Abstract
Magnetic resonance imaging (MRI) is gaining popularity in guiding radiation treatment for intrahepatic cancers due to its superior soft tissue contrast and potential of monitoring individual motion and liver function. This study investigates a deep learning-based method that generates synthetic CT volumes from T1-weighted MR Dixon images in support of MRI-based intrahepatic radiotherapy treatment planning. Training deep neutral networks for this purpose has been challenged by mismatches between CT and MR images due to motion and different organ filling status. This work proposes to resolve such challenge by generating ‘semi-synthetic’ CT images from rigidly aligned CT and MR image pairs. Contrasts within skeletal elements of the ‘semi-synthetic’ CT images were determined from CT images, while contrasts of soft tissue and air volumes were determined from voxel-wise intensity classification results on MR images. The resulting ‘semi-synthetic’ CT images were paired with their corresponding MR images and used to train a simple U-net model without adversarial components. MR and CT scans of 46 patients were investigated and the proposed method was evaluated for 31 patients with clinical radiotherapy plans, using 3-fold cross validation. The averaged mean absolute errors between synthetic CT and CT images across patients were 24.10 HU for liver, 28.62 HU for spleen, 47.05 HU for kidneys, 29.79 HU for spinal cord, 105.68 HU for lungs and 110.09 HU for vertebral bodies. VMAT and IMRT plans were optimized using CT-derived electron densities, and doses were recalculated using corresponding synthetic CT-derived density grids. Resulting dose differences to planning target volumes and various organs at risk were small, with the average difference less than 0.15 Gy for all dose metrics evaluated. The similarities in both image intensity and radiation dose distributions between CT and synthetic CT volumes demonstrate the accuracy of the method and its potential in supporting MRI-only radiotherapy treatment planning.
Keywords: MRI guided radiation therapy, synthetic CT, intrahepatic cancer, deep learning
1. Introduction
MRI is increasingly popular for use in planning treatments of intrahepatic cancers, most notably hepatocellular carcinoma. In addition to improved tumor visualization, MRI has been shown to produce very high quality reconstructions of breathing motion (Johansson et al 2018a, 2018b, Stemkens et al 2018) as well as maps of functional liver to aid in optimally sparing liver function with precision treatments such as modulated stereotactic radiation therapy (Wang et al 2016, Tsegmed et al 2017, Simeth et al 2018). Furthermore, the advent of MR-guided radiation therapy invites higher precision of intrahepatic treatments due to the availability of MR-based monitoring of and gating based on tumor and/or nearby surrogate tissue positions (Wojcieszynski et al 2016). Given this interest, investigators have pursued various means of synthetic CT generation to support MRI-only workflows for simulation of patients with intrahepatic tumors (Bredfeldt et al 2017, Liu et al 2019). While these methodologies have produced some useful results, they have been either challenged by the degeneracy of intensity mapping between air and bone from MR images (Bredfeldt et al 2017) or the complex changes in internal anatomy that routinely occur in the abdomen, which confounds direct correspondence of CT and MR voxels to aid Hounsfield Units (HU) determination from MR images (Liu et al 2019).
Among various technologies proposed in supporting synthetic CT generation, deep neural networks have shown significant value (Han 2017). However, deep neutral networks require a large amount of high quality data for training, which can be particularly challenging when applying deep learning for abdominal synthetic CT generation. Soft tissue in abdominal region exhibits significant deformation between scans due to respiration, physiological motion and different organ filling status. As a result, it is difficult to establish voxel-wise correspondence between CT and MR images of the same patient. Deformable registration may improve soft tissue alignment between CT and MR images but the accuracy is limited and there are usually residual mismatches between scans. Such mismatch, if unresolved, will degrade the quality of synthetic CT images generated and result in artifacts. Although several network structures with adversarial components have been proposed to handle such mismatch (Emami et al 2018, Maspero et al 2018, Nie et al 2018, Yang et al 2018), it adds on model complexity and may require more training samples.
In this work, we propose a novel scheme to train a simple U-net structure without any adversarial components for abdominal synthetic CT generation. Instead of using deformably aligned CT and MR image pairs for training, we use ‘semi-synthetic’ CT and MR image pairs. The justification for this method is based on two observations. First, MRI presents image contrast sufficient for soft tissue differentiation of organs for which HU values are stable across most patients. Second, although MRI has limited capability to generate bony contrast, skeletal tissues are rigid and can to first order be easily aligned rigidly in the upper abdomen between MRI and CT. We generate ‘semi-synthetic’ CT images with soft tissue contrast determined from voxel-wise soft tissue classification results on MRI, and bone contrast determined from CT images rigidly aligned to MRI. In this way the problem of soft tissue mismatch is completely avoided. We show that, by training with well-aligned image pairs, a simple network can produce synthetic CT images that present image qualities comparable or better to published results.
2. Methods and materials
2.1. Image acquisition and preprocessing
Under institution review board approval, 46 patients treated with radiation for intrahepatic carcinomas were investigated. All patients had both CT and MRI scans. CT scans were acquired on an in-house CT simulator (Brilliance, Philips, Amsterdam, Netherlands) with slice thickness of 3 mm, in-plane pixel size of 1 × 1 mm2 and tube currents ranging from 250 mAs to 400 mAs. Most CT scans were acquired with a tube voltage of 120 kVp except for four patients whose scans were acquired with 140 kVp photon spectra. Various breathing control schemes were used during CT scans, including voluntary breath hold (32 patients), free-breathing (9 patients) and voluntary breath hold guided with a visual feedback spirometry respiratory gating system (SDX, DYN’R medical systems, France, 5 patients). MR scans of relevance were acquired using an in-house 3 T MR scanner (Skyra, Siemens Medical Systems, Erlangen, Germany), under breath held conditions, using a 3D gradient echo sequence (VIBE Dixon). The imaging parameters were TEs (in/out-of-phase) 2.46/1.23 ms, TR 4.26 ms, flip angle 9°, and 1.5 × 1.5 × 3 mm3 voxel size. The in-phase (T1-weighted) images as well as fat and water images, computed from the in-phase and out-of-phase images were used in this study. Geometric distortions on the MR images were corrected using the scanner’s built-in 3-dimensional Gradient Non-Linearity Correction software. Intensity inhomogeneity correction was applied to MR images using an open source algorithm (N4) (Tustison et al 2010), implemented in a publicly available image analysis software environment (SLICER, surgical processing laboratory, Brigham and Women’s Hospital, Boston, MA). All image volumes were reformatted to axial cuts with voxel sizes interpolated to 1 × 1 × 1 mm3. Figure 1 shows example images from one subject. The axial field sizes of MR scans ranged from 380 × 380 mm2 to 500 × 500 mm2 across patients, and fields of view incorporated widely varying amounts of lung and background air. Figure 2 shows example MRI scans of two patients of different sizes and lung volumes.
Figure 1.
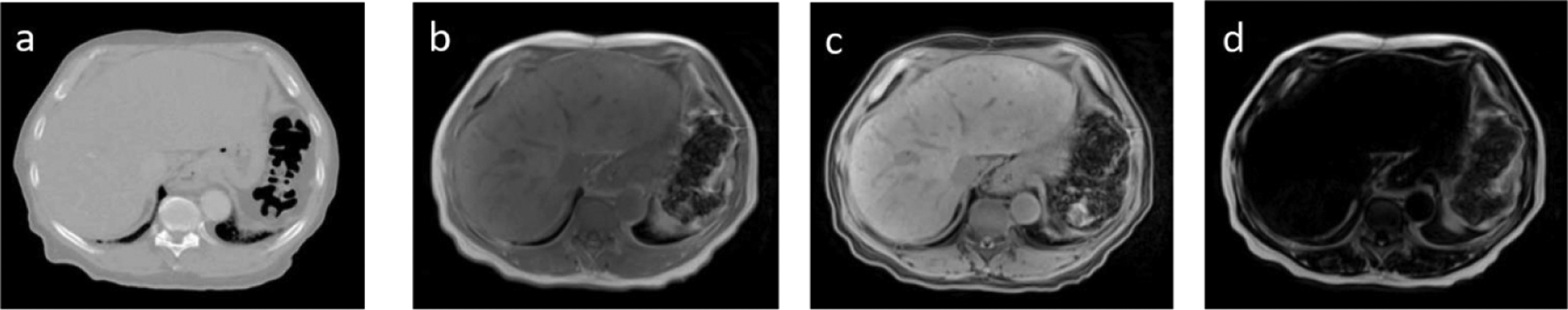
Example CT and MR images from one study subject. (a) CT (b) T1-weighted (in-phase), (c) water and (d) fat images.
Figure 2.
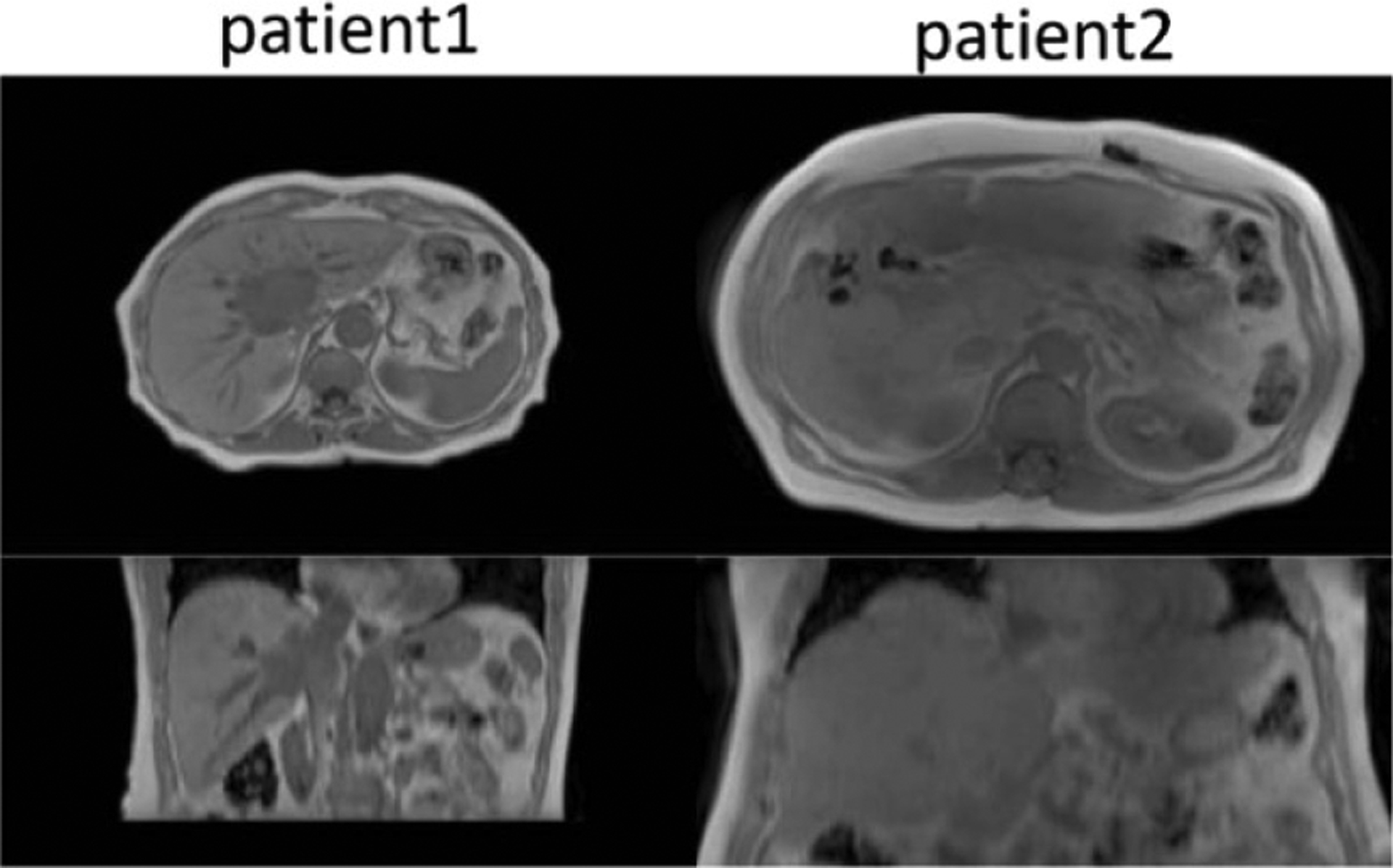
Example MR images of two patients of different sizes and lung volumes. Both images have been reformatted to axial cuts with voxel sizes interpolated to 1 × 1 × 1 mm3. Axial slices were padded to the same size of 512 × 512.
2.2. Training dataset construction
To avoid mismatches between MR and CT images, we constructed a training dataset that consists of MR and ‘semi-synthetic’ CT images, which have voxel-wise correspondence to MR images. Figure 3 shows the general workflow of ‘semi-synthetic’ CT generation.
Figure 3.
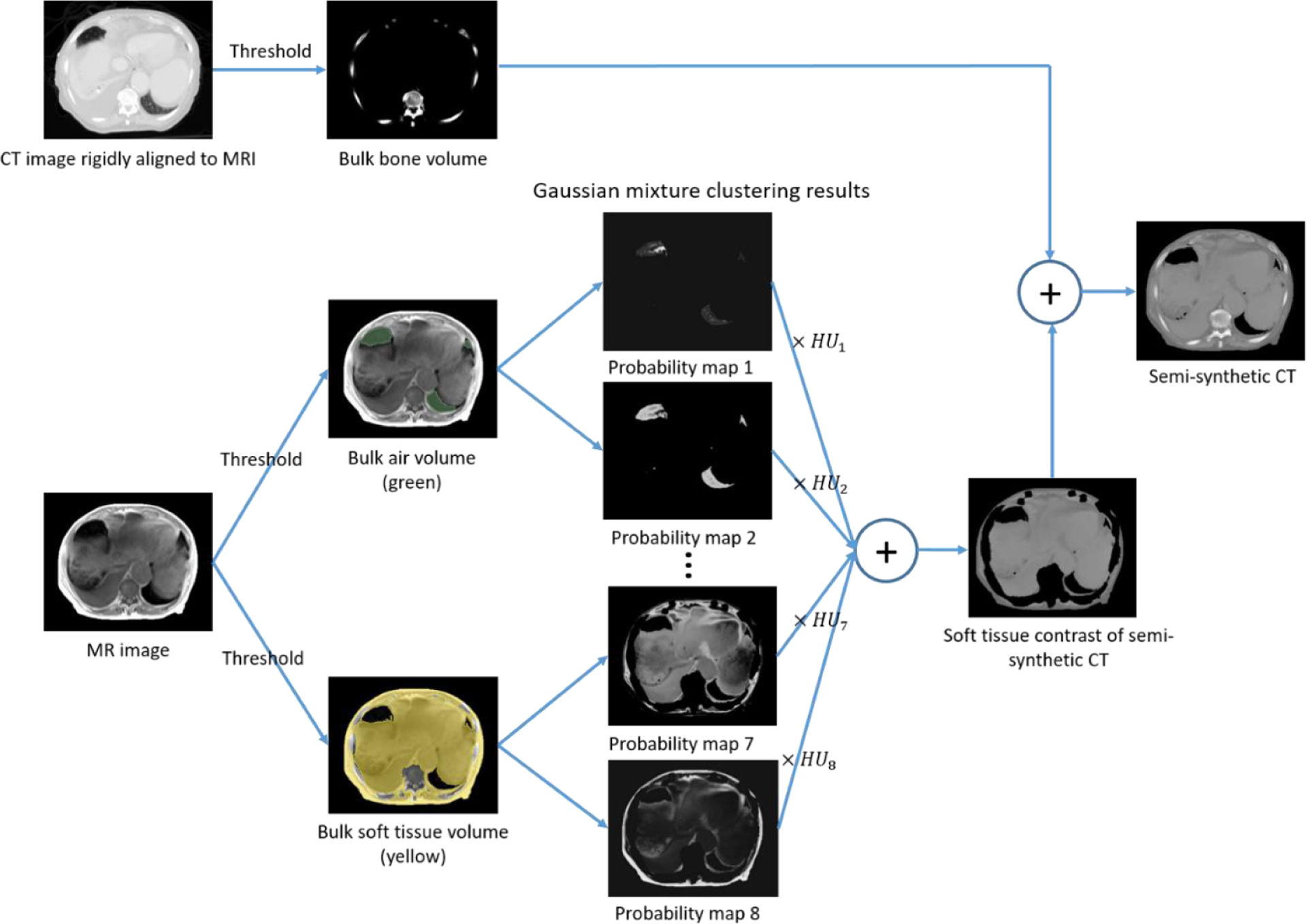
General workflow of ‘semi-synthetic’ CT generation for training dataset construction.
2.2.1. Bulk volume extraction
Three types of bulk volumes were extracted from MR and CT images: bone, soft tissue and air. A bone mask was extracted from CT images by applying a threshold of 150 HU. The CT-derived bone mask was rigidly aligned to the corresponding MR image volume using registration tools implemented in SLICER (Brigham and Women’s Hospital, Boston, MA). The calculated rigid transformation was also applied to the entire CT image volume to produce rigidly aligned CT and MR image pairs.
The soft tissue and air volumes were extracted from CT and MR images separately due to large deformation between scans. The external surface contour was first automatically delineated using intensity thresholds of −200 HU on CT images and 120 on T1-weighted MR images. Air volumes were determined as voxels with an intensity below −500 HU on CT images within the body contour. To determine air volumes on MR images, we first normalized image intensities by scaling so that the mean intensity of T1-weighted image voxels within the external body contour was 1000. This scale factor was also applied to the corresponding water images. Air volumes were then identified within the space occupied by the body contour but excluding the CT-based bone mask. Within this space, air voxels were identified as the intersection of two mask regions, generated by thresholding T1-weighted images at intensities below 150 and water images below 200, respectively. All remaining non-bone or air voxels within the body contour were defined to be soft tissue. All thresholding operations were followed by morphological operations (3 mm dilation, filling holes and 3 mm erosion) to clean up holes and gaps, using an in-house Functional Imaging Analysis Tool (FIAT). The second column in figure 3 shows an example of different bulk tissue volumes extracted from CT and MR images.
2.2.2. Voxel-wise intensity classification
To determine HU values for soft tissue and air voxels on MR images, we performed voxel-wise intensity classification on multi-spectral MR images (T1-weighted, water and fat) through Gaussian-mixture clustering using 5 classes for soft tissue and 3 for air. Gaussian-mixture models have been used for synthetic CT generation before (Johansson et al 2011), but suffered from degeneracies resulting from intensity overlaps between different tissues on MRI. The difference here is that we used Gaussian-mixture clustering for training data generation. Therefore, we can fully use corresponding rigidly aligned CT images to improve the accuracy of clustering. We first fitted a Gaussian-mixture model (GMM) to CT data x = [x1, x2, …, xN]T, where N is the total number of voxels, and xi is the HU number of voxel i. Using the expectation maximization (EM) algorithm implemented in MATLAB (Natick, MA), for each class j, we obtained the following model parameters: (class mean), (class variation), (class weight) and the probability of xi belonging to class . We then hardened soft clustering results to label maps LCT (i|j) where
Using such label maps, we constructed a GMM model on MRI data y = [y1, y2, …, yN]T, where yi is the MR intensity of voxel i. The model parameters were determined as
where 1 is the indicator function. In addition, to account for mismatches between MR and CT images, voxels that belonged to different bulk tissue volumes on CT and MR images (for example, voxels that belonged to soft tissue volume on CT but air on MRI) were excluded from the GMM model construction process above. In this case, were renormalized to ensure that . From the GMM model constructed, we calculated the probability map of voxel i belonging to class j on MRI as . Figure 4 shows the general workflow, with example Gaussian-mixture clustering results on CT images, model construction for MRI and final clustering results on MR images.
Figure 4.
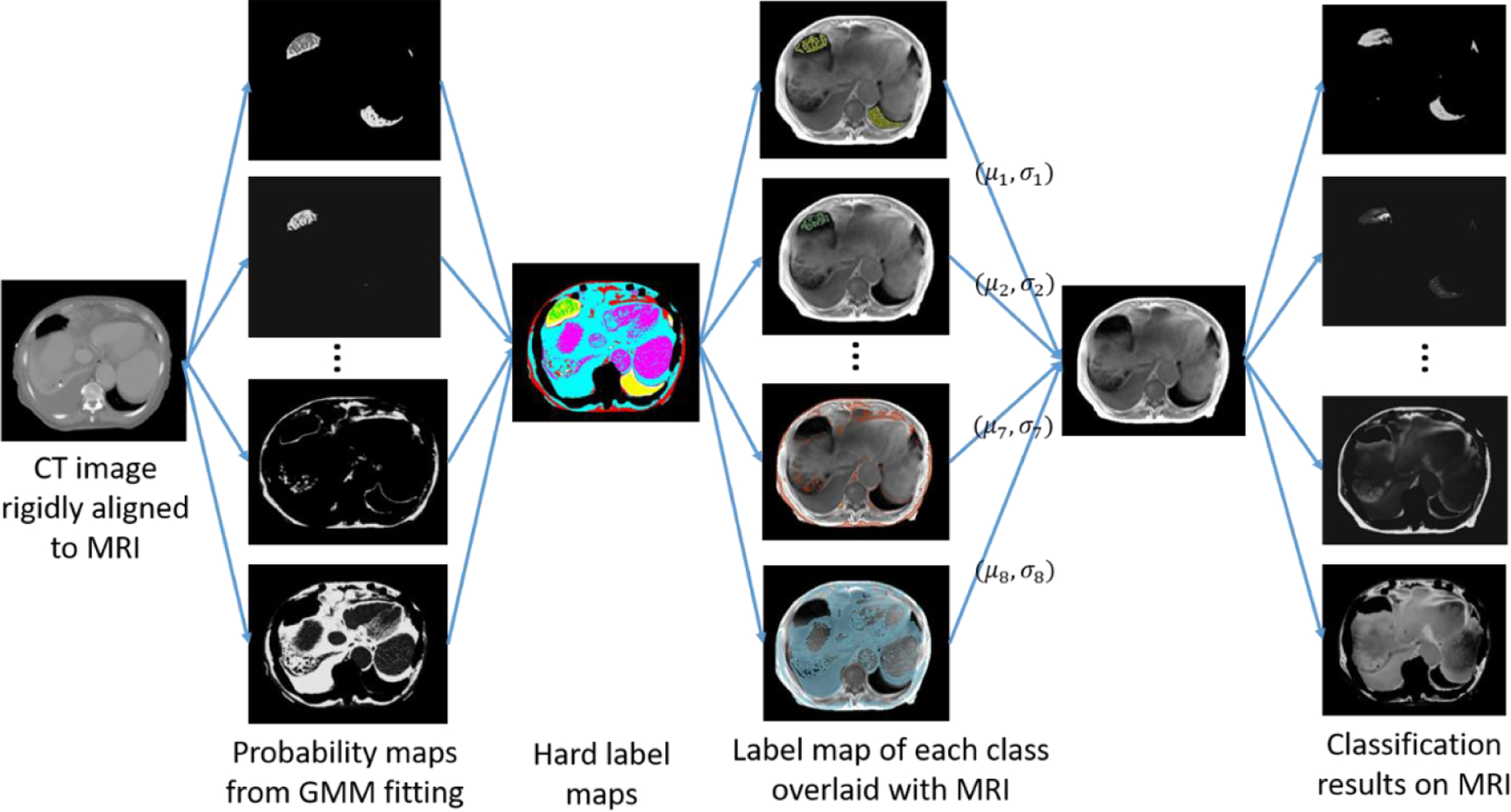
Voxel-wise classification of soft tissue and air volumes on MRI aided by classification results on rigidly aligned CT data.
2.2.3. ‘Semi-synthetic’ CT generation
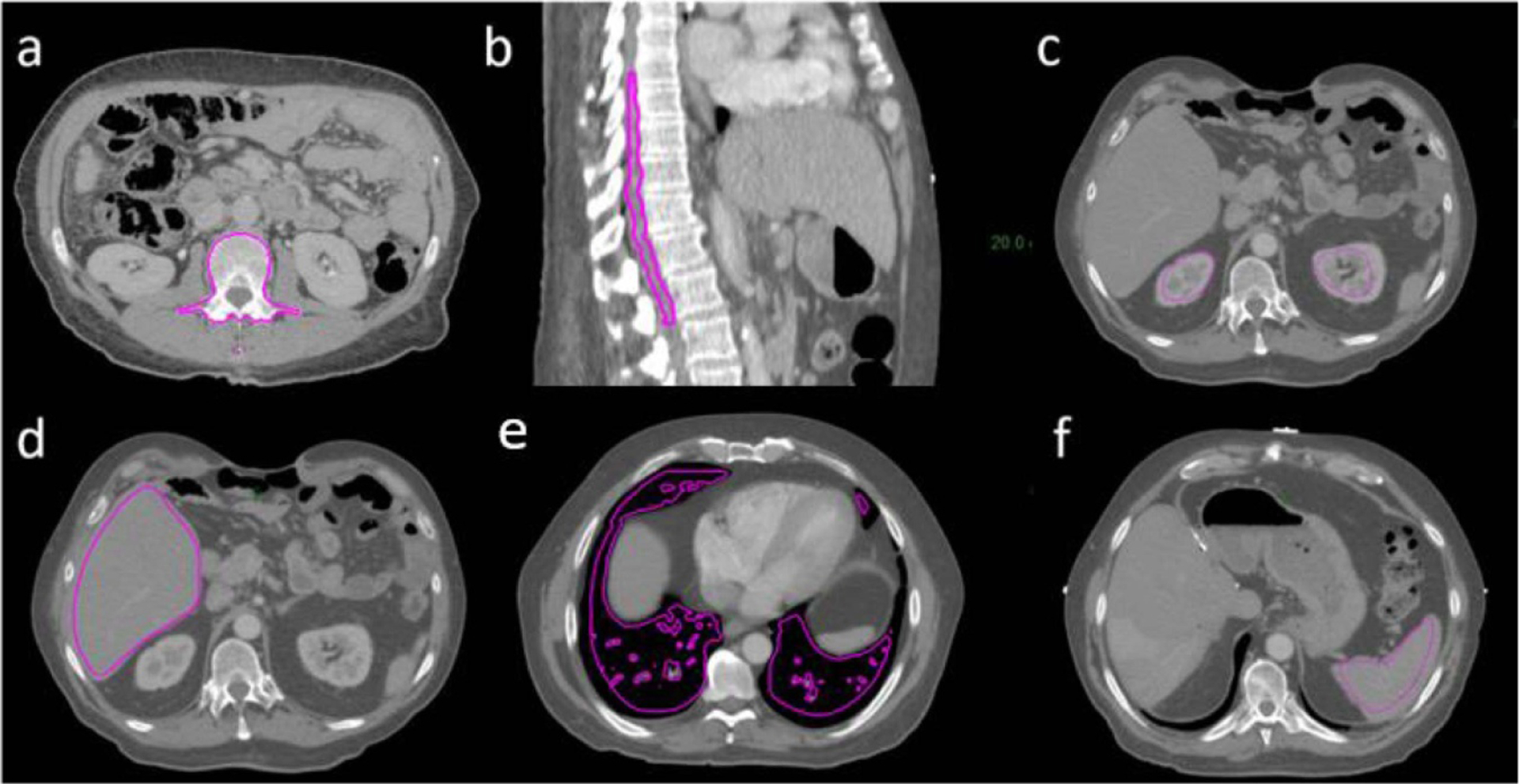
As illustrated in figure 3, soft tissue and air contrasts of ‘semi-synthetic’ CT were generated from voxel-wise tissue classification results on MRI by multiplying the probability of voxels belonging to different classes with the average HU value of respective classes and summing the results together. Specifically, the mean HU value for class j was calculated by averaging the HU values from CT images across all voxels that have the highest probability of belonging to class j on both MR and corresponding CT image volumes based on the GMM fitting results. Intensities of voxels within the bone mask were directly copied from CT image to fill their spaces within the ‘semi-synthetic’ CT images. Figure 5 shows example CT and MR images following skeletal alignment, as well as ‘semi-synthetic’ CT images generated using both MR and CT images. ‘Semi-synthetic’ CT images present contrast similar to CT images but with voxel-wise correspondence to MR images. The CT images, on the other hand, show considerable soft tissue deformation relative to MR images, as well as different amount of gas filling.
Figure 5.
Rigidly aligned (a) CT and (b) T1-weighted MR image pairs, and (c) ‘semi-synthetic’ CT image generated using both CT and MRI. Magenta contours indicate the bone mask within which ‘semi-synthetic’ CT voxel intensities were copied directly from CT.
2.3. U-net model for synthetic CT estimation
2.3.1. Network architecture
We used a simple U-net model to learn a mapping from MR images to ‘semi-synthetic’ CT images (figure 6). The input layers consisted of six 512 × 512 channels. The first three channels contained three contiguous T1-weighted image slices and the remaining three channels held their corresponding water image slices. By including neighboring slices, we constructed a 2.5D U-net with the goal of improving continuity between slices. The network structure is the same as the original U-net (Ronneberger et al 2015). It performs four downsampling steps in the contracting path and four corresponding upsampling steps in the expansive path. The final layer performs a 1 × 1 convolution with three channels of output, to map the input multi-contrast MR images to ‘semi-synthetic’ CT images of three contiguous slices.
Figure 6.
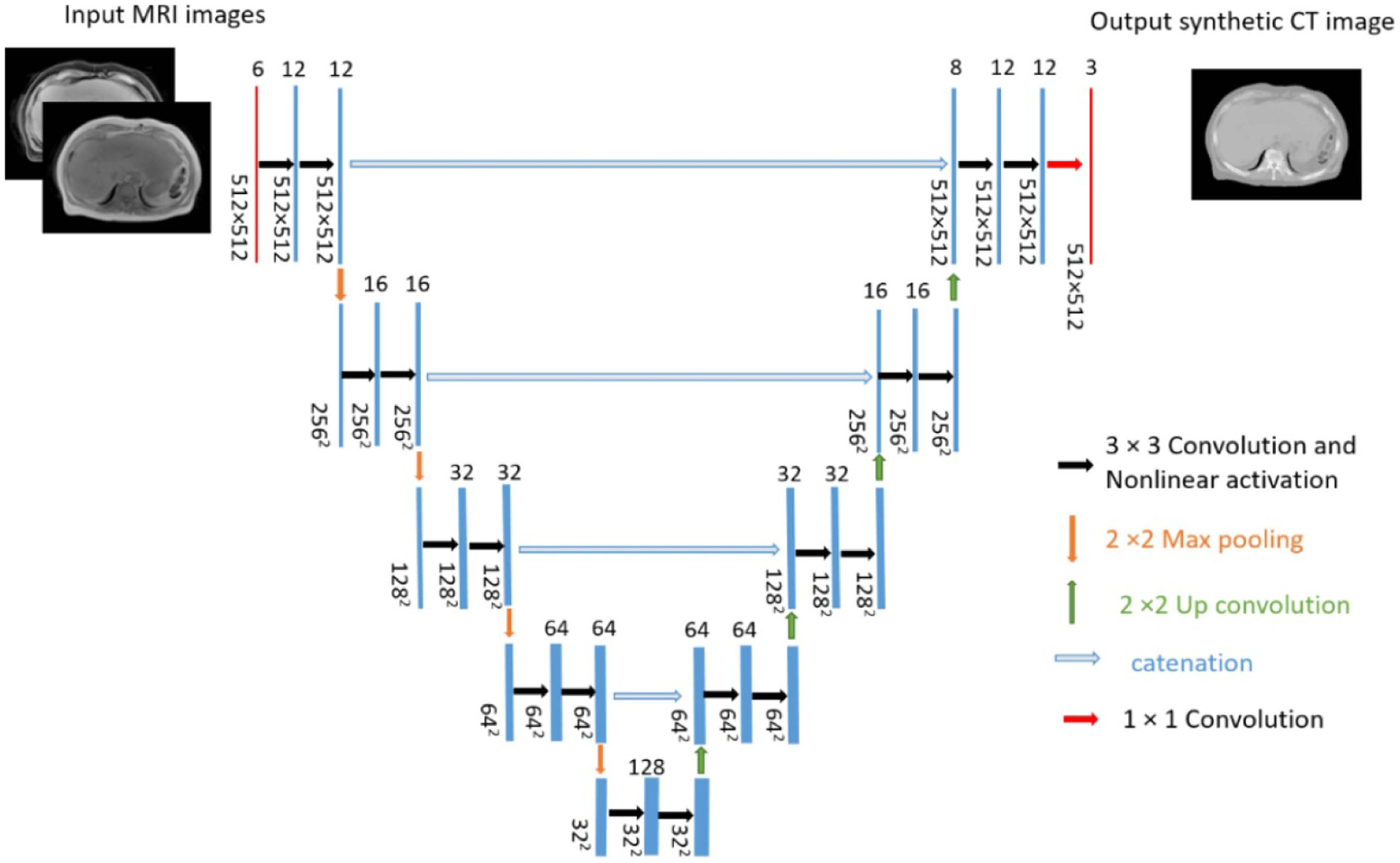
U-net architecture. For simplicity, the central of three contiguous input (T1-weighted in-phase and water images from Dixon MRI) and output (‘semi-synthetic’ CT image) channels are displayed.
2.3.2. Network training
Weighted-mean square error was chosen as the loss function, to offset potential bias in optimization due to unbalanced sizes of different bulk tissue volumes in the abdomen. For example, within the field of view of current MRI scans, the number of soft tissue voxels was on the order of 107, while the numbers of air voxels and bony voxels were less than 10% and 20% of soft tissue voxels respectively. The number of background voxels on the other hand, was four times of the number of soft tissue voxels. Based on such observations, we assigned lowest weights to background regions and highest weights to air pockets. Specifically, the weights associated with voxels that belong to background, soft tissue, bone and air were 0.01, 0.5, 1 and 2 respectively. Each 2D axial image slice, together with its two neighboring slices, were treated as one sample. The splitting ratio for training and validation was 0.9. Both MRI and ‘semi-synthetic’ CT data were normalized to an intensity range of [0,1] before training. We implemented the network using the publicly available Keras package and trained the network on two GPUs with a total memory of 22 GB. The batch size was 8 and the optimization method was Adam. Data augmentation, including random shift (up to 25 voxels), rotation (up to 30°), scaling (range from 0.9 to 1.1) and horizontal flip was applied to each batch of input. Training was stopped after 500 epochs.
2.4. Model evaluation
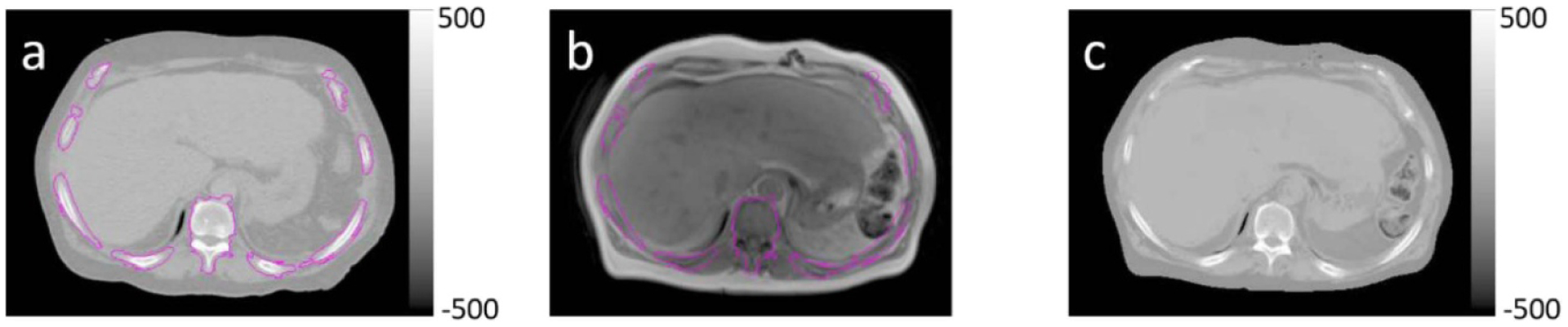
The trained model was used to generate synthetic CT images for patients whose data were not used during the training stage. The training process took 144 h. After the training was completed, generation of synthetic CT for one patient took less than 30 s on a CPU. A 3-fold cross validation was performed to evaluate the model on a subset of 31 of the 46 patients, specifically those that had treatment plans generated in the current clinical treatment planning system (Eclipse 15.6, Varian Medical Systems, Palo Alto, CA) with MRI scan ranges covering the entire treatment fields. Both intensity correlations between synthetic CT and corresponding CT image volumes, as well as accuracy of using synthetic CT images for treatment planning dose calculations were evaluated. We first aligned MR, synthetic CT and CT image volumes into the same coordinate system using rigid registration implemented in SLICER (Brigham and Women’s Hospital, Boston, MA). Similar with previous studies (Bredfeldt et al 2017), we calculated mean absolute error (MAE) between synthetic CT and CT images in 6 different organs (vertebral bodies, spinal cord, kidneys, liver, spleen and lungs). Regions of interest (ROIs) of vertebral bodies were generated from CT images by intensity thresholding at 150 HU. To avoid confounding factors introduced by soft tissue deformation, we generated ROIs on MR and CT images independently for the remaining 5 organs. MAE values were calculated within the spatial intersections of MR and CT ROIs between CT and synthetic CT images for each organ. Figure 7 shows example ROIs for each organ overlaid with CT images.
Figure 7.
Example ROIs (magenta contours) overlaid with CT images. (a) Vertebral body, (b) spinal cord, (c) kidneys, (d) liver, (e) lungs and (f) spleen.
For dosimetric evaluation purposes, we performed deformable registration using a commercial software package (VelocityAI 4.0, Varian Medical Systems, Palo Alto, CA) to improve alignment between synthetic CT and corresponding treatment planning CT images in soft tissue regions. The aligned synthetic CT and planning CT image volumes were then imported into the clinical treatment planning system. Among the 31 patients evaluated, 18 patients received 5 fractions with 10 to 12 Gy per fraction, 1 patient received 20 fractions with 2.8 Gy per fraction, 1 patient received 30 fractions with 1.8 Gy per fraction, and the remaining patients were from an adaptive therapy protocol that prescribes 3 fractions with 7 to 20 Gy per fraction. We copied clinical structure sets contoured on planning CT images to synthetic CT images. Due to the limited scan range of the MRI data used, contours of heart and spinal cord were edited to be within the body contour of synthetic CT volumes. Treatment plans (22 volumetric modulated arc therapy plans and 9 intensity modulated radiation therapy plans) were first optimized using CT images for electron density estimation with the goal of meeting clinical treatment planning directives. Beam fluences from each of the CT-optimized plans were then used to calculate dose using the associated synthetic CT-derived electron density grids. Clinically relevant dose metrics between CT optimizations and synthetic CT recalculations were compared for PTVs and various organs at risk (OARs) including liver, spinal cord, colon, duodenum, esophagus and heart.
3. Results
Figure 8 shows rigidly aligned CT and MR images of two example patients, as well as synthetic CT images generated from MR images using the trained network. Synthetic CT images present contrast similar to that of CT images and successfully preserve small bony structures such as ribs. Structural details, such as local soft tissue interfaces, are consistent between synthetic CT and source MR images, as indicated by red arrows in figure 8. Figure 9 shows bar plots of MAEs for different organs between CT and synthetic CT images. Table 1 summarizes the mean and standard deviation of MAEs across the 31 patients.
Figure 8.
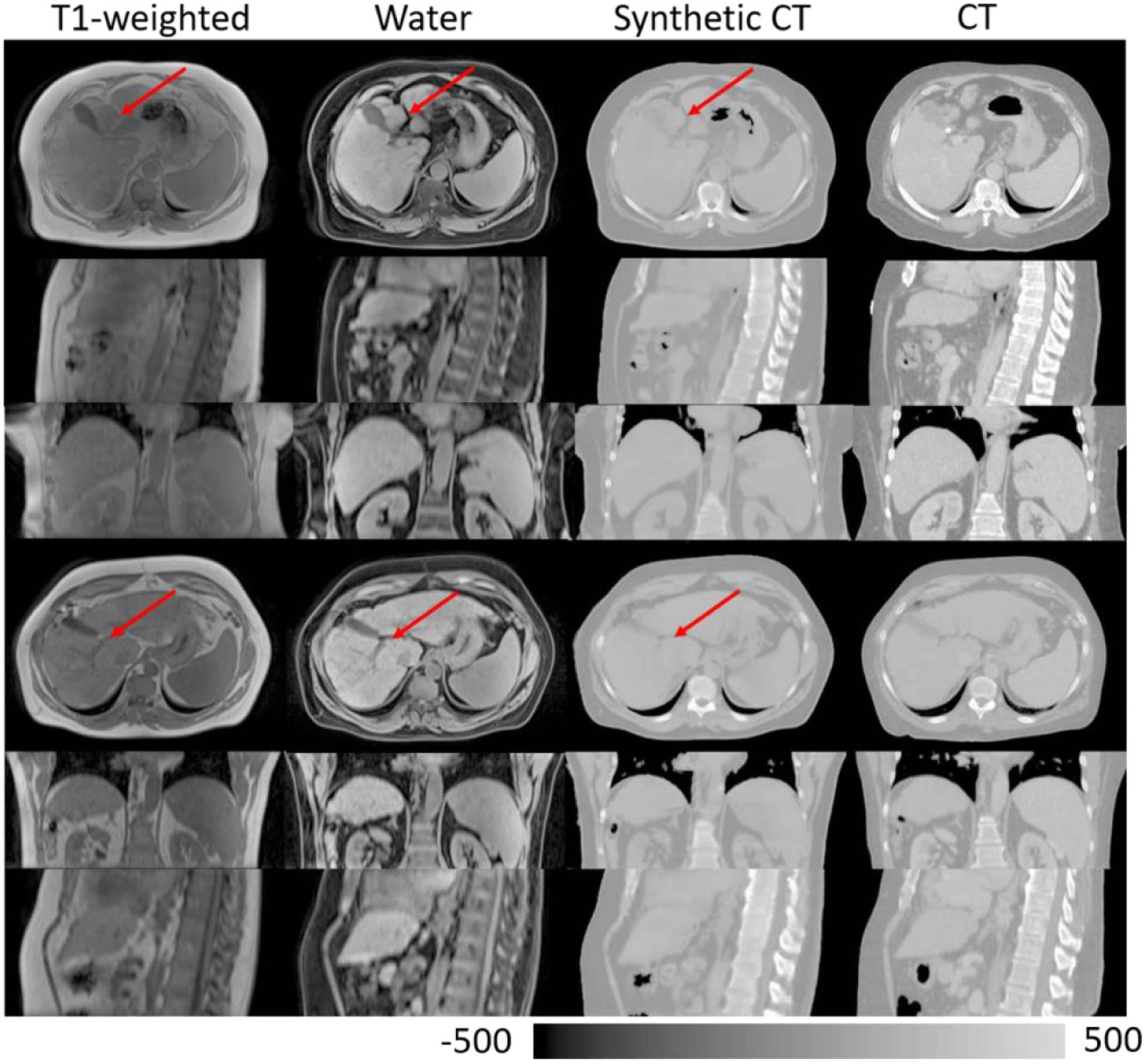
Rigidly aligned MR images, synthetic CT images and CT images of two example patients. Red arrows point to example structural details that are preserved between MR and synthetic CT images.
Figure 9.
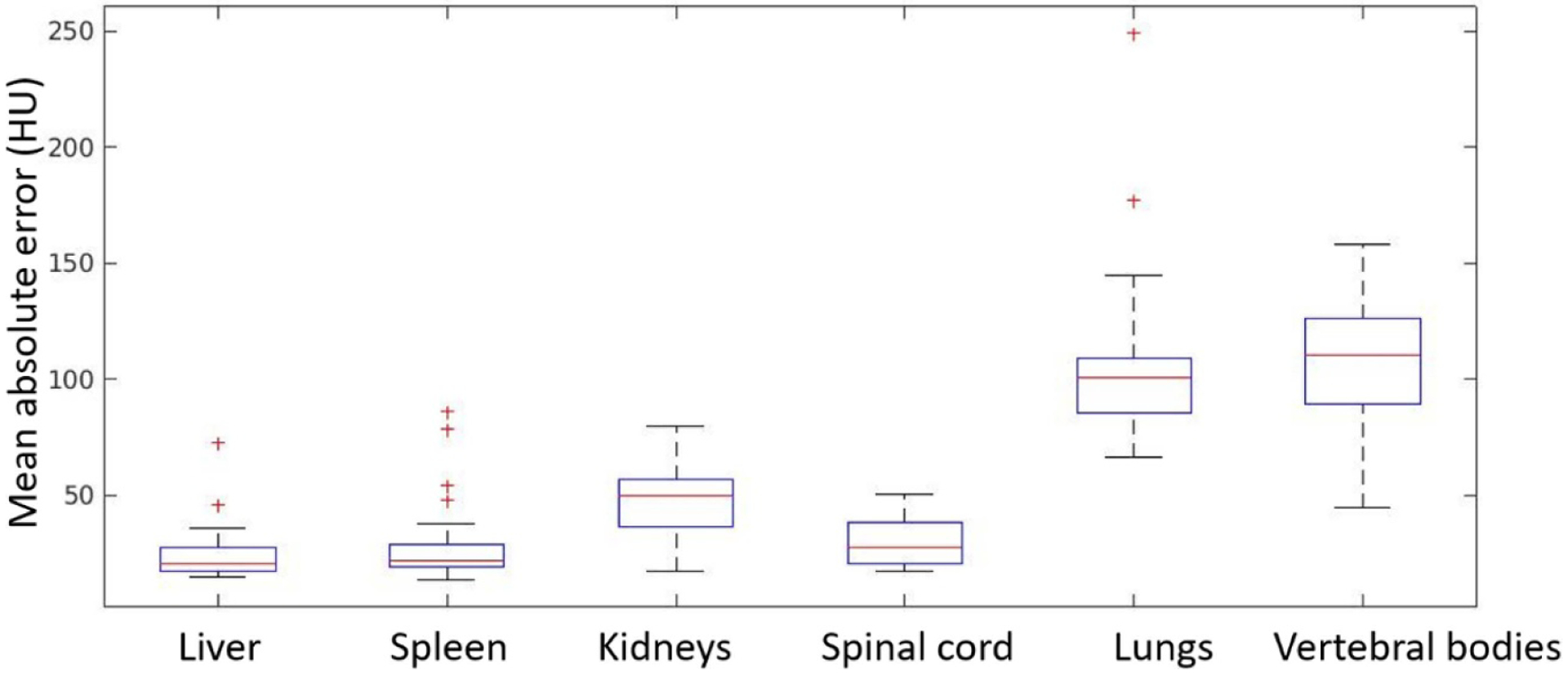
Mean absolute error (MAE) between synthetic CT and CT images within ROIs of various organs. Box and whisker plots show the median (center-line), 75th percentile (top of box), 25th percentile (bottom box), max and min (whisker ends) and outliers (‘+’ symbol).
Table 1.
Summary of MAEs across patients in different organs (HU).
| Liver | Spleen | Kidneys | Spinal cord | Lungs | Vertebral bodies | |
|---|---|---|---|---|---|---|
| Mean | 24.10 | 28.62 | 47.05 | 29.79 | 105.68 | 110.09 |
| Standard deviation | 11.54 | 17.48 | 14.98 | 9.43 | 35.00 | 29.23 |
Figure 10 shows example dose distributions on CT and synthetic CT images, as well as dose volume histograms (DVH) for PTVs, non-tumor bearing liver and spinal cord. The dose distributions are visually very similar between CT and synthetic CT images. Differences in DVH plots are also minimal. Figure 11 shows differences in clinically relevant dose metrics between CT plans and synthetic CT recalculations. The mean differences across patients were less than 0.15 Gy for all PTV and OAR dose metrics, and the maximum dose difference across patients was less than 1 Gy, with the noted exception of one patient where the maximum dose to PTV differed by more than 2 Gy. A closer look at the plan for this patient showed that the tumor was located at the superior border of the liver and the clinical PTV volume contained a significant amount of air. As shown in figure 12, deformable registration was not able to fully resolve air volume differences within PTV contours between CT and synthetic CT images, which resulted in a visible difference in PTV maximum doses.
Figure 10.
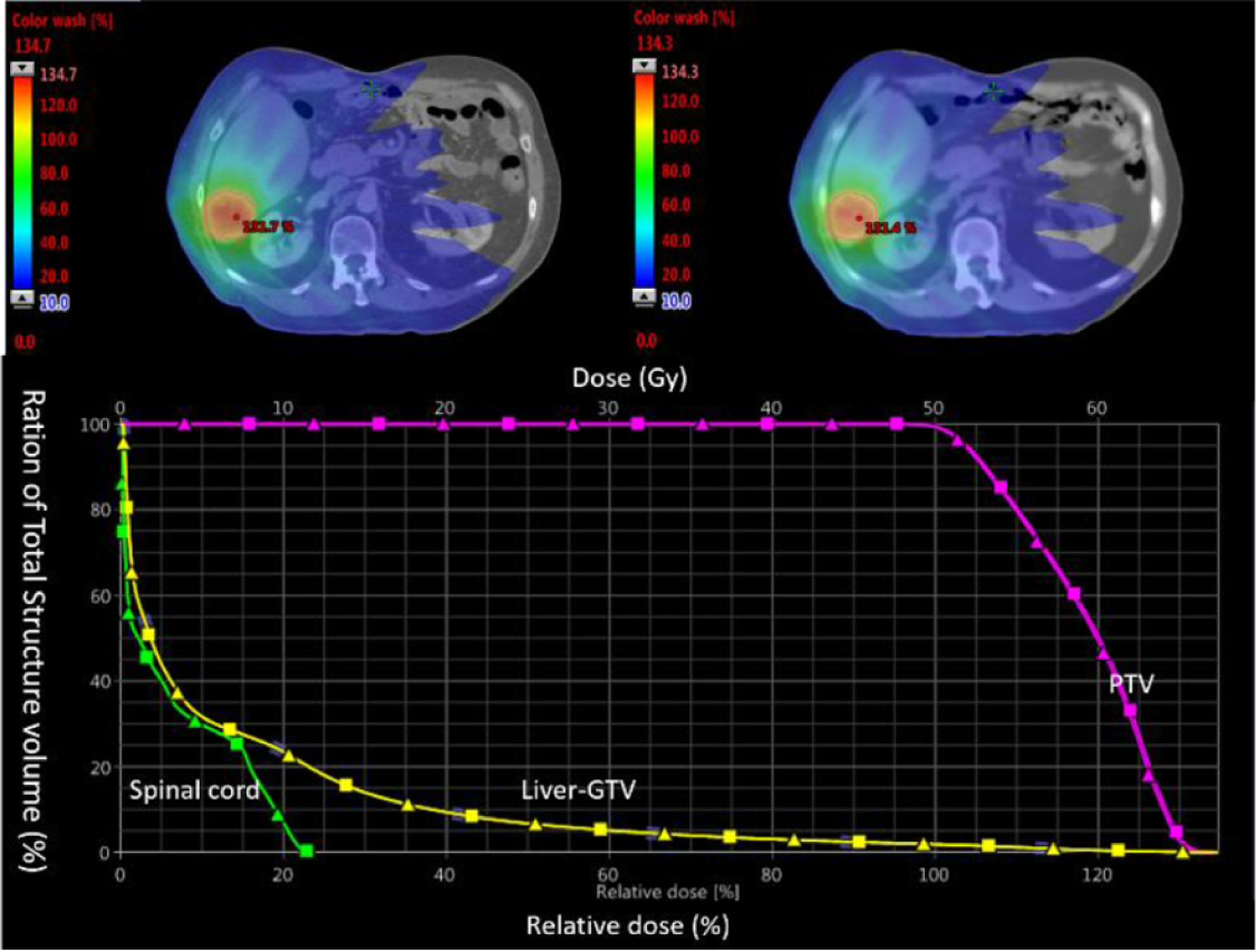
Top: dose distribution on CT (left) and synthetic CT (right) images. Bottom: dose volume histograms of PTV, liver (exclude gross tumor volume) and spinal cord from CT (squares) and synthetic CT (triangles) calculations.
Figure 11.
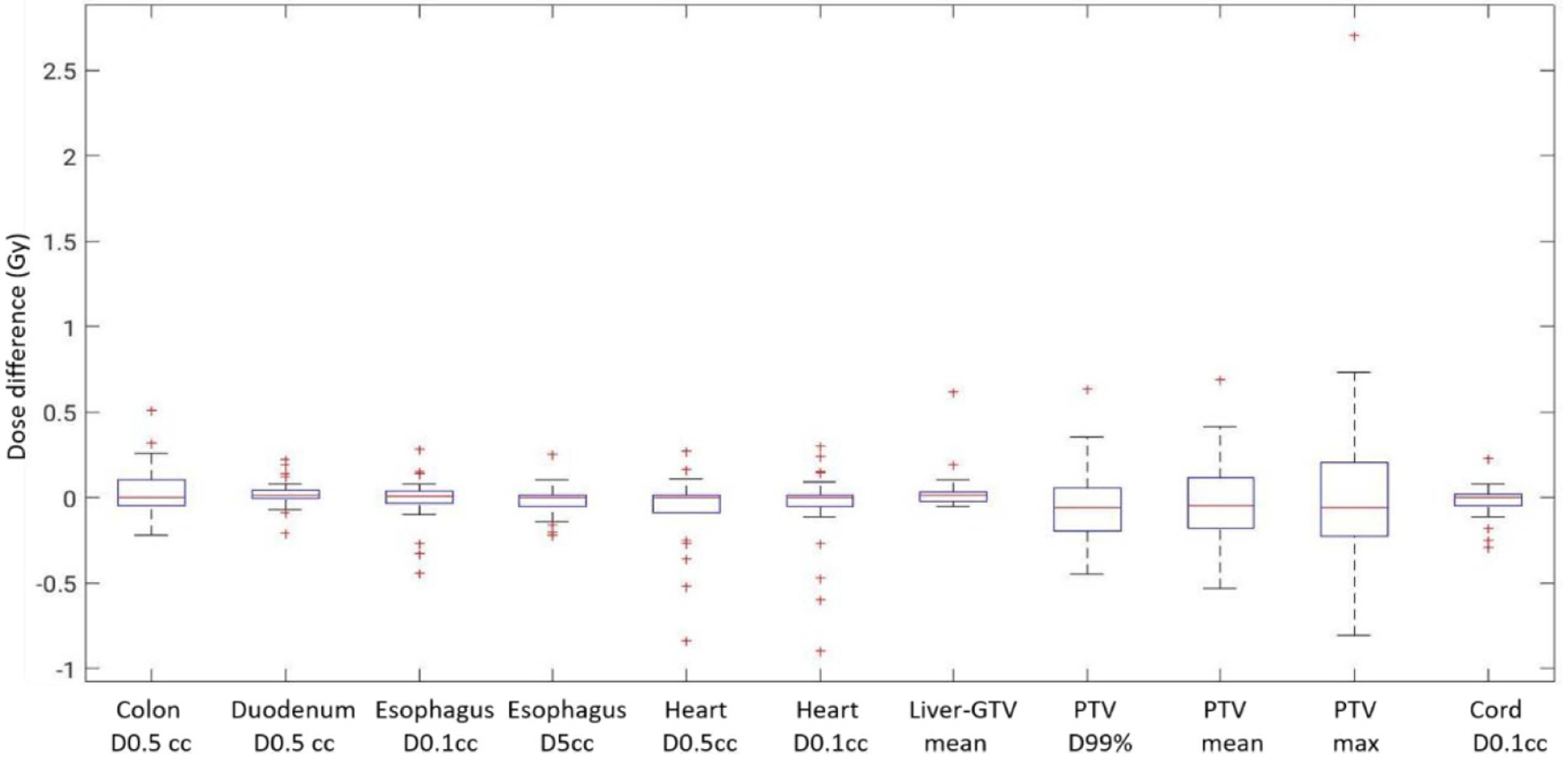
Dose differences between CT plans and synthetic CT recalculations for OARs and PTV. Box and whisker plots show the median (center-line), 75th percentile (top of box), 25th percentile (bottom box), max and min (whisker ends) and outliers (‘+’ symbol).
Figure 12.
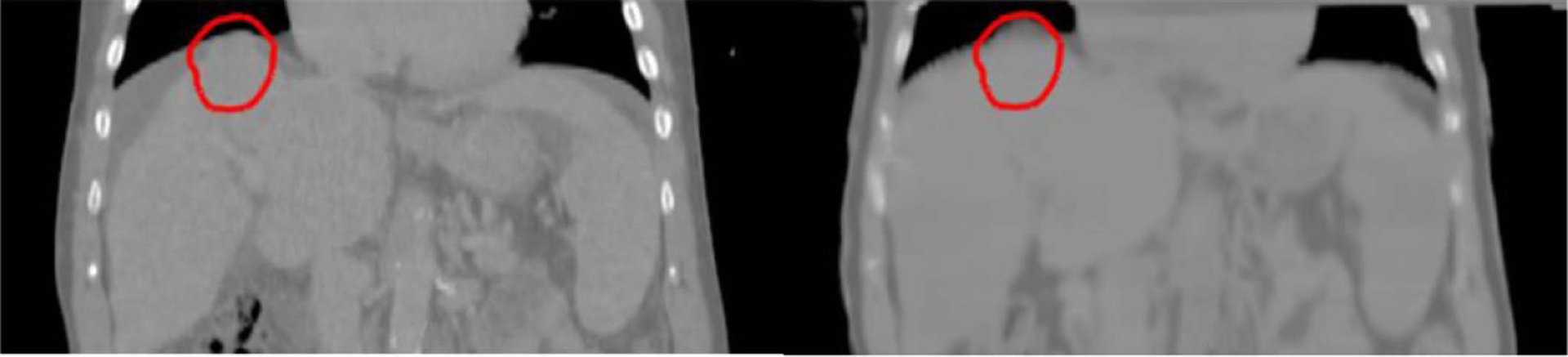
Outlier patient whose PTV volume (red contour) contained a significant amount of air. The amount of air was different between CT and synthetic CT images, which resulted in difference in maximum PTV dose.
4. Discussion
This study investigated a novel scheme to train deep neural networks for abdominal synthetic CT generation. The major challenge for this task has been the mismatch between MRI and CT scans due to various sources of patient motion. Instead of relying on deformable registration to resolve such mismatch, this study proposed to avoid this issue through an intermediate step, where ‘semi-synthetic’ CT images that have voxel-wise correspondence to MR images were generated and used for training. We trained a simple U-net model from such MR and ‘semi-synthetic’ CT image pairs. Synthetic CT images predicted by the trained model presented contrast sufficiently close to CT images to support radiation treatment planning, as validated by both direct intensity comparison and by performing treatment planning using CT image volumes and recalculating dose distributions on synthetic CT image volumes.
The presented method differs from previously published studies. First, it eliminates the need of deformable alignment of MRI and CT images for training. Deformable registration in the abdomen is still an open problem. Limited accuracy in registration has introduced blurry artifacts and local distortions in synthetic CT images generated (Liu et al 2019). Second, our network structure is simple without adversarial loss components. The goal of introducing adversarial blocks has been to deal with mismatches in the training dataset. We showed that by training from well aligned image pairs, the classic U-net model is capable of producing comparable or even better results. Comparing to the work by Bredfeldt et al (Bredfeldt et al 2017), the presented method is able to preserve small bony structures such as ribs in synthetic CT images generated. The MAEs in both bony tissues and air (lung volumes) were also lower than previously reported (Liu et al 2019). Although direct comparison between dose distributions on CT and synthetic CT was complicated by the challenge of deformable alignment of the two image volumes, treatment planning evaluation showed acceptably small variations. The network was trained/validated on a group of patients with varying sizes and lung volumes, suggesting the robustness of the proposed method to different patient configurations.
The MRI scans used in this study covered a reduced longitudinal scan range as compared to their corresponding CT scans. Future studies will extend the field of view of MRI scans through stitching of scans at 2 table positions. Geometric distortion and peripheral signal loss of MR images may also impact the quality of synthetic CT images and require further investigation. Due to memory limits, the current U-net model used very thin image slabs (3 slices). Including more neighboring slices or training a 3D U-net may have the potential of learning richer information and further improving the quality of synthetic CT images. Another alternative to speed up the training process and reduce the use of GPU memory may be splitting images into patches to reduce the size of input, which will be investigated in future studies. It will also be interesting to investigate the weighted loss function more thoroughly, by experimenting with different weighting schemes and evaluating the sensitivity of the network to different weight assignments. Finally, this study assumed that the entire bony structure in the abdomen is rigid, which is a simplified assumption. Although each separate skeletal element (a single vertebral body or rib) is rigid, different elements may move relative to each other. More sophisticated registration algorithms that attempt to preserve rigidity of each bony element (Kim et al 2013) may further improve the quality of ‘semi-synthetic’ CT images and thus the quality of network predicted synthetic CT images.
5. Conclusion
This study presented a novel scheme of training neural network for abdominal synthetic CT generation from MRI scans. By generating a training dataset that consisted of paired MR images and ‘semi-synthetic’ CT images, which were generated by combining MRI and CT information and had voxel-wise correspondence to MR images, the problem of mismatch between MRI and CT scans was avoided, and a simple network structure without adversarial loss components was able to predict synthetic CT images of sufficient integrity to support MRI-only radiotherapy treatment planning.
Acknowledgment
This work was supported by NIH R01 EB016079.
References
- Bredfeldt JS, Liu L, Feng M, Cao Y and Balter JM 2017. Synthetic CT for MRI-based liver stereotactic body radiotherapy treatment planning Phys. Med. Biol 62 2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami H, Dong M, Nejad-Davarani SP and Glide-Hurst CK 2018. Generating synthetic CTs from magnetic resonance images using generative adversarial networks Med. Phys 45 3627–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X 2017. MR-based synthetic CT generation using a deep convolutional neural network method Med. Phys 44 1408–19 [DOI] [PubMed] [Google Scholar]
- Johansson A, Balter J and Cao Y 2018a. Rigid-body motion correction of the liver in image reconstruction for golden-angle stack-of-stars DCE MRI Magn. Reson. Med 79 1345–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Balter JM and Cao Y 2018b. Abdominal DCE-MRI reconstruction with deformable motion correction for liver perfusion quantification Med. Phys 45 4529–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson A, Karlsson M and Nyholm T 2011. CT substitute derived from MRI sequences with ultrashort echo time Med. Phys 38 2708–14 [DOI] [PubMed] [Google Scholar]
- Kim J, Matuszak MM, Saitou K and Balter JM 2013. Distance-preserving rigidity penalty on deformable image registration of multiple skeletal components in the neck Med. Phys 40 121907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. et al. MRI-based treatment planning for proton radiotherapy: dosimetric validation of a deep learning-based liver synthetic CT generation method. Phys. Med. Biol. 2019;64:145015. doi: 10.1088/1361-6560/ab25bc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maspero M, Savenije MH, Dinkla AM, Seevinck PR, Intven MP, Jurgenliemk-Schulz IM, Kerkmeijer LG and van den Berg CA 2018. Dose evaluation of fast synthetic-CT generation using a generative adversarial network for general pelvis MR-only radiotherapy Phys. Med. Biol 63 185001. [DOI] [PubMed] [Google Scholar]
- Nie D, Trullo R, Lian J, Wang L, Petitjean C, Ruan S, Wang Q and Shen D 2018. Medical image synthesis with deep convolutional adversarial networks IEEE Trans. Biomed. Eng 65 2720–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronneberger O, Fischer P and Brox T 2015. U-net: convolutional networks for biomedical image segmentation Int. Conf. on Medical Image Computing and Computer-Assisted Intervention (Lecture Notes in Computer Science, vol 9351) (Berlin: Springer; ) pp 234–41 [Google Scholar]
- Simeth J, Johansson A, Owen D, Cuneo K, Mierzwa M, Feng M, Lawrence TS and Cao Y 2018. Quantification of liver function by linearization of a two-compartment model of gadoxetic acid uptake using dynamic contrast-enhanced magnetic resonance imaging NMR Biomed. 31 e3913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemkens B, Paulson ES and Tijssen RH 2018. Nuts and bolts of 4D-MRI for radiotherapy Phys. Med. Biol 63 21TR01. [DOI] [PubMed] [Google Scholar]
- Tsegmed U et al. 2017. Functional image-guided stereotactic body radiation therapy planning for patients with hepatocellular carcinoma Med. Dosim 42 97–103 [DOI] [PubMed] [Google Scholar]
- Tustison NJ, Avants BB, Cook PA, Zheng Y, Egan A, Yushkevich PA and Gee JC 2010. N4ITK: improved N3 bias correction IEEE Trans. Med. Imaging 29 1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Sun J, Carass A, Zhao C, Lee J, Xu Z and Prince J 2018. Unpaired brain MR-to-CT synthesis using a structure-constrained CycleGAN Deep Learning in Medical Image Analysis and Multimodal Learning for Clinical Decision Support DLMIA 2018, ML-CDS 2018 (Lecture Notes in Computer Science, vol 11045) (Berlin: Springer; ) pp 174–82 [Google Scholar]
- Wang H, Feng M, Jackson A, Ten Haken RK, Lawrence TS and Cao Y 2016. Local and global function model of the liver Int. J. Radiat. Oncol. Biol. Phys 94 181–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wojcieszynski AP et al. 2016. Gadoxetate for direct tumor therapy and tracking with real-time MRI-guided stereotactic body radiation therapy of the liver Radiother. Oncol 118 416–18 [DOI] [PubMed] [Google Scholar]


