Abstract
Sjögren’s syndrome (SS) is an autoimmune disease with no effective treatment options. Resolvin D1 (RvD1) belongs to a class of lipid‐based specialized pro‐resolving mediators that showed efficacy in preclinical models of SS. We developed a physiologically‐based pharmacokinetic (PBPK) model of RvD1 in mice and optimized the model using plasma and salivary gland pharmacokinetic (PK) studies performed in NOD/ShiLtJ mice with SS‐like features. The predictive performance of the PBPK model was also evaluated with two external datasets from the literature reporting RvD1 PKs. The PBPK model adequately captured the observed concentrations of RvD1 administered at different doses and in different species. The PKs of RvD1 in virtual humans were predicted using the verified PBPK model at various doses (0.01–10 mg/kg). The first‐in‐human predictions of RvD1 will be useful for the clinical trial design and translation of RvD1 as an effective treatment strategy for SS.
Study Highlights.
WHAT IS THE CURRENT KNOWLEDGE ON THE TOPIC?
☑ Sjögren’s syndrome (SS) is a complex inflammatory disorder with no clinically approved treatment options. Resolvin D1 (RvD1), a specialized pro‐resolving mediator with potent anti‐inflammatory effects, has shown promise in preclinical studies to restore the salivary gland function and promote tissue repair.
WHAT QUESTION DID THIS STUDY ADDRESS?
☑ We leveraged physiologically‐based pharmacokinetic (PBPK) modeling to predict the RvD1 pharmacokinetics (PKs) in humans by extrapolation from PK data obtained from in vivo experiments in a mouse model of SS.
WHAT DOES THIS STUDY ADD TO OUR KNOWLEDGE?
☑ The PBPK model provides an appropriate dose and estimates of clearance and other PK parameters for RvD1 in humans. These PK parameters and dose will inform the initial dosing of RvD1 for first‐in‐human clinical studies.
HOW MIGHT THIS CHANGE CLINICAL PHARMACOLOGY OR TRANSLATIONAL SCIENCE?
☑ This study significantly advances the potential for clinical translation of RvD1 into clinical trials for the treatment of SS.
Sjögren’s syndrome (SS) is a chronic inflammatory autoimmune disease characterized by the diminished secretory function of the exocrine glands. 1 It affects ~ 1% of the general population and up to 3% of people above the age of 50 years. 2 Women account for > 90% of diagnosed cases. 2 Diagnostic hallmarks for SS include xerostomia or dry mouth, impaired tear production, lymphocytic infiltration into salivary glands, and presence of anti‐Ro and anti‐La autoantibodies in plasma. 1 SS reduces salivary gland function and leads to oral diseases, such as gingivitis and caries, and candidiasis. 3 The advanced complications of reduced salivary gland function include recurrent infection of the parotid gland 4 and lymphomas. 5 Together, these issues impose significant physical, psychological, and economic burdens on patients with SS. 6 Furthermore, the cause of SS is still unknown, and current therapeutic interventions are ineffective and limited to the use of saliva substitutes and medications that provide only temporary relief. 7 Therefore, the development of alternative treatments to restore salivary gland function are an urgent and unmet medical need.
Viral and bacterial infections, in conjunction with the activation of susceptibility genes, stimulates chronic salivary gland inflammation. 8 A state of chronic inflammation causes tissue damage followed by cytokine and chemokine release. 8 When resolution mechanisms are working correctly, neutrophils and M2 macrophages can clear the site of injury or infection. 9 However, in SS, these resolution mechanisms are impaired, 10 and the dead cells involved in the process cannot be removed from the injury/infection site leading to the production of autoantigens and elevations in proinflammatory cytokine levels. 11 The increased production of autoantigens and proinflammatory cytokines stimulate peripheral lymphocytes to bind to and infiltrate across the vascular endothelium into the salivary gland. 12 Lymphocyte infiltration exacerbates the pathologic proinflammatory state, tissue damage, and hastens salivary gland dysfunction. 13 The cascade of events leading to inflammation resolution is an actively regulated process mediated in part by a family of endogenous lipid‐based specialized pro‐resolving mediators, which include resolvins, maresins, lipoxins, and protectins. 14 The administration of pro‐resolving lipid mediators have demonstrated efficacy in treating diseases having a pathologic proinflammatory basis, such as osteoarthritis, by terminating proinflammatory signaling, thereby enhancing tissue regeneration. 15 These pro‐resolving lipid mediators have been detected in animal models of infection and chronic inflammation. 16 Specific to SS, exogenous resolvin administration has the potential to reduce inflammation and restore salivary gland function. 17
Naturally occurring resolvin subtypes include the D series (derived from docosahexaenoic acid), the E series (derived from eicosapentaenoic acid), and six analogs of the D series, which are produced in response to aspirin. 18 Previously, we identified resolvin D1 (RvD1) as a therapeutic candidate for the treatment of SS. 19 RvD1 reduced inflammation and restored salivary gland tissue integrity in animal models of SS. 19 , 20 , 21 In addition, the progression of SS‐like features in NOD/ShiLtJ mice can be halted using an aspirin‐triggered form of RvD1. In this circumstance, mice treated systemically, prior to disease onset, displayed downregulation of proinflammatory cytokines, upregulation of anti‐inflammatory mediators, and intact saliva production. 22 , 23 Based on the preclinical safety and efficacy data of RvD1, we are preparing for phase I studies in humans. Therefore, the purpose of this study was to perform first‐in‐human predictions of RvD1 pharmacokinetics (PKs) in plasma and saliva using a verified whole‐body physiologically‐based pharmacokinetic (PBPK) model. The mouse PBPK model of RvD1 was developed using the physical, chemical, and biological properties of RvD1 and was optimized with plasma and salivary gland PK data obtained in SS‐like mice. The mouse PBPK model was then extrapolated to humans to generate appropriate first‐in‐human dose predictions.
METHODS
Drugs and chemicals
RvD1 was purchased from Cayman Chemicals, Ann Arbor, MI. RvD1 was formulated in a vehicle with 8.9% ethanol in phosphate‐buffered saline for intravenous injection. All chemicals were of analytical grade or better (ethanol was high‐performance liquid chromatography grade solvent) and were obtained from Merck (Darmstadt, Germany).
Preclinical pharmacokinetic study
Female NOD/ShiLtJ mice, 8–10 weeks old, were obtained from Jackson Laboratories (Bar Harbor, ME). Mice were injected intravenously in the tail vein with RvD1 (Cayman Chemicals, Ann Arbor, MI) at a dosage of 0.1 mg/kg. At the indicated experimental time points, the mice were euthanized via compressed CO2 inhalation. Animals were housed in cages in a room with a controlled environment (12‐hour day/night cycles) and fed with standard pellet diet and water. All animal management, anesthesia, and surgeries followed a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Utah.
Pharmacokinetic analysis
Blood was collected immediately upon euthanization via abdominal exsanguination into heparin‐coated tubes (BD Microtainer; BD Biosciences, San Jose, CA), and centrifuged at 14,000 revolutions per minute for 10 minutes at 4°C to obtain supernatant plasma. Submandibular glands (SMGs) were harvested and homogenized mechanically in stomacher bags (Nasco Whirl‐Pak; Nasco, Ft. Atkinson, WI) with 0.5 mL enzyme‐linked immunosorbent assay (ELISA) buffer. SMG is the largest of the three salivary glands in mice contributing to maximal saliva production. 24 SMG homogenate was centrifuged at 1,200 revolutions per minute for 5 minutes at 4°C to obtain supernatant SMG lysate. RvD1 concentrations in plasma and SMG lysate samples were measured using a commercially available RvD1 ELISA kit (Cayman Chemicals, Ann Arbor, MI) following the manufacturer’s instructions. The RvD1 ELISA method was validated according to manufacturer’s instructions. The percent recovery of RvD1 in plasma and tissue was 73% and the lower limit of quantification was 3.3 pg/mL. Baseline endogenous RvD1 concentrations were established by using plasma and submandibular concentrations of RvD1 at time 0 (“predose”). The PKs of baseline‐corrected RvD1 plasma and salivary gland concentrations were initially evaluated by noncompartmental PK analysis (NCA) using PKanalix version 2019R1 (Lixoft, Antony, France).
Model building workflow
A PBPK model was first developed in the NOD/ShiLtJ mouse population. The default PBPK model of mouse species in PK‐Sim version 8 does not have a salivary gland compartment. 25 Therefore, the salivary gland compartment was added to the model using MoBi version 8. 25 The NOD/ShiLtJ salivary gland PBPK model was optimized using the data obtained from our in vivo PK studies of RvD1 in NOD/ShiLtJ mice. We compared model predictions with independent datasets from the literature. 26 , 27 After model optimization, the NOD/ShiLtJ mouse PBPK model was scaled to humans, and first‐in‐human dosing predictions were performed. A schematic representing the NOD/ShiLtJ mouse RvD1 PBPK model is provided in Figure 1 .
Figure 1.
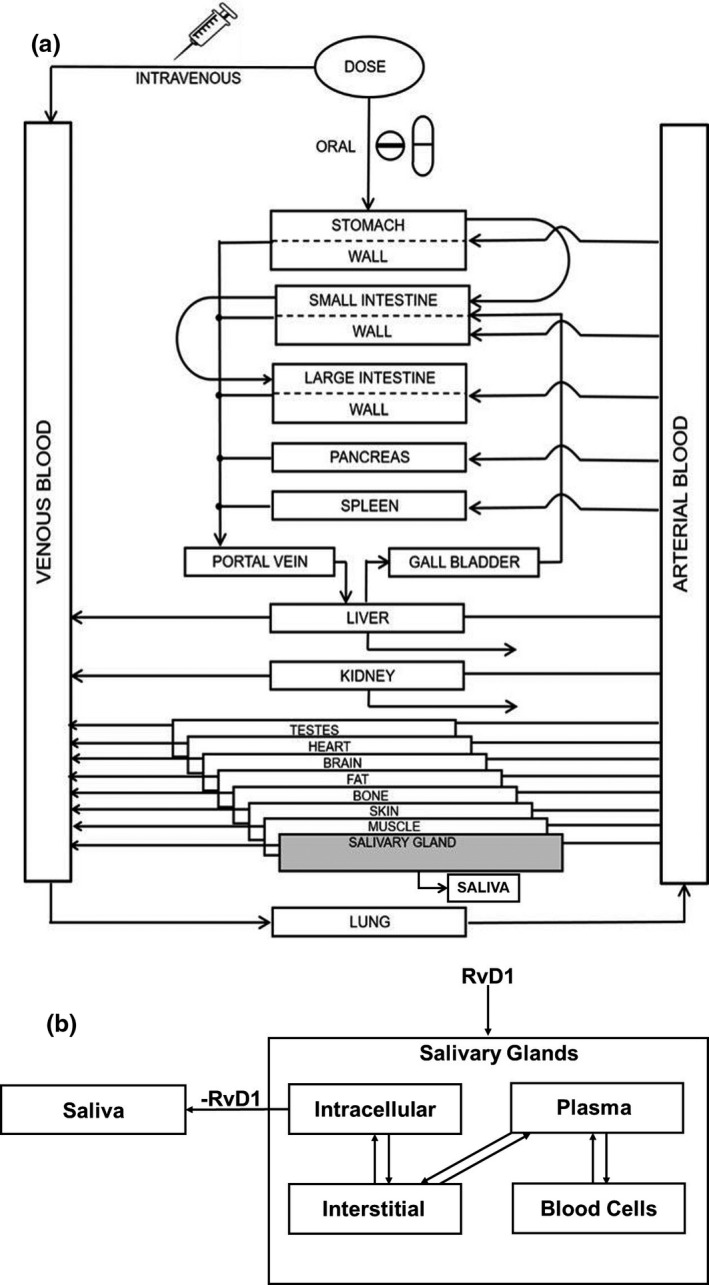
Schematic representation of physiologically‐based pharmacokinetic (PBPK) model for resolvin D1 (RvD1). (a) A whole‐body PBPK model of RvD1 with added salivary gland compartment. (b) Schematic of salivary gland compartment with subcompartments.
PBPK model development
The drug‐specific information for the model parameters was obtained from literature and quantitative structure‐property relationship calculations (Table 1 ). 28 The quantitative structure‐property relationship calculations were performed using the Chemicalize application by ChemAxon Company, Cambridge, MA. 29 Chemicalize application provided the structure‐based calculations and predictions of RvD1 when a chemical name of RvD1 was provided as an input. The parameters compound type (acidic or basic), pKa, and solubility at pH 7.4 were predicted by Chemicalize and used for the PBPK model development. The total clearance of RvD1 in NOD/ShiLtJ mice was calculated from NCA using the data obtained from our in vivo i.v. RvD1 PK study in NOD/ShiLtJ mice. The total clearance value was incorporated into the ADME tab of the RvD1 compound building block of the PK‐Sim as “total hepatic clearance.” The time points between 6 and 24 hours on the plasma concentration vs. time data were used for the calculation of total clearance of RvD1 in NOD/ ShiLtJ mice after i.v. administration.
Table 1.
Drug‐dependent parameters of RvD1
| Parameter | Reported value | Optimized value |
|---|---|---|
| Physicochemical properties | ||
| Lipophilicity (LogP) | 3.22 28 | 1.54 |
| Fraction unbound % | Not reported | 2.08 |
| Molecular weight | 376.5 28 | |
| Compound type/pKa | Acidic/4.47 29 | |
| Charge | Neutral | |
| Solubility at pH 7.4, mg/mL | 3.1 29 | |
| Metabolism and elimination | ||
| Plasma clearance, L/hour/kg | 2.65 a | |
RvD1, resolvin D1.
The total plasma clearance was calculated from the in vivo pharmacokinetic study of RvD1 in NODShiLtJ mice with Sjögren’s syndrome‐like features.
We developed a whole‐body NOD/ShiLtJ mouse PBPK model for RvD1 using PK‐Sim and MoBi, version 8 (Open Systems Pharmacology Suite, open‐systemspharmacology.com). The PBPK model was structured with 16 organs using mass balance differential equations describing drug entering and exiting the various organ compartments. The link between physiologic spaces was blood circulation. The NOD/ShiltJ mouse species model was developed in PK‐Sim by modifying the parameters, such as weight and age‐based growth of weight of the default mouse species in PK‐Sim, using the information obtained from Mouse Strain Datasheet‐001976 provided by the supplier, the Jackson Laboratory, Bar Harbor, ME. 30
The model for the distribution of RvD1 across each organ consisted of four subcompartments; plasma, red blood cells (together forming the vascular space), interstitial fluid, and cellular space. A permeation barrier exists between the vascular space and the interstitial fluid, and between the interstitial fluid and the cellular space. The organ‐to‐plasma partition coefficients were determined using drug physicochemical properties based on methods described by Rodgers et al., 31 and Rodgers and Rowland. 32
The salivary gland compartment was modeled using the following four default subcompartments: blood, plasma, interstitial fluid, and intracellular compartments. 25 These subcompartments were linked to the entire PBPK model through arterial and venous blood compartments. 25 The intracellular compartment was linked to saliva to represent the excretion of RvD1 from the salivary gland into the saliva. Additionally, salivary blood flow and volume were incorporated from the literature, whereas salivary gland weight was derived from in‐house laboratory data using NOD/ShiLtJ mice. Drug permeability into the salivary gland was determined by Eq. 1 below. Equation 1 was obtained from standard PK‐Sim model structure equation defining transport of drug from plasma to saliva. 33
| (1) |
where P is the specific organ permeability, fu is the fraction of RvD1 unbound in plasma, C_pls_art is the concentration of RvD1 in arterial blood plasma, C_slv is the concentration of RvD1 in salivary gland compartment, K_slv_pls in the partition coefficient of RvD1 across plasma and intracellular compartments, and Q_slv is the salivary flow rate. Excretion of RvD1 from salivary gland to saliva was determined by Eq. 2 below. Equation 2 is based on published literature reports indicating that passive transcellular diffusion is the predominant transport process of lipophilic compounds like RvD1 from salivary gland to saliva. 34
| (2) |
where dN/dt is rate of excretion of RvD1, K Saliva is the first‐order rate constant, and CRvD1 is the amount of RvD1 in salivary glands. The physiological and drug‐specific parameters used for the salivary gland compartment are obtained from either literature sources 33 , 35 , 36 or calculated using PK Sim 25 , 37 and provided in Table 2 .
Table 2.
Physiologic and drug‐specific parameters used for the salivary gland compartment in mice and humans
| Parameter | Mice | Humans | ||
|---|---|---|---|---|
| Value | Ref. | Value | Ref. | |
| Physiological parameters | ||||
| Blood flow rate, mL/minute | 3.6 × 10−4 | 33 | 160.56 | 41 |
| Tissue density, g/cm3 | 1 | 35 | 1 | 35 |
| Fluid recirculation flow rate, mL/minute | 2.2 × 10−7 | 33 | N/A | N/A |
| Fluid circulation flow rate scaling exponent | N/A | N/A | 0.67 | 33 |
| Weight of tissue, mg | 60 a | N/A | 48,000 | 40 |
| Organ volume, mL | 0.06 b | N/A | 48 b | N/A |
| Surface area, plasma/interstitial, cm2 | 5.7 × 10−5 | 33 | N/A | N/A |
| Surface area, plasma/interstitial, proportionality factor, cm−1 | N/A | N/A | 950 | 37 |
| Drug‐specific parameters | ||||
| Partition coefficient, cell/saliva | 0.5 | 25, 37 | 0.5 | 25, 37 |
| Partition coefficient, intracellular/plasma | 0.04 | 25, 37 | 0.04 | 25, 37 |
N/A, not applicable.
Weight of the salivary gland tissue was obtained from laboratory data.
Volume of the salivary gland tissue was calculated using density and mass values.
A global sensitivity analysis was performed following simulation of the NOD/ShiLtJ mouse PBPK model. The medial demographics were used to identify model input parameters, that when altered by 25%, caused a ≥ 10% change in simulated plasma area under the plasma concentration‐time curve from the time of the first dose to the time of the last dose (AUC_tEnd) or maximum plasma concentration (Cmax). 25 The parameters that were altered are: lipophilicity, hematocrit, organ‐specific blood flow rate, organ volume, organism‐plasma protein scale factor, fraction unbound, pKa, and solubility.
Model verification and optimization
The NOD/ShiLtJ mouse PBPK model was optimized using the RvD1 PK data from the in vivo PK experiment. A virtual population of 10,000 female NOD/ShiLtJ mice was simulated using PK‐Sim and incorporated characteristics of the NOD/ShiLtJ mice used for the in vivo RvD1 PK experiments. Several physiological parameters, such as blood flow rates to the organs, fluid recirculation flow rate, lymph flow rate, peripheral blood flow fraction, tissue/plasma—albumin and lipoprotein ratio, hematocrit, and endosomal clearance were varied by 30–40% in the virtual population. The predictive performance of the model was further evaluated using digitized RvD1 PK data from two studies published by Recchiuti et al. 26 and Krashia et al., 27 representing virtual populations of 10,000 C57BL/6N mice and 10,000 Sprague‐Dawley rats, respectively. RvD1 was administered orally in data reported by Recchiuti et al. and intraperitoneally in data reported by Krashia et al. As with the NOD/ShiLtJ mice virtual population, the physiological parameters of individuals comprising of virtual populations of C57BL/6N and Sprague‐Dawley rats were also varied. The parameters that were optimized in C57BL/6N and Sprague‐Dawley rat models are provided as Supplementary Data Table S1 . Based on these studies, the number of observed RvD1 concentrations outside of the 90% prediction interval was calculated. The accuracy of model predictions was explored by goodness‐of‐fit plots with 90% prediction intervals. The accuracy of model predictions was also explored by calculating the average fold error (AFE) of the observed to median simulated concentrations for all samples according to Eq. 3:
| (3) |
Model acceptance criteria were defined as simulated concentrations with AFE within twofold of observed values. The AFE values for concentrations and PK parameters for all PBPK models in this study are provided in Table 3 .
Table 3.
AFE in concentration, AUC0‐∞, and Cmax for all RvD1 PBPK models
| Animal model | AFE | ||
|---|---|---|---|
| Concentration | AUC0–∞ | Cmax | |
| NODShiLtJ mice—plasma | 0.52 | 0.42 | 0.22 |
| NODShiLtJ mice—salivary glands | 0.35 | 0.31 | 0.8 |
| C57BL/6N mice—plasma | 0.14 | 0.53 | 0.81 |
| Sprague‐Dawley rats—plasma | 0.69 | 0.7 | 1.7 |
AFE, average fold error; AUC0–∞, area under the concentration‐time curve from zero to infinity; Cmax, peak plasma concentration; PBPK, physiologically‐based pharmacokinetic; RvD1, resolvin D1.
First‐in‐human predictions
The human PBPK model was created by incorporating known human physiological data, including blood flow rate and variability in organ volumes from a 70‐kg individual. 25 , 38 The virtual human population consisted of a female to male ratio of 9:1 because of the predominance of SS in women. 10 Based on these anthropometric measurements, organ weights, volumes, and blood flows were generated using the International Commission on Radiological Protection database. 39 The physiological and drug‐specific parameters used for the human salivary gland compartment are either obtained from the literature 40 , 41 or calculated using PK Sim and are provided in Table 2 . The optimized NOD/ShiLtJ mouse PBPK model was extrapolated to the human model using PK Sim default human population database characteristics. 25 The simulations were performed at various doses (0.01–10 mg/kg), and the PK parameters of RvD1 in plasma and saliva compartments were obtained. The clearance of RvD1 from the mouse model was scaled to the human model using allometric scaling method. 42 The first‐in‐human dosing will be selected based on the Cmax of RvD1 in the saliva compartment reaching a threshold efficacy value of 100 ng/mL. The threshold efficacy value of 100 ng/mL was selected based on our previous in vitro cell culture experiments on salivary epithelial cells freshly isolated from mouse salivary glands. 43
RESULTS
Preclinical pharmacokinetics of RvD1
Concentrations of RvD1 in plasma and SMG declined to undetectable levels after 8 and 4 hours, respectively (Supplementary Data Figure S1 ). The concentration vs. time curve of RvD1 in plasma showed a biphasic response with an additional peak at 4 hours (Supplementary Data Figure S1 a). The PK analysis by NCA showed that after i.v. injection, the terminal half‐life of RvD1 in plasma and SMG was 4.77 hours and 3.41 hours, respectively. The time to reach maximum concentration (Tmax) for plasma and SMG was the same (0.0833 hours), indicating the rapid distribution of RvD1 to SMG after i.v. administration. The information on PK parameters of RvD1 in plasma and submandibular glands is provided in Supplementary Data Figure S1 a,b, respectively. The datasets containing plasma and SMG PK data of RvD1 in NOD/ShiLtJ mice are provided as supplementary Files , [Link] , [Link] respectively.
NOD/ShiLtJ mouse PBPK model
Lipophilicity, fraction unbound in plasma, and volume of saliva were optimized to 1.54, 2.08%, and 0.1 L, respectively (Table 1 ) using in vivo PK data. The optimized NOD/ShiLtJ PBPK model predictions vs. observed data, from the in vivo preclinical PK studies showed good agreement with ≥ 90% of the observed concentrations falling within the 90% prediction interval of the simulated population concentrations of RvD1 in both plasma (Figure 2a ) and salivary glands (Figure 2b ). Furthermore, the AFE values demonstrated that the median predicted concentrations generally fell within twofold of the observed values for plasma and salivary gland concentrations (Table 3 ).
Figure 2.
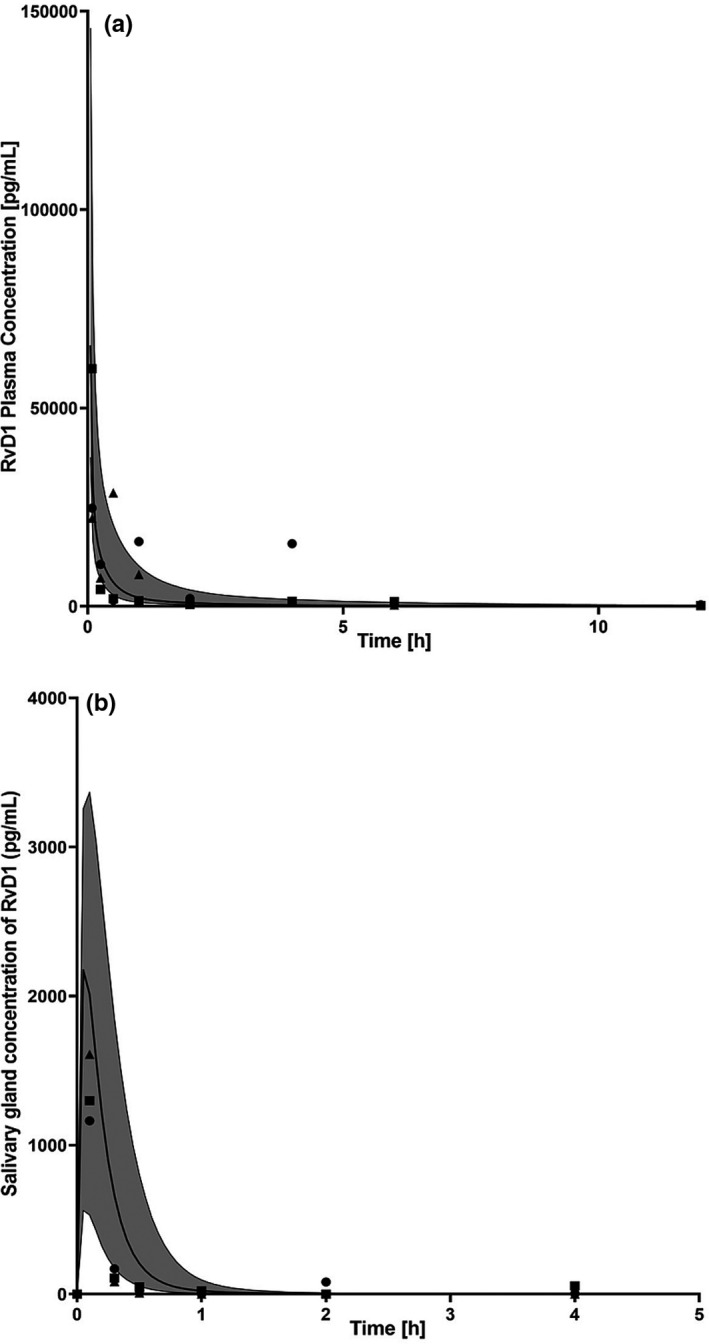
Observed and physiologically‐based pharmacokinetic (PBPK) model‐simulated concentrations of resolvin D1 (RvD1). Measurements were performed in (a) plasma and (b) submandibular glands following a dose of 0.1 mg/kg in NOD/ShiLtJ mice with Sjögren’s syndrome (SS) plasma. Black circle, square, and triangle symbols represent data from each individual animal of the observed data, and solid lines represent median RvD1 concentrations from the PBPK model. The shaded region represents the 90% prediction interval for the simulated RvD1 concentrations.
Additionally, the optimized NOD/ShiLtJ PBPK model adequately described RvD1 concentration‐time data from two external in vivo animal PK datasets. 26 , 27 The predicted vs. observed data from the two datasets showed good agreement, with over 90% of the observed data within the 90% prediction interval of the simulated population concentrations (Figure 3 a,b). Furthermore, the AFE values demonstrated that the median predicted concentrations generally fell within twofold of the observed values for all datasets (Table 3 ). The external datasets with PK data from Recchiuti et al. and Krashia et al. are provided as Supplementary Files [Link] , [Link] , respectively.
Figure 3.
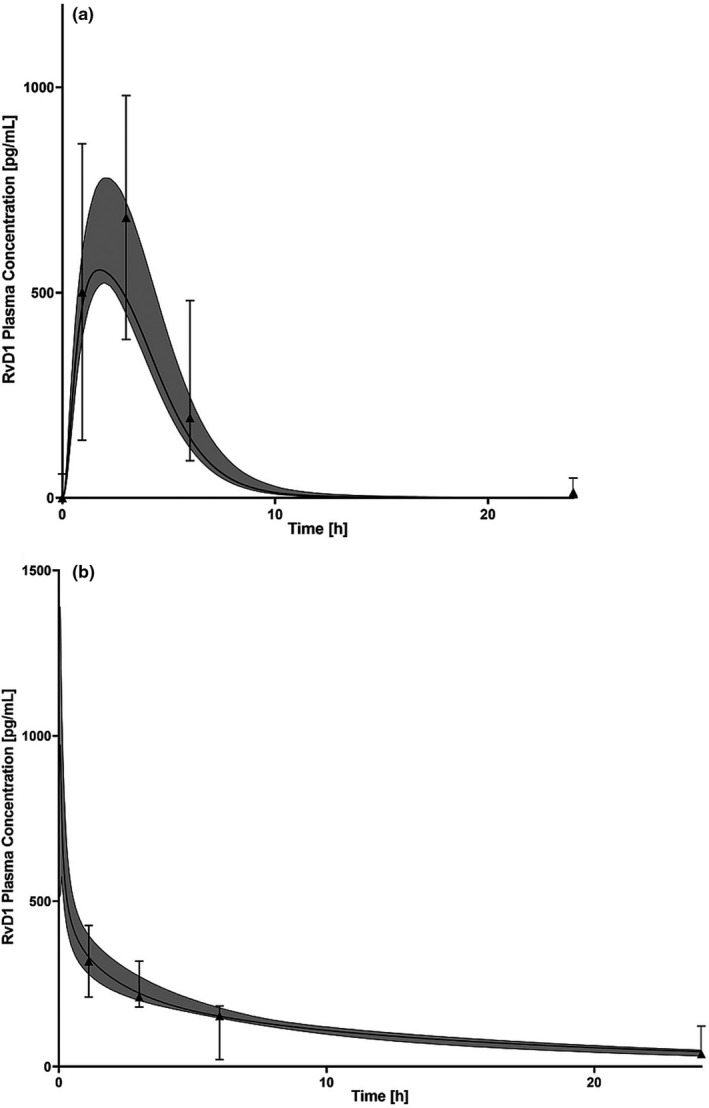
The physiologically‐based pharmacokinetic concentration‐time model predictions of resolvin D1 (RvD1). Modelling was performed (a) following administration of an oral dose of 0.005 mg per kg in C57BL/6N mice and (b) intraperitoneal dose 0.2 µg/kg in Sprague‐Dawley rats . Solid black lines represent the median predicted concentration; and black triangle symbols represent data from each individual animal of the observed data obtained from the animal study reported by Recchiuti et al. 26 (a) and Krashia et al. 27 (b). The shaded region represents the 90% prediction interval for the simulated RvD1 concentrations.
AUC and Cmax were sensitive to each of the parameters tested, with the greatest sensitivity associated with total hepatic clearance, followed by log P and hematocrit value. Hematocrit value may be an influential parameter affecting AUC and Cmax of RvD1 because of its ability to impact the free RvD1 plasma concentration. 44 Furthermore, the hematocrit value is a patient‐specific physiological parameter, which is highly variable due to patients’ age and disease state. 45
The first‐in‐human simulations were performed at a dose range of 0.01 to 10 mg/kg. Based on the simulations, 1 mg/kg dose was chosen as the appropriate first‐in‐human dose as > 90% of the individuals reached a Cmax of 119.36 ng/mL (box in Figure 4b ), which is greater than the efficacy threshold of 100 ng/mL of RvD1. 43 The concentration vs. time curves of RvD1 in plasma and saliva compartment with PK data is provided in Figure 4 a,b, respectively.
Figure 4.
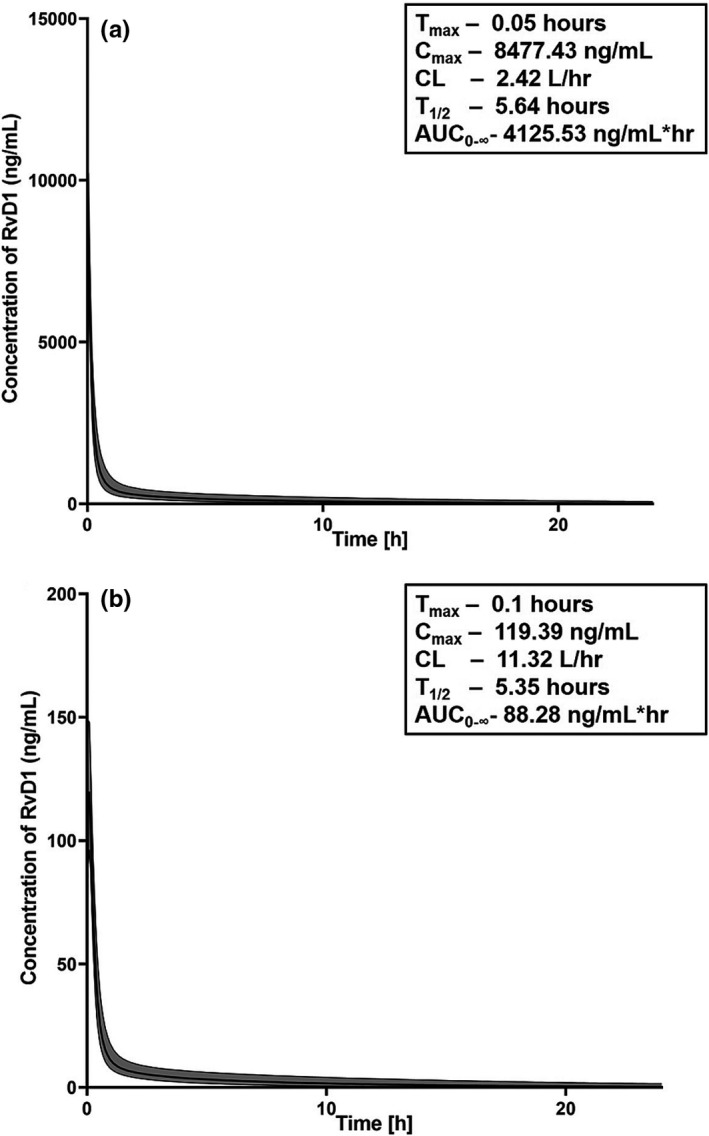
The physiologically‐based pharmacokinetic model concentration‐time model predictions of resolvin D1 (RvD1). Modeling was performed in (a) plasma and (b) saliva compartments of the virtual human population (n = 10,000) at 1 mg/kg I.V. dose. The shaded area represents 5th to 95th percentiles of the predicted concentrations. The pharmacokinetic (PK) parameters inside the boxes of each figure were calculated by noncompartmental analysis. AUC0–∞, area under the concentration‐time curve from zero to infinity; CL, clearance; Cmax, peak plasma concentration; T1/2, terminal half‐life; Tmax, time to peak plasma concentration.
DISCUSSION
SS has a profound negative impact on the quality of life, 6 and, unfortunately, no effective treatment exists. Symptomatic treatments for hyposalivation are limited to the use of saliva substitutes with modest effectiveness, and salivary secretory agonists that only provide temporary relief. 8 Due to the significant unmet medical need, and the limitations of current therapies, the development of novel treatments to decrease inflammation and restore salivary gland secretory function is essential. RvD1 is a novel therapeutic for the treatment of SS. RvD1 belongs to the resolvin class of potent lipid inflammatory mediators and has demonstrated efficacy in treating inflammatory‐related diseases 14 and promoting tissue repair in the salivary epithelium. 43 In this study, we utilized the PBPK modeling strategy for first‐in‐human PK predictions of RvD1.
PBPK modeling is well‐established in the pharmaceutical industry and is accepted by regulatory agencies for the prediction of drug‐drug interactions. 33 PBPK modeling can be extended to address a wide range of pharmaceutical applications that impact regulatory decisions. One such application is the translation of preclinical PK data to inform first‐in‐human predictions. 46 PBPK modeling is superior to empiric scaling for predicting first‐in‐human PKs and is routinely applied for first‐in‐human predictions within pharmaceutical companies. 46
In this study, a novel PBPK model of RvD1 was developed and optimized using in vivo PK data using NOD/ShiLtJ mice representing an SS‐like phenotype. The in vivo PK data of RvD1 in mice showed a biphasic response in plasma concentration with an increase in RvD1 concentration around 4 hours. This biphasic response may be attributed to the endogenous release of RvD1 and the SS‐like inflammatory phenotype of NOD/ShiLtJ. Previous studies have shown that RvD1 levels are dysregulated in patients with inflammatory diseases, such as arthritis and systemic lupus erythematosus, when compared with healthy subjects 47 , 48 and, likewise, it is possible that RvD1 concentrations in NOD/ShiLtJ mice may be dysregulated due to their inflammatory phenotype. Furthermore, local biosynthesis of specialized pro‐resolving mediators like RvD1 are likely to be controlled in part by circadian responses as similar lipid mediators, including glucocorticoids and prostaglandins, are influenced by circadian oscillations. 49 Successful clinical translation of RvD1 as a therapeutic to treat SS will necessitate a detailed understanding of RvD1 PKs at the salivary gland. The developed PBPK model adequately predicted the PKs of RvD1 in both the plasma and the salivary gland compartments. This model can be used to predict the RvD1 concentrations in salivary glands across different dose regimens and various species. RvD1 was rapidly absorbed into the salivary glands following i.v. injection and reaching maximum concentrations within 3–5 minutes. This rapid absorption into the salivary glands is expected due to the lipophilicity of RvD1. However, the model underpredicted the maximum concentration of RvD1 in salivary gland tissue. This may be due to an unaccounted mechanism for RvD1 uptake by salivary gland cells and will require further investigation.
The predictive performance of the PBPK model was further evaluated with two independent datasets to evaluate robustness in predicting oral RvD1 administration in mice and RvD1 PK in another rodent species (Sprague‐Dawley rats). The model was able to adequately predict the PK of RvD1 after oral administration when compared with the observed data. Although, the model had a tendency toward underprediction of in vivo RvD1 concentrations, it predicted an oral bioavailability of 31%. However, this prediction needs to be confirmed, along with a potential RvD1 transport mechanism, in future in vivo experiments.
The optimized mouse PBPK model was extrapolated to a human PBPK model population. Our predictions showed that after i.v. administration, the RvD1 concentrations decline rapidly after 4–6 hours with a half‐life under 3 hours. This result suggests to developing strategies to prolong the half‐life of RvD1, allowing less frequent dosing when used in humans.
In our previous in vitro cell culture experiments using freshly isolated salivary epithelial cells from mouse salivary glands, we reported that RvD1 at 100 ng/mL shows potent anti‐inflammatory activity by preventing tumor necrosis factor alpha mediated disruption of salivary epithelial formation. 43 Based on this anti‐inflammatory efficacy data of RvD1 we concluded that 1 mg/kg RvD1 is the appropriate dose for first‐in‐human studies as the Cmax of RvD1 in human simulations at 1 mg/kg was 119.36 ng/mL. The results from our study illustrate the value of PBPK modeling for the integration of physiologic and physicochemical data from multiple sources to refine and improve understanding of the PK properties of RvD1 in humans before first‐in‐human studies.
However, our model has limitations that need to be addressed in order to enhance the confidence of the predictions for future use. First, parameters, such as degree of protein binding, red blood cells to plasma partitioning, the contribution of specific enzymes to hepatic degradation/elimination, renal excretion process, and the role of potential transport mechanisms, need to be evaluated experimentally for RvD1 and incorporated into the model. Second, the first‐in‐human PK predictions need to be verified with observed human concentration data. Third, our model does not account for the endogenous synthesis and elimination of RvD1 due to lack of sufficient information. Due to these limitations, the proposed animal model can be considered preliminary and caution must be exercised before relying on the first‐in‐human predictions from this model. Overall, future studies will investigate PK of RvD1 in a phase I clinical trial.
In conclusion, a mouse PBPK model of RvD1 was extrapolated to humans to predict RvD1 PK. The model simulations provide valuable information on PK of RvD1 and appropriate dose in humans that can be used to inform and design first‐in‐human studies.
Funding
This project was supported by the National Institutes of Health Grant R01DE027884 awarded to Olga J. Baker.
Conflict of Interest
The authors declared no competing interests for this work.
Author Contributions
V.Y., K.W., and O.B. wrote the manuscript. V.Y., K.W., and O.B designed the research. V.Y., K.P., and S.D. performed the research. J.C. analyzed the data.
Supporting information
Figure S1
Table S1
Dataset S1
Dataset S2
Dataset S3
Dataset S4
References
- 1. Luciano, N. et al. One year in review 2015: Sjogren's syndrome. Clin. Exp. Rheumatol. 33, 259–271 (2015). [PubMed] [Google Scholar]
- 2. Gaubitz, M. Epidemiology of connective tissue disorders. Rheumatology (Oxford) 45(suppl. 3), iii3–iii4 (2006). [DOI] [PubMed] [Google Scholar]
- 3. Lopez‐Pintor, R.M. , Fernandez Castro, M. & Hernandez, G. Oral involvement in patients with primary Sjogren's syndrome. Multidisciplinary care by dentists and rheumatologists. Reumatol. Clin. 11, 387–394 (2015). [DOI] [PubMed] [Google Scholar]
- 4. Knopf, A. , Pickhard, A. , Stark, T. , Schulz, S. & Scherer, E.Q. Recurrent abcesses of the parotid gland in Sjogren's syndrome. HNO 57, 959–963 (2009). [DOI] [PubMed] [Google Scholar]
- 5. Awasthi, N.P. , Kashyap, R. , Kumari, N. & Krishnani, N. Sjogren's syndrome complicated by non‐Hodgkin's lymphoma: spectrum of lymphoid proliferations and associated hepatitis C virus infection. Indian J. Pathol. Microbiol. 54, 430–431 (2011). [DOI] [PubMed] [Google Scholar]
- 6. Segal, B. et al. Primary Sjogren's syndrome: health experiences and predictors of health quality among patients in the United States. Health Qual. Life Outcomes 7, 46 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ferro, F. et al. One year in review 2016: Sjogren's syndrome. Clin. Exp. Rheumatol. 34, 161–171 (2016). [PubMed] [Google Scholar]
- 8. Wang, J. , Zhou, L. & Liu, B. Update on disease pathogenesis, diagnosis, and management of primary Sjogren's syndrome. Int. J. Rheum. Dis. 23, 723–727 (2020). [DOI] [PubMed] [Google Scholar]
- 9. Park, G.T. et al. Formyl peptide receptor 2 activation ameliorates dermal fibrosis and inflammation in bleomycin‐induced scleroderma. Front. Immunol. 10, 2095 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Parashar, K. , Schulte, F. , Hardt, M. & Baker, O.J. Sex‐mediated elevation of the specialized pro‐resolving lipid mediator levels in a Sjogren's syndrome mouse model. FASEB J. 34, 7733–7744 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Munoz, L.E. et al. Autoimmunity and chronic inflammation ‐ two clearance‐related steps in the etiopathogenesis of SLE. Autoimmun. Rev. 10, 38–42 (2010). [DOI] [PubMed] [Google Scholar]
- 12. Saito, I. et al. Expression of cell adhesion molecules in the salivary and lacrimal glands of Sjogren's syndrome. J. Clin. Lab. Anal. 7, 180–187 (1993). [DOI] [PubMed] [Google Scholar]
- 13. Mueller, C.G. , Nayar, S. , Gardner, D. & Barone, F. Cellular and vascular components of tertiary lymphoid structures. Methods Mol. Biol. 1845, 17–30 (2018). [DOI] [PubMed] [Google Scholar]
- 14. Serhan, C.N. & Levy, B.D. Resolvins in inflammation: emergence of the pro‐resolving superfamily of mediators. J. Clin. Invest. 128, 2657–2669 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yacoubian, S. & Serhan, C.N. New endogenous anti‐inflammatory and proresolving lipid mediators: implications for rheumatic diseases. Nat. Clin. Pract. Rheumatol. 3, 570–579; quiz 571 page following 589 (2007). [DOI] [PubMed] [Google Scholar]
- 16. Serhan, C.N. Treating inflammation and infection in the 21st century: new hints from decoding resolution mediators and mechanisms. FASEB J. 31, 1273–1288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sommakia, S. & Baker, O.J. Regulation of inflammation by lipid mediators in oral diseases. Oral Dis. 23, 576–597 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Sun, Y.P. et al. Resolvin D1 and its aspirin‐triggered 17R epimer. Stereochemical assignments, anti‐inflammatory properties, and enzymatic inactivation. J. Biol. Chem. 282, 9323–9334 (2007). [DOI] [PubMed] [Google Scholar]
- 19. Leigh, N.J. , Nelson, J.W. , Mellas, R.E. , Aguirre, A. & Baker, O.J. Expression of resolvin D1 biosynthetic pathways in salivary epithelium. J. Dent. Res. 93, 300–305 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Keinan, D. , Leigh, N.J. , Nelson, J.W. , De Oleo, L. & Baker, O.J. Understanding resolvin signaling pathways to improve oral health. Int. J. Mol. Sci. 14, 5501–5518 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nelson, J.W. et al. ALX/FPR2 receptor for RvD1 is expressed and functional in salivary glands. Am. J. Physiol. Cell Physiol. 306, C178–C185 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Easley, J.T. , Maruyama, C.L. , Wang, C.S. & Baker, O.J. AT‐RvD1 combined with DEX is highly effective in treating TNF‐alpha‐mediated disruption of the salivary gland epithelium. Physiol. Rep. 4, e12990 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang, C.S. , Maruyama, C.L. , Easley, J.T. , Trump, B.G. & Baker, O.J. AT‐RvD1 promotes resolution of inflammation in NOD/ShiLtJ mice. Sci. Rep. 7, 45525 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maruyama, C.L. , Monroe, M.M. , Hunt, J.P. , Buchmann, L. & Baker, O.J. Comparing human and mouse salivary glands: a practice guide for salivary researchers. Oral Dis. 25, 403–415 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bayer Technology Services . Computational systems biology software suite. PKSim® and MOBI® manual. Version 8.0.0, SB Suite <http://www.systems‐biology.com/products/pk‐sim.html>. Accessed January 28, 2020.
- 26. Recchiuti, A. et al. Immunoresolving actions of oral resolvin D1 include selective regulation of the transcription machinery in resolution‐phase mouse macrophages. FASEB J. 28, 3090–3102 (2014). [DOI] [PubMed] [Google Scholar]
- 27. Krashia, P. et al. Blunting neuroinflammation with resolvin D1 prevents early pathology in a rat model of Parkinson's disease. Nat. Commun. 10, 3945 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. U.S. National Library of Medicine .Resolvin D1 Compound Summary, <https://pubchem.ncbi.nlm.nih.gov/compound/44251266>. Accessed October 1, 2020.
- 29. Chemicalize . Instant Cheminformatics Solutions, Chemaxon, MA, USA <https://chemaxon.com/products/chemicalize>.
- 30. Mouse strain datasheet‐001976. Jackson Laboratories <https://www.jax.org/strain/001976> (2018).
- 31. Rodgers, T. , Leahy, D. & Rowland, M. Physiologically based pharmacokinetic modeling 1: predicting the tissue distribution of moderate‐to‐strong bases. J. Pharm. Sci. 94, 1259–1276 (2005). [DOI] [PubMed] [Google Scholar]
- 32. Rodgers, T. & Rowland, M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J. Pharm. Sci. 95, 1238–1257 (2006). [DOI] [PubMed] [Google Scholar]
- 33. Niederalt, C. et al. A generic whole body physiologically based pharmacokinetic model for therapeutic proteins in PK‐Sim. J. Pharmacokinet. Pharmacodyn. 45, 235–257 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith, J.N. , Carver, Z.A. , Weber, T.J. & Timchalk, C. Predicting transport of 3,5,6‐trichloro‐2‐pyridinol into saliva using a combination experimental and computational approach. Toxicol. Sci. 157, 438–450 (2017). [DOI] [PubMed] [Google Scholar]
- 35. Rabito, S.F. , Orstavik, T.B. , Scicli, A.G. , Schork, A. & Carretero, O.A. Role of the autonomic nervous system in the release of rat submandibular gland kallikrein into the circulation. Circ. Res. 52, 635–641 (1983). [DOI] [PubMed] [Google Scholar]
- 36. Duck, F.A. Physical Properties of Tissue. A Comprehensive Reference Book (Academic Press Limited, San Diego, CA, 1990). [Google Scholar]
- 37. Schmitt, W. General approach for the calculation of tissue to plasma partition coefficients. Toxicol. In Vitro 22, 457–467 (2008). [DOI] [PubMed] [Google Scholar]
- 38. Statistics NCfH . Third National Health and Nutrition Examination Survey (NHANES III) (Statistics NCfH, Hyattsville, MD, 1997). [Google Scholar]
- 39. ICRP Basic anatomical and physiological data for use in radiological protection reference values. Ann. ICRP 32, 3–4 (2002). [PubMed] [Google Scholar]
- 40. de Paula, F. et al. Overview of human salivary glands: highlights of morphology and developing processes. Anat. Rec. 300, 1180–1188 (2017). [DOI] [PubMed] [Google Scholar]
- 41. Schwenzer, N.F. et al. MR measurement of blood flow in the parotid gland without contrast medium: a functional study before and after gustatory stimulation. NMR Biomed. 21, 598–605 (2008). [DOI] [PubMed] [Google Scholar]
- 42. Boxenbaum, H. Interspecies scaling, allometry, physiological time, and the ground plan of pharmacokinetics. J. Pharmacokinet. Biopharm. 10, 201–227 (1982). [DOI] [PubMed] [Google Scholar]
- 43. Odusanwo, O. , Chinthamani, S. , McCall, A. , Duffey, M.E. & Baker, O.J. Resolvin D1 prevents TNF‐alpha‐mediated disruption of salivary epithelial formation. Am. J. Physiol. Cell. Physiol. 302, C1331–C1345 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Emoto, C. et al. A Theoretical physiologically‐based pharmacokinetic approach to ascertain covariates explaining the large interpatient variability in tacrolimus disposition. CPT Pharmacometrics Syst. Pharmacol. 8, 273–284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yip, R. , Johnson, C. & Dallman, P.R. Age‐related changes in laboratory values used in the diagnosis of anemia and iron deficiency. Am. J. Clin. Nutr. 39, 427–436 (1984). [DOI] [PubMed] [Google Scholar]
- 46. Miller, N.A. , Reddy, M.B. , Heikkinen, A.T. , Lukacova, V. & Parrott, N. Physiologically based pharmacokinetic modelling for first‐in‐human predictions: an updated model building strategy illustrated with challenging industry case studies. Clin. Pharmacokinet. 58, 727–746 (2019). [DOI] [PubMed] [Google Scholar]
- 47. Barden, A.E. et al. Specialised pro‐resolving mediators of inflammation in inflammatory arthritis. Prostaglandins Leukot. Essent. Fatty Acids 107, 24–29 (2016). [DOI] [PubMed] [Google Scholar]
- 48. Navarini, L. et al. Role of the specialized proresolving mediator resolvin d1 in systemic lupus erythematosus: preliminary results. J. Immunol. Res. 2018, 5264195 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Claria, J. , Dalli, J. , Yacoubian, S. , Gao, F. & Serhan, C.N. Resolvin D1 and resolvin D2 govern local inflammatory tone in obese fat. J. Immunol. 189, 2597–2605 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1
Table S1
Dataset S1
Dataset S2
Dataset S3
Dataset S4


