Abstract
The emergence of Coronavirus disease 2019 as a global pandemic has increased popular concerns about diseases caused by viruses. Fermented foods containing high loads of viable fungi and bacteria are potential sources for virus contamination. The most common include viruses that infect bacteria (bacteriophage) and yeasts reported in fermented milks, sausages, vegetables, wine, sourdough, and cocoa beans. Recent molecular studies have also associated fermented foods as vehicles for pathogenic human viruses. Human noroviruses, rotavirus, and hepatitis virus have been identified in different fermented foods through multiple routes. No severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) virus or close members were found in fermented foods to date. However, the occurrence/persistence of other pathogenic viruses reveals a potential vulnerability of fermented foods to SARS-CoV-2 contamination. On the other side of the coin, some bacteriophages are being suggested for improving the fermentation process and food safety, as well as owing potential probiotic properties in modern fermented foods. This review will address the diversity and characteristics of viruses associated with fermented foods and what has been changed after a short introduction to the most common next-generation sequencing platforms. Also, the risk of SARS-CoV-2 transmission via fermented foods and preventive measures will be discussed.
Keywords: SARS-CoV-2, Rotavirus, Next-generation sequencing, Bacteriophage, Fermented milks
Abbreviations: BA, Biological amines; BLAST, Basic Local Alignment Search Tool; COVID-19, Coronavirus disease 2019; FAO, Food and Agriculture Organization of the United Nations; GIT, Gastrointestinal tract; HAV, Hepatitis A virus; HV, Hepatitis virus; HEV, Hepatitis E virus; HGT, Horizontal gene transfer; HTS, High-throughput screening; kb, Kilobases; LA, Linker amplification; LAB, Lactic acid bacteria; LASL, Linker amplification shotgun libraries; MERS-CoV, Middle East respiratory syndrome coronavirus; MG-RAST, MetaGenomic-Rapid Annotation using Subsystem Technology; NGS, Next-generation sequencing; NiV, Nipah virus; nm, Nanometer; NoV, Norovirus; MDA, Multiple displacement amplification; MLST, Multilocus sequence typing; ORF, Open reading frame; OUT, Operational taxonomic units; PCR, Polymerase chain reaction; PHACCS, PHAge Communities from Contig Spectrum; qPCR, Real-time quantitative PCR; RAPD, Random Amplified Polymorphic DNA; RFLP, Restriction Fragment Length Polymorphism; RT-PCR, Reverse transcription polymerase chain reaction; RV, Rotavirus; RVA, Rotavirus A; SARS-CoV-2, Severe Acute Respiratory Syndrome Coronavirus 2; TBEV, Tick-borne encephalitis virus; TEM, Transmission electron microscopy; UV, Ultraviolet; VACV, Vaccinia virus; VIROME, Viral Informatics Resource for Metagenome Exploration; VMGAP, Viral MetaGenome Annotation Pipeline; VP, Viral particles; WGA, Whole genome amplification; WHO, World Health Organization
1. Introduction
Viruses are ubiquitous in every ecosystem and infect all forms of life, from prokaryotes to eukaryotes (Hyman and Abedon, 2012). Food fermentation is driven by dense microbial consortia consisting mainly of bacteria and fungi (Pereira et al., 2020). This constitutes a rich reservoir for the development of many microorganism-infecting viruses. Studies have reported the presence of viruses that infect bacteria (bacteriophages) and yeast in plenty of fermented food products, including wine, meat, cheese, yoghurt, sourdough, sauerkraut, kimchi, soybean, and cocoa (Auad et al., 1997; Barrangou et al., 2002; Foschino et al., 2005; Illeghems et al., 2012; Kiliç et al., 1996; Kleppen et al., 2012a; Pringsulaka et al., 2011; Umene et al., 2009). In general, bacteriophages are considered harmful by decreasing the fermentative capacity of lactic acid bacteria (LAB) and yeasts, occasionally resulting in complete fermentation failure. The diversity of phages in fermented foods has been seen to vary according to geography, climate, environment, type of raw material, preparation methods, and microbial composition (Tamang et al., 2020). However, although there is evidence that bacteriophages can cause disease in humans (Tetz and Tetz, 2018), they are not associated with sanitary and public health concerns.
In addition to bacteriophages and yeast-associated viruses, pathogenic viruses have been reported in fermented foods causing injuries and even death (Cho et al., 2016; Colson et al., 2010; Holzmann et al., 2009; Hossain et al., 2016). The zoonotic Nipah virus (NiV), for example, was attributed as the probable cause of the death of eight victims who ingested a traditional fermented liquor in Bangladesh (Hossain et al., 2016). Viruses have high environmental resistance being able to survive against microorganisms elimination processes (Vasickova et al., 2010), using these products as vehicles to human contagion. Currently, human noroviruses (NoVs) are recognized as the main cause of viral foodborne outbreaks, followed by rotavirus (RV) and hepatitis virus (HV) (Leblanc et al., 2019). Contamination can occur in different ways, mainly through infected raw materials and improper food handling. RVs causes an estimated 111 thousand cases of diarrhea per year, two million hospitalizations, and 400 thousand fatalities in children under five years old, being more than 80% from undeveloped countries (Tamang et al., 2020, World Health Organization). The emergence of Coronavirus disease 2019 (COVID-19) as a global pandemic has increased popular concerns about diseases caused by viruses (Lai et al., 2020). Despite the principal form of spreading is human-to-human contact, the COVID-19 similar Middle East Respiratory Syndrome (MERS-CoV) virus remained infectious up to 60 min outside the body (Pyankov et al., 2018). Security authorities raised concern on viral transmission by food, as preparation and delivery can be critical step on transmission (Rizou et al., 2020).
The presence of viruses in fermented foods has traditionally been studied by culture-dependent methods. These methods are focused on singular bacteriophages that cause fermentative flaws and on pathogenic human viruses (Park et al., 2011). However, with the advancement of molecular techniques and the outgrowth of next-generation sequencing (NGS), a large body of metagenomic sequencing information was allowed, circumventing the need for gene cloning or cultivation (de Melo Pereira et al., 2020). Fermented food microbiome has been accessed, and what is known as “viromes” has emerged. However, the number of viromes studies still lags far behind that of bacteria and fungi publications; for each virome in 2019, there were 42 microbiome studies (Ledormand et al., 2020; Tamang et al., 2020).
The lack of a universal molecular marker, as the 16S and 18S rRNA in bacteria and fungi, respectively, can be considered an obstacle for virome studies in food matrices. Nevertheless, shotgun metagenomic, which does not depend on a target ribosomal marker, has successively characterized viral communities from freshwater, soil, ocean, mammalian gut and, to a lesser extent, fermented food products (Dugat-Bony et al., 2020; Hayes et al., 2017; Park et al., 2011). Recently, pyrosequencing and Illumina Miseq platforms have been used to characterize viral communities of different fermented foods (Dugat-Bony et al., 2020; Jung et al., 2018). NGS viromes confirmed the dominance of Caudovirales bacteriophages as on culture-dependent approaches (Dugat-Bony et al., 2020; Jung et al., 2018; Park et al., 2011).
NGS studies have enabled new applications and perspectives for bacteriophages and, thus, accessing the other side of the coin. It has been observed that some bacteriophages can modulate bacterial community succession during the fermentation process, positively affecting food quality and sensorial properties (Agyirifo et al., 2019). Quorum sensing studies have not yet been accomplished to better understand this modulation and relationship. Gastrointestinal tract (GIT) bacterial community is also shaped by bacteriophages contained in fermented foods, impacting host physiology and metabolism. They are able to trigger immune responses through direct contact with mucosal epithelial cells (locally) or with immune system components (systemically) (Sausset et al., 2020). These features enable bacteriophages to provide probiotic effects, as suggested by Pacini and Ruggiero (2019), through the ingestion of phage-containing fermented milk and colostrum.
This review will provide a general overview of viruses associated with fermented foods and what has been changed after a short introduction to the most common NGS platforms, as well as a critical discussion on the potential of fermented foods to deliver viruses with public health concerns. Additionally, NGS strategies and methods for describing food viromes will be addressed with the ultimate objective of assisting future evaluation studies.
2. Bacteriophages and yeast viruses
Bacteria comprise a highly diverse group, representing the second major biomass element on Earth (~15%), behind only plants (~80%). This abundant living mass is a huge reservoir for bacteriophage (or simply called phage) predation. As phages can be found repeatedly in different hosts, they represent on approximately 1031 particles, ten times the number of bacteria (1030 cells) (Breitbart and Rohwer, 2005). Bacteriophages are small in size (isometric heads are typically 45–170 nm in diameter) and are composed of a single type of nucleic acid with single or double-stranded (ssDNA, dsDNA, ssRNA, dsRNA) protected by a protein or lipoprotein capsid (Orlova, 2012). Phage genomes vary between families ranging from ~3.5 kb (e.g. Escherichia coli phage genome) to ~540 kb (Prevotella spp. phages genome) (Sausset et al., 2020). They do not have cellular machinery required for transcription, translation, and energy production, using from their hosts. When inside, phage particles are formed and, when bacterial lysis occurs, they are released (lytic cycle). The phages that use only the lytic cycle to propagate are called virulent.
The infection of LAB by bacteriophages is widely investigated being considered the primary cause of fermentation failure in the dairy industry (Garneau and Moineau, 2011). However, some phages have specific genes to direct their integration into the bacterial chromosome and remain dormant as a prophage until stresses or specific conditions induce the lytic cycle. A bacterial host carrying a prophage is called lysogenic. A temperate phage can form lysogens and initiate either a lytic cycle or a lysogenic cycle, whereas a virulent phage is obligately lytic (Samson and Moineau, 2013).
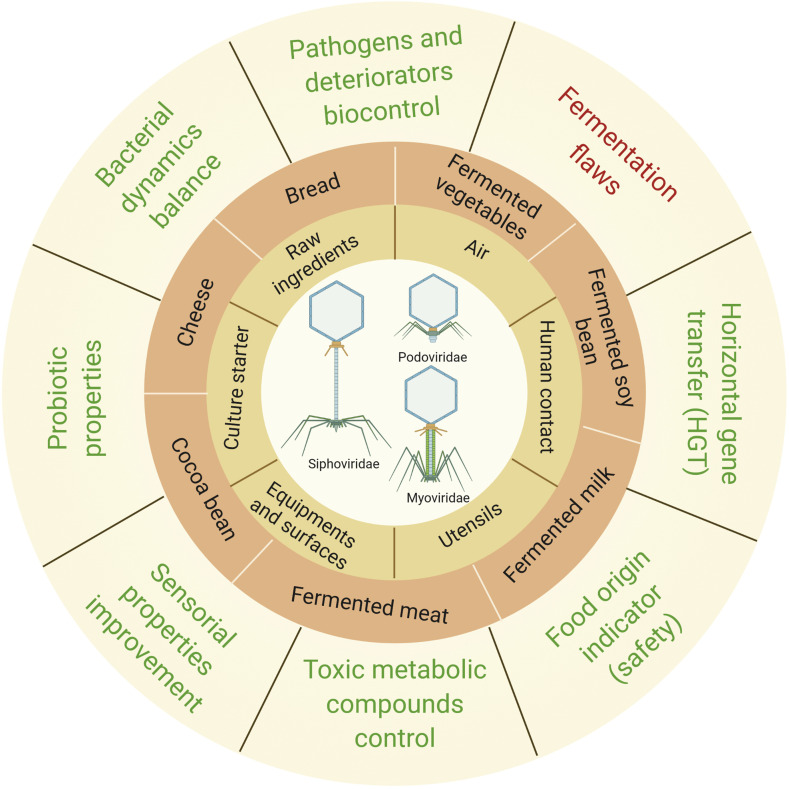
Phages have various contamination routes during the manufacture of fermented foods (Fig. 1 ). Eventually, LAB can naturally prevent phage invasion by evolving phage-resistance systems, or it can be genetically engineered to avoid culture devastation (e.g., origin-derived phage-encoded resistance, gene silencing, suicide system, and subunit poisoning) (Murphy et al., 2017). The use of physical-chemical methods on materials and industrial facilities also acts in phage contamination prevention. Examples include thermal treatment, use of biocidal agents (sodium hypochlorite and peracetic acid), UV photocatalysis, and high-pressure treatments (high hydrostatic pressure and high-pressure homogenization) (Murphy et al., 2017). However, the existence of resistant viruses allows contamination to still occur (Fig. 1). Phage-related issues are not restricted to foods, but also pharmaceutical, chemical, and pesticide industries (Pujato et al., 2019).
Fig. 1.
Main bacteriophages families and contagion routes associated with fermented foods (adapted from Samson and Moineau, 2013). Positive associations are written in green and negative in red on the outer part of the circle. This figure was created using BioRender (https://biorender.com/). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
A survey on bacteriophages and yeast virus's diversity in fermented foods is reported in Table 1 .They are mainly represented by LAB phages (Lactococcus, Lactobacillus, Leuconostoc, Enterococcus, Streptococcus, and Weissella), being Lactococcus, Lactobacillus, and Leuconostoc most commonly reported in fermented foods (Pereira et al., 2020). Bacteriophages infecting LAB are all members of the Caudovirales, an order known as the non-enveloped dsDNA tailed phages. Caudovirales use the tail section to bind to the receptor on the bacterial cell wall, and the genome passes down the tail into the bacteria cell. Once inside, the virus genome is replicated by overlapping bacterial DNA and replicating the complete viral genome. Three families within the Caudovirales order, i.e., Myoviridae, Siphoviridae, and Podoviridae, were reported in fermented foods (Fig. 1). Myoviridae are characterized by the presence of a long contractile tail, Siphoviridae has a long, non-contractile tail, and Podoviridae a short, non-contractile tail (Samson and Moineau, 2013). This classification, which is based on morphology instead of DNA sequences, was originated by Bradley in 1969 and has been extended to date.
Table 1.
Bacteriophages and yeast virus diversity in fermented food.
The first LAB phage report was published 85 years ago, a streptococci attack-phage in cheese (Whitehead and Hunter, 1947). Posteriorly, the presence of phage was widely reported during lactic fermentation (e.g., sauerkraut, yogurt, natto, and cucumber) (Barrangou et al., 2002; Brussow et al., 1994; Kiliç et al., 1996; Kleppen et al., 2012b; Lu et al., 2003b; McIntyre et al., 1991; Quiberoni et al., 2003; Saxelin et al., 1986; Umene and Shiraishi, 2013). Phage attack was frequently associated with fermentation failure until complete loss of the product batch due to unfeasibility of starter cultures (Quiberoni et al., 2003). Starter-destroying phages are not restricted to LAB. Other genera that compose starter cultures are also attacked by phages, such as Bacillus spp. in fermented soybean known as natto (Umene et al., 2009), Staphylococcus spp. in salami (Bruttin et al., 1992), and even on yeast (Saccharomyces cerevisiae) as called ‘yeast viruses’ in winemaking (Ramírez et al., 2015; Rodríguez-Cousiño et al., 2011). Differently from bacteriophages, S. cerevisiae viruses belong to the Totiviridae family (Ghabrivirales order) and infect not only yeasts but also protozoa, filamentous fungi, plants, invertebrates, and vertebrates (Rowley, 2017).
The presence of bacteriophages in microbial cultures is not itself sufficient for affecting fermentation processes. It depends on the microbial composition in the starter, being the more diverse the composition, the less chance of failure (Spus et al., 2015). This can be explained by the fact that bacteria have different levels of sensitivity to phages and, when an attack occurs, some bacteria are resistant and recover fermentation. Beyond that, contamination seems to be attributed to the consistency of the food matrix in which liquid matrices are more susceptible to allowing a rapid phage spread than solid or semi-solid. Bruttin et al. (1992) attributed the solid-state of meat in salami as the key factor to prevent attack by Staphylococcus carnosus bacteriophages, while liquid milk fermentation allows easiest phages propagation (Garneau and Moineau, 2011).
3. Human foodborne and zoonotic viruses
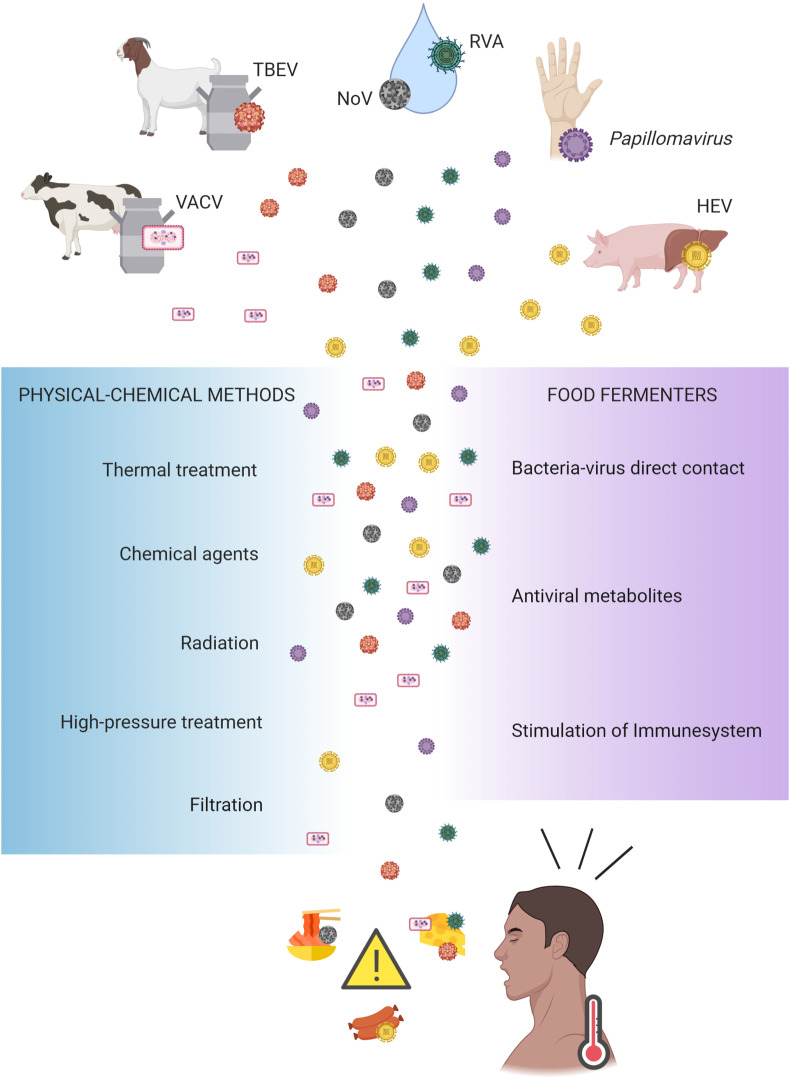
Food fermenters produce different end-metabolites having antiviral activity, including bacteriocins, hydrogen peroxide, ethanol, and lactic acid. However, some enteric viruses, including NoV, hepatitis A virus (HAV), hepatitis E virus (HEV), and Orthopoxvirus and Henipavirus genus, are allowed to remain viable in foods for periods from two days to four weeks (Hewitt and Greening, 2004), even in an environment with the lack of specific host cells to replicate. Contamination routes include (i) the raw material contaminated before food preparation, (ii) food preparation and processing, emphasizing the role of food handler, hygienic conditions, and the use of polluted materials, and (iii) food delivery and facilities routes where the food passes after it is finished (Fig. 3).
Fig. 3.
Main sources of pathogenic viruses and prevention measures associated with fermented foods. VACV (vaccinia virus); TBVE (tick-borne encephalitis virus); NoV (Norovirus); HEV (hepatitis E virus); RVA (rotavirus A). This figure was created using BioRender (https://biorender.com/).
Tick-borne encephalitis virus (TBEV) outbreaks were associated with cheese made with infected goat's milk in Europe (Brockmann et al., 2018; Holzmann et al., 2009; Markovinović et al., 2016). Contaminated milk with vaccinia virus (VACV) during cheese production in Brazil has also been reported (Rehfeld et al., 2017). VACV is considered a zoonosis affecting cows and humans. Artisanal cheese produced with not pasteurized milk increases the chance of infection. Rehfeld et al. (2017) found that VACV remains viable after 60 days of cheese ripening at 25 °C. Meat products are also potential targets for viral contamination, giving special attention to HEV due to its zoonotic potential (Colson et al., 2010). The target cells of the virus are hepatocytes; therefore, the greatest risk is the consumption of contaminated animals' liver. It was recently found that, even in pH 2, there were remaining infectious virus particles in fermented meat products (Wolff et al., 2020).
Fermented oyster and kimchi were associated with NoV produced with polluted water in South Korea (Park et al., 2015). NoV has high infectivity, having the ability to withstand a broad range of temperatures and high resistance to acidic conditions. These factors facilitate NoV transmission (Park et al., 2015). Bae et al. (2018) and Gagné et al. (2015) evaluated NoV survival in experimentally contaminated kimchi and sauerkraut, respectively. In extended periods of fermentation by 90 days, virus load has decreased; however, viable copies were still present at the end of the fermentation. Rotavirus A (RVA) also easily spread by polluted water or sewage. Recently, de Castro Carvalho et al. (2020) reported RVA in homemade Minas frescal cheese collected from local markets in the city of Mariana, Brazil. This area suffered an environmental disaster in 2015 when a rupture of an ore dam adversely affected water, soil, and air quality. Even today, Mariana and other nearby cities affected by this man-causes disaster have poor sanitation conditions and contaminated mudflow.
Colombo et al. (2018) characterized airborne VP isolated from two dairies in Italy. The authors raised some considerations about the safety of cheese ripening cellars, as human viruses belonging to the Papillomaviridae family showed a high abundance (17%) and identity higher than 90% with human papillomavirus. Although it is normally transmitted through direct skin-to-skin contact, the authors showed that even a slight human presence is sufficient for contamination, due to papillomavirus's high stability outside the host. Proper industrial environmental and hand workers sanitization are essential to avoid plant contamination. Although fermented foods pass through manufacturing processes that can inactivate viruses, many of them still carry infectious particles in the final product. The rapid identification of pathogens and monitoring sources of contamination are extremely important, especially for traditional fermented foods and minimally processed products (MASKE et al., 2020).
RV and HEV are members of the enteric viruses group that have zoonotic patterns (Leblanc et al., 2019). RV is distributed into ten groups or species (A to J) (International Committee on Taxonomy of Viruses, 2020) and are the most frequent species in human outbreaks. RV has segmented double-stranded RNA, which favors reassortments among human and zoonotic species. RV enteritis is frequently reported in calves and piglets in livestock (Martella et al., 2010). Contamination of food by RV occurs by primary source (water or infected animals) or along the food chain by food handlers. The WHO has recently estimated that 20 HEV infection occur annually worldwide (World Health Organization WHO, 2020.). HEV is a single-stranded RNA virus with four genotypes causing diseases in humans. Genotypes 1 and 2 infect only humans, mainly in undeveloped countries by contaminated water. Genotypes 3 and 4 have zoonotic patterns (infecting humans and other animals, mainly domestic pigs), and are associated with disease in industrialized countries after consumption of raw or undercooked meat from viremic animals. Genotype 3 is responsible for the majority of zoonotic episodes. Normally, HEV causes self-limited disease, but in chronic liver disease patients and pregnant women may occur fulminant hepatic failure. HEV has been detected in meat, liver, kidney, and heart, principally from domestic pigs and wild boars (Doceul et al., 2016).
Severe acute respiratory syndrome Coronavirus 2 (SARS-CoV-2) is the causative agent of the ongoing coronavirus disease pandemic, named Coronavirus Disease 2019 (COVID-19) by WHO (Sohrabi et al., 2020). The SARS-CoV-2 is primarily transmitted by human-to-human transference via droplets (Lai et al., 2020). Other transmission routes include direct contact with an infected person and indirect contact through hand-mediated viral transfer from contaminated fomites to the nose, eyes, and mouth (La Rosa et al., 2020). Although there is no evidence that SARS-CoV-2 can be transmitted by food and water ingestion, the lack of scientific evidence leads to public health concerns mainly because the virus can remain viable for days in favorable atmospheric conditions (Singhal, 2020).
Animal-to-human SARS-CoV transmission emerged through ingestion of Chinese ferret badgers, raccoon dogs, and Himalayan palm civets sold as food (Goli, 2020), representing the primary route of transmission. The most relevant animal reservoir of human MERS-CoV, for example, are dromedary camels that caused human–human infections as occurred in Saudi Arabia in 2012 (Park et al., 2018). Reusken et al. (2014) hypothesized a foodborne transmission through consumption of raw camel milk or rawmeat, as antibodies of MERS-CoV were detected in serum and milk of dromedary camel. It was demonstrated that MERS-CoV experimentally introduced in camel milk can survive for up to 72 h at 4 °C and 22 °C (van Doremalen et al., 2014). Human Coronavirus (HCoV) was also experimentally recovered from lettuce after 4 days at 4 °C with titer 1.2 × 106 (Yépiz-Gómez et al., 2013). The threat of secondary HCoV transmission through water used for food production is also hypothesized via aerosolization/fecal-oral route (Singhal, 2020).
Usually, thermal treatment at 60 °C for 30 min is sufficient to reduce SARS-CoV in free cell matrices (Goli, 2020). CoVs are also sensitiveto basic and acidic pHs (Rabenau et al., 2005), but seem to be stable at 4 °C. It was demonstrated that viral infectious level declines faster at ~24 °C than at 4 °C (La Rosa et al., 2020). In addition, CoVs seem to be susceptible to chemical agents (e.g., salt and nitrates) and physical treatments (Thippareddi et al., 2020). Fermented foods are non-thermally or chemically treated and constitute a potential route of virus transmission. On the other hand, in pH around 5, SARS-CoV nucleocapsid starts to unfold and is denatured at a pH 2.7, suggesting the sensibility of SARS-CoV to pH changes (Wang et al., 2004). Innumerous fermented foods reach low pH levels and, consequently, can play a fundamental role in the elimination of the virus. All of these questions are hypothetical and should be the subject to study for verification.
After production, contamination still can occur at processing and during handling and delivery of food products by infected personnel via respiratory droplets, aerosols, or from contaminated equipment (Thippareddi et al., 2020). Recent SARS-CoV-2 outbreaks in food processing and food stores are highlighting the potential for employees being a source of contamination (Rizou et al., 2020). Considering the persistence of CoV on fresh food at 4 °C, the survival of SARS-CoV-2 on food packages was evaluated (Malenovská, 2020). The loss of infectivity of the virus on plastic surfaces was, on average, 0.93 log10 (i.e. 83%) per day of storage at 4 °C. However, when using wipes saturated with a combination of disinfectant agents (hydrogen peroxide and didecyl-dimethyl-ammonium chloride), it decreased the viral titer still more efficiently, by 3.8 log10 (99.98%). The Centre for Disease Control and Prevention (CDC) from United States of America states that COVID-19 infection from handling contaminated food packages have low-risk, however, it is recommended cleaning and disinfection (Seymour et al., 2020).
4. Current virus detection methods
The recovery of viruses from fermented foods consists of their separation from other microorganisms and suspended solids (Barrangou et al., 2002). In liquid samples, bacteriophages separation process can be performed by ultracentrifugation, followed by a filtering step to eliminate contaminants. Solid samples must be previously added in a buffer or sterile culture medium and stirred to elute the phages from the food matrix (Foschino et al., 2005). If the sample contains a low phage titer, concentration by ultracentrifugation should be performed previously. For phage propagation, the filtrate is inoculated into a culture medium containing the host cell (Bandara et al., 2012, Lu et al., 2003a). After overnight incubation, the culture is ultracentrifuged and the supernatant is filtered to remove the remaining bacteria cells. Then, the presence of phages and host range can be stablished by spot testing, plaque testing, or culture lysis. For classification, the most used method is the direct observation of morphology through Transmission Electron Microscopy (TEM), which despite being old, is still widely used. To a lesser extent, the PCR-based methods (e.g. RAPD, Multiplex PCR, MLST, and qPCR) and DNA analysis (DNA sequencing and RFLP) are eventually performed (Samson and Moineau, 2013).
Pathogenic viruses, differently from bacteriophages, are rarely propagated in cell-culture assays, and its presence is based mainly on genome copies detection. Due to significance of viral foodborne diseases, validated methods for viral analysis in food are increasing over the years, such as international standard method ISO 15216–1:2017 for NoV and HAV detection. Surrogates' viruses are frequently used to on pathogenic viruses’ trials. Surrogate viruses share molecular characteristics with pathogenic viruses, such as size and chemical composition (Richards, 2012). Murine Norovirus-1, Feline Calicivirus, and some human virus strains, adapted to cell culture propagation (HAV strain HM-175, Human Adenovirus, RV, Enterovirus, and others), are the most used in cell culture-based assay or molecular techniques coupled to cell culture (Plaque Assay, Tissue Culture Infectious Dose (TCID) and Integrated Cell Culture Quantitative PCR (ICC-et-RT-qPCR)) (Cromeans et al., 2008). However, these trials are still not available in the routine of food analysis laboratories and the use of surrogate viruses is not fully elucidated to predict pathogenic viruses (Richards, 2012).
Pathogenic viruses were found in fermented foods samples through PCR techniques [Quantitative (qPCR) and reverse transcription (RT-PCR), or the combination of both (RT-qPCR)] (Table 2 ). They are prominent in sensitivity and specificity, being more employed than antigen detection and serology. In the PCR assay, DNA/RNA isolated from the target virus are amplified with specific primers. In the supplemental material, we compiled an extensive list of specific primers designed to detect viral pathogens (Table S1). However, PCR does not show virus viability and, to generate selectively amplifiable primers, the target sequence is required in advance (Sekse et al., 2017). This limitation is aggravated by the fact that novel viruses can arise and the symptoms of viral diseases are very similar, making it difficult to know which virus is involved. To overcome this, Lee et al. (2018) proposed a PCR multiplex reverse transcription using six primer sets to simultaneously detect and quantify NoV, HAV, RV, and astrovirus in food samples (lettuce, oysters, and vegetable products). Considering the aforementioned barriers, fast and more generalized techniques for identifying pathogenic viruses are required.
Table 2.
Human pathogenic viral outbreaks associated to fermented food ingestion.
| Virus | Classification | Fermented food | Location | Identification method | Disorder | Reference |
|---|---|---|---|---|---|---|
| TBEV | Flavivirus | Goat cheese | Germany | RT-qPCR | Meningitis, meningoencephalitis, or meningoencephalomyelitis | Brockmann et al. (2018) |
| TBEV | Flavivirus | Goat cheese | Austria | Sample no longer available | Meningitis, meningoencephalitis, or meningoencephalomyelitis | Holzmann et al. (2009) |
| TBEV | Flavivirus | Goat cheese | Croatia | Sample no longer available, however goats tested positive | Meningitis, meningoencephalitis, or meningoencephalomyelitis | Markovinović et al. (2016) |
| VACV | Orthopoxvirus | Minas cheese | Brazil | qPCR | Skin lesions | de Oliveira et al. (2018) |
| HEV | Orthohepevirus | Figatelli | Southeastern | RT-PCR | Acute and chronic hepatitis, death | Colson et al. (2010) |
| HEV | Orthohepevirus | Raw pork sausage | Netherlands | RT-qPCR | Acute and chronic hepatitis | Boxman et al. (2020) |
| NoV | Calciviridae | Fermented oyster | South Korea | RT-PCR | Acute gastroenteritis | Cho et al. (2016) |
| NoV | Calciviridae | Kimchi | South Korea | RT-PCR | Acute gastroenteritis | Park et al. (2015) |
| RVA | Rotavirus | Minas frescal cheese | Brazil | qPCR | Acute gastroenteritis | de Castro Carvalho et al. (2020) |
| NiV | Henipavirus | Tari (fermented palm sap liquor) | Bangladesh | Sample no longer available | Encephalitis and death | Hossain et al. (2016) |
Tick-borne encephalitis virus: TBEV; Vaccinia virus: VACV; Hepatitis E Virus: HEV; Norovirus: NoV; Rotavirus A: RVA; Bat Nipah virus: NiV.
5. Food virome
Great effort over the last years has been done to prevent virus contamination during fermentation processes (Park et al., 2011). However, surprisingly, viral presence started to be more noticed with the advancement of NGS technologies, and it was reframed in the fermented food niche. This transition was decelerated because food microbiome investigations are mainly focused on the characterization of bacterial and fungal communities, being virus largely neglected (Ledormand et al., 2020).
The challenges of using NGS to characterize viral content are numerous, including the absence of universal marker genes (Eric Wommack et al., 2012), the low concentration of viral DNA for preparation of genomic libraries (Garmaeva et al., 2019), the contamination of the sample with bacterial and fungal DNA (Kim and Bae, 2011), and the scarcity of databases for virus sequences as they remain largely unknown (Bikel et al., 2015; Garmaeva et al., 2019). Nevertheless, some food microbiome studies have detected, but not classified viruses in fermented food systems (Liu et al., 2020; Lyu et al., 2013). To a lesser extent, some studies have achieved a classification, even though it was focused on bacterial and fungal communities (Agyirifo et al., 2019; Illeghems et al., 2012; Kumar et al., 2019).
Kumar et al. (2019) analyzed kinema (traditional fermented soybean from Himalaya) samples using Illumina NGS technology, and found less than 1% of total reads belonging to bacteriophages, with Siphoviridae family being dominant. Illeghems et al. (2012) and Agyirifo et al. (2019) analyzed cocoa beans by Illumina and reported 0,25 and 1%, respectively, of the total metagenomics read sequences being bacteriophages. Both studies reported Lactobacillus phages of Siphoviridae family as the dominant, and the presence of phages infecting Enterobacter, Klebsiella, Pseudomonas, Bacillus, and Staphylococcus. Interestingly, Agyirifo et al. (2019) attributed Lactobacillus bacteriophages to positively influence the aroma formation during cocoa beans fermentation. The authors mentioned that bacterial cell lysis caused by phages releases intracellular enzymes in the food matrix, degrading the substrate, and stimulating aroma production.
Currently, there are two approaches for metagenomic studies; i) metagenomic amplification of the target gene, where only a specific region is sequenced, performed to characterize mainly bacterial and fungal communities, and ii) shotgun metagenomics which consists of the sequencing of random fragments of all microbial DNA present in a given sample, frequently used in viral communities’ studies (Sharpton, 2014). In any case, due to these advances, the terms “ome” and “omics” have been attributed to viruses.
The “virome” and the (meta) viromics refer to all viruses present in each sample and the study of their genomes, respectively (Garmaeva et al., 2019). However, since the vast majority of existing phages belong to the Caudovirales order, current studies on viral communities in fermented foods are strictly focused on the presence of dsDNA phages, unfortunately excluding the possibility of identifying other viruses and RNA-containing phages (Mokili et al., 2012). Most pathogenic viruses associated with fermented foods, due to their RNA genetic material content, cannot be identified by next-generation approaches. However, when associated with PCR, seems to be a promising tool for food safety monitoring. Metabarcoding strategy after RT-qPCR sample treatment has successfully been used, despite being limited to closely related viruses or viral families (Desdouits et al., 2020). If food is contaminated by innumerous viral strains belonging to different genotypes, PCR products are synthesized for each strain, and bioinformatics analyses each reading and classifies genotypes. Imamura et al. (2017) used PCR primers targeting the N-terminal area of the VP1 protein and analyzed the diversity of NoV genogroups I and II in naturally contaminated oysters with Illumina platform. Oshiki et al. (2018) combined a microfluidic tool allowing the use of multiplex PCR and Miseq sequencing to the detection of 11 different human RNA viruses from human feces, sewage, and oysters artificially contaminated. Even so, these techniques require more studies to attend to fermented foods, whereas they have peculiar characteristics affecting viral content access.
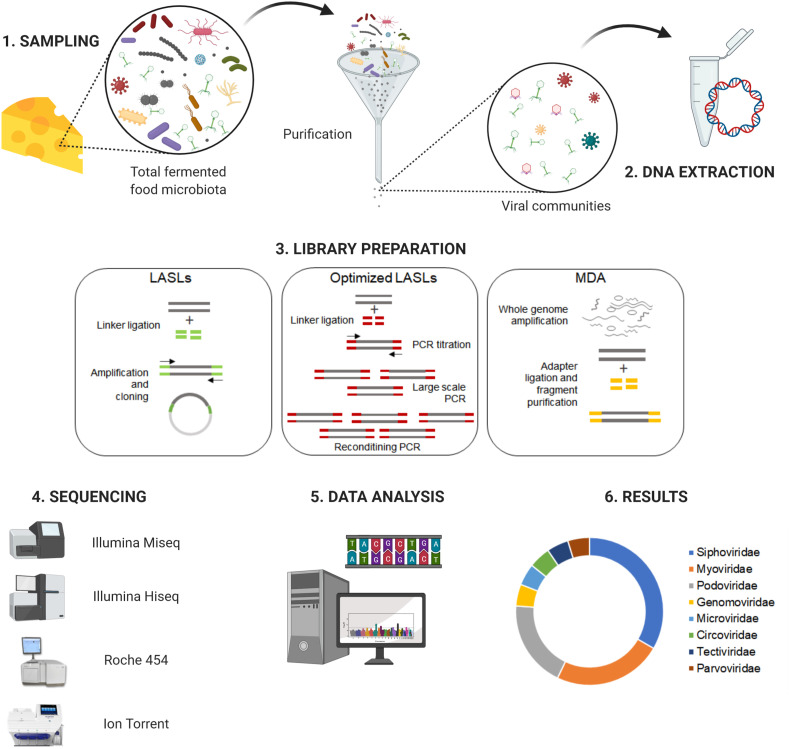
Alternatively to the term “virome”, studies of the viral community are called frequently by “phageome” (Ledormand et al., 2020). Fig. 2 illustrates the NGS standard workflow for viral community analysis in fermented foods, including (1) sampling, (2) DNA extraction, (3) library preparation, (4) sequencing, (5) data analysis, and (6) results.
Fig. 2.
Schematic workflow for analyzing virus communities in fermented foods by next-generation sequencing. LASLs (linker amplification shotgun libraries) and MDA (multiple displacement amplification). This figure was created using BioRender (https://biorender.com/).
5.1. Sampling and DNA extraction
All virome study starts with the step of sample purification. The purification method must be able to satisfactorily represent the original virus population (Hayes et al., 2017). Usually, the purification of the viral particles (VPs) has divided into three steps: i) VPs recovery, ii) VPs purification and concentration, and iii) an optional second purification via cesium chloride gradient (Park et al., 2011). Although there is no standardized sampling protocol for all fermented foods, recently, Dugat-Bony et al. (2020) optimized a method for extraction and purification of bacteriophages from feces to perform, for the first time, viral metagenomic analysis of cheese surface. They used a chloroform treatment and a filtration step to extract viral DNA prior to the Illumina Miseq sequencing.
Virome studies of other fermented foods (e.g., kimchi, sauerkraut, and fermented shrimp) have used different strategies, mainly in the VPs concentration step. While Park et al. (2011) used ultracentrifuged at 100,000×g for 4 h at 4°C, Jung et al. (2018) opted to precipitate the VPs with polyethylene glycol 8000 e NaCl 1 M. In addition, due to the high concentration of cellular microorganisms in fermented food, samples are prone to contamination by bacteria. An alternative is the treatment of concentrated VPs with lysozyme and chloroform, followed by incubation with DNase and RNase to remove the remaining genetic material (Dugat-Bony et al., 2020; Jung et al., 2018; Park et al., 2011). Regardless of which purification technique is employed, a step of eliminating bacteria and fungi also means eliminating prophages that are frequently inserted in the genome of these hosts, generating a bias for the technique, as it considers only the free phage (Sausset et al., 2020).
Nevertheless, subsequently, the extraction of DNA from the VPs is performed with commercial kits. It is important to note that the choice of extraction kit influences the composition of the microbial community produced by NGS and may generate inaccurate results. Therefore, the extraction method should be chosen according to the objective of the researcher. Also, it is recommended that after viral DNA extraction a small aliquot should be amplified by PCR using 16S/18S rRNA primers to ensure the absence of bacteria and fungi DNA contamination (Hurwitz et al., 2016).
5.2. Library preparation and sequencing
Most of the viromes that have been performed so far have used linker amplification shotgun libraries (LASLs) or whole (meta) genome amplification (WGA) methods [e.g., multiple displacement amplification (MDA)] (Willner and Hugenholtz, 2013). LASLs consist of the random fragmentation of genomic DNA and the binding of known linkers in these fragments that can be used for PCR amplification. At the time this method was developed, the fragments were cloned in plasmid vectors and sequenced by the Sanger method to generate the viral metagenomes. However, this strategy had the bias of large-scale cloning and sequencing (Breitbart et al., 2002).
NGS technologies eliminated the need for cloning vectors and significantly increased the sequencing capacity; however, the low viral DNA concentration remains a challenge since some NGS protocols require nucleic acid micrograms. Duhaime et al. (2012) optimized the LASLs technique with the addition of a titration step, significantly reducing the number of cycles and the DNA concentration required to build the library (only 1 pg), enabling large scale PCR (Willner and Hugenholtz, 2013). The optimized linker amplification (LA) technique has been adapted for 454 sequencing, but also can be used to build genomic libraries on other sequencing platforms, such as Illumina and Ion Torrent (Duhaime et al., 2012).
The MDA method uses the high efficiency of polymerase ϕ29 which synthesizes >70,000 nucleotides per cycle from small concentrations of DNA, allowing the amplification of complete viral genomes through adapter ligations followed by purification step (Bikel et al., 2015). LA and MDA are powerful tools for studying virome, however, MDA is prone to unevenly amplify linear genome fragments and might generate biases into the representation of ssDNA circular viruses (Kim and Bae, 2011), while A-LA provides a more reliable representation of ssDNA and dsDNA viruses (Roux et al., 2016).
After the construction of the genomic libraries, the sequencing is performed using mainly the Illumina, Roche 454, and Ion Torrent platforms (Hayes et al., 2017). The quality control steps of the generated sequences must be performed as reviewed by Hurwitz et al. (2016). Succinctly, quality control consists of ensuring optimal sequencing coverage for each sample and removing rare reads that may occur from sequencing errors or contamination of the sample, leading to overestimation of the virome diversity (Hurwitz et al., 2016).
5.3. Data analysis
The central challenge in bioinformatics analysis of viromes is the absence of universal genes, which makes diversity estimation difficult. Currently, taxonomic classification is often performed by aligning the generated sequences against a “generalist” database using the BLAST (Basic Local Alignment Search Tool) or BLAST-based programs, such as MetaPhyler (Liu et al., 2011), CARMA (Gerlach et al., 2009), and MG-RAST (MetaGenomic-Rapid Annotation using Subsystem Technology) (Glass et al., 2010). However, a large part (about 60–99%) of the sequences produced in viromes has no homology with viral sequences available in the databases.
To solve this problem, database focused on the taxonomic classification of viruses [ACLAME (Leplae et al., 2009) and Phage SEED (Overbeek et al., 2005)] and specific data analysis programs [VIROME (Viral Informatics Resource for Metagenome Exploration) (Eric Wommack et al., 2012), VMGAP (Viral MetaGenome Annotation Pipeline) (Lorenzi et al., 2011), and Metavir (Roux et al., 2014)], were developed. Usually, these pipelines use an ORF (open reading frame) localization algorithm and perform a comparison with a protein database (Hayes et al., 2017) creating a functional and taxonomic profile of the viral community (Bikel et al., 2015). In addition, an independent method of similarity PHACCS (PHAge Communities from Contig Spectrum) software was developed to better understand the structure of the viral community. It provides the estimation of diversity and uniformity revealing the most abundant viruses in a viral metagenome. This analysis is based on the principle that the most abundant virotypes in a VPs sample will more likely be assembled into large contigs (Reyes et al., 2012).
5.4. Results
The diversity revealed by virome studies in fermented foods agrees with previous findings described by culture-dependent methods. Most of the sequences belong to Caudovirales order of the families Siphoviridae, Myoviridae, and Podoviridae (Agyirifo et al., 2019; Cheng et al., 2018; Del Rio et al., 2019; Illeghems et al., 2012; Jung et al., 2018; Kot et al., 2014; Kumar et al., 2019; Park et al., 2011). Similar results are seen on the human gut virome (Garmaeva et al., 2019).
Park et al. (2011) applied pyrosequencing on fermented sauerkraut, kimchi, and shrimp samples and verified that Siphoviridae dominated sauerkraut (60.07%) and fermented shrimp (53.55%), differently from kimchi, which Podoviridae prevailed (52.82%). They also first identified Phyconaviridae in a fermented food, an attacker of harmful eukaryotic algae responsible for algal blooms, demonstrating that metagenomics studies contribute to the discovery of biocontrol agents for different fields other than food. Jung et al. (2018) described the dominance of ssDNA viruses in Korean kimchi, including Circoviridae (hosts: birds and pigs), Genomoviridae (plants and fungi) and Microviridae (bacteria), not previously described. They applied Illumina HiSeq to differentiate Chinese and Korean kimchi origins and found that viral clusters were more clearly distinguished than bacterial clusters by beta diversity analysis, making viruses more strongly associated with the geographic origins of fermented foods than the bacterial ones. The origin of traditional food reveals its quality and safety, being of great importance to consumers.
6. Beneficial viruses: the other side of the coin
Phages directly control bacterial dynamics, promoting their balance in fermentation. In the ecological study of sauerkraut fermentation, Lu et al. (2003b) observed that the succession of bacteria was associated with the content of the respective phages found in different stages of fermentation. At the beginning of the process, Leuconostoc spp. and Weissella spp. prevailed and the phages that infected these species were isolated. After seven days, Lactobacillus was the main genus, and phages infecting them were observed. Recently, Kumar et al. (2019) also noticed that phages seem to determine the abundance of bacterial communities during the fermentation of Kinema, acting in the biocontrol of Bacillus, Staphylococcus, Enterococcus, Lactococcus, and Streptococcus.
These results suggest that phage dynamics during fermentation is crucial to guarantee a good succession of bacteria. In addition, phages can move between different environments, elevating horizontal gene transfer (HGT) and, therefore, forcing bacteria to evolve (Breitbart and Rohwer, 2005). Understand the process of phage-bacteria evolution assists the selection of good strategies to control harmful phages and maintain beneficial ones, improving fermentation performance.
Phages can act as biocontrol agents combating pathogenic and deteriorating bacteria, as well as toxic metabolic components in fermented foods (García et al., 2008). Bandara et al. (2012) found two phages belonging to Myoviridae family were capable of eradicating Bacillus cereus quickly when supplemented with divalent cations (Ca2 +, Mg2 + or Mn2 +) in cheonggukjang, a product of fermented soybean mass. Philippe et al. (2018) found a phage that infects Gluconobacter cerinus, a spoilage acetic acid bacterium in the wine making process. G. cerinus produces ethyl alcohol and transform it into acetic acid, representing hazardous to the final product. Additionally, phages can also combat toxic metabolic components released by LAB in fermented foods (Del Rio et al., 2019; Ladero et al., 2016). Recently, Del Rio et al. (2019) proved the efficiency of an Enterococcus faecalis bacteriophage of Myoviridae family in reducing biological amines (BA) in an experimental model of cheese. Phage presence reduced BA and putrescine without devastation the starter culture due to its specificity for E. faecalis.
The discovery of other perspectives for phages that infect LAB was also enabled. Phages that attack Streptococcus thermophilus were associated for decades as the cause of defects and flaws in the yogurt fermentation process (Bendadis et al., 1990; Brussow et al., 1994; Ishlimova et al., 2012; Ma et al., 2014; Quiberoni et al., 2003). However, recently, Pacini and Ruggiero (2019) attributed a probiotic potential to fermented milk and colostrum after analyzing the genomes of innumerous Streptococcus and Lactococcus's phages contained in the product using Axiom Microbiome Array. When ingested with food, phages can influence the host in three ways: i) modulation of the gastrointestinal microbiota, as they can act against pathogens and, by promoting horizontal transfer of genetic material, it operates in the improvement and evolution of bacterial community diversity; ii) intestinal mucosa cell interaction, indirectly triggering immune system response; and iii) immune system components interaction, directly driving immune response, as they can overcome anatomical and physiological barriers, being found in compartments of the human body earlier considered sterile (Sausset et al., 2020). In this way, Lactococcus and Streptococcus phages can possess antimicrobial, antitumor, and antiviral effects on the host. In addition, phages may interact with the immune system, opening possibility to immunotherapies to treat diseases such as cancer and autism. Phages presence can enhance notable benefits, which are still untapped.
7. Final considerations
The worldwide emergence of COVID-19 resulted in abrupt awareness of the presence of viruses in all sectors of the economy. Cases of NoVs, RVs, and HV have been reported in association with cheeses, sausages, fermented vegetables, and fermented cereals. However, there is no evidence of COVID-19 transmission through fermented foods. The possibility of food transmission needs to be investigated and clarified since no food virome study has been conducted since the first case identified in China, in December 2019. Studies have reported that Coronaviruses can remain infectious in waters and are highly stable at 4 °C, the main raw material, and storage temperature, respectively, of fermented foods. While respiratory droplets are the main way the virus spreads, transmission via fermented foods is considered negligible. However, transmission appears to be possible if the virus is transferred from hands to food and the food itself to the mucous membranes of the mouth, throat, or eyes.
A recent study showed that physical contact and shared food during a conference in Singapore resulted in a cluster of COVID-19 patients (Pung et al., 2020). The major risk enhancing factors of fermented foods is the use of contaminated raw materials, the conduction of poorly controlled natural fermentation, and the lack of pasteurization. Thus, to minimize the risk of virus contagion, good hygiene practices and cleaning of the fermentation room (taps, door handles, fermentation vessels, and utensils) should be followed by hand washing or using hand sanitizer.
On the other side of the coin, emerging evidence of bacteriophages diversity in fermented foods by NGS has revealed an ongoing paradigm in understanding their role in this ecosystem. The positive influence of phage is ample, ranging from sensorial improvement of the fermentation process to probiotic potential by cell interaction. To overcome unexplored fermented food viromes ecosystems, current challenges, such as the expansion of databases for non-cultivable viruses, the optimization of isolation protocols, and new bioinformatics tools, demands to be faced. Establishing viromes of innumerous fermented foods, studying the long-term evolution of virus-bacterial interactions, analyzing the influence of external factors, and elucidating the interaction of viruses with consumer's gastrointestinal tract cells will be essential in keeping viruses as allies.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This work was financed by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) and the Conselho Nacional de Desenvolvimento Científico e Tecnológico do Brasil (CNPq).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fm.2021.103794.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- Agyirifo D.S., Wamalwa M., Otwe E.P., Galyuon I., Runo S., Takrama J., Ngeranwa J. Metagenomics analysis of cocoa bean fermentation microbiome identifying species diversity and putative functional capabilities. Heliyon. 2019;5 doi: 10.1016/j.heliyon.2019.e02170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali Y., Kot W., Atamer Z., Hinrichs J., Vogensen F.K., Heller K.J., Neve H. Classification of lytic bacteriophages attacking dairy leuconostoc starter strains. Appl. Environ. Microbiol. 2013;79:3628–3636. doi: 10.1128/AEM.00076-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamer Z., Ali Y., Neve H., Heller K.J., Hinrichs J. Thermal resistance of bacteriophages attacking flavour-producing dairy Leuconostoc starter cultures. Int. Dairy J. 2011;21:327–334. doi: 10.1016/j.idairyj.2010.11.005. [DOI] [Google Scholar]
- Auad L., De Ruiz Holgado A.A.P., Forsman P., Alatossava T., Raya R.R. Isolation and characterization of a new Lactobacillus delbrueckii ssp. bulgaricus temperate bacteriophage. J. Dairy Sci. 1997;80:2706–2712. doi: 10.3168/jds.s0022-0302(97)76231-3. [DOI] [Google Scholar]
- Bae G., Kim J., Kim H., Seok J.H., Lee D.B., Kim K.H., Chung M.S. Inactivation of norovirus surrogates by kimchi fermentation in the presence of black raspberry. Food Contr. 2018;91:390–396. doi: 10.1016/j.foodcont.2018.04.025. [DOI] [Google Scholar]
- Bandara N., Jo J., Ryu S., Kim K.P. Bacteriophages BCP1-1 and BCP8-2 require divalent cations for efficient control of Bacillus cereus in fermented foods. Food Microbiol. 2012;31:9–16. doi: 10.1016/j.fm.2012.02.003. [DOI] [PubMed] [Google Scholar]
- Barrangou R., Yoon S.S., Breidt F., Fleming H.P., Klaenhammer T.R. Characterization of six Leuconostoc fallax bacteriophages isolated from an industrial sauerkraut fermentation. Appl. Environ. Microbiol. 2002;68:5452–5458. doi: 10.1128/AEM.68.11.5452-5458.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendadis L., Faelen M., Slos P., Fazel A., Mercenier A. Characterization and comparison of virulent bacteriophages of Streptococcus thermophilus isolated from yogurt. Biochimie. 1990;72:855–862. doi: 10.1016/0300-9084(90)90002-X. [DOI] [PubMed] [Google Scholar]
- Bikel S., Valdez-Lara A., Cornejo-Granados F., Rico K., Canizales-Quinteros S., Soberón X., Del Pozo-Yauner L., Ochoa-Leyva A. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: towards a systems-level understanding of human microbiome. Comput. Struct. Biotechnol. J. 2015;13:390–401. doi: 10.1016/j.csbj.2015.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxman I.L.A., Jansen C.C.C., Zwartkruis-Nahuis A.J.T., Hägele G., Sosef N.P., Dirks R.A.M. Detection and quantification of hepatitis E virus RNA in ready to eat raw pork sausages in The Netherlands. Int. J. Food Microbiol. 2020;333:108791. doi: 10.1016/j.ijfoodmicro.2020.108791. [DOI] [PubMed] [Google Scholar]
- Breitbart M., Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Breitbart M., Salamon P., Andresen B., Mahaffy J.M., Segall A.M., Mead D., Azam F., Rohwer F. Genomic analysis of uncultured marine viral communities. Proc. Natl. Acad. Sci. U. S. A. 2002;99:14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann S.O., Oehme R., Buckenmaier T., Beer M., Jeffery-Smith A., Spannenkrebs M., Haag-Milz S., Wagner-Wiening C., Schlegel C., Fritz J., Zange S., Bestehorn M., Lindau A., Hoffmann D., Tiberi S., Mackenstedt U., Dobler G. A cluster of two human cases of tick-borne encephalitis (TBE) transmitted by unpasteurised goat milk and cheese in Germany, May 2016. Euro Surveill. 2018;23 doi: 10.2807/1560-7917.ES.2018.23.15.17-00336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brussow H., Fremont M., Bruttin A., Sidoti J., Constable A., Fryder V. Detection and classification of Streptococcus thermophilus bacteriophages isolated from industrial milk fermentation. Appl. Environ. Microbiol. 1994;60:4537–4543. doi: 10.1128/aem.60.12.4537-4543.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruttin A., Marchesinl B., Moreton R.S. 1992. Factories in Germany and Italy. [Google Scholar]
- Cheng L., Marinelli L.J., Grosset N., Fitz-Gibbon S.T., Bowman C.A., Dang B.Q., Russell D.A., Jacobs-Sera D., Shi B., Pellegrini M., Miller J.F., Gautier M., Hatfull G.F., Modlin R.L. Complete genomic sequences of Propionibacterium freudenreichii phages from Swiss cheese reveal greater diversity than Cutibacterium (formerly Propionibacterium) acnes phages. BMC Microbiol. 2018;18:1–13. doi: 10.1186/s12866-018-1159-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho H.G., Lee S.G., Lee M.Y., Hur E.S., Lee J.S., Park P.H., Park Y.B., Yoon M.H., Paik S.Y. An outbreak of norovirus infection associated with fermented oyster consumption in South Korea, 2013. Epidemiol. Infect. 2016;144:2759–2764. doi: 10.1017/S0950268816000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo S., Arioli S., Gargari G., Neri E., Della Scala G., Mora D. Characterization of airborne viromes in cheese production plants. J. Appl. Microbiol. 2018;125:1444–1454. doi: 10.1111/jam.14046. [DOI] [PubMed] [Google Scholar]
- Colson P., Borentain P., Queyriaux B., Kaba M., Moal V., Gallian P., Heyries L., Raoult D., Gerolami R. Pig liver sausage as a source of hepatitis E virus transmission to humans. J. Infect. Dis. 2010;202:825–834. doi: 10.1086/655898. [DOI] [PubMed] [Google Scholar]
- Cromeans T.L., Lu X., Erdman D.D., Humphrey C.D., Hill V.R. Development of plaque assays for adenoviruses 40 and 41. J. Virol. Methods. 2008;151:140–145. doi: 10.1016/j.jviromet.2008.03.007. [DOI] [PubMed] [Google Scholar]
- de Castro Carvalho S.V., Rogovski P., Cadamuro R.D., Viancelli A., Michelon W., dos Reis D.A., Santana das Chagas I.A., Assenço R., da Silva Lanna M.C., Treichel H., Fongaro G. Co-contamination of food products from family farms in an environmental disaster area in Southeast Brazil with pathogenic bacteria and enteric viruses. Arch. Virol. 2020;165:715–718. doi: 10.1007/s00705-019-04501-9. [DOI] [PubMed] [Google Scholar]
- de Melo Pereira G.V., de Carvalho Neto D.P., Maske B.L., De Dea Lindner J., Vale A.S., Favero G.R., Viesser J., de Carvalho J.C., Góes-Neto A., Soccol C.R. An updated review on bacterial community composition of traditional fermented milk products: what next-generation sequencing has revealed so far? Crit. Rev. Food Sci. Nutr. 2020:1–20. doi: 10.1080/10408398.2020.1848787. 0. [DOI] [PubMed] [Google Scholar]
- de Oliveira T.M.L., Guedes M.I.M.C., Rehfeld I.S., Matos A.C.D., Rivetti Júnior A.V., da Cunha A.F., Cerqueira M.M.O.P., Abrahão J.S., Lobato Z.I.P. Vaccinia virus detection in dairy products made with milk from experimentally infected cows. Transbound. Emerg. Dis. 2018;65:e40–e47. doi: 10.1111/tbed.12666. [DOI] [PubMed] [Google Scholar]
- Del Rio B., Sánchez-Llana E., Redruello B., Magadan A.H., Fernández M., Martin M.C., Ladero V., Alvarez M.A. Enterococcus faecalisbacteriophage 156 is an effective biotechnological tool for reducing the presence of tyramine and putrescine in an experimental cheese model. Front. Microbiol. 2019;10:1–10. doi: 10.3389/fmicb.2019.00566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desdouits M., Wacrenier C., Ollivier J., Schaeffer J., Le Guyader F.S. A targeted metagenomics approach to study the diversity of norovirus GII in shellfish implicated in outbreaks. Viruses. 2020;12:1–17. doi: 10.3390/v12090978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doceul V., Bagdassarian E., Demange A., Pavio N. Zoonotic hepatitis E virus: classification, animal reservoirs and transmission routes. Viruses. 2016;8:1–24. doi: 10.3390/v8100270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doria F., Napoli C., Costantini A., Berta G., Saiz J.C., Garcia-Moruno E. Development of a new method for detection and identification of Oenococcus oeni bacteriophages based on endolysin gene sequence and randomly amplified polymorphic DNA. Appl. Environ. Microbiol. 2013;79:4799–4805. doi: 10.1128/AEM.01307-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dugat-Bony E., Lossouarn J., De Paepe M., Sarthou A.S., Fedala Y., Petit M.A., Chaillou S. Viral metagenomic analysis of the cheese surface: a comparative study of rapid procedures for extracting viral particles. Food Microbiol. 2020;85:103278. doi: 10.1016/j.fm.2019.103278. [DOI] [PubMed] [Google Scholar]
- Duhaime M.B., Deng L., Poulos B.T., Sullivan M.B. Towards quantitative metagenomics of wild viruses and other ultra-low concentration DNA samples: a rigorous assessment and optimization of the linker amplification method. Environ. Microbiol. 2012;14:2526–2537. doi: 10.1111/j.1462-2920.2012.02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eric Wommack K., Bhavsar J., Polson S.W., Chen J., Dumas M., Srinivasiah S., Furman M., Jamindar S., Nasko D.J. VIROME: a standard operating procedure for analysis of viral metagenome sequences. Stand. Genomic Sci. 2012;6:427–439. doi: 10.4056/sigs.2945050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foschino R., Venturelli E., Picozzi C. Isolation and characterization of a virulent Lactobacillus sanfranciscensis bacteriophage and its impact on microbial population in sourdough. Curr. Microbiol. 2005;51:413–418. doi: 10.1007/s00284-005-0122-y. [DOI] [PubMed] [Google Scholar]
- Frantzen C.A., Holo H. Unprecedented diversity of lactococcal group 936 bacteriophages revealed by amplicon sequencing of the portal protein gene. 2019. Viruses 11. [DOI] [PMC free article] [PubMed]
- Gagné M.J., Barrette J., Savard T., Brassard J. Evaluation of survival of murine norovirus-1 during sauerkraut fermentation and storage under standard and low-sodium conditions. Food Microbiol. 2015;52:119–123. doi: 10.1016/j.fm.2015.07.009. [DOI] [PubMed] [Google Scholar]
- García P., Martínez B., Obeso J.M., Rodríguez A. Bacteriophages and their application in food safety. Lett. Appl. Microbiol. 2008;47:479–485. doi: 10.1111/j.1472-765X.2008.02458.x. [DOI] [PubMed] [Google Scholar]
- Garmaeva S., Sinha T., Kurilshikov A., Fu J., Wijmenga C., Zhernakova A. Studying the gut virome in the metagenomic era: challenges and perspectives. BMC Biol. 2019;17:1–14. doi: 10.1186/s12915-019-0704-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garneau J.E., Moineau S. Bacteriophages of lactic acid bacteria and their impact on milk fermentations. Microb. Cell Factories. 2011;10:1–10. doi: 10.1186/1475-2859-10-S1-S20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach W., Jünemann S., Tille F., Goesmann A., Stoye J. WebCARMA: a web application for the functional and taxonomic classification of unassembled metagenomic reads. BMC Bioinf. 2009;10:1–10. doi: 10.1186/1471-2105-10-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass E.M., Wilkening J., Wilke A., Antonopoulos D., Meyer F. Using the metagenomics RAST server (MG-RAST) for analyzing shotgun metagenomes. Cold Spring Harb. Protoc. 2010;5 doi: 10.1101/pdb.prot5368. [DOI] [PubMed] [Google Scholar]
- Goli M. Review of novel human β-coronavirus (2019-nCoV or SARS-CoV-2) from the food industry perspective—appropriate approaches to food production technology. Food Sci. Nutr. 2020;8:5228–5237. doi: 10.1002/fsn3.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greer G.G., Dilts B.D., Ackermann H.W. Characterization of a Leuconostoc gelidum bacteriophage from pork. Int. J. Food Microbiol. 2007;114:370–375. doi: 10.1016/j.ijfoodmicro.2006.09.021. [DOI] [PubMed] [Google Scholar]
- Hayes S., Mahony J., Nauta A., Van Sinderen D. Metagenomic approaches to assess bacteriophages in various environmental niches. Viruses. 2017;9:1–22. doi: 10.3390/v9060127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewitt J., Greening G.E. Survival and persistence of norovirus, hepatitis A virus, and feline calicivirus in marinated mussels. J. Food Protect. 2004;67:1743–1750. doi: 10.4315/0362-028X-67.8.1743. [DOI] [PubMed] [Google Scholar]
- Holzmann H., Aberle S.W., Stiasny K., Werner P., Mischak A., Zainer B., Netzer M., Koppi S., Bechter E., Heinz F.X. Tick-borne encephalitis from eating goat cheese in a mountain region of Austria. Emerg. Infect. Dis. 2009;15:1671–1673. doi: 10.3201/eid1510.090743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain M.J., Hasan M., Rahman M., Campbell S., Cannon D.L., Ströher U., Daszak P., Luby S.P., Gurley E.S. Nipah virus transmission from bats to humans associated with drinking traditional liquor made from date palm sap. Emerg. Infect. Dis. 2016;22:664–670. doi: 10.3201/eid2204.151747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurwitz B.L., U'Ren J.M., Youens-Clark K. Computational prospecting the great viral unknown. FEMS Microbiol. Lett. 2016;363:1–12. doi: 10.1093/femsle/fnw077. [DOI] [PubMed] [Google Scholar]
- Hyman P., Abedon S.T. Smaller fleas: viruses of microorganisms. Sci. Tech. Rep. 2012:1–23. doi: 10.6064/2012/734023. 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illeghems K., de Vuyst L., Papalexandratou Z., Weckx S. Phylogenetic analysis of a spontaneous cocoa bean fermentation metagenome reveals new insights into its bacterial and fungal community diversity. PloS One. 2012;7 doi: 10.1371/journal.pone.0038040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imamura S., Kanezashi H., Goshima T., Haruna M., Okada T., Inagaki N., Uema M., Noda M., Akimoto K. Next-generation sequencing analysis of the diversity of human noroviruses in Japanese oysters. Foodb. Pathog. Dis. 2017;14:465–471. doi: 10.1089/fpd.2017.2289. [DOI] [PubMed] [Google Scholar]
- International Committee on Taxonomy of Viruses . 2020. Rotavirus taxonomy. [WWW Document]. ICTV) [Google Scholar]
- Ishlimova D., Urshev Z., Alexandrov M., Doumanova L. DIVERSITY OF BACTERIOPHAGES INFECTING THE STREPTOCOCCUS THERMOPHILUS COMPONENT OF INDUSTRIAL YOGURT STARTERS IN. 2012. Diversity of bacteriophages infecting the Streptococcus thermophilus component of industrial yogurt starters in Bulgaria. [Google Scholar]
- Jung J.Y., Lee S.H., Kim J.M., Park M.S., Bae J.W., Hahn Y., Madsen E.L., Jeon C.O. Metagenomic analysis of kimchi, a Traditional Korean fermented food. Appl. Environ. Microbiol. 2011;77:2264–2274. doi: 10.1128/AEM.02157-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M.J., Kim M.S., Yun J.H., Lee J.Y., Kim P.S., Lee H.W., Ha J.H., Roh S.W., Bae J.W. Viral community predicts the geographical origin of fermented vegetable foods more precisely than bacterial community. Food Microbiol. 2018;76:319–327. doi: 10.1016/j.fm.2018.06.010. [DOI] [PubMed] [Google Scholar]
- Kiliç A.O., Pavlova S.I., Ma W.G.E., Tao L. Analysis of Lactobacillus phages and bacteriocins in American dairy products and characterization of a phage isolated from yogurt. Appl. Environ. Microbiol. 1996;62:2111–2116. doi: 10.1128/aem.62.6.2111-2116.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K.H., Bae J.W. Amplification methods bias metagenomic libraries of uncultured single-stranded and double-stranded DNA viruses. Appl. Environ. Microbiol. 2011;77:7663–7668. doi: 10.1128/AEM.00289-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppen H.P., Holo H., Jeon S.R., Nes I.F., Yoon S.S. Novel Podoviridae family bacteriophage infecting Weissella cibaria isolated from kimchi. Appl. Environ. Microbiol. 2012;78:7299–7308. doi: 10.1128/AEM.00031-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleppen H.P., Nes I.F., Holo H. Characterization of a Leuconostoc bacteriophage infecting flavor producers of cheese starter cultures. Appl. Environ. Microbiol. 2012;78:6769–6772. doi: 10.1128/AEM.00562-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kot W., Neve H., Heller K.J., Vogensen F.K. Bacteriophages of leuconostoc, oenococcus, and Weissella. Front. Microbiol. 2014;5:1–9. doi: 10.3389/fmicb.2014.00186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J., Sharma N., Kaushal G., Samurailatpam S., Sahoo D., Rai A.K., Singh S.P. Metagenomic insights into the taxonomic and functional features of kinema, a traditional fermented soybean product of Sikkim Himalaya. Front. Microbiol. 2019;10:1–17. doi: 10.3389/fmicb.2019.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Rosa G., Bonadonna L., Lucentini L., Kenmoe S., Suffredini E. Coronavirus in water environments: occurrence, persistence and concentration methods - a scoping review. Water Res. 2020;179:115899. doi: 10.1016/j.watres.2020.115899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladero V., Gómez-Sordo C., Sánchez-Llana E., del Rio B., Redruello B., Fernández M., Martín M.C., Alvarez M.A. Q69 (an E. faecalis-infecting bacteriophage) as a biocontrol agent for reducing tyramine in dairy products. Front. Microbiol. 2016;7:1–10. doi: 10.3389/fmicb.2016.00445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C.C., Shih T.P., Ko W.C., Tang H.J., Hsueh P.R. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and coronavirus disease-2019 (COVID-19): the epidemic and the challenges. Int. J. Antimicrob. Agents. 2020;55:105924. doi: 10.1016/j.ijantimicag.2020.105924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leblanc D., Gagné M.J., Poitras É., Brassard J. Persistence of murine norovirus, bovine rotavirus, and hepatitis A virus on stainless steel surfaces, in spring water, and on blueberries. Food Microbiol. 2019;84:103257. doi: 10.1016/j.fm.2019.103257. [DOI] [PubMed] [Google Scholar]
- Ledormand P., Desmasures N., Dalmasso M. Phage community involvement in fermented beverages: an open door to technological advances? Crit. Rev. Food Sci. Nutr. 2020:1–10. doi: 10.1080/10408398.2020.1790497. 0. [DOI] [PubMed] [Google Scholar]
- Lee S.Y., Kim M.J., Kim H.J., Jeong K.C.C., Kim H.Y. Simultaneous detection of four foodborne viruses in food samples using a one-step multiplex reverse transcription PCR. J. Microbiol. Biotechnol. 2018;28:210–217. doi: 10.4014/jmb.1710.10008. [DOI] [PubMed] [Google Scholar]
- Leplae R., Lima-Mendez G., Toussaint A. ACLAME: a CLAssification of mobile genetic elements, update 2010. Nucleic Acids Res. 2009;38:57–61. doi: 10.1093/nar/gkp938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B., Gibbons T., Ghodsi M., Treangen T., Pop M. Accurate and fast estimation of taxonomic profiles from metagenomic shotgun sequences. BMC Genom. 2011;12:1–10. doi: 10.1186/1471-2164-12-S2-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.F., Liu C.J., Zeng X.Q., Zhang H.Y., Luo Y.Y., Li X.R. Metagenomic and metatranscriptomic analysis of the microbial community structure and metabolic potential of fermented soybean in Yunnan Province. Food Sci. Technol. 2020;40:18–25. doi: 10.1590/fst.01718. [DOI] [Google Scholar]
- Lorenzi H.A., Hoover J., Inman J., Safford T., Murphy S., Kagan L., Williamson S.J. The viral metagenome annotation pipeline (VMGAP): an automated tool for the functional annotation of viral metagenomic shotgun sequencing data. Stand. Genomic Sci. 2011;4:418–429. doi: 10.4056/sigs.1694706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Altermann E., Breidt F., Kozyavkin S. Sequence analysis of Leuconostoc mesenteroides bacteriophage Φ1-A4 isolated from an industrial vegetable fermentation. Appl. Environ. Microbiol. 2010;76:1955–1966. doi: 10.1128/AEM.02126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Breidt F., Fleming H.P., Altermann E., Klaenhammer T.R. Isolation and characterization of a Lactobacillus plantarum bacteriophage, ΦJL-1, from a cucumber fermentation. Int. J. Food Microbiol. 2003;84:225–235. doi: 10.1016/S0168-1605(03)00111-9. [DOI] [PubMed] [Google Scholar]
- Lu Z., Breidt F., Plengvidhya V., Fleming H.P. Bacteriophage ecology in commercial sauerkraut fermentations. Appl. Environ. Microbiol. 2003;69:3192–3202. doi: 10.1128/AEM.69.6.3192-3202.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z., Pérez-Díaz I.M., Hayes J.S., Breidt F. Bacteriophage ecology in a commercial cucumber fermentation. Appl. Environ. Microbiol. 2012;78:8571–8578. doi: 10.1128/AEM.01914-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu C., Chen C., Ge F., Liu D., Zhao S., Chen D. A preliminary metagenomic study of puer tea during pile fermentation. J. Sci. Food Agric. 2013;93:3165–3174. doi: 10.1002/jsfa.6149. [DOI] [PubMed] [Google Scholar]
- Ma C., Pan N., Chen Z., Liu Z., Gong G., Ma A. Geographical diversity of Streptococcus thermophilus phages in Chinese yoghurt plants. Int. Dairy J. 2014;35:32–37. doi: 10.1016/j.idairyj.2013.10.007. [DOI] [Google Scholar]
- Mahony J., Moscarelli A., Kelleher P., Lugli G.A., Ventura M., Settanni L., van Sinderen D. Phage biodiversity in artisanal cheese wheys reflects the complexity of the fermentation process. Viruses. 2017;9:1–18. doi: 10.3390/v9030045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malenovská H. Coronavirus persistence on a plastic carrier under refrigeration conditions and its reduction using wet wiping technique, with respect to food safety. Food Environ. Virol. 2020 doi: 10.1007/s12560-020-09447-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markovinović L., Kosanović Ličina M.L., Tešić V., Vojvodić D., Vladušić Lucić I., Kniewald T., Vukas T., Kutleša M., Krajinović L.C. 2016. An Outbreak of Tick-Borne Encephalitis Associated with Raw Goat Milk and Cheese Consumption, Croatia, 2015. Infection. [DOI] [PubMed] [Google Scholar]
- Martella V., Bányai K., Matthijnssens J., Buonavoglia C., Ciarlet M. Zoonotic aspects of rotaviruses. Vet. Microbiol. 2010;140:246–255. doi: 10.1016/j.vetmic.2009.08.028. [DOI] [PubMed] [Google Scholar]
- Maske B.L., Pereira G.V. de M., Carvalho Neto D.P. de, Lindner J. de D., Letti L.A.J., Pagnoncelli M.G., Soccol C.R. Presence and persistence of Pseudomonas sp. during Caspian Sea-style spontaneous milk fermentation highlights the importance of safety and regulatory concerns for traditional and ethnic foods. Food Sci. Technol. 2020 doi: 10.1590/fst.15620. 2061. [DOI] [Google Scholar]
- McIntyre K., Heap H.A., Davey G.P., Limsowtin G.K.Y. The distribution of lactococcal bacteriophage in the environment of a cheese manufacturing plant. Int. Dairy J. 1991;1:183–197. doi: 10.1016/0958-6946(91)90010-6. [DOI] [Google Scholar]
- Mokili J.L., Rohwer F., Dutilh B.E. Metagenomics and future perspectives in virus discovery. Curr. Opin. Virol. 2012;2:63–77. doi: 10.1016/j.coviro.2011.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mudgal P., Breidt F., Lubkin S.R., Sandeep K.P. Quantifying the significance of phage attack on starter cultures: a mechanistic model for population dynamics of phage and their hosts isolated from fermenting sauerkraut. Appl. Environ. Microbiol. 2006;72:3908–3915. doi: 10.1128/AEM.02429-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J., Mahony J., Fitzgerald G.F., van Sinderen D. Cheese: Chemistry, Physics and Microbiology. Elsevier Ltd; 2017. Bacteriophages infecting lactic acid bacteria. fourth ed. fourth ed. [DOI] [Google Scholar]
- Murphy J., Royer B., Mahony J., Hoyles L., Heller K., Neve H., Bonestroo M., Nauta A., van Sinderen D. Biodiversity of lactococcal bacteriophages isolated from 3 Gouda-type cheese-producing plants. J. Dairy Sci. 2013;96:4945–4957. doi: 10.3168/jds.2013-6748. [DOI] [PubMed] [Google Scholar]
- Nagai T., Yamasaki F. Bacillus subtilis (natto) bacteriophages isolated in Japan. Food Sci. Technol. Res. 2009;15:293–298. doi: 10.3136/fstr.15.293. [DOI] [Google Scholar]
- Orlova E.V. Bacteriophages; 2012. Bacteriophages and Their Structural Organisation. [DOI] [Google Scholar]
- Oshiki M., Miura T., Kazama S., Segawa T., Ishii S., Hatamoto M., Yamaguchi T., Kubota K., Iguchi A., Tagawa T., Okubo T., Uemura S., Harada H., Kobayashi N., Araki N., Sano D. Microfluidic PCR amplification and MiSeq amplicon sequencing techniques for high-throughput detection and genotyping of human pathogenic RNA viruses in human feces, sewage, and oysters. Front. Microbiol. 2018;9:1–10. doi: 10.3389/fmicb.2018.00830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overbeek R., Begley T., Butler R.M., Choudhuri J.V., Chuang H.Y., Cohoon M., de Crécy-Lagard V., Diaz N., Disz T., Edwards R., Fonstein M., Frank E.D., Gerdes S., Glass E.M., Goesmann A., Hanson A., Iwata-Reuyl D., Jensen R., Jamshidi N., Krause L., Kubal M., Larsen N., Linke B., McHardy A.C., Meyer F., Neuweger H., Olsen G., Olson R., Osterman A., Portnoy V., Pusch G.D., Rodionov D.A., Rül. ckert C., Steiner J., Stevens R., Thiele I., Vassieva O., Ye Y., Zagnitko O., Vonstein V. The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res. 2005;33:5691–5702. doi: 10.1093/nar/gki866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E.J., Kim K.H., Abell G.C.J., Kim M.S., Roh S.W., Bae J.W. Metagenomic analysis of the viral communities in fermented foods. Appl. Environ. Microbiol. 2011;77:1284–1291. doi: 10.1128/AEM.01859-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacini S, Ruggiero M. Phage composition of a fermented milk and colostrum product assessed by microbiome array; putative role of open reading frames. bioRxiv. 2019 doi: 10.1101/714154. [DOI] [Google Scholar]
- Park J.E., Jung S., Kim A. MERS transmission and risk factors: a systematic review. BMC Publ. Health. 2018;18 doi: 10.1186/s12889-018-5484-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.H., Jung S., Shin J., Lee J.S., Joo I.S., Lee D.Y. Three gastroenteritis outbreaks in South Korea caused by the consumption of kimchi tainted by norovirus GI.4. Foodb. Pathog. Dis. 2015;12:221–227. doi: 10.1089/fpd.2014.1879. [DOI] [PubMed] [Google Scholar]
- Pereira G., De Carvalho Neto D.P., Junqueira A.C.D.O., Karp S.G., Letti L.A.J., Magalhães Júnior A.I., Soccol C.R. A review of selection criteria for starter culture development in the food fermentation industry. Food Rev. Int. 2020;36:135–167. doi: 10.1080/87559129.2019.1630636. [DOI] [Google Scholar]
- Philippe C., Krupovic M., Jaomanjaka F., Claisse O., Petrel M., le Marrec C. Bacteriophage GC1, a novel tectivirus infecting Gluconobacter cerinus, an acetic acid bacterium associated withwine-making. Viruses. 2018;10:1–16. doi: 10.3390/v10010039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phumkhachorn P. Isolation and characterization of lytic phage against Lactococcus lactis RP359, kem-buk-nud starter culture. Afr. J. Microbiol. Res. 2012;6:6678–6684. doi: 10.5897/ajmr12.1145. [DOI] [Google Scholar]
- Pringsulaka O., Patarasinpaiboon N., Suwannasai N., Atthakor W., Rangsiruji A. Isolation and characterisation of a novel Podoviridae-phage infecting Weissella cibaria N 22 from Nham, a Thai fermented pork sausage. Food Microbiol. 2011;28:518–525. doi: 10.1016/j.fm.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Pujato S.A., Quiberoni A., Mercanti D.J. Bacteriophages on dairy foods. J. Appl. Microbiol. 2019;126:14–30. doi: 10.1111/jam.14062. [DOI] [PubMed] [Google Scholar]
- Pung R., Chiew C.J., Young B.E., Chin S., Chen M.I.C., Clapham H.E., Cook A.R., Maurer-Stroh S., Toh M.P.H.S., Poh C., Low M., Lum J., Koh V.T.J., Mak T.M., Cui L., Lin R.V.T.P., Heng D., Leo Y.S., Lye D.C., Lee V.J.M., Kam K. qian, Kalimuddin S., Tan S.Y., Loh J., Thoon K.C., Vasoo S., Khong W.X., Suhaimi N.A., Chan S.J., Zhang E., Oh O., Ty A., Tow C., Chua Y.X., Chaw W.L., Ng Y., Abdul-Rahman F., Sahib S., Zhao Z., Tang C., Low C., Goh E.H., Lim G., Hou Y., Roshan I., Tan James, Foo K., Nandar K., Kurupatham L., Chan P.P., Raj P., Lin Y., Said Z., Lee A., See C., Markose J., Tan Joanna, Chan G., See W., Peh X., Cai V., Chen W.K., Li Z., Soo R., Chow A.L., Wei W., Farwin A., Ang L.W. Investigation of three clusters of COVID-19 in Singapore: implications for surveillance and response measures. Lancet. 2020;395:1039–1046. doi: 10.1016/S0140-6736(20)30528-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pyankov O.V., Bodnev S.A., Pyankova O.G., Agranovski I.E. Survival of aerosolized coronavirus in the ambient air. J. Aerosol Sci. 2018;115:158–163. doi: 10.1016/j.jaerosci.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiberoni A., Auad L., Binetti A.G., Suárez V.B., Reinheimer J.A., Raya R.R. Comparative analysis of Streptococcus thermophilus bacteriophages isolated from a yogurt industrial plant. Food Microbiol. 2003;20:461–469. doi: 10.1016/S0740-0020(02)00143-0. [DOI] [Google Scholar]
- Quiberoni A., Tremblay D., Ackermann H.W., Moineau S., Reinheimer J.A. Diversity of Streptococcus thermophilus phages in a large-production cheese factory in Argentina. J. Dairy Sci. 2006;89:3791–3799. doi: 10.3168/jds.S0022-0302(06)72420-1. [DOI] [PubMed] [Google Scholar]
- Rabenau H.F., Kampf G., Cinatl J., Doerr H.W. Efficacy of various disinfectants against SARS coronavirus. J. Hosp. Infect. 2005;61:107–111. doi: 10.1016/j.jhin.2004.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramírez M., Velázquez R., Maqueda M., López-Piñeiro A., Ribas J.C. A new wine Torulaspora delbrueckii killer strain with broad antifungal activity and its toxin-encoding double-stranded RNA virus. Front. Microbiol. 2015;6:1–12. doi: 10.3389/fmicb.2015.00983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rehfeld I.S., Fraiha A.L.S., Matos A.C.D., Guedes M.I.M.C., Costa E.A., de Souza M.R., Cavalcante L.F.L., Lobato Z.I.P. Short communication: survival of Vaccinia virus in inoculated cheeses during 60-day ripening. J. Dairy Sci. 2017;100:7051–7054. doi: 10.3168/jds.2017-12560. [DOI] [PubMed] [Google Scholar]
- Reusken C.B., Farag E.A., Jonges M., Godeke G.J., El-Sayed A.M., Pas S.D., Raj V.S., Mohran K.A., Moussa H.A., Ghobashy H., Alhajri F., Ibrahim A.K., Bosch B.J., Pasha S.K., Al-Romaihi H.E., Al-Thani M., Al-Marri S.A., Alhajri M.M., Haagmans B.L., Koopmans M.P. Middle east respiratory syndrome coronavirus (MERS-CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels. Euro Surveill. 2014;19:1–5. doi: 10.2807/1560-7917.ES2014.19.23.20829. Qatar, April 2014. [DOI] [PubMed] [Google Scholar]
- Reyes A., Semenkovich N.P., Whiteson K., Rohwer F., Gordon J.I. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat. Rev. Microbiol. 2012;10:607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards G.P. Critical review of norovirus surrogates in food safety research: rationale for considering volunteer studies. Food Environ. Virol. 2012;4:6–13. doi: 10.1007/s12560-011-9072-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizou M., Galanakis I.M., Aldawoud T.M.S., Galanakis C.M. Safety of foods, food supply chain and environment within the COVID-19 pandemic. Trends Food Sci. Technol. 2020;102:293–299. doi: 10.1016/j.tifs.2020.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cousiño N., Maqueda M., Ambrona J., Zamora E., Esteban R., Ramírez M. A new wine Saccharomyces cerevisiae killer toxin (Klus), encoded by a double-stranded RNA virus, with broad antifungal activity is evolutionarily related to a chromosomal host gene. Appl. Environ. Microbiol. 2011;77:1822–1832. doi: 10.1128/AEM.02501-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S., Solonenko N.E., Dang V.T., Poulos B.T., Schwenck S.M., Goldsmith D.B., Coleman M.L., Breitbart M., Sullivan M.B. Towards quantitative viromics for both double-stranded and single-stranded DNA viruses. PeerJ. 2016:1–17. doi: 10.7717/peerj.2777. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux S., Tournayre J., Mahul A., Debroas D., Enault F. Metavir 2: new tools for viral metagenome comparison and assembled virome analysis. BMC Bioinf. 2014;15 doi: 10.1186/1471-2105-15-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley P.A. The frenemies within: viruses, retrotransposons and plasmids that naturally infect Saccharomyces yeasts. Yeast. 2017;34:279–292. doi: 10.1002/yea.3234. [DOI] [PubMed] [Google Scholar]
- Samson J.E., Moineau S. Bacteriophages in food fermentations: new frontiers in a continuous arms race. Annu. Rev. Food Sci. Technol. 2013;4:347–368. doi: 10.1146/annurev-food-030212-182541. [DOI] [PubMed] [Google Scholar]
- Sausset R., Petit M.A., Gaboriau-Routhiau V., De Paepe M. New insights into intestinal phages. Mucosal Immunol. 2020;13:205–215. doi: 10.1038/s41385-019-0250-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxelin M.-L., Nurmiaho-Lassila E.-L., Meriläinen V.T., Forsén R.I. Ultrastructure and host specificity of bacteriophages of Streptococcus cremoris, Streptococcus lactis subsp. diacetylactis, and leuconostoc cremoris from Finnish fermented milk “viili. Appl. Environ. Microbiol. 1986;52:771–777. doi: 10.1128/aem.52.4.771-777.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekse C., Holst-Jensen A., Dobrindt U., Johannessen G.S., Li W., Spilsberg B., Shi J. High throughput sequencing for detection of foodborne pathogens. Front. Microbiol. 2017;8:1–26. doi: 10.3389/fmicb.2017.02029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour C.W., Bauchner H., Golub R.M. COVID-19 infection-preventing clinical deterioration. Jama. 2020;324:2020. doi: 10.1001/jama.2020.21720. [DOI] [PubMed] [Google Scholar]
- Sharpton T.J. An introduction to the analysis of shotgun metagenomic data. Front. Plant Sci. 2014;5:1–14. doi: 10.3389/fpls.2014.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Bandara N., Shin E., Ryu S., Kim K. pyo. Prevalence of Bacillus cereus bacteriophages in fermented foods and characterization of phage JBP901. Res. Microbiol. 2011;162:791–797. doi: 10.1016/j.resmic.2011.07.001. [DOI] [PubMed] [Google Scholar]
- Singhal T. A review of coronavirus disease-2019 (COVID-19) Indian J. Pediatr. 2020;87:281–286. doi: 10.1007/s12098-020-03263-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrabi C., Alsafi Z., O'Neill N., Khan M., Kerwan A., Al-Jabir A., Iosifidis C., Agha R. World Health Organization declares global emergency: a review of the 2019 novel coronavirus (COVID-19) Int. J. Surg. 2020;76:71–76. doi: 10.1016/j.ijsu.2020.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spus M., Li M., Alexeeva S., Wolkers-Rooijackers J.C.M., Zwietering M.H., Abee T., Smid E.J. Strain diversity and phage resistance in complex dairy starter cultures. J. Dairy Sci. 2015;98:5173–5182. doi: 10.3168/jds.2015-9535. [DOI] [PubMed] [Google Scholar]
- Tamang J.P., Cotter P.D., Endo A., Han N.S., Kort R., Liu S.Q., Mayo B., Westerik N., Hutkins R. Fermented foods in a global age: east meets West. Compr. Rev. Food Sci. Food Saf. 2020;19:184–217. doi: 10.1111/1541-4337.12520. [DOI] [PubMed] [Google Scholar]
- Tetz G., Tetz V. Bacteriophages as new human viral pathogens. Microorganisms. 2018;6:54. doi: 10.3390/microorganisms6020054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thippareddi H., Balamurugan S., Patel J., Singh M., Brassard J. Coronaviruses – potential human threat from foodborne transmission? LWT (Lebensm.-Wiss. & Technol.) 2020;134:110147. doi: 10.1016/j.lwt.2020.110147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trevors K.E., Holley R.A., Kempton A.G. Isolation and characterization of a Lactobacillus plantarum bacteriophage isolated from a meat starter culture*. J. Appl. Bacteriol. 1983;54:281–288. doi: 10.1111/j.1365-2672.1983.tb02618.x. [DOI] [Google Scholar]
- Umene K., Oohashi S., Yamanaka F., Shiraishi A. Molecular characterization of the genome of Bacillus subtilis (natto) bacteriophage PM1, a phage associated with disruption of food production. World J. Microbiol. Biotechnol. 2009;25:1877–1881. doi: 10.1007/s11274-009-0086-3. [DOI] [Google Scholar]
- Umene K., Shiraishi A. Complete nucleotide sequence of Bacillus subtilis (natto) bacteriophage PM1, a phage associated with disruption of food production. Virus Gene. 2013;46:524–534. doi: 10.1007/s11262-013-0876-4. [DOI] [PubMed] [Google Scholar]
- van Doremalen N., Bushmaker T., Karesh W.B., Munster V.J. Stability of middle east respiratory syndrome coronavirus in milk. Emerg. Infect. Dis. 2014;20:1263–1264. doi: 10.3201/eid2007.140500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasickova P., Pavlik I., Verani M., Carducci A. Issues concerning survival of viruses on surfaces. Food Environ. Virol. 2010;2:24–34. doi: 10.1007/s12560-010-9025-6. [DOI] [Google Scholar]
- Wang Yulong, Wu X., Wang Yihua, Li B., Zhou H., Yuan G., Fu Y., Luo Y. Low stability of nucleocapsid protein in SARS virus. Biochemistry. 2004;43:11103–11108. doi: 10.1021/bi049194b. [DOI] [PubMed] [Google Scholar]
- Whitehead H.R., Hunter G.J.E. 356. Bacteriophage in cheese manufacture: contamination from farm equipment. J. Dairy Res. 1947;15:112–119. doi: 10.1017/S0022029900004994. [DOI] [Google Scholar]
- Willner D., Hugenholtz P. From deep sequencing to viral tagging: recent advances in viral metagenomics. Bioessays. 2013;35:436–442. doi: 10.1002/bies.201200174. [DOI] [PubMed] [Google Scholar]
- Wolff A., Günther T., Albert T., Schilling-Loeffler K., Gadicherla A.K., Johne R. Stability of hepatitis E virus at different pH values. Int. J. Food Microbiol. 2020;325:108625. doi: 10.1016/j.ijfoodmicro.2020.108625. [DOI] [PubMed] [Google Scholar]
- Yépiz-Gómez M.S., Gerba C.P., Bright K.R. Survival of respiratory viruses on fresh produce. Food Environ. Virol. 2013;5:150–156. doi: 10.1007/s12560-013-9114-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinno P., Janzen T., Bennedsen M., Ercolini D., Mauriello G. Characterization of Streptococcus thermophilus lytic bacteriophages from mozzarella cheese plants. Int. J. Food Microbiol. 2010;138:137–144. doi: 10.1016/j.ijfoodmicro.2009.12.008. [DOI] [PubMed] [Google Scholar]
- World Health Organization (WHO) Hepatitis E Fact Sheets. 2020. (Accessed 1 november 2020) [Google Scholar]
- World Health Organization, (2009). Rotavirus vaccines: an update. Weekly Epidemiological Record = Relevé épidémiologique hebdomadaire, 84 (51-52), 533 - 540. https://apps.who.int/iris/handle/10665/241496. [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.