Abstract
Bread wheat is a major crop that has long been the focus of basic and breeding research. Assembly of its genome has been difficult because of its large size and allohexaploid nature (AABBDD genome). Following the first reported assembly of the genome of the experimental strain Chinese Spring (CS), the 10+ Wheat Genomes Project was launched to produce multiple assemblies of worldwide modern cultivars. The only Asian cultivar in the project is Norin 61, a representative Japanese cultivar adapted to grow across a broad latitudinal range, mostly characterized by a wet climate and a short growing season. Here, we characterize the key aspects of its chromosome-scale genome assembly spanning 15 Gb with a raw scaffold N50 of 22 Mb. Analysis of the repetitive elements identified chromosomal regions unique to Norin 61 that encompass a tandem array of the pathogenesis-related 13 family. We report novel copy-number variations in the B homeolog of the florigen gene FT1/VRN3, pseudogenization of its D homeolog and the association of its A homeologous alleles with the spring/winter growth habit. Furthermore, the Norin 61 genome carries typical East Asian functional variants different from CS, ranging from a single nucleotide to multi-Mb scale. Examples of such variation are the Fhb1 locus, which confers Fusarium head-blight resistance, Ppd-D1a, which confers early flowering, Glu-D1f for Asian noodle quality and Rht-D1b, which introduced semi-dwarfism during the green revolution. The adoption of Norin 61 as a reference assembly for functional and evolutionary studies will enable comprehensive characterization of the underexploited Asian bread wheat diversity.
Keywords: Adaptation, Asian germplasm, Bread wheat, Genome assembly, Norin 61, Polyploidy
Accession numbers: The nucleotide sequence reported in this paper has been submitted to EBI database under the accession number GCA_904066035.1.
Introduction
About 35% of land plant species are estimated to be recent polyploids (Wood et al. 2009, Mayrose et al. 2011), and polyploid species are common in crops (Leitch and Leitch 2008). However, genome-scale studies of polyploid species have been challenging because of their large genome size and the high sequence similarity among homeologs; i.e. duplicated genes derived from genome duplication. Recent advances in sequencing and assembly algorithms have drastically improved the quality of genome assemblies, enabling improved genome-wide polymorphism analyses and transcriptomic analysis of allopolyploid species (Paape et al. 2018, Hu et al. 2020, Kuo et al. 2020). Assemblies of many polyploid species were reported with high contiguity (N50) values in the order of megabases [Mb; Avni et al. 2017, Hatakeyama et al. 2018, The International Wheat Genome Sequencing Consortium (IWGSC) 2018, Schreiber et al. 2018].
Bread wheat (Triticum aestivum L.), together with rice and maize, is a major crop that has a long tradition of cytogenetic, evolutionary and agricultural research (Dubcovsky and Dvorak 2007, Matsuoka 2011). However, its genome assembly has been one of the most difficult, yet sought-after challenges in genomics [The International Wheat Genome Sequencing Consortium (IWGSC) 2014] because of its allohexaploid nature and large size of about 16 Gb (Doležel et al. 2018). Kihara (1944) and McFadden and Sears (1944) independently discovered that bread wheat (AABBDD genome) originated from the allopolyploidization of domesticated emmer wheat T. turgidum (tetraploid AABB) with the wild species Aegilops tauschii (diploid DD). This polyploid speciation was inferred to have occurred near the Caspian Sea during the Neolithic period, around 8,000 years ago (Dubcovsky and Dvorak 2007, Matsuoka 2011). Neolithic crop species may have dispersed either by demic diffusion (associated with the spread of people and language) or by cultural diffusion (horizontal knowledge transfer without gene flow between human groups) (Stoneking 2016). A demic diffusion of agriculture, including T. aestivum, was proposed to have occurred from the Middle East toward Europe (Ammerman and Cavalli-Sforza 1971) and to have eventually continued its spread to America and Australia by the migration of European people in modern times. In its move toward East Asia, wheat may have integrated with preexisting farming systems in China through cultural diffusion (Li et al. 2007, Dodson et al. 2013, Barton and An 2014, Stevens and Fuller 2017) and arrived at the eastern end, Japan, about 2,000 years ago (Ohta 2010, Fujita 2013).
Despite their significant contribution to the green revolution, recent studies suggest that traditional Asian bread wheat cultivars are still underutilized for breeding (Balfourier et al. 2019, Sansaloni et al. 2020). A principal component analysis of 188 worldwide accessions using DArTseq markers indicated that Japanese, West Asian, and European accessions were the furthest apart from each other, with the Chinese accessions lying on the edge between Japanese and West Asian accessions (Takenaka et al. 2018). The areas of wheat cultivation throughout Japan and China (except for the northern parts of both countries) are characterized by their wet environment and short growing seasons because of the rainy period in early summer and because of multiple cropping with rice (Yang et al. 2009). Early flowering cultivars with no or weak requirement for vernalization (‘spring wheat’) are typically sown in early winter to be harvested in early summer before the arrival of the rainy season and rice cropping. Important targets of wheat breeding in East Asia include early flowering (Fu et al. 2005, Tanio et al. 2005, Seki et al. 2011, Seki et al. 2013), semi-dwarfism (Kojima et al. 2017), resistance to preharvest sprouting (Himi et al. 2011), favorable quality for Asian type noodles and especially resistance to Fusarium head blight (FHB)—a severe threat in wet climates (Niwa et al. 2018).
The analysis of population genomic data and pangenomic analysis is critically important for demographic, evolutionary and breeding studies (Saisho and Purugganan 2007, Brohammer et al. 2018, Gutaker et al. 2020). In contrast to the extensive information from population genomic studies of rice and maize (van Heerwaarden et al. 2011, Wang et al. 2018, Gutaker et al. 2020), little is known about wheat, although it is one of the three species most widely grown and consumed. The first draft genome assembly of bread wheat released by the IWGSC was highly fragmented, with ≈10 kb of N50 statistics [The International Wheat Genome Sequencing Consortium (IWGSC) 2014]. In 2018, the first chromosome-scale assembly of the experimentally used landrace Chinese Spring (CS) [The International Wheat Genome Sequencing Consortium (IWGSC) 2018] was reported. The 10+ Wheat Genomes Project was launched to produce multiple high-quality de novo genome assemblies of worldwide modern cultivars, to be subsequently analyzed in a pangenomic context. The Japanese cultivar Komugi Norin 61 (‘Komugi’ means ‘wheat’ in Japanese, thus shortened to Norin 61) was the sole Asian cultivar represented in the project. The assembly data have been publicly released, and Walkowiak et al. (2020) analyzed the entire dataset.
Norin 61 is a representative Japanese cultivar of bread wheat that is characterized by broad adaptation and environmental robustness (Ishikawa 2010, Fujita 2013). It has been utilized for a broad range of molecular and physiological studies, including transgenic experiments and mutant screening (e.g. Taya et al. 1981, Ikeda et al. 2002, Matsuo et al. 2005, Kasajima et al. 2009, Shimada et al. 2009, Minoda 2010, Rikiishi and Maekawa 2010). It has also been used as a representative cultivar in studies of worldwide and regional variation (e.g. Fu et al. 2005, Tanio et al. 2005). Norin 61 was developed in 1935 and released in 1944. Its flour is mainly used as a soft red wheat for Japanese noodles, and between 1950 and 1980 Norin 61 was most widely cultivated in Japan. It was still the third most widely grown cultivar in 2010. Norin 61 is grown in a broad region spanning from Kyushu to Kanto and partly in the northern island of Hokkaido (Ishikawa 2010). When grown in Sudan, it also demonstrated heat tolerance (Elbashir et al. 2017). The paternal parent is the cultivar Shinchunaga, which originated by pure-line selection from the landrace Nakanaga (alternatively pronounced as Chunaga). Shinchunaga is characterized by FHB resistance, early flowering, and high yield but is not semi-dwarf: its name ‘naga’ means long. The semi-dwarfism in Norin 61 was introduced from the maternal parent Fukuokakomugi 18. Norin 61 is genetically related to a large number of Japanese cultivars, including Shiroganekomugi, that are grown from Kyushu to Honshu islands, partly because its paternal parent Shinchunaga was widely used in the breeding programs by crossing with Japanese and Western cultivars (Fukunaga and Inagaki 1985, Kobayashi et al. 2016, Takenaka et al. 2018). Overall, the history and features of Norin 61 suggest that it could be established as the reference genotype for adaptation and breeding research in modern East Asian cultivars. In this article, we support this conclusion by examining the quality of the genome assembly, studying unique chromosomal regions and investigating known and novel functional variants of Norin 61.
Results
Genome assembly and scaffolding
The Norin 61 genome was sequenced using the Illumina short-read sequencing technology, combining different library formats and insert sizes. Most of the coverage was contributed by paired-end (PE) and mate-pair (MP) whole-genome shotgun (WGS) libraries and 10x Genomics linked reads and Hi-C data were produced to connect adjacent regions to hundreds of Mb-sized pseudomolecules (Table 1). After assembling an approximately 323-fold coverage of the WGS and linked-read data, the sequences were inspected for misassemblies and later scaffolded to chromosome-level pseudomolecules using POPSEQ and the Hi-C data (as described in Walkowiak et al. 2020, Supplementary Fig. S1).
Table 1.
Genomic DNA libraries and sequencing strategy for Norin 61
| Library type | Insert size | Sequencing length | Platform | # of flow cells | Approximate coverage (×) |
|---|---|---|---|---|---|
| PCR-free PE470 | 450–470 bp | 2 × 266 bp | HiSeq 2500v2 | 17 | 92 |
| PCR-free PE700 | 700–800 bp | 2 × 150 bp | HiSeq 2500v4 | 7 | 56 |
| Nextera MP3kb | 2–4 kb | 2 × 150 bp | HiSeq 4000 | 4 | 43 |
| Nextera MP6kb | 5–7 kb | 2 × 150 bp | HiSeq 4000 | 4 | 41 |
| Nextera MP9kb | 8–10 kb | 2 × 150 bp | HiSeq 4000 | 4 | 63 |
| 10× Genomics Chromium | N/A | 2 × 150 bp | HiSeq 4000 | 4 | 28 |
| Hi-C | 300 bp | 2 × 100 bp | HiSeq 2500v2 | 4 | 5.2 |
Because the initial inspection of the assembly indicated that a key flowering gene (FT-B1/VRN-B3) was missing, the raw PE, MP and 10x Genomics linked-read data were reassembled. Two copies of the FT-B1 chromosomal region were identified in two short (≈19 kb each) contigs. Overall, approximately 755 Mb of sequences not present in the v1.0 assembly were added to chrUn (unplaced sequences). This updated assembly was labeled v1.1 and was used as the reference for all downstream analyses. The assembly spanned 14.9 Gb, having very high contiguity (L50 and N50, or half of the assembly, represented in 162 sequences spanning 21.9 Mb or more; Supplementary Table S1, Walkowiak et al. 2020). The vast majority of the assembly (14.1 Gb, 94.9% of the bases) was anchored to 21 chromosome pseudomolecules, leaving unanchored only 852 Mb of smaller sequences (96 kb N50). The data are available for download and BLAST search from IPK Gatersleben (https://wheat.ipk-gatersleben.de/) and from the National Institute of Genetics, Japan, (https://shigen.nig.ac.jp/wheat/komugi/about/norin61GenomeSequence.jsp), NCBI and EBI databases with the accession number GCA_904066035.1. Genes were annotated by projecting the CS annotation [The International Wheat Genome Sequencing Consortium (IWGSC) 2018], as described by Walkowiak et al. (2020).
Assembly validation
The completeness of the assembly was evaluated using three complementary methods. After alignment of six genome equivalents (part of PE700 data), 97.4% of the bases mapped to the assembly. Similarly, 97.4% of the read pairs mapped, and 86.2% were properly paired. Across the 14.9-Gb assembly, 98.5% of the positions were realigned to at least one read and 92.2% to at least four reads. We measured the degree of inclusion of the sequenced bases in the assembly using the comp function of the KAT tool (Mapleson et al. 2017). By comparing k-mers between the raw reads and the assembly, KAT estimated an overall assembly completeness of 97.8%. The k-mer distribution in the raw reads showed two peaks, at 21 and 40 (Fig. 1A). The copy numbers of the k-mers in the assembly (colored areas) and in the raw reads (curve profile) corresponded strongly. The homozygous and repeat peaks (k = 21 and 40, respectively) contained 96.3% and 70% of the read k-mers at the appropriate frequency in the assembly.
Fig. 1.
Norin 61 assembly validation. (A) KAT plot comparing the sequence content in the raw reads (curve profile) and in the assembly (colored areas). The peak at X ≈ 21× contains the single-copy regions that are assembled in one copy (red area). The shoulder at 40–60× contains duplicated genomic sequences that are mostly assembled (as expected) in two copies. (B) Presence and copy number of universal SCOs in the Norin 61 assembly and gene annotation.
Walkowiak et al. (2020) reported that in the 10 reference-quality pseudomolecule assemblies (RQAs), >97% of plant single-copy orthologs (SCOs) were always present, implying a high level of completeness of the gene fraction across all RQAs. We inspected in more detail the representation of SCOs in the Norin 61 assembly and the gene annotation. In the assembly, <4% of the ultraconserved SCOs were either fragmented or missing. Of the 1,383 complete SCOs, 1,090 (75.7%) were present in three copies. In the remaining 293 complete models, most were present in two copies (14.5% of the total) and only 5.3% in one copy (Fig. 1B). Similarly, the scan for SCOs on the gene annotation identified 1,421 of the 1,440 SCOs (98.7%), with the majority of genes being present three times. These data support that the majority of the SCOs are present in three copies, which is consistent with the genome allohexaploidy, and that some homeologs are absent.
The conserved, large-scale chromosomal structure of Norin 61
To inspect for possible large-scale rearrangements, we compared the karyotype of Norin 61 with that of CS after probing with three repetitive sequence probes. The hybridization showed a similar pattern in the two varieties, indicating a lack of any large-scale rearrangements in the genome of Norin 61 relative to CS. Six minor fluorescence in situ hybridization (FISH) polymorphisms were detected (Fig. 2): a gain of a terminal pSc119.2 signal on chromosome arm 1BS, a gain of terminal pSc119.2 signal on 1DS, a weaker terminal pTa535 signal on 2AS, a loss of terminal pSc119.2 signal on 3BS, a loss of terminal pSc119.2 signal on 4AL and a loss of terminal pTa713 signal on 6BL. Moreover, we did not identify any chromatin containing a large block of repetitive elements.
Fig. 2.
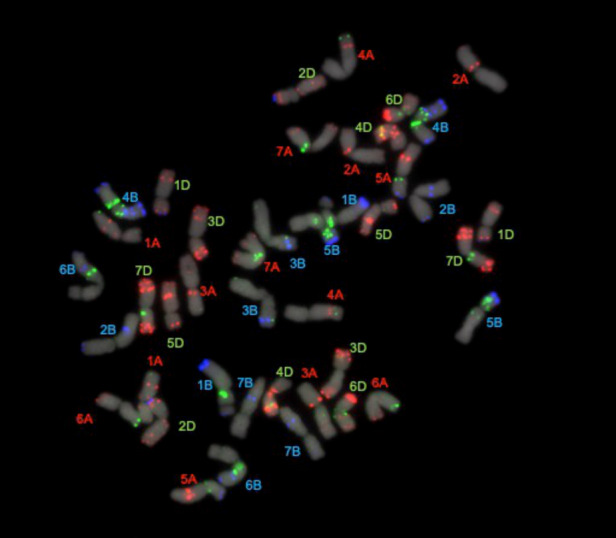
Nondenaturing fluorescence in situ hybridization of a metaphase spread of Norin 61. A pseudocolored image of the stacked pictures taken with red (Oligo-pSc119.2-1; shown in blue), green (Oligo-pTa535; shown in red), near-infrared (Oligo-pTa713; shown in green) and blue (counterstaining with DAPI; shown in gray) filters.
The uniform/consistent large-scale structure of Norin 61 and CS genomes was confirmed by genome-wide sequence alignment (Supplementary Fig. S2). The local inversions identified at the very end or in the middle of the chromosomes may be the result of the difficulty of assembling repetitive subtelomeric or centromeric regions. These analyses supported that the Norin 61 genome does not have the large-scale interchromosomal rearrangements observed in some other 10+ Wheat Genomes Project cultivars. We next investigated the genomic variation at a smaller scale.
Unique chromosomal regions identified by the patterns of repetitive elements
The analysis of repetitive elements in the 10 genomes identified unique chromosomal segments that were considered candidates for foreign introgressions (Walkowiak et al. 2020, Zhou et al. 2020). Norin 61 contains 45 such segments that are unique among the 10 RQAs, span between 20 and 169 Mb in size and together account for 1.9 Gb (or 13.5%) of the assembly. The candidate introgressed segments contain between 75 and 1,328 genes each (Supplementary Table S2). Overall, 20,225 wheat genes (17.0% of the 118,734 total genes) were annotated in the 45 regions. By comparing their coding sequences (CDSs) to the annotation for CS, we classified such genes as syntenic or private: 1,669 genes were private (i.e. lacked significant homology to genes in the corresponding syntenic region in CS—see Materials and Methods for a more complete definition of syntenyc or private genes).
Typically, 6–8% of the genes annotated in the candidate regions for containing an introgression event were private, with a moderate enrichment in distal and centromeric regions (Supplementary Fig. S3A). We identified 10 introgressions that contained over 10% private genes, in three of which the count reached 15% (Supplementary Table S2). Overall, the 45 regions contained a larger fraction of private genes than the genome-wide average (Supplementary Fig. S3B). This indicates that the gene content in the candidate regions for introgression differs considerably from that of the CS reference.
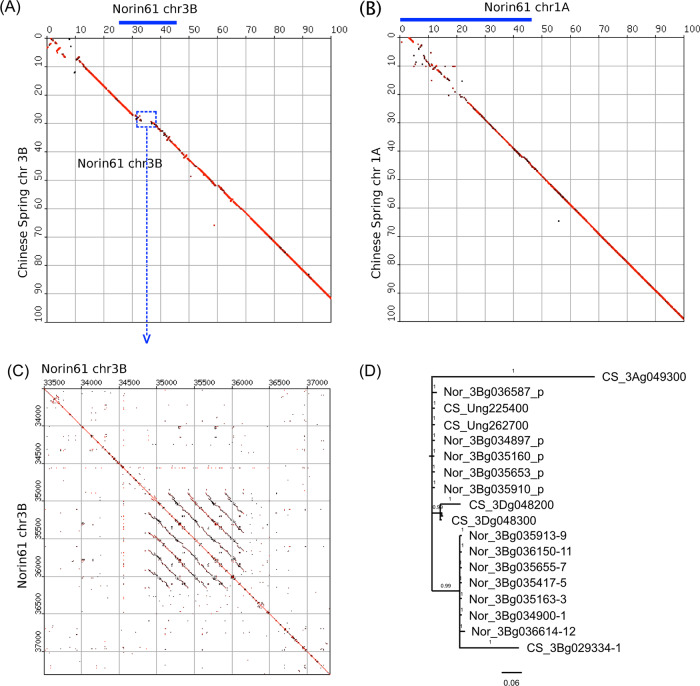
Two introgressions (chromosome 3B, 25–46.3 Mb, and chromosome 1A, 0–41.7 Mb) were of particular interest because their sequence conservation between Norin 61 and CS was very low (Fig. 3A, B). The 3B introgression spans about 21 Mb, contains 61 private genes and includes an ≈3.9 Mb unique segment that is completely absent from CS (Fig. 3A). Structurally, this segment is composed of five direct sequence repeats arrayed in tandem, each one ≈250 kb in size (Fig. 3C). The formation of this array is probably the result of recurrent unequal crossing-over events. The sequence identity between individual repeat units is >96%. Interestingly, 27 genes in the 3B introgression candidate belong to two large gene families—family 3B_25-46-1 and family 3B_25-46-2, which contain 12 and 15 members, respectively. All genes belonging to family 3B_25-46-1 are located on the unique 3.9 Mb, five of them inside the tandem repeat array. Because of repetitive nature of the region in Norin 61, genes cluster in two clades with high copy number, with most sequences within the clades being identical (Fig. 3D). Genes of this family encode short peptides of 84–88 amino acids that are rich in positively charged amino acids (Supplementary Fig. S4A). Because of their lack of homology to characterized sequences, their function is unclear. Family 3B_25-46-2 also encodes short sequences with cysteine-rich motifs and a signal peptide, named the pathogenesis-related 13 (PR-13) gene family or thionins (Goyal and Mattoo 2014). The clade highlighted in Supplementary Fig. S4B contains 12 genes in Norin 61 in contrast to the six copies present in the CS region carrying the introgression on 3B (Supplementary Fig. S4B). The even distribution of Norin 61 and CS genes in the clade suggests that there was no preferential amplification of one or a few founder copies. Similar to the 3B introgression described above, we identified a cluster of putative PR-13 genes on the 1A introgression, which in Norin 61 also comprised a large number of gene copies (Supplementary Fig. S4C). The shorter branch lengths may suggest that the Norin 61 PR-13 copies could have amplified more recently than the CS homologs.
Fig. 3.
Analysis of chromosomal introgression in Norin 61. (A) Dot plot comparison of the terminal 100 Mb of chromosome 3B from Norin 61 (horizontal) and CS. Norin 61 contains an introgression at approximately position 25–46 Mb (indicated with a blue bar). Discontinuity in the diagonal denotes lack of sequence conservation in that region, and Norin 61 contains a unique ≈3.9 Mb segment (shift to the right in the dashed blue rectangle). (B) Dot plot comparison of the terminal 100 Mb of chromosome 1A from Norin 61 (horizontal) and CS. The introgression was identified through the analysis of repetitive sequences and ranges between 0 and 47.7 Mb (indicated with a blue bar). Sequences are particularly diverse in the 5′ terminal 25 Mb. (C) Self dot plot alignment of the 3.9 Mb 3B segment that is unique to Norin 61. The alignment of the segment with itself identifies a large array of five tandem repeats (series of parallel lines). (D) Phylogenetic analysis of proteins encoded by gene family 3B_25-46-1 whose members are identified in the repeat array shown in (C). Note that the individual genes cluster in two clades that cannot be further resolved because of the high similarity of the protein sequences.
Identification of flowering-time-related genes and their variants
Flowering time is a major target of selection in wheat and other crop species (Eshed and Lippman 2019, Hyles et al. 2020). The gene family of FLOWERING LOCUS T (FT), which has been denoted as Vrn3 or Vrn-1 in wheat, plays a central role in its regulation (Kikuchi et al. 2009, Halliwell et al. 2016). By homology search, we mined Asian wheat homeologs of flowering-related genes (Table 2), identifying 45 and 62 FT homeologs in the Norin 61 and CS assemblies, respectively (Table 2). The FT family members clustered in 12 subfamilies (Fig. 4A, Supplementary Fig. S5). Among these, FT1 and FT3 play important roles in controlling flowering time in grasses (Kikuchi et al. 2009, Halliwell et al. 2016). In both Norin 61 and CS, all three homeologs in the FT3 subfamily are present in a single copy. Its ortholog in barley, HvFT3, is colocalized with its major flowering-time locus Ppd-H2 (Faure et al. 2007, Kikuchi et al. 2009). In the following section below, we will focus on the variation in the FT1 subfamily.
Table 2.
Flowering time-related gene homologs between Norin 61 and CS
| Accession | CS |
Norin 61 |
Notes on potential extra copies in Norin 61 | |||||
|---|---|---|---|---|---|---|---|---|
| Gene symbol | Reference | A | B | D | A | B | D | |
| TaFT1-1/VRN-3-1 | Halliwell et al. (2016) | TraesCS7A02G115400 | TraesCS7B02G013100 | TraesCS7D02G111600 | TraesNOR7A01G123700 | NA | TraesNOR7D01G129000 | See main text. Copy number variation of the B homoeologs in Norin 61 as well as in CS |
| TaFT2-1* | Halliwell et al. (2016) | TraesCS3A02G143100 | TraesCS3B02G162000 | TraesCS3D02G144500 | TraesNOR3A01G166100 | TraesNOR3B01G189800 | TraesNOR3D01G160200 | |
| TaFT3-1 | Halliwell et al. (2016) | TraesCS1A02G338600 | TraesCS1B02G351100 | TraesCS1D02G340800 | TraesNOR1A01G362800 | TraesNOR1B01G382200 | TraesNOR1D01G378400 | |
| TaFT4-1 | Halliwell et al. (2016) | TraesCS2A02G132300 | TraesCS2B02G154800 | TraesCS2D02G134200 | TraesNOR2A01G150300 | TraesNOR2B01G174600 | TraesNOR2D01G153400 | |
| TaFT5-1 | Halliwell et al. (2016) | TraesCS5A02G546900 | TraesCS4B02G379100 | TraesCSU02G130900 | TraesNOR5A01G583800 | TraesNOR4B01G414800 | TraesNORUn01G040600 | There might be multiple copies on the B genome of Norin 61 |
| TaFT5-2 | Halliwell et al. (2016) | TraesCS5A02G546800 | TraesCS4B02G380600 | TraesCSU02G130800 | TraesNOR5A01G583700 | TraesNOR4B01G416300 | TraesNORUn01G040500 | A second Norin 61D homoeolog was not annotated |
| TaFT5-4 | Halliwell et al. (2016) | NA | NA | TraesCSU02G130700 | NA | NA | TraesNORUn01G040400 | |
| TaFT6-1 | Halliwell et al. (2016) | TraesCS6A02G160200 | TraesCS6B02G191200 | TraesCS6D02G152500 | TraesNOR6A01G175800 | TraesNOR6B01G210900 | TraesNOR6D01G177500 | |
| TaFT7-1* | Halliwell et al. (2016) | TraesCS5A02G154600 | TraesCS5B02G152800 | TraesCS5D02G159500 | TraesNOR5A01G167400 | TraesNOR5B01G169100 | TraesNOR5D01G173200 | |
| TaFT8-1 | Halliwell et al. (2016) | TraesCS2A02G536600 | TraesCS2B02G511400 | TraesCS2D02G538000 | TraesNOR2A01G582200 | TraesNOR2B01G557400 | TraesNOR2D01G585300 | High copy number |
| TaFT8-2 | Halliwell et al. (2016) | TraesCS2A02G485000 | TraesCS2B02G567400 | TraesCS2D02G485100 | TraesNOR2A01G524400 | TraesNOR2B01G619000 | NA | High copy number |
| TaFT8-3 | Halliwell et al. (2016) | TraesCS2A02G536900 | TraesCS3B02G015800 | TraesCS2D02G538100 | NA | TraesNOR3B01G017500 | NA | |
| TaFT8-4 | Halliwell et al. (2016) | TraesCS2A02G536700 | TraesCSU02G045800 | TraesCS3D02G011600 | NA | TraesNOR3B01G017400 | NA | |
| TaFT9-1 | Halliwell et al. (2016) | TraesCS2A02G347000 | TraesCS2B02G365300 | TraesCS2D02G345700 | TraesNOR2A01G378700 | TraesNOR2B01G400600 | TraesNOR2D01G376200 | |
| New | Halliwell et al. (2016) | TraesCS2A02G346900 | TraesCS2B02G365200 | TraesCS2D02G345600 | TraesNOR2A01G378600 | TraesNOR2B01G400500 | TraesNOR2D01G376100 | |
| TaFT10-1 | Halliwell et al. (2016) | TraesCS5A02G297300 | TraesCS5B02G296600 | TraesCS5D02G304400 | TraesNOR5A01G314900 | TraesNOR5B01G321800 | TraesNOR5D01G327800 | |
| TaFT11-1 | Halliwell et al. (2016) | NA | TraesCS4B02G073800 | TraesCS4D02G072300 | NA | TraesNOR4B01G083000 | TraesNOR4D01G076400 | |
| TaFT12-1 | Halliwell et al. (2016) | TraesCS6A02G214400 | TraesCS6B02G244400 | TraesCS6D02G197100 | TraesNOR6A01G234900 | TraesNOR6B01G268400 | TraesNOR6D01G227500 | |
| TaMFT1 | Higuchi et al. (2013) | TraesCS7A02G229400 | TraesCS7B02G195200 | TraesCS7D02G230000 | TraesNOR7A01G248800 | TraesNOR7B01G154000 | TraesNOR7D01G258900 | |
| TaMFT2-1 | Higuchi et al. (2013) | TraesCS3A02G006600 | TraesCS3B02G010100 | TraesCS3D02G004100 | TraesNOR3A01G008900 | TraesNOR3B01G009400 | TraesNOR3D01G000600 | |
| TaMFT2-2 | Higuchi et al. (2013) | NA | TraesCS3B02G007400 | NA | NA | TraesNOR3B01G009500 | NA | |
| TaTFL1 | Faure et al. (2007) | TraesCS5A02G128600 | TraesCS5B02G127600 | TraesCS5D02G136300 | TraesNOR5B01G141700 | TraesNOR5B01G141700 | TraesNOR5D01G148800 | |
| TaTFL2 | Higuchi et al. (2013) | TraesCS4A02G409200 | TraesCS4B02G307600 | TraesCS4D02G305800 | TraesNOR4A01G399300 | TraesNOR4B01G335300 | TraesNOR4D01G327600 | |
| TaTFL3 | Higuchi et al. (2013) | TraesCSU02G202000 | TraesCS2B02G310700 | TraesCS2D02G292000 | TraesNOR2A01G323700 | TraesNOR2B01G343700 | TraesNOR2D01G320200 | |
| Vrn-1 | Yan et al. (2003), Danyluk et al. (2003) and Murai et al. (2003) | TraesCS5A02G391700 | TraesCS5B02G396600 | TraesCS5D02G401500 | TraesNOR5A01G416600 | TraesNOR5B01G433300 | TraesNOR5D01G434800 | |
| ZCCT-1 (Vrn2-1) | Yan et al. (2004) | TraesCS5A02G541300 | TraesCS4B02G372700 | TraesCS4D02G364500 | TraesNOR5A01G578600 | TraesNOR4B01G408300 | TraesNORUn01G034600 | |
| ZCCT-2 (Vrn2-2) | Yan et al. (2004) | TraesCS5A02G541200 | NA | TraesCS4D02G364400 | TraesNOR5A01G578700 | NA | TraesNORUn01G034500 | |
| TaGRP-2 | Xiao et al. (2014) | TraesCS4A02G293000 | TraesCS4B02G020300 | TraesCS4D02G018500 | TraesNOR4A01G307500 | TraesNOR4B01G023800 | TraesNOR4D01G020000 | |
| TaVRT-2 | Kane et al. (2005) | TraesCS7A02G175200 | TraesCS7B02G080300 | TraesCS7D02G176700 | TraesNOR7A01G188500 | TraesNOR7B01G090400 | TraesNOR7D01G200800 | |
| TaGI-1 | Zhao et al. (2005) | TraesCS3A02G116300 | TraesCS3B02G135400 | TraesCS3D02G118200 | TraesNOR3A01G128300 | TraesNOR3B01G162100 | TraesNOR3D01G131800 | |
| WSOC-1 | Shitsukawa et al. (2007) | TraesCS4D02G341700 | TraesCS4B02G346700 | TraesCS5A02G515500 | TraesNOR5A01G549300 | TraesNOR4B01G377200 | TraesNOR4D01G366500 | A second Norin 61D homoeolog was not annotated |
| WPCL-1 | Mizuno et al. (2012) and Mizuno et al. (2016) | TraesCS3A02G526600 | TraesCS3B02G594300 | TraesCS3D02G531900 | TraesNOR3A01G575200 | TraesNOR3B01G660800 | TraesNOR3D01G579900 | |
| WCO-1 | Shimada et al. (2009) | TraesCS1A02G220300 | TraesCS1B02G233600 | TraesCS1D02G221800 | TraesNOR1A01G240300 | TraesNOR1B01G254700 | TraesNOR1D01G249900 | |
| TaHD-1 (CO-2) | Nemoto et al. (2003) | TraesCS6A02G289400 | TraesCS6B02G319500 | TraesCS6D02G269500 | TraesNOR6A01G314800 | TraesNOR6B01G348100 | TraesNOR6D01G304400 | |
| TaPHYC | Mizuno et al. (2012) and Mizuno et al. (2016) | TraesCS5A02G391300 | TraesCS5B02G396200 | TraesCS5D02G401000 | TraesNOR5A01G416100 | TraesNOR5B01G432800 | TraesNOR5D01G434300 | |
| Ppd-1 (PRR) | Turner et al. (2005), and Beales et al. (2007) | TraesCS2A02G081900 | NA | TraesCS2D02G079600 | TraesNOR2A01G096300 | NA | TraesNOR2D01G091900 | There might be multiple copies of the B homoeologs in Norin 61 as well as in CS |
| TaAP1 | Murai et al. (2003) | TraesCS5A02G391700 | TraesCS5B02G396600 | TraesCS5D02G401500 | TraesNOR5A01G416600 | TraesNOR5B01G433300 | TraesNOR5D01G434800 | Splicing variant in CS. Splicing variant in Norin 61 was not annotated |
| TaAP2 | Ning et al. (2013) | TraesCS2A02G514200 | TraesCS2B02G542400 | TraesCS2D02G515800 | TraesNOR2A01G556000 | TraesNOR2B01G592300 | TraesNOR2D01G559400 | |
| TaAGL12 | Zhao et al. (2006) | TraesCS7A02G260900 | TraesCS7B02G158900 | TraesCS7D02G261900 | TraesNOR7A01G280100 | TraesNOR7B01G185700 | TraesNOR7D01G293000 | A second Norin 61D homoeolog was not annotated |
| TaAGL22 | Zhao et al. (2006) | TraesCS3A02G434900 | NA | NA | TraesNOR3A01G466500 | TraesNOR3B01G517700 | TraesNOR3D01G458800 | |
| TaAGL33 | Zhao et al. (2006) | TraesCS3A02G435000 | TraesCS3B02G470000 | TraesCS3D02G428000 | TraesNOR3A01G466400 | TraesNOR3B01G517500 | TraesNOR3D01G458600 | |
| TaAGL41 | Sharma et al. (2016) | TraesCS3A02G434400 | TraesCS3D02G427700 | TraesCS3B02G469700 | TraesNOR3A01G465900 | TraesNOR3B01G517500 | TraesNOR3D01G458500 | |
| TaAGL42 | Zhao et al. (2006) | TraesCS3A02G432900 | NA | TraesCS3D02G427900 | TraesNOR3A01G466500 | NA | TraesNOR3D01G457000 | |
| TaAG1 | Meguro et al. (2003) | TraesCS1A02G125800 | TraesCS1B02G144800 | TraesCS1D02G127700 | TraesNOR1A01G139600 | TraesNOR1B01G161700 | TraesNOR1D01G145300 | Potential another copies on chr Un |
| WAG2/TaAGL39 | Zhao et al. (2006) | TraesCS3A02G314300 | TraesCS3B02G157500 | TraesCS3D02G140200 | TraesNOR3A01G153000 | NA | TraesNOR3D01G155900 | |
Fig. 4.
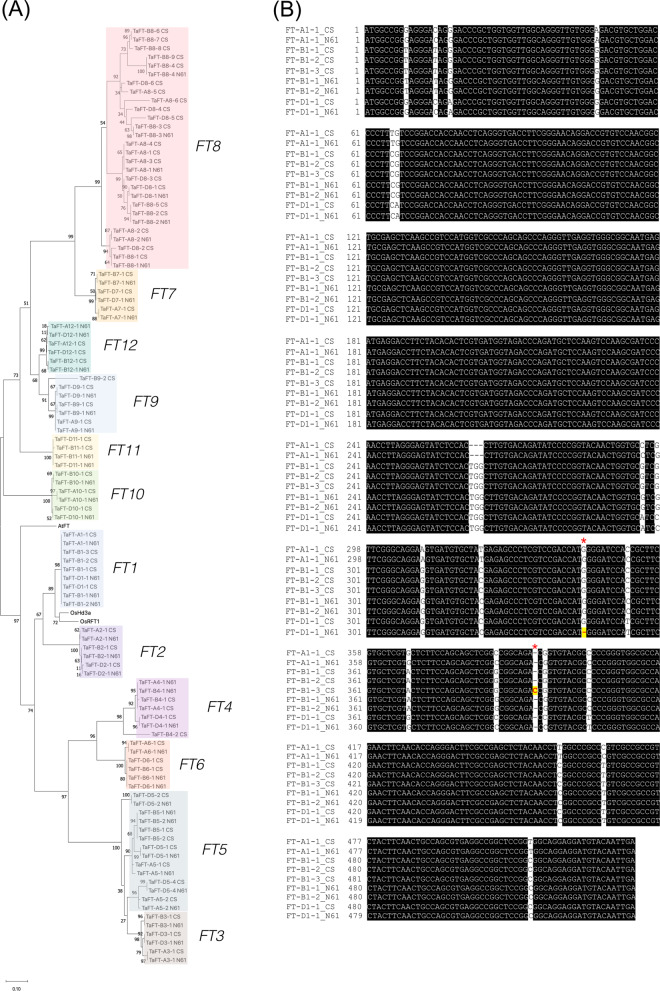
(A) Maximum-likelihood tree of 108 FT homologs based on amino acid sequences. The bootstrap values are shown on the branches. N61 and CS stand for Norin 61 and CS, respectively. (B) Multiple sequence alignment of FT1 genes on CS and N61. The red stars highlight the positions with the 1-bp deletion in FT-D1 of Norin 61 and the 1-bp insertion in FT-B1-3 of CS. The black boxes indicate the conserved sequences. The white regions show the SNPs or indels between sequences.
Norin 61 had two FT-B1 (the B-genome homeolog of FT1, synonymous to Vrn-B3) copies as described above. Nucleotide phylogeny of the FT1 subfamily demonstrated the close relatedness of the two copies, and we named FT-B1-1 and FT-B1-2 (Supplementary Fig. S6, Table 2). The two contigs containing the FT-B1 genes (contig 78046 and 91980, each about 19 kb) were highly similar, only differing by a 48-bp tandem duplication (two copies in 78046, three in 91980) and two mismatches outside of the coding regions. To confirm the existence of multiple copies, we compared the resequencing coverage values of the three FT1 homeologs with that for Adh-1, a single-copy gene. The coverage of FT-A1, FT-D1 and three Adh-1 homeologs was all comparable (Table 3). By contrast, increased coverage was found at both FT-B1 copies (Table 3), supporting its increased copy number.
Table 3.
Resequencing coverage of Adh-1 and FT1 loci on CS and Norin 61
| Genotype | Gene | Homeolog coverage |
||
|---|---|---|---|---|
| A | B | D | ||
| CS | Adh-1 | 7.8 | 7.4 | 9.4 |
| FT-B1 | 9.7 | 22.8 | 8.9 | |
| Norin 61 | Adh-1 | 9.1 | 9.7 | 8.0 |
| FT-B1 | 7.6 | 13.4 | 11.5 | |
| 14.3 | ||||
Compared with the Adh-1 gene, the mean coverage of the FT1 copies is less homogeneous (higher coverages are highlighted in bold), hinting at a possible collapse of multiple (even more than two) copies in the B homeolog in both the Norin and CS assemblies. The coverage values of the two Norin 61 FT-B1 copies detected in the assembly are represented on two separate lines.
Unexpectedly, we also noticed the higher coverage on the B homeolog of CS despite a single copy (TraesCS7B02G013100) was reported in the RefSeq v1.1 [The International Wheat Genome Sequencing Consortium (IWGSC) 2018]. When the improved CS assembly was interrogated (IWGSC RefSeq v2.0 and Alonge et al. 2020), we indeed identified three copies of FT-B1 on chromosome 7B (Supplementary Table S3), one of which showed a frameshift mutation (Fig. 4B). In the phylogenetic tree, the three CS FT-B1 copies clustered with the two Norin 61 copies of FT-B1 (Supplementary Fig. S6).
We next examined the Norin 61 FT-D1 (the D-genome homeolog of FT1, synonymous to Vrn-D3) and found that a 1-bp deletion introduced a frameshift in Norin 61. The predicted protein lacked the conserved segment B that is necessary for the function of FT proteins (Fig. 4B) (Ahn et al. 2006).
Several known haplotypes of the FT-A1 (the A-genome homeolog of FT1, synonymous to Vrn-A3) gene are suggested to be functional variations in tetraploid emmer wheat and in bread wheat (Nishimura et al. 2018, Chen et al. 2020). We examined the sequence variation among the 10+ Wheat Genomes Project wheat cultivars and found two alleles—Vrn-A3b and Vrn-A3c (Table 4). Both alleles were well represented (Vrn-A3c present in 4 of 11 RQAs and in 3 of 5 scaffold-level assemblies). This showed that these are common alleles in the cultivars used in the 10+ Wheat Genomes Project. Interestingly, there is an association of the Vrn-A3 alleles and growth habit: all surveyed spring wheat cultivars except Cadenza tend to have the deletion allele Vrn-A3b, and all winter wheats except SY Mattis have the non-deletion allele Vrn-A3c (χ2 = 6.1315, P = 0.01328, Pearson’s chi-squared test with Yates’ continuity correction).
Table 4.
Variation at FT-A1 (also denoted as Vrn-A3) and the growth habit
| Cultivar |
Vrn-A3
|
Habita | ||
|---|---|---|---|---|
| Size | Type | Allele | ||
| Norin 61 | 799 | del | Vrn-A3b | Facultative spring |
| ArinaLrFor | 830 | Ins | Vrn-A3c | Winter |
| CS | 799 | Del | Vrn-A3b | Spring |
| Jaggar | 830 | Ins | Vrn-A3c | Winter |
| Julius | 830 | Ins | Vrn-A3c | Winter |
| LongReach Lancer | 799 | Del | Vrn-A3b | Spring |
| CDC Landmark | 799 | Del | Vrn-A3b | Spring |
| Mace | 799 | Del | Vrn-A3b | Spring |
| CDC Stanley | 799 | Del | Vrn-A3b | Spring |
| SY Mattis | 799 | Del | Vrn-A3b | Winter |
| PI190962 (spelt wheat) | 830 | Ins | Vrn-A3c | Winter |
| Cadenza | 830 | Ins | Vrn-A3c | Spring |
| Paragon | 799 | Del | Vrn-A3b | Spring |
| Robigus | 830 | Ins | Vrn-A3c | Winter |
| Claire | 830 | Ins | Vrn-A3c | Winter |
| Weebill 1 | 799 | Del | Vrn-A3b | Spring |
Data from Walkowiak et al. (2020).
The photoperiod response is another key mechanism that regulates flowering. In wheat, it is mainly regulated by three homeologous genes: Ppd-D1 (previously designated Ppd1), Ppd-B1 (Ppd2) and Ppd-A1 (Ppd3), each on their respective group-2 chromosome (Welsh et al. 1973, Scarth and Law 1983). Ppd genes encode pseudoresponse regulator (PRR) proteins that control the circadian clock. Because of a 2,089-bp deletion in the promoter region that includes the repressor (Turner et al. 2005, Beales et al. 2007), the Ppd-D1a allele confers early flowering with high expression of Ppd-D1 itself and the downstream FT1/Vrn3 genes. This allele has been widely used for breeding in southern Europe and the southern part of East Asia (Worland 1996, Guo et al. 2010). Consistent with previous polymerase chain reaction (PCR) genotyping (Seki et al. 2011), we confirmed the deletion characteristic for Ppd-D1a in Norin 61 in contrast to CS (Fig. 5). We did not use the CS Ppd-B1 sequence for the comparison because of the increased copy number in CS (Díaz et al. 2012). Although both Norin 61 and CS show a sensitive phenotype with Ppd-A1b, the two lines displayed independent indels in intronic and promoter regions of the Ppd-A1 gene, as reported previously (Beales et al. 2007) (Fig. 5).
Fig. 5.
Sequence features of Ppd-1 genes with the 5-kb upstream region extracted from the CS and Norin 61 pseudomolecules. (A) Both CS and Norin 61 have the photoperiod-sensitive Ppd-A1b allele. Three insertions/deletions were detected. (B) The red region spans the 2,089-bp deletion of the Ppd-D1a allele of Norin 61 compared with the Ppd-D1b allele of CS. The boxes indicate CDSs. The red lines show the sequence polymorphisms between CS and Norin 61 assemblies.
Syntenic analysis at the FHB-resistant loci
FHB resistance is a major target of breeding in East Asia and other regions that have relatively high humidity at harvest season. With multiple quantitative trait loci (QTLs) contributing to the resistance, Niwa et al. (2018) reported that Norin 61, Sumai 3, Ning 7840 and other related cultivars had several FHB-resistant QTLs, including Fhb1. Although the TaPFT gene encoding a pore-forming toxin-like protein was first suggested to be responsible for Fhb1 (Rawat et al. 2016), the TaHRC gene encoding a nuclear protein was supported in subsequent studies (Li et al. 2019, Su et al. 2019). The CS chromosomal region containing Fhb1 falls within a region (0–32.5 Mb on chromosome 3B) identified as an introgression candidate (Walkowiak et al. 2020). A comparison of the CS Fhb1 region to the orthologous sequence in Norin 61 identified a ≈340-kb region with very little sequence homology (Fig. 6A). The Fhb1 region in the Norin 61 assembly showed very high sequence conservation with the bacterial artificial chromosome (BAC)-based contigs of CM-82036 derived from Sumai 3 (Schweiger et al. 2016) (Fig. 6B), providing at the same time an independent validation of the high quality of the Norin 61 assembly. The gene annotation of the Norin 61 orthologs (projected CS models, Walkowiak et al. 2020) identifies 20 protein-coding regions. However, given the remarkably low sequence conservation with CS at this locus, we annotated genes in the interval de novo and identified 50 additional genes (Fig. 6C). In contrast to the susceptible allele of CS and Clark, the Norin 61 assembly lacked the region, including the start codon in the third exon of the TaHRC gene (Fig. 6D, Supplementary Fig. S7), which is equivalent to resistant alleles of Ning 7840 and Sumai 3 (Li et al. 2019, Su et al. 2019). The occurrence of the deletion in the Norin 61 assembly, coupled with its FHB resistance, corroborates the role of the deletion in TaHRC for Fusarium resistance.
Fig. 6.
Characterization of the Fhb1 locus in CS and in FHB-resistant varieties. (A) Sequence dot plot of the region surrounding Fhb1 in CS (X-axis chromosome 3B: 7.99–8.88 Mb) and Norin 61 (Y-axis, chromosome 3B: 11.41–12.37 Mb). The gap in the bottom part of the diagonal denotes the region with different sequence composition (spanning about 340 kb). (B) Dot plot comparing CM-82036 derived from Sumai 3 (X-axis, from Schweiger et al. 2016; BAC assembly, GenBank accession: MK450312) and Norin 61 (Y-axis, chromosome 3B: 11.41–12.37 Mb) showing the almost complete identity of the sequence. (C) Annotation of the ≈390 kb Norin 61 region containing Fhb1 (chromosome 3B: 11.89–12.31 Mb). The newly annotated genes (red boxes) and the span of the Norin 61 region differing from CS are shown below the published annotation of Norin 61 (Walkowiak et al. 2020, first three tracks). The TaHRC gene is highlighted in purple. (D) Schematic structure of the deletion in the TaHRC gene. The boxes indicate the third exon. The red dashed line shows the sequence polymorphisms between CS and Norin 61 assemblies.
The short arm of chromosome 2D also harbors a QTL for FHB resistance and mycotoxin accumulation and is closely linked with the Reduced height 8 (Rht8) gene (Korzun et al. 1998, Chai et al. 2019, Supplementary Table S4). The gene for multidrug-resistant-associated protein (TaMRP-D1) was postulated as a candidate gene for this QTL (Handa et al. 2008). Because Norin 61 and CS as well as Sumai 3 have this allele of TaMRP-D1 (Niwa et al. 2018) and a susceptible haplotype, we surveyed for allelic variation at TaMRP-D1 and Rht8 loci in other cultivars sequenced in the 10+ Wheat Genomes Project (Supplementary Table S4). We found a tight link between one allele of TaMRP-D1 and the semi-dwarf and dense spike allele of Rht8 in the FHB-susceptible haplotype, which is consistent with a linkage drag potentially caused by the selection of the semi-dwarf allele (Niwa et al. 2018). By contrast, we observed that the TaMRP-D1 variants were linked to the FHB and mycotoxin resistance in the cultivars with the non-semi-dwarf allele of Rht8. The efficient use of this FHB-QTL on the 2DS region as breeding resource requires the breaking of the linkage drag between the FHB-QTL and semi-dwarfism (Basnet et al. 2012, Niwa et al. 2018, Fabre et al. 2020).
Other functional variations between Norin 61 and CS responsible for adaptation to East Asian environments
The three homeologous R-1 genes (on group 3 chromosomes) control grain color and preharvest sprouting (Warner et al. 2000, Flintham et al. 2002, Himi et al. 2002, Lin et al. 2016, Vetch et al. 2019). A plant homozygous for the recessive alleles of R-A1a, R-B1a and R-D1a (r2, r3 and r1, respectively, in a former notation) has white grain. The presence of at least one dominant allele, i.e. R-A1b, R-B1b or R-D1b (R2, R3 and R1, respectively), turns the grain color to red (McIntosh et al. 2013). The Tamyb10 genes, which encode R2R3-type MYB domain proteins, are candidates for R-1 and are used as a marker to detect dominant R-1 alleles (Himi and Noda 2005, Himi et al. 2011). Although both CS and Norin 61 are red grain varieties, their underlying genotypes are different (Himi et al. 2011, Kojima et al. 2017). The sequence analysis suggested that Norin 61 retains the functional (i.e. dominant) allele at all three homeologous loci (Fig. 7, Supplementary Fig. S8). By contrast, Tamyb10-A1 of CS contained a deletion of the first half of the R2 repeat, including the start codon (Fig. 7A), as well as a synonymous mutation and an intronic mutation. Tamyb10-B1 of CS contained a disrupting 19-bp deletion in the third exon (Fig. 7B). Tamyb10-D1 of both CS and Norin 61 were identical dominant alleles (Fig. 7C).
Fig. 7.
Schematic features of Tamyb10 sequences of CS and Norin 61. (A) Tamyb10-A1. CS has the recessive allele (Tamyb10-A1a), characterized by a large deletion, including the start codon. Norin 61 has the (wild type) dominant allele (Tamyb10-A1b). (B) Tamyb10-B1. The CS has the recessive allele (Tamyb10-B1a) has a 19-bp deletion and Norin 61 has the dominant Tamyb10-B1b allele. (C) Tamyb10-D1. Both CS and Norin 61 have the identical dominant allele (Tamyb10-D1b). The boxes indicate CDSs. The red dashed lines show the sequence polymorphisms between CS and Norin 61 assemblies.
Semi-dwarf mutations of wheat are thought to be advantageous to increase yield in short growing seasons and to confer lodging resistance (Miralles and Slafer 1995, Asplund et al. 2012, Jobson et al. 2019). Rht-B1b (formerly designated Rht1) and Rht-D1b (Rht2) genes are orthologous to the GIBBERELLIN INSENSITIVE (GAI) of Arabidopsis (Peng et al. 1999, Kojima et al. 2017) and confer semi-dwarfism in wheat. Each allele contains a single-nucleotide polymorphism (SNP) that results in a stop codon within the DELLA domain, and translational reinitiation after this polymorphism produces a gain-of-function protein that represses gibberellin signaling (Peng et al. 1999). Both variants are present in the Japanese cultivar Norin 10 and were used during the green revolution to confer lodging resistance to wheat grown in fertilized conditions (Peng et al. 1999). The genome analysis confirmed that Norin 61 possesses a semi-dwarf Rht-D1b allele with the SNP in contrast to CS (Supplementary Fig. S9). We further genotyped other cultivars in the next section.
Norin 61 carries the Glu-D1f allele that produces the 2.2 + 12 subunits of the high-molecular-weight (HMW) glutenin, first described by Payne et al. (1983) in Japanese hexaploid wheat varieties. HMW glutenin proteins are integral to end-use quality characteristics through the formation of the gluten matrix. The Glu-D1 locus contains two genes, each encodes for an x and y subunits, that together constitute the HMW glutenin types (i.e. 2.2x + 12y or 2x + 12y). The 2.2 + 12 allele is also referred to as the f allele, and 2 + 12 as the a allele (Payne and Lawrence 1983). The very low electrophoretic mobility of the 2.2x protein on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) suggested that the protein was much larger in size than any other hexaploid HMW glutenin subunits known at the time. It was then hypothesized that 2.2x arose from an unequal crossing over in the central repeat domain of 2x that increased the size of the gene and that this novel allele is very recent, within the modern breeding history. This is supported by failure to identify 2.2 + 12 in surveys of Glu-D1 SDS-PAGE mobilities in Ae. tauschii (Lagudah and Halloran 1988, William et al. 1993, Mackie et al. 1996), the D genome donor to hexaploid wheat, and that 2.2 + 12 is present primarily only in Japanese germplasm (Nakamura et al. 1999, Yanaka et al. 2016) indicated that 2.2x arose from recent mutation within hexaploid wheat. Further evidence for this hypothesis was provided by the full gene CDSs comparing 2.2x to 2x that showed 2.2x has a perfect 396 nucleotide duplication in the central repeat region (Wan et al. 2005).
Within the 257-kb region around the Glu-D1 locus, we surveyed SNPs. There exists only 10 SNPs differentiating 2.2 + 12 from 2 + 12 of CS and other wheat varieties (Long Reach Lancer, ArinaLrFor, Mace, SY Mattis) (Supplementary Fig. S10A). The number was much less than the 72 SNPs differentiating the 2 + 12 sequences among the wheat varieties CS, Long Reach Lancer, ArinaLrFor, Mace and SY Mattis in the same region. Analysis with MUMmer showed no large-scale structural differences between 2.2 + 12 and 2 + 12 of CS, aside from 11 gaps ranging between 33 and 5,381 bp. Gaps were likely assembly artifacts, evidenced by Norin 61 reads aligning to the CS ‘gap’ regions. Due to the highly repetitive nature of the central repeat domain and the limitations of short-read sequencing, we were unable to directly detect the 396 duplication in 2.2x relative to 2x. However, with the genome assembly of Norin 61, we support here that the unique 2.2 + 12 in Japanese germplasm arose from a recent mutation of 2 + 12, as well as identify several molecular variants that could be used as a high-throughput molecular marker in breeding to differentiated 2.2 + 12 from the common 2 + 12 (Supplementary Fig. S10B).
Functional variations in other Japanese cultivars
To examine whether the variants found in Norin 61 are shared with other Japanese cultivars, PCR testing for markers of eight loci was performed on three Japanese cultivars (Shinchunaga, Norin 26 and Shirasagi komugi), as well as Norin 61 and CS (Table 5). As described above, Shinchunaga is a pure-line selection from a landrace and was used as a parent of Norin 61. Norin 26 is also derived from Shinchunaga and was released in 1937. It was the second most cultivated variety after Norin 61 in the 1950s and 1960s and was popular for Japanese udon noodles. Shirasagi komugi is also a cultivar of Shinchunaga lineage (released in 1956) with earlier maturing than Norin 61. The analysis showed that the same alleles present in Norin 61 were found in these three Japanese cultivars at three Ppd1 and three Vrn1 loci (Table 5, Saisho et al. 2011, Nishida et al. 2013). The semi-dwarf allele of Rht-D1b (Kojima et al. 2017) of Norin 61 was not found in the other cultivars; instead, Norin 26 and Shirasagi komugi possessed another semi-dwarf allele of Rht-B1b. By contrast, CS possesses different genotypes at Ppd and Rht loci (Table 5).
Table 5.
Genotypes of Japanese cultivars at loci regulating flowering time and plant height
| Cultivar name | Status | Ppd-A1 | Ppd-B1 | Ppd-D1 | Vrn-A1 | Vrn-B1 | Vrn-D1 | Rht-B1 | Rht-D1 |
|---|---|---|---|---|---|---|---|---|---|
| Komugi Norin 61 | Breeders line | ppd-A1 | ppd-B1 | Ppd-D1 | vrn-A1 | vrn-B1 | Vrn-D1 | Rht-B1a | Rht-D1b |
| Shinchunaga | Pure selected line | ppd-A1 | ppd-B1 | Ppd-D1 | vrn-A1 | vrn-B1 | Vrn-D1 | Rht-B1a | Rht-D1a |
| Komugi Norin 26 | Breeders line | ppd-A1 | ppd-B1 | Ppd-D1 | vrn-A1 | vrn-B1 | Vrn-D1 | Rht-B1b | Rht-D1a |
| Shirasagi komugi | Breeders line | ppd-A1 | ppd-B1 | Ppd-D1 | vrn-A1 | NA | Vrn-D1 | Rht-B1b | Rht-D1a |
| CS | Standard line | ppd-A1 a | ppd-B1 | ppd-D1 | vrn-A1 | vrn-B1 | Vrn-D1 | Rht-B1a | Rht-D1a |
Ppd-D1 and ppd-D1 correspond to Ppd-D1a and Ppd-D1b in Fig. 5, respectively.
Data from Nishida et al. (2013).
Discussion
In this study, we analyzed the chromosome-scale assembly of the Japanese wheat Norin 61 as the sole Asian cultivar in the 10+ Wheat Genomes Project. In addition to using standard statistics, such as N50, remapping, KAT, and BUSCO analysis, we validated the quality of the assembly by showing synteny to more than an Mb of the BAC contigs encompassing the resistant Fhb1 haplotype on chromosome 3B. We were able to identify and confirm SNPs and indel mutations of up to kilobase scales of functional variants, including Ppd, TaHRC, Rht and Tamyb10. These data supported the high quality of the assembly for analysis of functional variations. Our manual inspection identified copy-number variations in FT-B1, suggesting that tandem duplications remain a major challenge in wheat genomics (Alonge et al. 2020).
Of the accessions whose karyotypes were compared, Norin 61 had the karyotype most similar to that of CS (Walkowiak et al. 2020). In contrast to the modern introgressions from different species that are seen in Western cultivars used in the 10+ Wheat Genomes Project, none of the known ancestors of Norin 61 are foreign species. The number of unique chromosomal segments in Norin 61 (45 regions) was less than the 54 observed in CS. These cytogenetic and repetitive element analyses suggested that Norin 61 as well as CS can serve as useful reference genomes for functional and evolutionary analysis.
Analysis of the distribution pattern of repetitive sequences on the chromosomes of the 10 wheat genomes identified hundreds of chromosomal segments with unique or rare retrotransposon insertions (>20 Mb) in A, B and D chromosomes that were hypothesized to be introgressions (Walkowiak et al. 2020). Among these are introgressions introduced during modern breeding of Western cultivars that originate from Triticum timopheevii, Aegilops ventricosa and Thinopyrum ponticum and that confer disease resistance (Walkowiak et al. 2020). Introgression from wild diploid and tetraploid species with A, B or D genomes may be common (Zhou et al. 2020). Norin 61 contains 45 unique chromosomal regions, and we analyzed two of these in detail. We found that these Norin 61 regions contain numerous genes that lack homology with CS genes. In both regions, Norin 61 contains tandem copies of the PR-13/Thionin gene family, which is well known to be toxic to plant pathogens (Stec 2006, Sels et al. 2008, Goyal and Mattoo 2014, Rogozhin et al. 2018). The low level of sequence conservation and presence or absence of large chromosomal segments indicates that these introgressions must have come from distant genetic backgrounds. One of the 54 unique regions of CS coincided with the Fhb1 chromosomal region responsible for resistance to Fusarium. Our de novo annotation of the region supported that the gene content and CDSs are highly divergent between CS and Norin 61. The haplotype of CS may confer an unknown different phenotype, such as resistance to other pathogens. We suggest that these unique chromosomal segments were introgressions from distinct genotypes and may have contributed to adaptive variations. These data also highlight the diversity present (and partially untapped) in East Asian cultivars.
In contrast to the three introgressions from distant species into Western cultivars discussed above (Walkowiak et al. 2020), the origin of the unique chromosomal regions in Norin 61 and CS is currently unclear. They are not likely introgressions from other species introduced during modern breeding because their known pedigrees do not include other species. In East Asia, the distribution of wild wheat relatives is limited, except for some populations of the DD genome species Ae. tauschii; thus, local introgression from wild relatives does not seem adequate to explain the large number of such regions distributed in the A, B and D chromosomes. We suggest that early bread wheat acquired these diverse chromosomal regions close to its origin in the Near East, and some of the polymorphisms were maintained throughout its dispersal to East Asia.
One way to address this question is to consider and unravel the demography of Asian wheat. Archaeological studies suggest that wheat arrived in China >4,000 years ago, reaching the islands of Japan >2,000 years ago (Kato 2000, Li et al. 2007, Fujita 2013, Barton and An 2014) The arrival of agriculture in Japan during the Yayoi period is considered a typical case of the farming/language dispersal hypothesis (Diamond and Bellwood 2003), but, compared with that of rice, the role of wheat in this process is not clear. Bioinformatic tools to analyze the genome-wide polymorphisms of polyploid species have been developed (Akama et al. 2014, Paape et al. 2018). The polymorphism analysis of Asian wheat together with a recently reported Tibetan wheat genome (Guo et al. 2020) and an updated version of CS (Alonge et al. 2020) will contribute to shed light on the origin of their high genetic diversity and selection.
Although both Norin 61 and CS originated in East Asia, our analysis of functional variations highlighted important differences between the two lines. CS is an experimental strain likely to be derived from a landrace in Sichuan, China (Liu et al. 2018). Norin 61 is a modern cultivar that has been widely cultivated in Japan for decades because of traits that confer high yield and superior grain quality in the Japanese climate. Our genome assembly of Norin 61 verified variations, including FHB resistance (Fhb1), grain quality (Glu-D1), semi-dwarfism (Rht-D1) and early flowering (Ppd-D1). In addition, our genotyping data together with previous genotyping of 12 genes of 96 cultivars (Kojima et al. 2017) supported that these functional variants are shared with many other Japanese cultivars, suggesting that the genome sequence of Norin 61 would be useful as a representative of modern Japanese cultivars.
In this analysis, we identified novel variations in the homeologs of the florigen gene FT1/Vrn3. Interestingly, all three homeologs of wheat showed copy number or indel variations. FT-D1 of Norin 61 is a pseudogene with a frameshift 1-bp deletion. By contrast, we found two copies of FT-B1 in Norin 61. The presence of at least two copies was supported both by the assembly sequence and by coverage analysis. Unexpectedly, although the RefSeq v1.1 of CS [The International Wheat Genome Sequencing Consortium (IWGSC) 2018] had only a single copy of FT-B1, our coverage analysis suggested the presence of additional copies also in CS, which were subsequently confirmed by the new assembly of CS (CS RefSeq v2.0). The presence of three or more copies in Norin 61 cannot be excluded from the current data. In addition, we found that allelic differences of FT-A1 were associated with the spring or winter habit. Although the data presented indicate only an association without considering population structure, they suggest that the Vrn-A3c allele contributes to the early flowering of winter wheat, not that of spring wheat, assuming a functional difference in tetraploid wheat. An obvious next step would be to test the functional significance of these mutations on flowering time because flowering time has been a major target in Japanese wheat breeding programs.
Given the underexploitation of Asian germplasm in the most recent worldwide varieties (Balfourier et al. 2019), the use of the material to identify new QTLs and the responsible mutations would be important. A nested association mapping line using Norin 61 as the common parent crossed with diverse Asian cultivars is being developed by the National BioResource Project-KOMUGI. For example, a resistant allele of the FHB-resistant gene, Fhb1, was found in East Asian cultivars (Niwa et al. 2018) and is now used for breeding worldwide. However, Fhb1 alone is not sufficient to control FHB and so FHB resistance from additional and more diverse sources should be integrated into new varieties. The molecular nature of many QTLs on the FHB-resistant gene is yet to be studied (Handa et al. 2008, Niwa et al. 2018). The use of FHB-QTL in 2DS, another gene resource for the FHB resistance of Asian germplasms (Basnet et al. 2012, Niwa et al. 2018, Fabre et al. 2020), was hampered by its linkage to the non-semi-dwarf allele of Rht8 and so will be facilitated by further studies to allow pyramiding the FHB-resistant alleles. We propose that the de novo genome assembly of Norin 61 will open the way to analysis of the functional variants and evolution of East Asian wheat.
Materials and Methods
Plant material
The accession of Norin 61 was provided by NBRP-Wheat, Japan (Accession Number LPGKU 2305). This genotype is originally in the collection of Dr. Takashi Endo, Kyoto University, Japan, who provided the seed to Dr. Koji Murai, Fukui Prefectural University. Shuhei Nasuda obtained the seed from Dr. Koji Murai in 2010 and self-pollinated by bagging more than five times. A bulk collection of seeds from a single Norin 61 plant was used for this study. The seed is accessible at NBRP-Wheat (https://shigen.nig.ac.jp/wheat/komugi/, last accessed on Dec. 15, 2020) and also from the John Innes Centre (https://www.seedstor.ac.uk/index.php, last accessed on Dec. 15, 2020).
Sequencing data
WGS PE and MP libraries were prepared at the Carver Biotechnology Center, University of Illinois, USA. 10× Genomics Chromium libraries were developed at HudsonAlpha, USA. The libraries were sequenced on Illumina HiSeq 2500 (v2 and v4) and HiSeq 4000 sequencers at the Functional Genomics Center Zurich (Switzerland), IPK (Germany), and Carver Biotechnology Center, University of Illinois (USA). Details about library features and sequencing coverage are reported in Table 1. The conformation capture library was prepared using a variant (TCC) of the original Hi-C method (Lieberman-Aiden et al. 2009, Tiang et al. 2012, Concia et al. 2020) as described previously (Mascher et al. 2017, Himmelbach et al. 2018).
Assembly strategy
WGS libraries and linked read data were assembled with NRGene’s (Ness-Ziona, Israel) DenovoMAGIC™ 3.0 pipeline and later scaffolded with Hi-C and POPSEQ data using TRITEX (Monat et al. 2019) (more details in Walkowiak et al. 2020). Assembly statistics were calculated with assembly-stats (https://github.com/sanger-pathogens/assembly-stats).
Assembly validation and coverage analysis
After including bread wheat chloroplast and mitochondrial genomes (NC_002762.1 and NC_036024.1, respectively), short reads of the PE700 library were aligned to the final assembly with minimap2 v2.17-r974-dirty (Li 2018) and statistics were calculated using the SAMtools v1.9 (Li et al. 2009) functions stats and flagstats. KAT v2.4.1 (Mapleson et al. 2017) was run in comp mode with –h –m 21 –H 30000000000 settings on the PE700 reads and the assembly. BUSCO v3.1.0 (Seppey et al. 2019) was run (with embryophyta_odb9 as a database and wheat as a species for Augustus) on the assembly and on the primary isoform of each predicted gene.
For the coverage analysis, a subset of CS and Norin 61 PE700 reads was aligned to the respective assemblies with HISAT2 v2.1.0 (Kim et al. 2019). Secondary and supplementary alignments were removed with SAMtools view and coverage functions. The coverage of the assembly region containing the Adh-1 and Vrn3 transcripts was calculated with BEDtools coverage (Quinlan and Hall 2010, v2.26.0) adopting the –mean option. Given that only primary alignments were retained to calculate coverage, there is no possibility that the counts could have originated from inflation of the coverage by counting alignments of the same read multiple times.
Cytogenetics
Chromosomes in the root tip meristem were prepared by the conventional acetocarmine-squash method (Murata et al. 2018). We performed nondenaturing FISH with three repetitive-sequence probes following the methodology described by Tang et al. (2014) and Zhao et al. (2016). The three oligo probes were: Oligo-pSc119.2-1 (Tamra-5′-CCGTT TTGTG GACTA TTACT CACCG CTTTG GGGTC CCATA GCTAT-3′), Oligo-pTa535 (AlexaFluor488-5′-GACGA GAACT CATCT GTTAC ATGGG CACTT CAATG TTTTT TAAAC TTATT TGAAC TCCA-3′), and Oligo-pTa713 (AlexaFluor647-5′-AGACG AGCAC GTGAC ACCAT TCCCA CCCTG TCTTA GCGTA ACGCG AGTCG-3′). Chromosomes were counterstained with 4′,6-diamidino-2-phenylindole. Chromosomes were viewed using a BX61 epifluorescence microscope (Olympus, Tokyo, Japan) and captured using a CCD camera DP80 (Olympus). Images were processed and pseudocolored using ImageJ 1.51n in the Fiji package (Schindelin et al. 2012). For karyotyping, at least four chromosomes per accession were examined with reference to the karyotype of CS reported by Komuro et al. (2013).
Whole-genome alignment
Whole-genome sequence alignment was performed using LAST v957 (Frith and Kawaguchi 2015) with the CS assembly as the reference and Norin 61 as query. The many-to-one Norin 61 to CS alignments were then parsed to keep only the one-to-one hits. We processed the alignments and created dot plot visualizations using the scripts included with LAST. First, we used last-postmask to discard low complexity alignments and alignments with error probability >10−5. Then, the resulting output files were converted into a tabular format using maf-convert from which the dot plots were created with last-dotplot.
Identification of flowering-related genes and phylogenetic analysis
Flowering-related genes in CS were identified by BLAST search (Altschul et al. 1997) of the sequence using known FT, MFT and FTL genes of Arabidopsis, rice and wheat as queries (Table 2) against CS RefSeq v1.1 [The International Wheat Genome Sequencing Consortium (IWGSC) 2018] with default parameters. Splicing variants of the FT genes described by Halliwell et al. (2016) were not used for this study. From Norin 61, the flowering-related genes were detected based on homology search comparing CS sequences with the Norin 61 pseudomolecules.
A total of 108 amino acid sequences of FT genes were aligned using MUSCLE (Edgar 2004). The alignments and the splicing boundaries of FT genes were manually curated according to BLASTN results. The nucleotide sequences of FT1 homeologs in CS and Norin 61 (Table 2) were used for phylogenetic analysis.
The extra copies of FT-B1 were identified based on the CDSs of FT-B1-1 in CS compared with the pseudomolecules of CS (RefSeq v2.0, https://www.wheatgenome.org/News2/IWGSC-RefSeq-v2.0-now-available-at-URGI (last accessed on Dec. 15, 2020), and Alonge et al. 2020) and Norin 61.
Phylogenetic trees were built using translated protein sequences of 108 FT homeologs of wheat with a few rice and Arabidopsis sequences identified. A neighbor-joining (NJ) tree and a maximum-likelihood (ML) tree were constructed using MEGA X (Kumar et al. 2018). All positions of gaps and missing data were removed, and bootstrap probability was calculated for 1,000 replications. Both NJ and ML phylogenetic analyses were performed. The phylogenetic analysis of all FT genes involved 111 amino acid positions. The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling 1965) for the NJ tree. The JTT matrix-based model (Jones et al. 1992) was used as the substitution model and the tree inference for the ML tree. The phylogenetic analysis of FT1 genes involved 11 nucleotide sequences. OsHd3a (GenBank accession: AB052941) and OsRFT1 (GenBank accession: AB062675.1) were used as the outgroups. The trees were constructed based on 530 nucleotide sequences, and the evolutionary distances were computed using the Kimura 2-parameter method (Kimura 1980).
Definition of syntenic and unique genes, annotation and phylogenetic analysis of the unique chromosomal regions
All CDSs of all genes annotated on Norin 61 chromosomes were used for BLASTN (Walkowiak et al. 2020) searches against the corresponding chromosomes in the CS reference genome. A Norin 61 gene was classified as syntenic if its closest homolog in CS was >95% identical at the DNA level and its chromosomal position differed by <5% of the total chromosome length from that of the CS homolog. A Norin 61 gene was classified as ‘private’ if no homeolog with an E-value below 10−6 was identified on the corresponding CS chromosome.
To obtain a genome-wide assessment of the level of collinearity, chromosome lengths were normalized to 100% and each gene assigned a relative position along the chromosome. Then, ratios of syntenic to private genes were calculated in running windows corresponding to 10% of the chromosome lengths. Data from individual subgenomes were compiled to assess the overall levels of synteny along the chromosomes of all three subgenomes (Supplementary Fig. S3A). It should be noted that the gene annotation of Norin 61 is projected from that of CS; thus, it could be biased by the gene content in CS. To compensate for this bias, we did bi-directional BLAST searches of gene families of interest (i.e. those on 1A and 3B candidate introgressions) and annotated genes by hand, if they were missed in the annotation either in CS or Norin61. The gene content of the genomic regions studied here will most likely change with an improved (de novo) gene annotation.
To identify possible genes that were missed during automated annotation, one representative protein sequence for each family was used in TBLASTN searches against the respective chromosomes of Norin 61 and CS. Protein sequences were extracted directly from BLAST outputs and segments corresponding to individual exons were connected, resulting in some cases in incomplete protein sequences. Nevertheless, the data contained enough sequence information to allow phylogenetic analyses. Newly identified genes were named using an index for the chromosome and the chromosome position divided by 1,000. In very dense gene clusters, an additional running number was added to the name. CS homeologs from other subgenomes were used as outgroups for the phylogenetic analysis.
Predicted protein sequences were then aligned with ClustalW using a gap-opening penalty of 5.0 and a gap-extension penalty of 0.01. Alignments were manually checked; sequences that were predicted to be much longer than the majority were trimmed and realigned. Phylogenetic trees were constructed with mrBayes, using the mcmc option. MrBayes was run for at least 100,000 generations, or until the average standard deviation of split frequencies fell below 0.01. Trees were visualized with FigTree.
Sequence analysis of functional variants
Four genes controlling important breeding traits (Ppd1, TaHRC, Rht1 and Tamyb10) were selected and identified by BLASTN search using default parameters against CS and Norin 61 pseudomolecules based on reported sequences (Supplementary Table S5). The CDSs were extracted from CDS fasta files of CS and Norin 61 based on the BLASTN results.
The region containing the Fhb1-resistant allele was annotated with MAKER v2.31.9 (Campbell et al. 2014) using the Norin 61 genomic interval, the Uniprot Liliopsida proteome (downloaded in July 2020) and the annotation from Walkowiak et al. (2020). Augustus was run with wheat as a species, and the output was parsed removing models that had >40% similarity over >50% of their length to TE proteins via BLAST. Dot plots of genomic regions were produced with Gepard v1.40 (Krumsiek et al. 2007). The sequence of the variety Clark was downloaded from NCBI (GenBank accession MK450309). The sequences of Rht-D1 in Norin 61, Tamyb10-A1 in CS and Norin 61 were manually extracted from the pseudomolecules using SAMtools (Li et al. 2009), because they were not annotated. The Ppd-1 sequences were extracted with 5 kb of 5′-UTR region from pseudomolecules using SAMtools. The obtained sequences were aligned using MUSCLE (Edgar 2004) in MEGA X (Kumar et al. 2018).
Glu-D1 variation
Nucleotide variation in the Glu-D1 locus of Norin 61 was determined by aligning 470-bp PE Illumina reads of all 10+ wheat pan genome varieties to the CS reference genome v1 with HISAT2 v2.1.0 using default parameters (Kim et al. 2019) and alignment sorting and indexing done with samtools v1.6 (Li et al. 2009). Variants within the Glu-D1 subunits, the 57 kb between subunits and 100 kb flanking were called with bcftools v1.11 ‘mpileup’ and ‘call’ with minimum alignment quality score of 20 and ‘--group-samples–’ option (Li 2011). Heterozygous calls were set to missing and missing variants exceeding a proportion of 0.1 filtered out. Structural variation between the 2.2 + 12 locus of Norin 61 genome assembly v1.1 and 2 + 12 locus of CS genome assembly v1 was assessed with MUMmer v3.23 using default parameters (Kurtz et al. 2004).
Availability
The data are also accessible at IPK, Germany, https://wheat.ipk-gatersleben.de/ (last accessed on Dec. 15, 2020), and BLAST server at the National Institute of Genetics, Japan, https://shigen.nig.ac.jp/wheat/komugi/about/norin61GenomeSequence.jsp (last accessed on Dec. 15, 2020). The gene annotation of the Fhb1 locus can be downloaded from https://de.cyverse.org/dl/d/6A85909D-942B-4C95-AEB8-7B5516680878/Fhb1_N61_340kbregion.tar.gz.
Supplementary Data
Supplementary data are available at PCP online.
Funding
JST CREST (JPMJCR16O3) to K.K.S., S.N., J.S. and T.B.; Swiss National Science Foundation 31003A_182318, CRSII5_183578, NCCR Evolving Language # 51NF40_180888 and European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie (MSC) grant agreement No 847585 to K.K.S.; MEXT KAKENHI The Birth of New Plant Species (JP16H06469, JP16H06464, JP16H06466, JP16K21727) to J.S., K.K.S., and H.T.; National Agriculture and Food Research Organization (NARO) Vice President Fund to H.H.; NBRP Genome Information Upgrading Project 2017, AMED to S.N., J.S., K.K. and K.K.S.; University of Zurich Research Priority Program Evolution in Action to K.K.S. and T.W.; German Federal Ministry of Food and Agriculture grant 2819103915 ‘WHEATSEQ’ to N.S.; and Canadian Triticum Applied Genomics research project (CTAG2) funded by Genome Canada, Genome Prairie, the Western Grains Research Foundation, Government of Saskatchewan, Saskatchewan Wheat Development Commission, Alberta Wheat Commission, Viterra and Manitoba Wheat and Barley Growers Association to C.P.
Supplementary Material
Acknowledgments
We would like to thank the IT Support Group D-HEST, ETH Zurich, the Functional Genomics Center Zurich and the International Wheat Genome Sequencing Consortium (IWGSC).
Disclosures
The authors declare no competing interests.
References
- Ahn J.H., Miller D., Winter V.J., Banfield M.J., Jeong H.L., So Y.Y., et al. (2006) A divergent external loop confers antagonistic activity on floral regulators FT and TFL1. EMBO J. 25: 605–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama S.Shimizu-Inatsugi R., Shimizu K.K., Sese J. (2014) Genome-wide quantification of homeolog expression ratio revealed nonstochastic gene regulation in synthetic allopolyploid Arabidopsis. Nucleic Acids Res. 42: e46–e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonge M., Shumate A., Puiu D., Zimin A.V., Salzberg S.L. (2020) Chromosome-scale assembly of the bread wheat genome reveals thousands of additional gene copies. Genetics 216: 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S., Madden T., Schäffer A., Zhang J., Zhang Z., Miller W., et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammerman A.J., Cavalli-Sforza L.L. (1971) Measuring the rate of spread of early farming in Europe. Man 6: 674. [Google Scholar]
- Asplund L., Leino M.W., Hagenblad J. (2012) Allelic variation at the Rht8 locus in a 19th century wheat collection. Sci. World J. 2012: 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R., Nave M., Barad O., Baruch K., Twardziok S.O., Gundlach H., et al. (2017) Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science 357: 93–97. [DOI] [PubMed] [Google Scholar]
- Balfourier F., Bouchet S., Robert S., De Oliveira R., Rimbert H., Kitt J., et al. ; International Wheat Genome Sequencing Consortium (2019) Worldwide phylogeography and history of wheat genetic diversity. Sci. Adv. 5: eaav0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton L., An C.-B. (2014) An evaluation of competing hypotheses for the early adoption of wheat in East Asia. World Archaeol. 46: 775–798. [Google Scholar]
- Basnet B.R., Glover K.D., Ibrahim A.M.H., Yen Y., Chao S. (2012) A QTL on chromosome 2DS of ‘Sumai 3’ increases susceptibility to Fusarium head blight in wheat. Euphytica 186: 91–101. [Google Scholar]
- Beales J., Turner A., Griffiths S., Snape J.W., Laurie D.A. (2007) A Pseudo-Response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L.). Theor. Appl. Genet. 115: 721–733. [DOI] [PubMed] [Google Scholar]
- Brohammer A.B., Kono T.J.Y., Hirsch C.N. (2018) The maize pan-genome. In The Maize Genome. Compendium of Plant Genomes. Edited by Bennetzen J., Flint-Garcia S., Hirsch C., Tuberosa R. Springer, Cham. 10.1007/978-3-319-97427-9_2. (Last accessed February 1, 2021) [DOI] [Google Scholar]
- Campbell M.S., Law M.Y., Holt C., Stein J.C., Moghe G.D., Hufnagel D.E., et al. (2014) MAKER-P: A tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol. 164: 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai L., Chen Z., Bian R., Zhai H., Cheng X., Peng H., et al. (2019) Dissection of two quantitative trait loci with pleiotropic effects on plant height and spike length linked in coupling phase on the short arm of chromosome 2D of common wheat (Triticum aestivum L.). Theor. Appl. Genet. 132: 1815–1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Cheng X., Chai L., Wang Z., Du D., Wang Z., et al. (2020) Pleiotropic QTL influencing spikelet number and heading date in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 133: 1825–1838. [DOI] [PubMed] [Google Scholar]
- Concia L., Veluchamy A., Ramirez-Prado J.S., Martin-Ramirez A., Huang Y., Perez M., et al. (2020) Wheat chromatin architecture is organized in genome territories and transcription factories. Genome Biol. 21: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danyluk J., Kane N.A., Breton G., Limin A.E., Fowler D.B., Sarhan F. (2003) TaVRT-1, a putative transcription factor associated with vegetative to reproductive transition in cereals. Plant Physiol. 132: 1849–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond J., Bellwood P. (2003) Farmers and their languages: the first expansions. Science 300: 597–603. [DOI] [PubMed] [Google Scholar]
- Díaz A., Zikhali M., Turner A.S., Isaac P., Laurie D.A. (2012) Copy number variation affecting the photoperiod-B1 and vernalization-A1 genes is associated with altered flowering time in wheat (Triticum aestivum). PLoS One 7: e33234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson J.R., Li X., Zhou X., Zhao K., Sun N., Atahan P. (2013) Origin and spread of wheat in China. Quat. Sci. Rev. 72: 108–111. [Google Scholar]
- Doležel J., Čížková J., Šimková H., Bartoš J. (2018) One major challenge of sequencing large plant genomes is to know how big they really are. Int. J. Mol. Sci. 19: 3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubcovsky J., Dvorak J. (2007) Genome plasticity a key factor in the success of polyploid wheat under domestication. Science 316: 1862–1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R.C. (2004) MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32: 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir A.A.E., Gorafi Y.S.A., Tahir I.S.A., Elhashimi A.M.A., Abdalla M.G.A., Tsujimoto H. (2017) Genetic variation in heat tolerance-related traits in a population of wheat multiple synthetic derivatives. Breed. Sci. 67: 483–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eshed Y., Lippman Z.B. (2019) Revolutions in agriculture chart a course for targeted breeding of old and new crops. Science 366: eaax0025. [DOI] [PubMed] [Google Scholar]
- Fabre F., Rocher F., Alouane T., Langin T., Bonhomme L. (2020) Searching for FHB resistances in bread wheat: susceptibility at the crossroad. Front. Plant Sci. 11. 10.3389/fpls.2020.00731 (Last accessed February 1, 2021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure S., Higgins J., Turner A., Laurie D. (2007) The FLOWERING LOCUS T-like gene family in barley (Hordeum vulgare). Genetics 176: 599–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flintham J., Adlam R., Bassoi M., Holdsworth M., Gale M. (2002) Mapping genes for resistance to sprouting damage in wheat. Euphytica 126: 39–45. [Google Scholar]
- Frith M.C., Kawaguchi R. (2015) Split-alignment of genomes finds orthologies more accurately. Genome Biol. 16: 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu D., Szűcs P., Yan L., Helguera M., Skinner J.S., von Zitzewitz J., et al. (2005) Large deletions within the first intron in VRN-1 are associated with spring growth habit in barley and wheat. Mol. Genet. Genomics 273: 54–65. [DOI] [PubMed] [Google Scholar]
- Fujita M. (2013) Wheat. In Japanese History of Breeding. Edited by Ukai Y., Osawa R. pp. 48–73. Yuushokan, Tokyo, Japan: (in Japanese). [Google Scholar]
- Fukunaga K., Inagaki M. (1985) Genealogical pedigrees of Japanese wheat cultivars. Jpn. J Breed. 35: 89–92 (in Japanese). [Google Scholar]
- Goyal R.K., Mattoo A.K. (2014) Multitasking antimicrobial peptides in plant development and host defense against biotic/abiotic stress. Plant Sci. 228: 135–149. [DOI] [PubMed] [Google Scholar]
- Guo Z., Song Y., Zhou R., Ren Z., Jia J. (2010) Discovery, evaluation and distribution of haplotypes of the wheat Ppd-D1 gene. New Phytol. 185: 841–851. [DOI] [PubMed] [Google Scholar]
- Guo W., Xin M., Wang Z., Yao Y., Hu Z., Song W., et al. (2020) Origin and adaptation to high altitude of Tibetan semi-wild wheat. Nat. Commun. 11: 5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutaker R.M., Groen S.C., Bellis E.S., Choi J.Y., Pires I.S., Bocinsky R.K., et al. (2020) Genomic history and ecology of the geographic spread of rice. Nat. Plants 6: 492–502. [DOI] [PubMed] [Google Scholar]
- Halliwell J., Borrill P., Gordon A., Kowalczyk R., Pagano M.L., Saccomanno B., et al. (2016) Systematic investigation of FLOWERING LOCUS T-like Poaceae gene families identifies the short-day expressed flowering pathway gene, TaFT3 in wheat (Triticum aestivum L.). Front. Plant Sci. 7: 857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa H., Namiki N., Xu D., Ban T. (2008) Dissecting of the FHB resistance QTL on the short arm of wheat chromosome 2D using a comparative genomic approach: From QTL to candidate gene. Mol. Breeding 22: 71–84. [Google Scholar]
- Hatakeyama M., Aluri S., Balachadran M.T., Sivarajan S.R., Patrignani A., Grüter S., et al. (2018) Multiple hybrid de novo genome assembly of finger millet, an orphan allotetraploid crop. DNA Res. 25: 39–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi Y., Narumi T., Oda A., Nakano Y., Sumitomo K., Fukai S., et al. (2013) The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA 110: 17137–17142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himi E., Maekawa M., Miura H., Noda K. (2011) Development of PCR markers for Tamyb10 related to R-1, red grain color gene in wheat. Theor. Appl. Genet. 122: 1561–1576. [DOI] [PubMed] [Google Scholar]
- Himi E., Mares D.J., Yanagisawa A., Noda K. (2002) Effect of grain colour gene (R) on grain dormancy and sensitivity of the embryo to abscisic acid (ABA) in wheat. J. Exp. Bot. 53: 1569–1574. [DOI] [PubMed] [Google Scholar]
- Himi E., Noda K. (2005) Red grain colour gene (R) of wheat is a Myb-type transcription factor. Euphytica 143: 239–242. [Google Scholar]
- Himmelbach A., Walde I., Mascher M., Stein N. (2018) Tethered chromosome conformation capture sequencing in Triticeae: a valuable tool for genome assembly. Bioprotocol 8: e2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G., Grover C.E., Arick M.A., Liu M., Peterson D.G., Wendel J.F. (2020) Homoeologous gene expression and co-expression network analyses and evolutionary inference in allopolyploids. Brief Bioinform. doi:10.1093/bib/bbaa035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyles J., Bloomfield M.T., Hunt J.R., Trethowan R.M., Trevaskis B. (2020) Phenology and related traits for wheat adaptation. Heredity 125: 417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda T.M., Nagamine T., Fukuoka H., Yano H. (2002) Identification of new low-molecular-weight glutenin subunit genes in wheat. Theor. Appl. Genet. 104: 680–687. [DOI] [PubMed] [Google Scholar]
- Ishikawa N. (2010) Recent progress and future tasks in wheat breeding at Kanto, Tokai and western Japan. J. Crop Res. 55: 1–8 (in Japanese with English summary). [Google Scholar]
- Jobson E.M., Johnston R.E., Oiestad A.J., Martin J.M., Giroux M.J. (2019) The impact of the wheat Rht-B1b semi-dwarfing allele on photosynthesis and seed development under field conditions. Front. Plant Sci. 10: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.T., Taylor W.R., Thornton J.M. (1992) The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8: 275–282. [DOI] [PubMed] [Google Scholar]
- Kane N.A., Danyluk J., Tardif G., Ouellet F., Laliberté J., Limin A.E., et al. (2005) TaVRT-2, a member of the StMADS-11 clade of flowering repressors, is regulated by vernalization and photoperiod in wheat. Plant Physiol. 138: 2354–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasajima S.-Y., Inoue N., Mahmud R. (2009) Response spectrum for green light-induced acceleration of heading in wheat cv. Norin 61. Plant Prod. Sci. 12: 54–57. [Google Scholar]
- Kato K. (2000) Komugi ga nihon ni kita michi. In Mugi No Shizenshi. Edited by Sato Y., Kato K. pp. 113–136 Hokkaido University Press, Sapporo: (in Japanese). [Google Scholar]
- Kihara H. (1944) Discovery of the DD-analyser, one of the ancestors of Triticum vulgare (abstr). Agric. Hortic. 19: 889–890 (in Japanese). [Google Scholar]
- Kikuchi R., Kawahigashi H., Ando T., Tonooka T., Handa H. (2009) Molecular and functional characterization of PEBP genes in barley reveal the diversification of their roles in flowering. Plant Physiol. 149: 1341–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Paggi J.M., Park C., Bennett C., Salzberg S.L. (2019) Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37: 907–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16: 111–120. [DOI] [PubMed] [Google Scholar]
- Kobayashi F., Tanaka T., Kanamori H., Wu J., Katayose Y., Handa H. (2016) Characterization of a mini core collection of Japanese wheat varieties using single-nucleotide polymorphisms generated by genotyping-by-sequencing. Breed. Sci. 66: 213–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima H., Fujita M., Matsunaka H., Seki M., Chono M., Kiribuchi-Otobe C., et al. (2017) Development and evaluation of the core collection of Japanese wheat varieties. Bull. NARO Crop Sci. 1: 1–13 (in Japanese with English summary). [Google Scholar]
- Komuro S., Endo R., Shikata K., Kato A. (2013) Genomic and chromosomal distribution patterns of various repeated DNA sequences in wheat revealed by a fluorescence in situ hybridization procedure. Genome 56: 131–137. [DOI] [PubMed] [Google Scholar]
- Korzun V., Röder M.S., Ganal M.W., Worland A.J., Law C.N. (1998) Genetic analysis of the dwarfing gene (Rht8) in wheat. Part I. Molecular mapping of Rht8 on the short arm of chromosome 2D of bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 96: 1104–1109. [Google Scholar]
- Krumsiek J., Arnold R., Rattei T. (2007) Gepard: A rapid and sensitive tool for creating dotplots on genome scale. Bioinformatics 23: 1026–1028. [DOI] [PubMed] [Google Scholar]
- Kumar S., Stecher G., Li M., Knyaz C., Tamura K. (2018) MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35: 1547–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo T.C.Y., Hatakeyama M., Tameshige T., Shimizu K.K., Sese J. (2020) Homeolog expression quantification methods for allopolyploids. Brief Bioinform. 21: 395–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurtz S., Phillippy A., Delcher A.L., Smoot M., Shumway M., Antonescu C., et al. (2004) Versatile and open software for comparing large genomes. Genome Biol. 5: R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagudah E.S., Halloran G.M. (1988) Phylogenetic relationships of Triticum tauschii the D genome donor to hexaploid wheat. Theor. Appl. Genet. 75: 592–598. [DOI] [PubMed] [Google Scholar]
- Leitch A.R., Leitch I.J. (2008) Genomic plasticity and the diversity of polyploid plants. Science 320: 481–483. [DOI] [PubMed] [Google Scholar]
- Li G., Zhou J., Jia H., Gao Z., Fan M., Luo Y., et al. (2019) Mutation of a histidine-rich calcium-binding-protein gene in wheat confers resistance to Fusarium head blight. Nat. Genet. 51: 1106–1112. [DOI] [PubMed] [Google Scholar]
- Li H. (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27: 2987–2993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. (2018) Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 34: 3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. ; 1000 Genome Project Data Processing Subgroup (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25: 2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Dodson J., Zhou X., Zhang H., Masutomoto R. (2007) Early cultivated wheat and broadening of agriculture in Neolithic China. Holocene 17: 555–560. [Google Scholar]
- Lieberman-Aiden E., Van Berkum N.L., Williams L., Imakaev M., Ragoczy T., Telling A., et al. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M., Zhang D., Liu S., Zhang G., Yu J., Fritz A.K., et al. (2016) Genome-wide association analysis on pre-harvest sprouting resistance and grain color in U.S. winter wheat. BMC Genomics 17: 794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Zhang L., Hao M., Ning S., Yuan Z., Dai S., et al. (2018) Wheat breeding in the hometown of Chinese Spring. Crop J. 6: 82–90. [Google Scholar]
- Mackie A.M., Lagudah E.S., Sharp P.J., Lafiandra D. (1996) Molecular and biochemical characterisation of HMW glutenin subunits from T. tauschii and the D genome of hexaploid wheat. J. Cereal Sci. 23: 213–225. [Google Scholar]
- Mapleson D., Accinelli G.G., Kettleborough G., Wright J., Clavijo B.J. (2017) KAT: A K-mer analysis toolkit to quality control NGS datasets and genome assemblies. Bioinformatics 33: 574–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mascher M., Gundlach H., Himmelbach A., Beier S., Twardziok S.O., Wicker T., et al. (2017) A chromosome conformation capture ordered sequence of the barley genome. Nature 544: 427–433. [DOI] [PubMed] [Google Scholar]
- Matsuo H., Kohno K., Morita E. (2005) Molecular cloning, recombinant expression and IgE-binding epitope of omega-5 gliadin, a major allergen in wheat-dependent exercise-induced anaphylaxis. FEBS J. 272: 4431–4438. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y. (2011) Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 52: 750–764. [DOI] [PubMed] [Google Scholar]
- Mayrose I., Zhan S.H., Rothfels C.J., Magnuson-Ford K., Barker M.S., Rieseberg L.H., et al. (2011) Recently formed polyploid plants diversify at lower rates. Science 333: 1257–1257. [DOI] [PubMed] [Google Scholar]
- McFadden E., Sears E. (1944) The artificial synthesis of Triticum spelta. Rec. Genet. Soc. Am. 13: 26–27. [Google Scholar]
- McIntosh R.A., Yamazaki Y., Dubcovsky J., Rogers J., Morris C., Apples R., et al. (2013) Catalogue of gene symbols for wheat. In Proceedings of the 12th International Wheat Genetics Symposium. pp. 74–75.
- Meguro A., Takumi S., Ogihara Y., Murai K. (2003) WAG, a wheat AGAMOUS homolgo, is associated with development of pistil-like stamens in alloplasmic wheat. Sex. Plant Reprod. 15: 221–230. [Google Scholar]
- Minoda T. (2010) Effects of climatic conditions on the yield of wheat variety ‘Norin 61’ in the experimental upland field in Saitama Prefecture. Jpn. J. Crop Sci. 79: 62–68 (in Japanese with English summary). [Google Scholar]
- Miralles D.J., Slafer G.A. (1995) Yield, biomass and yield components in dwarf, semi‐dwarf and tall isogenic lines of spring wheat under recommended and late sowing dates. Plant Breed. 114: 392. [Google Scholar]
- Mizuno N., Nitta M., Sato K., Nasuda S. (2012) A wheat homologue of PHYTOCLOCK1 is a candidate gene conferring the early heading ohenotype to einkorn wheat. Genes Genet. Syst. 87: 357–367. [DOI] [PubMed] [Google Scholar]
- Mizuno N., Kinoshita M., Kinoshita S., Nishida H., Fujita M., Kato K., et al. (2016) Loss-of-function mutations in three homoeologous PHYTOCLOCK1 genes in common wheat are associated with the extra-early flowering phenotype. PLoS One 11: e0165618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monat C., Padmarasu S., Lux T., Wicker T., Gundlach H., Himmelbach A., et al. (2019) TRITEX: chromosome-scale sequence assembly of Triticeae genomes with open-source tools. Genome Biol. 20: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murai K., Miyamae M., Kato H., Takumi S., Ogihara Y. (2003) WAP1, a wheat APETALA1 homolog, plays a central role in the phase transition from vegetative to reproductive growth. Plant Cell Physiol. 44: 1255–1265. [DOI] [PubMed] [Google Scholar]
- Murata K., Watanabe S., Tsujimoto H., Nasuda S. (2018) Cytological observation of chromosome breakage in wheat male gametophytes caused by gametocidal action of Aegilops triuncialis-derived chromosome 3Ct. Genes Genet. Syst. 93: 111–118. [DOI] [PubMed] [Google Scholar]
- Nakamura H., Inazu A., Hirano H. (1999) Allelic variation in high-molecular-weight glutenin subunit Loci of Glu-1 in Japanese common wheats. Euphytica 106: 131–138. [Google Scholar]
- Nemoto Y., Kisaka M., Fuse T., Yano M., Ogihara Y. (2003) Characterization and functional analysis of three wheat genes with homology to the CONSTANS flowering time gene in transgenic rice. Plant J. 36: 82–93. [DOI] [PubMed] [Google Scholar]
- Ning S., Wang N., Sakuma S., Pourkheirandish M., Wu J., Matsumoto T., et al. (2013) Structure, transcription and post-transcriptional regulation of the bread wheat orthologs of the barley cleistogamy gene Cly1. Theor. Appl. Genet. 126: 1273–1283. [DOI] [PubMed] [Google Scholar]
- Nishida H., Yoshida T., Kawakami K., Fujita M., Long B., Akashi Y., et al. (2013) Structural variation in the 5′ upstream region of photoperiod-insensitive alleles Ppd-A1a and Ppd-B1a identified in hexaploid wheat (Triticum aestivum L.), and their effect on heading time. Mol. Breed. 31: 27–37. [Google Scholar]
- Nishimura K., Moriyama R., Katsura K., Saito H., Takisawa R., Kitajima A., et al. (2018) The early flowering trait of an emmer wheat accession (Triticum turgidum L. ssp. dicoccum) is associated with the cis-element of the Vrn-A3 locus. Theor. Appl. Genet. 131: 2037–2053. [DOI] [PubMed] [Google Scholar]
- Niwa S., Kazama Y., Abe T., Ban T. (2018) Tracking haplotype for QTLs associated with Fusarium head blight resistance in Japanese wheat (Triticum aestivum L.) lineage. Agric. Food Secur. 7: 4. [Google Scholar]
- Ohta S. (2010) Wheat. In History of Crop Breeding. Edited by Ukai Y., Osawa R. pp. 41–66. Yuushokan, Tokyo, Japan: (in Japanese). [Google Scholar]
- Paape T., Briskine R.V., Halstead-Nussloch G., Lischer H.E.L., Shimizu-Inatsugi R., Hatakeyama M., et al. (2018) Patterns of polymorphism and selection in the subgenomes of the allopolyploid Arabidopsis kamchatica. Nat. Commun. 9: 3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne P.I., Holt L.M., Lawrence G.J. (1983) Detection of a novel high molecular weight subunit of glutenin in some Japanese hexaploid wheats. J. Cereal Sci. 1: 3–8. [Google Scholar]
- Payne P.I., Lawrence G.J. (1983) Catalogue of alleles for the complex gene loci, Glu-A1, Glu-B1, and Glu-D1 which code for high-molecular-weight subunits of glutenin in hexaploid wheat. Cereal Res. Commun. 11: 29–35. [Google Scholar]
- Peng J., Richards D.E., Hartley N.M., Murphy G.P., Devos K.M., Flintham J.E., et al. (1999) ‘ Green revolution’ genes encode mutant gibberellin response modulators. Nature 400: 256–261. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. (2010) BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawat N., Pumphrey M.O., Liu S., Zhang X., Tiwari V.K., Ando K., et al. (2016) Wheat Fhb1 encodes a chimeric lectin with agglutinin domains and a pore-forming toxin-like domain conferring resistance to Fusarium head blight. Nat. Genet. 48: 1576–1580. [DOI] [PubMed] [Google Scholar]
- Rikiishi K., Maekawa M. (2010) Characterization of a novel wheat (Triticum aestivum L.) mutant with reduced seed dormancy. J. Cereal Sci. 51: 292–298. [Google Scholar]
- Rogozhin E., Ryazantsev D., Smirnov A., Zavriev S. (2018) Primary structure analysis of antifungal peptides from cultivated and wild cereals. Plants 7: 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saisho D., Ishii M., Hori K., Sato K. (2011) Natural variation of barley vernalization requirements: Implication of quantitative variation of winter growth habit as an adaptive trait in East Asia. Plant Cell Physiol. 52: 775–784. [DOI] [PubMed] [Google Scholar]
- Saisho D., Purugganan M.D. (2007) Molecular phylogeography of domesticated barley traces expansion of agriculture in the old world. Genetics 177: 1765–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sansaloni C., Franco J., Santos B., Percival-Alwyn L., Singh S., Petroli C., et al. (2020) Diversity analysis of 80,000 wheat accessions reveals consequences and opportunities of selection footprints. Nat. Commun. 11: 4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarth R., Law C.N. (1983) The location of the photoperiod gene, Ppd2 and an additional genetic factor for ear-emergence time on chromosome 2B of wheat. Heredity 51: 607–619. [Google Scholar]
- Schindelin J., Arganda-Carreras I., Frise E., Kaynig V., Longair M., Pietzsch T., et al. (2012) Fiji: an open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M., Stein N., Mascher M. (2018) Genomic approaches for studying crop evolution. Genome Biol. 19: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweiger W., Steiner B., Vautrin S., Nussbaumer T., Siegwart G., Zamini M., et al. (2016) Suppressed recombination and unique candidate genes in the divergent haplotype encoding Fhb1, a major Fusarium head blight resistance locus in wheat. Theor. Appl. Genet. 129: 1607–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Chono M., Matsunaka H., Fujita M., Oda S., Kubo K., et al. (2011) Distribution of photoperiod-insensitive alleles Ppd-B1a and Ppd-D1a and their effect on heading time in Japanese wheat cultivars. Breed. Sci. 61: 405–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M., Chono M., Nishimura T., Sato M., Yoshimura Y., Matsunaka H., et al. (2013) Distribution of photoperiod-insensitive allele Ppd-A1a and its effect on heading time in Japanese wheat cultivars. Breed. Sci. 63: 309–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sels J., Mathys J., De Coninck B.M.A., Cammue B.P.A., De Bolle M.F.C. (2008) Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46: 941–950. [DOI] [PubMed] [Google Scholar]
- Seppey M., Manni M., Zdobnov E.M. (2019) BUSCO: assessing genome assembly and annotation completeness. Methods Mol. Biol . 1962: 227–245. [DOI] [PubMed] [Google Scholar]
- Sharma N., Ruelens P., D'hauw M., Maggen T., Dochy N., Torfs S., et al. (2017) A flowering locus C homolog is a vernalization-regulated repressor in Brachypodium and is cold regulated in wheat. Plant Physiol. 173: 1301–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada S., Ogawa T., Kitagawa S., Suzuki T., Ikari C., Shitsukawa N., et al. (2009) A genetic network of flowering-time genes in wheat leaves, in which an APETALA1/FRUITFULL-like gene, VRN1, is upstream of FLOWERING LOCUS T. Plant J. 58: 668–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shitsukawa N., Tahira C., Kassai K., Hirabayashi C., Shimizu T., Takumi S., et al. (2007) Genetic and epigenetic alteration among three homeologous genes of a class E MADS box gene in hexaploid wheat. Plant Cell 19: 1723–1737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stec B. (2006) Plant thionins—the structural perspective. Cell. Mol. Life Sci. 63: 1370–1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens C.J., Fuller D.Q. (2017) The spread of agriculture in eastern Asia. Lang. Dyn. Change 7: 152–186. [Google Scholar]
- Stoneking M. (2016) An Introduction to Molecular Anthropology. Wiley-Blackwell, Hoboken, NJ, USA. [Google Scholar]
- Su Z., Bernardo A., Tian B., Chen H., Wang S., Ma H., et al. (2019) A deletion mutation in TaHRC confers Fhb1 resistance to Fusarium head blight in wheat. Nat. Genet. 51: 1099–1105. [DOI] [PubMed] [Google Scholar]
- Takenaka S., Nitta M., Nasuda S. (2018) Population structure and association analyses of the core collection of hexaploid accessions conserved ex situ in the Japanese gene bank NBRP-Wheat. Genes Genet. Syst. 93: 237–254. [DOI] [PubMed] [Google Scholar]
- Tang Z., Yang Z., Fu S. (2014) Oligonucleotides replacing the roles of repetitive sequences pAs1, pSc119.2, pTa-535, pTa71, CCS1, and pAWRC.1 for FISH analysis. J. Appl. Genetics 55: 313–318. [DOI] [PubMed] [Google Scholar]
- Tanio M., Kato K., Ishikawa N., Tamura Y., Sato M., Takagi H., et al. (2005) Genetic analysis of photoperiod response in wheat and its relation with the earliness of heading in the southwestern part of Japan. Breed. Sci. 55: 327–334. [Google Scholar]
- Taya S., Araki H., Nonaka S. (1981) Effect of air temperature, sunshine and precipitation on the yield and other agronomic characteristics of wheat cultivar ‘Norin 61’. Nihon Sakumotsu Gakkai Kyushu Shibu Kaiho 48: 15–18 (in Japanese). [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC) (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345: 6194. [DOI] [PubMed] [Google Scholar]
- The International Wheat Genome Sequencing Consortium (IWGSC) (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 361: eaar7191. [DOI] [PubMed] [Google Scholar]
- Tiang C.L., He Y., Pawlowski W.P. (2012) Chromosome organization and dynamics during interphase, mitosis, and meiosis in plants. Plant Physiol. 158: 26–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner A., Beales J., Faure S., Dunford R.P., Laurie D.A. (2005) The pseudo-response regulator Ppd-H1 provides adaptation to photoperiod in barley. Science 310: 1031–1034. [DOI] [PubMed] [Google Scholar]
- van Heerwaarden J., Doebley J., Briggs W.H., Glaubitz J.C., Goodman M.M., Gonzalez J.D.J.S., et al. (2011) Genetic signals of origin, spread, and introgression in a large sample of maize landraces. Proc. Natl. Acad. Sci. USA 108: 1088–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetch J.M., Stougaard R.N., Martin J.M., Giroux M.J. (2019) Review: revealing the genetic mechanisms of pre-harvest sprouting in hexaploid wheat (Triticum aestivum L.). Plant Sci 281: 180–185. [DOI] [PubMed] [Google Scholar]
- Walkowiak S. Gao L. Monat C. Haberer G. Kassa M. T. Brinton J., et al. (2020) Multiple wheat genomes reveal global variation in modern breeding. Nature 588: 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Yan Z., Liu K., Zheng Y., D’Ovidio R., Shewry P.R., et al. (2005) Comparative analysis of the D genome-encoded high-molecular weight subunits of glutenin. Theor. Appl. Genet. 111: 1183–1190. [DOI] [PubMed] [Google Scholar]
- Wang W., Mauleon R., Hu Z., Chebotarov D., Tai S., Wu Z., et al. (2018) Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557: 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner R.L., Kudrna D.A., Spaeth S.C., Jones S.S. (2000) Dormancy in white-grain mutants of Chinese Spring wheat (Triticum aestivum L.). Seed Sci. Res. 10: 51–60. [Google Scholar]
- Welsh J., Keim D., Pirasteh B., Richards R.D. (1973) Genetic control of photoperiod response in wheat. In Proceedings of the 4th International Wheat Genetics Symposium. pp. 879–884.
- William M.D.H.M., Peña R.J., Mujeeb-Kazi A. (1993) Seed protein and isozyme variations in Triticum tauschii (Aegilops squarrosa). Theoret. Appl. Genetics 87: 257–263. [DOI] [PubMed] [Google Scholar]
- Wood T.E., Takebayashi N., Barker M.S., Mayrose I., Greenspoon P.B., Rieseberg L.H. (2009) The frequency of polyploid speciation in vascular plants. Proc. Natl. Acad. Sci. USA 106: 13875–13879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worland A.J. (1996) The influence of flowering time genes on environmental adaptability in European wheats. Euphytica 89: 49–57. [Google Scholar]
- Xiao J., Xu S., Li C., Xu Y., Xing L., Niu Y., et al. (2014) O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat. Commun. 5: 4572. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Yan L., Loukoianov A., Tranquilli G., Helguera M., Fahima T., Dubcovsky J. (2003) Positional cloning of the wheat vernalization gene VRN1. Proc. Natl. Acad. Sci. USA 100: 6263–6268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Louloianov A., Blechl A., Tranquilli G., Ramakrishna W., SanMiguel P., et al. (2004) The wheat VRN2 gene is a flowering repressor down-regulated by vernalization. Science 303: 1640–1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanaka M., Takata K., Ishikawa N., Takahashi T. (2016) Effects of the composition of glutenin subunits at Glu-A1 and Glu-D1 loci on the protein composition in Japanese soft wheat. Jpn. J. Crop Sci. 85: 403–410. [Google Scholar]
- Yang F.P., Zhang X.K., Xia X.C., Laurie D.A., Yang W.X., He Z.H. (2009) Distribution of the photoperiod insensitive Ppd-D1a allele in Chinese wheat cultivars. Euphytica 165: 445–452. [Google Scholar]
- Zhao X.Y., Liu M.S., Li J.R., Guan C.M., Zhang X.S. (2005) The wheat TaGI1, involved in ohotoperiodic flowering, encodes an Arabidopsis GI ortholog. Plant Mol. Biol. 58: 53–64. [DOI] [PubMed] [Google Scholar]
- Zhao L., Ning S., Yu J., Hao M., Zhang L., Yuan Z., et al. (2016) Cytological identification of an Aegilops variabilis chromosome carrying stripe rust resistance in wheat. Breed. Sci. 66: 522–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T., Ni Z., Dai Y., Yao Y., Nie X., Sun Q. (2006) Characterization and expression of 42 MADS-box genes in wheat (Triticum aestivum L.). Mol. Gen. Genomics 276: 334–350. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Zhao X., Li Y., Xu J., Bi A., Kang L., et al. (2020) Triticum population sequencing provides insights into wheat adaptation. Nat. Genet. 52: 1412–1422. [DOI] [PubMed] [Google Scholar]
- Zuckerkandl E., Pauling L. (1965) Molecules as documents of evolutionary history. J. Theor. Biol. 8: 357–366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.