Abstract
Antibiotics resistance is becoming increasingly common, involving almost all antibiotics on the market. Diseases caused by drug resistant bacteria, such as MRSA, have high mortality and negatively affect public health. The development of new drugs would be an effective means of solving this problem. Modifications based on bioactive natural products could greatly shorten drug development time and improve success rate. Pleuromutilin, a natural product from the basidiomycete bacterial species, is a promising antibiotic candidate. In this study, a series of novel pleuromutilin derivatives possessing piperazinyl urea linkage were efficiently synthesised, and their antibacterial activities and bactericidal properties were evaluated via MIC, MBC and Time-kill kinetics assays. The results showed that all compounds exhibited potent activities against tested strains, especially MRSA strains with MIC values as low as 0.125 μg/mL; 8 times lower than that of marketed antibiotic Tiamulin. Docking studies indicate substituted piperazinyl urea derivatives could provide hydrogen bonds and initiate π-π stacking between molecules and surrounding residues.
Keywords: Pleuromutilin, piperazinyl urea, synthesis, biological activities, MRSA
Introduction
Current available antibiotics are losing their effectiveness due to increasing bacterial resistance. Diseases caused by multi-drug resistant bacteria are particularly difficult to prevent and treat, and across the world, approximately 700,000 people die annually as a result of them1. Among drug-resistant strains, methicillin-resistant Staphylococcus aureus (MRSA) is one of the most problematic, with pathogens that spread rapidly and cause severe diseases such as septicaemia, pneumonia, osteomyelitis and endocarditis2,3. Average treatment time and MRSA patient mortality are, respectively, five and three times higher than those of patients infected by common pathogens. Antibiotics are generally used to control MRSA infections, however the majority of common antibiotics used in clinical treatment were ineffective against MRSA. Therefore, it is crucial to develop and discover novel antibacterial agents4.
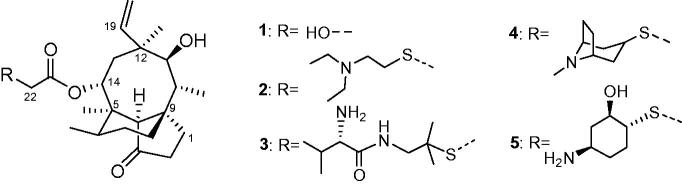
Natural products such as Penicillin, Erythromycin, Cephalosporin C and Kanamycin are often used as main compounds in drug discoveries due to their potent bioactivity. Corresponding semi-synthesised derivatives Amoxicillin, Azithromycin, Cefpirome and Amikacin all exhibit higher potency than natural products5–7. Pleuromutilin (Figure 1, 1), a diterpenoid natural product from the basidiomycete species, shows moderate activities against Gram-positive strains and mycoplasmas8,9. Four semi-synthesised marketed drugs based on its structure have already been developed. Tiamulin and valnemulin (Figure 1, 2 and 3) can effectively prevent and control swine dysentery, mycoplasmal diseases such as enzootic pneumonia and chronic respiratory disease in poultry10,11. Retapamulin (Figure 1, 4) is used for the treatment of skin impetigo12,13. Lefamulin (Figure 1, 5) is approved for treatment of community-acquired bacterial pneumonia (CABP)14. These derivatives can inhibit bacterial protein synthesis via specific interaction with 23S rRNA of the 50S bacterial ribosome unit15, which are unaffected by resistance to major antibiotic classes, such as beta-lactam antibiotics, tetracyclines, macrolides, fluoroquinolones, and others. Thus, pleuromutilin is a promising candidate for treating drug-resistant bacteria infections16, and many compounds derived from four marketed drugs using the thioether as the linkage at the C14 side chain were developed in recent years17–20. To increase side chain diversity and improve modification success rate, the bioactive moiety piperazinyl urea, displaying multiple biological activities such as antifungal21, analgesic22, antibacterial23 and antitumour24 effects was introduced. In this study, a series of pleuromutilin derivatives 6a∼z with piperazinyl urea (Scheme 1) were efficiently synthesised. Their activities were evaluated against MRSA and Gram-negative strains, and interactions were examined by molecular docking.
Figure 1.
Structures of pleuromutilin derivatives 2 ∼ 5 developed from natural Pleuromutilin 1.
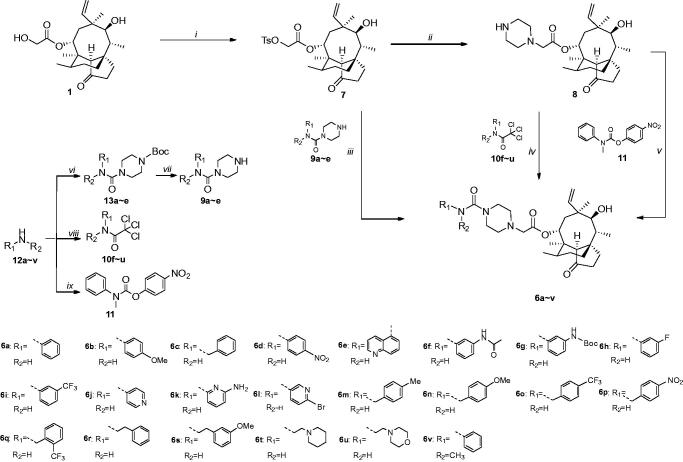
Scheme 1.
Synthesis route of target compounds 6a ∼ v. Reagents and conditions: (i) TsCl, triethylamine, DCM, 25 °C; (ii) Piperazine, K2CO3, NaI, THF, reflux; (iii) 9a ∼ e, NaI, K2CO3, MeCN, 70 °C; (iv) 10f ∼ u, K2CO3, DMF, 80 °C; (v) 11, DMAP, MeCN, 70 °C; (vi) CDI, triethylamine, N-Boc piperazine, MeCN and DMF, 25 °C for 13a and 13b, or BTC, triethylamine, N-Boc piperazine, THF and DCM, 25 °C for 13c ∼ e, (vii) TFA, DCM, 25 °C; (viii) Trichloroacetyl chloride, triethylamine, DCM, 25 °C; (ix) 4-Nitrophenyl carbonochloridate, NMM, DCM, 25 °C.
Results and discussion
Chemistry
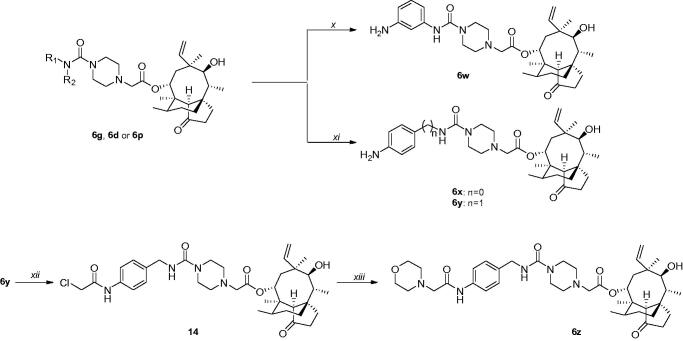
The piperazinyl urea linkage can be constructed with substituted amines via three strategies (Scheme 1). Pleuromutilin 1 was reacted with tosyl chloride and triethylamine in DCM to form pleuromutilin-22-O-tosylate 7 in 78.3% yield, which could convert to 22-(piperazine-1-yl)-22-deoxypleuromutilin 8 by treatment with NaI, piperazine and potassium carbonate in dry THF with a 75.2% yield. Compounds 6a ∼ e were synthesised by treating 7 with N-substituted-1-carboxamide 9a ∼ e in the presence of a catalytic amount of NaI and K2CO3 in MeCN with 78.3 ∼ 92.3% yield. Here, the primary amines 12a∼e reacted with bis(trichloromethyl)carbonate (BTC) or N, N'-carbonyldiimidazole (CDI) to form the corresponding isocyanate, which was used to form piperazinyl urea moiety by reacting with N-Boc-piperazine in the presence of triethylamine with 56.2 ∼ 82.3% yield. Afterwards, 9a ∼ e were obtained via removal of the above Boc protected group using trifluoroacetic acid (TFA) in 93.7 ∼ 97.2% yield. Compounds 6f ∼ u were synthesised in 45.6 ∼ 82.7% yield by treating 8 in the presence of potassium carbonate with substituted 2,2,2-trichloro-acetamide 10f∼u, which were acquired by reacting primary amines 12f∼u with trichloroacetyl chloride in DCM with 54.7 ∼ 97.2% yield. Moreover, in regard to the secondary amine, the expected product was rarely obtained using the above two strategies. N-methylaniline 12v was treated with 4-nitrophenyl carbonochloridate in the presence of N-methylmorpholine (NMM) in DCM to give 11, which was condensed without further purification and reacted with 22-(piperazine-1-yl)-22-deoxypleuromutilin 8 to form N-methyl derivative 6v in 40.5% yield. Additionally, 6w∼z were synthesised from corresponding Boc-protected amine and nitro molecules (Scheme 2). 6w was gained from deprotecting 6g in the presence of TFA in 95.6% yield, and two other amine derivatives 6x and 6y were attained by stannous chloride reduction of 6d and 6p in 92.5% and 75.5% yields, respectively. 6y was used to react with chloracetyl chloride to get 14 in the presence of triethylamine with 70.1% yield, which was treated with morpholine to produce 6z in 50.3% yield.
Scheme 2.
Synthesis route of target compounds 6w ∼ z. Reagents and conditions: (x) TFA, DCM, 25 °C from 6g; (xi) SnCl2, EtOH, reflux from 6d or 6p; (xii) Chloroacetyl chloride, triethylamine, THF, 25 °C; (xiii) Morpholine, K2CO3, THF, reflux.
Biological activities
MIC and MBC values of the synthesised compounds against three Gram-positive bacteria (i.e.: Staphylococcus aureus (S. aureus) ATCC 25923, MRSA ATCC 33591 and ATCC 43300) and one Gram-negative bacterium (i.e.: Escherichia coli (E. coli) ATCC 25922) were determined using the broth microdilution method with the marketed antibiotic Tiamulin as reference. The lipophilicities (ClogP) were predicted by ACD/Labs (https://www.acdlabs.com/resources/ilab/).
As Table 1 indicates, almost all the synthesised compounds exhibited broad-spectrum antibacterial activities and showed high potency against both Gram-positive and Gram-negative bacteria with MIC values of 0.5 ∼ 4 μg/mL. Moreover, the majority of synthesised compounds displayed bactericidal activities against MRSA ATCC 33591, as MBC/MIC values were less than 4. Among them, the antibacterial activities of 6d, 6o, 6p and 6q were at least 2 times stronger than those of Tiamulin. In particular, compound 6p had a 0.125 μg/mL MIC value (8 times lower than that of Tiamulin) and displayed the best performance against MRSA ATCC 33591. 6p could also be considered as a bactericide, since MBC/MIC values against four tested strains were less than 2. Furthermore, its activity against E. coli ATCC 25922 was 16 times stronger than those of Tiamulin.
Table 1.
MIC and MBC values of pleuromutilin derivatives possessing piperazinyl urea linkage.
| Cpd | MRSA ATCC33591 |
MRSA ATCC43300 |
S. aureus ATCC25923 |
E.coli ATCC25922 |
CLogP | ||||
|---|---|---|---|---|---|---|---|---|---|
| MIC | MBC | MIC | MBC | MIC | MBC | MIC | MBC | ||
| 6a | 1 | 2 | 1 | >4 | 1 | 2 | 1 | >4 | 4.94 |
| 6b | 1 | 2 | 0.5 | 2 | 1 | 2 | 1 | >4 | 4.78 |
| 6c | 0.5 | 1 | 0.5 | 2 | 1 | 4 | 0.5 | 2 | 5.28 |
| 6d | 0.5 | 1 | 0.5 | 2 | 0.5 | 2 | 0.5 | 2 | 5.47 |
| 6e | 1 | 2 | 1 | >4 | 1 | 2 | 1 | 4 | 4.56 |
| 6f | 2 | 4 | 2 | 4 | 2 | 4 | 4 | >16 | 4.35 |
| 6g | 1 | 4 | 1 | 4 | 1 | 2 | 1 | >4 | 6.26 |
| 6h | 1 | 2 | 0.5 | 1 | 1 | 1 | 1 | >4 | 5.33 |
| 6i | 1 | 4 | 1 | >4 | 1 | 2 | 4 | >16 | 6.32 |
| 6j | 4 | 8 | 4 | 16 | 4 | 16 | 32 | 32 | 4.42 |
| 6k | 1 | 2 | 0.5 | 2 | 1 | 4 | 1 | >4 | 3.68 |
| 6l | 1 | 2 | 1 | >4 | 1 | 4 | 1 | 4 | 5.22 |
| 6m | 2 | 4 | 1 | 4 | 0.5 | 2 | 1 | >4 | 5.74 |
| 6n | 0.5 | 2 | 1 | 2 | 0.5 | 2 | 0.5 | 2 | 5.19 |
| 6o | 0.5 | 2 | 0.5 | >2 | 0.5 | 0.5 | 0.5 | 2 | 5.85 |
| 6p | 0.125 | 0.125 | 0.25 | 0.25 | 0.125 | 0.25 | 0.125 | 0.125 | 5.01 |
| 6q | 0.5 | 2 | 0.25 | 0.25 | 0.125 | 0.5 | 1 | >4 | 5.85 |
| 6r | 1 | 4 | 2 | 4 | 1 | 4 | 2 | 8 | 6.02 |
| 6s | 1 | 1 | 1 | 2 | 1 | 4 | 1 | 4 | 5.93 |
| 6t | 2 | 2 | 2 | 8 | 2 | 4 | 2 | 4 | 5.24 |
| 6u | 4 | 8 | 4 | 4 | 4 | 4 | 4 | 8 | 3.59 |
| 6v | 2 | 8 | 4 | 16 | 4 | 16 | 4 | 16 | 4.94 |
| 6w | 2 | 4 | 1 | 2 | 2 | 8 | 2 | >8 | 3.85 |
| 6x | 1 | 4 | 1 | 4 | 1 | >4 | 1 | 4 | 3.25 |
| 6y | 1 | 1 | 1 | 2 | 1 | >4 | 1 | 4 | 4.00 |
| 6z | 1 | 1 | 1 | 4 | 1 | 4 | 1 | 4 | 4.30 |
| Tiamulin (2) | 1 | 4 | 1 | >4 | 1 | >4 | 2 | >8 | 5.93 |
The effects of terminal ring types (e.g. phenyl, pyridinyl, morpholinyl and piperidinyl), ring substituents and distance between rings and urea groups on changes in activities were investigated. MIC value of phenyl piperazinyl urea pleuromutilin 6a was 1 μg/mL against four strains and when substituents were introduced to the terminal benzene, activities varied within a small range. Compound 6d with electron-withdrawing 4-nitro groups was the most potent among all phenyl derivatives and exhibited an MIC value of 0.5 μg/mL. However, the introduction of N-methyl groups to the urea led to decreased activities, since actions of compound 6v was 1/4 or 1/2 of compound 6a. To render the side chain more flexible, a CH2 group was introduced to increase the distance between terminal rings and urea groups. As result, the benzyl derivatives with an additional methylene group could noticeably improve activity level. In addition, similar as the effect of substituents on phenyl piperazinyl urea pleuromutilin activity, compound 6p with the 4-nitro benzyl group exhibited the best performance among all the benzyl derivatives. However, continuously extending the space between the benzene and piperazinyl urea exerts a slight influence on activities. Activity declined when the phenyl group was replaced by 4-pyridnyl group, but phenyl group replacement by substituting 2-pyridinyl, 3-pyridinyl or 5-quinolinyl groups had nearly zero impact on activities. Activity level visibly decreased when aliphatic rings such as piperidine and morpholine were added. The majority of target molecules with an aromatic terminal ring were fairly lipophilic with a CLogP value of greater than 5. To enhance hydrophilic properties, compounds 6w∼y with amino groups were produced from corresponding Boc-protected amines and nitro compounds. Their ClogP values were approximately equal to or less than 4. Furthermore, a water-soluble morpholine ring was attached to the terminal of 6y through N-acetyl group. Although hydrophilic properties were clearly optimised via the above modification, activity levels were only slightly changed.
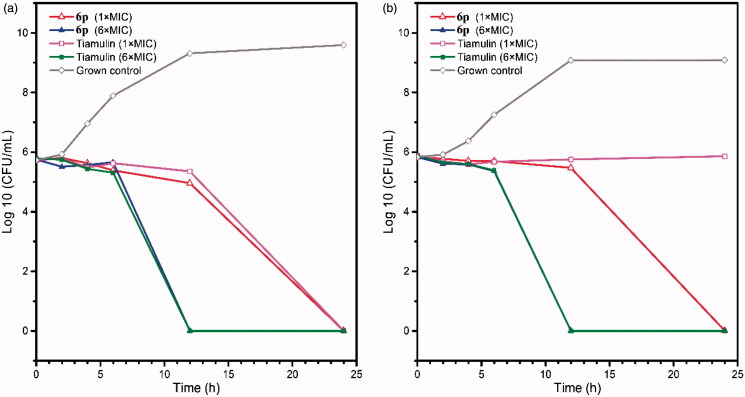
Since compound 6p showed the most potent antibacterial activities against MRSA ATCC 33591 and E. coli ATCC 25922, its bactericidal properties were examined by in vitro time-kill assay. Bactericidal properties of 6p against two strains were evaluated at concentrations of 1 × MIC and 6 × MIC, and two concentrations of Tiamulin were set as controls (Figure 2). The MRSA ATCC 33591 propagation-inhibition effect of 6p was visibly higher at the 1 × MIC concentration than that of the Grown control, which shows similarity to inhibition effects of Tiamulin at 1 × MIC. Meanwhile, at 6 × MIC, compound 6p achieved complete bactericidal effects in 12 h, a much faster rate than that under 1 × MIC (24 h). In addition, compound 6p displayed better bactericidal effects against E. coli ATCC 25922 compared with Tiamulin at 1 × MIC, and was capable of eliminating total bacteria colony in 24 h, while Tiamulin could only limit bacterial propagation speed. Outcomes were nearly identical at the 6 × MIC concentration.
Figure 2.
Time-kill kinetics of compound 6p against MRSA ATCC 33591(a) and E. coli ATCC 25922 (b).
Cytotoxicity evaluation
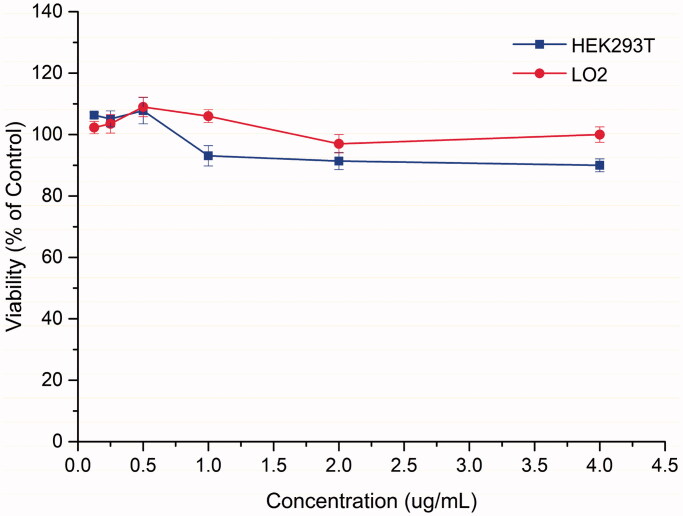
The cytotoxicity of the most potent compound 6p was investigated in normal human hepatic cell line LO2 and human embryo kidney cell line HEK293T using a cell counting kit-8 (CCK-8) assay25,26 (Figure 3). The CCK-8 assay is a colorimetric assay based on the reduction of dye WST-8 [2–(2-methoxy-4-nitrophenyl)-3–(4-nitrophenyl)-5–(2,4disulfophenyl)-2H-tetrazolium, monosodium salt] to a water-soluble orange-coloured formazan via dehydrogenase in viable cells. The amount of formazan dye produced by cellular dehydrogenase is correlated with the number of living cells. The result showed that 6p hardly affected cell viability at concentrations ranging from 0.125 μg/mL to 4 μg/mL in either of the two cell lines. More than 90% of cells remained alive at the 4 μg/mL concentration, indicating 6p exhibited almost no cytotoxicity even at the concentration of 32 × MIC.
Figure 3.
Effects of compound 6p on cell viability by CCK-8 assay.
Docking studies
Docking investigation was conducted to examine the binding mode between synthesised compounds and the peptidyl transferase centre (PTC) of the 50S ribosomal subunit. The method was confirmed by re-docking, revealing that the docking pose of the original ligand Tiamulin nearly coincided with the X-ray structure conformation of RMSD 0.9 Å. All of the title compounds were used for the docking evaluation (Figs. S36–39). Among them, phenyl derivative 6d and benzyl derivative 6p with the most potent antibacterial activity, and aliphatic rings derivative 6z with moderate activity were chosen for the binding mode study. The favourably docked molecules were ranked according to their Grid_Score, which consisted of two parameters: Grid_vdw_energy (mainly referring to hydrophobic interactions and π–π stacking) and Grid_es_energy (often containing hydrogen bonds and salt bridges). The results presented in Table 2 showed that the order of Grid_Scores went in decreasing order (6p > 6d > 6z>Tiamulin), which matched those found in the biological assay.
Table 2.
Molecular docking analysis of PTC with selected ligands (kcal/mol).
| Cpd | Grid_Score | Grid_vdw_energy | Grid_es_energy |
|---|---|---|---|
| 6d | −107.27 | −103.75 | −3.52 |
| 6p | −112.91 | −111.64 | −1.27 |
| 6z | −105.50 | −98.69 | −6.81 |
| Tiamulin | −95.34 | −91.14 | −4.20 |
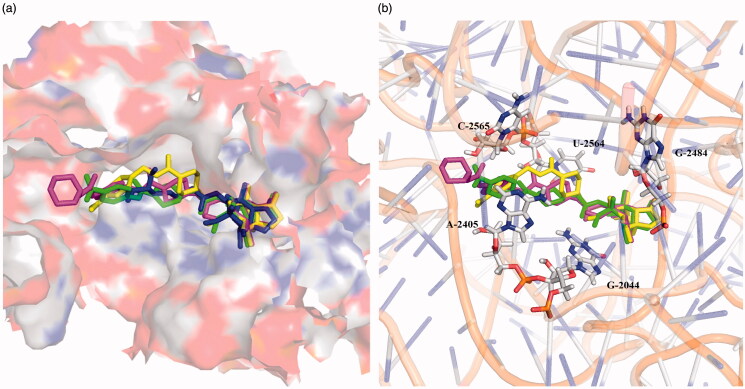
As shown by the superimposed poses in Figures 4(a,b), three synthesised ligands 6d (green), 6p (yellow) and 6z (magenta) were located at the same pocket of PTC as Tiamulin (blue). The tricyclic skeleton of the synthesised compounds was in good agreement with that of Tiamulin, while the side chain at C14 varied noticeably. Many active interactions occurred continuously between the above-mentioned ligands and residues C2565, U2564, G2484, G2044 and A2405.
Figure 4.
Superimposed poses of selected ligands in the PTC of the 50 s ribosomal subunit. (a) The interaction groove; (b) Residues involved in the interactions between ligands and PTC.
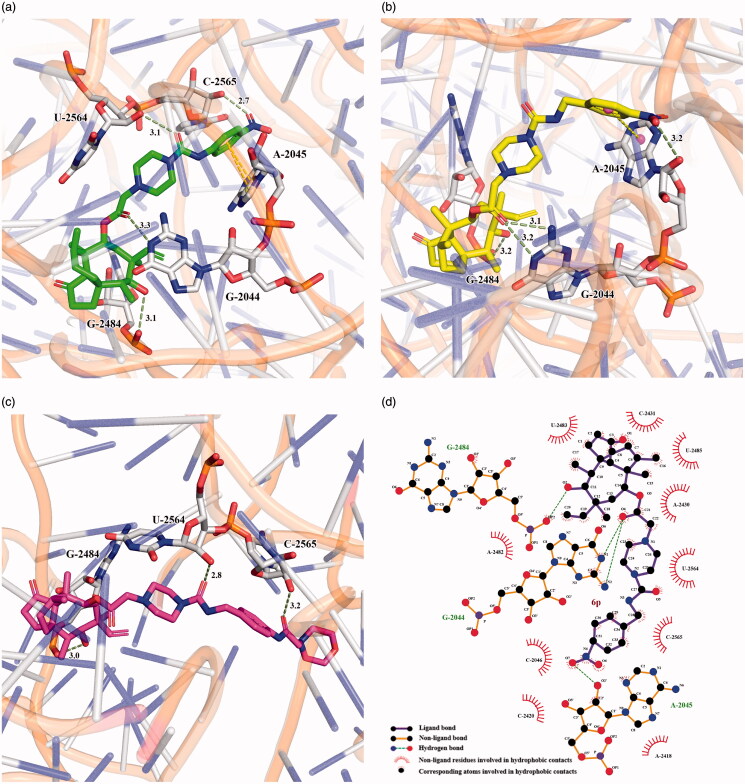
The important intermolecular PTC-ligand interactions are depicted in Figure 5. In addition to hydrogen bonds between the hydroxyl group on C11 and residue G2484, and between the carbonyl group on C21 and residue G2044, 6d (Figure 5(a)) and 6p (Figure 5(b)) were able to interact with A2045 by π-π stacking; the former adopting face-to-face stacking, and the latter displaying nearly edge-to-face stacking. Moreover, the terminal nitro group was able to form one additional hydrogen bond by interacting with C2565 or A2045. 6z (Figure 5(c)) was able to interact with residues G2484, U2564 and C2565 by three hydrogen bonds, however the π-π stacking was missing. This might explain why the Grid_vdw_energy of 6z (-98.69 kcal/mol) was worse than 6d (-103.75 kcal/mol) and 6p (-111.64 kcal/mol). The hydrophobic environment of the most potent compound 6p is shown in Figure 5(d). Residues G2484, U2483, C2431, U2485, A2430, U2564, C2565, A2045, A2418, C2420, C2046, G2044 and A2482 were involved in the hydrophobic interactions, which provided strong van der Waals forces in conjunction with the above mentioned π–π stacking of 6p.
Figure 5.
Interactions between ligands and residues of the 50 s ribosomal subunit. (a) 6d and site; (b) 6p and site; (c) 6z and site; (d) Hydrogen bonds and hydrophobic interactions.
Experimental
Synthesis
Analytical grade reagents were used and purchased from Energy Chemical (Shanghai, China) and Kelong Chemical (Chengdu, China). Melting points were determined on an Shenguang WRS-1B apparatus (Shanghai, China). 1H NMR and 13 C NMR spectra were measured on a Bruker AV400 spectrometer in CDCl3 or DMSO-d6. Mass spectra were recorded with a X500R mass spectrometer (AB SCIEX) using the electro spray ionisation (ESI) method.
Synthesis of compound 7 and 8
Tosyl chloride (1.91 g, 10.0 mmol) in DCM (10 ml) was added to a solution of pleuromutilin (3.56 g, 9.40 mmol) in DCM (10 ml) containing triethylamine (4.54 g, 12.0 mmol) at 0 °C, then the reaction was stirred at 25 °C for 20 h and the resultant was washed with water (50 ml). The organic phase was collected, dried over Na2SO4 and evaporated under vacuum to produce yellow oil, which could be purified by recrystallisation in ethanol to obtain white solid 7 (Yield: 78.3%). Spectral data of 7 were identical to those from reports in the literature27.
7 (533 mg, 1.0 mmol) and NaI (30.0 mg, 0.2 mmol) were stirred in dry THF (5 ml) at 25 °C for 0.5 h, then K2CO3 (276 mg, 2.0 mmol) and piperazine (172 mg, 2.0 mmol) were added to the aforementioned solution. The mixture was heated to reflux and stirred for 6 h. After cooling to room temperature, the resulting mixture was washed with water and condensed until dry. Compound 8 was collected after silica gel column chromatography (Yield: 75.2%). Spectral data results of 8 were identical to those from reports in the literature28.
Synthesis of compounds 9a ∼ e
Compounds 9a ∼ d were synthesised according to previous literature29,30, and 9e was synthesised as follows:
BTC (386 mg, 1.3 mmol) in dry THF (4 ml) was added dropwise to a mixture of triethylamine (526 mg, 5.2 mmol) and 12e (173 mg, 1.2 mmol) in dry THF (6 ml) solution under an ice-water bath. The mixture was then stirred for 0.5 h. After condensation, N-Boc piperazine (503 mg, 2.7 mmol), triethylamine (546 mg, 5.4 mmol) and DCM (10 ml) were added and stirred for 2 h at 25 °C. The solvent was then evaporated before crude product purification via silica gel column chromatography to produce compound 13e.
Compound 13e (356 mg, 1.0 mmol) was added to the mixture of 6 ml DCM and TFA (v/v = 10/1) and stirred at 25 °C until 13e completely converted. The resulting solution was washed with saturated aqueous NaHCO3 to neutralise TFA, then f the organic phase was collected and dried over anhydrous Na2SO4. After filtering, the filtrate was concentrated to acquire the crude product, which was purified via silica gel chromatography to produce compound 9e.
13e: white powder; yield: 56.2%; mp: 222.9–223.8 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.86 (dd, J = 4.0, 1.4 Hz, 1H), 8.18 − 8.05 (m, 1H), 7.92 (d, J = 8.4 Hz, 1H), 7.65 − 7.52 (m, 1H), 7.43 (d, J = 7.2 Hz, 1H), 7.33 (dd, J = 8.4, 4.0 Hz, 1H), 6.89 (s, 1H), 3.45 (s, 8H), 1.48 (s, 9H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 156.2, 154.7, 150.4, 148.8, 134.2, 131.0, 129.2, 127.3, 124.4, 122.4, 121.0, 80.5, 28.5; HRMS: calculated for C19H24N4O3 ([M + H]+): 357.1921, found 357.1929.
9e: yellow liquid; yield: 96.3%; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 9.08 (s, 1H), 8.87 (d, J = 4.0 Hz, 1H), 8.35 (d, J = 8.4 Hz, 1H), 7.84 (d, J = 8.4 Hz, 1H), 7.70 (t, J = 8.0 Hz, 1H), 7.54 − 7.44 (m, 2H), 3.80 − 3.72 (m, 4H), 3.24 − 3.17 (m, 4H); 13 C NMR (100 MHz, DMSO-d6): δ (ppm) 156.3, 150.7, 148.6, 136.1, 132.9, 129.4, 126.3, 124.8, 123.1, 121.2, 43.1, 41.5; HRMS: calculated for C14H16N4O ([M + H]+): 257.1391, found 257.1402.
Synthesis of compounds 10f ∼ u
Compounds 10f, 10h, 10i, 10j, 10l, 10m, 10o, 10r, 10t and 10u were synthesised as in literature31,32. The remaining compounds were synthesised as follows:
A mixture of trichloroacetyl chloride (218 mg, 1.2 mmol) in an appropriate amount of DCM was added to a DCM (3 ml) solution containing compounds 12g (208 mg, 1.0 mmol) and triethylamine (121 mg, 1.2 mmol) under an ice-water bath. Next, the mixture was stirred at 25 °C until 12g was completely dispersed. The reaction mixture was evaporated by vacuum and residue was dissolved in DCM. After washing using water and saturated aqueous ammonium chloride, the organic layer was separated and dried over anhydrous Na2SO4. The crude compound was attained after evaporating the solvent, which was recrystallised by ethanol/water to give 10g. Compounds 10k, 10n, 10p, 10q and 10s were synthesised in the same manner.
10g: white powder; yield: 97.2%; mp: 124.9–126.3 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.33 (s, 1H), 7.78 (t, J = 2.0 Hz, 1H), 7.42 − 7.35 (m, 1H), 7.29 (t, J = 8.0 Hz, 1H), 7.08 − 7.00 (m, 1H), 6.62 (s, 1H), 1.52 (s, 9H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 159.2, 152.7, 139.3, 136.7, 129.8, 115.7, 114.7, 110.2, 92.8, 81.0, 28.3; HRMS: calculated for C13H15Cl3N2NaO3 ([M + Na]+): 375.0040, found 375.0046.
10k: white powder; yield: 54.1%; mp: 148.1–149.3 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.57 (s, 1H), 7.56 − 7.45 (m, 2H), 6.35 (d, J = 7.6 Hz, 1H), 4.41 (s, 2H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 159.2, 157.5, 148.3, 140.5, 106.1, 103.3, 92.8; HRMS: calculated for C7H6Cl3N3O ([M + H]+): 253.9649, found 253.9652.
10n: white powder; yield: 88.1%; mp: 84.9–85.6 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.26 − 7.22 (m, 2H), 6.92 − 6.88 (m, 2H), 4.48 (d, J = 5.6 Hz, 2H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 161.8, 159.6, 129.3, 128.3, 114.4, 92.6, 55.4, 45.0; HRMS: calculated for C10H10Cl3NNaO2 ([M + Na]+): 303.9669, found 303.9685.
10p: light yellow powder; yield: 85.2%; mp: 87.2–88.4 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.24 (d, J = 8.8 Hz, 2H), 7.49 (d, J = 8.8 Hz, 2H), 4.67 (d, J = 6.0 Hz, 2H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 162.4, 147.7, 143.7, 128.3, 124.2, 92.2, 44.5; HRMS: calculated for C9H7Cl3N2O3 ([M + H]+): 296.9595, found 296.9601.
10q: white powder; yield: 87.5%; mp: 77.3–78.7 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.70 (d, J = 7.6 Hz, 1H), 7.59 − 7.57 (m, 2H), 7.48 − 7.44 (m, 1H), 4.73 (dd, J = 6.0, 0.8 Hz, 2H), 1.25 (s, 1H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 162.0, 134.5, 132.7, 130.8, 128.4, 128.3 (q, J = 31 Hz), 126.4 (q, J = 6 Hz), 124.3 (q, J = 272 Hz), 92.4, 42.1; HRMS: calculated for C10H7Cl3F3NO ([M + H]+): 319.9618, found 319.9624.
10s: white powder; yield: 86.8%; mp: 48.1–49.5 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.26 − 7.25 (m, 1H), 6.84 − 6.83 (m, 2H), 6.78 (t, J = 2.0 Hz, 1H), 3.83 (s, 3H), 3.65 (q, J = 6.8 Hz, 2H), 2.92 (d, J = 6.8 Hz, 2H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 161.8, 160.0, 139.4, 129.9, 121.1, 114.4, 112.4, 92.6, 55.2, 42.5, 35.1; HRMS: calculated for C11H12Cl3NO2 ([M + H]+): 296.0006, found 296.0022.
Synthesis of compound 1133
4-Nitrophenyl chloroformate (97 mg, 0.48 mmol) and NMM (143 mg, 1.41 mmol) were added to a DCM (5 ml) solution containing N-methylaniline 12v (51 mg, 0.47 mmol) at 0 °C. The subsequent mixture was stirred for 2 h before pouring into water (5 ml). The organic layer was separated and dried over anhydrous Na2SO4 After solvent evaporation, the residue was directly used for the next step (Yield: 97.5%).
Synthesis of compounds 6a ∼ e
Pleuromutilin 22-tosylate 7 (213 mg, 0.4 mmol) and NaI (14.9 mg, 0.1 mmol) in MeCN (5 ml) were stirred at 25 °C for 0.5 h, then compounds 9a∼e (0.5 mmol) and K2CO3 (111 mg, 0.8 mmol) were added. The mixture was stirred under 70 °C for 5 h. After solvent evaporation, the crude product was purified via silica gel column chromatography to obtain pure products 6a∼e.
6a: white powder; yield: 78.3%; mp: 98.6–100.4 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.39 − 7.11 (m, 4H), 6.95 (t, J = 6.8 Hz, 1H), 6.61 (s, 1H), 6.43 (dd, J = 17.2, 11.2 Hz, 1H), 5.73 (d, J = 8.0 Hz, 1H), 5.27 (d, J = 10.8 Hz, 1H), 5.13 (d, J = 17.2 Hz, 1H), 3.47 (s, 4H), 3.29 (s, 1H), 3.11 (ABq, J = 17.2 Hz, 2H), 2.70 − 2.38 (m, 4H), 2.36 − 1.92 (m, 5H), 1.81 − 1.26 (m, 11H), 1.10 (s, 3H), 1.07 − 0.98 (m, 1H), 0.82 (d, J = 6.8 Hz, 3H), 0.66 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.0, 155.1, 139.2, 139.1, 128.9, 123.2, 120.2, 117.3, 74.6, 68.6, 59.8, 58.3, 52.6, 45.5, 45.1, 44.0, 43.9, 41.8, 36.8, 36.1, 34.5, 30.5, 26.9, 26.5, 24.9, 16.8, 15.0, 11.6; HRMS: calculated for C33H47N3O5 ([M + H]+): 566.3588, found 566.3588.
6b: white powder; yield: 87.6%; mp: 121.6–122.4 °C; 1H NMR (400 MHz, DMSO-d6): δ (ppm) 8.33 (s, 1H, NH), 7.33 (d, J = 8.8 Hz, 2H), 6.81 (d, J = 8.8 Hz, 2H), 6.18 (dd, J = 17.2, 11.2 Hz, 1H), 5.60 (d, J = 8.0 Hz, 1H), 5.09 − 5.05 (m, 2H), 4.54 (d, J = 6.0 Hz, 1H), 3.70 (s, 3H), 3.53 − 3.34 (m, 5H), 3.30 − 3.01 (m, 2H), 2.45 − 2.35 (m, 4H), 2.28 − 2.01 (m, 5H), 1.75 − 1.21 (m, 11H), 1.07 (s, 3H), 1.03 − 0.95 (m, 1H), 0.83 (d, J = 6.8 Hz, 3H), 0.64 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, DMSO-d6): δ (ppm) 217.3, 155.2, 154.4, 141.0, 133.5, 121.5, 115.2, 113.5, 72.6, 68.3, 59.8, 58.9, 57.3, 55.1, 51.8, 45.0, 44.1, 43.7, 41.5, 36.5, 36.4, 34.0, 30.2, 28.6, 26.6, 24.5, 20.8, 16.0, 14.6, 11.6; HRMS: calculated for C34H49N3O6 ([M + H]+): 596.3694, found 596.3693.
6c: white powder; yield: 86.4%; mp: 107.9–109.1 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.42 − 7.13 (m, 5H), 6.46 (dd, J = 17.2, 11.2 Hz, 1H), 5.74 (d, J = 8.4 Hz, 1H), 5.29 (d, J = 11.2 Hz, 1H), 5.15 (d, J = 17.2 Hz, 1H), 4.75 (d, J = 5.2 Hz, 1H), 4.37 (d, J = 5.2 Hz, 2H), 3.39 (t, J = 4.8 Hz, 4H), 3.31 (s, 1H), 3.11 (ABq, J = 17.2 Hz, 2H), 2.71 − 2.40 (m, 4H), 2.40 − 1.96 (m, 5H), 1.83 − 1.29 (m, 11H), 1.12 (s, 3H), 1.10 − 1.02 (m, 1H), 0.84 (d, J = 6.8 Hz, 3H), 0.67 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.0, 157.6, 139.4, 139.2, 128.7, 127.9, 127.4, 117.4, 74.7, 68.5, 59.9, 58.3, 52.6, 45.6, 45.1, 44.1, 43.8, 41.9, 36.8, 36.2, 34.6, 30.5, 26.9, 26.5, 24.9, 16.8, 15.0, 11.6; HRMS: calculated for C34H49N3O5 ([M + H]+): 580.3745, found 580.3741.
6d: yellow powder; yield: 88.5%; mp: 87.6–88.9 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.19 − 8.09 (m, 2H), 7.55 − 7.46 (m, 2H), 6.94 (s, 1H), 6.49 (dd, J = 17.2, 11.2 Hz, 1H), 5.79 (d, J = 8.4 Hz, 1H), 5.33 (dd, J = 11.2, 1.2 Hz, 1H), 5.19 (dd, J = 17.2, 1.2 Hz, 1H), 3.57 (t, J = 4.8 Hz, 4H), 3.41 − 3.30 (m, 1H), 3.15 (ABq, J = 17.2 Hz, 2H), 2.71 − 2.53 (m, 4H), 2.39 − 2.02 (m, 5H), 1.93 − 1.32 (m, 11H), 1.16 (s, 3H), 1.14 − 1.07 (m, 1H), 0.87 (d, J = 6.8 Hz, 3H), 0.71 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.0, 153.8, 145.5, 142.6, 139.2, 125.2, 118.6, 117.4, 74.7, 68.7, 59.7, 58.3, 52.5, 45.6, 45.1, 44.2, 44.1, 41.9, 36.8, 36.2, 34.6, 30.5, 27.0, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C33H46N4O7 ([M + H]+): 611.3439, found 611.3440.
6e: white powder; yield: 92.3%; mp: 138.1–139.3 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.98 − 8.85 (m, 1H), 8.22 − 812 (m, 1H), 7.96 − 7.89 (m, 1H), 7.69 − 7.60 (m, 1H), 7.57 − 7.48 (m, 1H), 7.43 − 7.34 (m, 1H), 6.70 (d, J = 16.0 Hz, 1H), 6.59 − 6.43 (m, 1H), 5.80 (d, J = 8.0 Hz, 1H), 5.35 (d, J = 11.2 Hz, 1H), 5.21 (d, J = 17.2 Hz, 1H), 3.59 (d, J = 4.8 Hz, 4H), 3.42 − 3.30 (m, 1H), 3.15 (ABq, J = 17.2 Hz, 2H), 2.77 − 2.51 (m, 4H), 2.45 − 2.00 (m, 5H), 1.87 − 1.33 (m, 11H), 1.17 (s, 3H), 1.14 − 1.07 (m, 1H), 0.88 (d, J = 5.6 Hz, 3H), 0.73 (d, J = 5.6 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.1, 155.9, 150.4, 148.9, 139.2, 134.2, 130.9, 129.3, 127.2, 122.1, 121.0, 117.4, 74.7, 68.7, 59.9, 58.3, 52.6, 45.6, 45.2, 44.3, 44.1, 41.9, 36.8, 36.2, 34.6, 30.6, 27.0, 26.6, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C36H48N4O5 ([M + H]+): 617.3697, found 617.3695.
Synthesis of compounds 6f ∼ u
Compounds 10f ∼ u (1.0 mmol) and K2CO3 (276 mg, 2.0 mmol) were added to a stirred solution of compound 8 (402 mg, 0.9 mmol) in anhydrous DMF (10 ml) at 25 °C. The mixture was then heated to 80 °C until 8 was fully converted. Afterwards, the mixture was diluted by water and extracted via DCM (20 ml × 3). Subsequently, the combined organic layer was washed with brine and dried over anhydrous MgSO4. Solvent evaporation was completed before the crude product was purified by silica gel column chromatography to produce compounds 6f ∼ u.
6f: white powder; yield: 81.5%; mp: 110.9–113.2 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.65 (s, 1H), 7.59 (s, 1H), 7.20 − 7.03 (m, 3H), 6.65 (s, 1H), 6.50 (dd, J = 17.2, 11.2 Hz, 1H), 5.79 (d, J = 8.4 Hz, 1H), 5.33 (dd, J = 11.2, 1.2 Hz, 1H), 5.20 (dd, J = 17.2, 1.6 Hz, 1H), 3.55–3.50 (m, 4H), 3.36 (s, 1H), 3.15 (ABq, J = 17.2 Hz, 2H), 2.68 − 2.46 (m, 4H), 2.40 − 2.04 (m, 8H), 1.81 − 1.32 (m, 11H), 1.16 (s, 3H), 1.14 − 1.08 (m, 1H), 0.87 (d, J = 6.8 Hz, 3H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.1, 168.7, 155.1, 139.7, 139.2, 138.6, 129.4, 117.4, 116.1, 114.7, 111.9, 74.7, 68.7, 59.9, 58.3, 52.7, 45.6, 45.2, 44.1, 44.0, 41.9, 36.9, 36.2, 34.6, 30.6, 27.0, 26.6, 25.0, 24.6, 16.9, 15.0, 11.6; HRMS: calculated for C35H50N4O6 ([M + H]+): 623.3803, found 623.3804.
6g: white powder; yield: 77.4%; mp: 109.7–111.2 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.51 (s, 1H), 7.20 − 7.10 (m, 2H), 6.88 (dd, J = 7.6, 1.6 Hz, 1H), 6.63 − 6.36 (m, 3H), 5.79 (d, J = 8.4 Hz, 1H), 5.34 (dd, J = 11.2, 1.6 Hz, 1H), 5.20 (dd, J = 17.2, 1.6 Hz, 1H), 3.51 (t, J = 5.0 Hz, 4H), 3.35 (s, 1H), 3.13 (ABq, J = 17.2 Hz, 2H), 2.68 − 2.47 (m, 4H), 2.42 − 2.04 (m, 5H), 1.90 − 1.33 (m, 20H), 1.16 (s, 3H), 1.14 − 1.08 (m, 1H), 0.87 (d, J = 6.8 Hz, 3H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.1, 154.8, 152.9, 139.8, 139.2, 139.0, 129.5, 117.5, 114.5, 113.1, 109.8, 80.7, 74.7, 68.6, 60.5, 59.9, 58.3, 52.7, 45.6, 45.2, 44.1, 44.0, 41.9, 36.9, 36.2, 34.6, 30.6, 28.5, 27.0, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C38H56N4O7 ([M + H]+): 681.4222, found 681.4258.
6h: white powder; yield: 82.3%; mp: 103.2–104.4 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.30 − 7.27 (m, 1H),7.23 − 7.15 (m, 1H), 6.99 (dd, J = 8.0, 1.2 Hz, 1H), 6.71 (td, J = 8.4, 2.0 Hz, 1H), 6.56 − 6.43 (m, 2H), 5.79 (d, J = 8.4 Hz, 1H), 5.34 (dd, J = 11.2, 1.2 Hz, 1H), 5.20 (dd, J = 17.2, 1.6 Hz, 1H), 3.53 (t, J = 5.2 Hz, 4H), 3.35 (d, J = 5.6 Hz, 1H), 3.15 (ABq, J = 17.2 Hz, 2H), 2.71 − 2.50 (m, 4H), 2.39 − 2.04 (m, 5H), 1.85 − 1.33 (m, 11H), 1.16 (s, 3H), 1.14 − 1.07 (m, 1H), 0.88 (d, J = 6.8 Hz, 3H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.0, 163.2 (J = 244 Hz), 154.6, 140.8, 140.7, 139.2, 130.0 (J = 9.4 Hz), 117.4, 115.0 (J = 3.2 Hz), 114.9, 109.8 (J = 21 Hz), 107.4, 107.1, 74.7, 68.7, 60.5, 59.8, 58.3, 52.6, 45.6, 44.1, 41.9, 36.8, 36.2, 34.6, 30.6, 27.0, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C33H46FN3O5 ([M + H]+): 584.3494, found 584.3488.
6i: white powder; yield: 81.5%; mp: 116.3–117.7 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.62 (s, 1H), 7.55 (d, J = 8.0 Hz, 1H), 7.37 (t, J = 8.0 Hz, 1H), 7.30 − 7.26 (m, 1H), 6.61 (s, 1H), 6.50 (dd, J = 17.2, 11.2 Hz, 1H), 5.79 (d, J = 8.4 Hz, 1H), 5.34 (dd, J = 11.2, 1.2 Hz, 1H), 5.20 (dd, J = 17.2, 1.6 Hz, 1H), 3.55 (t, J = 5.2 Hz, 4H), 3.35 (s, 1H), 3.27 (ABq, J = 17.2 Hz, 2H), 2.73 − 2.51 (m, 4H), 2.39 − 2.04 (m, 5H), 1.83 − 1.33 (m, 11H), 1.16 (s, 3H), 1.14 − 1.08 (m, 1H), 0.88 (d, J = 6.8 Hz, 3H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.0, 154.6, 139.7, 139.2, 131.5, 131.2, 129.5, 123.0, 119.8, 117.4, 116.6, 74.7, 68.7, 60.5, 59.8, 58.3, 52.6, 45.6, 45.2, 44.1, 41.9, 36.9, 36.2, 34.6, 30.6, 27.0, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C34H46F3N3O5 ([M + H]+): 634.3462, found 634.3468.
6j: white powder; yield: 73.2%; mp: 127.9–129.5 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.33 (d, J = 6.6 Hz, 2H), 7.66 (d, J = 6.2 Hz, 2H), 6.50 (dd, J = 17.2, 11.2 Hz, 1H), 5.80 (d, J = 8.4 Hz, 1H), 5.34 (dd, J = 11.2, 1.6 Hz, 1H), 5.20 (dd, J = 17.2, 1.6 Hz, 1H), 3.64 (t, J = 4.8 Hz, 4H), 3.36 (d, J = 6.6 Hz, 1H), 3.16 (ABq, J = 17.2 Hz, 2H), 2.72 − 2.54 (m, 4H), 2.41 − 2.05 (m, 5H), 1.83 − 1.33 (m, 11H), 1.17 (s, 3H), 1.15 − 1.08 (m, 1H), 0.88 (d, J = 6.8 Hz, 3H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.0, 153.8, 150.4, 146.7, 139.2, 117.4, 113.4, 74.7, 68.7, 59.8, 58.3, 52.5, 45.6, 45.2, 44.2, 44.1, 41.9, 36.8, 36.2, 34.6, 30.6, 27.0, 26.6, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C32H46N4O5 ([M + H]+): 567.3541, found 567.3546.
6k: white powder; yield: 67.8%; mp: 129.6–130.9 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.43 − 7.35 (m, 1H), 7.31 (d, J = 8.0 Hz, 1H), 6.96 (s, 1H), 6.49 (dd, J = 17.2, 11.2 Hz, 1H), 6.14 (d, J = 7.6 Hz, 1H), 5.78 (d, J = 8.4 Hz, 1H), 5.33 (d, J = 11.2 Hz, 1H), 5.19 (dd, J = 17.2, 1.2 Hz, 1H), 4.25 (s, 2H), 3.54 (t, J = 4.8 Hz, 4H), 3.42 − 3.29 (m, 1H), 3.12 (ABq, J = 17.2 Hz, 2H), 2.72 − 2.39 (m, 4H), 2.39 − 2.01 (m, 5H), 1.82 − 1.31 (m, 11H), 1.15 (s, 3H), 1.13 − 1.06 (m, 1H), 0.86 (d, J = 6.8 Hz, 3H), 0.70 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.0, 157.0, 153.9, 151.2, 140.2, 139.1, 117.5, 102.9, 102.5, 74.7, 68.6, 59.8, 58.3, 52.6, 45.6, 45.1, 44.1, 43.9, 41.9, 36.8, 36.2, 34.6, 30.5, 26.9, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C32H47N5O5 ([M + H]+): 582.3650, found 582.3658.
6l: white powder; yield: 77.2%; mp: 149.9–151.8 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.20 (d, J = 2.4 Hz, 1H), 7.88 (dd, J = 8.8, 2.8 Hz, 1H), 7.38 (d, J = 8.8 Hz, 1H), 6.91 (s, 1H), 6.50 (dd, J = 17.2, 11.2 Hz, 1H), 5.80 (d, J = 8.4 Hz, 1H), 5.34 (dd, J = 11.2, 1.2 Hz, 1H), 5.21 (dd, J = 17.2, 1.2 Hz, 1H), 3.56 (t, J = 4.8 Hz, 4H), 3.45 − 3.31 (m, 1H), 3.15 (ABq, J = 17.2 Hz, 2H), 2.69 − 2.51 (m, 4H), 2.40 − 2.06 (m, 5H), 1.84 − 1.34 (m, 11H), 1.17 (s, 3H), 1.15 − 1.07 (m, 1H), 0.89 (d, J = 6.8 Hz, 3H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.0, 154.4, 141.1, 139.2, 135.9, 134.6, 130.3, 127.9, 117.4, 74.7, 68.7, 59.8, 58.3, 52.5, 45.6, 45.1, 44.1, 41.9, 36.8, 36.2, 34.6, 30.5, 26.9, 26.5, 24.9, 16.9, 15.0, 11.6; HRMS: calculated for C32H46BrN4O5 ([M + H]+): 645.2646, found 645.2648, 647.2638.
6m: white powder; yield: 56.2%; mp: 102.3–103.7 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.19 (d, J = 8.0 Hz, 2H), 7.13 (d, J = 8.0 Hz, 2H), 6.50 (dd, J = 17.2, 11.2 Hz, 1H), 5.79 (d, J = 8.4 Hz, 1H), 5.33 (dd, J = 11.2, 1.2 Hz, 1H), 5.19 (dd, J = 17.2, 1.2 Hz, 1H), 4.63 (t, J = 5.2 Hz, 1H), 4.37 (d, J = 5.2 Hz, 2H), 3.47 − 3.31 (m, 5H), 3.13 (ABq, J = 17.2 Hz, 2H), 2.57–2.50 (m, 4H), 2.33 (s, 3H), 2.25 − 2.04 (m, 5H), 1.80 − 1.74 (m, 1H), 1.70 − 1.42 (m, 10H), 1.16 (s, 3H), 1.13 − 1.08 (m, 1H), 0.87 (d, J = 6.8 Hz, 3H), 0.71 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.0, 157.4, 139.0, 137.1, 136.3, 129.3, 127.9, 117.4, 74.6, 68.4, 59.9, 58.2, 52.6, 45.5, 45.0, 44.8, 44.0, 43.6, 41.8, 36.7, 36.1, 34.5, 30.4, 26.8, 26.4, 24.9, 21.1, 16.8, 14.9, 11.6; HRMS: calculated for C35H51N3O5 ([M + H]+): 594.3901, found 594.3904.
6n: white powder; yield: 60.1%; mp: 104.2–105.8 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.22 (d, J = 8.4 Hz, 2H), 6.85 (d, J = 8.4 Hz, 2H), 6.49 (dd, J = 17.2, 11.2 Hz, 1H), 5.78 (d, J = 8.4 Hz, 1H), 5.33 (d, J = 11.2 Hz, 1H), 5.19 (d, J = 17.2 Hz, 1H), 4.61 (t, J = 5.6 Hz, 1H), 4.34 (d, J = 5.2 Hz, 2H), 3.79 (s, 3H), 3.45 − 3.41 (m, 4H), 3.35 (dd, J = 10.4, 6.4 Hz, 1H), 3.15 (ABq, J = 17.2 Hz, 2H), 2.62 − 2.46 (m, 4H), 2.36 − 2.03 (m, 5H), 1.81 − 1.40 (m, 11H), 1.16 (s, 3H), 1.13 − 1.08 (m, 1H), 0.87 (d, J = 6.8 Hz, 3H), 0.71 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.0, 157.4, 139.0, 131.4, 129.2, 117.3, 114.0, 74.6, 68.4, 59.9, 58.2, 55.3, 45.5, 45.0, 44.5, 44.0, 43.6, 41.8, 36.7, 36.1, 34.5, 30.4, 26.8, 26.4, 24.9, 16.8, 14.9, 11.5; HRMS: calculated for C35H51N3O6 ([M + H]+): 610.3851, found 610.3831.
6o: white powder; yield: 53.1%; mp: 109.6–110.7 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.58 (d, J = 8.4 Hz, 2H), 7.41 (d, J = 8.4 Hz, 2H), 6.50 (dd, J = 17.2, 11.2 Hz, 1H), 5.79 (d, J = 8.4 Hz, 1H), 5.34 (dd, J = 11.2, 1.2 Hz, 1H), 5.20 (d, J = 17.2 Hz, 1H), 4.83 (t, J = 5.6 Hz, 1H), 4.48 (d, J = 5.6 Hz, 2H), 3.48(t, J = 4.4 Hz, 4H), 3.35 (d, J = 6.2 Hz, 1H), 3.25 (ABq, J = 17.2 Hz, 2H), 2.67 − 2.45 (m, 4H), 2.38 − 2.03 (m, 5H), 1.81 − 1.37 (m, 11H), 1.16 (s, 3H), 1.14 − 1.09 (m, 1H), 0.88 (d, J = 6.4 Hz, 3H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.1, 168.3, 157.4, 143.6, 139.1, 129.7 (q, J = 33 Hz), 127.9, 125.6 (q, J = 4 Hz), 117.5, 74.7, 69.0, 59.3, 58.3, 52.7, 45.6, 45.1, 44.6, 44.1, 43.5, 41.9, 36.8, 36.2, 34.6, 30.5, 26.9, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C35H48F3N3O5 ([M + H]+): 648.3619, found 648.3597.
6p: light yellow powder; yield: 55.1%; mp: 115.2–116.5 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.14 (d, J = 8.4 Hz, 2H), 7.44 (d, J = 8.4 Hz, 2H), 6.49 (dd, J = 17.2, 11.2 Hz, 1H), 5.79 (d, J = 8.4 Hz, 1H), 5.33 (d, J = 11.2 Hz, 1H), 5.21–5.15 (m, 2H), 4.50 (d, J = 5.6 Hz, 2H), 3.46 (t, J = 4.4 Hz, 4H), 3.36 (s, 1H), 3.15 (ABq, J = 17.2 Hz, 2H), 2.62–2.50 (m, 4H), 2.38 − 2.05 (m, 5H), 1.82 − 1.41 (m, 11H), 1.16 (s, 3H), 1.11 (d, J = 13.6 Hz, 1H), 0.88 (d, J = 6.8 Hz, 3H), 0.71 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.1, 168.9, 157.3, 147.4, 147.1, 139.1, 128.0, 123.8, 117.2, 74.6, 68.5, 60.41, 59.7, 58.2, 52.4, 45.5, 45.0, 44.2, 44.0, 43.7, 41.8, 36.7, 36.1, 34.5, 30.4, 26.8, 26.5, 24.8, 16.7, 14.9, 11.5; HRMS: calculated for C34H48N4O7 ([M + H]+): 625.3596, found 625.3596.
6q: white powder; yield: 45.6%; mp: 107.6–108.7 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.61 (d, J = 7.6 Hz, 1H), 7.57 (d, J = 7.6 Hz, 1H), 7.49 (t, J = 7.6 Hz, 1H), 7.34 (t, J = 7.6 Hz, 1H), 6.47 (dd, J = 17.2, 11.2 Hz, 1H), 5.76 (d, J = 8.4 Hz, 1H), 5.31 (d, J = 11.2 Hz, 1H), 5.17 (d, J = 17.2 Hz, 1H), 4.93 (d, J = 5.6 Hz, 1H), 4.57 (d, J = 5.6 Hz, 2H), 3.40 (t, J = 4.8 Hz, 4H), 3.33 (dd, J = 10.4, 6.8 Hz, 1H), 3.11 (ABq, J = 17.2 Hz, 2H), 2.58 − 2.44 (m, 4H), 2.35 − 2.04 (m, 5H), 1.80 − 1.39 (m, 11H), 1.14 (s, 3H), 1.12 − 1.06 (m, 1H), 0.86 (d, J = 6.8 Hz, 3H), 0.69 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.1, 168.9, 157.2, 139.1, 137.9, 132.3, 130.6, 127.4, 125.9, 125.9, 117.3, 74.6, 68.5, 59.8, 58.2, 52.5, 45.5, 45.0, 44.0, 43.6, 41.8, 41.4, 36.7, 36.1, 34.5, 30.4, 26.8, 26.4, 24.8, 16.7, 14.9, 11.5; HRMS: calculated for C35H48F3N3O5 ([M + H]+): 648.3619, found 648.3620.
6r: white powder; yield: 65.1%; mp: 136.5–137.9 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.32 − 7.27 (m, 2H), 7.24 − 7.15 (m, 3H), 6.49 (dd, J = 17.2, 11.2 Hz), 5.78 (d, J = 8.4 Hz), 5.33 (dd, J = 11.2, 1.2 Hz), 5.19 (dd, J = 17.2, 1.2 Hz), 4.44 (t, 1H, J = 5.6 Hz), 3.47 (dd, J = 12.4, 6.8 Hz, 2H), 3.35 (t, J = 4.8 Hz, 4H), 3.10 (ABq, J = 17.2 Hz, 2H), 2.81 (t, J = 6.8 Hz, 2H), 2.58 − 2.42 (m, 4H), 2.37 − 2.02 (m, 5H), 1.79 − 1.24 (m, 11H), 1.15 (s, 3H), 1.13 − 1.07 (m, 1H), 0.87 (d, J = 6.4 Hz, 3H), 0.70 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.0, 157.6, 139.1, 139.5, 129.0, 128.7, 126.5, 117.5, 74.7, 68.5, 59.9, 58.3, 52.6, 45.6, 45.1, 44.1, 43.6, 42.1, 41.9, 36.8, 36.4, 36.2, 34.6, 30.5, 26.9, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C35H51N3O5 ([M + H]+): 594.3901[M + H]+, found 594.3905.
6s: white powder; yield: 61.7%; mp: 105.8–107.1 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.22 (t, J = 7.6 Hz, 1H), 6.78 − 6.73 (m, 3H), 6.50 (dd, J = 17.2, 11.2 Hz, 1H), 5.78 (d, J = 8.4 Hz, 1H), 5.33 (d, J = 11.2 Hz, 1H), 5.20 (d, J = 17.2 Hz, 1H), 4.42 (t, J = 5.6 Hz, 1H), 3.79 (s, 3H), 3.47 (dd, J = 12.4, 6.8 Hz, 2H), 3.36 (t, J = 4.8 Hz, 4H), 3.12 (ABq, J = 17.2 Hz, 2H), 2.79 (t, J = 6.8 Hz, 2H), 2.54 − 2.49 (m, 4H), 2.37 − 2.03 (m, 5H), 1.80 − 1.42 (m, 11H), 1.16 (s, 3H), 1.13 − 1.09 (m, 1H), 0.87 (d, J = 6.8 Hz, 3H), 0.71 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.1, 159.9, 157.6, 139.1, 141.1, 129.7, 121.3, 114.6, 111.9, 117.5, 74.7, 68.5, 59.9, 58.3, 55.3, 52.6, 45.6, 45.1, 44.1, 43.6, 42.0, 41.9, 36.8, 36.5, 36.2, 34.6, 30.5, 26.9, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C36H53N3O6 ([M + H]+): 624.4007, found 624.3995.
6t: white powder; yield: 82.7%; mp: 119.5–122.2 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 6.49 (dd, J = 17.2, 11.2 Hz, 1H), 5.77 (d, J = 8.4 Hz, 1H), 5.35 (s, 1H), 5.32 (dd, J = 11.2, 1.6 Hz, 1H), 5.18 (dd, J = 17.2, 1.6 Hz, 1H), 3.40 (t, J = 4.8 Hz, 4H), 3.36 − 3.24 (m, 3H), 3.12 (ABq, J = 17.2 Hz, 2H), 2.63 − 1.99 (m, 16H), 1.83 − 1.31 (m, 17H), 1.15 (s, 3H), 1.13 − 1.03 (m, 1H), 0.86 (d, J = 6.8 Hz, 3H), 0.70 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.1, 162.6, 157.9, 139.2, 117.4, 77.5, 77.2, 76.8, 74.7, 68.5, 60.0, 58.3, 57.6, 54.3, 52.7, 45.6, 45.1, 44.0, 43.6, 41.9, 37.3, 36.8, 36.6, 36.2, 34.6, 31.5, 30.5, 26.9, 26.5, 26.0, 25.0, 24.4, 16.8, 15.0, 11.6; HRMS: calculated for C34H56N4O5 ([M + H]+): 601.4323, found 601.4326.
6u: white powder; yield: 79.2%; mp: 92.1–93.8 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.98 (s, 1H), 6.47 (dd, J = 17.2, 11.2 Hz, 1H), 5.76 (d, J = 8.4 Hz, 1H), 5.30 (d, J = 11.2 Hz, 1H), 5.22 − 5.07 (m, 2H), 3.78 − 3.62 (m, 4H), 3.49 − 3.24 (m, 6H), 3.10 (ABq, J = 17.2 Hz, 2H), 2.63 − 2.37 (m, 10H), 2.36 − 1.96 (m, 5H), 1.81 − 1.29 (m, 11H), 1.13 (s, 3H), 1.11 − 1.04 (m, 1H), 0.85 (d, J = 6.8 Hz, 3H), 0.69 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 169.0, 157.8, 139.2, 117.3, 74.6, 68.5, 67.0, 59.9, 58.3, 57.6, 53.4, 52.6, 45.5, 45.1, 44.0, 43.6, 41.9, 37.0, 36.1, 34.5, 30.5, 26.9, 26.5, 24.9, 16.8, 15.0, 11.6; HRMS: calculated for C33H54N4O6 ([M + H]+): 603.4116, found 603.4144.
Synthesis of compound 6v
The solution of 11 (128 mg, 0.47 mmol) in acetonitrile (10 ml) was treated with 22-(piperazine-1-yl)-22-deoxypleuromutilin 8 (210 mg, 0.47 mmol) and DMAP (57 mg, 0.47 mmol), which was heated to reflux for 24 h. After solvent removal, the residue was dissolved in EtOAc (10 ml). The organic phase was washed with 1 N NaOH and brine, followed by drying over anhydrous Na2SO4. The filtrate obtained via filtration was concentrated until dry, and then purification by silica gel chromatography was conducted to obtain compound 6v. white powder; yield: 40.5%; mp: 105.1–106.3 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.33 − 7.31 (m, 2H), 7.11 − 7.06 (m, 3H), 6.47 (dd, J = 17.2, 11.2 Hz, 1H), 5.74 (d, J = 8.4 Hz, 1H), 5.30 (dd, J = 11.2, 1.2 Hz, 1H), 5.17(dd, J = 17.2, 1.2 Hz, 1H), 3.33 (dd, J = 10.4, 6.8 Hz, 1H), 3.25 (t, J = 4.9 Hz, 4H), 3.21 (s, 3H), 3.02 (ABq, J = 17.2 Hz, 2H), 2.40 − 2.23 (m, 7H), 2.26 − 1.99 (m, 2H), 1.80 − 1.38 (m, 11H), 1.14 (s, 3H), 1.12 − 1.05 (m, 1H), 0.85 (d, J = 7.2 Hz, 3H), 0.66 (d, J = 7.2 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.2, 168.9, 160.9, 146.7, 139.0, 129.5, 124.6, 123.8, 117.3, 74.6, 68.3, 59.8, 58.2, 52.5, 45.5, 45.4, 44.9, 43.9, 41.8, 39.6, 36.7, 36.0, 34.5, 30.4, 26.8, 26.4, 24.9, 16.7, 14.9, 11.5; HRMS: calculated for C34H49N3O5 ([M + H]+): 580.3745, found 580.3744.
Synthesis of compound 6w
Compound 6g (136 mg, 0.2 mmol) was added to a mixture of 6 ml DCM and TFA (v/v = 10/1) and stirred at 25 °C until completely converted. After neutralisation by saturated aqueous NaHCO3, the organic phase was collected and dried over anhydrous Na2SO4. Then, the filtrate obtained via filtration was concentrated to generate the crude product, which was purified by silica gel chromatography to produce compound 6w. white powder; yield: 95.6%; mp: 146.2–147.5 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.02 (t, J = 8.0 Hz, 1H), 6.95 (t, J = 2.0 Hz, 1H), 6.56 − 6.44 (m, 2H), 6.38 − 6.31 (m, 2H), 5.79 (d, J = 8.4 Hz, 1H), 5.34 (dd, J = 11.2, 1.6 Hz, 1H), 5.20 (dd, J = 17.2, 1.2 Hz, 1H), 3.52 (t, J = 4.8 Hz, 4H), 3.35 (d, J = 6.0 Hz, 1H), 3.12 (ABq, J = 17.2 Hz, 2H), 2.68 − 2.49 (m, 4H), 2.41 − 2.02 (m, 7H), 1.86 − 1.21 (m, 13H), 1.16 (s, 3H), 1.14 − 1.07 (m, 1H), 0.87 (d, J = 6.8 Hz, 4H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.1, 155.0, 147.3, 140.0, 139.2, 129.7, 117.5, 110.2, 109.9, 106.8, 74.7, 68.6, 59.9, 58.3, 52.7, 45.6, 45.2, 44.1, 44.0, 41.9, 36.8, 36.2, 34.6, 30.6, 27.0, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C33H48N4O5 ([M + H]+): 581.3697, found 581.3704.
Synthesis of compound 6x and 6y
A mixture of 6d (122 mg, 0.2 mmol), SnCl2 (474 mg, 2.5 mmol) and ethanol (5 ml) was stirred under reflux for 5 h. After cooling to 25 °C, 1 N NaOH was added. The resultant mixture was extracted with DCM (10 ml × 3) and separated. After the organic phase was dried over Na2SO4 and concentrated, the residue was purified via silica gel column chromatography to acquire 6x. 6y was synthesised by a similar method used for 6p.
6x: yellow powder; yield: 92.5%; mp: 137.4–138.7 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.08 (d, J = 8.4 Hz, 2H), 6.62 (d, J = 8.4 Hz, 2H), 6.50 (dd, J = 17.2, 11.2 Hz, 1H), 6.18 (s, 1H), 5.79 (d, J = 8.4 Hz, 1H), 5.34 (d, J = 11.2 Hz, 1H), 5.20 (d, J = 17.2 Hz, 1H), 3.50 (t, J = 4.8 Hz, 4H), 3.35 (d, J = 5.2 Hz, 1H), 3.14 (ABq, J = 17.2 Hz, 2H), 2.67 − 2.49 (m, 4H), 2.41 − 2.01 (m, 5H), 1.83 − 1.32 (m, 13H), 1.16 (s, 3H), 1.14 − 1.07 (m, 1H), 0.87 (d, J = 6.8 Hz, 3H), 0.72 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.3, 169.1, 155.8, 142.9, 139.2, 130.2, 123.1, 117.5, 115.7, 74.7, 68.6, 59.9, 58.3, 52.7, 45.6, 45.2, 44.1, 44.0, 41.9, 36.8, 36.2, 34.6, 30.6, 29.8, 27.0, 26.5, 25.0, 16.9, 15.0, 11.6; HRMS: calculated for C33H48N4O5 ([M + H]+): 581.3697, found 581.3705.
6y: light yellow powder; yield: 75.5%; mp: 118.5–119.3 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 7.08 (d, J = 8.4 Hz, 2H), 6.65 − 6.60 (m, 2H), 6.48 (dd, J = 17.2, 11.2 Hz, 1H), 5.77 (d, J = 8.4 Hz, 1H), 5.32 (dd, J = 11.2, 1.2 Hz, 1H), 5.18 (dd, J = 17.2, 1.2 Hz, 1H), 4.59 (t, J = 5.6 Hz, 1H), 4.27 (d, J = 5.2 Hz, 2H), 3.40 (t, J = 5.2 Hz, 4H), 3.35 (s, 1H), 3.11 (ABq, J = 17.2 Hz, 2H), 2.56 − 2.46 (m, 4H), 2.36 − 2.02 (m, 6H), 1.74– 1.27 (m, 10H), 1.15 (s, 3H), 1.12 − 1.06 (m, 1H), 0.86 (d, J = 6.8 Hz, 3H), 0.70 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3) δ (ppm) 217.1, 168.9, 157.4, 145.8, 139.1, 129.2, 129.1, 117.3, 115.2, 74.6, 68.4, 59.9, 58.2, 52.6, 45.5, 45.0, 44.7, 44.0, 43.6, 41.8, 36.7, 36.1, 34.5, 30.4, 26.8, 26.4, 24.9, 16.7, 14.9, 11.5; HRMS: calculated for C34H50N4O5 ([M + H]+): 595.3854, found 595.3860.
Synthesis of compound 6z
6y (190 mg, 0.32 mmol) and triethylamine (49 mg, 0.48 mmol) were dissolved in dry THF, and chloroacetyl chloride (54 mg, 0.48 mmol) was added under an ice bath condition to the above solution. The mixture was stirred at 25 °C for about 4 h before water was added. The resulting mixture was extracted via DCM, washed with brine, and dried over anhydrous Na2SO4. Then, the organic phase was concentrated to obtain the crude intermediate, which was purified by silica gel column chromatography to produce 14.
Compound 14 (201 mg, 0.30 mmol), morpholine (52 mg, 0.60 mmol), and K2CO3 (82 mg, 0.60 mmol) were dissolved in dry THF (5 ml) and maintained at reflux for about 6 h. After the solution was concentrated, the residue was purified by silica gel column chromatography to generate 6z.
14: white powder; yield: 70.1%; mp: 118.1–119.2 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 8.25 (s, 1H), 7.49 (d, J = 8.4 Hz, 2H), 7.29 (d, J = 8.4 Hz, 2H), 6.49 (dd, J = 17.2, 11.2 Hz, 1H), 5.79 (d, J = 8.4 Hz, 1H), 5.34 (d, J = 11.2 Hz, 1H), 5.20 (d, J = 17.2 Hz, 1H), 4.77 (s, 1H), 4.39 (d, J = 5.6 Hz, 2H), 4.18 (s, 2H), 3.48 (s, 4H), 3.39 − 3.32 (m, 1H), 3.30 − 3.07 (m, 2H), 2.64 (br, 4H), 2.38 − 2.15 (m, 3H), 2.08 (dd, J = 16.4, 8.0 Hz, 2H), 1.80 − 1.47 (m, 11H), 1.16 (s, 3H), 1.13 − 1.09 (m, 1H), 0.88 (d, J = 6.8 Hz, 3H), 0.71 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 216.7, 188.9, 164.0, 157.4, 139.0, 135.8, 128.6, 120.6, 117.4, 74.6, 58.2, 52.3, 45.5, 45.0, 44.5, 44.0, 42.9, 41.8, 36.7, 36.1, 34.5, 30.4, 26.8, 26.4, 24.9, 16.8, 14.9, 11.5; HRMS: calculated for C35H51ClN4O6 ([M + H]+): 671.3586, found 671.3570.
6z: white powder; yield: 50.3%; mp: 120.1–121.5 °C; 1H NMR (400 MHz, CDCl3): δ (ppm) 9.02 (s, 1H), 7.49 (d, J = 8.4 Hz, 2H), 7.26 (d, J = 8.4 Hz, 2H), 6.48 (dd, J = 17.2, 11.2 Hz, 1H), 5.77 (d, J = 8.4 Hz, 1H), 5.31 (dd, J = 11.2, 1.2 Hz, 1H), 5.18 (dd, J = 17.2, 1.2 Hz, 1H), 4.79 (t, J = 5.6 Hz, 1H), 4.35 (d, J = 5.6 Hz, 2H), 3.78 − 3.74 (m, 4H), 3.41 (t, J = 5.0 Hz, 4H), 3.34 (dd, J = 9.6, 6.8 Hz, 1H), 3.21 − 3.02 (m, 4H), 2.63 − 2.58 (m, 4H), 2.58 − 2.45 (m, 4H), 2.36 − 2.01 (m, 6H), 1.89 (s, 1H), 1.76 (dd, J = 14.2, 2.4 Hz, 1H), 1.69 − 1.51 (m, 5H), 1.49 − 1.43 (m, 1H), 1.38 − 1.31 (m, 1H), 1.15 (s, 3H), 1.12 − 1.06 (m, 1H), 0.86 (d, J = 6.8 Hz, 3H), 0.70 (d, J = 6.8 Hz, 3H); 13 C NMR (100 MHz, CDCl3): δ (ppm) 217.1, 168.9, 167.9, 157.4, 139.1, 136.6, 135.5, 128.5, 119.8, 117.3, 74.6, 68.5, 67.0, 59.8, 58.2, 53.8, 52.6, 45.5, 45.0, 44.5, 44.0, 43.7, 41.8, 36.7, 36.1, 34.5, 30.4, 29.7, 26.8, 26.5, 24.9, 16.7, 14.9, 11.5; HRMS: calculated for C40H59N5O7 ([M + H]+): 722.4487, found 722.4497.
Biological assay
Bacterial solution preparation
Upon overall recovery of S. aureus ATCC 25923, MRSA ATCC 33591, MRSA ATCC 43300 and E.coli ATCC 25922, the single selected colony was inoculated in sterile Luria-Bertani (LB) broth and incubated at 37 °C for 1 8 ∼ 20 h. The bacterial solution was adjusted to 0.5 McFarland standard with saline and subsequently diluted with LB broth to the approximate concentration of 106∼107 CFU·mL−1.
MIC And MBC assays
MIC values were established in vitro using the agar dilution method34,35. Moreover, tiamulin fumarate was selected as the reference drug. All compounds were dissolved in a small amount of ethanol and diluted in distilled water to a concentration of 1280 µg·mL−1. Next, two-fold serially diluted pleuromutilin compound solutions containing distilled water were produced until the final concentration reached 0.625 µg·mL−1. After ten-fold dilution with LB broth, the two-fold serially diluted solutions were successively added to plate wells containing 10 ml of the above four bacterial solutions; then, solutions were incubated at 37 °C for 1 6 ∼ 18 h and data was recorded. Each procedure was repeated 3 times. MBC was considered when the compounds killed over 99% of the tested bacterial culture. MIC-corresponding wells and three previous wells were homogenised, serially diluted, and plated on Mueller-Hinton (MH) agar to determine MBC. Data was recorded after plates underwent incubation at 37 °C for 18–24 h. Each procedure was repeated 3 times.
Time-kill kinetics assay
Compounds 6p and tiamulin fumarate were dissolved in MH broth to generate different solutions of 1 × MIC and 6 × MIC, followed by inoculation with the above bacteria suspension at 37 °C; After specified time intervals (0, 1, 2, 4, 6, 12 and 24 h), 10 µL of the above mixture was serially diluted 10-fold with saline and incubated at 37 °C for 24 h on MH agar plates. Following this, viable colonies were counted and expressed as log 10 CFU·mL−1. Each procedure was done a total of 3 times.
CCK-8 assay
All cell lines were cultured in Dulbecco’s modified Eagle’s medium (DMEM), with 10% foetal bovine serum (FBS) and supplemented with 1% penicillin-streptomycin (P/S). The cell counting kit-8 (CCK-8) was purchased from Dojindo Laboratories (Kumamoto, Japan).
Briefly, a 100 μL suspension of 10000 LO2 cells and 20000 HEK293T cells were seeded in 96-well plates, respectively. They were treated with six different concentrations of 6p (0.125 μg/mL, 0.25 μg/mL, 0.5 μg/mL, 1 μg/mL, 2 μg/mL and 4 μg/mL) and incubated at 37 °C in a CO2 incubator for 48 h. Afterwards, the CCK-8 solution (10 μL) was added to each well, and plates were incubated for an additional 1 h at 37 °C. The absorbance at 450 nm was then recorded using a SpectraMax-190 microplate reader (Molecular Devices, USA). Each procedure was done a total of 3 times.
Molecular docking
The binding of synthesised compound 6p was examined using the PTC ribosome complex and Tiamulin (PDB ID: 1XBP). In particular, the PTC ribosome model was constructed using residues within 30 Å from the binding site after extraction of the original ligand Tiamulin. In addition, both 6p and the above ribosome model were prepared via AutoDockTools by removing water molecules, adding polar hydrogens, and assigning Gasteiger charges. Finally, the box (52.791, 122.679, 114.289) was established using original ligand as the grid centre with box size (36.758, 44.448, 36.761). Docking was performed by DOCK 6.9, and the 2 D interaction diagrams was generated via LigPlot+ v1.436.
Conclusions
In summary, a series of pleuromutilin derivatives possessing the piperazinyl urea linkage were designed and synthesised, and their antibacterial activities against Gram-positive and Gram-negative strains were evaluated. Nearly all synthesised compounds exhibited broad-spectrum antibacterial activities, and particularly, more potent activities against MRSA strains. Activities of 6p were most effective with a MIC concentration of 0.1 2 5 ∼ 0.25 μg/mL, which was 8 ∼ 16 times that of Tiamulin. Time-kill kinetics indicated that at 1 × MIC, compound 6p and Tiamulin had similar bactericidal effects against MRSA ATCC 33591; however compound 6p was more effective than Tiamulin against E.coli ATCC 25922, since it eliminated total bacteria colony in 24 h while Tiamulin could not. Molecular docking demonstrated that aside from hydrophobic interactions, benzyl urea linkage and terminal nitro groups from the side chain could produce several hydrogen bonds and π-π stacking with surrounding residues.
Funding Statement
This research was funded by Chunhui Project of Ministry of Education of China [Z2016164], Scientific Research Foundation of the Education Department of Sichuan Province [12ZB128 and 18TD0023], National Undergraduate Training Program for Innovation and Entrepreneurship [202010650016], National Natural Science Foundation of China [81703355] and Scientific Research Project from Science and Technology Department of Yibin [2019GY007 and 2018ZGY013].
Author contributions
Y.Z., C.X., F.S. and R.S. designed and synthesised the compounds. Y.L. and D.L. performed the biological evaluation. C. W. implemented the cytotoxicity test. X. Y. performed the molecular docking. Y.Z., J.Y. and Z.W. interpreted the data and wrote the paper. All authors have read and agreed to the published version of the manuscript.
Disclosure statement
There are no conflicts to declare.
References
- 1.Rohde C, Wittmann J, Kutter E.. Bacteriophages: a therapy concept against multi-drug-resistant bacteria. Surg Infect (Larchmt) 2018;19:737–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hassoun A, Linden PK, Friedman B.. Incidence, prevalence, and management of MRSA bacteremia across patient populations-a review of recent developments in MRSA management and treatment. Crit Care 2017;21:211–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.David MZ, Dryden M, Gottlieb T, et al. Recently approved antibacterials for methicillin-resistant Staphylococcus aureus (MRSA) and other Gram-positive pathogens: the shock of the new. Int J Antimicrob Agents 2017;50:303–7. [DOI] [PubMed] [Google Scholar]
- 4.Theuretzbacher U, Outterson K, Engel A, et al. The global preclinical antibacterial pipeline. Nat Rev Microbiol 2020;18:275–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goyal V, Grimwood K, Byrnes CA, et al. Amoxicillin-clavulanate versus azithromycin for respiratory exacerbations in children with bronchiectasis (BEST-2): a multicentre, double-blind, non-inferiority, randomised controlled trial. Lancet 2018;392:1197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Arechiga-Alvarado NA, Medellin-Garibay SE, Milan-Segovia RC, et al. Population pharmacokinetics of amikacin administered once daily in patients with different renal functions. Antimicrob Agents Chemother 2020;64:e02178–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Newman DJ, Cragg GM.. Natural products as sources of new drugs over the nearly four decades from 01/1981 to 09/2019. J Nat Prod 2020;83:770–803. [DOI] [PubMed] [Google Scholar]
- 8.Sandargo B, Chepkirui C, Cheng T, et al. Biological and chemical diversity go hand in hand: Basidiomycota as source of new pharmaceuticals and agrochemicals. Biotechnol Adv 2019;37:107344. [DOI] [PubMed] [Google Scholar]
- 9.Gatadi S, Madhavi YV, Chopra S, et al. Promising antibacterial agents against multidrug resistant Staphylococcus aureus. Bioorg Chem 2019;92:103252. [DOI] [PubMed] [Google Scholar]
- 10.Garmyn A, Vereecken M, Degussem K, et al. Efficacy of tiamulin alone or in combination with chlortetracycline against experimental Mycoplasma gallisepticum infection in chickens. Poult Sci 2017;96:3367–74. [DOI] [PubMed] [Google Scholar]
- 11.Hata E, Harada T, Itoh M.. Relationship between antimicrobial susceptibility and multilocus sequence type of Mycoplasma bovis isolates and development of a method for rapid detection of point mutations involved in decreased susceptibility to macrolides, lincosamides, tetracyclines, and spectinomycin. Appl Environ Microbiol 2019;85:e00575–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun F, Zhang H, Gonzales GB, et al. Unraveling the metabolic routes of retapamulin: insights into drug development of pleuromutilins. Antimicrob Agents Chemother 2018;62:e02388–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williamson DA, Carter GP, Howden BP.. Current and emerging topical antibacterials and antiseptics: agents, action, and resistance patterns. Clin Microbiol Rev 2017;30:827–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Perry W, Golan Y.. Therapeutic potential of lefamulin in the treatment of community-acquired pneumonia. Future Microbiol 2019;14:927–39. [DOI] [PubMed] [Google Scholar]
- 15.Veve MP, Wagner JL.. Lefamulin: review of a promising novel pleuromutilin antibiotic. Pharmacotherapy 2018;38:935–46. [DOI] [PubMed] [Google Scholar]
- 16.Paukner S, Riedl R.. Pleuromutilins: potent drugs for resistant bugs-mode of action and resistance. Cold Spring Harb Perspect Med 2017;7:a027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shang R, Pu X, Xu X, et al. Synthesis and biological activities of novel pleuromutilin derivatives with a substituted thiadiazole moiety as potent drug-resistant bacteria inhibitors. J Med Chem 2014;57:5664–78. [DOI] [PubMed] [Google Scholar]
- 18.Yi Y, Xu X, Liu Y, et al. Synthesis and antibacterial activities of novel pleuromutilin derivatives with a substituted pyrimidine moiety. Eur J Med Chem 2017;126:687–95. [DOI] [PubMed] [Google Scholar]
- 19.Deng Y, Wang X-Z, Huang S-H, et al. Antibacterial activity evaluation of synthetic novel pleuromutilin derivatives in vitro and in experimental infection mice. Eur J Med Chem 2019;162:194–202. [DOI] [PubMed] [Google Scholar]
- 20.Ling C, Fu L, Gao S, et al. Design, synthesis, and structure-activity relationship studies of novel thioether pleuromutilin derivatives as potent antibacterial agents. J Med Chem 2014;57:4772–95. [DOI] [PubMed] [Google Scholar]
- 21.Wang BL, Shi YX, Zhang SJ, et al. Syntheses, biological activities and SAR studies of novel carboxamide compounds containing piperazine and arylsulfonyl moieties. Eur J Med Chem 2016;117:167–78. [DOI] [PubMed] [Google Scholar]
- 22.Keith JM, Apodaca R, Tichenor M, et al. Aryl piperazinyl ureas as inhibitors of fatty acid amide hydrolase (FAAH) in rat, dog, and primate. ACS Med Chem Lett 2012;3:823–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babu DS, Srinivasulu D, Kotakadi VS.. Synthesis of novel N-aryl-4- 6-(2-fluoropyridin-3-yl)quinazolin-2-yl -piperazine-1-carboxam ide or -carbothioamide derivatives and their antimicrobial activity. Chem Heterocycl Compd 2015;51:60–6. [Google Scholar]
- 24.Trivedi P, Adhikari N, Amin SA, et al. Design, synthesis, biological evaluation and molecular docking study of arylcarboxamido piperidine and piperazine-based hydroxamates as potential HDAC8 inhibitors with promising anticancer activity. Eur J Pharm Sci 2019;138:105046. [DOI] [PubMed] [Google Scholar]
- 25.Zhu L, Kalimuthu S, Gangadaran P, et al. Exosomes derived from natural killer cells exert therapeutic effect in melanoma. Theranostics 2017;7:2732–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yu S, Zhang X, Tan G, et al. A novel pH-induced thermosensitive hydrogel composed of carboxymethyl chitosan and poloxamer cross-linked by glutaraldehyde for ophthalmic drug delivery. Carbohydr Polym 2017;155:208–17. [DOI] [PubMed] [Google Scholar]
- 27.Gao M-L, Zeng J, Fang X, et al. Design, synthesis and antibacterial evaluation of novel pleuromutilin derivatives possessing piperazine linker. Eur J Med Chem 2017;127:286–95. [DOI] [PubMed] [Google Scholar]
- 28.Luo J, Yang Q-E, Yang Y-Y, et al. Design, synthesis, and structure-activity relationship studies of novel pleuromutilin derivatives having a piperazine ring. Chem Biol Drug Des 2016;88:699–709. [DOI] [PubMed] [Google Scholar]
- 29.Liao C, Liu Y, Liu C, et al. Phenylquinoline transient receptor potential vanilloid 1 antagonists for the treatment of pain: discovery of 1-(2-phenylquinoline-4-carbonyl)-N-(4-(trifluoromethyl)phenyl)pyrrolidine-3-carboxamide. Bioorg Med Chem 2018;26:845–54. [DOI] [PubMed] [Google Scholar]
- 30.Modranka J, Li J, Parchina A, et al. Synthesis and structure-activity relationship studies of benzo[b][1,4]oxazin-3(4H)-one analogues as inhibitors of mycobacterial thymidylate synthase X. ChemMedChem 2019;14:645–62. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Alsina LA, Murray JC, Buzon LM, et al. Spiropiperidine sultam and lactam templates: diastereoselective overman rearrangement and metathesis followed by NH arylation. J Org Chem 2017;82:12246–56. [DOI] [PubMed] [Google Scholar]
- 32.Zhang Y, Yang W, Xu K, et al. Synthesis and antibacterial activities of pleuromutilin derivatives containing aryl urea groups. Lett Drug Des Discov 2013;10:219–25. [Google Scholar]
- 33.Batey RA, Yoshina-Ishii C, Taylor SD, et al. A new protocol for the formation of carbamates and thiocarbamates using carbamoyl imidazolium salts. Tetrahedron Lett 1999;40:2669–72. [Google Scholar]
- 34.Shang R, Liu Y, Xin Z, et al. Synthesis and antibacterial evaluation of novel pleuromutilin derivatives. Eur J Med Chem 2013;63:231–8. [DOI] [PubMed] [Google Scholar]
- 35.Yi Y, Fu Y, Wang K, et al. Synthesis and antibacterial activities of novel pleuromutilin derivatives. Archiv Der Pharmazie 2018;351:e1800155. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, Swindells MB.. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 2011;51:2778–86. [DOI] [PubMed] [Google Scholar]