Abstract
Background
In-depth analysis of the HIV pandemic at its epicenter in the Congo basin has been hampered by 40 years of political unrest and lack of functional public health infrastructure. In recent surveillance studies (2017-18), we found that the prevalence of HIV in Kinshasa, Democratic Republic of Congo (11%) far exceeded previous estimates.
Methods
10,457 participants were screened in Kinshasa with rapid tests from 2017-2019. Individuals confirmed as reactive by the Abbott ARCHITECT HIV Ag/Ab Combo assay (n=1968) were measured by the Abbott RealTime HIV-1 viral load assay. Follow up characterization of samples was performed with alternate manufacturer viral load assays, qPCR for additional blood borne viruses, unbiased next generation sequencing, and HIV Western blotting.
Findings
Our data suggested the existence of a significant cohort (n=429) of HIV antibody positive/viral load negative individuals. We systematically eliminated collection site bias, sample integrity, and viral genetic diversity as alternative explanations for undetectable viral loads. Mass spectroscopy unexpectedly detected the presence of 3TC antiviral medication in approximately 60% of those tested (209/354), and negative Western blot results indicated false positive serology in 12% (49/404). From the remaining Western blot positives (n=53) and indeterminates (n=31) with reactive Combo and rapid test results, we estimate 2.7-4.3% of infections in DRC to be potential elite controllers. We also analyzed samples from the DRC collected in 1987 and 2001-03, when antiretroviral drugs were not available, and found similarly elevated trends.
Interpretation
Viral suppression to undetectable viral loads without therapy occurs infrequently in HIV-1 infected patients around the world. Mining of global data suggests a unique ability to control HIV infection arose early in central Africa and occurs in <1% of founder populations. Identification of this group of elite controllers presents a unique opportunity to study potentially novel genetic mechanisms of viral suppression.
Funding
Abbott Laboratories funded surveillance in DRC and subsequent research efforts. Additional funding was received from a MIZZOU Award from the University of Missouri. Research was supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH.
Keywords: HIV, Viral controller, Elite controller, Democratic Republic of Congo
Research in context.
Evidence before this study
Surveillance in Kinshasa has demonstrated extensive viral genetic diversity and indicated that HIV prevalence in Democratic Republic of Congo far exceeds most estimates. Further characterization of this population here revealed an unusually high number of HIV positive individuals with undetectable viral loads. The ability to naturally suppress HIV replication generally occurs in fewer than 1% of infections globally.
Added value of this study
Approximately 60% of these participants failed to disclose ongoing anti-retroviral therapy, highlighting the perils of reliance on self-reporting. Nevertheless, having eliminated other explanations, we report that 2.7-4.3% of all HIV-infected individuals from DRC are antibody positive/viral load negative and represent potential elite controllers.
Implications of all the available evidence
Just as the HIV epidemic is believed to have its origins in the Congo River basin, it suggests a unique ability to control HIV infection arose there as well. Studying this group of individuals may provide insight into novel genetic mechanisms of viral suppression.
Alt-text: Unlabelled box
Introduction
The introduction of antiretroviral therapy (cART) has been a major contributor to the decreasing incidence and prevalence of HIV-1 worldwide [1]. In order to reach the 90-90-90 goal outlined by the World Health Organization (WHO), individuals who do not know they are infected must be diagnosed and treated. Surveillance efforts in resource-limited settings like the Democratic Republic of the Congo (DRC) can provide this basic public health framework. Indeed, a recent investigation in Kinshasa estimated that the HIV prevalence in DRC (11%) far exceeds previous estimates (2.86%) and that a concerted response will be necessary to control the epidemic there and in neighboring countries [2]. The origin of the HIV pandemic has been traced to the Congo Basin and the oldest known strains, ZR59 (subtype D) and DRC60 (subtype A), were found in Kinshasa, DRC [[3], [4], [5]]. Habitats of non-human primates carrying SIV strains ancestral to HIV reside in close proximity to towns and villages, and thus countries like Cameroon and the DRC are home to every subtype and group of HIV-1 (M, N, O, P) [[6], [7], [8], [9], [10], [11]]. Surveillance in settings such as the DRC with a high degree of viral genetic diversity provides the opportunity to ensure that diagnostic tests reliably detect all infections [12].
It is well established that an exceptionally rare group of HIV-1 seropositive individuals can control their HIV viral loads to undetectable levels without antiviral therapy for 10 years or more [13]. These individuals, elite controllers (ECs), are defined by a durable threshold viral load of <50 copies/ml. Individuals with sustained viral loads (VL) from 50-2000 copies/ml are termed viral controllers (VCs) [14,15]. Elite controllers comprise approximately 0.1-2.5% of all HIV infections worldwide and are less likely to develop AIDS [16]. These patients resemble those infected with HIV-2, where lower VLs and more robust antibody and T cell responses delay progression to AIDS [[17], [18], [19], [20]]. Elite controllers and HIV-2 infections are ideal sources for identification of naturally occurring mechanisms evolved to restrict and possibly prevent HIV replication. Characterization of ECs has confirmed infection with replication-competent strains of HIV, and multiple host factors that contribute toward viral restriction have been identified, including specific HLA subtypes associated with slower disease progression, higher mRNA levels of T cell p21 expression, and lower CCR5 levels [[21], [22], [23]]. These key insights into the virology and immunology of HIV demonstrate some of the critical elements of immunity that would be required of a protective HIV vaccine [24,25].
Recent surveillance efforts (2017-2018) in the DRC have involved 48 sites within the greater Kinshasa city area, occupying approximately two hundred square miles, with sites located in multiple health zones controlled by different directors, and one remote site at the rural hospital in Vanga, 150 miles from Kinshasa [2]. We consistently observed an unusually high number of antibody-reactive, viral load-negative specimens. Follow-up experiments confirmed these findings but revealed that many individuals (~60%) were receiving antiretroviral therapy, unbeknownst to the surveillance team, and others were eliminated for false positive serology. Nevertheless, the unexplained and intriguing absence of viremia in the remaining individuals (n ≈ 84) prompted us to revisit historic surveillance data and analyze archival samples obtained more than thirty years ago from former Zaire. Each of these cohorts exhibited a similar trend, indicating that a sizeable percentage of individuals in the Congo basin are able to control HIV infection compared to what is observed globally.
Methods
Patient recruitment and specimen acquisition Patients seeking healthcare at any of 48 clinical sites in Kinshasa, Democratic Republic of Congo (DRC) from 2017-2018 (Fig. 1a) were recruited for this study [2]. One rural hospital at Vanga also participated and is included. Exclusions were children under 18 and HIV positive patients receiving cART; specimens were de-identified. Patients provided a plasma sample and received an HIV rapid test (RDT) free of charge and the health centers received a stipend. Seropositivity was confirmed following the national algorithm. Subsequent testing and sequencing occurred at Abbott Laboratories. De-identified specimens from the Democratic Republic of Congo were obtained in 1987 as part of Project SIDA by the US National Institute of Allergy and Infectious Diseases. DRC plasma specimens from 2001-2003 were collected at the Vanga Hospital, Bandundu Province and the Good Shepherd Hospital, located 12 km from Kananga, Kasai-Occidental Province. These were from participants seeking voluntary testing and pregnant women attending an HIV clinic [26]. Cameroon study participants (2004-2017) were recruited from blood bank donors, hospitals, and chest clinics in the urban centers of Douala and Yaoundé [6].
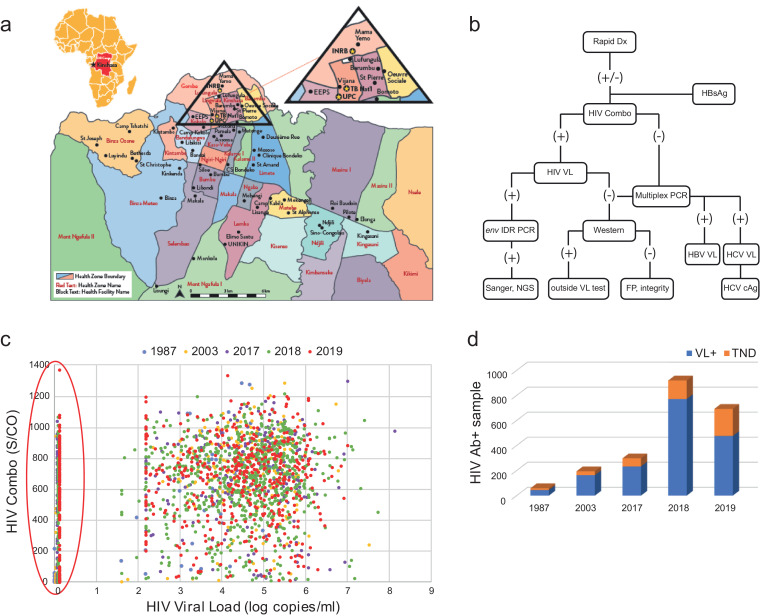
Fig. 1.
High percentage of viral load ‘TND’ in DRC. a) Map of collection sites in Kinshasa, DRC. b) Surveillance testing algorithm. FP = false positive, IDR = gp41 immunodominant region, outside test = alternate VL manufacturer. c) Plot of HIV Combo S/CO vs HIV-1 viral load. Legend indicates color for year sample was received. Samples on the y-axis with viral load results of ‘target not detected’ (TND) are circled. d) Histogram relating the number of tentative TNDs among HIV viral load positives for each cohort.
Serology testing Abbott on-market ARCHITECT assays were used for testing according to manufacturer instructions: HIV Ag/Ab Combo (2P36), HBV surface antigen (HBsAg) Qualitative (4P53), and HCV Ag (6L47).
HIV Viral load testing HIV viral loads were first determined on Ab+ samples using the Abbott m2000 HIV-1 RealTime assay (Abbott Laboratories, Des Plaines IL) using the 0.2 ml input volume (200 µl extraction and 300 µl dead-volume). 115 ‘TND’ samples (+ 3 VL positives as controls) were sent to Quest Diagnostics for independent evaluation (test code 40085). To conserve volume, patient plasma was diluted 1:1 in normal human plasma (e.g. 500 µl + 500 µl). The Roche Cobas 8800 HIV-1 RNA Viral Load quantitative RT-PCR was performed with a 0.7 ml input volume. Twenty-two samples with remaining volume and detectable VLs by Cobas were re-tested with the RealTime HIV-1 assay using the 0.6 ml sample input volume.
Western Blotting Specimens with discordant Architect HIV Combo 2P36 Reactive and HIV-1 RealTime viral load results (Target Not Detected; TND) were evaluated by Western Blot using the FUJIREBIO INNO-LIA HIV I/II Score line immunoassay. Test strips were submerged in individual troughs containing 20 µl of specimen or control diluted in 1 ml of sample diluent (e.g. 1:50) and incubated with rocking for 3 hr at room temperature. Primary antibodies were removed by aspiration and test strips were washed with 3 × 6 min incubations in 1 ml of wash solution. A 1 ml anti-human conjugate solution added to each trough was incubated for 30 min, aspirated, then followed by another 3 × 3 min series of washes. Finally, a 1 ml substrate solution was incubated for 30 minutes, aspirated, and then quenched with a 1 ml Stop Solution for 10 minutes. Strip were removed with tweezers, dried on absorbent paper, and photographed.
Multiplex qPCR Viral loads were approximated with a quantitative Multiplex PCR. This is a research-use only assay developed on the m2000sp (Abbott Molecular Diagnostics, Des Plaines, IL) and CFX96 Real Time (Bio-Rad, Hercules, CA) instruments that simultaneously quantifies HBV DNA, HCV RNA, HIV-1 RNA, and HIV-2 RNA with respective limits of detection of 10 IU/ml, 100 IU/ml, 500 cp/ml, and 100 IU/ml. Quantitation was extrapolated from relative Ct values of diluted standards. The assay includes probes for two distinct regions of the HIV-1 genome.
cDNA synthesis, Nextera XT, and HIV-xGen library production Viral RNA extracted from plasma on the m2000sp was concentrated to 10 µl with RNA Clean and Concentrator-5 spin columns (Zymo Research, CA). Metagenomic libraries were prepared with random primers using Superscript III (SSRTIII) 1st Strand reagents (Life Technologies) followed by 2nd strand synthesis with Sequenase V2.0 T7 DNA pol (Affymetrix). Double stranded cDNA was recovered with DNA Clean and Concentrator-5 spin columns (Zymo Research) and eluted in 7 μl. SSRTIII libraries were tagmented by Nextera XT and amplified for 24 cycles (Illumina, Carlsbad CA). Nextera libraries were purified with Agencourt AMPure XP beads (Beckman Coulter), visualized on a BioAnalyzer 2200TapeStation (Agilent), quantified on a Qubit instrument (Life Technologies) with dsDNA high sensitivity reagents (Molecular Diagnostics), and sequenced on a MiSeq instrument with V2 chemistry (2 × 150 bp). Where indicated, plasma samples were heated for 12 hrs at 50°C. HIV-xGen libraries were enriched from mNGS libraries essentially as described [27].
NGS analysis HIV mapping and consensus sequence generation was performed as previously described with CLC Genomics Workbench 9.0 software (CLC bio/Qiagen, Aarhus Denmark). Additionally, fastq data was processed through an automated, custom HIV pipeline. Metagenomic analysis was performed with SURPI [28,29]. To detect possible contaminating reads from barcode hopping, raw data from each sample was individually mapped to the consensus sequences of other samples sequenced on the same run, removing any with ≥99% identity. Unmapped reads (e.g. unique to the sample of interest) were collected and realigned to generate the final consensus and mapping metrics.
Mass Spectroscopy Analysis TND plasma samples were sent to PPD Laboratories (Madison, WI) for validated detection of Lamivudine (3TC) and Abacavir (ABC) (test #P1165) by LC/MS/MS. Values are reported between 2.50-2500 ng/ml.
Ethics The investigation protocol for DRC samples acquired in 2017-2019 conformed to local regulations and was approved by the Université Protestante au Congo ethics committee in Kinshasa; ethics approval was received by memorandum dated May 2017 via # CEUPC-0027. Informed verbal consent in the DRC was obtained from all participants in either French or Lingala and plasma samples were deidentified for analysis. IRB approval from the University of Missouri-Kansas City Research Board for protocol 16-411 was approved on 10/18/2016. Samples from Vanga 2001-2003 were acquired according to protocol 98-041e, approved by the University of Missouri-Kansas City Research Board IRB in 1998. Informed verbal consent was obtained from participants at Vanga after a full explanation of the goals of the study. The Cameroonian sample collection from 2004-2017 was approved by the Cameroon National Ethical Review Board, the Faculty of Medicine and Biomedical Science IRB at the University of Yaounde, and the Ministry of Health of Cameroon. In accordance with study protocols, written informed consent was acquired and plasma was collected and deidentified.
Statistical Analysis Values are primarily reported as percentages of total samples screened or total HIV Combo positives and did not require statistical analysis. Given our initial aim was to characterize viral genetic diversity, sample size was determined by the number of participants seeking healthcare in the given year and not computed by statistical power analysis. In Fig. 2b, the total N for each cohort and the mean values ± SD for HIV Combo results are reported. In Figure S3, the total N for males and females and the median values ± SD for age are reported.
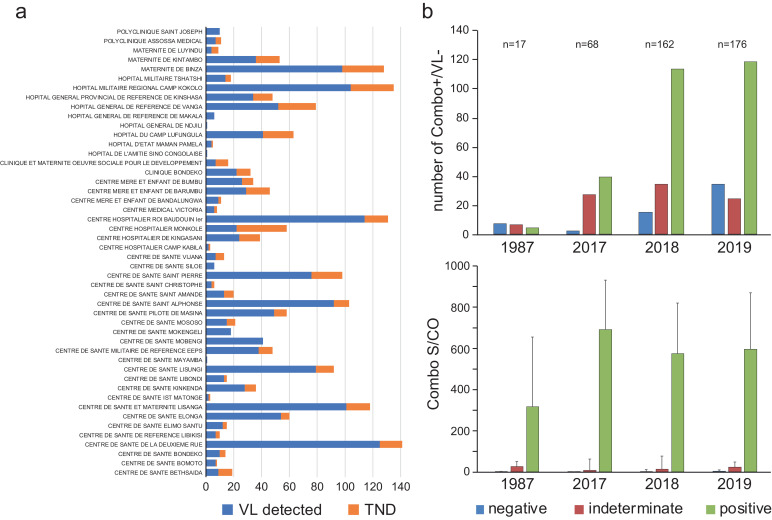
Fig. 2.
TND are confirmed sero-positives collected from numerous sites and observed over decades. a) Number of TND (orange) and VL+ (blue) collected at each site from 2017-2019. b) Western blot results according to year (top) and corresponding HIV Combo S/CO values (bottom).
Role of funding source Abbott Laboratories did not have any role in study design, data collection, data analyses, interpretation, writing of report, or decision to publish the study.
Results
An unusually high number of individuals in DRC without detectable viral loads. To inform individuals of their HIV status and in turn assess overall prevalence and viral genetic diversity among people in the region seeking healthcare, screening campaigns restricted to ART naïve subjects were conducted at 48 sites across Kinshasa and one rural hospital (Vanga) in Democratic Republic of Congo (Fig. 1a) [2]. Samples were collected in 2017-2018 but shipped and tested as three cohorts from 2017-2019. HIV rapid diagnostic tests (RDT) were administered to 10,457 participants and all samples were rescreened in the US for HIV on the Abbott ARCHITECT HIV Ag/Ab Combo assay (2P36), as well as for HBV surface antigen (HBsAg Qualitative, 4P53; Fig. 1b). In 2017, 2018, and 2019 cohorts, 300/2041, 934/3245, and 734/5171 respectively, were confirmed HIV antibody positives, with 3.45% (N=66/1911) meeting the criteria for a recent infection (<200 S/CO, >100 copies/ml RNA). Overall agreement between the RDT and HIV Combo was 94.5%, with 2.1% RDT false positives (n=169), and 3.0% RDT false negatives (n=239). All HIV Combo positives were then tested on the Abbott m2000 RealTime HIV-1 viral load assay. Values for each assay by individual and year spanned the entire measurable range, with most having serology S/CO values between 400-1000 and viral loads of 3 log10-6 log10 copies per ml, as would be expected for a population not receiving treatment (Fig. 1c) [30]. However, we observed a significant number of individuals with no detectable viral load (e.g. ‘target not detected’, TND; circled in red on y-axis in Fig. 1c). In specimens collected in 2017, there were 65/298 (22%), in 2018 there were 148/920 (16%), and in 2019 there were 216/693 (31%) TNDs (Fig. 1d).
We recently analyzed archival plasma samples from two collaborative studies exploring viral genetic diversity in the DRC, each dating back >15 and 30 years, respectively [26,27]. We applied a similar surveillance algorithm to that described in Fig. 1b. In the first study conducted at two rural hospitals (Vanga and Kananga) from 2001-2003, 262 individuals were screened for HIV by serology, and seropositive specimens (n=211) were later tested for viral RNA [26]. Samples from the second study (n=93) were collected in 1987 as part of Project SIDA in Kinshasa (in former Zaire), which was established by the United States Centers for Disease Control (US CDC), the US National Institutes of Health (US NIH), Belgium's Institute of Tropical Medicine (ITM), and the Mama Yemo Hospital in Kinshasa during the initial HIV/AIDS outbreak in the 1980s [31]. The effort was disbanded in 1991 due to civil unrest, and samples were air lifted out of the country for long-term storage at the US National Institutes of Health (NIH). A subset of these specimens were previously characterized at the molecular level to classify strains present, and in this study both HIV antibodies and viral load were assessed [27]. A comparison of ARCHITECT HIV Ag/Ab Combo (2P36) and the Abbott RealTime HIV-1 (6L18) results yielded similar profiles when compared to the 2017-2019 data (Fig. 1c). In the 2001-2003 cohort, there were 31/194 antibody positive specimens with undetectable viral loads, and in 1987 before anti-retrovirals (cART) were available, there were 17/59 (Fig. 1d). Given the abundance of samples (476/2164; ~22% of HIV-1+), the wide range and typically high Combo S/CO values, and the consistent historic trend observed in this region, we initially concluded these results were not sampling artifacts. Thus, we conducted further studies to eliminate possible technical reasons for the finding of so many samples with low viral loads.
Neither collection site bias nor antibody false positives explain TNDs. We reviewed 2017-2019 patient demographic data to determine whether TND samples were concentrated in one or a few collection sites. This was not the case: TND individuals were found at most of the 48 locations and were not overrepresented at any particular testing site (Fig. 2a, Table S1, Table S2). Therefore, a bias due to location, patient recruitment, sample procurement and/or storage, was absent.
Next, we excluded the possibility of false seropositivity. To confirm the reactive results in both the rapid test and ARCHITECT HIV Ag/Ab Combo, we performed Western blots (WB; Fujirebio Innolia HIV1/2) with patient plasma to determine if prior results were false positives or due to HAMA (human anti-mouse antibody) interference (Fig. 2b). Images of all WB strips sorted by year can be found in Supplemental Figure S1. A minority of samples from each year were WB negative (upper panel; blue), however, all of these had Combo S/CO values near the assay cut-off of 1.0 (lower panel, blue). A slightly larger percentage were ruled ‘indeterminate’, with reactivity to only the gp41 band, and once again, these also had low S/CO values (red). We ascertained that 55% of these ‘indeterminate’ blots had originally been rapid test-negative and that the remaining 45% rapid test-positives had comparatively greater HIV Combo S/CO values (median 6.74 vs 25.4). The majority of TND samples were therefore confirmed by Western blot (upper panel, green). Median S/CO ranges of 300-700 (lower panel, green) were consistent with the population at large (see Fig. 1c). Therefore, three separate antibody tests determined that these individuals were infected with HIV-1.
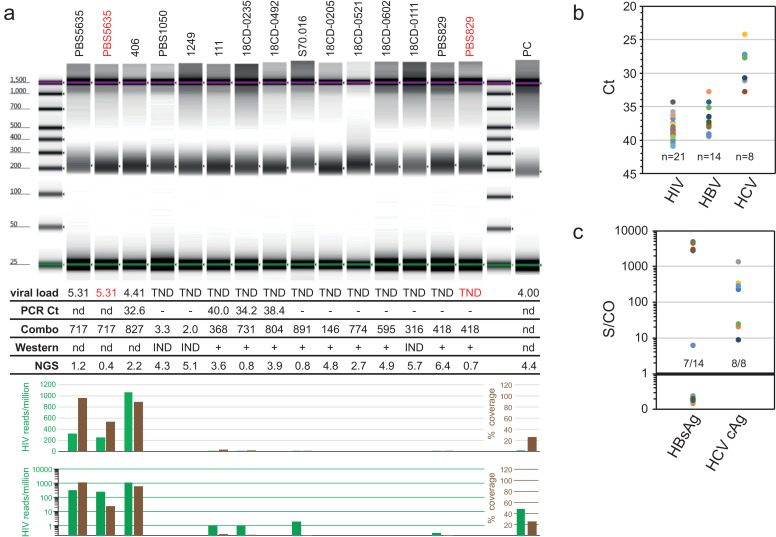
Sample integrity and genetic diversity do not explain TND results. While no individual site(s) was implicated as the source of most TND samples, the possibility remained that individual, unrelated samples from multiple sites could independently be degraded. We addressed specimen integrity by generating metagenomic next-generation sequencing (mNGS) libraries from plasma using randomly primed cDNA synthesis and Nextera tagmentation. Bioanalyzer profiles illustrate that not only was sufficient nucleic acid present to generate libraries for every sample, but also that TND samples (WB indeterminate and positive) were visually identical to those with detectable HIV viral loads (Fig. 3a). When VL+ control samples (red; lanes 2 & 15) were deliberately heat-treated (12hrs at 50°C), they still yielded comparable libraries. The breakdown of NGS read percentages reflects that most libraries consist of host background and once again that TND are indistinguishable from HIV RNA positives (Supplemental Figure S2) [29]. Viral load positive samples (e.g. PBS5635; 5.31 log10 cp/ml) yielded >300 HIV reads/million (green histograms) and >89% genome coverage (brown histograms). Even after prolonged heating, >50% of the PBS5635 genome was obtained (lane 2) from only 400,000 reads, demonstrating that ‘degraded’ samples still contain viral RNA templates. Likewise, despite the large drop in total reads, the abundance of GBV-C virus reads in PBS829 (lanes 14 &15) remained at 12% after heating (Supplemental Figure S2).
Fig. 3.
Sample integrity and sequence diversity do not explain absence of HIV RNA detection. a) Bioanalyzer profiles of mNGS libraries. PBS5635 (lane 2) and PBS829 (lane 15) samples (red) were heat-treated. The table beneath lists each sample's viral load (log copies/ml), Multiplex qPCR Ct, HIV Combo S/CO, Western blot, and total number of NGS reads (in millions). The histogram plots the # HIV reads/million (green) and the % HIV genome coverage (brown) on both linear and log scales (PC = positive control, an HIV+ sample of 4.0 log10 cp/ml; (-) = negative PCR; IND = indeterminate Western Blot; (+) = positive Western Blot; n.d.= not done). b) TNDs were reassessed by a multiplex qPCR that detects HIV-1/-2, HBV, and HCV. Ct values for each positive sample are plotted by virus. c) S/CO values of antigen assays for HBV and HCV qPCR positives shown in 3b.
The above mNGS data also addresses an altogether separate issue: sequence diversity. The DRC is home to a myriad of subtypes, CRFs, and URFs, all of which could avoid conventional molecular detection [11,26]. Viral loads could be under-quantified or infections go undetected if hybridization is impaired due to mismatches. Random priming followed by alignment of every read to the entire NT database eliminates the possibility that amplification and detection of HIV was unsuccessful due to lack of primer and probe specificity. Despite this unbiased approach, few if any HIV reads were detected in the TND samples, providing further evidence that titers were correctly determined as either non-existent or very low. Similarly, no divergent retroviral reads were detected by RAPsearch, eliminating the possibility that a novel virus with antibody cross-reactivity was present [32]. To be certain that mNGS sensitivity was not limiting, 32 libraries constructed from the 2017 panel were enriched by the HIV-xGen target capture approach [27]. The median genome coverage obtained was 10.6% (range: 1.3- 37%) with a depth ≤ 5 reads and included sequences expected from DRC such as subtype A, CRF02, CRF45, CRF63, etc. (Table S3). Thus, for randomly selected specimens, the ultra-sensitive NGS method was able to detect some HIV reads, but the low coverage was another indication that viral loads were extremely low.
Continuing with the premise that mismatches in target sequences relative to primers or probes may be the root cause, 115 TND samples from the 2017 and 2018 panels were sent to a contract laboratory for independent viral load measurements by an alternate method, the Roche Cobas 8800 HIV-1 assay [11,26]. This test targets a different region of the HIV genome compared to Abbott's m2000 HIV-1 RealTime. Three samples with detectable viral loads (2.31, 4.95, and 6.79 log10 cp/ml) by the Abbott assay were included as positive controls and resulted in nearly identical levels by the Roche Cobas 8800 HIV-1 assay (Table 1). We note that in order to preserve sample for follow up studies, all viral loads were initially determined on the Abbott Abbott RealTime HIV-1 assay using the 0.2 ml sample input volume, with an LOD of 150 cp/ml. With the Roche Cobas 8800 HIV-1 assay using a 0.7 ml sample input volume with an LOD of 20 cps/ml, 62 samples (54%) were also reported as ‘target not detected’, 19 (17%) were <1.3 log10 cp/ml (<20 copies/ml), 28 (24%) were between 1.3-2.3 log10 cp/ml (20-200 copies/ml), and 7 (6%) were between 2.3-2.8 log10 cp/ml (200-630 copies/ml). Samples with VL >20 copies/ml by Roche Cobas 8800 HIV-1 assay having sufficient volume (22/35) were retested with the Abbott RealTime HIV-1 assay using the 0.6 ml sample input volume (LOD of 40 copies/ml): 13/22 confirmed a low viral load was present. Therefore, using an alternate manufacturer's viral load assay, we substantiated that viral RNA was indeed either completely absent or present at extremely low levels, confirming that this observation was not due to under-quantitation.
Table 1.
Outside testing confirms viral loads and reveals many are on ART therapy. Results for TND samples with outside or additional viral load testing (n=115) are listed. HIV viral loads (log copies/ml) were determined by Abbott HIV-1 RealTime (0.2 ml, neat), Roche Cobas (0.7 ml, 1:1 dilution), and Abbott HIV-1 RealTime (0.6 ml, neat) assays. Concentrations (ng/ml) of 3TC (lamivudine) were determined by mass spectroscopy and WB determined by INNO-LIA.
| Year | Sample ID | Rapid test | Combo S/CO | HIV Real Time 0.2 ml log10 cp/ml | Roche Cobas 0.7 ml log10 cp/ml | HIV Real Time 0.6ml log10 cp/ml | Multiplex qPCR | Western Blot | Mass Spec 3TC (ng/mL) |
|---|---|---|---|---|---|---|---|---|---|
| CTRL1 | 25 | 1.96 | 2.31 | ||||||
| CTRL2 | 15 | 4.88 | 4.95 | ||||||
| CTRL3 | 51 | 6.65 | 6.79 | ||||||
| Potential Elite/Viral Controllers | |||||||||
| 2017 | 14 | NEG | 150.04 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2017 | 31 | POS | 154.61 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2017 | 36 | POS | 16.46 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2017 | 69 | POS | 14.67 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2017 | 135 | 811.78 | Not Detected | Not detected | HIV NEG | HIV-1 | <2.50 | ||
| 2017 | 207 | POS | 35.31 | Not Detected | Not detected | HIV GREY | IND | <2.50 | |
| 2017 | 240 | POS | 41.89 | Not Detected | Not detected | IND | <2.50 | ||
| 2017 | 243 | POS | 91.03 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2017 | 254 | POS | 117.12 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2017 | 712 | NEG | 191.66 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2017 | 2009 | NEG | 51.64 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0096 | POS | 40.41 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0111 | POS | 316.13 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0125 | POS | 295.1 | Not Detected | Not detected | HIV NEG | HIV-1 | <2.50 | |
| 2018 | 18CD-0146 | POS | 566.67 | Not Detected | Not detected | HIV NEG | HIV-1 | <2.50 | |
| 2018 | 18CD-0185 | POS | 38.84 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0205 | POS | 145.9 | Not Detected | 1.76 | HIV NEG | HIV-1 | <2.50 | |
| 2018 | 18CD-0220 | POS | 12.25 | Not Detected | Not detected | IND | <2.50 | ||
| 2018 | 18CD-0222 | POS | 12.59 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0235 | POS | 730.74 | Not Detected | Not detected | HIV-1 | <2.50 | ||
| 2018 | 18CD-0314 | POS | 437.61 | Not Detected | 1.73 | <1.60 | HIV NEG | HIV-1 | <2.50 |
| 2018 | 18CD-0381 | POS | 158.46 | Not Detected | 1.62 | 1.96 | HIV NEG | HIV-1 | <2.50 |
| 2018 | 18CD-0423 | POS | 760.43 | Not Detected | 2.71 | Not detected | HIV NEG | HIV-1 | <2.50 |
| 2018 | 18CD-0428 | POS | 382.51 | Not Detected | 1.72 | HIV-1 | <2.50 | ||
| 2018 | 18CD-0500 | POS | 807.14 | Not Detected | 1.34 | 1.67 | HIV NEG | HIV-1 | <2.50 |
| 2018 | 18CD-0513 | POS | 570.81 | Not Detected | Not detected | HIV NEG | HIV-1 | <2.50 | |
| 2018 | 18CD-0544 | POS | 55.4 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0558 | POS | 64.92 | Not Detected | <1.30 | Not detected | HIV NEG | HIV-1 | <2.50 |
| 2018 | 18CD-0572 | POS | 574.8 | Not Detected | Not detected | HIV NEG | HIV-1 | <2.50 | |
| 2018 | 18CD-0739 | POS | 473.89 | Not Detected | 2.82 | 3.14 | HIV NEG | HIV-1 | <2.50 |
| 2018 | 18CD-0750 | POS | 3.54 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0754 | POS | 143.32 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0763 | POS | 23.27 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0776 | POS | 599.96 | Not Detected | 2.25 | 2.16 | HIV NEG | HIV-1 | <2.50 |
| 2018 | 18CD-0893 | 407.3 | Not Detected | Not detected | HIV GREY | HIV-1 | <2.50 | ||
| 2018 | 18CD-0902 | POS | 27.6 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-0911 | POS | 607.59 | Not Detected | <1.30 | HIV NEG | HIV-1 | <2.50 | |
| 2018 | 18CD-1321 | NEG | 126.69 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 2018 | 18CD-2545 | NEG | 28.7 | Not Detected | Not detected | HIV NEG | IND | <2.50 | |
| 3TC Positives | |||||||||
| 2017 | 60 | POS | 710.38 | Not Detected | Not detected | HIV NEG | HIV-1 | 168 | |
| 2017 | 73 | POS | 829.98 | Not Detected | Not detected | HIV NEG | HIV-1 | 7.79 | |
| 2017 | 76 | POS | 723.37 | Not Detected | Not detected | HIV NEG | HIV-1 | 627 | |
| 2017 | 77 | 508.57 | Not Detected | Not detected | HIV NEG | HIV-1 | 329 | ||
| 2017 | 105 | POS | 845.04 | Not Detected | 1.4 | HIV POS | HIV-1 | 194 | |
| 2017 | 111 | POS | 367.65 | Not Detected | 1.3 | HIV POS | 115 | ||
| 2017 | 118 | POS | 660.07 | Not Detected | Not detected | HIV NEG | HIV-1 | 22.5 | |
| 2017 | 120 | POS | 687.67 | Not Detected | 1.91 | 2.4 | HIV POS | HIV-1 | 2460 |
| 2017 | 123 | POS | 776.07 | Not Detected | <1.30 | HIV NEG | HIV-1 | 649 | |
| 2017 | 124 | POS | 529.25 | Not Detected | Not detected | HIV NEG | HIV-1 | 1040 | |
| 2017 | 127 | 927.25 | Not Detected | Not detected | HIV NEG | HIV-1 | 2190 | ||
| 2017 | 137 | 279.01 | Not Detected | Not detected | HIV NEG | HIV-1 | 455 | ||
| 2017 | 141 | POS | 561.03 | Not Detected | Not detected | HIV NEG | HIV-1 | 889 | |
| 2017 | 148 | POS | 251.17 | Not Detected | Not detected | HIV NEG | HIV-1 | 28.7 | |
| 2017 | 195 | POS | 480.12 | Not Detected | <1.30 | HIV NEG | HIV-1 | 520 | |
| 2017 | 198 | NEG | 21.3 | Not Detected | Not detected | HIV NEG | IND | 86.9 | |
| 3TC Positives | |||||||||
| 2017 | 202 | POS | 873.79 | Not Detected | <1.30 | HIV GREY | HIV-1 | 58.5 | |
| 2017 | 217 | 691.57 | Not Detected | Not detected | HIV NEG | HIV-1 | 18.6 | ||
| 2017 | 219 | POS | 928.95 | Not Detected | <1.30 | HIV NEG | HIV-1 | 662 | |
| 2017 | 225 | 765.54 | Not Detected | Not detected | HIV NEG | HIV-1 | 905 | ||
| 2017 | 245 | POS | 674.91 | Not Detected | Not detected | HIV NEG | HIV-1 | 368 | |
| 2017 | 250 | POS | 385.13 | Not Detected | Not detected | HIV NEG | HIV-1 | 41.3 | |
| 2018 | 18CD-0045 | POS | 950.46 | Not Detected | 2.06 | HIV-1 | 1940 | ||
| 2018 | 18CD-0084 | POS | 953.23 | Not Detected | 2.55 | HIV NEG | HIV-1 | 2160 | |
| 2018 | 18CD-0137 | POS | 583.79 | Not Detected | <1.30 | Not detected | HIV NEG | HIV-1 | 386 |
| 2018 | 18CD-0138 | POS | 581.8 | Not Detected | Not detected | HIV NEG | HIV-1 | 442 | |
| 2018 | 18CD-0158 | POS | 632.85 | Not Detected | Not detected | HIV NEG | HIV-1 | 887 | |
| 2018 | 18CD-0164 | POS | 760.58 | Not Detected | 1.95 | HIV NEG | HIV-1 | 152 | |
| 2018 | 18CD-0182 | POS | 730.51 | Not Detected | Not detected | HIV NEG | HIV-1 | 495 | |
| 2018 | 18CD-0210 | POS | 451.86 | Not Detected | <1.30 | <1.60 | HIV NEG | HIV-1 | 297 |
| 2018 | 18CD-0263 | POS | 705.77 | Not Detected | 1.49 | HIV NEG | HIV-1 | 238 | |
| 2018 | 18CD-0275 | POS | 569.37 | Not Detected | 1.48 | <1.60 | HIV-1 | 99.5 | |
| 2018 | 18CD-0306 | POS | 693.24 | Not Detected | 1.99 | HIV NEG | HIV-1 | 10.5 | |
| 2018 | 18CD-0356 | POS | 620.59 | Not Detected | Not detected | HIV-1 | 585 | ||
| 2018 | 18CD-0379 | POS | 526.13 | Not Detected | 1.45 | 2.02 | HIV NEG | HIV-1 | 658 |
| 2018 | 18CD-0384 | POS | 427.02 | Not Detected | Not detected | HIV NEG | HIV-1 | 288 | |
| 2018 | 18CD-0391 | POS | 484.68 | Not Detected | Not detected | HIV NEG | HIV-1 | 95.7 | |
| 2018 | 18CD-0437 | POS | 557.83 | Not Detected | Not detected | HIV NEG | HIV-1 | 53.3 | |
| 2018 | 18CD-0443 | POS | 590.61 | Not Detected | Not detected | HIV NEG | HIV-1 | 889 | |
| 2018 | 18CD-0451 | POS | 795.28 | Not Detected | 1.68 | HIV NEG | HIV-1 | 209 | |
| 2018 | 18CD-0452 | POS | 907.85 | Not Detected | 1.3 | HIV NEG | HIV-1 | 335 | |
| 2018 | 18CD-0453 | POS | 378.33 | Not Detected | 1.83 | HIV NEG | HIV-1 | 1250 | |
| 2018 | 18CD-0471 | POS | 756.43 | Not Detected | <1.30 | HIV GREY | HIV-1 | 430 | |
| 2018 | 18CD-0484 | POS | 563.69 | Not Detected | 1.6 | HIV-1 | 6.26 | ||
| 2018 | 18CD-0492 | POS | 804 | Not Detected | 2.77 | HIV-1 | 159 | ||
| 2018 | 18CD-0503 | POS | 970.37 | Not Detected | Not detected | HIV NEG | HIV-1 | 1320 | |
| 2018 | 18CD-0509 | POS | 529.2 | Not Detected | 1.79 | HIV NEG | HIV-1 | 146 | |
| 2018 | 18CD-0510 | POS | 284.69 | Not Detected | <1.30 | Not detected | HIV-1 | 393 | |
| 2018 | 18CD-0521 | POS | 773.66 | Not Detected | 1.96 | HIV NEG | HIV-1 | 233 | |
| 2018 | 18CD-0535 | 1014.7 | Not Detected | 2.83 | HIV NEG | HIV-1 | 704 | ||
| 2018 | 18CD-0537 | 858.46 | Not Detected | <1.30 | 1.91 | HIV NEG | HIV-1 | 3070 | |
| 2018 | 18CD-0592 | POS | 571.44 | Not Detected | Not detected | HIV NEG | HIV-1 | 491 | |
| 2018 | 18CD-0602 | POS | 594.73 | Not Detected | <1.30 | <1.60 | HIV NEG | HIV-1 | 1570 |
| 2018 | 18CD-0603 | POS | 407.11 | Not Detected | Not detected | HIV NEG | HIV-1 | 261 | |
| 2018 | 18CD-0621 | POS | 676.08 | Not Detected | Not detected | HIV NEG | HIV-1 | 1270 | |
| 2018 | 18CD-0649 | POS | 282.38 | Not Detected | <1.30 | Not detected | HIV NEG | HIV-1 | 578 |
| 2018 | 18CD-0654 | POS | 210.47 | Not Detected | 1.58 | HIV NEG | HIV-1 | 210 | |
| 2018 | 18CD-0665 | POS | 976.37 | Not Detected | 1.85 | Not detected | HIV NEG | HIV-1 | 2980 |
| 2018 | 18CD-0669 | POS | 767.02 | Not Detected | 1.86 | 2.25 | HIV NEG | HIV-1 | 806 |
| 2018 | 18CD-0671 | 573.49 | Not Detected | <1.30 | HIV NEG | HIV-1 | 225 | ||
| 2018 | 18CD-0723 | POS | 736.27 | Not Detected | Not detected | HIV NEG | HIV-1 | 519 | |
| 2018 | 18CD-0728 | POS | 298.37 | Not Detected | <1.30 | HIV NEG | HIV-1 | 870 | |
| 2018 | 18CD-0738 | POS | 18.09 | Not Detected | Not detected | HIV NEG | IND | 4.76 | |
| 2018 | 18CD-0789 | POS | 56.05 | Not Detected | 1.51 | <1.60 | HIV NEG | HIV-1 | 3060 |
| 2018 | 18CD-0851 | POS | 942.66 | Not Detected | <1.30 | Not detected | HIV NEG | HIV-1 | 195 |
| 2018 | 18CD-0855 | POS | 776.73 | Not Detected | <1.30 | HIV NEG | HIV-1 | 1140 | |
| 2018 | 18CD-0861 | POS | 712.31 | Not Detected | Not detected | HIV NEG | HIV-1 | 567 | |
| 2018 | 18CD-0868 | POS | 220.64 | Not Detected | 1.36 | Not detected | HIV NEG | HIV-1 | 546 |
| 2018 | 18CD-0869 | POS | 225.09 | Not Detected | Not detected | HIV NEG | HIV-1 | 198 | |
| 2018 | 18CD-0901 | POS | 733.52 | Not Detected | 2.34 | HIV NEG | HIV-1 | 257 | |
| 2018 | 18CD-0912 | POS | 664.4 | Not Detected | 2.06 | HIV NEG | HIV-1 | 121 | |
| 2018 | 18CD-0913 | POS | 343.31 | Not Detected | <1.30 | HIV NEG | HIV-1 | 4.67 | |
| 2018 | 18CD-2316 | NEG | 445.84 | Not Detected | Not detected | HIV NEG | HIV-1 | 13.1 | |
| 2018 | 18CD-2414 | NEG | 483.74 | Not Detected | Not detected | HIV NEG | HIV-1 | 215 | |
| 2018 | 18CD-2721 | NEG | 1069.3 | Not Detected | <1.30 | HIV NEG | HIV-1 | 358 | |
To further address genetic diversity and sample integrity in one assay, a multiplex PCR was deployed that detects HIV-1 at two genome targets, while simultaneously detecting HIV-2, HCV, and HBV (Figs. 1b, 3b). From 175 TND in the 2018 panel, no samples with high HIV viral loads were present, however, HIV was detected in 21 instances, the majority with Cts >35 (< 100 cp/ml) (Fig. 3b, Table 1). Once again, potential mutations were not causing samples to be missed outright, but differential assay sensitivity confirmed that some of these have very low levels of RNA. Importantly, coinfections of HBV (n=14; 8.0%) and HCV (n=8; 4.6%) were observed in samples, once more indicating that (viral) nucleic acid was indeed intact. These results were bolstered by positive serology data for HBsAg Qualitative (4P53; 8/15) and HCV Ag (6L47; 10/10) assays (Fig. 3c), which again confirmed sample quality was maintained. Thus, we still believed that numerous individuals from the DRC were able to control HIV VL to (nearly) undetectable levels.
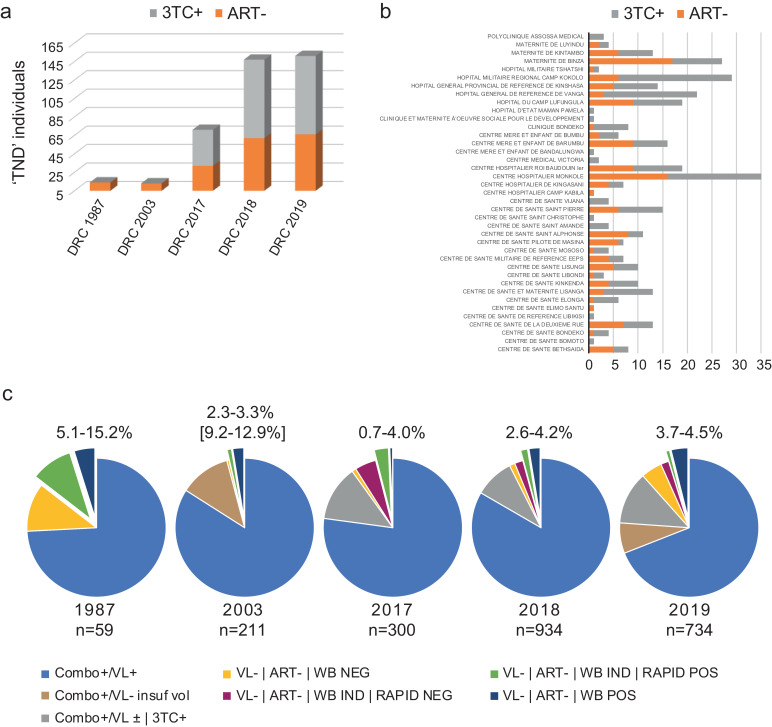
Increased percentages of elite controllers reside in the Congo Basin. Finally, given the “test and treat” policy, we considered that patients may not have acknowledged ongoing cART upon enrollment. With respect to older samples collected in 1987 and 2001-2003, this was impossible or highly unlikely, since anti-retroviral drugs either did not exist or were unavailable in the DRC. Nevertheless, 421 TND samples including individuals from the above early cohorts and later cohorts (2017-2019), as well as TND samples from Cameroonian blood donors collected between 2004-2017, were analyzed by mass spectroscopy for detection of lamivudine (3TC) and abacavir (ABC), the frontline antivirals prevalent in DRC. As expected, both drugs were absent in all 1987 and 2001-2003 samples tested (Table 1, Fig. 4, Table S1, Table S2, Figure S1). Similarly, cART positives were largely absent from Cameroonian samples (Table S4). However, approximately 60% of DRC plasma samples obtained during 2017-2019 were 3TC positive; only one DRC sample was dually reactive for abacavir (Table 1, Fig. 4a). cART+ individuals were found at nearly every location, once again indicating that no particular testing site was responsible for this trend (Fig. 4b). We were surprised to learn that so many patients in these recent cohorts had enrolled in our study that specifically required cART-naive patients: our approved protocol excluded HIV patients who had received drugs. However, there remained nearly 40% (144/353) of the 2017-2019 DRC cohort tested by mass spectroscopy that did not receive therapy and still had undetectable viral loads. With respect to the 32 samples in the 2017 panel sequenced by HIV-xGen, 31/32 samples tested positive for 3TC, indicating that cART had suppressed replication, but as would be expected, there was still detectable virus present (Table S3) [33]. By contrast, only one yielded sequence from among the true “TNDs” with <2.5 ng/ml of 3TC.
Fig. 4.
A sizeable population of potential elite controllers reside in the Democratic Republic of Congo. a) Breakdown by cohort of TND that were positive (grey) and negative (orange) for 3TC by mass spec. b) Breakdown by collection site of TND that were positive (grey) and negative (orange) for 3TC by mass spec. c). Percentage range by year of potential elite controllers in DRC are represented by the slices extending from the pie, which include VL-|ART-|WB IND|RAPID POS and VL-|ART-|WB POS samples. Total number of HIV Combo+ samples evaluated are listed beneath each year.
We performed an accounting of ‘TND’ (Combo+/VL-) individuals, excluding those on therapy, WB negatives, and WB indeterminates with negative rapid results (Table 2, Fig. 4c). Remaining cART- WB positives and indeterminates with positive rapid results were included to provide an estimate of the percentage of potential elite controllers in Kinshasa. Percentages in 1987 ranged from 5.1-15.2% (n=3-9/60) and in 2003 from as low as 2.3-3.3% to as high as 9.2-12.9%, since for many, volume was depleted and we were unable to test (n=31/194). Bear in mind there are comparatively fewer individuals in these cohorts and that long-term storage could have adversely affected these specimens. For the 2017 cohort, elite controllers are now projected at 0.7-4.0% (n=2-12/298), reduced from 22% before mass spectrometry testing. Roughly half of the remaining 3TC- individuals with an indeterminate WB and negative rapid test were excluded (Table 2). In 2018, we identified 24 WB positives and 15 WB IND/Rapid+ from 920 Combo reactives for a range of 2.5-4.2%, reduced from 17%. Similarly, in 2019 specimens we identified 27 WB positives and 6 WB IND/Rapid+ from 639 Combo reactives for a range of 3.7-4.5%, down from 28%. Combining all TND's identified over 2017-2019 and dividing by all HIV Combo Ab+ screened, we estimate that potential EC comprise 2.7-4.3% of the HIV+ Kinshasa, DRC population. Neither the age nor gender appear to be factors. The median age of men (TND/3TC-) was 38 years compared to 43 overall (VL+), while for women it was 36 years compared to 34 overall (Supplemental Figure S3).
Table 2.
Summary of potential elite controller data in DRC. An accounting of samples in each category is listed to show how percentages were determined.
| DRC Cohort | Combo tested | Combo+ | Combo+ VL+ | Combo+ VL- | ART tested | ART+ | ART- | WB tested | ART+ WB NEG | ART+ WB IND | ART+ WB POS | ART- WB NEG | ART- WB IND | ART- WB POS | RAPID+ ART- WB IND | RAPID- ART- WB IND | Potential ECs | Percentage Potential ECs |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1987 | 93 | 59 | 43 | 16 | 9 | 0 | 9 | 17 | 0 | 0 | 0 | 7 | 6 | 4 | N/A | N/A | 3-9 | 5.1-15.2% |
| 2003 | 262 | 211 | 163 | 31 | 8 | 0 | 8 | 8 | 0 | 0 | 0 | 1 | 2 | 5 | N/A | N/A | 2-7 [19-27] | 2.3-3.3% [9.2-12.9%] |
| 2017 | 2041 | 300 | 233 | 65 | 66 | 39 | 27 | 69 | 0 | 2 | 36 | 3 | 25 | 2 | 10 | 15 | 2-12 | 0.7-4.0% |
| 2018 | 3245 | 934 | 772 | 148 | 142 | 85 | 57 | 159 | 0 | 1 | 84 | 12 | 33 | 24 | 15 | 18 | 24-39 | 2.5-4.2% |
| 2019 | 5171 | 734 | 477 | 216 | 146 | 85 | 61 | 176 | 0 | 2 | 82 | 13 | 20 | 27 | 6 | 13 | 27-33 | 3.7-4.5% |
| 2017-2019 | 10457 | 1968 | 1482 | 429 | 354 | 209 | 145 | 404 | 0 | 5 | 202 | 28 | 78 | 53 | 31 | 46 | 53-84 | |
| Percentages | 19% | 75% | 22% | 59% | 41% | 0% | 1% | 50% | 7% | 19% | 13% | 40% | 59% | 2.7-4.3% |
In neighboring Cameroon, Ngo-Malabo et al. found 2.95% of participants were elite controllers, suggesting this phenomenon may not be unique to the DRC [34]. We mined our own historic surveillance data collected from 2004-2017 in Cameroon and found 1533 specimens with paired viral load and HIV serology (ARCHITECT HIV Combo 2P36 or HIV-1/2 3A77) data to compare. Of these with a Combo S/CO >1.0, 177 had an undetectable viral load. Extrapolating from our mass spectroscopy and WB results, wherein 94% (48/51) of Cameroonian blood donors were not on cART and only 28/51 of these were WB indeterminate or positive, we estimate potential elite controllers could represent 2.5-5.8% of individuals in Cameroon (Table S4, Supplemental Figures S4, S5). Collectively, the data suggests that a greater proportion of individuals from West Central Africa possess a natural ability to control HIV-1 infection.
Discussion
The majority of individuals infected with HIV-1 require antiretroviral therapy to block viral replication and prevent the development of HIV/AIDS. Opportunistic infections accompanying AIDS lead to eventual patient death. Elite controllers, on the other hand, are a unique sub-group of patients who are able to control HIV-1 replication to undetectable levels for many years. In this study, using plasma collected over a span of 30 years, we have identified numerous HIV+ patients with undetectable viral loads in DRC. We suggest they are potential elite controllers and excluded other explanations such as serological false positives, specimen degradation, genetic diversity, and cART. In most instances, we applied antibody tests produced by three different manufacturers to confirm infection and conducted viral load assays produced by two different manufacturers. In addition, a 3rd RUO qPCR was employed, once again confirming that extremely low or undetectable VLs were present in a sub-group of our cohort. Potential ECs were represented at most collection sites and had the same demographic make-up as other study participants.
Surprisingly, nearly half of the TND individuals in the recent patient pool from 2017-2019 failed to disclose they were receiving cART. As cART has become widely available in Africa with ‘test and treat’ policies, a troubling uptick in ‘false elite controllers’ has also been observed [35]. The unreliability of self-reported cART usage ranges from 11% (Uganda) to upwards of 42-46% in the KAIS (Kenya) and HPTN 052 (Malawi and Zimbabwe) trials, respectively [36], [37], [38]. However, an important distinction applies to our study and the other reports. For example, in Sykes, et al, out of 12,705 HIV+ blood donations from South Africa, 270 (2.12%) were Ab+/RNA-, which was reduced by 66% to only 0.7% of EC after retrospective cART detection [35]. In the DRC, we determined 2.7-4.3% of HIV patients have undetectable VL in the absence of cART, an approximately 5-fold increase in frequency. Importantly, we show these findings were present decades before anti-retroviral drugs became available. We also found a sizeable number of our potential ECs had indeterminate Western blots (Figure S1, Table 2, Fig. 4c). Anti-gp41 cross-reactivity may result from other infections, immunizations, pregnancy, or autoimmunity, and a study of 1020 known sero-positives from Nigeria found 20.1% to be Western blot indeterminate, demonstrating its limitation for confirmatory diagnosis [39]. Nevertheless, there are examples in the literature of LTNPs/ECs with indeterminate or weakly reactive Westerns [40], [41], [42]. By the same token, a recent survey of 85 HIV virus-controllers showed that 6% were negative by rapid test and 7% indeterminate by Western blot, with enzyme immunoassay ratios declining over time [43]. To be conservative, we only included those indeterminates who were positive for both HIV Combo and the rapid test.
Elite/viral controllers are indeed rare and most eventually lose immunological and clinical control of HIV infection to succumb to AIDS [44]. Worldwide, the percentage of EC (VL < 50 cp/ml)/VC (50-2000 cp/ml) reported in studies range from 0.1-2% and are frequently never detected (0%). For example, European cohorts examined in Sweden, Italy, and France all report fewer than 1%. Multinational cohorts like CASCADE (Europe, Australia, Canada) had 1.4%, while MERIT (S Africa, US, Belgium, Canada, Italy, UK, Australia) and studies in India, reported zero [45], [46], [47]. A survey conducted by the US Department of Defense found that among HIV+ military personnel, only 25/4586 (0.55%) were EC and 153/4586 (3.34%) were VC [15]. The landscape is similar in most of Africa, with cohorts in Uganda (0.26%), Botswana (0.3%), South Africa (1.25%), and Ethiopia (1.17%, 0.6%) all reporting very few controllers of either type [48], [49], [50], [51], [52]. Our own data (not shown) from the Abbott Global Surveillance program, characterizing specimens from over 60 countries for nearly 30 years, mirrors these trends. Interestingly, our numbers in Cameroon are elevated, in support of Ngo-Malabo et al., whose comparative study found 188/6355 (2.95%) patients with undetectable VLs. Of note, however, they did not actively exclude cART+ participants [34]. This is the first report estimating the prevalence of elite controllers from the DRC. The political situation in DRC has hampered public health research there for many decades. Accurate estimates of HIV prevalence have also been limited for a decade or more, and incidence data is estimated using computer modeling of small studies [2].
Mechanistically, it is widely accepted that the host immune response determines whether a patient becomes an elite controller or non-progressor [53]. Shortly after resolution of the acute phase and seroconversion, individuals reach an equilibrium, or viral set point, which for EC falls below the limit of detection in plasma. A multifunctional, HIV-specific CD8 T cell response is central to mediating virological control in these individuals [54]. Due to the rarity of this phenomenon and that heterogenous, unrelated ECs can be found globally, cellular restriction factors were originally not thought to play a major role [55]. More recently, higher levels of T cell p21, which blocks cyclin-dependent kinases supporting several HIV replication steps, have been found in EC/NPs compared to uninfected patients [21,22]. In addition, lower levels of CCR5 in EC/NPs compared to HIV progressors have been reported for HIV subtype C infections [23]. There are numerous other cellular factors such as RICH2 and CXCR6 that play a role in controlling HIV-1 replication and that distinguish ECs from VCs [56,57]. The notion of an attenuated or defective viral strain(s) whose replication is more easily suppressed has largely been dismissed, as has the inability of activated CD4(+) T cells to support HIV infection in ECs [58,59]. Virus recovered from 4/10 elite controllers was found to be replication competent and devoid of large deletions, yet the remaining 6 failed to culture, so arguably this is still an open question [60]. Given the numbers of potential EC we find in DRC and considering the origins of the HIV epidemic, the role of EC/NPs in sustaining the epidemic is a vital unanswered question.
With the early emergence of HIV in this region many decades ago, it is perhaps no accident that potential ECs could be found in larger numbers in the Congo Basin rather than any other part of the world. Another possibility for the higher percentages is the survival advantage of ECs in areas with no or less treatment, wherein susceptible individuals die and thereby proportionally increase the EC prevalence. CCR5Δ32 deletions can preclude transitions in viral tropism, and HLA restricted epitopes (HLA-B*57) can mediate a robust cellular response [61,62]. While this allele is not prevalent in African populations, there are several other genotypes of CCR5 involved in HIV-1 control, just as there are other protective HLA-B alleles with greater representation in Africa [63,64]. Is there another yet to be discovered genetic basis shared among this population that permits development of HIV control? Would fixation of these resistance alleles in the population be expected to develop in the span of an only 4 decades-long pandemic? In terms of the virus, the wide range of viral sequence diversity present in DRC and the Congo Basin recasts the intriguing possibility of an attenuated strain, perhaps like HIV-2 with a less severe phenotype, for which certain aspects intrinsic to the virus and independent of the host response contribute to its lesser phenotype [65]. The converse has been reported in CRF19 and subtype D, where these more fit viruses achieve higher viral loads and lead to a more rapid progression to AIDS [66], [67], [68]. A recent incidence study in Africa showed that infection with subtype A is associated with viral control (compared to subtype C), and subtype A along with recombinants thereof, are the predominant strains in DRC [69]. Perhaps this phenomenon was a factor in sustaining the epidemic, particularly in remote areas of DRC, otherwise all may have died. Determining the subtype of HIV cleared by ’TND’ patients in our study was not possible since the RNA was by definition absent in plasma: the only virus we recovered was from individuals subsequently found to be on therapy. Going back to lymphocytes of potential ECs to sequence integrated virus could prove useful if these were available. Continued exploration of sequence diversity in this region may begin to address some fundamental questions as well as the ‘age’ of strains. Basal branching patterns without evidence of recombination, the presumed most recent common ancestors, cover the spectrum in the DRC and include subtypes A, C, D [26]. Molecular clock dating of these and other diverse strains from the Congo Basin could reset the evolutionary timeline of HIV-1 [4,[70], [71], [72]].
The paucity of elite/viral controllers and the identification of a homogenous population achieving durable suppression in the absence of drugs has limited our understanding of immune mechanisms critical to develop a vaccine or functional cure. A primary limitation of our ‘observational’, cross-sectional study was that with only one timepoint it was not designed to monitor individuals or establish how many of these retained long-term HIV control (e.g. at timepoints >6 months apart) to meet the classical definition of ECs or VCs. Another consideration is the short half-life of 3TC or the possibility that other non-frontline drugs were used, in which case additional patients might have been excluded [73]. Countering this decrease is the possibility there are individuals who received cART that did not need it based on viral load. Nevertheless, with the cost of viral loads out of reach for most DRC citizens and ‘test and treat’ policies in place, indiscriminate use of cART+ is unavoidable. A controlled, prospective longitudinal study to follow a cohort of potential controllers in DRC and Cameroon could determine the true proportion of ECs and allow a comprehensive characterization of recoverable virus sequence and the immune responses in these patients. For example, genome wide association studies identifying genetic predisposition(s) for viral control (e.g. HLA alleles, cytokines, polymorphisms, restriction factors like APOBEC, Trim 5alpha, RICH2, etc.) as well as the targets/epitopes of humoral and cellular responses would now have the statistical power required as the number of affected subjects increases. These studies might inform vaccine design and suggest potential new targets of antivirals. They might also indicate whether viral control or pre-existing specific anti-HIV responses prevent reinfection with HIV. While antivirals have removed the death sentence that HIV once was, there are still 37 million individuals infected globally and incidence remains stubbornly stable (even in the US, and is rising in certain geographies). Harnessing what we learn from studying this unique population can hopefully stem this tide and get us closer to the 90-90-90 goals and “ending HIV in the US in 10 years”.
Data sharing statement
All data for this study has been included in manuscript figures, tables, and online supplemental information.
Figure S1. Western blot images for all TNDs in DRC. Western blot strips are shown for each DRC cohort with keys on sides indicating the identity of each HIV band.
Figure S2. Breakdown of metagenomic NGS reads. Stacked histogram representing the taxonomic classification of mNGS library reads in Figure 3a as determined by SURPI.
Figure S3. Potential EC breakdown by age and sex. Individuals with complete age and sex demographic data were plotted to compare VL-/3TC- (true TND) versus VL positives.
Figure S4. Western blot images for TNDs from Cameroon. Western blot strips are shown for Cameroonian samples with figure keys on sides indicating the identity of each HIV band.
Figure S5. Summary of Cameroon TND results. a) Number of Combo+/VL- (TND) individuals on 3TC (grey) or not on therapy (ART-, orange). b) Western blot results for TNDs in Cameroon. c) Pie chart showing distribution of TNDs and the percentage of potential elite controllers in Cameroon.
Table S1. Results and demographic data for ART- TND individuals.
Table S2. Results and demographic data for ART+ TND individuals.
Table S3. HIV-xGen enrichment indicates sequence is only present in ART+ TND individuals. NGS results for 32 ‘TND’ samples from the 2017 panel enriched by the HIV-xGen method. The final column indicates the concentration of 3TC determined by mass spectrometry.
Table S4. Results for Cameroon TNDs. Cohort years are listed for samples collected between 2004-2017. In some cases, multiple results were obtained for HIV Combo. Western blot, RealTime VLs, and 3TC levels determined by LC/MS are listed.
Author contributions
Michael G. Berg: conceptualization, methodology, project administration, supervision, data analysis, created figures, wrote manuscript, researched literature, verified data. Ana Olivo: experimental methodology, data analysis and organization, verified data. Barbara J. Harris: experimental methodology, data analysis and organization, verified data. Mary A. Rodgers: conceptualization, methodology, project administration, supervision, arranged collaboration and sourcing of samples, designed experiments, data analysis, wrote manuscript, researched literature, verified data. Linda James: Sample collection and testing DRC, supervision. Samuel Mampunza: Sample collection and testing DRC, supervision. Jonathan Niles: Sample collection and testing DRC, supervision. Franklin Baer: Sample collection and testing DRC, supervision. Julie Yamaguchi: experimental methodology, data analysis. Lazare Kaptue: Sample collection and testing Cameroon, supervision. Oliver Laeyendecker: resources, data analysis, researched literature. Thomas C. Quinn: resources, data analysis, researched literature. Carole McArthur: data curation, methodology, project administration, supervision, established collaboration, managed testing and collection sites in DRC, resources, data analysis, wrote manuscript, researched literature, verified data. Gavin A. Cloherty: conceptualization, project administration, supervision, resources, wrote manuscript, researched literature. All authors read and approved the final version of the manuscript.
Declaration of Competing Interests
MGB, AO, BJH, JY, MAR, and GAC are all employees and shareholders of Abbott Laboratories. The funder provided support in the form of salaries for authors MGB, AO, BJH, JY, MAR, GAC, but did not have any additional role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript. OL is an NIAID employee. LJ, SM, JN, FB, LK, TCQ, and CM do not have conflicts of interest to disclose. No patents have been applied for and no products are in development related to this research.
Acknowledgements
The authors acknowledge the following individuals for their role in sample collection: Gaëtan Bondo, Jonathan Fumunguya, Jeansy Mavinga, Médard Omakoy, Yves Tshangala and François Tshibaka at Université Protestante au Congo. We thank Matthew Frankel for guidance with the Multiplex qPCR assay and Ana Vallari for technical assistance with Western Blots. Abbott Laboratories funded surveillance in DRC and subsequent research efforts. Additional funding was received from a MIZZOU Award from the University of Missouri. Research was supported in part by the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, NIH.
Footnotes
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.ebiom.2021.103258.
Appendix. Supplementary materials
References
- 1.Williams BG, Lima V, Gouws E. Modelling the impact of antiretroviral therapy on the epidemic of HIV. Curr HIV Res. 2011;9(6):367–382. doi: 10.2174/157016211798038533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pour M, James L, Singh K, Mampunza S, Baer F, Scott J. Increased HIV in greater Kinshasa urban health zones: Democratic Republic of Congo (2017-2018) AIDS Res Ther. 2020;17(1):67. doi: 10.1186/s12981-020-00322-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hemelaar J. The origin and diversity of the HIV-1 pandemic. Trends Mol Med. 2012;18(3):182–192. doi: 10.1016/j.molmed.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Worobey M, Gemmel M, Teuwen DE, Haselkorn T, Kunstman K, Bunce M. Direct evidence of extensive diversity of HIV-1 in Kinshasa by 1960. Nature. 2008;455(7213):661–664. doi: 10.1038/nature07390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu T, Korber BT, Nahmias AJ, Hooper E, Sharp PM, Ho DD. An African HIV-1 sequence from 1959 and implications for the origin of the epidemic. Nature. 1998;391(6667):594–597. doi: 10.1038/35400. [DOI] [PubMed] [Google Scholar]
- 6.Brennan CA, Bodelle P, Coffey R, Devare SG, Golden A, Hackett J., Jr. The prevalence of diverse HIV-1 strains was stable in Cameroonian blood donors from 1996 to 2004. J Acquir Immune Defic Syndr. 2008;49(4):432–439. doi: 10.1097/QAI.0b013e31818a6561. [DOI] [PubMed] [Google Scholar]
- 7.Keele BF, Van Heuverswyn F, Li Y, Bailes E, Takehisa J, Santiago ML. Chimpanzee reservoirs of pandemic and nonpandemic HIV-1. Science. 2006;313(5786):523–526. doi: 10.1126/science.1126531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Peeters M, D'Arc M, Delaporte E. Origin and diversity of human retroviruses. AIDS Rev. 2014;16(1):23–34. [PMC free article] [PubMed] [Google Scholar]
- 9.Plantier JC, Leoz M, Dickerson JE, De Oliveira F, Cordonnier F, Lemee V. A new human immunodeficiency virus derived from gorillas. Nat Med. 2009;15(8):871–872. doi: 10.1038/nm.2016. [DOI] [PubMed] [Google Scholar]
- 10.Simon F, Mauclere P, Roques P, Loussert-Ajaka I, Muller-Trutwin MC, Saragosti S. Identification of a new human immunodeficiency virus type 1 distinct from group M and group O. Nat Med. 1998;4(9):1032–1037. doi: 10.1038/2017. [DOI] [PubMed] [Google Scholar]
- 11.Vidal N, Peeters M, Mulanga-Kabeya C, Nzilambi N, Robertson D, Ilunga W. Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) group M genetic diversity in the Democratic Republic of Congo suggests that the HIV-1 pandemic originated in Central Africa. J Virol. 2000;74(22):10498–10507. doi: 10.1128/jvi.74.22.10498-10507.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brennan CA, Bodelle P, Coffey R, Harris B, Holzmayer V, Luk KC. HIV global surveillance: foundation for retroviral discovery and assay development. J Med Virol. 2006;78(Suppl 1):S24–S29. doi: 10.1002/jmv.20603. [DOI] [PubMed] [Google Scholar]
- 13.Pantaleo G, Menzo S, Vaccarezza M, Graziosi C, Cohen OJ, Demarest JF. Studies in subjects with long-term nonprogressive human immunodeficiency virus infection. N Engl J Med. 1995;332(4):209–216. doi: 10.1056/NEJM199501263320402. [DOI] [PubMed] [Google Scholar]
- 14.Grabar S, Selinger-Leneman H, Abgrall S, Pialoux G, Weiss L, Costagliola D. Prevalence and comparative characteristics of long-term nonprogressors and HIV controller patients in the French Hospital Database on HIV. AIDS. 2009;23(9):1163–1169. doi: 10.1097/QAD.0b013e32832b44c8. [DOI] [PubMed] [Google Scholar]
- 15.Okulicz JF, Marconi VC, Landrum ML, Wegner S, Weintrob A, Ganesan A. Clinical outcomes of elite controllers, viremic controllers, and long-term nonprogressors in the US Department of Defense HIV natural history study. J Infect Dis. 2009;200(11):1714–1723. doi: 10.1086/646609. [DOI] [PubMed] [Google Scholar]
- 16.Olson AD, Meyer L, Prins M, Thiebaut R, Gurdasani D, Guiguet M. An evaluation of HIV elite controller definitions within a large seroconverter cohort collaboration. PLoS One. 2014;9(1):e86719. doi: 10.1371/journal.pone.0086719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Silva TI, Peng Y, Leligdowicz A, Zaidi I, Li L, Griffin H. Correlates of T-cell-mediated viral control and phenotype of CD8(+) T cells in HIV-2, a naturally contained human retroviral infection. Blood. 2013;121(21):4330–4339. doi: 10.1182/blood-2012-12-472787. [DOI] [PubMed] [Google Scholar]
- 18.Kong R, Li H, Bibollet-Ruche F, Decker JM, Zheng NN, Gottlieb GS. Broad and potent neutralizing antibody responses elicited in natural HIV-2 infection. J Virol. 2012;86(2):947–960. doi: 10.1128/JVI.06155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leligdowicz A, Yindom LM, Onyango C, Sarge-Njie R, Alabi A, Cotten M. Robust Gag-specific T cell responses characterize viremia control in HIV-2 infection. J Clin Invest. 2007;117(10):3067–3074. doi: 10.1172/JCI32380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Onyango CO, Leligdowicz A, Yokoyama M, Sato H, Song H, Nakayama EE. HIV-2 capsids distinguish high and low virus load patients in a West African community cohort. Vaccine. 2010;28(Suppl 2):B60–B67. doi: 10.1016/j.vaccine.2009.08.060. [DOI] [PubMed] [Google Scholar]
- 21.Leng J, Ho HP, Buzon MJ, Pereyra F, Walker BD, Yu XG. A cell-intrinsic inhibitor of HIV-1 reverse transcription in CD4(+) T cells from elite controllers. Cell Host Microbe. 2014;15(6):717–728. doi: 10.1016/j.chom.2014.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moosa Y, Tanko RF, Ramsuran V, Singh R, Madzivhandila M, Yende-Zuma N. Case report: mechanisms of HIV elite control in two African women. BMC Infect Dis. 2018;18(1):54. doi: 10.1186/s12879-018-2961-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ostrowski MA, Justement SJ, Catanzaro A, Hallahan CA, Ehler LA, Mizell SB. Expression of chemokine receptors CXCR4 and CCR5 in HIV-1-infected and uninfected individuals. J Immunol. 1998;161(6):3195–3201. [PubMed] [Google Scholar]
- 24.Baker BM, Block BL, Rothchild AC, Walker BD. Elite control of HIV infection: implications for vaccine design. Expert Opin Biol Ther. 2009;9(1):55–69. doi: 10.1517/14712590802571928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saag M, Deeks SG. How do HIV elite controllers do what they do? Clin Infect Dis. 2010;51(2):239–241. doi: 10.1086/653678. [DOI] [PubMed] [Google Scholar]
- 26.Rodgers MA, Wilkinson E, Vallari A, McArthur C, Sthreshley L, Brennan CA. Sensitive next-generation sequencing method reveals deep genetic diversity of HIV-1 in the democratic Republic of the Congo. J Virol. 2017;91(6) doi: 10.1128/JVI.01841-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamaguchi J, Olivo A, Laeyendecker O, Forberg K, Ndembi N, Mbanya D. Universal target capture of HIV sequences from NGS libraries. Front Microbiol. 2018;9:2150. doi: 10.3389/fmicb.2018.02150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berg MG, Yamaguchi J, Alessandri-Gradt E, Tell RW, Plantier JC, Brennan CA. A pan-HIV strategy for complete genome sequencing. J Clin Microbiol. 2016;54(4):868–882. doi: 10.1128/JCM.02479-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Naccache SN, Federman S, Veeraraghavan N, Zaharia M, Lee D, Samayoa E. A cloud-compatible bioinformatics pipeline for ultrarapid pathogen identification from next-generation sequencing of clinical samples. Genome Res. 2014;24(7):1180–1192. doi: 10.1101/gr.171934.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fraser C, Hollingsworth TD, Chapman R, de Wolf F, Hanage WP. Variation in HIV-1 set-point viral load: epidemiological analysis and an evolutionary hypothesis. Proc Natl Acad Sci U S A. 2007;104(44):17441–17446. doi: 10.1073/pnas.0708559104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen J. The rise and fall of Project SIDA. Science. 1997;278(5343):1565–1568. doi: 10.1126/science.278.5343.1565. [DOI] [PubMed] [Google Scholar]
- 32.Ye Y, Choi JH, Tang H. RAPSearch: a fast protein similarity search tool for short reads. BMC Bioinform. 2011;12:159. doi: 10.1186/1471-2105-12-159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dornadula G, Zhang H, VanUitert B, Stern J, Livornese L, Jr., Ingerman MJ. Residual HIV-1 RNA in blood plasma of patients taking suppressive highly active antiretroviral therapy. JAMA. 1999;282(17):1627–1632. doi: 10.1001/jama.282.17.1627. [DOI] [PubMed] [Google Scholar]
- 34.Ngo-Malabo ET, Ngoupo TP, Zekeng M, Ngono V, Ngono L, Sadeuh-Mba SA. A cheap and open HIV viral load technique applicable in routine analysis in a resource limited setting with a wide HIV genetic diversity. Virol J. 2017;14(1):224. doi: 10.1186/s12985-017-0893-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sykes W, Van den Berg K, Jacobs G, Jauregui A, Roubinian N, Wiesner L. Discovery of false elite controllers: HIV antibody-positive rna-negative blood donors found to be on antiretroviral therapy. J Infect Dis. 2019;220(4):643–647. doi: 10.1093/infdis/jiz145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fogel JM, Wang L, Parsons TL, Ou SS, Piwowar-Manning E, Chen Y. Undisclosed antiretroviral drug use in a multinational clinical trial (HIV Prevention Trials Network 052) J Infect Dis. 2013;208(10):1624–1628. doi: 10.1093/infdis/jit390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grabowski MK, Reynolds SJ, Kagaayi J, Gray RH, Clarke W, Chang LW. The validity of self-reported antiretroviral use in persons living with HIV: a population-based study. AIDS. 2018;32(3):363–369. doi: 10.1097/QAD.0000000000001706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim AA, Mukui I, Young PW, Mirjahangir J, Mwanyumba S, Wamicwe J. Undisclosed HIV infection and antiretroviral therapy use in the Kenya AIDS indicator survey 2012: relevance to national targets for HIV diagnosis and treatment. AIDS. 2016;30(17):2685–2695. doi: 10.1097/QAD.0000000000001227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Uneke CJ, Alo MN, Ogbu O, Ngwu BA. Western blot-indeterminate results in Nigerian patients HIV serodiagnosis: the clinical and public health implication. AIDS Patient Care STDS. 2007;21(3):169–176. doi: 10.1089/apc.2006.0089. [DOI] [PubMed] [Google Scholar]
- 40.Mendoza D, Johnson SA, Peterson BA, Natarajan V, Salgado M, Dewar RL. Comprehensive analysis of unique cases with extraordinary control over HIV replication. Blood. 2012;119(20):4645–4655. doi: 10.1182/blood-2011-10-381996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rhodes D, Solomon A, Bolton W, Wood J, Sullivan J, Learmont J. Identification of a new recipient in the Sydney Blood Bank Cohort: a long-term HIV type 1-infected seroindeterminate individual. AIDS Res Hum Retroviruses. 1999;15(16):1433–1439. doi: 10.1089/088922299309946. [DOI] [PubMed] [Google Scholar]
- 42.Zaunders J, Dyer WB, Churchill M, Munier CML, Cunningham PH, Suzuki K. Possible clearance of transfusion-acquired nef/LTR-deleted attenuated HIV-1 infection by an elite controller with CCR5 Delta32 heterozygous and HLA-B57 genotype. J Virus Erad. 2019;5(2):73–83. doi: 10.1016/S2055-6640(20)30696-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hage-Sleiman M, Tremeaux P, Fillion M, Boufassa F, Melard A, Gardiennet E. False-negative Results of Human Immunodeficiency Virus (HIV) Rapid Testing in HIV Controllers. Clin Infect Dis. 2020;70(8):1754–1757. doi: 10.1093/cid/ciz734. [DOI] [PubMed] [Google Scholar]
- 44.van der Helm JJ, Geskus R, Lodi S, Meyer L, Schuitemaker H, Gunsenheimer-Bartmeyer B. Characterisation of long-term non-progression of HIV-1 infection after seroconversion: a cohort study. Lancet HIV. 2014;1(1):e41–e48. doi: 10.1016/S2352-3018(14)70016-5. [DOI] [PubMed] [Google Scholar]
- 45.Cooper DA, Heera J, Ive P, Botes M, Dejesus E, Burnside R. Efficacy and safety of maraviroc vs. efavirenz in treatment-naive patients with HIV-1: 5-year findings. AIDS. 2014;28(5):717–725. doi: 10.1097/QAD.0000000000000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Madec Y, Boufassa F, Porter K, Prins M, Sabin C, d'Arminio Monforte A. Natural history of HIV-control since seroconversion. AIDS. 2013;27(15):2451–2460. doi: 10.1097/01.aids.0000431945.72365.01. [DOI] [PubMed] [Google Scholar]
- 47.Solomon SS, Solomon S, McFall AM, Srikrishnan AK, Anand S, Verma V. Integrated HIV testing, prevention, and treatment intervention for key populations in India: a cluster-randomised trial. Lancet HIV. 2019;6(5):e283–ee96. doi: 10.1016/S2352-3018(19)30034-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kayongo A, Gonzalo-Gil E, Gumusgoz E, Niwaha AJ, Semitala F, Kalyesubula R. Brief report: identification of elite and viremic controllers from a large urban HIV ambulatory center in Kampala, Uganda. J Acquir Immune Defic Syndr. 2018;79(3):394–398. doi: 10.1097/QAI.0000000000001825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiros YK, Elinav H, Gebreyesus A, Gebremeskel H, Azar J, Chemtob D. Identification and characterization of HIV positive Ethiopian elite controllers in both Africa and Israel. HIV Med. 2019;20(1):33–37. doi: 10.1111/hiv.12680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Moyo S, Gaseitsiwe S, Mohammed T, Pretorius Holme M, Wang R, Kotokwe KP. Cross-sectional estimates revealed high HIV incidence in Botswana rural communities in the era of successful ART scale-up in 2013-2015. PLoS One. 2018;13(10) doi: 10.1371/journal.pone.0204840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shoko C, Chikobvu D. A superiority of viral load over CD4 cell count when predicting mortality in HIV patients on therapy. BMC Infect Dis. 2019;19(1):169. doi: 10.1186/s12879-019-3781-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Telele NF, Kalu AW, Marrone G, Gebre-Selassie S, Fekade D, Tegbaru B. Baseline predictors of antiretroviral treatment failure and lost to follow up in a multicenter countrywide HIV-1 cohort study in Ethiopia. PLoS One. 2018;13(7) doi: 10.1371/journal.pone.0200505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalo-Gil E, Ikediobi U, Sutton RE. Mechanisms of virologic control and clinical characteristics of HIV+ Elite/Viremic controllers. Yale J Biol Med. 2017;90(2):245–259. [PMC free article] [PubMed] [Google Scholar]
- 54.Saez-Cirion A, Lacabaratz C, Lambotte O, Versmisse P, Urrutia A, Boufassa F. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A. 2007;104(16):6776–6781. doi: 10.1073/pnas.0611244104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okulicz JF, Lambotte O. Epidemiology and clinical characteristics of elite controllers. Curr Opin HIV AIDS. 2011;6(3):163–168. doi: 10.1097/COH.0b013e328344f35e. [DOI] [PubMed] [Google Scholar]
- 56.Limou S, Coulonges C, Herbeck JT, van Manen D, An P, Le Clerc S. Multiple-cohort genetic association study reveals CXCR6 as a new chemokine receptor involved in long-term nonprogression to AIDS. J Infect Dis. 2010;202(6):908–915. doi: 10.1086/655782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paximadis M, Ngqobe RN, Chaisson RE, Martinson NA, Tiemessen CT. RICH2 is implicated in viraemic control of HIV-1 in black South African individuals. Infect Genet Evol. 2017;49:78–87. doi: 10.1016/j.meegid.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Julg B, Pereyra F, Buzon MJ, Piechocka-Trocha A, Clark MJ, Baker BM. Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers. Clin Infect Dis. 2010;51(2):233–238. doi: 10.1086/653677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamine A, Caumont-Sarcos A, Chaix ML, Saez-Cirion A, Rouzioux C, Delfraissy JF. Replication-competent HIV strains infect HIV controllers despite undetectable viremia (ANRS EP36 study) AIDS. 2007;21(8):1043–1045. doi: 10.1097/QAD.0b013e3280d5a7ac. [DOI] [PubMed] [Google Scholar]
- 60.Blankson JN, Bailey JR, Thayil S, Yang HC, Lassen K, Lai J. Isolation and characterization of replication-competent human immunodeficiency virus type 1 from a subset of elite suppressors. J Virol. 2007;81(5):2508–2518. doi: 10.1128/JVI.02165-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Blanpain C, Libert F, Vassart G, Parmentier M. CCR5 and HIV infection. Receptors Channels. 2002;8(1):19–31. [PubMed] [Google Scholar]
- 62.Lecuroux C, Saez-Cirion A, Girault I, Versmisse P, Boufassa F, Avettand-Fenoel V. Both HLA-B*57 and plasma HIV RNA levels contribute to the HIV-specific CD8+ T cell response in HIV controllers. J Virol. 2014;88(1):176–187. doi: 10.1128/JVI.02098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Gonzalez E, Bamshad M, Sato N, Mummidi S, Dhanda R, Catano G. Race-specific HIV-1 disease-modifying effects associated with CCR5 haplotypes. Proc Natl Acad Sci U S A. 1999;96(21):12004–12009. doi: 10.1073/pnas.96.21.12004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiepiela P, Leslie AJ, Honeyborne I, Ramduth D, Thobakgale C, Chetty S. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432(7018):769–775. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 65.de Silva TI, Cotten M, SL Rowland-Jones. HIV-2: the forgotten AIDS virus. Trends Microbiol. 2008;16(12):588–595. doi: 10.1016/j.tim.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 66.Baeten JM, Chohan B, Lavreys L, Chohan V, McClelland RS, Certain L. HIV-1 subtype D infection is associated with faster disease progression than subtype A in spite of similar plasma HIV-1 loads. J Infect Dis. 2007;195(8):1177–1180. doi: 10.1086/512682. [DOI] [PubMed] [Google Scholar]
- 67.Kiwanuka N, Laeyendecker O, Robb M, Kigozi G, Arroyo M, McCutchan F. Effect of human immunodeficiency virus Type 1 (HIV-1) subtype on disease progression in persons from Rakai, Uganda, with incident HIV-1 infection. J Infect Dis. 2008;197(5):707–713. doi: 10.1086/527416. [DOI] [PubMed] [Google Scholar]
- 68.Kouri V, Khouri R, Aleman Y, Abrahantes Y, Vercauteren J, Pineda-Pena AC. CRF19_cpx is an Evolutionary fit HIV-1 Variant Strongly Associated With Rapid Progression to AIDS in Cuba. EBioMedicine. 2015;2(3):244–254. doi: 10.1016/j.ebiom.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Price MA, Rida W, Kilembe W, Karita E, Inambao M, Ruzagira E. Control of the HIV-1 load varies by viral subtype in a large cohort of African adults with incident HIV-1 infection. J Infect Dis. 2019;220(3):432–441. doi: 10.1093/infdis/jiz127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gryseels S, Watts TD, Kabongo Mpolesha JM, Larsen BB, Lemey P, Muyembe-Tamfum JJ. A near full-length HIV-1 genome from 1966 recovered from formalin-fixed paraffin-embedded tissue. Proc Natl Acad Sci U S A. 2020;117(22):12222–12229. doi: 10.1073/pnas.1913682117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tongo M, Dorfman JR, Abrahams MR, Mpoudi-Ngole E, Burgers WA, Martin DP. Near full-length HIV type 1M genomic sequences from Cameroon : Evidence of early diverging under-sampled lineages in the country. Evol Med Public Health. 2015;2015(1):254–265. doi: 10.1093/emph/eov022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tongo M, Dorfman JR, Martin DP. High Degree of HIV-1 Group M (HIV-1M) genetic diversity within circulating recombinant forms: insight into the early events of HIV-1M evolution. J Virol. 2015;90(5):2221–2229. doi: 10.1128/JVI.02302-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Else LJ, Jackson A, Puls R, Hill A, Fahey P, Lin E. Pharmacokinetics of lamivudine and lamivudine-triphosphate after administration of 300 milligrams and 150 milligrams once daily to healthy volunteers: results of the ENCORE 2 study. Antimicrob Agents Chemother. 2012;56(3):1427–1433. doi: 10.1128/AAC.05599-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.