SUMMARY
The peptide ghrelin targets the growth hormone secretagogue receptor 1a (GHSR) to signal changes in cell metabolism and is a sought-after therapeutic target, although no structure is known to date. To investigate the structural basis of ghrelin binding to GHSR, we used solid-state nuclear magnetic resonance (NMR) spectroscopy, site-directed mutagenesis, and Rosetta modeling. The use of saturation transfer difference NMR identified key residues in the peptide for receptor binding beyond the known motif. This information combined with assignment of the secondary structure of ghrelin in its receptor-bound state was incorporated into Rosetta using an approach that accounts for flexible binding partners. The NMR data and models revealed an extended binding surface that was confirmed via mutagenesis. Our results agree with a growing evidence of peptides interacting via two sites at G protein-coupled receptors.
Graphical Abstract
In Brief
Structural knowledge of peptide binding at GPCRs is relatively unknown. Bender et al. investigate ghrelin binding to its receptor using a combination of NMR spectroscopy, mutagenesis, and protein modeling that revealed an extended binding surface for this peptide-receptor interaction.
INTRODUCTION
Ghrelin is a 28-amino acid peptide that binds to the G protein-coupled receptor (GPCR) growth hormone secretagogue receptor 1a (GHSR) to induce a release of growth hormone (Kojima et al., 1999). Ghrelin is best characterized as an orexigenic peptide, as increased levels stimulate hunger via GHSR activation (Cummings et al., 2001). GHSR has high constitutive activity (Holst et al., 2004) and dynamism in the apo state (Schrottke et al., 2017). Mutational work has examined the structural basis for this basal activity (Goze et al., 2010; Holst et al., 2010; Valentin-Hansen et al., 2012) and identified ligand binding sites (Els et al., 2012; Holst et al., 2004, 2006, 2007, 2009; Liu et al., 2007; Mokrosinski et al., 2012). However, the majority of this work has focused on small-molecule ligands, while binding of the endogenous peptide remains unknown.
Ghrelin contains an n-octanoylation at its Ser3 residue (Kojima et al., 1999). While des-acyl ghrelin is found in high concentrations in the blood, only the lipidated form is active at GHSR (Bednarek et al., 2000). We previously showed that ghrelin interacts with the membrane via its lipid modification (Vortmeier et al., 2015). Further, membrane-bound ghrelin was found to possess a small α-helical core and disordered termini in contrast to free ghrelin, which displays no helical character. This demonstrated that the environment was crucial to the structure of ghrelin and suggested that the receptor-bound state may yet differ further.
Peptide-binding GPCRs comprise a large subset of the GPCR family and structural knowledge is growing (Wu et al., 2017). However, only a few of these structures were determined with their respective peptide bound. These include the receptors for neurotensin (White et al., 2012), endothelin (Shihoya et al., 2016), opioid peptides (Fenalti et al., 2015), C5a (Liu et al., 2018), apelin (Ma et al., 2017), and chemokines (Burg et al., 2015; Qin et al., 2015). Nuclear magnetic resonance (NMR) has added structural and dynamic studies of peptides binding their receptors including bradykinin (Joedicke et al., 2018; Lopez et al., 2008), NPY (Kaiser et al., 2015; Yang et al., 2018), and dynorphin (O’Connor et al., 2015). An emerging theme in these studies is the large binding surface areas of peptide ligands compared with small-molecule ligands over two sites. One site correlates with the canonical orthosteric site of rhodopsin-like GPCRs, while an additional site is located distally in the N terminus, as in chemokine receptors (Burg et al., 2015; Qin et al., 2015), or extracellular loop 2 (ECL2), as in NPY type 2 receptor (Kaiser et al., 2015).
Initial studies of ghrelin found that the N-terminus of ghrelin including the lipid-modified Ser3 residue was critical for receptor binding, and the first five residues were suggested to represent the minimal binding motif (Bednarek et al., 2000). However, this short peptide possessed a binding affinity two orders of magnitude lower than full-length ghrelin and could not activate GHSR in isolated membranes (Torsello et al., 2002). This suggested a role for residues outside of the first five amino acids. We report here an extension of this binding motif to at least nine residues at a site in GHSR distinct from the orthosteric binding pocket. We measured peptide secondary structure and receptor-proximal residues using isotopically labeled ghrelin in complex with GHSR. We incorporated this data into a modeling method that built the ligand-receptor complex while accounting for high flexibility. Resulting models were filtered against the experimental NMR data and were compared against existing and novel mutational analysis. The final ensemble of models identifies an extended binding pocket from the central transmembrane (TM) bundle out to GHSR’s extracellular loops.
RESULTS
Expression of Functional GHSR
GHSR was expressed in inclusion bodies in E. coli using batch-fed fermentation (Schrottke et al., 2017). GHSR was reconstituted into lipid bicelles formed from 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) and 1,2-diheptanoyl-sn-glycero-3-phosphocholine (DHPC). Either small bicelles (q = 0.25, q is the molar ratio of DMPC to DHPC) or large bicelles (q > 10) were used in solution and solid-state NMR experiments, respectively. We used bicelles as they provide the best lipid systems for the reconstitution of high amounts of functional GHSR. Receptor functionality was confirmed by saturation ligand binding using atto520-labeled ghrelin peptide as in Figure S1 (Schrottke et al., 2017).
Investigation of Ghrelin Partitioning at GHSR
Octanoylated ghrelin is lipophilic providing the basis for membrane partitioning (Vortmeier et al., 2015). Although the affinity of ghrelin to GHSR is much higher than to the membrane, the membrane area exceeds that of the receptor, which may lead to sizable unspecific membrane binding. To quantitatively determine this equilibrium, we measured the 2H NMR spectra of a deuterated octanoyl chain to ghrelin (ghrelin-d15) bound to empty lipid bicelles and bicelles containing GHSR (Figure S2). To compare data with the study of ghrelin bound to membranes in the absence of GHSR (Vortmeier et al., 2015), negatively charged DMPC/DMPS (5/1, mol/mol) were used to increase membrane binding. Two dominating contributions can be extracted from the spectral line shape: (1) a large isotropic NMR signal, indicative of high mobility or unbound ghrelin, and (2) a Pake spectrum with a dominating quadrupolar splitting of ~110 kHz for the CH2 groups and an order parameter of 0.88, indicative of largely immobilized receptor-bound ghrelin. However, the superposition of the isotropic signal and immobilized Pake spectrum do not provide a complete description of the experimental 2H NMR line shape. Therefore, a third contribution was used in the numerical line shape analysis, which is the 2H NMR spectrum of ghrelin bound to empty bicelles and is characterized by motionally averaged quadrupolar splitting (spectrum reproduced in Figure S2B [Vortmeier et al., 2015]). As 2H NMR spectra were excited by a direct pulse and acquired with sufficient relaxation delay, the individual contributions to the spectral intensity can be readily obtained from the experimental 2H NMR spectrum using a quantitative line shape analysis on the basis of numerical simulations (Stahlberg et al., 2015). Quantification of the peak fitting yielded (1) 63% isotropic contribution, indicative of free or fast exchanging ghrelin, (2) 15% rigid contribution, indicative of receptor-bound ghrelin, and (3) 22% liquid-crystalline contribution, indicative of membrane-bound ghrelin.
Determination of Ghrelin Residues in Close Contact with GHSR
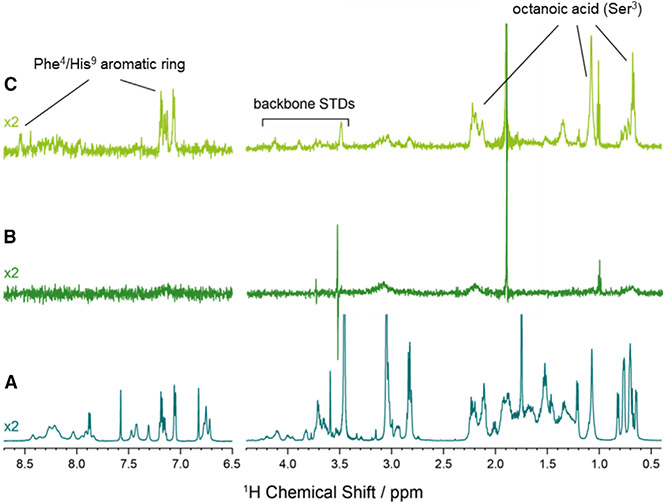
Given the high exchange between receptor-bound, membrane-bound, and free ghrelin seen in the 2H NMR results, saturation transfer difference (STD) NMR spectroscopy (Meyer and Peters, 2003) was suggested to determine ghrelin residues that are in close contact with GHSR. Figure 1 displays a 1H NMR spectrum of ghrelin in solution (A), ghrelin in the presence of empty bicelles (B), and ghrelin in the presence of bicelles containing GHSR (C). We used zwitterionic DMPC/DHPC bicelles in these experiments to reduce the affinity of ghrelin to the membrane (Vortmeier et al., 2015), allowing us to exclusively determine STD effects of ghrelin in interaction with GHSR. Clearly defined ghrelin signals that are observed in the STD NMR spectrum in the presence of GHSR can be assigned to the octanoyl chain at Ser3 and hydrophobic residues Phe4 and His9. The amplitude of signal in the STD spectrum is related to the proximity and rigidness of the binding residues with a greater signal indicating more tightly bound component. The STD effect is greatest for the octanoyl chain of Ser3 (60%–100% of saturation maximum) suggesting that this residue is tightly bound within the receptor (Table S1).
Figure 1. 1H NMR Saturation Transfer Difference Spectroscopy of Ghrelin at GHSR.
(A) 1H NMR spectrum of ghrelin.
(B) 1H STD NMR spectrum of ghrelin in the presence of 5 mM DMPC/DHPC bicelles.
(C) 1H STD NMR spectrum of ghrelin in the presence of 5 mM DMPC/DHPC bicelles with 25 μM GHSR. All NMR spectra were acquired in 50 mM NaP buffer (pH 7) at 20°C.
Structural Features of Receptor-Bound Ghrelin
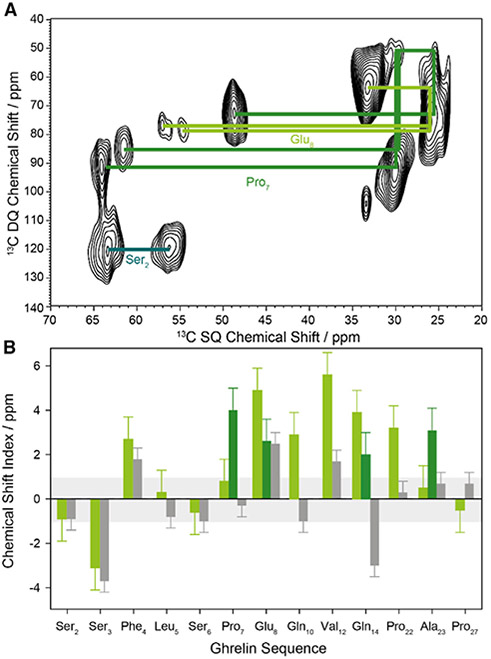
We next sought to characterize structural features of ghrelin in its receptor-bound form. We produced a library of peptides with three to four 13C-labeled residues each (Table 1). 13C magic-angle spinning NMR of the ghrelin/GHSR complex in large DMPC/DHPC bicelles (q > 10) was carried out at −30°C. Using 13C/13C single quantum/double quantum correlation NMR spectroscopy, we were able to assign chemical shifts for each labeled residue (Figure 2A). The cross-peaks were assigned according to their correlation schemes within the amino acid side chain. For Pro7, Glu8, Gln14, and Ala23, we detected two sets of cross-peaks, as reported in Table S2. The exact origin of the second contribution is unclear at the moment but points to slightly different backbone conformation and/or chemical environment. Pro7 cis/trans isomerization is unlikely as the measured chemical shifts do not support this. The chemical shift index (CSI) calculated by the difference in the Cα-Cβ chemical shift values relates information regarding the backbone geometry of each residue. Using the CSI across the collected residues we were able to measure the secondary structure of specific residues. We compared these results with our previously collected data on the membrane-bound structure (Vortmeier et al., 2015) and found that, while the peptide maintains a lack of secondary structure at the N terminus, the degree of α-helical nature in the central region becomes more defined (Figure 2B).
Table 1.
Peptides Used in this Study
| Peptide Sequence | Molecular Weight (Da) |
|---|---|
| GHR1: H2N- GSS(n-octanoyl-d15)FL SPEHQ RVQQR KESKK PPAKL QPR -OH | 3,398 |
| GHR2: H2N- GSS(n-octanoyl)FL SPEHQ RVQQR KESKK PPAKL QPR -OH | 3,384 |
| GHR3: H2N- GSS(n-octanoyl)FL SPEHQ RVQQR KESKK PPAKL QPR -OH | 3,385 |
| GHR4: H2N- GSS(n-octanoyl)FL SPEHQ RVQQR KESKK PPAKL QPR -OH | 3,388 |
| GHR5: H2N- GSS(n-octanoyl)FL SPEHQ RVQQR KESKK PPAKL QPR -OH | 3,379 |
| ∑: H2N- GSS(n-octanoyl-d15)FL SPEHQ RVQQR KESKK PPAKL QPR -OH |
Residues that are underlined were 13C/15N labeled. See also Figure S1.
Figure 2. Structural Data on GHSR-Bound Ghrelin.
(A) 13C/13C single-quantum/double-quantum (SQ/DQ) correlation NMR spectrum of a representative ghrelin peptide labeled in Ser2, Pro7, and Glu8, with the correlation pattern indicated by lines.
(B) Chemical shift index (CSI) for the investigated labeled amino acids of GHSR-bound ghrelin (light green bars). Positive values greater than 1 ppm indicate a tendency for α-helical structure, whereas values less than −1 ppm suggest β sheet character. For comparison, the CSI for membrane-bound ghrelin is given as gray bars (data adopted from Vortmeier et al., 2015). Two chemical shifts were found for Pro7, Glu8, Gln14, and Ala23 (dark green bars). Error bars are estimated from single repeats and the linewidths of the NMR signals.
See also Table S2.
Taken together, three types of structural information were obtained from the NMR analysis: (1) information on the binding equilibrium of ghrelin to GHSR and the membrane, (2) ghrelin residues in close contact with GHSR, and (3) the backbone conformation of ghrelin bound to GHSR.
Modeling of the Ghrelin/GHSR Complex
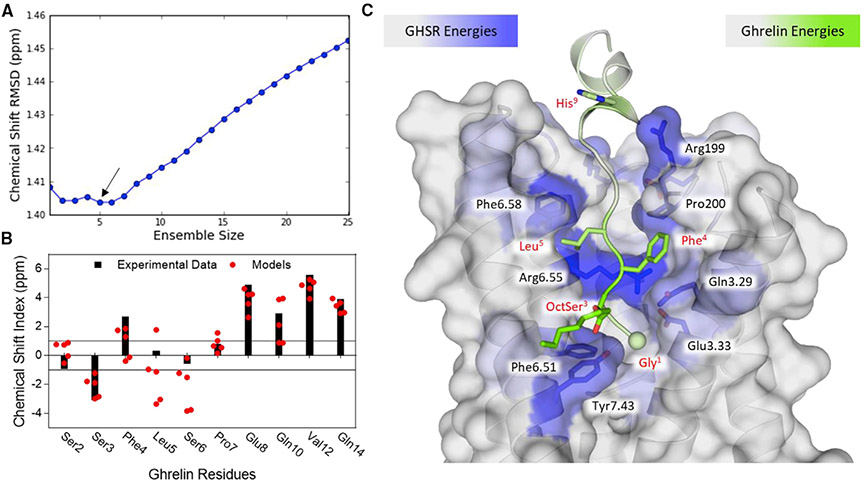
To better understand the structure of receptor-bound ghrelin, we used Rosetta to generate a model of the GHSR/ghrelin complex. An initial model of the receptor was generated with RosettaCM using related receptors (Vortmeier et al., 2015). Of note, these initial models were generated in the absence of a bound ligand and energetic minimization often collapsed the binding pocket. In addition, as the peptide was expected to extend out of the central binding pocket and make extensive contacts with the loop regions, it was anticipated that the loops would need to rearrange to accommodate the ligand. To generate models of the peptide-bound receptor, we removed all extracellular loops and rebuilt them in the presence of the peptide. We devised an approach to achieve this by alternating between docking ghrelin and rebuilding loops with a greater number of residues in each step (Figure S3). Each docking step oriented the ghrelin N terminus amino group toward Glu1243.33 (superscripts indicate Ballesteros-Weinstein numbering [Ballesteros and Weinstein, 1995]) as this residue has been previously suggested to balance its positive charge (Holst et al., 2009). By alternating between peptide docking and loop modeling, we densely sampled the conformational space available to these flexible partners. We modeled only the first 17 amino acids of ghrelin as the C terminus has no affinity for the receptor and does not enhance ligand binding over truncated variants (Bednarek et al., 2000). Following complex modeling, Cα and Cβ chemical shifts were calculated for ghrelin in the top 20% of models by energy. These values were compared against the experimentally determined chemical shifts and filtered to identify an ensemble of five models that collectively best represent the data (Figures 3A, 3B, and S3). Smaller ensemble sizes suffer from individual deviations that get averaged out in a larger ensemble, while larger ensembles suffer from additional noise. As such, the remaining analysis is performed on this ensemble of five ghrelin-GHSR complexes. Of note, the filtering step only addressed the conformation of the ghrelin peptide backbone and ambiguities remain, particularly in the GHSR loop regions and octanoyl chain (Figure S4).
Figure 3. Rosetta Modeling of Ghrelin at GHSR.
(A) Identification of ensemble by comparison with experimental chemical shift root-mean-square deviation (RMSD) in ppm. The minimum RMSD and smallest ensemble required five models (highlighted with an arrow).
(B) Comparison of the chemical shift index between the experimental data points and the five-model ensemble. Black bars correspond to the experimental CSI and red circles show the CSI for each model in the ensemble.
(C) A representative model of ghrelin binding at GHSR (TM 1 and 2 are removed for clarity). Residues are colored according to the predicted binding energies with green corresponding to strong energies in ghrelin and blue corresponding to strong energies in GHSR. The N-terminal nitrogen is shown as a sphere. The side chains shown represent those that contribute the most to binding energy.
See also Figures S3 and S4 and Table S3.
Characterization of the Ghrelin/GHSR Complex
The final ensemble of models converge with the N-terminus of the peptide extending down into the binding pocket of GHSR (Figures 3C and S3B). The depth is similar to what is observed for small-molecule-binding GPCRs. The N-terminus has no defined secondary structure propensity as seen in the membrane-bound and solutions states. Distinct from these states, however, a well-defined α helix extends from residue 8 or 9, depending on the model, through residue 17. This region of the peptide extends out of the central pocket and lies along ECL3. This is also the region where two sets of 13C chemical shifts are found, suggesting some plasticity in the conformation of the bound ligand. To better understand the molecular recognition between receptor and peptide, we calculated per-residue energetic contributions across the interface (Figure 3C; Table S3). As expected, the N-terminal binding motif of ghrelin was found to contribute significant energy to the binding interface. In addition, residues His9 and Arg11 were ranked highly in binding energy which agrees with the STD results. On the receptor, the strongest interaction partner for ghrelin was found to be Arg2836.55 (Table S3). This residue has been shown to be critical for ghrelin signaling at GHSR (Goze et al., 2010; Holst et al., 2004, 2007; Holst and Schwartz, 2006). In addition, residue Glu1243.33, the proposed charge balance for the positive amino terminus, was also highly ranked (Holst et al., 2007, 2009; Pantel et al., 2006). Further residues contributing significant binding energy were identified in the binding pocket along TMs 5, 6, and 7.
Validation of Binding Interface via Mutagenesis
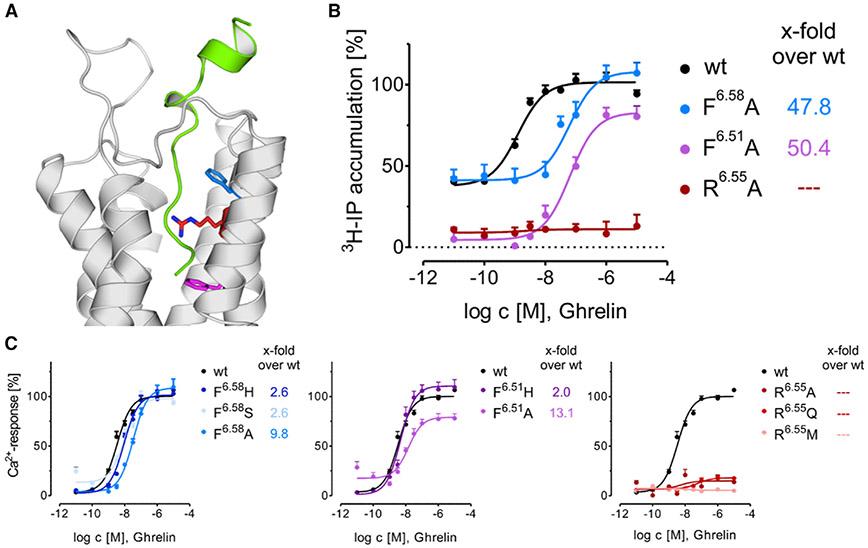
Many of the top 15 interacting residues in GHSR have been previously implicated in peptide binding, confirming our localization of the peptide within a central cavity (Table S4). In particular Glu1243.33 and Gln1203.29, which in our models show interactions with Gly1 and Phe4, displayed a dramatic loss of signal transduction after mutagenesis (Holst et al., 2004, 2006, 2007, 2009). To shed additional light on the structure of the binding complex, receptor mutants of the top 15 residues were tested in Ca2+ signaling and inositol triphosphate (IP3) accumulation assays. Alanine mutation of the top three interacting residues, Arg2836.55, Phe2866.58, and Phe2796.51, exhibited a strong impact on the receptor activation profiles (Figure 4). As seen before, Arg2836.55Ala and Phe2796.51Ala lost all constitutive activity (Holst et al., 2004), while Phe2866.58Ala retained basal activity. In addition, all of these mutations decreased activation via ghrelin stimulation. Phe2866.58 and Phe2796.51 could be partly rescued by a less-disruptive histidine mutation, suggesting that a bulky hydrophobic side chain is allowed. According to our model, these two residues pack alongside hydrophobic segments of ghrelin including octanoyl-Ser3 and Leu5. Another residue suggested to interact with the octanoyl chain of Ser3 is Tyr3137.43. However, mutation to alanine or phenylalanine resulted in loss of trafficking of the receptor to the plasma membrane, suggesting that this residue is critical to the structural integrity of the receptor (Figure S5). Ile1784.60, which may interact with Phe4 in ghrelin, displayed only a 2-fold loss of potency for alanine (Holst et al., 2004) but a 15-fold loss of potency for threonine (Table S4), indicating a hydrophobic interaction between receptor and ligand. Taken together, these mutations highlight the likelihood that our structural model represents a relevant binding pose of ghrelin at GHSR.
Figure 4. Signaling Impact of Residues with the Highest Rosetta Energy Score.
(A) Position of the residues with the highest Rosetta energy score in the ghrelin/GHSR complex. GHSR show in gray, ghrelin in green, and side chains tested colored according to (B and C).
(B) Concentration-response curves of GHSR mutants determined by IP accumulation assay in transiently transfected COS7 cells.
(C) Ca2+ mobilization assay of selected GHSR1a mutants in transiently transfected HEK293 cells. Data are means ± SEM of ≥2 independent experiments performed in duplicate.
DISCUSSION
Ghrelin Binds to GHSR via an Extended Binding Surface
Our models of ghrelin in complex with GHSR depict binding from the canonical orthosteric binding pocket outward along TMs 6 and 7 to ECL3. STD NMR identified Ser3 and Phe4 in the ghrelin N-terminus and His9 in the central helix to be closely interacting with GHSR. His9 is unique in that it is outside the first five residues that comprise the minimal signaling motif (Bednarek et al., 2000). This identifies a secondary region of the peptide that is needed for full signaling potential. Two-site binding modes have been seen in the few available peptide-bound GPCR crystal structures (Burg et al., 2015; Liu et al., 2018; Ma et al., 2017; Qin et al., 2015). This secondary site may explain why the minimal peptide could not compete full-length ghrelin in vivo (Torsello et al., 2002). Our modeling experiments also found His9 to be strongly interacting with the receptor as measured in our energy calculations. Interestingly, the adjacent residue Glu8 was found to lie in a fairly hydrophobic pocket at the top of TM 7. This agrees with previous studies that showed that mutation of negatively charged Glu8 to hydrophobic Ala or Tyr increases the affinity for the peptide to the receptor (Van Craenenbroeck et al., 2004). Overall, our model shows that the N-terminal motif binds within the central cavity in GHSR while the central α helix including His9 lies alongside and interacts with ECL3. There is increased flexibility in the ensemble as to the exact interaction of this α helix (Figure 4A), which may account for why we could not determine any high-specificity interactions between this α-helical region of ghrelin and the extracellular loops via mutagenesis. However, the sum total of interactions provides a strong signaling potential for the receptor as noted by enhanced activity for peptides from four up to ten residues in length (Matsumoto et al., 2001). This is similar to the endothelin-1/ET-B complex in which mutation of known crystal contacts resulted in maximal decreases of binding by only 10-fold (Shihoya et al., 2016). At present, the available data converge on a model in which this α helix extends between ECLs 2 and 3 and away from the receptor N terminus, unlike what has been seen in the chemokines (Wu et al., 2010). In addition, our use of an E. coli expression system suggests that no additional post-translational modifications are necessary beyond the ghrelin lipidation to achieve receptor-ligand binding. While additional data may become apparent in the future, we remain confident in the model given the current known experimental results.
The Binding Model of Ghrelin Adopts a Fairly Small Structural Ensemble
We filtered the models to generate an ensemble that best represents the experimental data. Five peptide models were identified that collectively satisfied the NMR restraints. This number was low compared with the ensemble of 22 models needed to represent the membrane-bound structure (Vortmeier et al., 2015). The five peptide models were fairly similar in conformation (Figure S4B), again contrasting with the membrane-bound ensemble with highly diverse backbone structures. For residues with multiple chemical shifts, we examined the secondary structure propensities individually. Glu8 and Gln14 display α-helical conformations in both populations and therefore we ensured that all models had α-helical character at these positions. Pro7 shows one population with distinct α-helical character and a second population of random coil. Our ensemble shows that this proline is important for initiating the α helix at the following position. Of note, we chose not to model the flexible C-terminus of ghrelin from residues 18 to 27 as these residues are unstructured and do not contribute to receptor binding (Bednarek et al., 2000). Addition of these residues would likely increase the ensemble size and conformational space, but provide little additional information on the binding interaction between the peptide and receptor. Compared with the membrane-bound state, the flexible N-terminus is highly converged to a single conformation in the receptor-bound state. The specific interaction between Gly1 and Glu1243.33 lock the N-terminus in place. This results in a well-defined pocket that can accommodate only small variations in the backbone structure. Evidence for this defined N-terminal conformation has been confirmed in complementary NMR studies (Ferré et al., 2018).
A Hierarchical Approach to Modeling Highly Flexible Protein-Ligand Complexes
We introduce here an approach for modeling protein-ligand complexes when both binding partners have a high degree of flexibility. A unique challenge of peptide ligands is the large degree of freedom associated with each amino acid. In addition, GPCRs possess a vast degree of internal flexibility, as seen in spectroscopic studies (Isogai et al., 2016; Kofuku et al., 2012; Manglik et al., 2015; Schafer and Farrens, 2015; Schrottke et al., 2017; Sounier et al., 2015; Ye et al., 2016). Flexibility within the receptor loops is likely to select for different ligand types, as seen in crystal structures of the C5a receptor, in which ECL2 adopted different conformations for small-molecule-bound versus peptide-bound states (Liu et al., 2018; Robertson et al., 2018). To account for these properties, we used an iterative approach in which we built and docked the peptide into the receptor. While no new code was developed for this method, the iterative use of homology modeling and flexible peptide docking of progressively longer segments of the target peptide is a unique technique. Each step in the protocol allowed for dense sampling of flexible components at the receptor binding pocket. Further, the use of homology modeling of loops ensures that we only build loops with structures relevant to known GPCRs. This approach was significantly guided by our understanding of GPCR biology and the ghrelin system. We believe the approach presented here will be useful for ligand-GPCR modeling in general with particular emphasis in complexes that require extensive remodeling of the extracellular loops for ligand binding to occur.
Concluding Remarks
Currently, there is a lack of knowledge of how endogenous peptides bind their GPCRs. While the rate of deposited structures is increasing for peptide-binding GPCRs, structures with a bound peptide remain sparse. The methodologies presented here use orthogonal structural methods to derive a model that best represents all available data. NMR allows the study of the receptor in its native state absent of fusion proteins and with full-length termini. Computational modeling rapidly eliminates unlikely solutions and allows researchers to focus on native-like structures. Use of these two methods resulted in a novel understanding of ghrelin binding to GHSR. Further, this model adds to the growing structural evidence of two-site binding modes in peptide-binding GPCRs.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Daniel Huster (daniel.huster@medizin.uni-leipzig.de).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Parts of this study were conducted in human and animal cell lines. HEK293 (human embryonic kidney) cells were cultured in Dulbecco’s modified Eagle’s medium with 4.5 g/L glucose and L-glutamine (DMEM) and HAM’s F-12 (1:1, v/v) Lonza, Basel, Switzerland) supplied with 15% (v/v) heat-inactivated FBS (Biochrom, Berlin, Germany). COS-7 (African green monkey, kidney) cells were maintained in DMEM with 10% (v/v) heat-inactivated FBS. All cell lines were cultured under humidified atmosphere at 37°C and 5% CO2.
HEK293 were authenticated using a PCR based multiplex assay based on the use of short tandem repeats (STR) (Anthentication of Human Cell Lines: Standardization of STR Profiling, ANSI/ATCC ASN-0002-2011). Identity of COS-7 was confirmed by DNA Barcoding by PCR amplification of 5′coding region of cytochrome c oxidase I. Cycle sequencing of respective PCR products revealed taxon assignment upon submission to BOLD (Ratnasingham and Hebert, 2007). All cell lines were routinely tested (negative) for mycoplasma contamination. HEK293 cells are derived from a female fetus and COS-7 cells are derived from a male adult monkey.
METHOD DETAILS
Expression and Purification of GHSR
The GHSR was prepared as described before (Schrottke et al., 2017). Briefly, the receptor was expressed with a C-terminal His8-tag (pET41b(+)-GHSR_M1-T366_LE_8xHis) in E. coli NiCo21(DE3) (NEB, Frankfurt am Main, Germany) in an insoluble form in inclusion bodies. Subsequently, the receptor was solubilized with 15 mM SDS and 50 mM DTT. After removal of DTT, it was purified using affinity chromatography. For reconstitution of the receptor into a lipid membrane the correct disulfide was formed at room temperature by dialysis in 50 mM sodium phosphate (pH 8), 2 mM SDS, 1 mM EDTA, 2 mM reduced and 1 mM oxidized glutathione at a receptor concentration of 0.5 mg/ml. After concentrating the receptor to 1 mg/ml it was added to preformed bicelles comprised of DMPC (or DMPC-d54) and DHPC with a ratio of receptor:DMPC:DHPC of 1:600:2400 (q=0.25) for solution NMR experiments, or 1 : 200 : 800 for solid-state NMR experiments. Three cycles of temperature jumps between 0°C and 42°C were performed. The solution NMR sample was then dialyzed against 50 mM sodium phosphate pH 7, 1.5 mM DHPC and it was concentrated with 20-30% (w/v) PEG20,000. For the solid-state NMR samples, BioBeads SM-2 (BioRad, Germany) were added to a concentration of 50 mg/ml to reduce the amount of detergent to yield samples with a q value higher than 10. The resulting turbid solution was incubated with a twofold molar excess of peptide for 1 h at 37°C. The sample was finally centrifuged and the pellet loaded into the MAS rotor.
Peptide Synthesis
Ghrelin was synthesized by Fmoc/tert-butyl strategy as described before (Park et al., 2015; Vortmeier et al., 2015). Briefly, 15 μmol of [Ser(Trt)3]-Ghrelin were synthesized automatically on R-Wang residue on a robot system (SyroI, MultiSynTech, Bochum, Germany). After selective deprotection of Ser3 by treatment of the resin with 10 times 1 ml of TFA/TIS/DCM (1:5:94, v/v/v), octanoic acid was coupled. First, free reactive hydroxyl groups at the resin were preactivated with 5 equivalents of DIC in DMF for 10 min. Subsequently, 5 equivalents of HOBt, 5 equivalents of octanoic acid, 5 equivalents of methylimidazole and 0.1 equivalents of DMAP in DMF were added and shaken overnight. 13C/15N-labeled peptides (Table 1) were synthesized on R-Wang resin as described for ghrelin (Vortmeier et al., 2015). 13C/15N-amino acids were coupled manually with 5 equivalents of Fmoc-protected amino acid, 5 equic HOBt and 5 equivalents of DIC in DMF overnight. Fluorescent tracer [Dpr3-Oct; Dpr16-atto520]-Ghrelin (Dpr, diaminopropionic acid; Oct, octanoic acid) for in vitro functionality assay was prepared as described (Schrottke et al., 2017). Briefly, 15 μmol of Boc-[Dpr(Mtt)3, Dpr(ivDde)16]-ghrelin was synthesized automatically on R-Wang resin. After selective deprotection of methyltrityl (Mtt) with 1% TFA/DCM (10x 1 min), octanoic acid was coupled using 1-hydroxybenzotriazole/diisopropylcarbodiimide (HOBT/DIC) over night (5 eq. each). Next, ivDde (1-(4,4-dimethyl-2,6-dioxocyclohex-1-ylidene)-3methylbutyl) protection group was selectively cleaved off using 2% hydrazine/DMF (10x 10min), and 1 eq. of the atto520 dye (free acid) was coupled using HOBT/DIC in DMF overnight. Peptides were cleaved off the resin using 90/5/5 (v/v/v) trifluoroacetic acid/H2O/triisopropylsilane for 3h and precipitated by ice cold diethyl ether. Peptide identity was confirmed by MALDI-ToF mass spectrometry (Ultraflex III MALDI ToF/ToF, Bruker), and purified to > 95% by reversed-phase HPLC applying linear gradients of H2O + 0.1% TFA and acetonitrile (ACN) + 0.08% TFA.
In Vitro Binding Assay
Functionality of GHSR reconstituted in DMPC/DHPC bicelles (with q=0.25) was tested by a fluorescence binding assay using [Dpr3-Oct; Dpr16-atto520]-ghrelin (ghrelin-atto520). Various concentrations of GHSR-containing bicelles in 50 mM sodium phosphate buffer pH 7 were incubated with 50 nM ghrelin-atto520 overnight. Samples were transferred into a 10 mm quartz cuvette and measurements were carried out on a FluoroMax-2 (JOBIN YVON) at 20°C with linear polarized light, an excitation wavelength of 500 nm, an emission wavelength of 540 nm, and 90° detection angle. Two independent measurements were carried out in duplicate and given data points are the mean values of the four experiments (with their standard deviation). Saturation binding data were fit with a sigmoidal dose-response curve using the Origin software. To assess the binding of ghrelin-atto520 to the membrane alone, the measurements were repeated with empty bicelles (a single measurement in duplicate).
2H NMR of Deuterated Ghrelin-d15
20 mg of GHSR embedded in large bicelles composed of DMPS/DMPC (1/5 mol/mol) was prepared as stated above. The pellet was resuspended three times in 5 ml of de-deuterated water and centrifuged 15 min at 5,000 rpm. 5.25 mg of ghrelin with a deuterated octanoyl chain (ghrelin-d15) was dissolved in 1 ml de-deuterated water and the pH was adjusted to pH 7 with NaOH. The peptide solution was added to the receptor sample and the complex incubated 1 h at 37°C. Unbound ghrelin was removed by centrifugation (15 min at 5,000 rpm).
Static 2H NMR spectra were acquired on a Bruker Avance I 750 MHz spectrometer with a 2H resonance frequency of 115 MHz at a temperature of 30°C. A quadrupolar echo pulse sequence was used. The 90° pulses had a length of 3.1 μs, the echo time was 60 μs and the relaxation delay 1 s. 2H NMR spectra were processed and simulated with a program written in Mathcad to estimate the amount of tightly bound, membrane associated and free ghrelin (Stahlberg et al., 2015).
STD NMR of Ghrelin
To investigate ligand-receptor contacts by saturation transfer difference (STD) NMR, GHSR was reconstituted in DMPC/DHPC bicelles (q=1) to achieve a receptor concentration of 25 μM. By dialysis with 25% PEG 20,000 the volume was adjusted to 1 ml. Ghrelin peptide was added in a 100-fold molar excess. A pseudo-2D version of the STD NMR sequence was used for the interleaved acquisition of on- and off-resonance spectra (Meyer and Peters, 2003). For selective saturation of the protein, a cascade of 50 E-Burpshaped pulses with a length of 50 ms each and an interpulse delay of 1 ms was used. The on-resonance frequency was set to −1 ppm and the off-resonance pulse was applied at 33.3 ppm. For suppression of the residual water signal, a Watergate sequence was used. Typically, STD NMR spectra were acquired with a 100-fold excess of ligand, a saturation time of 3 s, 5120 scans, and 8 K complex points. Prior to Fourier transformation FIDs were multiplied by an exponential line broadening function of 1 Hz.
13C/13C Single Quantum Double Quantum NMR
The experiments were carried out on a Bruker Avance III 600 NMR spectrometer with a 4 mm spinning module at a temperature of −30°C and an MAS frequency of 7 kHz. 1H as well as 13C 90° pulses had a length of 4 μs. For heteronuclear decoupling, the SPINAL64 was used with a 1H radio-frequency field strength of about 65 kHz. 13C magnetization was achieved with a cross polarization step with a contact time of 700 μs. The SQ/DQ correlation spectra were acquired using the SPC5 recoupling sequence for double quantum excitation and reconversion (set to 0.571 ms each). The relaxation delay was 2 s.
Modeling of the GHSR-Ghrelin Complex
We expanded on the structures generated in the previous study which were only used to define the membrane in Rosetta simulations but did not interact with the peptide. An ensemble of starting structures were generated previously used with varying backbone conformations (Vortmeier et al., 2015). The extracellular loops of the receptor were removed and the first 7 amino acids of ghrelin with an acetyl group on Ser3 were docked using FlexPepDock (Raveh et al., 2011). A constraint between the N-terminal amino head group and the oxygen atoms of Glu 1243.33 was used to filter the resulting models. Top models were passed through to a round of loop modeling guided by known crystal structures using RosettaCM (Song et al., 2013) in which most of ECL1 was built and the initial residues of ECL2 and ECL3 were built. A 12mer peptide was docked into these new extended models but were trimmed to a 10mer because of significant clashes with ECL3. A subsequent round of loop modeling completed ECL1 and ECL3 and built ECL2 through the β-sheet secondary structure region seen in all peptide binding crystal structures to date. The N-terminal 17 residues of ghrelin comprising the N-terminal flexible region and α-helical middle region were docked into this nearly complete structure. Lastly ECL2 was closed in a final loop modeling step. These last structures were modified such that the acetyl group on Ser3 was replaced by the full octanoyl chain followed by a last round of energetic minimization. These models were filtered by overall energy then analyzed by Proshift to compare calculated chemical shifts with the experimentally determined data. This resulted in a small ensemble size of 5 peptide conformations that had the lowest RMSD to the experimental data.
Mutagenesis of GHSR Plasmid
QuikChange™ site-directed mutagenesis (Agilent, Santa Clara, USA) was used to introduce mutations into the eukaryotic expression plasmid GHSR1a_EYFP_pVitro2 (Els et al., 2012). The correct sequences were confirmed by Sanger DNA sequencing.
Ca2+ Mobilization Assay
HEK293 cells were cultured in 25 cm2 flasks until 70-80 % confluency and subsequently transfected with 4 αg plasmid DNA and 15 αl Metafectene®Pro (Biontex, Munich, Germany) as a transfection reagent. One day after transfection, the cells were seeded into poly-D-lysine coated 96-well plates (black) and incubated overnight. Prior to stimulation, the medium was replaced by assay buffer (HBSS, 20 mM HEPES, 2.5 mM Probenecid (Sigma-Aldrich, St. Louis, USA)) containing 4 μM fluorescent dye Fluo-8 AM (Abcam, Cambridge, UK) and 0.1% Pluronic F127(Sigma-Aldrich, St. Louis, USA). After incubating the cells for 60 min at 37°C and 5% CO2, the dye solution was replaced by 150 μl assay buffer. Subsequently, fluorescence measurement (excitation 485 nm, emission 525 nm, cutoff 515 nm) was carried out with a Flexstation® 3 microplate reader (Molecular Devices, Sunnyvale, USA). Following a baseline recording for 20 s, 50 μl of serial dilutions of ghrelin (6-fold concentrated, 10−11 M to 10−5 M) were added automatically through a multi-channel pipetter and the fluorescence was detected for another 50 s. The experiments were repeated at least two times independently in technical triplicate.
Inositol Phosphate Accumulation Assay
COS7 cells were transiently transfected with 4 μg receptor plasmid in 25 cm2 culture flasks using 15 μl of MetafectenePro (Biontex, Munich, Germany) according to the manufacturer’s instructions. 24 h post-transfection, the cells were seeded into 48-well plates and grown to 90% confluency. The cells were labeled in DMEM supplied with 10% (v/v) FBS containing 2 μCi/ml myo-[2-3H]-inositol (Perkin Elmer, Waltham, USA) at 37°C in the presence of 5% CO2 for 16-18 h. After aspirating the labeling solution, the cells were washed once with DMEM containing 10 mM LiCl and stimulated for 2 h with increasing ghrelin concentrations, ranging from 10−11 M to 10−5 M in DMEM with 10 mM LiCl. Stimulation was stopped by aspiration of the medium followed by basic cell lysis with 0.1 M NaOH and subsequent neutralization with 0.13 M formic acid. After removal of the cell debris, radioactive inositol phosphate species were diluted and purified on an AG 1-X8 anion exchange resin (Bio-Rad, Hercules, USA) as described (Els et al., 2010). Radioactivity of the eluates was measured on a liquid scintillation counter. The experiments were repeated at least two times independently in technical triplicate.
Live Cell Microscopy
Membrane localization of the mutant GHSR variants was verified by fluorescence imaging. HEK293 cells were seeded onto microslide 8-wells (ibidiTreat, Martinsried, Germany) to 70-80% confluency and transiently transfected with 1 μg plasmid DNA and 1 μl Lipofectamine®2000 (Invitrogen, Carlsbad, USA) for 1 h. Following cultivation in DMEM and Ham’s F-12 (1:1, v/v) supplemented with 15% FBS overnight, the medium was replaced by Opti-MEM® (Life Technologies, Darmstadt, Germany) supplemented with 10 μM Hoechst33342 (Sigma-Aldrich, St. Louis, USA) and incubated at 37°C for 10 min. Membrane localization was documented in pure Opti-MEM® using an AxioObserver.Z1 microscope with an ApoTome Imaging System (Carl Zeiss AG, Oberkochen, Germany). Representative images of at least two independent experiments are shown.
QUANTIFICATION AND STATISTICAL ANALYSIS
Statistics from receptor activation assays were calculated in GraphPad Prism 5.03 (GraphPad Software, San Diego, USA) using standard non-linear regression (log(agonist) vs. response (three parameters)) and normalization to wild type response. All experiments were performed at least in duplicates. EC50 values are given as mean ± standard error of the mean (SEM). Error bars are displayed in Figure 4 and described in the caption.
DATA AND SOFTWARE AVAILABILITY
The 13C chemical shift data has been deposited in the BMRB under ID code 27600. The ensemble of ghrelin/GHSR complexes has been deposited in the PDB-Dev under ID code PDBDEV_00000024.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Library Efficiency™ DH5α™ Competent Cells | ThermoFisher Scientific | Cat# 18263012 |
| E. coli NiCo21(DE3) | BioRad | Cat# 1523920 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Atto520, free acid | Sigma-Aldrich | Cat# 70706-1MG-F |
| MetafectenePro | Biontex | Cat# T040-5.0 |
| Fluo-8 AM | Abcam | Cat# ab142773 |
| Pluronic F127 | Sigma-Aldrich | Cat# P2443-250G |
| Probenecid | Sigma-Aldrich | Cat# P8761-25G |
| myo-[2-3H]-inositol | Perkin Elmer | Cat# NET1156001MC |
| 1,2-diheptanoyl-sn-glycero-3-phosphocholine (DHPC) | Avanti Polar Lipids | Cat# 850306 |
| 1,2-dimyristoyl-sn-glycero-3-phophocholine (DMPC) | Avanti Polar Lipids | Cat# 850345 |
| 1,2-dimyristoyl(d54)-sn-glycero-3-phosphocholine (DMCP-d54) | Avanti Polar Lipids | Cat# 860345 |
| 1,2-dimyristoyl-sn-glycero-3-phophatidylserine (DMPS) | Avanti Polar Lipids | Cat# 840033 |
| BioBeads SM-2 | BioRad | Cat# 1523920 |
| Ghrelin and 13C variants | (Vortmeier et al., 2015) | NA |
| [Dpr3-Oct; Dpr16-atto520]-Ghrelin | (Schrottke et al., 2017) | NA |
| Deposited Data | ||
| Ensemble of Ghrelin-GHSR | PDB-Dev | PDBDEV_00000024 |
| Ghrelin Peptide 13C Chemical Shift Data | BMRB | 27600 |
| Experimental Models: Cell Lines | ||
| HEK293 | DSMZ | ACC305 |
| COS-7 | ATCC | CRL-1651 |
| Oligonucleotides | ||
| Primers for mutagenesis, see Table S5 | ||
| Recombinant DNA | ||
| Plasmid: pV2_GHSR1a_eYFP | (Els et al., 2012) | NA |
| Plasmid: pET41b(+)-GHSR_M1-T366_LE_8xHis (ATGTGGAACGCGACGCCCAGCGAAGAGCCGGGGTTCAACCTCACACTGGCCGACCTGGACTGGGATGCTTCCCCCGGCAACGACTCGCTGGGCGACGAGCTGCTGCAGCTCTTCCCCGCGCCGCTGCTGGCGGGCGTCACAGCCACCTGCGTGGCACTCTTCGTGGTGGGTATCGCTGGCAACCTGCTCACCATGCTGGTGGTGTCGCGCTTCCGCGAGCTGCGCACCACCACCAACCTCTACCTGTCCAGCATGGCCTTCTCCGATCTGCTCATCTTCCTCTGCATGCCCCTGGACCTCGTTCGCCTCTGGCAGTACCGGCCCTGGAACTTCGGCGACCTCCTCTGCAAACTCTTCCAATTCGTCAGTGAGAGCTGCACCTACGCCACGGTGCTCACCATCACAGCGCTGAGCGTCGAGCGCTACTTCGCCATCTGCTTCCCACTCCGGGCCAAGGTGGTGGTCACCAAGGGGCGGGTGAAGCTGGTCATCTTCGTCATCTGGGCCGTGGCCTTCTGCAGCGCCGGGCCCATCTTCGTGCTAGTCGGGGTGGAGCACGAGAACGGCACCGACCCTTGGGACACCAACGAGTGCCGCCCCACCGAGTTTGCGGTGCGCTCTGGACTGCTCACGGTCATGGTGTGGGTGTCCAGCATCTTCTTCTTCCTTCCTGTCTTCTGTCTCACGGTCCTCTACAGTCTCATCGGCAGGAAGCTGTGGCGGAGGAGGCGCGGCGATGCTGTCGTGGGTGCCTCGCTCAGGGACCAGAACCACAAGCAAACCGTGAAAATGCTGGCTGTAGTGGTGTTTGCCTTCATCCTCTGCTGGCTCCCCTTCCACGTAGGGCGATATTTATTTTCCAAATCCTTTGAGCCTGGCTCCTTGGAGATTGCTCAGATCAGCCAGTACTGCAACCTCGTGTCCTTTGTCCTCTTCTACCTCAGTGCTGCCATCAACCCCATTCTGTACAACATCATGTCCAAGAAGTACCGGGTGGCAGTGTTCAGACTTCTGGGATTCGAACCCTTCTCCCAGAGAAAGCTCTCCACTCTGAAAGATGAAAGTTCTCGGGCCTGGACAGAATCTAGTATTAATACACTCGAGCACCACCACCACCACCACCACCAC) | Modified from Schrottke et al. (2017) | N/A |
| Software and Algorithms | ||
| TopSpin | Bruker | https://www.bruker.com/products/mr/nmr/nmr-software/software/topspin/overview.html |
| Rosetta | (Leaver-Fay et al., 2011) | https://www.rosettacommons.org/ |
| GraphPad Prism | GraphPad Software, Inc. | www.graphpad.com/scientific-software/prism/ |
| ProShift | (Meiler, 2003) | http://www.meilerlab.org/index.php/servers/show?s_id=9 |
| PyMol | Schrödinger | http://www.pymol.org/ |
Highlights.
NMR spectroscopy and mutagenesis analysis provide insight on ghrelin binding mode
Residues outside of the known binding motif are identified
Ghrelin binds via an extended surface to its G protein-coupled receptor
Hybrid method of modeling peptide-binding receptors is described
ACKNOWLEDGMENTS
Parts of this study were supported by the EU and the Free State of Saxony (ESF 100148835 and 100227414). Work in the Meiler laboratory is supported by the NIH (R01 GM080403, R01 DK097376, and R01 HL122010) and NSF (CHE 1305874). B.J.B. is supported by the Pharmacology Training Grant at Vanderbilt University (T32 GM007628).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes five figures and five tables and can be found with this article online at https://doi.org/10.1016/j.str.2018.12.004.
REFERENCES
- Ballesteros JA, and Weinstein H (1995). Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. In Receptor Molecular Biology, Sealfon SC, ed. (Academic Press; ), pp. 366–428. [Google Scholar]
- Bednarek MA, Feighner SD, Pong SS, McKee KK, Hreniuk DL, Silva MV, Warren VA, Howard AD, Van Der Ploeg LH, and Heck JV (2000). Structure-function studies on the new growth hormone-releasing peptide, ghrelin: minimal sequence of ghrelin necessary for activation of growth hormone secretagogue receptor 1a. J. Med. Chem 43, 4370–4376. [DOI] [PubMed] [Google Scholar]
- Burg JS, Ingram JR, Venkatakrishnan AJ, Jude KM, Dukkipati A, Feinberg EN, Angelini A, Waghray D, Dror RO, Ploegh HL, et al. (2015). Structural basis for chemokine recognition and activation of a viral G protein-coupled receptor. Science 347, 1113–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings DE, Purnell JQ, Frayo RS, Schmidova K, Wisse BE, and Weigle DS (2001). A preprandial rise in plasma ghrelin levels suggests a role in meal initiation in humans. Diabetes 50, 1714–1719. [DOI] [PubMed] [Google Scholar]
- Els S, Beck-Sickinger AG, and Chollet C (2010). Ghrelin receptor: high constitutive activity and methods for developing inverse agonists. Methods Enzymol. 485, 103–121. [DOI] [PubMed] [Google Scholar]
- Els S, Schild E, Petersen PS, Kilian TM, Mokrosinski J, Frimurer TM, Chollet C, Schwartz TW, Holst B, and Beck-Sickinger AG (2012). An aromatic region to induce a switch between agonism and inverse agonism at the ghrelin receptor. J. Med. Chem 55, 7437–7449. [DOI] [PubMed] [Google Scholar]
- Fenalti G, Zatsepin NA, Betti C, Giguere P, Han GW, Ishchenko A, Liu W, Guillemyn K, Zhang H, James D, et al. (2015). Structural basis for bifunctional peptide recognition at human delta-opioid receptor. Nat. Struct. Mol. Biol 22, 265–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferré G, Damian M, M’Kadmi C, Saurel O, Czaplicki G, Demange P, Marie J, Floquet N, Fehrentz J-A, Milon A, et al. (2018). Structure and dynamics of the receptor-bound ghrelin lipopeptide. Biophys. J 114, 235a. [Google Scholar]
- Goze C, Berge G, M’Kadmi C, Floquet N, Gagne D, Galleyrand JC, Fehrentz JA, and Martinez J (2010). Involvement of tryptophan W276 and of two surrounding amino acid residues in the high constitutive activity of the ghrelin receptor GHS-R1a. Eur. J. Pharmacol 643, 153–161. [DOI] [PubMed] [Google Scholar]
- Holst B, Frimurer TM, Mokrosinski J, Halkjaer T, Cullberg KB, Underwood CR, and Schwartz TW (2009). Overlapping binding site for the endogenous agonist, small-molecule agonists, and ago-allosteric modulators on the ghrelin receptor. Mol. Pharmacol 75, 44–59. [DOI] [PubMed] [Google Scholar]
- Holst B, Holliday ND, Bach A, Elling CE, Cox HM, and Schwartz TW (2004). Common structural basis for constitutive activity of the ghrelin receptor family. J. Biol. Chem 279, 53806–53817. [DOI] [PubMed] [Google Scholar]
- Holst B, Lang M, Brandt E, Bach A, Howard A, Frimurer TM, Beck-Sickinger A, and Schwartz TW (2006). Ghrelin receptor inverse agonists: identification of an active peptide core and its interaction epitopes on the receptor. Mol. Pharmacol 70, 936–946. [DOI] [PubMed] [Google Scholar]
- Holst B, Mokrosinski J, Lang M, Brandt E, Nygaard R, Frimurer TM, Beck-Sickinger AG, and Schwartz TW (2007). Identification of an efficacy switch region in the ghrelin receptor responsible for interchange between agonism and inverse agonism. J. Biol. Chem 282, 15799–15811. [DOI] [PubMed] [Google Scholar]
- Holst B, Nygaard R, Valentin-Hansen L, Bach A, Engelstoft MS, Petersen PS, Frimurer TM, and Schwartz TW (2010). A conserved aromatic lock for the tryptophan rotameric switch in TM-VI of seven-transmembrane receptors. J. Biol. Chem 285, 3973–3985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holst B, and Schwartz TW (2006). Ghrelin receptor mutations – too little height and too much hunger. J. Clin. Invest 116, 637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai S, Deupi X, Opitz C, Heydenreich FM, Tsai CJ, Brueckner F, Schertler GF, Veprintsev DB, and Grzesiek S (2016). Backbone NMR reveals allosteric signal transduction networks in the beta1-adrenergic receptor. Nature 530, 237–241. [DOI] [PubMed] [Google Scholar]
- Joedicke L, Mao J, Kuenze G, Reinhart C, Kalavacherla T, Jonker HRA, Richter C, Schwalbe H, Meiler J, Preu J, et al. (2018). The molecular basis of subtype selectivity of human kinin G-protein-coupled receptors. Nat. Chem. Biol 14, 284–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser A, Muller P, Zellmann T, Scheidt HA, Thomas L, Bosse M, Meier R, Meiler J, Huster D, Beck-Sickinger AG, et al. (2015). Unwinding of the C-terminal residues of neuropeptide Y is critical for Y(2) receptor binding and activation. Angew. Chem. Int. Ed 54, 7446–7449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofuku Y, Ueda T, Okude J, Shiraishi Y, Kondo K, Maeda M, Tsujishita H, and Shimada I (2012). Efficacy of the beta(2)-adrenergic receptor is determined by conformational equilibrium in the transmembrane region. Nat. Commun 3, 1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, and Kangawa K (1999). Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature 402, 656–660. [DOI] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. (2011). ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Fortin JP, Beinborn M, and Kopin AS (2007). Four missense mutations in the ghrelin receptor result in distinct pharmacological abnormalities. J. Pharmacol. Exp. Ther 322, 1036–1043. [DOI] [PubMed] [Google Scholar]
- Liu H, Kim HR, Deepak R, Wang L, Chung KY, Fan H, Wei Z, and Zhang C (2018). Orthosteric and allosteric action of the C5a receptor antagonists. Nat. Struct. Mol. Biol 25, 472–481. [DOI] [PubMed] [Google Scholar]
- Lopez JJ, Shukla AK, Reinhart C, Schwalbe H, Michel H, and Glaubitz C (2008). The structure of the neuropeptide bradykinin bound to the human G-protein coupled receptor bradykinin B2 as determined by solid-state NMR spectroscopy. Angew. Chem. Int. Ed 47, 1668–1671. [DOI] [PubMed] [Google Scholar]
- Ma Y, Yue Y, Ma Y, Zhang Q, Zhou Q, Song Y, Shen Y, Li X, Ma X, Li C, et al. (2017). Structural basis for apelin control of the human apelin receptor. Structure 25, 858–866.e4. [DOI] [PubMed] [Google Scholar]
- Manglik A, Kim TH, Masureel M, Altenbach C, Yang Z, Hilger D, Lerch MT, Kobilka TS, Thian FS, Hubbell WL, et al. (2015). Structural insights into the dynamic process of beta2-adrenergic receptor signaling. Cell 161, 1101–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M, Hosoda H, Kitajima Y, Morozumi N, Minamitake Y, Tanaka S, Matsuo H, Kojima M, Hayashi Y, and Kangawa K (2001). Structure-activity relationship of ghrelin: pharmacological study of ghrelin peptides. Biochem. Biophys. Res. Commun 287, 142–146. [DOI] [PubMed] [Google Scholar]
- Meiler J (2003). PROSHIFT: protein chemical shift prediction using artificial neural networks. J. Biomol. NMR 26, 25–37. [DOI] [PubMed] [Google Scholar]
- Meyer B, and Peters T (2003). NMR spectroscopy techniques for screening and identifying ligand binding to protein receptors. Angew. Chem. Int. Ed 42, 864–890. [DOI] [PubMed] [Google Scholar]
- Mokrosinski J, Frimurer TM, Sivertsen B, Schwartz TW, and Holst B (2012). Modulation of constitutive activity and signaling bias of the ghrelin receptor by conformational constraint in the second extracellular loop. J. Biol. Chem 287, 33488–33502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor C, White KL, Doncescu N, Didenko T, Roth BL, Czaplicki G, Stevens RC, Wuthrich K, and Milon A (2015). NMR structure and dynamics of the agonist dynorphin peptide bound to the human kappa opioid receptor. Proc. Natl. Acad. Sci. U S A 112, 11852–11857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantel J, Legendre M, Cabrol S, Hilal L, Hajaji Y, Morisset S, Nivot S, Vie-Luton MP, Grouselle D, de Kerdanet M, et al. (2006). Loss of constitutive activity of the growth hormone secretagogue receptor in familial short stature. J. Clin. Invest 116, 760–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M, Sivertsen BB, Els-Heindl S, Huber T, Holst B, Beck-Sickinger AG, Schwartz TW, and Sakmar TP (2015). Bioorthogonal labeling of ghrelin receptor to facilitate studies of ligand-dependent conformational dynamics. Chem. Biol 22, 1431–1436. [DOI] [PubMed] [Google Scholar]
- Qin L, Kufareva I, Holden LG, Wang C, Zheng Y, Zhao C, Fenalti G, Wu H, Han GW, Cherezov V, et al. (2015). Crystal structure of the chemokine receptor CXCR4 in complex with a viral chemokine. Science 347, 1117–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratnasingham S, and Hebert PD (2007). bold: the Barcode of Life Data System (http://www.barcodinglife.org). Mol. Ecol. Notes 7, 355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raveh B, London N, Zimmerman L, and Schueler-Furman O (2011). Rosetta FlexPepDock ab-initio: simultaneous folding, docking and refinement of peptides onto their receptors. PLoS One 6, e18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson N, Rappas M, Dore AS, Brown J, Bottegoni G, Koglin M, Cansfield J, Jazayeri A, Cooke RM, and Marshall FH (2018). Structure of the complement C5a receptor bound to the extra-helical antagonist NDT9513727. Nature 553, 111–114. [DOI] [PubMed] [Google Scholar]
- Schafer CT, and Farrens DL (2015). Conformational selection and equilibrium governs the ability of retinals to bind opsin. J. Biol. Chem 290, 4304–4318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrottke S, Kaiser A, Vortmeier G, Els-Heindl S, Worm D, Bosse M, Schmidt P, Scheidt HA, Beck-Sickinger AG, and Huster D (2017). Expression, functional characterization, and solid-state NMR investigation of the G protein-coupled GHS receptor in bilayer membranes. Sci. Rep 7, 46128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shihoya W, Nishizawa T, Okuta A, Tani K, Dohmae N, Fujiyoshi Y, Nureki O, and Doi T (2016). Activation mechanism of endothelin ETB receptor by endothelin-1. Nature 537, 363–368. [DOI] [PubMed] [Google Scholar]
- Song Y, DiMaio F, Wang RY, Kim D, Miles C, Brunette T, Thompson J, and Baker D (2013). High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sounier R, Mas C, Steyaert J, Laeremans T, Manglik A, Huang W, Kobilka BK, Demene H, and Granier S (2015). Propagation of conformational changes during mu-opioid receptor activation. Nature 524, 375–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahlberg S, Skolova B, Madhu PK, Vogel A, Vavrova K, and Huster D (2015). Probing the role of the ceramide acyl chain length and sphingosine unsaturation in model skin barrier lipid mixtures by (2)H solid-state NMR spectroscopy. Langmuir 31, 4906–4915. [DOI] [PubMed] [Google Scholar]
- Torsello A, Ghe C, Bresciani E, Catapano F, Ghigo E, Deghenghi R, Locatelli V, and Muccioli G (2002). Short ghrelin peptides neither displace ghrelin binding in vitro nor stimulate GH release in vivo. Endocrinology 143, 1968–1971. [DOI] [PubMed] [Google Scholar]
- Valentin-Hansen L, Holst B, Frimurer TM, and Schwartz TW (2012). PheVI:09 (Phe6.44) as a sliding microswitch in seven-transmembrane (7TM) G protein-coupled receptor activation. J. Biol. Chem 287, 43516–43526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Craenenbroeck M, Gregoire F, De Neef P, Robberecht P, and Perret J (2004). Ala-scan of ghrelin (1-14): interaction with the recombinant human ghrelin receptor. Peptides 25, 959–965. [DOI] [PubMed] [Google Scholar]
- Vortmeier G, DeLuca SH, Els-Heindl S, Chollet C, Scheidt HA, Beck-Sickinger AG, Meiler J, and Huster D (2015). Integrating solid-state NMR and computational modeling to investigate the structure and dynamics of membrane-associated ghrelin. PLoS One 10, e0122444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JF, Noinaj N, Shibata Y, Love J, Kloss B, Xu F, Gvozdenovic-Jeremic J, Shah P, Shiloach J, Tate CG, et al. (2012). Structure of the agonist-bound neurotensin receptor. Nature 490, 508–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B, Chien EY, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, et al. (2010). Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F, Song G, de Graaf C, and Stevens RC (2017). Structure and function of peptide-binding G protein-coupled receptors. J. Mol. Biol 429, 2726–2745. [DOI] [PubMed] [Google Scholar]
- Yang Z, Han S, Keller M, Kaiser A, Bender BJ, Bosse M, Burkert K, Kogler LM, Wifling D, Bernhardt G, et al. (2018). Structural basis of ligand binding modes at the neuropeptide Y Y1 receptor. Nature 556, 520–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Van Eps N, Zimmer M, Ernst OP, and Prosser RS (2016). Activation of the A2A adenosine G-protein-coupled receptor by conformational selection. Nature 533, 265–268. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 13C chemical shift data has been deposited in the BMRB under ID code 27600. The ensemble of ghrelin/GHSR complexes has been deposited in the PDB-Dev under ID code PDBDEV_00000024.