Abstract
Background
Diffuse midline gliomas (DMGs), including diffuse intrinsic pontine gliomas (DIPGs), have a dismal prognosis, with less than 2% surviving 5 years postdiagnosis. The majority of DIPGs and all DMGs harbor mutations altering the epigenetic regulatory histone tail (H3 K27M). Investigations addressing DMG epigenetics have identified a few promising drugs, including the HDAC inhibitor (HDACi) panobinostat. Here, we use clinically relevant DMG models to identify and validate other effective HDACi and their biomarkers of response.
Methods
HDAC inhibitors were tested across biopsy-derived treatment-naïve in vitro and in vivo DMG models with biologically relevant radiation resistance. RNA sequencing was performed to define and compare drug efficacy and to map predictive biomarkers of response.
Results
Quisinostat and romidepsin showed efficacy with low nanomolar half-maximal inhibitory concentration (IC50) values (~50 and ~5 nM, respectively). Comparative transcriptome analyses across quisinostat, romidepsin, and panobinostat showed a greater degree of shared biological effects between quisinostat and panobinostat, and less overlap with romidepsin. However, some transcriptional changes were consistent across all 3 drugs at similar biologically effective doses, such as overexpression of troponin T1 slow skeletal type (TNNT1) and downregulation of collagen type 20 alpha 1 chain (COL20A1), identifying these as potential vulnerabilities or on-target biomarkers in DMG. Quisinostat and romidepsin significantly (P < 0.0001) inhibited in vivo tumor growth.
Conclusions
Our data highlight the utility of treatment-naïve biopsy-derived models; establishes quisinostat and romidepsin as effective in vivo; illuminates potential mechanisms and/or biomarkers of DMG cell lethality due to HDAC inhibition; and emphasizes the need for brain tumor–penetrant versions of potentially efficacious agents.
Keywords: diffuse intrinsic pontine glioma (DIPG), diffuse midline glioma, H3 K27M-mutant (DMG), histone deacetylase inhibitor (HDACi), quisinostat, romidepsin
Key Points.
1. Treatment-naïve biopsy-derived DMG models have biologically relevant radiation resistance and blood-brain-tumor barriers.
2. Quisinostat and romidepsin are cytotoxic HDAC inhibitors against DMG.
3. Upregulation of TNNT1 or downregulation of COL20A1 may serve as a clinical biomarker of HDACi-induced lethality.
Importance of the Study.
Clinical progress toward effective treatment of DMG has been hampered by limited preclinical identification of agents active at relevant biological concentrations. Here, we create biopsy-derived treatment-naïve DMG models and are the first to demonstrate the in vivo efficacy of the HDAC inhibitors quisinostat and romidepsin in DMG. Furthermore, transcriptional analysis revealed critical overlapping and distinct profiles between panobinostat and quisinostat versus romidepsin, suggestive of potential vulnerabilities within DMG and biomarkers of on-target cytotoxic HDAC inhibition.
Diffuse intrinsic pontine glioma (DIPG) is a universally fatal brainstem tumor diagnosed in more than 300 children per year in the US.1 The lack of clinical progress beyond the advent of focal radiation has stranded the median overall survival (OS) at 11 months for decades.2 Due to its critical location in the brainstem, DIPG cannot be surgically resected, so many patients are diagnosed radiographically.3 For many years, it was recommended not to perform DIPG biopsies, as pathology failed to alter therapy.4 The subsequent lack of available pathologic tissue hampered both the biologic understanding and the development of preclinical models. Only in the past decade have postmortem DIPG collections and a resurgence in DIPG biopsies provided a scaffolding on which biologic investigations of DIPG can be built.5,6 Subsequently, H3 K27M mutations, most often in genes encoding histone 3.3 (H3.3) or 3.1 (H3.1), were discovered to decrease epigenetically driven transcriptional repression in DIPG.7,8 Midline gliomas sharing these histone 3 mutations and fatal outcomes are now unified under the term “diffuse midline glioma, H3 K27M-mutant” (DMG).9 Expression of H3 K27M causes an increase in histone acetylation and reduction in H3 K27 methylation.10 Further increasing this pathological histone acetylation through the use of the histone deacetylase inhibitor (HDACi) panobinostat has shown efficacy in several preclinical DMG models. Panobinostat treatment causes metabolic dysfunction and transcriptome dysregulation, although the precise mechanism of cytotoxicity of many HDACi in DIPG has yet to be fully elucidated.11,12 With available models and deeper molecular understanding, epigenetic-regulating agents have been evaluated, including panobinostat, a pan-HDACi improving OS in some in vivo models.5,13,14 Considering its clinical availability and preclinical efficacy, panobinostat is being evaluated in several DIPG clinical trials (eg, NCT02717455, -03566199, -04341311).
Histone deacetylases (HDACs) regulate gene expression by enzymatically removing histone acetyl groups; therefore, HDAC inhibition can result in profound transcriptional effects especially with multiple, variably expressed HDACs among different cancers.15 While pan-HDACi are preclinically effective against a range of cancers, they have broad effects on normal tissue as well. In a phase I study of children with refractory solid tumors treated with panobinostat, one third of children experienced grades 3–4 thrombocytopenia.16 However, there are other HDACi that have demonstrated preclinical/clinical efficacy and may provide the same cytotoxicity of panobinostat with less toxicity, so they warrant further evaluations into their underlying mechanisms of cytotoxicity against DMG. Here, we utilize novel biopsy-derived treatment-naïve DIPG models to identify novel HDACi that are cytotoxic to DIPG. Specifically, we show in vivo efficacy of quisinostat and romidepsin. In addition, we use gene expression analysis to describe markers of optimal HDAC inhibition.
Materials and Methods
Histology
Paraffin sections were cut and placed on charged slides. Hematoxylin and eosin (H&E) staining was performed in the standard fashion. Immunohistochemical (IHC) staining was performed using a Ventana Benchmark Stainer. Sections were incubated with primary antibody to H3 K27M at 1:1200 (Millipore). Slides were incubated with biotinylated secondary antibodies, followed by incubation with the streptavidin and biotinylated peroxidase complex. Sections were counterstained with hematoxylin and mounted.
Human Specimen and Patient-Derived Cell Cultures
Human cell cultures were generated with informed consent in compliance with institutional review board approval at Seattle Children’s Hospital (#14449), Children’s National Medical Center (#1339), and University Children’s Hospital Zurich (#2019-00615). PBT-09FH, PBT-22FH, PBT-24FH, PBT-27FH, and DRIz-D105 were biopsy derived at diagnosis. For PBT-09FH, PBT-22FH, PBT-24FH, PBT-27FH, and MED-411 tumor tissue was obtained at Seattle Children’s Hospital and cell cultures were created at Fred Hutchinson Cancer Research Center (FHCRC). HSJD-DIPG007, from Dr Angel Montero Carcaboso (Hospital Sant Joan de Deu, Barcelona), and SU-DIPG48, from Dr Michelle Monje (Stanford University), were generously donated to University Children’s Hospital Zurich. Cells were maintained in NeuroCult NS-A Basal Medium with NS-A Proliferation Supplement (Stemcell Technologies), 1X Antibiotic/Antimycotic (ThermoFisher Scientific), 40 ng/mL epidermal growth factor (PeproTech), and 40 ng/mL fibroblast growth factor (PeproTech). All cell culture models were validated by DNA fingerprinting.
Drugs, Radiation, Cell Viability Assays, Apoptosis Assays
Panobinostat (LBH589), quisinostat (JNJ-26481585), vorinostat (MK0683), entinostat (MS-275), romidepsin (FK228), and CAY10603 were purchased from Selleck Chemicals Llc. For in vitro drug studies, cells were plated in 96-well plates at 15 000 cells per well and cultured 72 hours in the presence of drug in at least duplicate. Experiments were repeated for validation. Radiation studies were performed using an X-rad 320 Precision X-ray (Precision X-Ray) using stage position 4, 320 KV, and 12.50 mA on filter 1. For cell viability studies following radiation, 96-well plates were coated in 10 µg/mL of laminin (Sigma-Aldrich) in Dulbecco’s phosphate buffered saline and incubated for >4 hours, after which 15 000 cells per well of a single-cell suspension were plated, 24 hours later irradiated, then viability was measured 96 hours later. Cell viability was measured using CellTiter-Glo Luminescent Cell Viability Assay (Promega), and data were collected on a Synergy 2 plate reader (Bio-Tek). Flow cytometry was performed on a NovoCyte Flow Cytometer (ACEA Biosciences) using annexin V–fluorescein isothiocyanate (Biolegend) and data were analyzed using FlowJo software (Becton-Dickinson). Cell viability for CNMC-XD-760, DRIz-D105, CNMC-D967, CNMC-D1008, HSJD-DIPG007, and SU-DIPG48 cells was done by plating 5000 cells/well into 96-well plates, then, after 24 hours, cells were exposed to drug for 72 hours. Viability was measured using CellTiter-Glo assay (Promega) and data were collected on a Biotek Cytation 3 luminescence reader.
Antibodies and Western Blotting
Cells were lysed in M-PER lysis buffer (ThermoFisher) and supplemented with PhosSTOP and cOmplete inhibitors (Sigma-Aldrich), and the protein concentration was measured utilizing QuickStart Bradford 1X Dye Reagent (Bio-Rad). Samples were resolved on Bolt 4–12% Bis-Tris gels (ThermoFisher), transferred to 0.2 µm nitrocellulose, and blotted with either acetylated lysine antibody at 1:1000, acetyl α-tubulin (Lys40) antibody at 1:1000, β-actin (8H10D10) mouse monoclonal antibody (mAb) at 1:2000 (Cell Signaling Technologies), or anti-histone H3 (acetyl K9, K14, K18, K23, K27) antibody 47915 (Abcam) at 1:1000. Select lysates were stained for poly(ADP-ribose) polymerase (PARP) cleavage using a Cell Signal Technology antibody (D64E10). Secondary antibodies from Li-Cor Biosciences were used at 1:10 000 to detect primaries (IRDye 800CW donkey anti-mouse immunoglobulin [Ig]G and IRDye 680RD goat anti-rabbit IgG).
Surgical Procedure and In Vivo Treatment of Tumor-Bearing Mice
Mouse studies were conducted in accordance with the FHCRC Institutional Animal Care and Use Committee approved protocol #1457. Eight-week-old NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ (NSG) mice were provided by internal breeding. Athymic nude (Hsd:Athymic Nude-Foxn1nu) mice were obtained from Envigo. Intracranial xenografts were established in nonobese diabetic severe combined immunodeficient gamma (NSG) mice by injecting 100 000 tumor cells suspended in 2 µL phosphate buffered saline (PBS) at 2 mm lateral and 1 mm caudal to lambda. Symptomatic mice were euthanized, and tumors were resected for analysis and generation of flank xenografts. Flank xenografts were established by injection of ~2 × 106 dissociated PBT-09 cells into soft tissue flank of athymic nude mice. Quisinostat and romidepsin were dissolved in 2% Tween80, 2% DMSO, 48% Peg300, and 48% water and dosed intraperitoneally.
RNA Sequencing and Expression Analysis
DMG cells were treated in culture with HDACi for 72 hours before total RNA isolation using the RNeasy Plus Mini Kit (Qiagen). Multiplexed RNA-Seq was performed on libraries generated using the TruSeq Stranded Total RNA Library Prep kit (Illumina) and sequenced on a HiSeq 2500 (Illumina). Resultant reads passing Illumina’s quality threshold were aligned to hg38 using STAR v2.5.2a (2-pass mapping), counts per gene were generated using Subread featureCounts v1.6.0, and log2 ratios of normalized data were calculated for vehicle treatment versus the various drug treatments using edgeR v3.25.8. Normalized log2 FC were utilized to perform hierarchical clustering in edgeR on the full dataset, top 1000 most variable genes, and top 500 most variable genes. Quantitative PCR using Taqman probes (Life Technologies) was performed on FSTL5, ITIH5, and ACTB following generation of cDNA using SuperScript IV reverse transcriptase (Life Technologies) according to manufacturer’s instructions.
Statistical Analysis
To evaluate cell viability following treatment with panobinostat, quisinostat, or romidepsin compared with other HDACi, we used the one-sided Kolmogorov–Smirnov test, designed to test if 2 curves originate from the same distribution. P-values were calculated by the “ks.test” function from R “stats” package. To compare postradiation cell viability of MED411 versus our DMG models and to compare cell viability post-HDACi in our DMG models results to those from Zurich, we utilized a 2-sided Kolmogorov–Smirnov test. To evaluate if there was decreasing cell viability from 24 to 48 to 72 hours, we employed the 1-sided Kolmogorov–Smirnov test to compare the 48 with the 24 hour curve and the 72 to 48 hour curve. To compare western blot protein acetylation, we used a t-test. To evaluate the pairwise overlapping numbers in the Venn diagram, we performed a hypergeometric test using the “phyper” function in R “stats” package. To assign differentially expressed gene significance, we used EBSeq.17 To evaluate consistency between Taqman and RNA-seq results, we performed a paired t-test with degrees of freedom 5 (two-sided test) and calculated a Pearson correlation coefficient. Statistical data is provided in Supplementary Document 1.
Results
Treatment-Naïve DMG Model Development from Biopsy-Derived Tissue Is Feasible
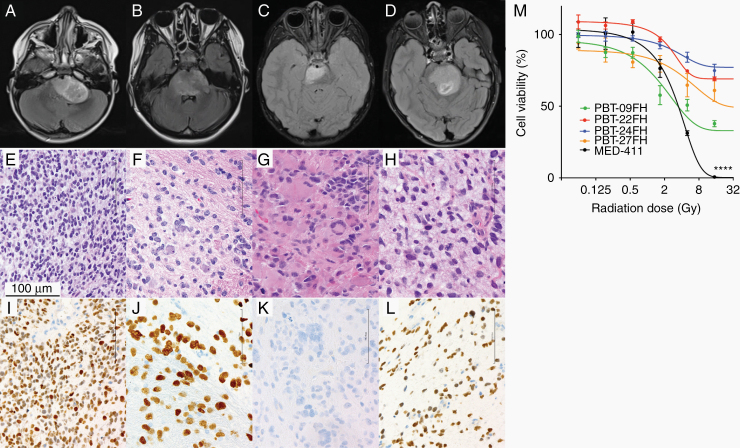
To investigate the feasibility of developing treatment-naïve biopsy-derived models, we processed tissue from diagnostic DIPG biopsies. We established cell cultures from 4 patients whose tumors were radiographically DIPG (Figure 1A–D). All showed histologic features of high-grade glioma (Figure 1E–H) and 3 met the criteria of DMG by H3 K27M IHC (Figure 1I–L). UW-OncoPlex, a clinical targeted DNA platform, was used to interrogate genome alterations, summarized in Table 1 along with clinical information of the patients from whom cultures were derived.18
Fig. 1.
Formation of treatment-naïve biopsy-derived DMG cell cultures. Clinical correlates for patients providing PBT-09FH, PBT-22FH, PBT-24FH, and PBT-27FH including MRI brain axial T2 fluid attenuated inversion recovery postcontrast (A–D), H&E immunohistochemistry (E–H), and H3 K27M IHC (I–L). Scale bar 100 µm. Cell viability following in vitro radiation treatment (****P < 0.0001) (M).
Table 1.
Characterization of the treatment-naïve biopsy-derived DIPG/DMG models
| ID | Genomic Mutations | Age at Diagnosis, y | Patient PFS, days | Patient OS, days | Median In Vitro Viability at 8 Gy (n = 3) |
|---|---|---|---|---|---|
| PBT-09FH | H3FA3, NF1, PI3KCA biallelic | 2 | 284 | 468 | 36% |
| PBT-22FH | H3F3A, TP53 | 5 | 121 | 190 | 81% |
| PBT-24FH | PMS2 (hypermutant) | 13 | 79 | 222 | 71% |
| PBT-27FH | HIST1H3B, TP53 biallelic, NTRK2 ITD | 6 | 288 | 544 | 58% |
To correlate radiation resistance in our patients and their corresponding cell cultures, we reviewed each patient’s progression-free survival (PFS) and OS. Two cultures (PBT-22FH and PBT-24FH) were generated from patients who experienced early progression despite standard ~54 Gy focal radiation: the former progressed at day 121 and died on day 190 postdiagnosis, while the latter progressed 79 days postdiagnosis and died on day 222 postdiagnosis. In contrast, the patient represented by PBT-09FH did not experience progression until 284 days postdiagnosis, and death occurred on day 468. The patient from whom PBT-27FH was derived experienced tumor progression on day 288 and death occurred on day 544. To assess the cultures’ radiation resistance, we treated them with 0–32 Gy and measured cell viability at 96 hours (Figure 1M). PBT-22FH and PBT-24FH showed 69.1% and 78.1% viability, respectively, after 32 Gy (Figure 1M). In contrast, PBT-09FH had only 32.6% viability under the same conditions. As medulloblastoma is another highly malignant pediatric central nervous system (CNS) tumor, we investigated the radiation response of our previously published medulloblastoma culture MED-411FH.19 As hypothesized, MED-411FH was more sensitive to radiation than the DMG models, evidenced by lower cell viability (P = 9.03e-6), emphasizing DMG’s radiation resistance compared with other aggressive, high-grade CNS tumors (Figure 1M).
Quisinostat and Romidepsin Are Effective Against DMG at Low Nanomolar Concentrations
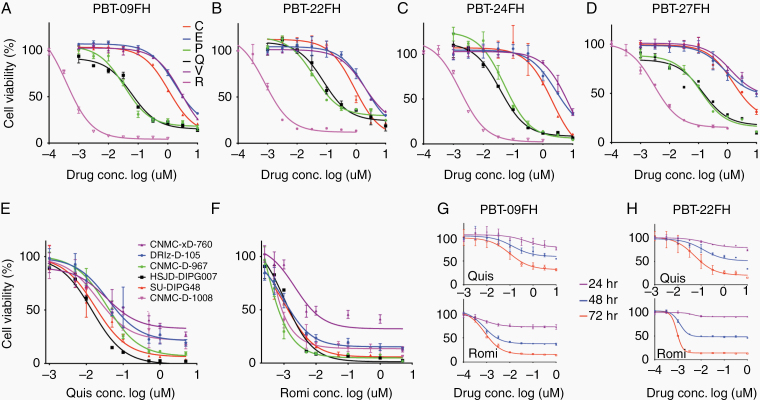
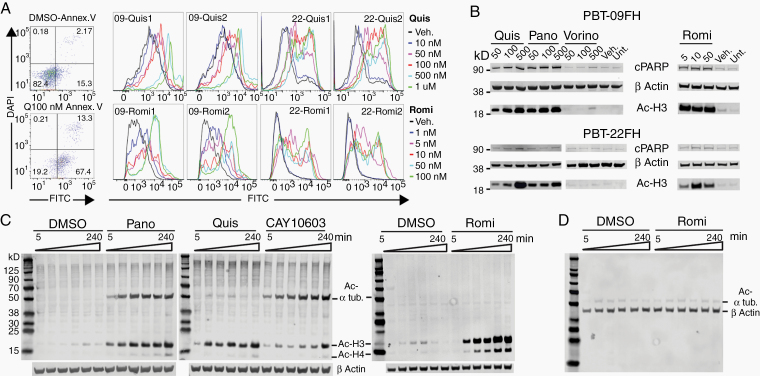
The HDACi panobinostat has demonstrated efficacy in some DMG models and has entered pediatric clinical trials.20 As HDACi comprise a broad class with variable effects that may induce cytotoxicity, we evaluated several HDACi, including CAY10603, entinostat (MS-275), panobinostat (LBH589), quisinostat (JNJ-26481585), romidepsin (FK288), and vorinostat (MK0683). We performed a 72-hour dose titration cell viability assay in PBT-09FH, PBT-22FH, PBT-24FH, and PBT-27FH cells, revealing that panobinostat, quisinostat, and romidepsin lowered cell viability at biologically relevant doses (ie, <~1µM) to a much greater extent than other HDACi (Figure 2A–D). For example, in PBT-09FH the half-maximal inhibitory concentration (IC50) values of panobinostat, quisinostat, and romidepsin were 34 nM, 60 nM, and 0.39 nM, significantly lower than the other HDACi (P = 0.008, 0.0001, and 6.1E-6, respectively) (Figure 2A). Statistical significance was seen across all models (Supplementary Document 1). Considering historical variability in drug responses in vitro due to drug stock potency, laboratory techniques, and models tested, we requested the research team at University Children’s Hospital Zurich evaluate quisinostat and romidepsin.21 With separately purchased drugs and across multiple DMG cultures (n = 6), mean IC50 values were 24.8 nM and 1.26 nM following treatment with quisinostat and romidepsin, respectively, which were consistent with our findings (P = 0.35, 0.47, respectively) (Figure 2E, F). Our timecourse tracking of quisinostat- and romidepsin-treated PBT-09FH and PBT-22FH cells showed significantly decreasing cell viability consistent with a cytocidal effect (eg, P-values of changing viability in quisinostat-treated PBT-09FH at 48 compared with 24 hours and at 72 compared with 48 hours were 0.029 and 0.029, respectively) (Figure 2G, H). Statistical significance was also seen in PBT-22FH (Supplementary Doc 1). To confirm apoptosis as the mechanism of cell death, flow cytometries of 100 nM quisinostat-treated and romidepsin-treated PBT-09FH and PBT-22FH cells were performed (Figure 3A). In contrast to only 15.3% of vehicle-treated cells, 67.4% percent of quisinostat-treated PBT-22FH cells stained positive for annexin-V and negative for 4′,6′-diamidino-2-phenylindole (DAPI), indicating early apoptosis. As validation, we treated PBT-09FH and PBT-22FH with panobinostat, quisinostat, romidepsin, and vorinostat (a negative control due to its relative ineffectiveness) and demonstrated a dose-dependent increase in apoptosis, as measured by PARP cleavage, following treatment with panobinostat and quisinostat to a much greater degree than after vorinostat (Figure 3B). While the HDACi panobinostat reduces DMG cell viability in vitro and in some models extends OS in vivo, dose-limiting hematologic toxicities remain a clinical challenge.16,22 As HDAC6 modulation of α-tubulin acetylation has been implicated in the development of cytopenias, we performed a western blot to assess protein expression.23–25 Panobinostat and CAY10603 (a positive control due to its HDAC6 specificity), elicited dose-dependent increases in the abundance of acetylated α-tubulin, while quisinostat and romidepsin did not (Figure 3C).
Fig. 2.
Quisinostat and romidepsin exhibit low nanomolar efficacy against DMG cultures. Cell viability assay of HDACi (72 hours) in (A) PBT-09FH, (B) PBT-22FH, (C) PBT-24FH, and (D) PBT-27FH. C = CAY10603, E = entinostat, P = panobinostat, Q = quisinostat, R = romidepsin, and V = vorinostat. (E) Cell viability assay following 72 hours of quisinostat and (F) romidepsin treatment performed at University Children’s Hospital Zurich. Cell viability timecourse assay following quisinostat and romidepsin treatment in (G) PBT-09FH and (H) PBT-22FH.
Fig. 3.
Quisinostat and romidepsin induce apoptosis in DMG cultures. (A) Flow cytometry of PBT-22FH stained with DAPI and FITC-annexin V following 72 hour treatment with 100 nM quisinostat (left) and duplicate histogram overlays for annexin V staining over concentrations of quisinostat- and romidepsin-treated PBT-09FH and PBT-22FH (right). (B) Western blot of cPARP and Ac-histone 3 in lysates generated from HDACi treated PBT-09FH and PBT-22FH (concentrations in nM). (C) Four-hour timecourse western blot showing decreased acetyl α-tubulin but very similar H3 acetylation by 500 nM quisinostat and 50 nM romidepsin compared with 500 nM panobinostat (HDAC6 inhibitor CAY10603 as a positive control of α-tubulin acetylation) in PBT-22FH. (D) Western blot of acetyl α-tubulin–specific antibody, demonstrating no change over the timecourse of treatment of PBT-22FH with 50 nM romidepsin.
Quisinostat and Romidepsin Are Effective in an In Vivo Treatment-Naïve DMG Flank Model
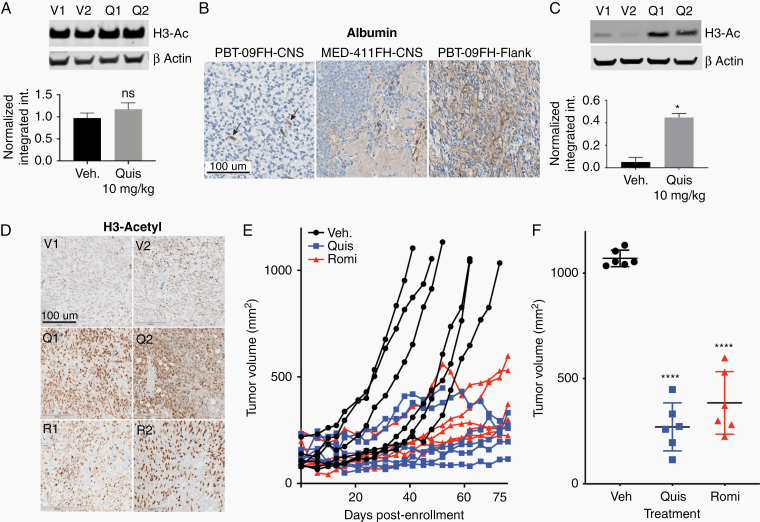
To evaluate blood-brain-tumor drug penetration, we treated orthotopic xenograft DMG tumor-bearing athymic mice with vehicle versus 1 week of intraperitoneal (IP) quisinostat (10 mg/kg Monday, Wednesday, Friday [MWF]) and euthanized mice 3 hours following the final dose to evaluate histone 3 acetylation as a marker of on-tumor effect. Intratumoral histone 3 acetylation did not increase following treatment with quisinostat (P = 0.39) (Figure 4A). To evaluate if this was due to lack of drug efficacy or tumor penetration, we assessed albumin staining of the orthotopic PBT-09FH model by IHC. Albumin staining is restricted to blood vessels in normal CNS, while in tumors causing blood–brain barrier disruption, such as our orthotopic xenograft MED-411FH model, albumin can be detected within the tumor (Figure 4B).26 In our orthotopic xenograft PBT-09FH model, CNS IHC revealed albumin staining restricted to blood vessels (Figure 4B). Given the concern that HDACi could not effectively penetrate the orthotopic tumors of our treatment-naïve DMG model, we implanted PBT-09FH cells into flanks of athymic mice to develop a DMG flank model as an alternative method to evaluate in vivo efficacy. Abundant albumin staining was observed in the flank tumor (Figure 4B).
Fig. 4.
Quisinostat and romidepsin induce prolonged tumor growth inhibition in an in vivo DMG flank model. (A) Western blots for H3 acetylation in vehicle and quisinostat-treated orthotopic xenograft PBT-09FH tumor lysate, with corresponding histograms of β-actin normalized intensities below. V = vehicle, Q = quisinostat, ns = no significant difference. (B) IHC of albumin in orthotopic xenograft PBT-09FH tumor compared with orthotopic xenograft MED-411FH and flank PBT-09FH tumors. Arrows indicate albumin-positive blood vessels. Scale bar 100 µm. (C) Western blots for H3 acetylation in vehicle and quisinostat-treated flank PBT-09FH tumors, with corresponding histograms of β-actin normalized intensities below (*P < 0.05). (D) H3-Ac IHC replicates of xenograft PBT-09FH flank tumors following systemic vehicle, quisinostat (10 mg/kg, MWF), or romidepsin (1 mg/kg, MF). V = vehicle, Q = quisinostat, R = romidepsin. (E) Tumor volume over time in flank xenograft cohorts treated with vehicle, quisinostat (10 mg/kg MWF), or romidepsin (1 mg/kg MF). (F) Boxplot of tumor volumes at study endpoint showing significantly decreased tumor volume in quisinostat and romidepsin-treated cohorts when compared with vehicle (****P < 0.0001).
To evaluate on-tumor drug effect, DMG flank tumors were formed over ~4 weeks and, after reaching a volume of 100 mm3, tumor-bearing mice were treated with vehicle, quisinostat, or romidepsin. Quisinostat was dosed at 10 mg/kg MWF. The romidepsin dose was chosen based on a pilot study in which cohorts of 3 mice were treated with either vehicle or romidepsin at 0.3 mg/kg, 1 mg/kg, or 3 mg/kg MF. Found to have unacceptable toxicity was 3 mg/kg within the first week of dosing, while 0.3 mg/kg and 1 mg/kg were tolerated with no significant weight loss (ie, >20%) or neurologic toxicity. Laboratory studies revealed anemia and aspartate aminotransferase elevations at 1 mg/kg dosing (without corresponding clinical changes); platelets were not significantly altered (Supplementary Table 1). Treatment with quisinostat (10 mg/kg MWF) or romidepsin (1 mg/kg MF), increased acetylation by western blot (P = 0.011; Figure 4C) and IHC (Figure 4D) within the flank DMG tumor, suggesting tumor penetration of quisinostat and romidepsin. Therefore, a larger 3-arm study was performed: vehicle control (n = 6), IP quisinostat (n = 6), and IP romidepsin (n = 6). Flank tumor-bearing mice (minimum volume = 100 mm3) were enrolled with median tumor volumes of 119.4, 134, and 135.3 mm3 for the vehicle, quisinostat, and romidepsin cohorts, respectively (Figure 4E). Study endpoints were tumor growth ≥1000 mm3 or treatment cutoff of 90 days, and tumor volume was measured with calipers. Due to COVID-related workplace restrictions, the 90-day treatment was amended to 75 days. While 6/6 vehicle-treated mice exited the study for tumor burden (≥1000 mm3), this did not occur in any quisinostat-treated or romidepsin-treated mice (Figure 4D). One quisinostat-treated mouse was euthanized on day 52 for gastrointestinal obstruction (without systemic toxicity). The mice in both the quisinostat-treated and romidepsin-treated cohorts demonstrated inhibited tumor growth compared with vehicle, with median tumor volume of the quisinostat- and romidepsin-treated cohorts 270.5 mm3 and 384.5 mm3, respectively, at study endpoint (P < 0.0001) (Figure 4F). This confirmed that systemically delivered quisinostat and romidepsin were tolerable and provided prolonged tumor control in vivo.
Cytotoxic HDAC Inhibitors Induce Shared RNA Expression Changes in DMG
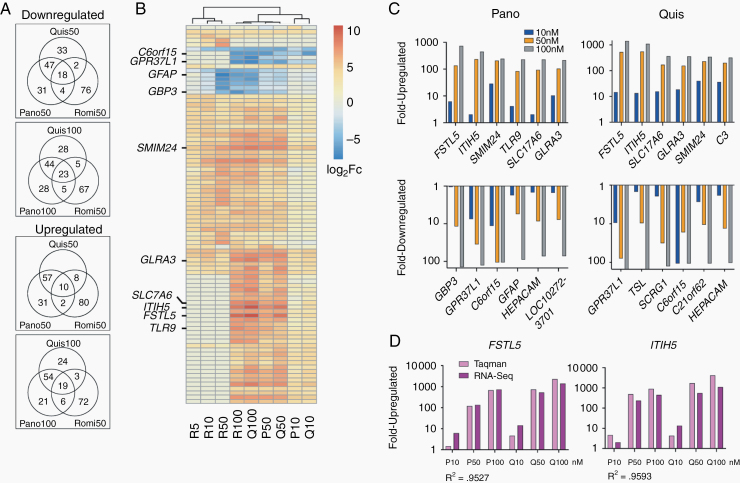
To investigate if panobinostat, quisinostat, and romidepsin induced similar transcriptomic changes, we performed RNA sequencing using PBT-22FH, the most common molecular subtype. To compare across doses inducing cytotoxicity, panobinostat, quisinostat, and romidepsin were tested at their approximate IC25, IC50, and IC75 values. Following 72-hour drug exposure, RNA was extracted and RNA-Seq differential expression analysis was performed (Supplementary Table 2). Unsupervised hierarchical clustering revealed similar gene expression changes between panobinostat and quisinostat, overlapping at each dose level. When including romidepsin, we took 2 approaches: testing romidepsin at the same dose as panobinostat and quisinostat (50 nM) and at closer biologically equivalent dose by in vitro cytotoxicity (100 nM panobinostat and quisinostat vs 50 nM romidepsin) and found statistically significantly overlapping genes (Figure 5A; Supplementary Doc 1). We performed hierarchical clustering across 3 drug concentrations and found high correlation between transcriptional changes at each dose of quisinostat and panobinostat, while all 3 doses of romidepsin clustered independently (Figure 5B). A smaller gene subset, including troponin T1 slow skeletal type (TNNT1) and collagen type 20 alpha 1 chain (COL20A1), correlated among all 3 drugs. In each approach, there was less overlap between the transcriptomic changes of romidepsin compared with the other 2 drugs. Analysis of the most differentially regulated genes by absolute fold-change revealed striking overlap between panobinostat-treated and quisinostat-treated samples, with 73/100 most upregulated genes, and 67/100 most downregulated genes in common at 100 nM. While less similar, romidepsin at 50 nM still demonstrated considerable overlap with the other 2 drugs at 100 nM (Figure 5A), with approximately one fifth of the top 100 affected genes overlapping (23 upregulated, 19 downregulated). Comparison of all 3 drugs at equimolar concentration (50 nM) only modestly reduced the correlative gene counts.
Fig. 5.
Transcriptomic studies reveal targets of cytotoxic HDAC inhibition. (A) Venn diagrams show the overlap between the 100 most up and downregulated genes for quisinostat, panobinostat, and romidepsin treated PBT-22FH cells relative to vehicle control. Comparisons are shown for equimolar treatment (50 nM) or 50 nM romidepsin versus 100 nM quisinostat and panobinostat. (B) Unsupervised hierarchical clustering of the union of the top 500 most differentially regulated genes, displaying union of top 20 for each treatment (87 genes total). (C) Modulation of expression levels with drug concentration for top 6 differentially up- and downregulated genes following panobinostat and quisinostat treatment. (D) TaqMan PCR validation of expression changes in FSTL5 and ITIH5, compared with RNA-seq (R2 = Pearson coefficient).
In all 3 drug treatments, small integral membrane protein 24 (SMIM24) and TNNT1 were upregulated, while COL20A1 and interferon induced transmembrane protein 3 (IFITM3) were downregulated. Five of the 6 most upregulated genes were shared between panobinostat and quisinostat treatments: follistatin-like 5 (FSTL5), inter-alpha-trypsin inhibitor heavy chain 5 (ITIH5), SMIM24, SLC17A6, and GLRA3 (Figure 5C). FSTL5 and ITIH5 (P < 0.001) were 2 of the most upregulated genes following both 50 nM and 100 nM panobinostat and quisinostat treatment. At the 100 nM quisinostat dosing, FSTL5 and ITIH5 expression increased over 1000×. Of note, an analysis of paired glioma tumor/normal brain samples found FSTL5 had the strongest expression correlation with other genes in glioma tissue.27 Also shared were 3/6 most downregulated genes following panobinostat and quisinostat treatment: GPR37L1, C6orf15, and HEPACAM (Figure 5C). GPR37L1 (P < 0.001) was the second most downregulated gene following panobinostat and quisinostat treatment, with ~90× lower expression and was the most downregulated gene following quisinostat treatment, with ~80× lower expression. Of note, GPR37L1 appears critical to sonic hedgehog (SHH) medulloblastoma, in which its ablation delays tumor development.28 Transcriptome changes identified in our RNA-seq data were validated by targeted TaqMan quantitative PCR and displayed high correlation (Pearson correlation coefficient = 0.976 and 0.979 for FSTL5 and ITIH5, respectively) (Figure 5D).
Discussion
The lack of clinical progress against DMG and the staggering number of failed trials necessitates better understanding of drugs advanced to the clinic. Here, we describe 4 novel treatment-naïve biopsy-derived DMG models whose radiation-naïve status may provide a valuable platform to evaluate novel therapeutics, including sequentially with radiation. We demonstrate low nanomolar efficacy of quisinostat and romidepsin in DMG (IC50 values of ~50 and ~5 nM, respectively), findings validated in 6 other models by international colleagues. We also demonstrate that quisinostat and romidepsin cause prolonged tumor growth inhibition in vivo in a xenograft DMG flank model. RNA sequencing posttreatment unveiled tight transcriptional overlap between the panobinostat- and quisinostat-induced changes. DMG cells treated with romidepsin had a more distinct transcriptome, though potentially important biological overlap between panobinostat, quisinostat, and romidepsin remain.
While panobinostat has shown preclinical efficacy against DMG, it should not be assumed to work similarly to other HDACi or to be the best-in-class agent. Considering the broad array of HDACi and diverse molecular targets including the metabolome, epigenome, and DNA damage response,29 the assumption should be that HDACi induce varying differential gene expression. While on-target acetylation and tumor permeability were not present in our orthotopic xenograft models, we established flank DIPG tumors in which quisinostat and romidepsin had a significant and prolonged effect of controlling tumor growth and are the first to show this in vivo benefit. As HDAC inhibition can affect megakaryocyte function, HDAC6 activity impacts lymphocyte chemotaxis, and α-tubulin acetylation plays a role in platelet formation, we investigated HDAC6-induced effects between quisinostat and romidepsin versus panobinostat as they were otherwise similar.30–32 We found less acetylation of α-tubulin by quisinostat and romidepsin, suggesting potential for lower hematopoietic toxicity than panobinostat. Based on early clinical experience, panobinostat may more commonly cause grades 3 and 4 hematologic toxicities compared with quisinostat and romidepsin.16,22,33–36 In particular, a 92-person phase I quisinostat study found only 5% of patients had grade 1 or 2 thrombocytopenia and no patients had grade 3 or 4.35 Our data, and their clinical tolerability, support their further investigation of quisinostat and romidepsin in other models, potentially as part of multi-agent regimens. While quisinostat and romidepsin were not tumor-penetrant in our orthotopic model, HDACi likely have variable on-target intratumoral activity across different in vivo model systems, so this should be considered as part of all laboratory CNS-related HDACi investigations. Considering the dearth of preclinically effective agents against DIPG and the decades of clinical trials failing to display efficacy, options for developing blood-brain-tumor penetrant versions of these agents is warranted. Locoregional delivery of HDACi and other epigenetic drugs either intrathecally or, as water-soluble formulations, via convention-enhanced delivery has already demonstrated in vivo efficacy and is advancing into clinical trials.37,38 Agents may also have superior intratumoral penetration when conjugated to blood-brain-tumor trafficking molecules, such as chlorotoxin.39,40 Preclinical glioma studies have already demonstrated that this approach is feasible and that chlorotoxin conjugation can lead to increased efficacy.41
Considering that many HDACi display limited cytotoxicity against DMG, evaluations for methods of action or shared effects among effective agents are critical. Our hierarchical analysis displayed panobinostat and quisinostat’s striking commonalities of the gene expression at each dose level. The tight transcriptional overlap suggests potential vulnerabilities or biomarkers, including upregulation of TNNT1, SMIM24, FSTL5, and ITIH5. Of note, TNNT1 promotes colorectal cancer progression and breast cancer cell proliferation,42,43 while COL20A1 is aberrantly expressed in adult gliomas.44FSTL5 was recently shown to have the strongest expression correlation with other genes in a coexpression network of gliomas, making its overexpression of post-cytotoxic HDACi a particular interest.27 In medulloblastoma, FSTL5 was found to be a negative prognostic marker by sequencing and by IHC.45 Also, FSTL5 modulation has been studied in a preclinical model of hepatocellular carcinoma, in which its overexpression decreased tumor size in vivo.46ITIH5 is a tumor suppressor associated with chemoradiotherapy response and metastatic dissemination.47,48 Several genes were also consistently downregulated, including COL20A1, IFITM3, and GPR37L1. In gliomas, IFITM3 has been implicated in tumor growth, migration, and invasion.49,50 In SHH medulloblastoma, knocking out GPR37L1 leads to a delayed, less aggressive tumor.28 Further biological assessment of how individual differential expression of the most highly ranked genes affects DMG cell viability may be valuable. Ultimately, these genes warrant further exploration as potentially clinically relevant biomarkers of HDACi on-target cytotoxic effect or avenues for narrower targeting.
In conclusion, our data support the biological relevance of our treatment-naïve DMG models and support the development of more blood-brain-tumor penetrating versions of quisinostat and romidepsin or enhanced locoregional delivery systems.
Supplementary Material
Acknowledgments
We thank our patients and their families who selflessly contributed to this study through tissue donation. We thank M. Monje for her pioneering work and mentorship. We thank F. Wu and R. Basom of the Fred Hutchinson Cancer Research Center Genomics Shared Resources for support in performing and interpreting our RNA sequencing. We thank J. Stevens, as well as the Seattle Children’s Hospital’s Department of Anatomic Pathology and TTS Brain Tumor Committee, for assistance in tissue collection and research coordination. We thank neuro-radiologists F. Perez and J. Wright for providing MR images. We are grateful for the clinical expertise of our neuro-oncology team, including N. Millard, A. Sato, E. Crotty, C. Hoeppner, S. Holtzclaw, S. Chaffee, A. Laurine, and W. Iwata. Portions of the in vitro HDACi testing previously have been presented (ISPNO; Denver, 2018).
Funding
We are grateful for generous funding from the National Cancer Institute (R01 CA114567; JMO), the Seattle Run of Hope (NAV, JMO), the Pediatric Brain Tumor Research Fund Guild of Seattle Children’s Hospital (NAV, JMO), the McKenna Claire Foundation (NAV), Unravel Pediatric Cancer (NAV, JMO), Team Cozzi Foundation (NAV), Love for Lucy (NAV), the Julianna Sayler Foundation (NAV), the Grousemont Foundation (NAV), Avery Huffman Defeat DIPG Foundation (NAV), Michael Mosier Defeat DIPG Foundation (NAV), the ChadTough Foundation (NAV), and the Kellen Joyce Heart of a Warrior Research Fund (MCB).
Conflict of interest statement. None.
Authorship interest. NAV, MCB, CM, EF, YZ, EJG, AN, JS, SMM, CC, NAPL, SM, JN, MYB, AJM, and JMO participated in the design or interpretation of the reported experiments or results. NAV, MCB, CM, EF, JMP, GP, FP, CAW, BLC, SESL, RG, and JMO participated in the acquisition or analysis of data. NAV, MCB, NAPL, and JMO wrote the manuscript with revisions and approval from all authors. JMO supervised all aspects of the research.
References
- 1. Ostrom QT, Gittleman H, Liao P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;16(Suppl 4):iv1–iv63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cooney T, Lane A, Bartels U, et al. Contemporary survival endpoints: an International Diffuse Intrinsic Pontine Glioma Registry study. Neuro Oncol. 2017;19(9):1279–1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Barkovich AJ, Krischer J, Kun LE, et al. Brain stem gliomas: a classification system based on magnetic resonance imaging. Pediatr Neurosurg. 1990;16(2):73–83. [DOI] [PubMed] [Google Scholar]
- 4. Albright AL, Packer RJ, Zimmerman R, Rorke LB, Boyett J, Hammond GD. Magnetic resonance scans should replace biopsies for the diagnosis of diffuse brain stem gliomas: a report from the Children’s Cancer Group. Neurosurgery. 1993;33(6):1026–1029; discussion 1029–1030. [DOI] [PubMed] [Google Scholar]
- 5. Grasso CS, Tang Y, Truffaux N, et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med. 2015;21(6):555–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gupta N, Goumnerova LC, Manley P, et al. Prospective feasibility and safety assessment of surgical biopsy for patients with newly diagnosed diffuse intrinsic pontine glioma. Neuro Oncol. 2018;20(11):1547–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Schwartzentruber J, Korshunov A, Liu XY, et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature. 2012;482(7384):226–231. [DOI] [PubMed] [Google Scholar]
- 8. Wu G, Broniscer A, McEachron TA, et al. ; St Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project . Somatic histone H3 alterations in pediatric diffuse intrinsic pontine gliomas and non-brainstem glioblastomas. Nat Genet. 2012;44(3):251–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803–820. [DOI] [PubMed] [Google Scholar]
- 10. Herz HM, Morgan M, Gao X, et al. Histone H3 lysine-to-methionine mutants as a paradigm to study chromatin signaling. Science. 2014;345(6200):1065–1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin GL, Wilson KM, Ceribelli M, et al. Therapeutic strategies for diffuse midline glioma from high-throughput combination drug screening. Sci Transl Med. 2019;11(519). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Srivatsan SR, McFaline-Figueroa JL, Ramani V, et al. Massively multiplex chemical transcriptomics at single-cell resolution. Science. 2020;367(6473):45–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Halvorson KG, Barton KL, Schroeder K, et al. A high-throughput in vitro drug screen in a genetically engineered mouse model of diffuse intrinsic pontine glioma identifies BMS-754807 as a promising therapeutic agent. PLoS One. 2015;10(3):e0118926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Nagaraja S, Vitanza NA, Woo PJ, et al. Transcriptional dependencies in diffuse intrinsic pontine glioma. Cancer Cell. 2017;31(5):635–652 e636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Seto E, Yoshida M. Erasers of histone acetylation: the histone deacetylase enzymes. Cold Spring Harb Perspect Biol. 2014;6(4):a018713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Wood PJ, Strong R, McArthur GA, et al. A phase I study of panobinostat in pediatric patients with refractory solid tumors, including CNS tumors. Cancer Chemother Pharmacol. 2018;82(3):493–503. [DOI] [PubMed] [Google Scholar]
- 17. Leng N, Dawson JA, Thomson JA, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pritchard CC, Salipante SJ, Koehler K, et al. Validation and implementation of targeted capture and sequencing for the detection of actionable mutation, copy number variation, and gene rearrangement in clinical cancer specimens. J Mol Diagn. 2014;16(1):56–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brabetz S, Leary SES, Gröbner SN, et al. A biobank of patient-derived pediatric brain tumor models. Nat Med. 2018;24(11):1752–1761. [DOI] [PubMed] [Google Scholar]
- 20. Vitanza NA, Monje Met al. Diffuse Intrinsic Pontine Glioma: from diagnosis to next-generation clinical trials. Curr Treatment Options Neurol. 2019;21(8):37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Larsson P, Engqvist H, Biermann J, et al. Optimization of cell viability assays to improve replicability and reproducibility of cancer drug sensitivity screens. Sci Rep. 2020;10(1):5798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ibrahim N, Buchbinder EI, Granter SR, et al. A phase I trial of panobinostat (LBH589) in patients with metastatic melanoma. Cancer Med. 2016;5(11):3041–3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cuenca-Zamora EJ, Ferrer-Marin F, Rivera J, Teruel-Montoya R. Tubulin in platelets: when the shape matters. Int J Mol Sci. 2019;20(14). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sadoul K, Wang J, Diagouraga B, et al. HDAC6 controls the kinetics of platelet activation. Blood. 2012;120(20):4215–4218. [DOI] [PubMed] [Google Scholar]
- 25. Aslan JE, Phillips KG, Healy LD, Itakura A, Pang J, McCarty OJ. Histone deacetylase 6-mediated deacetylation of alpha-tubulin coordinates cytoskeletal and signaling events during platelet activation. Am J Physiol Cell Physiol. 2013;305(12):C1230–C1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dziegielewska KM, Habgood MD, Møllgård K, Stagaard M, Saunders NR. Species-specific transfer of plasma albumin from blood into different cerebrospinal fluid compartments in the fetal sheep. J Physiol. 1991;439:215–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liang A, Zhou B, Sun W. Integrated genomic characterization of cancer genes in glioma. Cancer Cell Int. 2017;17:90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Di Pietro C, La Sala G, Matteoni R, Marazziti D, Tocchini-Valentini GP. Genetic ablation of Gpr37l1 delays tumor occurrence in Ptch1+/- mouse models of medulloblastoma. Exp Neurol. 2019;312:33–42. [DOI] [PubMed] [Google Scholar]
- 29. Marks PA, Xu WS. Histone deacetylase inhibitors: potential in cancer therapy. J Cell Biochem. 2009;107(4):600–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iancu-Rubin C, Gajzer D, Mosoyan G, Feller F, Mascarenhas J, Hoffman R. Panobinostat (LBH589)-induced acetylation of tubulin impairs megakaryocyte maturation and platelet formation. Exp Hematol. 2012;40(7):564–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Valenzuela-Fernandez A, Cabrero JR, Serrador JM, Sanchez-Madrid F. HDAC6: a key regulator of cytoskeleton, cell migration and cell-cell interactions. Trends Cell Biol. 2008;18(6):291–297. [DOI] [PubMed] [Google Scholar]
- 32. Bishton MJ, Harrison SJ, Martin BP, et al. Deciphering the molecular and biologic processes that mediate histone deacetylase inhibitor-induced thrombocytopenia. Blood. 2011;117(13):3658–3668. [DOI] [PubMed] [Google Scholar]
- 33. Zaja F, Salvi F, Rossi M, et al. Single-agent panobinostat for relapsed/refractory diffuse large B-cell lymphoma: clinical outcome and correlation with genomic data. A phase 2 study of the Fondazione Italiana Linfomi. Leuk Lymphoma. 2018:1–7. [DOI] [PubMed] [Google Scholar]
- 34. Maruyama D, Tobinai K, Ogura M, et al. Romidepsin in Japanese patients with relapsed or refractory peripheral T-cell lymphoma: a phase I/II and pharmacokinetics study. Int J Hematol. 2017;106(5):655–665. [DOI] [PubMed] [Google Scholar]
- 35. Venugopal B, Baird R, Kristeleit RS, et al. A phase I study of quisinostat (JNJ-26481585), an oral hydroxamate histone deacetylase inhibitor with evidence of target modulation and antitumor activity, in patients with advanced solid tumors. Clin Cancer Res. 2013;19(15):4262–4272. [DOI] [PubMed] [Google Scholar]
- 36. Otterson GA, Hodgson L, Pang H, Vokes EE; Cancer and Leukemia Group B . Phase II study of the histone deacetylase inhibitor Romidepsin in relapsed small cell lung cancer (Cancer and Leukemia Group B 30304). J Thorac Oncol. 2010;5(10):1644–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Anastas JN, Zee BM, Kalin JH, et al. Re-programing chromatin with a bifunctional LSD1/HDAC inhibitor induces therapeutic differentiation in DIPG. Cancer Cell. 2019;36(5):528–544 e510. [DOI] [PubMed] [Google Scholar]
- 38. Singleton WGB, Bienemann AS, Woolley M, et al. The distribution, clearance, and brainstem toxicity of panobinostat administered by convection-enhanced delivery. J Neurosurg Pediatr. 2018;22(3):288–296. [DOI] [PubMed] [Google Scholar]
- 39. Baik FM, Hansen S, Knoblaugh SE, et al. Fluorescence identification of head and neck squamous cell carcinoma and high-risk oral dysplasia with BLZ-100, a chlorotoxin-indocyanine green conjugate. JAMA Otolaryngol Head Neck Surg. 2016;142(4):330–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stroud MR, Hansen SJ, Olson JM. In vivo bio-imaging using chlorotoxin-based conjugates. Curr Pharm Des. 2011;17(38):4362–4371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang X, Guo Z. Anti-gliomas effect of chlorotoxin-conjugated onconase at high dose. Cell Biochem Biophys. 2015;73(2):389–392. [DOI] [PubMed] [Google Scholar]
- 42. Chen Y, Wang J, Wang D, et al. TNNT1, negatively regulated by miR-873, promotes the progression of colorectal cancer. J Gene Med. 2020;22(2):e3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Shi Y, Zhao Y, Zhang Y, et al. TNNT1 facilitates proliferation of breast cancer cells by promoting G1/S phase transition. Life Sci. 2018;208:161–166. [DOI] [PubMed] [Google Scholar]
- 44. Ishihara E, Takahashi S, Fukaya R, Ohta S, Yoshida K, Toda M. Identification of KLRC2 as a candidate marker for brain tumor-initiating cells. Neurol Res. 2019;41(11):1043–1049. [DOI] [PubMed] [Google Scholar]
- 45. Remke M, Hielscher T, Korshunov A, et al. FSTL5 is a marker of poor prognosis in non-WNT/non-SHH medulloblastoma. J Clin Oncol. 2011;29(29):3852–3861. [DOI] [PubMed] [Google Scholar]
- 46. Li C, Dai L, Zhang J, et al. Follistatin-like protein 5 inhibits hepatocellular carcinoma progression by inducing caspase-dependent apoptosis and regulating Bcl-2 family proteins. J Cell Mol Med. 2018;22(12):6190–6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rose M, Meurer SK, Kloten V, et al. ITIH5 induces a shift in TGF-β superfamily signaling involving Endoglin and reduces risk for breast cancer metastasis and tumor death. Mol Carcinog. 2018;57(2):167–181. [DOI] [PubMed] [Google Scholar]
- 48. Sasaki K, Kurahara H, Young ED, et al. Genome-wide in vivo RNAi screen identifies ITIH5 as a metastasis suppressor in pancreatic cancer. Clin Exp Metastasis. 2017;34(3–4):229–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zhao B, Wang H, Zong G, Li P. The role of IFITM3 in the growth and migration of human glioma cells. BMC Neurol. 2013;13:210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wang H, Tang F, Bian E, et al. IFITM3/STAT3 axis promotes glioma cells invasion and is modulated by TGF-β. Mol Biol Rep. 2020;47(1):433–441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.