Abstract
Objective
Identifying symptoms and characteristics highly specific to coronavirus disease 2019 (COVID-19) would improve the clinical and public health response to this pandemic challenge. Here, we describe a high-throughput approach – Concept-Wide Association Study (ConceptWAS) – that systematically scans a disease's clinical manifestations from clinical notes. We used this method to identify symptoms specific to COVID-19 early in the course of the pandemic.
Methods
We created a natural language processing pipeline to extract concepts from clinical notes in a local ER corresponding to the PCR testing date for patients who had a COVID-19 test and evaluated these concepts as predictors for developing COVID-19. We identified predictors from Firth's logistic regression adjusted by age, gender, and race. We also performed ConceptWAS using cumulative data every two weeks to identify the timeline for recognition of early COVID-19-specific symptoms.
Results
We processed 87,753 notes from 19,692 patients subjected to COVID-19 PCR testing between March 8, 2020, and May 27, 2020 (1,483 COVID-19-positive). We found 68 concepts significantly associated with a positive COVID-19 test. We identified symptoms associated with increasing risk of COVID-19, including “anosmia” (odds ratio [OR] = 4.97, 95% confidence interval [CI] = 3.21–7.50), “fever” (OR = 1.43, 95% CI = 1.28–1.59), “cough with fever” (OR = 2.29, 95% CI = 1.75–2.96), and “ageusia” (OR = 5.18, 95% CI = 3.02–8.58). Using ConceptWAS, we were able to detect loss of smell and loss of taste three weeks prior to their inclusion as symptoms of the disease by the Centers for Disease Control and Prevention (CDC).
Conclusion
ConceptWAS, a high-throughput approach for exploring specific symptoms and characteristics of a disease like COVID-19, offers a promise for enabling EHR-powered early disease manifestations identification.
Keywords: COVID-19, EHR, Natural language processing
1. Introduction
As of October 14, 2020, over 7.7 million people in the United States (U.S.) and 37 million worldwide have been infected with coronavirus SARS-CoV-2, the agent responsible for COVID-19 [1]. The virus's high transmissibility, lack of native immunity, high mutability, and the dearth of effective treatments make managing COVID-19 uniquely challenging. Hence, timely recognition of emerging symptoms specific to COVID-19 plays an essential role in the clinical and public health response, enabling rapid symptom screening, diagnostic testing, and contact tracing.
Early in the pandemic, physicians observed fever, cough, and shortness of breath as presenting symptoms of COVID-19; however, these symptoms are common to many viral and bacterial illnesses [2]. Subsequently, as new symptoms were reported, health departments and ministries updated the list of COVID-19 symptoms [3]; for example, the U.S. CDC and the Department of Health and Social Care in the United Kingdom added loss of smell and loss of taste, highly indicative symptoms [4], to the list in late April and mid-May, respectively [5], [6]. As demonstrated by the COVID-19 pandemic, identifying the specific disease symptoms early in the course of the pandemic is crucial to inform the public on when to present for testing and can potentially be used to reduce the size of the outbreak, lowering overall morbidity and mortality.
Recent efforts to track COVID-19 symptoms have used methods such as scanning scientific publications or Twitter [7], [8], deploying questionnaires [9], or releasing apps to self-report symptoms [10]. However, results from publications and questionnaires are often delayed; data from social media or self-reported apps do not always include proper controls and lack physiological assessments to determine COVID-19 status. Electronic Health Records (EHR) data has also been used to characterize COVID-19, due to the availability of routinely collected medical data. However, existing studies of EHRs have been mostly limited to structured data (e.g., coded diagnoses, procedures, or lab tests) [11], [12] and have lacked a portable and high-throughput approach [13].
Here, we present a high-throughput approach (ConceptWAS) for early identification of clinical manifestations of COVID-19 using natural language processing (NLP) on EHR clinical notes. ConceptWAS was modeled after the methodology of genome-wide association studies (GWAS) [14], which scan the genomes from different people to identify genetic markers that can be used to predict the presence of a disease, and phenome-wide association studies (PheWAS) [15], which operate in reverse to GWAS by screening thousands of diagnosis codes in EHR for a given genetic variant. Numerous studies have applied GWAS and PheWAS to reveal the inheritance patterns of various diseases [16]. However, unstructured EHR data, in particular clinical notes, are a rich but underutilized EHR resource, containing detailed descriptions of patients’ signs or symptoms, medical histories, and progression [17].Yet, using clinical notes to systematically identify the symptoms and clinical characteristics of a pandemic disease has been largely untapped.
In this study, we used ConceptWAS to identify the symptoms and clinical characteristics associated with COVID-19. In particular, we performed serial ConceptWAS analyses using every 2-week cumulative data to demonstrate the time course of emerging clinical manifestations. We also conducted a chart review to validate the significant associations.
2. Methods
2.1. Study setting
The study was performed at Vanderbilt University Medical Center (VUMC), one of the largest primary care and referral health systems serving over one million patients annually from middle Tennessee and the Southeast United States. We used data from patients represented in the VUMC EHR aged ≥ 18 years. The study was approved by the VUMC Institutional Review Board (IRB #200512).
2.2. Cohort definition
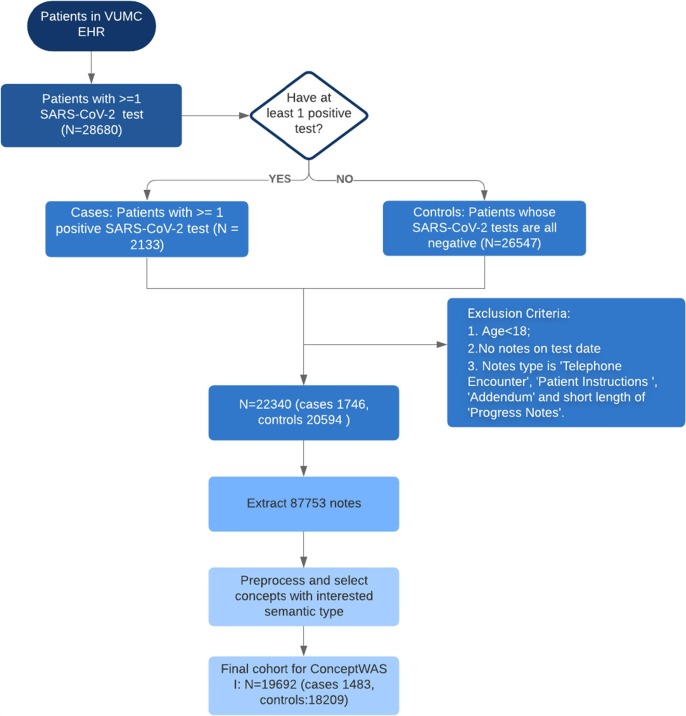
We identified patients who received at least one SARS-CoV-2 polymerase chain reaction (PCR) test between March 8 (when the first COVID-19 case emerged at VUMC) and May 27, 2020 (Fig. A1). The COVID-19 status was determined using the PCR test result. The case group (COVID-19-positive) was defined as patients who had >=1 PCR positive result, and the control group (COVID-19-negative) consisted of patients with only negative PCR tests. We excluded patients who had no clinical notes on the day when the PCR test was ordered.
Fig. A1.
Flowchart of study design for ConceptWAS between COVID-19-positive (case) and COVID-19 negatives (control).
2.3. Data collection
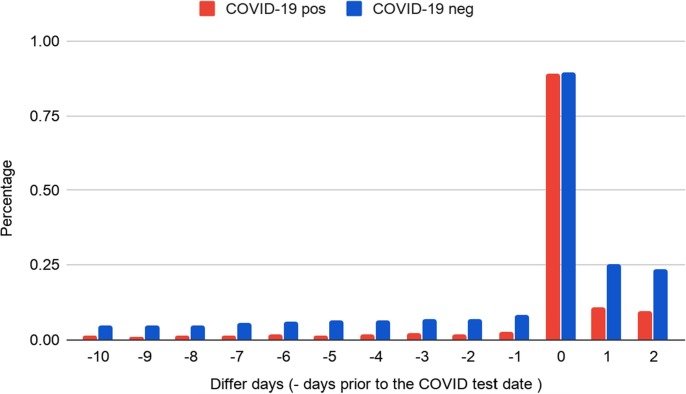
We extracted clinical notes from 24 h prior to PCR testing date (day0) for the cohort (>86% of patients had at least one note within the time window, see Fig. B1). If a patient first tested negative and then subsequently tested positive or if a patient tested positive more than once, we used the date of the first positive PCR test as day0. We also segmented the study period into a 2-week interval window and performed a temporal analysis using every 2-week cumulative data. The primary types of clinical notes that we extracted included progress notes, problem lists, Emergency Department (ED) provider notes, ED triage notes, imaging reports, social histories, etc., (full list is shown in Table B.1).
Fig. B1.
Proportion of cases/controls with clinical notes on the days around COVID-19 test date. The x-axis indicates the note day relative to the COVID-19 test date. > 86% patients who have a PCR test had a clinical note within 24 h before the test date.
2.4. Concept extraction
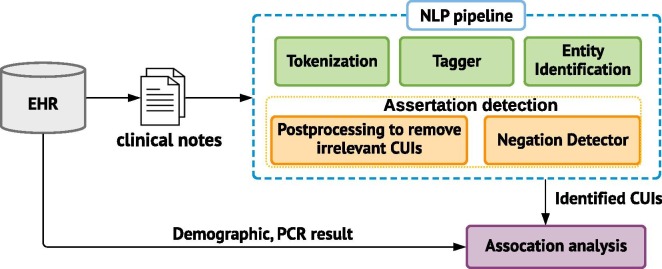
We used KnowledgeMap Concept Indexer (KMCI [18]) to extract concepts (Fig. C1). The KMCI is an NLP pipeline developed at VUMC for preprocessing medical notes and entity recognition, which has been used for several clinical and genomic studies [18], [19], [20]. The preprocessing includes sentence boundary detection, tokenization, part-of-speech tagging, section header identification. The concepts were represented as Unified Medical Language System concept unique identifiers (UMLS CUIs). Since we focused on capturing clinical manifestations of COVID-19, we restricted the concepts to SNOMED Clinical Terms and a specific range of semantic types, e.g., finding, sign or symptom, disease or syndrome, individual behaviors, or mental process (see full list in Table C.1).
Fig. C1.
Schematic framework of the ConceptWAS and NLP pipeline.
2.5. Assertion and negation detection
A main challenge of clinical NLP is to accurately detect the clinical entities' assertion modifier such as negated, uncertain, and hypothetical information (e.g. describe a future hypothetical or instruction for patients). We took the following steps to post-process the KMCI output to remove concepts that appear in sentences reflecting uncertainty and theoretical thoughts. We first excluded any concepts that arose from family history sections. Next, we removed any sentences with future tense or subjunctive mood (e.g. “should”, “could”, or “if”) that describe a hypothetical or instruction for patients. We excluded inquiry sentences that served as the template questions without a simple confirmed answer (e.g. “Yes”, “No”, or “None”) as well. For recognition of negated concepts (e.g. “patient denies having any fever”), we used NegEx, which was implemented in KMCI. NegEx is a widely-used algorithm to detect negations, but it still could miss post-negation triggers such as “Cough: No”. To enhance negation detection, we added regular expression rules based on our local note templates. The extended processing modules was implemented using Python 3.6. After processing, the extracted concepts served as the input for following ConceptWAS analysis.
2.6. ConceptWAS analysis
Similar to how GWAS and PheWAS scan genomic and phenomic data for discovery of disease associations [15], [21], ConceptWAS examines the clinical concepts retrieved from clinical notes to determine if any concept is associated with a disease. In this study, we applied ConceptWAS to identify associations between symptoms-related concepts and the presence of COVID-19.
We applied Firth's logistic regression to examine the association for each concept, adjusted by age, gender, and race. We chose Firth's logistic regression because it has become a standard approach for analyzing binary outcomes with small samples [22]. Negated and non-negated concepts are treated separately. Concepts were coded as binary variables for each patient. Firth's logistic regression was implemented using R version 3.4.3 and the logistf package. As we tested multiple hypotheses, we used a Bonferroni correction for the significance level. For each concept, we report the odds ratio (OR), p-values, and the prevalence in case and control groups. We used a volcano plot to show p-values and the odds ratio for all concepts. We also used a forest plot to show the significant concepts that were relevant to signs and symptoms.
2.7. Chart review
We performed a manual chart review to evaluate the clinical plausibility of identified signals. We reviewed a concept if 1) its p-value met Bonferroni-corrected significance, and 2) it was clinically meaningful (e.g., we excluded CUIs such as “finding [CUI C0243095]” in a sentence like “Findings are nonspecific.”). We randomly selected notes from which the CUI was identified. Two authors (M.E.G. and H.N.E.) with clinical background ascertained whether the identified CUI was a true signal or false positive.
3. Results
We identified 19,692 patients with COVID-19 PCR test results during the study period (Fig. A1). Of these, a total of 1,483 (7.5%) patients tested positive for COVID-19. Patients' mean age was 45 (44.6 ± 16.9) years. The COVID-19-positive group was younger (41.5 ± 16.2 vs. 44.9 ± 16.9), more often male (48.0% vs. 41.7%), less often white (49.6% vs. 66.7%), and newer to VUMC (EHR length 7.3 years ± 8.1 vs. 9.2 ± 8.5) compared to COVID-19-negative patients (Table 1 ).
Table 1.
Patient characteristics of the study cohort.
| Attribute | Cases: COVID-19-positive (n = 1,483) | Controls: COVID-19-negative (n = 18,209) | P- value |
|---|---|---|---|
| Age (mean years +/- stddev) | 41.5 (16.2) | 44.9 (16.9) | <0.0001 |
| Gender (% Male) | 48.0% | 41.7% | <0.0001* |
| Race (% White) | 49.6% | 66.7% | <0.0001* |
| Average EHR length (years, +/- stddev) | 7.3 (8.1) | 9.2 (8.5) | <0.0001 |
| Average CUIs (+/- stddev) | 46.1 (61.1) | 71.9 (96.3) | <0.0001 |
* 2-proportion z hypothesis test was performed. For age, EHR length, and average CUIs, a t-test was performed for comparing the mean and standard deviations.
3.1. Comparison of EHR-derived concepts between COVID-19 positive and negative patients
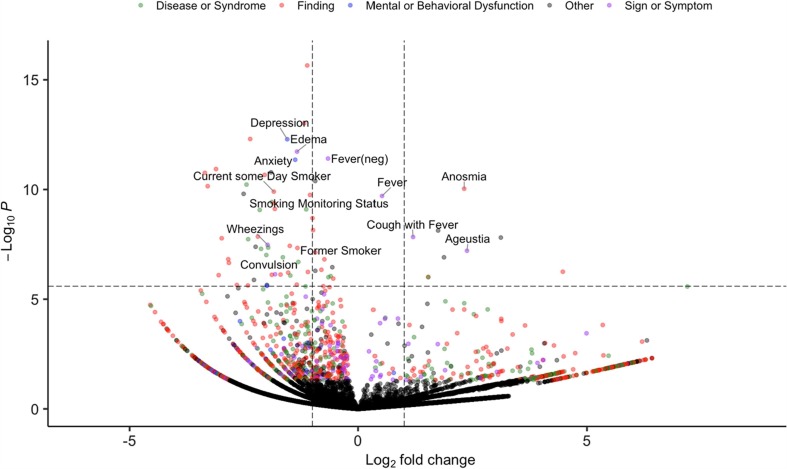
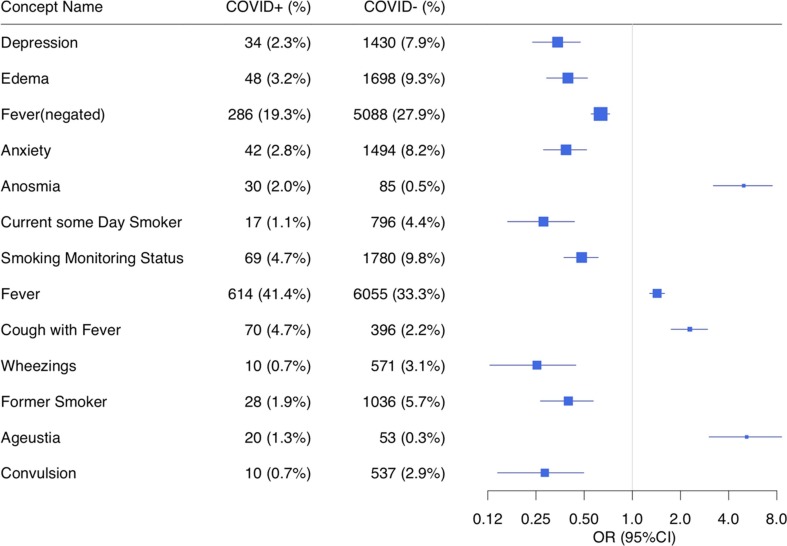
We extracted 87,753 clinical notes from the 19,692 patients. After using the NLP pipeline to process the notes, we recognized 19,595 unique concepts (including negated status) with semantic types of interests (Table B.1). Using ConceptWAS to compare EHR-derived concepts for COVID-19 positive and negative patients, 68 concepts were identified after adjusting for multiple testing (Bonferroni-corrected significance, P < 2.55E-06) (Fig. 1 , Table E.1). The top signals included “depression” (OR = 0.34, 95% CI = 0.24–0.47), “edema” (OR = 0.40, 95% CI = 0.29–0.53), “fever (negated)” (OR = 0.63, 95% CI = 0.55–0.72), and “anxiety” (OR = 0.39, 95% CI = 0.28–0.52). Specifically, symptoms concepts associated with COVID-19-positive patients included “anosmia” (loss of smell, OR = 4.97, 95% CI = 3.21–7.50), “fever” (OR = 1.43, 95% CI = 1.28–1.59), “cough with fever” (OR = 2.29, 95% CI = 1.75–2.96), and “ageusia” (loss of taste, OR = 5.18, 95% CI = 3.02–8.58) (Fig. 2 ).
Fig. 1.
Volcano plot of a ConceptWAS scan for 19, 692 patients that included COVID-19-positive group (cases) and negative group (controls). The points are colored by the semantic type of the concepts. Selected associations related to signs, symptoms, or diseases/syndromes are labeled. The volcano plot indicates -log10 (p-value) for association (y-axis) plotted against their respective log2 (fold change) (x-axis). The dashed line represents significance level using a Bonferroni correction.
Fig. 2.
Forest plot comparing individual concepts between COVID-19-positive (case) and COVID-19-negative (control) patients. Selected associations include the significant signals related to semantic types of symptoms that met Bonferroni-corrected significance (p-value < 2.55E-06). The odds ratio has been adjusted for age, gender, and race. The concepts are ordered by p-value.
Concepts related to smoking status such as “current some day smoker”, “former smoker”, and “smoking monitoring status” were more frequently reported in the COVID-negative group than in the COVID-positive group (OR < 1, P < 2.55E-06), suggesting more smokers in control group. To ascertain whether this signal was true or false positives due to wrongly assertion detection by NLP pipeline, we performed a chart review of 80 patients' notes that had smoking-related CUIs. We found that 79 of 80 patients confirmed an affirmative smoking status (see below chart review).
3.2. Temporal analysis
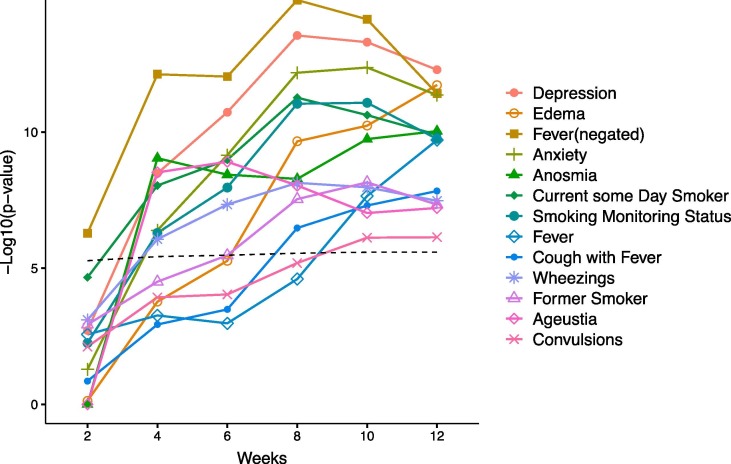
We performed ConceptWAS using the every 2-week cumulative data within the study period (Fig. 3 , Fig. D1). By week 4 (by April 5, 2020), “anosmia” (OR = 10.24; 95% CI = 5.18–20.06) and “ageusia” (loss of taste, OR = 11.79; 95% CI = 5.55–25.2) became significantly associated with increased risk of COVID-19 infection. These two signals remained significant through the subsequent weeks (Supplementary Data). Fever (negated) appeared (OR = 0.55; 95% CI = 0.43–0.71) at week 2 (between March 8 and 22, 2020), and “cough with fever” became significant (OR = 2.09; 95% CI = 1.60–2.70) from the 8th week (between March 8 and May 3, 2020). The “depression” and “anxiety” appeared significantly starting from week 4 (by April 5, 2020).
Fig. 3.
Temporal ConceptWAS using every 2-week cumulative data. For significant signals (related to signs, symptoms) using all data (labeled in Fig. 2), the plot indicates their -log 10 (p-value) for association (y-axis) against using the cumulative data started between March 8, 2020 to n weeks (x-axis). The dashed line indicates a significant association using a Bonferroni correction.
Fig. D1.
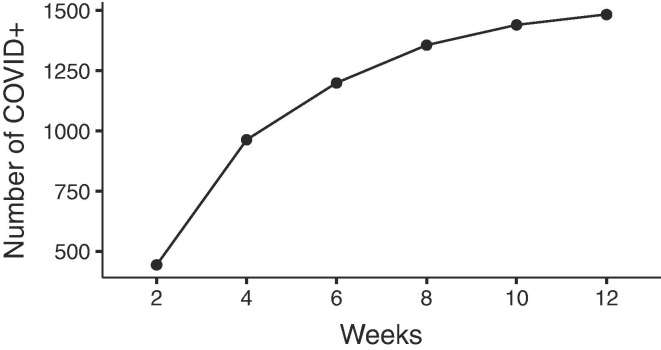
The cumulative number of COVID-19-positive(cases) along weeks.
3.3. Chart review
To validate the signals, we reviewed patient's charts for significant concepts. We randomly selected 10–20 notes for each concept to review whether the notes mentioned the symptoms in the expected attribute (e.g. affirmative or negated). Table 2 shows the results for significant concepts that with high clinical relevance (full list in supplementary material). The significant concepts such as “ anosmia”, “ageusia”, “depression”, and concepts related to smoking status (e.g. “current some day smoker”, “former smoker”, and “smoking monitoring status”) were consistent with the expected attribute based on chart review.
Table 2.
Results of chart reviews.
| Concepts | Reviewed samples | True signals | True signals percentage % | Examples of false positive |
|---|---|---|---|---|
| Anosmia | 20 | 19 | 95.00% | “(-) altered/loss of smell”, were wrongly recognized as an affirmative/ positive attribute. |
| Ageustia | 20 | 19 | 95.00% | “Symptoms, n/v, fever, cough, loss of taste or smell or around anyone + for Covid 19.” |
| Depression | 20 | 18 | 90.00% | One was recognized from a medical history title without any answers; the other came from a recommendation for further Psychosocial assessment. |
| Current some day smoker | 20 | 20 | 100.00% | |
| Smoking monitoring status | 20 | 19 | 95.00% | One is uncertain. “Smoking Status Not on file” . |
| Fever | 20 | 17 | 85.00% | Template issue. “The following ROS were reviewed and are negative, unless otherwise stated as + positive: 1 Constitutional: Fever; malaise” |
| Pericardial Fluid (neg) | 20 | 20 | 100.00% | |
| Hydrocephalus (neg) | 20 | 20 | 100.00% | |
| Hydronephrosis | 20 | 20 | 100.00% | |
| Blood group AB Rh(D) negative | 20 | 0 | 0.00% | From blood typing tests. This signal was not specific to blood type AB+, but generated by other ABO blood types and Rh-positive patients. |
| Allergy test positive | 20 | 5 | 25.00% | The false positives were wrongly mapped from a sentence like “He /She has been exposed to covid, family member or friends have tested positive.” |
| Laurin-Sandrow syndrome | 20 | 20 | 100.00% | |
| Cough nonproductive | 20 | 20 | 100.00% | |
| In total | 260 | 217 | 83.46% |
Although “smoking monitoring status” was generated by an inquiry term used in a template of a chart, after we post-processed the KMCI output to remove irrelevant concepts and refine negation, the smoking monitoring status followed by a negated answer was recognized as a negated attribute. We reviewed 20 notes that mentioned the “smoking monitoring status (affirmative/positive attribute)” and 19 were either current or former smokers.
We also found false positive concepts, mostly due to NLP entity recognition errors. For example, “additional information” was recognized as “adequate knowledge”. The concept “fever” with positive attribute has three false positives, mainly due to a few specific chart templates used for denoting the negation, which were not captured by NLP pipeline.
4. Discussion
We present a high-throughput and reproducible approach (ConceptWAS) that uses EHR notes to identify early emerging disease symptoms and investigate clinical manifestations for further hypothesis-driven study. Most of the previous studies used well-known tools such as GWAS and PheWAS for disease association discovery; however, these studies focused on structured EHR data, e.g., diagnosis codes and lab results. Few studies take advantage of the rich information within clinical notes to systematically detect relevant symptoms in the early stage of a disease that was not well characterized early in the pandemic. Singh et al. analyzed the nephrology notes of 4,013 patients, and identified 960 concepts to predict kidney failure. Their study demonstrated the feasibility of using clinical notes for a systematic analysis [17]. In this study, we developed a high-throughput pipeline to systematically scan clinical notes and detect unique concepts of COVID-19 in real-time.
We applied ConceptWAS to a cohort of patients who underwent COVID-19 PCR testing. We replicated several well-known symptoms of COVID-19, such as fever, loss of smell/taste, and cough with fever [23], [24], [25]. By performing temporal analysis on every 2-week cumulative data, we detected the signal of loss of smell and taste as early as April 5, 2020, nearly three weeks earlier than the date that they were listed as COVID-19 symptoms by the CDC [4]. Our results demonstrate the feasibility of using ConceptWAS for early detection of symptoms of an unknown disease.
We also observed several signals enriched in the COVID-19-negative group. For example, depression and anxiety have a higher prevalence among patients who tested negative. These signals first became significant starting from April 5, 2020, which may correspond to a period when the Governor of Tennessee issued a “safer at home” Executive Order and a “stay at home” order. It reflects the mental health issues that the shutdown and quarantine policies may bring to the people [26], [27]. We also find a higher percentage of smoking status concepts in the COVID-19-negative group. Earlier epidemiological studies found that fewer smokers are among COVID-19 patients or hospitalized COVID-19 patients [24], [28], which are consistent with our findings of the negative correlation between smoking and COVID-19. One explanation could be the impact of nicotine on ACE-2, as nicotine has been suggested to play a protective role against COVID-19 [29]. It is also possible that smokers are taking greater social precautions because of perceived higher risk for respiratory complications from COVID-19, thus reducing their risk of contracting the virus. Although these findings suggest that smoking may be a protective factor, lack of evidence and known adverse events associated with smoking dissuade continued smoking as a protective measure against COVID-19.
While our analysis was able to detect many of the known symptoms of COVID-19 included on the CDC’s list, including fever, loss of smell, and loss of taste, other symptoms present on the list were not found to be significant, including shortness of breath, muscle/body aches, and vomiting/diarrhea. Upon further review of 200 notes from 13 concepts that were on the list of symptoms maintained by the CDC but not significant in our analysis, we found the true positive percentage to be 77% (Supplemental Material Table 2).
ConceptWAS is open-source, portable, and reproducible. Researchers/users can choose other NLP pipelines (e.g. MetaMap, CLAMP, cTAKES) [26] for concept extraction and use the derived concepts as the input to ConceptWAS. Below we summarize the lessons that we learned in our proof-of-concept study applying NLP techniques to identify COVID-19 symptoms and characteristics, in the hope that these lessons may help others apply this method.
-
(1)
A high-throughput, lightweight, and reproducible method is important for an emerging pandemic disease. ConceptWAS enables a rapid scan of symptoms using clinical notes. These symptoms provided an initial hypothesis for further investigation and could alert clinicians to pay attention to patients who present with specific symptoms. Researchers can run ConceptWAS regularly (e.g. using weekly or 2-week cumulative data) to track changes in the identified symptoms of a pandemic disease.
-
(2)
Running ConceptWAS, one needs to be cautious about the distribution of different clinical note types. Clinical notes differ from each other due to their specific clinical usage. They may have variable templates and inconsistent lengths. Therefore, we recommend that researchers check the distribution of document types between cases and controls to avoid sampling bias.
-
(3)
Although NLP has been used in various medical fields to improve information processing and practice [30], [31], [32], [33], recognition of negative and uncertain concepts remains a challenge. We enhanced the detection of uncertain arguments and negated concepts by developing rule-based methods as wrappers for entity-identification generated results. Still, our manual chart review suggest that the outcome is not perfect. For example, some notes mentioned negative concepts such as “the following ROS were reviewed and are negative, unless otherwise stated as + positive: Constitutional: Fever; malaise.” Such scenarios are difficult for NLP tools to identify. A combination of machine learning and rule-based approaches may improve the detection.
-
(4)
To detect differential concepts at various levels of magnitude of change, the sample size needed for the study could be estimated beforehand. For example, a sample size calculation tool (e.g. https://vbiostatps.app.vumc.org/ps/dichot/1) could be used to generally estimate the minimum sample size given the input of desired odds ratio, type I Error (α), power (e.g. 80%), and probability of exposure in controls.
-
(5)
We also learned and recognized that our study had several limitations. First, the study was performed at a single institution with a limited number of COVID-19 patients. As the pandemic crisis evolves and more patients are tested for SARS-CoV-2 in our healthcare system, our ability to detect clinical concepts associated with COVID-19 will continue to improve. Second, this study used data from a limited time (before May 27, 2020). Third, ConceptWAS accepts concepts as input, which relies on an NLP pipeline and addon packages to identify. In this study, we used a locally developed NLP pipeline and customized several RegEx rules. A user may use different tools to extract the concepts, and the following step remains the same. However, the overall performance may vary.
-
(6)
In the future, we will extract notes from visits/calls before the test date to study symptom progression, and also extract notes after the test date to further explore the symptoms and their severity after the diagnosis. Lastly, as the performance of an NLP system may vary across institutions and databases [30], [34], further studies are necessary to assess the generalizability of our findings.
5. Conclusion
In this study, we describe a high-throughput approach (ConceptWAS) that systematically scans a disease's clinical manifestations from clinical notes. By applying ConceptWAS on EHR clinical notes from patients who received a COVID-19 PCR test, we detected loss of smell and taste three weeks prior to their inclusion as symptoms of the disease by the CDC. This study demonstrates the capability of EHR-based methods to enable early recognition of COVID-19-specific symptoms and to improve our response to such pandemic challenges.
Code availability
Up-to-date developments of ConceptWAS are available in GitHub (https://github.com/zhaojuanwendy/ConceptWAS).
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Dr. Vivian Siegel from Department of Biology at MIT Department of Medicine at Vanderbilt University for helpful suggestions on the study design and manuscript drafting.
Funding
The project was supported by the National Institutes of Health (NIH), National Institute of General Medical Studies (P50 GM115305), National Heart, Lung, and Blood Institute (R01 HL133786), National Library of Medicine (T15 LM007450, R01 LM010685), and the American Heart Association (18AMTG34280063), as well as the Vanderbilt Biomedical Informatics Training Program, Vanderbilt Faculty Research Scholar Fund, and the Vanderbilt Medical Scientist Training Program. The datasets used for the analyses described were obtained from Vanderbilt University Medical Center's resources and the Synthetic Derivative, which are supported by institutional funding and by the Vanderbilt National Center for Advancing Translational Science grant (UL1 TR000445) from NCATS/NIH. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Contributions
J.Z. and W.Q.W. developed the ConceptWAS concept and methodology. J.Z. and M.E.G. wrote the code, performed analysis, and plotted the figure. J.Z. and J.C.S. developed the ConceptWAS NLP pipelines. V.E.K. and W.Q.W. interpreted the clinical meaning of the results. M.E.G. and H.N.E. performed the chart review. V.E.K, Q.P., J.P., S.T.R. and K.B.J. provided the feedback. J.Z., M.E.G., V.E.K., J.C.S., H.N.E., Q.P., J.P., S.T.R., K.B.J., and W.Q.W. wrote and edited the manuscript.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jbi.2021.103748.
Appendix A. Study design
See Fig. A1
Appendix B. EHR notes distribution
The COVID-19-positive group had a distribution of clinical notes types similar to that of the COVID-19-negative group on the PCR test day (e.g. progress notes 81.36% versus [vs] 73.73%, social history 22.95% vs 28.41%, emergency department [ED] provider notes 6.26% vs 7.98%).
Table B1.
The notes types that were extracted in the study.
| Note type |
|---|
| Progress Notes |
| Social History |
| Imaging |
| ECG_IMPRESSION |
| ED Triage Notes |
| ED Provider Notes |
| H&P |
| Problem List |
| Assessment & Plan Note |
| Synopsis Sub-Note |
| Procedures |
| Subjective & Objective |
| History of Present Illness Sub-Note |
| Consults |
| Diagnostic Studies Sub-Note |
| Initial Assessments |
| ED Notes |
| Anesthesia Preprocedure Evaluation |
| Hospital Course Sub-Note |
| Clinical Update |
| Pathology And Cytology |
| Anesthesia Procedure Notes |
| Cardiac Services |
| Operative Report |
| Consult Reason Sub-Note |
| ED Progress Note |
| Anesthesia Postprocedure Evaluation |
| Technologist Note |
| Brief Op Note |
| Perioperative Nursing Note |
| Nursing Note |
| Neurology |
| Discharge Summary |
| Plan by Systems Sub-Note |
| ED Procedure Note |
| Echocardiography |
| Transthoracic Echocardiogram Report |
| Significant Event |
| Lactation Note |
| LAB |
| Post-operative Check |
| Research Informed Consent Note |
| Treatment Plan |
| Group Note |
| Pre-Procedure Note |
| Pre-Procedure Instructions |
| Plan of Care |
| Death Summary |
| Covering Surgeon |
| Research Coordinator Notes |
| Post-Procedure Note |
| Transition Plan Sub-Note |
| Interval H&P Note |
| Anesthesia Post-op Follow-up Note |
| Discharge Instr - Other Orders |
| Discharge Instr - Appointments |
| Interim Summary |
| Research Billing Note |
| External Transfer Orders |
| Declaration of Brain Death |
| Radiation oncology |
| Teleconsult |
| Discharge Instr - Activity |
| Discharge Instr - Diet |
| ACP (Advance Care Planning) |
| Code Documentation |
| Letter |
| Medical Student Progress Note |
| Onc Cost of Treatment |
| Transesophageal Echocardiogram Report |
Appendix C. Framework of the NLP pipeline and ConceptWAS
See Fig. C1
See Table C1
Table C1.
Semantic type of concepts that were included in the analysis.
| Semantic type |
|---|
| Sign or Symptom |
| Finding |
| Disease or Syndrome |
| Mental Process |
| Mental or Behavioral Dysfunction |
| Organism Function |
| Laboratory or Test Result |
| Individual Behavior |
| Social Behavior |
| Acquired Abnormality |
| Age Group |
| Population Group |
Appendix D. Temporal analysis
See Fig. D1
Appendix E. ConceptWAS results
See Table E1
Table E1.
ConceptWAS between COVID-19-positive (case) and COVID-19 negative(control). The table presents the significant concepts related to sign or symptom, disease or syndrome, or individual behaviors, which crossed Bonferroni p-value < 2.55E-06. “Neg” stands for negated attribute.
| Concept CUI (attribute) | Concept Name | Semantic type | Case Count (%) | Control Count(%) | OR (95%CI) | P-value |
|---|---|---|---|---|---|---|
| C1998726 | Adequate Knowledge | Finding | 109 (7.3%) | 3033 (16.7%) | 0.46 (0.38,0.56) | 2.22E-16 |
| C0243095 | Finding | Finding | 75 (5.1%) | 2309 (12.7%) | 0.44 (0.34,0.56) | 9.55E-14 |
| C0205400 | Increased thickness (Finding) | Finding | 11 (0.7%) | 854 (4.7%) | 0.19 (0.10,0.33) | 4.97E-13 |
| C0011570 | Depression | Mental or Behavioral Dysfunction | 34 (2.3%) | 1430 (7.9%) | 0.34 (0.24,0.47) | 5.12E-13 |
| C0013604 | Edema | Sign or Symptom | 48 (3.2%) | 1698 (9.3%) | 0.40 (0.29,0.53) | 1.88E-12 |
| C0015967 (neg) | Fever (neg) | Sign or Symptom | 286 (19.3%) | 5088 (27.9%) | 0.63 (0.55,0.72) | 3.87E-12 |
| C0003467 | Anxiety | Mental or Behavioral Dysfunction | 42 (2.8%) | 1494 (8.2%) | 0.39 (0.28,0.52) | 4.40E-12 |
| C1305863 | Anesthesia Type | Finding | 4 (0.3%) | 536 (2.9%) | 0.12 (0.04,0.26) | 1.17E-11 |
| C0554862 | Result, Lab.- General (Observable Entity) | Laboratory or Test Result | 17 (1.1%) | 891 (4.9%) | 0.27 (0.16,0.42) | 1.65E-11 |
| C0445088 | Neck Flexion (Finding) | Finding | 3 (0.2%) | 494 (2.7%) | 0.10 (0.03,0.24) | 1.74E-11 |
| C0231224 | Crisis | Finding | 14 (0.9%) | 850 (4.7%) | 0.24 (0.14,0.39) | 2.16E-11 |
| C0453996 | Tobacco smoking behavior | Individual Behavior | 99 (6.7%) | 2445 (13.4%) | 0.52 (0.42,0.64) | 4.11E-11 |
| C0020255 (neg) | Hydrocephalus (neg) | Disease or Syndrome | 8 (0.5%) | 662 (3.6%) | 0.18 (0.09,0.34) | 5.98E-11 |
| C0235195 (neg) | Sedated State(neg) | Finding | 3 (0.2%) | 445 (2.4%) | 0.10 (0.03,0.25) | 7.07E-11 |
| C0003126 | Anosmia | Finding | 30 (2.0%) | 85 (0.5%) | 4.97 (3.21,7.50) | 9.21E-11 |
| C1880200 | Current some day smoker | Finding | 17 (1.1%) | 796 (4.4%) | 0.28 (0.17,0.44) | 1.25E-10 |
| C0457318 | Blood Group Ab Rh(d) Negative | Laboratory or Test Result | 7 (0.5%) | 589 (3.2%) | 0.18 (0.08,0.34) | 1.58E-10 |
| C0586120 | Smoking monitoring status | Finding | 69 (4.7%) | 1780 (9.8%) | 0.48 (0.37,0.61) | 1.77E-10 |
| C0015967 | Fever | Sign or Symptom | 614 (41.4%) | 6055 (33.3%) | 1.43 (1.28,1.59) | 1.97E-10 |
| C0031039 (neg) | Pericardial Fluid(neg) | Disease or Syndrome | 15 (1.0%) | 819 (4.5%) | 0.27 (0.16,0.43) | 3.72E-10 |
| C1262869 | Body position | Finding | 16 (1.1%) | 821 (4.5%) | 0.28 (0.16,0.44) | 4.26E-10 |
| C0221198 | Lesion | Finding | 16 (1.1%) | 860 (4.7%) | 0.28 (0.17,0.45) | 7.68E-10 |
| C0521530 (neg) | Consolidation of Lung(neg) | Disease or Syndrome | 53 (3.6%) | 1604 (8.8%) | 0.46 (0.34,0.60) | 8.03E-10 |
| C0020295 (neg) | Hydronephroses(neg) | Disease or Syndrome | 10 (0.7%) | 667 (3.7%) | 0.22 (0.11,0.39) | 8.57E-10 |
| C0750426 | White Blood Cell Count Increased (Lab Result) | Finding | 70 (4.7%) | 1951 (10.7%) | 0.50 (0.39,0.64) | 2.02E-09 |
| C0455735 | Comments on own reading | Finding | 66 (4.5%) | 1850 (10.2%) | 0.51 (0.39,0.65) | 7.13E-09 |
| C0580359 | Allergy test positive | Laboratory or Test Result | 39 (2.6%) | 128 (0.7%) | 3.35 (2.29,4.79) | 7.42E-09 |
| C0231170 | Disability | Finding | 8 (0.5%) | 559 (3.1%) | 0.22 (0.10,0.40) | 1.40E-08 |
| C1277295 | Cough with fever | Sign or Symptom | 70 (4.7%) | 396 (2.2%) | 2.29 (1.75,2.96) | 1.46E-08 |
| C0427451 | Sickling test positive | Laboratory or Test Result | 15 (1.0%) | 22 (0.1%) | 8.66 (4.38,16.69) | 1.55E-08 |
| C0184763 | Patient condition unchanged | Finding | 3 (0.2%) | 403 (2.2%) | 0.13 (0.04,0.31) | 1.67E-08 |
| C0030554 (neg) | Paresthesias(neg) | Disease or Syndrome | 6 (0.4%) | 441 (2.4%) | 0.19 (0.08,0.38) | 1.84E-08 |
| C0043144 | Wheezings | Sign or Symptom | 10 (0.7%) | 571 (3.1%) | 0.25 (0.13,0.44) | 3.35E-08 |
| C3853152 | Does with Much Difficulty | Finding | 21 (1.4%) | 871 (4.8%) | 0.35 (0.22,0.53) | 3.71E-08 |
| C0028259 | Nodule | Acquired Abnormality | 7 (0.5%) | 552 (3.0%) | 0.21 (0.09,0.41) | 4.08E-08 |
| C0023518 | Leukocytosis | Disease or Syndrome | 10 (0.7%) | 607 (3.3%) | 0.26 (0.13,0.45) | 4.42E-08 |
| C0337671 | Former smoker | Finding | 28 (1.9%) | 1036 (5.7%) | 0.40 (0.27,0.57) | 4.62E-08 |
| C0014544 | Epilepsy | Disease or Syndrome | 8 (0.5%) | 556 (3.1%) | 0.23 (0.11,0.42) | 5.09E-08 |
| C2364111 | Ageustia | Sign or Symptom | 20 (1.3%) | 53 (0.3%) | 5.18 (3.02,8.58) | 6.16E-08 |
| C0444867 | both patent | Finding | 64 (4.3%) | 1673 (9.2%) | 0.52 (0.40,0.67) | 7.36E-08 |
| C0032227 | Pleural effusion disorder | Disease or Syndrome | 9 (0.6%) | 587 (3.2%) | 0.25 (0.12,0.45) | 9.69E-08 |
| C1851100 | Laurin-sandrow syndrome | Disease or Syndrome | 15 (1.0%) | 706 (3.9%) | 0.32 (0.18,0.51) | 1.23E-07 |
| C0086409 | Hispanics | Population Group | 30 (2.0%) | 71 (0.4%) | 3.66 (2.33,5.61) | 1.23E-07 |
| C1287298 | Urine volume finding | Finding | 3 (0.2%) | 374 (2.1%) | 0.14 (0.04,0.34) | 1.51E-07 |
| C0002871 | Anemia | Disease or Syndrome | 26 (1.8%) | 964 (5.3%) | 0.40 (0.27,0.59) | 2.00E-07 |
| C4081907 | Patient Identity Verified (Finding) | Finding | 3 (0.2%) | 311 (1.7%) | 0.14 (0.04,0.35) | 2.21E-07 |
| C0032074 | Planning | Mental Process | 195 (13.1%) | 3636 (20.0%) | 0.68 (0.58,0.79) | 3.48E-07 |
| C0455458 | Pmh - Past Medical History | Finding | 80 (5.4%) | 1887 (10.4%) | 0.57 (0.45,0.72) | 4.55E-07 |
| C0004048 | Inspiration Function | Organism Function | 54 (3.6%) | 1420 (7.8%) | 0.52 (0.39,0.68) | 5.06E-07 |
| C0442770 | Sees hand movements | Finding | 7 (0.5%) | 5 (0.0%) | 22.18 (7.26,71.96) | 5.60E-07 |
| C0332148 | Probable diagnosis | Finding | 37 (2.5%) | 1114 (6.1%) | 0.47 (0.33,0.64) | 5.73E-07 |
| C0700124 | Dilated | Finding | 16 (1.1%) | 691 (3.8%) | 0.35 (0.20,0.55) | 6.16E-07 |
| C0036572 | Convulsion | Sign or Symptom | 10 (0.7%) | 537 (2.9%) | 0.28 (0.14,0.50) | 7.28E-07 |
| C0233519 (neg) | Suspiciousness(neg) | Finding | 12 (0.8%) | 595 (3.3%) | 0.31 (0.17,0.52) | 7.50E-07 |
| C0332219 | Not Difficult at all | Finding | 9 (0.6%) | 501 (2.8%) | 0.27 (0.13,0.49) | 7.71E-07 |
| C0475269 | G1 Grade (Finding) | Finding | 2 (0.1%) | 309 (1.7%) | 0.12 (0.03,0.34) | 8.01E-07 |
| C0025517 | Disease, Metabolic | Disease or Syndrome | 17 (1.1%) | 746 (4.1%) | 0.36 (0.21,0.56) | 8.35E-07 |
| C0032326 (neg) | Pneumothorax (neg) | Disease or Syndrome | 161 (10.9%) | 3280 (18.0%) | 0.66 (0.55,0.78) | 8.87E-07 |
| C0277803 (neg) | Normal vital signs (neg) | Finding | 34 (2.3%) | 146 (0.8%) | 2.88 (1.94,4.17) | 9.60E-07 |
| C0032227 (neg) | Pleural effusion Disorder(neg) | Disease or Syndrome | 127 (8.6%) | 2674 (14.7%) | 0.64 (0.53,0.77) | 9.94E-07 |
| C0032310 | Pneumonias, Viral | Disease or Syndrome | 33 (2.2%) | 180 (1.0%) | 2.88 (1.94,4.15) | 1.00E-06 |
| C0033213 | Problem | Finding | 191 (12.9%) | 3525 (19.4%) | 0.69 (0.58,0.80) | 1.15E-06 |
| C1285647 | Characteristic of Perceptual Performance (Observable Entity) | Mental Process | 5 (0.3%) | 382 (2.1%) | 0.21 (0.08,0.43) | 1.32E-06 |
| C0574839 | Seen on arrival (Finding) | Finding | 20 (1.3%) | 729 (4.0%) | 0.39 (0.24,0.60) | 2.17E-06 |
| C0043157 | Caucasians | Population Group | 7 (0.5%) | 421 (2.3%) | 0.25 (0.11,0.48) | 2.25E-06 |
| C0449850 (neg) | Patient position finding (neg) | Finding | 3 (0.2%) | 269 (1.5%) | 0.16 (0.04,0.39) | 2.25E-06 |
| C0278061 | Altered Mental Status (Finding) | Mental or Behavioral Dysfunction | 7 (0.5%) | 469 (2.6%) | 0.25 (0.11,0.48) | 2.46E-06 |
Appendix F. Supplementary data
The following are the Supplementary data to this article:
References
- 1.WHO Coronavirus Disease (COVID-19) Dashboard, (n.d.). https://covid19.who.int/ (accessed May 26, 2020).
- 2.Guan W., Ni Z., Hu Y., Liang W., Ou C., He J., Liu L., Shan H., Lei C., Hui D.S.C., Du B., Li L., Zeng G., Yuen K.-Y., Chen R., Tang C., Wang T., Chen P., Xiang J., Li S., Wang J., Liang Z., Peng Y., Wei L., Liu Y., Hu Y., Peng P., Wang J., Liu J., Chen Z., Li G., Zheng Z., Qiu S., Luo J., Ye C., Zhu S., Zhong N. Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 2020 doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng X., Deng Y., Dai Z., Meng Z. COVID-19 and anosmia: A review based on up-to-date knowledge. Am J Otolaryngol. 2020;41 doi: 10.1016/j.amjoto.2020.102581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makaronidis J., Mok J., Balogun N., Magee C.G., Omar R.Z., Carnemolla A., Batterham R.L. Seroprevalence of SARS-CoV-2 antibodies in people with an acute loss in their sense of smell and/or taste in a community-based population in London, UK: An observational cohort study. PLoS Med. 2020;17 doi: 10.1371/journal.pmed.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.A. Fritz, M. Brice-Saddler, M. Judkis, CDC confirms six coronavirus symptoms showing up in patients over and over, Washington Post. (n.d.). https://www.washingtonpost.com/health/2020/04/27/six-new-coronavirus-symptoms/ (accessed September 25, 2020).
- 6.Statement from the UK Chief Medical Officers on an update to coronavirus symptoms: 18 May 2020, GOV.UK. (n.d.). https://www.gov.uk/government/news/statement-from-the-uk-chief-medical-officers-on-an-update-to-coronavirus-symptoms-18-may-2020 (accessed June 5, 2020).
- 7.R. Awasthi, R. Pal, P. Singh, A. Nagori, S. Reddy, A. Gulati, P. Kumaraguru, T. Sethi, CovidNLP: A Web Application for Distilling Systemic Implications of COVID-19 Pandemic with Natural Language Processing, MedRxiv. (2020) 2020.04.25.20079129. http://doi.org/10.1101/2020.04.25.20079129.
- 8.Mackey T., Purushothaman V., Li J., Shah N., Nali M., Bardier C., Liang B., Cai M., Cuomo R. Machine Learning to Detect Self-Reporting of Symptoms, Testing Access, and Recovery Associated With COVID-19 on Twitter: Retrospective Big Data Infoveillance Study. JMIR Public Health Surveill. 2020;6 doi: 10.2196/19509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R.M. Burke, Symptom Profiles of a Convenience Sample of Patients with COVID-19 — United States, January–April 2020, MMWR Morb Mortal Wkly Rep. 69 (2020). 10.15585/mmwr.mm6928a2. [DOI] [PMC free article] [PubMed]
- 10.Menni C., Valdes A.M., Freidin M.B., Sudre C.H., Nguyen L.H., Drew D.A., Ganesh S., Varsavsky T., Cardoso M.J., El-Sayed Moustafa J.S., Visconti A., Hysi P., Bowyer R.C.E., Mangino M., Falchi M., Wolf J., Ourselin S., Chan A.T., Steves C.J., Spector T.D. Real-time tracking of self-reported symptoms to predict potential COVID-19. Nat. Med. 2020;26:1037–1040. doi: 10.1038/s41591-020-0916-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richardson S., Hirsch J.S., Narasimhan M., Crawford J.M., McGinn T., Davidson K.W., Barnaby D.P., Becker L.B., Chelico J.D., Cohen S.L., Cookingham J., Coppa K., Diefenbach M.A., Dominello A.J., Duer-Hefele J., Falzon L., Gitlin J., Hajizadeh N., Harvin T.G., Hirschwerk D.A., Kim E.J., Kozel Z.M., Marrast L.M., Mogavero J.N., Osorio G.A., Qiu M., Zanos T.P. Presenting Characteristics, Comorbidities, and Outcomes Among 5700 Patients Hospitalized With COVID-19 in the New York City Area. JAMA. 2020;323:2052–2059. doi: 10.1001/jama.2020.6775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.G.A. Brat, G.M. Weber, N. Gehlenborg, P. Avillach, N.P. Palmer, L. Chiovato, J. Cimino, L.R. Waitman, G.S. Omenn, A. Malovini, J.H. Moore, B.K. Beaulieu-Jones, V. Tibollo, S.N. Murphy, S. L’Yi, M.S. Keller, R. Bellazzi, D.A. Hanauer, A. Serret-Larmande, A. Gutierrez-Sacristan, J.H. Holmes, D.S. Bell, K.D. Mandl, R.W. Follett, J.G. Klann, D.A. Murad, L. Scudeller, M. Bucalo, K. Kirchoff, J. Craig, J. Obeid, V. Jouhet, R. Griffier, S. Cossin, B. Moal, L.P. Patel, A. Bellasi, H.U. Prokosch, D. Kraska, P. Sliz, A.L. Tan, K.Y. Ngiam, A. Zambelli, D.L. Mowery, E. Schiver, B. Devkota, R.L. Bradford, M. Daniar, APHP/Universities/INSERM COVID-19 research collaboration, C. Daniel, V. Benoit, R. Bey, N. Paris, A.S. Jannot, P. Serre, N. Orlova, J. Dubiel, M. Hilka, A.S. Jannot, S. Breant, J. Leblanc, N. Griffon, A. Burgun, M. Bernaux, A. Sandrin, E. Salamanca, T. Ganslandt, T. Gradinger, J. Champ, M. Boeker, P. Martel, A. Gramfort, O. Grisel, D. Leprovost, T. Moreau, G. Varoquaux, J.-J. Vie, D. Wassermann, A. Mensch, C. Caucheteux, C. Haverkamp, G. Lemaitre, I.D. Krantz, S. Cormont, A. South, The Consortium for Clinical Characterization of COVID-19 by EHR (4CE), T. Cai, I.S. Kohane, International Electronic Health Record-Derived COVID-19 Clinical Course Profiles: The 4CE Consortium, Infectious Diseases (except HIV/AIDS), 2020. http://doi.org/10.1101/2020.04.13.20059691.
- 13.Wagner T., Shweta F., Murugadoss K., Awasthi S., Venkatakrishnan A., Bade S., Puranik A., Kang M., Pickering B.W., O’Horo J.C., Bauer P.R., Razonable R.R., Vergidis P., Temesgen Z., Rizza S., Mahmood M., Wilson W.R., Challener D., Anand P., Liebers M., Doctor Z., Silvert E., Solomon H., Anand A., Barve R., Gores G., Williams A.W., Morice W.G., II, Halamka J., Badley A., Soundararajan V. Augmented curation of clinical notes from a massive EHR system reveals symptoms of impending COVID-19 diagnosis. ELife. 2020;9 doi: 10.7554/eLife.58227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Welter D., MacArthur J., Morales J., Burdett T., Hall P., Junkins H., Klemm A., Flicek P., Manolio T., Hindorff L., Parkinson H. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42:D1001–D1006. doi: 10.1093/nar/gkt1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denny J.C., Ritchie M.D., Basford M.A., Pulley J.M., Bastarache L., Brown-Gentry K., Wang D., Masys D.R., Roden D.M., Crawford D.C. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene–disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verma A., Verma S.S., Pendergrass S.A., Crawford D.C., Crosslin D.R., Kuivaniemi H., Bush W.S., Bradford Y., Kullo I., Bielinski S.J., Li R., Denny J.C., Peissig P., Hebbring S., De Andrade M., Ritchie M.D., Tromp G. eMERGE Phenome-Wide Association Study (PheWAS) identifies clinical associations and pleiotropy for stop-gain variants. BMC Med. Genomics. 2016;9:32. doi: 10.1186/s12920-016-0191-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singh K., Betensky R.A., Wright A., Curhan G.C., Bates D.W., Waikar S.S. A Concept-Wide Association Study of Clinical Notes to Discover New Predictors of Kidney Failure. CJASN. 2016;11:2150–2158. doi: 10.2215/CJN.02420316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Denny J.C., Spickard A., Miller R.A., Schildcrout J., Darbar D., Rosenbloom S.T., Peterson J.F. Identifying UMLS concepts from ECG Impressions using KnowledgeMap. AMIA Annu Symp Proc. 2005;196–200 [PMC free article] [PubMed] [Google Scholar]
- 19.Denny J.C., Irani P.R., Wehbe F.H., Smithers J.D., Spickard A. The KnowledgeMap Project: Development of a Concept-Based Medical School Curriculum Database. AMIA Annu Symp Proc. 2003;2003:195–199. [PMC free article] [PubMed] [Google Scholar]
- 20.Denny J.C., Peterson J.F., Choma N.N., Xu H., Miller R.A., Bastarache L., Peterson N.B. Extracting timing and status descriptors for colonoscopy testing from electronic medical records. J Am Med Inform Assoc. 2010;17:383–388. doi: 10.1136/jamia.2010.004804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hindorff L.A., Sethupathy P., Junkins H.A., Ramos E.M., Mehta J.P., Collins F.S., Manolio T.A. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. PNAS. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Firth’s logistic regression with rare events: accurate effect estimates and predictions? - Puhr - 2017 - Statistics in Medicine - Wiley Online Library, (n.d.). 10.1002/sim.7273 (accessed June 7, 2020). [DOI] [PubMed]
- 23.Vaira L.A., Salzano G., Deiana G., De Riu G. Anosmia and Ageusia: Common Findings in COVID-19 Patients. Laryngoscope. 2020;130:1787. doi: 10.1002/lary.28692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.S.T. Moein, S.M. Hashemian, B. Mansourafshar, A. Khorram‐Tousi, P. Tabarsi, R.L. Doty, Smell dysfunction: a biomarker for COVID-19, International Forum of Allergy & Rhinology. n/a (n.d.). 10.1002/alr.22587. [DOI] [PMC free article] [PubMed]
- 25.Li L.-Q., Huang T., Wang Y.-Q., Wang Z.-P., Liang Y., Huang T.-B., Zhang H.-Y., Sun W., Wang Y. COVID-19 patients’ clinical characteristics, discharge rate, and fatality rate of meta-analysis. J Med Virol. 2020;92:577–583. doi: 10.1002/jmv.25757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pfefferbaum B., North C.S. Mental Health and the Covid-19 Pandemic. N. Engl. J. Med. 2020;383:510–512. doi: 10.1056/NEJMp2008017. [DOI] [PubMed] [Google Scholar]
- 27.W. Sturges, Gov. Bill Lee issues stay-at-home order through April 14, Impact. (2020). https://communityimpact.com/nashville/franklin-brentwood/coronavirus/2020/03/30/gov-bill-lee-issues-statewide-stay-at-home-order-for-tennesseans/ (accessed October 7, 2020).
- 28.A. Emami, F. Javanmardi, N. Pirbonyeh, A. Akbari, Prevalence of Underlying Diseases in Hospitalized Patients with COVID-19: a Systematic Review and Meta-Analysis, Arch Acad Emerg Med. 8 (2020). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7096724/ (accessed July 31, 2020). [PMC free article] [PubMed]
- 29.Farsalinos K., Niaura R., Le Houezec J., Barbouni A., Tsatsakis A., Kouretas D., Vantarakis A., Poulas K. Editorial: Nicotine and SARS-CoV-2: COVID-19 may be a disease of the nicotinic cholinergic system. Toxicol Rep. 2020;7:658–663. doi: 10.1016/j.toxrep.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Soysal E., Wang J., Jiang M., Wu Y., Pakhomov S., Liu H., Xu H. CLAMP - a toolkit for efficiently building customized clinical natural language processing pipelines. J Am Med Inform Assoc. 2018;25:331–336. doi: 10.1093/jamia/ocx132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yetisgen-Yildiz M., Gunn M.L., Xia F., Payne T.H. Automatic identification of critical follow-up recommendation sentences in radiology reports. AMIA Annu Symp Proc. 2011;2011:1593–1602. [PMC free article] [PubMed] [Google Scholar]
- 32.D. Mf, S. S, B. Ws, D. Jc, H. Jl, Automated extraction of clinical traits of multiple sclerosis in electronic medical records, Journal of the American Medical Informatics Association : JAMIA. 20 (2013). http://doi.org/10.1136/amiajnl-2013-001999. [DOI] [PMC free article] [PubMed]
- 33.Y. Ww, Y. M, H. Wp, K. Sw, Natural Language Processing in Oncology: A Review, JAMA Oncology. 2 (2016). http://doi.org/10.1001/jamaoncol.2016.0213. [DOI] [PubMed]
- 34.Wu S., Miller T., Masanz J., Coarr M., Halgrim S., Carrell D., Clark C. Negation’s Not Solved: Generalizability Versus Optimizability in Clinical Natural Language Processing. PLoS ONE. 2014;9 doi: 10.1371/journal.pone.0112774. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.