Abstract
People suspected of having COVID-19 need to know quickly if they are infected, so they can receive appropriate treatment, self-isolate, and inform those with whom they have been in close contact. Currently, the formal diagnosis of COVID-19 requires a laboratory test (RT-PCR) on samples taken from the nose and throat. The RT-PCR test requires specialized equipment and takes at least 24 h to produce a result. Chest imaging has demonstrated its valuable role in the development of this lung disease. Fast and accurate diagnosis of COVID-19 is possible with the chest X-ray (CXR) and computed tomography (CT) scan images. Our manuscript aims to compare the performances of chest imaging techniques in the diagnosis of COVID-19 infection using different convolutional neural networks (CNN). To do so, we have tested Resnet-18, InceptionV3, and MobileNetV2, for CT scan and CXR images. We found that the ResNet-18 has the best overall precision and sensitivity of 98.5% and 98.6%, respectively, the InceptionV3 model has achieved the best overall specificity of 97.4%, and the MobileNetV2 has obtained a perfect sensitivity for COVID-19 cases. All these performances have occurred with CT scan images.
Keywords: COVID-19, Chest imaging, CT scan, Chest X-ray, Convolutional neural network, Transfer learning
Graphical abstract
1. Introduction
SARS-CoV-2 belongs to the coronavirus (CoV) family, a name linked to the “crown” formed by certain proteins on the surface of these viruses. It was first identified in Wuhan, China, in December 2019. Several coronaviruses are already known to be capable of infecting humans and mammals: SARS-CoV responsible for the severe acute respiratory syndrome (SARS), and MERS-CoV responsible for Middle East Respiratory Syndrome (MERS). SARS-CoV-2 is the seventh coronavirus pathogenic to humans. He is responsible for the Covid-19 disease [1].
The Covid-2019 pandemic has subjected the world to a state of containment intending to limit contagion and the loss of lives. So far, 80% of infections are mild or asymptomatic, 15% are severe and require oxygen therapy, and 5% are critical and require respiratory support [2]. The most common symptoms of SARS-CoV2 are fever, asthenia, dry cough, and gastrointestinal symptoms including diarrhea, nausea, vomiting, and then anorexia. One in six people is subject to severe forms of the disease including pneumonia and acute respiratory distress syndrome [3]. The person suspected of having a Covid-19 infection needs to know quickly whether or not they are infected, in order to be well isolated, to receive the appropriate treatment, and to inform the people with whom she has been in contact. Often it is necessary to have two RT-PCR tests to confirm a diagnosis, which is time and resources consuming [4]. Also, the RT-PCR has high false-negative rates which cause sometimes COVID-19 patients can be assigned as healthy people [5], which has severe consequences. Therefore, a tool that can establish an earlier diagnosis would help decrease the prevalence of the disease.
Very similar to SARS and MERS, a wide variety of lung lesions have been described for COVID-19 on CT scan and CXR imaging [6]. The most common CT manifestations and features include frosted glass opacities (87%), bilateral lesion involvement (80%), peripheral distribution (75%), multilobar involvement (89%), posterior lesion topography (80%), and parenchymal condensations (33%) [7,8]. These ground-glass opacities have often been reported to be rounded, nodular, or with a crazy-paving pattern. The lower lobes are the most affected and the middle lobe is the least affected by this pneumonia. Pure frosted glass opacities or those associated with condensation were one of the most frequently found patterns [9].
Of the full range of imaging techniques available, the most widely used to examine patients with COVID-19 are CXR and CT scan [10,11]. These techniques help the clinicians identifying the effects of COVID-19 on different organs at various stages of the disease. They are used on the chest and lungs because respiratory symptoms are known to be among the first signs of COVID-19.
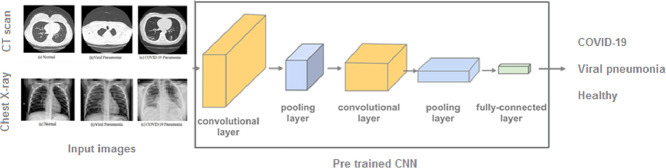
This study uses artificial intelligence (AI), in particular a deep learning approach, to build an automated system for mass screening, and early diagnosis of COVID-19. To achieve this objective, we have used the HUST-19 dataset containing 19,685 CT slices [12], and the COVIDx dataset comprising 5026 CXR images [13]. These datasets formed three architectures that represent different tiers of complexity: InceptionV3, ResNet-18, and MobileNetV2. This research offers the following contributions:
-
•
Developing three CNN models for mass screening, and accurate early diagnosis of COVID-19.
-
•
Detecting COVID-19 using CT scan and CXR imaging techniques.
-
•
Providing a detailed performance analysis of the proposed system in terms of the confusion matrix, precision, sensitivity, specificity, and F1-score.
-
•
Comparing the proposed method with the current RT-PCR, and highlighting the problems encountered in implementing AI-based models in real clinical usage.
2. Literature review
A range of deep learning models has been proposed to address the COVID-19 epidemic [10,11]. Based on clinical images, these models have shown promising results in the detection of COVID-19. Zhao et al. [14] have conducted a study to investigate the relationship between chest CT imaging and COVID-19 pneumonia. They have used a dataset collected from four institutions in Hunan, China. The results have shown typical imaging features for confirmed COVID-19 pneumonia cases, that can help in early screening and tracking the disease. Bernheim et al. [15] have analyzed the chest CTs images of 121 symptomatic coronavirus patients. The hallmarks of COVID-19 infection as seen on the CT scan images were bilateral and peripheral ground-glass and consolidative pulmonary opacities. Zhao et al. [16] have developed an AI-based CT scan image model to diagnose COVID-19, using an open-source dataset called COVID-CT, containing 812 CT images; 349 positive COVID-19, and 463 non-COVID-19 cases. Gozes et al. [17] have achieved an accuracy of 95%, by a model tested on 157 international coronavirus patients. The model is used to detect, quantify and track the evolution of the disease. Yasin et al. [18] have conducted a study to correlate patients’ age, sex, and outcome with COVID-19 disease evolution and severity using a CXR scoring system. Zheng et al. [19] have built an automatic software deep learning-based system to predict the COVID-19 infections, using a pre-trained UNet and a 3D deep neural network. Ai et al. [20] and Fang et al. [21] have shown a sensitivity superior to 97% of chest CT images in suggesting COVID-19, compared to 71% of the RT-PCR. Wang et al. [13] have proposed a COVID-NET convolutional neural network designed to detect COVID-19 cases from CXR images. They also introduced COVIDx, an open-access dataset that contains 13,975 CXR images from 13,870 COVID-19 patients. We have used the latest update COVIDx V7A from this dataset, by combining COVID-19 Radiography Database, Actualmed-COVID-chestxray-dataset, -COVID-chestxray-dataset, and covid-chestxray-dataset [22], and we have kept an equal number of observations for the three classes to avoid classification bias and improved the training performance by k-fold cross-validation technique.
3. Methods
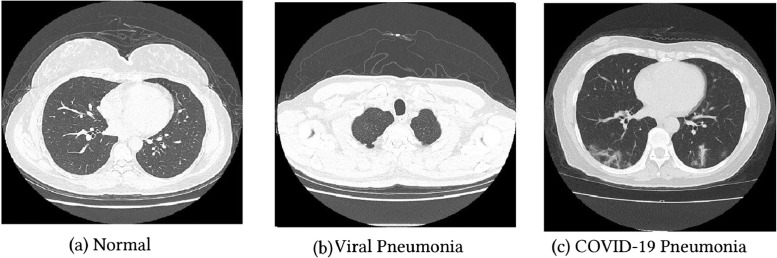
The images are collected from two datasets, HUST-19 for CT scan images, and the CXR images from the COVIDx dataset, the total number of images is 24,711 (19,685 CT scan, and 5026 CXR). The HUST-19 dataset has 5705 non-informative CT (NiCT), 4001 positive CT (pCT), and 9979 negative CT (nCT) slices, randomly selected from persons with and without COVID-19 pneumonia Figure 1 . 14,765 images are used for the models’ training, 2460 for the validation, and 2460 for the test, with a ratio of 75%, 12.5%, and 12.5%, respectively.
Fig. 1.
CT scan images.
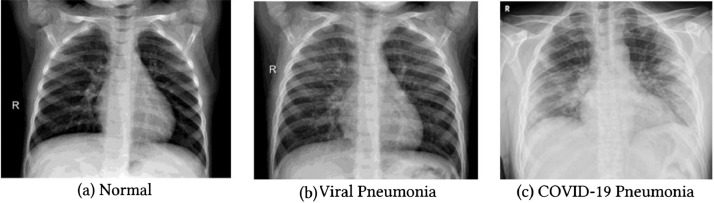
The COVIDx has 5026 CXR images: 1778 CXR COVID-19 patient images, 1530 who have no pneumonia (i.e., healthy), and 1718 non-COVID19 pneumonia patients Figure 2 . The dataset is divided into two sets: 70% of the data to build the models, and 30% to test them. The models are built with a 5-fold cross-validation technique: 70% of the dataset is divided into 5 folds roughly equal, alternately, 4 folds are used for the training, and the remaining one is used as a validation set. So, each observation is used 4 times in the training, and one time in the validation.
Fig. 2.
CXR images.
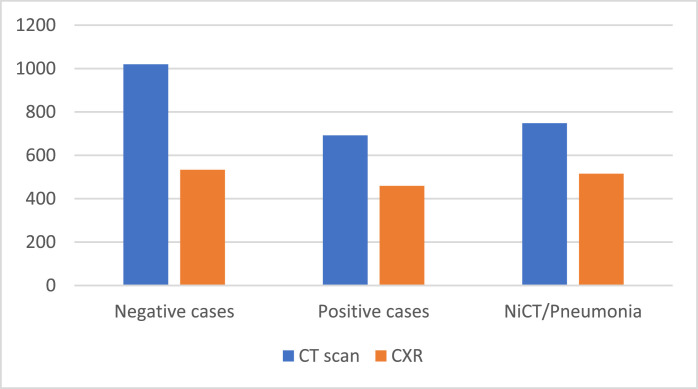
The CT scan test set has 692 positive, 1020 negative, and 748 NiCT observations. Whereas the CXR test set consists of 533 COVID-19, 515 viral pneumonia, and 459 healthy persons Figure 3 .
Fig. 3.
Test sets for the CXR and CT scan datasets.
The main task is to label CXR and CT images into three predefined categories. So, the models learn to classify CT slices as NiCT, pCT, and nCT. Or COVID-19, non-COVID-19 pneumonia, healthy persons, in the case of CXR images. Input images within specific categories are fed to the networks, and the softmax function is applied to the output to generate probabilities corresponding to these categories. The architectures used in this study belong to three tiers of complexity; InceptionV3 with 48 layers 29.3 million parameters, ResNet-18 with 18 layers deep and 11.7 million parameters, and MobileNetV2 with 53 layers and 3.5 million parameters. So, when comparing classification performances of our models, their complexity was taken into consideration, thus their ability to be implemented in mobile devices for real-world clinical practice.
Based on GoogleNet architecture, InceptionV3 was presented to reduce the computational cost by replacing large (5 × 5) and (7 × 7) filters with small asymmetric (1 × 5) and (1 × 7) filters, and using (1 × 1) convolution before the large filters as bottleneck [22]. This architecture has inception blocks that are stacked on top of each other, allowing the network to choose in each block between multiple filters with different sizes. In 2015, ResNet introduced the concept of residual units with shortcut connections, that skips one or more layers [23]. These residual connections improve the efficiency and reduce the effect of the vanishing gradient problem. The ResNet architecture was developed with different numbers of layers: 18, 34, 50, 101… We used ResNet-18 which represents a good compromise between the complexity and performance. Then we tried the MobileNetV2 [24]. It is an architecture that aims to perform well on mobile devices. This model uses depthwise separable convolution and has inverted residual building blocks, where the residual connections are between the bottleneck layers. We trained the models with 10 epochs using adam optimizer and a learning rate of (1* e − 3), mini-batch size of 16, L2 regularization was set to (1* e − 4), and we activated shuffling in every epoch.
4. Results and discussions
The models are evaluated with precision, sensitivity, specificity, and f1-score metrics. This allows us to compare the models’ performances within a dataset, and the effectiveness of the CXR and CT scan techniques in the detection of COVID-19, non-COVID-19 pneumonia, and healthy cases
Table 1 shows the resulted confusion matrix of the InceptionV3 model. For the CT scan images we have:
-
•
669 pCT were correctly classified, and 23 are considered nCT. The sensitivity of this model in detecting positive COVID-19 cases is 96.7%.
-
•
As for the 1020 nCT, 1002 were correctly detected, and the remaining 18 were wrongly classified, 3 as pCT and 15 as NiCT cases, resulting in a specificity of 98.2%.
-
•
And for the 748 NiCT cases, 723 were correctly classified, 20 cases are considered nCT, and 5 as pCT.
Table 1.
Confusion matrix for InceptionV3 model.
 |
As for the CXR images classification, the model has achieved:
-
•
In the case of the 533 COVID-19 cases, 530 were classified as positive, and 3 were wrongly classified, 2 as normal, and 1 as viral pneumonia. So, the model has achieved a sensitivity of 99.4% in detecting positive COVID-19 cases.
-
•
Out of 459 healthy persons, 456 persons were correctly classified, 1 person was diagnosed as COVID-19, and 2 were considered as pneumonia cases. Thus, the specificity is 99.3%.
-
•
And for the 515 viral pneumonia cases, 403 were correctly classified, 101 cases as COVID-19, and 11 were considered by the model as healthy persons.
Table 2 shows the ResNet-18 confusion matrix. The results of CT scan classification are:
-
•
685 images out of 692 pCT were correctly classified, 4 were considered as nCT, and 3 as NiCT. The sensitivity resulted is 99%.
-
•
999 nCT images were correctly detected, and 21 were wrongly classified as NiCT cases, meaning the model has achieved a specificity of 97.9%.
-
•
As for the 748 NiCT cases, 741 were correctly classified, 5 cases were diagnosed as nCT, and 2 cases as pCT.
Table 2.
Confusion matrix for ResNet-18 model.
 |
As for the CXR images classification, ResNet-18 has achieved:
-
•
506 COVID-19 cases were classified as positive, 5 cases were diagnosed as healthy, and 22 were considered other pneumonia, indicating a sensitivity of 94.9%.
-
•
455 healthy persons were correctly detected, 3 diagnosed positive and 1 case considered as other pneumonia. So, we have a specificity of 99.1% using this model
-
•
For the viral pneumonia cases, 408 were correctly classified, 100 cases as COVID-19, and 7 were considered by the model as healthy persons.
MobileNetV2 confusion matrix is illustrated in Table 3. By classifying CT scan images, we have:
-
•
All 692 COVID19 positive CT scan images (pCT) were correctly classified. So, we have perfect sensitivity.
-
•
As for the 1020 nCT images, 937 were correctly detected, and the remaining 47 were classified as 67 pCT and 16 NiCT cases. The specificity achieved is 91.9%.
-
•
And for the 748 NiCT cases, 731 were correctly classified, 9 cases are considered nCT, and 8 as pCT.
Table 3.
Confusion matrix for MobileNetV2 model.
 |
And by classifying CXR images, we obtain:
-
•
In the case of the COVID-19 class, 488 were correctly detected, 5 mislabeled as normal, and 22 were diagnosed as other non-covid-19 pneumonia. The sensitivity resulted using this model is 91.6%.
-
•
420 healthy CXR images were correctly classified, the remaining 39 are considered as 6 normal, and 33 other pneumonia. Leading to a specificity of 91.5%.
-
•
Finally, 409 other viral pneumonia cases were correctly classified, 102 diagnosed as COVID-19, and 4 were identified by the model as healthy persons.
To compare models’ performances within a dataset, and show the CXR and CT scan imaging techniques' effectiveness, we calculated the overall precision, sensitivity, specificity, and F1-score. The results are lustrated in Table 4.
Table 4.
The overall precision, sensitivity, specificity, and f1-score.
| Precision in% | Sensitivity in% | Specificity in% | F1-score in% | |||||
|---|---|---|---|---|---|---|---|---|
| CT scan | CXR | CT scan | CXR | CT scan | CXR | CT scan | CXR | |
| InceptionV3 | 97.5 | 93.4 | 97.2 | 92.3 | 97.4 | 88.8 | 97.3 | 92.8 |
| ResNet-18 | 98.5 | 91.7 | 98.6 | 91 | 97 | 89.1 | 98.5 | 91.4 |
| MobileNetV3 | 95.7 | 88.2 | 96.5 | 87.5 | 91.5 | 85.4 | 96 | 87.8 |
The highest precision using CXR dataset is 93.4% achieved by InceptionV3, and 98.5% by ResNet-18 using the CT scan dataset. With a low number of false positives, these models can correctly label the observations with the minimum overlapping between classes. As for sensitivity, the same models have achieved 92.3% and 98.6%, respectively. To calculate the specificity, we focused on non-COVID-19 true positive and COVID-19 false-positive, InceptionV3 and ResNet-18 have very close performances; for the CT scan dataset, we got 97.4% and 97% advantage InceptionV3, whereas, for the CXR images, we had 89.1% and 88.8% in favor of ResNet-18. Finally, we provide the F1-score, a metric that takes into account the precision and the sensitivity, ResNet-18 has obtained the highest F1-score of 98.5%, using the CT scan dataset, while InceptionV3 classifying CXR images has 92.8%.
For binary COVID-19 non-COVID-19 classification, the specificity resulted from CT scan InceptionV3, ResNet-18, MobileNetV2 is 99.5%, 99.8%, and 95.75%, respectively. Whereas the sensitivity is 96.7%, 99%, and 100%, respectively. ResNet-18 offers the best sensitivity/specificity compromise. MobileNetV2 model has a perfect sensitivity but sacrifices the specificity, in real-world usage, where the positive COVID-19 rate is lower and the resources are limited, having a high false alarm rate is time and resources consuming, meaning people with COVID-19 may not have the proper treatment.
These findings outperform saliva sample RT-PCR conducted by Pasomsub [25], where they get a sensitivity, specificity, and a precision of 84.2%, 98.9%, and 88.9%, respectively, and the findings of [26], where the authors got a 100% specificity of the diagnostic assay, and the sensitivity achieved was 90.7%. Compared to the current RT-PCR method, our results are provided quickly and accurately, suggesting that chest imaging could be a reliable method, allowing efficiency in recognizing and detecting COVID-19 patients.
From the confusion matrix, all three models, built by CXR images, misclassify approximately 20% (100 observations) of viral pneumonia cases as COVID-19, this consistency in false positive could be due to the quality of image compression (the COVIDx was not published in standard DICOM), so the models could not distinguish extracted features for both classes from the compressed images. Also, it could be that 100 viral pneumonia observations have close CXR symptoms to COVID-19, so the models have confused them both. Or there is a bias in the dataset since COVIDx is a combination of different datasets collected in completely different environments. This misclassification rises the problem of implementing CNN models in real clinical usage, where we could encounter a bias in merging multiple datasets with different contrast, resolution, and signal-to-noise ratio. While the results obtained in this study are encouraging, the occurred issue noted must be addressed before we could confidently use deep learning models in clinical routine.
4. Conclusion
Combining Chest imaging with deep learning models provides an accurate and efficient method to detect, quantify, and track the evolution of the COVID-19 disease. This study has revealed a positive outcome distinguishing COVID-19 patients from other pneumonia and negative cases. The method used is easy to implement by clinicians for mass screening of the patients. It will yield a faster rate as compared to the currently used RT-PCR method. Comparing CT scan and CXR images, CT scan has shown better performances detecting the positive cases using the same CNN models. This imaging technique offers better contrast and creates detailed quality images over CXR, which helps the models to extract pertinent information from the images. Although the findings encourage more chest imaging diagnosis for COVID-19, the downside is that image quality and merging multiple datasets could lead to poor classification performances.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Borghesi A., Zigliani A., Masciullo R., et al. Radiographic severity index in COVID-19 pneumonia: relationship to age and sex in 783 Italian patients. Nucl. Med. Med. Imaging. 2020;20 doi: 10.1007/s11547-020-01202-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grewal M., Srivastava M.M., Kumar P., Varadarajan S. Proceedings of the IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018) IEEE; 2018. pp. 281–284. [Google Scholar]
- 3.Song Q., Zhao L., Luo X., Dou X. Using deep learning for classification of lung nodules on computed tomography images. J Healthc Eng. 2017:2017. doi: 10.1155/2017/8314740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020 doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fang Y., et al. Sensitivity of chest CT for covid-19: comparison to RT-PCR. Radiology. 2020 doi: 10.1148/radiol.2020200432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wong H.Y.F., Lam H.Y.S., Fong A.H., Leung S.T., Chin T.W., Lo C.S.Y., et al. Frequency and distribution of chest radiographic findings in COVID-19 positive patients. Radiology. 2019 doi: 10.1148/radiol.2020201160. Mar 27:201160Epub ahead of print. PMID: 32216717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19) imaging reporting and data system (COVID-RADS) and common lexicon: a proposal based on the imaging data of 37 studies. Eur. Radiol. 2020:1–13. doi: 10.1007/s00330-020-06863-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye Z., Zhang Y., Wang Y., Huang Z., Song B. Chest CT manifestations of new coronavirus disease 2019 (COVID-19): a pictorial review. Eur. Radiol. 2020:1–9. doi: 10.1007/s00330-020-06801-0. Mar 19Epub ahead of print. PMID: 32193638PMCID: PMC7088323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Salehi S., Abedi A., Balakrishnan S., Gholamrezanezhad A. Coronavirus disease 2019 (COVID-19): a systematic review of imaging findings in 919 patients. AJR Am. J. Roentgenol. 2020;14:1–7. doi: 10.2214/AJR.20.23034. MarEpub ahead of print. PMID: 32174129. [DOI] [PubMed] [Google Scholar]
- 10.Rubin G.D., et al. The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the fleischner society. Radiology. 2020 doi: 10.1148/radiol.2020201365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen, J.P., Morrison, P. & Dao, L. COVID-19 image data collection. arXiv:2003.11597 (2020).
- 12.Ning W., Lei S., Yang J., Cao Y., Jiang P., Yang Q., Xiong L. Open resource of clinical data from patients with pneumonia for the prediction of COVID-19 outcomes via deep learning. Nat. Biomed. Eng. 2020:1–11. doi: 10.1038/s41551-020-00633-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L., Lin Z.Q., Wong A. Covid-net: a tailored deep convolutional neural network design for detection of covid-19 cases from chest x-ray images. Sci. Rep. 2020;10(1):1–12. doi: 10.1038/s41598-020-76550-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao W., Zhong Z., Xie X., Yu Q., Liu J. Relation between chest ct findings and clinical conditions of coronavirus disease (covid-19) pneumonia: a multicenter study. Am. J. Roentgenol. 2020;214(5):1072. doi: 10.2214/AJR.20.22976. [DOI] [PubMed] [Google Scholar]
- 15.Bernheim A., Mei X., Huang M., Yang Y., Fayad Z.A., Zhang N., Diao K., Lin B., Zhu X., Li K., et al. Chest ct findings in coronavirus disease19 (covid-19): relationship to duration of infection. Radiology. 2020 doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.J. Zhao, Y. Zhang, X. He, P. Xie, Covid-ct-dataset: a CT scan dataset about covid-19, arXiv:2003.13865 (2020).
- 17.O. Gozes, M. Frid-Adar, H. Greenspan, P.D. Browning, H. Zhang, W. Ji, A. Bernheim, E. Siegel, Rapid ai development cycle for the coronavirus (covid-19) pandemic: initial results for automated detection & patient monitoring using deep learning ct image analysis, arXiv:2003.05037 (2020).
- 18.Yasin R., Gouda W. Chest X-ray findings monitoring COVID-19 disease course and severity. Egypt. J. Radiol. Nucl. Med. 2020;51(1):1–18. [Google Scholar]
- 19.Zheng C., Deng X., Fu Q., Zhou Q., Feng J., Ma H., Liu W., Wang X. Deep learning-based detection for covid-19 from chest ct using weak label. medRxiv. 2020 [Google Scholar]
- 20.Ai T., Yang Z., Hou H., Zhan C., Chen C., Lv W., Tao Q., Sun Z., Xia L. Correlation of chest ct and RT-PCR testing in coronavirus disease 2019 (covid19) in China: a report of 1014 cases. Radiology. 2020 doi: 10.1148/radiol.2020200642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Y., Zhang H., Xie J., Lin M., Ying L., Pang P., Ji W. Sensitivity of chest ct for covid-19: comparison to RT-PCR. Radiology. 2020 doi: 10.1148/radiol.2020200432. p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Szegedy C., Vanhoucke V., Ioffe S., Shlens J., Wojna Z. Rethinking the inception architecture for computer vision. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Las Vegas, NV, USA; 2016. pp. 2818–2826. 26 June–1 July. [Google Scholar]
- 23.He K., Zhang X., Ren S., Sun J. Deep residual learning for image recognition. Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition; Las Vegas, NV, USA; 2016. pp. 770–778. 26 June–1 July. [Google Scholar]
- 24.Sandler M., Howard A., Zhu M., Zhmoginov A., Chen L. "MobileNetV2: inverted residuals and linear bottlenecks,". Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition; Salt Lake City, UT; 2018. pp. 4510–4520. [DOI] [Google Scholar]
- 25.Pasomsub E., Watcharananan S.P., Boonyawat K., Janchompoo P., Wongtabtim G., Suksuwan W., Phuphuakrat A. Saliva sample as a non-invasive specimen for the diagnosis of coronavirus disease 2019: a cross-sectional study. Clin. Microbiol. Infect. 2020 doi: 10.1016/j.cmi.2020.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanji J.N., Zelyas N., MacDonald C., et al. False negative rate of COVID-19 PCR testing: a discordant testing analysis. Virol J. 2021;18(13) doi: 10.1186/s12985-021-01489-0. [DOI] [PMC free article] [PubMed] [Google Scholar]