Chung et al. discuss the architectural design of CD8+ T cell responses during acute and chronic viral infection, emphasizing the parallelism of the differentiation pathways, the heterogeneity of the resultant T cell states, and the underlying transcriptional and epigenetic networks controlling these differentiation processes.
Abstract
In response to infection, T cells adopt a range of differentiation states, creating numerous heterogeneous subsets that exhibit different phenotypes, functions, and migration patterns. This T cell heterogeneity is a universal feature of T cell immunity, needed to effectively control pathogens in a context-dependent manner and generate long-lived immunity to those pathogens. Here, we review new insights into differentiation state dynamics and population heterogeneity of CD8+ T cells in acute and chronic viral infections and cancer and highlight the parallels and distinctions between acute and chronic antigen stimulation settings. We focus on transcriptional and epigenetic networks that modulate the plasticity and terminal differentiation of antigen-specific CD8+ T cells and generate functionally diverse T cell subsets with different roles to combat infection and cancer.
Heterogeneity is a universal feature of T cell differentiation, providing context-specific immunity
A T cell response to infection conceptually has two primary goals. The first is an immediate goal of generating large numbers of effector T cells to help eliminate the present infection. The second is a long-term goal of developing immunologic memory by endowing a portion of the antigen-experienced cells with enhanced longevity and regenerative capacity to protect against future encounters by the same pathogen. To accomplish these goals, T cells have evolved to differentiate into remarkably heterogeneous cellular subsets that differ based on phenotype, function, proliferative capacity, longevity, and anatomic location. Indeed, the ability of a single T cell, especially naive T cells, to differentiate into multiple types of T cells based on the environmental conditions experienced has made “plasticity” a signature T cell trademark (Fig. 1). “Cell plasticity” describes the ability of cells to readily transition from one differentiation state to another in response to environmental fluctuations, as opposed to “terminal differentiation,” which describes a cell in a more stable or fixed differentiation state that does not easily transition (see text box). The plasticity of CD4+ T cells to adopt distinct effector states was the first to be noted and has been a major focus in the field for >30 yr, fueled by the original discovery of the polarization of CD4+ T cells into T helper 1 (Th1) and Th2 subsets in 1986 (Mosmann et al., 1986). CD8+ T cells could also be functionally polarized in vitro into cells with more or less cytotoxic activity based on the types of cytokines present (Curtsinger et al., 1999). Subsequently, T cell plasticity was highlighted through in vivo studies showing that, upon infection, a single virus-specific CD4+ or CD8+ T cell could give rise to heterogeneous subsets of effector and memory cells (Gerlach et al., 2013; Buchholz et al., 2013; Plumlee et al., 2013; Tubo et al., 2013; Fig. 1 and Fig. 2 A). Multiple distinct memory T cell subsets that vary in their migratory and effector properties have been characterized: effector memory (TEM), central memory (TCM), stem-cell memory, tissue-resident memory (TRM), and peripheral memory (TPM) T cells (Schluns et al., 2000; Joshi et al., 2007; Kaech et al., 2002, 2003; Masopust et al., 2001; Wherry et al., 2003b; Gerlach et al., 2013; Buchholz et al., 2013; Sallusto et al., 1999; Gattinoni et al., 2011).
Figure 1.
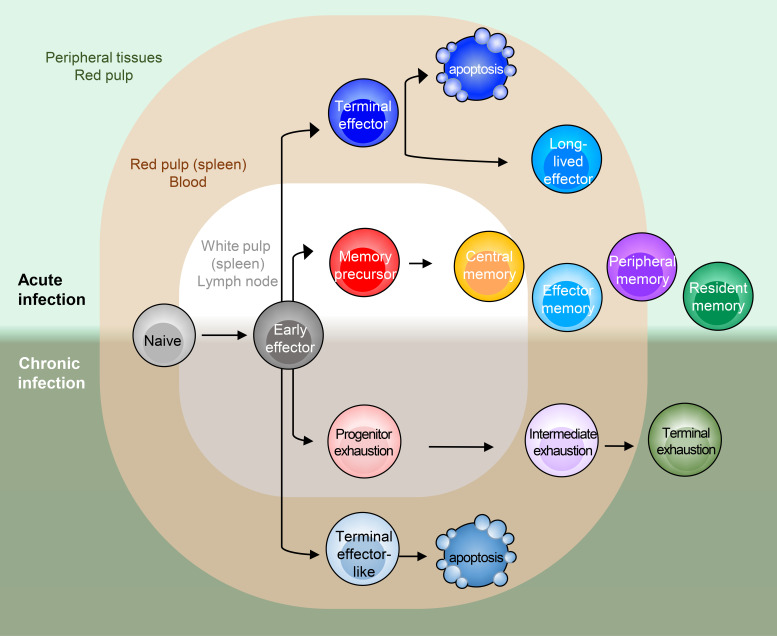
Heterogeneous immune populations in virus infection model. In both acute and chronic infection, early effector (EE) cells differentiate in a parallel manner into various CD8+ T cell subsets. Notable distinctions in trafficking patterns are signified by residence in lymphoid organs (gray), blood (red), or peripheral tissues (green). In acute infection, several effector and memory states are found. TE cells are typically found in the red pulp of spleen or blood, whereas MP cells are primarily found in white pulp or lymphoid structures, but they are also capable of recirculation. TCM and TEM cells both circulate in the blood, but TCM cells predominate in lymphoid organs, whereas TEM cells are also found in tissues. TPM cells are proposed to circulate throughout lymph, blood, and tissues. TRM cells do not circulate much and reside long-term in tissues. As in acute infection, heterogeneous states and distinct localization of CD8+ T cells are found in chronic infection. Texprog and MP cells are often observed in lymphoid structures, yet their circulation tendency might not be equivalent. Texint and Texterm cells are predominantly found in blood and peripheral tissue, respectively.
Figure 2.
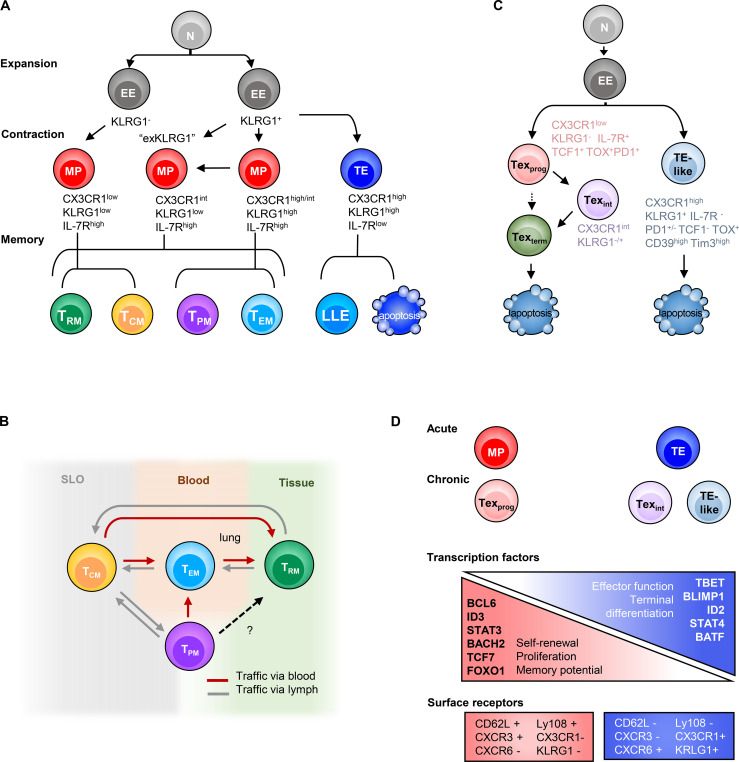
Heterogeneous T cell subsets in both acute and chronic infection are possibly driven by a similar mechanism. (A) In acute infection, depending on T cell activation and inflammation intensity, heterogeneous MP cells are formed with various levels of CX3CR1, which give rise to diverse memory subsets. KRLG1hi IL-7Rahi or exKLRG1 MP cells develop memory cells of higher cytotoxicity (Herndler-Brandstetter et al., 2018; Olson et al., 2013). (B) Several memory subsets maintain plasticity, which allows homeostatic (TRM to TCM; TEM or TPM to TCM) or antigen rechallenge–induced (TCM to TEM, TPM, or TRM) cell state conversion (Marzo et al., 2005, 2007; Beura et al., 2018b; Park et al., 2018; Osborn et al., 2019; Fonseca et al., 2020; Frizzell et al., 2020; Wherry et al., 2003b; Wherry and Ahmed, 2004; Bouneaud et al., 2005; Gattinoni et al., 2011; Gerlach et al., 2016). (C) Parallel differentiation ontology in chronic infection. Texprog cells diverge from EE due to TOX and Tcf7 activation, which suppresses TE-like cell formation. Texprog further differentiate in a linear manner, or also perhaps in a bifurcated manner, to Texint or Texterm cells. (D) Transcription gradient model for both acute and chronic infection. Shared TF gradients and surface receptors are found that drive the proliferative, stem cell–like cell states to more terminally differentiated cell states with effector function. KRLG1 expression is found in TE in acute and TE-like cells in chronic LCMV clone 13 and exhausted cells in HIV-infected patients. N, naive; SLO, secondary lymphoid organs.
Comparative analyses of the effector and memory CD8+ T cell populations over the course of immune response have revealed the complex diversity of cell types (or, as we prefer to call them, cell states; see text box) that arise in different settings of infection and disease. For example, there are notable distinctions in the quality of T cell response, the subtypes of T cells produced among different types of pathogens, and whether they cause acute infection (such as lymphocytic choriomeningitis virus [LCMV] Armstrong strain, Listeria monocytogenes, influenza virus, hepatitis A virus, vaccinia virus, and yellow fever vaccine), persistent chronic infection (HIV, hepatitis C virus [HCV], or hepatitis B virus), or latent infection with periods of virus reactivation (EBV or CMV; Boutboul et al., 2005; van Leeuwen et al., 2005; Wherry et al., 2006; Schulte et al., 2011; DeWitt et al., 2015). Likewise, cancer, autoimmunity, and transplantation can also be considered settings of chronic T cell activation. However, as we learn more about how T cells adapt to these settings of chronic stimulation, we also see a great deal of symmetry or parallelism in the overall “design” of the CD8+ T cell response between acute and chronic settings. This is the focus of this review.
Furthermore, with the advent of single-cell technologies to characterize the transcriptomes, epigenomes, and clonotypes (TCR repertoire) of T cells, we are operating in an unprecedented manner with exceptional speed to probe and deconvolute T cell diversity and heterogeneity and define T cell subsets or states (Pauken et al., 2016; Sen et al., 2016; Scott-Browne et al., 2016; Philip et al., 2017; Mognol et al., 2017; Scharer et al., 2017; Beltra et al., 2020; Brummelman et al., 2018; Bengsch et al., 2018; Yao et al., 2019; Chen et al., 2019b). As the determination of cell subsets or states using single-cell RNA-sequencing is based on the unbiased clustering of cells according to the genes expressed (of which the upper limit is 30,000, the number of genes in a cell), one may predict that this analysis would reveal infinite numbers of possible T cell differentiation states. However, as more data emerges from various contexts and disease settings, we are identifying a reasonably finite number of T cell subsets, on the order of dozens, not thousands (Tirosh et al., 2016; Zheng et al., 2017; Azizi et al., 2018; Guo et al., 2018; Sade-Feldman et al., 2018; Zhang et al., 2018, 2019; Li et al., 2019; Miller et al., 2019; Yost et al., 2019; Sandu et al., 2020). Thus, although each individual CD8+ T cell has the potential to adopt multiple cell fates, there is likely an upper limit to the number of possible cell states acquired, with some states being more stable than others (see text box). Needless to say, the sheer amount of transcriptional and epigenetic information being generated will undoubtedly lend critical insight into the vast heterogeneity of functional states, in addition to the identification of key regulators governing T cell differentiation and plasticity.
Defining cell types, cell states, and cell fates
A great number of phenotypic marker discoveries and advances in multiomics approaches have led to the identification of an extensive (but not infinite) number of T cell subsets that arise from naive CD4+ or CD8+ T cells. This subset classification is useful to understand T cell functionality based on the activity of distinct groups of cells. However, this classification raises the following questions: (a) Should different T cell subsets be considered distinct “cell types” or “cell fates”—or rather, “cell states”—based on their differentiation states? (b) How does the stability of a differentiation state (stable vs. quasi-stable vs. transient) influence these definitions? (c) How should differences in signature genes, epigenetic states, functions, and migratory patterns be applied to these definitions?
The term cell type has historically been used to define cells based on certain attributes, such as specialized cellular function, unique morphology, and cytoplasmic architecture. CD4+ and CD8+ T cells, for example, are well-recognized cell types, yet molecular dissection has revealed that they comprise numerous subtypes. Cellular properties can vary dramatically between subtypes. Alternatively, the difference is subtle and gradual rather than categorical, which makes the classification of cell subsets along developmental trajectories challenging. For example, CD4+ effector Th2 cells, Th1 cells, and Th17 cells have a clear difference in the pattern of gene expression, epigenetic states, and cytokine production, but the population of each effector type is inherently heterogeneous, containing multiple subsets that often display a continuum of differentiation states (Marshall et al., 2011; Pepper et al., 2011; Pepper and Jenkins, 2011; Cano-Gamez et al., 2020; Hale et al., 2013).
Cell subsets can be defined as epigenetically and transcriptionally similar groups of cells with a certain level of plasticity. In response to various stimuli, such as cell–cell interactions, signal transduction, and metabolic and mechanosensory cues, one subset could give rise to transcriptionally and epigenetically distinct states and to a new subset, but it is also possible that a subset could transcriptionally respond to stimuli without major epigenetic rewiring, giving rise to a transient cell state (Zheng et al., 2017; Guo et al., 2018; Sade-Feldman et al., 2018; Savas et al., 2018; Zhang et al., 2018, 2019; Satpathy et al., 2019; Yost et al., 2019; Low et al., 2020). Therefore, a cell subset may take on various cell states depending on the stimuli received. Thus, we propose the notion of referring to T cell subsets as cell states, knowing that each state has a variable degree of temporal stability and cell fate potential. For example, there are longer-lived and more stable states such as resting TCM or TRM cells, but there are more transient or conditional states such as transitional TEM cells that convert to TCM cells, “transitory” Texint cells, or EE cells that convert into KLRG1+ TE or IL-7R+ MP cells. Unless fate-mapping approaches are used, individual cells are interrogated at just a single point in time, making it difficult to assess the spatiotemporal changes in differentiation states experienced by a cell. Moreover, as we learn more about the epigenetic states of cells on a single-cell level, we will be able to better define cell subsets and their relative cell states.
The term cell fate is more restricted and describes the longer-term developmental destination of a cell as it relates to its natural environment. Cell fate is driven by a sequence of instructions: its genetic program, coupled with how the cell responds or adapts to its environment. The sequences of multiple cell states can end with a specified cell fate. However, as cell states can be plastic, one cell state is not necessarily destined to have a particular cell fate. Understanding of the heterogeneity and stability of T cell states and how they connect to cell fates is improving in T cells, but still faces challenges.
CD8 T cell responses to acute and chronic infection
The heterogeneity and plasticity of T cells have been revealed at every phase of the immune response: expansion and effector cell differentiation; resolution and effector cell contraction; and memory formation. Decades ago, it was noted in murine viral infections that several proteins are dynamically expressed by virus-specific CD8+ T cells over time, with peak expression of proteins that define highly cytotoxic CD8+ T cells during the first phase: granzymes (GZMBhi), perforinhi, IFNγhi, CD43 (IB11)hi, CD62Llo, and CD27lo (Tripp et al., 1995; Hamann et al., 1997; Harrington et al., 2000; Kaech et al., 2003). Similar properties distinguishing highly cytotoxic CD8+ T cells (CD45RAhi, CD62Llo, CD27lo, and CD28lo) could also be found in human virus-specific CD8+ T cells (Hamann et al., 1997; Akondy et al., 2017). Gradually, cells with this phenotype mostly decayed as memory cells formed throughout the contraction phase. However, for a long time, it was not possible to distinguish the cytotoxic effector cells that died during the contraction phase from those that simply down-regulated their protein expression as they matured into resting memory T cells. As flow cytometry expanded the number of proteins and parameters one could analyze simultaneously, it was then revealed that even greater heterogeneity existed within the pool of pathogen-specific CD8+ T cells, with numerous subsets developing, spanning a range of differentiation states, and displaying different long-term fates and degrees of plasticity (Kaech et al., 2003; Wherry et al., 2003b; Huster et al., 2004; Gerlach et al., 2013; Herndler-Brandstetter et al., 2018; Chang et al., 2014; Wherry et al., 2007; Sarkar et al., 2008). In this section, we compare how CD8+ T cell differentiation varies across time and within different contexts of acute and chronic infection.
Acute infection
Upon virus infection, CD8+ T cells differentiate into diverse T cell states. One way to explain the rise of various states is the asymmetric division of daughter cells as T cells proliferate (Chang et al., 2007). The first interaction between a CD8+ T cell and an antigen-presenting cell polarizes the T cell at the immunological synapse, the site where the T cell receives all of its most essential instructions from antigen:TCR (signal 1), costimulation (signal 2), and cytokines (signal 3). The coordination of these signals leads to polarized segregation of transcription factors (TFs) and cell fate determinants that influence its differentiation trajectory. Sufficient costimulation and inflammatory signals induce clonal expansion and expression of TFs such as RUNX3, T-bet, and Eomesodermin (Eomes) that trigger cytotoxic effector T cell differentiation (Banerjee et al., 2010; Intlekofer et al., 2005, 2007; Joshi et al., 2007). The effector cells start to produce granzymes, perforin, cytokines (such as IL-2, IFNγ, and TNF), chemokines (such as CCL5 and CCL3), and chemokine receptors (such as CXCR3, CX3CR1, CXCR6, and CCR5) to traffic to sites of inflammation. However, the effector molecules expressed by these canonical “killer” CD8+ T cells can vary across the pool of antigen-specific T cells according to their extent of differentiation. For instance, during an acute infection (Fig. 1, top half, and Fig. 2 A), a large fraction of effector CD8+ T cells terminally differentiate into highly specialized cytotoxic effector cells that coproduce IFNγ, GZMB, and perforin but lose the ability to produce IL-2 and often TNF. These cells express the highest levels of chemokine receptors CX3CR1 and S1PR5 and the inhibitory receptor KLRG1 (Joshi et al., 2007; Rao et al., 2010; Gerlach et al., 2016; Sarkar et al., 2008) and are commonly referred to as terminal effector (TE) cells (a.k.a., short-lived effector cells) because while they are highly functional killer T cells and pertinent to fighting the present infection, most TE cells commit to a terminal endpoint, literally, and die during the contraction phase (Joshi et al., 2007; Sarkar et al., 2008). As TE cells terminally differentiate, they induce effector programs and actively repress promemory genetic programs (including IL-7 receptor α [IL-7Ra] and CD27 expression); this diminishes their multipotency, proliferative capacity, and longevity (Kaech et al., 2003; Joshi et al., 2007; Herndler-Brandstetter et al., 2018). However, a small number of TE-like cells persist into the memory pool with some memory-like features, dwell in the blood, and contribute to secondary responses. Such cells are often referred to as long-lived effector (LLE) or terminal TEM cells (Renkema et al., 2020; Milner et al., 2020a; Olson et al., 2013).
As the activated T cells expand, most of the early effector (EE) cells down-regulate IL-7Ra, but then toward the end of the proliferative burst, a small portion of the cells begin to reexpress IL-7Ra and display enhanced longevity and stem-like properties. These cells are referred to as memory precursor (MP) cells, because they are multipotent and can develop into many types of memory cells (TCM, TEM, TRM, and TPM) or further differentiate into effector cells upon restimulation (Schluns et al., 2000; Joshi et al., 2007; Kaech et al., 2002, 2003; Masopust et al., 2001; Wherry et al., 2003b; Gerlach et al., 2013; Knell et al., 2013). MP cells are lesser differentiated cells typically defined by increased expression of IL-7Ra as well as Bcl-2, CD27, CD28, and CXCR3 (Badovinac and Harty, 2007; Hikono et al., 2007; Kaech et al., 2003; Wherry and Ahmed, 2004; Wherry et al., 2003b). Compared with TE cells, MP cells are more functionally heterogeneous and display greater proliferative capacity and cytokine polyfunctionality (IL-2+, IFNγ+, and TNF+), but lower GZMB. Some IL-7Rahi MPs also express high to intermediate amounts of CX3CR1 or KLRG1 concordant with greater amounts of cytotoxic molecules. These MPs are often referred to as double-positive (DP) cells, relating to their dual KLRG1hi IL-7Rahi expression (Obar et al., 2011; Rubinstein et al., 2008; Goldrath et al., 2002; Plumlee et al., 2013; Fig. 2 A). Indeed, many of the MP cells are quite plastic and can be seen converting from one state to another during the resolution and contraction phase (and even much later at memory stages; Kaech et al., 2003; Joshi et al., 2007; Gerlach et al., 2016; Huster et al., 2004; Böttcher et al., 2015; Wherry et al., 2003b; Marzo et al., 2007; Youngblood et al., 2017). This is especially evident in the effector cells that express intermediate amounts of CX3CR1int (Gerlach et al., 2016) or DP (IL-7Rhi KLRG1hi) cells (Herndler-Brandstetter et al., 2018; Olson et al., 2013) that convert to KLRG1lo (ex-KLRG1) cells during the contraction/resolution phase and become TRM, TPM, TCM, or TEM cells (Fig. 2 A).
As memory T cells mature following infection, they develop into subsets that play critical and diverse roles in mediating long-term protective immunity. TEM and TRM cells normally confer first-line defense at portals of pathogen entry in the blood or peripheral tissues, respectively, and can exert immediate effector responses, whereas TCM cells and stem-cell memory T cells, located in secondary lymphoid organs, focus their efforts on proliferation to resupply the host with large bursts of effector cells (Jabbari and Harty, 2006; Masopust et al., 2006; Marzo et al., 2007; Gattinoni et al., 2011). However, it is important to remember that these overly simple classifications do not accurately portray the broad phenotypic and functional heterogeneity that exists in the memory T cell population, as nicely reviewed by Jameson and Masopust (2018), and that the composition of memory CD8 T cells are not static. Rather, the composition of memory T cells is dynamic and changes with age, repeated infection, and environmental fluctuations (such as inflammation). For example, interconversions between TEM or TRM cells → TCM (Beura et al., 2018b; Osborn et al., 2019; Frizzell et al., 2020; Fonseca et al., 2020; Slütter et al., 2017; Van Braeckel-Budimir et al., 2018) or conversely TCM → TEM, have been described at rest (Marzo et al., 2005, 2007; Gattinoni et al., 2011; Wherry et al., 2003b; Bouneaud et al., 2005; Wherry and Ahmed, 2004; Gerlach et al., 2016). During a secondary challenge of skin infection, even circulating memory cells can convert to TRM (Osborn et al., 2019; Park et al., 2018; Kok et al., 2020; Beura et al., 2018a; Enamorado et al., 2017). Further, following reinfection or serial reinfections, the secondary and tertiary memory pools contain larger numbers of long-living TE- and TEM-like cells that have elevated cytotoxic properties (Jabbari and Harty, 2006; Masopust et al., 2006; Marzo et al., 2007; Gattinoni et al., 2011; Nolz and Harty, 2011; Wirth et al., 2010; Cui et al., 2011; Russell et al., 2017; Fraser et al., 2013). However, the properties of these “boosted” memory cells are largely influenced by the frequency of preexisting memory cells (Fraser et al., 2013; Joshi and Kaech, 2008). A larger number of preexisting memory cells encourage the formation of hybrid memory cells that display elevated cytotoxicity, IL-7Ra, and proliferative potential.
Persistent infections: Chronic and latent infections
Different types of chronic infections, persistent and latent, vary the duration of viremia and types of viral antigens produced. During a chronic infection with persistent viremia, such as infection with LCMV clone 13 in mice or HIV, HCV, or hepatitis B virus in humans, the virus-specific CD8+ T cells initially expand and contract, quite like during acute infection, albeit with different kinetics and outcomes (Badovinac et al., 2004, 2002; Wherry et al., 2003a; Zajac et al., 1998). In contrast to acute infection, which generates memory cell fates long-term, in persistent chronic infections and cancer, the long-term fate of most effector CD8+ T cells is to become dysfunctional, or what is more commonly referred to as “exhausted” (Fig. 1, bottom half; Schietinger et al., 2016; Philip et al., 2017; Willimsky and Blankenstein, 2005; Spranger et al., 2015; Alfei et al., 2019; Khan et al., 2019; Chen et al., 2019a; Seo et al., 2019; Thommen et al., 2018; Li et al., 2019). CD8+ T cell exhaustion is driven by chronic TCR activation and is characterized by (a) increased expression of multiple inhibitory receptors, such as PD1, TIM3, LAG3, CTLA4, and TIGIT, and (b) progressive loss of IL-2, TNF, and IFNγ secretion (Zajac et al., 1998; Paley et al., 2012; Barber et al., 2006; Khan et al., 2019; Wherry et al., 2003a). While the increase in inhibitory receptor expression represses TCR signaling and proinflammatory effector functions in CD8+ T cells, it also appears to protect the cells from activation-induced cell death, as PD-1–deficient T cells fare poorly and deteriorate quickly (Wei et al., 2019; Odorizzi et al., 2015). More recently, it was discovered that sustained Ca2+/nuclear factor of activated T cells (NFAT) signaling leads to induction of the HMG-box TF TOX, which directs a distinct exhaustion transcriptional and epigenetic developmental program (Khan et al., 2019; Seo et al., 2019; Yao et al., 2019; Scott et al., 2019).
Similar to acute infection, the CD8+ T cell response to chronic infection comprises multiple cell subsets that serve different roles in short- and long-term viral control. Akin to the multipotent MP cells in acute infection, in persistent chronic infection or tumors, a related stem-like exhaustion progenitor T cell type (Texprog) forms that is distinguished by increased SLAMF6 and CXCR5 and decreased TIM3 expression. Texprog cells have also been referred to as stem cell–like (Im et al., 2016), precursors of exhausted T cells (Utzschneider et al., 2020), or T memory-like exhausted cells (Utzschneider et al., 2016; Fig. 1, bottom half). Like MP cells, Texprog cells also display plasticity and proliferative capacity and can differentiate into transitory-intermediate (Texint) “effector-like” cells that up-regulate CX3CR1, T-bet, and effector molecules (GZMB, IFNγ, and TNF). Sustained production of the Texint subset is critical to control chronic virus infection or tumor (Hudson et al., 2019; Zander et al., 2019; Yan et al., 2018). Importantly, PD-1 blockade acts on the Texprog cells by releasing their proliferative restraints and generating new bursts of Texint cells (Im et al., 2016; Miller et al., 2019; Hudson et al., 2019; Zander et al., 2019; Utzschneider et al., 2016; Leong et al., 2016; Wu et al., 2016; Beltra et al., 2020; Im et al., 2020). Texint cells then further develop into terminally differentiated exhausted T cells (Texterm; Figs. 1 and 2 C) that express even higher amounts of PD-1, TIM3, and other inhibitory receptors including CD101, CD39, and CD160. Texterm cells show impaired expression of effector-related proteins (TNF, IFNγ, GZMB, and T-bet), and stemness and proliferation-related proteins (TCF1, MYB, MYC, and Ki-67; Beltra et al., 2020; Hudson et al., 2019). It is possible that some multipotent Texprog cells directly develop into Texterm cells, bypassing the Texint state, because the Texterm cell population did not seem greatly diminished several weeks after deletion of CX3CR1hi T cells (Zander et al., 2019; Hudson et al., 2019).
However, it is noteworthy that not all chronic infections drive CD8+ T cell exhaustion phenotypes. For example, during latent viral infections followed by episodes of viral reactivation such as commonly seen by adenovirus or herpes viruses (HSV, EBV, CMV, and murine CMV), a mixed population with a variety of effector and memory subsets is formed. This is likely driven mostly by the sporadic production of low-abundance antigen depots at sites of viral latency. Importantly, a distinctive feature is that the CD8+ T cells recognizing latently produced antigens gradually expand over time and develop into a so-called “inflationary” memory cell pool that contains mostly CX3CR1hi KLRG1hi TE-like cells (similar in many ways to memory cells formed after sequential acute virus immunizations; Gordon et al., 2018; Ouyang et al., 2003; Ibegbu et al., 2005). Even though CX3CR1hi inflationary memory cells show some similarity to Texint cells, unlike their exhaustion counterpart in LCMV clone 13, inflationary memory cells up-regulate KLRG1 (Gordon et al., 2018; Ouyang et al., 2003; Ibegbu et al., 2005) and express low levels of inhibitory receptors such as PD1, TIM3, and CTLA4 (Hertoghs et al., 2010; Sauce et al., 2007). We outline these series of events in comparison to those that occur in acute infection in the next section.
Comparative analysis of CD8+ T cell differentiation trajectories during acute and chronic infection
Even though T cell exhaustion is a progressive process that occurs over several weeks to months, the effector differentiation programs begin to diverge between acute and chronic infection within the first week of infection (Wherry et al., 2007; Ahmadzadeh et al., 2009; Sen et al., 2016; Chen et al., 2019b, 2021; Yao et al., 2019; Zhang et al., 2019). More specifically, the effector cells in both acute and chronic LCMV infection are transcriptionally similar up to day 4.5 after infection (Yao et al., 2019), but a significant transcriptional and epigenetic divergence begins a couple of days later and then continues for several more weeks (Wherry et al., 2007; Ahmadzadeh et al., 2009; Sen et al., 2016; Philip et al., 2017; Chen et al., 2019b, 2021; Yao et al., 2019; Zhang et al., 2019). This time period coincides exactly with changes in the expression of the TF TOX, as it dwindles in antigen-specific CD8+ T cells in acute infection but becomes amplified in chronic infection and tumors (Page et al., 2018; Yao et al., 2019; Khan et al., 2019). Indeed, TOX is critical for inducing chromatin remodeling and gene expression programs associated with T cell exhaustion and sustaining Texprog cells (Khan et al., 2019; Page et al., 2018; Yao et al., 2019; Scott et al., 2019). Yet, despite these clear divisions in the outcomes of CD8+ T cell fates during acute versus chronic infection, the underlying structure of the T cell differentiation programs appears quite similar between the two types of infections (Fig. 1; and Fig. 2, A and C).
Shared features of T cell differentiation in acute and chronic infection
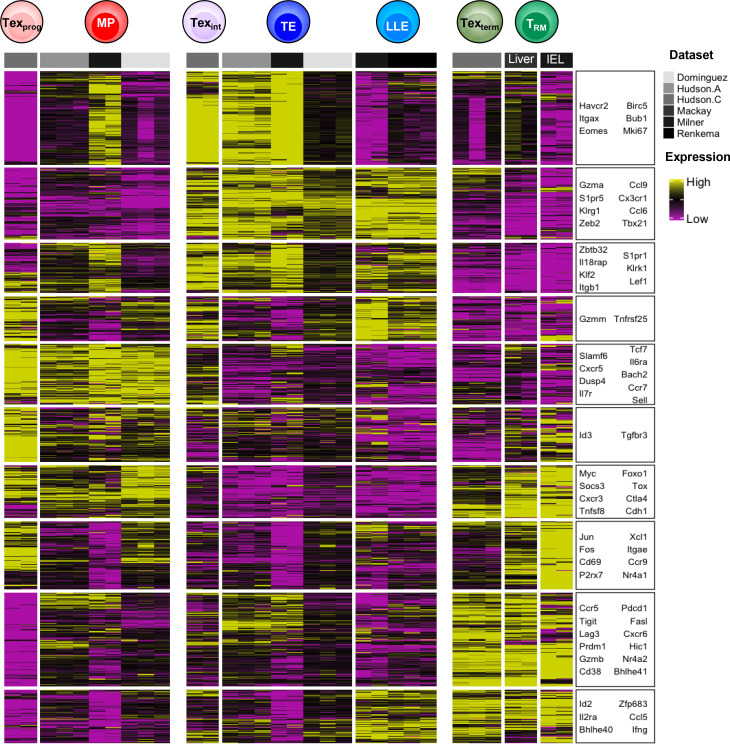
In spite of overt differences in CD8+ T cell phenotypes and effector functions, the core purpose of the developmental trajectories established in acute and chronic infection appears similar: to generate progenitor cells that sustain the pool of virus-specific CD8+ T cells long-term and can regenerate effector cells to battle viral-infected cells when present. Therefore, it is perhaps not surprising that both MP and Texprog subsets, as well as TE and Texint, have a great deal in common with regard to trafficking patterns and transcriptional programs (Fig. 1). For example, the precursor cells in both infection settings (Texprog and MP) prefer to home to lymphoid zones (e.g., white pulp in spleen, lymph nodes, or tertiary lymphoid organs), whereas the CX3CR1-expressing effector-like cells (TE and Texint) are predominantly found in the blood. Interestingly, Texterm and TRM also seem to have tissue homing in common, and both express CXCR6 and CD69. Regarding the shared transcriptional programs, both Texprog and MP express high Tcf7, Foxo1, and Bach2; TE and Texint express Tbx21 (encoding T-bet) and Zeb2; Texterm and TRM express Prdm1, Id2, Nr4a2, Bhlhe40, and Tox (Fig. 2 D and Fig. 3). Another common element between acute and chronic infection is that terminally differentiated cell types are produced in both acute and chronic infection as they develop into TE (KLRG1hi CX3CR1hi IL-7Ralo CD27lo) and Texterm (CD101hi TIM3hi SLAMF6lo) cells, respectively. This definition of terminal differentiation stems from experiments showing that, when adoptively transferred into naive or infection-matched hosts, these cells maintain their physical and genetic properties.
Figure 3.
Transcriptional meta-analysis of T cell subsets in both acute and chronic infection. Scaled and batch-corrected RNA-sequencing expression from sorted effector and exhausted cell populations (Dominguez et al., 2015; Mackay et al., 2016; Hudson et al., 2019; Milner et al., 2020a; Renkema et al., 2020). T cell states that arise during chronic and acute infection can be characterized by similar transcriptional archetypes, such as up-regulation of sets of stem-like genes (Texprog and MP cells), cytotoxicity and circulating potential (Texint, TE, and LLE cells), or inhibitory receptor genes (Texterm and TRM cells).
Distinctive features of T cell differentiation in acute and chronic infection
Before we understood the limits of terminal differentiation of effector cells, it seemed sensible to assume that CD8+ T cell exhaustion was simply an end-stage product of effector differentiation and that TE cells would convert into Texterm cells with prolonged antigenic stimulation. However, this is not the case, because KLRG1hi TE cells do not generate Texterm cells when restimulated (Chen et al., 2019b; Khan et al., 2019; Angelosanto et al., 2012). In fact, there are many similarities between the transcriptional states of TE and Texint cells found in acute and chronic infection, respectively (Fig. 2 D and Fig. 3). One of the most notable distinctions is that TE cells are terminally differentiated, whereas Texint cells are intermediary and not yet terminally differentiated. Another notable difference is the expression of KLRG1 itself. KLRG1 is expressed by LCMV-specific TE-like CD8+ T cells during the first week of chronic infection, but these KLRG1hi cells rapidly wane and are highly dependent on CD4+ T cell help (Chen et al., 2019b; Khan et al., 2019; Stelekati et al., 2018). The inability of KLRG1hi TE cells to persist in chronic infection may be because these cells are highly sensitive to activation-induced cell death (Joshi et al., 2007; Chen et al., 2019b) and/or because TOX antagonizes T-bet and impairs maximal T-bet induction and TE cell induction, which circumvents terminal differentiation into TE cells (Beltra et al., 2020; Khan et al., 2019). Even though exhausted HIV- or HCV-specific T cells in humans express KRLG1, they show significant reduction in T-bet expression (Wang et al., 2020; Bengsch et al., 2007). In this way, TOX may help to endow Texint cells with plasticity to continue differentiating into Texterm cells.
Transcriptional and epigenetic regulation of CD8+ T cell heterogeneity during acute and chronic infection
Active maintenance of plasticity and terminally differentiated states in CD8+ T cells
The spectrum of CD8+ T cell differentiation states that arise during infection depend on the coordinated actions of multiple TFs and chromatin remodeling complexes that can generate positive feed-forward circuits to promote one cell state while simultaneously opposing another. Indeed, some differentiation states seem more stable (e.g., naive, TCM, TEMRA [effector memory T cells re-expressing CD45RA], Texterm, and TE states) than others (e.g., MP, CX3CR1int, or Texint states). These stable states perhaps represent epigenetic and metabolic equilibrium, since the epigenetic landscape of a cell is directly influenced by its metabolic activities (Franco et al., 2020; Buck et al., 2017). In this section, we outline several transcriptional and epigenetic changes that occur to generate the subsets described above.
TE cell differentiation in acute infection is a stepwise process. In EE cells, TFs such as T-bet, Zeb2, Stat4, Rbpj, Irf4, Blimp-1 (Prdm1), and Id2 are induced and set up a counterregulatory network that leads to the transcriptional and epigenetic repression of critical promemory TFs (such as Tcf7, Bach2, Bcl6, Id3, Foxo1, Zeb1, and Stat3) active within MP cells to resist terminal differentiation and maintain plasticity, multipotency, and memory cell development (Fig. 2 D; Yang et al., 2011; Joshi et al., 2007; Cui et al., 2011; Ichii et al., 2002; Oestreich et al., 2012; Delpoux et al., 2017; Intlekofer et al., 2005; McLane et al., 2013; Kaech and Cui, 2012; Dominguez et al., 2015; Guan et al., 2018; Omilusik et al., 2015; Kallies et al., 2009; Rutishauser et al., 2009; Knell et al., 2013; Best et al., 2013). The developing TE cells epigenetically repress promemory genes via targeted deposition of H3K27me3 and H3K9me3 and DNA methylation on promemory genes by the enzymes EZH2, SUV39H1, and DNMT3a, respectively (Gray et al., 2017; Pace et al., 2018; Ladle et al., 2016; Youngblood et al., 2017). This epigenetic remodeling likely occurs several days after the initial transcriptional repression of promemory genes that begins as early as the first CD8+ T cell division (Arsenio et al., 2014). In other words, EE cells may begin to commit to TE cell differentiation (e.g., via transcriptional induction of TE genes [Klrg1 and Cx3cr1]), but their determination to a terminally differentiated TE state (as defined by epigenetic silencing of promemory genetic programs) is not achieved until several days later. These findings, nearly 20 yr later, provide a molecular framework for the decreasing-potential model to describe the contraction of effector cells and survival of memory cells originally put forth by Ahmed and Gray (1996). Evidence for a similar process occurring in chronic infection to promote terminally differentiated Texterm by restricting Texint and Texprog signature genes also exists but needs to be investigated more closely (Ghoneim et al., 2017; Schietinger et al., 2016; Beltra et al., 2020; Yao et al., 2019; Zander et al., 2019; Hudson et al., 2019).
MP cells, on the other hand, maintain promemory, prosurvival, and many TE-signature gene loci in active or permissive epigenetic states, allowing MP cells to simultaneously promote memory development and remain poised for the future expression of “effector” genes. Similarly, TCM and TEM cells maintain poised and bivalent epigenetic marks, activating H3K4me3 and repressive H3K27me3, on the promoter regions of Id2, Tbx21, Eomes, Irf4, Map3k1, Mlk4, and Mkx (Russ et al., 2014; Araki et al., 2009). Bivalency allows multipotent memory cells to epigenetically mark transcriptionally silenced genes that are destined for activation followed by T cell activation, facilitating a rapid transition of memory cells from resting to an activated state. The capacity of the transition can be stably maintained and has been observed in smallpox-specific T cells up to 83 yr after infection, suggesting that this effector inducibility is extremely durable (Hammarlund et al., 2010). The plasticity seen in MP and memory cells depends on the continued expression of promemory genes such as Foxo1 and exposure to cytokines via TGFβ receptor, because late deletion of these genes in resting memory CD8+ T cells leads to the spontaneous acquisition of TE-like states (Ma and Zhang, 2015; Utzschneider et al., 2018). Possibly, FOXO1 acts in MP cells to insulate promemory genes from (a) EZH2-mediated silencing by locally impairing H3K27me3 deposition at such loci (Gray et al., 2017) or (b) recruiting DNA-demethylase machinery because the selective loss of DNA methylation at promemory-associated loci such as Sell (CD62L) and Tcf7 was observed in MP cells, but not TE cells (Youngblood et al., 2017). These data provide mechanistic insight into how developmental plasticity is epigenetically wired in subsets of effector CD8+ T cells or lost in others as they are terminally differentiated.
Terminally differentiated states are usually associated with long-term stability; therefore, it was surprising to see that maintenance of TE states during the contraction and resolution phase depended on sustained ID2 expression (Omilusik et al., 2015). When ID2 was deleted after day 8 of acute virus infection, the KLRG1hi TE cells rapidly converted into MP-like cells, indicating that terminal differentiation depends on sustained expression of key TFs that prevent dedifferentiation. Coupled with the data above that sustained expression of Foxo1 is needed to preserve resting memory cell states, these data suggest that maintaining T cell differentiation states, even those that appear to be fairly stable like TE or TCM cells, is an active process and requires sustained genomic surveillance by “inducer” TFs. Natural perturbations in the expression of such inducer TFs and epigenetic modifiers such as demethylases likely account for the dynamic interconversions found between various differentiation states (e.g., TEM → TCM, DP → TRM, TRM → TCM) in CD8+ T cells after infection has resolved.
Imprinting CD8+ T cell exhaustion
Along the trajectory from Texprog → Texint → Texterm, there is extensive remodeling of the transcriptional and epigenetic landscape that occurs from one state to another (Philip et al., 2017; Beltra et al., 2020; Utzschneider et al., 2020; Miller et al., 2019). Chronic TCR signaling is proposed as a core mechanistic driver of functional exhaustion, especially via calcineurin-dependent TF, NFAT, and other NFAT-driven, TCR-responsive TFs such as IRF4, BATF, nuclear receptor subfamily 4 group A (NR4A), and TOX (Zajac et al., 1998; Paley et al., 2012; Barber et al., 2006; Seo et al., 2019; Chen et al., 2019a). The formation and maintenance of Texprog cells depend on TOX, but also on TFs found in memory cells and their precursors, such as TCF1 (encoded by Tcf7) and FOXO1 (Utzschneider et al., 2018). As discussed in the section Comparative analysis of CD8+ T cell differentiation trajectories…, TOX promotes Tex differentiation in part by inhibiting TE differentiation. TOX was found to bind to Kat7, the acetyl transferase component of the HBO1 complex, suggesting it may direct histone H4 and H3 acetylation in exhausted CD8+ T cells (Khan et al., 2019; Yao et al., 2019; Page et al., 2018). TOX may also have a role in regulating DNA methylation (Khan et al., 2019; Alfei et al., 2019). Now, an important next step is to understand how TOX specifically interacts with and influences the functions of promemory TFs to steer the cells toward T cell exhaustion in chronic infection and tumors.
The transition of Texprog to Texint cells is associated with the proliferation and induction of Tbx21, Zeb2, Cx3Cr1, and S1pr5 (similar to TE cells), but then these cells continue to convert into CD101hi Texterm cells and progressively methylate the DNA and silence Ifng, Tcf7, and Tbx21. In contrast, the promoters of Pdcd1, Lag3, and Havcr1 (encoding TIM3) and Cd101 are demethylated and further up-regulated (Scharer et al., 2013; Ghoneim et al., 2017; Scott-Browne et al., 2016; Sen et al., 2016). De novo methylation of effector loci by DNMT3a is critical for commitment to exhaustion by silencing effector- and memory-related gene loci (Ghoneim et al., 2017). Indeed, the epigenetic stability of the exhaustion program is remarkably robust. Even though greater numbers of functional Texint-like cells emerge with anti-PD-1 treatment, this therapy is unable to reprogram the epigenetic landscape of Tex cells to that of an effector or memory state (Pauken et al., 2016). Thus, drugs that inhibit epigenetic modifiers will likely be needed to redirect the differentiation of exhausted CD8+ T cells (Chiappinelli et al., 2016; Henning et al., 2018; Utzschneider et al., 2020; Alfei et al., 2019).
How does the same TF specify distinct differentiation states?
While we use differentially expressed genes to distinguish one CD8+ T cell differentiation state from another, there are also shared sets of coordinately expressed genes across multiple T cell subsets. This indicates that certain TFs are “multitaskers” and are used repeatedly to generate multiple types of differentiation states within T cells. For example, as mentioned above, Tcf7 and Foxo1 are involved in the generation of both MP cells in acute infection and Texprog cells in chronic infection (Fig. 2 D and Fig. 3). Similarly, T-bet and Zeb2 mRNA is common to both TE and Texint cells, and there is a great deal of overlap between TFs expressed in TRM and Texterm cells (e.g., Nr4a1, Nr4a2, Prdm1, Irf4, and Tox; Fig. 3; Hudson et al., 2019; Beltra et al., 2020; Mackay et al., 2016; Milner et al., 2017, 2020b). Eomes is particularly busy, because it is expressed the most in EE, TCM, and Texterm cells and plays a role in the development of each cell subtype. Interestingly, Eomes demonstrates cooperative behavior with T-bet in EE cells (Pearce et al., 2003) but develops a seemingly antagonistic relationship later in the balance between MP versus TE, TCM versus TEM, or Texint versus Texterm cell states. Collectively, this raises the all-important question: How does a CD8+ T cell commit to one cell fate over another if similar TFs are being used?
Another way to ask this question is, what are the context-dependent specifiers that differentially control the activity of the same TF in discrete differentiation states? One of the first mechanisms put forth by our laboratory was the use of TF gradients, wherein the expression level of a TF has one type of activity and regulates certain target genes at low amounts, but then regulates other target genes at higher amounts. Strong support for this mechanism has been observed with T-bet controlling the differentiation of MP → TE cells, Texprog → Texint cells, and TRM versus circulating TCM and TEM cells (Joshi et al., 2007; Wherry et al., 2003b; Mackay et al., 2016, 2015). Furthermore, T-bet nuclear localization is also used to regulate its function in different CD8 T cell subsets (McLane et al., 2013), but little is known about how T-bet functions to control gene expression at high and low amounts in the nucleus. Likewise, the levels of TOX can also influence Tex states (Khan et al., 2019; Scott et al., 2019; Seo et al., 2019). Other common mechanisms to alter the activity of a single TF in a context-dependent manner is via posttranslational modifications, such as phosphorylation, which allows the same TF to acquire multiple activities or binding partners. Shockingly, we know very little about the differences between TF posttranslational modifications, their genomic binding patterns, and partnerships with other TFs in acute versus chronic infection. This knowledge would go a long way toward understanding how the same TFs can be involved in overtly distinct differentiation programs.
Concluding remarks
T cell immunity has coevolved over millions of years, with pathogens adapting to diverse forms of acute and chronic infections. The result has been the creation of T cells that develop immense plasticity and heterogeneity to enable the generation of (a) expendable pools of effector cells to control the present infection and (b) longer-lived pools of multipotent cells to sustain immune responses to chronic infection or provide secondary “recall” responses at a later time of reinfection. Therefore, while the qualities of the CD8+ T cells between acute and chronic infection are overtly distinct, one can see core similarities in the underlying structure of the T cell responses between these two types of infections.
With recent technological advances, including sequencing at the single-cell level, we are now able to appreciate the heterogeneity and plasticity of antigen-specific T cells even more in various tissues, infections, cancer, autoimmunity, and other diseases. All things considered, we propose referring to T cell subsets as cell states (see text box) to convey the dynamic nature of T cell differentiation with different levels of plasticity. The current comprehensive understanding of T cell differentiation in multiple contexts raises outstanding questions: (a) Given the similarity in the transcriptional networks between Texprog versus MP, Texint versus TE or LLE, and Texterm versus TRM cells, will these states be capable of transdifferentiation to one another? What are the signaling and transcriptional pathways that drive the conversion from one cell state to another? Is it possible to reverse epigenetic imprinting of exhaustion and reprogram the state to effector or memory T cells? Can we exploit this plasticity that possibly allows transdifferentiation or reverse imprinting to synthetically create even more diverse states, aiming for better therapy for different diseases? (b) Other than TCR signaling, what are the environmental factors and their signaling pathways that divert the T cell differentiation trajectory from memory to exhaustion? (c) Which T cell states offer the most desirable therapeutic benefit in a tumor or chronic infection? Understanding these similarities and differences in T cell differentiation and programmability at different stages of the T cell response will provide insight into and new targets for novel therapeutic interventions to both rejuvenate dysfunctional, exhausted cells and to promote memory in virus infection and cancer.
Acknowledgments
We thank Drs. Jun Siong Low, Robert A. Amezquita, Hubert Tseng, Thomas H. Mann, and Anna-Maria Globig for their helpful discussion.
This work was supported by National Institutes of Health grants to S.M. Kaech (R37AI066232, R01AI123864, R21AI151986, and R01CA240909). H.K. Chung is a Damon Runyon Fellow supported by a Damon Runyon Cancer Research Foundation grant (DRG-2374-19).
Author contributions: S.M. Kaech and H.K. Chung contributed to the conceptualization and writing of the manuscript. H.K. Chung and B. McDonald contributed to the figures. S.M. Kaech provided supervision of the manuscript.
References
- Ahmadzadeh, M., Johnson L.A., Heemskerk B., Wunderlich J.R., Dudley M.E., White D.E., and Rosenberg S.A.. 2009. Tumor antigen-specific CD8 T cells infiltrating the tumor express high levels of PD-1 and are functionally impaired. Blood. 114:1537–1544. 10.1182/blood-2008-12-195792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed, R., and Gray D.. 1996. Immunological memory and protective immunity: understanding their relation. Science. 272:54–60. 10.1126/science.272.5258.54 [DOI] [PubMed] [Google Scholar]
- Akondy, R.S., Fitch M., Edupuganti S., Yang S., Kissick H.T., Li K.W., Youngblood B.A., Abdelsamed H.A., McGuire D.J., Cohen K.W., et al. 2017. Origin and differentiation of human memory CD8 T cells after vaccination. Nature. 552:362–367. 10.1038/nature24633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alfei, F., Kanev K., Hofmann M., Wu M., Ghoneim H.E., Roelli P., Utzschneider D.T., von Hoesslin M., Cullen J.G., Fan Y., et al. 2019. TOX reinforces the phenotype and longevity of exhausted T cells in chronic viral infection. Nature. 571:265–269. 10.1038/s41586-019-1326-9 [DOI] [PubMed] [Google Scholar]
- Angelosanto, J.M., Blackburn S.D., Crawford A., and Wherry E.J.. 2012. Progressive loss of memory T cell potential and commitment to exhaustion during chronic viral infection. J. Virol. 86:8161–8170. 10.1128/JVI.00889-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araki, Y., Wang Z., Zang C., Wood W.H. III, Schones D., Cui K., Roh T.-Y., Lhotsky B., Wersto R.P., Peng W., et al. 2009. Genome-wide analysis of histone methylation reveals chromatin state-based regulation of gene transcription and function of memory CD8+ T cells. Immunity. 30:912–925. 10.1016/j.immuni.2009.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arsenio, J., Kakaradov B., Metz P.J., Kim S.H., Yeo G.W., and Chang J.T.. 2014. Early specification of CD8+ T lymphocyte fates during adaptive immunity revealed by single-cell gene-expression analyses. Nat. Immunol. 15:365–372. 10.1038/ni.2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azizi, E., Carr A.J., Plitas G., Cornish A.E., Konopacki C., Prabhakaran S., Nainys J., Wu K., Kiseliovas V., Setty M., et al. 2018. Single-Cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment. Cell. 174:1293–1308.e36. 10.1016/j.cell.2018.05.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badovinac, V.P., and Harty J.T.. 2007. Manipulating the rate of memory CD8+ T cell generation after acute infection. J. Immunol. 179:53–63. 10.4049/jimmunol.179.1.53 [DOI] [PubMed] [Google Scholar]
- Badovinac, V.P., Porter B.B., and Harty J.T.. 2002. Programmed contraction of CD8(+) T cells after infection. Nat. Immunol. 3:619–626. 10.1038/ni804 [DOI] [PubMed] [Google Scholar]
- Badovinac, V.P., Porter B.B., and Harty J.T.. 2004. CD8+ T cell contraction is controlled by early inflammation. Nat. Immunol. 5:809–817. 10.1038/ni1098 [DOI] [PubMed] [Google Scholar]
- Banerjee, A., Gordon S.M., Intlekofer A.M., Paley M.A., Mooney E.C., Lindsten T., Wherry E.J., and Reiner S.L.. 2010. Cutting edge: The transcription factor eomesodermin enables CD8+ T cells to compete for the memory cell niche. J. Immunol. 185:4988–4992. 10.4049/jimmunol.1002042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barber, D.L., Wherry E.J., Masopust D., Zhu B., Allison J.P., Sharpe A.H., Freeman G.J., and Ahmed R.. 2006. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 439:682–687. 10.1038/nature04444 [DOI] [PubMed] [Google Scholar]
- Beltra, J.C., Manne S., Abdel-Hakeem M.S., Kurachi M., Giles J.R., Chen Z., Casella V., Ngiow S.F., Khan O., Huang Y.J., et al. 2020. Developmental Relationships of Four Exhausted CD8+ T Cell Subsets Reveals Underlying Transcriptional and Epigenetic Landscape Control Mechanisms. Immunity. 52:825–841.e8. 10.1016/j.immuni.2020.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengsch, B., Ohtani T., Khan O., Setty M., Manne S., O’Brien S., Gherardini P.F., Herati R.S., Huang A.C., Chang K.M., et al. 2018. Epigenomic-Guided Mass Cytometry Profiling Reveals Disease-Specific Features of Exhausted CD8 T Cells. Immunity. 48:1029–1045.e5. 10.1016/j.immuni.2018.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengsch, B., Spangenberg H.C., Kersting N., Neumann-Haefelin C., Panther E., von Weizsäcker F., Blum H.E., Pircher H., and Thimme R.. 2007. Analysis of CD127 and KLRG1 expression on hepatitis C virus-specific CD8+ T cells reveals the existence of different memory T-cell subsets in the peripheral blood and liver. J. Virol. 81:945–953. 10.1128/JVI.01354-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best, J.A., Blair D.A., Knell J., Yang E., Mayya V., Doedens A., Dustin M.L., Goldrath A.W., Monach P., and Shinton S.A.. Immunological Genome Project Consortium . 2013. Transcriptional insights into the CD8(+) T cell response to infection and memory T cell formation. Nat. Immunol. 14:404–412. 10.1038/ni.2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura, L.K., Mitchell J.S., Thompson E.A., Schenkel J.M., Mohammed J., Wijeyesinghe S., Fonseca R., Burbach B.J., Hickman H.D., Vezys V., et al. 2018a. Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19:173–182. 10.1038/s41590-017-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura, L.K., Wijeyesinghe S., Thompson E.A., Macchietto M.G., Rosato P.C., Pierson M.J., Schenkel J.M., Mitchell J.S., Vezys V., Fife B.T., et al. 2018b. T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity. 48:327–338.e5. 10.1016/j.immuni.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher, J.P., Beyer M., Meissner F., Abdullah Z., Sander J., Höchst B., Eickhoff S., Rieckmann J.C., Russo C., Bauer T., et al. 2015. Functional classification of memory CD8(+) T cells by CX3CR1 expression. Nat. Commun. 6:8306. 10.1038/ncomms9306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouneaud, C., Garcia Z., Kourilsky P., and Pannetier C.. 2005. Lineage relationships, homeostasis, and recall capacities of central- and effector-memory CD8 T cells in vivo. J. Exp. Med. 201:579–590. 10.1084/jem.20040876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutboul, F., Puthier D., Appay V., Pellé O., Ait-Mohand H., Combadière B., Carcelain G., Katlama C., Rowland-Jones S.L., Debré P., et al. 2005. Modulation of interleukin-7 receptor expression characterizes differentiation of CD8 T cells specific for HIV, EBV and CMV. AIDS. 19:1981–1986. 10.1097/01.aids.0000191919.24185.46 [DOI] [PubMed] [Google Scholar]
- Brummelman, J., Mazza E.M.C., Alvisi G., Colombo F.S., Grilli A., Mikulak J., Mavilio D., Alloisio M., Ferrari F., Lopci E., et al. 2018. High-dimensional single cell analysis identifies stem-like cytotoxic CD8+ T cells infiltrating human tumors. J. Exp. Med. 215:2520–2535. 10.1084/jem.20180684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchholz, V.R., Flossdorf M., Hensel I., Kretschmer L., Weissbrich B., Gräf P., Verschoor A., Schiemann M., Höfer T., and Busch D.H.. 2013. Disparate individual fates compose robust CD8+ T cell immunity. Science. 340:630–635. 10.1126/science.1235454 [DOI] [PubMed] [Google Scholar]
- Buck, M.D., Sowell R.T., Kaech S.M., and Pearce E.L.. 2017. Metabolic Instruction of Immunity. Cell. 169:570–586. 10.1016/j.cell.2017.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano-Gamez, E., Soskic B., Roumeliotis T.I., So E., Smyth D.J., Baldrighi M., Willé D., Nakic N., Esparza-Gordillo J., Larminie C.G.C., et al. 2020. Single-cell transcriptomics identifies an effectorness gradient shaping the response of CD4+ T cells to cytokines. Nat. Commun. 11:1801. 10.1038/s41467-020-15543-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang, J.T., Palanivel V.R., Kinjyo I., Schambach F., Intlekofer A.M., Banerjee A., Longworth S.A., Vinup K.E., Mrass P., Oliaro J., et al. 2007. Asymmetric T lymphocyte division in the initiation of adaptive immune responses. Science. 315:1687–1691. 10.1126/science.1139393 [DOI] [PubMed] [Google Scholar]
- Chang, J.T., Wherry E.J., and Goldrath A.W.. 2014. Molecular regulation of effector and memory T cell differentiation. Nat. Immunol. 15:1104–1115. 10.1038/ni.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, J., López-Moyado I.F., Seo H., Lio C.J., Hempleman L.J., Sekiya T., Yoshimura A., Scott-Browne J.P., and Rao A.. 2019a. NR4A transcription factors limit CAR T cell function in solid tumours. Nature. 567:530–534. 10.1038/s41586-019-0985-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Ji Z., Ngiow S.F., Manne S., Cai Z., Huang A.C., Johnson J., Staupe R.P., Bengsch B., Xu C., et al. 2019b. TCF-1-Centered Transcriptional Network Drives an Effector versus Exhausted CD8 T Cell-Fate Decision. Immunity. 51:840–855.e5. 10.1016/j.immuni.2019.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Z., Arai E., Khan O., Zhang Z., Ngiow S.F., He Y., Huang H., Manne S., Cao Z., Baxter A.E., et al. 2021. In vivo CD8+ T cell CRISPR screening reveals control by Fli1 in infection and cancer. Cell. 184:1262–1280.e22. 10.1016/j.cell.2021.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiappinelli, K.B., Zahnow C.A., Ahuja N., and Baylin S.B.. 2016. Combining epigenetic and immunotherapy to combat cancer. Cancer Res. 76:1683–1689. 10.1158/0008-5472.CAN-15-2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, W., Liu Y., Weinstein J.S., Craft J., and Kaech S.M.. 2011. An interleukin-21-interleukin-10-STAT3 pathway is critical for functional maturation of memory CD8+ T cells. Immunity. 35:792–805. 10.1016/j.immuni.2011.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtsinger, J.M., Schmidt C.S., Mondino A., Lins D.C., Kedl R.M., Jenkins M.K., and Mescher M.F.. 1999. Inflammatory cytokines provide a third signal for activation of naive CD4+ and CD8+ T cells. J. Immunol. 162:3256–3262. [PubMed] [Google Scholar]
- Delpoux, A., Lai C.-Y., Hedrick S.M., and Doedens A.L.. 2017. FOXO1 opposition of CD8+ T cell effector programming confers early memory properties and phenotypic diversity. Proc. Natl. Acad. Sci. USA. 114:E8865–E8874. 10.1073/pnas.1618916114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWitt, W.S., Emerson R.O., Lindau P., Vignali M., Snyder T.M., Desmarais C., Sanders C., Utsugi H., Warren E.H., McElrath J., et al. 2015. Dynamics of the cytotoxic T cell response to a model of acute viral infection. J. Virol. 89:4517–4526. 10.1128/JVI.03474-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez, C.X., Amezquita R.A., Guan T., Marshall H.D., Joshi N.S., Kleinstein S.H., and Kaech S.M.. 2015. The transcription factors ZEB2 and T-bet cooperate to program cytotoxic T cell terminal differentiation in response to LCMV viral infection. J. Exp. Med. 212:2041–2056. 10.1084/jem.20150186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enamorado, M., Iborra S., Priego E., Cueto F.J., Quintana J.A., Martínez-Cano S., Mejías-Pérez E., Esteban M., Melero I., Hidalgo A., and Sancho D.. 2017. Enhanced anti-tumour immunity requires the interplay between resident and circulating memory CD8+ T cells. Nat. Commun. 8:16073. 10.1038/ncomms16073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca, R., Beura L.K., Quarnstrom C.F., Ghoneim H.E., Fan Y., Zebley C.C., Scott M.C., Fares-Frederickson N.J., Wijeyesinghe S., Thompson E.A., et al. 2020. Developmental plasticity allows outside-in immune responses by resident memory T cells. Nat. Immunol. 21:412–421. 10.1038/s41590-020-0607-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco, F., Jaccard A., Romero P., Yu Y.-R., and Ho P.-C.. 2020. Metabolic and epigenetic regulation of T-cell exhaustion. Nat. Metab. 2:1001–1012. 10.1038/s42255-020-00280-9 [DOI] [PubMed] [Google Scholar]
- Fraser, K.A., Schenkel J.M., Jameson S.C., Vezys V., and Masopust D.. 2013. Preexisting high frequencies of memory CD8+ T cells favor rapid memory differentiation and preservation of proliferative potential upon boosting. Immunity. 39:171–183. 10.1016/j.immuni.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frizzell, H., Fonseca R., Christo S.N., Evrard M., Cruz-Gomez S., Zanluqui N.G., von Scheidt B., Freestone D., Park S.L., McWilliam H.E.G., et al. 2020. Organ-specific isoform selection of fatty acid-binding proteins in tissue-resident lymphocytes. Sci. Immunol. 5:eaay9283. 10.1126/sciimmunol.aay9283 [DOI] [PubMed] [Google Scholar]
- Gattinoni, L., Lugli E., Ji Y., Pos Z., Paulos C.M., Quigley M.F., Almeida J.R., Gostick E., Yu Z., Carpenito C., et al. 2011. A human memory T cell subset with stem cell-like properties. Nat. Med. 17:1290–1297. 10.1038/nm.2446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach, C., Moseman E.A., Loughhead S.M., Alvarez D., Zwijnenburg A.J., Waanders L., Garg R., de la Torre J.C., and von Andrian U.H.. 2016. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 45:1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerlach, C., Rohr J.C., Perié L., van Rooij N., van Heijst J.W.J., Velds A., Urbanus J., Naik S.H., Jacobs H., Beltman J.B., et al. 2013. Heterogeneous differentiation patterns of individual CD8+ T cells. Science. 340:635–639. 10.1126/science.1235487 [DOI] [PubMed] [Google Scholar]
- Ghoneim, H.E., Fan Y., Moustaki A., Abdelsamed H.A., Dash P., Dogra P., Carter R., Awad W., Neale G., Thomas P.G., and Youngblood B.. 2017. De novo epigenetic programs inhibit PD-1 blockade-mediated T cell rejuvenation. Cell. 170:142–157.e19. 10.1016/j.cell.2017.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldrath, A.W., Sivakumar P.V., Glaccum M., Kennedy M.K., Bevan M.J., Benoist C., Mathis D., and Butz E.A.. 2002. Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells. J. Exp. Med. 195:1515–1522. 10.1084/jem.20020033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, C.L., Lee L.N., Swadling L., Hutchings C., Zinser M., Highton A.J., Capone S., Folgori A., Barnes E., and Klenerman P.. 2018. Induction and Maintenance of CX3CR1-Intermediate Peripheral Memory CD8+ T Cells by Persistent Viruses and Vaccines. Cell Rep. 23:768–782. 10.1016/j.celrep.2018.03.074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, S.M., Amezquita R.A., Guan T., Kleinstein S.H., and Kaech S.M.. 2017. Polycomb Repressive Complex 2-Mediated Chromatin Repression Guides Effector CD8+ T Cell Terminal Differentiation and Loss of Multipotency. Immunity. 46:596–608. 10.1016/j.immuni.2017.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan, T., Dominguez C.X., Amezquita R.A., Laidlaw B.J., Cheng J., Henao-Mejia J., Williams A., Flavell R.A., Lu J., and Kaech S.M.. 2018. ZEB1, ZEB2, and the miR-200 family form a counterregulatory network to regulate CD8+ T cell fates. J. Exp. Med. 215:1153–1168. 10.1084/jem.20171352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X., Zhang Y., Zheng L., Zheng C., Song J., Zhang Q., Kang B., Liu Z., Jin L., Xing R., et al. 2018. Global characterization of T cells in non-small-cell lung cancer by single-cell sequencing. Nat. Med. 24:978–985. 10.1038/s41591-018-0045-3 [DOI] [PubMed] [Google Scholar]
- Hale, J.S., Youngblood B., Latner D.R., Mohammed A.U.R., Ye L., Akondy R.S., Wu T., Iyer S.S., and Ahmed R.. 2013. Distinct memory CD4+ T cells with commitment to T follicular helper- and T helper 1-cell lineages are generated after acute viral infection. Immunity. 38:805–817. 10.1016/j.immuni.2013.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamann, D., Baars P.A., Rep M.H.G., Hooibrink B., Kerkhof-Garde S.R., Klein M.R., and van Lier R.A.W.. 1997. Phenotypic and functional separation of memory and effector human CD8+ T cells. J. Exp. Med. 186:1407–1418. 10.1084/jem.186.9.1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund, E., Lewis M.W., Hanifin J.M., Mori M., Koudelka C.W., and Slifka M.K.. 2010. Antiviral immunity following smallpox virus infection: a case-control study. J. Virol. 84:12754–12760. 10.1128/JVI.01763-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrington, L.E., Galvan M., Baum L.G., Altman J.D., and Ahmed R.. 2000. Differentiating between memory and effector CD8 T cells by altered expression of cell surface O-glycans. J. Exp. Med. 191:1241–1246. 10.1084/jem.191.7.1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning, A.N., Roychoudhuri R., and Restifo N.P.. 2018. Epigenetic control of CD8+ T cell differentiation. Nat. Rev. Immunol. 18:340–356. 10.1038/nri.2017.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herndler-Brandstetter, D., Ishigame H., Shinnakasu R., Plajer V., Stecher C., Zhao J., Lietzenmayer M., Kroehling L., Takumi A., Kometani K., et al. 2018. KLRG1+ Effector CD8+ T Cells Lose KLRG1, Differentiate into All Memory T Cell Lineages, and Convey Enhanced Protective Immunity. Immunity. 48:716–729.e8. 10.1016/j.immuni.2018.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hertoghs, K.M.L., Moerland P.D., van Stijn A., Remmerswaal E.B.M., Yong S.L., van de Berg P.J.E.J., van Ham S.M., Baas F., ten Berge I.J.M., and van Lier R.A.W.. 2010. Molecular profiling of cytomegalovirus-induced human CD8+ T cell differentiation. J. Clin. Invest. 120:4077–4090. 10.1172/JCI42758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hikono, H., Kohlmeier J.E., Takamura S., Wittmer S.T., Roberts A.D., and Woodland D.L.. 2007. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J. Exp. Med. 204:1625–1636. 10.1084/jem.20070322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, W.H., Gensheimer J., Hashimoto M., Wieland A., Valanparambil R.M., Li P., Lin J.X., Konieczny B.T., Im S.J., Freeman G.J., et al. 2019. Proliferating Transitory T Cells with an Effector-like Transcriptional Signature Emerge from PD-1+ Stem-like CD8+ T Cells during Chronic Infection. Immunity. 51:1043–1058.e4. 10.1016/j.immuni.2019.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huster, K.M., Busch V., Schiemann M., Linkemann K., Kerksiek K.M., Wagner H., and Busch D.H.. 2004. Selective expression of IL-7 receptor on memory T cells identifies early CD40L-dependent generation of distinct CD8+ memory T cell subsets. Proc. Natl. Acad. Sci. USA. 101:5610–5615. 10.1073/pnas.0308054101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibegbu, C.C., Xu Y.-X., Harris W., Maggio D., Miller J.D., and Kourtis A.P.. 2005. Expression of killer cell lectin-like receptor G1 on antigen-specific human CD8+ T lymphocytes during active, latent, and resolved infection and its relation with CD57. J. Immunol. 174:6088–6094. 10.4049/jimmunol.174.10.6088 [DOI] [PubMed] [Google Scholar]
- Ichii, H., Sakamoto A., Hatano M., Okada S., Toyama H., Taki S., Arima M., Kuroda Y., and Tokuhisa T.. 2002. Role for Bcl-6 in the generation and maintenance of memory CD8+ T cells. Nat. Immunol. 3:558–563. 10.1038/ni802 [DOI] [PubMed] [Google Scholar]
- Im, S.J., Hashimoto M., Gerner M.Y., Lee J., Kissick H.T., Burger M.C., Shan Q., Hale J.S., Lee J., Nasti T.H., et al. 2016. Defining CD8+ T cells that provide the proliferative burst after PD-1 therapy. Nature. 537:417–421. 10.1038/nature19330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Im, S.J., Konieczny B.T., Hudson W.H., Masopust D., and Ahmed R.. 2020. PD-1+ stemlike CD8 T cells are resident in lymphoid tissues during persistent LCMV infection. Proc. Natl. Acad. Sci. USA. 117:4292–4299. 10.1073/pnas.1917298117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Intlekofer, A.M., Takemoto N., Wherry E.J., Longworth S.A., Northrup J.T., Palanivel V.R., Mullen A.C., Gasink C.R., Kaech S.M., Miller J.D., et al. 2005. Effector and memory CD8+ T cell fate coupled by T-bet and eomesodermin. Nat. Immunol. 6:1236–1244. 10.1038/ni1268 [DOI] [PubMed] [Google Scholar]
- Intlekofer, A.M., Takemoto N., Kao C., Banerjee A., Schambach F., Northrop J.K., Shen H., Wherry E.J., and Reiner S.L.. 2007. Requirement for T-bet in the aberrant differentiation of unhelped memory CD8+ T cells. J. Exp. Med. 204:2015–2021. 10.1084/jem.20070841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jabbari, A., and Harty J.T.. 2006. Secondary memory CD8+ T cells are more protective but slower to acquire a central-memory phenotype. J. Exp. Med. 203:919–932. 10.1084/jem.20052237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jameson, S.C., and Masopust D.. 2018. Understanding Subset Diversity in T Cell Memory. Immunity. 48:214–226. 10.1016/j.immuni.2018.02.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi, N.S., and Kaech S.M.. 2008. Effector CD8 T cell development: a balancing act between memory cell potential and terminal differentiation. J. Immunol. 180:1309–1315. 10.4049/jimmunol.180.3.1309 [DOI] [PubMed] [Google Scholar]
- Joshi, N.S., Cui W., Chandele A., Lee H.K., Urso D.R., Hagman J., Gapin L., and Kaech S.M.. 2007. Inflammation directs memory precursor and short-lived effector CD8(+) T cell fates via the graded expression of T-bet transcription factor. Immunity. 27:281–295. 10.1016/j.immuni.2007.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech, S.M., and Cui W.. 2012. Transcriptional control of effector and memory CD8+ T cell differentiation. Nat. Rev. Immunol. 12:749–761. 10.1038/nri3307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech, S.M., Hemby S., Kersh E., and Ahmed R.. 2002. Molecular and functional profiling of memory CD8 T cell differentiation. Cell. 111:837–851. 10.1016/S0092-8674(02)01139-X [DOI] [PubMed] [Google Scholar]
- Kaech, S.M., Tan J.T., Wherry E.J., Konieczny B.T., Surh C.D., and Ahmed R.. 2003. Selective expression of the interleukin 7 receptor identifies effector CD8 T cells that give rise to long-lived memory cells. Nat. Immunol. 4:1191–1198. 10.1038/ni1009 [DOI] [PubMed] [Google Scholar]
- Kallies, A., Xin A., Belz G.T., and Nutt S.L.. 2009. Blimp-1 transcription factor is required for the differentiation of effector CD8(+) T cells and memory responses. Immunity. 31:283–295. 10.1016/j.immuni.2009.06.021 [DOI] [PubMed] [Google Scholar]
- Khan, O., Giles J.R., McDonald S., Manne S., Ngiow S.F., Patel K.P., Werner M.T., Huang A.C., Alexander K.A., Wu J.E., et al. 2019. TOX transcriptionally and epigenetically programs CD8+ T cell exhaustion. Nature. 571:211–218. 10.1038/s41586-019-1325-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knell, J., Best J.A., Lind N.A., Yang E., D’Cruz L.M., and Goldrath A.W.. 2013. Id2 influences differentiation of killer cell lectin-like receptor G1(hi) short-lived CD8+ effector T cells. J. Immunol. 190:1501–1509. 10.4049/jimmunol.1200750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kok, L., Dijkgraaf F.E., Urbanus J., Bresser K., Vredevoogd D.W., Cardoso R.F., Perié L., Beltman J.B., and Schumacher T.N.. 2020. A committed tissue-resident memory T cell precursor within the circulating CD8+ effector T cell pool. J. Exp. Med. 217:e20191711. 10.1084/jem.20191711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ladle, B.H., Li K.P., Phillips M.J., Pucsek A.B., Haile A., Powell J.D., Jaffee E.M., Hildeman D.A., and Gamper C.J.. 2016. De novo DNA methylation by DNA methyltransferase 3a controls early effector CD8+ T-cell fate decisions following activation. Proc. Natl. Acad. Sci. USA. 113:10631–10636. 10.1073/pnas.1524490113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong, Y.A., Chen Y., Ong H.S., Wu D., Man K., Deleage C., Minnich M., Meckiff B.J., Wei Y., Hou Z., et al. 2016. CXCR5(+) follicular cytotoxic T cells control viral infection in B cell follicles. Nat. Immunol. 17:1187–1196. 10.1038/ni.3543 [DOI] [PubMed] [Google Scholar]
- Li, H., van der Leun A.M., Yofe I., Lubling Y., Gelbard-Solodkin D., van Akkooi A.C.J., van den Braber M., Rozeman E.A., Haanen J.B.A.G., Blank C.U., et al. 2019. Dysfunctional CD8 T Cells Form a Proliferative, Dynamically Regulated Compartment within Human Melanoma. Cell. 176:775–789.e18. 10.1016/j.cell.2018.11.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low, J.S., Farsakoglu Y., Amezcua Vesely M.C., Sefik E., Kelly J.B., Harman C.C.D., Jackson R., Shyer J.A., Jiang X., Cauley L.S., et al. 2020. Tissue-resident memory T cell reactivation by diverse antigen-presenting cells imparts distinct functional responses. J. Exp. Med. 217. 10.1084/jem.20192291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma, C., and Zhang N.. 2015. Transforming growth factor-β signaling is constantly shaping memory T-cell population. Proc. Natl. Acad. Sci. USA. 112:11013–11017. 10.1073/pnas.1510119112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay, L.K., Minnich M., Kragten N.A.M.M., Liao Y., Nota B., Seillet C., Zaid A., Man K., Preston S., Freestone D., et al. 2016. Hobit and Blimp1 instruct a universal transcriptional program of tissue residency in lymphocytes. Science. 352:459–463. 10.1126/science.aad2035 [DOI] [PubMed] [Google Scholar]
- Mackay, L.K., Wynne-Jones E., Freestone D., Pellicci D.G., Mielke L.A., Newman D.M., Braun A., Masson F., Kallies A., Belz G.T., and Carbone F.R.. 2015. T-box Transcription Factors Combine with the Cytokines TGF-β and IL-15 to Control Tissue-Resident Memory T Cell Fate. Immunity. 43:1101–1111. 10.1016/j.immuni.2015.11.008 [DOI] [PubMed] [Google Scholar]
- Marshall, H.D., Chandele A., Jung Y.W., Meng H., Poholek A.C., Parish I.A., Rutishauser R., Cui W., Kleinstein S.H., Craft J., and Kaech S.M.. 2011. Differential expression of Ly6C and T-bet distinguish effector and memory Th1 CD4(+) cell properties during viral infection. Immunity. 35:633–646. 10.1016/j.immuni.2011.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzo, A.L., Klonowski K.D., Le Bon A., Borrow P., Tough D.F., and Lefrançois L.. 2005. Initial T cell frequency dictates memory CD8+ T cell lineage commitment. Nat. Immunol. 6:793–799. 10.1038/ni1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzo, A.L., Yagita H., and Lefrançois L.. 2007. Cutting edge: migration to nonlymphoid tissues results in functional conversion of central to effector memory CD8 T cells. J. Immunol. 179:36–40. 10.4049/jimmunol.179.1.36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust, D., Ha S.J., Vezys V., and Ahmed R.. 2006. Stimulation history dictates memory CD8 T cell phenotype: implications for prime-boost vaccination. J. Immunol. 177:831–839. 10.4049/jimmunol.177.2.831 [DOI] [PubMed] [Google Scholar]
- Masopust, D., Vezys V., Marzo A.L., and Lefrançois L.. 2001. Preferential localization of effector memory cells in nonlymphoid tissue. Science. 291:2413–2417. 10.1126/science.1058867 [DOI] [PubMed] [Google Scholar]
- McLane, L.M., Banerjee P.P., Cosma G.L., Makedonas G., Wherry E.J., Orange J.S., and Betts M.R.. 2013. Differential localization of T-bet and Eomes in CD8 T cell memory populations. J. Immunol. 190:3207–3215. 10.4049/jimmunol.1201556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, B.C., Sen D.R., Al Abosy R., Bi K., Virkud Y.V., LaFleur M.W., Yates K.B., Lako A., Felt K., Naik G.S., et al. 2019. Subsets of exhausted CD8+ T cells differentially mediate tumor control and respond to checkpoint blockade. Nat. Immunol. 20:326–336. 10.1038/s41590-019-0312-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Nguyen H., Omilusik K., Reina-Campos M., Tsai M., Toma C., Delpoux A., Boland B.S., Hedrick S.M., Chang J.T., and Goldrath A.W.. 2020a. Delineation of a molecularly distinct terminally differentiated memory CD8 T cell population. Proc. Natl. Acad. Sci. USA. 117:25667–25678. 10.1073/pnas.2008571117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Toma C., Yu B., Zhang K., Omilusik K., Phan A.T., Wang D., Getzler A.J., Nguyen T., Crotty S., et al. 2017. Runx3 programs CD8+ T cell residency in non-lymphoid tissues and tumours. Nature. 552:253–257. 10.1038/nature24993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milner, J.J., Toma C., He Z., Kurd N.S., Nguyen Q.P., McDonald B., Quezada L., Widjaja C.E., Witherden D.A., Crowl J.T., et al. 2020b. Heterogenous Populations of Tissue-Resident CD8+ T Cells Are Generated in Response to Infection and Malignancy. Immunity. 52:808–824.e7. 10.1016/j.immuni.2020.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mognol, G.P., Spreafico R., Wong V., Scott-Browne J.P., Togher S., Hoffmann A., Hogan P.G., Rao A., and Trifari S.. 2017. Exhaustion-associated regulatory regions in CD8+ tumor-infiltrating T cells. Proc. Natl. Acad. Sci. USA. 114:E2776–E2785. 10.1073/pnas.1620498114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosmann, T.R., Cherwinski H., Bond M.W., Giedlin M.A., and Coffman R.L.. 1986. Two types of murine helper T cell clone. I. Definition according to profiles of lymphokine activities and secreted proteins. J. Immunol. 136:2348–2357. [PubMed] [Google Scholar]
- Nolz, J.C., and Harty J.T.. 2011. Protective capacity of memory CD8+ T cells is dictated by antigen exposure history and nature of the infection. Immunity. 34:781–793. 10.1016/j.immuni.2011.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obar, J.J., Jellison E.R., Sheridan B.S., Blair D.A., Pham Q.M., Zickovich J.M., and Lefrançois L.. 2011. Pathogen-induced inflammatory environment controls effector and memory CD8+ T cell differentiation. J. Immunol. 187:4967–4978. 10.4049/jimmunol.1102335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odorizzi, P.M., Pauken K.E., Paley M.A., Sharpe A., and Wherry E.J.. 2015. Genetic absence of PD-1 promotes accumulation of terminally differentiated exhausted CD8+ T cells. J. Exp. Med. 212:1125–1137. 10.1084/jem.20142237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oestreich, K.J., Mohn S.E., and Weinmann A.S.. 2012. Molecular mechanisms that control the expression and activity of Bcl-6 in TH1 cells to regulate flexibility with a TFH-like gene profile. Nat. Immunol. 13:405–411. 10.1038/ni.2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, J.A., McDonald-Hyman C., Jameson S.C., and Hamilton S.E.. 2013. Effector-like CD8+ T cells in the memory population mediate potent protective immunity. Immunity. 38:1250–1260. 10.1016/j.immuni.2013.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omilusik, K.D., Best J.A., Yu B., Goossens S., Weidemann A., Nguyen J.V., Seuntjens E., Stryjewska A., Zweier C., Roychoudhuri R., et al. 2015. Transcriptional repressor ZEB2 promotes terminal differentiation of CD8+ effector and memory T cell populations during infection. J. Exp. Med. 212:2027–2039. 10.1084/jem.20150194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborn, J.F., Hobbs S.J., Mooster J.L., Khan T.N., Kilgore A.M., Harbour J.C., and Nolz J.C.. 2019. Central memory CD8+ T cells become CD69+ tissue-residents during viral skin infection independent of CD62L-mediated lymph node surveillance. PLoS Pathog. 15:e1007633. 10.1371/journal.ppat.1007633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, Q., Wagner W.M., Voehringer D., Wikby A., Klatt T., Walter S., Müller C.A., Pircher H., and Pawelec G.. 2003. Age-associated accumulation of CMV-specific CD8+ T cells expressing the inhibitory killer cell lectin-like receptor G1 (KLRG1). Exp. Gerontol. 38:911–920. 10.1016/S0531-5565(03)00134-7 [DOI] [PubMed] [Google Scholar]
- Pace, L., Goudot C., Zueva E., Gueguen P., Burgdorf N., Waterfall J.J., Quivy J.P., Almouzni G., and Amigorena S.. 2018. The epigenetic control of stemness in CD8+ T cell fate commitment. Science. 359:177–186. 10.1126/science.aah6499 [DOI] [PubMed] [Google Scholar]
- Page, N., Klimek B., De Roo M., Steinbach K., Soldati H., Lemeille S., Wagner I., Kreutzfeldt M., Di Liberto G., Vincenti I., et al. 2018. Expression of the DNA-Binding Factor TOX Promotes the Encephalitogenic Potential of Microbe-Induced Autoreactive CD8+ T Cells. Immunity. 48:937–950.e8. 10.1016/j.immuni.2018.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paley, M.A., Kroy D.C., Odorizzi P.M., Johnnidis J.B., Dolfi D.V., Barnett B.E., Bikoff E.K., Robertson E.J., Lauer G.M., Reiner S.L., and Wherry E.J.. 2012. Progenitor and terminal subsets of CD8+ T cells cooperate to contain chronic viral infection. Science. 338:1220–1225. 10.1126/science.1229620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.L., Zaid A., Hor J.L., Christo S.N., Prier J.E., Davies B., Alexandre Y.O., Gregory J.L., Russell T.A., Gebhardt T., et al. 2018. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19:183–191. 10.1038/s41590-017-0027-5 [DOI] [PubMed] [Google Scholar]
- Pauken, K.E., Sammons M.A., Odorizzi P.M., Manne S., Godec J., Khan O., Drake A.M., Chen Z., Sen D.R., Kurachi M., et al. 2016. Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science. 354:1160–1165. 10.1126/science.aaf2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce, E.L., Mullen A.C., Martins G.A., Krawczyk C.M., Hutchins A.S., Zediak V.P., Banica M., DiCioccio C.B., Gross D.A., Mao C.A., et al. 2003. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science. 302:1041–1043. 10.1126/science.1090148 [DOI] [PubMed] [Google Scholar]
- Pepper, M., and Jenkins M.K.. 2011. Origins of CD4(+) effector and central memory T cells. Nat. Immunol. 12:467–471. 10.1038/ni.2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper, M., Pagán A.J., Igyártó B.Z., Taylor J.J., and Jenkins M.K.. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 35:583–595. 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philip, M., Fairchild L., Sun L., Horste E.L., Camara S., Shakiba M., Scott A.C., Viale A., Lauer P., Merghoub T., et al. 2017. Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature. 545:452–456. 10.1038/nature22367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumlee, C.R., Sheridan B.S., Cicek B.B., and Lefrançois L.. 2013. Environmental cues dictate the fate of individual CD8+ T cells responding to infection. Immunity. 39:347–356. 10.1016/j.immuni.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao, R.R., Li Q., Odunsi K., and Shrikant P.A.. 2010. The mTOR kinase determines effector versus memory CD8+ T cell fate by regulating the expression of transcription factors T-bet and Eomesodermin. Immunity. 32:67–78. 10.1016/j.immuni.2009.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renkema, K.R., Huggins M.A., Borges da Silva H., Knutson T.P., Henzler C.M., and Hamilton S.E.. 2020. KLRG1+ Memory CD8 T Cells Combine Properties of Short-Lived Effectors and Long-Lived Memory. J. Immunol. 205:1059–1069. 10.4049/jimmunol.1901512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinstein, M.P., Lind N.A., Purton J.F., Filippou P., Best J.A., McGhee P.A., Surh C.D., and Goldrath A.W.. 2008. IL-7 and IL-15 differentially regulate CD8+ T-cell subsets during contraction of the immune response. Blood. 112:3704–3712. 10.1182/blood-2008-06-160945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ, B.E., Olshanksy M., Smallwood H.S., Li J., Denton A.E., Prier J.E., Stock A.T., Croom H.A., Cullen J.G., Nguyen M.L., et al. 2014. Distinct epigenetic signatures delineate transcriptional programs during virus-specific CD8(+) T cell differentiation. Immunity. 41:853–865. 10.1016/j.immuni.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell, S., Bennett J., Wellman J.A., Chung D.C., Yu Z.-F., Tillman A., Wittes J., Pappas J., Elci O., McCague S., et al. 2017. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. Lancet. 390:849–860. 10.1016/S0140-6736(17)31868-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rutishauser, R.L., Martins G.A., Kalachikov S., Chandele A., Parish I.A., Meffre E., Jacob J., Calame K., and Kaech S.M.. 2009. Transcriptional repressor Blimp-1 promotes CD8(+) T cell terminal differentiation and represses the acquisition of central memory T cell properties. Immunity. 31:296–308. 10.1016/j.immuni.2009.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sade-Feldman, M., Yizhak K., Bjorgaard S.L., Ray J.P., de Boer C.G., Jenkins R.W., Lieb D.J., Chen J.H., Frederick D.T., Barzily-Rokni M., et al. 2018. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 175:998–1013.e20. 10.1016/j.cell.2018.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto, F., Lenig D., Förster R., Lipp M., and Lanzavecchia A.. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Sandu, I., Cerletti D., Oetiker N., Borsa M., Wagen F., Spadafora I., Welten S.P.M., Stolz U., Oxenius A., and Claassen M.. 2020. Landscape of Exhausted Virus-Specific CD8 T Cells in Chronic LCMV Infection. Cell Rep. 32:108078. 10.1016/j.celrep.2020.108078 [DOI] [PubMed] [Google Scholar]
- Sarkar, S., Kalia V., Haining W.N., Konieczny B.T., Subramaniam S., and Ahmed R.. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205:625–640. 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satpathy, A.T., Granja J.M., Yost K.E., Qi Y., Meschi F., McDermott G.P., Olsen B.N., Mumbach M.R., Pierce S.E., Corces M.R., et al. 2019. Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37:925–936. 10.1038/s41587-019-0206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauce, D., Almeida J.R., Larsen M., Haro L., Autran B., Freeman G.J., and Appay V.. 2007. PD-1 expression on human CD8 T cells depends on both state of differentiation and activation status. AIDS. 21:2005–2013. 10.1097/QAD.0b013e3282eee548 [DOI] [PubMed] [Google Scholar]
- Savas, P., Virassamy B., Ye C., Salim A., Mintoff C.P., Caramia F., Salgado R., Byrne D.J., Teo Z.L., Dushyanthen S., et al. Kathleen Cuningham Foundation Consortium for Research into Familial Breast Cancer (kConFab) . 2018. Single-cell profiling of breast cancer T cells reveals a tissue-resident memory subset associated with improved prognosis. Nat. Med. 24:986–993. 10.1038/s41591-018-0078-7 [DOI] [PubMed] [Google Scholar]
- Scharer, C.D., Bally A.P.R., Gandham B., and Boss J.M.. 2017. Cutting Edge: Chromatin Accessibility Programs CD8 T Cell Memory. J. Immunol. 198:2238–2243. 10.4049/jimmunol.1602086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharer, C.D., Barwick B.G., Youngblood B.A., Ahmed R., and Boss J.M.. 2013. Global DNA methylation remodeling accompanies CD8 T cell effector function. J. Immunol. 191:3419–3429. 10.4049/jimmunol.1301395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schietinger, A., Philip M., Krisnawan V.E., Chiu E.Y., Delrow J.J., Basom R.S., Lauer P., Brockstedt D.G., Knoblaugh S.E., Hämmerling G.J., et al. 2016. Tumor-specific T cell dysfunction is a dynamic antigen-driven differentiation program initiated early during tumorigenesis. Immunity. 45:389–401. 10.1016/j.immuni.2016.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluns, K.S., Kieper W.C., Jameson S.C., and Lefrançois L.. 2000. Interleukin-7 mediates the homeostasis of naïve and memory CD8 T cells in vivo. Nat. Immunol. 1:426–432. 10.1038/80868 [DOI] [PubMed] [Google Scholar]
- Schulte, I., Hitziger T., Giugliano S., Timm J., Gold H., Heinemann F.M., Khudyakov Y., Strasser M., König C., Castermans E., et al. 2011. Characterization of CD8+ T-cell response in acute and resolved hepatitis A virus infection. J. Hepatol. 54:201–208. 10.1016/j.jhep.2010.07.010 [DOI] [PubMed] [Google Scholar]
- Scott, A.C., Dündar F., Zumbo P., Chandran S.S., Klebanoff C.A., Shakiba M., Trivedi P., Menocal L., Appleby H., Camara S., et al. 2019. TOX is a critical regulator of tumour-specific T cell differentiation. Nature. 571:270–274. 10.1038/s41586-019-1324-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Browne, J.P., López-Moyado I.F., Trifari S., Wong V., Chavez L., Rao A., and Pereira R.M.. 2016. Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity. 45:1327–1340. 10.1016/j.immuni.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen, D.R., Kaminski J., Barnitz R.A., Kurachi M., Gerdemann U., Yates K.B., Tsao H.-W.W., Godec J., LaFleur M.W., Brown F.D., et al. 2016. The epigenetic landscape of T cell exhaustion. Science. 354:1165–1169. 10.1126/science.aae0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo, H., Chen J., González-Avalos E., Samaniego-Castruita D., Das A., Wang Y.H., López-Moyado I.F., Georges R.O., Zhang W., Onodera A., et al. 2019. TOX and TOX2 transcription factors cooperate with NR4A transcription factors to impose CD8+ T cell exhaustion. Proc. Natl. Acad. Sci. USA. 116:12410–12415. 10.1073/pnas.1905675116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slütter, B., Van Braeckel-Budimir N., Abboud G., Varga S.M., Salek-Ardakani S., and Harty J.T.. 2017. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2:eaag2031. 10.1126/sciimmunol.aag2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spranger, S., Bao R., and Gajewski T.F.. 2015. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 523:231–235. 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- Stelekati, E., Chen Z., Manne S., Kurachi M., Ali M.A., Lewy K., Cai Z., Nzingha K., McLane L.M., Hope J.L., et al. 2018. Long-Term Persistence of Exhausted CD8 T Cells in Chronic Infection Is Regulated by MicroRNA-155. Cell Rep. 23:2142–2156. 10.1016/j.celrep.2018.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thommen, D.S., Koelzer V.H., Herzig P., Roller A., Trefny M., Dimeloe S., Kiialainen A., Hanhart J., Schill C., Hess C., et al. 2018. A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 24:994–1004. 10.1038/s41591-018-0057-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tirosh, I., Izar B., Prakadan S.M., Wadsworth M.H. II, Treacy D., Trombetta J.J., Rotem A., Rodman C., Lian C., Murphy G., et al. 2016. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. 352:189–196. 10.1126/science.aad0501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripp, R.A., Hou S., and Doherty P.C.. 1995. Temporal loss of the activated L-selectin-low phenotype for virus-specific CD8+ memory T cells. J. Immunol. 154:5870–5875. [PubMed] [Google Scholar]
- Tubo, N.J., Pagán A.J., Taylor J.J., Nelson R.W., Linehan J.L., Ertelt J.M., Huseby E.S., Way S.S., and Jenkins M.K.. 2013. Single naive CD4+ T cells from a diverse repertoire produce different effector cell types during infection. Cell. 153:785–796. 10.1016/j.cell.2013.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider, D.T., Charmoy M., Chennupati V., Pousse L., Ferreira D.P., Calderon-Copete S., Danilo M., Alfei F., Hofmann M., Wieland D., et al. 2016. T cell factor 1-expressing memory-like CD8+ T cells sustain the immune response to chronic viral infections. Immunity. 45:415–427. 10.1016/j.immuni.2016.07.021 [DOI] [PubMed] [Google Scholar]
- Utzschneider, D.T., Delpoux A., Wieland D., Huang X., Lai C.Y., Hofmann M., Thimme R., and Hedrick S.M.. 2018. Active Maintenance of T Cell Memory in Acute and Chronic Viral Infection Depends on Continuous Expression of FOXO1. Cell Rep. 22:3454–3467. 10.1016/j.celrep.2018.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utzschneider, D.T., Gabriel S.S., Chisanga D., Gloury R., Gubser P.M., Vasanthakumar A., Shi W., and Kallies A.. 2020. Early precursor T cells establish and propagate T cell exhaustion in chronic infection. Nat. Immunol. 21:1256–1266. 10.1038/s41590-020-0760-z [DOI] [PubMed] [Google Scholar]
- Van Braeckel-Budimir, N., Varga S.M., Badovinac V.P., and Harty J.T.. 2018. Repeated Antigen Exposure Extends the Durability of Influenza-Specific Lung-Resident Memory CD8+ T Cells and Heterosubtypic Immunity. Cell Rep. 24:3374–3382.e3. 10.1016/j.celrep.2018.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen, E.M.M., de Bree G.J., Remmerswaal E.B.M., Yong S.L., Tesselaar K., ten Berge I.J.M., and van Lier R.A.W.. 2005. IL-7 receptor α chain expression distinguishes functional subsets of virus-specific human CD8+ T cells. Blood. 106:2091–2098. 10.1182/blood-2005-02-0449 [DOI] [PubMed] [Google Scholar]
- Wang, S., Zhang Q., Hui H., Agrawal K., Karris M.A.Y., and Rana T.M.. 2020. An atlas of immune cell exhaustion in HIV-infected individuals revealed by single-cell transcriptomics. Emerg. Microbes Infect. 9:2333–2347. 10.1080/22221751.2020.1826361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, J., Luo C., Wang Y., Guo Y., Dai H., Tong C., Ti D., Wu Z., and Han W.. 2019. PD-1 silencing impairs the anti-tumor function of chimeric antigen receptor modified T cells by inhibiting proliferation activity. J. Immunother. Cancer. 7:209. 10.1186/s40425-019-0685-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J., and Ahmed R.. 2004. Memory CD8 T-cell differentiation during viral infection. J. Virol. 78:5535–5545. 10.1128/JVI.78.11.5535-5545.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J., Blattman J.N., Murali-Krishna K., van der Most R., and Ahmed R.. 2003a. Viral persistence alters CD8 T-cell immunodominance and tissue distribution and results in distinct stages of functional impairment. J. Virol. 77:4911–4927. 10.1128/JVI.77.8.4911-4927.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J., Day C.L., Draenert R., Miller J.D., Kiepiela P., Woodberry T., Brander C., Addo M., Klenerman P., Ahmed R., and Walker B.D.. 2006. HIV-specific CD8 T cells express low levels of IL-7Ralpha: implications for HIV-specific T cell memory. Virology. 353:366–373. 10.1016/j.virol.2006.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wherry, E.J., Ha S.J., Kaech S.M., Haining W.N., Sarkar S., Kalia V., Subramaniam S., Blattman J.N., Barber D.L., and Ahmed R.. 2007. Molecular signature of CD8+ T cell exhaustion during chronic viral infection. Immunity. 27:670–684. 10.1016/j.immuni.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Wherry, E.J., Teichgräber V., Becker T.C., Masopust D., Kaech S.M., Antia R., von Andrian U.H., and Ahmed R.. 2003b. Lineage relationship and protective immunity of memory CD8 T cell subsets. Nat. Immunol. 4:225–234. 10.1038/ni889 [DOI] [PubMed] [Google Scholar]
- Willimsky, G., and Blankenstein T.. 2005. Sporadic immunogenic tumours avoid destruction by inducing T-cell tolerance. Nature. 437:141–146. 10.1038/nature03954 [DOI] [PubMed] [Google Scholar]
- Wirth, T.C., Xue H.H., Rai D., Sabel J.T., Bair T., Harty J.T., and Badovinac V.P.. 2010. Repetitive antigen stimulation induces stepwise transcriptome diversification but preserves a core signature of memory CD8(+) T cell differentiation. Immunity. 33:128–140. 10.1016/j.immuni.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, T., Ji Y., Moseman E.A., Xu H.C., Manglani M., Kirby M., Anderson S.M., Handon R., Kenyon E., Elkahloun A., et al. 2016. The TCF1-Bcl6 axis counteracts type I interferon to repress exhaustion and maintain T cell stemness. Sci. Immunol. 1:eaai8593. 10.1126/sciimmunol.aai8593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan, Y., Cao S., Liu X., Harrington S.M., Bindeman W.E., Adjei A.A., Jang J.S., Jen J., Li Y., Chanana P., et al. 2018. CX3CR1 identifies PD-1 therapy-responsive CD8+ T cells that withstand chemotherapy during cancer chemoimmunotherapy. JCI Insight. 3:e97828. 10.1172/jci.insight.97828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, C.Y., Best J.A., Knell J., Yang E., Sheridan A.D., Jesionek A.K., Li H.S., Rivera R.R., Lind K.C., D’Cruz L.M., et al. 2011. The transcriptional regulators Id2 and Id3 control the formation of distinct memory CD8+ T cell subsets. Nat. Immunol. 12:1221–1229. 10.1038/ni.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao, C., Sun H.-W., Lacey N.E., Ji Y., Moseman E.A., Shih H.-Y., Heuston E.F., Kirby M., Anderson S., Cheng J., et al. 2019. Single-cell RNA-seq reveals TOX as a key regulator of CD8+ T cell persistence in chronic infection. Nat. Immunol. 20:890–901. 10.1038/s41590-019-0403-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yost, K.E., Satpathy A.T., Wells D.K., Qi Y., Wang C., Kageyama R., McNamara K.L., Granja J.M., Sarin K.Y., Brown R.A., et al. 2019. Clonal replacement of tumor-specific T cells following PD-1 blockade. Nat. Med. 25:1251–1259. 10.1038/s41591-019-0522-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood, B., Hale J.S., Kissick H.T., Ahn E., Xu X., Wieland A., Araki K., West E.E., Ghoneim H.E., Fan Y., et al. 2017. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature. 552:404–409. 10.1038/nature25144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zajac, A.J., Blattman J.N., Murali-Krishna K., Sourdive D.J., Suresh M., Altman J.D., and Ahmed R.. 1998. Viral immune evasion due to persistence of activated T cells without effector function. J. Exp. Med. 188:2205–2213. 10.1084/jem.188.12.2205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zander, R., Schauder D., Xin G., Nguyen C., Wu X., Zajac A., and Cui W.. 2019. CD4+ T cell help is required for the formation of a cytolytic CD8+ T cell subset that protects against chronic infection and cancer. Immunity. 51:1028–1042.e4. 10.1016/j.immuni.2019.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, L., Yu X., Zheng L., Zhang Y., Li Y., Fang Q., Gao R., Kang B., Zhang Q., Huang J.Y., et al. 2018. Lineage tracking reveals dynamic relationships of T cells in colorectal cancer. Nature. 564:268–272. 10.1038/s41586-018-0694-x [DOI] [PubMed] [Google Scholar]
- Zhang, Q., He Y., Luo N., Patel S.J., Han Y., Gao R., Modak M., Carotta S., Haslinger C., Kind D., et al. 2019. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell. 179:829–845.e20. 10.1016/j.cell.2019.10.003 [DOI] [PubMed] [Google Scholar]
- Zheng, C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., et al. 2017. Landscape of Infiltrating T Cells in Liver Cancer Revealed by Single-Cell Sequencing. Cell. 169:1342–1356.e16. 10.1016/j.cell.2017.05.035 [DOI] [PubMed] [Google Scholar]