Abstract
Background
Plasmacytoid dendritic cells (pDCs) play a key role in the induction and maintenance of antitumor immunity. Conversely, they can act as tolerogenic DCs by inhibiting tumor-directed immune responses. Therefore, pDCs may profoundly influence tumor progression. To gain novel insights into the role of pDCs in colon cancer, we investigated the frequency and clinical relevance of pDCs in primary tumor tissues from patients with colon cancer with different clinicopathological characteristics.
Methods
Immunohistochemical stainings were performed to explore the frequency of tumor-infiltrating BDCA-2+ pDCs in patients with colon cancer. Statistical analyses were conducted to determine an association between the pDC density and clinicopathological characteristics of the patients. Furthermore, we used multiplex immunofluorescence stainings to evaluate the localization and phenotype of pDCs in stroma and tertiary lymphoid structures (TLS) of colon cancer tissues.
Results
An increased density of infiltrating pDCs was associated with lower Union for International Cancer Control (UICC) stages. Furthermore, a higher pDC frequency was significantly correlated with increased progression-free and overall survival of patients with colon cancer. Moreover, a lower number of coloncancer-infiltrating pDCs was significantly and independently linked to worse prognosis. In addition, we found that a proportion of pDCs shows a nuclear expression of the transcription factor interferon regulatory factor 7 (IRF7), which is characteristic for an activated phenotype. In various tumor stroma regions, IRF7+ pDCs were located in the neighborhood of granzyme B-expressing CD8+ T cells. Moreover, pDCs were identified as a novel component of the T cell zone of colon cancer-associated TLS, which are major regulators of adaptive antitumor immunity. A proportion of TLS-associated pDCs displayed a nuclear IRF7 expression and was preferentially located close to CD4+ T cells.
Conclusions
These results indicate that higher densities of tumor-infiltrating pDCs are associated with prolonged survival of patients with colon cancer. Moreover, colon cancer-infiltrating pDCs may represent a novel prognostic factor. The colocalization of activated pDCs and T cells in tumor stroma and within TLS may contribute to the correlation between higher pDC densities and better prognosis. In addition, our findings may have implications for the design of novel immunotherapeutic strategies that are based on targeting colon cancer-infiltrating pDCs.
Keywords: dendritic cells, gastrointestinal neoplasms, immunohistochemistry, tumor microenvironment
Background
Accumulating evidence indicates that the tumor immune contexture comprising the spatial organization, density, and functional orientation of tumor-infiltrating immune cells plays a critical role for the clinical outcome of patients with cancer.1–4 Thus, it has been reported that higher frequencies of CD45RO+ T helper (Th) 1 cells and CD8+ T cells are associated with prolonged survival of patients with cancer.1 3 In addition, recent studies provided evidence that higher densities of tumor-infiltrating CD8+ T cells at baseline correlate with an improved clinical efficacy of immune checkpoint inhibitor (CPI) therapy, which evolved as a very promising treatment modality for patients with cancer.5 6
Dendritic cells (DCs) play a key role in the orchestration of innate and adaptive antitumor immunity. Consequently, DCs may profoundly influence tumor progression and may have a significant impact on the clinical outcome of patients with cancer. When investigating the clinical impact of native human blood DC subsets, it has been demonstrated that a higher expression of specific gene signatures for myeloid DC1 and DC2 is associated with a higher probability for disease-free survival of patients with luminal breast cancer.7 Furthermore, a higher DC1-specific gene signature was significantly linked to improved survival in patients with cutaneous melanoma, breast invasive carcinoma, head and neck squamous cell carcinoma, and lung adenocarcinoma.8
Plasmacytoid DCs (pDCs) represent another major subset of human blood-circulating DCs that are characterized by their selective phenotype (BDCA-2+, BDCA-4+, CD123+, CD1c−, CD11c−, CD14−, CD141−) and by their extraordinary capacity to produce large amounts of type I interferon (IFN) upon stimulation.9 10 Functional studies revealed that activated pDCs efficiently enhance the antitumoral properties of natural killer and T cells.11 12 Moreover, it has been reported that pDCs kill tumor cells directly via tumor necrosis factor-related apoptosis-inducing ligand and granzyme B (GrzB)-dependent mechanisms.13 14 Based on these functional properties, stimulated pDCs can efficiently exhibit antitumor responses in vivo. Thus, intratumoral pDC administration led to tumor regression in a B16 melanoma mouse model.15 In addition, it has been shown in an orthotopic murine mammary tumor model that the intratumoral application of a toll-like receptor 7 (TLR7) ligand results in the activation of tumor-associated pDCs and tumor regression.16 In a clinical trial, intranodal injections of activated pDCs loaded with tumor antigen-associated peptides in patients with metastatic melanoma induced specific CD8+ and CD4+ T cell responses.17 However, tumor-infiltrating pDCs can also display tolerogenic properties contributing to tumor progression.9 10 For example, it has been demonstrated that pDCs activate regulatory T cells (Tregs) via indoleamine 2,3-dioxygenase.18 The presence of tolerogenic pDCs may explain previous findings, indicating that a higher density of tumor-infiltrating pDCs is associated with poor prognosis in various cancer types such as head and neck, breast, and ovarian cancer.19–21
So far, little is known about the potential role of pDCs in colon cancer and their impact on prognosis of patients. To gain novel insights into the clinical relevance of colon cancer-infiltrating pDCs, we determined the frequency of pDCs in primary tumor tissues and the association between pDC numbers and important clinicopathological characteristics of patients with colon cancer. In addition, we employed multiplex immunofluorescence stainings to explore the presence, localization, and phenotype of colon cancer-associated pDCs.
Materials and methods
Patients and study design
This is a retrospective study including 149 patients with Union for International Cancer Control (UICC) stage I–IV colon cancer treated with primary resection at the University Hospital Carl Gustav Carus of Dresden between 2002 and 2013. Each case was classified according to the tumor, node, metastases staging system of the UICC. The clinicopathological characteristics of the patients are outlined in online supplemental table S1.
jitc-2020-001813supp001.pdf (21.5KB, pdf)
Immunohistochemistry
Formalin-fixed paraffin-embedded (FFPE) tissue blocks were cut into 3–5 µm sections. Tissue preparation consisted of deparaffinization in xylene (2×8 min, VWR International, Fontenay-sous-Bois, France) and rehydration by washes in graded ethanol (Berkel AHK, Ludwigshafen, Germany) to water (B. Braun, Melsungen, Germany). For antigen retrieval, the slides were boiled in citrate buffer (Zytomed Systems GmbH, Berlin, Germany) at pH 6.0 for 20 min. To determine the presence and frequency of tumor-infiltrating pDCs, the tissue sections were stained overnight at 4°C with the polyclonal goat anti-BDCA-2 antibody (1:200, R&D Systems, Minneapolis, Minnesota, USA), followed by a 10 min incubation with a mouse anti-goat antibody solution (Thermo Fisher Scientific, Rockford, Illinois, USA). Subsequently, labeling with dextran-coupled anti-mouse antibody (Dako, Glostrup, Denmark) was performed for all sections. pDCs were visualized by the alkaline phosphatase-based EnVision detection system according to the manufacturer’s instructions (Dako). Finally, all tissue sections were counterstained with Mayer’s hematoxylin (Merck, Darmstadt, Germany). For quantification, 80 randomly selected regions of interest (size 0.0835 mm²) within every whole section were acquired (×400 magnification) using the Vectra 3 automated quantitative pathology imaging system (Akoya Biosciences, Menlo Park, California, USA). Positively stained pDCs were counted and the mean value was determined and converted into cell density.
Immunofluorescence staining
For additional experiments, we employed the Opal kit together with the Vectra 3 automated imaging system (both from Akoya Biosciences) to perform various multiplex immunofluorescence stainings. For the stainings conducted manually, tissues were deparaffinized and hydrated as described earlier. A microwave treatment in the AR6 or AR9 buffer (both from Akoya Biosciences) was applied for antigen retrieval. Subsequently, the Opal kit was used as described previously.22 Briefly, after a 10 min blocking step, tissues were incubated with the primary antibody for 1 hour and then with a horseradish peroxidase-conjugated secondary antibody (Akoya Biosciences) for 10 min. In the case of goat anti-human primary antibodies, a bridge step with a mouse anti-goat antibody (1:100, Thermo Fisher Scientific) preceded the application of the secondary antibody. Finally, one of the six TSA fluorophores available for Vectra 3 (Opal 520, 540, 570, 620, 650, and 690, all from Akoya Biosciences) was added to the tissue sections for 10 min. Subsequent stripping of the primary together with secondary antibodies was performed by microwave treatment. All the steps mentioned earlier, starting with blocking and ending with the microwave treatment, were repeated for each of the primary antibodies.
For the stainings performed on the Ventana Ultra Instrument (Ventana Medical Systems, Arizona, USA), the steps employed were similar. Tissues were deparaffinized and antigen retrieval was performed in the cell conditioning 1 solution (Ventana Medical Systems). Incubation of the primary antibody followed for 32 min at 36ᐤC. Then, the appropriate secondary antibody (OmniMap anti-mouse, anti-rabbit or anti-goat antibody, ready-to-use, all from Ventana Medical Systems) was applied for 12 min. Finally, one of the TSA fluorophores (Akoya Biosciences) was added to the tissue slides for 8 min. Stripping of the primary and secondary antibodies took place by heating the slides at 100ᐤC in the cell conditioning 2 solution (Ventana Medical Systems). This sequence of steps (except deparaffinization and antigen retrieval) was repeated for each primary antibody.
Finally, for both the manual and automatic approach, all tissue slides were counterstained with spectral DAPI (Akoya Biosciences) and coverslipped with fluoromount medium (SouthernBiotech, Birmingham, Alabama, USA). The primary antibodies used for the various multiplex immunofluorescence stainings are summarized in online supplemental table S2.
jitc-2020-001813supp002.pdf (84.3KB, pdf)
Acquisition of the multispectral images was performed with the Vectra 3 automated imaging system (Akoya Biosciences). Spectral unmixing was done in the inForm software (Akoya Biosciences) using a library built from single stained tissue slides for each primary antibody-TSA fluorophore combination.
Quantification of the density of tertiary lymphoid structures (TLS) was conducted using the whole scans of all tissue sections stained with a seven-color multiplex immunofluorescence protocol. TLS were defined as aggregates of CD20+ cells surrounded by CD8+ and CD4+ lymphocytes that were associated with the tumor. The number of TLS was determined by a blinded observer and converted into frequency for each tissue section.
Analysis of microsatellite stability
To determine whether the 149 colon cancers are microsatellite stable (MSS) or microsatellite instable (MSI), immunohistochemistry was performed on FFPE colon cancer tissue sections using an autostainer. The sections were deparaffinized and rehydrated, and heat-induced epitope retrieval was performed using the cell conditioning 1 solution (Ventana Medical Systems). Slides were incubated with primary antibodies for PMS2 (clone: EPR3947; diluted 1:1; Ventana Medical Systems) or MSH6 (clone: 44; diluted 1:1; Ventana Medical Systems). For visualization, the Roche OptiView DAB IHC Detection Kit (Ventana Medical Systems) was used and the samples were counterstained with hematoxylin. The expression of the mismatch repair (MMR) proteins (PMS2 and MSH6) was defined as absent (loss−MSI) when nuclear staining of tumor cells was not detectable despite positive staining in normal epithelial or stromal cells in at least one of the MMR proteins.
Analysis of public transcriptomic data
Publicly available data were downloaded from the NCBI Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/) using the R software packages Biobase and affy. The following gene expression datasets, determined on the Affymetrix U133 Plus 2.0 Array, were used for this analysis: GSE33113,23 GSE31595,24 GSE38832,25 GSE39582,26 and GSE17536.27 Raw gene expression data was first processed using the frozen robust multi-array analysis (fRMA). Based on the intratumoral C-type lectin domain family 4 member C gene (CLEC4C) expression the patients were split into two groups and assessed by Kaplan-Meier curves and log-rank test.
Statistical analysis
Statistical analysis was performed using unpaired Student’s t-test for the evaluation of pDC density among the groups of patients with different clinicopathological characteristics. Kaplan-Meier curves were used to visualize differences in progression-free survival (PFS) and overall survival (OS). Significance was determined using the log-rank test. Using a Cox proportional hazards regression model, we explored the HR of the pDC infiltration in combination with clinicopathological characteristics of the patients. We assessed the spatial distribution of tumor-infiltrating immune cell subsets by calculating a cross-type cumulative distribution of the nearest neighbor distances for each pair of phenotypes using Ripley’s K function. Thereby, we determined the fraction of pDCs located close (≤25 µm) to CD8+ T cells or Tregs. All statistical analyses were performed using the software R (survival, survminer package). In all analyses, values of p≤0.05 were considered significant.
Results
pDCs infiltrate human colon cancer
pDCs can essentially contribute to the induction and maintenance of antitumor immunity, resulting in tumor growth inhibition. However, they can also act as tolerogenic DCs by inhibiting tumor-directed immune responses, leading to tumor progression. In this context, it has been demonstrated that pDCs infiltrate a variety of human cancers and that a higher density is associated either with better7 or poor clinical outcome.19–21
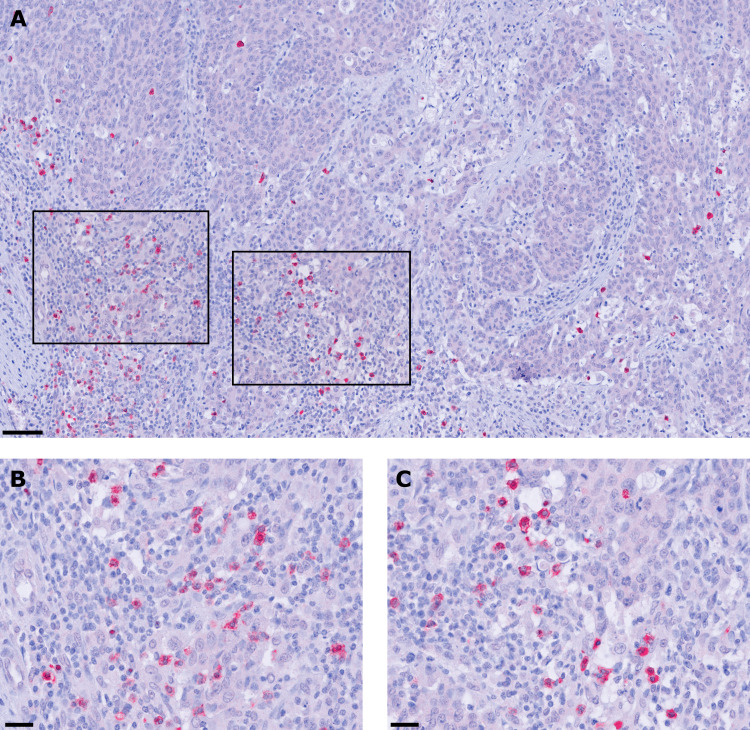
To gain novel insights into the potential role of pDCs in colon cancer, we explored the presence and frequency of BDCA-2-expressing pDCs in tissue specimens from 149 patients with colon cancer with different clinicopathological characteristics (online supplemental table S1). BDCA-2 is a protein encoded by the CLEC4C gene that is characteristically expressed by pDCs and is involved in ligand internalization and antigen presentation.10 28 As depicted in figure 1A–C, pDCs were detectable in all colon cancer samples at varying frequencies (12.2±14.1 pDCs/mm2). They were preferentially located in the tumor stroma and often accumulated along the invasive tumor margin. These results indicate that pDCs are a component of the colon cancer-associated immune contexture and may contribute to the regulation of antitumor immune responses.
Figure 1.
Plasmacytoid dendritic cells (pDCs) infiltrate the tumor microenvironment of patients with colon cancer. (A–C) Immunohistochemical stainings were performed to assess the frequency of tumor-infiltrating pDCs. As a representative example, the presence of pDCs in the stroma of a histologically confirmed primary colon cancer tissue is shown. Images were taken at (A) 100× and (B, C) 400× magnification. Scale bars indicate 50 µm and 25 µm, respectively.
Higher frequency of infiltrating pDCs is associated with better prognosis for patients with colon cancer
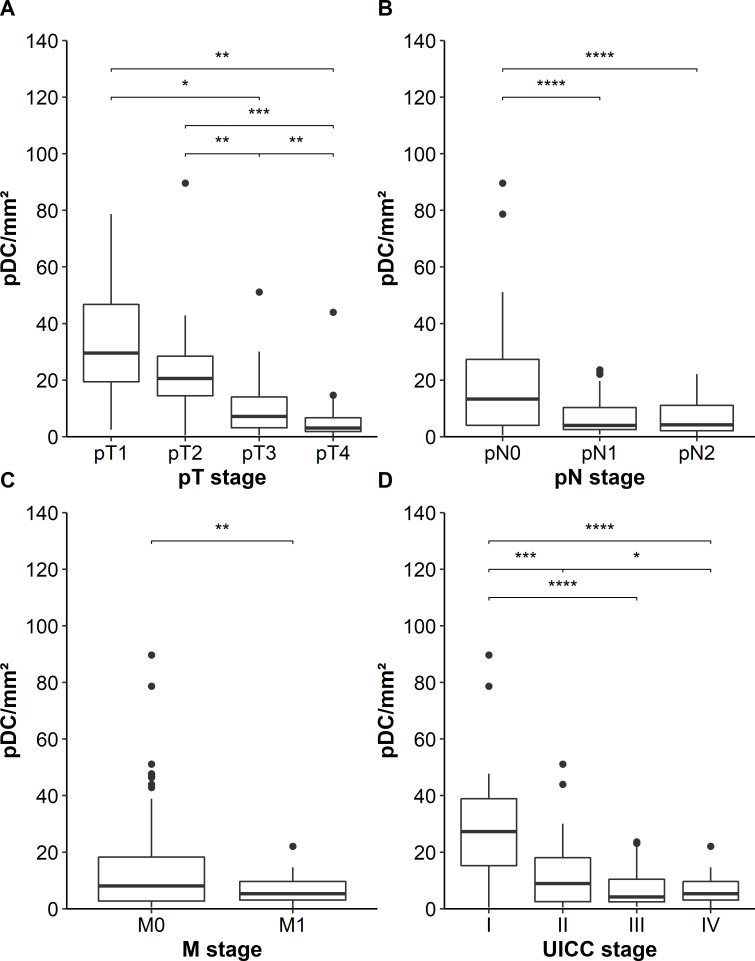
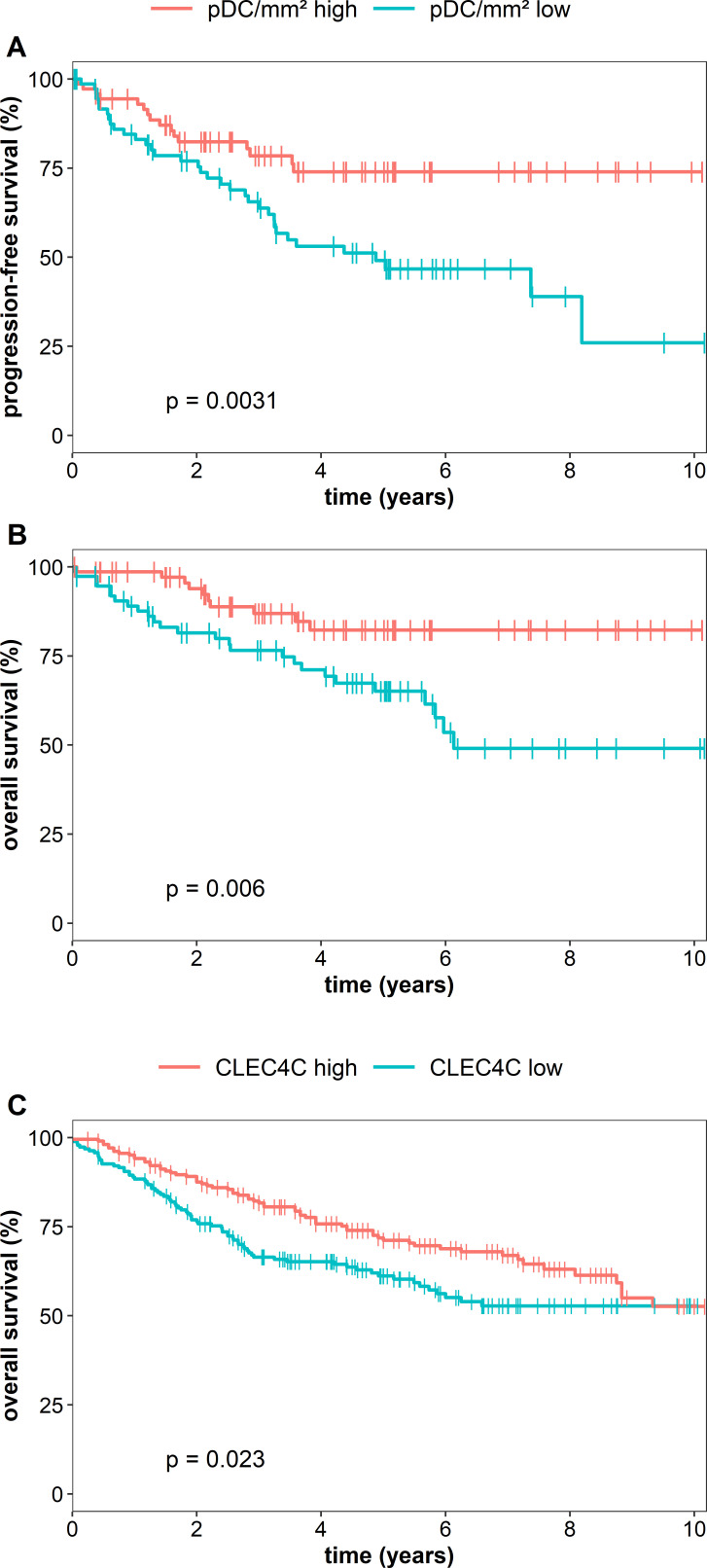
Next, we investigated whether the density of pDCs in primary tumor tissues is associated with relevant clinicopathological characteristics of patients with colon cancer. We found that a lower infiltration of pDCs is associated with locally more advanced tumors (figure 2A). Furthermore, tumors with lymph node or distant metastasis at the time of resection showed a lower density of infiltrating pDCs at the primary tumor site (figure 2B, C). A lower number of colon cancer-infiltrating pDCs correlated with an advanced UICC stage (figure 2D). Previously, it has been demonstrated that MSI-colorectal cancers are associated with a high density of activated CD8+ T cells and a favorable prognosis of patients.29 Following these findings, we explored a potential link between the pDC frequency and the MSI/MSS status of our cohort comprising 23 MSI- and 126 MSS-colon cancers. However, the density of colon cancer-infiltrating pDCs did not differ between these two groups (online supplemental figure S1). In further experiments, we analyzed a potential correlation between the intratumoral pDC density and survival of patients. Interestingly, we found that an increased number of infiltrating pDCs was associated with an improved PFS and OS of patients with colon cancer (figure 3A, B). The 10-year PFS for patients with high pDC densities was 74% compared with 26% for patients with low pDC counts. Moreover, the 10-year OS for patients with colon cancer with high pDC frequencies was 82% compared with 49% for the cohort with low pDC values. To validate our findings, we took advantage of publicly available gene expression datasets of colorectal cancer specimens. Here, we evaluated the data from 1011 patients with colorectal cancer for the expression of CLEC4C in tissue samples. Each gene expression set was normalized using the fRMA method. The datasets were ranked for CLEC4C-expression and the top (208 patients) versus the bottom quartile (191 patients) was assessed for OS. Consistent with our observation, a higher expression of CLEC4C in colorectal cancer samples was significantly linked to an increased OS (figure 3C). Following these findings, we conducted a Cox multivariable regression analysis to assess the relevance of pDC density compared with clinicopathological characteristics. For a low pDC infiltration, we calculated a HR of 2.29 (95% CI 1.22 to 4.3), which was significantly associated with shorter PFS of patients with colon cancer and independent of patients’ gender, age at resection, and UICC stage (table 1). Besides pDC infiltration, UICC IV stage was significantly associated with worse prognosis. These results indicate that a higher frequency of infiltrating pDCs is associated with improved clinical outcome and that tumor-infiltrating pDCs may represent a novel prognostic biomarker in colon cancer.
Figure 2.
Association between the frequency of plasmacytoid dendritic cells(pDCs) in tumor tissues and clinicopathological characteristics of patients with colon cancer. (A–D) Boxplots show the association between pDC infiltration and (A) local tumor progression state (pT stage), (B) lymph node metastases (pN stage), (C) distant metastases (M stage), and (D) combined Union for International Cancer Control (UICC) stage. Significant differences are indicated by asterisks (*p≤0.05, **p≤0.01, ***p≤0.001, ****p≤0.0001).
Figure 3.
Increased plasmacytoid dendritic cell (pDC) frequency is associated with better prognosis of patients with colon cancer. (A, B) Kaplan-Meier curves illustrate the association between the density of pDCs and (A) progression-free survival (PFS) and (B) overall survival (OS). (C) Kaplan-Meier curves demonstrate the correlation between CLEC4C expression in colorectal cancer tissues and OS of patients. The results were obtained from publicly available datasets of patients with colorectal cancer. Statistical comparison between groups of patients was conducted by using log-rank test.
Table 1.
pDC infiltration is an independent prognostic factor for patients with colon cancer
| HR | 95% CI | Wald p value | |
| pDC infiltration | |||
| Low vs high | 2.29 | 1.22 to 4.3 | 0.01* |
| UICC stage | |||
| II vs I | 2.06 | 0.56 to 7.54 | 0.27 |
| III vs I | 1.79 | 0.5 to 6.45 | 0.37 |
| IV vs I | 14.86 | 4.05 to 54.57 | <0.0001**** |
| Age at resection | 0.99 | 0.97 to 1.01 | 0.23 |
| Sex | |||
| Female vs male | 0.71 | 0.4 to 1.26 | 0.24 |
Results of a multivariable Cox proportional hazards analysis of progression-free survival for 149 patients with colon cancer, including patients’ gender, age at resection, UICC stage, and pDC infiltration. Significance was assessed using the Wald p value and is indicated by bold font in the table (*p≤0.05, ****p≤0.0001). When testing for proportionality, none of the parameters violated the proportional hazard assumption (all p values >0.05).
UICC, Union for International Cancer Control.
jitc-2020-001813supp003.pdf (89.5KB, pdf)
jitc-2020-001813supp004.pdf (28KB, pdf)
pDCs preferentially colocalize with CD8+ T cells in colon cancer stroma
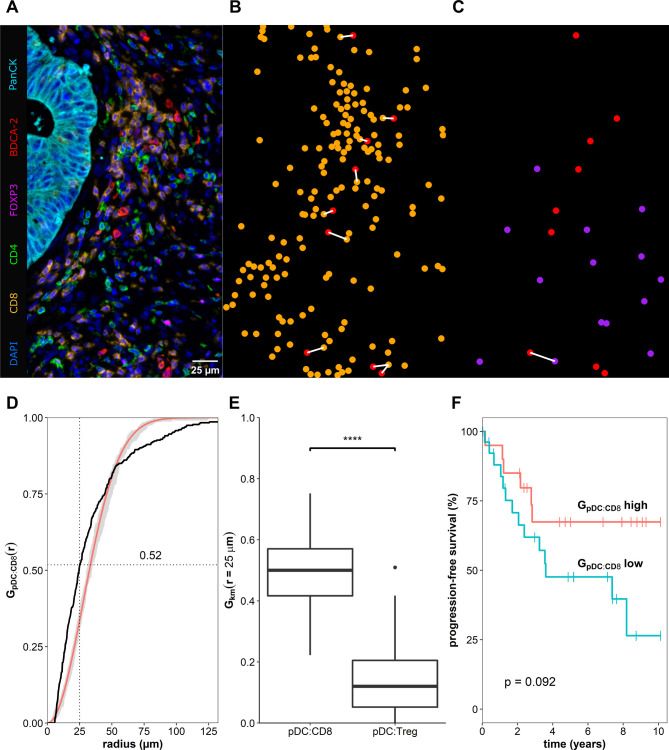
Recent reports revealed that cell-to-cell interactions, defined as neighboring cells located within a distance of 25–30 µm and measured in tissue samples stained with multiplex techniques, can be used as biomarkers for the clinical outcome of the patients.30 31 To identify potential underlying mechanisms for the correlation between increased numbers of colon cancer-associated pDCs and better prognosis, we explored the spatial distribution of various T cell subsets in the neighborhood of pDCs in colon cancer tissues by using multiplex immunofluorescence staining (figure 4A–C). For this, we determined the spatial organization of pDCs to CD8+ T cells or FoxP3+CD4+ Tregs by calculating a cross-type cumulative distribution of the nearest neighbor distances G(r) for each pair of phenotypes (figure 4D). More explicitly, we analyzed which of the phenotypes (CD8+ or FoxP3+CD4+) is preferentially located in proximity (≤25 µm) to pDCs. Interestingly, we found that 49.5%±13.1% of all pDCs across the whole tissue sections of 47 colon cancer samples were located in the neighborhood of a CD8+ T cell (figure 4E). In contrast, only 14.2%±11.5% were located close to a Treg (figure 4E). Our data further suggests a clustered spatial arrangement, meaning that CD8+ cells and pDCs tend to locate nearby (figure 4D). Additionally, we evaluated the clinical significance of the location of pDCs close to CD8+ T cells by dividing the group of patients based on the fraction of neighboring cells and performed survival analysis. Thus, we observed a trend towards prolonged PFS for patients with a higher frequency of colon cancer-infiltrating pDCs located close to CD8+ T cells (figure 4F).
Figure 4.
Plasmacytoid dendritic cells (pDCs) colocalize preferentially with CD8+ T cells. (A) Representative example of a tissue region from a whole slide stained with a seven-color multiplex immunofluorescence protocol. Samples were automatically segmented and phenotyped based on their fluorescence staining. (B, C) Localization of pDCs (red) and CD8+ T cells (orange) or FoxP3+CD4+ Tregs (purple) are illustrated. The white line indicates a distance ≤25 µm from a pDC to their closest T cell. (D) Cumulative distribution of the closest distance for pDCs to CD8+ T cells (black line) is shown in comparison to a theoretical distribution based on a Poisson distribution with its confidence envelope (red line, gray area). (E) Boxplot illustrating the averaged fraction of pDCs in close distance to a CD8+ T cell or a FoxP3+CD4+ Treg, respectively, based on 47 colon cancer tissue samples. Asterisks indicate a statistically significant difference (****p≤0.0001). (F) Kaplan-Meier curves demonstrate the correlation between the fraction of pDCs neighboring a CD8+ T cell and overall survival of patients with colon cancer. Significance was assessed using the log-rank test.
Proportion of colon cancer-infiltrating pDCs displays an activated phenotype
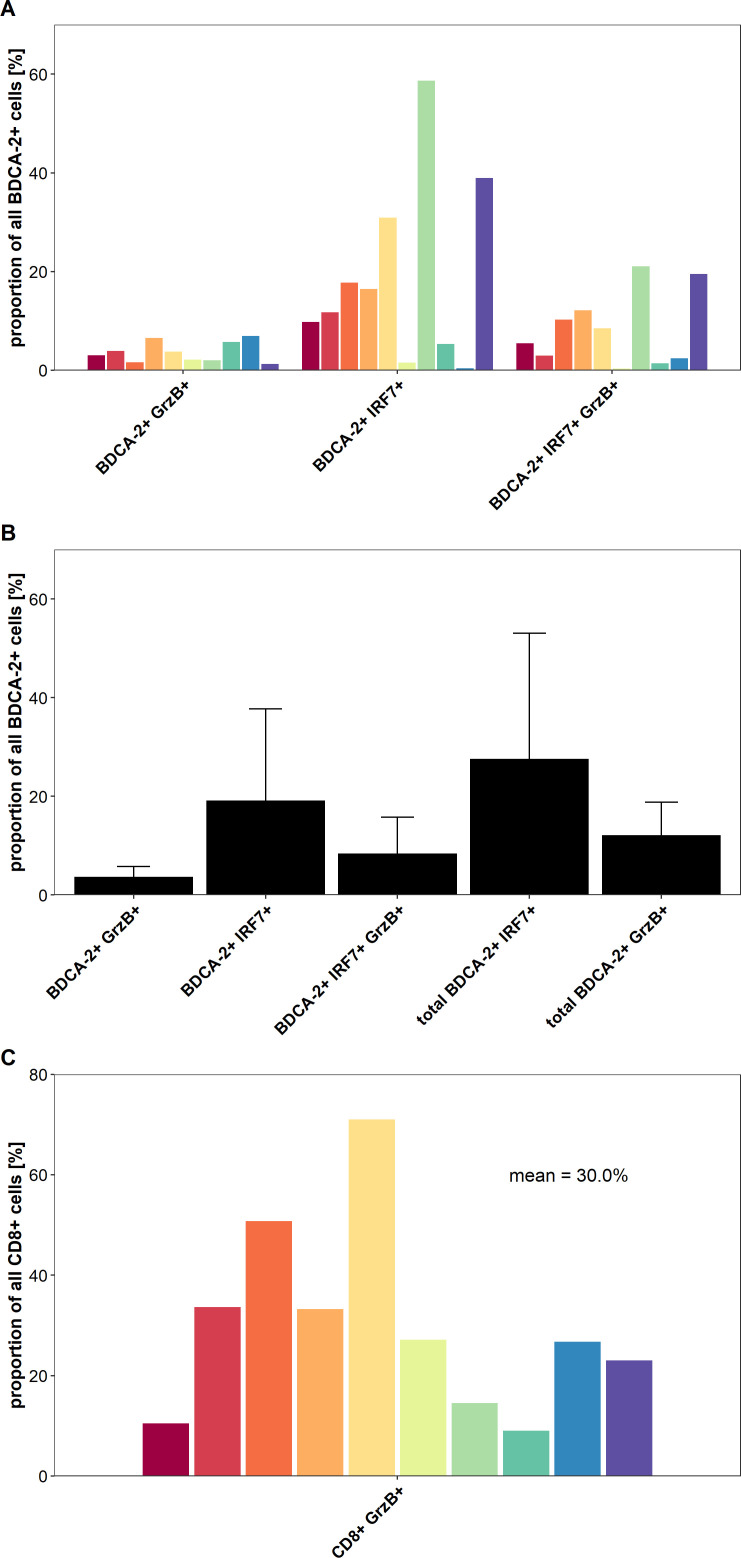
IFN regulatory factor 7 (IRF7) is a key transcriptional factor for the induction of type I IFN production and its nuclear expression is characteristic for activated pDCs, which are major producers of this cytokine.9 10 32 Activation of pDCs through TLR7 and TLR9 can trigger the phosphorylation and nuclear translocation of IRF7, resulting in the induction of large amounts of type I IFN. To gain novel insights into the functional status of colon cancer-infiltrating pDCs and of CD8+ T cells, we conducted a multiparametric immunofluorescence staining of colon cancer samples using antibodies against pan-cytokeratin, BDCA-2, IRF7, CD8, and GrzB. We selected 10 tissue samples containing a high frequency of pDCs from patients with colon cancer with a good prognosis. As demonstrated in online supplemental figure S2A–C and figure 5A, B, activated pDCs characterized by a nuclear expression of IRF7 were present in all colon cancer tissue specimens at varying frequencies (mean: 27.6%, range: 1.8%–79.7%). As depicted in online supplemental figure S2A–C and figure 5A, B, we also found varying percentages of pDCs expressing the cytotoxic effector molecule GrzB in the analyzed tissues (mean: 12.1%, range: 2.5%–23.1%). When investigating the functional status of infiltrating CD8+ T cells, we detected a significant proportion of colon cancer-infiltrating GrzB+ CD8+ T cells (mean: 30.0%, range: 9.0%–71.0%) as shown in online supplemental figure S2A–C and figure 5C. Tumor stroma regions with a colocalization of IRF7+ pDCs and GrzB+ CD8+ T cells were observed in all analyzed colon cancer tissues (online supplemental figure S2A–C). These results provide evidence that activated colon cancer-infiltrating pDCs may contribute to an efficient antitumor response by the production of soluble molecules such as type I IFN and GrzB as well as the stimulation of GrzB+ CD8+ T cells.
Figure 5.
A proportion of colon cancer-infiltrating plasmacytoid dendritic cells (pDCs) and CD8+ T cells displays an activated phenotype. (A) The percentage of BDCA-2+ pDCs expressing interferon regulatory factor 7 (IRF7) and/or granzyme B (GrzB) for each of the 10 analyzed tissues is demonstrated. (B) The results are presented as mean value±SD of the percentage of IRF7-expressing and/or GrzB-expressing pDCs in the 10 tissues. (C) The percentage of CD8+ T cells expressing GrzB for each of the 10 analyzed tissues is depicted.
jitc-2020-001813supp005.pdf (236.8KB, pdf)
pDCs are a component of colon cancer-associated TLS
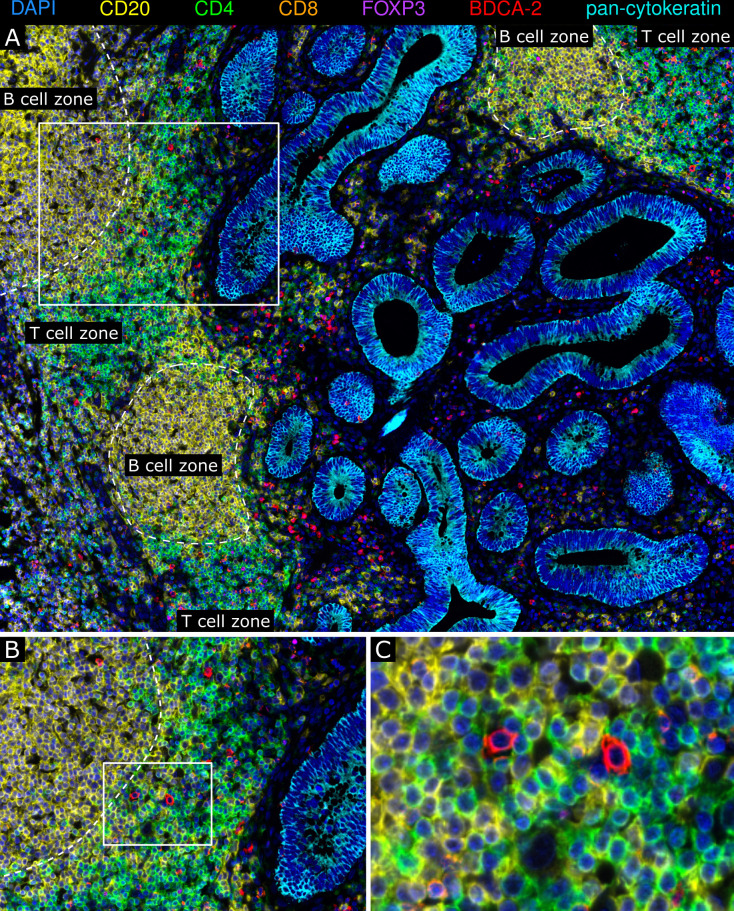
TLS are ectopic, organized lymphoid formations, which can occur in tumor tissues and are considered to play an important role in modulating adaptive antitumor immunity.33 34 Here, we identified colon cancer-associated TLS by multiplex immunofluorescence stainings based on the occurrence of CD4+ cells, CD8+ T cells, and CD20+ B cells and their organization in a lymph node-like structure. When investigating a potential correlation between the TLS density and clinical parameters of patients, we found that a higher TLS frequency is associated with improved PFS and OS of patients with colon cancer (online supplemental figure S3). Moreover, pDCs were detectable in colon cancer-associated TLS and were mainly located in the T cell zone (figure 6A–C). To explore the functional status of TLS-associated pDCs, we performed a multiparametric immunofluorescence staining of colon cancer samples using antibodies against BDCA-2, IRF7, CD3, CD4, and CD20. For this, tissue samples containing an appropriate number of TLS-associated pDCs from seven patients with colon cancer with a good prognosis were used. When analyzing 156 TLS across these tissues, we found that a relevant proportion (mean: 28.4%; range: 5.7%–55.1%) of TLS-associated pDCs shows a nuclear IRF7 expression and is preferentially located close to CD4+ T cells (online supplemental figure S4). These results demonstrate that pDCs are a cellular component of the TLS-associated T cell zone in colon cancer. Moreover, a proportion of these pDCs displays an activated phenotype, indicating that pDCs may participate in the induction and regulation of tumor-directed T cell responses.
Figure 6.
Plasmacytoid dendritic cells (pDCs) are a cellular component of colon cancer-associated tertiary lymphoid structures (TLS). (A–C) Multiplex immunofluorescence stainings of colon cancer tissues for CD4 (green), CD8 (orange), CD20 (yellow), FoxP3 (magenta), pan-cytokeratin (cyan), and BDCA-2 (red) were performed. (A) As a representative example, the spatial distribution of BDCA-2+ pDCs within colon cancer-associated TLS is shown. (B–C) The localization of BDCA-2+ pDCs in the TLS-associated T cell zone is depicted. Original magnification was ×200.
jitc-2020-001813supp006.pdf (161.5KB, pdf)
jitc-2020-001813supp007.pdf (2.9MB, pdf)
Discussion
Recent studies highlight the importance of the tumor immune contexture for the prognosis of patients with colorectal cancer.1–4 Thus, high densities of CD4+ Th1 cells and CD8+ T cells in the tumor center and the invasive margin were associated with improved survival of patients with colorectal cancer.35 36 Recently, a multi-center study has been initiated to assess the prognostic value of tumor-infiltrating T cell numbers in patients with colon cancer.37 Patients with a so-called high Immunoscore, which is characterized by a high frequency of CD3+ and CD8+ T cells in the tumor center and the invasive margin, had the longest survival and the lowest risk of recurrence. These results suggest that the Immunoscore may represent a reliable estimate of the risk of disease recurrence and support its implementation in the classification of colon cancer. In addition to T cells, the clinical impact of colorectal cancer-infiltrating macrophages has been explored. A pooled analysis of various studies has shown that a high density of colorectal cancer-infiltrating macrophages is associated with favorable OS.38
Previous studies revealed that DCs, as key regulators of innate and adaptive antitumor immunity, are a component of the immune architecture in colorectal cancer and may influence the clinical outcome of patients. Thus, it has been reported that S-100+ DCs are detectable in almost all colorectal specimens and are mainly located in the tumor stroma.39–41 Increased densities of tumor epithelium-infiltrating S-100+ DCs were associated with higher numbers of intraepithelial CD4+ and CD8+ T cells.40 When investigating the clinical impact of colorectal cancer-infiltrating DCs, it has been demonstrated that a higher frequency of S-100+ DCs is correlated with prolonged survival of patients.39 42 43 However, other studies did not find a statistically significant association between colorectal cancer-infiltrating S-100+ DCs and improved prognosis.40 41 It has also been reported that DCs expressing the maturation marker CD83 are detectable in colorectal cancer and are located predominantly in the invasive margin of the cancer stroma, often in clusters with lymphocytes.43 44 In addition, a lower number of CD83+ DCs was found in locally advanced tumors.43 Whereas all these findings rely on the detection of general marker molecules that are not selectively expressed by DCs, studies investigating the frequency and clinical impact of distinct human DC subsets in colon cancer tissues are rather limited.
In the present study, we explored the frequency of colon cancer-infiltrating BDCA-2+ (CLEC4C) pDCs and their correlation with clinicopathological characteristics of patients. We found that pDCs are detectable in all primary tumor tissues, indicating that pDCs represent an immune contexture component of colon cancer. Notably, a higher density of infiltrating pDCs was linked to lower tumor stages and to improved PFS and OS of patients with colon cancer. This was further substantiated by publicly available gene expression datasets of a large cohort of colorectal cancer specimens, demonstrating that the expression of CLEC4C was correlated with prolonged OS of patients. These findings are in line with a recent study, indicating that a higher z-score for a pDC subset-specific gene signature is linked to an increased probability of disease-free survival for patients with luminal breast cancer.7 Similar findings have been reported for blood-circulating pDCs in breast cancer, where higher levels of pDCs represented a positive prognostic indicator.45 In contrast, previous studies revealed that a higher density of pDCs is associated with poor clinical outcome for patients with head and neck, breast, hepatocellular, and ovarian cancer.19–21 46
When exploring the underlying mechanisms for this correlation, it has been shown that head and neck cancer can efficiently suppress the ability of infiltrating pDCs to produce IFN-α upon activation with a TLR9 agonist, which may be based on the tumor-induced downregulation of TLR9 expression.19 This finding was further substantiated by studies demonstrating a reduced capacity of ovarian cancer- and breast cancer-associated pDCs to produce IFN-α upon stimulation.21 47 Breast cancer-derived transforming growth factor β and tumor necrosis factor α were shown to impair the IFN-α production of pDCs by blocking IRF7 expression and nuclear translocation.48 In addition, it has been reported that breast cancer-associated pDCs efficiently promote the expansion of Tregs.47
To gain novel insights into the role of colon cancer-infiltrating pDCs that may explain the correlation between a higher pDC frequency and better survival, we explored a potential colocalization of pDCs with CD8+ T cells. Activated CD8+ T cells are characterized by the production of pro-inflammatory cytokines and an extraordinary cytotoxic potential. Due to these functional properties, CD8+ T cells play an essential role in antitumor immunity. We demonstrated that a higher number of pDCs is located close to CD8+ T cells in tumor stroma. When further analyzing the phenotype of colon cancer-infiltrating pDCs, we found a relevant proportion of activated pDCs that were characterized by the nuclear expression of IRF7, representing a master transcription factor for the induction of type I IFN production.9 10 32 A lower percentage of pDCs expressing the cytotoxic effector molecule GrzB was also detected. Furthermore, we observed a significant proportion of colon cancer-infiltrating GrzB+ CD8+ T cells. The colocalization of IRF7+ pDCs and GrzB+ CD8+ T cells in various tumor regions provides evidence that activated pDCs may contribute to the stimulation of tumor-reactive CD8+ cytotoxic T cells. These observations indicate that the tumor microenvironment plays an important role in shaping the functional properties (tolerogenic vs activated) of infiltrating pDCs that may influence the regulation of tumor growth and the prognosis of patients.
Tumor-associated TLS are characterized by features of lymph nodes including a T-cell zone with DCs and a germinal center with follicular DCs and proliferating B cells, indicating that these lymphoid structures may play an important role in modulating adaptive antitumor immunity.34 49 Previous studies have indicated that a higher frequency of TLS is associated with better prognosis of patients with cancer.1 3 In addition, accumulating evidence suggests that TLS essentially contribute to the therapeutic efficacy of CPI in patients with tumor.50 Thus, it has been demonstrated that higher numbers of tumor-associated TLS are detectable in treatment responders and are correlated with increased survival of patients with cancer receiving CPI therapy.51–53 When focusing on the localization and phenotype of TLS-associated DCs in patient-derived tumor tissues, a high frequency of TLS-associated DCs expressing the maturation marker lysosomal-associated membrane protein has been linked to a strong infiltration of T cells in non-small cell lung cancer.54 The density of mature DC was also associated with expression of genes related to T-cell activation, Th1 phenotype, and cytotoxic orientation. Moreover, a high number of TLS-associated DCs correlated with long-term survival of patients. Here, we observed that pDCs are present in colon cancer-associated TLS and are preferentially located close to CD4+ T cells within the T cell zone. When investigating the potential role of TLS-associated pDCs, we found that a significant proportion pDCs shows a nuclear IRF7 expression, which is characteristic for an activated phenotype. These findings indicate that TLS-associated pDCs may participate in the induction and regulation of CD4+ T cell responses. CD4+ effector T cells can essentially contribute to antitumor immunity by enhancing the capacity of DCs to induce CD8+ T cell responses and by providing help for the expansion of CD8+ T cells directly.55 In addition, they can kill tumor cells and promote the differentiation of B cells into antibody-producing plasma cells.
In summary, we found that pDCs are present in all primary tumor tissues and that a higher density of infiltrating pDCs is associated with lower tumor stages and improved PFS and OS of patients with colon cancer. Moreover, a lower pDC density was significantly and independently linked to worse prognosis, indicating that infiltrating pDCs may represent a novel prognostic factor for patients with colon cancer. In addition, we demonstrated that a proportion of colon cancer-associated pDCs displays a nuclear expression of IRF7, which is characteristic for an activated phenotype. In various tumor stroma regions, IRF7+ pDCs were located in the neighborhood of GrzB+ CD8+ T cells. Furthermore, our studies revealed that a higher number of colon cancer-associated TLS is linked to better prognosis and that pDCs are a novel component of the T cell zone of TLS. A proportion of the TLS-associated pDCs showed a nuclear IRF7 expression and was preferentially located close to CD4+ T cells. The colocalization of pDCs and T cells in tumor stroma and TLS may lead to the induction and maintenance of efficient T cell-mediated antitumor responses and may contribute to the association between high pDC frequencies and better prognosis for patients with colon cancer.
Acknowledgments
We thank Bärbel Löbel and Susanne Doms for excellent technical assistance.
Footnotes
MK and IP contributed equally.
Contributors: Conception and design: MK, IP, GB, MS. Acquisition of data: MK, IP, US, RW, FW, LM, AT, XL, KP. Analysis and interpretation of data: MK, IP, US, AR, AMS, LS, MB, DEA, GB, MS. Writing, review, and/or revision of the manuscript: MK, IP, US, AMS, LS, JW, SZ, MB, MB, DEA, GB, MS. Study supervision: MS.
Funding: This study was supported by grants from the Federal Ministry of Education and Research (01ZX1912C) and the Faculty of Medicine, TU Dresden to R. Wehner.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: This study was approved by the institutional review board of the Faculty of Medicine of the TU Dresden. Patients gave their written informed consent.
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplemental information.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
References
- 1.Fridman WH, Zitvogel L, Sautès-Fridman C, et al. The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 2017;14:717–34. 10.1038/nrclinonc.2017.101 [DOI] [PubMed] [Google Scholar]
- 2.Binnewies M, Roberts EW, Kersten K, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med 2018;24:541–50. 10.1038/s41591-018-0014-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galon J, Bruni D. Tumor immunology and tumor evolution: intertwined histories. Immunity 2020;52:55–81. 10.1016/j.immuni.2019.12.018 [DOI] [PubMed] [Google Scholar]
- 4.Bruni D, Angell HK, Galon J. The immune contexture and immunoscore in cancer prognosis and therapeutic efficacy. Nat Rev Cancer 2020;20:662–80. 10.1038/s41568-020-0285-7 [DOI] [PubMed] [Google Scholar]
- 5.Tumeh PC, Harview CL, Yearley JH, et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014;515:568–71. 10.1038/nature13954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havel JJ, Chowell D, Chan TA. The evolving landscape of biomarkers for checkpoint inhibitor immunotherapy. Nat Rev Cancer 2019;19:133–50. 10.1038/s41568-019-0116-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Michea P, Noël F, Zakine E, et al. Adjustment of dendritic cells to the breast-cancer microenvironment is subset specific. Nat Immunol 2018;19:885–97. 10.1038/s41590-018-0145-8 [DOI] [PubMed] [Google Scholar]
- 8.Böttcher JP, Bonavita E, Chakravarty P, et al. NK cells stimulate recruitment of cDC1 into the tumor microenvironment promoting cancer immune control. Cell 2018;172:1022–37. 10.1016/j.cell.2018.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Swiecki M, Colonna M. The multifaceted biology of plasmacytoid dendritic cells. Nat Rev Immunol 2015;15:471–85. 10.1038/nri3865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wculek SK, Cueto FJ, Mujal AM, et al. Dendritic cells in cancer immunology and immunotherapy. Nat Rev Immunol 2020;20:7–24. 10.1038/s41577-019-0210-z [DOI] [PubMed] [Google Scholar]
- 11.Salio M, Cella M, Vermi W, et al. Plasmacytoid dendritic cells prime IFN-gamma-secreting melanoma-specific CD8 lymphocytes and are found in primary melanoma lesions. Eur J Immunol 2003;33:1052–62. 10.1002/eji.200323676 [DOI] [PubMed] [Google Scholar]
- 12.Hanabuchi S, Watanabe N, Wang Y-H, et al. Human plasmacytoid predendritic cells activate NK cells through glucocorticoid-induced tumor necrosis factor receptor-ligand (GITRL). Blood 2006;107:3617–23. 10.1182/blood-2005-08-3419 [DOI] [PubMed] [Google Scholar]
- 13.Stary G, Bangert C, Tauber M, et al. Tumoricidal activity of TLR7/8-activated inflammatory dendritic cells. J Exp Med 2007;204:1441–51. 10.1084/jem.20070021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drobits B, Holcmann M, Amberg N, et al. Imiquimod clears tumors in mice independent of adaptive immunity by converting pDCs into tumor-killing effector cells. J Clin Invest 2012;122:575–85. 10.1172/JCI61034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Lou Y, Lizée G, et al. Plasmacytoid dendritic cells induce NK cell-dependent, tumor antigen-specific T cell cross-priming and tumor regression in mice. J Clin Invest 2008;118:1165–75. 10.1172/JCI33583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Mercier I, Poujol D, Sanlaville A, et al. Tumor promotion by intratumoral plasmacytoid dendritic cells is reversed by TLR7 ligand treatment. Cancer Res 2013;73:4629–40. 10.1158/0008-5472.CAN-12-3058 [DOI] [PubMed] [Google Scholar]
- 17.Tel J, Aarntzen EHJG, Baba T, et al. Natural human plasmacytoid dendritic cells induce antigen-specific T-cell responses in melanoma patients. Cancer Res 2013;73:1063–75. 10.1158/0008-5472.CAN-12-2583 [DOI] [PubMed] [Google Scholar]
- 18.Sharma MD, Baban B, Chandler P, et al. Plasmacytoid dendritic cells from mouse tumor-draining lymph nodes directly activate mature Tregs via indoleamine 2,3-dioxygenase. J Clin Invest 2007;117:2570–82. 10.1172/JCI31911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hartmann E, Wollenberg B, Rothenfusser S, et al. Identification and functional analysis of tumor-infiltrating plasmacytoid dendritic cells in head and neck cancer. Cancer Res 2003;63:6478–87. [PubMed] [Google Scholar]
- 20.Treilleux I, Blay J-Y, Bendriss-Vermare N, et al. Dendritic cell infiltration and prognosis of early stage breast cancer. Clin Cancer Res 2004;10:7466–74. 10.1158/1078-0432.CCR-04-0684 [DOI] [PubMed] [Google Scholar]
- 21.Labidi-Galy SI, Sisirak V, Meeus P, et al. Quantitative and functional alterations of plasmacytoid dendritic cells contribute to immune tolerance in ovarian cancer. Cancer Res 2011;71:5423–34. 10.1158/0008-5472.CAN-11-0367 [DOI] [PubMed] [Google Scholar]
- 22.Wagner F, Hölig U, Wilczkowski F, et al. Neoadjuvant radiochemotherapy significantly alters the phenotype of plasmacytoid dendritic cells and 6-sulfo LacNAc+ monocytes in rectal cancer. Front Immunol 2019;10:1–16. 10.3389/fimmu.2019.00602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.de Sousa E Melo F, Colak S, Buikhuisen J, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell 2011;9:476–85. 10.1016/j.stem.2011.10.008 [DOI] [PubMed] [Google Scholar]
- 24.Thorsteinsson M, Kirkeby LT, Hansen R, et al. Gene expression profiles in stages II and III colon cancers: application of a 128-gene signature. Int J Colorectal Dis 2012;27:1579–86. 10.1007/s00384-012-1517-4 [DOI] [PubMed] [Google Scholar]
- 25.Tripathi MK, Deane NG, Zhu J, et al. Nuclear factor of activated T-cell activity is associated with metastatic capacity in colon cancer. Cancer Res 2014;74:6947–57. 10.1158/0008-5472.CAN-14-1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marisa L, de Reyniès A, Duval A, et al. Gene expression classification of colon cancer into molecular subtypes: characterization, validation, and prognostic value. PLoS Med 2013;10:e1001453. 10.1371/journal.pmed.1001453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology 2010;138:958–68. 10.1053/j.gastro.2009.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dzionek A, Sohma Y, Nagafune J, et al. BDCA-2, a novel plasmacytoid dendritic cell-specific type II C-type lectin, mediates antigen capture and is a potent inhibitor of interferon alpha/beta induction. J Exp Med 2001;194:1823–34. 10.1084/jem.194.12.1823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guidoboni M, Gafà R, Viel A, et al. Microsatellite instability and high content of activated cytotoxic lymphocytes identify colon cancer patients with a favorable prognosis. Am J Pathol 2001;159:297–304. 10.1016/S0002-9440(10)61695-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feng Z, Bethmann D, Kappler M, et al. Multiparametric immune profiling in HPV– oral squamous cell cancer. JCI Insight 2017;2:1–17. 10.1172/jci.insight.93652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Angelova M, Mlecnik B, Vasaturo A, et al. Evolution of metastases in space and time under immune selection. Cell 2018;175:751–65. 10.1016/j.cell.2018.09.018 [DOI] [PubMed] [Google Scholar]
- 32.Honda K, Yanai H, Negishi H, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature 2005;434:772–7. 10.1038/nature03464 [DOI] [PubMed] [Google Scholar]
- 33.Goc J, Fridman W-H, Sautès-Fridman C, et al. Characteristics of tertiary lymphoid structures in primary cancers. Oncoimmunology 2013;2:e26836. 10.4161/onci.26836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sautès-Fridman C, Petitprez F, Calderaro J, et al. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer 2019;19:307–25. 10.1038/s41568-019-0144-6 [DOI] [PubMed] [Google Scholar]
- 35.Galon J, Costes A, Sanchez-Cabo F, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 2006;313:1960–4. 10.1126/science.1129139 [DOI] [PubMed] [Google Scholar]
- 36.Pagès F, Kirilovsky A, Mlecnik B, et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J Clin Oncol 2009;27:5944–51. 10.1200/JCO.2008.19.6147 [DOI] [PubMed] [Google Scholar]
- 37.Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus immunoscore for the classification of colon cancer: a prognostic and accuracy study. Lancet 2018;391:2128–39. 10.1016/S0140-6736(18)30789-X [DOI] [PubMed] [Google Scholar]
- 38.Li J, Li L, Li Y, et al. Tumor-Associated macrophage infiltration and prognosis in colorectal cancer: systematic review and meta-analysis. Int J Colorectal Dis 2020;35:1203–10. 10.1007/s00384-020-03593-z [DOI] [PubMed] [Google Scholar]
- 39.Ambe K, Mori M, Enjoji M. S-100 protein-positive dendritic cells in colorectal adenocarcinomas. Distribution and relation to the clinical prognosis. Cancer 1989;63:496–503. [DOI] [PubMed] [Google Scholar]
- 40.Dadabayev AR, Sandel MH, Menon AG, et al. Dendritic cells in colorectal cancer correlate with other tumor-infiltrating immune cells. Cancer Immunol Immunother 2004;53:978–86. 10.1007/s00262-004-0548-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sandel MH, Dadabayev AR, Menon AG, et al. Prognostic value of tumor-infiltrating dendritic cells in colorectal cancer: role of maturation status and intratumoral localization. Clin Cancer Res 2005;11:2576–82. 10.1158/1078-0432.CCR-04-1448 [DOI] [PubMed] [Google Scholar]
- 42.Nagorsen D, Voigt S, Berg E, et al. Tumor-infiltrating macrophages and dendritic cells in human colorectal cancer: relation to local regulatory T cells, systemic T-cell response against tumor-associated antigens and survival. J Transl Med 2007;5:1–8. 10.1186/1479-5876-5-62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gulubova MV, Ananiev JR, Vlaykova TI, et al. Role of dendritic cells in progression and clinical outcome of colon cancer. Int J Colorectal Dis 2012;27:159–69. 10.1007/s00384-011-1334-1 [DOI] [PubMed] [Google Scholar]
- 44.Suzuki A, Masuda A, Nagata H, et al. Mature dendritic cells make clusters with T cells in the invasive margin of colorectal carcinoma. J Pathol 2002;196:37–43. 10.1002/path.1018 [DOI] [PubMed] [Google Scholar]
- 45.Kini Bailur J, Gueckel B, Pawelec G. Prognostic impact of high levels of circulating plasmacytoid dendritic cells in breast cancer. J Transl Med 2016;14:1–10. 10.1186/s12967-016-0905-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou Z-J, Xin H-Y, Li J, et al. Intratumoral plasmacytoid dendritic cells as a poor prognostic factor for hepatocellular carcinoma following curative resection. Cancer Immunol Immunother 2019;68:1223–33. 10.1007/s00262-019-02355-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sisirak V, Faget J, Gobert M, et al. Impaired IFN-α production by plasmacytoid dendritic cells favors regulatory T-cell expansion that may contribute to breast cancer progression. Cancer Res 2012;72:5188–97. 10.1158/0008-5472.CAN-11-3468 [DOI] [PubMed] [Google Scholar]
- 48.Sisirak V, Vey N, Goutagny N, et al. Breast cancer-derived transforming growth factor-β and tumor necrosis factor-α compromise interferon-α production by tumor-associated plasmacytoid dendritic cells. Int J Cancer 2013;133:771–8. 10.1002/ijc.28072 [DOI] [PubMed] [Google Scholar]
- 49.Dieu-Nosjean M-C, Giraldo NA, Kaplon H, et al. Tertiary lymphoid structures, drivers of the anti-tumor responses in human cancers. Immunol Rev 2016;271:260–75. 10.1111/imr.12405 [DOI] [PubMed] [Google Scholar]
- 50.Plesca I, Tunger A, Müller L, et al. Characteristics of tumor-infiltrating lymphocytes prior to and during immune checkpoint inhibitor therapy. Front Immunol 2020;11:1–8. 10.3389/fimmu.2020.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petitprez F, de Reyniès A, Keung EZ, et al. B cells are associated with survival and immunotherapy response in sarcoma. Nature 2020;577:556–60. 10.1038/s41586-019-1906-8 [DOI] [PubMed] [Google Scholar]
- 52.Helmink BA, Reddy SM, Gao J, et al. B cells and tertiary lymphoid structures promote immunotherapy response. Nature 2020;577:549–55. 10.1038/s41586-019-1922-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cabrita R, Lauss M, Sanna A, et al. Tertiary lymphoid structures improve immunotherapy and survival in melanoma. Nature 2020;577:561–5. 10.1038/s41586-019-1914-8 [DOI] [PubMed] [Google Scholar]
- 54.Goc J, Germain C, Vo-Bourgais TKD, et al. Dendritic cells in tumor-associated tertiary lymphoid structures signal a Th1 cytotoxic immune contexture and license the positive prognostic value of infiltrating CD8+ T cells. Cancer Res 2014;74:705–15. 10.1158/0008-5472.CAN-13-1342 [DOI] [PubMed] [Google Scholar]
- 55.Borst J, Ahrends T, Bąbała N, et al. CD4+ T cell help in cancer immunology and immunotherapy. Nat Rev Immunol 2018;18:635–47. 10.1038/s41577-018-0044-0 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jitc-2020-001813supp001.pdf (21.5KB, pdf)
jitc-2020-001813supp002.pdf (84.3KB, pdf)
jitc-2020-001813supp003.pdf (89.5KB, pdf)
jitc-2020-001813supp004.pdf (28KB, pdf)
jitc-2020-001813supp005.pdf (236.8KB, pdf)
jitc-2020-001813supp006.pdf (161.5KB, pdf)
jitc-2020-001813supp007.pdf (2.9MB, pdf)