Abstract
Coronavirus disease 2019 (COVID-19) is a worldwide pandemic. To understand the changes in plasma proteomics upon SARS-CoV-2 infection, we analyzed the protein profiles of plasma samples from 10 COVID-19 patients and 10 healthy volunteers by using the DIA quantitative proteomics technology. We compared and identified differential proteins whose abundance changed upon SARS-CoV-2 infection. Bioinformatic analyses were then conducted for these identified differential proteins. The GO/KEEG database was used for functional annotation and enrichment analysis. The interaction relationship of differential proteins was evaluated with the STRING database, and Cytoscape software was used to conduct network analysis of the obtained data. A total of 323 proteins were detected in all samples. Difference between patients and healthy donors was found in 44 plasma proteins, among which 36 proteins were up-regulated and 8 proteins were down-regulated. GO functional annotation showed that these proteins mostly composed of cellular anatomical entities and proteins involved in biological regulation, cellular processes, transport, and other processes. KEEG functional annotation further showed that these proteins were mainly involved in complement system activation and infectious disease processes. Importantly, a KEEG pathway (natural killer cell-mediated cytotoxicity) was enriched, with three important activators of this pathway, ICAM1/2 and IgG, being up-regulated. Protein–protein interaction (PPI) statistics indicated that, among these 44 proteins, 6 were the most significantly up-regulated (DBH, SHGB, TF, ICAM2, THBS1, and C1RL) while 2 were the most significantly down-regulated (APCS and ORM1). Results from this study showed that a few proteins associated with immune activation were up-regulated in patient plasma. In addition, this study established a method for extraction and quantitative determination of plasma components in convalescent plasma from COVID-19 patients.
1. Introduction
Coronavirus disease 2019 (COVID-19) is an epidemic disease characterized by acute respiratory distress syndrome caused by SARS-CoV-2. Since its first discovery in Wuhan, it has rapidly spread to 212 countries and regions around the world. Being the first pandemic in the 21st century, by August 4, 2020, the total number of confirmed cases worldwide has exceeded 18 million, with a crude mortality rate of about 3.7%. Based on the severity of the disease, symptoms were divided into mild and severe. Eighty percent of patients presented with mild symptoms such as fever, cough, loss of appetite, dizziness, fatigue, etc. Conventional antiviral therapy and other supportive treatments could effectively alleviate the conditions. Patients with severe symptoms required life-sustaining treatment such as intensive care and mechanical ventilation. At present, a specific antiviral treatment for COVID-19 is very limited. To effectively control the spread of the epidemic, tremendous efforts are being made all over the world in vaccine and medicine research and development for prevention and treatment of this disease.
SARS-CoV-2 and SARS-CoV share 79.5% sequence homology, and the mechanisms of infection and pathogenesis of them are similar.1 SARS-CoV-2 binds to host cells using the spike protein (S protein) on the surface of the virus envelope and mediates the fusion of the host cell and virus during the infection process.2 SARS-CoV-2 has one polyprotein (Orf1ab), four structural proteins (S, E, M, and N), and six accessory proteins (Orf3a, Orf6, Orf7a, Orf7b, Orf8, and Orf10) involved in virus replication and transcription. The interaction between the S protein receptor binding domain (RBD) of SARS-CoV-2 and the ACE2 (angiotensin converting enzyme 2) molecule was a necessary process for SARS-CoV-2 to invade host cells.3,4
Some studies have investigated the vulnerability of medical staff in the COVID-19 epidemic. Korth et al. detected SARS-CoV-2-IgG antibodies in 1.6% of 316 health care workers, suggesting that the medical staff were vulnerable to SARS-CoV-2 infection.5 Another study in the United States found that 1.5% of 3500 medical workers were positive for SARS-CoV-2 antibodies.6 In addition to conventional antiviral therapy and basic supportive therapy, convalescent plasma therapy was considered a promising treatment approach. Historically, this approach has been adopted for a variety of infectious diseases. The Spanish flu in the 1920s was the first viral infection to be treated effectively with convalescent plasma. Convalescent plasma therapy was found to be effective in treating SARS in 2003, the Middle East respiratory syndrome in 2014, Ebola in 2015, and some other viral infectious diseases.7−9 Convalescent plasma contains neutralizing antibodies, natural antibodies, and anti-inflammatory factors, clotting factors, and other proteins of uncertainty.10 As such, convalescent plasma not only directly neutralizes the virus but also reduces inflammatory response, which mediates inflammatory storm in COVID-19 patients.11 Analyzing the plasma protein components of COVID-19 patients may provide insights into body response to SARS-CoV-2 infection and will have profound implication to vaccine and therapeutic research and development against COVID-19. In this study, we quantitatively analyzed the plasma protein profiles of COVID-19 patients and healthy subjects and identified protein components, which showed different abundance between the two groups. Results from this work demonstrated that multiple factors involved in humoral and cellular immune responses were increased in patient plasma, suggesting that both humoral and cellular immune responses may be required for fighting SARS-CoV-2 infection.
2. Material and Methods
2.1. Subjects
Inclusion criteria: (i) aged over 18 years; (ii) SARS-CoV-2 infection was confirmed as respiratory tract specimens by real-time reverse transcription polymerase chain reaction (RT-PCR); (iii) the severity of the disease is classified as mild according to the COVID-19 guidelines (Trial Version 7).12
Exclusion criteria: (i) under the age of 18; (ii) patients with severe cardiopulmonary disease, immunodeficiency syndrome, malignant tumor, or mental abnormalities who were unable to cooperate.
Ten patients diagnosed with COVID-19 in the Union Hospital and Liyuan Hospital of Tongji Medical College of Huazhong University of Science and Technology from January to April 2020 were included in the case group, and 10 healthy subjects were included in the control group. All investigators were informed of the purpose of the study and precautions that should be taken. All participants in this study signed written informed consent forms. This study was approved by the ethics committee of the respective hospitals.
2.2. Preparation of Plasma Samples
Fasting venous blood of patients and healthy subjects was collected in the morning. Reaction solution (1% SDC, 10 mM TCEP, 40 mM CAA) was added to the plasma samples and incubated at 60 °C for 30 min for protein denaturation disulfide bond reduction as well as sulfhydryl alkylation. Protein concentration in samples was determined using the Bradford assay. To conduct enzymatic digestion for the samples, the concentration of SDC was diluted to less than 0.5%, trypsin was added to reach a mass ratio of 1:50 between the enzyme and protein, and the samples were incubated at 37 °C overnight. On the following day, TFA was added to terminate the enzymatic digestion, and the pH value of the solution was adjusted to about 6.0. The solution was centrifuged at 12,000g for 15 min, and the supernatant was taken for desalting. After desalination, the peptide solution was filtered by using centrifugal concentrators and stored at −20 °C until being analyzed.
2.3. Data-Independent Acquisition (DIA) Analysis
DIA technology was used for mass spectrometry analysis. The analysis was conducted on an Ultimate3000 (capillary flow) combined with a Q Exactive HF-X. While conducting a SWATH scan, the scan of each cycle contained one MS1 scan (scan range, 350–1250 m/z; resolution, 60 K; AGC, 3 × 106; max interval time (IT), 20 ms) and 40 variable window MS2 scans (resolution, 30 K; AGC, 1 × 106; max IT, auto). Secondary mass spectrometry files obtained from the DIA scan were processed by DIA-Umpire, which could be used for database retrieval. The MSFragger software was used for database retrieval of the secondary mass spectrogram, and the results obtained were used as the spectrogram library. Quantitative algorithm DIA-NN was used for subsequent DIA-targeted extraction. Quantitative information for components obtained by DIA analysis was screened by a 1% false discovery rate (FDR), the normalized data was converted to log2 data, which was then used for difference comparison and t test analysis, and the fold change ratio and P value were used to screen for protein components, which showed different abundance between patients and the control group. Differential protein was screened by the ratio and P value. Screening criteria: ① if fold change ≥ 1.2 and P ≤ 0.05, then the protein was over-represented; ② if fold change ≤ 0.833 and P ≤ 0.05, then the protein was under-represented.
2.4. Bioinformatics Analysis
The GO/KEEG database was used for functional annotation and enrichment analysis. The interaction relationship of differential proteins was evaluated with the STRING database, and Cytoscape software was used to conduct network analysis of the obtained data.
2.5. Statistical Analysis
Continuous variables were represented by mean ± standard deviation. The percentage of categorical variables was used to compare the difference between case and control groups. The chi-square test or Fisher test was used to compare categorical variables between the two groups. SPSS Version 24 (Chicago, IL, USA) was used for all statistical analyses. P < 0.05 was considered statistically significant.
3. Results
3.1. Basic Patient Information
Basic data for all patients and healthy individuals included in the study are shown in Table 1. There were four males in the case group. The average age of this group was 62.8 ± 14.597, and six patients in this group had basic diseases including diabetes, hypertension, and kidney stones. All 10 patients had mild symptoms. There were four males in the control group. The average age of the control group was 50.7 ± 14.103. There was a statistically significant difference in the percentage of lymphocytes between the two groups (P = 0.02). No significant difference in age, gender, and other blood test results was found between the two groups (P > 0.05).
Table 1. Baseline Characteristics of Patients Included in the Study.
| subject/group | case group (mean ± SD) | control group (mean ± SD) | P value |
|---|---|---|---|
| average age | 62.8 ± 14.597 | 50.7 ± 14.103 | 0.076 |
| sex/men/n | 4 | 5 | 0.395 |
| complications/n | 6 | ||
| severity (light/severe) | light | ||
| WBC × 109 | 6.44 ± 1.810 | 6.53 ± 1.443 | 0.909 |
| Lym % | 26.07 ± 11.393 | 36.64 ± 6.337 | 0.02 |
| Neu % | 61.08 ± 13.376 | 60.05 ± 8.268 | 0.838 |
| RBC × 1012 | 3.79 ± 0.907 | 4.42 ± 0.546 | 0.078 |
| PLT × 109 | 187.7 ± 77.424 | 227.6 ± 41.078 | 0.167 |
| ALT (u/L) | 83.01 ± 192.283 | 26.98 ± 15.589 | 0.371 |
| AST (μ/L) | 58.84 ± 105.445 | 26.1 ± 7.790 | 0.34 |
| TBIL (μmol/L) | 19.26 ± 14.527 | 9.6 ± 5.039 | 0.064 |
| BUN (μmol/L) | 7.4 ± 5.609 | 5.8 ± 1.755 | 0.41 |
| Crea (μmol/L) | 101.39 ± 108.967 | 63.16 ± 16.568 | 0.287 |
| PCT (ng/mL) | 0.49 ± 1.206 | 0.03 ± 0.026 | 0.258 |
| LDH (μ/L) | 245.6 ± 91.504 | 201.3 ± 46.070 | 0.188 |
3.2. Proteomics Analysis
A total of 323 plasma proteins were detected in both groups, and the quantitative values for all proteins are shown in Table 1. Proteins with fold change ≥ 1.2 and P ≤ 0.05 were classified as up-regulated proteins, and proteins with fold change ≤ 0.833 and P ≤ 0.05 were classified as down-regulated proteins. The quantitative correlation for proteins in each sample was good, and the quantitative correlation coefficient R2 values of different samples were all greater than 0.9 (Figure 1). The ratio distribution of the case group is roughly normal (Figure 2). A total of 44 differential proteins were detected between the two groups, of which 36 were up-regulated and 8 were down-regulated (Table 2 and Figures 3 and 4).
Figure 1.

Quantitative correlation analysis of sample protein.
Figure 2.
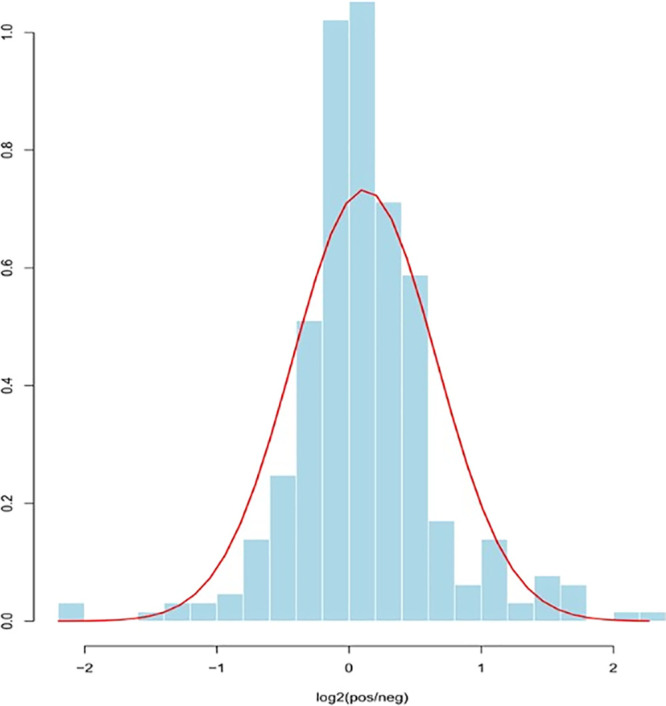
Histone ratio distribution of cases.
Table 2. Differential Protein Information.
| protein names | protein description | FC (pos/neg) | log2FC (pos/neg) | P value | state |
|---|---|---|---|---|---|
| HV5X1_HUMAN | immunoglobulin heavy variable 5-10-1 | 4.82 | 2.26872 | 0.005737 | up |
| GPV_HUMAN | platelet glycoprotein V | 4.17 | 2.05876 | 0.001849 | up |
| SHBG_HUMAN | sex hormone-binding globulin | 3.32 | 1.7328 | 0.006005 | up |
| TSP1_HUMAN | thrombospondin-1 | 3.11 | 1.63661 | 0.015421 | up |
| DOPO_HUMAN | dopamine β-hydroxylase | 2.82 | 1.49417 | 0.003681 | up |
| ICAM2_HUMAN | intercellular adhesion molecule 2 | 2.66 | 1.40923 | 0.005953 | up |
| KV621_HUMAN | immunoglobulin kappa variable 6-21 | 2.34 | 1.22735 | 0.02518 | up |
| LV469_HUMAN | immunoglobulin lambda variable 4-69 | 2.26 | 1.17911 | 0.02029 | up |
| ARAP1_HUMAN | Arf-GAP with Rho-GAP domain, ANK repeat and PH domain-containing protein 1 | 2.17 | 1.11569 | 0.041246 | up |
| S10AC_HUMAN | protein S100-A12 | 2.16 | 1.11386 | 0.005249 | up |
| HV205_HUMAN | immunoglobulin heavy variable 2-5 | 2.06 | 1.04224 | 0.024872 | up |
| LV325_HUMAN | immunoglobulin lambda variable 3-25 | 2.02 | 1.01292 | 0.008503 | up |
| HV43D_HUMAN | immunoglobulin heavy variable 3-43D | 1.93 | 0.94639 | 0.012763 | up |
| TRFE_HUMAN | serotransferrin | 1.79 | 0.83607 | 0.046888 | up |
| CRAC1_HUMAN | cartilage acidic protein 1 | 1.63 | 0.70302 | 0.010019 | up |
| KVD15_HUMAN | immunoglobulin kappa variable 3D-15 | 1.61 | 0.69054 | 0.007783 | up |
| KV320_HUMAN | immunoglobulin kappa variable 3-20 | 1.57 | 0.651 | 0.012032 | up |
| HV323_HUMAN | 1.57 | 0.64824 | 0.012064 | up | |
| C1RL_HUMAN | complement C1r subcomponent-like protein | 1.56 | 0.64178 | 0.002045 | up |
| HVD82_HUMAN | immunoglobulin heavy variable 4-38-2 | 1.54 | 0.62082 | 0.017265 | up |
| HV118_HUMAN | immunoglobulin heavy variable 1-18 | 1.53 | 0.61393 | 0.046497 | up |
| CETP_HUMAN | cholesteryl ester transfer protein | 1.47 | 0.55667 | 0.029024 | up |
| A2MG_HUMAN | alpha-2-macroglobulin | 1.44 | 0.52321 | 0.015519 | up |
| IGHG2_HUMAN | immunoglobulin heavy constant gamma 2 | 1.43 | 0.51655 | 0.022341 | up |
| KV108_HUMAN | immunoglobulin kappa variable 1-8 | 1.40 | 0.48435 | 0.02486 | up |
| AFAM_HUMAN | afamin | 1.39 | 0.47767 | 0.041463 | up |
| HV428_HUMAN | immunoglobulin heavy variable 4-28 | 1.38 | 0.46815 | 0.034601 | up |
| PLTP_HUMAN | phospholipid transfer protein | 1.38 | 0.46297 | 0.00479 | up |
| TENX_HUMAN | tenascin-X | 1.38 | 0.46195 | 0.031929 | up |
| KV106_HUMAN | 1.35 | 0.43012 | 0.037791 | up | |
| KV37_HUMAN | probable non-functional immunoglobulin kappa variable 3-7 | 1.33 | 0.4118 | 0.013726 | up |
| IGG1_HUMAN | 1.31 | 0.3898 | 0.008494 | up | |
| PGRP2_HUMAN | N-acetylmuramoyl-l-alanine amidase | 1.30 | 0.37717 | 0.011929 | up |
| APOA1_HUMAN | apolipoprotein A-I | 1.29 | 0.36524 | 0.02888 | up |
| IGKC_HUMAN | immunoglobulin kappa constant | 1.28 | 0.35081 | 0.011717 | up |
| C1QA_HUMAN | complement C1q subcomponent subunit A | 1.27 | 0.34757 | 0.025015 | up |
| C4BPA_HUMAN | C4b-binding protein alpha chain | 0.77 | –0.38011 | 0.00728 | down |
| CERU_HUMAN | ceruloplasmin | 0.76 | –0.38873 | 0.009802 | down |
| SAA4_HUMAN | serum amyloid A-4 protein | 0.70 | –0.51451 | 0.000425 | down |
| IC1_HUMAN | plasma protease C1 inhibitor | 0.68 | –0.56084 | 0.007387 | down |
| LG3BP_HUMAN | galectin-3-binding protein | 0.66 | –0.60309 | 0.018873 | down |
| LBP_HUMAN | lipopolysaccharide-binding protein | 0.59 | –0.77189 | 0.029039 | down |
| A1AG1_HUMAN | alpha-1-acid glycoprotein 1 | 0.58 | –0.78207 | 0.018007 | down |
| SAMP_HUMAN | serum amyloid P component | 0.48 | –1.06577 | 0.001006 | down |
Figure 3.
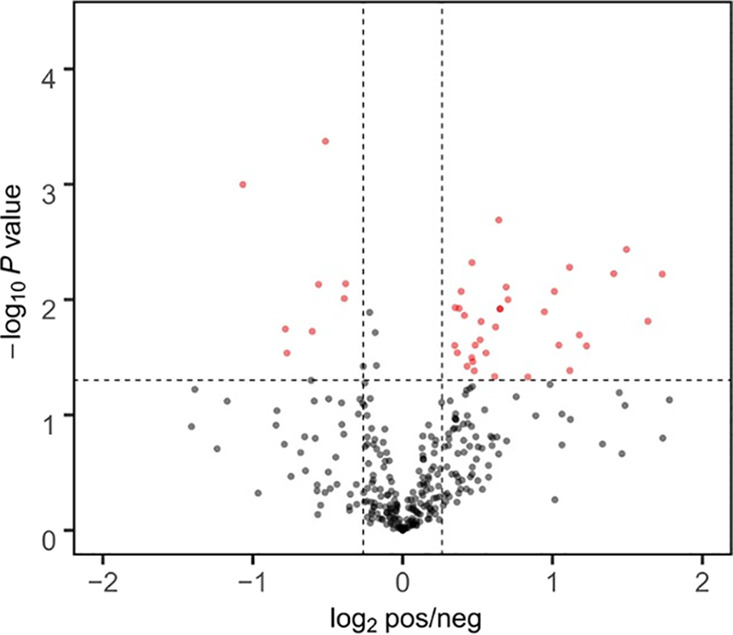
Volcano figure.
Figure 4.
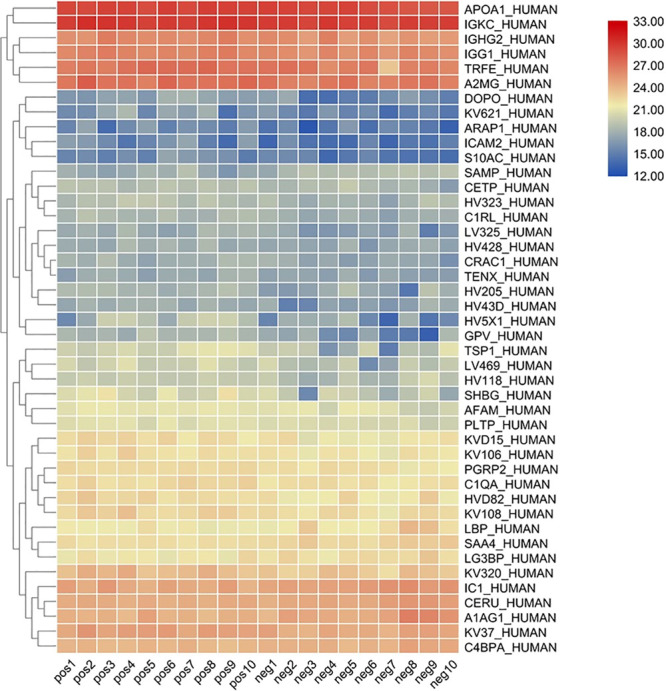
Clustering heat map of all proteins.
3.3. GO Functional Annotation and Enrichment Analysis
Differential proteins identified were annotated as three GO levels and three categories, namely, the biological process (BP), cellular component (CC), and molecular function (MF) (Figure 5A–C, respectively). In the BP category, differential proteins in the patient group were mainly involved in biological regulation (13.06%), cellular processes (12.16%), and the response to stimulation (10.36%). In the CC category, 39.24% belonged to the anatomical entity of the cell, 27.85% belonged to the cell, and 18.3% belonged to the intracellular. In the MF category, 53.49% had a binding function, 18.6% were molecular function regulators, and 11.63% had catalytic activity. The hypergeometric test was used to analyze the functional classification or pathway in which differentially represented proteins were significantly enriched compared with the background proteins (the total number of proteins with quantitative and annotated information) (P ≤ 0.05). This case group did not have the GO classification information enriched.
Figure 5.
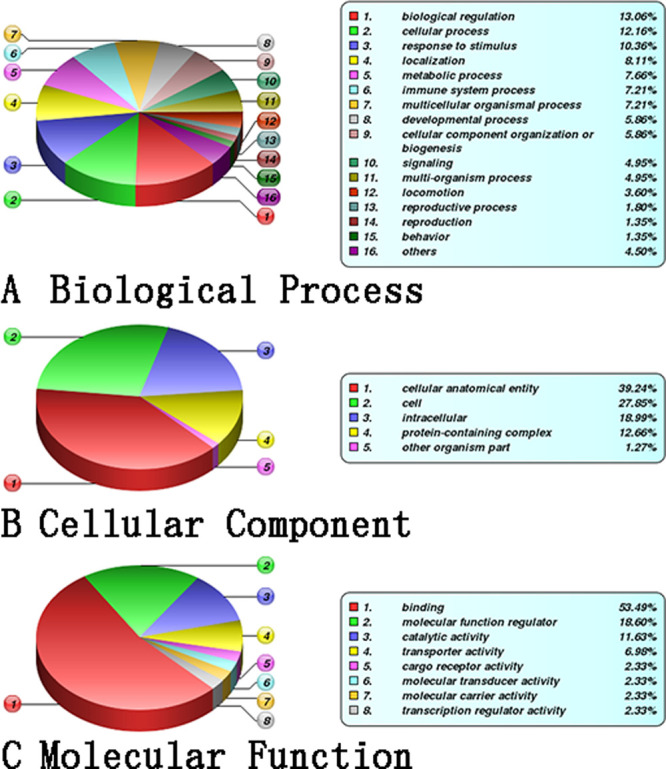
GO annotation and enrichment analysis. (A) Biological process, (B) cellular component, and (C) molecular function.
3.4. KEEG Functional Annotation and Enrichment Analysis
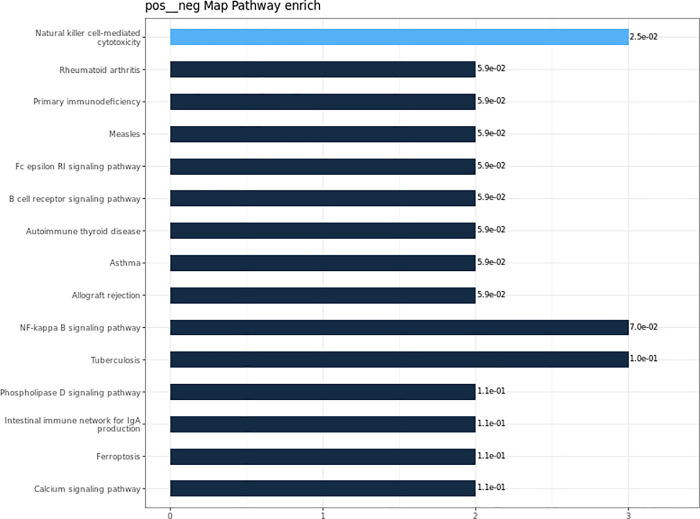
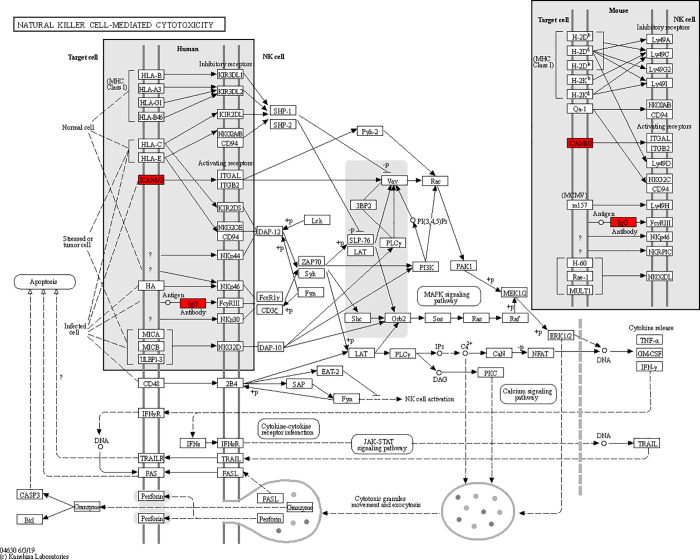
To better understand the functions of the identified differential proteins in physiological or pathological activities and to determine the metabolic and signaling pathways involved, we carried out KEEG pathway analysis for the differential proteins. Our analyses showed that the differential proteins were mainly involved in the following biological process: NK cell-mediated primary immunodeficiency NF-κB signaling pathway tuberculosis in rheumatoid arthritis (Figure 6). The pathway with the highest KEEG enrichment was natural killer cell-mediated cytotoxicity (Figure 7).
Figure 6.
KEGG annotation and enrichment analysis.
Figure 7.
Enrichment KEGG pathway diagram.
3.5. PPI Network Construction
The interaction relationship of differential proteins was evaluated with the STRING database, and Cytoscape software was used to conduct network analysis of the obtained data (Figure 8). Each node in the figure represented a protein, and the attachment between nodes represented the interaction between proteins. Red represented up-regulation, blue represented down-regulation, and the darker the color, the larger the variation multiple of the protein. As shown in the figure, over-represented proteins with significant changes included DBH (dopamine β-hydroxylase), SHBG (sex hormone-binding globulin), TF (serum transferrin), THBS1 (thrombospondin-1), ICAM2 (intercellular adhesion molecule 2), and C1RL (complement C1r subcomponent-like protein). Two proteins, namely, APCS (serum amyloid P component) and ORM1 (alpha-1-acid glycoprotein 1) were significantly under-represented.
Figure 8.

Protein interaction network analysis.
4. Discussion
With the spread of COVID-19 and the urgent need for treatment and prevention options, a systematic study of virus–host interactions is critically important. In a recent study, Gordon et al. reported the SARS-CoV-2 protein interaction map and identified 66 human proteins or cytokines as potential therapeutic targets for COVID-19.13 As is well known, after viruses invade host cells, the body undergoes a series of antiviral reactions, including inflammatory cell aggregation, cytokine release, humoral immunity, and cellular immune activation. Among these factors, antiviral antibodies play a very critical role. By detecting IgM and IgG in the sera of 29 COVID-19 patients during convalescence, Jiang et al.14 constructed a proteomics microarray containing 18 predicted viral proteins and characterized the profile of SARS-CoV-2-specific serum antibodies, especially antibodies against S1 and N proteins.15 Long et al. analyzed the acute antibody response in 285 COVID-19 patients, and the results showed that 100% of patients tested positive for SARS-CoV-2 virus IgG within 19 days from the symptom onset.16 These studies characterized direct interactions between viral and host proteins and the antibody response to viral infection. However, a systematic investigation of change in protein components in patient serum as a response to SARS-CoV-2 infection was never reported yet. In this study, a total of 44 differential proteins were identified in plasma samples from COVID-19 patients with 36 being over-represented and 8 under-represented in patient samples in comparison with control samples.
Through GO functional annotation, it was found that these proteins were mostly cellular anatomical entities and factors involved in biological regulation, cell process, and transportation. KEEG functional annotation further revealed that these differential proteins were mainly involved in the complement system activation, pertussis, and other infectious diseases. Notably, members of the pathway of natural killer cell-mediated cytotoxicity were enriched, as evidenced by the finding that ICAM1/2 and IgG were all over-represented in patient samples. ICAM1/2 participates in the NF-κB signaling pathway, natural killer cell-mediated cytotoxicity, the TNF signaling pathway, the interaction between signaling molecules, leukocyte migration across the endothelium, etc.17,18 PPI interaction analysis suggested that SHBG participates in receptor-mediated processes; TF may play a role in stimulating cell proliferation; THBS1 mediates intracellular and intercellular interactions. APCS can interact with DNA and histones. ORM1 may play a role in regulating the activity of the immune system. These factors can directly or indirectly block viral invasion and replication and other processes, which are of great significance for virus defense.19
Plasma phospholipid transfer protein (PLTP), a member of the lipid transfer/lipopolysaccharide binding protein gene family, is a major regulator of lipoprotein metabolism and also has the function of regulating inflammation and immune response. PLTP has a specific effect on Th1/Th2 polarization. Lack of PLTP will reduce the expression of IL-18, and antigen-presenting cells cannot induce Th1 cell differentiation due to lack of IL-18, thereby affecting the immune function of the body.20 Ceruloplasmin is a protein that maintains iron and copper homeostasis in the body. It also has the functions of copper transport, blood coagulation, angiogenesis, and oxidative stress. Down-regulation of this protein has been observed in many viral infections.21 Cholesteryl ester transfer protein (CETP) can mediate the exchange of CE in HDL to TG in other lipoproteins.22 It is speculated that the up-regulation of CETP may be due to the important role of lipids in virus infection. Lipids participate in membrane fusion, envelope, and transformation as direct or indirect virus receptors, fusion cofactors, and entrance cofactors to help viruses invade host cells.23,24 Study has suggested that the lipid may play important roles in the virus life cycle and can be potential targets for therapeutic intervention of COVID-19.25
In this study, 44 differential proteins were identified, among which 15 belonged to the immunoglobulin superfamily.
There are some limitations in this study. On the one hand, the sample size of the study was relatively small, which might lead to selection bias in the results. On the other hand, the differential proteins identified were not confirmed with alternative assays. Therefore, in the future, studies with larger sample sizes may be conducted if conditions allow, and differential proteins identified will need to be confirmed by other analytical methods.
Acknowledgments
We are grateful to all the medical staff who worked on the front line to fight the epidemic.
Author Contributions
# X.L. and Y.C. contributed equally to this work.
The authors declare no competing financial interest.
References
- Zhou P.; Yang X.; Wang X.; Hu B.; Zhang L.; Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020, 579, 270–273. 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F. Structure, Function, and Evolution of Coronavirus Spike Proteins. Annu. Rev. Virol. 2016, 3, 237–261. 10.1146/annurev-virology-110615-042301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang J.; Ye G.; Shi K.; Wan Y.; Luo C.; Aihara H. Structural basis of receptor recognition by SARS-CoV-2. Nature 2020, 581, 221–224. 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S.; Cui H.; Gao Z.; Liu M.; Lu S.; Mkandawire W. Structural Genomics of SARS-CoV-2 Indicates Evolutionary Conserved Functional Regions of Viral Proteins. Viruses 2020, 12, 360. 10.3390/v12040360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth J.; Wilde B.; Dolff S.; Anastasiou O. E.; Krawczyk A.; Jahn M. SARS-CoV-2-specific antibody detection in healthcare workers in Germany with direct contact to COVID-19 patients. J. Clin. Virol. 2020, 128, 104437. 10.1016/j.jcv.2020.104437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendavid E.; Mulaney B.; Sood N.; Shah S.; Ling E.; Bromley-Dulfano R. COVID-19 Antibody Seroprevalence in Santa Clara County, California. medRxiv 2020, 10.1101/2020.04.14.20062463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo Y. O. Y.; Cheng Y.; Wong R.; Hui D. S.; Lee C. K.; Tsang K. K. S. Retrospective comparison of convalescent plasma with continuing high-dose methylprednisolone treatment in SARS patients. Clin. Microbiol. Infect. 2004, 10, 676–678. 10.1111/j.1469-0691.2004.00956.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi Y. M.; Hajeer A. H.; Luke T.; Raviprakash K.; Balkhy H.; Johani S. Feasibility of Using Convalescent Plasma Immunotherapy for MERS-CoV Infection, Saudi Arabia. Emerging Infect. Dis. 2016, 22, 1554–1561. 10.3201/eid2209.151164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Griensven J.; Edwards T.; de Lamballerie X.; Semple M. G.; Gallian P.; Baize S. Evaluation of Convalescent Plasma for Ebola Virus Disease in Guinea. N. Engl. J. Med. 2016, 374, 33–42. 10.1056/NEJMoa1511812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraud O.; Heshmati F.; Pozzetto B.; Lefrere F.; Girot R.; Saillol A. Plasma therapy against infectious pathogens, as of yesterday, today and tomorrow. Clin. Biol. Trans. 2016, 23, 39–44. 10.1016/j.tracli.2015.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lünemann J. D.; Nimmerjahn F.; Dalakas M. C. Intravenous immunoglobulin in neurology—mode of action and clinical efficacy. Nat. Rev. Neurol. 2015, 11, 80–89. 10.1038/nrneurol.2014.253. [DOI] [PubMed] [Google Scholar]
- ″New Coronavirus Pneumonia Diagnosis and Treatment Plan (Trial Seventh Edition)″ National Health Commission. Journal of Practical Traditional Chinese Medicine.2020;34:3.
- Gordon D. E.; Jang G. M.; Bouhaddou M.; Xu J.; Obernier K.; White K. M. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468. 10.1038/s41586-020-2286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H.; Li Y.; Zhang H.; Wang W.; Yang X.; Qi H. SARS-CoV-2 proteome microarray for global profiling of COVID-19 specific IgG and IgM responses. Nat. Commun. 2020, 11, 3581. 10.1038/s41467-020-17488-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S.; Hillyer C.; Du L. Neutralizing Antibodies against SARS-CoV-2 and Other Human Coronaviruses. Trends Immunol. 2020, 41, 355–359. 10.1016/j.it.2020.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q.; Liu B.; Deng H.; Wu G.; Deng K.; Chen Y. Antibody responses to SARS-CoV-2 in patients with COVID-19. Nat. Med. 2020, 26, 845–848. 10.1038/s41591-020-0897-1. [DOI] [PubMed] [Google Scholar]
- Lyck R.; Enzmann G. The physiological roles of ICAM-1 and ICAM-2 in neutrophil migration into tissues. Curr. Opin. Hematol. 2015, 22, 53–59. 10.1097/MOH.0000000000000103. [DOI] [PubMed] [Google Scholar]
- Akella R.; Hall R. E. Expression of the adhesion molecules ICAM-1 and ICAM-2 on tumor cell lines does not correlate with their susceptibility to natural killer cell-mediated cytolysis: evidence for additional ligands for effector cell beta integrins. Eur. J. Immunol. 1992, 22, 1069. 10.1002/eji.1830220429. [DOI] [PubMed] [Google Scholar]
- Wack A.; Openshaw P.; Garra O. A. Contribution of cytokines to pathology and protection in virus infection. Curr. Opin. Virol. 2011, 1, 184–195. 10.1016/j.coviro.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Desrumaux C.; Lemaire-Ewing S.; Ogier N.; Yessoufou A.; Hammann A.; Sequeira-Le Grand A. Plasma phospholipid transfer protein (PLTP) modulates adaptive immune functions through alternation of T helper cell polarization. Cell. Mol. Immunol. 2016, 13, 795–804. 10.1038/cmi.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallianpur A. R.; Gittleman H.; Letendre S.; Ellis R.; Barnholtz-Sloan J. S.; Bush W. S. Cerebrospinal Fluid Ceruloplasmin, Haptoglobin, and Vascular Endothelial Growth Factor Are Associated with Neurocognitive Impairment in Adults with HIV Infection. Mol. Neurobiol. 2019, 56, 3808–3818. 10.1007/s12035-018-1329-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh K. High level of serum cholesteryl ester transfer protein in active hepatitis C virus infection. World J. Hepatol. 2016, 8, 291. 10.4254/wjh.v8.i5.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo A.; Vera-Estrella R.; Barkla B. J.; Méndez E.; Arias C. F. Identification of Host Cell Factors Associated with Astrovirus Replication in Caco-2 Cells. J. Virol. 2015, 89, 10359–10370. 10.1128/JVI.01225-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorizate M.; Krausslich H. G. Role of Lipids in Virus Replication. Cold Spring Harbor Perspect. Biol. 2011, 3, a004820. 10.1101/cshperspect.a004820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abu-Farha M.; Thanaraj T. A.; Qaddoumi M. G.; Hashem A.; Abubaker J.; Al-Mulla F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int. J. Mol. Sci. 2020, 21, 3544. 10.3390/ijms21103544. [DOI] [PMC free article] [PubMed] [Google Scholar]