resistance of mouse mammary progenitor cells. Proc Natl Acad Sci USA. 2007;104:618–623. doi: 10.1073/pnas.0606599104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C, Chen L, Jin S, Li H. Glutaminase inhibitors: a patent review. Expert Opin Ther Pat. 2018;28:823–835. doi: 10.1080/13543776.2018.1530759. [DOI] [PubMed] [Google Scholar]
- Xie J, Xiao Y, Zhu X-y, Ning Z-y, Xu H-f, Wu H-m. Hypoxia regulates stemness of breast cancer MDA-MB-231 cells. Med Oncol. 2016;33:42. doi: 10.1007/s12032-016-0755-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Meng Y, Li L, Xu P, Wang J, Li Z, Bian J. Overview of the development of glutaminase inhibitors: achievements and future directions. J Med Chem. 2018;62:1096–1115. doi: 10.1021/acs.jmedchem.8b00961. [DOI] [PubMed] [Google Scholar]
- Ye F, Qiu Y, Li L, Yang L, Cheng F, Zhang H, Wei B, Zhang Z, Sun L, Bu H, Ye F, et al. The presence of EpCAM−/CD49f+ cells in breast cancer is associated with a poor clinical outcome. J Breast Cancer. 2015;18:242–248. doi: 10.4048/jbc.2015.18.3.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information 1 (DOCX 2162 kb)