Abstract
X-ray-based analytics are routinely applied in many fields, including physics, chemistry, materials science, and engineering. The full potential of such techniques in the life sciences and medicine, however, has not yet been fully exploited. We highlight current and upcoming advances in this direction. We describe different X-ray-based methodologies (including those performed at synchrotron light sources and X-ray free-electron lasers) and their potentials for application to investigate the nano–bio interface. The discussion is predominantly guided by asking how such methods could better help to understand and to improve nanoparticle-based drug delivery, though the concepts also apply to nano–bio interactions in general. We discuss current limitations and how they might be overcome, particularly for future use in vivo.
Keywords: nano−bio interface, X-ray techniques, synchrotron radiation, imaging, nanoparticles, delivery, degradation, spectroscopy
Several drugs on the market consist of more than just a homogeneous pill for oral delivery or the injection of a solution for intravenous administration, but are intrinsically heterogeneous, comprising multiple compounds. In the case of nanoparticle (NP)-based drugs, they usually involve a particulate delivery vehicle and a pharmaceutically active compound, which is to be delivered to and released at the target site. This concept reaches back decades,1 well before the actual term “nanomedicine”, under which such NP-based drugs nowadays are referred, was coined. Examples include Abraxane, where the pharmaceutical therapeutic paclitaxel is delivered via protein carriers,2 or Doxil, where doxorubicin (DOX) is delivered via liposomes.3 However, before reaching acceptance for clinical use, such NP-based systems (as with any drug) have to undergo rigorous clinical trials. Apart from probing the biological effects of the drug (i.e., pharmacodynamics), testing also involves characterization of the drug itself and the NP vehicle, specifically, its pharmacokinetics.
Independent of the route of administration (e.g., oral, intravenous, etc.), drugs are formulated with numerous pharmaceutical excipients to ensure the desired pharmacological function. Conventionally, absorption enhancers, emulsifiers, diluents, preservatives, solvents, sustained release matrices, etc., are used to shape the pharmacokinetic profile of a drug.4 At the dawn of nanotechnology, novel and fascinating opportunities for drug formulations emerge. For instance, it has been demonstrated that the therapeutic window of approved drugs will dramatically expand, if dissipated into a nanocrystalline formulation.5 Far beyond established methodologies, nanotechnology enhances the targeted delivery and controlled release onto the cellular and even subcellular levels, hence emulating fundamental biological mechanisms, such as cell–cell or cell-messenger interactions. Albeit much more sophisticated, nanoformulations have to meet the same regulatory requirements of pharmacovigilance as conventional drugs with respect to effectiveness, efficacy, safety, and benefit.6 However, due to the cardinal differences compared to conventional drugs, improved methods are urgently needed to monitor the adsorption, distribution, metabolism, excretion-toxicity (ADME-T) profile of a NP-based drug.
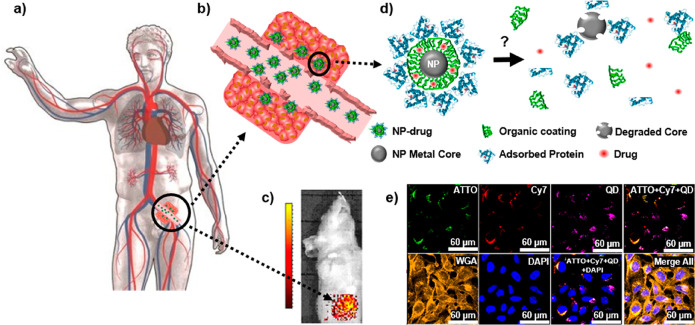
Figure 1 depicts a scenario in which a hypothetical patient is being treated with an intravenously injected NP-based drug. We describe the NP-based drug with a model system composed of the NP carrier matrix/vehicle, which is modified on its surface with a coating that provides colloidal stability, could help to minimize immune response, and might also carry targeting ligands, and an encapsulated or appended pharmaceutical compound.7,8 However, such a NP-based drug is not a static assembly and, after its administration to the human body, may undergo changes in composition.9 The fate of the NP-based drug over time thus becomes an important consideration. As expected, initially, a part of the NP-based drugs will be cleared from circulation by the immune system and end up in the liver and spleen, while some fraction will arrive at the actual target site, such as a tumor.10,11 On the route to reach the target site, however, there are several scenarios that can render delivery less efficient. For example, the surfaces of NP-based drugs may be overcoated due to the formation of a protein corona12 or may suffer enzymatic degradation of their targeting moieties.9 Ultimately, these surface changes may worsen the capability of the NP-based drug to achieve active targeting, which depends on ligand density.13 If all targeting ligands are cleaved, there will no longer be active targeting. Even partial cleavage may have dramatic effects on targeting, if multivalent interactions play roles in target recognition.14 Furthermore, degradation of the surface coating15 may also lead to the agglomeration of the carrier NPs and thus potentially induce clogging of the blood vessels.16 Besides agglomeration, degradation can lead to significantly altered chemical surface structures with respect to polarity or charge. These changes, in turn, will impact in the biodistribution of a NP-based drug by altering its solubility, ability for serum protein binding, and membrane permeability. Finally, degradation of the carrier NP vehicle in the blood may also lead to the loss of the pharmaceutical compound before it actually reaches the target site. In this case, there would be no biological effects as a result of treatment with the NP-based drug, as only the empty NP carrier, but not the embedded pharmaceutical compound, would reach the target site. Thus, as different parts of NP-based drugs degrade over time, following in vivo the fate of their individual components is important to understand what is exactly delivered to the target site. Equally important is understanding the efficacy of the targeting strategy chosen to enrich the NP-based drug in its target and the delivery efficiency of the drug from the NP carrier once it has reached it. Again, if the intact NP carrier is not capable of accumulating in the areas that need to be treated (e.g., organs or tumors), or the embedded therapeutic drug cannot be released when required, the overall therapeutic activity will be compromised.
Figure 1.
(a) Sketch of a hypothetical scenario in which nanoparticle (NP)-based drugs are administered intravenously for the purpose of cancer treatment. (b) In leaky tumor tissue, some NP-based drugs may be retained by passive targeting.10,11 But do these drugs penetrate effectively into the tumor tissue or remain at the tumor surface? (c) Fluorescently labeled NP-based drugs can be imaged in vivo in animal models, but spatial resolution is typically too low and does not allow conclusions about the distribution of the NPs in the tumor tissue. Reprinted with permission from ref (13) under a CC BY-NC-ND 4.0 International License. Copyright 2016 Nature Research. (d) Inside the human body, the NP-based drugs, comprising a NP carrier/vehicle (gray), an organic surface coating including ligands for targeting (green), the drug to be delivered (red), and a corona of adsorbed proteins (blue) may degrade, which can completely change the NP properties. (e) Example of in vitro degradation of a model for a NP-based drug after endocytosis by cells. Here, the NP-based model drug used can be identified in different compartments by their fluorescence (shown in false colors in the overlay of different fluorescence channels): CdSe/ZnS quantum dots (QDs) (purple) as NP carriers, ATTO-labeled polymer shell around the QDs (green), representing the ligands on the NP surface, and preadsorbed Cy7-labeled proteins (red) symbolizing a drug to be delivered. As shown in the image, after exposure, the location of the different compounds of the NP-based model drug can be mapped by fluorescence imaging. Data from Carrillo-Carrion et al. demonstrate that the three components of the NP (QD core, polymer shell, protein corona) disintegrate over time, as colocalization is partly lost.17 However, fluorescence imaging is not ideal to follow all the different parts of the NP-based drugs. For example, Cd ions released from the CdSe/Zn core cannot be directly imaged based on fluorescence. Reprinted with permission from ref (17). Copyright 2019 American Chemical Society.
Moreover, once at the target site, NP-based drugs need to penetrate into the tissue and typically get internalized by cells (usually through endosomal pathways), where the pharmaceutical component is supposed to be released into the cytosol. Therefore, targeted drug delivery and controlled drug release represent important parts of drug delivery. Keeping in mind that only a small fraction of the drug or the NP carrier will reach the target and that the drug may cause side effects in other organs, ensuring stable encapsulation that only releases the toxic drug/pharmaceutical agent at the target site would be advantageous. Following this approach, the drug would be released from the NP carrier once the carrier reaches the target tissue, and thus the therapeutic effect would be limited to the immediate vicinity of the target cells, with maximum efficacy. Several strategies have been developed for controlled release, including site-activated release, for example, based on environment sensing mechanisms or by triggering heat dissipation from magnetic NPs embedded in liposomes. For environment-sensing drug release, several approaches based on pH-dependent release have been reported, premised on the observation that pH levels can be different in pathological (e.g., inflamed or infected) tissues or cells. Lipid structures can be made responsive to pH changes, so they are ideal for environment-sensing delivery systems, both for extracellular as well as intracellular release. To understand the release of the pharmacological agent from the NP carrier vehicle, the localization of each of the components needs to be mapped with subcellular resolution. The localization and transformations of the different units of the NP-based drug are important for its biological activity. For example, pharmaceutical compounds can lead to completely different cellular effects depending on their intracellular location (e.g., therapeutic agents acting at the mRNA level would be ineffective if they are confined to endosomes/lysosomes and not released to the cytosol, where the mRNA is located).18 Also, metal cores (e.g., for NP carrier vehicles) may undergo a variety of transformations once internalized by cells/tissue, which might be vital to understanding the activity or potential risks of NP-based drugs due to NP-induced toxicity (e.g., release of Ag+ when using Ag-based carriers).19−24 Thus, in order to understand the effects of NP-based drugs, biological responses need to be correlated to the intracellular locations,25 speciation, and behavior of the different compounds of the NP-based drug.
However, it is difficult to obtain information about the structural rearrangements in the delivery system while in a liquid environment. Small-angle X-ray scattering (SAXS) has been used to study these processes and is now an essential tool to gather information and to fine-tune lipid drug release systems.26−29 Alternatively, drugs can be released from liposomes that incorporate sphingomyelin in their membranes. If such sphingomyelin-liposomes are in contact with the enzyme sphingomyelinase (expressed by stressed cells in tumors or inflammatory tissue), the lipid molecule is broken down into its constituents, ceramide and phosphorylcholine. These molecules no longer have an optimal fit, so the liposome membrane becomes more permeable, and the contents of the liposomes are released. X-ray-based techniques can be used to study these microdomain changes for optimized drug release. External activation methods using magnetic fields have also been developed as an additional means of controlled drug release.30 Here, ferric NP-containing liposomes are used. Once enough drug carriers have accumulated at the target site, an external alternating magnetic field is directed at the desired location and membrane disruption induced from local heating or by mechanical actuation results in a leaky liposome membrane and subsequent drug release. The synergistic effect of the enzyme and the applied magnetic field induces a more potent and selective release than a single approach alone.31,32 Again, for the selection of optimized ferric NPs and monitoring conformal changes leading to the release of the drug, X-ray-based analyses are most valuable. In order to study the process, different markers can be attached to the carrier (to monitor delivery) and the drug (to monitor release).
The examples presented here illustrate the potential of NP carriers with mechanisms for controlled release. Related alternative approaches have been reviewed recently.33 The development of improved luminescent NPs is of interest for targeted drug delivery as well. Photoactive NPs that respond to near-infrared (NIR) excitation could be used for spatially targeted delivery, as NIR light penetrates tissue to greater depths than shorter wavelengths with considerably less phototoxicity.34,35 Upconverting NPs (UCNPs) make use of energy-transfer upconversion between neighboring lanthanide ions to convert tissue-penetrating NIR light efficiently to visible or UV light. Targeted UCNPs can be used to release therapeutics through UV-based uncaging strategies, creating highly localized light sources of wavelengths that might otherwise be cytotoxic.36−38 The stable optical response of these NPs also enables extended tracking39,40 as well as sensitive detection of cellular fluctuations in response to temperature and pressure.41−43 Moreover, UCNPs have been shown to excite retinal opsins to endow mice with NIR vision44 as well as to power optogenetic switches in awake mouse brains,45 suggesting that even more complex therapeutic strategies are possible with these NIR-responsive NPs. For each of these NP-based approaches, understanding the fate of NP-based drugs in biological organisms is of vital importance to improve their potential medical applications.
All of these aspects require efficient imaging or spectroscopic tools to study the behavior of NP-based drugs in situ in biological environments or whole biological samples such as small animals, allowing direct detection of the different components of the NP-based drugs (to overcome the loss of information as a result of their degradation) and, more importantly, providing high spatial resolution. In mapping the fate of NPs, it is desirable to access different size scales, ranging from subcellular resolution to full organisms (i.e., humans or small animals). Therefore, if a single analytical technique is to be used, it should be capable of changing magnification, to provide optimal spatial resolution according to the requirements of the sample to be analyzed.
The functionalization of NP surfaces is critical for targeting therapeutics to organs and cells of interest and for minimizing toxicity and off-target effects. Intrinsically hydrophobic NPs must be transformed into being hydrophilic by surface functionalization, and this combination of aqueous surface passivation and biomolecule targeting determines the interactions of the NPs with living systems. While many NP–biomolecule complexes have been assembled by nonspecific or reversible linkages, the challenge of maintaining the stability of these complexes within tissue has spawned a number of bioconjugation strategies to attach proteins and organic ligands covalently to NP surfaces, with controlled stoichiometry.46−50 Identifying a suitable labeling approach for deciphering the degradation processes occurring in the NP-coatings while keeping the efficiency of targeting or delivery of the drug is not straightforward. The label needs to be stable (to limit degradation throughout the course of the study) and detectable with analytical techniques of high sensitivity and resolution. But most of all, it needs to be modular, in the sense that it should allow chemoselective conjugation to the region of interest, for example, by immobilization on metallic NPs or covalently linked to organic coatings on the surface of the NPs (which helps to understand degradation and targeting) or to the drug embedded (which provides vital information about delivery to the target).51 In this context, bifunctional metal chelators such as 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid and its derivatives enable the introduction of various labels as tracers into the coating of NPs and may thus be used for detailed analysis of degradation processes.52 This includes radiolabels compatible with positron emission tomography (PET) or single photon emission computed tomography (SPECT) imaging,53 nonradioactive heavy metals compatible with magnetic resonance imaging (MRI) and NIR fluorescence, or elemental labels with high contrast in inductively coupled plasma mass spectrometry (ICP-MS) and X-ray detection (such as X-ray fluorescence imaging, XFI).54−57 Nevertheless, any modification of the structure of a drug might alter its therapeutic activity and biological behavior, an important consideration that must be understood and considered before deciding upon one or more labels.
In addition to using NPs alone as drug delivery vehicles, it is also possible to integrate NPs within different types of materials or as components of gels. Integration of NPs within hydrogels allows improving the mechanical, biological, and chemical properties of the material. One example of this strategy is the integration of nanosilicates, such as Laponite, within polymers that form shear-thinning hydrogels. Such nanosilicates have charge distributions that allow them to interact with gelatin polymers by electrostatic interactions forming bonds that can be broken upon application of force. The incorporation of Laponite within gelatin has been shown to form hydrogels that can be used for various regenerative applications as well as medical devices for embolization.58 These hydrogels have shown great potential; one advantage is their ability to be delivered through catheters and utilized in minimally invasive manners. Unfortunately, some of these materials are not radio-opaque by themselves. In such cases, the addition of contrast agents such as tantalum can make these hydrogels visible to X-rays and thus suitable for computed tomography (CT) scans and other X-ray techniques. Integration of nanomaterials with non-invasive imaging approaches provides opportunities to address a number of issues such as internal bleeding, aneurysms, and other medical problems. Ultimately in situ and in vivo analyses will be most valuable.
While with current technologies, in situ analyses in humans are not yet feasible, we discuss in this Review current developments toward this goal and obstacles still to be solved. We start first with a description of the state-of-the-art techniques, followed by a forward-looking analysis of the possible improvements in the near term.
Selected Current Techniques to Study Nanoparticle-Based Drugs in Biological Environments
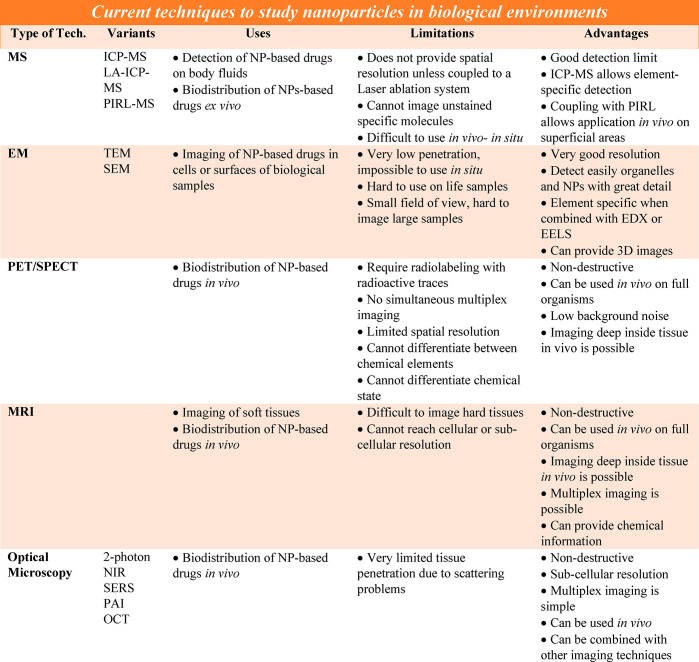
Over the past century, analytical chemistry has developed a large set of methodologies for the detection of molecular species in biological settings (Table 1). From samples of body fluids, in particular blood and urine, degradation and clearance of the different components of a drug can be tracked, using, for example, mass spectrometry (MS).59−62 Though not in situ, body fluids can be extracted at different time points from the organism, providing important details on the pharmacokinetics of drugs. However, as detection is performed in blood, urine, etc., spatial resolution is lost. This strategy results in it not being possible to know the concentrations of drugs and their metabolites in the specific organs where a drug is being degraded. Often, to acquire spatial information, animals must be sacrificed, and analytics performed ex vivo on the dissected pieces, which is essentially the opposite of in situ detection. Still, MS is a convenient technique to record the biodistribution of drugs ex vivo. Different components of NPs and loaded pharmaceutical agents can be labeled to enable element-specific detection with ICP-MS,52,63 and different organic ligands and pharmaceutical compounds can also be directly detected by MS.64 This strategy has been applied in models of NP-based drugs, in which biodistributions of metal-containing NPs have been recorded ex vivo.15,65,66 Recent progress now also allows applying MS in vivo, by using picosecond-infrared-lasers (PIRL) for scar-free minimally invasive surgery and MS analyses of the ablated tissue.67,68 This advance has led to the development of MS microscopes.69,70 While MS microscopes enable detailed analyses of molecular species, and thus imaging of the different parts of NP-based drugs, in situ-in vivo analyses of NP-based drugs deep inside tissue remain complicated. For example, it is straightforward to probe the NPs with the surface region of tissue fragments using PIRL-coupled MS methods, but reaching tumors deep inside a tissue with the laser, followed by in situ extraction of tissue fragments for mass spectrometry analyses (of the presence of NPs), has not thus far been demonstrated.
Table 1. List of Current Techniques Commonly Used to Study the Biodistribution or Fate of Nanoparticle (NP)-Drugs in Biological Samples.
Alternatively, electron microscopy (EM) techniques such as transmission electron microscopy (TEM) and scanning electron microscopy (SEM) allow imaging cells with extremely high resolution. Moreover, experimental approaches such as cryo-electron tomography coupled with focused ion-beam milling provide three-dimensional (3D) images of organelles and large protein structures inside cells.71−73 While the development of liquid-phase supports for EM allows studying samples over time in biologically relevant environments,74 it is not yet clear whether imaging live cells will be possible.75−77 Finally, EM techniques also permit direct detection of metals with adequate sensitivity through spectroscopic methods such as energy dispersive X-ray analysis (EDX) and electron energy loss spectroscopy (EELS).63 However, EM is not capable of imaging deep inside tissues and cells. Although it can be possible to image thin parts of intact whole cells (i.e., 500 nm) using cryoEM (at a voltage of 300 kV), EM traditionally requires extensive sample preparation (mostly using thin sections of fixed cells), making in situ usage impossible.
Visualization of NP-based drugs can be accomplished in situ via different standard medical imaging techniques. A comprehensive overview of techniques suitable for imaging of NP carriers is beyond the scope of this Review, but a number of relevant recent reviews can be recommended.78,79 In particular, MRI and PET/SPECT enable performing analyses on small animals and humans. Both PET and SPECT require radio-labeling of the different NP components with different radioactive traces (e.g., one for the pharmaceutical agent and one for the matrix of the carrier vehicle) in order to follow the fates of the different entities by performing different (multiplexed) imaging experiments. Imaging deep inside tissue in vivo is possible, but spatial resolution is limited. Nevertheless, only information about the location of the radio-labeled compounds can be obtained, but not about their molecular state (e.g., 64Cu or 64Cu2+ cannot be distinguished, excluding for example the identification of changes in the surface chemistry of Cu NPs). Imaging deep inside tissue with up to near-cellular-level resolution (below 10 μm) can be achieved with MRI.80−82 Element-based MRI diagnostics (e.g., Gd3+, Mn2+)83,84 and NP-based MRI diagnostics (e.g., MnO, Fe3O4)85,86 can avoid the use of radioactive elements and have been used to monitor signal changes of NPs within tumor mass. By using functional MRI, local chemical information can be provided in some cases.87 Multiplexed imaging is possible by using different elements (1H, 13C, 19F, etc.). However, the diagnostic outcome of MRI is also closely dependent on the resolution of MRI equipment. In addition, there can be nonlinear signal responses to concentration of agent and endogenous changes in contrast that can create uncertainty in the measurements, and signal intensity and imaging resolution are not yet sufficient to reach subcellular levels.88
While scarcely used in clinics, optical imaging (fluorescence and bioluminescence) is one of the most commonly used imaging techniques in preclinical settings, in particular for in vitro imaging of cell/tissues (both fixed or live) and for in vivo imaging of small animals, due to its low cost, rapid throughput, and multiplexing ability. However, we note that it is not highly accurate for determining biodistribution. Other than the localization of fluorescently labeled molecules/structures, it can also provide information about the local microenvironment by using analyte-sensitive fluorophores.89 Multifunctional NPs have been developed as biocompatible probes of external stimuli, such as force sensors. For example, ceramic NPs doped with lanthanide ions have been widely used as temperature, electric field, and pressure sensors for MRI (with gadolinium) and for biomarker detection using their upconversion.90,91 In principle, fluorescence imaging is also possible in humans, though there are many limitations, primarily that it is limited to imaging structures <1 cm from an endothelial surface.92 Fluorescence allows tracking of pharmaceutical agents and NP carriers by measuring organ distribution and subcellular localization. Commonly used NIR fluorophores (e.g., IR780, chlorin e6) enable monitoring the changes of NP-based drugs in tumors over time and can also be combined with photothermal or photodynamic therapy to integrate cancer diagnosis and treatment.93−95 The above-mentioned lanthanide dopants, besides facilitating upconversion (i.e., when irradiated with NIR light, emitting in the visible), can also be used as markers for X-ray fluorescence or nanoscintillator-driven photodynamic therapies.96−98
Fluorescence can be detected with spatial resolution (i.e., recording images) down to the level of single molecules and also with temporal resolution (i.e., enabling fluctuation-based correlation analyses such as fluorescence correlation spectroscopy, or in combination with spatial resolution, enabling the recording of movies). Although the standard diffraction-based resolution limit of light is a few hundreds of nanometers, super-resolution and near-field approaches have pushed recordings to spot sizes of only a few nanometers.99−102 Exceptional spectral resolution permits a multiplexed recording of the fluorescence originating from different fluorophores,17 see Figure 2. Finally, changes in the local environment of the imaged drug can also be detected, both qualitatively and quantitatively, by using analyte-sensitive reporter fluorophores. The use of fluorescence-based analytics is therefore a powerful methodology, but not without its practical limitations. Maybe the greatest obstacle for its use in many “real” samples is light scattering, see Figure 3.
Figure 2.
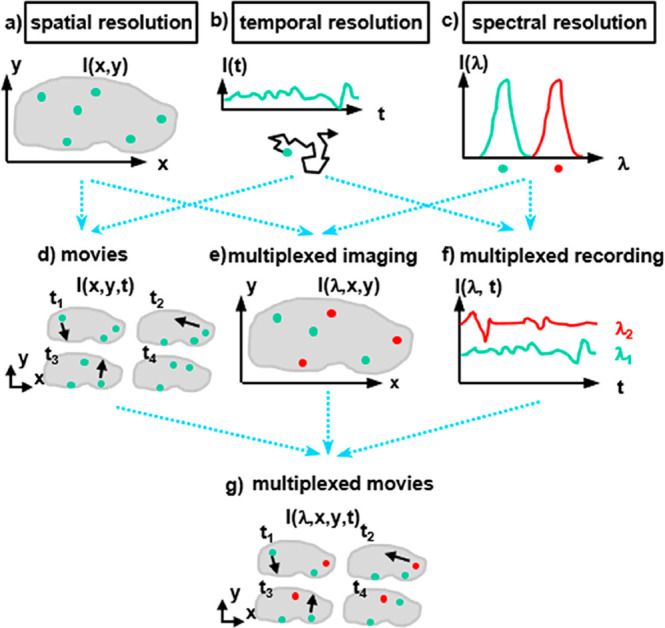
(a) Spatial resolution allows recording monochromatic images, such as the distribution I(x,y) of one type of fluorophore within one cell. (b) Temporal resolution allows recording intensity fluctuations I(t) at one point, which with correlation analysis enables diffusion measurements of fluorophores. (c) Spectral resolution permits discrimination of the fluorescence I(λ) of fluorophores emitting at different wavelengths, λ. (d) Spatial and temporal resolution taken together allow recording of monochromatic movies, such as the movement of one type of fluorophores within one cell. (e) Spatial and spectral resolution taken together allow recording multicolor images, such as the distribution of different fluorophores in one cell. (f) Temporal and spectral resolution together enable multiplexed recording of intensity variations of multiple fluorophores. (g) Taking spatial, temporal, and spectral resolution together makes it possible to record multicolor movies, such as recording the movement of multiple different fluorophores in one cell.
Figure 3.
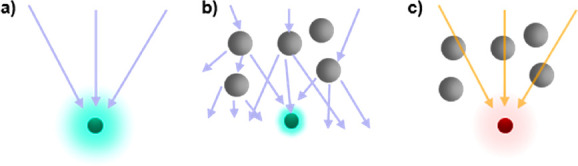
(a) Visible light can be focused to a fluorophore. (b) In the case of scatterers in the path, the focus is diffused, limiting spatial resolution as well as the intensity that arrives at the fluorophore. (c) In case wavelengths are used at which no scattering occurs with the intermediate material, the illumination path would remain unaffected.
For fluorescence measurements, a fluorophore needs to be optically excited, and the resulting fluorescent emission needs to be recorded with a detector. To achieve spatial resolution and to maximize the excitation probability, the stimulating radiation should be focused as tightly as possible to the position of the fluorophore (see Figure 3a). However, in many “real” samples, the incoming beam encounters obstacles in the form of light scattering. This scattering limits focus, leading to reductions in the spatial resolution and effective excitation probability at the target (see Figure 3b). Consider the fluorescence of a fluorophore in water versus milk. It is relatively simple to record the emission of a standard fluorophore in water; however, milk is an emulsion and scatters light, thereby appearing opaque to visible light. This scattering affects both the excitation and emission, hampering fluorescence measurements. The interiors of cells growing as two-dimensional (2D) monolayer cultures can be probed using fluorescence imaging. These measurements can be used, for example, to detect specific molecules using immunostaining103 or to track individual molecules.104 However, the same measurements would no longer be possible if the cell to be probed lay 1 mm deep inside three-dimensional (3D) tissue. As such, light scattering is a fundamental problem to acquire fluorescence measurements in tissue and full organisms efficiently. Visible light is strongly scattered by both biological tissues and inhomogeneous fluids, such as blood. The overall absorption of the light used for irradiation as well as the emission from the fluorophores by the different components of the sample hampers the useful penetration depth of optical microscopy. Penetration limits of optical imaging can be avoided by combining it with intraoperative or endoscopic procedures, yielding imaging methods that have already been translated for use in humans, but are highly invasive, as most require surgical intervention. Alternatively, tissue penetration issues caused by absorption and scattering can be partially solved by working in the so-called spectral “biological window”,105 which designates the reduced scattering of light by tissue in the NIR region. Therefore, scattering and absorption effects can be reduced by shifting the wavelength of the beam used for the irradiation of a sample from visible to NIR light.106 Alternatively, two-photon microscopy uses longer wavelength photons to excite fluorophores, achieving deeper penetration. As such, two-photon intravital microscopy enables important in vivo and in situ insight into fundamental biological processes (e.g., immunological responses) occurring 100–200 μm deep within animals.107−109 While two-photon imaging techniques also suffer from optical scattering, advances in modern image analysis, applying effective and adaptive Fourier filtering algorithms (FFA), promise to improve image quality and analyses.110,111 Despite these advances, acquisition of high-resolution optical recordings deep within tissue is not possible using existing methodologies. One could dissect the specimen, but that would exclude in situ and longitudinal detection. While such ex vivo analyses can be done in animal experiments, it is ruled out for most clinical applications in humans. Thus, tissue effectively remains nontransparent to analytics based on visual light optics.
Raman microscopy presents an alternative and a complement to the methods discussed above. Although Raman microspectroscopy has been used frequently for the characterization of tissues and cells,112 the intrinsic low Raman cross sections of NPs, especially with infrared light, limit its applicability in bioimaging. The intensity of conventional Raman scattering, however, can be enhanced by many orders of magnitude when the target molecules are located close to plasmonic surfaces, giving rise to the so-called surface-enhanced Raman scattering (SERS).113 Whereas most applications of SERS have been directed to the analytical detection of molecules at low concentrations, direct SERS spectroscopy has also been used in the classification and characterization of tissues114 and cells.115,116 The intrinsic complexity of these biological samples makes the spectral output of the direct use of SERS difficult to interpret. Thus, as an alternative to direct SERS, labeled particles known as SERS tags or SERS-encoded NPs have been developed specifically for imaging. Such particles typically comprise a metal (Au or Ag) NP core, on which molecules with high SERS cross-section are adsorbed. Then, the particle is protected with an oxide or a polymer material that, in turn, can be functionalized with targeting biomolecules.117 The SERS tags can be used for bioimaging, with the advantage that the signal can be readily excited with NIR lasers, circumventing the photodegradation common to visible lasers.118 Applications in vivo have been reported using fiber optics-based illumination and signal collection, which enables acquiring images as deep as a few millimeters (or even centimeters) within the subject, but only with moderate spatial resolution.119,120 Although high-resolution imaging is more challenging, recent reports demonstrate the possibility of using confocal SERS to classify cell types, both in 2D121 and in 3D122 cell co-cultures. Additionally, as SERS is a surface active methodology, changes in the local environment of the metal NP can be detected, such as conformation of adsorbed proteins.123−125 Thus, SERS potentially enables one to follow the fate and degradation of NP-based drugs, also in vivo.126 Nonetheless, resolution and penetration depth are hindered by the same issues discussed for fluorescence imaging above, while acquisition times for SERS images are still typically much slower than those for fluorescence.
Photoacoustic imaging (PAI) is another optical imaging modality to monitor NP-based drugs, labeled macromolecules and/or cells, combining the sensitivity of fluorescence imaging with the high spatial resolution of ultrasound imaging. This method measures the echo waves initiated from the heat generated by a laser beam and subsequently thermo-elastic expansion of the tissue and is capable of greater spatial resolution when imaging NPs deep within tissues compared to fluorescence imaging.127 For instance, PAI has been widely utilized to investigate the distribution of different Au NPs.128 The strong localized surface plasmon resonance (LSPR) effect in Au NPs enables tunable photoacoustic absorption in vivo. Alternatively, a series of protein nanostructures filled with gas generated by some microorganisms can be used as PAI contrast agents and enable to probe macrophage phagocytosis and lysosomal degradation in the liver of living animals.129−131 Single-walled carbon nanotubes (SWNTs) can also be monitored in vivo with a wide PA absorption spectrum without specific peaks. Other suitable candidates for PAI include NPs loaded or labeled with organic photoacoustic contrast agents, such as cyanine-based dyes, melanin, and porphyrin.132 Similar to fluorescence imaging, the NIR window (780–900 nm) and the second NIR window (900–1700 nm) are optimal for in vivo applications in order to avoid laser absorption from endogenous agents such as hemoglobin.133 Multiplexed PAI enables the quantification of signals from NPs, oxygenated hemoglobin, and deoxygenated hemoglobin separately, but remains a limitation for PAI. Thus, PAI has great potential for use in investigating pharmacokinetics, biodistribution, stem cell homing, metastasis dynamics, etc. Presently, PAI is used mostly in research laboratories to follow blood flow, plaque formation in blood vessels, and blood vessel elasticity. Instruments approved for clinical application are not yet available, limiting the widespread use of this method.
Optical coherence tomography (OCT) is a non-invasive optical imaging technique that may be used in combination with PAI to measure the time delay from photons backscattered by samples irradiated with low-coherence NIR or visible light.134 This method is widely used in vivo, especially in the eye, and measures the morphology of tissues with millimeter penetration depth and micrometer resolution.135−137 NPs can be used as contrast agents for OCT.138−141 Therefore, OCT might also help to detect NP-based drugs in biological environments and can be used to probe the biodistribution and/or behavior of such nanomaterials in vivo.142
Due to the noted limitations of the above-discussed imaging techniques, especially with respect to penetration depth, there are clear needs for the development of further methodologies for the in situ and in vivo analyses of NP-based drugs. In the following section, the use of X-ray-based analytics to characterize and to image NP-based drugs will be discussed as methodologies with significant potential in this context. Capabilities for such measurements in situ in complex biological environments such as blood, in vitro in cells and tissue, and ultimately in vivo in animals and humans will be outlined. While this Review focuses on NP-based drugs, the concepts discussed herein apply to the characterization of nano–bio interactions in general.
X-ray-Based Techniques As an Alternative to Study Nanoparticle-Based Drugs in Biological Environments
Visible light is an electromagnetic wave within a specific range of wavelengths λ. Scattering imposes limits to visible light-based imaging, but scattering is highly wavelength dependent (λ–4) and therefore can be reduced by shifting the optical excitation from the visible to the NIR.143−145 Alternatively, it is possible to reduce the scattering of the incident light further and to achieve deeper tissue penetration by shifting fluorescence-based methodologies to a different spectral range, such as X-rays. Standard X-ray projection imaging in a physician’s office allows visualizing bones deep inside the body in contrast to surrounding soft tissues and is also used clinically in hospitals in the form of computed tomography (CT).146 X-ray-based medical imaging remains by far the most commonly used method, exceeding the use of all other imaging techniques combined. This technique is based on the differential X-rays attenuation in different organs, which depends on the elemental composition (mostly O, C, H, and N for biological tissue, but bone contains high quantities of heavier Ca) and tissue density.147
Such dramatic changes in wavelength alter the fundamental interactions of light with matter, as many phenomena, such as absorption, fluorescence emission, and scattering, are wavelength dependent. For example, X-rays can carry enough energy to excite electrons located in orbitals at the inner shells of heavy elements, whereas optical techniques normally study electronic transitions only between valence orbitals. Therefore, X-ray-based techniques such as X-ray fluorescence (XRF),148,149 or for in situ/in vivo, also called X-ray fluorescence imaging (XFI), or X-ray absorption spectroscopy (XAS)148,150,151 enable direct detection of the different components in NP-based drugs (Table 2). Due to the characteristic discrete electronic levels of different elements, XRF is element specific, and thus multiplexed detection is possible.148,149,152 Furthermore, as the chemical environment also changes the electronic levels, information about the electronic and chemical states of the elements under study can be obtained by using XAS.150,151,153 As X-ray scattering occurs on the basis of the electron shells of atoms, the sizes of labels reduce to the sizes of individual atoms. However, working with biological samples requires consideration of radiation-induced toxicity, which is a particular concern with X-ray radiation.
Table 2. List of X-ray Imaging, Spectroscopy, and Scattering Techniques Commonly Used to Study the Biodistribution or Fate of Nanoparticle (NP) Drugs in Biological Samples.
X-ray-based techniques may fully or partially solve some of the problems of fluorescence-based analytics and constitute a valuable alternative to optical imaging for the study of NP-based drugs in situ (Table 2). This set includes a number of spectroscopic techniques such as XRF, XAS, and X-ray emission spectroscopy (XES), among others. There are also scattering techniques, such as powder X-ray diffraction (PXRD), total X-ray scattering, or SAXS imaging,154 X-ray phase contrast imaging techniques,155−157 and X-ray photon correlation spectroscopy (XPCS).158−162 These techniques could be used to probe nanomaterials directly, including NP-based drugs, and to provide important information about their location, quantification, state, and supramolecular arrangements.
In fact, X-ray-based techniques are frequently used to characterize NP syntheses and properties. We first start with a short summary on how such techniques are used to characterize NPs under laboratory conditions (e.g., dissolved in water), for example, to monitor their synthesis and assessing their materials’ properties. Based on this information, prospects for extending such approaches to “biological” environments will be discussed.
Detailed analyses of PXRD data for NPs can provide useful insights into size, shape, crystal structure, crystallinity, and sample purity.163 Amorphous or poorly crystallized components of a sample are not detectable by PXRD, so NPs that do not have a crystalline core will not be observed with diffraction. The most interesting properties of NPs are caused by quantum confinement as a result of their small sizes. But it is precisely these small sizes that make it challenging to obtain a solid and reliable characterization of the synthesis of NPs in situ. The peaks in PXRD patterns of NPs get considerably broader as NP size decreases, leading to lower signal-to-noise ratios and making it challenging to identify phases and to evaluate purity. TEM and SEM, optical measurements, nuclear magnetic resonance, or MS techniques are generally used to obtain information about size, composition, and chemical environment of NPs, and these techniques can be used in conjunction with PXRD to provide additional insights into the characteristics of samples. However, despite their suitability to study NP samples, there remain considerable limitations with regard to their applicability to in situ studies (i.e., including extensive sample preparation requirements, of specific solvent requirements or vacuum conditions, low sensitivity, or incapacity of separating between the different populations found within the growing particles). A wide variety of X-ray-based techniques have been used to characterize the structure, composition, size, or aggregation state of NPs (e.g., SAXS, wide-angle X-ray scattering (WAXS), XRF, XAS, PXRD, pair distribution function (PDF), and X-ray photoelectron spectroscopy (XPS)).164 In most cases, the small X-ray scattering cross sections of NPs impose the need for synchrotron-radiation-based techniques. Despite the possibility of characterizing NPs of different types, the dynamic character of synthetic processes leads to the need for in situ studies. Control of the syntheses of NPs, from their nucleation and growth to the attachment to other NPs or conjugation with biological ligands/pharmaceutical agents, would benefit heavily from monitoring the reactions involved in situ. These measurements would enable not only observing the evolution of the NPs in real time but also obtaining data without any disruption of the initial structure. The breakthroughs in this field will come from the possibility to combine different characterization techniques to study the synthesis and properties of NPs in their different stages. Indeed, the use of microfluidic devices to perform both synthesis and in situ characterization offers enormous potential. At this point, it is critical to consider the interface of the reaction container. Inexpensive, X-ray-transparent polyimide windows are suitable for in situ X-ray characterization, for example, by means of SAXS, WAXS, XAS, or PDF. However, other promising techniques, such as XPS or MS, require vacuum conditions. Therefore, while the use of flow reactors is a reality, combining multiple interfaces that allow the successful application of several X-ray-based techniques at different time windows of the reaction represent real challenges in this context.
These techniques could also be applied to a wide variety of NP-based drugs. Conversely, the identification of ideal drug delivery systems to study under X-ray techniques can facilitate the use of existing infrastructure to observe nano–bio interactions that previously could only be inferred. For example, crystalline materials such as liposomes, metal NPs, and metal–organic frameworks (MOFs)-based NPs have distinct X-rays scattering profiles. In particular, MOFs can also be loaded with therapeutics and engineered to disassemble in acidic microenvironments, meaning that dissolution of the MOF carrier system could be monitored by SAXS. This process would not require any labels and would also give information on whether the NPs fracture to release cargo, to disassemble into crystallites, or to dissolve completely during internalization. The reverse time course (formation instead of dissolution) has been performed by looking at the formation of MOFs on the cell walls/membranes of microorganisms.165 Further, SAXS has recently been used to monitor the cellular uptake and interactions of cubic liposomes (cubosomes) in a microfluidic setup. Roughly 16 min after interacting with the cells, the cubosomes demonstrated a phase transition and evolved into hexasomes, a phenomenon that would not have been observable without X-ray techniques.166 Theoretically, X-ray absorption near-edge structure (XANES) spectroscopy and XRF can similarly be used to monitor whether the metal state changes and how the loaded drug is released, due to its proximity to the metal center. In particular, the protein corona will also dictate whether changes in the microenvironment can be observed after internalization, as demonstrated by in situ extended X-ray absorption fine structure (EXAFS) measurements of TiO2 NPs, which revealed no fine structure change upon internalization into cells from cell media.167 Other EXAFS work has also confirmed that small molecules may stay bound to NPs (maghemite) during internalization.168
The question is now how such in situ methods could be extended to in situ–in vivo measurements, that is, requiring the observation of NP samples not under test condition, but ultimately deep inside tissue. Modern development of synchrotron radiation sources allows for advanced beam properties, such as foci down to the range of a few nm,169−177 excellent coherence, and brilliance exceeding 1021 photons/(s·mm2·mrad2) at 0.1% bandwidth.178 As such, measurements deep inside tissue with subcellular resolution are potentially possible. Therefore, these X-ray techniques constitute an exciting alternative to study the behavior and fate of NP-based drugs in biological systems at different length scales. Particularly, X-ray techniques can replicate, or might be able eventually to replicate, the following types of measurements which are standard for fluorescence-based analytics:
Imaging (i.e., spatial resolution) (see Figure 2a) based on X-ray transmission, fluorescence, (coherent) scattering, diffraction, or phase contrast techniques has been demonstrated using synchrotron radiation.57,149,154,179−182 Furthermore, recently developed nanoprobe beamlines169−176,183 possess advanced X-ray optics which are capable of focusing synchrotron radiation below 50 nm, allowing the use of X-ray imaging techniques to study biological samples with subcellular resolution.
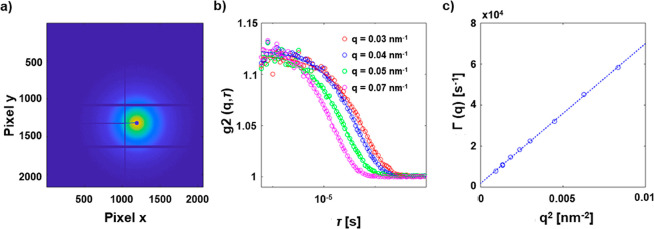
Temporal resolution is possible (see Figure 2b), in particular when using a fast 2D pixel detector running at kHz or even MHz frame rates, ideally synchronized with the bunch pattern of the synchrotron radiation source.184,185 For example, intensity fluctuation-based studies of the dynamics of objects scattering X-rays have been performed with XPCS using synchrotron radiation with highly coherent X-ray beams.186,187 The scattering pattern is modulated by an interference pattern. Changes in this pattern are correlated to the motion of the scattering and thus to its diffusion coefficient (which provides information on the size and shape of the object).158−160 The application of time-resolved techniques also allows studying the dynamics of metal centers upon photoexcitation and photoactivation.188
Spectral resolution (see Figure 2c) with X-rays is possible due to the characteristic discrete electronic levels of different elements. As such, X-ray absorption, fluorescence, and some X-ray inelastic scattering techniques (i.e., resonant inelastic X-ray scattering, RIXS) are element specific. Multiplexed measurementsof different elements such as Au, Cu, Fe, Ag, Pt, Os, etc. are thus possible.148,149,152,189−191 Spectral resolution is even more important when studying the mechanisms of action of NP-based drugs, as it permits following various species of the same element that can be generated once the NP has been administered.192 From the detector side, arrays of microcalorimeters hold potential for microspectroscopy applications using X-rays.193−196
Movies (see Figure 2d) are conceived as the combination of spatial and temporal resolution. Such experiments can be recorded using synchrotron radiation to probe biological samples.197,198 However, in general, high spatial resolution or large image areas and high temporal resolution are still mutually exclusive. This limit is mainly due to two primary factors. First, biological samples normally contain low concentrations of the NPs of interest, making it necessary to use relatively long acquisition times on instruments providing high photon fluxes to obtain high-quality images. Furthermore, many X-ray imaging techniques use scanning-based approaches needing mechanical translation of the sample during acquisition, which takes longer for higher resolution images. This limitation holds true even for full-field imaging techniques, as the size of the field of view normally affects the final resolution of the image (i.e., imaging larger fields of view leads to lower resolutions). Therefore, scanning approaches are still needed to image large areas of the sample with high resolution.
Multiplexed imaging (see Figure 2e) is possible, as different elements can be spectrally resolved using different techniques, which also allow achieving spatial resolution.199 For example, the simultaneous acquisition of maps of different elements within a single XRF scan with synchrotron radiation is a clear example of well-established multiplexed imaging.149 Furthermore, in multimodal imaging, XRF can be coupled also with other techniques such as XAS and XRD (at fluorescence microprobes), ptychography, transmission, and XRD simultaneously. Moreover, collecting images at different energies around an X-ray absorption edge permits imaging chemical states and electronic states which are compound specific. Similar to spectral resolution, detectors based on microcalorimeters may hold potential for multiplexed imaging as well.
Multiplexed recording might be possible with XRF (see Figure 2f). Parallel fluctuation analysis as recorded by XRF from different elements could be achieved using an energy-discriminating detector with high temporal resolution (kHz or more). Alternatively, XES should allow probing two orbitals within the same element with subnanosecond temporal resolution, by combining a van Hamos spectrometer200 with fast detectors.
Multiplexed movies would involve the combination of spatial, temporal, and spectral resolution (see Figure 2g). This kind of measurement using X-ray-based techniques is currently at the technically possible limit, due to multiple technical restrictions (as described above) and by the maximum biologically tolerable dose. Nevertheless, there is no fundamental physical principle that would rule out multiplexed movies, although there are tremendous practical hurdles, and at the state of the art, multiplexed movies are not yet possible.
While the aforementioned examples are structured on a conceptual basis, in the following sections we will discuss the current aspects of employing X-ray-based techniques for their potential use in analyzing NP-based drugs in situ in more practical terms.
X-ray Imaging of Nanoparticle-Based Drugs (and Related Systems) from the Subcellular to the Animal Level
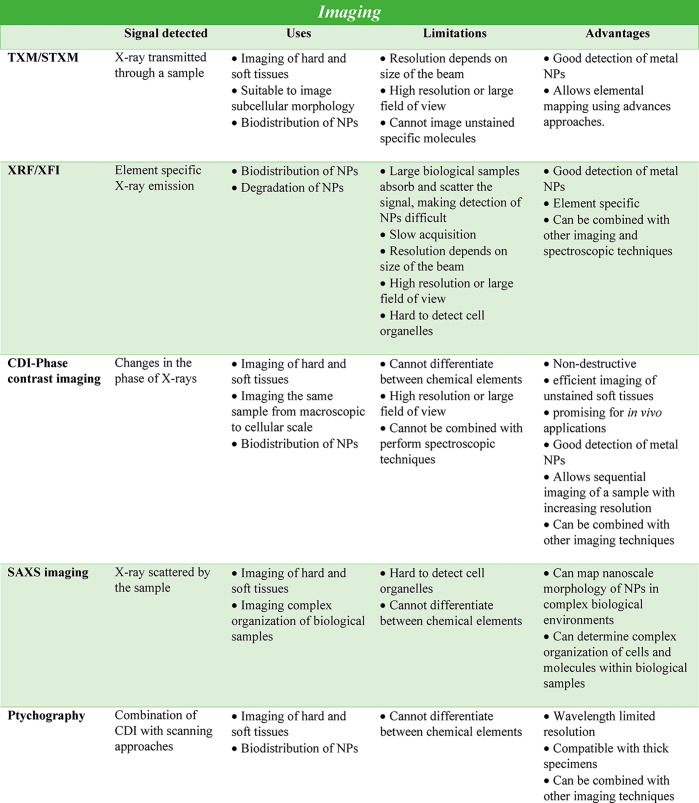
X-ray imaging has the potential to overcome the penetration depth limitation of EM and to image cellular components in fully intact cells with high spatial resolution and minimal sample preparation, i.e., imaging biological material in a native or near-native state. Different from fluorescence imaging with visible/NIR light, synchrotron radiation-based imaging methods are capable of providing element-specific and precise distribution information on NPs at subtissue, cellular, and even organelle levels as well as the morphology information on biological specimens. The key factor here is the short wavelength of the X-rays. Whereas visible/NIR light suffers from low spatial resolution of a few 100 nm due to Abbe’s diffraction limit and even super-resolution techniques thus far typically achieve only a few tens of nm spatial resolution in cells99,201,202 (or 5 nm spatial resolution in nonbiological synthetic samples),203 X-rays enable single nm spatial resolution, which is more than enough for the imaging of NP-based drugs. Also, by choosing the X-ray wavelength selectively, scattering effects by tissue can be minimized. This high resolution, however, comes with the price of potentially higher radiation damage than that caused by optical imaging techniques, which will be discussed in a separate section. Strategies for deep-tissue recordings will be discussed at the end of this section. For the majority of X-ray-based microscopies suitable for biological specimens, soft, tender, and hard X-ray wavelength ranges (which are discussed below) can be used as light sources, and each method has its own advantages and drawbacks. Thus, the applicable imaging method should be chosen depending on the desired penetration depth, spatial resolution, and contrast mechanism.204 In the case of inorganic NPs, X-ray-based imaging technologies can visualize the position and distribution of the NPs inside the cell/tissue in situ, without any further functionalization or labeling. Also, released heavy-element ions can be directly imaged. Loaded pharmaceutical compounds, however, may require tagging with atoms that provide enough contrast for the respective imaging method (unless they contain certain atoms such as Pt in the case of cisplatin). There are a variety of different methods for X-ray imaging, such as analyzing the phase contrast, absorption, fluorescence emission, or diffraction signals, etc., of different elements/NPs. From these, several X-ray-based microscopies have been developed, which enable imaging in the field of nanobiotechnology,205−207 both using soft and/or hard X-rays as light sources.181,182 Examples include transmission X-ray microscopy (TXM), scanning transmission X-ray microscopy (STXM), micro- or submicro-focused XFI, and coherent diffraction imaging microscopy (CDI)/X-ray ptychography.
X-rays are ionizing radiation that can be divided into low penetrating soft X-ray (with energies from 100 eV to 1 keV, penetrating up to a few μm), tender X-rays (energies from 1 to 5 keV), and high penetrating hard X-rays (wavelengths below 2 Å and energies above 5 keV). The interactions of photons with soft matter are dominated in the soft energy X-ray range by the photoelectric effect and in the hard X-ray energy range above 50 keV by Compton scattering. X-rays are orders of magnitude more penetrating than charged particles. The attenuation of the beam increases exponentially with the thickness of the sample and decreases with increasing X-ray energy. The intensity of the beam is attenuated by 1/e (attenuation length) after transmission of soft tissue, for example, through 30 μm (cell), 300 μm (cell spheroid), 3 mm (tumor), and 30 mm (organ) at energies of 2.4, 5.4, 11.8, and 33 keV, respectively.208 For radiography of the human chest in clinics, X-ray beams with maximum energies of 50–150 kV are used (i.e., a beam composed of X-rays with a range of energies from a minimum of about 25 kV, depending on the filtering used, up to the maximum selected). The high penetration depth of X-rays can be employed to obtain real 3D imaging by tomographic methods, usually by computational reconstruction of virtual slices from a series of projections recorded at various angles. Being tiny and sparse, NPs in a tissue can only be investigated in 3D if the local tomographic resolution approaches the size of the NP itself, which requires coherent scattering techniques such as X-ray holotomography or ptychographic tomography and small sample volumes. The sensitivity for NPs can be increased by selecting XRF contrast. When composed of heavy elements inside a light matrix, NPs generate an XRF signal that can be efficiently separated from the background signal by energy dispersive detectors. XRF tomography enables measuring intrinsic trace element distributions with parts-per-million sensitivity in cells without the need to add or to encode genetically-specific fluorescent labels. However, the spatial resolution is limited by the X-ray optics used, to about 60 nm in 3D. Indeed, XRF tomography is a raster scanning technique utilizing a pencil beam, and as a consequence, this technique is comparatively slow. The measurement of a mega voxel 3D image can easily take several hours. On the other hand, XRF tomography offers free spatial scalability. The size of the scanned volume is generally limited by the available measurement time once a suitable beam size is selected. For thick samples, the “over absorption” of the X-ray fluorescence radiation emitted from the NP inside the sample matrix is important. For Au NPs, for example, the Lα radiation (9.7 keV) would transmit soft matter of 15, 150, and 1500 μm at 99.1%, 91.5%, and 41% intensity, respectively. A sample of 15 mm would be transmitted only at 0.0015% intensity. Using the high-energetic Au–Kα1 line (68.8 keV) would allow 75% and 5.6% transmission through 15 mm and 150 mm soft tissue, respectively.208 The application of high energies for XRF microscopy requires dedicated sources, X-ray optics and detectors, and is discussed in a separate section below. At high-resolution conditions including coherent illumination, XRF tomography can be combined with ptychography, a scanning coherent X-ray diffraction imaging technique, to image the internal structures simultaneously, including organelles and distributions of trace elements within cells.209 In ptychography, the effective numerical aperture of the imaging system can be increased, resulting in higher resolution than the size of the beam and imaging the natural contrast arising from internal electron density. Larger XRF tomographies may be combined with X-ray holotomography or phase contrast tomography to locate the NP position precisely relative to the tissue structure.
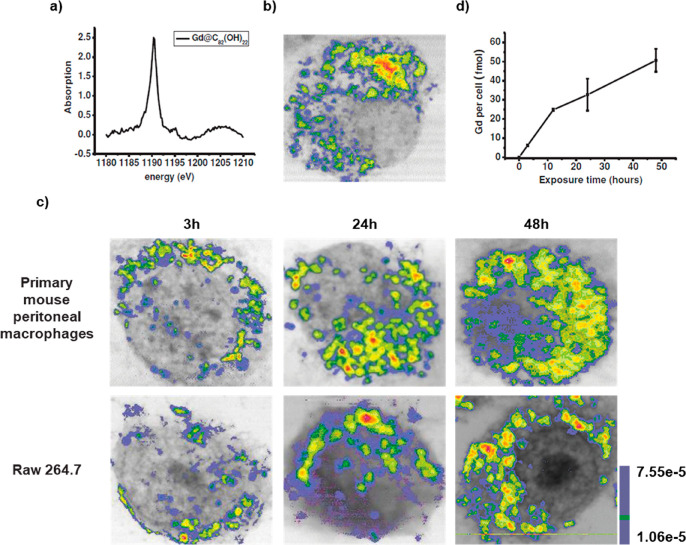
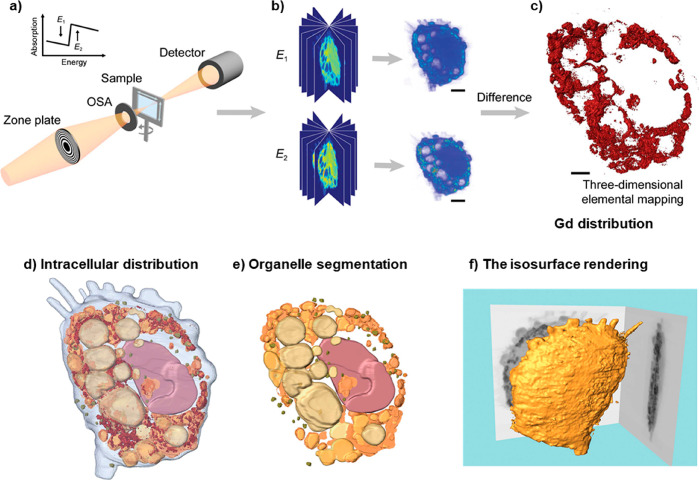
Soft X-ray-based microscopy is suitable for imaging the subcellular morphology together with the distribution of NPs/pharmaceutical agents in cells. As an example, Chen et al. used STXM to observe the continuous uptake and subcellular distribution of metallofullerenols in macrophages with 2D spatial resolution of 30 nm (Figure 4).210 Taking images below and above the absorption edge provides elemental contrast in STXM imaging (dual-energy STXM). Cells were scanned at two energies, E1 (1189 eV) and E2 (1185 eV) just above and below the M5 absorption edge of the Gd atoms from Gd@C82(OH)22. The result showed that the Gd@C82(OH)22 NPs were taken up by primary mouse peritoneal macrophages and RAW264.7 after 3 h exposure, and the content of Gd@C82(OH)22 kept increasing over 48 h. The internalized Gd@C82(OH)22 NPs were mainly located in the cytoplasm, but almost never entered into the nucleus (Figure 4).210 Being element specific, this method is suitable for studying the distributions of elemental Gd, even when Gd is integrated with other NPs, such as in the case of Gd-hybridized Au@SiO2NPs (Au@SiO2(Gd)). With this method, both uptake and intracellular distribution of NP-based drugs were investigated. Hyaluronic acid (HA) and DOX were added to Au@SiO2 (Gd) carrier NPs as pharmaceutical agents. With dual-energy STXM, the cellular uptake of these NP-based drugs was imaged in MDA-MB-231 cells.211 Data showed a time-dependent uptake and how the intracellular localization of the Au@SiO2 (Gd) NPs moved from the membrane to around the cell nuclei. Reduced cellular uptake was detected when cells were pretreated with HA, verifying that the HA targeting modification efficiently enhances cellular uptake. These results on the internalization of Au@SiO2 (Gd) NPs were in accord with data obtained with laser confocal scanning microscopy and TEM.211
Figure 4.
Internalization of metallofullerenol by macrophages in vivo and in vitro. (a) A Gd M5-edge XANES spectrum of Gd@C82(OH)22 NPs. (b) Soft X-ray dual-energy contrast STXM images of Gd@C82(OH)22 in a primary mouse peritoneal macrophage in vivo. (c) Soft X-ray dual-energy contrast STXM images of the time-dependent uptake of Gd@C82(OH)22 NPs by primary mouse peritoneal macrophages and RAW 264.7 cells in vitro. (d) ICP-MS quantification of the time-dependent uptake of the NPs in macrophages of primary mouse peritoneal macrophages. Reprinted with permission from ref (210). Copyright 2014 John Wiley and Sons, Inc.
In the aforementioned example, besides the 2D distribution, 3D ultrastructural imaging of the Gd@C82(OH)22NPs inside the cell was performed by combining dual-energy contrast STXM and equally sloped tomography (EST; Figure 5a,b). This method is a type of tomography where projections are acquired using a constant slope increment (instead of more common angle increments). This technique facilitates the use of iterative image reconstruction algorithms based on pseudopolar fast Fourier transform,212 producing high-quality images with reduced exposure to radiation.213 In this experiment, the detailed distribution of Gd@C82(OH)22NPs in macrophages was obtained (Figure 5c).214 A large number of NPs were found to be aggregated within cells, and they were mainly located in phagosomes. No NPs were observed in the nuclei, which is in agreement with 2D imaging results. Based on the morphologies and the linear attenuation coefficients, μ, of the organelles,215 the 3D images were segmented into subvolume regions, and the lysosomes, mitochondria, and nuclei could be segmented (Figure 5d). The quantitative analysis results of the segmentation suggest that the majority of aggregated NPs were only located in phagocytic vesicles, instead of other organelles, including the nuclei (Figure 5e). This method also can show the characteristic morphological features of macrophages, for example, the pseudopods, rough surfaces, and flat shapes (Figure 5f).
Figure 5.
(a) Schematic layout of the dual-energy STXM imaging technique. Two sets of projections are acquired from various angles by STXM at energies below and above the absorption edge of the observed element, which in the reported work was Gd. (b) Tomographic data sets for both energies were separately reconstructed using the EST algorithm. (c) From this the quantitative 3D distribution of the specific element, here Gd, was obtained. (d) Intracellular distribution of Gd@C82(OH)22NPs. (e) Organelle segmentation based on differences in the linear attenuation coefficient and specific morphology of the different organelles. (f) Isosurface rendering of the macrophage at 1189 eV. Reprinted with permission from ref (214) under a CC-BY License. Copyright 2018 International Union of Crystallography.
Recently, soft X-ray based TXM nano-CT has been applied to visualize Escherichia coli (AMR) cells. Data indicated that La@graphene oxide (GO) NPs are able to insert perpendicularly into the cell membrane, causing a number of irregularly shaped perforations, leading to disruption of the bacterial membrane and thus ultimately to killing the bacteria (Figure 6).216 There are also examples of using soft X-ray CDI to obtain morphological information on some bacteria, green algae, viruses, chromosomes, etc.217−219 The development of XRF tomography enabled the visualization of internal chemical elemental structure nondestructively, initially demonstrated in investigations of the freshwater diatom Cyclotella meneghiniana and later extended to the model organism Caenorhabditis elegans and others.220,221 The chemical coordination of Cu within an intact organism was revealed using the four-dimensional combination of XRF XANES tomography, by mapping the distribution of cuprous and cupric complexes within Drosophila melanogaster.222 The lack of sensitivity of XRF to lighter elements leads to a symbiotic correlation with ptychography, the latter technique particularly suited to revealing the structures of lighter elements.223,224 STXM tomography, optical fluorescence, and 2D ptychography have also been correlated.189
Figure 6.
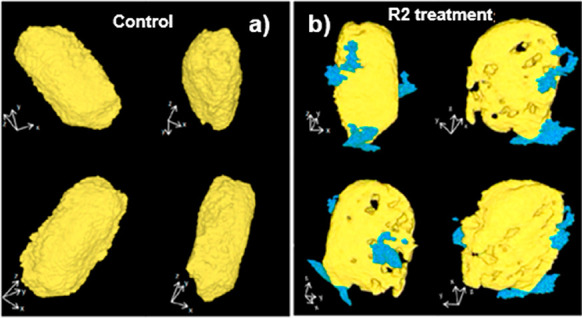
Nanocomputed tomography images of Escherichia coli: (a) untreated or (b) upon exposure to La@GO nanocomposites to decipher the bactericidal mechanism. Reprinted with permission from ref (216). Copyright 2019 American Chemical Society.
Soft X-ray microscopy (XRM) is also suitable for recording tomography of heterogeneous atmospheric particulate matter. The morphology and the distribution of elemental Fe can be observed, which can help to understand biological phenomena caused by atmospheric particulate matter after entering the biological environment, which is of relevance for ecotoxicology.225 Soft X-rays have strong interactions with organic materials, which limits the cell penetration to ∼10–15 μm, depending on different cell types226 and the incident photon energies used.
As illustrated with the above examples, in addition to high spatial resolution, an important advantage of using soft X-rays for imaging is that cell membranes and intracellular structures can be imaged without the use of contrasting methods. This is a significant advantage over laboratory-based optical fluorescence microscopy imaging and OCT in the visible/NIR, where fluorescence staining of such structures is required, in order to correlate the location of NPs to intracellular organelles.103 Standard thin-section TEM yields high-resolution images, but visualization of intracellular organelles requires staining,227 and 3D tomography typically needs to be done by reconstruction of images obtained from different slices. Cryo-EM can image these organelles in 3D without staining, but only in thin parts of the cells (i.e., 500 nm). Nevertheless, presently, soft X-ray techniques do not allow imaging different types of individual biological molecules in a cell directly, which would be important in investigating intracellular nano–bio interactions. Biomolecules such as proteins are small when compared with organelles or the complex structures found within them (i.e., a few nm versus hundreds of nm respectively) and normally provide a low contrast independent of their type (unless they contain large quantities of heavier elements such as Fe). This issue makes it challenging to determine the location of a type of protein or to discriminate between different types of individual proteins using soft X-rays. However, the use of staining strategies might help to solve this problem. For example, by using immuno-gold, it was possible to stain cellular components such as microtubules228 or mitochondria.229 Kong et al. reported a genetically encoded method for in situ labeling of intracellular proteins.230 Analogous to green fluorescent protein for fluorescence imaging, the genetically encoded tags provided a means for site-specific labeling of proteins of interest in mammalian cells with high-contrast elements, which enabled imaging of protein locations using STXM with 30 nm resolution (Figure 7). This ability to image multiple proteins holds promise for multimodal imaging to understand the biological effects and mechanisms of how NP-based drugs interact with cells at the molecular level.231
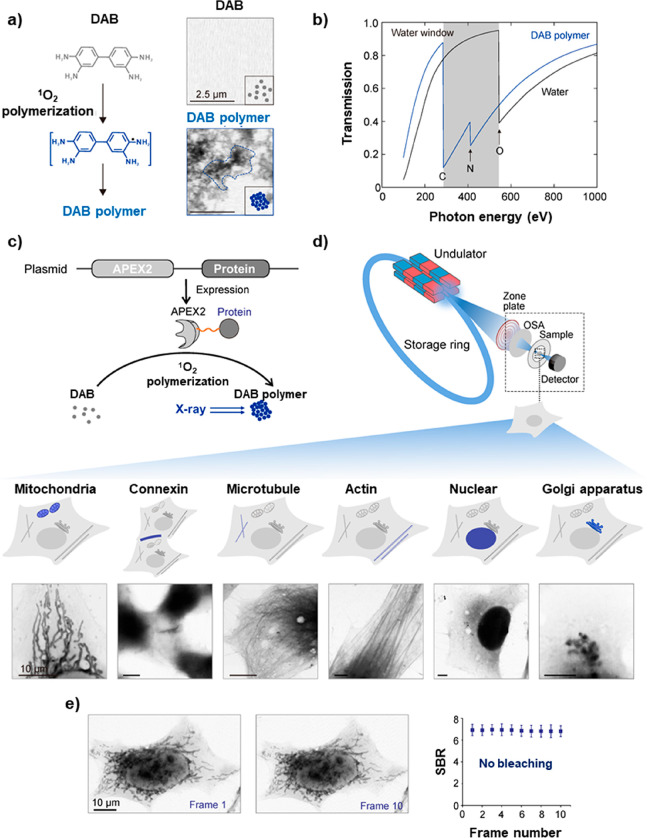
Figure 7.
Repurpose engineered peroxidase as genetically encoded tags for protein localization with XRM. (a) Schematics showing the catalytic polymerization of 3,3′-diaminobenzidine (DAB) into DAB polymer (left) and X-ray imaging of DAB polymer (right). (b) X-ray absorption spectra of water and DAB polymer. In the “water window”, absorption by carbon and nitrogen is much stronger than by oxygen. (c) Schematics showing APEX2 as a genetically encoded tag for protein localization with XRM. By using fusion expression plasmids including APEX2 and biotargets, these tags are highly specific and can polymerize DAB into localized X-ray-visible dense DAB polymers. This strategy enables localizing and imaging various cellular targets with high resolution. (d) STXM images of cellular proteins and specific amino acid sequences: COX4 (mitochondrial), Cx43, α-tubulin, β-actin, NLS, and GalT. Scale bars: 10 μm. (e) Photostability characterization of the genetically encoded tag for protein localization with XRM. No photobleaching occurred after 10 frames of STXM scans (for each STXM scan, the signal-to-background ratio of 10 loci was calculated and averaged to obtain a single value). Scale bars: 10 μm. Reprinted with permission from ref (230) under a CC-BY License. Copyright 2020 Oxford University Press.
Overall, soft X-ray-based imaging methods make it possible to obtain morphological information from intact samples, quantitative mass information (e.g., of the internalized NPs), and localization information about the different NP parts to study nano–bio interactions. Accompanied with advances on X-ray monochromator technology, optics, X-ray detectors, experiment control, and the quality of the X-ray light source, the diversity of imaging modalities based on synchrotron and X-ray free-electron lasers light sources are on the way toward achieving efficient imaging of cells. The combination of different modalities of X-ray microscopy to build multimodal instruments can succeed in achieving correlative imaging on the same cell, providing complementary information from each method.
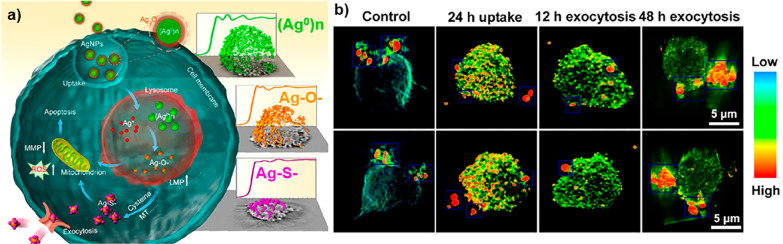
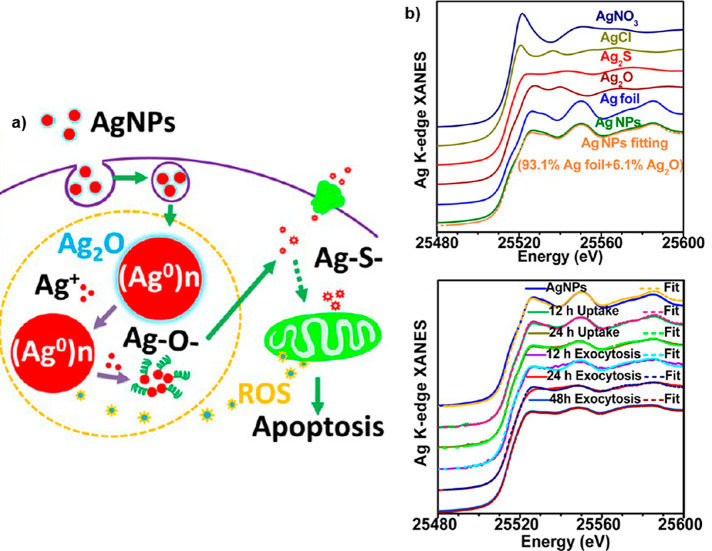
In contrast to soft X-rays, the greater biological penetration depth of hard X-rays enables imaging of larger cells, tissues, and organisms. Furthermore, the morphology of tissues and cells can be visualized with hard X-rays through use of chemical staining to enhance the signal of organic structures of cells and tissue,232 but also by using phase contrast data acquisition protocols,155−157 including holotomography,233 or coherent diffraction-based ptychography.234 The morphology and organelle localization can also be assessed on cellular samples by using experimental approaches based on the correlative acquisition of optical and hard X-ray microscopy images.235 Additionally, hard X-ray tomography can provide the high-resolution 3D distribution of metal NPs in cells,236,237 as an important alternative to methodologies based on visible fluorescence and OCT.238 Due to their pH independence, these methods also avoid the photobleaching considerations inherent to working with fluorophores. For example, in Figure 8, the 3D distribution of 20 nm Ag NPs inside a single human monocyte (THP-1) at different time points is shown, using hard X-ray TXM with Zernike phase contrast imaging at high spatial resolution of 60 nm at 8 keV.22 These images directly demonstrate the cellular accumulation and exclusion processes of Ag NPs in THP-1 cells. The different content, 3D distribution, and aggregation states of the Ag NPs elucidated the time-dependent interactions of cells with the NPs. The Ag NPs were internalized by cells, trafficked from engulfed vesicles to the lysosomes, disrupted the lysosomal membranes, decreased matrix metalloproteinases, generated reactive oxygen species (ROS), and finally caused apoptosis of the cell.22 While these findings could have been also determined with fluorescence imaging in the visible by using fluorescently labeled NPs, in this work, additional information was extracted that would not have been possible with fluorescence spectroscopy in the visible. The trick was to combine imaging with XANES, which also allows observing the oxidation states of Ag atoms. The cytotoxicity of Ag NPs is largely due to the chemical transformation from elemental Ag into particulate Ag, as (Ag0)n, to Ag+ ions and Ag–O– and then Ag–S– species (see Figure 8a).22 The same method has also been applied to visualize the distributions of TiO2 NPs and nano-MoS2 in cells.239,240 Further possibilities of spectroscopic analyses are discussed in greater detail in a separate section, below.
Figure 8.
(a) Schematic diagram of the chemical mechanism of Ag NP toxicity to human monocytes (THP-1), showing also the XANES spectra of Ag atoms in different oxidation states/chemical environments. (b) Single-cell imaging with 3D hard X-ray tomography (NanoCT) to observe the spatial distribution of Ag NPs in a single THP-1 cell. Reprinted with permission from ref (22). Copyright (2015) American Chemical Society.
Phase contrast approaches are more efficient than absorption-based techniques for imaging low-absorbing samples,233 such as soft biological tissues without using staining procedures (and, therefore, promising for in situ analysis). Phase contrast imaging follows alterations on the phase of X-rays as they go through an object. Such changes are related to the electron densities of the components of the sample, meaning the different cells or tissues in biological specimens. Imaging fine tissue structures or individual cells in hydrated samples is challenging, as differences in electron density between such biological structures and water are small (especially at the micro- or nanoscale). However, good contrast images with cellular or subcellular resolution can be obtained by using the correct experimental set-ups and phase retrieval algorithms. For example, full-field propagation-based phase contrast tomography has shown promising results for 3D imaging of weakly absorbing specimens such as biological samples with good resolution.241−244 Apart from being applied to probe nanomaterials inside single cells, X-rays have already been used for imaging tissues or organisms. Current developments in benchtop X-ray sources make it possible to extend the same experimental approach (with submicrometer resolution) to clinical and biomedical research within a laboratory environment.243,245,246 As such, the application of full-field propagation-based phase contrast tomography has enabled acquisition of a variety of data, from structural information on full or large sections of organs with μm resolution243,245−248 to mapping in 3D the cellular organization of large areas of brains or lungs (from mice or human origin) with outstanding resolution241−244 and collecting images from isolated cells with subcellular resolution.237,249,250 Furthermore, as metal-based NP-based drugs would show much higher electron densities than the soft elements normally found in tissues or organs, it will be possible to detect them easily in biological samples using phase contrast techniques. Again, propagation-based phase contrast tomography has enabled scientists to discriminate individual barium-based NPs (used as contrast agents) and to map their locations within isolated macrophages.237 It has also been used to determine the distributions of barium-labeled macrophages in lungs of healthy and asthmatic mice (showing preferential localization of macrophages within the alveoli and their ability to penetrate epithelial layers within lungs, Figure 9).241 Interestingly, propagation-based phase contrast is highly dose efficient, and the size of the volume analyzed can be easily controlled by changing the relative distances between source, sample, and detector.241,243,246,251 Yet, the analysis of larger volumes normally leads to images with lower spatial resolution. Nevertheless, as the technique is nondestructive, it is possible to implement sequential analytical strategies. For example, images of full organs can be initially acquired to identify interesting areas, which can then be analyzed at higher resolution.241,243,246,251 Thus, hard X-ray propagation phase contrast tomography is a promising approach to study NP-based drugs in situ at different levels (i.e., from the cellular level to full organs or small animals) with a single technique. There is however concern that X-ray phase contrast imaging may suffer from increased required radiation doses.252
Figure 9.

Example of propagation-based phase contrast tomography of a lung section from a healthy mouse where macrophages labeled with barium NPs were instilled, showing the barium NPs (green), blood vessel (purple), bronchial area (yellow), and the contours of macrophages (blue). (a) Lung section mounted on the sample holder. (b) 3D rendering of the reconstructed volume of a large field of view of the lung section. (c) 3D rendering of the reconstructed volume obtained from tomographic data zooming on the bronchial area in (b). (d) Detail of barium-labeled macrophage highlighted in (c) from two orientations showing the internal distribution of the NP. Reprinted with permission from ref (241) under a CC BY-NC-ND 4.0 International License. Copyright 2015 Nature Research.
As shown in Figure 9, there is potential for in situ studies in vivo, at least in animals. Au NPs are attractive as CT contrast agent because of their strong X-ray attenuation, flexibility for surface functionalization, and biocompatibility. Wen et al.253 studied kinetics of Gd-loaded dendrimer-entrapped Au NPs and monitored the accumulation of contrast in several organs over 45 min post-intravenous injection in rats using micro-CT. Zhang et al.254 demonstrated an accumulation of a contrast agent in tumor tissue. They used a multimodal imaging nanoprobe by co-loading an aggregation-induced fluorescent dye (NPAPF) and Au NPs into FDA-approved micelles. The combination of fluorescence and micro-CT results in a probe with high sensitivity (fluorescence) and high spatial resolution (micro-CT). Of course, it does not yet overcome the limitation of restricted penetration depth of fluorescence imaging, but for small animal studies, this is not a crucial problem. Use of such a probe may be useful for testing strategies for enhanced sensitivity using synchrotron radiation. For translation to human imaging, the fluorescent marker could be replaced for example, by a SPECT label, as demonstrated in a study by Xu et al.255 Their Au/99mTc-PEG-RGD dendrimer entrapped NPs can be used as a nanoprobe for targeted SPECT/CT dual mode imaging of cancer cells in vitro and subcutaneous tumor models in vivo.
A form of CT imaging, known as spectral photon counting CT (SPCCT), has recently emerged as a tool for both preclinical and clinical studies.256 Conventional CT uses energy-integrating detectors, whereas SPCCT uses photon-counting detectors. This approach allows characterization of the energy profile of the beam that has exited the subject as well as higher spatial resolution and lower radiation dose.257 Of particular relevance to this Review, SPCCT allows “K-edge imaging” of elements ranging approximately from cerium to bismuth. K-edge imaging provides maps of elemental distributions within the subject and can image more than one element at a time.258,259 This technique has been used with cerium, tantalum, ytterbium, gold, and bismuth NPs and for applications such as blood vessel imaging, targeted imaging, and cell tracking.260−263 For example, Si-Mohamed et al. reported the use of SPCCT for tracking poly(ethylene glycol) (PEG)-coated Au NPs (Figure 10).264 While conventional CT images suggested liver and spleen uptake, it was found by Au “K-edge” images that Au NPs were also in the bone marrow. Other applications of Au NPs for drug delivery, targeting, and imaging have been reviewed by Kong et al.265
Figure 10.
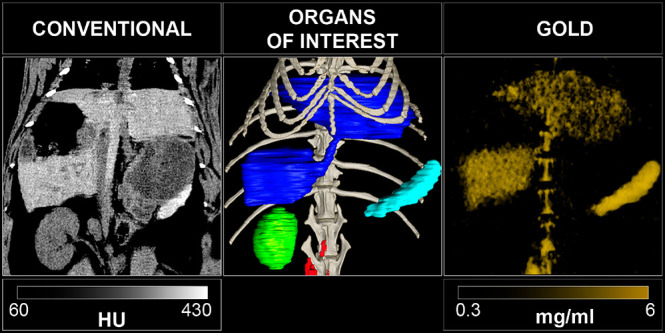
SPCCT images of a rabbit injected with PEG-coated, 15 nm core Au NPs, at 6 months post-injection. From left to right: A conventional CT image, segmentation of organs of interest (dark blue: liver, light blue: spleen, green: right kidney, red: lymph nodes, light gray: bone structure) and a Au “K-edge” image. Reprinted with permission from ref (264). Copyright 2017 Royal Society of Chemistry.
Ex vivo–in situ studies can provide important information when linked to complementary studies in vivo. Study designs featuring multimodal longitudinal imaging in vivo with imaging methods permit good soft tissue contrast (e.g., MRI) and high sensitivity for monitoring drug kinetics (e.g., fluorescence or photoacoustics). These methods can be combined with synchrotron radiation-based detailed ex vivo assessment on intact animals or excised organs. Labels such as Au NPs are good candidates for this approach (not for MRI, which still provides the high-resolution anatomical background information). For drug-delivery systems, one needs to decide whether to label the carrier, the drug, or both. While drug labels may alter its efficacy, there are a few drugs that show intrinsic CT contrast, such as cisplatin and other metal-based drugs. Oxaliplatin is a second-generation platinum anticancer drug that has potent therapeutic effects against several gastrointestinal cancers, and platinum (but also other metals) can be imaged, albeit at this time point only ex vivo, with synchrotron-based XRF.266,267 The latter was used to study drug distributions within tumor tissue. This ex vivo approach was subsequently also applied to human cancer tissue specimens. Higher platinum concentrations in the tumor stroma were an independent predictive factor of limited histologic response.268 These results suggest that XRF analysis may contribute to predicting the therapeutic effect of l-OHP-based chemotherapy by quantifying the distribution of platinum. This result is an example of X-ray-based 3D histology that can complement standard histological approaches. XRF tomography was also used to determine the 3D accumulation of LaF3:Ce nanoscintillators in spheroid tumor models with micron resolution (Figure 11), helping to assess their capacity to act as radiotherapy agents against solid tumors.269 In addition, XRF maps collected using nanofocused synchrotron radiation have enabled following not only the cellular internalization and degradation of labile Ag particles23,24 and nanowires270 but also other nanomaterials such as Au and Ti NPs236 and Pt-based NP-drugs.271 In fact, XRF permits multiplexed imaging of the distributions of different elements269,272 (Figure 11) and provides lower limits of detection down to ultratrace elemental sensitivity for high-Z elements. In principle, the smallest units providing signal are individual atoms (in contrast to fluorescence in the visible/NIR, where the smallest unit providing signal are small molecules, i.e., fluorophores). Elemental mapping by XRF has been also coupled to XAS to determine the speciation of nanomaterials in tissue or cell samples.273
Figure 11.
X-ray fluorescence 3D elemental maps showing the distributions of Zn and La in F98 spheroids (a,b) untreated or (c,d) treated with LaF3:Ce NPs for anticancer radiotherapy. Axes are shown in μm. Adapted with permission from ref (269) under a CC-BY International License. Copyright 2020 Wiley-VCH GmbH.
In addition, XRF can be used for imaging at scales above the cellular level. For example, synchrotron-based XFI has helped to study possible systemic toxicity caused by exposure to NPs found in pollution or consumer products by assessing ex vivo their accumulation in animal or plant tissues.152,274−277 Furthermore, it was shown that XFI computed tomography permits detection of single cells in a Au-loaded tumor implanted in the head of an adult rat, thus demonstrating the suitability of this technique for ex vivo studies on brain-tumor cell migration.278 Yet, for quite some time, XFI was seen as essentially unusable for objects of human size. This problem was overcome by a variant called “spatial filtering”.57 The broad background exhibited by XFI in the X-ray spectra of photons emerging from the irradiated object originate predominantly from multiple Compton scattering. For example, from about 1000 measured photons within the detector-resolved X-ray fluorescence lines’ energy range, only 1 photon is a fluorescence photon, while the other 999 photons come from Compton scattering. In such a case, no XFI signal can be recorded. However, especially for larger objects, the XFI background can show strong anisotropy if the incident photon energy is close to and just above the K-edge of the element excited.57 With the help of a computer algorithm, only such pixels from a large-area pixelated detector are taken into account, yielding the highest information density in terms of XFI signal versus background noise. The subsets of selected pixels (hence “spatial filtering”) show maximum imaging sensitivity, that is, statistical significance, respectively, whereas if all pixels are taken into account, the 1:999 ratio renders the signal unobservable (Figure 12). Thus, future clinical XFI applications are within reach, and this finding should trigger additional research starting with small-animal XFI. As a first example in this direction, in situ imaging of the natural iodine concentration in a mouse is shown in Figure 13. This XFI method, however, depends on the use of pencil X-ray beams with monochromatic spectra, as provided at synchrotron-based beamlines. Although one can perform basic research at such facilities, translation into clinical application needs ultracompact X-ray sources, such as laser-driven inverse Compton sources.279
Figure 12.
Simulated XFI spectra showing XFI signals obtained from a 30 cm-diameter water-filled sphere. (a) X-ray spectrum from the full solid angle (4π) with X-ray fluorescence (red) Au NPs (peaks around 67 and 69 keV) are undetectable within a “sea of background photons” from multiple Compton scattering: of 1000 measured photons, only 1 is from fluorescence, the other 999 arise from Compton scattering (the color indicates how often a photon is Compton scattered). (b) Spectrum for the same situation, but after performing optimized “spatial filtering”, which enables the detection of X-ray fluorescence signals from the Au NPs. Reprinted with permission from ref (57) under a CC BY-NC-ND 4.0 International License. Copyright 2018 Nature Research.
Figure 13.
X-ray fluorescence imaging full-body (a) and fine scan of the thyroid region (b) of a mouse. The euthanized mouse was placed sideways with the X-ray beam impinging perpendicular to the figure plane (mouse head on the right side). The left map depicts the number of Compton-scattered photons for each scan position. As seen, the only visible iodine concentration is the natural one found in the thyroid with the local iodine mass in the beam volume as retrieved from data analysis (each pixel of the fine scan covers an area of 0.25 mm2 and shows the amount of iodine Kα fluorescence photons). These data were recorded at Deutsches Elektronen-Synchrotron (DESY) by C. Körnig, O. Schmutzler, Y. Liu, T. Staufer, A. Machicote, Beibei Liu, W. J. Parak, N. Feliu, S. Huber, F. Grüner and have not been published previously. Experiments involving animals were carried out in accordance with the Institutional Review Board “Behörde für Soziales, Familie, Gesundheit und Verbraucherschutz” (Hamburg, Germany).
Finally, to go from images to movies, time-lapse in vivo recordings are at the borderline of what is possible today with existing synchrotron technology. As biodistributions are dynamic and the effects of drugs are described by their pharmacokinetics, such capabilities are tantalizing and widely anticipated. Examples were reported in living cells using low-energy synchrotron-based Fourier-transformed infrared (FTIR) spectroscopy.280−284 A successful transition to working in the X-ray regime will essentially need the development of methodologies to keep radiation damage at tolerable levels. However, this transition might be possible to achieve, and synchrotron-based hard X-ray phase contrast microtomography has been used to acquire time-lapse images of a living embryo.197,198 Furthermore, the delivery of respiratory treatments in the lungs of mice has been recently studied in vivo by using time-lapse phase contrast imaging with hard X-rays produced by a compact synchrotron light source (CLS).285−287
Scattering Experiments for Monitoring the Time-Dependent Structure/Composition of Nanoparticles and Their Assemblies
In this section, X-ray scattering techniques with applications in the characterization of NPs and NP-based drugs are highlighted. While some of these techniques have only been applied thus far to NP suspensions, their possible applications for studying intracellular NPs etc. are also discussed. In this direction, SAXS has been widely used and elucidates a wide range of properties. Before describing more complex systems, we give an overview of what SAXS can do in the analysis of NP properties. With regard to NP size distribution, SAXS can be applied to study NP formation and dissolution by making use of micro- and then nanofluidic devices.288,289 For designing reproducible experiments that yield in situ time-resolved structural information at fast time scales, it is necessary to build X-ray compatible microfluidic devices. Polyimide/Kapton-only devices enable the ex vivo investigation of structural dynamics and phase transitions of a wide range of colloidal NPs and soft matter samples down to millisecond time scales. Such devices then can be used to follow structural evolution in situ at millisecond time scales using on-chip time-resolved SAXS under continuous-flow conditions. In combination with other techniques such as ultrafast Coulter counters,290 this approach can have major impact on the design and formulation of amphiphilic polymer NPs for drug-delivery systems in medicine. More sophisticated reciprocal space mapping enables determining NP atomic-scale shape evolution in situ and in operando as a function of externally changing conditions.291−293 Concerning shape changes, X-ray diffraction can be used to investigate NP degradation systematically in the form of oxidation or deactivation during catalytic reactions.294,295 Imaging techniques such as CDI can provide detailed information on the shape and shape changes of single-metal NP in the size range of 100 nm diameter, as demonstrated previously in gas environments.296 Ptychography can be used to follow changes in the size and shape of NPs in liquid phase, both ex situ and under operando conditions.183 Similar responses of the NPs can be expected from other stimuli in the biological, wet chemical environment. Single PtRh alloy NPs and PdRh NP ensembles were found to dealloy by the formation of Rh oxide when switching from reducing to oxidizing conditions.297,298 Such processes may also occur in cells electrochemically driven at room temperature when changes in pH take place. This change has potential relevance to biological applications, as surface-based catalysis is one origin of NP toxicity.299,300 These technical possibilities also have potential for the development of applications concerning in situ observation of NP-based drugs. Simulations of in situ CDI data of the fusion of glioblastoma cells were already reported301 and also considered radiation damage. Once CDI permits in situ imaging of NPs and NP-based drugs in biological environments, these experiments will provide important information about the states of the NP-based drugs such as changes in size and shape and also their effective size increases due to agglomeration. Dissolution of the NP carrier, for example, could be observed by reductions in size and also by changes in its shape. Time-resolved data may offer important information, as discussed below. Loss of the surface coating of the carrier NPs, involving, for example, attached pharmaceutical agents and ligands, might be detected by the onset of agglomeration. While such ideas (i.e., to investigate the state of NPs in biological environments by measuring their effective hydrodynamic diameters) are also explored with other techniques,302 a fundamental understanding of the interpretation remains lacking.
At the next level, SAXS is helpful in investigating NP assemblies. For “uncontrolled” assemblies, aggregated NPs give rise to diverging SAXS intensity in the direct beam direction, while distributed NPs show a plateau, which allows model-free extraction of the NP diameter. The exact length scale that can be probed by SAXS depends on X-ray collimation and X-ray wavelength. Conservative values for standard university lab sources using Mo X-ray radiation range from ca. 1 to ca. 50 nm.303 The exact limits for a given sample depend on signal-to-noise ratio and the brilliance of the X-ray source. Synchrotrons can reach much more extreme values, that is, in ultrasmall-angle X-ray scattering, in which length scales of several microns have been probed in dental composites,304 bridging the gap all the way to optical microscopy. An interesting recent development from the medical point of view is the use of high-energy X-rays of 50 keV and higher. In this regime, absorption due to the photoelectric effect is small. Since the Compton effect at higher X-ray energies affects mainly back scattering, there should be little influence of Compton scattering on the SAXS pattern, as previously shown for X-ray reflectometry.305 Thus, it seems possible to apply highly collimated high-energy X-ray beams for medical SAXS diagnosis of rather large tissue sections of 10 cm thickness (i.e., full organ size). Nevertheless, this measurement would require developing methods capable of dealing with the low coherent scattering signals obtained from soft tissues at such high energies, which would be further attenuated by multiple scattering in thick samples.
Even more information can be obtained from “controlled” NP assemblies. Biomimetic NP assemblies can be conveniently investigated with synchrotron SAXS (Figure 14).306 For instance, Xia et al. self-assembled self-limiting monodisperse supraparticles (SP) from polydisperse NPs.307 For the majority of the NP assemblies forming spontaneously in bulk solution, self-organization occurs continuously until the components are exhausted and the NPs form a dry crystal, complex solid, or precipitate. A self-limiting self-assembly process would be conceptually different from currently known self-organization reactions. Because self-limiting structures are common in biological systems, the realization that using inorganic NPs might lead to unexpected parallels between the world of inorganic colloids and biomacromolecules. By conducting synchrotron SAXS, a distinct scattering pattern was observed and confirmed monodispersity in the large ensemble of SPs in solution. The corresponding diameters and dispersibility of the SPs calculated from SAXS data matched the TEM, SEM, and dynamic light scattering (DLS) data. Together, they determine the sizes of SPs both in solution and in dry state and observed dense packing of the NPs. Data fitting using three different form factor models revealed a core–shell sphere as the most likely possibility. Fitting SAXS curves yielded the number of NPs in a single SP and the thickness of the shell, along with a loosely packed core and more densely packed outer shell. All these SAXS results give insight into the potential effects of different forces in self-limiting assembly.307 Related work by Merkens et al. addressed formation of Au NP clusters within a microfluidic chip, driven by hydrophobic interactions.308
Figure 14.
Example SAXS experiment to study the structure of a microporous nongraphitic carbon material. (a) Schematic showing the normal setup of a SAXS instrument. (b) Intensity versus scattering vector curve plot (log–log scale), highlighting morphological (low Q ranges), microstructural (intermediate Q ranges), and structural (large Q ranges) features of the material probed by the technique. (c) Intensity versus scattering angle 2θ plot (linear scale) of the same spectrum, which is normally used for PXRD. Reprinted with permission from ref (306) under a CC BY-NC-ND 4.0 International License. Copyright 2019 Elsevier.
X-ray techniques are popular in investigating principles in biomimetic nanocomposites. In layer-by-layer (LbL) assembly, Podsiadlo et al. investigated the effects of combining polymers and NPs (clay nanosheets), both components with strong tendencies toward self-organization, into a single LbL assembly.309 They found diffusional self-organization in exponential LbL films on the micro- and nanoscale. SAXS was employed to reveal the morphologies of the LbL films. In films that did not contain clay nanosheets, only diffuse scattering from the polymer was observed. Clay nanosheets spontaneously adsorb almost exclusively in orientations parallel to the substrate.310 In the film containing clay, a sharp peak corresponding to basal spacing for Na+-montmorillonite was shown, and a less prominent peak to a larger basal spacing indicated significant intercalation of polymer between the clay sheets. No distinct peak was observed in another film with clay nanosheets, demonstrating intercalated basal spacing or exfoliation of clay platelets. All these results were helpful in determining the morphologies between the multilayers.309 Another interesting case study was done by Zhang et al. in fabricating fibers with high toughness.311 The materials architecture with alternating layers of hard inorganic components and soft organic polymers effectively arrests the propagation of cracks. Further improvement of toughness in biomimetic nanocomposites is restricted by the low strains of composite materials. The combination of two structural motifs at different scales (nanoscale and microscale) was designed to increase both the stretchability and toughness simultaneously. They transformed flat nacre films into fibers that combine layered nanoscale and spiral microscale structural motifs. Synchrotron SAXS was used to check the enhanced alignment. Since polymers scatter X-rays weakly, the diffraction peaks originated from the graphene nanosheets. The belt-like fibers without sharp scattering peaks indicated poor alignment of the nanosheets. The nearly perfect nacre-like layering in the transverse direction of the fiber yielded sharp peaks after initial twisting. Further twisted coiled fiber resulted in an absence of sharp peaks. All three fibers showed monotonic intensity drops, indicating the uniform dispersion of graphene in the poly(vinyl alcohol) (PVA) matrix.311
Chiral NPs or assemblies with intrinsic geometries that lack inversion symmetry, or with imprinted optical activity attributed to chiral ligands, have recently garnered significant attention for controlling biorecognition and optical sensing.312−314 Chirality determines many structure–function relationships in nature at many levels of biological organization. Omnipresent chiral properties in biology inspire and necessitate further studies of chirality of nanoscale materials due to many structural parallels between nano- and biomaterials as well as multiple biomedical applications of NPs. Typically, electronic circular dichroism (CD) in molecules ranging from individual amino acids to hierarchically more complex peptides, proteins, and oligonucleotides is analyzed using UV light, which is incompatible with living cells. However, chiral NP systems have been shown to exhibit significantly enhanced CD at optical and near-IR wavelengths, offering more sensitive and selective routes toward driving enantioselective intermolecular reactions,315 facilitating higher NP–biomolecule affinities and tuning polarization-dependent light–matter interactions.316 Indeed, the emergence of chirality in NPs increases their tendency to interact with specific biomolecules featuring similar chirality, a critical condition in drug discovery and delivery, and can also promote further remodeling of NPs by these species. While chiral discrimination can be applied with in vitro systems,317−319in vivo will be substantially more challenging, but engineered orders-of-magnitude enhancements in achievable optical electromagnetic density of chirality in plasmonic- and dielectric-based NP systems may enable discernible chiral hotspots to be detectable in imaging and spectroscopy.320,321
Experiments including synchrotron SAXS and XRD can be employed in many chiral nanostructure studies. For direct resolution using X-ray techniques, inversion asymmetric crystallinity of NPs or surface reconstruction with chiral ligands may show polarization-selective scattering, enabling tracking of enantioselective chiral drug delivery, NP toxicity, and protein corona formation.322,323 In another study, Yan et al. self-assembled chiral NP pyramids with strong R/S optical activity.324 They applied synchrotron SAXS combined with DLS data to give evidence for the actual space occupied by the NPs and their assemblies in “wet” states. This insight can help elucidate the degree of expansion upon hydration as well as the ensemble composition of the dispersions. By looking at SAXS features located at different scattering vectors, one can obtain information about different nanoscale superstructures in terms of size, shape, and conformation.324 Jana et al. twisted stacking nanoplatelets to self-assemble into chiral ribbons.325 All SAXS patterns of the dispersion at all three different steps of self-assembly displayed two scattering peaks whose position does not change over the whole process, confirming that the nanoplatelets remain stacked and the stacking period keeps constant.325 Jiang et al. self-assembled hierarchically organized particles with greater complexity; SAXS served as a significant tool to substantiate the atomic structure of Au-Cys nanoribbons along with density functional theory calculations and XRD data.326 Ahn et al. investigated the effects of chemical modification on the chirality of organic inorganic hybrid perovskites.327 They used XRD to show the shift of a peak at the smallest 2θ angle (interlayer spacing of lead halide layers), ascribed to the phase transition of the crystalline structure due to the modulation of halide anion mixing ratio.327
As discussed above, such local scattering can be involved in X-ray imaging. Originally, X-ray-based scattering techniques were developed for bulk samples. However, by using low-emittance synchrotron sources with diffraction-limited storage rings, both from the current third-generation (i.e., DESY or NSLS)328,329 and increasingly from the fourth-generation instruments,330−333 nanofocused beams providing increased photon flux and coherence are possible, and thus scattering can be carried out locally with high spatial resolution, enabling improved imaging modalities.
SAXS is particularly useful for studying nanoscale morphology in complex environments.334 Examples range from plant and bone structures up to brain and breast tumors and cardiac tissue.335−338 By combining scanning approaches along the x–y plane, rotation around the tomographic axis (y), and tilting around the x axis, SAXS serves as a tomography technique.339−342 SAXS-computed tomography (SAXS-CT) bridges the gap between information retrieved from high-resolution local techniques and information from low-resolution, large field-of-view imaging techniques when some hierarchical structure is present. Figure 15 shows the schematic procedure for SAXS-CT from the data acquisition to the volume-resolved architectural nanostructure, passing by the choice of the q-value for reconstruction. Detection of NPs in soft tissues by SAXS is efficient if the electron density of embedded NPs is higher than the hydrocarbon matrix, that is, oxide, semiconductor, or metal NPs are easy to detect in soft tissue matrices. Information that can be readily extracted from such measurements includes the NP size distributions, NP shapes, pair distances between interacting NPs due to molecular interactions,343,344 and aggregation states.345 These structural parameters can also be spatially resolved by applying the reverse analysis approach to the SAXS tomograms.342
Figure 15.
(a) Phantom for SAXS tomography: Dried collagen (anisotropic scattering) and 120 nm SiO2 nanospheres (isotropic scattering) inserted in a homogeneous matrix of RW3 solid water (PTW Freiburg, Freiburg, Germany), which simulates water. (b) Tomographic data set for pencil-beam geometry. (c) Depending on the q-value choice, some structure is highlighted. In this case, reconstructing at q = 0.23 nm–1 emphasizes the SiO2 nanospheres, while if the signal from collagen is desired, the reconstruction is carried out at the q = 0.59 nm–1. (d) 3D view of the SAXS-CT phantom from the top and the side. These data were recorded at the beamline BL40B2 at the Spring-8 synchrotron source for this work by Andre L. C. Conceição and have not been published previously.
A number of X-ray diffraction or scattering techniques can also generate cell images capable of showing the structure of different organelles with high resolution. Such imaging techniques have been extensively discussed in a recent review.154 Combinations between those and other X-ray imaging techniques can help probing NPs in cells. In particular, correlative imaging of chemically fixed HeLa cancer cells by fluorescence (optical) microscopy, diffraction-based ptychography, and STXM using X-rays tuned to the Fe L-edge were effective in determining the specific cellular localization of individual Fe NPs, showing that particles were internalized by cells in <30 min.189
It should also be possible to observe the interactions of NPs with cellular membranes directly. X-ray reflectivity (XRR) can be utilized to analyze the adsorption behavior and conformational arrangements of proteins on biomaterials surfaces.346 XRR furthermore permits the characterization of the lipid-induced fibrillation at the molecular level.347 The assembly of ligands on NP surfaces can be tested in comparatively simple experiments on planar surfaces using XRR. The XRR technique enables us to probe the attachment and penetration of NP-based drugs through model cell membranes containing lipids and proteins and, in turn, may aid our understanding regarding various physiological functions such as cellular transport, signaling, membrane trafficking, and molecular recognition. For these studies, a Langmuir monolayer of lipids is formed on the water surface, and high-energy synchrotron X-rays are used for XRR and related measurements to cover a larger momentum-transfer (q) space (Figure 16).348 Apart from measurements on the liquid surface, one can also collect XRR data from solid surfaces immersed in liquid using high-energy X-rays. This method becomes particularly helpful as X-rays, which can penetrate through the thick water bath over the membrane.349 The electron density profile (EDP) obtained from XRR measurements and pressure molecular area data of Langmuir monolayers over water surfaces have demonstrated membrane localization of heme.348 Heme and its analog hemin, are among the most biologically relevant planar organic molecules. Therefore, it is important to understand the molecular mechanism of intercalation and adsorption of this cytotoxic molecule after its dissociation from proteins such as hemoglobin. Continuous hemin uptake from the subphase and intercalation into and/or adsorption on to the membrane surface have been witnessed in a strong membrane surface packing-specific manner. Competitive interactions between hemin–membrane and hemin–hemin are proposed to be responsible for the critical hemin concentration. Systematic studies of the EDP showed that up to the limit, continuous hemin uptake is possible and beyond that the hemin–hemin interactions dominate, effectively reducing the hemin intercalation into the membrane. The technique developed could be easily adopted for NP-based drugs by putting them in the subphase to study uptake and orientation-specific attachment in the Langmuir monolayer of bioengineered membranes. Two lipids, namely 1,2-dimyristoyl-sn-glycero-3-phosphocholine and 1,2-dimyristoyl-sn-glycero-3-phosphoethanolamine, with differences only in their head groups, were studied to understand the specificity of a model protein spectrin for zwitterionic lipids,349 which constitute the major part of the physiological membrane. Spectrin is a high-molecular-weight, ca. 100 nm-long, flexible rod-like protein composed of two subunits. Similar measurements could be carried out to model NP-based drug attachment and penetration of appropriately engineered biomembranes. X-ray reflectivity is a powerful non-invasive technique to determine the buried structure of thin films along the depth,350 and the extracted EDP from the reflectivity analysis showed that spectrin chains form a uniform layer on top of the phosphocholine-containing bilayer, whereas spectrin gets adsorbed into the phosphoethanolamine-containing membrane, possibly through one or two permanent binding sites with the rest of the chains projected out of the membrane.
Figure 16.
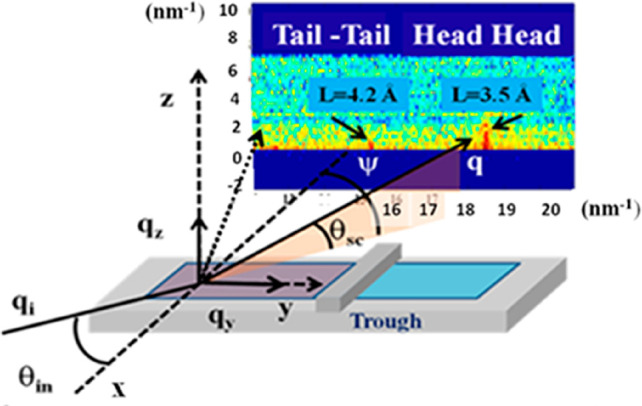
Typical arrangement for X-ray reflectivity and related measurements in a synchrotron experiment. Reprinted with permission from ref (351). Copyright 2019 American Chemical Society.
Spectroscopies to Investigate Biotransformation of Nanoparticles in Biological Environment
Spectroscopies also provide information regarding the chemical states of NPs. Exposure of nanomaterials to biological fluids can affect the surface properties of NPs. Such effects often occur at the outer surface (e.g., loss or rearrangement of capping ligands) and may affect solubility, aggregation, and interactions with their environment. These effects can also happen at the cores of the NPs (e.g., dissolution, remodeling). These changes are especially significant for small NPs having size- and shape-dependent properties, since dissolution of even a few outer layers of the NP core and modifications to the faceting or shapes of plasmonic NPs can dramatically alter their behavior. Ex situ XRD and TEM can provide information about changes to the NP cores that are permanent and therefore do not revert back to their original state. Since NPs are rarely employed in their pristine forms, dynamic exchange of corona proteins,352 or the ligands that serve in a protection role with the surrounding medium, is expected for almost all types of NPs. Continuous interplay with biomolecules in biological environments gives NPs different bioidentities.352 Some biomolecules also induce variations in surface properties (charge, hydrophobicity, etc.), roughness, and local chemical environment of NPs, cause dissolution/degradation, and ion leaching of NPs, subsequently generating ROS and oxidative stress, which eventually can cause toxicity to cells.353,354 To reveal such processes, imaging surface plasmon resonance (ISPR) is frequently leveraged,355,356 in which the loss of previous ligands indirectly causes shifts in the optical resonance peaks of the NPs through remodeling the local refractive index. Kinetics of ligand exchange or loss can further be derived from time-dependent ISPR profiles to give insight into the interplay of coated NPs with the surrounding biological fluids.357 Under dark-field illumination, ISPR has a resolution down to the level of an individual NP.358 Major limitations of ISPR characterization include the inability to reflect the kinetics of multiple ligand loss and dynamic processes, as witnessed during the establishment of the soft protein corona. In the presence of persistent ligand loss, there stands a chance for the NP itself to be influenced, in which the remodeling of surface roughness is likely to be observed. Under such conditions, under-coordinated atoms (referred to as adatoms) are more prone to oxidation or atom-exchange with respect to their fully coordinated counterparts.359 The dynamic processes of surface roughness variation can be visualized by using in situ liquid-TEM imaging. An updated modality of a gas-supplement equipment might even enable this system to image living cells at nanometer resolution.360 Beyond projections, reconstruction of the whole NP through tomographic TEM (3D TEM) permits the view from varying directions to reach unbiased conclusions.361
The interactions between X-rays and matter are determined by the elemental composition of the matter and the energies of the photons. Hard X-ray spectroscopy involves promoting a core electron of an atom to unoccupied levels by absorption of the incident X-ray photon, followed by emission of a lower energy photon during electron decay to fill the hole. Both XANES (also called NEXAFS at energies below 1 keV) and EXAFS can provide quantitative information about chemical bonding, the oxidation state of the absorbing atom, the local atomic environment such as coordination number, type, and length of the metal–ligand bond, independent of the state of aggregation (NPs, ions, clusters).153 Thus, they are not only used to study the molecular reactions preceding the nucleation of colloidal NPs but also can be applied to investigate NP assembly and NP uptake by cells and related toxicity.
Supramolecular assemblies have been characterized using XANES. For example, Kenji et al. were inspired by coordination assemblies of organic building blocks, which occurred in many biotic systems, and these systems attained exemplary performance in redox and photonic reactions optimized for the cellular environment.362 Synchrotron EXAFS was used to help establish the coordination pattern of Zn2+ and the supramolecular geometry of the interparticle bridges. Fourier-transformed XAFS plots of Zn2+ in the nanoscale sheets were found to be nearly identical to that of [Zn4(μ4-O)]6+SP clusters in zinc stearate, indicating that the coordination clusters in nanosheets have the same coordination geometry with Zn2+ stearate. These results, along with additional complementary experimental data sets (XRD, EDX, etc.), demonstrate that the assembly was driven by coordination bonds rather than other intermolecular forces.362
Nanoparticles can be related to their catalytic properties, such as in the case of metal NPs.363,364 Metal clusters and metal complexes show a number of chemical and catalytic activities that can be explored by a combination of optical and X-ray spectroscopies. Examples are entatic state model complexes showing catalytic activity due to charge and electron transfer between the metal center and its ligand sphere. Studies involve the activation of catalytic activity by optically exciting metal complexes and probing them by means of EXAFS and XANES, raising the possibility of studying photoactivated complexes in operando.188,365 The photocatalytic activity of Cu-based metal clusters has been studied through such combinations of optical and X-ray spectroscopies.366 These studies involved the activation of catalytic activity by optically exciting metal complexes and probing them by means of EXAFS and XANES. The XAS showed shifts in the XANES edges due to changes in the metal oxidation state, while EXAFS tracked local changes of the structural environment surrounding the metal site. The complementary use of optical and X-ray techniques enabled the study of a wide range of different time scales.188 Such studies could also be carried out in cells. For example, the genotoxicity of Ag NPs has been investigated by testing the DNA and chromosomal damage to CHO-K1 cells according to Organization for Economic Cooperation and Development guidelines, and a ROS and Ag+-releasing mechanism was proposed (Figure 17a).367 Further, as discussed above in a different context, Chen et al. (and others) reported the intracellular stability and chemical state of Ag NPs with the help of XANES spectroscopy (Figure 17b), together with a degradation study, proved the quick dissolution of Ag NPs inside cells and that the released Ag+ was oxidized to Ag–O– species, subsequently stabilized by thiol groups, forming Ag–S bonds within the cells. The degradation of Ag NPs inside cells increased ROS production, decreased cell viability, and decreased the ultimate toxicity to cells.20,23,24 This ion-releasing and redox-related mechanism of toxicity has also been found in ZnO2, CeO2, and other metal oxide NPs.368,369 Thus, XANES spectroscopy is a powerful method to study the oxidation states of NPs inside cells, ion release, and surface redox and oxidative stress-related mechanisms of toxicities. This information can aid the safe design of NPs by inhibiting the dissolution of metal ions of NPs or to modulate oxidative stress.370−372
Figure 17.
(a) Schematic illustration of the mechanism of toxicity of Ag NPs. Adapted from ref (22). Copyright 2015 American Chemical Society. (b) Different chemical species of Ag as indicated in normalized Ag L3-edge XANES. Reprinted with permission from ref (22).
In another example, Gong et al.373 applied XANES and EXAFS to investigate the behavior of dispersed Au atoms in distinct carbon-dot-supported Au NPs. They found that dispersed Au atoms in NPs, compared with small Au clusters, enable to react efficiently with glutathione (GSH) to form Au–S bonds and to reduce the GSH levels in cells. Taking advantage of XANES and EXAFS, researchers observed the dispersion of Au0 as a peak at ∼2.5 Å and confirmed that the GSH depletion is due to the atomic-level dispersed Au, which highlights the potential of atomic economy in NP design. Thus, by deciphering the chemical mechanism of NP–bio interactions, X-ray-based spectroscopy could provide insights into cellular regulation with NPs.
Today’s questions in NP research and particularly in toxicity in human tissue go far beyond studying oxidation state. In this context, studying the electronic structure of the central metal atom (M) and differentiating between various metal ligands (e.g., carbon, nitrogen, oxygen) during NP uptake is important. However, conventional hard X-ray spectroscopy is not able to distinguish between M–C, M–N, and M–O ligands. This limitation could be circumvented by the recent implementation of high-energy resolution spectrometers, which provide a superb opportunity to access the types, protonation states, and ionization energies of ligands bound to metals.374−376 Moreover, with the same experimental setup, high-energy-resolution fluorescence detected-XANES and X-ray emission spectroscopy (XES) can provide complementary information about the highest occupied and lowest unoccupied electronic states. Thus far, the method was successfully applied to shed light on weak noncovalent bonding of CO2 to NPs, the roles of a central carbon in the nitrogenase iron–molybdenum cofactor, and photocatalysis.377−379 Furthermore, the implementation of high-energy-resolution spectrometers facilitates measurements of hard X-ray magnetic circular dichroism combined with resonant inelastic X-ray scattering (RIXS-MCD) on magnetic NPs relevant in biomedical applications as contrast agents. The RIXS-MCD method enables the determination of the size distributions of superparamagnetic iron oxide NPs in frozen samples and in concentrated solutions, which are below the detection limits of light-scattering probes.380−382
An alternative method to monitoring oxidation and spin-state changes is XES, which spectrally disperses the X-ray fluorescence beyond elemental sensitivity and records spectral emission line-shapes of Kα and Kβ emission. These shapes are characteristic of the oxidation and spin states and of the chemical environment of the NPs.383−386 The high resolution of XES enables elemental determination, and XES greatly benefits from not requiring monochromatic X-ray radiation, as needed for XANES spectroscopy (the latter typically operating at bandwidths, ΔE, relative to the photon energy, E, of ΔE/E ≈ 10–4), thereby exploiting the full flux of many X-ray sources, such as undulators at synchrotron radiation facilities. Indeed, XES is particularly well suited for heavier elements in solution and cellular phases because the total fluorescence yield greatly increases from ∼1% for light elements (C, N, O, etc.) versus ∼30% for 3d transition metals (Mn, Fe, Co, etc.) to >80% for 4d and 5d transition metals.387 Spectral filters can further reduce fluorescent background from organic matter. X-ray emission is widely used in analytical techniques such as proton-induced X-ray emission388 and is often combined with monochromatic X-ray sources to study NPs with resonant RIXS, statically and dynamically.383−386 With advances in relatively simple high-resolution spectrometers and fast line and area detectors, pink-beam sources can provide a high chemical specificity if required or increased flux at lower spectral resolution, providing sufficient elemental sensitivity to distinguish heavier elements in NPs from organic matter. The development of microcalorimeter pixel arrays with high spectral resolution is relatively recent in the field of X-ray spectroscopy and has great potential for combining X-ray emission spectroscopy, multiplexed recording, and even imaging.
Finally, classical X-ray-based spectroscopies, such as XPS, provide information about the surfaces of NPs. XPS has been used to probe the degradation of labile and stable NPs (Ag and Pt, respectively), once internalized by cancer cells.389 The results showed that in 48 h, only 30% of the stable Pt internalized was oxidized (to PtII or PtIV), while all of the labile Ag NPs were degraded in the same time (forming inside cells nanoclusters, AgO, AgS, or AgCl species).
In parallel to the advantages outlined in the previous section on scattering techniques, through the use of synchrotron sources (especially from the fourth generation),330−333 X-ray spectroscopy can be carried out locally with high spatial resolution, giving way to improved imaging modalities.
Methods for Observing the Colloidal Properties of Nanoparticles in Biological Environments
When coherent X-rays are scattered from disordered samples, such as an assembly of NPs or proteins, the photons scattered by the individual objects interfere and give rise to characteristic modulation of the scattered intensities. These patterns, usually referred to as speckle patterns, contain information about the exact spatial arrangement of the individual sample objects. Movements of the sample lead to corresponding changes of the scattering pattern. These changes can be quantified by calculating the time intensity autocorrelation function g(q,τ) = ⟨I(q,t) × I(q,t + τ)⟩/⟨I(q)⟩2 for a given point of the scattering pattern, a technique known as XPCS,158,390 the X-ray analogue of DLS. As an ensemble-averaging technique, XPCS allows for measuring the dynamics of all objects scattering at a particular length scale. In the simplest case of unhindered Brownian motion of a colloidal NP, the hydrodynamic diameter can be directly obtained, which changes depending on the state and surrounding medium of the colloidal NPs,161 as shown in Figure 18. It thus offers the possibility of monitoring colloidal stability or the size of a shell of adsorbed proteins, which slows down the free diffusion of a single NP. Moreover, XPCS gives access to dynamics that are directly associated with a certain length scale in a sample by the momentum transfer wave vector q. In the case of NP internalization by endocytosis/phagocytosis, hindered NP movement can be observed by q- and thus length-scale-dependent measurements.
Figure 18.
(a) SAXS pattern of an aqueous dispersion of 50 nm-diameter Au NPs. (b) Autocorrelation functions g(q,τ) for different scattering vectors q. The autocorrelation function was fitted by a diffusion model g(q,τ) ∝ exp(−2 × Γ(q)) with Γ(q) = D(q) × q2 + c). The diffusion coefficient D was fitted from the Γ(q) data, leading with the Stokes–Einstein equation (assuming 22 °C and using the viscosity of water) to an effective hydrodynamic diameter of 66 nm. These data were recorded at Deutsches Elektronen-Synchrotron (DESY) for this work by X. Sun, F. Otto, C. Sanchez-Cano, N. Feliu, F. Westermeier, and W. J. Parak and have not been published previously.
The advent of fast 2D X-ray detectors with framing rates in the milli- and even microsecond time ranges offers the possibility of observing direction-dependent dynamics by XPCS,391,392 which is beneficial in flowing systems or in directionally ordered structures as can be ubiquitously found in biological specimens, starting from individual cell organelles like the Golgi apparatus, individual cells like myocytes, up to complete organs like the heart. Changes in the dynamics, due, for example, to colloidal disintegration or the adsorption of proteins, can be tracked by calculating the two-time intensity correlation function g(q,t1,t2), which gives access to nonstationary dynamics and thereby enables following the temporal evolution of sample systems.393 One major challenge of most X-ray-based techniques, and in particular XPCS experiments, when applied to biological samples systems, is beam damage (see below for details about this topic). One possibility for minimizing damage is to employ XPCS based on speckle visibility, where the degradation of a speckle pattern is observed as a function of illumination time. As each measurement can be performed on a fresh sample spot, this greatly reduces the dose to the sample.394,395 In principle, measurements in this direction could offer in situ monitoring of NP degradation (which would leads reduced diameters) or changes in surface chemistry, which would change the state of agglomeration and thus alter the effective hydrodynamic diameter. In particular, colloidal stability of NP-based drugs in biological environment containing salt and large amounts of proteins (e.g., in blood) could be monitored.
Until now, XPCS has been used primarily to study the dynamics of colloidal glasses, gels, and polymers.396 In analogy to such systems, biological soft matter exhibits complex dynamics over many time and length scales, including nondiffusive, anisotropic, and spatially and temporally heterogeneous dynamics.397 Studying and understanding these dynamics, ranging from localized fast rattling-like motions of, for example, spatially confined NPs to slow network dynamics, could help to assign characteristic dynamic “signatures” to different cellular compartments and binding states. Beyond the state of the NP and its ligand shell in response to the complex environment mentioned above, analyses on the ensemble level are possible that quantify and characterize the fraction of localized NPs, as has been shown for gel-forming Au NP suspensions.398 Although the underlying questions in studying colloidal glasses and gels versus biological soft matter are quite different, the capability of XPCS to develop a complex microscopic picture of the system under study offers exciting possibilities. The increasing quality of photon sources, in particular with the advent of the fourth generation of synchrotron sources, in terms of coherent flux and coherence lengths helps to improve the signal-to-noise ratios, thus allowing XPCS studies of weakly scattering samples, as demonstrated recently for concentrated lysozyme solutions.399 Modern free-electron lasers (FELs) have superior coherence properties and high-repetition rates that result in outstanding temporal resolution.399 However, the high brilliance and ultrashort pulse lengths lead to radiation damage being a severe problem beyond single-shot studies. Nevertheless, FEL-XPCS studies of radiation-sensitive samples have been demonstrated and are therefore also conceivable for biological systems.401
First steps using related techniques have already been applied for monitoring the release of drugs from carrier NPs. In recent years it has been realized that the cellular distribution and delivery of NPs to cells under in vitro and in vivo conditions can be very different, primarily due to shear stresses generated by the fluid flow inside the body.402−406 The shear stress generated by the blood flow through a healthy artery is ∼1 Pa. As the stress generated is inversely proportional to r3 (r being radius of the vessel), the shear stress is enhanced by almost 1 order of magnitude (∼10–20 Pa) in stenosed vessels. The fluid shear stress contributes to regulating specific cellular processes and determines the efficacy of intracellular drug delivery. The interactions of NPs with cells depend on NP composition, charge, concentration, and shape as well as on the types of cells. In general, cationic NPs interact more strongly with cells due to negatively charged groups on the cell membrane407 (although this may not be true for bigger particles),238 and shear stress can further stimulate this interaction. The shear stress can affect ligand–receptor adhesion for NPs to cells, resulting in altered cellular uptake of the NPs and hence varied drug delivery. In recent years, mechano-sensitive drug delivery systems have been demonstrated where payload is released under enhanced stress. It has been suggested that shear-based drug delivery can be more powerful than biological and chemical methods, especially in conditions such as atherosclerosis where disease-specific markers are not well identified. Two routes have been demonstrated: (i) Shear-activated nanotherapeutics, where supraparticulate NPs (of the size of natural platelets) are composites of smaller NPs (with drugs), which are stable under normal blood flow, but break into individual NPs under high local shear stress in an obstructed region, thus delivering the drug or drugs.406 (ii) Vesicles loaded with drugs change their shape from spherical to lenticular shapes at elevated shear stresses in semiclogged portions, thus releasing drugs either due to pore formation or disruption of the vesicles (Figure 19).403 The exact mechanism can vary with the nature of the instabilities of the vesicles under shear stress. This discussion highlights the importance of fluidic shear stress both in tuning the cell-NP interaction as well as in targeted drug delivery. X-ray-based techniques like SAXS, XPCS, and X-ray fluorescence correlation spectroscopy in microfluidic devices, which emulate in vivo environments by mimicking fluid flow conditions of the body (including flexible channels and pulsating flows), will provide real-time structure and dynamics information on the interaction of the NPs carrying drugs to cells. Another exciting possibility is to use in situ grazing incidence X-ray diffraction from a monolayer consisting of NPs and cells at the air–water interface undergoing shear in an interfacial rheology setup. An experiment of this type has recently been demonstrated, wherein changes in the lipid–protein monolayers were quantitatively studied as a function of shear stress.408
Figure 19.
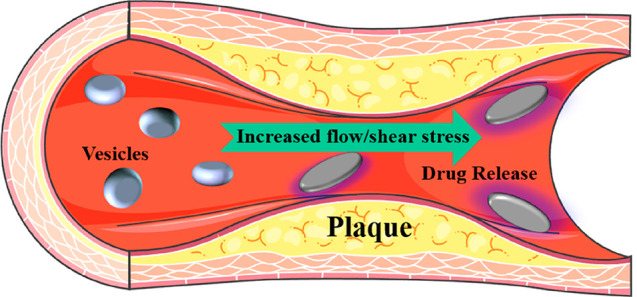
Schematic of shear-stress-induced drug delivery using shape changes of the drug-loaded vesicles. Cartoon created using images modified from Servier Medical Art (Servier, www.servier.com), under a CC-BY 3.0 International License.
Challenges for the Application of X-ray-Based Techniques to Biological Samples: Radiation Damage
Although X-ray-based techniques are promising tools to study NP-drugs and other nanomaterials in situ, there are still issues that must be addressed before they can be used to their full potential. For example, there is a lack of suitable labeling methods that enable detecting simultaneously the different components of NP-drugs. Moreover, while probing the variations in the chemical properties of metal nanomaterials using XAS with nm resolution has been possible in vitro,148,409−413 its application to biological samples has been limited due to concentration and sensitivity issues. Using longer acquisition times to obtain meaningful spectra is not always possible, as it normally causes unwanted radiation damage. Also, to achieve time-lapse in vivo recordings using X-rays, the challenge is to keep organisms alive; prolonged excitation time can lead to significant beam damage to biological samples. In particular, it remains a great challenge to track the fate and degradation/transformation of NPs in vivo, as the pristine NPs may break down and corrode into smaller NPs, clusters, molecules, or ions, due to the cellular and biomolecular interactions under the biological settings. Thus, concerning long-term tissue penetration, sometimes only some debris from the original NPs might be able to be translocated into an individual cell, or cells only would retain few NPs after a long-term therapy or upon a low-dose administration. These circumstances demand extremely high detection sensitivity and resolution of X-ray-based techniques.
Most synchrotron-based X-ray techniques are not capable of analyzing large numbers of samples in a short time. Therefore, they cannot provide data from significant cell populations, limiting the strength of results obtained to some extent. Most of these issues are due to the experimental approaches or set-ups currently used to acquire data using X-ray-based methods. As such, these limitations might be partially or totally overcome by a series of technical improvements currently available or in the process of being implemented. For example, the XFM beamline at the Australian Synchrotron can perform high-throughput analyses, and one study analyzed the uptake of ZnO NPs with roughly 1000 cells.414 Also, large-area and solid-angle XRF detector arrays are capable of achieving high-count rates in extremely short times, enabling the collection of on-the-fly, real-time elemental images or high-resolution XAS image stacks minimizing irradiation times,415−417 while CCD-based energy dispersive 2D detectors can be used to acquire full-field XRF images (both 2D and 3D).418 Developments in silicon drift detector array chips will increase throughput of the large-area, XRF detector arrays by orders of magnitude.419 Additionally, the fourth generation of synchrotron radiation sources (ESRF, MAX IV, and upcoming implementation of APS-U and PETRA IV at DESY) will provide access to much brighter and more coherent X-ray photon beams.330−333 These advances will help to increase the sensitivity of X-ray-based analytic techniques (e.g, sub-ppm for XRF imaging currently).149 Moreover, such improvements can dramatically reduce the irradiation time needed to obtain good quality data, enabling information collecting from larger populations and minimizing the damage to samples.
In fact, potential beam damage is one of the significant hurdles in advancing X-ray-based analysis toward more in vivo applications. Ideally, X-rays should be only an interrogator, leaving the sample (e.g., the NP-based drug) and its environment (e.g., tissue) unaffected. However, if exposure to X-rays is too severe, it can destroy biological molecules and damage tissues. Any radiation would be harmful, especially in terms of cumulative dose, which accounts for its own long-term health risks. Classified as a Group 1 carcinogen by the World Health Organization,420 any exposure to X-rays can cause DNA mutations, genetic damage, and further the consequent occurrence of cancers. For instance, leukemias have long been known to occur after detrimental radiation of several hundreds of mSv,421 but according to the widely accepted linear no threshold model, the risk to develop this type of cancer is assumed to increase linearly with dose from zero, and significant effects have recently been reported with doses as low as 50 mSv;422 although recent reports suggest that the role of radiation on cancer risk is far more complex.423 In contrast, for general clinical diagnosis in hospitals, the emerging risks of low-dose X-ray radiation below 10 mGy (or 10 mSv considering RBE = 1) are rather low, whereas the cumulative risk of cancer from diagnostic X-ray exposure was estimated to be approximately 0.6–1.8% to the age of 75 years.424 Nevertheless, radiation risk cannot be excluded from the young and occupational populations.425,426 Therefore, damage caused by X-rays is an important issue concerning its use in biomedical applications.427 The mechanism of radiation injury depends predominantly on the changes of biological macromolecules with exposure to X-rays. Radiation can directly interact with biological macromolecules and induce their ionization and excitation, resulting in molecular structure changes and loss of biological activity. Free radicals formed by X-rays can, in turn, damage biomolecules and, in particular, DNA. Meanwhile, the ionized and excited molecules are unstable, and the electronic structure within the molecules can be changed. This process can induce decomposition of molecules and changes in their structure, leading to the loss of biological function, especially when chromosomal DNA is affected.
Importantly, in the study of living organisms, radiative injury depends on many factors, including exposure-related factors (e.g., the irradiation time, dosage, fractionation, the size of the exposed area, and its site) and biological factors (e.g., physical structure, hormonal status, oxygen status, tissue renewal rate, and capillary density). Among them, acute high-dose irradiation may cause more tissue damage, including acute and chronic injuries, than long-term low-dose exposure with the same total dose. The extent of radiation damage often differs between different biological tissues and organs (e.g., skin, bone marrow, etc.). For instance, when the entire or a part of the body is exposed to X-rays, the skin, as an external organ, is first damaged to initiate and to promote skin radiation injury, including acute skin burns, chronic skin fibrosis, and, rarely, skin cancer.428−430 Bone marrow is one of the main sensitive target tissues following ionizing radiation exposure.431,432 Various hematopoietic stem and progenitor cells, naive hematopoietic cells in the bone marrow, and mature blood cells in lymphatic tissues are sensitive to radiation. Particularly when the bone marrow is exposed to large doses of X-rays, hematopoietic stem cells suffer a greater degree of radiation damage, and their self-renewal, proliferation, and differentiation will appear unbalanced, manifested as weakened self-renewal, resulting in serious reductions or even depletions of hematopoietic stem cells, which ultimately leads to bone marrow hematopoietic failure and loss of immune function. Although tremendous progress has been made in the prevention and treatment of radiation damage, there remains major scientific issues that need to be studied and overcome. These issues include not only the differences in radiosensitivity of different tissues and organs and the physiological damage caused to sensitive tissues and organs under high-dose irradiation but also the development of countermeasures against normal tissue radiation injury. The cumulative radiation dose should also be considered in preclinical animal models. In order to reach an optimal signal-to-noise ratio and high spatial resolution in X-ray-based analyses, relatively high radiation doses are necessary. Such high doses will not allow repeated measurements since high total body doses are lethal because of the bone marrow suppression. Second, high radiation doses certainly affect the experimental animal models and outcomes, that is, immune responses, tumor microenvironment, and/or tumor growth. Thus, the fate and therapeutic effect of NPs measured by X-ray-based methods should be re-evaluated using multimodality methods considering the effects of cumulative radiation doses on the animals.
For in vivo X-ray imaging of living organisms, radiation parameters such as dose limits need to be established. Here, a molecular understanding of radiation damage in model systems, such as NPs co-crystallized with proteins,433 might help to determine these parameters. X-rays can interact with water in the cells, causing water molecules to ionize or to be excited, to form highly active free radicals and peroxides after a series of reactions. These species actions on biological macromolecules can lead to changes in molecular structure and function, causing dysfunction and systemic lesions. In this context, X-ray protein crystallography may provide important insights into radiation damage at the molecular level, because it has a strong background on studying the effects of irradiating hydrated biological macromolecules with X-rays.434 In protein crystals, prolonged exposure to X-rays may alter structural features such as side chains or oxidation states of metal ions within the protein or affect global parameters such as unit cell dimensions.435 Strategies to overcome the site-specific and global radiation damage are, for example, low-temperature data collection436 and the determination of dose limits.437 Typically, synchrotron-based characterization of biological samples has been conducted through the use of microfluidics or using frozen or crystallized samples. Recent advances in near-ambient X-ray spectroscopy open the door to studying the behavior of NPs in biological samples under near-physiological conditions.438 With the advent of serial crystallography methods, including experiments with XFEL sources, most studies are carried out at room temperature. Here, radiation damage is mitigated by using a large number of irradiated species. Either the radiation is distributed over a large number of crystals or, in the case of XFEL studies, the diffraction outruns the destruction of the crystals. Thus, only minimal radiation damage is observed in the final data.439 For investigation of nano–bio interactions, these serial approaches might be suitable for minimizing radiation damage in cellular studies. Here, a high throughput of cellular material could ensure limited radiation damage after data processing. Such an approach also requires further development of serial X-ray-based imaging techniques, as discussed above.
There are strategies to reduce the impact of radiation damage. While large biological objects can readily be investigated by X-rays, microscopy of small biological objects, such as cells with high spatial resolution, risks potential radiation damage.440 Due to the ionizing nature of X-rays, radicals can be formed that lead to the cleavage of chemical bonds and cause structural changes,441 which become particularly obvious when nonconductive biological objects are analyzed.442 According to the empirical Rose criterion, the dose required for imaging an object reliably against the background noise increases with the desired resolution.443 Thus, the achievable resolution is ultimately limited by the X-ray dose that can be applied before radiation damage occurs.444,445 The impact of radiation damage on biological samples critically depends on their environment and sample preparation. For many mammalian cells already, ≈10 Gy is deadly.446 Elemental redistributions in XRF of unfixed vanadocytes and mass loss in dried chromosomes were observed for doses >105 Gy.447,448 As the resistance against radiation damage rises exponentially with decreasing temperature,449 structure determination by X-ray-based biocrystallography could be greatly improved when done at cryogenic temperatures, as much higher radiation doses can be applied than at room temperature.450 When investigated under cryogenic conditions, the diffraction signal of lysozyme crystals remained visible up to radiation doses of 107 Gy.451 In X-ray microscopy, the tolerable radiation dose for imaging before artifacts due to radiation damage occur was calculated to be 108–109 Gy.452 An additional advantage of cryogenic sample preparation, particularly for X-ray fluorescence analysis, is the preservation of the location of ions as close to the natural, hydrated state as possible. This feature was demonstrated by imaging the elemental distribution within duckweed roots,453 green algae,454 and fibroblast cells.455 While cryo-preparation of the samples is usually performed in the biological laboratories, transfer systems are required to deliver the frozen samples free of contaminants to the experiment. During measurements, the samples are kept in the frozen state either by a cryo-stream453 or in a vacuum chamber using cooled sample stages.454
Another possibility would be to harness the molecular machinery of cells. Consider that in addition to the direct and indirect effects discussed above, X-ray radiation can trigger a series of biochemical and molecular signaling events together forming or providing the radiation response of the organism (Figure 20) that may either repair the radiation-induced damage or result in long-term physiological changes or cell death. DNA damage can be caused even by irradiation as low as 1 mGy, and DNA repair proteins are recruited to these damaged sites.456 Important factors in radiation response are DNA repair,456,457 antioxidant defense systems,458 and immune and inflammatory responses.459−461 These systems provide the organism with highly effective protection against radiation-induced ROS and DNA damage, but they can also trigger long-term adverse effects. The efficiency of these radioprotective systems is reduced, and even plays a reverse role in the case of high-dose irradiation. This response is due to (i) abundant generation of radiation-induced ROS that affects the oxidant–antioxidant balance of the organism and causes potentially lethal DNA damage and (ii) radiation-induced mutations caused by erroneous repair. Consequently, high-dose irradiation can result in the alteration of enzyme activities and trigger uncontrolled inflammatory responses,462 which in turn can cause secondary vascular dysfunction and tissue damage with the subsequent activation of a variety of cell death mechanisms, such as apoptosis, necrosis, fibrosis, etc.463
Figure 20.
Mechanisms involved in radiation damage and subsequent radiation response.
To reduce radiation damage during X-ray-based in vivo research and in clinical radiotherapy, different natural or synthetic radioprotective agents have been shown to moderate radiation-induced molecular and cellular damage and/or to restore the physiological balance of the organism in vitro, in vivo, or in human randomized controlled trials. The radioprotective effect of such compounds is provided mostly by their antioxidative and immunomodulatory features, as they are able to suppress free radical production, remove already generated free radicals, and reduce radiation-induced inflammatory response.464 Another approach to mitigating radiation damage is the use of local radioenhancers, which would achieve higher on-site radiation at tumor cells or at the region of interest by applying lower overall radiation doses.465 Despite the large number of radioenhancers synthesized and studied even at the level of clinical trials,466 they are not largely used in clinical practice. Moreover, their application in NP research is also challenging, as it requires additional studies on whether the particular radioenhancer interacts with the NPs directly or has any indirect effects on the NP-based drug or NP carrier delivery.
When considering the applications of X-rays applied to biological samples, the use of the above-discussed imaging and spectroscopic techniques, especially to large biological specimens (such as whole mice or humans) will also need to address the issue of the trade-off between X-ray tissue penetration and the quality of the signals obtained. As described above, X-ray radiation can be divided into low-penetrating soft and tender X-rays (100 eV to 1 keV and 1 to 5 keV, respectively, penetrating up to a few μm) and high-penetrating hard X-rays (5 keV and higher). However, the penetration of hard X-rays is controlled by their energy. As such, X-rays with energies of ca. 15 keV are needed to penetrate over 1 cm of tissues, while much higher energies are required to analyze whole organisms. Up to 50 keV, the interactions of hard X-rays with tissue are dominated by photoabsorption events, which enable acquisition of multiple images and spectra with low background. It is thus possible to obtain easily high-quality XRF maps, XAS spectra, or scattering images (among others) in cell samples or ex vivo tissue samples (up to a couple of cm thick). However, at energies over 50 keV, the interaction between X-rays and tissue is dominated by inelastic Compton scattering. Although not critical for medical imaging (which uses X-rays in the energy range 10–150 keV), such scattering events can lead to the generation of large background noise and make it difficult to acquire maps of most elements, and other images and spectroscopic data. Thus, it will be necessary to develop improved acquisition and/or data analysis methods to reduce the background created by Compton scattering before certain X-ray-based analytics (i.e., XRF, or XAS-based techniques) can be properly applied in situ. There is progress in this direction, as described above. In a recent report, the localization of Au NPs on both tumor models and objects with human-size scales was determined with XFI by applying a spatial filtering scheme for background reduction, applying a local dose of only 10 mGy within the scanning X-ray beam volume, resulting in an even lower effective organ dose, as only parts of the body were scanned by XFI.57
Finally, most X-ray techniques that can be used to study NP-drugs in organisms currently require the use of synchrotron facilities. Therefore, the methodologies developed for those synchrotron-based tools will need to be transferred to benchtop environments if we want to apply X-ray-based analytics to clinically relevant situations. Although XRF elemental maps can be obtained from mice using current benchtop X-ray sources,467 they cannot reach the flux, coherence, or subcellular resolution achieved by synchrotron-based nanoprobe beamlines and still need to be improved significantly to reach adequate performance.
Perspectives and Outlook
Tissue is nontransparent to probes used in many analytical methods. However, for future biomedical applications concerning delivery, imaging, and diagnostics, the use of in situ analytics that include monitoring what is happening inside tissue at the (sub-) cellular level will be important. In biological media, NPs may interact with cells, organelles, and molecules,353 resulting in variations in aggregation, distribution, surface properties, and chemical environment and even in the structures of the NPs. Meanwhile, the biological functions and structures of the biological components are also affected by nano–bio interactions. For example, many metal NPs have been designed for nanomedicine. Compared to traditional pharmaceuticals, NP-based drug delivery may exhibit distinct pharmacokinetic and pharmacodynamics properties, which rely on step-by-step interactions between the NPs and the biological targets. The intracellular localization and the chemical transformation (valence state or chemical environment variation) of the NPs are critical to their biological functions and may help us to understand the degradation of NPs and the toxicological mechanisms of NPs. When NPs are treated with external stimuli to react with biological systems, in situ analyses could provide direct and visual details on physiological and pathological development. These details would offer better understanding of dynamic regulation in biological homeostasis. However, it is not currently possible to obtain detailed and comprehensive understanding of the (biological or molecular) events that affect NPs once inside the body of animals and humans. Detailed analytics can be achieved by testing blood samples, which, unfortunately, is not a local technique. It is possible to perform local analytics ex vivo on dissected organs, which is neither in situ nor applicable to humans. Thus, comprehensive in situ analytics at the molecular/cellular level are needed that make the body “transparent”, in order to observe the site of action. Therefore, the aim of this Review lies in highlighting the need for developing X-ray-based methods that are suitable for studying nanomaterials in complicated biological environments. Applications to humans and translation to the clinic of the techniques discussed here will often not be possible within a short time frame, but in vivo work on animals is already in the exploration stage.
This methodology would apply generally to all optically nontransparent samples and could also be used for other applications, such as environmental analysis and toxicology. Nanomedicine needs to look to other fields dealing with nano–bio interactions for inspiration. In this respect, toxicology has been using X-rays to track and to monitor the distributions of metal and metal–organic NPs in plants and animals for decades, and similar approaches could be straightforwardly adapted for drug delivery. XRF and STXM can be combined with XANES to identify the intracellular fate of ZnO NPs, with which it was found that toxicity can arise due to the dissolution of Zn and its complexation with molecules in the cell.468 In animals, the biodistribution of copper NPs in earthworms was monitored by XRF and the speciation of the copper by EXAFS.469 Additionally, the woody tissue of plants makes them ideal for testing X-ray based characterization techniques. SAXS has been used to monitor the formation and trafficking of Zn-based MOFs in plants.470 In another experiment, after uptake by algae, EXAFS was used to determine that Ag NPs can dissolve into Ag+, but then reaccumulate into different cellular compartments.471 By combining, X-ray microscopy, XANES, and electron microscopy, it was found that a wide size range of Au NPs can be taken up into the vasculatures of land plants, but only the smallest (∼3.5 nm) can then be internalized into the plant cells.472
For breakthroughs in applications in humans, it will be necessary to address not only the issues of potential radiation damage but also the development of improved laboratory X-ray sources and table-top or compact synchrotrons for clinical applications with low flux. Current laboratory X-ray sources can generate stable (both in emission and position) and reasonably brilliant (107–108 ph/s in the focus) micro- and nanofocused hard X-ray beams with certain levels of coherence that enable imaging on small animals and ex vivo tissue samples.243,248 Still, they normally produce polychromatic or broadband radiation that cannot be tuned and with much lower overall brilliance (at least 1000 times lower) and coherence than synchrotron sources, which makes it difficult to translate many synchrotron-based X-ray techniques to a laboratory or clinical environment. It is possible to produce highly monochromatic and collimated hard X-ray beams (i.e., with energies between 15 and 35 keV) with a brilliance (about 1010 ph/s) intermediate between that of laboratory sources and synchrotron facilities using CLS based on inverse Compton scattering.473,474 CLS can be installed in biomedical research institutions or hospitals and produces radiation that is stable enough for the acquisition of X-ray imaging,475 including in vivo experiments,285−287 but also can be used to perform X-ray spectroscopy experiments (i.e., XAS).476 Still, NP-based drugs are normally found at very low concentrations inside patients. Therefore, CLS would need to improve greatly before we can start thinking about applying such techniques to study NP-based drugs in situ, if possible at all.
The technological requirements for moving from synchrotron to conventional sources required for any successful translation to human patients already seems possible, in certain cases. One example of this opportunity is the application of gratings-based set-ups for the diagnosis of lung conditions using dark-field imaging.477−480 Gratings-based methods were demonstrated for phase-contrast imaging using synchrotron sources almost 20 years ago.481,482 The same approach was later used to acquire both phase-contrast483 and dark-field images484 with conventional X-ray sources, helping to bring them closer to the clinic. Gratings-based dark-field imaging proved to be promising and has been employed successfully in in vivo preclinical studies with animals of different sizes, from mice477−479 to pigs.480 Current efforts are directed to optimize the technique for use in humans, by testing it on cadavers,485,486 and will hopefully translate into clinical practice in the near future. Nevertheless, analytical methodologies that can be used to study NP-based drugs with laboratory instruments must be developed and optimized, which can be done now using state-of-the-art synchrotron facilities.
Next, we consider other potential medical applications of X-ray-based imaging for possible future clinical use. A number of pilot studies described above have illustrated the potential for imaging small animals with synchrotron X-ray-based methods. No single method yields all relevant information, and therefore, multimodal imaging should be considered, complementing the assessment by synchrotron X-rays with other non-invasive imaging approaches, preferably employing multimodal labels such as Au NPs. This strategy would enable combining longitudinal animal studies with synchrotron X-rays in in vivo–in situ assessments of intact animals or excised tissue for final examinations ex vivo. Another exciting opportunity would be to collect human tissue, for example, from organ transplantation or tumor resection surgery, to keep that tissue functional, and to study these specimens with synchrotron X-ray technologies.487 Similarly, bioreactors containing and preserving large tissue constructs and 3D in vitro tumor models for drug evaluation could also be used.488,489 Analyses of the perfused tissue could provide valuable insight into cellular uptake of NPs and subsequent responses. For example, for tomography with resolution below 1 μm, one could study 3D receptor distributions on tumors or other cells and thus tumor heterogeneity. It may be possible to detect single receptor-scale events using Au NPs and high-affinity antibodies, thus elucidating the targeting process. This insight could help to achieve accurate molecular imaging where still living tissue could be imaged and investigated with single-receptor resolution and sensitivity to understand and to improve drug delivery by nanocarriers.
Cancer is the leading cause of death worldwide. It is estimated that by 2030, the number of cancer cases will increase by more than 50% to 22 million per year.490 Early detection and optimal therapy of cancer largely depend on patho-anatomical information provided by imaging. Nearly all aspects of patient care require precise visualization of spatial information, that is, the tumor site, its anatomic relation to adjacent structures including displacement and/or infiltration of healthy organs as well as probable spreading to distant organs. Treatment decisions are made by multidisciplinary teams consisting of medical specialists of various disciplines, who collect history, clinical information, family and genetic data, laboratory data, and imaging data about the patient. Spatial information provided by imaging is an essential pillar of many diagnostic as well as therapeutic interventions, such as biopsy, surgery, radiation therapy, and minimally invasive focal as well as systemic therapies. Therapy planning and response are not only based on precise information on the localization, extension, and spreading of the tumor but also on the individual anatomy of the healthy structures. The overall quality of patient care depends on the quality of cancer imaging and repeated imaging over the course of years of patient care. Over the last five decades, medical imaging technologies have been improved by the invention of cross-sectional imaging technologies providing unforeseen opportunities for patient care. Various methods of ultrasound, CT, MRI, and positron emission tomography (PET/CT and PET/MRI), along with advances in computer science, provide high-resolution 3D visualization of anatomical and functional tumor features. While macroscopic medical imaging has improved impressively, the final diagnoses of cancer and the definite decisions on therapeutic strategies inevitably require microscopic information about tumor pathobiology on the cellular, subcellular, and molecular levels. Medical imaging often reaches its intrinsic limits with ca. 0.5–0.1 mm spatial resolution. This limit is not only technological but also due to human anatomical and physiological restrictions related to the size of the human body, tissue movement resulting from, for example, a beating heart, breathing, and bowel peristalsis, as well as the need for the lowest possible radiation exposure to the patient and limited examination time. To bridge this gap of the “nano–bio interface”, tissue often has to be sampled for further examination outside of the body. It is important to realize that small tissue samples must represent the disease characteristics and consequently critically depend on the right selection and extraction of tissue out of a heterogeneous tumor and peritumoral tissue identified by macroscopic imaging. A pathway of optimum cancer diagnostics in the clinical setting should follow a stepwise approach from macro- to microscale, bridging the nano–bio interface: (i) detection and localization of suspicious, potentially cancerous lesions by whole-body imaging (CT, MR, PET/CT, PET/MR); (ii) visualization of local tumor extensions and tumor heterogeneity by local high-resolution multiparametric imaging; (iii) high-precision sampling of representative tumor tissue by sophisticated image-guided biopsy methods; (iv) microstructural characterization of cellular, subcellular, and molecular features using ultrahigh-resolution synchrotron imaging; and finally (v) integration and evaluation of the entire set of multiplexed data exploiting deep-learning bioinformatic methods, leading to clinically relevant information on diagnosis, potential treatment, and prognosis. Possible fields of application are widely spread and include a variety of cancers, for example, prostate, breast, lung, pancreas, etc.
Another potential medical application would be XFI in vivo cell tracking in live animals, for example, tracking T cells (labeled with metal NPs or molecules suitable for XFI detection, such as iohexol) in immune-mediated inflammatory diseases (IMIDs). These diseases are a group of seemingly unrelated medical conditions affecting multiple organs, such as autoimmune hepatitis, nephritis, multiple sclerosis, and inflammatory bowel disease (IBD). All of these diseases are characterized by dysregulated immune response and nonhealing tissue damage, which promote a vicious cycle leading to chronic disease.491−493 Furthermore, chronic inflammation can promote the development of cancer. IBD, for example, is associated with colorectal cancer, especially in patients suffering from chronic intestinal inflammation. IMIDs are already among the leading causes of mortality in developed countries, and their prevalence is increasing.491−493 However, in most cases, current therapies are palliative and do not offer cures. Indeed, most therapies are based on immune-suppressive drugs, but are not able to reestablish homeostasis between the immune system, the tissue, and the microbiota. The resulting problems are relapsing flares and opportunistic infections that occur as a consequence of immune suppression. Thus, there is a major need for improved targeted therapies. Of note, CD4+ T cells are characteristic of the inflammation seen in IMIDs, and recent genome-wide association studies indicate that they do play key roles in the etiology of IMIDs and especially IBD.494 These data are further supported by murine studies, which have shown that an imbalance of effector T helper (TH) subsets and regulatory T cells, such as Foxp3+ T regulatory (TREG) and IL-10-producing Foxp3Neg type 1 regulatory T cells (TR1), plays important roles in IMIDs.495−500 CD4+ T cells are central players in adaptive immune responses. Naïve CD4+ T cells differentiate into a plethora of TH subsets, including TH1, TH2, TH17, and TH22 effector T cells with exquisite levels of functional specialization. Accordingly, one problem to understand is the spatiotemporal dynamics of these T-helper cell subsets during IMIDs and cancer. Note that inflammatory responses are different in distinct IMIDs and cancer. Intravital microscopy could be useful for this task, as it has been used successfully to explore in vivo and in situ immunological responses in surface tissues of animals (100–200 μm deep).107−109 However, until now, there has been no in vivo imaging method that would allow tracking several different T cell subsets at the same time in an entire animal with sufficient spatial and temporal resolution. There are trade-offs between increasing spatial versus temporal resolution, but detection of a small local number of marked T cells should be possible. Furthermore, current therapies are known to modulate T-helper cell responses. Therefore, a suitable in vivo imaging technology would have the potential not only to answer basic scientific questions but also to identify biomarkers for diagnoses of different IMIDs and to track the responses to specific therapies. By using different XFI tracers (either molecular or NP-based) that have similar sensitivities, several different types of immune cells could be tracked in a single measurement. The different T cells could be preloaded with different contrast, providing molecular agents or NPs via endocytosis, as has been demonstrated for stem cells and macrophages.501 By spatial filtering, the imaging sensitivity, in terms of the minimum local amount of XRF tracers, could be minimized, meaning that a small number of XFI-labeled immune cells could be visible via XFI in living animals.
Radiation damage has been discussed as a looming hurdle concerning the safety of in vivo X-ray analytics. On the other hand, such radiation damage to tissue via NPs could also be used for treatment, in which X-rays are intentionally used to destroy malignant tissue. Here, X-ray-based imaging techniques for detecting metallic NPs have important potential roles in radiation therapy. Ideally targeted delivery of metallic NPs to a tumor can be leveraged to increase the efficacy of radiation therapy.502−504 On one hand, high-resolution imaging of NPs could provide more accurate, single-cell localization of the tumor prior to irradiation, thus largely sparing surrounding healthy tissue.505,506 X-rays interacting with strongly absorbing metallic NPs deposit greater fractions of incident photon energy, thereby releasing higher amounts of low-energy secondary electrons, fluorescence X-rays, and ROS where needed. Both secondary electrons and ROS can further increase cellular damage, so that metallic NPs selectively accumulated in tumor tissue could be used as radiation sensitizers for cancer therapy. While the physical processes involved in the mechanisms of NP-mediated radiosensitization are being modeled with increasing accuracy,507,508 the roles of related chemical and biological processes are not fully understood.505,508 Since measurements of radiation-induced free radicals are quite complex, only a few experiments have performed to date. For example, it was found that Au NPs directly and indirectly increase hydroxyl and superoxide production in water.509−511 Experiments with macrophages incubated with Au and FeOx NPs could visualize the spatial patterns of hydroxyl radicals and superoxide anions produced by fluorescence X-rays and Auger electrons that were emitted upon cell irradiation with a scanning polychromatic synchrotron microbeam. These experiments showed that, while enhanced radical production from Auger electrons is limited to a range of ca. 100 μm from the NP-loaded cell, fluorescence X-rays can increase this production up to a distance of 1.5 mm for FeOx NPs and to 2 mm for Au NPs, respectively. Therefore, for enhancing radiation-induced cell damage for cancer treatment, in situ spatial distributions of specific NP systems, as well as biokinetics, possible toxicity, and tumor-targeting efficiencies will need to be thoroughly investigated prior to clinical implementation.
While this Review has predominantly addressed synchrotron-based X-ray analytics, the potential of XFELs should also be highlighted. XFELs, offering mJ pulse energies at hard X-ray energies and delivered in subhundred fs pulses, provide additional opportunities for structural investigations of NPs in both the physical and life sciences. These studies are typically performed either by exploiting the short pulse duration for time-resolved investigations512 or by leveraging the extreme pulse intensity to “outrun” radiation damage (caused by the X-ray–matter interactions) to determine the structures of noncrystalline (bio) materials at high resolution.513 Indeed, ultrafast pump–probe capabilities with XFELs have been demonstrated using solution-based samples.514 Pump–probe methods enable powerful means of data collection, including fs X-ray crystallography and temporal resolution of chemical processes spanning charge transfer and bond cleavage. Moreover, ultrafast pump–probe capabilities are superbly positioned to access NP dynamics in biological samples and are crucial in assessing the viability of some nanomaterials-based therapeutic approaches such as highly localized pulsed photothermal heating.515
In general, using coherent single-particle imaging (SPI) with free electron lasers, the 3D structures of biological samples can be investigated. SPI has progressed from observations of single cells,516,517 cellular substructures,518 and large viruses519 to the determination of the conformational landscape of smaller viruses.520 Simulations now suggest that the structures of single protein molecules can be determined at sub-nm length scales during experimental time allocated at high-repetition rate XFELs.521 Such high rates of data collection have been shown to be feasible at, for example, European XFEL (EuXFEL).522 For inorganic NPs, a large number of X-ray structural investigations are reported from diverse materials such as iron/silica,523 soot,524 gold/palladium,525 thiol/gold,526 and others, including from heterogeneous populations.527 These investigations inform both the development of methods applicable to the life sciences and, as outlined earlier, biofunctionalized inorganic NPs (bio–NPs) may be used as carriers to transport biologically active compounds (BAC) such as small molecules, peptides, and drugs. One concept is to release BACs (or “drugs”) from their light-sensitive NP carriers by application of an optical laser. Phytochromes are light sensitive and were originally identified in plants and then subsequently found in many other organisms.528 The large structural changes of the phytochromes upon red light illumination are unmatched (Figure 21). Red light is relatively harmless and, most importantly, penetrates deep into soft tissue. It is conceivable to engineer phytochrome constructs that are specifically optimized for uptake and release of BACs. An advantage would be that intense red light could be applied simultaneously and localized to multiple positions allowing for the treatment of multiple sites at the same time. To investigate the mechanism of BAC uptake and release by these light controlled bioinorganic nanomachines, their structures must be determined to high enough resolution that is roughly equivalent to the amplitude of the structural changes after light illumination.529−531 Resolution between 1 and 2 nm should be sufficient (Figure 21). These experiments are likely to become feasible in the near future with existing free electron lasers.522 We anticipate that such SPI experiments are extremely “photon hungry”, both in photons per X-ray pulse and in the number of pulses (and hence diffraction images) collected.532 The EuXFEL is presently the world’s highest repetition rate XFEL, offering up to 3520 measured images per sec—more than an order of magnitude more than the next XFEL source. This repetition rate enables the collection of large data sets in reasonable measurement times and bodes well for the applicability of this method to both organic and inorganic NPs.533
Figure 21.
Structural changes in phytochrome photosensory core modules. (a) The Stigmatella aurantiaca photochrome P2 in the Pr state.530 The central biliverdin (BV) chromophore is marked. The chromophore absorbs 700 nm red light. Upon light absorption, it isomerizes from a Z configuration to an E configuration. The configurational change triggers a large conformational change. Centroid distance of the movable domain: 30 Å. The sensory tongue (red dashed box) adopts a β-sheet structure. The BAC (green star) is bound. (b) Reaction product after light absorption as depicted by the Deinococcus radiodurans phytochrome in the Pfr state.531 The chromophore absorbs at 750 nm in the far red. The centroid distance of the movable domain is 55 Å. The sensory tongue (red box) then adopts an α-helical structure. The BAC (green star) is free to leave. The reaction between Pr and Pfr states is reversible to facilitate uptake and release of the BAC.
Improvements in the development of X-ray light sources will enable their use for biological/medical experiments that have not yet been feasible. Inorganic NPs, due to their high X-ray absorption cross sections and their potential conjugation with BACs/drugs, are an important part of the molecular and nanoscience toolkits for exploiting such developments. We anticipate that there will be increases in the uses of X-ray techniques to explore the fates and mechanisms of therapeutic nanomaterials once administered to animal models and patients. We further expect applications to experiments in laboratory, preclinical, and clinical environments, as large efforts are currently directed to developing improved compact X-ray sources. We foresee that X-rays will have important, fundamental roles in the advance of nanomedicine toward its maturity over the next decades.
Acknowledgments
Parts of this work were supported by the Cluster of Excellence ’Advanced Imaging of Matter’ of the Deutsche Forschungsgemeinschaft (DFG) - EXC 2056 - project ID 390715994. Part of this work was performed under the Maria de Maeztu Units of Excellence Programme - grant no. MDM-2017-0720 Ministry of Science, Innovation and Universities. C.S.C. thanks Gipuzkoa Foru Aldundia (Gipuzkoa Fellows program; grant no. 2019-FELL-000018-01/62/2019) for financial support. R.A.A.P. acknowledges the Ministerio de Economia y Competitividad (CTQ2017-88648R), the Generalitat de Cataluña (2017SGR883), and the Universitat Rovira i Virgili (2019PFR-URV-B2-02) for financial support. F.C. acknowledges the award of a National Health and Medical Research Council Senior Principal Research Fellowship (GNT1135806) and support from the Australian Research Council Centre of Excellence in Convergent Bio-Nano Science and Technology (project no. CE140100036). F.G., C.K., Y.L., and O.S. acknowledge DESY (Hamburg, Germany), a member of the Helmholtz Association HGF, for the provision of experimental facilities. Parts of this research were carried out at PETRA III, and we would like to thank the beamline team for assistance in using P21.1. C.F. and Y.Z. thank the National Key R&D Program of China (2016YFA0400900) for financial support. Y.Z. also acknowledges the Key Research Program of Frontier Sciences (QYZDJ-SSW-SLH031) and the LU Jiaxi International Team of the Chinese Academy of Sciences. Y.H., Y.K., Y.L., Z.L., B.Q., X.S., and H.Y. acknowledge the Chinese Scholarship Council for funding. N.K. thanks the Alexander von Humboldt Foundation for a Visiting Professorship. L.M.L.-M. acknowledges funding by the European Research Council (ERC AdG 4DbioSERS, no. 787510). A.K.S. and M.K.S. thank the Department of Science and Technology, India, for support. K.R. acknowledges support by BMBF grant nos. 02NUK032 and 02NUK035B. D.M.S. and P.S.W. thank the U.S. National Science Foundation, grant no. CHE-2004238, for support. Part of the phytochrome work is supported by BioXFEL STC grant no. NSF-1231306.
Glossary
Vocabulary
- Synchrotron radiation
electromagnetic radiation emitted by electrons (or other charged particles) traveling at near the speed of light when their direction is altered by an external magnetic field
- synchrotron brilliance
indicates the quality of a synchrotron source and can be defined as the number of photons within a bandwidth of 0.1% of the central wavelength with the same angular divergence found per unit area of the beam every second
- synchrotron emittance
average distribution of the relative position and momentum of the electron beam of the synchrotron. Low synchrotron emittances normally lead to smaller X-ray beams and higher brilliance
- diffraction limited storage ring
a synchrotron storage ring that maintains an electron beam with similar or lower emittance than the smaller X-ray photon beam that it produces
- coherent X-rays
X-ray radiation with a fixed relationship between their properties, normally referred to as X-rays with a constant difference between their phases
- collimated X-ray beam
X-ray beam with high spatial coherence. This means that its photons follow parallel or almost parallel trajectories and will not get dispersed with distance
- free electron laser
fourth-generation synchrotron radiation sources that produce short pulses of highly coherent and extremely brilliant radiation.
The authors declare no competing financial interest.
Dedication
This article is dedicated to our co-author, colleague, and friend Dr. Theo Schotten, who sadly passed away the day the galley proofs of our article were received, and who will be sadly missed.
References
- Holding J. D.; Lindup W. E.; Laer C. v.; Vreeburg G. C.; Schilling V.; Wilson J. A.; Stell P. M. Phase I Trial of a Cisplatin-Albumin Complex for the Treatment of Cancer of the Head and Neck. Br. J. Clin. Pharmacol. 1992, 33, 75–81. 10.1111/j.1365-2125.1992.tb04003.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins M. J.; Soon-Shiong P.; Desai N. Protein Nanoparticles as Drug Carriers in Clinical Medicine. Adv. Drug Delivery Rev. 2008, 60, 876–885. 10.1016/j.addr.2007.08.044. [DOI] [PubMed] [Google Scholar]
- Barenholz Y. Doxil® — The First FDA-Approved Nano-Drug: Lessons Learned. J. Controlled Release 2012, 160, 117–134. 10.1016/j.jconrel.2012.03.020. [DOI] [PubMed] [Google Scholar]
- Abrantes C. G.; Duarte D.; Reis C. P. An Overview of Pharmaceutical Excipients: Safe or Not Safe?. J. Pharm. Sci. 2016, 105, 2019–2026. 10.1016/j.xphs.2016.03.019. [DOI] [PubMed] [Google Scholar]
- Junghanns J.-U. A. H.; Müller R. H. Nanocrystal Technology, Drug Delivery and Clinical Applications. Int. J. Nanomed. 2008, 3, 295–309. 10.2147/IJN.S595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limaye V.; Fortwengel G.; Limaye D. Regulatory Roadmap for Nanotechnology Based Medicines. Int. J. Drug Regul. Aff. 2014, 2, 33–41. 10.22270/ijdra.v2i4.151. [DOI] [Google Scholar]
- Roy S.; Liu Z.; Sun X.; Gharib M.; Yan H.; Huang Y.; Megahed S.; Schnabel M.; Zhu D.; Feliu N.; Chakraborty I.; Sanchez-Cano C.; Alkilany A. M.; Parak W. J. Assembly and Degradation of Inorganic Nanoparticles in Biological Environments. Bioconjugate Chem. 2019, 30, 2751–2762. 10.1021/acs.bioconjchem.9b00645. [DOI] [PubMed] [Google Scholar]
- Pelaz B.; Alexiou C.; Alvarez-Puebla R. A. A.; Alves F.; Andrews A. M.; Ashraf S.; Balogh L. P.; Ballerini L.; Bestetti A.; Brendel C.; Bosi S.; Carril M.; Chan W. C. W.; Chen C.; Chen X.; Chen X.; Cheng Z.; Cui D.; Du J.; Dullin C.; et al. Diverse Applications of Nanomedicine. ACS Nano 2017, 11, 2313–2381. 10.1021/acsnano.6b06040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliu N.; Docter D.; Heine M.; Del Pino P.; Ashraf S.; Kolosnjaj-Tabi J.; Macchiarini P.; Nielsen P.; Alloyeau D.; Gazeau F.; Stauber R. H.; Parak W. J. In Vivo Degeneration and the Fate of Inorganic Nanoparticles. Chem. Soc. Rev. 2016, 45, 2440–2457. 10.1039/C5CS00699F. [DOI] [PubMed] [Google Scholar]
- Rosenblum D.; Joshi N.; Tao W.; Karp J. M.; Peer D. Progress and Challenges towards Targeted Delivery of Cancer Therapeutics. Nat. Commun. 2018, 9, 1410. 10.1038/s41467-018-03705-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narum S. M.; Le T.; Le D. P.; Lee J. C.; Donahue N. D.; Yang W.; Wilhelm S.. Chapter 4 - Passive Targeting in Nanomedicine: Fundamental Concepts, Body Interactions, and Clinical Potential. In Nanoparticles for Biomedical Applications; Chung E. J., Leon L., Rinaldi C., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp 37–53. [Google Scholar]
- Salvati A.; Pitek A. S.; Monopoli M. P.; Prapainop K.; Bombelli F. B.; Hristov D. R.; Kelly P. M.; Aberg C.; Mahon E.; Dawson K. A. Transferrin-Functionalized Nanoparticles Lose Their Targeting Capabilities When a Biomolecule Corona Adsorbs on the Surface. Nat. Nanotechnol. 2013, 8, 137–143. 10.1038/nnano.2012.237. [DOI] [PubMed] [Google Scholar]
- Colombo M.; Fiandra L.; Alessio G.; Mazzucchelli S.; Nebuloni M.; De Palma C.; Kantner K.; Pelaz B.; Rotem R.; Corsi F.; Parak W. J.; Prosperi D. Tumour Homing and Therapeutic Effect of Colloidal Nanoparticles Depend on the Number of Attached Antibodies. Nat. Commun. 2016, 7, 13818. 10.1038/ncomms13818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasting C.; Schalley C. A.; Weber M.; Seitz O.; Hecht S.; Koksch B.; Dernedde J.; Graf C.; Knapp E.-W.; Haag R. Multivalency as a Chemical Organization and Action Principle. Angew. Chem., Int. Ed. 2012, 51, 10472–10498. 10.1002/anie.201201114. [DOI] [PubMed] [Google Scholar]
- Kreyling W. G.; Abdelmonem A. M.; Ali Z.; Alves F.; Geiser M.; Haberl N.; Hartmann R.; Hirn S.; de Aberasturi D. J.; Kantner K.; Khadem-Saba G.; Montenegro J. M.; Rejman J.; Rojo T.; de Larramendi I. R.; Ufartes R.; Wenk A.; Parak W. J. In Vivo Integrity of Polymer-Coated Gold Nanoparticles. Nat. Nanotechnol. 2015, 10, 619–623. 10.1038/nnano.2015.111. [DOI] [PubMed] [Google Scholar]
- Bruinink A.; Wang J.; Wick P. Effect of Particle Agglomeration in Nanotoxicology. Arch. Toxicol. 2015, 89, 659–675. 10.1007/s00204-015-1460-6. [DOI] [PubMed] [Google Scholar]
- Carrillo-Carrion C.; Bocanegra A. I.; Arnaiz B.; Feliu N.; Zhu D.; Parak W. J. Triple-Labeling of Polymer-Coated Quantum Dots and Adsorbed Proteins for Tracing Their Fate in Cell Cultures. ACS Nano 2019, 13, 4631–4639. 10.1021/acsnano.9b00728. [DOI] [PubMed] [Google Scholar]
- Guan S.; Rosenecker J. Nanotechnologies in Delivery of mRNA Therapeutics Using Nonviral Vector-Based Delivery Systems. Gene Ther. 2017, 24, 133–143. 10.1038/gt.2017.5. [DOI] [PubMed] [Google Scholar]
- Sharma V. K.; Siskova K. M.; Zboril R.; Gardea-Torresdey J. L. Organic-Coated Silver Nanoparticles in Biological and Environmental Conditions: Fate, Stability and Toxicity. Adv. Colloid Interface Sci. 2014, 204, 15–34. 10.1016/j.cis.2013.12.002. [DOI] [PubMed] [Google Scholar]
- Jiang X. M.; Miclaus T.; Wang L. M.; Foldbjerg R.; Sutherland D. S.; Autrup H.; Chen C. Y.; Beer C. Fast Intracellular Dissolution and Persistent Cellular Uptake of Silver Nanoparticles in CHO-K1 Cells: Implication for Cytotoxicity. Nanotoxicology 2015, 9, 181–189. 10.3109/17435390.2014.907457. [DOI] [PubMed] [Google Scholar]
- Manshian B. B.; Pfeiffer C.; Pelaz B.; Heimerl T.; Gallego M.; Moller M.; del Pino P.; Himmelreich U.; Parak W. J.; Soenen S. J. High-Content Imaging and Gene Expression Approaches to Unravel the Effect of Surface Functionality on Cellular Interactions of Silver Nanoparticles. ACS Nano 2015, 9, 10431–10444. 10.1021/acsnano.5b04661. [DOI] [PubMed] [Google Scholar]
- Wang L. M.; Zhang T. L.; Li P. Y.; Huang W. X.; Tang J. L.; Wang P. Y.; Liu J.; Yuan Q. X.; Bai R.; Li B.; Zhang K.; Zhao Y. L.; Chen C. Y. Use of Synchrotron Radiation-Analytical Techniques to Reveal Chemical Origin of Silver-Nanoparticle Cytotoxicity. ACS Nano 2015, 9, 6532–6547. 10.1021/acsnano.5b02483. [DOI] [PubMed] [Google Scholar]
- Veronesi G.; Aude-Garcia C.; Kieffer I.; Gallon T.; Delangle P.; Herlin-Boime N.; Rabilloud T.; Carrière M. Exposure-Dependent Ag+ Release from Silver Nanoparticles and Its Complexation in AgS2 Sites in Primary Murine Macrophages. Nanoscale 2015, 7, 7323–7330. 10.1039/C5NR00353A. [DOI] [PubMed] [Google Scholar]
- Veronesi G.; Deniaud A.; Gallon T.; Jouneau P.-H.; Villanova J.; Delangle P.; Carrière M.; Kieffer I.; Charbonnier P.; Mintz E.; Michaud-Soret I. Visualization, Quantification and Coordination of Ag+ Ions Released from Silver Nanoparticles in Hepatocytes. Nanoscale 2016, 8, 17012–17021. 10.1039/C6NR04381J. [DOI] [PubMed] [Google Scholar]
- Rejman J.; Nazarenus M.; Jimenez de Aberasturi D.; Said A. H.; Feliu N.; Parak W. J. Some Thoughts about the Intracellular Location of Nanoparticles and the Resulting Consequences. J. Colloid Interface Sci. 2016, 482, 260–266. 10.1016/j.jcis.2016.07.065. [DOI] [PubMed] [Google Scholar]
- Angelov B.; Angelova A.; Mutafchieva R.; Lesieur S.; Vainio U.; Garamus V. M.; Jensen G. V.; Pedersen J. S. SAXS Investigation of a Cubic to a Sponge (L3) Phase Transition in Self-Assembled Lipid Nanocarriers. Phys. Chem. Chem. Phys. 2011, 13, 3073–3081. 10.1039/C0CP01029D. [DOI] [PubMed] [Google Scholar]
- Mertins O.; Mathews P. D.; Angelova A. Advances in the Design of pH-Sensitive Cubosome Liquid Crystalline Nanocarriers for Drug Delivery Applications. Nanomaterials 2020, 10, 963. 10.3390/nano10050963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salentinig S.; Tangso K. J.; Hawley A.; Boyd B. J. pH-Driven Colloidal Transformations Based on the Vasoactive Drug Nicergoline. Langmuir 2014, 30, 14776–14781. 10.1021/la503824z. [DOI] [PubMed] [Google Scholar]
- Angelov B.; Angelova A.; Garamus V. M.; Drechsler M.; Willumeit R.; Mutafchieva R.; Štĕpánek P.; Lesieur S. Earliest Stage of the Tetrahedral Nanochannel Formation in Cubosome Particles from Unilamellar Nanovesicles. Langmuir 2012, 28, 16647–16655. 10.1021/la302721n. [DOI] [PubMed] [Google Scholar]
- Reimhult E. Nanoparticle-Triggered Release from Lipid Membrane Vesicles. New Biotechnol. 2015, 32, 665–672. 10.1016/j.nbt.2014.12.002. [DOI] [PubMed] [Google Scholar]
- Peñate-Medina O.; Peñate-Medina T.; Humbert J.; Qi B.; Baum W.; Will O.; Damm T.; Glüer C. Using Alendronic Acid Coupled Fluorescently Labelled SM Liposomes as a Vehicle for Bone Targeting. Curr. Pharm. Des. 2020, 26, 6021–6027. 10.2174/1381612826666200614175905. [DOI] [PubMed] [Google Scholar]
- Peñate-Medina T.; Kraas E.; Luo K.; Humbert J.; Zhu H.; Mertens F.; Gerle M.; Rochweder A.; Damoah C.; Will O.; Acil Y.; Kairemo K.; Wiltfang J.; Glüer C. C.; Scherließ R.; Sebens S.; Peñate-Medina O. Utilizing ICG Spectroscopical Properties for Real-Time Nanoparticle Release Quantification in Vitro and in Vivo in Imaging Setups. Curr. Pharm. Des. 2020, 26, 3828–3833. 10.2174/1381612826666200318170849. [DOI] [PubMed] [Google Scholar]
- Mi P. Stimuli-Responsive Nanocarriers for Drug Delivery, Tumor Imaging, Therapy and Theranostics. Theranostics 2020, 10, 4557–4588. 10.7150/thno.38069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian B.; Fernandez-Bravo A.; Najafiaghdam H.; Torquato N. A.; Altoe M. V. P.; Teitelboim A.; Tajon C. A.; Tian Y.; Borys N. J.; Barnard E. S.; Anwar M.; Chan E. M.; Schuck P. J.; Cohen B. E. Low Irradiance Multiphoton Imaging with Alloyed Lanthanide Nanocrystals. Nat. Commun. 2018, 9, 3082. 10.1038/s41467-018-05577-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy E. S.; Tajon C. A.; Bischof T. S.; Iafrati J.; Fernandez-Bravo A.; Garfield D. J.; Chamanzar M.; Maharbiz M. M.; Sohal V. S.; Schuck P. J.; Cohen B. E.; Chan E. M. Energy-Looping Nanoparticles: Harnessing Excited-State Absorption for Deep-Tissue Imaging. ACS Nano 2016, 10, 8423–8433. 10.1021/acsnano.6b03288. [DOI] [PubMed] [Google Scholar]
- Chien Y.-H.; Chou Y.-L.; Wang S.-W.; Hung S.-T.; Liau M.-C.; Chao Y.-J.; Su C.-H.; Yeh C.-S. Near-Infrared Light Photocontrolled Targeting, Bioimaging, and Chemotherapy with Caged Upconversion Nanoparticles in Vitro and in Vivo. ACS Nano 2013, 7, 8516–8528. 10.1021/nn402399m. [DOI] [PubMed] [Google Scholar]
- Jalani G.; Tam V.; Vetrone F.; Cerruti M. Seeing, Targeting and Delivering with Upconverting Nanoparticles. J. Am. Chem. Soc. 2018, 140, 10923–10931. 10.1021/jacs.8b03977. [DOI] [PubMed] [Google Scholar]
- Lai J.; Shah B. P.; Zhang Y.; Yang L.; Lee K.-B. Real-Time Monitoring of ATP-Responsive Drug Release Using Mesoporous-Silica-Coated Multicolor Upconversion Nanoparticles. ACS Nano 2015, 9, 5234–5245. 10.1021/acsnano.5b00641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam S. H.; Bae Y. M.; Park Y. I.; Kim J. H.; Kim H. M.; Choi J. S.; Lee K. T.; Hyeon T.; Suh Y. D. Long-Term Real-Time Tracking of Lanthanide Ion Doped Upconverting Nanoparticles in Living Cells. Angew. Chem., Int. Ed. 2011, 50, 6093–6097. 10.1002/anie.201007979. [DOI] [PubMed] [Google Scholar]
- Wu S.; Han G.; Milliron D. J.; Aloni S.; Altoe V.; Talapin D. V.; Cohen B. E.; Schuck P. J. Non-Blinking and Photostable Upconverted Luminescence from Single Lanthanide-Doped Nanocrystals. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 10917–10921. 10.1073/pnas.0904792106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetrone F.; Naccache R.; Zamarron A.; Juarranz de la Fuente A.; Sanz-Rodriguez F.; Martinez Maestro L.; Martin Rodriguez E.; Jaque D.; Garcia Sole J.; Capobianco J. A. Temperature Sensing Using Fluorescent Nanothermometers. ACS Nano 2010, 4, 3254–3258. 10.1021/nn100244a. [DOI] [PubMed] [Google Scholar]
- Ortgies D. H.; Tan M.; Ximendes E. C.; del Rosal B.; Hu J.; Xu L.; Wang X.; Martín Rodríguez E.; Jacinto C.; Fernandez N.; Chen G.; Jaque D. Lifetime-Encoded Infrared-Emitting Nanoparticles for in Vivo Multiplexed Imaging. ACS Nano 2018, 12, 4362–4368. 10.1021/acsnano.7b09189. [DOI] [PubMed] [Google Scholar]
- Lay A.; Wang D. S.; Wisser M. D.; Mehlenbacher R. D.; Lin Y.; Goodman M. B.; Mao W. L.; Dionne J. A. Upconverting Nanoparticles as Optical Sensors of Nano- to Micro-Newton Forces. Nano Lett. 2017, 17, 4172–4177. 10.1021/acs.nanolett.7b00963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y.; Bao J.; Zhang Y.; Li Z.; Zhou X.; Wan C.; Huang L.; Zhao Y.; Han G.; Xue T. Mammalian Near-Infrared Image Vision through Injectable and Self-Powered Retinal Nanoantennae. Cell 2019, 177, 243–255. 10.1016/j.cell.2019.01.038. [DOI] [PubMed] [Google Scholar]
- Chen S.; Weitemier A. Z.; Zeng X.; He L.; Wang X.; Tao Y.; Huang A. J. Y.; Hashimotodani Y.; Kano M.; Iwasaki H.; Parajuli L. K.; Okabe S.; Teh D. B. L.; All A. H.; Tsutsui-Kimura I.; Tanaka K. F.; Liu X.; McHugh T. J. Near-Infrared Deep Brain Stimulation via Upconversion Nanoparticle Mediated Optogenetics. Science 2018, 359, 679–684. 10.1126/science.aaq1144. [DOI] [PubMed] [Google Scholar]
- Mann V. R.; Powers A. S.; Tilley D. C.; Sack J. T.; Cohen B. E. Azide–Alkyne Click Conjugation on Quantum Dots by Selective Copper Coordination. ACS Nano 2018, 12, 4469–4477. 10.1021/acsnano.8b00575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thal L. B.; Mann V. R.; Sprinzen D.; McBride J. R.; Reid K. R.; Tomlinson I. D.; McMahon D. G.; Cohen B. E.; Rosenthal S. J. Ligand-Conjugated Quantum Dots for Fast Sub-Diffraction Protein Tracking in Acute Brain Slices. Biomater. Sci. 2020, 8, 837–845. 10.1039/C9BM01629E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichner S. M.; Mann V. R.; Powers A. S.; Segal M. A.; Mir M.; Bandaria J. N.; DeWitt M. A.; Darzacq X.; Yildiz A.; Cohen B. E. Covalent Protein Labeling and Improved Single-Molecule Optical Properties of Aqueous CdSe/CdS Quantum Dots. ACS Nano 2017, 11, 6773–6781. 10.1021/acsnano.7b01470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo M.; Mazzucchelli S.; Montenegro J. M.; Galbiati E.; Corsi F.; Parak W. J.; Prosperi D. Protein Oriented Ligation on Nanoparticles Exploiting O6-Alkylguanine-DNA Transferase (SNAP) Genetically Encoded Fusion. Small 2012, 8, 1492–1497. 10.1002/smll.201102284. [DOI] [PubMed] [Google Scholar]
- Alkilany A. M.; Zhu L.; Weller H.; Mews A.; Parak W.; Barz M.; Feliu N. Ligand Density on Nanoparticles: A Parameter with Critical Impact on Nanomedicine. Adv. Drug Delivery Rev. 2019, 143, 22–36. 10.1016/j.addr.2019.05.010. [DOI] [PubMed] [Google Scholar]
- Kriemen E.; Holzapfel M.; Ruf E.; Rehbein J.; Maison W. Synthesis and Structural Analysis of 1,4,7,10-Tetraazacyclododecane-1,4,7,10--Tetraazidoethylacetic Acid (DOTAZA) Complexes. Eur. J. Inorg. Chem. 2015, 2015, 5368–5378. 10.1002/ejic.201500789. [DOI] [Google Scholar]
- Ali Z.; Abbasi A. Z.; Zhang F.; Arosio P.; Lascialfari A.; Casula M. F.; Wenk A.; Kreyling W.; Plapper R.; Seidel M.; Niessner R.; Knoll J.; Seubert A.; Parak W. J. Multifunctional Nanoparticles for Dual Imaging. Anal. Chem. 2011, 83, 2877–2882. 10.1021/ac103261y. [DOI] [PubMed] [Google Scholar]
- Price E. W.; Orvig C. Matching Chelators to Radiometals for Radiopharmaceuticals. Chem. Soc. Rev. 2014, 43, 260–290. 10.1039/C3CS60304K. [DOI] [PubMed] [Google Scholar]
- Holzapfel M.; Mutas M.; Chandralingam S.; von Salisch C.; Peric N.; Segelke T.; Fischer M.; Chakraborty I.; Parak W. J.; Frangioni J. V.; Maison W. Nonradioactive Cell Assay for the Evaluation of Modular Prostate-Specific Membrane Antigen Targeting Ligands via Inductively Coupled Plasma Mass Spectrometry. J. Med. Chem. 2019, 62, 10912–10918. 10.1021/acs.jmedchem.9b01606. [DOI] [PubMed] [Google Scholar]
- Wahsner J.; Gale E. M.; Rodríguez-Rodríguez A.; Caravan P. Chemistry of MRI Contrast Agents: Current Challenges and New Frontiers. Chem. Rev. 2019, 119, 957–1057. 10.1021/acs.chemrev.8b00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S.; Song J.; Qu J.; Cheng Z. Crucial Breakthrough of Second Near-Infrared Biological Window Fluorophores: Design and Synthesis toward Multimodal Imaging and Theranostics. Chem. Soc. Rev. 2018, 47, 4258–4278. 10.1039/C8CS00234G. [DOI] [PubMed] [Google Scholar]
- Grüner F.; Blumendorf F.; Schmutzler O.; Staufer T.; Bradbury M.; Wiesner U.; Rosentreter T.; Loers G.; Lutz D.; Richter B.; Fischer M.; Schulz F.; Steiner S.; Warmer M.; Burkhardt A.; Meents A.; Kupinski M.; Hoeschen C. Localising Functionalised Gold-Nanoparticles in Murine Spinal Cords by X-Ray Fluorescence Imaging and Background-Reduction Through Spatial Filtering for Human-Sized Objects. Sci. Rep. 2018, 8, 16561. 10.1038/s41598-018-34925-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avery R. K.; Albadawi H.; Akbari M.; Zhang Y. S.; Duggan M. J.; Sahani D. V.; Olsen B. D.; Khademhosseini A.; Oklu R. An Injectable Shear-Thinning Biomaterial for Endovascular Embolization. Sci. Transl. Med. 2016, 8, 365ra156 10.1126/scitranslmed.aah5533. [DOI] [PubMed] [Google Scholar]
- Carrillo-Carrion C.; Carril M.; Parak W. J. Techniques for the Experimental Investigation of the Protein Corona. Curr. Opin. Biotechnol. 2017, 46, 106–113. 10.1016/j.copbio.2017.02.009. [DOI] [PubMed] [Google Scholar]
- Witzler M.; Küllmer F.; Günther K. Validating a Single-Particle ICP-MS Method to Measure Nanoparticles in Human Whole Blood for Nanotoxicology. Anal. Lett. 2018, 51, 587–599. 10.1080/00032719.2017.1327538. [DOI] [Google Scholar]
- Wang W.-Y.; Yao C.; Shao Y.-F.; Mu H.-J.; Sun K.-X. Determination of Puerarin in Rabbit Aqueous Humor by Liquid Chromatography Tandem Mass Spectrometry Using Microdialysis Sampling after Topical Administration of Puerarin PAMAM Dendrimer Complex. J. Pharm. Biomed. Anal. 2011, 56, 825–829. 10.1016/j.jpba.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Epemolu O.; Mayer I.; Hope F.; Scullion P.; Desmond P. Liquid Chromatography/Mass Spectrometric Bioanalysis of a Modified γ-Cyclodextrin (Org 25969) and Rocuronium Bromide (Org 9426) in Guinea Pig Plasma and Urine: Its Application to Determine the Plasma Pharmacokinetics of Org 25969. Rapid Commun. Mass Spectrom. 2002, 16, 1946–1952. 10.1002/rcm.812. [DOI] [PubMed] [Google Scholar]
- Liu Z.; Escudero A.; Carrillo-Carrion C.; Chakraborty I.; Zhu D.; Gallego M.; Parak W. J.; Feliu N. Biodegradation of Bi-Labeled Polymer-Coated Rare-Earth Nanoparticles in Adherent Cell Cultures. Chem. Mater. 2020, 32, 245–254. 10.1021/acs.chemmater.9b03673. [DOI] [Google Scholar]
- Gowda G. A. N.; Djukovic D.. Overview of Mass Spectrometry-Based Metabolomics: Opportunities and Challenges. In Mass Spectrometry in Metabolomics: Methods and Protocols; Raftery D., Ed.; Springer: New York, 2014; pp 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talamini L.; Violatto M. B.; Cai Q.; Monopoli M. P.; Kantner K.; Krpetić Z. e.; Perez-Potti A.; Cookman J.; Garry D.; Silveira C. P.; Boselli L.; Pelaz B.; Serchi T.; Cambier S. b.; Gutleb A. C.; Feliu N.; Yan Y.; Salmona M.; Parak W. J.; Dawson K. A.; Bigini P. Influence of Size and Shape on the Anatomical Distribution of Endotoxin-Free Gold Nanoparticles. ACS Nano 2017, 11, 5519–5529. 10.1021/acsnano.7b00497. [DOI] [PubMed] [Google Scholar]
- Xu M.; Soliman M. G.; Sun X.; Pelaz B.; Feliu N.; Parak W. J.; Liu S. How Entanglement of Different Physicochemical Properties Complicates the Prediction of in Vitro and in Vivo Interactions of Gold Nanoparticles. ACS Nano 2018, 12, 10104–10113. 10.1021/acsnano.8b04906. [DOI] [PubMed] [Google Scholar]
- Amini-Nik S.; Kraemer D.; Cowan M. L.; Gunaratne K.; Nadesan P.; Alman B. A.; Miller R. J. Ultrafast Mid-IR Laser Scalpel: Protein Signals of the Fundamental Limits to Minimally Invasive Surgery. PLoS One 2010, 5, e13053. 10.1371/journal.pone.0013053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwiatkowski M.; Wurlitzer M.; Krutilin A.; Kiani P.; Nimer R.; Omidi M.; Mannaa A.; Bussmann T.; Bartkowiak K.; Kruber S.; Uschold S.; Steffen P.; Lubberstedt J.; Kupker N.; Petersen H.; Knecht R.; Hansen N. O.; Zarrine-Afsar A.; Robertson W. D.; Miller R. J. D.; Schluter H. Homogenization of Tissues via Picosecond-Infrared Laser (PIRL) Ablation: Giving a Closer View on the in-Vivo Composition of Protein Species as Compared to Mechanical Homogenization. J. Proteomics 2016, 134, 193–202. 10.1016/j.jprot.2015.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rompp A.; Guenther S.; Takats Z.; Spengler B. Mass Spectrometry Imaging with High Resolution in Mass and Space (HR(2) MSI) for Reliable Investigation of Drug Compound Distributions on the Cellular Level. Anal. Bioanal. Chem. 2011, 401, 65–73. 10.1007/s00216-011-4990-7. [DOI] [PubMed] [Google Scholar]
- Abramowski P.; Kraus O.; Rohn S.; Riecken K.; Fehse B.; Schluter H. Combined Application of RGB Marking and Mass Spectrometric Imaging Facilitates Detection of Tumor Heterogeneity. Cancer Genomics Proteomics 2015, 12, 179–187. [PubMed] [Google Scholar]
- Cui Y.; Cao W.; He Y.; Zhao Q.; Wakazaki M.; Zhuang X.; Gao J.; Zeng Y.; Gao C.; Ding Y.; Wong H. Y.; Wong W. S.; Lam H. K.; Wang P.; Ueda T.; Rojas-Pierce M.; Toyooka K.; Kang B.-H.; Jiang L. A Whole-Cell Electron Tomography Model of Vacuole Biogenesis in Arabidopsis Root Cells. Nat. Plants 2019, 5, 95–105. 10.1038/s41477-018-0328-1. [DOI] [PubMed] [Google Scholar]
- Zachs T.; Schertel A.; Medeiros J.; Weiss G. L.; Hugener J.; Matos J.; Pilhofer M. Fully Automated, Sequential Focused Ion Beam Milling for Cryo-Electron Tomography. eLife 2020, 9, e52286 10.7554/eLife.52286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lučić V.; Rigort A.; Baumeister W. Cryo-Electron Tomography: The Challenge of Doing Structural Biology in Situ. J. Cell Biol. 2013, 202, 407–419. 10.1083/jcb.201304193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Friedrich H.; Patterson J. P.; Sommerdijk N. A. J. M.; de Jonge N. Liquid-Phase Electron Microscopy for Soft Matter Science and Biology. Adv. Mater. 2020, 32, 2001582. 10.1002/adma.202001582. [DOI] [PubMed] [Google Scholar]
- Koo K.; Dae K. S.; Hahn Y. K.; Yuk J. M. Live Cell Electron Microscopy Using Graphene Veils. Nano Lett. 2020, 20, 4708–4713. 10.1021/acs.nanolett.0c00715. [DOI] [PubMed] [Google Scholar]
- Kennedy E.; Nelson E. M.; Damiano J.; Timp G. Gene Expression in Electron-Beam-Irradiated Bacteria in Reply to “Live Cell Electron Microscopy Is Probably Impossible. ACS Nano 2017, 11, 3–7. 10.1021/acsnano.6b06616. [DOI] [PubMed] [Google Scholar]
- de Jonge N.; Peckys D. B. Live Cell Electron Microscopy Is Probably Impossible. ACS Nano 2016, 10, 9061–9063. 10.1021/acsnano.6b02809. [DOI] [PubMed] [Google Scholar]
- Key J.; Leary J. F. Nanoparticles for Multimodal in Vivo Imaging in Nanomedicine. Int. J. Nanomed. 2014, 9, 711–726. 10.2147/IJN.S53717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leary J. F. Design of Sophisticated Shaped, Multilayered, and Multifunctional Nanoparticles for Combined in-Vivo Imaging and Advanced Drug Delivery. Proc. SPIE 2019, 10891, 108910R. 10.1117/12.2511821. [DOI] [Google Scholar]
- van Schadewijk R.; Krug J. R.; Shen D.; Sankar Gupta K. B. S.; Vergeldt F. J.; Bisseling T.; Webb A. G.; Van As H.; Velders A. H.; de Groot H. J. M.; Alia A. Magnetic Resonance Microscopy at Cellular Resolution and Localised Spectroscopy of Medicago truncatula at 22.3 T. Sci. Rep. 2020, 10, 971. 10.1038/s41598-020-57861-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H.; Bengtsson N.; Chrzanowski S. M.; Flint J. J.; Walter G. A.; Blackband S. J. Magnetic Resonance Microscopy (MRM) of Single Mammalian Myofibers and Myonuclei. Sci. Rep. 2017, 7, 39496. 10.1038/srep39496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. H.; Flint J. J.; Hansen B.; Blackband S. J. Investigation of the Subcellular Architecture of L7 Neurons of Aplysia californica Using Magnetic Resonance Microscopy (MRM) at 7.8 Microns. Sci. Rep. 2015, 5, 11147. 10.1038/srep11147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T.; Zhou J.; Wang L.; Zhang H.; Song C.; de la Fuente J. M.; Pan Y.; Song J.; Zhang C.; Cui D. Photo-Fenton-Like Metal–Protein Self-Assemblies as Multifunctional Tumor Theranostic Agent. Adv. Healthcare Mater. 2019, 8, 1900192. 10.1002/adhm.201900192. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Pan Y.; Cao W.; Xia F.; Liu B.; Niu J.; Alfranca G.; Sun X.; Ma L.; Fuente J. M. d. l.; Song J.; Ni J.; Cui D. A Tumor Microenvironment Responsive Biodegradable CaCO3/MnO2- Based Nanoplatform for the Enhanced Photodynamic Therapy and Improved PD-L1 Immunotherapy. Theranostics 2019, 9, 6867–6884. 10.7150/thno.37586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding B.; Zheng P.; Ma P. a.; Lin J. Manganese Oxide Nanomaterials: Synthesis, Properties, and Theranostic Applications. Adv. Mater. 2020, 32, 1905823. 10.1002/adma.201905823. [DOI] [PubMed] [Google Scholar]
- Shen Z.; Chen T.; Ma X.; Ren W.; Zhou Z.; Zhu G.; Zhang A.; Liu Y.; Song J.; Li Z.; Ruan H.; Fan W.; Lin L.; Munasinghe J.; Chen X.; Wu A. Multifunctional Theranostic Nanoparticles Based on Exceedingly Small Magnetic Iron Oxide Nanoparticles for T1-Weighted Magnetic Resonance Imaging and Chemotherapy. ACS Nano 2017, 11, 10992–11004. 10.1021/acsnano.7b04924. [DOI] [PubMed] [Google Scholar]
- Tassa C.; Shaw S. Y.; Weissleder R. Dextran-Coated Iron Oxide Nanoparticles: A Versatile Platform for Targeted Molecular Imaging, Molecular Diagnostics, and Therapy. Acc. Chem. Res. 2011, 44, 842–852. 10.1021/ar200084x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladd M. E.; Bachert P.; Meyerspeer M.; Moser E.; Nagel A. M.; Norris D. G.; Schmitter S.; Speck O.; Straub S.; Zaiss M. Pros and Cons of Ultra-High-Field MRI/MRS for Human Application. Prog. Nucl. Magn. Reson. Spectrosc. 2018, 109, 1–50. 10.1016/j.pnmrs.2018.06.001. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Ali Z.; Amin F.; Feltz A.; Oheim M.; Parak W. J. Ion and pH Sensing with Colloidal Nanoparticles: Influence of Surface Charge on Sensing and Colloidal Properties. ChemPhysChem 2010, 11, 730–735. 10.1002/cphc.200900849. [DOI] [PubMed] [Google Scholar]
- Lay A.; Sheppard O. H.; Siefe C.; McLellan C. A.; Mehlenbacher R. D.; Fischer S.; Goodman M. B.; Dionne J. A. Optically Robust and Biocompatible Mechanosensitive Upconverting Nanoparticles. ACS Cent. Sci. 2019, 5, 1211–1222. 10.1021/acscentsci.9b00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehlenbacher R. D.; Kolbl R.; Lay A.; Dionne J. A. Nanomaterials for in Vivo Imaging of Mechanical Forces and Electrical Fields. Nature Rev. Mater. 2018, 3, 17080. 10.1038/natrevmats.2017.80. [DOI] [Google Scholar]
- Rasmussen J. C.; Tan I. C.; Marshall M. V.; Fife C. E.; Sevick-Muraca E. M. Lymphatic Imaging in Humans with Near-Infrared Fluorescence. Curr. Opin. Biotechnol. 2009, 20, 74–82. 10.1016/j.copbio.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song C.; Li Y.; Li T.; Yang Y.; Huang Z.; de la Fuente J. M.; Ni J.; Cui D. Long-Circulating Drug-Dye-Based Micelles with Ultrahigh pH-Sensitivity for Deep Tumor Penetration and Superior Chemo-Photothermal Therapy. Adv. Funct. Mater. 2020, 30, 1906309. 10.1002/adfm.201906309. [DOI] [Google Scholar]
- Pan S.; Pei L.; Zhang A.; Zhang Y.; Zhang C.; Huang M.; Huang Z.; Liu B.; Wang L.; Ma L.; Zhang Q.; Cui D. Passion Fruit-Like Exosome-PMA/Au-BSA@Ce6 Nanovehicles for Real-Time Fluorescence Imaging and Enhanced Targeted Photodynamic Therapy with Deep Penetration and Superior Retention Behavior in Tumor. Biomaterials 2020, 230, 119606. 10.1016/j.biomaterials.2019.119606. [DOI] [PubMed] [Google Scholar]
- Zhang A.; Pan S.; Zhang Y.; Chang J.; Cheng J.; Huang Z.; Li T.; Zhang C.; Fuente J. M. d. l.; Zhang Q.; Cui D. Carbon-Gold Hybrid Nanoprobes for Real-Time Imaging, Photothermal/Photodynamic and Nanozyme Oxidative Therapy. Theranostics 2019, 9, 3443–3458. 10.7150/thno.33266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudheendra L.; Das G. K.; Li C.; Stark D.; Cena J.; Cherry S.; Kennedy I. M. NaGdF4:Eu3+ Nanoparticles for Enhanced X-Ray Excited Optical Imaging. Chem. Mater. 2014, 26, 1881–1888. 10.1021/cm404044n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C.-C.; Lin S.-L.; Chang C. A. Lanthanide-Doped Core–Shell–Shell Nanocomposite for Dual Photodynamic Therapy and Luminescence Imaging by a Single X-Ray Excitation Source. ACS Appl. Mater. Interfaces 2018, 10, 7859–7870. 10.1021/acsami.8b00015. [DOI] [PubMed] [Google Scholar]
- Ma J.; Huang P.; He M.; Pan L.; Zhou Z.; Feng L.; Gao G.; Cui D. Folic Acid-Conjugated LaF3:Yb,Tm@SiO2 Nanoprobes for Targeting Dual-Modality Imaging of Upconversion Luminescence and X-Ray Computed Tomography. J. Phys. Chem. B 2012, 116, 14062–14070. 10.1021/jp309059u. [DOI] [PubMed] [Google Scholar]
- Pujals S.; Feiner-Gracia N.; Delcanale P.; Voets I.; Albertazzi L. Super-Resolution Microscopy as a Powerful Tool to Study Complex Synthetic Materials. Nat. Rev. Chem. 2019, 3, 68–84. 10.1038/s41570-018-0070-2. [DOI] [Google Scholar]
- Möckl L.; Lamb D. C.; Bräuchle C. Super-Resolved Fluorescence Microscopy: Nobel Prize in Chemistry 2014 for Eric Betzig, Stefan Hell, and William E. Moerner. Angew. Chem., Int. Ed. 2014, 53, 13972–13977. 10.1002/anie.201410265. [DOI] [PubMed] [Google Scholar]
- Schermelleh L.; Heintzmann R.; Leonhardt H. A Guide to Super-Resolution Fluorescence Microscopy. J. Cell Biol. 2010, 190, 165–175. 10.1083/jcb.201002018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang B.; Bates M.; Zhuang X. Super-Resolution Fluorescence Microscopy. Annu. Rev. Biochem. 2009, 78, 993–1016. 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X.; Hartmann R.; Jimenez de Aberasturi D.; Yang F.; Soenen S. J. H.; Manshian B. B.; Franz J.; Valdeperez D.; Pelaz B.; Feliu N.; Hampp N.; Riethmuller C.; Vieker H.; Frese N.; Golzhauser A.; Simonich M.; Tanguay R. L.; Liang X.-J.; Parak W. J. Colloidal Gold Nanoparticles Induce Changes in Cellular and Subcellular Morphology. ACS Nano 2017, 11, 7807–7820. 10.1021/acsnano.7b01760. [DOI] [PubMed] [Google Scholar]
- Itano M. S.; Neumann A. K.; Liu P.; Zhang F.; Gratton E.; Parak W. J.; Thompson N. L.; Jacobson K. DC-SIGN and Influenza Hemagglutinin Dynamics in Plasma Membrane Microdomains are Markedly Different. Biophys. J. 2011, 100, 2662–2670. 10.1016/j.bpj.2011.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A. M.; Mancini M. C.; Nie S. M. BIOIMAGING Second Window for in Vivo Imaging. Nat. Nanotechnol. 2009, 4, 710–711. 10.1038/nnano.2009.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong N.-N.; Pedroni M.; Piccinelli F.; Conti G.; Sbarbati A.; Ramirez-Hernandez J. E.; Maestro L. M.; Iglesias-De La Cruz M. C.; Sanz-Rodriguez F.; Juarranz A.; Chen F.; Vetrone F.; Capobianco J. A.; Sole J. G.; Bettinelli M.; Jaque D.; Speghini A. NIR-to-NIR Two-Photon Excited CaF2:Tm3+,Yb3+ Nanoparticles: Multifunctional Nanoprobes for Highly Penetrating Fluorescence Bio-Imaging. ACS Nano 2011, 5, 8665. 10.1021/nn202490m. [DOI] [PubMed] [Google Scholar]
- Lopez M. J.; Seyed-Razavi Y.; Yamaguchi T.; Ortiz G.; Sendra V. G.; Harris D. L.; Jamali A.; Hamrah P. Multiphoton Intravital Microscopy of Mandibular Draining Lymph Nodes: A Mouse Model to Study Corneal Immune Responses. Front. Immunol. 2020, 11, 39. 10.3389/fimmu.2020.00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii M. Intravital Imaging Technology Reveals Immune System Dynamics in Vivo. Allergol. Int. 2016, 65, 225–227. 10.1016/j.alit.2016.05.001. [DOI] [PubMed] [Google Scholar]
- Pittet M. J.; Garris C. S.; Arlauckas S. P.; Weissleder R. Recording the Wild Lives of Immune Cells. Sci. Immunol. 2018, 3, eaaq0491 10.1126/sciimmunol.aaq0491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witte M.; Jaspers S.; Wenck H.; Rübhausen M.; Fischer F. Noise Reduction and Quantification of Fiber Orientations in Greyscale Images. PLoS One 2020, 15, e0227534. 10.1371/journal.pone.0227534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellem D.; Sattler M.; Pagel-Wolff S.; Jaspers S.; Wenck H.; Rübhausen M. A.; Fischer F. Fragmentation of the Mitochondrial Network in Skin in Vivo. PLoS One 2017, 12, e0174469. 10.1371/journal.pone.0174469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huser T.; Chan J. Raman Spectroscopy for Physiological Investigations of Tissues and Cells. Adv. Drug Delivery Rev. 2015, 89, 57–70. 10.1016/j.addr.2015.06.011. [DOI] [PubMed] [Google Scholar]
- Langer J.; Jimenez de Aberasturi D.; Aizpurua J.; Alvarez-Puebla R. A.; Auguié B.; Baumberg J. J.; Bazan G. C.; Bell S. E. J.; Boisen A.; Brolo A. G.; Choo J.; Cialla-May D.; Deckert V.; Fabris L.; Faulds K.; García de Abajo F. J.; Goodacre R.; Graham D.; Haes A. J.; Haynes C. L.; et al. Present and Future of Surface-Enhanced Raman Scattering. ACS Nano 2020, 14, 28–117. 10.1021/acsnano.9b04224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin O.; Altas M.; Kahraman M.; Bayrak O. F.; Culha M. Differentiation of Healthy Brain Tissue and Tumors Using Surface-Enhanced Raman Scattering. Appl. Spectrosc. 2009, 63, 1095–1100. 10.1366/000370209789553219. [DOI] [PubMed] [Google Scholar]
- Rivera Gil P.; Vazquez C. V.; Giannini V.; Callao M. P.; Parak W. J.; Duarte M. A. C.; Alvarez-Puebla R. A. Plasmonic Nanoprobes for Real-Time Optical Monitoring of Nitric Oxide inside Living Cells. Angew. Chem., Int. Ed. 2013, 52, 13694–13698. 10.1002/anie.201306390. [DOI] [PubMed] [Google Scholar]
- Köker T.; Tang N.; Tian C.; Zhang W.; Wang X.; Martel R.; Pinaud F. Cellular Imaging by Targeted Assembly of Hot-Spot SERS and Photoacoustic Nanoprobes Using Split-Fluorescent Protein Scaffolds. Nat. Commun. 2018, 9, 607. 10.1038/s41467-018-03046-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzi E.; Jimenez de Aberasturi D.; Liz-Marzán L. M. Surface-Enhanced Raman Scattering Tags for Three-Dimensional Bioimaging and Biomarker Detection. ACS Sensors 2019, 4, 1126–1137. 10.1021/acssensors.9b00321. [DOI] [PubMed] [Google Scholar]
- Álvarez-Puebla R. A. Effects of the Excitation Wavelength on the SERS Spectrum. J. Phys. Chem. Lett. 2012, 3, 857–866. 10.1021/jz201625j. [DOI] [PubMed] [Google Scholar]
- Smith B. R.; Gambhir S. S. Nanomaterials for in Vivo Imaging. Chem. Rev. 2017, 117, 901–986. 10.1021/acs.chemrev.6b00073. [DOI] [PubMed] [Google Scholar]
- Zavaleta C. L.; Smith B. R.; Walton I.; Doering W.; Davis G.; Shojaei B.; Natan M. J.; Gambhir S. S. Multiplexed Imaging of Surface Enhanced Raman Scattering Nanotags in Living Mice Using Noninvasive Raman Spectroscopy. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 13511. 10.1073/pnas.0813327106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez de Aberasturi D.; Serrano-Montes A. B.; Langer J.; Henriksen-Lacey M.; Parak W. J.; Liz-Marzan L. M. Encoded Gold Nanostars for Multiplexed SERS Cell Differentiation. Chem. Mater. 2016, 28, 6779–6790. 10.1021/acs.chemmater.6b03349. [DOI] [Google Scholar]
- Jimenez de Aberasturi D.; Henriksen-Lacey M.; Litti L.; Langer J.; Liz-Marzán L. M. Using SERS Tags to Image the Three-Dimensional Structure of Complex Cell Models. Adv. Funct. Mater. 2020, 30, 1909655. 10.1002/adfm.201909655. [DOI] [Google Scholar]
- Alvarez-Puebla R. A.; Zubarev E. R.; Kotov N. A.; Liz-Marzan L. M. Self-Assembled Nanorod Supercrystals for Ultrasensitive SERS Diagnostics. Nano Today 2012, 7, 6–9. 10.1016/j.nantod.2011.11.001. [DOI] [Google Scholar]
- Alvarez-Puebla R. A.; Agarwal A.; Manna P.; Khanal B. P.; Aldeanueva-Potel P.; Carbo-Argibay E.; Pazos-Perez N.; Vigderman L.; Zubarev E. R.; Kotov N. A.; Liz-Marzan L. M. Gold Nanorods 3D-Supercrystals as Surface Enhanced Raman Scattering Spectroscopy Substrates for the Rapid Detection of Scrambled Prions. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 8157–8161. 10.1073/pnas.1016530108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feliu N.; Hassan M.; Garcia Rico E.; Cui D.; Parak W.; Alvarez-Puebla R. SERS Quantification and Characterization of Proteins and Other Biomolecules. Langmuir 2017, 33, 9711–9730. 10.1021/acs.langmuir.7b01567. [DOI] [PubMed] [Google Scholar]
- Carrillo-Carrion C.; Martinez R.; Navarro Poupard M. F.; Pelaz B.; Polo E.; Arenas-Vivo A.; Olgiati A.; Taboada P.; Soliman M. G.; Catalan U.; Fernandez-Castillejo S.; Sola R.; Parak W. J.; Horcajada P.; Alvarez-Puebla R. A.; del Pino P. Aqueous Stable Gold Nanostar/ZIF-8 Nanocomposites for Light-Triggered Release of Active Cargo inside Living Cells. Angew. Chem., Int. Ed. 2019, 58, 7078–7082. 10.1002/anie.201902817. [DOI] [PubMed] [Google Scholar]
- Gerling M.; Zhao Y.; Nania S.; Norberg K. J.; Verbeke C. S.; Englert B.; Kuiper R. V.; Bergstrom A.; Hassan M.; Neesse A.; Lohr J. M.; Heuchel R. L. Real-Time Assessment of Tissue Hypoxia in Vivo with Combined Photoacoustics and High-Frequency Ultrasound. Theranostics 2014, 4, 604–613. 10.7150/thno.7996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Zheng W.; Hassan M.. Chapter 4 - Nanoparticles for Imaging Application. In Frontiers of Nanoscience- Colloids for Nanobiotechnology. Synthesis, Characterization and Potential Applications; Parak W. J., Feliu N., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; Vol. 16, pp 67–88. [Google Scholar]
- Lakshmanan A.; Farhadi A.; Nety S. P.; Lee-Gosselin A.; Bourdeau R. W.; Maresca D.; Shapiro M. G. Molecular Engineering of Acoustic Protein Nanostructures. ACS Nano 2016, 10, 7314–7322. 10.1021/acsnano.6b03364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling B.; Lee J.; Maresca D.; Lee-Gosselin A.; Malounda D.; Swift M. B.; Shapiro M. G. Biomolecular Ultrasound Imaging of Phagolysosomal Function. ACS Nano 2020, 14, 12210–12221. 10.1021/acsnano.0c05912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro M. G.; Goodwill P. W.; Neogy A.; Yin M.; Foster F. S.; Schaffer D. V.; Conolly S. M. Biogenic Gas Nanostructures as Ultrasonic Molecular Reporters. Nat. Nanotechnol. 2014, 9, 311–316. 10.1038/nnano.2014.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q.; Zhu R.; Song J.; Yang H.; Chen X. Photoacoustic Imaging: Contrast Agents and Their Biomedical Applications. Adv. Mater. 2019, 31, 1805875. 10.1002/adma.201805875. [DOI] [PubMed] [Google Scholar]
- Cai Y.; Wei Z.; Song C.; Tang C.; Han W.; Dong X. Optical Nano-Agents in the Second Near-Infrared Window for Biomedical Applications. Chem. Soc. Rev. 2019, 48, 22–37. 10.1039/C8CS00494C. [DOI] [PubMed] [Google Scholar]
- Shu X.; Beckmann L.; Zhang H. F. Visible-Light Optical Coherence Tomography: A Review. J. Biomed. Opt. 2017, 22, 1–14. 10.1117/1.JBO.22.12.121707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan A. C. S.; Tan G. S.; Denniston A. K.; Keane P. A.; Ang M.; Milea D.; Chakravarthy U.; Cheung C. M. G. An Overview of the Clinical Applications of Optical Coherence Tomography Angiography. Eye 2018, 32, 262–286. 10.1038/eye.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pahlevaninezhad H.; Khorasaninejad M.; Huang Y.-W.; Shi Z.; Hariri L. P.; Adams D. C.; Ding V.; Zhu A.; Qiu C.-W.; Capasso F.; Suter M. J. Nano-Optic Endoscope for High-Resolution Optical Coherence Tomography in Vivo. Nat. Photonics 2018, 12, 540–547. 10.1038/s41566-018-0224-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimoto J. G. Optical Coherence Tomography for Ultrahigh Resolution in Vivo Imaging. Nat. Biotechnol. 2003, 21, 1361–1367. 10.1038/nbt892. [DOI] [PubMed] [Google Scholar]
- Li Y. L.; Seekell K.; Yuan H.; Robles F. E.; Wax A. Multispectral Nanoparticle Contrast Agents for True-Color Spectroscopic Optical Coherence Tomography. Biomed. Opt. Express 2012, 3, 1914–1923. 10.1364/BOE.3.001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liba O.; SoRelle E. D.; Sen D.; de la Zerda A. Contrast-Enhanced Optical Coherence Tomography with Picomolar Sensitivity for Functional in Vivo Imaging. Sci. Rep. 2016, 6, 23337. 10.1038/srep23337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen V. P.; Li Y.; Qian W.; Liu B.; Tian C.; Zhang W.; Huang Z.; Ponduri A.; Tarnowski M.; Wang X.; Paulus Y. M. Contrast Agent Enhanced Multimodal Photoacoustic Microscopy and Optical Coherence Tomography for Imaging of Rabbit Choroidal and Retinal Vessels in Vivo. Sci. Rep. 2019, 9, 5945. 10.1038/s41598-019-42324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si P.; Yuan E.; Liba O.; Winetraub Y.; Yousefi S.; SoRelle E. D.; Yecies D. W.; Dutta R.; de la Zerda A. Gold Nanoprisms as Optical Coherence Tomography Contrast Agents in the Second Near-Infrared Window for Enhanced Angiography in Live Animals. ACS Nano 2018, 12, 11986–11994. 10.1021/acsnano.8b03862. [DOI] [PubMed] [Google Scholar]
- Tucker-Schwartz J. M.; Beavers K. R.; Sit W. W.; Shah A. T.; Duvall C. L.; Skala M. C. In Vivo Imaging of Nanoparticle Delivery and Tumor Microvasculature with Multimodal Optical Coherence Tomography. Biomed. Opt. Express 2014, 5, 1731–1743. 10.1364/BOE.5.001731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R. A Clearer Vision for in Vivo Imaging. Nat. Biotechnol. 2001, 19, 316–317. 10.1038/86684. [DOI] [PubMed] [Google Scholar]
- Smith A. M.; Mancini M. C.; Nie S. Second Window for in Vivo Imaging. Nat. Nanotechnol. 2009, 4, 710–711. 10.1038/nnano.2009.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenry; Duan Y.; Liu B. Recent Advances of Optical Imaging in the Second Near-Infrared Window. Adv. Mater. 2018, 30, 1802394. 10.1002/adma.201802394. [DOI] [PubMed] [Google Scholar]
- Liguori C.; Frauenfelder G.; Massaroni C.; Saccomandi P.; Giurazza F.; Pitocco F.; Marano R.; Schena E. Emerging Clinical Applications of Computed Tomography. Med. Devices: Evidence Res. 2015, 8, 265–278. 10.2147/MDER.S70630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momose A.; Takeda T.; Itai Y.; Hirano K. Phase-Contrast X-Ray Computed Tomography for Observing Biological Soft Tissues. Nat. Med. 1996, 2, 473–475. 10.1038/nm0496-473. [DOI] [PubMed] [Google Scholar]
- Martínez-Criado G.; Segura-Ruiz J.; Alén B.; Eymery J.; Rogalev A.; Tucoulou R.; Homs A. Exploring Single Semiconductor Nanowires with a Multimodal Hard X-ray Nanoprobe. Adv. Mater. 2014, 26, 7873–7879. 10.1002/adma.201304345. [DOI] [PubMed] [Google Scholar]
- Decelle J.; Veronesi G.; Gallet B.; Stryhanyuk H.; Benettoni P.; Schmidt M.; Tucoulou R.; Passarelli M.; Bohic S.; Clode P.; Musat N. Subcellular Chemical Imaging: New Avenues in Cell Biology. Trends Cell Biol. 2020, 30, 173–188. 10.1016/j.tcb.2019.12.007. [DOI] [PubMed] [Google Scholar]
- Yano J.; Yachandra V. K. X-Ray Absorption Spectroscopy. Photosynth. Res. 2009, 102, 241. 10.1007/s11120-009-9473-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziej D. Revealing Complexity of Nanoparticle Synthesis in Solution by in Situ Hard X-Ray Spectroscopy—Today and Beyond. Chem. Mater. 2016, 28, 2478–2490. 10.1021/acs.chemmater.6b00486. [DOI] [Google Scholar]
- Pushie M. J.; Pickering I. J.; Korbas M.; Hackett M. J.; George G. N. Elemental and Chemically Specific X-Ray Fluorescence Imaging of Biological Systems. Chem. Rev. 2014, 114, 8499–8541. 10.1021/cr4007297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamberti C.; Bordiga S.; Bonino F.; Prestipino C.; Berlier G.; Capello L.; D’Acapito F.; Llabrés i Xamena F. X.; Zecchina A. Determination of the Oxidation and Coordination State of Copper on Different Cu-Based Catalysts by XANES Spectroscopy in Situ or in Operando Conditions. Phys. Chem. Chem. Phys. 2003, 5, 4502–4509. 10.1039/B305810G. [DOI] [Google Scholar]
- Hémonnot C. Y. J.; Koster S. Imaging of Biological Materials and Cells by X-Ray Scattering and Diffraction. ACS Nano 2017, 11, 8542–8559. 10.1021/acsnano.7b03447. [DOI] [PubMed] [Google Scholar]
- Morrison G. R.; Niemann B.. Differential Phase Contrast X-Ray Microscopy. In X-Ray Microscopy and Spectromicroscopy: Status Report from the Fifth International Conference, Würzburg, August 19–23, 1996; Thieme J., Schmahl G., Rudolph D., Umbach E., Eds.; Springer: Berlin, Heidelberg, 1998; pp 85–94.
- Chen H.; Wang Z.; Gao K.; Hou Q.; Wang D.; Wu Z. Quantitative Phase Retrieval in X-Ray Zernike Phase Contrast Microscopy. J. Synchrotron Radiat. 2015, 22, 1056–1061. 10.1107/S1600577515007699. [DOI] [PubMed] [Google Scholar]
- Yang Y.; Cheng Y.; Heine R.; Baumbach T. Contrast Transfer Functions for Zernike Phase Contrast in Full-Field Transmission Fard X-Ray Microscopy. Opt. Express 2016, 24, 6063–70. 10.1364/OE.24.006063. [DOI] [PubMed] [Google Scholar]
- Grübel G.; Zontone F. Correlation Spectroscopy with Coherent X-Rays. J. Alloys Compd. 2004, 362, 3–11. 10.1016/S0925-8388(03)00555-3. [DOI] [Google Scholar]
- Holmqvist P.; Meester V.; Westermeier F.; Kleshchanok D. Rotational Diffusion in Concentrated Platelet Systems Measured with X-Ray Photon Correlation Spectroscopy. J. Chem. Phys. 2013, 139, 084905. 10.1063/1.4818532. [DOI] [PubMed] [Google Scholar]
- Westermeier F.; Pennicard D.; Hirsemann H.; Wagner U. H.; Rau C.; Graafsma H.; Schall P.; Lettinga M. P.; Struth B. Connecting Structure, Dynamics and Viscosity in Sheared Soft Colloidal Liquids: A Medley of Anisotropic Fluctuations. Soft Matter 2016, 12, 171–180. 10.1039/C5SM01707F. [DOI] [PubMed] [Google Scholar]
- Caronna C.; Chushkin Y.; Madsen A.; Cupane A. Dynamics of Nanoparticles in a Supercooled Liquid. Phys. Rev. Lett. 2008, 100, 055702. 10.1103/PhysRevLett.100.055702. [DOI] [PubMed] [Google Scholar]
- Pearson A. R.; von Stetten D.; Huse N. If You Can Get a Crystal Structure, Why Bother with Anything Else?. Synchrotron Radiat. News. 2015, 28, 10–14. 10.1080/08940886.2015.1101321. [DOI] [Google Scholar]
- Holder C. F.; Schaak R. E. Tutorial on Powder X-Ray Diffraction for Characterizing Nanoscale Materials. ACS Nano 2019, 13, 7359–7365. 10.1021/acsnano.9b05157. [DOI] [PubMed] [Google Scholar]
- Mourdikoudis S.; Pallares R. M.; Thanh N. T. K. Characterization Techniques for Nanoparticles: Comparison and Complementarity upon Studying Nanoparticle Properties. Nanoscale 2018, 10, 12871–12934. 10.1039/C8NR02278J. [DOI] [PubMed] [Google Scholar]
- Liang K.; Richardson J. J.; Cui J.; Caruso F.; Doonan C. J.; Falcaro P. Metal–Organic Framework Coatings as Cytoprotective Exoskeletons for Living Cells. Adv. Mater. 2016, 28, 7910–7914. 10.1002/adma.201602335. [DOI] [PubMed] [Google Scholar]
- Lam Y. Y.; Hawley A.; Tan A.; Boyd B. J. Coupling in Vitro Cell Culture with Synchrotron SAXS to Understand the Bio-Interaction of Lipid-Based Liquid Crystalline Nanoparticles with Vascular Endothelial Cells. Drug Delivery Transl. Res. 2020, 10, 610–620. 10.1007/s13346-020-00718-3. [DOI] [PubMed] [Google Scholar]
- Veronesi G.; Brun E.; Fayard B.; Cotte M.; Carrière M. Structural Properties of Rutile TiO2 Nanoparticles Accumulated in a Model of Gastrointestinal Epithelium Elucidated by Micro-Beam X-Ray Absorption Fine Structure Spectroscopy. Appl. Phys. Lett. 2012, 100, 214101. 10.1063/1.4720172. [DOI] [Google Scholar]
- Auffan M.; Decome L.; Rose J.; Orsiere T.; De Meo M.; Briois V.; Chaneac C.; Olivi L.; Berge-Lefranc J.-l.; Botta A.; Wiesner M. R.; Bottero J.-y. In Vitro Interactions between DMSA-Coated Maghemite Nanoparticles and Human Fibroblasts: A Physicochemical and Cyto-Genotoxical Study. Environ. Sci. Technol. 2006, 40, 4367–4373. 10.1021/es060691k. [DOI] [PubMed] [Google Scholar]
- Martínez-Criado G.; Villanova J.; Tucoulou R.; Salomon D.; Suuronen J.-P.; Labouré S.; Guilloud C.; Valls V.; Barrett R.; Gagliardini E.; Dabin Y.; Baker R.; Bohic S.; Cohen C.; Morse J. ID16B: A Hard X-Ray Nanoprobe Beamline at the ESRF for Nano-Analysis. J. Synchrotron Radiat. 2016, 23, 344–352. 10.1107/S1600577515019839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cesar da Silva J.; Pacureanu A.; Yang Y.; Bohic S.; Morawe C.; Barrett R.; Cloetens P. Efficient Concentration of High-Energy X-Rays for Diffraction-Limited Imaging Resolution. Optica 2017, 4, 492–495. 10.1364/OPTICA.4.000492. [DOI] [Google Scholar]
- Welcome to B24: Full Field Cryo-X-Ray Microscopy for the Life Sciences; Diamond Light Source Ltd.: Oxfordshire, UK. http://www.diamond.ac.uk/Instruments/Biological-Cryo-Imaging/B24.html (accessed 2020-11-01).
- Welcome to I08: The Diamond Scanning X-Ray Microscopy Beamline; Diamond Light Source Ltd.: Oxfordshire, UK. http://www.diamond.ac.uk/Instruments/Imaging-and-Microscopy/I08.html (accessed 2020-11-01).
- Welcome to I14: Hard X-Ray Nanoprobe; Diamond Light Source Ltd.: Oxfordshire, UK. https://www.diamond.ac.uk/Instruments/Imaging-and-Microscopy/I14.html (accessed 2020-11-01).
- Schroer C. G.; Boye P.; Feldkamp J. M.; Patommel J.; Samberg D.; Schropp A.; Schwab A.; Stephan S.; Falkenberg G.; Wellenreuther G.; Reimers N. Hard X-Ray Nanoprobe at Beamline P06 at PETRA III. Nucl. Instrum. Methods Phys. Res., Sect. A 2010, 616, 93–97. 10.1016/j.nima.2009.10.094. [DOI] [Google Scholar]
- Pereiro E.; Nicolás J.; Ferrer S.; Howells M. R. A Soft X-Ray Beamline for Transmission X-Ray Microscopy at ALBA. J. Synchrotron Radiat. 2009, 16, 505–512. 10.1107/S0909049509019396. [DOI] [PubMed] [Google Scholar]
- Somogyi A.; Medjoubi K.; Baranton G.; Le Roux V.; Ribbens M.; Polack F.; Philippot P.; Samama J.-P. Optical Design and Multi-Length-Scale Scanning Spectro-Microscopy Possibilities at the Nanoscopium Beamline of Synchrotron Soleil. J. Synchrotron Radiat. 2015, 22, 1118–1129. 10.1107/S1600577515009364. [DOI] [PubMed] [Google Scholar]
- Kalbfleisch S.; Neubauer H.; Krüger S. P.; Bartels M.; Osterhoff M.; Mai D. D.; Giewekemeyer K.; Hartmann B.; Sprung M.; Salditt T.; et al. The Göttingen Holography Endstation of Beamline P10 at PETRA III/DESY. AIP Conf. Proc. 2010, 1365, 96–99. 10.1063/1.3625313. [DOI] [Google Scholar]
- PETRA III - Facility Information:; DESY: Hamburg, Germany. http://photon-science.desy.de/facilities/petra_iii/facility_information/index_eng.html. (accessed 2020-11-01).
- Dullin C.; dal Monego S.; Larsson E.; Mohammadi S.; Krenkel M.; Garrovo C.; Biffi S.; Lorenzon A.; Markus A.; Napp J.; Salditt T.; Accardo A.; Alves F.; Tromba G. Functionalized Synchrotron in-Line Phase-Contrast Computed Tomography: A Novel Approach for Simultaneous Quantification of Structural Alterations and Localization of Barium-Labelled Alveolar Macrophages within Mouse Lung Samples. J. Synchrotron Radiat. 2015, 22, 143–155. 10.1107/S1600577514021730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien C.-C.; Chen H.-H.; Lai S.-F.; Wu K.-C.; Cai X.; Hwu Y.; Petibois C.; Chu Y.; Margaritondo G. Gold Nanoparticles as High-Resolution X-Ray Imaging Contrast Agents for the Analysis of Tumor-Related Micro-Vasculature. J. Nanobiotechnol. 2012, 10, 10. 10.1186/1477-3155-10-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinhardt V.; Chen J. H.; Ekman A.; McDermott G.; Le Gros M. A.; Larabell C. Imaging Cell Morphology and Physiology Using X-Rays. Biochem. Soc. Trans. 2019, 47, 489–508. 10.1042/BST20180036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Midgley P. A.; Ward E. P.; Hungria A. B.; Thomas J. M. Nanotomography in the Chemical, Biological and Materials Sciences. Chem. Soc. Rev. 2007, 36, 1477–1494. 10.1039/b701569k. [DOI] [PubMed] [Google Scholar]
- Schroer C. G.; Seyrich M.; Schropp A.; Dohrmann R.; Botta S.; Wiljes P.; Bruckner D.; Kahnt M.; Wittwer F.; Grote L.; Koziej D.; Garrevoet J.; Falkenberg G. Ptychographic Nano-Analytical Microscope (PtyNAMi) at PETRA III: Signal-to-Background Optimization for Imaging with High Sensitivity. Proc. SPIE 2019, 11112, 111120D. 10.1117/12.2529096. [DOI] [Google Scholar]
- Shavorskiy A.; Neppl S.; Slaughter D. S.; Cryan J. P.; Siefermann K. R.; Weise F.; Lin M.-F.; Bacellar C.; Ziemkiewicz M. P.; Zegkinoglou I.; Fraund M. W.; Khurmi C.; Hertlein M. P.; Wright T. W.; Huse N.; Schoenlein R. W.; Tyliszczak T.; Coslovich G.; Robinson J.; Kaindl R. A.; et al. Sub-Nanosecond Time-Resolved Ambient-Pressure X-Ray Photoelectron Spectroscopy Setup for Pulsed and Constant Wave X-Ray Light Sources. Rev. Sci. Instrum. 2014, 85, 093102. 10.1063/1.4894208. [DOI] [PubMed] [Google Scholar]
- Neppl S.; Mahl J.; Tremsin A. S.; Rude B.; Qiao R.; Yang W.; Guo J.; Gessner O. Towards Efficient Time-Resolved X-Ray Absorption Studies of Electron Dynamics at Photocatalytic Interfaces. Faraday Discuss. 2016, 194, 659–682. 10.1039/C6FD00125D. [DOI] [PubMed] [Google Scholar]
- Allahgholi A.; Becker J.; Delfs A.; Dinapoli R.; Goettlicher P.; Greiffenberg D.; Henrich B.; Hirsemann H.; Kuhn M.; Klanner R.; Klyuev A.; Krueger H.; Lange S.; Laurus T.; Marras A.; Mezza D.; Mozzanica A.; Niemann M.; Poehlsen J.; Schwandt J.; et al. The Adaptive Gain Integrating Pixel Detector at the European XFEL. J. Synchrotron Radiat. 2019, 26, 74–82. 10.1107/S1600577518016077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q.; Bahadur D.; Dufresne E. M.; Grybos P.; Kmon P.; Leheny R. L.; Maj P.; Narayanan S.; Szczygiel R.; Ramakrishnan S.; Sandy A. Dynamic Scaling of Colloidal Gel Formation at Intermediate Concentrations. Phys. Rev. Lett. 2017, 119, 178006. 10.1103/PhysRevLett.119.178006. [DOI] [PubMed] [Google Scholar]
- Dicke B.; Hoffmann A.; Stanek J.; Rampp M. S.; Grimm-Lebsanft B.; Biebl F.; Rukser D.; Maerz B.; Göries D.; Naumova M.; Biednov M.; Neuber G.; Wetzel A.; Hofmann S. M.; Roedig P.; Meents A.; Bielecki J.; Andreasson J.; Beyerlein K. R.; Chapman H. N.; et al. Transferring the Entatic-State Principle to Copper Photochemistry. Nat. Chem. 2018, 10, 355–362. 10.1038/nchem.2916. [DOI] [PubMed] [Google Scholar]
- Gallagher-Jones M.; Dias C. S. B.; Pryor A. Jr.; Bouchmella K.; Zhao L.; Lo Y. H.; Cardoso M. B.; Shapiro D.; Rodriguez J.; Miao J. Correlative Cellular Ptychography with Functionalized Nanoparticles at the Fe L-Edge. Sci. Rep. 2017, 7, 4757. 10.1038/s41598-017-04784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A.; Citek C.; Binder S.; Goos A.; Rübhausen M.; Troeppner O.; Ivanović-Burmazović I.; Wasinger E. C.; Stack T. D. P.; Herres-Pawlis S. Catalytic Phenol Hydroxylation with Dioxygen: Extension of the Tyrosinase Mechanism Beyond the Protein Matrix. Angew. Chem., Int. Ed. 2013, 52, 5398–5401. 10.1002/anie.201301249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler H. D.; Drinkel E. E.; Orzechovicz B.; Leopoldino E. C.; Souza F. D.; Almerindo G. I.; Perdona C.; Nome F. Simultaneous Nondestructive Analysis of Palladium, Rhodium, Platinum, and Gold Nanoparticles Using Energy Dispersive X-Ray Fluorescence. Anal. Chem. 2013, 85, 10142–10148. 10.1021/ac402419r. [DOI] [PubMed] [Google Scholar]
- Zheng W.; He R.; Boada R.; Subirana M. A.; Ginman T.; Ottosson H.; Valiente M.; Zhao Y.; Hassan M. A General Covalent Binding Model between Cytotoxic Selenocompounds and Albumin Revealed by Mass Spectrometry and X-Ray Absorption Spectroscopy. Sci. Rep. 2020, 10, 1274. 10.1038/s41598-020-57983-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlig J.; Doriese W. B.; Fowler J. W.; Swetz D. S.; Jaye C.; Fischer D. A.; Reintsema C. D.; Bennett D. A.; Vale L. R.; Mandal U.; O’Neil G. C.; Miaja-Avila L.; Joe Y. I.; El Nahhas A.; Fullagar W.; Parnefjord Gustafsson F.; Sundstrom V.; Kurunthu D.; Hilton G. C.; Schmidt D. R.; Ullom J. N. High-Resolution X-Ray Emission Spectroscopy with Transition-Edge Sensors: Present Performance and Future Potential. J. Synchrotron Radiat. 2015, 22, 766–775. 10.1107/S1600577515004312. [DOI] [PubMed] [Google Scholar]
- Bandler S. R.; Adams J. S.; et al. Development of X-Ray Microcalorimeter Imaging Spectrometers for the X-Ray Surveyor Mission Concept. Proc. SPIE 2016, 9905, 99050Q. 10.1117/12.2232156. [DOI] [Google Scholar]
- Doriese W. B.; Abbamonte P.; Alpert B. K.; Bennett D. A.; Denison E. V.; Fang Y.; Fischer D. A.; Fitzgerald C. P.; Fowler J. W.; Gard J. D.; Hays-Wehle J. P.; Hilton G. C.; Jaye C.; McChesney J. L.; Miaja-Avila L.; Morgan K. M.; Joe Y. I.; O’Neil G. C.; Reintsema C. D.; Rodolakis F.; et al. A Practical Superconducting-Microcalorimeter X-Ray Spectrometer for Beamline and Laboratory Science. Rev. Sci. Instrum. 2017, 88, 053108. 10.1063/1.4983316. [DOI] [PubMed] [Google Scholar]
- den Herder J. W.; Kelley R. L.; Mitsuda K.; Piro L.; Bandler S. R.; Bastia P.; Boyce K. R.; Bruin M.; Chervenak J. A.; Colasanti L.; Doriese W. B.; DiPirro M.; Eckart M. E.; Ezoe Y.; Figueroa-Feliciano E.; Ferrari L.; Fujimoto R.; Gatti F.; Gendreau K. C.; Gottardi L.; et al. The X-Ray Microcalorimeter Spectrometer Onboard of IXO. Proc. SPIE 2010, 7732, 77321H. 10.1117/12.856018. [DOI] [Google Scholar]
- Moosmann J.; Ershov A.; Altapova V.; Baumbach T.; Prasad M. S.; LaBonne C.; Xiao X.; Kashef J.; Hofmann R. X-Ray Phase-Contrast in Vivo Microtomography Probes New Aspects of Xenopus Gastrulation. Nature 2013, 497, 374–377. 10.1038/nature12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moosmann J.; Ershov A.; Weinhardt V.; Baumbach T.; Prasad M. S.; LaBonne C.; Xiao X.; Kashef J.; Hofmann R. Time-Lapse X-Ray Phase-Contrast Microtomography for in Vivo Imaging and Analysis of Morphogenesis. Nat. Protoc. 2014, 9, 294–304. 10.1038/nprot.2014.033. [DOI] [PubMed] [Google Scholar]
- Chen H.; Rogalski M. M.; Anker J. N. Advances in Functional X-Ray Imaging Techniques and Contrast Agents. Phys. Chem. Chem. Phys. 2012, 14, 13469–13486. 10.1039/c2cp41858d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinko A.; Caliebe W. A.; Schoch R.; Bauer M. A Von Hamos-Type Hard X-Ray Spectrometer at the PETRA III Beamline P64. J. Synchrotron Radiat. 2020, 27, 31–36. 10.1107/S1600577519013638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. A.; Shim S.-H.; He J.; Zhuang X. Fast, Three-Dimensional Super-Resolution Imaging of Live Cells. Nat. Methods 2011, 8, 499–505. 10.1038/nmeth.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl S. J.; Hell S. W.; Jakobs S. Fluorescence Nanoscopy in Cell Biology. Nat. Rev. Mol. Cell Biol. 2017, 18, 685–701. 10.1038/nrm.2017.71. [DOI] [PubMed] [Google Scholar]
- Dai M.; Jungmann R.; Yin P. Optical Imaging of Individual Biomolecules in Densely Packed Clusters. Nat. Nanotechnol. 2016, 11, 798–807. 10.1038/nnano.2016.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaulich B.; Thibault P.; Gianoncelli A.; Kiskinova M. Transmission and Emission X-Ray Microscopy: Operation Modes, Contrast Mechanisms and Applications. J. Phys.: Condens. Matter 2011, 23, 083002. 10.1088/0953-8984/23/8/083002. [DOI] [PubMed] [Google Scholar]
- Miao J.; Ishikawa T.; Robinson I. K.; Murnane M. M. Beyond Crystallography: Diffractive Imaging Using Coherent X-Ray Light Sources. Science 2015, 348, 530–535. 10.1126/science.aaa1394. [DOI] [PubMed] [Google Scholar]
- Hanssen E.; Knoechel C.; Dearnley M.; Dixon M. W.; Le Gros M.; Larabell C.; Tilley L. Soft X-Ray Microscopy Analysis of Cell Volume and Hemoglobin Content in Erythrocytes Infected with Asexual and Sexual Stages of Plasmodium falciparum. J. Struct. Biol. 2012, 177, 224–232. 10.1016/j.jsb.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.; Li W.; Liu G.; Zhang X.; Chen J.; Wu W.; Guan Y.; Xiong Y.; Tian Y.; Wu Z. 3D Visualization of Subcellular Structures of Schizosaccharomyces pombe by Hard X-Ray Tomography. J. Microsc. 2010, 240, 14–20. 10.1111/j.1365-2818.2010.03379.x. [DOI] [PubMed] [Google Scholar]
- Hubbell J.; Seltzer S.. X-Ray Mass Attenuation Coefficients. Tables of X-Ray Mass Attenuation Coefficients and Mass Energy-Absorption Coefficients from 1 keV to 20 MeV for Elements Z = 1 to 92 and 48 Additional Substances of Dosimetric Interest. NIST 5632; Physics Laboratory, NIST: Gaithersburg, MD, 2004.
- Deng J.; Lo Y. H.; Gallagher-Jones M.; Chen S.; Pryor A.; Jin Q.; Hong Y. P.; Nashed Y. S. G.; Vogt S.; Miao J.; Jacobsen C. Correlative 3D X-Ray Fluorescence and Ptychographic Tomography of Frozen-Hydrated Green Algae. Sci. Adv. 2018, 4, eaau4548 10.1126/sciadv.aau4548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z. Y.; Liu Y.; Sun B. Y.; Li H.; Dong J. Q.; Zhang L. J.; Wang L. M.; Wang P.; Zhao Y. L.; Chen C. Y. Polyhydroxylated Metallofullerenols Stimulate IL-1 Beta Secretion of Macrophage through TLRs/MyD88/NF-Kappa B Pathway and NLRP3 Inflammasome Activation. Small 2014, 10, 2362–2372. 10.1002/smll.201302825. [DOI] [PubMed] [Google Scholar]
- Wang J.; Liu J.; Liu Y.; Wang L.; Cao M.; Ji Y.; Wu X.; Xu Y.; Bai B.; Miao Q.; Chen C.; Zhao Y. Gd-Hybridized Plasmonic Au-Nanocomposites Enhanced Tumor-Interior Drug Permeability in Multimodal Imaging-Guided Therapy. Adv. Mater. 2016, 28, 8950–8958. 10.1002/adma.201603114. [DOI] [PubMed] [Google Scholar]
- Miao J.; Förster F.; Levi O. Equally Sloped Tomography with Oversampling Reconstruction. Phys. Rev. B: Condens. Matter Mater. Phys. 2005, 72, 052103. 10.1103/PhysRevB.72.052103. [DOI] [Google Scholar]
- Lee E.; Fahimian B. P.; Iancu C. V.; Suloway C.; Murphy G. E.; Wright E. R.; Castaño-Díez D.; Jensen G. J.; Miao J. Radiation Dose Reduction and Image Enhancement in Biological Imaging through Equally-Sloped Tomography. J. Struct. Biol. 2008, 164, 221–227. 10.1016/j.jsb.2008.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao S.; Fan J.; Chen Z.; Zong Y.; Zhang J.; Sun Z.; Zhang L.; Tai R.; Liu Z.; Chen C.; Jiang H. Three-Dimensional Ultrastructural Imaging Reveals the Nanoscale Architecture of Mammalian Cells. IUCrJ 2018, 5, 141–149. 10.1107/S2052252517017912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Do M.; Isaacson S. A.; McDermott G.; Le Gros M. A.; Larabell C. A. Imaging and Characterizing Cells Using Tomography. Arch. Biochem. Biophys. 2015, 581, 111–121. 10.1016/j.abb.2015.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H.; Ji Z.; Roy K. R.; Gao M.; Pan Y.; Cai X.; Wang L.; Li W.; Chang C. H.; Kaweeteerawat C.; Chen C.; Xia T.; Zhao Y.; Li R. Engineered Graphene Oxide Nanocomposite Capable of Preventing the Evolution of Antimicrobial Resistance. ACS Nano 2019, 13, 11488–11499. 10.1021/acsnano.9b04970. [DOI] [PubMed] [Google Scholar]
- Jiang H.; Song C.; Chen C. C.; Xu R.; Raines K. S.; Fahimian B. P.; Lu C. H.; Lee T. K.; Nakashima A.; Urano J.; Ishikawa T.; Tamanoi F.; Miao J. Quantitative 3D Imaging of Whole, Unstained Cells by Using X-Ray Diffraction Microscopy. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 11234–11239. 10.1073/pnas.1000156107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishino Y.; Takahashi Y.; Imamoto N.; Ishikawa T.; Maeshima K. Three-Dimensional Visualization of a Human Chromosome Using Coherent X-Ray Diffraction. Phys. Rev. Lett. 2009, 102, 018101. 10.1103/PhysRevLett.102.018101. [DOI] [PubMed] [Google Scholar]
- Nelson J.; Huang X.; Steinbrener J.; Shapiro D.; Kirz J.; Marchesini S.; Neiman A. M.; Turner J. J.; Jacobsen C. High-Resolution X-Ray Diffraction Microscopy of Specifically Labeled Yeast Cells. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 7235–7239. 10.1073/pnas.0910874107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge M. D.; Holzner C.; Baines S. B.; Twining B. S.; Ignatyev K.; Diaz J.; Howard D. L.; Legnini D.; Miceli A.; McNulty I.; Jacobsen C. J.; Vogt S. Quantitative 3D Elemental Microtomography of Cyclotella meneghiniana at 400-nm Resolution. Proc. Natl. Acad. Sci. U. S. A. 2010, 107, 15676–15680. 10.1073/pnas.1001469107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cagno S.; Brede D. A.; Nuyts G.; Vanmeert F.; Pacureanu A.; Tucoulou R.; Cloetens P.; Falkenberg G.; Janssens K.; Salbu B.; Lind O. C. Combined Computed Nanotomography and Nanoscopic X-Ray Fluorescence Imaging of Cobalt Nanoparticles in Caenorhabditis elegans. Anal. Chem. 2017, 89, 11435–11442. 10.1021/acs.analchem.7b02554. [DOI] [PubMed] [Google Scholar]
- James S. A.; Burke R.; Howard D. L.; Spiers K. M.; Paterson D. J.; Murphy S.; Ramm G.; Kirkham R.; Ryan C. G.; de Jonge M. D. Visualising Coordination Chemistry: Fluorescence X-Ray Absorption near Edge Structure Tomography. Chem. Commun. 2016, 52, 11834–11837. 10.1039/C6CC06747F. [DOI] [PubMed] [Google Scholar]
- Deng J.; Vine D. J.; Chen S.; Nashed Y. S.; Jin Q.; Phillips N. W.; Peterka T.; Ross R.; Vogt S.; Jacobsen C. J. Simultaneous Cryo X-Ray Ptychographic and Fluorescence Microscopy of Green Algae. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 2314–2319. 10.1073/pnas.1413003112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. W.; Elgass K. D.; Junker M. D.; de Jonge M. D.; van Riessen G. A. Molar Concentration from Sequential 2-D Water-Window X-Ray Ptychography and X-Ray Fluorescence in Hydrated Cells. Sci. Rep. 2016, 6, 24280. 10.1038/srep24280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J.; Guan Y.; Cong Y.; Chen L.; Li Y. F.; Zhang L.; Zhang L.; Wang J.; Bai R.; Zhao Y.; Chen C.; Wang L. Single-Particle Analysis for Structure and Iron Chemistry of Atmospheric Particulate Matter. Anal. Chem. 2020, 92, 975–982. 10.1021/acs.analchem.9b03913. [DOI] [PubMed] [Google Scholar]
- Le Gros M. A.; McDermott G.; Larabell C. A. X-Ray Tomography of Whole Cells. Curr. Opin. Struct. Biol. 2005, 15, 593–600. 10.1016/j.sbi.2005.08.008. [DOI] [PubMed] [Google Scholar]
- Brandenberger C.; Mühlfeld C.; Ali Z.; Lenz A.-G.; Schmid O.; Parak W. J.; Gehr P.; Rothen-Rutishauser B. Quantitative Evaluation of Cellular Uptake and Trafficking of Plain and Polyethylene Glycol-Coated Gold Nanoparticles. Small 2010, 6, 1669–1678. 10.1002/smll.201000528. [DOI] [PubMed] [Google Scholar]
- Chapman H. N.; Fu J.; Jacobsen C.; Williams S. Dark-Field X-Ray Microscopy of Immunogold-Labeled Cells. Microsc. Microanal. 1996, 2, 53–62. 10.1017/S1431927696210530. [DOI] [Google Scholar]
- Matsuyama S.; Shimura M.; Mimura H.; Fujii M.; Yumoto H.; Sano Y.; Yabashi M.; Nishino Y.; Tamasaku K.; Ishikawa T.; Yamauchi K. Trace Element Mapping of a Single Cell Using a Hard X-Ray Nanobeam Focused by a Kirkpatrick-Baez Mirror System. X-Ray Spectrom. 2009, 38, 89–94. 10.1002/xrs.1123. [DOI] [Google Scholar]
- Kong H.; Zhang J.; Li J.; Wang J.; Shin H.-J.; Tai R.; Yan Q.; Xia K.; Hu J.; Wang L.; Zhu Y.; Fan C. Genetically Encoded X-Ray Cellular Imaging for Nanoscale Protein Localization. Natl. Sci. Rev. 2020, 7, 1218–1227. 10.1093/nsr/nwaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J.; Wang Y.; Xu C.; Zheng L.; Wang M.; Feng W.; Gao L.; Zhao L.; Liu R.; Gao F.; Zhao Y.; Chai Z.; Gao X. Facile Approach to Observe and Quantify the αIIbβ3 Integrin on a Single-Cell. Anal. Chem. 2015, 87, 2546–2549. 10.1021/ac504639u. [DOI] [PubMed] [Google Scholar]
- Metscher B. D. MicroCT for Comparative Morphology: Simple Staining Methods Allow High-Contrast 3D Imaging of Diverse Non-Mineralized Animal Tissues. BMC Physiol. 2009, 9, 11. 10.1186/1472-6793-9-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloetens P.; Barrett R.; Baruchel J.; Guigay J.-P.; Schlenker M. Phase Objects in Synchrotron Radiation Hard X-Ray Imaging. J. Phys. D: Appl. Phys. 1996, 29 (1), 133–146. 10.1088/0022-3727/29/1/023. [DOI] [Google Scholar]
- Pfeiffer F. X-Ray Ptychography. Nat. Photonics 2018, 12, 9–17. 10.1038/s41566-017-0072-5. [DOI] [Google Scholar]
- Carmona A.; Zogzas C. E.; Roudeau S.; Porcaro F.; Garrevoet J.; Spiers K. M.; Salomé M.; Cloetens P.; Mukhopadhyay S.; Ortega R. SLC30A10 Mutation Involved in Parkinsonism Results in Manganese Accumulation within Nanovesicles of the Golgi Apparatus. ACS Chem. Neurosci. 2019, 10, 599–609. 10.1021/acschemneuro.8b00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y.; Chen S.; Paunesku T.; Gleber S. C.; Liu W. C.; Doty C. B.; Mak R.; Deng J.; Jin Q.; Lai B.; Brister K.; Flachenecker C.; Jacobsen C.; Vogt S.; Woloschak G. E. Epidermal Growth Factor Receptor Targeted Nuclear Delivery and High-Resolution Whole Cell X-Ray Imaging of Fe3O4@TiO2 Nanoparticles in Cancer Cells. ACS Nano 2013, 7, 10502–10517. 10.1021/nn4033294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krenkel M.; Toepperwien M.; Alves F.; Salditt T. Three-Dimensional Single-Cell Imaging with X-Ray Waveguides in the Holographic Regime. Acta Crystallogr., Sect. A: Found. Adv. 2017, 73, 282–292. 10.1107/S2053273317007902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashraf S.; Said A. H.; Hartmann R.; Assmann M.-A.; Feliu N.; Lenz P.; Parak W. J. Quantitative Particle Uptake by Cells as Analyzed by Different Methods. Angew. Chem., Int. Ed. 2020, 59, 5438–5453. 10.1002/anie.201906303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Cai X.; Zhang Y.; Li X.; Li W.; Tian Y.; Li A.; Yu X.; Fan C.; Huang Q. Imaging Cellular Uptake and Intracellular Distribution of TiO2 Nanoparticles. Anal. Methods 2013, 5, 6611–6616. 10.1039/c3ay41121d. [DOI] [Google Scholar]
- Mei L.; Zhang X.; Yin W.; Dong X.; Guo Z.; Fu W.; Su C.; Gu Z.; Zhao Y. Translocation, Biotransformation-Related Degradation, and Toxicity Assessment of Polyvinylpyrrolidone-Modified 2H-Phase Mano-MoS2. Nanoscale 2019, 11, 4767–4780. 10.1039/C8NR10319D. [DOI] [PubMed] [Google Scholar]
- Krenkel M.; Markus A.; Bartels M.; Dullin C.; Alves F.; Salditt T. Phase-Contrast Zoom Tomography Reveals Precise Locations of Macrophages in Mouse Lungs. Sci. Rep. 2015, 5, 9973. 10.1038/srep09973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cedola A.; Bravin A.; Bukreeva I.; Fratini M.; Pacureanu A.; Mittone A.; Massimi L.; Cloetens P.; Coan P.; Campi G.; Spanò R.; Brun F.; Grigoryev V.; Petrosino V.; Venturi C.; Mastrogiacomo M.; Kerlero de Rosbo N.; Uccelli A. X-Ray Phase Contrast Tomography Reveals Early Vascular Alterations and Neuronal Loss in a Multiple Sclerosis Model. Sci. Rep. 2017, 7, 5890. 10.1038/s41598-017-06251-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpperwien M.; Krenkel M.; Vincenz D.; Stöber F.; Oelschlegel A. M.; Goldschmidt J.; Salditt T. Three-Dimensional Mouse Brain Cytoarchitecture Revealed by Laboratory-Based X-Ray Phase-Contrast Tomography. Sci. Rep. 2017, 7, 42847. 10.1038/srep42847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpperwien M.; van der Meer F.; Stadelmann C.; Salditt T. Three-Dimensional Virtual Histology of Human Cerebellum by X-Ray Phase-Contrast Tomography. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 6940. 10.1073/pnas.1801678115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reichardt M.; Töpperwien M.; Khan A.; Alves F.; Salditt T. Fiber Orientation in a Whole Mouse Heart Reconstructed by Laboratory Phase-Contrast Micro-CT. J. Med. Imaging. 2020, 7, 023501. 10.1117/1.JMI.7.2.023501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpperwien M.; Doeppner T. R.; Zechmeister B.; Bähr M.; Salditt T. Multiscale X-Ray Phase-Contrast Tomography in a Mouse Model of Transient Focal Cerebral Ischemia. Biomed. Opt. Express 2019, 10, 92–103. 10.1364/BOE.10.000092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels M.; Hernandez V. H.; Krenkel M.; Moser T.; Salditt T. Phase Contrast Tomography of the Mouse Cochlea at Microfocus X-Ray Sources. Appl. Phys. Lett. 2013, 103, 083703. 10.1063/1.4818737. [DOI] [Google Scholar]
- Krenkel M.; Töpperwien M.; Dullin C.; Alves F.; Salditt T. Propagation-Based Phase-Contrast Tomography for High-Resolution Lung Imaging with Laboratory Sources. AIP Adv. 2016, 6, 035007. 10.1063/1.4943898. [DOI] [Google Scholar]
- Bartels M.; Priebe M.; Wilke R. N.; Krüger S. P.; Giewekemeyer K.; Kalbfleisch S.; Olendrowitz C.; Sprung M.; Salditt T. Low-Dose Three-Dimensional Hard X-Ray Imaging of Bacterial Cells. Opt. Nanoscopy 2012, 1, 10. 10.1186/2192-2853-1-10. [DOI] [Google Scholar]
- Nicolas J.-D.; Bernhardt M.; Krenkel M.; Richter C.; Luther S.; Salditt T. Combined Scanning X-Ray Diffraction and Holographic Imaging of Cardiomyocytes. J. Appl. Crystallogr. 2017, 50, 612–620. 10.1107/S1600576717003351. [DOI] [Google Scholar]
- Bartels M.; Krenkel M.; Cloetens P.; Möbius W.; Salditt T. Myelinated Mouse Nerves Studied by X-Ray Phase Contrast Zoom Tomography. J. Struct. Biol. 2015, 192, 561–568. 10.1016/j.jsb.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Raupach R.; Flohr T. G. Analytical Evaluation of the Signal and Noise Propagation in X-Ray Differential Phase-Contrast Computed Tomography. Phys. Med. Biol. 2011, 56, 2219–2244. 10.1088/0031-9155/56/7/020. [DOI] [PubMed] [Google Scholar]
- Wen S.; Li K.; Cai H.; Chen Q.; Shen M.; Huang Y.; Peng C.; Hou W.; Zhu M.; Zhang G.; Shi X. Multifunctional Dendrimer-Entrapped Gold Nanoparticles for Dual Mode CT/MR Imaging Applications. Biomaterials 2013, 34, 1570–1580. 10.1016/j.biomaterials.2012.11.010. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Li C.; Zhang X.; Huo S.; Jin S.; An F.-F.; Wang X.; Xue X.; Okeke C. I.; Duan G.; Guo F.; Zhang X.; Hao J.; Wang P. C.; Zhang J.; Liang X.-J. In Vivo Tumor-Targeted Dual-Modal Fluorescence/CT Imaging Using a Nanoprobe Co-Loaded with an Aggregation-Induced Emission Dye and Gold Nanoparticles. Biomaterials 2015, 42, 103–111. 10.1016/j.biomaterials.2014.11.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Zhao L.; Li X.; Wang P.; Zhao J.; Shi X.; Shen M. Targeted Tumor SPECT/CT Dual Mode Imaging Using Multifunctional RGD-Modified Low Generation Dendrimer-Entrapped Gold Nanoparticles. Biomater. Sci. 2017, 5, 2393–2397. 10.1039/C7BM00826K. [DOI] [PubMed] [Google Scholar]
- Willemink M. J.; Persson M.; Pourmorteza A.; Pelc N. J.; Fleischmann D. Photon-Counting CT: Technical Principles and Clinical Prospects. Radiology 2018, 289, 293–312. 10.1148/radiol.2018172656. [DOI] [PubMed] [Google Scholar]
- Si-Mohamed S.; Bar-Ness D.; Sigovan M.; Cormode D. P.; Coulon P.; Coche E.; Vlassenbroek A.; Normand G.; Boussel L.; Douek P. Review of an Initial Experience with an Experimental Spectral Photon-Counting Computed Tomography System. Nucl. Instrum. Methods Phys. Res., Sect. A 2017, 873, 27–35. 10.1016/j.nima.2017.04.014. [DOI] [Google Scholar]
- Symons R.; Krauss B.; Sahbaee P.; Cork T. E.; Lakshmanan M. N.; Bluemke D. A.; Pourmorteza A. Photon-Counting CT for Simultaneous Imaging of Multiple Contrast Agents in the Abdomen: An in Vivo Study. Med. Phys. 2017, 44, 5120–5127. 10.1002/mp.12301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormode D. P.; Si-Mohamed S.; Bar-Ness D.; Sigovan M.; Naha P. C.; Balegamire J.; Lavenne F.; Coulon P.; Roessl E.; Bartels M.; Rokni M.; Blevis I.; Boussel L.; Douek P. Multicolor Spectral Photon-Counting Computed Tomography: In Vivo Dual Contrast Imaging with a High Count Rate Scanner. Sci. Rep. 2017, 7, 4784. 10.1038/s41598-017-04659-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D.; Schirra C. O.; Senpan A.; Schmieder A. H.; Stacy A. J.; Roessl E.; Thran A.; Wickline S. A.; Proska R.; Lanza G. M. An Early Investigation of Ytterbium Nanocolloids for Selective and Quantitative “Multicolor” Spectral CT Imaging. ACS Nano 2012, 6, 3364–3370. 10.1021/nn300392x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. C.; Nieves L. M.; Betzer O.; Sadan T.; Noël P. B.; Popovtzer R.; Cormode D. P. Nanoparticle Contrast Agents for X-Ray Imaging Applications. Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol. 2020, 12, e1642 10.1002/wnan.1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan D.; Roessl E.; Schlomka J.-P.; Caruthers S. D.; Senpan A.; Scott M. J.; Allen J. S.; Zhang H.; Hu G.; Gaffney P. J.; Choi E. T.; Rasche V.; Wickline S. A.; Proksa R.; Lanza G. M. Computed Tomography in Color: NanoK-Enhanced Spectral CT Molecular Imaging. Angew. Chem., Int. Ed. 2010, 49, 9635–9639. 10.1002/anie.201005657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naha P. C.; Hsu J. C.; Kim J.; Shah S.; Bouché M.; Si-Mohamed S.; Rosario-Berrios D. N.; Douek P.; Hajfathalian M.; Yasini P.; Singh S.; Rosen M. A.; Morgan M. A.; Cormode D. P. Dextran-Coated Cerium Oxide Nanoparticles: A Computed Tomography Contrast Agent for Imaging the Gastrointestinal Tract and Inflammatory Bowel Disease. ACS Nano 2020, 14, 10187–10197. 10.1021/acsnano.0c03457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si-Mohamed S.; Cormode D. P.; Bar-Ness D.; Sigovan M.; Naha P. C.; Langlois J.-B.; Chalabreysse L.; Coulon P.; Blevis I.; Roessl E.; Erhard K.; Boussel L.; Douek P. Evaluation of Spectral Photon Counting Computed Tomography K-Edge Imaging for Determination of Gold Nanoparticle Biodistribution in Vivo. Nanoscale 2017, 9, 18246–18257. 10.1039/C7NR01153A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong F.-Y.; Zhang J.-W.; Li R.-F.; Wang Z.-X.; Wang W.-J.; Wang W. Unique Roles of Gold Nanoparticles in Drug Delivery, Targeting and Imaging Applications. Molecules 2017, 22, 1445. 10.3390/molecules22091445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando H.; Abu Lila A. S.; Tanaka M.; Doi Y.; Terada Y.; Yagi N.; Shimizu T.; Okuhira K.; Ishima Y.; Ishida T. Intratumoral Visualization of Oxaliplatin within a Liposomal Formulation Using X-Ray Fluorescence Spectrometry. Mol. Pharmaceutics 2018, 15, 403–409. 10.1021/acs.molpharmaceut.7b00762. [DOI] [PubMed] [Google Scholar]
- Sanchez-Cano C.; Romero-Canelón I.; Geraki K.; Sadler P. J. Microfocus X-Ray Fluorescence Mapping of Tumour Penetration by an Organoosmium Anticancer Complex. J. Inorg. Biochem. 2018, 185, 26–29. 10.1016/j.jinorgbio.2018.04.014. [DOI] [PubMed] [Google Scholar]
- Koba R.; Fujita H.; Nishibori M.; Saeki K.; Nagayoshi K.; Sadakari Y.; Nagai S.; Sekizawa O.; Nitta K.; Manabe T.; Ueki T.; Ishida T.; Oda Y.; Nakamura M. Quantitative Evaluation of the Intratumoral Distribution of Platinum in Oxaliplatin-Treated Rectal Cancer: In Situ Visualization of Platinum via Synchrotron Radiation X-Ray Fluorescence Spectrometry. Int. J. Cancer 2020, 146, 2498–2509. 10.1002/ijc.32592. [DOI] [PubMed] [Google Scholar]
- Bulin A.-L.; Broekgaarden M.; Chaput F.; Baisamy V.; Garrevoet J.; Busser B.; Brueckner D.; Youssef A.; Ravanat J.-L.; Dujardin C.; Motto-Ros V.; Lerouge F.; Bohic S.; Sancey L.; Elleaume H. Radiation Dose-Enhancement Is a Potent Radiotherapeutic Effect of Rare-Earth Composite Nanoscintillators in Preclinical Models of Glioblastoma. Adv. Sci. 2020, 7, 2001675. 10.1002/advs.202001675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann S. G.; Toybou D.; Pradas Del Real A. E.; Arndt D.; Tagmount A.; Viau M.; Safi M.; Pacureanu A.; Cloetens P.; Bohic S.; Salome M.; Castillo-Michel H.; Omana-Sanz B.; Hofmann A.; Vulpe C.; Simonato J. P.; Celle C.; Charlet L.; Gilbert B. Crumpling of Silver Nanowires by Endolysosomes Strongly Reduces Toxicity. Proc. Natl. Acad. Sci. U. S. A. 2019, 116, 14893–14898. 10.1073/pnas.1820041116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitol E. A.; Rozhkova E. A.; Rose V.; Stripe B. D.; Young N. R.; Cohen E. E. W.; Leoni L.; Novosad V. Efficient Cisplatin Pro-Drug Delivery Visualized with Sub-100 nm Resolution: Interfacing Engineered Thermosensitive Magnetomicelles with a Living System. Adv. Mater. Interfaces 2014, 1, 1400182. 10.1002/admi.201400182. [DOI] [Google Scholar]
- Sanchez-Cano C.; Romero-Canelón I.; Yang Y.; Hands-Portman I. J.; Bohic S.; Cloetens P.; Sadler P. J. Synchrotron X-Ray Fluorescence Nanoprobe Reveals Target Sites for Organo-Osmium Complex in Human Ovarian Cancer Cells. Chem. - Eur. J. 2017, 23, 2512–2516. 10.1002/chem.201605911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reith F.; Etschmann B.; Grosse C.; Moors H.; Benotmane M. A.; Monsieurs P.; Grass G.; Doonan C.; Vogt S.; Lai B.; Martinez-Criado G.; George G. N.; Nies D. H.; Mergeay M.; Pring A.; Southam G.; Brugger J. Mechanisms of Gold Biomineralization in the Bacterium Cupriavidus metallidurans. Proc. Natl. Acad. Sci. U. S. A. 2009, 106, 17757–17762. 10.1073/pnas.0904583106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korbas M.; Blechinger S. R.; Krone P. H.; Pickering I. J.; George G. N. Localizing Organomercury Uptake and Accumulation in Zebrafish Larvae at the Tissue and Cellular Level. Proc. Natl. Acad. Sci. U. S. A. 2008, 105, 12108. 10.1073/pnas.0803147105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiver I.; Hesse B.; Seim C.; Castillo-Michel H.; Villanova J.; Laux P.; Dreiack N.; Penning R.; Tucoulou R.; Cotte M.; Luch A. Synchrotron-Based ν-XRF Mapping and μ-FTIR Microscopy Enable to Look into the Fate and Effects of Tattoo Pigments in Human Skin. Sci. Rep. 2017, 7, 11395. 10.1038/s41598-017-11721-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Servin A. D.; Castillo-Michel H.; Hernandez-Viezcas J. A.; Diaz B. C.; Peralta-Videa J. R.; Gardea-Torresdey J. L. Synchrotron Micro-XRF and Micro-XANES Confirmation of the Uptake and Translocation of TiO2 Nanoparticles in Cucumber (Cucumis sativus) Plants. Environ. Sci. Technol. 2012, 46, 7637–7643. 10.1021/es300955b. [DOI] [PubMed] [Google Scholar]
- Castillo-Michel H. A.; Larue C.; Pradas del Real A. E.; Cotte M.; Sarret G. Practical Review on the Use of Synchrotron Based Micro- and Nano- X-Ray Fluorescence Mapping and X-Ray Absorption Spectroscopy to Investigate the Interactions between Plants and Engineered Nanomaterials. Plant Physiol. Biochem. 2017, 110, 13–32. 10.1016/j.plaphy.2016.07.018. [DOI] [PubMed] [Google Scholar]
- Schultke E.; Menk R.; Pinzer B.; Astolfo A.; Stampanoni M.; Arfelli F.; Harsan L. A.; Nikkhah G. Single-Cell Resolution in High-Resolution Synchrotron X-Ray CT Imaging with Gold Nanoparticles. J. Synchrotron Radiat. 2014, 21, 242–250. 10.1107/S1600577513029007. [DOI] [PubMed] [Google Scholar]
- Brümmer T.; Debus A.; Pausch R.; Osterhoff J.; Grüner F. Design Study for a Compact Laser-Driven Source for Medical X-Ray Fluorescence Imaging. Phys. Rev. Accel. Beams. 2020, 23, 031601. 10.1103/PhysRevAccelBeams.23.031601. [DOI] [Google Scholar]
- Chan K. L. A.; Fale P. L. V.; Atharawi A.; Wehbe K.; Cinque G. Subcellular Mapping of Living Cells via Synchrotron MicroFTIR and ZnS Hemispheres. Anal. Bioanal. Chem. 2018, 410, 6477–6487. 10.1007/s00216-018-1245-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holman H.-Y. N.; Miles R.; Hao Z.; Wozei E.; Anderson L. M.; Yang H. Real-Time Chemical Imaging of Bacterial Activity in Biofilms Using Open-Channel Microfluidics and Synchrotron FTIR Spectromicroscopy. Anal. Chem. 2009, 81, 8564–8570. 10.1021/ac9015424. [DOI] [PubMed] [Google Scholar]
- Doherty J.; Raoof A.; Hussain A.; Wolna M.; Cinque G.; Brown M.; Gardner P.; Denbigh J. Live Single Cell Analysis Using Synchrotron FTIR Microspectroscopy: Development of a Simple Dynamic Flow System for Prolonged Sample Viability. Analyst 2019, 144, 997–1007. 10.1039/C8AN01566J. [DOI] [PubMed] [Google Scholar]
- Miller L. M.; Bourassa M. W.; Smith R. J. FTIR Spectroscopic Imaging of Protein Aggregation in Living Cells. Biochim. Biophys. Acta, Biomembr. 2013, 1828, 2339–2346. 10.1016/j.bbamem.2013.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pijanka J.; Sockalingum G. D.; Kohler A.; Yang Y.; Draux F.; Parkes G.; Lam K. P.; Collins D.; Dumas P.; Sandt C.; van Pittius D. G.; Douce G.; Manfait M.; Untereiner V.; Sule-Suso J. Synchrotron-Based FTIR Spectra of Stained Single Cells. Towards a Clinical Application in Pathology. Lab. Invest. 2010, 90, 797–807. 10.1038/labinvest.2010.8. [DOI] [PubMed] [Google Scholar]
- Morgan K. S.; Parsons D.; Cmielewski P.; McCarron A.; Gradl R.; Farrow N.; Siu K.; Takeuchi A.; Suzuki Y.; Uesugi K.; Uesugi M.; Yagi N.; Hall C.; Klein M.; Maksimenko A.; Stevenson A.; Hausermann D.; Dierolf M.; Pfeiffer F.; Donnelley M. Methods for Dynamic Synchrotron X-Ray Respiratory Imaging in Live Animals. J. Synchrotron Radiat. 2020, 27, 164–175. 10.1107/S1600577519014863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradl R.; Dierolf M.; Yang L.; Hehn L.; Günther B.; Möller W.; Kutschke D.; Stoeger T.; Gleich B.; Achterhold K.; Donnelley M.; Pfeiffer F.; Schmid O.; Morgan K. S. Visualizing Treatment Delivery and Deposition in Mouse Lungs Using in Vivo X-Ray Imaging. J. Controlled Release 2019, 307, 282–291. 10.1016/j.jconrel.2019.06.035. [DOI] [PubMed] [Google Scholar]
- Gradl R.; Dierolf M.; Günther B.; Hehn L.; Möller W.; Kutschke D.; Yang L.; Donnelley M.; Murrie R.; Erl A.; Stoeger T.; Gleich B.; Achterhold K.; Schmid O.; Pfeiffer F.; Morgan K. S. In Vivo Dynamic Phase-Contrast X-Ray Imaging Using a Compact Light Source. Sci. Rep. 2018, 8, 6788. 10.1038/s41598-018-24763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vakili M.; Merkens S.; Gao Y.; Gwozdz P. V.; Vasireddi R.; Sharpnack L.; Meyer A.; Blick R. H.; Trebbin M. 3D Micromachined Polyimide Mixing Devices for in Situ X-Ray Imaging of Solution-Based Block Copolymer Phase Transitions. Langmuir 2019, 35, 10435–10445. 10.1021/acs.langmuir.9b00728. [DOI] [PubMed] [Google Scholar]
- Merkens S.; Vakili M.; Sanchez-Iglesias A.; Litti L.; Gao Y.; Gwozdz P. V.; Sharpnack L.; Blick R. H.; Liz-Marzan L. M.; Grzelczak M.; Trebbin M. Time-Resolved Analysis of the Structural Dynamics of Assembling Gold Nanoparticles. ACS Nano 2019, 13, 6596–6604. 10.1021/acsnano.9b00575. [DOI] [PubMed] [Google Scholar]
- Bhat A.; Gwozdz P. V.; Seshadri A.; Hoeft M.; Blick R. H. Tank Circuit for Ultrafast Single-Particle Detection in Micropores. Phys. Rev. Lett. 2018, 121, 078102. 10.1103/PhysRevLett.121.078102. [DOI] [PubMed] [Google Scholar]
- Nolte P.; Stierle A.; Jin-Phillipp N. Y.; Kasper N.; Schulli T. U.; Dosch H. Shape Changes of Supported Rh Nanoparticles during Oxidation and Reduction Cycles. Science 2008, 321, 1654–1658. 10.1126/science.1160845. [DOI] [PubMed] [Google Scholar]
- Nolte P.; Stierle A.; Kasper N.; Jin-Phillipp N. Y.; Jeutter N.; Dosch H. Reversible Shape Changes of Pd Nanoparticles on MgO(100). Nano Lett. 2011, 11, 4697–4700. 10.1021/nl2023564. [DOI] [PubMed] [Google Scholar]
- Hejral U.; Franz D.; Volkov S.; Francoual S.; Strempfer J.; Stierle A. Identification of a Catalytically Highly Active Surface Phase for CO Oxidation over PtRh Nanoparticles under Operando Reaction Conditions. Phys. Rev. Lett. 2018, 120, 126101. 10.1103/PhysRevLett.120.126101. [DOI] [PubMed] [Google Scholar]
- Nolte P.; Stierle A.; Kasper N.; Jin-Phillipp N. Y.; Reichert H.; Rühm A.; Okasinski J.; Dosch H.; Schöder S. Combinatorial High-Energy X-Ray Microbeam Study of the Size-Dependent Oxidation of Pd Nanoparticles on MgO(100). Phys. Rev. B: Condens. Matter Mater. Phys. 2008, 77, 115444. 10.1103/PhysRevB.77.115444. [DOI] [Google Scholar]
- Hejral U.; Müller P.; Balmes O.; Pontoni D.; Stierle A. Tracking the Shape-Dependent Sintering of Platinum-Rhodium Model Catalysts under Operando Conditions. Nat. Commun. 2016, 7, 10964. 10.1038/ncomms10964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuin M.; Kim Y. Y.; Runge H.; Kulkarni S.; Maier S.; Dzhigaev D.; Lazarev S.; Gelisio L.; Seitz C.; Richard M.-I.; Zhou T.; Vonk V.; Keller T. F.; Vartanyants I. A.; Stierle A. Coherent X-Ray Imaging of CO-Adsorption-Induced Structural Changes in Pt Nanoparticles: Implications for Catalysis. ACS Appl. Nano Mater. 2019, 2, 4818–4824. 10.1021/acsanm.9b00764. [DOI] [Google Scholar]
- Kawaguchi T.; Keller T. F.; Runge H.; Gelisio L.; Seitz C.; Kim Y. Y.; Maxey E. R.; Cha W.; Ulvestad A.; Hruszkewycz S. O.; Harder R.; Vartanyants I. A.; Stierle A.; You H. Gas-Induced Segregation in Pt-Rh Alloy Nanoparticles Observed by in Situ Bragg Coherent Diffraction Imaging. Phys. Rev. Lett. 2019, 123, 246001. 10.1103/PhysRevLett.123.246001. [DOI] [PubMed] [Google Scholar]
- Müller P.; Hejral U.; Rütt U.; Stierle A. In Situ Oxidation Study of Pd-Rh Nanoparticles on MgAl2O4(001). Phys. Chem. Chem. Phys. 2014, 16, 13866–13874. 10.1039/C4CP01271B. [DOI] [PubMed] [Google Scholar]
- Sayes C. M.; Wahi R.; Kurian P. A.; Liu Y. P.; West J. L.; Ausman K. D.; Warheit D. B.; Colvin V. L. Correlating Nanoscale Titania Structure with Toxicity: A Cytotoxicity and Inflammatory Response Study with Human Dermal Fibroblasts and Human Lung Epithelial Cells. Toxicol. Sci. 2006, 92, 174–185. 10.1093/toxsci/kfj197. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Lai W.; Yin T.; Zhang C.; Yue C.; Cheng J.; Wang K.; Yang Y.; Cui D.; Parak W. J. Investigation of the Viability of Cells upon Co-Exposure to Gold and Iron Oxide Nanoparticles. Bioconjugate Chem. 2018, 29, 2120–2125. 10.1021/acs.bioconjchem.8b00349. [DOI] [PubMed] [Google Scholar]
- Lo Y. H.; Zhao L.; Gallagher-Jones M.; Rana A.; Lodico J. J.; Xiao W.; Regan B. C.; Miao J. In Situ Coherent Diffractive Imaging. Nat. Commun. 2018, 9, 1826. 10.1038/s41467-018-04259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carril M.; Padro D.; del Pino P.; Carrillo-Carrion C.; Gallego M.; Parak W. J. In Situ Detection of the Protein Corona in Complex Environments. Nat. Commun. 2017, 8, 1542. 10.1038/s41467-017-01826-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruetzel L. K.; Fischer S.; Salditt A.; Sedlak S. M.; Nickel B.; Lipfert J. A Mo-Anode-Based In-House Source for Small-Angle X-Ray Scattering Measurements of Biological Macromolecules. Rev. Sci. Instrum. 2016, 87, 025103. 10.1063/1.4940936. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Allen A. J.; Levine L. E.; Espinal L.; Antonucci J. M.; Skrtic D.; O’Donnell J. N.; Ilavsky J. Ultra-Small-Angle X-Ray Scattering-X-Ray Photon Correlation Spectroscopy Studies of Incipient Structural Changes in Amorphous Calcium Phosphate-Based Dental Composites. J. Biomed. Mater. Res., Part A 2012, 100, 1293–1306. 10.1002/jbm.a.34018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich C.; Hochrein M. B.; Krause B.; Nickel B. A Microfluidic Setup for Studies of Solid-Liquid Interfaces Using X-Ray Reflectivity and Fluorescence Microscopy. Rev. Sci. Instrum. 2005, 76, 095103. 10.1063/1.2040187. [DOI] [Google Scholar]
- Saurel D.; Segalini J.; Jauregui M.; Pendashteh A.; Daffos B.; Simon P.; Casas-Cabanas M. A SAXS Outlook on Disordered Carbonaceous Materials for Electrochemical Energy Storage. Energy Stor. Mater. 2019, 21, 162–173. 10.1016/j.ensm.2019.05.007. [DOI] [Google Scholar]
- Xia Y.; Nguyen T. D.; Yang M.; Lee B.; Santos A.; Podsiadlo P.; Tang Z.; Glotzer S. C.; Kotov N. A. Self-Assembly of Self-Limiting Monodisperse Supraparticles from Polydisperse Nanoparticles. Nat. Nanotechnol. 2011, 6, 580–587. 10.1038/nnano.2011.121. [DOI] [PubMed] [Google Scholar]
- Merkens S.; Vakili M.; Sánchez-Iglesias A.; Litti L.; Gao Y.; Gwozdz P. V.; Sharpnack L.; Blick R. H.; Liz-Marzán L. M.; Grzelczak M.; Trebbin M. Time-Resolved Analysis of the Structural Dynamics of Assembling Gold Nanoparticles. ACS Nano 2019, 13, 6596–6604. 10.1021/acsnano.9b00575. [DOI] [PubMed] [Google Scholar]
- Podsiadlo P.; Michel M.; Critchley K.; Srivastava S.; Qin M.; Lee J. W.; Verploegen E.; Hart A. J.; Qi Y.; Kotov N. A. Diffusional Self-Organization in Exponential Layer-by-Layer Films with Micro- and Nanoscale Periodicity. Angew. Chem., Int. Ed. 2009, 48, 7073–7077. 10.1002/anie.200901720. [DOI] [PubMed] [Google Scholar]
- Tang Z.; Kotov N. A.; Magonov S.; Ozturk B. Nanostructured Artificial Nacre. Nat. Mater. 2003, 2, 413–418. 10.1038/nmat906. [DOI] [PubMed] [Google Scholar]
- Zhang J.; Feng W.; Zhang H.; Wang Z.; Calcaterra H. A.; Yeom B.; Hu P. A.; Kotov N. A. Multiscale Deformations Lead to High Toughness and Circularly Polarized Emission in Helical Nacre-Like Fibres. Nat. Commun. 2016, 7, 10701. 10.1038/ncomms10701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slocik J. M.; Govorov A. O.; Naik R. R. Plasmonic Circular Dichroism of Peptide-Functionalized Gold Nanoparticles. Nano Lett. 2011, 11, 701–705. 10.1021/nl1038242. [DOI] [PubMed] [Google Scholar]
- Karst J.; Cho N. H.; Kim H.; Lee H.-E.; Nam K. T.; Giessen H.; Hentschel M. Chiral Scatterometry on Chemically Synthesized Single Plasmonic Nanoparticles. ACS Nano 2019, 13, 8659–8668. 10.1021/acsnano.9b04046. [DOI] [PubMed] [Google Scholar]
- Huang Y.; Fu Y.; Li M.; Jiang D.; Kutyreff C. J.; Engle J. W.; Lan X.; Cai W.; Chen T. Chirality-Driven Transportation and Oxidation Prevention by Chiral Selenium Nanoparticles. Angew. Chem., Int. Ed. 2020, 59, 4406–4414. 10.1002/anie.201910615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Liu J.; Ramesar N. S.; Heinz H.; Xu L.; Xu C.; Kotov N. A. Single- and Multi-Component Chiral Supraparticles as Modular Enantioselective Catalysts. Nat. Commun. 2019, 10, 4826. 10.1038/s41467-019-12134-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W.; Kim J.-Y.; Wang X.; Calcaterra H. A.; Qu Z.; Meshi L.; Kotov N. A. Assembly of Mesoscale Helices with Near-Unity Enantiomeric Excess and Light-Matter Interactions for Chiral Semiconductors. Sci. Adv. 2017, 3, e1601159 10.1126/sciadv.1601159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun M.; Xu L.; Bahng J. H.; Kuang H.; Alben S.; Kotov N. A.; Xu C. Intracellular Localization of Nanoparticle Dimers by Chirality Reversal. Nat. Commun. 2017, 8, 1847. 10.1038/s41467-017-01337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.; Xu L.; Ma W.; Wu X.; Sun M.; Kuang H.; Wang L.; Kotov N. A.; Xu C. Dual-Mode Ultrasensitive Quantification of MicroRNA in Living Cells by Chiroplasmonic Nanopyramids Self-Assembled from Gold and Upconversion Nanoparticles. J. Am. Chem. Soc. 2016, 138, 306–312. 10.1021/jacs.5b10309. [DOI] [PubMed] [Google Scholar]
- Sun M.; Hao T.; Li X.; Qu A.; Xu L.; Hao C.; Xu C.; Kuang H. Direct Observation of Selective Autophagy Induction in Cells and Tissues by Self-Assembled Chiral Nanodevice. Nat. Commun. 2018, 9, 4494. 10.1038/s41467-018-06946-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Y.; Cohen A. E. Optical Chirality and Its Interaction with Matter. Phys. Rev. Lett. 2010, 104, 163901. 10.1103/PhysRevLett.104.163901. [DOI] [PubMed] [Google Scholar]
- Solomon M. L.; Saleh A. A. E.; Poulikakos L. V.; Abendroth J. M.; Tadesse L. F.; Dionne J. A. Nanophotonic Platforms for Chiral Sensing and Separation. Acc. Chem. Res. 2020, 53, 588–598. 10.1021/acs.accounts.9b00460. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zhou Y.; Wang H. Y.; Perrett S.; Zhao Y.; Tang Z.; Nie G. Chirality of Glutathione Surface Coating Affects the Cytotoxicity of Quantum Dots. Angew. Chem., Int. Ed. 2011, 50, 5860–5864. 10.1002/anie.201008206. [DOI] [PubMed] [Google Scholar]
- González-Rubio G.; Mosquera J.; Kumar V.; Pedrazo-Tardajos A.; Llombart P.; Solís D. M.; Lobato I.; Noya E. G.; Guerrero-Martínez A.; Taboada J. M.; Obelleiro F.; MacDowell L. G.; Bals S.; Liz-Marzán L. M. Micelle-Directed Chiral Seeded Growth on Anisotropic Gold Nanocrystals. Science 2020, 368, 1472–1477. 10.1126/science.aba0980. [DOI] [PubMed] [Google Scholar]
- Yan W.; Xu L.; Xu C.; Ma W.; Kuang H.; Wang L.; Kotov N. A. Self-Assembly of Chiral Nanoparticle Pyramids with Strong R/S Optical Activity. J. Am. Chem. Soc. 2012, 134, 15114–15121. 10.1021/ja3066336. [DOI] [PubMed] [Google Scholar]
- Jana S.; de Frutos M.; Davidson P.; Abécassis B. Ligand-Induced Twisting of Nanoplatelets and Their Self-Assembly into Chiral Ribbons. Sci. Adv. 2017, 3, e1701483 10.1126/sciadv.1701483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang W.; Qu Z.-b.; Kumar P.; Vecchio D.; Wang Y.; Ma Y.; Bahng J. H.; Bernardino K.; Gomes W. R.; Colombari F. M.; Lozada-Blanco A.; Veksler M.; Marino E.; Simon A.; Murray C.; Muniz S. R.; de Moura A. F.; Kotov N. A. Emergence of Complexity in Hierarchically Organized Chiral Particles. Science 2020, 368, 642–648. 10.1126/science.aaz7949. [DOI] [PubMed] [Google Scholar]
- Ahn J.; Ma S.; Kim J.-Y.; Kyhm J.; Yang W.; Lim J. A.; Kotov N. A.; Moon J. Chiral 2D Organic Inorganic Hybrid Perovskite with Circular Dichroism Tunable over Wide Wavelength Range. J. Am. Chem. Soc. 2020, 142, 4206–4212. 10.1021/jacs.9b11453. [DOI] [PubMed] [Google Scholar]
- Schroer C. G.; Falkenberg G. Hard X-Ray Nanofocusing at Low-Emittance Synchrotron Radiation Sources. J. Synchrotron Radiat. 2014, 21, 996–1005. 10.1107/S1600577514016269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weckert E. The Potential of Future Light Sources to Explore the Structure and Function of Matter. IUCrJ 2015, 2, 230–245. 10.1107/S2052252514024269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroer C. G.; Agapov I.; Brefeld W.; Brinkmann R.; Chae Y.-C.; Chao H.-C.; Eriksson M.; Keil J.; Nuel Gavalda X.; Rohlsberger R.; Seeck O. H.; Sprung M.; Tischer M.; Wanzenberg R.; Weckert E. PETRA IV: The Ultralow-Emittance Source Project at DESY. J. Synchrotron Radiat. 2018, 25, 1277–1290. 10.1107/S1600577518008858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Mitri S. One Way Only to Synchrotron Light Sources Upgrade?. J. Synchrotron Radiat. 2018, 25, 1323–1334. 10.1107/S160057751800810X. [DOI] [PubMed] [Google Scholar]
- Hettel R. DLSR Design and Plans: An International Overview. J. Synchrotron Radiat. 2014, 21, 843–855. 10.1107/S1600577514011515. [DOI] [PubMed] [Google Scholar]
- Eriksson M.; van der Veen J. F.; Quitmann C. Diffraction-Limited Storage Rings - A Window to the Science of Tomorrow. J. Synchrotron Radiat. 2014, 21, 837–842. 10.1107/S1600577514019286. [DOI] [PubMed] [Google Scholar]
- Boldon L.; Laliberte F.; Liu L. Review of the Fundamental Theories Behind Small Angle X-Ray Scattering, Molecular Dynamics Simulations, and Relevant Integrated Application. Nano Rev. 2015, 6, 25661. 10.3402/nano.v6.25661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fratzl P.; Jakob H. F.; Rinnerthaler S.; Roschger P.; Klaushofer K. Position-Resolved Small-Angle X-Ray Scattering of Complex Biological Materials. J. Appl. Crystallogr. 1997, 30, 765–769. 10.1107/S0021889897001775. [DOI] [Google Scholar]
- He W. X.; Rajasekharan A. K.; Tehrani-Bagha A. R.; Andersson M. Mesoscopically Ordered Bone-Mimetic Nanocomposites. Adv. Mater. 2015, 27, 2260–2264. 10.1002/adma.201404926. [DOI] [PubMed] [Google Scholar]
- Nicolas J. D.; Bernhardt M.; Markus A.; Alves F.; Burghammer M.; Salditt T. Scanning X-Ray Diffraction on Cardiac Tissue: Automatized Data Analysis and Processing. J. Synchrotron Radiat. 2017, 24, 1163–1172. 10.1107/S1600577517011936. [DOI] [PubMed] [Google Scholar]
- Jensen T. H.; Bech M.; Bunk O.; Thomsen M.; Menzel A.; Bouchet A.; Le Duc G.; Feidenhans’l R.; Pfeiffer F. Brain Tumor Imaging Using Small-Angle X-Ray Scattering Tomography. Phys. Med. Biol. 2011, 56, 1717–1726. 10.1088/0031-9155/56/6/012. [DOI] [PubMed] [Google Scholar]
- Schaff F.; Bech M.; Zaslansky P.; Jud C.; Liebi M.; Guizar-Sicairos M.; Pfeiffer F. Six-Dimensional Real and Reciprocal Space Small-Angle X-Ray Scattering Tomography. Nature 2015, 527, 353–356. 10.1038/nature16060. [DOI] [PubMed] [Google Scholar]
- Liebi M.; Georgiadis M.; Menzel A.; Schneider P.; Kohlbrecher J.; Bunk O.; Guizar-Sicairos M. Nanostructure Surveys of Macroscopic Specimens by Small-Angle Scattering Tensor Tomography. Nature 2015, 527, 349–352. 10.1038/nature16056. [DOI] [PubMed] [Google Scholar]
- Liebi M.; Georgiadis M.; Kohlbrecher J.; Holler M.; Raabe J.; Usov I.; Menzel A.; Schneider P.; Bunk O.; Guizar-Sicairos M. Small-Angle X-Ray Scattering Tensor Tomography: Model of the Three-Dimensional Reciprocal-Space Map, Reconstruction Algorithm and Angular Sampling Requirements. Acta Crystallogr., Sect. A: Found. Adv. 2018, 74, 12–24. 10.1107/S205327331701614X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conceição A. L. C.; Perlich J.; Haas S.; Funari S. S. SAXS-CT: A Nanostructure Resolving Microscopy for Macroscopic Biologic Specimens. Biomed. Phys. Eng. Express 2020, 6, 035012. 10.1088/2057-1976/ab7cad. [DOI] [PubMed] [Google Scholar]
- Allec N.; Choi M.; Yesupriya N.; Szychowski B.; White M. R.; Kann M. G.; Garcin E. D.; Daniel M. C.; Badano A. Small-Angle X-Ray Scattering Method to Characterize Molecular Interactions: Proof of Concept. Sci. Rep. 2015, 5, 12085. 10.1038/srep12085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl C.; Frank K.; Amenitsch H.; Fischer S.; Liedl T.; Nickel B. Position Accuracy of Gold Nanoparticles on DNA Origami Structures Studied with Small-Angle X-Ray Scattering. Nano Lett. 2018, 18, 2609–2615. 10.1021/acs.nanolett.8b00412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberdisse J. Aggregation of Colloidal Nanoparticles in Polymer Matrices. Soft Matter 2006, 2, 29–36. 10.1039/B511959F. [DOI] [PubMed] [Google Scholar]
- Kiesel I.; Paulus M.; Nase J.; Tiemeyer S.; Sternemann C.; Rüster K.; Wirkert F. J.; Mende K.; Büning T.; Tolan M. Temperature-Driven Adsorption and Desorption of Proteins at Solid–Liquid Interfaces. Langmuir 2014, 30, 2077–2083. 10.1021/la404884a. [DOI] [PubMed] [Google Scholar]
- Evers F.; Jeworrek C.; Tiemeyer S.; Weise K.; Sellin D.; Paulus M.; Struth B.; Tolan M.; Winter R. Elucidating the Mechanism of Lipid Membrane-Induced IAPP Fibrillogenesis and Its Inhibition by the Red Wine Compound Resveratrol: A Synchrotron X-Ray Reflectivity Study. J. Am. Chem. Soc. 2009, 131, 9516–9521. 10.1021/ja8097417. [DOI] [PubMed] [Google Scholar]
- Giri R. P.; Mukhopadhyay M. K.; Basak U. K.; Chakrabarti A.; Sanyal M. K.; Runge B.; Murphy B. M. Continuous Uptake or Saturation—Investigation of Concentration and Surface-Packing-Specific Hemin Interaction with Lipid Membranes. J. Phys. Chem. B 2018, 122, 7547–7554. 10.1021/acs.jpcb.8b03327. [DOI] [PubMed] [Google Scholar]
- Giri R. P.; Mukhopadhyay M. K.; Mitra M.; Chakrabarti A.; Sanyal M. K.; Ghosh S. K.; Bera S.; Lurio L. B.; Ma Y.; Sinha S. K. Differential Adsorption of a Membrane Skeletal Protein, Spectrin, in Phospholipid Membranes. EPL 2017, 118, 58002. 10.1209/0295-5075/118/58002. [DOI] [Google Scholar]
- Basu J. K.; Sanyal M. K. Ordering and Growth of Langmuir–Blodgett films: X-Ray Scattering Studies. Phys. Rep. 2002, 363, 1–84. 10.1016/S0370-1573(01)00083-7. [DOI] [Google Scholar]
- Bhattacharyya A.; Sanyal M. K.; Mogera U.; George S. J.; Dhiman S.; Kulkarni G. U.; Fontaine P. Formation of Two-Dimensional Network of Organic Charge-Transfer Complexes at the Air–Water Interface. Langmuir 2019, 35, 12630–12635. 10.1021/acs.langmuir.9b01635. [DOI] [PubMed] [Google Scholar]
- Ke P. C.; Lin S.; Parak W. J.; Davis T. P.; Caruso F. A Decade of the Protein Corona. ACS Nano 2017, 11, 11773–11776. 10.1021/acsnano.7b08008. [DOI] [PubMed] [Google Scholar]
- Wang Y.; Cai R.; Chen C. The Nano–Bio Interactions of Nanomedicines: Understanding the Biochemical Driving Forces and Redox Reactions. Acc. Chem. Res. 2019, 52, 1507–1518. 10.1021/acs.accounts.9b00126. [DOI] [PubMed] [Google Scholar]
- Nel A. E.; Madler L.; Velegol D.; Xia T.; Hoek E. M. V.; Somasundaran P.; Klaessig F.; Castranova V.; Thompson M. Understanding Biophysicochemical Interactions at the Nano-Bio Interface. Nat. Mater. 2009, 8, 543–557. 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- Fang Y.; Wang H.; Yu H.; Liu X.; Wang W.; Chen H. Y.; Tao N. J. Plasmonic Imaging of Electrochemical Reactions of Single Nanoparticles. Acc. Chem. Res. 2016, 49, 2614–2624. 10.1021/acs.accounts.6b00348. [DOI] [PubMed] [Google Scholar]
- Fang Y.; Li Z.; Jiang Y.; Wang X.; Chen H. Y.; Tao N.; Wang W. Intermittent Photocatalytic Activity of Single CdS Nanoparticles. Proc. Natl. Acad. Sci. U. S. A. 2017, 114, 10566–10571. 10.1073/pnas.1708617114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y.; Wang W.; Wo X.; Luo Y.; Yin S.; Wang Y.; Shan X.; Tao N. Plasmonic Imaging of Electrochemical Oxidation of Single Nanoparticles. J. Am. Chem. Soc. 2014, 136, 12584–12587. 10.1021/ja507097y. [DOI] [PubMed] [Google Scholar]
- Chen Z.; Li J.; Chen X.; Cao J.; Zhang J.; Min Q.; Zhu J. J. Single Gold@Silver Nanoprobes for Real-Time Tracing the Entire Autophagy Process at Single-Cell Level. J. Am. Chem. Soc. 2015, 137, 1903–1908. 10.1021/ja5112628. [DOI] [PubMed] [Google Scholar]
- Xia Y.; Xia X.; Peng H. C. Shape-Controlled Synthesis of Colloidal Metal Nanocrystals: Thermodynamic versus Kinetic Products. J. Am. Chem. Soc. 2015, 137, 7947–7966. 10.1021/jacs.5b04641. [DOI] [PubMed] [Google Scholar]
- Peckys D. B.; de Jonge N. Visualizing Gold Nanoparticle Uptake in Live Cells with Liquid Scanning Transmission Electron Microscopy. Nano Lett. 2011, 11, 1733–1738. 10.1021/nl200285r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liz-Marzan L. M.; Grzelczak M. Growing Anisotropic Crystals at the Nanoscale. Science 2017, 356, 1120–1121. 10.1126/science.aam8774. [DOI] [PubMed] [Google Scholar]
- Hirai K.; Yeom B.; Chang S.-H.; Chi H.; Mansfield J. F.; Lee B.; Lee S.; Uher C.; Kotov N. A. Coordination Assembly of Discoid Nanoparticles. Angew. Chem., Int. Ed. 2015, 54, 8966–8970. 10.1002/anie.201502057. [DOI] [PubMed] [Google Scholar]
- Choi S.-J.; Choy J.-H. Effect of Physico-Chemical Parameters on the Toxicity of Inorganic Nanoparticles. J. Mater. Chem. 2011, 21, 5547–5554. 10.1039/c1jm10167f. [DOI] [Google Scholar]
- Limbach L. K.; Wick P.; Manser P.; Grass R. N.; Bruinink A.; Stark W. J. Exposure of Engineered Nanoparticles to Human Lung Epithelial Cells: Influence of Chemical Composition and Catalytic Activity on Oxidative Stress. Environ. Sci. Technol. 2007, 41, 4158–4163. 10.1021/es062629t. [DOI] [PubMed] [Google Scholar]
- Schön F.; Biebl F.; Greb L.; Leingang S.; Grimm-Lebsanft B.; Teubner M.; Buchenau S.; Kaifer E.; Rübhausen M. A.; Himmel H.-J. On the Metal Cooperativity in a Dinuclear Copper–Guanidine Complex for Aliphatic C-H Bond Cleavage by Dioxygen. Chem. - Eur. J. 2019, 25, 11257–11268. 10.1002/chem.201901906. [DOI] [PubMed] [Google Scholar]
- Naumova M.; Khakhulin D.; Rebarz M.; Rohrmüller M.; Dicke B.; Biednov M.; Britz A.; Espinoza S.; Grimm-Lebsanft B.; Kloz M.; Kretzschmar N.; Neuba A.; Ortmeyer J.; Schoch R.; Andreasson J.; Bauer M.; Bressler C.; Gero Schmidt W.; Henkel G.; Rübhausen M. Structural Dynamics upon Photoexcitation-Induced Charge Transfer in a Dicopper(i)–Disulfide Complex. Phys. Chem. Chem. Phys. 2018, 20, 6274–6286. 10.1039/C7CP04880G. [DOI] [PubMed] [Google Scholar]
- Jiang X.; Foldbjerg R.; Miclaus T.; Wang L.; Singh R.; Hayashi Y.; Sutherland D.; Chen C.; Autrup H.; Beer C. Multi-Platform Genotoxicity Analysis of Silver Nanoparticles in the Model Cell Line CHO-K1. Toxicol. Lett. 2013, 222, 55–63. 10.1016/j.toxlet.2013.07.011. [DOI] [PubMed] [Google Scholar]
- Xia T.; Kovochich M.; Liong M.; Madler L.; Gilbert B.; Shi H.; Yeh J. I.; Zink J. I.; Nel A. E. Comparison of the Mechanism of Toxicity of Zinc Oxide and Cerium Oxide Nanoparticles Based on Dissolution and Oxidative Stress Properties. ACS Nano 2008, 2, 2121–2134. 10.1021/nn800511k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirshafiee V.; Sun B.; Chang C. H.; Liao Y. P.; Jiang W.; Jiang J.; Liu X.; Wang X.; Xia T.; Nel A. E. Toxicological Profiling of Metal Oxide Nanoparticles in Liver Context Reveals Pyroptosis in Kupffer Cells and Macrophages versus Apoptosis in Hepatocytes. ACS Nano 2018, 12, 3836–3852. 10.1021/acsnano.8b01086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naatz H.; Lin S.; Li R.; Jiang W.; Ji Z.; Chang C. H.; Koser J.; Thoming J.; Xia T.; Nel A. E.; Madler L.; Pokhrel S. Safe-by-Design CuO Nanoparticles via Fe-Doping, Cu-O Bond Length Variation, and Biological Assessment in Cells and Zebrafish Embryos. ACS Nano 2017, 11, 501–515. 10.1021/acsnano.6b06495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J.; Wang P.; Zhang X.; Wang L.; Wang D.; Gu Z.; Tang J.; Guo M.; Cao M.; Zhou H.; Liu Y.; Chen C. Rapid Degradation and High Renal Clearance of Cu3BiS3 Nanodots for Efficient Cancer Diagnosis and Photothermal Therapy in Vivo. ACS Nano 2016, 10, 4587–4598. 10.1021/acsnano.6b00745. [DOI] [PubMed] [Google Scholar]
- Wang L.; Yan L.; Liu J.; Chen C.; Zhao Y. Quantification of Nanomaterial/Nanomedicine Trafficking in Vivo. Anal. Chem. 2018, 90, 589–614. 10.1021/acs.analchem.7b04765. [DOI] [PubMed] [Google Scholar]
- Gong N.; Ma X.; Ye X.; Zhou Q.; Chen X.; Tan X.; Yao S.; Huo S.; Zhang T.; Chen S.; Teng X.; Hu X.; Yu J.; Gan Y.; Jiang H.; Li J.; Liang X.-J. Carbon-Dot-Supported Atomically Dispersed Gold as a Mitochondrial Oxidative Stress Amplifier for Cancer Treatment. Nat. Nanotechnol. 2019, 14, 379–387. 10.1038/s41565-019-0373-6. [DOI] [PubMed] [Google Scholar]
- Bauer M. HERFD-XAS and Valence-to-Core-XES: New Tools to Push the Limits in Research with Hard X-Rays?. Phys. Chem. Chem. Phys. 2014, 16, 13827–13837. 10.1039/C4CP00904E. [DOI] [PubMed] [Google Scholar]
- Gallo E.; Glatzel P. Valence to Core X-Ray Emission Spectroscopy. Adv. Mater. 2014, 26, 7730–7746. 10.1002/adma.201304994. [DOI] [PubMed] [Google Scholar]
- Szlachetko J.; Nachtegaal M.; de Boni E.; Willimann M.; Safonova O.; Sa J.; Smolentsev G.; Szlachetko M.; van Bokhoven J. A.; Dousse J. C.; Hoszowska J.; Kayser Y.; Jagodzinski P.; Bergamaschi A.; Schmitt B.; David C.; Lücke A. A von Hamos X-Ray Spectrometer Based on a Segmented-Type Diffraction Crystal for Single-Shot X-Ray Emission Spectroscopy and Time-Resolved Resonant Inelastic X-Ray Scattering Studies. Rev. Sci. Instrum. 2012, 83, 103105. 10.1063/1.4756691. [DOI] [PubMed] [Google Scholar]
- Hirsch O.; Kvashnina K. O.; Luo L.; Süess M. J.; Glatzel P.; Koziej D. High-Energy Resolution X-Ray Absorption and Emission Spectroscopy Reveals Insight into Unique Selectivity of La-Based Nanoparticles for CO2. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 15803. 10.1073/pnas.1516192113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lancaster K. M.; Roemelt M.; Ettenhuber P.; Hu Y.; Ribbe M. W.; Neese F.; Bergmann U.; DeBeer S. X-Ray Emission Spectroscopy Evidences a Central Carbon in the Nitrogenase Iron-Molybdenum Cofactor. Science 2011, 334, 974–977. 10.1126/science.1206445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressler C.; Milne C.; Pham V. T.; ElNahhas A.; van der Veen R. M.; Gawelda W.; Johnson S.; Beaud P.; Grolimund D.; Kaiser M.; Borca C. N.; Ingold G.; Abela R.; Chergui M. Femtosecond XANES Study of the Light-Induced Spin Crossover Dynamics in an Iron(II) Complex. Science 2009, 323, 489–492. 10.1126/science.1165733. [DOI] [PubMed] [Google Scholar]
- Sikora M.; Juhin A.; Weng T.-C.; Sainctavit P.; Detlefs C.; de Groot F.; Glatzel P. Strong K-Edge Magnetic Circular Dichroism Observed in Photon-In--Photon-Out Spectroscopy. Phys. Rev. Lett. 2010, 105, 037202. 10.1103/PhysRevLett.105.037202. [DOI] [PubMed] [Google Scholar]
- Daffé N.; Sikora M.; Rovezzi M.; Bouldi N.; Gavrilov V.; Neveu S.; Choueikani F.; Ohresser P.; Dupuis V.; Taverna D.; Gloter A.; Arrio M.-A.; Sainctavit P.; Juhin A. Nanoscale Distribution of Magnetic Anisotropies in Bimagnetic Soft Core–Hard Shell MnFe2O4@CoFe2O4 Nanoparticles. Adv. Mater. Interfaces 2017, 4, 1700599. 10.1002/admi.201700599. [DOI] [Google Scholar]
- Kuciakowski J.; Kmita A.; Lachowicz D.; Wytrwal-Sarna M.; Pitala K.; Lafuerza S.; Koziej D.; Juhin A.; Sikora M. Selective Magnetometry of Superparamagnetic Iron Oxide Nanoparticles in Liquids. Nanoscale 2020, 12, 16420–16426. 10.1039/D0NR02866E. [DOI] [PubMed] [Google Scholar]
- Liu B.; van Schooneveld M. M.; Cui Y.-T.; Miyawaki J.; Harada Y.; Eschemann T. O.; de Jong K. P.; Delgado-Jaime M. U.; de Groot F. M. F. In-Situ 2p3d Resonant Inelastic X-Ray Scattering Tracking Cobalt Nanoparticle Reduction. J. Phys. Chem. C 2017, 121, 17450–17456. 10.1021/acs.jpcc.7b04325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfold T. J.; Szlachetko J.; Santomauro F. G.; Britz A.; Gawelda W.; Doumy G.; March A. M.; Southworth S. H.; Rittmann J.; Abela R.; Chergui M.; Milne C. J. Revealing Hole Trapping in Zinc Oxide Nanoparticles by Time-Resolved X-Ray Spectroscopy. Nat. Commun. 2018, 9, 478. 10.1038/s41467-018-02870-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szlachetko J.; Kubas A.; Cieślak A. M.; Sokołowski K.; Ma̧kolski Ł.; Czapla-Masztafiak J.; Sá J.; Lewiński J. Hidden Gapless States during Thermal Transformations of Preorganized Zinc Alkoxides to Zinc Oxide Nanocrystals. Mater. Horiz. 2018, 5, 905–911. 10.1039/C8MH00106E. [DOI] [Google Scholar]
- Kayser Y.; Milne C.; Juranić P.; Sala L.; Czapla-Masztafiak J.; Follath R.; Kavčič M.; Knopp G.; Rehanek J.; Błachucki W.; Delcey M. G.; Lundberg M.; Tyrała K.; Zhu D.; Alonso-Mori R.; Abela R.; Sá J.; Szlachetko J. Core-Level Nonlinear Spectroscopy Triggered by Stochastic X-Ray Pulses. Nat. Commun. 2019, 10, 4761. 10.1038/s41467-019-12717-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M. O. Atomic Radiative and Radiationless Yields for K and L Shells. J. Phys. Chem. Ref. Data 1979, 8, 307–327. 10.1063/1.555594. [DOI] [Google Scholar]
- Veith L.; Böttner J.; Vennemann A.; Breitenstein D.; Engelhard C.; Meijer J.; Estrela-Lopis I.; Wiemann M.; Hagenhoff B. Detection of ZrO2 Nanoparticles in Lung Tissue Sections by Time-of-Flight Secondary Ion Mass Spectrometry and Ion Beam Microscopy. Nanomaterials 2018, 8, 44. 10.3390/nano8010044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turco A.; Moglianetti M.; Corvaglia S.; Rella S.; Catelani T.; Marotta R.; Malitesta C.; Pompa P. P. Sputtering-Enabled Intracellular X-Ray Photoelectron Spectroscopy: A Versatile Method to Analyze the Biological Fate of Metal Nanoparticles. ACS Nano 2018, 12, 7731–7740. 10.1021/acsnano.8b01612. [DOI] [PubMed] [Google Scholar]
- Sutton M.; Mochrie S. G. J.; Greytak T.; Nagler S. E.; Berman L. E.; Held G. A.; Stephenson G. B. Observation of Speckle by Diffraction with Coherent X-Rays. Nature 1991, 352, 608–610. 10.1038/352608a0. [DOI] [Google Scholar]
- Westermeier F.; Autenrieth T.; Gutt C.; Leupold O.; Duri A.; Menzel A.; Johnson I.; Broennimann C.; Grubel G. Fast Two-Dimensional Detection for X-Ray Photon Correlation Spectroscopy Using the PILATUS Detector. J. Synchrotron Radiat. 2009, 16, 687–689. 10.1107/S0909049509023280. [DOI] [PubMed] [Google Scholar]
- Fluerasu A.; Moussaid A.; Falus P.; Gleyzolle H.; Madsen A. X-Ray Photon Correlation Spectroscopy under Flow. J. Synchrotron Radiat. 2008, 15, 378–384. 10.1107/S0909049508006420. [DOI] [PubMed] [Google Scholar]
- Busch S.; Jensen T. H.; Chushkin Y.; Fluerasu A. Dynamics in Shear Flow Studied by X-Ray Photon Correlation Spectroscopy. Eur. Phys. J. E: Soft Matter Biol. Phys. 2008, 26, 55–62. 10.1140/epje/i2007-10305-2. [DOI] [PubMed] [Google Scholar]
- Hruszkewycz S. O.; Sutton M.; Fuoss P. H.; Adams B.; Rosenkranz S.; Ludwig K. F. Jr.; Roseker W.; Fritz D.; Cammarata M.; Zhu D.; Lee S.; Lemke H.; Gutt C.; Robert A.; Grubel G.; Stephenson G. B. High Contrast X-Ray Speckle from Atomic-Scale Order in Liquids and Glasses. Phys. Rev. Lett. 2012, 109, 185502. 10.1103/PhysRevLett.109.185502. [DOI] [PubMed] [Google Scholar]
- Verwohlt J.; Reiser M.; Randolph L.; Matic A.; Medina L. A.; Madsen A.; Sprung M.; Zozulya A.; Gutt C. Low Dose X-Ray Speckle Visibility Spectroscopy Reveals Nanoscale Dynamics in Radiation Sensitive Ionic Liquids. Phys. Rev. Lett. 2018, 120, 168001. 10.1103/PhysRevLett.120.168001. [DOI] [PubMed] [Google Scholar]
- Shpyrko O. G. X-Ray Photon Correlation Spectroscopy. J. Synchrotron Radiat. 2014, 21, 1057–1064. 10.1107/S1600577514018232. [DOI] [PubMed] [Google Scholar]
- Bandyopadhyay R.; Liang D.; Harden J. L.; Leheny R. L. Slow Dynamics, Aging, and Glassy Rheology in Soft and Living Matter. Solid State Commun. 2006, 139, 589–598. 10.1016/j.ssc.2006.06.023. [DOI] [Google Scholar]
- Jain A.; Schulz F.; Lokteva I.; Frenzel L.; Grübel G.; Lehmkühler F. Anisotropic and Heterogeneous Dynamics in an Aging Colloidal Gel. Soft Matter 2020, 16, 2864–2872. 10.1039/C9SM02230A. [DOI] [PubMed] [Google Scholar]
- Möller J.; Sprung M.; Madsen A.; Gutt C. X-Ray Photon Correlation Spectroscopy of Protein Dynamics at Nearly Diffraction-Limited Storage Rings. IUCrJ 2019, 6, 794–803. 10.1107/S2052252519008273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmkühler F.; Valerio J.; Sheyfer D.; Roseker W.; Schroer M. A.; Fischer B.; Tono K.; Yabashi M.; Ishikawa T.; Grübel G. Dynamics of Soft Nanoparticle Suspensions at Hard X-Ray FEL Sources below the Radiation-Damage Threshold. IUCrJ 2018, 5, 801–807. 10.1107/S2052252518013696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T.; Tran T. T.-T.; Park C.; Lee B.-J. Biomimetic Shear Stress and Nanoparticulate Drug Delivery. J. Pharm. Invest. 2017, 47, 133–139. 10.1007/s40005-017-0313-0. [DOI] [Google Scholar]
- Holme M. N.; Fedotenko I. A.; Abegg D.; Althaus J.; Babel L.; Favarger F.; Reiter R.; Tanasescu R.; Zaffalon P.-L.; Ziegler A.; Müller B.; Saxer T.; Zumbuehl A. Shear-Stress Sensitive Lenticular Vesicles for Targeted Drug Delivery. Nat. Nanotechnol. 2012, 7, 536–543. 10.1038/nnano.2012.84. [DOI] [PubMed] [Google Scholar]
- Saxer T.; Zumbuehl A.; Müller B. The Use of Shear Stress for Targeted Drug Delivery. Cardiovasc. Res. 2013, 99, 328–333. 10.1093/cvr/cvt102. [DOI] [PubMed] [Google Scholar]
- Epshtein M.; Korin N. Shear Targeted Drug Delivery to Stenotic Blood Vessels. J. Biomech. 2017, 50, 217–221. 10.1016/j.jbiomech.2016.11.015. [DOI] [PubMed] [Google Scholar]
- Korin N.; Kanapathipillai M.; Matthews B. D.; Crescente M.; Brill A.; Mammoto T.; Ghosh K.; Jurek S.; Bencherif S. A.; Bhatta D.; Coskun A. U.; Feldman C. L.; Wagner D. D.; Ingber D. E. Shear-Activated Nanotherapeutics for Drug Targeting to Obstructed Blood Vessels. Science 2012, 337, 738–742. 10.1126/science.1217815. [DOI] [PubMed] [Google Scholar]
- Nazarenus M.; Zhang Q.; Soliman M. G.; del Pino P.; Pelaz B.; Carregal-Romero S.; Rejman J.; Rothen-Ruthishauser B.; Clift M. J. D.; Zellner R.; Nienhaus G. U.; Delehanty J. B.; Medintz I. L.; Parak W. J. In Vitro Interaction of Colloidal Nanoparticles with Mammalian Cells: What Have We Learned Thus Far?. Beilstein J. Nanotechnol. 2014, 5, 1477–1490. 10.3762/bjnano.5.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bera P. K.; Kandar A. K.; Krishnaswamy R.; Fontaine P.; Impéror-Clerc M.; Pansu B.; Constantin D.; Maiti S.; Sanyal M. K.; Sood A. K. Grazing Incidence X-Ray Diffraction Studies of Lipid–Peptide Mixed Monolayers during Shear Flow. ACS Omega 2020, 5, 14555–14563. 10.1021/acsomega.0c01261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura-Ruiz J.; Martínez-Criado G.; Chu M. H.; Geburt S.; Ronning C. Nano-X-Ray Absorption Spectroscopy of Single Co-Implanted ZnO Nanowires. Nano Lett. 2011, 11, 5322–5326. 10.1021/nl202799e. [DOI] [PubMed] [Google Scholar]
- Segura-Ruiz J.; Martinez-Criado G.; Chu M. H.; Denker C.; Malindretos J.; Rizzi A. Synchrotron Nanoimaging of Single In-Rich InGaN nanowires. J. Appl. Phys. 2013, 113, 136511. 10.1063/1.4795544. [DOI] [Google Scholar]
- Kuzmin A.; Chaboy J. EXAFS and XANES Analysis of Oxides at the Nanoscale. IUCrJ 2014, 1, 571–589. 10.1107/S2052252514021101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Criado G.; Homs A.; Alén B.; Sans J. A.; Segura-Ruiz J.; Molina-Sánchez A.; Susini J.; Yoo J.; Yi G.-C. Probing Quantum Confinement within Single Core–Multishell Nanowires. Nano Lett. 2012, 12, 5829–5834. 10.1021/nl303178u. [DOI] [PubMed] [Google Scholar]
- Segura-Ruiz J.; Martínez-Criado G.; Denker C.; Malindretos J.; Rizzi A. Phase Separation in Single InxGa1–xN Nanowires Revealed through a Hard X-Ray Synchrotron Nanoprobe. Nano Lett. 2014, 14, 1300–1305. 10.1021/nl4042752. [DOI] [PubMed] [Google Scholar]
- James S. A.; Feltis B. N.; de Jonge M. D.; Sridhar M.; Kimpton J. A.; Altissimo M.; Mayo S.; Zheng C.; Hastings A.; Howard D. L.; Paterson D. J.; Wright P. F. A.; Moorhead G. F.; Turney T. W.; Fu J. Quantification of ZnO Nanoparticle Uptake, Distribution, and Dissolution within Individual Human Macrophages. ACS Nano 2013, 7, 10621–10635. 10.1021/nn403118u. [DOI] [PubMed] [Google Scholar]
- Boesenberg U.; Ryan C. G.; Kirkham R.; Siddons D. P.; Alfeld M.; Garrevoet J.; Nunez T.; Claussen T.; Kracht T.; Falkenberg G. Fast X-Ray Microfluorescence Imaging with Submicrometer-Resolution Integrating a Maia Detector at Beamline P06 at PETRA III. J. Synchrotron Radiat. 2016, 23, 1550–1560. 10.1107/S1600577516015289. [DOI] [PubMed] [Google Scholar]
- Etschmann B. E.; Ryan C. G.; Brugger J.; Kirkham R.; Hough R. M.; Moorhead G.; Siddons D. P.; De Geronimo G.; Kuczewski A.; Dunn P.; Paterson D.; de Jonge M. D.; Howard D. L.; Davey P.; Jensen M. Reduced As Components in Highly Oxidized Environments: Evidence from Full Spectral XANES Imaging Using the Maia Massively Parallel Detector. Am. Mineral. 2010, 95, 884–887. 10.2138/am.2010.3469. [DOI] [Google Scholar]
- Ryan C. G.; Siddons D. P.; Kirkham R.; Dunn P. A.; Kuczewski A.; Moorhead G.; De Geronimo G.; Paterson D. J.; de Jonge M. D.; Hough R. M.; Lintern M. J.; Howard D. L.; Kappen P.; Cleverley J.; et al. The New Maia Detector System: Methods for High Definition Trace Element Imaging of Natural Material. AIP Conf. Proc. 2009, 1221, 9–17. 10.1063/1.3399266. [DOI] [Google Scholar]
- De Samber B.; Scharf O.; Buzanich G.; Garrevoet J.; Tack P.; Radtke M.; Riesemeier H.; Reinholz U.; Evens R.; De Schamphelaere K.; Falkenberg G.; Janssen C.; Vincze L. Three-Dimensional X-Ray Fluorescence Imaging Modes for Biological Specimens Using a Full-Field Energy Dispersive CCD Camera. J. Anal. At. Spectrom. 2019, 34, 2083–2093. 10.1039/C9JA00198K. [DOI] [Google Scholar]
- Chen W.; DeGeronimo G.; Elliott D.; Giacomini G.; Kuczewski A. J.; Mead J.; Pinelli D.; Rumaiz A. K.; Siddons D. P.; Smith G.; Vernon E. O.. A New Prototype X-Ray Fluorescence Detector System with Silicon Drift Detector Array. Proceedings from the 2016 IEEE Nuclear Science Symposium, Medical Imaging Conference and Room-Temperature Semiconductor Detector Workshop (NSS/MIC/RTSD), October 29–November 6, 2016, Strasbourg, France; IEEE: New York, 2016; pp 1–3.
- IARC Classifies Radiofrequency Electromagnetic Fields as Possibly Carcinogenic to Humans. Press Release No. 208; International Agency for Research on Cancer: Lyon, France, 2011. https://www.iarc.fr/wp-content/uploads/2018/07/pr208_E.pdf (accessed 2020-06-16).
- Cuttler J. M. Evidence of a Dose Threshold for Radiation-Induced Leukemia. Dose-Response 2018, 16, 1559325818811537. 10.1177/1559325818811537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little M. P.; Wakeford R.; Borrego D.; French B.; Zablotska L. B.; Adams M. J.; Allodji R.; de Vathaire F.; Lee C.; Brenner A. V.; Miller J. S.; Campbell D.; Pearce M. S.; Doody M. M.; Holmberg E.; Lundell M.; Sadetzki S.; Linet M. S.; Berrington de González A. Leukaemia and Myeloid Malignancy among People Exposed to Low Doses (<100 mSv) of Ionising Radiation during Childhood: A Pooled Analysis of Nine Historical Cohort Studies. Lancet Haematol. 2018, 5, e346–e358. 10.1016/S2352-3026(18)30092-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks A. L.; Hoel D. G.; Preston R. J. The Role of Dose Rate in Radiation Cancer Risk: Evaluating the Effect of Dose Rate at the Molecular, Cellular and Tissue Levels Using Key Events in Critical Pathways Following Exposure to Low LET Radiation. Int. J. Radiat. Biol. 2016, 92, 405–426. 10.1080/09553002.2016.1186301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de González A. B.; Darby S. Risk of Cancer from Diagnostic X-Rays: Estimates for the UK and 14 Other Countries. Lancet 2004, 363, 345–351. 10.1016/S0140-6736(04)15433-0. [DOI] [PubMed] [Google Scholar]
- Pearce M. S.; Salotti J. A.; Little M. P.; McHugh K.; Lee C.; Kim K. P.; Howe N. L.; Ronckers C. M.; Rajaraman P.; Craft A. W.; Parker L.; Berrington de González A. Radiation Exposure from CT Scans in Childhood and Subsequent Risk of Leukaemia and Brain Tumours: A Retrospective Cohort Study. Lancet 2012, 380, 499–505. 10.1016/S0140-6736(12)60815-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boffetta P.; Mannetje A. t.; Zaridze D.; Szeszenia-Dabrowska N.; Rudnai P.; Lissowska J.; Fabiánová E.; Mates D.; Bencko V.; Navratilova M.; Janout V.; Cardis E.; Fevotte J.; Fletcher T.; Brennan P. Occupational X-Ray Examinations and Lung Cancer Risk. Int. J. Cancer 2005, 115, 263–267. 10.1002/ijc.20854. [DOI] [PubMed] [Google Scholar]
- McBride W. H.; Schaue D. Radiation-Induced Tissue Damage and Response. J. Pathol. 2020, 250, 647–655. 10.1002/path.5389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geleijns J.; Wondergem J. X-Ray Imaging and the Skin: Radiation Biology, Patient Dosimetry and Observed Effects. Radiat. Prot. Dosim. 2005, 114, 121–125. 10.1093/rpd/nch544. [DOI] [PubMed] [Google Scholar]
- Koike M.; Sugasawa J.; Koike A.; Kohno Y. p53 Phosphorylation in Mouse Skin and in Vitro Human Skin Model by High-Dose-Radiation Exposure. J. Radiat. Res. 2005, 46, 461–468. 10.1269/jrr.46.461. [DOI] [PubMed] [Google Scholar]
- Kumar S.; Kolozsvary A.; Kohl R.; Lu M.; Brown S.; Kim J. H. Radiation-Induced Skin Injury in the Animal Model of Scleroderma: Implications for Post-Radiotherapy Fibrosis. Radiat. Oncol. 2008, 3, 40. 10.1186/1748-717X-3-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai J.; He F.; Wang J.; Chen J.; Tong L.; Zhu G. Influence of Radiation Exposure Pattern on the Bone Injury and Osteoclastogenesis in a Rat Model. Int. J. Mol. Med. 2019, 44, 2265–2275. 10.3892/ijmm.2019.4369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.; Jiang J.; Huang R.; Wang Y.; Nie X.; Gui R. Circular RNA Expression Profiles Are Significantly Altered in Mice Bone Marrow Stromal Cells After Total Body Irradiation. Leuk. Res. 2018, 70, 67–73. 10.1016/j.leukres.2018.05.010. [DOI] [PubMed] [Google Scholar]
- Künzle M.; Eckert T.; Beck T. Binary Protein Crystals for the Assembly of Inorganic Nanoparticle Superlattices. J. Am. Chem. Soc. 2016, 138, 12731–12734. 10.1021/jacs.6b07260. [DOI] [PubMed] [Google Scholar]
- Garman E. F.; Weik M. X-Ray Radiation Damage to Biological Samples: Recent Progress. J. Synchrotron Radiat. 2019, 26, 907–911. 10.1107/S1600577519009408. [DOI] [PubMed] [Google Scholar]
- Garman E. Radiation Damage in Macromolecular Crystallography: What Is It and Why Should We Care?. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2010, 66, 339–351. 10.1107/S0907444910008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton J. A Beginner’s Guide to Radiation Damage. J. Synchrotron Radiat. 2009, 16, 133–142. 10.1107/S0909049509004361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen R. L.; Rudiño-Piñera E.; Garman E. F. Experimental Determination of the Radiation Dose Limit for Cryocooled Protein Crystals. Proc. Natl. Acad. Sci. U. S. A. 2006, 103, 4912–4917. 10.1073/pnas.0600973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axnanda S.; Crumlin E. J.; Mao B.; Rani S.; Chang R.; Karlsson P. G.; Edwards M. O. M.; Lundqvist M.; Moberg R.; Ross P.; Hussain Z.; Liu Z. Using “Tender” X-Ray Ambient Pressure X-Ray Photoelectron Spectroscopy as a Direct Probe of Solid-Liquid Interface. Sci. Rep. 2015, 5, 9788. 10.1038/srep09788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahim A.; Moreno-Chicano T.; Appleby M. V.; Chaplin A. K.; Beale J. H.; Sherrell D. A.; Duyvesteyn H. M. E.; Owada S.; Tono K.; Sugimoto H.; Strange R. W.; Worrall J. A. R.; Axford D.; Owen R. L.; Hough M. A. Dose-Resolved Serial Synchrotron and XFEL Structures of Radiation-Sensitive Metalloproteins. IUCrJ 2019, 6, 543–551. 10.1107/S2052252519003956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirz J.; Jacobsen C.; Howells M. Soft X-Ray Microscopes and Their Biological Applications. Q. Rev. Biophys. 1995, 28, 33–130. 10.1017/S0033583500003139. [DOI] [PubMed] [Google Scholar]
- Cazaux J. A Physical Approach to the Radiation Damage Mechanisms Induced by X-Rays in X-Ray Microscopy and Related Techniques. J. Microsc. 1997, 188, 106–124. 10.1046/j.1365-2818.1997.2550812.x. [DOI] [Google Scholar]
- Teng T.-Y.; Moffat K. Radiation Damage of Protein Crystals at Cryogenic Temperatures between 40 and 150 K. J. Synchrotron Radiat. 2002, 9, 198–201. 10.1107/S0909049502008579. [DOI] [PubMed] [Google Scholar]
- Rose A. A Unified Approach to the Performance of Photographic Film, Television Pickup Tubes, and the Human Eye. J. Soc. Motion Pict. Eng. 1946, 47, 273–294. 10.5594/J12772. [DOI] [Google Scholar]
- Howells M. R.; Beetz T.; Chapman H. N.; Cui C.; Holton J. M.; Jacobsen C. J.; Kirz J.; Lima E.; Marchesini S.; Miao H.; Sayre D.; Shapiro D. A.; Spence J. C. H.; Starodub D. M. An Assessment of the Resolution Limitation Due to Radiation-Damage in X-Ray Diffraction Microscopy. J. Electron Spectrosc. Relat. Phenom. 2009, 170, 4–12. 10.1016/j.elspec.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson R. The Potential and Limitations of Neutrons, Electrons and X-Rays for Atomic Resolution Microscopy of Unstained Biological Molecules. Q. Rev. Biophys. 1995, 28, 171–193. 10.1017/S003358350000305X. [DOI] [PubMed] [Google Scholar]
- Skarsgard L. D.; Harrison I.; Durand R. E. The Radiation Response of Asynchronous Cells at Low Dose: Evidence of Substructure. Radiat. Res. 1991, 127, 248–56. 10.2307/3577938. [DOI] [PubMed] [Google Scholar]
- Fayard B.; Salomé M.; Takemoto K.; Kihara H.; Susini J. Some Practical Considerations About the Effects of Radiation Damage on Hydrated Cells Imaged by X-Ray Fluorescence Microscopy. J. Electron Spectrosc. Relat. Phenom. 2009, 170, 19–24. 10.1016/j.elspec.2008.11.003. [DOI] [Google Scholar]
- Williams S.; Zhang X.; Jacobsen C.; Kirz J.; Lindaas S.; Van’T Hof J.; Lamm S. S. Measurements of Wet Metaphase Chromosomes in the Scanning Rransmission X-Ray Microscope. J. Microsc. 1993, 170, 155–165. 10.1111/j.1365-2818.1993.tb03335.x. [DOI] [Google Scholar]
- Kempner E. S.; Wood R.; Salovey R. The Temperature Dependence of Radiation Sensitivity of Large Molecules. J. Polym. Sci., Part B: Polym. Phys. 1986, 24, 2337–2343. 10.1002/polb.1986.090241015. [DOI] [Google Scholar]
- Hope H. Crystallography of Biological Macromolecules at Ultra-Low Temperature. Annu. Rev. Biophys. Biophys. Chem. 1990, 19, 107–126. 10.1146/annurev.bb.19.060190.000543. [DOI] [PubMed] [Google Scholar]
- Teng T.-y.; Moffat K. Primary Radiation Damage of Protein Crystals by an Intense Synchrotron X-Ray Beam. J. Synchrotron Radiat. 2000, 7, 313–317. 10.1107/S0909049500008694. [DOI] [PubMed] [Google Scholar]
- Schneider G. Cryo X-Ray Microscopy with High Spatial Resolution in Amplitude and Phase Contrast. Ultramicroscopy 1998, 75, 85–104. 10.1016/S0304-3991(98)00054-0. [DOI] [PubMed] [Google Scholar]
- Kanngießer B.; Malzer W.; Pagels M.; Lühl L.; Weseloh G. Three-Dimensional Micro-XRF under Cryogenic Conditions: A Pilot Experiment for Spatially Resolved Trace Analysis in Biological Specimens. Anal. Bioanal. Chem. 2007, 389, 1171–1176. 10.1007/s00216-007-1494-6. [DOI] [PubMed] [Google Scholar]
- Chen S.; Deng J.; Yuan Y.; Flachenecker C.; Mak R.; Hornberger B.; Jin Q.; Shu D.; Lai B.; Maser J.; Roehrig C.; Paunesku T.; Gleber S. C.; Vine D. J.; Finney L.; VonOsinski J.; Bolbat M.; Spink I.; Chen Z.; Steele J.; et al. The Bionanoprobe: Hard X-Ray Fluorescence Nanoprobe with Cryogenic Capabilities. J. Synchrotron Radiat. 2014, 21, 66–75. 10.1107/S1600577513029676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Samber B.; Meul E.; Laforce B.; De Paepe B.; Smet J.; De Bruyne M.; De Rycke R.; Bohic S.; Cloetens P.; Van Coster R.; Vandenabeele P.; Vanden Berghe T. Nanoscopic X-Ray Fluorescence Imaging and Quantification of Intracellular Key-Elements in Cryofrozen Friedreich’s Ataxia Fibroblasts. PLoS One 2018, 13, e0190495. 10.1371/journal.pone.0190495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothkamm K.; Löbrich M. Evidence for a Lack of DNA Double-Strand Break Repair in Human Cells Exposed to Very Kow X-Ray Doses. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 5057–5062. 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Zhong R.; Sun L.; Jia J.; Ma S.; Liu X. Ionizing Radiation-Induced Adaptive Response in Fibroblasts under Both Monolayer and 3-Dimensional Conditions. PLoS One 2015, 10, e0121289. 10.1371/journal.pone.0121289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray D.; Mirzayans R.; McBride W. H. Defenses Against Pro-Oxidant Forces - Maintenance of Cellular and Genomic Integrity and Longevity. Radiat. Res. 2018, 190, 331–349. 10.1667/RR15101.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaue D.; Micewicz E. D.; Ratikan J. A.; Xie M. W.; Cheng G.; McBride W. H. Radiation and Inflammation. Semin. Radiat. Oncol. 2015, 25, 4–10. 10.1016/j.semradonc.2014.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Ruiz M. E.; Vitale I.; Harrington K. J.; Melero I.; Galluzzi L. Immunological Impact of Cell Death Signaling Driven by Radiation on the Tumor Microenvironment. Nat. Immunol. 2020, 21, 120–134. 10.1038/s41590-019-0561-4. [DOI] [PubMed] [Google Scholar]
- McKelvey K. J.; Hudson A. L.; Back M.; Eade T.; Diakos C. I. Radiation, Inflammation and the Immune Response in Cancer. Mamm. Genome 2018, 29, 843–865. 10.1007/s00335-018-9777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azzam E. I.; Jay-Gerin J.-P.; Pain D. Ionizing Radiation-Induced Metabolic Oxidative Stress and Prolonged Cell Injury. Cancer Lett. 2012, 327, 48–60. 10.1016/j.canlet.2011.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesulu B. P.; Mahadevan L. S.; Aliru M. L.; Yang X.; Bodd M. H.; Singh P. K.; Yusuf S. W.; Abe J.-i.; Krishnan S. Radiation-Induced Endothelial Vascular Injury: A Review of Possible Mechanisms. J. Am. Coll. Cardiol. Basic Trans. Science 2018, 3, 563–572. 10.1016/j.jacbts.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith T. A.; Kirkpatrick D. R.; Smith S.; Smith T. K.; Pearson T.; Kailasam A.; Herrmann K. Z.; Schubert J.; Agrawal D. K. Radioprotective Agents to Prevent Cellular Damage Due to Ionizing Radiation. J. Transl. Med. 2017, 15, 232. 10.1186/s12967-017-1338-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Zhang P.; Li F.; Jin X.; Li J.; Chen W.; Li Q. Metal-Based NanoEnhancers for Future Radiotherapy: Radiosensitizing and Synergistic Effects on Tumor Cells. Theranostics 2018, 8, 1824–1849. 10.7150/thno.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonvalot S.; Rutkowski P. L.; Thariat J.; Carrère S.; Ducassou A.; Sunyach M.-P.; Agoston P.; Hong A.; Mervoyer A.; Rastrelli M.; Moreno V.; Li R. K.; Tiangco B.; Herraez A. C.; Gronchi A.; Mangel L.; Sy-Ortin T.; Hohenberger P.; de Baère T.; Le Cesne A.; et al. NBTXR3, a First-in-Class Radioenhancer Hafnium Oxide Nanoparticle, Plus Radiotherapy versus Radiotherapy Alone in Patients with Locally Advanced Soft-Tissue Sarcoma (Act. In. Sarc): A Multicentre, Phase 2–3, Randomised, Controlled Trial. Lancet Oncol. 2019, 20, 1148–1159. 10.1016/S1470-2045(19)30326-2. [DOI] [PubMed] [Google Scholar]
- Manohar N.; Reynoso F. J.; Diagaradjane P.; Krishnan S.; Cho S. H. Quantitative Imaging of Gold Nanoparticle Distribution in a Tumor-Bearing Mouse Using Benchtop X-Ray Fluorescence Computed Tomography. Sci. Rep. 2016, 6, 22079. 10.1038/srep22079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert B.; Fakra S. C.; Xia T.; Pokhrel S.; Madler L.; Nel A. E. The Fate of ZnO Nanoparticles Administered to Human Bronchial Epithelial Cells. ACS Nano 2012, 6, 4921–4930. 10.1021/nn300425a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unrine J. M.; Tsyusko O. V.; Hunyadi S. E.; Judy J. D.; Bertsch P. M. Effects of Particle Size on Chemical Speciation and Bioavailability of Copper to Earthworms (Eisenia fetida) Exposed to Copper Nanoparticles. J. Environ. Qual. 2010, 39, 1942–1953. 10.2134/jeq2009.0387. [DOI] [PubMed] [Google Scholar]
- Richardson J. J.; Liang K. Nano-Biohybrids: In Vivo Synthesis of Metal–Organic Frameworks inside Living Plants. Small 2018, 14, 1702958. 10.1002/smll.201702958. [DOI] [PubMed] [Google Scholar]
- Wang S.; Lv J.; Ma J.; Zhang S. Cellular Internalization and Intracellular Biotransformation of Silver Nanoparticles in Chlamydomonas reinhardtii. Nanotoxicology 2016, 10, 1129–1135. 10.1080/17435390.2016.1179809. [DOI] [PubMed] [Google Scholar]
- Sabo-Attwood T.; Unrine J. M.; Stone J. W.; Murphy C. J.; Ghoshroy S.; Blom D.; Bertsch P. M.; Newman L. A. Uptake, Distribution and Toxicity of Gold Nanoparticles in Tobacco (Nicotiana xanthi) Seedlings. Nanotoxicology 2012, 6, 353–360. 10.3109/17435390.2011.579631. [DOI] [PubMed] [Google Scholar]
- Eggl E.; Dierolf M.; Achterhold K.; Jud C.; Gunther B.; Braig E.; Gleich B.; Pfeiffer F. The Munich Compact Light Source: Initial Performance Measures. J. Synchrotron Radiat. 2016, 23, 1137–1142. 10.1107/S160057751600967X. [DOI] [PubMed] [Google Scholar]
- Hornberger B.; Kasahara J.; Gifford M.; Ruth R.; Loewen R. A Compact Light Source Providing High-Flux, Quasi-Monochromatic, Tunable X-Rays in the Laboratory. Proc. SPIE 2019, 11110, 1111003. 10.1117/12.2527356. [DOI] [Google Scholar]
- Eggl E.; Schleede S.; Bech M.; Achterhold K.; Loewen R.; Ruth R. D.; Pfeiffer F. X-Ray Phase-Contrast Tomography with a Compact Laser-Driven Synchrotron Source. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 5567–5572. 10.1073/pnas.1500938112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.; Günther B.; Achterhold K.; Cui Y.-t.; Gleich B.; Dierolf M.; Pfeiffer F. Energy-Dispersive X-Ray Absorption Spectroscopy with an Inverse Compton Source. Sci. Rep. 2020, 10, 8772. 10.1038/s41598-020-65225-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaroshenko A.; Pritzke T.; Koschlig M.; Kamgari N.; Willer K.; Gromann L.; Auweter S.; Hellbach K.; Reiser M.; Eickelberg O.; Pfeiffer F.; Hilgendorff A. Visualization of Neonatal Lung Injury Associated with Mechanical Ventilation Using X-Ray Dark-Field Radiography. Sci. Rep. 2016, 6, 24269. 10.1038/srep24269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaroshenko A.; Hellbach K.; Yildirim A. Ö.; Conlon T. M.; Fernandez I. E.; Bech M.; Velroyen A.; Meinel F. G.; Auweter S.; Reiser M.; Eickelberg O.; Pfeiffer F. Improved in Vivo Assessment of Pulmonary Fibrosis in Mice Using X-Ray Dark-Field Radiography. Sci. Rep. 2015, 5, 17492. 10.1038/srep17492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bech M.; Tapfer A.; Velroyen A.; Yaroshenko A.; Pauwels B.; Hostens J.; Bruyndonckx P.; Sasov A.; Pfeiffer F. In-Vivo Dark-Field and Phase-Contrast X-Ray Imaging. Sci. Rep. 2013, 3, 3209. 10.1038/srep03209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gromann L. B.; De Marco F.; Willer K.; Noël P. B.; Scherer K.; Renger B.; Gleich B.; Achterhold K.; Fingerle A. A.; Muenzel D.; Auweter S.; Hellbach K.; Reiser M.; Baehr A.; Dmochewitz M.; Schroeter T. J.; Koch F. J.; Meyer P.; Kunka D.; Mohr J.; et al. In Vivo X-Ray Dark-Field Chest Radiography of a Pig. Sci. Rep. 2017, 7, 4807. 10.1038/s41598-017-05101-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C.; Nöhammer B.; Solak H. H.; Ziegler E. Differential X-Ray Phase Contrast Imaging Using a Shearing Interferometer. Appl. Phys. Lett. 2002, 81, 3287–3289. 10.1063/1.1516611. [DOI] [Google Scholar]
- Momose A.; Kawamoto S.; Koyama I.; Hamaishi Y.; Takai K.; Suzuki Y. Demonstration of X-Ray Talbot Interferometry. Jpn. J. Appl. Phys. 2003, 42, L866–L868. 10.1143/JJAP.42.L866. [DOI] [Google Scholar]
- Pfeiffer F.; Weitkamp T.; Bunk O.; David C. Phase Retrieval and Differential Phase-Contrast Imaging with Low-Brilliance X-Ray Sources. Nat. Phys. 2006, 2, 258–261. 10.1038/nphys265. [DOI] [Google Scholar]
- Pfeiffer F.; Bech M.; Bunk O.; Kraft P.; Eikenberry E. F.; Brönnimann C.; Grünzweig C.; David C. Hard-X-Ray Dark-Field Imaging Using a Grating Interferometer. Nat. Mater. 2008, 7, 134–137. 10.1038/nmat2096. [DOI] [PubMed] [Google Scholar]
- Willer K.; Fingerle A. A.; Gromann L. B.; De Marco F.; Herzen J.; Achterhold K.; Gleich B.; Muenzel D.; Scherer K.; Renz M.; Renger B.; Kopp F.; Kriner F.; Fischer F.; Braun C.; Auweter S.; Hellbach K.; Reiser M. F.; Schroeter T.; Mohr J. X-Ray Dark-Field Imaging of the Human Lung—A Feasibility Study on a Deceased Body. PLoS One 2018, 13, e0204565. 10.1371/journal.pone.0204565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauter A. P.; Andrejewski J.; De Marco F.; Willer K.; Gromann L. B.; Noichl W.; Kriner F.; Fischer F.; Braun C.; Koehler T.; Meurer F.; Fingerle A. A.; Pfeiffer D.; Rummeny E.; Herzen J.; Pfeiffer F. Optimization of Tube Voltage in X-Ray Dark-Field Radiography. Sci. Rep. 2019, 9, 8699. 10.1038/s41598-019-45256-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köcher S.; Beyer B.; Lange T.; Nordquist L.; Volquardsen J.; Burdak-Rothkamm S.; Schlomm T.; Petersen C.; Rothkamm K.; Mansour W. Y. A Functional ex Vivo Assay to Detect PARP1-EJ Repair and Radiosensitization by PARP-Inhibitor in Prostate Cancer. Int. J. Cancer 2019, 144, 1685–1696. 10.1002/ijc.32018. [DOI] [PubMed] [Google Scholar]
- Xu X.; Farach-Carson M. C.; Jia X. Three-Dimensional in Vitro Tumor Models for Cancer Research and Drug Evaluation. Biotechnol. Adv. 2014, 32, 1256–1268. 10.1016/j.biotechadv.2014.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peroglio M.; Gaspar D.; Zeugolis D. I.; Alini M. Relevance of Bioreactors and Whole Tissue Cultures for the Translation of New Therapies to Humans. J. Orthop. Res. 2018, 36, 10–21. 10.1002/jor.23655. [DOI] [PubMed] [Google Scholar]
- Stewart B.; Wild C.. World Cancer Report 2014; International Agency for Research on Cancer: Lyon, France, 2014.
- El-Gabalawy H.; Guenther L. C.; Bernstein C. N. Epidemiology of Immune-Mediated Inflammatory Diseases: Incidence, Prevalence, Natural History, and Comorbidities. J. Rheumatol., Suppl. 2010, 85, 2–10. 10.3899/jrheum.091461. [DOI] [PubMed] [Google Scholar]
- Molodecky N. A.; Soon I. S.; Rabi D. M.; Ghali W. A.; Ferris M.; Chernoff G.; Benchimol E. I.; Panaccione R.; Ghosh S.; Barkema H. W.; Kaplan G. G. Increasing Incidence and Prevalence of the Inflammatory Bowel Diseases with Time, Based on Systematic Review. Gastroenterology 2012, 142, 46–54. 10.1053/j.gastro.2011.10.001. [DOI] [PubMed] [Google Scholar]
- Teng M. W. L.; Bowman E. P.; McElwee J. J.; Smyth M. J.; Casanova J.-L.; Cooper A. M.; Cua D. J. IL-12 and IL-23 Cytokines: From Discovery to Targeted Therapies for Immune-Mediated Inflammatory Diseases. Nat. Med. 2015, 21, 719–729. 10.1038/nm.3895. [DOI] [PubMed] [Google Scholar]
- Farh K. K.-H.; Marson A.; Zhu J.; Kleinewietfeld M.; Housley W. J.; Beik S.; Shoresh N.; Whitton H.; Ryan R. J. H.; Shishkin A. A.; Hatan M.; Carrasco-Alfonso M. J.; Mayer D.; Luckey C. J.; Patsopoulos N. A.; De Jager P. L.; Kuchroo V. K.; Epstein C. B.; Daly M. J.; Hafler D. A.; Bernstein B. E. Genetic and Epigenetic Fine Mapping of Causal Autoimmune Disease Variants. Nature 2015, 518, 337–343. 10.1038/nature13835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber S.; Gagliani N.; Esplugues E.; O’Connor W. Jr.; Huber F. J.; Chaudhry A.; Kamanaka M.; Kobayashi Y.; Booth C. J.; Rudensky A. Y.; Roncarolo M. G.; Battaglia M.; Flavell R. A. Th17 Cells Express Interleukin-10 Receptor and Are Controlled by Foxp3(−) and Foxp3+ Regulatory CD4+ T Cells in an Interleukin-10-Dependent Manner. Immunity 2011, 34, 554–65. 10.1016/j.immuni.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhry A.; Samstein R. M.; Treuting P.; Liang Y.; Pils M. C.; Heinrich J. M.; Jack R. S.; Wunderlich F. T.; Bruning J. C.; Muller W.; Rudensky A. Y. Interleukin-10 Signaling in Regulatory T C Is Required for Suppression of Th17 Cell-Mediated Inflammation. Immunity 2011, 34, 566–578. 10.1016/j.immuni.2011.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glocker E. O.; Kotlarz D.; Boztug K.; Gertz E. M.; Schaffer A. A.; Noyan F.; Perro M.; Diestelhorst J.; Allroth A.; Murugan D.; Hatscher N.; Pfeifer D.; Sykora K. W.; Sauer M.; Kreipe H.; Lacher M.; Nustede R.; Woellner C.; Baumann U.; Salzer U.; et al. Inflammatory Bowel Disease and Mutations Affecting the Interleukin-10 Receptor. N. Engl. J. Med. 2009, 361, 2033–2045. 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelhardt K. R.; Grimbacher B. IL-10 in Humans: Lessons from the Gut, IL-10/IL-10 Receptor Deficiencies, and IL-10 Polymorphisms. Curr. Top. Microbiol. Immunol. 2014, 380, 1–18. 10.1007/978-3-662-43492-5_1. [DOI] [PubMed] [Google Scholar]
- Bennett C. L.; Christie J.; Ramsdell F.; Brunkow M. E.; Ferguson P. J.; Whitesell L.; Kelly T. E.; Saulsbury F. T.; Chance P. F.; Ochs H. D. The Immune Dysregulation, Polyendocrinopathy, Enteropathy, X-Linked Syndrome (IPEX) Is Caused by Mutations of FOXP3. Nat. Genet. 2001, 27, 20–21. 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- Wildin R. S.; Ramsdell F.; Peake J.; Faravelli F.; Casanova J.-L.; Buist N.; Levy-Lahad E.; Mazzella M.; Goulet O.; Perroni L.; Dagna Bricarelli F.; Byrne G.; McEuen M.; Proll S.; Appleby M.; Brunkow M. E. X-Linked Neonatal Diabetes Mellitus, Enteropathy and Endocrinopathy Syndrome Is the Human Equivalent of Mouse Scurfy. Nat. Genet. 2001, 27, 18–20. 10.1038/83707. [DOI] [PubMed] [Google Scholar]
- Sun X.; Gamal M.; Nold P.; Said A.; Chakraborty I.; Pelaz B.; Schmied F.; Pückler K. v.; Figiel J.; Zhao Y.; Brendel C.; Hassan M.; Parak W. J.; Feliu N. Tracking Stem Cells and Macrophages with Gold and Iron Oxide Nanoparticles – The Choice of the Best Suited Particles. Appl. Mater. Today 2019, 15, 267–279. 10.1016/j.apmt.2018.12.006. [DOI] [Google Scholar]
- Chen H.; Wang G. D.; Chuang Y.-J.; Zhen Z.; Chen X.; Biddinger P.; Hao Z.; Liu F.; Shen B.; Pan Z.; Xie J. Nanoscintillator-Mediated X-Ray Inducible Photodynamic Therapy for in Vivo Cancer Treatment. Nano Lett. 2015, 15, 2249–2256. 10.1021/nl504044p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W.; Shi T.; Luo L.; Chen X.; Lv P.; Lv Y.; Zhuang Y.; Zhu J.; Liu G.; Chen X.; Chen H. Monodisperse and Uniform Mesoporous Silicate Nanosensitizers Achieve Low-Dose X-Ray-Induced Deep-Penetrating Photodynamic Therapy. Adv. Mater. 2019, 31, 1808024. 10.1002/adma.201808024. [DOI] [PubMed] [Google Scholar]
- Kamkaew A.; Chen F.; Zhan Y.; Majewski R. L.; Cai W. Scintillating Nanoparticles as Energy Mediators for Enhanced Photodynamic Therapy. ACS Nano 2016, 10, 3918–3935. 10.1021/acsnano.6b01401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuncic Z.; Lacombe S. Nanoparticle Radio-Enhancement: Principles, Progress and Application to Cancer Treatment. Phys. Med. Biol. 2018, 63, 02TR01. 10.1088/1361-6560/aa99ce. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Zhang P.; Li F.; Jin X.; Li J.; Chen W.; Li Q. Metal-Based NanoEnhancers for Future Radiotherapy: Radiosensitizing and Synergistic Effects on Tumor Cells. Theranostics 2018, 8, 1824–1849. 10.7150/thno.22172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus H.; Gargioni E.; Li W. B.; Nettelbeck H.; Villagrasa C. Determining Dose Enhancement Factors of High-Z Nanoparticles from Simulations where Lateral Secondary Particle Disequilibrium Exists. Phys. Med. Biol. 2019, 64, 155016. 10.1088/1361-6560/ab31d4. [DOI] [PubMed] [Google Scholar]
- Rudek B.; McNamara A.; Ramos-Mendez J.; Byrne H.; Kuncic Z.; Schuemann J. Radio-Enhancement by Gold Nanoparticles and Their Impact on Water Radiolysis for X-Ray, Proton and Carbon-Ion Beams. Phys. Med. Biol. 2019, 64, 175005. 10.1088/1361-6560/ab314c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misawa M.; Takahashi J. Generation of Reactive Oxygen Species Induced by Gold Nanoparticles Under X-Ray and UV Irradiations. Nanomedicine 2011, 7, 604–614. 10.1016/j.nano.2011.01.014. [DOI] [PubMed] [Google Scholar]
- Sicard-Roselli C.; Brun E.; Gilles M.; Baldacchino G.; Kelsey C.; McQuaid H.; Polin C.; Wardlow N.; Currell F. A New Mechanism for Hydroxyl Radical Production in Irradiated Nanoparticle Solutions. Small 2014, 10, 3338–3346. 10.1002/smll.201400110. [DOI] [PubMed] [Google Scholar]
- Seo S. J.; Jeon J. K.; Han S. M.; Kim J. K. Reactive Oxygen Species-Based Measurement of the Dependence of the Coulomb Nanoradiator Effect on Proton Energy and Atomic Z Value. Int. J. Radiat. Biol. 2017, 93, 1239–1247. 10.1080/09553002.2017.1361556. [DOI] [PubMed] [Google Scholar]
- Clark J. N.; Beitra L.; Xiong G.; Fritz D. M.; Lemke H. T.; Zhu D.; Chollet M.; Williams G. J.; Messerschmidt M. M.; Abbey B.; Harder R. J.; Korsunsky A. M.; Wark J. S.; Reis D. A.; Robinson I. K. Imaging Transient Melting of a Nanocrystal Using an X-Ray Laser. Proc. Natl. Acad. Sci. U. S. A. 2015, 112, 7444–7448. 10.1073/pnas.1417678112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neutze R.; Wouts R.; Van der Spoel D.; Weckert E.; Hajdu J. Potential for Biomolecular Imaging with Femtosecond X-Ray Pulses. Nature 2000, 406, 752–757. 10.1038/35021099. [DOI] [PubMed] [Google Scholar]
- Wernet P.; Kunnus K.; Josefsson I.; Rajkovic I.; Quevedo W.; Beye M.; Schreck S.; Grübel S.; Scholz M.; Nordlund D.; Zhang W.; Hartsock R. W.; Schlotter W. F.; Turner J. J.; Kennedy B.; Hennies F.; de Groot F. M. F.; Gaffney K. J.; Techert S.; Odelius M.; Föhlisch A. Orbital-Specific Mapping of the Ligand Exchange Dynamics of Fe(CO)5 in Solution. Nature 2015, 520, 78–81. 10.1038/nature14296. [DOI] [PubMed] [Google Scholar]
- Spence J. XFELs for Structure and Dynamics in Biology. IUCrJ 2017, 4, 322–339. 10.1107/S2052252517005760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso A. P.; Yefanov O. M.; Vartanyants I. A. Coherent Diffractive Imaging of Biological Samples at Synchrotron and Free Electron Laser Facilities. J. Biotechnol. 2010, 149, 229–237. 10.1016/j.jbiotec.2010.01.024. [DOI] [PubMed] [Google Scholar]
- van der Schot G.; Svenda M.; Maia F. R. N. C.; Hantke M.; DePonte D. P.; Seibert M. M.; Aquila A.; Schulz J.; Kirian R.; Liang M.; Stellato F.; Iwan B.; Andreasson J.; Timneanu N.; Westphal D.; Almeida F. N.; Odic D.; Hasse D.; Carlsson G. H.; Larsson D. S. D.; et al. Imaging Single Cells in a Beam of Live Cyanobacteria with an X-Ray Laser. Nat. Commun. 2015, 6, 5704. 10.1038/ncomms6704. [DOI] [PubMed] [Google Scholar]
- Hantke M. F.; Hasse D.; Maia F. R. N. C.; Ekeberg T.; John K.; Svenda M.; Loh N. D.; Martin A. V.; Timneanu N.; Larsson D. S. D.; van der Schot G.; Carlsson G. H.; Ingelman M.; Andreasson J.; Westphal D.; Liang M.; Stellato F.; DePonte D. P.; Hartmann R.; Kimmel N.; et al. High-Throughput Imaging of Heterogeneous Cell Organelles with an X-Ray Laser. Nat. Photonics 2014, 8, 943–949. 10.1038/nphoton.2014.270. [DOI] [Google Scholar]
- Seibert M. M.; Ekeberg T.; Maia F. R. N. C.; Svenda M.; Andreasson J.; Jönsson O.; Odić D.; Iwan B.; Rocker A.; Westphal D.; Hantke M.; DePonte D. P.; Barty A.; Schulz J.; Gumprecht L.; Coppola N.; Aquila A.; Liang M.; White T. A.; Martin A.; et al. Single Mimivirus Particles Intercepted and Imaged with an X-Ray Laser. Nature 2011, 470, 78–81. 10.1038/nature09748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseinizadeh A.; Mashayekhi G.; Copperman J.; Schwander P.; Dashti A.; Sepehr R.; Fung R.; Schmidt M.; Yoon C. H.; Hogue B. G.; Williams G. J.; Aquila A.; Ourmazd A. Conformational Landscape of a Virus by Single-Particle X-Ray Scattering. Nat. Methods 2017, 14, 877–881. 10.1038/nmeth.4395. [DOI] [PubMed] [Google Scholar]
- Poudyal I.; Schmidt M.; Schwander P. Single-Particle Imaging by X-Ray Free-Electron Lasers—How Many Snapshots Are Needed?. Struct. Dyn. 2020, 7, 024102. 10.1063/1.5144516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobolev E.; Zolotarev S.; Giewekemeyer K.; Bielecki J.; Okamoto K.; Reddy H. K. N.; Andreasson J.; Ayyer K.; Barak I.; Bari S.; Barty A.; Bean R.; Bobkov S.; Chapman H. N.; Chojnowski G.; Daurer B. J.; Dörner K.; Ekeberg T.; Flückiger L.; Galzitskaya O.; et al. Megahertz Single-Particle Imaging at the European XFEL. Commun. Phys. 2020, 3, 97. 10.1038/s42005-020-0362-y. [DOI] [Google Scholar]
- Malik V.; Petukhov A. V.; He L.; Yin Y.; Schmidt M. Colloidal Crystallization and Structural Changes in Suspensions of Silica/Magnetite Core–Shell Nanoparticles. Langmuir 2012, 28, 14777–14783. 10.1021/la301942t. [DOI] [PubMed] [Google Scholar]
- Martin A. V.; Loh N. D.; Hampton C. Y.; Sierra R. G.; Wang F.; Aquila A.; Bajt S.; Barthelmess M.; Bostedt C.; Bozek J. D.; Coppola N.; Epp S. W.; Erk B.; Fleckenstein H.; Foucar L.; Frank M.; Graafsma H.; Gumprecht L.; Hartmann A.; Hartmann R.; et al. Femtosecond Dark-Field Imaging with an X-Ray Free Electron Laser. Opt. Express 2012, 20, 13501–13512. 10.1364/OE.20.013501. [DOI] [PubMed] [Google Scholar]
- Li X.; Chiu C.-Y.; Wang H.-J.; Kassemeyer S.; Botha S.; Shoeman R. L.; Lawrence R. M.; Kupitz C.; Kirian R.; James D.; Wang D.; Nelson G.; Messerschmidt M.; Boutet S.; Williams G. J.; Hartmann E.; Jafarpour A.; Foucar L. M.; Barty A.; Chapman H.; et al. Diffraction Data of Core-Shell Nanoparticles from an X-Ray Free Electron Laser. Sci. Data 2017, 4, 170048. 10.1038/sdata.2017.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadzinsky P. D.; Calero G.; Ackerson C. J.; Bushnell D. A.; Kornberg R. D. Structure of a Thiol Monolayer-Protected Gold Nanoparticle at 1.1 A Resolution. Science 2007, 318, 430–433. 10.1126/science.1148624. [DOI] [PubMed] [Google Scholar]
- Ayyer K.; Xavier P. L.; Bielecki J.; Shen Z.; Daurer B. J.; Samanta A. K.; Awel S.; Bean R.; Barty A.; Bergemann M.; Ekeberg T.; Estillore A. D.; Fangohr H.; Giewekemeyer K.; Hunter M. S.; Kirian R.; Karnevskiy M.; Kirian R. A.; Kirkwood H.; Kim Y.; et al. 3D Diffractive Imaging of Nanoparticle Ensembles Using an X-Ray Laser. Optica 2021, 8, 15–23. 10.1364/OPTICA.410851. [DOI] [Google Scholar]
- Auldridge M. E.; Forest K. T. Bacterial Phytochromes: More than Meets the Light. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 67–88. 10.3109/10409238.2010.546389. [DOI] [PubMed] [Google Scholar]
- Burgie E. S.; Zhang J.; Vierstra R. D. Crystal Structure of Deinococcus Phytochrome in the Photoactivated State Reveals a Cascade of Structural Rearrangements during Photoconversion. Structure 2016, 24, 448–457. 10.1016/j.str.2016.01.001. [DOI] [PubMed] [Google Scholar]
- Sanchez J. C.; Carrillo M.; Pandey S.; Noda M.; Aldama L.; Feliz D.; Claesson E.; Wahlgren W. Y.; Tracy G.; Duong P.; Nugent A. C.; Field A.; Šrajer V.; Kupitz C.; Iwata S.; Nango E.; Tanaka R.; Tanaka T.; Fangjia L.; Tono K.; et al. High-Resolution Crystal Structures of a Myxobacterial Phytochrome at Cryo and Room Temperatures. Struct. Dyn. 2019, 6, 054701. 10.1063/1.5120527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takala H.; Björling A.; Berntsson O.; Lehtivuori H.; Niebling S.; Hoernke M.; Kosheleva I.; Henning R.; Menzel A.; Ihalainen J. A.; Westenhoff S. Signal Amplification and Transduction in Phytochrome Photosensors. Nature 2014, 509, 245–248. 10.1038/nature13310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giewekemeyer K.; Aquila A.; Loh N.-T. D.; Chushkin Y.; Shanks K. S.; Weiss J. T.; Tate M. W.; Philipp H. T.; Stern S.; Vagovic P.; Mehrjoo M.; Teo C.; Barthelmess M.; Zontone F.; Chang C.; Tiberio R. C.; Sakdinawat A.; Williams G. J.; Gruner S. M.; Mancuso A. P. Experimental 3D Coherent Diffractive Imaging from Photon-Sparse Random Projections. IUCrJ 2019, 6, 357–365. 10.1107/S2052252519002781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielecki J.; Maia F. R. N. C.; Mancuso A. P. Perspectives on Single Particle Imaging with X- Rays at the Advent of High Repetition Rate X-Ray Free Electron Laser Sources. Struct. Dyn. 2020, 7, 040901. 10.1063/4.0000024. [DOI] [PMC free article] [PubMed] [Google Scholar]