Abstract

Therapeutic proteins such as enzymes, hormones, and cytokines suffer from poor stability, inefficient cellular penetration, and rapid clearance from circulation. Conjugation with polymers (such as poly(ethylene glycol)) and fusion with long-acting proteins (such as albumin and Fc fragments) have been utilized to partially address the delivery issues, but these strategies require the introduction of new macromolecular substances, resulting in potential immunogenicity and toxicity. Herein, we report an easy strategy to increase the intracellular delivery efficiency and stability of proteins by combining of sortase-mediated protein cyclization and cell-penetrating peptide (CPP)-mediated intracellular delivery. We, for the first time, genetically constructed a green fluorescence protein (GFP) fused with a CPP, a transacting activator of transcription (TAT) peptide, at its C-terminus for intracellular internalization, and two sortase recognition sequences, pentaglycine and LPETG, at its N- and C-termini for cyclization. Notably, the cyclized GFP-TAT (cGFP-TAT) not only highly retained the photophysical properties of the protein but also significantly improved the in vitro stability compared with the native linear GFP (lGFP) and linear TAT peptide-fused GFP (lGFP-TAT).Moreover, cGFP-TAT showed better cellular internalization ability compared with lGFP. In C26 tumor-inoculated mice, cGFP-TAT exhibited enhanced in vivo tumor retention, with increases of 7.79- and 6.52-fold relative to lGFP and lGFP-TAT in tumor retention 3 h after intratumor administration. This proof-of-concept study has provided an easy strategy to increase the in vitro stability, intracellular delivery efficiency, and in vivo tumor retention of GFP, which would be applicable to numerous therapeutic proteins and peptides for clinical practice.
1. Introduction
Proteins have been marked as potential therapeutics in clinical practice due to their high biological activity. However, there are many therapeutic proteins, such as enzymes and especially cytokines, suffering from conspicuous limitations including rapid clearance from blood circulation, inefficient cellular penetration, quick enzymatic degradation, and poor stability.1−3 Numerous approaches have been implemented to address the issues described above.1−4 For example, conjugating poly(ethylene glycol) (PEG) at the reactive sites (such as lysine or cysteine residues) of proteins, known as PEGylation,5,6 can effectively extend the circulating half-life of a protein, and many PEGylated proteins have been approved by the Food and Drug Administration (FDA) for clinical use.7,8 Another widely used strategy for improving pharmaceutical profiles is to fuse biomolecules with long-acting human serum albumin or Fc fragments using protein engineering technology.9−11 However, these strategies require the introduction of new macromolecular substances, and the potential immunogenicity and toxicity effects induced by the extra moieties still remain of a concern.12−14
Covalent conjugation of the N- and C-termini of a protein is often used as a tool to enhance conformational stability during protein delivery.15−17 There are two main reasons: (1) backbone cyclization of proteins can reduce the conformational entropy in the unfolding state with improved melting temperature (Tm);17,18 and (2) the cyclized proteins do not present free termini, thus greatly decreasing enzymes’ and other substances’ attack.19−21 Previously, various proteins such as dihydrofolate reductase,18,22 maltose-binding protein,23 green fluorescent protein,19,24,25 ubiquitin carboxyl-terminal hydrolase isozyme L3,25 erythropoietin,26 and interferon26 have been cyclized via disulfide bond formation, SpyTag/SpyCatcher system and intein- or sortase-mediated ligation with well-retained bioactivity, better thermal stability, and enhanced pharmaceutical profiles.
The plasma membrane is impenetrable and can exclude biomolecules, which greatly limits the potential of proteins for pharmaceutical applications.27−30 Cell-penetrating peptides (CPPs) are a class of peptides that have the ability to cross the plasma membrane and have been applied to efficiently assist molecular cargoes in internalizing in living cells in a manner termed “protein transduction”.30−36 Fusion or chemical conjugation of these short peptides has also been proven to be an effective way to deliver large proteins into cells, and numerous “CPP–protein complexes” have been reported, thus providing an easy alternative to increase the delivery efficiency of therapeutic proteins and peptides.28,29,37−42
In this study, we report a system to increase the stability and intracellular delivery efficiency of proteins via a combination of sortase-mediated cyclization and CPP-mediated delivery. We chose a TAT peptide as the cellular penetrant carrier. The TAT peptide is a nine amino acid sequence (RKKRRQRRR) derived from a motif (residues 49–57) of the human immunodeficiency virus-1 (HIV-1) transacting activator of transcription (TAT) protein; and it can transfer proteins across plasma membranes and mediate nuclear localization.31,39,43,44 We adopted sortase-mediated protein ligation (SPL) for the backbone cyclization of a green fluorescent protein (GFP). There are two reasons for our choice: (1) the reaction condition of SPL is mild, and the reaction efficiency is high;19,45 and (2) the N- and C-termini of GFP stay at a relatively close distance to facilitate effective ligation.24,46 Herein, as the first case, we demonstrate that cyclized GFP fused with TAT (cGFP-TAT) significantly improved not only in vitro stability and cellular internalization ability but also in vivo tumor retention compared with native linear GFP (lGFP) and linear TAT-fused GFP (lGFP-TAT).
2. Results and Discussion
2.1. Biosynthesis and Physicochemical Characterization
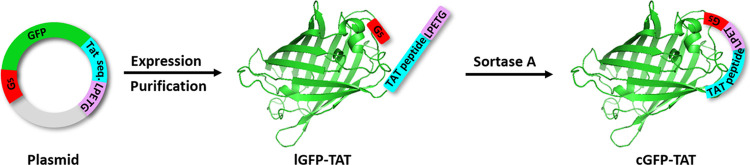
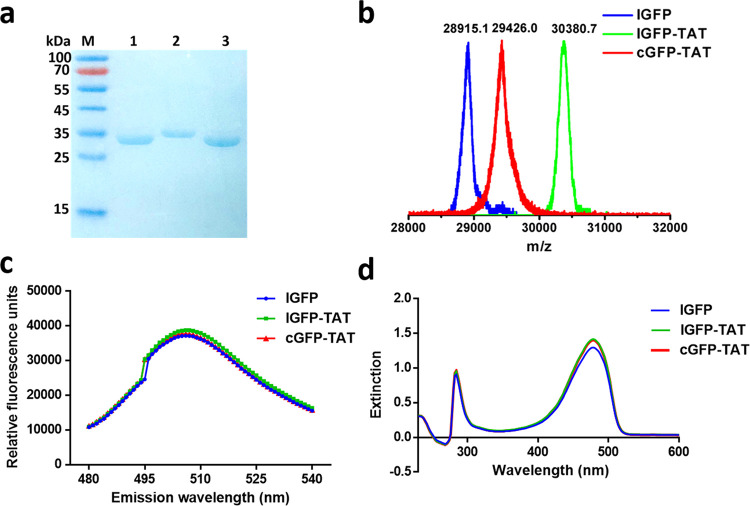
To obtain the cGFP-TAT protein, an lGFP-TAT protein was first synthesized by protein engineering technology, followed by SPL for cyclization (Scheme 1). The sequence of the lGFP-TAT recombinant plasmid consisted of the following important motifs: the nine amino acid TAT peptide was fused at the C-terminus of the lGFP; while two sortase A recognition motifs, G5 and LPETG, were introduced at the N-terminus of lGFP and the C-terminus of TAT peptide, respectively; the 6× His tag was located at the C-terminus of the LPETG sequence for nickel-nitrilotriacetic acid immobilized metal affinity chromatography (Ni-NTA IMAC) purification. The lGFP-TAT fusion protein was successfully overexpressed in Escherichia coli (E. coli) with isopropyl-β-d-1-thiogalactopyranoside (IPTG) induction and purified by Ni-NTA IMAC with high yield (∼100 mg/L) (Figure S1). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) showed a single band around the molecular weight (MW) position of ∼30.4 kDa corresponding to lGFP-TAT (Figure 1a). The theoretical MW of lGFP-TAT is 30381.1 Da, which was in accordance with the value (30380.7 Da) confirmed by matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) for accurate mass (Figure 1b). Similarly, we designed and obtained sortase A (with the 6× His at the N-terminus) of high quality for the cyclization reaction via the same protein purification method (Figure S2).
Scheme 1. Schematic Illustration of the Biosynthesis Process of cGFP-TAT. First, lGFP Was Selectively Fused with a CPP, the TAT Peptide, at Its C-Terminus Using Protein Engineering Technology to Form lGFP-TAT. Second, lGFP-TAT Was Cyclized to Produce cGFP-TAT for Subsequent Sortase A-Catalyzed Ligation.
Figure 1.
Synthesis and characterization of cGFP-TAT, lGFP-TAT, and lGFP. (a) SDS-PAGE analysis. Lane 1: lGFP; lane 2: lGFP-TAT; lane 3: cGFP-TAT. (b) MALDI-TOF spectra of proteins. (c) Fluorescence spectra of proteins from 480 to 540 nm, excited at 460 nm. (d) UV–vis absorption spectra of proteins from 230 to 600 nm.
The addition of sortase A to lGFP-TAT resulted in cGFP-TAT. Sortase A recognizes the substrate motif LPETG at the C-terminus of lGFP-TAT and cleaves the amide bond between Thr and Gly, utilizing an active site cysteine.19,47 The generating covalent acyl enzyme intermediate then undergoes nucleophilic attack by oligoglycine at the N-terminus, yielding the cyclized protein, cGFP-TAT. SDS-PAGE analysis of the reaction mixture demonstrated the complete disappearance of the band for lGFP-TAT at approximately 30.4 kDa and the appearance of a lower band corresponding to cGFP-TAT at approximately 29.4 kDa, indicating that the efficacy of cyclization was almost quantitative and that few oligomers of GFP were formed during the reaction (Figure S3). cGFP-TAT was then separated from the reaction mixture by anion exchange (AEX) chromatography, yielding products at a purity of ∼95% (Figure 1a). Purified cGFP-TAT was characterized by MALDI-TOF for MW determination, and the mass detected was 29426.0 Da, closely agreeing with the theoretical value of 29426.1 Da (Figure 1b). We also overexpressed and purified native lGFP (with the 6× His at the C-terminus) as a control for further assays (Figure S4). The purity (∼95%) and MW (theoretical value of 28915.3 Da) were confirmed by SDS-PAGE (Figure 1a) and MALDI-TOF (Figure 1b), respectively. The data show that the backbone of lGFP-TAT can be efficiently conjugated using SPL and successfully yield cGFP-TAT with high purity.
The hydrodynamic radii (Rh) of cGFP-TAT, lGFP-TAT, and lGFP analyzed by dynamic light scattering (DLS) were similar, approximately 3.5 nm (Figure S5). Circular dichroism (CD) spectroscopy showed that cGFP-TAT displayed an identical signature of β-sheet barrel, with a positive peak at 195–198 nm and a negative peak at 217–218 nm (Figure S6). The spectra of cGFP-TAT were consistent with those of lGFP-TAT and lGFP, suggesting that sortase A-catalyzed cyclization has no obvious influence on the secondary structure of GFP.
To characterize the photophysical properties and determine the functional activities of proteins, the fluorescence and UV absorption spectra were quantified at similar concentrations. The GFP concentration was confirmed by UV–vis absorption at 280 nm (Figure S7). The fluorescence spectra of cGFP-TAT were consistent with those of lGFP and lGFP-TAT, with emission maxima at 507 nm (Figure 1c and Table S1) and absorbance maxima at 478 nm (Figure 1d and Table S2). These results indicate that the photophysical properties of cGFP-TAT are almost completely retained, and the covalent cyclization has no influence on the function of GFP.
2.2. In Vitro Stability
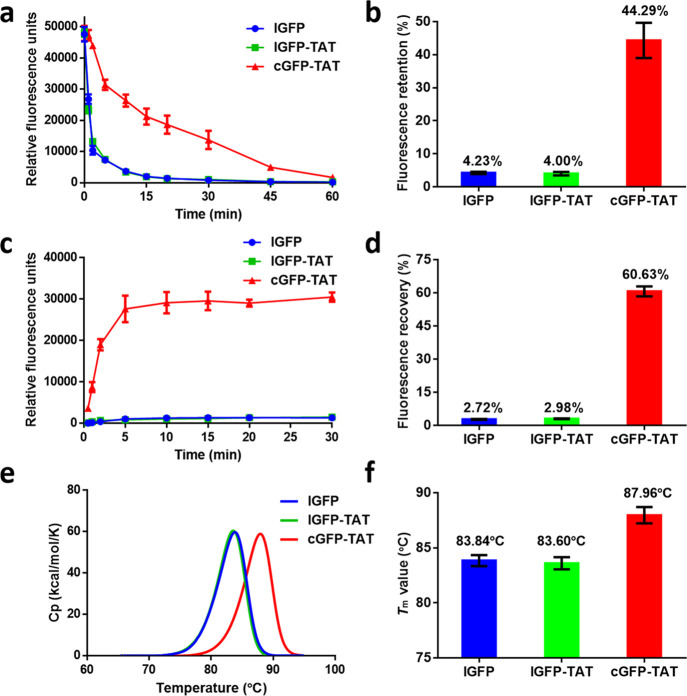
Because cyclization of the backbone of a protein can increase the thermal stability compared with its linear form, we investigated the fluorescence retention of cGFP-TAT, lGFP-TAT, and lGFP after incubation at 80 °C for the indicated times. lGFP and lGFP-TAT dramatically lost their fluorescence within a few minutes; in contrast, cGFP-TAT slowly lost its fluorescence within 1 h (Figure 2a and Figure S8). After 15 min, the fluorescence retention percentage of cGFP-TAT (44.29%) was over 10-fold than those of lGFP (4.23%) and lGFP-TAT (4.00%) (Figure 2b). We also assessed the fluorescence recovery of cGFP-TAT by simple thermal denaturation at 90 °C for 5 min, followed by an ice bath. The fluorescence of cGFP-TAT quickly regained and increased in 5 min, while the fluorescence of lGFP and lGFP-TAT could not be recovered for a long time (Figure 2c and Figure S9). Thirty minutes after denaturation, the fluorescence recovery percentage of cGFP-TAT was 60.63%, over 20-fold higher than those of lGFP (2.72%) and lGFP-TAT (2.98%) (Figure 2d). Because GFP should be diluted in a buffer containing NaCl (at least at a concentration of 50 mM) to maintain relative stable during detection, we chose differential scanning calorimetry (DSC) over CD to measure the Tm of proteins.48−50 As the DSC curve showed, the Tm value of lGFP (83.84 °C) was in accordance with the reported value (∼357 K at a heating rate of 1 K/min).50 lGFP-TAT had a similar Tm value compared with lGFP, whereas cGFP-TAT had an improved Tm value of ∼4 °C (Figure 2e,f), which further explained the increased thermal stability of GFP after cyclization.
Figure 2.
In vitro thermal stability of cGFP-TAT, lGFP-TAT, and lGFP. (a) Fluorescence retention of proteins heated at 80 °C for the indicated times. P < 0.005 for cGFP-TAT vs lGFP and lGFP-TAT. (b) Fluorescence retention percentage of proteins heated at 80 °C for 15 min. P < 0.001 for cGFP-TAT vs lGFP and lGFP-TAT. (c) Fluorescence recovery of proteins following thermal denaturation at 90 °C for 5 min. P < 0.0001 for cGFP-TAT vs lGFP and lGFP-TAT. (d) Fluorescence recovery percentage of proteins 30 min after thermal denaturation. P < 0.0001 for cGFP-TAT vs lGFP and lGFP-TAT. (e) DSC curve of proteins. (f) Tm value of proteins. P < 0.01 for cGFP-TAT vs lGFP and lGFP-TAT. Data are shown as the mean ± standard deviation (n = 3).
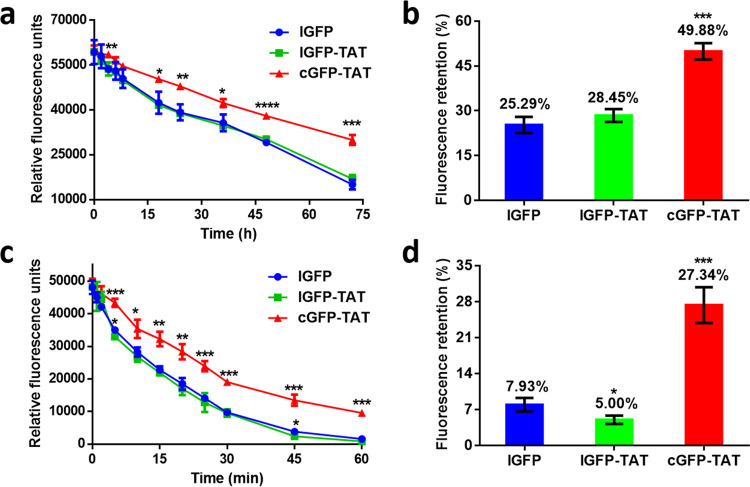
As proteins are susceptible to enzymatic degradation, we tested the proteolytic stability of cGFP-TAT against papain.Papain is a kind of sulfhydryl protease with broad substrate specificity; and it can hydrolyze the carboxyl end of arginine and lysine distribution on proteins. lGFP and lGFP-TAT showed a rapid decrease in fluorescence upon the addition of papain, while cGFP-TAT exhibited a slower fluorescence decrease than both lGFP and lGFP-TAT (Figure 3a). After 72 h, the fluorescence retention percentages of lGFP, lGFP-TAT, and cGFP-TAT were 25.29, 28.45, and 49.88%, respectively (Figure 3b). We also investigated the chemical denaturation of cGFP-TAT against guanidine hydrochloride (GdnHCl), a common chemical denaturant that has been widely used in protein inclusion body purification. It was obviously visible that cGFP-TAT unfolded more slowly than lGFP-TAT and lGFP in a 6 M GdnHCl solution (Figure 3c). The fluorescence retentions of cGFP-TAT (27.34%) were 5.47- and 3.45-fold higher than those of lGFP-TAT (5.00%) and lGFP (7.93%), respectively (Figure 3d). The data suggest that cGFP-TAT exhibits more resistance to enzymatic degradation and chemical denaturation than lGFP and lGFP-TAT.
Figure 3.
In vitro proteolytic and chemical stability of cGFP-TAT, lGFP-TAT, and lGFP. (a) Fluorescence retention of proteins against papain as a function of time. (b) Fluorescence retention percentage of proteins after incubating with papain for 72 h. (c) Fluorescence retention of proteins after incubating in a 6 M GdnHCl solution as a function of time. (d) Fluorescence retention percentage of proteins after incubating in a GdnHCl solution for 45 min. Data are shown as the mean ± standard deviation (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 for cGFP-TAT and lGFP-TAT vs lGFP.
2.3. Intracellular Delivery
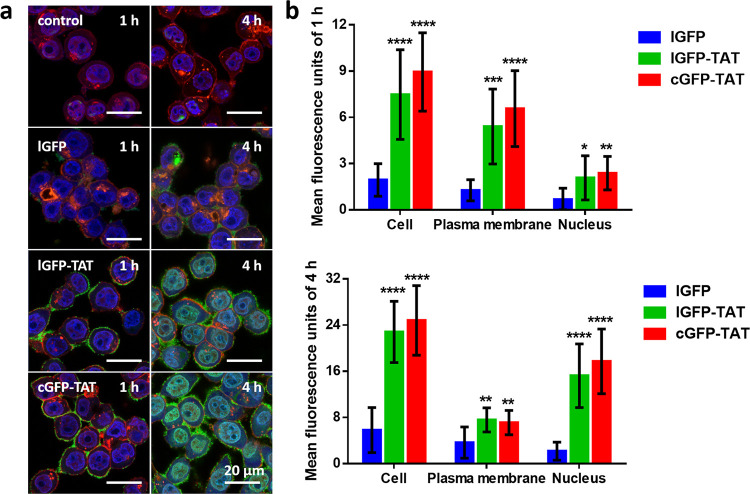
Next, we investigated the cellular internalization and distribution of cGFP-TAT, lGFP-TAT, and lGFP in a C26 cell (murine colon carcinoma) because it is a cell line that has been widely applied in assessing the intracellular drug delivery.51 We first tested the cellular internalization of proteins at different concentrations using confocal laser scanning microscopy (CLSM). After incubating with 10, 30, and 50 μM proteins for 1 and 4 h at 37 °C in a fresh RPMI-1640 medium, C26 cells were directly imaged without fixation to decrease the artifactual redistribution of the TAT peptide-fused GFP in the nucleus.52 We found the efficient and similar cellular uptake of lGFP-TAT and cGFP-TAT into cells at high concentrations of 30 and 50 μM (Figure 4a and Figure S10). Most proteins were enriched in the plasma membrane and nucleus, and some were homogeneously distributed throughout the cytoplasm; and no obvious endocytic vesicles were observed in the cytoplasm after incubating 30 and 50 μM proteins for 4 h (Figure S10).37 However, proteins were heterogeneously distributed, and some vesicles can be observed in the cytoplasm after incubating at a low concentration of 10 μM for 4 h, indicating partial endosomal delivery (Figure S11).52,53 Herein, in the intracellular delivery experiments, we chose to fix the protein concentration at 30 μM, as the TAT-fused proteins were internalized mainly in a non-endosomal mode.
Figure 4.
Intracellular uptake of cGFP-TAT, lGFP-TAT, and lGFP. (a) Transduction of proteins to C26 cells. C26 cells incubated without GFP were used as the control. The cell nucleus was stained with Hoechst 33342 in blue; the plasma membrane was stained with wheat germ agglutinin in red; and GFP was shown in green with a final incubation concentration of 30 μM at 37 °C in the fresh RPMI-1640 medium. (b) Quantification of the mean fluorescence units in the cell, plasma membrane, and nucleus after taken up by C26 for 1 h (up) and 4 h (below). Data are shown as the mean ± standard deviation (n = 9). *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001 for lGFP-TAT and cGFP-TAT vs lGFP in the cell, plasma membrane, and nucleus, respectively.
From the results, we found that cGFP-TAT rapidly bound to the plasma membrane after incubation for 1 h, and the green fluorescence was well distributed in the nucleus after internalization for 4 h (Figure 4a and Figure S10), which was in accordance with the fluorescence quantification (Figure 4b and Tables S3–S4), suggesting that cGFP-TAT was successfully taken up by cells and translocated in the nucleus. However, when cells were treated with lGFP, green fluorescence could only be clearly observed on the plasma membrane until 4 h, a much longer incubation period than that of cGFP-TAT (Figure 4a and Figure S12); lGFP also accumulated in the nucleus at a low level (Figure 4b and Tables S3 and S4). In particular, the mean fluorescence unit of cGFP-TAT in the nucleus was 3.54 times higher than that of lGFP at 1 h, and the ratio increased to 8.20 times after incubation for 4 h (Figure 4b and Tables S3 and S4). In addition, the cellular delivery is temperature-independent because lGFP-TAT and cGFP-TAT exhibited a similar intracellular distribution throughout the cytosol and nucleus after incubating at 4 °C (Figure S13), which further proved that the entry mode is mainly non-endosomal.29,53 We have also tested the uptake of proteins into cells in the serum, and the proteins exhibited the same internalized behaviors as in the fresh medium (Figure S14). The data demonstrate that the TAT peptide can be utilized to efficiently transport biomolecules into cells.
2.4. In Vivo Tumor Retention
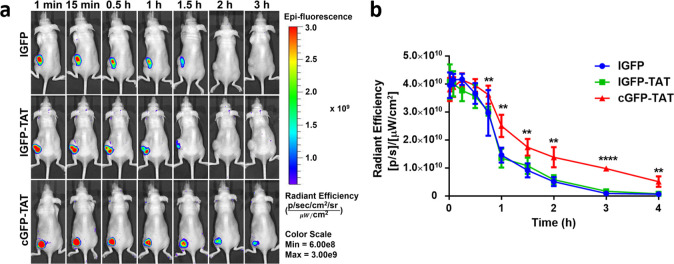
One of the major problems in the delivery of therapeutic proteins is their poor in vivo retention time. Herein, we evaluated the retention of cGFP-TAT, lGFP-TAT, and lGFP in tumors in C26-inoculated mice via in vivo imaging. Mice with a mean tumor volume of 100 mm3 were intratumorally injected with cGFP-TAT, lGFP-TAT, and lGFP. Upon administration, the fluorescence of lGFP, as well as lGFP-TAT, in tumors quickly decreased and was completely undetectable within 2 h (Figure 5a). In contrast, the fluorescence of cGFP-TAT exhibited a relatively slower decrease and could still be visible even after 3 h (Figure 5a). Notably, the fluorescence of cGFP-TAT (1.74 × 1010 photons) in tumors was 1.91- and 1.61-fold higher than those of lGFP (9.11 × 109 photons) and lGFP-TAT (1.08 × 1010 photons) at 1.5 h; and the fluorescence of cGFP-TAT (5.13 × 109 photons) displayed 7.79- and 6.52-fold higher relative to those of lGFP (6.58 × 108 photons) and lGFP-TAT (7.86 × 108 photons) after administration for 4 h (Figure 5b). This slower clearance in the tumor might be attributed to the better thermal, proteolytic, and chemical stability of cGFP-TAT compared with lGFP-TAT and lGFP, as the data we achieved above. These results indicate that the backbone ligation of cGFP-TAT can remarkably improve the retention of GFP in tumors.
Figure 5.
In vivo tumor retention of cGFP-TAT, lGFP-TAT, and lGFP. (a) In vivo fluorescence imaging of proteins in tumor-bearing mice after intratumor injection from 1 min to 3 h. From top to bottom: lGFP, lGFP-TAT, and cGFP-TAT. (b) Fluorescence of proteins in tumors as a function of time. Data are shown as the mean ± standard deviation (n = 4). **P < 0.01 and ****P < 0.001 for cGFP-TAT vs lGFP and lGFP-TAT.
3. Conclusions
In summary, we have demonstrated an easy strategy to improve the in vitro stability, intracellular delivery efficiency, and in vivo tumor retention of GFP. We, for the first time, genetically constructed a TAT peptide and sortase recognition motifs at the N- and C-termini of GFP for CPP-mediated delivery and sortase-mediated cyclization, respectively. Compared with lGFP and lGFP-TAT, cGFP-TAT displays three major advantages that are vital for the design of advanced therapeutic protein agents: (i) cGFP-TAT has superior resistance to heat inactivation, enzymatic degradation, and chemical denaturation because the N- and C-termini are not exposed to external stressors; (ii) cGFP-TAT can efficiently be taken up into living cells with the assistance of fused CPPs on the protein; and (iii) cGFP-TAT exhibits enhanced tumor retention of GFP because of the increased thermal, proteolytic, and chemical stability. We believe that this strategy would be applicable to various therapeutic proteins and peptides for clinical treatments.
4. Experimental Section
4.1. Materials
All biological reagents were purchased from New England Biolabs or Thermo Scientific and used as received, unless otherwise stated. All chemical reagents were purchased from Sigma-Aldrich or J&K Scientific and used as received, unless otherwise stated. All cell culture-related reagents were obtained from Gibco or Corning, unless otherwise stated. C26 cells were obtained from the cell bank of the Chinese Academy of Medical Sciences. Female BALB/c-nude mice were purchased from Vital River Laboratories (Beijing, China) and accommodated in the Laboratory Animal Center of Peking Union Medical College Hospital accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC), and all animal methods used in this study were approved by the Institutional Animal Care and Use Committee (IACUC).
4.2. Expression and Production of lGFP-TAT
The gene encoding the green fluorescent protein (GFP) with a TAT peptide (lGFP-TAT) was designed with a sortase A recognition motif (GGGGG) at the N-terminus and a TAT peptide (RKKRRQRRR), sortase A recognition motif (LPETG) and 6× His tag at the C-terminus. The lGFP-TAT gene synthesized by Sangon Biotech (Shanghai, China) was cloned into the pET-25b (+) vector (Novagen) using a standard molecular cloning technique and verified by DNA sequencing. The constructed plasmid was then transformed into E. coli BL21(DE3) competent cells (TransGen Biotech, Beijing, China) and cultivated in Luria Bertani media containing 100 μg/mL of ampicillin at 37 °C with shaking at 220 rpm. Once the optical density at 600 nm (OD600) reached 0.5, isopropyl-β-d -thiogalactopyranoside (IPTG) (with a final concentration of 500 μM) was added to the cell culture for induction. After overnight expression at 22 °C, cells were harvested by centrifugation at 4500× g for 15 min. Cell pellets from 1 L of culture were resuspended in 50 mL of lysis buffer (50 mM Tris·HCl, 150 mM NaCl, pH 8.0) and lysed by a continuous flow cell disrupter (JNBIO, Guangzhou, China) twice with an ultrahigh pressure of 1200 bar. After centrifugation, the supernatant was collected and applied to a 5 mL HisTrap column (GE Healthcare, USA) on an AKTApurifier chromatographic system with a UV detector at 280 nm. The protein-loaded column was first equilibrated with an equilibration buffer (50 mM Tris·HCl, 150 mM NaCl, and 5 mM imidazole) and then washed with a washing buffer (50 mM Tris·HCl, 150 mM NaCl, and 20 mM imidazole). After eluted by an eluent buffer (50 mM Tris·HCl, 150 mM NaCl, and 200 mM imidazole), lGFP-TAT was further purified by running through a HiPrep 26/10 desalting column (GE Healthcare, USA) to change the buffer to 50 mM Tris·HCl, 150 mM NaCl, pH 7.4 and stored at −80 °C until use. The concentration of lGFP-TAT was estimated by bicinchoninic acid (BCA) assay according to the instructions of the BCA kit (Beyotime Biotech, Shanghai, China). The purification process and purity of protein were assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
As a control, native GFP with a 6× His tag at the C-terminus (lGFP) was produced and purified via the same method.
Sortase A was expressed and purified as previously described.19
The amino
acid sequence of lGFP-TAT is listed as follows: The amino acids
with dotted underline are the sortase A
recognition motifs; the amino acids with solid underline are the sequence
of GFP; the amino acids with wavy underline are the sequence of TAT
peptide; and the starting amino acid M can be cleaved automatically
during the expression process.
The amino acids
with dotted underline are the sortase A
recognition motifs; the amino acids with solid underline are the sequence
of GFP; the amino acids with wavy underline are the sequence of TAT
peptide; and the starting amino acid M can be cleaved automatically
during the expression process.
4.3. Synthesis and Purification of cGFP-TAT
lGFP-TAT (50 μM) was mixed with sortase A (25 μM) in 50 mM Tris·HCl, 150 mM NaCl, 10 mM CaCl2, pH 7.4 for 6 h at 25 °C without agitation, resulting in cGFP-TAT. The reaction mixture was diluted into 4 volumes of 20 mM Tris·HCl, pH 7.4, and purified by anion exchange (AEX) chromatography on a HiTrap Capto Q column (GE Healthcare, USA) performed on an AKTApurifier chromatographic system. The column was eluted with 20 mM Tris·HCl, pH 7.4, and the concentration of NaCl in the buffer was increased from 0 to 1 M using a linear flow velocity gradient with time. The peak containing the desired cGFP-TAT protein was collected; and the contents were buffer exchanged into 50 mM Tris·HCl, 150 mM NaCl, pH 7.4 with a desalting column and finally concentrated via ultrafiltration.
4.4. Physicochemical Characterization
4.4.1. Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE)
Samples were diluted in a loading buffer containing 5% (v/v) β-mercaptoethanol and 0.5% (m/v) bromophenol blue and boiled at 95 °C for 10 min before loading onto a precast 12.5% SDS-PAGE gel (GenScript, Nanjing, China). The gel was applied to a Mini-Protein gel apparatus (Bio-Rad) at 130 V for 50 min using 25 mM Tris, 250 mM glycine, and 0.1% SDS as the running buffer and stained with Coomassie blue. Protein markers with a broad range of 10–170 kDa were used as the standard.
4.4.2. Bicinchoninic Acid (BCA)
The concentrations of cGFP-TAT, lGFP-TAT, and lGFP were assessed via a protein BCA assay kit following the directions of the manufacturer. Bovine serum albumin (BSA) was used as the protein standard.
4.4.3. Matrix-Assisted Laser Desorption/Ionization Time-of-Flight Mass Spectrometry (MALDI-TOF MS)
The molecular weights of cGFP-TAT, lGFP-TAT, and lGFP were detected by a 4800 Plus MALDI-TOF/TOF analyzer (Applied Biosystems, USA) with a nitrogen laser. Samples were diluted in 2 mM PBS, pH 7.4 and mixed with the matrix solution (a saturated solution of sinapic acid in a 1:1 mixture of acetonitrile and water containing 0.1% trifluoroacetic acid) at a volume ratio of 1:1, and then 1 μL of the mixture was spotted onto the sample plate and air dried. The spectra were acquired in a linear mode (1.5 m) and calibrated with proteins of known masses.
4.4.4. Dynamic Light Scattering (DLS)
Samples were filtered and then measured on a Malvern Zetasizer Nano ZS90 using a laser wavelength of 633 nm and a scattering angle of 90° at 25 °C. The data were analyzed with Zetasizer software 6.32.
4.4.5. Circular Dichroism (CD) Spectra
Samples were diluted in H2O at a concentration of 0.2 mg/mL and applied on Pistar π-180 (Applied Photophysics Ltd., UK) with a wavelength range from 195 to 250 nm.
4.4.6. Fluorescence Spectra Assay
Fluorescence spectra were recorded on a SpectraMax M3 microplate reader (Molecular Devices, USA) at room temperature, with an excitation wavelength of 460 nm. The concentrations of cGFP-TAT, lGFP-TAT, and lGFP were 0.3 mg/mL in 50 mM Tris·HCl, 150 mM NaCl, pH 7.4.
4.4.7. UV–vis Absorption Spectra Assay
UV–vis absorption spectra were measured on a SpectraMax M3 microplate reader (Molecular Devices, USA) at room temperature. The concentrations of cGFP-TAT, lGFP-TAT, and lGFP were 1.0 mg/mL in 50 mM Tris·HCl, 150 mM NaCl, pH 7.4.
4.5. In Vitro Stability
4.5.1. Thermal Stability
In order to assess the fluorescence retention of proteins in the thermal environment, the samples with a final protein concentration of 0.4 mg/mL were incubated at 80 °C for given times. The fluorescence of the samples was measured as described above, with excitation and emission wavelengths of 460 and 507 nm, respectively.
To estimate the fluorescence recovery of cGFP-TAT, the samples were denatured at 90 °C for 5 min, followed by an ice bath for activity recovery.
4.5.2. Differential Scanning Calorimetry (DSC) Assay
Samples were diluted in 50 mM Tris·HCl, 150 mM NaCl, pH 7.4 at a concentration of 1 mg/mL and applied on MicroCal PEAQ-DSC (Malvern Instruments Ltd., UK). The temperature was ranged from 65 to 100 °C with an increase in rate of 1 °C/min. The melting temperature (Tm) was calculated using MicroCal PEAQ-DSC software 1.52.
4.5.3. Proteolytic Stability
A mixture containing 0.5 mg/mL protein and 1.0 mg/mL papain in 50 mM sodium phosphate buffer, 10 mM EDTA, 10 mM cysteine, pH 6.5 was incubated at 55 °C for the indicated times. The fluorescence of the reaction mixtures was measured as described above.
4.5.4. Chemical Denaturation
The samples were diluted at a final concentration of 0.4 mg/mL in 6 M guanidine hydrochloride (GdnHCl), 50 mM Tris·HCl, 150 mM NaCl, 1 mM EDTA, 1 mM DTT, pH 8.0 at room temperature. The fluorescence of the reaction mixtures was measured at selected times.
4.6. Intracellular Delivery
C26 cells were cultured in the RPMI-1640 medium containing 10% (v/v) fetal bovine serum (FBS), 1% penicillin/streptomycin, 10 mM HEPES, 1 mM sodium pyruvate, 1 mM nonessential amino acid, and 4.5 g/L d-glucose at 37 °C in a 5% CO2 humidified atmosphere. The cells were grown in a T-75 flask and passaged every 2 days. After incubating with 2 mL of 0.1% trypsin, 0.5 mM EDTA for 3 min, cells were harvested by centrifugation at 300× g. The cells were then resuspended in the fresh medium, seeded in a confocal dish at a density of 5 × 104 per well, and cultured overnight at 37 °C for attachment. Samples (diluted to a final concentration of 30 μM in the fresh RPMI-1640 medium, unless otherwise indicated) were added into the dish for cellular uptake. After incubation for 1 and 4 h (at 37 °C, unless otherwise indicated), the cells were washed with the medium to remove the remaining samples. The plasma membrane and nucleus were dyed with wheat germ agglutinin (WGA) Alexa Fluor 594 (5 μg/mL) and bisBenzimide H33342 trihydrochloride (Hoechest 33342) (1 μg/mL) for 10 min. The cells were then washed with 0.01 M PBS, 150 mM NaCl, pH 7.4 containing 2% heparan sulfate for three times and imaged using an LSM710 laser scanning confocal microscope (Carl Zeiss, Germany). Cells incubated without a GFP were used as the negative control. For the uptake experiment in the serum, proteins were diluted in the serum at a ratio of 1:1 before added into C26 cells for attachment. RPMI-1640 media were replaced with a serum buffer (1:1 mixture of serum and 0.01 M PBS, pH 7.4) during the incubation period. The serum samples were obtained from the orbit of rats under standard protocols. The excitation and emission wavelengths of Hoechest, GFP, and WGA were 346/460, 485/590, and 590/617 nm, respectively. Images were analyzed by Zen_2012 (blue edition) software, and the fluorescence value was calculated with ImageJ.
4.7. In Vivo Tumor Retention
The protocols of in vivo tumor retention assessment were employed as described previously with slight changes.19 Eight-week-old female BALB/c nude mice were implanted subcutaneously with 5 × 107 cells in the left flank. The tumor volume was calculated as length × width2 × 0.5. When the tumor size grew to 100–150 mm3, the mice were randomly divided into 3 groups (4 mice per group) and intratumorally injected with 15 μg of cGFP-TAT, lGFP-TAT, and lGFP. The fluorescence retention of tumors in mice was recorded on an IVIS Lumina II in vivo imaging system (Caliper Life Sciences, USA) at selected times (1, 5, 15, 30, and 45 min and 1, 1.5, 2, 3, and 4 h), and the images were analyzed by Living Image 4.2 software.
4.8. Statistical Analysis
Data were analyzed using GraphPad Prism software 5.0 and are shown as the mean ± standard deviation. Comparisons of the data including in vitro stability, cellular uptake, and in vivo tumor retention were carried out using student’s t test and one-way ANOVA. A P value less than 0.05 was considered significant. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
Acknowledgments
This work was financially supported by grants from the National Natural Science Foundation of China (grant nos. 21805311 to J.H. and 81803419 to B.Z.).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.1c00532.
SDS-PAGE analysis of lGFP, lGFP-TAT, sortase A, and cGFP-TAT purification; DLS analysis of cGFP-TAT, lGFP-TAT, and lGFP; circular dichroism spectra of cGFP-TAT, lGFP-TAT, and lGFP; UV–vis absorption at 280 nm of cGFP-TAT, lGFP-TAT, and lGFP to verify similar protein concentrations; fluorescence retention of cGFP-TAT, lGFP-TAT, and lGFP after heating at 80 °C; fluorescence recovery of cGFP-TAT, lGFP-TAT, and lGFP following thermal denaturation for 5 min at 90 °C; intracellular uptake of cGFP-TAT, lGFP-TAT, and lGFP under different conditions; individual fluorescent channels of intracellular uptake of cGFP-TAT, lGFP-TAT, and lGFP to C26 cells; fluorescence data of proteins from 480 to 540 nm; UV–vis absorption data of proteins from 230 to 600 nm; and fluorescence values (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Torchilin V. P.; Lukyanov A. N. Peptide and Protein Drug Delivery to and into Tumors: Challenges and Solutions. Drug Discovery Today 2003, 8, 259–266. 10.1016/S1359-6446(03)02623-0. [DOI] [PubMed] [Google Scholar]
- Yu M.; Wu J.; Shi J.; Farokhzad O. C. Nanotechnology for Protein Delivery: Overview and Perspectives. J. Controlled Release 2016, 240, 24–37. 10.1016/j.jconrel.2015.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. M.; Gran M. P.; Peppas N. A. Designing the New Generation of Intelligent Biocompatible Carriers for Protein and Peptide Delivery. Acta Pharm. Sin. B 2018, 8, 147–164. 10.1016/j.apsb.2018.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J.; Kamaly N.; Shi J.; Zhao L.; Xiao Z.; Hollett G.; John R.; Ray S.; Xu X.; Zhang X.; Kantoff P. W.; Farokhzad O. C. Development of Multinuclear Polymeric Nanoparticles as Robust Protein Nanocarriers. Angew. Chem. Int. Edit. 2014, 53, 8975–8979. 10.1002/anie.201404766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X.; Hu J.; Wang Z.; Xu Z.; Gao W. Polypeptides and Engineered Proteins. Acs. Sym. Ser. 2017, 1252, 93–127. 10.1021/bk-2017-1252.ch006. [DOI] [Google Scholar]
- Grigoletto A.; Mero A.; Yoshioka H.; Schiavon O.; Pasut G. Covalent Immobilisation of Transglutaminase: Stability and Applications in Protein PEGylation. J. Drug Targeting 2017, 25, 856–864. 10.1080/1061186X.2017.1363211. [DOI] [PubMed] [Google Scholar]
- Veronese F. M.; Pasut G. PEGylation, Successful Approach to Drug Delivery. Drug Discovery Today 2005, 10, 1451–1458. 10.1016/S1359-6446(05)03575-0. [DOI] [PubMed] [Google Scholar]
- Alconcel S. N. S.; Baas A. S.; Maynard H. D. FDA-approved Poly(ethylene glycol)-Protein Conjugate Drugs. Polym. Chem. 2011, 2, 1442–1448. 10.1039/c1py00034a. [DOI] [Google Scholar]
- Subramanian G. M.; Fiscella M.; Lamousé-Smith A.; Zeuzem S.; McHutchison J. G. Albinterferon Alpha-2b: a Genetic Fusion Protein for the Treatment of Chronic Hepatitis C. Nat. Biotechnol. 2007, 25, 1411–1419. 10.1038/nbt1364. [DOI] [PubMed] [Google Scholar]
- Rampotas A.; Desborough M. J. R.; Raza-Burton S.; Taylor S.; Wilkinson A.; Hall G. W.; Shapiro S.; Curry N. A Single Centre Retrospective Study of Low Dose Prophylaxis with Extended Half-life Factor IX for Severe Haemophilia B. Haemophilia 2020, 26, 278–281. 10.1111/hae.13936. [DOI] [PubMed] [Google Scholar]
- Chung H. S.; Oh J. Y.; Yoo S. B.; Lee S. M.; Cho H. S. The N-terminal Alanine-Extended GLP-1/IgG-Fc Fusion Protein Confers Resistance to DPP-IV and Reduces Serum Glucose Level in db/db Mice. Regul. Pept. 2011, 170, 1–3. 10.1016/j.regpep.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Knop K.; Hoogenboom R.; Fischer D.; Schubert U. S. Poly(ethylene glycol) in Drug Delivery: Pros and Cons as Well as Potential Alternatives. Angew. Chem. Int. Edit. 2010, 49, 6288–6308. 10.1002/anie.200902672. [DOI] [PubMed] [Google Scholar]
- Pasut G.; Sergi M.; Veronese F. M. Anti-cancer PEG-enzymes: 30 Years Old, but Still a Current Approach. Adv. Drug Deliver. Rev. 2008, 60, 69–78. 10.1016/j.addr.2007.04.018. [DOI] [PubMed] [Google Scholar]
- Sroda K.; Rydlewski J.; Langner M.; Kozubek A.; Grzybek M.; Sikorski A. F. Repeated Injections of PEG-PE Liposomes Generate Anti-PEG Antibodies. Cell Mol. Biol. Lett. 2005, 10, 37–47. [PubMed] [Google Scholar]
- Williams N. K.; Liepinsh E.; Watt S. J.; Prosselkov P.; Matthews J. M.; Attard P.; Beck J. L.; Dixon N. E.; Otting G. Stabilization of Native Protein Fold by Intein-mediated Covalent Cyclization. J. Mol. Biol. 2005, 346, 1095–1108. 10.1016/j.jmb.2004.12.037. [DOI] [PubMed] [Google Scholar]
- Sun X. B.; Cao J. W.; Wang J. K.; Lin H. Z.; Gao D. Y.; Qian G. Y.; Park Y. D.; Chen Z. F.; Wang Q. SpyTag/SpyCatcher Molecular Cyclization Confers Protein Stability and Resilience to Aggregation. New Biotechnol. 2019, 49, 28–36. 10.1016/j.nbt.2018.12.003. [DOI] [PubMed] [Google Scholar]
- Camarero J. A.; Fushman D.; Sato S.; Giriat I.; Cowburn D.; Raleigh D. P.; Muir T. W. Rescuing a Destabilized Protein Fold through Backbone Cyclization. J. Mol. Biol. 2001, 308, 1045–1062. 10.1006/jmbi.2001.4631. [DOI] [PubMed] [Google Scholar]
- Wu X. L.; Liu Y.; Liu D.; Sun F.; Zhang W. B. An Intrinsically Disordered Peptide-Peptide Stapler for Highly Efficient Protein Ligation Both in Vivo and in Vitro. J. Am. Chem. Soc. 2018, 140, 17474–17483. 10.1021/jacs.8b08250. [DOI] [PubMed] [Google Scholar]
- Hu J.; Zhao W.; Gao Y.; Sun M.; Wei Y.; Deng H.; Gao W. Site-specific in Situ Growth of a Cyclized Protein-polymer Conjugate with Improved Stability and Tumor Retention. Biomaterials 2015, 47, 13–19. 10.1016/j.biomaterials.2015.01.002. [DOI] [PubMed] [Google Scholar]
- Malde A. K.; Hill T. A.; Iyer A.; Fairlie D. P. Crystal Structures of Protein-Bound Cyclic Peptides. Chem. Rev. 2019, 119, 9861–9914. 10.1021/acs.chemrev.8b00807. [DOI] [PubMed] [Google Scholar]
- Pelay-Gimeno M.; Bange T.; Hennig S.; Grossmann T. N. In Situ Cyclization of Native Proteins: Structure-Based Design of a Bicyclic Enzyme. Angew. Chem. Int. Edit. 2018, 57, 11164–11170. 10.1002/anie.201804506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott C. P.; Abel-Santos E.; Wall M.; Wahnon D. C.; Benkovic S. J. Production of Cyclic Peptides and Proteins in vivo. Proc. Natl. Acad. Sci. U. S. A. 1999, 96, 13638–13643. 10.1073/pnas.96.24.13638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panne D.; Liu X. Q.; Evans T. C. Jr.; Martin D.; Kolly R.; Sun L.; Ghosh I.; Chen L.; Benner J.; Liu X. Q.; Xu M. Q. Protein Trans-splicing and Cyclization by a Naturally Split Intein from the DnaE Gene of Synechocystis Species PCC6803. J. Biol. Chem. 2000, 275, 9091–9094. 10.1074/jbc.275.13.9091. [DOI] [PubMed] [Google Scholar]
- Iwai H.; Lingel A.; Plückthun A. Cyclic Green Fluorescent Protein Produced in vivo Using an Artificially Split PI-PfuI Intein from Pyrococcus Furiosus. J. Biol. Chem. 2001, 276, 16548–16554. 10.1074/jbc.M011639200. [DOI] [PubMed] [Google Scholar]
- Antos J. M.; Popp M. W. L.; Ernst R.; Chew G. L.; Spooner E.; Ploegh H. L. A Straight Path to Circular Proteins. J. Biol. Chem. 2009, 284, 16028–16036. 10.1074/jbc.M901752200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp M. W.; Dougan S. K.; Chuang T. Y.; Spooner E.; Ploegh H. L. Sortase-catalyzed Transformations that Improve the Properties of Cytokines. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 3169–3174. 10.1073/pnas.1016863108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.; Saw P. E.; Tao W.; Li Y.; Ji X.; Yu M.; Mahmoudi M.; Rasmussen J.; Ayyash D.; Zhou Y.; Farokhzad O. C.; Shi J. Tumor Microenvironment-Responsive Multistaged Nanoplatform for Systemic RNAi and Cancer Therapy. Nano Lett. 2017, 17, 4427–4435. 10.1021/acs.nanolett.7b01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S. G.; Sayers E. J.; He L.; Narayan R.; Williams T. L.; Mills E. M.; Allemann R. K.; Luk L. Y. P.; Jones A. T.; Tsai Y. H. Cell-penetrating Peptide Sequence and Modification Dependent Uptake and Subcellular Distribution of Green Fluorescent Protein in Different Cell Lines. Sci. Rep. 2019, 9, 6298. 10.1038/s41598-019-42456-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herce H. D.; Schumacher D.; Schneider A. F. L.; Ludwig A. K.; Mann F. A.; Fillies M.; Kasper M. A.; Reinke S.; Krause E.; Leonhardt H.; Cardoso M. C.; Hackenberger C. P. R. Cell-permeable Nanobodies for Targeted Immunolabelling and Antigen Manipulation in Living Cells. Nat. Chem. 2017, 9, 762–771. 10.1038/nchem.2811. [DOI] [PubMed] [Google Scholar]
- Allolio C.; Magarkar A.; Jurkiewicz P.; Baxová K.; Javanainen M.; Mason P. E.; Šachl R.; Cebecauer M.; Hof M.; Horinek D.; Heinz V.; Rachel R.; Ziegler C. M.; Schröfel A.; Jungwirth P. Arginine-rich Cell-penetrating Peptides Induce Membrane Multilamellarity and Subsequently Enter via Formation of a Fusion Pore. Proc. Natl. Acad. Sci. U. S. A. 2018, 115, 11923–11928. 10.1073/pnas.1811520115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel A. D.; Pabo C. O. Cellular Uptake of the Tat Protein from Human Immunodeficiency Virus. Cell 1988, 55, 1189–1193. 10.1016/0092-8674(88)90263-2. [DOI] [PubMed] [Google Scholar]
- Schneider A. F. L.; Wallabregue A. L. D.; Franz L.; Hackenberger C. P. R. Targeted Subcellular Protein Delivery Using Cleavable Cyclic Cell-Penetrating Peptides. Bioconjugate Chem. 2019, 30, 400–404. 10.1021/acs.bioconjchem.8b00855. [DOI] [PubMed] [Google Scholar]
- Fukuoka Y.; Khafagy E. S.; Goto T.; Kamei N.; Takayama K.; Peppas N. A.; Takeda-Morishita M. Combination Strategy with Complexation Hydrogels and Cell-Penetrating Peptides for Oral Delivery of Insulin. Biol. Pharm. Bull. 2018, 41, 811–814. 10.1248/bpb.b17-00951. [DOI] [PubMed] [Google Scholar]
- Wu L. P.; Ahmadvand D.; Su J.; Hall A.; Tan X.; Farhangrazi Z. S.; Moghimi S. M. Crossing the Blood-brain-Barrier with Nanoligand Drug Carriers Self-assembled from a Phage Display Peptide. Nat. Commun. 2019, 10, 4635. 10.1038/s41467-019-12554-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashina T.; Koyama T.; Nohara S.; Hasegawa M.; Ishiguro A.; Iijima K.; Lu J.; Shimura M.; Okamura T.; Sakuma T.; Yamamoto T.; Ishizaka Y. Identification of a Cell-penetrating Peptide Applicable to a Protein-based Transcription Activator-like Effector Expression System for Cell Engineering. Biomaterials 2018, 173, 11–21. 10.1016/j.biomaterials.2018.04.040. [DOI] [PubMed] [Google Scholar]
- Zhang P.; Moreno R.; Lambert P. F.; DiMaio D. Cell-penetrating Peptide Inhibits Retromer-mediated Human Papillomavirus Trafficking During Virus Entry. Proc. Natl. Acad. Sci. U. S. A. 2020, 117, 6121–6128. 10.1073/pnas.1917748117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nischan N.; Herce H. D.; Natale F.; Bohlke N.; Budisa N.; Cardoso M. C.; Hackenberger C. P. R. Covalent Attachment of Cyclic TAT Peptides to GFP Results in Protein Delivery into Live Cells with Immediate Bioavailability. Angew. Chem. Int. Ed. Engl. 2015, 54, 1950–1953. 10.1002/anie.201410006. [DOI] [PubMed] [Google Scholar]
- Kadkhodayan S.; Sadat S. M.; Irani S.; Fotouhi F.; Bolhassani A. Generation of GFP Native Protein for Detection of Its Intracellular Uptake by Cell-Penetrating Peptides. Folia Biol. 2016, 62, 103–109. [DOI] [PubMed] [Google Scholar]
- Wang F.; Wang Y.; Zhang X.; Zhang W.; Guo S.; Jin F. Recent Progress of Cell-penetrating Peptides as New Carriers for Intracellular Cargo Delivery. J Control Release 2014, 174, 126–136. 10.1016/j.jconrel.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Sun L.; Xie S.; Qi J.; Liu E.; Liu D.; Liu Q.; Chen S.; He H.; Yang V. C. Cell-Permeable, MMP-2 Activatable, Nickel Ferrite and His-Tagged Fusion Protein Self-Assembled Fluorescent Nanoprobe for Tumor Magnetic-Targeting and Imaging. ACS Appl. Mater. Interfaces 2017, 9, 39209–39222. 10.1021/acsami.7b12918. [DOI] [PubMed] [Google Scholar]
- Gao G.; Jiang Y. W.; Jia H. R.; Sun W.; Guo Y.; Yu X. W.; Liu X.; Wu F. G. From Perinuclear to Intranuclear Localization: A Cell-penetrating Peptide Modification Strategy to Modulate Cancer Cell Migration Under Mild Laser Irradiation and Improve Photothermal Therapeutic Performance. Biomaterials 2019, 223, 119443. 10.1016/j.biomaterials.2019.119443. [DOI] [PubMed] [Google Scholar]
- Ma D.; Zou Y.; Chu Y.; Liu Z.; Liu G.; Chu J.; Li M.; Wang J.; Sun S. Y.; Chang Z. A Cell-permeable Peptide-based PROTAC Against the Oncoprotein CREPT Proficiently Inhibits Pancreatic Cancer. Theranostics 2020, 10, 3708–3721. 10.7150/thno.41677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo H.; Xu C.; Liu Z.; Yang L.; Hong Y.; Liu G.; Zhong H.; Cai X.; Lin X.; Chen X.; Wang C.; Zhang N.; Xu W. Neural Differentiation of Bone Marrow Mesenchymal Stem Cells with Human Brain-derived Neurotrophic Factor Gene-modified in Functionalized Self-assembling Peptide Hydrogel in vitro. J. Cell. Biochem. 2019, 120, 2828–2835. 10.1002/jcb.26408. [DOI] [PubMed] [Google Scholar]
- Quan X.; Sun D.; Zhou J. Molecular Mechanism of HIV-1 TAT Peptide and its Conjugated Gold Nanoparticles Translocating across Lipid Membranes. Phys. Chem. Chem. Phys. 2019, 21, 10300–10310. 10.1039/C9CP01543D. [DOI] [PubMed] [Google Scholar]
- Baumann A. L.; Hackenberger C. P. R. Modern Ligation Methods to Access Natural and Modified Proteins. Chimia 2018, 72, 802–808. 10.2533/chimia.2018.802. [DOI] [PubMed] [Google Scholar]
- Yang F.; Moss L. G.; Phillips G. N. Jr. The Molecular Structure of Green Fluorescent Protein. Nat. Biotechnol. 1996, 14, 1246–1251. 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- Tsukiji S.; Nagamune T. Sortase-mediated Ligation: a Gift from Gram-positive Bacteria to Protein Engineering. ChemBioChem 2009, 10, 787–798. 10.1002/cbic.200800724. [DOI] [PubMed] [Google Scholar]
- Nicholls S. B.; Hardy J. A. Structural basis of fluorescence quenching in caspase activatable-GFP. Protein Sci. 2013, 22, 247–257. 10.1002/pro.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy G.; Liu Z.; Thirumalai D. Denaturant-dependent folding of GFP. Proc. Natl. Acad. Sci. U. S. A. 2012, 109, 17832–17838. 10.1073/pnas.1201808109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik T. N.; Povarnitsyna T. V.; Glukhov A. S.; Uversky V. N.; Melnik B. S. Sequential melting of two hydrophobic clusters within the green fluorescent protein GFP-cycle3. Biochemistry 2011, 50, 7735–7744. 10.1021/bi2006674. [DOI] [PubMed] [Google Scholar]
- Zhang Q.; Tang J.; Fu L.; Ran R.; Liu Y.; Yuan M.; He Q. A pH-responsive Alpha-helical Cell Penetrating Peptide-mediated Liposomal Delivery System. Biomaterials 2013, 34, 7980–7993. 10.1016/j.biomaterials.2013.07.014. [DOI] [PubMed] [Google Scholar]
- Richard J. P.; Melikov K.; Vives E.; Ramos C.; Verbeure B.; Gait M. J.; Chernomordik L. V.; Lebleu B. Cell-penetrating Peptides, a Reevaluation of the Mechanism of Cellular Uptake. J. Biol. Chem. 2003, 278, 585–590. 10.1074/jbc.M209548200. [DOI] [PubMed] [Google Scholar]
- Chen K.; Pei D. Engineering Cell-Permeable Proteins through Insertion of Cell-Penetrating Motifs into Surface Loops. ACS Chem. Biol. 2020, 15, 2568–2576. 10.1021/acschembio.0c00593. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.