Abstract
Aging is associated with loss of muscle mass and strength that leads to a condition termed sarcopenia. Impaired conditions, morbidity, and malnutrition are the factors of devaluation of muscle fibers in aged animals. Satellite cells play an important role in maintaining muscle homeostasis during tissue regeneration and repair. Proteomic profiling on the skeletal muscle tissues of different age group rats helps to determine the differentially expressed (DE) proteins, which may eventually lead to the development of biomarkers in treating the conditions of sarcopenia. In this study, nanoscale liquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS) analysis was implemented in the calf tissues of young and old groups of rats. The mass spectrometry (MS) analysis revealed the presence of 335 differentially expressed proteins between the two different age conditions, among which those based on log-fold change 25 proteins were upregulated and 77 were downregulated. The protein–protein interaction network analysis revealed 18 upregulated proteins with three distinct interconnected networks and 57 downregulated proteins with two networks. Further, gene ontology (GO) enrichment analysis showed the biological process, cellular component, and molecular function of the differential proteins. Pathway enrichment analysis of the DE proteins identified nine significantly enriched pathways with a list of eight significant genes (Cryab, Hspb2, Acat1, Ak1, Adssl1, Anxa5, Gys1, Ogdh, Gc, and Adssl1). Quantification of significant genes by quantitative real-time polymerase chain reaction (qRT-PCR) confirmed the downregulation at the mRNA level. Western blot analysis of their protein expression showed concordant results on two candidate proteins (Ogdh and annexin 5) confirming their differential regulation between the two age groups of rats. Thus, these proteomic approaches on young and aged rats provide insights into the development of protein targets in the treatment of sarcopenia (muscle loss).
Introduction
Aging in humans is strongly associated with a decrease in skeletal muscle mass progressively leading to weakness of muscle strength.1 Muscular dystrophy owing to muscle weakness eventually leads to a condition called sarcopenia, which is characterized by muscle mass loss and increased fibrosis of the intramuscular.2 In fact, the mobility-impaired muscle loss is related to reduced quality of life and increased health needs among older adults.3 Satellite cells play an important role in maintaining the muscle homeostasis by its stimulation during regeneration and repair.4 With increased treatment requirements, researchers are focusing on sarcopenia and muscle dystrophy that contribute to the production of biomarkers using satellite cells in animal models.5
Proteomics is a highly complex platform for understanding the protein expression and function on a wide scale with a rapid improvement in clinical research.6 Interest in the proteome research is increased as transcriptome analysis has shown that only half of the variations found in proteins are expected.7 Thus, the proteomics-based strategy is considered to be one of the complex and innovative methods that provide more comprehensive knowledge on the disease beyond that provided from other high-throughput approaches.8
Proteomic approaches provide exclusive detection of proteins and peptides that are currently suitable for the extensive biomarker research devoid of the complex traditional methods.9 Several technologies for protein profiling have been developed that enable quantitative analysis from a single cell, tissue, or other analytic samples of the living system in a specific health condition.10 Proteomics based on mass spectrometry (MS) is an innovative practice that can identify proteins with exquisite precision and sensitivity. The mass spectrometry method coupled with liquid chromatography aids in the quantification of thousands of protein expressions and helps to analyze the dynamic proteome changes occurring in a particular condition.11 The use of nano-LC-MS to achieve the differential protein profiling of a diseased state is an emerging approach in the development of biomarkers at present.12
Differential expression based on proteomics technologies to detect, identify, and characterize proteins on a wide scale makes it a highly promising biomarker discovery tool for various diseases.13 Biomarkers are any substances that include mainly proteins, metabolites, or electrolytes that can be measured to evaluate any physiological processes for therapeutic outcomes.14 The identification of protein biomarkers from biological samples can be rarely detected through traditional methods like two-dimensional (2D) gel electrophoresis due to their low abundance.15 Therefore, proteomics techniques based on gel-free or mass spectrometry approaches are considered to be the best of choice for measuring protein levels with increased sustainability and reproducibility16
Proteomics can be a more challenging method when paired with MS to analyze complex molecular events of diseases and classify essential biomarkers through differential expression analysis.17 The study of gene expression shows the origin and condition of cells involving the pharmacological reaction, disease development, and biological mechanism. They provide a group of genes and enriched pathways associated with the diagnosis of the disease. Thus, identification of differentially expressed (DE) gene signatures is an important method for disease detection and drug development.18 Expression analysis through omics platforms aids the comparison of different marker proteins and signaling pathways between experimental and control groups.19 Thus, molecular expression studies involving quantitative differential patterns may serve as the basis for identifying targets for therapy.20
In this study, we aim to report the protein profiling between two groups of Sprague-Dawley (SD) rat models, namely, young and aged, by performing the nanoscale liquid chromatography coupled to tandem mass spectrometry (nano-LC-MS/MS) technique. Also, we have analyzed the differential protein profile between the two conditions by Bioinformatics tools, leading to the identification of gene enrichment and significant pathways. Further, based on the significantly enriched pathways, the relative proteins and genes were identified between the two conditions followed by in vitro validations.
Results
Identification of the Differentially Expressed Proteins
The expression patterns of the entire protein profile between the young and old group samples were obtained by preprocessing the raw tandem mass spectrometry data using ProteinPilot 5.0.1 software and UniProt database. The results of the database search were curated using Scaffold (version Scaffold_4.10.0, Proteome Software, Inc.) software. The protein identification was considered accepted if they could be established at 99% protein threshold; 95% peptide threshold, and contained at least one identified peptide. The differentially expressed proteins between young and old conditions were grouped individually. A total of 335 differentially expressed proteins were identified and filtered based on species “Rattus norvegicus”. The number of upregulated and downregulated proteins were distinctly identified based on the log-fold change criteria between the two groups. The log-fold change values were set between (0.5 and 2.0), the proteins expressed with values less than 0.5 were upregulated and the proteins expressed with values greater than 2.0 were downregulated. By considering the fold-change range, a total of 25 proteins were found to be upregulated and 77 proteins were found to be downregulated, as shown in Tables 1 and 2.
Table 1. List of Proteins Identified as Upregulated from the Expression Profile of 503 Proteins between the Young and Old Categoriesa.
| protein name | accession no. | fold change |
|---|---|---|
| proteasome subunit β | G3V7Q6 | 0.08 |
| glyoxylate and hydroxypyruvate reductase | B0BN46 | 0.1 |
| phosphoglycerate mutase | Q6P6G4 | 0.2 |
| peroxiredoxin-6 | O35244 | 0.2 |
| lactoylglutathione lyase | Q6P7Q4 | 0.2 |
| heat shock protein β-2 | O35878 | 0.2 |
| glycerol-3-phosphate dehydrogenase [NAD(+)], cytoplasmic | O35077 | 0.2 |
| myozenin 3 | A0A0H2UHR2 | 0.2 |
| protein-l-isoaspartate (d-aspartate) O-methyltransferase | P22062 | 0.3 |
| myosin light chain kinase 2, skeletal muscle | G3V731 | 0.3 |
| myosin-binding protein C, fast-type (predicted) | D3ZA38 | 0.3 |
| myomesin 2 | G3V7K1 | 0.3 |
| glycogenin-1 | A0A0G2JXP1 | 0.3 |
| aspartate-β-hydroxylase | A0A096MIY8 | 0.3 |
| adenylate kinase isoenzyme 1 | P39069 | 0.3 |
| AMP deaminase 1 | P10759 | 0.3 |
| ADP-ribose glycohydrolase MACROD1 | A0A0G2K5F1 | 0.3 |
| Sh3bgr protein (fragment) | B0K040 | 0.3 |
| truncated α-actinin | Q8K551 | 0.4 |
| protein/nucleic acid deglycase DJ-1 | O88767 | 0.4 |
| proteasome subunit α type | A0A0G2K0W9 | 0.4 |
| glutathione S-transferase pi | B6DYQ7 | 0.4 |
| annexin | Q5U362 | 0.4 |
| amylo-1,6-glucosidase,4-α-glucanotransferase | D4AEH9 | 0.4 |
The DE genes are grouped based on log-fold change between 0.5 and 2.0. Fold change <0.5 were upregulated.
Table 2. List of Proteins Identified as Downregulated from the Expression Profile of 503 Proteins between the Young and Old Categoriesa.
| protein name | accession no | fold change |
|---|---|---|
| succinate–CoA ligase [ADP/GDP-forming] subunit α, mitochondrial | A0A0H2UHE1 | 2.1 |
| citrate synthase | G3V936 | 2.1 |
| voltage-dependent anion-selective channel protein 3 | A0A0G2JSR0 | 2.2 |
| keratin, type II cytoskeletal 2 epidermal | A0A0G2JWX4 | 2.2 |
| glutathione peroxidase | M0RAM5 | 2.2 |
| four and a half LIM domains 1 | Q6P792 | 2.2 |
| myosin-binding protein C, slow-type | A0A0G2JUE5 | 2.2 |
| succinate–CoA ligase subunit β (fragment) | B2RZ24 | 2.3 |
| phosphate carrier protein, mitochondrial | Q6IRH6 | 2.3 |
| 2-oxoglutarate dehydrogenase, mitochondrial | Q5XI78 | 2.3 |
| Rab GDP dissociation inhibitor β | P50399 | 2.4 |
| NADH dehydrogenase (ubiquinone) Fe–S protein 7 | Q5RJN0 | 2.4 |
| mitochondrial 2-oxoglutarate/malate carrier protein | G3V6H5 | 2.4 |
| glycogen [starch] synthase, muscle | A2RRU1 | 2.4 |
| serotransferrin | P12346 | 2.5 |
| prohibitin-2 | A0A0G2KB63 | 2.5 |
| electron transfer flavoprotein subunit β | Q68FU3 | 2.5 |
| uncharacterized protein | F1LM19 | 2.6 |
| serum albumin | P02770 | 2.6 |
| carbonic anhydrase 1 | B0BNN3 | 2.6 |
| serine protease inhibitor A3N | A0A0G2JXK5 | 2.7 |
| protein disulfide isomerase | P04785 | 2.7 |
| NADH-ubiquinone oxidoreductase 75 kDa subunit, mitochondrial | Q66HF1 | 2.7 |
| isocitrate dehydrogenase [NADP], mitochondrial | P56574 | 2.7 |
| heat shock protein β-6 | P97541 | 2.7 |
| acetyl-CoA acetyltransferase, mitochondrial | P17764 | 2.7 |
| serum albumin | A0A0G2JSH5 | 2.8 |
| trifunctional enzyme subunit β, mitochondrial | Q60587 | 2.9 |
| α-1-macroglobulin | Q63041 | 2.9 |
| Gup1, glycerol uptake/transporter homologue (yeast) (predicted) | D4A9P9 | 3 |
| vinculin | A0A0G2K8V2 | 3.1 |
| kelch-like protein 41 | Q9ER30 | 3.1 |
| electron transfer flavoprotein subunit α, mitochondrial | P13803 | 3.1 |
| Ckmt2 protein | B0BNC0 | 3.1 |
| dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex, mitochondrial | P08461 | 3.2 |
| 3-mercaptopyruvate sulfurtransferase | P97532 | 3.2 |
| cadherin 13 | F1M7X3 | 3.4 |
| α-crystallin B chain | P23928 | 3.4 |
| α-1-antiproteinase | A0A0G2JY31 | 3.4 |
| aldehyde dehydrogenase, mitochondrial | F1LN88 | 3.4 |
| AHNAK nucleoprotein | A0A0G2JUA5 | 3.4 |
| l-lactate dehydrogenase B chain | P42123 | 3.5 |
| NADH dehydrogenase [ubiquinone] 1 α subcomplex subunit 9, mitochondrial | Q5BK63 | 3.6 |
| calreticulin | P18418 | 3.6 |
| WD repeat-containing protein 1 | Q5RKI0 | 3.8 |
| trifunctional enzyme subunit α, mitochondrial | Q64428 | 3.9 |
| elongation factor 1-delta | Q68FR9 | 3.9 |
| succinate dehydrogenase [ubiquinone] flavoprotein subunit, mitochondrial | Q920L2 | 4 |
| osteoglycin | D3ZVB7 | 4.1 |
| NADH dehydrogenase [ubiquinone] iron–sulfur protein 2, mitochondrial | Q641Y2 | 4.4 |
| hemopexin | P20059 | 4.4 |
| annexin A2 | Q07936 | 4.4 |
| ubiquinone biosynthesis protein COQ9, mitochondrial | Q68FT1 | 4.4 |
| apolipoprotein H | Q5I0M1 | 4.8 |
| adenosylhomocysteinase | Q6P743 | 5.1 |
| NADH:ubiquinone oxidoreductase subunit B10 | D4A0T0 | 5.7 |
| complement C3 | M0RBF1 | 5.7 |
| dysferlin | A0A0G2K7B6 | 6.1 |
| annexin A5 | P14668 | 6.1 |
| vitamin D-binding protein | P04276 | 6.8 |
| decorin | Q01129 | 6.8 |
| acyl-coenzyme A dehydrogenase, very long chain | Q5M9H2 | 7 |
| 3-ketoacyl-CoA thiolase, mitochondrial | A0A0G2K642 | 7.2 |
| carbonic anhydrase 2 | P27139 | 7.9 |
| muscle carnitine palmitoyltransferase I | O70253 | 8.2 |
| long-chain-specific acyl-CoA dehydrogenase, mitochondrial | P15650 | 8.4 |
| long-chain fatty-acid–CoA ligase 1 | P18163 | 8.8 |
| apolipoprotein E | A0A0G2K151 | 8.9 |
| plectin | A0A0G2K1J5 | 8.9 |
| hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | Q9WVK7 | 9.3 |
| acetyl-coenzyme A dehydrogenase, medium chain | G3V796 | 9.4 |
| NADH dehydrogenase [ubiquinone] 1 α subcomplex subunit 10, mitochondrial | A0A1W2Q6F8 | 9.6 |
| prolargin | Q9EQP5 | 11 |
| cathepsin D | P24268 | 11 |
| carnitine O-palmitoyltransferase 2, mitochondrial | P18886 | 11 |
| nicotinamide nucleotide transhydrogenase | Q5BJZ3 | 13 |
The DE genes are grouped based on log-fold change between 0.5 and 2.0. Fold change >2.0 were downregulated.
PPI Network Analysis of the Differentially Expressed Proteins
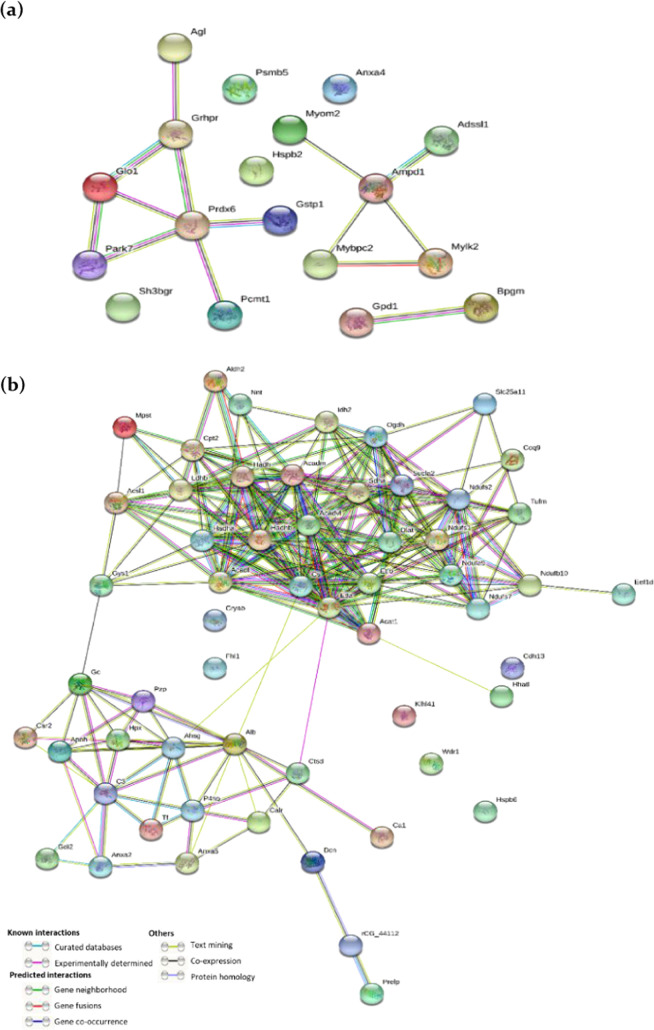
The protein–protein interaction network analysis was constructed using the STRING database for the upregulated and downregulated categories of proteins. A total of 18 differentially expressed proteins in the upregulated group and 57 differentially expressed proteins in the downregulated group were found to show interaction in the matched PPI networks. In the upregulated group, three distinct interactions were found, among which seven proteins (UniProt: B0BN46, O35244, Q6P7Q4, P22062, O88767, B6DYQ7, and D4AEH9) presented the highest degree of connectivity as shown in Figure 1a. In the downregulated group, two distinct interactive networks were found, with 31 proteins (UniProt: F1LN88, Q5BJZ3, P56574, Q5XI78, G3V6H5, P97532, P18886, Q9WVK7, G3V796, Q920L2, B2RZ24, Q641Y2, Q68FT, P18163, P42123, Q64428, Q60587, Q5M9H2, P08461, Q66HF1, P85834, A2RRU1, P15650, G3V936, P13803, Q68FU3, Q5BK63, Q5RJN0, D4A0T0, Q68FR9, and P17764) presented the highest degree of connectivity, as shown in Figure 1b.
Figure 1.
STRING interaction analysis of the differentially expressed genes. (a) Upregulated gene interaction network and (b) downregulated gene interaction network. (The line color indicates the type of interaction evidence, and the line thickness indicates the strength of data support).
Gene Ontology (GO) Enrichment Analysis of the Differentially Expressed Proteins
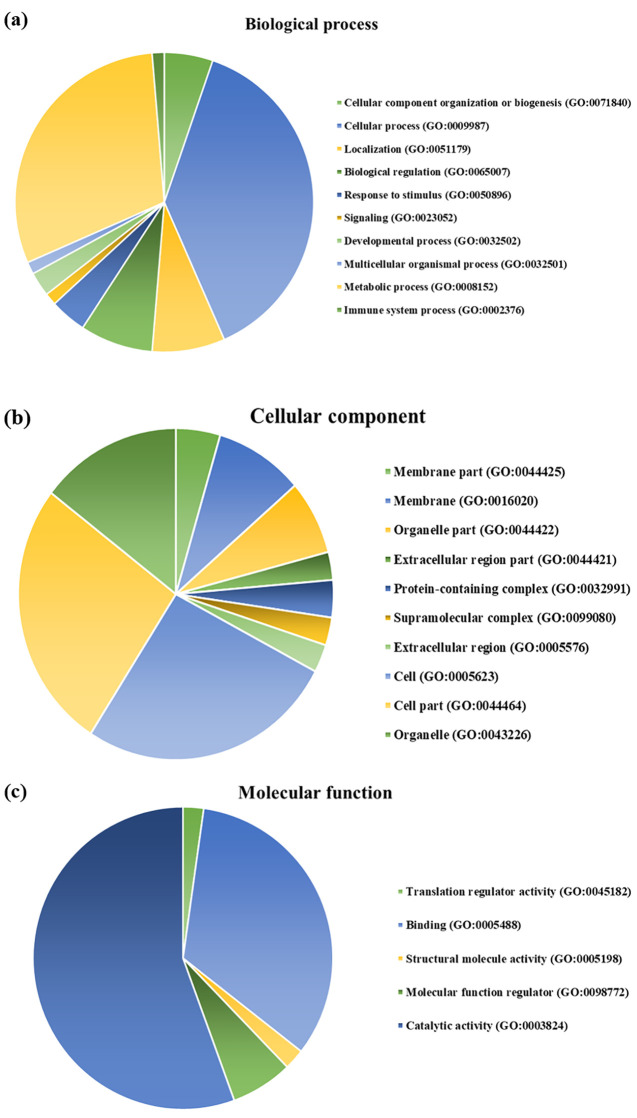
GO enrichment analysis is widely used to identify the biological roles and functions of specific gene and its products. All of the differentially expressed proteins were mapped to their enriched GO terms based on the functional annotation from the PANTHER database. The differentially expressed proteins were categorized into three classes, including biological process, molecular function, and cellular component, as shown in Figure 2. In terms of biological process, majority of the protein class are associated with cellular process (29) and metabolic process (23), as represented in Figure 2a. For the cellular component, most proteins were distributed in the cellular part (29) and the organelle part (16), as represented in Figure 2b. The proteins of molecular function were mainly involved in catalytic activity (25) and binding (15), as represented in Figure 2c.
Figure 2.
Gene ontology of the differentially expressed genes. (a) The hub genes expressed in terms of biological process, (b) the hub genes expressed in terms of cellular component, and (c) the hub genes expressed in terms of molecular function.
Pathway Analysis of the Differentially Expressed Proteins
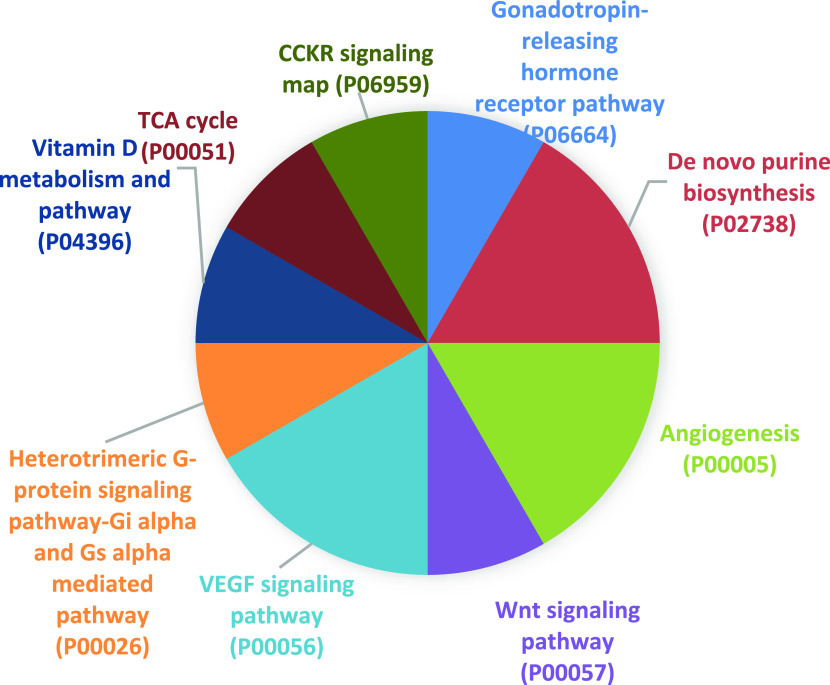
The PANTHER database provides a collection of various biological pathways, which represent the molecular interaction network between each protein/gene function. To identify the enriched pathways among the differentially expressed proteins, PANTHER pathway analysis was performed. Nine significantly enriched pathways were identified in the differentially expressed proteins of young and old groups, as shown in Figure 3. The enriched pathways include angiogenesis (P00005); CCKR signaling map (P06959); de novo purine biosynthesis (P02738); gonadotropin-releasing hormone receptor pathway (P06664); heterotrimeric G-protein signaling pathway—Gi α- and Gs α-mediated pathway (P00026); tricarboxylic acid (TCA) cycle (P00051); vascular endothelial growth factor (VEGF) signaling pathway (P00056); vitamin D metabolism and pathway (P04396); and Wnt signaling pathway (P00057). All of the significantly enriched pathways with their differentially expressed proteins mapped to their genes pertaining to indicated pathway are represented in Table 3.
Figure 3.
Pathway analysis of the differentially expressed genes. A total of nine significantly enriched pathways were identified using PANTHER database.
Table 3. List of Significantly Enriched Genes and Proteins between the Young and Old Groups Obtained from the Enriched Pathways by PANTHER.
| enriched pathways | proteins | genes |
|---|---|---|
| angiogenesis (P00005) | α-crystallin B chain, heat shock protein β-2 | Cryab, Hspb2 |
| CCKR signaling map (P06959) | acetyl-CoA acetyltransferase | Acat1 |
| de novo purine biosynthesis (P02738) | adenylate kinase isoenzyme 1, adenylosuccinate synthetase isozyme 1 | Ak1, Adssl1 |
| gonadotropin-releasing hormone receptor pathway (P06664) | annexin A5 | Anxa5 |
| heterotrimeric G-protein signaling pathway—Gi α and Gs α mediated pathway (P00026) | glycogen synthase | Gys1 |
| TCA cycle (P00051) | 2-oxoglutarate dehydrogenase | Ogdh |
| VEGF signaling pathway (P00056) | α-crystallin B chain, heat shock protein β-2 | Cryab, Hspb2 |
| vitamin D metabolism and pathway (P04396) | vitamin D-binding protein | Gc |
| Wnt signaling pathway (P00057) | adenylosuccinate synthetase isozyme 1 | Adssl1 |
Quantification of the Differentially Expressed Genes through Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR) Analysis
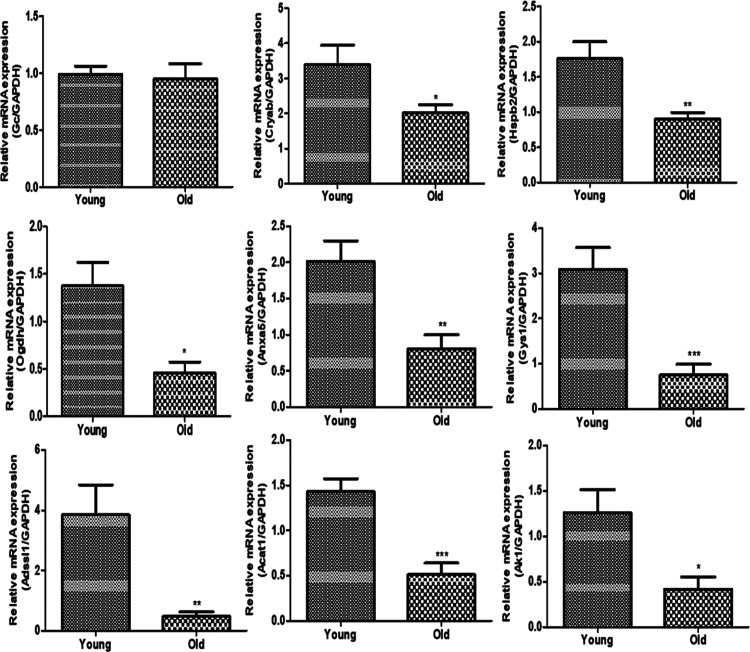
To experimentally refine the identified significantly enriched genes based on the enriched pathways, qRT-PCR analysis was performed on the nine genes (Cryab, Hspb2, Acat1, Ak1, Adss1, Anxa5, Gys1, Ogdh, and Gc). The relative mRNA expression levels of the identified genes were comparatively analyzed between the young and old groups. The results obtained indicated that the identified significant genes were found to be suppressed on the old group compared to the young group, as shown in Figure 4.
Figure 4.
mRNA quantification by qRT-PCR analysis. The mRNA expression of the significantly enriched genes between the young and old groups was measured. Nine genes were quantified (Cryab, Hspb2, Acat1, Ak1, Adssl1, Anxa5, Gys1, Ogdh, Gc, and Adssl1). The data are analyzed statistically and represented graphically.
Validation of Protein Expression through Western Blot Analysis
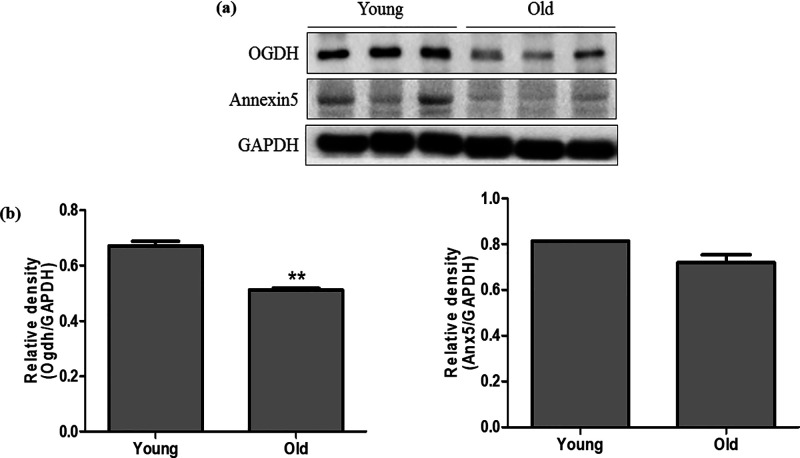
Further, western blot analysis of the validated differentially expressed proteins was performed to confirm the results of their mRNA expression. The protein levels of the relative proteins (Cryab, Hspb2, Acat1, Ak1, Adss1, AnxA5, Gys1, Ogdh, and Gc) were analyzed comparatively between the young and old groups. The results indicated that seven proteins—Cryab, Hspb2, Actat1, Ak1, Adss1, Gys1, and Gc—demonstrate no significant difference between the young and old groups (data not shown). However, two proteins Ogdh and AnxA5 showed a significant difference between the two groups with suppressed expression in the old group, as shown in Figure 5. The protein expression data are consistent with the relative mRNA expression data of the respective genes.
Figure 5.
Analysis of protein expression by Western blot. (a) Western blot analysis of the significant proteins (Ogdh and annexin 5 (AnxA5)). (b) Expressions of the protein levels are represented graphically based on its densitometry. Values are given as mean ± standard error of the mean (SEM) of three independent experiments.
Discussion
Proteomic techniques have become a strong approach to obtain insights into the mechanisms of biological process by characterizing the composition of cellular proteins and functional linkages between protein molecules.21 It provides extensive knowledge of the proteins expressed differentially in a particular condition exposing the signaling changes uniquely.22 The analysis of differential expression helps to provide useful information about the molecular behavior of a particular condition of the disease and aids in biomarker development.23 Reported studies used the proteomic approach to study the changes in different rat tissues in a diseased state or upon treated conditions identified novel insights with tandem mass tags.24
Our previous study on cultured muscle cells of young and old rats by flavonoid sinensetin showed regulation on age-related sarcopenia.25 In the current study, we adopted the proteomics-based tandem mass spectrometry approach to identify the differentially expressed proteins between the two different age groups of young and old rats. Herein, we explored the complete proteome profile between the two age group samples to elucidate the molecular mechanism and significantly enriched pathways of the differentially expressed proteins. In addition, the expressions of the proteins and respective genes related to the significantly enriched pathways were validated quantitatively.
A comprehensive study of the biochemical profiling and characterization of the wide ranging changes in the proteome profile enables the understanding of the role of metabolic networks inside the cell and will lead to the identification of the signature proteins.26 The nano-LC-MS/MS analysis of the samples of young and old groups of rats resulted in a set of protein spectral data with 503 proteins. The proteins were filtered based on the sample organism R. norvegicus, resulting in 335 proteins that were identified to be differentially expressed between the two sample groups (young and old). Among the 335 differentially expressed proteins, we identified 25 upregulated and 77 downregulated proteins based on log-fold change between 0.5 and 2.0. STRING network analysis performed between the upregulated proteins showed interaction between 18 differentially expressed proteins with three distinct networks and downregulated proteins showed interaction among 57 differentially expressed proteins with two distinct networks.
The functional enrichment of the proteins was identified in terms of GO involving the biological process, cellular component, and molecular functions. The most significantly enriched biological processes were associated with cellular process (GO: 0009987), with 29 genes and metabolic process (GO: 0008152) involving 23 genes. In terms of molecular function, the enriched categories were catalytic activity (GO: 0003824) and binding (GO: 0005488) with 25 and 15 representative genes, respectively. In the case of cellular component, most of the differential genes were found to be localized at the cellular part (GO: 0044464) comprising 29 hub genes, respectively.
The evaluation of biological significance in enriched pathways of the differentially expressed proteins was detected using PANTHER pathways. A total of nine significantly enriched pathways were identified, namely, angiogenesis (P00005); CCKR signaling map (P06959); de novo purine biosynthesis (P02738); gonadotropin-releasing hormone receptor pathway (P06664); heterotrimeric G-protein signaling pathway—Gi α- and Gs α-mediated pathway (P00026); TCA cycle (P00051); VEGF signaling pathway (P00056); vitamin D metabolism and pathway (P04396); and Wnt signaling pathway (P00057). The top three pathway hits that were highly enriched were found to be angiogenesis, de novo purine biosynthesis, and the VEGF signaling pathway. Sarcopenia is highly related with reduced capillary density and determined by the levels of various angiogenic factors, which decline during muscle aging. The induction of angiogenesis can be a promising therapeutic approach to improve the vascular function in aging people with muscle loss.27 The purine biosynthesis pathway has been shown to be highly related with the skeletal muscle fiber variations. The degradation of adenine nucleosides prone to be elevated during physical activity and the rates of de novo synthesis must be maintained to regulate the adenine pool. Observations on the level of purine salvage in male Sprague-Dawley rat muscles were observed to be in higher rise in terms of adenine levels.28,29 Vascular endothelial growth factor (VEGF) is a vital regulatory factor that has been reported to be involved in the process of muscle and blood vessel formation during embryogenesis. In vitro and in vivo studies on skeletal muscles and mouse models have successfully shown the upregulated expression of VEGF receptors (VEGF-1 and VEGF-2) promoting the growth of myogenic fibers, which employs a therapeutic approach in musculoskeletal disorders.30
The relative genes for each of the significantly enriched pathway hits were identified as Cryab, Hspb2, Acat1, Ak1, Adssl1, AnxA5, Gys1, Ogdh, and Gc. The validation of the significant hits in terms of mRNA and protein expression was quantified using real-time PCR and Western blot analysis to explore the consistency with that of the determined proteomics data. The relative mRNA expression of the nine genes was found to be downregulated in the aged muscle tissue group compared with that of the young group. The in vitro protein validation of the significant hits with their respective antibodies has identified two proteins (Ogdh and annexin 5) that were downregulated in the old group compared with the young group.
Ogdh is a mitochondrial protein also known as 2-oxoglutarate dehydrogenase, which is functionally active in the decarboxylation of α-ketoglutarate in the metabolic citric acid cycle.31 Proteomic profile of skeletal muscles from rodents and humans has identified about 42% of metabolic enzymes that brings about changes in the muscle mass loss.32 Structural and functional analogues of metabolic enzymes like Ogdh protect against the oxidative stress by increasing the antioxidant enzyme activity.33 Aging effect on the rate of metabolic enzyme Ogdh measured in older skeletal muscles has shown decreased activity compared with the middle-aged group.34
Annexins are a group of Ca2+-related membrane-binding proteins that are involved in the process of repair, and several annexin subtypes have been implicated in muscle injury disorders.35 Annexin 5 (AnxA5) is well known as an anticoagulant that functions as an indirect inhibitor of the thromboplastin complex involved in the cascades of blood coagulation.36 The inventory reports on AnxA5 2-D arrays that are shown to encourage the repair of membrane in protecting cells against the damage effects and membrane disorders in mouse perivascular cells.37 The facilitation of AnxA5 by forming a membrane band at the broken points of muscle wound in resealing of membrane repair in human skeletal muscle cells.38
Sarcopenia, the condition of loss in skeletal muscle mass during aging, is a major contributing factor that leads to fragility and disability in the elderly.33 Our data on validation of protein expressions (Ogdh and AnxA5) showed downregulation in the old group compared to the young group. It suggests the contribution of muscle loss and metabolic alterations caused during aging, and these can be specific targets in treatment strategies. This result has also attributed to the relevance of the proteins in promoting muscle loss or other membrane-related disorders in old skeletal muscles with earlier reported evidence. This proteomic profile on the muscle tissues of young and old groups has identified distinctive target markers that may aid in the development of therapeutics in muscle-related disorders with further extensive research.
Conclusions
In conclusion, in this study, we conducted a comprehensive proteomic analysis on the young and old muscle tissues in SD rats. As such, we identified the differentially expressed proteins in both the conditions followed by analysis of its gene ontology terms and significantly enriched pathways based on Bioinformatics tools. Further, in vitro validations on the differential expressed proteins related to the enriched pathways were analyzed by mRNA and protein expression. Notably, two important proteins (Ogdh and annexin 5) were found to be confirmed between the young and old groups, suggesting the differential pattern in terms of aging. Taken together, these findings have important clinical significance as they can improve our understanding on the biological and molecular mechanism of age-related sarcopenia. Moreover, these results may also lead to provide insights into the potential biomarker targets for the prediction of prognosis in sarcopenia with future validations on the identified proteins.
Materials and Methods
Animal Experiments and Handing
The experiment used tissues of Sprague-Dawley rats of two different age groups (6 months) and (21 months) that were obtained from Aging Tissue Bank, Pusan National University, Busan, Republic of Korea. The animals were housed and handled under maintenance of controlled environment (24 °C; 50–60% humidified atmosphere) with a 12 h dark/light cycles interval followed by the Animal Care Committee of Pusan National University, Busan, Republic of Korea.
Preparation of Protein Samples for Proteomics Analysis
To extract the calf tissue proteins from Sprague-Dawley rats’ (rat) young (6 months) and old (21 months) muscles, the tissues were homogenized using radioimmunoprecipitation assay (RIPA) buffer (iNtRON Biotechnology, Gyeonggi, Republic of Korea; 50 mM Tris–HCl, pH 7.5, 150 mM sodium chloride, 0.5% sodium deoxycholate, 1% Triton X-100, 0.1% sodium dodecyl sulfate (SDS), and 2 mM ethylenediaminetetraacetic acid (EDTA)) containing a protease inhibitor cocktail (Thermo Fisher Scientific). Then, it was kept on ice for 30 min and centrifuged at 10 000 rpm at 4°C for 10 min. The protein concentration present in the extracted samples was quantified using the bicinchoninic acid (BCA) assay (Thermo Fisher Scientific) method. The samples with equal protein concentration were subjected to proteomic analysis.
Identification of Proteins Using MS/MS Analysis
One-Dimensional Gel Electrophoresis (Nanomas)
For one-dimensional SDS-PAGE (acrylamide concentration of 4% for the stacking and 12% for the resolving), 50 μg of rat’s total protein was diluted with denaturing sample buffer (0.5M Tris–HCl pH 8.8, 10% SDS, 20% glycerol, 1% bromophenol blue, 0.2% dithiothreitol (DTT)) and heated at 95 °C for 5 min. The samples were subjected to SDS-PAGE using 12% polyacrylamide gel. Electrophoresis was performed at 100 V for 60 min, followed by 200 V for 2 h. The gels were then stained with Coomassie Brilliant R250 (Sigma-Aldrich, St. Louis, MO) and destained with distilled water.
In-Gel Digestion
A one-dimensional SDS-PAGE lane containing all protein bands was excised using a razor blade from top to bottom, and the excised gel slices were washed twice with 100 μL of distilled water for 15 min at RT. The excised gel bands were destained with 50% acetonitrile and shrunk with 100% acetonitrile. The proteins in the gel were then soaked in 500 μL of 50 mM ammonium bicarbonate for 5 min at RT. The ammonium bicarbonate solution was removed by pipetting, and 500 μL of acetonitrile was then added and incubated for 5 min at RT. After acetonitrile incubation, the gel slices were dried completely in vacuum. The gel slices were incubated in 10 mM DTT/0.1 M ammonium bicarbonate for 45 min at 56 °C and then alkylated by incubation with 55 mM iodoacetamide/0.1 M ammonium bicarbonate for 30 min at RT in the dark. After alkylation, the gel slices were dried again and the dried gel slices were swollen in 5 μL of digestion buffer that contained 25 mM ammonium bicarbonate, 0.1% n-octyl glucoside, and 50 ng/mL of sequencing grade trypsin (Promega, Madison, MI) for rehydration. After rehydration, the band slices were incubated overnight at 37 °C in 50 μL of digestion buffer (without trypsin) to allow enzymatic cleavage in siliconized tube. About 0.1 μg of protease was used for one gel band. Peptides were extracted from the gel slices with 66% acetonitrile, 33% water, and 0.1% trifluoroacetic acid (TFA). After centrifugation, the peptides were transferred into a new tube and dried with a SpeedVac (Hanil, Korea). The dried peptides form the gel slices stored at −80 °C before analysis.
Nano-Flow Liquid Chromatography
The dried peptides from the gel slices were redissolved in 20 μL of 5% formic acid and analyzed via on-line nanoflow LC-MS/MS. All nano-LC-MS/MS experiments were performed on an Eksigent nanoLC415 system (Eksigent, Dublin) connected to a triple time-of-flight (TOF) 6600 mass spectrometry system (SCIEX, Redwood City, CA) with a nano-electrospray ion source (New Objective, Woburn, MA). The tryptic digested peptides were separated in a 15 cm analytical column (ChromXP C18, 75 μm × 15 cm, 3 μm, 120 Å, Eksigent, Dublin) with a 90 min gradient from 5 to 60% acetonitrile in 0.1% formic acid. The effluent from nano-LC was directly sprayed into the mass spectrometer through electrospraying.
LC-MS/MS Analysis and Data Search
For gel-free proteomic analysis, protein samples were resuspended in water/formic acid solution (water in 0.1% formic acid). On-line nano-high-performance liquid chromatography (nanoHPLC) was conducted by Eksigent nanoLC415 system (Eksigent, Dublin). After injection, each peptide was transferred onto a C18 nonoLC trap column (ChromXP, 350 μm × 0.5 mm, 3 μm, 120 Å) and washed at 300 nL/min for 10 min with loading solvent (water/0.1% formic acid) for desalting. Subsequently, each peptide was transferred to an analytical ChromXP C18 column (75 μm × 15 cm, 3 μm, 120 Å) and eluted at a flow rate 300 nL/min using a 90 min gradient using analysis solvent (water/0.1% formic acid). The samples were loaded on a C18 nonoLC trap column (ChromXP, 350 μm × 0.5 mm, 3 μm, 120 Å) and washed with nanoHPLC buffer A at a rate of 300 nL/min for 10 min. An elution gradient of 5–60% water (0.1% formic acid) over 90 min gradient period was used on an analytical ChromXP C18 column (75 μm × 15 cm, 3 μm, 120 Å) with a nanospray tip. Data acquisition was performed with a triple time-of-flight (TOF) 6600 system (SCIEX, Redwood City, CA) coupled with nonospray source (New Objective, Woburn, MA), with a pulled 10 μm fused silica emitter, 360/20 μm (New Objective, Woburn, MA). Data were acquired using an ionspray voltage floating (ISVF) of 2.1 kV, curtain gas of 20 psi, ion source gas (GS1) of 15 psi, and interface heater temperature of 150 °C. For information-dependent acquisition (IDA), the survey scans range was set between 250 and 2000 m/z (250 ms accumulation time) followed by dependent MS/MS scan with a mass range set between 100 and 2000 m/z (100 ms accumulation time) of the 20 most intense ion in the high-sensitivity mode with a 2+ to 5+ charge state. Dynamic exclusion was carried out for a period of 15 s, and a tolerance of 50 ppm was adopted. Rolling collision energy was used throughout the process.
After MS/MS analysis, data files were processed using UniProt and ProteinPilot 5.0.1 (SCIEX, Redwood City, CA) database software. Based on the combined MS and MS/MS spectra, proteins were successfully identified at a 95% or higher confidence interval, using their scores in the MASCOT v2.6 search engine (Matrix Science Ltd., London, U.K.), and the following search parameters were adopted: rat database, trypsin as the digestion enzyme, single missed cleavage sites, fixed modifications of carbamidomethyl (C) and oxidation of methionine, ±0.1 Da precursor ion tolerance, and ±0.1 Da MS/MS fragment ion tolerance.
Criteria for Protein Identification
Protein identification was undertaken using a “through” search effort with ProteinPilot 5.0.1 software with the Paragon algorithm. The search parameters were defined as iodoacetamide modified for cysteine alkylation and trypsin as the digestion enzyme. Tandem mass spectrometry data were searched against a database comprising UniProt-rat (version 2019/20) and rat peptide sequences (sequence downloaded in November 2019; 215 352 sequences). The database search results were manually curated to achieve protein identification using 1% global false discovery rate (FDR) determined by the in-built FDR tool within ProteinPilot software, and Scaffold (version Scaffold_4.10.0, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at a greater than 96.0% probability to achieve an FDR less than 1.0% by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at a greater than 99.0% probability and contained at least one identified peptide. Scaffold (version Scaffold_4.10.0, Proteome Software, Inc., Portland, OR) was used to validate the MS/MS-based peptide and protein identification.
Peptide Clean-Up Progress
After drying the resulting peptide mixture, samples were reconstituted by 1% trifluoroacetic acid (TFA) in water and applied to Sep-Pak C18 cartridges (Waters, Milford, MA). After washing the column with 1 mL of 0.1% trifluoroacetic acid (TFA) in water, digested peptides were twice eluted with 0.5 mL of 50% acetonitrile in water. Eluted peptides were dried in a SpeedVac (Hanil, Korea), and the dried peptides were stored at −80 °C before analysis.
Differential Gene Expression Analysis
The proteomics mass spectra data obtained from MS/MS analysis were visualized using Scaffold (version Scaffold_4.10.0, Proteome Software, Inc., Portland, OR). The differential expressions of proteins between the young and old groups were identified and grouped individually. The protein identification was considered accepted if they could be established at 99% protein threshold; 95% peptide threshold and contained at least one identified peptide. The total differentially expressed proteins obtained based on the peptide and protein thresholds were scrutinized by species selection (R. norvegicus). Further, the differentially expressed proteins were segmented into upregulated and downregulated categories based on the log-fold change criteria between 0.5 and 2.0.
Gene Ontology and Pathway Enrichment Analysis
Upon protein identification into upregulated and downregulated categories, the interaction of the proteins between the two classes was viewed using STRING interaction network construction (https://string-db.org/). The differentially expressed genes were analyzed to know their annotations through evolutionary relationship in functional pathways and classification of proteins with gene ontology terms were studied. The gene ontology categories in terms of biological process, molecular function, and cellular component were individually sorted using PANTHER GO terms. Similarly, all of the functionally enriched pathways of the significantly expressed proteins were determined with PANTHER pathways. Gene ontology and pathway analysis was carried out using PANTHER database (http://www.pantherdb.org/). From the enriched pathway terms, each protein was expressed and its subjective genes were chosen for further quantification studies.
mRNA Quantification by RT-qPCR Analysis
Real-time PCR was employed to validate the differentially expressed proteins obtained from the proteomic analysis. To extract mRNA from the calf muscles in young and old Sprague-Dawley rats, the tissues were homogenized in TRIzol reagent (Thermo Fisher Scientific, Inc.). The 1 μg of RNA was reverse-transcribed to cDNA synthesis using iScript cDNA synthesis kit (Bio-Rad Laboratories, Inc.), according to the manufacturer’s instructions, and cDNA templates were conducted in quantitative real-time PCR using AccuPower 2× Greenstar qPCR Master (Bioneer, Daejeon, Republic of Korea). To know each gene primet, we commissioned primer design service in Bioneer Corp. (Bioneer, Daejeon, Republic of Korea). The sequence of primers is listed in Table 4. Thermocycling conditions were organized of a predenaturation for 2 min at 95 °C, followed by 40 cycles at 95 °C for 5 s, 58 °C for 30 s, and 95 °C for 5 s. qPCR was performed with a CFX384 Real-Time PCR Detection System (Bio-Rad Laboratories, Inc.). All data were analyzed with the Bio-Rad CFX Manager version 3.1 software using the difference method (ΔΔCq). The mRNA expression levels were normalized using the expression of GAPDH as control.
Table 4. Primer Sequences of the Nine Genes Used in qRT-PCR Analysis.
| pimer name | primer sequence (5′–3′) |
|---|---|
| Acat1 | F: TGCTACACGAACTCCCATTG |
| R: ATGACGTTGCCCATGTAGAC | |
| Adssl1 | F: ACCTTGTGTTCGACTTCCAC |
| R: TGGTGGTGCCGATATTCTTC | |
| Ak1 | F: GAAGTCAGCTCTGGATCGTC |
| R: TCAAGAAGCCGTTGGAAGAG | |
| Anxa5 | F: CATCACCATCCTTGGGACAC |
| R: CGACAGCCAGGAGTAAGTTC | |
| Cryab | F: TCTCTACAGCCACTTCCCTG |
| R: TGTCCTTCTCCATACGCATC | |
| Gc | F: GTTTGCCAGGAACTCTCCAC |
| R: TTCACAAGCTGACTGACCTG | |
| Gys1 | F: CAGACCTGCTGGATTGGAAG |
| R: GGCTCATAGGTGAAGTGGTC | |
| Hspb2 | F: CGAGTACGAATTTGCCAACC |
| R: AGTAGCCATGGTAGAGGGTG | |
| Ogdh | F: CTTCACTCCCAAATCCCTCC |
| R: TTTGTCTGGGTTCTGAGCTG |
Western Blot Analysis
The protein levels of the differentially expressed proteins were measured by western blot to validate the differentially expressed proteins identified by mass spectrometry. The extracted young and old calf muscle protein was quantified using BCA assay (Thermo Fisher Scientific) method. Proteins were separated by 8–15% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a poly(vinylidene fluoride) (PVDF) membrane (immunobilon-P, 0.45 mm; Millipore, Billerica, MA) using the semidry transfer system (Atto Corp., Tokyo, Japan). The membranes were blocked with 5% skim milk or bovine serum albumin (BSA) in Tris-buffered saline containing 1% Tween 20 (TBS-T, pH 7.4) at RT for 1 h and incubated overnight at 4 °C with a 1:1000 dilution of the respective primary antibody. The membranes were washed five times with TBS-T for 10 min each at RT and incubated with a 1:1000–1:5000 dilution of horseradish peroxidase (HRP)-conjugated secondary antibody for 3 h at RT. The membranes were then washed again five times with TBS-T. The proteins were detected by ECL reagent (Bio-Rad, Hercules, CA) and analyzed using Image Lab 4.1 (Bio-Rad) program. The densitometry readings of the bands were normalized according to GAPDH expression as control.
Statistical Analysis
All experimental results are expressed as ±standard error of the mean (SEM) of at least triplicate samples using GraphPad Prism software (version 5.02; GraphPad Software, Inc.). Significant differences between the two groups were calculated by one-way factorial analysis of variance (ANOVA), followed by Dunnett’s test. P-values <0.05 were considered statistically significant.
Acknowledgments
The authors thank Aging Tissue Bank for providing research materials for this study (NRF-2015M3A9B8029074).
Glossary
Abbreviations
- nano LC-MS/MS
nanoscale liquid chromatography coupled to tandem mass spectrometry
- DE
differentially expressed
- GO
gene ontology
- SD
Sprague-Dawley
- SDS-PAGE
SDS-polyacrylamide gel electrophoresis
- PVDF
poly(vinylidene fluoride)
Author Contributions
# J.A.K. and P.V. contributed equally to this work.
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (no. NRF-2020R1F1A1069319).
The authors declare no competing financial interest.
References
- Bross R.; Storer T.; Bhasin S. Aging and Muscle Loss. Trends Endocrinol. Metab. 1999, 10, 194–198. 10.1016/S1043-2760(98)00143-X. [DOI] [PubMed] [Google Scholar]
- Merlini L.; Bonaldo P.; Marzetti E. Editorial: Pathophysiological Mechanisms of Sarcopenia in Aging and in Muscular Dystrophy: A Translational Approach. Front. Aging Neurosci. 2015, 7, 153 10.3389/fnagi.2015.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzetti E.; Lees H. A.; Wohlgemuth S. E.; Leeuwenburgh C. Sarcopenia of aging: underlying cellular mechanisms and protection by calorie restriction. BioFactors 2009, 35, 28–35. 10.1002/biof.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber L.; Scicchitano B. M.; Musaro A. Molecular and Cellular Mechanisms of Muscle Aging and Sarcopenia and Effects of Electrical Stimulation in Seniors. Eur. J. Transl. Myol. 2015, 25, 231–236. 10.4081/ejtm.2015.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alway S. E.; Myers M. J.; Mohamed J. S. Regulation of satellite cell function in sarcopenia. Front. Aging Neurosci. 2014, 6, 246 10.3389/fnagi.2014.00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedl T. J.; San Gil R.; Cheng F.; Rayner S. L.; Davidson J. M.; De Luca A.; Villalva M. D.; Ecroyd H.; Walker A. K.; Lee A. Proteomics Approaches for Biomarker and Drug Target Discovery in ALS and FTD. Front. Aging Neurosci. 2019, 13, 548 10.3389/fnins.2019.00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas S.; Hao L.; Ricke W. A.; Li L. Biomarker discovery in mass spectrometry-based urinary proteomics. Proteomics: Clin. Appl. 2016, 10, 358–370. 10.1002/prca.201500102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallam R. M. Proteomics in cancer biomarkers discovery: challenges and applications. Dis. Markers 2015, 2015, 321370 10.1155/2015/321370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier E. F.; Ducamp S.; Leduc M.; Salnot V.; Guillonneau F.; Dussiot M.; Hale J.; Giarratana M. C.; Raimbault A.; Douay L.; Lacombe C.; Mohandas N.; Verdier F.; Zermati Y.; Mayeux P. Comprehensive Proteomic Analysis of Human Erythropoiesis. Cell Rep. 2016, 16, 1470–1484. 10.1016/j.celrep.2016.06.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuli L.; Ressom H. W. LC-MS Based Detection of Differential Protein Expression. J. Proteomics Bioinf. 2009, 2, 416–438. 10.4172/jpb.1000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W.; Petricoin E. F. 3rd; Longo C. Mass Spectrometry-Based Biomarker Discovery. Methods Mol. Biol. 2017, 1606, 297–311. 10.1007/978-1-4939-6990-6_19. [DOI] [PubMed] [Google Scholar]
- Toms S. A.; Weil R. J.. Nanoproteomics: Methods and Protocols; Humana Press: New York, 2011; Vol. xiv, pp 322. [Google Scholar]
- Vaiopoulou A.; Gazouli M.; Papadopoulou A.; Anagnostopoulos A. K.; Karamanolis G.; Theodoropoulos G. E.; M’Koma A.; Tsangaris G. T. Serum protein profiling of adults and children with Crohn disease. J. Pediatr. Gastroenterol. Nutr. 2015, 60, 42–47. 10.1097/MPG.0000000000000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner J. A.; Williams S. A.; Webster C. J. Biomarkers and surrogate end points for fit-for-purpose development and regulatory evaluation of new drugs. Clin. Pharmacol. Ther. 2007, 81, 104–107. 10.1038/sj.clpt.6100017. [DOI] [PubMed] [Google Scholar]
- Rifai N.; Gillette M. A.; Carr S. A. Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol. 2006, 24, 971–983. 10.1038/nbt1235. [DOI] [PubMed] [Google Scholar]
- Zhao Y.; Lee W. N.; Xiao G. G. Quantitative proteomics and biomarker discovery in human cancer. Expert Rev. Proteomics 2009, 6, 115–118. 10.1586/epr.09.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralhan R.; Desouza L. V.; Matta A.; Tripathi S. C.; Ghanny S.; Datta Gupta S.; Bahadur S.; Siu K. W. Discovery and verification of head-and-neck cancer biomarkers by differential protein expression analysis using iTRAQ labeling, multidimensional liquid chromatography, and tandem mass spectrometry. Mol. Cell. Proteomics 2008, 7, 1162–1173. 10.1074/mcp.M700500-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monforte J.; McPhail S. Strategy for gene expression-based biomarker discovery. BioTechniques 2005, 38, 25–29. 10.2144/05384SU05. [DOI] [PubMed] [Google Scholar]
- Theofilatos K.; Korfiati A.; Mavroudi S.; Cowperthwaite M. C.; Shpak M. Discovery of stroke-related blood biomarkers from gene expression network models. BMC Med. Genomics 2019, 12, 118118 10.1186/s12920-019-0566-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siu K. W.; DeSouza L. V.; Scorilas A.; Romaschin A. D.; Honey R. J.; Stewart R.; Pace K.; Youssef Y.; Chow T. F.; Yousef G. M. Differential protein expressions in renal cell carcinoma: new biomarker discovery by mass spectrometry. J. Proteome Res. 2009, 8, 3797–3807. 10.1021/pr800389e. [DOI] [PubMed] [Google Scholar]
- Malkawi A. K.; Masood A.; Shinwari Z.; Jacob M.; Benabdelkamel H.; Matic G.; Almuhanna F.; Dasouki M.; Alaiya A. A.; Rahman A. M. A. Proteomic Analysis of Morphologically Changed Tissues after Prolonged Dexamethasone Treatment. Int. J. Mol. Sci. 2019, 20, 3122 10.3390/ijms20133122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn J.; Ferluga S.; Sharma V.; Futschik M.; Hilton D. A.; Adams C. L.; Lasonder E.; Hanemann C. O. Proteomic analysis discovers the differential expression of novel proteins and phosphoproteins in meningioma including NEK9, HK2 and SET and deregulation of RNA metabolism. EBioMedicine 2019, 40, 77–91. 10.1016/j.ebiom.2018.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi A.; Kaufman K. L.; Ali M.; Arthur J.; Kossard S.; Fernandez-Penas P. Differential proteomic analysis of actinic keratosis, Bowen’s disease and cutaneous squamous cell carcinoma by label-free LC-MS/MS. J. Dermatol. Sci. 2018, 91, 69–78. 10.1016/j.jdermsci.2018.04.006. [DOI] [PubMed] [Google Scholar]
- Chen R.; Vendrell I.; Chen C. P.; Cash D.; O’Toole K. G.; Williams S. A.; Jones C.; Preston J. E.; Wheeler J. X. Proteomic analysis of rat plasma following transient focal cerebral ischemia. Biomarkers Med. 2011, 5, 837–846. 10.2217/bmm.11.89. [DOI] [PubMed] [Google Scholar]
- Kim J. A.; Kim S. M.; Ha S. E.; Vetrivel P.; Saralamma V. V. G.; Kim E. H.; Kim G. S. Sinensetin regulates age-related sarcopenia in cultured primary thigh and calf muscle cells. BMC Complementary Altern. Med. 2019, 19, 287 10.1186/s12906-019-2714-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Delgado J.; Lazaro A.; Osende J. I.; Barderas M. G.; Blanco-Colio L. M.; Duran M. C.; Martin-Ventura J. L.; Vivanco F.; Egido J. Proteomic approach in the search of new cardiovascular biomarkers. Kidney Int. 2005, 68, S103–S107. 10.1111/j.1523-1755.2005.09919.x. [DOI] [PubMed] [Google Scholar]
- Ambrose C. Muscle weakness during aging: a deficiency state involving declining angiogenesis. Ageing Res. Rev. 2015, 23, 139–153. 10.1016/j.arr.2015.03.005. [DOI] [PubMed] [Google Scholar]
- Mielcarek M.; Smolenski R. T.; Isalan M. Transcriptional Signature of an Altered Purine Metabolism in the Skeletal Muscle of a Huntington’s Disease Mouse Model. Front. Physiol. 2017, 8, 127 10.3389/fphys.2017.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brault J. J.; Terjung R. L. Purine salvage to adenine nucleotides in different skeletal muscle fiber types. J. Appl. Physiol. 2001, 91, 231–238. 10.1152/jappl.2001.91.1.231. [DOI] [PubMed] [Google Scholar]
- Arsic N.; Zacchigna S.; Zentilin L.; Ramirez-Correa G.; Pattarini L.; Salvi A.; Sinagra G.; Giacca M. Vascular endothelial growth factor stimulates skeletal muscle regeneration in vivo. Mol. Ther. 2004, 10, 844–854. 10.1016/j.ymthe.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Araújo W. L.; Martins A. O.; Fernie A. R.; Tohge T. 2-Oxoglutarate: linking TCA cycle function with amino acid, glucosinolate, flavonoid, alkaloid, and gibberellin biosynthesis. Front. Plant Sci. 2014, 5, 552 10.3389/fpls.2014.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio D.; Fania C.; Torretta E.; Vigano A.; Moriggi M.; Bravata V.; Caretti A.; Levett D. Z. H.; Grocott M. P. W.; Samaja M.; Cerretelli P.; Gelfi C. TCA cycle rewiring fosters metabolic adaptation to oxygen restriction in skeletal muscle from rodents and humans. Sci. Rep. 2017, 7, 9723 10.1038/s41598-017-10097-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh M. E.; Bhattacharya A.; Sataranatarajan K.; Qaisar R.; Sloane L.; Rahman M. M.; Kinter M.; Van Remmen H. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell 2015, 14, 957–970. 10.1111/acel.12387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaczor J. J.; Ziolkowski W.; Antosiewicz J.; Hac S.; Tarnopolsky M. A.; Popinigis J. The effect of aging on anaerobic and aerobic enzyme activities in human skeletal muscle. J. Gerontol., Ser. A 2006, 61, 339–344. 10.1093/gerona/61.4.339. [DOI] [PubMed] [Google Scholar]
- Demonbreun A. R.; Fallon K. S.; Oosterbaan C. C.; Bogdanovic E.; Warner J. L.; Sell J. J.; Page P. G.; Quattrocelli M.; Barefield D. Y.; McNally E. M. Recombinant annexin A6 promotes membrane repair and protects against muscle injury. J. Clin. Invest. 2019, 129, 4657–4670. 10.1172/JCI128840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rescher U.; Gerke V. Annexins--unique membrane binding proteins with diverse functions. J. Cell Sci. 2004, 117, 2631–2639. 10.1242/jcs.01245. [DOI] [PubMed] [Google Scholar]
- Bouter A.; Gounou C.; Berat R.; Tan S.; Gallois B.; Granier T.; d’Estaintot B. L.; Poschl E.; Brachvogel B.; Brisson A. R. Annexin-A5 assembled into two-dimensional arrays promotes cell membrane repair. Nat. Commun. 2011, 2, 270 10.1038/ncomms1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeille R.; Bouvet F.; Tan S.; Croissant C.; Gounou C.; Mamchaoui K.; Mouly V.; Brisson A. R.; Bouter A. Membrane repair of human skeletal muscle cells requires Annexin-A5. Biochim. Biophys. Acta, Mol. Cell Res. 2016, 1863, 2267–2279. 10.1016/j.bbamcr.2016.06.003. [DOI] [PubMed] [Google Scholar]